Abstract
Immune checkpoint inhibitor (ICI) therapies used to treat cancer, such as anti-PD-1 antibodies, can induce autoimmune conditions in some individuals. The T cell mechanisms mediating such iatrogenic autoimmunity and their overlap with spontaneous autoimmune diseases remain unclear. Here we compared T cells from the joints of twenty patients with an inflammatory arthritis induced by ICI therapy (ICI-arthritis) to two archetypal autoimmune arthritides, rheumatoid arthritis (RA) and psoriatic arthritis (PsA). Single cell transcriptomic and antigen receptor repertoire analyses highlighted clonal expansion of an activated effector CD8 T cell population in the joints and blood of ICI-arthritis patients. These cells were identified as CD38hiCD127− CD8 T cells, and were uniquely enriched in ICI-arthritis joints compared to RA and PsA and also displayed an elevated interferon signature. In vitro, type I interferon induced CD8 T cells to acquire the ICI-associated CD38hi phenotype and enhanced cytotoxic function. In a cohort of patients with advanced melanoma, ICI therapy dramatically expanded circulating CD38hiCD127− T cells, which were frequently bound by the therapeutic anti-PD-1 drug. In ICI-arthritis patients, drug-bound CD8 T cells in circulation showed marked clonal overlap with drug-bound CD8 T cells from synovial fluid. These results suggest that ICI therapy directly targets CD8 T cells in patients that develop ICI-arthritis and induces an autoimmune pathology that is distinct from prototypical spontaneous autoimmune arthritides.
One-sentence summary:
Checkpoint inhibitor-associated arthritis involves a CD8 T cell response that differs from that seen in common autoimmune arthritides.
Introduction
Immune checkpoint inhibitor (ICI) therapies have markedly improved treatment of advanced cancers through their ability to promote activation of tumor-reactive T cells (1). Prior to this clinical application, targets of ICI therapies, such as PD-1 and CTLA-4, were studied for their critical function in preventing autoimmune disease pathogenesis (2–4). It is now clear that ICI therapies used to treat cancers also unleash autoimmune responses, known as immune related adverse events (irAEs), in >80% of treated patients (1, 5–11). These iatrogenic autoimmune events allow examination of human autoimmunity from an unprecedented vantage point of a defined inciting event, ICI therapy. Identifying the cells directly activated by ICI, and the downstream autoimmune cellular cascades in target tissues, may provide mechanistic insights into human autoimmune diseases and immune tolerance.
Inflammatory arthritis induced by ICI therapy occurs in ~4% of patients treated with antibodies targeting PD-1/PD-L1 or CTLA-4 (12). Clinically, ICI-associated arthritis (ICI-arthritis) resembles the classic inflammatory arthritides, rheumatoid arthritis (RA) and psoriatic arthritis (PsA), with small and large joint pain, swelling, and inflammatory effusions (1, 6–10). ICI-arthritis presents within days to months after ICI initiation and—unlike most irAEs—often persists for months to years, even after ICI discontinuation (7). The affected joints limit mobility and can incur permanent damage that necessitates joint replacement surgery. To subvert this trajectory, ICI-arthritis is commonly treated with immunosuppressive medications used for RA and PsA, despite their potential to counteract the therapeutic anti-tumor response.
Recent analyses of RA synovial joint tissue have defined characteristic T cell features, including high frequencies of PD-1hi CD4 T peripheral helper (Tph) cells and granzyme K+ CD8 T cells (13–15). Whether similar or distinct T cell phenotypes accumulate in ICI-arthritis is unknown. Further, it is not clear whether dysfunctional T cell states, prominent in tumors, also populate in irAE target tissues (16–20). Identifying distinct features of the autoimmune response in target tissues from patients with irAEs following ICI therapy may reveal opportunities for tailored immunosuppression.
Here, we leveraged the clinical experience of a patient with severe, destructive ICI-arthritis who required bilateral knee replacements, which enabled study of tissue-infiltrating T cells across two joints in the same individual. We further studied synovial fluid T cells from an additional 23 patients using transcriptomic, cytometric, and functional analyses to define an expanded, oligoclonal, cytotoxic CD8 T cell population in ICI-arthritis. Comparison of CD8 T cells from ICI-arthritis and common inflammatory arthritides, RA and PsA, demonstrated that this CD8 T cell population, marked by high CD38, HLA-DR and granzyme expression, represents a unique effector T cell phenotype in ICI-arthritis, unlike those found in spontaneous autoimmune arthritides. We identified type I interferon (IFN) as an inducer of this prominent CD8 T cell phenotype in ICI-arthritis joints. We demonstrated that these cells are dramatically expanded in the circulation of ICI-treated cancer patients and circulate with anti-PD-1 drug bound to them. T cell receptor (TCR) repertoire studies further linked drug-bound CD8 T cells in the circulation to those in joints of ICI-arthritis patients. These findings define a predominant T cell phenotype in the target tissue of ICI-induced arthritis and highlight the unique immunologic features of this newly recognized, inducible autoimmune disease.
Results
Activated CD8 T cells accumulate in synovial tissue in ICI-arthritis
We evaluated a patient who developed a severe, destructive inflammatory arthritis following anti-PD-1 therapy for metastatic melanoma. The patient had no known autoimmune conditions and had an excellent tumor response to anti-PD-1 therapy. Two years after initiating ICI therapy, the patient developed persistent joint pain and swelling, without RA-associated autoantibodies. Anti-PD-1 therapy was discontinued, and the patient was treated with methotrexate and glucocorticoids, but arthritis symptoms progressed. One year after arthritis symptom onset, radiographs of the knees showed tricompartmental joint space narrowing and periarticular osteopenia, consistent with an aggressive inflammatory arthritis (Fig. 1a). The patient underwent total knee replacement of the left knee, followed by replacement of the right knee six months later, without any change in immunosuppressive treatment between the two surgeries. Synovial tissue samples from both joints were collected for histologic and cellular analyses.
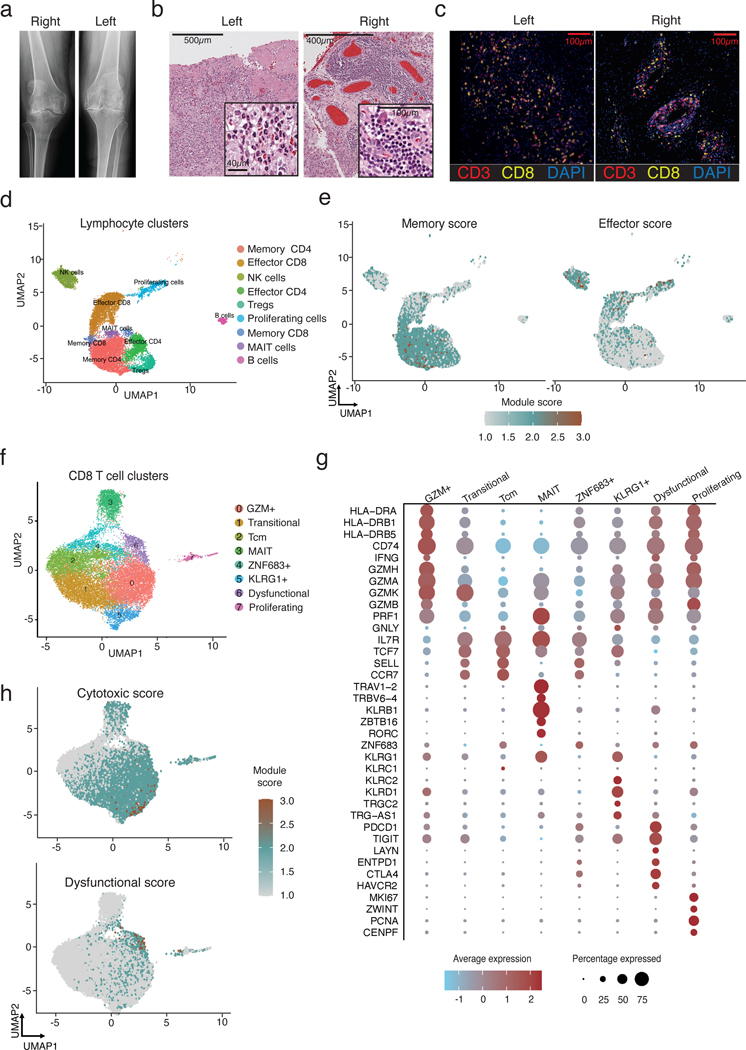
Figure 1. Activated CD8 T cells accumulate in the synovium in ICI-arthritis.
a) Left and right knee radiographs prior to arthroplasty. b) Left and right knee synovial tissue H&E histology. c) Multiplexed immunofluorescence images showing diffuse CD8 T cell infiltration in left knee and aggregate-like CD8 T cell infiltration in right knee. CD3 staining is shown in red, CD8 in yellow, nuclear stain DAPI in blue. d) Synovial tissue (bilateral knee explants from n=1) and synovial fluid (n=5) lymphocytes resolve into 9 distinct clusters using scRNA-seq. e) UMAP visualization of T cell effector and memory signature scores for all lymphocytes in synovial samples. f) Synovial fluid (n=4) and tissue (bilateral knee explants from n=1) CD8 T cells analyzed by scRNA-seq and plotted in clusters in UMAP space. g) Dotplot visualization of differentially expressed genes that distinguish CD8 T cell clusters. Color of the dot represents the average expression of the gene across cells in the cluster. Size of the dot represents the percentage of cells h) UMAP visualization of T cell cytotoxic and dysfunctional signature scores for all CD8 T cells in synovial samples.
Histologic analysis of synovium from the left knee demonstrated features of acute and chronic inflammation, with mononuclear cell and neutrophil infiltrates and fibrin exudate into the synovial cavity (Fig. 1b). Six months later, synovium from the right knee showed extensive lymphocyte aggregates, plasma cells, and Russel bodies; a pattern indistinguishable from RA (Fig. 1b) (21). Immunofluorescence microscopy demonstrated extensive CD3+ T cell infiltration in both knees, although with different patterns, involving a diffuse infiltrate in the left knee and more organized lymphoid aggregates in the right knee (Fig. 1c). Both PD-1 and the proliferation marker Ki67 were detected on the synovial T cells (Fig. S1).
To further evaluate the cellular infiltrate, we performed single cell RNA-sequencing (scRNA-seq) on synovial cells from both knees of this patient, as well as on cells from synovial fluid of an additional 4 patients with ICI-arthritis (Fig. 1d, Fig. S2a-b, Supplementary table 1). Across all ICI-arthritis patients included in the study, the mean age was 61 (range 42–79), with 37 % female, and the mean duration of ICI-treatment of 77 weeks (range 1–333) (Supplementary table 1). The combined single-cell transcriptomic yield across the initial five patients (6 joints) included 14,974 synovial lymphocytes that passed quality filtering (<4,000 detectable genes, <20% mitochondrial genes), which distributed into distinct CD8+ T, CD4+ T, NK, and B cell populations (Fig. S2c-e). A sizable population of effector CD8 T cells were found in both synovial tissue and fluid and exhibited elevated activation and effector function signatures with lower levels of a resting memory signature (Fig. 1d-e, S2f, Supplementary table 2). In contrast, the majority of CD4 T cells expressed a memory phenotype (Fig. 1d,e). Proliferating cells (MKI67+) formed a discrete cluster in which ~50% of the T cells (CD3D+CD3E+) expressed CD8A, 35% expressed CD4 and the remaining 15% were absent for both markers. Thus, synovial tissue and fluid from patients with ICI-arthritis contain a large population of activated CD8 T cells.
Synovial CD8 T cells are clonally expanded and actively proliferating in ICI-arthritis joints
To examine CD8 T cells from ICI-arthritis joints more thoroughly, we sorted CD8 T cells and performed paired single-cell antigen receptor and transcriptomic sequencing. The samples included the two bilateral knee tissues, as well as synovial fluid from four patients (three of which were analyzed in Fig. 1d) (Fig. S3a). From 18,472 synovial CD8 T cells that passed quality filtering, we defined eight transcriptionally distinct clusters, which were comparably represented across all samples (Fig. 1f, S3b,c). These included effector, memory and dysfunctional CD8 T cells. The largest cluster, Cluster 0 (GZM+), featured high expression of HLA-DRA, CD74, IFNG, and multiple granzymes including GZMA, GZMB, GZMH, and GZMK and scored highest for a cytotoxic signature (Fig. 1f-h, Supplementary table 2). In contrast, Clusters 1, 2 and 4 showed higher expression of central memory markers including IL7R (encoding the CD127 protein), TCF7, SELL, and CCR7. Cluster 1 shared features with Cluster 0, such as GZMK, and Cluster 2, a central memory like (Tcm) cluster with TCF7, SELL, and CCR7 and appeared as a transitional population between these two phenotypes. Cluster 4 (ZNF683) was further distinguished by expression of ZNF683, a tissue-resident memory marker. Cluster 3, consistent with a mucosal-associated invariant T (MAIT) cell phenotype, contained cells with high levels of KLRB1, RORC, ZBTB16, and the TCR chains TRAV1–2 and TRBV6–4. Cluster 5 (KLRG1+) contained a mixture of γδ and CD8 T cells expressing the effector markers KLRG1, KLRC1/2, KLRD1 and XCL1/2. Cluster 6 (Dysfunctional) was distinguished by high expression of genes associated with dysfunction, including PCDC1, ENTPD1, CTLA4, HAVCR2, and TIGIT. These cells also scored highest for a gene module defined from dysfunctional tumor-infiltrating lymphocytes, further suggesting that the small Cluster 6 represented dysfunctional T cells in ICI-arthritis joints (Fig. 1h, Supplementary table 2) (22). Cluster 7 (Proliferating) captured proliferating cells, marked by strong expression of cell-cycle genes including MKI67, ZWINT, CENPF, and PCNA (Fig. 1f,g).
We then used the T cell receptor (TCR) sequences to examine clonal distributions within and across the transcriptionally defined CD8 T cell clusters. The Tcm cluster contained the highest number and frequency of cells with a unique TCR (~1,500, 60%). In contrast, despite having almost twice as many total cells, the GZM+ cluster contained fewer cells with a unique TCR (~900) and instead exhibited extensive clonal expansion with 80% of the cells sharing their TCR with other cells (Fig. 2a). In this cluster, the six largest clonotypes were populated by over 100 T cell clones. Across the dataset, the largest clonotypes (accounting for >5% of all CD8 T cells from a patient) were concentrated in the GZM+ cluster, with representation in the Transitional cluster as well (Fig. 2a,b). Consistent with these observations, the GZM+ and MAIT clusters exhibited the highest clonality and therein the lowest repertoire diversity score by the Simpson Index and by the Shannon Equitability metric, which accounts for variability in abundance and evenness (Fig. S3d).
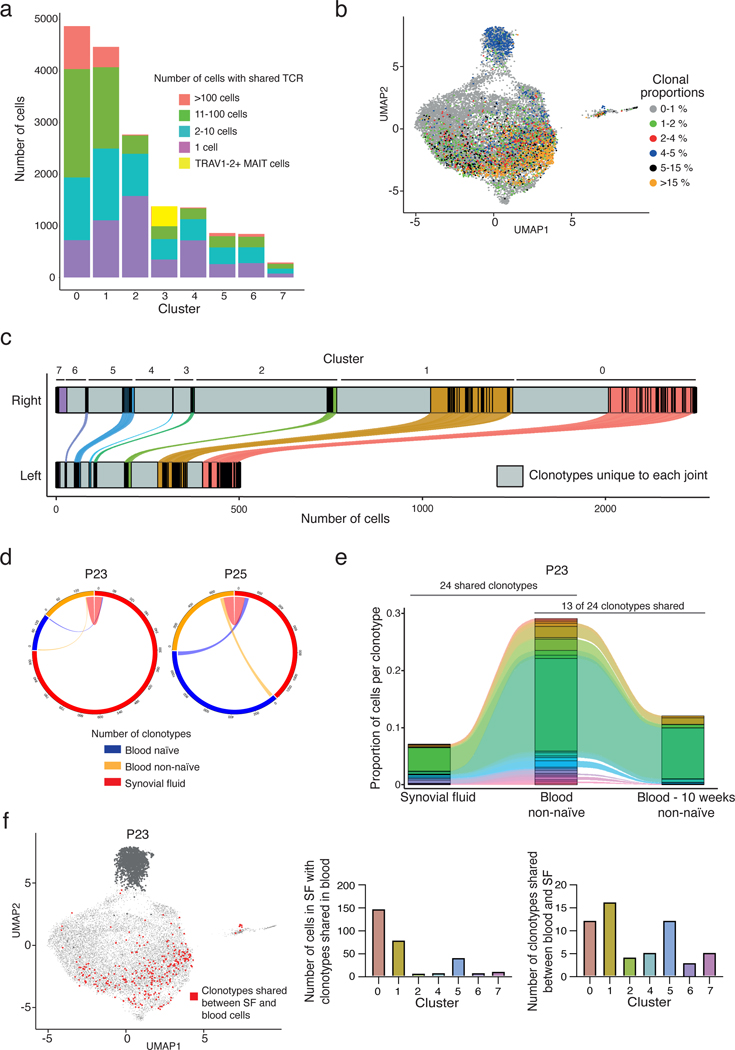
Figure 2. Extensive clonal expansion and proliferation of synovial CD8 T cells in ICI-arthritis.
a) The number of cells in each synovial CD8 T cell cluster that either contains a unique TCR (purple) or a shared TCR (all other colors, for which the total number of clones from the sample is represented by the specific color). Yellow in Cluster 3 represents TRAV1–2+ invariant MAIT cells. b) Expanded TCR clones (>1% of T cells from a patient) depicted onto the transcriptionally defined UMAP clustering. c) Number of distinct (gray) and shared CD8 TCR clonotypes in left and right knee from each transcriptionally defined cluster. Colored bars represent clonotypes in each cluster that can be found in both joints. Distance between black lines in the colored segment represents the size (number of cells) of shared clonotypes. d) Circle plot showing the number of clonotypes expressed in and shared between paired synovial fluid and blood CD8 T cells (n=2 patients). e) Representative alluvial plot of the proportion of shared clonotypes between paired synovial fluid and non-naïve blood CD8 T cells further compared to non-naïve blood CD8 T cells collected 10 weeks later for P23. f) UMAP representation of clonotypes shared between paired synovial fluid CD8 T cells and blood naïve and non-naïve CD8 T cells in P23. MAIT cells were excluded in the analysis (dark gray dots). Bar graphs show the number of cells contained in the shared clonotypes and the number of shared clonotypes in each cluster. Cluster IDs for figures (a), (c) and (f): 0 - GZM+, 1 - Transitional, 2 - Tcm, 3 - MAIT, 4 - ZNF683+, 5 - KLRG1+, 6 - Dysfunctional, and 7 - Proliferating.
Further assessment of TCR overlap between clusters indicated that the GZM+ cells were most highly related to transitional, KLRG1+, and proliferating clusters, with ~30% of their TCR clonotypes shared with each of these clusters (Fig. S3e). The proliferating cluster was largely populated by TCR clonotypes from the GZM+ and KLRG1+ clusters (7.2% and 6.7%, respectively). Together, these data suggest that the large GZM+ population results from extensive clonal expansion of locally proliferating cells in the ICI-arthritis joint.
The bilateral knee synovial tissue allowed us to evaluate the extent of clonal sharing between two joints in the same patient. Strikingly, T cells from the left and right knees demonstrated considerable sharing of TCR clonotypes, particularly among T cells within the GZM+ and transitional clusters, suggesting that the expanded effector clones are present systemically and not restricted to a single site (Fig. 2c, Fig. S3f).
Considering the abundant sharing across joints, we then investigated whether ICI-arthritis synovial clonotypes could also be found in the circulation. Using blood from ICI-arthritis patients with pair-matched synovial fluid, we sorted naïve (CD45RA+CCR7+) and non-naïve (CD45RA− and CD45RA+CCR7−) CD8 T cells and performed single-cell transcriptomic and TCR-sequencing (Fig. S4a). MAIT cells were excluded from this clonal sharing analysis as TCR relatedness is established during development via invariant TCR sequences and does not necessarily indicate clonal expansion. Across synovial fluid and blood from ICI-arthritis patients, we identified considerable sharing of TCR clonotypes particularly between the joint and the circulating non-naïve subset (27 joint-blood shared clonotypes for P23 and 106 for P25) (Fig. 2d-f, S4b). Further, for P23, ~50% (13 of 24) of the joint-blood shared clonotypes identified during an initial visit persisted in the blood during a follow-up visit 10 weeks later (Fig. 2e).
The shared joint-blood clonotypes were largely populated by clones from three synovial CD8 T cell clusters: the GZM+, transitional, and proliferating. This included sharing of 50% and 40% of the GZM+ cells from P23 and P25, respectively, plus 27% and 41% from Transitional clusters (Fig. 2f, S4c). Using the McPAS-TCR database of virus-specific TCRs, we found no known antiviral TCRs among the shared clonotypes between blood and synovial fluid (23). These results indicate that CD8 T cell clones with extensive clonal expansion in ICI-arthritis joints can be found in the circulation in a state consistent with previous activation and that they persist over time.
Activated CD38hi CD8 T cells distinguish ICI-arthritis from spontaneous autoimmune arthritis
The extent to which immune responses in irAEs resemble spontaneous autoimmune diseases remains unclear; therefore, we compared the synovial fluid T cell phenotypes in ICI-arthritis patients to those in RA and PsA in a complementary approach using mass cytometry of an independent cohort of patients. Synovial fluid mononuclear cells from six ICI-arthritis patients, alongside five RA and five PsA patients (Supplementary table 1), were analyzed by mass cytometry using a panel that included multiple T cell activation markers (Supplementary table 3). For PD-1 detection, we used an antibody that binds regardless of a bound therapeutic anti-PD-1 antibody (Fig. S5a). Among synovial fluid mononuclear cells, T cells were the largest population accounting for ~50% of cells, followed by monocytes, NK, and then B cells. Across the three diseases, these proportions were comparable, as were the frequencies of CD4 and CD8 T cells (Fig. S5b,c). The RA samples contained a PD-1hiCXCR5− CD4 T peripheral helper (Tph) cell population (~30% of CD4 T cells), consistent with prior observations (Fig. S5d) (13). In ICI-arthritis samples, the frequency of PD-1hi Tph cells was significantly lower and more comparable to PsA samples (~10% of CD4 T cells, p<0.05) (Fig. S5d).
Visualization of the mass cytometry profiles of synovial fluid CD8 T cells revealed multiple populations with heterogeneous expression of markers for activation (CD38, HLA-DR) and memory states (CD127, encoded by IL7R), as was observed by scRNA-seq (Fig. 3a, S6a,b). An unbiased comparison of the CD8 T cell phenotypes in ICI-arthritis, RA, and PsA by FlowSOM revealed a significant expansion of three CD8 T cell populations (metaclusters 1–3), with 5- and 3-fold higher levels than RA and PsA joints, respectively (p<0.05 vs PsA, p<0.001 vs RA) (Fig. 3a,b). Cells within the three metaclusters shared a profile of activation, with high expression of CD38 and HLA-DR and low expression of CD127, and together comprised ~33% of the CD8 T cells in ICI-arthritis samples (Fig. 3b-d). Biaxial gating validated the unbiased clustering result, demonstrating marked expansion of CD38hiCD127− CD8 cells in ICI-arthritis samples compared to RA and PsA (Fig. 3e,f). Metacluster 5 was significantly reduced in ICI-arthritis samples compared to RA (p<0.001); this cluster showed few distinguishing features and few markers of activation, suggesting an unactivated status.
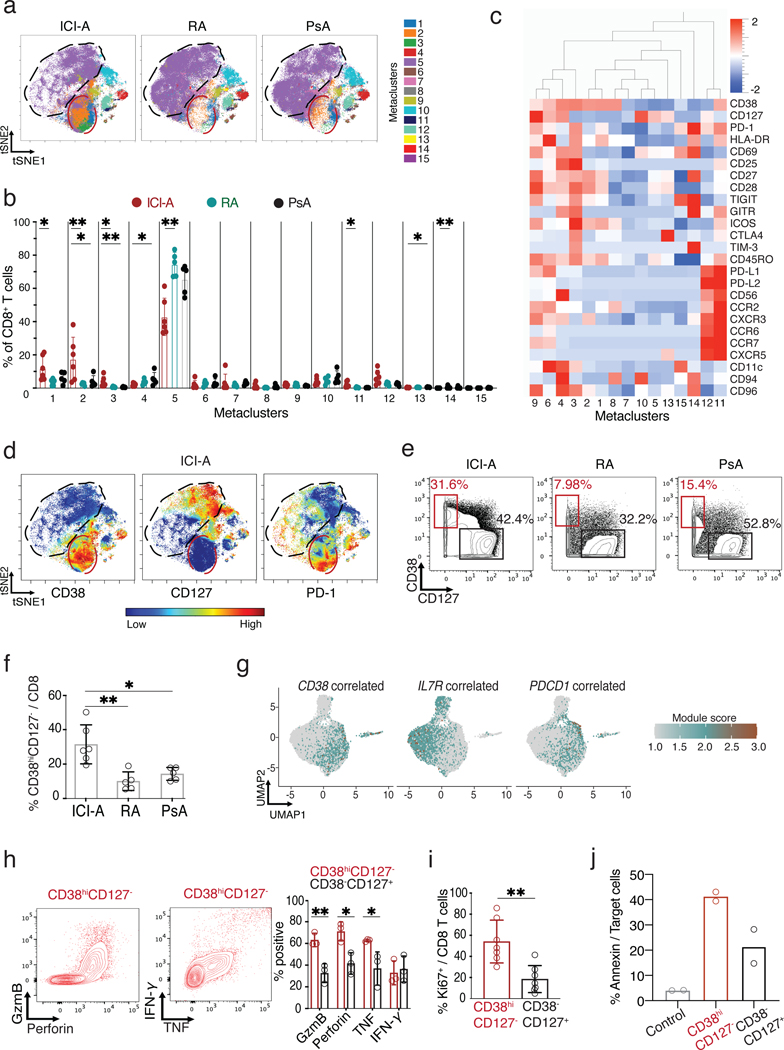
Figure 3. Expansion of CD38hiCD127− CD8 T cells in ICI-arthritis (ICI-A).
a) tSNE visualization of FlowSOM metaclusters of CD8 T cells from ICI-A (n=6), RA (n=5) and PsA (n=5) synovial fluid. The red circle indicates area that includes MC1–3 with increased density in ICI-A. Black circle indicates area of MC5 with lower density in ICI-A. b) Frequency of FlowSOM metaclusters of CD8 T cells from ICI-A, RA and PsA synovial fluid. c) Heatmap of marker expression in CD8 T cell metaclusters. d) tSNE plots of mass cytometry data showing expression of indicated markers on CD8 T cells from ICI-A, RA and PsA synovial fluid. e,f) Biaxial gating (e) and quantification (f) of CD38hiCD127- cells among CD8 T cells from ICI-A, RA and PsA synovial fluid detected by mass cytometry. g) UMAP overlay of signature scores for gene sets that correlate with CD38, IL7R (CD127) or PDCD1 (PD-1) expression derived from bulk RNA-seq data. h) Representative flow cytometric plots and summarized frequency of intracellular granzyme B, perforin, IFN-γ, and TNF in CD38hiCD127− and CD38−CD127+ populations from ICI-A synovial fluid, detected after PMA/ionomycin stimulation (n=3 donors). i) Frequency of intracellular Ki67 in sorted CD38hiCD127− and CD38−CD127+ populations from ICI-A synovial fluid (n=7). Mean ± SD shown. j) Cytotoxicity assay of sorted CD38hiCD127− or CD38−CD127+ CD8 T cells from synovial fluid. Control condition contains only target cells without CD8 T cells. Data represents ≥6 individual experiments. *p<0.05, **p<0.001, ***p<0.0001 by Kruskal-Wallis test in (b), (f), (h) and (i).
The high expression of HLA-DR and absence of CD127 on the CD38hi cells suggested that these cells might represent the same population as the GZM+ cluster in the scRNA-seq analyses, which also expressed little IL7R (Fig. 1f,g). CD38 gene expression was poorly detected in the scRNA-seq data, even though ~30% of these synovial CD8 T cells expressed CD38 protein by flow cytometry, precluding direct assessment of CD38 expression in the scRNA-seq clusters (Fig. S3a). Therefore, we determined the transcriptional program of the CD38hiCD127− CD8 T cells in ICI-arthritis joints in more detail by sorting synovial fluid T cell subsets from an independent cohort (n=6–7 patients each for ICI-arthritis, RA and PsA) for bulk RNA-sequencing (Fig. S7a,b, Supplementary table 1). We sorted five CD8 T cell subsets based on patterns from the mass cytometry profiles: CD38hiCD127−PD-1int, two CD38−CD127+ subsets with little or no PD-1, PD-1hi (dysfunctional), and CD38−CD127−KLRG1+ (cytotoxic). Consistent with mass cytometry results, flow cytometry sorting validated CD38hiCD127− cells as the largest CD8+ subset in ICI-arthritis joints (Fig. S7b).
To demonstrate a relationship between CD38hiCD127− cells and the scRNA-seq clusters, we identified a set of genes that strongly correlated with CD38 expression across all populations in the bulk RNA-seq dataset (Pearson correlation coefficient r > 0.6) (Supplementary table 4 and 5) and visualized expression of this gene set in the scRNA-seq clusters. scRNA-seq GZM+ cluster showed the highest expression of CD38-correlated genes (Fig. 3g). Conversely, the transitional and Tcm displayed the opposite pattern, with high expression of IL7R and high scores for a set of genes correlated with IL7R expression in the bulk RNA-seq dataset (Fig. 3g, Supplementary table 5). Cells from the dysfunctional cluster had the highest PDCD1 expression and scored highest for genes associated with PD-1 in T cells (Fig. 3g, Supplementary table 5). We conclude that CD38hiCD127− cells identified by mass cytometry correspond to GZM+ cluster cells visualized by scRNA-seq.
Analysis of the deeper bulk RNA-seq confirmed the transcriptomic patterns indicated by scRNA-seq. Hierarchical clustering with the 100 most variable genes clearly separated CD38hiCD127− and PD-1hi cells from the CD38−CD127+ cells (Fig. S7c and Supplementary table 4). ICI-arthritis CD38hiCD127− transcriptomes expressed elevated levels of genes associated with effector, cytotoxic, dysfunctional and proliferation gene modules (Fig. S7c,d, Supplementary table 6) (17–20, 24–27). PD-1hi T cells showed a similar pattern, except for a lower score for cytotoxicity. In contrast, CD38−CD127+ cells preferentially expressed genes associated with memory states (Fig. S7d). These populations also exhibited differential expression of chemokines and chemokine receptors, indicating potential differences in migratory capacity or localization (Fig. S7e).
We next used immunofluorescence microscopy to confirm the presence of CD38+ CD8 T cells ICI-arthritis synovial tissue. CD38+ CD8 T cells could be observed in synovial tissue from both the left and right knee of the ICI-arthritis patient described above (Fig. S8). To demonstrate effector functions of these cells at the protein level, we sorted CD8 T cell subpopulations based on CD38 and CD127 expression from ICI-arthritis synovial fluid samples and quantified granzyme B, perforin, IFN-γ and TNF by flow cytometry after PMA/ionomycin stimulation. Compared to CD38−CD127+ cells, CD38hiCD127− cells more frequently expressed granzyme B, perforin, and TNF, consistent with transcriptomic results (Fig. 3h). CD38hiCD127− cells were also more likely to express the Ki67 protein, consistent with the proliferative gene signature (Fig. 3i). We also assessed the cytotoxic function of CD38hiCD127− CD8 T cells in ICI-arthritis synovial fluid. We developed a cytotoxicity assay using a murine fibroblast cell line that expresses the human Fc receptor CD32 and can be loaded with an agonist anti-CD3 antibody (28). Upon co-culturing, CD8 T cells become stimulated to kill the target fibroblasts, measurable by annexin V positivity. We sorted CD38hiCD127− and CD38−CD127+ CD8 T cells out of the synovial fluid of two separate patients and assessed cytotoxicity (Fig. 3j). CD38hiCD127− CD8 T cells killed target cells more so than CD38−CD127+ CD8 T cells in both patients confirming that the observed higher expression of granzyme B and perforin results in increased cytotoxic capacity. Thus scRNA-seq and mass cytometry/bulk RNA-seq analyses converged on an abundant CD38hiCD127− CD8 T cell population, distinct from dysfunctional PD-1hi cells, that is activated, cytotoxic, and specifically expanded in ICI-arthritis joints.
IFN-inducible genes are upregulated in ICI-arthritis synovial T cells
We next sought to identify transcriptomic features that distinguish ICI-arthritis T cells from RA or PsA T cells. Principal component analysis (PCA) on highly variable genes (ANOVA with q-value <0.05) separated ICI-arthritis CD8 T cell subsets from RA and PsA T cells along principal component (PC) 1, suggesting substantial transcriptomic differences (Fig. 4a). Pathway analyses identified four reactome pathways enriched in ICI-arthritis cells, including IFN response pathways (Supplementary table 7). Differential expression analyses comparing the three arthritides also highlighted higher levels of IFN-inducible genes across ICI-arthritis T cells, including OAS1/3, STAT1/2, IFIT1/3, CXCL9/10/11, IFI35/44/44L, IRF1/7/9, ISG15, and MX1 (Supplementary table 8). Direct assessment of 106 IFN-inducible genes showed marked upregulation in ICI-arthritis (vs. RA p=0.0004 and PsA, p<0.0001) (Fig. 4b, Supplementary table 8).
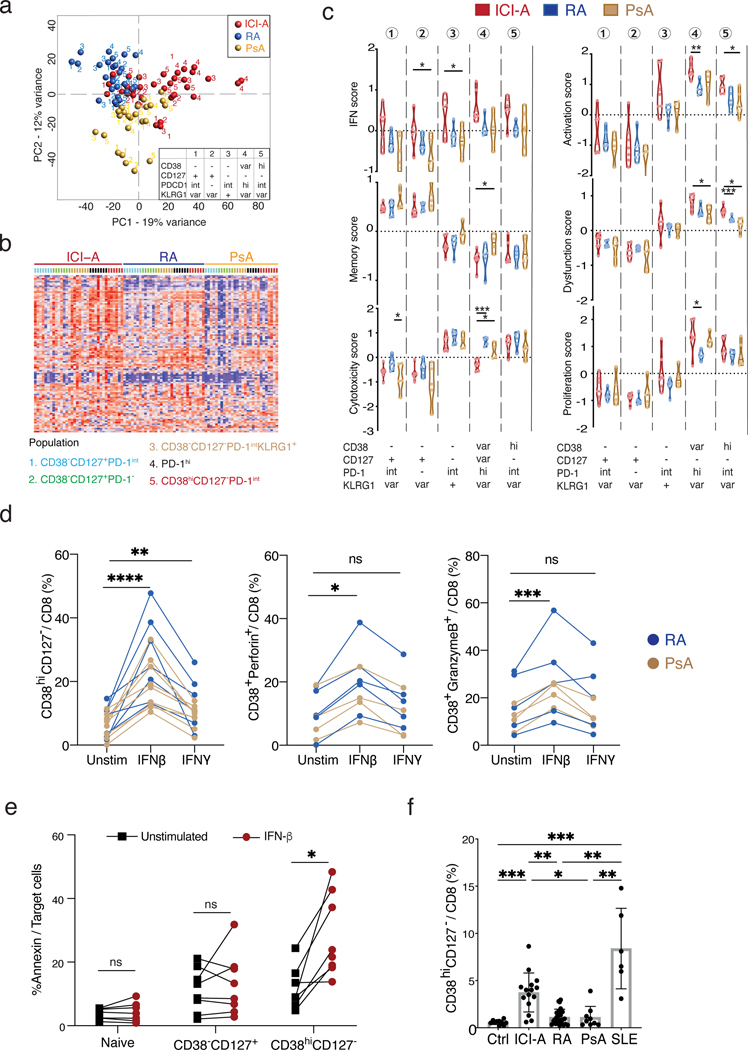
Figure 4. Type I IFN drives the expansion of synovial CD8 T cells in ICI-arthritis.
a) PCA plot showing the expression of 106 IFN-inducible genes across the sorted populations ordered by diseases. b) Heatmap showing clusters of CD8 T cell populations using genes differentially expressed by ICI-A, RA and PsA, with numbers indicating five sorted populations colored by disease. c) Gene module scores of the CD8 T cell populations as in (b) from ICI-A, PsA and RA synovial fluid, calculated based on differentially expressed genes (int = intermediate, var = variable). d) Frequency of CD38hiCD127− cells, CD38hiperforin+ cells and CD38+granzyme B+ cells in CD8 T cells from RA or PsA SFMC cultured with IFN-β or IFN-γ for indicated times. Lines link the same patient sample under the different conditions. Data represents ≥5 individual experiments. e) Cytotoxicity assay of sorted naïve, CD38−CD127+, or CD38hiCD127− CD8 T cells from healthy blood. Data represents 2 individual experiments. f) Frequency of CD38hiCD127− cells among CD8 T cells from PBMC from control (n=10), ICI-A (n=15), RA (n=22), PsA (n=9), and SLE (n=6) patients. Mean ± SD shown. *p<0.05, **p<0.001, ***p<0.0001 by Kruskal-Wallis test in (c) and (f), Wilcoxon matched-pair test in (d) paired T test in (e).
Relative to RA and PsA, the IFN gene module scores appeared higher in ICI-arthritis across all CD8 T cell populations, with the highest levels in CD38hiCD127− and PD-1hi cells (Fig. 4c, Supplementary table 6). The ICI-arthritis CD38hiCD127− cells also demonstrated elevated T cell activation and dysfunctional scores, with similar levels of cytotoxic, proliferative, and cytokine-response pathways compared to RA or PsA counterparts (Fig. 4c). A focused analysis on cytokines demonstrated high expression of IFNG and IL10 in ICI-arthritis CD38hiCD127− and PD-1hi subsets, with similar levels for most other cytokines across the diseases (Fig. S9a). Further, an analysis examining more than 1000 transcription factors found only eight that were upregulated in the ICI-arthritis CD38hiCD127− T cells compared to RA/PsA. Of these eight transcription factors, three are the classical constituents (STAT1, STAT2 and IRF9) of the IFN-stimulated gene factor 3 (ISGF3) complex that is known to mediate type I IFN signaling responses (Fig. S9b and Supplementary table 8) (29).
Based on the elevation of interferon inducible genes and the ISGF3 complex, we hypothesized that type I IFN may induce the CD38hi T cell phenotype that is expanded in ICI-arthritis. To test this, we treated synovial fluid mononuclear cells from RA or PsA patients with IFN-β (type I IFN) or IFN-γ in vitro. Treatment with IFN-β, more so than IFN-γ, induced RA and PsA synovial fluid T cells to acquire a CD38hiCD127− profile. These CD38hiCD127− cells expressed more perforin, granzyme B and Ki67 than did CD38−CD127+ cells (Fig. S10a,b). This trend was consistent, even at maximal concentrations of each cytokine (Fig. 4d, S10c). IFN-β increased CD38hiperforin+ cells and CD38higranzymeB+ cells and increased the frequency of perforin in both CD38hiCD127− and CD38−CD127+ cells. Additionally, IFN-β increased the frequency of granzyme B in total CD8 T cells (Fig. 4d, S10a). This appeared to be a direct effect of IFN-β on CD8 T cells, as treatment of purified CD8 T cells sorted from RA or PsA synovial fluid with IFN-β similarly increased the frequency of CD38hiCD127−, CD38hiperforin+, and CD38higranzymeB+ CD8 T cells (Fig. S10b). In contrast, treatment of synovial fluid mononuclear cells from ICI-arthritis patients with IFN-β or IFN-γ resulted in only a modest increase in the frequency of CD38hiCD127− and CD38hiPerforin+ CD8 T cells (Fig. S10a), which may be consistent with their prior exposure to type I IFN in vivo. These results implicate type I IFN in the induction of the cytotoxic CD38hiCD127− CD8 T cell phenotype prominent in ICI-arthritis.
We next assessed the effect of IFN-β on the cytotoxic function of CD38hiCD127− CD8 T cells. We sorted naïve as well as non-naïve CD8 T cells from healthy control PBMCs based on expression of CD38 and CD127 and assessed cytotoxic function after 3 days of culture with and without IFN-β (Fig. 4e, S10d). As expected, non-naive T cells killed target cells more effectively than naive T cells. IFN-β substantially increased the cytotoxic function of CD38hiCD127− cells but did not significantly increase the cytotoxic function of CD38−CD127+ cells (Fig. 4e, n = 7 donors; p<0.05). Thus IFN-β can increase the cytotoxic function of CD38hi CD8 T cells.
CD38hiCD127− CD8 T cells are expanded in the circulation of ICI-treated patients
We next used flow cytometry to compare the frequency of CD38hiCD127− CD8 T cells in the circulation of ICI-arthritis patients to that of healthy controls or patients with spontaneous autoimmune diseases (30). CD38hiCD127− CD8 T cells were increased in the blood of patients with systemic lupus erythematosus (SLE), a disease characterized by high type I IFN production (Fig. 4f, S10e,f) (16). ICI-arthritis patients had a significantly higher frequency of circulating CD38hiCD127− CD8 T cells relative to RA and PsA patients (p<0.001, p<0.05, respectively) (Fig. 4f). However, despite the elevated circulating CD38hiCD127− cell frequency in the blood of ICI-arthritis patients, an increased IFN signature was not consistently detected, suggesting that the IFN activity may be locally restricted to the joints, while the cellular phenotype is detectable systemically (Fig. S10f).
Since CD38hiCD127− CD8 T cells were expanded in the circulation of ICI-arthritis patients, we addressed whether this is a phenomenon common to patients initiating ICI therapy. Blood samples were obtained from patients with advanced melanoma enrolled in a clinical trial, both before and 6 weeks after initiating combination ICI therapy (anti-PD-1 nivolumab plus anti-CTLA-4 ipilimumab) (n = 48; 31 with paired pre/post samples) (31). Of patients in this cohort, 94% developed an irAE (57% high grade), and 60% of these irAEs had occurred by the time of the 6-week blood sample collection. PBMCs from these patients were analyzed by mass cytometry using a T cell-focused panel (Fig. S11a). Unsupervised clustering of the CD8 T cells resulted in 11 distinct clusters (Fig. 5a).
Figure 5. Expansion of CD38hiCD127− CD8 T cells following ICI-therapy.
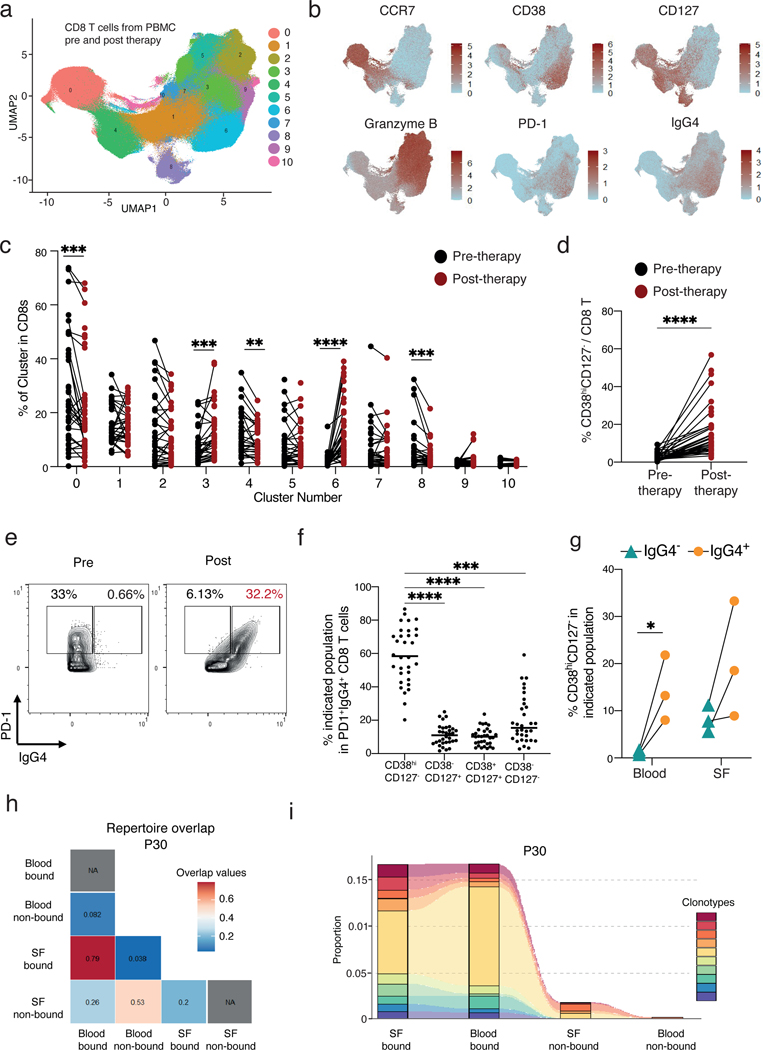
a) UMAP visualization of CD8 T cells plotted in clusters from mass cytometry analysis of melanoma patient PBMC before and after anti-PD-1 and anti-CTLA-4 therapy. b) Expression of markers present in the CD8 T clusters shown in the UMAP space. c) Distribution of CD8 T cells in the clusters depicted in (a). Black lines connect paired patients before and 6-weeks post initiation of therapy. d) Frequency of CD38hiCD127− CD8 T cells among total CD8 T cells pre and post initiation of therapy. e) Example of the gating strategy of the population divided in (f). f) PD-1+IgG4+ CD8 T cells are divided by their expression of CD38 and CD127. Statistical significance as compared to CD38hiCD127− is depicted. g) Frequency of CD38hiCD127− CD8 T cells in IgG4+ or IgG4− CD8 T cells in either blood or synovial fluid. h) Plot denoting the Morisita index value for each comparison demonstrating the repertoire overlap. Representative plot of three patients. See Fig S.12. i) Alluvial plot demonstrating the location and overlap of the top 10 most abundant TCR clones in anti-PD-1 bound and not bound blood and synovial fluid in one of the three patients. *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001 by Wilcoxon in (c), (d), and (f) and by Two-tailed Student’s t test (g).
Cluster 6 showed the largest and most consistent increase post-therapy (fold change =10.6, p<0.0001). Cells in this cluster were characterized by high levels of CD38, HLA-DR, and Ki67 and low levels of CD127; thereby closely resembling the CD38hiCD127− CD8 T cells that we observed in ICI-arthritis joints (Fig. 5b, S11b). Additionally, Cluster 6 had high expression of CD39, which has been used as a marker of tumor-reactive CD8 T cells in melanoma (32, 33). A portion of cells in Cluster 6 expressed granzyme B, granzyme A, and perforin while others expressed granzyme K. In addition to Cluster 6, we also observed a significant but smaller increase in the frequency Cluster 3, which was composed of granzyme B expressing cells (p<0.001). There was a significant decrease in the naïve, CD127hi memory, and MAIT cell clusters (Cluster 0, p<0.001; Cluster 4, p<0.001; Cluster 8, p<0.01 respectively) after ICI treatment (Fig. 5c). Biaxial gating for CD38hiCD127- CD8 T cells confirmed a significant increase in this population post ICI therapy (fold change = 12.8, p <0.0001; Fig. 5d). The frequency of CD8 T cells in total did not change post-ICI therapy (Fig. S11c).
We then sought to identify and evaluate cells in circulation that have the therapeutic anti-PD-1 antibody bound to them. To detect these cells, we included in the mass cytometry panel an anti-IgG4 antibody, which can detect either pembrolizumab or nivolumab, two common anti-PD-1 therapeutic antibodies, bound to the cell surface. The anti-IgG4 antibody does not detect ipilimumab, which is an IgG1 antibody. At the 6-week time point, Cluster 6 had the highest level of IgG4, indicating the presence of nivolumab on these cells (34). We again included in the mass cytometry panel an anti-PD-1 antibody that can bind to cell surface PD-1 even in the presence of nivolumab or pembrolizumab and observed that the IgG4+ cells had high levels of PD-1 (Fig. 5b). Biaxial gating of non-naïve PD-1+ CD8 T cells bound by nivolumab (IgG4+) demonstrated that the majority of drug-bound cells in circulation displayed a CD38hiCD127− surface phenotype (59%) (Fig. 5e, f).
Anti-PD-1 drug-bound CD8 T cells in circulation and synovial fluid are clonally linked in ICI arthritis patients
To evaluate the relationship between anti-PD-1 drug-bound cells (IgG4+) in the circulation and in the joints of patients with ICI-arthritis, we assessed anti-PD-1 drug-bound cells in paired synovial fluid and blood from 3 ICI-arthritis patients. The proportion of anti-PD-1 drug-bound CD8 T cells was substantially higher in synovial fluid than in the blood (Fig. 5g). In both synovial fluid and blood samples, the proportion of CD38hiCD127− CD8 T cells was higher among anti-IgG4+ drug-bound cells than among non-drug-bound cells (Fig. 5g).
We hypothesized that the drug-bound CD8 T cells in circulation would be clonally related to the cells that are expanded in ICI-arthritis joints. To evaluate this, we sorted CD8 T cells from these same 3 paired blood and synovial fluid samples, dividing CD8 T cells into anti-IgG4+ (drug-bound) or anti-IgG4- (non-drug-bound) populations, and performed bulk TCR sequencing. We observed a range of clonality in both the drug-bound and non-drug-bound CD8 T cells across the samples (Fig. S12a). Analysis of repertoire similarity using the Morisita index revealed high overlap of TCR sequences between the drug-bound cells in synovial fluid and drug-bound cells in blood (0.79, 0.22, 0.47; for P30, P14, P31 respectively; Fig. 5h, S12b). In contrast, almost no overlap was observed between drug-bound cells in the synovial fluid and non-drug-bound cells in blood (0.04, 0.03, and 0.01). There was also less overlap between drug-bound cells in synovial fluid and non-drug-bound cells in synovial fluid in the 3 patients (0.2, 0.14, and 0.14). Tracking the 10 most abundant clonotypes in the drug-bound synovial fluid CD8 T cells also demonstrated substantial overlap in these large clones between drug-bound cells in synovial fluid and blood, and much less sharing with non-drug-bound cells in all 3 patients (Fig. 5i, S12c). Together, these results demonstrate that anti-PD-1 drug-bound CD8 T cells in the circulation of patients with ICI arthritis are clonally and developmentally related to those active in the irAE target tissue.
Discussion
Here we define the T cell infiltrate within the target organ system of a human autoimmune condition with a known causal mechanism—administration of an ICI therapy that is designed to activate T cells. Cytometric, transcriptomic, TCR repertoire, and functional studies implicate CD38hiCD127− CD8 T cells as cytotoxic effector cells that are clonally expanded within affected joints in ICI-arthritis and are induced in response to type I IFN. These CD8 T cells are often direct targets of anti-PD1 antibodies and are expanded in joints long after ICI-therapy initiation. The accumulation of cytotoxic T cells is emerging as a shared feature in tissues affected by irAEs, with apparent detection in intestine (35), liver (36), skin (37), heart (38, 39), and thyroid (40, 41). We demonstrate cytotoxic function of expanded CD38hiCD127− CD8 T cells as one means by which CD38hiCD127− cells may mediate tissue injury in ICI-arthritis joints. While tumors often contain large populations of dysfunctional T cells (16–20), we found relatively few in ICI-arthritis joints, with a far larger number of CD38hiCD127− effectors. We further demonstrate that the CD38hiCD127− CD8 T cells expanded in ICI-arthritis joints are profoundly expanded in blood following combination anti-PD-1 and anti-CTLA-4 therapy. CD38+ T cells have also been identified in tumors, with varying levels of function (42–44). Further comparisons of the functional and clonal relationships of T cells across tumors and ICI affected joints may help determine the extent to which the anti-tumor and autoimmune responses are linked
Our results are consistent with recent observations of a clonally expanded and activated CD8 T cell population in ICI-arthritis patients (45) and now define the specific T cell phenotype expanded in both synovial tissue and fluid. In ICI-associated colitis, activated CD8 T cells were clonally related to tissue resident T cells suggestive of local activation (46). Our detection of shared expanded T cell clones across two joints in the same patient in addition to our data demonstrating clonal overlap between drug bound CD8 T cells in blood and synovial fluid, suggests these T cells travel between blood and joints. This is consistent with the clonal expansion of CD8 T cells observed in blood from ipilimumab-treated patients prior to the development of severe irAEs (47). The fact that we detected the expanded TCR clonotypes identified in the joint circulating in the blood several months later may provide an immunologic mechanism for the clinical observation that ICI-arthritis can persist for months or years after cessation of ICI therapy (7).
Our comparison of T cells from ICI-arthritis to two common spontaneous arthritides, RA and PsA, highlights key differences in the predominant T cell responses across these conditions. Surprisingly, anti-PD-1 ICI therapy did not substantially expand PD-1hi Tph cells, a population of CD4 T cells with B cell-helper function that is highly enriched in seropositive RA synovium(13). Analyses of T cells from PsA synovium highlighted clonally expanded CD8 T cells with expression of CXCR3. Our analyses of ICI-arthritis similarly detected clonally expanded CD8 T cells but with a clearly distinct phenotype, CD38hiCD127−, suggesting that the predominant T cell effector types differ in these clinical conditions. These differences raise the possibility that therapies developed to treat pathologic T cell responses in RA and PsA may not optimally target the active effector pathways in ICI-arthritis or other irAEs. It is likely that clinical factors may affect the extent of expansion of CD38hiCD127- cells in ICI-arthritis patients; however, our study was not powered to detect these effects.
Transcriptomic comparisons across the diseases highlighted the expression of IFN-inducible genes selectively in ICI-arthritis T cells, and type I IFN was sufficient to induce the CD38hiCD127− perforin- and granzyme B-producing phenotype in synovial T cells and significantly boost T cell cytotoxic function (p<0.05). These results implicate a role for type I IFN in ICI-arthritis and draw further distinction from RA and PsA. Type I IFN has generally not been implicated in PsA pathogenesis and is only modestly elevated in a subset of RA patients (48–50). It is interesting to note that while type I IFN induces strong signatures across the immune cell types in SLE, ICI therapy rarely results in SLE-like disease (9). Further research will be required to determine the source of type I IFN as well as better determine the systemic nature of the type I IFN signal following ICI treatment. Blocking type I IFN responses, as used in treating SLE (51), could be considered for ICI-arthritis. However, this approach involves the risk in compromising the anti-tumor response of ICI therapy where IFNs have a potential role (52–55).
Our study has the following limitations. Our observations in synovial fluid and tissue are made from a small cohort of patients; a larger cohort is required to assess potential heterogeneity in T cell phenotypes and associations with clinical outcome and other irAEs. Further, our study includes patients on anti-PD-1 monotherapy and patients on dual anti-PD-1/anti-CTLA-4 and is underpowered to detect differences between the treatment regimens. Our identification of the almost universal expansion of CD38hiCD127− CD8 T cells following dual ICI therapy suggests that additional factors, including temporal dynamics, need to be considered to utilize CD38hi T cell expansion as a circulating biomarker of irAEs. We have shown that CD38hiCD127− CD8 T cells are abundant within the joints of patients with ICI-arthritis; however, the pathologic function of the cells in vivo has not yet been demonstrated. We provide several plausible mechanisms by which these cells may contribute to joint inflammation, including through direct cytotoxicity and production of inflammatory cytokines, yet further studies are required to assess the clinical effect of disrupting the function of these cells.
In summary, amidst a resurgent appreciation for CD8 T cell pathologic mechanisms in autoimmune diseases (56–59), our study identifies a specific CD8 T effector phenotype that characterizes an autoimmune disease induced by T cell activating ICI therapy.
Materials and Methods
Study Design
Human subjects research was performed according to the Institutional Review Boards at Mass General Brigham (IRB protocol 2014P002558) or Hospital for Special Surgery (IRB protocols 2017–1898 and 2014–233) via approved protocols with informed consent as required. Synovial fluid samples were collected from patients with ICI-arthritis, RA or PsA as discarded fluid from clinically indicated arthrocentesis. Patients with ICI-arthritis developed arthritis after receiving checkpoint inhibitor therapy to treat a malignancy and were diagnosed by experienced rheumatologists. Type and status of cancer, type and duration of CI therapy, tender or swollen joint counts (mono-, oligo- or poly- arthritis), other irAE, prior history of rheumatologic conditions, serum rheumatoid factor (RF) and anti-CCP antibody status, C-reactive protein level, and medication usage were obtained by review of electronic medical records. Seropositive (RF+ and/or anti-CCP+) RA patients fulfilled 2010 ACR/EULAR classification criteria. PsA patients were diagnosed with PsA by their treating rheumatologist. Blood samples were obtained from individuals with ICI-arthritis, ICI-thyroiditis, seropositive RA, PsA, SLE as well as individuals without inflammatory arthritis. Mononuclear cells from synovial fluid and peripheral blood were isolated by density centrifugation using Ficoll-Paque Plus (GE healthcare) and cryopreserved in FBS + 10% DMSO by slow freeze, followed by storage in liquid nitrogen for batched analyses. For experimental analyses, cryopreserved samples were thawed into RPMI medium + 10% FBS.
Multiplexed Immunofluorescence (mIF)
Formalin Fixed Paraffin sections were made from synovial tissue explants from right and left knees of an ICI-arthritis patient. The staining shown in Fig. 1 and Fig. S1 was performed at the Molecular Cytology Core Facility at Memorial Sloan Kettering Cancer Research Center. Antibodies used included anti-CD8A (clone SP57, Ventana), anti-CD3 (polyclonal, Dako), anti-Ki67 (clone D2H10, Cell Signaling Technology) and anti-PD-1 (clone NAT105, Ventana). mIF slides were analyzed using SlideViewer (3DHISTECH v2.5) and the final images were rendered using ImageJ (v2.1.0). The staining shown in Fig. S8 included antibodies anti-CD8 (clone SP16, Invitrogen), anti-CD3 (clone CD3–12, Abcam) and anti-CD38 (clone 38C03 (SPC32), Invitrogen). Secondary antibodies were raised in goat and included anti-rabbit IgG AlexaFluor 488 (Invitrogen) for detection of CD8, anti-rat IgG AlexaFluor 647 (Invitrogen) for detection of CD3 and anti-mouse IgG AlexaFluor 568 (Invitrogen) for detection of CD38. Mounting Medium with DAPI (Abcam) was used for nuclei identification and mounting. The slides for Fig. S8 were imaged using a Zeiss LSM 780 confocal microscope and analyzed using Zen Lite software (Zeiss).
Sample preparation for single cell RNA-seq
Synovial tissue: Fresh synovial tissues were cut into 3-mm3 fragments and preserved in Cryostor (BioLife). For experimental processing, tissues were thawed into RPMI++. Following two subsequent washes in RPMI, the tissues were finely chopped and transferred into 5ml polystyrene tubes containing digestion buffer (5ml/sample) made with RPMI medium + Liberase TL (100μg/ml; Roche) + DNaseI (100μg/ml; Roche). The tubes were placed securely in a MACSmix tube rotator (Miltenyi Biotec) and placed in an incubator at 37°C, 5% CO2 for 30 minutes. The digested tissue was filtered over 70μM cell strainer (BD) and subjected to further mechanical dissociation using a syringe plunger. The eluate was washed with RMPI++ and centrifuged at 1500 rpm for 4 minutes at 4°C. The cells were resuspended in RPMI++ and passed through a second filtration using 40μM cell strainer (BD). Cells were then counted and used for downstream applications.
Synovial fluid and peripheral blood: Mononuclear cells from synovial fluid and blood were isolated using Ficoll-Paque Plus (GE healthcare) and cryopreserved in Cryostor10 (BioLife) in liquid nitrogen. For experimental analyses, cryopreserved samples were thawed into RPMI medium (Corning) + 10% FBS (HyClone) + 1% L-glutamine (Gibco), referred to as RPMI++.
Single cell suspensions from fluid and tissue were subjected to Human TruStain FcX (Biolegend) in FACS buffer (PBS supplemented with 5% FBS (HyClone) prior to staining. The antibodies used for identification of CD8 T-cells were anti-CD3 (APC, clone UCHT1, Biolegend) and CD8 (PerCP/Cyanine5.5, clone RPA-T8, Biolegend). Cells were sorted on a three-laser BD FACS Aria Fusion cell sorter at the Flow Cytometry Core Facility at Weill Cornell Medicine (WCM). Intact cells were gated according to forward scatter and side scatter area (FSC-A and SSC-A). Doublets were excluded by serial FSC-H/FSC-W and SSC-H/SSC-W gates (H, height; W, width). Non-viable cells were excluded based on DAPI uptake. Cells were further selected for CD3 and CD8 surface expression and sorted through a 70μM nozzle at 70 psi. Flow cytometric quantification of cell populations was performed using FlowJo v.10.0.7.
Single cell RNA-seq library preparation
3’ gene expression (GEX) of sorted live synovial cells was prepared using Chromium Single Cell 3’ v3 Kit reagents and protocols provided by 10X genomics. The pooled libraries at 10nM concentration were sequenced using NovaSeq6000 S2 Flow Cell by the Illumina platform at Genomics Research Core Facility at WCM.
5’ gene expression (GEX) and paired TCR libraries for sorted CD8 T cells from synovial fluid and blood were prepared using the 5’ Chromium Next GEM Single Cell v2 (Dual Index) reagents and protocol provided by 10X Genomics. The pooled 5’ GEX and TCR libraries at 10nM concentration were sequenced using NovaSeq6000 S1 Flow Cell by the Illumina platform at Genomics Research Core Facility at WCM.
scRNA-seq data analysis
scRNA-seq data processing and alignment: 10x FASTQ files were processed with the Cellranger count 4.0 pipeline with default parameters. Reads were aligned to the human reference sequence GRCh38. Seurat package (v.4.0.0) was used to perform unbiased clustering of the 5’ GEX of sorted CD8+ T cells and 3’ GEX of sorted synovial cells from our patient samples. QC was performed on the 5’ GEX sorted T cells dataset to remove cells that had less than 50 genes, more than 3000 genes, or >10% mitochondrial gene expression, resulting in a total of 18,472 cells and 19,406 genes (Fig. S3b). The 3’ GEX dataset of sorted live synovial cells had QC performed to also remove cells with less than 150 genes, more than 7500 genes, or > 25% mitochondrial gene expression, resulting in a total of 44,959 cells and 26,424 genes. Both datasets were then log-normalized using a scale factor of 10,000. Potential confounders such as percent mitochondrial gene expression and number of UMI per cell was regressed out during scaling (mean of 0 and variance of 1) for 3’ GEX dataset, while the 5’ GEX dataset had additional batch parameter included to consider samples being from different sequencing runs. Principal component analysis was used with the top 2000 highly variable genes for both datasets. Elbow plot was used to determine the statistically significant principal components, where then for the 5’ and 3’ datasets used the first 21 PCs and 17 PCs, respectively, for follow-up analysis. Harmony (v1.0) was performed to improve integration and correct for batch effects on our samples, with parameters of max.iter.cluster = 30, and max.iter.harmony = 20 and sample as the only covariate. Eight clusters for the 5’ dataset were found at 0.5 resolution and eleven clusters for the 5’ dataset at 0.2 resolution were found, and their identity were annotated based on the expression of differentially expressed genes (DEG) using FindAllMarkers function using default parameters, where only genes detected in at least 25% of the cells in the two comparison sets are used. The AddModuleScore function in Seurat was used to calculate module scores, where the average expression level of genes of interest are subtracted from a randomly aggregated expression of control features, to distinguish clusters. Additional grouping of the 3’ GEX dataset of clusters 1, 3, 6, 10 and 15 at resolution 0.3 was used to define only lymphocytes. These lymphocyte clusters contained 15,562 cells and nine clusters after going through linear dimensional reduction of the top 17 PCs with Harmony as described above.
scRNA sequencing TCR analyses
Cellranger vdj 4.0 pipeline was used to generate clonotype information from 10× 5’ VDJ FASTQ files, with default parameters. Reads were aligned to the human reference sequence GRCh38. Clonotype information was then manually inputted into our Seurat object as metadata information, using the cell barcodes to match the clonotype information to our cells. The integration resulted in 6843 unique clonotypes, where all cells within a clonotype share the same CDR3 alpha and CDR3 beta sequences. This allowed us to explore the relationship between TCR sequence and its phenotype. The immune receptor database McPAS-TCR was used to annotate our clonotypes and match them to condition-associated receptors. As most of the TCR sequences were from bulk sequencing, only the beta-chain of our clonotypes were used in the analysis and we allowed for 1 amino acid mismatch. Additionally, we only included human virus specific TCR sequences from the database, such as CMV and EBV.
Simpson’s diversity index:
where ni is the number of cells within the ith clonotype, c is the total number of unique clonotypes in our cluster, and n is the total number of cells with clonotypes within our cluster. Simpson Diversity Index ranges from 0 to 1, with 0 being a community with no diversity and 1 being a community of the highest diversity.
Shannon Equitability:
E = H / S; where H is the Shannon’s diversity index value and S is the number of clonotypes in that sample. Shannon Equitability ranges from 0 to 100 and is based on the Shannon Diversity Index but takes into account the actual number of unique clonotypes present within each cluster to assign a value that can be directly compared across all clusters. As the value approaches 100, the community is approaching maximum diversity.
Mass cytometry staining of ICI-arthritis synovial fluid cells
Cryopreserved synovial fluid cells were thawed and trypan blue negative viable cells were counted by hemocytometry. Approximately 1 million live cells per sample were used for mass cytometry staining. All antibodies were obtained from the Longwood Medical Area CyTOF Core except for anti-PD-1 antibody (MIH4), which was conjugated with heavy-metal isotope (Fluidigm kit) and validated in house. Buffers were from Fluidigm unless otherwise specified. Cells were stained with cisplatin (Fluidigm) for viability then washed. Surface antibody cocktail was prepared in cell staining buffer (Fluidigm) and added to all samples equally after Fc block (BD). Cells were washed then fixed and permeabilized using eBioscience Transcription Factor Fix/Perm Buffer followed by barcoding (Fluidigm). Barcoded samples were pooled together and stained with the intracellular antibody cocktail in an intracellular staining buffer (Fluidigm). Cells were re-fixed in 4% formalin (Sigma-Aldrich). Intercalator-Ir was diluted in CyTOF PBS and applied to cells. Cells were then washed and resuspended with cell acquisition solution (Fluidigm) containing 1:10 diluted EQ beads (Fluidigm). Acquisition was performed on a CyTOF-Helios mass cytometer (Fluidigm).
Mass cytometry staining of PBMC from ICI-treated melanoma cohort
PBMC were made available from the ADAPT-IT Study from melanoma patients that had received 2 doses of both nivolumab and ipilimumab. PBMC were prepared in a similar manner to the ICI-arthritis synovial fluid samples. Cryopreserved PBMC were thawed, washed, and counted. 2 million live PBMC were stained from each sample. Antibodies were obtained from the Longwood Medical Area CyTOF Core and Fluidigm. Cells were incubated with 103Rh for viability, Fc blocked, surface stained, barcoded, combined, and IC stained as before. Acquisition was performed on a CyTOF-XT (Fluidigm).
Mass cytometry data analysis of ICI-arthritis synovial fluid cells
Cytometry data were normalized and debarcoded as previously described (58). Live cells (DNA+ 195_Pt− 140_Beads−) were first gated prior to gating for specific cell populations using the following scheme: monocytes (CD3−CD14+), B cells (CD14−CD19+), and T cells (CD3+CD14−). Gated populations from 6 ICI-arthritis, 5 RA and 5 PsA were concatenated for high-dimensional analyses using the implementations on Cytobank (www.cytobank.org). For donors with more than 10,000 cells, we randomly selected 10,000 cells to ensure that samples were equally represented. In this way, we created downsampled datasets of 160,000 viable cells or 42,144 CD8+ T cells from 16 samples for analysis. Dimensional reduction was performed using viSNE algorithm (59) using 2000 iterations with a perplexity = 30 and a theta = 0.5. Hierarchical consensus clustering was performed using FlowSOM algorithm to generate 15 metaclusters containing 225 clusters using 100 iterations (60). A metacluster represents an aggregation of cytometrically similar clusters and can be considered a cell ‘population. Antibody channels excluding gating markers were used for analyses. Heatmaps of row-normalized median expression of representing markers in the metaclusters are shown, in which the metaclusters were arranged by hierarchical consensus clustering. Metaclusters and markers of interest were overlaid on tSNE plot for visualization. Manual biaxial gating was performed using FlowJo v.10.4.2 for quality control and independent examination of the expression of markers and frequencies of populations.
Mass cytometry data analysis of PBMC from ICI-treated melanoma cohort
Cytometry data were normalized and debarcoded as before. Live cells were gated as DNA+103Rh-Beads- using FlowJo (BD). Live cells were then corrected for batch effects using CyCombine (61). Batch-corrected total live cells, as well as CD14-CD19-CD3+CD8+ live batch-corrected cells were then analyzed using Seurat for principal component analysis, neighborhood analysis, and clustering visualized by UMAP (62).
Flow cytometry staining
Cryopreserved cells were thawed, washed, and counted. Cells were stained in PBS with Aqua fixable live/dead dye (Invitrogen) for 15 minutes at room temperature and washed.
For surface staining, cells were then stained in PBS with 1% BSA with the following antibodies for 30 minutes at 4°C: anti-CD14-BV510-dump (M5E2), anti-CD25-FITC (M-A251), anti-CD8-BUV395 (RPA-T8), anti-CD4-BV605 (RPA-T4), anti-CD38-PercpCy5.5 (HIT2), anti-CD45RA-BV711 (HI100), anti-PD-1-PE-Cy7 (MIH4), anti-CD3-AF700 (UCHT1), anti-CD127-APC (A019D5) and anti-KLRG1-BV421 (SA231A2) from BioLegend. Cells were washed in cold PBS, passed through a 70μM filter, and data acquired on a BD Fortessa analyzer using FACSDiva software. Data were analyzed using FlowJo 10.4.2. For intracellular staining of transcription factors, cells were processed and stained for viability and indicated surface markers as described above. Cells were washed and incubated with 1x eBioscience Transcription Factor Fixation/Permeabilization Buffer at room temperature for 1 hour. Cells were then washed in 1x eBioscience Permeabilization Buffer twice and incubated with indicated intracellular antibodies including anti-T-bet-BV785 (4B10, BD Biosciences), anti-EOMES-PerCP-ef710 (X4–83, BD Biosciences), anti-granzyme B-AF647 (GB11, Invitrogen), anti-granzyme K-FITC (GM26E7, Invitrogen), anti-perforin-BV421 (dG9, Invitrogen) and anti-Ki-67-BV605 (Ki67, BioLegend) at room temperature for 1 hour. Cells were washed twice in 1x eBioscience Permeabilization Buffer, passed through a 70μM filter, and data acquired on a BD Fortessa analyzer. For intracellular detection of TNF and IFN-γ, sorted cells were stimulated with 1x PMA/ionomycin + Brefeldin A/Monesin (Invitrogen) at 37°C for 2 hours. Cells were stained for viability and indicated surface markers as described above. Cells were washed and incubated with 1x eBioscience Transcription Factor Fixation/Permeabilization Buffer at room temperature for 1 hour. Cells were then washed in 1x eBioscience Permeabilization Buffer twice and incubated with indicated intracellular antibodies including anti-TNF-PE (MAb1) and anti-IFN-γ-APC (4S.B3) from BioLegend at room temperature for 1 hour. Cells were washed twice in 1x eBioscience Permeabilization Buffer, passed through a 70μM filter, and data acquired on a BD Fortessa analyzer.
Flow cytometric cell sorting for bulk RNA-seq
An 8-color flow cytometry panel was developed to identify CD8 T cell populations within SFMC. Antibodies include anti-CD14-BV510 (M5E2), anti-3-Alexa Fluor 700 (UCHT1), anti-CD8-BV510 (RPA-T8), anti-PD-1-PE-Cy7 (MIH4), anti-CD127-APC (A019D5), anti-CD38-PE (HIT2), anti-KLRG1-BV421 (SA231A2) and propidium iodide (all from BioLegend). SFMC were incubated at 4°C with antibodies in HBSS/1% BSA for 30 min. Cells were washed once in HBSS/1% BSA, centrifuged and passed through a 70μM filter and propidium iodide was added immediately prior to sorting. Cells were sorted on a 4-laser BD FACSAria Fusion cell sorter. Intact cells were gated according to forward scatter and side scatter area (FSC-A and SSC-A). Doublets were excluded by serial FSC-H/FSC-W and SSC-H/SSC-W gates (H, height; W, width). Non-viable cells were excluded based on propidium iodide uptake. Cells were sorted through a 70μM nozzle at 70 psi. Cell purity was routinely >98%. For functional analyses, approximately 200,000 cells were sorted from each population into cold RPMI/10% FBS. For RNA-seq, up to 1,500 cells were collected from each cell subset directly into buffer TCL (Qiagen) with 1% β-mercaptoethanol (Sigma). Flow cytometric quantification of cell populations was performed using FlowJo v.10.0.7.
Sample preparation for low-input bulk RNA-seq
RNA was isolated from 1,500 cells from sorted T cell subpopulations. 5μL of total RNA were placed in wells of a 96-well plate and RNA-seq libraries were prepared at Broad Technology Labs at the Broad Institute of Harvard and MIT using the Illumina SmartSeq2 platform. Libraries were sequenced to generate 38 base paired end reads.
Low-input bulk RNA-seq analysis
Sequencing samples were examined with FastQC for quality control and trimmed with trimmomatic. Samples with low sequencing quality were removed and 86 samples were used for downstream analysis. Reads were mapped to GRCh38 (Ensembl release 96) using STAR alignment software. Lowly expressed genes (log2 FPKM<2) were filtered, and the expression of 17,779 genes were used for downstream analysis. TMM normalized read counts were generated and data was log2 transformed, batched corrected and scaling was applied (mean=0, variation=1) when appropriate. Differential expression analysis and gene set enrichment analysis was performed in Qlucore software. In comparisons between two specific cell populations by t-test, genes with log fold change >1.5 and FDR <5% were considered differentially expressed. In comparisons among more than two cell populations by ANOVA, genes with an FDR <1% were considered differentially expressed.
Analysis of gene module, IFN response, and therapeutic pathway scores
To broadly score for transcriptional evidence of cell states, we compiled gene lists summarizing modules of memory and effector states, cytotoxicity, dysfunction, and proliferation from published studies and calculated scores based on the average expression for each cell population (Supplementary table 5) (17–20, 24–27). These 5 modules were examined by 2 approaches. First, the full list was used for scoring to evaluate broad expression patterns (module score, Fig. 4c, S7d). Second, the differentially expressed genes identified by ANOVA were assigned to these modules and used for scoring to evaluate the difference in expression among populations (DEG score).
Module scores based on specific gene sets (memory, effector, proliferation, IFN-inducible genes etc.) were calculated for each population as the average of the scaled (Z-normalized) expression of the genes in the list. A similar approach was used when calculating gene set-based scores in bulk RNA-seq data. Module gene lists were compiled from published studies and molecular signature databases. (MSigDB v7.4 from UC San Diego and Broad Institute). Memory module genes (Tcm, Tem, Trm and Temra) were from Zhang et al., Nature 2018 (17). Dysfunction module genes were from Zhang et al., Nature 2018 (17) and Li et al., Cell 2019 (18). Cytotoxicity module genes were from Li et al., Cell 2019 (18), and MSigDB gene sets M16355 (BIOCARTA_NK cells pathway), M13247 (BIOCARTA_ T Cytotoxic Cell Surface Molecules) and M5669 (KEGG_ Natural killer cell mediated cytotoxicity) Proliferation module genes were from Gene Ontology gene sets (GO_G1_S_TRANSITION_OF_MITOTIC_CELL_CYCLE, GO_G2_M_TRANSITION_OF_MITOTIC_CELL_CYCLE) and Reactome MSigDB gene sets M1017 (DNA_REPLICATION), M1080 (G2_M_DNA_DAMAGE_CHECKPOINT), M27662 (M_PHASE), M17283 (MITOTIC_G1_PHASE_AND_G1_S_TRANSITION), M27673 (MITOTIC_SPINDLE_CHECKPOINT) and M3158 (S_PHASE) Effector module genes were from MSigDB gene sets M3044 (GOLDRATH_EFF_VS_MEMORY_CD8_TCELL_DN), M3041 (GOLDRATH_EFF_VS_MEMORY_CD8_TCELL_UP), M5834 (GSE9650_EFFECTOR_VS_EXHAUSTED_CD8_TCELL_DN), M5837 (GSE9650_EFFECTOR_VS_MEMORY_CD8_TCELL_DN), M3073 (GSE10239_MEMORY_VS_DAY4.5_EFF_CD8_TCELL_UP), M4407 (GSE22886_NAIVE_CD8_TCELL_VS_MEMORY_TCELL_DN), M8435 (GSE23321_CENTRAL_MEMORY_VS_NAIVE_CD8_TCELL_UP), M9490 (GSE41867_DAY6_EFFECTOR_VS_DAY30_EXHAUSTED_CD8_TCELL_LCMV_CLONE13_DN), M9492 (GSE41867_DAY8_EFFECTOR_VS_DAY30_EXHAUSTED_CD8_TCELL_LCMV_CLONE13_DN), M9480 (GSE41867_MEMORY_VS_EXHAUSTED_CD8_TCELL_DAY30_LCMV_UP) and M3027 (KAECH_DAY8_EFF_VS_MEMORY_CD8_TCELL_UP) IFN-inducible genes were from Arazi, et al., Nat Immunol, 2019 (26). IL-6 pathway genes were from Brocke-Heidrich et al., Blood 2004 (63). TNF pathway genes were from Zhou et al., Oncogene 2003 (64). IL-1 beta pathway genes were from Wu, et al., Cancer Res. 2018 (65). We compiled the gene lists using 2 strategies: 1) we took the union of genes from gene sets; 2) we reduced the overlap in the memory-effector, memory-dysfunction, and effector-dysfunction modules by assigning the genes shared by these lists to the effector module. Differentially expressed module genes were then obtained from the intersection of each of the above module genes and the ANOVA-tested, batch-corrected DEGs (q < 0.01) among the sorted bulk populations (Supplementary table 4).
RNA extraction, reverse transcription, and real-time PCR (qPCR)
RNA isolated using RNeasy Micro Kits (Qiagen). cDNA was prepared using Quantitect RT- PCR (Qiagen) and PCR performed with Brilliant III SYBRGreen on a Stratagene Mx3000. Primers used were as follows: 18S forward: GGGAGCCTGAGAAACGGC, reverse: GGGTCGGGAGTGGGTAATTT. IFI27 forward: GAATCGCCTCGTCCTCCATAG, reverse: CGCCAGGATACTTACCCAGTG. IFI44 forward: CTGAGACGAATGCTATGGGCT, reverse: GACAGAGAGCTGCCAGGTATT. IFI44L forward: CAGATTTGGAACTGGACCCCA, reverse: AGGGCCAGATTACCAGTTTCC. ISG15 forward: CGCAGATCACCCAGAAGATCG, reverse: TTCGTCGCATTTGTCCACCA. IFIT1 forward: TTGATGACGATGAAATGCCTGA, reverse: CAGGTCACCAGACTCCTCAC. IFIT3 forward: TCAGAAGTCTAGTCACTTGGGG, reverse: ACACCTTCGCCCTTTCATTTC. IRF7 forward: CCCACGCTATACCATCTACCT, reverse: GATGTCGTCATAGAGGCTGTTG. RASD2 forward: GAGCGCCACAAAGAAGTGTC, reverse: ACAATGTGTGGGGTCCTTGG.
Cell culture with IFN stimulation
Cryopreserved mononuclear cells from synovial fluid or peripheral blood were thawed and counted as usual. Cells were plated at 2.5×106 cells/mL in 1mL per well in a 12-well plate and cultured with or without IFN-β (1kU/mL) or IFN-γ (50ng/mL) for 72 hours. For analysis of isolated CD8 T cells from synovial fluid, thawed synovial fluid CD8 T cells were isolated from synovial fluid cells by MACS total CD8 isolation kit (Miltenyi). At the time of harvest, cells were washed and stained with surface markers and intracellular markers as described above.
Cytotoxicity Assays
CD8 T cells were isolated from leukoreduction collars with MACS total CD8 isolation kit (Miltenyi) and subsequently sorted with a 5-laser BD FACSAria Fusion cell sorter for naïve and non-naive CD38hiCD127− cells using anti-CD14 FITC (63D3), anti-CD8 BV711 (RPA-T8), anti-CD45RA BV510 (HI100), anti-CCR7 Pe-Cy7 (G043H7), anti-CD38 PerCP Cy5.5 (HIT2), and anti-CD127 APC (A019D5) (all from Biolegend). Sorted CD8 T cells were then cultured with or without IFN-β (1kU/mL) for 3 days before the cytotoxicity assay.
CD32-expressing murine fibroblast L cells were kindly gifted to Deepak Rao by Megan Levings(28). CD32 expressing L cells between passage number 4 and 20 were incubated with an agonist anti-CD3 antibody (OKT3, BioXcell) for 30 min on ice and then plated with sorted CD8 T cells at a ratio of 5:1 CD8 T cells:L cells. After 3.5 hours, the T cells and L cells were removed from culture and stained with AnnexinV APC and 7-AAD in Annexin Binding Buffer (all from Biolegend) and analyzed on a BD CantoII analyzer.
Synovial fluid cell cytotoxicity assays were performed in a similar manner. Cryopreserved SFMCs were thawed, washed, and counted before staining with Aqua fixable live/dead dye (Invitrogen) in PBS. Subsequently the cells were stained in PBS+ 1% FBS with the following antibodies anti-CD14 FITC (63D3), anti-CD8 BV711 (RPA-T8), anti-CD4 APC-Cy7 (RPA-T4), anti-CD45RA BV605 (HI100), anti-CCR7 Pe-Cy7 (G043H7), anti-CD38 Percp Cy5.5 (HIT2), anti-CD127 APC (A019D5), and anti-PD-1 BV421 (EH12.2H7) (all from Biolegend) and sorted by a 5-laser BD FACSAria Fusion cell sorter. The sorted SF T cells were then rested at 37°C for 2 hours before plating with L cells, incubated as before with anti-CD3 antibody for 30 min, co-cultured for 3.5 hrs and then analyzed on a BD Fortessa analyzer.
TCR sequencing
3 cryopreserved paired blood and synovial fluid cell samples were thawed, washed, and stained with viability dye in PBS. Subsequently cells were stained with the following flow cytometry antibodies anti-IgG4 PE (abcam), anti-CD8 BV711 (RPA-T8), anti-CD45RA BV605, anti-CCR7 Pe-Cy7, anti-CD38 Percp Cy5.5 (HIT2), and anti-CD127 APC (A019D5) (all but IgG4 from Biolegend) for 20 minutes in PBS + 1% FBS. Live non-naive CD8 T cells were sorted by IgG4 positivity on a 5-laser BD FACSAria Fusion cell sorter. Following the sort, cells were processed by a DNA extraction kit (Qiagen). Obtained DNA was quantified bulk TCR sequencing performed at Adaptive Biotechnologies. Data analysis was performed using the immunoSEQ software (Adaptive) and the immunarch R package(66). Morisita overlap index was utilized to measure overlap between samples. This analysis produces unitless values ranging from 0 to 1 with 0 being completely dissimilar. The productive Simpson clonality was calculated as the square root of Simpson’s Diversity index of productive clonotypes. This analysis also produces values ranging from 0 to 1, with 0 being more polyclonal.
Statistics
Statistical analysis was performed as described in each section and figure legends using Prism 8 software. Unless otherwise stated, data are presented as mean ± SD from data obtained from at least two independent experiments. Parametric and non-parametric analyses were used where appropriate based on testing for a normal distribution using the D’Agostino–Pearson normality test or Shapiro-Wilk normality test. Two-tailed Student’s t test was used for two-group comparisons (Mann-Whitney test was used for nonparametric data). One-way analysis of variation (ANOVA) followed by the Holm-Sidak test was used for multiple comparisons (Kruskal-Wallis test was used for nonparametric data). P-values < 0.05 were considered significant after adjusting for multiple testing where appropriate.
Supplementary Material
Acknowledgements
We thank the BWH Center for Cellular Profiling for cell sorting assistance.
Funding
This work has been supported in part by funding from the Rheumatology Research Foundation (to DAR, LTD, ARB), Burroughs Wellcome Fund Career Award in Medical Sciences, NIAMS K08 AR072791, P30 AR070253 (to DAR), NIAMS R01 (to MBB), NIAID R01 AI148435, Carson Foundation, Ambrose Monell Foundation (to LTD), HSS Department of Medicine (to ARB).
Competing interests
D.A. Rao reports personal fees from Pfizer, Janssen, Merck, GlaxoSmithKline, and Bristol-Myers Squibb and grant support from Janssen and Bristol-Myers Squibb, outside the submitted work. J. A. Sparks is funded by NIH/NIAMS (grant numbers R01 AR080659, R01 AR077607, P30 AR070253, and P30 AR072577), the R. Bruce and Joan M. Mickey Research Scholar Fund, and the Llura Gund Award for Rheumatoid Arthritis Research and Care. J. A. Sparks reports research support from Bristol Myers Squibb; consulting fees from AbbVie, Amgen, Boehringer Ingelheim, Bristol Myers Squibb, Gilead, Inova Diagnostics, Janssen, Optum, and Pfizer, all unrelated to this work. The funders had no role in the decision to publish or preparation of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard University, its affiliated academic healthcare centers, or the National Institutes of Health. M.B. Brenner reports consulting fees from GlaxoSmithKline, 4FO Ventures, Third Rock Ventures and is consultant and founder of Mestag Therapeutics. M. A. Postow reports consulting fees from BMS, Merck, Novartis, Eisai, Pfizer, Chugai and institutional support from RGenix, Infinity, BMS, Merck, Novartis. A.R. Bass reports being a Member of the American College of Rheumatology, Board of Directors from 2019–2022.
Data Availability
Bulk and single cell RNA-seq data will be shared through Synapse upon publication. Project SynID: syn51534310 DOI: https://doi.org/10.7303/syn51534310
References
- 1.Dougan M, Luoma AM, Dougan SK, Wucherpfennig KW, Understanding and treating the inflammatory adverse events of cancer immunotherapy. Cell 184, 1575–1588 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leach DR, Krummel MF, Allison JP, Enhancement of antitumor immunity by CTLA-4 blockade. Science 271, 1734–1736 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Nishimura H, Nose M, Hiai H, Minato N, Honjo T, Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity 11, 141–151 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Sharpe AH, Pauken KE, The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol 18, 153–167 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Michot JM, Bigenwald C, Champiat S, Collins M, Carbonnel F, Postel-Vinay S, Berdelou A, Varga A, Bahleda R, Hollebecque A, Massard C, Fuerea A, Ribrag V, Gazzah A, Armand JP, Amellal N, Angevin E, Noel N, Boutros C, Mateus C, Robert C, Soria JC, Marabelle A, Lambotte O, Immune-related adverse events with immune checkpoint blockade: a comprehensive review. Eur J Cancer 54, 139–148 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Pundole X, Abdel-Wahab N, Suarez-Almazor ME, Arthritis risk with immune checkpoint inhibitor therapy for cancer. Curr Opin Rheumatol 31, 293–299 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Braaten TJ, Brahmer JR, Forde PM, Le D, Lipson EJ, Naidoo J, Schollenberger M, Zheng L, Bingham CO, Shah AA, Cappelli LC, Immune checkpoint inhibitor-induced inflammatory arthritis persists after immunotherapy cessation. Ann Rheum Dis 79, 332–338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cappelli LC, Thomas MA, Bingham CO 3rd, Shah AA, Darrah E, Immune checkpoint inhibitor-induced inflammatory arthritis as a model of autoimmune arthritis. Immunol Rev 294, 106–123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan KK, Bass AR, Autoimmune complications of immunotherapy: pathophysiology and management. BMJ 369, m736 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Jeurling S, Cappelli LC, Treatment of immune checkpoint inhibitor-induced inflammatory arthritis. Curr Opin Rheumatol 32, 315–320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Postow MA, Sidlow R, Hellmann MD, Immune-Related Adverse Events Associated with Immune Checkpoint Blockade. N Engl J Med 378, 158–168 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Kostine M, Rouxel L, Barnetche T, Veillon R, Martin F, Dutriaux C, Dousset L, Pham-Ledard A, Prey S, Beylot-Barry M, Daste A, Gross-Goupil M, Lallier J, Ravaud A, Forcade E, Bannwarth B, Truchetet ME, Richez C, Mehsen N, Schaeverbeke T, ACRONIM F, Rheumatic disorders associated with immune checkpoint inhibitors in patients with cancer-clinical aspects and relationship with tumour response: a single-centre prospective cohort study. Ann Rheum Dis 77, 393–398 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Rao DA, Gurish MF, Marshall JL, Slowikowski K, Fonseka CY, Liu Y, Donlin LT, Henderson LA, Wei K, Mizoguchi F, Teslovich NC, Weinblatt ME, Massarotti EM, Coblyn JS, Helfgott SM, Lee YC, Todd DJ, Bykerk VP, Goodman SM, Pernis AB, Ivashkiv LB, Karlson EW, Nigrovic PA, Filer A, Buckley CD, Lederer JA, Raychaudhuri S, Brenner MB, Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 542, 110–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang F, Wei K, Slowikowski K, Fonseka CY, Rao DA, Kelly S, Goodman SM, Tabechian D, Hughes LB, Salomon-Escoto K, Watts GFM, Jonsson AH, Rangel-Moreno J, Meednu N, Rozo C, Apruzzese W, Eisenhaure TM, Lieb DJ, Boyle DL, Mandelin AM 2nd, Accelerating Medicines Partnership Rheumatoid A, Systemic Lupus Erythematosus C, Boyce BF, DiCarlo E, Gravallese EM, Gregersen PK, Moreland L, Firestein GS, Hacohen N, Nusbaum C, Lederer JA, Perlman H, Pitzalis C, Filer A, Holers VM, Bykerk VP, Donlin LT, Anolik JH, Brenner MB, Raychaudhuri S, Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol 20, 928–942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephenson W, Donlin LT, Butler A, Rozo C, Bracken B, Rashidfarrokhi A, Goodman SM, Ivashkiv LB, Bykerk VP, Orange DE, Darnell RB, Swerdlow HP, Satija R, Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat Commun 9, 791 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, Freeman SS, Reuben A, Hoover PJ, Villani AC, Ivanova E, Portell A, Lizotte PH, Aref AR, Eliane JP, Hammond MR, Vitzthum H, Blackmon SM, Li B, Gopalakrishnan V, Reddy SM, Cooper ZA, Paweletz CP, Barbie DA, Stemmer-Rachamimov A, Flaherty KT, Wargo JA, Boland GM, Sullivan RJ, Getz G, Hacohen N, Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 175, 998–1013 e1020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, Gao R, Kang B, Zhang Q, Huang JY, Konno H, Guo X, Ye Y, Gao S, Wang S, Hu X, Ren X, Shen Z, Ouyang W, Zhang Z, Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564, 268–272 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, van den Braber M, Rozeman EA, Haanen J, Blank CU, Horlings HM, David E, Baran Y, Bercovich A, Lifshitz A, Schumacher TN, Tanay A, Amit I, Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 176, 775–789 e718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, Modak M, Carotta S, Haslinger C, Kind D, Peet GW, Zhong G, Lu S, Zhu W, Mao Y, Xiao M, Bergmann M, Hu X, Kerkar SP, Vogt AB, Pflanz S, Liu K, Peng J, Ren X, Zhang Z, Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell 179, 829–845 e820 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, Yates KB, Lako A, Felt K, Naik GS, Manos M, Gjini E, Kuchroo JR, Ishizuka JJ, Collier JL, Griffin GK, Maleri S, Comstock DE, Weiss SA, Brown FD, Panda A, Zimmer MD, Manguso RT, Hodi FS, Rodig SJ, Sharpe AH, Haining WN, Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 20, 326–336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orange DE, Agius P, DiCarlo EF, Robine N, Geiger H, Szymonifka J, McNamara M, Cummings R, Andersen KM, Mirza S, Figgie M, Ivashkiv LB, Pernis AB, Jiang CS, Frank MO, Darnell RB, Lingampali N, Robinson WH, Gravallese E, Accelerating A. Medicines Partnership in Rheumatoid, Lupus N, Bykerk VP, Goodman SM, Donlin LT, Identification of Three Rheumatoid Arthritis Disease Subtypes by Machine Learning Integration of Synovial Histologic Features and RNA Sequencing Data. Arthritis Rheumatol 70, 690–701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, Kang B, Liu Z, Jin L, Xing R, Gao R, Zhang L, Dong M, Hu X, Ren X, Kirchhoff D, Roider HG, Yan T, Zhang Z, Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med 24, 978–985 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Tickotsky N, Sagiv T, Prilusky J, Shifrut E, Friedman N, McPAS-TCR: a manually curated catalogue of pathology-associated T cell receptor sequences. Bioinformatics 33, 2924–2929 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z, Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep 18, 248–262 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Savas P, Virassamy B, Ye C, Salim A, Mintoff CP, Caramia F, Salgado R, Byrne DJ, Teo ZL, Dushyanthen S, Byrne A, Wein L, Luen SJ, Poliness C, Nightingale SS, Skandarajah AS, Gyorki DE, Thornton CM, Beavis PA, Fox SB, Kathleen C. Cuningham Foundation Consortium for Research into Familial Breast, Darcy PK, Speed TP, Mackay LK, Neeson PJ, Loi S, Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med 24, 986–993 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Arazi A, Rao DA, Berthier CC, Davidson A, Liu Y, Hoover PJ, Chicoine A, Eisenhaure TM, Jonsson AH, Li S, Lieb DJ, Zhang F, Slowikowski K, Browne EP, Noma A, Sutherby D, Steelman S, Smilek DE, Tosta P, Apruzzese W, Massarotti E, Dall’Era M, Park M, Kamen DL, Furie RA, Payan-Schober F, Pendergraft WF 3rd, McInnis EA, Buyon JP, Petri MA, Putterman C, Kalunian KC, Woodle ES, Lederer JA, Hildeman DA, Nusbaum C, Raychaudhuri S, Kretzler M, Anolik JH, Brenner MB, Wofsy D, Hacohen N, Diamond B, S. L. E. n. Accelerating Medicines Partnership in, The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol 20, 902–914 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jing Y, Liu J, Ye Y, Pan L, Deng H, Wang Y, Yang Y, Diao L, Lin SH, Mills GB, Zhuang G, Xue X, Han L, Multi-omics prediction of immune-related adverse events during checkpoint immunotherapy. Nat Commun 11, 4946 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boardman DA, Wong MQ, Rees WD, Wu D, Himmel ME, Orban PC, Vent-Schmidt J, Zachos NC, Steiner TS, Levings MK, Flagellin-specific human CAR Tregs for immune regulation in IBD. J Autoimmun 134, 102961 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivashkiv LB, Donlin LT, Regulation of type I interferon responses. Nat Rev Immunol 14, 36–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katsuyama E, Suarez-Fueyo A, Bradley SJ, Mizui M, Marin AV, Mulki L, Krishfield S, Malavasi F, Yoon J, Sui SJH, Kyttaris VC, Tsokos GC, The CD38/NAD/SIRTUIN1/EZH2 Axis Mitigates Cytotoxic CD8 T Cell Function and Identifies Patients with SLE Prone to Infections. Cell Rep 30, 112–123 e114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Postow MA, Goldman DA, Shoushtari AN, Betof Warner A, Callahan MK, Momtaz P, Smithy JW, Naito E, Cugliari MK, Raber V, Eton O, Nair SG, Panageas KS, Wolchok JD, Chapman PB, Adaptive Dosing of Nivolumab + Ipilimumab Immunotherapy Based Upon Early, Interim Radiographic Assessment in Advanced Melanoma (The ADAPT-IT Study). J Clin Oncol 40, 1059–1067 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliveira G, Stromhaug K, Klaeger S, Kula T, Frederick DT, Le PM, Forman J, Huang T, Li S, Zhang W, Xu Q, Cieri N, Clauser KR, Shukla SA, Neuberg D, Justesen S, MacBeath G, Carr SA, Fritsch EF, Hacohen N, Sade-Feldman M, Livak KJ, Boland GM, Ott PA, Keskin DB, Wu CJ, Phenotype, specificity and avidity of antitumour CD8. Nature 596, 119–125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duhen T, Duhen R, Montler R, Moses J, Moudgil T, de Miranda NF, Goodall CP, Blair TC, Fox BA, McDermott JE, Chang SC, Grunkemeier G, Leidner R, Bell RB, Weinberg AD, Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat Commun 9, 2724 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osa A, Uenami T, Koyama S, Fujimoto K, Okuzaki D, Takimoto T, Hirata H, Yano Y, Yokota S, Kinehara Y, Naito Y, Otsuka T, Kanazu M, Kuroyama M, Hamaguchi M, Koba T, Futami Y, Ishijima M, Suga Y, Akazawa Y, Machiyama H, Iwahori K, Takamatsu H, Nagatomo I, Takeda Y, Kida H, Akbay EA, Hammerman PS, Wong KK, Dranoff G, Mori M, Kijima T, Kumanogoh A, Clinical implications of monitoring nivolumab immunokinetics in non-small cell lung cancer patients. JCI Insight 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luoma AM, Suo S, Williams HL, Sharova T, Sullivan K, Manos M, Bowling P, Hodi FS, Rahma O, Sullivan RJ, Boland GM, Nowak JA, Dougan SK, Dougan M, Yuan GC, Wucherpfennig KW, Molecular Pathways of Colon Inflammation Induced by Cancer Immunotherapy. Cell 182, 655–671 e622 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zen Y, Yeh MM, Checkpoint inhibitor-induced liver injury: A novel form of liver disease emerging in the era of cancer immunotherapy. Semin Diagn Pathol 36, 434–440 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Berner F, Bomze D, Diem S, Ali OH, Fassler M, Ring S, Niederer R, Ackermann CJ, Baumgaertner P, Pikor N, Cruz CG, van de Veen W, Akdis M, Nikolaev S, Laubli H, Zippelius A, Hartmann F, Cheng HW, Honger G, Recher M, Goldman J, Cozzio A, Fruh M, Neefjes J, Driessen C, Ludewig B, Hegazy AN, Jochum W, Speiser DE, Flatz L, Association of Checkpoint Inhibitor-Induced Toxic Effects With Shared Cancer and Tissue Antigens in Non-Small Cell Lung Cancer. JAMA Oncol 5, 1043–1047 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tadokoro T, Keshino E, Makiyama A, Sasaguri T, Ohshima K, Katano H, Mohri M, Acute Lymphocytic Myocarditis With Anti-PD-1 Antibody Nivolumab. Circ Heart Fail 9, (2016). [DOI] [PubMed] [Google Scholar]
- 39.Johnson DB, Balko JM, Compton ML, Chalkias S, Gorham J, Xu Y, Hicks M, Puzanov I, Alexander MR, Bloomer TL, Becker JR, Slosky DA, Phillips EJ, Pilkinton MA, Craig-Owens L, Kola N, Plautz G, Reshef DS, Deutsch JS, Deering RP, Olenchock BA, Lichtman AH, Roden DM, Seidman CE, Koralnik IJ, Seidman JG, Hoffman RD, Taube JM, Diaz LA Jr., Anders RA, Sosman JA, Moslehi JJ, Fulminant Myocarditis with Combination Immune Checkpoint Blockade. N Engl J Med 375, 1749–1755 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kotwal A, Gustafson MP, Bornschlegl S, Kottschade L, Delivanis DA, Dietz AB, Gandhi M, Ryder M, Immune Checkpoint Inhibitor-Induced Thyroiditis Is Associated with Increased Intrathyroidal T Lymphocyte Subpopulations. Thyroid 30, 1440–1450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yasuda Y, Iwama S, Sugiyama D, Okuji T, Kobayashi T, Ito M, Okada N, Enomoto A, Ito S, Yan Y, Sugiyama M, Onoue T, Tsunekawa T, Ito Y, Takagi H, Hagiwara D, Goto M, Suga H, Banno R, Takahashi M, Nishikawa H, Arima H, CD4(+) T cells are essential for the development of destructive thyroiditis induced by anti-PD-1 antibody in thyroglobulin-immunized mice. Sci Transl Med 13, (2021). [DOI] [PubMed] [Google Scholar]
- 42.Wagner J, Rapsomaniki MA, Chevrier S, Anzeneder T, Langwieder C, Dykgers A, Rees M, Ramaswamy A, Muenst S, Soysal SD, Jacobs A, Windhager J, Silina K, van den Broek M, Dedes KJ, Rodriguez Martinez M, Weber WP, Bodenmiller B, A Single-Cell Atlas of the Tumor and Immune Ecosystem of Human Breast Cancer. Cell 177, 1330–1345 e1318 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verma V, Shrimali RK, Ahmad S, Dai W, Wang H, Lu S, Nandre R, Gaur P, Lopez J, Sade-Feldman M, Yizhak K, Bjorgaard SL, Flaherty KT, Wargo JA, Boland GM, Sullivan RJ, Getz G, Hammond SA, Tan M, Qi J, Wong P, Merghoub T, Wolchok J, Hacohen N, Janik JE, Mkrtichyan M, Gupta S, Khleif SN, PD-1 blockade in subprimed CD8 cells induces dysfunctional PD-1(+)CD38(hi) cells and anti-PD-1 resistance. Nat Immunol 20, 1231–1243 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen L, Diao L, Yang Y, Yi X, Rodriguez BL, Li Y, Villalobos PA, Cascone T, Liu X, Tan L, Lorenzi PL, Huang A, Zhao Q, Peng D, Fradette JJ, Peng DH, Ungewiss C, Roybal J, Tong P, Oba J, Skoulidis F, Peng W, Carter BW, Gay CM, Fan Y, Class CA, Zhu J, Rodriguez-Canales J, Kawakami M, Byers LA, Woodman SE, Papadimitrakopoulou VA, Dmitrovsky E, Wang J, Ullrich SE, Wistuba II, Heymach JV, Qin FX, Gibbons DL, CD38-Mediated Immunosuppression as a Mechanism of Tumor Cell Escape from PD-1/PD-L1 Blockade. Cancer Discov 8, 1156–1175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim ST, Chu Y, Misoi M, Suarez-Almazor ME, Tayar JH, Lu H, Buni M, Kramer J, Rodriguez E, Hussain Z, Neelapu SS, Wang J, Shah AY, Tannir NM, Campbell MT, Gibbons DL, Cascone T, Lu C, Blumenschein GR, Altan M, Lim B, Valero V, Loghin ME, Tu J, Westin SN, Naing A, Garcia-Manero G, Abdel-Wahab N, Tawbi HA, Hwu P, Oliva ICG, Davies MA, Patel SP, Zou J, Futreal A, Diab A, Wang L, Nurieva R, Distinct molecular and immune hallmarks of inflammatory arthritis induced by immune checkpoint inhibitors for cancer therapy. Nat Commun 13, 1970 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luoma AM, Suo S, Williams HL, Sharova T, Sullivan K, Manos M, Bowling P, Hodi FS, Rahma O, Sullivan RJ, Boland GM, Nowak JA, Dougan SK, Dougan M, Yuan GC, Wucherpfennig KW, Molecular Pathways of Colon Inflammation Induced by Cancer Immunotherapy. Cell 182, 655–671.e622 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subudhi SK, Aparicio A, Gao J, Zurita AJ, Araujo JC, Logothetis CJ, Tahir SA, Korivi BR, Slack RS, Vence L, Emerson RO, Yusko E, Vignali M, Robins HS, Sun J, Allison JP, Sharma P, Clonal expansion of CD8 T cells in the systemic circulation precedes development of ipilimumab-induced toxicities. Proc Natl Acad Sci U S A 113, 11919–11924 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez-Carrio J, Alperi-Lopez M, Lopez P, Ballina-Garcia FJ, Suarez A, Heterogeneity of the Type I Interferon Signature in Rheumatoid Arthritis: A Potential Limitation for Its Use As a Clinical Biomarker. Front Immunol 8, 2007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Pouw Kraan TC, Wijbrandts CA, van Baarsen LG, Voskuyl AE, Rustenburg F, Baggen JM, Ibrahim SM, Fero M, Dijkmans BA, Tak PP, Verweij CL, Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: assignment of a type I interferon signature in a subpopulation of patients. Ann Rheum Dis 66, 1008–1014 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veale DJ, Fearon U, The pathogenesis of psoriatic arthritis. Lancet 391, 2273–2284 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Morand EF, Furie R, Tanaka Y, Bruce IN, Askanase AD, Richez C, Bae SC, Brohawn PZ, Pineda L, Berglind A, Tummala R, Investigators T-T, Trial of Anifrolumab in Active Systemic Lupus Erythematosus. N Engl J Med 382, 211–221 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, Archambault JM, Lee H, Arthur CD, White JM, Kalinke U, Murphy KM, Schreiber RD, Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med 208, 1989–2003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dunn GP, Bruce AT, Sheehan KC, Shankaran V, Uppaluri R, Bui JD, Diamond MS, Koebel CM, Arthur C, White JM, Schreiber RD, A critical function for type I interferons in cancer immunoediting. Nat Immunol 6, 722–729 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Lu C, Klement JD, Ibrahim ML, Xiao W, Redd PS, Nayak-Kapoor A, Zhou G, Liu K, Type I interferon suppresses tumor growth through activating the STAT3-granzyme B pathway in tumor-infiltrating cytotoxic T lymphocytes. J Immunother Cancer 7, 157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li W, Lu L, Lu J, Wang X, Yang C, Jin J, Wu L, Hong X, Li F, Cao D, Yang Y, Wu M, Su B, Cheng J, Yang X, Di W, Deng L, cGAS-STING-mediated DNA sensing maintains CD8(+) T cell stemness and promotes antitumor T cell therapy. Sci Transl Med 12, (2020). [DOI] [PubMed] [Google Scholar]
- 56.McKinney EF, Lee JC, Jayne DR, Lyons PA, Smith KG, T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature 523, 612–616 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korem Kohanim Y, Tendler A, Mayo A, Friedman N, Alon U, Endocrine Autoimmune Disease as a Fragility of Immune Surveillance against Hypersecreting Mutants. Immunity 52, 872–884.e875 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bocharnikov AV, Keegan J, Wacleche VS, Cao Y, Fonseka CY, Wang G, Muise ES, Zhang KX, Arazi A, Keras G, Li ZJ, Qu Y, Gurish MF, Petri M, Buyon JP, Putterman C, Wofsy D, James JA, Guthridge JM, Diamond B, Anolik JH, Mackey MF, Alves SE, Nigrovic PA, Costenbader KH, Brenner MB, Lederer JA, Rao DA, Accelerating Medicines Partnership RASLEN, PD-1hiCXCR5- T peripheral helper cells promote B cell responses in lupus via MAF and IL-21. JCI Insight 4, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amir el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe’er D, viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 31, 545–552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, Saeys Y, FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Pedersen CB, Dam SH, Barnkob MB, Leipold MD, Purroy N, Rassenti LZ, Kipps TJ, Nguyen J, Lederer JA, Gohil SH, Wu CJ, Olsen LR, cyCombine allows for robust integration of single-cell cytometry datasets within and across technologies. Nat Commun 13, 1698 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brocke-Heidrich K, Kretzschmar AK, Pfeifer G, Henze C, Löffler D, Koczan D, Thiesen HJ, Burger R, Gramatzki M, Horn F, Interleukin-6-dependent gene expression profiles in multiple myeloma INA-6 cells reveal a Bcl-2 family-independent survival pathway closely associated with Stat3 activation. Blood 103, 242–251 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Zhou A, Scoggin S, Gaynor RB, Williams NS, Identification of NF-kappa B-regulated genes induced by TNFalpha utilizing expression profiling and RNA interference. Oncogene 22, 2054–2064 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Wu TC, Xu K, Martinek J, Young RR, Banchereau R, George J, Turner J, Kim KI, Zurawski S, Wang X, Blankenship D, Brookes HM, Marches F, Obermoser G, Lavecchio E, Levin MK, Bae S, Chung CH, Smith JL, Cepika AM, Oxley KL, Snipes GJ, Banchereau J, Pascual V, O’Shaughnessy J, Palucka AK, IL1 Receptor Antagonist Controls Transcriptional Signature of Inflammation in Patients with Metastatic Breast Cancer. Cancer Res 78, 5243–5258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Team I, immunarch: An R Package for Painless Bioinformatics Analysis of T-Cell and B-Cell Immune Repertoires. Zenodo, (2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bulk and single cell RNA-seq data will be shared through Synapse upon publication. Project SynID: syn51534310 DOI: https://doi.org/10.7303/syn51534310