Abstract
The mass production of the graphics processing unit and the coronavirus disease 2019 (COVID-19) pandemic have provided the means and the motivation, respectively, for rapid developments in artificial intelligence (AI) and medical imaging techniques. This has led to new opportunities to improve patient care but also new challenges that must be overcome before these techniques are put into practice. In particular, early AI models reported high performances but failed to perform as well on new data. However, these mistakes motivated further innovation focused on developing models that were not only accurate but also stable and generalizable to new data. The recent developments in AI in response to the COVID-19 pandemic will reap future dividends by facilitating, expediting, and informing other medical AI applications and educating the broad academic audience on the topic. Furthermore, AI research on imaging animal models of infectious diseases offers a unique problem space that can fill in evidence gaps that exist in clinical infectious disease research. Here, we aim to provide a focused assessment of the AI techniques leveraged in the infectious disease imaging research space, highlight the unique challenges, and discuss burgeoning solutions.
Keywords: artificial intelligence, AI, imaging, infectious disease
Artificial intelligence has shown great potential to accelerate infectious disease imaging research and improve clinical care. However significant challenges must be solved to ensure that these emerging tools are used responsibly, prioritizing patient well-being and scientific rigor.
The global burden of the coronavirus disease 2019 (COVID-19) pandemic has led to an unprecedented acceleration of data science research focused on digital clinical data and medical imaging. Patients with COVID-19 develop a spectrum of lung abnormalities, ranging from ground-glass opacities and crazy paving (interlobular septal thickening) to lung consolidation leading to acute respiratory distress syndrome. In addition to radiography and computed tomography, acute and chronic cardiopulmonary manifestations of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection may also be imaged with cardiac magnetic resonance (MR) imaging and ultrasonography. In the context of the rapidly changing pandemic, the data science community broadly applied emerging artificial intelligence (AI) tools toward quantitative and semiautomated measurement, classification, and interpretation of medical images. This commonly included fundamental models for deep-learning-based image segmentation and machine learning (ML) classification of disease.
Unfortunately, early fervor and broad speculation outpaced practical and validated deployment of impactful models that were generalizable and not overfit to a geographic area, demographic group, disease stage, or genetic variant. The number of research papers and models in this field grew quickly, and a growing need emerged to review and assess the current status and impact of efforts. AI tools have been proposed for assisting radiologist workflows in resource-challenged settings, optimizing triage, or quantifying disease severity. However, it is generally agreed in retrospect that reported model performances may be misrepresented and may not generalize to larger more diverse populations, disease settings, and geographies.
Although the application of AI methods to imaging patients with COVID-19 may prove important in the current pandemic, the major scientific impact of this acceleration and lessons learned may well be felt indirectly in future broader applications to other infectious diseases, each with their own disease phenotype. It is in this space that AI methods have been less successfully applied in the past. The common translational paradigm, from bench to preclinical model to the patient, does not always apply to the high-consequence infectious disease setting. Whereas the human outbreak setting informs epidemiology, transmission, infection dynamics, and early characterization of human disease, preclinical models are often required to interrogate pathophysiology, mechanisms of disease, and the early development of therapeutic countermeasures.
The application of AI methods to preclinical and clinical imaging shares some common features and challenges, though the unique properties and often varying goals present specific challenges that will be discussed in this review article. ML methods require high-quality training data sets and often need time-intensive and human resource–intensive manual definition of disease abnormalities or features in medical images. Preclinical modeling and imaging present a unique opportunity for rigorously controlled experiments, with preinfection baseline followed by serial imaging uniquely enabling longitudinal assessment of disease. This enables a focus on specific hypotheses and can restrict the influence of confounding factors.
SCOPE AND APPROACH
We aim to provide an overview and assessment of the AI techniques leveraged in the infectious disease imaging research space. We also aim to highlight the unique challenges of this research space as it applies to humans and animal models of disease. This review will focus on organ–scale imaging (eg, chest radiography, computed tomography [CT], MR imaging, and positron emission tomography [PET]) in the context of infectious disease research. Literature searches were conducted using PubMed and Google Scholar. with attention to both preclinical and clinical applications and models. Key terms used throughout the article are defined in Table 1.
Table 1.
Definitions of Key Terms
| Term | Definition |
|---|---|
| AI | Simulation of human intelligence in machines |
| Artificial neural networks | Algorithm inspired by biological neural networks in which interconnected neurons process information |
| Convolutional neural networks | Type of neural network that uses a series of learnable convolutional layers to distill spatial features from imaging data |
| Deep learning | A subset of ML, algorithms that use an artificial neural network to extract high-level features from data; these methods can be used to distill complex data types, such as images and text for predictive tasks |
| Labeled data | Data that include class labels; for example, if the task is to predict the fruit name (class label) given the color and shape, the labeled data would include the fruit name, color, and shape |
| Low/poor-quality labeled data | A labeled data set wherein the label is not accurate for some data points |
| ML | A subset of AI, algorithms that learn without explicit instructions |
| Model generalizability | The ability for a model to perform well on new data it has not been trained on |
| Partially labeled data | A data set that includes some combination of labeled and unlabeled data |
| Preclinical model | Nonhuman (typically animal) model of a disease in humans |
| Self-supervised learning | A type of ML in which the algorithm learns from unlabeled data to form representations; the representations can be used later to better complete a more useful downstream task |
| Supervised learning | A type of ML in which the algorithm learns from labeled data to produce the label for new data |
| Traditional/classic ML | A subset of ML, algorithms that learn from structured (tabular) data |
| Unlabeled data | Data that do not include class labels; for example, if the task is to predict the fruit name (class label) given the color and shape, the unlabeled data would include only the fruit color and shape |
| Weakly supervised or semi-supervised learning | A type of ML that falls between self-supervised and supervised learning, in which a small amount of labeled and a large amount of unlabeled data are used for model training |
Abbreviations: AI, artificial intelligence; ML, machine learning.
EARLY APPLICATIONS OF AI IN INFECTIOUS DISEASE RESEARCH
One of the earliest attempts at a computer-based clinical decision support system for infectious diseases was MYCIN, which used >500 rules to determine the bacterial species responsible for an infection, provide a diagnosis, and recommend an antibody regimen [1]. However, it eventually became clear that the decision process clinicians make is far too complex to encode in explicitly defined rules. Furthermore, inputting a long series of answers to questions was not easily integrated into clinical practice [2]. ML has the potential to solve many of these issues by using large amounts of data to automatically derive a logic system for providing clinical decision support. Initial applications of AI to aid in clinical decision making on infectious diseases focused on structured and easily accessible types of medical data, including vital signs, laboratory measures, demographics, medical history, and physical examination data [2]. More complex (unstructured) data types, such as images, particularly 3-dimensional (3D) and time–series images, are difficult to quantify and thus require specialized AI methods to take full advantage of the wealth of information hidden within.
CURRENT AI APPROACHES FOR THE STUDY OF INFECTIOUS DISEASES USING IMAGING
As AI computer vision techniques have grown, so have their applications to infectious disease imaging. Imaging tasks that have been automated using AI have been directed toward 2 fundamental questions: “Where is it?” (segmentation) and “What is it?” (classification). Segmentation is the process of identifying the pixels in an image that correspond with a region of interest. Once an image is segmented, further analyses can be focused on specific organs or tissues. Manual segmentation of images is a time-consuming process, particularly for high-field-of-view and high-resolution 3D images commonly produced by modern medical imaging modalities, such as CT and MR imaging. Manual segmentation is even more time-consuming for modalities that image over time, such as functional MR imaging and PET. Therefore, the development of reliable automated segmentation methods is critical to advancing infectious disease imaging research and improving clinical practice.
With the advent of deep learning and enhanced computing resources, AI techniques have grown in popularity as an effective method to automatically segment images and have been applied across a wide range of infectious diseases. A probabilistic information method [3] was used to segment different regions of the whole brain to map abnormal subcortical brain morphometry in a human immunodeficiency virus (HIV) study [4]. A deep-learning-based method for tuberculosis detection and segmentation in chest radiographs was also used [5]. More recently, a deep-learning-based method was developed to segment the liver in CT images of animal models of Ebola and Marburg virus, Lassa virus, and Nipah virus infections [6]. In this study, Reza and colleagues [6] found that a feature pyramid network model could segment the liver from a CT scan with a dice score of 95%. During the COVID-19 pandemic, many automated lung lesions segmentation methods of CT scans and radiographs have been proposed, including the dual-branch combination network [7], semisupervised Inf-Net [8], slice-based 2-dimensional UNet [9], Dense-UNet [10], encoder-decoder-based attention network [11], dual-sampling attention network [12], and many similar convolutional neural network (CNN)–based methods [13].
In the context of infectious disease imaging, classification commonly involves predicting the infection status, disease severity or stage, or response to therapeutic intervention. Classification of images can be performed in segmented regions or directly on the original image. Traditional ML algorithms, such as logistic regression, support vector machine, k-nearest neighbors, naive Bayes, linear discriminant analysis, and tree-based algorithms, can be used to classify images, but the unstructured data must first be converted into a structured data format (ie, a table) through the calculation of descriptive features. Image features can be quantified using simple metrics, such as volume or mean intensity (eg, volume and mean intensity of a lesion). In addition, more complex metrics, such as radiomic [14] features, can be calculated to quantify shapes and textures found within the image.
Studies of SARS-CoV-2 [15–19] and other infectious lung diseases [20] have successfully deployed traditional ML algorithms for classifying images using radiomic features. AI approaches that have been applied to the imaging of infectious diseases have ranged in complexity from simple algorithms applied to structured data sets to the more complex deep-learning algorithms that excel in making predictions from unstructured data sets, including large imaging data sets. Deep-learning algorithms, such as CNNs, have been widely used in infectious disease research, including for the classification (ie, detection) of COVID-19, pneumonia, and pulmonary tuberculosis [21–23].
CNNs have proved useful in a diverse array of studies related to infectious disease research and medical imaging; however, the introduction of the vision transformer model, has had a great impact in these domains and seems to suggest a new era in deep learning [24,25]. This model has been shown to outperform CNN models and ensemble approaches trained on binary and multiclass scan data sets [26]. Similarly, a multitask vision transformer model was trained to perform both diagnostics and severity prediction of patients with COVID-19 [27]. When tasked with classifying radiographs as normal, COVID-19, or other infection, the model performed with areas under the curve of 0.932, 0.947, and 0.928; sensitivities of 83.4%, 88.4%, and 85.4%; and accuracies of 83.8%, 84.9%, and 86.9% on a set of 3 external data sets [26].
Challenge: Disease Specificity
Imaging techniques, such as CT and MR imaging, can produce high-resolution 3D images with intensities that relate to the atomic density of the tissue and the nuclear (usually hydrogen, used as a proxy for water) density within the tissue, respectively. Typically, viral infection either causes an inflammatory immune response (leading to an increase in tissue density) or causes cell death (leading to a reduction in tissue density). As a result, ML models have performed well when trained to detect infections from medical images in areas such as the lungs [28–30]. However, changes in tissue density (captured by CT, structural MR imaging, and ultrasonography), brain blood flow (captured by functional MR imaging), and metabolic activity (captured by [18F]Fluorodeoxyglucose PET) are secondary effects of infection and thus not pathogen specific. As a result, AI models trained only on in vivo imaging modalities are limited in their potential performance as biomarkers of infectious disease progression. Of note, some PET tracers have been developed that aim to be bacteria specific, but they have been largely restricted to preclinical applications [31–34]. Building AI models that are trained on data that contain both physical and biological properties (eg, CT and blood biomarkers, or MR imaging and PET) would enable more specific models of disease that better generalize across the spectrum of infectious pathogens.
Challenge: Data Scarcity
AI models using medical images, particularly neural networks, are large and complex and thus require a large number of labeled samples to train adequately. This is a prominent challenge, particularly in the infectious disease imaging research space. Data scarcity is a function of multiple factors, including the prevalence of the disease, severity of the disease, duration of the disease, difficulty of the labeling task, prevalence of experts that can perform labeling, data-sharing hurdles, and privacy regulations. As a result, human data are more available for some diseases (eg, COVID-19), while in other diseases, data from animal models of disease are more available (eg, Ebola virus disease, Nipah virus disease, and Lassa fever). Methods such as self-supervised, semisupervised, and weakly supervised learning aim to provide robust models trained on unlabeled, partially labeled, or low-quality labeled data. Self-supervised learning, which has been used to pretrain a range of different attention models, has proved especially effective at leveraging unlabeled data toward significant improvements in model generalization and transfer learning [35].
Self-supervision, wherein a model learns a robust representation of a domain (eg, the English language), rather than task-specific information, has yielded significant improvements in model performance on complex downstream tasks involving small annotated data sets [36,37]. After the pretraining stage is complete, the models can be fine-tuned on the domain-specific data set, and the learned representation can be shifted slightly to facilitate performance on a complex task, while preserving the robust features learned from the large unlabeled training set [38–40] (Figure 1). Infectious disease researchers can deploy various self-supervision techniques to use the large quantities of unlabeled data sets (including those from other, related domains) that are often readily available. Both vision transformers and deep CNN models used for COVID-19 classification or detection are very often pretrained on large public data sets that include medical images as well as images from across many different categories (ie, ImageNet) [26,27,41–43]. Without this pretraining, the models would likely be overfitted to task-specific data sets owing to the scarcity of data.
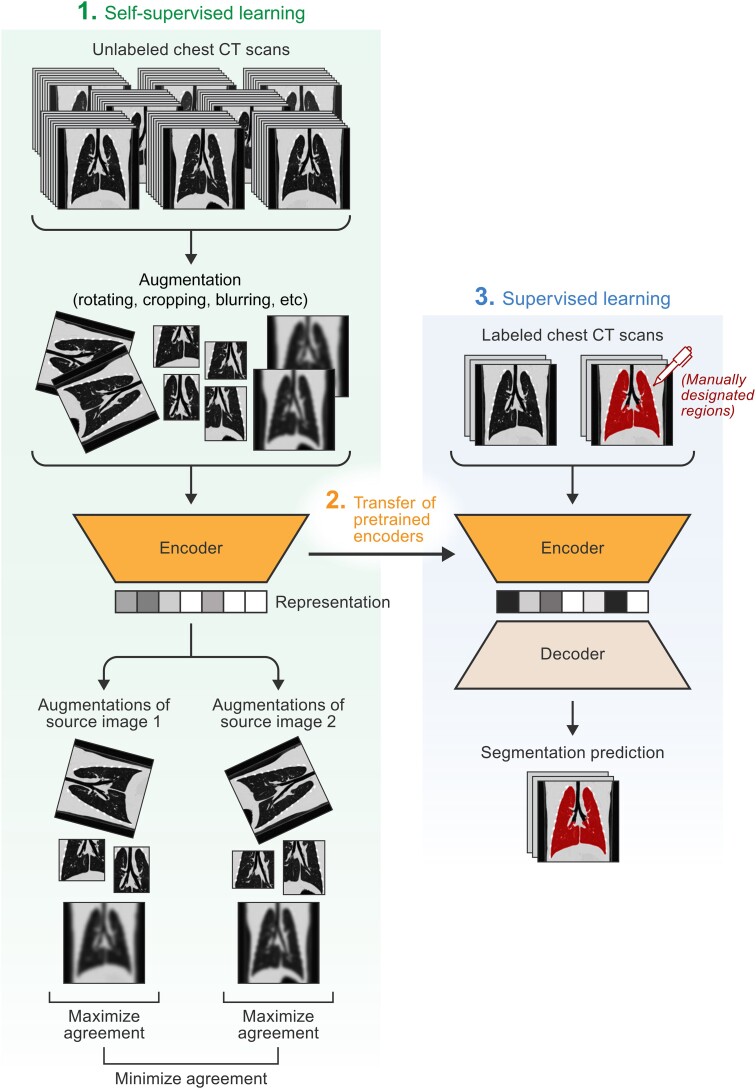
Figure 1.
Self-supervised learning for medical image segmentation. The diagram describes an implementation of how unlabeled computed tomographic (CT) scans and self-supervised learning (specifically contrastive learning) can be used to enhance the performance of a supervised learning segmentation model. First, unlabeled scans are augmented using simple transformations, such as cropping, rotation, and blurring. These augmented scans are inputted into the self-supervised model, and the model is tasked with distinguishing augmented images that come from the same source image from augmented images that come from different images (ie, pretask). After training, the pretrained encoders can be transferred to a supervised learning model, which is given a small batch of labeled scans and tasked with producing the segmentation masks. Pretraining with a self-supervised learning task has been shown to enhance the performance of supervised learning models.
In addition to algorithmic improvements to accomplish higher performance with less training data, some have turned their focus on improving the quality of the data used to train models. This approach, termed data-centric AI, focuses on using domain knowledge and systematic processes to remove poor-quality data points and design the input features to guide the model to be more robust and generalizable. DataPerf is a recently developed benchmark suite for ML data sets that aims to implement data-centric AI principles [44].
Challenge: Explainable AI
Most deep-learning models do not explain their predictions in a way that humans can understand [45]. For example, in CNNs, the convolutional layers are adjusted through the training process to deconstruct the image into relevant features. However, the trained weights of the convolutional layers are not organized into human-interpretable concepts (eg, shapes and textures) making the inner logic of the model unknown (Figure 2). Such black-box models may not be safe to use in high-stake applications, such as medical image diagnosis [46,47]. It has been demonstrated that current AI systems can easily be fooled: a small, carefully designed change in how inputs are presented to an AI system can completely change diagnostic performance (eg, from a benign to a malignant diagnostic decision when rotating the input image a few degrees or putting a small amount of noise into the image) [48].
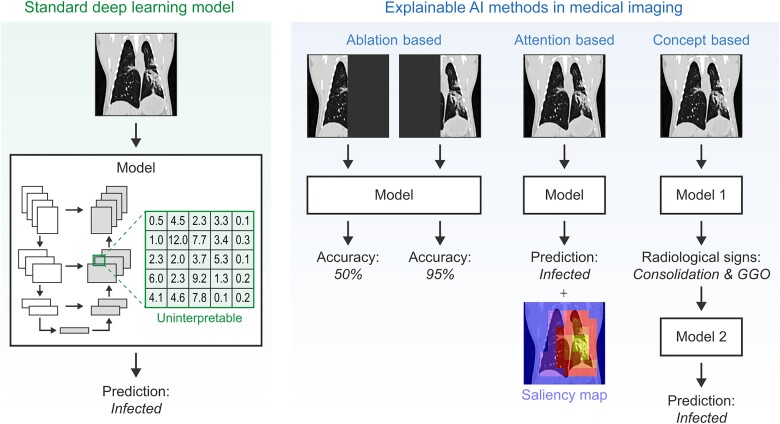
Figure 2.
Explainable artificial intelligence (AI) methods in medical imaging. Standard deep learning models are uninterpretable and therefore work as a “black box.” Explainable AI methods, such as ablation-based, attention-based, and concept-based methods, provide clinicians and researchers with additional information about how the model forms its predictions. Abbreviation: GGO, ground-glass opacities.
Without the ability to understand the reasoning behind a radiological prediction, radiologists are unlikely to trust and adopt deep-learning models. This interpretability barrier is a critical challenge that AI researchers must overcome before these predictive models can be applied responsibly and adopted into clinical practice. Uninterpretable algorithms are still useful in some applications, such as the knowledge discovery process and the creation of baselines for performance comparison. However, uninterpretable AI models could have catastrophic consequences, such as severe impediments in therapy planning, intervention, and healthcare costs, [46,49,50].
It should be noted that there is no all-purpose definition for explainability or interpretability because the proper application of the concept is domain specific [47,51,52]. A large number of interpretable prediction studies exist in the literature [47,53–111], but most provide explanations that are not faithful to what the original model computes. As agreed by pioneers in the field of deep learning, including Rudin, G. Marcus, Schölkopf, Doshi-Velez, and several others [112–118], the currently available methods in the literature tend to present “interpretable” AI models in a misleading way such that the underlying mechanisms are not faithfully revealed.
Studies exploring how CNNs make predictions are typically done through post hoc interpretation techniques, but these do not provide a true (fully transparent) explanation. For instance, some studies remove parts of an image (pixels or regions) to determine their impact on the final prediction (called perturbation-based or ablation-based methods) [119–121]. These methods do not reflect built-in explainability, and their interpretations fail for several reasons. For instance, perturbation-based methods assume that the model trained on the ablated data set follows a similar process to the model trained on the full data set; however, deep-learning models are known to vary widely as a result of subtle changes in the training data set [48].
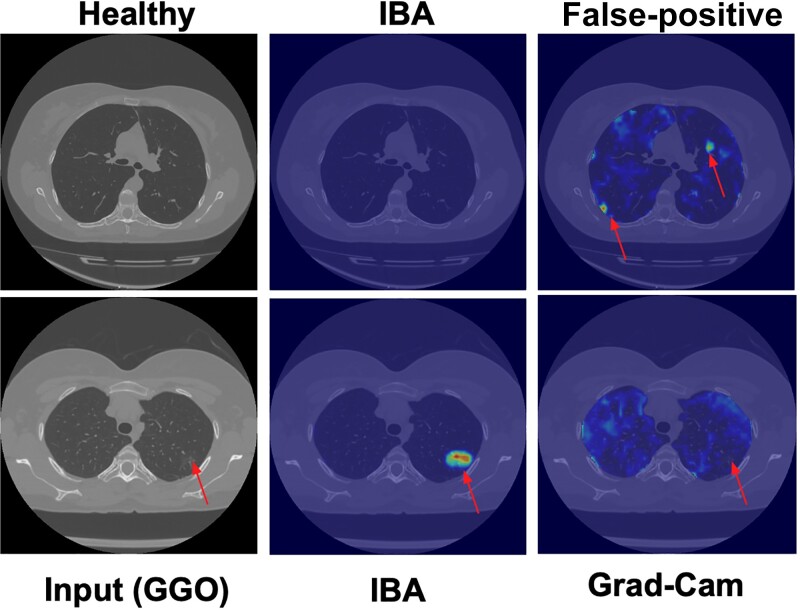
A second major group of post hoc interpretation techniques uses neuron activation maps to discover attention (eg, CAM and Grad-CAM) (called localization-based or attention-based methods) [122,123] or looks at the interpretability of individual neurons [56,124]. It has been shown that attention and gradient information are often uncorrelated, with many different attention maps yielding identical results, while others have shown that removing visually interpretable neurons versus uninterpretable ones had no measurable effect on network prediction accuracy [125]. Although Grad-CAM is becoming the de facto visualization method, it has been noted by several researchers that Grad-CAM is very sensitive to noise and is not a completely reliable technique. It has been shown that an alternative approach, based on information bottleneck attribution (IBA) [126], is far superior to the widely adopted Grad-CAM approach [123]. The study tested >1100 CT scans of patients with varying levels of COVID-19 severity and without dense annotations and found that IBA had minimal false-positive regions and was superior to Grad-CAM in >95% of the visual evaluations. Figure 3 shows the accurate localization of the IBA approach and failures of Grad-CAM.
Figure 3.
Grad-CAM compared with information bottleneck attribution (IBA) attention maps. Left, Computed tomographic (CT) scan with subtle ground-glass opacities (GGO) pattern. The proposed IBA shows the exact location of pathology without false-positives and precisely, while Grad-CAM fails. Arrows on the first row are pointing to areas within the image that are not true lesions but false positives predicted by Grad-Cam. Arrows on the bottom row point to true lesions not detected by Grad-Cam.
In medical imaging of infectious diseases, many studies applied “explainable” models with varying levels of explainability. Methods such as Grad-CAM [123] and other direct gradient approaches have been applied to infectious disease imaging [127,128]. For example, one study developed a robust deep-learning model for COVID-19 detection/characterization on CT scans from a diverse multinational cohort [129]. The model achieved 92.4% accuracy in the validation set and 90.8% in the independent test set for the COVID-19 diagnosis. The Grad-CAM algorithm was used to highlight the pathological regions that the algorithm learns from. While the accuracy was high, the Grad-CAM saliency maps did not reveal intuitive attention trends.
Beyond commonly considered visual interpretation methods, some researchers have proposed improving the interpretability of deep-learning models by embedding radiographic interpretations (concept-based explainable AI). The idea is to learn the radiographic explanations of the object of interest (eg, pathological region) from radiologists and train the deep-learning architecture to learn these features in addition to the main outcome (eg, classification). For instance, the explainable capsule network (X-Caps) study [130] describes a novel multitask capsule network providing radiographic explainability for prediction. Visual explanations (called “attributes”) are encoded as tasks and defined by radiologists using standard guidelines. For example, to detect infectious lung disease in a CT scan, the attributes may be set up as the existence of the following patterns: ground-glass opacities, consolidation, traction bronchiectasis, cysts, centrilobular nodules, reticulations, honeycombing, and subpleural lines.
Challenge: Spectrum Bias
The clinical utility of AI models is highly dependent on their ability to perform well across the spectrum of cases they are anticipated to analyze. AI researchers use techniques such as stratified cross-validation to ensure that the distribution of sub-classes in the testing set matches the training set. However, if the full data set does not replicate the range of cases in the population, then the calculated performance metrics may not represent the model's performance on the population (termed spectrum bias). In clinical studies, spectrum bias may affect model performance on ethnic, age, or sex minorities. Spectrum bias can also occur as a result of recruiting methods and can lead to models biased toward those financially able to take time off to participate in a research study.
In preclinical research, much more control is maintained over animal demographics. However, differences in disease presentation across species can lead to another form of spectrum bias. In nonhuman primate models of SARS-CoV-2 infection, only a mild form of the disease has been replicated [131,132]. However, in crab-eating (cynomolgus) macaque models of Ebola virus disease, infection leads to death faster than that seen in humans (macaques, 5–8 days from exposure to death; humans, 4–10 days incubation period, followed by death at 6–16 days after onset of symptoms) [133,134]. Thus, animal models of infectious diseases sometimes overrepresent and sometimes underrepresent the disease's pace and severity in humans. More work is needed to fine-tune these animal models to replicate human disease more accurately. In the meantime, AI researchers must keep this in mind while training their models on animal data for predictions on human data.
The primary method to improve the generalizability of a biased data set is to collect more data from the underrepresented group. If this is not an available option, other methods that have been developed to compensate for class imbalances may be applied. For example, designing the loss function to increase the penalty for incorrect predictions of the minority group draws the model's “focus” to that group, potentially balancing performance across the minority and majority groups [135]. Other techniques involve inflating the weight of minority samples by reducing the number of majority samples (undersampling), creating duplicates of the minority samples (oversampling), and creating synthetic minority samples. In medical imaging, synthetic minority images can be created using generative adversarial neural networks, which have shown success in alleviating class imbalances [136]. In a study by Waheed and colleagues, it was found that COVID-19 detection in radiographs could be improved from 85% to 95% accuracy using generative adversarial neural networks [137].
UNIQUE GOALS AND CHALLENGES OF AI APPLICATIONS TO IMAGING ANIMAL MODELS OF INFECTIOUS DISEASES
Animal models of infectious diseases provide unique opportunities to study infectious diseases in a highly controlled environment typically not possible in the clinic, especially in most outbreak settings. With these opportunities also comes unique challenges in collecting and analyzing data in this field (Table 2). Ultimately, AI research on animal models of infectious diseases must be designed to build toward improving patient care, and thus goals must be carefully crafted to ensure this outcome.
Table 2.
Major Challenges for Humans and Animal Artificial Intelligence Infectious Disease Imaging Research and Corresponding Solutions
| Challenges | Solutions | Animal Research | Human Research |
|---|---|---|---|
| Data scarcity | Self-supervised learning, semisupervised learning, and data-centric AI | X | X |
| Model interpretability | Explainable AI techniques | X | X |
| Disease specificity | Multimodal models | X | X |
| Spectrum bias | Improvement of recruiting methods and animal models, bias loss term, oversampling or undersampling, and synthetic data | X | X |
| Development of imaging tools | Additional funding for research | X | … |
| Control over environmental variables | Careful design of input features and explainable AI techniques | … | X |
| Cost per image | Additional funding for research | X | … |
| Privacy | Federated and swarm learning | … | X |
Abbreviation: AI, artificial intelligence;
Goals of AI Applications to Imaging Animal Models of Infectious Diseases
There are 3 main goals of AI in the research space of animal models of infectious disease imaging. One goal is to directly translate predictive models to humans. For diseases in which high-quality labeled data are more easily produced in animals than collected in humans (eg, rare and severe diseases); predictive models can be trained on animal models and applied in humans. However, this is challenging because the animal model of disease must replicate all key features of the disease in humans. An example of a model trained on animal data and likely to perform equally well on human data is deep learning for lung-lesion phenotyping in nonhuman primates. This is because lung lesion types (eg, ground-glass opacities, crazy-paving, and consolidation) are defined by common radiological patterns and are not specific to anatomy, disease, or species [138].
The second major goal is to build biomarkers of disease progression for application in animal models. Such biomarkers could be used as a benchmark for assessing the effectiveness of emerging therapies. Imaging biomarkers (and AI-empowered imaging biomarkers) can be collected in vivo, enabling longitudinal studies and eliminating the need for serial sacrifice. Imaging biomarkers are well positioned to fill this role because all major imaging modalities (ie, CT, MR imaging, PET, and ultrasonography) quantify disease characteristics downstream of the initial infection. For example, imaging biomarkers of neurological impairment from HIV infection (termed HIV/neuroAIDS) have been developed using MR spectroscopy, diffusion tensor imaging, and functional MR imaging; these modalities measure changes in neurochemicals, brain tissue structure, and brain function, respectively. All of these measures do not detect HIV but instead detect a downstream consequence of HIV infection and can be correlated with clinical symptoms and signs. Imaging biomarkers bridge the gap between virus detection (too detached from symptoms) and symptom detection (too late in the disease process), thus filling an important role in therapy development.
The third major goal is to enhance our understanding of underlying disease mechanisms that are common between animal models of disease and the disease in humans. Particularly, models with high interpretability (eg, feature weights in logistic regression, feature importance in decision trees, and saliency maps in CNNs and graph-based data mining) provide information on the structure of predictive models and consequently a window into the system being modeled. Feature-importance measures have been used to determine the most important features in applications such as the prediction of COVID-19 disease progression [139], detection of influenza [140], and prediction of HIV therapy potency [141]. In a study examining the use of laboratory data for predicting the disease progression of COVID-19, feature importance was used to identify D-dimer, C-reactive protein, and age as the top 3 features used by the ML model [139]. Unlike linear statistical models, feature importance in a nonlinear ML model can highlight strong nonlinear relationships between features and the classification task. It should be noted that, although high feature importance suggests a relationship between a predictor and a classification task, there are no currently agreed-on conventions for determining how accurate a model must be and how important a feature must be to be considered “significant” (eg, the 95% confidence interval convention). Further work is needed in this area to maximize the knowledge gained from feature-importance calculations; until then, AI researchers must be cautious in their interpretation.
Challenges of AI Applications to Imaging Animal Models of Infectious Diseases
Animal imaging studies of infectious diseases with sample sizes typical of AI research are rare, primarily owing to the costs associated with such studies. In vivo modeling of highly infectious diseases imposes significant logistical and financial challenges. In a typical animal imaging experiment, specialized scanners (eg, high-field MR imaging, micro-CT, and small-animal PET) are needed to image the small organs found in rodent models of disease. Furthermore, infectious disease imaging requires specialized infrastructure to protect researchers during the scanning procedure [142]. To study highly infectious and high-consequence biological agents (eg, Marburg, Ebola, Lassa, Hendra, and Nipah viruses) entire facilities must be designed for maximum contaminant (biosafety level 4) conditions that encompass the animal care sections as well as imaging suites [143]. Although imaging animal models of infectious diseases is associated with a high cost per sample, experiments can be precisely designed to improve the value of each sample for training a predictive model.
A core tenet of the data-centric AI approach is that a predictive model trained on a small but well-designed data set may learn the generalizable predictive features better than a model trained on a large and noisy data set [44]. Finely tuned experimental parameters and highly controlled environmental factors can be used to create higher-quality training data compared to what is possible in humans. Parameters such as exposure dose and time from exposure can be precisely controlled in animal studies. Furthermore, preexposure data points and long-term follow-up data points are more easily collected in animal studies. Environmental factors, such as diet, physical activity, and comorbid conditions, vary widely in the human population but are easily standardized in animal studies. By keeping environmental factors consistent and using preexposure-corrected data, researchers can be more confident that high-performing predictive models are learning generalizable concepts and are not biased by confounding factors.
Image preprocessing, such as normalization, registration, and segmentation, can have an immense impact on the performance of a predictive model. Classic image-analysis techniques provide an opportunity for imaging scientists to leverage decades of previous research to minimize variability in imaging data unrelated to the infectious agent. Importantly, many of these techniques can significantly improve model performances without acquiring more data. Unfortunately, processing tools for animal images are less refined than those for human images. For example, brain researchers use registration tools to align brains across scans and control for slight differences in brain shape and size to focus predictive models on changes in intensity within common regions of the brain. While well-tuned for application in human imaging, the necessary templates and tools for preprocessing are not as developed for animal imaging, increasing the noise that predictive models must work around.
Unique Challenges of AI Applications to Imaging Infectious Diseases in Humans
Compared with animal models of infectious diseases, human imaging data are much noisier and more complicated by uncontrolled and confounding factors. These factors range across the full spectrum of human environmental and genetic variability, and each factor can modulate the response to infection directly or indirectly. Human data are also more often collected across multiple sites, introducing another layer of confounding factors. These confounders can distort model performance in ways that are not readily apparent. For example, it has been found that some imaging-based predictive models were detecting medical interventions in response to diseases [144,145] rather than biological markers of diseases. This can be particularly dangerous for clinical applications, as untreated patients with a disease are at the greatest risk for harm (compared with treated patients and those without disease). Great care must be taken to ensure that data fed into AI models do not contain features that have spurious correlations with the response variable. In addition, explainable AI techniques must become common practice to ensure that models are using appropriate features to form predictions.
CONCLUSIONS
Infectious disease imaging during the pandemic has proven a fertile ground for the development and application of classic ML, deep learning, and AI data science. AI tools have been used for both preclinical and clinical purposes but need to be fine-tuned. As AI researchers tackle increasingly complex problems, increasingly complex solutions have been developed. When patients’ well-being is at stake, an equal, if not greater, effort must be dedicated to developing methods to ensure that new AI techniques are adequately generalizable, explainable, and unbiased. An AI roadway laid on the foundation of COVID-19 imaging and data acquisition attempts may facilitate subsequent passage toward robust and generalizable models. Certainly, the long-term impact on the data-science research community is broad.
Contributor Information
Winston T Chu, Center for Infectious Disease Imaging, Radiology and Imaging Sciences, Clinical Center, National Institutes of Health, Bethesda, Maryland, USA; Integrated Research Facility at Fort Detrick, Division of Clinical Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Frederick, Maryland, USA.
Syed M S Reza, Center for Infectious Disease Imaging, Radiology and Imaging Sciences, Clinical Center, National Institutes of Health, Bethesda, Maryland, USA.
James T Anibal, Center for Interventional Oncology, Clinical Center, National Institutes of Health, Bethesda, Maryland, USA.
Adam Landa, Center for Interventional Oncology, Clinical Center, National Institutes of Health, Bethesda, Maryland, USA.
Ian Crozier, Clinical Monitoring Research Program Directorate, Frederick National Laboratory for Cancer Research sponsored by the National Cancer Institute, Frederick, Maryland, USA.
Ulaş Bağci, Department of Radiology, Feinberg School of Medicine, Northwestern University, Chicago, Illinois, USA.
Bradford J Wood, Center for Interventional Oncology, Clinical Center, National Institutes of Health, Bethesda, Maryland, USA; Center for Interventional Oncology, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Jeffrey Solomon, Clinical Monitoring Research Program Directorate, Frederick National Laboratory for Cancer Research sponsored by the National Cancer Institute, Frederick, Maryland, USA.
Notes
Acknowledgments. We thank Maryam Homayounieh for her consultation in the planning of this manuscript and Anya Crane (the Integrated Research Facility at Fort Detrick [IRF-Frederick]) for critically editing the manuscript. We thank Jiro Wada (IRF-Frederick) for preparing the figures and Linh Shinguyen for providing diagrams used for the figures.
Disclaimer. The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication. The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the US Department of Health and Human Services) or of the institutions and companies affiliated with the authors, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Financial support. This work was supported by the National Cancer Institute, National Institutes of Health (NIH; contract 75N910D00024 [task order no. 75N91019F00130, with Leidos Biomedical Research] and support to Leidos Biomedical Research through the Clinical Monitoring Research Program Directorate, Frederick National Laboratory for Cancer Research; I. C. and J. S. performed this work as employees of Leidos Biomedical Research); the National Institutes of Health Clinical Center Radiology and Imaging Sciences Center for Infectious Disease Imaging (W. T. C. and S. M. S. R.); the National Institute of Allergy and Infectious Diseases (contract HHSN272201800013C to Laulima Government Solutions; W. T. C. performed this work as an employee of Tunnell Government Services, a subcontractor of Laulima Government Solutions); the NIH Center for Interventional Oncology, the NIH Intramural Research Program, the NIH Clinical Center, the National Cancer Institute, and the National Institute of Biomedical Imaging and Bioengineering (intramural NIH grants Z1A CL040015 and 1ZIDBC011242); and the NIH Intramural Targeted Anti-COVID-19 Program, funded by the National Institute of Allergy and Infectious Diseases.
Supplement sponsorship. This article appears as part of the supplement “Seeing Is Believing: The Potential of Molecular Imaging Approaches for Infectious Diseases,” sponsored by the Center for Infectious Disease Imaging, Clinical Center, NIH (Intramural Research Program), and the Center for Infection and Inflammation Imaging Research, Johns Hopkins University School of Medicine, with individual contributions from Long Island Jewish Medical Center; University of California, San Francisco; and University of Maryland School of Pharmacy.
References
- 1. Shortliffe EH. Design considerations for MYCIN. In: Shortliffe EH, ed. Computer-based medical consultations: MYCIN. Amsterdam: Elsevier, 1976:63–78. [Google Scholar]
- 2. Peiffer-Smadja N, Rawson TM, Ahmad R, et al. Machine learning for clinical decision support in infectious diseases: a narrative review of current applications. Clin Microbiol Infect 2020; 26:584–95. [DOI] [PubMed] [Google Scholar]
- 3. Fischl B, Salat DH, Busa E, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 2002; 33:341–55. [DOI] [PubMed] [Google Scholar]
- 4. Wade BS, Valcour VG, Wendelken-Riegelhaupt L, et al. Mapping abnormal subcortical brain morphometry in an elderly HIV+ cohort. NeuroImage Clin 2015; 9:564–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stirenko SK, Kochura YP, Alienin O, et al. Chest X-ray analysis of tuberculosis by deep learning with segmentation and augmentation. In: 2018 IEEE 38th International Conference on Electronics and Nanotechnology (ELNANO). Kyiv, Ukraine. 2018:422–8.
- 6. Reza SMS, Bradley D, Aiosa N, et al. Deep learning for automated liver segmentation to aid in the study of infectious diseases in nonhuman primates. Acad Radiol 2021; 28:S37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gao K, Su J, Jiang Z, et al. Dual-branch combination network (DCN): towards accurate diagnosis and lesion segmentation of COVID-19 using CT images. Med Image Anal 2021; 67:101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fan DP, Zhou T, Ji GP, et al. Inf-Net: automatic COVID-19 lung infection segmentation from CT images. IEEE Trans Med Imaging 2020; 39:2626–37. [DOI] [PubMed] [Google Scholar]
- 9. Hofmanninger J, Prayer F, Pan J, Röhrich S, Prosch H, Langs G. Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem. Eur Radiol Exp 2020; 4:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chaganti S, Grenier P, Balachandran A, et al. Automated quantification of CT patterns associated with COVID-19 from chest CT. Radiol Artif Intell 2020; 2:e200048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu YH, Gao SH, Mei J, et al. JCS: an explainable COVID-19 diagnosis system by joint classification and segmentation. IEEE Trans Image Process 2021; 30:3113–26. [DOI] [PubMed] [Google Scholar]
- 12. Ouyang X, Huo J, Xia L, et al. Dual-Sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans Med Imaging 2020; 39:2595–605. [DOI] [PubMed] [Google Scholar]
- 13. Ranjbarzadeh R, Jafarzadeh Ghoushchi S, Bendechache M, et al. Lung infection segmentation for COVID-19 pneumonia based on a cascade convolutional network from CT images. Biomed Res Int 2021; 2021:5544742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res 2017; 77:e104–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fang X, Li X, Bian Y, Ji X, Lu J. Radiomics nomogram for the prediction of 2019 novel coronavirus pneumonia caused by SARS-CoV-2. Eur Radiol 2020; 30:6888–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fu L, Li Y, Cheng A, Pang P, Shu Z. A novel machine learning-derived radiomic signature of the whole lung differentiates stable from progressive COVID-19 infection: a retrospective cohort study. J Thorac Imaging 2020; 35:361–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu H, Ren H, Wu Z, et al. CT radiomics facilitates more accurate diagnosis of COVID-19 pneumonia: compared with CO-RADS. J Transl Med 2021; 19:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zeng QQ, Zheng KI, Chen J, et al. Radiomics-based model for accurately distinguishing between severe acute respiratory syndrome associated coronavirus 2 (SARS-CoV-2) and influenza A infected pneumonia. MedComm (2020) 2020; 1:240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yue H, Yu Q, Liu C, et al. Machine learning-based CT radiomics method for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection: a multicenter study. Ann Transl Med 2020; 8:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bağci U, Bray M, Caban J, Yao J, Mollura DJ. Computer-assisted detection of infectious lung diseases: a review. Comput Med Imaging Graph 2012; 36:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Varshni D, Thakral K, Agarwal L, Nijhawan R, Mittal A. Pneumonia detection using CNN based feature extraction. In: 2019 IEEE International Conference on Electrical, Computer and Communication Technologies (ICECCT), Coimbatore, India, 2019:1–7.
- 22. Irmak E. Implementation of convolutional neural network approach for COVID-19 disease detection. Physiol Genomics 2020; 52:590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li L, Huang H, Jin X. AE-CNN classification of pulmonary tuberculosis based on CT images. In: 2018 9th International Conference on Information Technology in Medicine and Education (ITME). Hangzhou, China: IEEE, 2018:39–42. [Google Scholar]
- 24. Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. In: 31st Conference on Neural Information Processing Systems (NIPS 2017). Long Beach, CA: 2017:39–42. [Google Scholar]
- 25. Dosovitskiy A, Beyer L, Kolesnikov A, et al. An image is worth 16 × 16 words: transformers for image recognition at scale. ICLR 2021. arXiv [Preprint: not peer reviewed]. 22 October 2020. Available from: 10.48550/arXiv.2010.11929. [DOI] [Google Scholar]
- 26. Mehboob F, Rauf A, Jiang R, et al. Towards robust diagnosis of COVID-19 using vision self-attention transformer. Sci Rep 2022; 12:8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park S, Kim G, Oh Y, et al. Vision transformer for COVID-19 CXR diagnosis using chest X-ray feature corpus. arXiv [Preprint: not peer reviewed]. 12 March 2021. Available from: 10.48550/arXiv.2103.07055. [DOI] [Google Scholar]
- 28. Cao Y, Zhang C, Peng C, et al. A convolutional neural network-based COVID-19 detection method using chest CT images. Ann Transl Med 2022; 10:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aslan MF, Sabanci K, Durdu A, Unlersen MF. COVID-19 diagnosis using state-of-the-art CNN architecture features and Bayesian optimization. Comput Biol Med 2022; 142:105244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen YM, Chen YJ, Ho WH, Tsai JT. Classifying chest CT images as COVID-19 positive/negative using a convolutional neural network ensemble model and uniform experimental design method. BMC Bioinformatics 2021; 22:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Auletta S, Varani M, Horvat R, Galli F, Signore A, Hess S. PET radiopharmaceuticals for specific Bacteria imaging: a systematic review. J Clin Med 2019; 8:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ordonez AA, Weinstein EA, Bambarger LE, et al. A systematic approach for developing bacteria-specific imaging tracers. J Nucl Med 2017; 58:144–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cho SY, Rowe SP, Jain SK, et al. Evaluation of musculoskeletal and pulmonary bacterial infections with [124I]FIAU PET/CT. Mol Imaging 2020; 19:1536012120936876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mota F, De Jesus P, Jain SK. Kit-based synthesis of 2-deoxy-2-[18F]-fluoro-D-sorbitol for bacterial imaging. Nat Protoc 2021; 16:5274–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Devlin J, Chang MW, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers). Minneapolis, MN: Association for Computational Linguistics, 2018:4171–86. [Google Scholar]
- 36. Raffel C, Shazeer N, Roberts A, et al. Exploring the limits of transfer learning with a unified text-to-text transformer. arXiv [Preprint: not peer reviewed]. 23 October 2019. Available from: 10.48550/arXiv.1910.10683. [DOI] [Google Scholar]
- 37. Nguyen H, Phan L, Anibal J, Peltekian A, Tran H. Viesum: how robust are transformer-based models on Vietnamese summarization? arXiv [Preprint: not peer reviewed]. 8 October 2021. Available from: 10.48550/arXiv.2110.04257. [DOI] [Google Scholar]
- 38. Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2019; 36:1234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang K, Altosaar J, Ranganath R. ClinicalBERT: modeling clinical notes and predicting hospital readmission. arXiv [Preprint: not peer reviewed]. 10 April 2019. Available from: 10.48550/arXiv.1904.05342. [DOI] [Google Scholar]
- 40. Phan LN, Anibal JT, Tran H, et al. Scifive: a text-to-text transformer model for biomedical literature. arXiv [Preprint: not peer reviewed]. 28 May 2021. Available from: 10.48550/arXiv.2106.03598. [DOI]
- 41. Han J, Xia T, Spathis D, et al. Sounds of COVID-19: exploring realistic performance of audio-based digital testing. NPJ Digit Med 2022; 5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abbasian Ardakani A, Acharya UR, Habibollahi S, Mohammadi A. COVIDiag: a clinical CAD system to diagnose COVID-19 pneumonia based on CT findings. Eur Radiol 2021; 31:121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pham TD. A comprehensive study on classification of COVID-19 on computed tomography with pretrained convolutional neural networks. Sci Rep 2020; 10:16942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mazumder M, Banbury C, Yao X. DataPerf: benchmarks for data-centric AI development. arXiv [Preprint: not peer reviewed]. 20 July 2022. Available from: 10.48550/arXiv.2207.10062. [DOI] [Google Scholar]
- 45. Geirhos R, Temme CR, Rauber J, Schütt HH, Bethge M, Wichmann FA. Generalisation in humans and deep neural networks. In: Advances in neural information processing systems, Red Hook, NY, 7538–50.
- 46. Knight W. The dark secret at the heart of AI.MIT Technology Rev 2017. Available from: https://www.technologyreview.com/2017/04/11/5113/the-dark-secret-at-the-heart-of-ai/
- 47. Rudin C. Please stop explaining black box models for high stakes decisions. arXiv [Preprint: not peer reviewed]. 26 November 2018. Available from: 10.48550/arXiv.1811.10154. [DOI] [Google Scholar]
- 48. Finlayson SG, Bowers JD, Ito J, Zittrain JL, Beam AL, Kohane IS. Adversarial attacks on medical machine learning. Science 2019; 363:1287–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rotemberg V, Halpern A. Towards ‘interpretable’ artificial intelligence for dermatology. Br J Dermatol 2019; 181:5–6. [DOI] [PubMed] [Google Scholar]
- 50. Lamy JB, Sekar B, Guezennec G, Bouaud J, Séroussi B. Explainable artificial intelligence for breast cancer: a visual case-based reasoning approach. Artif Intell Med 2019; 94:42–53. [DOI] [PubMed] [Google Scholar]
- 51. Freitas AA. Comprehensible classification models: a position paper. ACM SIGKDD Explorations Newsl 2014; 15:1–10. [Google Scholar]
- 52. Huysmans J, Dejaeger K, Mues C, Vanthienen J, Baesens B. An empirical evaluation of the comprehensibility of decision table, tree and rule based predictive models. Decis Support Syst 2011; 51:141–54. [Google Scholar]
- 53. Dieng A, Liu Y, Roy S, Rudin C, Volfovsky A. Interpretable almost-exact matching for causal inference. Proc Mach Learn Res 2019; 89:2445–53. [PMC free article] [PubMed] [Google Scholar]
- 54. Fong R, Vedaldi A. Net2vec: quantifying and explaining how concepts are encoded by filters in deep neural networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Salt Lake City, UT: 2018; 8730–8. [Google Scholar]
- 55. Morcos AS, Barrett DG, Rabinowitz NC, Botvinick M. On the importance of single directions for generalization. arXiv [Preprint: not peer reviewed]. 19 March 2018. Available from: 10.48550/arXiv.1803.06959. [DOI] [Google Scholar]
- 56. Zhou B, Sun Y, Bau D, Torralba A. Revisiting the importance of individual units in CNNs via ablation. arXiv [Preprint: not peer reviewed]. 7 June 2018. Available from: 10.48550/arXiv.1806.02891. [DOI] [Google Scholar]
- 57. Vellido A, Martín-Guerrero JD, Lisboa PJ. Making machine learning models interpretable. In: The European Symposium on Artificial Neural Networks. Bruges, Belgium: Citeseer, 2012:163–72. [Google Scholar]
- 58. Hainmueller J, Hazlett C. Kernel regularized least squares: reducing misspecification bias with a flexible and interpretable machine learning approach. Political Analysis 2014; 22:143–68. [Google Scholar]
- 59. Nemati S, Holder A, Razmi F, Stanley MD, Clifford GD, Buchman TG. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit Care Med 2018; 46:547–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen X, Duan Y, Houthooft R, Schulman J, Sutskever I, Abbeel P. InfoGAN: interpretable representation learning by information maximizing generative adversarial nets. Barcelona, Spain: ACM, 2016:2172–80 [Google Scholar]
- 61. Ponte P, Melko RG. Kernel methods for interpretable machine learning of order parameters. Phys Rev B 2017; 96:205146. [Google Scholar]
- 62. Du M, Liu N, Hu X. Techniques for interpretable machine learning. arXiv [Preprint: not peer reviewed]. 31 July 2018. Available from: 10.48550/arXiv.1808.00033. [DOI] [Google Scholar]
- 63. Kim B. Interactive and interpretable machine learning models for human machine collaboration. USA. Massachusetts Institute of Technology, 2015:135–43. [Google Scholar]
- 64. Van Belle V, Lisboa P. Research directions in interpretable machine learning models. In: Proceedings of the European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning. Bruges, Belgium: ESANN, 2013:533–41. [Google Scholar]
- 65. Otte C. Safe and interpretable machine learning: a methodological review. In: Computational intelligence in intelligent data analysis. Berlin, Heidelberg: Springer, 2013:111–22. [Google Scholar]
- 66. Ribeiro MT, Singh S, Guestrin C. Model-agnostic interpretability of machine learning. arXiv [Preprint: not peer reviewed]. 26 June 2016. Available from: 10.48550/arXiv.1606.05386. [DOI] [Google Scholar]
- 67. Murphy B, Talukdar P, Mitchell T. Learning effective and interpretable semantic models using non-negative sparse embedding. In: International Conference on Computational Linguistics (COLING 2012), Mumbai, India. Association for Computational Linguistics, 2012:1933–50. [Google Scholar]
- 68. Murdoch WJ, Singh C, Kumbier K, Abbasi-Asl R, Yu B. Interpretable machine learning: definitions, methods, and applications. arXiv [Preprint: not peer reviewed]. 14 January 2019. Available from: 10.48550/arXiv.1901.04592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ridgeway G, Madigan D, Richardson T, O'Kane J. Interpretable boosted naïve Bayes classification. In: Proceedings of the Fourth International Conference on Knowledge Discovery and Data Mining. New York, NY, USA. ACM, 1998:101–4. [Google Scholar]
- 70. Li Y, Song J, Ermon S. InfoGAIL: interpretable imitation learning from visual demonstrations. In: Advances in neural information processing systems. Long Beach, CA, USA. 3812–22.
- 71. Lisboa PJ. Interpretability in machine learning–principles and practice. In: International Workshop on Fuzzy Logic and Applications. Genoa, Italy: Springer, 2013:15–21. [Google Scholar]
- 72. Tao L, Vidal R. Moving poselets: a discriminative and interpretable skeletal motion representation for action recognition. In: Proceedings of the IEEE International Conference on Computer Vision Workshops. Santiago, Chile. 61–9.
- 73. Huang GM, Huang KY, Lee TY, Weng JTY. An interpretable rule-based diagnostic classification of diabetic nephropathy among type 2 diabetes patients. BMC Bioinformatics 2015; 16 (suppl 1):S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Worrall DE, Garbin SJ, Turmukhambetov D, Brostow GJ. Interpretable transformations with encoder-decoder networks. In: Proceedings of the IEEE International Conference on Computer Vision. Venice, Italy. 5726–35.
- 75. Chen C, Li O, Tao C, Barnett AJ, Su J, Rudin C. This looks like that: deep learning for interpretable image recognition. arXiv [Preprint: not peer reviewed]. 27 June 2018. Available from: 10.48550/arXiv.1806.10574. [DOI] [Google Scholar]
- 76. Bengio Y. Learning deep architectures for AI. Found Trends Mach Learn 2009; 2:1–127. [Google Scholar]
- 77. Ehsan U, Harrison B, Chan L, Riedl MO. Rationalization: a neural machine translation approach to generating natural language explanations. In: Proceedings of the 2018 AAAI/ACM Conference on AI, Ethics, and Society. New York, NY, USA. ACM: 2018:81–7. [Google Scholar]
- 78. Zhang Z, Xie Y, Xing F, McGough M, Yang L. MDNet: a semantically and visually interpretable medical image diagnosis network. In: Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition. Honolulu, HI, USA. 2017:6428–36.
- 79. Vilamala A, Madsen KH, Hansen LK. Deep convolutional neural networks for interpretable analysis of EEG sleep stage scoring. In: 2017 IEEE 27th International Workshop on Machine Learning for Signal Processing (MLSP). Tokyo, Japan: IEEE. 2017:1–6. [Google Scholar]
- 80. Kim B, Shah JA, Doshi-Velez F. Mind the gap: a generative approach to interpretable feature selection and extraction. In: Advances in neural information processing systems. Montreal Canada. 2015:2260–8.
- 81. Min MR, Ning X, Cheng C, Gerstein M. Interpretable sparse high-order Boltzmann machines. In: Artificial intelligence and statistics. Reykjavik, Iceland. 614–22.
- 82. Martin-Barragan B, Lillo R, Romo J. Interpretable support vector machines for functional data. Eur J Oper Res 2014; 232:146–55. [Google Scholar]
- 83. Pelleg D, Moore A. Mixtures of rectangles: interpretable soft clustering. In: International Conference on Machine Learning. Williamstown, MA, USA. 2001:401–8.
- 84. Ahmad MA, Eckert C, Teredesai A. Interpretable machine learning in healthcare. In: Proceedings of the 2018 ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics. Washington DC, USA: ACM, 2018:559–60. [Google Scholar]
- 85. Seo S, Huang J, Yang H, Liu Y. Interpretable convolutional neural networks with dual local and global attention for review rating prediction. In: Proceedings of the Eleventh ACM Conference on Recommender Systems. Como, Italy: ACM, 2017:297–305. [Google Scholar]
- 86. Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects. Science 2015; 349:255–60. [DOI] [PubMed] [Google Scholar]
- 87. Fyshe A, Wehbe L, Talukdar PP, Murphy B, Mitchell TM. A compositional and interpretable semantic space. In: Proceedings of the 2015 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Denver, CO: Association for Computational Linguistics, 32–41. [Google Scholar]
- 88. Cano A, Zafra A, Ventura S. An EP algorithm for learning highly interpretable classifiers. In: 2011 11th International Conference on Intelligent Systems Design and Applications. Cordoba, Spain: IEEE, 2011:325–30. [Google Scholar]
- 89. Letham B, Rudin C, McCormick TH, Madigan D. Building interpretable classifiers with rules using Bayesian analysis. Department of Statistics technical report no. 609. Seattle, WA, USA: University of Washington, 2012. [Google Scholar]
- 90. Rudin C. Algorithms for interpretable machine learning. In: Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. New York, NY, USA: ACM, 2014:1519. [Google Scholar]
- 91. Weiss SM, Indurkhya N. Rule-based machine learning methods for functional prediction. J Artif Intell Res 1995; 3:383–403. [Google Scholar]
- 92. Verma A, Murali V, Singh R, Kohli P, Chaudhuri S. Programmatically interpretable reinforcement learning. arXiv [Preprint: not peer reviewed]. 6 April 2018. Available from: 10.48550/arXiv.1804.02477. [DOI] [Google Scholar]
- 93. Burrell J. How the machine ‘thinks’: understanding opacity in machine learning algorithms. Big Data Soc 2016; 3:2053951715622512. [Google Scholar]
- 94. Brinkrolf J, Hammer B. Interpretable machine learning with reject option. at-Automatisierungstechnik 2018; 66:283–90. [Google Scholar]
- 95. Lakkaraju H, Kamar E, Caruana R, Leskovec J. Interpretable & explorable approximations of black box models. arXiv [Preprint: not peer reviewed]. 4 July 2017. Available from: 10.48550/arXiv.1707.01154. [DOI] [Google Scholar]
- 96. Caywood MS, Roberts DM, Colombe JB, Greenwald HS, Weiland MZ. Gaussian Process regression for predictive but interpretable machine learning models: an example of predicting mental workload across tasks. Front Hum Neurosci 2017; 10:647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wang T, Rudin C, Doshi-Velez F, Liu Y, Klampfl E, MacNeille P. A Bayesian framework for learning rule sets for interpretable classification. J Mach Learn Res 2017; 18:2357–93. [Google Scholar]
- 98. Papernot N, McDaniel P. Deep k-nearest neighbors: towards confident, interpretable and robust deep learning. arXiv [Preprint: not peer reviewed]. 13 March 2018. Available from: 10.48550/arXiv.1803.04765. [DOI] [Google Scholar]
- 99. Murphy KP. Machine learning: a probabilistic perspective. MIT press, Cambridge, MA, USA. 2012. [Google Scholar]
- 100. Grosenick L, Greer S, Knutson B. Interpretable classifiers for FMRI improve prediction of purchases. IEEE Trans Neural Syst Rehabil Eng 2008; 16:539–48. [DOI] [PubMed] [Google Scholar]
- 101. Rüping S. Learning interpretable models. University Dortmund. Dortmund, North Rhine-Westphalia, Germany. 2006.
- 102. Bien J, Tibshirani R. Prototype selection for interpretable classification. Ann Appl Stat 2011; 5:2403–24. [Google Scholar]
- 103. Bibal A, Frénay B. Interpretability of machine learning models and representations: an introduction. In: 2016 European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning. Bruges, Belgium: ESANN.2016. [Google Scholar]
- 104. Kim J, Canny J. Interpretable learning for self-driving cars by visualizing causal attention. In: Proceedings of the 2017 IEEE International Conference on Computer Vision. Venice, Italy. 2942–50, 2017.
- 105. Biran O, Cotton C. Explanation and justification in machine learning: a survey. In: IJCAI-17 Workshop on Explainable AI (XAI). Melbourne, VIC, Australia. 2017.
- 106. Tomsett R, Braines D, Harborne D, Preece A, Chakraborty S. Interpretable to whom? a role-based model for analyzing interpretable machine learning systems. arXiv [Preprint: not peer reviewed]. 20 June 2018. Available from: 10.48550/arXiv.1806.07552. [DOI] [Google Scholar]
- 107. Hsu WN, Zhang Y, Glass J. Unsupervised learning of disentangled and interpretable representations from sequential data. In: Advances in neural information processing systems. Long Beach, CA, USA. 2017:1878–89.
- 108. Lakkaraju H, Bach SH, Leskovec J. Interpretable decision sets: a joint framework for description and prediction. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. San Francisco, CA: ACM, 2016:1675–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Narayanan M, Chen E, He J, Kim B, Gershman S, Doshi-Velez F. How do humans understand explanations from machine learning systems? An evaluation of the human-interpretability of explanation. arXiv [Preprint: not peer reviewed]. 2 February 2018. Available from: 10.48550/arXiv.1802.00682. [DOI] [Google Scholar]
- 110. Choi E, Bahadori MT, Sun J, Kulas J, Schuetz A, Stewart W. Retain: an interpretable predictive model for healthcare using reverse time attention mechanism. In: Advances in neural information processing systems. Barcelona, Spain: ACM, 2016:3504–12. [Google Scholar]
- 111. Molnar C. Interpretable machine learning: a guide for making black box models explainable. 2nd ed. Germany. Independently published, 2022. [Google Scholar]
- 112. Doshi-Velez F, Kim B. Towards a rigorous science of interpretable machine learning. arXiv [Preprint: not peer reviewed]. 28 February 2017. Available from: 10.48550/arXiv.1702.08608. [DOI] [Google Scholar]
- 113. Letham B, Rudin C, McCormick TH, Madigan D. Interpretable classifiers using rules and Bayesian analysis: building a better stroke prediction model. Ann Appl Stat 2015; 9:1350–71. [Google Scholar]
- 114. Wang T, Rudin C, Velez-Doshi F, Liu Y, Klampfl E, MacNeille P. Bayesian rule sets for interpretable classification. In: 2016 IEEE 16th International Conference on Data Mining (ICDM). Barcelona, Spain. IEEE, 2016:1269–74. [Google Scholar]
- 115. Marcus GF. The algebraic mind: integrating connectionism and cognitive science. MIT Press, Cambridge, MA, USA. 2018. [Google Scholar]
- 116. von Kügelgen J, Loog M, Mey A, Schölkopf B. Semi-supervised learning, causality and the conditional cluster assumption. arXiv [Preprint: not peer reviewed]. 28 May 2019. Available from: 10.48550/arXiv.1905.12081. [DOI] [Google Scholar]
- 117. BakIr G, Hofmann T, Schölkopf B, Smola AJ, Taskar B. Predicting structured data. MIT Press, Cambridge, MA, USA. 2007. [Google Scholar]
- 118. Holzinger A, Malle B, Kieseberg P, et al. Towards the augmented pathologist: challenges of explainable-AI in digital pathology. arXiv [Preprint: not peer reviewed]. 18 December 2017. Available from: 10.48550/arXiv.1712.06657. [DOI] [Google Scholar]
- 119. Shrikumar A, Greenside P, Kundaje A. Learning important features through propagating activation differences. In: Proceedings of the 34th International Conference on Machine Learning. Vol 70. Sydney, Australia: ACM, 2017:3145–53. [Google Scholar]
- 120. Simonyan K, Vedaldi A, Zisserman A. Deep inside convolutional networks: visualising image classification models and saliency maps. arXiv [Preprint: not peer reviewed]. 20 December 2013. Available from: 10.48550/arXiv.1312.6034. [DOI] [Google Scholar]
- 121. Zeiler MD, Fergus R. Visualizing and understanding convolutional networks. In: 2014 European Conference on Computer Vision. Zurich, Switzerland: Springer, 2014:818–33. [Google Scholar]
- 122. Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning deep features for discriminative localization. In: Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition. Las Vegas, NV, USA: IEEE, 2016:2921–9 [Google Scholar]
- 123. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. In: Proceedings of the 2017 IEEE International Conference on Computer Vision. Venice, Italy: IEEE, 2017:618–26. [Google Scholar]
- 124. Yosinski J, Clune J, Nguyen A, Fuchs T, Lipson H. Understanding neural networks through deep visualization. arXiv [Preprint: not peer reviewed]. 22 June 2015. Available from: 10.48550/arXiv.1506.06579. [DOI] [Google Scholar]
- 125. Jain S, Wallace BC. Attention is not explanation. arXiv [Preprint: not peer reviewed]. 26 February 2019. Available from: 10.48550/arXiv.1902.10186. [DOI] [Google Scholar]
- 126. Demir U, Irmakci I, Keles E, et al. Information bottleneck attribution for visual explanations of diagnosis and prognosis. In: Machine learning in medical imaging. Virtual. MLMI, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Pennisi M, Kavasidis I, Spampinato C, et al. An explainable AI system for automated COVID-19 assessment and lesion categorization from CT-scans. Artif Intell Med 2021; 118:102114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Palatnik de Sousa I, Vellasco M, Costa da Silva E. Explainable artificial intelligence for bias detection in COVID CT-scan classifiers. Sensors (Basel) 2021; 21:5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Harmon SA, Sanford TH, Xu S, et al. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat Commun 2020; 11:4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. LaLonde R, Torigian D, Bagci U. Encoding visual attributes in capsules for explainable medical diagnoses. In: Medical image computing and computer assisted intervention—MICCAI 2020. Cham: Springer International Publishing, 2020:294–304. [Google Scholar]
- 131. Munster VJ, Feldmann F, Williamson BN, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 2020; 585:268–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Finch CL, Crozier I, Lee JH, et al. Characteristic and quantifiable COVID-19-like abnormalities in CT- and PET/CT-imaged lungs of SARS-CoV-2-infected crab-eating macaques (Macaca fascicularis). bioRxiv [Preprint: not peer reviewed]. 14 May 2020. Available from: 10.1101/2020.05.14.096727. [DOI] [Google Scholar]
- 133. Zawilinska B, Kosz-Vnenchak M. General introduction into the Ebola virus biology and disease. Folia Med Cracov 2014; 54:57–65. [PubMed] [Google Scholar]
- 134. Klenk HD, Feldmann H. Ebola and Marburg viruses: molecular and cellular biology. Horizon Bioscience, Wymondham, Norfolk, UK. 2004. [Google Scholar]
- 135. Thai-Nghe N, Gantner Z, Schmidt-Thieme L. Cost-sensitive learning methods for imbalanced data. In: The 2010 International Joint Conference on Neural Networks (IJCNN). Barcelona, Spain: IEEE, 2010:1–8. [Google Scholar]
- 136. Sampath V, Maurtua I, Aguilar Martín JJ, Gutierrez A. A survey on generative adversarial networks for imbalance problems in computer vision tasks. J Big Data 2021; 8:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Waheed A, Goyal M, Gupta D, Khanna A, Al-Turjman F, Pinheiro PR. CovidGAN: data augmentation using auxiliary classifier GAN for improved COVID-19 detection. IEEE Access 2020; 8:91916–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Walker C, Chung J. Muller's imaging of the chest. Elsevier, Philadelphia, PA, USA. 2018. [Google Scholar]
- 139. Xu F, Chen X, Yin X, et al. Prediction of disease progression of COVID-19 based upon machine learning. Int J Gen Med 2021; 14:1589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Hogan CA, Rajpurkar P, Sowrirajan H, et al. Nasopharyngeal metabolomics and machine learning approach for the diagnosis of influenza. EBioMedicine 2021; 71:103546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Leidner F, Kurt Yilmaz N, Schiffer CA. Target-specific prediction of ligand affinity with structure-based interaction fingerprints. J Chem Inf Model 2019; 59:3679–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Jahrling PB, Keith L, St Claire M, et al. The NIAID integrated research facility at Frederick, Maryland: a unique international resource to facilitate medical countermeasure development for BSL-4 pathogens. Pathog Dis 2014; 71:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Chosewood LC. Biosafety in microbiological and biomedical laboratories. US Department of Health and Human Services, Public Health Service, Centers for Disease Control and Prevention, National Institutes of Health, Atlanta, GA, USA. 2010. [Google Scholar]
- 144. Oakden-Rayner L. Exploring large-scale public medical image datasets. Acad Radiol 2020; 27:106–12. [DOI] [PubMed] [Google Scholar]
- 145. Winkler JK, Fink C, Toberer F, et al. Association between surgical skin markings in dermoscopic images and diagnostic performance of a deep learning convolutional neural network for melanoma recognition. JAMA Dermatol 2019; 155:1135–41. [DOI] [PMC free article] [PubMed] [Google Scholar]