Abstract
Objectives
To evaluate a deep learning model for automated and interpretable classification of central canal stenosis, neural foraminal stenosis, and facet arthropathy from lumbar spine MRI.
Methods
T2-weighted axial MRI studies of the lumbar spine acquired between 2008 and 2019 were retrospectively selected (n = 200) and graded for central canal stenosis, neural foraminal stenosis, and facet arthropathy. Studies were partitioned into patient-level train (n = 150), validation (n = 20), and test (n = 30) splits. V-Net models were first trained to segment the dural sac and the intervertebral disk, and localize facet and foramen using geometric rules. Subsequently, Big Transfer (BiT) models were trained for downstream classification tasks. An interpretable model for central canal stenosis was also trained using a decision tree classifier. Evaluation metrics included linearly weighted Cohen’s kappa score for multi-grade classification and area under the receiver operator characteristic curve (AUROC) for binarized classification.
Results
Segmentation of the dural sac and intervertebral disk achieved Dice scores of 0.93 and 0.94. Localization of foramen and facet achieved intersection over union of 0.72 and 0.83. Multi-class grading of central canal stenosis achieved a kappa score of 0.54. The interpretable decision tree classifier had a kappa score of 0.80. Pairwise agreement between readers (R1, R2), (R1, R3), and (R2, R3) was 0.86, 0.80, and 0.74. Binary classification of neural foraminal stenosis and facet arthropathy achieved AUROCs of 0.92 and 0.93.
Conclusion
Deep learning systems can be performant as well as interpretable for automated evaluation of lumbar spine MRI including classification of central canal stenosis, neural foraminal stenosis, and facet arthropathy.
Keywords: Deep learning, MRI, Stenosis, Arthropathy
Introduction
Lumbar spinal stenosis (LSS) is a common cause for spinal surgery in patients older than 65 years with a predominantly degenerative etiology [1]. Degenerative narrowing of the central and foraminal canal can be secondary to changes that include disk protrusion, extrusion; ligamentum flavum hypertrophy; or facet joint arthropathy [2]. Accurate classification of LSS is therefore essential for subsequent patient management [3].
Clinical symptoms, examination, and radiologic findings contribute to the diagnosis of symptomatic LSS. MRI is the preferred imaging modality due to superior softtissue contrast [4, 5], and a number of grading systems have been proposed for diagnosing LSS from MRI [6–8].
Detailed interpretation of lumbar spine MRI can be time consuming; recent advances in deep learning have led to unprecedented systems that augment the radiology workflow, ranging from applications in thoracic imaging to musculoskeletal radiology [9–13]. Notwithstanding the apparent advantages, adoption of deep learning in clinical settings is limited due to concerns regarding the black-box nature of the algorithms and lack of clinical interpretability [11, 14, 15].
A number of deep learning systems have been recently proposed for automated evaluation of lumbar spine MRI [16]. This includes detection of vertebral bodies [17], disk degeneration [18], central canal [19–21], lateral recess [21], and neural foraminal stenosis [20, 21]. In SpineNet [19], a multi-task approach was used to classify conditions of the lumbar spine, including central canal stenosis. The grades were binarized and the model relied on only sagittal images. Deep Spine [20] proposed an end-to-end approach for detection and classification of central canal and neural foraminal stenosis using both sagittal and axial images; however, their ground-truth labels were extracted from radiology reports in a weakly supervised fashion. Most recently in 2021, Hallinan et al [21] reported an end-to-end approach to automatically detect and classify central canal, lateral recess, and neural foraminal stenosis using both axial and sagittal MRI sequences. To our knowledge, none of the prior approaches include lumbar facet arthropathy in their evaluation.
Moreover, previously described systems rely on two-staged black-box deep learning: localization (either segmentation or object detection) followed by convolutional neural network (CNN) for classification, making it difficult for a radiologist to interpret and explain model predictions.
In this paper, we evaluate a deep learning system to detect and classify central canal stenosis, neural foraminal stenosis, and facet arthropathy from T2-weighted axial MRI slices of the lumbar spine. We present an interpretable approach for classifying lumbar spinal stenosis that may be more data efficient, generalizable, and amenable to clinical adoption compared to prior systems.
Materials and methods
This study was compliant with the Health Insurance Portability and Accountability Act (HIPAA) and approved by our institutional review board (IRB). Informed consent was waived due to the retrospective nature of the study.
Study cohort
Lumbar spine MRIs were collected retrospectively using the following inclusion and exclusion criteria: age 19 years or older with MR imaging of the lumbar spine acquired between 2008 and 2019. Patients with transitional anatomy, fractures, post-operative changes, extensive hardware, infection, primary tumors, and widespread metastatic disease to the spine were excluded; studies with absence of a T2-weighted axial sequence or poor image quality were also excluded. A total of 30,619 patients were identified, of which a subset of randomly selected patients (n = 200) was included in the study. The cohort was partitioned into random patient-level splits of training data (n = 150, 75%), validation data (n = 20, 10%), and test data (n = 30, 15%).
Image acquisition
All T2-weighted axial MRIs used in this study were fast spin echo (FSE) sequences acquired within our institution as part of routine clinical lumbar spine MRI imaging studies using a Discovery MR750 scanner (GE Healthcare). Acquisition parameters are reported in Table 1.
Table 1.
Acquisition parameters of the clinically acquired MRI sequences
| Sequence | T2 |
|---|---|
|
| |
| Orientation | Axial, Sagittal |
| Field strength (T) | 1.5, 3.0 |
| Matrix (pixels) | 256×256–512×512 |
| Field of view (cm) | 24.0–37.0 |
| Slice thickness (mm) | 3.0–4.0 |
| Pixel bandwidth (Hz) | 81.4–325.5 |
| Repetition time (ms) | 2430–6307 |
| Echo time (ms) | 26.1–107.8 |
| Flip angle (°) | 90–160 |
Data labeling
A board-certified neuroradiologist (R1) with 25 years of experience qualitatively graded MRIs from the study cohort (n = 200) for central canal stenosis as normal, mild, moderate, or severe with one grade per spinal level (L1/L2, L2/L3, L3/L4, L4/L5, L5/S1) based on a previously published qualitative grading system (Schizas system) [8]. The neuroradiologist also qualitatively classified each exam for lumbar foraminal stenosis as normal (graded normal or mild) vs stenosed (graded moderate or severe) based on the Park system [22], and classified lumbar facet arthropathy as normal vs arthropathy based on Pathria’s criteria [23]. While subsequent deep-learning models were trained on T2-weighted axial sequences alone, R1’s grades were based on the evaluation of all clinical sequences—T1-weighted as well as T2-weighted sagittal, axial, and coronal sequences—necessitated by each grading system. In the case of facet arthropathy, CT imaging was not available and R1’s assessment was based on a modification of Pathria’s criteria [23] to MRI. Imaging examples from the study sample are provided in Fig. 1.
Fig. 1.
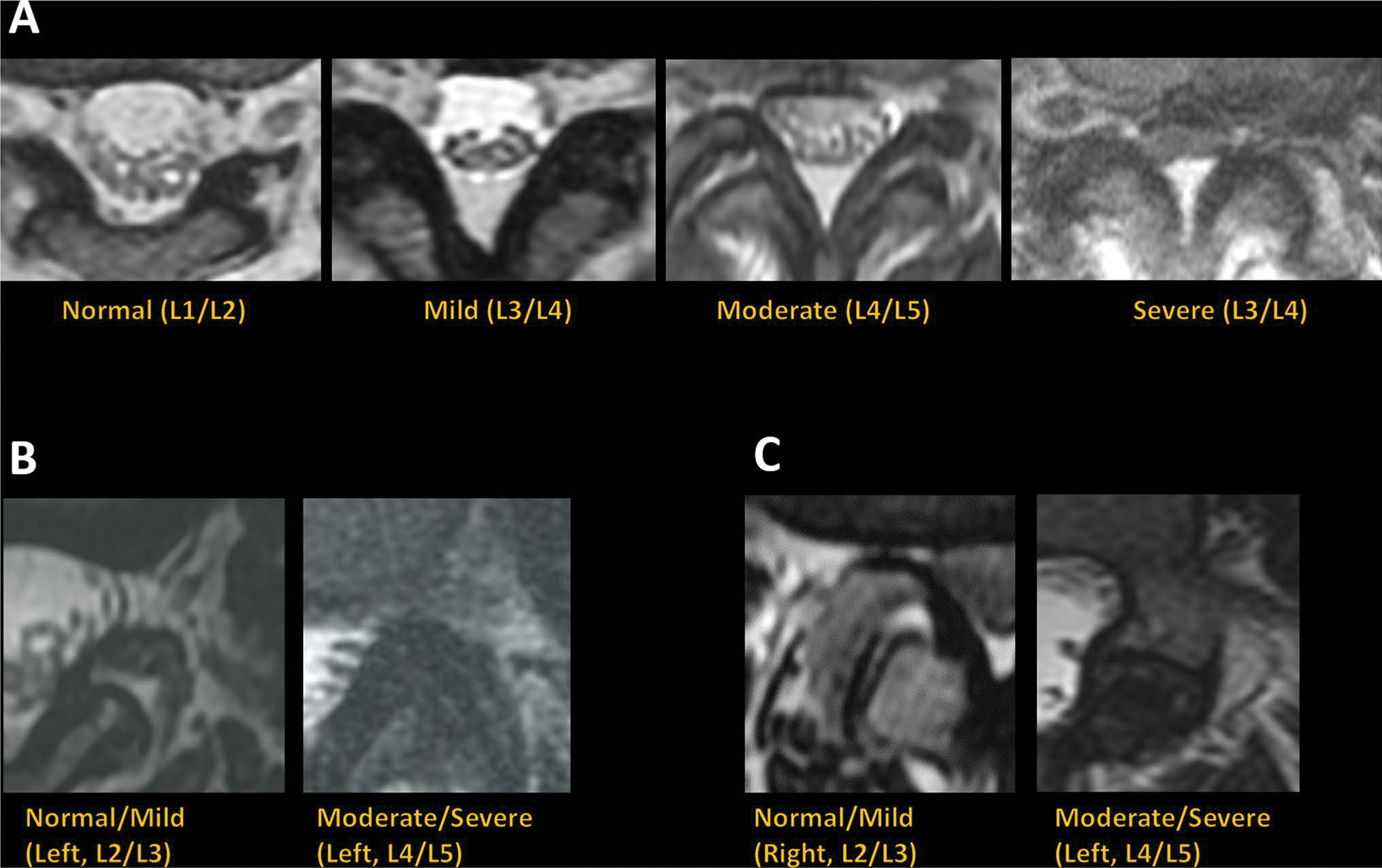
Imaging examples of T2-weighted axial MRI graded as normal, mild, moderate, or severe for central canal stenosis (A), normal/mild or moderate/severe for neural foraminal stenosis (B), and normal/mild or moderate/severe for lumbar facet arthropathy (C)
Using a research annotation platform (MD.ai), a trained researcher and a radiology trainee (R2) annotated the T2-weighted axial slices with freeform masks of the dural sac and intervertebral disk as well as landmark coordinates denoting the location of each facet and foramen on the same slice. All image annotations were subsequently verified by a board-certified neuroradiologist (R1).
Deep learning models
A two-staged system was developed for automated grading and classification of central canal stenosis, foraminal stenosis, and facet arthropathy: region localization in the first stage, followed by classification in the second stage. Fig. 2 provides an overview of the system.
Fig. 2.
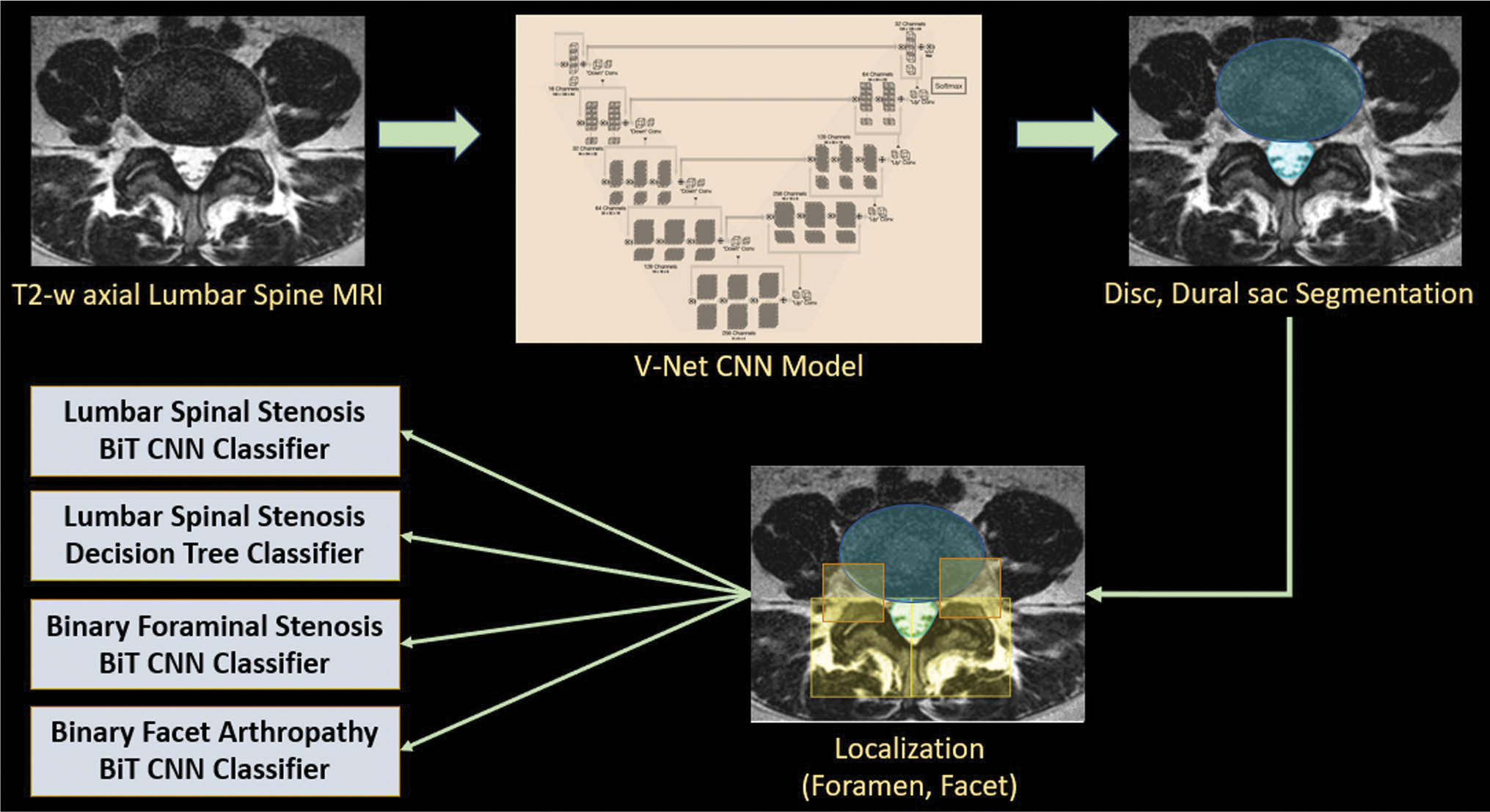
Overview of the deep learning pipeline. T2-weighted axial slices are passed into V-Net segmentation models to obtain masks for the intervertebral disc and dural sac. Geometric rules based on the disc and dural sac are used to localize bounding boxes around foramen and facet. Each localized region is passed into its corresponding classifier: Big Transfer (BiT) convolutional neural network (CNN) for classification of lumbar spinal stenosis, foraminal stenosis, and facet arthropathy. Interpretable classification (decision tree) of lumbar spinal stenosis relies on additional quantitative metrics extracted from the disc and dural sac segmentations
In the first stage, convolutional neural networks (CNN) based on the 2D V-Net architecture [24] were used to segment the dural sac and the intervertebral disk, one independently trained model for each anatomical region. Right and left facets were localized using a rule-based algorithm: 36 mm × 36 mm bounding boxes originating from the top-most point of the predicted dural sac, with the centroid partitioning the image into left and right were extracted. A similar approach was used to also localize bounding boxes around the left and right foramina.
In the second stage, a Big Transfer (BiT) ResNet-50 CNN pretrained on ImageNet-21 k images [25] was fine-tuned for each classification task: binary as well as multi-class grading of central canal stenosis, binary classification of foraminal stenosis, and binary classification of facet arthropathy on their respective localized patches. All segmentation and classification CNNs were trained on a 32 GB Nvidia Tesla V100 GPU in mixed precision using TensorFlow 1.15 [26].
A landmark coordinate regression model [27] with 5 coordinates was trained on T2-weighted mid-sagittal slice to identify the axial slice centered at the disk level (L1/L2, L2/L3, L3/L4, L4/L5) for inference. Sagittal sequences were not used anywhere else in the model development pipeline.
Interpretable classification
In addition to the CNN, an interpretable decision tree classifier was also trained to classify central canal stenosis using a quantitative metric of the dural sac and intervertebral disk extracted from segmentations obtained in the first stage. The metric, based on previously published biomarkers, is the ratio between the cross-sectional areas of the dural sac and the intervertebral disk [28–30].
Decision trees have been previously used to obtain thresholds for lumbar spinal stenosis and offer the advantage of clinically interpretable rules [31]. The Scikitlearn Python library version 0.24.2 DecisionTreeClassifier module was used with the max_depth parameter set to 3, and max_leaves parameter set to 4 to avoid overfitting [32].
Statistical analysis
Segmentation of the dural sac and intervertebral disk was evaluated using the Sørensen-Dice coefficient [33], which measures spatial overlap between ground-truth and model-predicted masks. Localization of foramen and facets was characterized using intersection over union (IoU) between ground-truth and model-predicted bounding boxes [34].
All binary classifiers were evaluated using area under the receiver operating characteristic curve (AUROC). Multi-grade classification of central canal stenosis was evaluated using model accuracy, multi-class AUROC with the one-v-one criterion, and agreement with R1’s grades using linearly weighted Cohen’s kappa coefficient.
Landmark coordinate regression was evaluated using the mean absolute error.
Reproducibility
A board-certified musculoskeletal radiologist (R3) with 23 years of experience also graded the test set (n = 30) for central canal stenosis qualitatively as normal, mild, moderate, and severe. Agreement scores between R1, R2, and R3 were characterized using linearly weighted Cohen’s kappa coefficient. A kappa score of 0.2 or higher was considered fair agreement, 0.4 or higher was considered moderate agreement, and 0.6 or higher was considered substantial [35].
The SciPy v1.6.0 Python library and its stats module were used for all statistical analyses [36].
Results
Study cohort
The study cohort (n = 200) consisted of 100 female and 100 male patients with a mean age 56.7 [19.0, 96.0] years and mean BMI of 26.9 [15.3, 58.8] kg/m2. Patients presented with either low back pain (n = 45), radicular pain (n = 20), or both low back pain and radicular pain (n = 105), as well as other symptoms (n = 30) that include numbness, tingling, weakness, dysesthesia, and tightness. Patients had an average low back pain score of 5.8 ± 2.6 and a radicular pain score of 5.9 ± 2.6 on an 11-point qualitative numerical pain rating scale. The distribution of demographics and symptoms across train (n = 150), validation (n = 20), and test (n = 30) had no significant differences.
Grading of central canal stenosis, neural foraminal stenosis, and facet arthropathy
A total of 987 slices across lumbar levels L1/L2, L2/L3, L3/L4, L4/L5, and L5/S1, one slice per lumbar level, were annotated in the study cohort with the following distribution of grades for central canal stenosis: normal (n = 487), mild (n = 351), moderate (n = 72), and severe (n = 77); neural foraminal stenosis: normal/mild (n = 809) and moderate/severe (n = 178); and facet arthropathy normal/mild (n = 671) and moderate/severe (n = 316).
Localization of dural sac, intervertebral disk, foramen, and facets
On the test set (n = 30), V-Net-based segmentation of the dural sac achieved a volumetric Dice score of 0.93 (95% CI: [0.92, 0.95]); segmentation of the intervertebral disk achieved a volumetric Dice score of 0.94 (95% CI: [0.92, 0.94]).
Localization of foramen and facet based on predicted segmentations achieved intersection over union of 0.72 (95% CI: [0.70, 0.74]) and 0.83 (95% CI: [0.82, 0.84]), respectively.
Visual examples of model-predicted segmentations of the dural sac and intervertebral disk along with generated bounding boxes for foramen and facet are shown in Fig. 3.
Fig. 3.
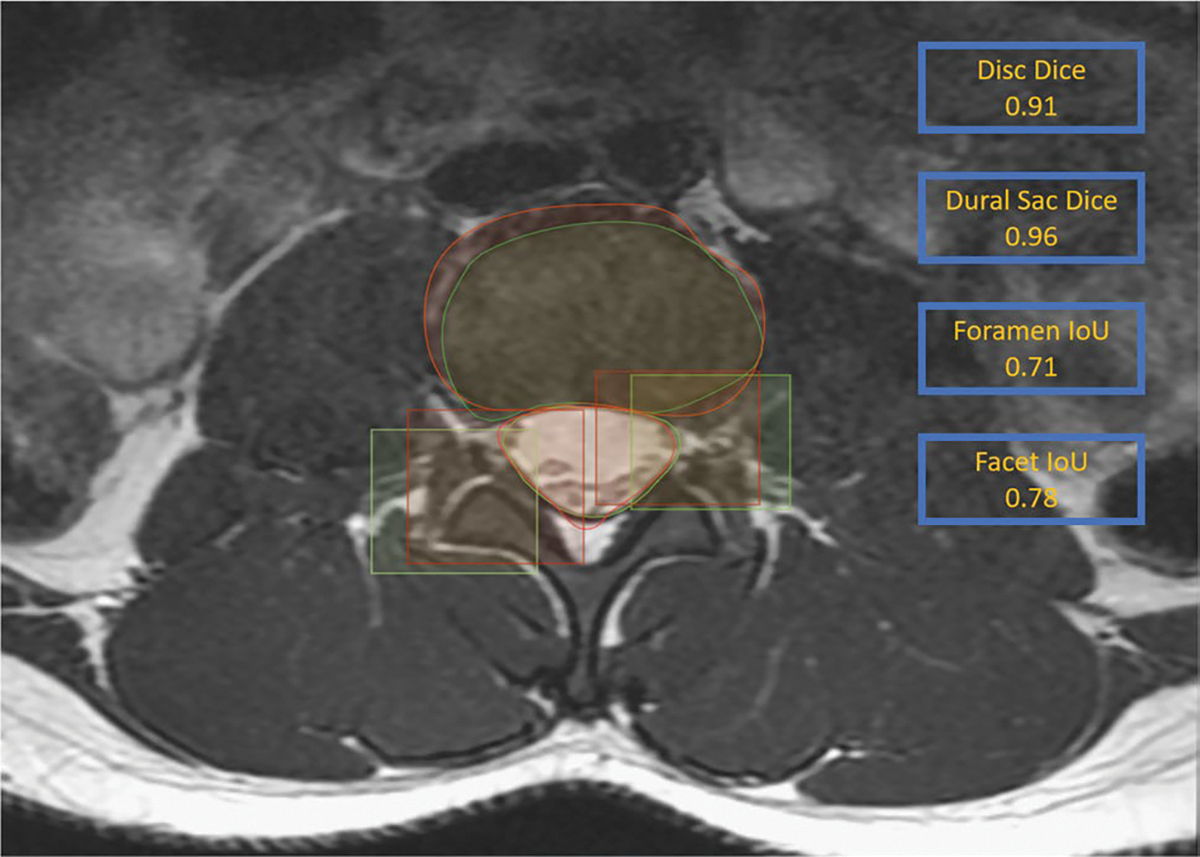
Results of the V-Net segmentation model on a T2-weighted MR axial slice at L2/L3 from the test set. Model predictions (red) and ground-truth annotations (green) show significant overlap with Dice scores of 0.91 and 0.96 for the intervertebral disc and the dural sac, respectively. Also illustrated are the generated bounding boxes (red) compared to ground truth (green) for the right facet and left foramen
Classification of central canal stenosis
Binary classification (normal/mild vs moderate/severe) of central canal stenosis using the BiT CNN classifier in the second stage achieved an AUROC of 0.94 (95% CI: [0.93, 0.95]) on the test set (n = 30). The decision tree classifier using interpretable biomarkers in the second stage achieved an AUROC of 0.95 (95% CI: [0.93, 0.96]). A receiver operator characteristic (ROC) curve comparing the two models is shown in Fig. 4.
Fig. 4.
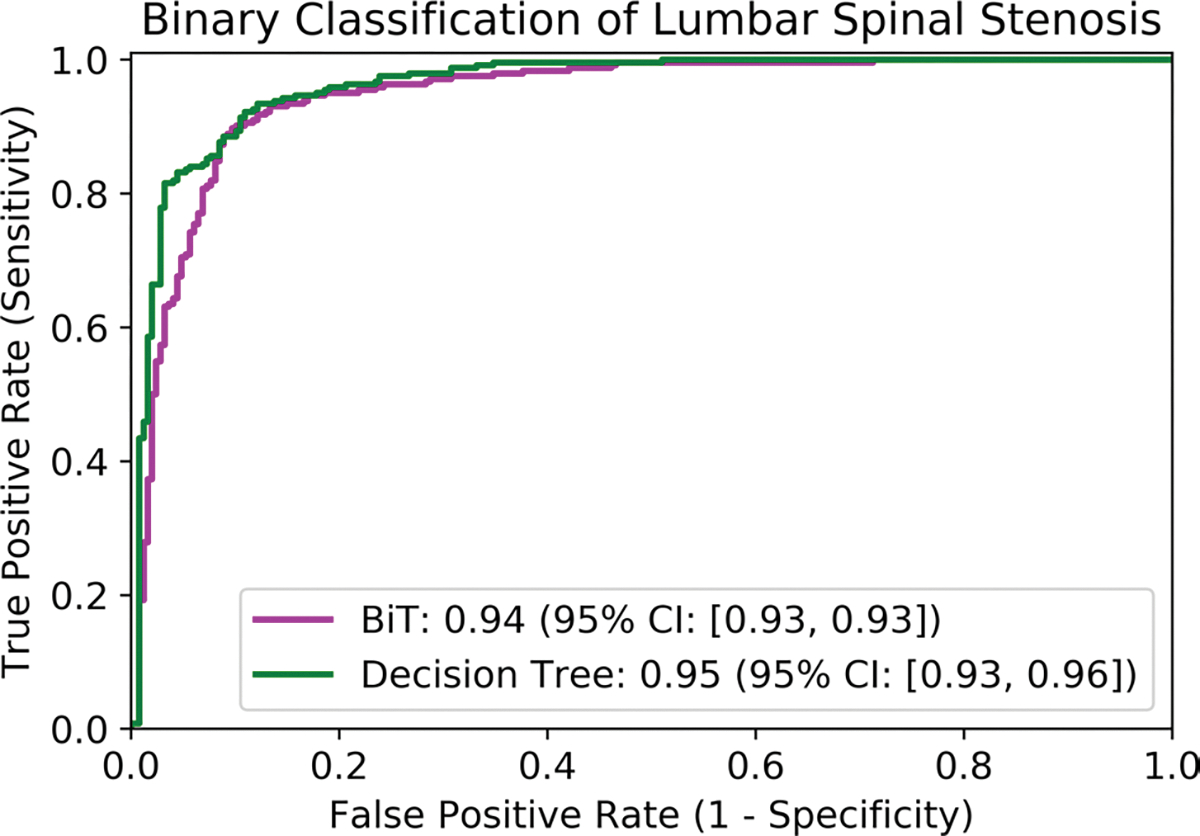
Receiver operator characteristic curve (ROC) of the BiT convolutional neural network (magenta) and the interpretable decision tree classifier (green) for binary classification of lumbar spinal stenosis with their respective area under the ROC (AUROC) values reported in the legend. The difference in AUROC was statistically significant (p < 0.01) based on DeLong’s paired test for AUROC
Multi-class (normal, mild, moderate, severe) grading of central canal stenosis using the BiT CNN classifier achieved a linearly weighted Cohen’s kappa score of 0.54 (95% CI: [0.51, 0.60]) with respect to R1’s grades. The decision tree classifier with interpretable quantitative metric in the second stage achieved a linearly weighted Cohen’s kappa score of 0.80 (95% CI: [0.76, 0.82]). Detailed results are presented in Table 2.
Table 2.
Multi-class grading of central canal stenosis using the Big Transfer (BiT) convolutional neural network (CNN) and the interpretable decision tree based on a clinical quantitative metric. Reported are multi-class accuracy, multi-class area under the receiver operator characteristic curve (AUROC) using the one-v-one criterion, and linearly weighted Cohen’s kappa
| Model | Accuracy | AUROC | Cohen’s kappa | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| Accuracy | 95% CI | AUROC | 95% CI | Kappa | 95% CI | |
|
| ||||||
| BiT CNN | 63.8 | [59.7, 68.1] | 75.4 | [72.6, 78.3] | 0.54 | [0.51, 0.60] |
| Decision Tree | 72.7 | [67.8, 76.3] | 81.9 | [78.5, 83.8] | 0.80 | [076, 0.82] |
Classification of neural foraminal stenosis and lumbar facet arthropathy
Binary classification (normal/mild vs moderate/severe) of neural foraminal stenosis using the BiT CNN classifier in the second stage achieved an AUROC of 0.92 (95% CI: [0.91, 0.93]) on the test set (n = 30). Classification (normal/mild vs moderate/severe) of facet arthropathy achieved an AUROC of 0.93 (95% CI: [0.91, 0.94]). ROC curves for binary classification of foraminal stenosis and facet arthropathy are shown in Figs. 5A and B, respectively.
Fig. 5.
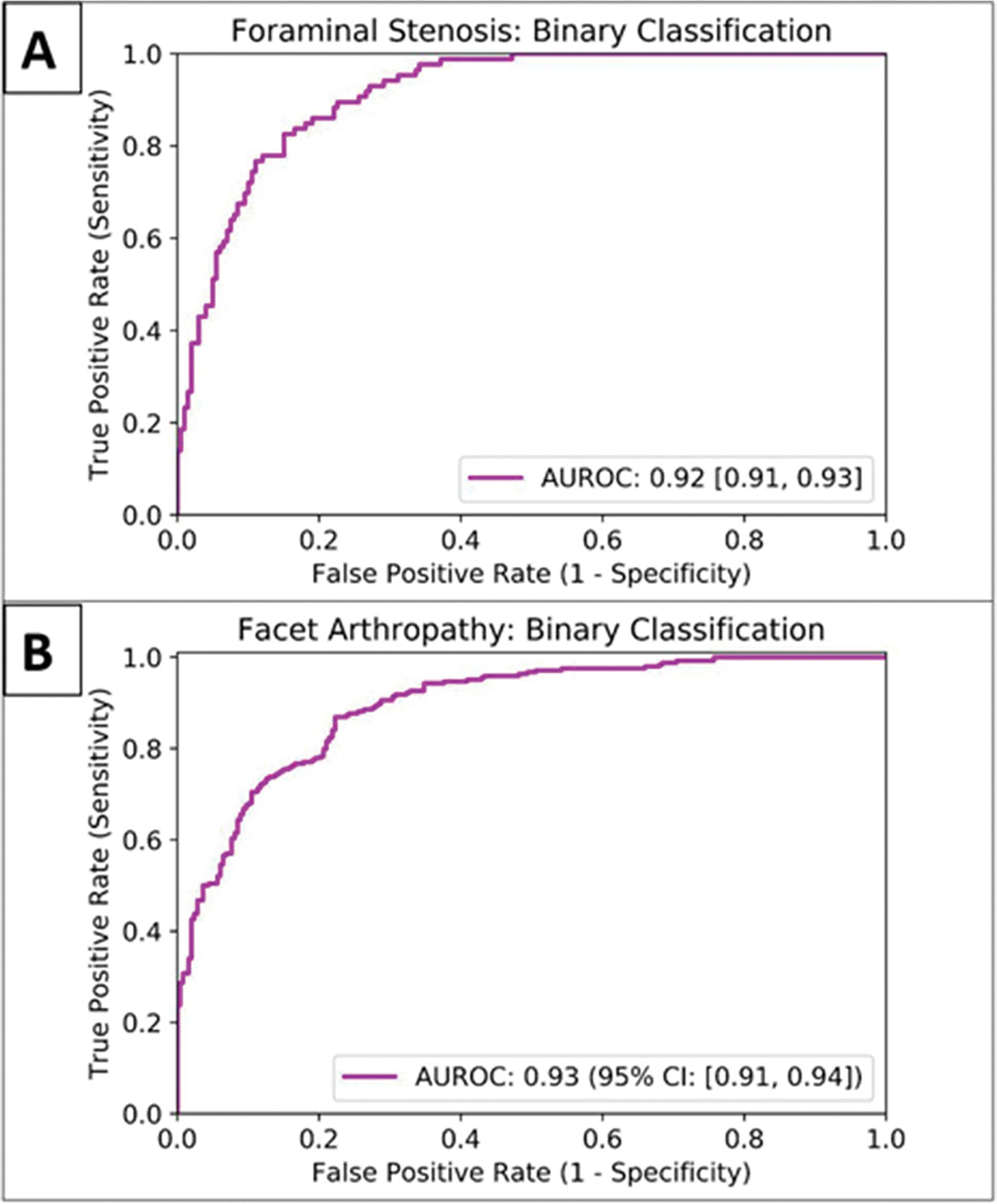
Receiver operator characteristic curve (ROC) of the BiT convolutional neural network for binary classification of foraminal stenosis (A) and facet arthropathy (B)
Interpretable classification
The decision tree classifier was of depth 3, and amenable to grading central canal stenosis in an interpretable manner with cut-off thresholds derived as follows for the ratio between the cross-sectional areas of the dural sac and intervertebral disk: greater than 0.31 is normal, greater than 0.23 is mild, greater than 0.19 is moderate, and anything less than 0.19 is severe stenosis.
Landmark coordinate regression of vertebral levels
Landmark coordinate regression of the vertebral levels on the mid-sagittal slice, used during inference to identify the corresponding axial slices, achieved a mean absolute error of 2.14 [0.97, 3.22] mm. Predicted coordinates are visualized in Fig. 6.
Fig. 6.
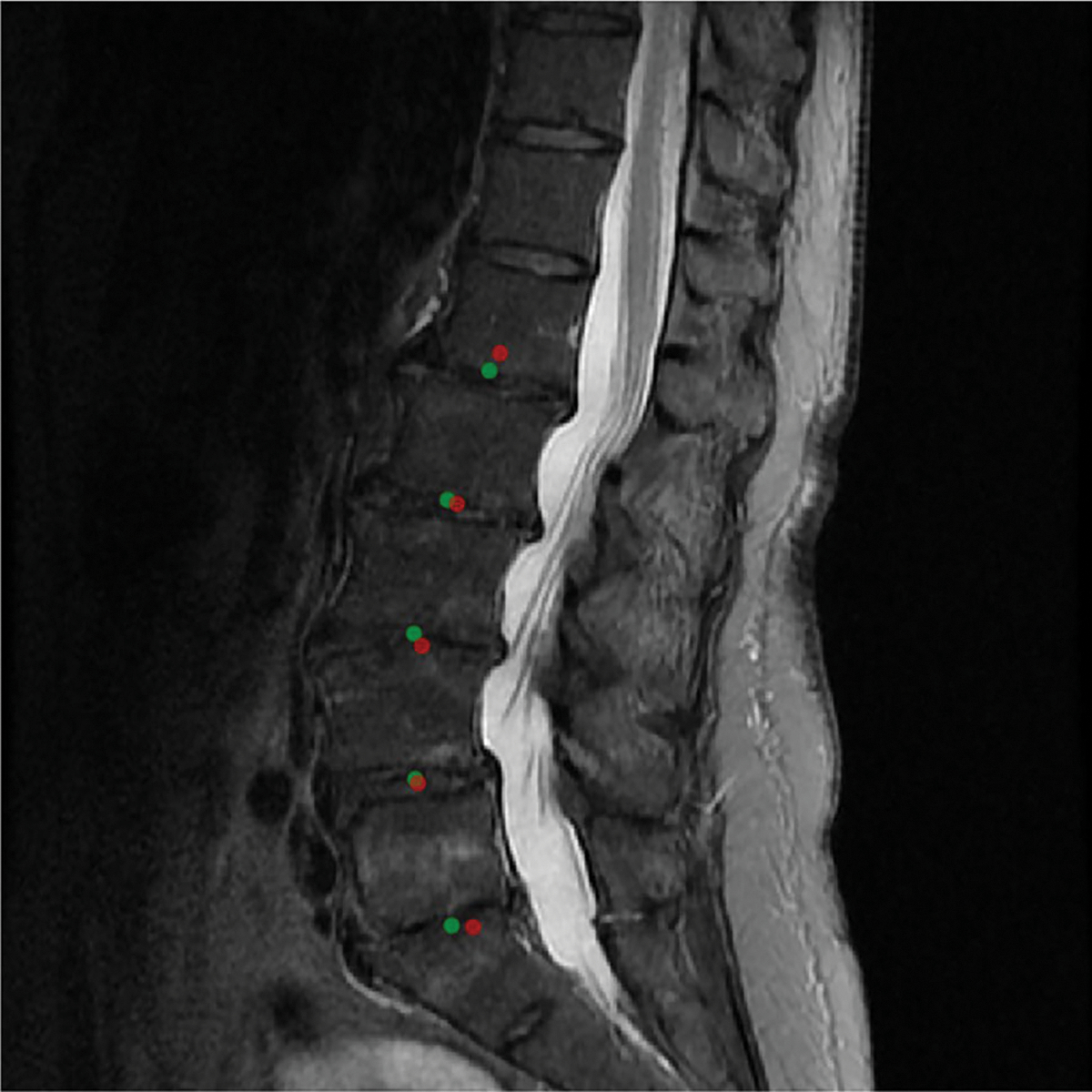
Visualization of the predictions of the landmark coordinate regression model on the mid-sagittal slice of the T2 sequence. Green (ground-truth landmark) and red (model-predicted landmark) had a mean absolute error of 1.14 mm in this example. Since the model is constrained to generate exactly 5 landmarks, they correspond to L1/L2, L2/L3, L3/L4, L4/L5, and L5/S1
Reproducibility
Pairwise agreement between readers (R1, R2), (R1, R3), and (R2, R3), measured with linearly weighted Cohen’s kappa score, were 0.86, 0.80, and 0.74, respectively. Agreement between R1 and readers (R1, R2) as well as models (BiT CNN and Decision Tree) is presented in Table 3. Pairwise agreement scores between readers, measured with linearly weighted Cohen’s kappa, were all 1.0 for binarized grading of lumbar foraminal stenosis as well as facet arthropathy.
Table 3.
Agreement between R1 and R2, R3, Big Transfer (BiT) convolutional neural network (CNN) model, and the interpretable decision tree model based on dural-sac to disc ratios (DDR) for multi-class grading of central canal stenosis evaluated using linearly weighted Cohen’s kappa coefficient on the held-out test set (n = 30)
| Reader 2 | Reader 3 | BiT CNN model | Decision Tree model |
|---|---|---|---|
|
| |||
| 0.86 | 0.80 | 0.54 | 0.80 |
Discussion
We proposed a two-staged learning system that can be used to automatically evaluate T2-weighted axial MRI of the lumbar spine for classification of central canal stenosis, neural foraminal stenosis, and facet arthropathy. The first stage—localization of anatomical regions—was performant with excellent volumetric Dice scores (well above 0.90) for the dural sac and intervertebral disk, and with no additional training or fine-tuning, localization of the foramen and facet was also favorable. In the second stage, our interpretable approach to multi-class grading (normal, mild, moderate, severe) of central canal stenosis was in line with pairwise agreements between three radiologists, and significantly outperformed a black-box convolutional neural network. While no systematic trends were observed in differences between radiologists’ grades as well as model predictions, nearly all disagreements were borderline within 1 grade, and mild-moderate or moderate-severe discrepancies were most prevalent. Our models also showed accurate binary classification (normal/mild vs moderate/severe) of both neural foraminal stenosis and facet arthropathy with AUROC values greater than 0.90.
Our approach targets a more comprehensive evaluation of lumbar spine MRI and assessment of features associated with low back pain. Facet arthropathy, prevalent in 15 to 45% of patients presenting with chronic low back pain, is a very important yet underdiagnosed etiology for low back pain [37]. In clinical practice, MRI is used to identify causes of low back pain such as disk herniation, foraminal stenosis, and central canal stenosis—whereas facet arthropathy is often omitted as a descriptor in radiology reports [38]. Our model therefore provides additional insights during assessment of low back pain on MRI that may influence subsequent management of pain.
In the multi-class setting, our interpretable decision tree classifier outperformed black-box CNN classification of central canal stenosis. Performant results reported in prior literature [18–21] rely on significantly more training data, which is essential for training accurate deep learning systems. To obtain the most performant deep learning model using our dataset, we utilize the BiT classification model which was originally designed for few-shot learning—i.e., training with limited labeled data [25]. The performance delta between the decision tree classifier and CNN suggests that not all deep learning models are highly performant, and efficient alternatives may be preferred in settings where data and annotation resources can be limited.
While it is difficult to compare deep learning models across different datasets, our approach is as performant as previously published results for both binary as well as multiclass grading of central canal stenosis [19–21]. Our cohort consists of 987 axial slices on which the models have been trained, validated, and tested, which is 10 × fewer than all previously published models: SpineNet [19] was trained on more than 10,000 slices, Deep Spine [20] on 16,000 slices, and most recently the work by Hallinan et al [21] on over 10,000 slices. A detailed summary of our results in comparison to prior publications is presented in Table 4.
Table 4.
Summary of our results compared to previously published results. Values reported are not directly comparable since they are based on heterogeneous datasets with potentially different grading systems, methodologies, and evaluation
| SpineNet [19] | DeepSpine [20] | Hallinan et al [21] | Our study | |
|---|---|---|---|---|
|
| ||||
| Training data (number of slices) | 10,836 | 15,957 | ~10,000 | 750 |
| Sequences | T2 sagittal, axial | T2 sagittal | T2 sagittal, axial | T2 axial |
| Interpretability | Localization | Localization | Localization | Localization + quantitative metrics |
| Anomalies | Binary central canal stenosis | Central canal and foraminal stenosis | Central canal, lateral recess, and foraminal stenosis | Central canal and foraminal stenosis, facet arthropathy |
| Central canal stenosis (multi-class grading) | – | 78.6% accuracy | 0.82 kappa | 0.80 kappa 81.9% AUROC |
| Central canal stenosis (binary grading) | 0.95 AUROC | 0.97 AUROC | 0.98 AUROC | 0.95 AUROC |
| Foraminal stenosis (binary grading) | – | 0.94 AUROC | 0.96 AUROC | 0.92 AUROC |
| Facet arthropathy (binary grading) | – | – | – | 0.93 AUROC |
We believe our approach also mitigates some effects of “black-box” deep learning. In the first stage of our system, we emphasize precise segmentations of the dural sac and intervertebral disk so that downstream results can be interpreted in an anatomical context, instead of more economical alternatives such as coarse-grained region-of-interest identification as discussed in Hallinan et al [21]. In the second stage, we rely on a simple quantitative metric from prior literature coupled with a decision tree classifier from which meaningful thresholds can be derived for each grade.
In addition to enhanced interpretability, each stage of the pipeline is very amenable to human intervention: in case of disagreement, radiologists can update segmentations or downstream quantitative metrics without altering the pipeline—providing more control over the level of automation and alignment with clinical deployment goals.
Our approach also generalizes to detection of the foramen and facet in a sample-efficient manner; compared to previous approaches such as Deep Spine [20] and Hallinan et al [21], where a separate localization model was trained for each region of interest, we use geometric principles to localize additional structures from the dural sac segmentations. New deep learning applications require task-specific data; however, annotation costs for obtaining high-quality masks or bounding boxes can be exorbitant; we address this gap by leveraging a precise dural sac segmentation model and localizing the foramen and facet in a zero-shot manner, with no additional training or fine-tuning.
This study has a few limitations. Small sample size is indeed a double-edged sword; while the leading tenet of this paper is to demonstrate that accurate classification models can be trained even in under-resourced settings, our model evaluation may suffer from the lack of generalizability to more heterogeneous datasets across multiple vendors, institutions, and patient demographics. A follow-up study on a large, diverse, clinical cohort would provide additional insights into the generalizability of our approach. Although we rely on a published grading system for central canal stenosis, there is significant variability among radiologists in clinical practice. We attempt to capture some of this variability by including three independent readers for the test set; with more resources, a consensus-based approach for the entire cohort including training and validation data may have resulted in cleaner labels. We also simplified neural foraminal stenosis and facet arthropathy to a binarized setting (normal/mild vs moderate/severe) in the interest of labeling resources, hoping that subsequent management may be similar for moderate and severe cases; however, granular grading is certainly more clinically relevant. Lastly with the notable exception of identifying axial slices at each lumbar level, for which we use the mid-sagittal slice, our pipeline relies entirely on axial sequences. While more complex deep learning systems that utilize both axial as well as sagittal sequences can be developed, we believe our results provide an upper bound on what is feasible using axial sequences alone.
In conclusion, we demonstrated that our deep learning system is performant for automated evaluation of lumbar spine MRI including classification of central canal stenosis, neural foraminal stenosis, and facet arthropathy; it is also one of the first systems designed to be clinically interpretable, sample efficient, and generalizable to other applications; hence, it is amenable to clinical deployment in various automated and semi-automated settings.
Key Points.
Interpretable deep-learning systems can be developed for the evaluation of clinical lumbar spine MRI. Multi-grade classification of central canal stenosis with a kappa of 0.80 was comparable to inter-reader agreement scores (0.74, 0.80, 0.86). Binary classification of neural foraminal stenosis and facet arthropathy achieved favorable and accurate AUROCs of 0.92 and 0.93, respectively.
While existing deep-learning systems are opaque, leading to clinical deployment challenges, the proposed system is accurate as well as interpretable, providing valuable information to a radiologist in clinical practice.
Acknowledgements
We would like to thank Jeremy Huang and Alma Rechav Ben-Natan of the University of California, San Francisco, for their valuable contributions to data collection.
Funding
This study was funded by the National Institute of Arthritis and Musculoskeletal and Skin Diseases, Grant/Award Number: UH2AR076724–01. Additionally, Upasana Upadhyay Bharadwaj was supported by the NIH T32 grant for research training in biomedical imaging at the University of California, San Francisco and Cynthia T. Chin was supported by the Edward A. Dickinson Professorship Award at the University of California, San Francisco.
Abbreviations
- AUROC
Area under the receiver operating characteristic curve
- BiT
Big Transfer
- CI
Confidence interval
- CNN
Convolutional neural network
- HIPAA
Health Insurance Portability and Accountability Act
- IoU
Intersection over union
- IRB
Institutional review board
- LSS
Lumbar spinal stenosis
Footnotes
Conflict of interest Author Thomas M. Link is a member of the European Radiology Advisory Editorial Board. He has not taken part in the review or selection process of this article. The remaining authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Guarantor The scientific guarantor of this publication is Dr. Sharmila Majumdar, PhD, University of California, San Francisco, San Francisco, CA, USA. Email: Sharmila.Majumdar@ucsf.edu.
Statistics and biometry No complex statistical methods were necessary for this paper.
Informed consent Written informed consent was waived by the institutional review board.
Ethical approval Institutional review board approval was obtained.
Methodology
• retrospective
• experimental
• performed at one institution
References
- 1.Deyo RA, Gray D, Kreuter W, Mirza S, Martin BI (2005) United States trends in lumbar fusion surgery for degenerative conditions. (Phila Pa 1976) 30:1441–1445 [DOI] [PubMed] [Google Scholar]
- 2.Cowley P (2016) Neuroimaging of spinal canal stenosis. Magn Reson Imaging Clin N Am 24:523–529 [DOI] [PubMed] [Google Scholar]
- 3.Lurie J, Tomkins-Lane C (2016) Management of lumbar spinal stenosis. BMJ 352:h6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morita M, Miyauchi A, Okuda S, Oda T, Iwasaki M (2011) Comparison between MRI and myelography in lumbar spinal canal stenosis for the decision of levels of decompression surgery. J Spinal Disord Tech 24:31–36 [DOI] [PubMed] [Google Scholar]
- 5.Alsaleh K, Ho D, Rosas-Arellano MP, Stewart TC, Gurr KR, Bailey CS (2017) Radiographic assessment of degenerative lumbar spinal stenosis: is MRI superior to CT? Eur Spine J 26:362–367 [DOI] [PubMed] [Google Scholar]
- 6.Arana E, Royuela A, Kovacs FM et al. (2010) Lumbar spine: agreement in the interpretation of 1.5-T MR images by using the Nordic Modic Consensus Group Classification Form. Radiology 254(3):809–817 [DOI] [PubMed] [Google Scholar]
- 7.Guen YL, Joon WL, Hee SC, Kyoung-Jin O, Heung SK (2011) A new grading system of lumbar central canal stenosis on MRI: an easy and reliable method. Skeletal Radiol 40:1033–1039 [DOI] [PubMed] [Google Scholar]
- 8.Schizas C, Theumann N, Burn A et al. (2010) Qualitative grading of severity of lumbar spinal stenosis based on the morphology of the dural sac on magnetic resonance images. (Phila Pa 1976) 35:1919–1924 [DOI] [PubMed] [Google Scholar]
- 9.Aggarwal R, Sounderajah V, Martin G et al. (2021) Diagnostic accuracy of deep learning in medical imaging: a systematic review and meta-analysis. NPJ Digital Medicine 4(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazurowski MA, Buda M, Saha A, Bashir MR (2018) Deep learning in radiology: an overview of the concepts and a survey of the state of the art with focus on MRI. J Magn Reson Imaging 49:939–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montagnon E, Cerny M, Cadrin-Chênevert A et al. (2020) Deep learning workflow in radiology: a primer. Insights Imaging 11:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng PM, Montagnon E, Yamashita R et al. (2021) Deep learning: an update for radiologists. Radiographics 41:1427–1445 [DOI] [PubMed] [Google Scholar]
- 13.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL (2018) Artificial intelligence in radiology. Nat Rev Cancer 18:500–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh A, Sengupta S, Lakshminarayanan V (2020) Explainable deep learning models in medical image analysis. J Imaging 6:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.England JR, Cheng PM (2019) Artificial Intelligence for medical image analysis: a guide for authors and reviewers. AJR Am J Roentgenol 212:513–519 [DOI] [PubMed] [Google Scholar]
- 16.Galbusera F, Casaroli G, Bassani T (2019) Artificial intelligence and machine learning in spine research. JOR Spine 2:e1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Liu Y, Chen Q, Gu G, Sui X (2019) Automatic lumbar MRI detection and identification based on deep learning. J Digit Imaging 32:513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jamaludin A, Lootus M, Kadir T et al. (2017) ISSLS PRIZE IN BIOENGINEERING SCIENCE 2017: automation of reading of radiological features from magnetic resonance images (MRIs) of the lumbar spine without human intervention is comparable with an expert radiologist. Eur Spine J 26:1374–1383 [DOI] [PubMed] [Google Scholar]
- 19.Jamaludin A, Kadir T, Zisserman A (2017) SpineNet: automated classification and evidence visualization in spinal MRIs. Med Image Anal 41:73. [DOI] [PubMed] [Google Scholar]
- 20.Lu J-T, Pedemonte S, Bizzo BC et al. (2018) Deep spine: automated lumbar vertebral segmentation, disc-level designation, and spinal stenosis grading using deep learning. Proceedings of Machine Learning Research 85:1–16 [Google Scholar]
- 21.Hallinan JTPD, Zhu L, Yang K et al. (2021) Deep learning model for automated detection and classification of central canal, lateral recess, and neural foraminal stenosis at lumbar spine MRI. Radiology 300:130–138 [DOI] [PubMed] [Google Scholar]
- 22.Park H-J, Kim SS, Lee S-Y et al. (2012) Clinical correlation of a new MR imaging method for assessing lumbar foraminal stenosis. AJNR Am J Neuroradiol 33:818–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pathria M, Sartoris DJ, Resnick D (1987) Osteoarthritis of the facet joints: accuracy of oblique radiographic assessment. Radiology 164:227–230 [DOI] [PubMed] [Google Scholar]
- 24.Milletari F, Navab N, Ahmadi S-A (2016) V-Net: fully convolutional neural networks for volumetric medical image segmentation. 2016 Fourth International Conference on 3D Vision (3DV):565–571 [Google Scholar]
- 25.Kolesnikov A, Beyer L, Zhai X et al. (2019) Big Transfer (BiT): general visual representation learning. arXiv:1912.11370 [Google Scholar]
- 26.Abadi M, Agarwal A, Barham P et al. (2015) TensorFlow: large-scale machine learning on heterogeneous systems. arXiv:1603.04467 [Google Scholar]
- 27.Nibali A, He Z, Morgan S, Prendergast LA (2018) Numerical coordinate regression with convolutional neural networks. CoRR abs/ 1801.07372 [Google Scholar]
- 28.Steurer J, Roner S, Gnannt R, Hodle J (2011) Quantitative radiologic criteria for the diagnosis of lumbar spinal stenosis: a systematic literature review. BMC Musculoskelet Disord 12:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laurencin CT, Lipson SJ, Senatus P et al. (1999) The stenosis ratio: a new tool for the diagnosis of degenerative spinal stenosis. Int J Surg Investig 1:127–131 [PubMed] [Google Scholar]
- 30.Hamanishi C, Matukura N, Fujita M, Tomihara M, Tanaka S (1994) Cross-sectional area of the stenotic lumbar dural tube measured from the transverse views of magnetic resonance imaging. J Spinal Disord 7:388–393 [PubMed] [Google Scholar]
- 31.Huber FA, Stutz S, Martini IVd et al. (2019) Qualitative versus quantitative lumbar spinal stenosis grading by machine learning supported texture analysis—experience from the LSOS study cohort. Eur J Radiol 114:45–50 [DOI] [PubMed] [Google Scholar]
- 32.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O (2011) Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830 [Google Scholar]
- 33.Zou KH, Warfield SK, Bharatha A et al. (2004) Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol 11:178–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rezatofighi SH, Tsoi N, Gwak J, Sadeghian A, Reid ID, Savarese S (2019) Generalized intersection over union: a metric and a loss for bounding box regression. (2019) IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR):658–666 [Google Scholar]
- 35.Landis JR, Koch GG (1977) The measurement of observer agreement for categorical data. Biometrics 33:159–174 [PubMed] [Google Scholar]
- 36.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D (2020) SciPy 1.0: fundamental algo rithms for scientific computing in Python. Nat Methods 17:261–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen SP, Raja SN (2007) Pathogenesis, diagnosis, and treatment of lumbar zygapophysial (facet) joint pain. Anesthesiology 106:591–614 [DOI] [PubMed] [Google Scholar]
- 38.Berg L, Thoresen H, Neckelmann G, Furunes H, Hellum C, Espeland A (2019) Facet arthropathy evaluation: CT or MRI? Eur Radiol 29:4990–4998 [DOI] [PubMed] [Google Scholar]


