Abstract
Ecotropic viral integration site 1 (EVI1), encoded at the MECOM locus, is an oncogenic zinc finger transcription factor with diverse roles in normal and malignant cells, most extensively studied in the context of hematopoiesis. EVI1 interacts with other transcription factors in a context-dependent manner and regulates transcription and chromatin remodeling, thereby influencing the proliferation, differentiation, and survival of cells. Interestingly, it can act both as a transcriptional activator as well as a transcriptional repressor. EVI1 is expressed, and fulfills important functions, during the development of different tissues, including the nervous system and hematopoiesis, demonstrating a rigid spatial and temporal expression pattern. However, EVI1 is regularly overexpressed in a variety of cancer entities, including epithelial cancers such as ovarian and pancreatic cancer, as well as in hematologic malignancies like myeloid leukemias. Importantly, EVI1 overexpression is generally associated with a very poor clinical outcome and therapy-resistance. Thus, EVI1 is an interesting candidate to study to improve the prognosis and treatment of high-risk patients with “EVI1high” hematopoietic malignancies.
EVI1 PROTEIN IS ENCODED AT THE MECOM LOCUS
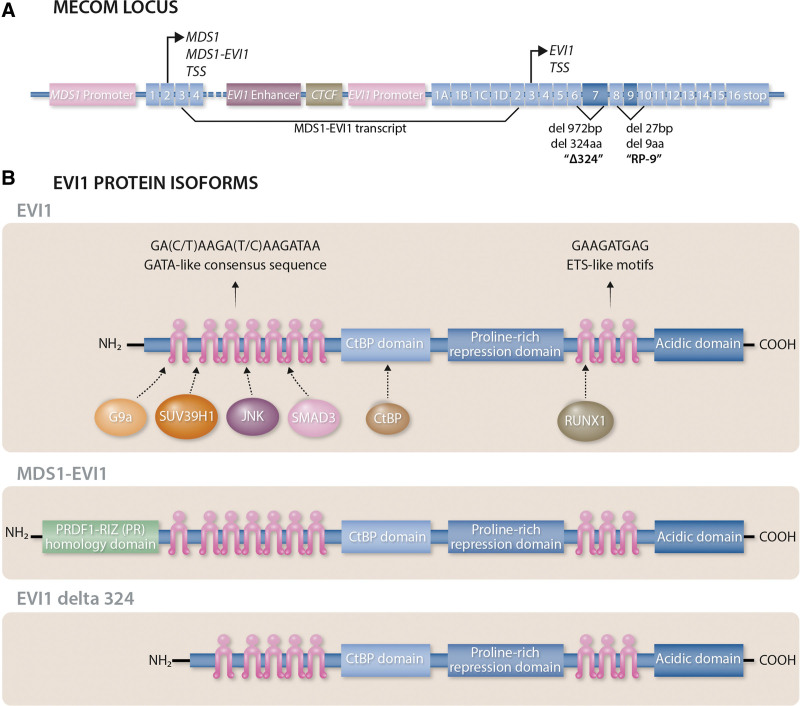
The proto-oncogene ecotropic viral integration site 1 (EVI1) is a zinc finger transcription factor with chromatin remodeling activity that is encoded at the MDS1 and EVI1 complex locus (MECOM), located at chromosomal band 3q26.2.1,2 The MECOM locus encodes several transcripts and protein isoforms (Figure 1). The divergent transcripts are generated as a combined effect of differential splicing and the presence of 2 distinct transcription start sites (TSS) located either upstream of the MDS1 gene or upstream of the EVI1 gene (Figure 1A). The MDS1 TSS generates 2 major protein isoforms: the MDS1 protein and the MDS1-EVI1 protein, while the EVI1 TSS generates transcripts that only encode EVI1. The MDS1-EVI1 protein is generated as a consequence of alternative splicing of exon 3 of MDS1 to exon 2 of EVI1 (Figure 1A), resulting in the N-terminal addition of a PRDF1-RIZ (PR) homology domain to the entire coding region of the EVI1 protein (Figure 1B). The EVI1 protein has 2 distinct zinc finger domains that mediate DNA binding: ZF1 at the N-terminus of the protein with 7 zinc finger repeats that binds to GATA-like consensus sequences1; and the more distal ZF2 with 3 more zinc finger repeats that binds to ETS-like motifs3 (Figure 1B). Alternative splicing of EVI1 includes an exon skipping event of the 27bp long exon 9, resulting in the so-called RP-9 isoform, and a deletion of 972bp within exon 7 of EVI1, resulting in the so-called EVI1 delta 324 isoform (Figure 1A), lacking 324 amino acids and thus zinc finger motifs 6 and 7 within ZF1 (Figure 1B). These different protein isoforms, encoded from the different TSS of the MECOM locus, possess alternative functional characteristics that apparently directly oppose each other in some contexts and are also independently expressed and (de-)regulated, both in the setting of normal hematopoiesis as well as malignancies.4–6 These differences can at least in part be explained by the ability of MDS1-EVI1 and EVI1 to engage different interaction partners.7 While an oncogenic role for the MDS1 and MDS1-EVI1 protein isoforms has not been described, the EVI1 protein isoforms are associated with hematologic8 as well as solid cancers.7 Furthermore, the EVI1 protein is also regulated at the post-translational level, for example via (de-)phosphorylation through casein kinase II and protein phosphatase-1α.9 While phosphorylation at the serine residue 436 (S436) has been shown to impact the association of EVI1 with protein partners,10 phosphorylation at S538 and S858 was shown to influence DNA binding via ZF2.9
Figure 1.
The MECOM locus and EVI1 protein isoforms. (A) The MECOM locus: The MDS1 and EVI1 complex locus (MECOM) encodes several transcripts and protein isoforms through differential splicing and the presence of 2 distinct transcription start sites (TSS), each with their own promoter region, located either upstream of the MDS1 cDNA or upstream of the EVI1 cDNA. The MDS1 TSS generates the MDS1 protein and the MDS1-EVI1 protein via alternative splicing of exon 3 of MDS1 to exon 2 of EVI1. Alternative splicing events of EVI1 include a deletion of 972bp within exon 7, resulting in the so-called EVI1 delta 324 isoform, as well as an exon skipping event of the 27bp long exon 9, resulting in the so-called RP-9 isoform. (B) EVI1 protein isoforms: The EVI1 protein has 2 distinct zinc finger domains. ZF1, containing 7 zinc finger repeats, is located at the N-terminus of the protein and binds to GATA-like consensus sequences. The more distal ZF2, containing 3 additional zinc finger repeats, binds to ETS-like motifs. EVI1 can interact with the histone methyltransferases G9a and SUV39H1 via ZF1, as well as the protein kinase JNK and the transcription factor SMAD3. EVI1 additionally possesses domains to directly interact with the transcriptional corepressor CtBP and the transcription factor RUNX1. Alternative splicing of exon 3 of MDS1 to exon 2 of EVI1 results in the N-terminal addition of the PRDF1-RIZ (PR) homology domain in the MDS1-EVI1 protein. The deletion of 972 bp within exon 7 of EVI1 leads to a deletion of 324 amino acids and the so-called EVI1 delta 324 protein isoform lacking zinc finger motifs 6 and 7 within ZF1.
EVI1 IN NORMAL HEMATOPOIESIS
EVI1 plays an essential role during embryonic development and is important for hematopoiesis, angiogenesis, and the development of the heart and nervous system.11–13 Homozygous knockout of EVI1 is embryonic lethal in mice at day E10.5, with knockout embryos failing to develop and expand definitive HSCs.13 A mouse model incorporating a GFP knock-in into the endogenous Evi1 locus revealed that, in normal hematopoiesis, Evi1 expression is tightly restricted to the hematopoietic stem cell (HSC) compartment, being highest in the most primitive long-term HSCs (LT-HSCs) and strictly downregulated before the generation of lineage-restricted progenitors.14 Interestingly, it has recently been shown that during hemogenic endothelial to hematopoietic transition in the fetal liver during embryonic development, it is the EVI1high cells that give rise to HSCs.15 Thus, this HSC-specific expression pattern also holds true in the setting of developmental hematopoiesis, meaning that EVI1 expression can be considered to act as a molecular marker of the most primitive HSCs in normal adult and embryonic hematopoiesis.16 In this context, it has been reported that Evi1 functions as a major positive regulator of HSC self-renewal divisions,14 increasing HSC proliferation and blocking myeloid differentiation.17,18 Haploinsufficiency of Evi1 leads to reduced self-renewal of LT-HSCs in mice, which correlates with the clinical observation of rare early-onset bone marrow failure syndromes associated with near-complete depletion of HSCs.19 These syndromes have been observed neonatally or early in childhood in humans who carry heterozygous loss of function mutations in the MECOM locus.20–22 In engineered models of this disease, haploinsufficiency of EVI1 results in the collapse of an HSC-specific transcriptional network consisting of hundreds of genes, suggesting that EVI1 is a master epigenetic regulator of functional identity in these cells.19
Evi1 gain of function models similarly illustrates an important role for Evi1 in HSC biology, with lentiviral overexpression of Evi1 in primary murine hematopoietic progenitor cells leading to a block in differentiation and prolonged survival of myeloblasts in long-term culture.23 However, in this study, the authors also observed a cytostatic effect caused by the inhibition of cell cycle progression, which the authors interpret as being consistent with Evi1 enforcing the important feature of quiescence upon HSCs.23 However, it must be noted that this does not appear consistent with the role EVI1 plays in driving malignant transformation. This could potentially be explained by context or expression level-dependent activities of EVI1 that are different when EVI1 is lentivirally overexpressed in vitro versus a leukemia setting in patients.
CLINICAL RELEVANCE OF EVI1 IN CANCER
As noted above, EVI1 expression is tightly restricted to the most primitive HSC compartment in normal hematopoiesis. However, approximately 10% of acute myeloid leukemia (AML) patients display abnormally high expression of the EVI1 transcript, but not the MDS1-EVI1 mRNA.24–27 Elevated EVI1 expression can be a result of 3q rearrangements, which comprise a high-risk AML entity as defined by the World Health Organization (WHO),28 but can also occur outside the context of such translocations, often in co-occurrence with mixed-lineage leukemia (MLL) rearrangements or chromosome 7 deletions.26 In Fanconi anemia patients suffering from MDS or AML, approximately every second patient shows EVI1 overexpression. Importantly, regardless of the presence of a 3q rearrangement and the exact mechanism of aberrant expression, all EVI1 overexpressing (EVI1high) leukemias are associated with a low rate of disease remission, high rates of relapse and chemotherapy resistance and very poor overall survival.26,29–31 A significant proportion of the EVI1high AML patients remains refractory, and of those who achieve complete remission,25 many relapse after a short time. Indeed, the median survival of 3q-rearranged AML patients is only 10 months.32 EVI1 is overexpressed in around 10%–28% of pediatric AML cases, where it predicts a poor prognosis and is associated with MLL rearrangements in around one-third of the cases, while 3q rearrangements are very rare.29,33
In addition to AML, EVI1 apparently plays an important causal role in the evolution of other myeloid malignancies. EVI1 is overexpressed in 10% of patients with myelodysplastic syndrome (MDS),34–36 and in 30% of patients with chronic myeloid leukemia (CML) in blast crisis.37–39 Moreover, EVI1 is elevated in around 10% of chronic phase CML patients who demonstrate therapy resistance to the thymidine kinase inhibitor imatinib, with these patients not surprisingly exhibiting a particularly poor prognosis.38 In mouse models of both chronic phase CML, as well as blast crisis, Evi1 overexpression enhanced the proliferation and leukemogenic potential of these cells and conferred resistance to tyrosine kinase inhibitors.40
While EVI1 is most frequently associated with myeloid malignancies, it is also dysregulated in some cases of adult and pediatric acute lymphoblastic leukemia (ALL) and chronic lymphocytic leukemia, most commonly in B-ALL.41 As in AML, CML, and MDS, high EVI1 expression in pediatric ALL may serve as a predictive factor for adverse clinical outcome. 3q rearrangements are rare in ALL and an association of high EVI1 expression with cytogenetic subgroups was not found, but elevated levels of EVI1 were again linked with poor prognosis in patients and enhanced cell survival, increased growth, and elevated leukemia initiation capacity in an experimental setting.41,42{Stevens, 2014 #151}
Interestingly, aberrantly high levels of EVI1 have also been observed across a range of non-hematologic malignancies including ovarian cancer,43,44 breast cancer,45 pancreatic cancer,46 colorectal cancer,47 and prostate cancer.48 While transcription factors like EVI1 might have cell type-specific targets and functions resulting from a context-dependent remodeling of the epigenetic landscape and gene expression, it would be of interest to investigate the commonalities in the mechanism of action of EVI1 across these diverse cancer entities.
DOWNSTREAM CONSEQUENCES OF EVI1 OVEREXPRESSION
Although EVI1 is overexpressed in a variety of cancers and is associated with therapy resistance and poor clinical outcome, the exact oncogenic mechanism of action downstream of EVI1 overexpression is still the subject of extensive investigation. Broadly speaking, EVI1 is canonically thought of as a classical transcription factor that can both positively and negatively modulate the expression of target genes via the recruitment of the transcriptional machinery to specific genomic loci, or by the alteration of local chromatin structure into an active or repressive state as a result of interactions with proteins with epigenetic remodeling activity. However, some literature also suggests that EVI1 may elicit effects that are not directly related to the regulation of transcription.
EVI1 as a transcriptional co-activator and co-repressor
EVI1 has been proposed to act as a transcriptional regulator of several important mediators of gene expression in hematopoiesis. For example, several groups have confirmed that EVI1 directly binds to the GATA2 promoter and regulates its expression, providing a clear link to regulation of HSC biology given the well-characterized role of GATA2 in both adult and developmental hematopoiesis at the stem cell/progenitor level (Figure 2A, left panel).13 Indeed, the proliferation of definitive HSCs in mouse embryos was heavily impaired in Evi1-/- mice, presumably due to the decrease in Gata2 expression.13 EVI1 also binds to an upstream regulatory element of Spi1, which encodes the pioneer transcription factor PU.1.49 Binding to this site, approximately 14kb upstream of Spi1, requires both zinc finger domains and increases Spi1 transcription. As a consequence, erythropoiesis and lymphopoiesis are suppressed while myelopoiesis is enhanced. Aberrant overexpression of EVI1 can thus result in aberrant myeloid expansion and, ultimately, in leukemic transformation.49 A recent study found the ETS transcription factor ERG to be a direct transcriptional target of EVI1.50 EVI1 can drive ERG expression by binding to an intragenic +85 enhancer that is conserved between humans and mice. This region is occupied by EVI1 in AML cell lines and also in primary AML patient samples, leading to aberrant ERG expression that maintains cells in an immature differentiation stage and constitutes a dependency in EVI1-driven AML.50,51 Furthermore, EVI1 can also directly bind to the promoter of the proto-oncogenic transcription factor PBX1 and activate its transcription52 (Figure 2A, left panel). In line with this data, EVI1 and PBX1 expression levels are found to positively correlate in AML patients. Knockdown of Pbx1 in Evi1-overexpressing primary murine hematopoietic stem and progenitor cells reduced the cell’s capacity to self-renew and be re-plated in a colony-forming unit assay.52 Moreover, EVI1 increases the activity of the c-fos promoter and cooperates functionally with the FOS protein to increase cell proliferation.53
Figure 2.
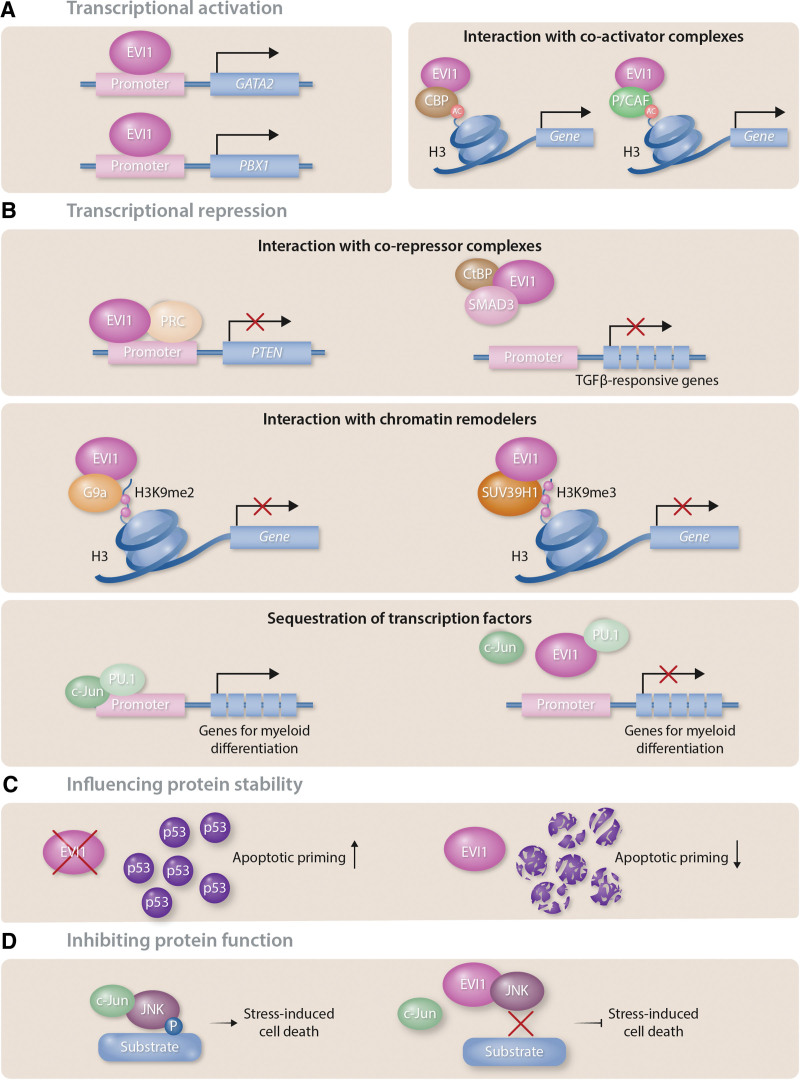
EVI1 protein functions. (A) Transcriptional activation: Left panel: EVI1 can act as a transcriptional activator, initiating transcription by directly binding to promoters such as those of GATA2 and the PBX1. Right panel: CBP and P/CAF can interact with EVI1, leading to histone acetylation. (B) Transcriptional repression: Upper panel: EVI1 can act as a transcriptional repressor via interaction with corepressor complexes. EVI1 binds both PRC1 and PRC2/3/4 and recruits them to the PTEN promoter. EVI1 also forms a co-repressor complex with CtBP and prevents SMAD3 binding to DNA. As a consequence, TGFβ-mediated transcription is inhibited. Middle panel: EVI1 interacts with chromatin remodelers such as the histone methyltransferases G9a and SUV39H1, that methylate histone H3 at lysine 9 (H3K9) resulting in transcriptional repression. Lower panel: EVI1 binds PU.1 and blocks its interaction with the coactivator c-Jun, thereby preventing PU.1 binding to promoters of genes involved in myeloid differentiation. (C) Influencing protein stability: EVI1 decreases p53 protein abundance via a proteasome-independent mechanism, resulting in decreased apoptotic priming in EVI1high cells. (D) Inhibiting protein function: EVI1 inhibits JNK substrate phosphorylation via direct association with JNK, thus blocking JNK-mediated stress-induced cell death.
In its role as both a transcriptional activator as well as a transcriptional repressor, EVI1 also interacts with a range of proteins with chromatin remodeling activities. In bone marrow hematopoietic cells, Evi1 directly binds to several members of the polycomb repressive complex (PRC), including PRC1 and PRC2/3/4, facilitating their recruitment to the promoter of the phosphatase and tensin homologue deleted on chromosome 10 (Pten) gene (Figure 2B, upper panel).54 This leads to the repression of Pten transcription and decreased Pten protein levels, which in turn results in the activation of the AKT/mammalian target of rapamycin (mTOR) signaling. Treatment of murine Evi1-driven leukemias with the mTOR inhibitor rapamycin significantly prolonged the survival of recipient mice.54 This sensitivity towards inhibition of mTOR using rapamycin might also be an attractive therapeutic strategy for human EVI1high leukemia. EVI1 also recruits other transcriptional co-repressors to chromatin. Evi1 can form a co-repressor complex with C-terminal binding protein (CtBP)55 and phosphorylation at S436 of the EVI1 protein seems to influence the association with CtBP1.10 Moreover, it was shown that Evi1 can bind directly to the MH2 domain of Smad3, an intracellular mediator of TGFβ signaling,56 via its first zinc finger domain.57 In this context, through the interaction with the EVI1/CtBP co-repressor complex, binding of the Smad3 complex to DNA is prevented (Figure 2B, upper panel). As a consequence, Evi1 inhibits TGFβ-mediated transcription resulting in a release of TGFβ-mediated growth arrest.56 Using the histone deacetylase (HDAC) inhibitor trichostatin A, the transcriptional repression of TGFβ signaling could be prevented, suggesting an involvement of HDAC in this EVI1-mediated process.55 In addition to Smad3, Evi1 has also been shown to interact with, and thereby repress the transcription of, other proteins of the Smad protein family, including Smad1 and Smad2 which are targets of bone morphogenetic protein (BMP) and activin signaling. Moreover, following stimulation with TGFβ, Evi1 binds to the promoter of Smad7 in a complex with CtBP to inhibit Smad7 transcription.57 In this context, the deletion of the CtBP1 binding site in EVI1 prevented an increase in proliferation observed in 32Dcl3 cells upon EVI1 overexpression, demonstrating the functional relevance of this interaction.58 Taken together, EVI1 mediates signaling of several TGFβ family ligands, including TGFβ, BMP, and activin, that are all implicated in embryogenesis and are often deregulated during malignant transformation, resulting in de-regulation of cellular functions including proliferation, differentiation, and cell survival.
EVI1 can also directly interact with the histone methyltransferases (HMTs) G9a and SUV39H1 (Figure 2B, middle panel).59 These HMTs methylate histone H3 at lysine 9 (H3K9) which is generally associated with the silencing of genes. Inactivation of SUV39H1 prevented the transcriptional repression of TGFβ-responsive elements by Evi1 and knockdown of either of these 2 HMTs decreased the colony-forming potential of Evi1-overexpressing hematopoietic stem and progenitor cells in a CFU assay and thus restricted their self-renewal capacity.59
Immuno-precipitation experiments have also identified that several HDACs interact with EVI1 via its proximal zinc finger domain.9 These include both class I and class II HDACs, with the strongest interaction being with HDAC1 and HDAC4.58 This adds a further class of transcriptional repressors that EVI1 can potentially recruit to its target genes.
Moreover, EVI1 forms complexes with the 2 de novo DNA methyltransferases (DNMTs) DNMT3A and DNMT3B via its proximal zinc finger domain and mediates de novo DNA methylation of target regions.60 The phosphorylation site at S436 of the EVI1 protein seems to be important for the interaction with DNMT3A.10 With regards to this interaction with DNMTs, it is interesting to note that high expression of EVI1 in AML correlates with a distinct hypermethylation signature.61 This signature is characterized by hypermethylated CpG-rich promoter regions that are enriched for EVI1 binding sites. Whether the interaction with DNMTs is also responsible for the aberrant DNA hypermethylation observed in these AML cases still needs to be determined. Nonetheless, it has been shown that one of the targets of the EVI1/DNMT3 protein complex is a regulatory region of the microRNA-124-3. The methylation of CpG dinucleotides in this regulatory region through the complex leads to silencing of the microRNA-124-3 locus which in turn results in increased self-renewal and cell cycling.
With regards to EVI1’s ability to act as an activator of gene expression, it can also associate with the 2 co-activators: cAMP-responsive element-binding protein-binding protein (CBP); and p300/CBP-associated factor (P/CAF), which are both known to exhibit acetyltransferase activity, acetylating both transcription factors and histone lysine residues58 (Figure 2A, right panel). This interaction leads to the acetylation of EVI1, the consequences of which still need to be investigated.
Interactions with other transcription factors
In addition to its association with transcriptional co-activators, co-repressors, and chromatin remodeling enzymes that cannot directly bind DNA, EVI1 forms complexes with other transcription factors, resulting in a range of functional consequences. EVI1 binds to PU.1 and therefore interferes with its function by specifically blocking its interaction with the coactivator c-Jun (Figure 2B, lower panel), preventing PU.1 binding to promoters of genes important for myeloid differentiation.62 EVI1 also binds to GATA1 via 2 zinc fingers in the proximal zinc finger domain, preventing DNA binding and repressing GATA1 function, resulting in a block in erythroid differentiation.63 The distal zinc finger domain of EVI1 is able to mediate interaction with AP1, modulating its transcriptional regulatory activity.64 Indeed, the ChIPseq experiment in human ovarian carcinoma cells revealed an approximate 25% overlap in EVI1 and AP1 binding sites, suggesting a synergistic interaction of these transcription factors. The eighth zinc finger within the distal zinc finger domain of EVI1 mediates interaction with the DNA-binding Runt domain of RUNX1, thereby reducing the binding of RUNX1 to DNA.65 Consequences of this interaction include a block in differentiation and an increase in cell death.65 Thus, overexpression of EVI1 effectively leads to a functional repression of RUNX1. One consequence of this inhibition is the upregulation of the creatine kinase CKMT1, which in turn modulates WNT and GSK3 signaling.66
Regulation of signal transduction via non-transcriptional mechanisms
EVI1 can also influence processes other than gene transcription via interaction with a range of different classes of proteins. Thus, EVI1 directly associated with the c-Jun N-terminal kinase (JNK), which plays a crucial role in signal transduction related to stress-induced cell death67 (Figure 2D). As a consequence, EVI1 interferes with the interaction of JNK with its substrates, effectively resulting in a block of UV/ TNF-α-induced JNK kinase activity and cell death.67 More recently, it has also been shown that EVI1 can modulate p53 at the protein level, by influencing its stability and thus protein abundance68 (Figure 2C), which has been linked with resistance of EVI1high AML cells to chemotherapy. Although the authors present data to suggest that this is not as a result of EVI1-mediated alterations in gene expression, the exact mechanism of action still needs to be investigated. Mass spectrometry and co-immunoprecipitation experiments have also shown association of EVI1 with several different proteins involved in DNA repair, including the DNA helicases XRCC5 and XRCC6, RAD50 that is involved in double-strand break repair and the DNA mismatch repair protein MSH2.9
In summary, the EVI1 protein is able to directly interact with a wide range of additional proteins, including transcription factors, transcriptional co-activators, transcriptional co-repressors, chromatin remodelers, and other signaling proteins such as protein kinases. These diverse interactions may in part explain the apparent “master regulator” effect that EVI1 appears to play in both normal and malignant HSC biology and how it is able to coordinately impact upon several cellular processes that are directly implicated in malignant disease, including differentiation, cell proliferation, and cell death.
MECHANISMS OF EVI1 DEREGULATION IN MALIGNANT CELLS
The downstream functional activity of EVI1 is frequently deregulated in a variety of different cancer entities and is often associated with a particularly poor prognosis. This oncogenic deregulation of EVI1 frequently relates to a simple alteration in mRNA expression which mediates elevated protein levels but can additionally be a consequence of changes in protein function.
EVI1 fusion proteins
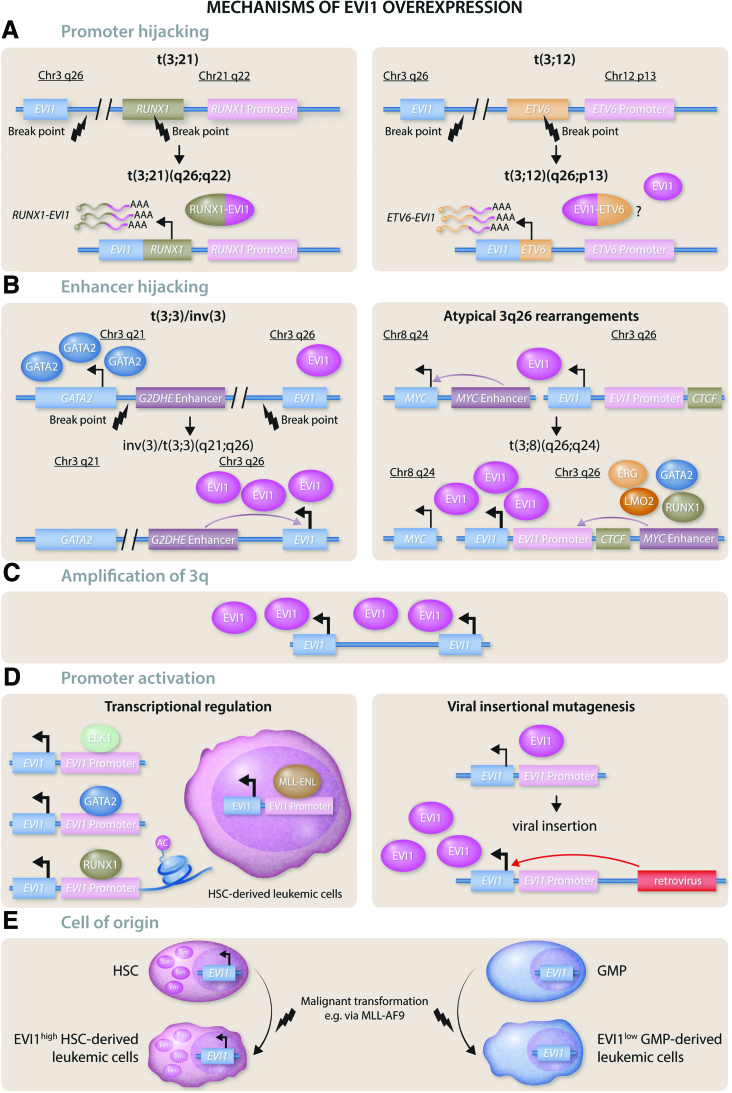
While many translocations involving the MECOM locus exert their oncogenicity via aberrant overexpression of EVI1 while leaving the protein structure intact, the translocation t(3;21)(q26;q22) results in a fusion of the EVI1 protein with another transcription factor required for normal hematopoiesis: RUNX1 (Figure 3A, left panel). This translocation results in the first 5 or 6 exons of RUNX1, including the DNA-binding RUNT domain, being fused to EVI1, while the RUNX1 transactivation domain is lost. The RUNX1-EVI1 fusion occurs as a secondary mutation in patients suffering from CML in blast crisis,69 MDS and AML,70 and is associated with a poor outcome. The prognosis is even worse than that of patients having an EVI1 mutation alone and the 5-year event-free survival rate of t(3;21) patients is only 14%.32 In a chimeric knock-in mouse model, Runx1-Evi1 expression alone was sufficient to induce a megakaryoblastic leukemia in adult mice,71 while Runx1-Evi1 expression early during hematopoietic development was embryonically lethal around day E13.5. There are several molecular consequences resulting from expression of this transcription factor fusion: transcriptional activation and repression mediated by wild-type RUNX1 and EVI1 are disrupted; and the fusion protein gains new oncogenic functions and novel transcriptional targets,72 resulting in extensive epigenetic reprogramming of hematopoietic progenitor cells undergoing lineage fate decisions.73 In the t(3;21) SKH-1 cell line, knockdown of the RUNX1-EVI1 fusion gene resulted in myeloid differentiation, indicating a role of the oncogenic protein in blocking the normal differentiation process.72 The effect of the RUNX1-EVI1 fusion was also studied in an in vitro differentiation model for hematopoietic specification using mouse embryonic stem cells. Combined analysis of transcription factor binding and gene expression in these cells revealed that the cell fate decision of multipotent hematopoietic progenitor cells was disrupted by RUNX1-EVI1, and a multi-lineage gene expression pattern was induced. Both the RUNX1- and the EVI1-mediated gene expression programs were disturbed by RUNX1-EVI1 expression. In addition to the observed changes in the differentiation and lineage specification of the cells, the fusion also led to a block in the cell cycle, decreased colony-forming potential and increased apoptosis.73 In summary, the RUNX1-EVI1 fusion both disrupts the normal RUNX1 and EVI1-mediated gene expression programs and regulates its own unique targets. This results in alterations of the cell cycle and the failure of hematopoietic progenitor cells to undergo cell fate commitment.73
Figure 3.
Mechanisms of EVI1 overexpression. (A) Promoter hijacking: EVI1 can be aberrantly expressed through the translocation of other genes’ promoters to the MECOM locus. Left panel: In t(3;21)(q26;q22), parts of the RUNX1 gene and the RUNX1 promoter are dislocated to the MECOM locus, resulting in a RUNX1-EVI1 fusion that also encodes a RUNX1-EVI1 fusion protein. Right panel: In t(3;12)(q26;p13), parts of the ETV6 gene and the ETV6 promoter are dislocated to the MECOM locus, resulting in an ETV6-EVI1 fusion. Whether this fusion also leads to the expression of an ETV6-EVI1 fusion protein is still subject of current research. (B) Enhancer hijacking: A recurrent mechanism of EVI1 overexpression is through translocation of a potent cellular enhancer to the MECOM locus. Left panel: In t(3;3)(q21;q26) and inv(3)(q21;q26), the GATA2 distal hematopoietic enhancer (G2DHE) is dislocated to the MECOM locus, resulting in increased EVI1 levels and decreased levels of GATA2, while the expression of the MDS1-EVI1 isoform is disrupted. Right panel: The translocation t(3;8)(q26;q24 is an atypical 3q26 rearrangement resulting in the relocation of a MYC “super-enhancer” to the MECOM locus, serving as a binding site for ERG, GATA2, LMO2 and RUNX1. Upstream of the EVI1 promoter there is a CTCF enhancer-docking site that mediates promoter-enhancer looping. (C) Amplification of 3q: Amplification of 3q26q29 is a frequent event in myeloid malignancies of Fanconi anemia (FA) patients. (D) Promoter activation: Left panel: Transcriptional activation of the EVI1 locus is mediated by other transcription factors including ELK1, GATA2, and RUNX1, that bind the EVI1 promoter directly. RUNX1 also mediates acetylation of histone H3 in the EVI1 promoter region. In cells with a translocation t(11;19) (q23;p13.3), the MLL-ENL oncoprotein activates EVI1 transcription in HSC-derived leukemic cells. Right panel: The MECOM locus is a recurrent site of retroviral insertion and viral insertional mutagenesis can result in increased EVI1 expression through insertion of viral regulatory elements that aberrantly activate the EVI1 promoter. (E) Cell of origin: The epigenetic state of the cell of origin can be preserved after malignant transformation. Normal HSCs physiologically express high levels of EVI1, and leukemias arising from HSCs, for example after transformation with the MLL-AF9 oncogene, retain these high levels of EVI1. While MLL-AF9 is capable of transforming more mature myeloid progenitor cells, such progenitor-derived leukemias retain the low EVI1 expression levels present in the cell of origin.
In addition to RUNX1-EVI1 fusions, several cases of a t(3;12)(q26;p13) translocation have been reported in poor prognosis AML, MDS, and blast crisis MDS.74 This translocation leads to a fusion of ETV6 to the MECOM locus (Figure 3A, right panel). However, whether this translocation leads to a novel oncogenic fusion protein or whether the oncogenic potential stems from the ETV6 promoter increasing EVI1 transcription is still not clear.
t(3;3)/inv(3)
The best-characterized mechanisms leading to EVI1 overexpression are the translocations t(3;3)(q21;q26) and inv(3)(q21;q26), occurring in approximately 2.5% of all AML cases.32 Indeed, these rearrangements are defining features of a distinct AML entity recognized in the WHO classification system28 and the more recently established International Consensus Classification (ICC) of myeloid neoplasms and acute leukemias,75 and can also be observed in CML in blast crisis.76 In AML with t(3;3)(q21;q26) or inv(3)(q21;q26), an enhancer of GATA2 at 3q21, also called the GATA2 distal hematopoietic enhancer (G2DHE), is relocated proximal to the MECOM locus at 3q26 (Figure 3B, left panel).77,78 The following 3 recurrent changes in gene expression can be seen as hallmarks of this entity:
Since the breakpoint of the translocation often lies between the MDS1 gene and the EVI1 promoter, the expression of the full-length MECOM transcript MDS1-EVI1 is disrupted.
In contrast, EVI1, now under control of the GATA2 enhancer, is overexpressed.
Conversely, GATA2 is only expressed from the remaining normal allele, leading to a decrease in GATA2 abundance.
In mice, it was shown that GATA2 haploinsufficiency cooperates with Evi1 overexpression and accelerates the onset of Evi1-induced leukemia,79 which appears to correlate with the observation that in humans, inherited GATA2 haploinsufficiency constitutes a predisposition to develop MDS or AML.80 Thus, this specific translocation can be considered to result in a “double hit” leading to malignant transformation.
Mutations in the splicing factor SF3B1 have been found to co-occur in >30% of t(3;3)/inv(3) cases.81 The mutated SF3B1 causes alternative splicing of the EVI1 transcript, leading to a novel oncogenic isoform with an addition of 18 base pairs/6 amino acids in the distal zinc finger domain.82 This EVI1+18 isoform exhibits altered DNA binding and increased oncogenicity.
Atypical 3q26 rearrangements
In addition to t(3;3)/inv(3), other translocations can also lead to EVI1 overexpression, with enhancer hijacking operating as a recurrent mechanism of dysregulation. In AML with t(3;8)(q26;q24), an MYC “super-enhancer” comprised of several enhancer modules is translocated to the MECOM locus (Figure 3B, right panel).83 One of these modules serves as a binding site for key transcription factors of early hematopoiesis, including ERG, FLI1, GATA2, LMO2, LYL1, and RUNX1.83,84 This enhancer module facilitates the interaction of the enhancer with the EVI1 promoter via promoter-enhancer looping, and is essential for the overexpression of EVI1. This looping interaction is mediated by the CCCTC-binding factor CTCF and a CTCF enhancer-docking site that lies upstream of the EVI1 promoter and is preserved in 3q26 rearranged AMLs.
There are also other atypical 3q26 translocations that involve super-enhancers of genes that play an important role in myeloid development, including CD133 (PROM1), CD164, and CDK6 (through t(3;7)(q26;q21)).85,86 The mechanism of EVI1 overexpression as well as the pathophysiology of these leukemias resembles that of the classical inv(3)/t(3;3)(q21q26) AML entity in that EVI1 expression is high, the expression of MDS-EVI1 is low or absent and in half of the cases, levels of GATA2 are also reduced.85 Since the translocation in these leukemias does not involve the GATA2 enhancer, the mechanism behind the low GATA2 expression in these cases is unclear. Interestingly, some of the transcription factors known to bind to the GATA2 enhancer at 3q21, for example, ERG, GATA2, LMO2, and RUNX1, are reported to also interact with the loci involved in these atypical 3q26 translocations.87 All in all, it is possible that atypical 3q26 rearrangements and the classical inv(3)/t(3;3)(q21;q26) constitute a single AML entity that is characterized by elevated EVI1 levels through enhancer hijacking, absent MDS-EVI1 and a reduction in GATA2, while clinically sharing a poor prognosis and the occurrence of resistance to chemotherapy.
While EVI1 overexpression was accompanied by an insufficiency of GATA2 as a result of the translocation events described above, other translocations leading to high levels of EVI1 in the absence of altered GATA2 expression have also been described. Cases of t(3;21)(q26;q11), as reported by Haferlach et al88 and D’Angió et al,89 relocate the EVI1 gene proximal to regulatory elements of the Nuclear Receptor Interacting Protein 1 (NRIP1) gene. As is the case for the t(3;21)(q26;q22) RUNX1-EVI1 translocation, there is no evidence that t(3;21)(q26;q11) results in the expression of a novel fusion protein.90 However, the NRIP1 promoter and associated enhancers contain multiple elements that are responsive to retinoic acid and NRIP1 transcription can be stimulated by treatment with all-trans-retinoic acid (ATRA). It is therefore likely, that ATRA treatment will also lead to the upregulation of EVI1 in cases with t(3;21)(q26;q11).
In myeloid malignancies with t(3;17)(q26;q22), EVI1 is translocated to the MSI2 locus.91 While no fusion protein can be detected, the translocation increases the expression of EVI1.91 Also cases of translocations involving the short arm of chromosome 2, t(2;3)(p15-23;q26), have been reported in AML and CML in blast crisis92 with poor clinical outcome, leading to a >20-fold overexpression of EVI1.93
Amplification of 3q
Amplification of the EVI1 locus at 3q, for example spanning the region of 3q26q29, is a frequent early event in myeloid malignancies that occur in Fanconi anemia (FA) patients,94,95 with the EVI1 locus being contained in the minimally amplified region (Figure 3C). Almost half of FA patients with MDS or AML exhibit this genomic abnormality that leads to a >10-fold increase of the EVI1 transcript and constitutes an adverse risk factor.94–97 In contrast, gain of 3q is a very rare event in non-FA patients. It is tempting to speculate that this type of mutation is more likely to occur in the setting of a defective DNA repair pathway. Moreover, the fact that EVI1 overexpression is strongly selected for in the context of the molecular defect in DNA repair that is characteristic of this disease, suggests that EVI1 may either directly modulate the cellular DNA damage response (DDR) or the growth-restrictive cell fate outcomes that result from activation of the DDR. It is tempting to speculate that such a relationship may be more broadly relevant to the poor outcome and therapy resistance observed in EVI1high leukemias in non-FA patients.
Transcriptional (de)regulation of EVI1
The majority of cases of EVI1 overexpression in hematologic malignancies do not occur in the context of translocations involving the MECOM locus, suggesting that aberrant epigenetic programming of the EVI1 promoter/enhancer elements is a more frequent driver of this oncogenic event. Indeed, known upstream regulators of EVI1 transcription include GATA1, ELK1, and RUNX1, meaning that altered expression or activity of any of these transcription factors could modulate EVI1 activity (Figure 3D, left panel).98 Indeed, these transcription factors directly bind to a 318bp region that has been characterized as the minimal promoter region of EVI1. Knockdown of ELK1 or RUNX1 results in decreased EVI1 levels, while their overexpression led to an increase. EVI1 and RUNX1 levels also seem to positively correlate in AML patient samples, indicating a causal relationship which appears to relate to RUNX1 exerting a regulatory role on EVI1 expression via mediating acetylation of histone H3 in the EVI1 promoter region.98 Whether translocations involving the RUNX1 gene influence its ability to bind to the EVI1 promoter has yet to be determined.
In addition to these normal transcription factors, MLL fusion oncoprotein MLL-ENL directly binds to the promoter of EVI1 and elevates its transcription selectively in leukemic HSCs (Figure 3D, left panel).99 Interestingly, this association cannot be observed in the more differentiated leukemic myeloid progenitor populations, demonstrating a surprising context-specific effect of the fusion protein within different hierarchically organized sub-sets of leukemic cells. Knockdown of EVI1 in MLL-ENL leukemic cells decreases their potential to form colonies.18 Overexpression of EVI1 is also observed in other MLL-rearranged leukemias, including those with a MLL-AF9 fusion25 and EVI1 expression serves as an independent and adverse prognostic factor within MLL-rearranged leukemias.30 Nevertheless, EVI1low leukemias also exist among the MLL-rearranged cases, possibly relating to the cell of origin of these leukemias, as discussed below.
Dysregulation of the MECOM locus due to retro/lentiviral insertional mutagenesis
An aberrant increase of EVI1 transcript levels through non-physiologic EVI1 promoter activation can also result from viral insertional mutagenesis when murine or human HSCs have been transduced with integrating retro- or lentiviral vectors (Figure 3D, right panel). In fact, the oncogene EVI1 was first identified as a murine retroviral insertion site causing myeloid leukemia in mice as a result of Evi1 overexpression.100–103 In this particular setting, the recombinant vector integrated within the vicinity of the Evi1 promoter, resulting in the powerful enhancer elements encoded in the viral long terminal repeats driving expression of the Evi1 transcript. The clinical relevance of this phenomenon became apparent later since the MECOM locus was also found as an insertional mutagenesis site in a gene therapy clinical trial treating patients with X-linked granulomatous disease. During the attempt to correct mutations of gp91PHOX in hematopoietic progenitor cells, the vector integration resulted in EVI1 overexpression and the competitive outgrowth of mutated clones in 2 patients, eventually contributing to the development of myeloid malignancies.104 These cases formally illustrate the capacity of the EVI1 gene to initiate myeloid malignant transformation when aberrantly expressed in primary HSCs.
Capturing endogenous high EVI1 levels in the leukemic cell of origin
Leukemias can arise from a range of different cells of origin depending on the initial driver mutations present. For example, while certain oncogenic drivers, such as MLL-AF9, are competent at mediating the transformation of both HSC and more differentiated progenitor cells, others, such as HOXA9 and MEIS1 are only capable of transforming HSCs.105 It is widely accepted that the cell of origin can also influence clinical features, such as sensitivity toward chemotherapy.106,107 Often, some features of the epigenetic state of the cell of origin are preserved after malignant transformation.108,109 This phenomenon has been demonstrated to comprise a route via which Evi1 can be aberrantly expressed in murine models of leukemia. Thus, using the MLL-AF9 transformation model referenced above, the introduction of this fusion oncoprotein into murine HSCs, which express high levels of Evi1, resulted in the generation of an AML-like disease that retained these high Evi1 expression levels106 and retained epigenetic marks at the MECOM locus that were consistent with those found in HSCs (Figure 3E). In contrast, the introduction of MLL-AF9 into more differentiated granulocyte-macrophage progenitors resulted in an Evi1low AML (Figure 3E). Intriguingly, despite the identical nature of the leukemic driver in these 2 AMLs, the Evi1high disease was found to be inherently more resistant to chemotherapy and demonstrated lower levels of apoptotic priming, which could be ascribed to retaining high expression levels of Evi1 after the transformation process.68 It remains to be investigated whether this capturing of the EVI1high epigenetic state present in HSCs can also be achieved in non-MLL-AF9 leukemias.
POSSIBLE THERAPEUTIC INTERVENTIONS TO TREAT EVI1high MALIGNANCIES
With more than 10% of all AMLs demonstrating EVI1 overexpression, and this phenomenon correlating with poor prognosis, it would clearly be desirable to develop targeted therapies to effectively treat these EVI1high leukemias; both in the setting of 3q rearrangements, as well as the more frequent non-3q rearranged EVI1high AMLs. Three therapeutic approaches to treat EVI1high cancers are conceptually plausible:
Prevent or revert the aberrant EVI1 overexpression at the transcriptional level.
Prevent the association of the EVI1 protein with its protein interaction partners.
Modulate downstream effects of EVI1.
Inhibition of super-enhancers
As described previously, a recurrent mechanism of EVI1 overexpression in AML is the rearrangement of (super) enhancers to the MECOM locus via translocations at the 3q locus. Thus, an obvious therapeutic strategy in 3q-rearranged AML could be to prevent the aberrant activation of the EVI1 promoter by inhibition of such super-enhancers. One way that this could be achieved by pharmacologic BET bromodomain inhibition.110,111 Indeed, in both 3q-rearranged cell lines as well as primary t(3;3) AML patient samples, treatment with the BET bromodomain inhibitor JQ1 led to a decrease of EVI1 and growth arrest of the AML cells.77 Other studies have specifically investigated the binding of important myeloid transcription factors to the GATA2 distal hematopoietic enhancer that is often relocated to the EVI1 locus, including CEBPA and RUNX1. Proteome analyses have identified PARP1 and IKZF1 among the colocalizing proteins at this locus, thereby identifying novel therapeutic targets to interfere with the oncogenic super-enhancer.112 Inhibition of PARP1, both pharmacologically with olaparib or talazoparib, and shRNA-mediated, was able to reduce the interaction of the super-enhancer with the EVI1 promoter and decreased the expression of EVI1. As a consequence, differentiation and apoptosis in 3q-rearranged AML cell lines was increased.112
Inhibition of EVI1 as a transcriptional co-repressor
The EVI1 protein contains several different functional domains, allowing it to associate with a range of proteins that can impact on target gene expression, including chromatin remodelers like HMTs and HADCs. With regard to the latter, HDAC inhibitors, such as trichostatin A, already exist. It has already been shown in vitro that trichostatin A treatment is able to revert EVI1-mediated transcriptional repression of TGFβ signaling.55 Thus, selective inhibition of the enzymatic activity of proteins that act as co-regulators of the EVI1 transcription factors may well comprise a feasible targeted approach for the treatment of EVI1high malignancies.
Modulating downstream effectors
Retinoic acid receptor signaling
As described previously, the t(3;21)(q26;q11) translocation leads to the EVI1 gene falling under the control of regulatory elements of the retinoid acid-responsive gene NRIP1.90 Upregulation of NRIP1 can be observed in AML cases with elevated EVI1 levels, presumably due to binding of EVI1 to a NRIP1 enhancer. Knockdown of NRIP1 in 3q-rearranged cell lines reduced the proliferation and viability of these AML cells and was also able to increase their sensitivity towards treatment with all-trans-retinoic acid (ATRA),90 making NRIP1 a potential therapeutic target in EVI1high AML cases. Whether EVI1high AML cases might profit from treatment with ATRA is still under continued investigation.
More generally, following on from the clinical success of treating acute promyelocytic leukemia (APL) with ATRA, this so-called “differentiation therapy” has been broadly tested against a range of hematologic malignancies, regardless of whether they harbored the APL-specific translocation between the retinoic acid receptor-α (RARA) and APL genes that specifically confers sensitivity to ATRA. Some of these studies have encompassed 3q-rearranged/EVI1high AML samples. Verhagen et al observed in vitro induction of differentiation upon ATRA treatment in 9 out of 13 EVI1high AML cases, coupled with a reduced viability of the AML blasts in some cases.113 The pretreatment with ATRA also resulted in decreased survival of these cells after treatment with doxorubicin. In 2 out of 3 cases, ATRA treatment also resulted in reduced leukemic engraftment of primary AML blasts in NOD SCID-IL2g knockout (NSG) mice. Interestingly, a second study has shown a synergistic effect of combining the bromodomain inhibitor JQ1 with ATRA to induce differentiation and apoptosis in vitro in the non-APL AML cell lines HL-60 and MV4-11.114 In a clinical study by Paubelle et al, 13 high-risk EVI1high AML cases were treated with ATRA, of which 7 achieved complete remission.115 While these results are promising, it is also clear that the response of EVI1high AML cells to ATRA is very heterogenous and warrants further investigation with larger cohort/sample sizes to determine whether it is possible to identify criteria that robustly sub-classify patients with EVI1high leukemias who would respond at ATRA treatment as a combined or mono-therapy.
Creatine kinase inhibition
The EVI1-mediated inhibition of RUNX1 results in the upregulation of the mitochondrial creatine kinase CKMT1,66,116 which has been reported to constitute a metabolic vulnerability of EVI1high leukemic cells. Both the shRNA-mediated knockdown of CKMT1, as well as pharmacological inhibition using the small molecule cyclocreatine, reduced the growth of EVI1high AML cells and induced cell cycle arrest and apoptosis in human cell lines, orthotopic xenograft models and mouse models of primary AML. At a mechanistic level, cyclocreatine treatment interfered with the cell’s mitochondrial respiration and ATP production and altered both WNT signaling and GSK3 signaling.66,116 Given the important role of these signaling pathways in normal HSCs, it has to be clarified whether a therapeutic window exists to facilitate the successful treatment of EVI1high leukemias.
ERG inhibition
Two independent studies have demonstrated a therapeutic vulnerability of EVI1high AMLs towards the inhibition of ERG signaling.50,51 In the EVI1high leukemic cells, knockdown of ERG led to a decrease in proliferation and an increase in apoptosis, and also induced differentiation.50 However, achieving a pharmacological inhibition of ERG in AML patients, as is the case for many transcription factors, remains a challenge and is currently still a subject of research and development.117
Apoptotic priming
We have previously discussed how the epigenetic state of the EVI1 locus and corresponding transcript levels can be preserved after the malignant transformation of HSCs with the MLL-AF9 oncogenic fusion gene.68 In an MLL-AF9 AML mouse model, the GMP-derived EVI1low murine leukemic cells showed a higher degree of apoptotic priming and were more sensitive to treatment with doxorubicin and to treatment with the LSD1 inhibitors IMG-7289 and IMG-98 than the HSC/multipotent-progenitor (MPP)-derived EVI1high malignant cells. Knockdown of Evi1 in HSC/MPP-derived MLL-AF9 leukemic cells sensitized the cells to treatment with several chemotherapeutic agents, including doxorubicin, ionizing radiation, and combination treatment with venetoclax and LSD1 inhibitors.68 The authors could ascribe this EVI1-mediated reduction in apoptotic priming to decreased p53 protein abundance via a proteasome-independent mechanism that is as yet uncharacterized68 (Figure 2C). One possible treatment strategy for EVI1high leukemias with non-mutated TP53 could therefore be the use of an MDM2 inhibitor, for example, RG7388, to increase p53 protein levels and sensitize cells to subsequent therapy. Potentially, this effect could be enhanced with dual inhibition of both MDM2 and MDMX, for example using a stapled α-helical peptide called ALRN-6924, which has been previously shown to be very effective in xenograft models with p53 wildtype AML.118 Another strategy to increase the apoptotic priming of EVI1high leukemias could be treatment with BH3 mimetics such as venetoclax, that target anti-apoptotic BCL2 family members.
CONCLUSIONS AND FUTURE DIRECTIONS
EVI1 is a multifunctional transcription factor with diverse interaction partners. It can influence several hallmarks of cancer, including cell proliferation, differentiation, and apoptosis. It has already been acknowledged that expression levels of EVI1 are of prognostic value in different cancer types, including hematologic malignancies like AML32 and CML,38 as well as solid cancers such as prostate48 and ovarian cancer,44 suggesting that EVI1 expression levels might also be informative to navigate and improve treatment strategies of EVI1high malignancies in the future.
Many groups have already investigated the mechanisms responsible for EVI1 overexpression, including several translocations that result in an increase in EVI1 levels. However, while translocations involving the MECOM locus are only found in around 2.5% of AML patients, EVI1 overexpression can be detected in around 10% of all AMLs and for many patients, the mechanism behind this is not clear. Since the exact etiology underlying EVI1 overexpression might comprise a potential novel therapeutic target to treat the patient, further characterization of the epigenetic mechanisms promoting this phenomenon in non-3q rearranged EVI1high AMLs appears warranted.
While significant traction has been gained in dissecting some of the mechanisms via which EVI1 expression is deregulated in leukemia, so far there appears to be little consensus in the literature on the critical downstream effects of EVI1 that relate to the evolution of therapy-resistant hematologic malignancies. Several studies suggest functional attributes for EVI1 that extend beyond its canonically defined role as a transcription factor, the most recent of which is the modulation of p53 protein levels. To further interrogate this phenomenon, and potentially uncover other important roles of this protein that may not relate to the regulation of transcription, it will be necessary to take alternative approaches that extend beyond the classical analysis of gene expression and chromatin conformation/modifications. Given the context-specificity of EVI1 function, it might well be of importance to investigate whether therapies to treat EVI1high malignant cells are only effective in a specific cellular setting while also bearing in mind potential confounding effects of concomitant mutations.
Despite the fact that EVI1 was identified as a powerful pro-leukemic factor more than 3 decades ago, many of the details of its biological function remain enigmatic. While EVI1 clearly plays an important role in tissue development and homeostasis, clearly one of the main motivational forces underlying the desire to elucidate its role is its presumed causal role in a significant proportion of high-risk hematologic malignancies, as well as a range of solid cancers. We would predict that any gains that are made in this endeavor could have direct translational application toward the unmet goal of treating such diseases.
AUTHOR CONTRIBUTIONS
SL and MDM interpreted the relevant literature and wrote the article.
DISCLOSURES
The authors have no conflicts of interest to disclose.
SOURCES OF FUNDING
This work was supported by the DKFZ Postdoctoral Fellowship Program (SL) and the Dietmar Hopp Stiftung (MDM).
REFERENCES
- 1.Delwel R, Funabiki T, Kreider B, et al. Four of the seven zinc fingers of the Evi-1 myeloid-transforming gene are required for sequence-specific binding to GA (C/T) AAGA (T/C) AAGATAA. Mol Cell Biol. 1993;13:4291–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morishita K, Parganas E, Bartholomew C, et al. The human Evi-1 gene is located on chromosome 3q24-q28 but is not rearranged in three cases of acute nonlymphocytic leukemias containing t (3; 5)(q25; q34) translocations. Oncogene Res. 1990;5:221–231. [PubMed] [Google Scholar]
- 3.Morishita K, Suzukawa K, Taki T, et al. EVI-1 zinc finger protein works as a transcriptional activator via binding to a consensus sequence of GACAAGATAAGATAAN1-28 CTCATCTTC. Oncogene. 1995;10:1961–1967. [PubMed] [Google Scholar]
- 4.Soderholm J, Kobayashi H, Mathieu C, et al. The leukemia-associated gene MDS1/EVI1 is a new type of GATA-binding transactivator. Leukemia. 1997;11:352–358. [DOI] [PubMed] [Google Scholar]
- 5.Sitailo S, Sood R, Barton K, et al. Forced expression of the leukemia-associated gene EVI1 in ES cells: a model for myeloid leukemia with 3q26 rearrangements. Leukemia. 1999;13:1639–1645. [DOI] [PubMed] [Google Scholar]
- 6.Sood R, Talwar-Trikha A, Chakrabarti S, et al. MDS1/EVI1 enhances TGF-β1 signaling and strengthens its growth-inhibitory effect, but the leukemia-associated fusion protein AML1/MDS1/EVI1, product of the t (3; 21), abrogates growth-inhibition in response to TGF-β1. Leukemia. 1999;13:348–357. [DOI] [PubMed] [Google Scholar]
- 7.Ivanochko D, Halabelian L, Henderson E, et al. Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex. Nucleic Acids Res. 2019;47:1225–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinai AA, Valk PJ. Aberrant EVI 1 expression in acute myeloid leukaemia. Br J Haematol. 2016;172:870–878. [DOI] [PubMed] [Google Scholar]
- 9.Bard-Chapeau EA, Gunaratne J, Kumar P, et al. EVI1 oncoprotein interacts with a large and complex network of proteins and integrates signals through protein phosphorylation. Proc Natl Acad Sci USA. 2013;110:E2885–E2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paredes R, Kelly JR, Geary B, et al. EVI1 phosphorylation at S436 regulates interactions with CtBP1 and DNMT3A and promotes self-renewal. Cell Death Dis. 2020;11:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoyt PR, Bartholomew C, Davis AJ, et al. The Evil proto-oncogene is required at midgestation for neural, heart, and paraxial mesenchyme development. Mech Dev. 1997;65:55–70. [DOI] [PubMed] [Google Scholar]
- 12.Bard-Chapeau EA, Szumska D, Jacob B, et al. Mice carrying a hypomorphic Evi1 allele are embryonic viable but exhibit severe congenital heart defects. PLoS One. 2014;9:e89397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuasa H, Oike Y, Iwama A, et al. Oncogenic transcription factor Evi1 regulates hematopoietic stem cell proliferation through GATA-2 expression. EMBO J. 2005;24:1976–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kataoka K, Sato T, Yoshimi A, et al. Evi1 is essential for hematopoietic stem cell self-renewal, and its expression marks hematopoietic cells with long-term multilineage repopulating activity. J Exp Med. 2011;208:2403–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokomizo T, Ideue T, Morino-Koga S, et al. Independent origins of fetal liver haematopoietic stem and progenitor cells. Nature. 2022;609:779–784. [DOI] [PubMed] [Google Scholar]
- 16.Calvanese V, Capellera-Garcia S, Ma F, et al. Mapping human haematopoietic stem cells from haemogenic endothelium to birth. Nature. 2022;604:534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laricchia-Robbio L, Nucifora G. Significant increase of self-renewal in hematopoietic cells after forced expression of EVI1. Blood Cells Mol Dis. 2008;40:141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goyama S, Yamamoto G, Shimabe M, et al. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell. 2008;3:207–220. [DOI] [PubMed] [Google Scholar]
- 19.Voit RA, Tao L, Yu F, et al. A genetic disorder reveals a hematopoietic stem cell regulatory network co-opted in leukemia. Nat Immunol. 2023;24:69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Germeshausen M, Ancliff P, Estrada J, et al. MECOM-associated syndrome: a heterogeneous inherited bone marrow failure syndrome with amegakaryocytic thrombocytopenia. Blood Adv. 2018;2:586–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niihori T, Ouchi-Uchiyama M, Sasahara Y, et al. Mutations in MECOM, encoding oncoprotein EVI1, cause radioulnar synostosis with amegakaryocytic thrombocytopenia. Am J Hum Genet. 2015;97:848–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bluteau O, Sebert M, Leblanc T, et al. A landscape of germ line mutations in a cohort of inherited bone marrow failure patients. Blood. 2018;131:717–732. [DOI] [PubMed] [Google Scholar]
- 23.Kustikova O, Schwarzer A, Stahlhut M, et al. Activation of Evi1 inhibits cell cycle progression and differentiation of hematopoietic progenitor cells. Leukemia. 2013;27:1127–1138. [DOI] [PubMed] [Google Scholar]
- 24.Haas K, Kundi M, Sperr WR, et al. Expression and prognostic significance of different mRNA 5′-end variants of the oncogene EVI1 in 266 patients with de novo AML: EVI1 and MDS1/EVI1 overexpression both predict short remission duration. Genes Chromosomes Cancer. 2008;47:288–298. [DOI] [PubMed] [Google Scholar]
- 25.Lugthart S, van Drunen E, van Norden Y, et al. High EVI1 levels predict adverse outcome in acute myeloid leukemia: prevalence of EVI1 overexpression and chromosome 3q26 abnormalities underestimated. Blood. 2008;111:4329–4337. [DOI] [PubMed] [Google Scholar]
- 26.Groeschel S, Lugthart S, Schlenk RF, et al. High EVI1 expression predicts outcome in younger adult patients with acute myeloid leukemia and is associated with distinct cytogenetic abnormalities. J Clin Oncol. 2010;28:2101–2107. [DOI] [PubMed] [Google Scholar]
- 27.Vázquez I, Maicas M, Cervera J, et al. Down-regulation of EVI1 is associated with epigenetic alterations and good prognosis in patients with acute myeloid leukemia. Haematologica. 2011;96:1448–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. [DOI] [PubMed] [Google Scholar]
- 29.Ho PA, Alonzo TA, Gerbing RB, et al. High EVI 1 expression is associated with MLL rearrangements and predicts decreased survival in paediatric acute myeloid leukaemia: a report from the children’s oncology group. Br J Haematol. 2013;162:670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groeschel S, Schlenk RF, Engelmann J, et al. Deregulated expression of EVI1 defines a poor prognostic subset of MLL-rearranged acute myeloid leukemias: a study of the German-Austrian Acute Myeloid Leukemia Study Group and the Dutch-Belgian-Swiss HOVON/SAKK Cooperative Group. J Clin Oncol. 2013;31:95–103. [DOI] [PubMed] [Google Scholar]
- 31.Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, van Putten WL, et al. High EVI1 expression predicts poor survival in acute myeloid leukemia: a study of 319 de novo AML patients. Blood. 2003;101:837–845. [DOI] [PubMed] [Google Scholar]
- 32.Lugthart S, Groschel S, Beverloo HB, et al. Clinical, molecular, and prognostic significance of WHO type inv (3)(q21q26. 2)/t (3; 3)(q21; q26. 2) and various other 3q abnormalities in acute myeloid leukemia. J Clin Oncol. 2010;28:3890–3898. [DOI] [PubMed] [Google Scholar]
- 33.Balgobind B, Lugthart S, Hollink I, et al. EVI1 overexpression in distinct subtypes of pediatric acute myeloid leukemia. Leukemia. 2010;24:942–949. [DOI] [PubMed] [Google Scholar]
- 34.Russell M, List A, Greenberg P, et al. Expression of EVI1 in myelodysplastic syndromes and other hematologic malignancies without 3q26 translocations. Blood. 1994;84:1243–1248. [PubMed] [Google Scholar]
- 35.Dreyfus F, Bouscary D, Melle J, et al. Expression of the Evi-1 gene in myelodysplastic syndromes. Leukemia. 1995;9:203–205. [PubMed] [Google Scholar]
- 36.Thol F, Yun H, Sonntag AK, et al. Prognostic significance of combined MN1, ERG, BAALC, and EVI1 (MEBE) expression in patients with myelodysplastic syndromes. Ann Hematol. 2012;91:1221–1233. [DOI] [PubMed] [Google Scholar]
- 37.Ogawa S, Kurokawa M, Tanaka T, et al. Increased Evi-1 expression is frequently observed in blastic crisis of chronic myelocytic leukemia. Leukemia. 1996;10:788–794. [PubMed] [Google Scholar]
- 38.Daghistani M, Marin D, Khorashad JS, et al. EVI-1 oncogene expression predicts survival in chronic-phase CML patients resistant to imatinib treated with second-generation tyrosine kinase inhibitors. Blood. 2010;116:6014–6017. [DOI] [PubMed] [Google Scholar]
- 39.Paquette RL, Nicoll J, Chalukya M, et al. Frequent EVI1 translocations in myeloid blast crisis CML that evolves through tyrosine kinase inhibitors. Cancer Genet. 2011;204:392–397. [DOI] [PubMed] [Google Scholar]
- 40.Sato T, Goyama S, Kataoka K, et al. Evi1 defines leukemia-initiating capacity and tyrosine kinase inhibitor resistance in chronic myeloid leukemia. Oncogene. 2014;33:5028–5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konantz M, Andre M, Ebinger M, et al. EVI-1 modulates leukemogenic potential and apoptosis sensitivity in human acute lymphoblastic leukemia. Leukemia. 2013;27:56–65. [DOI] [PubMed] [Google Scholar]
- 42.Stevens A, Hanson D, De Leonibus C, et al. EVI1 expression in childhood acute lymphoblastic leukaemia is not restricted to MLL and BCR/ABL rearrangements and is influenced by age. Blood Cancer J. 2014;4:e179–e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nanjundan M, Nakayama Y, Cheng KW, et al. Amplification of MDS1/EVI1 and EVI1, located in the 3q26. 2 amplicon, is associated with favorable patient prognosis in ovarian cancer. Cancer Res. 2007;67:3074–3084. [DOI] [PubMed] [Google Scholar]
- 44.Österberg L, Levan K, Partheen K, et al. Potential predictive markers of chemotherapy resistance in stage III ovarian serous carcinomas. BMC Cancer. 2009;9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel J, Appaiah H, Burnett R, et al. Control of EVI-1 oncogene expression in metastatic breast cancer cells through microRNA miR-22. Oncogene. 2011;30:1290–1301. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka M, Suzuki H, Shibahara J, et al. EVI1 oncogene promotes KRAS pathway through suppression of microRNA-96 in pancreatic carcinogenesis. Oncogene. 2014;33:2454–2463. [DOI] [PubMed] [Google Scholar]
- 47.Deng X, Cao Y, Liu Y, et al. Overexpression of Evi-1 oncoprotein represses TGF-β signaling in colorectal cancer. Mol Carcinog. 2013;52:255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Queisser A, Hagedorn S, Wang H, et al. Ecotropic viral integration site 1, a novel oncogene in prostate cancer. Oncogene. 2017;36:1573–1584. [DOI] [PubMed] [Google Scholar]
- 49.Ayoub E, Wilson MP, McGrath KE, et al. EVI1 overexpression reprograms hematopoiesis via upregulation of Spi1 transcription. Nat Commun. 2018;9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmoellerl J, Barbosa IA, Minnich M, et al. EVI1 drives leukemogenesis through aberrant ERG activation. Blood. 2023;141:453–466. [DOI] [PubMed] [Google Scholar]
- 51.Masamoto Y, Chiba A, Mizuno H, et al. EVI1 exerts distinct roles in AML via ERG and cyclin D1 promoting a chemoresistant and immune-suppressive environment. Blood Adv. 2023;7:1577–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimabe M, Goyama S, Watanabe-Okochi N, et al. Pbx1 is a downstream target of Evi-1 in hematopoietic stem/progenitors and leukemic cells. Oncogene. 2009;28:4364–4374. [DOI] [PubMed] [Google Scholar]
- 53.Bard-Chapeau EA, Jeyakani J, Kok CH, et al. Ecotopic viral integration site 1 (EVI1) regulates multiple cellular processes important for cancer and is a synergistic partner for FOS protein in invasive tumors. Proc Natl Acad Sci USA. 2012;109:2168–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshimi A, Goyama S, Watanabe-Okochi N, et al. Evi1 represses PTEN expression and activates PI3K/AKT/mTOR via interactions with polycomb proteins. Blood. 2011;117:3617–3628. [DOI] [PubMed] [Google Scholar]
- 55.Izutsu K, Kurokawa M, Imai Y, et al. The corepressor CtBP interacts with Evi-1 to repress transforming growth factor β signaling. Blood. 2001;97:2815–2822. [DOI] [PubMed] [Google Scholar]
- 56.Kurokawa M, Mitani K, Irie K, et al. The oncoprotein Evi-1 represses TGF-β signalling by inhibiting Smad3. Nature. 1998;394:92–96. [DOI] [PubMed] [Google Scholar]
- 57.Alliston T, Ko TC, Cao Y, et al. Repression of bone morphogenetic protein and activin-inducible transcription by Evi-1. J Biol Chem. 2005;280:24227–24237. [DOI] [PubMed] [Google Scholar]
- 58.Chakraborty S, Senyuk V, Sitailo S, et al. Interaction of EVI1 with cAMP-responsive element-binding protein-binding protein (CBP) and p300/CBP-associated factor (P/CAF) results in reversible acetylation of EVI1 and in co-localization in nuclear speckles. J Biol Chem. 2001;276:44936–44943. [DOI] [PubMed] [Google Scholar]
- 59.Goyama S, Nitta E, Yoshino T, et al. EVI-1 interacts with histone methyltransferases SUV39H1 and G9a for transcriptional repression and bone marrow immortalization. Leukemia. 2010;24:81–88. [DOI] [PubMed] [Google Scholar]
- 60.Senyuk V, Premanand K, Xu P, et al. The oncoprotein EVI1 and the DNA methyltransferase Dnmt3 co-operate in binding and de novo methylation of target DNA. PLoS One. 2011;6:e20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lugthart S, Figueroa ME, Bindels E, et al. Aberrant DNA hypermethylation signature in acute myeloid leukemia directed by EVI1. Blood. 2011;117:234–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laricchia-Robbio L, Premanand K, Rinaldi CR, et al. EVI1 Impairs myelopoiesis by deregulation of PU. 1 function. Cancer Res. 2009;69:1633–1642. [DOI] [PubMed] [Google Scholar]
- 63.Laricchia-Robbio L, Fazzina R, Li D, et al. Point mutations in two EVI1 Zn fingers abolish EVI1-GATA1 interaction and allow erythroid differentiation of murine bone marrow cells. Mol Cell Biol. 2006;26:7658–7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanaka T, Nishida J, Mitani K, et al. Evi-1 raises AP-1 activity and stimulates c-fos promoter transactivation with dependence on the second zinc finger domain. J Biol Chem. 1994;269:24020–24026. [PubMed] [Google Scholar]
- 65.Senyuk V, Sinha KK, Li D, et al. Repression of RUNX1 activity by EVI1: a new role of EVI1 in leukemogenesis. Cancer Res. 2007;67:5658–5666. [DOI] [PubMed] [Google Scholar]
- 66.Benajiba L, Alexe G, Su A, et al. Creatine kinase pathway inhibition alters GSK3 and WNT signaling in EVI1-positive AML. Leukemia. 2019;33:800–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurokawa M, Mitani K, Yamagata T, et al. The evi-1 oncoprotein inhibits c-Jun N-terminal kinase and prevents stress-induced cell death. EMBO J. 2000;19:2958–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cai SF, Chu SH, Goldberg AD, et al. Leukemia cell of origin influences apoptotic priming and sensitivity to LSD1 inhibition. Cancer Discov. 2020;10:1500–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rubin CM, Larson RA, Bitter MA, et al. Association of a chromosomal 3; 21 translocation with the blast phase of chronic myelogenous leukemia. Blood. 1987;70:1338–1342. [PubMed] [Google Scholar]
- 70.Rubin CM, Larson RA, Anastasi J, et al. t (3; 21)(q26; q22): a recurring chromosomal abnormality in therapy-related myelodysplastic syndrome and acute myeloid leukemia. Blood. 1990;76:2594–2598. [PubMed] [Google Scholar]
- 71.Maki K, Yamagata T, Yamazaki I, et al. Development of megakaryoblastic leukaemia in Runx1-Evi1 knock-in chimaeric mouse. Leukemia. 2006;20:1458–1460. [DOI] [PubMed] [Google Scholar]
- 72.Loke J, Assi SA, Imperato MR, et al. RUNX1-ETO and RUNX1-EVI1 differentially reprogram the chromatin landscape in t (8; 21) and t (3; 21) AML. Cell Rep. 2017;19:1654–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kellaway SG, Keane P, Kennett E, et al. RUNX1-EVI1 disrupts lineage determination and the cell cycle by interfering with RUNX1 and EVI1 driven gene regulatory networks. Haematologica. 2021;106:1569–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peeters P, Wlodarska I, Baens M, et al. Fusion of ETV6 to MDS1/EVI1 as a result of t (3; 12)(q26; p13) in myeloproliferative disorders. Cancer Res. 1997;57:564–569. [PubMed] [Google Scholar]
- 75.Arber DA, Orazi A, Hasserjian RP, et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: integrating morphologic, clinical, and genomic data. Blood. 2022;140:1200–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bernstein R, Bagg A, Pinto M, et al. Chromosome 3q21 abnormalities associated with hyperactive thrombopoiesis in acute blastic transformation of chronic myeloid leukemia. Blood. 1986;68:652–657. [PubMed] [Google Scholar]
- 77.Gröschel S, Sanders MA, Hoogenboezem R, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–381. [DOI] [PubMed] [Google Scholar]
- 78.Yamazaki H, Suzuki M, Otsuki A, et al. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv (3)(q21; q26) by activating EVI1 expression. Cancer Cell. 2014;25:415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Katayama S, Suzuki M, Yamaoka A, et al. GATA2 haploinsufficiency accelerates EVI1-driven leukemogenesis. Blood. 2017;130:908–919. [DOI] [PubMed] [Google Scholar]
- 80.Hahn CN, Chong C-E, Carmichael CL, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43:1012–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huber S, Haferlach T, Meggendorfer M, et al. SF3B1 mutations in AML are strongly associated with MECOM rearrangements and may be indicative of an MDS pre-phase. Leukemia. 2022;36:2927–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tanaka A, Nakano TA, Nomura M, et al. Aberrant EVI1 splicing contributes to EVI1-rearranged leukemia. Blood. 2022;140:875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ottema S, Mulet-Lazaro R, Erpelinck-Verschueren C, et al. The leukemic oncogene EVI1 hijacks a MYC super-enhancer by CTCF-facilitated loops. Nat Commun. 2021;12:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Beck D, Thoms JA, Perera D, et al. Genome-wide analysis of transcriptional regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. Blood. 2013;122:e12–e22. [DOI] [PubMed] [Google Scholar]
- 85.Ottema S, Mulet-Lazaro R, Beverloo HB, et al. Atypical 3q26/MECOM rearrangements genocopy inv (3)/t (3; 3) in acute myeloid leukemia. Blood. 2020;136:224–234. [DOI] [PubMed] [Google Scholar]
- 86.Storlazzi C, Anelli L, Albano F, et al. A novel chromosomal translocation t (3; 7)(q26; q21) in myeloid leukemia resulting in overexpression of EVI1. Ann Hematol. 2004;83:78–83. [DOI] [PubMed] [Google Scholar]
- 87.Chacon D, Beck D, Perera D, et al. BloodChIP: a database of comparative genome-wide transcription factor binding profiles in human blood cells. Nucleic Acids Res. 2014;42(D1):D172–D177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haferlach C, Bacher U, Grossmann V, et al. Three novel cytogenetically cryptic EVI1 rearrangements associated with increased EVI1 expression and poor prognosis identified in 27 acute myeloid leukemia cases. Genes Chromosomes Cancer. 2012;51:1079–1085. [DOI] [PubMed] [Google Scholar]
- 89.D’Angiò M, Fazio G, Grioni A, et al. High EVI1 expression due to NRIP1/EVI1 fusion in therapy-related acute myeloid leukemia: description of the first pediatric case. HemaSphere. 2020;4:e471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grasedieck S, Cabantog A, MacPhee L, et al. The retinoic acid receptor co-factor NRIP1 is uniquely upregulated and represents a therapeutic target in acute myeloid leukemia with chromosome 3q rearrangements. Haematologica. 2020;107:1758–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.De Weer A, Speleman F, Cauwelier B, et al. EVI1 overexpression in t (3; 17) positive myeloid malignancies results from juxtaposition of EVI1 to the MSI2 locus at 17q22. Haematologica. 2008;93:1903–1907. [DOI] [PubMed] [Google Scholar]
- 92.Stevens-Kroef M, Poppe B, van Zelderen-Bhola S, et al. Translocation t (2; 3)(p15–23; q26–27) in myeloid malignancies: report of 21 new cases, clinical, cytogenetic and molecular genetic features. Leukemia. 2004;18:1108–1114. [DOI] [PubMed] [Google Scholar]
- 93.Trubia M, Albano F, Cavazzini F, et al. Characterization of a recurrent translocation t (2; 3)(p15–22; q26) occurring in acute myeloid leukaemia. Leukemia. 2006;20:48–54. [DOI] [PubMed] [Google Scholar]
- 94.Meyer S, Bristow C, Wappett M, et al. Fanconi anemia (FA)–associated 3q gains in leukemic transformation consistently target EVI1, but do not affect low TERC expression in FA. Blood. 2011;117:6047–6050. [DOI] [PubMed] [Google Scholar]
- 95.Quentin S, Cuccuini W, Ceccaldi R, et al. Myelodysplasia and leukemia of Fanconi anemia are associated with a specific pattern of genomic abnormalities that includes cryptic RUNX1/AML1 lesions. Blood. 2011;117:e161–e170. [DOI] [PubMed] [Google Scholar]
- 96.Cioc AM, Wagner JE, MacMillan ML, et al. Diagnosis of myelodysplastic syndrome among a cohort of 119 patients with Fanconi anemia: morphologic and cytogenetic characteristics. Am J Clin Pathol. 2010;133:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tönnies H, Huber S, Kühl JS, et al. Clonal chromosomal aberrations in bone marrow cells of Fanconi anemia patients: gains of the chromosomal segment 3q26q29 as an adverse risk factor. Blood. 2003;101:3872–3874. [DOI] [PubMed] [Google Scholar]
- 98.Maicas M, Vázquez I, Vicente C, et al. Functional characterization of the promoter region of the human EVI1 gene in acute myeloid leukemia: RUNX1 and ELK1 directly regulate its transcription. Oncogene. 2013;32:2069–2078. [DOI] [PubMed] [Google Scholar]
- 99.Arai S, Yoshimi A, Shimabe M, et al. Evi-1 is a transcriptional target of mixed-lineage leukemia oncoproteins in hematopoietic stem cells. Blood. 2011;117:6304–6314. [DOI] [PubMed] [Google Scholar]
- 100.Mucenski M, Taylor B, Ihle J, et al. Identification of a common ecotropic viral integration site, Evi-1, in the DNA of AKXD murine myeloid tumors. Mol Cell Biol. 1988;8:301–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Morishita K, Parker DS, Mucenski ML, et al. Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell. 1988;54:831–840. [DOI] [PubMed] [Google Scholar]
- 102.Modlich U, Schambach A, Brugman M, et al. Leukemia induction after a single retroviral vector insertion in Evi1 or Prdm16. Leukemia. 2008;22:1519–1528. [DOI] [PubMed] [Google Scholar]
- 103.Kustikova O, Fehse B, Modlich U, et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. [DOI] [PubMed] [Google Scholar]
- 104.Stein S, Ott MG, Schultze-Strasser S, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med. 2010;16:198–204. [DOI] [PubMed] [Google Scholar]
- 105.Wang Y, Krivtsov AV, Sinha AU, et al. The Wnt/β-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Krivtsov AV, Figueroa ME, Sinha AU, et al. Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia. 2013;27:852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pandolfi A, Barreyro L, Steidl U. Concise review: preleukemic stem cells: molecular biology and clinical implications of the precursors to leukemia stem cells. Stem Cells Transl Med. 2013;2:143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wierzbinska JA, Toth R, Ishaque N, et al. Methylome-based cell-of-origin modeling (Methyl-COOM) identifies aberrant expression of immune regulatory molecules in CLL. Genome Med. 2020;12:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oakes CC, Seifert M, Assenov Y, et al. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat Genet. 2016;48:253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lovén J, Hoke HA, Lin CY, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kiehlmeier S, Rafiee M-R, Bakr A, et al. Identification of therapeutic targets of the hijacked super-enhancer complex in EVI1-rearranged leukemia. Leukemia. 2021;35:3127–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Verhagen HJ, Smit MA, Rutten A, et al. Primary acute myeloid leukemia cells with overexpression of EVI-1 are sensitive to all-trans retinoic acid. Blood. 2016;127:458–463. [DOI] [PubMed] [Google Scholar]
- 114.Kang C, Kim C-Y, Kim HS, et al. The bromodomain inhibitor JQ1 enhances the responses to all-trans retinoic acid in HL-60 and MV4-11 leukemia cells. Int J Stem Cells. 2018;11:131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Paubelle E, Plesa A, Hayette S, et al. Efficacy of All-Trans-Retinoic Acid in high-risk acute myeloid leukemia with overexpression of EVI1. Oncol Ther. 2019;7:121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fenouille N, Bassil CF, Ben-Sahra I, et al. The creatine kinase pathway is a metabolic vulnerability in EVI1-positive acute myeloid leukemia. Nat Med. 2017;23:301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang X, Qiao Y, Asangani IA, et al. Development of peptidomimetic inhibitors of the ERG gene fusion product in prostate cancer. Cancer Cell. 2017;31:532–548.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Carvajal LA, Neriah DB, Senecal A, et al. Dual inhibition of MDMX and MDM2 as a therapeutic strategy in leukemia. Sci Transl Med. 2018;10:eaao3003. [DOI] [PMC free article] [PubMed] [Google Scholar]