Abstract
Previous study has demonstrated that the WNK kinases 1 and 3 are direct osmosensors consistent with their established role in cell-volume control. WNK kinases may also be regulated by hydrostatic pressure. Hydrostatic pressure applied to cells in culture with N2 gas or to Drosophila Malpighian tubules by centrifugation induces phosphorylation of downstream effectors of endogenous WNKs. In vitro, the autophosphorylation and activity of the unphosphorylated kinase domain of WNK3 (uWNK3) is enhanced to a lesser extent than in cells by 190 kPa applied with N2 gas. Hydrostatic pressure measurably alters the structure of uWNK3. Data from size exclusion chromatography in line with multi-angle light scattering (SEC-MALS), SEC alone at different back pressures, analytical ultracentrifugation (AUC), NMR, and chemical crosslinking indicate a change in oligomeric structure in the presence of hydrostatic pressure from a WNK3 dimer to a monomer. The effects on the structure are related to those seen with osmolytes. Potential mechanisms of hydrostatic pressure activation of uWNK3 and the relationships of pressure activation to WNK osmosensing are discussed.
WNK kinases are intracellular protein kinases associated with familial hypertension. This association caused us to ask whether WNKs are directly activated by hydrostatic pressure.
We show the kinase domain of WNK3 is activated by hydrostatic pressure both in vivo and in vitro. Hydrostatic pressure induced modest ~30%-50% enhanced autophosphorylation or substrate phosphorylation. Biophysical characterization exposed a dimer to monomer equilibrium promoted by pressure.
This reveals that an intracellular protein kinase is activated by hydrostatic pressure.
INTRODUCTION
Cells respond to hydrostatic pressure to maintain cellular, organ, and organism level functions including cell volume control, blood pressure regulation, and myogenic responses. Integral membrane proteins are the best characterized pressure sensors (Volkers et al., 2015; Zeng et al., 2018). However, there are suggestions from the literature that WNK kinases, a family of soluble-intracellular proteins first identified in 2000 (Xu et al., 2000), may also be involved in hydrostatic-pressure responses. WNK kinases are genetically linked both to hypertension (Wilson et al., 2001) and hypotension (Tobin et al., 2008). Mice heterozygous for Wnk1 have low-blood pressure (Zambrowicz et al., 2003) and reduced-myogenic responses (Bergaya et al., 2011). The linkage between WNKs and familial hypertension showed that WNKs regulate the sodium chloride cotransporter (NCC), the target of the antihypertensive reagent used to treat familial hyperkalemic hypertension (Farfel et al., 1978). WNKs also control the activity of the related cotransporters NKCCs and KCCs (Piechotta et al., 2002; Xu et al., 2002; Vitari et al., 2005; Richardson and Alessi, 2008), which have been well-studied for their roles in transepithelial transport and cell volume recovery (Parker, 1993; Russell, 2000). The same pathways that regulate cell volume (osmotic effects) also function in blood pressure control (hydrostatic pressure effects; Richardson et al., 2008).
WNKs are directly activated by osmotic pressure in cells (Xu et al., 2000) inducing Activation Loop (AL) phosphorylation (Xu et al., 2002; Zagorska et al., 2007). WNKs were identified as potential direct osmosensors in a Caenorhabditis elegans RNAi screen for proteins involved in volume control (Choe and Strange, 2007). Recently, WNK1 knockout in the OVLT neuron-blunts vasopressin release in response to water restriction (Jin et al., 2023). This suggests that WNK1 acts neuronally as an osmosensor. We have demonstrated recently that WNKs 1 and 3 autophosphorylate in response to osmolytes in vitro (Akella et al., 2021). We further showed through a combination of biophysical methods, crystallography, and assays, that osmolytes affect a conformational equilibrium between a chloride-binding and inhibited dimer and an autophosphorylation-competent monomer. In this equilibrium, water functions as an allosteric inhibitor by binding extensively to the inhibited dimer (Akella et al., 2021). Thus, WNKs can function as direct osmosensors.
Here we explore whether in addition to osmotic pressure, hydrostatic pressure also activates WNKs. HEK293 cells expressing endogenous WNKs, and Drosophila melanogaster Malpighian tubules expressing full-length WNK3, both exhibited pressure-induced phosphorylation of WNK effectors. Further, the uWNK3 (kinase domain) exhibits modest enhancements of autophosphorylation and activity under 190 kPa (∼28 PSI) pressure. Effects of hydrostatic pressure on uWNK, measured by multiple modalities, indicate a role for de-dimerization in the action of hydrostatic pressure to activate uWNKs. Most of our analysis was carried out on uWNK3, the more pressure sensitive WNK of the two isoforms we tested (uWNK3 and uWNK1). Apparently, in addition to integral-membrane sensors, a soluble-cytosolic kinase is pressure sensitive. Here we probe the mechanism of hydrostatic-pressure effects on WNKs, and discuss potential origins for differences between in vitro and in vivo responses.
RESULTS
In vivo pressure-induced phosphorylation of the WNK effector KCC2
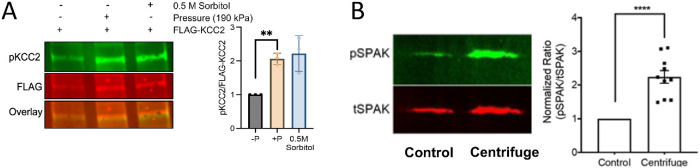
To test whether the endogenous WNK-OSR/SPAK-CCC pathway is activated by hydrostatic pressure in cells, pressure was applied to MDAMB231 cancer cells (Rodriguez et al., 2022). Modest pressure (190 kPa) was applied in an Amicon concentrator using N2 gas (schematic in Supplemental Figure S1A). Phosphorylation of transiently overexpressed KCC2, a downstream effector of WNKs, was used as a readout. Endogenous WNK activity was quantified as the ratio of phosphorylated KCC2 (pKCC2) to total flag-tag KCC2 (pKCC2/tKCC2) by Western blots (Figure 1A, left, blots; right, quantitation, and Supplemental Table S1). A known cellular activator of WNKs, sorbitol, was used as a positive control (Lenertz et al., 2005; Anselmo et al., 2006; Zagorska et al., 2007). A ∼two-fold increase in the pKCC2/tKCC2 ratio was induced by 0.5 M Sorbitol. Hydrostatic pressure (190 kPa) induced a similar two-fold increase of pKCC2/tKCC2 ratio compared with the benchtop control (no added pressure) with a p value of 0.008. Thus, the WNK-OSR/SPAK-CCC pathway in MDAMB231 cells is activated by modest-hydrostatic pressure, and the activation is similar to that induced by sorbitol under the conditions used.
FIGURE 1:
Pressure effects on cellular assays of endogenous WNKs and full length WNK3. (A) Left, MDAMB231 cells transiently transfected with PCMV-FLAG-KCC2, lysed and immunoblotted with anti-pKCC2 antibodies and quantification of normalized pKCC2 infrared signal. Mean ± SEM, n = 3 (biological replicates). Significance measured using one sample t and Wilcoxon test in Graphpad Prism 9 (right). (B) Malpighian tubules in which Drosophila WNK was knocked down and fl-human WNK3 was expressed with SPAKD219A (w; UAS-DmWNKRNAi UAS-WNK3/UAS-SPAKD219A; c42-GAL4/+) were subjected to pressure by centrifugation, or to a similar 0.75°C temperature change (control). WNK3 activity was estimated from the ratio of pSPAK to tSPAK. The ratio from the centrifuged sample was normalized to control. Mean ± SEM with individual data points for n = 10 independent replicates shown. ****, p < 0.0001, one-sample t test to theoretical mean of one. A sample Western blot is shown.
In vivo hydrostatic-pressure activation of full-length WNK3 in Malpighian tubules
We further tested whether WNK3 is affected by pressure in vivo in Drosophila. We previously used the GAL-UAS modified Drosophila to replace the single Drosophila WNK (dWNK) with full-length human WNK3 in Malpighian (renal) tubules (Pleinis et al., 2021). Using this system, we showed that WNK-SPAK/OSR1 signaling regulates ion transport in the Malpighian tubule (Wu et al., 2014; Sun et al., 2018). To measure SPAK phosphorylation, flies transgenically expressing kinase-dead rat SPAKD219A were crossed with the human WNK3 flies (Supplemental Figure S1B, as described previously [Sun et al., 2018]). WNK3 activity was quantified as the ratio of phosphorylated SPAK (pSPAK)/total SPAK (tSPAK) by Western blots (Figure 1B). Isolated Malpighian tubules were subjected to 81 kPa hydrostatic pressure by centrifugation and SPAK phosphorylation compared with unpressurized control tubules. Each experiment involved 15 sets of pressurized and unpressurized tubules, and the experiment was replicated 10 times. Phosphorylation of SPAKD219A increased by about a factor of two in the presence of applied pressure. Apparently, WNK3 is activated by hydrostatic pressure as well as osmotic pressure in cells.
In vitro hydrostatic-pressure activation of uWNK3
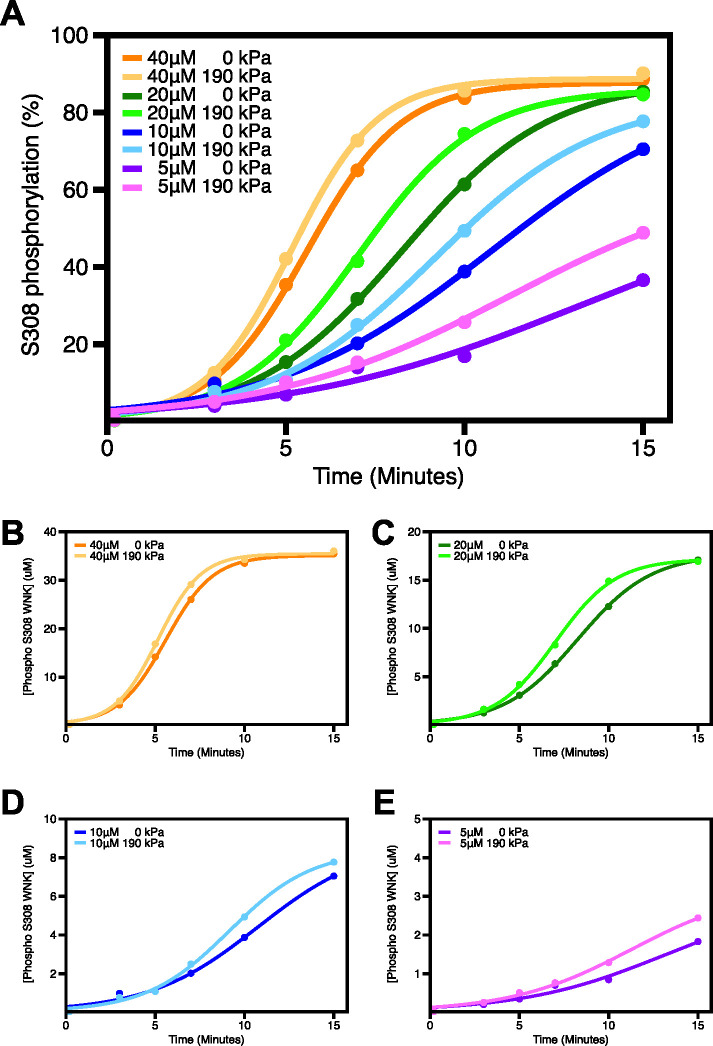
To probe the biochemical and biophysical basis for the effects of hydrostatic pressure on WNKs, we focused on the kinase domains of WNK1 and WNK3. The kinase domains express well, are highly conserved and likely house essential activities of WNKs, and are amenable for biophysical studies. WNK AL phosphorylation is required for WNK activity (Xu et al., 2002). We hypothesized that pressure effects should be on autophosphorylation of uWNKs to form active pWNKs. To obtain uWNK3, bacterially expressed pWNK3 was dephosphorylated with phosphatases to make unphosphorylated and inactive WNK3 (uWNK3) (Akella et al., 2021). Autophosphorylation reactions require relatively high ATP concentrations (Supplemental Figure S2A) and 5 mM ATP was used in these analyses. Different readouts of autophosphorylation and substrate phosphorylation were used, each revealing small, 20–30% enhancements of phosphorylation. 190 kPa of pressure was applied for 15 min using 4 μM uWNK3 and 40 μM GST-OSR1 (gOSR1) in 150 mM total Cl–. ProQ Diamond ® phosphoprotein stain offers a visual of autophosphorylation and activity (Figure 2A). Triplicate measurements revealed ∼15% in vitro enhancements of uWNK3 autophosphorylation (p < 0.01) and gOSR1 substrate phosphorylation (p < 0.001; Figure 2A. The reagent ADP-Glo® tracks the disappearance of ATP, and showed slightly larger effects at 10 and 15 min (40–50%; Figure 2B) than observed with ProQ- Diamond. A third readout was 32P radiography of phosphate incorporation into WNK3. Autophosphorylation reactions were carried out at higher-protein concentrations (40 μM uWNK3) above the ∼20 μM dimer equilibrium (0.8 mg/mL; Akella et al., 2021), and higher total chloride (400 mM) was used to slow the reaction (Figure 2C). Modest pressure enhancement of autophosphorylation was again observed.
FIGURE 2:
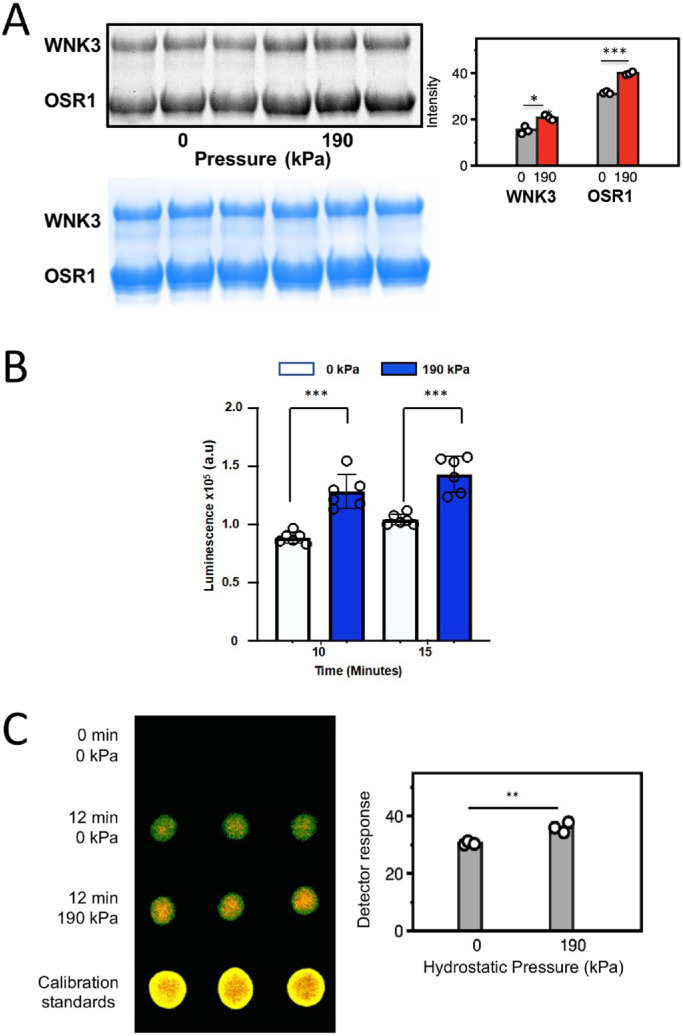
Pressure effects on WNK3 autophosphorylation and activity in vitro. (A) using pWNK and gOSR. in the absence or presence of added hydrostatic pressure. Pro-Q® stain (back) of phospho-proteins pWNK3 and phopho-gOSR. Triplicated 15-min reactions –/+ 190 kPa using 4 mM uWNK3 and 40 mM gOSR1 in 150 mM Cl–, run at 25°C. The blue bands are Coomassie staining as loading controls. In the bar graph, gray corresponds to unpressurized pWNK3 and phospho-gOSR; red, under pressure. Two-sided t test was conducted for each protein giving * p < 0.01, *** p < 0.0001. (B) ADP-Glo ® measuring ATP consumption using uWNK3 (4 mM) and gOSR1 (40 mM). Reactions were run at 30°C. Appearance of ADP without (white) and with (blue) 190 kPa measured at 10 min and 15 min (background from reagents subtracted). (C) g32P ATP spot blots imaged by false color autoradiography of autophosphorylation of 40 mM uWNK3 at 0 and 190 kPa hydrostatic pressure induced with N2 gas. p = 0.007.
Tracking autophosphorylation by LC/MS
LC/MS was used to observe autophosphorylation of specific known activating phosphorylation sites (Xu et al., 2002). Multiple serine residues in WNK3 are phosphorylated as expressed (Akella et al., 2021). The four most prominent sites were tracked in autophosphorylation assays (Supplemental Figure S2B). The known primary Activation-Loop site (Ser308) was fully autophosphorylated under the conditions used, and the secondary Activation-Loop site (Ser304) became 70% autophosphorylated. The minor sites did not change in phosphorylation over the 20-min reaction time course.
Hydrostatic pressure sensitivity of WNK3 AL autophosphorylation
Autophosphorylation reactions were carried out in 5 to 40 μM uWNK3, spanning a range of activities from about 25 to 85% phosphorylation of S308 over the 15-min time course (darker colors in Figure 3A). Application of hydrostatic pressure in an Amicon concentrator induced ∼20–45% (Δ%phos/%phos) increased autophosphorylation (lighter colors in Figure 3A). In general, larger effects were observed at lower levels of basal autophosphorylation. The progress curves were fit to an autocatalytic model in DynaFit (Kuzmic, 2009; Kuzmic et al., 2009; Figure 3B–E). The DynaFit script used is presented in Supplemental Table S2. Table 1 lists the computed reaction rate at each [uWNK3] with and without added pressure. At each [uWNK3], added pressure increases the observed rate of autophosphorylation. It is also interesting that the rate increases as the [uWNK3] drops. This enhanced activity is consistent with data described below that pressure dissociates dimeric uWNK3, and a model that the monomer is autophosphorylation competent.
FIGURE 3:
Tracking pressure effects on individual autophosphorylation reactions measured by LC/MS. (A) Time-course of uWNK3 autophosphorylation of Ser308 as a function of [uWNK3], ±190 kPa added hydrostatic pressure at each [uWNK3]. Lighter colors represent assays conducted under 190 kPa, darker colors, bench (no added pressure) at concentrations indicated (points are % pWNK3). (B–E; pWNK3) +in μM fit to autocatalytic mechanism using DynaFit.
TABLE 1:
DynaFit modeling of autophosphorylation in Figure 3B.
| [uWNK3] μM | 0 added kPa | 190 kPa | ||
|---|---|---|---|---|
| rate μM/min | R2 | rate μM/min | R2 | |
| 40 | 0.022 ± 0.0013 | 1.0 | 0.023 ± 0.0015 | 1.0 |
| 20 | 0.028 ± 0.0015 | 1.0 | 0.035 ± 0.0024 | 1.0 |
| 10 | 0.037 ± 0.0097 | 0.99 | 0.052 ± 0.0058 | 1.0 |
| 5 | 0.085 ± 0.078 | 0.98 | 0.092 ± 0.027 | 0.99 |
The pressure effects on autophosphorylation at 10 μM uWNK3 was triplicated using a different uWNK3 preparation (Supplemental Figure S3). This WNK3 sample was less active, but again showed significant enhancement of WNK3/S308 autophosphorylation with pressure (p = 0.0043 at 30 min). In contrast, pressure did not increase the activity of pWNK3 toward a peptide substrate (p = 0.33 at 15 min; Supplemental Figure S4A) as measured using the Kinase-Glo® reagent. As a further control, and to assure that the small effects observed are not due to sample handling, a mock reaction was run in the Amicon concentrator, without adding pressure (Supplemental Figure S4B). The mock Amicon autophosphorylation progress curve superimposed with the benchtop control.
De-dimerization of uWNK3 under pressure
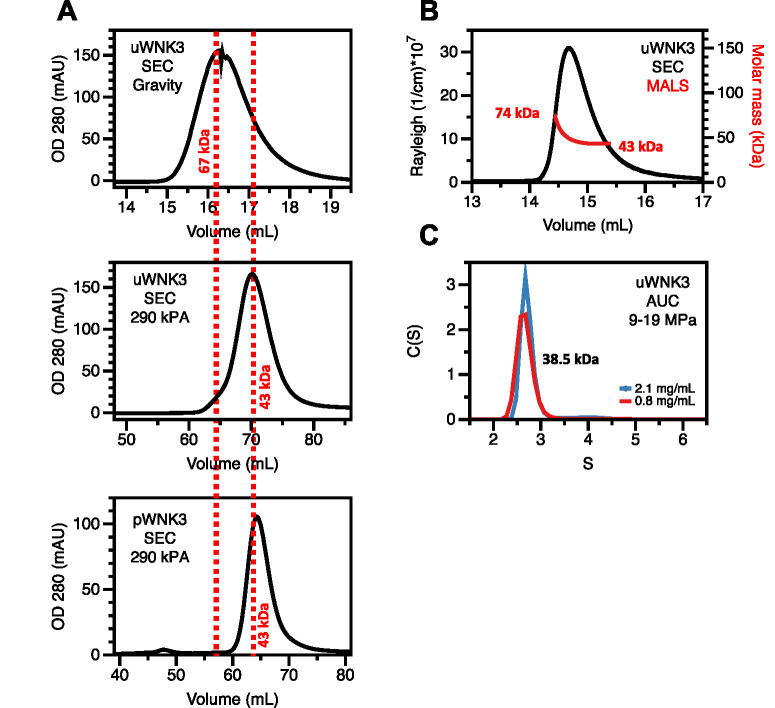
In our previous study of WNK osmotic activation, we found that osmolytes affected the WNK oligomeric state as well as activity. We showed previously by static light scattering (SLS) that uWNK3 is dimeric in solution above 20 μM (0.8 mg/mL). Further, we showed by SLS that osmolytes promote the monomeric state (Akella et al., 2021). Here we tested whether hydrostatic-pressure activation of WNKs is also associated with changes in oligomeric state. Figure 4A shows gel filtration elution traces for WNK3 at different back pressures. The elution volumes for the 67 kDa and 43 kDa standards are indicated by dashed red lines. Using gravity-driven gel filtration on Sephadex 75, uWNK3 eluted as a single peak at a volume close to the 67 kDa standard. This elution volume is intermediate between a uWNK3 monomer (39.3 kDa) and a dimer (78.6 kDa), suggesting a mixture in rapid equilibrium (Stevens, 1989). In contrast, the elution volume of uWNK3 gel-filtered on an FPLC at a backpressure of 280 kPa, also using SuperdexTM 75, was close to that of the 43 kDa standard and an elution volume similar to that of pWNK3, indicating a monomer. Elution traces for the albumin and ovalbumin standards are given in Supplemental Figures S5, A and B.
FIGURE 4:
Apparent molecular weight of uWNK3 as a function of back-pressure. (A) Gravity gel filtration of uWNK3 on SephadexTM 75 resin in 150 mM NaCl and 25°C. Elution volumes of standard albumin (67 kDa, 16.0 mL) and ovalbumin (43 kDa, 17.0 mL; red vertical dashed lines). Gel filtration of uWNK3 as run on the Acta FPLC at 290 kPa SephadexTM 75 column. Albumin standard (63.4 mL); ovalbumin (69.6 mL). Gel Filtration of pWNK3 in the same buffer (standard, ovalbumin [43 kDa, 64.3 mL]). (B) SEC-MALS data collected with a backpressure of 250 kPA. (C) Analytical ultracentrifugation (9–19 MPa) at two uWNK3 concentrations, 2.1 mg/mL (52 mM, black) and 0.8 mg/mL (19.2 mM, red). The calculated apparent calculated molecular weight was 38.5 kg/mol.
Further support for the idea that uWNK3 is in a pressure-sensitive oligomeric equilibrium comes from SEC-MALS analysis at a back pressure of 250 kPa: uWNK3 eluted as a single peak that exhibits a range of molecular weights (Figure 4B). The MALS molecular weight estimates for the leading and trailing edge of the eluant were 74 kDa and 43 kDa, respectively. These data again suggest a mixture in rapid equilibrium (Stevens, 1989).
To access a different pressure regime and in the absence of column interactions, sedimentation velocity analytical ultracentrifugation (AUC) of uWNK3 was conducted. A dominant sedimentation boundary was observed at 2.7S for the two uWNK3 concentrations analyzed (Figure 4C). The SEDFIT calculated mass was 38.5 kDa, close to the 39.3 kDa WNK3 monomer mass. The AUC experiment generated pressures ranging from 9–19 MPa, depending on radius. This pressure is much higher than the gravity or 280 kPa used in the gel-filtration experiments. Apparently, uWNK3 is fully monomeric at these pressures. The molar masses observed here and measured previously by SLS, SEC-MALS, AUC, and gel filtration are presented in (Supplemental Tables S3 and S4). Thus, the apparent oligomeric state as measured under filtration of uWNK3 is affected by hydrostatic pressure. The range of molecular weights from dimer to monomer are consistent with crystallographic analyses of WNK kinase domains as seen in PDB file 6CN9 and 5W7T.
NMR of uWNK3 under applied pressure
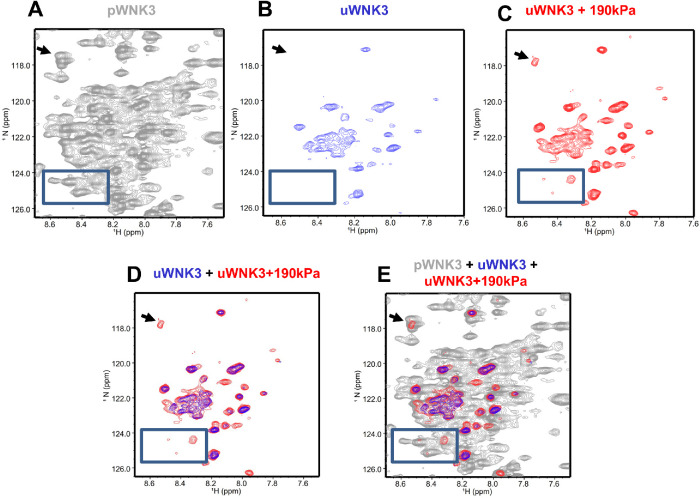
As another measure of the effects of hydrostatic pressure on uWNK3, we used NMR spectroscopy. First, we obtained 1H,15N TROSY-HSQC spectra for uWNK3 and pWNK3 at 50 μM WNK3, at 37°C. The time frame for data collection was 11 h for pWNK3 and 24 h for uWNK3. The two forms show very distinct spectra as exemplified by the boxes shown in Figures 5, A and B. The pWNK3 spectrum (Figure 5A) exhibited reasonable quality, as expected for a monomeric protein of 38.5 kDa. In contrast, considerably fewer cross-peaks were observed in the uWNK3 spectrum (Figure 5B) indicative of a higher-molecular weight (Takeuchi et al., 2015). To test the effects of hydrostatic pressure on uWNK3, an 1H,15N TROSY-HSQC spectrum was collected on uWNK3 in a valved-NMR pressure tube charged to 190 kPA with N2 gas. Data on the pressurized uWNK3 were collected for 12 h. These data (red in Figure 5C) show new cross-peaks appearing not present in the unpressurized uWNK3 sample (blue; Figure 5D–E). Most peaks appearing in the pressurized uWNK3 sample overlay with pWNK3 cross-peaks (indicated by arrows). These data suggest a reduction in molecular weight under pressure, potentially due to monomerization. Thus, hydrostatic-pressure affects the structure of uWNK3 without any other changes in the solution environment.
FIGURE 5:
uWNK3 and pWNK3 1H,15N TROSY-HSQC. (A) pWNK3 at 50 mM chloride and 37°C. (B) uWNK3 as in (A). (C) uWNK3 with 190 kPa applied with N2. (D) Overlay of (B) and (C). (E) Overlay of (A), (B), and (C). Comparable contour levels were used for uWNK3 and uWNK3+190 kPa applied pressure. pWNK3 spectra was plotted at higher contour levels for clarity. Arrow and box indicate peaks in the uWNK3 spectrum appearing under pressure in panel (C).
DSS crosslinking of uWNK3 under applied pressure
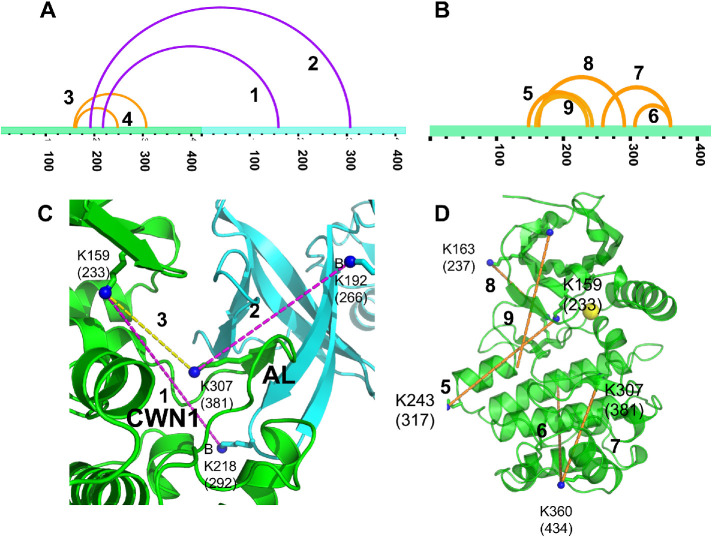
Another measure of protein—protein interactions in solution is crosslinking. We used the lysine-directed crosslinking reagent disuccinylsuberate (DSS) that covalently links lysines with Cα—Cα distances < 28 Å (Rappsilber, 2011). Crosslinking reactions were carried out in the absence or presence of 190 kPa hydrostatic pressure applied in an Amicon chamber, as above. Reactions were quenched with Tris-HCl, and proteolyzed using chymotrypsin. Crosslinked uWNK3 peptides were analyzed by LC–MS/MS. Crosslinked peptides found in unpressurized versus pressurized uWNK3 reactions are presented in Supplemental Tables S5 and S6, respectively. As expected, many crosslinks were common to both unpressurized and pressurized reactions or arose from the nonnative pET29b C-terminal tag. Crosslinks were mapped onto the dimer structure of uWNK1 (PDB file 6CN9). Table 2 lists crosslinks that are unique to unpressurized or pressurized uWNK3 together with an interpretation of the lysine Cα–Cα distances as arising from monomer or dimer. We found more unique crosslinks in the pressurized sample, and more crosslinks likely arising from the monomer. Two of the crosslinks unique to the unpressurized uWNK3 are most easily interpreted as arising from the dimer (magenta arches) in xVis (Grimm et al., 2015; Figure 6A). Orange arches show unique crosslinks we interpret as arising from the monomer. Figure 6B shows pressure-unique crosslinks (all orange, from the monomer). Figure 6C show the unpressurized-unique crosslinks mapped onto the uWNK1 dimer, and Figure 6D shows the pressurized-unique crosslinks mapped onto the uWNK1 monomer. Overall, the crosslinking data lend support the existence of a uWNK3 dimer as the 80 kDa species in SLS and gel filtration ([Akella et al., 2021] and Figure 4, A and B). The data are further consistent with the idea that this dimer dissociates under pressure as observed above by gel filtration and NMR.
TABLE 2:
Distance between crosslinked residues calculated from a monomer vs. a dimer of uWNK3 based on the WNK1-KDm dimer (6CN9) or Subunit A of 6CN9
| X-LINK | A. Crosslinks in uWNK3 only without added pressure ζ | |||||
|---|---|---|---|---|---|---|
| 1 WNK3 (WNK1 #) | 2 WNK3 (WNK1) | Score | Monomer (Å) | Dimer (Å) | ||
| 1 | 218 (292) | 159 (233) | 15 | 18 | 25 | β3 to inside barrel |
| 2 | 307 (381) | 192 (266) | 6 | 25 | 19 | Act. Loop to Helix C |
| 3 | 159 (233) | 307 (381) | 4 | 14 | 43 | β3 to Act. Loop |
| 4 | 248 (322) | 163 (237) | 10 | 33 | 53 | β3 to Helix E |
| 5 | 222 | 333 | 9 | 38 | 54 | β1 to Helix E |
| B. Crosslinks in uWNK3 occurring only under 190 kPa ζ | ||||||
| 6 | 192 (266) | 159 (233) | 13 | 16 | 38 | Helix C to β3 |
| 7 | 159 (233) | 243 (317) | 8 | 24 | 52 | β3 to DE |
| 8 | 236 (310) | 259 (333) | 7 | 20 | 56 | Helix D to Helix E |
| 9 | 185 (259) | 192 (266) | 7 | 11 | 33 | BC to Helix C |
| 360 (434) | 259 (333) | 6 | 30 | 51 | Helix E to Helix G | |
| 307 (381) | 360 (434) | 6 | 25 | 51 | Act. Loop to Helix G | |
| 291 (365) | 163 (237) | 4 | 19 | 41 | β7 to β3 | |
| 236 (310) | 148 (222) | 4 | 25 | 31 | β1 to Helix D | |
| 236 (310) | 159 (233) | 3 | 15 | 40 | Helix D to β3 | |
| 221 (295) | 259 (333) | 6 | 36 | 42 | Inside barrel to helix E | |
ζCalculated distances between residue pairs (Resn1-Resn2) in the uWNK1/SA structure. Blue indicates distances typical for lysine–lysine DSS crosslinking. Gray indicates distances outside the DSS crosslinking limits.
FIGURE 6:
DSS crosslinks of wild-type uWNK3 formed without and with added hydrostatic pressure. Crosslinks unique to (A) unpressurized and (B) pressurized WNK3 visualized as a function of sequence in xVis (Grimm et al., 2015). Orange, intramolecular crosslinks, magenta, inter-subunit crosslinks. (C) Crosslinks in unpressurized uWNK3 mapped onto the crystallographic dimer of uWNK1 (PDB file 6CN9). Subunit A, green; B, cyan. The AL from Subunit A, which forms intimate contacts with Subunit B, and the position of the CWN1 are indicated. Intermolecular crosslinks are in magenta, and intramolecular crosslinks are yellow. Crosslink 1 links WNK3/A/K159 to B/K218 (WNK1 A/K233 to B/K292), 2 links WNK3/B/K192 to A/K307 (WNK1/B/K266 to A/K381), and intrachain crosslink 3 links WNK3/A/K159 to A/K307 (WNK1/A/K233 to A/K381). Crosslink 4 is not shown. (D) Selected crosslinks observed in the presence of 190 kPa mapped onto a monomer of uWNK1. Residues crosslinked are indicated in Table 2.
DISCUSSION
The data presented here show that a soluble-protein kinase, WNK3, is modestly activated by hydrostatic pressure in vivo and in vitro. Cellular assays of endogenous WNKs or full-length WNK3 show 50–100% activity enhancement, as observed by phosphorylation of downstream effectors. In vitro assays show 20–45% (Δ%phos/%phos) enhancement of autophosphorylation and substrate phosphorylation. Here we applied 190 kPa pressure to uWNK3 in cells in culture and in vitro, and 80 kPa to Malpighian tubules. These pressures are higher than the 16 kPa (120 mm Hg) typical of systolic blood pressure, but orders of magnitude below pressures used (200 MPa and more) in studies of effects on soluble proteins (Collins et al., 2011; Dellarole and Royer, 2014). Our observed pressure effects are on a smaller scale than those recorded on integral membrane proteins such as Piezo (Coste et al., 2010). We do observe larger effects in vivo than in vitro. Potential origins for these discrepancies could include effects from phase transitions exhibited by full-length WNKs in cells (Sengupta et al., 2012; Boyd-Shiwarski et al., 2022). Alternatively, other cellular components might be involved in WNK pressure activation. The hydrostatic pressure experienced by tissues varies from pascals to megapascals (Liu et al., 2019). The pressure we used in our study, 190 kPa, is within this range, suggesting potential physiological relevance of direct pressure sensing by WNKs.
Concerning the effects of hydrostatic pressure on uWNK3 kinase domains, it is interesting that the same changes in uWNK structure, de-dimerization, are observed with both osmotic and hydrostatic pressure (Akella et al., 2021). Osmotic pressure, a demand on solvent, disfavors the dimer, promoting an alternative, autophosphorylation-competent conformation. We posited that the dimer is disfavored under osmotic pressure because of a conserved water network (CWN1 in Figure 6C) in the dimer, as discussed previously (Akella et al., 2021). Does hydrostatic pressure work in the same way? Hydrostatic pressure, a force to change volume, may affect the WNK conformation similarly. Nonhydrogen-bonded water in contact with the surface of proteins is lower density than bulk water, as is clear from radial distribution analysis of waters in the Protein Data Bank (Carugo, 2017) and high-resolution crystallography (Teeter, 1984). Thus, hydrostatic pressure, like osmotic pressure, should disfavor a WNK conformation with more bound water. This potential similarity in mechanism raises the possibility that some pressure sensitivity should be expected where there is osmosensitivity. Further structural and mutational studies are needed to probe mechanisms of hydrostatic activation of WNKs, especially to evaluate effects on full-length WNKs, and whether hydrostatic effects and osmotic effects are coupled in other pressure sensors. More data is required also to probe the extent to which WNKs are activated by pressure in cells, and how these effects on WNKs are integrated with other cellular-pressure sensors. We look forward also to identification and analysis of other intracellular-pressure sensors to determine whether molecular mechanisms related to those observed in WNKs are involved.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Clones and reagents
The clone WNK3 (118-409) has been described (Akella et al., 2021). GST-OSR1 peptide (314-344) encompassing the phosphorylation site at 325 of OSR1 was a gift from Clinton Taylor and Melanie Cobb. A peptide containing the OSR1/S325 site, OSR-tide (RVPGSSGRLHK), was obtained from Biomatik (Ontario, CANADA). Kinase-Glo ® (Promega), ADP Glo ® (Promega), ProQ Diamond ® (Thermo Fisher Scientific), 15N ammonia (Cambridge Isotope Laboratories), Terrific Broth (Fisher). and tris carboxyethyl phosphene (TCEP) (Hampton, CA) were used. Polyclonal anti-total KCC2 antibodies were from Sigma-Aldrich (Cat. no. 07-432) and polyclonal anti-KCC2/pT2007 antibodies were from Thermo Fisher Scientific (Cat. no. PA5-95677). The plasmid for human WNK3 kinase domain (118-409) expression and the phosphatases used have been described (Akella et al., 2021). MDAMB231 cells have been used previously (Jaykumar et al., 2021) and were obtained from ATCC (MDA-MB-231-HTB-26). Monoclonal ANTI-FLAG® M2 antibody was from Sigma-Aldrich (Cat. No. F1804);
IRDye® 800CW Goat anti-Rabbit IgG Secondary Antibody (Cat. no. 926-32211) and IRDye® 680RD Goat anti-Mouse IgG Secondary Antibody (926-68070) were from Fisher Scientific; Dulbecco’s modified Eagle’s medium (DMEM) from Life Technologies (Cat. no. 11965-092); fetal bovine serum (FBS) from Genesee Scientific, (Cat. no. 25-514); and Pierce™ IP Lysis Buffer is from Thermo Fisher Scientific (Cat. no. 87787) Protease and phosphatase inhibitors were from Fisher Science. (Cat. nos. PIA32963 and PIA32965). Precast gels used were Mini-PROTEAN gels (4568096) LI-COR Intercept (TBS) blocking buffer was from Fisher Science. (Cat. No. NC22420).
Bacterial expression and purification of pWNK1, pWNK3, and GST-OSR1 (314-344)
Protein expression was carried out in the Escherichia coli strain Bl21 DE3, and purification of (pWNK3) and (pWNK1) followed published protocols (Min et al., 2004; Piala et al., 2014) and were stored in 50 mM hydroxyethyl piperazine ethanesulfonic acid (HEPES) pH 7.4, 50 mM NaCl. The purification of pWNK3 includes Ni affinity (NTA agarose, Qiagen), MonoQ (10/10; GE), and gel filtration on Sephadex 75 [GE]). The GST-OSR1(314-344; gOSR1) encompassing the phosphorylation site S325 was a gift of Clinton Taylor and Melanie Cobb (Taylor et al., 2018). The gOSR1 was expressed similarly and purified on a glutathione Sepharose (GE Healthcare) and buffer-exchanged into 20 mM HEPES, pH 7.4 and 50 mM NaCl, with the addition of 5% glycerol.
Dephosphorylation methods
The kinase domains of WNK1 and WNK3 are phosphorylated as expressed in E. coli (pWNK1 and pWNK3). The pWNKs were dephosphorylated using phosphatases to make uWNK1 and uWNK3. WNK1 was dephosphorylated using λ−phosphatase and shrimp alkaline phosphatase in 10:1 and 5:1 (kinase/phosphatase) ratios respectively. WNK3 was dephosphorylated using PP1cγ and λ-phosphatase in a 10:1 ratio, in 0.5 mM MnCl2. were removed using Ni-NTA and gel filtration chromatography. The state of phosphorylation was confirmed by mass-spectrometry. Residual activity of phosphatase was checked using p-nitrophenyl phosphate (G-Biosciences, St. Louis, MO) following published protocols (Zhou and Zhang, 1999). The kinases were buffer exchanged into 50 mM HEPES, pH 7.4, 150 mM NaCl. The phosphatase inhibitor orthovanadate prepared as described (Zhou and Zhang, 1999) was added to the protein solutions to 2 mM. The GST-OSR1(314-344; gOSR1) encompassing the phosphorylation site S325 was a gift of Clinton Taylor and Melanie Cobb (Taylor et al., 2018). The gOSR1 was expressed similarly and purified on a glutathione Sepharose (GE Healthcare) and buffer-exchanged as for uWNK1 with the addition of 5% glycerol.
Preparation of 15N enriched proteins for NMR
BL21-DE3 cells are grown initially in 5 mL of TB. 800 μL of culture is transferred to 50 mL of M9 medium (described below) and inoculated at 37°C and shaken at 210 rpm. 16 mL of culture is transferred to 1L of M9. Cells grew for 18 h, at 20°C and 200 rpm.1L of minimal medium M9 is composed of 100 mL 10x M9 salts (30 g/L KH2PO4, 67.8 g/L Na2HPO4 and 5g/L NaCl), 10 mg biotin, 10 mg thiamine, 25 mg/L MgSO4.7H2O, 11 mg/L CaCl2, 3 mg/L glucose as carbon source and 1 g/L 15NH4Cl as nitrogen source.
In cell-assay methods
MDAMB231 cells were incubated in DMEM high glucose medium (Thermo Fisher Scientific). Cells were plated at 37°C in a CO2 incubator to an initial density of 2.5 × 106 in10 cm tissue culture plates. Cells were transfected with 8 μg of the KCC2 plasmid and further incubated at 37°C. After 24 h, cells were split into three 6-cm plates. The following day, fresh medium or medium containing to 0.5 M sorbitol was introduced. One 6-cm plate was pressurized to 190 kPa using N2 in an Amicon concentrator for 30 min (Supplemental Figure S1A). The unpressurized negative control plate and the 0.5 M Sorbitol positive control plate were incubated on lab bench. Media was removed and cells were washed three times with ice-cold 1X phosphate buffered saline (13.7 mM NaCl, 0.27 mM KCl, 0.8 mM Na2HPO4, and 0.2 mM K2H2PO4). All plates were simultaneously flash frozen in liquid N2, and stored at 80°C.
Cells were thawed in 400 μL Laemmli lysis buffer for 30 min. Lysed cells were transferred to 1.5 mL Eppendorf tubes and centrifuged at 14,000 rpm for 10 min. 30 μL of supernatant from each of the three conditions were loaded onto 5–15% acrylamide precast gels. Proteins were transferred to nitrocellulose membrane using 8 V for 40 min in a semidry transfer apparatus. The nitrocellulose membrane was blocked with 5% bovine serum albumin for 1 h. Primary ANTI-FLAG® M2 antibodies (1/1000) were applied for 11 h. The membrane was washed three times in tris-buffered saline (5 mins each). Incubation with secondary antibody (IR conjugate) (1/1000) was conducted overnight. Protein bands were visualized using a LI-COR Odessey imager and quantified in Image Studio. The pKCC2/tKCC2 ratio was set to one for the negative control, and the pKCC2/tKCC2 ratio in the pressurized and positive control samples were normalized to the negative control ratio. P values were calculated using a one-sample t and Wilcoxon in Graphpad Prism.
Measurement of in vivo WNK3 activity in D. melanogaster
Endogenous Drosophila WNK was knocked down using UAS-DmWNKRNAi (Bloomington Drosophila Stock Center, stock #42521 (Sun et al., 2018), and full-length human WNK3 (Stenesen et al., 2019) was expressed in the principal cell of the Drosophila renal tubule, together with the WNK substrate, kinase-dead rat SPAKD219A (Sun et al., 2018), using c42-GAL4 (Julian Dow, Glasgow, UK; Rosay et al., 1997). The genotype used was w; UAS-DmWNKRNAiUAS-HsWNK3WT/UAS-SPAKD219A; c42-GAL4/+0.15 pairs of tubules were dissected from 4- to 6-day-old adult females, transferred to 300 μL of bathing solution for 1 h, and then centrifuged at relative centrifugal force = 250 for 10 min (Eppendorf Centrifuge 5424, 1632 rpm; estimated pressure, 81 kPa), or were subjected to an equivalent temperature gradient in a PTC-100 Programmable Thermal Controller (MJ Research, Inc). The effect of temperature was controlled for by imposing similar temperature increase to control tubules in a PTC-100 Programmable Thermal Controller. The temperature gradient was determined by centrifuging two separate sets of tubules and measuring temperature at different timepoints, which increased by a total of 0.75°C over the first 2 min, beginning at room temperature (∼23°C). Bathing solution was the normal potassium/low intracellular chloride solution described in (Pleinis et al., 2021). After centrifugation, tubules were lysed in 2X Laemmli buffer. 50 μL lysate was used to detect pSPAK or tSPAK by Western blotting using 1:1000 dilution of antibodies to pSPAK (Ser373)/pOSR1 (Ser325; Millipore, Cat. #07-2273), total-SPAK (GeneTex anti-STK39 [2E10], Cat.#GTX83543) and actin (JLA20-s; DSHB).These antibodies do not react to antigen in the absence of transgenically expressed SPAK (Sun et al., 2018). Secondary antibodies, used at 1:2500 dilution, were fluorophore-conjugated anti-rabbit AzureSpectra 800 (VWR, Cat. #AC2134, Lot #170124-06) or anti-mouse AzureSpectra 700 (VWR, Cat.#AC2129). Protein bands were visualized using a c600 (Azure Biosystems) and quantified in ImageJ as previously described (Pleinis et al., 2021). The pSPAK/tSPAK ratio was set to one in each experiment, and the pSPAK/tSPAK ratio of the centrifuged sample was normalized to the control sample. Ratios were compared using a two-sided one-sample t test to a theoretical mean of one using GraphPad Prism, version 9.
Calculation of tubule pressure
Tubule pressure was calculated using the following equation:
Force was calculated using the following equation: Force (Newtons or kg × m/s2) = mass (kg) × acceleration (m/s2). The mass of 80 anterior tubules was determined to be 0.0003 g (using AB54-S/FACT Mettler Toledo Balance). Converting this mass to kg per tubule gives 3.75 × 10–9 kg/tubule. Acceleration was calculated by the following equation (per centrifuge specifications): Acceleration (m/s2) = relative centrifuge speed (RCF) × g (m/s2) × 1000, where RCF = 250. Thus, the force was determined to be 9.20 × 10–3 Newtons. To determine the surface area, we assumed tubules to have a cylindrical shape, and that the potential space of the lumen is obliterated under pressure. The outer diameter of the tubule is 35 μm, the inner (luminal) diameter is 17 μm (Dow et al., 1994), and therefore, under pressure, tubule diameter is 18 μm. Surface area was calculated by the following equation: (2×π×r2 + 2×π×r×h), where r = radius = 0.000009 m for the tubule, and h = height = 0.002 m for anterior tubules (Dow et al., 1994). Thus, the calculated pressure = 80.9 kPa.
Generic assay conditions
Autophosphorylation assays were carried out in in 20 mM HEPES, pH 7.4, 20 mM MgCl2, 5 mM ATP, and usually in 150 mM Cl– at room temperature (unless otherwise stated). Reactions were initiated by the addition of 4 μM uWNK3 or uWNK1. Assays containing gOSR1 were 40 μM in gOSR1. Reactions run at (uWNK3) above the dimer equilibrium were run at either 40 μM (1.6 mg/mL) or 20 μM (0.8 mg/mL) and in 400 mM NaCl. Some reactions were conducted at 30°C in a water bath.
Pressure chamber reactions
Hydrostatic pressure was applied in an Amicon concentrator connected to a N2 tank fitted with a Boclow low pressure regulator (BOC Corp. UK). Samples were subject to 190 kPa (schematic in Supplemental Figure S1A).
ProQ Diamond ® Gel-based phosphorylation assays
To visualize phospho-proteins, 12% precast BioRad gels were run with 30 μL of the reaction aliquot. Pro-Q Diamond® (Life Technologies Inc.) stain was applied to the gels using the manufacturer’s protocol which includes fixing, staining, and wash steps. The Pro-Q Diamond stain reflects total phosphorylation of the uWNK3 and gOSR1. Gels were imaged using a ChemiDoc MP (Biorad) and volumes calculated in ImageLab (Biorad). The gels were then stained with Coomassie (Biorad) to obtain loading controls.
ATP depletion assays with ADP-Glo ®
ADP-Glo® (Promega Inc.) was one assay method tested to track WNK autophosphorylation and substrate phosphorylation. ADP-Glo® allows the use of the high-ATP concentrations (2–5 mM) necessary for uWNK autophosphorylation, in contrast to Kinase-Glo® that requires no more than 0.1 mM ATP. 50 µL reactions contained 40 mM HEPES (pH 7.0), 10 mM MgCl2, 4 µM uWNK3, and 40 µM gOSR1. pH 7.0 was necessary to eliminate an ATPase activity. Reactions were run at 30°C. The chloride concentration was maintained at 150 mM (unless otherwise stated). The reaction was started by the addition of 2 mM ATP. Reactions were stopped after 15 min by addition of a pan-WNK inhibitor WNK463 (Yamada et al., 2016). The manufacturer’s protocol was followed for the remaining steps, except that the ATP depletion step (conversion to cyclic AMP) was conducted for 2 h to reduce the background. 100 μL aliquots from each reaction were transferred to a 96-well plate which was centrifuged for 2 min. at 800 rpm. Luminescence was read on a CLARIOstar plate reader and data analyzed using MARS software (both reader and software, BMG Labtech, Ortenberg, GER). The background fluorescence in the absence of added proteins, enzyme uWNK3 and substrate gOSR1 was subtracted. Data were analyzed in Excel to obtain standard deviations and t tests. Data were plotted in DataGraph (Visual Data Tools, Inc.)
32P Assays of WNK3 autophosphorylation
Reaction conditions were the same as stated above, with exceptions. Reactions were initiated by the addition of uWNK3 (final concentration of 40 µM). Assay termination and phosphorylation was accomplished using protocols described by Racker (Racker, 1991). Reactions were terminated at timepoints by spotting aliquots on 3MM Whatman filter paper followed by immediate immersion in termination solution (10% trichloroacetic acid and 10 mM pyrophosphate). Extensive washing with fresh termination solution to remove unincorporated radioactivity was followed by drying the filter paper. 32P-incorporation was measured by autoradiography using a phosphor screen (Kodak) and Typhoon 9450 (General Electric). Additional aliquots of the reaction mixtures were diluted then spotted on unwashed filter paper to establish the ratio of CPM to cold ATP concentration. ImageQuant software (General Electric) was used to quantify the autoradiograms.
Mass spectrometry to track specific serine auto-phosphorylation in WNK3
Proteins were digested in an 8:1 M ratio with sequence-grade chymotrypsin (Roche) in the presence of 100 mM Tris pH 8.0, and 25 mM CaCl2 (100 μL total reaction volume) at 30°C overnight. Following digestion, the peptide mixture was separated by HPLC on a RP-C18 column (Phenomenex Aeris Widepore 150 × 2.1 mm) using an acetonitrile-water gradient from 4 to 28% acetonitrile containing 0.2% formic acid throughout. Mass spectrometric analysis was performed on a Finnigan LTQ ion-trap mass spectrometer (Thermo Finnigan) with the Shimadzu HPLC (10ATvp) coupled inline to an IonMAX electrospray ionization source. MS detector responses were obtained by integration under ion traces corresponding to m/z ranges for AL peptides. Synthetic isotopically labeled (+10 amu) peptide standard (21st Century Biochemicals) was used to confirm HPLC elution time and for quantitation of the AKSVIGTPEFMAPEMY and AKpSVIGTPEFMAPEMY (pS phosphorylated on serine corresponding to Ser308 in WNK3) peptides. The phosphopeptide standards gave volumes that differed by (∼10%), with the phosphopeptide slightly underrepresented. MS/MS spectra were acquired in a data dependent mode and analyzed using Mass Matrix and MASCOT software (Koenig et al., 2008; Xu et al., 2010).
Kinetic Models of Autophosphorylation Data
Autophosphorylation progress curves were modeled in DynaFit (Kuzmic, 2009; Kuzmic et al., 2009), as we have done previously (Humphreys et al., 2013). DynaFit performs nonlinear least squares regression analysis of progress curve data. Global minimization was achieved by a differential evolution of trial parameters. A very simple model (DynaFit script in Supplemental Table S2), uWNK + pWNK → pWNK + pWNK was found sufficient to fit the data with high coefficients of determination (R2).
Kinase-Glo ® reactions of pWNK3 peptide phosphorylation
Kinase-Glo assays were performed as described previously (Akella et al., 2021). Fifty μL reactions were comprised of 50 mM HEPES, pH 7.4, 5 mM MgCl2, 150 mM NaCl, 4 μM pWNK3, 400 μM OSR-tide (RVPGSSGRLHK; Biomatic Corp. Kitchener, Ontario), and 100 μM ATP. Time zero reactions established maximum luminescence. Assays were started by ATP addition, then aliquoted into tubes that were incubated either on the benchtop or under 190 kPa (∼28 PSI) nitrogen in an Amicon pressure chamber. Assays were stopped at specified times by the addition of 30 μL Kinase-Glo® with Amicon pressure momentarily disrupted for tube removal. All stopped reactions were transferred to a 96-well plate and centrifuged, then luminescence was read on a CLARIOstar plate reader (BMG Labtech, Ortenberg, Germany). The data were processed using MARS data analysis software (BMG Labtech).
Gel filtration and SEC-MALS methods
The uWNK3 was concentrated in spin columns (Millipore, Germany) to 5 mg/mL. Gel filtration was conducted under gravity using Hi Load SuperdexTM 75 (GE Healthcare) resin packed in a 20 mm × 240 mm column (25 mL bed volume). The column void volume, measured with Blue Dextran 2000 (Sigma-Aldrich), was 4.5 mL. The column was calibrated using standards from the GE Healthcare calibration kit (albumin [67 kDa] and ovalbumin [43 kDa]). 0.4 mL of 5 mg/mL uWNK3 was eluted at 0.2 mL/min. Gel filtration of uWNK3 and pWNK3 was conducted at a back pressure of 290 kPa using a Hi LoadTM 16/600 SuperdexTM -75 column with pressure applied by an AKTA FPLC (GE Healthcare).
SEC-MALS was measured with a Wyatt TREOS II light scattering detector in line with a Wyatt Optilab t-REX differential refractive index detector. Before SEC-MALS analysis, 250 mL of 5 mg/mL uWNK3 was centrifuged through a 0.22 μm filter. The SEC was performed on a SephadexTM 200 10/300 mm column at a back pressure of 250 kPa (flow rate 0.5 mL/min). The instrumentation was calibrated with BSA. Integration of the differential-refractive index signal gave a starting concentration of 2.4 mg/mL uWNK3. MALS data were analyzed in ASTRA 7.3.0.11 (Wyatt Technologies).
AUC Methods
Sedimentation velocity experiments were conducted in an Optima XL-I ultracentrifuge (Beckman-Coulter, Indianapolis, IN). 400 μL of uWNK3 at two concentrations (2.1 mM and 7.1 mM, calculated post hoc from the c(s) distribution described below) in a buffer containing 50 mM HEPES pH 7.4 and 150 mM NaCl were introduced into these cells with buffer alone in reference cells. The An50-Ti rotor with samples was incubated under vacuum at 20°C for 2.5 h before centrifugation at 50,000 rpm for 16 h. Absorbance data at 280 nm were analyzed using the continuous sedimentation coefficient distribution c(s) methodology in SEDFIT (Schuck, 2000; Schuck et al., 2002). SEDNTERP (Laue et al., 1992) was used to estimate the partial-specific volume, density and viscosity based on the protein sequence and the buffer conditions. The s-range was 0.2 to 10s in 100 steps. Time-invariant noise modeling, and a regularization confidence level of 0.68 were used. Figures were rendered in GUSSI (Brautigam, 2015) and DataGraph (Visual Data Tools, Inc., Chapel Hill NC).
NMR Methods
NMR spectra were acquired at 37°C on an Agilent DD2 spectrometer operating at 800 MHz. The [WNK3], either pWNK3 or uWNK3, was 50 μM (based on the monomer MW of 39.3 kDa). Pressure was introduced using N2 gas in a valved-NMR pressure tube (Norell, N.C. Cat. No. S-5-600-MW-IPB-8). NMR spectra were processed with NMRPipe (Delaglio et al., 1995) and analyzed with NMRview (Johnson, 2004) was used to superimpose and scale spectra.
DSS crosslinking methods
DSS crosslinks-lysine residues (Rappsilber, 2011). Samples were unpressurized or pressurized at 190 kPa in the Amicon concentrator/N2 chamber. Reaction solutions contained 4μM WNK3-KDm, 150 mM NaCl, 10 mM HEPES pH 8.0 and 1 mM DSS. 25 μL reactions were run for 20 mins at 25°C. Reactions were quenched to 25 mM Tris-HCl, pH 7.4. The samples were treated with 6M guanidine-HCl, to a final concentration of 1M and then proteolyzed with chymotrypsin overnight. Peptides were observed by mass spectrometry on an Orbitrap Fusion Lumos (Thermo Finnigan). Crosslinks were identified using the xQuest/xProphet software (Leitner et al., 2010). Only crosslinks with a score above 3 (from xQuest) were considered. DSS crosslinks were depicted as a function of sequence in xVis (Grimm et al., 2015).
Supplementary Material
Acknowledgments
We thank the American Heart Association (14GRNT20500035 [EJG] and 16CSA28530002 [EJG and ARR]), NIH DK110358 (ARR and EJG), CPRIT RP190421 (EJG), Mary Kay Ash Charitable Foundation International Postdoctoral Scholars in Cancer Research Fellowship (LRT), and the Welch Foundation I-2100-20220331 (EJG) and I1243 (MHC) for support. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. We thank Julian Dow for the c42-GAL4 fly stock. We thank Steven McKnight for discussion of this work.
Abbreviations used:
- AUC
analytical ultra-centrifugation
- CCC
cation chloride cotransporter
- DSS
disuccinylsuberate
- GST
glutathione S-transferase
- HEPES
hydroxyethyl piperazine ethanesulfonic acid
- KCC
K + Cl – co-transporter
- LC/MS
liquid chromatography/mass spectrometry
- NCC
Na + Cl – co-transporter
- NKCC
Na + K + 2Cl – co-transporter
- OSR1/SPAK
oxidative stress responsive 1/Ste20-related proline/alanine-rich kinase
- SEC
size exclusion chromatography
- SEC-MALS
size exclusion chromatography multiangle light scattering
- SLS
static light scattering
- TCEP
Tris(2-carboxyethyl)phosphine
- TROSY-HSQC
Transverse relaxation optimized spectroscopy- heteronuclear single-quantum correlation spectroscopy
- WNK
With No Lysine (K).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E23-03-0113) on August 16, 2023.
REFERENCES
- Akella R, Humphreys JM, Sekulski K, He H, Durbacz M, Chakravarthy S, Liwocha J, Mohammed ZJ, Brautigam CA, Goldsmith EJ (2021). Osmosensing by WNK Kinases. Mol Biol Cell 32, 1614–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselmo AN, Earnest S, Chen W, Juang YC, Kim SC, Zhao YM, Cobb MH (2006). WNK1 and OSR1 regulate the Na+, K+, 2Cl(–) cotransporter in HeLa cells. Proc Natl Acad Sci USA 103, 10883–10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergaya S, Faure S, Baudrie V, Rio M, Escoubet B, Bonnin P, Henrion D, Loirand G, Achard JM, Jeunemaitre X, Hadchouel J (2011). WNK1 regulates vasoconstriction and blood pressure response to alpha 1-adrenergic stimulation in mice. Hypertension 58, 439–445. [DOI] [PubMed] [Google Scholar]
- Boyd-Shiwarski CR, Shiwarski DJ, Griffiths SE, Beacham RT, Norrell L, Morrison DE, Wang J, Mann J, Tennant W, Anderson EN, et al. (2022). WNK kinases sense molecular crowding and rescue cell volume via phase separation. Cell 185, 4488–4506 e4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam CA (2015). Calculations and Publication-Quality Illustrations for Analytical Ultracentrifugation Data. Methods Enzymol 562, 109–133. [DOI] [PubMed] [Google Scholar]
- Carugo O (2017). Protein hydration: Investigation of globular protein crystal structures. Int J Biol Macromol 99, 160–165. [DOI] [PubMed] [Google Scholar]
- Choe KP, Strange K (2007). Molecular and genetic characterization of osmosensing and signal transduction in the nematode Caenorhabditis elegans. FEBS J 274, 5782–5789. [DOI] [PubMed] [Google Scholar]
- Collins MD, Kim CU, Gruner SM (2011). High-pressure protein crystallography and NMR to explore protein conformations. Annu Rev Biophys 40, 81–98. [DOI] [PubMed] [Google Scholar]
- Coste B, Mathur J, Schmidt M, Earley TJ, Ranade S, Petrus MJ, Dubin AE, Patapoutian A (2010). Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995). NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Dellarole M, Royer CA (2014). High-pressure fluorescence applications. In: Fluorescence Spectroscopy and Microscopy, New Jersey, NJ: Springer, 53–74. [DOI] [PubMed] [Google Scholar]
- Dow JA, Maddrell SH, Gortz A, Skaer NJ, Brogan S, Kaiser K (1994). The malpighian tubules of Drosophila melanogaster: A novel phenotype for studies of fluid secretion and its control. J Exp Biol 197, 421–428. [DOI] [PubMed] [Google Scholar]
- Farfel Z, Iaina A, Rosenthal T, Waks U, Shibolet S, Gafni J (1978). Familial hyperpotassemia and hypertension accompanied by normal plasma aldosterone levels: Possible hereditary cell membrane defect. Arch Intern Med 138, 1828–1832. [PubMed] [Google Scholar]
- Grimm M, Zimniak T, Kahraman A, Herzog F (2015). xVis: A web server for the schematic visualization and interpretation of crosslink-derived spatial restraints. Nucleic Acids Res 43, W362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys JM, Piala AT, Akella R, He H, Goldsmith EJ (2013). Precisely ordered phosphorylation reactions in the p38 mitogen-activated protein (MAP) kinase cascade. J Biol Chem 288, 23322–23330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaykumar AB, Jung JU, Parida PK, Dang TT, Wichaidit C, Kannangara AR, Earnest S, Goldsmith EJ, Pearson GW, Malladi S, Cobb MH (2021). WNK1 Enhances Migration and Invasion in Breast Cancer Models. Mol Cancer Ther 20, 1800–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Xie J, Yeh CW, Chen JC, Cheng CJ, Lien CC, Huang CL (2023). WNK1 promotes water homeostasis by acting as a central osmolality sensor for arginine vasopressin release. J Clin Invest 133, e164222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA (2004). Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol 278, 313–352. [DOI] [PubMed] [Google Scholar]
- Koenig T, Menze BH, Kirchner M, Monigatti F, Parker KC, Patterson T, Steen JJ, Hamprecht FA, Steen H (2008). Robust prediction of the MASCOT score for an improved quality assessment in mass spectrometric proteomics. J Proteome Res 7, 3708–3717. [DOI] [PubMed] [Google Scholar]
- Kuzmic P (2009). DynaFit–a software package for enzymology. Methods Enzymol 467, 247–280. [DOI] [PubMed] [Google Scholar]
- Kuzmic P, Lorenz T, Reinstein J (2009). Analysis of residuals from enzyme kinetic and protein folding experiments in the presence of correlated experimental noise. Anal Biochem 395, 1–7. [DOI] [PubMed] [Google Scholar]
- Laue TM, Shah BD, Ridgeway TM, Pelletier SL (1992). Sednterp. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science, ed. Harding, S.E., Row, A.J., Horton, J.C., Cambridge, UK: The Royal Society of Chemistry, 90–125. [Google Scholar]
- Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R (2010). Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteomics 9, 1634–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenertz LY, Lee BH, Min X, Xu BE, Wedin K, Earnest S, Goldsmith EJ, Cobb MH (2005). Properties of WNK1 and implications for other family members. J Biol Chem 280, 26653–26658. [DOI] [PubMed] [Google Scholar]
- Liu S, Tao R, Wang M, Tian J, Genin GM, Lu TJ, Xu F (2019). Regulation of Cell Behavior by Hydrostatic Pressure. Appl Mech Rev 71, 0408031–4080313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min XS, Lee BH, Cobb MH, Goldsmith EJ (2004). Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure 12, 1303–1311. [DOI] [PubMed] [Google Scholar]
- Parker JC (1993). In defense of cell volume? Am J Physiol 265, C1191–1200. [DOI] [PubMed] [Google Scholar]
- Piala AT, Moon TM, Akella R, He H, Cobb MH, Goldsmith EJ (2014). Chloride sensing by WNK1 involves inhibition of autophosphorylation. Sci Signal 7, ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechotta K, Lu J, Delpire E (2002). Cation chloride cotransporters interact with the stress-related kinases Ste20-related proline-alanine-rich kinase (SPAK) and oxidative stress response 1 (OSR1). J Biol Chem 277, 50812–50819. [DOI] [PubMed] [Google Scholar]
- Pleinis JM, Norrell L, Akella R, Humphreys JM, He H, Sun Q, Zhang F, Sosa-Pagan J, Morrison DE, Schellinger JN, et al. (2021). WNKs are potassium-sensitive kinases. Am J Physiol Cell Physiol 320, C703–C721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racker E (1991). Use of synthetic amino acid polymers for assay of protein-tyrosine and protein-serine kinases. Methods Enzymol 200, 107–111. [DOI] [PubMed] [Google Scholar]
- Rappsilber J (2011). The beginning of a beautiful friendship: Cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J Struct Biol 173, 530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C, Alessi DR (2008). The regulation of salt transport and blood pressure by the WNK-SPAK/OSR1 signalling pathway. J Cell Sci 121, 3293–3304. [DOI] [PubMed] [Google Scholar]
- Richardson C, Rafiqi FH, Karlsson HKR, Moleleki N, Vandewalle A, Campbell DG, Morrice NA, Alessi DR (2008). Activation of the thiazide-sensitive Na+-Cl– cotransporter by the WNK-regulated kinases SPAK and OSR1. J Cell Sci 121, 675–684. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, Kannangara A, Chlebowicz J, Akella R, He H, Tambar UK, Goldsmith EJ (2022). Synthesis and Structural Characterization of Novel Trihalo-sulfone Inhibitors of WNK1. ACS Med Chem Lett 13, 1678–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosay P, Davies SA, Yu Y, Sozen MA, Kaiser K, Dow JA (1997). Cell-type specific calcium signalling in a Drosophila epithelium. J Cell Sci 110, 1683–1692. [DOI] [PubMed] [Google Scholar]
- Russell JM (2000). Sodium-potassium-chloride cotransport. Physiol Rev 80, 211–276. [DOI] [PubMed] [Google Scholar]
- Schuck P (2000). Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J 78, 1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D (2002). Size-distribution analysis of proteins by analytical ultracentrifugation: Strategies and application to model systems. Biophys J 82, 1096–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Tu SW, Wedin K, Earnest S, Stippec S, Luby-Phelps K, Cobb MH (2012). Interactions with WNK (with no lysine) family members regulate oxidative stress response 1 and ion co-transporter activity. J Biol Chem 287, 37868–37879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenesen D, Moehlman AT, Schellinger JN, Rodan AR, Kramer H (2019). The glial sodium-potassium-2-chloride cotransporter is required for synaptic transmission in the Drosophila visual system. Sci Rep 9, 2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens FJ (1989). Analysis of protein-protein interaction by simulation of small-zone size exclusion chromatography. Stochastic formulation of kinetic rate contributions to observed high-performance liquid chromatography elution characteristics. Biophys J 55, 1155–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Wu Y, Jonusaite S, Pleinis JM, Humphreys JM, He H, Schellinger JN, Akella R, Stenesen D, Kramer H, et al. (2018). Intracellular Chloride and Scaffold Protein Mo25 Cooperatively Regulate Transepithelial Ion Transport through WNK Signaling in the Malpighian Tubule. J Am Soc Nephrol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Arthanari H, Shimada I, Wagner G (2015). Nitrogen detected TROSY at high field yields high resolution and sensitivity for protein NMR. J Biomol NMR 63, 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CA IV, An SW, Kankanamalage SG, Stippec S, Earnest S, Trivedi AT, Yang JZ, Mirzaei H, Huang CL, Cobb MH (2018). OSR1 regulates a subset of inward rectifier potassium channels via a binding motif variant. Proc Natl Acad Sci USA 115, 3840–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter MM (1984). Water structure of a hydrophobic protein at atomic resolution: Pentagon rings of water molecules in crystals of crambin. Proc Natl Acad Sci USA 81, 6014–6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin MD, Tomaszewski M, Braund PS, Hajat C, Raleigh SM, Palmer TM, Caulfield M, Burton PR, Samani NJ (2008). Common variants in genes underlying monogenic hypertension and hypotension and blood pressure in the general population. Hypertension 51, 1658–1664. [DOI] [PubMed] [Google Scholar]
- Vitari AC, Deak M, Morrice NA, Alessi DR (2005). The WNK1 and WNK4 protein kinases that are mutated in Gordon’s hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem J 391, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkers L, Mechioukhi Y, Coste B (2015). Piezo channels: from structure to function. Pflugers Arch 467, 95–99. [DOI] [PubMed] [Google Scholar]
- Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, et al. (2001). Human hypertension caused by mutations in WNK kinases. Science 293, 1107–1112. [DOI] [PubMed] [Google Scholar]
- Wu Y, Schellinger JN, Huang CL, Rodan AR (2014). Hypotonicity stimulates potassium flux through the WNK-SPAK/OSR1 kinase cascade and the Ncc69 sodium-potassium-2-chloride cotransporter in the Drosophila renal tubule. J Biol Chem 289, 26131–26142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, Cobb MH (2000). WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem 275, 16795–16801. [DOI] [PubMed] [Google Scholar]
- Xu BE, Min XS, Stippec S, Lee BH, Goldsmith EJ, Cobb MH (2002). Regulation of WNK1 by an autoinhibitory domain and autophosphorylation. J Biol Chem 277, 48456–48462. [DOI] [PubMed] [Google Scholar]
- Xu H, Hsu PH, Zhang L, Tsai MD, Freitas MA (2010). Database search algorithm for identification of intact cross-links in proteins and peptides using tandem mass spectrometry. J Proteome Res 9, 3384–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Park HM, Rigel DF, DiPetrillo K, Whalen EJ, Anisowicz A, Beil M, Berstler J, Brocklehurst CE, Burdick DA, et al. (2016). Small-molecule WNK inhibition regulates cardiovascular and renal function. Nat Chem Biol 12, 896–898. [DOI] [PubMed] [Google Scholar]
- Zagorska A, Pozo-Guisado E, Boudeau J, Vitari AC, Rafiqi FH, Thastrup J, Deak M, Campbell DG, Morrice NA, Prescott AR, Alessi DR (2007). Regulation of activity and localization of the WNK1 protein kinase by hyperosmotic stress. J Cell Biol 176, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz BP, Abuin A, Ramirez-Solis R, Richter LJ, Piggott J, Person C, Sands AT (2003). Wnk1 kinase deficiency lowers blood pressure in mice: A gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci USA 100, 14109–14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng WZ, Marshall KL, Min S, Daou I, Chapleau MW, Abboud FM, Liberles SD, Patapoutian A (2018). PIEZOs mediate neuronal sensing of blood pressure and the baroreceptor reflex. Science 362, 464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Zhang ZY (1999). Mechanism of mitogen-activated protein kinase phosphatase-3 activation by ERK2. J Biol Chem 274, 35526–35534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.