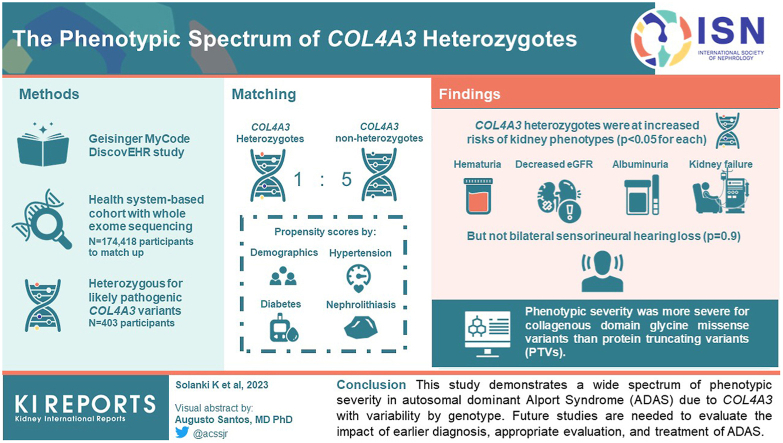
Abstract
Introduction
The penetrance and phenotypic spectrum of autosomal dominant Alport Syndrome (ADAS), affecting 1 in 106, remains understudied.
Methods
Using data from 174,418 participants in the Geisinger MyCode/DiscovEHR study, an unselected health system-based cohort with whole exome sequencing, we identified 403 participants who were heterozygous for likely pathogenic COL4A3 variants. Phenotypic data was evaluated using International Classification of Diseases (ICD) codes, laboratory data, and chart review. To evaluate the phenotypic spectrum of genetically-determined ADAS, we matched COL4A3 heterozygotes 1:5 to nonheterozygotes using propensity scores by demographics, hypertension, diabetes, and nephrolithiasis.
Results
COL4A3 heterozygotes were at significantly increased risks of hematuria, decreased estimated glomerular filtration rate (eGFR), albuminuria, and kidney failure (P < 0.05 for all comparisons) but not bilateral sensorineural hearing loss (P = 0.9). Phenotypic severity was more severe for collagenous domain glycine missense variants than protein truncating variants (PTVs). For example, patients with Gly695Arg (n = 161) had markedly increased risk of dipstick hematuria (odds ratio [OR] 9.50; 95% confidence interval [CI]: 6.32, 14.28) and kidney failure (OR 7.02; 95% CI: 3.48, 14.16) whereas those with PTVs (n = 119) had moderately increased risks of dipstick hematuria (OR 1.64; 95% CI: 1.03, 2.59) and kidney failure (OR 3.44; 95% CI: 1.28, 9.22). Less than a third of patients had albuminuria screening completed, and fewer than 1 of 3 were taking inhibitors of the renin-angiotensin-aldosterone system.
Conclusion
This study demonstrates a wide spectrum of phenotypic severity in ADAS due to COL4A3 with phenotypic variability by genotype. Future studies are needed to evaluate the impact of earlier diagnosis, appropriate evaluation, and treatment of ADAS.
Keywords: Alport Syndrome, Chronic Kidney Disease, Clinical Epidemiology, Genetic Renal Disease, Kidney Failure
Graphical abstract
Alport Syndrome (AS), the second most common cause of monogenic kidney failure, is characterized by hematuria, sensorineural deafness, and ocular abnormalities. Historically, about 85% of AS cases have been due to X-linked COL4A5, with the remainder mostly due to autosomal recessive disease from COL4A3 and COL4A4 variants.1,2 AS develops because the COL4A3, COL4A4, and COL4A5 genes encode the alpha-3, alpha-4, and alpha-5 subunits of type IV collagen, which is the major structural component of glomerular basement membrane (GBM). Heterozygous carriers for autosomal recessive and X-linked conditions were often mistakenly thought to be asymptomatic. However, recent data have shown that heterozygous carriers for COL4A3, COL4A4, and COL4A5 (females only) often present with at least microscopic hematuria, and can have a wide spectrum of renal manifestations, including thin basement membrane nephropathy (TBMN), focal segmental glomerulosclerosis (FSGS) and kidney failure.3 Up to 40% of patients with TBMN and 10% of FSGS cases have pathogenic or likely pathogenic heterozygous variants in COL4A3, COL4A4, or COL4A5.4,5 Because most studies include cohorts enriched with the most severely affected individuals, the prevalence, penetrance and full phenotypic spectrum of AS remain unknown.
Advances in molecular genetics have led to the use of genetic testing in confirming the diagnosis of AS, and expert opinions suggest that genetic testing for COL4A3, COL4A4, and COL4A5 variants could take precedence over renal biopsy when AS is suspected or if there is significant family history of AS-related kidney disease.6 Genetic testing with next generation sequencing gene panels is also recommended in patients with steroid-resistant FSGS.7 What remains less clear is the role of earlier genetic diagnosis of ADAS, especially as costs of next generation sequencing decrease and availability of testing increases. More information is needed on individuals with undiagnosed disease who could potentially receive early treatment with angiotensin converting enzyme inhibitors (ACEis) to reduce future risk of kidney failure.8, 9, 10
In this study, we examine the penetrance and phenotypic spectrum of heterozygous COL4A3 pathogenic variants using data from GeisingerMyCode-DiscovEHR study, an unselected health system-based cohort. We hypothesized that heterozygous carriers of COL4A3 pathogenic variants would be at increased risks of hematuria, albuminuria, FSGS, and kidney failure.
Methods
Study Population
The Geisinger Institutional Review Board approved this study. Informed consent was waived because participants were previously consented in the MyCode Community Health Initiative as part of the Geisinger-Regeneron DiscovEHR collaboration.11 We adhered to the Strengthening the Reporting of Observational Studies in Epidemiology guidelines (see Supplementary Materials; STROBE Statement). For this study, we included 174,361 participants in MyCode who had whole exome sequencing data available, and a COL4A3 variant that was classified as pathogenic (P) or likely pathogenic (LP) previously in ClinVar.12
Exome Sequencing and Variant Calling
Exome sequencing was performed in collaboration with Regeneron Genetics Center, as previously described.11 A modified version of the xGEN probe from Integrated DNA Technologies were used for target sequence capture (Supplementary Methods).13 We included COL4A3 variants that were listed in ClinVar at least once as P/LP (accessed May 25, 2021), with any number of ClinVar stars. To examine potential genotype-phenotype comparisons, we categorized P/LP variants into 4 separate categories as follows: (i) Gly695Arg, which was the most common P/LP variant; (ii) other glycine missense variants located within the collagenous domain (between the end of exon 2 and the beginning of exon 48)14 because glycine variants are critical in the intermediate collagenous domain; (iii) PTVs; and (iv) other missense variants or inframe deletions. Varsome (www.varsome.com accessed February 23, 2023), a human genomic variant search engine that provides automated classification of pathogenicity using American College of Medical Genetics criteria was used to report predicted pathogenicity. We also documented whether COL4A3 P/LP heterozygotes had any additional rare (allele frequency <0.001) variants in COL4A3, COL4A4, or COL4A5.
Phenotyping
Electronic health record (EHR) data, including ICD 9 and 10 diagnosis codes and laboratory data were extracted. We linked the data to the United States Renal Data System (USRDS) to ascertain kidney failure status and presumed cause of kidney failure,15 and used a definition including ICD diagnosis codes for dialysis or kidney transplant. Due to USRDS data restrictions, we report only data using kidney failure per ICD code for subgroups <11. ICD diagnosis codes used for kidney failure, hematuria, bilateral sensorineural hearing loss, and FSGS are listed in Supplementary Table S1.
Laboratory data included serum creatinine, urinalysis, urine albumin-to-creatinine ratio (ACR), and urine protein-to-creatinine ratio. To minimize false positive hematuria tests that could be due to urinary tract infection, we included only urinalyses negative for leukocyte esterase and nitrites. Urinalysis-based hematuria outcomes were categorized as trace or greater blood and 1+ or greater if present on >50% of urinalyses. Urinalysis-based proteinuria outcomes were categorized as 1+ or greater or 2+ or greater if on at least 2 urinalyses. Albuminuria was defined as having moderate albuminuria (ACR 30–299 mg/g or urine protein-to-creatinine ratio 150–499 mg/g) or severe albuminuria (ACR 300+ mg/g or urine protein-to-creatinine ratio 500+ mg/g). We calculated eGFR using the Chronic Kidney Disease Epidemiology Collaboration 2021 formula.16
To provide comprehensive phenotypic data as available in the EHR, chart review was performed on the 402 COL4A3 carriers by author KVS with additional review of patients with kidney biopsies by authors ARC and IDB. We searched for additional data on AS-related phenotypic features, family history of AS and TBMN, audiometry, treatment with ACEis or angiotensin receptor blockers, kidney biopsies, urologic workup of hematuria, and genetic testing.
Outcomes
The primary outcome was having any phenotypic feature of AS (hematuria on urinalysis or ICD code, dipstick proteinuria 1+, moderate albuminuria, severe albuminuria, eGFR <60 and <30 ml/min per 1.73 m2, FSGS, kidney failure, or bilateral sensorineural hearing loss). Secondary outcomes included each of these outcomes separately, as well as Kidney Disease Improving Global Outcomes (KDIGO) risk categories,17 which included no chronic kidney disease (CKD), hematuria alone, moderately increased risk, high risk, very high risk, and extremely high risk. Only individuals with eGFR and urinalysis data were included in the KDIGO risk category analysis; if quantitative ACR was unavailable, we classified dipstick protein 1+ twice as ACR 30–299 mg/g, and dipstick protein 2+ or greater twice as ACR 300+ mg/g.
Statistical Analysis
We first compared phenotypic features between COL4A3 P/LP heterozygotes and the rest of the MyCode cohort. Considering that urinalysis testing was done more frequently in COL4A3 P/LP heterozygotes, we performed propensity score matching to reduce potential selection bias because hematuria is a key feature of AS. We used a combination of propensity score based on age, Black race, Hispanic ethnicity, and exactly-matched categories (sex, hypertension, diabetes, nephrolithiasis, and year of first outpatient encounter) to match COL4A3 P/LP variant heterozygotes 1:5 to individuals without COL4A3/A4/A5 P/LP variants in ClinVar, using RStudio (Version 2022.02.3). The standardized mean difference was calculated to measure the balance of covariates between COL4A3 P/LP heterozygotes and the matched control cohort. Categorical and continuous outcomes were compared between heterozygotes and nonheterozygotes using χ2 and t-tests, respectively. Logistic regression was used to estimate the adjusted ORs for AS-related phenotypes using STATA/MP 15.1. We also examined whether outcomes differed between the 4 COL4A3 subgroups, using chi-square tests. Logistic regression analyses comparing subgroups to the overall control group were adjusted for age, sex, and race. We used complete case analyses.
Results
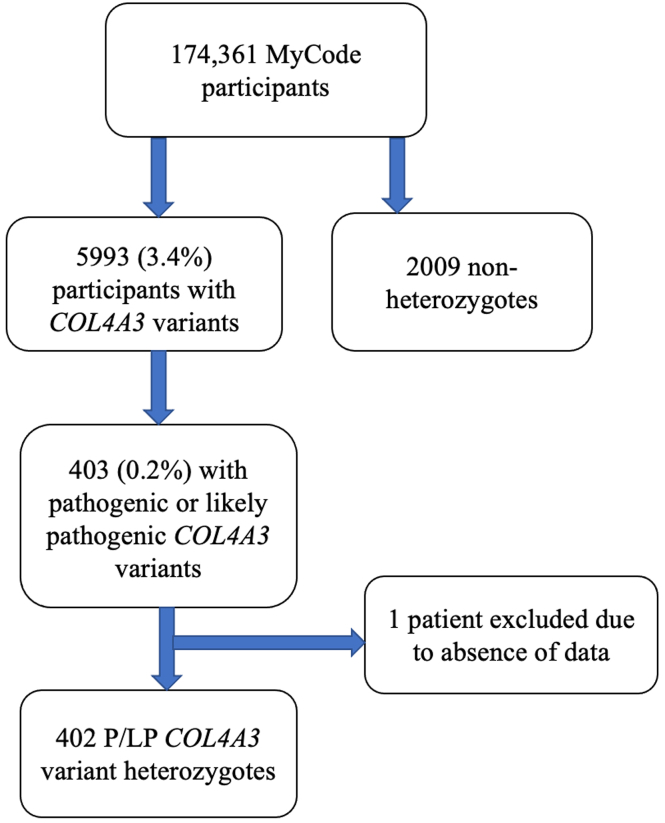
Out of 174,361 MyCode participants, 5993 (3.4%) had COL4A3 variants at minor allele frequency <1%, including 403 (0.2%) who had a COL4A3 variant previously reported in ClinVar as P/LP at least once. After excluding 1 COL4A3 P/LP heterozygote because of missing data, we examined 402 heterozygotes in detail (Figure 1). The most common P/LP COL4A3 variant was Gly695Arg (n = 161). There were 47 patients with other glycine missense variants located within the collagenous domain, 119 with PTVs and 75 with other missense variants or inframe deletions (n = 73) (Supplementary Table S2 includes additional details by variant).
Figure 1.
Flowchart. This figure does not use United States Renal Data System data.
Compared to the rest of the MyCode cohort, COL4A3 P/LP heterozygotes were more likely to have undergone urinalysis evaluation (81.1% vs. 75.7%; P = 0.01) but had similar rates of evaluation for eGFR (89.1% vs. 88.8%; P = 0.9) and ACR (29.4% vs. 28.1%; P = 0.6) and similar prevalence of hypertension, diabetes, and nephrolithiasis (Table 1). After successfully matching 402 COL4A3 P/LP heterozygotes to 2009 nonheterozygote controls using a 1:5 ratio, mean age was 59.1 years and 64% were female, with similar urinalysis availability (81.1% vs. 77.7%; P = 0.2) and median follow-up time (15 years, interquartile interval 7.8–18.6). Compared to controls, COL4A3 P/LP heterozygotes were more likely to have at least 1 phenotypic feature (64.4% vs. 45.7%; P < 0.001) than controls with significant differences by variant group (χ2 P < 0.001) (Table 2, Supplementary Table S3). All AS phenotypic features (hematuria, albuminuria, decreased eGFR, FSGS, and kidney failure) were significantly higher in COL4A3 P/LP heterozygotes compared to controls (P < 0.05 for all comparisons; Supplementary Table S3), except for bilateral sensorineural hearing loss.
Table 1.
Characteristics of 402 COL4A3 P/LP heterozygotes and nonheterozygotes before and after matching
| Characteristic | Before matching (N = 174,361) |
P-value | SMD | After propensity score matching (N = 2411) |
P-value | SMD | ||
|---|---|---|---|---|---|---|---|---|
| COL4A3 P/LP heterozygotes (n = 402) | Nonheterozygotes (n = 173,959) | COL4A3 P/LP heterozygotes (n = 402) | Nonheterozygotes (n = 2009) | |||||
| Age, Mean (SD) | 59.0 (18.6) | 57.5 (18.9) | 0.104 | 0.082 | 59.1 (18.7) | 59.1 (18.5) | 0.986 | 0.001 |
| Female (%) | 257 (63.9) | 105,441 (60.6) | 0.19 | 0.069 | 257 (63.9) | 1284 (63.9) | 1 | <0.001 |
| Black, n (%) | 9 (2.2) | 3951 (2.3) | 1 | 0.002 | 9 (2.2) | 32 (1.6) | 0.482 | 0.047 |
| Hispanic, n (%) | 4 (1.0) | 4613 (2.7) | 0.056 | 0.124 | 4 (1.0) | 47 (2.3) | 0.128 | 0.105 |
| Year of first outpatient visit, median (IQR) | 2003 (2001–2011) | 2004 (2001–2011) | 0.12 | 0.079 | 2003 (2001–2011) | 2003 (2001–2011) | 0.939 | 0.004 |
| Hypertension (%) | 226 (56.2) | 92,964 (53.3) | 0.259 | 0.059 | 226 (56.2) | 1129 (56.2) | 1 | <0.001 |
| Diabetes (%) | 90 (22.4) | 41,056 (23.6) | 0.608 | 0.029 | 90 (22.4) | 449 (22.3) | 1 | 0.001 |
| Nephrolithiasis (%) | 22 (5.5) | 10,155 (5.8) | 0.838 | 0.016 | 22 (5.5) | 109 (5.4) | 1 | 0.002 |
| eGFR available, n (%) | 358 (89.1) | 154,420 (88.8) | 0.918 | 0.009 | 358 (89.1) | 1815 (90.3) | 0.484 | 0.042 |
| Urinalysis available, n (%) | 326 (81.1) | 131,667 (75.7) | 0.014 | 0.132 | 326 (81.1) | 1561 (77.7) | 0.15 | 0.084 |
| ACR available, n (%) | 118 (29.4) | 48,836 (28.1) | 0.607 | 0.028 | 118 (29.4) | 539 (26.8) | 0.329 | 0.056 |
ACR, urine albumin-to-creatinine ratio; eGFR, estimated glomerular filtration rate; LP, likely pathogenic; P, pathogenic; SMD, standardized mean difference.
To reduce potential selection bias since hematuria is an important phenotypic feature of Alport Syndrome, COL4A3 P/LP heterozygotes were matched 1:5 to controls without COL4A3 variants using a combination of propensity score based on age, Black race, Hispanic ethnicity, and exact-matched categories (sex, hypertension, diabetes, nephrolithiasis, year of first outpatient encounter).
The cells with <11 individuals do not present USRDS data.
Table 2.
Phenotypic features of COL4A3 P/LP variant groups and controls
| Gly695Arg (n = 161) | Other collagenous domain glycine variants (n = 47) | PTVs (n = 119) | Other missense variants or inframe deletions (n = 75) | P-value | Controls (n = 2009) | |
|---|---|---|---|---|---|---|
| Any phenotypic feature below | 125 (77.6) | 38 (80.9) | 62 (52.1) | 34 (45.3) | <0.001 | 949 (47.2) |
| ICD Code-based diagnoses | ||||||
| Hematuria ICD code (%) | 57 (35.4) | 22 (46.8) | 23 (19.3) | 8 (10.7) | <0.001 | 279 (13.9) |
| FSGS ICD (%) | 5 (3.1) | 0 | 1 (0.8) | 1 (1.4) | 0.4 | 4 (0.2) |
| Kidney failure per ICD code (%) | 13 (8.1) | 3 (6.1) | 5 (4.2) | 2 (2.7) | 0.3 | 25 (1.2) |
| Bilateral sensorineural hearing loss ICD (%) | 7 (4.4) | 3 (6.4) | 3 (2.5) | 10 (13.3) | 0.01 | 109 (5.4) |
| Lab-based diagnoses | ||||||
| Trace blood or greater on UAa (%) | 90/128 (70.3) | 31/41 (75.6) | 29/97 (29.9) | 8/60 (13.3) | <0.001 | 327/1561 (21.0) |
| 1+ blood or greater on UAa (%) | 71/128 (55.5) | 25/41 (61.0) | 17/97 (17.5) | 6/60 (10.0) | <0.001 | 184/1561 (11.8) |
| 1+ protein twice on UA | 53/128 (41.4) | 16/40 (40.0) | 22/97 (22.7) | 12/59 (20.3) | 0.008 | 357/1560 (22.9) |
| 2+ protein twice on UA | 31/128 (24.2) | 12/40 (30.0) | 18/97 (18.6) | 5/59 (8.5) | 0.03 | 171/1560 (11.0) |
| ACR ≥ 30 mg/g (%) | 25/42 (59.5) | 9/22 (40.9) | 11/32 (34.4) | 5/22 (22.7) | 0.02 | 117 (24.6) |
| ACR ≥ 300 mg/g (%) | 11/42 (26.2) | 5/22 (22.7) | 3/32 (9.4) | 1/22 (4.6) | 0.08 | 26 (5.5) |
| eGFR < 60 (%) | 46/145 (31.7) | 15/45 (33.3) | 25/101 (24.5) | 11/68 (16.2) | 0.07 | 388 (21.4) |
| eGFR < 30 (%) | 18/145 (12.4) | 4/45 (8.9) | 6/102 (5.9) | 2/68 (2.9) | 0.09 | 59 (3.3) |
| KDIGO CKD risk categorya | 122 | 39 | 85 | 54 | <0.001 | |
| N with eGFR and UA data available | 17 (13.9) | 7 (18.0) | 42 (49.4) | 28 (51.9) | 1439 | |
| No CKD (%) | 40 (32.8) | 15 (38.5) | 8 (9.4) | 3 (5.6) | 752 (52.3) | |
| Hematuria alone (%) | 22 (18.0) | 6 (15.4) | 15 (17.7) | 13 (24.1) | 154 (10.7) | |
| Moderately increased risk (%) | 19 (15.6) | 3 (7.7) | 12 (12.1) | 6 (11.1) | 280 (19.5) | |
| High risk (%) | 9 (7.4) | 4 (10.3) | 4 (4.7) | 2 (3.7) | 153 (10.6) | |
| Very high risk (%) | 15 (12.3) | 4 (10.3) | 4 (4.7) | 2 (3.7) | 65 (4.5) | |
| Extremely high risk (%) | 34 (2.4) |
ACR, urine albumin-to-creatinine ratio; eGFR, estimated glomerular filtration rate; FSGS, focal segmental glomerulosclerosis; ICD, International Classification of Disease; KDIGO, Kidney Disease Improving Global Outcomes; LP, likely pathogenic; P, pathogenic; PTV, protein truncating variant; UA, urinalysis.
If ACR data was unavailable, urinalysis data was used with dipstick 1+ twice classified as ACR 30–299 mg/g and dipstick 2+ twice or greater classified as ACR 300+ mg/g.
P-value for comparisons across COL4A3 variant groups using ANOVA.
This table does not include any USRDS data.
KDIGO CKD Risk Categories: Hematuria only category had trace hematuria but eGFR >60 and ACR <30; moderately increased risk (eGFR >60 with ACR 30–300 or eGFR 45–59 with ACR <30); high risk (eGFR >60 with ACR >300, eGFR 45–59 with ACR 30–300, eGFR 30–44 with ACR <30); very high risk (eGFR 15–29 with ACR <30, eGFR 15–44 with ACR 30–300, eGFR 30–59 with ACR >300), extremely high risk (eGFR <15 or eGFR 15–29 with ACR>300).
Phenotypic Variability by P/LP COL4A3 Genotype
There was significant variability between the 4 genotype subgroups for having any phenotypic feature present as well as the number of phenotypic features present (Tables 2 and 3). Penetrance and phenotypic severity was highest in glycine missense variants located within the collagenous domain: Gly695Arg (n = 161; 78% any phenotypic feature, 70% trace blood on dipstick, 32% eGFR <60, 12% eGFR <30, 3% FSGS, 8% kidney failure per ICD code); other glycine collagenous domain variants (n = 47; 81% any phenotypic feature, 76% trace blood on dipstick, 33% eGFR<60, 9% eGFR<30, and 6% kidney failure per ICD code).
Table 3.
Risk of Alport syndrome phenotypic features
| Phenotypic feature | Gly695Arg (n = 161) |
Other collagenous domain glycine variants (n = 47) |
PTVs (n = 119) |
Other missense variants or inframe deletions (n = 75) |
|---|---|---|---|---|
| OR (95%) | OR (95%) | OR (95%) | OR (95%) | |
| Any phenotypic feature below | 4.47 (2.99, 6.67)a | 5.17 (2.44, 10.93)a | 1.17 (0.80, 1.73) | 0.99 (0.61, 1.61) |
| ICD diagnoses | ||||
| Hematuria ICD code | 3.44 (2.42, 4.88)a | 5.63 (3.11, 10.18)a | 1.46 (0.91, 2.35) | 0.77 (0.36, 1.62) |
| FSGS ICD code | 15.74 (4.16, 59.50)a | - | 4.22 (0.45, 39.25) | 7.64 (0.82, 70.89) |
| Kidney failure per ICD code | 7.02 (3.48, 14.16)a | 5.88 (1.68, 20.60)b | 3.44 (1.28, 9.22)c | 2.29 (0.53, 9.94) |
| Kidney failure per USRDS | 9.01 (4.23, 19.20)a | 5.05 (1.13, 22.67)c | 2.94 (0.85, 10.19) | 3.04 (0.69, 13.51) |
| Bilateral sensorineural hearing loss ICD code | 0.77 (0.35, 1.70) | 1.22 (0.37, 4.03) | 0.44 (0.14, 1.40) | 2.91 (1.44, 5.91)b |
| Lab-based diagnoses | ||||
| Trace blood or greater on UAc | 9.50 (6.32, 14.28)a | 13.06 (6.23, 27.38)a | 1.64 (1.03, 2.59)c | 0.63 (0.29, 1.35) |
| 1+ blood or greater on UAc | 9.52 (6.47, 14.02)a | 12.48 (6.47, 24.07)a | 1.61 (0.93, 2.80) | 0.89 (0.38, 2.12) |
| 1+ protein twice on UA | 2.21 (1.53, 3.19)a | 2.40 (1.28, 4.50)b | 1.05 (0.65, 1.70) | 0.90 (0.47, 1.72) |
| 2+ protein twice on UA | 2.42 (1.57, 3.72)a | 3.82 (1.94, 7.51)a | 1.98 (1.17, 3.36)c | 0.77 (0.30, 1.94) |
| ACR ≥30 mg/g | 4.79 (2.49, 9.22)a | 2.31 (0.95, 5.59) | 1.66 (0.78, 3.55) | 0.89 (0.32, 2.46) |
| ACR ≥300 mg/g | 6.20 (2.81, 13.69)a | 4.92 (1.67, 14.46)b | 1.80 (0.51, 6.28) | 0.84 (0.11, 6.48) |
| eGFR <60 | 1.89 (1.21, 2.95)b | 2.79 (1.32, 5.91)b | 1.09 (0.64, 1.85) | 0.75 (0.35, 1.58) |
| eGFR <30 | 4.35 (2.42, 7.82)a | 3.42 (1.12, 10.41)c | 1.76 (0.73, 4.29) | 0.96 (0.22, 4.11) |
ACR, albumin-to-/creatinine ratio; eGFR, estimated glomerular filtration rate; FSGS, focal segmental glomerulosclerosis; ICD, International Classification of Disease; PTV, protein truncating variant; UA, urinalysis; USRDS, United States Renal Data System.
Logistic regression analyses comparing each variant subgroup to control group, adjusted for age, sex, and race.
P < 0.001.
P < 0.01.
P < 0.05.
Penetrance was lower in PTVs (n = 119; 52% any phenotypic feature, 30% trace blood on dipstick, 25% eGFR<60, 6% eGFR<30, and 4% kidney failure per ICD code). Results were similar among PTVs at earlier versus later exons, including the most common PTV Gly1602AlafsTer13 (2/32 [6%] kidney failure per ICD code) (Supplementary Table S3). Patients with other missense or inframe deletions (n = 75) did not have significantly higher risk of most AS phenotypes (45% any phenotypic feature, 13% trace blood on dipstick, 16% eGFR <60, 3% eGFR<30, 3% kidney failure per ICD code) (Tables 2 and 3), other than bilateral sensorineural hearing loss (13%). In terms of patients with multiple COL4A variants, there was 1 patient with COL4A3 p.Gly695Arg, a LP COL4A3 Arg1661Cys, and a COL4A4 p.Gly1465Asp variant of unknown significance (in the carboxy noncollagenous domain) who lacked eGFR and urinalysis data; 1 patient with COL4A3 p.Gly695Arg and a LP COL4A4 Gly774Arg who had hematuria, severe albuminuria, and eGFR >60 ml/min per 1.73 m2. There were 24 additional patients who had a rare variant (per Varsome: 18 benign or likely benign, 6 unknown significance) in COL4A3, COL4A4, or COL4A5 with 4 having kidney failure ICD diagnosis (full details in Supplementary Table S4).
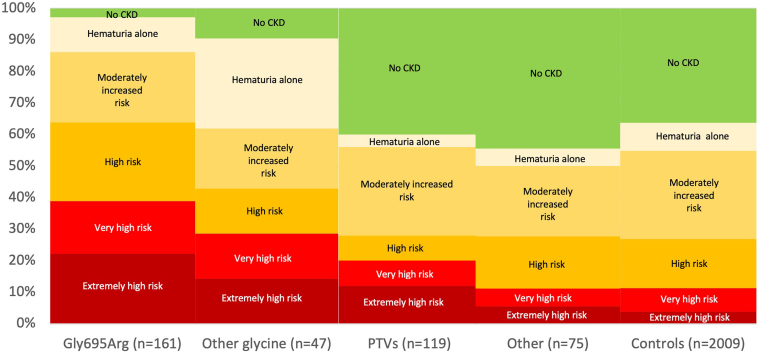
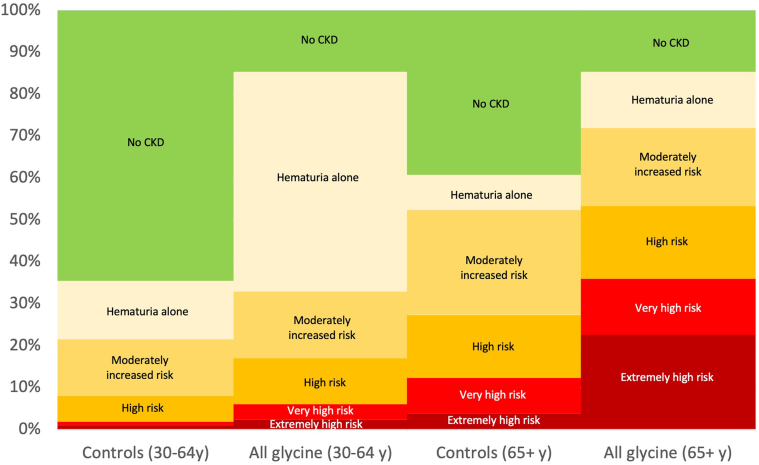
ORs of having any phenotypic feature were increased for Gly695Arg (OR 4.47; 95% CI: 2.99, 6.67; P < 0.001) and other collagenous domain glycine variant groups (OR 5.17; 95% CI: 2.44, 10.93; P < 0.001) but not the PTV group (OR 1.17; 95% CI: 0.80, 1.73; P = 0.4) or the other missense variants or inframe deletion group (OR 0.99; 95% CI: 0.61, 1.61; P = 1.0). Risk of kidney failure per ICD was higher for Gly695Arg (OR 7.02; 95% CI: 3.48, 14.16; P < 0.001), other collagenous domain glycine variants (OR 5.88; 95% CI: 1.68, 20.60; P = 0.006), and PTVs (OR 3.44; 95% CI: 1.28, 9.22; P = 0.01), compared to controls. Dipstick hematuria (trace blood or greater on at least 50% of urinalyses) was markedly increased for Gly695Arg (OR 9.50; 95% CI: 6.32, 14.28; P < 0.001) and other glycine variants (OR 13.06; 95% CI: 6.23, 27.38; P < 0.001), intermediate for PTVs (OR 1.64; 95% CI: 1.03, 2.59; P = 0.04), and not increased for other missense and inframe deletions (OR 0.63; 95% CI: 0.29, 1.35; P = 0.2), compared to controls. The glycine collagenous domain variant groups also had worse KDIGO CKD risk categories than controls (Figure 2, Table 2, Supplementary Figure S1). Among patients with Gly695Arg, prevalence of high risk or greater KDIGO CKD risk category was 17.1% for those age 30 to <65 years and 53.3% for those 65 years and older (Figure 3, Supplementary Figure S2).
Figure 2.
KDIGO CKD risk categories by variant groups. KDIGO CKD Risk Categories: Hematuria only category had trace hematuria but eGFR >60 and ACR <30; moderately increased risk (eGFR >60 with ACR 30–300 or eGFR 45–59 with ACR <30); high risk (eGFR >60 with ACR >300, eGFR 45–59 with ACR 30–300, eGFR 30–44 with ACR <30); very high risk (eGFR 15–29 with ACR <30, eGFR 15–44 with ACR 30–300, eGFR 30–59 with ACR >300), extremely high risk (eGFR <15 or eGFR 15–29 with ACR >300, or kidney failure by ICD code). If ACR data was unavailable, urinalysis data was used with dipstick 1+ twice classified as ACR 30–299 mg/g and dipstick 2+ twice or greater classified as ACR 300+ mg/g. This figure does not include any United States Renal Data System data. ACR, urine albumin-to-creatinine ratio; CKD, chronic kidney disease; eGFR, estimated glomerular filtration rate; ICD, International Classification of Diseases; KDIGO, Kidney Disease Improving Global Outcomes.
Figure 3.
KDIGO CKD Risk Categories for Glycine Collagenous Domain Variants versus Controls, by Age Group. This figure includes 169 patients heterozygous for glycine variants in the collagenous domain (including 129 Gly695Arg), and 1475 controls with eGFR and urinalysis data. KDIGO CKD Risk Categories: Hematuria only category had trace hematuria but eGFR >60 and ACR <30; moderately increased risk (eGFR >60 with ACR 30–300 or eGFR 45–59 with ACR <30); high risk (eGFR >60 with ACR >300, eGFR 45–59 with ACR 30–300, eGFR 30–44 with ACR <30); very high risk (eGFR 15–29 with ACR <30, eGFR 15–44 with ACR 30–300, eGFR 30–59 with ACR >300), extremely high risk (eGFR <15 or eGFR 15–29 with ACR >300, or kidney failure by ICD code). If ACR data was unavailable, urinalysis data was used with dipstick 1+ twice classified as ACR 30–299 mg/g and dipstick 2+ twice or greater classified as ACR 300+ mg/g. This figure does not include any United States Renal Data System data. ACR, urine albumin-to-creatinine ratio; CKD, chronic kidney disease; eGFR, estimated glomerular filtration rate; ICD, International Classification of Diseases; KDIGO, Kidney Disease Improving Global Outcomes.
Chart Review of All COL4A3 P/LP Heterozygotes
Only 4 patients had been diagnosed with AS or TBMN, and only 30% of patients were receiving ACEis or angiotensin receptor blockers, the mainstay for treatment of AS (Supplementary Table S5). Among those on ACEis or angiotensin receptor blockers, 18 of 52 (34.6%) had albuminuria per ACR and 33 of 101 (32.7%) had albuminuria per ACR or urinalysis. Nearly one-third of patients with glycine collagenous variants had undergone urologic workup for hematuria. Eleven patients underwent kidney biopsy; all had glycine collagenous domain variants and 4 (36.4%) had at least 1 other rare variant in a COL4A gene (Table 4). Diagnoses included FSGS (n = 6), TBMN (n = 1), IgA nephropathy (n = 2), light chain deposition disease (n = 1), advanced glomerulosclerosis (n = 1), and glomerulomegaly with severe tubular atrophy (n = 1). Presence of thin GBM on kidney biopsy, a feature of ADAS was noted on 3 biopsies; 1 other biopsy noted variable GBM thickness ranging from 125 to 375 nm.
Table 4.
Characteristics of COL4A3 P/LP heterozygotes with kidney biopsies
| Study ID, variant(s)a | Age | CKD stage, microscopic hematuria | Extra-renal features | Biopsy lab (yr), diagnosis | Thin GBM | IFTA | Global GS |
|---|---|---|---|---|---|---|---|
| Study_122123 COL4A3 p.Gly695Arg |
70–75 | G3bA3, Y |
N | Arkana (2017–2021): Light chain deposition disease, kappa type | NR | Severe | 14/23 |
| Study_14567 COL4A3 p.Gly883Arg, COL4A4 p.Gly774Arg, COL4A4 p.Gly1465Asp |
40–45 | G2A3, Y | N | Arkana (2017–2021): EM notes thin GBM; Severe IFTA, Global and segmental glomerulosclerosis 5/12 | Y | Severe | 5/12 |
| Study_148551 COL4A3 p.Gly695Arg |
45–50 | G3A3, Y | N | #1 - Brigham (2012–2016): Widespread endothelial cell injury with early glomerular capillary wall remodeling, suggesting primary vascular injury. She had thin BM and there was no evidence of immune complex disease or primary podocytopathy. #2 Arkana (2017–2021): FSGS (secondary favored). Moderate foot process effacement in the relatively well-preserved areas. | Y (2010–2015), NR 2016–2021) | Mild | 2/13 (most recent bx) |
| Study_49318 COL4A3 p.Gly336Cys COL4A4 p.Lys17Arg |
50–55 | G3bA3, Y | N | Arkana (2016–2021): Advanced glomerulosclerosis w/moderate-severe interstitial fibrosis, moderate arteriosclerosis. EM shows focal epithelial foot process effacement, rare small intramembranous electron dense materials seen. | NR | Moderate | 23/28 |
| Study_45371 COL4A3 c.Gly695Arg |
50–55 | G1A3, Y | N | GMC (2007–2011): FSGS w segmental foot process effacement | N | Mild | 0 |
| Study_6481 COL4A3 c.Gly695Arg |
25–30 | G1A3, Y | SNHL | #1 GMC (2002-2006)-MCD; #2 GMC (2011–2015): FSGS. Focal foot process effacement noted on EM. No thinning of GBM in two different kidney biopsy reports | NR | Moderate | 14/41 |
| Study_65222 COL4A3 c.Gly695Arg |
55–60 | G3A3, Y | N | GMC (2012–2016): FSGS and acute and chronic interstitial nephritis. EM showed variable GBM thickness ranging from 125–375 nm, averaging 260 nm thickness. Swollen and effaced podocyte foot processes noted | variable thickness 125–375 nm | Moderate | 1/6 |
| Study_79006 COL4A3 c.Gly695Arg |
50–55 | G3bA3, Y | N | GMC (2002–2006): FSGS, diffuse fusion of foot processes with segmental sclerosis. No thin GBM or foot process effacement mentioned | NR | Moderate | 1/5 |
| Study_89428 COL4A3 c.Gly695Arg COL4A3 Leu1474Pro |
45–50 | Not available; ESKD now | N | GMC (2007–2011): thin basement membrane nephropathy. EM with diffuse and uniform thinning of GBM. | Y | Minimal | 1/12 |
| Study_110766 COL4A3 c.Gly1167Arg |
55–60 | G3bA3, Y | N | Arkana (2016–2021): mild glomerulomegaly with severe tubular atrophy noted. No thinning of GBM on EM and mild epithelial foot process effacement noted | N | Severe | 10/16 |
| Study_80355 COL4A3 c.Gly695Arg COL4A3 c.Arg1661Cys |
40–45 | G2A0, Y | N | UPenn (1982–1986): no biopsy report available. PCP note-IgA nephropathy. No additional details available. | NR | NR | NR |
Arkana, Arkana Laboratories; CKD, chronic kidney disease; CR, complete response; EM, electronic microscopy; FSGS, focal segmental glomerulosclersoossis; GBM, glomerular basement membrane; GMC, Geisinger Medical Center; GS, glomerulosclerosis; IFTA, interstitial fibrosis and tubular atrophy; MM, multiple myeloma; NR, not reported; PCP, primary care provider; SRNS, steroid resistant nephrotic syndrome; UPenn, University of Pennsylvania.
Patient characteristics at the time of biopsy are shown.
Sample/Patient IDs used in Table 4 are not known to anyone outside of the research group.
Among the 19 patients with kidney failure per USRDS, the mean age at kidney failure was 58.3 (13.4) years. Causes of kidney failure listed on USRDS 2728 forms included AS (n = 2) nephrotic syndrome with FSGS (n = 3), hypertension (n = 2), renal artery stenosis (n = 2), unspecified with renal failure (n = 2), cholesterol emboli (n = 2), type 2 diabetes, other vasculitis, IgA nephropathy, chronic interstitial nephritis, ADPKD, and unspecified injury.
Discussion
In this study examining the phenotypic spectrum of pathogenic COL4A3 variants in an unselected population with a median of 15 years of follow-up time, we demonstrate a wide spectrum of phenotypic severity in genetically-determined ADAS due to COL4A3 with phenotypic variability by genotype. Our results expand our knowledge of heterozygotes beyond previous studies which mostly focused on family members of patients. Overall, rare P/LP COL4A3 variant heterozygotes were at increased risks of hematuria, albuminuria, FSGS, CKD, and kidney failure though penetrance and phenotypic severity varied by genotype group with glycine missense variants located in the collagenous domain being particularly penetrant with at least 80% of individuals having any phenotypic feature, including 70% with hematuria on urinalysis, and 8% with kidney failure per ICD code. We found that PTVs were less severe than glycine missense variants with only 52% PTV heterozygotes having any phenotypic feature, 30% having hematuria on urinalysis, and 4% having kidney failure per ICD code. Among individuals with glycine variants in the collagenous domain and available data, approximately 1 of 6 adults aged between 30 and 65 years were KDIGO high risk or greater and more than 1 of 2 adults aged 65+ years were KDIGO high risk or greater. Over a median follow-up period of 15 years, the majority (70%) of the COL4A3 heterozygotes had never had ACR testing done before, highlighting the potential importance of early genetic diagnosis to inform appropriate management.
Previous studies have not been able to comprehensively examine penetrance of genetically-defined ADAS, with varied reports depending on the study population (population-based vs. disease-specific) and the comprehensiveness of phenotyping. In case series of patients with ADAS or TBMN, the vast majority have hematuria, and most have proteinuria.18, 19, 20, 21 In a systematic review of literature on TBMN due to COL4A3 or COL4A4,20 phenotypic spectrum of 777 patients with heterozygous COL4A3/COL4A4 variants from 258 families included the following: 95% hematuria, 29% CKD, 15% kidney failure (mean age ∼53 years), 16% hearing loss, and 3% ocular lesions. However, the study noted lack of clear, uniform definitions for proteinuria, CKD, and hearing loss in the included studies, and did not compare these phenotypic features by COL4A3 variant groups. Results from research studies such as the Genomics England 100,000 Genomes Project report much lower prevalence of hematuria of 17.1% in collagenous domain glycine variants, due to reliance on defining hematuria from medical records.14 In our study with detailed phenotyping using extensive longitudinal EHR data including urinalyses, prevalence of hematuria was 70.3% using urinalysis data and 35.4% using ICD codes among patients heterozygous for COL4A3 Gly695Arg, a top hit for hematuria in genome-wide association studies.22,23
We also confirmed a strong association between COL4A3 and FSGS24 and found a prevalence of diagnosed FSGS of approximately 2% in COL4A3 heterozygotes and approximately 3% in Gly695Arg variant heterozygotes. This likely represents an underestimate because kidney biopsies are not done universally, and many were not screened for albuminuria. In a study by Wang et al.24 examining rare variants in FSGS genes in 363 FSGS cases and 363 ancestry-matched controls, the top 3 contributors in a dominant or X-linked model were COL4A5, WT1, and COL4A4. The study also reported 8 COL4A3 variants in FSGS cases versus 4 variants of unknown significance in matched controls; when restricted to glycine variants located in the collagenous domain, there were 6 in FSGS cases versus 0 in the controls. In a Toronto cohort of 193 individuals with FSGS from 174 families, 11% had a genetic diagnosis, including 28% in families with kidney disease, and 8% in sporadic cases. Heterozygous and hemizygous COL4A variants accounted for 55% of cases, including 3 COL4A3 (all glycine variants located within the collagenous domain), 1 COL4A4, and 8 COL4A5.5
Among previously described COL4A3 P/LP variants, collagenous domain glycine missense variants had the highest risk whereas PTVs had intermediate severity. These findings are consistent with previous literature that has shown that heterozygous COL4A3/COL4A4 pathogenic variants resulting in substitution of glycine residues in the collagenous domain with highly destabilizing residues (Arg, Val, Glu, and Asp) were associated with increased risk of kidney failure and hematuria in comparison to those adjacent to noncollagenous domains.14 To our knowledge, the weaker association of heterozygous PTVs compared to glycine collagenous domain variants has not been previously described and deserves further investigation. Type IV collagen proteins form protomers in the GBM, where α3(IV) forms multimers with α4(IV) and α5(IV).25 We hypothesize that disruption of the collagenous domain with glycine substitutions in 1 copy of COL4A3 will disrupt a significant proportion of heteromeric protein complexes, resulting in GBM defects and kidney phenotypes. In contrast, because the noncollagenous domains in the C-terminus of α3 chain of type IV collagen are essential to specificity and initiation of protomer assembly,26,27 PTVs of COL4A3 are likely impaired in heteromer formation. We hypothesize that for PTV carriers, the reference (nontruncated) allele predominantly produces protein to form functional heteromers, which may be sufficient for basement membrane function; therefore, PTV carriers manifest a milder phenotype than carriers of glycine substitutions in the collagenous domain. In addition, we found no association between other missense or inframe deletions that were not in the collagenous domain with kidney phenotypes, although this group had higher prevalence of bilateral sensorineural hearing loss. Computational prediction programs to assign COL4A3/4/5 variant pathogenicity have been found to have limited value.28 Interestingly, out of 26 patients in our COL4A3 P/LP cohort who had an additional rare variant in a COL4A3/4/5 gene, 4 (15.4%) had kidney failure, suggesting these second variants may contribute to AS severity. Further research is needed to fully evaluate genotype-phenotype correlations and individual variants’ pathogenicity.
Numerous opportunities for improved management of AS were identified, which may impact considerations for returning secondary findings of heterozygous COL4A3 P/LP variants to patients for earlier diagnosis and management.29 First, few patients with genetic ADAS had been diagnosed. Even among the patients with kidney failure, only 2 were recognized as due to AS. Early knowledge could be important for ensuring ACR testing is done because patients at risk of CKD should undergo screening,17 and less than a third of COL4A3 heterozygotes had ACR testing. Three-quarters of patients were not on ACEis or angiotensin receptor blockers, which have been shown to reduce the risk of CKD progression in autosomal recessive AS with similar suggestive evidence in ADAS.10,30,31 In addition, knowledge of ADAS could be useful in minimizing unnecessary testing for hematuria because approximately a third of glycine collagenous domain variant heterozygotes underwent urologic workup of hematuria.
The major strengths of our study are the use of an unselected patient population with careful phenotyping and use of propensity-matched controls over a median follow-up of 15 years, allowing us to provide insights on penetrance and phenotypic spectrum of ADAS. We were able to look at multiple phenotypic features of AS using a combination of ICD codes, longitudinal laboratory, USRDS, and kidney biopsy data. Although only 29% had ACR data available, 81% had urinalysis data available, and we examined albuminuria defined by ACR alone and by incorporating urinalysis data. There were limitations in our study. We focused on known P/LP variants in ClinVar as a starting point to establish our electronic phenotyping strategy before expanding research efforts to novel variants in COL4A3 as well as COL4A4 and COL4A5. We plan on expanding efforts to examine novel variants and variants of unknown significance in all 3 COL4A genes in the future. Data were collected from the EHR rather than rigorously collected research data, which limits our ability to describe more subtle phenotypic features such as ocular manifestations. Our study population was mostly European in ancestry, and additional studies in other cohorts, including more diverse populations are needed. In conclusion, we provide a strong foundation of evidence for the clinical significance and phenotypic spectrum of genetically-determined COL4A3 ADAS in an unselected population using carefully phenotyped EHR data and for demonstrating variability of phenotypic severity by genotype. Future studies are needed to evaluate the impact of earlier diagnosis, appropriate evaluation, and treatment of ADAS.
Appendix
List of Members of Regeneron Genetics Center
RGC Management and Leadership Team
Goncalo Abecasis, Ph.D., Aris Baras, M.D., Michael Cantor, M.D., Giovanni Coppola, M.D., Andrew Deubler, Aris Economides, Ph.D., Katia Karalis, Ph.D., Luca A. Lotta, M.D., Ph.D., John D. Overton, Ph.D., Jeffrey G. Reid, Ph.D., Katherine Siminovitch, M.D., Alan Shuldiner, M.D.
Sequencing and Lab Operations
Christina Beechert, Caitlin Forsythe, M.S., Erin D. Fuller, Zhenhua Gu, M.S., Michael Lattari, Alexander Lopez, M.S., John D. Overton, Ph.D., Maria Sotiropoulos Padilla, M.S., Manasi Pradhan, M.S., Kia Manoochehri, B.S., Thomas D. Schleicher, M.S., Louis Widom, Sarah E. Wolf, M.S., Ricardo H. Ulloa, B.S.
Clinical Informatics
Amelia Averitt, Ph.D., Nilanjana Banerjee, Ph.D., Michael Cantor, M.D., Dadong Li, Ph.D., Sameer Malhotra, M.D., Deepika Sharma, M.H.I., Jeffrey Staples, Ph.D.
Genome Informatics
Xiaodong Bai, Ph.D., Suganthi Balasubramanian, Ph.D., Suying Bao, Ph.D., Boris Boutkov, Ph.D., Siying Chen, Ph.D., Gisu Eom, B.S., Lukas Habegger, Ph.D., Alicia Hawes, B.S., Shareef Khalid, Olga Krasheninina, M.S., Rouel Lanche, B.S., Adam J. Mansfield, B.A., Evan K. Maxwell, Ph.D., George Mitra, B.A., Mona Nafde, M.S., Sean O’Keeffe, Ph.D., Max Orelus, B.B.A., Razvan Panea, Ph.D., Tommy Polanco, B.A., Ayesha Rasool, M.S., Jeffrey G. Reid, Ph.D., William Salerno, Ph.D., Jeffrey C. Staples, Ph.D., Kathie Sun, Ph.D., Jiwen Xin, Ph.D.
Analytical Genomics and Data Science
Goncalo Abecasis, D.Phil., Joshua Backman, Ph.D., Amy Damask, Ph.D., Lee Dobbyn, Ph.D., Manuel Allen Revez Ferreira, Ph.D., Arkopravo Ghosh, M.S., Christopher Gillies, Ph.D., Lauren Gurski, B.S., Eric Jorgenson, Ph.D., Hyun Min Kang, Ph.D., Michael Kessler, Ph.D., Jack Kosmicki, Ph.D., Alexander Li, Ph.D., Nan Lin, Ph.D., Daren Liu, M.S., Adam Locke, Ph.D., Jonathan Marchini, Ph.D., Anthony Marcketta, M.S., Joelle Mbatchou, Ph.D., Arden Moscati, Ph.D., Charles Paulding, Ph.D., Carlo Sidore, Ph.D., Eli Stahl, Ph.D., Kyoko Watanabe, Ph.D., Bin Ye, Ph.D., Blair Zhang, Ph.D., Andrey Ziyatdinov, Ph.D.
Research Program Management & Strategic Initiatives
Marcus B. Jones, Ph.D., Lyndon J. Mitnaul, Ph.D.
Disclosure
ARC has served as a consultant for Novartis, Reata, and Amgen, and has received research funding from Novartis, Novo Nordisk, and Bayer. All the other authors declared no competing interests.
Acknowledgments
The authors are grateful to the many patients who contributed to the MyCode Community Health Initiative by providing genomic and electronic health information. The authors thank the Regeneron Genetics Center for providing funding for patient enrollment and exome sequencing for the DiscovEHR study, and the authors would like to acknowledge the Geisinger-Regeneron DiscovEHR Collaboration for the genotypic and phenotypic data. This work was supported by NIGMS grant GM111913 to TM.
The authors thank the United States Renal Data System (USRDS) for their support. The data reported here have been supplied by the USRDS. The interpretation and reporting of these data are the responsibility of the author(s) and in no way should be seen as an official policy or interpretation of the US government.
Data Availability
The data supporting the findings of this study are available within the article and its Supplementary Data files. Additional information for reproducing the results described in the article is available upon reasonable request and subject to a data use agreement.
Ethics Declaration
This research was approved by the Geisinger Clinic Institutional Review Board and included participants in the MyCode Health Initiative who have exome sequencing data obtained as part of the Geisinger-Regeneron DiscovEHR collaboration. All participants provided written informed consent, and all experiments were performed in accordance with relevant guidelines and regulations.
Author Contributions
Study was designed by ARC and NTS; data collection was done by KVS, BSM, TM, VAb, and VAv; analysis was performed by YH and ARC; interpretation of results was conducted by KVS, NTS, IDB, TM, and ARC; manuscript was drafted by KVS and ARC; and the manuscript was revised critically for important intellectual content by all authors. All authors have approved the version to be published.
Footnotes
Supplementary Methods–Whole Exome Sequencing.
Figure S1. KDIGO Risk Categories by COL4A3 P/LP Variant Group and Controls.
Figure S2. KDIGO Risk Categories in Glycine collagenous domain variants versus controls, by age group.
Table S1. Coding definitions for comorbidities.
Table S2. Characteristics by individual variants.
Table S3. Characteristics of COL4A3 P/LP Heterozygotes and nonheterozygotes before and after matching.
Table S4.COL4A3 P/LP heterozygotes with an additional rare variant in COL4A3/4/5.
Table S5. Chart Review data of COL4A3 AD Alport Syndrome-related Diagnosis and Management.
STROBE Statement—checklist of items that should be included in reports of observational studies.
Supplementary Material
Supplementary Methods–Whole Exome Sequencing.
Figure S1. KDIGO Risk Categories by COL4A3 P/LP Variant Group and Controls.
Figure S2. KDIGO Risk Categories in Glycine collagenous domain variants versus controls, by age group.
Table S1. Coding definitions for comorbidities.
Table S2. Characteristics by individual variants.
Table S3. Characteristics of COL4A3 P/LP Heterozygotes and nonheterozygotes before and after matching.
Table S4.COL4A3 P/LP heterozygotes with an additional rare variant in COL4A3/4/5.
Table S5. Chart Review data of COL4A3 AD Alport Syndrome-related Diagnosis and Management.
STROBE Statement—checklist of items that should be included in reports of observational studies.
References
- 1.Kashtan C.E., Ding J., Garosi G., et al. Alport syndrome: a unified classification of genetic disorders of collagen IV α345: a position paper of the Alport Syndrome Classification Working Group. Kidney Int. 2018;93:1045–1051. doi: 10.1016/j.kint.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 2.Storey H., Savige J., Sivakumar V., Abbs S., Flinter F.A. COL4A3/COL4A4 mutations and features in individuals with autosomal recessive Alport syndrome. J Am Soc Nephrol. 2013;24:1945–1954. doi: 10.1681/ASN.2012100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savige J., Gregory M., Gross O., Kashtan C., Ding J., Flinter F. Expert guidelines for the management of Alport syndrome and thin basement membrane nephropathy. J Am Soc Nephrol. 2013;24:364–375. doi: 10.1681/ASN.2012020148. [DOI] [PubMed] [Google Scholar]
- 4.Voskarides K., Damianou L., Neocleous V., et al. COL4A3/COL4A4 mutations producing focal segmental glomerulosclerosis and renal failure in thin basement membrane nephropathy. J Am Soc Nephrol. 2007;18:3004–3016. doi: 10.1681/ASN.2007040444. [DOI] [PubMed] [Google Scholar]
- 5.Yao T., Udwan K., John R., et al. Integration of genetic testing and pathology for the diagnosis of adults with FSGS. Clin J Am Soc Nephrol. 2019;14:213–223. doi: 10.2215/CJN.08750718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adam J., Connor T.M.F., Wood K., et al. Genetic testing can resolve diagnostic confusion in Alport syndrome. Clin Kidney J. 2014;7:197–200. doi: 10.1093/ckj/sft144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sen E.S., Dean P., Yarram-Smith L., et al. Clinical genetic testing using a custom-designed steroid-resistant nephrotic syndrome gene panel: analysis and recommendations. J Med Genet. 2017;54:795–804. doi: 10.1136/jmedgenet-2017-104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rheault M.N., Smoyer W.E. Long-term ACE inhibition in Alport syndrome: are the benefits worth the risks? Kidney Int. 2020;97:1104–1106. doi: 10.1016/j.kint.2020.01.030. [DOI] [PubMed] [Google Scholar]
- 9.Kashtan C.E., Gross O. Clinical practice recommendations for the diagnosis and management of Alport syndrome in children, adolescents, and young adults-an update for 2020. Pediatr Nephrol. 2021;36:711–719. doi: 10.1007/S00467-020-04819-6. [DOI] [PubMed] [Google Scholar]
- 10.Gross O., Tönshoff B., Weber L.T., et al. A multicenter, randomized, placebo-controlled, double-blind phase 3 trial with open-arm comparison indicates safety and efficacy of nephroprotective therapy with ramipril in children with Alport’s syndrome. Kidney Int. 2020;97:1275–1286. doi: 10.1016/J.KINT.2019.12.015. [DOI] [PubMed] [Google Scholar]
- 11.Carey D.J., Fetterolf S.N., Davis F.D., et al. The Geisinger MyCode community health initiative: an electronic health record–linked biobank for precision medicine research. Genet Med. 2016;18:906–913. doi: 10.1038/gim.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landrum M.J., Lee J.M., Benson M., et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dewey F.E., Murray M.F., Overton J.D., et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016;354(6314):aaf6814. doi: 10.1126/science.aaf6814. [DOI] [PubMed] [Google Scholar]
- 14.Gibson J.T., Huang M., Shenelli Croos Dabrera M., et al. Genotype–phenotype correlations for COL4A3–COL4A5 variants resulting in Gly substitutions in Alport syndrome. Sci Rep. 2022;12:2722. doi: 10.1038/s41598-022-06525-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.U.S. Renal Data System. 2022 Researcher’s Guide to the USRDS Database. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases: Bethesda, MD, 2022.
- 16.Inker L.A., Eneanya N.D., Coresh J., et al. Collaboration Chronic Kidney Disease Epidemiology New creatinine- and cystatin C-based equations to estimate GFR without race. N Engl J Med. 2021;385:1737–1749. doi: 10.1056/NEJMoa2102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevens P.E., Levin A. Kidney Disease: Improving Global Outcomes Chronic Kidney Disease Guideline Development Work Group Members. Evaluation and management of chronic kidney disease: synopsis of the kidney disease: improving global outcomes 2012 clinical practice guideline. Ann Intern Med. 2013;158:825–830. doi: 10.7326/0003-4819-158-11-201306040-00007. [DOI] [PubMed] [Google Scholar]
- 18.Kamiyoshi N., Nozu K., Fu X.J., et al. Genetic, clinical, and pathologic backgrounds of patients with autosomal dominant Alport syndrome. Clin J Am Soc Nephrol. 2016;11:1441–1449. doi: 10.2215/CJN.01000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groen in ’t Woud S., Rood I.M., Steenbergen E., et al. Kidney disease associated with mono-allelic COL4A3 and COL4A4 variants: a case series of 17 families. Kidney Med. 2023;5 doi: 10.1016/J.XKME.2023.100607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthaiou A., Poulli T., Deltas C. Prevalence of clinical, pathological and molecular features of glomerular basement membrane nephropathy caused by COL4A3 or COL4A4 mutations: a systematic review. Clin Kidney J. 2020;13:1025–1036. doi: 10.1093/CKJ/SFZ176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savige J., Rana K., Tonna S., Buzza M., Dagher H., Wang Y.Y. Thin basement membrane nephropathy. Kidney Int. 2003;64:1169–1178. doi: 10.1046/j.1523-1755.2003.00234.x. [DOI] [PubMed] [Google Scholar]
- 22.Benonisdottir S., Kristjansson R.P., Oddsson A., et al. Sequence variants associating with urinary biomarkers. Hum Mol Genet. 2019;28:1199–1211. doi: 10.1093/hmg/ddy409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gagliano Taliun S.A., Sulem P., Sveinbjornsson G., et al. GWAS of hematuria. Clin J Am Soc Nephrol. 2022;17:672–683. doi: 10.2215/CJN.13711021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M., Chun J., Genovese G., et al. Contributions of rare gene variants to familial and sporadic FSGS. J Am Soc Nephrol. 2019;30:1625–1640. doi: 10.1681/ASN.2019020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khoshnoodi J., Cartailler J.P., Alvares K., Veis A., Hudson B.G. Molecular recognition in the assembly of collagens: terminal noncollagenous domains are key recognition modules in the formation of triple helical protomers. J Biol Chem. 2006;281:38117–38121. doi: 10.1074/JBC.R600025200. [DOI] [PubMed] [Google Scholar]
- 26.Söder S., Pöschl E. The NC1 domain of human collagen IV is necessary to initiate triple helix formation. Biochem Biophys Res Commun. 2004;325:276–280. doi: 10.1016/J.BBRC.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 27.Boutaud A., Borza D.B., Bondar O., et al. Type IV collagen of the glomerular basement membrane. Evidence that the chain specificity of network assembly is encoded by the noncollagenous NC1 domains. J Biol Chem. 2000;275:30716–30724. doi: 10.1074/JBC.M004569200. [DOI] [PubMed] [Google Scholar]
- 28.Shulman C., Liang E., Kamura M., et al. Type IV collagen variants in CKD: performance of computational predictions for identifying pathogenic variants. Kidney Med. 2021;3:257–266. doi: 10.1016/j.xkme.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller D.T., Lee K., Gordon A.S., et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2021 update: a policy statement of the American College of Medical Genetics and Genomics (ACMG) Genet Med. 2021;23:1391–1398. doi: 10.1038/s41436-021-01171-4. [DOI] [PubMed] [Google Scholar]
- 30.Boeckhaus J., Hoefele J., Riedhammer K.M., et al. Lifelong effect of therapy in young patients with the COL4A5 Alport missense variant p.(Gly624Asp): a prospective cohort study. Nephrol Dial Transplant. 2022;37:2496–2504. doi: 10.1093/NDT/GFAC006. [DOI] [PubMed] [Google Scholar]
- 31.Stock J., Kuenanz J., Glonke N., et al. Prospective study on the potential of RAAS blockade to halt renal disease in Alport syndrome patients with heterozygous mutations. Pediatr Nephrol. 2017;32:131–137. doi: 10.1007/S00467-016-3452-Z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the article and its Supplementary Data files. Additional information for reproducing the results described in the article is available upon reasonable request and subject to a data use agreement.