Abstract
CRISPR diagnostics have recently emerged as powerful diagnostic tools for the rapid detection of infections. The ultimate goal is to develop these diagnostics for the point of care, where patients quickly receive and easily interpret results. Although they are in their infancy, the COVID-19 pandemic has accelerated innovation of CRISPR diagnostics and led to an explosion of improvements to these systems. Challenges that have impeded the implementation at the point of care have been addressed, and CRISPR diagnostics have been dramatically simplified. Here we outline recent developments and advancements in CRISPR diagnostics that have pushed these technologies to the point of care.
Graphical Abstract
Rapid diagnostics are crucial in the battle against highly transmissible diseases such as COVID-19. Point-of-care (POC) diagnostics ideally require intuitive laboratory equipment and rapidly produce easily interpretable results.1 For the detection of infectious diseases, POC diagnostics have primarily relied on the detection of antigens to identify a pathogen in an infected individual.2 Antigen tests produce rapid results; however, their accuracy can vary, and successful detection is dependent on virus maturation and high viral titers.3 On the contrary, nucleic acid tests can detect an infection prior to a patient becoming transmissive.4,5 Recently, CRISPR-based diagnostics (CRISPR-Dx) have emerged as revolutionary nucleic acid tests with the potential to operate in POC settings.
CRISPR-Dx generally combine amplification technologies with CRISPR-Cas (Cas) enzymes and exploit their sequence-specific recognition properties to detect pathogens. CRISPR-Dx have primarily utilized two main CRISPR effector types to accomplish nucleic acid detection: type V (Cas12) DNA-targeting systems6 and type VI (Cas13) RNA-targeting systems7,8 (Table 1). Both Cas12 and Cas13 effectors exhibit collateral cleavage activity, or subsequent cleavage of bystander nucleic acid sequences, where Cas12 cleaves secondary ssDNA molecules9,10 and Cas13 cleaves secondary ssRNA molecules.7,8,11,12 For CRISPR-Dx, the collateral cleavage activity has been harnessed as the primary mode of nucleic acid detection9,13–17 (Figure 1). This is accomplished by introducing a modified secondary probe that, upon cleavage, provides readouts by fluorescence or visually by lateral flow. Further investigation into this phenomenon identified distinct nucleic acid sequence preferences of collateral probes for various Cas13 orthologs and subtypes, ultimately demonstrating multiplexing capabilities of CRISPR-Dx.14,17 Cas effectors have improved clinical diagnostic systems by providing highly specific sequence recognition; however, the sensitivity of CRISPR-Dx is primarily dependent on the amplification method applied.
Table 1.
Summary of CRISPR-Dx Technologies Using Collateral Cleavage
| diagnostic tool | Cas effector | amplification method | readout method | ref |
|---|---|---|---|---|
| SHERLOCK | Cas13a, Cas13b, Cas12a, Csm6 | RPA | fluorescence/lateral flow | 13, 14 |
| DETECTR | Cas12a | RPA, LAMP | fluorescence/lateral flow | 9, 16 |
| STOP | Cas12b | LAMP | fluorescence/lateral flow | 24 |
| SENSR | Cas13d | RPA | fluorescence/lateral flow | 17 |
| amplification-free detection | Cas13a, Csm6 | N/A | fluorescence | 8, 26–, 28 |
Figure 1.
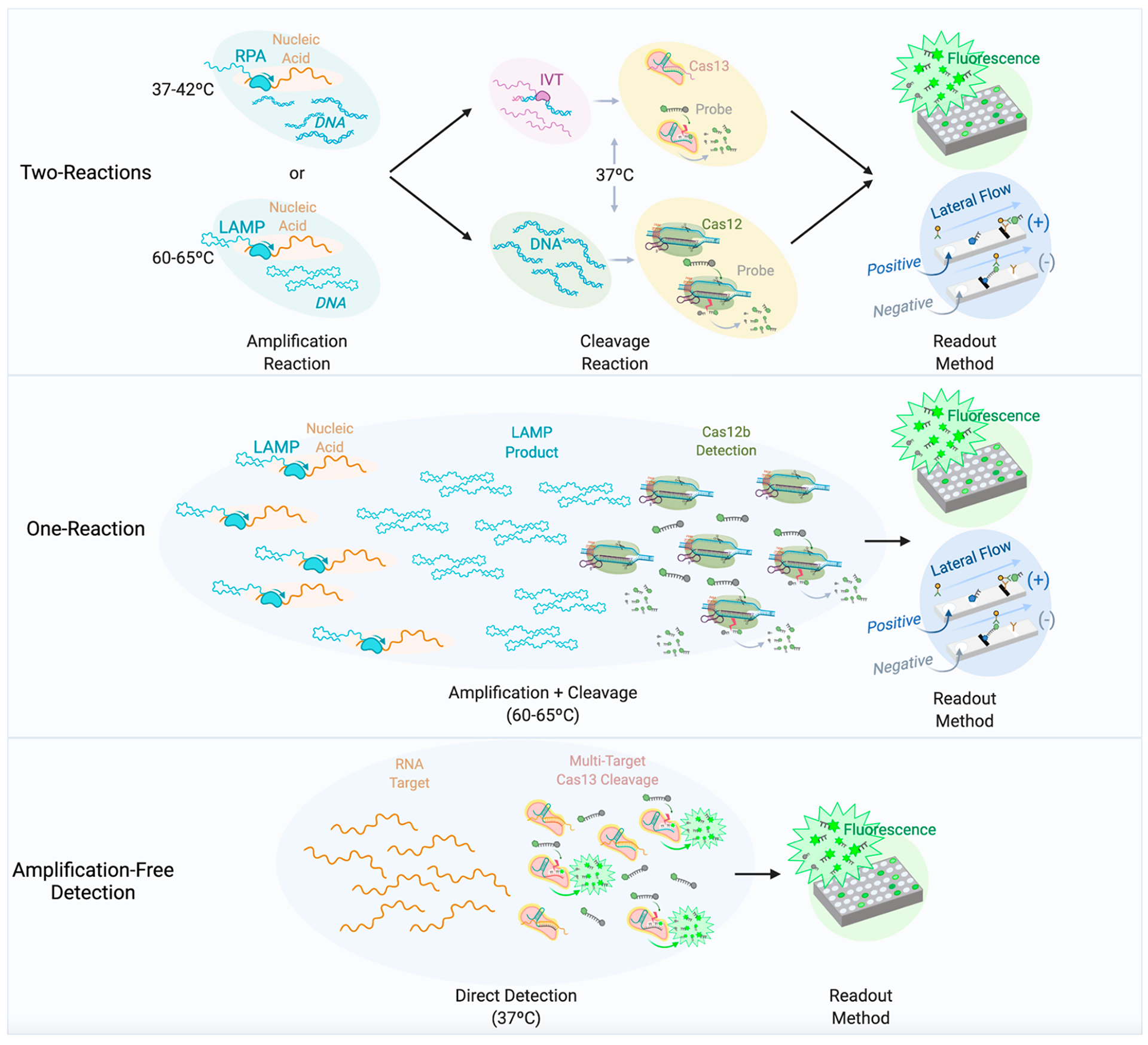
CRISPR-Dx formats for detection of nucleic acids. Schematic representation of different formats of CRISPR-Dx systems. Two-reaction diagnostics represent the original formats established for CRISPR-Dx. One-reaction diagnostics involve the combination of isothermal amplification methods and Cas detection into one reaction, most notably represented by STOP, which uses LAMP and Cas12b for detection. Amplification-free diagnostics exploit direct detection of nucleic acids using Cas enzymes and utilize multiple targets to amplify the detection signal. Only amplification-based detection methods have been established for both fluorescence and lateral flow detection, while amplification-free systems have been established only for detection via fluorescence.
The concept of harnessing collateral cleavage for nucleic acid detection was first established in 2016.8 Subsequently, the addition of an amplification method to the SHERLOCK system dramatically improved the sensitivity of CRISPR-Dx.13 To simplify these diagnostic tools, the first iterations of CRISPR-Dx utilized an isothermal amplification method for targeted sequence amplification termed recombinase polymerase amplification (RPA).18 Successive versions of these diagnostics swapped RPA for an alternative isothermal amplification method with greater specificity and sensitivity termed loop-mediated isothermal amplification (LAMP).16,19–21 The use of isothermal amplification methods simplifies the technical handling and equipment requirements of CRISPR-Dx and thus makes the transition to the POC setting smooth.
CRISPR-Dx have the potential to be developed as effective POC diagnostics; however, notable incompatibilities between isothermal amplification methods and Cas enzymes have limited this progress. For example, the viscosity of RPA has been reported to decrease the sensitivity and diminish the reliability in single-reaction SHERLOCK diagnostics,22 and LAMP reactions operate at high temperatures (60–65 °C) beyond the optimal operation temperature of most Cas effectors (37–42 °C).6–8,12,23 These incompatibilities have limited the development of single-reaction CRISPR-Dx and led researchers to develop two-reaction diagnostics that are technically complex and prone to contamination. However, through discovery and innovation, significant progress has been made to further the development of CRISPR-Dx for POC settings.
SINGLE-REACTION CRISPR-DX
The transfer steps required in earlier versions of CRISPR-Dx were the main drawbacks for implementing these tests in POC settings. Recently, significant progress has been made in the development of single-reaction CRISPR-Dx technologies. For example, the SHERLOCK group recently demonstrated a system for detection of SARS-CoV-2 termed STOP (SHERLOCK testing in one pot).24 To develop this assay, the group identified a thermostable Cas12 effector (AapCas12b) that demonstrates robust cleavage activity at higher temperatures.25 The thermostable property of this Cas12 enzyme enabled the integration of this unique effector with LAMP into a single reaction. Furthermore, this test utilized a simplified extraction protocol that allowed the entire sample to be used as input leading to improved detection compared with the CDC standard RT-qPCR protocol.
STOP is a significant advance for CRISPR-Dx and demonstrates the robustness of these CRISPR-Dx tools; however, this development also highlights significant obstacles these technologies face. For instance, the use of AapCas12b underscores the need to discover more thermostable Cas effectors to enable multiplexing capabilities previously demonstrated in two-reaction systems.14 In addition, although STOP requires only a single temperature for detection, the heat requirement of this reaction increases the technological requirements for this tool, whereas POC diagnostics ideally operate at room temperature.
While much effort has been focused on integrating CRISPR-mediated detection with isothermal amplification into a single reaction, other researchers have focused solely on exploiting the robust cleavage activity of Cas effectors for direct detection of nucleic acids. Upon removal of the amplification methods, CRISPR-Dx systems can be simplified into a single reaction with a focus on improving the effect of collateral cleavage activity. Recently, groups have developed amplification-free CRISPR-Dx for the detection of SARS-CoV-2, by exploiting the robust collateral cleavage activity of Cas13a effectors.26–28 To achieve sensitive detection comparable to amplification-based CRISPR-Dx, the groups screened multiple targets and combined the best targets together to improve detection. The use of multiple targets in one reaction dramatically improved the sensitivity of these systems; however, considerable variation was observed between orthologs. Using LwaCas13a, detection of 3400 copies per microliter was achieved,27 whereas detection of 100 copies per microliter was achieved using LbuCas13a.26 In a later study, direct detection with LbuCas13a was enhanced by leveraging a type III CRISPR effector, Csm6,29 and modifying the collateral probe to contain a Csm6 activator resulting in improved sensitivity (31 copies per microliter). Although the sensitivity was significantly improved, this method required eight targets as opposed to three targets used in the previously mentioned studies.28
Amplification-free CRISPR-Dx that exploit the targeting activities of Cas effectors are very attractive for their simplicity; however, notable drawbacks hinder these applications. One considerable drawback is the requirement for multiple target sites, which prevents detection of single-nucleotide polymorphisms and subjects the accuracy of these diagnostics to sequence variations. Amplification-free methods also require RNA extraction to achieve detection, and thus until direct detection is achieved without extraction, application of these tests in the POC setting remains impractical. Furthermore, recent evidence has suggested a disagreement between published results and theoretical limits of direct detection with Cas effectors, presenting a clear need to evaluate these technologies further.30 Finally, Cas effectors preferentially target one nucleic acid species, and therefore, the flexibility of amplification-free CRISPR-Dx is limited to one nucleic acid species that depends on the Cas effector chosen.
As groups work to innovate the CRISPR-Dx reactions, others have decided to engineer novel devices and take advantage of the mass use of smartphones. Two innovative methods were developed on the basis of the established SHERLOCK methods using Cas13 and Cas12 (SHINE and miSHERLOCK, respectively).31,32 SHINE created a streamlined detection method by improving HUSDON,33 a previously developed lysis method, and developing an app-based readout of fluorescence to minimize sample contamination and user bias. miSHERLOCK engineered a unique lysis method and device to enable simplified lysis of viral particles and interpretation of results. The device is built to allow users to capture fluorescence results on a smartphone app that automatically quantifies and determines results, similar to the app used in SHINE. Alternatively, another group engineered a smartphone-compatible device in which numerous Cas12 detection reactions run on a chip in parallel.34 This diagnostic, termed CRISPR-FDS, enables user-friendly assay setup and interpretation, does not require an extraction step, and provides rapid turnover of results with demonstrated sensitivity comparable to that of RT-PCR. These innovations provide promise for engineering novel devices or developing smartphone applications to simplify CRISPR-Dx reactions and interpretation that will further streamline these diagnostic tools.
CONCLUSION
CRISPR-Dx remain in their nascency, yet innovation is progressing rapidly. Although still early in development, advances toward simplified CRISPR-Dx are happening rapidly and, due to the COVID-19 pandemic, have been dramatically accelerated. Future developments of CRISPR-Dx are likely to address concerns related to technical handling, sensitivity, and multiplexing capabilities necessary for POC diagnostics. With these rapid advances, CRISPR-Dx present promising nucleic acid detection technologies that are likely to shape the future of POC diagnostics.
Funding
This work was supported in part by a Directors New Innovator award from the National Institute of Allergy and Infectious Diseases (DP2 AI152071-01) to O.S.A.
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.biochem.2c00051
The authors declare no competing financial interest.
Contributor Information
Daniel J. Brogan, Division of Biological Sciences, Section of Cell and Developmental Biology, University of California, San Diego, La Jolla, California 92093, United States;
Omar S. Akbari, Division of Biological Sciences, Section of Cell and Developmental Biology, University of California, San Diego, La Jolla, California 92093, United States;
REFERENCES
- (1).Valera E; Jankelow A; Lim J; Kindratenko V; Ganguli A; White K; Kumar J; Bashir R COVID-19 Point-of-Care Diagnostics: Present and Future. ACS Nano 2021, 15, 7899–7906. [DOI] [PubMed] [Google Scholar]
- (2).Pavia CS; Plummer MM The evolution of rapid antigen detection systems and their application for COVID-19 and other serious respiratory infectious diseases. J. Microbiol. Immunol. Infect 2021, 54, 776–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Pilarowski G; Lebel P; Sunshine S; Liu J; Crawford E; Marquez C; Rubio L; Chamie G; Martinez J; Peng J; Black D; Wu W; Pak J; Laurie MT; Jones D; Miller S; Jacobo J; Rojas S; Rojas S; Nakamura R; Tulier-Laiwa V; Petersen M; Havlir DV; DeRisi J Performance Characteristics of a Rapid Severe Acute Respiratory Syndrome Coronavirus 2 Antigen Detection Assay at a Public Plaza Testing Site in San Francisco. J. Infect. Dis 2021, 223, 1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Pan Y; Zhang D; Yang P; Poon LLM; Wang Q Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis 2020, 20, 411–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Byrne AW; McEvoy D; Collins AB; Hunt K; Casey M; Barber A; Butler F; Griffin J; Lane EA; McAloon C; O’Brien K; Wall P; Walsh KA; More SJ Inferred duration of infectious period of SARS-CoV-2: rapid scoping review and analysis of available evidence for asymptomatic and symptomatic COVID-19 cases. BMJ. Open 2020, 10, No. e039856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zetsche B; Gootenberg JS; Abudayyeh OO; Slaymaker IM; Makarova KS; Essletzbichler P; Volz SE; Joung J; van der Oost J; Regev A; Koonin EV; Zhang F Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Abudayyeh OO; Gootenberg JS; Konermann S; Joung J; Slaymaker IM; Cox DBT; Shmakov S; Makarova KS; Semenova E; Minakhin L; Severinov K; Regev A; Lander ES; Koonin EV; Zhang F C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).East-Seletsky A; O’Connell MR; Knight SC; Burstein D; Cate JHD; Tjian R; Doudna JA Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Chen JS; Ma E; Harrington LB; Da Costa M; Tian X; Palefsky JM; Doudna JA CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Li S-Y; Cheng Q-X; Liu J-K; Nie X-Q; Zhao G-P; Wang J CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018, 28, 491–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Smargon AA; Cox DBT; Pyzocha NK; Zheng K; Slaymaker IM; Gootenberg JS; Abudayyeh OA; Essletzbichler P; Shmakov S; Makarova KS; Koonin EV; Zhang F aCas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol. Cell 2017, 65, 618–630.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Konermann S; Lotfy P; Brideau NJ; Oki J; Shokhirev MN; Hsu PD Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 2018, 173, 665–676.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Gootenberg JS; Abudayyeh OO; Lee JW; Essletzbichler P; Dy AJ; Joung J; Verdine V; Donghia N; Daringer NM; Freije CA; Myhrvold C; Bhattacharyya RP; Livny J; Regev A; Koonin EV; Hung DT; Sabeti PC; Collins JJ; Zhang F Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Gootenberg JS; Abudayyeh OO; Kellner MJ; Joung J; Collins JJ; Zhang F Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ackerman CM; Myhrvold C; Thakku SG; Freije CA; Metsky HC; Yang DK; Ye SH; Boehm CK; Kosoko-Thoroddsen T-SF; Kehe J; Nguyen TG; Carter A; Kulesa A; Barnes JR; Dugan VG; Hung DT; Blainey PC; Sabeti PC Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Broughton JP; Deng X; Yu G; Fasching CL; Servellita V; Singh J; Miao X; Streithorst JA; Granados A; Sotomayor-Gonzalez A; Zorn K; Gopez A; Hsu E; Gu W; Miller S; Pan C-Y; Guevara H; Wadford DA; Chen JS; Chiu CY CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol 2020, 38, 870–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Brogan DJ; Chaverra-Rodriguez D; Lin CP; Smidler AL; Yang T; Alcantara LM; Antoshechkin I; Liu J; Raban RR; Belda-Ferre P; Knight R; Komives EA; Akbari OS Development of a Rapid and Sensitive CasRx-Based Diagnostic Assay for SARS-CoV-2. ACS Sens. 2021, 6, 3957–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Piepenburg O; Williams CH; Stemple DL; Armes NA DNA detection using recombination proteins. PLoS Biol. 2006, 4, No. e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Notomi T; Okayama H; Masubuchi H; Yonekawa T; Watanabe K; Amino N; Hase T Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, No. 63e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Howson ELA; Kurosaki Y; Yasuda J; Takahashi M; Goto H; Gray AR; Mioulet V; King DP; Fowler VL Defining the relative performance of isothermal assays that can be used for rapid and sensitive detection of foot-and-mouth disease virus. J. Virol. Methods 2017, 249, 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Agrawal S; Fanton A; Chandrasekaran SS; Prywes N; Lukarska M; Biering SB; Smock DCJ; Mok A; Knott GJ; Van E; Dugan E; Kim S; Liu TY; Harris E; Stanley SA; Lareau LF; Doudna JA; Savage DF; Hsu PD Rapid detection of SARS-CoV-2 with Cas13. medRxiv 2020, DOI: 10.1101/2020.12.14.20247874. [DOI] [Google Scholar]
- (22).Kellner MJ; Koob JG; Gootenberg JS; Abudayyeh OO; Zhang F SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc 2019, 14, 2986–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Yan WX; Chong S; Zhang H; Makarova KS; Koonin EV; Cheng DR; Scott DA Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Mol. Cell 2018, 70, 327–339.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Joung J; Ladha A; Saito M; Kim N-G; Woolley AE; Segel M; Barretto RPJ; Ranu A; Macrae RK; Faure G; Ioannidi EI; Krajeski RN; Bruneau R; Huang M-LW; Yu XG; Li JZ; Walker BD; Hung DT; Greninger AL; Jerome KR; Gootenberg JS; Abudayyeh OO; Zhang F Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N. Engl. J. Med 2020, 383, 1492–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Teng F; Cui T; Feng G; Guo L; Xu K; Gao Q; Li T; Li J; Zhou Q; Li W Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discovery 2018, 4, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Fozouni P; Son S; Díaz de León Derby M; Knott GJ; Gray CN; D’Ambrosio MV; Zhao C; Switz NA; Kumar GR; Stephens SI; Boehm D; Tsou C-L; Shu J; Bhuiya A; Armstrong M; Harris AR; Chen P-Y; Osterloh JM; Meyer-Franke A; Joehnk B; Walcott K; Sil A; Langelier C; Pollard KS; Crawford ED; Puschnik AS; Phelps M; Kistler A; DeRisi JL; Doudna JA; Fletcher DA; Ott M Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2021, 184, 323–333.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Shinoda H; Taguchi Y; Nakagawa R; Makino A; Okazaki S; Nakano M; Muramoto Y; Takahashi C; Takahashi I; Ando J; Noda T; Nureki O; Nishimasu H; Watanabe R Amplification-free RNA detection with CRISPR-Cas13. Commun. Biol 2021, 4, 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Liu TY; Knott GJ; Smock DCJ; Desmarais JJ; Son S; Bhuiya A; Jakhanwal S; Prywes N; Agrawal S; Díaz de León Derby M; Switz NA; Armstrong M; Harris AR; Charles EJ; Thornton BW; Fozouni P; Shu J; Stephens SI; Kumar GR; Zhao C; Mok A; Iavarone AT; Escajeda AM; McIntosh R; Kim S; Dugan EJ; IGI Testing Consortium; Pollard KS; Tan MX; Ott M; Fletcher DA; Lareau LF; Hsu PD; Savage DF; Doudna JA; et al. Accelerated RNA detection using tandem CRISPR nucleases. Nat. Chem. Biol 2021, 17, 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Jiang W; Samai P; Marraffini LA Degradation of Phage Transcripts by CRISPR-Associated RNases Enables Type III CRISPR-Cas Immunity. Cell 2016, 164, 710–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ramachandran A; Santiago JG CRISPR Enzyme Kinetics for Molecular Diagnostics. Anal. Chem 2021, 93, 7456–7464. [DOI] [PubMed] [Google Scholar]
- (31).Arizti-Sanz J; Freije CA; Stanton AC; Petros BA; Boehm CK; Siddiqui S; Shaw BM; Adams G; Kosoko-Thoroddsen T-SF; Kemball ME; Uwanibe JN; Ajogbasile FV; Eromon PE; Gross R; Wronka L; Caviness K; Hensley LE; Bergman NH; MacInnis BL; Happi CT; Lemieux JE; Sabeti PC; Myhrvold C Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun 2020, 11, 5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).de Puig H; Lee RA; Najjar D; Tan X; Soenksen LR; Angenent-Mari NM; Donghia NM; Weckman NE; Ory A; Ng CF; Nguyen PQ; Mao AS; Ferrante TC; Lansberry G; Sallum H; Niemi J; Collins JJ Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci. Adv 2021, 7, eabh2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Myhrvold C; Freije CA; Gootenberg JS; Abudayyeh OO; Metsky HC; Durbin AF; Kellner MJ; Tan AL; Paul LM; Parham LA; Garcia KF; Barnes KG; Chak B; Mondini A; Nogueira ML; Isern S; Michael SF; Lorenzana I; Yozwiak NL; MacInnis BL; Bosch I; Gehrke L; Zhang F; Sabeti PC Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ning B; Yu T; Zhang S; Huang Z; Tian D; Lin Z; Niu A; Golden N; Hensley K; Threeton B; Lyon CJ; Yin X-M; Roy CJ; Saba NS; Rappaport J; Wei Q; Hu TY A smartphone-read ultrasensitive and quantitative saliva test for COVID-19. Sci. Adv 2021, 7, eabe3703. [DOI] [PMC free article] [PubMed] [Google Scholar]


