Abstract
Three-dimensional (3D) chromatin structure has been shown to play a role in regulating gene transcription during biological transitions. Although our understanding of loop formation and maintenance is rapidly improving, much less is known about the mechanisms driving changes in looping and the impact of differential looping on gene transcription. One limitation has been a lack of well-powered differential looping data sets. To address this, we conducted a deeply sequenced Hi-C time course of megakaryocyte development comprising four biological replicates and 6 billion reads per time point. Statistical analysis revealed 1503 differential loops. Gained loop anchors were enriched for AP-1 occupancy and were characterized by large increases in histone H3K27ac (over 11-fold) but relatively small increases in CTCF and RAD21 binding (1.26- and 1.23-fold, respectively). Linear modeling revealed that changes in histone H3K27ac, chromatin accessibility, and JUN binding were better correlated with changes in looping than RAD21 and almost as well correlated as CTCF. Changes to epigenetic features between—rather than at—boundaries were highly predictive of changes in looping. Together these data suggest that although CTCF and RAD21 may be the core machinery dictating where loops form, other features (both at the anchors and within the loop boundaries) may play a larger role than previously anticipated in determining the relative loop strength across cell types and conditions.
The three-dimensional (3D) organization of chromatin is thought to play an important role in transcriptional regulation and has been implicated in many biological processes, including cellular differentiation and response to external stimuli (Dixon et al. 2015). Whereas several types of 3D chromatin structures exist, chromatin loops are of particular interest as they are thought to regulate gene expression by bringing distal regulatory elements (e.g., enhancers) into close physical proximity with gene promoters via point to point interactions. Loop anchors are typically enriched for enhancers and promoters and correlate with differences in gene expression (Kagey et al. 2010; Kim and Dean 2012; Grubert et al. 2020). Aberrations to chromatin looping are associated with a variety of human diseases and developmental disorders such as Cornelia de Lange syndrome (Panarotto et al. 2022), polydactyly (Lettice et al. 2003; Paliou et al. 2019), and cancer (Kon et al. 2013; Ahn et al. 2021). Whereas the basic mechanisms of loop formation have been established, major questions remain regarding the mechanisms driving differential looping during biological development and their functional impact.
The majority of chromatin loops are thought to form through a process called loop extrusion, in which the cohesin complex is loaded onto DNA and reels in chromatin until it reaches convergently bound CTCF proteins (Sanborn et al. 2015). In rare circumstances, loops can form through noncanonical mechanisms including phase separation (Krivega and Dean 2017; Monahan et al. 2019; Ahn et al. 2021) or binding of lineage-specific factors like LDB1 (Krivega and Dean 2017; Monahan et al. 2019; Ahn et al. 2021). Although chromatin loops have been shown to change over cellular transitions, the mechanisms that govern these structural changes, and the impact of these changes on gene expression, remain poorly understood (Rao et al. 2017; Jeppsson et al. 2022).
The relationship between looping and gene expression is even less clear. Cell type–specific loops correlate with differential expression patterns, supporting a role for loops in gene regulation (Rao et al. 2014; Hu et al. 2021; Sey et al. 2022). Moreover, forced looping between enhancers and promoters at select loci has been shown to activate transcription (Rupon et al. 2013; Deng et al. 2014). However, several recent studies have called the role of loops in regulating gene expression into question. Live cell imaging of looping between SOX2 and an enhancer known to regulate its expression showed no correlation between enhancer-promoter proximity and gene transcription (Alexander et al. 2019). In another study, rapid and thorough degradation of cohesin led to a complete removal of cohesin-driven loops in human cancer cells with only a minor impact on gene expression (Rao et al. 2017). In summary, the degree to which chromatin looping regulates gene expression is still unresolved.
One impediment to answering these questions is that identifying differential loops between cells and conditions remains challenging. Because of the depth of sequencing required for Hi-C data sets, the statistical requirements of differential analysis, and the cost of DNA sequencing, most existing differential looping studies lack the statistical power to adequately identify differential loops. And without comprehensive and rigorously defined sets of differential loops, it is challenging to determine what mechanisms drive differential looping and what transcriptional impact they have.
To address this gap, we generated a deeply sequenced Hi-C data set characterizing the differentiation of K562 cells into a megakaryocyte-like state. By sequencing over 18 billion reads across three time points and four biological replicates, we achieved a statistical power of roughly 0.932 and identified 1503 differential loops. Generation and intersection with accompanying maps of chromatin accessibility, histone acetylation, transcription factor (TF) binding, and gene expression revealed insights into both the mechanisms and the functional impacts of differential looping during cellular differentiation. We find that regulatory features both at and between loop anchors correlate with changes in loop strength. Finally, we show that incorporating H3K27 acetylation and chromatin looping dynamics into linear models in addition to promoter acetylation improves predictions of changes in gene expression.
Results
Differentiation of K562 cells induces large-scale changes to 3D chromatin structure across multiple scales
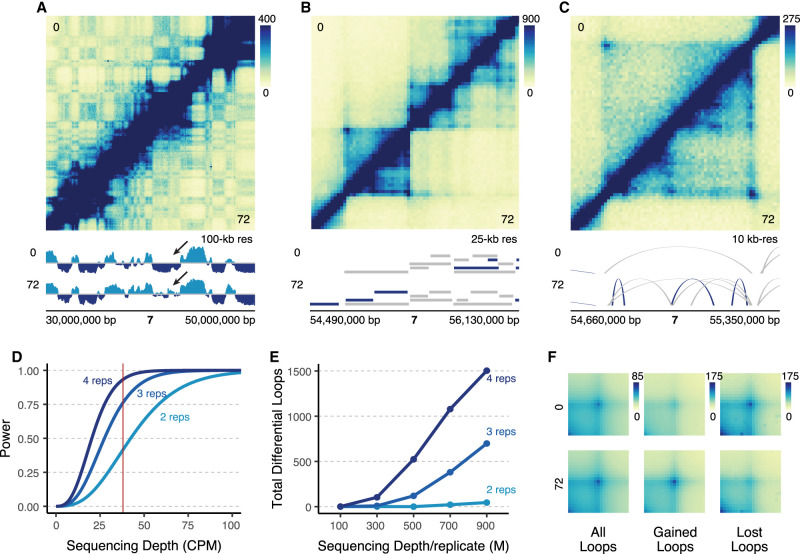
To understand how 3D chromatin structures change over cellular differentiation, we performed a deeply sequenced, three-time-point Hi-C time course tracking the differentiation of K562 cells into a megakaryocyte-like state (Huang et al. 2014). We treated K562 cells with phorbol 12-myrisate 13-acetate (PMA), which has been shown to induce a megakaryocyte-like phenotype (Whalen et al. 1997; Huang et al. 2014), for 0, 6, and 72 h. We confirmed differentiation using qPCR for ITGB3, a megakaryocyte marker (Supplemental Fig. S1A; Pencovich et al. 2011). As K562 cells differentiate into a megakaryocyte-like state, they lose their potential to differentiate into erythroid cells. We confirmed this with qPCR for KLF1, an erythroid marker, which decreases in expression over differentiation (Supplemental Fig. S1B; Kuvardina et al. 2015). We then performed in situ Hi-C on four biological replicates and sequenced them to a depth of roughly 6 billion reads per time point (Supplemental Table S1). We generated Hi-C contact maps using the Juicer pipeline (Rao et al. 2014), identified compartments using the EigenVector package (https://github.com/moshe-olshansky/EigenVector), topologically associating domains (TADs) using arrowhead (Rao et al. 2014), and chromatin loops using SIP (Supplemental Fig. S1C; Rowley et al. 2020). Replicates showed high similarity as measured by principal component analysis (PCA) (Supplemental Fig. S1D). We identified a total of 33,914 loops merged across all samples (Supplemental Table S2). Our loop calls agreed well with existing 3D chromatin and functional data. For example, 98% of the loops identified by Belaghzal were identified in our study (Supplemental Fig. S2A,B; Belaghzal et al. 2021). In addition, we found that 32% of the Perturb-seq pairs were supported by a loop in our data, and 69% of the CRISPRi pairs were supported by a loop (Supplemental Table S3; Fulco et al. 2019; Gasperini et al. 2019).
Visual inspection of the data revealed clear changes at multiple scales including nuclear compartments, TADs, and chromatin loops (Fig. 1A–C). To assess the sequencing depth and replicates required to achieve sufficient statistical power, we analyzed our data set using the RNASeqPower package (Hart et al. 2013). Using our dispersion of 0.0019, median sequencing depth of 38 counts per million (CPM) per loop, and an alpha value of 0.05 divided by 33,914 loops to account for multiple hypothesis testing, the statistical power to detect twofold changes was 0.932, which is generally considered to be well-powered (Fig. 1D; Cohen 1992). We used the dispersion from our Hi-C data to model predicted statistical power across multiple different sequencing depths and numbers of replicates (Fig. 1D). Holding sequencing depth per replicate constant, we found that decreasing to three or two replicates reduced the power estimates to 0.762 and 0.416, respectively. To determine how this increase in power translated into the number of differential loops, we subsampled our data to multiple depths and analyzed it with two, three, or all four replicates (Fig. 1E). Using our full data set of four biological replicates (∼900 million reads per replicate), we identified 1503 differential loops using DESeq2 (Love et al. 2014) with a log2(fold-change) (LFC) greater than 1.5 (adjusted P-value < 0.05) compared to only 698 and 45 identified with three or two replicates, respectively (Fig. 1E). This underscores how critical sequencing depth is in the sensitivity to detect differential loops. Finally, we confirmed the quality of these differential loops through aggregate peak analysis (Rao et al. 2014) (APA), showing that gained loops have higher contact at 72 versus 0 h, and lost loops have higher contact at 0 versus 72 h (Fig. 1F). We also identified differential loops with the alternative approach of using HiC-DC+ (Sahin et al. 2021), and found that our differential loop calls overlapped very strongly with HiC-DC+ (Supplemental Fig. S2C). The differential loops that were unique to either DESeq2 or HiC-DC+ had overall weaker P-values when compared to the differential loops that were identified by both methods (Supplemental Fig. S2D).
Figure 1.
Deeply sequenced Hi-C experiments provide sensitive detection of differential chromatin loops. (A) 20 Mb region on Chromosome 7 at 100-kb resolution comparing K562s at 0 h (top) to differentiated megakaryocytes at 72 h (bottom). Signal tracks show compartmental eigenvector calls (light blue = compartment A, dark blue = compartment B). The arrow points to qualitative changes in compartmentalization. (B) Zoom-in of a 1.6 Mb region of Chromosome 7 at 25-kb resolution. TAD calls are indicated by ranges below the Hi-C map for each cell type (dark blue = cell type–specific, gray = shared across cell types). (C) Zoom-in of a 690 kb region in B at 10-kb resolution showing a region with differential loops. Arches indicate loop calls (dark blue = cell type–specific, gray = shared across cell types). (D) Statistical power modeled across various theoretical sequencing depths at two, three, and four biological replicates. Red line indicates the median sequencing depth per loop (CPM: counts per million). (E) Actual number of differential loops called at multiple different subsampled sequencing depths for different numbers of replicates (M: millions). (F) Aggregate peak analysis for all loops, gained loops, and lost loops.
Most loops were bound by CTCF at least at one anchor, yet 26.61% of loops were not bound by CTCF. We detected an equal proportion of CTCF and non-CTCF loops at each of our time points, and in the merged map. Differential loops were more enriched for CTCF than static loops (Supplemental Fig. S3A–C). We compared the differential looping events observed between the 6 and 72 h treatments. Only 118 loops were differential between 0 and 6 h, and the majority of these were also differential between 0 and 72 h (Supplemental Fig. S3D,E). We found no significant differences in their anchor CTCF enrichment (Supplemental Fig. S3F), their significance in our DESeq2 analysis (Supplemental Fig. S3G), or their size distributions (Supplemental Fig. S3H). There was a slight increase in absolute log2(fold-change) for 72 h differential loops (Supplemental Fig. S3I). Given these mostly nonsignificant differences between 6 h and 72 h differential loops, we decided to focus our comparisons on 72 h versus 0 h.
Genes at the anchors of gained, but not lost, loops show concordant changes in expression
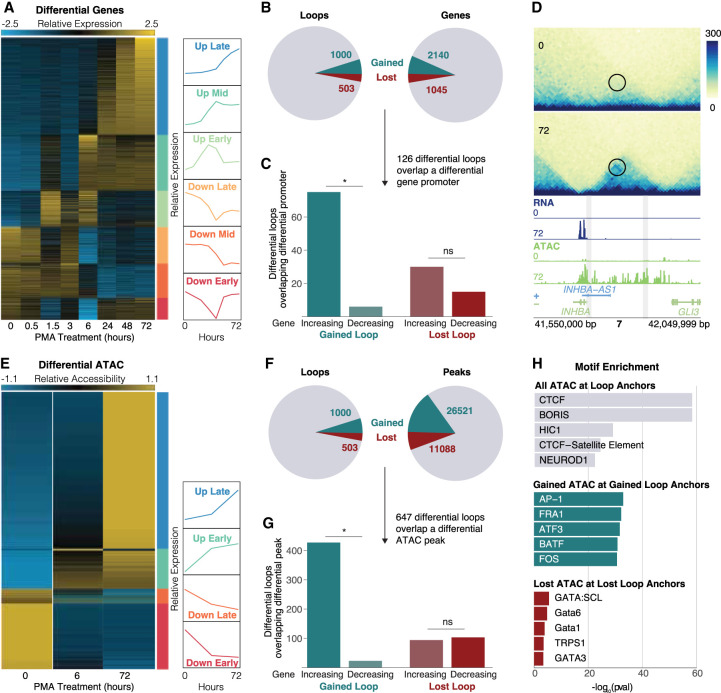
To assess the potential transcriptional impacts of differential loops, we performed RNA-seq across eight time points (0, 0.5, 1.5, 3, 6, 24, 48, and 72 h) in K562 cells treated with PMA. Gene expression levels of biological replicates showed very strong similarities (Supplemental Fig. S4A–D). DESeq2 analysis identified 3190 differential genes (adjusted P-value < 0.05, LFC > 2) which were grouped into six clusters using k-means clustering (Fig. 2A). The biggest and most unique changes were observed at 6 and 72 h (1619 differential, 527 unique genes after 6 h; 1925 differential, 236 unique genes after 72 h), which coincide exactly with our selected Hi-C time points. Consistent with the differentiation of these cells into a megakaryocyte-like state, the up-regulated genes were enriched for Gene Ontology (GO) terms relating to cell differentiation, cell adhesion, and morphogenesis, and pathways including focal adhesion, hematopoietic cell lineage, and regulation of actin cytoskeleton (Supplemental Fig. S4E,F). Up-regulated genes include megakaryocyte markers VWF, FLI1, and ITGB3, further supporting acquisition of a megakaryocyte-like phenotype.
Figure 2.
Gene expression and chromatin accessibility changes at differential loops. (A) RNA-seq normalized counts for all differential genes. Clusters are indicated by bars on the right side of the heatmap. Line plots show the mean expression per cluster. (B) Pie charts showing the proportion of differential loops (left) and genes (right). (C) Concordance analysis for the 126 differential loops that had a differential gene promoter at an anchor. Asterisk represents P < 0.05 (binomial test). (D) Example region of a gained loop with increased gene expression and chromatin accessibility at the INHBA locus. (E) ATAC-seq normalized counts for all differential peaks. Clusters indicated by bars on the right side of the heatmap. Line plots show the mean expression per cluster. (F) Pie charts showing the proportion of differential loops (left) and ATAC peaks (right). (G) Concordance analysis for the 647 differential loops that had a differential ATAC peak at a promoter. Asterisk represents P < 0.05 (binomial test). (H) TF motif enrichment analysis on all ATAC peaks at all loops (top), concordant gained ATAC peaks at gained loop anchors (middle), and concordant lost ATAC peaks at lost loop anchors (bottom).
To determine the relationship between differential loops and gene transcription, we intersected the promoters of differential genes with differential loop calls (Fig. 2B). We found that 9% (126 of 1503) of differential loop anchors overlapped a differential gene promoter. We also found that 93% (75 out of 81) of gained loops that overlapped a differential gene promoter showed the same direction of change as the gene (P = 2.91 × 10−16, binomial test) (Fig. 2C). This is consistent with our previous work in macrophages (Phanstiel et al. 2017; Reed et al. 2022) and suggests that gained loops are relevant in increasing transcription of genes at their anchors. Up-regulated genes found at the anchors of gained loops include TGFB1 and THBS1, both of which are megakaryocyte-related (Villeval and Vainchenker 2020; Wang et al. 2021). An example of a gained loop with a concordant increase in gene expression and chromatin accessibility is present at the INHBA locus (Fig. 2D).
In contrast, lost loops that overlapped a differential gene promoter showed no such concordant behavior, with only 33% (15 out of 45) showing the same directional change as the gene (P = 0.04, binomial test) (Fig. 2C). These findings again agree with our previous work in macrophages (Phanstiel et al. 2017; Reed et al. 2022), where we did not see a significant decrease in expression of genes at lost loop anchors, suggesting that loss of looping is not sufficient to decrease transcriptional output (Phanstiel et al. 2017; Reed et al. 2022). This is also consistent with work by Rao et al. that found almost no change in gene transcription following virtually complete abrogation of loop extrusion (Rao et al. 2017). Taken together, this suggests that loss of looping is generally not sufficient to induce a decrease in transcriptional output of genes at loop anchors. The anticoncordance we observed at loop anchors was also observed when looking at genes found between loop anchors; 45% of lost loops (228 out of 503) had a differential gene between their anchors, 64% of which (146 out of 228) overlapped a gene that was increasing (P = 2.69 × 10−5, binomial test) (Supplemental Fig. S4G). Genes that overlapped the interior of lost loops were expressed at significantly higher levels compared to genes within gained loops (P = 1.85 × 10−23, Wilcoxon rank-sum test) (Supplemental Fig. S4H). This also agrees with our work in macrophages (Phanstiel et al. 2017; Reed et al. 2022) showing that extremely high expression of genes within loop boundaries was associated with a weakening of the loop, suggesting that high transcription at loop interiors might antagonize loop extrusion. Indeed, several other studies provide evidence that transcription can serve as a barrier to and/or interfere with loop extrusion (Heinz et al. 2018; Brandão et al. 2019; Gu et al. 2020).
Gained loops are associated with increased accessibility at AP-1 motifs
To assess which TFs were involved in loop-based regulation, we mapped chromatin accessibility using ATAC-seq in K562 cells treated with PMA for 0, 6, and 72 h. Differential chromatin accessibility analysis with DESeq2 revealed 37,609 differential peaks (adjusted P-value, LFC > 2), which we grouped into four clusters based on whether the peak reached maximal or minimal normalized counts at 6 or 72 h (Fig. 2E). As was the case with gene expression, differential chromatin accessibility peaks were highly concordant at the anchors of gained, but not lost, loops (Fig. 2F,G).
To understand the mechanisms driving differential looping, we performed TF motif enrichment on various sets of ATAC peaks at loop anchors (Fig. 2H). As expected, CTCF was the most enriched motif at ATAC peaks overlapping all loop anchors, which is consistent with its known role in loop formation and maintenance. In contrast, peaks of gained chromatin accessibility at gained loop anchors were highly enriched for Activator Protein 1 (AP-1) family members. This is consistent with our previous work showing the enrichment of AP-1 at the anchors of gained loops during macrophage development (Phanstiel et al. 2017). In agreement with these findings, members of the AP-1 transcription factor family tended to be up-regulated early/midway during the time course (Supplemental Fig. S5A). Peaks of decreased chromatin accessibility at lost loop anchors were enriched for GATA family members, albeit to a far lesser degree compared to the enrichments observed at all or gained loops. GATA TFs have a well-established role in mediating cell fate decisions in myeloid cell development (Stachura et al. 2006; Tijssen and Ghevaert 2013; Noh et al. 2015). AP-1 family members were also found to be enriched more at proximal (promoter) than distal loop anchors. Instead, members of NF-kB were more enriched at distal loop anchors (Supplemental Fig. S5B,C).
Differential looping is associated with chromatin features both at and between loop anchors
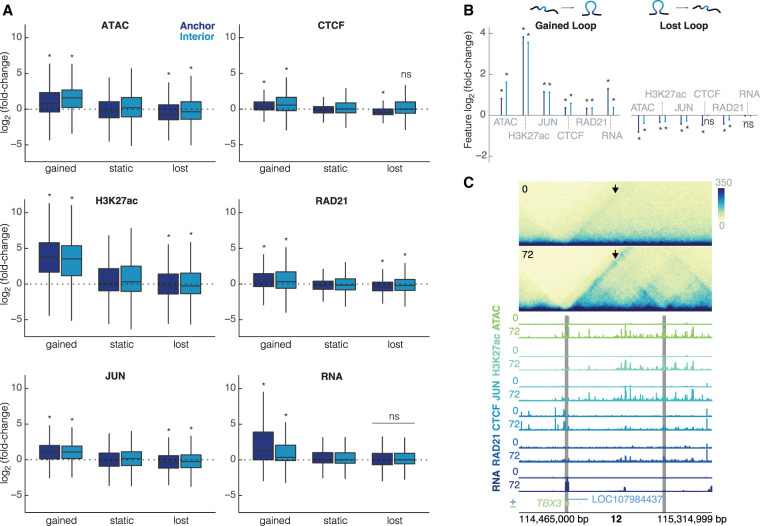
To gain further insight into the mechanisms driving differential looping, we generated matched CUT&RUN data sets for multiple TFs and histone modifications. Because of the enrichment of AP-1 motifs at the anchors of gained loops, we performed CUT&RUN for JUN, a member of the AP-1 family. We also targeted histone H3K27 acetylation, which is commonly used to identify putative enhancers, and CTCF and RAD21, proteins with known roles in DNA looping. For each assay, we performed differential analysis and compared how each differential feature corresponds to differential looping (Supplemental Fig. S6A–D). For all features, we observed statistically significant concordance at gained loop anchors (Fig. 3A,B). With the notable exception of RNA, all features were also concordant at lost loop anchors, albeit to a lesser degree.
Figure 3.
Changes in chromatin features are correlated with changes in looping. (A) Intersections of each feature at the loop anchors and interior for gained, static, and lost loops (dark blue = anchor, light blue = interior). All plots are on the same scale for the y-axis, showing log2(fold-change). Wilcoxon rank-sum test was performed for each feature to compare gained/lost anchors to static anchors and gained/lost interiors to static interiors; asterisks represent P < 0.05. (B) Median unshrunken log2(fold-change) of each data set at gained loops (left), and lost loops (right). Asterisks represent P < 0.05, dots represent P > 0.05. (C) 600-kb region around a loop at the TBX3 locus at 10-kb resolution. The arrow is pointing to the gained loop. Signal tracks for ATAC-seq, H3K27ac, JUN, CTCF, RAD21, and RNA for K562s at 0 h and differentiated megakaryocytes at 72 h show increased occupancy for all features. Gray bars indicate 10-kb loop anchors.
Investigating these trends more closely revealed two prominent features. First, although all features increased at gained loop anchors, some showed bigger changes than others. For instance, the canonical mediators of chromatin looping, CTCF and RAD21, increased by only 1.26- and 1.23-fold, respectively, at gained loop anchors. In contrast, histone H3K27ac increased nearly 14-fold at gained loop anchors (median = 13.86-fold). Chromatin accessibility and JUN binding increased by modest amounts, 1.72- and 2.16-fold, respectively. Second, although increased signals were observed at gained loop anchors, they were also observed between loop anchors. For example, chromatin accessibility and histone H3K27ac peaks within the boundaries of gained loops increased by 2.99- and 11.52-fold, respectively.
Examples of these trends are evident at the TBX3 locus (Fig. 3C). Gained enhancer-promoter looping is associated with increases in CTCF and RAD21 occupancy at loop anchors as well as large gains in chromatin accessibility and JUN occupancy at, between, and even beyond loop boundaries. Expression of TBX3 itself, a TF known to regulate developmental transitions (Weidgang et al. 2013), increases by over 50-fold. Taken together, these results suggest that differential looping may involve more than just alterations to CTCF and RAD21 occupancy. They may also be mediated by chromatin-modifying proteins and condition-specific TF binding events that act both at loop anchors and within the loop interior.
Changes in chromatin features predict changes in chromatin looping
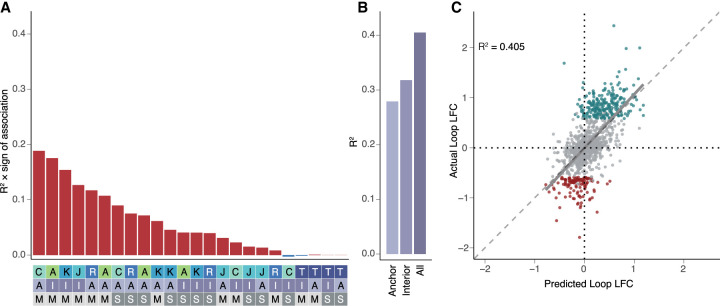
To explore this further and determine which chromatin features are the most predictive of changes in chromatin looping, we investigated how changes in each feature correlated with changes in looping (Fig. 4A). Despite the relatively small fold change of CTCF peaks at differential loop anchors, we found that CTCF occupancy changes had the highest correlation (R2 = 0.189) with loop log2(fold-change) (Fig. 4A), consistent with the role of CTCF in the formation and maintenance of loops. However, changes to multiple features in the loop “interior”—the region in between the two anchors—also strongly showed strong correlations with differential looping. Interior chromatin accessibility, histone H3K27 acetylation, and JUN occupancy had correlations of 0.175, 0.154, 0.127, followed by anchor RAD21 occupancy (R2 = 0.117). Several features showed very slight negative correlations including interior gene expression and CTCF occupancy, which is consistent with each of these having the ability to antagonize loop extrusions as previously described (Brandão et al. 2019; Jeppsson et al. 2022; Banigan et al. 2023). In addition to correlations with changes in looping, many of the features are highly correlated with each other (Supplemental Fig. S7A).
Figure 4.
Changes in chromatin features predict changes in chromatin looping. (A) R2 multiplied by the sign of association for all possible features correlated individually against changes in all loops in the model (top). Heatmap (bottom) is a legend in which the feature, position, and measure for each bar are reported (features: A = ATAC, K = H3K27ac, J = JUN, C = CTCF, R = RAD21, T = RNA; positions: A = anchor, I = interior; measures: M = max, S = sum). (B) R2 values calculated for the anchor only, interior only, or all feature models. (C) Scatterplot showing the predicted loop fold-change versus actual loop fold-change for the testing data set for looping. (Gray = static loops, teal = gained loops, maroon = lost loops, R2 calculated for all loops included in the testing data set.)
Given the relationships between changes in looping and multiple features both at and between loop anchors, we next asked if we could build a better model to predict differential looping using multiple features simultaneously. We used the caret package to perform LASSO regression (Kuhn 2008; Friedman et al. 2010). We selected 1127 differential loops and 2254 nondifferential loops that were matched for distance and contact frequency as our training set and held out 376 differential loops and 752 nondifferential loops as a testing set to evaluate our model. We then used LASSO with 10 cross-validations to generate a predictive model using various sets of features (Fig. 4A) and evaluated it by applying it to our test set. Using all anchor features improved the correlation to R2 = 0.279, far higher than using the best single feature alone (CTCF anchor max, R2 = 0.189; Fig. 4B). Using only interior features yielded an even more accurate model with an R2 of 0.317 (Fig. 4B). Combining all features at both the anchors and interiors produced the strongest predictions with an R2 of 0.405 (Fig. 4B,C). Further, the model accurately predicted the signs of 91% of gained loops and 72% of lost loops. Our predictions are much stronger for CTCF loops than non-CTCF loops, where CTCF loops had a correlation of R2 = 0.413 (Supplemental Fig. S7B) and non-CTCF loops had a much weaker correlation of R2 = 0.28 (Supplemental Fig. S7C). Additionally we used a random forest regression model (Breiman 2001) to predict changes in looping and found similar results (Supplemental Fig. S7D). Whereas there were some differences in the importance of each of the individual features, the three most important features from both the linear models and random forest were the same (anchor CTCF, interior ATAC, and interior H3K27ac) (Supplemental Fig. S7E). The predictive power of histone H3K27 acetylation, chromatin accessibility, and JUN occupancy is surprising, and especially so given that the highest correlations were observed within loop boundaries rather than at anchors themselves. This may suggest that epigenetic changes between anchor sites play a significant role in modulating loop strength. However, we cannot rule out the participation of other TFs and architectural proteins that we did not directly measure with CUT&RUN.
Changes in histone acetylation and chromatin structure predict changes in gene expression
Predicting gene expression patterns from chromatin features is a long-standing and difficult problem in the field of gene regulation. Recent advances have been made by incorporating both 2D (e.g., histone H3K27 acetylation) and 3D (e.g., Hi-C contacts) features into the activity-by-contact model of gene regulation (Fulco et al. 2019). This model has been successful in assigning enhancers to their target genes but has typically been applied to resting cells rather than biological transitions. A notable exception is the work of Beagan et al. that correlated changes to ABC score to changes in gene expression (Beagan et al. 2020) albeit at only a handful of genomic loci. We leveraged this approach to determine if chromatin dynamics could help predict changes in gene expression in a genome-wide fashion.
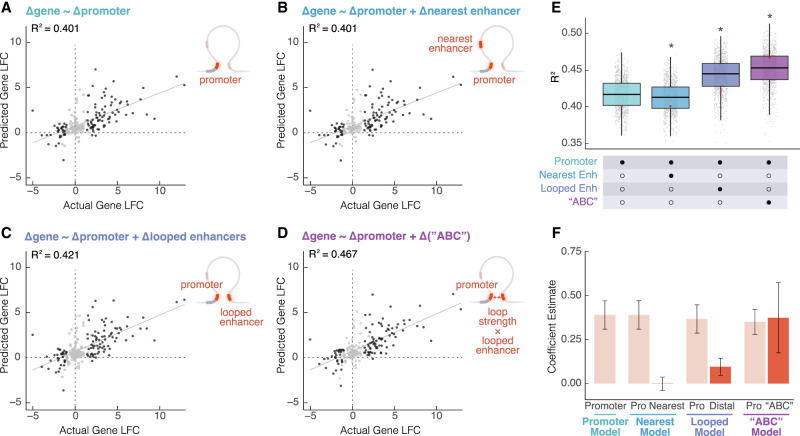
To evaluate our ability to predict changes in gene expression, we built and evaluated four linear models over 1000 permutations. In the first model, changes in gene expression were predicted based solely on changes in promoter H3K27ac, which performed well with a median R2 of 0.417 (Fig. 5A). In the second model, adding information from the nearest enhancer slightly decreased the predictive power (Wilcoxon rank-sum test on permutations, P = 1.99 × 10−6; median R2 = 0.412; Fig. 5B). In contrast, in the third model, by combining H3K27ac information from both the promoter and enhancers that were physically looped to the promoter, the median R2 increased significantly to 0.444 (Wilcoxon rank-sum test on permutations, P = 1.99 × 10−122; Fig. 5C). We then calculated a modified ABC score (see Methods) by taking the product of loop strength and distal enhancer activity for each looped enhancer-promoter pair, summing across all enhancers that were looped to each gene, and then calculating the fold change. Finally, the last linear model built using both promoter histone H3K27ac and change in ABC score increased the median R2 even further to 0.453 (Fig. 5D), a significant improvement compared to all other models (Wilcoxon rank-sum test on permutations, P = 1.95 × 10−182; Fig. 5E,F). The improvement was even more drastic when building and applying the model to specifically differential genes and differential loops. For the models built on differential loops and differential genes, R2 values improve from 0.674 (promoter only model) to 0.763 for the promoter plus ABC score model (Supplemental Fig. S8A–E). These findings suggest that alterations to both enhancer activity and contact frequency can tune transcriptional programs during cellular differentiation. At the same time, this does not rule out the participation of other TFs and their abilities to tune gene expression.
Figure 5.
Changes in gene expression are explained by combined proximal and distal enhancer activity and loop strength. Scatterplots showing predicted gene fold change versus actual gene log2(fold-change) (LFC) based on one permutation of (A) promoter H3K27ac LFC alone, (B) promoter H3K27ac LFC and the nearest enhancer to the promoter's LFC, (C) promoter H3K27ac LFC and distal looped H3K27ac, and (D) promoter H3K27ac LFC and the modified ABC LFC (dark gray = differential gene, light gray = static gene). (E) R2 for each model calculated based on 1000 permutations of splitting the data into training and testing sets. A Wilcoxon rank-sum test was performed to compare each group to the promoter only model; asterisks represent P < 0.05. Red dots represent the single permutation from A–D. (F) Estimates for each term in each model were calculated based on 1000 permutations of splitting the data into training and testing sets.
Discussion
Collection and integration of deeply sequenced Hi-C and other genomic data characterizing the differentiation of K562 cells into a megakaryocyte-like state strengthened previous findings and provided novel observations into the mechanisms and impacts of changes to 3D chromatin structure. Our results confirmed canonical roles for CTCF and RAD21 in loop establishment and were consistent with chromatin looping playing a role in transcriptional activation and/or enhancement; however, these results also revealed strong correlations between differential looping and other regulatory features including chromatin accessibility, histone H3K27 acetylation, and AP-1 occupancy, and found a lack and/or anticorrelation of gene expression at lost loops. Differential looping correlated with transcriptional and regulatory features both at and between loop boundaries. In many cases, the correlations with internal features were stronger than the correlation with anchor signals. These results raise further questions regarding the mechanisms driving differential looping and point to previously underappreciated roles of molecules other than CTCF and cohesin.
The correlation between gained loops and the expression genes at their anchors agrees with many previous studies and supports the role of chromatin looping in gene activation. The increased predictive power when incorporating acetylation dynamics of looped enhancers and the dynamics of loop strength itself further emphasize this point. In contrast, loss of looping does not coincide with decreased expression of anchor genes. In fact, more genes were increasing in expression than decreasing at the anchors of lost loops. This is consistent with our previous studies of macrophage development and activation, neither of which identified a decrease in expression at lost loop anchors (Phanstiel et al. 2017; Reed et al. 2022). Taken together, this suggests that although loops may be involved in transcriptional activation, loss of looping alone may not be sufficient for transcriptional repression or decreased expression. This agrees well with previous studies involving rapid depletion of cohesin and subsequent global loss of loop extrusion-driven loops. Rao et al. 2017 found that loop elimination did not substantially alter gene transcription—as measured directly using PRO-seq—in human colorectal cancer (HCT-116) cells. In contrast, loop disruption does inhibit activation of proinflammatory transcription in macrophages treated with LPS (Takeuchi and Akira 2010) (a bacterial cell wall component commonly used as a model for inflammatory activation), again suggesting that loops do play a role in gene activation. While it is difficult to fully reconcile these findings, the evidence seems to be mounting that although increased looping can play a role in gene activation, events beyond loop disruption are required to decrease expression levels. One possible explanation is that looping imparts some sort of regulatory memory that is not erased as soon as the loop is disrupted. Similar mechanisms (e.g., the kiss-and-run mechanism) (Karr et al. 2022) have been suggested before. While these trends are coming into focus, more functional studies are required to understand the exact role of chromatin looping in gene activation and repression.
Our data also support the theory that loop loss may be a result, rather than cause, of changes in gene transcription. Differential genes within lost loops are significantly biased toward increased expression. And those increased genes tend to be fairly highly expressed. We observed these exact same phenomena in macrophages responding to LPS (Reed et al. 2022). This is consistent with transcription occurring at a high level between loop boundaries being antagonistic to loop extrusion, something that is easy to imagine given that both involve fairly large molecular complexes traversing the same stretch of DNA. Indeed several previous studies have suggested that transcription can act as a molecular barrier to the loop extrusion process (Heinz et al. 2018; Brandão et al. 2019; Gu et al. 2020).
We revealed that changes to multiple other chromatin features (i.e., chromatin accessibility, histone H3K27 acetylation, and AP-1 occupancy) are roughly as predictive as changes to known loop extrusion-related proteins CTCF and RAD21. The correlation between looping and these other features was strongest for features within the loop interior, which may provide clues into the nature of this relationship. One possibility is that increased accessibility, histone H3K27 acetylation, and AP-1 binding might play a role in increasing the efficiency or rate of cohesin loading. This seems consistent with previous findings that suggested that cohesin loading takes place preferentially at active gene promoters (Kagey et al. 2010), which are also associated with histone acetylation, TF binding, and chromatin accessibility. It is important to acknowledge that we cannot rule out the possibility that all of these internal changes are the result, rather than cause, of differential looping events.
Despite the findings presented here, several limitations of this study must be considered when interpreting the data and speculating about their meaning. First, this study is largely correlative. Although intersecting the results of this study can provide important mechanistic insights, several of the results raise new questions that must be addressed by future functional experiments. Second, in contrast to our previous work (Reed et al. 2022), this time course lacked the temporal resolution to put regulatory and transcriptional events in temporal order, which makes it difficult to infer the direction of causality between any two features. This was a conscious decision as the current cost of sequencing makes it unfeasible to acquire deeply sequenced data sets across deeply sampled time courses; however, this is likely to change soon as sequencing costs continue to decrease (November 2018; Almogy et al. 2022). The recent development of Micro-C will also greatly benefit the resolution of future studies, thereby allowing for even greater detection of differential looping with reduced sequencing costs (Krietenstein et al. 2020). Finally, although this study encompassed a broad number of regulatory features—including ATAC-seq, which when combined with motif analysis can provide insights into the binding patterns of hundreds of TFs—there are a vast number of other features that may influence looping (e.g., DNA methylation and other architectural proteins) for which we are not measuring nor explicitly accounting for.
Despite these limitations, these findings improve our understanding of how different trans regulators and epigenetic features govern changes in looping, as well as our understanding of the relationship between looping and gene expression. The deeply sequenced nature of this differential Hi-C analysis offers a uniquely well-powered data set with which to explore a pressing number of biological questions. Moreover, these data were acquired in one of the most widely studied human cell lines (K562) for which hundreds of publicly available genome-wide data sets are already available. As such, this study provides a valuable new resource for future studies of chromatin biology.
Methods
K562 culture and differentiation
K562s were cultured in RPMI media (Corning 10-040-CV) with 10% fetal bovine serum (FBS) (Gibco 26140079) and 1% penicillin-streptomycin (PS) (Gibco 15140122). For megakaryocyte differentiation, K562s were plated in either 6-well plates (RNA-seq, ATAC-seq) or T-175 flasks (Hi-C, CUT&RUN) at a density of 1 × 105 cells/mL and treated with 25 nM PMA (Sigma-Aldrich P1585-1MG). After 24 h, the cells become semi-adherent. Cells were provided with fresh media and PMA after 24 h and 48 h. Cells were collected without treatment or after 6 or 72 h. For all treatments and library preparations, K562s were thawed and immediately split into two T-25 flasks to create biological replicates.
Genomic library preparation
Hi-C libraries were prepared according to the protocol as described in Rao et al. (2014). RNA-seq libraries were prepared using the KAPA RNA HyperPrep kit with RiboErase (HMR). ATAC-seq libraries were prepared according to the Omni ATAC-seq protocol described in Corces et al. (2017). CUT&RUN libraries were prepared following existing protocols (Skene and Henikoff 2017) modified for use with cross-linked cells. Complete descriptions of all genomic library preparations can be found in the Supplemental Material.
Genomic library processing
All genomic data were processed using in-house pipelines, utilizing Juicer, FastQC (version 0.11.5, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), MultiQC (Ewels et al. 2016), Trim Galore! (version 0.4.3, https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), Salmon (Patro et al. 2017), HISAT2 (Kim et al. 2019), SAMtools (Danecek et al. 2021), deepTools2 (Ramírez et al. 2016), txImport (version 1.2.0) (Soneson et al. 2015), BWA-MEM (Li and Durbin 2009), MACS2 (Zhang et al. 2008), and BEDTools (Quinlan and Hall 2010). Complete descriptions of all genomic library processing can be found in the Supplemental Material.
Differential analysis
Differential analysis of loops, genes, and peaks were performed with DESeq2. All significant loops/genes/peaks had an adjusted P-value < 0.05. A log2(fold-change) > 2 was used for differential genes/peaks and a log2(fold-change) > 1.5 was used for differential loops. Complete descriptions of all differential analysis can be found in the Supplemental Material.
Linear models
LASSO regression within the caret package was used to build the chromatin looping linear model. The base R function lm was used to build the gene expression linear models. Complete descriptions of all linear modeling can be found in the Supplemental Material.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under SuperSeries GSE213909. The Hi-C data are available under accession number GSE214123. The RNA-seq data are available under accession number GSE214123. The ATAC-seq data are available under accession number GSE213295. The CUT&RUN data are available under accession number GSE213908. The code to process and analyze these data is available at GitHub (https://github.com/mbond0718/MEGA2023) and as Supplemental Code.
Supplementary Material
Acknowledgments
We thank Erika Deoudes for graphic design of figures and preparation of the manuscript. We thank Sam Pattenden for allowing us to use the Covaris LE220 instrument. We thank Katie Reed for assistance with project design and intellectual discussions. We thank Nicole Kramer for the development of plotgardener and help with generating figures and data visualization. This work was supported by National Institutes of Health (NIH) Grant No. R35-GM128645 to D.H.P. M.L.B. was supported by NIH training Grant No. T32-GM135128. E.S.D. was supported by NIH Training Grant No. T32-GM067553. I.Y.Q. was supported by the BrightFocus Postdoctoral Fellowship 911831. A.D. was supported by the Institute of Eminence fellowship UoH-IoE-RC2-21-012 from the University of Hyderabad. M.K. was supported by the Science and Engineering Research Board (SERB) Start-up Research Grant No. SRG/2020/002146 and SERB International Research Experience fellowship SIR/2022/000458. M.I.L. was supported by NIH Grant No. R01-MH118349. H.W. was supported by NIH Grant No. DP2MH122403.
Author contributions: M.L.B. designed and performed most of the experiments, performed all of the computational analysis, and wrote the manuscript. E.S.D. developed some of the software used and assisted with computational analysis. I.Y.Q. prepared ATAC-seq libraries. A.D. performed random forest modeling. M.K. performed differential loop analysis with HiC-DC+ and oversaw random forest modeling. M.I.L. oversaw data analysis and linear modeling. H.W. acquired funding, supervised data analysis, and helped write the manuscript. D.H.P. acquired funding, conceptualized the project, supervised experiments and data analysis, and helped write the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.277397.122.
Competing interest statement
The authors declare no competing interests.
References
- Ahn JH, Davis ES, Daugird TA, Zhao S, Quiroga IY, Uryu H, Li J, Storey AJ, Tsai Y-H, Keeley DP, et al. 2021. Phase separation drives aberrant chromatin looping and cancer development. Nature 595: 591–595. 10.1038/s41586-021-03662-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander JM, Guan J, Li B, Maliskova L, Song M, Shen Y, Huang B, Lomvardas S, Weiner OD. 2019. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife 8: e41769. 10.7554/eLife.41769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almogy G, Pratt M, Oberstrass F, Lee L, Mazur D, Beckett N, Barad O, Soifer I, Perelman E, Etzioni Y, et al. 2022. Cost-efficient whole genome-sequencing using novel mostly natural sequencing-by-synthesis chemistry and open fluidics platform. bioRxiv 10.1101/2022.05.29.493900 [DOI]
- Banigan EJ, Tang W, van den Berg AA, Stocsits RR, Wutz G, Brandão HB, Busslinger GA, Peters J-M, Mirny LA. 2023. Transcription shapes 3D chromatin organization by interacting with loop extrusion. Proc Natl Acad Sci 120: e2210480120. 10.1073/pnas.2210480120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagan JA, Pastuzyn ED, Fernandez LR, Guo MH, Feng K, Titus KR, Chandrashekar H, Shepherd JD, Phillips-Cremins JE. 2020. Three-dimensional genome restructuring across timescales of activity-induced neuronal gene expression. Nat Neurosci 23: 707–717. 10.1038/s41593-020-0634-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaghzal H, Borrman T, Stephens AD, Lafontaine DL, Venev SV, Weng Z, Marko JF, Dekker J. 2021. Liquid chromatin Hi-C characterizes compartment-dependent chromatin interaction dynamics. Nat Genet 53: 367–378. 10.1038/s41588-021-00784-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão HB, Paul P, van den Berg AA, Rudner DZ, Wang X, Mirny LA. 2019. RNA polymerases as moving barriers to condensin loop extrusion. Proc Natl Acad Sci 116: 20489–20499. 10.1073/pnas.1907009116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. 2001. Random forests. Mach Learn 45: 5–32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- Cohen J. 1992. A power primer. Psychol Bull 112: 155–159. 10.1037/0033-2909.112.1.155 [DOI] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. 2017. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14: 959–962. 10.1038/nmeth.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. 2021. Twelve years of SAMtools and BCFtools. GigaScience 10: giab008. 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Rupon JW, Krivega I, Breda L, Motta I, Jahn KS, Reik A, Gregory PD, Rivella S, Dean A, et al. 2014. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell 158: 849–860. 10.1016/j.cell.2014.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331–336. 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, Käller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32: 3047–3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. 2010. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33: 1–22. 10.18637/jss.v033.i01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco CP, Nasser J, Jones TR, Munson G, Bergman DT, Subramanian V, Grossman SR, Anyoha R, Doughty BR, Patwardhan TA, et al. 2019. Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nat Genet 51: 1664–1669. 10.1038/s41588-019-0538-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini M, Hill AJ, McFaline-Figueroa JL, Martin B, Kim S, Zhang MD, Jackson D, Leith A, Schreiber J, Noble WS, et al. 2019. A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell 176: 377–390.e19. 10.1016/j.cell.2018.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubert F, Srivas R, Spacek DV, Kasowski M, Ruiz-Velasco M, Sinnott-Armstrong N, Greenside P, Narasimha A, Liu Q, Geller B, et al. 2020. Landscape of cohesin-mediated chromatin loops in the human genome. Nature 583: 737–743. 10.1038/s41586-020-2151-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Comerci CJ, McCarthy DG, Saurabh S, Moerner WE, Wysocka J. 2020. Opposing effects of cohesin and transcription on CTCF organization revealed by super-resolution imaging. Mol Cell 80: 699–711.e7. 10.1016/j.molcel.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart SN, Therneau TM, Zhang Y, Poland GA, Kocher J-P. 2013. Calculating sample size estimates for RNA sequencing data. J Comput Biol 20: 970–978. 10.1089/cmb.2012.0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Texari L, Hayes MGB, Urbanowski M, Chang MW, Givarkes N, Rialdi A, White KM, Albrecht RA, Pache L, et al. 2018. Transcription elongation can affect genome 3D structure. Cell 174: 1522–1536.e22. 10.1016/j.cell.2018.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Won H, Mah W, Park RB, Kassim B, Spiess K, Kozlenkov A, Crowley CA, Pochareddy S, Ashley-Koch AE, et al. 2021. Neuronal and glial 3D chromatin architecture informs the cellular etiology of brain disorders. Nat Commun 12: 3968. 10.1038/s41467-021-24243-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Zhao L, Chen H, Yin R-H, Li C-Y, Zhan Y-Q, Zhang J-H, Ge C-H, Yu M, Yang X-M. 2014. Megakaryocytic differentiation of K562 cells induced by PMA reduced the activity of respiratory chain complex IV. PLoS One 9: e96246. 10.1371/journal.pone.0096246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppsson K, Sakata T, Nakato R, Milanova S, Shirahige K, Björkegren C. 2022. Cohesin-dependent chromosome loop extrusion is limited by transcription and stalled replication forks. Sci Adv 8: eabn7063. 10.1126/sciadv.abn7063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. 10.1038/nature09380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karr JP, Ferrie JJ, Tjian R, Darzacq X. 2022. The transcription factor activity gradient (TAG) model: contemplating a contact-independent mechanism for enhancer-promoter communication. Genes Dev 36: 7–16. 10.1101/gad.349160.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Dean A. 2012. Chromatin loop formation in the β-globin locus and its role in globin gene transcription. Mol Cells 34: 1–5. 10.1007/s10059-012-0048-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon A, Shih L-Y, Minamino M, Sanada M, Shiraishi Y, Nagata Y, Yoshida K, Okuno Y, Bando M, Nakato R, et al. 2013. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet 45: 1232–1237. 10.1038/ng.2731 [DOI] [PubMed] [Google Scholar]
- Krietenstein N, Abraham S, Venev SV, Abdennur N, Gibcus J, Hsieh T-HS, Parsi KM, Yang L, Maehr R, Mirny LA, et al. 2020. Ultrastructural details of mammalian chromosome architecture. Mol Cell 78: 554–565.e7. 10.1016/j.molcel.2020.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivega I, Dean A. 2017. LDB1-mediated enhancer looping can be established independent of mediator and cohesin. Nucleic Acids Res 45: 8255–8268. 10.1093/nar/gkx433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M. 2008. Building predictive models in R using the caret package. J Stat Softw 28: 1–26. 10.18637/jss.v028.i0527774042 [DOI] [Google Scholar]
- Kuvardina ON, Herglotz J, Kolodziej S, Kohrs N, Herkt S, Wojcik B, Oellerich T, Corso J, Behrens K, Kumar A, et al. 2015. RUNX1 represses the erythroid gene expression program during megakaryocytic differentiation. Blood 125: 3570–3579. 10.1182/blood-2014-11-610519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJH, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. 2003. A long-range Shh enhancer regulates expression in the developing limb and Fin and is associated with preaxial polydactyly. Hum Mol Genet 12: 1725–1735. 10.1093/hmg/ddg180 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short-read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan K, Horta A, Lomvardas S. 2019. LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice. Nature 565: 448–453. 10.1038/s41586-018-0845-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh J-Y, Gandre-Babbe S, Wang Y, Hayes V, Yao Y, Gadue P, Sullivan SK, Chou ST, Machlus KR, Italiano JE, et al. 2015. Inducible Gata1 suppression expands megakaryocyte-erythroid progenitors from embryonic stem cells. J Clin Invest 125: 2369–2374. 10.1172/JCI77670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- November J. 2018. More than Moore's mores: computers, genomics, and the embrace of innovation. J Hist Biol 51: 807–840. 10.1007/s10739-018-9539-6 [DOI] [PubMed] [Google Scholar]
- Paliou C, Guckelberger P, Schöpflin R, Heinrich V, Esposito A, Chiariello AM, Bianco S, Annunziatella C, Helmuth J, Haas S, et al. 2019. Preformed chromatin topology assists transcriptional robustness of Shh during limb development. Proc Natl Acad Sci USA 116: 12390–12399. 10.1073/pnas.1900672116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panarotto M, Davidson IF, Litos G, Schleiffer A, Peters J-M. 2022. Cornelia de Lange syndrome mutations in NIPBL can impair cohesin-mediated DNA loop extrusion. Proc Natl Acad Sci 119: e2201029119. 10.1073/pnas.2201029119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencovich N, Jaschek R, Tanay A, Groner Y. 2011. Dynamic combinatorial interactions of RUNX1 and cooperating partners regulates megakaryocytic differentiation in cell line models. Blood 117: e1–e14. 10.1182/blood-2010-07-295113 [DOI] [PubMed] [Google Scholar]
- Phanstiel DH, Van Bortle K, Spacek D, Hess GT, Shamim MS, Machol I, Love MI, Aiden EL, Bassik MC, Snyder MP. 2017. Static and dynamic DNA loops form AP-1-bound activation hubs during macrophage development. Mol Cell 67: 1037–1048.e6. 10.1016/j.molcel.2017.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang S-C, St Hilaire BG, Engreitz JM, Perez EM, Kieffer-Kwon K-R, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. 2017. Cohesin loss eliminates all loop domains. Cell 171: 305–320.e24. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed KSM, Davis ES, Bond ML, Cabrera A, Thulson E, Quiroga IY, Cassel S, Woolery KT, Hilton I, Won H, et al. 2022. Temporal analysis suggests a reciprocal relationship between 3D chromatin structure and transcription. Cell Rep 41: 111567. 10.1016/j.celrep.2022.111567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Poulet A, Nichols MH, Bixler BJ, Sanborn AL, Brouhard EA, Hermetz K, Linsenbaum H, Csankovszki G, Lieberman Aiden E, et al. 2020. Analysis of Hi-C data using SIP effectively identifies loops in organisms from C. elegans to mammals. Genome Res 30: 447–458. 10.1101/gr.257832.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupon JW, Deng W, Wang H, Gregory PD, Reik A, Dean A, Blobel GA. 2013. Using forced chromatin looping to overcome developmental silencing of embryonic and fetal β-type globin genes in adult erythroid cells. Blood 122: 433. 10.1182/blood.V122.21.433.433 [DOI] [Google Scholar]
- Sahin M, Wong W, Zhan Y, Van Deynze K, Koche R, Leslie CS. 2021. HiC-DC+ enables systematic 3D interaction calls and differential analysis for Hi-C and HiChIP. Nat Commun 12: 3366. 10.1038/s41467-021-23749-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. 2015. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci 112: E6456–E6465. 10.1073/pnas.1518552112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sey NYA, Hu B, Iskhakova M, Lee S, Sun H, Shokrian N, Hutta GB, Marks JA, Quach BC, Johnson EO, et al. 2022. Chromatin architecture in addiction circuitry identifies risk genes and potential biological mechanisms underlying cigarette smoking and alcohol use traits. Mol Psychiatry 27: 3085–3094. 10.1038/s41380-022-01558-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6: e21856. 10.7554/eLife.21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI, Robinson MD. 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4: 1521. 10.12688/f1000research.7563.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachura DL, Chou ST, Weiss MJ. 2006. Early block to erythromegakaryocytic development conferred by loss of transcription factor GATA-1. Blood 107: 87–97. 10.1182/blood-2005-07-2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140: 805–820. 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- Tijssen MR, Ghevaert C. 2013. Transcription factors in late megakaryopoiesis and related platelet disorders. J Thromb Haemost 11: 593–604. 10.1111/jth.12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeval J-L, Vainchenker W. 2020. Megakaryocytes tame erythropoiesis with TGFβ1. Blood 136: 1016–1017. 10.1182/blood.2020006906 [DOI] [PubMed] [Google Scholar]
- Wang H, He J, Xu C, Chen X, Yang H, Shi S, Liu C, Zeng Y, Wu D, Bai Z, et al. 2021. Decoding human megakaryocyte development. Cell Stem Cell 28: 535–549.e8. 10.1016/j.stem.2020.11.006 [DOI] [PubMed] [Google Scholar]
- Weidgang CE, Russell R, Tata PR, Kühl SJ, Illing A, Müller M, Lin Q, Brunner C, Boeckers TM, Bauer K, et al. 2013. TBX3 directs cell-fate decision toward mesendoderm. Stem Cell Reports 1: 248–265. 10.1016/j.stemcr.2013.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen AM, Galasinski SC, Shapiro PS, Nahreini TS, Ahn NG. 1997. Megakaryocytic differentiation induced by constitutive activation of mitogen-activated protein kinase kinase. Mol Cell Biol 17: 1947–1958. 10.1128/MCB.17.4.1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.