Abstract
Despite advances in diagnosis and treatment, gastric cancer (GC) remains a refractory disease, which limits overall survival. Therefore, it is key to identify novel targets to develop more effective and precise treatment. Circular RNAs (circRNAs) serve essential roles in the process of various human cancers. Through analyzing GSE83521 dataset, the present study identified a novel circRNA derived from ribosomal protein S19 (circRPS19), which was considered a potential treatment target for GC. Results of RT-qPCR indicated that circRPS19 was upregulated in GC compared with normal gastric epithelial cells. Loss-of function assays revealed that silencing of circRPS19 suppressed proliferation and aerobic glycolysis but increased apoptosis of GC cells. circRPS19 upregulated ubiquitin-specific processing protease 7 (USP7) expression by sponging microRNA (miR)-125a-5p. circRPS19 stabilized hexokinase 2 (HK2) protein by USP7-mediated deubiquitination of HK2. In vivo experiments confirmed that circRPS19 promoted GC progression and aerobic glycolysis. Taken together, circRPS19 induced aerobic glycolysis of GC cells by stabilizing HK2 protein via the miR-125a-5p/USP7 axis and thus promoting the progression of GC. These findings suggested that circRPS19 served a critical role in the progression of GC and may be a novel therapeutic target for GC.
Keywords: circRPS19, hexokinase 2, deubiquitination, aerobic glycolysis, gastric cancer
Introduction
As a malignancy of the digestive system, gastric cancer (GC) originates in the gastric mucosal epithelium (1). Based on global cancer statistics in 2020, more than one million patients were newly diagnosed with GC and ~769,000 patients died from GC; GC ranks fifth in incidence and fourth in cancer-associated mortality of all types of cancer (2). The occurrence and development of GC are influenced by multiple factors, such as diet, family history, consumption of alcohol and tobacco and infection of Helicobacter pylori and Epstein-Barr virus (3). Endoscopic ultrasound is the most efficient diagnostic methods for early-stage GC, however, a majority of patients are diagnosed at advanced stages (4). Surgical resection is mainly applied to treat early-stage patients, while chemotherapy proves most efficient in advanced-staged patients (5). In spite of improvement in GC diagnosis and treatment, the prognosis of GC still remains poor and the 5-year survival rate is <30% (6). Hence, it is of great importance to identify novel molecular mechanisms in GC progression.
Circular RNA (circRNA) is a type of RNA with covalently closed structure, generated by back-splicing of precursor mRNA (7,8). Former studies have revealed the effect of circRNAs in GC progression: For examples, circRNA_100290 modulated by eukaryotic translation initiation factor 4A3 (EIF4A3) can interact with microRNA (miRNA or miR)-29b-3p to target integrin subunit alpha 11 (ITGA11), thus facilitating proliferation and invasion of GC cells (9); circRNA nuclear receptor interacting protein 1 targets the Akt1/mTOR pathway by sequestering miR-149-5p to promote GC progression (10); circRNA ELAV like RNA binding protein 1 (circ-HuR) downregulates HuR expression by binding to CCHC-type zinc finger nucleic acid binding protein (CNBP), thereby suppressing metastasis, proliferation and invasion of GC cells (11); the circ_0004872/miR-224/Smad4/ADAR1 axis forms a negative feedback loop to hinder GC progression (12). A recent study showed that Helicobacter pylori infection-induced circRNA mannosidase alpha class 1A member 2 (circMAN1A2) promotes GC progression via sponging miR-1236-3p (13). The aforementioned findings indicate that circRNAs are crucial regulators in GC development, highlighting the research value of circRNAs in GC. The present study investigated the role of a new-found GC-associated circRPS19 selected from Gene Expression Omnibus (GEO) database in GC cell proliferation.
Aerobic glycolysis, or the Warburg effect, was discovered by Otto Warburg in the 1920s. Tumor cells, despite sufficient oxygen, prefer the conversion of glucose into lactate to generate energy (14). As reported by previous studies, aerobic glycolysis serves a key role in a variety of carcinomas, including GC: For example, long non-coding RNA H19 (imprinted maternally expressed transcript interacts with miR-519d-3p to regulate lactate dehydrogenase A (LDHA), thus facilitating aerobic glycolysis and proliferative ability of GC cells (15); Opa interacting protein 5 antisense RNA 1 (OIP5-AS1) sequesters miR-186 to enhance proliferation and aerobic glycolysis in GC cells (16); pyruvate kinase M2 (PKM2) serves an oncogenic role in GC cells by facilitating aerobic glycolysis (17); Hexokinase 2 (HK2), as a rate-limiting enzyme, catalyzes glucose metabolism and cancer cells commonly exhibit increased glucose metabolism (18). Previous research has demonstrated that circRNA functions as a regulator of HK2 in GC cells to facilitate aerobic glycolysis (19). Nevertheless, the association between HK2 and circRPS19 in GC cells needs to be further investigated.
The present current study aimed to investigate the functions of circRPS19 in GC cell proliferation and aerobic glycolysis and its downstream molecular mechanism.
Materials and methods
Cell culture and treatment
Human GC cell lines (AGS, HGC-27, NCI-N87 and SNU-1) were obtained from China Center for Type Culture Collection and normal human gastric epithelial cell line (GES-1) was obtained from American Type Culture Collection (ATCC). All cell lines were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). Cell lines were incubated in a humid atmosphere with 5% CO2 at 37°C.
For RNA stability analysis, GC cells were treated with 3 U/µg RNase R (9001-99-4, Sigma-Aldrich; Merck KGaA) for 24 h at room temperature or treated with 2 µg/ml ActD (50-76-0, Merck) for different time intervals (0, 6, 12, 18, 24 h) at room temperature. RNA stability was determined by using RT-qPCR as following description.
To analyze protein stability, GC cells were treated with 5 µM cycloheximide (CHX, HY-12320, MedChemExpress) for 0, 3, 6, 9 h or treated with 20 µM MG132 (HY-13259, MedChemExpress) for 4 h. Cells were treated with 10 µM HBX19818 (HY-17540, MedChemExpress) to inhibit USP7 expression. Protein levels were detected using western blot as following description.
Data collection and bioinformatics analysis
circRNAs significantly overexpressed in GC tissues (under the condition of P≤0.05) were screened out from GSE83521 dataset in Gene Expression Omnibus (GEO; ncbi.nlm.nih.gov/gds). starBase database (rnasysu.com/encori/) was used to screen miRNAs that potentially interact with both circRPS19 and USP7.
Cell transfection
Small interfering RNAs (siRNAs) targeting circRPS19 or ubiquitin-specific processing protease 7 (USP7) sequence were synthesized for knockdown with non-targeted siRNA as the negative control (NC). For overexpression of USP7, the full-length sequence of USP7 was inserted into pcDNA3.1 vector to establish USP7 overexpression vector, with empty vector as NC. miR-125a-5p mimics was synthesized and used to upregulate miR-125a-5p expression, with mimics-NC as NC. All plasmids and vectors were synthesized and constructed by Guangzhou RiboBio Co., Ltd. Cell transfection was performed at 37°C using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After being transfected with siRNAs and pcDNA3.1 vectors for 48 h or transfected with for miRNA mimics or inhibitor for 24 h, cells were harvested for subsequent experiments. The duration between end of transfection of siRNAs and pcDNA3.1 vectors and subsequent experiments was 48 h. The duration between end of transfection of miRNA mimics or inhibitor and subsequent experiments is 24 h. siRNA and pcDNA3.1 expression vector (0.8/50 µl) were treated with Lipofectamine 2000 for transfection at 24 well plates. miRNA mimics were transfected at a final concentration of 100 nM. The sequences were as follows: si-NC, 5′-CCG GCG GCC GCA AAT TAC GAA TGA CTC GAG TCA TTC GTA ATT TGC GGC CGT TTT T G-3′; si-circRPS19-1: 5′-CCG GCT CAT TCG TAA CGG CCG CAA ACT CGA GGA GTA AGC ATT GCC GGC GTT TTT TTT G-3′; si-circRPS19-2: 5′-CCG GAT TCG TAA CGG CCG CAA ACT GCT CGA GTA AGC ATT GCC GGC GTT TGA CTT TTT G-3′; si-circRPS19-3: 5′-CCG GGC CTC ATT CGT AAC GGC CGC ACT CGA GCG GAG TAA GCA TTG CCG GCG TTT TTT G-3′; mimics-NC: 5′-UCU CCG AGA UCU UAC CAC CUG GUA-3′; miR-125a-5p mimics: 5′-UCC CUG AGA CCC UUU AAC CUG UGA-3′; inhibitor NC: 5′-UCA UCU CAG GAA GGG UAU CGA GGA-3′; miR-125a-5p inhibitor: 5′-UCA CAG GUU AAA GGG UCU CAG GGA-3′; si-NC: 5′-CCG GGT TAA TTT ATT ATC CAG CAT TCT CGA GAA TGC TGG ATA ATA A AT TAA CTT TTT G-3′; si-USP7-1, 5′-CAC CGT GTA ATT TAT TAT CTT CAG CAC TCG AGT GCT GAA GAT AAT AAA T TA CA-3′; si-USP7-2, 5′-CAC CAT TCA TAG AAC ACT CTT TGT ACT CGA GTA CAA AGA GTG TTC TAT GAA-3′; si-USP7-3, 5′-CAC CAG TTG TAG ATG TAA CAC TGG TCT CGA GAC CAG TGT TAC ATC TAC AAC-3′.
Reverse transcription-quantitative (RT-q) PCR
Total RNA was extracted from AGS, HGC-27, NCI-N87, SNU-1 or GES-1 cells using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). RT was performed using the PrimeScript 1st Strand cDNA Synthesis kit (Takara Bio, Inc.) according to the manufacturer's protocol. RT-qPCR was performed on ABI 7500 fast PCR System with SYBR green PCR Master Mix (both Applied Biosystems; Thermo Fisher Scientific, Inc.). The qRT-PCR reaction was performed 95°C for 5 min, followed by 40 cycles of 95°C for 10 sec and a primer-specific annealing temperature of 60°C for 30 sec. GAPDH and U6 were used as internal controls. Relative RNA expression was calculated using 2−ΔΔCq method, as previously described (20). Additionally, gel electrophoresis was performed to identify the circular structure of circRPS19. Briefly, circular structure of circRPS19 was validated by PCR reactions using the HotStar HiFidelity Polymerase Kit (#202602; Qiagen) by using pairs of both convergent and divergent primers to amplify circRPS19. Convergent primers were all designed within the same exon. Both cDNA and genomic DNA (gDNA) were applied as the template for PCR analysis. The sequences for primers were as follows (5′~3′): divergent primer for circRPS19 (forward): CAG CTG CCA ACA AGA AGC AT; divergent primer for circRPS19 (reverse): CTG TCC GGC GAT TCT GTC C; convergent primer for circRPS19 (forward): ACT GAC ACC TCA GGG ACA AAG; convergent primer for circRPS19 (reverse): ATG AGG CAA TTT ATT AAC CCA GCA; RPS19 (forward): GAA CCA GCA GGA GTT CGT CA and reverse): AGA ACC AGT TCT CAT CGT AGG G; miR-125a-5p forward): GCCGAG TCC CTG AGA CCC TTT A; miR-125a-5p (reverse): CTC AAC TGG TGT CGT GGA; USP7 (forward): CCG AGG ACA TGG AGA TGG AA and reverse, TTT CGC ACA AAA CAC GGA GG; GAPDH (forward): GAC AGT CAG CCG CAT CTT CT; GAPDH (reverse): GCG CCC AAT ACG ACC AAA TC; U6 (forward): TCC CTT CGG GGA CAT CCG; U6 (reverse): AAT TTT GGA CCA TTT CTC GAT TTG T.
Western blotting
Total protein was isolated from AGS cells with RIPA lysis buffer (Sigma-Aldrich; Merck KGaA), Protein concentration was measured by using a bicinchoninic acid (BCA) Kit (Pierce Biotechnology, Rockford, IL, USA). Protein samples (30 ng/lane) were then separated by 10% SDS-PAGE (Bio-Rad Laboratories, Inc.). Afterwards, the samples were transferred onto PVDF membranes, which were blocked with 5% defatted milk in 5% TBST at room temperature for 2 h. Subsequently, membranes were incubated with primary antibodies, including anti-HK2 (1/1,000, ab209847, Abcam, MA, USA), anti-6-phosphofructokinase subunit alpha (anti-PFK1, 1/1,000, ab154804, Abcam), anti-PKM2 (1/1,000, ab85555, Abcam), anti-USP7 (1/1,000, ab264422, Abcam) and the internal reference anti-β-actin (1:1,000, ab8227, Abcam) at 4°C overnight. After being washed in TBST buffer, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (1/2,000, ab154804, Abcam) at room temperature for 1 h. Pierce ECL Plus Substrate (Pierce; Thermo Fisher Scientific, Inc.) was then used to visualize blots. Image J software v1.8.0 (National Institutes of Health, USA) was used to quantify protein blots.
Cell Counting Kit-8 (CCK-8) assay
Transfected AGS or HGC-27 cells were inoculated into 96-well culture plates at a density of 3×103 cells/well. GC cells were incubated with 10 µl CCK-8 solution (Dojindo Laboratories, Inc.) for 2 h. After incubation, the absorbance at 450 nm was detected at different time points (0, 24, 48 and 72 h) by a spectrophotometer (Thermo Fisher Scientific, Inc.).
EdU assay
Transfected AGS or HGC-27 cells were plated in 12-well plates at a density of 1×105 cells/well. GC cells were treated with EdU reagent (Guangzhou RiboBio Co., Ltd.). Following incubation at 37°C for 2 h, cells were washed in PBS and fixed in 4% paraformaldehyde (Sangon Biotech Co., Ltd.) at 4°C for 30 min. The nuclei of GC cells were stained by 400 µl DAPI (Sigma-Aldrich; Merck KGaA) at room temperature for 30 min. GC cell proliferation was observed under a Leica GmbH fluorescence microscope (×200) and quantified by calculating the percentage of EdU-positive cells in five randomly selected view fields using Image J software v1.8.0 (National Institutes of Health, Bethesda, MD, USA).
TUNEL assay
The apoptosis of transfected AGS or HGC-27 cells was measured by TUNEL assay. GC cells were fixed in 4% paraformaldehyde (Sangon Biotech Co., Ltd.) at 4°C for 30 min. The fixed cells were then washed with 5 ml cold 70% ethanol, and the dehydrated cells were incubated at -20°C for 4 h. Cells were permeabilized with 20 µg/ml Proteinase K in PBS for 5 min. Cells were then treated with the ProLong Antifade Kit (P-7481, Molecular Probes) and incubated with TUNEL reaction mixture (Roche Diagnostics) containing 200 U/ml TdT and 4 µM dUTP at room temperature for 1 h. After removing the reaction buffer, cells were washed with PBS for 3 times. GC cell nuclei were stained by 400 µl DAPI (Sigma-Aldrich; Merck KGaA) that was added to the media (1:10) at room temperature for 30 min. The percentage of TUNEL-positive cells in five randomly selected view fields was observed under a Leica GmbH fluorescence microscope (×200) and calculated using Image J software v1.8.0 (National Institutes of Health, Bethesda, MD, USA).
Flow cytometry
Flow cytometry analysis was used to detect cell apoptosis. Briefly, cells were collected and washed twice in cold PBS. Cells were then fixed with 4% paraformaldehyde (Sangon Biotech Co., Ltd.) at -20°C overnight. GC cells were suspended in 600 µl eBioscience™ flow cytometry binding buffer (Invitrogen; Thermo Fisher Scientific, Inc.) at a concentration of 106 cells/ml and then stained with 5 µl Annexin V/FITC and 5 µl propidium iodide (both BD Biosciences) in dark for 15 min. Samples were analyzed using a FACSAria flow cytometry (BD Biosciences, San Jose, CA, USA). Results were analyzed using FlowJo 7.6 software (Tree Star, Inc.).
Measurement of glucose consumption and lactate production
The glucose consumption and lactate production of transfected AGS or HGC-27 cells were detected by using a Glucose Assay kit (CBA086) and a Lactate Assay Kit (MAK064; both Sigma-Aldrich) as previously described (22).
Examination of extracellular acidification rate (ECAR) and oxygen consumption rate (OCR)
ECAR and OCR were examined as previously reported (22). Seahorse XF96 (cat. no. 102601-100, Seahorse Bioscience) was applied for the analysis of ECAR and OCR in AGS or HGC-27 cells. For ECAR analysis, GC cells were exposed to 1 µM oligomycin for elimination of mitochondria ATP production and 150 mM 2-Deoxy-D-glucose (2-DG) at 37°C for 1 h for inhibition of aerobic glycolysis (both Sigma-Aldrich; Merck KGaA) successively, followed by assessment using Glycolytic Stress Test kit (Seahorse Bioscience). For OCR analysis, GC cells were exposed to 1.0 µM oligomycin, 0.5 µM carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP; a protonophore uncoupler) and 0.5 µM antimycin A at 37°C for 1 h for inhibition of electron transfer chain (all Sigma-Aldrich; Merck KGaA) successively, then measured by XF Cell Mito Stress kit (Seahorse Bioscience). ECAR and OCR were measured by normalizing to total protein content.
Fluorescence in situ hybridization (FISH)
In brief, AGS cells were washed in PBS, followed by fixation in 4% paraformaldehyde (Sangon Biotech Co., Ltd.) at 4°C for 30 min and permeabilization in 0.5% Triton X-100 (Beijing Leigen Biotechnology Co., Ltd.). Cells were blocked with 50 µl goat serum (Sigma-Aldrich; Merck KGaA) for 1 h at room temperature. Primer sets were designed based on the sequence of circRPS19 and USP7 in the pGEM-T Easy vector (Promega). Digoxigenin-labeled DNA probes targeting circRPS19 (genomic position: chr19:42373768-42375484; accession no. NM_001022) and USP7 mRNA (genomic position: chr16:8985951-9057341 accession no. NM_003470.3) or control probes synthesized by Guangzhou RiboBio Co., Ltd. DNA probes were denatured at 75°C for 5 min. Afterwards, the probes were incubated with GC cells in 10 µl hybridization buffer (30% formamide, 0.9 M NaCl, 20 mM Tris-HCl at pH 8.0, 0.01% SDS, a 1-µM probe, and MQ water) at 55°C for 4 h. To remove excess probes, samples was washed in 50 ml tubes of washing buffer without salt [20 mM Tris-HCl, 5 mM EDTA and 0.01% (w/v) SDS] or with 1.8 M NaCl at 47°C for 3 min.
DAPI (Sigma-Aldrich; Merck KGaA) was applied to counterstain nuclei of GC cells at room temperature for 30 min. A confocal microscope (×200, Zeiss AG) was employed to obtain images. The probe sequence for circRPS19: TTT GCG GCC GTT ACG AAT GA.
To identify the co-location of USP7 and HK2 proteins, we used the anti-HK2 (1/1,000, ab209847, Abcam, MA, USA), and anti-USP7 (1/1,000, ab264422, Abcam) to incubated cell lysates at 4°C overnight, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (1/2,000, ab154804, Abcam) at room temperature for 1 h. Results were analyzed using Image J software v1.8.0 (National Institutes of Health).
Co-immunoprecipitation (Co-IP) assay and mass spectrometry
Co-IP assay followed by mass spectrometry was used to identify the interacting protein of HK2. Cells were washed twice with pre-cold PBS. Cell lysates were obtained with RIPA lysis buffer (Sigma-Aldrich; Merck KGaA). For each IP reaction, 50 µl lysates were used. After being centrifuged at 14,000 g for 15 min at 4°C, supernatants were collected. The supernatants were incubated with 50 µg of the negative control anti-Flag (1/200, 66008-3-Ig; Proteintech) or 50 µg Flag-labeled anti-HK2 (1/200, ab209847, Abcam) at 4°C overnight, then with protein A/G magnetic beads (Thermo Fisher Scientific) in accordance with manufacturer's protocol. After being centrifuged at 3,000 × g for 3 min at 4°C, the magnetic beads were deposited at the bottom of the tube. Removing the supernatant, the magnetic beads were washed three times in 1 ml lysis buffer. After being added 15 µl 2xSDS loading buffer, the magnetic beads were boiled for 5 min. The gel bands were excised for in-gel trypsin digestion. Subsequently, extracted peptides were analyzed using nano-LC-MS/MS (AB SCIEX TripleT OF 5600) for mass spectrometry. Mass spectrometry weas conducted on an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific, Inc.) equipped with an Easynano UHPLC, using a 75 µm x 250 mm Pepmap RSLC reverse phase column with a PepMap 1000 trapping column (Thermo Fisher Scientific). The mass spectrometer was equipped with an electrospray ionization (ESI) and operated in the positive ion mode to monitor the m/z transitions for all peptides and IS. Two multiple reaction monitorings were simultaneously monitored for each peptide using manually optimized declustering potentials (DP), collision energies (CE), collision cell entrance (CEP) and exit potentials (CXP). Nitrogen gas was desorbed from the SPME fibre in the injector port at 200°C, under splitless injection mode. The spray voltage was set as 5,000 V, the atomization pressure was set as 45 psi, the auxiliary gas pressure was set as 10 psi. Nitrogen gas with high purity (≥99%) was used as the carrier gas (constant flow of 1.1 ml/min; average velocity 37 cm/s).
IP
To detect ubiquitination of HK2, transfected AGS cells were cultured for 24 h at room temperature in PBS with 0.1% formaldehyde. Cells were treated with the proteasome inhibitor MG132 (10 µM) at room temperature for 8 h. GC cells were lysed in IP buffer (Beyotime Institute of Biotechnology, Shanghai, China) with protease and protein phosphatase inhibitors (Roche Applied Science). Cell lysates (100 ml) were then immunoprecipitated with anti-β-actin (1/1,000, ab8227, Abcam) and anti-HK2 (1/1,000, ab209847, Abcam) at 4°C overnight. To measure HK2 ubiquitination, the samples were incubated with anti-ubiquitin (1/1,000, 10201-2-AP, Proteintech Group, Inc.) 4°C overnight and then subjected to western blot analysis as described above.
Luciferase reporter assay
The wild-type or mutant (MUT) sequences of circRPS19/USP7 3′ untranslated region (UTR) were inserted into pmirGLO vectors (Promega Corporation) to construct pmirGLO-circRPS19/USP7-3′ UTR or pmirGLO-circRPS19/USP7-3′ UTR (MUT). pmirGLO-circRPS19 and pmirGLO-circRPS19 (MUT) were co-transfected into AGS or HGC-27 cells with miR-125a-5p mimics or mimics-NC using Lipofectamine 2000 Transcription Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 36 h. pmirGLO-USP7-3′ UTR or pmirGLO-USP7-3′ UTR (MUT) were co-transfected into AGS or HGC-27 cells with miR-125a-5p mimics or mimics-NC using Lipofectamine 2000 Transcription Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 48 h. The duration between end of transfectioin and activity measurement is 48 h. After that, relative luciferase activity was measured using a Dual-Lucy Assay kit (Progema) with Renilla luciferase activity as the internal reference. The sequence of miR-125a-5p mimics: 5′-UCC CUG AGA CCC UUU AAC CUG UGA-3′; the sequence of NC-mimics: 5′-UCU CCG AGA UCU UAC CAC CUG GUA-3′.
RNA pulldown assay
RNA pulldown assay was performed in AGS or HGC-27 cells to demonstrate RNA interaction. Briefly, 1 µg biotin-labeled circRPS19/USP7-3′ UTR synthesized by Guangzhou RiboBio Co., Ltd. was added to 1% Triton X-100, followed by heating at 95°C for 2 min and cooling in an ice bath for 3 min. Next, the denatured biotinylated RNA was mixed with 15 µl streptavidin beads (Thermo Fisher Scientific, Inc.). RNA-bead complexes were incubated with 100 ml cell lysate obtained using RIPA lysis buffer. After purification, RNA in pulldown products was analyzed by RT-qPCR as described above.
RNA-binding protein IP (RIP) assay
AGS cells were lysed in RIPA buffer for RIP assay using Magna RIP kit (MilliporeSigma). Briefly, cell lysate was treated with RIP buffer at room temperature for 3 h and added to 10 mg/ml magnetic beads (Thermo Fisher Scientific, Inc.) conjugated with anti-argonaute 2 (Ago2, 1/200, 04-642, MilliporeSigma) or NC anti-IgG (1/200, AP113, MilliporeSigma). After washing the beads with 500 µl RIP Wash Buffer, the complexes were incubated with 150 µl proteinase K at 55°C for 30 min to remove protein. The enrichment of circRPS19 or USP7 was analyzed by RT-qPCR as described above.
Animal experiments
A total of 20 male BALB/C nude mice aged 4-6 weeks and weighed 20±2 g were obtained from Shanghai Lab Animal Research Center. All mice were raised in specific pathogen free (SPF) condition at 22-24°C with a stable humidity (55±15%) with free access to food/water in a 12/12 h light/dark cycle, according to the guidelines of the local Animal Care and Use Committee at Affiliated Hospital of Nanjing University of Chinese Medicine. Animal experiments were approved by Ethics Committee of Affiliated Hospital of Nanjing University of Chinese Medicine (approval NO. 2021DW-35-01, Nanjing, China). All nude mice were randomly classified into two groups (n=10). Each mouse was subcutaneously injected with 2×106 AGS cells transfected with si-NC or si-circRPS19-1 (5 nmol/injection). Tumors were monitored on day 7 after injection and measured every three days. On day 28, all nude mice were subjected to euthanasia via cervical dislocation following intravenous injection with 30 mg/kg pentobarbital sodium (23) and tumors were resected and weighed. Death was confirmed by lack of spontaneous breathing for 2-3 min and loss of eye blink response. Body weight decrease of 15-20% was taken as the humane endpoint. No animals were prematurely sacrificed according to this endpoint. Tumors were cut into 4-µm thick small pieces. The tissues were fixed with 10% formalin for 24 h at room temperature, and then embedded in paraffin at 58-62°C for 30 min. The sections were deparaffinized in xylene and rehydrated in a graded series of alcohol. A steamer was used for 20 min in an EDTA buffer for antigen retrieval at 92°C. Next, samples were treated with 3% hydrogen peroxide in methanol at room temparature for 20 min to block endogenous peroxidase activity and the reaction was blocked by incubating in a 5% solution of non-fat milk for 1 h. Slides were then incubated with polyclonal rabbit antibodies against Ki67 (1:200; cat. no. ab16667, Abcam), HK2 (1/500; cat. no. ab209847, Abcam), or USP7 (1/200, ab239936, Abcam) at room temperature for 1 h, followed by incubation with the secondary antibody HRP-polymer anti-rabbit (1/1,000, ab6721, Abcam) at 37°C for 15 min. The samples were counterstained with DAB) at room temperature for 5 min. Images of five typical visual fields of each section were obtained under a light microscope (×200, DMI1, Leica, Wetzlar, Germany) and analyzed using software (Image-Pro Plus 6.0; Media Cybernetics, Inc.).
Statistical analysis
Unpaired Student's t test was used to analyze differences between two groups. Data from three independent assays are presented as the mean ± SD. SPSS v22.0 software (IBM Corp.) was used for data analysis. One-way ANOVA followed by LSD (one control group) or Tukey (more than one control group) post hoc test was used for comparing >2 groups. P<0.05 was considered to indicate a statistically significant difference.
Results
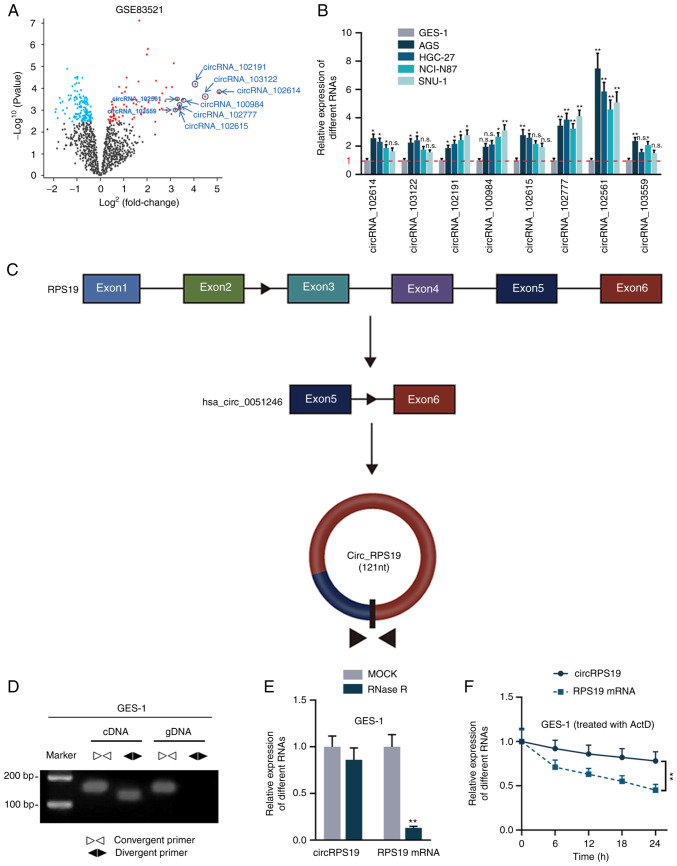
CircRPS19 has circular characteristics and is upregulated in GC cells
circRNAs significantly overexpressed in GC tissue were screened from GEO database. According to the GSE83521 dataset, under the condition of P≤0.05, the top eight circRNAs with the highest logFC values were screened, including circRNA_102614, circRNA_103122, circRNA_102191, circRNA_100984, circRNA_102615, circRNA_102777, circRNA_102561 and circRNA_103559 (Fig. 1A). Of these, hsa_circRNA_103122 (24), hsa_circRNA_102191 (25) and hsa_circRNA_102615 (26) have been reported in GC and were excluded from subsequent study. RT-qPCR was used to probe the expression levels of the other five candidates in human GC (AGS, HGC-27, NCI-N87 and SNU-1) and normal gastric epithelial cell lines (GES-1). circRNA_102561 was most significantly upregulated in all four GC cell lines compared with GES-1 (Fig. 1B) and was therefore selected for subsequent experiments. circRNA_102561 (circRPS19) originates from exons 5 to 6 of its host gene RPS19 (Fig. 1C). PCR-agarose gel electrophoresis and RT-qPCR were performed in GES-1 cells to validate the circular structure of circRPS19. As shown by PCR-agarose gel, circRPS19 was amplified by convergent and divergent primer in cDNA but only amplified by convergent primer in gDNA (Fig. 1D). Moreover, after the treatment of RNase R, circRPS19 expression was almost unchanged, while linear RPS19 mRNA was nearly completely digested. Expression levels of circRPS19 and RPS19 mRNA at 0, 6, 12, 18, 24 h in GES-1 cells treated with Actinomycin D (ActD) were assessed. circRPS19 expression remained steady, while RPS19 mRNA level decreased with time (Fig. 1F). Collectively, the aforementioned results showed that circRPS19 had a circular structure and was associated with GC.
Figure 1.
circRPS19 has circular characteristics and is upregulated in GC cells. (A) Differentially expressed circRNAs in GC tissue obtained from GSE83521 dataset. (B) Expression of five circRNAs previously unreported in GC were detected by RT-qPCR in AGS, HGC-27, NCI-N87, SNU-1 and GES-1 cells. (C) circRPS19 structure. (D) PCR-agarose gel electrophoresis verified the circular structure of circRPS19 in GES-1 cells. cDNA was created by RT of circRPS19; gDNA was the genome sequence of circRPS19. circRPS19 and RPS19 mRNA levels were detected by RT-qPCR in GES-1 cells following (E) RNase R and (F) ActD treatment. *P<0.05, **P<0.01 vs. control group. circRPS19, circRNA ribosomal protein S19; GC, gastric cancer; RT-q, reverse transcription-quantitative; n.s., not significant.
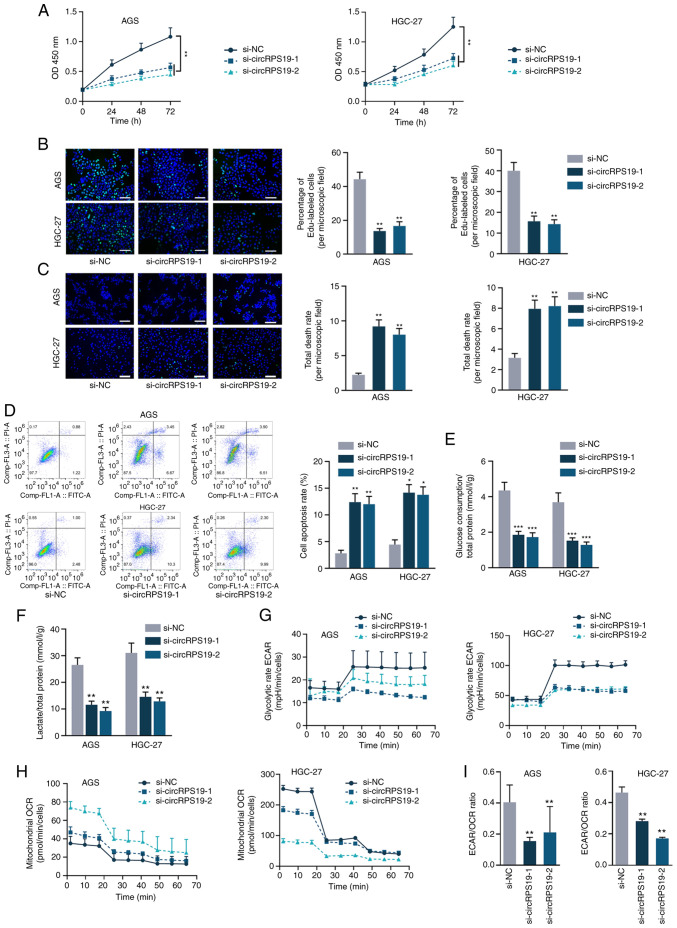
circRPS19 knockdown inhibits GC cell proliferation and aerobic glycolysis
As aforementioned, circRPS19 was upregulated in GC cell lines but its specific effects on the malignant phenotype of GC cells is unknown. It was investigated whether knockdown of circRPS19 alters biological processes of GC cells. RT-qPCR was used to determine the efficiency of circRPS19-1-specific siRNAs on silencing circRPS19 in AGS and HGC-27 cells (Fig. S1A). si-circRPS19-1 and si-circRPS19-2 were selected for subsequent assays owing to their higher efficiency. CCK-8 and EdU assay showed that GC cell proliferation decreased following circRPS19 knockdown (Fig. 2A and B). TUNEL assay and flow cytometry analysis of GC cells showed that GC cell apoptosis was enhanced following circRPS19 depletion (Fig. 2C and D). circRPS19 facilitated GC cell proliferation but prevented GC cell apoptosis (Fig. 2A-D). Furthermore, the effect of circRPS19 knockdown on glucose consumption and lactate production of GC cells was assessed. Glucose consumption and lactate production were decreased following circRPS19 knockdown (Fig. 2E and F). circRPS19 ablation led to a decrease in ECAR but an increase in OCR (Fig. 2G and H). ECAR/OCR ratio was decreased following circRPS19 interference (Fig. 2I). Taken together, these data indicated that circRPS19 facilitated GC cell proliferation and aerobic glycolysis.
Figure 2.
circRPS19 knockdown inhibits GC cell proliferation and aerobic glycolysis. (A) Proliferation of AGS and HGC-27 cells following circRPS19 ablation was detected by Cell Counting Kit-8. (B) Proliferation of AGS and HGC-27 cells following circRPS19 ablation was detected by EdU assay. (C) TUNEL assay showed GC cell apoptosis following circRPS19 ablation (scale bar, 200 µm). (D) Flow cytometry assay showed GC cell apoptosis following circRPS19 ablation. (E) Glucose consumption, (F) lactate production, (G) ECAR and (H) OCR were determined in GC cells following circRPS19 silencing. (I) ECAR/OCR ratio was assessed in GC cells following circRPS19 silencing. *P<0.05, **P<0.01, ***P<0.001 vs. control group. circRPS19, circRNA ribosomal protein S19; GC, gastric cancer; ECAR, extracellular acidification rate; OCR, oxygen consumption rate; OD, optical density; si, small interfering; NC, negative control.
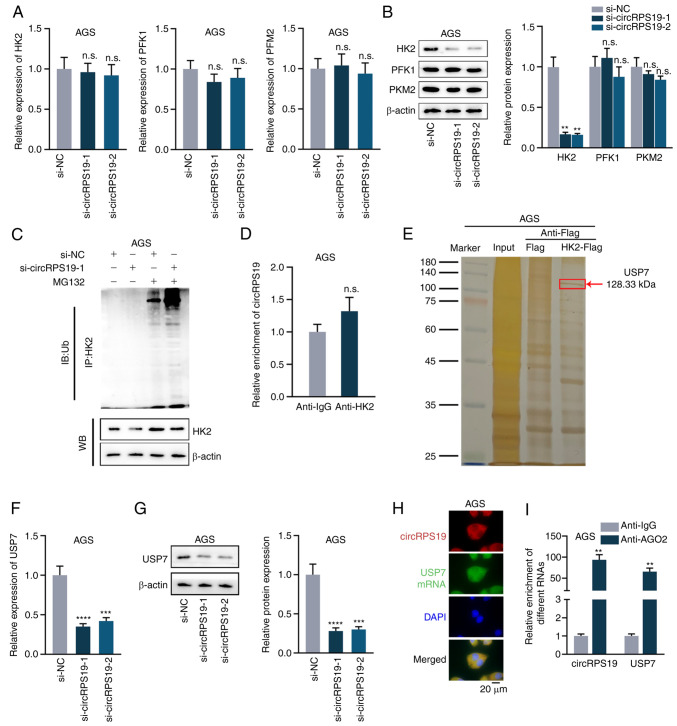
circRPS19 upregulates HK2 protein expression via USP7
A previous study identified three rate-limiting enzymes in the regulation of aerobic glycolysis, including pyruvate kinases type M2 (PKM2), HK2 and phosphofructokinase 1 (PFK1) (14). Thus, the present study investigated which rate-limiting enzyme was regulated by circRPS19. RT-qPCR and western blotting showed that knockdown of circRPS19 had no effect on mRNA levels of these three enzymes (Fig. 3A) but effectively decreased protein levels of HK2 (Fig. 3B). Therefore, it was investigated whether circRPS19 influenced ubiquitination of HK2 protein. IP showed ubiquitination of HK2 was notably increased by circRPS19 silencing (Fig. 3C). It was hypothesized circRPS19 may stabilize HK2 by directly interacting with it. However, RIP showed that circRPS19 did not directly bind to HK2 protein (Fig. 3D). The protein interacting with HK2 and modulating ubiquitination of HK2 protein was assessed by Co-IP followed by mass spectrometry. Deubiquitinating enzyme USP7 interacted with HK2 (Fig. 3E). RT-qPCR and western blotting were performed in AGS cells to investigate if circRPS19 affected USP7 mRNA and protein expression. mRNA and protein levels of USP7 were significantly decreased by circRPS19 depletion (Fig. 3F and G). FISH assay was used to investigate co-localization of circRPS19 and USP7 mRNA in AGS cells. circRPS19 and USP7 mRNA were co-localized in the cytoplasm (Fig. 3H). RIP showed that circRPS19 and USP7 mRNA were more abundant in AGO2 immunoprecipitate compared with that in IgG immunoprecipitate (Fig. 3I). In summary, circRPS19 upregulated USP7 expression to enhance HK2 protein levels and promote aerobic glycolysis of GC cells.
Figure 3.
circRPS19 upregulates HK2 protein expression via USP7. (A) mRNA and (B) protein levels of PKM2, HK2 and PFK1 were assessed by RT-qPCR and WB in AGS cells following circRPS19 ablation. (C) IP showed HK2 ubiquitination in AGS cells following circRPS19 depletion. (D) RIP assay was used to examine the interaction between circRPS19 and HK2 protein in AGS cells. (E) Co-IP followed by mass spectrometry analysis was used in AGS cells to determine enzymes affecting ubiquitination of HK2 protein. (F) mRNA and (G) protein levels of USP7 were assessed by RT-qPCR and WB in AGS cells following circRPS19 ablation. (H) FISH assay was used to determine co-localization of circRPS19 and USP7 mRNA in AGS cells. (I) RIP assay was used to detect abundance of circRPS19 and USP7 mRNA in RISC. **P<0.01, ***P<0.001, ****P<0.0001 compared with control group. circRPS19, circRNA ribosomal protein S19; HK2, hexokinase 2; USP7, ubiquitin-specific processing protease 7; PKM2, pyruvate kinase M2; PFK1, 6-phosphofructokinase subunit alpha; RT-q, reverse transcription-quantitative; IP, immunoprecipitation; FISH, Fluorescence in situ hybridization; RISC, RNA-induced silencing complex; n.s., not significant; si, small interfering; NC, negative control; WB, western blot; IB, immunoblot; Ub, ubiquitin.
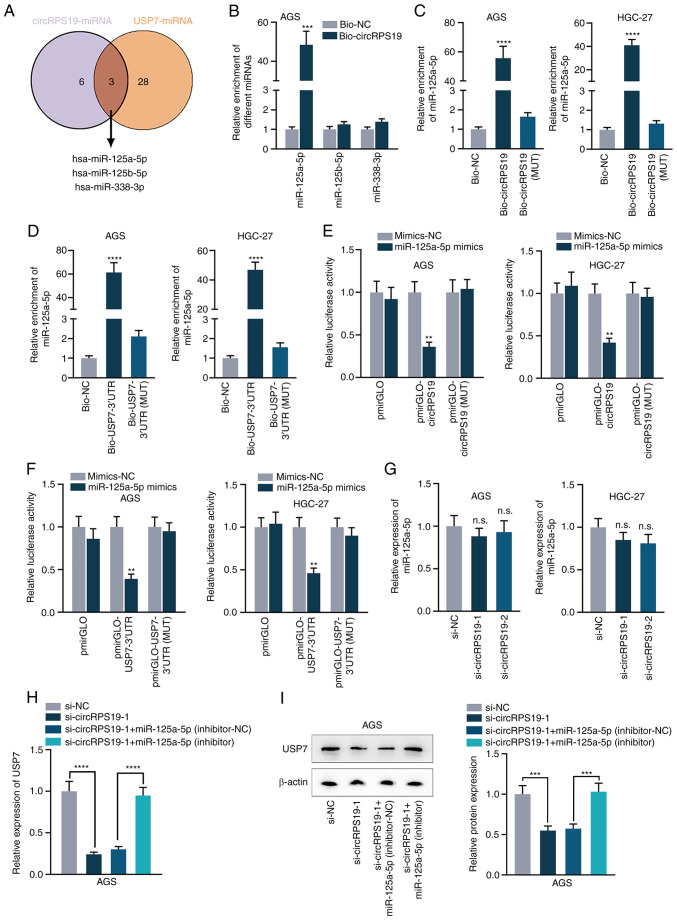
CircRPS19 upregulates USP7 expression via competitively binding to miR-125a-5p
It was hypothesized that circRPS19 might modulate USP7 via competitive endogenous RNA (ceRNA) network. starBase database was used to screen miRNAs that potentially interact with both circRPS19 and USP7. As a result, three common miRNAs (hsa-miR-125a-5p, hsa-miR-125b-5p and hsa-miR-338-3p) were obtained (Figs. S2A and B and 4A). RNA pulldown assay was used for screening in AGS cells and binding affinity of miR-125a-5p with circRPS19 was the highest compared with other miRNAs (Fig. 4B). RNA pulldown assay verified the interaction of miR-125a-5p with circRPS19 (Fig. 4C). RNA pulldown assay in AGS and HGC-27 cells showed that miR-125a-5p could combine with USP7 3′ UTR (Fig. 4D). Next, miR-125a-5p was overexpressed by transfection with miR-125a-5p mimics into AGS and HGC-27 cells and the overexpression efficiency were detected by RT-qPCR (Fig. S1B). As indicated by luciferase reporter assay, upregulation of miR-125a-5p decreased luciferase activity of pmirGLO-circRPS19/USP7-3′ UTR, while no obvious changes were observed in MUT groups. miR-125a-5p could bind with both circRPS19 and USP7 3′ UTR (Fig. 4E and F). RT-qPCR results indicated that circRPS19 silencing had no effects on miR-125a-5p expression (Fig. 4G), indicating that circRPS19 exerted its function by competitively binding to miR-125a-5p. mRNA and protein levels of USP7 suppressed by circRPS19 knockdown were completely reversed by miR-125a-5p inhibition (Fig. 4H and I). circRPS19 upregulated USP7 expression via competitively binding to miR-125a-5p.
Figure 4.
circRPS19 upregulates USP7 expression via competitively binding to miR-125a-5p. (A) Common miRs interacting with USP7 and circRPS19 predicted by starBase database (starbase.sysu.edu.cn/). (B) RNA pulldown assay showed the combination of circRPS19 with miR-125a-5p, miR-125b-5p and miR-338-3p in AGS cells. (C) RNA pulldown assay revealed the interaction of miR-125a-5p with circRPS19 in AGS and HGC-27 cells. (D) RNA pulldown assay revealed the interaction of miR-125a-5p with USP7 3′-UTR in AGS and HGC-27 cells. (E) Luciferase reporter assay revealed the interaction of miR-125a-5p with circRPS19 in AGS and HGC-27 cells. (F) Luciferase reporter assay revealed the interaction of miR-125a-5p with USP7 3′UTR in AGS and HGC-27 cells. (G) miR-125a-5p expression was probed by RT-qPCR in AGS and HGC-27 cells subsequent to circRPS19 knockdown. (H) mRNA and (I) protein levels of USP7 were assessed by RT-qPCR and western blot in AGS cells transfected with si-NC, si-circRPS19-1, si-circRPS19-1 + inhibitor-NC or si-circRPS19-1 + miR-125a-5p inhibitor. **P<0.01, ***P<0.001, ****P<0.0001 compared with control group. circRPS19, circRNA ribosomal protein S19; USP7, ubiquitin-specific processing protease 7; miR, microRNA; UTR, untranslated region; RT-q, reverse transcription-quantitative; NC, negative control; bio, biotinylated; MUT, mutant; si, small interfering; n.s., not significant.
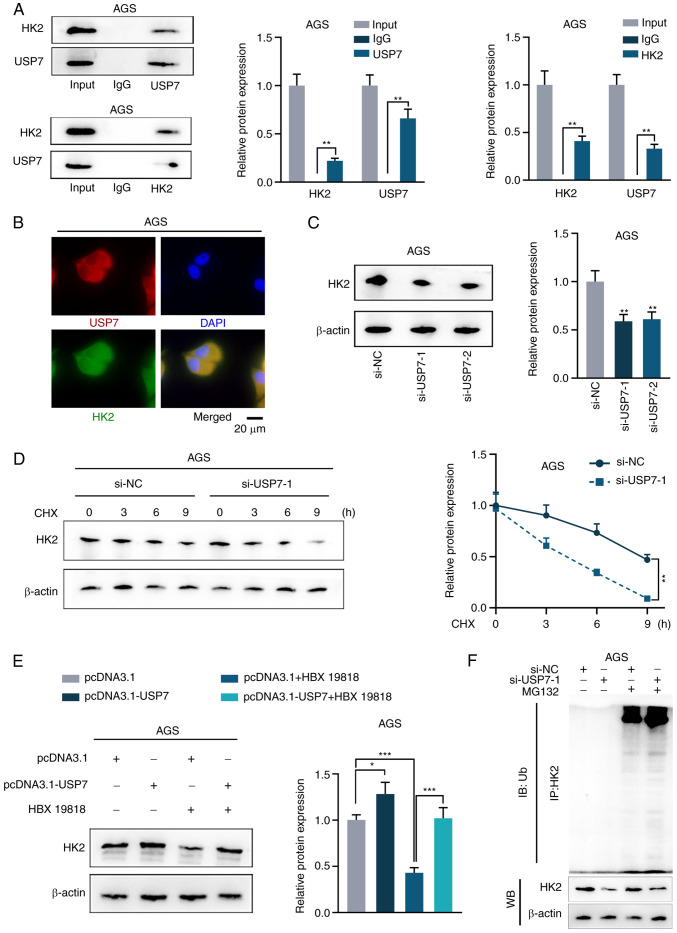
circRPS19 upregulates H2K protein through USP7-mediated deubiquitination
Based on the aforementioned interaction between USP7 and HK2, USP7-mediated HK2 deubiquitination was investigated. Co-IP in AGS cells demonstrated an interaction between USP7 and HK2 (Fig. 5A). Immunofluorescence assay revealed that USP7 and HK2 were co-localized in the cytoplasm of AGS cells (Fig. 5B). To validate the regulatory effect of USP7 on HK2, USP7 expression was knocked down and efficiency was assessed by RT-qPCR (Fig. S1C). HK2 protein was downregulated after USP7 interference (Fig. 5C). Degradation rate of HK2 protein was accelerated after interference with USP7 in CHX-treated cells (Fig. 5D). The overexpression efficiency of pcDNA3.1-USP7 was detected by RT-qPCR in AGS and HGC-27 cells (Fig. S1D). The western blot results showed that increased HK2 protein expression induced by USP7 overexpression was reversed by USP7 inhibitor HBX19818, indicating that USP7 mediated deubiquitination of HK2 (Fig. 5E). IP showed that HK2 ubiquitination was notably decreased by USP7 knockdown (Fig. 5F). circRPS19 promoted USP7-mediated deubiquitination of HK2.
Figure 5.
circRNA ribosomal protein S19 upregulates HK2 protein via USP7-mediated deubiquitination. (A) Co-IP analysis of the interaction between USP7 and HK2 in AGS cells. (B) Immunofluorescence assay detected the co-localization of USP7 and HK2 in AGS cells. WB of (C) HK2 protein expression in AGS cells and (D) degradation rate of HK2 protein in CHX-treated AGS cells following USP7 ablation and (E) HK2 protein expression following USP7 overexpression and HBX19818 treatment in AGS cells. (F) IP showed HK2 ubiquitination in MG132-treated AGS cells following USP7 depletion. *P<0.05, **P<0.01, ***P<0.001 vs. control group. HK2, hexokinase 2; USP7, ubiquitin-specific processing protease 7; IP, immunoprecipitation; CHX, cycloheximide; HBX19818, a specific inhibitor of USP7; si, small interfering; NC, negative control; WB, western blotting; Ub, ubiquitin.
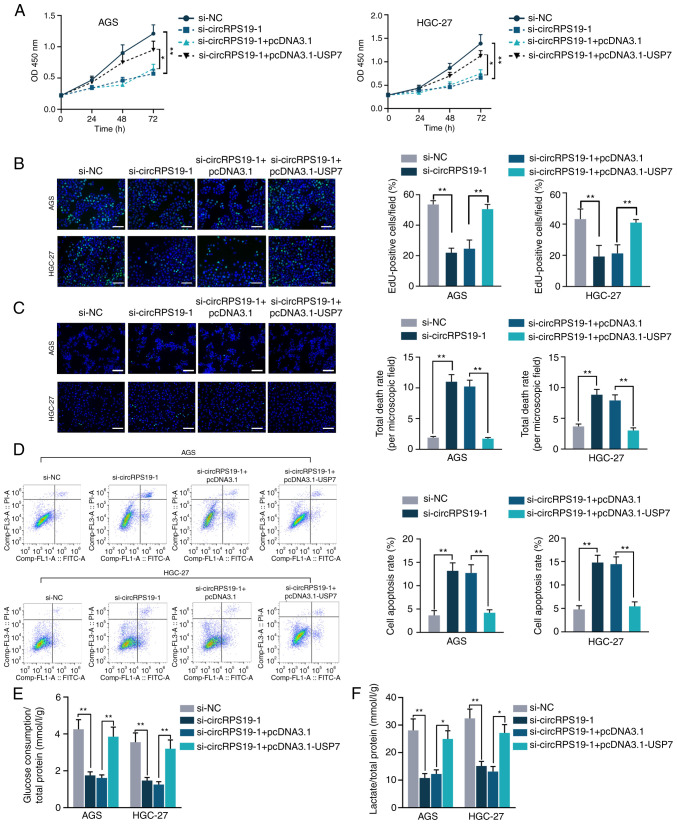
circRPS19 facilitates GC cell proliferation and aerobic glycolysis by modulating USP7
In vitro rescue assays were performed to investigate whether circRPS19 promotes aerobic glycolysis of GC cells by regulating USP7, thus facilitating GC cell progression. As indicated by CCK-8 and EdU assay, decreased GC cell proliferation induced by circRPS19 depletion was counteracted by USP7 overexpression (Fig. 6A and B). TUNEL and flow cytometry showed that the effect of circRPS19 knockdown on GC cell apoptosis was counteracted by USP7 overexpression (Fig. 6C and D). The present study investigated the roles of circRPS19 and USP7 in aerobic glycolysis of GC cells. Glucose consumption and lactate production suppressed by circRPS19 silencing was restored by USP7 overexpression (Fig. 6E and F). To summarize, circRPS19 facilitated proliferation and aerobic glycolysis but inhibited apoptosis of GC cells by modulating USP7.
Figure 6.
circRPS19 facilitates GC cell proliferation and aerobic glycolysis by modulating USP7. (A) Cell Counting Kit-8 and (B) EdU assays showed proliferation in AGS and HGC-27 cells (scale bar=200 µm). (C) TUNEL assay (scale bar=200 µm) and (D) flow cytometry showed GC cell apoptosis. (E) Glucose consumption and (F) lactate production of GC cells. *P<0.05, **P<0.01 vs. control. circRPS19, circRNA ribosomal protein S19; GC, gastric cancer; OD, optical density; si, small interfering; NC, negative control.
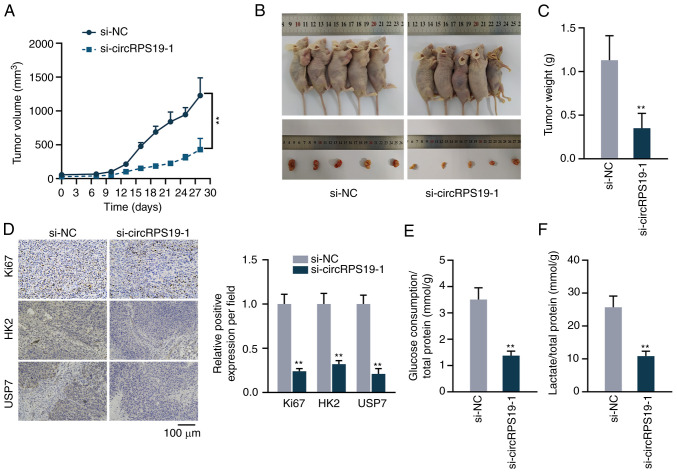
circRPS19 promotes GC cell proliferation and aerobic glycolysis in vivo
The present study performed animal experiments to detect the effect of circRPS19 on GC in vivo. Tumor volume in nude mice was measured every 3 days beginning on day 7. The growth curve showed that circRPS19 knockdown decreased tumor growth (Fig. 7A). On day 28, tumors were resected. Size and volume of excised tumors were reduced in si-circRPS19-1 compared with si-NC group (Fig. 7B and C). Immunohistochemistry showed that the expression of Ki67, HK2, and USP7 was decreased in si-circRPS19-1 group compared with si-NC group (Fig. 7D). The effects of circRPS19 on aerobic glycolysis of tumor cells were analyzed. Glucose consumption and lactate production of tumor cells were decreased in si-circRPS19-1 group in comparison with si-NC group (Fig. 7E and F). Therefore, circRPS19 promoted GC cell proliferation and aerobic glycolysis in vivo.
Figure 7.
circRPS19 promotes GC cell proliferation and aerobic glycolysis in vivo. (A) Tumor volume was recorded every 3 days. (B) Representative image and (C) weight of excised tumors. (D) Immunohistochemistry was implemented to investigate the expression of Ki-67, HK2 and USP7 in tumor tissues. (E) Glucose consumption and (F) lactate production were examined in tumor tissues. **P<0.01 compared with control group. circRPS19, circRNA ribosomal protein S19; GC, gastric cancer; HK2, hexokinase 2; USP7, ubiquitin-specific processing protease 7; si, small interfering; NC, negative control.
Discussion
Previous studies have demonstrated that circRNAs play either a tumor-suppressing or -promoting role in GC: For example, circRNA circ_001988 can sequester miR-197-3p to hamper progression of GC both in vitro and in vivo (27); circRNA circ_0004104 enhances GC progression by sequestering miR-539-3p and regulating ring finger protein 2 (RNF2) (28). In addition, circRNA Ran GTPase activating protein 1 (circ-RanGAP1) targets miR-877-3p to modulate VEGFA expression, thereby promoting GC cell migration and invasion (29). The present study identified a novel circRNA, circRPS19, that was notably upregulated in GC cell lines. Furthermore, circRPS19 knockdown attenuated GC cell proliferation both in vitro and in vivo. FISH results demonstrated that circRPS19 and USP7 mRNA localized in the cytoplasm of GC cells. The cytoplasmic localization is indicative of the regulation of circRNA on mRNA at the post-transcriptional level (30). As indicated by previous studies, circRNAs serve as ceRNAs by competitively interacting with miRNAs to affect levels of target mRNAs: For example, circRNA DNA topoisomerase IIA as a ceRNA increases sushi domain containing 2 (SUSD2) expression via sequestering miR-346 in glioma cells (31); circ_0058357 competitively combines with miR-24-3p to increase AVL9 cell migration associated (AVL9) expression in non-small cell lung cancer cells (32); circRNA itchy E3 ubiquitin protein ligase (circ-ITCH) acts as a sponge for miR-199a-5p to enhance Klotho expression in GC cells (33). It was hypothesized that circRPS19 as a ceRNA binds to a certain miRNA to regulate USP7 mRNA. Through bioinformatics analysis, miR-125a-5p was screened. The regulatory influence of miR-125a-5p on GC progression has been studied previously (34). Additionally, tyrosine kinase non receptor 2 antisense RNA 1 (TNK2-AS1) targets miR-125a-5p to modulate the PI3K/Akt pathway, thereby facilitating epithelial-mesenchymal transition, invasion and migration of GC cells (35). The present study showed that miR-125a-5p bound to both circRPS19 and USP7 mRNA. Rescue assays proved that circRPS19 upregulated USP7 mRNA expression by competitively binding to miR-125a-5p.
Rate-limiting enzyme HK2 is a key factor in glycolysis of GC cells (36). According to previous studies, circRNA cullin 3 (circCUL3) upregulates transcription factor STAT3, which facilitates HK2 transcription to increase aerobic glycolysis of GC cells (19); circRNF20 increases aerobic glycolysis of breast cancer cells by upregulating hypoxia inducible factor-1α, a transcription factor for HK2 (37). The present study showed that circRPS19 promoted aerobic glycolysis of GC cells by modulating HK2. Nevertheless, gene expression analysis showed that circRPS19 could only affect HK2 protein, excluding the possibility of ceRNA network and transcription activation. It was hypothesized that circRPS19 influences ubiquitination of HK2 protein via deubiquitinating enzyme USP7. USP7 mediates ubiquitination of target protein in cancer cells (38). The present study showed that circRPS19 can upregulate USP7 to mediate deubiquitination of HK2.
The present study showed that circRPS19 promotes GC cell proliferation via aerobic glycolysis. Furthermore, circRPS19 mediated deubiquitination of HK2 by USP7, which was also regulated by circRPS19 via ceRNA network. The present findings provide understanding of the molecular mechanisms underlying GC, which may facilitate development of treatment for GC.
Supplementary Data
Acknowledgments
Not applicable.
Funding Statement
The present study was supported by National Traditional Chinese Medicine Inheritance and Innovation Platform Construction Project by National Administration of Traditional Chinese Medicine (grant no. Y2020CX570) and College Project of Jiangsu Province Hospital of Chinese Medicine (No. Y2021ZR12).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SL and JQ conceived and designed the study. XZ and JS performed experiments. XZ analyzed and interpreted data. XZ and JQ wrote the manuscript. XZ and JS confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was reviewed and approved by the Ethics Committee of Affiliated Hospital of Nanjing University of Chinese Medicine (approval no. 2021DW-35-01; Nanjing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Liu AG, Zhong JC, Chen G, He RQ, He YQ, Ma J, Yang LH, Wu XJ, Huang JT, Li JJ, et al. Upregulated expression of SAC3D1 is associated with progression in gastric cancer. Int J Oncol. 2020;57:122–138. doi: 10.3892/ijo.2020.5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric cancer: Epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int J Mol Sci. 2020;21:4012. doi: 10.3390/ijms21114012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA Cancer J Clin. 2021;71:264–279. doi: 10.3322/caac.21657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen W, Zhang K, Yang Y, Guo Z, Wang X, Teng B, Zhao Q, Huang C, Qiu Z. MEF2A-mediated lncRNA HCP5 inhibits gastric cancer progression via MiR-106b-5p/p21 axis. Int J Biol Sci. 2021;17:623–634. doi: 10.7150/ijbs.55020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang B, Zhang Y, Qing T, Xing K, Li J, Zhen T, Zhu S, Zhan X. Comprehensive analysis of metastatic gastric cancer tumour cells using single-cell RNA-seq. Sci Rep. 2021;11:1141. doi: 10.1038/s41598-020-80881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang X, Ren H, Guo M, Qian J, Yang Y, Gu C. Review on circular RNAs and new insights into their roles in cancer. Comput Struct Biotechnol J. 2021;19:910–928. doi: 10.1016/j.csbj.2021.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu T, Sun Y, Sun Z, Li S, Wang W, Yu B, Wang G. Hsa_circ_0042823 accelerates cancer progression via miR-877-5p/FOXM1 axis in laryngeal squamous cell carcinoma. Ann Med. 2021;53:960–970. doi: 10.1080/07853890.2021.1934725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G, Sun D, Li W, Xin Y. CircRNA_100290 promotes GC cell proliferation and invasion via the miR-29b-3p/ITGA11 axis and is regulated by EIF4A3. Cancer Cell Int. 2021;21:324. doi: 10.1186/s12935-021-01964-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, Xu P, Sun G, Xu J, Lv J, Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang F, Hu A, Li D, Wang J, Guo Y, Liu Y, Li H, Chen Y, Wang X, Huang K, et al. Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol Cancer. 2019;18:158. doi: 10.1186/s12943-019-1094-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma C, Wang X, Yang F, Zang Y, Liu J, Wang X, Xu X, Li W, Jia J, Liu Z. Circular RNA hsa_circ_0004872 inhibits gastric cancer progression via the miR-224/Smad4/ADAR1 successive regulatory circuit. Mol Cancer. 2020;19:157. doi: 10.1186/s12943-020-01268-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo R, Cui X, Li X, Zang W, Chang M, Sun Z, Liu Z, Sun Y, Jia J, Li W. CircMAN1A2 is upregulated by Helicobacter pylori and promotes development of gastric cancer. Cell Death Dis. 2022;13:409. doi: 10.1038/s41419-022-04811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng J, Li J, Wu L, Yu Q, Ji J, Wu J, Dai W, Guo C. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39:126. doi: 10.1186/s13046-020-01629-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, Li J, Yan W, Yao Z, Wang R, Zhou X, Wu H, Zhang G, Shi T, Chen W. H19 promotes aerobic glycolysis, proliferation, and immune escape of gastric cancer cells through the microRNA-519d-3p/lactate dehydrogenase A axis. Cancer Sci. 2021;112:2245–2259. doi: 10.1111/cas.14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J, Hou S, Xu J, Wu J, Yin J. Long non-coding RNA OIP5-AS1 promotes cell proliferation and aerobic glycolysis in gastric cancer through sponging miR-186. Arch Med Sci. 2021;17:1742–1751. doi: 10.5114/aoms.2019.87213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Xu H, Xing R, Pan Y, Li W, Cui J, Lu Y. Pyruvate kinase M2 contributes to cell growth in gastric cancer via aerobic glycolysis. Pathol Res Pract. 2019;215:152409. doi: 10.1016/j.prp.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Zheng M, Wu C, Yang K, Yang Y, Liu Y, Gao S, Wang Q, Li C, Chen L, Li H. Novel selective hexokinase 2 inhibitor Benitrobenrazide blocks cancer cells growth by targeting glycolysis. Pharmacol Res. 2021;164:105367. doi: 10.1016/j.phrs.2020.105367. [DOI] [PubMed] [Google Scholar]
- 19.Pu Z, Xu M, Yuan X, Xie H, Zhao J. Circular RNA circCUL3 accelerates the warburg effect progression of gastric cancer through regulating the STAT3/HK2 axis. Mol Ther Nucleic Acids. 2020;22:310–318. doi: 10.1016/j.omtn.2020.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Cheng D, Zhu M, Yu H, Pan Z, Liu L, Geng Q, Pan H, Yan M, Yao M. OTUB2 stabilizes U2AF2 to promote the Warburg effect and tumorigenesis via the AKT/mTOR signaling pathway in non-small cell lung cancer. Theranostics. 2019;9:179–195. doi: 10.7150/thno.29545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Peng Y, Li J, Chen Z, Chen F, Tu J, Lin S, Wang H. N(6)-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat Commun. 2020;11:2578. doi: 10.1038/s41467-020-16306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joo DT, Xiong Z, MacDonald JF, Jia Z, Roder J, Sonner J, Orser BA. Blockade of glutamate receptors and barbiturate anesthesia: Increased sensitivity to pentobarbital-induced anesthesia despite reduced inhibition of AMPA receptors in GluR2 null mutant mice. Anesthesiology. 1999;91:1329–1341. doi: 10.1097/00000542-199911000-00025. [DOI] [PubMed] [Google Scholar]
- 24.Ding L, Zhao Y, Dang S, Wang Y, Li X, Yu X, Li Z, Wei J, Liu M, Li G. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol Cancer. 2019;18:45. doi: 10.1186/s12943-019-1006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang D, Jiang X, Liu Y, Cao G, Zhang X, Kuang Y. Circular RNA circ_HN1 facilitates gastric cancer progression through modulation of the miR-302b-3p/ROCK2 axis. Mol Cell Biochem. 2021;476:199–212. doi: 10.1007/s11010-020-03897-2. [DOI] [PubMed] [Google Scholar]
- 26.Chen B, Ji F, Wen X, Jin Z. Circular RNA circ_ASAP2 promotes cell viability, migration, and invasion of gastric cancer cells by regulating the miR-770-5p/CDK6 axis. Int J Clin Exp Pathol. 2020;13:2806–2819. [PMC free article] [PubMed] [Google Scholar]
- 27.Sun D, Wang G, Xiao C, Xin Y. Hsa_circ_001988 attenuates GC progression in vitro and in vivo via sponging miR-197-3p. J Cell Physiol. 2021;236:612–624. doi: 10.1002/jcp.29888. [DOI] [PubMed] [Google Scholar]
- 28.Yue F, Peng K, Zhang L, Zhang J. Circ_0004104 accelerates the progression of gastric cancer by regulating the miR-539-3p/RNF2 axis. Dig Dis Sci. 2021;66:4290–4301. doi: 10.1007/s10620-020-06802-5. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW, Wang JB, Lin JX, Chen QY, Cao LL, et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38–48. doi: 10.1016/j.canlet.2019.11.038. [DOI] [PubMed] [Google Scholar]
- 30.Zhou J, Zhang S, Chen Z, He Z, Xu Y, Li Z. CircRNA-ENO1 promoted glycolysis and tumor progression in lung adenocarcinoma through upregulating its host gene ENO1. Cell Death Dis. 2019;10:885. doi: 10.1038/s41419-019-2127-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sang J, Li X, Lv L, Zhang C, Zhang X, Li G. Circ-TOP2A acts as a ceRNA for miR-346 and contributes to glioma progression via the modulation of sushi domain-containing 2. Mol Med Rep. 2021;23:255. doi: 10.3892/mmr.2021.11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei D, Sun L, Feng W. hsa_circ_0058357 acts as a ceRNA to promote non-small cell lung cancer progression via the hsa-miR-24-3p/AVL9 axis. Mol Med Rep. 2021;23:470. doi: 10.3892/mmr.2021.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Wang H, Zheng R, Wu P, Sun Z, Chen J, Zhang L, Zhang C, Qian H, Jiang J, Xu W. Circular RNA ITCH suppresses metastasis of gastric cancer via regulating miR-199a-5p/Klotho axis. Cell Cycle. 2021;20:522–536. doi: 10.1080/15384101.2021.1878327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li R, Hu Z, Wang Z, Zhu T, Wang G, Gao B, Wang J, Deng X. miR-125a-5p promotes gastric cancer growth and invasion by regulating the Hippo pathway. J Clin Lab Anal. 2021;35:e24078. doi: 10.1002/jcla.24078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo L, Ma H, Kong Y, Leng G, Liu G, Zhang Y. Long non-coding RNA TNK2 AS1/microRNA-125a-5p axis promotes tumor growth and modulated phosphatidylinositol 3 kinase/AKT pathway. J Gastroenterol Hepatol. 2022;37:124–133. doi: 10.1111/jgh.15683. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Cao B, Zhao R, Li H, Wei B, Dai G. Knockdown of circBFAR inhibits proliferation and glycolysis in gastric cancer by sponging miR-513a-3p/hexokinase 2 axis. Biochem Biophys Res Commun. 2021;560:80–86. doi: 10.1016/j.bbrc.2021.04.131. [DOI] [PubMed] [Google Scholar]
- 37.Cao L, Wang M, Dong Y, Xu B, Chen J, Ding Y, Qiu S, Li L, Zaharieva EK, Zhou X, Xu Y. Circular RNA circRNF20 promotes breast cancer tumorigenesis and Warburg effect through miR-487a/HIF-1α/HK2. Cell Death Dis. 2020;11:145. doi: 10.1038/s41419-020-2336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JE, Park CM, Kim JH. USP7 deubiquitinates and stabilizes EZH2 in prostate cancer cells. Gene Mol Biol. 2020;43:e20190338. doi: 10.1590/1678-4685-gmb-2019-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.