Abstract
To develop reproducible results, it is critical that all reagents used in an experiment be validated in an alternative or independent method. We present two such independent methods for determining the specificity of antibodies: (1) “MILKSHAKE,” which can be used to validate the liability and specificity of antibodies directed against post-translationally-modified epitopes, and (2) “Sundae,” which is a more complete alanine-like scanning method that can be used to better understand the binding and bioactivity of specific residues of a protein. We apply both of these methods to the interaction between an antibody and its antigen.
Keywords: Alanine scanning, Antibody specificity, Antibody validation, ELISA, Western
1. Introduction
A current cause for concern in the scientific literature is the poor reproducibility of published experimental results because of a lack of rigor in reagent validation and inter-experimental consistency. This is especially true for antibodies (Abs) used in experiments and a topic of several excellent reviews and conferences [1-13]. As a result of these concerns, many detailed recommendations for Ab validation before beginning experimental work have been proposed [3-6, 13].
Even beyond affinity reagent-antigen binding, the reproducibility issue drastically increases in complexity when validating Abs against post-translationally modified (PTM) targets. Often Abs are sold as being post-translational modification specific, however, when tested under the recommended protocol appear to have some affinity to the non-modified state of the epitope (Fig. 1). Thus a real difficulty in validating any anti-PTM Abs is access to a validated antigen or cell lysate. It is especially difficult to generate cell lysates and purified antigen wherein the modified amino acid in the protein is present at either 100% (fully modified) or 0% (fully non-modified) in the sample [14]. Even when a particularly specific and targeted treatment is known for a particular PTM, other variables such as batch to batch variation, cell health, modifying-enzyme activity, storage conditions, and researcher technique can still persist to allow for errors in an appropriate validation.
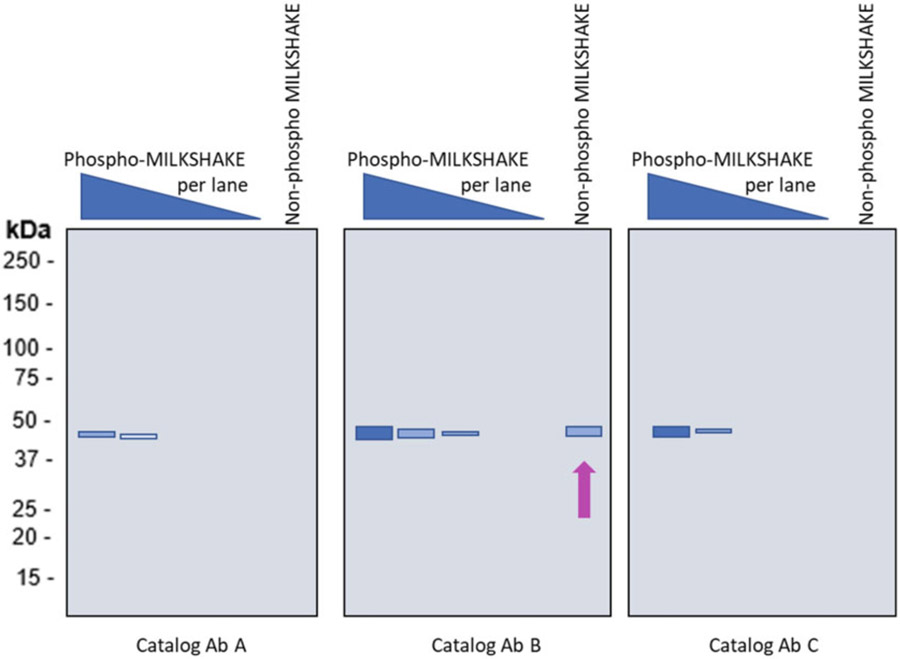
Fig. 1.
Commercially available phospho-specific Abs show varying degrees of specificity to phospho epitopes in MILKSHAKE. Simulated MILKSHAKE Western blots tested with catalog Abs from three different vendors. Abs A and C show binding specific to the phosphorylated epitope. Catalog Ab B, however, binds to the phosphorylated as well as the non-phosphorylated MILKSHAKE residue (arrow). For data and additional Ab validation examples see Ref. 14
Alanine scanning is a technique that is often used to determine the contribution of a specific amino acid residue to the stability or function of a given protein [15, 16]. Alanine is used because it is non-bulky and chemically inert. The alanine methyl group can be used to mimic the secondary structure preferences that the other amino acids possess. Sometimes bulky amino acids such as the branched-chain amino acids valine and leucine are used where the conservation of the size of mutated residues is needed.
Alanine scanning is further used to determine whether a specific amino acid residue plays a significant role in bioactivity by replacing the naturally occurring amino acid with alanine. This can often be performed without the requirement for highly purified protein and often uses the method of site-directed mutagenesis to change the residues in question to alanine [17, 18]. Once the change is made, the protein can be retested for bioactivity with the alanine replaced. Often these tests can yield a quantitative measurement. However, one limitation of alanine scanning is that it only reveals whether alanine as a replacement at a specific site retains bioactivity. It reveals nothing about the effect of the other 18 amino acids (i.e., the set of 20 amino acids minus the native and alanine residues). So a true picture of the importance of a specific residue for interaction or bioactivity cannot be painted if only alanine is used for scanning (Fig. 2).
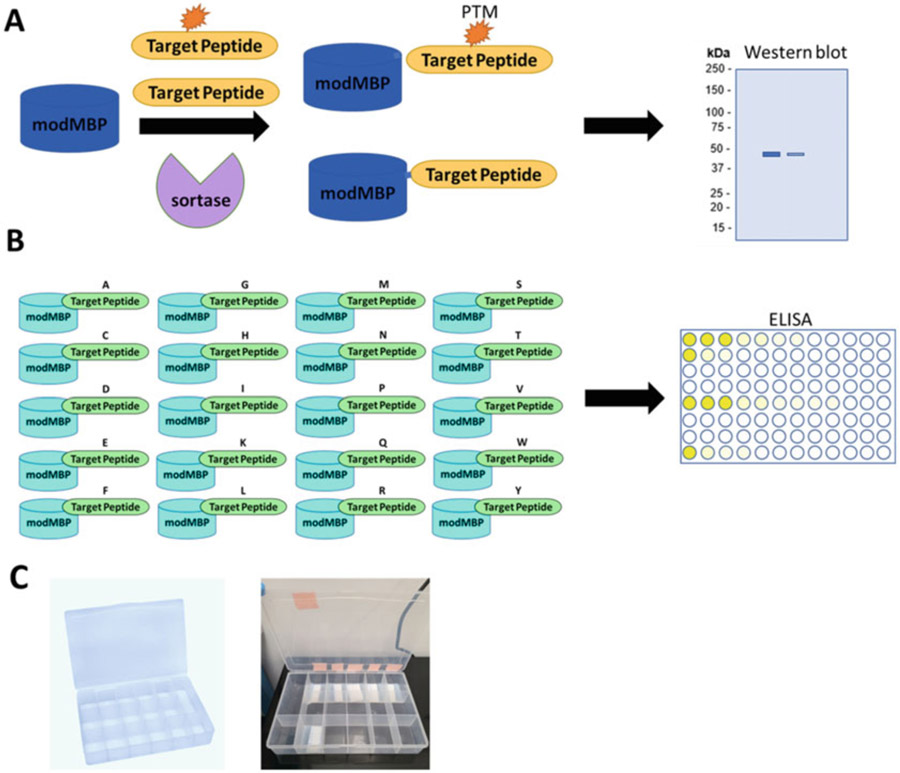
Fig. 2.
MILKSHAKE and Sundae workflow (a) MILKSHAKE proteins used in Western blot result from a conjugation reaction between a target peptide (with or without a PTM) and a modified MBP using the sortase enzyme. These proteins can then be tested to determine the specificity of PTM Abs. (b) Sundae proteins contain genetically encoded target sequences in which one residue has been replaced by all 20 amino acids. These proteins can be used in ELISA to determine binding affinity of Abs when this single residue is replaced, (c) Trays containing multiple small compartments (sold as bead organizers at hobby stores) facilitate testing a variety of different primary Abs in MILKSHAKE Western blot
To summarize, the most reliable method for validation of anti-PTM-specific Abs is one in which each antigen sample contains the target sequence with either 100% modification or 100% non-modification at the residue or epitope of interest. The MILKSHAKE protocol provides that certainty for a Western blot assay. This protocol uses a modified maltose binding protein (MBP) and the sortase enzyme to produce surrogate proteins containing the modified or non-modified epitope sequence (Fig. 3). These MILKSHAKE proteins can then be used in Western blot to evaluate the PTM specificity of catalog Abs (Fig. 4).
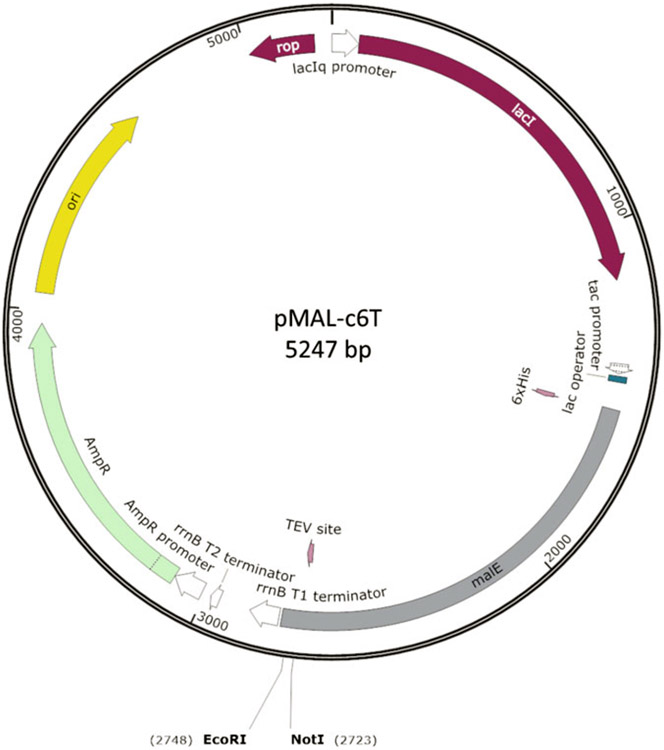
Fig. 3.
Plasmid map of pMAL-c6T. This vector [New England Biolabs] was modified to construct both the sortase acceptor plasmid for MILKSHAKE and the Sundae proteins
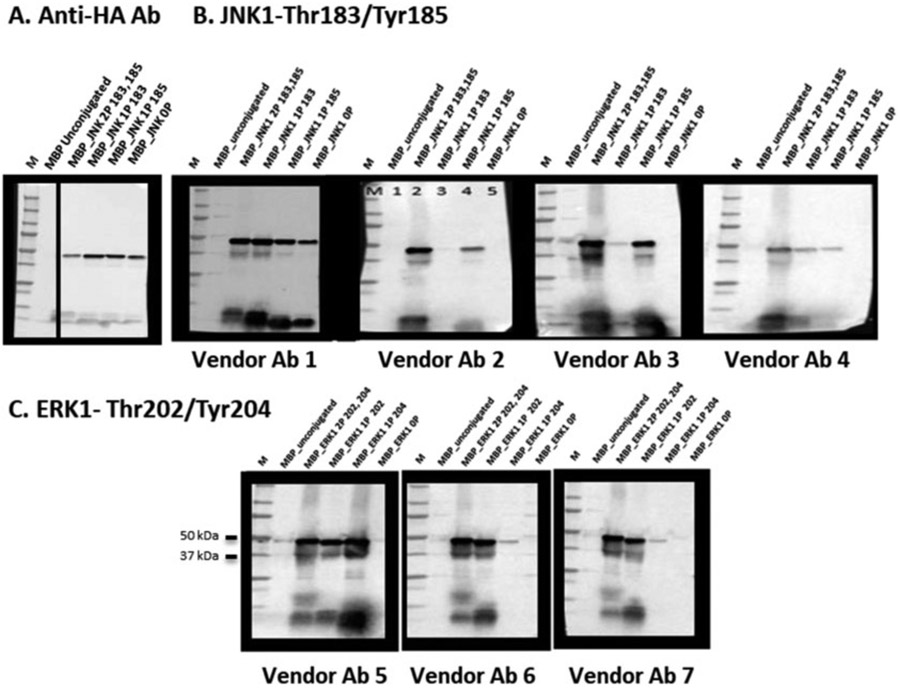
Fig. 4.
MILKSHAKE Validation of Vendor catalog Abs. (a) Blot loaded with unconjugated MILKSHAKE and four versions of MILKSHAKE protein containing the target sequence for JNK1 with or without phosphorylation at Thr183 and Tyr185. Blot probed with anti-HA Ab. Each lane containing conjugated MILKSHAKE protein shows reactivity with anti-HA and the unconjugated sample shows no reactivity, (b) Blot loaded with unconjugated MILKSHAKE and four versions of MILKSHAKE protein containing the target sequence for JNK1 with or without phosphorylation at Thr183 and Tyr185. Blots probed with vendor Abs labeled for sale as specific binders to the JNK1 protein, only when phosphorylated at both Thr183 and Tyr185. Vendor Ab 1 binds non-specifically to phospho as well as the non-phospho_JNK MILKSHAKE proteins. Vendor Abs 2 and 3 recognize MILKSHAKE when both sites are phosphorylated and phospho_Tyr185 but are not able to bind when Thr183 alone is phosphorylated. Vendor Ab 4 is able to bind all three phosphorylated versions of MILKSHAKE, (c) Loading as in (b) for Abs against ERK1 phospho-positions 202 (phospho-threonine) and 204 (phospho-tyrosine). Vendor Ab 5 is able to bind when either residue is phosphorylated. Vendor Abs 6 and 7 are able to bind only when position 202 is phosphorylated. None of the three vendor Abs bind unphosphorylated residues. In all blots, M = size marker
To determine more completely the binding contribution of a specific amino acid at a specific site, we have developed a protocol we term “Sundae” (because we are using multiple ‘flavors’ of amino acids at the site of interest, not just alanine). Using Sundae, we replace the targeted site with up to 20 different amino acids (Fig. 5) and retest for bioactivity. The Sundae method is able to distinguish varying reactivity to each amino acid in ELISA (Fig. 6).
Fig. 5.
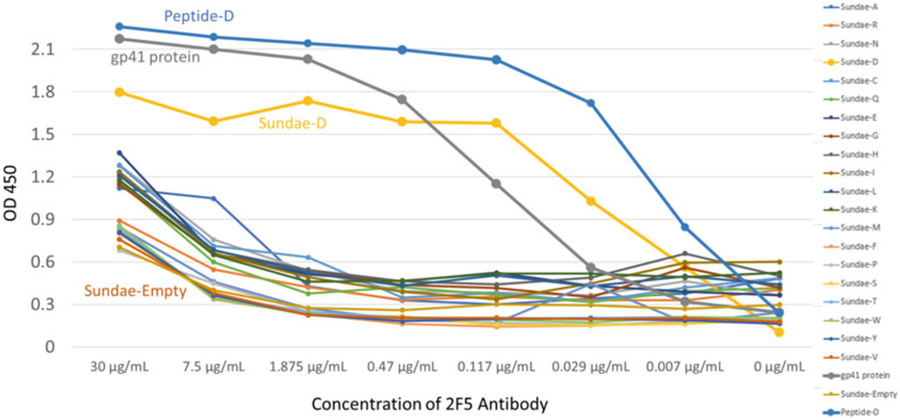
Sundae proteins shed light on which residues are important for Ab binding. 2F5 is a broadly neutralizing human monoclonal Ab known to bind the sequence 657-EQELLELdKWASLW-670 found in the human immunodeficiency virus (HIV). Sundae proteins were constructed with an insert of this HIV sequence in which the underlined residue (EQELLELxKWASLW) was replaced with each of the naturally occurring amino acids (Sundae-A for alanine etc.). These Sundae proteins were then tested in ELISA for binding to 2F5. ELISA plates were coated with 1 μg mL−1 of Sundae antigens and primary Ab 2F5 [Polymun Scientific] was tested using four-fold dilutions in 2% BSA. The 2F5 Ab binds strongly to a full-length gp41 HIV protein [Abcam], a native HIV sequence peptide as well as the Sundae-D protein containing the sequence EQELLELdKWASLW. Binding is reduced when tested against the Sundae proteins containing each of the other 19 amino acids as well the negative control, Sundae-Empty (MBP with no HIV sequence present). These data suggest that the D at position 664 is critical for 2F5 binding
Fig. 6.
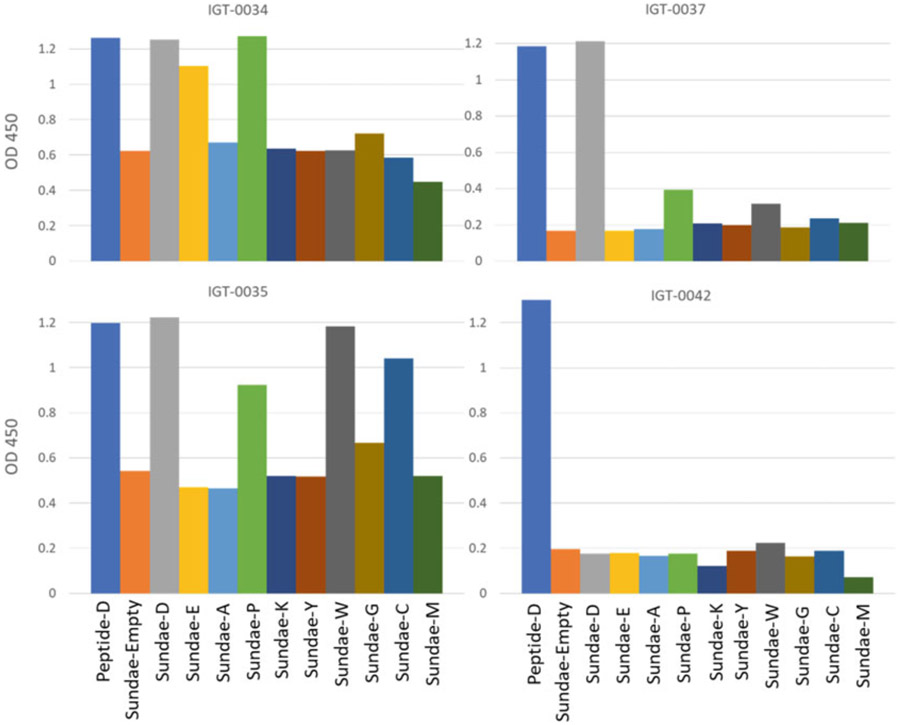
Ab reactivity varies depending on which residue is present in Sundae proteins. Different rabbit IgG clones (measured at 5 μg mL−1) show different reactivity when a single residue in the Ab’s epitope (an aspartic acid in the native sequence) is replaced with other amino acids (Sundae-A, Sundae-E, etc.). IGT-0037 is able to bind the original amino acid, aspartic acid as well as a peptide version containing aspartic acid (D). However, IGT-0034 is able to bind both those antigens as well as Sundae proteins with glutamic acid and proline substitutions (Sundae-E and Sundae-P). IGT-0035 recognizes tryptophan and cysteine (Sundae-W and Sundae-C) as well at that same position in the epitope sequence but not glutamic acid. IGT-0042 is only able to recognize the peptide version of the antigen sequence and not any of the Sundae proteins. None of these Abs were able to bind alanine at this position. Unlike alanine scanning, the Sundae method can fully interrogate the contribution of a single residue to Ab binding
2. Materials and Reagents
Materials
Prepare all solutions using ultrapure water (prepared by purifying deionized water to attain a sensitivity of 18 MΩ-cm at 25 °C). Prepare and store all reagents at room temperature unless indicated otherwise. Follow all waste disposal regulations when disposing of waste materials.
Media and Solutions
Rich Broth: deionized water, to 950 mL, Tryptone, 10 g, NaCl, 5 g, Yeast extract, 5 g. Combine the above reagents and mix until the solutes have dissolved. Adjust the volume of the solution to 1 L with deionized water and autoclave. Before use, add glucose, 2 g.
Column buffer: 20 mM Tris–HCL, pH 7.4, 200 mM NaCl, 1 mM EDTA, 1 mM DTT.
Elution buffer: column buffer with maltose to 10 mM (final).
10× Tris-NaCl buffered saline (TBS): 1.5 M Sodium Chloride, and 0.25 M Tris, pH 7.2-7.5.
1× TBST: 1 × TBS with final concentration of 0.1% Tween 20.
3% MTBST: 1 × TBST with 3% nonfat dry milk powder.
5% BSA-TBST: 1× TBST with 5% bovine serum albumin.
10× Sortase Buffer: 200 mM Tris–HCl, 1.5 M NaCl, 50 mM CaCl2, 2 mM beta-mercaptoethanol prepared in sterile water.
2× Laemmeli + DTT: 2× Laemmli sample buffer [BioRad], 200 mM DTT
1 × PBST: 1 × PBS with final concentration of 0.1% Tween 20.
3. Methods
Carry out all procedures at room temperature unless otherwise specified.
3.1. Construction of Sortase-Acceptor Plasmid
To begin construction of a sortase-acceptor plasmid (pAT27 or equivalent), the commercially available plasmid pMAL-c6t can be purchased from New England BioLabs (NEB catalog #: N0378S, see Note 1). A sortase conjugation site (LPETG, encoded in the plasmid DNA as: TTACCGGAAACTGGT) with a preceding Gly4Ser peptide linker (GGGGS encoded in the DNA as: GGTGGCGGTGGCTCG) are cloned into the parent vector in-frame with the MBP into the multiple cloning site using EcoRI and NotI. The resulting plasmid pAT27 expresses a modified MBP (MILKSHAKE) containing the sortase conjugation site in its C-terminus (Fig. 3). The recombinant vector can be transformed into NEB Express cells and frozen in 15% glycerol stock solution and stored at −80 °C. Purified plasmid DNA can be stored at −20 °C in Tris-EDTA solution.
3.2. Preparation of MILKSHAKE Protein Inoculant
Inoculate 5 mL of LB media supplemented with ampicillin (final concentration of 100 μg mL−1) in a 14 mL snapcap culture tube with a scraping of a glycerol stock of the sortase acceptor plasmid in NEB Express cells. (see Note 2).
Incubate tube with shaking at 37 °C overnight.
3.3. Expression of MILKSHAKE Protein
Inoculate 75 mL of Rich Broth media supplemented with ampicillin (final concentration of 100 μg mL−1) in a 250 mL baffled glass flask with 750 μL of the overnight culture.
Incubate 250 mL flask shaking at 37 °C until an OD600 = 0.5 is reached (approximately 1–3 h). Determine OD600 using a spectrophotometer.
Add 15 μL of 1 MIPTG stock to each flask (final concentration 0.3 mM).
Incubate flasks with shaking at 37 °C for an additional 2 h.
Pour culture from one 250 mL flask into one 250 mL centrifuge tube. (see Note 3).
Centrifuge tubes in a high-speed floor centrifuge at 5000 × g for 20 min at 4 °C.
Discard supernatant in biohazardous liquid waste container.
Freeze pellet overnight at −80 °C.
3.4. Purification of MILKSHAKE Protein
Thaw pellets at room temperature on a benchtop. Add 2 mL of B-PER II Reagent [Thermo] or up to 4 mL per gram of pellet. Incubate for 15 min at room temperature on a nutator or equivalent piece of equipment.
Transfer sample to a 45 mL centrifuge tube and place on ice.
Centrifuge sample at 15,000 × g for 15 min at 4 °C in a high-speed floor centrifuge.
While the sample is spinning, prepare one 2 mL purification column in a cold room or deli case refrigerator for each sample. Snap off the bottom of the column and set the column up in a rack above a waste container to catch the flow through.
Add 2 mL of amylose resin [New England BioLabs] to each column and allow to drain via gravity. Be sure to shake resin stock bottle to resuspend resin before pipetting into column.
Wash each column once with 2 mL of ultra-pure water followed by 2 mL of column buffer.
Load the clarified sample onto the washed column by pouring, being careful not to disturb the cell pellet. Discard pellet. (see Note 4).
Allow sample to flow by gravity. Collect the flow through in a clean tube and pour back over column a second time without collecting the flow through.
Wash each column with 12 mL column buffer.
Place a new sterile tube under the column.
Add 1 mL of elution buffer 3 times. Make sure to allow buffer to flow through completely before adding the next 1 mL. Collect each elution fraction in a separate tube.
Place eluted protein on ice. Add glycerol to a final concentration of 15% (333 μL of 45% glycerol stock per 1 mL eluate).
Quantitate the amount of protein in the sample using a Biotek plate reader or equivalent. Use elution buffer with 15% glycerol as your blank. (see Note 5).
Label each tube with the concentration of MILKSHAKE protein in mg mL−1.
Store MILKSHAKE protein at 4 °C for short term. For long-term storage, flash freeze, the sample in liquid nitrogen and store at −80 °C. Be sure to wear appropriate personal protective equipment when working with liquid nitrogen.
3.5. Modified Maltose Binding Protein Conjugation to Peptide
3.5.1. Conjugation Controls
Positive control: Use anti-HA as a secondary Ab [Abcam] in a separate blot to confirm conjugation reaction was successful.
Negative control: Load unconjugated MILKSHAKE protein in one lane per blot to confirm primary Ab binding is specific to conjugated peptide and not to the MILKSHAKE protein itself (see Note 6).
3.5.2. Peptide Design
Design and order two peptides per target being tested: (i) one peptide should be the modified peptide containing the N-terminal sortase compatible site and a C-terminal HA tag and (ii) the other peptide will be the non-modified version containing both an N-terminal sortase compatible site and a C-terminal HA tag (see Note 7).
3.5.3. Modified Maltose Binding Protein Conjugation to Peptides
Reactions will be arranged in sets of three in a 96-well deep-well plate: (i) unconjugated MILKSHAKE protein negative control, (ii) MILKSHAKE protein conjugated to the modified peptide, and (iii) MILKSHAKE protein conjugated to the non-modified peptide.
For wells containing unconjugated MILKSHAKE protein, mix the following per well: 10× Sortase buffer (11 μL), MILKSHAKE protein (0.145 mg), 1× PBS to a final volume of 113 μL.
For wells containing “MILKSHAKE protein conjugated to the modified peptide” mix the following per well: modified peptide, 4 μL of a 10 mg mL−1 stock (in DMSO), 10× Sortase Buffer (11 μL), MILKSHAKE protein (0.145 mg), Sortase Enzyme (1.5 μL) [Moradec LLC] and 1× PBS to a final well volume of 113 μL.
For wells containing “MILKSHAKE protein conjugated to the non-modified peptide” mix the following per well: non-modified peptide, 4 μL of a 10 mg mL−1 stock (in DMSO), 10× Sortase Buffer (11 μL), MILKSHAKE protein (0.145 mg), Sortase Enzyme (1.5 μL) and 1× PBS to a final well volume of 113 μL (see Note 8).
Seal the reactions with plate tape and incubate with shaking at 250 RPM at 37 °C for 2 h.
After incubation, centrifuge plate for 5 min in tabletop centrifuge at 2100× g to make sure reaction volume is at the bottom of the well.
Add 113 μL of 2× Laemmli + DTT buffer to each well. Transfer each mixture to separate and labeled 1.7 × g Eppendorf tubes.
Incubate tubes containing conjugation reactions at 95 °C for 10 min. Reaction is now ready to load in a gel for Western blot (see Note 9).
3.6. Ab Validation Using Western Blot
3.6.1. Protein Electrophoresis and Membrane Transfer
Use the lanes described below for each Ab to be tested. Load up to 15 μL (or 5 μg) of sample (unconjugated MILKSHAKE protein or conjugated MILKSHAKE protein) per lane. Load 5 μl of a protein standard [Precision Plus from BioRad or equivalent] per lane (see Note 10): (Lane 1) Protein standard, (Lane 2) unconjugated MILKSHAKE protein, (Lane 3) modified peptide conjugated to MILKSHAKE protein, and (Lane 4) non-modified peptide conjugated to MILKSHAKE protein.
Run the gel(s) until the dye front reaches the bottom of the gel. If using a Criterion apparatus [BioRad], we typically run the gel (s) at 180 V for 40 min.
Transfer the proteins in the gel to a nitrocellulose membrane according to the Western blot supplier’s protocol (BioRad Protocol 10019593).
When transfer is complete, cut the membranes between the molecular weight standard lanes to generate separate blots.
Place membranes in separate compartments in a tray and wash with 1× TBS for 5 min on a platform shaker. We use a compartmentalized plastic tray such as one purchased from a hobby shop (Fig. 2).
Discard the 1× TBS and add 3% MTBST (enough volume to cover the membrane) to each blot. Incubate at room temperature for 1 h on a platform shaker with gentle shaking to block the membranes.
Discard blocking solution and wash membranes with 1× TBST 3 times for 5 min each on a platform shaker at room temperature.
During washes, dilute Ab(s) to be tested in 5% BSA-TBST at a concentration of 1 μg mL−1 (or according to vendor recommendations). Remove final 1× TBST wash from blots and add primary Ab to the corresponding blot(s). For secondary only or anti-HA blots, add 5% BSA-TBST only.
Incubate blots overnight at 4 °C on a platform shaker with gentle shaking.
3.6.2. Developing and Exposing the Membrane
Discard primary Abs and wash membranes with 1× TBST 3 times for 5 min each on a platform shaker at room temperature.
Discard TBST and add secondary Ab diluted in 3% MTBST. For starting conditions, try a 1:5000 dilution. Incubate at room temperature for 1 h on a platform shaker with gentle shaking.
Remove secondary Ab and wash membranes with 1× TBST 3 times for 5 min each at room temperature on a platform shaker with gentle shaking.
Develop the membranes with ECL Reagent [BioRad] or equivalent for at least 10 s. Mix together equal volumes of the Clarity Western ECL Substrates [BioRad] and then add a small volume of the mixture to each membrane (just enough to cover the surface of the membrane). It is important to note not to mix the substrates together until immediately before use.
Image the membranes using an appropriate available imaging system.
3.6.3. Interpretation of Data
- Expected band size of MILKSHAKE protein is 42 kDa (Fig. 4).
- In the anti-HA blot, verify the presence of the HA tag on all of the conjugation reactions but NOT on the unconjugated reactions.
- Verify PTM-specific binding for your positive control Abs (if available). Most general Abs will not bind the peptide. Check the vendor’s catalog page to determine whether you should expect binding to your conjugated MILKSHAKE protein.
- As an example, a phospho-specific Ab should only bind the phospho-conjugated MILKSHAKE protein. Phospho-independent Abs should bind to both phospho- and to the non-phospho-conjugated MILKSHAKE proteins. Non-phospho specific Abs should bind only to the nonphospho-conjugated MILKSHAKE proteins. No Abs should bind to the unconjugated MILKSHAKE proteins (Fig. 4).
3.7. Construction of Genetically Encoded Target Sequence Plasmid (Sundae)
To begin construction of a Sundae expression plasmid (pAT28 or equivalent), the commercially available plasmid pMAL-c6T was purchased from NEB (catalog #: N8108S, see Note 1). A sequence from the target of interest (10–14 amino acids) preceded by a GSGS linker was cloned into the parent vector in-frame with the MBP into the multiple cloning site using NotI and EcoRI (see Note 11). The resulting plasmid pAT28 expresses a modified MBP containing a short sequence of the target of interest in its C-terminus (termed “Sundae”). In order to further probe Ab binding to a specific residue in a target sequence, a set of 20 Sundae plasmids should be constructed, each with a different amino acid at the site of interest (Fig. 2). The recombinant vector(s) can be transformed into NEB Express cells and frozen in 15% glycerol stock solution and stored at −80 °C. Purified plasmid DNA can be stored at −20 °C in Tris-EDTA solution.
3.8. Sundae Protocol—Preparation of Inoculant
Follow the same steps as Subheading 3.2 for each Sundae plasmid.
3.9. Expression of Sundae Protein
Follow the same procedure as Subheading 3.3 for each Sundae plasmid.
3.10. Purification of Sundae Protein
Follow the same procedure as Subheading 3.4 for each Sundae plasmid.
3.11. Ab Validation Using Sundae ELISA
Coat wells of a 96-well microtiter plate with 50 μL of the Sundae protein(s) (antigen) diluted to 1 μg mL−1 in 1× PBS and incubate, sealed, overnight at 4 °C.
Following day, wash plates 3 times with PBS and block the wells with 50 μL of 2% BSA diluted in 1× PBS. Incubate for 1 h. Wash 3 times with 1 × PBS.
Add 50 μL per well of primary Ab using four-fold dilutions in 2% BSA beginning at 30 μg mL−1. Incubate for 1 h. Wash 3 times with 1 × PBST.
Add 50 μL of anti-Protein A HRP conjugated Ab using 1:5000 dilution (or other appropriate secondary Ab) to each well and incubate for 1 h. Wash 3 times with 1 × PBST.
Add 50 μL of room temperature TMB to each well and allow color to develop for 5–10 min.
Add 50 μL of 0.16 M H2SO4 to each well to quench the reaction. Be sure to wear PPE and dispose of acidic solutions in a hazardous waste stream.
Read absorbance at 450 nm in an appropriate available plate reader.
4. Notes
The more recent pMAL-c6T vector is a replacement for NEB #N8108, pMAL™-c5X Vector. Like the pMAL-c5x vector, the pMAL-c6T vector is designed to produce maltose-binding protein (MBP) fusions in the cytoplasm. The MBP has been engineered for tighter binding to amylose resin. The vector expresses an N-terminal his-tagged malE gene followed by a multiple cloning site containing a TEV protease recognition sequence, allowing MBP to be cleaved from the protein of interest after purification.
This plasmid contains a modified maltose binding protein (MILKSHAKE protein) with a C-terminal sortase site. The protein is expressed in NEB Express Cells (New England BioLabs Catalog # C2523H). Genotype: fhuA2 [lon] ompT gal sulA11 R(mcr-73::miniTn10--TetS)2 [dcm] R(zgb-210::Tn10--TetS) end A1 delta (mcrC-mrr)114::IS10. See Fig. 3 for details on this plasmid.
Do not overfill centrifuge bottles or tubes to avoid spillage and damage to the centrifuge. Make sure tubes are weighed and balanced to prevent faulting.
If the sample volume is large and it will require multiple additions.
When using the Biotek plate reader, use a protein quantification setting for a percent extinction coefficient of 1.4.
The anti-HA Ab recognizes the C-terminal HA tag on the peptide. Confirm the presence of the HA tag in a conjugated reaction and the absence of HA tag in the unconjugated sample.
When ordering peptides, request 2 mg of peptide with 90% purity. When peptides arrive (depending on the vendor this takes approximately 4–6 weeks), resuspend them in DMSO to a concentration of 10 mg mL−1. An example of one peptide set for the target akt1_473 (a PTM phospho-site) would consist of the following two peptides (sortase compatible site, target sequence, HA tag); Phospho_akt1_473_HA: GGGSGSS ERRPHFPQF{pSer}YSASGTA YPYDVPDYA and Non-Phospo_akt1_473_HA GGGSGSS ERRPHFPQFSYSASGTA YPYDVPDYA
Determine how many reactions you will need to test all of your Abs for each target. Each conjugation reaction will allow you to load approximately 11 lanes using 15 μL per lane in Western blot. If you have additional Abs to test, set up additional reactions in the plate.
The denatured reactions may be stored at 4 °C for up to 1 month before use in a Western blot. Settling may occur during 4 °C storage, so be sure to vortex these samples before use.
As an added control, include one additional lane per blot containing 15 μL MILKSHAKE protein conjugated to an irrelevant modified-HA peptide. This control can confirm that binding of the primary Ab is specific to the target peptide.
-
An example of two Sundae plasmids for the MPER target (found in the human immunodeficiency virus HIV) would consist of the following (pMAL-c6T sequence, GS linker, target sequence):
MPER-Sundae-D: MLMGGR GSGS EQELLELDKWASLW
MPER-Sundae-E: MLMGGR GSGS EQELLELEKWASLW
Acknowledgments
The authors would like to thank Emily P. Fuller and Brian K. Kay for insightful discussions.
This research was funded by NIH Appl. ID 1R44GM148998-01 “Platform for the High Throughput Generation and Validation of Affinity Reagents” and NIH Appl. ID 1R43GM146473-01 “Method for the validation by Western analysis of affinity reagents against post-translationally modified proteins. I. survey of existing Abs, and II. development of method improvements.”
References
- 1.Adhikari S, Nice EC, Deutsch EW et al. (2020) A high-stringency blueprint of the human proteome. Nat Commun 11(1):5301. 10.1038/s41467-020-19045-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebersold R, Agar JN, Amster IJ et al. (2018) How many human proteoforms are there? Nat Chem Biol 14(3):206–214. 10.1038/nchembio.2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bordeaux J, Welsh A, Agarwal S et al. (2010) Antibody validation. BioTechniques 48(3):197–209. 10.2144/000113382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourbeillon J, Orchard S, Benhar I et al. (2010) Minimum information about a protein affinity reagent (MIAPAR). Nat Biotechnol 28(7):650–653. 10.1038/nbt0710-650 [DOI] [PubMed] [Google Scholar]
- 5.Bradbury A, Plückthun A (2015) Reproducibility: standardize antibodies used in research. Nature 518(7537):27–29. 10.1038/518027a [DOI] [PubMed] [Google Scholar]
- 6.Bradbury AM, Plückthun A (2015) Antibodies: validate recombinants once. Nature 520(7547):295. 10.1038/520295b [DOI] [PubMed] [Google Scholar]
- 7.Bradbury AR, Plückthun A (2015) Getting to reproducible antibodies: the rationale for sequenced recombinant characterized reagents. Protein Eng Des Sel 28(10):303–305. 10.1093/protein/gzv051 [DOI] [PubMed] [Google Scholar]
- 8.Jonasson K, Berglund L, Uhlen M (2010) The 6th HUPO Ab Initiative (HAI) workshop: sharing data about affinity reagents and other recent developments September 2009, Toronto, Canada. Proteomics 10(11):2066–2068. 10.1002/pmic.201090040 [DOI] [PubMed] [Google Scholar]
- 9.Shankar G, Devanarayan V, Anaravadi L et al. (2008) Recommendations for the validation of immunoassays used for detection of host antibodies against biotechnology products. J Pharm Biomed Anal 48(5):1267–1281. 10.1016/j.jpba.2008.09.020 [DOI] [PubMed] [Google Scholar]
- 10.Taussig MJ, Fonseca C, Trimmer JS (2018) Antibody validation: a view from the mountains. New Biotechnol 45:1–8. 10.1016/j.nbt.2018.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taussig MJ, Stoevesandt O, Borrebaeck CA et al. (2007) ProteomeBinders: planning a European resource of affinity reagents for analysis of the human proteome. Nat Methods 4(1):13–17. 10.1038/nmeth0107-13 [DOI] [PubMed] [Google Scholar]
- 12.Uhlen M, Bandrowski A, Carr S et al. (2016) A proposal for validation of antibodies. Nat Methods 13(10):823–827. 10.1038/nmeth.3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voskuil JLA, Bandrowski A, Begley CG et al. (2020) The antibody society’s antibody validation webinar series. MAbs 12(1):1794421. 10.1080/19420862.2020.1794421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones KS, Chapman AE, Driscoll HA et al. (2022) MILKSHAKE: novel validation method for antibodies to post-translationally modified targets by surrogate Western blot. BioTechniques 72(1):11–20. 10.2144/btn-2021-0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrison KL, Weiss GA (2001) Combinatorial alanine-scanning. Curr Opin Chem Biol 5(3):302–307. 10.1016/s1367-5931(00)00206-4 [DOI] [PubMed] [Google Scholar]
- 16.Weiss GA, Watanabe CK, Zhong A et al. (2000) Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc Natl Acad Sci U S A 97(16):8950–8954. 10.1073/pnas.160252097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costa GL, Bauer JC, McGowan B et al. (1996) Site-directed mutagenesis using a rapid PCR-based method. Methods Mol Biol 57:239–248. 10.1385/0-89603-332-5:239 [DOI] [PubMed] [Google Scholar]
- 18.Costa GL, Weiner MP (2006) Rapid PCR site-directed mutagenesis. CSH Protoc 2006:pdb.prot4144. 10.1101/pdb.prot4144 [DOI] [PubMed] [Google Scholar]