Abstract
The molecular mechanisms and evolutionary changes accompanying synapse development are still poorly understood1,2. Here we generate a cross-species proteomic map of synapse development in the human, macaque and mouse neocortex. By tracking the changes of more than 1,000 postsynaptic density (PSD) proteins from midgestation to young adulthood, we find that PSD maturation in humans separates into three major phases that are dominated by distinct pathways. Cross-species comparisons reveal that human PSDs mature about two to three times slower than those of other species and contain higher levels of Rho guanine nucleotide exchange factors (RhoGEFs) in the perinatal period. Enhancement of RhoGEF signalling in human neurons delays morphological maturation of dendritic spines and functional maturation of synapses, potentially contributing to the neotenic traits of human brain development. In addition, PSD proteins can be divided into four modules that exert stage- and cell-type-specific functions, possibly explaining their differential associations with cognitive functions and diseases. Our proteomic map of synapse development provides a blueprint for studying the molecular basis and evolutionary changes of synapse maturation.
Synapses establish the neuronal networks that mediate information processing in the brain. Synaptic dysfunction is implicated in most brain diseases affecting individuals from childhood to adulthood3-5. Therefore, understanding synapse formation, maturation and specification is crucial for understanding human cognition and mental disorders.
The brain contains two main types of chemical synapse: excitatory glutamatergic and inhibitory GABAergic synapses. They differ in neurotransmitters, morphology, molecular composition and postsynaptic organization6-8. Excitatory synapses possess a specialized structure underneath the postsynaptic membrane, the PSD, which is more prominent than its counterpart in inhibitory synapses. Acting as an organizing centre and a signalling apparatus within synapses, the PSD is a sophisticated protein complex consisting of more than 1,000 proteins9. Alterations in these proteins are linked to more than 130 brain diseases10.
During brain development, excitatory synapses and their associated PSDs undergo substantial morphological and compositional changes11-13. For instance, the developmental shifts from the glutamate receptor subunits GRIN2B to GRIN2A, and from the scaffolding proteins DLG3 to DLG4, are critical for synapse maturation14-16. However, studies of these developmental changes have been limited to dozens of well-known PSD proteins typically identified in the adult brain11,12,17. Unbiased, systematic characterization has been limited, especially in humans18. In addition, synapse density, composition and maturation rates vary among species, potentially contributing to the evolutionary variation of neurotransmission properties, cognitive ability and behavioural repertoires19-24. For example, prolonged maturation or neoteny of human synapses has been suggested as a possible explanation for the emergence of human-specific cognitive traits2,25,26. Nevertheless, we still know little about the underlying molecular mechanisms.
Here we generate a cross-species proteomic map of synapse development in the neocortex. This map identifies the dynamics of more than 1,000 PSD proteins and the molecular pathways governing synapse maturation. A comparison of the maturing PSDs in humans to those in macaques and mice reveals that PSD maturation in humans is two to three times slower. This neotenic trait could be attributed to the higher abundance of RhoGEFs in human PSDs during the perinatal period. Integrating these data with transcriptomic and genetic data allows the determination of gene regulatory networks, cell type specificity and disease vulnerability during synapse maturation. Our data provide a temporal map of synapse development in the neocortex and offer insight into the evolutionary mechanisms of synaptic neoteny in humans.
PSD composition during human cortical development
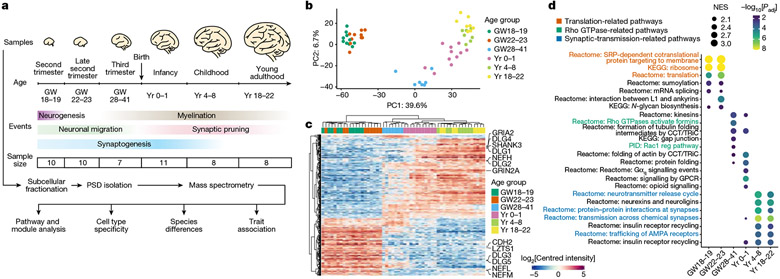
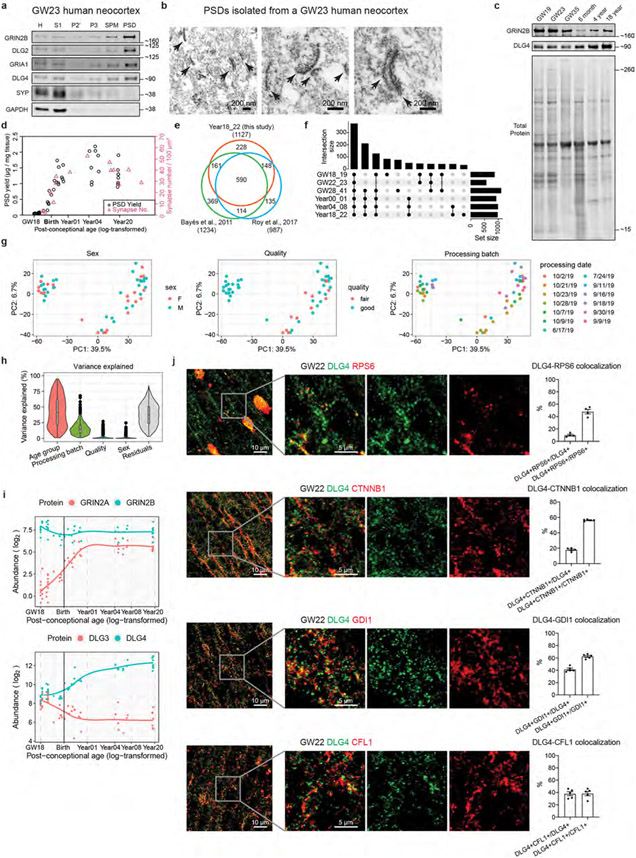
To understand the molecular changes of the PSD in developing human neocortex, we obtained neocortical samples across six main developmental stages ranging from the second trimester to young adulthood, covering key events such as neuronal migration, synaptogenesis, myelination and synaptic pruning (Fig. 1a and Supplementary Table 1). To reduce the confounding effect of cortical areas, we used samples from the prefrontal cortex (PFC), except for second-trimester samples that lacked area information owing to limited availability. PSDs were isolated from each sample as described previously27. Isolation of the PSD, including from immature human brain samples, was successful, as indicated by the following quality control metrics. Integral components of the PSD, but not presynaptic (SYP) or cytoplasmic (GAPDH) proteins, were enriched in the PSD fraction of early-stage samples (Extended Data Fig. 1a). In addition, electron microscopy revealed typical PSD-like electron-dense structures in the PSD fraction (Extended Data Fig. 1b). Furthermore, GRIN2B and DLG4, two PSD proteins that change during PSD maturation12, exhibited expected temporal abundance patterns in isolated PSDs (Extended Data Fig. 1c). Finally, PSD yield correlated well with estimated synapse numbers (Extended Data Fig. 1d).
Fig. 1 ∣. Changes in PSD composition during human neocortical development.
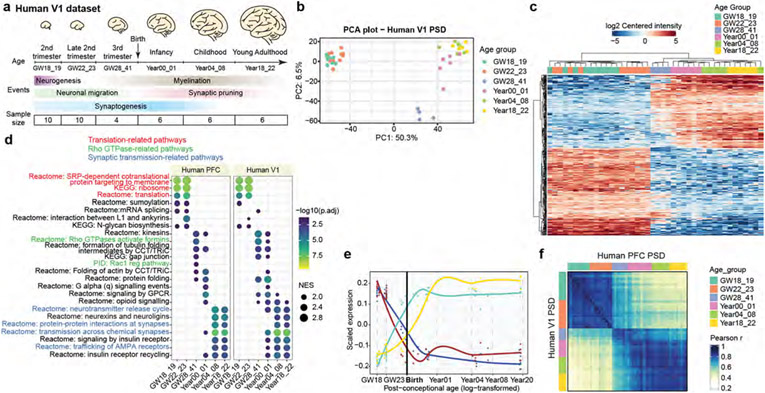
a, Flow chart of the overall approach. GW, gestational week. b, Principal component (PC) analysis plots of samples coloured by their age groups. c, Hierarchical clustering of samples based on proteins with differential abundance. d, GSEA for individual age groups. NES, normalized enrichment score; KEGG, Kyoto Encyclopedia of Genes and Genomes; PID, Pathway Interaction Database; reg, regulatory; GPCR, G-protein-coupled receptor. Nominal P values were adjusted (adj) by the Benjamini–Hochberg method.
We carried out liquid chromatography with tandem mass spectrometry analysis and label-free quantification of 54 PSD samples, each from a different neurotypical individual, and screened for synaptic proteome preservation28. The identified proteins significantly overlapped with previously reported PSD proteins at comparable stages10,29 (Extended Data Fig. 1e). After removing potential contaminants, we found 1,765 PSD proteins, including some that were stage specific (Extended Data Fig. 1f and Supplementary Table 2). Principal component analysis revealed that samples from the same age group were closely clustered (Fig. 1b). Principal component 1, accounting for 39.5% of the variability, strongly correlated with sample age but not with other potential confounding factors such as sex or processing batch (Extended Data Fig. 1g). Variance across age groups explained a median of 41.7% of the dataset’s variation, after correcting for processing batch, PSD quality and sex (Extended Data Fig. 1h). Hierarchical clustering also indicated age-related sample grouping (Fig. 1c). Key PSD proteins (GRIN2A, GRIN2B, DLG3 and DLG4) exhibited expected abundance patterns during PSD maturation, in agreement with western blotting data (Fig. 1c and Extended Data Fig. 1c,i). Immunostaining of proteins enriched at midgestation, including RPS6, β-catenin (CTNNB1), GDI1 and cofilin (CFL1), showed colocalization with the canonical PSD marker DLG4 in a subset of synapses (Extended Data Fig. 1j).
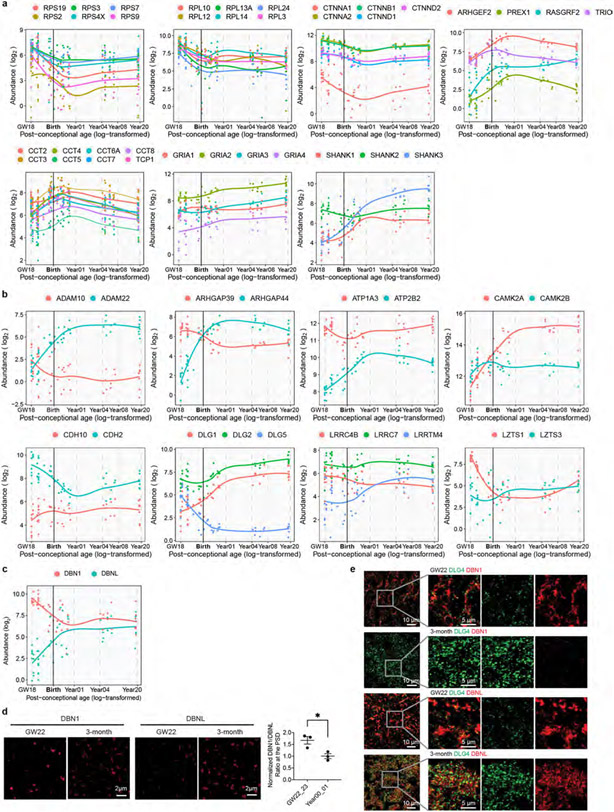
We carried out gene set enrichment analysis (GSEA) to identify molecular pathways with higher activity at different developmental stages. The three major phases observed were a midgestational phase (gestational week 18 to 23) enriched for translation-related pathways, a perinatal phase (third trimester to 1 year of age) enriched for Rho GTPase and protein folding pathways, and a postnatal phase (above 4 years of age) enriched for synaptic-transmission-related and neurexin- and neuroligin-associated pathways (Fig. 1d and Extended Data Fig. 2a). These findings indicate sequential activation of local protein synthesis, actin cytoskeleton reorganization and synaptic efficacy enhancement during PSD development. Although proteins from the same complex or pathway typically exhibited similar abundance changes during development (Extended Data Fig. 2a), it is known that relative changes in the abundance of homologous proteins such as GRIN2A and GRIN2B, and DLG3 and DLG4, are critical for synapse maturation14-16. As another example, GRIA2 increased steadily during development, whereas GRIA1 remained relatively constant (Extended Data Fig. 2a), consistent with the essential role of calcium-permeable α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors in early synaptic function30. Additionally, we found reciprocal pattern changes in many other homologous proteins (Extended Data Fig. 2b), including drebrin and drebrin-like proteins (Extended Data Fig. 2c-e), suggesting their potential significance for PSD maturation.
Protein modules and their functions in the PSD
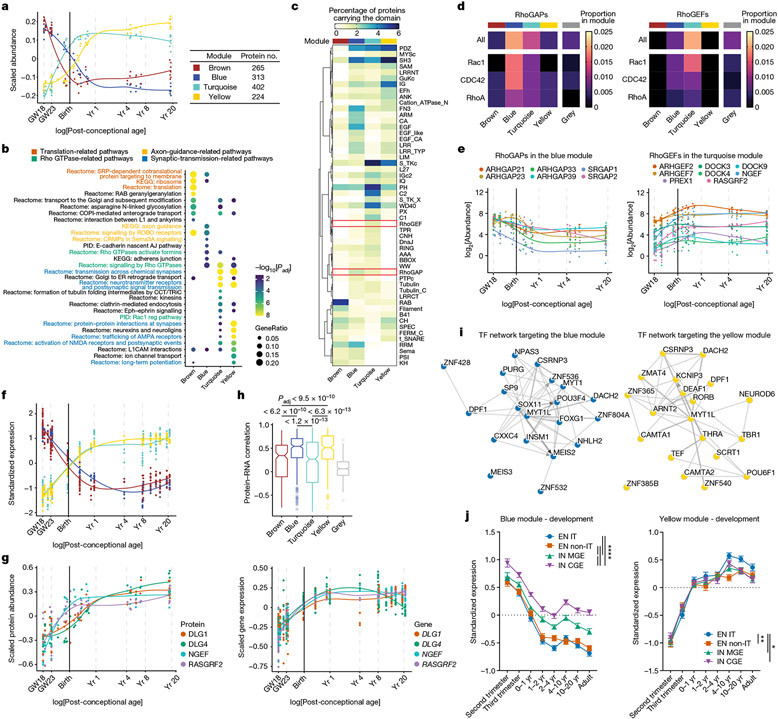
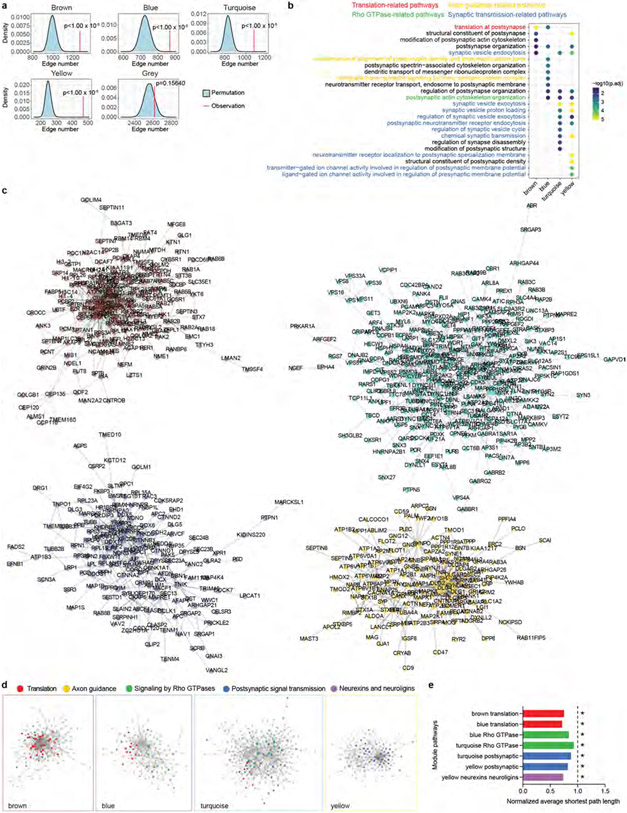
Proteins with similar abundance patterns during PSD maturation could represent protein modules with specific molecular functions. We identified four protein modules by weighted gene co-expression network analysis31 (WGCNA; Fig. 2a). All four modules were significantly enriched for protein–protein interactions, whereas no enrichment was found for proteins with no module assignment (the grey module; Extended Data Fig. 3a), indicating that proteins of the same module work synergistically by forming protein complexes. Pathway overrepresentation analysis revealed that the brown, blue, turquoise and yellow modules, ranked by their timing of peak abundance, were enriched with translation-, axon-guidance-, Rho GTPase- and synaptic-transmission-related pathways, respectively (Fig. 2b and Supplementary Table 3). Similar results were obtained by synaptic gene ontology (using SynGO) enrichment analysis32 (Extended Data Fig. 3b and Supplementary Table 3). In summary, abundance patterns and molecular functions of the PSD protein modules align with the GSEA results at individual developmental stages.
Fig. 2 ∣. Protein modules of the developing human PSD.
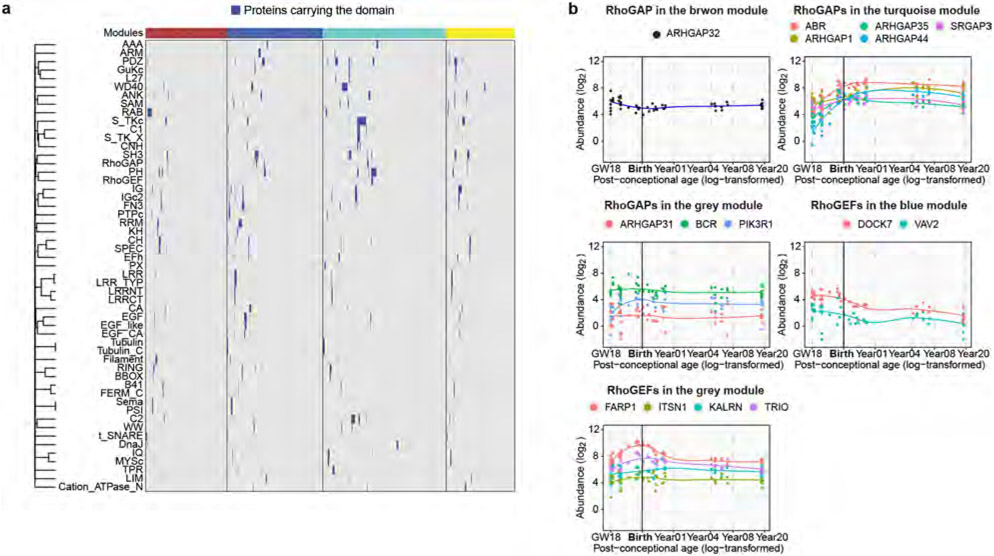
a, Scaled abundance patterns (module eigengene values) of four protein modules of the human PSD identified by WGCNA. b, Pathway enrichment analysis of each module. GeneRatio, proportion of genes in the pathway that are present in the module; AJ, adhesion junction. Nominal P values from hypergeometric test were adjusted by the Benjamini–Hochberg method. c, Distribution of protein domains in each module. d, Proportions of RhoGAPs (left) and RhoGEFs (right) and their subtypes in each module. e, Abundance patterns of RhoGAPs in the blue module (left) and RhoGEFs in the turquoise module (right). f, Standardized median expression values of genes encoding proteins of the four PSD modules in the BrainSpan data. g, Scaled protein abundance (left) and gene expression (right) patterns of DLG1, DLG4, NGEF and RASGRF2. h, Spearman correlation coefficients between protein abundance and gene expression of PSD proteins in each module (left to right: n = 236, 283, 371, 212 and 504 proteins). Box plot centre, median; hinges, the 25th and 75th percentiles; whiskers, 1.5 × interquartile range. The P values were obtained from Kruskal–Wallis test with Dunn’s multiple comparisons test. i, Transcription factor (TF) networks that regulate genes in the blue (left) and yellow (right) modules. j, Standardized expression values of genes in the blue (n = 298 genes; left) and yellow (n = 217 genes; right) modules in individual neuronal subtypes of developing human neocortex. EN IT, excitatory intratelencephalic neuron; EN non-IT, excitatory non-intratelencephalic neuron; IN MGE, inhibitory neuron derived from the medial ganglionic eminence; IN CGE, inhibitory neuron derived from the caudal ganglionic eminence. Data are presented as mean values s.e.m. Nominal P values from two-way analysis of variance (ANOVA) were adjusted by the Benjamini–Hochberg method; *P < 0.05, **P < 0.01, ****P < 0.0001.
To visualize potential protein complexes and interactions in the PSD, we generated networks of protein–protein interactions and co-abundance in each module (Methods and Extended Data Fig. 3c). As expected, proteins within the same pathway clustered closely together with shorter average path lengths, highlighting their functional associations (Extended Data Fig. 3d,e). Protein–protein interactions and biological functions of a protein are often mediated by protein domains. We determined the distribution of protein domains in PSD modules (Fig. 2c) and individual proteins (Extended Data Fig. 4a and Supplementary Table 4). Domains involved in vesicle trafficking (RAB and t_SNARE), cell adhesion (LRR, CA and ARM), signal transduction (S_TKc, C1 and C2) and adult PSD scaffolds (PDZ, SH3 and GuKc)17 were enriched in the brown, blue, turquoise and yellow modules, respectively (Fig. 2c). Notably, although both the blue and turquoise modules were involved in Rho GTPase signalling, RhoGAP and RhoGEF domains were selectively enriched in each of them (Fig. 2c). Indeed, Rho GTPase-activating proteins (RhoGAPs), particularly those specific for Rac1 and Cdc42, were enriched in the blue module, whereas RhoGEFs, particularly those specific for Rac1, were enriched in the turquoise module (Fig. 2d,e, Extended Data Fig. 4b and Supplementary Table 4). These results indicate that synaptic Rho GTPases gradually shift towards a more active state during PSD maturation, facilitating stage-specific cytoskeleton reorganization and morphological changes.
Generalization to other cortical regions
Our initial analysis focused on PFC. To assess the applicability of our findings to other cortical regions, we conducted a similar analysis on PSD samples from human primary visual cortex (V1), a sensory cortical area located on the opposite pole of the rostral–caudal axis from PFC (Extended Data Fig. 5a and Supplementary Tables 1 and 5). Similar to PFC, V1 samples were separated into three clusters corresponding to the midgestational, perinatal and postnatal phases (Extended Data Fig. 5b,c). Additionally, we observed comparable pathway enrichment patterns with translation-, Rho GTPase- and synaptic-transmission-related pathways activated sequentially from midgestation to adulthood (Extended Data Fig. 5d and Supplementary Table 5). Further, the four PSD modules showed similar abundance patterns in V1 (Extended Data Fig. 5e). Quantitative comparison of PSD samples between the two regions indicated a strong correlation, with a Pearson r > 0.75 for counterparts in the same phase (Extended Data Fig. 5f). Thus, our findings in the PFC dataset are generalizable to other regions of the human neocortex, including V1.
Cell-type-specific transcription of PSD proteins
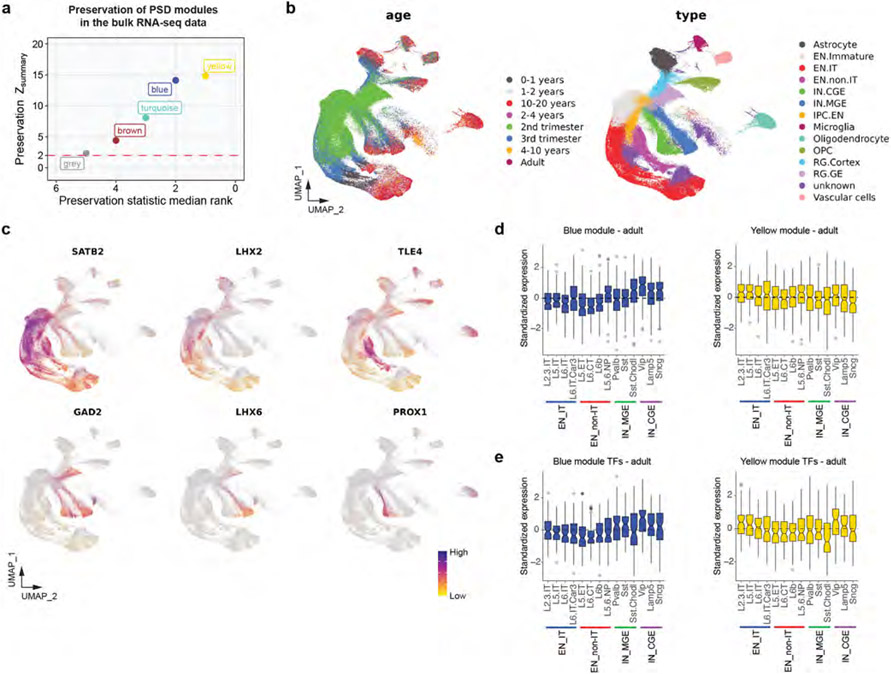
To understand the role of transcription in PSD development, we compared the RNA levels of PSD modules with their abundance patterns. Integrating BrainSpan and PsychENCODE transcriptomic data33 with our proteomic data, we found that general trends were preserved. However, the differences between the brown and blue and between the turquoise and yellow modules diminished (Fig. 2f and Supplementary Table 6). For example, Rho GTPase regulators and PSD scaffolding proteins from the turquoise and yellow modules, respectively, showed different abundance patterns in proteomic data, but had similar expression patterns in transcriptomic data (Fig. 2g). We quantified the RNA–protein concordance by calculating the Spearman’s rank coefficient of correlation for all PSD proteins (Supplementary Table 6). The blue and yellow modules showed high concordance (median Spearman r > 0.5), whereas the brown and turquoise modules had lower concordance, suggesting that post-transcriptional and/or translational regulation affects the PSD abundance of the latter (Fig. 2h). In alignment with these findings, the results of module density and connectivity preservation analysis34 showed that although all four modules were at least moderately preserved in transcriptomic data, the brown and turquoise modules were among the least preserved (Extended Data Fig. 6a).
The high RNA–protein concordance of proteins in the blue and yellow modules suggests that their PSD abundance is mainly regulated by transcription. Therefore, we focused on these two modules to study the transcriptional regulatory mechanisms of PSD development. Transcription factor enrichment analysis by ChEA3 revealed core transcription factors targeting the two modules, including FOXG1, MEIS2, MYT1L and RORB, known critical regulators of neuronal differentiation and synapse development (Fig. 2i and Supplementary Table 6). To investigate the transcription of PSD proteins in a cell-type-specific manner, we integrated PSD proteomic data with single-cell RNA-sequencing data from developing and adult human neocortex35,36 (Extended Data Fig. 6b,c and Supplementary Table 7). We found that both excitatory neurons and inhibitory neurons had a reduction in blue module and an increase in yellow module gene expression during development. However, the reduction of blue module genes in inhibitory neurons, particularly those from the caudal ganglionic eminence, was slower than that of excitatory neurons (Fig. 2j). Instead, yellow module gene expression increased significantly faster in excitatory intratelencephalic neurons (Fig. 2j). Similar patterns were seen in the adult brain (Extended Data Fig. 6d). This difference in expression can be attributed to the differential expression of upstream transcription factors (Extended Data Fig. 6e). Differences in transcription and abundance of PSD proteins may contribute to distinct excitatory postsynaptic responses observed in these cortical neurons37.
Species differences in PSD development
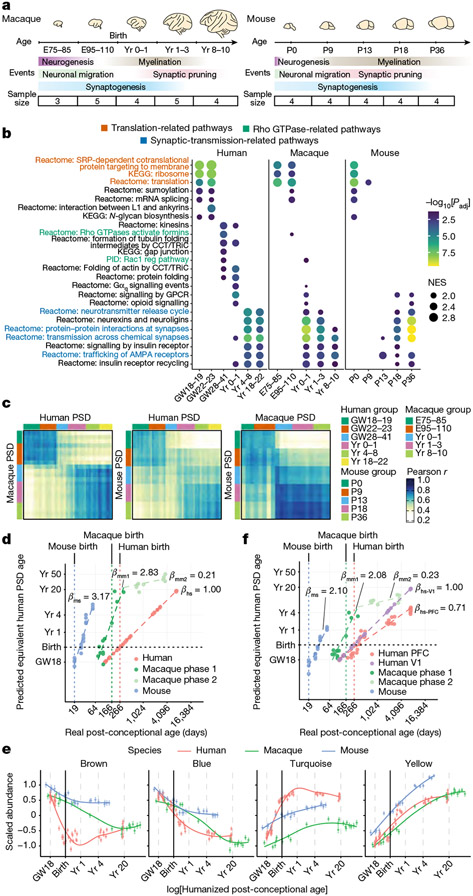
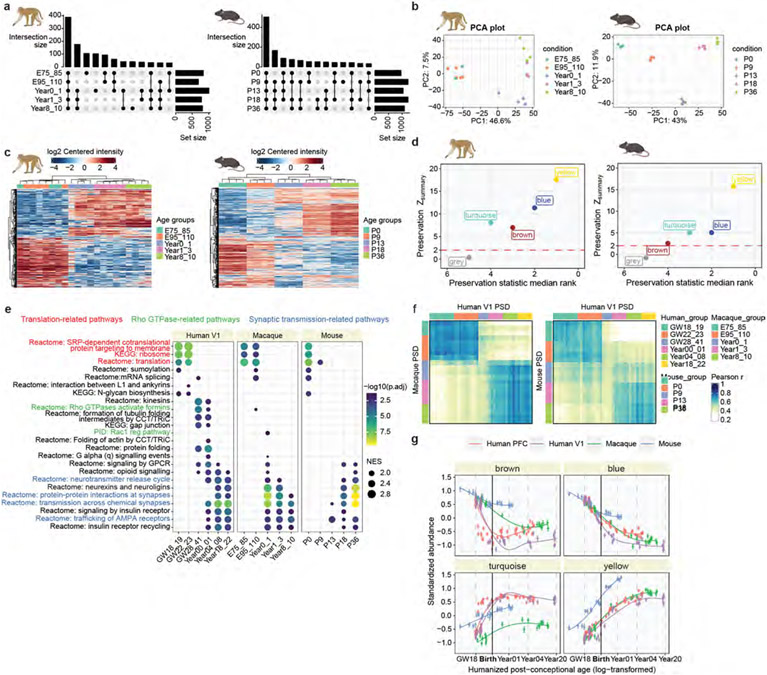
We investigated PSD development in humans, macaques and mice to understand the differences across these species. Macaque and mouse neocortical samples were collected at five time points (Fig. 3a and Supplementary Table 8), roughly corresponding to the developmental stages of our human samples38. Most of the macaque samples were collected from PFC, similar to our human samples. As mice lack a granular PFC39, and our human PFC findings can be applied to other cortical areas, we collected whole mouse neocortex instead.
Fig. 3 ∣. Comparison of PSD development across humans, macaques and mice.
a, Schematic of developmental stages of macaque (left) and mouse (right) samples. E, embryonic day; P, postnatal day. b, GSEA for individual age groups across species. Nominal P values were adjusted by the Benjamini–Hochberg method. c, Similarity matrices representing pairwise Pearson correlations between human, macaque and mouse samples. d, Predicted equivalent human PSD ages. β indicates the slope coefficients of the linear regression models in each species. ms, mouse; mm1, macaque phase 1; mm2, macaque phase 2; hs, human, e, Standardized abundance patterns of proteins in the four PSD modules (left to right: n = 127, 169, 159 and 178 proteins) across species. Data are presented as mean values ± s.e.m. f, Predicted equivalent human PSD ages based on the human V1 dataset. β indicates the slope coefficients of the linear regression models in each region and species; hs-V1, human V1; hs-PFC, human PFC.
Proteomic analysis identified 1,572 proteins in developing macaque PSDs and 1,572 proteins in mouse PSDs (Supplementary Tables 9 and 10), with some proteins being stage specific (Extended Data Fig. 7a). Both principal component analysis and hierarchical clustering revealed age-specific clustering of samples for both species (Extended Data Fig. 7b,c). GSEA showed that translation- and synaptic-transmission-related pathways were more active during early and late PSD development in all three species (Fig. 3b). However, enrichment of pathways in the human perinatal phase, including Rho GTPase signalling and protein folding, was largely diminished in macaques and mice (Fig. 3b). Cross-species similarity analysis showed that human samples in the second trimester and above 4 years of age correlated well with macaque and mouse samples at the corresponding stages (Pearson r > 0.6; Fig. 3c). Nevertheless, human samples between the third trimester and 1 year of age (the perinatal phase) exhibited lower correlations with all age groups in other species, prompting us to investigate the underlying differences that led to this variation.
To make a fair comparison of PSD maturation among species, we first applied a regularized linear approach to predict the equivalent human PSD ages for all samples (Fig. 3d, Methods and Supplementary Table 11). We found that multiplicative changes in real age were approximately linearly associated with multiplicative changes in the predicted equivalent human PSD age, except that macaque samples exhibited two different stages separated by 1 year of age (Fig. 3d). We thus regressed the log-transformed humanized ages against the real log-transformed ages using a linear model (or a linear spline model for macaque samples) and obtained the slope coefficients as an estimator of PSD maturation rate normalized to the developmental timescale of individual species. This analysis revealed that PSD maturation was about three times slower in humans than in mice and macaques (<l year; Fig. 3d).
On the basis of the equivalent PSD ages, we compared the abundance patterns of human PSD modules in all three species. Whereas the patterns of the blue and yellow modules were similar across all species, the brown and turquoise modules exhibited species-specific differences (Fig. 3e). Specifically, the brown module was less abundant, and the turquoise module was more abundant in humans during the perinatal phase, which probably contributed to the low correlation observed in the similarity analysis. Module density and connectivity preservation analysis confirmed that these two modules were among the least preserved in macaques and mice (Extended Data Fig. 7d). Given the low RNA–protein concordance of these two modules (Fig. 2h), our results also highlight the role of post-transcriptional and/or translational regulation in shaping the distinctive features of human synapses.
Comparing the macaque and mouse datasets to the human V1 dataset yielded similar results (Extended Data Fig. 7e-g). One minor but notable difference is that the predicted PSD maturation rate in human V1 was about two times slower than in mice and macaques (<1 year) and about 40% faster than in human PFC (Fig. 3f). This aligns with previous findings that sensory cortical areas, such as V1, tend to mature faster than association areas such as PFC40,41. In summary, human PSD matures at a slower rate, and the perinatal phase of its development is less represented in macaques and mice.
RhoGEFs promote neoteny of human synapses
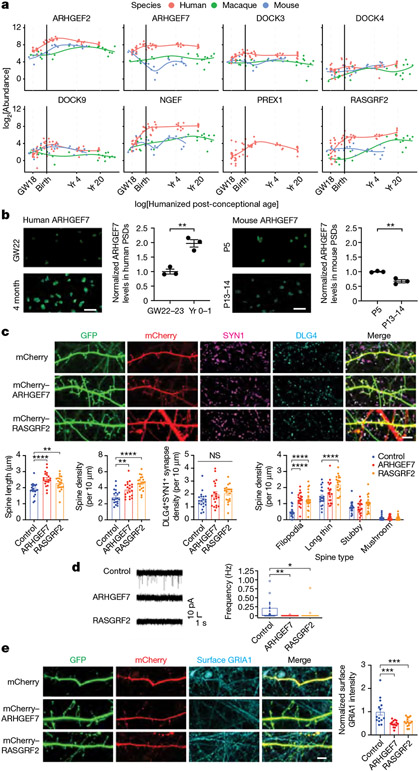
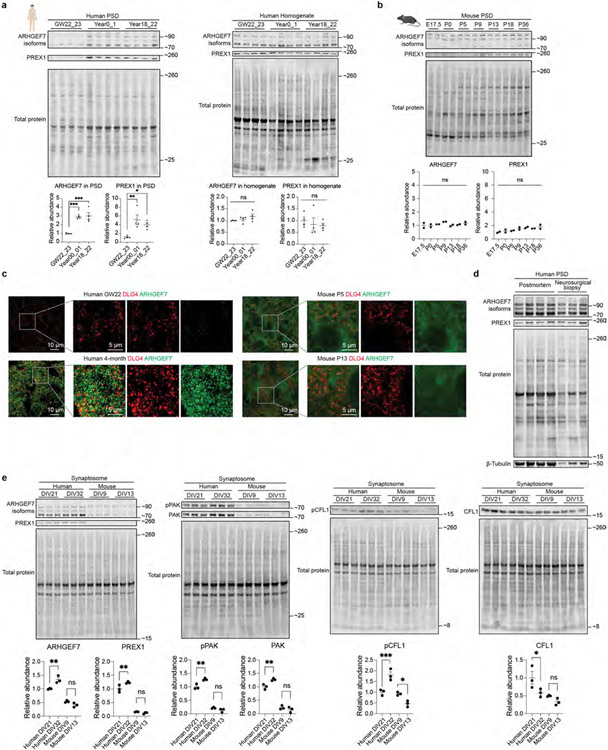
The slower human PSD maturation rate may be due to increased turquoise module proteins and Rho GTPase regulators during the perinatal phase. To test this hypothesis, we further investigated the increase of RhoGEF signalling in human PSDs. Indeed, most RhoGEF proteins in the turquoise module were greatly increased at the perinatal phase in humans and remained more abundant compared to those in other species (Fig. 4a). Western blotting and immunostaining further validated these findings (Fig. 4b and Extended Data Fig. 8a-c). No significant changes were observed in the total homogenate over time, supporting the role of protein trafficking in regulating turquoise module protein abundance (Extended Data Fig. 8a). Postmortem accumulation could lead to an artificial increase in PSD proteins, as has been reported for tubulins42. To rule out the possibility that postmortem accumulation caused the observed increase in RhoGEF proteins, we compared adult PSDs prepared from postmortem and neurosurgical biopsy samples and found similar RhoGEF levels (Extended Data Fig. 8d). Most RhoGEF proteins in the turquoise module target Rac1. We observed that phosphorylation of PAK and CFL1, both downstream effectors of Rac1, increased in human synaptosomes along with RhoGEFs during synapse maturation (Extended Data Fig. 8e). However, no such increases were observed in mouse neurons (Extended Data Fig. 8e). Together, these results validated the enhancement of RhoGEF signalling in human PSDs.
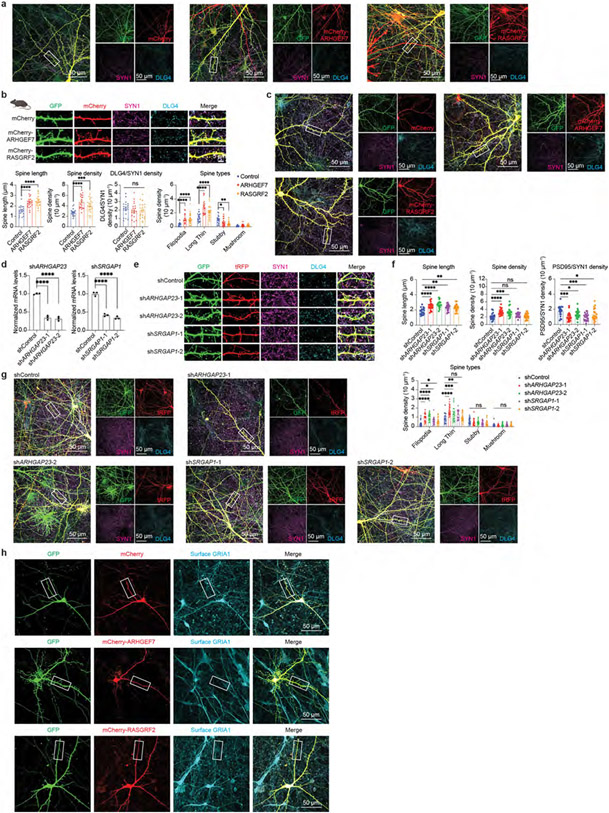
Fig. 4 ∣. Increase in RhoGEF proteins promotes neoteny of human synapses.
a, Abundance patterns of RhoGEFs in the turquoise module across species. b, Immunofluorescence intensity of ARHGEF7 at DLG4 loci in developing human and mouse neocortex (n = 3, 3, 3 and 3 samples; scale bars, 2 μm). Data are presented as mean values. The P values were obtained from unpaired two-tailed t-test; **P < 0.01. c, Immunostaining of dendrites from primary human cortical neurons cultured 6 weeks in vitro. Neurons were transfected with mEGFP-C1 and vectors expressing mCherry, mCherry–ARHGEF7 or mCherry–RASGRF2 (n = 20, 20 or 20 neurons, respectively, from 4 cultures; scale bars, 5 μm). Data are presented as mean values ± s.e.m. The P values were obtained from one-way ANOVA with Holm–Sidak’s multiple comparisons test; **P < 0.01, ****P < 0.0001; NS, not significant. d, Miniature excitatory postsynaptic current recording of primary human cortical neurons cultured 6 weeks in vitro. Neurons were transfected with mEGFP-C1 and vectors expressing mCherry, mCherry–ARHGEF7 or mCherry–RASGRF2 (n = 20, 17 or 18 neurons, respectively, from 8 cultures). Box plot centre, median; hinges, the 25th and 75th percentiles; whiskers, 1.5 × interquartile range. The P values were obtained from Kruskal–Wallis test with Dunn’s multiple comparisons test; *P < 0.05, **P < 0.01. e, Immunostaining against surface GRIA1 of dendrites from primary human cortical neurons cultured 6 weeks in vitro. Neurons were transfected with mEGFP-C1 and vectors expressing mCherry, mCherry–ARHGEF7 or mCherry-RASGRF2 (n = 14, 15 or 15 neurons, respectively, from 3 cultures; scale bar, 5 μm). Data are presented as mean values ± s.e.m. The P values were obtained from one-way ANOVA with Holm–Sidak’s multiple comparisons test; ***P < 0.001.
Next we manipulated Rho GTPase regulators in neurons to understand their role in synapse maturation. Individually overexpressing two RhoGEF proteins from the turquoise module, ARHGEF7 and RASGRF2, in human cortical neurons increased spine length and density, and promoted more immature spine types without affecting the number of DLG4+SYN1+ synapses (Fig. 4c and Extended Data Fig. 9a). Similar results were observed in mouse cortical neurons (Extended Data Fig. 9b,c). Conversely, individually knocking down two RhoGAP proteins from the blue module, ARHGAP23 and SRGAP1, partially phenocopied RhoGEF overexpression (Extended Data Fig. 9d-g and Supplementary Table 12). These morphological changes also translate into functional consequences, as the miniature excitatory postsynaptic current frequency decreased in neurons overexpressing ARHGEF7 or RASGRF2 (Fig. 4d), along with a reduction in AMPA receptor GRIA1 surface levels (Fig. 4e and Extended Data Fig. 9h). These data suggest that overexpression of specific RhoGEF proteins increases the number of filopodia and inhibits the maturation of synapses. Altogether, our results indicate that the human-specific increase in selective RhoGEF proteins delays maturation and promotes neoteny of human synapses.
PSD modules in human cognition and brain disorders
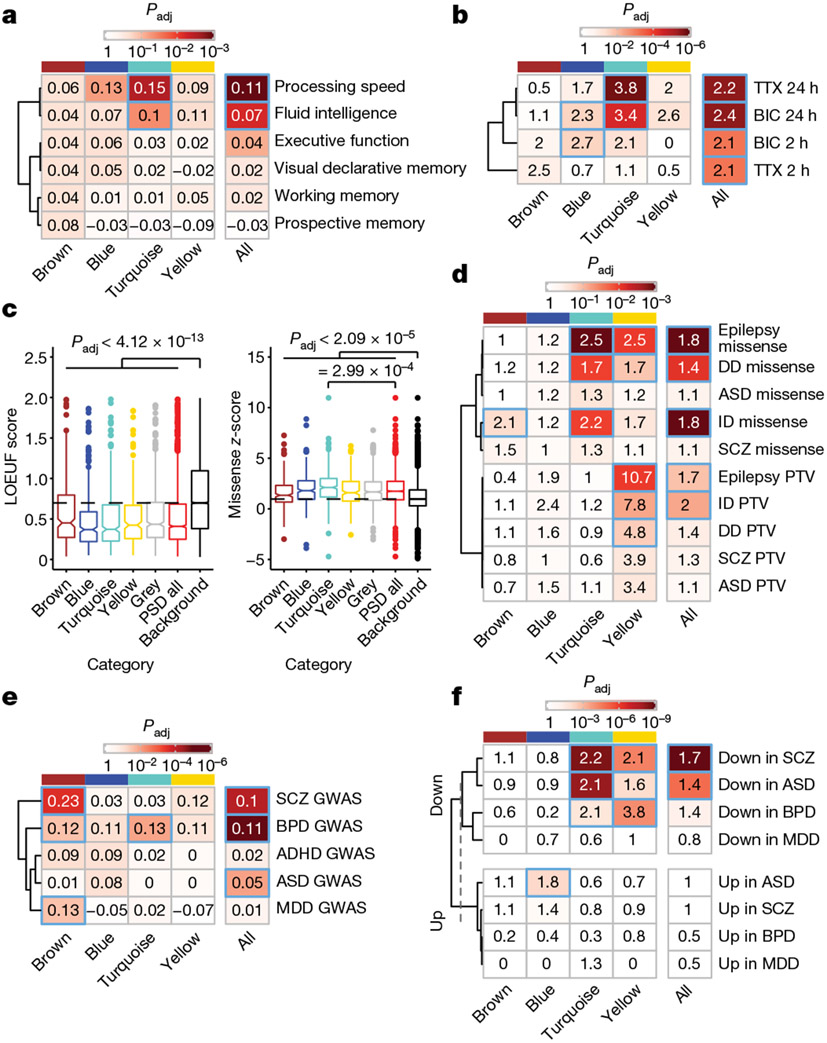
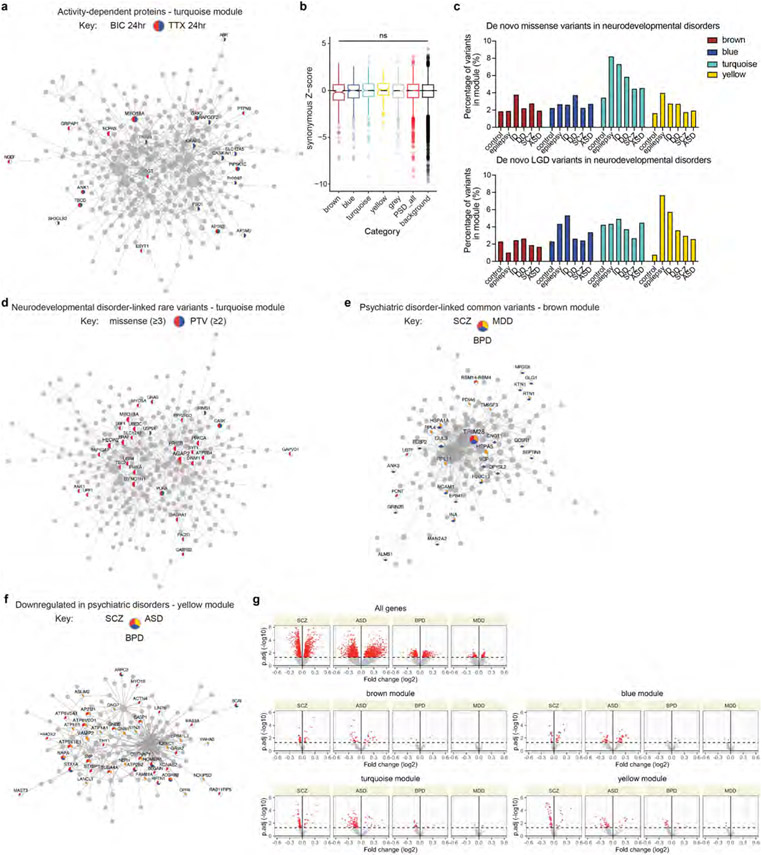
We next investigated whether genetic variants associated with human cognition converge onto human PSD modules, identifying that the turquoise module was enriched for common variants associated with processing speed and fluid intelligence (Fig. 5a). The turquoise module has its peak expression shortly after birth, at which time infants perceive a wealth of external stimuli. We posited that proteins in this module might be important for activity-dependent synaptic remodelling. Indeed, the turquoise module was highly enriched for activity-dependent proteins found in rat neurons43 (odds ratio > 3; Fig. 5b, Extended Data Fig. 10a and Supplementary Table 13). Combined with the fact that the turquoise module is more abundant in human PSDs and that RhoGEF proteins in this module promote synaptic neoteny, these results highlight the possible significance of this module in the evolutionary enhancement of human cognitive function.
Fig. 5 ∣. Association of human PSD modules with cognitive functions and brain disorders.
a, Enrichment of common variants associated with human cognitive functions in PSD modules. The numbers indicate the MAGMA linear regression coefficient β. The P values were obtained from MAGMA analysis on genome-wide association study (GWAS) summary statistics; the blue borders denote that the Benjamini–Hochberg-adjusted P value is <0.05. b, Enrichment of neuronal activity-dependent proteins in PSD modules. The numbers indicate the odds ratio. TTX, tetrodotoxin; BIC, bicuculine. The P values were obtained from hypergeometric test; the blue borders denote that the Benjamini–Hochberg-adjusted P value is <0.05. c, Distribution of gnomAD LOEUF scores and missense z-scores of genes in each category (n = 265, 313, 402, 224, 561, 1,765 and 12,892 genes). Box plot centre, median; hinges, the 25th and 75th percentiles; whiskers, 1.5 × interquartile range. The P values were obtained from Kruskal–Wallis test with Dunn’s multiple comparisons test. d, Enrichment of de novo variants associated with neurodevelopmental disorders in PSD modules. The numbers indicate the odds ratio. PTV, protein-truncating variant. The P values were obtained from hypergeometric test; the blue borders denote that the Benjamini–Hochberg-adjusted P value is <0.05. e, Enrichment of common variants associated with psychiatric disorders in PSD modules. The numbers indicate the MAGMA linear regression coefficient β. The P values were obtained from MAGMA analysis on GWAS summary statistics; the blue borders denote that the Benjamini–Hochberg-adjusted P value is <0.05. f, Enrichment of misexpressed genes after the onset of psychiatric disorders in PSD modules. The numbers indicate the odds ratio. The P values were obtained from hypergeometric test; the blue borders denote that the Benjamini–Hochberg-adjusted P value is <0.05. DD, developmental delay; ASD, autism spectrum disorder; ID, intellectual disability; SCZ, schizophrenia; BPD, bipolar disorder; ADHD, attention-deficit hyperactivity disorder; MDD, major depressive disorder.
Synaptic dysfunction contributes to both neurodevelopmental and psychiatric disorders, often caused by de novo and common variants, respectively. Regarding de novo variants, genes encoding PSD proteins were more intolerant of protein-truncating variants (lower loss-of-function observed over expected upper bound fraction (LOEUF) scores) and missense variants (higher missense z-scores) compared with background genes (Fig. 5c). Remarkably, the turquoise module was particularly intolerant of missense variants (Fig. 5c). No difference was observed for synonymous mutations (Extended Data Fig. 10b). Accordingly, we found that genes encoding PSD proteins were enriched for de novo nonsynonymous variants associated with neurodevelopmental disorders including epilepsy, developmental delay and intellectual disability (Fig. 5d, Extended Data Fig. 10c,d and Supplementary Table 13). Notably, the turquoise module had excessive missense variants, whereas the yellow module was enriched for both missense variants and protein-truncating variants. Turquoise module genes with disease-associated missense variants included ion channel genes such as KCNQ2 and SCN2A, and molecular motor genes such as DYNC1H1 and KIF1A. By contrast, many yellow module genes with protein-truncating variants encoded enzymes regulating PSD organization and postsynaptic receptor trafficking, such as SYNGAP1, IQSEC2 and CDKL5. Hence, the two modules contribute to neurodevelopmental disorders through distinct mechanisms, targeting specific stages of synapse maturation.
For psychiatric disorders, the brown module was enriched for common variants associated with schizophrenia, bipolar disorder and major depressive disorder (Fig. 5e, Extended Data Fig. 10e and Supplementary Table 13). Proteins in the brown module had peak abundance at midgestation, indicating an early aetiology involving synapse development for these adolescence- or adult-onset disorders. However, after the onset of these psychiatric disorders, genes encoding PSD proteins in the late modules (turquoise and yellow) were downregulated in brain samples from patients compared with those from unaffected individuals (Fig. 5f, Extended Data Fig. 10f,g and Supplementary Table 13). This is probably the consequence of a downstream cascade of biological events following earlier-acting genetic risk factors that disrupt synapse development.
Discussion
In this study, we generated a cross-species proteomic map of synapse development and revealed the temporal dynamics of more than 1,000 PSD proteins. We demonstrated that human PSDs undergo three major phases of maturation, with four protein modules exerting stage-, cell-type- and species-specific functions. Furthermore, we found that human PSDs develop two to three times slower than in other species, possibly owing to the higher abundance of RhoGEF proteins in the turquoise module. The turquoise module is also associated with synaptic plasticity, human cognitive function and mental disorders. Together, these data provide a blueprint for studying the molecular and evolutionary mechanisms of synapse maturation in humans.
Synapse development is regulated at both RNA and protein levels. We found that different PSD modules have distinct RNA–protein concordance, suggesting module-specific post-transcriptional and/or translational regulation. Focusing on the modules with high RNA–protein concordance, we inferred neuronal-subtype-specific PSD signatures from single-cell RNA-sequencing data. Our analysis revealed major differences in the PSD between excitatory and inhibitory neuron subtypes in the neocortex. This is consistent with previous studies showing that the composition of the PSD is diverse among neuronal subtypes44,45. Moreover, inhibitory neurons have higher levels of blue module genes, including those encoding RhoGAPs, potentially contributing to spine density differences observed between excitatory neurons and inhibitory neurons46. One limitation of this inference is that single-cell RNA sequencing does not include RNAs in dendrites. Future studies to determine the proteomic profiles of neuronal-subtype-specific PSDs will help expand these findings.
Previous studies identified a critical role of SRGAP2, a RhoGAP protein, in the human-specific delay in synapse maturation and increase in synaptic density47,48. It has been shown that the Rac1-GAP activity of the ancestral protein SRGAP2A limits spine neck length and density in neocortical neurons. Human-specific partial duplications of SRGAP2 inhibited the function of SRGAP2A, resulting in longer spine necks and higher spine density in humans. We found increased RhoGEF proteins in human PSDs and that overexpression of RhoGEFs in neurons increased spine density and delayed synapse maturation. Given the antagonistic roles of RhoGAP and RhoGEF proteins, our results are consistent with previous findings and suggest that increased synaptic Rho GTPase activity contributes to the neoteny of human synapses. Moreover, RhoGEF proteins are enriched in the turquoise module associated with human cognitive function. Thus, our analysis provides molecular evidence that links synaptic neoteny to the evolution of human cognition.
There are several limitations to this study. The isolation of PSDs by subcellular fractionation may include contaminants. We therefore applied rigorous data filtering to minimize this effect. Additional independent validation will help confirm new PSD components. Additionally, isoforms and alternative splicing play a role in synapse development, but our analysis did not quantify isoforms owing to technical constraints. Furthermore, PSDs are heterogeneous and probably different between individual synapses, yet our data represent the averaged profiles at the bulk tissue level. Last, the enrichment of activity-dependent proteins in the turquoise module was based on experimental data from rat neurons, not human43. With the development of new methods, future studies determining developmental and activity-dependent changes of the synaptic proteome at the isoform level across different brain regions, cell types and species will provide further insight into the mechanisms of brain development, evolution and disease.
Methods
Brain tissue samples
Acquisition of all second-trimester primary human tissue samples (Supplementary Table 1a) was approved by the Human Gamete, Embryo and Stem Cell Research Committee at the University of California San Francisco (study number 10-05113). All experiments were carried out in accordance with protocol guidelines. Informed consent was obtained before sample collection and use for this study. After tissue acquisition, the cortical plate and subplate were dissected and frozen at −80 °C.
Thirty-four de-identified snap-frozen postmortem PFC tissue samples without known neurological disorders were obtained from the University of Maryland Brain and Tissue Bank through NIH NeuroBioBank and the Pediatric Brain Tissue Bank at UCSF (Supplementary Table 1a).
Twenty-two de-identified snap-frozen postmortem V1 tissue samples without known neurological disorders were obtained from the University of Maryland Brain and Tissue Bank through NIH NeuroBioBank (Supplementary Table 1b).
Three adult cortical samples were obtained from neurosurgical operations (Supplementary Table 1c). In two cases, the reason for surgery was tumour resection and in one, focal cortical dysplasia type 2b treatment. In all cases, the tissue samples were collected from peripheral regions removed together with the affected regions. The samples were snap-frozen in liquid nitrogen and stored at −80 °C before further processing.
Macaque samples were obtained from Oregon National Primate Research Center, Southwest National Primate Research Center, Wisconsin National Primate Research Center, Dr Alex Pollen at UCSF and Dr Nenad Sestan at Yale University (Supplementary Table 8a).
Mouse experiments were approved by UCSF Institutional Animal Care and Use Committee and carried out in accordance with relevant institutional guidelines. Mice were housed under a standard 12:12 light/dark cycle with humidity of 30–70% and temperature of 68–79 °F. Neocortices from C57BL/6J mice were dissected and frozen at −80 °C (Supplementary Table 8b).
Subcellular fractionation and PSD isolation
Subcellular fractionation and PSD isolation were carried out as previously described27. Briefly, about 300 to 2,000 mg of human brain tissue (dependent on the age group) was thawed on ice and cut into small pieces. It was then homogenized using a 10-ml tissue grinder in 3 ml homogenization buffer (0.32 M sucrose, 2 mM EDTA and 4 mM HEPES, pH 7.4) with freshly added protease and phosphatase inhibitors (Roche). Postnuclear supernatants were obtained through a 1,000g spin. The supernatant (S1) was spun at 10,000g for 15 min, and the pelleted membranes were resuspended in 10 ml homogenization buffer and spun again to obtain P2′. This pellet was lysed by 10 ml hypoosmotic shock in 4 mM HEPES (pH 7.4) buffer and incubated for 30 min at 4 °C. This was followed by a spin at 25,000g for 20 min. The pellet (P3) was layered onto a 0.8/1.0/1.2 M discontinuous sucrose gradient and spun at 150,000g for 2 h. Synaptic plasma membranes were recovered from the 1.0/1.2 M interface and pelleted. These membranes were extracted with 3 ml 0.5% Triton X-100 for 15 min and pelleted with a 32,000g spin for 20 min (PSD I). For mass spectrometry and western blot analysis, fractions obtained by subcellular fractionation were lysed by sonication. Protein concentrations were determined using the BCA assay (Pierce).
Macaque and mouse PSD samples were prepared in the same way.
Western blot
LDS sample buffer (Invitrogen) with a reducing agent was added to each protein lysate, followed by a 10-min incubation at 95 °C. Samples were spun down and electrophoresed on a 4–12% Bis-Tris gel and transferred to a nitrocellulose membrane. Total protein was quantified by Revert Total Protein Stains (LI-COR). The membrane was then blocked for 1 h with Intercept (TBS) blocking buffer (LI-COR) before primary antibody incubation overnight. After secondary antibody incubation, the membrane was imaged using Odyssey Classic or Odyssey CLx (LI-COR).
The following antibodies were used: GRIN2B (Abcam, ab93610, 1:1,000), DLG2 (NeuroMab, 75–284, 1:1,000), GRIA1 (Millipore, AB1504, 1:1,000), DLG4 (NeuroMab, 75-028, 1:1,000), SYP (Sigma, S5768, 1:1,000), GAPDH (Cell Signaling Technology, 2118S, 1:1,000), ARHGEF7 (Sigma, 07-1450-I, 1:1,000), PREX1 (Sigma, HPA001927-100UL, 1:1,000), β-tubulin (Cell Signaling Technology, 2146S, 1:1,000), Phospho-PAK (Cell Signaling Technology, 2606S, 1:1,000), PAK (Cell Signaling Technology, 2604S, 1:1,000), phospho-CFL (Cell Signaling Technology, 3311S, 1:1,000) and CFL (Cell Signaling Technology, 5175S, 1:1,000).
Transmission electron microscopy
Human PSD I pellets prepared from one GW23 sample were fixed with 3% glutaraldehyde (Electron Microscopy Sciences (EMS)) for 10 min at 4 °C, washed three times with 0.1 M phosphate buffer and post-fixed with 2% osmium tetroxide (EMS) in 0.1 M phosphate buffer for 1 h at room temperature. Dehydration in graded series of ethanol (30, 50 and 70%) was followed by 2% uranyl acetate (EMS) incubation for 2.5 h. Samples were rinsed with 70%, 96% and 100% ethanol, washed two times with propylene oxide (EMS), embedded in Durcupan resin (Sigma-Aldrich) and allowed to polymerize at 69 °C for 72 h. Ultrathin sections (60–70 nm) were sectioned with a diamond knife (DiATOME) in a UC7 ultramicrotome (Leica), stained with lead citrate and examined under a transmission electron microscope (Tecnai Spirit G2, FEI) using a digital camera (Morada, Soft Imaging System, Olympus).
Sample quality control, digestion and liquid chromatography with tandem mass spectrometry analysis
The quality of sample synaptic proteomes was checked before mass spectrometry analysis using the human synapse proteome integrity ratio (HUSPIR)28. PSD samples with HUSPIR > 2 were deemed good, and those with 1 < HUSPIR < 2 were deemed fair. Samples with HUSPIR ≤ 1 were excluded from further analysis.
Five micrograms of isolated PSD samples was electrophoresed on a 4–12% Bis-Tris gel for 20 min. Proteins were visualized by Bio-Safe Coomassie stain (BIO-RAD) and excised from the gel. An individual gel piece was subjected to in-gel digestion using trypsin (Promega). The resulting dried peptides were analysed in technical duplicate on a ThermoFisher Orbitrap Fusion Lumos Tribrid mass spectrometry system equipped with an EASY-nLC 1200 ultrahigh-pressure liquid chromatography system interfaced via a Nanospray Flex nanoelectrospray source. Samples were injected into a C18 reverse-phase column (25 cm × 75 μm packed with ReprosilPur C18 AQ 1.9 μm particles). Peptides were separated by an organic gradient from 5 to 30% ACN in 0.02% heptafluorobutyric acid over 180 min at a flow rate of 300 nl min−1 for the phosphorylated peptides or unmodified peptides for global abundance. Spectra were continuously acquired in a data-dependent manner throughout the gradient, acquiring a full scan in the Orbitrap (at 120,000 resolution with an AGC target of 400,000 and a maximum injection time of 50 ms) followed by as many tandem mass spectrometry scans as could be acquired on the most abundant ions in 3 s in the dual linear ion trap (rapid scan type with an intensity threshold of 5,000, HCD collision energy of 32%, AGC target of 10,000, maximum injection time of 30 ms and isolation width of 0.7 m/z). Single and unassigned charge states were rejected. Dynamic exclusion was enabled with a repeat count of 2, an exclusion duration of 20 s and an exclusion mass width of ±10 ppm.
Protein identification, quantification and potential contaminant removal
Protein identification and quantification were carried out using MaxQuant v1.6.11.0 (ref. 49). Spectra from the human, macaque and mouse raw files were matched to the reference proteomes from UniProt (Homo sapiens UP000005640_9606, Macaca mulatta UP000006718_9544 and Mus musculus UP000000589_10090, respectively). Default settings of MaxQuant with FDR = 0.01 were used except that “Match between runs” was enabled to improve proteome coverage with an alignment window of 20 min and a match time window of 0.7 min. The intensity-based absolute quantification (iBAQ) value for each protein group was calculated50.
Potential external contaminants including keratins and proteins known to be localized at mitochondria51, a principal contaminant in PSD preparation52, were excluded. Moreover, because some identified proteins could be non-PSD proteins artificially bound to the PSD in the postmortem condition42, we curated a list of proteins that have been identified in non-postmortem brain tissues by combining our data and data from ref. 10. This list was used to filter the identified PSD proteins so that those present only in postmortem brain samples were excluded. After contaminant filtration, the remaining proteins were considered present in an age group if more than half of the samples in that group had the protein identified by tandem mass spectrometry.
Data normalization, imputation and quality effect correction
Our initial analysis focused on the human PFC dataset consisting of the 20 cortical samples without area information in the second trimester and 34 PFC samples from the third trimester to young adulthood. After filtering out potential contaminants, the iBAQ values of the remaining proteins were used to calculate the normalized molar intensity relative iBAQ (riBAQ)53:
The riBAQ values were multiplied by a scale factor of 106 and log2-transformed to obtain the abundance values for each protein. After log-transformation, 1,765 human PSD proteins missing in fewer than three samples in any age group (either by tandem mass spectrometry or by matching) were included for further analysis. Next, abundance values were normalized by variance-stabilizing transformation, and missing values were imputed using the MinProb method (), both of which were implemented using the R package DEP54.
Linear models combined with empirical Bayes methods were used for the differential abundance analysis of the human samples. We accounted for the fact that synaptic proteome integrity could affect the abundance values. The abundance , for a given protein in sample was modelled using the following linear model:
where is the intercept value, is sample ’s age group, is sample ’s quality (HUSPIR category good or fair), and are protein ’s estimated regression coefficients of the respective covariates, and is the model error. This was carried out using the R package limma55. After the model was fitted, a pairwise comparison was made to identify proteins with differential abundance between two age groups. Proteins with a log2[fold change] of at least one and an adjusted P value56 of the moderated t-test of less than 0.05 were selected as proteins with differential abundance. The effect of quality was removed using the removeBatchEffect function in limma. The final corrected abundance matrix can be found in Supplementary Table 2a.
A similar differential abundance analysis was carried out with the macaque and mouse samples, except that the quality covariate was removed in the linear model. The final abundance matrix can be found in Supplementary Tables 9a and 10a.
A similar differential abundance analysis was carried out with the human V1 dataset consisting of the same 20 cortical samples without area information in the second trimester as the PFC dataset and 22 V1 samples from the third trimester to young adulthood. Variance-stabilizing transformation was carried out using the same model from the PFC dataset. The final corrected abundance matrix after applying the removeBatchEffect function can be found in Supplementary Table 5a.
Dimensionality reduction and clustering
Dimensionality reduction was carried out by the principal component analysis. The first two principal components were used for the principal component analysis plots. Heat maps were generated using the R package ComplexHeatmap57. Samples and proteins were clustered on the basis of the Spearman correlation distance.
Variance partitioning
Variance partitioning was carried out using the R package variance-Partition58. Age group, processing batch, PSD quality and sex of the sample donor were included in the formula.
GSEA using MSigDB gene sets
Age specificity for each protein was calculated by comparing samples within an age group to all other samples outside the age group using linear models combined with empirical Bayes methods implemented in limma. The resulting moderated t statistics of each protein were ranked and used as input for GSEA59 using the R package clusterProfiler60. GSEA was carried out against the MSigDB C2 canonical pathways, which contain curated gene sets representing different molecular pathways61. Only pathway sets with gene numbers between 10 and 500 were used for the analysis.
Immunohistochemistry and confocal imaging
Prenatal human tissue samples and mouse tissue samples were fixed in 4% paraformaldehyde in PBS at 4 °C overnight. The samples were cryoprotected in 15% and 30% sucrose in PBS and frozen in OCT. Samples were sectioned at a thickness of 15 μm, air dried and rehydrated in PBS. Postnatal unfixed frozen human tissue samples were sectioned at a thickness of 15 μm, air dried, fixed in 4% paraformaldehyde in PBS for 10 min and washed three times with PBS. Antigen retrieval was carried out using citrate-based Antigen Unmasking Solution (Vector Laboratory) at 95 °C for 10 min. The slides were then washed in PBS and blocked in PBS-based blocking buffer containing 10% donkey serum, 0.2% gelatin and 0.1% Triton X-100 at room temperature for 1 h. After blocking, slides were incubated with primary antibodies in the blocking buffer at 4 °C overnight. The slides were washed in PBS three times and incubated with secondary antibodies in the blocking buffer at room temperature for 2 h. The slides were then washed in PBS twice, counterstained with DAPI and washed in PBS once more. Slides were mounted with coverslips with ProLong Gold (Invitrogen). Confocal images were acquired with a Leica TCS SP8 using a 63× oil immersion objective. Acquired images were processed using Imaris v9.7 (Oxford Instruments) and Fiji/ImageJ v1.54 (ref. 62). The following antibodies were used: DLG4 (NeuroMab, 75-028, 1:250), DLG4 (Synaptic Systems, N3702-At488-L, 1:250), RPS6 (Cell Signaling Technology, 2217S, 1:200), CTNNB1 (Cell Signaling Technology, 8480S, 1:100), GDI1 (Proteintech, 10249-1-AP, 1:100), CFL1 (Cell Signaling Technology, 5175S, 1:250), DBN1 (Abcam, ab11068, 1:200), DNBL (Proteintech, 13015-1-AP, 1:100) and ARHGEF7 (Sigma, 07-1450-I, 1:200).
Colocalization analysis was carried out using Imaris. Protein puncta were identified on the basis of signal intensity and local contrast. Puncta of two different proteins within 0.5 μm were considered colocalized. DBN1, DBNL and ARHGEF7 abundances at the PSD were quantified using Imaris. The DLG4 puncta were identified on the basis of signal intensity and local contrast. These DLG4 puncta, deemed as the PSD loci, were used to create a mask channel. Intensities of DBN1, DBNL or ARHGEF7 were then quantified within the mask channel.
Weighted gene coexpression network analysis
WGCNA was carried out using the R package WGCNA31.
For the human samples, the blockwiseModules function (power = 20, corType = “pearson”, networkType = “signed”, deepSplit = 2, minModuleSize = 45, reassignThreshold =1 × 10−6, mergeCutHeight = 0.15, min-KMEtoStay = 0.3, numericLabels = F, pamRespectsDendro = F) was used to build a signed weighted correlation network. In total, four modules plus the grey module for unassigned proteins were identified. Module memberships and module eigengene values of the samples are available in Supplementary Table 3.
Protein–protein interaction enrichment analysis by Monte Carlo permutation tests
The protein–protein interaction (PPI) network consisting of interactions between human proteins and those between mouse proteins was downloaded from the BioGRID database (https://thebiogrid.org)63. We used the PPI network consisting of all human PSD proteins as the background PPI network. For each module, we constructed the module protein interaction network by extracting the interactions connecting all of the proteins in the module and calculated the observed number of interactions. We then randomly sampled the same number of proteins with similar degree distribution from the background PPI network and calculated the number of interactions in the random network. We repeated this randomization process 100,000 times and calculated the P value as the fraction of the random numbers of interactions that are greater than the observed number of interactions.
Statistical overrepresentation test using MSigDB gene sets
The one-sided hypergeometric test implemented in clusterProfiler was used to identify overrepresented pathways in each protein module. The MSigDB C2 canonical pathways were used as input gene sets. The union of genes encoding PSD proteins and those expressed in the human neocortex curated using the BrainSpan RNA-sequencing (RNA-seq) data was used as the background. To define genes expressed in the human neocortex, we downloaded the developing human brain RNA-seq data from the BrainSpan database (http://www.brainspan.org). The neocortical samples of the BrainSpan data that have RNA integrity numbers ≥8 were temporally divided into prenatal and postnatal stages. Genes with RPKM ≥ 1 in at least half of the neocortical samples at either stage were defined as genes expressed in the human neocortex. Only pathway sets with gene numbers between 10 and 500 were used for the analysis.
SynGO enrichment analysis
SynGO (release 1.1) enrichment analysis was carried out using the online tool at https://syngoportal.org (ref. 32). We used the same background gene list as the above statistical overrepresentation test.
PPI–co-abundance network analysis and visualization
The pairwise topological overlap similarity scores of all human PSD protein pairs were calculated by WGCNA. Protein pairs with topological overlap similarity scores in the top 10% were considered coexpressed. Protein pairs that are coexpressed and interacting with each other in the BioGRID database were deemed connected in the PPI–co-abundance network. Subnetworks were generated within each module. All network plots were constructed using Cytoscape 3.8.2 (ref. 64). The shortest path lengths between proteins were calculated using igraph65. The average length within pathway proteins was compared to that between the pathway proteins and non-pathway proteins in the network by the one-sided Wilcoxon rank-sum test.
Protein domain analysis
Protein domain information of all PSD proteins was downloaded from the SMART database (https://smart.embl.de)66 and is summarized in Supplementary Table 4a. Domains present in more than six PSD proteins were included in Fig. 2c and Extended Data Fig. 4a. Domains were clustered on the basis of the Pearson correlation distance in Fig. 2c and Jaccard distance in Extended Data Fig. 4a.
RhoGAP and RhoGEF specificity
The target specificity of individual RhoGAPs and RhoGEFs was hand-annotated on the basis of a literature review. The annotation results are summarized in Supplementary Table 4b,c.
Comparison between PSD proteomic data and bulk transcriptomic data
The BrainSpan human brain bulk RNA-seq data were downloaded from the PsychENCODE website (http://evolution.psychencode.org)67. Neocortical samples older than GW16 were included for analysis. Transcripts were filtered to include only those encoding PSD proteins and normalized to obtain the TPM values. log2(TPM + 1) values were used for downstream analysis.
Module preservation analysis was carried out using the module-Preservation function in WGCNA34. We carried out the permutation 500 times to obtain the zsummary statistics and composite module preservation statistic medianRank.
To estimate the correlation at the individual PSD protein level between the proteomic data and transcriptomic data, we first imputed the transcriptomic profiles of human cortical samples used to generate our proteomic data. To do so, we trained a generalized additive model by regressing the transcriptome of human cortical samples against their ages using the R package mgcv 1.8-40 (ref. 68). We then predicted the corresponding transcriptomic profiles of our PSD samples on the basis of their ages and calculated the Spearman correlation coefficients between RNA and protein of individual PSD proteins (Supplementary Table 6). The Kruskal–Wallis rank-sum test with post hoc Dunn’s test was used to compare differences in correlation coefficients between PSD modules.
ChEA3 transcription factor enrichment analysis
ChEA3 analysis was carried out using the online tool at https://maayanlab.cloud/chea3 (ref. 69). Results are listed in Supplementary Table 6d,e.
Developing human brain single-nucleus RNA-seq data analysis
Single-nucleus RNA-seq data from developing human neocortex35 were downloaded from the UCSC Cell Browser (https://cells.ucsc.edu/?ds=human-cortical-dev+all)70. Uniform manifold approximation and projection coordinates from the original authors were used. The identity of specific lineages and cell types was reannotated on the basis of the expression of known marker genes (Extended Data Fig. 6). Only excitatory intratelencephalic neurons, excitatory non-intratelencephalic neurons, inhibitory neurons derived from the medial ganglionic eminence and inhibitory neurons derived from the caudal ganglionic eminence were included for further analysis. Pseudobulk samples were constructed by aggregating the raw counts of all the cells within the same cell type. Counts for genes encoding PSD proteins were extracted from the peudobulk data, normalized for sequencing depth and log-transformed to obtain the log2[CPM + 1] values (Supplementary Table 7a). Gene expression levels were standardized by z-transformation across pseudobulk samples and are summarized in Fig. 2j.
Adult human brain single-cell RNA-seq data analysis
Single-cell RNA-seq data from the adult human motor cortex36 were downloaded from the UCSC Cell Browser (https://cells.ucsc.edu/?ds=allen-celltypes+human-cortex+m1)70. Only neocortical neurons were included for further analysis. Pseudobulk samples were constructed by aggregating the raw counts of all the cells within the same neuronal subtype annotated by the original authors. Counts for genes encoding PSD proteins were extracted from the peudobulk data, normalized for sequencing depth, and log-transformed to obtain the log2[CPM + 1] values (Supplementary Table 7b). Counts for transcription factors regulating PSD modules were extracted from the original pseudobulk data without renormalizing for sequencing depth (Supplementary Table 7c,d). Gene expression levels were standardized by z-transformation across pseudobulk samples and are summarized in Extended Data Fig. 6d,e.
Correlation analysis between species and regions
The orthologues of genes encoding human PSD proteins were obtained from Ensembl. Only PSD proteins with one-on-one orthologues present in the PSD of all three species (854 proteins) were included for further analysis. The riBAQ values of the filtered data were renormalized to obtain updated riBAQ values comparable between all species and regions (Supplementary Table 11a). Pairwise correlations between samples were obtained by calculating the Pearson correlation coefficients. The detailed results can be found in Supplementary Table 11.
Prediction of equivalent human PSD ages and comparison of PSD maturation rates
To predict equivalent human PSD ages, we log-transformed the age (post-conceptional days) of all samples in the human PFC dataset and then trained a regularized linear model by regressing the transformed ages against the riBAQ values of all PSD proteins. Specifically, ridge regression was carried out using the R package glmnet71 with λ (equals 0.4466836) selected by tenfold cross-validation. The equivalent human PSD ages for macaque and mouse samples were then predicted using the trained model and are listed in Supplementary Table 11b.
The predicted PSD ages of human, macaque and mouse samples were regressed against the real post-conceptional ages using a linear regression model to obtain the slope coefficients as an estimator of the PSD maturation rate. For the macaque samples, because the PSD maturation rate seems to be different before and after the age of 1 year, the predicted PSD ages were regressed against the real post-conceptional ages using a linear spline model with the knot set at post-conceptional day 330 using the R package lspline.
Similar predictions were made based on the human V1 dataset. For ridge regression, λ (equals 0.7943282) was selected by tenfold cross-validation. The equivalent human PSD ages based on the human V1 dataset are listed in Supplementary Table 11c.
Primary neuronal culture
Primary human cortical neurons were prepared from GW21 to GW23 human dorsal cortical tissue samples. The cortical plate and subplate were dissected and dissociated using the Papain Dissociation System (Worthington Biochemical). Dissociated neurons were resuspended in a plating medium (Neurobasal medium supplemented with 1× B27, 2 mM GlutaMAX and antibiotics) and plated into tissue culture plates coated with PEI–laminin or containing a 12-mm coverslip precoated with PDL and laminin (Corning 354087) at the density of 100,000 cells cm−2. Cells were cultured in a humidified incubator with 5% CO2 and 8% O2. On day in vitro (DIV) 1, the medium was changed to maturation medium (BrainPhys medium supplemented with 1× B27, 1× N2, 20 ng ml−1 BDNF, 20 ng ml−1 GDNF, 1 mM dibutyryl-cAMP, 200 nM ascorbic acid, 1 μg ml−1 laminin and antibiotics). Half of the medium was changed with fresh medium every 3–4 days until collection.
Primary mouse cortical neurons were prepared from P0 C57BL6/J mice. The neocortices were dissected, dissociated and plated using the same procedure as for primary human neurons.
Synaptosome preparation from cultured primary neurons
About two million neurons were collected at the indicated DIV in 2 ml homogenization buffer (0.32 M sucrose, 2 mM EDTA and 4 mM HEPES, pH 7.4) with freshly added protease and phosphatase inhibitors (Roche) and homogenized using a 10-ml tissue grinder. Postnuclear supernatants were obtained through a 1,000g spin. The supernatant (S1) was spun at 10,000g for 15 min, and the pelleted membranes were resuspended in 1 ml of 0.8 M sucrose buffer and spun at 20,000g for 30 min. The pellet (synaptosomes) was resuspended in the homogenization buffer and stored at −80 °C before western blot analysis. Protein concentrations were determined using the BCA assay (Pierce).
Plasmid cloning
pmCherry-C1 (632524, Clontech) was used as the backbone and control vector for the overexpression experiments. mCherry–ARHGEF7 was a gift from Dorus Gadella (Addgene #129611). The cDNA of RASGRF2 was cloned into pmCherry-C1 from R777-E241 Hs.RASGRF2, a gift from Dominic Esposito (Addgene #70525). mEGFP-C1 was a gift from M. Davidson (Addgene #54759). pLKO-RFP-shCntrl, a gift from William Kaelin (Addgene #69040), was used as the backbone and control vector for the knockdown experiments. Sequences of short hairpin RNAs (shRNAs) against ARHGAP23 or SRGAP1 can be found in Supplementary Table 12a.
Validation of shRNA knockdown efficiency
HEK293T (ATCC CRL-3216) cells cultured in Dulbecco’s modified Eagle medium (Corning) containing 10% fetal bovine serum (Hyclone) and antibiotics (penicillin–streptomycin) were plated in 12-well plates. The next day, 1 μg of corresponding shRNA vectors was transfected into the cells using Lipofectamine 3000 (Invitrogen). Cells were collected 48 h after transfection for mRNA extraction using the RNAeasy mini plus kit (Qiagen). RNA quantity and quality were checked using NanoDrop 1000 (Thermo Scientific). Quantitative PCR with reverse transcription was carried out using the ViiA 7 Real-time PCR System with PowerUp SYBR Green Master Mix and analysed with the comparative Ct method normalized against the housekeeping gene GAPDH. Primers used are listed in Supplementary Table 12b. HEK293T cells are sometimes contaminated by HeLa cells. As our purpose is to validate shRNA efficiency, the identity of the line is not critical. HEK293T cells used in this study were not authenticated or tested for mycoplasma contamination.
Plasmid DNA transfection into primary neurons and immunocytochemistry
Human neurons were transfected on DIV28 with mEGFP-C1 (0.3 μg per well) plus vectors expressing mCherry, mCherry–ARHGEF7, mCherry–RASGRF2, tRFP–shControl, tRFP–shARHGAP23–1, tRFP–shARHGAP23–2, tRFP–shSRGAP1–1 or tRFP–shSRGAP1–2 (0.7 μg per well) with Lipofectamine 2000. On DIV42, human neurons were fixed with 4% formaldehyde and 4% sucrose in PBS and permeabilized and blocked with PBS-based blocking buffer containing 10% donkey serum, 0.2% gelatin and 0.1% Triton X-100 at room temperature for 1 h. Samples were then incubated with primary antibodies at 4 °C overnight. The next day, samples were washed in PBS three times and incubated with secondary antibodies in the blocking buffer at room temperature for 1 h. Samples were then washed in PBS twice, counterstained with DAPI and washed in PBS once more. z-stack images were acquired with a Leica TCS SP8 using a 63× oil immersion objective. Dendritic spine analysis was carried out using Imaris (Oxford Instruments). Spine density and morphology were measured from secondary or tertiary dendrites. Automatic spine classification was carried out using the Classify Spines Xtension in Imaris with the following rules: Stubby–length(spine) < 1 μm; Mushroom–length(spine) < 3 μm and max_width(head) > mean_width(neck) × 2; Long Thin–length(spine) < 3 μ m but not Mushroom; Filopodia–length(spine) ≥3 μm. Mouse neurons were transfected on DIV5 and analysed on DIV8 in a similar manner.
For surface GRIA1 staining, after fixation, neurons were blocked by PBS-based blocking buffer containing 10% donkey serum and 0.2% gelatin at room temperature for 1 h. Samples were then incubated with primary antibodies against extracellular GRIA1 under the non-permeabilized condition at room temperature for 1 h. Next, samples were washed in PBS three times and permeabilized and blocked with blocking buffer containing 0.1% Triton X-100 at room temperature for 1 h. Samples were then incubated with primary antibodies against intracellular proteins at 4 °C overnight. The rest of the steps of the procedure were the same as mentioned above.
The following antibodies were used: GFP (Aves, GFP-1020, 1:1,000), mCherry (Invitrogen, M11217, 1:500), SYN1 (Cell Signaling Technology, 5297S, 1:250), DLG4 (Synaptic Systems, N3702-AF647-L, 1:250), tRFP (OriGene, TA150061, 1:250), GRIA1 (Alomone, AGC-004, 1:25).
Whole-cell recording of primary human cortical neurons
Human neurons transfected with pEGFP-C1 plus vectors expressing mCherry, mCherry–ARHGEF7 or mCherry–RASGRF2 were recorded between DIV42 and DIV49. Whole-cell recordings were carried out in maturation medium supplemented with 1 mM tetrodotoxin at room temperature. The voltage was set at −60 mV. Recording pipettes (4–10 mOhm) were filled with intrapipette solution containing 140 mM K-gluconate, 2 mM MgCl2, 10 mM HEPES, 0.2 mM EGTA, 4 mM MgATP, 0.3 mM NaGTP, 10 mM phosphocreatine d-tris and 0.25% biocytin. Signals were collected at a sampling rate of 10 kHz, using a 10K Bessel filter (MultiClamp 700B, Axon Instruments, Molecular Devices) and digitized (Digidata, Axon Instruments, Molecular Devices). Putative miniature excitatory postsynaptic current events were extracted in the open-source software Stimfit 0.15.8 (https://github.com/neurodroid/stimfit).
MAGMA analysis of PSD modules using GWAS data of human cognitive functions
MAGMA v1.09 (ref. 72) was used to determine whether human PSD modules are enriched for common variants associated with human cognitive function.
GWAS summary statistics for human cognitive function studies conducted by the UK Biobank were downloaded from the GWAS ATLAS resource (https://atlas.ctglab.nl)73. We analysed GWAS data of Reaction Time Test for processing speed, Fluid Intelligence Test for fluid intelligence, Trail Making Test Part B for executive function, Pairs Matching Test for visual declarative memory, Numeric Memory Test for working memory, and Prospective Memory Test for prospective memory.
For gene analysis in MAGMA, the 1000 Genomes European panel was used to estimate linkage disequilibrium between single nucleotide polymorphisms, and the single nucleotide polymorphism-wise mean model was used as the gene analysis model. For gene-set analysis, several common technical confounders are included in the linear regression model as covariates. These confounders include gene size, gene density, the inverse of the mean minor allele count of variants, and their log-transformed values. The resulting nominal P values were adjusted using the Benjamini–Hochberg method56. The detailed results can be found in Supplementary Table 13a.
Module enrichment of activity-dependent proteins
Lists of proteins whose abundances depend on neuronal activities were obtained from ref. 43. The rat protein list was converted to human genes using orthologue data obtained from the Alliance of Genome Resources (https://www.alliancegenome.org). The one-sided hypergeometric test was used to determine whether there was a significant overlap with individual PSD modules. The union of genes encoding PSD proteins and those expressed in the human neocortex was used as the background. The resulting nominal P values were adjusted using the Benjamini–Hochberg method. The detailed results can be found in Supplementary Table 13b.
Gene constraint analysis
LOEUF scores, missense z-scores and synonymous z-scores for all human genes were downloaded from gnomAD v2.1.1 (https://gnomad.broadinstitute.org)74. The union of genes encoding PSD proteins and those expressed in the human neocortex was used as the background. Data were plotted using the geom_boxplot() function from the R package ggplot2 with default settings for elements. The Kruskal–Wallis rank-sum test with post hoc Dunn’s test was used to compare differences in scores between each category of genes.
Module enrichment of de novo variants associated with neurodevelopmental disorders
De novo variants in neurodevelopmental disorders, including epilepsy, developmental delay, autism spectrum disorder, intellectual disability and schizophrenia, were obtained from denovo-db v.1.6.1 (https://denovo-db.gs.washington.edu/denovo-db)75. We also included one additional epilepsy dataset76 and two additional schizophrenia datasets77,78 not included in the denovo-db database. We defined “stop-gained”, “start-lost”, “stop-gained-near-splice”, “frameshift”, “frameshift-near-splice”, “splice-donor” and “splice-acceptor” mutations as PTVs and “missense” and “missense-near-splice” mutations as missense variants. The number of PTVs or missense variants for a gene in a disorder was defined as the number of individuals with the disorder harbouring PTVs or missense variants in the gene. The one-sided Fisher’s exact test was used to determine whether there is a significant enrichment of de novo variants in genes within individual PSD modules compared with those outside the module. The resulting nominal P values were adjusted using the Benjamini–Hochberg method. The summarized results can be found in Supplementary Table 13c.
MAGMA analysis of PSD modules using GWAS data of human psychiatric disorders
GWAS summary statistics for schizophrenia (June 2018 release), bipolar disorder (June 2018 release), autism spectrum disorder (November 2017 release), major depressive disorder (mdd2019edinburgh) and attention-deficit hyperactivity disorder (January 2022 release) were downloaded from The Psychiatric Genomics Consortium database (https://www.med.unc.edu/pgc)79-82. MAGMA analysis was carried out in the same way as described above. The detailed results can be found in Supplementary Table 13d.
Module enrichment of misexpressed genes after the onset of psychiatric disorders
Gene expression data of brain samples from patients with schizophrenia, bipolar disorder, autism spectrum disorder or major depressive disorder and unaffected individuals were obtained from ref. 83. Genes with an adjusted P value less than 0.05 were considered misexpressed genes in a psychiatric disorder. The summary can be found in Supplementary Table 13e. The one-sided hypergeometric test was used to determine whether there was a significant overlap with individual PSD modules. The union of genes encoding PSD proteins and those expressed in the human neocortex was used as the background. The resulting nominal P values were adjusted using the Benjamini–Hochberg method.
Extended Data
Extended Data Fig. 1 ∣. Isolation of PSDs from immature and mature human cortices.
a, Western blot analysis of different subcellular fractions of a GW23 sample demonstrating enrichment of PSD proteins and depletion of presynaptic SYP and cytoplasmic GAPDH in the PSD fraction. Enrichment of PSD proteins after fractionation was validated for all samples subjected to mass spectrometry analysis. b, Electron micrographs of the PSD fraction isolated from a GW23 sample (scale bar: 200 nm). Arrows denote structures resembling the PSD. The experiment was performed on one sample. c, Western blot analysis of purified PSDs from different age groups demonstrating changes in GRIN2B and DLG4 during development. The experiment was performed once. d, Correlation between PSD yield and synapse number of developing human prefrontal cortex. e, Venn diagram showing the overlap between Year18_22 samples in this study and the human PSD proteomes published in Roy et al., 2017 and Bayés et al., 2011. f, UpSet plot describing the number of identified proteins and their overlaps at each age group. g, PCA plots of the samples colored by various covariates. h, Variance explained by individual covariates (n = 1765 proteins). Boxplot center: median; hinges: the 25th and 75th percentiles; whiskers: 1.5 × inter-quartile range. i, Abundance patterns of GRIN2A, GRIN2B, DLG3, and DLG4. j, Colocalization of RPS6, CTNNB1, GDI1, or CFL1 with DLG4 in second-trimester human neocortex (n = 5, 5, 5 and 5 samples, scale bar: 10 μm or 5 μm as indicated in the figure). Data are presented as mean values ± s.e.m.
Extended Data Fig. 2 ∣. Examples of PSD protein abundance patterns in human cortical development.
a, Abundance patterns of representative PSD proteins. b, Examples of PSD protein paralogs that undergo reciprocal developmental changes. c, Abundance patterns of DBN1 and DBNL. d, Immunofluorescent intensity of DBN1 and DBNL at DLG4 loci in human neocortex (n = 3 and 3 samples, scale bar: 2 μm). Data are presented as mean values ± s.e.m. The P value was obtained from unpaired two-tailed t test; *P < 0.05. e, Original large field of view images for producing processed images in panel d (scale bar: 10 μm or 5 μm as indicated in the figure).
Extended Data Fig. 3 ∣. Protein modules of the developing human PSD.
a, Kernel density estimation of the null distributions of protein-protein interaction (PPI) numbers assuming no enrichment of PPI in individual modules; the vertical red lines indicate the observed PPI numbers in each module. The exact P value was derived from the one-tailed permutation test. b, SynGO biological pathway enrichment analysis of each module. Nominal P values from Fisher’s exact test were adjusted by the Benjamini and Hochberg method. c, PPI-co-abundance network of the four PSD modules. d, PPI-co-abundance network of each module highlighting proteins in enriched pathways. e, The normalized average shortest path lengths of pathways in individual modules. The asterisks denote that the average shortest path length is significantly shorter within pathway proteins than between pathway and non-pathway proteins. The P values were obtained from one-tailed Wilcoxon rank sum test; *P < 0.01.
Extended Data Fig. 4 ∣. Protein domains in PSD proteins.
a, Protein domains in individual PSD proteins. The rows are clustered based on the Jaccard distance. b, Abundance patterns of RhoGAPs and RhoGEFs not listed in Fig. 2e.
Extended Data Fig. 5 ∣. Changes in PSD composition during human V1 development.
a, Schematic illustrating the developmental stages of samples in the human V1 dataset. b, PCA plots of samples in the human V1 dataset colored by their age groups. c, Hierarchical clustering of the samples in the human V1 dataset based on proteins with differential abundance. d, Gene set enrichment analysis (GSEA) for individual age groups across species. NES, normalized enrichment score. Nominal P values were adjusted by the Benjamini and Hochberg method. e, Scaled abundance patterns (module eigengene values) of four protein modules in the human V1 dataset. f, Similarity matrices representing pairwise Pearson correlations between human PFC and human V1 samples.
Extended Data Fig. 6 ∣. Transcription of PSD proteins.
a, Preservation of human PSD modules in the bulk RNA-seq data. b, Uniform manifold approximation and projection (UMAP) plots showing the distribution of age groups and cell types in the single-nucleus RNA-seq data from developing human neocortex. c, UMAP plots showing the expression patterns of neuronal subtype-specific markers in the single-nucleus RNA-seq data from developing human neocortex. d, Standardized expression values of genes in the blue (n = 301 genes) and yellow (n = 218 genes) modules in individual neuronal subtypes of the adult human neocortex. Boxplot center: median; hinges: the 25th and 75th percentiles; whiskers: 1.5 × inter-quartile range. e, Standardized expression values of top TFs predicted to regulate the blue (95 genes) and yellow (97 genes) modules in individual neuronal subtypes of the adult human neocortex. Boxplot center: median; hinges: the 25th and 75th percentiles; whiskers: 1.5 × inter-quartile range. EN_IT, excitatory intratelencephalic neuron; EN_non-IT, excitatory non-intratelencephalic neuron; IN_MGE, inhibitory neuron derived from the medial ganglionic eminence; IN_CGE, inhibitory neuron derived from the caudal ganglionic eminence.
Extended Data Fig. 7 ∣. Changes in PSD composition during macaque and mouse neocortical development.
a, UpSet plot describing the number of identified proteins and their overlaps at each age group of macaque and mouse datasets. b, PCA plots of the macaque and mouse samples colored by their age groups. c, Hierarchical clustering of macaque and mouse samples based on proteins with differential abundance. d, Preservation of Human PSD modules in macaque and mouse PSD proteomic data. e, Gene set enrichment analysis (GSEA) for individual age groups across human V1, macaque, and mouse samples. NES, normalized enrichment score. Nominal P values were adjusted by the Benjamini and Hochberg method. f, Similarity matrices representing pairwise Pearson correlations between human V1, macaque, and mouse samples. g, Standardized abundance patterns of proteins in the four PSD modules (n = 127, 169, 159 and 178 proteins) across regions and species along the humanized age based on the human V1 dataset. Data are presented as mean values ± s.e.m. The macaque and mouse illustrations were created with BioRender.com.
Extended Data Fig. 8 ∣. RhoGEF levels and activities during synapse development.
a, Immunoblots and quantification of ARHGEF7 and PREX1 in the PSD and homogenate of developing human cortex (n = 4, 4 and 4 samples). Data are presented as mean values ± s.e.m. The P values were obtained from one-way ANOVA with Holm-Sidak’s multiple comparisons test; *P < 0.05, **P < 0.01, ***P < 0.001. b, Immunoblots and quantification of ARHGEF7 and PREX1 in the PSD of developing mouse cortex (n = 2, 2, 2, 2, 2, 2 and 2 samples). Data are presented as mean values ± s.e.m. The P values were obtained from one-way ANOVA; ns, not significant. c, Original large field of view images for producing processed images in Fig. 4b (scale bar: 10 μm or 5 μm as indicated in the figure). d, Comparison of adult human PSD samples from postmortem brain tissues and neurosurgical biopsy tissues by western blot analysis (n = 4 and 3 samples). e, Immunoblots and quantification of ARHGEF7, PREX1, phospho-PAK, PAK, phospho-CFL1, and CFL1 in the synaptosomes of cultured primary human (n = 3 and 3 samples) and mouse cortical neurons (n = 3 and 3 samples) at indicated days in vitro (DIV). Data are presented as mean values ± s.e.m. The P values were obtained from one-way ANOVA with Holm-Sidak’s multiple comparisons test; *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant. The human and mouse illustrations were created with BioRender.com.
Extended Data Fig. 9 ∣. Manipulation of Rho GTPase regulators alters synapse development in human and mouse cortical neurons.
a, Original large field of view images for producing processed images in Fig. 4c (scale bar: 50 μm). b, Immunostaining of dendrites from primary mouse cortical neurons cultured 8 days in vitro. Neurons were transfected with mEGFP-C1 and vectors expressing mCherry, mCherry-ARHGEF7, or mCherry-RASGRF2 (n = 20, 20 and 20 neurons from 4 cultures, scale bar: 5 μm). Data are presented as mean values ± s.e.m. The P values were obtained from one-way ANOVA with Holm-Sidak’s multiple comparisons test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. c, Original large field of view images for producing processed images in panel b (scale bar: 50 μm). d, Quantification of mRNA levels of ARHGAP23 and SRGAP1 in HEK293T cells transfected with control shRNAs (shControl), two shRNAs targeting ARHGAP23 (shARGGAP23–1 and shARGGAP23–2), or two shRNAs targeting SRGAP1 (shSRGAP1–1 and shSRGAP1–2) (n = 3 cultures for all conditions). Data are presented as mean values ± s.e.m. The P values were obtained from one-way ANOVA with Holm-Sidak’s multiple comparisons test; ****P < 0.0001. e,f, Immunostaining of dendrites from primary human cortical neurons cultured six weeks in vitro. Neurons were transfected with mEGFP-C1 and vectors co-expressing turbo-RFP (tRGP) and shControl, shARGGAP23–1, shARGGAP23–2, shSRGAP1–1 or shSRGAP1–2 (n = 20, 22, 20, 20 and 20 neurons from 4 cultures, scale bar: 5 μm). Data are presented as mean values ± s.e.m. The P values were obtained from one-way ANOVA with Holm-Sidak’s multiple comparisons test; *P< 0.05, **P < 0.01, ***P < 0.001, ****P< 0.0001. g, Original large field of view images for producing processed images in panel e (scale bar: 50 μm). h, Original large field of view images for producing processed images in Fig. 4e (scale bar: 50 μm).
Extended Data Fig. 10 ∣. Association of PSD modules with cognitive functions and brain disorders.
a, PPI-co-abundance network of the turquoise module with activity-dependent proteins highlighted. b, Distribution of gnomAD synonymous Z-scores of genes in each category (n = 265, 313, 402, 224, 561, 1765 and 12892 genes). Boxplot center: median; hinges: the 25th and 75th percentiles; whiskers: 1.5 × inter-quartile range. The P values were obtained from Kruskal–Wallis test with Dunn’s multiple comparisons test; ns, not significant. c, Percentage of rare variants located at PSD module genes in subjects with or without neurodevelopmental disorders. d, PPI-co-abundance network of the turquoise module with genes carrying neurodevelopmental disorder-linked (at least 3) de novo missense variants or (at least 2) PTVs highlighted. e, PPI-co-abundance network of the brown module with genes carrying psychiatric disorder-linked common variants highlighted. f, PPI-co-abundance network of the yellow module with genes downregulated in psychiatric disorders highlighted. g, Volcano plots for misexpressed genes after the onset of psychiatric disorders in PSD modules. Red dots indicate differentially expressed genes with the Benjamini–Hochberg adjusted P values < 0.05.
Supplementary Material
Acknowledgements
We thank NIH NeuroBioBank, the University of Maryland School of Medicine Brain and Tissue Bank for providing postmortem brain tissue samples; L. Kohlstaedt for help in mass spectrometry data analysis; and S. Sanders, R. Nicoll, N. Parikshak, T. Mukhtar, M. Song and H. Zoghbi for comments on the manuscript. This work used the Vincent J. Proteomics/Mass Spectrometry Laboratory at UC Berkeley, supported in part by NIH S10 Instrumentation grant S10RR025622. This study was supported by National Institute of Neurological Disorders and Stroke grant R35NS097305 to A.R.K., National Institute of Mental Health grant K99MH131832 to L.W., National Human Genome Research Institute grant HG010898 to N.S. and National Institute of Mental Health grant MH124619 to N.S.
Footnotes
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-023-06542-2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Competing interests A.R.K. is a co-founder, consultant and director of Neurona Therapeutics. The remaining authors declare no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-023-06542-2.
Data availability
All raw proteomic data were deposited to ProteomeXchange through MassIVE (human PFC dataset: MSV000091887 and PXD042067; human V1 dataset: MSV000091888 and PXD042068; macaque dataset: MSV000091889 and PXD042069; mouse dataset: MSV000091890 and PXD042071). All processed data are available in Supplementary Tables 1-13 and at an online interactive portal (https://liwang.shinyapps.io/PSD_development_explorer). The reference proteomes (Homo sapiens UP000005640_9606, Macaca mulatta UP000006718_9544 and Mus musculus UP000000589_10090) can be downloaded from UniProt (https://www.uniprot.org/proteomes). The PPI data can be downloaded from the BioGRID database (https://thebiogrid.org/). Bulk-level RNA-seq data of developing human brain can be downloaded from the BrainSpan database (http://www.brainspan.org) or from the PsychENCODE website (http://evolution.psychencode.org/). Protein domain information can be downloaded from the SMART database (https://smart.embl.de/). Single-cell and single-nucleus RNA-seq data from developing and adult human neocortex can be downloaded from the UCSC Cell Browser (https://cells.ucsc.edu). GWAS summary statistics for human cognitive function can be downloaded from the GWAS ATLAS resource (https://atlas.ctglab.nl). LOEUF scores, missense z-scores and synonymous z-scores for all human genes can be downloaded from the gnomAD database (https://gnomad.broadinstitute.org). De novo variants in neurodevelopmental disorders can be obtained from denovo-db (https://denovo-db.gs.washington.edu/denovo-db). GWAS summary statistics for psychiatric disorders can be downloaded from The Psychiatric Genomics Consortium database (https://www.med.unc.edu/pgc).
Code availability
Code used for data analysis in this manuscript is available at GitHub (https://github.com/alexwang1001/PSD_development).
References
- 1.Südhof TC Towards an understanding of synapse formation. Neuron 100, 276–293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt ERE & Polleux F Genetic mechanisms underlying the evolution of connectivity in the human cortex. Front. Neural Circuits 15, 787164 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zoghbi HY & Bear MF Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb. Perspect. Biol 4, a009886 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frankle WG, Lerma J & Laruelle M The synaptic hypothesis of schizophrenia. Neuron 39, 205–216 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Shankar GM & Walsh DM Alzheimer’s disease: synaptic dysfunction and Aβ. Mol. Neurodegener 4, 964075 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig AM & Boudin H Molecular heterogeneity of central synapses: afferent and target regulation. Nat. Neurosci 4, 569–578 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Harris KM & Weinberg RJ Ultrastructure of synapses in the mammalian brain. Cold Spring Harb. Perspect. Biol 4, 7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uezu A. et al. Identification of an elaborate complex mediating postsynaptic inhibition. Science 353, 1123–1129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheng M & Kim E The postsynaptic organization of synapses. Cold Spring Harb. Perspect. Biol 3, a005678 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayés A. et al. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat. Neurosci 14, 19–21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swulius MT, Kubota Y, Forest A & Waxham MN Structure and composition of the postsynaptic density during development. J. Comp. Neurol 518, 4243–4260 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petralia RS, Sans N, Wang YX & Wenthold RJ Ontogeny of postsynaptic density proteins at glutamatergic synapses. Mol. Cell. Neurosci 29, 436–452 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris KM, Jensen FE & Tsao B Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. J. Neurosci 12, 2685–2705 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Zundert B, Yoshii A & Constantine-Paton M Receptor compartmentalization and trafficking at glutamate synapses: a developmental proposal. Trends Neurosci. 27, 428–437 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Elias GM, Elias LAB, Apostolides PF, Kriegstein AR & Nicoll RA Differential trafficking of AMPA and NMDA receptors by SAP102 and PSD-95 underlies synapse development. Proc. Natl Acad. Sci. USA 105, 20953–20958 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray JA et al. Distinct modes of AMPA receptor suppression at developing synapses by GluN2A and GluN2B: single-cell NMDA receptor subunit deletion in vivo. Neuron 71, 1085–1101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J. et al. Spatiotemporal profile of postsynaptic interactomes integrates components of complex brain disorders. Nat. Neurosci 20, 1150–1161 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaizuka T. et al. Developmental dynamics of the postsynaptic proteome to understand synaptic maturation and dysmaturation. Preprint at bioRxiv 10.1101/2022.05.05.490828 (2022). [DOI] [Google Scholar]
- 19.Elston GN, Benavides-Piccione R & DeFelipe J The pyramidal cell in cognition: a comparative study in human and monkey. J. Neurosci 21, RC163 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayés À. et al. Comparative study of human and mouse postsynaptic proteomes finds high compositional conservation and abundance differences for key synaptic proteins. PLoS ONE 7, e46683 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emes RD et al. Evolutionary expansion and anatomical specialization of synapse proteome complexity. Nat. Neurosci 11, 799–806 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benavides-Piccione R, Ballesteros-Yáñez I, DeFelipe J & Yuste R Cortical area and species differences in dendritic spine morphology. J. Neurocytol 31, 337–346 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Shibata M. et al. Hominini-specific regulation of CBLN2 increases prefrontal spinogenesis. Nature 598, 489–494 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loomba S. et al. Connectomic comparison of mouse and human cortex. Science 377, eabo0924 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Petanjek Z. et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc. Natl Acad. Sci. USA 108, 13281–13286 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X. et al. Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res. 22, 611–622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L. et al. An autism-linked missense mutation in SHANK3 reveals the modularity of Shank3 function. Mol. Psychiatry 25, 2534–2555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayés À. et al. Human post-mortem synapse proteome integrity screening for proteomic studies of postsynaptic complexes. Mol. Brain 7, 88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy M. et al. Proteomic analysis of postsynaptic proteins in regions of the human neocortex. Nat. Neurosci 21, 130–141 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Pickard L, Noel J, Henley JM, Collingridge GL & Molnar E Developmental changes in synaptic AMPA and NMDA receptor distribution and AMPA receptor subunit composition in living hippocampal neurons. J. Neurosci 20, 7922–7931 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langfelder P & Horvath S WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koopmans F. et al. SynGO: an evidence-based, expert-curated knowledge base for the synapse. Neuron 103, 217–234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li M. et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362, eaat7615 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langfelder P, Luo R, Oldham MC & Horvath S Is my network module preserved and reproducible? PLoS Comput. Biol 7, e1001057 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Velmeshev D. et al. Single-cell analysis of prenatal and postnatal human cortical development. Preprint at bioRxiv 10.1101/2022.10.24.513555 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakken TE et al. Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu LJ, Li X, Chen T, Ren M & Zhuo M Characterization of intracortical synaptic connections in the mouse anterior cingulate cortex using dual patch clamp recording. Mol. Brain 2, 32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Workman AD, Charvet CJ, Clancy B, Darlington RB & Finlay BL Modeling transformations of neurodevelopmental sequences across mammalian species. J. Neurosci 33, 7368–7383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laubach M, Amarante LM, Swanson K & White SR What, if anything, is rodent prefrontal cortex? eNeuro 5, 315–333 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huttenlocher PR & Dabholkar AS Regional differences in synaptogenesis in human cerebral cortex. J. Comp. Neurol 387, 167–178 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Sydnor VJ et al. Neurodevelopment of the association cortices: patterns, mechanisms, and implications for psychopathology. Neuron 109, 2820–2846 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlin RK, Grab DJ & Siekevitz P Postmortem accumulation of tubulin in postsynaptic density preparations. J. Neurochem 38, 94–100 (1982). [DOI] [PubMed] [Google Scholar]
- 43.Schanzenbächer CT, Langer JD & Schuman EM Time- and polarity-dependent proteomic changes associated with homeostatic scaling at central synapses. eLife 7, e33322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu F. et al. Cell-type-specific visualisation and biochemical isolation of endogenous synaptic proteins in mice. Eur. J. Neurosci 51, 793–805 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bygrave AM et al. Btbd11 supports cell-type-specific synaptic function. Cell Rep. 42, 112591 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawaguchi Y, Karube F & Kubota Y Dendritic branch typing and spine expression patterns in cortical nonpyramidal cells. Cereb. Cortex 16, 696–711 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Charrier C. et al. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell 149, 923–935 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fossati M. et al. SRGAP2 and its human-specific paralog co-regulate the development of excitatory and inhibitory synapses. Neuron 91, 356–369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Schwanhüusser B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Rath S. et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49, D1541–D1547 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walikonis RS et al. Identification of proteins in the postsynaptic density fraction by mass spectrometry. J. Neurosci 20, 4069–4080 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shin JB et al. Molecular architecture of the chick vestibular hair bundle. Nat. Neurosci 16, 365–374 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X. et al. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc 13, 530–550 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Ritchie ME et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benjamini Y & Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc 57, 289–300 (1995). [Google Scholar]
- 57.Gu Z, Eils R & Schlesner M Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Hoffman GE & Schadt EE variancePartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics 17, 17–22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu G, Wang LG, Han Y & He QY ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liberzon A. et al. The Molecular Signatures Database hallmark gene set Collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schindelin J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stark C. et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34, 535–539 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shannon P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Csardi G & Nepusz T The igraph software package for complex network research. InterJ. Complex Syst (2006). [Google Scholar]
- 66.Letunic I, Khedkar S & Bork P SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu Y. et al. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362, eaat8077 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wood SN Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J. R. Stat. Soc. B 73, 3–36 (2011). [Google Scholar]
- 69.Keenan AB et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 47, W212–W224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Speir ML et al. UCSC Cell Browser: visualize your single-cell data. Bioinformatics 37, 4578–4580 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Friedman J, Hastie T & Tibshirani R Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw 33, 1–22 (2010). [PMC free article] [PubMed] [Google Scholar]
- 72.de Leeuw CA, Mooij JM, Heskes T & Posthuma D MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol 11, 1–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watanabe K. et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet 51, 1339–1348 (2019). [DOI] [PubMed] [Google Scholar]
- 74.Karczewski KJ et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turner TN et al. denovo-db: a compendium of human de novo variants. Nucleic Acids Res. 45, D804–D811 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heyne HO et al. De novo variants in neurodevelopmental disorders with epilepsy. Nat. Genet 50, 1048–1053 (2018). [DOI] [PubMed] [Google Scholar]
- 77.Rees E. et al. De novo mutations identified by exome sequencing implicate rare missense variants in SLC6A1 in schizophrenia. Nat. Neurosci 23, 179–184 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Howrigan DP et al. Exome sequencing in schizophrenia-affected parent–offspring trios reveals risk conferred by protein-coding de novo mutations. Nat. Neurosci 23, 185–193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ruderfer DM et al. Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell 173, 1705–1715 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grove J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet 51, 431–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Howard DM et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci 22, 343–352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Demontis D. et al. Genome-wide analyses of ADHD identify 27 risk loci, refine the genetic architecture and implicate several cognitive domains. Nat. Genet 55, 198–208 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gandal MJ et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362, eaat8127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw proteomic data were deposited to ProteomeXchange through MassIVE (human PFC dataset: MSV000091887 and PXD042067; human V1 dataset: MSV000091888 and PXD042068; macaque dataset: MSV000091889 and PXD042069; mouse dataset: MSV000091890 and PXD042071). All processed data are available in Supplementary Tables 1-13 and at an online interactive portal (https://liwang.shinyapps.io/PSD_development_explorer). The reference proteomes (Homo sapiens UP000005640_9606, Macaca mulatta UP000006718_9544 and Mus musculus UP000000589_10090) can be downloaded from UniProt (https://www.uniprot.org/proteomes). The PPI data can be downloaded from the BioGRID database (https://thebiogrid.org/). Bulk-level RNA-seq data of developing human brain can be downloaded from the BrainSpan database (http://www.brainspan.org) or from the PsychENCODE website (http://evolution.psychencode.org/). Protein domain information can be downloaded from the SMART database (https://smart.embl.de/). Single-cell and single-nucleus RNA-seq data from developing and adult human neocortex can be downloaded from the UCSC Cell Browser (https://cells.ucsc.edu). GWAS summary statistics for human cognitive function can be downloaded from the GWAS ATLAS resource (https://atlas.ctglab.nl). LOEUF scores, missense z-scores and synonymous z-scores for all human genes can be downloaded from the gnomAD database (https://gnomad.broadinstitute.org). De novo variants in neurodevelopmental disorders can be obtained from denovo-db (https://denovo-db.gs.washington.edu/denovo-db). GWAS summary statistics for psychiatric disorders can be downloaded from The Psychiatric Genomics Consortium database (https://www.med.unc.edu/pgc).
Code used for data analysis in this manuscript is available at GitHub (https://github.com/alexwang1001/PSD_development).