Abstract
Oxidative stress can damage DNA and thereby contribute to genome instability. To avoid an imbalance or overaccumulation of reactive oxygen species (ROS), cells are equipped with antioxidant enzymes that scavenge excess ROS. Cells lacking the RecQ-family DNA helicase Sgs1, which contributes to homology-dependent DNA break repair and chromosome stability, are known to accumulate ROS, but the origin and consequences of this oxidative stress phenotype are not fully understood. Here, we show that the sgs1 mutant exhibits elevated mitochondrial superoxide, increased mitochondrial mass, and accumulation of recombinogenic DNA lesions that can be suppressed by antioxidants. Increased mitochondrial mass in the sgs1Δ mutant is accompanied by increased mitochondrial branching, which was also inducible in wildtype cells by replication stress. Superoxide dismutase Sod2 genetically interacts with Sgs1 in the suppression of nuclear chromosomal rearrangements under paraquat (PQ)-induced oxidative stress. PQ-induced chromosome rearrangements in the absence of Sod2 are promoted by Rad51 recombinase and the polymerase subunit Pol32. Finally, the dependence of chromosomal rearrangements on the Rev1/Pol ζ mutasome suggests that under oxidative stress successful DNA synthesis during DNA break repair depends on translesion DNA synthesis.
Keywords: Sgs1, Sod2, translesion DNA synthesis, homologous recombination, reactive oxygen species (ROS), mitochondria, chromosome instability
Vidushi, Campos, and Schmidt report that the mitochondrial superoxide dismutase Sod2 suppresses nuclear mutagenesis and chromosome rearrangements under oxidative stress in Saccharomyces cerevisiae. Notably, these chromosome rearrangements not only depend on homologous recombination factors, but also translesion DNA synthesis. Further, the authors identify novel genetic interactions between SOD2 and the RecQ-helicase gene SGS1 in the suppression of chromosome rearrangements and mitochondrial abnormalities. They discuss possible causes and consequences of oxidative stress in cell lacking Sgs1.
Introduction
Preserving genome integrity is essential for the proper biological functioning of organisms. To this end, eukaryotes have evolved multiple mechanisms of recognizing DNA lesions and repairing them to prevent the accumulation of mutations. One source of genome instability is oxidative stress, which causes major injury to cells by damaging DNA, proteins, and lipids (Carter et al. 2005) and is caused by a considerable increase in reactive oxygen species (ROS) (Schrader and Fahimi 2006). ROS include superoxide anions (O2·−) formed by the reduction of molecular oxygen, hydrogen peroxide (H2O2) formed by dismutation of O2·−, and the highly reactive, and, therefore, most toxic, hydroxyl free radicals (OH−) that can be formed by decomposition of H2O2 (Schrader and Fahimi 2006). The interest in ROS stems from its elevated levels in several neurodegenerative pathologies as well as cancer (Barnham et al. 2004; Lloret et al. 2008). Although still an ongoing debate, ROS has also been implicated as a cause of ageing (Laun et al. 2001; Fabrizio et al. 2004). Besides being produced by NADPH oxidases and peroxisomes (Salmon et al. 2004), endogenous ROS are mainly generated as a natural consequence of aerobic metabolism (Finkel and Holbrook 2000). To prevent ROS from reaching toxic levels cells are naturally equipped to employ antioxidant enzymes such as catalases, superoxide dismutases, and peroxidases to scavenge ROS.
Saccharomyces cerevisiae has 2 major superoxide dismutases that are highly conserved and convert superoxide to H2O2 and oxygen (Fridovich 1974; Miao and Clair 2009). The cytoplasmic Sod1, which also exists in the mitochondrial intermembrane space (Goscin and Fridovich 1972; Sturtz et al. 2001), comprises the majority of superoxide dismutase (SOD) activity in yeast (Das et al. 2018). The mitochondrial matrix protein Sod2 (Weisiger and Fridovich 1973; Ravindranath and Fridovich 1975; Vögtle et al. 2017) promotes chronological life span extension and its loss results in a gradual mitochondrial decline (Longo et al. 1996, 1999). Additionally, SOD2 deletion, like SOD1 deletion, renders cells hypersensitive to oxygen toxicity (Van Loon et al. 1986). H2O2 can further be reduced by enzymes like Tsa1, which is the most potent scavenger of H2O2. Belonging to a family of thioredoxin-dependent peroxidases, it reduces H2O2 and alkyl hydroperoxides by using electrons from NADPH and localizes predominantly to the cytoplasm (Chae et al. 1993; Park et al. 2000; Irokawa et al. 2016).
Some ROS-scavenging enzymes are important for maintaining genome stability. Mutations in SOD1, for example, produce an increase in the spontaneous mutation rate (Gralla and Valentine 1991; Huang et al. 2003), whereas mutations in TSA1 have been associated with an increase in spontaneous mutation rate as well as an increase in chromosomal rearrangements (Huang et al. 2003; Smith et al. 2004). Loss of SOD1 or TSA1 also results in increased DNA damage (Ragu et al. 2007; Choi et al. 2018), underscoring their significant contribution to the maintenance of nuclear genome integrity. Although Sod2 has been shown to prevent mitochondrial genomic instability (Doudican et al. 2005), its influence on nuclear genome integrity is unclear. While Sod1 takes up the mantle of being the primary superoxide scavenger and protector of genome stability in yeast, it is Sod2 (MnSOD) that is exceptionally important in higher eukaryotes as SOD2 disruption is lethal in mice and Drosophila (Lebovitz et al. 1996; Huang et al. 1998; Duttaroy et al. 2003), thereby representing the evolutionary selectivity and greater physiological relevance of MnSOD. Moreover, SOD2 is a putative tumor-suppressor gene and its epigenetic silencing results in cancer cell proliferation (Bravard et al. 1992; Li et al. 1998).
Oxidative damage to DNA occurs primarily in the form of base modifications and the formation of apurinic/apyrimidinic sites that are mainly repaired by the base excision repair (BER) pathway (Salmon et al. 2004). However, recombinational repair of DNA breaks has been associated with the repair of oxidative DNA damage (Slupphaug et al. 2003). Sgs1 is a non-replicative RecQ-like DNA helicase in yeast (Sun et al. 1999) and a homolog of the human Bloom syndrome (BLM) helicase, the inactivation of which causes a rare genetic disorder called Bloom syndrome (BS), which is characterized by aberrant recombination, chromosome instability, increased predisposition to cancer, short stature, and immunodeficiency (Ellis et al. 1995; Mirzaei and Schmidt 2012; Cunniff et al. 2017). The role of Sgs1 in homologous-recombination (HR)-mediated double-strand break (DSB) repair, maintenance of genome stability, and repair of stalled or damaged replication forks has been thoroughly investigated (Gangloff et al. 1994; Watt et al. 1995; Yamagata et al. 1998; Kusano et al. 1999; Frei and Gasser 2000; Myung et al. 2001; Cobb et al. 2003; Zhu et al. 2008; Cejka et al. 2010; Campos-Doerfler et al. 2018). In contrast, while mutants lacking Sgs1 are known to accumulate higher ROS than wildtype cells (Ringvoll et al. 2007), the cause of elevated ROS, the response of the sgs1Δ mutant to elevated ROS, and the contribution of ROS to sgs1Δ mutant phenotypes are less well-understood. Reports of an oxidative stress phenotype of human cells lacking BLM (Nicotera et al. 1989, 1993; Poot et al. 1989; Lloret et al. 2008; Subramanian et al. 2021) indicate that the role of Sgs1 in mitigating oxidative stress is conserved.
Evidence for a functional interaction between ROS signaling and DSB repair pathways is provided by a genetic study that shows a fitness defect and elevated levels of DNA damage and genome instability in cells lacking both SOD1 and the recombination factor RAD51 (Choi et al. 2018). Notably, mammalian SOD1 has been explored as a cancer therapeutic target for the selective killing of cancer cells with defective HR genes, such as BLM or RAD54B (Sajesh et al. 2013; Sajesh and McManus 2015). Moreover, deletion of RAD51 or RAD52 in the tsa1Δ mutant is synthetically lethal, and this lethality is dependent on the accumulation of ROS-related DNA damage (Huang and Kolodner 2005; Ragu et al. 2007). The report of a fitness defect and increased chromosomal rearrangements in the sgs1Δ tsa1Δ mutant (Huang and Kolodner 2005) highlights the importance of ROS scavenging enzymes in maintaining genome stability in the sgs1Δ mutant.
Here, we used SILAC-based quantitative proteomics to evaluate how cells respond to the lack of Sgs1, revealing mitochondrial changes, and investigated the cause and consequences of oxidative stress in the sgs1Δ mutant. We report a novel contribution of Sod2 to nuclear genome stability and identify functional interactions between SOD2 and SGS1 and other genome maintenance genes, including translesion DNA synthesis (TLS) genes, in the formation of chromosomal rearrangements.
Materials and methods
Yeast strains and media
Yeast strains for SILAC labeling, chromatin fractionation, and subsequent mass spectrometry were derived from KHSY5036 (MATa, ura3-52, trp1Δ63, his3Δ200). Yeast strains for all other experiments were derived from KHSY802 (MATa, ura3-52, trp1Δ63, his3Δ200, leu2Δ1, lys2Bgl, hom3-10, ade2Δ1, ade8, hxt13::URA3). SGS1 mutant alleles sgs1-K706A (sgs1-hd) and sgs1-F1192D (sgs1-FD), and EXO1 mutant allele exo1-E150D/D173A (exo1-ND) were previously described (Doerfler and Schmidt 2014; Campos-Doerfler et al. 2018). Gene deletions and C-terminal epitope tagging were carried out via recombination-mediated integration of selectable marker cassettes (Longtine et al. 1998) at the chromosomal loci by LiAc-mediated transformation (Gietz and Woods 2006). Haploids with multiple mutations were obtained by sporulating diploid heterozygous for the desired mutations and selection by random spore analysis on selective media or by PCR as previously described (Rockmill et al. 1991; Mirzaei et al. 2011). Yeast was grown at 30° in yeast extract (10 g/L), peptone (20 g/L), and dextrose (20 g/L) (YPD) or synthetic complete media with or without agar (20 g/l). All yeast strains used in this study are listed in Supplementary Table 1 and are available upon request.
Spot assay
Sensitivity of exponentially growing cell cultures to DNA damaging and oxidizing agents was examined with spot assays. Cultures from single colonies were grown overnight at 30° in liquid YPD, adjusted to an OD600 of 0.2, and grown to an OD600 of 0.5, followed by spotting 10-fold serial dilutions on YPD and YPD containing indicated concentrations of paraquat (PQ; Sigma), hydrogen peroxide (H2O2; Santa Cruz Biotechnology), hydroxyurea (HU; US Biological), and methyl methanesulfonate (MMS; Acros Organics). Growth was monitored at 30° and images were acquired every 12–24 hours for 2–4 days with a GelDoc IT Imaging system.
Chromatin enrichment for SILAC-based mass spectrometry
Isotope labeling of arginine and lysine was carried out according to (Syed et al. 2016). Briefly, KHSY5144 (lys2Δ arg4Δ) was grown at 30° in “light” medium containing 15 mg/L L-arginine and 30 mg/L L-lysine whereas KHSY5226 (lys2Δ arg4Δ sgs1Δ) was grown in “heavy” medium containing 15 mg/L L-arginine (13C6) and 30 mg/L L-lysine (13C6). Chromatin isolation was carried out as previously described (Kubota et al. 2012; Syed et al. 2016). Briefly, cells were suspended in pre-spheroplast buffer (100 mM PIPES/KOH, pH 9.4, 10 mM dithiothreitol (DTT), 0.1% sodium azide), followed by a 60-minute incubation in spheroplast buffer (50 mM KH2PO4/K2HPO4, pH 7.4; 0.6 M sorbitol, 10 mM DTT) containing Zymolase-100 T. Spheroplasts were washed, resuspended in wash buffer (20 mM KH2PO4/K2HPO4, pH 6.5; 0.6 M sorbitol, 1 mM MgCl2, 1 mM DTT, 20 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride (PMSF)), and overlaid onto 7.5% Ficoll-sorbitol cushion buffer (7.5% Ficoll, 20 mM KH2PO4/K2HPO4, pH 6.5; 0.6 M sorbitol, 1 mM MgCl2, 1 mM DTT, 20 mM β-glycerophosphate, 1 mM PMSF, Protease inhibitor cocktail (EDTA free, Thermo Scientific)), followed by centrifugation. Spheroplasts were resuspended and dropped onto 18% Ficoll, followed by homogenization and removal of unbroken cells by centrifugation at 5000×g for 10 minutes twice. Nuclei were pelleted and cytoplasmic fraction was collected, followed by nuclear lysis with 0.25% Triton X-100. Lysate was overlaid onto a buffer containing 30% sucrose, centrifuged, and the chromatin pellet collected.
Sample preparation and LC-MS/MS
Chromatin pellets were prepared for mass spectrometry using filter-aided sample preparation (FASP) as previously described (Syed et al. 2016). Briefly, proteins were alkylated with iodoacetamide (IAA), buffer exchanged with urea followed by ammonium bicarbonate, and finally digested with Trypsin/Lys-C overnight at 37°. Peptides were eluted and subsequently desalted using C18 solid-phase extraction cartridges (Waters) with a vacuum manifold. Desalted peptides were lyophilized in a vacuum concentrator. Peptides were resuspended in 0.1% formic acid for liquid chromatography (LC)-MS/MS analysis. Peptides were separated using a 75 µm × 50 cm C18 reversed-phase-HPLC column (Thermo Scientific) on an Ultimate 3000 UHPLC (Thermo Scientific) with a 120-minute gradient (2–32% acetonitrile with 0.1% formic acid) and analyzed on a hybrid quadrupole-Orbitrap instrument (Q Exactive Plus, Thermo Fisher Scientific). Full MS survey scans were acquired at 70,000 resolution and the top 10 most abundant ions were selected for MS/MS analysis. Raw data files were processed in MaxQuant (Cox and Mann 2008) and searched against the Saccharomyces genome database (SGD). Search parameters included constant modification of cysteine by carbamidomethylation and the variable modification of methionine oxidation. Proteins were identified using the filtering criteria of 1% protein/peptide false discovery rate (Hochberg and Benjamini 1990; Cox and Mann 2008). SILAC (H/L) ratios obtained for each replicate (n = 3) were analyzed by an outlier test (SigA test) in Perseus (Tyanova et al. 2016). Our focus for this analysis was statistical significance related to magnitude fold-change (SigA test, P < 0.05); however, we also addressed variance across replicates through additional filtering. Specifically, proteins were only included in the dataset if at least 2 of the 3 replicates had a measured stable isotope labeling by amino acids in cell culture (SILAC) ratio (Chaput et al. 2016). Additionally, proteins were only included in the final dataset if a coefficient of variation (CV) of less than 30% related to measured SILAC ratios across replicates was achieved. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2022) partner repository with the dataset identifier PXD040745.
Protein extraction and western blot analysis
Whole-cell, cytoplasmic, nucleoplasm, and chromatin extracts from yeast expressing Myc-tagged Sod2 in the absence or presence of Sgs1 were prepared as previously described (Syed et al. 2016). Samples were separated on 10% SDS-PAGE, transferred to PVDF membrane, and incubated with c-myc antibody (Santa Cruz Biotechnology) to detect myc epitope-tagged Sod2, with histone H3 (Abcam) and Adh1 (Abcam) antibodies to verify chromatin enrichment, and with α-tubulin (Santa Cruz Biotechnology) and replication factor A (Agrisera) antibodies as loading controls.
Fluctuation assay
Cells with gross-chromosomal rearrangements (GCRs) were identified by their resistance to both canavanine (canr) and 5-fluoro-orotic acid (5-FOAr) due to simultaneous inactivation of the CAN1 and URA3 genes, which are present within the nonessential end of the left arm of chromosome V (Schmidt et al. 2006a). Ten-milliliter cultures from single colonies were grown for 2 days at 30° with or without PQ, followed by plating appropriate dilutions on YPD to obtain the viable cell count and plating the remaining culture on synthetic media lacking arginine and uracil, supplemented with 60 mg/L canavanine (Sigma) and 1 g/L 5-FOA (US Biological) to select for cells with GCRs after incubation for 4 days at 30°. The rate of accumulating GCRs was determined by fluctuation analysis, and the median GCR rate from 15 cultures from 3 isolates per genotype was reported with 95% confidence intervals (Nair 1940; Lea and Coulson 1949; Schmidt et al. 2006a). The rate of accumulating mutations at the CAN1 locus was determined by fluctuation analysis by the method of the median (Nair 1940; Lea and Coulson 1949; Reenan and Kolodner 1992). Briefly, 6-ml cultures were grown for 2 days at 30° in liquid YPD with or without PQ and aliquots plated on synthetic media lacking arginine and supplemented with 60 µg/mL canavanine while appropriate dilutions were plated on YPD to obtain viable cell count. Fifteen cultures from 3 isolates per genotype were analyzed. The CAN1 ORF was amplified from canr clones by PCR and PCR products were analyzed by agarose gel electrophoresis.
Determination of ROS and mitochondrial superoxide content
Intracellular ROS levels were measured with 2′,7′-dichlorodihydrofluorescein (DCFH; λexc 485 nm λem 538 nm; Sigma) while mitochondrial superoxide levels were determined using the dihydroethidium (DHE) derivative MitoSOX Red (Invitrogen; λexc 488–510 nm λem 580 nm) (Johnson-Cadwell et al. 2007; Quaranta et al. 2011; Ramirez et al. 2014). Exponentially growing cells were stained with 10 µM DCFH or 5 µM MitoSOX Red for 30 minutes at 30° with gentle rocking as described (Ringvoll et al. 2007; Sariki et al. 2016), followed by washing and resuspension in PBS, and single-cell analysis of 100,000 cells by flow cytometry. Data was analyzed using Becton Dickinson (BD) FACSDiva software and is presented as the average of the median fluorescence values from 3 biological replicates, after adjusting for autofluorescence from unstained cells.
Determination of mitochondrial mass
Mitochondrial mass was determined using nonyl acridine orange (NAO; λex = 490 nm, λem = 518 nm; Invitrogen), which binds to cardiolipin independently of the energetic status of mitochondria (Benel et al. 1989). Exponentially growing cells were incubated in the dark with 1 µg/mL NAO for 20 minutes at 30°, followed by washing, resuspension in PBS, and flow cytometry of 100,000 cells (Lai et al. 2002).
Fluorescence microscopy
To assess accumulation of recombinogenic DNA lesions in yeast nuclei, the recombination factor Rad52 was tagged with GFP, cell cultures grown to an OD600 of 0.5–0.6, and the percentage of cells with Rad52-GFP foci determined by fluorescence microscopy (Lisby et al. 2001) on a BZ-X710 (Keyence); cells were cultured in the presence of 0.5 mM H2O2 for 30 mins or varying concentrations of N-acetyl-L cysteine (NAC) for 2 hours. To assess mitochondrial morphology, yeast expressing GFP-tagged Aco1 (Klinger et al. 2010) were grown to an OD600 of 0.8 in YPD before preparing cells for microscopy or incubated in 0.03 mM PQ for 18 hours, followed by dilution to OD600 of 0.2 and culturing to an OD600 of 0.8 in 0.03 mM PQ, followed by fluorescence microscopy on a BZ-X170 (Keyence) with a 60 × oil immersion objective. For visualizing mitochondrial morphology under replication stress, cells were grown to an OD600 of 0.5 and treated with 10 mM or 200 mM HU (US Biological) for 3 hours, and images were acquired on a BZ-X170 fluorescence microscope (Keyence). To assess the intracellular localization of red-fluorescent-protein-tagged Sod2 (Sod2-RFP), cells were grown for fluorescence microscopy to OD600 of 0.8 in the absence of PQ or 1 mM PQ for 1 hour. For 24-hour PQ treatment, cells were grown in 1 mM PQ for 24 hours, diluted to an OD600 of 0.2, and cultured to an OD600 of 0.8 in 1 mM PQ before harvesting cells for microscopy. Cells were fixed in 3.7% formaldehyde for 1 hour and stained with 50 ng/mL 4′,6-diamidino-2-phenylindole in an antifade mounting medium (DAPI; Vector Laboratories) to visualize nuclei. Following washes with PBS, cell suspensions were mounted on agarose pads over glass slides and imaged using a BZ-X170 fluorescence microscope (Keyence) with a 60 × oil immersion objective. One DIC image and 10–20 fluorescent images at 0.3 µm intervals along the z-axis were acquired to allow inspection of all focal planes. At least 200–250 cells were scored for Rad52-GFP foci and Sod2-RFP localization and 100 cells for mitochondrial morphology. Data is presented as the average of 3 biological replicates per genotype.
Results
Evaluation of the cellular response to SGS1 deletion by SILAC-based mass spectrometry
Cells lacking the RecQ family helicase Sgs1 exhibit DNA recombination and replication defects (Frei and Gasser 2000; Schmidt and Kolodner 2004; Branzei et al. 2006; Mankouri et al. 2007; Mimitou and Symington 2010; Nielsen et al. 2013; Fasching et al. 2015). To better understand the cellular response to the absence of Sgs1 and identify novel functional interactors of Sgs1, we performed SILAC-based quantitative mass spectrometry of chromatin-enriched fractions isolated from a mixture of light-labeled wildtype cells and heavy-labeled cells with an SGS1 deletion (sgs1Δ) (Fig. 1a, Supplementary Fig. 1). We identified 68 proteins that were significantly changed in the chromatin-enriched fraction of the sgs1Δ mutant (Fig. 1a, Supplementary Table 2). We were particularly interested in the proteins whose levels increased as they may include new genetic interactions with the sgs1Δ mutation. Notable increases were observed for Rnr2 and Rnr4, subunits of ribonucleotide reductase, which catalyzes balanced dNTP production throughout the cell cycle and during DNA damage (Elledge et al. 1993), and could be a response to elevated DNA lesions and genome instability in the sgs1Δ mutant (Myung et al. 2001; Chang et al. 2005). The ATP-dependent RNA helicase Nam7/Upf1, which functions in nonsense-mediated mRNA decay but is also required for telomere length maintenance (Leeds et al. 1992; Lew et al. 1998; Gatbonton et al. 2006), was also significantly upregulated (P < 0.05); its human homolog UPF1 also helps preserve telomere stability and is a potential tumor suppressor (Azzalin et al. 2007; Chen et al. 2021). Sgs1's functions in telomere maintenance involving telomeric end processing and telomere length maintenance (Bryan et al. 1997; Cohen and Sinclair 2001; Johnson et al. 2001; Cesare and Reddel 2008; Bonetti et al. 2009) raise the possibility of a functional interaction with Nam7/Upf1. Interestingly, GO Slim Term Mapping of the upregulated proteins revealed mitochondrial proteins as the largest fraction (13/37 mapped proteins; Supplementary Fig. 1c and Supplementary Table 2), including the superoxide dismutase Sod2 (P < 0.01), which eliminates superoxide radicals in mitochondria (Whittaker and Whittaker 2012). Although increased ROS has been detected in cells lacking Sgs1, and SGS1 was identified in a screen for genes important for the survival and chromosome stability of cells lacking the cytoplasmic peroxiredoxin Tsa1 (Huang and Kolodner 2005; Ringvoll et al. 2007), the oxidative stress phenotype in the sgs1Δ mutant and the link between oxidative stress and chromosome instability are poorly understood. Therefore, this study focused on understanding the importance of the mitochondrial antioxidant Sod2 in maintaining nuclear chromosome stability and its functional interaction with Sgs1.
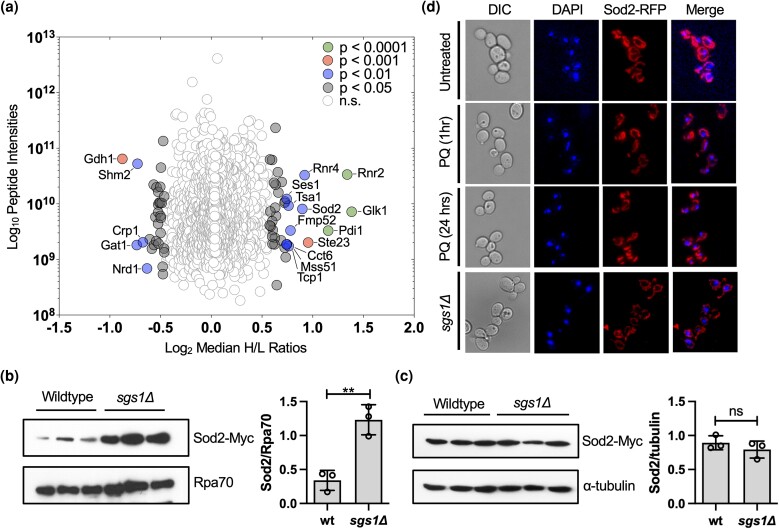
Fig. 1.
Cellular response to SGS1 deletion. a) SILAC-based quantitative proteomic analysis comparing chromatin-enriched fractions of wild-type cells and the sgs1Δ mutant. SILAC analysis was performed in triplicate with the median ratios (H/L) of heavy-labeled (sgs1Δ) to light-labeled (wildtype) proteins relative to protein abundance (estimated by summed peptide intensities) represented in this plot. The overall median ratios obtained from the entire dataset were used to demonstrate statistical significance based on magnitude fold-change (SigA test, P < 0.05). Details of additional statistical filtering are provided in Materials and Methods section. For a list of all proteins that significantly changed in the sgs1Δ mutant, see Supplementary Table 2 b) Western blot and quantification of Sod2-myc levels in 3 independent chromatin-enriched fractions from wild-type cells and the sgs1Δ mutant. Rpa70 was used as a loading control. The ratio between Sod2-myc and RPA70 is reported with standard deviation. c) Western blot and quantification of Sod2-myc expression levels in 3 independent whole-cell extracts from wild-type cells and the sgs1Δ mutant. α-tubulin was used as a loading control. The ratio between Sod2 and tubulin is reported with a standard deviation. d) Fluorescence microscopy images of RFP-tagged Sod2 and DAPI staining in wildtype cells in the absence and presence of PQ and in the absence of Sgs1 (sgs1Δ). The statistical significance of differences in b) and c) was determined with a Student's t-test and reported as **P ≤ 0.01; ns, not significant.
First, we verified Sod2 expression levels in the sgs1Δ mutant by Western blotting. This confirmed enrichment of Sod2 in the chromatin fraction of the sgs1Δ mutant (Fig. 1b, Supplementary Fig. 1b) but revealed no change in Sod2 levels in whole cell extracts (Fig. 1c), indicating that Sod2 enrichment in the chromatin fraction was not due to increased Sod2 expression in the sgs1Δ mutant. Therefore, we considered the possibility that the increase of Sod2 in the chromatin-enriched fraction of the sgs1Δ mutant was due to relocalization of Sod2 from the mitochondria to the nucleus as had previously been shown for Sod1 in cells under oxidative stress where Sod1 demonstrated cytoplasmic-to-nuclear relocalization (Tsang et al. 2014). Using RFP-tagged Sod2, we confirmed that Sod2-RFP was distributed throughout mitochondria (Vögtle et al. 2017) but did not colocalize with DAPI-stained nuclei during PQ-induced oxidative stress or in the sgs1Δ mutant (Fig. 1d). Only a small fraction of cells (1.6% ± 0.5) did show some overlap between Sod2-RFP and DAPI stained nuclei (Supplementary Fig. 2) whereas Sod1 localized to the nucleus in the majority of oxidatively stressed cells (Tsang et al. 2014). Thus, the enrichment of Sod2 in the chromatin fraction of the sgs1Δ mutant could not be explained by Sod2 relocalization to the nucleus. Considering the enrichment of several other mitochondrial proteins in the chromatin fraction of the sgs1Δ mutant besides Sod2 (Supplementary Table 2) and co-purification of mitochondrial DNA-associated proteins with nuclear chromatin (Tan et al. 2007; Kim et al. 2011; Cuevas-Bermúdez et al. 2019) we considered that mitochondrial network changes in the sgs1Δ mutant could explain the elevated levels of Sod2 and other mitochondrial proteins in the chromatin fraction of the sgs1Δ mutant. Therefore, we next evaluated mitochondrial morphology in the sgs1Δ mutant.
Absence of Sgs1 or HU-induced replication stress induces branched mitochondrial morphology
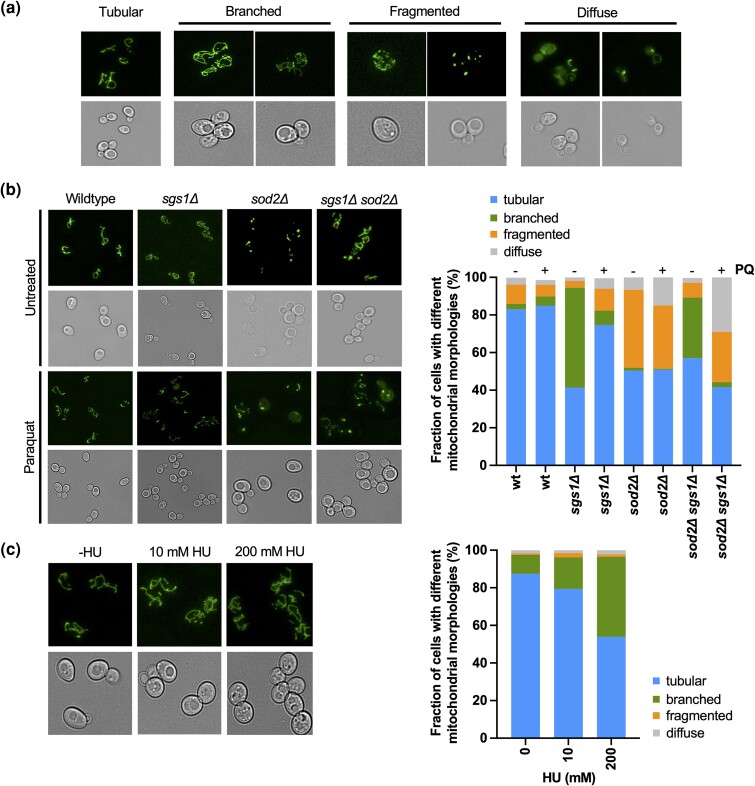
Mitochondria provide cells with energy via oxidative phosphorylation and are the predominant source of cellular ROS. The mitochondria in wildtype yeast cells appear as long, continuous tubules, which are the result of balanced fission and fusion dictated, among others, by metabolic requirements and life cycle events (Shaw and Nunnari 2002; Jakobs et al. 2003). Continuous tubules and branched tubules are associated with functional mitochondria whereas fragmented morphology indicates mitochondrial dysfunction (Aerts et al. 2009; Rudan et al. 2018). To characterize morphological states of mitochondria in cells lacking Sgs1 and/or Sod2, we tagged the mitochondrial matrix marker protein Aco1 with GFP (Aco1-GFP) and observed the cells by fluorescence microscopy, revealing 4 distinct morphological states that were easily scorable: long tubules, branched tubules, fragmented mitochondria, and diffuse mitochondria (Fig. 2a). Based on these 4 categories, we categorized mitochondrial morphology in at least 100–200 cells per genotype in the absence of exogenous stress and during PQ-induced oxidative stress (Fig. 2, b and c, Supplementary Table 3). Surprisingly, the sgs1Δ mutant displayed a large fraction of branched mitochondria (47%±1.7%) signifying a change from the continuous tubules that are characteristic of wildtype cells (82%±3.2). In contrast, many mitochondria in the sod2Δ mutant were fragmented (41%±5.8), indicative of dysfunctional mitochondria. When we introduced the sgs1Δ mutation into the sod2Δ mutant the branched mitochondrial morphology (32%±5.8), characteristic of the sgs1Δ mutant, dominated over the fragmented morphology (8%±1.7), characteristic of the sod2Δ mutant, indicating that mitochondrial morphology in the sgs1Δ sod2Δ mutant was more similar to the sgs1Δ mutant than the sod2Δ mutant. Although cell cycle analysis revealed a slight increase in the fraction of sod2Δ sgs1Δ cells in G2/M compared to the single mutants, mitochondrial branching was not limited to a particular cell cycle phase (Supplementary Fig. 3), suggesting that the small change in cell cycle distribution is unlikely to be responsible for the observed changes in mitochondrial morphology.
Fig. 2.
Changes in mitochondrial morphology upon deletion of SGS1 and/or SOD2 in the presence or absence of PQ. a) Overview of the 4 major mitochondrial morphologies (tubular, branched, fragmented, diffuse) we observed in wildtype cells or the sgs1Δ and sod2Δ mutants in the presence or absence of PQ (0.03 mM). Mitochondria were visualized by tagging mitochondrial aconitase Aco1-GFP, followed by fluorescence microscopy. b) Left panel: Representative images of different mitochondrial morphologies in wildtype and sgs1Δ and sod2Δ mutants in the presence and absence of PQ (0.03 mM). Mitochondria were visualized by GFP-tagging Aco1, followed by fluorescence microscopy. Right panel: The fraction of cells with either tubular, branched, fragmented, or diffuse mitochondrial morphology was determined from 3 experiments and a minimum of 100 cells of each strain and treatment condition. The mean is presented; for mean ± SD see Supplementary Table 3. c) Left panel: Representative images of mitochondrial morphology in wildtype cells treated with a low dose (10 mM) or high dose (200 mM) of HU. Mitochondria were visualized by GFP-tagging Aco1, followed by fluorescence microscopy. Right panel: The fraction of cells with either tubular, branched, fragmented, or diffuse mitochondrial morphology was determined from a minimum of 100 cells from 3 experiments performed in the absence of HU and for each HU concentration. The mean is presented; for mean ± SD see Supplementary Table 4.
Based on previous studies (Rudan et al. 2018), our findings of increased ROS and branched mitochondrial morphology suggest that the mitochondria in the sgs1Δ mutant are functional. Moreover, chronological and replicative lifespans of the sgs1Δ mutant growing on glycerol, a non-fermentable carbon source, are similar to those of wildtype cells (Ringvoll et al. 2007), further ruling out mitochondrial dysfunction in the sgs1Δ mutant. Furthermore, since there was no difference in mitochondrial morphology between the sgs1Δ mutant growing on 0.5 and 2% glucose (Supplementary Fig. 4), the observed branching appears to be independent of respiratory activity. Notably, there is evidence that high energy demands in a cell can result from increased DNA damage, which requires more ATP production to help repair DNA (Gafter-Gvili et al. 2011; Kulkarni et al. 2011). Since mitochondrial branching indicates increased mitochondrial activity (Rudan et al. 2018), we suspected that the increase in mitochondrial branching in the sgs1Δ mutant could be due to an increased need for ATP to respond to DNA lesions. Indeed, upon exposure to HU, which induces replication stress, wildtype cells underwent increased mitochondrial branching in a dose-dependent manner (Fig. 2c, Supplementary Table 4), providing evidence that the cellular response to replication stress is a cause for mitochondrial branching.
After treatment with PQ, which induces mitochondria-dependent superoxide production, the mitochondrial morphology in wildtype cells did not undergo a major change (Fig. 2b). In the sgs1Δ mutant, cells with branched mitochondria decreased sharply while those with tubular morphology increased, further supporting that the branched morphology in the sgs1Δ mutant is not caused by the increased ROS characteristic of sgs1Δ mutants but is due to DNA lesions and replication stress caused by the absence of Sgs1. Since branched and tubular mitochondria are considered functional this change in their proportions likely did not affect mitochondrial function in the sgs1Δ mutant. Fragmented or diffuse mitochondria, indicative of mitochondrial dysfunction, were still only seen in a small fraction of sgs1Δ cells (fragmented: 3.6% ± 3.4 vs 11.7% ± 4.9 with PQ, P = 0.079; diffuse: 1.9% ± 0.86 vs 5.6% ± 3 with PQ, P = 0.11), indicating that the vast majority of sgs1Δ cells in the presence or absence of induced oxidative stress had functional mitochondria. In the sod2Δ mutant, PQ exposure did not change the fraction of cells with functional mitochondria and only caused a small increase in cells with diffuse mitochondria (6.6% ± 3.5 to 14.9% ± 3.5; P < 0.05), indicative of a mild increase in dysfunction or cell death. Fragmented and diffuse mitochondrial morphologies spiked, however, in the sgs1Δ sod2Δ mutant after PQ exposure (fragmented: 7.9% ± 1.7 to 26.8% ± 3.9; P < 0.01; diffuse: 2.5% ± 0.9 to 29% ± 3.2, P < 0.001), raising the possibility of a negative genetic interaction between sgs1Δ and sod2Δ.
Causes of the oxidative stress phenotype of the sgs1Δ mutant
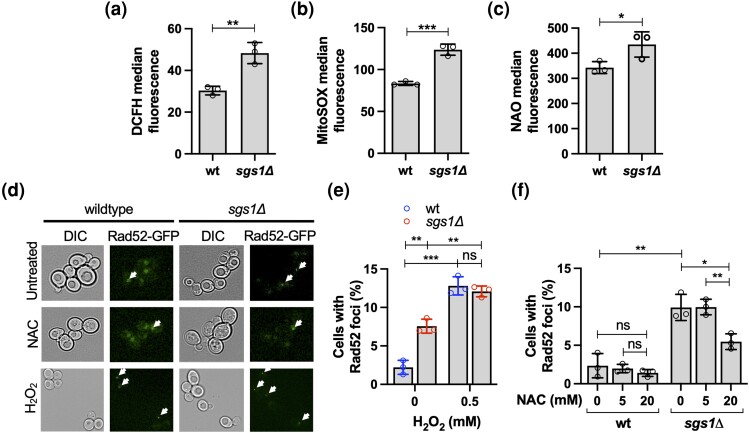
To better understand the oxidative stress phenotype of the sgs1Δ mutant (Ringvoll et al. 2007), we measured endogenous ROS levels by DCFH staining and MitoSox staining and found a 40% increase in overall ROS levels (Fig. 3a) and a 50% increase in mitochondrial superoxide content (Fig. 3b) in the sgs1Δ mutant. Since mitochondria are the predominant sources of cellular ROS (Rossignol and Frye 2012), we asked if this increase in ROS in the sgs1Δ mutant was due to an increase in mitochondrial mass. Performing flow cytometry using NAO, which stains mitochondria independently of their energetic status (Benel et al. 1989; Lai et al. 2002), we found a significant 26% increase in mitochondrial mass in the sgs1Δ mutant (Fig. 3c). Taken together with the increased mitochondrial branching in the sgs1Δ mutant (Fig. 2b) and the inducibility of mitochondrial branching by replication stress (Fig. 2c), these results suggest that the oxidative stress phenotype of the sgs1Δ mutant could be due to genome instability that induces increased mitochondrial mass and branching, which in turn causes increases in cellular ROS.
Fig. 3.
Endogenous ROS, mitochondrial mass, and recombinogenic DNA lesion formation in the sgs1Δ mutant. a) Measurement of ROS in the sgs1Δ mutant by flow cytometry using the fluorescent dye DCFH diacetate. b) Measurement of mitochondrial superoxide content in the sgs1Δ mutant by flow cytometry using MitoSOX Red fluorescent dye. c) Measurement of mitochondrial mass in the sgs1Δ mutant by flow cytometry using the fluorescent dye NAO. d) To detect recombinogenic DNA lesions, homologous recombination factor Rad52 was tagged with GFP (Rad52-GFP), and foci formation detected by fluorescence microscopy (BZ-X170, Keyence) of exponentially growing cell cultures either untreated or treated with the antioxidant NAC (20 mM) for 2 hours or H2O2 (0.5 mM) for 30 minutes. Representative images are shown with arrows pointing to nuclei with Rad52-GFP foci. In nuclei without Rad52-GFP foci, the GFP signal appears diffused throughout the nucleus. e) Percentage of cells with Rad52-GFP foci in cultures of wildtype cells and the sgs1Δ mutant in the presence or absence of oxidative stress induced by H2O2. Exponentially growing cell cultures were incubated at 0.5 mM H2O2 for 30 minutes and imaged by fluorescence microscopy (BZ-X170, Keyence). f) Effect of the antioxidant NAC (5 mM, 20 mM) on the percentage of cells with Rad52-GFP foci in cultures of wildtype cells and the sgs1Δ mutant. Exponentially growing cell cultures were incubated at the indicated concentration of NAC for 2 hours and imaged by fluorescence microscopy (BZ-X170, Keyence). Experiments were performed in triplicate and the mean ± SD is reported. Statistical significance of differences was determined with a Student's t-test and reported as *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001; ns, not significant.
To determine the effect of increased ROS in the sgs1Δ mutant on nuclear DNA integrity, we measured the percentage of cells that exhibited foci of the nuclear homologous recombination factor Rad52. Using fluorescence microscopy, we observed more spontaneous Rad52-GFP foci in the sgs1Δ mutant than in the wildtype (Fig. 3, d and e), consistent with previous reports (Chang et al. 2005). Upon treatment with H2O2, however, Rad52 foci increased 6-fold in wildtype cells, but only 1.7-fold in the sgs1Δ mutant (Fig. 3, d and e), raising the possibility that endogenous oxidative stress caused by the absence of Sgs1 is a source of Rad52-GFP foci. To test this hypothesis, we treated the sgs1Δ mutant with varying amounts of the antioxidant NAC, a well-established ROS scavenger that reacts with H2O2 and hydroxyl radicals (Aruoma et al. 1989; Zafarullah et al. 2003; Kim et al. 2013). Indeed, NAC supplementation resulted in a nearly 50% reduction of sgs1Δ cells with Rad52-GFP foci, but had no significant effect on spontaneous Rad52-GFP foci formation in wildtype cells (Fig. 3, d and f), suggesting that the increase in endogenous ROS significantly contributes to the formation of recombinogenic DNA lesions in cells lacking Sgs1.
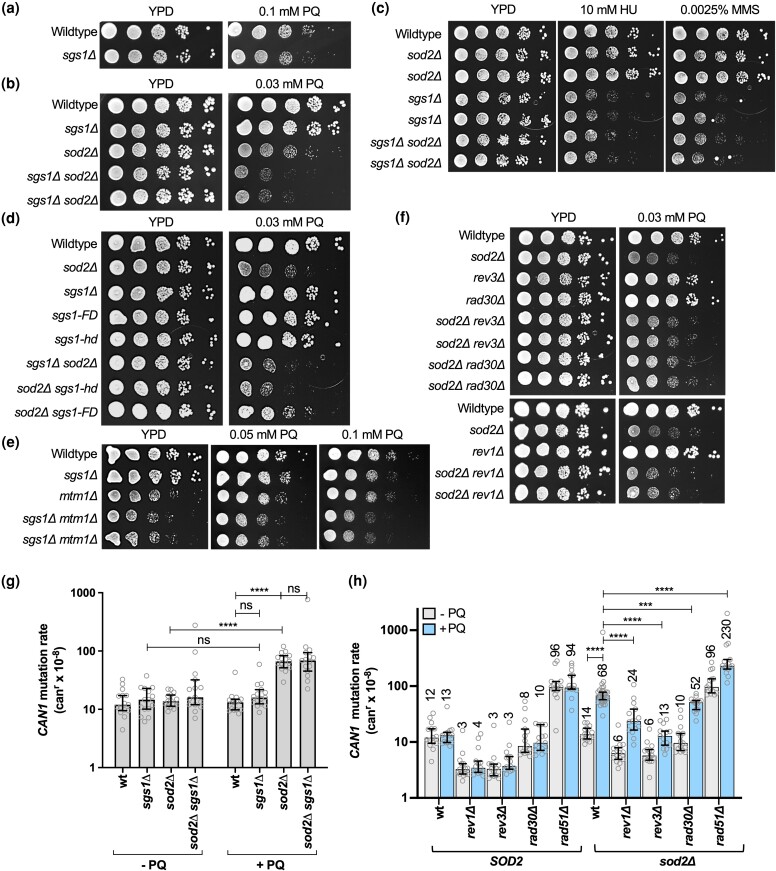
Sod2 suppresses nuclear genome instability under oxidative stress and genetically interacts with Sgs1
To better understand ROS as a source of nuclear genome instability we investigated the effect of the lack of a mitochondrial antioxidant on the formation of small-scale mutations (CAN1 mutation assay) and GCR assay in the sod2Δ mutant and a possible genetic interaction with the sgs1Δ mutation, which caused mild sensitivity to PQ (Fig. 4a). Notably, we observed a synergistic increase in sensitivity of the sod2Δ sgs1Δ mutant to PQ but not to HU and MMS, which result in replication stress and DNA damage (Fig. 4, b and c), suggesting a PQ-specific functional interaction between SGS1 and SOD2. This functional interaction is dependent on Sgs1 helicase activity since the sod2Δ sgs1-hd mutant was as sensitive to PQ as the sgs1Δ sod2Δ mutant (Fig. 4d). On the other hand, an sgs1-FD mutation, which disrupts Sgs1 binding to Rad51 and causes a hypo-recombination phenotype (Campos-Doerfler et al. 2018), had no effect on the growth of the sod2Δ mutant on PQ (Fig. 4d).
Fig. 4.
Sensitivity to DNA damage, replication stress, oxidative stress, and level of genome instability of cells lacking Sgs1 and Sod2 activities. a) Spot assay to determine the sensitivity of the sgs1Δ mutant to PQ. Ten-fold-dilutions of exponentially growing cell cultures were spotted on YPD or on YPD supplemented with 0.1 mM PQ and incubated at 30°. b) Spot assay to determine the sensitivity of the sgs1Δ sod2Δ mutant to PQ. Ten-fold-dilutions of exponentially growing cell cultures were spotted on YPD or YPD supplemented with 0.03 mM PQ and incubated at 30°. c) Spot assay to determine the sensitivity of the sgs1Δ sod2Δ mutant to HU and MMS. Ten-fold-dilutions of exponentially growing cell cultures were spotted on YPD or YPD supplemented with the indicated concentrations of HU or MMS and incubated at 30°. d) Spot assay to determine PQ-sensitivity of the sod2Δ mutant harboring sgs1 mutations that interrupt helicase activity (sgs1-hd) or Rad51-binding (sgs1-FD). Ten-fold-dilutions of exponentially growing cell cultures were spotted on YPD or YPD supplemented with 0.03 mM PQ and incubated at 30°. e) Spot assay to determine PQ-sensitivity of the sgs1Δ mutant with a deletion of MTM1. Ten-fold-dilutions of exponentially growing cell cultures were spotted on YPD or on YPD supplemented with 0.1 mM or 0.05 mM PQ and incubated at 30°. f) Spot assay to determine the effect of rev1Δ, rev3Δ, or rad30Δ mutations on PQ sensitivity of the sod2Δ mutant. Ten-fold-dilutions of exponentially growing cell cultures were spotted on YPD or YPD supplemented with 0.03 mM PQ and incubated at 30°. g) Rates of accumulating inactivating mutations in CAN1 in cells harboring sgs1Δ and/or sod2Δ mutations in the presence or absence of PQ. Rates of accumulating canavanine-resistant (canr) clones were determined from at least 15 cell cultures (gray dots) for each yeast strain in the presence or absence of 0.03 mM PQ. The statistical significance of differences was determined by a Mann–Whitney test and reported as **** P ≤ 0.0001; ns, not significant. Numbers above the columns in the graph indicate the median rate of accumulating mutations conferring resistance to canavanine (per cell/generation, canr×10−8). For a complete list of all canr rates with 95% confidence intervals see also Supplementary Table 5. h) Rates of accumulating inactivating mutations in the CAN1 gene (canr) of sod2Δ mutants harboring deletions of TLS genes (rev1Δ, rev3Δ, rad30Δ) or a homologous recombination gene (rad51Δ) in the presence or absence of PQ. At least 15 cell cultures (gray dots) from 3 isolates for each yeast strain were analyzed. The median rate with 95% confidence intervals is shown. Numbers above the columns in the graph indicate the median rate of accumulating mutations conferring resistance to canavanine (per cell/generation, canr×10−8). For a complete list of all canr rates with 95% confidence intervals see Supplementary Table 5. Statistical significance of differences was determined by a Mann–Whitney U-test and reported as ***P ≤ 0.001; ****P ≤ 0.0001.
Yeast Sod2 activation is dependent on intracellular manganese and iron levels, which are regulated by Smf2 and Mtm1, respectively (Edward et al. 2001; Luk et al. 2003; Yang et al. 2006). To determine if the genetic interaction between SOD2 and SGS1 in the suppression of PQ toxicity was indeed dependent on Sod2 function of redox regulation, we performed PQ-hypersensitivity spot assays for the sgs1Δ mutant carrying a deletion of MTM1 instead of SOD2 and observed increased PQ-sensitivity (Fig. 4e). Although this increase in PQ-sensitivity of the mtm1Δ sgs1Δ mutant was not as strong as that of the sod2Δ sgs1Δ mutant, probably due to residual Sod2 activity in the mtm1Δ mutant, it suggests that the ROS-scavenging function of Sod2 contributes to PQ-tolerance in the sgs1Δ mutant.
Using the CAN1 forward mutation assay (Fig. 4g, Supplementary Table 5), we found that PQ had no effect on the CAN1 mutation (canr) rate of wildtype cells. While the untreated sod2Δ mutant had a wildtype canr rate, the rate increased 5-fold upon PQ treatment, suggesting that the pro-oxidant PQ and loss of the mitochondrial antioxidant Sod2 synergize to induce point mutations in the nuclear genome of budding yeast. Introducing TLS mutations, such as deletions of the catalytic subunit of polymerase zeta (rev3Δ) or polymerase eta (rad30Δ), did not affect the PQ-hypersensitivity of the sod2Δ mutant (Fig. 4f), but significantly reduced its high canr rate, with rev3Δ and rev1Δ mutation having more substantial effects than rad30Δ, reducing the PQ-induced canr rate of the sod2Δ mutant to wildtype levels (Fig. 4h and Supplementary Table 5). These findings demonstrate that PQ-induced mutagenesis in the sod2Δ mutant is primarily controlled by the Rev1/Polζ (Rev3) mutasome.
Despite a strong synergistic interaction between sod1Δ and rad51Δ in the CAN1 assay (Choi et al. 2018), combining rad51Δ with the sod2Δ mutation did not cause an increase in the canr rate in the absence of PQ and only a slight increase above additive in the presence of PQ (Fig. 4h). This lack of a genetic interaction between sod2Δ and rad51Δ in the CAN1 assay suggests that, unlike in the sod1Δ mutant (Choi et al. 2018), HR does not suppress small-scale mutations in the sod2Δ mutant.
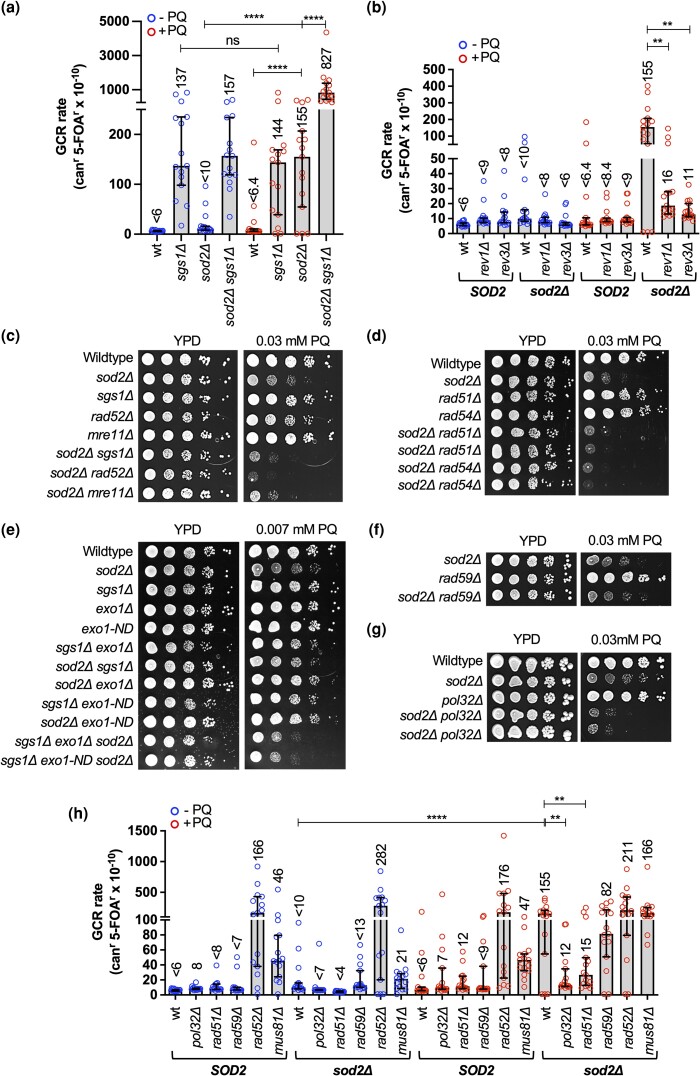
SGS1 deletion did not further increase the canr rate of the PQ-treated sod2Δ mutant (Fig. 4g and Supplementary Table 5), indicating that accumulation of point mutations and other small-scale mutations was not a major contributor to a decreased fitness of the sod2Δ sgs1Δ mutant on PQ. Therefore, we measured the rate of GCR formation in the sod2Δ and the sod2Δ sgs1Δ mutants. The GCR assay measures the rate of functional disruption of 2 counter-selectable marker genes, CAN1 and URA3, in the same cell as the result of a chromosome break (Myung et al. 2001; Schmidt et al. 2006a). We observed a significant (24-fold) increase in the GCR rate of the sod2Δ mutant upon treatment with PQ (Fig. 5a and Supplementary Table 6). This rate of PQ-induced GCRs in the sod2Δ mutant increased further, by more than 5-fold, upon deletion of SGS1 (Fig. 5a and Supplementary Table 6), suggesting a novel role for the mitochondrial antioxidant Sod2 in preventing nuclear chromosome rearrangements during exogenous oxidative stress and implicating Sgs1 in suppressing these sod2Δ-associated chromosome rearrangements.
Fig. 5.
Effect of TLS and the recombination factor Rad51 on PQ-sensitivity and PQ-induced mutator phenotype of the sod2Δ mutant. a) Rate of accumulating GCRs in cells harboring sgs1Δ and/or sod2Δ mutations in the presence or absence of PQ. The GCR rate is the rate of accumulating clones resistant to both canavanine (canr) and 5-FOAr, which was determined in least 15 cell cultures (open circles) for each yeast strain in the presence and absence of 0.03 mM PQ. Statistical significance of differences was determined by a Mann–Whitney test and reported as ****P ≤ 0.0001; ns, not significant. Numbers above the columns in the graph indicate the median rate of accumulating mutations conferring resistance to canavanine and 5-fluoro-orotic-acid (per cell/generation, canr 5-FOAr×10−10). For a complete list of all GCR rates with 95% confidence intervals see also Supplementary Table 6. b) Rate of accumulating GCRs in the presence or absence of PQ in the sod2Δ mutant harboring a deletion of the TLS genes REV1 or REV3. At least 15 cell cultures from 3 isolates for each yeast strain and each condition were analyzed (indicated by open circles). Numbers above the columns in the graph indicate the median rate of accumulating mutations conferring resistance to canavanine and 5-fluoro-orotic-acid (per cell/generation, canr 5-FOAr×10−10). Statistical significance of differences was determined by a Mann–Whitney test and reported as **P ≤ 0.01. For a complete list of GCR rates with 95% confidence intervals see Supplementary Table 6. c–g) Spot assays to determine PQ sensitivity of the sod2Δ mutant with rad52Δ, mre11Δ, rad51Δ, rad59Δ, rad54Δ, or pol32Δ mutations, or exo1 mutations in the presence or absence of Sgs1. Exo1-ND, nuclease-deficient allele of EXO1. Ten-fold-dilutions of exponentially growing cell cultures were spotted on YPD or YPD supplemented with PQ as indicated and incubated at 30°. h) Rate of accumulating GCRs in the presence or absence of PQ in the sod2Δ mutant with deletions of RAD51, RAD52, RAD59, MUS81, or POL32. Numbers above the columns in the graph indicate the median rate of accumulating mutations conferring resistance to canavanine and 5-fluoro-orotic-acid (per cell/generation, canr 5-FOAr×10−10). At least 15 cell cultures for each yeast strain (indicated by dots) and each condition (±PQ) were analyzed. Statistical significance of differences was determined by a Mann–Whitney test and reported as **P ≤ 0.01, ****P ≤ 0.0001. For a complete list of GCR rates with 95% confidence intervals see Supplementary Table 6.
Notably, these increases in PQ-induced GCRs in the sod2Δ and sod2Δ sgs1Δ mutants were not accompanied by increases in Rad52-marked recombinogenic DNA lesions (Supplementary Fig. 5). Considering this lack of Rad52-GFP foci accumulation in the sod2Δ mutant and its increased CAN1 point mutation rate, we reasoned that similar to mismatch-repair-deficient cells (Myung et al. 2001) the increased GCR rate in Sod2-deficient cells could be the result of pseudo-GCRs that arise by independent, inactivating point mutations in the CAN1 and URA3 genes in the same cell rather than a chromosome rearrangement where a DNA break leads to loss of the nonessential region that contains both CAN1 and URA3 (Chen and Kolodner 1999; Schmidt et al. 2006a). To distinguish between pseudo-GCRs and classic GCRs, we tested twelve independent GCR clones of the PQ-treated sod2Δ mutant for the presence of the CAN1 and URA3 genes, revealing that both genes had been lost in 11 clones (Supplementary Fig. 6, a and b), indicative of classic, chromosome-break-associated GCR formation in the PQ-treated sod2Δ mutant. Similarly, all fifteen GCR clones obtained from the PQ-treated sgs1Δ sod2Δ mutant had lost the CAN1 and URA3 genes (Supplementary Fig. 6a), indicating classic GCR formation. These findings support a novel role for the mitochondrial antioxidant Sod2 in the prevention of nuclear chromosome rearrangements during oxidative stress wherein it functionally interacts with Sgs1.
Chromosomal rearrangements in oxidatively stressed cells depend on TLS and HR
To better understand the mechanism of PQ-induced GCR formation in the sod2Δ mutant we tested the effect of disrupting DNA damage repair and tolerance pathways. Disruption of the Rev1/Polζ mutasome (rev1Δ, rev3Δ) led to major (10-fold) reductions in the GCR rate of the PQ-treated sod2Δ mutant (Fig. 5b, Supplementary Table 6). This was unexpected as the rev3Δ mutation previously had no effect on the elevated GCR rate of cells lacking the cytoplasmic antioxidant Tsa1 (Ragu et al. 2007), indicating a TLS-dependent mechanism of GCR formation in the PQ-treated sod2Δ mutant and a TLS-independent pathway in the tsa1Δ mutant. POL32, which codes for the noncatalytic subunit of Polδ that links the polymerase to PCNA, is also a component of Polζ and involved in TLS (Gibbs et al. 2005; Pages et al. 2008; Baranovskiy et al. 2012; Makarova and Burgers 2015). Its deletion also significantly reduced (13-fold) the GCR rate of the PQ-treated sod2Δ mutant (Fig. 5h, Supplementary Table 6). However, in contrast to rev1Δ and rev3Δ mutations, the pol32Δ mutation increased the PQ-sensitivity of the sod2Δ mutant (Fig. 5g), indicating that Pol32 is also required for increased oxidative stress tolerance of the sod2Δ mutant. Besides its role as a subunit of the Polζ mutasome, Pol32 is essential for break-induced replication (BIR) (Lydeard et al. 2007), a mutagenic recombination pathway that initiates at one-ended double-strand breaks (DSBs) that arise from collapsed replication forks (Huang et al. 2000; Lydeard et al. 2007), whereas Pol32 is dispensable for error-free HR pathways and for DNA replication. Together with the observation that SGS1 deletion increased the PQ-sensitivity of the sod2Δ mutant (Fig. 4a) and led to a synergistic increase in the GCR rate of the PQ-treated sod2Δ mutant (Fig. 5a), this prompted us to evaluate genetic interactions of the sod2Δ mutation with other HR genes.
Mre11 and Rad52 are important HR factors for DSB end resection (Mimitou and Symington 2010) and facilitate Rad51 filament formation (New et al. 1998; Song and Sung 2000), respectively. Rad51 and the Rad52 paralog Rad59 are involved in different HR events: Rad51 is involved in synthesis-dependent strand annealing where it performs homology search and strand invasion into a homologous duplex facilitated by Rad54 whereas Rad59 is involved in single strand annealing (SSA), wherein the resected DSB ends anneal at direct repeat sequences, often resulting in interstitial deletions, as well as in some minor pathways of DNA break repair, such as Rad51-independent BIR (Sung 1994; Petukhova et al. 1998, 1999; Davis and Symington 2001; Signon et al. 2001; Sugawara et al. 2003). We observed increased PQ sensitivity of the sod2Δ mutant when combined with rad52Δ, mre11Δ, rad51Δ, and rad54Δ mutations, but not with rad59Δ (Fig. 5, c, d and f), indicating that Rad51-dependent but not Rad59-dependent HR events are required for normal growth of the oxidatively stressed sod2Δ mutant. During HR, Sgs1 and Exo1 are both capable of performing long-range resection of DSB ends, but loss of both Sgs1 and Exo1 results in severe resection defects and extremely elevated GCR rates (Gravel et al. 2008; Mimitou and Symington 2008; Zhu et al. 2008; Doerfler and Schmidt 2014). We found that, unlike an SGS1 deletion, an EXO1 deletion had no effect on the growth of the sod2Δ mutant on PQ (Fig. 5e), even at higher concentrations (Supplementary Fig. 7a). Only when loss of SGS1 was combined with loss of EXO1 or the nuclease-deficient exo1-ND mutation did the sod2Δ mutant suffer from a severe fitness defect on PQ (Fig. 5e). These findings indicate that Rad51-dependent HR, including steps such as DSB end resection (Sgs1, Exo1), Rad51 filament formation (Rad51, Rad52) and duplex invasion (Rad51, Rad54) contribute to survival of the PQ-treated sod2Δ mutant whereas Rad59-dependent events are dispensable.
Since RAD52 deletion caused GCR accumulation comparable to that of the PQ-treated sod2Δ mutant (Fig. 5h, Supplementary Table 6) it was not possible to use Rad52 to further evaluate the role of HR for GCR formation in the PQ-treated sod2Δ mutant. However, deletions of RAD51 or RAD59 were suitable since they did not cause GCR rate increases in the absence or presence of PQ (Fig. 5h, Supplementary Table 6). RAD51 deletion significantly lowered the GCR rate of the PQ-treated sod2Δ mutant whereas RAD59 deletion did not (Fig. 5h, Supplementary Table 6), suggesting that Rad51-mediated HR events drive the formation of GCRs in the oxidatively stressed sod2Δ mutant. Mus81, a structure-specific endonuclease that cleaves HR intermediates and is important for genome stability (Boddy et al. 2001; Hwang et al. 2005; Ho et al. 2010; Agmon et al. 2011; Onaka et al. 2020), was dispensable for growth of the sod2Δ mutant on PQ (Supplementary Fig. 7b). Notably, despite the established role of Mus81 in suppressing GCRs (Hwang et al. 2005; Onaka et al. 2020; Muellner and Schmidt 2023), MUS81 deletion did not increase the GCR rate of PQ-treated cells or the PQ-treated sod2Δ mutant (Fig. 5h, Supplementary Table 6). That Mus81 was not required for preventing GCRs in these oxidatively stressed cells suggests that the HR intermediates Mus81 acts on during error-free DNA lesion repair do not arise in these cells. Instead, the GCR analysis suggests that PQ-induced lesions in cells lacking Sod2 are substrates for mutagenic repair mechanisms that require Rad51, Pol32, and the Rev1/Polζ (Rev3) mutasome.
Discussion
In this study, we have identified a novel function of the mitochondrial antioxidant Sod2 in the suppression of nuclear small-scale mutations as well as GCRs in cells under oxidative stress. Notably, these HR-driven chromosomal rearrangements were dependent on the TLS polymerase Polζ. Sod2 functionally interacts with Sgs1 in the suppression of these GCRs and in tolerating oxidative stress induced by PQ. We also show that cells lacking Sgs1 are hypersensitive to PQ-induced oxidative stress and exhibit elevated ROS levels, increased mitochondrial mass, and increased mitochondrial branching, the latter of which was also inducible by HU-induced replication stress in wildtype cells. Rad52-marked DNA repair centers induced by the absence of Sgs1 were significantly reduced by antioxidant treatment, suggesting that elevated ROS is a source of endogenous DNA lesions in the sgs1Δ mutant.
Although Sod2 prevents mitochondrial genome instability (Doudican et al. 2005), it was unknown whether this mitochondrial antioxidant contributes to the maintenance of nuclear genome integrity. Unlike Sod1, which relocalizes to the nucleus to act as a transcription factor during oxidative stress (Tsang et al. 2014), we show here that Sod2 does not relocalize to the nucleus under oxidative stress, yet suppresses nuclear GCRs under these conditions. The positive (REV1, REV3, POL32, RAD51) and negative regulators (SGS1) of GCR formation in the PQ-treated sod2Δ mutant give us some clues as to the nature of the underlying mechanisms. For example, Sgs1 promotes HR by resecting DSBs and through its physical interaction with Rad51, and it inhibits crossover formation and potentially mutagenic HR events, such as homeologous recombination and BIR (Myung et al. 2001; Ira et al. 2003; Wu and Hickson 2003; Spell and Jinks-Robertson 2004; Sugawara et al. 2004; Lo et al. 2006; Gravel et al. 2008; Mimitou and Symington 2008; Zhu et al. 2008; Jain et al. 2009; Lydeard et al. 2010; Campos-Doerfler et al. 2018). The downregulation of such mutagenic HR events by Sgs1 is likely responsible for the suppression of GCRs by Sgs1 in the PQ-treated sod2Δ mutant and would be consistent with Sgs1's known role as a suppressor of GCRs in other contexts (Myung et al. 2001; Schmidt et al. 2006b). In contrast to Sgs1, Rad51 is required for most HR events, including those that are mutagenic, such as BIR, providing an explanation for the decrease of the GCR rate of the PQ-treated sod2Δ mutant upon deletion of RAD51.
In addition to HR, we identified TLS polymerases, most notably the Rev1/Polζ mutasome, as a driver of GCR formation in the oxidatively stressed sod2Δ mutant. In contrast, GCR formation in cells lacking the antioxidant Tsa1 is not dependent on TLS polymerases (Ragu et al. 2007).
Analysis of the critical chromosome V region showed that the GCR clones had not arisen by a high rate of inactivating point mutations in the CAN1 and URA3 genes due to PQ treatment of the sod2Δ mutant as >90% of the GCRs had lost both ORFs, indicative of chromosome break involvement. While a mechanism by which TLS promotes genome rearrangements is not immediately evident, one possibility is that TLS is needed during DNA synthesis steps of GCR formation, such as those associated with HR, to bypass Polδ-stalling DNA lesions that arise in the PQ-treated sod2Δ mutant. Recently, TLS polymerases were shown to drive the formation of complex genome rearrangements when BIR is defective due to the absence of Pif1 (Sakofsky et al. 2015). The authors propose a model wherein extension of the invading strand in the BIR bubble stalls, leading to its dissociation and reannealing at a nearby microhomology where DNA synthesis depends on TLS polymerases. An interesting possibility is that the DNA lesions in the PQ-treated sod2Δ mutant can be similarly disruptive to DNA synthesis during GCR formation, necessitating an obligatory switch to TLS polymerases before the rearrangement can be resolved into a viable GCR.
The exact nature of the mutagenic damage that contributes to chromosome instability in the PQ-treated sod2Δ mutant is not known. PQ induces superoxide radicals and PQ-induced DNA damage includes oxidized bases, especially at pyrimidines, and DNA breaks (Keyer and Imlay 1996; Szabó and Ohshima 1997; Petrovska and Dusinska 1999; Tajai et al. 2018), which could be aggravated in the absence of a cellular antioxidant, such as Sod2, leading to increased GCR formation. Through lipid peroxidation, direct or indirect damage to protein structure or activity, or metabolic changes (Fukushima et al. 2002; Li et al. 2021) PQ could further threaten genome stability indirectly, compounding any direct effects of PQ-induced superoxide radicals on genome integrity.
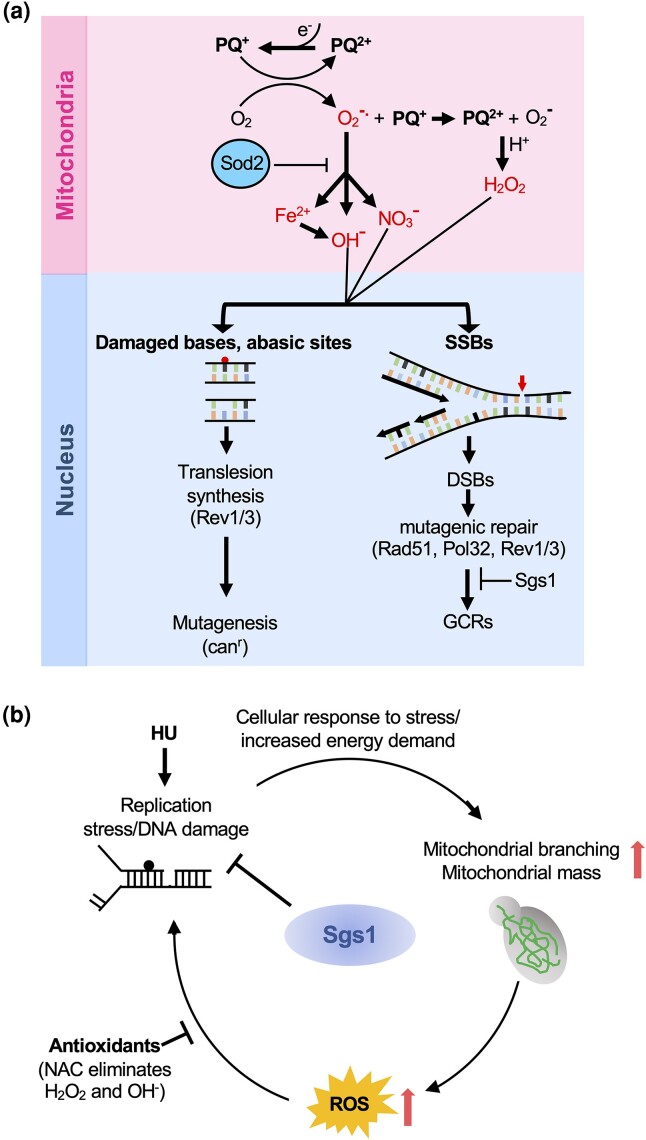
Based on the new functional interactions of sod2Δ in the suppression of nuclear genome instability, we propose the following working model (Fig. 6a): PQ-induced production of superoxide radicals in the mitochondrial matrix can lead to the formation of powerful oxidants (e.g. peroxynitrite, hydroxyl radicals) that can cause oxidative DNA damage in the nucleus that is normally prevented by Sod2, including base damage and DNA single-strand breaks (SSBs). First, increased base damage can stall DNA polymerases, requiring TLS to bypass the lesion, albeit in an error-prone manner, leading to increased mutagenesis. Second, hydroxyl radicals, peroxy-nitrite anions, and hydrogen peroxide can also directly damage the DNA backbone resulting in SSBs. SSBs can be converted into DSBs when a replication fork encounters them and collapses. Mutagenic repair of DNA breaks can result in GCRs whose formation in the sod2Δ mutant appears to be homology-directed, as suggested by their promotion by Rad51 and their suppression by Sgs1. Under conditions of elevated oxidative stress in the PQ-exposed sod2Δ mutant, the successful completion of DNA synthesis steps of homology-directed repair pathways (Pol32) depends on the successful bypass of oxidative-stress-induced DNA lesions by the Rev1/Polζ mutasome.
Fig. 6.
Working models for the role of Sod2 and Sgs1 in the suppression of oxidative stress and genome instability. a) Effects of PQ-induced oxidative stress on genome stability in cells lacking the mitochondrial superoxide dismutase Sod2. The redox cycling drug PQ continuously produces superoxide radicals and additional PQ ions in the mitochondrial matrix, among other cellular locations (Cochemé and Murphy 2008). This uncontrolled superoxide production in the mitochondria can be kept in check by Sod2. However, in the absence of Sod2, superoxide can generate powerful oxidants, such as hydroxyl radicals, peroxynitrite anions, and hydrogen peroxide, and can release free iron from proteins, which can react with other electron donors and produce more ROS like hydroxyl radicals (Keyer and Imlay 1996; Szabó and Ohshima 1997). These ROS can leak out of the mitochondria and result in oxidative DNA damage in the nucleus. Oxidative DNA damage, such as base modifications and abasic sites, the latter of which can be generated spontaneously or as intermediates during BER, can stall the DNA polymerase, requiring TLS to bypass the DNA lesion in an error-prone manner, leading to increased mutagenesis, which can be detected by the CAN1 forward mutation assay. Hydroxyl radicals and peroxy-nitrite anions directly damage the DNA backbone resulting in DNA SSBs, which can lead to one-ended DSBs. Mutagenic repair of DNA lesions in the PQ-exposed sod2Δ mutant by mechanisms dependent on Rad51 and Pol32 can give rise to GCRs. Completion of DNA synthesis steps during GCR formation appears to depend on the bypass of DNA lesions by TLS. In the absence of TLS, replication forks may stall at PQ-induced DNA lesions and abort the mutagenic repair event, thereby suppressing GCRs. These GCR events are suppressed by the antirecombinase Sgs1. b) Hypothesis for the causes and consequences of ROS in cells lacking Sgs1. Loss of Sgs1 results in replication stress and an increased need for DNA repair whose increased demand for ATP induces mitochondrial branching and an increase in mitochondrial mass. This proposed link between the need to respond to the disruption of normal DNA metabolism and the induction of mitochondrial branching is supported by our finding that replication stress induced by HU induces mitochondrial branching similar to the sgs1Δ mutant. Increased mitochondrial mass and activity result in an increase in endogenous ROS production and induce additional DNA damage in the sgs1Δ mutant, including recombinogenic DNA lesions that can be averted by treatment with the antioxidant NAC.
In this study, we have also characterized a novel mitochondrial branching phenotype for the sgs1Δ mutant based on which we propose a model for the oxidative stress phenotype of cells lacking Sgs1 (Fig. 6b): To help with the response to and repair of DNA lesions induced by the absence of Sgs1, cells induce mitochondrial branching to satisfy their increased energy needs. Mitochondrial branching is associated with increased mitochondrial activity and ATP production (Rudan et al. 2018). Notably, replication stress induced by chronic exposure to HU induced the same extensive mitochondrial branching as deletion of SGS1, suggesting that increased mitochondrial branching may be a general response of yeast cells with certain DNA metabolic defects or stressors whereby they produce more ATP for increased genome maintenance needs. However, increased mitochondrial activity resulting from increased mitochondrial mass and branching, such as in the sgs1Δ mutant, yields increased ROS as a mutagenic byproduct, leading to increased DNA repair need, thus, creating a vicious cycle (Fig. 6b). That mitochondrial branching may be the cause of increased ROS rather than its consequence is supported by our finding that neither the sod2Δ mutation nor PQ induced mitochondrial branching. Previous reports of a correlation between recombinogenic DNA lesions mediated by genotoxic stress and an increase in intracellular ROS levels (Yi et al. 2016; Choi et al. 2018) are also consistent with this model.
While the molecular events that lead to mitochondrial branching in the sgs1Δ mutant are unknown, they likely involve a shift from balanced fusion and fission, which generates the tubular mitochondria typical of wildtype cells, toward a state where fusion exceeds fission. Such a shift toward excessive fusion in the sgs1Δ mutant could explain why the sgs1Δ mutation appears to suppress mitochondrial fragmentation, which is a result of excessive fission, in the sod2Δ mutant (Fig. 2b).
In conclusion, we have demonstrated that the mitochondrial antioxidant Sod2 contributes to the maintenance of nuclear genome stability in oxidatively stressed cells by suppressing point mutations and chromosomal rearrangements. That Sod2 did not localize to the nucleus under oxidative stress, unlike Sod1 (Tsang et al. 2014), suggests that its contribution to nuclear genome stability may be mediated indirectly through mitochondria-to-nucleus signaling or directly by preventing the accumulation of mitochondrial ROS that could spread to the nucleus, mutate the nucleotide pool, and change the overall steady-state ROS balance in the cell. Since Sod2 is known to suppress mitochondrial genome instability (Doudican et al. 2005), mitochondrial DNA abnormalities could also promote nuclear DNA mutagenesis. Indeed, loss of the mitochondrial genome and mitochondrial DNA mutations have been shown to aggravate nuclear genome instability (Rasmussen et al. 2003; Veatch et al. 2009).
In human cells, pro- and anti-tumorigenic roles of Sod2 have been identified, which appear to be determined by the stage of cancer progression, the type of cancer, and the tumor environment. On one hand, Sod2 is considered a tumor suppressor (Li et al. 1995; Zhong et al. 1997; Weydert et al. 2003; Ough et al. 2004; Venkataraman et al. 2005; Weydert et al. 2006; Thomas and Sharifi 2012), most likely by preventing ROS-induced DNA damage and thereby suppressing tumor initiation, while on the other, pro-metastatic roles of Sod2 have also emerged, for instance in ROS-stress-responsive cancers such as ovarian clear cell carcinoma (Hemachandra et al. 2015), where increased Sod2 levels have been found to promote cancer cell migration and invasion and are thought to support the high metabolic activity of cancer cells (Liu et al. 2012; Hemachandra et al. 2015; Miar et al. 2015; Chang et al. 2016). Given the oxidative stress phenotype of cells lacking the Bloom's syndrome helicase BLM (Nicotera et al. 1989, 1993; Poot et al. 1989; Lloret et al. 2008; Subramanian et al. 2021), the human homolog of yeast Sgs1, increased ROS could contribute to both the initiation and progression of cancers in this highly cancer-prone syndrome (German and Ellis 1998), which would be further enhanced by any changes in Sod2 expression levels.
Supplementary Material
Acknowledgments
We would like to thank Dr. Dale Chaput and Charles Szekeres for their assistance with mass spectrometry and flow cytometry experiments, respectively, as well as Dr. Stanley Stevens for help with proteomics data analysis.
Contributor Information
Sonia Vidushi Gupta, Department of Molecular Biosciences, University of South Florida, 4202 East Fowler Avenue, Tampa, FL 33620, USA.
Lillian Campos, Department of Molecular Biosciences, University of South Florida, 4202 East Fowler Avenue, Tampa, FL 33620, USA.
Kristina Hildegard Schmidt, Department of Molecular Biosciences, University of South Florida, 4202 East Fowler Avenue, Tampa, FL 33620, USA; Cancer Biology and Evolution Program, H. Lee Moffitt Cancer Center and Research Institute, 12902 USF Magnolia Drive, Tampa, FL 33612, USA.
Data availability
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Yeast strains and their genotypes are listed in Supplementary Table 1 and are available upon request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2022) partner repository with the dataset identifier PXD040745. Supplementary Table 2 lists all proteins with significant changes identified by mass spectrometry proteomics.
Supplemental material available at GENETICS online.
Funding
This work was supported in part by National Institutes of Health grant R01GM018245 to K.H.S.
Author contributions
L.C. performed SILAC-MS, K.H.S. and S.V.G. performed data analysis, and S.V.G. verified the results of the SILAC-MS screen. S.V.G. and K.H.S. planned the study, and S.V.G. performed all experiments. S.V.G. and K.H.S. wrote the manuscript.
Literature cited
- Aerts AM, Zabrocki P, Govaert G, Mathys J, Carmona-Gutierrez D, Madeo F, Winderickx J, Cammue BPA, Thevissen K. Mitochondrial dysfunction leads to reduced chronological lifespan and increased apoptosis in yeast. FEBS Lett. 2009;583(1):113–117. doi: 10.1016/j.febslet.2008.11.028. [DOI] [PubMed] [Google Scholar]
- Agmon N, Yovel M, Harari Y, Liefshitz B, Kupiec M. The role of Holliday junction resolvases in the repair of spontaneous and induced DNA damage. Nucleic Acids Res. 2011;39(16):7009–7019. doi: 10.1093/nar/gkr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruoma OI, Halliwell B, Hoey BM, Butler J. The antioxidant action of N-acetylcysteine: its reaction with hydrogen peroxide, hydroxyl radical, superoxide, and hypochlorous acid. Free Radic Biol Med. 1989;6(6):593–597. doi: 10.1016/0891-5849(89)90066-X. [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat–containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318(5851):798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- Baranovskiy AG, Lada AG, Siebler HM, Zhang Y, Pavlov YI, Tahirov TH. DNA polymerase delta and zeta switch by sharing accessory subunits of DNA polymerase delta. J Biol Chem. 2012;287(21):17281–17287. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnham KJ, Masters CL, Bush AI. Neurodegenerative diseases and oxidative stress. Nat Rev Drug Discov. 2004;3(3):205–214. doi: 10.1038/nrd1330. [DOI] [PubMed] [Google Scholar]
- Benel L, Ronot X, Mounolou J, Gaudemer F, Adolphe M. Compared flow cytometric analysis of mitochondria using 10-n-nonyl acridine orange and rhodamine 123. Basic Appl Histochem. 1989;33(2):71–80. [PubMed] [Google Scholar]
- Boddy MN, Gaillard PHL, McDonald WH, Shanahan P, Yates JR III, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107(4):537–548. doi: 10.1016/S0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP. Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol Cell. 2009;35(1):70–81. doi: 10.1016/j.molcel.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M. Ubc9-and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127(3):509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- Bravard A, Sabatier L, Hoffschir F, Ricoul M, Luccioni C, Dutrillaux B. SOD2: a new type of tumor-suppressor gene? Int J Cancer. 1992;51(3):476–480. doi: 10.1002/ijc.2910510323. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3(11):1271. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- Campos-Doerfler L, Syed S, Schmidt KH. Sgs1 binding to Rad51 stimulates homology-directed DNA repair in Saccharomyces cerevisiae. Genetics. 2018;208(1):125–138. doi: 10.1534/genetics.117.300545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CD, Kitchen LE, Au W-C, Babic CM, Basrai MA. Loss of SOD1 and LYS7 sensitizes Saccharomyces cerevisiae to hydroxyurea and DNA damage agents and downregulates MEC1 pathway effectors. Mol Cell Biol. 2005;25(23):10273–10285. doi: 10.1128/MCB.25.23.10273-10285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1–Top3. Nat Struct Mol Biol. 2010;17(11):1377. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Reddel RR. Telomere uncapping and alternative lengthening of telomeres. Mech Ageing Dev. 2008;129(1–2):99–108. doi: 10.1016/j.mad.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Chae HZ, Kim I, Kim K, Rhee SG. Cloning, sequencing, and mutation of thiol-specific antioxidant gene of Saccharomyces cerevisiae. J Biol Chem. 1993;268(22):16815–16821. doi: 10.1016/S0021-9258(19)85489-3. [DOI] [PubMed] [Google Scholar]
- Chang B, et al. SOD2 deregulation enhances migration, invasion and has poor prognosis in salivary adenoid cystic carcinoma. Sci Rep. 2016;6:25918. doi: 10.1038/srep25918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Bellaoui M, Zhang C, Desai R, Morozov P, Delgado-Cruzata L, Rothstein R, Freyer GA, Boone C, Brown GW. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/topo III complex. EMBO J. 2005;24(11):2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput D, Kirouac L, Stevens SM Jr, Padmanabhan J. Potential role of PCTAIRE-2, PCTAIRE-3 and P-Histone H4 in amyloid precursor protein-dependent Alzheimer pathology. Oncotarget. 2016;7(8):8481–8497. doi: 10.18632/oncotarget.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 1999;23(1):81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- Chen B-L, Wang H-M, Lin X-S, Zeng Y-M. UPF1: a potential biomarker in human cancers. Front Biosci (Landmark Ed). 2021;26(5):76–84. doi: 10.52586/4925. [DOI] [PubMed] [Google Scholar]
- Choi JE, Heo S-H, Kim MJ, Chung W-H. Lack of superoxide dismutase in a rad51 mutant exacerbates genomic instability and oxidative stress-mediated cytotoxicity in Saccharomyces cerevisiae. Free Radic Biol Med. 2018;129:97–106. doi: 10.1016/j.freeradbiomed.2018.09.015. [DOI] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003;22(16):4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochemé HM, Murphy MP. Complex I is the major site of mitochondrial superoxide production by paraquat. J Biol Chem. 2008;283(4):1786–1798. doi: 10.1074/jbc.M708597200. [DOI] [PubMed] [Google Scholar]
- Cohen H, Sinclair DA. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc Natl Acad Sci U S A. 2001;98(6):3174–3179. doi: 10.1073/pnas.061579598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. Maxquant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cuevas-Bermúdez A, Garrido-Godino AI, Navarro F. A novel yeast chromatin-enriched fractions purification approach, yChEFs, for the chromatin-associated protein analysis used for chromatin-associated and RNA-dependent chromatin-associated proteome studies from Saccharomyces cerevisiae. Gene Rep. 2019;16(1):100450. doi: 10.1016/j.genrep.2019.100450. [DOI] [Google Scholar]
- Cunniff C, Bassetti JA, Ellis NA. Bloom’s syndrome: clinical spectrum, molecular pathogenesis, and cancer predisposition. Mol Syndromol. 2017;8(1):4–23. doi: 10.1159/000452082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AB, Sadowska-Bartosz I, Königstorfer A, Kettle AJ, Winterbourn CC. Superoxide dismutase protects ribonucleotide reductase from inactivation in yeast. Free Radic Biol Med. 2018;116:114–122. doi: 10.1016/j.freeradbiomed.2018.01.001. [DOI] [PubMed] [Google Scholar]
- Davis AP, Symington LS. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics. 2001;159(2):515–525. doi: 10.1093/genetics/159.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler L, Schmidt KH. Exo1 phosphorylation status controls the hydroxyurea sensitivity of cells lacking the Pol32 subunit of DNA polymerases delta and zeta. DNA Repair (Amst). 2014;24:26–36. doi: 10.1016/j.dnarep.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudican NA, Song B, Shadel GS, Doetsch PW. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25(12):5196–5204. doi: 10.1128/MCB.25.12.5196-5204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttaroy A, Paul A, Kundu M, Belton A. A Sod2 null mutation confers severely reduced adult life span in Drosophila. Genetics. 2003;165(4):2295–2299. doi: 10.1093/genetics/165.4.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edward E, Luk C, Culotta VC. Manganese superoxide dismutase in Saccharomyces cerevisiae acquires its metal co-factor through a pathway involving the Nramp metal transporter, Smf2p. J. Biol. Chem. 2001;276(50):47556–47562. doi: 10.1074/jbc.M108923200. [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Zhou Z, Allen JB, Navas TA. DNA Damage and cell cycle regulation of ribonucleotide reductase. Bioessays. 1993;15(5):333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- Ellis NA, Groden J, Ye T-Z, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83(4):655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Battistella L, Vardavas R, Gattazzo C, Liou L-L, Diaspro A, Dossen JW, Gralla EB, Longo VD. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J Cell Biol. 2004;166(7):1055–1067. doi: 10.1083/jcb.200404002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasching CL, Cejka P, Kowalczykowski SC, Heyer W-D. Top3-Rmi1 dissolve Rad51-mediated D loops by a topoisomerase-based mechanism. Mol Cell. 2015;57(4):595–606. doi: 10.1016/j.molcel.2015.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408(6809):239. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Frei C, Gasser SM. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14(1):81–96. doi: 10.1101/gad.14.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridovich I. Advances in Enzymology and Related Areas of Molecular Biology, Vol. 41. New York (NY): John Wiley; 1974. [Google Scholar]
- Fukushima T, Tanaka K, Lim H, Moriyama M. Mechanism of cytotoxicity of paraquat. Environ Health Prev Med. 2002;7(3):89–94. doi: 10.1265/ehpm.2002.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafter-Gvili A, Herman M, Ori Y, Korzets A, Chagnac A, Zingerman B, Rozen-Zvi B, Gafter U, Malachi T. Inhibition of mitochondrial function reduces DNA repair in human mononuclear cells. Leuk Res. 2011;35(2):219–225. doi: 10.1016/j.leukres.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14(12):8391–8398. doi: 10.1128/mcb.14.12.8391-8398.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatbonton T, Imbesi M, Nelson M, Akey JM, Ruderfer DM, Kruglyak L, Simon JA, Bedalov A. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2006;2(3):e35. doi: 10.1371/journal.pgen.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German J, Ellis NA. Bloom syndrome. In: Vogelstein B, Kinzler KW, editors. The Genetic Basis of Human Cancer. New York: McGraw-Hill, Health Professions Division; 1998. p. 301–315. [Google Scholar]
- Gibbs PE, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169(2):575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. In: Xiao W. editor. Yeast Protocol. Methods in Molecular Biology, Vol. 313. Totowa (NJ): Humana Press; 2006. p. 107–120. [Google Scholar]
- Goscin SA, Fridovich I. The purification and properties of superoxide dismutase from Saccharomyces cerevisiae. Biochim Biophys Acta. 1972;289(2):276–283. doi: 10.1016/0005-2744(72)90078-2. [DOI] [PubMed] [Google Scholar]
- Gralla EB, Valentine JS. Null mutants of Saccharomyces cerevisiae Cu, Zn superoxide dismutase: characterization and spontaneous mutation rates. J Bacteriol. 1991;173(18):5918–5920. doi: 10.1128/jb.173.18.5918-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP. DNA Helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22(20):2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemachandra LMP, Shin D-H, Dier U, Iuliano JN, Engelberth SA, Uusitalo LM, Murphy SK, Hempel N. Mitochondrial superoxide dismutase has a protumorigenic role in ovarian clear cell carcinoma. Cancer Res. 2015;75(22):4973–4984. doi: 10.1158/0008-5472.CAN-14-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Mazon G, Lam AF, Symington LS. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol Cell. 2010;40(6):988–1000. doi: 10.1016/j.molcel.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9(7):811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Huang T-T, Carlson EJ, Gillespie AM, Epstein CJ. Genetic modification of the dilated cardiomyopathy and neonatal lethality phenotype of mice lacking manganese superoxide dismutase. Age (Omaha). 1998;21(2):83. doi: 10.1007/s11357-998-0011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M-E, de Calignon A, Nicolas A, Galibert F. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase δ, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr Genet. 2000;38(4):178–187. doi: 10.1007/s002940000149. [DOI] [PubMed] [Google Scholar]
- Huang M-E, Kolodner RD. A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage. Mol Cell. 2005;17(5):709–720. doi: 10.1016/j.molcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Huang M-E, Rio A-G, Nicolas A, Kolodner RD. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc Natl Acad Sci U S A. 2003;100(20):11529–11534. doi: 10.1073/pnas.2035018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JY, Smith S, Myung K. The rad1-Rad10 complex promotes the production of gross chromosomal rearrangements from spontaneous DNA damage in Saccharomyces cerevisiae. Genetics. 2005;169(4):1927–1937. doi: 10.1534/genetics.104.039768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115(4):401–411. doi: 10.1016/S0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irokawa H, Tachibana T, Watanabe T, Matsuyama Y, Motohashi H, Ogasawara A, Iwai K, Naganuma A, Kuge S. Redox-dependent regulation of gluconeogenesis by a novel mechanism mediated by a peroxidatic cysteine of peroxiredoxin. Sci Rep. 2016;6(1):33536. doi: 10.1038/srep33536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Sugawara N, Lydeard J, Vaze M, Tanguy Le Gac N, Haber JE. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev. 2009;23(3):291–303. doi: 10.1101/gad.1751209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs S, Martini N, Schauss AC, Egner A, Westermann B, Hell SW. Spatial and temporal dynamics of budding yeast mitochondria lacking the division component Fis1p. J Cell Sci. 2003;116(10):2005–2014. doi: 10.1242/jcs.00423. [DOI] [PubMed] [Google Scholar]
- Johnson-Cadwell L, Jekabsons M, Wang A, Polster B, Nicholls D. ‘Mild uncoupling’ does not decrease mitochondrial superoxide levels in cultured cerebellar granule neurons but decreases spare respiratory capacity and increases toxicity to glutamate and oxidative stress. J Neurochem. 2007;101(6):1619–1631. doi: 10.1111/j.1471-4159.2007.04516.x. [DOI] [PubMed] [Google Scholar]
- Johnson FB, Marciniak RA, McVey M, Stewart SA, Hahn WC, Guarente L. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 2001;20(4):905–913. doi: 10.1093/emboj/20.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyer K, Imlay JA. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci U S A. 1996;93(24):13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DR, Gidvani RD, Ingalls BP, Duncker BP, McConkey BJ. Differential chromatin proteomics of the MMS-induced DNA damage response in yeast. Proteome Sci. 2011;9(1):1–14. doi: 10.1186/1477-5956-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Park J, Ryu TH, Nili M. Effect of N-acetyl-l-cysteine on Saccharomyces cerevisiae irradiated with gamma-rays. Chemosphere. 2013;92(5):512–516. doi: 10.1016/j.chemosphere.2013.02.035. [DOI] [PubMed] [Google Scholar]
- Klinger H, Rinnerthaler M, Lam YT, Laun P, Heeren G, Klocker A, Simon-Nobbe B, Dickinson JR, Dawes IW, Breitenbach M. Quantitation of (a) symmetric inheritance of functional and of oxidatively damaged mitochondrial aconitase in the cell division of old yeast mother cells. Exp Gerontol. 2010;45(7–8):533–542. doi: 10.1016/j.exger.2010.03.016. [DOI] [PubMed] [Google Scholar]
- Kubota T, Stead DA, Hiraga S-I, ten Have S, Donaldson AD. Quantitative proteomic analysis of yeast DNA replication proteins. Methods. 2012;57(2):196–202. doi: 10.1016/j.ymeth.2012.03.012. [DOI] [PubMed] [Google Scholar]
- Kulkarni R, Thomas RA, Tucker JD. Expression of DNA repair and apoptosis genes in mitochondrial mutant and normal cells following exposure to ionizing radiation. Environ Mol Mutagen. 2011;52(3):229–237. doi: 10.1002/em.20605. [DOI] [PubMed] [Google Scholar]
- Kusano K, Berres ME, Engels WR. Evolution of the RECQ family of helicases: a Drosophila homolog, dmblm, is similar to the human bloom syndrome gene. Genetics. 1999;151(3):1027–1039. doi: 10.1093/genetics/151.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C-Y, Jaruga E, Borghouts C, Jazwinski SM. A mutation in the ATP2 gene abrogates the age asymmetry between mother and daughter cells of the yeast Saccharomyces cerevisiae. Genetics. 2002;162(1):73–87. doi: 10.1093/genetics/162.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laun P, Pichova A, Madeo F, Fuchs J, Ellinger A, Kohlwein S, Dawes I, Fröhlich K-U, Breitenbach M. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol Microbiol. 2001;39(5):1166–1173. doi: 10.1111/j.1365-2958.2001.02317.x. [DOI] [PubMed] [Google Scholar]
- Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49(3):264. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- Lebovitz RM, Zhang H, Vogel H, Cartwright J, Dionne L, Lu N, Huang S, Matzuk MM. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc Natl Acad Sci U S A. 1996;93(18):9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12(5):2165–2177. doi: 10.1128/mcb.12.5.2165-2177.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew JE, Enomoto S, Berman J. Telomere length regulation and telomeric chromatin require the nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1998;18(10):6121–6130. doi: 10.1128/MCB.18.10.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Oberley TD, Oberley LW, Zhong W. Overexpression of manganese superoxide dismutase in DU145 human prostate carcinoma cells has multiple effects on cell phenotype. Prostate. 1998;35(3):221–233. doi:. [DOI] [PubMed] [Google Scholar]
- Li J-J, Oberley LW, St Clair DK, Ridnour LA, Oberley TD. Phenotypic changes induced in human breast cancer cells by overexpression of manganese-containing superoxide dismutase. Oncogene. 1995;10(10):1989–2000. [PubMed] [Google Scholar]
- Li Y, Zhong X, Ye J, Guo H, Long Y. Proteome of Saccharomyces cerevisiae under paraquat stress regulated by therapeutic concentration of copper ions. Ecotoxicol Environ Saf. 2021;217:112245. doi: 10.1016/j.ecoenv.2021.112245. [DOI] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci U S A. 2001;98(15):8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Li S, Cai Y, Wang A, He Q, Zheng C, Zhao T, Ding X, Zhou X. Manganese superoxide dismutase induces migration and invasion of tongue squamous cell carcinoma via H2O2-dependent Snail signaling. Free Radic Biol Med. 2012;53(1):44–50. doi: 10.1016/j.freeradbiomed.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloret A, Calzone R, Dunster C, Manini P, d'Ischia M, Degan P, Kelly FJ, Pallardó FV, Zatterale A, Pagano G. Different patterns of in vivo pro-oxidant states in a set of cancer-or aging-related genetic diseases. Free Radic Biol Med. 2008;44(4):495–503. doi: 10.1016/j.freeradbiomed.2007.10.046. [DOI] [PubMed] [Google Scholar]
- Lo YC, Paffett KS, Amit O, Clikeman JA, Sterk R, Brenneman MA, Nickoloff JA. Sgs1 regulates gene conversion tract lengths and crossovers independently of its helicase activity. Mol Cell Biol. 2006;26(11):4086–4094. doi: 10.1128/MCB.00136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Gralla EB, Valentine JS. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J Biol Chem. 1996;271(21):12275–12280. doi: 10.1074/jbc.271.21.12275. [DOI] [PubMed] [Google Scholar]
- Longo VD, Liou L-L, Valentine JS, Gralla EB. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch Biochem Biophys. 1999;365(1):131–142. doi: 10.1006/abbi.1999.1158. [DOI] [PubMed] [Google Scholar]
- Longtine MS, Mckenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–961. doi: [DOI] [PubMed] [Google Scholar]
- Luk E, Carroll M, Baker M, Culotta VC. Manganese activation of superoxide dismutase 2 in Saccharomyces cerevisiae requires MTM1, a member of the mitochondrial carrier family. Proc Natl Acad Sci U S A. 2003;100(18):10353–10357. doi: 10.1073/pnas.1632471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448(7155):820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Lipkin-Moore Z, Jain S, Eapen VV, Haber JE. Sgs1 and exo1 redundantly inhibit break-induced replication and de novo telomere addition at broken chromosome ends. PLoS Genet. 2010;6(5):e1000973. doi: 10.1371/journal.pgen.1000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova AV, Burgers PM. Eukaryotic DNA polymerase zeta. DNA Repair (Amst). 2015;29:47–55. doi: 10.1016/j.dnarep.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri HW, Ngo H-P, Hickson ID. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol Biol Cell. 2007;18(10):4062–4073. doi: 10.1091/mbc.e07-05-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L, Clair DKS. Regulation of superoxide dismutase genes: implications in disease. Free Radic Biol Med. 2009;47(4):344–356. doi: 10.1016/j.freeradbiomed.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miar A, Hevia D, Muñoz-Cimadevilla H, Astudillo A, Velasco J, Sainz RM, Mayo JC. Manganese superoxide dismutase (SOD2/MnSOD)/catalase and SOD2/GPx1 ratios as biomarkers for tumor progression and metastasis in prostate, colon, and lung cancer. Free Radic Biol Med. 2015;85:45–55. doi: 10.1016/j.freeradbiomed.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455(7214):770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29(19):3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei H, Schmidt KH. Non-bloom syndrome-associated partial and total loss-of-function variants of BLM helicase. Proc Natl Acad Sci U S A. 2012;109(47):19357–19362. doi: 10.1073/pnas.1210304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei H, Syed S, Kennedy J, Schmidt KH. Sgs1 truncations induce genome rearrangements but suppress detrimental effects of BLM overexpression in Saccharomyces cerevisiae. J Mol Biol. 2011;405(4):877–891. doi: 10.1016/j.jmb.2010.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muellner J, Schmidt KH. Helicase activities of Rad5 and Rrm3 genetically interact in the prevention of recombinogenic DNA lesions in Saccharomyces cerevisiae. DNA Repair (Amst). 2023;126:103488. doi: 10.1016/j.dnarep.2023.103488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Datta A, Chen C, Kolodner RD. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat Genet. 2001;27(1):113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- Nair K. Table of confidence interval for the median in samples from any continuous population. Sankhyā. 1940;4(4):551–558. [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391(6665):407. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- Nicotera TM, Notaro J, Notaro S, Schumer J, Sandberg AA. Elevated superoxide dismutase in Bloom’s syndrome: a genetic condition of oxidative stress. Cancer Res. 1989;49(19):5239–5243. [PubMed] [Google Scholar]
- Nicotera T, Thusu K, Dandona P. Elevated production of active oxygen in Bloom’s syndrome cell lines. Cancer Res. 1993;53(21):5104–5107. [PubMed] [Google Scholar]
- Nielsen I, Bentsen IB, Andersen AH, Gasser SM, Bjergbaek L. A Rad53 independent function of Rad9 becomes crucial for genome maintenance in the absence of the Recq helicase Sgs1. PLoS One. 2013;8(11):e81015. doi: 10.1371/journal.pone.0081015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaka AT, Su J, Katahira Y, Tang C, Zafar F, Aoki K, Kagawa W, Niki H, Iwasaki H, Nakagawa T. DNA Replication machinery prevents Rad52-dependent single-strand annealing that leads to gross chromosomal rearrangements at centromeres. Commun Biol. 2020;3(1):1–14. doi: 10.1038/s42003-020-0934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ough M, Lewis A, Zhang Y, Hinkhouse MM, Ritchie JM, Oberley LW, Cullen JJ. Inhibition of cell growth by overexpression of manganese superoxide dismutase (MnSOD) in human pancreatic carcinoma. Free Radic Res. 2004;38(11):1223–1233. doi: 10.1080/10715760400017376. [DOI] [PubMed] [Google Scholar]
- Pages V, Johnson RE, Prakash L, Prakash S. Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc Natl Acad Sci U S A. 2008;105(4):1170–1175. doi: 10.1073/pnas.0711227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SG, Cha M-K, Jeong W, Kim I-H. Distinct physiological functions of thiol peroxidase isoenzymes in Saccharomyces cerevisiae. J Biol Chem. 2000;275(8):5723–5732. doi: 10.1074/jbc.275.8.5723. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y, Bai J, Bandla C, Garcia-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu DJ, Prakash A, Frericks-Zipper A, Eisenacher M, et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50(D1):D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovska H, Dusinska M. Oxidative DNA damage in human cells induced by paraquat. Altern Lab Anim. 1999;27(3):387–395. doi: 10.1177/026119299902700314. [DOI] [PubMed] [Google Scholar]
- Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393(6680):91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- Petukhova G, Stratton SA, Sung P. Single strand DNA binding and annealing activities in the yeast recombination factor Rad59. J Biol Chem. 1999;274(48):33839–33842. doi: 10.1074/jbc.274.48.33839. [DOI] [PubMed] [Google Scholar]
- Poot M, Hoehn H, Nicotera TM, Rüdiger HW. Cell kinetic evidence suggests elevated oxidative stress in cultured cells of Bloom’s syndrome. Free Radic Res Commun. 1989;7(3–6):179–187. doi: 10.3109/10715768909087940. [DOI] [PubMed] [Google Scholar]
- Quaranta D, Krans T, Santo CE, Elowsky CG, Domaille DW, Chang CJ, Grass G. Mechanisms of contact-mediated killing of yeast cells on dry metallic copper surfaces. Appl Environ Microbiol. 2011;77(2):416–426. doi: 10.1128/AEM.01704-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragu S, Faye G, Iraqui I, Masurel-Heneman A, Kolodner RD, Huang ME. Oxygen metabolism and reactive oxygen species cause chromosomal rearrangements and cell death. Proc Natl Acad Sci U S A. 2007;104(23):9747–9752. doi: 10.1073/pnas.0703192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez O, Motta-Mena LB, Cordova A, Garza KM. A small library of synthetic di-substituted 1, 4-naphthoquinones induces ROS-mediated cell death in murine fibroblasts. PLoS One. 2014;9(9):e106828. doi: 10.1371/journal.pone.0106828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen AK, Chatterjee A, Rasmussen LJ, Singh KK. Mitochondria-mediated nuclear mutator phenotype in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31(14):3909–3917. doi: 10.1093/nar/gkg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindranath S, Fridovich I. Isolation and characterization of a manganese-containing superoxide dismutase from yeast. J Biol Chem. 1975;250(15):6107–6112. doi: 10.1016/S0021-9258(19)41165-4. [DOI] [PubMed] [Google Scholar]
- Reenan R, Kolodner RD. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics. 1992;132(4):975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringvoll J, Uldal L, Roed MA, Reite K, Baynton K, Klungland A, Eide L. Mutations in the RAD27 and SGS1 genes differentially affect the chronological and replicative lifespan of yeast cells growing on glucose and glycerol. FEMS Yeast Res. 2007;7(6):848–859. doi: 10.1111/j.1567-1364.2007.00248.x. [DOI] [PubMed] [Google Scholar]
- Rockmill B, Lambie EJ, Roeder GS. Spore enrichment. Methods Enzymol. 1991;194:146–149. doi: 10.1016/0076-6879(91)94012-2. [DOI] [PubMed] [Google Scholar]
- Rossignol D, Frye R. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry. 2012;17(3):290. doi: 10.1038/mp.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudan M, et al. Normal mitochondrial function in Saccharomyces cerevisiae has become dependent on inefficient splicing. Elife. 2018;7:e35330. doi: 10.7554/eLife.35330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajesh BV, Bailey M, Lichtensztejn Z, Hieter P, McManus KJ. Synthetic lethal targeting of superoxide dismutase 1 selectively kills RAD54B-deficient colorectal cancer cells. Genetics. 2013;195(3):757–767. doi: 10.1534/genetics.113.156836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajesh BV, McManus KJ. Targeting SOD1 induces synthetic lethal killing in BLM-and CHEK2-deficient colorectal cancer cells. Oncotarget. 2015;6(29):27907. doi: 10.18632/oncotarget.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakofsky CJ, Ayyar S, Deem AK, Chung WH, Ira G, Malkova A. Translesion polymerases drive microhomology-mediated break-induced replication leading to complex chromosomal rearrangements. Mol Cell. 2015;60(6):860–872. doi: 10.1016/j.molcel.2015.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon TB, Evert BA, Song B, Doetsch PW. Biological consequences of oxidative stress-induced DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32(12):3712–3723. doi: 10.1093/nar/gkh696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariki SK, Sahu PK, Golla U, Singh V, Azad GK, Tomar RS. Sen1, the homolog of human Senataxin, is critical for cell survival through regulation of redox homeostasis, mitochondrial function, and the TOR pathway in Saccharomyces cerevisiae. FEBS J. 2016;283(22):4056–4083. doi: 10.1111/febs.13917. [DOI] [PubMed] [Google Scholar]
- Schmidt KH, Kolodner RD. Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol Cell Biol. 2004;24(8):3213–3226. doi: 10.1128/MCB.24.8.3213-3226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KH, Pennaneach V, Putnam CD, Kolodner RD. Analysis of gross-chromosomal rearrangements in Saccharomyces cerevisiae. Methods Enzymol. 2006a;409:462–476. doi: 10.1016/S0076-6879(05)09027-0. [DOI] [PubMed] [Google Scholar]
- Schmidt KH, Wu J, Kolodner RD. Control of translocations between highly diverged genes by Sgs1, the Saccharomyces cerevisiae Homolog of the Bloom's Syndrome protein. Mol Cell Biol. 2006b;26(14):5406–5420. doi: 10.1128/MCB.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochim Biophys Acta. 2006;1763(12):1755–1766. doi: 10.1016/j.bbamcr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Shaw JM, Nunnari J. Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 2002;12(4):178–184. doi: 10.1016/S0962-8924(01)02246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signon L, Malkova A, Naylor ML, Klein H, Haber JE. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol Cell Biol. 2001;21(6):2048–2056. doi: 10.1128/MCB.21.6.2048-2056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupphaug G, Kavli B, Krokan HE. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat Res. 2003;531(1–2):231–251. doi: 10.1016/j.mrfmmm.2003.06.002. [DOI] [PubMed] [Google Scholar]
- Smith S, Hwang J-Y, Banerjee S, Majeed A, Gupta A, Myung K. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101(24):9039–9044. doi: 10.1073/pnas.0403093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Sung P. Functional interactions among yeast Rad51 recombinase, Rad52 mediator, and replication protein A in DNA strand exchange. J Biol Chem. 2000;275(21):15895–15904. doi: 10.1074/jbc.M910244199. [DOI] [PubMed] [Google Scholar]
- Spell RM, Jinks-Robertson S. Examination of the Roles of Sgs1 and Srs2 helicases in the enforcement of recombination fidelity in Saccharomyces cerevisiae. Genetics. 2004;168(4):1855–1865. doi: 10.1534/genetics.104.032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC. A fraction of yeast Cu, Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria: a physiological role for SOD1 in guarding against mitochondrial oxidative damage. J Biol Chem. 2001;276(41):38084–38089. doi: 10.1074/jbc.M105296200. [DOI] [PubMed] [Google Scholar]
- Subramanian V, Rodemoyer B, Shastri V, Rasmussen LJ, Desler C, Schmidt KH. Bloom syndrome DNA helicase deficiency is associated with oxidative stress and mitochondrial network changes. Sci Rep. 2021;11(1):1–18. doi: 10.1038/s41598-021-81075-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Goldfarb T, Studamire B, Alani E, Haber JE. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc Natl Acad Sci U S A. 2004;101(25):9315–9320. doi: 10.1073/pnas.0305749101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell. 2003;12(1):209–219. doi: 10.1016/S1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- Sun H, Bennett RJ, Maizels N. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds GG paired DNAs. Nucleic Acids Res. 1999;27(9):1978–1984. doi: 10.1093/nar/27.9.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265(5176):1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Syed S, Desler C, Rasmussen LJ, Schmidt KH. A novel Rrm3 function in restricting DNA replication via an Orc5-binding domain is genetically separable from Rrm3 function as an ATPase/helicase in facilitating fork progression. PLoS Genet. 2016;12(12):e1006451. doi: 10.1371/journal.pgen.1006451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó C, Ohshima H. DNA damage induced by peroxynitrite: subsequent biological effects. Nitric Oxide. 1997;1(5):373–385. doi: 10.1006/niox.1997.0143. [DOI] [PubMed] [Google Scholar]
- Tajai P, Fedeles BI, Suriyo T, Navasumrit P, Kanitwithayanun J, Essigmann JM, Satayavivad J. An engineered cell line lacking OGG1 and MUTYH glycosylases implicates the accumulation of genomic 8-oxoguanine as the basis for paraquat mutagenicity. Free Radic Biol Med. 2018;116:64–72. doi: 10.1016/j.freeradbiomed.2017.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan F, Li G, Chitteti BR, Peng Z. Proteome and phosphoproteome analysis of chromatin associated proteins in rice (Oryza sativa). Proteomics. 2007;7(24):4511–4527. doi: 10.1002/pmic.200700580. [DOI] [PubMed] [Google Scholar]
- Thomas R, Sharifi N. SOD Mimetics: a novel class of androgen receptor inhibitors that suppresses castration-resistant growth of prostate cancer. Mol Cancer Ther. 2012;11(1):87–97. doi: 10.1158/1535-7163.MCT-11-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang CK, Liu Y, Thomas J, Zhang Y, Zheng XS. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat Commun. 2014;5(1):3446. doi: 10.1038/ncomms4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat Methods. 2016;13(9):731. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Van Loon A, Pesold-Hurt B, Schatz G. A yeast mutant lacking mitochondrial manganese-superoxide dismutase is hypersensitive to oxygen. Proc Natl Acad Sci U S A. 1986;83(11):3820–3824. doi: 10.1073/pnas.83.11.3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137(7):1247–1258. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman S, Jiang X, Weydert C, Zhang Y, Zhang HJ, Goswami PC, Ritchie JM, Oberley LW, Buettner GR. Manganese superoxide dismutase overexpression inhibits the growth of androgen-independent prostate cancer cells. Oncogene. 2005;24(1):77–89. doi: 10.1038/sj.onc.1208145. [DOI] [PubMed] [Google Scholar]
- Vögtle F-N, Burkhart JM, Gonczarowska-Jorge H, Kücükköse C, Taskin AA, Kopczynski D, Ahrends R, Mossmann D, Sickmann A, Zahedi RP, et al. Landscape of submitochondrial protein distribution. Nat Commun. 2017;8(1):1–10. doi: 10.1038/s41467-017-00359-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt PM, Louis EJ, Borts RH, Hickson ID. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81(2):253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- Weisiger RA, Fridovich I. Mitochondrial superoxide dismutase. Site of synthesis and intramitochondrial localization. J Biol Chem. 1973;248(13):4793–4796. doi: 10.1016/S0021-9258(19)43735-6. [DOI] [PubMed] [Google Scholar]
- Weydert C, Roling B, Liu J, Hinkhouse MM, Ritchie JM, Oberley LW, Cullen JJ. Suppression of the malignant phenotype in human pancreatic cancer cells by the overexpression of manganese superoxide dismutase. Mol Cancer Ther. 2003;2(4):361–369. [PubMed] [Google Scholar]
- Weydert CJ, Waugh TA, Ritchie JM, Iyer KS, Smith JL, Li L, Spitz DR, Oberley LW. Overexpression of manganese or copper–zinc superoxide dismutase inhibits breast cancer growth. Free Radic Biol Med. 2006;41(2):226–237. doi: 10.1016/j.freeradbiomed.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Whittaker MM, Whittaker JW. Metallation state of human manganese superoxide dismutase expressed in Saccharomyces cerevisiae. Arch Biochem Biophys. 2012;523(2):191–197. doi: 10.1016/j.abb.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom’s Syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426(6968):870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Kato J-I, Shimamoto A, Goto M, Furuichi Y, Ikeda H. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc Natl Acad Sci U S A. 1998;95(15):8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Cobine PA, Molik S, Naranuntarat A, Lill R, Winge DR, Culotta VC. The effects of mitochondrial iron homeostasis on cofactor specificity of superoxide dismutase 2. EMBO J. 2006;25(8):1775–1783. doi: 10.1038/sj.emboj.7601064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi DG, Kim MJ, Choi JE, Lee J, Jung J, Huh W-K, Chung W-H. Yap1 and Skn7 genetically interact with Rad51 in response to oxidative stress and DNA double-strand break in Saccharomyces cerevisiae. Free Radic Biol Med. 2016;101:424–433. doi: 10.1016/j.freeradbiomed.2016.11.005. [DOI] [PubMed] [Google Scholar]
- Zafarullah M, Li W, Sylvester J, Ahmad M. Molecular mechanisms of N-acetylcysteine actions. Cell Mol Life Sci. 2003;60(1):6–20. doi: 10.1007/s000180300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Oberley LW, Oberley TD, St Clair DK. Suppression of the malignant phenotype of human glioma cells by overexpression of manganese superoxide dismutase. Oncogene. 1997;14(4):481–490. doi: 10.1038/sj.onc.1200852. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134(6):981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Yeast strains and their genotypes are listed in Supplementary Table 1 and are available upon request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2022) partner repository with the dataset identifier PXD040745. Supplementary Table 2 lists all proteins with significant changes identified by mass spectrometry proteomics.
Supplemental material available at GENETICS online.