Abstract
A clinical incident is typically manifested by several molecular events; therefore, it seems logical that a successful diagnosis, prognosis, or stratification of a clinical landmark require multiple biomarkers. In this report, we presented a machine learning pipeline, namely “Biomarker discovery process at binomial decision point” (2BDP) that took an integrative approach in systematically curating independent variables (e.g., multiple molecular markers) to explain an output variable (e.g., clinical landmark) of binary in nature. In a logical sequence, 2BDP includes feature selection, unsupervised model development and cross validation. In the present work, the efficiency of 2BDP was demonstrated by finding three biomarker panels that independently explained three stages of Alzheimer’s disease (AD) marked as Braak stages I, II and III, respectively. We designed three assortments from the entire cohort based on these Braak stages; subsequently, each assortment was split into two populations at Braak score I, II or III. 2BDP systematically integrated random forest and logistic regression fitting model to find biomarker panels with minimum features that explained these three assortments, e.g., significantly differentiated two populations segregated by Braak stage I, II or III, respectively. Thereafter, the efficacies of these panels were measured by the area under the curve (AUC) values of the receiver operating characteristic (ROC) plot. The AUC-ROC was calculated by two cross-validation methods. Final set of gene markers was a mix of novel and a priori established AD signatures. These markers were weighted by unique coefficients and linearly connected in a group of 2–10 to explain Braak stage I, II or III by AUC ≥ 0.8. Small sample size and a lack of distinctly recruited Training and Test sets were the limitations of the present undertaking; yet 2BDP demonstrated its capability to curate a panel of optimum numbers of biomarkers to describe the outcome variable with high efficacy.
Keywords: Algorithm pipeline, Artificial intelligence, Machine learning, Biomarker discovery, Logistic Regression Model, Alzheimer's Disease, Braak Stages, Diagnostic biomarker
Graphical Abstract
1. Introduction
A biomarker is defined as “an indicator of normal biological processes, pathogenic processes, or responses to an exposure or intervention, including therapeutic interventions” [1], [2], [3]. The biomarkers’ efficacy in indicating disease pathophysiology depends on the current advancements in assay and analysis capabilities. In particular, the recent advents of high throughput technologies have provided unprecedented resolution in the evaluating the host’s molecular, histologic, radiographic and physiologic underpinnings linked to disease pathophysiology [3]. Thereby, the overall performances of biomarkers have improved.
Improved aptitude for rapid handling of prospective and longitudinal samples extended an evidence-based support to the premise that every stage of disease pathophysiology is associated with multiple molecules with various degrees of involvements [4], [5]. Hence, an early marker of disease, which is by definition linked to the immediate effects of disease onset [2], [6] could be characteristically different from the late marker of disease, which is linked to the final phase of disease pathophysiology [6]. Likewise, multiple, and partially exclusive sets of molecules are expected to be associated with different degrees of disease severity. Recent trends of biomarker discovery are driven by the desire to identify signatures linked to certain clinical landmarks, such as the precise stratification of illness, the immediate vs. delayed characteristics of disease onset or acute vs. chronic phases of illness [3], [7], [8], [9], [10]. Early stratification, diagnostic or prognostic markers are of high demand in therapeutic studies since such tools can offer a golden time window to jumpstart the treatment.
In this context, we developed an analytical pipeline named Biomarker Discovery Process at Binomial Decision Point (2BDP), which is trained to identify the biomarkers associated with user-defined clinical landmarks, which could be binary or dichotomous in character. To explain further, a user-defined clinical landmark could be well-defined biological actions or clinical symptoms, such as the time since the disease onset (e.g., early vs, late stage of illness), dose of the drug or stimulants (e.g., lethal vs. sub-lethal dose) or any disease stage (e.g., mild vs. moderate state of illness). 2BDP presents a stepwise integrative pipeline that can guide the users starting from feature selection and performance-based ranking using Random Forest, an algorithm with proven success [11], [12], [13]. Logistic Regression model, a machine learning (ML) algorithm with strong theoretical background and well-understood assumptions was used to model these selected features to best explain the clinical variable. In this respect, we also considered Support vector machines (SVM) [14], and rather complex the Neuronal Network (NN) [15]; we finally selected Logistic Regression model for our purpose due its implementation simplicity, wide applicability and successful history [16], [17], [18]. Majority of comparative studies reported similar efficacies among these algorithms, namely Logistic Regression model, SVM and NN [19], [20], [21]. Applying the method described by Sullivan et al. [22], the coefficients of the logistic regression model generate risk scores associated with each feature and sum to provide a total risk score. 2BDP adapts this template and integrated it with two cross-validation routines, namely k-fold [23], an established routine and a novel routine to explain a binary clinical landmark. Overall, 2BDP presents a novel pipeline that takes a comprehensive approach for biomarker discovery from a high throughput data set to deliver panel of biomarkers with customizable feature counts. Supplementary section presents a table to compare 2BDP with other available routines and algorithms.
To demonstrate the performance of the present algorithm, we selected a study that investigated brain tissue samples collected from subjects suffering from Alzheimer’s disease (AD) of gradually increasing severity [24]. Here the user-defined clinical landmarks were the AD’s severity as stepwise graded by the Braak scale starting from 0 or no AD to VI or maximum severity [25], [26]. To note, we selected the work of Marttinen, M. et al. [24] following a curation of GEO database and this process is described in the Supplementary data.
Present tenet is that a successful model should be able to explain an outcome variable or a used-defined clinical landmark by a set of independent variables or molecular entities. In the present AD study design, the set of independent variables and used-defined clinical landmarks were the differentially expressed brain transcripts from the AD patients [24] and the Braak stages of AD [6], [24], respectively. Our objective was to identify those independent variables or transcriptomic signatures that were shifted in correlation with different Braak stages. To provide the best parametric model for 2BDP, the outcome variables was manipulated to become binary or dichotomous in nature. Hence, we selected each of the Braak stages, namely I, II or II as the cut-off marks to hypothetically separate the AD cohorts into either above or below any given cutoffs. For instance, the first cohort named as assortment 1 used Braak stage I as the singular cutoff. The cohort with Braak scores equal or less than I was named Under the Cutoff (UCo) population or UCo≤Braak_I and remaining cohort was considered Above the Cutoff (ACo) population or ACo≤Braak_I. Likewise, the second cohort named as assortment 2 used Braak stage II as the singular cutoff. The cohort with Braak scores equal or less than II was named UCo≤Braak_II and remaining cohort was considered ACo≤Braak_II. Similarly, the third cohort named as assortment 3 used Braak stage III as the singular cutoff. The cohort with Braak scores equal or less than III was named UCo≤Braak_III population and remaining cohort was considered ACo≤Braak_III. The goal was to identify a minimum number of transcriptomic markers that can differentiate UCo from ACo in each of the three assortments. It is also to note that these three Braak stages have significant biological implications [24]. Braak stage II is the clinical landmark of AD onset, where neuronal loss co-occurs with a surge of inflammation. The subclinical threshold or Braak I could be an appropriate instant to detect early AD markers, where the mitochondrial dysfunction begins. Braak stage III marks the post-threshold landmark, where extracellular matrix organization and inflammation surge meet their peaks [24].
The selection of the clinical landmark, and therefore the choice of parametric model is the primary criterion of any biomarker selection processes, including 2BDP. In addition, there are few secondary criteria to take in account. For instance, the capabilities or limitation of the downstream tool or technology to detect the biomarkers often governs the biomarker selection process. These secondary criteria deem further clarification. The molecular biomarkers, particularly transcriptomic biomarkers are fast becoming the preferred type of target to interrogate [27] and its reasons are manifold. Modern day’s high throughput arrays, Next Gen sequencing and quantitative polymerized chain reaction technology (qPCR) provide time-efficient, reproducible, and statistically powerful tools to screen the transcripts, which gives the scientists an unprecedented opportunity to down select high performing multi-gene biomarker panel from a reasonably large pool of putative candidates. Above mentioned multi-gene detection platforms are emerging as popular in vitro diagnostic tools [28], [29], [30], [31], [32] for multiple reasons including its highly sensitive and specific detection capability and relatively easy and hands-free operations. Nevertheless, biomarker detection processes are bound to select a certain number of target probes to maximize the detection capability of the platform of interest. For instance, qPCR-based platforms, which are gaining much tractions [33] due to its small footprint yet high sensitivity and specificity present a limited throughput capability; therefore this technology prefers a small number of transcripts to probe [34], [35]. These technological restrictions also play major role in determining the size of the biomarker panel.
Hence, a biomarker selection process needs to find an optimum solution. The constraints of the preferred detection technology should be accounted before down selecting the independent variables and the fitting model should aim to identify a realistic number of biomarkers. Our algorithm pipeline, 2BDP can optimize such constraints in the process to deliver most fitting model.
2. Materials and methods
We processed a dataset from public domain that was reported by Marttinen et al. [24]. Briefly, the authors collected 71 autopsied tissue samples from the temporal cortex of individuals with varying degrees of AD-related neurofibrillary pathology, and 60 samples of this entire cohort were used for the transcriptomic study, which was the focus of present work. These 60 samples were sorted based on the severity of AD-related neurofibrillary pathology, which was scored by Braak stages (Braak 0-VI). Transcriptomic analysis of the autopsied tissues was performed on the expression array (Agilent 8x60K Custom Exon array) [36]. Differential gene expression analysis across the seven Braak stages were analyzed using the ANOVA for Braak stage specific changes with the cut-off at Benjamini-Hochberg False Discovery Rate (FDR), and p-FDR < 0.05 [24].
2.1. Gene expression and sample data
Gene expression analysis was carried out by Marttinen et al. [24]. Microarray data were obtained from the NCBI’s GEO Expression Omnibus database series GSE106241 using the GEO Query package from Bioconductor [37]. The data were log2 transformed and quantile-normalized using limma [38], [39]. Samples were binned based on their annotated Braak score as a threshold. Braak scores were obtained from the sample metadata on GEO.
2.2. Outcome variables
We evaluated 2BDP’s performance in justifying the cohort’s AD severity as defined by Braak stages using a sliding window of cutoffs of I, II and III, respectively. These 60 samples were rearranged based on the relative grading on the Braak scale; hence three assortments were created. Each assortment had its unique set of UCo≤ Ω and ACo> Ω, where Ω is Braak I or II or II.
Assortment 1
By setting cut-off at Braak I, we binned all the samples at Braak stage of 0 and I under UCo≤Braak_I, and rest of the samples at Braak stage of II-V as ACo>Braak_I. The outcome variable of assortment 1 was to differentiate UCo≤Braak_I from ACo>Braak_I.
Assortment 2
By setting Braak II as the cut-off, all samples at Braak stage of 0, I, and II were binned under UCo≤Braak_II, while rest of the samples were binned under ACo>Braak_II. The outcome variable of assortment 2 was to differentiate UCo≤Braak_II from ACo>Braak_II.
Assortment 3
By setting Braak III as the cut-off, all samples at Braak stage of 0, I, II and III were binned under UCo≤Braak_III, while rest of the samples were binned under ACo>Braak_III. The outcome variable of assortment 3 was to differentiate UCo≤Braak_III from ACo>Braak_III.
2.3. Feature selection and model construction
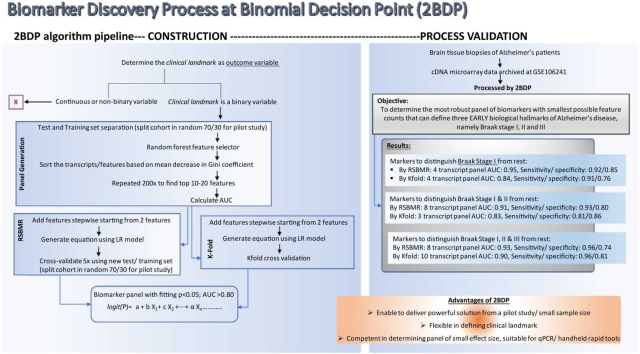
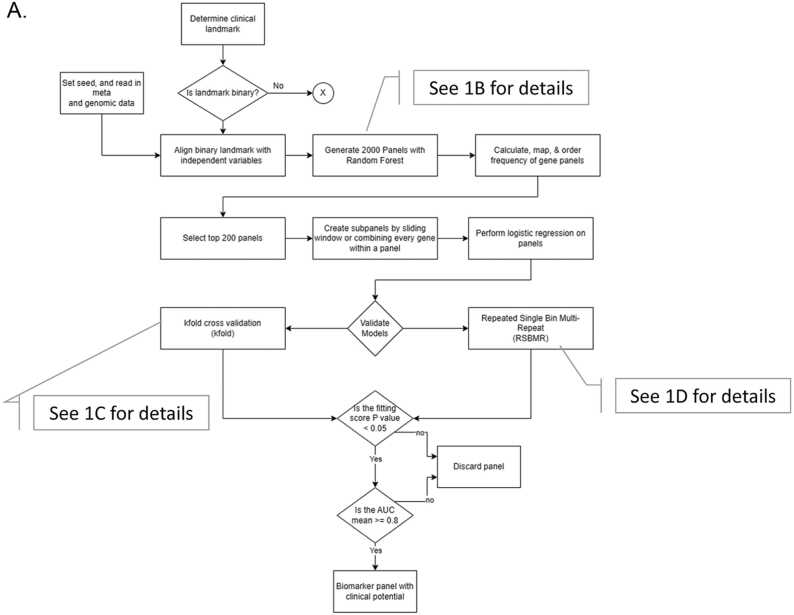
Marttinen et al. [24] probed a total of ∼30,000 gene transcripts to find 19,367 genes as differently expressed by the Braak Stages, and 2BDP pipeline used these genes as the independent features (Fig. 1). Following pipeline operated independently within each of the three assortments. The script of this work is documented in the supplementary materials.
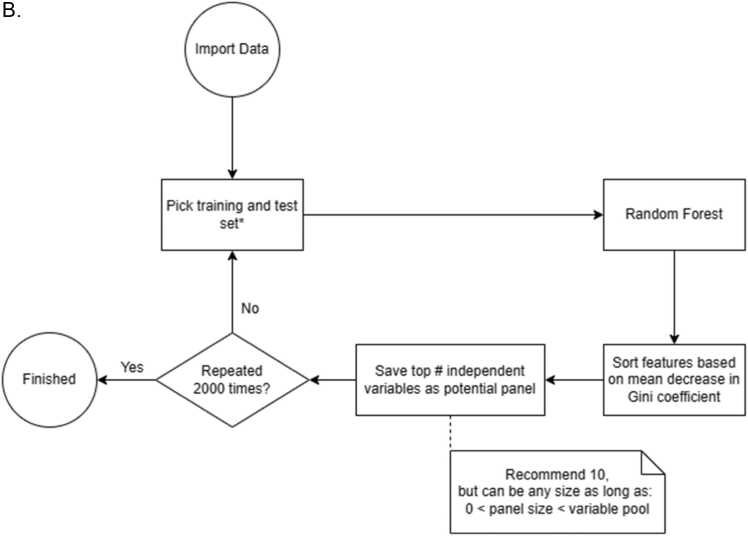
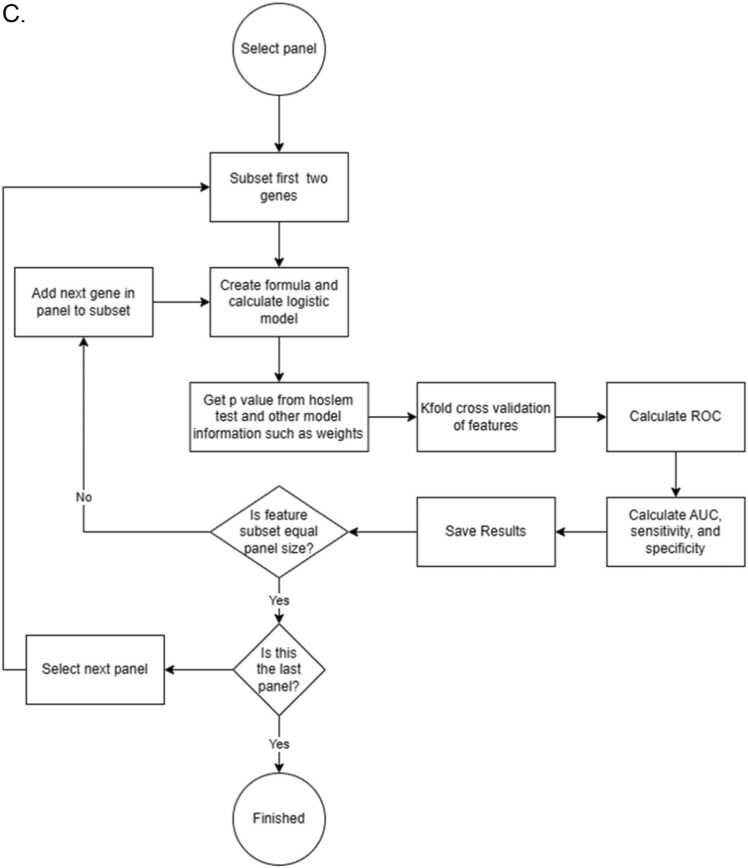
Fig. 1.
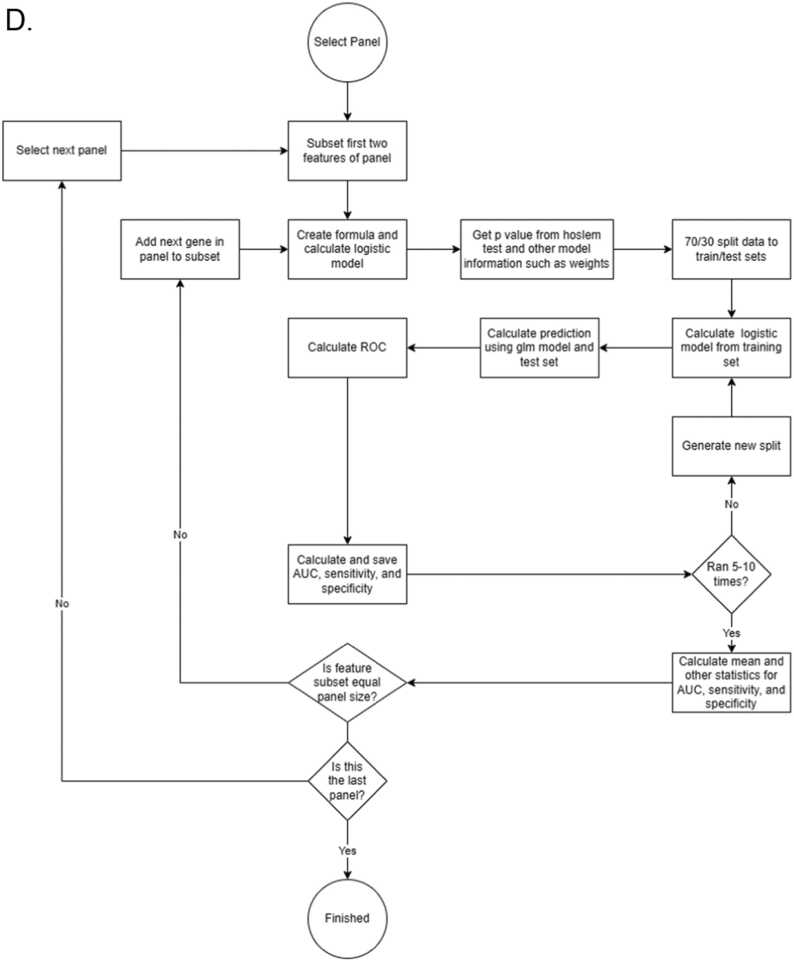
Flow chart showing the decision tree of 2BDP algorithm. (A) Complete flow diagram (B) Operational workflow to crease 2000 panels by Random Forest. (C) K-fold cross validation pipeline and (D) RSBMR cross validation pipeline. X: discard this path. AUC: Area under curve. ROC: receiver operating characteristic; RSBMR: Random Single Bin Multiple Repeats.
2.3.1. Feature ranking
Whole cohort was randomly divided into Training and Test set by 70:30 ratio. Fig. 2 displayed the cohort size at each Braak stage. For instance, there were 17 samples Braak Stage ≤ 1, which were segregated into 12 Training and 5 Test samples. Likewise, remaining 43 samples with Braak stage >1 were segregated into 31 Training and 12 Test samples. Now, total sizes of Training and Test sets became 43 and 17, respectively. Computed on the Training set, Random Forest curated high performing feature enabled to segregate the samples at Braak stage cutoff 1 and validated their performances in the Test set. Random Forest was used as our ML algorithm as it handles classification and regression problems. The independent features were ranked in the descending order of their Gini scores, top ten features were noted, and this Training and Test was discarded. A fresh randomization recreated Training and Test set split in 70:30, and new Gini scores of the top ten high performing independent features were calculated. This iteration was continued 2000 times. Deliverables include 10 independent features sorted by their descending Gini score as reported by each of 2000 iterations.
Fig. 2.
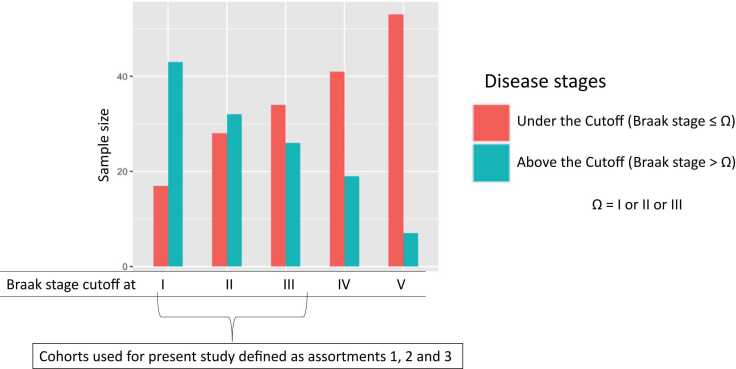
Bar chart showing the sample size distribution between Under the Cutoff (UCo) and Above the Cutoff (ACo) populations as determined by individual Braak stages. Thereby five sets of cohorts are shown here as defined by Braak stages I to V. For instances, the first set of UCo and ACo was differentiated by Braak stage I; UCo≤Braak_I (red colored bar) included those, which were graded 0 (i.e. no Alzheimer’s disease) and I; and ACo≤Braak_I (green colored bar) included those, which were graded from II to VI, and so on. Among these five sets of cohorts, first three sets were named as assortment 1, 2 and 3, respectively and used in this study. These groups were selected for two primary reasons. (a) Sample sizes of UCo and ACo became non-comparable beyond Braak stage III, so we preclude the sets determined by Braak stage IV and V. (b) Although, the sample sizes between ACo and UCo was not comparable at Braak Stage I, we still investigated these two sets, because the diagnosis at the sub-clinical level can enable the caregivers an extra time to intervene.
2.3.2. Feature selection
Objective of the next pipeline was to curate 200 independent feature panels that appeared most frequently in the previous set of deliverables, namely 10 features x 2000 iteration. The frequency of each unique feature was calculated from the summation of occurrences of a feature at the same position across 2000 panels. As logistic regression outputs are independent from the order of input variables, frequency by position was done to reduce duplicates of subset panels generated by sliding window over the 200 panels. The rationale behind this undertaking is the following: by calculating the frequency of a feature based on its position, the frequency of a feature is deflated and distributed across the positions. The intended result of this would allow more diverse panels to be discovered and reduce the duplicated number of subpanels created from every combination of features from a given panel. A panel’s rank was determined from the sum of calculated frequency of each feature within that panel. The top 200 deliverables from decreasing order by panel rank were selected as the potential list of biomarkers to determine the outcome variable of each of the three Assortments, as described above.
2.3.3. Cross-validation
In the next few steps, these biomarkers were linked by the logistic regression model as described in Eq. 1.
| (1) |
where logit() is the log odds function of a value, P is the probability of successful determination of outcome variable, a is the intercept of the equation, b through n are coefficient estimates of the independent variables, and X1 through Xn are the expression values of the transcript 1 to transcript n, respectively. The fitting criteria of these probe combinations were measured by multiple R2, adjusted R2 and p values (Chi-square).
This mathematical operation used to assess the efficacy of a biomarker panel in determining the outcome variables. The panel defined a group of independent features that are linearly associated with each other via unique weight factors or coefficients. This efficacy of individual panel was quantitively measured by the area under the curve (AUC) of receiver operating characteristic (ROC) curve. We used two methods to measure the ROC curve.
2.3.3.1. Random Single Bin Multiple Repeats (RSBMR)
Here the independent feature panels were stepwise added from 2 t to 10 to construct a series of unique subpanels. Hence, the maximum count of features in one panel should not exceed 10. Next, the whole cohort was randomly sorted into Training and Test set by 70:30 ratio. A unique panel of independent features was fitted onto the Training set to construct linear regression model as shown in Eq. 1. Afterwards, this linear regression model was operated on the Test set to calculate the AUC and sensitivity/ specificity. These deliverables were retained while this set of Training and Test set was discarded. This cycle was repeated 10 times for each subpanel and finally, the mean values of AUCs and sensitivity/ specificity calculated over these 10 iterations were reported. This iterative process continued until all 200 independent panels were exhausted.
2.3.3.2. k-fold method
The independent features were stepwise added from 1 to 10 to construct a series of unique panels. For any given unique panel, the entire cohort was segregated into 10 (k = 10) groups, and one randomly selected group was chosen as the Test set. Remaining groups were taken as Training set, where the unique panel was fitted to construct linear regression model as shown in Eq. 1. Next, the AUC was calculated using the Test set and duely reported. This iterative process continued until all possible combinations of the 200 independent features were exhausted.
3. Panel delivery
All panels curated by RSBMR and k-fold methods were screened to find those which had (a) significantly high fitting score, p < 0.05 and (b) AUC > 0.8.
3.1. Software
Training and Test set data partitions were generated using the “caret” package in R. Random forest models were generated using the caret package in R [40]. Logistic regression models were trained using the glm() function in base R for RSBMR and train() function in “caret” package for k-fold [27], [28]. Receiver operator curves were plotted using the pROC package in R [41]. Network analysis was carried out using Ingenuity Pathway Analysis (IPA), QIAGEN, Inc.
3.2. Parameters
The choices on the parameters for this algorithm were chosen based on a runtime of a few days. The parameters that we used herein are listed below, which should be considered as the guideline for future use:
(i) Size of seed:100 (To note, seed = 100 was used in the present run; however, this value only contributes to the reproducibility of results for a given run and can be any value allowed within R.).
(ii) Training: Test split ratio of cohort 70:30).
(iii) Metric: Accuracy (Note: However, “Kappa” is an acceptable option for metric as evaluation of selected classifiers are done separately from the random forest using AUC validation methods.).
(iv) Size of a panel for and selected from random forest: 10 * (please see its explanation in the trailing paragraph starting with *).
(v) Total trials of random forest: 2000 * (please see its explanation in the trailing paragraph starting with *).
(vi) Total panels to select: 200^ (please see its explanation in the trailing paragraph starting with ^).
(vii) AUC validation type: either k-fold or RSBMR‡ (please see its explanation in the trailing paragraph starting with ‡).
(viii) Number of repeats for RSBMR: 5‡ (please see its explanation in the trailing paragraph starting with ‡).
(ix) The train control for k-fold: method = “repeatedcv”, number = 10, repeats = 3‡ ((please see its explanation in the trailing paragraph starting with ‡).
*Explanation of Parameters iv and v- There are 2 cutoffs of features applied before, & after random forest. First is the cutoff of features used in the random forest, and second are the total amount of panels. The first cutoff should be less than the total features. The second cutoff is to limit the number of times random forest is ran. The recommend values for these cutoffs are 10 & 2000 for cutoffs 1 and 2 respectively. The first cutoff determines the size panel of a panel to generate and should be set to the maxed sized panel of features you are looking for. The second cutoff is the number of times random forest ran which affects frequency calculations of features. 2000 was selected as any more runs had little effect on the order of frequency of genes.
^Explanation of Parameter vi: The total panels to select after the frequency mapping is recommend at 200 with panels at size 10. With 200 10 gene panels, sliding window will expand the total number of panels to validate to 1800. The total panels to validate can be calculated by: Total panels x [sum(from 2 to the panel size)]. Increasing this limit resulted in more duplicated sub panels that were discarded with little increase in unique panels, while lowering this limit reduced both duplicated & unique panels. Its only recommended to increase this limit if the panel size of interest in approximately near the maxed panel size.
‡Explanation of Parameter vii-ix: Currently only one validation method can be selected per run, either RSBMR or k-fold. For RSBMR, 5 is the recommended minimum number of repeats; however, the repeats can be between 5 and 10 to prevent over or under reporting of AUC. This allows for a moderate calculation time while increasing the accuracy of the AUC score for a given panel. Under k-fold, the recommended values should be method = ”repeatedcv”, number = 10, repeats = 3. Like RSBMR, this produces a relatively more accurate AUC score within a moderate runtime. The method can be cv or repeatedcv, with the number between 5 and 10 to prevent over and under fitting of the data. The repeats allow for higher accuracy of the AUC calculation and is recommend between 1 and 3 for moderate runtimes.
3.3. Network Analysis
The genes of interest were uploaded to Ingenuity Pathway Analysis (IPA, QIAGEN, Inc.) for pathway analysis. We used human specific database for network building. The gene nodes were connected among themselves and those biofunctions, which are relevant to AD and met hypergeometric t-test p < 0.01.
4. Results
There were 60 samples in the study reported by Marttinen et al. [24]. Our objective was to screen these samples by 2BDP algorithm to determine the most robust panel of biomarkers with smallest possible feature count that can define three biological hallmarks of AD [24]. Here, we set up three assortments of the whole cohort that were defined by three Braak stage I, II or III, respectively; and we seek three sets of biomarker panels that can differentiate the following three pairs. Assortment 1: UCo≤Braak_I, vs. ACo>Braak_I; there were 17 and 43 samples in UCo≤Braak_I and ACo≥Braak_I, respectively. Assortment 2: UCo≤Braak_II vs. ACo>Braak_II; there were 28 and 32 samples in UCo≤Braak_II and ACo ≥Braak_II, respectively. Assortment 3: UCo≤Braak_III vs. ACo>Braak_III. there were 34 and 26 samples in UCo≤Braak_III and ACo≥Braak_III, respectively.
4.1. Biomarker panels for assortment 1
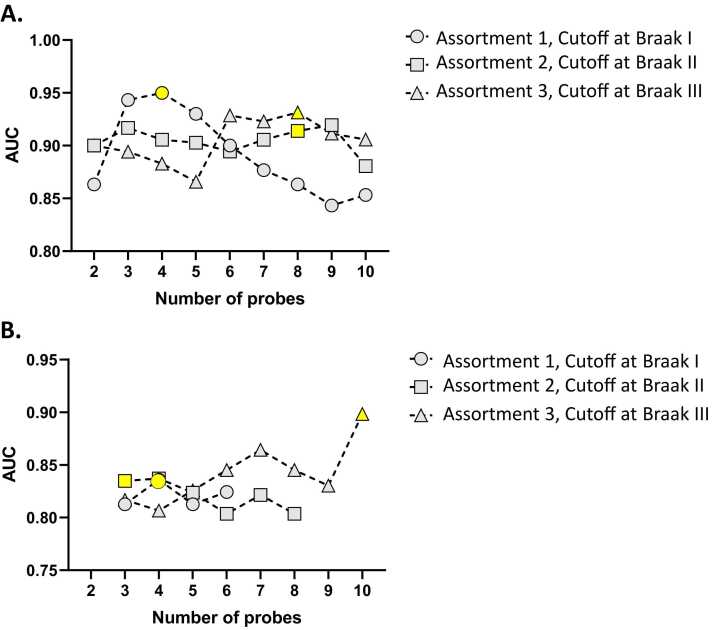
RSBMR and k-fold method separately reported putative panels of biomarkers linked to assortment 1. Table S1 listed those biomarker panels, which were curated by RSBMR. There were 159 putative panels that significantly well-fitted in a linear regression model (p < 0.0001) and can potentially distinguish UCo≤Braak_I, from ACo>Braak_I with AUC ≥ 0.8. In Fig. 3A, the circles with dotted line showed the maximum AUC scored by the biomarker panels with increasing feature count from 2 to 10. Maximum AUC was achieved by a 4-gene panel that was yellow highlighted in Fig. 3. This panel was marked as most potential candidate and reported in Table 1A. This panel included four genes, namely CRH, DLEC1, GSTT1 and HBD; together this panel scored mean AUC = 0.95, mean Sensitivity = 0.92 and mean Specificity = 0.85.
Fig. 3.
Distribution of AUC values generated by 2BDP for highest performing biomarker panels with increasing feature count. Here, the line curves stood for three assortments: assortment 1 (UCo≤Braak_I vs. ACo≤Braak_I) denoted by circles with dotted lines; assortment 2 (UCo≤Braak_II vs. ACo≤Braak_II) denoted by squares with dotted lines; assortment 3 (UCo≤Braak_III vs. ACo≤Braak_III) denoted by triangle with dotted lines. The top performing panel of individual assortments were highlighted yellow. (A) AUCs measured by RSBMR; (B) AUCs measured by k-fold.
Table 1A.
Top performing biomarker panels curated by RSBMR.
| Assortment/ Braak Stage cut-off | Panel.Size | Genes | R.Squared | Adjusted.R.Square | Validation.Error | AUC (Mean) | Sensitivity (Mean) | Specificity (Mean) | Intercept | Features' weighing factors |
Features' Expression valuesin in log base 2 |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene1 | Gene2 | Gene3 | Gene4 | Gene5 | Gene6 | Gene7 | Gene8 | Gene1 | Gene2 | Gene3 | Gene4 | Gene5 | Gene6 | Gene7 | Gene8 | ||||||||||
| Assortment 1/ Braak I | 4 | CRH,DLEC1,GSTT1,HBD | 0.48 | 0.44 | 0.14 | 0.95 | 0.92 | 0.85 | -6.67 | 1.93 | -0.75 | -0.36 | 0.64 | -0.105 | -0.184 | 0.2182 | 0.062 | ||||||||
| Assortment 2/ Braak II | 8 | CRH,DDX3Y,HBB,HBD,RPS4Y2,TSIX,USP9Y,VGF | 0.48 | 0.4 | 0.19 | 0.91 | 0.93 | 0.8 | 19.27 | -1.24 | 1.51 | -0.19 | -2.34 | -0.61 | -1.16 | 1.95 | 0.34 | -0.105 | -0.309 | -0.04 | 0.063 | -0.091 | -0.029 | 0.013 | 0.24 |
| Assortment 3/ Braak III | 8 | C4B,CARTPT,DDX3Y,HBB,HBD,RPS4Y2,TSIX,USP9Y | 0.79 | 0.75 | 0.18 | 0.93 | 0.96 | 0.74 | -85.43 | -3.06 | 5.42 | -9.81 | 10.97 | -13.76 | 13.46 | 1.02 | 5.43 | 0.003 | -0.04 | -0.309 | -0.04 | 0.063 | -0.091 | -0.029 | 0.013 |
Likewise, Table S2 listed those biomarker panels, which were curated by k-fold. There were 8 putative panels that significantly well-fitted in a linear regression model (p < 0.0001) and can potentially distinguish UCo≤Braak_I, from ACo>Braak_I with AUC ≥ 0.8. In Fig. 3B, the circles with dotted line showed the maximum AUC scored by biomarker panel with increasing feature count from 2 to 10. High AUC was achieved by two biomarker panels. A 4-gene panel included DLEC1, GSTT1, HBB and HBD; together this panel scored AUC = 0.83, Sensitivity = 0.90 and Specificity = 0.76. A similar scoring 6-gene panel included ALG2, DLEC1, GSTT1, HBB, HBD and USP9Y; together this panel scored AUC = 0.82, Sensitivity = 0.88 and Specificity = 0.76. Finally, we chose the 4-gene panel as the best candidate, primarily because of its smaller feature count. This panel was highlighted in Fig. 3B and reported in Table 1B.
Table 1B.
Top performing biomarker panels curated by k-fold.
| Assortment/ Braak Stage cut-off | Panel.Size | Genes | R.Squared | Adjusted.R.Square | Validation.Error | AUC | Sensitivity | Specificity | Intercept | Features' weighing factors |
Features' Expression values in log base 2 |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene1 | Gene2 | Gene3 | Gene4 | Gene5 | Gene6 | Gene7 | Gene8 | Gene9 | Gene10 | Gene1 | Gene2 | Gene3 | Gene4 | Gene5 | Gene6 | Gene7 | Gene8 | Gene9 | Gene10 | ||||||||||
| Assortment 1/ Braak I | 4 | DLEC1,GSTT1,HBB,HBD | 0.51 | 0.47 | 0.12 | 0.84 | 0.91 | 0.76 | -21.28 | 2.48 | -4.99 | 0.56 | 4.26 | -0.184 | 0.218 | -0.04 | 0.063 | ||||||||||||
| Assortment 2/ Braak II | 3 | HBB,SLC47A2,VGF | 0.5 | 0.47 | 0.13 | 0.83 | 0.81 | 0.86 | 22.42 | -0.83 | -1.26 | 1.08 | -0.04 | -0.024 | 0.24 | ||||||||||||||
| Assortment 3/ Braak III | 10 | C4B,CARTPT,DDX3Y,HBB,HBD,RPS4Y1,RPS4Y2,TSIX,USP9Y,XIST | 1 | 1 | 0.22 | 0.9 | 0.88 | 0.91 | -3191.51 | 185.87 | -66.68 | 259.3 | -320.64 | 63.84 | -484.2 | -146.26 | 419.78 | 409.83 | 122.79 | 0.003 | -0.04 | -0.309 | -0.04 | 0.063 | 0.052 | -0.091 | -0.029 | 0.013 | -0.056 |
4.2. Biomarker panels for assortment 2
RSBMR and k-fold method separately reported putative panels of biomarkers linked to assortment 2. Table S1 listed those biomarker panels, which were curated by RSBMR. There were 104 putative panels that significantly well-fitted in a linear regression model (p < 0.0001) and can potentially distinguish UCo≤Braak_II, from ACo>Braak_II with AUC ≥ 0.8. In Fig. 3A, the boxes with dotted line showed the maximum AUC scored by the biomarker panels with increasing feature count from 2 to 10. High AUC was achieved by three biomarker panels. A 9-gene panel included CARTPT, DDX3Y, HBB, HBD, PCSK1, RPS4Y2, USP9Y, VGF and XIST; together this panel scored mean AUC = 0.92, mean Sensitivity = 0.85 and mean Specificity = 0.80. There were two similarly scoring 8-gene panels. One of these two candidates included CRH, DDX3Y, HBB, HBD, PCSK1, RPS4Y2, USP9Y and VGF; together this panel scored AUC = 0.90, Sensitivity = 0.9 and Specificity = 0.8. Second of these two candidates included CRH, DDX3Y, HBB, HBD, RPS4Y2, TSIX, USP9Y and VGF; together this panel scored mean AUC = 0.91, mean Sensitivity = 0.92 and mean Specificity = 0.8. Finally, we selected the second 8-gene panel as the best candidate, primarily because of its smaller feature count that the 10-gene panela and little higher AUC value from the other 8-gene panel. This panel was highlighted in Fig. 3A and reported in Table 1A.
Likewise, Table S2 listed those biomarker panels, which were curated by k-fold. There were 19 putative panels that significantly well-fitted in a linear regression model (p < 0.0001) and can potentially distinguish UCo≤Braak_II, from ACo>Braak_II with AUC ≥ 0.8. In Fig. 3B, the boxes with dotted line showed the maximum AUC scored by biomarker panel with increasing feature count from 2 to 10. Maximum AUC was achieved by a 3-gene panel that included HBB, SLC47A2 and VGF; together this panel scored AUC = 0.83, Sensitivity = 0.81 and Specificity = 0.86.
4.3. Biomarker panels for assortment 3
RSBMR and k-fold method separately reported putative panels of biomarkers linked to assortment 3. Table S1 listed those biomarker panels, which were curated by RSBMR. There were 204 putative panels that significantly well-fitted in a linear regression model (p < 0.0001) and can potentially distinguish UCo≤Braak_III, from ACo>Braak_III with AUC ≥ 0.8. In Fig. 3A, the triangles with dotted line showed the maximum AUC scored by the biomarker panels with increasing feature count from 2 to 10. High AUC was achieved by five biomarker panels. A 7-gene panel included C4B, DDX3Y, HBB, HBD, NEUROD6, RPS4Y2, USP9Y; together this panel scored AUC = 0.92, mean Sensitivity = 0.88 and mean Specificity = 0.74. There were three similarly scoring 8-gene panels. One of these three candidates included C4B, CARTPT, DDX3Y, HBB, HBD, RPS4Y2, TSIX and USP9Y; together this panel scored mean AUC = 0.93, mean Sensitivity = 0.96 and mean Specificity = 0.74. Second of these three candidates included C4B, CARTPT, DDX3Y, HBB, HBD, RPS4Y2, USP9Y and XIST; together this panel scored mean AUC = 0.92, mean Sensitivity = 0.94 and mean Specificity = 0.71. Third candidates with 8 genes included CARTPT, DDX3Y, HBB, HBD, RPH3A, RPS4Y2, TSIX and USP9Y; together this panel scored mean AUC = 0.91, mean Sensitivity = 0.84 and mean Specificity = 0.77. In addition, there were a 9-gene biomarker panel candidate that included CARTPT, DDX3Y, HBB, HBD, RPH3A, RPS4Y2, TSIX, USP9Y; together this panel scored mean AUC = 0.91, mean Sensitivity = 0.96 and mean Specificity = 0.74. From these five candidates, we selected the first 8-gene panel that included C4B, CARTPT, DDX3Y, HBB, HBD, RPS4Y2, TSIX and USP9Y as the best candidate, primarily because of its little higher AUC value from the rest. This panel was highlighted in Fig. 3A and reported in Table 1A.
Likewise, Table S2 listed those biomarker panels, which were curated by k-fold. There were 74 putative panels that significantly well-fitted in a linear regression model (p < 0.0001) and can potentially distinguish UCo≤Braak_III, from ACo>Braak_III with AUC ≥ 0.8. In Fig. 3B, the triangles with dotted line showed the maximum AUC scored by biomarker panel with increasing feature count from 2 to 10. Maximum AUC was achieved by a 10-gene panel that included C4B, CARTPT, DDX3Y, HBB, HBD, RPS4Y1, RPS4Y2, TSIX, USP9Y and XIST; together this panel scored AUC = 0.90, Sensitivity = 0.88 and Specificity = 0.91 Fig. 4, Fig. 5.
Fig. 4.
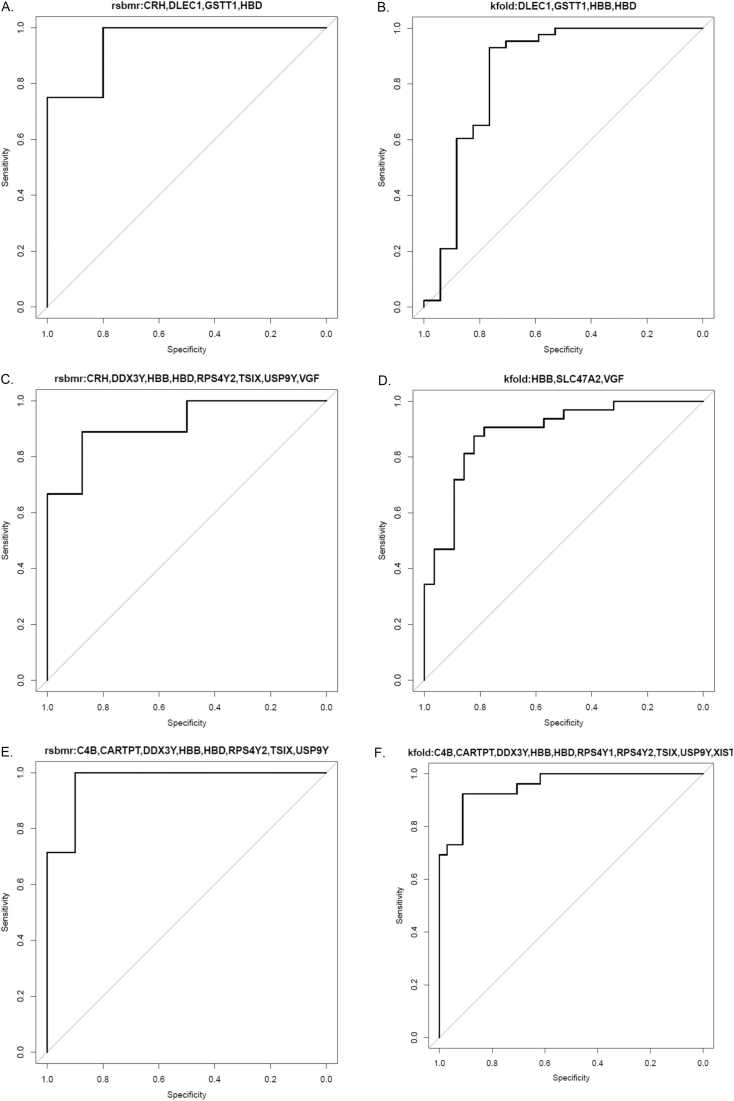
The AUC curves of top performing panels: (A) Assortment 1 (UCo≤Braak_I vs. BCo≤Braak_I measured by RSBMR. (B) Assortment 1 (UCo≤Braak_I vs. BCo≤Braak_I measured by k-fold. (C) Assortment 2 (UCo≤Braak_II vs. BCo≤Braak_II measured by RSBMR. (D) Assortment 2 (UCo≤Braak_II vs. BCo≤Braak_II measured by k-fold. (E) Assortment 3 (UCo≤Braak_III vs. BCo≤Braak_III measured by RSBMR. (F) Assortment 3 (UCo≤Braak_III vs. BCo≤Braak_III measured by k-fold.
Fig. 5.
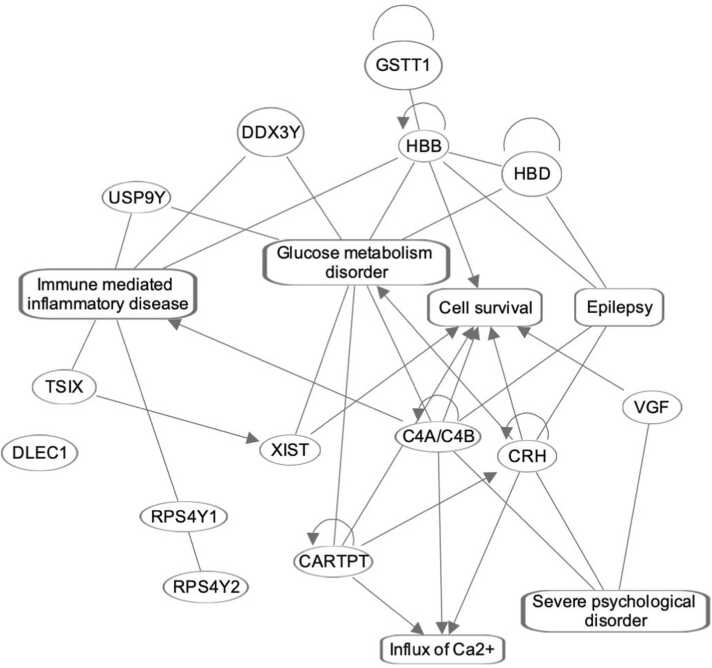
The gene network enriched by those genes, which were featured at least one time in the most promising set of biomarker panels listed in Table 1A, Table 1B. Here the oval and rectangular shaped nodes encircle the gene symbols and biofunctions, respectively. The edges represent the relationship between two interconnecting nodes. The solid lines represent their associations, and pointed arrowheads denote the activating relationships between the two connecting nodes. There is one node, which is not connected to any of the nodes in this network.
Final deliverables were listed in Table 1A, Table 1B that were the best possible candidate of biomarker panels with high translational potential. In this table, we reported coefficient estimates of individual genes of the panels. For instance, there were 4 genes in the biomarker panel of assortment 1 that was determined by RSBMR. Hence, the corresponding liner regression model would be the following.
Where X was the expression values of the genes noted in its subscript. The equations of rest of the biomarker panels would be similar.
There were fifteen unique genes that featured at least once in the top performing biomarker panels listed in Table 1A, Table 1B. The transcripts of all these genes were annotated in Table 2 reporting the topologies and sequences of these transcripts. Furthermore, these fifteen genes were functionally annotated. The genes were linked via either their significant functional attributes or their functional interactions. All annotations were curated from the existing literature.
Table 2.
| Gene Symbol | Gene Name | Chromosome | Starting | Ending | Transcriptomic sequence |
|---|---|---|---|---|---|
| C4B | Complement C4B | chr6 | 3.2E+ 07 | 3.2E+ 07 | GCTTTCCGCCTCTTTGAGACCAAGATCACCCAAGTCCTGCACTTCACCAAGGATGTCAAG |
| CARTPT | Cocaine And Amphetamine Regulated Transcript | chr5 | 7.1E+ 07 | 7.1E+ 07 | TTCCTCTGAAGGGAAAGGGCTCTTTTCCTGCTGTTTCAAAAATAAAAGAACACATTAGAT |
| CRH | Corticotropin Releasing Hormone | chr8 | 6.7E+ 07 | 6.7E+ 07 | AGAAGTCACTCAATTGTTTTTGTTGTGGTCTGAGCCAAAGAGAATGCCATTCTCTTGGGT |
| DDX3Y | DEAD-Box Helicase 3 Y-Linked | chrY | 1.5E+ 07 | 1.5E+ 07 | GCAGTATTCTTCAGTAAATAAAGAATGGAATTGCTGAATGTAATCATTGAACCTCGAGTC |
| DLEC1 | Cilia And Flagella Associated Protein | chr3 | 3.8E+ 07 | 3.8E+ 07 | CACATTGAGATCACTACTCAGTGCATAGCGAAGACCAGTATGGCAAAATTAGTCTTGGAA |
| GSTT1 | Glutathione S-Transferase Theta 1 | chr22 | 2.4E+ 07 | 2.4E+ 07 | AGCAGTCCACAAAGCATTTTCATTTCTAATGGCCCATGGGAGCCAGGCCCAGAAAGCAGG |
| HBB | Hemoglobin Subunit Beta | chr11 | 5246777 | 5246718 | GTCCAACTACTAAACTGGGGGATATTATGAAGGGCCTTGAGCATCTGGATTCTGCCTAAT |
| HBD | Hemoglobin Subunit Delta | chr11 | 5255317 | 5255258 | TGATGGCCTGGCTCACCTGGACAACCTCAAGGGCACTTTTTCTCAGCTGAGTGAGCTGCA |
| RPS4Y1 | Ribosomal Protein S4 Y-Linked 1 | chrY | 2722790 | 2733165 | CAACTTTATCAAATTTGATACAGGCAATTTGTGTATGGTGATTGGTGGAGCCAACCTCGG |
| RPS4Y2 | Ribosomal Protein S4 Y-Linked 2 | chrY | 2.3E+ 07 | 2.3E+ 07 | TGTTGGTGTGATCACAAACAGGGAAAGACATCCTGGTTCTTGCGATGTGGTACATGTGAA |
| SLC47A2 | Solute Carrier Family 47 Member 2 | chr17 | 2E+ 07 | 2E+ 07 | GGGACACTGCAGATAAAATCACAAAAACCACTGTTATATTAAAGATTACACATTTCCTGG |
| TSIX | X (Inactive)-Specific Transcript, Antisense | chrX | 7.3E+ 07 | 7.3E+ 07 | TCAGCTCTCTGCACTGCTTGTAGGAAGTATAATGATTTGGCAGATAGGAACAATGAAGAG |
| USP9Y | Ubiquitin Specific Peptidase 9 Y-Linked | chrY | 1.5E+ 07 | 1.5E+ 07 | GTACCATTGCACCAAGATGTCTGACTGAATTCATAGTCACACTTTTATTTGAAAGAAAGA |
| VGF | Nerve Growth Factor | chr7 | 1E+ 08 | 1E+ 08 | CTCTGTTGTAAATACCCCTCACGGAGGAAATAGTTTTGCTAAGAAATAAAAGTGACTATT |
| XIST | X Inactive Specific Transcript | chrX | 7.3E+ 07 | 7.3E+ 07 | GCCATCTAGATGTCACAATTGAAACAAACTGGGGAGTTGGTTGCTATTGTAAAATAAAAT |
5. Discussion
High throughput and high-resolution multi-omics readout made a paradigm shift in our understanding about the complex interconnectivity among groups of molecules to manifest clinical symptoms. Expectedly, this knowledge made a significant impact on the biomarker discovery. Present trend of biomarker discovery is shifting from finding a unique signature to a group of biomarkers that can collectively define a clinical event [3], [4], [42]. For example, Mamaprint is a 70-gene panel that was approved by the FDA as a prognostic marker for breast cancer relapse [43], [44]. The current hypothesis is that a systematic integration of a group of biomarkers can potentially demonstrate a higher efficacy than a single candidate biomarker [45].
Towards this objective, we presented a biomarker discovery pipeline that systematically integrated random forest and logistic regression fitting model in a computationally inexpensive fashion; its goal was to objectively define a clinically relevant binomial incident, where the decision was expressed by a ‘yes’ or ‘no’ answer. The decision was quantified by AUC, sensitivity, and specificity of individual biomarker panel, which were computed by RSBMR and k-fold. K-fold cross validation method performed well with small number of replications [23]. Following a similar template but applying a little relaxed and more flexible position, we formulated another cross-validation routine, namely RSBMR. By allowing the users to find the potential range of performance metrics, RSBMR addressed a limitation of k-fold [46], [47]. In RSBMR, the outlier panels with good performance becomes visible to be considered later. Similar solutions were offered via feedforward NN approach [48], and here we presented a sample pipeline integrating Random Forest and Logistic Regression model.
To evaluate the efficacy of 2BDP model, we used a publicly available transcriptomic study that performed gene microarray of autopsied human temporal cortical samples with variable severity of AD [24]. As discussed in supplementary section, this database met four exclusion-inclusion criteria set by us, namely, (1) The data should be untargeted microarray readouts; (2) the study should be focused on a clinically explainable endpoints, such as disease severity; (3) the clinically explainable endpoint should be binary in nature, if the sample size small and (4) data should be available in GEO dataset in an understandable format for public use.
Early diagnostic or subtyping markers of any disease are of high demand in clinical fraternity, since such markers can provide a golden time window allowing the clinicians to initiate therapy. Hence, we trained 2BDP to find markers to differentiate biological stages of AD. By definition, the Braak stages paralleled the progression of hyperphosphorylation status of aggregated-tau, an established histopathological signature of Alzheimer’s disease [25], [26]. The multi-omics study of Marttinen et al. [24] further associated Braak stage II and III with the disease threshold and the post-threshold onset of inflammation surge, respectively. The disease threshold is an important clinical landmark that technically pronounce the onset of Alzheimer’s disease. On the other hand, the onset of inflammation is a clear sign of discomfort and potentially triggers comorbidities, such as migraine. Clearly, these Braak stages are critical clinical landmarks and objective diagnosis of these stages would have significant diagnostic and therapeutic potential. Taken together, 2BDP identified biomarkers of following clinical values: (i) panels that explained Braak I could be the early markers of AD. These panels potentially can predict AD’s onset during its asymptomatic condition. (ii) The panels that explained Braak II could objectively determine the first onset of AD’s symptoms and could be used to monitor AD pathogenesis and perform disease triage along with the 3rd panel of biomarkers. (iii) The 3rd panel that explained Braak III could be a late marker of AD onset, which could be used for disease monitoring or to triage AD patients. It is also to note that all these markers were derived from post-mortem brain autopsies, hence it is warranted to check their expression levels in blood in order to assess their full clinical potential.
In addition to finding the appropriate panel of biomarkers, the number of features in the panel are other factors to consider. It is important that the panel size should meet the capabilities of available high throughput in vitro diagnostic (IVD) platforms. The limited multiplexing capability, prolonged hands-on time and the kits’ short shelf-lives typically reduce the approval of IVD devices. PCR platform is an increasingly popular IVD device despite of its limited high throughput capability; PCR presents high sensitivity and specificity, short hands-on time, and small footprint. Although, there are few incidents, where the PCR platform was used to detect panels with more than 20 genes [49], [50]; the market trend suggests that the protocol complexity could be greatly reduced should we able to keep the panel size less than10 probes [34], [51], [52], [53]. Hence, we presented a comparative performance analysis with panels enriched with variable number of transcripts. One major conclusion of this work is that unbiased and systematic evaluation can determine a panel of small sample size with high performance.
2BDP used an unbiased approach to find AD biomarkers; yet the list of features includes some of the established biomarkers of AD along with few novel markers. It is an essential signature of any unsupervised approach, as reported earlier by the Mammaprint biomarker discovery study, where 9 out of the 70-gene panel had no prior link to carcinoma or any biofunctions [54]. Some of the gene markers of 2BDP have already linked to AD, for instance, the gene copy number of C4B [55] and genetic variants on GSTT1 [56] are dependent variables of AD. Likewise, the risk of AD onset is linked to the instability of X-chromosome, which is typically governed by the interactions between TSIX and XIST [57] and both genes were featured in Table 1A, Table 1B. Atypical activity of HPA axis, and pertinent molecular expression of CRH are linked to AD [58]. Furthermore, this list included two transcripts encoding structural constituents of hemoglobin; this observation highlights the close connection between hemoglobin and AD onset [59]. Additional independent features with a priori association with AD included CARTPT [60] and, transcripts encoding ribosomal proteins [61] and solute carrier family [62]. Overall, present algorithm delivered a set of biomarkers, which was a mix of novel and established signatures of AD.
It is important to note that we had access to a single cohort; thus, we randomly sorted this single cohort into Training and Test set to meet our purpose. Braak stages were the only phenotypes accessible to the present analysis; therefore, it is beyond the scope of present analysis to check whether any other comorbidity plays role in driving the biomarker profile. Since, we only used a dataset from public domain, there was no scope to validate of the markers by different assay platforms or using independent cohort. The focus of this study was to present a novel pipeline for biomarker discovery and this AD data set was used for the proof-of-principal approach. To best assess the capability of 2BDP in handling large dataset, this study focused on high throughput array data; hence no other omics data was not considered herein. Nevertheless, we can posit that 2BDP is potentially flexible to handle omics data from other high throughput platforms, such as sequencers and spectroscopy machines. Furthermore, we did not attempt to find the biomarkers linked to Braak IV and V, since the sample size above IV and V thresholds were too small to make any meaningful analysis. Indeed, we customized our analysis to best fit the present sample size. For instance, the split of samples between training and validation was set to 70:30 as there were only 60 samples. With the purpose of the program to identify outcome variables related to diseases, a larger test set was used as performance estimates were more important than parameter estimates with the recommendation for splits on data as 70:30. If the difference between Training and Test sizes increases, then the variances in the validations will increase. Decreasing the split to 80:20 or 90:10 is only recommended when more than a few thousand samples are available. It is also important to note that 2BDP is designed to explain a binary clinical endpoint. More than one clinical endpoint will require further splitting of the cohort. For instance, a pursuit for biomarker panel to identify the cohort, who are between Braak I and II would need three-way splitting of 60-samples’ cohort, which would weaken the confidence on outcome. Indeed, 2BDP could be a valuable tool to explain non-binary clinical endpoints; however, present routine was not suitable to validate this claim.
In conclusion, we presented an algorithm that uses an machine learning-driven analytical tool to find biomarkers linked to a clinical variable. Our results produced a series of panels enriched by a different number of transcriptomic candidates that can explain the Braak stage I, II and II with high accuracy. This stage is of high clinical significance, due to its association with the initiation of synaptic modifications, onset of neuronal loss and escalation of mitochondrial dysfunctions. Although these promising results of 2BDP were limited by the absence of any independent validation cohort, our results underscored the capability of 2BDP in finding appropriate markers and pertinent scoring algorithms linked to clinical landmarks. Biomarker panels enable to diagnosis, prognosis and stratify disease pathogenies can be identified using 2BDP pipeline from high throughput big dataset.
Disclaimer
Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense.
CRediT authorship contribution statement
Conceptualization, NC, RY, RH; Methodology NC, AL, RC and RY, Supervision NC and RH, Data acquisition, curation and analysis: NC, AL, RC and RY, Drafting of the manuscript, NC and AL, Revision of manuscript content: NC, AL, RH, Funding acquisition, RH. All authors have read and approved the final submitted manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.09.025.
Appendix A. Supplementary material
Supplementary material.
.
Figure S1. Gini Scores of the top thirty independent features linked to (a) Assortment 1, (b) Assortment 2 and (c) Assortment 3.
.
Supplementary material.
.
Data Availability
Present work used the data that was shared here GSE106241.
References
- 1.Califf R.M. Biomarker definitions and their applications. Exp Biol Med (Maywood) 2018;243:213–221. doi: 10.1177/1535370217750088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Group, F.-N.B.W. BEST (Biomarkers, endpoints, and other tools) resource [Internet]. (2016). [PubMed]
- 3.Vincent J.-L., Bogossian E., Menozzi M.J.C. c c. The future of biomarkers. Future Biomark. 2020;36:177–187. doi: 10.1016/j.ccc.2019.08.014. [DOI] [PubMed] [Google Scholar]
- 4.Simon, R. (Oxford University Press, 2005).
- 5.Zaim, S.R., Li, Q., Schissler, A.G. , Lussier, Y.A. Emergence of pathway-level composite biomarkers from converging gene set signals of heterogeneous transcriptomic responses. (2018). [PMC free article] [PubMed]
- 6.Kamarudin A.N., Cox T., Kolamunnage-Dona R.J.B. m r m. Time-dependent ROC curve analysis in medical research: current methods and applications. BMC Med Res Methodol. 2017;17:1–19. doi: 10.1186/s12874-017-0332-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sengupta S., Parikh N.D. Biomarker development for hepatocellular carcinoma early detection: current and future perspectives. Hepatic Oncol. 2017;4:111–122. doi: 10.2217/hep-2017-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartwell M.J., Özbek U., Holler E., Renteria A.S., Major-Monfried H., Reddy P., Aziz M., Hogan W.J., Ayuk F., Efebera Y.A., Hexner E.O., Bunworasate U., Qayed M., Ordemann R., Wölfl M., Mielke S., Pawarode A., Chen Y.B., Devine S., Harris A.C., Jagasia M., Kitko C.L., Litzow M.R., Kröger N., Locatelli F., Morales G., Nakamura R., Reshef R., Rösler W., Weber D., Wudhikarn K., Yanik G.A., Levine J.E., Ferrara J.L. An early-biomarker algorithm predicts lethal graft-versus-host disease and survival. JCI Insight. 2017;2:89798. doi: 10.1172/jci.insight.89798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzikas, S., Vassilikos, V. , Keller, T. (Elsevier, 2019). [DOI] [PubMed]
- 10.Mahajan, K., Chand Negi P., Ganju N., Asotra S.. Cardiac biomarker-based risk stratification algorithm in patients with severe COVID-19. 14, 929–931 (2020). [DOI] [PMC free article] [PubMed]
- 11.Forghani R., Savadjiev P., Chatterjee A., Muthukrishnan N., Reinhold C., Forghani B. Radiomics and artificial intelligence for biomarker and prediction model development in oncology. Comput Struct Biotechnol J. 2019;17:995–1008. doi: 10.1016/j.csbj.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung H., Jo Y., Ryu D., Jeong C., Choe S., Lee J. Artificial-intelligence-driven discovery of prognostic biomarker for sarcopenia. J Cachex - Sarcopenia Muscle. 2021;12:2220–2230. doi: 10.1002/jcsm.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavanya C., Pooja S., Kashyap A., Rahaman A., Niranjan S., Niranjan V. Novel biomarker prediction for lung cancer using random forest classifiers. Cancer Inf. 2023;22 doi: 10.1177/11769351231167992. 11769351231167992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guyon I., Weston J., Barnhill S., Vapnik V. Gene selection for cancer classification using support vector machines. Mach Learn. 2002;46:389–422. [Google Scholar]
- 15.RA D. John Wiley & Sons; 2007. A statistical approach to neural networks for pattern recognition. [Google Scholar]
- 16.Liem Y., Judge A., Kirwan J., Ourradi K., Li Y., Sharif M. Multivariable logistic and linear regression models for identification of clinically useful biomarkers for osteoarthritis. Sci Rep. 2020;10 doi: 10.1038/s41598-020-68077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Mendonca E.B., Schmaltz C.A., Sant'Anna F.M., Vizzoni A.G., Mendes-de-Almeida D.P., de Oliverira R.V.C., et al. Anemia in tuberculosis cases: a biomarker of severity? PLoS One. 2021;16 doi: 10.1371/journal.pone.0245458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z., Yang D., Gao J., Xiang X., Hu X., Li S., et al. Discovery and validation of miR-452 as an effective biomarker for acute kidney injury in sepsis. Theranostics. 2020;10:11963–11975. doi: 10.7150/thno.50093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minarno, A.E., Kusuma, W.A., Wibowo, H. Performance comparisson activity recognition using logistic regression and support vector machine. 2020 3rd International conference on intelligent autonomous systems (ICoIAS), IEEE February, 19–24 (2020).
- 20.Westreich D., Lessler J., Funk M.J. Propensity score estimation: neural networks, support vector machines, decision trees (CART), and meta-classifiers as alternatives to logistic regression. J Clin Epidemiol. 2010;63:826–833. doi: 10.1016/j.jclinepi.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amini P., Ahmadinia H., Poorolajal J., Moqaddasi Amiri M. Evaluating the high risk groups for suicide: a comparison of logistic regression, support vector machine, decision tree and artificial neural network. Iran J Public Health. 2016;45:1179–1187. [PMC free article] [PubMed] [Google Scholar]
- 22.Sullivan L.M., Massaro J.M., D'Agostino R.B. Sr. Presentation of multivariate data for clinical use: the framingham study risk score functions. Stat Med. 2004;23:1631–1660. doi: 10.1002/sim.1742. [DOI] [PubMed] [Google Scholar]
- 23.Wong T.T., Yeh P.Y. Reliable accuracy estimates from k-fold cross validation. IEEE Trans Knowl Data Eng. 2019;32:1586–1594. [Google Scholar]
- 24.Marttinen M., Paananen J., Neme A., Mitra V., Takalo M., Natunen T., Paldanius K., Mäkinen P., Bremang M., Kurki M.I., Rauramaa T., Leinonen V., Soininen H., Haapasalo A., Pike I., Hiltunen M. A multiomic approach to characterize the temporal sequence in Alzheimer's disease-related pathology. Neurobiol Dis. 2019;124:454–468. doi: 10.1016/j.nbd.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 25.Braak H., Alafuzoff I., Arzberger T., Kretzschmar H., Del Tredici K.J.A. n. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 2006;112:389–404. doi: 10.1007/s00401-006-0127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Šimić G., Babić Leko M., Wray S., Harrington C., Delalle I., Jovanov-Milošević N., Bažadona D., Buée L., de Silva R., Di Giovanni G., Wischik C., Hof P.R. Tau protein hyperphosphorylation and aggregation in Alzheimer’s disease and other tauopathies, and possible neuroprotective strategies. Biomolecules. 2016;6:6. doi: 10.3390/biom6010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong H., Goodsaid F., Shi L., Tong W.J.B. i m. Molecular biomarkers: a US FDA effort. Biomark Med. 2010;4:215–225. doi: 10.2217/bmm.09.81. [DOI] [PubMed] [Google Scholar]
- 28.Paik S., Shak S., Tang G., Kim C., Baker J., Cronin M., Baehner F.L., Walker M.G., Watson D., Park T., Hiller W., Fisher E.R., Wickerham D.L., Bryant J., Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 29.Van der Hoeven J.J.N. t v g. 70-Gene signature as an aid to treatment decisions in early-stage breast cancer. Ned Tijdschr voor Geneeskd. 2017;161 D1369-D1369. D1369-D1369. [PubMed] [Google Scholar]
- 30.Wallden B., Storhoff J., Nielsen T., Dowidar N., Schaper C. Ferre S.,et al. Development and verification of the PAM50-based Prosigna breast cancer gene. signature assay. 2015;8:1–14. doi: 10.1186/s12920-015-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Easton D.F., Pharoah P.D., Antoniou A.C., Tischkowitz M., Tavtigian S.V., Nathanson K.L., Devilee P., Meindl A., Couch F.J., Southey M., Goldgar D.E., Evans D.G., Chenevix-Trench G., Rahman N., Robson M., Domchek S.M., Foulkes W.D. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372:2243–2257. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chong H.K., Wang T., Lu H.M., Seidler S., Lu H., Keiles S., Chao E.C., Stuenkel A.J., Li X., Elliott A.M. The validation and clinical implementation of BRCAplus: a comprehensive high-risk breast cancer diagnostic assay. PloS One. 2014;9 doi: 10.1371/journal.pone.0097408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jørgensen J.T. The current landscape of the FDA approved companion diagnostics. Transl Oncol. 2021;14 doi: 10.1016/j.tranon.2021.101063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verboom D.M., Koster-Brouwer M.E., Varkila M.R., Bonten M.J., Cremer O.L. Profile of the SeptiCyteTM LAB gene expression assay to diagnose infection in critically ill patients. Expert Rev Mol Diagn. 2019;19:95–108. doi: 10.1080/14737159.2019.1567333. [DOI] [PubMed] [Google Scholar]
- 35.FDA, U.J.U. List of cleared or approved companion diagnostic devices (in vitro and imaging tools). (2020).
- 36.Martiskainen H., Viswanathan J., Nykänen N.P., Kurki M., Helisalmi., Natunen T., et al. Transcriptomics and mechanistic elucidation of Alzheimer’s disease risk genes in the brain and in vitro models. Neurobiol Aging. 2015;36:e1215–1228. doi: 10.1016/j.neurobiolaging.2014.09.003. 1221. [DOI] [PubMed] [Google Scholar]
- 37.Davis S., Meltzer P.S.J.B. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinforma (Oxf, Engl) 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 38.Ritchie M.E., Silver J., Oshlack A., Holmes M., Diyagama D., Holloway A., Smyth G.K. A comparison of background correction methods for two-colour microarrays. Bioinforma (Oxf, Engl) 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 39.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv007. e47-e47. e47-e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liaw A., Wiener M.J.R. n. Classif Regres Random. 2002;2:18–22. [Google Scholar]
- 41.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinforma. 2011;12:1–8. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaim, S.R., Li, Q., Schissler, A.G. & Lussier, Y.A.Pacific symposium on biocomputing 2018: Proceedings of the Pacific Symposium. 484–495 (World Scientific). [PMC free article] [PubMed]
- 43.Slodkowska E.A., Ross J.S. MammaPrintTM 70-gene signature: another milestone in personalized medical care for breast cancer patients. Expert Rev Mol Diagn. 2009;9:417–422. doi: 10.1586/erm.09.32. [DOI] [PubMed] [Google Scholar]
- 44.Audeh W., Blumencranz L., Kling H., Trivedi H., Srkalovic G.J. Prospective validation of a genomic assay in breast cancer: the 70-gene MammaPrint Assay and the MINDACT Trial. Acta Med Acad. 2019;48:18–34. doi: 10.5644/ama2006-124.239. [DOI] [PubMed] [Google Scholar]
- 45.Dessi N., Pascariello E., Pes B. A comparative analysis of biomarker selection techniques. Biomed Res Int. 2013;2013 doi: 10.1155/2013/387673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiong Z., C.Y., Liu Z., Zhao Y., Hu M., Hu J. Evaluating explorative prediction power of machine learning algorithms for materials discovery using k-fold forward cross-validation. Comput Mater Sci. 2020;171 Jan 1. [Google Scholar]
- 47.Meredig B., Antono E., Church C., Hutchinson M., Ling J., Paradiso S., Blaiszik B., Foster I., Gibbons B., Hattrick-Simpers J., Mehta A. Can machine learning identify the next high-temperature superconductor? Examining extrapolation performance for materials discovery. Mol Syst Des Eng. 2018;3:819–825. [Google Scholar]
- 48.Martius G. a L., C.H. Extrapolation and learning equations. arXiv Prepr arXiv:1610 02995. 2016 [Google Scholar]
- 49.Paik S., Shak S., Tang G., Kim C., Baker J., Cronin M., et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 50.Clark-Langone K.M., Wu J.Y., Sangli C., Chen A., Snable J.L., Nguyen A., et al. Biomarker discovery for colon cancer using a 761 gene RT-PCR assay. BMC Genom. 2007;8:279. doi: 10.1186/1471-2164-8-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Byun J.M., Kim S.S., Kim K.T., Kang M.S., Jeong D.H., Lee D.S., Jung E.J., Kim Y.N., Han J., Song I.S., Lee K.B., Sung M.S. Overexpression of peroxiredoxin-3 and-5 is a potential biomarker for prognosis in endometrial cancer. Oncol Lett. 2018;15:5111–5118. doi: 10.3892/ol.2018.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li T., Shao Y., Fu L., Xie Y., Zhu L., Sun W., Yu R., Xiao B., Guo J. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl, Ger) 2018;96:85–96. doi: 10.1007/s00109-017-1600-y. [DOI] [PubMed] [Google Scholar]
- 53.Wang L., Zhang M., Zhu H., Sun L., Yu B., Cui X. Combined identification of lncRNA NONHSAG004550 and NONHSAT125420 as a potential diagnostic biomarker of perinatal depression. J Clin Lab Anal. 2021;35 doi: 10.1002/jcla.23890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tian S., et al. Biological functions of the genes in the mammaprint breast cancer profile reflect the hallmarks of cancer. Biomark Insights. 2010;5:129–138. doi: 10.4137/BMI.S6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zorzetto M., Datturi F., Divizia L., Pistono C., Campo I., Silvestri A.D., et al. Complement C4A and C4B gene copy number study in Alzheimer’s disease patients. Curr Alzheimer Res. 2017;14:303–308. doi: 10.2174/1567205013666161013091934. [DOI] [PubMed] [Google Scholar]
- 56.Wang T. Glutathione S-transferases variants as risk factors in Alzheimer's disease. Neurol Sci. 2015;36:1785–1792. doi: 10.1007/s10072-015-2245-7. [DOI] [PubMed] [Google Scholar]
- 57.Bajic V.P., Essack M., Zivkovic L., Stewart A., Zafirovic S., Bajic V., et al. The X Files: "The mystery of X chromosome instability in Alzheimer’s disease. Front Genet. 2019;10:1368. doi: 10.3389/fgene.2019.01368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rehman H.U. Role of CRH in the pathogenesis of dementia of Alzheimer's type and other dementias. Curr Opin Invest Drugs. 2002;3:1637–1642. [PubMed] [Google Scholar]
- 59.Arioz B.I., Tufekci K.U., Olcum M., Durur D.Y., Akarlar B.A., Ozlu N., et al. Proteome profiling of neuron-derived exosomes in Alzheimer’s disease reveals hemoglobin as a potential biomarker. Neurosci Lett. 2021;755:135914. doi: 10.1016/j.neulet.2021.135914. [DOI] [PubMed] [Google Scholar]
- 60.Huang C., Wen X., Xie H., Hu D., Li K. Identification and experimental validation of marker genes between diabetes and Alzheimer's disease. Oxid Med Cell Longev. 2022;2022 doi: 10.1155/2022/8122532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cohen D., Pilozzi A., Huang X. Network medicine approach for analysis of Alzheimer's disease gene expression data. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21010332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ayka A., Sehirli A.O. The role of the SLC transporters protein in the neurodegenerative disorders. Clin Psychopharmacol Neurosci. 2020;18:174–187. doi: 10.9758/cpn.2020.18.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Figure S1. Gini Scores of the top thirty independent features linked to (a) Assortment 1, (b) Assortment 2 and (c) Assortment 3.
Supplementary material.
Data Availability Statement
Present work used the data that was shared here GSE106241.