Abstract
Aims
Heart failure (HF) is a prevalent age‐related cardiovascular disease with poor prognosis in the elderly population. This study aimed to establish the causal relationship between ageing and HF by conducting a bidirectional Mendelian randomization (MR) analysis on epigenetic age (a marker of ageing) and HF.
Methods and results
Genome‐wide association study data for epigenetic age (GrimAge, HorvathAge, HannumAge, and PhenoAge) and HF were collected and assessed for significant genetic variables. A bidirectional MR analysis was carried out using the random‐effects inverse–variance weighted (IVW) method as the primary approach, while other methods (MR–Egger, weighted median, simple mode, and weighted mode) and multiple sensitivity analyses (heterogeneity analysis, leave‐one‐out sensitivity analysis, and horizontal pleiotropy analysis) were employed to evaluate the impact of epigenetic age on HF and vice versa. Bidirectional MR analysis of two samples revealed that the epigenetic PhenoAge clock increased the risk of HF [IVW odds ratio (OR) 1.015, 95% confidence interval (CI) 1.002–1.028, P = 0.028 and weighted median OR 1.020, 95% CI 1.001–1.038, P = 0.039]. Other results were not statistically significant.
Conclusions
The bidirectional MR analysis demonstrated a causal link between genetically predicted epigenetic age and HF in individuals of European descent. Further research into epigenetic age in other populations and additional genetic information related to HF is warranted.
Keywords: Epigenetic age, Heart failure (HF), Causality, Bidirectional Mendelian randomization (MR) study, Genome‐wide association study (GWAS)
Introduction
Heart failure (HF) is a common clinical syndrome that results in myocardial injury or overload during the pathogenesis of several diseases, ultimately causing decreased myocardial function. 1 HF is often accompanied by various inflammatory and metabolic disease complications with poor clinical prognosis, including diabetes, atrial fibrillation, and chronic kidney disease. 2 The prevalence of HF is around 26 million people worldwide, 3 with the elderly accounting for 80% of the population, and morbidity and mortality increase with ageing, 4 leading to significant socioeconomic and nursing burdens. 5 , 6 Cardiovascular ageing is an important risk factor for HF among all risk factors. 7
The study of ageing‐related biomarkers has recently been a hot topic in HF research. While epigenetic modifications are fundamental mechanisms of biological ageing, 7 these are increasingly implicated in the development of cardiovascular diseases. 8 , 9 Because there is significant heterogeneity in health outcomes in the elderly, 10 epigenetic age based on DNA methylation (DNAm) is more accurate than chronological age in predicting physiological ageing status in the elderly. 11 , 12 Each epigenetic clock measures the DNA methylation levels at specific CpG loci to capture the epigenetic ageing profiles. With the advancement of genome‐wide methylation research, epigenetic clocks have developed two generations of quantitative models: the first generation (HannumAge 13 and HorvathAge 14 ) and the second generation (PhenoAge 15 and GrimAge 16 ). Epigenetic age has been shown to be closely associated with atrial fibrillation when factors, such as actual age, are regarded as mediating factors. 17 However, there is a lack of current research on epigenetic age and HF.
Xu et al. 18 studied the susceptibility and severity of epigenetic age, such as PhenoAge, GrimAge, HannumAge, and HorvathAge, to novel coronavirus using inverse–variance weighted (IVW), Mendelian randomization (MR)–Egger, weighted median, simple mode, and weighted mode. Similarly, this analysis was used to investigate the causality between epigenetics and HF. To summarize, a bidirectional MR study was conducted using genome‐wide association study (GWAS) data for HF and epigenetic clocks to evaluate the causality between them and thus reduce the influence of confounding factors on outcomes through a large sample of clinical genetic data and rule out reverse causality.
Research design and methods
Research design
The research was designed in accordance with the STROBE‐MR guideline. 19 In this research, four epigenetic clocks (GrimAge, 16 HannumAge, 13 HorvathAge, 14 and PhenoAge 15 ) were included as exposures and HF as outcomes to determine the instrumental variables for bidirectional MR analysis. Subsequently, MR–Egger, Cochran's Q analysis, horizontal pleiotropic analysis, and leave‐one‐out analysis were performed to validate the reliability of causality, with the first two analyses used for heterogeneity analysis and the rest for sensitivity analysis confirmation. MR was then used to examine HF as an exposure factor and epigenetic clock as an outcome factor. In summary, MR studies were required to meet the following three criteria: (i) association hypothesis: instrumental variables closely associated with exposure factors; (ii) independence hypothesis: instrumental variables unrelated to confounding factors associated with exposure and outcome factors; and (iii) exclusivity hypothesis: instrumental variables influenced outcomes through exposure factors only. In this study, bidirectional MR studies were used to examine the bidirectional causality between epigenetic age and HF (Figure 1 ).
Figure 1.
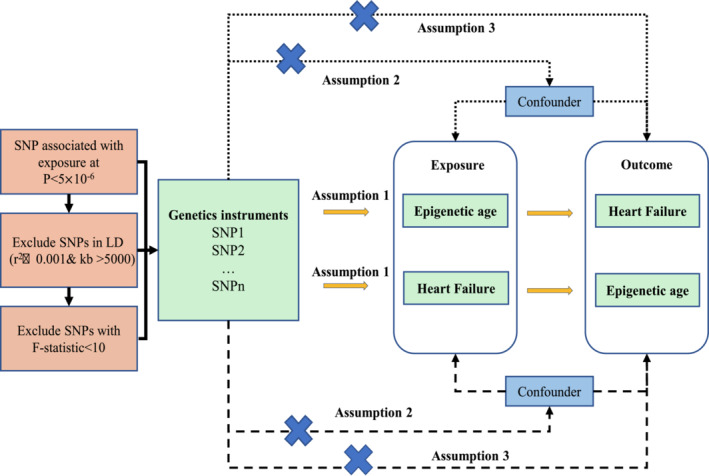
The flow chart of this study. LD, linkage disequilibrium; SNPs, single–nucleotide polymorphisms.
Data source
In this research, four epigenetic age data sets, GrimAge, 16 HannumAge, 13 HorvathAge, 14 and PhenoAge, 15 were obtained as GWAS data based on 28 cohort studies of 34 710 European ancestry investigators (https://doi.org/10.7488/ds/2834). These data identified 137 ageing‐related gene loci. 20 This research provided publicly available large GWAS summary data 21 for HF from the Molecular Epidemiology for Therapeutic Targets (HERMES) Consortium for HF, involving 26 HF cases from 47 309 and 930 014 HF cases from European ancestry and control groups, respectively, stored at the Cardiovascular Disease Knowledge Portal (https://cvd.hugeamp.org/). Details of these studies were presented in Table 1 .
Table 1.
Data description of epigenetic age and heart failure
| Type | Phenotype | Population | SNP | Sample size | n_cases | n_controls | Release date | Access address | DOI |
|---|---|---|---|---|---|---|---|---|---|
| Epigenetic age | PhenoAge | European | PhenoAge (7567585) | 34 710 | — | — | 1 July 2020 | https://datashare.ed.ac.uk/handle/10283/3645 | https://doi.org/10.7488/ds/2834 |
| GrimAge | GrimAge (7567701) | 34 710 | — | — | 2 July 2020 | https://datashare.ed.ac.uk/handle/10283/3646 | https://doi.org/10.7488/ds/2835 | ||
| HannumAge | HannumAge (7565045) | 34 710 | — | — | 3 July 2020 | https://datashare.ed.ac.uk/handle/10283/3647 | https://doi.org/10.7488/ds/2836 | ||
| HorvathAge | HorvathAge (7567532) | 34 710 | — | — | 4 July 2020 | https://datashare.ed.ac.uk/handle/10283/3648 | https://doi.org/10.7488/ds/2837 | ||
| Heart failure | Heart failure | European | Heart failure (8281262) | 964 057 | 47 309 | 930 014 | 9 January 2020 | https://cvd.hugeamp.org/downloads.html | https://doi.org/10.1038/s41467-019-13690-5 |
DOI, digital object identifier; SNP, single‐nucleotide polymorphism.
Tool variable acquisition
A series of parameter controls were performed on gene‐associated data for epigenetic age and HF to screen for eligible single‐nucleotide polymorphisms (SNPs) in a bidirectional MR analysis of two samples. First, epigenetic age (as the SNP threshold for exposure), kilobase pair (kb), and parameter r 2 were set at a loose threshold P < 5 × 10−6, 5000, and 0.001, respectively. Significant epigenetic age‐related SNPs obtained from these thresholds were used to match SNPs for HF outcome data. Moreover, the outcome SNPs that did not meet the threshold were removed. Similarly, according to the above principles, we also set HF as the threshold for significant SNP for exposure. Afterward, the most recent and stringent calculation methods for variance (R 2) and F‐statistic were performed to avoid weak shifts in instrumental variables. F‐statistic >10 is considered to avoid shifts caused by weak instrumental variables on MR results. 22 The formula is as follows:
where MAF is the minor allele frequency, β is the effect size, SE is the standard error, N is the sample size, and k is the number of tool variables.
Subsequently, exposed SNP from previous thresholds was extracted, integrated, and combined with outcome data, such that the effect value of the effector allele of the outcome was consistent with that of the exposure.
Statistical analysis
First, based on the results of the MR analysis, Cochran's Q test <0.05 was considered heterogeneous and a random‐effects model was used; otherwise, a fixed‐effects model was used. IVW was used as the primary method 23 for MR analysis and could reach robust conclusions. Meanwhile, MR–Egger, 24 weighted median, 25 simple mode, and weighted mode were used to assess the robustness of MR results. Second, the MR–Egger intercept was calculated to assess heterogeneity and horizontal pleiotropy, and the MR–Egger intercept <0.05 was considered horizontal pleiotropy. Third, a ‘leave‐one‐out’ sensitivity analysis was used to examine whether a single SNP affected MR‐level pleiotropy. Subsequently, forest plots and funnel plots were generated directly for horizontal pleiotropy testing. Finally, causal estimates (i.e. beta coefficients) were assessed and converted into random numbers [odds ratio (OR)]. The above approaches would provide the highest statistical effect based on the MR analysis of exposure to outcomes satisfying three key assumptions in the methodology section. Overall, these methods ensure the reliability of the causality between exposure and outcome. For multiple testing, the false discovery rate (FDR) is considered to be effective and robust. 26 , 27 In this study, FDR is implemented based on the R package ‘fdrtool’.
All statistical analyses were performed using the TwoSampleMR (Version 0.5.6) package of R software 4.2.1. P < 0.05 was considered statistically significant.
Results
Tool variable extraction
When utilizing epigenetic age as the exposure factor, extracted epigenetic age with GWAS data was significantly correlated SNP (P < 5 × 10−6), and linkage disequilibrium (LD) was removed (r 2 < 0.001, 5000 kb). Parallelly, we also eliminated palindromic SNPs (SNPs whose alleles consisted of one base and its complementary base). Eventually, an epigenetic age of SNP (instrumental variable: GrimAge = 26, R 2 = 1.89%, F‐statistic = 25.73; instrumental variable: HannumAge = 42, R 2 = 3.54%, F‐statistic = 30.25; instrumental variable: HorvathAge = 59, R 2 = 6.25%, F‐statistic = 39.14; and instrumental variable: PhenoAge = 37, R 2 = 3.34%, F‐statistic = 32.35) (Supporting Information, Table S1 ) satisfying the genome >2 for MR analysis was obtained. Correspondingly, when HF was the exposure factor, the number of instrumental variables was 54, R 2 = 0.15%, and F‐statistic = 27.00. The number of instrumental variables also met the requirements for MR analysis (Supporting Information, Table S2 ). In MR studies, F‐statistic was employed to evaluate the strength of instrumental variables. Generally, an F‐statistic >10 can rule out shifts caused by weak instrumental variables on MR results. In this study, the bidirectional MR had an F‐statistic >10 (range 20.73–239.73), and no weak instrumental variables were present. Therefore, these instrumental variables help determine the causality of exposure to outcomes.
Mendelian randomization analysis of epigenetic age on heart failure
Cochran's Q test (P > 0.05) indicated no heterogeneity, so IVW analysis was performed using a fixed‐effects model (Table 2 ). The IVW model suggested a causality between PhenoAge and HF [IVW OR 1.015, 95% confidence interval (CI) 1.001–1.028, P = 0.028]. And similar results were seen with the weighted median method (OR 1.020, 95% CI 1.001–1.038, P = 0.039). The OR and 95% CI of other epigenetic clocks and HF were not statistically significant (Figure 2 ) (GrimAge and HF: OR 1.013, 95% CI 0.992–1.034, P = 0.215; HannumAge and HF: OR 0.986, 95% CI 0.970–1.002, P = 0.081; HorvathAge and HF: OR 1.004, 95% CI 0.992–1.017, P = 0.519). The generated scatter plots were used to demonstrate the genetic visualization estimates of epigenetic age on HF (Figure 3 ). Results from other analytical methods and forest plots of MR analyses of individual SNPs were located in Supporting Information, Figure S1 . IVW (P = 0.212) and MR–Egger regression (P = 0.185) showed no significant heterogeneity between epigenetic age and MR analysis of HF. Egger_intercept of MR–Egger and zero were not statistically significant (P = 0.664). SNPs were not horizontally pleiotropic (Table 2 ). There were no SNPs in the study data that had a significant impact on the results, so the results have a high level of confidence (Supporting Information, Figure S2 ). Therefore, we can also draw the conclusion that the results were robust.
Table 2.
Heterogeneity and pleiotropy of epigenetic age and heart failure
| Exposure | Outcome | Q‐statistics | Pleiotropic test | ||
|---|---|---|---|---|---|
| MR–Egger | IVW | Egger_intercept | P value | ||
| GrimAge | HF | Q = 1.74E + 01, P = 6.88E − 01 | Q = 1.75E + 01, P = 7.37E − 01 | −2.12E − 03 | 7.65E − 01 |
| HannumAge | HF | Q = 3.23E + 01, P = 5.01E − 01 | Q = 3.24E + 01, P = 5.44E − 01 | −1.73E − 03 | 7.35E − 01 |
| HorvathAge | HF | Q = 5.50E + 01, P = 2.28E − 01 | Q = 5.50E + 01, P = 2.59E − 01 | 6.33E − 05 | 9.90E − 01 |
| PhenoAge | HF | Q = 3.20E + 01, P = 1.85E − 01 | Q = 3.92E + 01, P = 2.12E − 01 | 2.05E − 03 | 6.64E − 01 |
| HF | GrimAge | Q = 1.47E + 01, P = 1.18E − 02 | Q = 1.93E + 01, P = 3.64E − 03 | 1.67E − 01 | 2.65E − 01 |
| HF | HannumAge | Q = 4.18E + 01, P = 1.74E − 05 | Q = 4.44E + 01, P = 1.32E − 05 | −5.58E − 02 | 4.31E − 01 |
| HF | HorvathAge | Q = 1.35E + 01, P = 3.67E − 03 | Q = 1.42E + 01, P = 6.62E − 03 | 7.04E − 02 | 7.16E − 01 |
| HF | PhenoAge | Q = 9.95E + 00, P = 6.91E − 03 | Q = 1.03E + 01, P = 1.65E − 02 | −9.67E − 02 | 8.28E − 01 |
IVW, inverse‐variance weighted; MR, Mendelian randomization.
Figure 2.
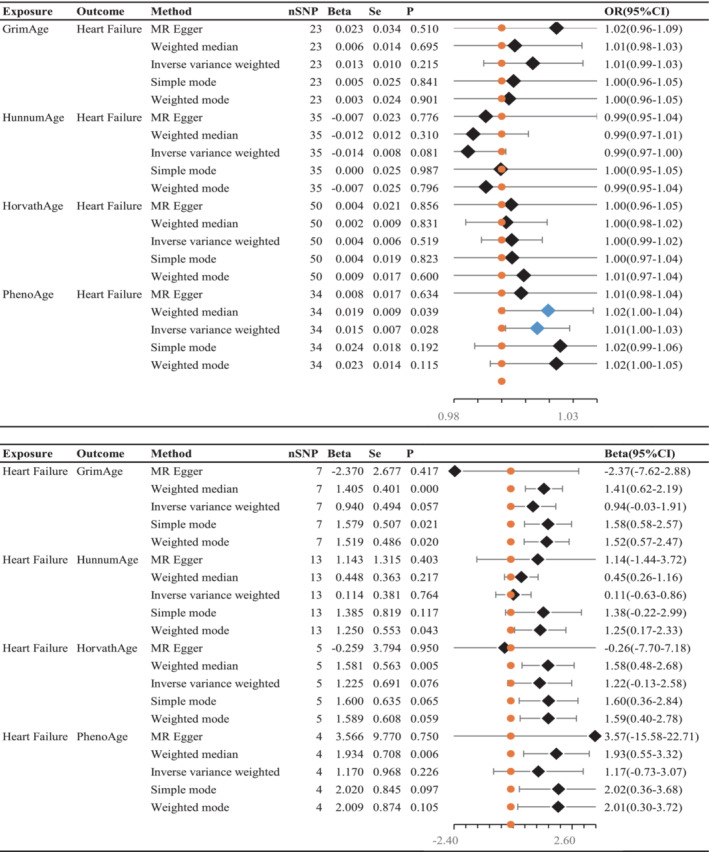
Inverse–variance weighted (IVW) was used as the main method to analyse the two‐way causal relationship between epigenetic age (GrimAge, HannumAge, HorvathAge, and PhenoAge) and heart failure. The forest map visualizes the causal effect of exposure on outcome risk by IVW method [when the outcome is heart failure, i.e. the dichotomy variable, the standard line is the ‘X = 1’ line (orange dashed line); when the outcome is epigenetic age, i.e. the continuity variable, the standard line is the ‘X = 0’ line (orange dashed line)], and the blue markers represent positive results with P < 0.05. Beta, risk index; CI, confidence interval; OR, odds ratio; Se, standard error; SNPs, single–nucleotide polymorphisms.
Figure 3.
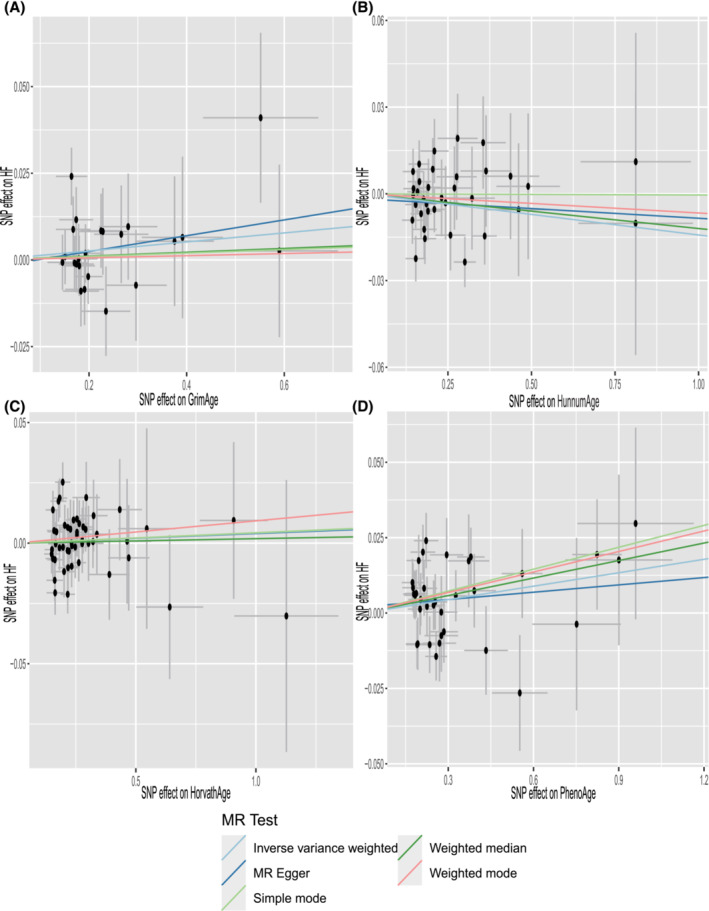
Scatter plot of epigenetic age and HF. Horizontal ordinate: single–nucleotide polymorphisms (SNPs) effect on ‘exposure’; vertical coordinates: SNP effect on ‘outcome’. (A) Exposure: GrimAge; outcome: HF. (B) Exposure: HannumAge; outcome: HF. (C) Exposure: HorvathAge; outcome: HF. (D) Exposure: PhenoAge; outcome: HF. MR, Mendelian randomization.
Mendelian randomization analysis of heart failure on epigenetic age
There is no clear evidence of causality between HF and epigenetic age (GrimAge IVW beta = 0.940, 95% CI −0.029 to 1.908, P = 0.057; HannumAge IVW beta = 0.114, 95% CI −0.632 to 0.861, P = 0.764; HorvathAge IVW beta = 0.200, 95% CI −0.129 to 2.579, P = 0.076; and PhenoAge IVW beta = 1.170, 95% CI −0.726 to 3.607, P = 0.226) (Figure 2 ). Visual estimation plots of the genetic variance are displayed in Figure 4 and forest plots of individual SNPs for reverse MR in Supporting Information, Figure S3 . The diagram of HF and epigenetic age was shown in Supporting Information, Figure S4 .
Figure 4.
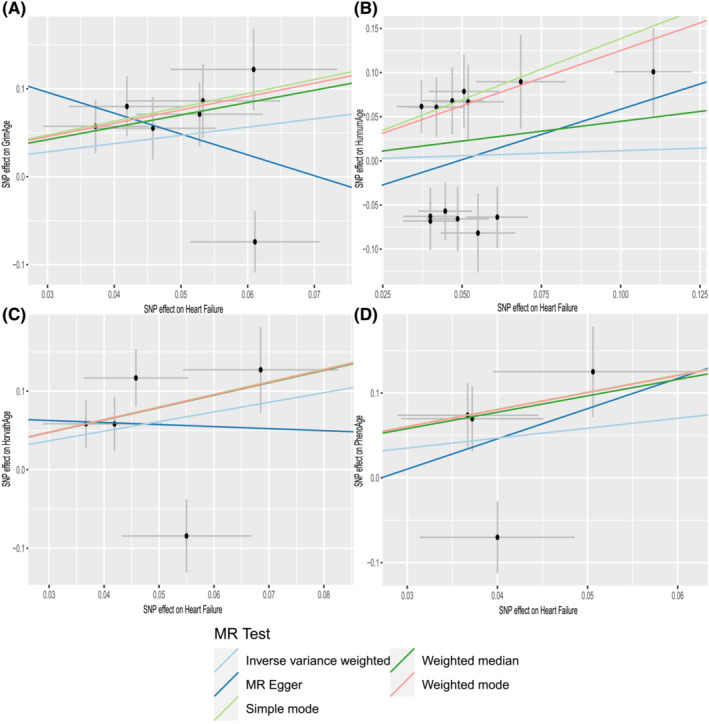
Scatter plots of HF and epigenetic age. Horizontal ordinate: single–nucleotide polymorphisms (SNPs) effect on ‘exposure’; vertical coordinates: SNP effect on ‘outcome’. (A) Exposure: HF; outcome: GrimAge. (B) Exposure: HF; outcome: HannumAge. (C) Exposure: HF; outcome: HorvathAge. (D) Exposure: HF; outcome: PhenoAge. MR, Mendelian randomization.
Discussion
Ageing is a risk factor in an ageing society, causing increased morbidity and mortality in various diseases. The mechanism of ageing is the focus of human physiological and pathological mechanism research. 28 Although the epigenetic age is not exactly comparable with the traditional age, epigenetic clocks can help researchers better understand the biological mechanisms of human health and ageing using different training samples and populations. 12 Investigating the causality between epigenetic age and cardiovascular disease is a novel and challenging research topic.
To the best of our knowledge, this is the first study to look into the bidirectional causality between epigenetic age and HF. In this research, the epigenetic clock PhenoAge was found to increase the risk of HF. On the other hand, HF also increased the risk of the epigenetic HorvathAge clock. In our study, no causality between other epigenetic clocks and HF was found.
We investigated the mechanism by which the epigenetic clock PhenoAge increases the risk of HF. As an epigenetic biomarker, PhenoAge can effectively combine blood DNAm indicating that epigenetic age, with a correlation coefficient with the heart of 0.66, can specifically assess the ageing characteristics of cardiovascular disease 15 and can better capture ‘preclinical ageing’. Second, it has been demonstrated that pathway enrichment of genes involved in PhenoAge positively correlates with the activation of pro‐inflammatory pathways such as response to lipopolysaccharide and nuclear factor‐κB (NF‐κB). Lipopolysaccharide is a toxic component produced by Gram‐negative cocci with immunostimulatory and immunomodulatory effects. 29 Increased plasma concentrations of lipopolysaccharide have been reported in patients with chronic HF. 30 They may reach the blood and cause the release of pro‐inflammatory factors such as tumour necrosis factor‐α (TNF‐α) by altering the intestinal barrier. 31 Lipopolysaccharide reactivity has also been demonstrated to be an independent predictor of mortality in HF. 32 NF‐κB activates myocardial senescence 33 and is closely related to cardiomyocyte survival and inflammatory regulation. 34 Prolonged activation of NF‐κB promotes inflammatory pathway signalling causing HF. Several studies 35 , 36 , 37 have shown that inhibiting NF‐κB pathway activity helps delay cardiomyocyte senescence and reduce age‐related HF, with the underlying mechanism possibly associated with apoptosis 38 and mitochondrial oxidative stress. 39 The findings of the preceding studies are consistent with the findings of this study regarding the causality of HF and epigenetic age. 40 We investigated why HF can accelerate the ageing of epigenetic clock (HorvathAge) malfunction due to HF's effect on telomere length and DNA methylation modifications. 41
However, our study does have inevitable limitations. First, the data in our study were primarily obtained from the European populations, and despite a large number of populations, genetic studies on epigenetic age and HF between ethnic groups are still lacking. This may cause discrepancies between observations and the actual situation. Second, because epigenetic age is intrinsically associated with environmental exposures rather than genetic factors, this highlights the limitations of MR in this context. Finally, the OR values of our findings were low; thus, they should be interpreted cautiously. We look forward to future research that will explain the relationship between epigenetic age and HF.
Conclusions
In this study, epigenetic age had a bidirectional causality with HF. PhenoAge, an epigenetic clock, increased the risk of HF. Moreover, the underlying mechanism may be related to inflammatory pathways. HF accelerated the epigenetic clock HorvathAge. This research explored the causality between epigenetic age and HF via MR analysis. However, more research into the mechanisms between epigenetic age and HF in different ethnic groups is needed.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This study was supported by the Natural Science Foundation of Shandong Province (ZR2020QH306) and the Department of Science and Technology of Sichuan Province of China (23QYCX0040).
Supporting information
Figure S1. Mendelian randomization effects forest plots for individual SNPs with epigenetic age as the exposure, Heart Failure as the outcome. (A) exposure: GrimAge, outcome: Heart Failure; (B) exposure: HannumAge, outcome: Heart Failure; (C) exposure: HovarthAge, outcome: Heart Failure; (D) exposure: PhenoAge, outcome: Heart Failure; The red marker points indicated All‐MR Egger and All‐IVW.
Figure S2. MR leave‐one‐out sensitivity analysis “outcome” on “Heart Failure”. (A) exposure: GrimAge, outcome: Heart Failure; (B) exposure: HannumAge, outcome: Heart Failure; (C) exposure: HovarthAge, outcome: Heart Failure; (D) exposure: PhenoAge, outcome: Heart Failure.
Figure S3. Mendelian randomization effects forest plots for individual SNPs with heart failure as the exposure, epigenetic age as the outcome. (A) exposure: Heart Failure, outcome: GrimAge; (B) exposure: Heart Failure, outcome: HannumAge; (C) exposure: Heart Failure, outcome: HorvathAge; (D) exposure: Heart Failure, outcome: PhenoAge. The red marker points indicated All‐MR Egger and All‐IVW.
Figure S4. MR leave‐one‐out sensitivity analysis “outcome” on “Epigenetic age”. (A) exposure: Heart Failure, outcome: GrimAge; (B) exposure: Heart Failure, outcome: HannumAge; (C) exposure: Heart Failure, outcome: HovarthAge; (D) exposure: Heart Failure, outcome: PhenoAge.
Table S1. Epigenetic age instrumental variables and R2 and F‐statistics.
Table S2. Heart failue instrumental variables and R2 and F‐statistics.
Data S1. Supporting Information.
Acknowledgements
We thank statisticians and GWAS data providers for their efforts in this study. Meanwhile, we thank Dr Guangli Sun, Chief Physician, Department of Traditional Chinese Medicine, Laixi City Hospital, for her financial assistance with this study. We thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript.
Zhang, F. , Deng, S. , Zhang, J. , Xu, W. , Xian, D. , Wang, Y. , Zhao, Q. , Liu, Y. , Zhu, X. , Peng, M. , and Zhang, L. (2023) Causality between heart failure and epigenetic age: a bidirectional Mendelian randomization study. ESC Heart Failure, 10: 2903–2913. 10.1002/ehf2.14446.
Contributor Information
Xiuli Zhu, Email: 15562592002@163.com.
Min Peng, Email: pengmin186@126.com.
Lin Zhang, Email: zhanglinfudan@zju.edu.cn.
References
- 1. McDonagh TA, Metra M, Adamo M, Gardner RS, Baumbach A, Böhm M, Burri H, Butler J, Čelutkienė J, Chioncel O, Cleland JGF, Coats AJS, Crespo‐Leiro MG, Farmakis D, Gilard M, Heymans S, Hoes AW, Jaarsma T, Jankowska EA, Lainscak M, Lam CSP, Lyon AR, McMurray JJV, Mebazaa A, Mindham R, Muneretto C, Francesco Piepoli M, Price S, Rosano GMC, Ruschitzka F, Kathrine Skibelund A. 2021 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure: developed by the Task Force for the Diagnosis and Treatment of Acute and Chronic Heart Failure of the European Society of Cardiology (ESC). With the special contribution of the Heart Failure Association (HFA) of the ESC. Eur J Heart Fail. 2022; 24: 4901. [DOI] [PubMed] [Google Scholar]
- 2. Li Z, Zhao H, Wang J. Metabolism and chronic inflammation: the links between chronic heart failure and comorbidities. Front Cardiovasc Med. 2021; 8: 650278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rajadurai J, Tse HF, Wang CH, Yang NI, Zhou J, Sim D. Understanding the epidemiology of heart failure to improve management practices: an Asia‐Pacific perspective. J Card Fail. 2017; 23: 327–339. [DOI] [PubMed] [Google Scholar]
- 4. Shah KS, Xu H, Matsouaka RA, Bhatt DL, Heidenreich PA, Hernandez AF, Devore AD, Yancy CW, Fonarow GC. Heart failure with preserved, borderline, and reduced ejection fraction: 5‐year outcomes. J Am Coll Cardiol. 2017; 70: 2476–2486. [DOI] [PubMed] [Google Scholar]
- 5. Conrad N, Judge A, Tran J, Mohseni H, Hedgecott D, Crespillo AP, Allison M, Hemingway H, Cleland JG, McMurray JJV, Rahimi K. Temporal trends and patterns in heart failure incidence: a population‐based study of 4 million individuals. Lancet. 2018; 391: 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ziaeian B, Fonarow GC. Epidemiology and aetiology of heart failure. Nat Rev Cardiol. 2016; 13: 368–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Triposkiadis F, Xanthopoulos A, Butler J. Cardiovascular aging and heart failure: JACC review topic of the week. J Am Coll Cardiol. 2019; 74: 804–813. [DOI] [PubMed] [Google Scholar]
- 8. Papait R, Greco C, Kunderfranco P, Latronico MVG, Condorelli G. Epigenetics: a new mechanism of regulation of heart failure? Basic Res Cardiol. 2013; 108: 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Raftopoulos L, Katsi V, Makris T, Tousoulis D, Stefanadis C, Kallikazaros I. Epigenetics, the missing link in hypertension. Life Sci. 2015; 129: 22–26. [DOI] [PubMed] [Google Scholar]
- 10. Lowsky DJ, Olshansky SJ, Bhattacharya J, Goldman DP. Heterogeneity in healthy aging. J Gerontol A Biol Sci Med Sci. 2014; 69: 640–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jylhävä J, Pedersen NL, Hägg S. Biological age predictors. EBioMedicine. 2017; 21: 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Z, Leung D, Thrush K, Zhao W, Ratliff S, Tanaka T, Schmitz LL, Smith JA, Ferrucci L, Levine ME. Underlying features of epigenetic aging clocks in vivo and in vitro. Aging Cell. 2020; 19: e13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T, Zhang K. Genome‐wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013; 49: 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013; 14: R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, Hou L, Baccarelli AA, Stewart JD, Li Y, Whitsel EA, Wilson JG, Reiner AP, Aviv A, Lohman K, Liu Y, Ferrucci L, Horvath S. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Milano). 2018; 10: 573–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, Hou L, Baccarelli AA, Li Y, Stewart JD, Whitsel EA, Assimes TL, Ferrucci L, Horvath S. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Milano). 2019; 11: 303–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roberts JD, Vittinghoff E, Lu AT, Alonso A, Wang B, Sitlani CM, Mohammadi‐Shemirani P, Fornage M, Kornej J, Brody JA, Arking DE, Lin H, Heckbert SR, Prokic I, Ghanbari M, Skanes AC, Bartz TM, Perez MV, Taylor KD, Lubitz SA, Ellinor PT, Lunetta KL, Pankow JS, Paré G, Sotoodehnia N, Benjamin EJ, Horvath S, Marcus GM. Epigenetic age and the risk of incident atrial fibrillation. Circulation. 2021; 144: 1899–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu W, Zhang F, Shi Y, Chen Y, Shi B, Yu G. Causal association of epigenetic aging and COVID‐19 severity and susceptibility: a bidirectional Mendelian randomization study. Front Med (Lausanne). 2022; 9: 989950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, VanderWeele TJ, Higgins JPT, Timpson NJ, Dimou N, Langenberg C, Golub RM, Loder EW, Gallo V, Tybjaerg‐Hansen A, Davey Smith G, Egger M, Richards JB. Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: the STROBE‐MR statement. JAMA. 2021; 326: 1614–1621. [DOI] [PubMed] [Google Scholar]
- 20. McCartney DL, Min JL, Richmond RC, Lu AT, Sobczyk MK, Davies G, Broer L, Guo X, Jeong A, Jung J, Kasela S, Katrinli S, Kuo PL, Matias‐Garcia PR, Mishra PP, Nygaard M, Palviainen T, Patki A, Raffield LM, Ratliff SM, Richardson TG, Robinson O, Soerensen M, Sun D, Tsai PC, van der Zee MD, Walker RM, Wang X, Wang Y, Xia R, Xu Z, Yao J, Zhao W, Correa A, Boerwinkle E, Dugue PA, Durda P, Elliott HR, Gieger C, The Genetics of DNA Methylation Consortium , de Geus EJC, Harris SE, Hemani G, Imboden M, Kahonen M, Kardia SLR, Kresovich JK, Li S, Lunetta KL, Mangino M, Mason D, McIntosh AM, Mengel‐From J, Moore AZ, Murabito JM, NHLBI Trans‐Omics for Precision Medicine (TOPMed) Consortium , Ollikainen M, Pankow JS, Pedersen NL, Peters A, Polidoro S, Porteous DJ, Raitakari O, Rich SS, Sandler DP, Sillanpaa E, Smith AK, Southey MC, Strauch K, Tiwari H, Tanaka T, Tillin T, Uitterlinden AG, Van Den Berg DJ, van Dongen J, Wilson JG, Wright J, Yet I, Arnett D, Bandinelli S, Bell JT, Binder AM, Boomsma DI, Chen W, Christensen K, Conneely KN, Elliott P, Ferrucci L, Fornage M, Hagg S, Hayward C, Irvin M, Kaprio J, Lawlor DA, Lehtimaki T, Lohoff FW, Milani L, Milne RL, Probst‐Hensch N, Reiner AP, Ritz B, Rotter JI, Smith JA, Taylor JA, van Meurs JBJ, Vineis P, Waldenberger M, Deary IJ, Relton CL, Horvath S, Marioni RE. Genome‐wide association studies identify 137 genetic loci for DNA methylation biomarkers of aging. Genome Biol. 2021; 22: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shah S, Henry A, Roselli C, Lin H, Sveinbjörnsson G, Fatemifar G, Hedman ÅK, Wilk JB, Morley MP, Chaffin MD, Helgadottir A, Verweij N, Dehghan A, Almgren P, Andersson C, Aragam KG, Ärnlöv J, Backman JD, Biggs ML, Bloom HL, Brandimarto J, Brown MR, Buckbinder L, Carey DJ, Chasman DI, Chen X, Chen X, Chung J, Chutkow W, Cook JP, Delgado GE, Denaxas S, Doney AS, Dörr M, Dudley SC, Dunn ME, Engström G, Esko T, Felix SB, Finan C, Ford I, Ghanbari M, Ghasemi S, Giedraitis V, Giulianini F, Gottdiener JS, Gross S, Guðbjartsson DF, Gutmann R, Haggerty CM, van der Harst P, Hyde CL, Ingelsson E, Jukema JW, Kavousi M, Khaw K‐T, Kleber ME, Køber L, Koekemoer A, Langenberg C, Lind L, Lindgren CM, London B, Lotta LA, Lovering RC, Ja L, Magnusson P, Mahajan A, Margulies KB, März W, Melander O, Mordi IR, Morgan T, Morris AD, Morris AP, Morrison AC, Nagle MW, Nelson CP, Niessner A, Niiranen T, O'Donoghue ML, Owens AT, Palmer CNA, Parry HM, Perola M, Portilla‐Fernandez E, Psaty BM, Rice KM, Ridker PM, Romaine SPR, Rotter JI, Salo P, Salomaa V, van Setten J, Shalaby AA, Smelser DT, Smith NL, Stender S, Stott DJ, Svensson P, Tammesoo M‐L, Taylor KD, Teder‐Laving M, Teumer A, Thorgeirsson G, Thorsteinsdottir U, Torp‐Pedersen C, Trompet S, Tyl B, Uitterlinden AG, Veluchamy A, Völker U, Voors AA, Wang X, Wareham NJ, Waterworth D, Weeke PE, Weiss R, Wiggins KL, Xing H, Yerges‐Armstrong LM, Yu B, Zannad F, Zhao JH, Hemingway H, Samani NJ, McMurray JJV, Yang J, Visscher PM, Newton‐Cheh C, Malarstig A, Holm H, Lubitz SA, Sattar N, Holmes MV, Cappola TP, Asselbergs FW, Hingorani AD, Kuchenbaecker K, Ellinor PT, Lang CC, Stefansson K, Smith JG, Vasan RS, Swerdlow DI, Lumbers RT. Genome‐wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun. 2020; 11: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011; 40: 740–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lawlor DA, Harbord RM, Sterne JAC, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008; 27: 1133–1163. [DOI] [PubMed] [Google Scholar]
- 24. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015; 44: 512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016; 40: 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Strimmer K. fdrtool: a versatile R package for estimating local and tail area‐based false discovery rates. Bioinformatics (Oxford, England). 2008; 24: 1461–1462. [DOI] [PubMed] [Google Scholar]
- 27. Strimmer K. A unified approach to false discovery rate estimation. BMC Bioinformatics. 2008; 9: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kennedy BK, Berger SL, Brunet A, Campisi J, Cuervo AM, Epel ES, Franceschi C, Lithgow GJ, Morimoto RI, Pessin JE, Rando TA, Richardson A, Schadt EE, Wyss‐Coray T, Sierra F. Geroscience: linking aging to chronic disease. Cell. 2014; 159: 709–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002; 71: 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Niebauer J, Volk HD, Kemp M, Dominguez M, Schumann RR, Rauchhaus M, Poole‐Wilson PA, Coats AJ, Anker SD. Endotoxin and immune activation in chronic heart failure: a prospective cohort study. Lancet (London, England). 1999; 353: 1838–1842. [DOI] [PubMed] [Google Scholar]
- 31. von Haehling S, Genth‐Zotz S, Sharma R, Bolger AP, Doehner W, Barnes PJ, Coats AJ, Anker SD. The relationship between age and production of tumour necrosis factor‐α in healthy volunteers and patients with chronic heart failure. Int J Cardiol. 2003; 90: 197–204. [DOI] [PubMed] [Google Scholar]
- 32. Ebner N, Földes G, Schomburg L, Renko K, Springer J, Jankowska EA, Sharma R, Genth‐Zotz S, Doehner W, Anker SD, von Haehling S. Lipopolysaccharide responsiveness is an independent predictor of death in patients with chronic heart failure. J Mol Cell Cardiol. 2015; 87: 48–53. [DOI] [PubMed] [Google Scholar]
- 33. Helenius M, Hänninen M, Lehtinen SK, Salminen A. Aging‐induced up‐regulation of nuclear binding activities of oxidative stress responsive NF‐κB transcription factor in mouse cardiac muscle. J Mol Cell Cardiol. 1996; 28: 487–498. [DOI] [PubMed] [Google Scholar]
- 34. Gordon JW, Shaw JA, Kirshenbaum LA. Multiple facets of NF‐κB in the heart: to be or not to NF‐κB. Circ Res. 2011; 108: 1122–1132. [DOI] [PubMed] [Google Scholar]
- 35. Wang X, Li X, Ong H, Tan T, Park KH, Bian Z, Zou X, Haggard E, Janssen PM, Merritt RE, Pawlik TM, Whitson BA, Mokadam NA, Cao L, Zhu H, Cai C, Ma J. MG53 suppresses NF‐κB activation to mitigate age‐related heart failure. JCI Insight. 2021; 6:e148375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dong X, Jiang J, Lin Z, Wen R, Zou L, Luo T, Guan Z, Li X, Wang L, Lu L, Li H, Huang Y, Yang Z, Wang J, Ye X, Hong X, Wang L, Xian S, Chen Z. Nuanxinkang protects against ischemia/reperfusion‐induced heart failure through regulating IKKβ/IκBα/NF‐κB‐mediated macrophage polarization. Phytomedicine. 2022; 101: 154093. [DOI] [PubMed] [Google Scholar]
- 37. Wang M, Luo W, Yu T, Liang S, Sun J, Zhang Y, Han X, Long X, Liang G, Li G. Corynoline protects Ang II‐induced hypertensive heart failure by increasing PPARα and inhibiting NF‐κB pathway. Biomed Pharmacother. 2022; 150: 113075. [DOI] [PubMed] [Google Scholar]
- 38. Hamid T, Guo SZ, Kingery JR, Xiang X, Dawn B, Prabhu SD. Cardiomyocyte NF‐κB p65 promotes adverse remodelling, apoptosis, and endoplasmic reticulum stress in heart failure. Cardiovasc Res. 2011; 89: 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dai D‐F, Chen T, Wanagat J, Laflamme M, Marcinek DJ, Emond MJ, Ngo CP, Prolla TA, Rabinovitch PS. Age‐dependent cardiomyopathy in mitochondrial mutator mice is attenuated by overexpression of catalase targeted to mitochondria. Aging Cell. 2010; 9: 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu AT, Seeboth A, Tsai P‐C, Sun D, Quach A, Reiner AP, Kooperberg C, Ferrucci L, Hou L, Baccarelli AA, Li Y, Harris SE, Corley J, Taylor A, Deary IJ, Stewart JD, Whitsel EA, Assimes TL, Chen W, Li S, Mangino M, Bell JT, Wilson JG, Aviv A, Marioni RE, Raj K, Horvath S. DNA methylation‐based estimator of telomere length. Aging (Milano). 2019; 11: 5895–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Duygu B, Poels EM, da Costa Martins PA. Genetics and epigenetics of arrhythmia and heart failure. Front Genet. 2013; 4: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Mendelian randomization effects forest plots for individual SNPs with epigenetic age as the exposure, Heart Failure as the outcome. (A) exposure: GrimAge, outcome: Heart Failure; (B) exposure: HannumAge, outcome: Heart Failure; (C) exposure: HovarthAge, outcome: Heart Failure; (D) exposure: PhenoAge, outcome: Heart Failure; The red marker points indicated All‐MR Egger and All‐IVW.
Figure S2. MR leave‐one‐out sensitivity analysis “outcome” on “Heart Failure”. (A) exposure: GrimAge, outcome: Heart Failure; (B) exposure: HannumAge, outcome: Heart Failure; (C) exposure: HovarthAge, outcome: Heart Failure; (D) exposure: PhenoAge, outcome: Heart Failure.
Figure S3. Mendelian randomization effects forest plots for individual SNPs with heart failure as the exposure, epigenetic age as the outcome. (A) exposure: Heart Failure, outcome: GrimAge; (B) exposure: Heart Failure, outcome: HannumAge; (C) exposure: Heart Failure, outcome: HorvathAge; (D) exposure: Heart Failure, outcome: PhenoAge. The red marker points indicated All‐MR Egger and All‐IVW.
Figure S4. MR leave‐one‐out sensitivity analysis “outcome” on “Epigenetic age”. (A) exposure: Heart Failure, outcome: GrimAge; (B) exposure: Heart Failure, outcome: HannumAge; (C) exposure: Heart Failure, outcome: HovarthAge; (D) exposure: Heart Failure, outcome: PhenoAge.
Table S1. Epigenetic age instrumental variables and R2 and F‐statistics.
Table S2. Heart failue instrumental variables and R2 and F‐statistics.
Data S1. Supporting Information.


