Abstract
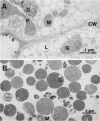
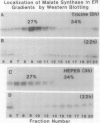
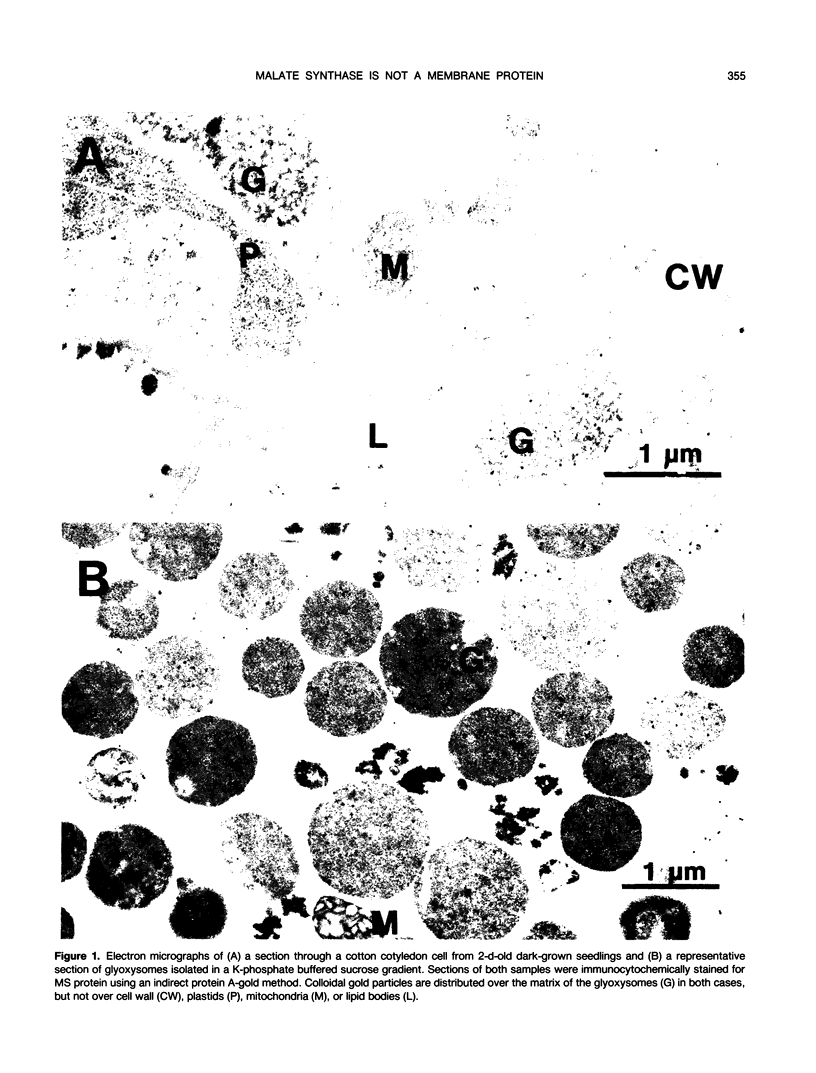
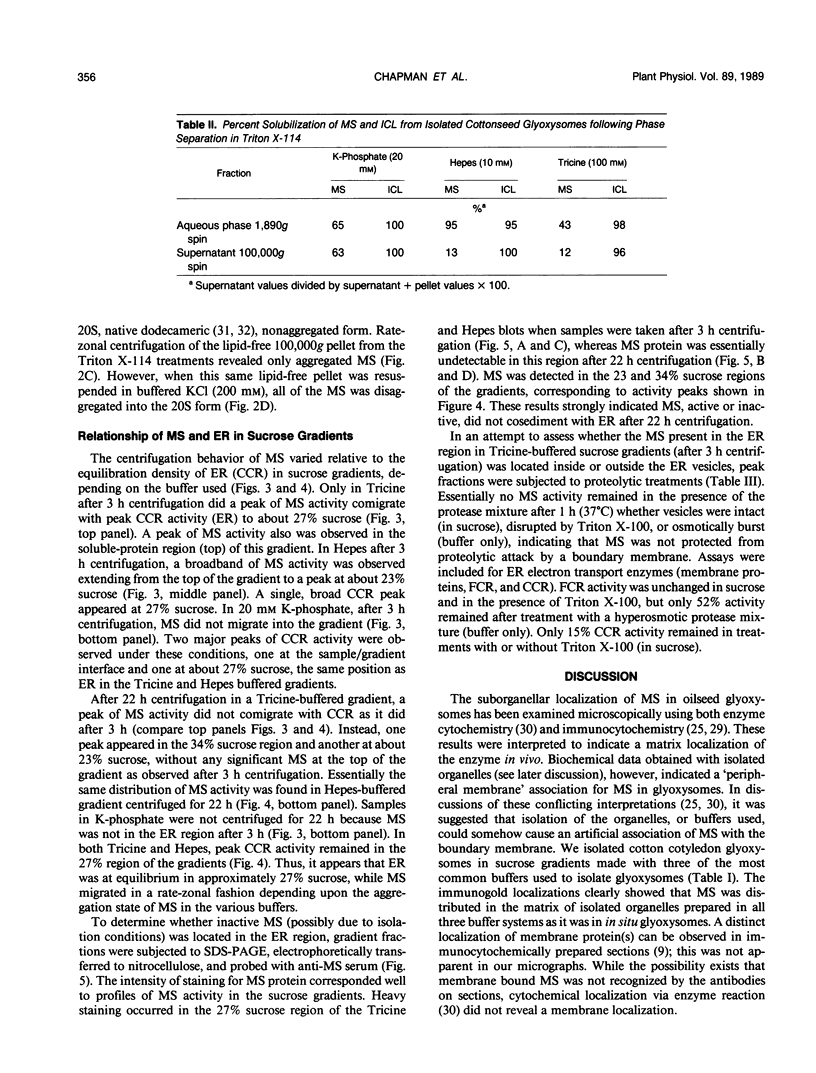
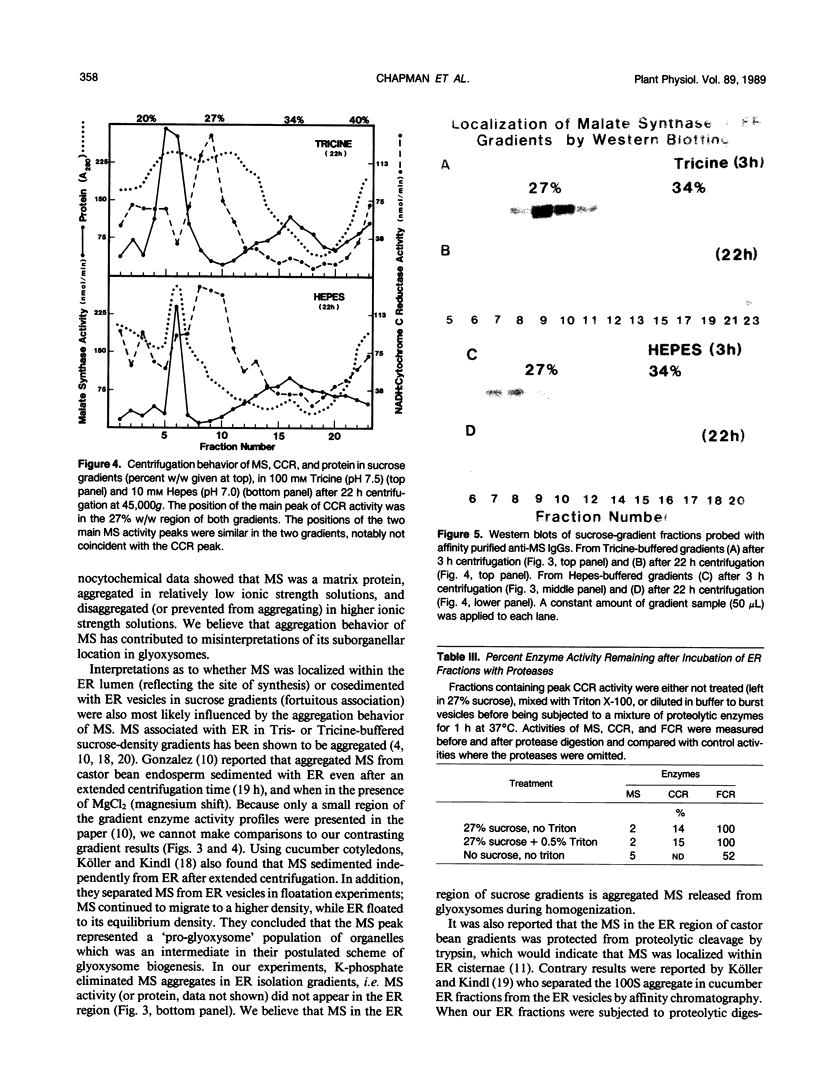
Malate synthase (EC 4.1.3.2) (MS), an enzyme unique to the glyoxylate cycle, was studied in cotyledons of dark-grown cotton (Gossypium hirsutum, L.) seedlings. MS has generally been regarded as a peripheral membrane protein in glyoxysomes and believed by some to be synthesized on rough ER. Immunocyto-chemical localization of MS in both in situ and isolated cottonseed glyoxysomes, however, showed that MS was located throughout the matrix of glyoxysomes, not specifically associated with their membranes. Biochemical data also supported matrix localization. Isolated glyoxysomes were diluted in variously-buffered salt solutions (200 millimolar KCl or 100 millimolar K-phosphate) or detergents (0.1% Triton X-100, 10 millimolar deoxycholate, or 1.0% Triton X-114) and centrifuged to pellet membranes. Greater than 70% of the MS was recovered in supernatants after treatment with salt solutions, whereas generally less than 30% was released following detergent treatments. MS in pellets derived from glyoxysomes burst in low ionic strength buffer solutions was aggregated (observed on rate-zonal gradients). MS released following salt treatments was the 20S nonaggregated form indicating that salt solutions either disaggregated (or prevented aggregation of) glyoxysomal MS rather than releasing it from membranes. We confirmed reports by others that MS comigrated with ER (NADH: cytochrome c reductase) in sucrose (20-40% w/w) gradients buffered with 100 millimolar Tricine (pH 7.5) after 3 hours centrifugation. However, cottonseed MS did not comigrate with ER in gradients buffered with 10 millimolar Hepes (pH 7.0) or 20 millimolar K-phosphate (pH 7.2) after 3 hours centrifugation, or after 22 hours centrifugation in Tricine or Hepes. Collectively, our data with cotton seeds indicate that MS is not a peripheral membrane protein, and that the aggregation behavior of MS (in various buffers) very likely has led to misinterpretations of its putative associations with ER and glyoxysomal membranes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981 Feb 25;256(4):1604–1607. [PubMed] [Google Scholar]
- Bortman S. J., Trelease R. N., Miernyk J. A. Enzyme development and glyoxysome characterization in cotyledons of cotton seeds. Plant Physiol. 1981 Jul;68(1):82–87. doi: 10.1104/pp.68.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden L., Lord J. M. Purification and comparative properties of microsomal and glyoxysomal malate synthase from castor bean endosperm. Plant Physiol. 1978 Feb;61(2):259–265. doi: 10.1104/pp.61.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. H., Lord J. M., Merrett M. J. Fractionation of the proteins of plant microbodies. Biochem J. 1974 Dec;144(3):559–566. doi: 10.1042/bj1440559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez D. E., Qu R., Huang A. H., Staehelin L. A. Immunogold Localization of the L3 Protein of Maize Lipid Bodies during Germination and Seedling Growth. Plant Physiol. 1988 Jan;86(1):270–274. doi: 10.1104/pp.86.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E. Aggregated Forms of Malate and Citrate Synthase are Localized in Endoplasmic Reticulum of Endosperm of Germinating Castor Bean. Plant Physiol. 1982 Jan;69(1):83–87. doi: 10.1104/pp.69.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks D. B., Donaldson R. P. Electron transport in glyoxysomal membranes. Arch Biochem Biophys. 1982 Apr 15;215(1):280–288. doi: 10.1016/0003-9861(82)90306-x. [DOI] [PubMed] [Google Scholar]
- Huang A. H., Beevers H. Localization of enzymes within microbodies. J Cell Biol. 1973 Aug;58(2):379–389. doi: 10.1083/jcb.58.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse C., Kindl H. Malate synthase: aggregation, deaggregation, and binding of phospholipids. Arch Biochem Biophys. 1983 Jun;223(2):618–628. doi: 10.1016/0003-9861(83)90626-4. [DOI] [PubMed] [Google Scholar]
- Kunce C. M., Trelease R. N. Heterogeneity of catalase in maturing and germinated cotton seeds. Plant Physiol. 1986 Aug;81(4):1134–1139. doi: 10.1104/pp.81.4.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köller W., Kindl H. 19S cytosolic malate synthase. A small pool characterized by rapid turnover. Hoppe Seylers Z Physiol Chem. 1980 Sep;361(9):1437–1444. doi: 10.1515/bchm2.1980.361.2.1437. [DOI] [PubMed] [Google Scholar]
- Köller W., Kindl H. Glyoxylate cycle enzymes of the glyoxysomal membrane from cucumber cotyledons. Arch Biochem Biophys. 1977 May;181(1):236–248. doi: 10.1016/0003-9861(77)90502-1. [DOI] [PubMed] [Google Scholar]
- Longo G. P., Bernasconi E., Longo C. P. Solubilization of enzymes from glyoxysomes of maize scutellum. Plant Physiol. 1975 Jun;55(6):1115–1119. doi: 10.1104/pp.55.6.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord J. M., Bowden L. Evidence that glyoxysomal malate synthase is segregated by the endoplasmic reticulum. Plant Physiol. 1978 Feb;61(2):266–270. doi: 10.1104/pp.61.2.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luster D. G., Donaldson R. P. Orientation of electron transport activities in the membrane of intact glyoxysomes isolated from castor bean endosperm. Plant Physiol. 1987 Nov;85(3):796–800. doi: 10.1104/pp.85.3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H., Ueda M., Tanaka A. Purification of peroxisomal malate synthase from alkane-grown Candida tropicalis and some properties of the purified enzyme. Arch Microbiol. 1986 Mar;144(2):137–141. doi: 10.1007/BF00414723. [DOI] [PubMed] [Google Scholar]
- Sandalio L. M., Del Río L. A. Intraorganellar distribution of superoxide dismutase in plant peroxisomes (glyoxysomes and leaf peroxisomes). Plant Physiol. 1988 Dec;88(4):1215–1218. doi: 10.1104/pp.88.4.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus D. E., Becker W. M. Investigation of the glyoxysome-peroxisome transition in germinating cucumber cotyledons using double-label immunoelectron microscopy. J Cell Biol. 1985 Oct;101(4):1288–1299. doi: 10.1083/jcb.101.4.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelease R. N., Hermerath C. A., Turley R. B., Kunce C. M. Cottonseed malate synthase : purification and immunochemical characterization. Plant Physiol. 1987 Aug;84(4):1343–1349. doi: 10.1104/pp.84.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley R. B., Trelease R. N. Cottonseed malate synthase : biogenesis in maturing and germinated seeds. Plant Physiol. 1987 Aug;84(4):1350–1356. doi: 10.1104/pp.84.4.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]