Abstract
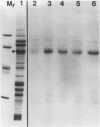
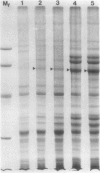
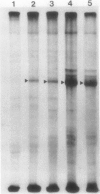
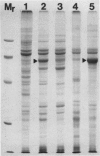
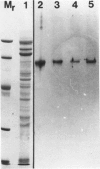
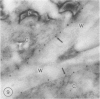
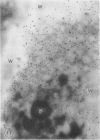
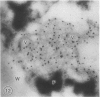
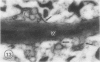
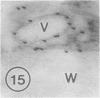
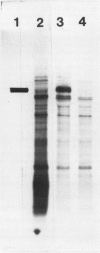
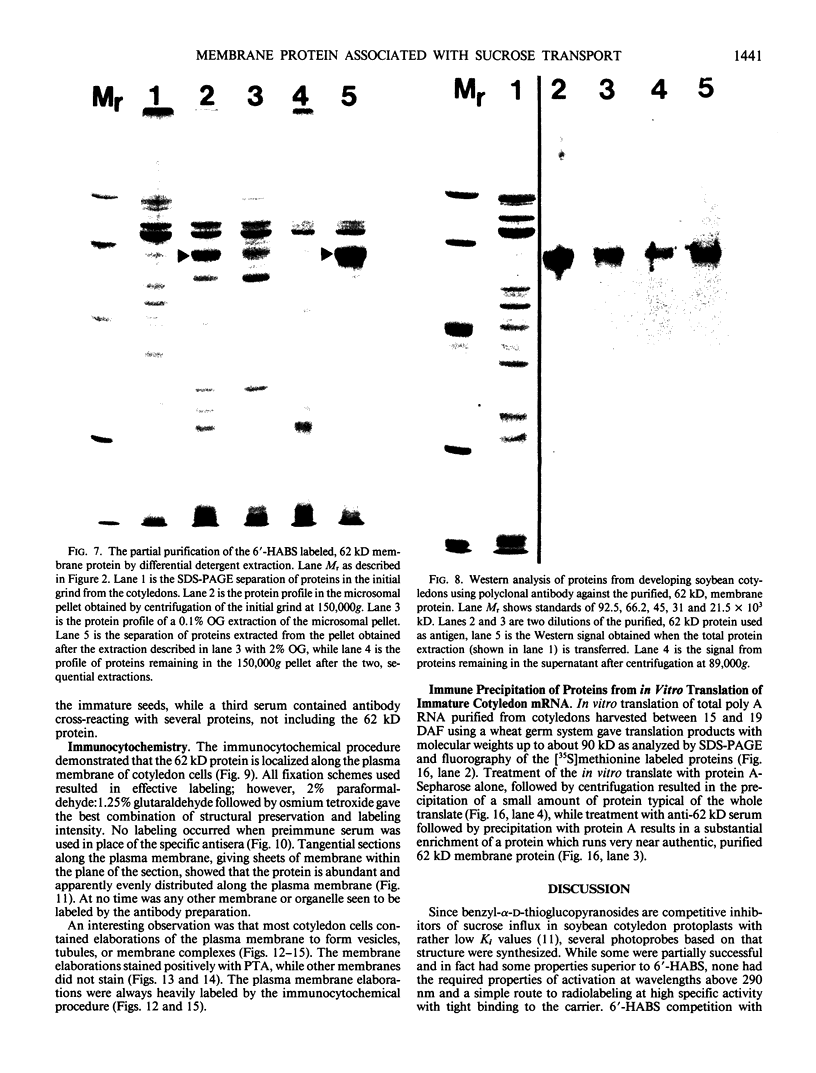
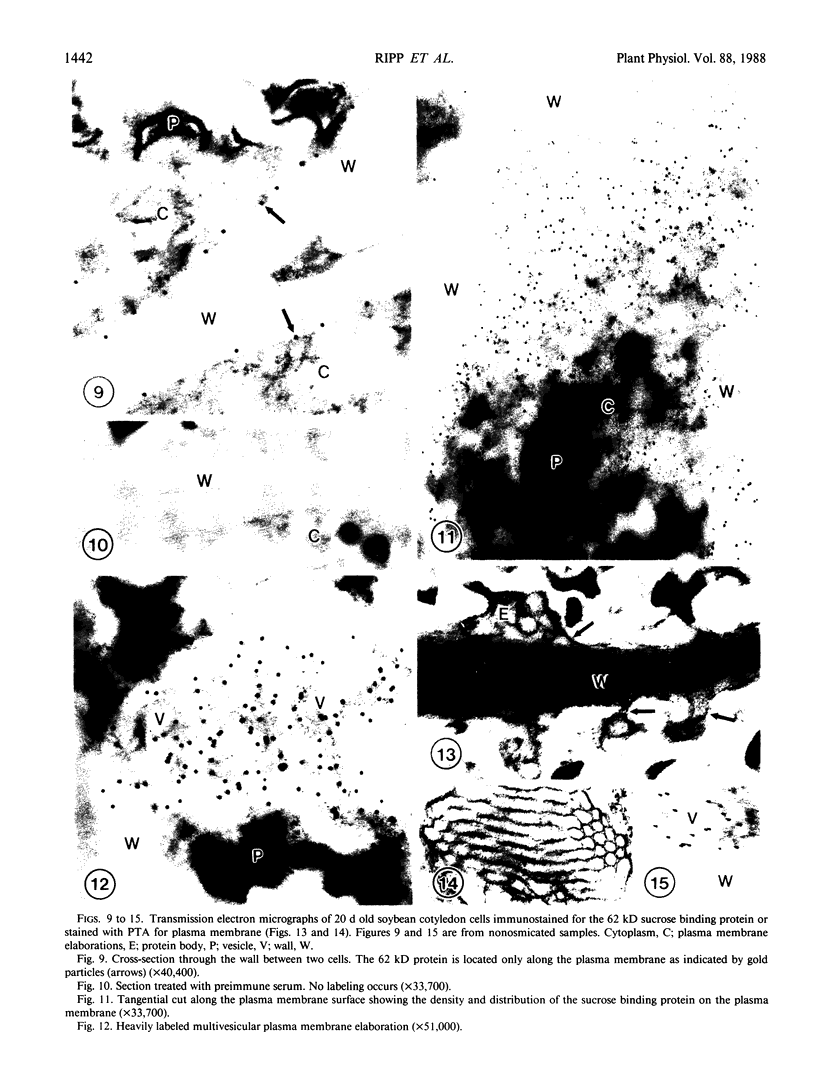
The photolyzable sucrose derivative 6′-deoxy-6′-(4-azido-2-hydroxy)-benzamidosucrose (6′-HABS), competitively inhibited the influx of [14C] sucrose into protoplasts from developing soybean (Glycine max L. Merr cv Wye) cotyledons. Photolysis of 125I-labeled 6′-HABS in the presence of 10 millimolar dithiothreitol and microsomal preparations from developing soybean cotyledons led to label incorporation into a moderately abundant membrane protein with an apparent molecular mass of about 62 kilodalton (kD) by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The 62 kD protein was partially protected from labeling by the inclusion of 100 millimolar sucrose in the photolysis medium and also by the inclusion of 10 millimolar phenyl α-d-thioglucopyranoside. Glucose, raffinose, or phenyl α-d-3-deoxy-3-fluoroglucopyranoside did not afford even partial protection from labeling. When the photolyzable moiety of 6′-HABS was attached to 6-deoxy-6-aminoglucose and 125I labeled, the resulting photoprobe did not label the 62 kD protein above background. The labeled protein at 62 kD is therefore apparently a specific, sucrose binding protein. Sucrose influx into cotlyedons of less than 25 milligrams fresh weight (approximately 10 days after flowering) occurred by passive processes, but metabolically dependent uptake became dominant over the next 5 to 7 days of development. Both the Coomassie staining protein at 62 kD and label incorporation at that position in analysis of membrane proteins appeared concomitant with the onset of active sucrose influx. Polyclonal antibodies to the purified 62 kD protein bound specifically to a protein in the plasmalemma of thin sections prepared from cotyledons and density stained with colloidal gold-protein A. The results suggest that the 62 kD membrane protein is associated with sucrose transport and may be the plasmalemma sucrose transporter.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Delrot S. Proton Fluxes Associated with Sugar Uptake in Vicia faba Leaf Tissues. Plant Physiol. 1981 Sep;68(3):706–711. doi: 10.1104/pp.68.3.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaquinta R. T. Phloem loading of sucrose: involvement of membrane ATPase and proton transport. Plant Physiol. 1979 Apr;63(4):744–748. doi: 10.1104/pp.63.4.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman H. M., MacDonald R. J. Cloning of hormone genes from a mixture of cDNA molecules. Methods Enzymol. 1979;68:75–90. doi: 10.1016/0076-6879(79)68007-2. [DOI] [PubMed] [Google Scholar]
- Henderson P. J. A linear equation that describes the steady-state kinetics of enzymes and subcellular particles interacting with tightly bound inhibitors. Biochem J. 1972 Apr;127(2):321–333. doi: 10.1042/bj1270321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitz W. D., Card P. J., Ripp K. G. Substrate recognition by a sucrose transporting protein. J Biol Chem. 1986 Sep 15;261(26):11986–11991. [PubMed] [Google Scholar]
- Ji T. H., Ji I. Macromolecular photoaffinity labeling with radioactive photoactivable heterobifunctional reagents. Anal Biochem. 1982 Apr;121(2):286–289. doi: 10.1016/0003-2697(82)90481-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lichtner F. T., Spanswick R. M. Sucrose uptake by developing soybean cotyledons. Plant Physiol. 1981 Sep;68(3):693–698. doi: 10.1104/pp.68.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W. Energetics of sucrose transport into protoplasts from developing soybean cotyledons. Plant Physiol. 1985 May;78(1):41–45. doi: 10.1104/pp.78.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Schmitt M. R., Hitz W. D., Giaquinta R. T. Sugar transport into protoplasts isolated from developing soybean cotyledons : I. Protoplast isolation and general characteristics of sugar transport. Plant Physiol. 1984 Aug;75(4):936–940. doi: 10.1104/pp.75.4.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden M. C., Davis E. O., Baldwin S. A., Moore D. C., Henderson P. J. Mammalian and bacterial sugar transport proteins are homologous. Nature. 1987 Feb 12;325(6105):641–643. doi: 10.1038/325641a0. [DOI] [PubMed] [Google Scholar]
- Marinetti T. D., Okamura M. Y., Feher G. Localization of the primary quinone binding site in reaction centers from Rhodopseudomonas sphaeroides R-26 by photoaffinity labeling. Biochemistry. 1979 Jul 10;18(14):3126–3133. doi: 10.1021/bi00581a033. [DOI] [PubMed] [Google Scholar]
- Schmitt M. R., Hitz W. D., Lin W., Giaquinta R. T. Sugar Transport into Protoplasts Isolated from Developing Soybean Cotyledons : II. Sucrose Transport Kinetics, Selectivity, and Modeling Studies. Plant Physiol. 1984 Aug;75(4):941–946. doi: 10.1104/pp.75.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne J. H. Morphology and ultrastructure of maternal seed tissues of soybean in relation to the import of photosynthate. Plant Physiol. 1981 May;67(5):1016–1025. doi: 10.1104/pp.67.5.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viitanen P. V., Doran E. R., Dunsmuir P. What is the role of the transit peptide in thylakoid integration of the light-harvesting chlorophyll a/b protein? J Biol Chem. 1988 Oct 15;263(29):15000–15007. [PubMed] [Google Scholar]