Abstract
Incomplete sampling of species’ geographic distributions has challenged biogeographers for many years to precisely quantify global-scale biodiversity patterns. After correcting for the spatial inequality of sample completeness, we generated a global species diversity map for woody angiosperms (82,974 species, 13,959,780 occurrence records). The standardized diversity estimated more pronounced latitudinal and longitudinal diversity gradients than the raw data and improved the spatial prediction of diversity based on environmental factors. We identified areas with potentially high species richness and rarity that are poorly explored, unprotected, and threatened by increasing human pressure: They are distributed mostly at low latitudes across central South America, Central Africa, subtropical China, and Indomalayan islands. These priority areas for botanical exploration can help to efficiently fill spatial knowledge gaps for better describing the status of biodiversity and improve the effectiveness of the protected area network for global woody plant conservation.
Bias-corrected diversity reveals latent peaks of underexplored species richness, detecting prospective sampling priorities
INTRODUCTION
The accumulation of species occurrence data is a fundamental basis for biodiversity science, providing rising opportunities toward addressing major challenges in ecology and conservation (1, 2). Occurrence records have been widely used to model species distribution (3) and to estimate diversity at given localities (4). However, occurrence records notoriously suffer from incompleteness and biases (5), where observed species diversity is statistically influenced by sample size (6). As most occurrence records stem from collections taken for purposes other than estimating diversity patterns, their coverage is usually not geographically systematic nor comprehensive, resulting in a dominance of omission errors (7, 8); to complicate matters, species absence is scale-dependent, and its information is usually unavailable (9). This so-called Wallacean shortfall in biodiversity knowledge (10) potentially precludes a solid understanding of geographical biodiversity patterns (11, 12) and implementation of spatial conservation planning (13).
To correctly capture species diversity patterns, knowing the geographic variation in sample completeness of species occurrence data is critical (14). The explicit link between sample size, completeness, and diversity enables standardization of an observed diversity using rarefaction or extrapolation based on sample completeness (15). This allows fair comparisons of species diversity across multiple assemblages measured at unequal sample completeness without necessarily knowing their true diversity (14). Notably, latitudinal and longitudinal diversity gradients have recently been revisited in this manner, especially in marine ecosystems (16, 17), and revealed unexpected diversity patterns (e.g., bi- or multimodality). Thus, diversity estimation theory challenges the generality of macroecological patterns that often suffer from serious sampling bias (18).
To achieve the global goals and milestones to counteract the current biodiversity crisis [e.g., the post-2020 Biodiversity Framework; (19)], the spatial allocation of conservation resources (e.g., land areas) is a key issue. For effective avoidance or mitigation of negative human impacts, spatial planning based on reliable information of biodiversity distribution is essential (20). However, spatial planning analyses implicitly assume that biodiversity patterns are accurately described, hereunder equally so inside and outside existing conservation areas; the validity of this assumption has not been examined at a global scale yet.
In this study, we focused on the species diversity of woody angiosperms. Woody angiosperms play a crucial role as ecosystem engineers, shaping most terrestrial biomes and supporting ecosystem functions and services on Earth (21). A recent study applied diversity estimation theory to a global occurrence record dataset and estimated the continental-level tree species richness, correcting for uneven sample completeness (4). However, their analysis did not include all woody angiosperms and only estimated diversity at the level of bioregions (biomes on continents). Global patterns of woody plant diversity at finer resolutions remain to be estimated from occurrence records and compared to previous studies using different data sources such as floristic checklists (22, 23) and plot surveys (24).
Here, we generated a global diversity map for woody angiosperms using 13,959,780 occurrence records for 82,974 species. We computed sample completeness and standardized species diversities using a Hill number–based approach to examine bias-corrected geographical patterns of species diversity. Hill numbers (or the effective number of species) (25) have been increasingly used to quantify the species diversity of assemblages. In particular, we evaluated the impact of sample completeness on the description of species diversity and ecological inferences, especially of latitudinal and longitudinal diversity patterns related to spatial resolution, and identified predominant environmental drivers of species diversity at the global and regional scales. We also examined the spatial congruence of the species diversity and sample completeness with the global protected area network and changes in the human pressure. Last, we identified spatial priority areas for allocation of future sampling effort to effectively fill knowledge gaps.
RESULTS AND DISCUSSION
Observed diversity and sample completeness
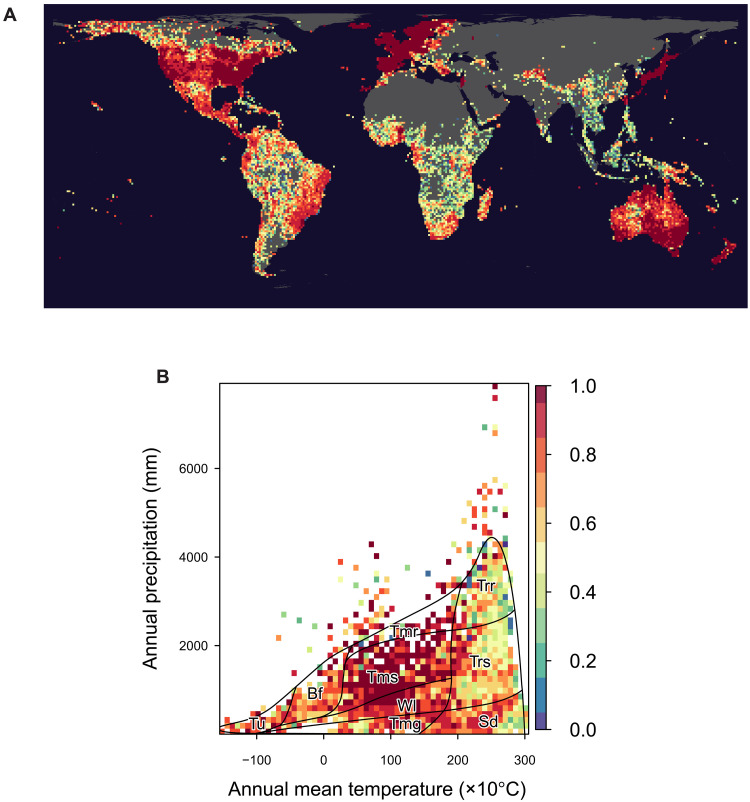
Sample completeness measured by sample coverage, a concept originally developed by Alan Turing in his cryptographic analysis during World War II, greatly varied globally for the occurrence records of woody angiosperms (Fig. 1) (5, 26). Sample coverage is defined as the proportion of the total number of incidences (counted by the 10 km–by–10 km subcells) belonging to detected species to the entire incidences including detected and undetected species. It tended to be high in temperate regions, including North America, Europe, Japan, Australia, and New Zealand. This trend likely reflects the sociopolitical histories of botanical collections rather than climatic conditions (27). Such geographical inequality of sampling effort distorts the description, interpretation, and prediction of biodiversity patterns (26, 28) because the observed diversity patterns reflect both multiple gradients of true diversity and the spatial bias of sampling efforts (29). The observed number of species showed a strong spatial congruence with the total number of occurrences (fig. S1). Expectedly, sample coverage was lower and more variable at the finest spatial resolution (100 km by 100 km) than at coarse resolution (~800 km by 800 km) (fig. S2). Such positive scale dependency of sample completeness has been reported previously in a regional-scale study of plants (30) and a global-scale study of stony corals (17).
Fig. 1. Sample completeness (sample coverage) of species occurrence records of woody angiosperms at global scale.
(A) Geographical map at the 100 km–by–100 km equal-area grids (n = 8427) and (B) the distribution on Whittaker’s biome plot: tundra (Tu), boreal forest (Bf), temperate grassland/desert (Tmg), woodland/shrubland (Wl), temperate seasonal forest (Tms), temperate rain forest (Tmr), tropical rain forest (Trr), tropical seasonal forest/savanna (Trs), and subtropical desert (Sd). In (B), the sample coverage values were aggregated to the median values in pixels divided 60 × 60 of the climate space.
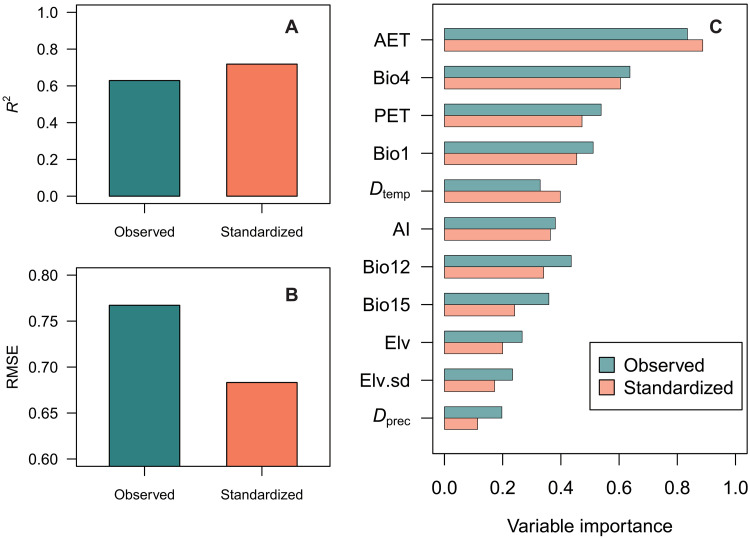
The sample coverage–based standardization of species diversity successfully mitigated the effect of uneven sample completeness (14), improving the description of relative geographical diversity patterns (Figs. 2 to 4 and figs. S3 to S9). Notably, the improvement is quantitatively clear from the increase in the predictive performance of macroecological models to explain species richness patterns after standardization (Fig. 5 and figs. S10 and S11). Specifically, the best-performing random forest model explained ~75% of the total variance of the sample coverage–based standardized species richness pattern, while, for the observed species richness, the model explained only 63% of the total variance (Fig. 5A and fig. S10). This is comparable to previously reported values (70 to 85%) in studies of macroscale plant diversity (23, 24, 31–33). These improved estimates of species richness showed deviations from the geographical patterns of observed species richness (Figs. 2 to 4 and fig. S12) especially for the latitudinal gradients in the Asia-Oceania region (around the Tropic of Cancer; Fig. 3C) and in the Africa-Europe region (around equator; Fig. 3B), likely reflecting that the observed species diversity is severely affected by undersampling. Furthermore, the deviations in geographic patterns between observed and standardized diversities were scale-dependent and tended to be greater at finer spatial resolutions (figs. S13 to S17).
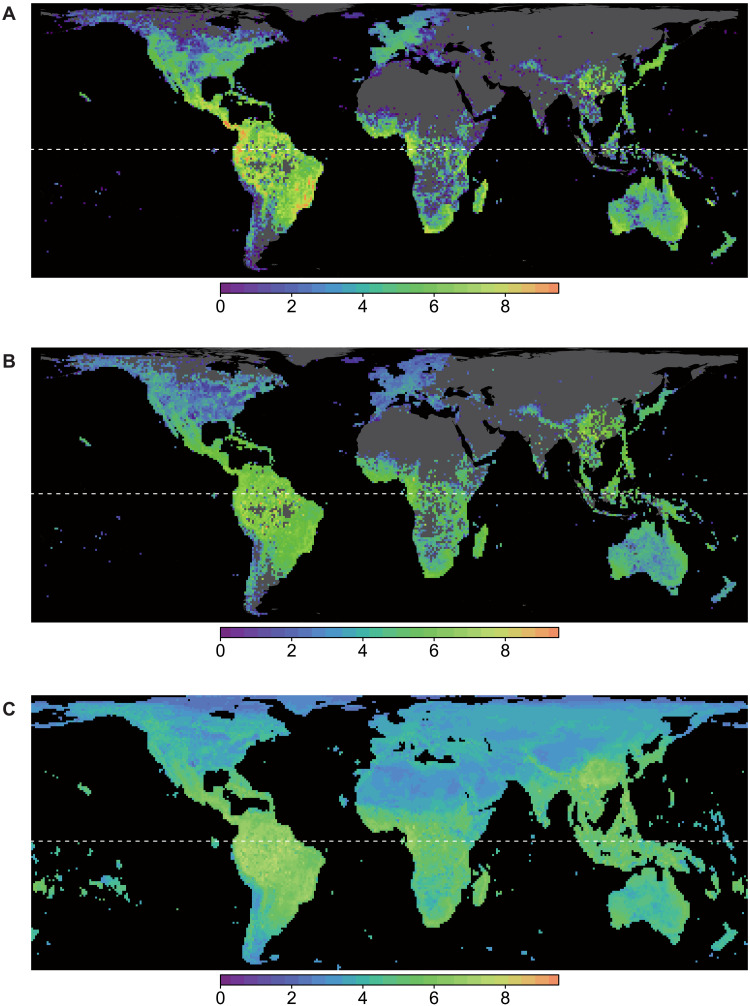
Fig. 2. Geographical distribution of species richness (Hill number q = 0) for 100 km–by–100 km grid cells.
(A) Observed species richness, (B) sample coverage–based standardized species richness (sample coverage = 0.82), and (C) spatial projection of random forest model for standardized richness. The values are log-scaled. Patterns for diversity at higher orders (q = 1 and 2) are given in fig. S18.
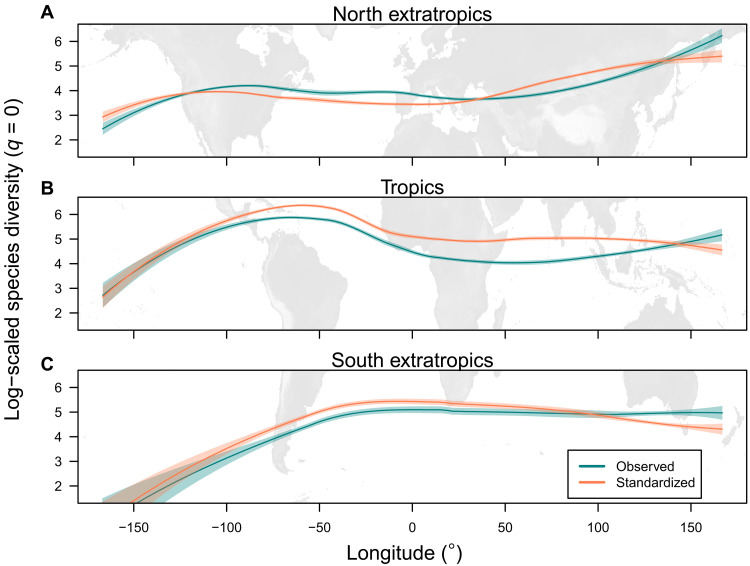
Fig. 4. Longitudinal pattern of species richness in three latitudinal zones for the 100 km–by–100 km grid cells.
The globe was subdivided into (A) northern extratropics, (B) tropics, and (C) southern extratropics: the observed species richness and the standardized species richness (species diversity at the order q = 0) based on sample coverage (0.82). The diversity values are log-scaled. Loess curve (scaling parameter α = 0.6) with 95% confidence interval is shown (green and red lines). Patterns for diversity at higher orders (q = 1 and 2) are given in fig. S20.
Fig. 5. Outputs of the random forest model explaining the observed and the sample coverage–based standardized species richness (sample coverage = 0.82) evaluated at the 100 km–by–100 km grid cell level.
(A) Explanatory power (R2), (B) root mean square error (RMSE) of prediction, and (C) the relative importance of environmental variables. The environmental explanatory variables are mean annual temperature (Bio1), temperature seasonality (Bio4), annual precipitation (Bio12), precipitation seasonality (Bio15), actual evapotranspiration (AET), potential evapotranspiration (PET), aridity index (AI), average elevation (Elv), SD of elevation (Elv.sd), and differences in temperature (Dtemp) and precipitation (Dprec) between the Last Glacial Maximum (LGM) and the present. Results for diversity at higher orders (q = 1 and 2) are given in figs. S21 and S22.
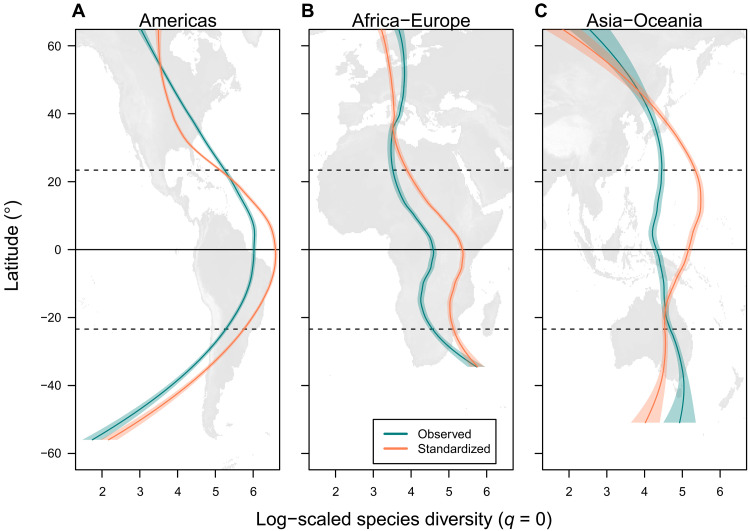
Fig. 3. Latitudinal pattern of species richness in three longitudinal zones for the 100 km–by–100 km grid cells.
The globe was subdivided into (A) Americas, (B) Africa-Europe, and (C) Asia-Oceania: the observed species richness and the standardized species richness (species diversity at the order q = 0) based on sample coverage (0.82). Loess (locally estimated scatterplot smoothing) curve (scaling parameter α = 0.6) with 95% confidence interval is shown (green and red lines). Thick horizontal line indicates the equator, and dashed lines represents the Tropics of Capricorn and Cancer. Patterns for diversity at higher orders (q = 1 and 2) are given in fig. S19.
After applying standardization, the different orders of diversities (q = 0, species richness; q = 1, Shannon diversity; and q = 2, Simpson diversity) showed similar geographical pattern (figs. S18 to S20). Because the rarefaction/extrapolation estimators for higher orders (q = 1 and 2) are nearly unbiased and valid for a wide range of prediction (15), the consistent results among different orders of diversities suggest the standardization of species diversity improved descriptions of geographical diversity patterns.
Geographical patterns of estimated diversity
The standardized (bias-corrected) species richness showed global latitudinal trends with regional diversity hot spots (areas characterized by high species diversity) mainly scattered in the tropics (Figs. 2 and 3). The highest diversity was identified in central South America followed by western tropical Africa and Indomalayan-Australasian region including subtropical China; these are generally in line with the estimation for tree species at continental and biome levels by Gatti et al. (4), while our analysis also captured finer-scale variation in species richness within continents, including a species diversity peak in the northern midlatitudes of Asia.
The sample coverage–based standardization changed the shapes of the latitudinal diversity gradient in the three longitudinal zones (Americas, Africa-Europe, and Asia-Oceania) (Fig. 3 and fig. S3), indicating that the biased raw data lead to mischaracterization of the latitudinal diversity gradient (18). After bias correction, the latitudinal diversity gradients showed regional differences (Fig. 3). The latitudinal diversity gradient in the Americas showed a typical symmetric shape with a peak at the equator and decline toward both poles, in line with the finding of meta-analyses (34). In contrast, the latitudinal diversity gradient in the Africa-Europe and Asia-Oceania regions showed complex patterns with asymmetric unimodality or bimodality. Specifically, the latitudinal diversity gradient in the Africa-Europe region showed a peak at the equator and decline toward higher latitude in the Northern Hemisphere and up to middle (~20°) latitude in the Southern Hemisphere; however, an extraordinary high species diversity at the Cape Floristic Region in South Africa (35) resulted in a distinctive bimodal diversity pattern (Fig. 3B). The latitudinal diversity gradient in the Asia-Oceania region showed a peak at northern central latitudes (around the Tropic of Cancer). These regional anomalies of the latitudinal diversity gradients have been pointed out for all vascular plants using different data sources (36).
These region-specific latitudinal diversity gradients can be understood in the context of a biodiversity anomaly among regions with similar climatic conditions (37), e.g., the diversity depression of African tropical rain forests in comparison with other tropical biomes (38) and the “Asian bias” in species diversity of temperate floras (39). Such regional diversity anomalies among continents were reflected in the longitudinal diversity gradients globally within the tropics and extratropics (Fig. 4 and fig. S4). Uni- and bimodal longitudinal diversity gradients were identified in the three latitudinal zones (northern extratropics, tropics, and southern extratropics). A tropical diversity peak in South America and relatively lower diversity in the rest of tropics (sub-Saharan Africa, Southeast Asia, Australia, and Oceania) was evident, as reported in a continental-level estimation (4) and community-based studies (40, 41). Furthermore, temperate East Asia was identified as a diversity peak on the longitudinal diversity gradient in the northern extratropics, as argued in previous studies (37, 41), although the diversity pattern was scale-dependent and assumed a bimodal shape at coarse spatial resolutions (≥400 km by 400 km; fig. S14). We noted that the longitudinal diversity gradients were not steep (except for oceanic gaps) and were relatively stable irrespective of the sampling bias (Fig. 4 and fig. S4) compared with the latitudinal diversity gradients. A possible explanation of this is relatively smaller variation of sample coverage within the latitudinal zones compared with the longitudinal zones (Fig. 1A).
Environmental drivers of woody species diversity
Within the 11 environmental (mostly climatic, see Materials and Methods for details) factors considered, actual evapotranspiration (AET; a measure that is high in warm and humid climate) was the most important factor to explain the geographical patterns of species richness at the global scale (Fig. 5C), regardless of the modeling framework and spatial resolution (figs. S8 and S9): AET was consistently positively associated with species richness (figs. S5 to S7), as reported in previous global-scale studies using different types of data (23, 24, 33, 35). The other climatic variables also were of relatively high importance. Temperature change from the Last Glacial Maximum (LGM) had a strong negative effect on species diversity, especially at coarser spatial resolutions (fig. S9), suggesting that geographic variation in late-Quaternary paleoclimate instability has had a regional effect on macroscale diversity patterns in woody angiosperms through extinction and dispersal limited range dynamics (42, 43). This is in line with the findings for local plant communities on global geographical gradients by Sabatini et al. (44) where the geohistorical effects on species diversity were greater at coarser grain sizes. In addition, the relative importance of environmental drivers, especially for the top four variables, was stable across the tested spatial resolutions (fig. S9). Such a grain-size–independent relationship of environmental variables with species diversity, which contrasts with the findings of Keil and Chase (24) covering a wider range of grain size across local communities (~10−3 km2) to regional species pools (~106 km2), suggests the predominance of environmental species sorting at the regional species pool level (>104 km2).
The latitudinal and longitudinal diversity gradients had region-specific links to different environmental variables assessed at the level of 100 km–by–100 km grid cells (figs. S23 and S24). Temperature seasonality exhibited a negative correlation with species diversity and outperformed AET in the Americas and the Africa-Europe region, but not in the Asia-Oceania region (fig. S24), suggestive of region-specific species sorting associated with latitudinal climatic seasonality gradients (45). A possible explanation could be that less temperature seasonality has facilitated the accumulation and/or diversification of species with varied strategies in reproduction and biological interactions (46). Although this effect might also exist in the Asia-Oceania region, the high species richness in the midlatitudes was not be explained by temperature seasonality (fig. S23). In addition, historical temperature change since the LGM contributed substantially to shaping the latitudinal diversity gradient in the Americas. This suggests the long-lasting impact of glacial disturbance and climatic disequilibrium in North America (47, 48): Seliger et al. (47) found that the potential ranges of North American trees and shrubs are largely unfilled due to dispersal lags in response to postglacial warming. Our result is also consistent with the finding of a strong correlation between historical climatic stability and number of restricted-range vertebrates in Meso- to South America (49). In contrast, historical climatic changes from the LGM showed minor importance in the Asia-Oceania region. Older historical imprints (e.g., those of the Paleo- and Neogene) might have played a pivotal role in shaping the heightened species richness observed in subtropical China (50). To gain more profound insights into these likely complex dynamics, it would be essential to conduct further investigations that integrate both phylogenetic and fossil information.
Across all continents, the longitudinal diversity gradients were mainly driven by energy and/or water variables (fig. S24). In particular, in the tropics, potential evapotranspiration (PET) and aridity index (AI) were of particularly high importance, indicating that low energy and water availability, i.e., a harsh environment, depress species diversity in particular tropical regions (51). In contrast to the latitudinal diversity gradients, historical temperature changes since the LGM played minor roles in explaining the longitudinal diversity gradients (fig. S24).
Spatial priority areas for future sampling
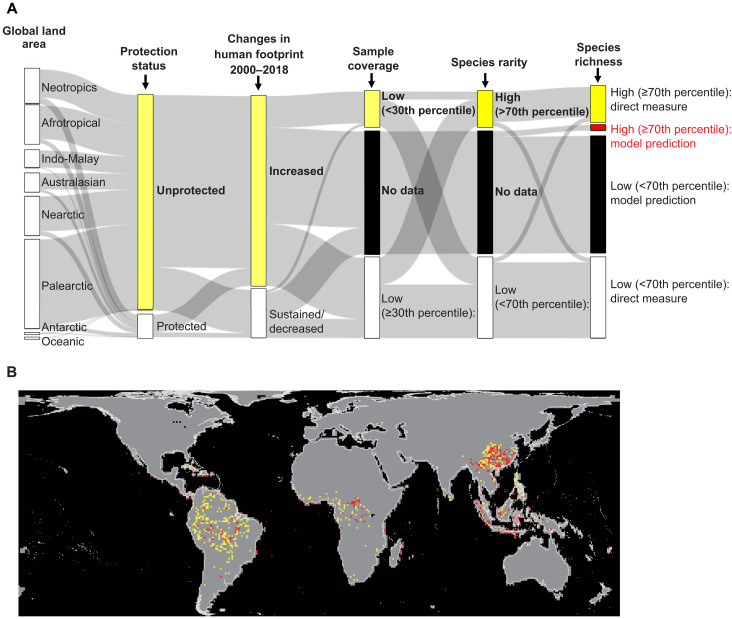
We found that woody angiosperm diversity had been explored with similar sample completeness inside and outside protected areas in general, except for the Eastern Palearctic region where sample coverage was much lower in unprotected areas (fig. S26). The areas with low sample coverage partly overlapped with high species richness and rarity areas that were distributed inside and outside the protected areas (Fig. 6 and fig. S26, A and B). In addition, the spatial prediction of the random forest model (see above) demonstrated that some sites with no occurrence data (where the species diversity estimation was impossible) could contain areas with high species diversity (Fig. 6). Those less or not explored sites are potentially suffering from escalating human pressure (Fig. 6A and fig. S26D). The threat of human pressure is likely to be more prominent in unprotected areas than in protected ones because of its absence of legal restrictions and spillover effects of human pressure from surrounding protected areas (52, 53). Our recommendation would be that, in such data-deficit areas, activities involving the destruction or removal of vegetation should be preceded by botanical surveys to avoid unexpectedly large biodiversity loss, including the extinction of scientifically undescribed species (54).
Fig. 6. Spatial priority areas for improving sample completeness of species occurrence records of woody angiosperms.
(A) Composition of the attributes used to define the priority areas. (B) The geographical map. Yellow color represents the priority areas defined as low sample coverage (<30th percentile of sample coverage values among the grid cells), high species rarity (≥70th percentile of species rarity values among the grid cells), high species richness (≥70th percentile of species richness values among the grid cells), unprotected, and increased human footprint from 2000 to 2018. Red color represents the grid cells with similar values except no occurrence data but predicted as potentially high species richness (≥70th percentile of species richness) by the random forest model.
To visualize the urgent needs for botanical exploration, we selected the grid cells (100 km by 100 km) with lower sample coverage (<30th percentile among the grid cells), higher species richness (≥70th percentile among the grid cells), and rarity (≥70th percentile among the grid cells) from the now unprotected areas experiencing escalating human pressure (Fig. 6 and fig. S27). Those areas represent spatial priority areas of future inventories where immediate assessments and evidence-based conservation decisions would be needed. We found the priority areas in South America, Central and part of West Africa, subtropical China, and Indomalayan islands (Fig. 6). These areas are spatially consistent with botanically unexplored areas (55) with poor mobilization of existing occurrence data (56) in the known global biodiversity hot spots (57). Notably, the priority map reflected not only the information gaps as reported by previous studies (5) but also the needs for preferential sampling efforts (including mobilization of existing records) that are informative to effectively fill the knowledge gap in plant biodiversity (58). Given that the spatial precision of species occurrence records in this study was restricted to less than 100 km (see Materials and Methods), the prioritization for sampling was carried out at the highest feasible resolution within that scope (i.e., 100 km–by–100 km grid cells). Nevertheless, for practical purposes in conservation and management, it would be imperative to perform a similar evaluation in individual regions at a resolution more aligned with actual management units (e.g., 1 km) to devise an effective sampling strategy.
Concluding remarks
This study evaluated the patterns of global woody angiosperm species diversities (Hill numbers at the order 0, 1, and 2) using rarefaction/extrapolation based on biodiversity estimation theory. The estimated diversity patterns and assessment of environmental drivers demonstrated that climatic factors mostly shape global-scale diversity gradients and anomalies through species sorting with latitude and longitude (38, 59). These diversity gradients and anomalies were also influenced by the combination of spatial extent and grain size in the diversity estimation, which is conceptually linked to local/regional species pool size. This study confirmed the nonlinear latitudinal and longitudinal diversity gradients that were determined by the relative effects of different climatic variables, including historical components, as recently argued in marine biodiversity patterns, e.g., bimodality with a tropical decline or truncated bimodality in response to paleo-/modern climatic changes (60, 61). The geographical arrangement of habitats characterized by historically warm and humid climates, coupled with less seasonality, emerges as key determinants of the symmetric unimodal diversity pattern peaking at the equator in the Americas, bimodality peaking at both the equator and southern high latitudes in Africa-Europe, and asymmetric unimodality peaking at northern midlatitudes in Asia-Oceania. Moreover, longitudinal diversity patterns of woody angiosperms peaking at South America and East Asia support historical imprints of the origin of tropical and temperate biome and their expansion across continents (62).
Our results revealed that there are areas, both inside and outside the global protected area network, where botanical sampling efforts have been inadequate or largely nonexistent. This incomplete data can result in spatial disparities between taxonomic diversity and protected area networks (63). Our findings demonstrated that locations with a potentially high number of rare and diverse woody angiosperms are present in the data-deficient areas, suggesting a conservation gap of the current protected areas. Prioritizing the inventory of biodiversity in threatened and unprotected locations is a pressing issue in enhancing the effectiveness of protected area expansion (64) in the post-2020 biodiversity framework.
MATERIALS AND METHODS
Woody angiosperm species data
We prepared a candidate species list of woody angiosperms (here defined as species having lignified stem tissues including trees, shrubs, lianas, bamboos, palms, and cacti) comprising all angiosperm species except exclusively herbaceous families such as Orchidaceae or Cyperaceae. The candidate species list contained 223,724 species. On the basis of information in the national floras, botanical literature, and various databases (see table S1 for the source list), we judged woody species by checking whether the original botanical literature include the following words: woody, tree, shrub, trunk, undershrub, semi-shrub, palm, or culm. We standardized the species names following the World Checklist of Vascular Plants (https://wcvp.science.kew.org/) and integrated subspecies and varieties into the parental binomials. The final list of confirmed woody angiosperms comprised 123,878 species from 6,844 genera and 296 families (table S2). On the basis of the species list, we retrieved 40,770,307 occurrence records from existing databases and literature (tables S1 and S3). We retained only records with precise geolocations (longitude/latitude coordinates or locality names with <100-km precision) with spatial uncertainty small enough to match to a 100 km–by–100 km grid cell. We removed records that were suspected to be a result of artificial introduction by verification in national floristic lists. After the data cleaning processes, the dataset comprised 13,959,780 occurrence records for 82,974 species (67% of known woody angiosperms).
The raw occurrence records comprise a heterogeneous assemblage of data, encompassing spot sampling, botanical expeditions, and standardized plot surveys, which may potentially result in the overrepresentation of rare species and the underrepresentation of common species. To avoid this problem, we converted the species occurrence records into species incidence data at the scale of approximately 10 km–by–10 km cells in the Behrmann projection (PROJ.4: + proj = cea + lat_ts = 30 + lon_0 = 0 + x_0 = 0 + y_0 = 0 + datum = WGS84 + units = m + no_defs). To calculate sample completeness and estimate diversity, we defined four coarser grids, with cells of 100 km–by–100 km, 200 km–by–200 km, 400 km–by–400 km, and 800 km–by–800 km resolution, respectively. Within each cell of those coarser grids, we counted the frequency of species incidence across 10 km–by–10 km equal area subcells. Note that uncertainty in the locations of subcells within a grid cell does not affect the species diversity estimation analysis. Because of the known limitations of the analytical framework of diversity estimation (65), we excluded the cells for which the sample size was deemed inadequate: The observed number of species was less than 6, the number of the subcells where at least one incidence was recorded was less than 6, or the total number of species incidence was equal to the number of singletons (i.e., there were no species recorded in more than one 10 km–by–10 km subcell).
Environmental data
To assess environmental drivers of species diversity, we selected environmental factors that reflect energy or water availability (66), environmental harshness [i.e., dryness or coldness; (50, 67)], climatic seasonality (45), historical climate stability (68), and topographic heterogeneity (69), which have been shown to affect plant diversity in past studies. We obtained climatic information for the present day and the LGM from WorldClim (70). As surrogates of historical climatic stability, we calculated absolute differences in annual mean temperature and annual precipitation between the present day and the LGM (68). Data for AET, PET, and AI were obtained from the Global Aridity and PET Database and Global High-Resolution Soil-Water Balance dataset (71, 72): PET was modeled using the WorldClim dataset; AI was calculated as the ratio between annual precipitation and PET; AET was modeled using PET and vegetation indices (71, 72). Elevation data at 15–arc sec resolution were obtained from the Geospatial Information Authority of Japan (www.gsi.go.jp/kankyochiri/gm_global_e.html).
Diversity estimation
We assessed the sample coverage and species diversities of the grid cells (100 km by 100 km, 200 km by 200 km, 400 km by 400 km, and 800 km by 800 km) by using the 10 km–by–10 km subcell as the fundamental unit for incidence counting. We calculated incidence-based species diversities (diversity at q = 0, 1, and 2, corresponding to species richness, Shannon diversity, and Simpson diversity, respectively) (15) in each coarse grid cell. Given that the relationships between diversity and the number of incidences were often nonsaturated and that empirical diversity depends on sampling effort and sample completeness, we estimated diversity using a combination of rarefaction and extrapolation based on standardizing sample completeness (15). We used sample coverage in each grid cell as a measure of sample completeness; the sample coverage based on incidence data is defined as the proportion of the total number of incidences belonging to detected species to the entire incidences including detected and undetected species in the grid cell (14). The sample coverage was calculated by following formulae (14)
Here, B is defined as
where U is the total number of species incidences in the data; T is the number of subcells where at least one incidence was recorded; and Q1 and Q2 are the number of uniques (those that are detected only in one subcell) and the number of duplicates (those that are detected only in two subcells), respectively.
The basic assumptions for inferring sample coverage and coverage-based diversity estimation are the following (73): (i) The detection probability of a species in any subcell is affected by two sources of heterogeneity: a species effect and a subcell effect. The species effect accounts for the fact that each species may have its own unique occurrence rate; the subcell effect may arise from different surveys, expeditions, mixed types of data collections, etc. All subcell effects are modeled as random effects taken from an unknown probability density function. Thus, species detection probability is allowed to be heterogeneous across subcells. (ii) Because the sample coverage estimate is mainly based on the incidence counts of rare species (i.e., uniques and duplicates), rare species should be correctly identified. One advantage of using incidence data is that only species detection/nondetection records in a subcell are required, regardless of species abundance. Thus, the under-recording of abundant species and over-recording of rare species do not affect our analysis.
We used sample coverage values of doubled reference sample size to capture a wide range of species diversities as possible within a reliable extrapolation range (74), while we slightly mitigated this limitation for extrapolation by using the percentiles of sample coverage values among the grid cells to standardize diversity (17). This means, if a percentile is 40th, then the standardized diversity of each grid cell was obtained by extrapolation to more than double its reference sample size for 40% of the grid cells; for the other 60% of the grid cells, the standardized diversity of each grid cell was obtained either by rarefaction or by extrapolation to less than double its reference sample size. By testing several levels of standardization (see Supplementary Text), we confirmed that the choice of level did not change the outcome of geographical pattern analyses (see below) unless the levels of standardization that were too low (first percentile) or too large (e.g., 100th percentile or asymptotic diversity) were selected. The former is because, when data in all grids were rarefied to a low coverage value, only a few species would be involved, and, thus, the true geographical pattern could not be detected. The reason for the latter is that asymptotic diversity (especially for q = 0) is typically subject to severe negative bias. As the level of standardization had only marginal influence on the outcome of geographical pattern analysis (see Supplementary Text), we presented the results of standardized species richness based on the 40th percentile of sample coverage at the 100 km–by–100 km grid cell level in Results and Discussion.
Geographical pattern analyses
We mapped global woody plant diversity and drew latitudinal and longitudinal diversity gradients in three longitudinal (Americas, Africa-Europe, and Asia-Oceania) and three latitudinal (northern extratropics, southern extratropics, and tropics) zones where previous studies have documented interzonal variations in species diversity patterns (36). For each latitudinal/longitudinal zone, we fitted a Loess (locally estimated scatterplot smoothing) regression curve and then compared the shape of the curves. In a preliminary analysis, we examined various values of smoothing parameter α ranging from 0.4 to 0.9 and verified that the global patterns of latitudinal and longitudinal diversity gradients were generally robust to the smoothing parameter configurations. Therefore, we present the findings for the alpha value of 0.6, which enables the delineation of major regional minima and maxima of species diversity, along with the overall global trends.
To detect the predominant environmental drivers of species diversity at the global and regional scales, we conducted regression analyses using three modeling approaches: ordinary least squares (OLS), generalized additive model [GAM; (75)], and random forest (76). In all models, we set log-scaled species diversity as the response variable. As the explanatory variables, we used annual mean temperature (Bio1), temperature seasonality (Bio4), annual precipitation (Bio12), precipitation seasonality (Bio15), AET, PET, AI, climatic difference between the LGM and present day for temperature (Dtemp) and precipitation (Dprec), average elevation (Elv), and SD of elevation (Elv.sd). In selecting explanatory variables, we excluded variables exhibiting exceedingly high correlations (|r| > 0.9). The maximum correlation coefficients and variance inflation factors (VIFs) among the 11 chosen variables were 0.86 and 20.8, respectively (table S5). Although the observed level of collinearity was a cause for concern (77), we decided to retain all 11 variables for the following reasons: the two variables with notably high VIFs (Bio1 and Bio12) have traditionary served as important drivers for species diversity, each variable contributed to improving the predictive power of the regression models, and the magnitudes and directions of their regression parameters were interpretable.
Individual relationships between species diversity and the environmental variables were visualized by plotting predictive lines (curves) using partial residuals. The relative importance of the explanatory variables was evaluated for OLS and random forest, based on the coefficients of partial determination for OLS, and the mean square error in out-of-bag data (permutation importance) for random forest. We refrained from the relative importance evaluation for GAM because, to the best of our knowledge, there is no established methodology for determining variable importance for GAM. The explanatory power of each model was evaluated using the coefficient of determination (R2). The predictive power was evaluated using root mean square errors computed by 10-fold cross-validation. In addition, we assessed spatial autocorrelation of diversity values and regression residuals for observed and standardized species richness (at sample coverage = 0.82) at the spatial resolution of 100 km by 100 km using the spatial correlograms of Moran’s I (78): This revealed a substantial reduction in spatial autocorrelation within the residuals, especially in the random forest model (fig. S25). In “Environmental drivers of woody species diversity,” we present the results for the random forest model, which showed the best overall performances among the three modeling approaches (see the Supplementary Materials for the results of the OLS and GAM approaches).
Priority map for future sampling
We visualize priority areas for future inventory as strategic sampling efforts that enable us to effectively fill knowledge gaps of woody angiosperm diversity. Specifically, we superimposed sample coverage, standardized species richness (at sample coverage = 0.82), species rarity, human footprint trend, and protected areas at the scale of 100 km–by–100 km grid cells. We defined the species rarity as the total number of unique (those that are detected only once in a grid cell) and duplicate (those that are detected twice in a grid cell) species (14). The information on protected areas was downloaded from the World Database on Protected Areas (www.protectedplanet.net/en), and, then, a grid cell was considered protected if it overlapped with the polygons belonging to the International Union for Conservation of Nature protected area categories I to VI. We regarded grid cells devoid of these categories as unprotected areas, representing locations with relatively high susceptibility to species loss by human pressure and, consequently, warranting higher priority for sampling. We used the global dataset of annual terrestrial human footprint, constructed upon human pressure variables encompassing human population density, agricultural land uses, built environment, and transportation infrastructure (79). We computed the difference in the human footprint between 2000 and 2018 in each grid cell. Last, we identified the priority areas as the grid cells (100 km by 100 km) with lower sample coverage (<0.52 or the 30th percentile among the grid cells) and higher species rarity (≥118 species or the 70th percentile among the grid cells): These priority areas were selected within now unprotected grid cells experiencing increasing human footprint (fig. S27), which would be more threatened by habitat loss or degradation (52).
Acknowledgments
We are grateful to members of the data management team of the Kubota lab, the University of the Ryukyus, for support with data compilation. We thank our colleagues at the Science Directorate of the Royal Botanic Gardens, Kew, for insights and comments.
Funding: This work was supported by the Program for Advancing Strategic International Networks to Accelerate the Circulation of Talented Researchers by the Japan Society for the Promotion of Science (Y.K., B.K., and T.S.); VILLUM FONDEN, grant 00025354 (W.L.E.); VILLUM Investigator project “Biodiversity Dynamics in a Changing World” funded by VILLUM FONDEN, grant 16549 (J.-C.S.); and Danish National Research Foundation “Center for Ecological Dynamics in a Novel Biosphere (ECONOVO),” grant DNRF173 (J.-C.S.).
Author contributions: Conceptualization: Y.K. and B.K. Methodology: B.K., A.C., and T.S. Data compilation: T.S. Visualization: B.K. and T.S. Writing—original draft: B.K. and Y.K. Writing—review and editing: B.K., Y.K., A.C., W.L.E., J.-C.S., and T.S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The original sources of species occurrence records were listed in the Supplementary Materials. The datasets and relevant R codes to reproduce all figures and results are available at Zenodo (DOI: 10.5281/zenodo.6462669).
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S27
Tables S1, S4, and S5
Legends for tables S2 and S3
References
Other Supplementary Material for this manuscript includes the following:
Tables S2 and S3
REFERENCES AND NOTES
- 1.J. Soberón, T. Peterson, Biodiversity informatics: Managing and applying primary biodiversity data. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 359, 689–698 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.J. M. Serra-Diaz, B. J. Enquist, B. Maitner, C. Merow, J.-C. Svenning, Big data of tree species distributions: How big and how good? For. Ecosyst. 4, 30 (2017). [Google Scholar]
- 3.A. Guisan, W. Thuiller, N. E. Zimmermann, Habitat Suitability and Distribution Models: With Applications in R (Cambridge Univ. Press, 2017). [Google Scholar]
- 4.R. Cazzolla Gatti, P. B. Reich, J. G. P. Gamarra, T. Crowther, C. Hui, A. Morera, J.-F. Bastin, S. De-Miguel, G.-J. Nabuurs, J.-C. Svenning, J. M. Serra-Diaz, C. Merow, B. Enquist, M. Kamenetsky, J. Lee, J. Zhu, J. Fang, D. F. Jacobs, B. Pijanowski, A. Banerjee, R. A. Giaquinto, G. Alberti, A. M. A. Zambrano, E. Alvarez-Davila, A. Araujo-Murakami, V. Avitabile, G. A. Aymard, R. Balazy, C. Baraloto, J. G. Barroso, M. L. Bastian, P. Birnbaum, R. Bitariho, J. Bogaert, F. Bongers, O. Bouriaud, P. H. S. Brancalion, F. Q. Brearley, E. N. Broadbent, F. Bussotti, W. C. da Silva, R. G. César, G. Češljar, V. Chama Moscoso, H. Y. H. Chen, E. Cienciala, C. J. Clark, D. A. Coomes, S. Dayanandan, M. Decuyper, L. E. Dee, J. Del Aguila Pasquel, G. Derroire, M. N. K. Djuikouo, T. Van Do, J. Dolezal, I. Đ. Đorđević, J. Engel, T. M. Fayle, T. R. Feldpausch, J. K. Fridman, D. J. Harris, A. Hemp, G. Hengeveld, B. Herault, M. Herold, T. Ibanez, A. M. Jagodzinski, B. Jaroszewicz, K. J. Jeffery, V. K. Johannsen, T. Jucker, A. Kangur, V. N. Karminov, K. Kartawinata, D. K. Kennard, S. Kepfer-Rojas, G. Keppel, M. L. Khan, P. K. Khare, T. J. Kileen, H. S. Kim, H. Korjus, A. Kumar, A. Kumar, D. Laarmann, N. Labrière, M. Lang, S. L. Lewis, N. Lukina, B. S. Maitner, Y. Malhi, A. R. Marshall, O. V. Martynenko, A. L. Monteagudo Mendoza, P. V. Ontikov, E. Ortiz-Malavasi, N. C. P. Camacho, A. Paquette, M. Park, N. Parthasarathy, P. L. Peri, P. Petronelli, S. Pfautsch, O. L. Phillips, N. Picard, D. Piotto, L. Poorter, J. R. Poulsen, H. Pretzsch, H. Ramírez-Angulo, Z. Restrepo Correa, M. Rodeghiero, R. D. P. Rojas Gonzáles, S. G. Rolim, F. Rovero, E. Rutishauser, P. Saikia, C. Salas-Eljatib, D. Schepaschenko, M. Scherer-Lorenzen, V. Šebeň, M. Silveira, F. Slik, B. Sonké, A. F. Souza, K. J. Stereńczak, M. Svoboda, H. Taedoumg, N. Tchebakova, J. Terborgh, E. Tikhonova, A. Torres-Lezama, F. van der Plas, R. Vásquez, H. Viana, A. C. Vibrans, E. Vilanova, V. A. Vos, H.-F. Wang, B. Westerlund, L. J. T. White, S. K. Wiser, T. Zawiła-Niedźwiecki, L. Zemagho, Z.-X. Zhu, I. C. Zo-Bi, J. Liang, The number of tree species on Earth. Proc. Natl. Acad. Sci. U.S.A. 119, e2115329119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.C. Meyer, P. Weigelt, H. Kreft, Multidimensional biases, gaps and uncertainties in global plant occurrence information. Ecol. Lett. 19, 992–1006 (2016). [DOI] [PubMed] [Google Scholar]
- 6.A. Chao, L. Jost, Coverage-based rarefaction and extrapolation: Standardizing samples by completeness rather than size. Ecology 93, 2533–2547 (2012). [DOI] [PubMed] [Google Scholar]
- 7.C. Rondinini, K. A. Wilson, L. Boitani, H. Grantham, H. P. Possingham, Tradeoffs of different types of species occurrence data for use in systematic conservation planning. Ecol. Lett. 9, 1136–1145 (2006). [DOI] [PubMed] [Google Scholar]
- 8.W. Jetz, M. A. McGeoch, R. Guralnick, S. Ferrier, J. Beck, M. J. Costello, M. Fernandez, G. N. Geller, P. Keil, C. Merow, C. Meyer, F. E. Muller-Karger, H. M. Pereira, E. C. Regan, D. S. Schmeller, E. Turak, Essential biodiversity variables for mapping and monitoring species populations. Nat. Ecol. Evol. 3, 539–551 (2019). [DOI] [PubMed] [Google Scholar]
- 9.P. Keil, A. M. Wilson, W. Jetz, Uncertainty, priors, autocorrelation and disparate data in downscaling of species distributions. Divers. Distrib. 20, 797–812 (2014). [Google Scholar]
- 10.J. Hortal, F. de Bello, J. A. F. Diniz-Filho, T. M. Lewinsohn, J. M. Lobo, R. J. Ladle, Seven shortfalls that beset large-scale knowledge of biodiversity. Annu. Rev. Ecol. Evol. Syst. 46, 523–549 (2015). [Google Scholar]
- 11.E. H. Boakes, P. J. K. McGowan, R. A. Fuller, D. Chang-qing, N. E. Clark, K. O’Connor, G. M. Mace, Distorted views of biodiversity: Spatial and temporal bias in species occurrence data. PLOS Biol. 8, e1000385 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.C. Maldonado, C. I. Molina, A. Zizka, C. Persson, C. M. Taylor, J. Albán, E. Chilquillo, N. Rønsted, A. Antonelli, Estimating species diversity and distribution in the era of Big Data: To what extent can we trust public databases? Glob. Ecol. Biogeogr. 24, 973–984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.J. Grand, M. P. Cummings, T. G. Rebelo, T. H. Ricketts, M. C. Neel, Biased data reduce efficiency and effectiveness of conservation reserve networks. Ecol. Lett. 10, 364–374 (2007). [DOI] [PubMed] [Google Scholar]
- 14.A. Chao, Y. Kubota, D. Zelený, C.-H. Chiu, C.-F. Li, B. Kusumoto, M. Yasuhara, S. Thorn, C.-L. Wei, M. J. Costello, R. K. Colwell, Quantifying sample completeness and comparing diversities among assemblages. Ecol. Res. 35, 292–314 (2020). [Google Scholar]
- 15.A. Chao, N. J. Gotelli, T. C. Hsieh, E. L. Sander, K. H. Ma, R. K. Colwell, A. M. Ellison, Rarefaction and extrapolation with Hill numbers: A framework for sampling and estimation in species diversity studies. Ecol Monogr. 84, 45–67 (2014). [Google Scholar]
- 16.C. Chaudhary, H. Saeedi, M. J. Costello, Bimodality of latitudinal gradients in marine species richness. Trends Ecol. Evol. 31, 670–676 (2016). [DOI] [PubMed] [Google Scholar]
- 17.B. Kusumoto, M. J. Costello, Y. Kubota, T. Shiono, C. Wei, M. Yasuhara, A. Chao, Global distribution of coral diversity: Biodiversity knowledge gradients related to spatial resolution. Ecol. Res. 35, 315–326 (2020). [Google Scholar]
- 18.A. Menegotto, T. F. Rangel, Mapping knowledge gaps in marine diversity reveals a latitudinal gradient of missing species richness. Nat. Commun. 9, 4713 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CBD Secretariat, Final text of Kunming-Montreal Global Biodiversity Framework; published 22 December 2022; https://prod.drupal.www.infra.cbd.int/sites/default/files/2022-12/221222-CBD-PressRelease-COP15-Final.pdf.
- 20.T. Shiono, Y. Kubota, B. Kusumoto, Area-based conservation planning in Japan: The importance of OECMs in the post-2020 Global Biodiversity Framework. Global Ecol. Conserv. 30, e01783 (2021). [Google Scholar]
- 21.R. T. Corlett, Plant diversity in a changing world: Status, trends, and conservation needs. Plant Divers. 38, 10–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.W. Barthlott, A. Hostert, G. Kier, W. Kueper, H. Kreft, J. Mutke, M. D. Rafiqpoor, J. H. Sommer, Geographic patterns of vascular plant diversity at continental to global scales. Erdkunde 61, 305–315 (2007). [Google Scholar]
- 23.H. Kreft, W. Jetz, Global patterns and determinants of vascular plant diversity. Proc. Natl. Acad. Sci. U.S.A. 104, 5925–5930 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.P. Keil, J. M. Chase, Global patterns and drivers of tree diversity integrated across a continuum of spatial grains. Nat Ecol Evol. 3, 390–399 (2019). [DOI] [PubMed] [Google Scholar]
- 25.M. O. Hill, Diversity and evenness: A unifying notation and its consequences. Ecology 54, 427–432 (1973). [Google Scholar]
- 26.W. Yang, K. Ma, H. Kreft, Geographical sampling bias in a large distributional database and its effects on species richness-environment models. J. Biogeogr. 40, 1415–1426 (2013). [Google Scholar]
- 27.A. Zizka, O. Rydén, D. Edler, J. Klein, A. Perrigo, D. Silvestro, S. C. Jagers, S. I. Lindberg, A. Antonelli, Bio-Dem, a tool to explore the relationship between biodiversity data availability and socio-political conditions in time and space. J. Biogeogr. 48, 2715–2726 (2021). [Google Scholar]
- 28.D. M. C. C. Alves, A. A. Eduardo, E. V. da Silva Oliveira, F. Villalobos, R. Dobrovolski, T. C. Pereira, A. de Souza Ribeiro, J. Stropp, J. F. M. Rodrigues, J. A. F. Diniz-Filho, S. F. Gouveia, Unveiling geographical gradients of species richness from scant occurrence data. Glob. Ecol. Biogeogr. 29, 748–759 (2020). [Google Scholar]
- 29.R. K. Colwell, J. A. Coddington, Estimating terrestrial biodiversity through extrapolation. Philos. Trans. R Soc. Lond. B Biol. Sci. 345, 101–118 (1994). [DOI] [PubMed] [Google Scholar]
- 30.M. S. Sousa-Baena, L. C. Garcia, A. T. Peterson, Completeness of digital accessible knowledge of the plants of Brazil and priorities for survey and inventory. Divers. Distrib. 20, 369–381 (2014). [Google Scholar]
- 31.R. Field, E. M. O’Brien, R. J. Whittaker, Global models for predicting woody plant richness from climate: Development and evaluation. Ecology 86, 2263–2277 (2005). [Google Scholar]
- 32.E. M. O’Brien, R. Field, R. J. Whittaker, Climatic gradients in woody plant (tree and shrub) diversity: Water-energy dynamics, residual variation, and topography. Oikos 89, 588–600 (2000). [Google Scholar]
- 33.A. P. Francis, D. J. Currie, A globally consistent richness-climate relationship for angiosperms. Am. Nat. 161, 523–536 (2003). [DOI] [PubMed] [Google Scholar]
- 34.N. L. Kinlock, L. Prowant, E. M. Herstoff, C. M. Foley, M. Akin-Fajiye, N. Bender, M. Umarani, H. Y. Ryu, B. Şen, J. Gurevitch, J. Pither, Explaining global variation in the latitudinal diversity gradient: Meta-analysis confirms known patterns and uncovers new ones. Glob. Ecol. Biogeogr. 27, 125–141 (2018). [Google Scholar]
- 35.J. F. Colville, C. M. Beale, F. Forest, R. Altwegg, B. Huntley, R. M. Cowling, Plant richness, turnover, and evolutionary diversity track gradients of stability and ecological opportunity in a megadiversity center. Proc. Natl. Acad. Sci. U.S.A. 117, 20027–20037 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.J. Mutke, W. Barthlott, Patterns of vascular plant diversity at continental to global scales. Biol. Skr. 55, 305–315 (2005). [Google Scholar]
- 37.R. E. Ricklefs, H. Qian, P. S. White, The region effect on mesoscale plant species richness between eastern Asia and eastern North America. Ecography (Cop.). 27, 129–136 (2004). [Google Scholar]
- 38.T. L. P. Couvreur, Odd man out: Why are there fewer plant species in African rain forests? Plant Syst. Evol. 301, 1299–1313 (2015). [Google Scholar]
- 39.H. Qian, R. E. Ricklefs, Large-scale processes and the Asian bias in species diversity of temperate plants. Nature 407, 180–182 (2000). [DOI] [PubMed] [Google Scholar]
- 40.A. H. Gentry, Changes in plant community diversity and floristic composition on environmental and geographical gradients. Ann. Missouri Bot. Gard. 75, 1–34 (1988). [Google Scholar]
- 41.Y. Kubota, B. Kusumoto, T. Shiono, W. Ulrich, Environmental filters shaping angiosperm tree assembly along climatic and geographic gradients. J. Veg. Sci. 29, 607–618 (2018). [Google Scholar]
- 42.G. Feng, Z. Ma, B. Sandel, L. Mao, S. Normand, A. Ordonez, J. Svenning, Species and phylogenetic endemism in angiosperm trees across the Northern Hemisphere are jointly shaped by modern climate and glacial–interglacial climate change. Glob. Ecol. Biogeogr. 28, 1393–1402 (2019). [Google Scholar]
- 43.T. Shiono, B. Kusumoto, M. Yasuhara, Y. Kubota, Roles of climate niche conservatism and range dynamics in woody plant diversity patterns through the Cenozoic. Glob. Ecol. Biogeogr. 27, 865–874 (2018). [Google Scholar]
- 44.F. M. Sabatini, B. Jiménez-Alfaro, U. Jandt, M. Chytrý, R. Field, M. Kessler, J. Lenoir, F. Schrodt, S. K. Wiser, M. A. S. Arfin Khan, F. Attorre, L. Cayuela, M. De Sanctis, J. Dengler, S. Haider, M. Z. Hatim, A. Indreica, F. Jansen, A. Pauchard, R. K. Peet, P. Petřík, V. D. Pillar, B. Sandel, M. Schmidt, Z. Tang, P. van Bodegom, K. Vassilev, C. Violle, E. Alvarez-Davila, P. Davidar, J. Dolezal, B. Hérault, A. Galán-de-Mera, J. Jiménez, S. Kambach, S. Kepfer-Rojas, H. Kreft, F. Lezama, R. Linares-Palomino, A. M. Mendoza, J. K. N’Dja, O. L. Phillips, G. Rivas-Torres, P. Sklenář, K. Speziale, B. J. Strohbach, R. V. Martínez, H.-F. Wang, K. Wesche, H. Bruelheide, Global patterns of vascular plant alpha diversity. Nature Comm. 13, 4683 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.N. V. Page, K. Shanker, Climatic stability drives latitudinal trends in range size and richness of woody plants in the Western Ghats, India. PLOS ONE 15, e0235733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.P. V. A. Fine, Ecological and evolutionary drivers of geographic variation in species diversity. Ann. Rev. Ecol. Evol. Syst. 46, 369–392 (2015). [Google Scholar]
- 47.B. J. Seliger, B. J. McGill, J. Svenning, J. L. Gill, Widespread underfilling of the potential ranges of North American trees. J. Biogeogr. 48, 359–371 (2020). [Google Scholar]
- 48.D. Montoya, M. A. Rodríguez, M. A. Zavala, B. A. Hawkins, Contemporary richness of holarctic trees and the historical pattern of glacial retreat. Ecography (Cop.). 30, 173–182 (2007). [Google Scholar]
- 49.B. Sandel, L. Arge, B. Dalsgaard, R. Davies, K. Gaston, W. Sutherland, J.-C. Svenning, The influence of Late Quaternary climate-change velocity on species endemism. Science 334, 660–664 (2011). [DOI] [PubMed] [Google Scholar]
- 50.R. I. Milne, R. J. Abbott, The origin and evolution of tertiary relict floras. Advance. Bot. Res. 38, 281–314 (2002). [Google Scholar]
- 51.M. Berdugo, M. Delgado-Baquerizo, S. Soliveres, R. Hernández-Clemente, Y. Zhao, J. J. Gaitán, N. Gross, H. Saiz, V. Maire, A. Lehmann, M. C. Rillig, R. V. Solé, F. T. Maestre, Global ecosystem thresholds driven by aridity. Science 367, 787–790 (2020). [DOI] [PubMed] [Google Scholar]
- 52.S. A. Ford, M. R. Jepsen, N. Kingston, E. Lewis, T. M. Brooks, B. MacSharry, O. Mertz, Deforestation leakage undermines conservation value of tropical and subtropical forest protected areas. Glob. Ecol. Biogeogr. 29, 2014–2024 (2020). [Google Scholar]
- 53.W. Li, W.-Y. Guo, M. Pasgaard, Z. Niu, L. Wang, F. Chen, Y. Qin, J.-C. Svenning, Human fingerprint on structural density of forests globally. Nat Sust. 6, 368–379 (2023). [Google Scholar]
- 54.M. M. A. Boehm, Q. C. B. Cronk, Dark extinction: The problem of unknown historical extinctions. Biol. Lett. 17, 20210007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.G. T. Prance, Floristic Inventory of the tropics: Where do we stand? Ann. Missouri Bot. Gard. 64, 659 (1977). [Google Scholar]
- 56.C. Meyer, H. Kreft, R. Guralnick, W. Jetz, Global priorities for an effective information basis of biodiversity distributions. Nat. Commun. 6, 8221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.R. A. Mittermeier, W. R. Turner, F. W. Larsen, T. M. Brooks, C. Gascon, in Biodiversity Hotspots (Springer Berlin, 2011), pp. 3–22. [Google Scholar]
- 58.A. H. Ariño, V. Chavan, N. King, The biodiversity informatics potential index. BMC Bioinformatics 12, S4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.I. Jiménez, R. E. Ricklefs, Diversity anomalies and spatial climate heterogeneity. Glob. Ecol. Biogeogr. 23, 988–999 (2014). [Google Scholar]
- 60.M. Yasuhara, C.-L. Wei, M. Kucera, M. J. Costello, D. P. Tittensor, W. Kiessling, T. C. Bonebrake, C. R. Tabor, R. Feng, A. Baselga, K. Kretschmer, B. Kusumoto, Y. Kubota, Past and future decline of tropical pelagic biodiversity. Proc. Natl. Acad. Sci. U.S.A. 117, 12891–12896 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.A. Zacaï, C. Monnet, A. Pohl, G. Beaugrand, G. Mullins, D. M. Kroeck, T. Servais, Truncated bimodal latitudinal diversity gradient in early Paleozoic phytoplankton. Sci. Adv. 7, eabd6709 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.M. J. Donoghue, A phylogenetic perspective on the distribution of plant diversity. Proc. Natl. Acad. Sci. U.S.A. 105, 11549–11555 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.F. Grattarola, J. A. Martínez-Lanfranco, G. Botto, D. E. Naya, R. Maneyro, P. Mai, D. Hernández, G. Laufer, L. Ziegler, E. M. González, I. da Rosa, N. Gobel, A. González, J. González, A. L. Rodales, D. Pincheira-Donoso, Multiple forms of hotspots of tetrapod biodiversity and the challenges of open-access data scarcity. Sci. Rep. 10, 22045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.H. Gaisberger, T. Fremout, C. J. Kettle, B. Vinceti, D. Kemalasari, T. Kanchanarak, E. Thomas, J. M. Serra-Diaz, J.-C. Svenning, F. Slik, W. Eiadthong, K. Palanisamy, G. Ravikanth, V. Bodos, J. Sang, R. R. Warrier, A. K. S. Wee, C. Elloran, L. T. Ramos, M. Henry, M. A. Hossain, I. Theilade, S. Laegaard, K. M. A. Bandara, D. P. Weerasinghe, S. Changtragoon, V. Yuskianti, P. Wilkie, N. H. Nghia, S. Elliott, G. Pakkad, P. Tiansawat, C. Maycock, C. Bounithiphonh, R. Mohamed, M. Nazre, B. N. Siddiqui, S.-L. Lee, C.-T. Lee, N. F. Zakaria, I. Hartvig, L. Lehmann, D. B. D. David, J.-P. B. Lillesø, C. Phourin, Z. Yongqi, H. Ping, H. A. Volkaert, L. Graudal, A. Hamidi, S. Thea, S. Sreng, D. Boshier, E. Tolentino Jr., W. Ratnam, M. M. Aung, M. Galante, S. F. M. Isa, N. Q. Dung, T. T. Hoa, T. C. Le, M. D. Miah, A. L. M. Zuhry, D. Alawathugoda, A. Azman, G. Pushpakumara, N. Sumedi, I. Z. Siregar, H. K. Nak, J. Linsky, M. Barstow, L. P. Koh, R. Jalonen, Tropical and subtropical Asia’s valued tree species under threat. Conserv. Biol. 36, e13873 (2022). [DOI] [PubMed] [Google Scholar]
- 65.A. Chao, S.-M. Lee, Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87, 210–217 (1992). [Google Scholar]
- 66.D. J. Currie, G. G. Mittelbach, H. V. Cornell, R. Field, J.-F. Guégan, B. A. Hawkins, D. M. Kaufman, J. T. Kerr, T. Oberdorff, E. O’Brien, J. R. G. Turner, Predictions and tests of climate-based hypotheses of broad-scale variation in taxonomic richness. Ecol. Lett. 7, 1121–1134 (2004). [Google Scholar]
- 67.F. I. Woodward, M. R. Lomas, C. K. Kelly, Global climate and the distribution of plant biomes. Phil. Trans. R. Soc. B. 359, 1465–1476 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.M. B. Araújo, D. Nogués-Bravo, J. A. F. Diniz-Filho, A. M. Haywood, P. J. Valdes, C. Rahbek, Quaternary climate changes explain diversity among reptiles and amphibians. Ecography 31, 8–15 (2008). [Google Scholar]
- 69.A. Stein, K. Gerstner, H. Kreft, Environmental heterogeneity as a universal driver of species richness across taxa, biomes and spatial scales. Ecol. Lett. 17, 866–880 (2014). [DOI] [PubMed] [Google Scholar]
- 70.S. E. Fick, R. J. Hijmans, WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315 (2017). [Google Scholar]
- 71.A. Trabucco, R. J. Zomer, Global high-resolution soil-water balance (2019); doi: 10.6084/m9.figshare.7707605.v3. [DOI]
- 72.R. J. Zomer, A. Trabucco, D. A. Bossio, L. V. Verchot, Climate change mitigation: A spatial analysis of global land suitability for clean development mechanism afforestation and reforestation. Agric. Ecosyst. Environ. 126, 67–80 (2008). [Google Scholar]
- 73.A. Chao, R. K. Colwell, C.-H. Chiu, D. Townsend, Seen once or more than once: Applying Good–Turing theory to estimate species richness using only unique observations and a species list. Method. Ecol. Evol. 8, 1221–1232 (2017). [Google Scholar]
- 74.R. K. Colwell, A. Chao, N. J. Gotelli, S.-Y. Lin, C. X. Mao, R. L. Chazdon, J. T. Longino, Models and estimators linking individual-based and sample-based rarefaction, extrapolation and comparison of assemblages. J. Plant Ecol. 5, 3–21 (2012). [Google Scholar]
- 75.S. N. Wood, Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J. R. Stat. Soc. B. 73, 3–36 (2011). [Google Scholar]
- 76.L. Breiman, Random forest. Machine Learn. 45, 5–32 (2001). [Google Scholar]
- 77.C. F. Dormann, J. Elith, S. Bacher, C. Buchmann, G. Carl, G. Carré, J. R. G. Marquéz, B. Gruber, B. Lafourcade, P. J. Leitão, T. Münkemüller, C. McClean, P. E. Osborne, B. Reineking, B. Schröder, A. K. Skidmore, D. Zurell, S. Lautenbach, Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography 36, 27–46 (2013). [Google Scholar]
- 78.P. A. P. Moran, Notes on continuous stochastic phenomena. Biometrika 37, 17–23 (1950). [PubMed] [Google Scholar]
- 79.H. Mu, X. Li, Y. Wen, J. Huang, P. Du, W. Su, S. Miao, M. Geng, A global record of annual terrestrial Human Footprint dataset from 2000 to 2018. Sci. Data 9, 176 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.M. A. Babyak, What you see may not be what you get: A brief, nontechnical introduction to overfitting in regression-type models. Psychosom. Med. 66, 411–421 (2004). [DOI] [PubMed] [Google Scholar]
- 81.R Core Team, R: A language and environment for statistical computing (2021); www.r-project.org/.
- 82.M. Dowle, A. Srinivasan, Data.table: Extension of ‘data.frame’. R package version 1.14.8 (2023); https://CRAN.R-project.org/package=data.table.
- 83.H. Wickham, R. Francois, L. Henry, K. Muller, D. Vaughan, dplyr: A Grammar of Data Manipulation. R package version 1.1.2 (2023); https://CRAN.R-project.org/package=dplyr.
- 84.H. Wickham, D. Vaughan, M. Girlich, tidyr: Tidy Messy Data. R package version 1.3.0 (2023); https://CRAN.R-project.org/package=tidyr.
- 85.Microsoft Corporation, S. Weston, doParallel: Foreach Parallel Adaptor for the ‘parallel’ Package. R package version 1.0.17 (2022); https://CRAN.R-project.org/package=doParallel.
- 86.Microsoft Corporation, S. Weston, foreach: Provides Foreach Looping Construct. R package version 1.5.2 (2022); https://CRAN.R-project.org/package=foreach.
- 87.R. J. Hijmans, M. Barbosa, A. Ghosh, A. Mandel, geodata: Download Geographic Data. R package version 0.5-8 (2023); https://CRAN.R-project.org/package=geodata.
- 88.Original S code by R. A. Becker, A. R. Wilks. R version by R. Brownrigg. Enhancements by T. P. Minka, A. Deckmyn, maps: Draw Geographical Maps. R package version 3.4.1 (2022); https://CRAN.R-project.org/package=maps.
- 89.R. Bivand, N. Lewin-Koh, maptools: Tools for handling spatial objects. R package version 1.1-6 (2022); https://CRAN.R-project.org/package=maptools.
- 90.R. J. Hijmans, raster: Geographic data analysis and modeling. R package version 3.6-20 (2023); https://CRAN.R-project.org/package=raster.
- 91.O. P. Lamigueiro, R. Hijmans, rasterVis. R package version 0.51.5 (2023); https://oscarperpinan.github.io/rastervis/.
- 92.R. Bivand, T. Keitt, B. Rowlingson, rgdal: Bindings for the ‘Geospatial’ data abstraction library. R package version 1.6-6 (2023); https://CRAN.R-project.org/package=rgdal.
- 93.R. Bivand, C. Rundel, rgeos: Interface to Geometry Engine - Open Source (‘GEOS’). R package version 0.6-2 (2023); https://CRAN.R-project.org/package=rgeos.
- 94.E. Pebesma, Simple features for R: standardized support for spatial vector data. R. J. 10, 439–446 (2018). [Google Scholar]
- 95.E. Pebesma, R. Bivand, Spatial Data Science: With Applications in R (Chapman and Hall/CRC, 2023). [Google Scholar]
- 96.R. J. Hijmans, terra: Spatial Data Analysis. R package version 1.7-23 (2023); https://CRAN.R-project.org/package=terra.
- 97.H. Wickham, ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2016). [Google Scholar]
- 98.T. Keitt, colorRamps: Builds Color Tables. R package version 2.3.1 (2022); https://CRAN.R-project.org/package=colorRamps.
- 99.K. Wright, pals: Color Palettes, Colormaps, and Tools to Evaluate Them. R package version 1.8 (2023); https://CRAN.R-project.org/package=pals.
- 100.E. Neuwirth, RColorBrewer: ColorBrewer Palettes. R package version 1.1-3 (2022); https://CRAN.R-project.org/package=RColorBrewer.
- 101.A. W. Bowman, A. Azzalini, R package ‘sm’: Nonparametric smoothing methods. R package version 2.2-5.7 (2021); www.stats.gla.ac.uk/~adrian/sm.
- 102.G. Snow, TeachingDemos: Demonstrations for Teaching and Learning. R package version 2.12 (2020); https://CRAN.R-project.org/package=TeachingDemos.
- 103.S. Chamberlain, K. Ram, V. Barve, Package ‘rgbif’ Interface to the Global “Biodiversity” Information Facility “API” (2016).
- 104.T. C. Hsieh, K. H. Ma, A. Chao, iNEXT: An R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol. 7, 1451–1456 (2016). [Google Scholar]
- 105.M. N. Wright, A. Ziegler, ranger: A fast implementation of random forests for high dimensional data in C++ and R. J. Stat. Softw. 77, 1–17 (2017). [Google Scholar]
- 106.B. M. Greenwell, pdp: An R package for constructing partial dependence plots. R. J. 9, 421–436 (2017). [Google Scholar]
- 107.J. Li, spm: Spatial Predictive Modeling. R package version 1.2.0 (2019); https://cran.r-project.org/package=spm.
- 108.S. V Levin, _plotbiomes: Plot Whittaker biomes with ggplot2_. R package version 0.0.0.9001 (2023); https://valentinitnelav.github.io/plotbiomes/.
- 109.J. Fox, S. Weisberg, An R Companion to Applied Regression (Sage Publications, ed. 3, 2019); https://socialsciences.mcmaster.ca/jfox/Books/Companion/.
- 110.P. Giraudoux, pgirmess: Spatial Analysis and Data Mining for Field Ecologists. R package version 2.0.2 (2023); https://CRAN.R-project.org/package=pgirmess.
- 111.R. Vallejos, F. Osorio, M. Bevilacqua, Spatial Relationships Between Two Georeferenced Variables: With Applications in R (Springer, 2020).
- 112.R. Vaidyanathan, Y. Xie, J. J. Allaire, J. Cheng, C. Sievert, K. Russell htmlwidgets: HTML Widgets for R. R package version 1.6.2 (2023); https://CRAN.R-project.org/package=htmlwidgets.
- 113.J. J. Allaire, C. Gandrud, K. Russell, C. J. Yetman, networkD3: D3 JavaScript Network Graphs from R. R package version 0.4 (2017); https://cran.r-project.org/web/packages/networkD3/index.html.
- 114.W. Chang, webshot: Take Screenshots of Web Pages. R package version 0.5.5 (2023); https://CRAN.R-project.org/package=webshot.
- 115.A.-W. A. Al-Khulaidi, Flora of Yemen. The Sustainable Natural Resource, Management Project (SNRMP II), EPA and UNDP, Republic of Yemen (2013).
- 116.Z. Al-Kuwari, Bahrain First National Report to the Convention on Biological Diversity (Public Commission for the Protection of Marine Resources, Environment and Wildlife, General Directorate for Environment and Wildlife Protection, 2006).
- 117.M. Arechavaleta, N. Zurita, M. C. Marrero, J. L. Martín, Lista preliminar de especies silvestres de Cabo Verde (hongos, plantas y animales terrestres) (Consejería de Medio Ambiente y Ordenación Territorial, Gobierno de Canarias, Santa Cruz de Tenerife, 2005).
- 118.J. Audru, J. Cesar, J. P. Lebrun, Les plantes vasculaires de la république de Djibouti: flore illustrée (CIRAD, Département d’Élevage et de Médicine Vétérinaire, Djibouti, 1994).
- 119.Australian Biological Resources Study, Flora of Australia Online (2011); www.awe.gov.au/science-research/abrs/online-resources/flora-of-australia-online.
- 120.BGCI, GlobalTreeSearch online database (2017); https://tools.bgci.org/global_tree_search.php.
- 121.M. L. Baker, M. F. Duretto, A Census of the Vascular Plants of Tasmania (Tasmanian Herbarium, Tasmanian Museum and Art Gallery, 2011).
- 122.M. J. Balick, M. H. Nee, D. E. Atha, Checklist of the Vascular Plants of Belize, with Common Names and Uses (The New York Botanical Garden Press, 2000). [Google Scholar]
- 123.P. Bamps, Flore d’Afrique Centrale (Zaïre-Rwanda-Burundi): Répertoire des lieux de récolte (Jardin Botanique National de Belgique, 1982).
- 124.H. J. Beentje, C. M. Whitehouse, Flora of Tropical East Africa (Routledge, 2017); 10.1201/9780203755761. [DOI]
- 125.M. Belov, Chileflora (2005–2012); www.chileflora.com/.
- 126.W. G. Berendsohn, A. K. Gruber, J. A. Monterrosa Salomón, N. S. Cuscatlanica, Árboles nativos e introducidos de El Salvador. Parte 1: Angiospermae—Familias A a L. Englera 29, 1–438 (2009). [Google Scholar]
- 127.W. G. Berendsohn, A. K. Gruber, J. A. Monterrosa Salomón, N. S. Cuscatlanica, Árboles nativos e introducidos de El Salvador. Parte 2: Angiospermae - Familias M a P y Pteridophyta. Englera 29, 1–300 (2012). [Google Scholar]
- 128.R. Bernal, S. R. Gradstein, M. Celis, Catálogo de plantas y líquenes de Colombia (2016); http://catalogoplantasdecolombia.unal.edu.co/en/.
- 129.Bhutan Biodiversity Portal, Bhutan Biodiversity Portal (2013); https://biodiversity.bt/.
- 130.M. G. Bingham, A. Willemen, B. T. Wursten, P. Ballings, M. A. Hyde, Flora of Zambia (2018); www.zambiaflora.com.
- 131.J. K. Boggan, V. A. Funk, C. L. Kelloff, M. Hoff, G. Cremers, C. Feuillet, Checklist of the Plants of the Guianas (Guyana, Surinam, French Guiana) (National Museum of Natural History Smithsonian Institution, 1997).
- 132.M. Bou Dagher-Kharrat, Lebanon FLORA (2013); www.lebanon-flora.org/default.php.
- 133.L. Boulos, M. Al-Dosari, Checklist of the flora of Kuwait. J. Univ. Kuwait 21, 203–217 (1994). [Google Scholar]
- 134.Brazil Flora Group, Brazilian Flora 2020 project - Projeto Flora do Brasil 2020. Version 393 (Instituto de Pesquisas Jardim Botanico do Rio de Janeiro, 2021).
- 135.I. Breitwieser, P. J. Brownsey, W. A. Nelson, A. D. Wilton, Eds., Flora of New Zealand Online (2010); www.nzflora.info/.
- 136.G. Brundu, I. Camarda, The Flora of Chad: A checklist and brief analysis. PhytoKeys 23, 1–18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.L. Catarino, E. S. Martins, M. F. Basto, M. A. Diniz, An annotated checklist of the vascular flora of Guinea-Bissau (West Africa). Blumea-Biodiversity, Evol. Biogeogr. Plant. 53, 1–222 (2008). [Google Scholar]
- 138.C.-S. Chang, H. Kim, K. Chang, Provisional Checklist of the Vascular Plants for the Korea Peninsular Flora (KPF) (Designpost, 2014).
- 139.K. Chayamarit, R. Pooma, N. Pattharahirantricin, A Checklist of Plants in Thailand, volume 1 (Office of Natural Resources and Environmental Policy and Planning, 2014).
- 140.K. Y. Chong, H. T. Tan, R. T. Corlett, A Checklist of the Total Vascular Plant Flora of Singapore: Native, Naturalised and Cultivated Species (Raffles Museum of Biodiversity Research, 2009).
- 141.B. J. Conn, R. Banka, L. L. Lee, Plants of Papua New Guinea (2006); www.pngplants.org.
- 142.Conservatoire et Jardin botaniques, South African National Biodiversity Institute, African Plant Database (2012); www.ville-ge.ch/musinfo/bd/cjb/africa/recherche.php.
- 143.P. Craven, H. Kolberg, Plants of Namibia (BRAHMS online, 2018); https://herbaria.plants.ox.ac.uk/bol/namibia.
- 144.I. Darbyshire, M. Kordofani, I. Farag, R. Candiga, H. A. Pickering, Eds., The Plants of Sudan and South Sudan: An Annotated Checklist (Royal Botanic Gardens, 2015).
- 145.G. Dauby, R. Zaiss, A. Blach-Overgaard, L. Catarino, T. Damen, V. Deblauwe, S. Dessein, J. Dransfield, V. Droissart, M. C. Duarte, H. Engledow, G. Fadeur, R. Figueira, R. E. Gereau, O. J. Hardy, D. J. Harris, J. de Heij, S. Janssens, Y. Klomberg, A. C. Ley, B. A. MacKinder, P. Meerts, J. L. van de Poel, B. Sonké, M. S. M. Sosef, T. Stévart, P. Stoffelen, J.-C. Svenning, P. Sepulchre, X. van der Burgt, J. J. Wieringa, T. L. P. Couvreur, RAINBIO: A mega-database of tropical African vascular plants distributions. PhytoKeys 74, 1–18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.A. Deomurari, M. Jani, K. Matieda, J. KaPatel, IBIS-Flora Beta Version—Angiosperm Flora of India (2014); https://flora.indianbiodiversity.org/.
- 147.Editorial Committee of the Flora of Taiwan, Ed., Flora of Taiwan (Department of Botany, National Taiwan University, Taipei, 1975).
- 148.Endemia, Faune et Flore de Nouvelle-Calédonie (2016); https://endemia.nc/.
- 149.Eswatini National Trust Commission, Eswatini’s Flora Database (2020); http://eswatininaturereserves.com/flora/index.asp.
- 150.Euro+Med, Euro+Med PlantBase: The information resource for Euro-Mediterranean plant diversity (2006); www.emplantbase.org/home.html.
- 151.E. Figueiredo, G. F. Smith, Plants of Angola/Plantas de Angola (South African National Biodiversity Institute, SANBI Publishing, 2008). [Google Scholar]
- 152.E. Figueiredo, J. Paiva, T. Stevart, F. Oliveira, G. F. Smith, Annotated catalogue of the flowering plants of São Tomé and Príncipe. Bothalia 41, 41–82 (2011). [Google Scholar]
- 153.Flora do Brasil, Brazilian Flora 2020 (2016); http://floradobrasil.jbrj.gov.br/.
- 154.Flora of China Editorial Committee, Flora of China (Science Press, 1994). [Google Scholar]
- 155.A. Ghahreman, F. Attar, Biodiversity of Plant Species in Iran: The vegetation of Iran, plant species, red data of Iran, endemic species, rare species, species threatened by extinction (vol. 1) (Central Herbarium of Tehran University, Faculty of Science, Tehran, 1999).
- 156.J. F. Gillet, J. L. Doucet, A commented checklist of woody plants in the Northern Republic of Congo. Plant. Ecol. Evol. 145, 258–271 (2012). [Google Scholar]
- 157.M. Hassler, “World plants: Synonymic checklists of the vascular plants of the world (version Nov 2018)” in Species 2000 & ITIS Catalogue of Life, Y. Roskov, L. Abucay, T. Orrell, D. Nicolson, N. Bailly, P. Kirk, T. Bourgoin, R. E. DeWalt, W. Decock, E. van Nieukerken, J. Zarucchi, L. Penev, Eds. (Species 2000: Naturalis, 2018), ISSN 2405-884X; www.catalogueoflife.org/annual-checklist/2019.
- 158.J. Hutchinson, J. M. Dalziel, Flora of West Tropical Africa (Crown Agents For Oversea Governments and Administrations, ed. 2, 1954).
- 159.M. A. Hyde, B. T. Wursten, P. Ballings, M. Coates Palgrave, Flora of Malawi (2016); www.malawiflora.com/index.php.
- 160.M. A. Hyde, B. T. Wursten, P. Ballings, M. Coates Palgrave, Flora of Mozambique (2016); www.mozambiqueflora.com/.
- 161.M. A. Hyde, B. T. Wursten, P. Ballings, M. Coates Palgrave, Flora of Zimbabwe (2018); www.zimbabweflora.co.zw/index.php.
- 162.J. Kellermann, Ed., Flora of South Australia (State Herbarium of South Australia, 2011).
- 163.A. H. Kitalong, R. DeMeo, T. Holm, Native Trees of Palau, A Field Guide (The Environment, Koror, 2008).
- 164.K. Kobisi, Preliminary checklist of the plants of Lesotho. SABONET 34, 1–84 (2005). [Google Scholar]
- 165.G. A. Lazkov, B. A. Sultanova, Checklist of Vascular Plants of Kyrgyzstan. Norrlinia, vol. 24. (Botanical Museum, Finnish Museum of Natural History, 2011).
- 166.Missouri Botanical Garden, Catalogue of the vascular plants of Madagascar (2013); http://legacy.tropicos.org/Project/Madagascar.
- 167.Missouri Botanical Garden, Pakistan Plant Database (1981); http://legacy.tropicos.org/project/Pakistan.
- 168.Missouri Botanical Garden, Bolivia Catalogue (2017); http://legacy.tropicos.org/Project/BC.
- 169.Missouri Botanical Garden, Catalogue of the vascular plants of Ecuador (2017); http://legacy.tropicos.org/Project/CE.
- 170.Missouri Botanical Garden, Flora de Nicaragua (2017); http://legacy.tropicos.org/Project/FC.
- 171.Missouri Botanical Garden, Manual de Plantas de Costa Rica (2017); http://legacy.tropicos.org/Project/Costa%20Rica.
- 172.Missouri Botanical Garden, Panama Checklist (2022); http://legacy.tropicos.org/Project/PAC.
- 173.Missouri Botanical Garden, Peru Checklist (2022); http://legacy.tropicos.org/Project/PEC.
- 174.M. Modzelevich, Flowers in Israel (2005–2021); www.flowersinisrael.com/.
- 175.National Museum of Natural History (U.S.), Botanical Exploration in Myanmar (Smithsonian, National Museum of Natural History, 2003).
- 176.National Museum of Natural Science, Flora of Solomon Islands (2022); http://siflora.nmns.edu.tw/.
- 177.New Zealand Plant Conservation Network, New Zealand’s Flora (2011); www.nzpcn.org.nz/flora/.
- 178.M. Newman, S. Ketphanh, B. Svengsuksa, P. Thomas, K. Sengdala, V. Lamxay, K. Armstrong, A Checklist of the Vascular Plants of Lao PDR (Royal Botanic Garden Edinburgh, 2007).
- 179.J. Norton, S. A. Majid, D. Allan, M. Al Safran, B. Böer, R. Richer, An Illustrated Checklist of the Flora of Qatar (Browndown Publications Gosport, 2009). [Google Scholar]
- 180.PVNH, The herbarium of Vanuatu (PVNH) (2015); http://publish.plantnet-project.org/project/vanuaflora_en.
- 181.P. B. Pelser, J. F. Barcelona, D. L. Nickrent, Co’s digital flora of the Philippines (2011); www.philippineplants.org/.
- 182.PlantNET, New South Wales Flora Online (2017); http://plantnet.rbgsyd.nsw.gov.au/.
- 183.D. Podlech, Checklist of the Flowering Plants of Afghanistan (Ludwig-Maximilians Universität Publ, 2012).
- 184.J. R. Press, K. K. Shrestha, D. A. Sutton, Annotated Checklist of the Flowering Plants of Nepal (Natural History Museum Publications, 2000). [Google Scholar]
- 185.Queensland Government, Census of the Queensland flora 2014 (2014); https://data.qld.gov.au/dataset/census-of-the-queensland-flora-2014.
- 186.RBG Kew, World checklist of selected plant families (2016); http://wcsp.science.kew.org/home.do.
- 187.RMI Office of Environmental Planning and Policy Coordination (OEPPC), Native plants of the Marshalls (2008); https://web.archive.org/web/20150619131813/http://biormi.org/index.shtml?en%2Fnative_plants.shtml.
- 188.S. A. Robertson, Flowering Plants of Seychelles (Royal Botanic Gardens, 1989).
- 189.R. Rolland, V. Boullet, J.-P. Quod, Eds., Mayotte: Biodiversité et Evaluation Patrimoniale (DAF Mayotte et CBN Mascarin (coord.), 2005), pp. 115–187.
- 190.V. E. Selvam, Trees and Shrubs of the Maldives (RAP Publication, 2007). [Google Scholar]
- 191.L. K. Senaratne, A Check List of the Flowering Plants of Sri Lanka (National Science Foundation, 2001).
- 192.M. Setshogo, Preliminary Checklist of the Plants of Botswana (Southern African Botanical Diversity Network Report, 2005).
- 193.P. S. Short, D. E. Albrecht, I. D. Cowie, D. L. Lewis, B. M. Stuckey, Checklist of the Vascular Plants of the Northern Territory (Department of Natural Resources, Environment, The Arts and Sport, 2011).
- 194.K. U. Siddiqui, M. A. Islam, Z. U. Ahmed, S. M. H. Kabir, Eds., Encyclopedia of flora and fauna of Bangladesh (Asiatic Society of Bangladesh, 2007).
- 195.Smithsonian Institution, Flora of the West Indies (2007); https://naturalhistory2.si.edu/botany/WestIndies/.
- 196.South African National Biodiversity Institute, Botanical Database of Southern Africa (BODATSA) (2016); https://posa.sanbi.org/.
- 197.W. R. Sykes, Contributions to the Flora of Niue (New Zealand Department of Scientific and Industrial Research Bulletin 200, 1970), pp. 1–321.
- 198.R. R. Thaman, The Flora of Tuvalu: Lakau Mo Mouku o Tuvalu. Atoll. Research Bulletin 611, 1–129 (2016). [Google Scholar]
- 199.R. R. Thaman, F. R. Fosberg, H. I. Manner, D. C. Hassall, The flora of Nauru. Atoll Research Bulletin 392, 1–223 (1994). [Google Scholar]
- 200.J. Thomas, Plant diversity of Saudi Arabia: Flora checklist (2011); www.plantdiversityofsaudiarabia.info/Biodiversity-Saudi-Arabia/Flora/Checklist/Cheklist.htm.
- 201.M. Thulin, Flora of Somalia (Royal Botanic Gardens, 2006).
- 202.USDA, NRCS, The PLANTS database (2017); https://plants.usda.gov/home.
- 203.University of Greifswald, Virtual Guide to the Flora of Mongolia (2010); https://floragreif.uni-greifswald.de/floragreif/.
- 204.Vietnam Plant Data Center, Plant Database (2007–2018); www.botanyvn.com/?lg=en.
- 205.J. L. Villaseñor, Checklist of the native vascular plants of Mexico. Revista Mexicana de Biodiversidad 87, 559–902 (2016). [Google Scholar]
- 206.W. L. Wagner, D. H. Lorence, Flora of the Marquesas Islands (2002–2018); https://naturalhistory2.si.edu/botany/marquesasflora/index.htm.
- 207.W. L. Wagner, D. R. Herbst, M. W. Tornabene, A. Weitzman, D. H. Lorence, Flora of Micronesia website (2012); http://botany.si.edu/pacificislandbiodiversity/micronesia/index.htm.
- 208.Western Australian Herbarium, FloraBase – the Western Australian Flora (2017); https://florabase.dpaw.wa.gov.au/.
- 209.K. Yonekura, T. Kajita, BG plants wamei-gakumei index (YList) (2003); http://ylist.info/.
- 210.M. Zohary, N. Feinbrun-Dothan, Flora Palaestina (Israel Academy of Sciences and Humanities, 1966).
- 211.F. O. Zuloaga, O. Morrone, M. Belgrano, Catálogo de las Plantas Vasculares del Cono Sur (2014); www.floraargentina.edu.ar/.
- 212.BioScripts, Flora vascular (2014); www.floravascular.com/.
- 213.Botanical Research Institute of Texas & San Diego Zoo Global, Atrium Biodiversity Information System (2005–2017); https://atrium.andesamazon.org/index.php.
- 214.California Native Plant Society, Rare Plant Program, Inventory of Rare and Endangered Plants of California (online edition, v9-01 1.0) (2021); www.rareplants.cnps.org.
- 215.E. Castro, La selva florula digital website (2013); https://sura.ots.ac.cr/florula4/.
- 216.W. D. Clayton, M. S. Vorontsova, K. T. Harman, H. Williamson, GrassBase-the online world grass flora (2016); www.kew.org/data/grasses-db.html.
- 217.Dave’s Garden, Dave’s Garden (2017); https://davesgarden.com/..
- 218.Department of the Environment, Australia, Species profile and threats database (2016); www.environment.gov.au/cgi-bin/sprat/public/sprat.pl.
- 219.K. Fern, A. Fern, R. Morris, Useful tropical plants database (2017); http://tropical.theferns.info/.
- 220.Flora Malesiana, Flora Malesiana (2017); http://portal.cybertaxonomy.org/flora-malesiana/.
- 221.Flora of North America Editorial Committee, Eds., Flora of North America North of Mexico, 20+ vols (Oxford Univ. Press, 1993). [Google Scholar]
- 222.T. Girija, Flowers of India (2016); www.flowersofindia.net.
- 223.W. H. Harvey, O. W. Sonder, Flora Capensis (Hodges Smith & Co., 1865).
- 224.Hortipedia, Hortipedia (2015); https://en.hortipedia.com/Main_page.
- 225.IUCN, The IUCN Red List of Threatened Species, version 2017 (2017); www.iucnredlist.org.
- 226.K. Iwatsuki, T. Yamazaki, D. E. Boufford, H. Ohba, Flora of Japan, vols. 1–4 (Kodansha, 1993–2020).
- 227.Missouri Botanical Garden, Flora of Pakistan (2017); http://legacy.tropicos.org/Project/Pakistan.
- 228.D. Oliver, Flora of Tropical Africa, vols. 1–4) (Lovell Reeve and Co. Publishers, 1904). [Google Scholar]
- 229.Plant Lust, Plant Lust (2007); https://plantlust.com/.
- 230.RBG Kew, Seed information database (SID) (2016); http://data.kew.org/sid/. The webpage was retired in May 2022. The information will be migrated to several of RBG Kew’s other databases; www.kew.org/about-us/reports-and-policies/website-accessibility-science.
- 231.J.P. Roux, Flora of Southern Africa (South African National Biodiversity Institute, 2003).
- 232.Smithsonian Institution’s National Museum of Natural History, Encyclopedia of Life (2018); https://eol.org/.
- 233.South Australian Seed Conservation Centre, Seeds of South Australia, (2017); www.saseedbank.com.au/.
- 234.State Herbarium of South Australia, eFloraSA: Electronic flora of South Australia (2007); www.flora.sa.gov.au/census.shtml.
- 235.T. Vattakaven, R. George, D. Balasubramanian, M. Réjou-Méchain, G. Muthusankar, B. Ramesh, R. Prabhakar, India biodiversity portal: An integrated, interactive and participatory biodiversity informatics platform. Biodivers. Data J. 4, e10279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Xycol database, Xycol (2021); https://xycol.net/.
- 237.CRIA (Centro de Referência em Informação Ambiental), SpeciesLink (2019); https://specieslink.net/.
- 238.Y. Kubota, B. Kusumoto, T. Shiono, W. Ulrich, F. Jabot, Non-neutrality in forest communities: Evolutionary and ecological determinants of tree species abundance distributions. Oikos 125, 237–244 (2016). [Google Scholar]
- 239.Y. Kubota, T. Shiono, B. Kusumoto, Role of climate and geohistorical factors in driving plant richness patterns and endemicity on the east Asian continental islands. Ecography 38, 639–648 (2015). [Google Scholar]
- 240.B. S. Maitner, B. Boyle, N. Casler, R. Condit, J. Donoghue, S. M. Durán, D. Guaderrama, C. E. Hinchliff, P. M. Jørgensen, N. J. B. Kraft, B. McGill, C. Merow, N. Morueta-Holme, R. K. Peet, B. Sandel, M. Schildhauer, S. A. Smith, J. C. Svenning, B. Thiers, C. Violle, S. Wiser, B. J. Enquist, Thebien rpackage: A tool to access the Botanical Information and Ecology Network (BIEN) database. Method. Ecol. Evol. 9, 373–379 (2018). [Google Scholar]
- 241.D. L. Nickrent, M. Costea, J. F. Barcelona, P. B. Pelser, K. Nixon, PhytoImages (2006); www.phytoimages.siu.edu.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S27
Tables S1, S4, and S5
Legends for tables S2 and S3
References
Tables S2 and S3