Abstract
Lentiviral vector (LV)–based gene therapy holds promise for a broad range of diseases. Analyzing more than 280,000 vector integration sites (VISs) in 273 samples from 10 patients with X-linked severe combined immunodeficiency (SCID-X1), we discovered shared LV integrome signatures in 9 of 10 patients in relation to the genomics, epigenomics, and 3D structure of the human genome. VISs were enriched in the nuclear subcompartment A1 and integrated into super-enhancers close to nuclear pore complexes. These signatures were validated in T cells transduced with an LV encoding a CD19-specific chimeric antigen receptor. Intriguingly, the one patient whose VISs deviated from the identified integrome signatures had a distinct clinical course. Comparison of LV and gamma retrovirus integromes regarding their 3D genome signatures identified differences that might explain the lower risk of insertional mutagenesis in LV-based gene therapy. Our findings suggest that LV integrome signatures, shaped by common features such as genome organization, may affect the efficacy of LV-based cellular therapies.
Signatures characterized by SCID-X1 patients treated by LV gene therapy shed light on genome organization and efficacy.
INTRODUCTION
Lentiviral vectors (LVs) are widely being used to deliver genes into hematopoietic stem cells (HSCs) or immune cells for therapeutic intent (1). Examples include gene therapy approaches for monogenic diseases such as immunodeficiencies and sickle cell disease (2–4) as well as the adoptive immunotherapy with T cells expressing chimeric antigen receptors (CARs) (5, 6). Lentiviral integration is mediated by the viral preintegration complex, in which the reverse-transcribed viral genome is associated with the host chromatin and integrated into the host genome via the viral integrase. Despite recent advances (7), fundamental questions regarding the underlying molecular mechanisms of viral integration remain elusive including the role of local chromatin organization and the global three-dimensional (3D) genome structure. While the precise location of a vector integration site (VIS) can serve as a marker for monitoring the corresponding clone and its subsequent evolution in longitudinal analysis (8–10), a systematic mapping of the integrome can provide further insights into integration site selection and may have important implications in terms of biosafety and efficacy for gene therapy.
Early studies of lentiviral VISs focused on the integration pattern of the HIV (11–13). Subsequently, VISs were analyzed in patients who received LV-transduced autologous HSCs as part of gene therapy studies (14–20). However, only a small number of samples were analyzed, resulting in a rather limited number of VISs, which prohibited a systematic analysis of the LV integrome to gain deeper insights into the genomics, epigenomics, and 3D genome signatures. To address this limitation, we now took advantage of our ongoing early-phase clinical study for infants with newly diagnosed X-linked severe combined immunodeficiency [SCID-X1; NCT01512888 (21)]. SCID-X1 is a rare, life-threatening disorder caused by mutations in the gene IL2RG, which is shared by multiple cytokine receptors necessary for the proper development and function of lymphocytes. Affected infants therefore present with severe opportunistic infections during the first months of life. On this ongoing clinical study, we have shown that this SCID-X1 gene therapy approach is well tolerated and results in the development of a functional immune system without evidence of malignant transformation with a median follow-up of >2.5 years (21, 22). Only 1 of our 23 patients (patient 1) required a second infusion of genetically modified stem cells 1 year after his initial infusion and since then has developed a functional immune system with a follow-up of >4.5 years. We compiled 273 samples from the first 10 patients enrolled on this study, and we have profiled more than 280,000 VISs. The unprecedented number of VISs provided enough statistical power to investigate genomic features at a high resolution. By integrating VISs with recent functional genomics datasets generated in big data consortia (23, 24), we characterized here a set of genomics, epigenomics, and 3D genome signatures that distinguish the LV integrome and confirmed our findings using clinical-grade CD19-specific CAR (CD19-CAR) T cell products. Beyond the performed analysis, our cohort presents a unique resource to examine LV integration site selection and its potential clinical implications for a broad range of LV-based gene therapy as well as immunotherapy approaches. In particular, the recent report of a single patient, who developed myelodysplastic syndrome (MDS) on a LV-based gene therapy study for adrenoleukodystrophy (ALD) (25), underscores the importance and relevance of our performed analysis.
RESULTS
Profiling LV integration sites of SCID-X1 patients
In our clinical study, infants received autologous HSCs transduced with a LV encoding the IL2RG gene after low-dose, targeted busulfan conditioning (21). Nine of analyzed 10 patients received a single dose of LV-transduced autologous CD34-positive Hematopoietic Stem and Progenitor Cells (HSPCs) with a vector copy number (VCN) of 0.16 to 1.13 except for one patient (patient 1), who received two LV-transduced HSC infusions, 1 year apart. We investigated the lentiviral integrations using patients’ samples, including unsorted peripheral blood mononuclear cells (PBMCs) and sorted PBMC populations, CD14+/CD15+ myeloid cells, CD19+ B cells, CD3+ T cells, and CD3−/CD56+ natural killer (NK) cells, for all patients, and unsorted and sorted bone marrow cells for patient 1 (fig. S1A). VISs were determined by quantitative shearing linear amplification (qsLAM) polymerase chain reaction (PCR) (26), and for this study, we built a VIS analysis pipeline as outlined in fig. S1B (see the “Profiling and quantification of LV integration sites” section for details, including signal and noise separation). By integrating samples from different cell types and time points, we compiled a list of VISs for each patient, with the total number of sites ranging from 770 to 72,000 (fig. S1C). Mapping VISs to the genome revealed VIS patterns that were consistent between patients except of patient 1 as judged by pairwise Pearson correlation coefficient analysis (fig. S2A). For individual patients, the locations of VISs within cell lineages were consistent; however, sharing of a specific VIS between cell lineages, defined by individual base-pair integration sites, was uncommon (fig. S2B) and similarly for VISs of individual patients across multiple time points (fig. S2C). Clonal diversity, as defined by VISs, correlated with VCN when multiple diversity metrics were used [UC50, oligoclonality index (OCI), Shannon diversity index, Chao estimator] (fig. S2D).
Genomic and epigenomic signatures of LV VIS
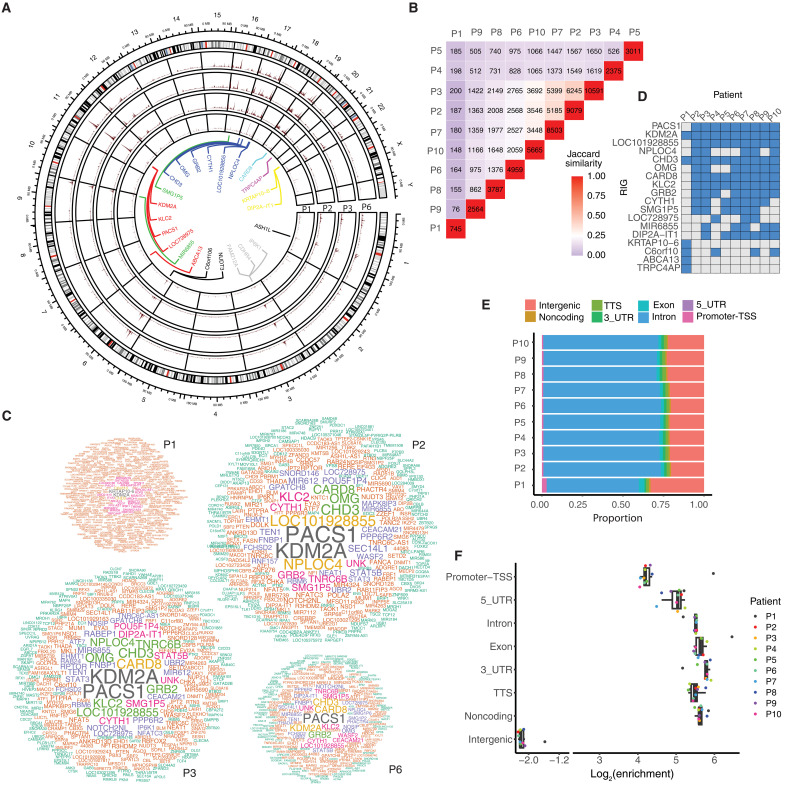
To gain insight into the observed similarities and differences between VIS patterns, we embarked on a detailed genomic and epigenomic analysis. One of the most notable features was the existence of genomic regions with a high density of integration sites, so-called hotspots (Fig. 1A) (27). More specifically, hotspots were identified as 10-kb regions in which vector integrations occurred at a greater frequency than expected. We found that these hotspots overlapped in patients 2 through 10 (Fig. 1B) and identified recurrent integration genes (RIGs) (Fig. 1C and table S1) with seven RIGs (KDM2A, PACS1, LOC101928855, CHD3, CARD8, GRB2, and KLC2) being shared by eight or nine patients (Fig. 1D). Apart from hotspots and RIGs, LVs integrated predominantly into introns and, after normalization, were strongly depleted in intergenic regions (Fig. 1, E and F).
Fig. 1. Genomic signatures of lentiviral VIS.
(A) Circular projection of the human genome with integration sites from patients 1, 2, 3, and 6. Gene names near integration site hotspots are listed in the inner circle. (B) Overlap of VIS hotspots in patients. The numbers along the diagonal represent the number of hotspots in individual patients, and the other numbers indicate hotspots common between pairs of patients. (C) RIGs in patients 1, 2, 3, and 6. Word clouds show the frequency of integration site clustering at each of the genes. Genes with more VISs within them are shown in larger font. Patients 2, 3, and 6 share similar RIGs. (D) Top RIGs among patients. Many RIGs were shared by patients 2 through 10; patient 1 had a set of unique RIGs. (E) The proportion of VISs located in various genomic regions. Most VISs fall in introns. (F) Enrichment of VISs in various genomic regions. The enrichment is calculated by normalizing the number of VIS fall upon a region by a background, in which is the number of VIS merely scales with the length of the region.
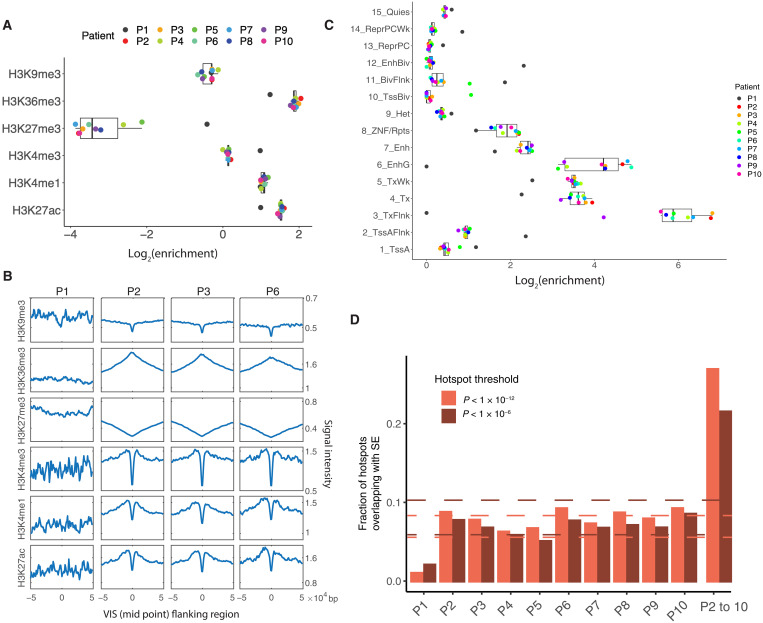
To investigate and extend the finding that lentiviral VISs are enriched in transcriptional units and active gene bodies (7, 28, 29), we took advantage of Chromatin immunoprecipitation followed by sequencing (ChIP-seq) data of CD34+ HSPCs from the Roadmap Epigenomics project (table S2) (23) and estimated the presence of VISs in histone modification marks. Despite not all integrations happening in bone marrow repopulating HSCs, but rather in a mixed population of HSCs and committed progenitors, VISs of patients 2 through 10 were enriched (log2 enrichment > 1) in active promoter H3K4me1, enhancer H3K27ac, and gene body H3K36me3 marks while being depleted in the repressive H3K27me3 mark (log2 enrichment < −1) (Fig. 2A). In contrast, for patient 1, VISs were not strongly associated with active or repressive histone marks (−1 < log2 enrichment < 1). Given the high number of VISs analyzed, we determined the signal intensity of the same histone modifications mapped in CD34+ HSPCs in a 100-kb window flanking the identified VIS. Patients 2 through 10 showed the same signatures, with active marks having the strongest signal and repressive marks the weakest signal (Fig. 2B; results are shown for patients 1, 2, 3, and 6). These results were further confirmed using the 15 chromatin states specific for CD34+ HSPCs as defined by the ChromHMM algorithm (Fig. 2C) (30). In all patients except for patient 1, VISs were strongly enriched in actively transcribed regions and enhancers (states 3, 4, 5, and 6) and depleted in repressive regions and heterochromatins (states 9, 13, 14, and 15).
Fig. 2. Epigenomic signatures of lentiviral VIS.
(A) Enrichment of VISs in six histone marks. (B) Aggregation plots showing the signal intensity of six histone marks at the VIS and its flanking regions. (C) Enrichment of VISs in 15 chromatin states. (D) Overlap of hotspots and SEs in all patients. Two sets of hotspots were called on the basis of different thresholds. The overlap was greater with the more stringent threshold (except for patient 1). Dashed lines mark the median ± 1.5 interquartile range. The overlap values corresponding to patient 1 were outliers with both thresholds. Bars are shown for individual patients and for hotspots shared by patients 2 through 10. The overlap between hotspots and SE in patient 1 is significantly different from that of patients 2 to 10 (P = 3.26 × 10−9 for threshold 10−6 and P = 1.07 × 10−7 for threshold 10−12, Fisher exact test). Note that the reported values are still higher than the expected overlap by chance, which are less than 0.005.
Although we demonstrated for patients 2 through 10 a twofold enrichment in active gene bodies and enhancer regions, this did not explain the observed 10- to 100-fold enrichment of VISs in RIGs. Examination of RIGs of HIV by DNA fluorescence in situ hybridization has demonstrated that these genes are located in proximity to the nuclear pores (31), which suggests that the viral genome preferentially integrates into active chromatin close to the nuclear pore complex (NPC) (32). Motivated by the recent finding that NPC proteins (e.g., NUP93 and NUP153) bind super-enhancers (SEs) (33), we compiled a list of SEs in HSPCs from the literature (34, 35) to determine whether VIS hotspots in our patient cohort were in proximity to NPCs. For patients 2 through 10, 8% of VIS hotspots overlapped with SEs (Fig. 2D). When we limited the analysis to shared VIS hotspots, this overlap increased to 20%. In contrast, the SE/VIS hotspot overlap was about 1 to 2% for patient 1 (P = 3.26 × 10−9 for threshold 10−6 and P = 1.07 × 10−7 for threshold 10−12, Fisher exact test). In general, hotspots were highly enriched in SEs in patients 2 to 10 (P = 0, randomization test using randomly sampled regions as hotspots) but not the case for patient 1 (P < 10−7 for threshold 10−6 and not significant for threshold 10−12; randomization test).
LV VIS signatures in 3D genome conformation
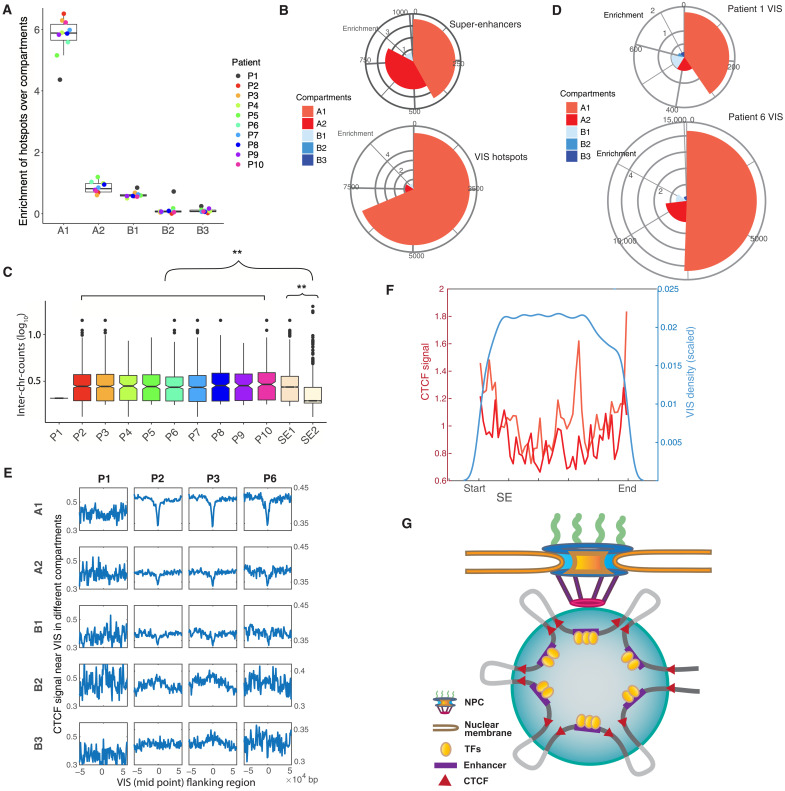
For additional insight into the relationship between VISs and the 3D architecture of the human genome, we analyzed the presence of VISs in the different nuclear compartments, which can be broadly divided into (i) an accessible and active compartment A and (ii) a compartment B that comprises repressed genes (36). More recently, these compartments have been further subdivided into A1, A2, B1, B2, and B3 based on clustering and on differences in multiple histone marks (37, 38). Using Hi-C data for HSPCs and the corresponding compartment predictions (38, 39), we found that hotspots of patients 2 through 10 showed notable enrichment in compartment A1 (Fig. 3A). Together, these results indicate that compartment A1 is in close proximity to NPCs, in contrast to compartment A2. This finding is consistent with a previous study that mapped compartment A2 in close proximity to speckles, which are located within the nucleus and not in its periphery (40). Whereas more than 60% of VIS hotspots were mapped to compartment A1, SEs were equally distributed, overall, between compartments A1 and A2 (Fig. 3B). Therefore, SEs located in compartment A1 (SE1) are more likely to overlap with VISs than are SEs in compartment A2 (SE2). Likewise, similar to VIS hotspots, SE1s are spatially proximal to one another by comparison with SE2s (Fig. 3C). Furthermore, although the HSC products of patients 1 and 6 had similar VCNs, 0.16 and 0.17, respectively (fig. S1C), 74% of VISs of patient 6 resided in the active compartments A1 and A2, as compared to only 51% of VISs of patient 1, excluding VCN bias as a potential explanation (Fig. 3D).
Fig. 3. 3D genome signatures of lentiviral VIS.
(A) Enrichment of hotspots in five compartments. (B) Mapping of SE and VIS hotspots to genome subcompartments. The pies indicate the fraction of VISs mapped to the compartments, with the radii representing the enrichment. The list of hotspots is compiled from all patients except patient 1. (C) Number of interchromosomal read counts between SEs. Only SEs overlapping with hotspots are considered. No boxplot for patient 1 because the number of SEs overlapping with hotspots was low. The number of interchromosomal reads is used as a proxy for spatial proximity. SE1s are in closer proximity than SE2s (P < 2.2 × 10−16, two-sided Wilcoxon test). (D) Mapping of VISs in patients 1 and 6 to the subcompartments. The distribution of VISs in P1 significantly differs from the distribution in P6 (P = 0.0175, Multinomial test, P value was estimated by a Monte Carlo approach). (E) Aggregation plots showing the intensity of CTCF signals at the VISs and their flanking regions at different subcompartments. For patients 2, 3, and 6, VISs were depleted at CTCF sites in compartments A1 and A2 but showed minor enrichment in compartments B2 and B3. (F) VIS density and CTCF signal within SEs. VISs were depleted near the two ends. A strong CTCF signal was observed near the ends of both SE1 and 2. (G) Model of LV integration. After entry into the nucleus through the nuclear pore, the LV integrates into/near SEs interacting with the NPC. Clusters of enhancers are brought together with the binding of multiple transcription factors (TFs). CTCFs at the ends are responsible for forming the whole SE, together with multiple CTCFs forming enhancer-promoter loops. LV integrations occur within SEs, being depleted near the CTCF binding sites but more likely in the regions in between (lightly colored intervals).
We then investigated how LVs integrated into SEs by examining VIS locations relative to CTCF binding sites. We found that depending on the corresponding compartments, VIS could be either depleted (in A1 and A2) or enriched (in B2 and B3) near CTCF sites (Fig. 3E). VIS density was low near the ends of SE (mostly in compartments A1 and A2), where the CTCF signal was particularly strong (Fig. 3F). This nonuniform distribution of VISs within a SE is most likely explained by the fact that CTCFs are responsible for forming multiple enhancer-promoter loops inside a SE (41). Connecting with the emerging picture on how SEs mediate the formation of coactivator condensation (42), our findings support a model in which LVs preferentially target SEs proximal to NPCs and integrate into chromatin loops created by CTCF and interacting proteins (Fig. 3G).
The HSC LV integrome signature is present in LV-transduced T cells
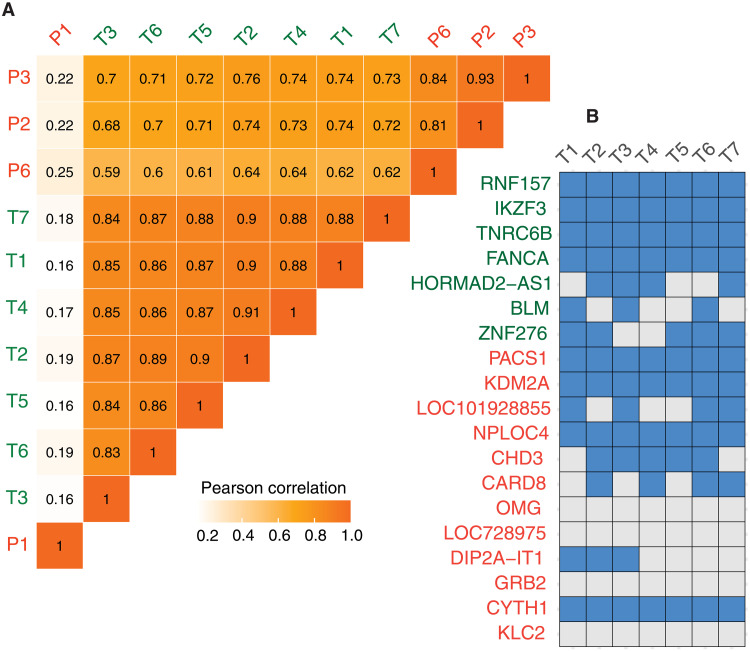
To determine the generalizability of the LV integrome signature identified in HSCs of patients with SCID-X1, we took advantage of T cells that were transduced with a clinical-grade LV encoding a CD19-CAR. Despite the differences in the underlying cell type, the LV integrome in CAR T cells exhibited (i) the same set of signatures, including the existence of hotspots (fig. S3A); (ii) the same preference for histone marks (fig. S3, B and C); (iii) overlap between hotspots and SEs (fig. S3D); and (iv) strong enrichment for the nuclear compartment A1 (fig. S3E). The VIS pattern in CAR T cells was similar to that in SCID-X1 patients 2 through 10, as judged by Pearson correlation coefficients, with SCID-X1 patient 1 again being an outlier (Fig. 4A). Although multiple RIGs identified post-HSC transduction in patients with SCID-X1 were shared by CAR T cells (e.g., PACS1, KDM2A, and CHD3), RIGs unique to each cell type were also identified (e.g., IKZF3) (Fig. 4B). The presence of specific RIGs in different hematopoietic cell types is not unexpected because histone modifications and 3D genome structure are, to a certain extent, cell type specific (43).
Fig. 4. The HSC lentiviral integrome signatures are present in lentiviral-transduced T cells.
(A) Correlation coefficients for the density profiles of integration sites across all CD19-CAR T cell samples and four patients with SCID-X1. (B) Top RIGs in CD19-CAR T cell samples. The list comprises RIGs identified in SCID-X1 (red), as shown in Fig. 1D, and RIGs specific for CD19-CAR T cells (green). SCID-X1 and CD19-CAR T cell RIGs are partially shared.
LV integrome signature is distinct from the γ-retroviral integrome
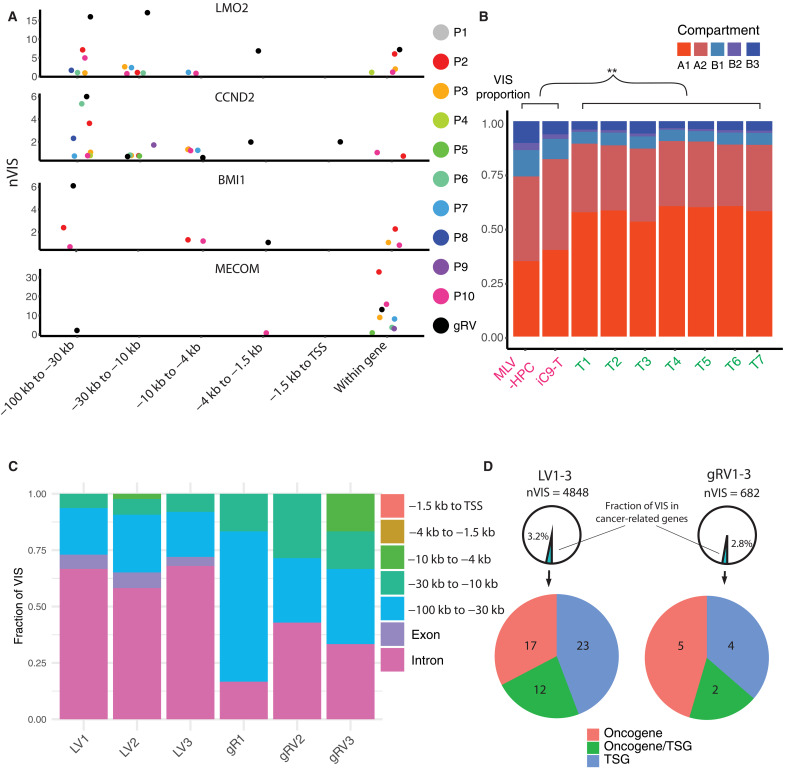
The distributions of lentiviral VISs have been compared with γ-retroviral (gRV) integration sites in the past (12, 44, 45). By integrating epigenetic data, it is widely established that LV integration sites are spread along active gene bodies, whereas gRV prefer to target promoter regions (7). This difference is of clinical importance because in early gRV therapy trials for SCID-X1 and Wiskott-Aldrich syndrome, patients later developed T cell acute lymphoblastic leukemia (T-ALL) with vector integration near the LMO2, CCND2, BMI1, and/or MECOM gene loci (46–48). In addition, two patients on a clinical study with X-linked chronic granulomatous disease developed MDS with monosomy 7 secondary to gRV insertions near the MECOM gene locus (49). We examined VISs of our patients near these genes, and while integration sites were located within introns, we did not identify integration sites at the promoter regions of these genes in the analyzed patients’ samples (Fig. 5A). We further investigated the discrepancy with respect to 3D genome organization. Using compartment prediction for CD3-positive T cells (38), we compared the LV integrome in our CAR T cells with the gRV integrome obtained from two studies in which hematopoietic progenitor cells (HPCs) or T cells were transduced with gRVs encoding different transgenes (29, 50). While gRVs also integrated into compartment A1, the frequency was significantly lower compared to LVs (Fig. 5B). This was mirrored by a higher frequency of gRV integration sites in compartments A2 and B. Intriguingly, the LMO2, BMI1, and MECOM gene loci are all found in compartment A2.
Fig. 5. LV integration sites near cancer genes and LV signatures are distinct from the gRV integrome.
(A) Distribution of VISs near and within known VIS-mediated mutagenesis genes. gRV integrome are profiled in murine leukemia virus–derived gRV transduced HPCs (MLV-HPC) and inducible caspase 9 gRV-encoded transduced T cells (iC9-T), as shown in (B). Unlike gRV, VISs identified in the patients’ cohort are rarely found in promoters (−1.5 kb to TSS). nVIS, number of VISs (B) Distribution of VISs in five genome compartments in LV and gRV integrome. gRV integrome signatures exhibit more VISs in compartments A2 and B, and fewer in compartment A1, as compared to LV signatures. The comparisons are all statistically significant (P = 0, Binomial test). (C and D) Comparison of the location of vector integration sites between LV and gRV in T cells from three healthy donors: 1, 2, and 3. In (C), VISs are categorized on the basis of the locations within or near the cancer-related genes relative to TSS. VISs are rarely found in the promoters. In (D), the number of VIS near cancer-related genes is displayed.
To compare LV and gRV integration sites in cells that were transduced in parallel, we transduced T cells using LV and gRV encoding the same transgene (IL2RG) used in our clinical trial. Integration sites were profiled, paying particular attention to VISs near a set of cancer-related genes that are associated with T cell acute lymphoblastic leukemia (T-ALL), acute myeloid leukemia (AML), B cell acute lymphoblastic leukemia (B-ALL), or MDS (51). Again, gRV integration sites were more likely upstream of transcriptional start site (TSS) (Fig. 5C). While only a small fraction of VISs were close to cancer-related genes, LV VISs were enriched near tumor suppressor genes as opposed to oncogenes based on annotation by Catalogue of Somatic Mutation in Cancer (COSMIC) (Fig. 5D) (52). We found a similar pattern when we examined the patients’ integration sites near the same set of cancer-related genes (fig. S4A). Of note, VISs were located in introns or 10 or 100 kb upstream of TSS and rarely in promoter regions (fig. S4B), and the relative abundance of the corresponding VIS clones in all samples was low (fig. S4C). VISs exhibited a polyclonal pattern in all profiled lineages except for patient 1 at all time points evaluated, consistent with our previous publication in which we had presented shorter follow-up data for the first eight patients (fig. S4D) (21). Likewise, while we found integration sites within in the HMGA2 gene locus (fig. S5A), the relative proportion of the corresponding clones were low, and there was no evidence of clonal selection (fig. S5B) as reported for one patient on a LV gene therapy study for β-thalassemia (15).
Single-cell profiling provides insight into distinct VIS signature of patient 1
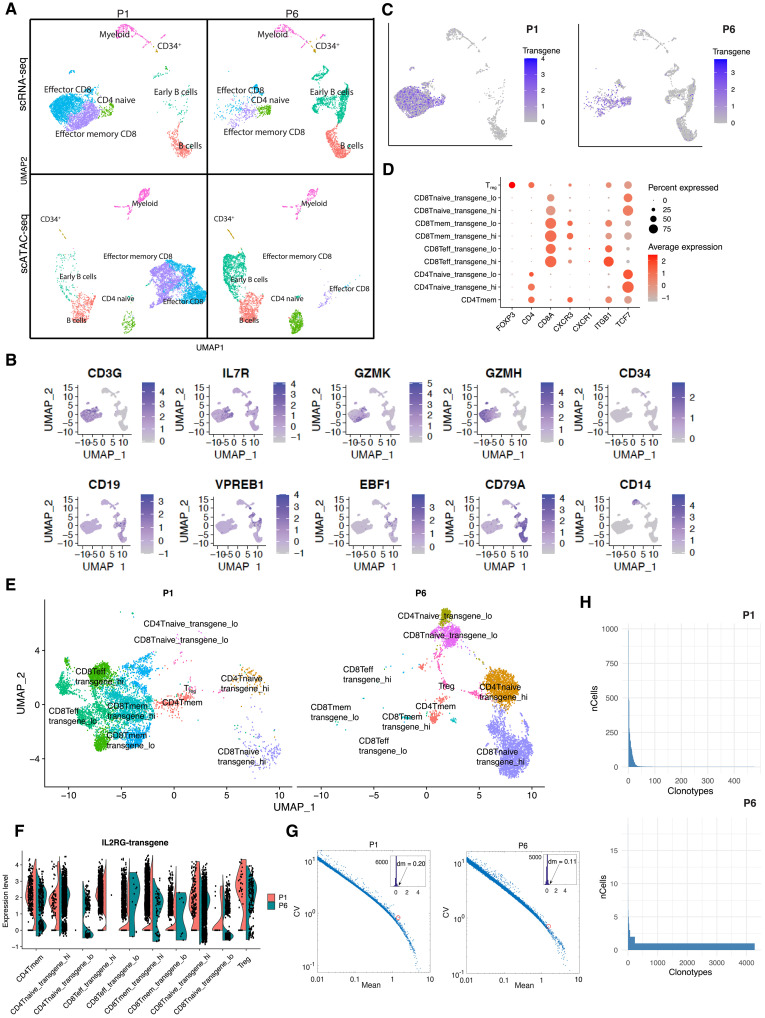
Why did the LV integrome differ in patient 1? Given the close link between 3D genome architecture and the LV integrome, we performed single-cell RNA-sequencing (scRNA-seq) and single-cell ATAC-Seq (scATAC-seq) profiling of bone marrow samples from patients 1 and 6 collected 18 months after gene therapy, to decipher whether patient 1 had an altered 3D genome architecture (Fig. 6A). Single-cell transcriptomic profiling revealed that major hematopoietic cell types like T cells, B cells, and myeloid cells were present in both patients (Fig. 6B) and that the LV-encoded IL2RG transgene was expressed in T cells (Fig. 6C). The 5′ scRNA-seq in sorted T cells further revealed the presence of different T cell subsets in both patients; however, the frequency varied between both patients (Fig. 6, D to F). Of particular interest is the emergence of independent clusters with high and low transgene expression in multiple types of T cells such as naïve CD4 and CD8 subsets. Gene sets, including cytoplasmic translation and ribosome, as well as immune gene sets like major histocompatibility complex class proteins, were enriched in T cells expressing high levels of the IL2RG transgene (fig. S6, A and B). As compared to genes with similar expression, IL2RG transgene expression varied to a greater extent, highlighting the positional effects of random integration on gene expression levels (53). Expression variation was higher in patient 1 than patient 6 (Fig. 6G). T cell receptor sequencing (TCR-seq) identified greater TCR diversity in patient 6 than in patient 1 (Fig. 6H), which is consistent with the observed difference in VIS diversity (fig. S2D). Last, we profiled VISs for a small portion of cells at a single-cell level using the scATAC-seq data and the EpiVIA method (fig. S7, A and B) (54) and confirmed the presence of two integration sites within a single cell, as suggested by the relative frequency of individual VISs (fig. S7C).
Fig. 6. Single-cell profiling of bone marrow samples from patients 1 and 6.
(A) Uniform Manifold Approximation and Projection (UMAP) plots showing the major cell types identified in patients 1 and 6 by using scRNA-seq and scATAC-seq. (B) Expression of markers of hematopoietic cell lineages. (C) Expression of the transgene IL2RG in patients 1 and 6. (D) Major markers for various T cell clusters. (E) UMAP plots showing several types of T cell in patients 1 and 6 profiled using 5′ scRNA-seq. Both patients have regulatory T cells (Tregs) and CD4 memory (mem) T cells; P6 has more CD4 and CD8 naïve T cells; P1 has more CD8 mem and effector T cells. (F) Expression of IL2RG transgene in patients 1 and 6 by 5′ scRNA-seq profiling of T cells. (G) Expression variation of IL2RG transgene (circled in red) in cells of patients 1 and 6. Expression variation is quantified by coefficient of variation (CV). Compared to other genes with similar mean expression, IL2RG expression variation (also known as expression noise) is high. Insets show the histogram of distance-to-median (dm) for all genes, which is defined as the difference between the gene’s CV and the running median of 11 neighboring genes. The dm values of the IL2RG gene are 0.20 for patient 1 (ranked top 5% of genes displayed) and 0.11 for patient 6 (ranked top 16% of displayed genes). (H) TCR clonotypes in patients 1 and 6.
On the basis of single-cell profiling, another discrepancy between P1 and P6 is the composition of their immune cells. Specifically, P1 has a large population of T cells and a small population of B cells, whereas B cells dominate in B6. To examine whether the immune cell composition in P6 is the norm just like all the VIS-related features, we analyzed available flow cytometric data of bone marrow samples from all 10 patients (fig. S8A). We found that the flow cytometry data are consistent with the single-cell profiling data. Moreover, unlike P1 in whom T cells are most abundant, the composition in most patients mirrors P6, with B cells being more abundant than T cells (fig. S8B).
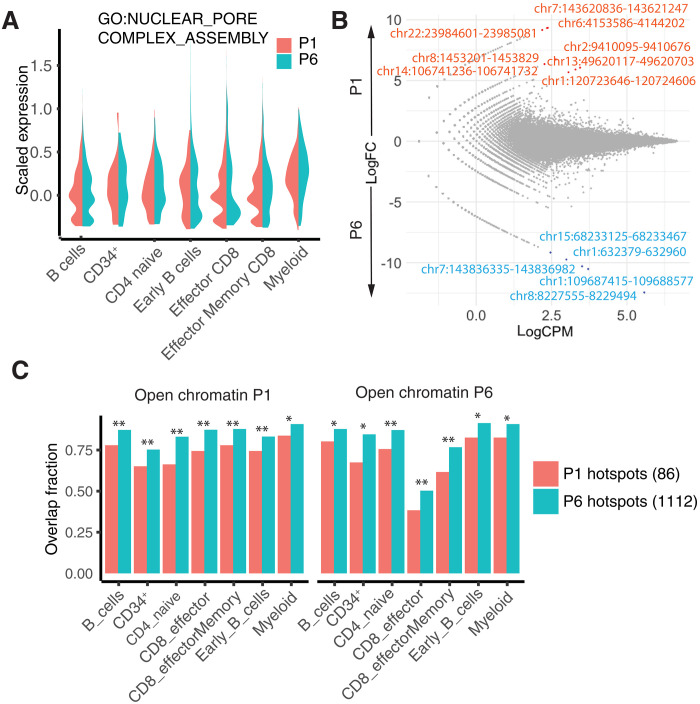
Given the close proximity of the LV integrome to the NPC, we next analyzed the expression of NPC-related genes and found no difference between the two patients, making an intrinsic NPC defect in patient 1 unlikely (Fig. 7A). Likewise, scATAC-seq data revealed no significant differential accessibility in these patients, indicating similar chromatin organization (Fig. 7B). In particular, the open chromatin regions of patient 1 overlapped more with the hotspots patient 6 than with patient 1’s own hotspots (Fig. 7C). Therefore, our results suggest that patient 1 had only a transient change, rather than a permanent change, in his 3D genome structure during LV transduction.
Fig. 7. Single-cell profiling provides insight into distinct lentiviral integrome of patient 1.
(A) Average expression of genes related to NPC assembly (TMEM170A, NUP205, AHCTF1, NUP98, NDC1, NUP107, RTN4, NUP93, and NUP153). (B) Differentially accessible regions (DARs) in B cells in patients 1 and 6. Accessibility is quantified by counts per million (CPM) and compared by fold change (FC). DARs in red were more accessible in patient 1 (log2FC > 0), and DARs in blue were more accessible in patient 6 (log2FC < 0). DARs were rare in all cell types [false discovery rate < 0.05, exact test with Benjamini-Hochberg (BH) correction]. (C) Overlap of hotspots with open chromatin regions identified by scATAC-seq. Open chromatin regions in both patients had a greater overlap with hotspots in patient 6 than with those in patient 1. There were 86 hotspots in patient 1 and 1112 hotspots in patient 6. Statistical significance was estimated by sampling two sets of random hotspots 500 times. All the differences reported were statistically significant (*P < 10−3, **P < 10−6).
Integrome signatures have limited predictive power on clonal selection
To determine whether VISs confer a selection advantage, we took advantage of the estimated relative VIS frequency (clonal abundance) in longitudinal samples from each patient and developed a classification model to predict clonal repopulation by using VISs. Abundant clones were more likely to have integrome signatures that (i) overlapped with SEs and (ii) histone modification signals such as H3K36me3. In contrast, clones with integrome signatures that included (i) VISs located within compartment B and (ii) a strong H3K27me3 signal tended to be rare (fig. S9A). However, the overall predictive power of the integrome signatures on future clonal population was rather limited, as evidenced by training a classification model that integrates all signatures (fig. S9B).
DISCUSSION
In this study, by incorporating the high number of VISs in samples from infant patients with SCID-X1, we have deciphered a set of LV integrome signatures in HSCs, which are shaped by local chromatin structure, the locations of SE, as well as the global 3D genome organization. Because of common underlying mechanisms in LV integration, these signatures are shared by CAR T cells, and some of them have been previously reported in vitro post-HIV infection, as well as in other HSC-based gene therapy studies in older patients (55–57). As compared to the study of HIV-1 integration in T cells by Lucic et al. (57), our study of lentiviral VISs confirms the generalizability of the integration of different lentiviruses and suggests a model in which LVs integrate into the first open chromatin region encountered (i.e., compartment A1) once they enter the nucleus through NPCs, which also function as scaffolds for SEs, thus influencing 3D genomic structure (58–61). While evolution shapes natural viruses to use this strategy for maximizing their survival, gene therapy has adopted the strategy to maximize the chance of inserting transgene in the open chromatin and thus optimizing transgene expression. The strategy could be a double-edged sword, as integration into active sites might increase the risk of inferring with existing gene regulation.
One of the most consistent observations in our VIS analysis is the unusual distribution of P1. scATAC-seq suggested that a potential cause is a transient change in the 3D genome structure of his HSCs during LV transduction. Intriguingly, patient 1 had disseminated CMV infection secondary to SCID-X1 when his HSCs were collected, and nuclear egress of CMV capsids in infected cells is associated with nuclear lamina disruption (62–64). It has been reported that the perturbation of NPC influences HIV integration. Specifically, translocated promoter region–depleted cells exhibited less integration mapping to regions enriched in deoxyribonuclease I hypersensitive sites or histone modification H3K36me3 (32). Their observations resemble the case of P1. Future studies are needed to determine whether there is CMV changes the VIS pattern of LVs.
Besides being used as a proxy to understand chromatin organization, the integrome signatures have potential implications in terms of safety and efficacy for gene therapy. Insertional mutagenesis remains a major concern of LV- and gRV-based gene therapy. For example, subsets of patients enrolled on gRV-based gene therapy trials for SCID-X1, Wiskott-Aldrich Syndrome, or chronic granulomatous disease developed malignancies (46–49). However, the risk of insertional mutagenesis seems to be lower with self-inactivating gRVs (65), suggesting the importance of the strength and long-range activity of gRV promoters, and moderate cellular promoters are less genotoxic (66, 67). While vector design plays a major role to insertional oncogenesis, we explored the possible influences of VISs on insertional mutagenesis in terms of their statistical integration preferences. Although LVs and gRVs have distinct preferences for histone modifications (7), gRV integration occurs during mitosis, in which compartmentalization disappears across the genome (68, 69). Our analysis showed that LV integration sites exhibit a strong bias toward compartment A1, whereas gRV sites are more uniform. Thus, genome compartmentalization might prevent LV integration close to oncogenes that are linked to gRV VIS-mediated mutagenesis, since these are located in compartment A2. In this regard, little is known currently about the LV integrome signature of the ALD patient, who developed MDS postgene therapy, and it would be of great interest to analyze it with our developed pipeline.
Apart from the integration signatures, we examined the integration sites near key cancer-related genes and used their frequency to monitor the possibility of clonal dominance. Being consistent with our observed signatures, we did not find VIS at the promoter of oncogenes like LMO2 (45). While we focused solely on the genomic location of VIS with respect to the cancer-related genes, it could be further examined in terms of the chromatin states. Given an enrichment of VIS in enhancer regions, the possibility of vector integrations causing global rearrangement such as disruption of topologically associating domain boundaries cannot be ruled out (70). In general, our data suggest that LV integration sites are more commonly found in tumor suppressor genes rather than oncogenes, presenting a rather different kind of safety concern because LV could inactivate the targets by insertion inside exons. Of course, the absence of VIS at certain problematic loci could be a consequence of the limitations of our VIS profiling assay. The overlap of integration sites identified across different time points or cell types showed no strong evidence of saturation, leading to the possibility of finding problematic integration sites by increasing the number of samples or sequencing depth. Fortunately, proliferating clones would be abundant and thus will be likely to be profiled even there is a lack of saturation. Nevertheless, as the VIS signatures reported are statistical in nature, the lack of saturation does not affect the related conclusions.
A related question of interest is whether integration sites drive clonal kinetics. Our model demonstrated that VISs do not appear to be key drivers for clonal kinetics. The hypothesis comes with a caveat that the promoter used in the vector is not particularly strong (71). Nevertheless, as LV signature regions tend to be in the open chromatin that is transcriptionally active, the abundant clones tend to express high levels of the LV-encoded transgene. High levels of IL2RG expression might be advantageous for SCID-X1 gene therapy since it could potentially promote the development of functional T, NK, and B cells. Since for other gene therapy applications, levels of transgene expression might not confer an advantage, correlations between VISs and outcome most likely dependent on the targeted disease. In addition, the influence of integrations sites on outcome might also be vector dependent based on recently performed VIS analyses on CD19-CAR T cell products (72, 73).
Here, we performed single-cell profiling on patients’ samples to examine transgene expression on a single-cell resolution. We identified the expected lineages and particularly the emergence of clusters with high and low transgene expression in multiple types of T cells. As compared to T cells with low transgene expression, T cells with high transgene expression expressed gene sets suggestive of a highly functional state. It is tempting to speculate that low and high transgene expression on a single-cell level is a consequence of the location of VISs, and future studies are needed to explore this possibility. The effects on gene expression by factors like local chromatin structures or spatial positioning of genes within the nucleus have previously been studied by high-throughput reporter assays (53). However, chromatin position effects have not been carefully explored in the context of LV-based gene therapy. Our single-cell profiling showed that IL2RG transgene expression varied to a greater degree (also known as expression noise) than genes with similar expression levels. While the consequences of positional effects remain unknown, the observed expression noise is likely due to the random spatial distribution of the transgene. Future studies that simultaneously profile VISs and transcriptomes on a single-cell level might be able to provide additional insights.
In conclusion, our study has uncovered previously unknown ways in which the 3D structure of the human genome influences lentiviral integration and highlights the significance of dynamic nuclear organization in lentiviral gene therapy. In addition to the biology, our data so far do not show evidence of clonal evolution consistent with insertional mutagenesis. Our findings have translational relevance because the identified integrome signatures could be used as biomarkers for LV-based gene therapy and should be applicable to a broad range of cellular therapies that are being developed to treat human diseases.
MATERIALS AND METHODS
Study design
The objective of this study was to characterize the canonical genomics and epigenomics properties of the LV insertion sites using samples from 10 patients currently enrolled in our SCID-X1 clinical study (NCT01512888). As integrome signatures are statistical properties, unsorted and sorted PBMC samples of a patient at different time points were integrated to increase statistical significance. To characterize the signatures, we integrated VISs with a variety of genomics and functional genomics including epigenomics and Hi-C data. To determine whether the signatures are independent of the vector but universal for a wide range of lentiviral therapy, samples from CD19-CAR T cell therapy study (NCT03573700) were analyzed. Single-cell profiling of bone marrow samples was performed to provide additional insight on the VISs of patient 1, which are farther away from the integrome signatures as compared to the other patients.
Human samples
Human samples were obtained from two clinical studies. The SCID-X1 clinical study (NCT01512888) (21) was approved by the US Food and Drug Administration and by the institutional review boards at St. Jude Children’s Research Hospital (St. Jude) and the University of California San Francisco (UCSF) Benioff Children’s Hospital. Ten consecutive patients who lacked a matched sibling donor (six at St. Jude Children’s Research Hospital and four at UCSF Benioff Children’s Hospital) received their stem cell product. Written informed consent was obtained from the legal guardians of the patients. The CD19-CAR T cell therapy study (NCT03573700) was approved by the US Food and Drug Administration and by the institutional review board at St. Jude.
LV and generation of genetically modified cell products
The replication-incompetent VSVG-pseudotyped LVs used for the clinical studies have been described previously (21, 71, 74, 75). Briefly, the vectors are self-inactivating third-generation lentiviruses that are devoid of any viral transcriptional enhancers or promoters. The 3′ partially deleted viral long terminal repeat (LTR) includes a 400-bp fragment from the chicken hypersensitive site 4 chromatin insulator element. The generation of clinical-grade LV-transduced CD34+ progenitors cells from patients with SCID-X1 was described previously (21). Figure S10 shows a schematic of the vector. LV-transduced CD19-CAR T cells from healthy donors and patients were generated from leukapheresis products by using a protocol established at our center (76).
Profiling and quantification of LV integration sites
Generation of a Jurkat clone with 20 defined VISs
For testing and optimizing the performance of our VIS identification pipeline, we used a previously generated Jurkat clone as described (26). The Jurkat clone has 20 VISs (it was originally reported of having 19 VIS, and since then, one additional VIS in chr X with a weaker signal was identified). In short, we spiked-in 5 or 10 ng of DNA from the 20-VIS clone to 1000 ng of DNA from wild-type Jurkat cell line and test the qsLAM PCR assay and the bioinformatics pipeline (see below).
Jurkat cells were transduced with the CL20-4i-EF1ahccOPT vector at a multiplicity of infection (MOI) of 100 and were subsequently sorted into 96-well plates at 1 cell per well. Cell clones were expanded, and genomic DNA was extracted from selected clones. VCN was measured using the quantitative PCR method. One clone with an estimated VCN of 19 was chosen for further experiments. VCN was verified by Southern blot analysis and fluorescence in situ hybridization using a vector-specific probe. The purified plasmid containing 4458 bp of the provirus form of the integrated vector was labeled with a red deoxyuridine triphosphate (dUTP) (AF594; Molecular Probes) by nick translation. The labeled probe was combined with sheared human DNA and hybridized to metaphase chromosomes or interphase chromatin derived from the 20 copy Jurkat cell clone using routine cytogenetic harvest methods (4′,6-diamidino-2-phenylindole) and analyzed. The locations of all 20 VISs are listed in fig. S11.
qsLAM PCR assay
To identify the genomic locations of lentiviral insertion sites and quantify their abundance, a qsLAM PCR assay was developed (26). Here, we summarize the procedures as previously described in (26). qsLAM PCR is based on random sonication so that the average DNA fragment size can be controlled to average about 250 to 800 bp per sonicated sample. While some shearing sites can still be too close to the VIS, the random shearing does allow fragments with appropriate length to be generated for a given clone. An advantage of random shearing is that each individual shear site arising from a specific VIS will be unique, allowing the clonal abundance to be estimated by counting the number of unique shear sites associated with each VIS (77, 78). Sheared DNA ends were repaired to form blunt-ended DNA, and 3′ deoxyadenosine monophosphate (dA) tails were then added to the blunt ends. Adapter ligation was performed using the commercially available NEBNext Multiplex Oligos for Illumina kit that allows direct compatibility with the Illumina MiSeq system. After adaptor ligation, 50 cycles of linear PCR were performed, which is then followed by streptavidin capture of a biotinylated primer homologous to the U5 LTR region of the CL20 LV (fig. S10, from 1 to 650 bp). Last, 12 cycles of nested PCR were performed using a second set of primers that bind 23 bp from the end of U5 region and to the adaptor. The sequences that are required for Illumina MiSeq have been incorporated into the nested LTR-U5 primer, so that the only exponential PCR amplification used in qsLAM PCR protocol is a single round of 12 cycles of PCR. The final PCR products were mixed and then size-selected for the 350- to 800-bp fragments on a 2% E-gel. The resulting amplified templates are then directly processed according to the protocol provided with the Illumina MiSeq instrument, and libraries were sequenced by Illumina MiSeq with a 150-bp read length in paired run.
Bioinformatics pipeline
A VIS calling pipeline was developed together with the qsLAM assay. Figure S12A shows an overview of the bioinformatics strategy. The pipeline begins by parsing the raw Illumina fastq files. The 5′ end of the chimeric reads is the 3′ LTR of the LV, whereas the 3′ end is human DNA with the PCR adaptor. Therefore, the primer sequences were first trimmed, specifically ATCCCTCAGACCCTTTTAGTCAGTGTGGAAAATCTC from the forward read and GACTGCGTATCAGT from reverse. The former sequence is part of the 3′ LTR of the LV, and the latter is the adapter sequence. An error rate (−e) of 0.1 was allowed, and a minimal trimmed length of 30 was required. Trimmed reads were aligned to the hg19 reference genome using Burrows-Wheeler Aligner (BWA). Postmapping processing was then performed, including the filtering of singleton reads, pairs mapped onto different chromosomes, as well as pairs with insertion >1000 bp. Figure S12B shows the distribution of the length of pair-end reads after postmapping processing in Jurkat clone samples. Vector integration to repetitive elements in the genome might present a technical issue. In general, less than 0.2% of reads are mapped to multiple locations in the genome.
As the LV is found at the 5′ end of a read, the genomic starting coordinate of a read mapped to the human genome corresponds to the location of a VIS (fig. S12C). We assume two reads with identical start, and end coordinates are the results of PCR amplification from a piece of DNA came in the same cell. On the other hand, two reads with the same start sites but different end sites came from two cells of the same clone (therefore with the same VIS), and the discrepancy in the end sites is a consequence of random sonication. The number of total reads and the number of unique reads (nonduplicated) are two ways to quantify the relative abundance of clones in a sample. Counting the number of unique reads associated with a VIS is essentially counting the number of shear sites in the 3′ end.
Although in theory the starting coordinate of a read corresponds to the location of a VIS, due to potential issues like PCR artifacts, a few base pairs might be truncated, and the starting position might offset from the actual integration site. To investigate the resolution limit of our assay, we made use of the Jurkat clone with known VISs. We aggregated the reads whose starting coordinates are ±50 bp near the 20 known sites. We found that the starting coordinates of most reads are within a few base pairs to the known sites, with the majority of those located precisely at the known sites (fig. S12D). We concluded that the setup has a resolution limit for identifying a VIS, and reads whose starting coordinates within the limit were originated from the integration site. Therefore, in the last step of our pipeline, a distance parameter “d” was introduced for merging. Two sites could only be merged if they are on the same strand. On the basis of fig. S12D, d was chosen to be 8.
Last, to reduce the number of false positive, we previously kept only the insertion sites with unique reads number of ≥2 or total reads number of ≥5 (21). Such a relaxed criterion was used because the primary concern was to detect clones that might cause clonal proliferation at the later stages of the clinical trial. For the purpose of this study, more stringent criteria were used (see the next sections). For each sample analyzed, our bioinformatic pipeline provides a list of VISs including genomic coordinates, strand, whether it is located at introns or intergenic regions, information of the nearest genes, etc. The source code of the pipeline and a step-by-step tutorial can be found in https://zenodo.org/record/8147727.
Separating signal and noise
We have run the qsLAM assay and the VIS bioinformatics pipeline for the Jurkat cell clone with 20 known insertion sites. We found a strong signal of all the 20 sites in all six biological replicates. Besides the 20 known sites, hundreds of extremely rare sites (presumably false positive) were detected. In all the replicates, we found that all combined these rare sites represented roughly 1% of the total number of reads (fig. S13A). As every cell contains the same 20 sites, the relative abundance of each site should theoretically equal to 5%. We found that the relative abundance of the sites was roughly 5%, in which the deviations observed are presumably due to the efficiency of PCR for different DNA sequences (fig. S13B).
The observation that extremely low abundance false discoveries compose of roughly 1% of all the reads suggests the noise level of the qsLAM assay. We therefore used the same approach to remove potential false discoveries in patient samples.
Effects of VCN, read coverage, and GC content
As the qSLAM PCR assay amplifies the regions near VISs, the number of reads generated at the end is determined by the VCN in a sample. We performed the qsLAM assay for Jurkat cell line with different amount of 20-VIS clone DNA (5 and 10 ng). Figure S14A shows the total number of reads in the raw fastq files and the number of reads containing the viral LTR used for PCR amplification. In general, samples with 10 ng of spike-in DNA (higher VCN) have higher fraction of reads with viral LTR (fig. S14B). Assume that the mass of a human haploid genome is 3.59 pg, the total VCN of a sample with 10 ng of spike-in DNA is given by
Samples with 5.6 × 104 total VCN lead to 1.2 to 2.5 million reads, and in which 90% of the reads have the LTR sequences. In general, such coverages lead to high-quality estimates. Coverages are reduced for samples with only 5 ng of spike-in DNA. Since in each patient sample, 1000 ng of DNA is extracted. If we assume the VCN per cell is 0.2, then
In other words, patient samples with average VCN 0.2 have comparable coverages with the Jurkat samples with 10 ng of spike-in DNA. In general, the read coverage of a sample is strongly determined by sample VCN, but in principle, a polyclonal sample and a clonal dominant sample could have the same VCN.
We then examined to what extent the read coverage determines the number of insertion sites identified in patient samples. Unlike the Jurkat samples, the effect of patient VCN on sample read coverage is not strong, but the number of VISs for patients with similar VCN could be very different. Nevertheless, for individual patients, we do not see a positive correlation between the number of reads in a sample and the number of integration sites (fig. S14C).
We further examined the effect of GC content. For Jurkat samples with spike-in DNA from the 20-VIS clone, we did find a strong correlation between the number of reads corresponding to a VIS and the GC content nearby (±100 bp) the site (fig. S15A). Apparently, the correlation is merely a result of a particular VIS (chrX:123484310-123484311) with very low GC content. For patient samples, we did not observe any strong dependence (fig. S15B).
Filtering and integration of VISs in patient samples
Peripheral blood and bone marrow samples were sorted using flow cytometry into CD34+ progenitors and myeloid cell (CD14+/CD15+), B cell (CD19+), T cell (CD3+), and NK cell (CD3− CD56+) lineages, and genomic DNA was extracted. DNA samples were analyzed by quantitative PCR with the use of a standard curve derived from a single-copy cell clone. The mean VCN per cell in the pre-infusion transduced CD34+ cells and bone marrow CD34+ cells was measured in pooled myeloid colony-forming unit assays after 14 days of culture. The VISs of individual patient samples were profiled using the same qsLAM PCR assay and the bioinformatics described above. To minimize the number of false discoveries, we used a rationale based on the observation found in the Jurkat clone analysis (fig. S13A). Essentially, any chimeric read with viral and human DNA could potentially correspond to a VIS. Nevertheless, some of these potential VIS have extremely low read counts. Therefore, we filtered out the bottom 1.5% of reads that correspond to the potential VIS with the lowest clonal abundance (the number of reads). As found in the Jurkat clone analysis, the fraction of reads corresponding to false discovery (everything other than the 20 known sites) is 0.77 ± 0.32%, the cutoff 1.5% is a rather stringent cutoff. Figure S16 shows the effect on the number insertion sites in each patient by varying the cutoff.
To compile a complete list of integration sites for a patient, integration sites across samples were matched. In short, overlapping integration sites (defined from the start position to the end position in the VIS calling output) from different samples were regarded to be the same site, although the procedure might potentially merge a few integration sites that were close to one another. On the basis of the compiled list, a frequency matrix storing the total number of reads mapped to each of the integration sites across samples from different cell types and time points was constructed for quantifying clonal populations.
It is worthwhile to mention that unlike the real integration sites, the noise/false discoveries in general do not follow the integrome signatures (fig. S17). Signatures like overlap with active histone marks H3K36me3 and H3K27ac are enriched in real VISs but depleted in false discoveries. On the other hand, false discoveries are likely to be found on regions like compartment B, overlap with repressive marks H3K27me3, as compared to real VISs.
Profiling integration sites of gRV and LV in T cells
CD4+ and CD8+ T cells were isolated from PBMCs from three healthy donors using CD4 and CD8 MicroBeads (Miltenyi Biotec #130-045-101 and #130-045-201, $680) according to the manufacturer’s recommendations and activated with T Cell TransAct (Miltenyi Biotec #130-111-160) in RPMI media supplemented with 10% fetal calf serum, 1% GlutaMAX (Thermo Fisher Scientific, #35050061), interleukin-17 (IL-7; 10 ng/ml; Peprotech, #200-07), and IL-15 (Peprotech, #200-15). After 48 hours, 5 × 105 cells were transduced on retronectin-coated plates (RetroNectin Recombinant Human Fibronectin Fragment, Takara #T100B) in the presence of protamin sulfate at a MOI of 25 with either LVs or gRVs encoding the IL2RG transgene. The day after transduced cells was transferred to G-Rex 6 Well Plate (WilsonWolf Superior Cell Culture Devices, #80240M). Half media was changed after 4 days in culture, and cells were grown in G-Rex for a total of 7 days before pelleting and cryopreservation. DNA from 5 × 106 cells was extracted using the Maxwell RSC Blood DNA Kit (Promega, #AS1400) according to manufacturer’s recommendation. VCN and VIS determination are described elsewhere in the “Profiling and quantification of LV integration sites” section above. New primers were designed for the gRV: IMC-Biot-gRV-LinPCR: /5BiotinTEG/AACCCTCTTGCAGTTGCATC and IMC-il-1gtta-gRV-LTR: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNagttaCTCCTCTGAGTGATTGACTACC.
Quantifying clonal diversity
Different measures of diversity were used to quantify the diversification of integration sites in each sample. The number of unique integration sites in a sample simply measures the number of unique clones without considering their sizes. Therefore, the UC50 was used; this is defined as the number of unique clones that constitute the top 50% of the sample’s abundance (79). The OCI, based on the Gini coefficient, ranges between 0 and 1, with a value of 0 representing a distribution in which each clone has the same population and a value of 1 representing an upper bound whereby a single clone makes up the entire population (77). The Chao estimator (80), which is often used in metagenomics for estimating the true number of species because of a lack of sequencing depth, was used as an alternative metric. Shannon diversity index (entropy) is another widely used measure of diversity that accounts for both the abundance and evenness of clones. For polyclonal samples, all these metrics are high.
Determining hotspots and RIGs
VIS density for a sample was calculated by binning the genome and counting the number of VISs in each bin. The density of VISs in Fig. 1A was generated using the circlize tool (81). The density was calculated by binning the genome into 100-kb bins with a sliding window of size 50 kb. Correlation coefficients shown in fig. S2 were Pearson coefficients between the density profiles across patients or samples. For hotspot calculation, a reduced bin size of 10 kb was used. The number of VISs inside a bin was compared to a Poisson distribution with an expectation equal to the total number of VISs normalized by the size of the genome. Bins with P values less than the cutoff of 10−6 with Bonferroni correction were defined as hotspots. A more stringent threshold of 10−12 was also used. The analysis of RIGs is performed by integrating all samples and counting the number of VISs falling upon a gene. Results in table S1, as well as the word clouds, did not depend on a particular threshold. The analysis like Figs. 1D and 4B showed the top RIGs, which are essentially for each patient, the genes with the highest number of VIS.
Genome annotation, functional genomics, and cancer data
Gencode (release 19) on the reference chromosomes was used for annotation. The mapping of VISs to genome annotation was performed using Homer (82). Histone modification data for CD34+ cells were downloaded from the ENCODE project (https://encodeproject.org). These data included bed files for peaks and bigWig files for signal tracks. The signal at a VIS and its flanking regions was extracted using bwtool (83). All file accession numbers and related information are listed in table S2. SEs were downloaded from dbSuper (35), but the data originated from reference (34). Lists of SEs in CD34+ samples were merged to form a final list for subsequent analysis. The overlap between VISs or hotspots and various genomic features such as SEs was calculated using bedtools (84). ChIP-seq data for HSPCs (CD34+), as well as Hi-C data for HSPCs and CD3+ T cells, were obtained in GSE104579 (39). The contact maps downloaded are of quality MAPQ30. Reads were extracted using the tool Straw (85) for interchromosomal read counts, at 100-kb resolution with Knight-Ruiz (KR) normalization. The compartment assignment of the HSPC contact map was downloaded from the Supplementary data of (38). All datasets used in the study were based on the hg19 annotation. The list of T-ALL–related genes was downloaded from the Bushman Lab website (http://bushmanlab.org/links/genelists).
Single-cell profiling of bone marrow samples
Bone marrow samples from patient 1 and patient 6 at the 18-month time points were used. These two patients were chosen because their VCN values were similar, but their clonal structures and clinical outcomes were different. Flow-sorted CD34/CD45-positive cells were captured with the Chromium Controller (10x Genomics). For scRNA-seq, libraries were prepared with Chromium Single Cell Gene Expression 3′ v3 Kits (10x Genomics) and sequenced on NovaSeq 6000 systems (Illumina). Sequencing data were run on Cell Ranger version 3.1.0 to generate feature barcode matrices. The resultant output was preprocessed using Seurat (86), and analysis for such features as cell type identification and differential expression was performed using Seurat and an in-house pipeline. For scATAC-seq, libraries were prepared with the Chromium Single Cell ATAC Kit v1.1 (10x Genomics) and sequenced on a NovaSeq 6000 system with an S1 flow cell to obtain more than 400 million read pairs per sample. Sequencing data were processed by Cell Ranger ATAC version 1.2.0. Analysis of scATAC-seq data, e.g., to detect clustering and identify differentially accessible regions (DARs), was performed using the SNAPATAC tool (87), with built-in specific tools MACS (88) for peak calling and chromVAR (89) for calculating the motif score. For 5′ scRNA-seq and TCR-seq, flow-sorted CD45+ CD3+ T cells were used to generate gene expression libraries according to the Chromium Next GEM Single Cell 5′ Kit v2 and TCR libraries according to the Chromium Single Cell Human TCR Amplification Kit (10x Genomics). The combined libraries were sequenced on Illumina NovaSeq 6000 system (Illumina).
The statistical significance of the difference in the overlap between hotspots and open chromatin in patients 1 and 6 shown in Fig. 6C was determined by sampling two sets of random genomic regions of the same size as the two sets of hotspots. The overlap between a set of open chromatin regions and the set of hotspots for patient 1 was denoted as X1 and that for patient 6 as X6. X1 and X6 were approximated by normal distributions in which their means μX and variances were estimated on the basis of the 500 trials. Under the null model, the difference X1 − X6 followed a normal distribution with mean μX1 − μX6 and variance P value was estimated by the resultant normal distribution.
Quantification of transgene expression at the single-cell level
The provirus sequence of length 4619 bp (fig. S10) was added as an additional chromosome to the human genome hg19. Pair-end reads were mapped to the host + virus genome by Cell Ranger. To quantify transgene expression in a single cell, the number of reads mapped to the provirus sequence, from the starting position of IL2RG (2758 bp) to the 3′ end, was counted.
Identification of VISs in single-cell ATAC-seq
The EpiVIA tool (54) was used to identify VISs from scATAC-seq data. Pair-end reads were mapped to the host + virus genome (the provirus sequence was added as an additional chromosome in hg19) by bwa. Chimeric reads were identified from the bam files, including cases (i) reads in which one end mapped to the viral genome and the other end to the host genome, (ii) reads in which both ends mapped to the host genome but a small soft-clipped fragment at one end matched the provirus sequence, (iii) reads in which both ends mapped to the viral genome but a small soft-clipped fragment at one end matched the human genome. Integration sites were then identified from the chimeric reads, and the cellular barcodes were extracted separately; thus, VISs were profiled at a single-cell level. Depending on the types of chimeric reads, a VIS may not be precisely located but rather specified by the start and end positions. For simplicity, only the potential starting positions were displayed. Because of factors such as the sequencing depth, chimeric reads were rare. VISs were identified in only a small fraction of the cells. Step-by-step instructions for 10× scATAC-seq can be found in our Zenodo/GitHub repository.
Classification model to predict clonal abundance
The location of a VIS was used to train a classification model to determine whether the abundance of the corresponding clone was high or low or, more specifically, to determine the average abundance of CD3 samples across time points. The VIS location was written into a set of features that could be 3D conformational (the compartment in which the VIS was located), epigenetic (the signals for various histone marks at/near the VIS), or genomic (annotation, DNA strand, etc.). A dataset was compiled from the VISs of nine patients (P2 to P10) because the clonal abundance in these patients exhibited clear near-Gaussian distributions. For each of the nine patients, the clonal abundance was fitted using a Gaussian model. The positive training set (high abundance) and the negative set (low abundance) were defined as clones with abundance of (≥μi + 2.8σi) and (≤μi − 2σi) respectively, where μi and σi are the mean and variance of the Gaussian model for patient i.
The classification model was constructed using CatBoost (90) with built-in categorical features support. Eighty percent of the VISs from the dataset were used for training the model and tuning the parameters; the remaining 20% were used for validation and for area under the curve calculations. Fivefold cross-validation was adopted for confirming the performance of the model on the entire dataset. The feature importance, whose total value was normalized to 100, shows by how much, on average, the prediction changed if the feature value changed.
Acknowledgments
We would like to thank S. McKinney-Freeman and G. Kang for critical reading of the manuscript, K. A. Laycock for scientific editing, and M. Weiss and J. Crispino for helpful suggestions; the staff of the St. Jude Regulatory Affairs Office and the Department of Bone Marrow Transplantation and Cellular Therapy, who assisted with the clinical studies from which the samples were obtained; and the staff of Children’s GMP, LLC, who assisted in generating clinical-grade cell products. We would like to thank E. Chang (Baylor College of Medicine, Houston) and G. Dotti (University of North Carolina, Chapel Hill) for providing integration site data of gRV-transduced T cells. We would like to thank C. Burdyshaw and M. Brewer from the Information Services for setting up the infrastructure to build the machine learning model. We would also like to thank the patients who participated in both clinical studies and the parents who entrusted the care of their children to us.
Funding: This work was supported by National Institutes of Health grant R01-GM13482 (J.Y.), National Institutes of Health grant P01-HL053749 (S.G.), National Institutes of Health grant U54-AI082973 (J.M.P.), National Institute of Allergy and Infectious Diseases grant U54-AI082973 (J.M.P. and M.J.C.), California Institute of Regenerative Medicine grant CLIN2-09504 (S.G., J.M.P., M.J.C., and E.M.), California Institute of Regenerative Medicine grant CLIN2-10830 (J.M.P. and M.J.C.), NIH/National Cancer Institute (NCI) Comprehensive Cancer Center grant P30CA021765, Assisi Foundation of Memphis (S.G. and E.M.), and American Lebanese Syrian Associated Charities (ALSAC) (S.G. and J.Y.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author contributions: Conception and design: K.-K.Y., S.G., and J.Y. Development of methodology: K.-K.Y., S.Z., L.E.P., and J.Y. Acquisition of data (e.g., acquired and managed patients and provided facilities): K.-K.Y., J.C., Z.M., J.-Y.M., F.F., R.T., B.J., L.E.P., D.M.L., J.-Y.M., S.B., B.S., T.D.L., M.M.M., J.E., A.C.T., J.M.P., M.J.C., S.Z., E.M., S.G., and J.Y. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, and computational analysis): K.-K.Y., S.Z., L.D., Y.D., L.E.P., H.S., I.R., S.G., and J.Y. Writing of the manuscript: K.-K.Y., S.G., and J.Y. Review and/or revision of the manuscript: J.C., Z.M., B.J., S.Z., L.D., Y.D., L.E.P., D.M.L., T.D.L., M.M.M., A.C.T., H.S., I.R., J.E., J.M.P., M.J.C., and EM. Administrative, technical, or material support (i.e., reporting or organizing data and constructing databases): K.-K.Y., J.C., Z.M., L.E.P., T.D.L., M.M.M., J.M.P., M.J.C., E.M., S.G., and J.Y. Study supervision: S.G. and J.Y.
Competing interests: St. Jude Children’s Research Hospital has an existing exclusive license and ongoing partnership with Mustang Bio for the further clinical development and commercialization of this SCID-X1 gene therapy. J.M.P. receives royalties from UpToDate, and J.M.P.’s spouse is employed by and holds stock in Invitae, a DNA sequencing company. M.J.C. is a member of the Data Safety Monitoring Board (DSMB) of Bluebird Bio, Orchard, Rocket Pharma, and Leadiant Biosciences. He serves on the Scientific Advisory Board for Homology Medicine and receives royalties from UpToDate. S.G. has patent applications in the fields of T cell and/or gene therapy for cancer. He has a research collaboration with TESSA Therapeutics, is a DSMB member of Immatics, and serves on the Scientific Advisory Board of Tidal. The authors declare that they have no other competing interests.
Data and materials availability: The VIS data for the 10 patients with SCID-X1 and the seven CAR T cell samples are provided in data S1. The data, together with tables S1 and S2, are deposited in Zenodo (https://zenodo.org/record/8147763). The scRNA-seq and scATAC-seq data for bone marrow samples from patients 1 and 6 have been deposited in the GEO database (accession number: GSE163083). The bioinformatics pipeline for quantifying VISs, the code for VIS analysis and visualization used in this study, the code for the classification model, and more detailed descriptions on using EpiVIA are available at Zenodo (https://zenodo.org/record/8147727), where future updates could be found in GitHub (https://github.com/jyyulab/LVIS_pipeline). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S17
Legends for tables S1 and S2
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
Data S1
REFERENCES AND NOTES
- 1.M. C. Milone, U. O’Doherty, Clinical use of lentiviral vectors. Leukemia 32, 1529–1541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.M. Cavazzana, F. D. Bushman, A. Miccio, I. André-Schmutz, E. Six, Gene therapy targeting haematopoietic stem cells for inherited diseases: Progress and challenges. Nat. Rev. Drug Discov. 18, 447–462 (2019). [DOI] [PubMed] [Google Scholar]
- 3.R. A. Morgan, D. Gray, A. Lomova, D. B. Kohn, Hematopoietic stem cell gene therapy: Progress and lessons learned. Cell Stem Cell 21, 574–590 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.C. E. Dunbar, K. A. High, J. K. Joung, D. B. Kohn, K. Ozawa, M. Sadelain, Gene therapy comes of age. Science 359, eaan6472 (2018). [DOI] [PubMed] [Google Scholar]
- 5.M. V. Maus, J. A. Fraietta, B. L. Levine, M. Kalos, Y. Zhao, C. H. June, Adoptive immunotherapy for cancer or viruses. Annu. Rev. Immunol. 32, 189–225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.C. H. June, M. Sadelain, Chimeric antigen receptor therapy. N. Engl. J. Med. 379, 64–73 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.V. Poletti, F. Mavilio, Interactions between retroviruses and the host cell genome. Mol. Ther. Methods Clin. Dev. 8, 31–41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.L. Biasco, D. Pellin, S. Scala, F. Dionisio, L. Basso-Ricci, L. Leonardelli, S. Scaramuzza, C. Baricordi, F. Ferrua, M. P. Cicalese, S. Giannelli, V. Neduva, D. J. Dow, M. Schmidt, C. Von Kalle, M. G. Roncarolo, F. Ciceri, P. Vicard, E. Wit, C. Di Serio, L. Naldini, A. Aiuti, In vivo tracking of human hematopoiesis reveals patterns of clonal dynamics during early and steady-state reconstitution phases. Cell Stem Cell 19, 107–119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.S. Scala, L. Basso-Ricci, F. Dionisio, D. Pellin, S. Giannelli, F. A. Salerio, L. Leonardelli, M. P. Cicalese, F. Ferrua, A. Aiuti, L. Biasco, Dynamics of genetically engineered hematopoietic stem and progenitor cells after autologous transplantation in humans. Nat. Med. 24, 1683–1690 (2018). [DOI] [PubMed] [Google Scholar]
- 10.E. Six, A. Guilloux, A. Denis, A. Lecoules, A. Magnani, R. Vilette, F. Male, N. Cagnard, M. Delville, E. Magrin, L. Caccavelli, C. Roudaut, C. Plantier, S. Sobrino, J. Gregg, C. L. Nobles, J. K. Everett, S. Hacein-Bey-Abina, A. Galy, A. Fischer, A. J. Thrasher, I. André, M. Cavazzana, F. D. Bushman, Clonal tracking in gene therapy patients reveals a diversity of human hematopoietic differentiation programs. Blood 135, 1219–1231 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A. R. Schröder, P. Shinn, H. Chen, C. Berry, J. R. Ecker, F. Bushman, HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110, 521–529 (2002). [DOI] [PubMed] [Google Scholar]
- 12.R. S. Mitchell, B. F. Beitzel, A. R. Schroder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, F. D. Bushman, Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLOS Biol. 2, E234 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.G. P. Wang, A. Ciuffi, J. Leipzig, C. C. Berry, F. D. Bushman, HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 17, 1186–1194 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.A. Aiuti, F. Cattaneo, S. Galimberti, U. Benninghoff, B. Cassani, L. Callegaro, S. Scaramuzza, G. Andolfi, M. Mirolo, I. Brigida, A. Tabucchi, F. Carlucci, M. Eibl, M. Aker, S. Slavin, H. Al-Mousa, A. Al Ghonaium, A. Ferster, A. Duppenthaler, L. Notarangelo, U. Wintergerst, R. H. Buckley, M. Bregni, S. Marktel, M. G. Valsecchi, P. Rossi, F. Ciceri, R. Miniero, C. Bordignon, M. G. Roncarolo, Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N. Engl. J. Med. 360, 447–458 (2009). [DOI] [PubMed] [Google Scholar]
- 15.M. Cavazzana-Calvo, E. Payen, O. Negre, G. Wang, K. Hehir, F. Fusil, J. Down, M. Denaro, T. Brady, K. Westerman, R. Cavallesco, B. Gillet-Legrand, L. Caccavelli, R. Sgarra, L. Maouche-Chrétien, F. Bernaudin, R. Girot, R. Dorazio, G.-J. Mulder, A. Polack, A. Bank, J. Soulier, J. Larghero, N. Kabbara, B. Dalle, B. Gourmel, G. Socie, S. Chrétien, N. Cartier, P. Aubourg, A. Fischer, K. Cornetta, F. Galacteros, Y. Beuzard, E. Gluckman, F. Bushman, S. Hacein-Bey-Abina, P. Leboulch, Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature 467, 318–322 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. Biffi, E. Montini, L. Lorioli, M. Cesani, F. Fumagalli, T. Plati, C. Baldoli, S. Martino, A. Calabria, S. Canale, F. Benedicenti, G. Vallanti, L. Biasco, S. Leo, N. Kabbara, G. Zanetti, W. B. Rizzo, N. A. L. Mehta, M. P. Cicalese, M. Casiraghi, J. J. Boelens, U. D. Carro, D. J. Dow, M. Schmidt, A. Assanelli, V. Neduva, C. D. Serio, E. Stupka, J. Gardner, C. V. Kalle, C. Bordignon, F. Ciceri, A. Rovelli, M. G. Roncarolo, A. Aiuti, M. Sessa, L. Naldini, Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 341, 1233158 (2013). [DOI] [PubMed] [Google Scholar]
- 17.A. Aiuti, L. Biasco, S. Scaramuzza, F. Ferrua, M. P. Cicalese, C. Baricordi, F. Dionisio, A. Calabria, S. Giannelli, M. C. Castiello, M. Bosticardo, C. Evangelio, A. Assanelli, M. Casiraghi, S. D. Nunzio, L. Callegaro, C. Benati, P. Rizzardi, D. Pellin, C. D. Serio, M. Schmidt, C. V. Kalle, J. Gardner, N. Mehta, V. Neduva, D. J. Dow, A. Galy, R. Miniero, A. Finocchi, A. Metin, P. P. Banerjee, J. S. Orange, S. Galimberti, M. G. Valsecchi, A. Biffi, E. Montini, A. Villa, F. Ciceri, M. G. Roncarolo, L. Naldini, Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science 341, 1233151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.S. S. De Ravin, X. Wu, S. Moir, S. Anaya-O’Brien, N. Kwatemaa, P. Littel, N. Theobald, U. Choi, L. Su, M. Marquesen, D. Hilligoss, J. Lee, C. M. Buckner, K. A. Zarember, G. O’Connor, D. McVicar, D. Kuhns, R. E. Throm, S. Zhou, L. D. Notarangelo, I. C. Hanson, M. J. Cowan, E. Kang, C. Hadigan, M. Meagher, J. T. Gray, B. P. Sorrentino, H. L. Malech, L. Kardava, Lentiviral hematopoietic stem cell gene therapy for X-linked severe combined immunodeficiency. Sci. Transl. Med. 8, 335ra357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.J. A. Ribeil, S. Hacein-Bey-Abina, E. Payen, A. Magnani, M. Semeraro, E. Magrin, L. Caccavelli, B. Neven, P. Bourget, W. El Nemer, P. Bartolucci, L. Weber, H. Puy, J. F. Meritet, D. Grevent, Y. Beuzard, S. Chrétien, T. Lefebvre, R. W. Ross, O. Negre, G. Veres, L. Sandler, S. Soni, M. de Montalembert, S. Blanche, P. Leboulch, M. Cavazzana, Gene therapy in a patient with sickle cell disease. N. Engl. J. Med. 376, 848–855 (2017). [DOI] [PubMed] [Google Scholar]
- 20.A. Khan, D. L. Barber, J. Huang, C. A. Rupar, J. W. Rip, C. Auray-Blais, M. Boutin, P. O'Hoski, K. Gargulak, W. M. McKillop, G. Fraser, S. Wasim, K. LeMoine, S. Jelinski, A. Chaudhry, N. Prokopishyn, C. F. Morel, S. Couban, P. R. Duggan, D. H. Fowler, A. Keating, M. L. West, R. Foley, J. A. Medin, Lentivirus-mediated gene therapy for Fabry disease. Nat. Commun. 12, 1178 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.E. Mamcarz, S. Zhou, T. Lockey, H. Abdelsamed, S. J. Cross, G. Kang, Z. Ma, J. Condori, J. Dowdy, B. Triplett, C. Li, G. Maron, J. C. Aldave Becerra, J. A. Church, E. Dokmeci, J. T. Love, A. C. da Matta Ain, H. van der Watt, X. Tang, W. Janssen, B. Y. Ryu, S. S. De Ravin, M. J. Weiss, B. Youngblood, J. R. Long-Boyle, S. Gottschalk, M. M. Meagher, H. L. Malech, J. M. Puck, M. J. Cowan, B. P. Sorrentino, Lentiviral gene therapy combined with low-dose busulfan in infants with SCID-X1. N. Engl. J. Med. 380, 1525–1534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.E. Mamcarz, S. Zhou, T. Lockey, Z. Ma, K.-K. Yan, J.-Y. Metais, D. Langfitt, S. J. Cross, G. Maron, G. Kang, in Molecular Therapy. (CELL PRESS 50 HAMPSHIRE ST, FLOOR 5, CAMBRIDGE, MA 02139 USA, 2022), vol. 30, pp. 552–552. [Google Scholar]
- 23.Roadmap Epigenomics Consortium, A. Kundaje, W. Meuleman, J. Ernst, M. Bilenky, A. Yen, A. Heravi-Moussavi, P. Kheradpour, Z. Zhang, J. Wang, M. J. Ziller, V. Amin, J. W. Whitaker, M. D. Schultz, L. D. Ward, A. Sarkar, G. Quon, R. S. Sandstrom, M. L. Eaton, Y. C. Wu, A. R. Pfenning, X. Wang, M. Claussnitzer, Y. Liu, C. Coarfa, R. A. Harris, N. Shoresh, C. B. Epstein, E. Gjoneska, D. Leung, W. Xie, R. D. Hawkins, R. Lister, C. Hong, P. Gascard, A. J. Mungall, R. Moore, E. Chuah, A. Tam, T. K. Canfield, R. S. Hansen, R. Kaul, P. J. Sabo, M. S. Bansal, A. Carles, J. R. Dixon, K. H. Farh, S. Feizi, R. Karlic, A. R. Kim, A. Kulkarni, D. Li, R. Lowdon, G. Elliott, T. R. Mercer, S. J. Neph, V. Onuchic, P. Polak, N. Rajagopal, P. Ray, R. C. Sallari, K. T. Siebenthall, N. A. Sinnott-Armstrong, M. Stevens, R. E. Thurman, J. Wu, B. Zhang, X. Zhou, A. E. Beaudet, L. A. Boyer, P. L. De Jager, P. J. Farnham, S. J. Fisher, D. Haussler, S. J. Jones, W. Li, M. A. Marra, M. T. McManus, S. Sunyaev, J. A. Thomson, T. D. Tlsty, L. H. Tsai, W. Wang, R. A. Waterland, M. Q. Zhang, L. H. Chadwick, B. E. Bernstein, J. F. Costello, J. R. Ecker, M. Hirst, A. Meissner, A. Milosavljevic, B. Ren, J. A. Stamatoyannopoulos, T. Wang, M. Kellis, Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ENCODE Project Consortium, J. E. Moore, M. J. Purcaro, H. E. Pratt, C. B. Epstein, N. Shoresh, J. Adrian, T. Kawli, C. A. Davis, A. Dobin, R. Kaul, J. Halow, E. L. Van Nostrand, P. Freese, D. U. Gorkin, Y. Shen, Y. He, M. Mackiewicz, F. Pauli-Behn, B. A. Williams, A. Mortazavi, C. A. Keller, X. O. Zhang, S. I. Elhajjajy, J. Huey, D. E. Dickel, V. Snetkova, X. Wei, X. Wang, J. C. Rivera-Mulia, J. Rozowsky, J. Zhang, S. B. Chhetri, J. Zhang, A. Victorsen, K. P. White, A. Visel, G. W. Yeo, C. B. Burge, E. Lécuyer, D. M. Gilbert, J. Dekker, J. Rinn, E. M. Mendenhall, J. R. Ecker, M. Kellis, R. J. Klein, W. S. Noble, A. Kundaje, R. Guigó, P. J. Farnham, J. M. Cherry, R. M. Myers, B. Ren, B. R. Graveley, M. B. Gerstein, L. A. Pennacchio, M. P. Snyder, B. E. Bernstein, B. Wold, R. C. Hardison, T. R. Gingeras, J. A. Stamatoyannopoulos, Z. Weng, Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.K. Servick, Gene therapy clinical trial halted as cancer risk surfaces. Science, (2021). [Google Scholar]
- 26.S. Zhou, M. A. Bonner, Y.-D. Wang, S. Rapp, S. S. De Ravin, H. L. Malech, B. P. Sorrentino, Quantitative shearing linear amplification polymerase chain reaction: An improved method for quantifying lentiviral vector insertion sites in transplanted hematopoietic cell systems. Hum. Gene Ther. Methods 26, 4–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.A. Biffi, C. C. Bartolomae, D. Cesana, N. Cartier, P. Aubourg, M. Ranzani, M. Cesani, F. Benedicenti, T. Plati, E. Rubagotti, S. Merella, A. Capotondo, J. Sgualdino, G. Zanetti, C. von Kalle, M. Schmidt, L. Naldini, E. Montini, Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood 117, 5332–5339 (2011). [DOI] [PubMed] [Google Scholar]
- 28.A. Ciuffi, M. Llano, E. Poeschla, C. Hoffmann, J. Leipzig, P. Shinn, J. R. Ecker, F. Bushman, A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11, 1287–1289 (2005). [DOI] [PubMed] [Google Scholar]
- 29.C. Cattoglio, G. Maruggi, C. Bartholomae, N. Malani, D. Pellin, F. Cocchiarella, Z. Magnani, F. Ciceri, A. Ambrosi, C. V. Kalle, F. D. Bushman, C. Bonini, M. Schmidt, F. Mavilio, A. Recchia, High-definition mapping of retroviral integration sites defines the fate of allogeneic T cells after donor lymphocyte infusion. PLOS ONE 5, e15688 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.J. Ernst, M. Kellis, ChromHMM: Automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.B. Marini, A. Kertesz-Farkas, H. Ali, B. Lucic, K. Lisek, L. Manganaro, S. Pongor, R. Luzzati, A. Recchia, F. Mavilio, M. Giacca, M. Lusic, Nuclear architecture dictates HIV-1 integration site selection. Nature 521, 227–231 (2015). [DOI] [PubMed] [Google Scholar]
- 32.M. Lelek, N. Casartelli, D. Pellin, E. Rizzi, P. Souque, M. Severgnini, C. Di Serio, T. Fricke, F. Diaz-Griffero, C. Zimmer, P. Charneau, F. Di Nunzio, Chromatin organization at the nuclear pore favours HIV replication. Nat. Commun. 6, 6483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.A. Ibarra, C. Benner, S. Tyagi, J. Cool, M. W. Hetzer, Nucleoporin-mediated regulation of cell identity genes. Genes Dev. 30, 2253–2258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D. Hnisz, B. J. Abraham, T. I. Lee, A. Lau, V. Saint-André, A. A. Sigova, H. A. Hoke, R. A. Young, Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.A. Khan, X. Zhang, dbSUPER: A database of super-enhancers in mouse and human genome. Nucleic Acids Res. 44, D164–D171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.E. Lieberman-Aiden, N. L. van Berkum, L. Williams, M. Imakaev, T. Ragoczy, A. Telling, I. Amit, B. R. Lajoie, P. J. Sabo, M. O. Dorschner, R. Sandstrom, B. Bernstein, M. A. Bender, M. Groudine, A. Gnirke, J. Stamatoyannopoulos, L. A. Mirny, E. S. Lander, J. Dekker, Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.S. S. P. Rao, M. H. Huntley, N. C. Durand, E. K. Stamenova, I. D. Bochkov, J. T. Robinson, A. L. Sanborn, I. Machol, A. D. Omer, E. S. Lander, E. L. Aiden, A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.K. Xiong, J. Ma, Revealing Hi-C subcompartments by imputing inter-chromosomal chromatin interactions. Nat. Commun. 10, 5069 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.X. Zhang, M. Jeong, X. Huang, X. Q. Wang, X. Wang, W. Zhou, M. S. Shamim, H. Gore, P. Himadewi, Y. Liu, I. D. Bochkov, J. Reyes, M. Doty, Y. H. Huang, H. Jung, E. Heikamp, A. P. Aiden, W. Li, J. Su, E. L. Aiden, M. A. Goodell, Large DNA methylation nadirs anchor chromatin loops maintaining hematopoietic stem cell identity. Mol. Cell 78, 506–521.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Y. Chen, Y. Zhang, Y. Wang, L. Zhang, E. K. Brinkman, S. A. Adam, R. Goldman, B. van Steensel, J. Ma, A. S. Belmont, Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J. Cell Biol. 217, 4025–4048 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.C. T. Ong, V. G. Corces, CTCF: An architectural protein bridging genome topology and function. Nat. Rev. Genet. 15, 234–246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.B. R. Sabari, A. Dall'Agnese, A. Boija, I. A. Klein, E. L. Coffey, K. Shrinivas, B. J. Abraham, N. M. Hannett, A. V. Zamudio, J. C. Manteiga, C. H. Li, Y. E. Guo, D. S. Day, J. Schuijers, E. Vasile, S. Malik, D. Hnisz, T. I. Lee, I. I. Cisse, R. G. Roeder, P. A. Sharp, A. K. Chakraborty, R. A. Young, Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.A. Kloetgen, P. Thandapani, A. Tsirigos, I. Aifantis, 3D chromosomal landscapes in hematopoiesis and immunity. Trends Immunol. 40, 809–824 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.P. Hematti, B. K. Hong, C. Ferguson, R. Adler, H. Hanawa, S. Sellers, I. E. Holt, C. E. Eckfeldt, Y. Sharma, M. Schmidt, C. von Kalle, D. A. Persons, E. M. Billings, C. M. Verfaillie, A. W. Nienhuis, T. G. Wolfsberg, C. E. Dunbar, B. Calmels, Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLOS Biol. 2, e423 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.B. C. Beard, D. Dickerson, K. Beebe, C. Gooch, J. Fletcher, T. Okbinoglu, D. G. Miller, M. A. Jacobs, R. Kaul, H. P. Kiem, G. D. Trobridge, Comparison of HIV-derived lentiviral and MLV-based gammaretroviral vector integration sites in primate repopulating cells. Mol. Ther. 15, 1356–1365 (2007). [DOI] [PubMed] [Google Scholar]
- 46.S. Hacein-Bey-Abina, C. Von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, M. Cavazzana-Calvo, LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302, 415–419 (2003). [DOI] [PubMed] [Google Scholar]
- 47.S. Hacein-Bey-Abina, A. Garrigue, G. P. Wang, J. Soulier, A. Lim, E. Morillon, E. Clappier, L. Caccavelli, E. Delabesse, K. Beldjord, V. Asnafi, E. MacIntyre, L. Dal Cortivo, I. Radford, N. Brousse, F. Sigaux, D. Moshous, J. Hauer, A. Borkhardt, B. H. Belohradsky, U. Wintergerst, M. C. Velez, L. Leiva, R. Sorensen, N. Wulffraat, S. Blanche, F. D. Bushman, A. Fischer, M. Cavazzana-Calvo, Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 118, 3132–3142 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.C. J. Braun, K. Boztug, A. Paruzynski, M. Witzel, A. Schwarzer, M. Rothe, U. Modlich, R. Beier, G. Göhring, D. Steinemann, R. Fronza, C. R. Ball, R. Haemmerle, S. Naundorf, K. Kühlcke, M. Rose, C. Fraser, L. Mathias, R. Ferrari, M. R. Abboud, W. Al-Herz, I. Kondratenko, L. Maródi, H. Glimm, B. Schlegelberger, A. Schambach, M. H. Albert, M. Schmidt, C. von Kalle, C. Klein, Gene therapy for Wiskott-Aldrich syndrome—long-term efficacy and genotoxicity. Sci. Transl. Med. 6, 227ra233 (2014). [DOI] [PubMed] [Google Scholar]
- 49.S. Stein, M. G. Ott, S. Schultze-Strasser, A. Jauch, B. Burwinkel, A. Kinner, M. Schmidt, A. Krämer, J. Schwäble, H. Glimm, U. Koehl, C. Preiss, C. Ball, H. Martin, G. Göhring, K. Schwarzwaelder, W. K. Hofmann, K. Karakaya, S. Tchatchou, R. Yang, P. Reinecke, K. Kühlcke, B. Schlegelberger, A. J. Thrasher, D. Hoelzer, R. Seger, C. von Kalle, M. Grez, Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 16, 198–204 (2010). [DOI] [PubMed] [Google Scholar]
- 50.E. C. Chang, H. Liu, J. A. West, X. Zhou, O. Dakhova, D. A. Wheeler, H. E. Heslop, M. K. Brenner, G. Dotti, Clonal dynamics in vivo of virus integration sites of T cells expressing a safety switch. Mol. Ther. 24, 736–745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.X. Ma, Y. Liu, Y. Liu, L. B. Alexandrov, M. N. Edmonson, C. Gawad, X. Zhou, Y. Li, M. C. Rusch, J. Easton, R. Huether, V. Gonzalez-Pena, M. R. Wilkinson, L. C. Hermida, S. Davis, E. Sioson, S. Pounds, X. Cao, R. E. Ries, Z. Wang, X. Chen, L. Dong, S. J. Diskin, M. A. Smith, J. M. Guidry Auvil, P. S. Meltzer, C. C. Lau, E. J. Perlman, J. M. Maris, S. Meshinchi, S. P. Hunger, D. S. Gerhard, J. Zhang, Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 555, 371–376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.J. G. Tate, S. Bamford, H. C. Jubb, Z. Sondka, D. M. Beare, N. Bindal, H. Boutselakis, C. G. Cole, C. Creatore, E. Dawson, P. Fish, B. Harsha, C. Hathaway, S. C. Jupe, C. Y. Kok, K. Noble, L. Ponting, C. C. Ramshaw, C. E. Rye, H. E. Speedy, R. Stefancsik, S. L. Thompson, S. Wang, S. Ward, P. J. Campbell, S. A. Forbes, COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–d947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.W. Akhtar, J. de Jong, A. V. Pindyurin, L. Pagie, W. Meuleman, J. de Ridder, A. Berns, L. F. Wessels, M. van Lohuizen, B. van Steensel, Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell 154, 914–927 (2013). [DOI] [PubMed] [Google Scholar]
- 54.W. Wang, M. Fasolino, B. Cattau, N. Goldman, W. Kong, M. A. Frederick, S. J. McCright, K. Kiani, J. A. Fraietta, G. Vahedi, Joint profiling of chromatin accessibility and CAR-T integration site analysis at population and single-cell levels. Proc. Natl. Acad. Sci. U.S.A. 117, 5442–5452 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.S. S. De Ravin, L. Su, N. Theobald, U. Choi, J. L. MacPherson, M. Poidinger, G. Symonds, S. M. Pond, A. L. Ferris, S. H. Hughes, H. L. Malech, X. Wu, Enhancers are major targets for murine leukemia virus vector integration. J. Virol. 88, 4504–4513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.C. Cattoglio, D. Pellin, E. Rizzi, G. Maruggi, G. Corti, F. Miselli, D. Sartori, A. Guffanti, C. Di Serio, A. Ambrosi, G. De Bellis, F. Mavilio, High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood 116, 5507–5517 (2010). [DOI] [PubMed] [Google Scholar]
- 57.B. Lucic, H.-C. Chen, M. Kuzman, E. Zorita, J. Wegner, V. Minneker, W. Wang, R. Fronza, S. Laufs, M. Schmidt, R. Stadhouders, V. Roukos, K. Vlahovicek, G. J. Filion, M. Lusic, Spatially clustered loci with multiple enhancers are frequent targets of HIV-1 integration. Nat. Commun. 10, 4059 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.K. E. Knockenhauer, T. U. Schwartz, The nuclear pore complex as a flexible and dynamic gate. Cell 164, 1162–1171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.M. Beck, E. Hurt, The nuclear pore complex: Understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 18, 73–89 (2017). [DOI] [PubMed] [Google Scholar]
- 60.C. Ptak, R. W. Wozniak, Nucleoporins and chromatin metabolism. Curr. Opin. Cell Biol. 40, 153–160 (2016). [DOI] [PubMed] [Google Scholar]
- 61.J. Demeulemeester, J. De Rijck, R. Gijsbers, Z. Debyser, Retroviral integration: Site matters: Mechanisms and consequences of retroviral integration site selection. Bioessays 37, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.D. Camozzi, S. Pignatelli, C. Valvo, G. Lattanzi, C. Capanni, P. Dal Monte, M. P. Landini, Remodelling of the nuclear lamina during human cytomegalovirus infection: Role of the viral proteins pUL50 and pUL53. J. Gen. Virol. 89, 731–740 (2008). [DOI] [PubMed] [Google Scholar]
- 63.V. Aho, M. Myllys, V. Ruokolainen, S. Hakanen, E. Mäntylä, J. Virtanen, V. Hukkanen, T. Kühn, J. Timonen, K. Mattila, C. A. Larabell, M. Vihinen-Ranta, Chromatin organization regulates viral egress dynamics. Sci. Rep. 7, 3692 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.M. F. Lye, A. R. Wilkie, D. J. Filman, J. M. Hogle, D. M. Coen, Getting to and through the inner nuclear membrane during herpesvirus nuclear egress. Curr. Opin. Cell Biol. 46, 9–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.S. Hacein-Bey-Abina, S. Y. Pai, H. B. Gaspar, M. Armant, C. C. Berry, S. Blanche, J. Bleesing, J. Blondeau, H. de Boer, K. F. Buckland, L. Caccavelli, G. Cros, S. De Oliveira, K. S. Fernández, D. Guo, C. E. Harris, G. Hopkins, L. E. Lehmann, A. Lim, W. B. London, J. C. van der Loo, N. Malani, F. Male, P. Malik, M. A. Marinovic, A. M. McNicol, D. Moshous, B. Neven, M. Oleastro, C. Picard, J. Ritz, C. Rivat, A. Schambach, K. L. Shaw, E. A. Sherman, L. E. Silberstein, E. Six, F. Touzot, A. Tsytsykova, J. Xu-Bayford, C. Baum, F. D. Bushman, A. Fischer, D. B. Kohn, A. H. Filipovich, L. D. Notarangelo, M. Cavazzana, D. A. Williams, A. J. Thrasher, A modified γ-retrovirus vector for X-linked severe combined immunodeficiency. N. Engl. J. Med. 371, 1407–1417 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.E. Montini, D. Cesana, M. Schmidt, F. Sanvito, C. C. Bartholomae, M. Ranzani, F. Benedicenti, L. S. Sergi, A. Ambrosi, M. Ponzoni, C. Doglioni, C. Di Serio, C. von Kalle, L. Naldini, The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J. Clin. Invest. 119, 964–975 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.D. Cesana, M. Ranzani, M. Volpin, C. Bartholomae, C. Duros, A. Artus, S. Merella, F. Benedicenti, L. Sergi Sergi, F. Sanvito, C. Brombin, A. Nonis, C. D. Serio, C. Doglioni, C. von Kalle, M. Schmidt, O. Cohen-Haguenauer, L. Naldini, E. Montini, Uncovering and dissecting the genotoxicity of self-inactivating lentiviral vectors in vivo. Mol. Ther. 22, 774–785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.N. Naumova, M. Imakaev, G. Fudenberg, Y. Zhan, B. R. Lajoie, L. A. Mirny, J. Dekker, Organization of the mitotic chromosome. Science 342, 948–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.J. H. Gibcus, K. Samejima, A. Goloborodko, I. Samejima, N. Naumova, J. Nuebler, M. T. Kanemaki, L. Xie, J. R. Paulson, W. C. Earnshaw, L. A. Mirny, J. Dekker, A pathway for mitotic chromosome formation. Science 359, eaao6135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.A. L. Valton, J. Dekker, TAD disruption as oncogenic driver. Curr. Opin. Genet. Dev. 36, 34–40 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.S. Zhou, D. Mody, S. S. DeRavin, J. Hauer, T. Lu, Z. Ma, S. Hacein-Bey Abina, J. T. Gray, M. R. Greene, M. Cavazzana-Calvo, H. L. Malech, B. P. Sorrentino, A self-inactivating lentiviral vector for SCID-X1 gene therapy that does not activate LMO2 expression in human T cells. Blood 116, 900–908 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.C. L. Nobles, S. Sherrill-Mix, J. K. Everett, S. Reddy, J. A. Fraietta, D. L. Porter, N. Frey, S. I. Gill, S. A. Grupp, S. L. Maude, D. L. Siegel, B. L. Levine, C. H. June, S. F. Lacey, J. J. Melenhorst, F. D. Bushman, CD19-targeting CAR T cell immunotherapy outcomes correlate with genomic modification by vector integration. J. Clin. Invest. 130, 673–685 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.A. Sheih, V. Voillet, L. A. Hanafi, H. A. DeBerg, M. Yajima, R. Hawkins, V. Gersuk, S. R. Riddell, D. G. Maloney, M. E. Wohlfahrt, D. Pande, M. R. Enstrom, H. P. Kiem, J. E. Adair, R. Gottardo, P. S. Linsley, C. J. Turtle, Clonal kinetics and single-cell transcriptional profiling of CAR-T cells in patients undergoing CD19 CAR-T immunotherapy. Nat. Commun. 11, 219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.R. E. Throm, A. A. Ouma, S. Zhou, A. Chandrasekaran, T. Lockey, M. Greene, S. S. De Ravin, M. Moayeri, H. L. Malech, B. P. Sorrentino, J. T. Gray, Efficient construction of producer cell lines for a SIN lentiviral vector for SCID-X1 gene therapy by concatemeric array transfection. Blood 113, 5104–5110 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.W. K. Chan, D. Suwannasaen, R. E. Throm, Y. Li, P. W. Eldridge, J. Houston, J. T. Gray, C. H. Pui, W. Leung, Chimeric antigen receptor-redirected CD45RA-negative T cells have potent antileukemia and pathogen memory response without graft-versus-host activity. Leukemia 29, 387–395 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.J. M. Riberdy, S. Zhou, F. Zheng, Y. I. Kim, J. Moore, A. Vaidya, R. E. Throm, A. Sykes, N. Sahr, C. L. Bonifant, B. Ryu, S. Gottschalk, M. P. Velasquez, The art and science of selecting a CD123-specific chimeric antigen receptor for clinical testing. Mol. Ther. Methods Clin. Dev. 18, 571–581 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.N. A. Gillet, N. Malani, A. Melamed, N. Gormley, R. Carter, D. Bentley, C. Berry, F. D. Bushman, G. P. Taylor, C. R. Bangham, The host genomic environment of the provirus determines the abundance of HTLV-1-infected T-cell clones. Blood 117, 3113–3122 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.B. C. Beard, J. E. Adair, G. D. Trobridge, H. P. Kiem, High-throughput genomic mapping of vector integration sites in gene therapy studies. Methods Mol. Biol. 1185, 321–344 (2014). [DOI] [PubMed] [Google Scholar]
- 79.C. C. Berry, C. Nobles, E. Six, Y. Wu, N. Malani, E. Sherman, A. Dryga, J. K. Everett, F. Male, A. Bailey, K. Bittinger, M. J. Drake, L. Caccavelli, P. Bates, S. Hacein-Bey-Abina, M. Cavazzana, F. D. Bushman, INSPIIRED: Quantification and visualization tools for analyzing integration site distributions. Mol. Ther. Methods Clin. Dev. 4, 17–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.A. Chao, Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270 (1984). [Google Scholar]
- 81.Z. Gu, L. Gu, R. Eils, M. Schlesner, B. Brors, circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 (2014). [DOI] [PubMed] [Google Scholar]
- 82.S. Heinz, C. Benner, N. Spann, E. Bertolino, Y. C. Lin, P. Laslo, J. X. Cheng, C. Murre, H. Singh, C. K. Glass, Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.A. Pohl, M. Beato, bwtool: A tool for bigWig files. Bioinformatics 30, 1618–1619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.A. R. Quinlan, I. M. Hall, BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.N. C. Durand, J. T. Robinson, M. S. Shamim, I. Machol, J. P. Mesirov, E. S. Lander, E. L. Aiden, Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.A. Butler, P. Hoffman, P. Smibert, E. Papalexi, R. Satija, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.R. Fang, S. Preissl, Y. Li, X. Hou, J. Lucero, X. Wang, A. Motamedi, A. K. Shiau, X. Zhou, F. Xie, E. A. Mukamel, K. Zhang, Y. Zhang, M. M. Behrens, J. Ecker, B. Ren, Comprehensive analysis of single cell ATAC-seq data with SnapATAC. Nat. Commun. 12, 1337 (2021) 10.1101/615179 10.1101/615179. [DOI] [PMC free article] [PubMed]
- 88.Y. Zhang, T. Liu, C. A. Meyer, J. Eeckhoute, D. S. Johnson, B. E. Bernstein, C. Nusbaum, R. M. Myers, M. Brown, W. Li, X. S. Liu, Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.A. N. Schep, B. Wu, J. D. Buenrostro, W. J. Greenleaf, chromVAR: Inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.A. V. Dorogush, V. Ershov, A. Gulin, CatBoost: Gradient boosting with categorical features support. arXiv:1810.11363 [cs.LG] (2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S17
Legends for tables S1 and S2
Legend for data S1
Tables S1 and S2
Data S1