Abstract
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), causative agent of the coronavirus disease-2019 (COVID-19) has resulted in several deaths and severe economic losses throughout the world. The spike protein in the virus binds to the human ACE-2 receptor in order to mediate virus-host interactions required for the viral transmission. Since first report of the SARS-CoV-2 sequence during December 2019 from patient infected with the virus in Wuhan, China, the virus has undergone rapid changes leading to mutations comprising substitutions, deletions and insertions in the sequence resulting in several variants of the virus that were more virulent and transmissible or less virulent but highly transmissible. The timely intervention with COVID-19 vaccines proved to be effective in controlling the number of infections. However, rapid mutations in the virus led to the lowering of vaccine efficacies being administered to people. In May 2023, the World Health Organization declared COVID-19 was not a public health emergency of international concern anymore. In order to take stock of mutations in the virus from early days to nearly end of COVID-19 pandemic, sequence analyses of the SARS-CoV-2 spike proteins available in the NCBI Virus database was carried out. The mutations and invariant residues in the SARS-CoV-2 spike protein sequences relative to the reference sequence were analysed. The location of the invariant residues and residues at interface of the protein chains in the spike protein trimer complex structure were examined. A total of 111,298 non-redundant SARS-CoV-2 spike protein sequences representing 2,345,585 spike proteins in the NCBI Virus database showed mutations at 1252 of the 1273 positions in the amino acid sequence. The mutations represented 6129 different mutation types in the sequences analysed. Besides, some sequences also contained insertion mutations. The SARS-CoV-2 spike protein sequences represented 1435 lineages. In addition, several spike protein sequences with mutations whose lineages were either ‘not classified’ or were ‘unclassifiable’ indicated the virus could still be evolving.
Keywords: Human SARS-CoV-2, Spike protein, Invariant residues, Mutations, Sequence analyses, Three-dimensional structure analyses
Graphical abstract
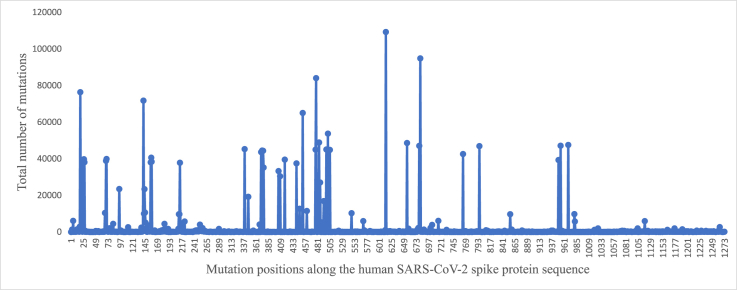
Total number of mutations observed at individual positions along the human SARS-CoV-2 spike protein reference sequence among 111,298 sequences representing 2,345,585 sequences obtained from the NCBI Virus database and corresponding to the period December 1, 2019 to April 14, 2023.
Highlights
-
•
Amino acid mutations among 111,298 human SARS-CoV-2 spike protein sequences representing a total of 2,345,585 sequences during the period Dec 1, 2019 to April 14, 2023 with respect to the first reported sequence from Wuhan, China were analysed.
-
•
1252 of the 1273 positions along the sequence were mutated in one or the other human SARS-CoV-2 spike proteins. The number of mutations ranged between 1 and 55 per spike protein sequence with upto 14 different mutation types at each position
-
•
Mutations in more than 40% of the sequences were associated with the following residues; D614, P681, T478, T19, G142, L452, N501, E484, H655, N969. The maximum mutation propensities were associated with the protease cleavage site, S1B, S1A domains and the least mutation propensities with C-terminal domain and linker regions.
-
•
The invariant residues were; M1, R319, K386, C391, F400, G416, N422, F559, G601, C662, C671, C749, P897, Q901, L962, S974, L996, R1000, G1044, F1256, K1269.
-
•
The spike proteins were represented by 1435 PANGO lineages and the prominent lineages were; Omicron (BA.2, BA.5.2.1, BA.1.1 and BA.2.12.1), Delta (AY.103, AY.44, AY.3, AY.25), Alpha (B.1.1.7) and clade G20 (B.1.2). 383 lineages represented by only single sequences including some of the recent spike proteins, 9657 lineages that were 'not classified' and 313 lineages that were 'unclassifiable', collectively indicate the virus could still be evolving.
1. Introduction
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) first reported in Wuhan, China during December 2019 is known to cause the coronavirus disease 2019 (COVID-19) (Wu et al., 2020). The virus since then had spread rapidly to various countries and the World Health Organization (WHO) made declarations of COVID-19 as a public health emergency of international concern (PHEIC) on 30th January 2020 and as a global pandemic on the 11th March 2020. This led to a total lockdown in most countries across the world. Although, it was initially thought COVID-19 would only cause mild to moderate respiratory illness, it subsequently became evident during the pandemic that COVID-19 caused severe respiratory illness leading to hospitalization and mortality, especially in individuals with cardiovascular disease, diabetes, chronic respiratory disease or with any immune-suppressed conditions (Bhaskaran et al., 2021). However, during the past 9 months or so, the number of COVID-19 infections decreased drastically with reports of causing only mild symptoms often not requiring hospitalization. Thereby, on 5th May 2023, WHO Director-General announced COVID-19 to be now an established and ongoing health issue and that it no longer constituted a PHEIC (https://www.who.int/news/item/05-05-2023-statement-on-the-fifteenth-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-coronavirus-disease-(covid-19)-pandemic?gclid=Cj0KCQjwpPKiBhDvARIsACn-gzAX5YkiaFCe6trdsmA6nRkiGfwV4OmPE7TCtvQ8uUQgBdBbybsChzUaArY3EALw_wcB). According to reports of September 6th, 2023 there were 770,437,327 confirmed cases of COVID-19 infections throughout the world and 6,956,900 deaths reported (https://covid19.who.int/).
During early COVID-19 infections, the WHO provided guidelines for individuals to maintain safe social distancing, personal hygiene, use of face masks to protect the spread of infections. The pharma and biotech companies also simultaneously worked towards drugs to treat COVID-19 by modifying known antiviral drugs (Fan et al., 2022), discover new drugs (Halford, 2022), diagnostic kits for the rapid detection of viral infections (Peeling et al., 2022) and vaccines to provide immunoprotection from SARS-CoV-2 (Rudan et al., 2022). As of 31st August 2023, a total of 13,500,122,024 vaccine doses were globally administered (https://covid19.who.int/). The timely discovery of COVID-19 vaccines (GeurtsvanKessel and de Vries, 2022) and their rapid dissemination and administration facilitated in lowering the number of deaths and hospitalization cases. However, rapid mutations led to the generation of new variants of the virus and individuals vaccinated for COVID-19 were prone to further SARS-CoV-2 infections (Chudzik et al., 2022; da Silva et al., 2022; Marks et al., 2023), thereby posing a serious challenge to deal with the situation and vaccine efficacy.
The SARS-CoV-2 comprises a single stranded RNA genome with ∼30 kb base pairs that is translated into structural and non-structural proteins (Chen et al., 2020). Single stranded RNA viruses have a high mutation rate as they lack the error correction mechanism of their genetic material and therefore can evolve to become more virulent strains or locally extinct (Duffy, 2018) During the virus transmission, SARS-CoV-2 has accumulated a variety of mutations that resulted in the virus either becoming more transmissible (Huai Luo et al., 2022; W. B. Wang et al., 2021), evade immune-protection resulting from the administration of vaccines (Jangra et al., 2021; Liu et al., 2021; Augusto et al., 2022), bind to the host with increased affinity (Ramanathan et al., 2021) or become less infective than previous variants (Uraki et al., 2022; Suzuki et al., 2022). The variants of SARS-CoV-2 are competitive in their co-transmission among human populations that ultimately decide fate of the virus (Plante et al., 2021; Chen et al., 2023). Evolutionary studies based on the spike protein sequences from SARS-CoV to SARS-CoV-2 and host receptor recognition were earlier studied (Guruprasad, 2020). The D614G was the first reported rapidly mutated substitution mutation among the SARS-CoV-2 spike proteins (Korber et al., 2020). The technical advisory group on SARS-CoV-2 virus evolution (https://www.who.int/groups/technical-advisory-group-on-sars-cov-2-virus-evolution) and the Phylogenetic Assignment of Named Global Outbreak (PANGO) lineage variants classification system (O'Toole et al., 2021) have independently classified variants of the virus. Accordingly, the SARS-CoV-2 variants have been classified (https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html) as: alpha, beta, gamma, delta, epsilon, eta, iota, kappa, omicron, zeta and mu (Rudan et al., 2022; Hebbani et al., 2022).
The SARS-CoV-2 spike protein is a structural protein that assembles to form a trimeric complex on the surface of the virus and it has a C-terminal membrane spanning region. The protein binds to the extracellular region of human ACE-2 receptor leading to fusion of the viral and host cellular membranes enabling the transfer of viral nucleocapsid into host cells to cause the infection (Zhang et al., 2020). The spike protein is heavily glycosylated. Based upon the nature of acceptor amino acid, protein glycosylation is classified as N- and O-linked glycosylation. The N-linked glycosylation occurs on the asparagine amide nitrogen side chain in the NxS and NxT sequence motif, where ‘x’ is any amino acid excepting proline. The O-linked glycosylation occurs on the side chain oxygen of serine or threonine residues. Both, N-linked (Shajahan et al., 2020; Walls et al., 2020) and O-linked glycosylations (Gong et al., 2021; Shajahan et al., 2023) are reported in SARS-CoV-2 spike proteins. The presence of extensive mutations in SARS-CoV-2 spike protein led to modifications in the glycosylation patterns. As spike protein is the first-line viral protein responsible for host infection, we intended to study the extent of changes in SARS-CoV-2 spike protein sequences of varying length by comparing with the first reported sequence from Wuhan, China during December 2019.
2. Data and analyses methods
The human SARS-CoV-2 spike protein sequences of varying length corresponding to the virus samples collected between December 1, 2019 until April 14, 2023 were obtained from the NCBI Virus database (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/). The redundant sequences were excluded using CD-HIT 2.0 program and representative sequences were selected where no two sequences share 100% identity (Li and Godzik, 2006) The partial sequences and sequences comprising ambiguous residues ‘X’ were excluded. The first reported SARS-CoV-2 spike protein sequence with NCBI code: YP_00972439019 was selected as reference sequence (RefSeq) for the comparative analyses (Wu et al., 2020). The alignment of sequences to the RefSeq was generated using the NGPhylogeny 2.1 program (Lemoine et al., 2019). Data management and identification of mutations with respect to the RefSeq based on the alignments and further analyses that included: identification of different mutation types at individual positions, calculation of the mutation percentages, evaluation of the domain-wise mutation propensities, identification of lineages representing the spike protein sequences and inter-chain interacting amino acid residues defined by a distance cut-off value ≤ 3.2 Å according to the three-dimensional structure of the SARS-CoV-2 spike protein trimer complex were carried out using the software suite of programs developed at ABREAST™ (https://www.abreast.in). The location of invariant residues in the spike protein three-dimensional electron microscopy structure available in the Protein Data Bank (PDB) (Berman et al., 2000) (PDB code: 7KRQ) (Zhang et al., 2021) were examined using UCSF Chimera24 (Pettersen et al., 2004). Also, residues at the interface of inter-chain interactions were examined to analyse whether they remained invariant or mutated.
3. Results and discussions
A total of 2,345,585 human SARS-CoV-2 spike protein sequences were obtained from the NCBI Virus database corresponding to the period December 1, 2019 to April 14, 2023 and after excluding the sequences comprising ambiguous residues marked ‘X’. Later, with the use of CD-HIT program and removal of 148 ‘partial’ sequences, 111,298 representative sequences were obtained that were used for identifying the mutations based on comparison to the reference human SARS-CoV-2 spike protein sequence (NCBI code: YP_009724390). The total number of mutations observed among the sequences were 2,294,482 including the deletion mutations and the mutations were associated with 1252 of the 1273 amino acid positions in RefSeq. In addition, certain insertion mutants were also identified for some of the spike protein sequences. For instance, sequences with the codes; BCN86413.1 and BCN86425.1 have extra 9 amino residues towards the N-terminal sequence with respect to the RefSeq and sequence with code; UJJ92015.1 has a “VV” dipeptide extension towards the C-terminus. The amino acid residues at twenty-one positions remained invariant among all the sequences analysed.
3.1. Mutations
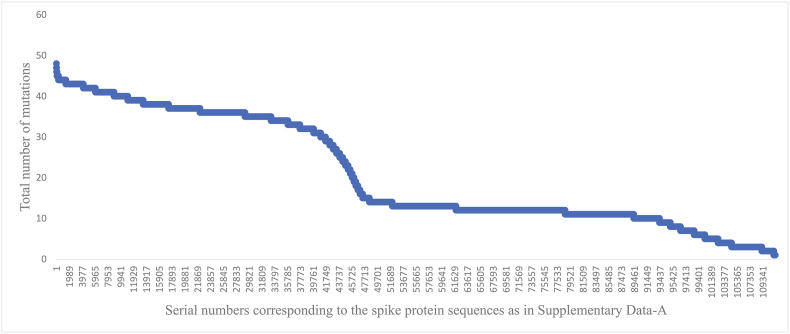
The total number of mutations (arranged in the decreasing order) observed among the spike protein sequences is shown in Fig. 1. The number of mutations ranged between 1 and 55 per spike protein sequence. For instance, proteins with 50 or more mutations were observed in the following sequences identified by their NCBI codes: UWH83352.1, WAU36028.1, WCM02064.1, URK41055.1, UZC40142.1, UPM00457.1, WFG71457.1, UPE42917.1, WCO18046.1, WCO18730.1, WDZ70513.1, UJN19575.1, UUZ31923.1, UNN34410.1, WBS32330.1. These represented the SARS-CoV-2 spike proteins from Omicron and sub-variants of virus samples collected January 2022 onwards from the USA. A relatively steep rise in the number of mutations comprising anywhere between 15 and 30 mutations per protein sequence were observed among some of the human SARS-CoV-2 spike proteins. The total number of mutations identified in each protein along with the corresponding protein codes are listed in the attached Supplementary Data-A.
Fig. 1.
Distribution of the total number of mutations (arranged in decreasing order) observed among 111,298 representative human SARS-CoV-2 spike protein sequences compared to the reference sequence.
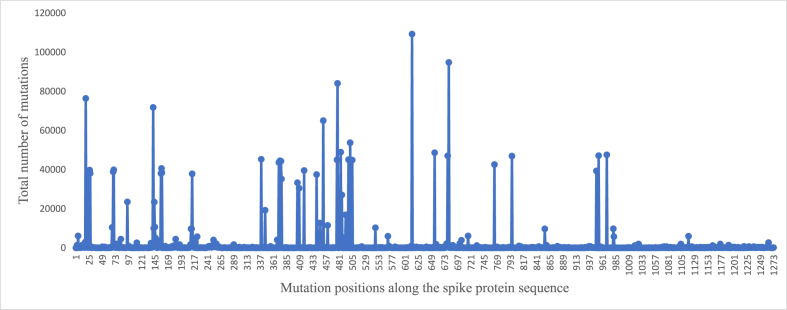
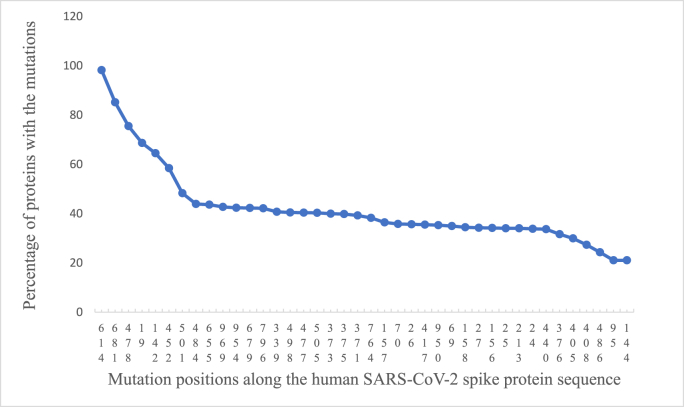
Fig. 2 shows the distribution of total number of mutations observed at individual positions along the protein sequence. Majority of the mutations were clustered around five to six regions and the C-terminal stretch comprised relatively few mutations. Fig. 3 shows the top 40 mutated positions. The position-wise mutation percentages are listed in the attached Supplementary Data-B. Accordingly, a higher percentage of mutations were observed along the SARS-CoV-2 spike protein sequence at the following positions; 614 (98.2%), 681 (85.2%), 478 (75.5%), 19 (68.6%), 142 (64.5%), 452 (58.4%), 501 (48.3%), 484 (453.93%), 655 (43.67%) and 969 (42.73%). The most frequent D614G mutation previously reported (Korber et al., 2020) remained the most frequent mutation observed in the present dataset too, since the outbreak of COVID-19 pandemic more than 3.5 years ago.
Fig. 2.
Total number of mutations at individual positions along the protein sequence observed among 111,298 representative human SARS-CoV-2 spike protein sequences relative to the reference sequence.
Fig. 3.
Top 40 mutations among the 111,298 human SARS-CoV-2 spike proteins relative to the reference sequence.
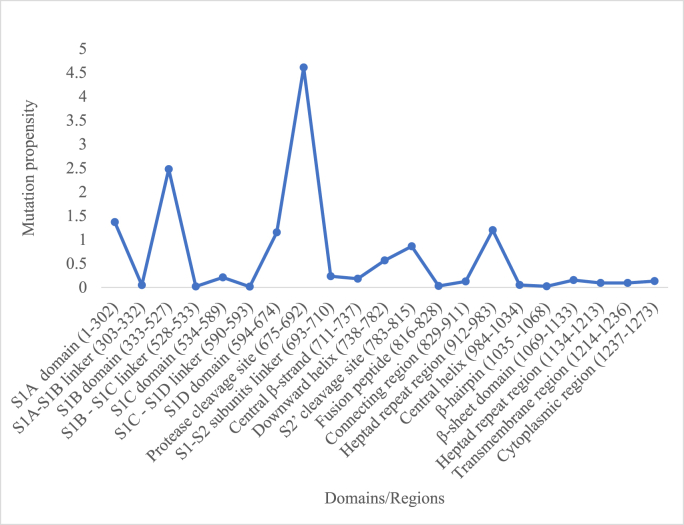
3.2. Domain-wise mutation propensities
The human SARS-CoV-2 spike protein comprises different domains/regions as previously described (Guruprasad, 2021a). The distribution of mutation propensities according to the different domains/regions is shown in Fig. 4. The relatively larger number of mutations were associated with the following domains/regions: S1A domain (residues 1–302), S1B domain (333–527), S1D (594–674), protease cleavage site (675–692), downward helix (738–782), S2’ cleavage site (783–815) and the first heptad repeat region (912–983). The maximum mutation propensity associated with the protease cleavage site among SARS-CoV-2 spike proteins previously reported (Guruprasad, 2021b) remained consistent in the present dataset too, comprising 111,298 sequences of variable length. The S1A domain involved in sialic acid recognition (Baker et al., 2020; Xu et al., 2021; Y. Wang et al., 2021) is associated with mutations and mutations in this domain are known to evade vaccines (Hebbani et al., 2022; Ahmadi et al., 2023). The S1B domain also known as receptor binding domain (RBD) comprises the receptor binding motifs (RBMs) that interact with human ACE-2 receptor leading to the human-host infection. Several non-bonding interactions inferred from the crystal structure of the protein (PDB code: 6LZG) stabilise the RBD-ACE-2 complex: Asn487(RBD)-Gln24(ACE-2), Gln493-Lys31, Gln493-Glu35, Tyr449-Asp38, Gln498-Gln42, Thr500-Tyr41, Asn487-Tyr83, Asn501-Lys353, Gly496-Lys353, Gln498-Lys353, Asn501-Lys353, Gly502-Lys353; salt-bridge; Lys417-Asp30 and hydrophobic interactions; Tyr489-Phe28, Tyr489-Leu79, Tyr489-Tyr83. Therefore, mutations associated with the residues above in SARS-CoV-2 spike proteins are likely to affect the virus-host interactions.
Fig. 4.
Mutation propensities corresponding to the different domains among 111,298 representative human SARS-CoV-2 spike protein sequences relative to the reference sequence.
Fourteen out of twenty-one domains/regions comprised relatively low mutation propensities. These correspond to the sites mainly associated with linker regions, fusion peptide, connecting regions and the central β-strand as in: S1A-S1B linker region (303–332), S1B–S1C linker region (528–533), S1C–S1D linker region (590–593), fusion peptide (816–828), connecting region (829–911) and the central β-strand (711–737). Thereby, there appears to be restrictions in flexibility of certain regions in the spike protein despite overall dynamical structure. Further, the entire stretch of C-terminal domains/regions between 984 and 1273 amino acid residue positions were associated with low mutation propensities. These regions of the SARS-CoV-2 spike protein could be potential regions for inhibitor design or targeted vaccines.
3.3. Mutation types
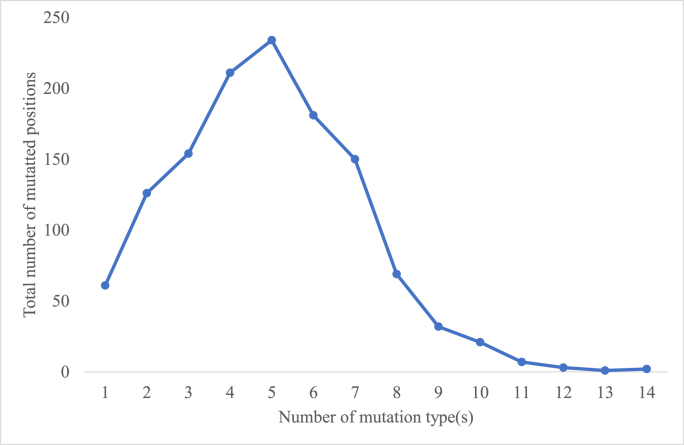
Multiple mutations occur at the same position in human SARS-CoV-2 spike protein (Guruprasad, 2021b). In the present dataset analysed, a total of 6129 different mutation types were observed. These represented one or more than one mutation type at each of the 1252 mutated positions. The total number of the different mutation types observed at each of the mutated positions is shown (arranged in the decreasing order) in Supplementary Data-C. The distribution of the total number of mutated positions associated with the different numbers of mutations among the human SARS-CoV-2 spike proteins relative to the reference sequence is shown in Fig. 5. These mutations were associated with 1252 amino acid positions among the 111,298 human SARS-CoV-2 spike proteins relative to the reference sequence. Accordingly, there were anywhere between 1 and 14 number of different mutation type(s) observed at the mutated positions and the amino acid residue positions comprising five different mutation types were observed to be more frequent, i.e., 234 out of 1252 mutated sites were associated with five different mutation types. The positions; 145 and 339 in the SARS-CoV-2 spike protein comprised a maximum number of 14 different mutation types. The corresponding residues; Y145 located in the β-hairpin in S1A domain and G339 in helical turn in S1B domain are exposed to the solvent according to the protein three-dimensional structure. These positions; 145 and 339 in the SARS-CoV-2 spike protein associated with the maximum number of diverse mutation types have been the most vulnerable positions at which mutations have occurred during COVID in the past 40 months. In Supplementary Data-D, the different mutation types observed at each of the mutated positions are listed. For instance, at position 1273, six different mutation types were observed; T1273S, T1273P, T1273A, T1273I, T1273K, T1273R. Likewise, at position 614, five different mutation types were observed; D614G, D614S, D614V, D614N, D614A. Multiple mutations were also observed within the RBD (i.e., between amino acid residues; 333–527) that interacts with the human ACE-2 receptor to cause the viral infection. In fact, it is reported that ACE-2 binding to the RBD is associated with ACE-2-RBD continuous swing motions and that the T470-T478 loop and Y505 are viral determinants for specific recognition of SARS-CoV-2 RBD by ACE-2 (Xu et al., 2021). In the present study, it was observed that different mutations were associated with all residues in the 470–478 loop and at the position 505. The residues at positions; 477 and 478 were associated with 8 different mutation types (refer Supplementary Data-C) that included a deletion mutation. The different mutation types can be referred in Supplementary Data-D. The mutations described may contribute to the inter-molecular interactions facilitating the ACE-2-RBD continuous swing motions. Likewise, it was observed that in the RBM too, the residue E484 was associated with 13 different mutation types which also included deletion mutation. The residue E484 is within a loop and does not directly interact with human ACE-2 (PDB code: 6LZG). The E484K mutation is shown to cause conformational rearrangements of the loop region in RBM leading to a tighter binding with ACE-2 and formation of hydrogen bonds (Li et al., 2020). The other mutations observed at position 484 are listed in the Supplementary Data-D that also includes a deletion mutant in three proteins identified by their NCBI codes; UTA82152.1, UYG26858.1 and WDZ70513.1. Previous results from our laboratory reported E484K mutant protein to have increased solvent interaction energy compared to the wild-type protein in binding to ACE-2, where the ionic interactions in the mutant protein contributed to the enhanced interactions (Mishra et al., 2022, Naresh and Guruprasad, 2023). Likewise, positions; 69, 88, 158, 213, 214 with 11 different mutation types and positions; 19, 144, 248 with 12 different mutations types are associated with the S1A domain. The protease cleavage site comprised 11 different mutation types located at positions; 679 and 681. All 2,329,012 mutations among the 111,298 human SARS-CoV-2 spike protein sequences relative to the RefSeq arranged protein code-wise are listed in the attached Supplementary Data-E.
Fig. 5.
Distribution of the total number of mutated positions associated with the different numbers of mutation type(s) among the human SARS-CoV-2 spike proteins relative to the reference sequence.
The variants of spike protein are known to have led to decreased or increased infectivity and therefore viral fitness, sensitivity to neutralizing monoclonal antibodies, sensitivity to convalescent sera and the deletion of glycosylation is known to have led to reduced infectivity (Li et al., 2020; Mishra et al., 2022; Kumar et al., 2023; Mittal et al., 2022; Kim et al., 2022; Harvey et al., 2021; Carabelli et al., 2023; Fan et al., 2022). The mutations in human SARS-CoV-2 spike proteins of sequence length 1273 amino acids and their implications on the potential drug development and epitope sites for vaccine design have been earlier reported (Carabelli et al., 2023; Guruprasad, 2022; Halford, 2022; Huai Luo et al., 2022). The spike protein is heavily glycosylated and large number of mutations are likely to affect the glycosylation patterns i.e., mutation can either introduce a glycosylation site or withdraw an existing glycosylation that could have functional implications on the protein.
3.4. Invariant residues
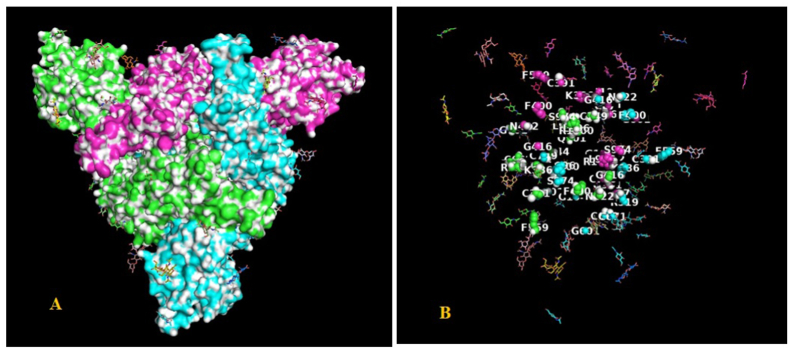
Among the 111,298 representative human SARS-CoV-2 spike protein sequences of variable length analysed, twenty-one amino acid residues were observed to be invariant. These residues were spread across the entire length of the spike protein sequence from the N- to the C-terminal region: M1, R319, K386, C391, F400, G416, N422, F559, G601, C662, C671, C749, P897, Q901, L962, S974, L996, R1000, G1044, F1256, K1269. The location of these residues was mapped onto the three-dimensional structure of the SARS-CoV-2 spike protein trimer complex comprising three chains; A, B, C (PDB code: 7KRQ) as shown in Fig. 6A. This can be more explicitly seen in Fig. 6B where only the invariant residues are displayed as spheres. The surface representation indicates that all the invariant residues lie within the protein ‘core’ and this may be important for maintaining the overall structure of the protein trimer complex. An animation showing the invariant residues is attached in the (Supplementary Video-1). The invariant residues; K386, C391, F400, G416, N422 are in the RBD domain important for the virus-host transmission. Among the four conserved cystines, C662 and C671 form a disulfide bridge and is present in the S1D domain. The invariant glycine residues; G416 and G601 and the proline residue; P897 in trans configuration (PDB_ID: 7KRQ) are possibly important structural determinants in the SARS-CoV-2 spike protein. Further, none of the invariant residues are involved in inter-chain interactions at a distance ≤3.2 Å, excepting the amino acid residue K386. The side-chain of K386 on the B-chain makes interactions with main chain carbonyl oxygen C=O of L981 on the A-chain and the equivalent interaction is present between all interacting partner chains in the protein trimer complex.
Fig. 6.
(A). The surface representation of the three-dimensional structure of human SARS-CoV-2 spike protein (PDB code: 7KRQ) showing; A-chain (green), B-chain (cyan), C-chain (magenta) and with the invariant residues mapped on to it. (B). Only invariant residues are shown as spheres and labelled. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. Glycosylation
Glycosylation in the SARS-CoV-2 spike protein is known to increase infectivity of the virus (Huang et al., 2021; Chawla et al., 2022; Shajahan et al., 2023). The invariant N422 residue, however, is not likely to be a potential glycosylation site as the residue at position two residues downstream, i.e., at position 424 has not been observed to be mutated in any of the spike protein sequences analysed as either S or T (refer Supplementary Data-D), in order to form the NxS or NxT sequence motif typical of N-glycosylation. Rather, the residue K424 in the spike protein is observed to be mutated only to either amino acid N (NCBI code: QOE90912.1) or R (NCBI code: UHS15838.1, UHT70778.1, UHT74898.1, UHE62980.1) among the spike proteins analysed (refer Supplementary Data-E).
3.6. Inter-chain interacting residues
The following residues lie at the interface of inter-chain interactions defined by a distance of ≤3.2 Å as deduced from the human SARS-CoV-2 spike protein electron microscopy structure (PDB code: 7KRQ_A-chain): F43, K113, D198, G232, S383, K386, D427, R466, I468, S469, E471, G545, N556, F565, R567, D571, D586, P589, C590, S591, F592, G614, A668, G669, L699, A701, E702, N703, Y707, N709, A713, Q755, Y756, Q787, I788, K790, D796, Q836, Y837, G842, D843, K854, P863, L864, Y873, Q895, F898, Y904, N907, Q913, N914, E918, Q965, F970, S975, L981, S982, R983, D985, K986, E988, Q1002, Q1005, E1017, E1031, R1039, N1074, P1090, V1094, R1107, S1123. All residues above, except the invariant amino acid K386, were observed to be mutated in one or the other SARS-CoV-2 spike proteins analysed. The mutations involving most residues that lie at the inter-chain interfaces within the trimer complex may facilitate the flexibility and dynamic nature of the spike protein.
3.7. Insertions in spike protein sequences relative to the RefSeq
In order to examine the positions of insertions in the sequences relative to the RefSeq and in order to get manageable number of sequences for examining the alignment, another multiple sequence alignment of the human SARS-CoV-2 spike protein sequences was carried out but only for sequences that shared 99% sequence identity. The sequence alignment obtained is shown in the Supplementary Data-F. Several short and long insertion regions with respect to the RefSeq were observed in some of the spike proteins as illustrated in Supplementary Video-2 in the spike protein structure. These insertions were mainly located on the surface of the protein trimer complex. The S1A domain comprises number of insertion mutants of variable length. Also, the region 453 to 458 close to the RBMs in the S1B domain that interacts with human ACE-2 receptor is associated with insertion mutants. The sequence regions: 640–643 (in S1D domain), 699–709 (S1–S2 subunits linker region) and 728–733 (loop region connecting the central β-strands) comprise insertions of variable length. The β-strand K786–K790 in C-chain forms parallel β-sheet with β-strand Gly700-Ser704 in B-chain. Extensive insertions comprising variable number of residues were also observed between L699 to N709 region.
3.8. PANGO lineages
The representative SARS-CoV-2 spike proteins constituted 1435 characterised PANGO lineages (O'Toole et al., 2021) according to the HEADER records for sequence ID entries in the NCBI Virus database. For instance, the protein ID UWH83352.1 from Georgia, U.S.A., with sequence entry date in the NCBI Virus database; 23-08-2022 corresponds to lineage BA.1.1. The prominent (top 10) lineages were; AY.103 (Delta variant according to WHO classification), B.1.1.7 (Alpha), AY.44 (Delta), BA.1.1 (Omicron), AY.3 (Delta), BA.2.12.1 (Omicron), AY.25 (Delta), B.1.2 (clade G20), BA.2 (Omicron) and BA.5.2.1 (Omicron). There were 9657 protein sequences with lineages that were ‘not classified’ and 313 sequences with lineage entries marked ‘unclassifiable’. Further, 383 lineages were represented only once in the NCBI Virus database, for instance, the lineage B.1.36.18 corresponding to the SARS-CoV-2 spike protein sequence entry from Hong Kong (NCBI code: UHM09432.1). The percentage of different lineages constituting the 111,298 human SARS-CoV-2 spike proteins analysed is shown in the Supplementary Data-G.
In summary, the present study demonstrates that a variety of mutations have been accommodated in the human SARS-CoV-2 spike protein, since the outbreak of COVID-19 pandemic more than 40 months ago. Twenty-one residues were observed to be invariant. The amino acid residues within interacting distance of ≤3.2 Å between the protein chains in the trimer complex are all known to be mutated, except K386. The presence of 9657 spike proteins whose lineages were ‘not classified’ and 313 spike proteins with ‘unclassifiable’ lineages and lineages corresponding to some of the recent spike proteins (Feb–April 2023), for instance, DS.1 (USA/WEQ44582.1), EC.1 (USA/WEQ45862.1), XBB.1.16 (USA/WFG70695.1, India/WEW81554.1) suggest the virus could still be evolving.
4. Conclusions
A total of 111,298 human SARS-CoV-2 spike protein sequences of variable length represented 2,345,585 sequences available in the NCBI Virus database corresponding to the period 1st Dec 2019 until 14th April 2023. Twenty-one residues were observed to be invariant as compared to the first reported human SARS-CoV-2 spike protein sequence from Wuhan, China comprising 1273 amino acid residues. These are possibly important for maintaining overall three-dimensional structure of the protein trimer complex. The mutations that include deletion mutants were associated with 1252 of the 1273 residue positions and correspond to 6129 different mutation types. Besides, there were sequences also comprising insertion mutations. A single SARS-CoV-2 spike protein comprised anywhere between 1 and 55 mutations. The number of different mutations observed at each position ranged between 1 and 14 with the positions associated with relatively higher number of mutations indicating vulnerability to accommodate the sequence changes in SARS-CoV-2 spike proteins. The conserved Cys662-Cys671 disulfide bridge may be an important structural constraint in the protein. The maximum mutations in the SARS-CoV-2 spike protein were associated with the protease cleavage site, whereas, the least number of mutations were associated with the C-terminal domain/regions. The structural plasticity of the SARS-CoV-2 spike protein and therefore promiscuous interactions with human ACE-2 receptor could be resulting from mutations in RBD, particularly, RBM. The prominent lineages represented were; Omicron (BA.2, BA.5.2.1, BA.1.1 and BA.2.12.1), Delta (AY.103, AY.44, AY.3, AY.25), Alpha (B.1.1.7) and clade G20 (B.1.2). The mutations described for the human SARS-CoV-2 spike proteins may have contributed to the increased transmissibility of the virus, or its escape from antibodies gradually during the past ∼3.5 years that may have resulted in the COVID-19 disease progressing from pandemic to the current endemic stage. However, mutations among human SARS-CoV-2 spike protein sequences constituting ‘unclassifiable’ lineages or lineages ‘not classified’ including some of the rare lineages that have recently emerged indicate the virus could still be evolving!
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Lalitha Guruprasad designed the project, carried out analyses and wrote the manuscript., Gatta KRS Naresh collected data and assisted in the analyses., Ganesh Boggarapu prepared the representative data set and provided technical assistance.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank School of Chemistry, University of Hyderabad for research facilities and ABREAST™ (https://www.abreast.in) for making available the computer programs used in this work for identification and analyses of the mutations. Gatta KRS Naresh thanks IOE, University of Hyderabad for research fellowship. Ganesh Boggarapu thanks UGC for research fellowship.
Handling Editor: Dr A Wlodawer
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2023.100107.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
All data are available in the submitted manuscript
References
- Ahmadi S., Bazargan M., Elahi R., Esmaeilzadeh A. Immune evasion of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2); molecular approaches. Mol. Immunol. 2023;156:10–19. doi: 10.1016/j.molimm.2022.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augusto G., Mohsen M.O., Zinkhan S., Liu X., Vogel M., Bachmann M.F. In vitro data suggest that Indian delta variant b.1.617 of SARS-CoV-2 escapes neutralization by both receptor affinity and immune evasion. Allergy. 2022;77(1):111–117. doi: 10.1111/all.15065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A.N., Richards S.J., Guy C.S., Congdon T.R., Hasan M., Zwetsloot A.J., Gallo A., Lewandowski J.R., Stansfeld P.J., Straube A., Walker M., Chessa S., Pergolizzi G., Dedola S., Field R.A., Gibson M.I. The SARS-CoV-2 spike protein binds sialic acids and enables rapid detection in a lateral flow point of care diagnostic device. ACS Cent. Sci. 2020;6(11):2046–2052. doi: 10.1021/acscentsci.0c00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskaran K., Bacon S., Evans S.J., Bates C.J., Rentsch C.T., MacKenna B., Tomlinson L., Walker A.J., Schultze A., Morton C.E., Grint D., Mehrkar A., Eggo R.M., Inglesby P., Douglas I.J., McDonald H.I., Cockburn J., Williamson E.J., Evans D., Curtis H.J., Hulme W.J., Parry J., Hester F., Harper S., Spiegelhalter D., Smeeth L., Goldacre B. Factors associated with deaths due to covid-19 versus other causes: population-based cohort analysis of UK primary care data and linked national death registrations within the opensafely platform. Lancet Reg. Health Eur. 2021;6:100–109. doi: 10.1016/j.lanepe.2021.100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli A.M., Peacock T.P., Thorne L.G., Harvey W.T., Hughes J., Consortium C.-G.U., Peacock S.J., Barclay W.S., de Silva T.I., Towers G.J., Robertson D.L. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023;21(3):162–177. doi: 10.1038/s41579-022-00841-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla H., Fadda E., Crispin M. Principles of SARS-CoV-2 glycosylation. Curr. Opin. Struct. Biol. 2022;75 doi: 10.1016/j.sbi.2022.102402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Gu C., Ruan Z., Tang M. Competition of SARS-CoV-2 variants on the pandemic transmission dynamics. Chaos, Solit. Fractals. 2023;169:113–193. doi: 10.1016/j.chaos.2023.113193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudzik M., Babicki M., Kapusta J., Kołat D., Kałuzińska Ż., Mastalerz-Migas A., Jankowski P. Do the successive waves of SARS-CoV-2, vaccination status and place of infection influence the clinical picture and covid-19 severity among patients with persistent clinical symptoms? The retrospective study of patients from the stop-covid registry of the polocov-study. J. Pers. Med. 2022;12(5) doi: 10.3390/jpm12050706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva S.J.R., do Nascimento J.C.F., Germano, Mendes R.P., Guarines K.M., Targino Alves, da Silva C., da Silva P.G., de Magalhaes J.J.F., Vigar J.R.J., Silva-Junior A., Kohl A., Pardee K., Pena L. Two years into the covid-19 pandemic: lessons learned. ACS Infect. Dis. 2022;8(9):1758–1814. doi: 10.1021/acsinfecdis.2c00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16(8) doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Lou F., Fan J., Li M., Tong Y. The emergence of powerful oral anti-covid-19 drugs in the post-vaccine era. Lancet Microbe. 2022;3(2):e91. doi: 10.1016/S2666-5247(21)00278-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., de Vries R.D. Evaluating novel covid-19 vaccines in the current chapter of the pandemic. Lancet Infect. Dis. 2022;22(12):1652–1654. doi: 10.1016/S1473-3099(22)00517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y., Qin S., Dai L., Tian Z. The glycosylation in SARS-CoV-2 and its receptor ACE2. Signal Transduct. Targeted Ther. 2021;6(1):396. doi: 10.1038/s41392-021-00809-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruprasad K. Mutations in human SARS-CoV-2 spike proteins, potential drug binding and epitope sites for covid-19 therapeutics development. Curr. Res. Struct. Biol. 2022;4:41–50. doi: 10.1016/j.crstbi.2022.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruprasad L. Evolutionary relationships and sequence‐structure determinants in human SARS coronavirus‐2 spike proteins for host receptor recognition. Proteins. 2020;88(11):1387–1393. doi: 10.1002/prot.25967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruprasad L. Human coronavirus spike protein-host receptor recognition. Prog. Biophys. Mol. Biol. 2021;161:39–53. doi: 10.1016/j.pbiomolbio.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruprasad L. Human SARS CoV-2 spike protein mutations. Proteins. 2021;89(5):569–576. doi: 10.1002/prot.26042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford B. The path to paxlovid. ACS Cent. Sci. 2022;8(4):405–407. doi: 10.1021/acscentsci.2c00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Consortium C.-G.U., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbani A.V., Pulakuntla S., Pannuru P., Aramgam S., Badri K.R., Reddy V.D. Covid-19: comprehensive review on mutations and current vaccines. Arch. Microbiol. 2022;204(1):8. doi: 10.1007/s00203-021-02606-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huai Luo C., Paul, Morris C., Sachithanandham J., Amadi A., Gaston D.C., Li M., Swanson N.J., Schwartz M., Klein E.Y., Pekosz A., Mostafa H.H. Infection with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) delta variant is associated with higher recovery of infectious virus compared to the alpha variant in both unvaccinated and vaccinated individuals. Clin. Infect. Dis. 2022;75(1):e715–e725. doi: 10.1093/cid/ciab986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.C., Lai Y.J., Liao C.C., Yang W.F., Huang K.B., Lee I.J., Chou W.C., Wang S.H., Wang L.H., Hsu J.M., Sun C.P., Kuo C.T., Wang J., Hsiao T.C., Yang P.J., Lee T.A., Huang W., Li F.A., Shen C.Y., Lin Y.L., Tao M.H., Li C.W. Targeting conserved n-glycosylation blocks SARS-CoV-2 variant infection in vitro. EBioMedicine. 2021;74 doi: 10.1016/j.ebiom.2021.103712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra S., Ye C., Rathnasinghe R., Stadlbauer D., Personalized Virology Initiative study g, Krammer F., Simon V., Martinez-Sobrido L., Garcia-Sastre A., Schotsaert M. SARS-CoV-2 spike e484k mutation reduces antibody neutralisation. Lancet Microbe. 2021;2(7):e283–e284. doi: 10.1016/S2666-5247(21)00068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Gaudreault N.N., Meekins D.A., Perera K.D., Bold D., Trujillo J.D., Morozov I., McDowell C.D., Chang K.O., Richt J.A. Effects of spike mutations in SARS-CoV-2 variants of concern on human or animal ACE2-mediated virus entry and neutralization. Microbiol. Spectr. 2022;10(3) doi: 10.1128/spectrum.01789-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield C.-G.G., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that d614g increases infectivity of the covid-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Srivastava Y., Muthuramalingam P., Singh S.K., Verma G., Tiwari S., Tandel N., Beura S.K., Panigrahi A.R., Maji S., Sharma P., Rai P.K., Prajapati D.K., Shin H., Tyagi R.K. Understanding mutations in human SARS-CoV-2 spike glycoprotein: a systematic review amp; meta-analysis. Viruses. 2023;15(4):856. doi: 10.3390/v15040856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine F., Correia D., Lefort V., Doppelt-Azeroual O., Mareuil F., Cohen-Boulakia S., Gascuel O. Ngphylogeny.Fr: new generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019;47(W1):W260–W265. doi: 10.1093/nar/gkz303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Zhang L., Li X., Huang W., Wang Y. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Liu Z., VanBlargan L.A., Bloyet L.M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., Alford B., Buchser W.J., Ellebedy A.H., Fremont D.H., Diamond M.S., Whelan S.P.J. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29(3):477–488 e474. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks P.W., Gruppuso P.A., Adashi E.Y. Urgent need for next-generation covid-19 vaccines. JAMA. 2023;329(1):19–20. doi: 10.1001/jama.2022.22759. [DOI] [PubMed] [Google Scholar]
- Mishra T., Dalavi R., Joshi G., Kumar A., Pandey P., Shukla S., Mishra R.K., Chande A. SARS-CoV-2 spike e156g/delta157-158 mutations contribute to increased infectivity and immune escape. Life Sci. Alliance. 2022;5(7) doi: 10.26508/lsa.202201415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal A., Khattri A., Verma V. Structural and antigenic variations in the spike protein of emerging SARS-CoV-2 variants. PLoS Pathog. 2022;18(2) doi: 10.1371/journal.ppat.1010260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naresh G., Guruprasad L. Mutations in the receptor-binding domain of human SARS CoV-2 spike protein increases its affinity to bind human ACE-2 receptor. J. Biomol. Struct. Dyn. 2023;41(6):2368–2381. doi: 10.1080/07391102.2022.2032354. [DOI] [PubMed] [Google Scholar]
- O'Toole A., Scher E., Underwood A., Jackson B., Hill V., McCrone J.T., Colquhoun R., Ruis C., Abu-Dahab K., Taylor B., Yeats C., du Plessis L., Maloney D., Medd N., Attwood S.W., Aanensen D.M., Holmes E.C., Pybus O.G., Rambaut A. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021;7(2):veab064. doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeling R.W., Heymann D.L., Teo Y.Y., Garcia P.J. Diagnostics for covid-19: moving from pandemic response to control. Lancet. 2022;399(10326):757–768. doi: 10.1016/S0140-6736(21)02346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Plante J.A., Mitchell B.M., K. S A., Debbink K., Weaver S.C., Menachery V.D. The variant gambit: covid-19's next move. Cell Host Microbe. 2021;29(4):508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan M., Ferguson I.D., Miao W., Khavari P.A. SARS-CoV-2 b.1.1.7 and b.1.351 spike variants bind human ACE2 with increased affinity. Lancet Infect. Dis. 2021;21(8):1070. doi: 10.1016/S1473-3099(21)00262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudan I., Adeloye D., Sheikh A. COVID-19: vaccines, efficacy and effects on variants. Curr. Opin. Pulm. Med. 2022;28(3):180–191. doi: 10.1097/MCP.0000000000000868. [DOI] [PubMed] [Google Scholar]
- Shajahan A., Supekar N.T., Gleinich A.S., Azadi P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;30(12):981–988. doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajahan A., Pepi L.E., Kumar B., Murray N.B., Azadi P. Site specific N- and O-glycosylation mapping of the spike proteins of SARS-CoV-2 variants of concern. Sci. Rep. 2023;13(1) doi: 10.1038/s41598-023-33088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R., Yamasoba D., Kimura I., Wang L., Kishimoto M., Ito J., Morioka Y., Nao N., Nasser H., Uriu K., Kosugi Y., Tsuda M., Orba Y., Sasaki M., Shimizu R., Kawabata R., Yoshimatsu K., Asakura H., Nagashima M., Sadamasu K., Yoshimura K., Genotype to Phenotype Japan C., Sawa H., Ikeda T., Irie T., Matsuno K., Tanaka S., Fukuhara T., Sato K. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 omicron variant. Nature. 2022;603(7902):700–705. doi: 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraki R., Kiso M., Iida S., Imai M., Takashita E., Kuroda M., Halfmann P.J., Loeber S., Maemura T., Yamayoshi S., Fujisaki S., Wang Z., Ito M., Ujie M., Iwatsuki-Horimoto K., Furusawa Y., Wright R., Chong Z., Ozono S., Yasuhara A., Ueki H., Sakai-Tagawa Y., Li R., Liu Y., Larson D., Koga M., Tsutsumi T., Adachi E., Saito M., Yamamoto S., Hagihara M., Mitamura K., Sato T., Hojo M., Hattori S.I., Maeda K., Valdez R., team I.s., Okuda M., Murakami J., Duong C., Godbole S., Douek D.C., Maeda K., Watanabe S., Gordon A., Ohmagari N., Yotsuyanagi H., Diamond M.S., Hasegawa H., Mitsuya H., Suzuki T., Kawaoka Y. Characterization and antiviral susceptibility of SARS-CoV-2 Omicron BA.2. Nature. 2022;607(7917):119–127. doi: 10.1038/s41586-022-04856-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292 e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.B., Liang Y., Jin Y.Q., Zhang J., Su J.G., Li Q.M. E484K mutation in SARS-CoV-2 RBD enhances binding affinity with hACE2 but reduces interactions with neutralizing antibodies and nanobodies: binding free energy calculation studies. J. Mol. Graph. Model. 2021;109 doi: 10.1016/j.jmgm.2021.108035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chen R., Hu F., Lan Y., Yang Z., Zhan C., Shi J., Deng X., Jiang M., Zhong S., Liao B., Deng K., Tang J., Guo L., Jiang M., Fan Q., Li M., Liu J., Shi Y., Deng X., Xiao X., Kang M., Li Y., Guan W., Li Y., Li S., Li F., Zhong N., Tang X. Transmission, viral kinetics and clinical characteristics of the emergent SARS-CoV-2 delta voc in guangzhou, China. eClinicalMedicine. 2021;40:101–129. doi: 10.1016/j.eclinm.2021.101129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Wang Y., Liu C., Zhang C., Han W., Hong X., Wang Y., Hong Q., Wang S., Zhao Q., Wang Y., Yang Y., Chen K., Zheng W., Kong L., Wang F., Zuo Q., Huang Z., Cong Y. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci. Adv. 2021;7(1) doi: 10.1126/sciadv.abe5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-COV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Cai Y., Xiao T., Lu J., Peng H., Sterling S.M., Walsh R.M., Jr., Rits-Volloch S., Zhu H., Woosley A.N., Yang W., Sliz P., Chen B. Structural impact on SARS-CoV-2 spike protein by d614g substitution. Science. 2021;372(6541):525–530. doi: 10.1126/science.abf2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the submitted manuscript