Abstract
The neurovascular unit, comprised of vascular cell types that collectively regulate cerebral blood flow to meet the needs of coupled neurons, is paramount for the proper function of the central nervous system. The neurovascular unit gatekeeps blood-brain barrier properties, which experiences impairment in several central nervous system diseases associated with neuroinflammation and contributes to pathogenesis. To better understand function and dysfunction at the neurovascular unit and how it may confer inflammatory processes within the brain, isolation and characterization of the neurovascular unit is needed. Here, we describe a singular, standardized protocol to enrich and isolate microvessels from archived snap-frozen human and frozen mouse cerebral cortex using mechanical homogenization and centrifugation-separation that preserves the structural integrity and multicellular composition of microvessel fragments. For the first time, microvessels are isolated from postmortem ventromedial prefrontal cortex tissue and are comprehensively investigated as a structural unit using both RNA sequencing and Liquid Chromatography with tandem mass spectrometry (LC-MS/MS). Both the transcriptome and proteome are obtained and compared, demonstrating that the isolated brain microvessel is a robust model for the NVU and can be used to generate highly informative datasets in both physiological and disease contexts.
Keywords: Neurovascular unit, Blood-Brain Barrier, Microvessels, Postmortem, RNA sequencing, LC-MS/MS
Highlights
-
•
Method presents singular protocol to isolate microvessels from postmortem and frozen mouse cortex.
-
•
High yield of microvessels with preserved integrity were isolated, and compatible with high-throughput techniques.
-
•
The transcriptome and proteome of isolated microvessels from human vmPFC tissue were characterized.
-
•
Limitations of past isolation methods are overcome, and standardized cross-species comparisons are made possible.
1. Introduction
To meet the metabolic needs of the 86 billion neurons in the human brain, an elaborate 400 mile-long microvascular network (Iadecola, 2017; Kisler et al., 2017) supplies blood flow to the deep structures of the cerebral hemispheres and gatekeeps blood-brain barrier (BBB) properties. Extensive research efforts have underscored the BBB as a highly selective cellular system. Ultrastructurally, the BBB consists of continuous non-fenestrated brain microvascular endothelial cells (BMECs) that precisely regulate movement between the blood and brain interface through the expression of specialized solute carriers and efflux transporters (Daneman, 2012; Betz and Goldstein, 1978; Betz et al., 1980; Cordon-Cardo et al., 1989; Thiebaut et al., 1989; Loscher and Potschka, 2005a; Mittapalli et al., 2010; Zlokovic, 2008). BMECs, along with astrocytic endfeet and mural cells (pericytes or smooth muscle cells), positioned at the vascular basement membrane, are the cell types that comprise the neurovascular unit (NVU) (McConnell et al., 2017) and work in concert to implement coordinated vascular responses to central and peripheral signals. Such responses include the continuous delivery of oxygen and glucose to neurons (Mintun et al., 2001; Hoge et al., 1999; Fox et al., 1988; De Vivo et al., 1991; Farrell and Pardridge, 1991; Gerhart et al., 1989), homeostatic maintenance of the brain (Jeong et al., 2006; Gendelman et al., 2009; Wilhelm et al., 2007; Mokgokong et al., 2014; Smith and Rapoport, 1986), the regulation of cerebral blood flow (Mathiesen et al., 1998; Caesar et al., 2003; Iadecola, 1993; Roy and Sherrington, 1890; Fergus and Lee, 1997) and clearance of interstitial fluid (Verheggen et al., 2018; Deane et al., 2009; Iliff et al., 2012; Iliff et al.). An underappreciated lens through which to investigate disease, NVU dysfunction contributes to cognitive decline in Aβ and tau pathology (Iturria-Medina et al., 2016; Montagne et al., 2015; Sweeney et al., 2015; Arvanitakis et al., 2016; Toledo et al., 2013; Rosenberg, 2014), traumatic brain injury (Stein et al., 2002; Schwarzmaier et al., 2010; Dietrich et al., 1994; del Zoppo and Mabuchi, 2003; Schroder et al., 1998; von Oettingen et al., 2002; Shapira et al., 1993; Baldwin et al., 1996; Hicks et al., 1997; Baskaya et al., 1997), perivenous myelin lesions presented in multiple sclerosis (Gaitan et al., 2013; Buch et al., 2021; Geraldes et al., 2020; Al-Louzi et al., 2022; Khan et al., 2010; Doepp et al., 2011), as well as multiphasic changes in BBB permeability after stroke (Liu et al., 2018; Lin et al., 2008; Strbian et al., 2008; Durukan and Tatlisumak, 2009; Pillai et al., 2009). Disease states may arise when BBB function can no longer match the needs of the central nervous system (CNS), which confers dire consequences for the ability of the BBB to communicate both with cells within the brain parenchyma and with cells in the periphery. Indeed, the BBB acts as the interface between the brain and peripheral systems through which neuroimmune interactions occur (Quan and Banks, 2007)and is highly responsive to immune activity encroaching the brain. Such interactions include, but are not limited to: regulation of major efflux transporter P-glycoprotein 1 (ABCB1), endothelial Toll-like receptor and NOD-like recepto activation by TNF-α signaling (Erickson and Banks, 2018; Nagyoszi et al., 2010, 2015), as well as modulation of Na–K–Cl cotransporter, which is critical for cerebral ionic homeostasis, by IL-6 secretion from astrocytes (Sun et al., 1997). The BBB itself secretes substances that interact with the neuroimmune system, including cytokines, prostaglandins, and nitric oxide (Fabry et al., 1993; Mandi et al., 1998; McGuire et al., 2003; Reyes et al., 1999). These substances may be constitutively expressed or pathologically induced, as shown by interleukin-8 in HIV infection (Hofman et al., 1999), invasion of autoreactive CD4+ T cells in multiple sclerosis (Traugott et al., 1983), microglial release of TNFα with cocaine exposure (Lewitus et al., 2016), and nitric oxide release in Alzheimer's disease (Dorheim et al., 1994). Because of its bipolar nature, dysfunction of the BBB can arise from, and be further aggravated by, either the CNS or peripheral compartments. Evidence of neurovascular dysfunction has similarly been observed in bipolar disorder (Kamintsky et al., 2020), schizophrenia (Kirkpatrick and Miller, 2013; Axelsson et al., 1982; Campana et al., 2023; Goldwaser et al., 2022), major depressive disorder (Torres-Platas et al., 2014; Gal et al., 2023; Najjar et al., 2013), and Parkinson's disease (Al-Bachari et al., 2020; Fowler et al., 2021).
Our understanding of neurovascular development and function has been advanced largely by mouse models. Functional characteristics of the BBB are regulated at the transcriptomic level and, in recent years, different methodologies have been employed to investigate the neurovascular transcriptome. Single-cell sequencing studies have leveraged transgenic-reporter claudin-5-GFP (Vanlandewijck et al., 2018; He et al., 2018), Tie2–eGFP (Zhang et al., 2014), and Pdgfrb-eGFP (He et al., 2016a) mouse lines in conjunction with fluorescence-activated cell sorting (FACS) to generate highly informative transcriptomic datasets of mouse BMECs and other vascular cell types, whereas other studies have carried out RNA sequencing of BMECs isolated from Rosa-tdTomato; VE-Cadherin-CreERT2 mouse models of stroke, multiple sclerosis, traumatic brain injury and seizure (Munji et al., 2019). While past mouse data have provided precious insight into defining core NVU gene expression and underscores the relevance of transcriptomic profiling for better understanding neurovascular function (and dysfunction), recent breakthroughs demonstrate that there are numerous species-specific differences between mouse and human neurovasculature, including solute carrier and efflux transporter expression (Munji et al., 2019; Garcia et al., 2022; Yang et al., 2022). Such findings reveal the partial utility of animal models for studying disease of the human CNS. Due to the scarcity of well-preserved human brain tissue available for research, transcriptomic profiling in human brain samples has been considerably more limited, leaving the investigation of vascular cells neglected in favour of non-vascular cell types, such as neurons and oligodendrocytes. In addition, single-cell or single-nucleus sequencing used to profile expression in all cell populations yield very low populations of endothelial cells and pericytes from human adult and embryonic cortex samples depiste vessel density ranging between 361 and 811 vessels/mm2 (Klein et al., 1986; Wu et al., 2004; Weber et al., 2008) at an overall endothelial cell density of 4504 ± 2666 cells/mm2 (Ventura-Antunes et al., 2022). Such techniques seem to deplete vascular cells/nuclei for reasons that are not understood and have impeded analysis of human neurovascular transcriptomes (Nagy et al., 2020; Velmeshev et al., 2019; Mathys et al., 2019; Grubman et al., 2019; Jakel et al., 2019). Recently, detailed transcriptome-wide atlases of human and mouse brain vascular nuclei were generated by two independent groups (Garcia et al., 2022; Yang et al., 2022), both addressing the underrepresentation of vascular cell types and elaborating on species-specific differences in NVU gene expression. Such progress and tools deepen our understanding of human NVU function, yet certain limitations that persist challenge further progress in the field: single-cell and single-nucleus sequencing remains inaccessible to many due to high costs and lack of bioinformatic expertise. Moreover, there is heavy reliance on brain banks for well-characterized frozen human brain tissue, which also requires considerable adaptation of techniques initially optimized for fresh tissue and creates even further disparity in how mouse and human brain tissue are utilized, even with the same experimental question in mind. The biological and bioinformatic biases these experimental decisions create and their extent are unknown. Understanding the molecular mechanisms of NVU dysfunction can be achieved by examining gene expression changes in brain microvessels in different disease contexts; and the limited number of existing human neurovascular datasets motivates transcriptomic characterization of more human samples. While it is understood that BMECs perform the BBB function and that other vascular-associated cell types critically regulate that function, the study of microvessels as a preserved unit provides greater insight into the neurovasculature in a manner that dissociated cell types cannot. To this aim, we use a singular, standardized protocol to enrich and isolate microvessels from archived snap-frozen human and frozen mouse cerebral cortex using mechanical homogenization and centrifugation-separation that is gentle enough to dissociate brain tissue while preserving the structural integrity and multicellular composition of microvessel fragments.
The common issue of multiple or contaminating cell types in samples used for tissue-derived RNA sequencing has been largely eliminated by single-cell workflows. However, single-cell and single-nucleus workflows introduce other significant challenges: measurements typically suffer from large fractions of observed zeros, possibly due to technical limitations or randomness (Hicks et al., 2018; Bacher and Kendziorski, 2016). Moreover, tissue dissociation and storage biases can induce unwanted transcriptomic alterations and cell type composition differences (Denisenko et al., 2020). Because of this, bulk and single-cell sequencing are complementary strategies in which the former approach warrants a versatile and effective method for isolating the NVU from human brain. Several approaches attempting to investigate the isolated NVU have been developed in the past, albeit with major drawbacks: FACS-sorting with antibodies against PECAM1/CD31 and CD13 (targeting the endothelial cell membrane and pericyte cell membrane, respectively) require fresh brain tissue (Yang et al., 2022), as do other iterations of microvessel isolation for cell culture expansion (Navone et al., 2013; van Beijnum et al., 2008). Opting instead for selective capture of endothelial and other vascular-associated cells from frozen human brain by laser capture microdissection (LCM) (Kinnecom and Pachter, 2005; Mojsilovic-Petrovic et al., 2004; Harris et al., 2008; Song et al., 2020a) demands considerable optimization if microvessels are to be used for high-throughput applications downstream (Almeida and Turecki, 2022). Finally, past attempts at microvessel isolation from frozen brain homogenates have only yielded samples suitable for qPCR and Western blot (Bourassa et al., 2019), and more comprehensive knowledge obtained from high-throughput techniques is currently lacking from such studies. Critically, microvessels isolated using the described method are in high yield, possess all major vascular-associated cell types, and maintain their in situ cellular structure, making them suitable for characterization using high-throughput techniques. The advantages of this simple protocol are manifold: it does not require the experimental setup needed by single-nucleus sorting, nor does it require transgenic mice (Lee et al., 2019), enzymatic dissociation (Crouch and Doetsch, 2018; Lee et al., 2019; Spitzer et al., 2023), or fresh brain tissue (Crouch and Doetsch, 2018; Lee et al., 2019; Spitzer et al., 2023). Importantly, this is the first standardized microvessel isolation method demonstrated to work with snap-frozen brain tissue and that is compatible across high-throughput downstream applications, removing unknown biases introduced by the use of varied isolation methods.
We have successfully applied the described protocol to postmortem ventromedial prefrontal cortex (vmPFC) tissue from individuals having died suddenly with no neurological or psychiatric disorder as well as mouse forebrain tissue, as proof of concept that the same procedure can be used in both species. To demonstrate the utility of microvessels isolated from postmortem vmPFC tissue, we processed 5 samples of extracted total RNA and 3 samples of extracted total protein using RNA sequencing and liquid chromatography with tandem mass spectrometry (LC-MS/MS), respectively. Moreover, we sorted endothelial nuclei from isolated microvessels using fluorescence-activated nuclei sorting (FANS) as proof of concept that specific neurovascular cell types may be further purified if needed. Bioinformatic processing and analysis of human transcriptomic and proteomic data indicated that isolated samples showed major enrichment for BMEC, pericyte, SMC, and astrocytic endfeet components at both the mRNA and protein level, generating the first multi-omic datasets from human brain microvessels.
2. Materials
2.1. Biological materials
2.1.1. Human cortex
This study was approved by the Douglas Hospital Research Ethics Board and written informed consent from next of kin was obtained for each individual included in this study. For each individual, the cause of death was determined by the Quebec Coroner's Office and medical records were obtained. Samples were obtained from Caucasian individuals having died suddenly with no neurological or psychiatric disorder (Table 1). Postmortem brain tissues were provided by the Douglas–Bell Canada Brain Bank (www.douglasbrainbank.ca). Frozen grey matter samples were dissected from the vmPFC (Brodmann area 11) by expert brain bank staff stored at −80 °C. The postmortem interval (PMI) is a metric for the delay between an individual's death, the collection and processing of the brain. To assess RNA quality, RNA integrity number (RIN) was measured for brain samples using tissue homogenates, with an average value of 5.34. A total of 5 subjects were subjected to RNA-sequencing and 3 subjects were subjected to Liquid Chromatography with tandem mass spectrometry (LC-MS/MS).
Table 1.
Background information on human subjects whose vmPFC tissue was studied. Information includes sex, age, cause of death, postmortem interval (PMI), and tissue pH.
| Experiment type | Brain ID | Sex | Age (years) | Cause of death | PMI (hours) | Tissue pH |
|---|---|---|---|---|---|---|
| RNA seq | 1 | Female | 76 | Polytrauma (accident) | 26.5 | 6.5 |
| RNA seq | 2 | Female | 51 | Pulmonary embolism | 111.3 | 6.5 |
| RNA seq | 3 | Male | 26 | Polytrauma (car accident) | 12.0 | 6.8 |
| RNA seq | 4 | Female | 28 | Undetermined | 80.0 | 6.5 |
| RNA seq | 5 | Female | 45 | Pulmonary embolism | 39.5 | 5.7 |
| Mass spec | 6 | Male | 45 | Gun wound | 20.5 | 6.6 |
| Mass spec | 7 | Male | 54 | Cardiac arrest | 25.3 | 6.6 |
| Mass spec | 8 | Male | 63 | Fall from several metres | 13.0 | 6.8 |
Note: All experiments involving the use of human brain samples must be performed in accordance with the relevant institutional and national regulations. Use of postmortem tissues was approved by the Institutional Review Board of the Douglas Hospital.
2.1.2. Mouse cortex
Male C57BL/6J mice (n = 2) aged between 120 and 126 days of age were bred, housed, and cared for in accordance with the Canadian Council on Animal Care guidelines (CCAC; http://ccac.ca/en_/standards/guidelines), and all methods were approved by the Animal Care Committee from the Douglas Institute Research Center under protocol number 5570. Mice were housed in standard conditions at 22 ± 1 °C with 60% relative humidity, and a 12-h light-dark cycle with food and water available ad libitum (Isingrini et al., 2017). Following pertinent guidelines and regulations, the mice were anesthetized via intraperitoneal injection of ketamine (10 mg/ml)/xylazine (1 mg/ml) and transcardially perfused with cold PBS 1X. The frontal cortices were removed and immediately frozen in liquid nitrogen and then stored at −80 °C.
2.2. Reagents
-
•
Sucrose (Fisher Scientific, cat. no. S5-500)
-
•
Bovine serum albumin (Sigma-Aldrich, cat. no. A3912)
-
•
DEPC-treated water (Invitrogen™, cat. no. 46–2224)
-
•
SIGMAFAST™ BCIP®/NBT tablets (Sigma-Aldrich, cat. no. B5655-25TAB)
-
•
Ethanol 70% (vol/vol) and 100% (Sigma-Aldrich)
-
•
Methanol 100% (Sigma-Aldrich)
-
•
1X Phosphate buffered saline (Wisent Bioproducts, D-PBS, cat. no. 311-425-CL)
-
•
Single Cell RNA Purification Kit (Norgen Biotek Corp., cat. no. 51800)
-
•
TapeStation RNA ScreenTape (Agilent, cat. no. 5067–5576) or TapeStation High Sensitivity RNA ScreenTape (Agilent, cat. no. 5067–5579)
-
•
TapeStation RNA ScreenTape sample buffer (Agilent, cat. no. 5067–5577) or TapeStation High Sensitivity RNA ScreenTape sample buffer (Agilent, cat. no. 5067–5580)
-
•
SMARTer Stranded Total RNA-Seq Kit v3 - Pico Input Mammalian (Takara Bio Inc., cat. no. 634485)
-
•
NucleoMag NGS Clean-up and Size Select beads (Takara Bio Inc., cat. no. 744970.50)
-
•
0.5% Triton X dissolved in PBS (Triton X-100 from Thermo Fisher Scientific, cat. no. AAA16046AE)
-
•
Normal donkey serum (Jackson ImmunoResearch Laboratories Inc., cat. no. 017-000-121)
-
•
Recombinant Alexa Fluor® 647 Anti-ERG antibody [EPR3864] (Abcam, cat. no. ab196149)
-
•
Anti-Aquaporin 4 antibody [4/18] (Abcam, cat. no. ab9512)
-
•
Anti-Claudin 5 antibody (Abcam, cat. no. ab15106)
-
•
Anti-Laminin antibody (Sigma-Aldrich, cat. no. L9393)
-
•
Anti-Myelin Basic Protein antibody (BioLegend, cat. no. SMI-99P)
-
•
Anti-NeuN antibody, clone A60 (Sigma-Aldrich, cat. no. MAB377)
-
•
Anti-PDGFRB monoclonal antibody (G.290.3) (Thermo Fisher Scientific, cat. no. PIMA515143)
-
•
Anti- PECAM-1 antibody (JC70) (Santa Cruz Biotechnology Inc., cat. no. sc-53411)
-
•
Anti-Vimentin antibody [RV202] (Abcam, cat. no. ab8978)
-
•
VECTASHIELD® Antifade Mounting Medium with DAPI (Vector Laboratories, cat. no. H-1200-10)
-
•
VECTASHIELD® Vibrance Antifade Mounting Media (Vector Laboratories, cat. no. H-1700-10)
-
•
Single Cell RNA Purification Kit (Norgen Biotek Corp., cat. no. 51800)
-
•
Hoechst 33342 (Invitrogen™, cat. no. H3570)
-
•
cOmplete™ EDTA-free Protease Inhibitor Cocktail (Roche, cat. no. 04693132001)
-
•
Phosphatase Inhibitor Cocktail 2 (Sigma-Aldrich, cat. no. P5726)
-
•
Pierce™ BCA Protein Assay Kit (Pierce Biotechnology Inc., cat. no. 23225)
-
•
Sodium dodecyl sulfate (SDS; Sigma-Aldrich)
-
•
100 mM TRIS (pH 7.8; Sigma-Aldrich)
2.3. Equipment
-
•
Vortex mixer
-
•
Lite High Speed Centrifuge Tubes, 15 mL (FroggaBio, cat. no. TL15-500B)
-
•
Lite High Speed Centrifuge Tubes, 50 mL (FroggaBio, cat. no. TL50-500B)
-
•
Costar® Stripette® serological pipettes (10 ml capacity, Corning Inc., cat. no. CLS4488)
-
•
GentleMACS™ C Tubes (Miltenyi Biotec, cat. no. 130-093-237)
-
•
GentleMACS™ Dissociator (Miltenyi Biotec, cat. no. 130-093-235)
-
•
Point-tip forceps
-
•
Razor blades
-
•
Flat-ended spatula
-
•
Weighing boats
-
•
Analytical Weighing scale
-
•
Low Protein Binding Microcentrifuge Tubes, 1.5 ml (Thermo Fisher Scientific, cat. no. 90410)
-
•
Fisherbrand™ Sure One™ Low Retention Non-Filtered Pipette Tips, 1000 μL (Fisher Scientific, cat. no. 02-707-026)
-
•
Dry ice
-
•
Wet ice
-
•
Refrigerated benchtop centrifuge for 15 mL tubes (Beckman Coulter, model Allegra X–14R)
-
•
Refrigerated benchtop centrifuge for 1.5 mL tubes (Eppendorf, model 5430)
-
•
Vacuum-aspiration system
-
•
FlowTubes™ with strainer cap (Canada Peptide, cat. no. FCT-9005)
-
•
Oven (37 °C) (Fisher Scientific, model Fisherbrand™ Isotemp™)
-
•
Multipurpose Digital Shaker (Mandel Scientific Inc., model Labnet Orbit 1000)
-
•
Nunc™ Lab-Tek™ II Chamber Slide™ System (Thermo Fisher Scientific, cat. no. 154453)
-
•
Fisherbrand™ Premium Cover Glasses (Fisher Scientific, cat. no. 12-548-5P)
-
•
Agilent TapeStation (Agilent Technologies, model 2200 TapeStation)
-
•
Flow Cytometer (BD Biosciences, model BD FACSAria™ Fusion)
-
•
Thermocycler (Applied Biosystems Corporation, model ProFlex PCR)
-
•
Probe-based sonicator (Fisher Scientific, model Thermo Sonic Dismembrator)
-
•
Heat block with tube rack
2.4. Reagent setup
2.4.1. Homogenization Buffer preparation
Prepare fresh Homogenization Buffer (1M sucrose + 1% BSA dissolved in DEPC-treated water) and keep at 4 °C until thoroughly chilled. If transcriptomic techniques are to be used downstream to microvessel isolation, it is recommended to use DEPC-treated water as opposed to Millipore water, as early work done by our group has demonstrated that DEPC treatment preserves an approximate 10% of transcripts that are otherwise lost when processing samples for RNA sequencing (Supplementary Table 1). DEPC treatment functions as a cost-effective RNAse inhibitor when homogenizing brain tissue in large volumes of buffer.
2.4.2. BCIP/NBT substrate preparation
If microvessel detection parallel to isolation is desired, BCIP/NBT (5-bromo-4-chloro-3'-indolyphosphate and nitro-blue tetrazolium) can be used as a chromogenic substrate for endothelial enzyme alkaline phosphatase. SIGMAFAST™ BCIP®/NBT tablet (Sigma-Aldrich, Missouri, United States) should be crushed and then dissolved in the fresh Homogenization Buffer according to the manufacturer's instructions (1 tablet per 10 ml of solution). Crushing the tablet first encourages faster dissolution in the highly viscous buffer. Within the brain, alkaline phosphatase is localized to cerebral blood vessels (Shimizu, 1950; Leduc and Wislocki, 1952; Bourne, 1958; Becker et al., 1960; Bannister and Romanul, 1963; Romanul and Bannister, 1962; Ball et al., 2002). BCIP is hydrolyzed by the alkaline phosphatase expressed exclusively in endothelial cells to form a blue intermediate that is then oxidized by NBT to produce a dimer, leaving an intense insoluble purple dye.
2.4.3. Protein extraction buffer
Prepare 100 mM TRIS at pH 7.8 with 5% final volume of sodium dodecyl sulfate (SDS).
3. Method
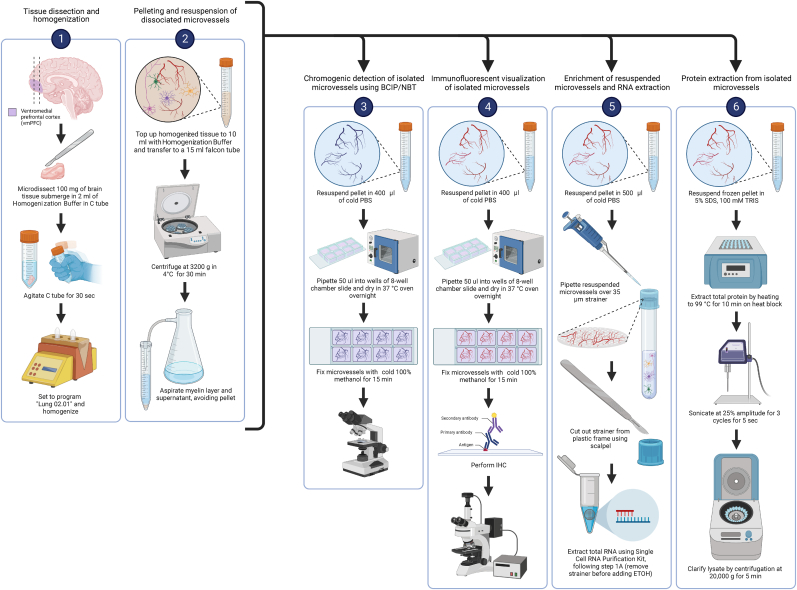
By virtue of a preserved basement membrane, the structures isolated using the described method contain all neurovascular-associated cell types and, therefore, are referred to as “microvessels”. The following protocol describes the specific steps used to isolate and enrich microvessels with retained in situ morphology, which is achieved by using semi-automated dissociation of microdissected brain tissue into a homogenate followed by low-speed centrifugation (schematic overview in Fig. 1). These steps have been optimized to isolate microvessels from 100 mg of frozen brain tissue, processing a maximum of 4 samples at a time.
Fig. 1.
Stage 1: After the vmPFC is dissected from a coronal slice containing the frontal lobe, 100 mg of vmPFC tissue is more precisely microdissected using a sterile blade or scalpel. Afterwards, the vmPFC tissue sample is dissociated in Homogenization Buffer using the GentleMACS™ Dissociator. Stage 2: Following dissociation, the sample is centrifuged at 3200 g for 30 mins in order to pellet dissociated microvessels. Additional downstream applications can be applied using microvessel-enriched pellets as shown in stages 3–6 by BCIP/NBT detection, immunofluorescent visualization, total RNA extraction, and total protein extraction, respectively. Image generated using BioRender.
3.1. Tissue microdissection
Timing ∼5 mins per sample
-
1.
Set up dissection station adjacent to an analytical balance needed to weigh microdissected brain tissue. Wash all dissection tools (point-tip forceps, razor blades, flat-ended spatula), bench, and any surface used in 70% (vol/vol) ethanol prior to microdissection. Sufficiently chill a fresh weigh boat, 1.5 ml Eppendorf tubes, and point-tip forceps on dry ice while leaving razor blades at room temperature.
-
2.
Cut tissue using a razor blade and weigh 100 mg of frozen tissue per sample using an analytical scale. Keep tissue on dry ice while microdissecting to minimize degradation and transfer to a chilled 1.5 ml Eppendorf tube. Clean razor, forceps, and spatula with 70% ethanol (vol/vol) and use a fresh weigh boat between samples.
Note: Postmortem human tissue can contain transmissible pathogens. Take appropriate precautions, including wearing PPE, and seek medical attention if the scalpel breaks skin.
3.2. Tissue homogenization and cellular fractionation
Timing 45 mins – 1h
-
3.
Set up homogenization station adjacent to benchtop gentleMACS™ Dissociator. Place gentleMACS™ C Tubes, 15 ml falcon tubes, and Homogenization Buffer on ice while keeping microdissected brain tissue samples on dry ice.
Note: Experimental objective must be decided at this step. If either transcriptomic, proteomic investigation, or immunofluorescent visualization is desired, then prepare Homogenization Buffer without BCIP/NBT tablet. If detection of microvessels is to be performed using BCIP/NBT substrate, BCIP/NBT must be prepared in the Homogenization Buffer, as described above.
-
4.
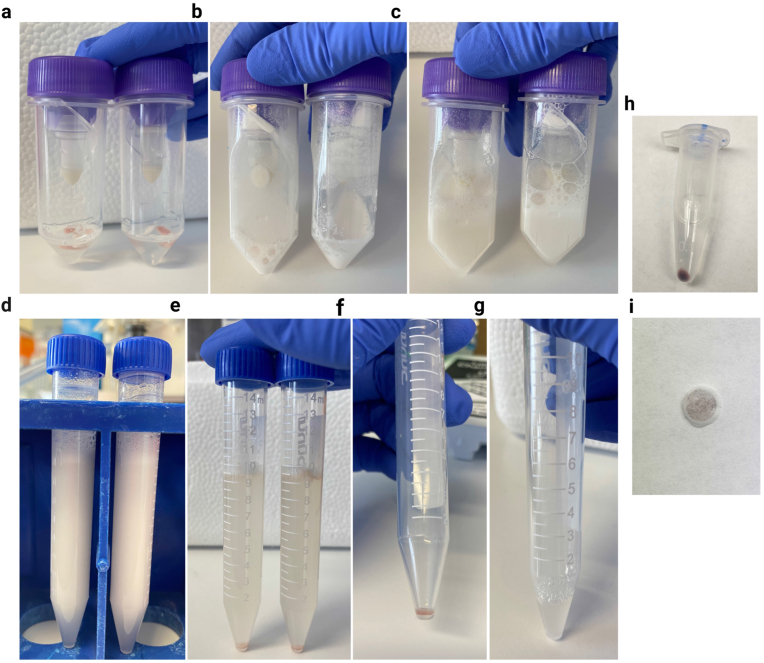
Using a serological pipette, transfer 2 ml of cold Homogenization Buffer to a gentleMACS™ C Tube and then transfer 100 mg brain tissue from 1.5 ml Eppendorf tube to gentleMACS™ C Tube. Ensure that tissue is fully submerged in Homogenization Buffer while gently agitating the tube for 30 seconds to encourage thawing and osmotic equilibrium (Fig. 2a). See Troubleshooting Step 1 in Supplementary Table 3.
Fig. 2.
a-g) Overview of experimental steps taken to collect and resuspend pelleted microvessels (BCIP/NBT staining is omitted). a) 100 mg of vmPFC tissue submerged in 2 ml of Homogenization Buffer. b) 100 mg of vmPFC tissue after homogenization. c) Lysate topped-up to 10 ml with Homogenization Buffer. d) Lysate transferred to 15 ml falcon tube. e-f) After centrifugation at 3200g for 30 min, a microvessel-enriched pellet forms at the bottom of the tube. g) microvessel-enriched pellet is resuspended in 500 ul of PBS. h-i) Output of experimental steps when BCIP/NBT staining is used. h) Example of BCIP/NBT-stained microvessel pellet, which lends a purple colour to the pellet. i) BCIP/NBT-stained microvessels trapped within the meshwork of a cellular strainer.
-
5.
Snap gentleMACS™ C Tube into the Dissociator and run the rotating paddle on program Lung 02.01 (Fig. 1). For the first half of the Lung 02.01 program, the speed of the rotating paddle is gradually increased to its maximum in a clockwise direction, and then in an anti-clockwise direction for the second half of the program. The duration of the program is 37 seconds.
-
6.
After tissue homogenization is complete, return gentleMACS™ C Tube to ice and pipette an additional 8 ml of cold Homogenization Buffer into the tube, topping up the homogenate to 10 ml. Gently invert to mix and collect homogenate (Fig. 2b–c).
-
7.
Using a serological pipette, transfer the 10 ml of homogenate to a chilled 15 ml falcon tube, including any foam produced during paddle rotation (Fig. 2d).
-
8.
Gently invert to mix homogenate one more time before centrifugation in order to prevent the formation of a foamy seal atop the homogenate when left sitting. Centrifuge homogenates at 3200 g for 30 mins at 4 °C. Once centrifugation is complete, a microvessel-enriched pellet will form at the bottom of the falcon tube (Fig. 2e). See Troubleshooting Step 2 in Supplementary Table 3.
-
9.
Carefully vacuum-aspirate the supernatant (which may include an upper layer of clumped dissociated myelin, similar to milk skin on top of boiled milk) without disturbing the microvessel-enriched pellet (Fig. 2e–f).
-
10.
Gently resuspend the pellet in 400μl of cold PBS and pipette 50 ul of resuspended pellet into each well of an 8-well chamber slide (Nunc™ Lab-Tek™ II Chamber Slide™ System, Thermo Scientific™, Massachusetts, United States) (Fig. 2g). Leave the chamber slide open-faced in a 37 °C oven overnight. Once the PBS has evaporated, microvessels will dry flush to the surface of the slide.
3.3. Detection of microvessels from the enriched pellet using BCIP/NBT substrate
Timing 35–40 mins
Yield and stability of microvessel isolation can be assessed using chromogenic substrate BCIP/NBT to visualize endothelial enzyme alkaline phosphatase. When detecting microvessels with BCIP/NBT, the process of staining microvessels occurs throughout homogenization and centrifugation steps (steps 5–11) (Fig. 2h).
-
11.
After resuspension of the pellet (section 3.2, step 10), fix microvessels by covering them to a depth of 2–3 cm with ice-cold 100% methanol. Cover chamber slide with lid and allow cells to fix for 15 min on ice or at 4 °C.
-
12.
Aspirate 100% methanol and wash wells in 1X PBS three times for 5 min.
-
13.
Remove the media chamber carefully with the provided chamber removal tool, according to the manufacturer's instructions.
-
14.
Mount the microvessels using VECTASHIELD Vibrance Antifade Mounting Media (California, United States) and place coverslip.
3.4. Detection of microvessels from the enriched pellet using immunohistochemistry
Timing 4–4.5 hours, spread across 3 days (2 overnight incubations)
Greater immunophenotypic characterization of isolated microvessels can be carried out following the resuspension of additional microvessel-enriched pellets. Primary antibodies raised against canonical expression markers for BMECs (Laminin and PECAM1), tight junctions (CLDN5), pericytes (PDGFRβ), smooth muscle cells (Vimentin), and astrocytic endfeet (AQP4) are utilized, along with appropriate fluorophore-conjugated secondary antibodies to characterize collected microvessels thoroughly.
-
15.
After resuspension of the pellet (section 3.2, step 10), fix microvessels by covering them to a depth of 2–3 cm with ice-cold 100% methanol. Cover chamber slide with lid and allow cells to fix for 15 min on ice or at 4 °C.
Note: Immunofluorescent visualization must omit any steps in which the microvessel-enriched pellet is filtered through a cellular strainer because microvessels cannot be released from the strainer and, therefore, cannot be mounted onto a microscope slide.
-
16.
Aspirate 100% methanol and wash wells in 1X PBS three times for 5 min.
-
17.
Incubate microvessels in blocking buffer (1% BSA + 0.5% Triton X dissolved in PBS) under agitation for 60 min at 4 °C.
-
18.
Aspirate blocking solution and pipette into each well 500 ul of primary antibody dilution (1:500 in 1% BSA + 2% normal donkey serum + 0.5% Triton X dissolved in PBS) and incubate under agitation overnight at 4 °C.
-
19.
Aspirate primary antibody dilution and wash wells in 1X PBS three times for 5 min.
-
20.
Incubate microvessels in fluorophore-conjugated secondary antibody dilution (1:500–1000 in 1% BSA + 2% normal donkey serum + 0.5% Triton X dissolved in PBS) and incubate under agitation for 2 hours at room temperature, protected from all light.
-
21.
Aspirate secondary antibody dilution and wash wells in 1X PBS three times for 5 min.
-
22.
Remove the media chamber carefully with the provided chamber removal tool, according to the manufacturer's instructions.
-
23.
Mount the microvessels using VECTASHIELD Antifade Mounting Medium with DAPI (California, United States) and place coverslip.
To expand upon experimental applications possible with isolated human brain microvessels, we adapted several downstream techniques including: total RNA extraction, RNA library construction for downstream RNA sequencing, fluorescence-activated nuclei sorting (FANS) of endothelial nuclei, and total protein extraction for downstream Liquid Chromatography with tandem mass spectrometry (LC-MS/MS).
3.5. RNA extraction from isolated microvessels
Timing 30–40 mins
Total RNA extraction from isolated microvessels begins with resuspension of the microvessel-enriched pellet (section 3.2, step 10), followed by filtration through a 35 μm cellular strainer in which microvessels become trapped. From these entrapped microvessels, total RNA is immediately extracted using the Single Cell RNA Purification Kit (Norgen Biotek Corp., Ontario, Canada). After total RNA is successfully extracted, a stopping point is possible during which extracted RNA is frozen and stored at −80 °C.
-
24.
Gently resuspend the pellet (section 3.2, step 10) in 500 μl of cold PBS and gradually pipette through a 35 μm Strainer Cap for FlowTubes™ (Canada Peptide, Quebec, Canada) using vacuum-aspiration underneath to encourage filtration. The result is intact microvessels trapped within the strainer mesh, where smaller cellular debris and free-floating nuclei have passed through. See Troubleshooting Step 3 in Supplementary Table 3.
-
25.
Using a flat-ended spatula and point-tip forceps, swiftly push and pull out the strainer mesh, removing it from its plastic frame (Fig. 2i).
-
26.
Immediately submerge the mesh into 100 μl of RL buffer in a 1.5 ml Eppendorf tube, according to step 1A of the manufacturer's protocol.
-
27.
Transfer 100 μl of fresh 70% ETOH, pipette 10 times to wash through the mesh (you may briefly vortex for good measure).
-
28.
Discard the mesh and follow steps according to the manufacturer's instructions, including on-column DNase digestion.
-
29.
Quantify RNA using the Agilent TapeStation 2200 or other quantification system of choice (for total RNA concentration and RIN, see Supplementary Table 2). Freeze and store RNA sample at −80 °C.
STOPPING POINT: tubes of total RNA should be frozen and stored at −80 °C.
3.6. Protein extraction from microvessel-enriched pellet or strained microvessels
Timing 1 hour 10 mins–1 hour 15 mins
Total protein has been successfully extracted from either the microvessel-enriched pellet or microvessels stained through a cellular sieve, with one modification to Homogenization Buffer preparation required in which cOmplete™ EDTA-free Protease Inhibitor Cocktail (Roche, Basel, Switzerland) and Phosphatase Inhibitor Cocktail 2 (Sigma-Aldrich, Missouri, United States) are added according to the manufacturer's instructions and based on final volume prepared. With the procedure identical to when using strained microvessels (kept on a strainer), we elaborate protein extraction from frozen microvessel-enriched pellets (pellets stored at −80 °C after section 3.2, step 10). After total protein is successfully extracted, a stopping point is possible during which extracted protein is frozen and stored at −80 °C.
-
30.
Resuspend frozen pellet in 5% sodium dodecyl sulfate (SDS), 100 mM TRIS (pH 7.8), transfer to a 1.5 ml Eppendorf tube and extract protein by heating to 99 °C for 10 minutes on a heat block with tube rack.
-
31.
Subject resuspended pellets to probe-based sonication using a Thermo Sonic Dismembrator at 25% amplitude for 3 cycles for 5 seconds.
-
32.
Clarify lysates by centrifugation at 20,000 g for 5 minutes.
-
33.
For estimation of protein concentration, aliquot approximately 10% of the sample and dilute to <1% SDS and use for estimation of protein concentration by Pierce™ bicinchoninic acid assay (BCA) Protein Assay Kit (Pierce Biotechnology Inc., Massachusetts, United States).
STOPPING POINT: tubes of total protein should be frozen and stored at −80 °C
Note: Total protein and total RNA extraction from the same microvessel-enriched pellet has not been validated. Two microvessel-enriched pellets per sample should be prepared, one for RNA and one for protein extraction.
3.7. FANS-sorting of endothelial nuclei from microvessel-enriched pellet
Timing 2 hours 5 mins preparation, ∼1–2 hours sorting
If further isolation of BMECs from the microvessel-enriched pellet is desired, FANS-sorting of ETS-related gene (ERG)+ BMECs from the obtained microvessel-enriched pellet can be performed. ERG is a transcription factor whose expression in normal physiological conditions is found exclusively in endothelial nuclei, making it a highly specific pan-endothelial nuclear marker (Nikolova-Krstevski et al., 2009; Miettinen et al., 2011; He et al., 2018; Garcia et al., 2022; Yang et al., 2022). FANS of BMECs can be carried out following the resuspension of the microvessel-enriched pellet (section 3.2, step 10).
-
34.
Resuspend the microvessel-enriched pellet in 500 ul of cold PBS and centrifuge a second time at 500 g for 3 min at 4 °C. Aspirate supernatant, removing any remaining sucrose.
-
35.
For a second time, resuspend the washed pellet in 250 ul of the following antibody solution: recombinant Alexa Fluor® 647 anti-ERG (1:100 dilution; ab196149, Abcam, Massachusetts, United States) in 0.5% BSA + 0.1% Triton in PBS. Transfer to a 1.5 ml Eppendorf tube and incubate under gentle agitation for 2 hours at 4 °C.
-
36.
In the last 10 minutes of the 2-h incubation, add 1 ul of Hoechst 33342 dye (Invitrogen, Massachusetts, United States) to stain nuclear DNA.
-
37.
Filter the suspension through a 35 μm FlowTubes™ with strainer cap and transfer flow-through into a FACS tube.
-
38.
The BD FACSAria™ Fusion Flow Cytometer (BD Biosciences, California, United States) was used to sort our ERG+ population. The gating strategy used for sorting was as follows: doublet discrimination was achieved by gating Hoechst 33342 stained singlets in a FSC-A versus Hoechst-A plot using a 350 nm UV laser and a 450/50 filter. The subsequent ERG+ population was gated in an Alexa Fluor 647-A vs. FSC-A plot using a 640 nm laser in combination with a 730/45 filter. Gating was applied to filter singlets using physical parameters and violet fluorescence (405-nm laser, 525/50 filter). Nonoverlapping gates were adjusted to collect endothelial nuclei based on Alexa Fluor® 647 anti-ERG immunoreactivity (640-nm laser, 730/45 filter). This approach was chosen due to the well-known observation that forward scatter is proportional to size.
4. High-throughput applications of isolated microvessels
4.1. Library construction and RNA-sequencing
Microvessel-enriched pellets yielded an average of 8.54 ug/ul of total RNA per sample (Supplementary Table 2). Libraries were then constructed using the SMARTer Stranded Total RNA-Seq Kit v3 - Pico Input Mammalian (Takara Bio Inc., Shiga, Japan), which features integration of unique molecular identifiers (UMIs). Libraries were constructed using 10 ng of RNA as input, 2 minutes of fragmentation at 94 °C (Applied Biosystems Corporation, model ProFlex PCR), 5 cycles of amplification at PCR1 (addition of Illumina adapters and indexes), 12 cycles of amplification at PCR2 (final RNA-seq library amplification) and clean-up of final library using NucleoMag NGS Clean-up and Size Select beads (Takara Bio Inc., Shiga, Japan). Libraries were then quantified at the Genome Quebec Innovation Centre (Montreal, Quebec) using a KAPA Library Quantification kit (Kapa Biosystems, USA), and average fragment size was determined using a LabChip GX (PerkinElmer, USA) instrument. Libraries were sequenced on the NovaSeq 6000 system (Illumina, Inc., California, United States) using S4 flow cells with 100bp PE sequencing kits.
4.2. Bioinformatic pipeline and analyses of RNA sequencing data
4.2.1. UMI extraction, alignment, de-duplication, metrics and gene counting
RNA sequencing of microvessel libraries yielded an average of ∼72 million reads per library, which were then processed following our in-house bioinformatic pipeline. Briefly, UMI extraction based on fastq files was performed using the module extract of umi_tools (v.1.1.2) (Smith et al., 2017). Reads were then aligned to the Human Reference Genome (GRCh38) using the STAR software v2.5.4b (Dobin et al., 2013) with Ensembl v90 as the annotation file and using the parameters: -twopassMode Basic --outSAMprimaryFlag AllBestScore --outFilterIntronMotifs RemoveNoncanonical --outSAMtype BAM SortedByCoordinate --quantMode TranscriptomeSAM GeneCounts. Resultant bam files were then sorted and indexed using SAMtools (v.1.3.1) (Li et al., 2009), and duplicate reads with the same UMI were removed using the dedup module of umi_tools (v.1.1.2) (Smith et al., 2017). Different metrics,including the fraction of the exonic, intronic, and intergenic reads were calculated using the CollectRnaSeqMetrics module of Picard (version 1.129; Picard2019toolkit, Broad Institute, GitHub repository). The expected counts and the transcripts per million (TPMs) were generated using RSEM (v1.3.3; reverse strand mode) (Li and Dewey, 2011). The number of reads, alignment percentages, genomic contamination, and duplication rate for each sample are shown in the Supplementary data.
4.2.2. Computational deconvolution of RNA sequencing data
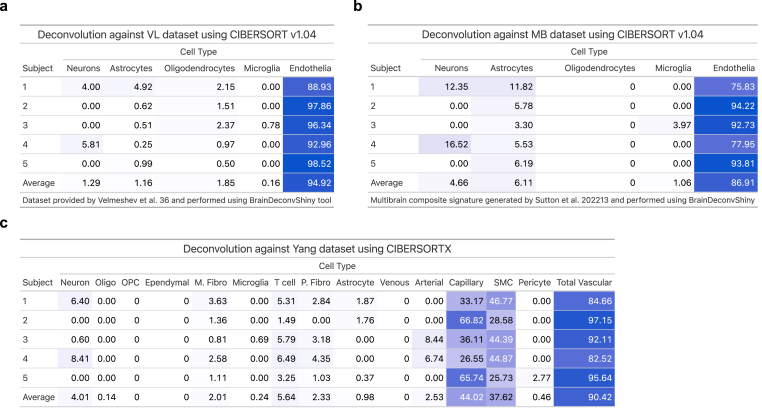
Two approaches were used for computational deconvolution of RNA sequencing data. The first approach was performed using the web tool BrainDeconvShiny (https://voineagulab.shinyapps.io/BrainDeconvShiny/), which implements the best-performing algorithms and all cell type signatures for brain, as well as goodness-of-fit calculations based on benchmark work conducted by Sutton et al. (2022) (Sutton et al., 2022). UMI counts for each gene were converted to transcripts per million (TPMs) to account for the varying length of gene and sequencing depth of each sample, facilitating comparisons across samples. Genes with zero TPMs were removed; 34370 genes from the original 58303 passed this QC criteria and were then used as input into the BrainDeconvShiny tool. Deconvolution was performed twice: the first approach used average expression in control samples from the Velmeshev et al. (2019) (Velmeshev et al., 2019) single nuclei dataset (raw data available through the Sequence Read Archive, accession number PRJNA434002; analyzed data available at https://autism.cells.ucsc.edu) as the reference signature for annotated cell types and CIBERSORT v1.04 algorithm to deconvolute sample profiles and estimate cell type composition. The second approach used the MultiBrain (MB) composite signature (Sutton et al., 2022) generated by averaging the expression signatures of five datasets for five cell types (neurons, astrocytes, oligodendrocytes, microglia, and endothelia). MB was used as cell type signatures for deconvolution of our dataset using CIBERSORT v1.04.
The second approach to deconvolution was performed in-house. Postmortem NVU single-nucleus data generated on the 10X Genomics Chromium system was accessed from Yang et al. (2022; raw sequencing data are accessible on GEO using the accession code GSE163577) (Yang et al., 2022) and used as the reference signature. Seurat (Stuart et al., 2019) was used to pre-process raw count expression data, removing genes with less than 3 cells or cells with less than 200 expressed genes. 23054 genes from a total of 23537 and 141468 nuclei from a total of 143793 passed these QC criteria. Counts per million (CPM) values were averaged across nuclei of each cell type to generate the Yang signature input for the CIBERSORTX tool (Newman et al., 2019).
4.3. Gas Chromatography−Mass spectrometry (LC-MS) with tandem mass tag fractionation
4.3.1. Protein digestion and TMT labelling
Extracted proteins were reduced with 20 mM tris(2-carboxyethyl)phosphine (TCEP) at 60 °C prior to alkylation with 25 mM iodoacetamide at room temperature for 30 minutes in the dark. An equivalent of 10 μg of total protein was used for proteolytic digestion using suspension trapping (S-TRAP). Briefly, samples were acidified with phosphoric (1.3% final concentration) and then diluted 6-fold in STRAP loading buffer (9:1 methanol:water in 100 mM TEAB, pH 8.5). Samples were loaded onto S-TRAP Micro cartridges (Protifi LLC, Huntington, NY) prior to centrifugation at 2000g for 2 minutes and washed three times with 50 μl of STRAP loading buffer. Proteins were digested with trypsin (Sigma Corporation, Kanagawa, Japan) at a 1:10 enzyme to substrate enzyme-to-substrate ratio for 2 hours at 47 °C. Peptides were sequentially eluted in 100 mM TEAB, 0.1% formic acid in water, and 50% acetonitrile and lyophilized to dryness prior to labelling with TMT 10plex reagents according to the vendor's specifications (Thermo Fisher Scientific, Massachusetts, United States).
4.3.2. Offline high-pH reversed-phase fractionation
Labelled peptides were pooled and again lyophilized to dryness, and then reconstituted in 5 mM ammonium formate and fractionated offline by high pH reversed-phase separation using a Waters Xbridge Peptide BEH C18 column (2.1 × 150mm, 2.5 μm) (Waters Corp., Massachusetts, United States) and an Agilent 1290 LC system (Agilent Technologies, California, United States). Binary gradient elution was performed at a flow rate of 400 μL/minute using mobile phase A) 5 mM ammonium formate adjusted with ammonium hydroxide to pH 10, and B) 100% acetonitrile using the following program: 0 min, 0% B; 2min, 0% B; 2.1min, 5% B; 25min, 30% B; 30min, 80% B; 32min, 80% B; 2 min post-run, 0% B. Fractions were collected every 30 seconds and the first and last 7 fractions were concatenated such that even and odd samples were pooled separately, resulting in 20 fractions in total.
4.3.3. LC-MS/MS
Samples were analyzed by data- dependent acquisition (DDA) using an Easy-nLC 1200 online coupled to a Q Exactive Plus (both Thermo Fisher Scientific, Massachusetts, United States). Samples were first loaded onto a pre-column (Acclaim PepMap 100 C18, 3 μm particle size, 75 μm inner diameter x 2 cm length) in 0.1% formic acid (buffer A). Peptides were then separated using a 60-min binary gradient ranging from 3 to 40% B (84% acetonitrile, 0.1% formic acid) on the analytical column (Acclaim PepMap 100 C18, 2 μm particle size, 75 μm inner diameter x 25 cm length) at 300 nL/min. MS spectra were acquired from m/z 350-1500 at a resolution of 70,000, with an automatic gain control (AGC) target of 1 × 106 ions and a maximum injection time of 50 ms. The 15 most intense ions (charge states +2 to +4) were isolated with a window of m/z 1.2, an AGC target of 2 × 104, and a maximum injection time of 64 ms and fragmented using a normalized higher-energy collisional dissociation (HCD) energy of 28. MS/MS spectra were acquired at a resolution of 17,500 and the dynamic exclusion was set to 30 s.
4.3.4. Bioinformatic pipeline and analyses of MS data
DDA MS raw data was processed with Proteome Discoverer 2.5 (Thermo Scientific, Massachusetts, United States) and searched using Sequest HT against a FASTA file containing all reviewed protein sequences of the canonical human proteome without isoforms downloaded from Uniprot (https://www.uniprot.org). The enzyme specificity was set to trypsin with a maximum of 2 missed cleavages. Carbamidomethylation of cysteine was set as static modification and methionine oxidation as variable modification. The precursor ion mass tolerance was set to 10 ppm, and the product ion mass tolerance was set to 0.02 Da. The percolator node was used, and the data was filtered using a false discovery rate (FDR) cut-off of 1% at both the peptide and protein level. The Minora feature detector node of Proteome Discoverer was used for precursor-based label-free quantitation.
5. Results
5.1. Immunophenotypic characterization reveals isolated brain microvessels have preserved morphology and expression integrity
Human brain microvessels were isolated from 5 frozen vmPFC grey matter samples microdissected from healthy individuals who died from peripheral diseases or natural events (Table 1). Because the same basement membrane that maintains the integrity of the endothelium also ensheaths astrocytic endfeet as well as pericytes or smooth muscle cells, it is impossible to isolate solely BMECs and, therefore, isolated microvessels also contain microvessel-associated cell types. Notably, the described method allows for isolation of microvessels from other brain regions (data not shown) and has additionally been performed using the dorsolateral prefrontal cortex (dlPFC, Brodmann area 8/9), the primary visual cortex (Brodmann area 17), as well as the hippocampus (Brodmann area 28).
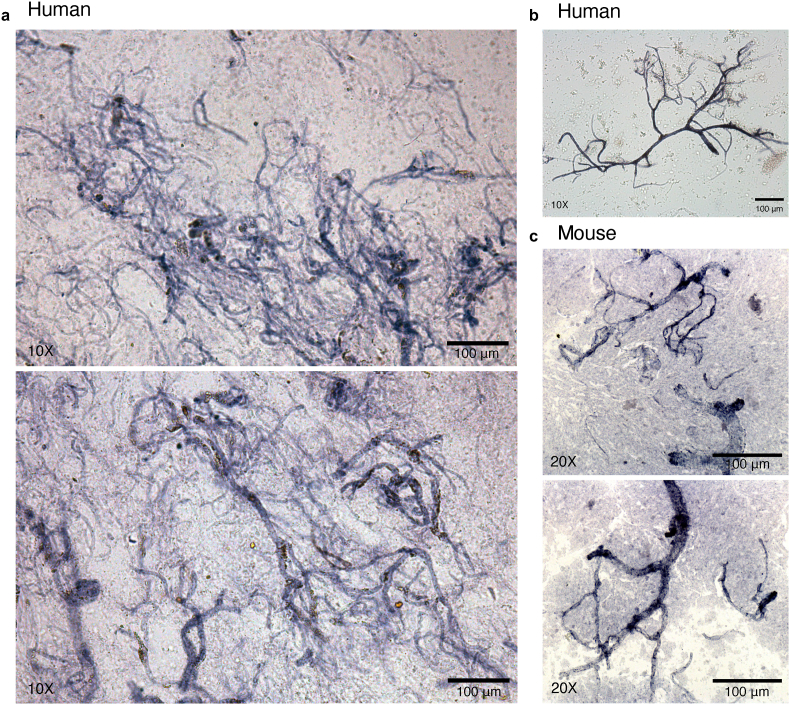
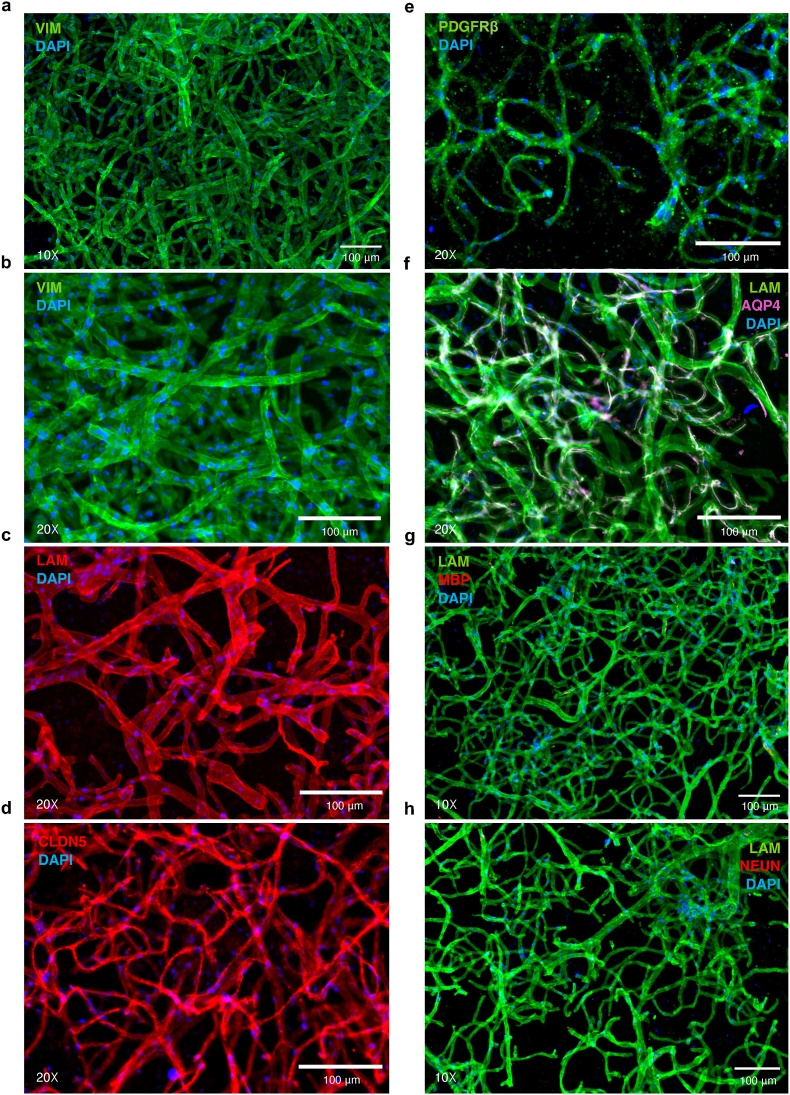
Following isolation of human brain microvessels, chromogenic staining of resuspended pellets using BCIP/NBT (5-bromo-4-chloro-3'-indolyphosphate and nitro-blue tetrazolium), a substrate for endothelial enzyme alkaline phosphatase, demonstrated isolation and enrichment of predominantly microvessels from vmPFC tissue samples (Fig. 3a–b), with similar success when isolating microvessels from mouse cortex (Fig. 3c). The cytoarchitecture of brain microvessels is both complex and comprised of several cell types; thus, following the isolation of brain microvessels, we aimed to characterize the structure and morphological integrity of isolated microvessels. Immunophenotypic characterization of several NVU markers revealed expression of vimentin (VIM; anti-Vimentin antibody RV202, Abcam), laminins (LAM; anti-Laminin antibody L9393, Sigma-Aldrich), claudin 5 (CLDN5, anti-Claudin 5 antibody ab15106, Abcam), platelet-derived growth factor receptor beta (PDGFRβ, anti-PDGFRβ monoclonal antibody G.290.3, Thermo Fisher Scientific) and aquaporin 4 (AQP4, anti-Aquaporin 4 antibody [4/18], Abcam). More precisely, vimentin (Fig. 4a–b), a regulator of actin cytoskeleton primarily in smooth muscle (Chang and Goldman, 2004) and to a lesser extent endothelial cells (Boraas and Ahsan, 2016) and pericytes (Bandopadhyay et al., 2001), as well as laminins (Fig. 4c), the major basement membrane component responsible for signal transduction via interaction with cell surface receptors (Aumailley and Smyth, 1998), are expressed continuously and homogeneously across the entire length of the endothelial surface. CLDN5, a major functional constituent of tight junctions (Greene et al., 2019), was also stained with no apparent discontinuity, suggesting that endothelial tight junctions were preserved (Fig. 4d). Pericyte coverage of BMECs, immunolabeled by regulator of angiogenesis and vascular stability PDGFRβ (Winkler et al., 2010), was detected adhering to the surface of microvessels (Fig. 4e). These results suggest that the overall in situ brain microvessel structure is conserved after isolation.
Fig. 3.
a-c) Isolated microvessels are enriched from brain tissue following the described protocol. a-c) Brightfield images of chromogenically stained microvessels using BCIP/NBT substrate demonstrated isolation and enrichment of predominantly microvessels from postmortem vmPFC tissue samples. b) example of preserved microvessel morphology and integrity isolated from postmortem vmPFC tissue. c) Similar microvessel enrichment was observed when the same protocol was carried out using mouse cortex.
Fig. 4.
a-b) Endothelial and smooth muscle cells immunolabelled for vimentin (green). c) Extracellular matrix laminins expressed by vascular-related cell types are immunolabelled (red). d) Endothelial cell tight junctions immunolabelled for CLDN5 (red). e) Pericytes immunolabelled for PDGFRβ (green). f) AQP4 expressed by the vessel-facing astrocyte membrane is immunolabelled (magenta) and laminins (green). g) Oligodendrocyte marker MBP (red) shows an absence of immunoreactivity in microvessel preparations; similarly, h) neuronal marker NeuN (red) shows an absence of immunoreactivity in microvessel preparations. Nuclei were stained with DAPI (blue) in all micrographs.
Astrocytes serve multiple essential functions in supporting normal brain physiology (Kimelberg and Nedergaard, 2010). This is, in part, due to the extension of astrocytic endfeet that surround approximately 99% of the cerebrovascular surface (Mathiisen et al., 2010) which, in conjunction with pericytes (Winkler et al., 2011), regulate expression of molecules that form the BBB including: tight junction, enzymatic, and transporter proteins (Abbott et al., 2006, 2010; Wolburg et al., 2009). Although astrocytes were not co-isolated with microvessels, their perivascular endfeet are ensheathed within the same vascular basement membrane as endothelial cells and pericytes, making it possible that astrocytic endfeet remained attached after tissue homogenization. Because of this, we further observed astrocyte vascular coverage of isolated microvessels. AQP4, a water channel protein essential for the maintenance of osmotic composition and volume within the interstitial, glial, and neuronal compartments (Nagelhus and Ottersen, 2013; Papadopoulos and Verkman, 2013), is expressed at the vessel-facing astrocytic membrane and superimposes the walls of isolated microvessels (Fig. 4f).
Finally, we examined possible contamination of our microvessel preparations by other cell types found within the brain. Immunolabelling for myelin basic protein (MBP; anti-Myelin Basic Protein antibody, BioLegend) was not detected (Fig. 4g), nor did immunolabelling for neuronal nuclear protein (NeuN; anti-NeuN antibody clone A60, Sigma-Aldrich) reveal the presence of neuronal elements (Fig. 4h). Thus, neurons and oligodendrocytes were consistently not observed to be co-isolated with microvessels. Together, these results indicate that the described protocol allows for the isolation and enrichment of structurally preserved brain microvessel fragments that are comprised of BMECs, astrocytic endfeet, pericytes, and tight junction proteins.
5.2. Computational deconvolution and characterization of transcriptomic data indicates high microvessel yield after isolation
To estimate the enrichment of our microvessel preparations, we performed computational deconvolution using the BrainDeconvShiny tool (https://voineagulab.shinyapps.io/BrainDeconvShiny/) and calculated TPMs as input (Fig. 5a–b) . To demonstrate stability of outcome regardless of resources used, different iterations of deconvolution were performed using the CIBERSORT v1.04 algorithm to estimate cell type composition and either the single-nucleus dataset from Velmeshev et al. (2019; VL) (Velmeshev et al., 2019) or the MultiBrain (MB) dataset from Sutton et al. (2022) (Sutton et al., 2022) as the reference signature. Regardless of the approach used, majority endothelial gene expression was estimated, with an average of 94.92% using the VL dataset (Fig. 5a) and an impressive 86.91% using the MB dataset (Fig. 5b), which is a composite signature generated by quantile-normalising and averaging five previously published datasets. Some contamination from neurons (1.29% and 4.66%, respectively), as well as negligible contamination from oligodendrocytes, was observed. When using either dataset as the reference signature, a limited presence of astrocytic genes was observed (1.16% and 6.11%, respectively), which may represent the contribution of astrocytic endfeet that cover the length of the neurovasculature. To estimate the multicellular composition of our microvessels at a finer resolution, computational deconvolution was performed a third time using the reference single-nucleus data generated by Yang et al. (2022), in which the different neurovascular cell type signatures were determined (Fig. 5c). Averaged CPMs across nuclei of each cell type were used as input for the CIBERSORTX tool (Newman et al., 2019), which estimated an average composition of 44.02% capillary and 37.62% SMC, along with much lower estimations for pericyte, arterial, venous, astrocyte and perivascular fibroblast genes (Fig. 5c). Although differentially distributed along the arteriovenous axis, both SMCs and pericytes are embedded within the vascular basement membrane (McConnell et al., 2017) and, therefore, it is unlikely that our isolated method preferentially selects one cell type over the other. Because of this, and the known similarity in molecular signature between SMCs and pericytes (Chasseigneaux et al., 2018; Muhl et al., 2020), as well as the high percentage of captured capillary segments (in which pericytes are predominantly observed) (Sweeney et al., 2016; Gonzales et al., 2020; Hartmann et al., 2021; Alarcon-Martinez et al., 2018), the surprisingly low estimation of pericyte genes may represent a limitation in comparing single-nucleus and bulk tissue datasets to one another. Critically, an average of 90.4% of the total TPMs across samples were assigned to NVU-constituent cell types.
Fig. 5.
a-b) Using the BrainDeconvShiny tool, different iterations of computational deconvolution were performed to demonstrate stability of outcome when using our microvessel isolation preparations. a) The average expression from every control sample from the VL dataset was calculated and used as cell type signatures for deconvolution of our dataset using CIBERSORT v1.04. b) The MB (MultiBrain) is a composite signature generated by Sutton et al. (2022) by averaging the expression signatures of five datasets for five cell types (neurons, astrocytes, oligodendrocytes, microglia, and endothelia). MB was used as cell type signatures for deconvolution of our dataset using CIBERSORT v1.04. c) In-house analysis where the average expression from every control sample from the Yang dataset was calculated and used as cell type signatures for deconvolution of our dataset using CIBERSORTX. Heattables were generated using the gt package in R.
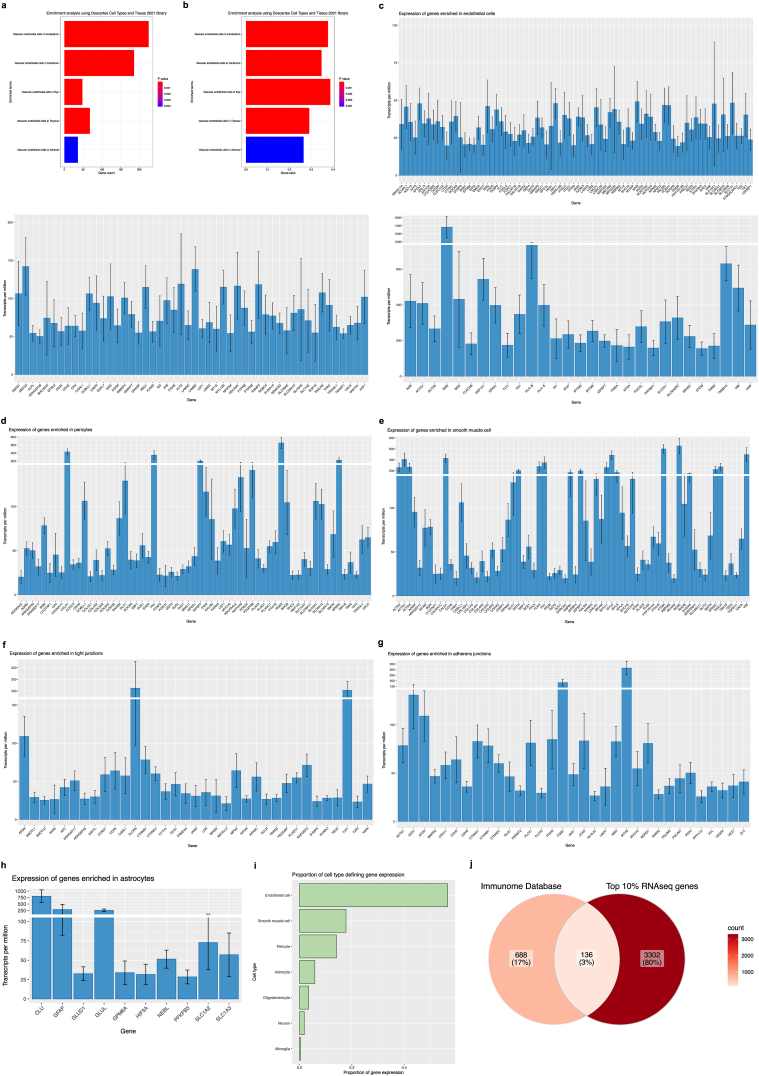
To explore our transcriptomic data, TPMs were averaged across the 5 subjects, and the top 10% of most highly expressed genes were designated (a total of 3437 genes) and used as input for over-representation analysis (ORA) using the enrichR package in R, selecting the “Descartes Cell Types and Tissue 2021” database to identify gene sets that are statistically over-represented (Fig. 6a–b and Supplementary Table 4). The threshold value of enrichment was selected by a p-value <0.05 and, as shown, over-represented genes were dramatically enriched for vascular-related terms, such as “Vascular endothelial cells in Cerebellum” and “Vascular endothelial cells in Cerebrum”. Genes behind enriched terms were extracted and the expression of a subset of known brain endothelial, pericyte, astrocytic, and smooth muscle genes in isolated microvessels were examined (Fig. 6c–i). As expected, microvessels had increased expression of canonical endothelial genes such as CLDN5, CDH5, SLC2A1, ABCB1, VWF, and MFSD2A, with the highest expression in endothelial genes ACTG1, B2M, BSG, EEF1A1, HLA-B, HLA-E, SPARCL1, TMSB10, and VIM (Fig. 6a).
Fig. 6.
a-b) Enrichment analysis of averaged 10% of most highly expressed genes (3437 genes) returns predominantly vascular-related terms. Over-representation analysis (ORA) using the enrichR package in R and the “Descartes Cell Types and Tissue 2021” database was used to identify gene sets that are statistically over-represented. The threshold value of enrichment was selected by a p-value <0.05, indicating that over-represented genes were significantly enriched for vascular-related terms. a) Count for genes in our dataset that are present in returned gene sets. b) Ratio for genes in our dataset that are present in returned gene sets, determined by the total number of genes in each set. c-i) Isolated microvessels have increased expression of canonical neurovascular-related genes. c) Bar plots showing TPMs for endothelial-defining genes, displayed according to general expression range. Highest expression found in endothelial genes B2M, BSG, FLT1, IFITM3, MT2A, SLC2A1, VIM, and VWF. d) Bar plot showing TPMs for pericyte-defining genes. Highest expression was detected in pericyte genes CALD1, FN1, IGFBP7, RGS5, and SPARCL1. e) Bar plot showing TPMs for smooth muscle cell-defining genes. Highest expression was detected in smooth muscle genes ACTG1, ACTN4, MYL6, PTMA, and TAGLN. f) Bar plot showing TPMs for tight junction-defining genes and g) Bar plot showing TPMs for adherens junction-defining genes. Several genes encoding for junctional proteins are found in the top 10% of most highly expressed genes, including CLDN5, CTNNB1, CTNND1, OCLN, JAM1, TJP1, and TJP2. h) Bar plot showing TPMs for astrocyte-defining genes. Astrocytic gene expression was predominantly limited to markers of astrocytic processes or endfeet, namely CLU, GFAP, and GLUL. i) TPMs from neurovascular-related genes were summarized according to cell type expression, demonstrating an overrepresentation of endothelial, smooth muscle cell, and pericyte genes. j) Overlap between top 10% of most highly expressed genes from our RNA sequencing data and immune genes found within the Immunome Database. Bar plots were generated using the ggplot package and Venn diagram was generated using the ggVennDiagram package in R.
Additionally, there was enrichment for other neurovascular cell types, as suggested by canonical pericyte genes PDGFRβ, MCAM, RGS5, AGRN, and NOTCH3 (Fig. 6b), as well as canonical smooth muscle genes ACTA2, MYL6, MYL9, TAGLN, and LGALS1 (Fig. 6c). Highest expression was detected in pericyte genes CALD1, FN1, IFGBP7, RGS5, SPARC, and SPARCL1 (Fig. 6b); as well as smooth muscle genes ACTG1, ACTN4, CALD1, MYL6, and PTMA (Fig. 6c). Complimentary to immunophenotypic characterization of isolated microvessels, several genes whose products are involved in junctional complex maintenance and organization (tight junctions, Fig. 6f; adherens junctions, Fig. 6g) are found in the top 10% of most highly expressed genes, for e.g., CLDN5, CTNNB1, CTNND1, ITGB1, JAM1, OCLN, TJP1, and TJP2; with additional vascular makers expressed at lower transcripts per million present throughout the entire dataset. Expression of astrocytic genes was observed to a lesser extent, several which have been previously validated as markers of astrocytic processes or endfeet (Boulay et al., 2017; Derouiche and Geiger, 2019; Sakers et al., 2017), namely EZR, GJA1, RDX, SLC1A2, and SLC1A3 (Fig. 6h). To better visualize the proportion of expression contributed by these genes, TPMs were summarized by cell type over the total number of TPMs (Fig. 6i), demonstrating an overrepresentation of endothelial, smooth muscle cell, and pericyte genes in samples.
Intriguingly, results from ORA reveal genes that are of interest in numerous disease contexts. Enriched in the described dataset is gene CLDN5, an indispensable junctional protein for the correct organization of tight junctions and maintenance of BMEC integrity (Greene et al., 2019), which was previously reported to be downregulated in the nucleus accumbens of depressed suicides (Menard et al., 2017) by altered epigenetic regulation via histone deacetylase 1 (HDAC1) (Dudek et al., 2020). Enriched pericyte genes TXNIP, RUNX1T1, ITGA1 and, DOCK9 are also reported to be differentially expressed in Schizophrenia (Puvogel et al., 2022). Similarly enriched in this dataset are angiogenic growth factors EGFL7, FLT1, VWF, and antigen-presentation machinery B2M and HLA-E, all of which are upregulated in a subpopulation of angiogenic BMECs from subjects with Alzheimer's disease (Lau et al., 2020), suggesting a compensatory angiogenic and immune response in AD pathogenesis. Likewise, endothelial genes PICALM, INPP5D, ADAMTS1, and PLCG2 that are found in the top 10% of the microvessel dataset are differentially expressed in Alzheimer's disease (Yang et al., 2022). Recent breakthroughs in deciphering the underlying etiology of Huntington's disease reveal aberrant downregulation of endothelial ABCB1, ABCG2, SLC2A1, and MFSD2A as well as mural PDGFRB, SLC20A2 and FTH1 (Garcia et al., 2022) – mutations in which are known to cause HD-like syndromes with primary pathology localized in the basal ganglia (Chinnery et al., 2007; Tadic et al., 2015). Indeed, human brain microvessel datasets integrated with others, like those from experimental models, will expedite our understanding of neurovascular contributions to mood disorders and neurodegenerative disease, and even further propagate hypothesis generation for established vascular diseases, such as white matter vascular dementia in which each cell type of the NVU exhibits a specific disease-associated expression signature (Mitroi et al., 2022).
During neuroinflammation, the BBB endothelium reconfigures its landscape of adhesion molecules, cytokines, chemokines, and reactive oxygen species, combined with reduced expression of junctional molecules. These changes prime for bidirectional interaction between the neuroimmune and peripheral immune compartments and enable increased recruitment of circulating leukocytes across the BBB. The top 10% of most highly expressed genes were also assessed for overlap with a curated immune-related gene list (Immunome Database accessed at https://www.innatedb.com/redirect.do?go=resourcesGeneLists, data originally provided by http://structure.bmc.lu.se/idbase/immunome/) (Breuer et al., 2013; Ortutay et al., 2007), sharing 145 out of 824 validated genes involved in immunological processes (Fig. 6j), including notable immune factors CX3CL1, IFNGR1, CD74, IL4R, CXCL2, CXCL12, CD81, IRF1, MIF, as well as HLAs (HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, HLA-DRA, HLA-DPA1, HLA-DPB1). Moreover, adhesion molecules known to mediate cell-cell adhesion at the BBB are also present, including: CD44, previously implicated in monocyte transmigration (He et al., 2016b) and T-cell-endothelial cell interaction (Flynn et al., 2013), MCAM, which mediates recruitment of pathogenic CD4+ T lymphocytes (Charabati et al., 2023) as well as T helper (TH) 1 cells (Breuer et al., 2018), and ICAM2, which is also critical for T helper (TH) 1 cell diapedesis (Laschinger et al., 2002). Other well-characterized immune, adhesion, and trafficking molecules in our data are listed in the Supplementary data.
5.3. High correspondence between generated transcriptomic data and published neurovascular dataset
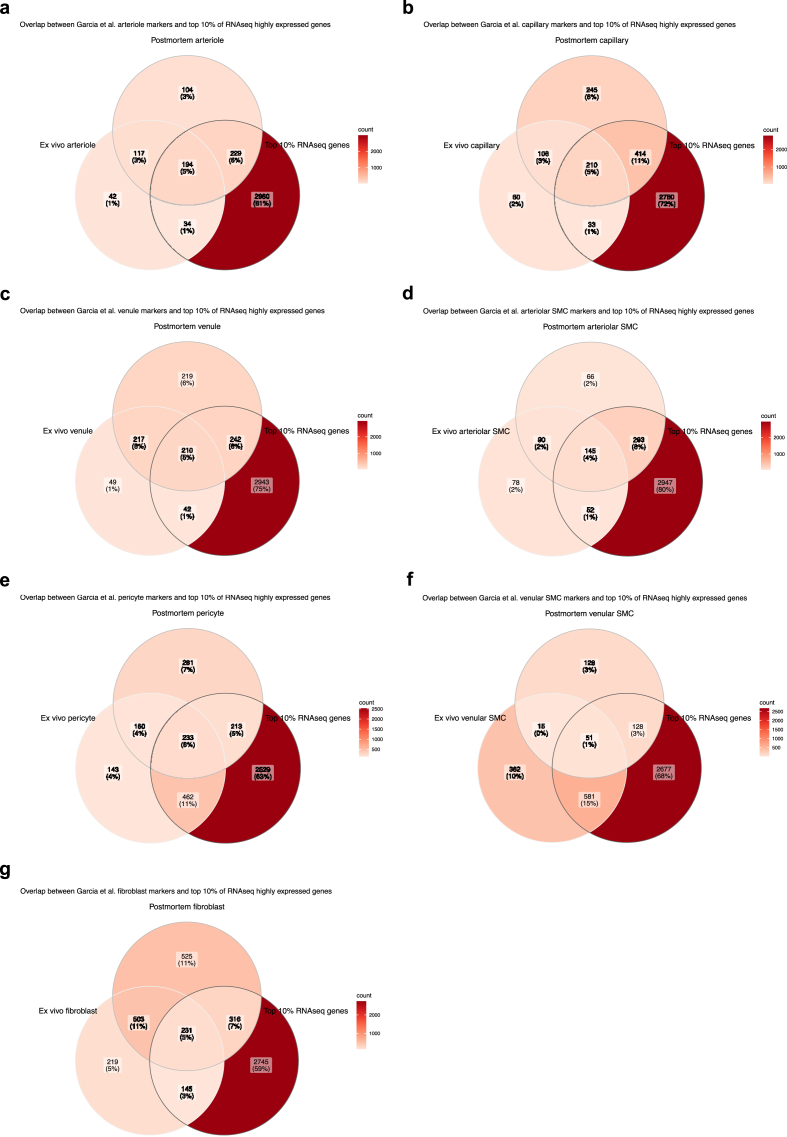
As a means to assess our isolation method, our top 10% of most highly expressed genes were juxtaposed to validated neurovascular cell type-defining markers, as designated by Garcia et al. (2022) (Garcia et al., 2022) based on sequencing of 4992 and 11,689 vascular nuclei from ex vivo and postmortem brain tissue, respectively. Results indicated substantial overlap between the three datasets for all NVU-constituent cell types (Fig. 7), including arteriole-defining genes (Fig. 7a), capillary-defining genes (Fig. 7b), venule-defining genes (Fig. 7c), arteriolar SMC-defining genes (Fig. 7d), pericyte-defining genes (Fig. 7e), venular SMC-defining genes (Fig. 7f), and perivascular fibroblast-defining genes (Fig. 7g). Interestingly, overlap was consistently greater between the two postmortem datasets.
Fig. 7.
a-g) Overlap between the top 10% of most highly expressed genes from our RNA sequencing data and published NVU dataset. Validated neurovascular cell type markers were obtained from Garcia et al. (2022) postmortem and human ex vivo single-nucleus sequencing datasets (found under Supplementary Table 2) and compared for potential overlap with the top 10% of most highly expressed genes from our data. Results indicate a significant overlap between the three datasets for all NVU-related cell types. a) Arteriole-defining genes. b) Capillary-defining genes. c) Venule-defining genes. d) Arteriolar SMC-defining genes. e) Pericyte-defining genes. f) Venular SMC-defining genes. g) Fibroblast-defining genes. Venn diagrams were generated using the ggVennDiagram package in R.
5.4. Proteomic characterization of isolated brain microvessels
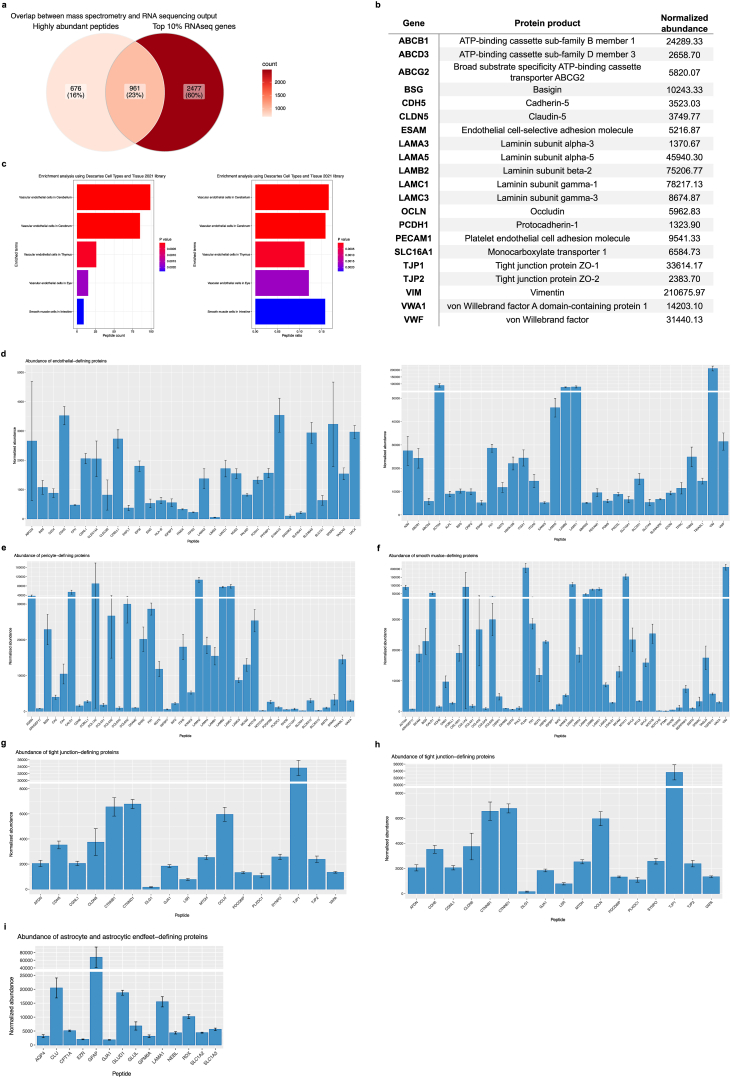
While transcript level may show positive correlations with protein level, protein abundance should not necessarily be inferred from RNA sequencing counts (Nie et al., 2006; Vogel and Marcotte, 2012); hence, interrogation of the highly dynamic proteome is needed to speculate the functional consequences of changes in protein expression. We sought to interrogate NVU-specific proteomic signatures using total protein extracted from isolated microvessels. Microvessel-enriched pellets were prepared from 3 frozen vmPFC grey matter samples microdissected from healthy individuals who died of peripheral diseases or natural events (Table 1). Although resuspended and strained pellets yield sufficient protein needed to perform LC-MS/MS (validated, data not shown), straining was omitted in favour of protein extraction directly from pellets to maximize microvessel material input for proteomic interrogation. Using Tandem Mass Tag (TMT) isobaric labeling and sample fractionation of peptides, global, relative quantitation of a total of 1638 individual proteins were detected from microvessel-enriched pellets (quality parametres shown in Supplementary Fig. 2). Importantly, there was significant overlap between transcriptomic and proteomic output, with 1635/1638 (99.8%) of detected proteins likewise identified in the transcriptomics data (albeit no corresponding transcripts were detected for proteins F9, DCD, and SERPINB12); and with 961/1638 (58.7%) proteins found in the top 10% of most highly expressed transcripts, resulting in an overall 23% overlap between high expressors in both datasets (Fig. 8a).
Fig. 8.
a-i) There is substantial overlap between transcriptomic and proteomic data output. a) 1635/1637 (99.9%) of proteins likewise identified in the transcriptomics data, and with 1024/1637 (62.6%) proteins found in the top 10% of most highly expressed genes. b) Several canonical BMEC markers were detected with high normalized average abundance. c) Over-representation analysis using the enrichR package in R and the “Descartes Cell Types and Tissue 2021” database was used to identify statistically over-represented sets. Genes corresponding to proteins detected during LC-MS/MS were used as input and the threshold value of enrichment was selected by a p-value <0.05. Output indicated that over-represented proteins were significantly enriched for vascular-related terms. a) Count for peptides in our dataset that are present in returned sets. b) Ratio for peptides in our dataset that are present in returned sets, determined by the total number in each set. d) Bar plot showing normalized abundance for endothelial cell-defining proteins. e) Bar plot showing normalized abundance for pericyte-defining proteins. f) Bar plot showing normalized abundance for smooth muscle cell defining-proteins. g) Bar plot showing normalized abundance for tight junction defining-proteins. h) Bar plot showing normalized abundance for adherens junction-defining proteins. i) Bar plot showing normalized abundance for astrocyte and astrocytic endfeet-defining proteins. Bar plots generated using the ggplot package and Venn diagram generated using the ggVennDiagram package in R.
Proteins known to be expressed by vascular-associated cell types and perivascular extracellular matrix were positively identified in all 3 samples. Several canonical endothelial markers were detected with high peptide abundance, including vimentin, several protein subunits of laminin (LAMA3, LAMA5, LAMB2, LAMC1, LAMC3), OCLN, TJP1, TJP2, VWF, VWA1, BSG, PECAM1, Monocarboxylate transporter 1 (SLC16A1), broad substrate specificity ATP-binding cassette transporter (ABCG2), ESAM, CLDN5, CDH5, ATP-binding cassette sub-family B member 1 (ABCB1), ATP-binding cassette sub-family D member 3 (ABCD3), and Protocadherin-1 (PCDH1) (normalized average abundance for these proteins are listed in Fig. 8b). As expected, enrichment terms returned by ORA (using corresponding gene names as input) were predominantly vascular-associated (Fig. 8c and Supplementary Table 5), and normalized abundance of known brain endothelial, pericyte, astrocytic and smooth muscle cell proteins were high (Fig. 8d–i).
The BBB poses a major pharmacological barrier as BMECs express a vast array of enzymes and transport systems that facilitate brain uptake processes of essential nutrients and neuroactive agents across the BBB (Hediger et al., 2004), controlling the rate and extent to which drugs are able to reach the brain parenchyma via the transcellular pathway (Ballabh et al., 2004). There is a pressing need for improved knowledge surrounding the expression and functionality of these systems at the human BBB as the majority of data comes from either in vitro cell culture or animal studies, making in vitro to in vivo or interspecies scaling less reliable. Analyses revealed high abundance of SLC2A1/GLUT1, a transmembrane protein responsible for the facilitated diffusion of glucose (Mueckler and Thorens, 2013), and the two glutamate transporters SLC1A2/EAAT2 and SLC1A3/EAAT1 in brain microvessels. Transporters SLC7A5/LAT1 and SLC3A2/4F2hc, which supply the brain with large neutral amino acids (Yanagida et al., 2001; Nakamura et al., 1999; Nicklin et al., 2009), and SLC16A1/MCT1 and SLC16A2/MCT8, which are involved in the transport of monocarboxylates (Vijay and Morris, 2014) and T3 thyroid hormone (Trajkovic et al., 2007) at the BBB, respectively, are also found in the described proteomic dataset. Also found are ABCB1/P-glycoprotein and breast cancer-related protein ABCG2/BCRP, the principal ABC efflux transporters (Begley, 2004; Loscher and Potschka, 2005b; Sun et al., 2003) that limit entry of drug candidates, toxic compounds, as well as xenobiotics from the central nervous system (Agarwal et al., 2010, 2011a, 2011b; Chen et al., 2009; de Vries et al., 2007a; Polli et al., 2009). Due to major species-specific differences in transporter expression profiles, there is utility in obtaining absolute protein amounts as they may elucidate the contribution of each transporter in facilitating the entry of endogenous substances and nutrients like glucose, glutamate, and amino and fatty acids into the brain, in addition to drugs and other xenobiotics that exploit these mechanisms (Hindle et al., 2017).
5.5. Further deconstruction of isolated brain microvessels using FANS
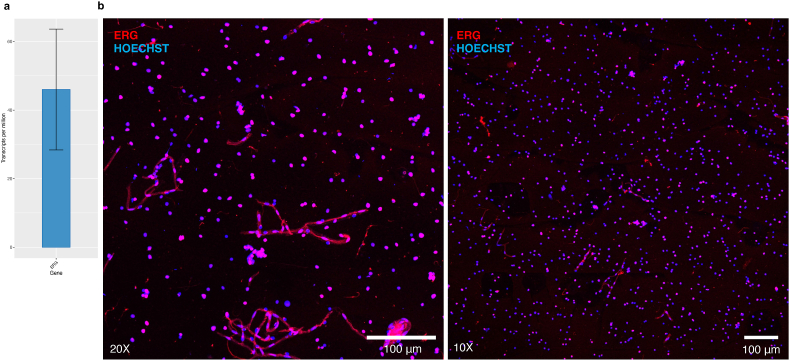
The use of frozen brain tissue demands sorting of target nuclei as opposed to intact cells and, therefore, requires the use of nuclear fluorescent tags to facilitate the isolation of endothelial nuclei. Expression of transcription factor ETS-related gene (ERG) has been previously validated as an endothelial-specific nuclear marker (Miettinen et al., 2011; Haber et al., 2015; Kim et al., 2013; Birdsey et al., 2015) and is found in the top 10% most highly expressed genes from isolated microvessels (TPM shown in Fig. 9a). As an additional testament to the versatility in downstream use of the described method, resuspended microvessels subjected to a 2-h incubation with anti-ERG antibody under rotation is sufficient for vascular disassembly and to expose the ERG epitope for endothelial nuclei immunolabelling (Fig. 9b) without the need for enzymatic digestion as previously described (Crouch and Doetsch, 2018; Pastrana et al., 2009; Codega et al., 2014). Critically, a 2-h incubation in combination with the appropriate gating strategy adjusted to collect endothelial nuclei, is sufficient for the purification of endothelial nuclei from frozen postmortem brain tissue (data not shown). The potential to isolate ERG+ endothelial nuclei from postmortem microvessels under different physiological conditions (Plane et al., 2010; Ohab et al., 2006; Yamashita et al., 2006; Kojima et al., 2010; Bardehle et al., 2013; Greenberg, 2014; Paul et al., 2012) is an advantage of our approach and addresses a growing interest in the use of primary BMECs, circumventing phenotypical or gene expression changes induced in primary BMEC cultures by prolonged adherence steps used in other isolation protocols (Durafourt et al., 2013; De Groot et al., 2000; Goldeman et al., 2020; Lyck et al., 2009).
Fig. 9.
Isolated microvessels can be used to further sort endothelial nuclei. a) ERG is a top 10% most highly expressed gene in isolated microvessels. b) Dissociated endothelial nuclei immunolabelled with ERG conjugated to Alexa-647 antibody prior to FANS protocol. Resuspended microvessels subjected to a 2-h incubation with anti-ERG antibody under rotation is sufficient for immunolabelling of endothelial nuclei prior to FANS sorting.
6. Discussion
We describe a singular, standardized protocol to enrich and isolate microvessels from archived snap-frozen human brain tissue with the ability to apply the same protocol to frozen mouse cerebral cortex. Integral to this method are three factors that confer gentility and simplicity: 1) the correct molarity of the sucrose-based buffer to separate and cushion microvessels during centrifugation-separation, 2) centrifugation-separation of microvessels from the rest of the tissue homogenate occurs in a 10 ml volume, which facilitates the formation of a microvessel-enriched pellet as heavier structures reach the bottom of the 15 ml falcon tube, and 3) the limited number of wash steps that minimize eventual damage done to the integrity of microvessel fragments. Microvessel enrichment and stability were assessed by chromogenic staining of microvessels using BCIP/NBT substrate and, through immunophenotypic characterization, we show that isolated microvessel fragments are comprised of NVU cellular components including BMECs, astrocytic endfeet, pericytes, as well as tight junction protein complexes. The demonstrated gentility and simplicity of the approach, in turn, confer versatility in the high-throughput techniques that can be utilized downstream to microvessel isolation, as demonstrated here with RNA sequencing and LC-MS/MS.
This protocol should have excellent reproducibility in isolating intact microvessels from any region of the adult human brain. It should be noted, however, that the current version of the protocol may need adjustments if myelin content of brain samples used is high, as dissociated myelin (which travels to the top of the homogenate; section 2.2, step 10) may interfere with the formation of a microvessel-enriched pellet (which travels to the bottom of the homogenate; section 2.2, step 10). As reported, microvessels isolated using the described method are in high yield wihout other contaminating cell types. However, the protocol enriches for but does not purify microvessels, so a limited proportion of contamination from non-vascular cell types might be found in collected pellets. Limited expression of neuronal markers is observed during computational deconvolution of RNA sequencing data; however, whether a minority of neurons are indeed co-enriched with microvessels remains unclear, as co-enrichment of neurons was not observed during immunophenotypic characterization of isolated microvessels. While some co-enrichment is consistent with the technically unavoidable capture of some parenchymal cells when utilizing a method suitable for frozen postmortem brain tissue (which necessitates as few and as gentle steps as possible), the human BBB atlases generated by Yang et al. (2022) and Garcia et al. (2022) reveal that “canonically neuronal” genes are also expressed in vascular-associated cell types. Therefore, it is possible that counts for such genes indeed originate from vascular-associated cells and not neurons. Additionally, representation of all vascular-associated cell types might not be uniform, as different cell types may vary across brain regions and even brain subregions, and different cell types may be differentially susceptible to steps taken during RNA or protein extraction. Nonetheless, in the absence of harsh experimental steps, it is reasonable to assume that the proportions of vascular-associated cell types found within isolated microvessels represent the natural multicellular composition of vasculature within the brain. During the development of this protocol, several brain tissue samples were used possessing a range of values for PMI, RIN, pH, and other metrics, including age at death, manner of death, freezer storage time, and prior medication exposure. Although not systematically assessed, success in isolating and enriching microvessels from different tissue samples was consistent and robust across variations in such quality metrics (as demonstrated here by the wide range of age and PMI across subjects shown in Table 1). We attribute such reproducibility to the protective nature of the vascular basement membrane that may protect the vessel structure and its contents against changes experienced by the brain parenchyma during the postmortem interval. Despite this, it is recommended to utilize postmortem samples with quality metrics PMI, RIN, and pH that indicate moderate to good quality, which is a fundamental prerequisite to investigating complex brain disorders using molecular profiling techniques. Inevitably, studies that make use of isolated human microvessels will encounter high inter-subject variation in both RNA and protein yield as well as experimental read-out. Large cohorts with matched subjects (if studying disease and/or sex differences) would be required to power a study investigating how brain vascular gene or proteomic expression varies with factors such as brain region, age, or disease. Although sensitivity of mass spectrometers has been greatly improved over the years (Hahne et al., 2013; Hebert et al., 2018; Shishkova et al., 2016, 2018; Timp and Timp, 2020), as well as peptide separation for untargeted proteome analysis (Shishkova et al., 2016, 2018; Toth et al., 2019), the literature consistently shows that high output from mass spectrometry relies on a balance between sample quantity as well as sample complexity. Because protein extracted frommicrovessels, as opposed to bulk tissue, could be considered a sample lacking complexity, it is reasonable to assume that this relays to lower output, which may be further impacted by several low signal-to-noise events that result in unidentified peptides (Griss et al., 2016) and a more limited dynamic range of peptide detection (Timp and Timp, 2020).
Our successfully generated datasets promise a more complete approach to investigating BBB function and disease, where one arm of a study may leverage experimental animal or cell culture models to identify pertinent biological mechanisms, and the other arm may utilize microvessels isolated from human brain to provide insight into species-specific differences and other limitations of experimental models. Species differences in disease-affected neuronal and myeloid gene expression have been characterized by single-cell or single-nucleus sequencing (Cosacak et al., 2022; Friedman et al., 2018; Kamath et al., 2022), but such differences in neurovascular gene expression have only just begun to be comprehensively analyzed, and solely in Huntington's disease and Alzheimer's disease (Garcia et al., 2022; Yang et al., 2022). Despite limited data, it has become evident there are striking differences, with one study reporting 142 mouse-enriched genes, including Vtn, Slco1c1, Slc6a20a, Atp13a5, Slc22a8, and 211 human-enriched genes, including SLCO2A1, GIMAP7, and A2M (Song et al., 2020b). Yang et al. (2022) (Yang et al., 2022) further reported hundreds of species-enriched genes in BMECs and pericytes (Yang et al., 2022), finding that BMECs and pericytes exhibit the greatest transcriptional divergence in several vascular solute transporters (for e.g., GABA transporter SLC6A12) and genes of disease and pharmacological importance (Yang et al., 2022). This observation was corroborated by Garcia et al. (2022) (Garcia et al., 2022), who further detailed that species-specific DEGs were strongly enriched for marker genes of vascular-associated cell types, indicating that cell type identity markers were among those that vary the most between species (Garcia et al., 2022). Breakthroughs can similarly be made using the described method, with the added benefit that microvessels kept as a structurally intact unit provide insight into the neurovasculature in a manner that nuclei or dissociated cells cannot: not only is unwanted technical-related depletion of vascular-associated cells avoided, but cytoplasm, which carries much higher amounts of mRNA and protein, as well as interstitium are also preserved. This is not trivial, as work done by others comparing microglia single cells versus single nuclei from postmortem tissue reveals that certain populations of genes are depleted in nuclei compared to whole cells. Those depleted were previously implicated in microglial activation, such as APOE, SPP1, CST3, and CD74, totalling 18% of previously identified microglial-disease-associated genes (Thrupp et al., 2020). While there is undeniable benefit in characterizing disease-related changes at the cell type level, the neurovasculature is so interconnected that dysfunction is typically observed in all vascular-associated cells, conferring whole-unit dysfunction that becomes difficult to parse when looking at its individual nuclei. We put forward the ATP-binding cassette (ABC) transporter family as an interesting candidate that can be further explored using microvessels isolated from postmortem brain. The human genome carries 48 different ABC transporters (Gil-Martins et al., 2020; Morris et al., 2017; Robey et al., 2018), several of which are expressed in the CNS, primarily at the BBB (Gil-Martins et al., 2020). Dysfunction of ABC transporters, at expression and/or activity level, has been repeatedly associated with neurological disease (Gil-Martins et al., 2020); and of note are the functionally important yet redundant ABCB1 and ABCG2, both of which have been observed in our transcriptomic and proteomic datasets. For example, ABCB1 is culpable in the neuroinflammatory mechanisms of mutliple sclerosis, where ABCB1 expression and activity are significantly decreased by mechanisms involving CD4+ T cells (Kooij et al., 2010), which is just one aspect of the broad barrier impairment that permits lymphocytes activated in the periphery to infiltrate the CNS (Cashion et al., 2023; Ortiz et al., 2014). Moreover, numerous drug candidates show potential anticancer effects against different brain cancer cell lines in vitro and yet, their efficacy in vivo and in clinical trials has been considerably more modest, in large part due to ABCB1/ABCG2-mediated efflux at the BBB. Both ABCB1 and ABCG2, which are expressed on the luminal membrane of BMECs (Biegel et al., 1995; Cooray et al., 2002; Eisenblatter and Galla, 2002; Zhang et al., 2003), present a double-edged sword at the blood-brain barrier: on the one hand, ABCB1/ABCG2-mediated efflux is vital for protecting the brain; on the other hand, several anticancer drugs have been identified as substrates of ABCB1 and/or ABCG2 (de Vries et al., 2007b; Agarwal and Elmquist, 2012; Traxl et al., 2019), and their function restricts brain uptake of anticancer drugs, significantly limiting their efficacy in the treatment of primary and metastatic brain tumors (de Vries et al., 2007b; Agarwal and Elmquist, 2012; Juliano and Ling, 1976; Doyle et al., 1998; Marchetti et al., 2008; Agarwal et al., 2011c; de Gooijer et al., 2018; Sorf et al., 2018; Schinkel et al., 1994). Efforts to better understand ABCB1/ABCG2 expression and circumvent the restriction of drug uptake have been considerable. Previous studies point to a lower ABCB1/ABCG2 ratio (Dehouck et al., 2022; Uchida et al., 2011), whereas our findings exhibit a higher ABCB1/ABCG2 ratio. This discrepancy may be an insight into the interindividual differences that give rise to variable drug responses, a common clinical challenge. Several synonymous single nucleotide polymorphisms (SNPs) that affect function and substrate binding have been identified in both transporters (Kimchi-Sarfaty et al., 2007; Fung and Gottesman, 2009; Dickens et al., 2013; Furukawa et al., 2009; Delord et al., 2013; El Biali et al., 2021). The individuals studied here were all of French-Canadian origin, a group with a distinct genotype, and so certain SNPs that affect mRNA stability or transporter activity may, in turn, modulate translational output of ABCB1 relative to ABCG2, as has been observed with ABCB1 SNPs (Wang et al., 2005). It should be noted, however, that these studies (including ours) comprised of small cohorts and warrant further investigations with larger sample sizes.
In sum, the isolated brain microvessel is a robust model for the NVU and can be used to generate a variety of highly dimensional datasets. The availability of characterized human neurovascular transcriptomes and proteomes can aid in identifying potential roles for BMECs and pericytes in the pathogenesis of neurological and psychiatric disorders and additionally aid in assessing the expression of molecules with potential relevance to drug delivery and novel therapies (e.g., SLCs, ABCs, and large molecule receptors) across the human brain vasculature.
Declaration of competing interest
None.
Acknowledgements
The authors have no competing interests to declare. This study was funded by an ERA NET Neuron grant and a CIHR Project grant to N.M. M.W. and R.R. respectively received scholarship and fellowship support from the FRQ-S. The Douglas-Bell Canada Brain Bank is supported in part by platform support grants from the Réseau Québécois sur le Suicide, les Troubles de l’Humeur et les Troubles Associés (FRQ-S), Healthy Brains for Healthy Lives (CFREF), and Brain Canada. The present study used the services of the Molecular and Cellular Microscopy Platform (MCMP) at the Douglas Research Centre. The authors would like to thank the expert help of Douglas-Bell Canada Brain Bank staff members (J. Prud'homme, M. Bouchouka, D. Mirault, V. Lariviere, A. Baccichet), the technology development team at the McGill University and Genome Quebec Innovation Centre, and the Segal Cancer Proteomics Centre at the Lady Davis Institute. The authors would also like to thank Dr. Ghazal Fakhfouri for rearing and providing the mice used in this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbih.2023.100684.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Abbott N.J., Ronnback L., Hansson E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat. Rev. Neurosci. 2006;7(1):41–53. doi: 10.1038/nrn1824. [DOI] [PubMed] [Google Scholar]
- Abbott N.J., Patabendige A.A., Dolman D.E., Yusof S.R., Begley D.J. Structure and function of the blood-brain barrier. Neurobiol. Dis. 2010;37(1):13–25. doi: 10.1016/j.nbd.2009.07.030. [DOI] [PubMed] [Google Scholar]
- Agarwal S., Elmquist W.F. Insight into the cooperation of P-glycoprotein (ABCB1) and breast cancer resistance protein (ABCG2) at the blood-brain barrier: a case study examining sorafenib efflux clearance. Mol. Pharm. 2012;9(3):678–684. doi: 10.1021/mp200465c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S., Sane R., Gallardo J.L., Ohlfest J.R., Elmquist W.F. Distribution of gefitinib to the brain is limited by P-glycoprotein (ABCB1) and breast cancer resistance protein (ABCG2)-mediated active efflux. J. Pharmacol. Exp. Therapeut. 2010;334:147–155. doi: 10.1124/jpet.110.167601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S., Hartz A.M., Elmquist W.F., Bauer B. Breast cancer resistance protein and P-glycoprotein in brain cancer: two gatekeepers team up. Curr. Pharmaceut. Des. 2011;17:2793–2802. doi: 10.2174/138161211797440186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S., Sane R., Ohlfest J.R., Elmquist W.F. The role of the breast cancer resistance protein (ABCG2) in the distribution of sorafenib to the brain. J. Pharmacol. Exp. Therapeut. 2011;336:223–233. doi: 10.1124/jpet.110.175034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S., Sane R., Ohlfest J.R., Elmquist W.F. The role of the breast cancer resistance protein (ABCG2) in the distribution of sorafenib to the brain. J. Pharmacol. Exp. Therapeut. 2011;336(1):223–233. doi: 10.1124/jpet.110.175034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bachari S., Naish J.H., Parker G.J.M., Emsley H.C.A., Parkes L.M. Blood-brain barrier leakage is increased in Parkinson's disease. Front. Physiol. 2020;11 doi: 10.3389/fphys.2020.593026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Louzi O., Govindarajan S.T., Tauhid S., et al. Central vein sign profile of newly developing lesions in multiple sclerosis: a 3-year longitudinal study. Neurol Neuroimmunol. Neuroinflamm. 2022;9(1) doi: 10.1212/NXI.0000000000001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon-Martinez L., et al. Capillary pericytes express alpha-smooth muscle actin, which requires prevention of filamentous-actin depolymerization for detection. Elife. 2018;7 doi: 10.7554/eLife.34861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida D., Turecki G. Profiling cell-type specific gene expression in post-mortem human brain samples through laser capture microdissection. Methods. 2022;207:3–10. doi: 10.1016/j.ymeth.2022.08.013. [DOI] [PubMed] [Google Scholar]
- Arvanitakis Z., Capuano A.W., Leurgans S.E., Bennett D.A., Schneider J.A. Relation of cerebral vessel disease to Alzheimer's disease dementia and cognitive function in elderly people: a cross-sectional study. Lancet Neurol. 2016;15(9):934–943. doi: 10.1016/S1474-4422(16)30029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumailley M., Smyth N. The role of laminins in basement membrane function. J. Anat. 1998;193(Pt 1):1–21. doi: 10.1046/j.1469-7580.1998.19310001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson R., Martensson E., Alling C. Impairment of the blood-brain barrier as an aetiological factor in paranoid psychosis. Br. J. Psychiatry. 1982;141:273–281. doi: 10.1192/bjp.141.3.273. [DOI] [PubMed] [Google Scholar]
- Bacher R., Kendziorski C. Design and computational analysis of single-cell RNA-sequencing experiments. Genome Biol. 2016;17:63. doi: 10.1186/s13059-016-0927-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin S.A., Fugaccia I., Brown D.R., Brown L.V., Scheff S.W. Blood-brain barrier breach following cortical contusion in the rat. J. Neurosurg. 1996;85(3):476–481. doi: 10.3171/jns.1996.85.3.0476. [DOI] [PubMed] [Google Scholar]
- Ball H.J., McParland B., Driussi C., Hunt N.H. Isolating vessels from the mouse brain for gene expression analysis using laser capture microdissection. Brain Res. Brain Res. Protoc. 2002;9(4):206–213. doi: 10.1016/s1385-299x(02)00147-2. [DOI] [PubMed] [Google Scholar]
- Ballabh P., Braun A., Nedergaard M. The blood-brain barrier: an overview: structure, regulation, and clinical implications. Neurobiol. Dis. 2004;16:1–13. doi: 10.1016/j.nbd.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Bandopadhyay R., Orte C., Lawrenson J.G., Reid A.R., De Silva S., Allt G. Contractile proteins in pericytes at the blood-brain and blood-retinal barriers. J. Neurocytol. 2001;30(1):35–44. doi: 10.1023/a:1011965307612. [DOI] [PubMed] [Google Scholar]
- Bannister R.G., Romanul F.C. The localization of alkaline phosphatase activity in cerebral blood vessels. J. Neurol. Neurosurg. Psychiatry. 1963;26(4):333–340. doi: 10.1136/jnnp.26.4.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardehle S., et al. Live imaging of astrocyte responses to acute injury reveals selective juxtavascular proliferation. Nat. Neurosci. 2013;16:580–586. doi: 10.1038/nn.3371. [DOI] [PubMed] [Google Scholar]
- Baskaya M.K., Rao A.M., Dogan A., Donaldson D., Dempsey R.J. The biphasic opening of the blood-brain barrier in the cortex and hippocampus after traumatic brain injury in rats. Neurosci. Lett. 1997;226(1):33–36. doi: 10.1016/s0304-3940(97)00239-5. [DOI] [PubMed] [Google Scholar]
- Becker N.H., Goldfischer S., Shin W.Y., Novikoff A.B. The localization of enzyme activities in the rat brain. J. Biophys. Biochem. Cytol. 1960;8(3):649–663. doi: 10.1083/jcb.8.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley D.J. ABC transporters and the blood-brain barrier. Curr. Pharmaceut. Des. 2004;10:1295–1312. doi: 10.2174/1381612043384844. [DOI] [PubMed] [Google Scholar]
- Betz A.L., Goldstein G.W. Polarity of the blood-brain barrier: neutral amino acid transport into isolated brain capillaries. Science. 1978;202:225–227. doi: 10.1126/science.211586. [DOI] [PubMed] [Google Scholar]
- Betz A.L., Firth J.A., Goldstein G.W. Polarity of the blood-brain barrier: distribution of enzymes between the luminal and antiluminal membranes of brain capillary endothelial cells. Brain Res. 1980;192:17–28. doi: 10.1016/0006-8993(80)91004-5. [DOI] [PubMed] [Google Scholar]
- Biegel D., Spencer D.D., Pachter J.S. Isolation and culture of human brain microvessel endothelial cells for the study of blood-brain barrier properties in vitro. Brain Res. 1995;692(1–2):183–189. doi: 10.1016/0006-8993(95)00511-n. [DOI] [PubMed] [Google Scholar]
- Birdsey G.M., et al. The endothelial transcription factor ERG promotes vascular stability and growth through Wnt/beta-catenin signaling. Dev. Cell. 2015;32:82–96. doi: 10.1016/j.devcel.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boraas L.C., Ahsan T. Lack of vimentin impairs endothelial differentiation of embryonic stem cells. Sci. Rep. 2016;6 doi: 10.1038/srep30814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay A.C., et al. Translation in astrocyte distal processes sets molecular heterogeneity at the gliovascular interface. Cell Discov. 2017;3 doi: 10.1038/celldisc.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourassa P., Tremblay C., Schneider J.A., Bennett D.A., Calon F. Beta-amyloid pathology in human brain microvessel extracts from the parietal cortex: relation with cerebral amyloid angiopathy and Alzheimer's disease. Acta Neuropathol. 2019;137(5):801–823. doi: 10.1007/s00401-019-01967-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne G.H. Histochemical demonstration of phosphatases in the central nervous system of the rat. Exp. Cell Res. 1958;14(1):101–117. [PubMed] [Google Scholar]
- Breuer K., Foroushani A.K., Laird M.R., Chen C., Sribnaia A., Lo R., et al. InnateDB: systems biology of innate immunity and beyond--recent updates and continuing curation. Nucleic Acids Res. 2013;41(Database issue):D1228–D1233. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer J., Korpos E., Hannocks M.J., Schneider-Hohendorf T., Song J., Zondler L., et al. Blockade of MCAM/CD146 impedes CNS infiltration of T cells over the choroid plexus. J. Neuroinflammation. 2018;15(1):236. doi: 10.1186/s12974-018-1276-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch S., Saville J.W., Schifitto G., et al. Revealing vascular abnormalities and measuring small vessel density in multiple sclerosis lesions using USPIO. Neuroimag.Clin. 2021;29 doi: 10.1016/j.nicl.2020.102525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar K., Thomsen K., Lauritzen M. Dissociation of spikes, synaptic activity, and activity-dependent increments in rat cerebellar blood flow by tonic synaptic inhibition. Proc. Natl. Acad. Sci. U. S. A. 2003;100(26):16000–16005. doi: 10.1073/pnas.2635195100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campana M., Lohrs L., Strauss J., Munz S., Oviedo-Salcedo T., Fernando P., et al. Blood-brain barrier dysfunction and folate and vitamin B12 levels in first-episode schizophrenia-spectrum psychosis: a retrospective chart review. Eur. Arch. Psychiatr. Clin. Neurosci. 2023 doi: 10.1007/s00406-023-01572-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashion J.M., Young K.M., Sutherland B.A. How does neurovascular unit dysfunction contribute to multiple sclerosis? Neurobiol. Dis. 2023;178 doi: 10.1016/j.nbd.2023.106028. [DOI] [PubMed] [Google Scholar]
- Chang L., Goldman R.D. Intermediate filaments mediate cytoskeletal crosstalk. Nat. Rev. Mol. Cell Biol. 2004;5(8):601–613. doi: 10.1038/nrm1438. [DOI] [PubMed] [Google Scholar]
- Charabati M., Zandee S., Fournier A.P., Tastet O., Thai K., Zaminpeyma R., et al. MCAM+ brain endothelial cells contribute to neuroinflammation by recruiting pathogenic CD4+ T lymphocytes. Brain. 2023;146(4):1483–1495. doi: 10.1093/brain/awac389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasseigneaux S., et al. Isolation and differential transcriptome of vascular smooth muscle cells and mid-capillary pericytes from the rat brain. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-30739-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., et al. P-glycoprotein and breast cancer resistance protein influence brain distribution of dasatinib. J. Pharmacol. Exp. Therapeut. 2009;330:956–963. doi: 10.1124/jpet.109.154781. [DOI] [PubMed] [Google Scholar]
- Chinnery P.F., et al. Clinical features and natural history of neuroferritinopathy caused by the FTL1 460InsA mutation. Brain. 2007;130(Pt 1):110–119. doi: 10.1093/brain/awl319. [DOI] [PubMed] [Google Scholar]
- Codega P., et al. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron. 2014;82:545–559. doi: 10.1016/j.neuron.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooray H.C., Blackmore C.G., Maskell L., Barrand M.A. Localisation of breast cancer resistance protein in microvessel endothelium of human brain. Neuroreport. 2002;13(16):2059–2063. doi: 10.1097/00001756-200211150-00014. [DOI] [PubMed] [Google Scholar]
- Cordon-Cardo C., O'Brien J.P., Boccia J., Casals D., Bertino J.R., Melamed M.R. Multidrug-resistance gene (P-glycoprotein) is expressed by endothelial cells at blood-brain barrier sites. Proc. Natl. Acad. Sci. U.S.A. 1989;86:695–698. doi: 10.1073/pnas.86.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosacak M.I., et al. Single cell/nucleus transcriptomics comparison in zebrafish and humans reveals common and distinct molecular responses to Alzheimer's disease. Cells. 2022;11 doi: 10.3390/cells11111807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch E.E., Doetsch F. FACS isolation of endothelial cells and pericytes from mouse brain microregions. Nat. Protoc. 2018;13(4):738–751. doi: 10.1038/nprot.2017.158. [DOI] [PubMed] [Google Scholar]
- Daneman R. The blood-brain barrier in health and disease. Ann. Neurol. 2012;72:648–672. doi: 10.1002/ana.23648. [DOI] [PubMed] [Google Scholar]
- de Gooijer M.C., Buil L.C.M., Citirikkaya C.H., Hermans J., Beijnen J.H., van Tellingen O. ABCB1 attenuates the brain penetration of the PARP inhibitor AZD2461. Mol. Pharm. 2018;15(11):5236–5243. doi: 10.1021/acs.molpharmaceut.8b00742. [DOI] [PubMed] [Google Scholar]
- De Groot C.J., et al. Isolation and characterization of adult microglial cells and oligodendrocytes derived from postmortem human brain tissue. Brain Res. Brain Res. Protoc. 2000;5:85–94. doi: 10.1016/s1385-299x(99)00059-8. [DOI] [PubMed] [Google Scholar]
- De Vivo D.C., Trifiletti R.R., Jacobson R.I., Ronen G.M., Behmand R.A., Harik S.I. Defective glucose transport across the blood-brain barrier as a cause of persistent hypoglycorrhachia, seizures, and developmental delay. N. Engl. J. Med. 1991;325(11):703–709. doi: 10.1056/NEJM199109053251006. [DOI] [PubMed] [Google Scholar]
- de Vries N.A., et al. P-glycoprotein and breast cancer resistance protein: two dominant transporters working together in limiting the brain penetration of topotecan. Clin. Cancer Res. 2007;13:6440–6449. doi: 10.1158/1078-0432.CCR-07-1335. [DOI] [PubMed] [Google Scholar]
- de Vries N.A., Zhao J., Kroon E., Buckle T., Beijnen J.H., van Tellingen O. P-glycoprotein and breast cancer resistance protein: two dominant transporters working together in limiting the brain penetration of topotecan. Clin. Cancer Res. 2007;13(21):6440–6449. doi: 10.1158/1078-0432.CCR-07-1335. [DOI] [PubMed] [Google Scholar]
- Deane R., Bell R.D., Sagare A., Zlokovic B.V. Clearance of amyloid-beta peptide across the blood-brain barrier: implication for therapies in Alzheimer's disease. CNS Neurol. Disord.: Drug Targets. 2009;8(1):16–30. doi: 10.2174/187152709787601867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehouck M.P., Tachikawa M., Hoshi Y., Omori K., Maurage C.A., Strecker G., et al. Quantitative targeted absolute proteomics for better characterization of an in vitro human blood-brain barrier model derived from hematopoietic stem cells. Cells. 2022;11(24) doi: 10.3390/cells11243963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Zoppo G.J., Mabuchi T. Cerebral microvessel responses to focal ischemia. J. Cerebr. Blood Flow Metabol. 2003;23(8):879–894. doi: 10.1097/01.WCB.0000078322.96027.78. [DOI] [PubMed] [Google Scholar]
- Delord M., Rousselot P., Cayuela J.M., Sigaux F., Guilhot J., Preudhomme C., et al. High imatinib dose overcomes insufficient response associated with ABCG2 haplotype in chronic myelogenous leukemia patients. Oncotarget. 2013;4(10):1582–1591. doi: 10.18632/oncotarget.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisenko E., Guo B.B., Jones M., et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 2020;21(1):130. doi: 10.1186/s13059-020-02048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derouiche A., Geiger K.D. Perspectives for ezrin and radixin in astrocytes: kinases, functions and pathology. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20153776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickens D., Owen A., Alfirevic A., Pirmohamed M. ABCB1 single nucleotide polymorphisms (1236C>T, 2677G>T, and 3435C>T) do not affect transport activity of human P-glycoprotein. Pharmacogenetics Genom. 2013;23(6):314–323. doi: 10.1097/FPC.0b013e328360d10c. [DOI] [PubMed] [Google Scholar]
- Dietrich W.D., Alonso O., Halley M. Early microvascular and neuronal consequences of traumatic brain injury: a light and electron microscopic study in rats. J. Neurotrauma. 1994;11(3):289–301. doi: 10.1089/neu.1994.11.289. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doepp F., Paul F., Valdueza J.M., Schmierer K., Schreiber S.J. Venous drainage in multiple sclerosis: a combined MRI and ultrasound study. Neurology. 2011;77(18):1745–1751. doi: 10.1212/WNL.0b013e318236f0ea. [DOI] [PubMed] [Google Scholar]
- Dorheim M.A., Tracey W.R., Pollock J.S., Grammas P. Nitric oxide synthase activity is elevated in brain microvessels in Alzheimer's disease. Biochem. Biophys. Res. Commun. 1994;205(1):659–665. doi: 10.1006/bbrc.1994.2716. [DOI] [PubMed] [Google Scholar]
- Doyle L.A., Yang W., Abruzzo L.V., Krogmann T., Gao Y., Rishi A.K., et al. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc. Natl. Acad. Sci. U. S. A. 1998;95(26):15665–15670. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek K.A., et al. Molecular adaptations of the blood-brain barrier promote stress resilience vs. depression. Proc. Natl. Acad. Sci. U. S. A. 2020;117(6):3326–3336. doi: 10.1073/pnas.1914655117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durafourt B.A., Moore C.S., Blain M., Antel J.P. Isolating, culturing, and polarizing primary human adult and fetal microglia. Methods Mol. Biol. 2013;1041:199–211. doi: 10.1007/978-1-62703-520-0_19. [DOI] [PubMed] [Google Scholar]
- Durukan A., Tatlisumak T. Post-ischemic blood-brain barrier leakage in rats: one-week follow-up by MRI. Brain Res. 2009;1280:158–165. doi: 10.1016/j.brainres.2009.05.025. [DOI] [PubMed] [Google Scholar]
- Eisenblatter T., Galla H.J. A new multidrug resistance protein at the blood-brain barrier. Biochem. Biophys. Res. Commun. 2002;293(4):1273–1278. doi: 10.1016/S0006-291X(02)00376-5. [DOI] [PubMed] [Google Scholar]
- El Biali M., Karch R., Philippe C., Haslacher H., Tournier N., Hacker M., et al. ABCB1 and ABCG2 together limit the distribution of ABCB1/ABCG2 substrates to the human retina and the ABCG2 single nucleotide polymorphism Q141K (c.421C> A) may lead to increased drug exposure. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.698966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson M.A., Banks W.A. Neuroimmune axes of the blood-brain barriers and blood-brain interfaces: bases for physiological regulation, disease states, and pharmacological interventions. Pharmacol. Rev. 2018;70(2):278–314. doi: 10.1124/pr.117.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabry Z., Fitzsimmons K.M., Herlein J.A., Moninger T.O., Dobbs M.B., Hart M.N. Production of the cytokines interleukin 1 and 6 by murine brain microvessel endothelium and smooth muscle pericytes. J. Neuroimmunol. 1993;47(1):23–34. doi: 10.1016/0165-5728(93)90281-3. [DOI] [PubMed] [Google Scholar]
- Farrell C.L., Pardridge W.M. Blood-brain barrier glucose transporter is asymmetrically distributed on brain capillary endothelial lumenal and ablumenal membranes: an electron microscopic immunogold study. Proc. Natl. Acad. Sci. U. S. A. 1991;88(13):5779–5783. doi: 10.1073/pnas.88.13.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergus A., Lee K.S. GABAergic regulation of cerebral microvascular tone in the rat. J. Cerebr. Blood Flow Metabol. 1997;17(9):992–1003. doi: 10.1097/00004647-199709000-00009. [DOI] [PubMed] [Google Scholar]
- Flynn K.M., Michaud M., Madri J.A. CD44 deficiency contributes to enhanced experimental autoimmune encephalomyelitis: a role in immune cells and vascular cells of the blood-brain barrier. Am. J. Pathol. 2013;182(4):1322–1336. doi: 10.1016/j.ajpath.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler A.J., Ahn J., Hebron M., Chiu T., Ayoub R., Mulki S., et al. CSF MicroRNAs reveal impairment of angiogenesis and autophagy in Parkinson disease. Neurol.Genet. 2021;7(6):e633. doi: 10.1212/NXG.0000000000000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox P.T., Raichle M.E., Mintun M.A., Dence C. Nonoxidative glucose consumption during focal physiologic neural activity. Science. 1988;241(4864):462–464. doi: 10.1126/science.3260686. [DOI] [PubMed] [Google Scholar]
- Friedman B.A., et al. Diverse brain myeloid expression profiles reveal distinct microglial activation states and aspects of Alzheimer's disease not evident in mouse models. Cell Rep. 2018;22:832–847. doi: 10.1016/j.celrep.2017.12.066. [DOI] [PubMed] [Google Scholar]
- Fung K.L., Gottesman M.M. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim. Biophys. Acta. 2009;1794(5):860–871. doi: 10.1016/j.bbapap.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T., Wakabayashi K., Tamura A., Nakagawa H., Morishima Y., Osawa Y., et al. Major SNP (Q141K) variant of human ABC transporter ABCG2 undergoes lysosomal and proteasomal degradations. Pharm. Res. (N. Y.) 2009;26(2):469–479. doi: 10.1007/s11095-008-9752-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitan M.I., de Alwis M.P., Sati P., Nair G., Reich D.S. Multiple sclerosis shrinks intralesional, and enlarges extralesional, brain parenchymal veins. Neurology. 2013;80(2):145–151. doi: 10.1212/WNL.0b013e31827b916f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal Z., Torok D., Gonda X., Eszlari N., Anderson I.M., Deakin B., et al. Inflammation and blood-brain barrier in depression: interaction of CLDN5 and IL6 gene variants in stress-induced depression. Int. J. Neuropsychopharmacol. 2023;26(3):189–197. doi: 10.1093/ijnp/pyac079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia F.J., Patel K., Costa A.S.H., et al. Single-cell dissection of the human brain vasculature. Nature. 2022;603(7897):893–899. doi: 10.1038/s41586-022-04521-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendelman H.E., Ding S., Gong N., et al. Monocyte chemotactic protein-1 regulates voltage-gated K+ channels and macrophage transmigration. J. Neuroimmune Pharmacol. 2009;4(1):47–59. doi: 10.1007/s11481-008-9135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes R., Esiri M.M., DeLuca G.C., Palace J. Vascular disease and multiple sclerosis: a post-mortem study exploring their relationships. Brain. 2020;143(10):2998–3012. doi: 10.1093/brain/awaa255. [DOI] [PubMed] [Google Scholar]
- Gerhart D.Z., LeVasseur R.J., Broderius M.A., Drewes L.R. Glucose transporter localization in brain using light and electron immunocytochemistry. J. Neurosci. Res. 1989;22(4):464–472. doi: 10.1002/jnr.490220413. [DOI] [PubMed] [Google Scholar]
- Gil-Martins E., Barbosa D.J., Silva V., Remiao F., Silva R. Dysfunction of ABC transporters at the blood-brain barrier: role in neurological disorders. Pharmacol. Ther. 2020;213 doi: 10.1016/j.pharmthera.2020.107554. [DOI] [PubMed] [Google Scholar]
- Goldeman C., Ozgur B., Brodin B. Culture-induced changes in mRNA expression levels of efflux and SLC-transporters in brain endothelial cells. Fluids Barriers CNS. 2020;17:32. doi: 10.1186/s12987-020-00193-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldwaser E.L., Swanson R.L., 2nd, Arroyo E.J., Venkataraman V., Kosciuk M.C., Nagele R.G., et al. A preliminary report: the Hippocampus and surrounding temporal cortex of patients with schizophrenia have impaired blood-brain barrier. Front. Hum. Neurosci. 2022;16 doi: 10.3389/fnhum.2022.836980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales A.L., et al. Contractile pericytes determine the direction of blood flow at capillary junctions. Proc. Natl. Acad. Sci. U. S. A. 2020;117(45):27022–27033. doi: 10.1073/pnas.1922755117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg D.A. Cerebral angiogenesis: a realistic therapy for ischemic disease? Methods Mol. Biol. 2014;1135:21–24. doi: 10.1007/978-1-4939-0320-7_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene C., Hanley N., Campbell M. Claudin-5: gatekeeper of neurological function. Fluids Barriers CNS. 2019;16(1):3. doi: 10.1186/s12987-019-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griss J., et al. Recognizing millions of consistently unidentified spectra across hundreds of shotgun proteomics datasets. Nat. Methods. 2016;13:651–656. doi: 10.1038/nmeth.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubman A., Chew G., Ouyang J.F., et al. A single-cell atlas of entorhinal cortex from individuals with Alzheimer's disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 2019;22(12):2087–2097. doi: 10.1038/s41593-019-0539-4. [DOI] [PubMed] [Google Scholar]
- Haber M.A., et al. ERG is a novel and reliable marker for endothelial cells in central nervous system tumors. Clin. Neuropathol. 2015;34:117–127. doi: 10.5414/NP300817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne H., et al. DMSO enhances electrospray response, boosting sensitivity of proteomic experiments. Nat. Methods. 2013;10:989–991. doi: 10.1038/nmeth.2610. [DOI] [PubMed] [Google Scholar]
- Harris L.W., Wayland M., Lan M., et al. The cerebral microvasculature in schizophrenia: a laser capture microdissection study. PLoS One. 2008;3(12) doi: 10.1371/journal.pone.0003964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann D.A., et al. Brain capillary pericytes exert a substantial but slow influence on blood flow. Nat. Neurosci. 2021;24(4):633–645. doi: 10.1038/s41593-020-00793-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Vanlandewijck M., Raschperger E., et al. Analysis of the brain mural cell transcriptome. Sci. Rep. 2016;6 doi: 10.1038/srep35108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Shi X., Puthiyakunnon S., Zhang L., Zeng Q., Li Y., et al. CD44-mediated monocyte transmigration across Cryptococcus neoformans-infected brain microvascular endothelial cells is enhanced by HIV-1 gp41-I90 ectodomain. J. Biomed. Sci. 2016;23:28. doi: 10.1186/s12929-016-0247-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Vanlandewijck M., Raschperger E., et al. Single-cell RNA sequencing of mouse brain and lung vascular and vessel-associated cell types. Sci. Data. 2018;5 doi: 10.1038/sdata.2018.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert A.S., et al. Improved precursor characterization for data-dependent mass spectrometry. Anal. Chem. 2018;90:2333–2340. doi: 10.1021/acs.analchem.7b04808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger M.A., et al. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins. Pflugers. Arch. 2004;447(5):465–468. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- Hicks R.R., Baldwin S.A., Scheff S.W. Serum extravasation and cytoskeletal alterations following traumatic brain injury in rats. Comparison of lateral fluid percussion and cortical impact models. Mol. Chem. Neuropathol. 1997;32(1):1–16. doi: 10.1007/BF02815164. [DOI] [PubMed] [Google Scholar]
- Hicks S.C., Townes F.W., Teng M., Irizarry R.A. Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics. 2018;19(4):562–578. doi: 10.1093/biostatistics/kxx053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindle S.J., et al. Evolutionarily conserved roles for blood-brain barrier xenobiotic transporters in endogenous steroid partitioning and behavior. Cell Rep. 2017;21:1304–1316. doi: 10.1016/j.celrep.2017.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman F.M., Chen P., Incardona F., Zidovetzki R., Hinton D.R. HIV-1 tat protein induces the production of interleukin-8 by human brain-derived endothelial cells. J. Neuroimmunol. 1999;94(1–2):28–39. doi: 10.1016/s0165-5728(98)00198-2. [DOI] [PubMed] [Google Scholar]
- Hoge R.D., Atkinson J., Gill B., Crelier G.R., Marrett S., Pike G.B. Linear coupling between cerebral blood flow and oxygen consumption in activated human cortex. Proc. Natl. Acad. Sci. U. S. A. 1999;96(16):9403–9408. doi: 10.1073/pnas.96.16.9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadecola C. Regulation of the cerebral microcirculation during neural activity: is nitric oxide the missing link? Trends Neurosci. 1993;16(6):206–214. doi: 10.1016/0166-2236(93)90156-g. [DOI] [PubMed] [Google Scholar]
- Iadecola C. The neurovascular unit coming of age: a journey through neurovascular coupling in health and disease. Neuron. 2017;96:17–42. doi: 10.1016/j.neuron.2017.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliff JJ, Wang M, Zeppenfeld DM, et al. Cerebral arterial pulsation drives paravascular CSF-interstitial fluid exchange in the murine brain. J. Neurosci.. [DOI] [PMC free article] [PubMed]
- Iliff J.J., Wang M., Liao Y., et al. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid β. Sci. Transl. Med. 2012;4(147):147ra111. doi: 10.1126/scitranslmed.3003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isingrini E., Perret L., Rainer Q., et al. Genetic elimination of dopamine vesicular stocks in the nigrostriatal pathway replicates Parkinson's disease motor symptoms without neuronal degeneration in adult mice. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-12810-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturria-Medina Y., Sotero R.C., Toussaint P.J., et al. Early role of vascular dysregulation on late-onset Alzheimer's disease based on multifactorial data-driven analysis. Nat. Commun. 2016;7 doi: 10.1038/ncomms11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel S., Agirre E., Mendanha Falcão A., et al. Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature. 2019;566(7745):543–547. doi: 10.1038/s41586-019-0903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S.M., Park S.W., Kim H.J., Kim N.H., Park J.W. Changes in magnesium concentration in the serum and cerebrospinal fluid of neuropathic rats. Acta Anaesthesiol. Scand. 2006;50(2):211–216. doi: 10.1111/j.1399-6576.2006.00925.x. [DOI] [PubMed] [Google Scholar]
- Juliano R.L., Ling V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim. Biophys. Acta. 1976;455(1):152–162. doi: 10.1016/0005-2736(76)90160-7. [DOI] [PubMed] [Google Scholar]
- Kamath T., et al. Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson's disease. Nat. Neurosci. 2022;25:588–595. doi: 10.1038/s41593-022-01061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamintsky L., Cairns K.A., Veksler R., Bowen C., Beyea S.D., Friedman A., et al. Blood-brain barrier imaging as a potential biomarker for bipolar disorder progression. Neuroimag.Clin. 2020;26 doi: 10.1016/j.nicl.2019.102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan O., Filippi M., Freedman M.S., et al. Chronic cerebrospinal venous insufficiency and multiple sclerosis. Ann. Neurol. 2010;67(3):286–290. doi: 10.1002/ana.22001. [DOI] [PubMed] [Google Scholar]
- Kim S., et al. ERG immunohistochemistry as an endothelial marker for assessing lymphovascular invasion. Kor. J. Pathol. 2013;47:355–364. doi: 10.4132/KoreanJPathol.2013.47.4.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., et al. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315(5811):525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Kimelberg H.K., Nedergaard M. Functions of astrocytes and their potential as therapeutic targets. Neurotherapeutics. 2010;7(4):338–353. doi: 10.1016/j.nurt.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnecom K., Pachter J.S. Selective capture of endothelial and perivascular cells from brain microvessels using laser capture microdissection. Brain Res. Brain Res. Protoc. 2005;16(1–3):1–9. doi: 10.1016/j.brainresprot.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick B., Miller B.J. Inflammation and schizophrenia. Schizophr. Bull. 2013;39(6):1174–1179. doi: 10.1093/schbul/sbt141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisler K., Nelson A.R., Montagne A., Zlokovic B.V. Cerebral blood flow regulation and neurovascular dysfunction in Alzheimer disease. Nat. Rev. Neurosci. 2017;18:419–434. doi: 10.1038/nrn.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein B., Kuschinsky W., Schrock H., Vetterlein F. Interdependency of local capillary density, blood flow, and metabolism in rat brains. Am. J. Physiol. 1986;251(6 Pt 2):H1333–H1340. doi: 10.1152/ajpheart.1986.251.6.H1333. [DOI] [PubMed] [Google Scholar]
- Kojima T., et al. Subventricular zone-derived neural progenitor cells migrate along a blood vessel scaffold toward the post-stroke striatum. Stem Cell. 2010;28:545–554. doi: 10.1002/stem.306. [DOI] [PubMed] [Google Scholar]
- Kooij G., van Horssen J., de Lange E.C., Reijerkerk A., van der Pol S.M., van Het Hof B., et al. T lymphocytes impair P-glycoprotein function during neuroinflammation. J. Autoimmun. 2010;34(4):416–425. doi: 10.1016/j.jaut.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Laschinger M., Vajkoczy P., Engelhardt B. Encephalitogenic T cells use LFA-1 for transendothelial migration but not during capture and initial adhesion strengthening in healthy spinal cord microvessels in vivo. Eur. J. Immunol. 2002;32(12):3598–3606. doi: 10.1002/1521-4141(200212)32:12<3598::AID-IMMU3598>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Lau S.F., Cao H., Fu A.K.Y., Ip N.Y. Single-nucleus transcriptome analysis reveals dysregulation of angiogenic endothelial cells and neuroprotective glia in Alzheimer's disease. Proc. Natl. Acad. Sci. U. S. A. 2020;117(47):25800–25809. doi: 10.1073/pnas.2008762117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc E.H., Wislocki G.B. The histochemical localization of acid and alkaline phosphatases, non-specific esterase and succinic dehydrogenase in the structures comprising the hemato-encephalic barrier of the rat. J. Comp. Neurol. 1952;97(2):241–279. doi: 10.1002/cne.900970203. [DOI] [PubMed] [Google Scholar]
- Lee Y.K., Uchida H., Smith H., Ito A., Sanchez T. The isolation and molecular characterization of cerebral microvessels. Nat. Protoc. 2019;14(10):3059–3081. doi: 10.1038/s41596-019-0212-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewitus G.M., Konefal S.C., Greenhalgh A.D., Pribiag H., Augereau K., Stellwagen D. Microglial TNF-alpha suppresses cocaine-induced plasticity and behavioral sensitization. Neuron. 2016;90(3):483–491. doi: 10.1016/j.neuron.2016.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.Y., Chang C., Cheah J.H., et al. Dynamic changes in vascular permeability, cerebral blood volume, vascular density, and size after transient focal cerebral ischemia in rats: evaluation with contrast-enhanced magnetic resonance imaging. J. Cerebr. Blood Flow Metabol. 2008;28(7):1491–1501. doi: 10.1038/jcbfm.2008.42. [DOI] [PubMed] [Google Scholar]
- Liu P., Wu H., Yang H., et al. Time-course investigation of blood-brain barrier permeability and tight junction protein changes in a rat model of permanent focal ischemia. J. Physiol. Sci. 2018;68(2):121–127. doi: 10.1007/s12576-016-0516-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscher W., Potschka H. Blood-brain barrier active efflux transporters: ATP-binding cassette gene family. NeuroRx. 2005;2:86–98. doi: 10.1602/neurorx.2.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscher W., Potschka H. Blood-brain barrier active efflux transporters: ATP-binding cassette gene family. NeuroRx. 2005;2:86–98. doi: 10.1602/neurorx.2.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyck R., et al. Culture-induced changes in blood-brain barrier transcriptome: implications for amino-acid transporters in vivo. J. Cerebr. Blood Flow Metabol. 2009;29:1491–1502. doi: 10.1038/jcbfm.2009.72. [DOI] [PubMed] [Google Scholar]
- Mandi Y., Ocsovszki I., Szabo D., Nagy Z., Nelson J., Molnar J. Nitric oxide production and MDR expression by human brain endothelial cells. Anticancer Res. 1998;18(4C):3049–3052. [PubMed] [Google Scholar]
- Marchetti S., de Vries N.A., Buckle T., Bolijn M.J., van Eijndhoven M.A., Beijnen J.H., et al. Effect of the ATP-binding cassette drug transporters ABCB1, ABCG2, and ABCC2 on erlotinib hydrochloride (Tarceva) disposition in in vitro and in vivo pharmacokinetic studies employing Bcrp1-/-/Mdr1a/1b-/- (triple-knockout) and wild-type mice. Mol. Cancer Therapeut. 2008;7(8):2280–2287. doi: 10.1158/1535-7163.MCT-07-2250. [DOI] [PubMed] [Google Scholar]
- Mathiesen C., Caesar K., Akgören N., Lauritzen M. Modification of activity-dependent increases of cerebral blood flow by excitatory synaptic activity and spikes in rat cerebellar cortex. J. Physiol. 1998;512(Pt 2):555–566. doi: 10.1111/j.1469-7793.1998.555be.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiisen T.M., Lehre K.P., Danbolt N.C., Ottersen O.P. The perivascular astroglial sheath provides a complete covering of the brain microvessels: an electron microscopic 3D reconstruction. Glia. 2010;58(9):1094–1103. doi: 10.1002/glia.20990. [DOI] [PubMed] [Google Scholar]
- Mathys H., Davila-Velderrain J., Peng Z., et al. Single-cell transcriptomic analysis of Alzheimer's disease. Nature. 2019;570(7761):332–337. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell H.L., Kersch C.N., Woltjer R.L., Neuwelt E.A. The translational significance of the neurovascular unit. J. Biol. Chem. 2017;292(3):762–770. doi: 10.1074/jbc.R116.760215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire T.R., Trickler W.J., Hock L., Vrana A., Hoie E.B., Miller D.W. Release of prostaglandin E-2 in bovine brain endothelial cells after exposure to three unique forms of the antifungal drug amphotericin-B: role of COX-2 in amphotericin-B induced fever. Life Sci. 2003;72(23):2581–2590. doi: 10.1016/s0024-3205(03)00172-3. [DOI] [PubMed] [Google Scholar]
- Menard C., et al. Social stress induces neurovascular pathology promoting depression. Nat. Neurosci. 2017;20(12):1752–1760. doi: 10.1038/s41593-017-0010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen M., Wang Z.F., Paetau A., et al. ERG transcription factor as an immunohistochemical marker for vascular endothelial tumors and prostatic carcinoma. Am. J. Surg. Pathol. 2011;35(3):432–441. doi: 10.1097/PAS.0b013e318206b67b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintun M.A., Raichle M.E., Kilbourn M.R., Wooten G.F., Welch M.J. Blood flow and oxygen delivery to human brain during functional activity: theoretical modeling and experimental data. Proc. Natl. Acad. Sci. U. S. A. 2001;98(12):6859–6864. doi: 10.1073/pnas.111164398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitroi D.N., Tian M., Kawaguchi R., Lowry W.E., Carmichael S.T. Single-nucleus transcriptome analysis reveals disease- and regeneration-associated endothelial cells in white matter vascular dementia. J. Cell Mol. Med. 2022;26(6):3183–3195. doi: 10.1111/jcmm.17315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittapalli R.K., Manda V.K., Adkins C.E., Geldenhuys W.J., Lockman P.R. Exploiting nutrient transporters at the blood-brain barrier to improve brain distribution of small molecules. Ther. Deliv. 2010;1:775–784. doi: 10.4155/tde.10.76. [DOI] [PubMed] [Google Scholar]
- Mojsilovic-Petrovic J., Nesic M., Pen A., Zhang W., Stanimirovic D. Development of rapid staining protocols for laser-capture microdissection of brain vessels from human and rat coupled to gene expression analyses. J. Neurosci. Methods. 2004;133(1–2):39–48. doi: 10.1016/j.jneumeth.2003.09.026. [DOI] [PubMed] [Google Scholar]
- Mokgokong R., Wang S., Taylor C.J., Barrand M.A., Hladky S.B. Ion transporters in brain endothelial cells that contribute to formation of brain interstitial fluid. Pflugers. Arch. 2014;466(5):887–901. doi: 10.1007/s00424-013-1342-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne A., Barnes S.R., Sweeney M.D., et al. Blood-brain barrier breakdown in the aging human hippocampus. Neuron. 2015;85(2):296–302. doi: 10.1016/j.neuron.2014.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M.E., Rodriguez-Cruz V., Felmlee M.A. SLC and ABC transporters: expression, localization, and species differences at the blood-brain and the blood-cerebrospinal fluid barriers. AAPS J. 2017;19(5):1317–1331. doi: 10.1208/s12248-017-0110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueckler M., Thorens B. The SLC2 (GLUT) family of membrane transporters. Mol. Aspect. Med. 2013;34:121–138. doi: 10.1016/j.mam.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhl L., et al. Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat. Commun. 2020;11:3953. doi: 10.1038/s41467-020-17740-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munji R.N., Soung A.L., Weiner G.A., Sohet F., Semple B.D., Trivedi A., et al. Profiling the mouse brain endothelial transcriptome in health and disease models reveals a core blood-brain barrier dysfunction module. Nat. Neurosci. 2019;22(11):1892–1902. doi: 10.1038/s41593-019-0497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagelhus E.A., Ottersen O.P. Physiological roles of aquaporin-4 in brain. Physiol. Rev. 2013;93(4):1543–1562. doi: 10.1152/physrev.00011.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy C., Torres-Platas S.G., Mechawar N., Turecki G. Single-nucleus transcriptomics of the prefrontal cortex in major depressive disorder implicates oligodendrocyte precursor cells and excitatory neurons. Nat. Neurosci. 2020;23(6):771–781. doi: 10.1038/s41593-020-0621-y. [DOI] [PubMed] [Google Scholar]
- Nagyoszi P., Wilhelm I., Farkas A.E., Fazakas C., Dung N.T., Hasko J., et al. Expression and regulation of toll-like receptors in cerebral endothelial cells. Neurochem. Int. 2010;57(5):556–564. doi: 10.1016/j.neuint.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Nagyoszi P., Nyul-Toth A., Fazakas C., Wilhelm I., Kozma M., Molnar J., et al. Regulation of NOD-like receptors and inflammasome activation in cerebral endothelial cells. J. Neurochem. 2015;135(3):551–564. doi: 10.1111/jnc.13197. [DOI] [PubMed] [Google Scholar]
- Najjar S., Pearlman D.M., Devinsky O., Najjar A., Zagzag D. Neurovascular unit dysfunction with blood-brain barrier hyperpermeability contributes to major depressive disorder: a review of clinical and experimental evidence. J. Neuroinflammation. 2013;10:142. doi: 10.1186/1742-2094-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura E., et al. 4F2 (CD98) heavy chain is associated covalently with an amino acid transporter and controls intracellular trafficking and membrane topology of 4F2 heterodimer. J. Biol. Chem. 1999;274:3009–3016. doi: 10.1074/jbc.274.5.3009. [DOI] [PubMed] [Google Scholar]
- Navone S.E., Marfia G., Invernici G., et al. Isolation and expansion of human and mouse brain microvascular endothelial cells. Nat. Protoc. 2013;8(9):1680–1693. doi: 10.1038/nprot.2013.107. [DOI] [PubMed] [Google Scholar]
- Newman A.M., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37(7):773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P., et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie L., Wu G., Zhang W. Correlation of mRNA expression and protein abundance affected by multiple sequence features related to translational efficiency in Desulfovibrio vulgaris: a quantitative analysis. Genetics. 2006;174(4):2229–2243. doi: 10.1534/genetics.106.065862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova-Krstevski V., Wang X., Yu J., et al. ERG is required for the differentiation of embryonic stem cells along the endothelial lineage. BMC Dev. Biol. 2009;9:72. doi: 10.1186/1471-213X-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohab J.J., Fleming S., Blesch A., Carmichael S.T. A neurovascular niche for neurogenesis after stroke. J. Neurosci. 2006;26:13007–13016. doi: 10.1523/JNEUROSCI.4323-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz G.G., Pacheco-Moises F.P., Macias-Islas M.A., Flores-Alvarado L.J., Mireles-Ramirez M.A., Gonzalez-Renovato E.D., et al. Role of the blood-brain barrier in multiple sclerosis. Arch. Med. Res. 2014;45(8):687–697. doi: 10.1016/j.arcmed.2014.11.013. [DOI] [PubMed] [Google Scholar]
- Ortutay C., Siermala M., Vihinen M. Molecular characterization of the immune system: emergence of proteins, processes, and domains. Immunogenetics. 2007;59(5):333–348. doi: 10.1007/s00251-007-0191-0. [DOI] [PubMed] [Google Scholar]
- Papadopoulos M.C., Verkman A.S. Aquaporin water channels in the nervous system. Nat. Rev. Neurosci. 2013;14(4):265–277. doi: 10.1038/nrn3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana E., Cheng L.C., Doetsch F. Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc. Natl. Acad. Sci. U. S. A. 2009;106:6387–6392. doi: 10.1073/pnas.0810407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul G., et al. The adult human brain harbors multipotent perivascular mesenchymal stem cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai D.R., Dittmar M.S., Baldaranov D., et al. Cerebral ischemia-reperfusion injury in rats--a 3 T MRI study on biphasic blood-brain barrier opening and the dynamics of edema formation. J. Cerebr. Blood Flow Metabol. 2009;29(10):1846–1855. doi: 10.1038/jcbfm.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plane J.M., Andjelkovic A.V., Keep R.F., Parent J.M. Intact and injured endothelial cells differentially modulate postnatal murine forebrain neural stem cells. Neurobiol. Dis. 2010;37:218–227. doi: 10.1016/j.nbd.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polli J.W., et al. An unexpected synergist role of P-glycoprotein and breast cancer resistance protein on the central nervous system penetration of the tyrosine kinase inhibitor lapatinib (N-3-chloro-4-[(3-fluorobenzyl)oxy]phenyl-6-[5-([2-(methylsulfonyl)ethyl]aminomethyl)-2-furyl]-4-quinazolinamine; GW572016) Drug Metab. Dispos. 2009;37:439–442. doi: 10.1124/dmd.108.024646. [DOI] [PubMed] [Google Scholar]
- Puvogel S., et al. Single-nucleus RNA sequencing of midbrain blood-brain barrier cells in schizophrenia reveals subtle transcriptional changes with overall preservation of cellular proportions and phenotypes. Mol. Psychiatr. 2022;27(9):4731–4740. doi: 10.1038/s41380-022-01796-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan N., Banks W.A. Brain-immune communication pathways. Brain Behav. Immun. 2007;21(6):727–735. doi: 10.1016/j.bbi.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Reyes T.M., Fabry Z., Coe C.L. Brain endothelial cell production of a neuroprotective cytokine, interleukin-6, in response to noxious stimuli. Brain Res. 1999;851(1–2):215–220. doi: 10.1016/s0006-8993(99)02189-7. [DOI] [PubMed] [Google Scholar]
- Robey R.W., Pluchino K.M., Hall M.D., Fojo A.T., Bates S.E., Gottesman M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer. 2018;18(7):452–464. doi: 10.1038/s41568-018-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanul F.C., Bannister R.G. Localized areas of high alkaline phosphatase activity in the terminal arterial tree. J. Cell Biol. 1962;15(1):73–84. doi: 10.1083/jcb.15.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg G.A. Blood-brain barrier permeability in aging and Alzheimer's disease. J. Prev. Alzheimers Dis. 2014;1(3):138–139. doi: 10.14283/jpad.2014.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy C.S., Sherrington C.S. On the regulation of the blood-supply of the brain. J. Physiol. 1890;11(1–2):85–158. doi: 10.1113/jphysiol.1890.sp000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakers K., et al. Astrocytes locally translate transcripts in their peripheral processes. Proc. Natl. Acad. Sci. U. S. A. 2017;114(17):E3830–E3838. doi: 10.1073/pnas.1617782114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinkel A.H., Smit J.J., van Tellingen O., Beijnen J.H., Wagenaar E., van Deemter L., et al. Disruption of the mouse mdr1a P-glycoprotein gene leads to a deficiency in the blood-brain barrier and to increased sensitivity to drugs. Cell. 1994;77(4):491–502. doi: 10.1016/0092-8674(94)90212-7. [DOI] [PubMed] [Google Scholar]
- Schroder M.L., Muizelaar J.P., Fatouros P.P., Kuta A.J., Choi S.C. Regional cerebral blood volume after severe head injury in patients with regional cerebral ischemia. Neurosurgery. 1998;42(6):1276–1280. doi: 10.1097/00006123-199806000-00042. ; discussion 1280-1271. [DOI] [PubMed] [Google Scholar]
- Schwarzmaier S.M., Kim S.W., Trabold R., Plesnila N. Temporal profile of thrombogenesis in the cerebral microcirculation after traumatic brain injury in mice. J. Neurotrauma. 2010;27(1):121–130. doi: 10.1089/neu.2009.1114. [DOI] [PubMed] [Google Scholar]
- Shapira Y., Setton D., Artru A.A., Shohami E. Blood-brain barrier permeability, cerebral edema, and neurologic function after closed head injury in rats. Anesth. Analg. 1993;77(1):141–148. doi: 10.1213/00000539-199307000-00028. [DOI] [PubMed] [Google Scholar]
- Shimizu N. Histochemical studies on the phosphatase of the nervous system. J. Comp. Neurol. 1950;93(2):201–217. doi: 10.1002/cne.900930203. [DOI] [PubMed] [Google Scholar]
- Shishkova E., Hebert A.S., Coon J.J. Now, more than ever, proteomics needs better chromatography. Cell Syst. 2016;3:321–324. doi: 10.1016/j.cels.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishkova E., et al. Ultra-high pressure (>30,000 psi) packing of capillary columns enhancing depth of shotgun proteomic analyses. Anal. Chem. 2018;90:11503–11508. doi: 10.1021/acs.analchem.8b02766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Q.R., Rapoport S.I. Cerebrovascular permeability coefficients to sodium, potassium, and chloride. J. Neurochem. 1986;46(6):1732–1742. doi: 10.1111/j.1471-4159.1986.tb08491.x. [DOI] [PubMed] [Google Scholar]
- Smith T., Heger A., Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017;27(3):491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.W., Castrillon-Rodriguez E., Armstrong E.A., et al. Transcriptomic comparison of human and mouse brain microvessels. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-69096-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.W., et al. Transcriptomic comparison of human and mouse brain microvessels. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-69096-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorf A., Hofman J., Kucera R., Staud F., Ceckova M. Ribociclib shows potential for pharmacokinetic drug-drug interactions being a substrate of ABCB1 and potent inhibitor of ABCB1, ABCG2 and CYP450 isoforms in vitro. Biochem. Pharmacol. 2018;154:10–17. doi: 10.1016/j.bcp.2018.04.013. [DOI] [PubMed] [Google Scholar]
- Spitzer D., Combes F., Colonna M. A flow cytometry-based protocol for syngenic isolation of neurovascular unit cells from mouse and human tissues. Nat. Protoc. 2023 doi: 10.1038/s41596-023-00805-y. [DOI] [PubMed] [Google Scholar]
- Stein S.C., Chen X.H., Sinson G.P., Smith D.H. Intravascular coagulation: a major secondary insult in nonfatal traumatic brain injury. J. Neurosurg. 2002;97(6):1373–1377. doi: 10.3171/jns.2002.97.6.1373. [DOI] [PubMed] [Google Scholar]
- Strbian D., Durukan A., Pitkonen M., et al. The blood-brain barrier is continuously open for several weeks following transient focal cerebral ischemia. Neuroscience. 2008;153(1):175–181. doi: 10.1016/j.neuroscience.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Stuart T., et al. Comprehensive integration of single-cell data. Cell. 2019;177(7) doi: 10.1016/j.cell.2019.05.031. 1888-1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Lytle C., O'Donnell M.E. IL-6 secreted by astroglial cells regulates Na-K-Cl cotransport in brain microvessel endothelial cells. Am. J. Physiol. 1997;272(6 Pt 1):C1829–C1835. doi: 10.1152/ajpcell.1997.272.6.C1829. [DOI] [PubMed] [Google Scholar]
- Sun H., Dai H., Shaik N., Elmquist W.F. Drug efflux transporters in the CNS. Adv. Drug Deliv. Rev. 2003;55:83–105. doi: 10.1016/s0169-409x(02)00172-2. [DOI] [PubMed] [Google Scholar]
- Sutton G.J., et al. Comprehensive evaluation of deconvolution methods for human brain gene expression. Nat. Commun. 2022;13(1):1358. doi: 10.1038/s41467-022-28655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M.D., Sagare A.P., Zlokovic B.V. Cerebrospinal fluid biomarkers of neurovascular dysfunction in mild dementia and Alzheimer's disease. J. Cerebr. Blood Flow Metabol. 2015;35(6):1055–1068. doi: 10.1038/jcbfm.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M.D., Ayyadurai S., Zlokovic B.V. Pericytes of the neurovascular unit: key functions and signaling pathways. Nat. Neurosci. 2016;19(6):771–783. doi: 10.1038/nn.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadic V., et al. Primary familial brain calcification with known gene mutations: a systematic review and challenges of phenotypic characterization. JAMA Neurol. 2015;72(4):460–467. doi: 10.1001/jamaneurol.2014.3889. [DOI] [PubMed] [Google Scholar]
- Thiebaut F., Tsuruo T., Hamada H., Gottesman M.M. Immunohistochemical localization in normal tissues of different epitopes in the multidrug transport protein P170: evidence for localization in brain capillaries and crossreactivity of one antibody with a muscle protein. J. Histochem. Cytochem. 1989;37:159–164. doi: 10.1177/37.2.2463300. [DOI] [PubMed] [Google Scholar]
- Thrupp N., Sala Frigerio C., Wolfs L., Skene N.G., Fattorelli N., Poovathingal S., et al. Single-nucleus RNA-seq is not suitable for detection of microglial activation genes in humans. Cell Rep. 2020;32(13) doi: 10.1016/j.celrep.2020.108189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timp W., Timp G. Beyond mass spectrometry, the next step in proteomics. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aax8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo J.B., Arnold S.E., Raible K., et al. Contribution of cerebrovascular disease in autopsy confirmed neurodegenerative disease cases in the National Alzheimer's Coordinating Centre. Brain. 2013;136(9):2697–2706. doi: 10.1093/brain/awt188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Platas S.G., Cruceanu C., Chen G.G., Turecki G., Mechawar N. Evidence for increased microglial priming and macrophage recruitment in the dorsal anterior cingulate white matter of depressed suicides. Brain Behav. Immun. 2014;42:50–59. doi: 10.1016/j.bbi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Toth G., Panic-Jankovic T., Mitulovic G. Pillar array columns for peptide separations in nanoscale reversed-phase chromatography. J. Chromatogr. A. 2019;1603:426–432. doi: 10.1016/j.chroma.2019.06.067. [DOI] [PubMed] [Google Scholar]
- Trajkovic M., et al. Abnormal thyroid hormone metabolism in mice lacking the monocarboxylate transporter 8. J. Clin. Invest. 2007;117:627–635. doi: 10.1172/JCI28253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traugott U., Reinherz E.L., Raine C.S. Multiple sclerosis. Distribution of T cells, T cell subsets and Ia-positive macrophages in lesions of different ages. J. Neuroimmunol. 1983;4(3):201–221. doi: 10.1016/0165-5728(83)90036-x. [DOI] [PubMed] [Google Scholar]
- Traxl A., Mairinger S., Filip T., Sauberer M., Stanek J., Poschner S., et al. Inhibition of ABCB1 and ABCG2 at the mouse blood-brain barrier with marketed drugs to improve brain delivery of the model ABCB1/ABCG2 substrate [(11)C]erlotinib. Mol. Pharm. 2019;16(3):1282–1293. doi: 10.1021/acs.molpharmaceut.8b01217. [DOI] [PubMed] [Google Scholar]
- Uchida Y., Ohtsuki S., Katsukura Y., Ikeda C., Suzuki T., Kamiie J., et al. Quantitative targeted absolute proteomics of human blood-brain barrier transporters and receptors. J. Neurochem. 2011;117(2):333–345. doi: 10.1111/j.1471-4159.2011.07208.x. [DOI] [PubMed] [Google Scholar]
- van Beijnum J.R., Rousch M., Castermans K., van der Linden E., Griffioen A.W. Isolation of endothelial cells from fresh tissues. Nat. Protoc. 2008;3(7):1085–1091. doi: 10.1038/nprot.2008.71. [DOI] [PubMed] [Google Scholar]
- Vanlandewijck M., He L., Mäe M.A., et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554(7693):475–480. doi: 10.1038/s41586-018-0232-x. [DOI] [PubMed] [Google Scholar]
- Velmeshev D., Schirmer L., Jung D., et al. Single-cell genomics identifies cell type-specific molecular changes in autism. Science. 2019;364(6441):685–689. doi: 10.1126/science.aav8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura-Antunes L., Dasgupta O.M., Herculano-Houzel S. Resting rates of blood flow and glucose use per neuron are proportional to number of endothelial cells available per neuron across sites in the rat brain. Front. Integr. Neurosci. 2022;16 doi: 10.3389/fnint.2022.821850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen I.C.M., Van Boxtel M.P.J., Verhey F.R.J., Jansen J.F.A., Backes W.H. Interaction between blood-brain barrier and glymphatic system in solute clearance. Neurosci. Biobehav. Rev. 2018;90:26–33. doi: 10.1016/j.neubiorev.2018.03.028. [DOI] [PubMed] [Google Scholar]
- Vijay N., Morris M.E. Role of monocarboxylate transporters in drug delivery to the brain. Curr. Pharmaceut. Des. 2014;20:1487–1498. doi: 10.2174/13816128113199990462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C., Marcotte E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13(4):227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Oettingen G., Bergholt B., Gyldensted C., Astrup J. Blood flow and ischemia within traumatic cerebral contusions. Neurosurgery. 2002;50(4):781–788. doi: 10.1097/00006123-200204000-00019. discussion 788-790. [DOI] [PubMed] [Google Scholar]
- Wang D., Johnson A.D., Papp A.C., Kroetz D.L., Sadee W. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenetics Genom. 2005;15(10):693–704. [PubMed] [Google Scholar]
- Weber B., Keller A.L., Reichold J., Logothetis N.K. The microvascular system of the striate and extrastriate visual cortex of the macaque. Cerebr. Cortex. 2008;18(10):2318–2330. doi: 10.1093/cercor/bhm259. [DOI] [PubMed] [Google Scholar]
- Wilhelm I., Fazakas C., Molnár J., et al. Regulation of cerebral endothelial cell morphology by extracellular calcium. Phys. Med. Biol. 2007;52(20):6261–6274. doi: 10.1088/0031-9155/52/20/012. [DOI] [PubMed] [Google Scholar]
- Winkler E.A., Bell R.D., Zlokovic B.V. Pericyte-specific expression of PDGF beta receptor in mouse models with normal and deficient PDGF beta receptor signaling. Mol. Neurodegener. 2010;5:32. doi: 10.1186/1750-1326-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.A., Bell R.D., Zlokovic B.V. Central nervous system pericytes in health and disease. Nat. Neurosci. 2011;14(11):1398–1405. doi: 10.1038/nn.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolburg H., Noell S., Mack A., Wolburg-Buchholz K., Fallier-Becker P. Brain endothelial cells and the glio-vascular complex. Cell Tissue Res. 2009;335(1):75–96. doi: 10.1007/s00441-008-0658-9. [DOI] [PubMed] [Google Scholar]
- Wu E.X., Tang H., Jensen J.H. High-resolution MR imaging of mouse brain microvasculature using the relaxation rate shift index Q. NMR Biomed. 2004;17(8):507–512. doi: 10.1002/nbm.921. [DOI] [PubMed] [Google Scholar]
- Yamashita T., et al. Subventricular zone-derived neuroblasts migrate and differentiate into mature neurons in the post-stroke adult striatum. J. Neurosci. 2006;26:6627–6636. doi: 10.1523/JNEUROSCI.0149-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida O., et al. Human L-type amino acid transporter 1 (LAT1): characterization of function and expression in tumor cell lines. Biochim. Biophys. Acta. 2001;1514:291–302. doi: 10.1016/s0005-2736(01)00384-4. [DOI] [PubMed] [Google Scholar]
- Yang A.C., Stevens M.Y., Chen M.B., et al. A human brain vascular atlas reveals diverse mediators of Alzheimer's risk. Nature. 2022;603(7897):885–892. doi: 10.1038/s41586-021-04369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Mojsilovic-Petrovic J., Andrade M.F., Zhang H., Ball M., Stanimirovic D.B. The expression and functional characterization of ABCG2 in brain endothelial cells and vessels. Faseb. J. 2003;17(14):2085–2087. doi: 10.1096/fj.02-1131fje. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S.A., et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34(36):11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlokovic B.V. The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron. 2008;57:178–201. doi: 10.1016/j.neuron.2008.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.