Summary
Background
Routine whole genome sequencing of Mycobacterium tuberculosis has been implemented with increasing frequency. However, its value for tuberculosis (TB) control programs beyond individual case management and enhanced drug resistance detection has not yet been explored.
Methods
We analysed routine sequencing data of culture-confirmed TB cases notified between 1st January 2017 and 31st December 2021 in New South Wales (NSW), Australia. Genomic surveillance included evidence of local TB transmission, defined by single nucleotide polymorphism (SNP) clustering over a variable (0–25) SNP threshold, and drug resistance conferring mutations.
Findings
M. tuberculosis sequences from 1831 patients were examined, representing 64.8% of all notified TB cases and 96.2% of culture-confirmed cases. Applying a traditional 5-SNP cluster threshold identified 62 transmission clusters with 183 clustered cases; 101/183 (55.2%) had 0 SNP differences. Cluster assessment over a 5-year period, using a 5-SNP threshold, provided a comprehensive overview of likely recent transmission within NSW, Australia, as an indicator of local TB control. Genotypic drug susceptibility testing (DST) was highly concordant with phenotypic DST and provided a 6.8% increase in antimycobacterial resistance detection. Importantly, it detected mutations missed by routine molecular tests. Lineage 2 strains were more likely to be drug resistant (p < 0.0001) and locally transmitted if drug resistant (p < 0.0001).
Interpretation
Performing routine prospective WGS in a low incidence country like Australia, provides genomically informed programmatic indicators of local TB control. A rolling 5-year cluster assessment reflects epidemic containment and progress towards ‘zero TB transmission’. Genomic DST also provides valuable information for clinical care and drug resistance surveillance.
Funding
NHMRC Centre for Research Excellence in Tuberculosis (www.tbcre.org.au) and NSW Health Prevention Research Support Program.
Keywords: Tuberculosis, Tuberculosis control, Genomic surveillance, Mycobacterium tuberculosis, Transmission, Drug resistance
Research in context.
Evidence before this study
M. tuberculosis culture-based whole genome sequencing (WGS) has been used with increasing frequency in low incidence settings for tuberculosis (TB) drug resistance surveillance and transmission tracking. However, the value of routine prospective WGS at the TB control program level, particularly in informing enhanced programmatic indicators of local TB control, have not been assessed.
Added value of this study
This is the first study to explore programmatic indicators of local TB control derived from routine WGS. A variable pairwise SNPs distance (0-25 SNPs) cluster definition was used to explore the most appropriate SNP cut-off for TB control monitoring and proactive transmission tracking. A rolling 5-year cluster assessment, using a 5-SNP cut off, seemed to offer a useful TB control program indicator of local TB transmission. A particular focus on large and/or protracted clusters identified instances of failed disease containment and opportunities for TB control improvement. Despite routine phenotypic drug susceptibility testing (DST), genomic DST provided an overall 6.8% increase in detecting drug resistance mutations with likely clinical relevance.
Implications of all the available evidence
Routine WGS provides high-resolution transmission surveillance that can enhance TB control efforts by guiding targeted public health interventions. A ‘rolling 5-year review’ of large and protracted transmission clusters offers a pragmatic indicator to monitor local TB transmission and TB control program performance. Assessment of TB cases representing likely local TB transmission is useful to inform progress towards ‘zero TB transmission’ in low incidence countries. Routine WGS also provides ongoing surveillance of drug resistance mutations with accurate drug resistance prediction guiding personalised TB treatment.
Introduction
Tuberculosis (TB) remains a leading infectious disease threat worldwide with more than 10 million people developing TB and ∼1.5 million deaths attributed to TB every year.1 The emergence of highly drug-resistant Mycobacterium tuberculosis strains poses additional challenges.1,2 Globally, 3–4% of new infections and 18–21% of previously treated TB cases have rifampicin- or multidrug-resistant (RR/MDR) TB.1 Before 2020, global TB incidence and mortality trends were declining, but not enough to meet World Health Organization (WHO) End TB targets.1 However, the health system disruption caused by the COVID-19 inflicted major setbacks to global TB control efforts.1
Australia has a low burden of TB with an annual incidence of ∼6 cases per 100,000 population and ∼2–3% of RR/MDR-TB cases per year.3 New South Wales (NSW) has the highest number of TB notifications, with ∼90% of TB cases occurring in people born in high TB incidence countries.4 Although TB case numbers in NSW remain relatively small compared to high incidence settings, TB control is a constitutionally mandated public health priority in Australia and the country has made a formal commitment to pursue local TB elimination.
Recent advances in whole genome sequencing (WGS) have accelerated the integration of pathogen genomics into public health surveillance and control efforts.5 WGS provides major advantages over conventional testing methods, particularly with regards to rapid drug resistance assessment and high-resolution surveillance, which allows better targeted public health responses.6,7 The benefits of routine genotypic drug susceptibility testing (DST) over phenotypic DST presents an opportunity to standardise techniques across laboratories and reduce time to drug resistance detection, which is important for clinical case management.7
Given these advantages, routine WGS has been increasingly applied in low TB incidence settings to improve personalised case management and disease control efforts.8,9 NSW began routine prospective WGS of all culture-confirmed TB cases in 2016 and standardised processes were implemented from 2017. The utility of WGS for timely recognition of drug resistance and accurate transmission tracking for individual cases and TB outbreaks has been recognised in many settings,10, 11, 12, 13 but the use of routine WGS in advising and evaluating the overall performance of TB control programs has yet to be established. One suggested metric is the calculation of “locally transmitted TB incidence” to reflect local TB control in low TB incidence settings.9 The calculation of ‘locally transmitted TB incidence’, based on standardized cluster definitions and epidemiological verification, presents an opportunity to establish programmatic indicators of local TB control that evaluate progress towards the aspirational goal of “zero TB transmission”. In this study, we aimed to explore the potential for routine WGS to provide genomically informed programmatic indicators, which can be used to evaluate TB control program performance.
Methods
TB is a notifiable condition under the NSW Public Health Act (2010). We used the STREGA reporting guidelines14 to perform an analysis of all TB cases with cultured isolates referred to the NSW Mycobacterium Reference Laboratory (MRL) at the Institute of Clinical Pathology and Medical Research (ICPMR) NSW Health Pathology, with a specimen collection date between 1st January 2017 and 31st December 2021. All positive M. tuberculosis cultures were routinely referred to the NSW MRL by public and private pathology providers for confirmatory identification, phenotypic DST, and prospective WGS, usually from primary cultures. M. tuberculosis isolates were cultured either in BACTEC MGIT 960 (Becton Dickinson) or on Lowenstein Jensen (LJ) slopes; confirmatory identification was performed using real-time PCR targeting IS6110 and regions of deletion analysis.15
Phenotypic drug susceptibility testing (pDST)
Phenotypic DST was conducted using the modified microdilution method in the BACTEC MGIT 960 system with WHO recommended critical concentrations.15 In brief, we tested first line TB drugs at the following critical concentrations; 0.1 μg/mL (low) and 0.4 μg/mL (high) for isoniazid (INH), 1 μg/mL for rifampicin (RIF), 100 μg/mL for pyrazinamide (PZA), and 5 μg/mL for ethambutol (EMB). Isolates with discrepant PZA resistance received further testing using the Wayne assay. Isolates resistant to rifampicin or more than one first-line drug, or upon request from the clinician, were tested for resistance against second-line TB drugs; including amikacin (1 μg/mL), and moxifloxacin (0.25 μg/mL).
Whole genome sequencing (WGS) and lineage determination
All M. tuberculosis isolates were prospectively sequenced in house. WGS libraries were prepared with DNA extracted from MGIT or LJ cultures using the Nextera XT kit (Illumina, San Diego, California). Sequencing was performed on an IIlumina NextSeq500 (Illumina, San Diego, California) instrument using 2 × 150 bp paired-end chemistry as previously described. The quality of short reads was checked using FastQC v0.1 (https://github.com/s-andrews/FastQC). Sequenced reads were trimmed using Trimmomatic v0.36 (https://github.com/usadellab/Trimmomatic). Trimmed reads were mapped to the M. tuberculosis H37Rv reference genome (NCBI GenBank accession NC_000962.3) using Mykrobe predict/master to classify isolates into major M. tuberculosis lineages.16
Genotypic drug susceptibility testing (gDST)
Single-nucleotide polymorphisms (SNPs) and small insertion/deletions (indels) were called using Snippy v3.1 (https://github.com/tseemann/snippy) with a minimum coverage of five reads and a minimum fraction of variant bases of 10%, using the M. tuberculosis H37Rv as reference genome. SNPs identified by Snippy were manually cross-checked against known TB drug resistance conferring mutations listed in the 2021 WHO Catalogue.17,18 Large indels associated with drug resistance were detected using RedDog v1beta.8 (https://github.com/katholt/RedDog). Mutations associated with resistance to any first-line drugs (isoniazid, rifampicin, pyrazinamide, ethambutol) were included in the analysis, as well as those associated with resistance to second-line injectables (amikacin, capreomycin, kanamycin) and fluoroquinolones.
The perceived added value of gDST for first-line drug resistance detection, defined as increased sensitivity to detect high confidence mutations missed by other routine tests, was assessed by considering the presence of drug resistance conferring mutations detected in strains that tested susceptible on pDST or in whom pDST was unsuccessful. We only considered the drug resistance conferring mutations classified as group 1 (Assoc W R) and group 2 (Assoc w R – Interim) in the 2021 WHO Catalogue.17,18
Genomic clusters suggestive of recent TB transmission
Sequence reads were mapped to the M. tuberculosis H37Rv genome using RedDog v1beta.8 (https://github.com/katholt/RedDog) with default settings. The quality filter for SNP alignment in cluster and phylogenetic analysis was defined as reads covering over 98% of the reference genome with >15x coverage of all isolates, and minimum depth of five in at least 95% of reads. PE/PPE and mobile genomic regions were excluded from SNP alignment.19 Pairwise SNP distances were calculated using Snp-dists v0.6 (https://github.com/tseemann/snp-dists). SNP clusters were identified using the hierarchical single-linkage agglomerative clustering algorithm in python package. A lineage specific transmission overview was obtained by incorporating a 5-SNP cut-off together with the case collection date and assumed molecular clock of 0.5 SNP per year per genome, using the Transcluster package (https://github.com/JamesStimson/transcluster). The estimated rate of recent transmission (RRT) was used to assess likely local transmission. The RRT was calculated using the formula (N-C)/T∗100, where N is the number of clustered isolates (using a 5-SNP cut off), C the number of clusters and T the total number of isolates analysed.20
Statistical analysis and ethics
Prism GraphPad v9.4.1 was used for statistical analyses. The accuracy of gDST was calculated using descriptive statistics and reported with 95% confidence intervals. Genomic markers of drug resistance and differences in transmission clusters between lineages were assessed using the Fisher's exact and paired-t tests with a significance level of p < 0.05. The study received ethics approval from the Western Sydney Local Health District (WSLHD) Human Research Ethics Committee (approval no. 2019/PID14240).
Role of the funding source
The funders of this study did not have any role in study design, data collection, data analysis, data interpretation or writing of this manuscript.
Results
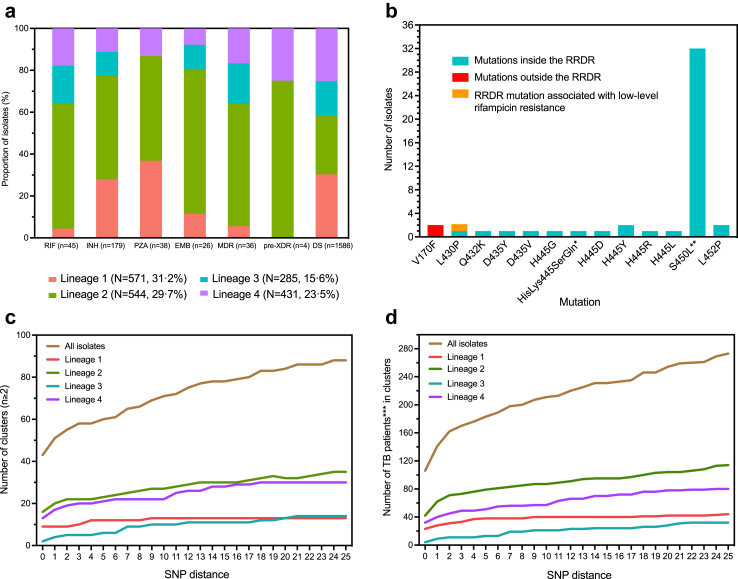
In total, M. tuberculosis strains from 1831 patients were sequenced, representing 64.8% of all notified TB cases in NSW during the study period; 84.8% of all bacteriologically confirmed cases and 96.2% of all culture confirmed cases (Figure S1). Patients with successfully sequenced M. tuberculosis strains were highly representative of the microbiologically-confirmed TB patient cohort in NSW (Table 1). The gender, age group and disease type (∼33.3% of sequences obtained from non-respiratory specimens) associated with sequenced isolates were also highly comparable to ratios observed among all notified TB cases. Children <15 years were the only under-represented group, representing 1.9% of all TB cases and only 0.8% of those sequenced, but numbers were very small and all 14 cases who had microbiological confirmation were successfully sequenced. All four major M. tuberculosis strain lineages were represented, with Lineage 1 (n = 571, 31.2%) and Lineage 2 (n = 544, 29.7%) strains being most common.
Table 1.
Overview of all notified TB cases and those whose M. tuberculosis isolates were successfully sequenced.
| All notified TB cases |
Microbiologically confirmeda |
Culture confirmedb |
Successfully sequenced |
With pDST |
|
|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | n (%) | |
| Gender | |||||
| Male | 1563 (55.3) | 1192 (55.2) | 1067 (56.0) | 1016 (55.5) | 1016 (55.5) |
| Female | 1258 (44.5) | 964 (44.7) | 835 (43.9) | 813 (44.4) | 811 (44.3) |
| Not stated | <5 (<0.1) | <5 (<0.1) | <5 (<0.1) | <5 (<0.1) | <5 (<0.1) |
| Transgender | <5 (<0.1) | <5 (<0.1) | <5 (<0.1) | <5 (<0.1) | <5 (<0.1) |
| Total | 2824 (100%) | 2158 (100%) | 1904 (100%) | 1831 (100%) | 1829 (100%) |
| Age (years) | |||||
| <15 | 55 (1.9) | 14 (0.6) | 14 (0.6) | 14 (0.8) | 14 (0.8) |
| 15–29 | 897 (31.8) | 699 (32.4) | 626 (32.9) | 582 (31.8) | 581 (31.8) |
| 30–44 | 793 (28.1) | 589 (27.3) | 510 (26.8) | 494 (27.0) | 494 (27.0) |
| 45–64 | 570 (20.2) | 432 (20.0) | 383 (20.1) | 375 (20.5) | 375 (20.5) |
| ≥65 | 509 (18.0) | 424 (19.6) | 373 (19.6) | 366 (20.0) | 365 (19.9) |
| Total | 2824 (100%) | 2158 (100%) | 1906 (100%) | 1831 (100%) | 1829 (100%) |
| Disease type | |||||
| PTB | 1521 (53.9) | 1218 (56.4) | 1113 (58.5) | NA | NA |
| PTB + EPTB | 305 (10.8) | 280 (13.0) | 260 (13.7) | NA | NA |
| EPTB only | 997 (35.3) | 660 (30.6) | 531 (27.9) | NA | NA |
| Total | 2824 (100%) | 2158 (100%) | 1904 (100%) | 1831c (100%) | 1829 (100%) |
EPTB: extrapulmonary tuberculosis; NA: not available; PCR: Polymerase Chain Reaction; pDST: phenotypic drug susceptibility testing; PTB: pulmonary tuberculosis; TB: tuberculosis; WGS: whole genome sequencing.
Culture and/or PCR positive.
254 cases were confirmed by PCR only; 96.2% of culture confirmed cases were successfully sequenced.
Successfully sequenced isolates represented 64.8% of all notified TB cases, 84.8% of all bacteriologically confirmed cases, and 96.2% of all culture confirmed cases.
Phenotypic and genotypic drug resistance
Routine pDST data were available for 1829/1831 (99.9%) isolates, with 198 (198/1829, 10.8%) resistant to one or more first-line TB drug (Table S1). Among drug resistant isolates, five (2.5%) were RR and 40 (20.2%) were MDR (defined as resistance to both isoniazid and rifampicin); four (10.0%) had additional fluoroquinolone resistance. Among strains with any drug resistance, 179 were resistant to isoniazid; 133 (74.3%) were mono-resistant to isoniazid, 40 (22.3%) were MDR and 6 (3.4%) were poly-resistant (isoniazid resistance with additional resistance to any drug other than rifampicin). Mono-resistance against pyrazinamide (13/198, 6.6%) and ethambutol (1/198, 0.5%) was uncommon. Second-line pDST data were available for RR/MDR strains; four were resistant to fluoroquinolones and none to injectables (amikacin, capreomycin, kanamycin). Drug resistant strains were detected in all lineages (Fig. 1a) but were significantly over-represented in Lineage 2 compared to other phylogenetic lineages combined (odds ratio 2.1, 95% CI 1.7–2.8, p < 0.0001).
Fig. 1.
Overview of M. tuberculosis drug resistance profiles, and transmission clusters identified over the 5-year study period. a): M. tuberculosis lineage-specific pDST profiles. Numbers (n) in brackets next to drugs (X axis) indicate the total number of isolates with phenotypic resistance against the specified drug. Numbers (N) in brackets next to lineages (legend) indicate the total number of isolates per lineage. b): Frequency of mutations inside or outside the RRDR and its association with phenotypic rifampicin resistance. All mutations identified for rifampicin resistance were group 1 (Assoc w R) mutations according to the 2021 WHO drug resistant TB mutation catalogue. Mutations outside RRDR (indicated by the red bar) would be missed by Gene Xpert. c): Number of M. tuberculosis transmission clusters identified. ‘SNP distance’ defined as the number of SNP differences from 0 to 25 and ‘SNP cluster’ defined using variable pairwise SNP distance cluster definition. d): Number of TB cases contained in these clusters. EMB: ethambutol; INH: isoniazid; MDR: multidrug-resistant; PZA: pyrazinamide; pDST: phenotypic drug susceptibility testing; RIF: rifampicin; RRDR: Rifampicin Resistance Determining Region used for rifampicin resistance detection by Xpert MTB/RIF®/ULTRA® and Hain GenoType MTBDRplus V2; SNP: single nucleotide polymorphism; TB: tuberculosis; XDR: extensively drug resistant. ∗delCACAinsTCCC p.HisLys445SerGln; ∗∗with compensatory mutations in rpoC P1040R (n = 3, MDR-TB), I491T (n = 3, 2 MDR-TB and 1 RR-TB); ∗∗∗Number of isolates reflects the number of TB patients in identified transmission clusters.
Among sequenced isolates 12.2% (223/1831) contained a mutation predicting likely resistance to at least one first-line drug.17,18 Mutations in the Rifampicin Resistance Determining Region (RRDR) of the rpoB gene (detected by Xpert MTB/RIF®) were documented in 97.8% (44/45) of strains with phenotypic rifampicin resistance (Fig. 1b). However, two strains had a rpoB V170F mutation outside the RRDR that was associated with phenotypic resistance but not detected by Xpert MTB/RIF®. In addition, one phenotypically susceptible strain had a rpoB L430P mutation located within the RRDR, which has been associated with low-level rifampicin resistance.21 Monitoring the local frequency of these clinically relevant mutations has important surveillance value.
Mutations in genes conferring resistance to INH were detected in 82.1% (197/240) of isolates with any gDST resistance (Table S3). The most common mutations were katG S315T (n = 113) and fabG1-inhA (−15 c > t, n = 45; −8 t > c, n = 4; −8 t>a, n = 5) promoter mutations, which are included in the Genotype MTBDRplus v2® kit (HAIN LifeSciences). katG mutations were always associated with high-level (>0.4 μg/mL) isoniazid resistance, except for a 275-bp deletion in the katG-furA intergenic region in one isolate that was phenotypically susceptible, as well as katG A106V and M105I mutations. katG A106V were detected in eight isolates, and M105I in one isolate, showing low-level (0.1–0.4 μg/mL) isoniazid resistance. In contrast, fabG1-inhA mutations, found in 27.0% of isolates, were mostly (80.8%) associated with low-level (0.1–0.4 μg/mL) isoniazid resistance.
Mutations in genes conferring resistance to PZA were detected in 84.2% (32/38) of phenotypically resistant isolates and in 16 susceptible isolates (Figure S2). All 26 isolates phenotypically resistant to ethambutol, as well as five phenotypically susceptible, had mutations in the embB gene (Figure S3). Mutations associated with fluroquinolone resistance were detected in six additional isolates that tested susceptible to all first-line TB drugs by pDST. Additionally, two MDR isolates were found to have mutations in genes conferring resistance to second-line injectables agents (amikacin, capreomycin, kanamycin).
Compared to pDST, gDST demonstrated high accuracy with 96.5% (95% CI 92.9%–98.3%) sensitivity and 98.2% (95% CI 97.4%–98.7%) specificity in predicting first-line drug resistance, with the reference standard defined by pDST using specified critical concentrations (Table 2). gDST detected all MDR and phenotypically drug resistant cases. In the study setting, where routine pDST was performed, gDST detected an additional 6.8% of cases with likely drug resistance (Table 2). The number of discordant pairs observed included six isolates that tested PZA resistant by pDST, but had no recognised resistance conferring mutations detected by gDST. These strains were considered to be ‘PZA resistanc missed by gDST’ and were subtracted from the perceived ‘added value of gDST’ calculation (198−6 = 192).
Table 2.
Overview of gDSTa accuracy for first-line drug resistance detection and consideration of its ‘perceived added value’ in the study context.
| pDST n (%) | gDST n (%) | Sensitivity % (95% CI) | Specificity % (95% CI) | PPV % (95% CI) | NPV % (95% CI) | Perceived added value of gDSTd |
|||
|---|---|---|---|---|---|---|---|---|---|
| All discordante | gDST onlyf | Frequency of perceived value addg | |||||||
| RIF | 45 (2.5%) | 46 (2.5%) | 100 (92.1–100) | 99.9 (99.7–100) | 97.8 (88.7–99.9) | 100 (99.8–100) | 1 | 0 | 2.2% (1/45) |
| INH | 179 (9.8%) | 195 (10.6%) | 99.4 (96.9–100) | 99.0 (98.4–99.4) | 91.3 (86.5–94.5) | 99.9 (99.7–100) | 11 | 1 | 6.7% (12/179) |
| PZAb | 38 (2.1%) | 48 (2.6%) | 84.2 (69.6–92.6) | 99.1 (98.6–99.5) | 66.7 (52.5–78.3) | 99.7 (99.3–99.9) | 16c | 0 | 0% (0/38) |
| EMBb | 26 (1.4%) | 31 (1.7%) | 100 (87.1–100) | 99.7 (99.4–99.9) | 83.9 (67.4–92.9) | 100 (99.8–100) | 5 | 0 | 19.2% (5/26) |
| MDR | 40 (2.2%) | 42 (2.3%) | 100 (91.2–100) | 99.9 (99.6–100) | 95.2 (84.2–99.2) | 100 (99.8–100) | 2 | 0 | 5.0% (2/40) |
| Total | 198/1829 (10.8%) | 221/1829 (12.1%) | 96.5 (92.9–98.3) | 98.2(97.4–98.7) | 86.4 (81.3–90.3) | 99.6 (99.1–99.8) | 12 | 1 | 6.8% (13/192g) |
CI: confidence interval; DST: drug susceptibility testing; gDST: genotypic DST; pDST: phenotypic DST; EMB: ethambutol; INH: isoniazid; PZA: pyrazinamide; RIF: rifampicin; NPV: Negative Predictive Value; PPV: Positive Predictive Value.
Compared to pDST (regarded as the reference standard).
pDST result was unavailable for 1 isolate.
Overall poor confidence in individual mutations associated with PZA resistance.
Reflects gDST ‘value add’ in the presence of routine pDST; would be greatly increased in settings without routine pDST. The ‘value add’ include discordant results with mutations that are graded as “Assoc w R″ in 2021 WHO catalogue and specimens with failed pDST.
All discordant’ indicate strains with resistance mutations that are graded as “Assoc W R″ and “Assoc w R -Interim” in 2021 WHO catalogue on gDST and that tested susceptible on pDST.
pDST failed.
Fraction with resistance mutations not detected by routine pDST. The number of discordant pairs observed included six isolates that tested PZA resistant by pDST, but had no recognised resistance conferring mutations detected by gDST. These resistant strains ‘missed by gDST’ were subtracted from the perceived added value calculation (198−6 = 192). The mutations that contributed to the 6.8% ‘value add’ estimate for first-line drug resistance detection, included rpoB L430P (n = 1), katG S315T (n = 1), fabG1 -15 c > t (n = 5), fabG1 -8 t>a (n = 5), and fabG1 -8 t > c (n = 1).
Transmission surveillance & examination of SNP thresholds for cluster assessment
Of 1831 sequenced isolates (median coverage of 83x; range 15–751x), 1821 (99.5%) passed the quality filter for SNP alignment and were included in transmission cluster analysis. Fig. 1 illustrates the number of clusters (1c), and the number of TB patients within these clusters (1d), identified using an increasing (0–25) SNP distance cut-off for cluster identification. Previous studies used variable (0, 5, or 12) SNP cut-offs to identify genomic clusters in comparison with identified epidemiological clusters.5 The application of a large and inclusive 25 SNP threshold that is not highly reflective of likely recent transmission, identified 88 clusters representing 15.0% (273/1821) of all sequenced TB cases, with an estimated RRT of 10.2% (25-SNP RRT). Application of a traditional 5-SNP threshold identified 62 clusters with 183 clustered TB cases and an estimated RRT of 6.8% (5-SNP RRT). A more conservative 2-SNP cut-off identified 55 clusters and 160 clustered TB cases, with an estimated RRT of 5.8% (2-SNP RRT). Of these clustered cases 101/160 (63.1%) had 0 SNP differences, which is highly indicative of recent person-to-person transmission within Australia.
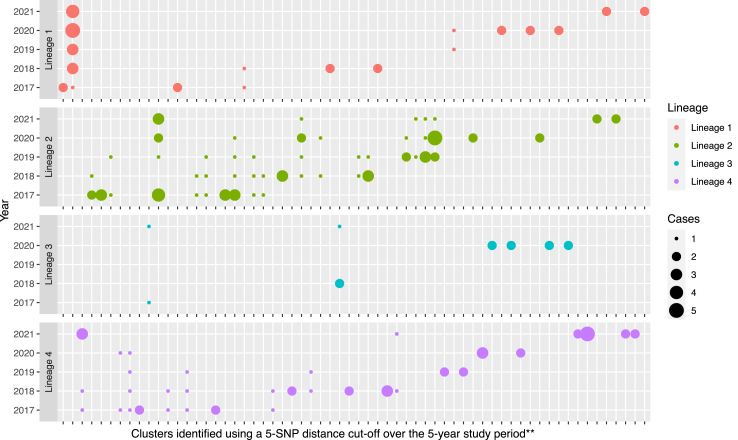
We assessed clusters by lineage using 0 SNP (Figure S4), 2 SNP (Table S1), 5 SNP (Fig. 2), and 12 SNP (Figure S5) cut-offs over the 5-year study period. A 5-SNP cut-off identified eight large clusters (defined as ≥5 cases per cluster) that included a total of 60 TB cases. Lineage 2 strains had the largest proportion of clustered isolates (79/543, 14.5%); significantly more than the other lineages combined (odds ratio 1.9, 95% CI 1.4–2.6, p < 0.0001) (Table S4). Lineage 2 strains also had a higher proportion of TB cases in large clusters (34/79, 43.0%) compared to the other lineages combined (odds ratio 2.8, 95% CI 1.7–4.6, p < 0.0001) (Table S4). We performed expanded cluster assessment, using 0, 2 and 5 SNP cut-offs, over a rolling 2-year period (Table S5, Figure S6). The proportion of TB isolates in large clusters were identical using a 2-SNP or 5-SNP cut-off. Overall we believe that use of a 5-SNP cut-off for cluster identification, with analysis over a rolling 5-year review period, should provide a standard and highly informative overview of likely local transmission in low incidence settings, like Australia.
Fig. 2.
All M. tuberculosis transmission clusters∗ identified over the 5-year study period. Y-axis indicates the study year (2017–2021) and the X-axis the sequential clusters identified in that year. ∗Using a five SNP distance cut-off; programmatically the focus should be on larger and/or multi-year clusters that indicate failed containment. ∗∗The clusters on the X-axis are presented in the order they were identified, based on the isolate in cluster with the earliest collection date. Single cases (small dots) included were part of multi-year clusters dispersed over the period indicated. SNP: single nucleotide polymorphism.
We also examined protracted multi-year clusters as an indication of sub-optimal local TB control. Such clusters included cases detected in at least three out of five consecutive years. In total, 29.0% (18/62) of all clusters (using a 5-SNP cut-off) were multi-year clusters, containing 83 TB cases (Table S4). Lineage 2 strains were significantly more likely to have a higher percentage of cases in multi-year clusters compared to the other lineages combined (odds ratio 1.6, 95% CI 1.2–2.3, p = 0.003). Among 18 multi-year clusters, seven were large clusters including 55 TB cases. Lineage 2 strains were more likely to be included in large multi-year clusters (odds ratio 2.1, 95% CI 1.4–3.4, p = 0.001).
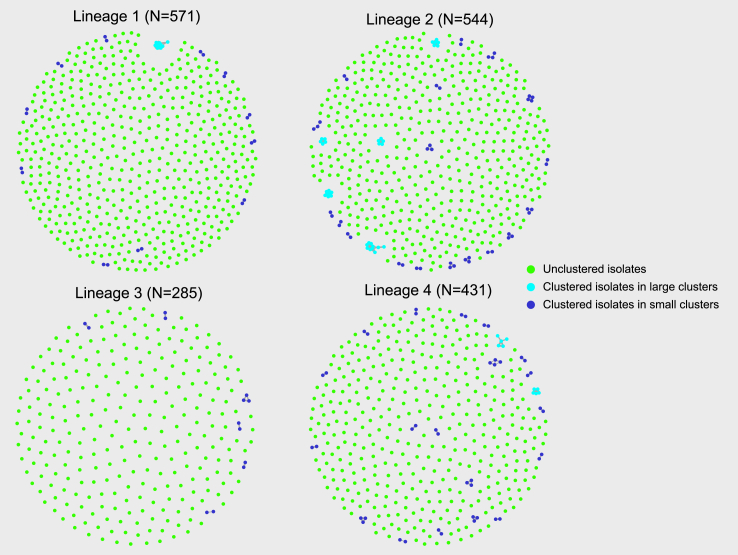
Fig. 3 provides a ‘birds eye’ overview of lineage-specific clusters suggestive of local TB transmission over the study period, including 35 (35/183, 19.1%) drug resistant isolates; one was MDR and 34 were isoniazid mono-resistant (Table S4). Among drug resistant isolates, Lineage 2 strains were more frequently associated with local transmission clusters than the other phylogenetic lineages combined (odds ratio 9.5, 95% CI 3.7–23.2, p < 0.0001) (Table S4).
Fig. 3.
‘Birds eye’ overview of lineage-specific tuberculosis cases and likely local M. tuberculosis transmission∗ observed in New South Wales, Australia, over the 5-year study period (2017–2021). Single dots within each lineage circle represents one sequenced M. tuberculosis isolate (TB patient). The varying density of dots reflects variation in the number of lineage specific TB cases (Table S4). For clustered cases the size of the cluster is explained in the legend with large clusters representing ≥5 cases/isolates and small clusters <5 cases/isolates. ∗Assessment of M. tuberculosis transmission clustering using a 5 SNPs cluster cut-off. SNP: single nucleotide polymorphism; TB: tuberculosis.
Discussion
This study employed data from routine M. tuberculosis cultures that were prospectively sequenced by the NSW TB control program to explore genomically-informed programmatic indicators of local TB control. Routine WGS can provide unique insight into local TB transmission dynamics, particularly in low TB incidence settings like Australia.6,7 During the study period, most microbiologically-confirmed TB cases were successfully sequenced with sufficient quality for high resolution genomic surveillance. We assessed pairwise SNP clustering over a wide range (0–25) of SNP differences, and over two different rolling time frames to ascertain the most useful metrics for objective programmatic performance review and comparison.
SNP-based clustering has been used to track transmission chains with high fidelity,22 but the use of standard SNP thresholds for transmission inference has been questioned.5 In our study, we applied a 5-SNP cut-off for cluster identification, since it is aligned with the most widely accepted international practice, which is important for data comparison. However, use of more stringent 0–2 SNP cut-offs also require consideration, since it may prove beneficial in settings with a high number of imported TB cases from specific high-incidence regions. A stringent SNP cut-off may also provide stronger impetus for detailed epidemiological investigation to verify local TB transmission. A stronger focus on stringent (0–2) SNP clusters may be warranted, since they are more representative of recent local transmission and justify targeted public health interventions. In NSW all clusters with ≤5 SNPs are currently investigated, although the majority of these clusters have 0 SNP differences indicating a high likelihood of recent transmission within NSW, Australia.
As a standard and reproducible TB control metric we propose a rolling 5-year cluster assessment, using a 5-SNP cluster threshold, alongside classical epidemiological investigation to monitor and track likely local transmission. Given molecular clock estimates for M. tuberculosis of ∼0.5 SNP/year this should broadly reflect transmission that occurred within the past decade,23,24 although it must be acknowledged that the ‘molecular clock’ of M. tuberculosis is highly variable and influenced by multiple factors.25 Progression from infection to disease usually occurs within five years of infection/re-infection, but can be delayed for many decades or occur within one year of infection as seen in young children.26 Australian data indicate that the majority of TB cases in migrants occur within 5-years of arrival from countries with a high TB incidence,27 although the contribution of reinfection following return visits to their country of origin in subsequent years remains uncertain. Incorporating a rolling 5-year cluster assessment as a standardised metric into annual TB control reports could provide an important programmatic indicator of local TB control.
We believe a 5-year time window represents a reasonable compromise between practicality and comprehensiveness within a low incidence setting. However, assessments over different rolling time frames, such as annual or 2-yearly reviews, may provide additional perspectives and valuable interim insights, particularly in the initial phases of routine WGS surveillance or in higher incidence settings. In our experience the use of a rolling 2-year period limited multi-year cluster recognition and showed a fluctuating trend, which may make it more difficult to recognise shifts in transmission patterns. We recommend a rolling 5-year time frame as a starting point for standardised cluster assessment, with the flexibility to review and modify it as additional data becomes available. The large and/or multi-year clusters identified during our study represent missed opportunities for disease prevention with delayed transmission control that can be improved by better targeted public health responses. In practice, a public health containment strategy for each cluster of concern should be developed in close collaboration with the TB control program, considering epidemiological data, as well as sociocultural context and workforce considerations.
Assessment of local transmission is particularly important for drug resistant TB,28 since the transmission of drug resistant strains adds urgency to containment efforts. Rapid drug resistance detection assists treatment optimisation, which is important to improve patient outcomes and to minimize community transmission. Active surveillance of the full repertoire of drug resistance mutations is also useful to guide empiric treatment when patient-specific drug resistance results are lacking. An important WGS surveillance function is the ability to detect resistance mutations missed by more selective methods. ‘Diagnostic selection’ of drug resistant strains with mutations outside the RRDR that are missed by Xpert MTB/RIF® has been demonstrated in Eswatini.29 From a programmatic perspective it is important to monitor the frequency of these mutations, since misdiagnosis of RR/MDR TB compromises patient outcomes and facilitates ongoing transmission of drug resistant strains within communities. Our initial findings also supported observations of potential ‘false positive’ RR/MDR-TB detection by Xpert MTB/RIF®.30 However, discordant results observed prior to August 2019 could be explained by changes in the critical concentration used to define in vitro rifampicin resistance. The one ‘false positive’ rifampicin resistant strain detected by gDST, would have tested phenotypically resistant using the new 0.5 μg/mL critical concentration.31 This highlights the difficulty of uncertain critical concentrations, especially for novel TB drugs where phenotypic methods are still in development.
Apart from mutation surveillance, gDST also provides clinically useful information. More than 15.0% of strains had genomic markers of isoniazid resistance that were undetectable by commercial molecular tests, including Gene Xpert XDR (Cepheid) and Genotype MTBDRplus v2 (HAIN LifeSciences). These included atypical katG-furA (including a large deletion), fabG1-inhA and ahpC mutations. WGS based gDST assesses the complete genetic repertoire, including uncommon isoniazid resistance conferring mutations such as katG S315N and fabG1 L203L that may be associated with low- or high-level resistance. The full diversity of mutations associated with pyrazinamide resistance have not been established and global mutation databases correlate poorly with pDST results.17,18 Discrepancies observed between pDST and gDST results require careful consideration. Routine gDST identified drug resistance mutations against first-line TB drugs in an additional 6.8% of M. tuberculosis isolates compared to routine pDST and identified genomic markers of fluoroquinolone resistance in cases that were phenotypically susceptible to all first-line drugs and therefore not phenotypically tested for second-line drug resistance. The limitations of pDST as reference standard are well recognised, given instances of suboptimal accuracy.1,18 The gDST ‘added value’ calculated in this study assumed routine pDST against first-line TB drugs, which is not available in most settings and may not be worthwhile to continue once routine gDST is well established.21 Replacing routine pDST with gDST is cost saving and will reduce ‘time to drug resistance detection’, with better tailored treatment providing clinical and programmatic benefits.6
Some important study limitations have to be acknowledged. We used laboratory data that lacked relevant epidemiological and clinical data, such as whether the cases were newly diagnosed or previously treated. In NSW the proportion of TB cases with a history of previous treatment is consistently below 5.0%,4 but in settings where re-treatment is more common further analysis of this subgroup may offer interesting insights. Like all non-metagenomic studies, our study only sequenced culture positive TB cases and sequenced strains might have been affected by culture selection. The fact that 64.8% of all notified TB cases were sequenced and that sequenced cases had a similar age, gender and disease profile as all notified cases, gives us confidence in the representativeness of our cohort and is a major study strength. However, it is important to consider that certain groups, such as children with low culture yields were underrepresented. Lastly, although close collaboration occurs between the laboratory and the NSW TB Program to integrate epidemiological data in day-to-day assessment of identified transmission clusters, this study could not integrate epidemiological or individual patient data into the analyses. We acknowledge the important added value of such data and the need for close collaboration with the TB control programme to assist critical assessment and interpretation of sequencing data.
In conclusion, routine WGS of M. tuberculosis offered important benefits for transmission and drug resistance surveillance. Monitoring of genomic case clusters was useful to guide public health interventions and could provide a useful programmatic indicator for enhanced TB control. It may also assist to measure progress towards ‘zero TB transmission’ in low TB incidence countries like Australia.9 As a programmatic tool we propose a standard rolling 5-year review of transmission clusters, using a 5-SNP threshold, with additional assessment as relevant to monitor local TB transmission and guide public health interventions. In addition, routine gDST provided valuable information to assist patient management and could serve an important drug resistance surveillance function.
Contributors
BM and VS designed the study and guided the data analysis. Data collection was done by TC, EM, CL, ES, and ED. XZ performed bioinformatic analysis and drafted the manuscript. All authors participated in manuscript revision and approved the final version.
Data sharing statement
The study was approved by the Western Sydney Local Health District (WSLHD) Human Research Ethics Committee (approval no. 2019/PID14240). Raw de-identified pathogen WGS data was deposited in the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) under BioProject number PRJNA899911.
Declaration of interests
No conflict.
Acknowledgements
The authors thank the Sydney Informatics Hub and the University of Sydney's high-performance computing cluster, Artemis. The first author (XZ) is funded by NHMRC Centre for Research Excellence in Tuberculosis.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.lanwpc.2023.100910.
Appendix A. Supplementary data
References
- 1.World Health Organization . World Health Organization; Geneva: 2022. Global tuberculosis report 2022. [Google Scholar]
- 2.McBryde E.S., Meehan M.T., Doan T.N., et al. The risk of global epidemic replacement with drug-resistant Mycobacterium tuberculosis strains. Int J Infect Dis. 2017;56:14–20. doi: 10.1016/j.ijid.2017.01.031. [DOI] [PubMed] [Google Scholar]
- 3.Bright A., Denholm J., Coulter C., Waring J., Stapledon R. Tuberculosis notifications in Australia, 2015-2018. Commun Dis Intell. 2020;44 doi: 10.33321/cdi.2020.44.88. [DOI] [PubMed] [Google Scholar]
- 4.NSW Tuberculosis Program CDB . Health Protection NSW; Sydney: 2023. Tuberculosis in NSW—surveillance report 2021. [Google Scholar]
- 5.Hatherell H.A., Colijn C., Stagg H.R., Jackson C., Winter J.R., Abubakar I. Interpreting whole genome sequencing for investigating tuberculosis transmission: a systematic review. BMC Med. 2016;14:21. doi: 10.1186/s12916-016-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kizny Gordon A., Marais B., Walker T.M., Sintchenko V. Clinical and public health utility of Mycobacterium tuberculosis whole genome sequencing. Int J Infect Dis. 2021;113(Suppl 1):S40–S42. doi: 10.1016/j.ijid.2021.02.114. [DOI] [PubMed] [Google Scholar]
- 7.de Vries G., van Dissel J., van Soolingen D. Measuring tuberculosis transmission in low-incidence countries. Lancet Respir Med. 2018;6(4):e13. doi: 10.1016/S2213-2600(18)30042-0. [DOI] [PubMed] [Google Scholar]
- 8.Cabibbe A.M., Trovato A., De Filippo M.R., et al. Countrywide implementation of whole genome sequencing: an opportunity to improve tuberculosis management, surveillance and contact tracing in low incidence countries. Eur Respir J. 2018;51(6) doi: 10.1183/13993003.00387-2018. [DOI] [PubMed] [Google Scholar]
- 9.Marais B.J., Walker T.M., Cirillo D.M., et al. Aiming for zero tuberculosis transmission in low-burden countries. Lancet Respir Med. 2017;5(11):846–848. doi: 10.1016/S2213-2600(17)30382-X. [DOI] [PubMed] [Google Scholar]
- 10.Alvarez G.G., Zwerling A.A., Duncan C., et al. Molecular epidemiology of Mycobacterium tuberculosis to describe the transmission dynamics among inuit residing in iqaluit nunavut using whole-genome sequencing. Clin Infect Dis. 2021;72(12):2187–2195. doi: 10.1093/cid/ciaa420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dale K., Globan M., Horan K., et al. Whole genome sequencing for tuberculosis in Victoria, Australia: a genomic implementation study from 2017 to 2020. Lancet Reg Health West Pac. 2022;28 doi: 10.1016/j.lanwpc.2022.100556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu D., Huang F., Zhang G., et al. Whole-genome sequencing for surveillance of tuberculosis drug resistance and determination of resistance level in China. Clin Microbiol Infect. 2022;28(5):731.e9–731.e15. doi: 10.1016/j.cmi.2021.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Walker T.M., Choisy M., Dedicoat M., et al. Mycobacterium tuberculosis transmission in Birmingham, UK, 2009-19: an observational study. Lancet Reg Health Eur. 2022;17 doi: 10.1016/j.lanepe.2022.100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Little J., Higgins J.P., Ioannidis J.P., et al. STrengthening the REporting of genetic association studies (STREGA)--an extension of the STROBE statement. Genet Epidemiol. 2009;33(7):581–598. doi: 10.1002/gepi.20410. [DOI] [PubMed] [Google Scholar]
- 15.Lam C., Martinez E., Crighton T., et al. Value of routine whole genome sequencing for Mycobacterium tuberculosis drug resistance detection. Int J Infect Dis. 2021;113(Suppl 1):48–54. doi: 10.1016/j.ijid.2021.03.033. [DOI] [PubMed] [Google Scholar]
- 16.Hunt M., Bradley P., Lapierre S.G., et al. Antibiotic resistance prediction for Mycobacterium tuberculosis from genome sequence data with Mykrobe. Wellcome Open Res. 2019;4:191. doi: 10.12688/wellcomeopenres.15603.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization . 2021. Catalogue of mutations in Mycobacterium tuberculosis complex and their association with drug resistance. [Google Scholar]
- 18.Walker T.M., Miotto P., Köser C.U., et al. The 2021 WHO catalogue of Mycobacterium tuberculosis complex mutations associated with drug resistance: a genotypic analysis. Lancet Microbe. 2022;3(4):e265–e273. doi: 10.1016/S2666-5247(21)00301-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eldholm V., Monteserin J., Rieux A., et al. Four decades of transmission of a multidrug-resistant Mycobacterium tuberculosis outbreak strain. Nat Commun. 2015;6:7119. doi: 10.1038/ncomms8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu G., Mao X., Wang J., Pan H. Clustering and recent transmission of Mycobacterium tuberculosis in a Chinese population. Infect Drug Resist. 2018;11:323–330. doi: 10.2147/IDR.S156534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allix-Béguec C., Arandjelovic I., Bi L., Beckert P., Bonnet M., Bradley P. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N Engl J Med. 2018;379(15):1403–1415. doi: 10.1056/NEJMoa1800474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meehan C.J., Goig G.A., Kohl T.A., et al. Whole genome sequencing of Mycobacterium tuberculosis: current standards and open issues. Nat Rev Microbiol. 2019;17(9):533–545. doi: 10.1038/s41579-019-0214-5. [DOI] [PubMed] [Google Scholar]
- 23.Meehan C.J., Moris P., Kohl T.A., et al. The relationship between transmission time and clustering methods in Mycobacterium tuberculosis epidemiology. eBioMedicine. 2018;37:410–416. doi: 10.1016/j.ebiom.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker T.M., Ip C.L., Harrell R.H., et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13(2):137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menardo F. Understanding drivers of phylogenetic clustering and terminal branch lengths distribution in epidemics of Mycobacterium tuberculosis. Elife. 2022;11 doi: 10.7554/eLife.76780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marais B.J., Gie R.P., Schaaf H.S., et al. The natural history of childhood intra-thoracic tuberculosis: a critical review of literature from the pre-chemotherapy era. Int J Tuberc Lung Dis. 2004;8(4):392–402. [PubMed] [Google Scholar]
- 27.Moyo N., Tay E.L., Trauer J.M., et al. Tuberculosis notifications in regional Victoria, Australia: implications for public health care in a low incidence setting. PLoS One. 2023;18(3) doi: 10.1371/journal.pone.0282884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marais B.J., Mlambo C.K., Rastogi N., et al. Epidemic spread of multidrug-resistant tuberculosis in Johannesburg, South Africa. J Clin Microbiol. 2013;51(6):1818–1825. doi: 10.1128/JCM.00200-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ardizzoni E., Ariza E., Mulengwa D., et al. Thin-layer-agar-based direct phenotypic drug susceptibility testing on sputum in Eswatini rapidly detects Mycobacterium tuberculosis growth and rifampicin resistance otherwise missed by WHO-endorsed diagnostic tests. Antimicrob Agents Chemother. 2021;65(6) doi: 10.1128/AAC.02263-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miotto P., Cabibbe A.M., Borroni E., Degano M., Cirillo D.M. Role of disputed mutations in the rpoB gene in interpretation of automated liquid MGIT culture results for rifampin susceptibility testing of Mycobacterium tuberculosis. J Clin Microbiol. 2018;56(5) doi: 10.1128/JCM.01599-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ajbani K., Comas I., Coulter C., et al. 2021. Technical Report on critical concentrations for drug susceptibility testing of isoniazid and the rifamycins (rifampicin, rifabutin and rifapentine) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.