Abstract
Lysyl hydroxylase 2 (LH2) catalyzes the formation of highly stable hydroxylysine aldehyde-derived collagen cross-links (HLCCs), thus promoting lung cancer metastasis through its capacity to modulate specific types of collagen cross-links within the tumor stroma. Using 1 and 2 from our previous high-throughput screening (HTS) as lead probes, we prepared a series of 1,3-diketone analogues, 1–18, and identified 12 and 13 that inhibit LH2 with IC50’s of approximately 300 and 500 nM, respectively. Compounds 12 and 13 demonstrate selectivity for LH2 over LH1 and LH3. Quantum mechanics/molecular mechanics (QM/MM) modeling indicates that the selectivity of 12 and 13 may stem from noncovalent interactions like hydrogen bonding between the morpholine/piperazine rings with the LH2-specific Arg661. Treatment of 344SQ WT cells with 13 resulted in a dose-dependent reduction in their migration potential, whereas the compound did not impede the migration of the same cell line with an LH2 knockout (LH2KO).
Keywords: Lysyl hydroxylase 2, LH2, α-Ketoglutarate, Hydroxylysine aldehyde-derived collagen cross-links, HLCC, Collagen, Inhibitor
Lysyl hydroxylase 2 (LH2)-mediated telopeptidyl hydroxylysines form structurally rigid hydroxylysine aldehyde-derived collagen cross-links (HLCCs) resistant to cleavage by collagenases.1 It is well-known that reduced lysine hydroxylation is a cause of genetic connective tissue disorders such as Bruck Syndrome,2−6 while abnormally elevated HLCC formation is associated with fibrosis and cancer metastasis.7−11
Three lysyl hydroxylase isoforms (LH1–3) play essential roles in collagen modifications. Each isoform possesses two functionally distinct enzymatically active domains.12 LH2 can hydroxylate Lys residues in the telopeptides, while all three LH isoforms hydroxylate lysine residues in the helical domain.2 In fibrotic diseases of the lung13 and liver,14 HLCCs are the predominant types of cross-links attributable to excessive hydroxylation of the telopeptidyl Lys, owing to the overexpression of LH2 in fibroblasts.8,15 A similar switch is associated with tumor stroma, where an increased level of LH2 leads to metastasis and is correlated with lower survival.7,9 Heretofore, LH2 was thought to be localized in the ER, where it hydroxylates procollagen Lys residues before the triple helix formation.16 However, it was recently found to modify collagen in the extracellular space as it is secreted by carcinoma cells.17 It is shown to be expressed in cancer-associated fibroblasts and contributes to a switch toward a high-HLCC, low-LCC state in the tumor stroma.18
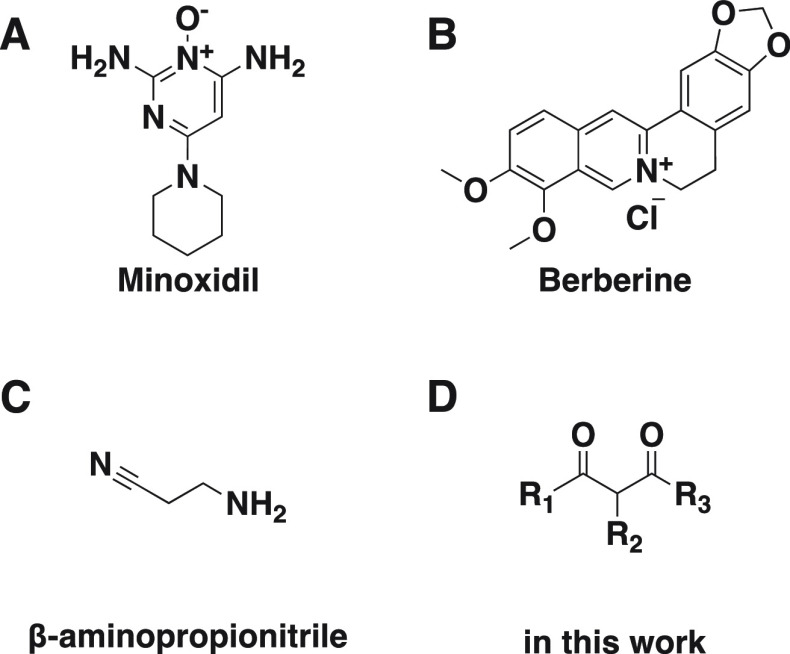
Minoxidil (Figure 1A), a widely recognized antifibrotic agent, was believed to reduce LH expression, thereby decreasing hydroxylysylpyridinoline cross-linking.19 However, Pfeffer et al. found that minoxidil does not significantly impact LH activity during postnatal mouse lung development.20 Berberine (Figure 1B) inhibits LH2 expression in TNBC cells, reducing the proliferation, motility, and glycolysis. This is achieved by suppressing interleukin-6 (IL-6) secretion, indirectly regulating LH2 activity.21 β-Aminopropionitrile (Figure 1C) has been observed to induce downregulation of LH2 expression and impede the formation of a stable matrix, likely by direct inhibition of LOX family members.22
Figure 1.
Compounds reported as LH2 inhibitors.
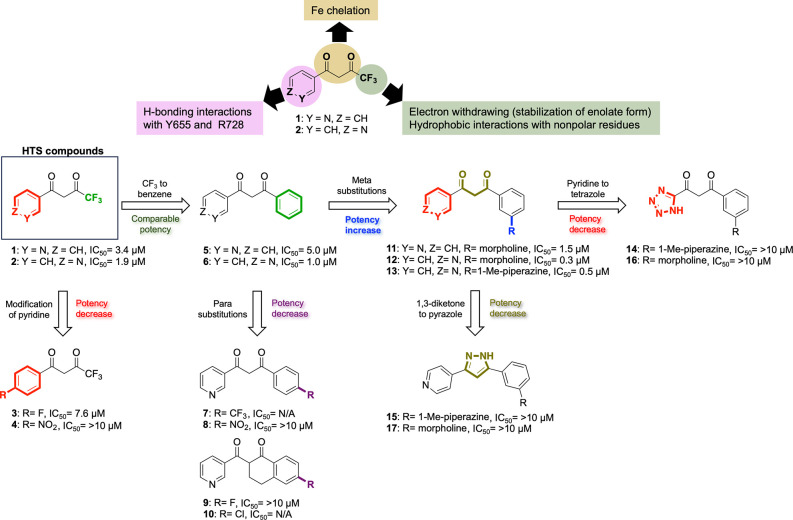
Our previous high-throughput screening utilizing luminescence-based lysyl hydroxylase activity assay identified two 1,3-diketone analogues (Figure 1D and compounds 1 and 2 in Figure 2) with IC50 values of 3.4 and 1.9 μM, respectively.31 The same assay conducted in the presence of an excess of Fe2+ demonstrated that the activity of 1 arises from its direct interaction with the enzyme rather than any interference with the assay components (Figure S48). While 1,3-diketone analogues are widely distributed in natural products and exhibit various pharmacological activities, including antibacterial, antioxidant, antiviral, insecticidal, and antifungal properties,23−27 their inhibition activity toward LH2 has not been reported. Notably, owing to their inherent tautomerism, these compounds are uniquely poised to engage in metal chelation and hydrogen bonding, endowing them with considerable potential as a molecular scaffold for intermediates in various organic reactions, metal–organic hybrid materials, and novel drug design.28,29 Indeed, our molecular docking studies confirmed the chelation of Fe(II) by the dicarbonyl moiety and hydrogen bonding of the pyridine nitrogen with LH2 active-site residues (Y655 and R728, human PLOD2), as illustrated in Figure 3A,B, providing a foundation for compounds 1 and 2 as leading probes for further optimization.
Figure 2.
Rational design flowcharts of LH2 inhibitors.
Figure 3.
Binding interactions between HTS hit compounds 1 (A and C), 2 (B), and 5 (C) and the LH2 active site predicted by molecular docking studies and observed in MD and QM/MM simulations. (A) Calculated distance between the phenol hydrogen (Y655) and the pyridine nitrogen of 1 is 2.1 Å. (B) Calculated distance between the guanidine hydrogen (R728) and the pyridine nitrogen of 2 is 1.9 Å. (C) The entry of the active site of LH2 associated with 1 shows the hydrophobic environment in proximity to trifluoromethyl (−CF3) group of 1. (D) Docked model of 5 presenting the surface environment of the benzene ring of the compound.
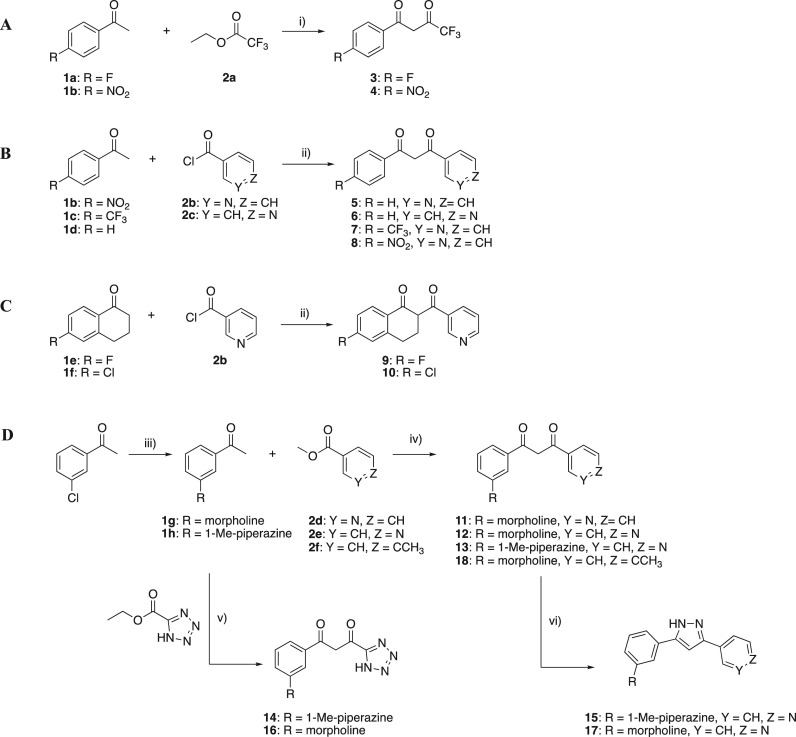
Here we report a structure-based rational design strategy of 1,3-diketone analogues (compounds 1–18) as LH2 inhibitors along with their inhibitory activity and computation investigation. Although compounds 1–8 were previously known, their biological activity against LH2 has not been studied. Compounds 9–18 were newly designed specifically for this study. The 1,3-diketone analogues were synthesized as depicted in Scheme 1. Compounds 3 and 4 were prepared via modified Claisen condensation.29 Compounds 5–10 were synthesized from acyl chlorides with enolated ketones by soft enolization30 in the presence of magnesium bromide diethyl etherate and Hünig’s base. 11–13, and 18 were synthesized from acylation of meta-substituted acetophenones 1g–1h with esters 2d–2f in the presence of lithium bis(trimethylsilyl) amide. The meta-substituted acetophenones also underwent an acylation reaction with 2-methoxypropyl-protected ethyl tetrazole-5-carboxylate to afford tetrazole-substituted variants 14 and 16. Pyrazole-substituted derivatives 15 and 17 were prepared by reacting 12 or 13 with hydrazine monohydrate in ethanol.
Scheme 1. Synthesis of Inhibitors 3–18.
Reagents and reaction conditions: (i) NaH, THF, 0 °C to RT, 12 h; (ii) MgBr2·OEt, DIPEA, DCM, RT, 12 h; (iii) 3-chloroacetopheone, morpholine or 1-methyl-piperazine, Pd2 (dba)3, DavePhos, K3PO4, DME, Ar, 100 °C, 12 h; (iv) LiN(SiMe3)2, Ar, THF, −40 °C, 6 h; (v) (a) 2-methoxypropene, p-TSA, THF, RT, 1 h. (b) LiN(SiMe3)2, Ar, THF, −40 °C, 6 h, (c) HCl; vi) NH2NH2·H2O, ethanol, RT, 12 h.
Previously, we conducted an exhaustive computational investigation on forty-four molecules,32 which included most of the compounds discussed in this paper. Based on polarizable molecular dynamics (MD) simulation and coupled quantum mechanics/molecular mechanics (QM/MM) optimizations, we found that the enolate forms of 1 and 2 have considerably strong interactions with the active site (−169.4 and −139.7 kcal mol–1, respectively) as well as several NCIs (noncovalent interactions) with the residues of the active site including Y655 and R728.
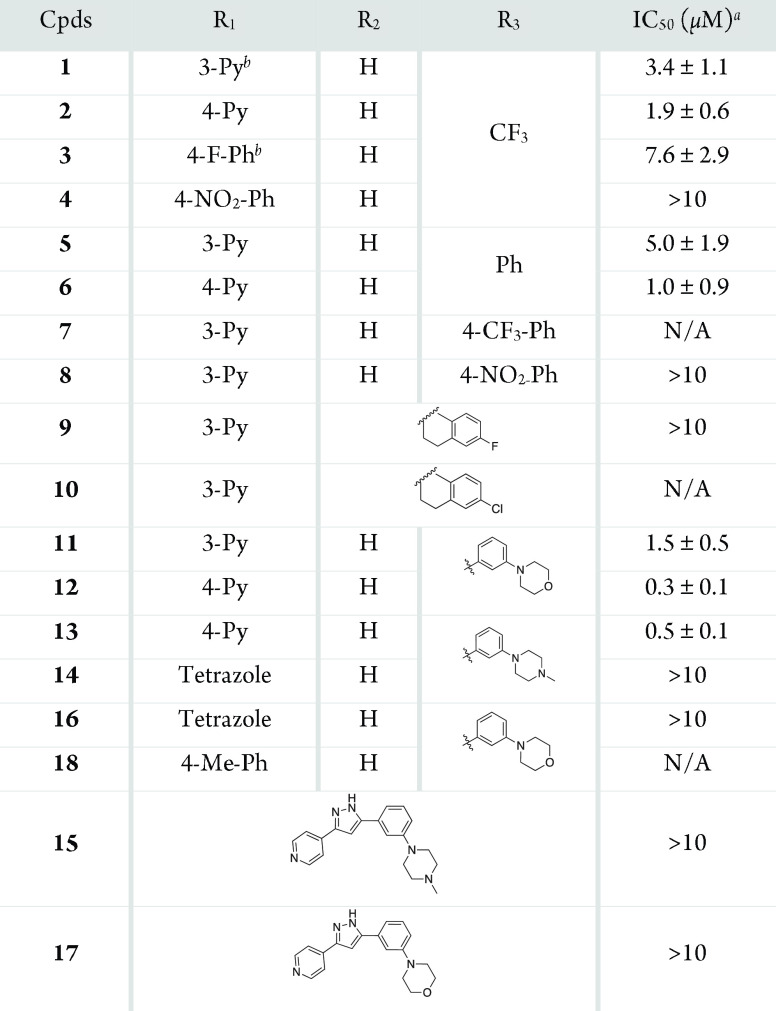
First, compounds 3 and 4 featuring para-substituted benzene rings in the R1 position were prepared to investigate the involvement of pyridine in the interaction with inner residues Y655 and R728. These compounds displayed diminished inhibitory activity relative to 1 and 2 and weaker calculated polarizable QM/MM interaction with LH2 (−50.4 and −38.7 kcal mol–1, respectively),32 underscoring the necessity of pyridine in mediating the interaction with the LH2 active site.
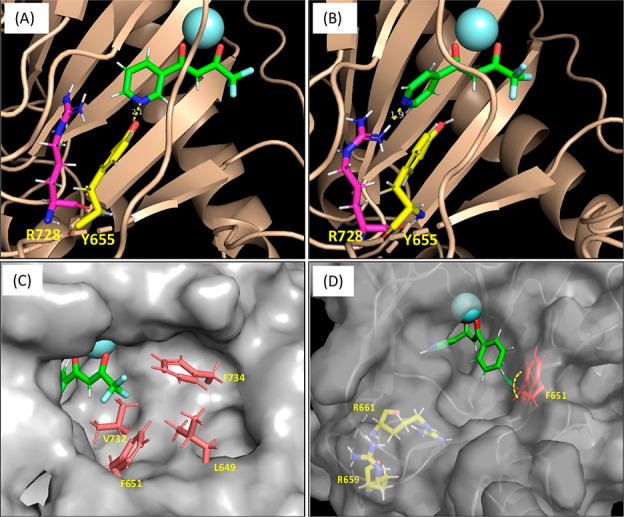
We then implemented a modification to the R3 position. We hypothesized that the trifluoromethyl (−CF3) group plays a role not only in withdrawing electron density, which stabilizes the formation of an enolate but also interacts with nonpolar residues situated at the entrance of the LH2 active site via hydrophobic interactions, as shown in Figure 3C. Thus, we opted to substitute the trifluoromethyl group with a benzene ring, which is considered to possess similar characteristics, along with better accessibility for functionalization. Notably, 5 (IC50 = 5.0 μM) and 6 (IC50 = 1.0 μM) retained inhibitory activity relative to the original hits from HTS. Compared with compounds 1 and 2, compounds 5(32) and 6 (Table S2) show considerably stronger interactions with the LH2 (−185.2 and −200.1 kcal mol–1, respectively).
We further functionalized compound 5 by introducing additional groups at the para position of the benzene ring (7 and 8) and at the 6-position of the tetralin group, which is linked to the R2 position (9 and 10). However, including functional groups at the para position of the benzene led to a decrease in the potency of the compounds (7; IC50 = N/A, 8; IC50 = > 10 μM, 9; IC50 = > 10 μM, 10; IC50 = N/A). This implies that the para position of the benzene ring may offer restricted space for functional groups to occupy, which is further corroborated by the molecular docking analysis and MD simulation of compound 5 onto the LH2 homology model, as depicted in Figure 3D. The calculated QM/MM interaction energies (IEQM/MM) for compounds 7–10 were −114.2, −190.5, −212.6, and −212.9 kcal mol–1, respectively.32 This suggests that the presence of −CF3 in the para position of the benzene ring in compound 7 led to a significant decrease in the interaction energies. At the same time, it was slightly increased for the other three compounds compared to compound 5. Interestingly, the number of residues having noncovalent interactions with these four compounds showed a slight decrease compared to compound 5, especially in the case of compounds 7 and 9.32 This suggested that our modifications at the para position of the benzene ring do not improve interaction between the inhibitor and the residues at the binding pocket. Given the LH2–inhibitor interaction energies, noncovalent interactions, and distinct hindrance by F651 on the para-modification, we investigated whether a meta-substitution could enhance binding to LH2.
The docked model of 5 revealed possible additional recognition sites, R661 and R659, proximal to the meta position of the benzene of 5 (Figure 3D). This was later supported by MD simulation of the ligand-bound system of this compound.32 These two basic amino acid residues adjacent to the LH2 active site are specific to this member of the LH family. (The corresponding residues in LH1 and LH3 are Glu and Pro, respectively.) These residues are thought to play a role in the electrostatic interactions with acidic aspartate and glutamate residues positioned adjacent to the Lys residues on fibrillar collagens.12 We postulated that LH2’s unique telopeptidyl lysyl hydroxylase activity and selectivity are determined partly by electrostatic interactions between the basic domain of LH2 and the acidic domain of collagen telopeptides. To achieve favorable interactions with the basic residues, morpholine (11 and 12) and 1-methyl piperazine (13) rings, which possess the intrinsic capability to participate in hydrogen bonding33,34 were incorporated at the meta-position of the benzene of compounds 5 and 6. Introducing the meta-substituted morpholine and methyl piperazine rings boosts the potencies (11; IC50 = 1.5 μM, 12; IC50 = 0.3 μM, and 13; IC50 = 0.5 μM) compared with the parent inhibitors 5 and 6. Notably, the weaker potency of 11 compared to that of 12 indicates that the pyridine nitrogen atom in the 4-position is optimal for interacting with inner residues in the active site.
The results of IEQM/MM between compounds 12 and 13 with LH2 are −259.9 and −216.3 kcal mol–1, respectively (see Table S2). The low values of IEQM/MM suggest a strong interaction between these two analogues and the enzyme. Table S2 also shows that the calculated interactions due to the QM region in compounds 12 and 13 are similar, −197.6 and −197.4 kcal mol–1, respectively, but the interactions due to the MM region in compound 12 are about three times greater than those in compound 13 (−62.3 and −18.9 kcal mol–1, respectively). In the case of 6, the interaction is weaker than the other two but the MM region stability is enhanced compared with the other two ligands.
A closer look into the NCI plots shows that compound 12 has two hydrogen bonds with R728 and R661 (Figure 4A-middle), and compound 13 has weak interactions with R661 (Figure 4A-right). In comparison, compound 6 only has two hydrogen bonds with two water molecules in the active site (Figure 4A-left). The interactions of 12 and 13, particularly with the R661 residue, suggest that their potencies might derive from the engagement of morpholine and methyl piperazine rings with LH2-specific R661.
Figure 4.
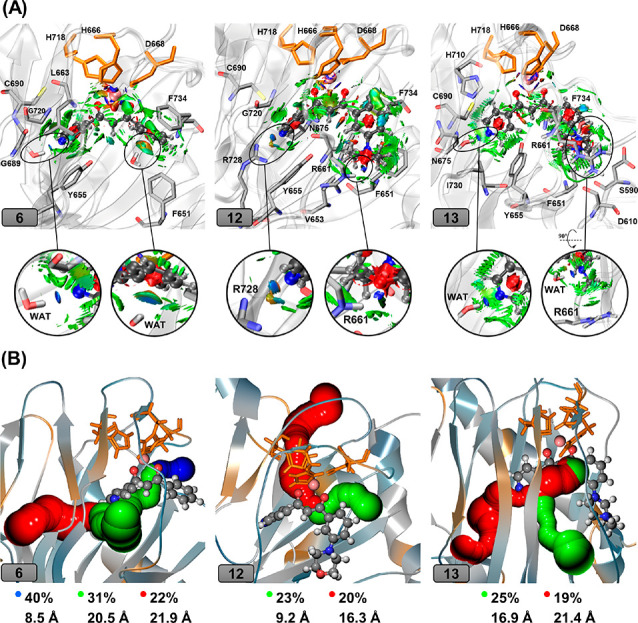
(A) The plot of the noncovalent interactions between compounds 6, 12, and 13 and the surrounding amino acid residues. The inhibitors are given in ball-and-sticks, and Fe(II) is shown in pink sphere. Residues forming noncovalent interactions with the inhibitor are shown in sticks. Red surfaces are related to repulsive interactions, while green surfaces denote weak interactions like van der Waals, and blue ones show strong attractive interactions such as hydrogen bonds. (B) O2-transporting tunnels with largest calculated percentages along the trajectory observed in compounds 6, 12, and 13. Calculated tunnels are colored in blue, green, and red, respectively, based on the tunnel’s length (Å) (blue: shortest, red: longest).
Our previous study suggested that three major tunnels exist in the apo-LH2, which can transport molecular oxygen to the active site, with tunnel availability probabilities of 63%, 51%, and 32% for blue, green, and red tunnels, respectively.32 Our results also showed that compounds 7 and 8 and some other studied molecules in that study considerably decreased the availability of these tunnels and elongated the tunnels’ lengths. As seen in Figure 4B, the tunneling results for compounds 6, 12, and 13 showed the same trend in which the availability/length of the tunnels decreased/increased. Interestingly, in the case of compounds 12 and 13, only two major putative tunnels are available for oxygen transportation.
A final set of modifications were performed, based on 12 and 13, by replacing the pyridine moiety with tetrazole (16; IC50 = > 10 μM, 14; IC50 = > 10 μM) and the 1,3-diketone with pyrazole (17; IC50 = > 10 μM, 15; IC50 = > 10 μM). The results indicated that pyridine and 1,3-diketone scaffolds are essential for the activity of the studied compounds. In addition, 18 with the toluene group on the R1 position shows no activity, supporting the importance of pyridine.
In light of the initial investigation of the structure–activity relationship (SAR) using a luminescence-based LH activity assay (Table 1) and further supported by computational studies, the HTS hit compounds (1 and 2) and the four most potent compounds (6, 11, 12, and 13) were further evaluated using the same activity assay against all three LH isoforms for selectivity profiling (Table 2). Compounds 12 and 13 exhibited the most promising LH2 selectivity among the six inhibitors tested. Specifically, the inhibition potency of compound 12 against LH2 was approximately 8-fold and 9-fold stronger than those against LH1 and LH3, respectively. In the case of 13, the LH2 selectivity over LH3 was observed as 9-fold, while it displayed a relatively lower selectivity (3-fold) over LH1. These results indicate that the engagement of the morpholine and methyl piperazine rings with the LH2-specific residue R661 is responsible for the selective binding to LH2.
Table 1. SAR of 1,3-Diketone Analogues.
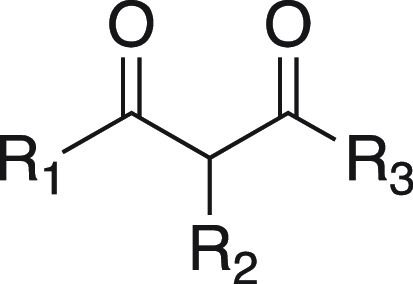
IC50 values were measured using a luminescence-based LH activity assay against LH2. See Supporting Information for a detailed description of the assay conditions.
Py = pyridyl, Ph = phenyl
Table 2. Selectivity of Inhibitors toward All Three LH Isoforms.
| IC50 (μM)a |
|||
|---|---|---|---|
| Cpds | LH2 | LH1 | LH3 |
| 1 | 3.4 ± 1.1 | 4.3 ± 1.3 | 5.2 ± 4.0 |
| 2 | 1.9 ± 0.6 | 4.8 ± 0.9 | 4.8 ± 0.8 |
| 6 | 1.0 ± 0.9 | 6.4 ± 8.0 | 5.2 ± 6.0 |
| 11 | 1.5 ± 0.5 | 1.7 ± 0.8 | 4.8 ± 1.9 |
| 12 | 0.3 ± 0.1 | 2.3 ± 0.9 | 2.7 ± 2.9 |
| 13 | 0.5 ± 0.1 | 1.6 ± 0.6 | 4.7 ± 8.8 |
IC50 values were measured using a luminescence-based LH activity assay. See Supporting Information for a detailed description of the assay conditions.
Compound 6 demonstrated modest selectivity toward LH2, with 6-fold and 5-fold more potency than LH1 and LH3, respectively, while compounds 1, 2, and 11 showed less than 3-fold or no selectivity. The moderate selectivity of compound 6 can be attributed to the hydrophobic interactions between the benzene ring of 6 and hydrophobic residues (F651 and F734) at the entry of the active site, which aids in the stable binding to LH2, as shown in Figure 4A.
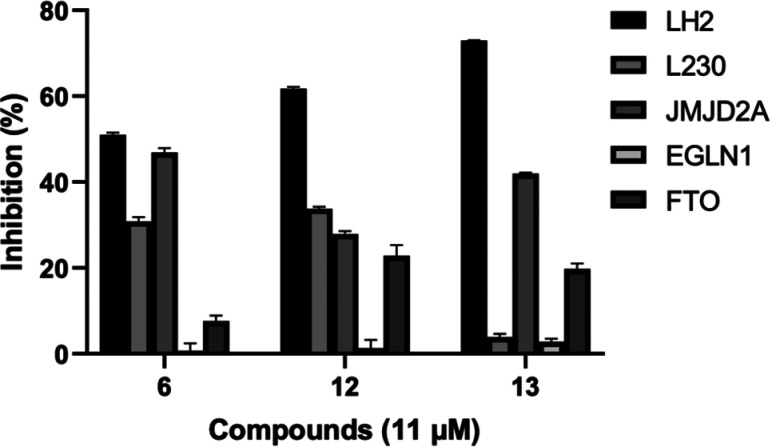
To further assess the selectivity of compounds 6, 12, and 13, we subjected them to screening against other Fe(II)/2OG-dependent enzymes. These enzymes included FTO (fat mass and obesity-associated protein, which preferentially demethylates N6-methyladenosine in RNA35), JMJD2A (the histone demethylase Jumonji domain-containing protein 2A36), EGLN1 (Egl-9 family hypoxia-inducible factor 1, also called prolyl hydroxylase domain-containing protein 2 or PHD237), and mimivirus L230, which hydroxylates lysine and glycosylates hydroxylysine residues on collagen.38 The percent inhibition of each of these enzymes at a dose of 11 μM compound (6, 12, or 13) is shown in Figure 5. The results indicate that all three compounds display a more pronounced inhibitory effect toward LH2 than other enzymes. Compound 6 shows lower selectivity for LH2 than the other compounds as it inhibits JMJD2A similarly to LH2.
Figure 5.
Selectivity profile of compounds against other Fe(II)/2OG-dependent enzymes. Compounds 6, 12, and 13 were tested at 11 μM for inhibition of LH2 and other Fe(II)/2OG-dependent enzymes L230, JMJD2A, EGLN1, and FTO. Inhibition (%) is the percent decrease in the luminescence signal relative to DMSO controls in the luminescence-based selectivity assay. All samples were performed in duplicate, and error bars indicate standard error.
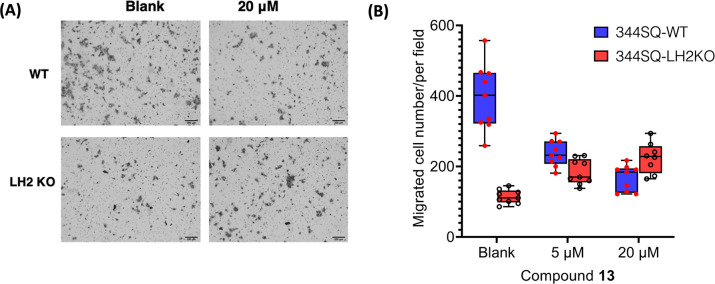
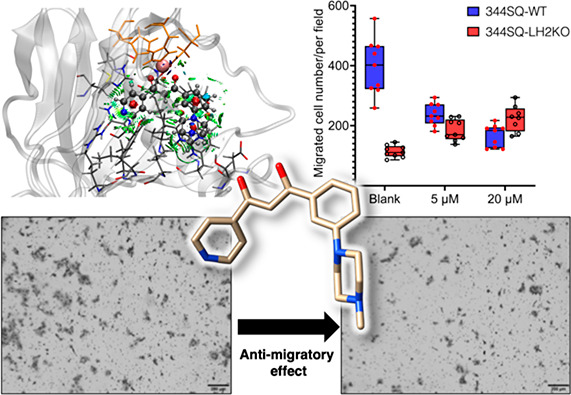
High expression of LH2 has been shown to drive cell migration in multiple cancer types.12,39,40 To determine if the most potent compounds identified in our study have selective activity against LH2 in cells, we evaluated the inhibitory effect of representative compound 13 on metastatic murine NSCLC (nonsmall cell lung cancer) cell line 344SQ that harbor mutations in Kras and p53.41 We used the parental 344SQ, Crispr-Cas9 edited LH2 KO 344SQ cell lines for the proliferation and migration assays.
Figure 6 shows that compound 13 demonstrated a dose-dependent inhibition on the parental 344SQ cells. In contrast, it was observed that compound 13 did not exert any obvious impact on the proliferation of parental 344SQ cells at the concentrations that were tested (Figure S47). Importantly, the compound did not hamper the migration of LH2KO cells, as depicted in Figures 6A and B, strongly suggesting its discerning activity for LH2 in both in vitro and cellular settings.
Figure 6.
Cell migration assay. Compound 13 suppresses cell migration of 344SQ lung adenocarcinoma cells (LH2b is highly expressed). A) Migrated 344SQ cells (WT and LH2 KO) in Boyden chambers were imaged after treatment with vehicle control (blank) and 20 μM of 13. B) Migrated 344SQ cells were counted after treatment with vehicle control (blank), 5 μM, and 20 μM of 13. Triplicates were performed for each condition, and the migration assay was repeated twice independently. When compared to 344SQ-WT cells treated with vehicle control (blank), 5 μM, and 20 μM of 13, a two-sided Student’s t-test analysis gave values of p = 0.000006, 0.001, and 0.07 for the corresponding concentrations of 13 in 344SQ LH2KO cells, respectively.
Notably, LH2KO cells were employed as a reference control to investigate whether the inhibitory effect of compound 13 is solely due to its action on LH2 or whether it interferes with other targets. If compound 13 targets additional molecules that promote cell migration, then it would still exhibit an inhibitory effect. Although there is no statistically significant difference between the migrations of WT and LH2KO cells treated with 20 μM compound 13 (p-value = 0.07 based on nine readings from a two-sided Student’s t test analysis), the observation that compound 13 induces a slight increase in the migration of LH2KO cells is intriguing. One plausible explanation for this observation is that compound 13 shows preferential binding to LH2 in the presence of LH2. In contrast, the compound interacts with alternative targets in its absence, resulting in the observed effect. Further experiments are underway to explore the potential alternative targets of compound 13 and to elucidate the mechanisms underlying its effect on cell migration in LH2KO cells.
In conclusion, the current study delineates the synthesis, design, and evaluation of a series of 18 1,3-diketone analogues (compounds 1–18) with respect to their potential to inhibit the LH2 enzymatic activity through luciferase-based biochemical and cell migration assays. The pyridine moiety at R1 was found to be a prerequisite for efficient hydrogen-bonding interactions with LH2’s active site residues. R3 was tailored to accommodate a benzene ring, replacing the trifluoromethyl group in the HTS compounds. Meta-substitution of the benzene ring conveys an advantageous effect on the LH2 inhibitory activity, whereas para-modification attenuates potency due to hindrance by F651. Notably, compounds 12 and 13, featuring morpholine and 1-methyl piperazine, respectively, on the meta position of the benzene moiety, exhibit improved potencies and selectivity for LH2 compared with the other isoforms and Fe(II)/2OG-dependent enzymes.
Further investigations by MD and QM/MM calculations suggest that 12 and 13 can firmly interact with the enzyme, corroborating their inhibitory activities and selectivity. Migration assays in the 344SQ lung adenocarcinoma cell line reveal that compound 13 demonstrates a dose-dependent antimigratory effect. While we acknowledge that there is still much to be done in the field of LH2 inhibition for the treatment of cancer metastasis, the findings presented in this study may represent a significant step forward in developing potential LH2 inhibitors, particularly given the limited availability of effective LH2 antagonists.
Acknowledgments
This work was supported by NIH CA070907, CPRIT RP210088, WELCH F-1390 (K.N.D); NIH R00CA225633, University of Kentucky Center for Cancer and Metabolism, funded through the NIH/NIGMS COBRE program under grant number P20 GM121327 (H.G.); NIH R01GM106137, R01GM114237 (P.R); NIH CA251067 (J.M.K); R01GM108583, computing from XSEDE through allocation TG-CHE160044, UNT CASCaM (partially funded by the NSF Grant Nos. CHE1531468 and OAC-2117247) and UT-Dallas’ Cyberinfrastructure and Research Services (G.A.C.).
Glossary
ABBREVIATIONS
- THF
tetrahydrofuran
- DIPEA
N,N-diisopropylethylamine
- DCM
dichloromethane
- DME
dimethoxyethane
- p-TSA
p-toluenesulfonic acid
- QM/MM
quantum mechanics/molecular mechanics
- NCI
noncovalent interactions
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.3c00305.
Author Contributions
The manuscript was written through contributions of all authors. All authors have approved the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Yamauchi M.; Sricholpech M. Lysine post-translational modifications of collagen. Essays Biochem 2012, 52, 113–33. 10.1042/bse0520113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scietti L.; Campioni M.; Forneris F. SiMPLOD, a Structure-Integrated Database of Collagen Lysyl Hydroxylase (LH/PLOD) Enzyme Variants. J. Bone Miner Res. 2019, 34 (7), 1376–1382. 10.1002/jbmr.3692. [DOI] [PubMed] [Google Scholar]
- Hyland J.; Ala-Kokko L.; Royce P.; Steinmann B.; Kivirikko K. I.; Myllyla R. A homozygous stop codon in the lysyl hydroxylase gene in two siblings with Ehlers-Danlos syndrome type VI. Nat. Genet. 1992, 2 (3), 228–31. 10.1038/ng1192-228. [DOI] [PubMed] [Google Scholar]
- Ha-Vinh R.; Alanay Y.; Bank R. A.; Campos-Xavier A. B.; Zankl A.; Superti-Furga A.; Bonafe L. Phenotypic and molecular characterization of Bruck syndrome (osteogenesis imperfecta with contractures of the large joints) caused by a recessive mutation in PLOD2. Am. J. Med. Genet A 2004, 131A (2), 115–120. 10.1002/ajmg.a.30231. [DOI] [PubMed] [Google Scholar]
- Vahidnezhad H.; Youssefian L.; Saeidian A. H.; Touati A.; Pajouhanfar S.; Baghdadi T.; Shadmehri A. A.; Giunta C.; Kraenzlin M.; Syx D.; Malfait F.; Has C.; Lwin S. M.; Karamzadeh R.; Liu L.; Guy A.; Hamid M.; Kariminejad A.; Zeinali S.; McGrath J. A.; Uitto J. Mutations in PLOD3, encoding lysyl hydroxylase 3, cause a complex connective tissue disorder including recessive dystrophic epidermolysis bullosa-like blistering phenotype with abnormal anchoring fibrils and type VII collagen deficiency. Matrix Biol. 2019, 81, 91–106. 10.1016/j.matbio.2018.11.006. [DOI] [PubMed] [Google Scholar]
- Salo A. M.; Cox H.; Farndon P.; Moss C.; Grindulis H.; Risteli M.; Robins S. P.; Myllyla R. A connective tissue disorder caused by mutations of the lysyl hydroxylase 3 gene. Am. J. Hum. Genet. 2008, 83 (4), 495–503. 10.1016/j.ajhg.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Terajima M.; Yang Y.; Sun L.; Ahn Y. H.; Pankova D.; Puperi D. S.; Watanabe T.; Kim M. P.; Blackmon S. H.; Rodriguez J.; Liu H.; Behrens C.; Wistuba I. I.; Minelli R.; Scott K. L.; Sanchez-Adams J.; Guilak F.; Pati D.; Thilaganathan N.; Burns A. R.; Creighton C. J.; Martinez E. D.; Zal T.; Grande-Allen K. J.; Yamauchi M.; Kurie J. M. Lysyl hydroxylase 2 induces a collagen cross-link switch in tumor stroma. J. Clin Invest 2015, 125 (3), 1147–62. 10.1172/JCI74725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Slot A. J.; Zuurmond A. M.; van den Bogaerdt A. J.; Ulrich M. M.; Middelkoop E.; Boers W.; Karel Ronday H.; DeGroot J.; Huizinga T. W.; Bank R. A. Increased formation of pyridinoline cross-links due to higher telopeptide lysyl hydroxylase levels is a general fibrotic phenomenon. Matrix Biol. 2004, 23 (4), 251–7. 10.1016/j.matbio.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Eisinger-Mathason T. S.; Zhang M.; Qiu Q.; Skuli N.; Nakazawa M. S.; Karakasheva T.; Mucaj V.; Shay J. E.; Stangenberg L.; Sadri N.; Pure E.; Yoon S. S.; Kirsch D. G.; Simon M. C. Hypoxia-dependent modification of collagen networks promotes sarcoma metastasis. Cancer Discov 2013, 3 (10), 1190–205. 10.1158/2159-8290.CD-13-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi M.; Barker T. H.; Gibbons D. L.; Kurie J. M. The fibrotic tumor stroma. J. Clin Invest 2018, 128 (1), 16–25. 10.1172/JCI93554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piersma B.; Bank R. A. Collagen cross-linking mediated by lysyl hydroxylase 2: an enzymatic battlefield to combat fibrosis. Essays. Biochem 2019, 63 (3), 377–387. 10.1042/EBC20180051. [DOI] [PubMed] [Google Scholar]
- Guo H. F.; Bota-Rabassedas N.; Terajima M.; Leticia Rodriguez B.; Gibbons D. L.; Chen Y.; Banerjee P.; Tsai C. L.; Tan X.; Liu X.; Yu J.; Tokmina-Roszyk M.; Stawikowska R.; Fields G. B.; Miller M. D.; Wang X.; Lee J.; Dalby K. N.; Creighton C. J.; Phillips G. N. Jr; Tainer J. A.; Yamauchi M.; Kurie J. M. A collagen glucosyltransferase drives lung adenocarcinoma progression in mice. Commun. Biol. 2021, 4 (1), 482. 10.1038/s42003-021-01982-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last J. A.; King T. E. Jr; Nerlich A. G.; Reiser K. M. Collagen cross-linking in adult patients with acute and chronic fibrotic lung disease. Molecular markers for fibrotic collagen. Am. Rev. Respir. Dis. 1990, 141 (2), 307–13. 10.1164/ajrccm/141.2.307. [DOI] [PubMed] [Google Scholar]
- Perepelyuk M.; Terajima M.; Wang A. Y.; Georges P. C.; Janmey P. A.; Yamauchi M.; Wells R. G. Hepatic stellate cells and portal fibroblasts are the major cellular sources of collagens and lysyl oxidases in normal liver and early after injury. Am. J. Physiol Gastrointest Liver Physiol 2013, 304 (6), G605–14. 10.1152/ajpgi.00222.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Slot A. J.; Zuurmond A. M.; Bardoel A. F.; Wijmenga C.; Pruijs H. E.; Sillence D. O.; Brinckmann J.; Abraham D. J.; Black C. M.; Verzijl N.; DeGroot J.; Hanemaaijer R.; TeKoppele J. M.; Huizinga T. W.; Bank R. A. Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. J. Biol. Chem. 2003, 278 (42), 40967–72. 10.1074/jbc.M307380200. [DOI] [PubMed] [Google Scholar]
- Myllyharju J.; Kivirikko K. I. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet 2004, 20 (1), 33–43. 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Guo H.; Terajima M.; Banerjee P.; Liu X.; Yu J.; Momin A. A.; Katayama H.; Hanash S. M.; Burns A. R.; Fields G. B.; Yamauchi M.; Kurie J. M. Lysyl Hydroxylase 2 Is Secreted by Tumor Cells and Can Modify Collagen in the Extracellular Space. J. Biol. Chem. 2016, 291 (50), 25799–25808. 10.1074/jbc.M116.759803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankova D.; Chen Y.; Terajima M.; Schliekelman M. J.; Baird B. N.; Fahrenholtz M.; Sun L.; Gill B. J.; Vadakkan T. J.; Kim M. P.; Ahn Y. H.; Roybal J. D.; Liu X.; Parra Cuentas E. R.; Rodriguez J.; Wistuba I. I.; Creighton C. J.; Gibbons D. L.; Hicks J. M.; Dickinson M. E.; West J. L.; Grande-Allen K. J.; Hanash S. M.; Yamauchi M.; Kurie J. M. Cancer-Associated Fibroblasts Induce a Collagen Cross-link Switch in Tumor Stroma. Mol. Cancer Res. 2016, 14 (3), 287–95. 10.1158/1541-7786.MCR-15-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuurmond A.-M; van der Slot-Verhoeven A. J.; van Dura E. A.; De Groot J.; Bank R. A. Minoxidil exerts different inhibitory effects on gene expression of lysyl hydroxylase 1, 2, and 3: implications for collagen cross-linking and treatment of fibrosis. Matrix Biol. 2005, 24, 261–270. 10.1016/j.matbio.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Pfeffer T.; Lignelli E.; Inoue H.; Mizikova I.; Surate Solaligue D. E.; Steenbock H.; Myti D.; Vadasz I.; Herold S.; Seeger W.; Brinckmann J.; Morty R. E. Minoxidil Cannot Be Used To Target Lysyl Hydroxylases during Postnatal Mouse Lung Development: A Cautionary Note. J. Pharmacol Exp Ther. 2020, 375, 478–487. 10.1124/jpet.120.000138. [DOI] [PubMed] [Google Scholar]
- Du Y.; Khan M.; Fang N.; Ma F.; Du H.; Tan Z.; Wang H.; Yin S.; Wei X. Berberine Attenuates Cell Motility via Inhibiting Inflammation-Mediated Lysyl Hydroxylase-2 and Glycolysis. Front. Pharmacol. 2022, 13, 856777. 10.3389/fphar.2022.856777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler R.; Spitzer S.; Rumpler M.; Fratzl-Zelman N.; Klaushofer K.; Paschalis E. P.; Varga F. Differential effects of homocysteine and beta aminopropionitrile on preosteoblastic MC3T3-E1 cells. Bone 2010, 46, 703–709. 10.1016/j.bone.2009.10.038. [DOI] [PubMed] [Google Scholar]
- Bennett I.; Broom N. J. P.; Cassels R.; Elder J. S.; Masson N. D.; O’Hanlon P. J. Synthesis and antibacterial properties of b-diketone acrylate bioisosteres of pseudomonic acid A. Bioorg. Med. Chem. Lett. 1999, 9, 1847–1852. 10.1016/S0960-894X(99)00296-6. [DOI] [PubMed] [Google Scholar]
- Sheikh J.; Parvez A.; Juneja H.; Ingle V.; Chohan Z.; Youssoufi M.; Ben Hadda T. Synthesis, biopharmaceutical characterization, antimicrobial and antioxidant activities of 1-(40-O-b-D-glucopyranosyloxy-20 -hydroxyphenyl)-3-aryl-propane-1,3-diones. Eur. J. Med. Chem. 2011, 46, 1390–1399. 10.1016/j.ejmech.2011.01.068. [DOI] [PubMed] [Google Scholar]
- Diana G. D.; Carabateas P. M.; Johnson R. E.; Williams G. L.; Pancic F.; Collins J. C. Antiviral activity of some b-diketones. 4. Benzyldiketones. In vitro activity against both RNA and DNA viruses. J. Med. Chem. 1978, 21, 889–894. 10.1021/jm00207a010. [DOI] [PubMed] [Google Scholar]
- Crouse G. D.; McGowan M. J.; Boisvenue R. J. Polyflouro 1,3- diketones as systemic insecticides. J. Med. Chem. 1989, 32, 2148–2151. 10.1021/jm00129a021. [DOI] [PubMed] [Google Scholar]
- Nishiyama T.; Shiotsu S.; Tsujita H. Antioxidative activity and active site of 1,3-indanediones with the b-diketone moiety. Polym. Degrad. Stab. 2002, 76, 435–439. 10.1016/S0141-3910(02)00046-0. [DOI] [Google Scholar]
- Pivarcsik T.; Toth G.; Szemeredi N.; Bogdanov A.; Spengler G.; Kljun J.; Kladnik J.; Turel I.; Enyedy E. A. Comparison of Solution Chemical Properties and Biological Activity of Ruthenium Complexes of Selected β-Diketone, 8-Hydroxyquinoline and Pyrithione Ligands. Pharmaceuticals 2021, 14, 518. 10.3390/ph14060518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel’in A. V. Recent Advances in the Synthesis of 1,3-Diketones. Curr. Org. Chem. 2003, 7, 1691–1711. 10.2174/1385272033486233. [DOI] [Google Scholar]
- Lim D.; Fang F.; Zhou G.; Coltart D. M. Direct carbon-carbon bond formation via soft enolization: a facile and efficient synthesis of 1,3-diketones. Org. Lett. 2007, 9 (21), 4139–42. 10.1021/ol701599v. [DOI] [PubMed] [Google Scholar]
- Devkota A. K.; Veloria J. R.; Guo H. F.; Kurie J. M.; Cho E. J.; Dalby K. N. Development of a High-Throughput Lysyl Hydroxylase (LH) Assay and Identification of Small-Molecule Inhibitors against LH2. SLAS Discov 2019, 24 (4), 484–491. 10.1177/2472555218817057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maghsoud Y.; Vázquez-Montelongo E. A.; Yang X.; Liu C.; Jing Z.; Lee J.; Harger M.; Smith A. K.; Espinoza M.; Guo H.-F.; Kurie J. M.; Dalby K. N.; Ren P.; Cisneros G. A. Computational Investigation of a Series of Small Molecules as Potential Compounds for Lysyl hydroxylase-2 (LH2) Inhibition. J. Chem. Inf. Model. 2023, 63 (3), 986–1001. 10.1021/acs.jcim.2c01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savastano M.; García-Gallarín C.; Giorgi C.; Gratteri P.; López de la Torre M. D.; Bazzicalupi C.; Bianchi A.; Melguizo M. Solid State and Solution Study on the Formation of Inorganic Anion Complexes with a Series of Tetrazine-Based Ligands. Molecules 2019, 24, 2247. 10.3390/molecules24122247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin A.; Oswald I. D. H.; Parsons S. Structures of piperazine, piperidine and morpholine. Acta Crystallogr. 2004, B60, 219–227. 10.1107/S0108768104003672. [DOI] [PubMed] [Google Scholar]
- Lan N.; Lu Y.; Zhang Y.; Pu S.; Xi H.; Nie X.; Liu J.; Yuan W. FTO - A Common Genetic Basis for Obesity and Cancer. Front Genet 2020, 11, 559138. 10.3389/fgene.2020.559138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture J. F.; Collazo E.; Ortiz-Tello P. A.; Brunzelle J. S.; Trievel R. C. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat. Struct Mol. Biol. 2007, 14 (8), 689–95. 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- Fong G. H.; Takeda K. Role and regulation of prolyl hydroxylase domain proteins. Cell Death Differ. 2008, 15 (4), 635–41. 10.1038/cdd.2008.10. [DOI] [PubMed] [Google Scholar]
- Luther K. B.; Hulsmeier A. J.; Schegg B.; Deuber S. A.; Raoult D.; Hennet T. Mimivirus collagen is modified by bifunctional lysyl hydroxylase and glycosyltransferase enzyme. J. Biol. Chem. 2011, 286 (51), 43701–43709. 10.1074/jbc.M111.309096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.; Zhang T.; Guo L.; Wang Y.; Yang Y. Lysyl hydroxylases are transcription targets for GATA3 driving lung cancer cell metastasis. Sci. Rep 2018, 8, 11905. 10.1038/s41598-018-30309-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Fan G.; Zhu H.; Yu L.; She D.; Wei Y.; Huang J.; Li T.; Zhan S.; Zhou S.; Zhu Y.; Wang Y.; Chen X.; Zhao J.; Zhou G. PLOD2 high expression associates with immune infiltration and facilitates cancer progression in osteosarcoma. Front. Oncol. 2022, 12, 980390. 10.3389/fonc.2022.980390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons D. L.; Lin W.; Creighton C. J.; Rizvi Z. H.; Gregory P. A.; Goodall G. J.; Thilaganathan N.; Du L.; Zhang Y.; Pertsemlidis A.; Kurie J. M. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009, 23, 2140–2151. 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.