Abstract
Summary
Computational methods for the quantification and visualization of the relative contribution of molecular interactions to the stability of biomolecular structures and complexes are fundamental to understand, modulate and engineer biological processes. Here, we present Surfaces, an easy to use, fast and customizable software for quantification and visualization of molecular interactions based on the calculation of surface areas in contact. Surfaces calculations shows equivalent or better correlations with experimental data as computationally expensive methods based on molecular dynamics.
Availability and implementation
All scripts are available at https://github.com/NRGLab/Surfaces. Surface’s documentation is available at https://surfaces-tutorial.readthedocs.io/en/latest/index.html.
1 Introduction
Molecular interactions determine all aspects of biological processes. Understanding the relative contributions of individual molecular interactions can guide the design of effective drugs to modulate biological processes as well as help understand the effect of mutations in natural processes or guide their introduction in protein engineering.
Protein engineering is an important tool in biotechnology that can produce proteins with improved therapeutic and industrial profiles (Chica 2015, Tobin et al. 2015, Bojar and Fussenegger 2020). In recent years, the field has made giant strides forward, but it still has the potential for exponential growth—as seen for many fields that benefit from high-throughput technologies and powerful new computational tools. However, a random search for all possible sequence configurations and respective structures and functions might reveal an enormous searchable space to explore. This is, even today, one of the biggest challenges of protein engineering, and one that makes the rational design necessary. Techniques that provide insights into the partial contribution of each atom or residue to protein interactions are critical for understanding the intricacies of the interaction interface. By identifying the specific residues and atoms that play a crucial role in the interaction, these techniques can aid in the development of rational protein engineering strategies. Unfortunately, these techniques, mostly based on ab initio approaches, are often computationally expensive, limiting the extent to which they can be used to explore this expansive searchable space (e.g. through simulating deep mutational scans or in large datasets).
With the decreasing cost of computational power, it is possible to simulate biological systems considering more realistically the underlying physical processes at the core of molecular interactions. Molecular dynamics (MD) simulations are at the forefront of such efforts, but such methods are computationally expensive and often also difficult to implement and thus remain impractical for high-throughput applications or broad adoption. Among these methods, we can highlight well known methods for modeling and analysis of protein structures, FoldX (Buß et al. 2018) and Rosetta (Kortemme et al. 2003, Leaver-Fay et al. 2011), free-energy perturbation (FEP) methods (McCarrick and Kollman 1999, Zhu et al. 2022, Sergeeva et al. 2023), molecular mechanics (MM/PBSA and MM/GBSA) methods (Genheden and Ryde 2015) that aim at predicting the ΔΔG of binding for different mutations relative to wild-type or the binding ΔG for biomolecular complexes, and gRINN (Serçinoglu and Ozbek 2018), a tool to breakdown the per-residue energetic contribution of molecular interactions based on MD simulations.
To address this challenge, simplified techniques using atomic surface areas in contact to estimate binding energy can be employed. This approach has previously been introduced in methods such as LPC (Ligand-Protein Contacts) and CSU (Contacts of Structural Units) (Sobolev et al. 1999) as well as STC (Structure-based Thermodynamic Calculations) (Lavigne et al. 2000). While these techniques are currently limited by server availability, as well as over-simplified atom type definitions and energetic pairwise matrices, they provide a faster alternative to evaluating energetic decompositions and served as the basis upon which many other methods that use atomic surfaces areas in contact were built (Gaudreault and Najmanovich 2015, Frappier et al. 2017, Ribeiro et al. 2019, Olechnovič and Venclovas 2021). Therefore, it is worth revisiting and improving these techniques by employing new libraries and visualization tools, making them customizable and user-friendly to broaden their general applicability. By doing so, we can enhance our understanding of the per-residue contribution to protein interactions and facilitate more efficient and effective protein engineering.
In this application note, we present Surfaces, a fast method that utilizes atomic surface areas in contact, user-defined atom-type definitions, and pairwise pseudo-energetic matrices as a proxy for highlighting favorable and unfavorable interactions within and between proteins, as well as between proteins and other biomolecules.
2 Methods
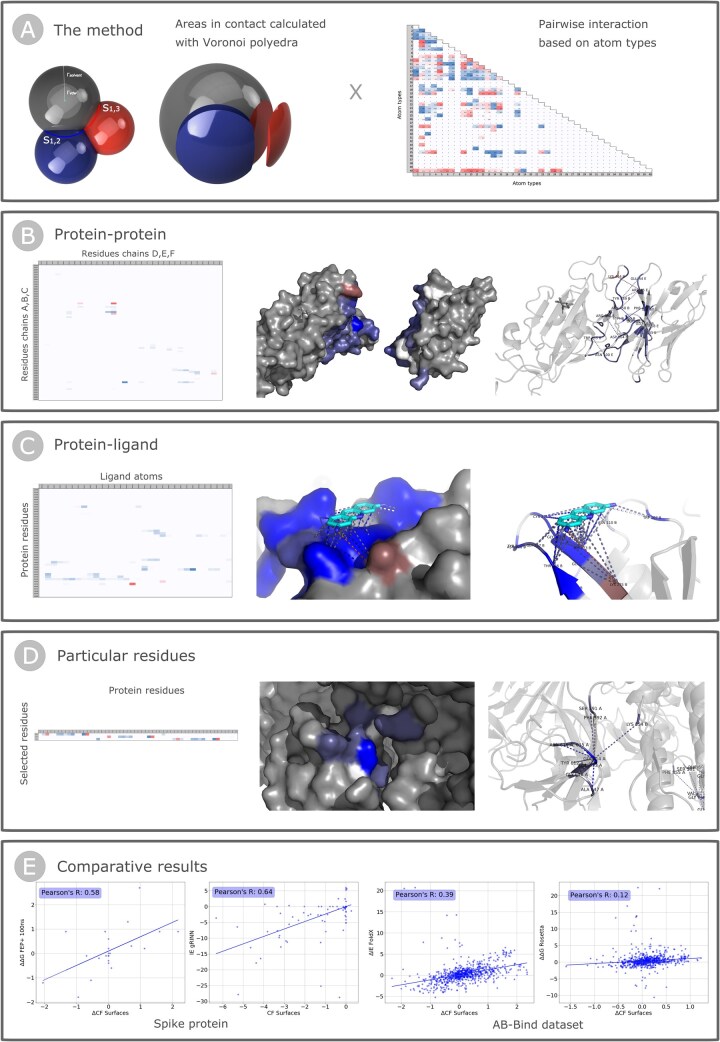
Surfaces quantifies atomic interactions using two measures. The first measure is the area in contact between atoms as described and calculated by Vcontacts using a Voronoi procedure (McConkey et al. 2002). This method restricts the evaluation of interactions to atoms within close proximity. The second measure is a pairwise pseudo-energetic matrix that assigns an interaction pseudo-energy based on the atom types (Fig. 1A).
Figure 1.
Schematic representation of the method and applications of Surfaces scripts. (A) The method: Representation of the calculated areas in contact between atoms 1 (gray), 2 (blue), and 3 (red), considering the expanded radii—that accounts for the van der Waals radii () and the radius of a water molecule ()—and leading to the definition of the surfaces in contact and , represented as exploded spherical caps, and the default matrix of pairwise interactions based on 40 SYBYL atom types. (B) Protein–protein: Example of the protein–protein application using the PDB structure 7VQ0 (Maeda et al. 2022). Representation of the. CSV output and of the visual output colored from red (unfavorable) to blue (favorable) showing the net value of interactions mapped into the surface of the chains, as well as the most relevant interactions highlighted with dash lines. (C) Protein–ligand: Example of the protein–ligand application using the PDB structure 7NT4 (Napolitano et al. 2022). Representation of the. CSV output and of the visual output colored from red (unfavorable) to blue (favorable) showing the net value of interactions mapped into the surface of the protein, as well as the interactions with the atoms of the ligand highlighted with dash lines. (D) Particular residues: Example of the particular residues application using the PDB structure 7EAZ (Yang et al. 2021) and selecting the residues GLY614. Representation of the. CSV output and of the visual output colored from red (unfavorable) to blue (favorable) showing the net value of interactions mapped into the surface of the protein, as well as the interactions involving the residues of interest highlighted with dash lines. (E) Comparative results of Surfaces with the calculations performed using FEP + 100 ns MD simulations for the interaction of 23 mutants of the Spike protein and the receptor ACE2 (Sergeeva et al. 2023), and with the pairwise Interaction Energies calculated with gRINN (Serçinoglu and Ozbek 2018) for a MD simulation of the Spike protein Receptor-Binding Domain of the Delta variant and the receptor ACE2 (Cheng et al. 2022), as well as the comparison with published results obtained using FoldX (Schymkowitz et al. 2005) and Rosetta (Kortemme et al. 2003) for the prediction of the effects on binding of 1101 mutations that constitute the AB-Bind dataset (Sirin et al. 2016).
| (1) |
The complementarity function (CF) that accounts for these two measures is described by Equation (1), in which N is the total of atoms of unit 1, M is the total of atoms of unit 2, is the pseudo-energy for the interaction between atoms i and j, and is the surface area in contact between atoms i and j. Units can correspond to residues, ligands or ligand atoms.
The CF calculation is based on the scoring function utilized by FlexAID (Gaudreault and Najmanovich 2015), a ligand docking tool, that also provides the SYBYL definition of atom types and the respective matrix of pairwise interaction energies optimized for the prediction of ligand poses using the PDBbind dataset (Wang et al. 2005). The 40 SYBYL atom types and the respective pairwise atom-type pseudo-energetic matrix from FlexAID are used as the default in Surfaces scripts. However, the Surfaces scripts are designed to be customizable, enabling the incorporation of modified residues or ligands, as well as the use of alternative atom type classifications and energetic matrices.
To visually represent the results of interaction analysis, Surfaces offers functions created using the PyMOL API (pymol.org). These functions are also customizable and allow for the visualization of the overall contribution of residues to interaction surfaces, as well as specific interactions between structural units, as PyMOL sessions.
3 Results and discussion
Surfaces was built as a set of python scripts designed to evaluate various types of interactions in proteins. The scripts are user-friendly, fast, and can be easily customized and automated, offering a scriptable way of generating data and visual representations for the interactions of interest.
3.1 Applications of surfaces
The specific types of interactions that can be analyzed by Surfaces are protein-protein, protein-ligand and residue interactions. In all cases, the input is a .PDB formatted file while the output of the quantitative analysis is saved as a .CSV file. The output for visualization is saved as a .PSE PyMOL session file. The visual output shows the surfaces of the relevant units of interest colored according to the net value of interactions and the specific pairwise interactions to the net total are represented as colored dash lines with a customizable color scale. In all cases, the input .PDB structure needs to be cleaned of any non-defined atoms in a pre-processing step, which can be done using scripts that are provided with the Surfaces software. The pre-processing is not done automatically to offer the use the possibility to customize. For example, if the user wishes to ignore specific hetero-atoms, these need to be removed but otherwise, defined. Surfaces offers a general script for atom type assignment mapping element types to the default SYBYL 40 atom types.
Protein–protein interfaces: For this application, two groups of protein chains are required as input, Surfaces calculates the interactions between residues of the first group and residues of the second group (Fig. 1B). This can also be used for protein–ligand evaluations by assigning a chain ID for the ligand’s atoms, if the user intends to evaluate the complete interaction of the ligand with each residue without a per-atom decomposition (see next paragraph).
Protein–ligand interactions: This application requires the PDB assigned three-letter code of the ligand of interest as input and calculates the interaction between residues of the structure and each atom of every instance of the ligand within the structure (Fig. 1C). Before running the application, two pre-processing steps are required: first, the assignment of atom types to the atoms of the ligand into the .DEF file, and second, cleaning the input .PDB structure of any non-defined atoms.
Particular residue interactions: This application requires a list of residues of interest as input and calculates all interactions (inter- or intra-chain) involving those residues (Fig. 1D).
3.2 Validation of surfaces
Common methods for analyzing protein-protein interactions and the per-residue energetic decomposition of the contributions of such interactions to the overall free energy are based on MD simulations (Kollman et al. 2000, Homeyer and Gohlke 2012, Serçinoglu and Ozbek 2018). MD simulations can provide detailed information on the structural changes and energetics associated with protein-protein interactions, including the binding free energy and per-residue energetic contributions. However, MD simulations are computationally intensive (Bopp et al. 2008, Ciccotti et al. 2022) making them less suitable for large-scale studies including protein engineering. Furthermore, MD simulations require expert knowledge to set up, which creates an additional obstacle for broad utilization.
The examples in Fig. 1 to illustrate the utilization of Surfaces to the three applications of Surfaces were taken from different datasets. The protein-protein interface analyzed in Fig. 1A was taken from a dataset of 738 structures of one or more chains of the SARS-CoV-2 Spike protein in complex with one or more chains of antibodies (Gowthaman et al. 2020). Such analysis required an average of 11.38 ± 9.62 CPU-seconds per structure including the time required for the preprocessing step. The analysis of protein-ligand interactions is derived from a dataset of 709 non-redundant structures and 669 non-redundant ligands, totaling 831 experimentally solved protein-ligand complexes of SARS-CoV-2 proteins and small molecules (Harrison et al. 2021). This analysis required an average of 3.97 ± 2.32 CPU-seconds per structure (including pre-processing).
To demonstrate the reliability of Surfaces, we performed two validations. The first one was performed using the AB-Bind dataset (Sirin et al. 2016), comprising 1101 mutations in 32 different protein complexes and associated experimentally determined ΔΔG changes. This dataset was published along with an extensive evaluation of the performance of seven different binding affinity prediction methods, performed using as input mutant structures generated with three different modeling methodologies (Supplementary Fig. S1A). By modeling the structures with two of these methods, FoldX (Buß et al. 2018) and Rosetta (Kortemme et al. 2003, Leaver-Fay et al. 2011), we can decouple the modeling quality from the prediction ability. Irrespective of the modelling method, Surfaces performance is similar to that of the best performing methods (Supplementary Fig. S1). Specifically, using FoldX mutants, Surfaces’ results showed a Pearson’s R of 0.32 and AUC |ΔΔG| > 0 of 0.65, compared to respective values of 0.34 and 0.70 obtained through FoldX for binding evaluation. For structures modelled with Rosetta, Surfaces predictions exhibited a Pearson’s R of 0.14, marginally lower than the correlation of 0.16 observed during interface assessment with Rosetta (Supplementary Fig. S1). Nonetheless, Surfaces shows slightly superior performance in ROC curve analysis compared to Rosetta, with an AUC for |ΔΔG| > 0 value of 0.61 compared to Rosetta’s 0.60 (Supplementary Fig. S1) (Detailed data available in the supplementary material). Other methods with equivalent or inferior performance to Surfaces are bASA (Hubbard and Thornton 1993, Sirin et al. 2016), dDFIRE (Yang and Zhou 2008), DFIRE (Zhou and Zhou 2002), and STATIUM (DeBartolo et al. 2012, 2014). Discovery Studio (Spassov and Yan 2013) shows slightly superior performance with Pearson’s R of 0.45 but in this as a paid software, we cannot test it to determine if the additional performance is due to a more accurate modelling of mutants or more accurate calculation of ΔΔG (Supplementary Fig. S1).
A second validation involved the comparison of binding affinity prediction performance for a highly curated dataset of 23 SARS-CoV-2 Spike Receptor Binding Domain (RBD)/ACE2 binding ΔΔG values measured by Surface Plasmon Resonance (Sergeeva et al. 2023). Surfaces shows a Pearson’s correlation of 0.556 with the experimental data, comparable to the performance of FEP + 100 ns MD, which obtains a correlation of 0.598, and considerably higher than all other methods tested, including state-of-the-art techniques such as MM/PBSA as MM/GBSA (Table S1 and Supplementary Fig. S2). Surfaces and FEP + 100 ns also obtained very similar ΔΔG root mean square errors (RMSE) and Pearson’s phi, showing an equivalent ability to classify mutations as stabilizing or destabilizing (detailed data available in the Supplementary Material).
Lastly, gRINN (Serçinoglu and Ozbek 2018) is a widely used method for residue-based energy decomposition that uses MD trajectories to calculate binding energy means and distributions. We used gRINN to calculate interface interactions of the Delta SARS-CoV-2 Spike in complex with the receptor ACE2 (Cheng et al. 2022) with MD trajectories available at the COVID-19 Molecular Structure and Therapeutics Hub (covid.molssi.org). The Pearson’s R obtained comparing Surfaces and gRINN is 0.64 (Fig. 1E and Supplementary Fig. S3). As additional validation of Surfaces (and comparison with gRINN) with experimental validation of per-residue contributions to binding, the two software were used to guide the selection of mutations to disrupt the binding interface between the Ebola protein VP35 and Ubiquitin (Rodríguez-Salazar et al. 2023). The two software highlighted a particular residue among the top contributing residues to the protein–protein interaction interface. Mutating this residue disrupted the interaction (Rodríguez-Salazar et al. 2023). The analysis of binding energy decomposition can help analyze interactions between proteins and ligands, potentially as a guide in rational drug design drug design (Supplementary Fig. S4).
In order to consider structural variations, captured when using MD-based approaches, Surfaces can be used to analyze protein ensembles, generated with tools such as the NRGTEN package (Mailhot and Najmanovich 2021). Such a procedure would permit to detect transient interactions utilizing the ensemble as a sample of the partition function near equilibrium by associating probabilities for each interaction.
4 Conclusions
Surfaces provides a simple, fast, and easy to use method to analyze and visualize biomolecular interactions. Surfaces performs equivalently or better than widely used methods such as Rosetta, FoldX, MM/GBSA and MM/PBSA, and, for specific datasets, comparable even to computationally expensive and cumbersome to implement methods such as gRINN and FEP. The use of variations in solvent accessible surface areas or those in contact to estimate free-energy contributions has been established previously (with LPC/CSU, STC) but somehow fell out of use over the past 20 years within the broader community. However, this work shows that such calculation can provide data of the same quality as more computationally expensive methods based on molecular dynamics, currently available methods based on machine learning, or other mean-field based statistical approaches.
As far as small-molecule protein interactions are concerned, docking simulations can be used to predict the binding pose, the relative ranking of molecules in virtual screening and the actual binding affinity. Until recently, no docking scoring function was able to tackle all three applications at once (Huang et al. 2010) and we believe this is still the case. The CF function parameters used here in the validations were optimized for the accurate detection of docking poses and tested for the relative ranking of ligands in virtual screening (Gaudreault and Najmanovich 2015). The CF function was not tested for the prediction of small-molecule binding affinities. As such, we do not believe Surfaces results correlate highly with binding affinities for small-molecules but this remains to be tested. Therefore, we recommend that Surfaces be used only to detect frustrated interactions between ligand and protein atom to guide rational drug design.
The scriptable and customizable nature of Surfaces makes it a valuable tool for researchers seeking to analyze large structural datasets, such as in virtual screening or protein engineering, thus permitting a broader exploration of search space. Lastly, the ease of use of Surfaces and low demand on computational resources makes this type of analysis accessible to a broader audience.
Supplementary Material
Acknowledgements
We would like to thank Nicolas DesCôteaux for the thorough testing of the software and identification of initial bugs. This work is dedicated to Vladimir Sobolev Z”L (1947-2023), creator of LIGIN and LPC/CSU and a pioneer in the utilization of surface areas in contact as a fundamental component of the analysis of biomolecular interactions.
Contributor Information
Natália Teruel, Department of Pharmacology and Physiology, Faculty of Medicine, Université de Montréal, Montreal H3T 1J4, Canada.
Vinicius Magalhães Borges, Department of Biomedical Sciences, Joan C. Edwards School of Medicine, Marshall University, Huntington, WV, USA.
Rafael Najmanovich, Department of Pharmacology and Physiology, Faculty of Medicine, Université de Montréal, Montreal H3T 1J4, Canada.
Supplementary data
Supplementary data are available at Bioinformatics online.
Conflict of interest
None declared.
Funding
This work was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery program grants; Genome Canada and Genome Quebec; Compute Canada.
Data availability
All data is available in the supplementary data file and within the documentation site.
References
- Bojar D, Fussenegger M. The role of protein engineering in biomedical applications of mammalian synthetic biology. Small 2020;16:e1903093. [DOI] [PubMed] [Google Scholar]
- Bopp PA, Buhn JB, Maier HA et al. Scope and limits of molecular simulations. Chem Eng Commun 2008;195:1437–56. [Google Scholar]
- Buß O, Rudat J, Ochsenreither K et al. FoldX as protein engineering tool: better than random based approaches? Comput Struct Biotechnol J 2018;16:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MH, Krieger JM, Banerjee A et al. Impact of new variants on SARS-CoV-2 infectivity and neutralization: a molecular assessment of the alterations in the spike-host protein interactions. Iscience 2022;25:103939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chica RA. Protein engineering in the 21st century. Protein Sci 2015;24:431–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccotti G, Dellago C, Ferrario M et al. Molecular simulations: past, present, and future (a topical issue in EPJB). Eur Phys J B 2022;95:3. [Google Scholar]
- DeBartolo J, Taipale M, Keating AE et al. Genome-wide prediction and validation of peptides that bind human prosurvival Bcl-2 proteins. PLoS Comput Biol 2014;10:e1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBartolo J, Dutta S, Reich L et al. Predictive Bcl-2 family binding models rooted in experiment or structure. J Mol Biol 2012;422:124–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier V, Chartier M, Najmanovich R. Applications of normal mode analysis methods in computational protein design. Methods Mol Biol 2017;1529:203–14. [DOI] [PubMed] [Google Scholar]
- Gaudreault F, Najmanovich RJ. FlexAID: revisiting docking on non-native-complex structures. J Chem Inf Model 2015;55:1323–36. [DOI] [PubMed] [Google Scholar]
- Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov 2015;10:449–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowthaman R, Guest JD, Yin R et al. CoV3D: a database of high resolution coronavirus protein structures. Nucleic Acids Res 2020;49:gkaa731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PW, Lopez R, Rahman N. et al. The COVID-19 data portal: accelerating SARS-CoV-2 and COVID-19 research through rapid open access data sharing. Nucleic Acids Res 2021;49:gkab417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homeyer N, Gohlke H. Free energy calculations by the molecular mechanics poisson−boltzmann surface area method. Mol Inform 2012;31:114–22. [DOI] [PubMed] [Google Scholar]
- Huang S-Y, Grinter SZ, Zou X et al. Scoring functions and their evaluation methods for protein-ligand docking: recent advances and future directions. Phys Chem Phys 2010;12:12899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard S, Thornton J. Naccess Dept. of Biochemistry and Molecular Biology. London: University College, 1993. [Google Scholar]
- Kollman PA, Massova I, Reyes C et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 2000;33:889–97. [DOI] [PubMed] [Google Scholar]
- Kortemme T, Morozov AV, Baker D et al. An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein-protein complexes. J Mol Biol 2003;326:1239–59. [DOI] [PubMed] [Google Scholar]
- Lavigne P, Bagu JR, Boyko R et al. Structure-based thermodynamic analysis of the dissociation of protein phosphatase-1 catalytic subunit and microcystin-LR docked complexes. Protein Sci 2000;9:252–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol 2011;487:545–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda R, Fujita J, Konishi Y et al. A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including omicron. Commun Biol 2022;5:669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailhot O, Najmanovich R. The NRGTEN python package: an extensible toolkit for coarse-grained normal mode analysis of proteins, nucleic acids, small molecules and their complexes. Bioinformatics 2021;37:3369–71. [DOI] [PubMed] [Google Scholar]
- McCarrick M, Kollman P. Predicting relative binding affinities of non-peptide HIV protease inhibitors with free energy perturbation calculations. J Comput Aided Mol Des 1999;13:109–21. [DOI] [PubMed] [Google Scholar]
- McConkey BJ, Sobolev V, Edelman M et al. Quantification of protein surfaces, volumes and atom-atom contacts using a constrained voronoi procedure. Bioinformatics 2002;18:1365–73. [DOI] [PubMed] [Google Scholar]
- Napolitano V, Dabrowska A, Schorpp K et al. Acriflavine, a clinically approved drug, inhibits SARS-CoV-2 and other betacoronaviruses. Cell Chem Biol 2022;29:774–84.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olechnovič K, Venclovas Č. VoroContacts: a tool for the analysis of interatomic contacts in macromolecular structures. Bioinformatics 2021;37:4873–5. [DOI] [PubMed] [Google Scholar]
- Ribeiro J, Ríos-Vera C, Melo F et al. Calculation of accurate interatomic contact surface areas for the quantitative analysis of non-bonded molecular interactions. Bioinformatics 2019;35:3499–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Salazar CA, van Tol S, Mailhot O et al. Ebola virus VP35 interacts non-covalently with ubiquitin chains to promote viral replication creating new therapeutic opportunities. bioRxiv 2023; 2023.07.14.549057, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz J, Borg J, Stricher F et al. The FoldX web server: an online force field. Nucleic Acids Res 2005;33:W382–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serçinoglu O, Ozbek P. gRINN: a tool for calculation of residue interaction energies and protein energy network analysis of molecular dynamics simulations. Nucleic Acids Res 2018;46:W554–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva AP, Katsamba PS, Liao J et al. Free energy perturbation calculations of mutation effects on SARS-CoV-2 RBD::ACE2 binding affinity. J Mol Biol 2023;435:168187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirin S, Apgar JR, Bennett EM et al. AB‐bind: antibody binding mutational database for computational affinity predictions. Protein Sci 2016;25:393–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev V, Sorokine A, Prilusky J et al. Automated analysis of interatomic contacts in proteins. Bioinformatics 1999;15:327–32. [DOI] [PubMed] [Google Scholar]
- Spassov VZ, Yan L. pH‐selective mutagenesis of protein–protein interfaces: in silico design of therapeutic antibodies with prolonged half‐life. Proteins Struct Funct Bioinform 2013;81:704–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin PH, Richards DH, Callender RA et al. Protein engineering: a new frontier for biological therapeutics. Curr Drug Metab 2015;15:743–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Fang X, Lu Y et al. The PDBbind database: methodologies and updates. J Med Chem 2005;48:4111–9. [DOI] [PubMed] [Google Scholar]
- Yang T-J, Yu P-Y, Chang Y-C et al. D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation. J Biol Chem 2021;297:101238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhou Y. Specific interactions for ab initio folding of protein terminal regions with secondary structures. Proteins Struct Funct Bioinform 2008;72:793–803. [DOI] [PubMed] [Google Scholar]
- Zhou H, Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci 2002;11:2714–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Bourguet FA, Bennett WFD et al. Large-scale application of free energy perturbation calculations for antibody design. Sci Rep 2022;12:12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available in the supplementary data file and within the documentation site.