Abstract
Engineered living materials (ELMs) combine living cells with polymeric matrices to yield unique materials with programmable functions. While the cellular platform and the polymer network determine the material properties and applications, there are still gaps in our ability to seamlessly integrate the biotic (cellular) and abiotic (polymer) components into singular material, then assemble them into devices and machines. Herein, we demonstrated the additive-manufacturing of ELMs wherein bioproduction of metabolites from the encapsulated cells enhanced the properties of the surrounding matrix. First, we developed aqueous resins comprising bovine serum albumin (BSA) and poly(ethylene glycol diacrylate) (PEGDA) with engineered microbes for vat photopolymerization to create objects with a wide array of 3D form factors. The BSA-PEGDA matrix afforded hydrogels that were mechanically stiff and tough for use in load-bearing applications. Second, we demonstrated the continuous in situ production of L-DOPA, naringenin, and betaxanthins from the engineered cells encapsulated within the BSA-PEGDA matrix. These microbial metabolites bioaugmented the properties of the BSA-PEGDA matrix by enhancing the stiffness (L-DOPA) or resistance to enzymatic degradation (betaxanthin). Finally, we demonstrated the assembly of the 3D printed ELM components into mechanically functional bolts and gears to showcase the potential to create functional ELMs for synthetic living machines.
Keywords: Engineered living materials, Additive Manufacturing, Bioaugmentation, Hydrogels, Microorganisms
Graphical abstract
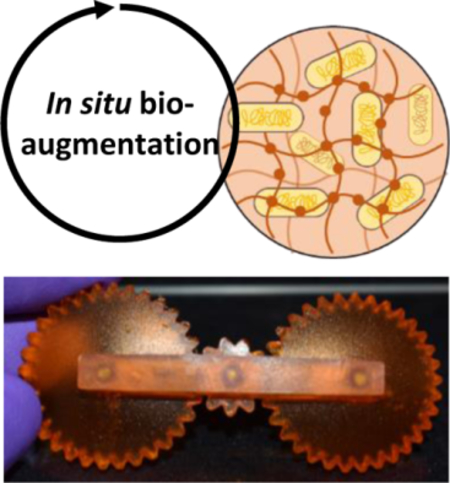
The additive manufacturing of sustainable engineered living materials (ELM) with arbitrary 3D form factors is further enhanced via bioaugmentation and affords operational machines: engineered cells produce chemical agents in situ to introduce a new function or capability to the material, and also contribute to material properties in manner that is complementary to the existing polymer matrix.
1. Introduction
The convergence of synthetic biology and polymer science has led to the rapid emergence of engineered living materials (ELMs), which are biocomposite materials comprised of engineered cells embedded within polymeric matrices. ELMs have the potential to attain a level of precision, control, responsiveness, and chemical recyclability unachievable with traditional, abiotic materials.[1–6] The synergistic effect of coupling living cells with abiotic material components enables bio-augmentation of material properties and function.[1,3] Bio-augmentation occurs when in-situ production of chemical agents introduces a new function or capability to the material, and/or contribute to material properties in manner that is complementary to the existing polymer matrix. In both cases, the polymeric material does not serve simply as a passive containment matrix for cells, but rather a chemical canvas upon which synergy between the cells and polymer matrix can be exerted. While the minimum requirements for fully living, bio-augmented ELMs are cell viability and metabolic activity, the implementation of ELMs at their fullest potential requires advanced manufacturing processes to fabricate desired three-dimensional (3D) form factors, in addition to establishing a synergy between the embedded cells and polymer matrix for material performance.
Hydrogel ELMs have been successfully fabricated via advanced manufacturing processes such as 3D-bioprinting.[1,2] Specifically, direct-write extrusion 3D-printing has been successful in the fabrication of sensors, bioreactors, actuators, drug screening kits and microfluidic devices.[3–9] However, this method of 3D-printing is limited in the complexity of the geometrical form factors that can be produced. Alternatively, vat photopolymerization is an additive manufacturing technique that allows for a greater range of geometrical designs that can vary in shape and connectivity.[10,11,12] In vat photopolymerization processes, objects are produced from a liquid resin in an additive fashion using visible light in a pattern-wise manner. Importantly, liquid resins must have both low viscosity (<10 Pa⸱s) and fast photocuring (at 405 nm) to achieve successful 3D-printing with high resolution.[13,14,15] Vat photopolymerization resins that can further incorporate and sustain microbial activity could provide an opportunity to create ELMs with greater geometrical complexity.
In this work, we posit that bovine serum albumin (BSA) is an ideal component for developing ELM resins for vat photopolymerization 3D printing as it affords mechanically robust hydrogels that exhibit high moduli and toughness (compressive modulus of 6.3 MPa,[13] which is comparable to articular cartilage[16]). These protein-based materials are also enzymatically degradable, which creates a route to a closed-loop life cycle for these materials. Recently, two different strategies have been developed to synthesize BSA-based resins for vat photopolymerization. These strategies include functionalization of the BSA surface lysines with methacrylic anhydride[13] or the conjugation of BSA with polyethylene glycol diacrylate (BSA-PEGDA).[14]
While these BSA-PEGDA materials can be synthetically manipulated to alter physical properties, we hypothesized that engineered microbes could be incorporated into these protein-based matrices to produce bio-augmented ELMs endowed with new functionality that is not inherent to the protein-polymer matrix or exceed the traits of BSA-PEGDA matrix alone. Previous studies have used microbial metabolites as the product of ELM bioreactors or to improve the mechanical properties of ELMs with the production of microbial-biopolymers such as genetically engineered protein nanofibers,[1] bacterial cellulose,[17,18] microbial pellet,[19] bacterial biofilm.[20] On the other hand, there are still opportunities to select metabolites that can specifically interact with the surrounding polymer matrix to affect the mechanical properties.
Herein, we demonstrate the additive manufacturing of ELM hydrogels wherein bioproduction of metabolites from the encapsulated cells bio-augment the innate properties of the surrounding BSA-PEGDA matrix (Fig. 1). Vat photopolymerization was used to fabricate ELM hydrogels that were mechanically stiff and a range of object geometries. We engineered E. coli for the in-situ production of L-DOPA to enhance the mechanical stiffness of an ELM (ELM-EC-LDOPA); naringenin production was used to demonstrate an ELM (ELM-EC-NGN) for continuous production of a therapeutic compound (as a comparative example of bio-augmentation); and S. cerevisiae was engineered to produce betaxanthins to afford an ELM (ELM-SC-BXN) that could resist microbial degradation. As a final proof of concept, we demonstrate the future potential for this field by 3D-printing robust ELM hydrogels as machine parts that can be assembled into mechanically functional ELM devices.
Figure 1.
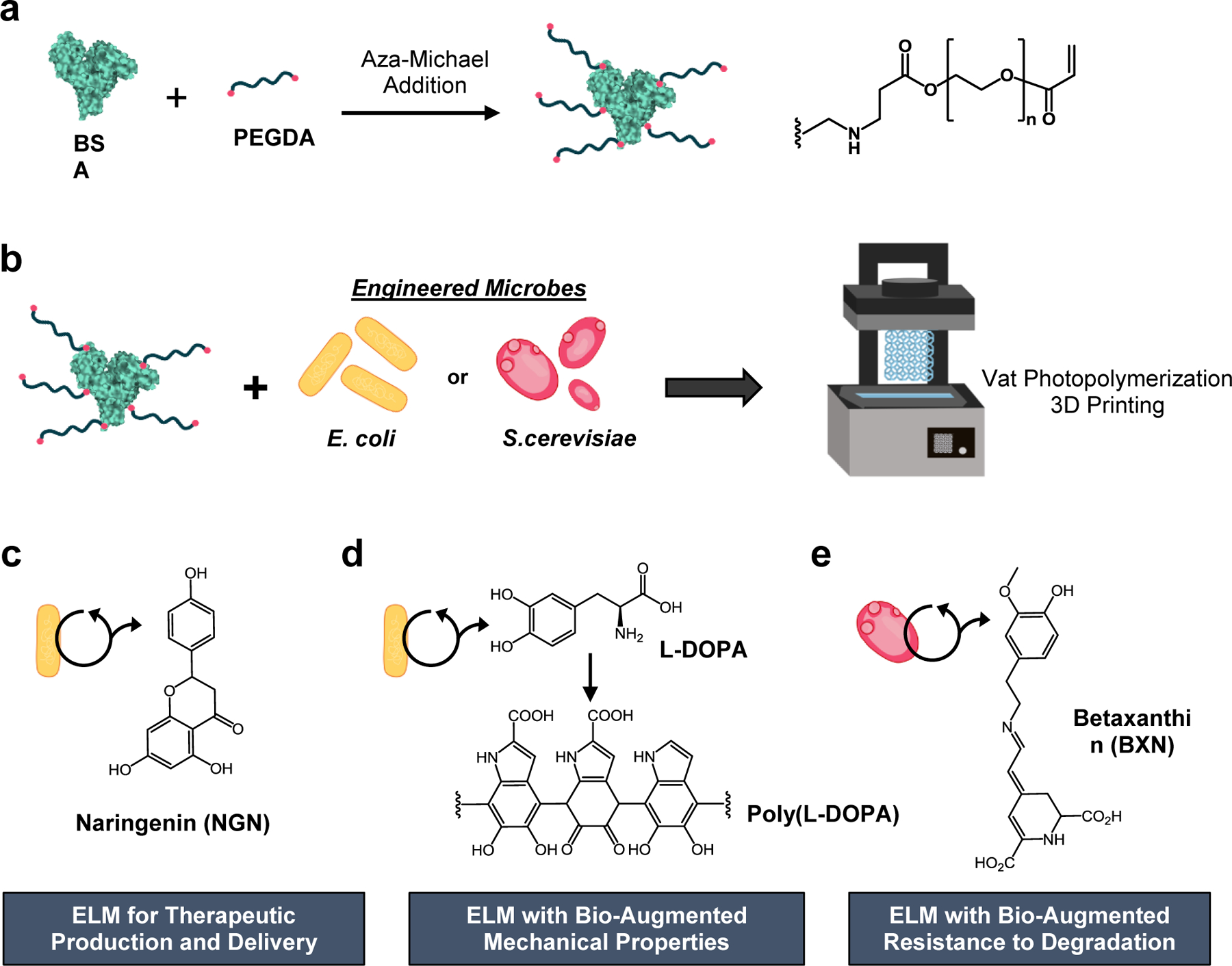
Additive manufacturing of ELMs with bio-augmented functionality and mechanical properties. a) BSA was functionalized via aza-Michael addition to afford pendant PEG-acrylate groups. b) Engineered E. coli or S. cerevisiae was added to the PEG-functionalized BSA to create an aqueous based resin for vat photopolymerization. The microbes were entrapped within the polymerized BSA-PEGDA hydrogel network. E. coli were engineered to produce c) naringenin or d) L-DOPA (the L-DOPA polymerized to afford poly(L-DOPA), and e) S. cerevisiae was engineered to produce betaxanthins. The 3D printed ELMs are referred to as ELM-EC-NGN, ELM-EC-LDOPA, or ELM-SC-BXN, respectively. (EC = E. coli, SC = S. cerevisiae, LDOPA = L-DOPA, BXN = betaxanthins, and NGN = naringenin).
2. Results and Discussion
2.1. ELM resins for vat photopolymerization 3D-printing.
ELM resins were created by adding the engineered microorganisms to an aqueous solution of BSA-PEGDA conjugates (Fig. 1). The resins in this study were optimized for their printability based on their rheometrical characterization. We first evaluated the viscosity of the resin formulations to ensure that the viscosity was <10 Pa⸱s. We, and others, have observed that this is ideal to ensure sufficient reflow during vat photopolymerization and to resolve finer features.[13,14,21] All the aqueous resins developed in this study were comprised of 30 wt% BSA with 0.075 wt% Ru(bpy)3Cl2 and 0.24 wt% sodium persulfate as the photoinitiating system. PEGDA was added to the formulation for the conjugation of PEGDA to BSA via an aza-Michael addition reaction.[14] We investigated resin formulations that contained 3, 5, or 10 wt% PEGDA to identify the minimum amount of PEGDA required for printability. All three of the resin formulations had viscosities that were <1 Pa⸱s. (Fig. 2a).
Figure 2.
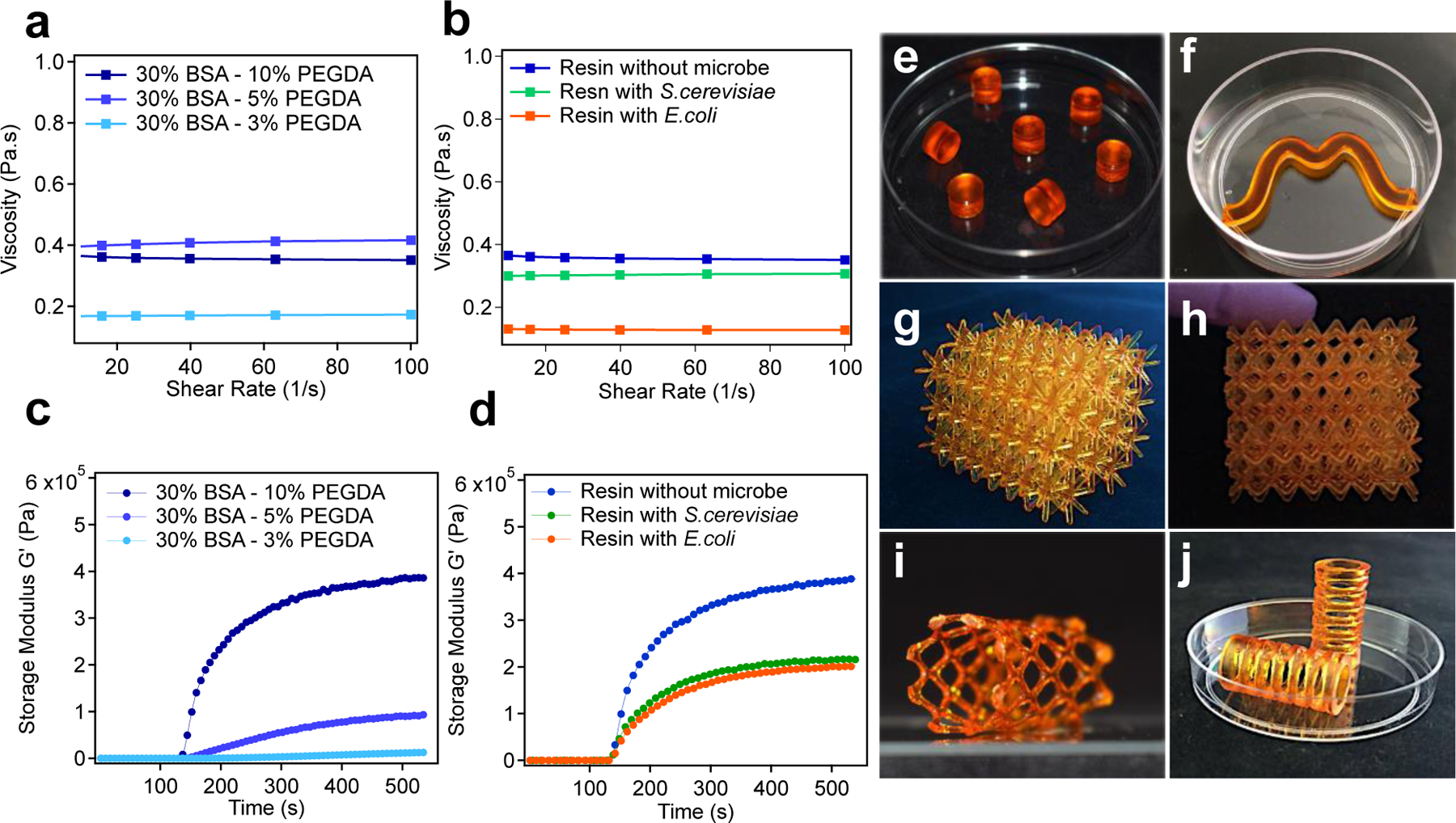
Rheological characterization and representative examples of ELMs 3D printed via vat photopolymerization. Graphs show minimal changes to the viscosity of the BSA-PEGDA solutions with a) different PEGDA concentrations, and b) in the presence and absence of microorganisms. Photorheometry experiments were performed to evaluate the change in the storage modulus (G’, Pa) over time upon irradiation with light, which was turned on after 120 s. The resulting graphs show the increase in the storage modulus for c) different PEGDA concentrations, and d) in the presence and absence of microorganisms. Optical images of 3D printed ELMs: e) 7 small cylinder-shaped constructs (in a 50 mm petri dish as a frame of reference); f) a wave-shaped construct (in a 50 mm petri dish); g) and h) lattice-shaped constructs (x=39.1 mm, y=33.6 mm, z=34.21 mm); i) stent-shape lattice (57.8 mm, y=33.4 mm, z=30.6 mm) and j) spring-shaped construct (in a 50 mm petri dish).
We next evaluated the time to reach gel-point (time at which storage modulus (G’) = loss modulus (G”)) and the rate of photocuring for each of the cell-free resins with different PEGDA concentrations in response to 405 nm light exposure. Photorheometry was used to evaluate the changes to G’ and G” over time as the sample was irradiated with light. Although all the formulations were low viscosity fluids to start, their G’ increased upon irradiation. The formulations with 3 wt% and 5 wt% PEGDA were insufficient to be used as SLA printable resins and failed to give successful prints (Supplementary Fig. 1a). The gel-point was reached after 47.5 s and 20 s for the two resins, respectively (Table S4). An increase in the PEGDA concentration to 10 wt% further decreased the time required to reach gel-point to 4 s, which is suitable for a commercial Formlabs Form 2 printer. Similarly, the 10 wt% formulation exhibited the fastest rate of photocuring (Fig. 2c, Supplementary Table 4). We found that 30 wt% BSA 10 wt% PEGDA showed the optimum concentration to formulate SLA printable resin for ELMs. To determine whether the addition of E. coli or S. cerevisiae to the resin formulations affected the viscosity and rate of photo curing, we added these cells (1 ml of cell culture, 1×109 cells/mL for S. cerevisiae and 1×1010 cells/mL for E. coli, to 20 g of resin) to the resin formulation containing 30 wt% BSA and 10 wt% PEGDA. We observed that the addition of cells did not significantly alter the rheometrical properties of the resin (Figs. 2b, 2d, Supplementary Table 4). The optimized aqueous ELM resins were 3D-printed to afford a range of cell-laden object geometries that included cylinders, lattices, and tubular stents (Figs. 2e–j). These hydrogels were mechanically stiff, and therefore, could sustain their printed shape.
2.2. Cell distribution and morphology of cells in 3D-printed ELMs
One of the lesser-studied aspects of ELMs is cell distribution and proliferation in 3D-printed networks. This distribution plays an important role in understanding the optimal geometries and dimensions of ELMs as well as the potential for homogenous, in situ production of bio-augmented compounds. Micro computed tomography (μCT) imaging showed the growth and distribution of the cells within the hydrogels in the hydrated state. After first printing, the cells were not visible in these scans owing to an initial low cell seeding density. As the cells proliferated within the hydrogels, the cell populations have been starting to image after 4 d (Fig 3a). The light-orange regions in dark orange are represents the S. cerevisiae colonies. When the samples cultured over 21 d, high cell density were observed in μCT (Fig 3b), which showed the S. cerevisiae cells were able to growth in BSA-PEGDA network, similar in time but different in spatial organization from another ELM formulation we previously studied.[22]
Figure 3.
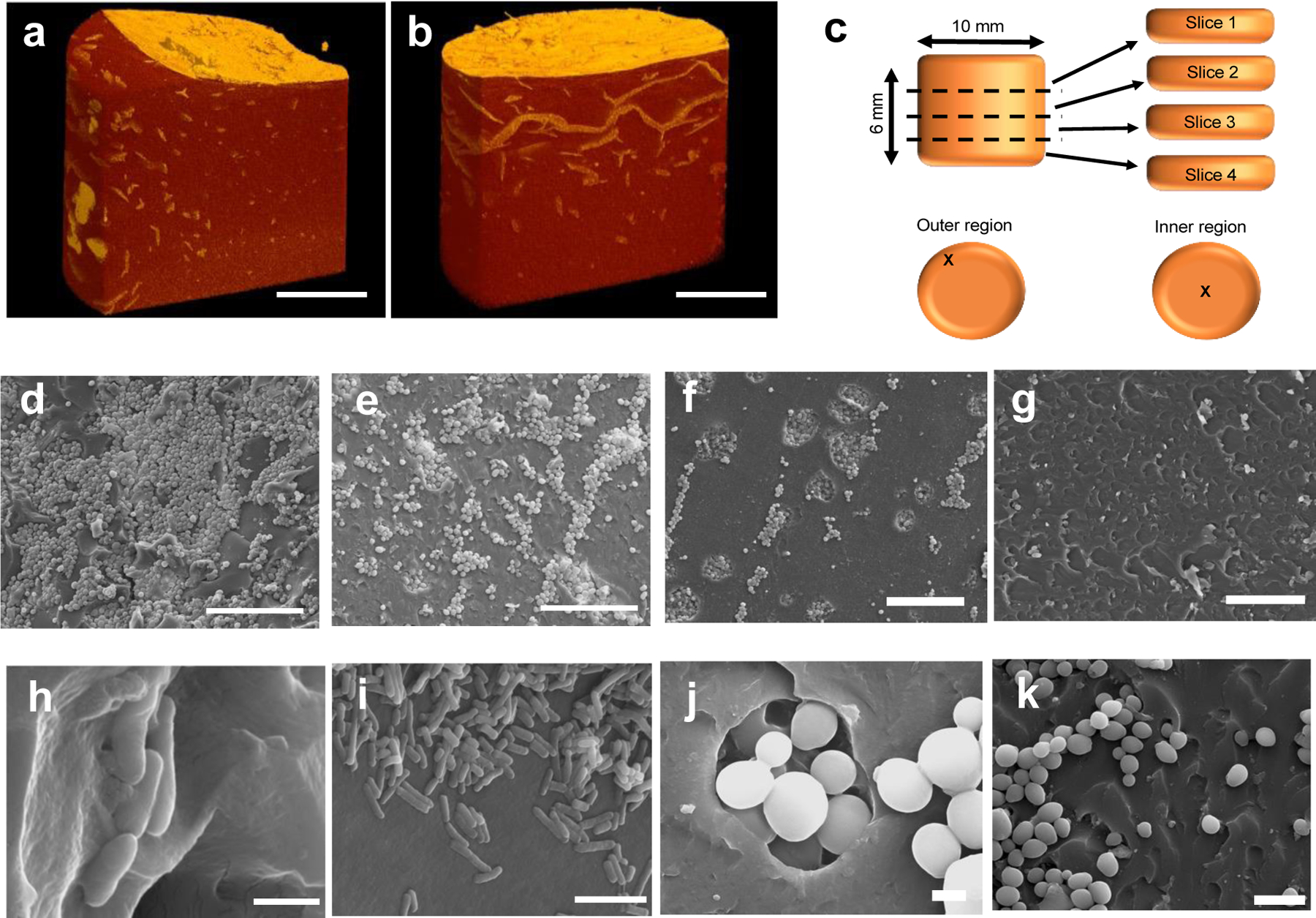
Cell growth and distribution were observed using μCT and SEM imaging of the ELMs. μCT image of S. cerevisiae in ELM-SC-BXN hydrogel after a) 4 d and b) 21 d of culturing (scale bar = 50 μm). c) Illustration showing the sample sectioning that was performed to evaluate the distribution of the cells within different regions of a 3D printed cylindrical construct. SEM images of S. cerevisiae in a 3D printed cylindrical ELM-SC-BXN construct in: d) the outer region of slice 1 at 500x magnification (scale bar = 50 μm); e) the inner region of slice 1 at 500x magnification (scale bar = 50 μm); f) the outer region of slice 2 at 500x magnification (scale bar = 50 μm); and g) the inner region of slice 2 at 500x magnification (scale bar = 50 μm). SEM images of E. coli in a 3D printed cylindrical ELM-EC-LDOPA construct at h) 20000x magnification (scale bar = 1 μm) and i, 5000x (scale bar = 5 μm). SEM images of S. cerevisiae in a 3D printed cylindrical ELM-SC-BXN construct at j) 7000x magnification (scale bar = 2 μm) and k) at 2500x magnification (scale bar = 10 μm).
Scanning electron microscopy (SEM) analysis of the dehydrated cylindrical samples were additionally used to assess the cell distribution and proliferation throughout the printed structure. Cross-sectional images were taken from different regions of a printed cylinder (Fig. 3c) of ELM-SC-BXN and ELM-EC-LDOPA after 3 d of culturing, which showed the presence of spherical-shaped yeast cells[23] and rod-shaped bacterial cells,[24] respectively (Fig. 3). The outer regions of the cylinder possessed the highest cell density (Figs. 3d–f) relative to the inner core regions (Fig. 3e–g), a phenomenon consistent with the 3D-reconstructed μCT images (Figs. 3a, b, Supplementary Videos 1 and 2). The difference in cell density between the surface and core of the printed cylinder is likely attributed to the gradient of available nutrients that may exist, with highest nutrient concentration at the outer surface in contact with the culturing media. The SEM images also proved the cell growth process in 3D printed BSA-PEGDA constructs. S. cerevisiae forms colonies process named budding.[23] In SEM images, both globose and ellipsoid in shape single S. cerevisiae cells (Figure 3j, Supplementary Figure 4b) and small, medium, and large types of S. cerevisiae buds were observed (Figure 3j, Supplementary Figure 4b). The observation of budding forms of S. cerevisiae showed that entrapped cells were able to proliferate across the BSA-PEGDA network. In SEM, the cellular surface of entrapped cells (either E. coli or S. cerevisiae) were visualized as smooth and non-damaged. Thus, it can be concluded that resin components (BSA-PEGDA conjugates, photoinitiator system), and photopolymerization of ELM resin during SLA 3D printing did not show remarkable damage to the morphology of entrapped cells.
2.3. Continuous production and effect of bio-augmentation on ELM material properties
As a first demonstration of bio-augmented manufacturing, we sought to enhance the ELM mechanical properties through incorporation of an engineered E. coli that produced L-DOPA (we will refer to this ELM as ELM-EC-LDOPA). L-DOPA can undergo oxidative coupling in the presence of oxygen to afford poly(L-DOPA)[25] (Fig. 1c) in a manner that can help cross-link and further strengthen the ELM matrix. Bio-adhesive poly(catecholamine)s including poly(dopamine), poly(norepinephrine), poly(epinephrine), and poly(L-DOPA) have been used as surface coating agents in applications such blood contact devices,[26] wound dressing gels[27] and electro-biosensors.[28] The poly(catecholamine) coatings can form conjugates with molecules containing sulfhydryl or amino groups via Michael-addition or Schiff-base reaction based on phenolic hydroxyl or quinone groups[25,26,29]. ELM-EC-LDOPA was 3D-printed, and we confirmed in situ L-DOPA production over the course of 5 d where the titer reached 199.0±3.2 mg/L (Fig. 4a). Interestingly, while ELM-EC-LDOPA initially had an orange-brown color (primarily from the ruthenium photocatalyst), the structure turned black after 5 d of culture (Fig. 4f). L-DOPA production and subsequent oxidation can form dark intermediate products such as melanin-like compounds and poly(L-DOPA) derivatives.[26] UV-vis and CD spectroscopies confirmed that L-DOPA can bind directly to BSA. We observed an increase in the strong UV-vis absorption band (280 nm) of BSA[30,31] upon the addition of L-DOPA, which was greater than the sum of the individual absorbance intensities of BSA and L-DOPA (Supplementary Fig. 5), thus indicating L-DOPA interactions with BSA. A small shift was also observed in the CD spectrum for BSA upon the addition of L-DOPA (Fig. 4c) corresponding to changes in the α-helical structure of the protein.[31,32] The obtained spectroscopic results provide evidence for L-DOPA binding with BSA.
Figure 4.
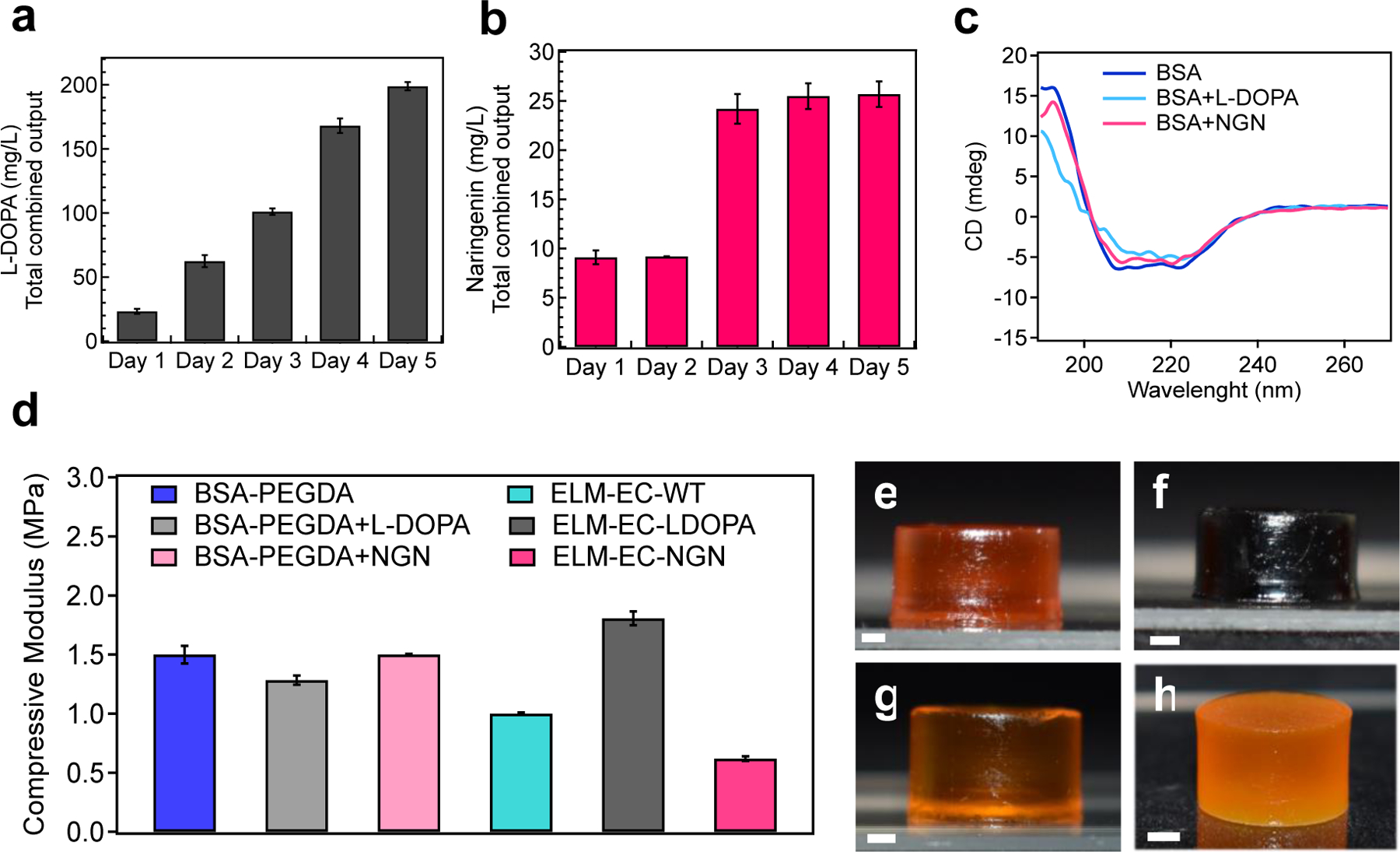
The continuous production of naringenin and L-DOPA were monitored over time and their effect on ELM moduli were characterized. a) Graph showing continuous L-DOPA production from ELM-EC-LDOPA over 5 d. b) Graph showing continuous naringenin production from ELM-EC-NGN over 5 d. c) CD spectra of BSA, BSA+L-DOPA (wherein L-DOPA was exogenously added to a solution of BSA), and BSA+NGN (wherein naringenin was exogenously added to a solution of BSA). BSA showed a negative peak that reach a maximum between 222 to 208 nm, which corresponds to α–helical structure. In the BSA+L-DOPA sample, the maximum peak was observed between 222 to 217 nm which suggests that the α–helical structure of BSA was altered by the presence of L-DOPA, but not naringenin. d) Uniaxial compression experiments were performed to obtain the compressive moduli for BSA-PEGDA, BSA-PEGDA+L-DOPA, BSA-PEGDA+NGN, ELM-EC-WT, ELM-EC-LDOPA and ELM-EC-NGN.. Optical images of 3D printed cylindrical constructs after 5 d of culturing: e) BSA-PEGDA; f) ELM-EC-LDOPA; g) ELM-EC-NGN; h) ELM-SC-BXN. Statistical analysis: one-way ANOVA followed by Welch Test. Each data point represents the mean (n = 3) and error bars represent standard deviation.
Uniaxial compression experiments were used to evaluate the mechanical properties of ELM-EC-LDOPA and showed that the bio-augmented ELM enabled by production of L-DOPA afforded enhanced stiffness. Specifically, the compressive modulus of BSA-PEGDA network (without cells) increased from 1.5 ± 0.08 MPa to 1.8 ± 0.06 MPa (p = 0.017) with the in-situ production of L-DOPA in ELM-EC-LDOPA (Fig. 4d). This increase in stiffness was not observed in any other instantiation of materials or ELMs that were often marked by decreases in stiffness when cells were present. For example, ELMs that were produced from wild type E. coli (ELM-EC-WT) without the production of L-DOPA, exhibited a decrease in elastic modulus of the BSA-PEGDA network (compared to the condition without cells) from 1.5 ± 0.08 MPa to 1.0 ± 0.01 MPa (p = 0.013) after 5 d of culturing (Fig. 4d). As another comparison, BSA-PEGDA (without cells) was exposed to L-DOPA exogenously by soaking the printed construct in a solution of L-DOPA (200 mg/L) for 5 d. The elastic modulus of these samples was 1.3 ± 0.04 MPa, which was lower than that of the ELMs with in-situ L-DOPA production (Fig. 4d). The difference (p = 0.0015) is attributed to the difference in the distribution of L-DOPA throughout the material. With the exogenous introduction of L-DOPA to printed samples, incorporation of L-DOPA can be limited by its diffusion into the matrix and likely more pronounced in the outer surface. In contrast, in-situ production offers the advantage of localized production and distribution of L-DOPA for enhanced mechanical properties via bio-augmentation.
To show the specificity of the effect of L-DOPA-BSA interaction on mechanical enhancement, we tested the mechanical properties of ELM-EC-NGN. First, we evaluated the metabolic activity of an ELM to produce the flavonoid structure therapeutic, naringenin.[33,34,35] In-situ naringenin production was confirmed by LC-MS (Fig. 4b) over 5 d. The naringenin titer was 9.1 ± 0.7 mg/L after 1 d of culturing in media and reached 25.7 ± 4.9 mg/L after 5 d. The instability of naringenin in liquid culture media may affect the titered amount in LCMS. The binding of naringenin to BSA was confirmed by UV-vis and CD spectroscopy. Although naringenin can interact with albumin protein[36,37] (Supplementary Fig. 6), this interaction did not cause conformational changes on BSA. The CD experiments confirmed that naringenin did not change the secondary structure of BSA, as both BSA and BSA+NGN samples showed the same CD spectrum (Fig. 4c). As expected, the in-situ production of naringenin did not increase the compressive modulus of the ELMs (Fig. 4d). This trend indicates that ELM bio-augmentation is strongly dependent on the choice of bioactive compound produced and its interaction with the surrounding polymer matrix.
As a second demonstration of bio-augmented manufacturing of ELMs, we sought to control degradation through a protective metabolite production. Microorganisms will exogenously secrete enzymes to degrade or remodel an extracellular matrix,[38] especially protein derived materials. Interestingly, we discovered that ELMs that produced betaxanthins from engineered S. cerevisiae (ELM-SC-BXN) resisted microbial degradation compared to ELMs that contained wild-type S. cerevisiae (ELM-SC-WT) (Fig. 5). CD spectroscopy of BSA in the presence of betaxanthins shows a significant change in the secondary structure of the protein (unlike other molecules such as L-DOPA and naringenin). This difference suggests that betaxanthins is specifically binding to the BSA and disrupting its folded conformation (Fig. 5b). Binding of betaxanthins to BSA also confirmed by UV-vis data (Supplementary Fig. 7). After a few days of culturing, there was a noticeable difference between the stiffness (p = 0.073) of ELM-SC-BXN and ELM-SC-WT, wherein the betaxanthin-producing ELM had a compressive modulus that was 2-fold higher than the ELM with wild-type cells (Fig. 5c). On the other hand, we did not find any significant difference (p = 0.07) between BSA-PEGDA and ELM-SC-BXN in terms of their elastic modulus (Figure 5 c). Therefore, we assumed that together with binding to BSA, betaxanthins may also bind to microbial enzyme as well which can contribute the resistance to microbial degradation in ELM-SC-BXN samples. Proteinase-K was selected as a model enzyme to show binding of betaxanthins to microbial enzymes which are responsible to protein degradation. Both UV-vis and CD spectroscopy data showed that betaxanthin interacted with Proteinase-K (Supplementary Fig. 8) and change the secondary structure of it (Supplementary Fig. 9).
Figure 5.
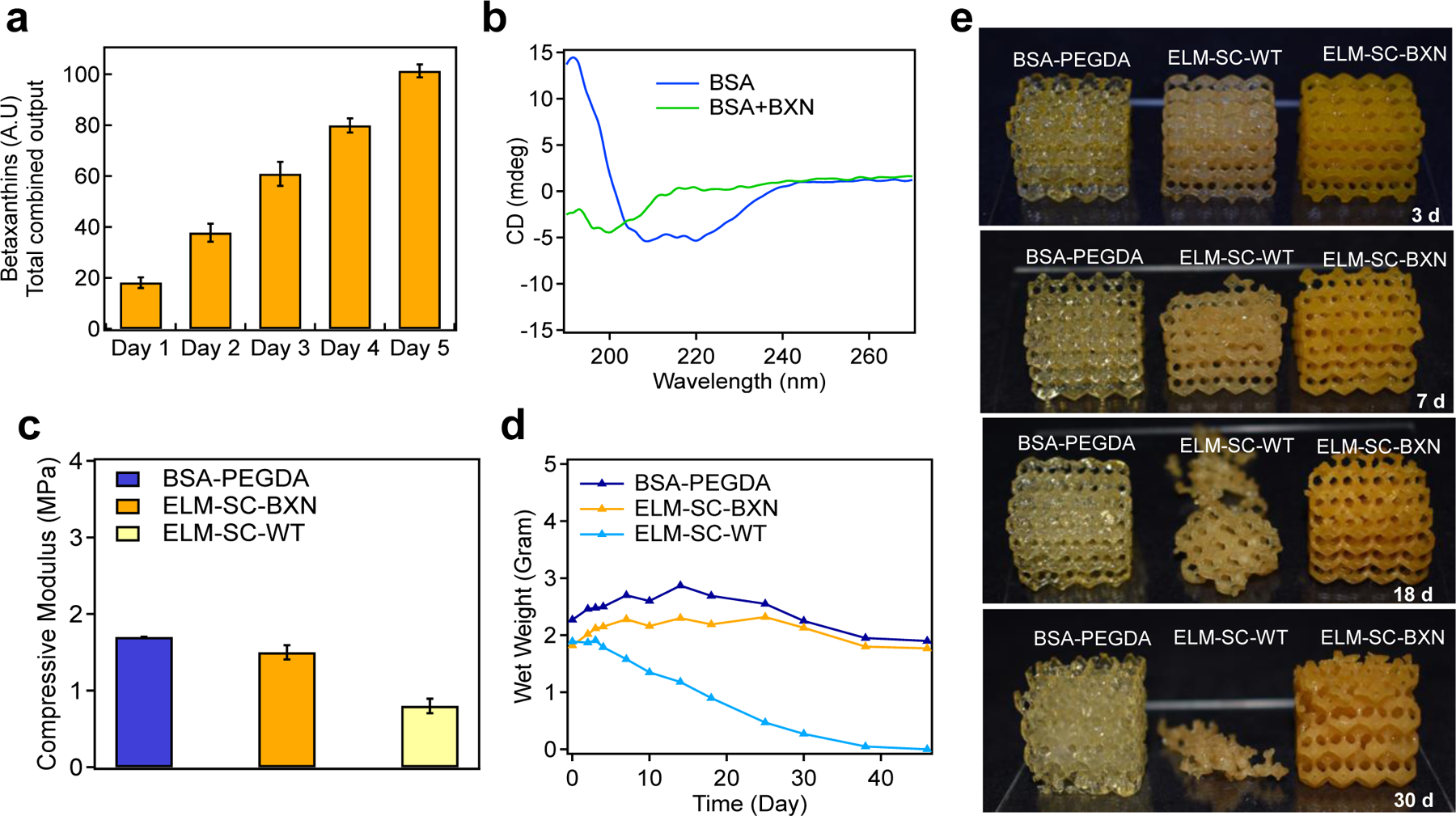
The effect of continuous betaxanthins production on microbial degradation of ELM constructs. a) Continuous betaxanthin production from ELM-SC-BXN over 5 d. b) CD spectra of BSA and BSA+BXN (wherein betaxanthin was exogenously added to a solution of BSA). c) Uniaxial compression experiments were performed to obtain the moduli for BSA-PEGDA, ELM-SC-BXN and ELM-SC-WT. d) Mass change of BSA-PEGDA, ELM-SC-BXN and ELM-SC-WT during degradation and e) optical images of BSA-PEGDA, ELM-SC-BXN and ELM-SC-WT over 30 d degradation period. Statistical analysis: one-way ANOVA followed by Welch Test. Each data point represents the mean (n = 3) and error bars represent standard deviation.
We compared the degradation of lattices printed from BSA-PEGDA, ELM-SC-WT, and ELM-SC-BXN over 45 d culture period (Figs. 5d–e, Supplementary Fig. 10). ELM-SC-WT showed an onset of mass loss after 3 d, which continued until complete microbial degradation after 38 d. In contrast, ELM-SC-BXN resisted microbial degradation (its performance was similar to BSA-PEGDA (without cells) control sample) and only exhibited a small degree of mass loss over the same period of time. At the end of the degradation period, the viability of ELM-SC-BXN samples was confirmed in agar plating experiments wherein viable cells were successfully cultured (Supplementary Fig. 11). Notably, ELM-SC-BXN remained intact throughout the culturing period, demonstrating potential for bio-augmented ELMs to be utilized for sustained microbe delivery and bioproduction applications. Thus, our findings demonstrate that in-situ betaxanthins production can prevent the microbial degradation. Betaxanthins can accomplish this by binding to degradation enzymes and showing enzyme inhibitor effects, or by binding to the BSA molecule and making it resistant to degradation by enzymes.
2.4. Mechanically functional robust ELM hydrogel constructs
The additive manufacturing of mechanically robust ELMs can enable the future fabrication of soft “living machines” that can be assembled from the printed parts (Fig. 6). A nut and bolt were individually printed using the ELM-EC-LDOPA resin. These two hydrogel components were mechanically stiff, and the nut was successfully screwed onto the bolt (Figs. 6a–b, Supplementary Video 3).
Figure 6.
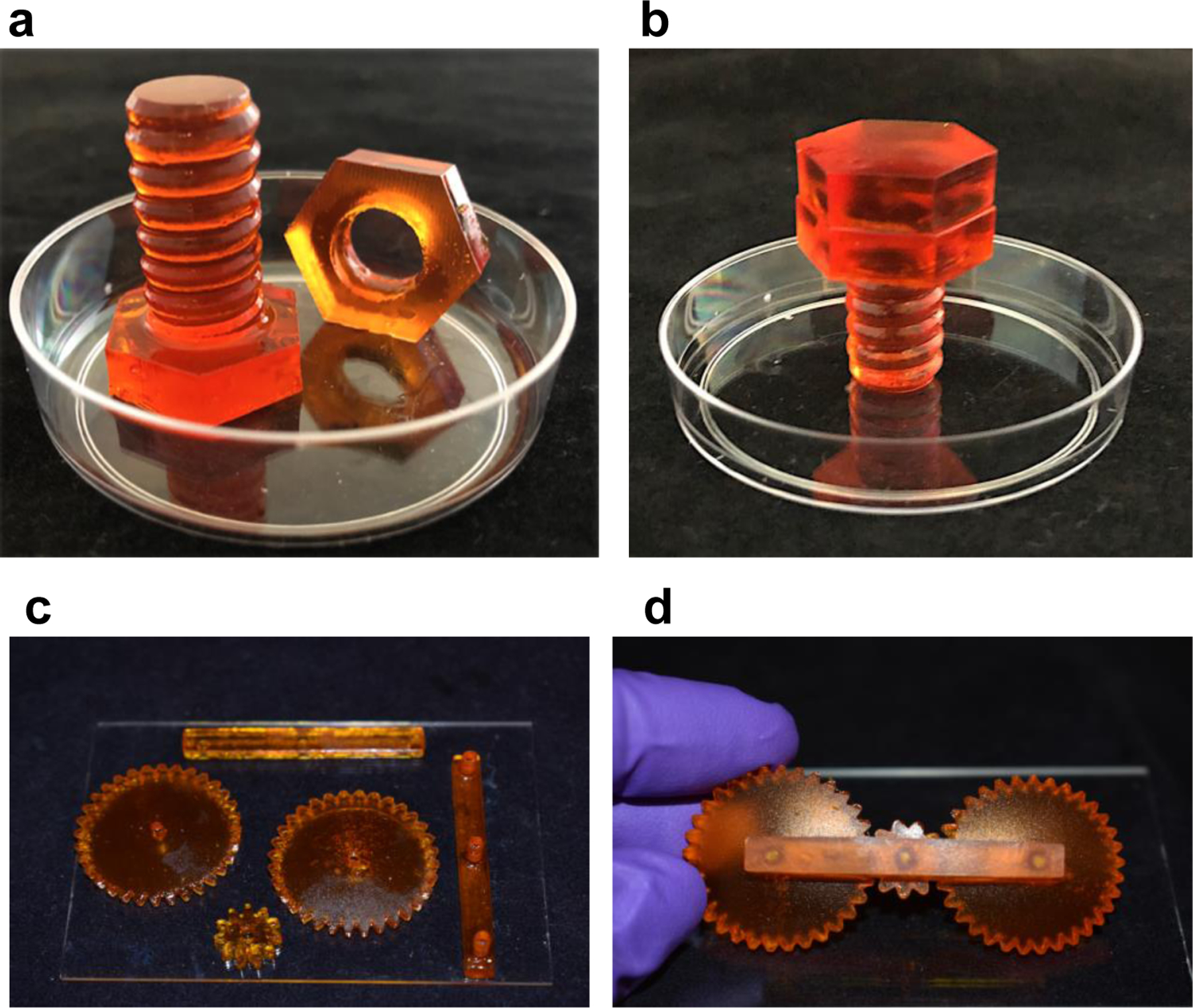
Mechanically functional ELM systems. Optical images of a) 3D printed ELM-EC-LDOPA nut and bolt (in a petri dish 50 mm diameter for reference), and b) after their assembly. c) Optical image of 3D printed ELM-SC-BXN gear components (placed on a glass slide with 114 × 159 mm). d) Optical image of the assembled 3D printed ELM-SC-BXN gear system.
As another demonstration, a set of three gears were printed from the ELM-SC-BXN resin and assembled (Figs. 6c–d). As expected for this device, the rotational motion of one gear was translated to adjacent gears (Supplementary Video 4). Our demonstrations show the potential for additive manufacturing as an advanced form of fabrication that will enable future users to print fully functional machines with arbitrary designs using ELMs and benefiting from the bio-augmented function of the cells within them.
3. Conclusion
The burgeoning field of ELMs is rapidly expanding the scope of synergistic, bio-augmented manufacturing to yield novel materials deployable in diverse settings ranging from on-demand production for natural disaster relief to in-situ therapeutic delivery. Successful production and scale-up of these materials requires an understanding of design principles for formulation and production of mechanically robust, bio-augmented systems. To this end, we fabricated ELMs via the additive manufacturing of photocurable BSA-PEGDA conjugates in the presence of engineered microorganisms. 3D-printed ELM constructs with complex geometries such as lattices, stents, bolts, and gears were successfully produced. Furthermore, μCT and SEM images indicated that the cells were viable within the BSA-PEGDA matrices. Despite the heterogeneous distribution of the cells within the cultured constructs, the presence of these cells introduced bio-augmented material functionality and properties. In particular, the production of L-DOPA and betaxanthin demonstrated the important role that those chemical agents can have upon the properties of ELM. Additional studies are underway to fully understand the factors that control cellular and chemical agent distribution within 3D-printed ELMs.
4. Experimental Section
Materials.
Bovine serum albumin was purchased from Nova Biologics, poly(ethylene glycol diacrylate), sodium persulfate, glutaraldehyde, naringenin, L-DOPA, spectinomycin, kanamycin, chloramphenicol, and ampicillin were obtained from Sigma-Aldrich. Tris(2,2′-bipyridyl)ruthenium(II) Chloride Hexahydrate, LB broth powder, LB agar powder, yeast extract, peptone, and glucose were purchased from Thermo Fisher Scientific.
Preparation of the resin formulation.
Bovine serum albumin (BSA) and poly (ethylene glycol) diacrylate (PEGDA, Mw = 700) were used to synthesize PEGDA modified BSA conjugates. To optimize the PEGDA ratio of BSA-PEGDA conjugates, 3 wt%, 5 wt % and 10 wt% of PEGDA were used against 30 wt% BSA. To synthesize the BSA-PEGDA conjugates, first PEGDA (Mw = 700) was dissolved in DI water. Then, the desired amount of BSA powder was slowly added to PEGDA solution while gently mixed and let them react overnight at 4 °C. Ru(bpy)3Cl (0.748 mg/g of resin) and sodium persulfate (2.38 mg/g of resin) were added to BSA-PEGDA formulation before SLA 3D printing. To prepare ELM resin formulations, microorganisms, 1×109 cells/mL for S. cerevisiae strains and 1×1010 cells/mL for E. coli strains, were added to resin formulation (2 wt%). Desired cell concentration was obtained by OD600 measurements.
Strains, plasmids, media.
All strains, plasmids, and primers used in this study are listed in Supplementary Table 2 and 3. NEB10β was used for gene cloning or propagation of expression vectors. It was cultivated in LB medium supplemented with appropriate antibiotics (100 μg mL−1 ampicillin, 50 μg mL−1 kanamycin, 30 μg mL−1 chloramphenicol, or 50 μg mL−1 spectinomycin (Sigma-Aldrich)) with 225 rpm orbital shaking at 37 °C.
Oligonucleotide primers that were used for PCR amplification were purchased from Integrated DNA Technologies (Coralville, IA). Ligated or Gibson-assembled DNA39 were electroporated (2 mm Electroporation Cuvettes, Bioexpress) into E. coli electrocompetent cells with a BioRad Genepulser Xcell at 2.5 kV. The Frozen EZ Yeast Transformation II Kit (Zymo Research) was used to transform integrative cassettes into the yeast. Plasmids utilized for L-DOPA and Naringenin production were constructed as previously described.[4] For Betaxanthins cassette generation, the DNA fragment amplified from plasmid pCMC0759[40] with primers P1 and P2 was cloned into the amplicon generated from the vector p415-TDH3-dCas9-VPRADH1t-UraInt[41] with primers P3 and P4 via Gibson Assembly, followed by NotI-HF digestion.
The L-DOPA generating strain was created by transforming pCDF-pLPP-B30rbshpaB-hpaC-T7t into a Tyrosine overproducing strain as previously described.[4] The Naringenin generating strain was created by transforming E. coli BL21(DE3) with plasmids pETM-PUTRtrxA-TAL-PUTRtalB-4CL, pCDM-PssrA-UTRrpsT-CHS-PUTRglpD-CHI, pACM-PfdeR-mut-FdeR-PfdeA-mut-acpH-asacpT-asacpS, and pRSM-PcspA-mut-PadR-acs-ACC as previously described.[40] To create the Betaxanthins-producing strain, the NotI-HF digested Betaxanthins cassette was gel extracted and transformed into BY4741 yeast, followed by selection of transformants on dropout media lacking leucine. Correct integrants were screened by colony PCR using primers P5 and P6 and confirmed via Sanger Sequencing. L-DOPA and naringenin can be secreted by the cells without any engineering efforts. Betaxanthins can be produced both intra and extracellularly.[42]
Culturing of metabolically engineered microorganism and ELM constructs.
S. cerevisiae BY01 and wild type S. cerevisiae BY4741 were grown in YPD (10 g L−1 yeast extract, 20 g L−1 peptone and 20 g L−1 glucose) media with 225 rpm at 30 °C. Wild type E. coli was grown in LB medium at 37 °C. 50 μg mL−1 spectinomycin and 50 μg mL−1 kanamycin was added to LB media to culture E. coli eBL0430D at 37 °C. To calculate L-dopa production from E. coli eBL0430D, antibiotic included LB media was enriched with 10 mM Vitamin C. E. coli BL21(DE3) was cultured in LB media contained 30 μg mL−1 chloramphenicol, 100 μg mL−1 ampicillin, 50 μg mL−1 spectinomycin and 50 μg mL−1 kanamycin at 30 °C. All strains were incubated in a shaking incubator at 225 rpm.
Rheology measurements.
TA Instruments Discovery Hybrid Rheometer-2 was used in rheology measurements. Viscosity versus shear rate experiments was performed at a shear rate increasing from 1 to 100 s−1. A 40 mm cone and plate geometry with a cone angle of 1.019°, a solvent trap, and a gap height of 26 μm were used during the analysis. For the photo rheology analysis, the rheometer was outfitted with a collimated light source (λ = 400 nm, 10 mW cm−2, Thorlabs) that was turned on 120 s after the start of the experiment. The storage and loss moduli were monitored for a total of 150 s at 1% strain and 6.28 rad/s by using a 20 mm parallel plate and a gap height of 1000 μm. Data collection and analysis were carried out with TRIOS software (TA Instruments).
SLA 3D Printing.
A Formlabs Form 2 printer was used to fabricate SLA 3D printed ELM constructs in the Open Mode with a layer height of 100 μm. 3D models were designed in Autodesk Fusion 360 or downloaded from Thingiverse. After 3D printing, samples were removed from the build plate, rinsed in DI water to remove any uncured resin, and post-cured in a custom photocuring chamber (Quans, 400 nm, 1 mW/ cm2) for 3 min. ELM constructs were cultured in appropriate liquid culture media and temperature according to the microbial strains present.
Micro computed tomography (μCT) imaging of ELM constructs.
ELM constructs were cultured for in YPD prior to sampling for μCT imaging in which ELMs were cultured for 4 d, and 21 d for comparison. After culturing, samples were removed from culture media and washing with Dulbecco’s phosphate-buffered saline (DPBS, 0.0009 M CaCl2, 0.0027 M KCl, 0.005 M MgCl2.6H2O, 0.1368 M NaCl, 0.0152 M Na2HPO4). Washed constructs were cut into cubical slices with side lengths ~2 mm with a razor blade. Samples were mounted and allowed to sit overnight (~12 h before imaging). Samples were scanned with a Zeiss Versa 620 Scanner with the following scan parameters: Zeiss. 20X, 70kV, 8.5 W, 2 s acquisition time, detector 6.015 mm, source −10.119 mm, XYZ [−65, −19373, 469], camera bin 2, 180+fan, 1201views, no filter, dithering. Multi reference and secondary reference with LE1 filter. Reconstructed with center shift 3.224, beam hardening 0, theta 0, byte scaling [−0.1, 0.5], binning 1, recon filter smooth (kernel size = 0.7). Scans were reconstructed by Xradia Reconstructor with 0.85 micron voxels. Images were processed using ImageJ.
Scanning Electron Microscopy (SEM) imaging of ELM constructs.
ELM constructs were cultured for 3 d prior to sampling for SEM analysis. After 3 d of culturing, constructs were removed from culture media and washed with Dulbecco’s phosphate-buffered saline (DPBS, 0.0009 M CaCl2, 0.0027 M KCl, 0.005 M MgCl2.6H2O, 0.1368 M NaCl, 0.0152 M Na2HPO4). Washed constructs were fixed with 8% glutaraldehyde (Sigma-Aldrich) solution for 48 h at room temperature. Cross-section samples were collected by cutting the samples 1.5 mm thickness after fixation. These samples were dried by stepwise ethanol dehydration with concentration range of 10, 20, 30, 40, 50, 60, 70, 80, 90 and 100 % were performed for 10 min in turn. Prior to SEM imaging, samples were coated with approximately 4 nm platinum by Leica EM ACE600 to maintain the conductivity. The Sirion XL30 scanning electron microscope was performed to image the microorganism in cross section area of ELM constructs. The SEM was operated at a high vacuum with an accelerating voltage of 15 kV.
Betaxanthins production.
Thermo Labsystems Fluoroskan Ascent FL Fluorescence Microplate Reader was used to determine the continuous betaxanthins production capacity of ELM-SC-BXN constructs. After SLA 3D printing, cylinder-shaped ELM-SC-BXN constructs (x = 10.5 mm, y = 10.5 mm, z = 7 mm) were placed in 15 mL sterile culture tubes and cultured in 3 mL YPD media (10 g L−1 yeast extract, 20 g L−1 peptone and 20 g L−1 glucose) with 225 rpm at 30 °C for 5 days. In each day, samples were collected from cultured media, centrifuged at 4400 rpm for 10 min, supernatants were collected and placed in a microplate. Fluorescence intensity of samples were measured at excitation: 485 nm / emission: 520 nm. ELM-SC-BXN constructs were washed with YPD and placed in new culture tubes and cultured in fresh YPD each day. Cylinder-shaped BSA-PEGDA constructs (x = 10.5 mm, y = 10.5 mm, z = 7 mm) were used as negative control and cylinder-shaped ELM-SC-WT constructs (x=10.5 mm, y = 10.5 mm, z = 7 mm) were used as positive control which were cultured in the same conditions for 5 d.
L-DOPA production.
Cylinder-shaped ELM-EC-LDOPA constructs (x = 10.5 mm, y = 10.5 mm, z = 7 mm) were placed in 15 mL sterile culture tubes after 3D printing. 3 mL of LB media (50 μg mL−1 spectinomycin, 50 μg mL−1 kanamycin) were added and constructs were cultured in shaking incubator with 225 rpm, at 37 °C for 5 days. To monitor the continuous production of L-DOPA, 10 mM Vitamin C was added the culture media to eliminate the oxidation of L-DOPA. In each day samples were collected from cultured media, centrifuged at 4400 rpm for 10 min, filtered with 0.2-μm nylon syringe filters (Wheaton Science) then analyzed. Continuous L-DOPA production was determined by Bruker Esquire LC-Ion Trap Mass Spectrometer with Agilent HPLC. Mass spectrometer was operated in positive mode, scan range was between 50.00 m/z to 1100.00 m/z, skim1 was 15.0 volt and capillary exit was 55.0 volt. In HPLC analysis column oven was held at 25 °C with 1 % acetic acid in water or acetonitrile as the mobile phase over the course of the 20-min sequence under the following conditions: 5–15% organic (v/v) for 5 min, 15 to 100% organic (v/v) for 8 min, 100% organic (v/v) for 2 min, 100 to 5% organic for 2 min followed by 5% organic for 3 min. The constant flow rate was set at 0.2 mL min−1. A standard curve was prepared using 98.0% purity L-DOPA (3,4-dihydroxy-L-phenylalanine) from Sigma-Aldrich. ELM-EC-LDOPA constructs were washed with LB, placed in new culture tubes, and cultured in fresh LB media (50 μg mL−1 spectinomycin, 50 μg mL−1 kanamycin, 10 mM Vitamin C) in each day. Cylinder-shaped BSA-PEGDA constructs (x=10.5 mm, y=10.5 mm, z=7 mm) were used as negative control and cylinder-shaped ELM-EC-WT constructs (x=10.5 mm, y=10.5 mm, z=7 mm) were used as positive control which were cultured in the same conditions for 5 days.
Naringenin production.
Cylinder-shaped ELM-EC-NGN constructs (x=10.5 mm, y=10.5 mm, z=7 mm) constructs were placed in 15 mL sterile culture tubes after 3D printing. 3mL of LB media (30 μg mL−1 chloramphenicol, 100 μg mL−1 ampicillin, 50 μg mL−1 spectinomycin and 50 μg mL−1 kanamycin) were added and constructs were cultured in a shaking incubator with 225 rpm, at 30 °C for 5 days. In each day samples were collected from cultured media, centrifuged at 4400 rpm for 10 min, filtered with 0.2-μm nylon syringe filters (Wheaton Science). Filtered samples were mixed with 100% ethanol (1:1 v/v) to dissolve the naringenin. Ethanol treated samples were analyzed in Bruker Esquire LC - Ion Trap Mass Spectrometer with Agilent HPLC. Mass spectrometer was operated in positive mode, scan range was between 50.00 m/z to 1100.00 m/z, skim1 was 15.0 volt and capillary exit was 55.0 volt. To identify naringenin in HPLC, same parameters were used that are described in the L-DOPA production section. A standard curve was prepared using ≥ 95% purity naringenin ( (±) - 2, 3 – Dihydro - 5, 7 – dihydroxy – 2 - (4 - hydroxyphenyl) - 4H – 1 – benzopyran – 4 - one, 4′, 5, 7 -Trihydroxyflavanone) from Sigma-Aldrich. ELM-EC-NGN constructs were washed with LB, placed in new culture tubes, and cultured in fresh LB media (30 μg mL−1 chloramphenicol, 100 μg mL−1 ampicillin, 50 μg mL−1 spectinomycin and 50 μg mL−1 kanamycin) in each day. Cylinder-shaped BSA-PEGDA constructs (x = 10.5 mm, y = 10.5 mm, z = 7 mm) were used as negative control and cylinder-shaped ELM-EC-WT constructs (x = 10.5 mm, y = 10.5 mm, z = 7 mm) were used as positive control which were cultured in the same conditions for 5 days.
Degree of swelling (q) and Mechanical Characterization.
Cylindrical shaped ELM constructs (x = 10.5 mm, y = 10.5 mm, z = 7 mm) were cultured for 5 d in the appropriate liquid culture media and conditions according to the microbial strain that they have. Constructions were removed from culture media after culturing and vacuum-dried overnight. Dried constructs were rehydrated in DI water overnight. Degree of swelling was calculated according to following equation:
These cultured, dried, and rehydrated samples were subjected to the compression test. The dimension of rehydrated samples (Table S5) was used to perform a compression test. For compression test, Newton Test Machine electromechanical test frame with 1 kN load cells and crosshead rates of 1.3 mm/min was used. The compressive modulus was calculated by the slope of the stress (kPa)- strain(mm/mm) curve. We formulated three control groups to examine the effects of in-situ L-DOPA production on mechanical properties of BSA-PEGDA networks. BSA-PEGDA constructs (without cell) were used as a negative control. ELM-EC-WT was studied to understand the effect of metabolic activity of incorporated cells on the BSA-PEGDA network. Finally, we treated the BSA-PEGDA constructs with the commercially available standard of L-DOPA (positive control-2) to compare the effect of L-DOPA production within the BSA-PEGDA network with the effect of providing this compound externally. To do that, 3D printed BSA-PEGDA constructs were treated with 200 mg/ml L-DOPA solution which is the amount that was produced from ELM-EC-LDOPA constructs after 5 d culturing (Figure 3b). All control samples were printed in cylinder-shape (x = 10.5 mm, y = 10.5 mm, z = 7 mm).
Biodegradation of ELM constructs.
Biodegradation of ELM constructs were followed by measuring the wet mass over time. The structural changes of constructs were determined by taking pictures of samples over time.
UV-vis Absorption and CD-Spectroscopy.
Varian Cary 5000 UV-Vis-NIR Spectrophotometer was used to measure UV-vis spectroscopy in a quartz cuvette at room temperature. Circular dichroism (CD) spectroscopy was carried out on a Jasco 720 Circular Dichroism Spectrophotometer by using a quartz cell of path length 0.1 cm.
Statistical analyses.
To compare the statistical difference Igor Pro (Version 8.04) software was used. Differences were analyzed by One-way ANOVA, Welch Test. For all tests, a p value < 0.05 was chosen to determine significant differences. Data are shown as the mean ± standard deviation (± s.d.). The standard deviations (± s.d) of replicates were calculated in Excel.
Supplementary Material
Acknowledgements
This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs through the CDMRP under Award No. (W81XWH-21–1-0167). Research reported in this publication was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under award number R21EB031256–01A1.
Supporting Information
Supporting information is available on the Wiley Online Library or from the author.
Footnotes
Competing interests
The authors declare no competing interests.
Data availability statement
Data generated and analyzed from this study are provided as source data in this publication and Supplementary Information. Additional data are available from the corresponding authors upon request.
Literature Cited
- [1].Duraj-Thatte AM, Manjula-Basavanna A, Rutledge J, Xia J, Hassan S, Sourlis A, Rubio AG, Lesha A, Zenkl M, Kan A, Weitz DA, Zhang YS, Joshi NS, Nat. Commun 2021, 12, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Decante G, Costa JB, Silva-Correia J, Collins MN, Reis RL, Oliveira JM, Biofabrication 2021, 13, 032001. [DOI] [PubMed] [Google Scholar]
- [3].Chen B, Kang W, Sun J, Zhu R, Yu Y, Xia A, Yu M, Wang M, Han J, Chen Y, Teng L, Tian Q, Yu Y, Li G, You L, Liu Z, Dai Z, Nat. Chem. Biol 2022, 18, 289–294. [DOI] [PubMed] [Google Scholar]
- [4].Johnston TG, Yuan SF, Wagner JM, Yi X, Saha A, Smith P, Nelson A, Alper HS, Nat. Commun 2020, 11, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rivera-Tarazona LK, Campbell ZT, Ware TH, Soft Matter 2021, 17, 785–809. [DOI] [PubMed] [Google Scholar]
- [6].Liu X, Inda ME, Lai Y, Lu TK, Zhao X, Adv. Mater 2022, 2201326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jin Y, Kim J, Lee JS, Min S, Kim S, Ahn DH, Kim Y, Cho SW, Adv. Funct. Mater 2018, 28, 1801954. [Google Scholar]
- [8].Yuan S, Brooks SM, Nguyen AW, Lin W, Johnston TG, Maynard JA, Nelson A, Alper HS, Bioact. Mater 2021, 6, 2390–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhang P, Shao N, & Qin L, Adv. Mater 2021, 33, 2005944. [DOI] [PubMed] [Google Scholar]
- [10].Chartrain NA, Williams CB, Whittington AR, Acta Biomater 2018, 74, 90–111. [DOI] [PubMed] [Google Scholar]
- [11].Andreu A, Su PC, Kim JH, Ng CS, Kim S, Kim I, Lee J, Noh J, Subramanian AS, Yoon YJ, Addit. Manuf 2021, 44, 102024. [Google Scholar]
- [12].Narupai B, Nelson A, ACS Macro Lett 2020, 9, 627–638. [DOI] [PubMed] [Google Scholar]
- [13].Smith PT, Narupai B, Tsui JH, Millik SC, Shafranek RT, Kim DH, Nelson A, Biomacromolecules 2019, 21, 484–492. [DOI] [PubMed] [Google Scholar]
- [14].Sanchez-Rexach E, Smith PT, Gomez-Lopez A, Fernandez M, Cortajarena AL, Sardon H, Nelson A, ACS Appl. Mater. Interfaces 2021, 13, 19193–19199. [DOI] [PubMed] [Google Scholar]
- [15].Smith PT, Altin G, Millik SC, Narupai B, Sietz C, Park JO, Nelson A, ACS Appl. Mater. Interfaces 2022, 18, 21418–21425. [DOI] [PubMed] [Google Scholar]
- [16].Beck EC, Barragan M, Tadros MH, Gehrke SH, Detamore MS, Acta Biomater 2016, 38, 94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yu K, Spiesz EM, Balasubramanian S, Schmieden DT, Meyer AS, Aubin-Tam ME, Cell Rep. Phys. Sci 2021, 2, 100464. [Google Scholar]
- [18].Gilbert C, Tang TC, Ott W, Dorr BA, Shaw WM, Sun GL, Lu TK, Ellis T, Nat. Mater 2021, 20, 691–700. [DOI] [PubMed] [Google Scholar]
- [19].Manjula‐Basavanna A, Duraj‐Thatte AM, Joshi NS, Adv. Funct. Mater 2021, 31, 2010784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Huang J, Liu S, Zhang C, Wang X, Pu J, Ba F, Zhong C, Nat. Chem. Biol 2019, 15, 34–41. [DOI] [PubMed] [Google Scholar]
- [21].Melchels FP, Feijen J, Grijpma DW, Biomaterials 2009, 30, 3801–3809. [DOI] [PubMed] [Google Scholar]
- [22].Brooks SM, Reed KB, Yuan SF, Altin‐Yavuzarslan G, Shafranek R, Nelson A, Alper HS, Biotechnol. Bioeng 2022, 1–11. [DOI] [PubMed] [Google Scholar]
- [23].Duina AA, Miller ME, Keeney JB, Genetics 2014, 197, 33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Xue R, Liu Y, Zhang Q, Liang C, Qin H, Liu P, Wei Y, Microbiol 2016, 82, 4663–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gaeta M, Randazzo R, Villari V, Micali N, Pezzella A, Purrello R, D’Urso A, Front. Chem 2020, 8, 1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Moulay S, Polym. Rev 2014, 54, 436–513. [Google Scholar]
- [27].Michalicha A, Roguska A, Przekora A, Budzyńska B, Belcarz A, Carbohydr. Polym 2021, 272, 118485. [DOI] [PubMed] [Google Scholar]
- [28].Kajisa T, Yanagimoto Y, Saito A, Sakata T, ACS Sens 2018, 3, 476–483. [DOI] [PubMed] [Google Scholar]
- [29].Park J, Brust TF, Lee HJ, Lee SC, Watts VJ, Yeo Y, ACS Nano 2014, 8, 3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yang Q, Liang J, Han H, J. Phys. Chem. B 2009, 113, 10454–10458. [DOI] [PubMed] [Google Scholar]
- [31].Siddiqui GA, Siddiqi MK, Khan RH, Naeem A, Spectrochim. Acta-A: Mol. Biomol. Spectrosc 2018, 203, 40–47. [DOI] [PubMed] [Google Scholar]
- [32].Sun C, Yang J, Wu X, Huang X, Wang F, Liu S, Biophys. J 2005, 88, 3518–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Salehi B, Fokou PVT, Sharifi-Rad M, Zucca P, Pezzani R, Martins N, Sharifi-Rad J, Pharmaceut 2019, 12, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zeng W, Jin L, Zhang F, Zhang C, Liang W, Pharmacol. Res 2018, 135, 122–126. [DOI] [PubMed] [Google Scholar]
- [35].Hernández-Aquino E, Muriel P, World J Gastroenterol 2018, 24, 1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hu YJ, Wang Y, Ou-Yang Y, Zhou J, Liu Y, J. Lumin 2010, 130, 1394–1399. [Google Scholar]
- [37].Zhang Y, Li Y, Dong L, Li J, He W, Chen X, Hu Z, J. Mol. Struct 2008, 875, 1–8. [Google Scholar]
- [38].Karygianni L, Ren Z, Koo H, Thurnheer T, Trends Microbiol 2020, 28, 668–681. [DOI] [PubMed] [Google Scholar]
- [39].Gibson DG, Young L, Venter JC, Hutchison CA, Smith HO, Nat. Methods 2009, 6, 343. [DOI] [PubMed] [Google Scholar]
- [40].Zhou S, Yuan S, Nair PH, Alper HS, Deng Y, Zhou J, Metab. Eng 2021, 67, 41–52. [DOI] [PubMed] [Google Scholar]
- [41].Morse NJ, Wagner JM, Reed KB, Gopal MR, Lauffer LH, Alper HS, ACS Synth. Biol 2018, 7, 1075–1084. [DOI] [PubMed] [Google Scholar]
- [42].Yuan SF, Nair PH, Borbon D, Coleman SM, Fan PH, Lin WL, & Alper HS Metab. Eng 2022, 74, 24–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated and analyzed from this study are provided as source data in this publication and Supplementary Information. Additional data are available from the corresponding authors upon request.


