Abstract
Respiratory viruses are a common cause of morbidity and mortality around the world. Viruses like influenza, RSV, and most recently SARS-CoV-2 can rapidly spread through a population, causing acute infection and, in vulnerable populations, severe or chronic disease. Developing effective treatment and prevention strategies often becomes a race against ever-evolving viruses that develop resistance, leaving therapy efficacy either short lived or relevant for specific viral strains. On June 29 to July 2, 2022, researchers met for the Keystone symposium “Respiratory Viruses: New Frontiers”. The meeting was held jointly with the symposium “Viral Immunity: Basic Mechanisms and Therapeutic Applications.” Researchers presented new insights into viral biology and virus-host interactions to understand the mechanisms of disease and identify novel treatment and prevention approaches that are effective, durable, and broad.
Keywords: adaptive immunity, COVID-19, influenza virus, innate immunity, respiratory virus, RSV
Graphical Abstract

Respiratory viruses are a common cause of morbidity and mortality around the world. Viruses like influenza, RSV, and most recently SARS-CoV-2 can rapidly spread through a population, causing acute infection and, in vulnerable populations, severe or chronic disease. On June 29 to July 2, 2022, researchers met for the Keystone symposium “Respiratory Viruses: New Frontiers”. The meeting was held jointly with the symposium “Viral Immunity: Basic Mechanisms and Therapeutic Applications.” Researchers presented new insights into viral biology and virus-host interactions to understand the mechanisms of disease and identify novel treatment and prevention approaches that are effective, durable, and broad.
Introduction
Respiratory viruses represent a significant cause of morbidity and mortality around the world. Air-borne transmission enables these viruses to spread quickly throughout a population, and while many infections cause acute illness, chronic disease and persistent lung damage are not uncommon, particularly in vulnerable populations. Despite intensive research, treatments and durable vaccination approaches remain limited.
On June 29 to July 2, 2022, researchers met for the Keystone symposium “Respiratory Viruses: New Frontiers”. The meeting was held jointly with the symposium “Viral Immunity: Basic Mechanisms and Therapeutic Applications.” The symposium brought together researchers studying diverse respiratory viruses, including SARS-CoV-2, influenza virus, and RSV. In light of the COVID-19 pandemic, presenters also discussed approaches to surveilling for and identifying emerging viruses with pandemic potential. Speakers focused on novel insights into respiratory virus biology, including host-virus interactions that enable viruses to evade the host immune response and cause chronic lung damage, and presented new data on host factors that render some populations more susceptible to severe infection and disease. The meeting ended with a session on novel prevention and treatment efforts, highlighting the continuous work to create effective therapies with broad, durable activity.
Beyond Acute Disease: Consequences of Respiratory Virus Infections
While respiratory viruses are known to cause acute disease and lung damage, they can also cause long-term sequelae both in the lung and other organ systems. These chronic sequelae may be a result of immune hyperresponsiveness and unchecked inflammation that persist after the virus is cleared and/or of residual low-level viral replication.1–3 Chronic sequelae of acute viral pneumonia are exacerbated during aging.4,5 Speakers presented research on understanding the host factors that contribute to long-term lung damage following influenza virus infection, evidence for persistent SARS-CoV-2 throughout the body, as well as viral features that contribute to persistent infection.
Persistent inflammation and chronic lung damage in the elderly
Jie Sun from the University of Virginia discussed the immune determinants of long-term influenza virus-mediated lung damage during aging. In mice, older mice experience higher mortality, more severe disease, and delayed recovery as a result of influenza infection. Sun showed that this is not due to persistent viral replication—viral titers in the lung are similar among young and aged mice. Instead, aged mice demonstrate persistent upregulation of pro-inflammatory genes and an increase in CD8+ resident memory T cells that have increased fibrogenic potential. Sun’s work shows that this inability to return to immune homeostasis likely contributes to the increased risk of chronic sequelae observed in the elderly after acute infection.6 Sun showed that CD8+ resident memory T cells may also contribute to the long-term lung sequelae of COVID-19. Elderly patients who have recovered from COVID-19 show signs of inflammation and fibrosis months after the infection is cleared. In those with persistent lung dysfunction, the lungs were enriched with memory CD8+ T cells. Further characterization of these T cells by single-cell RNA sequencing (scRNAseq), revealed three subpopulations, one of which had the characteristics of traditional tissue-resident memory T cells while the others may be pathogenic and promote chronic lung damage. Additional work in mice confirmed a causal role for CD8+ T cells in lung damage post-acute viral infection.7 Sun showed unpublished data supporting a role for the hyperactive CD8+ T cell response in promoting dysplastic lung repair following severe viral pneumonia.
The role of CD8+ and CD4+ T cells in persistent COVID-19-related lung damage
In Su Cheon from Sun’s group expanded on the role of CD8+ T cells in lung damage following acute COVID-19. Cheon assessed lung pathology and function in a cohort of convalescent individuals over 60 years of age. She showed that individuals experienced persistent lung pathology and impaired lung function after recovering from COVID-19, the severity of which correlated with disease severity. While peripheral immune markers were similar between the convalescent and uninfected cohorts, convalescent individuals had higher levels of CD8+ T cells and B cells in the lungs. Levels of tissue-resident CD8+ T cells negatively correlated with lung function and likely contribute to the chronic inflammation and fibrotic sequelae following acute COVID-19.7 Cheon showed unpublished work on the impact of tissue-resident helper (TRH)1 CD4+ cells on maintaining this pathologic CD8+ T cell population.
Mechanisms of dysplastic lung repair following viral infection
Andrew Vaughan from the University of Pennsylvania presented additional work on the mechanisms of dysplastic lung repair following viral pneumonia. During acute viral infection, many alveolar type 2 (AT2) cells are killed. As a result of lung damage, these bipotent cells can restore the alveolar epithelium by regenerating both themselves as well as AT1 cells, which are responsible for gas exchange. If too many AT2 cells are killed during infection, ectopic Krt5+ basal cells participate in the re-epithelization of the alveolus. While these cells can differentiate into many types of epithelial cells, they rarely became AT2 or AT1 cells. This epithelial dysplasia is essentially permanent and correlates with long-term loss of lung function. Vaughan’s group is interested in understanding why Krt5+ cells have such a low propensity to differentiate into AT2 cells. They showed that deletion of p63 increases the conversion of Krt5+ cells into AT2 cells.8 They have also looked at the impact of chronic inflammation that persists even after the virus is cleared. Vaughan focused on the impact of tuft cells, which are implicated in intestinal damage and repair, where they play a role in Type 2 mucosal immunity, characterized by IL-13, IL-25, and metaplasia.9 Tuft cells have also been shown to expand in the lung in response to influenza virus and SARS-CoV-2.10,11 Vaughan’s lab showed that in the lung, tuft cells arise from Krt5+ progenitors. Unlike in the intestine, lung tuft cells arise independent of Type II cytokines and have no obvious impact on the development of epithelial dysplasia.10 Vaughan’s group is continuing to work on understanding the signals for injury-induced lung tufT cell development as well as the role of these cells in lung dysplasia.
Persistent paramyxovirus infection and chronic lung disease
Italo Castro from Carolina López’s lab at the Washington University School of Medicine presented his work on the impact of persistent paramyxovirus infection on chronic lung pathology. López’s group had previously demonstrated the presence of chronic lung disease in mice after infection with Sendai virus or influenza virus. This disease was characterized by airway thickening, bronchiolization of the parenchyma, and type II inflammation. While viral RNA was detectable long term, lung disease persisted in the absence of replicating virus.12 Castro presented unpublished data using this mouse model to better understand whether Sendai virus can persist and replicate long term and whether persistent infection may contribute to chronic lung pathology.
Distribution and persistence of SARS-CoV-2
Sydney Stein from the National Institutes of Health (NIH) presented results from an unpublished study on the distribution of SARS-CoV-2 across the body from an autopsy cohort of individuals who died with (but not necessarily from) COVID-19. Results from 44 unvaccinated cases during the first year of the pandemic shed light on which tissues and cells the virus infects, how long viral RNA can persist in the body, and which cell types and anatomic compartments support viral replication.
Impact of nonstandard viral genomes in RSV infection
Carolina López from the Washington University School of Medicine in St. Louis presented work on how nonstandard viral genomes (nsVG) impact clinical outcomes in RSV infection. Nonstandard viral genomes are truncated genomes that lack essential genes for replication but can nonetheless impact viral infection by interfering with the replication of standard viral genomes and/or activating host immunity.13,14 López’s work has shown that standard and nonstandard genomes of RSV and Sendai virus preferentially accumulate in different cells.14,15 Cells with standard viral genomes undergo lipid stress and activate cell death pathways while cells with nsVGs upregulate antiviral and pro-survival pathways.15 López’s work has also shown that nsVGs impact the clinical outcome of viral infection. In a cohort of 56 adults inoculated with RSV, individuals with high levels of nsVGs early in infection had less severe disease. Conversely, the persistent presence of nsVGs late in infection was associated with more severe disease. Consistent with this, patients hospitalized with RSV also have higher levels of nsVGs.16 López ended by presenting unpublished work on characterizing the diversity of nsVGs using a tool developed in her lab dubbed viral open-source DVG key algorithm (VODKA).
Preclinical immunogenicity of a novel measles virus vaccine
Jessica Rubens from Diane Griffin’s lab at Johns Hopkins Bloomberg School of Public Health presented unpublished data on the immunogenicity and efficacy of a novel recombinant measles virus vaccine in nonhuman primates (NHP). The current measles vaccine included in the measles, mumps, and rubella (MMR) shot is a live virus vaccine. While it provides strong, lifelong protection in most recipients, several high-risk populations, including infants, pregnant women, and immunocompromised individuals, are ineligible to receive it. In addition, immunity can wane over time in some individuals. The Griffin lab is collaborating with the National Institutes of Health (NIH) and MEVOX to develop and evaluate a recombinant measles vaccine that contains the measles hemagglutinin (HA) dimer in its prefusion conformation. Such a vaccine could be administered to infants, may boost MMR-primed immunity, and is anticipated to be more thermostable than MMR.
Immunity and Respiratory Viruses
The immune system is continually exposed to and shaped by respiratory viruses. Speakers discussed how prior exposure to evolving viruses, like influenza virus, impacts protection when a similar virus is encountered, efforts to identify immune-mediated and other markers of disease severity during pandemics with novel viruses, tracking B cell maturation after vaccination, and elucidating the role of interferons in viral infections.
Impact of immune imprinting on protection
Katelyn Gostic from Sarah Cobey’s lab at the University of Chicago gave an overview of how early antigenic exposures can impact the adaptive immune response and protection. Immunologic data for respiratory viruses such as influenza virus and SARS-CoV-2 demonstrate that repeated infections with evolving pathogens reinforce the memory of past antigens, cause conserved epitopes to become immunodominant, and can interfere with responses against de novo antigens.17–20 Gostic’s work has demonstrated the impact of this immune imprinting on protection from avian influenza viruses. Before 1968, the majority of circulating influenza viruses were Group 1 subtypes; Group 2 subtypes dominated after 1968. When H5N1 influenza virus—a Group 1 subtype—emerged, elderly individuals (those born before 1968) were preferentially protected against infection. Likewise, when H7N9 influenza virus—a Group 2 subtype—emerged, those born after 1968 were largely protected. These data demonstrate that people maintain the strongest immune memory against strains that they encounter during childhood and that different birth cohorts may therefore have different levels of protection against viral strains.21 Immune imprinting has also been observed with seasonal influenza with varying breadth22–25 and has been shown to impact vaccine efficacy.23,26,27 However, there are also examples in which the host immune history has no discernible impact on vaccine effectiveness or susceptibility.26 As pathogens evolve, prior immunity may impact different populations differently. For example, those whose immune repertoire is skewed toward an antigen that mutates may lose protection while those whose immune repertoire is skewed toward an antigen that stays the same may maintain protection.25,28 Gostic stressed that to better understand these cohort effects, it is necessary to characterize immunologic diversity in hosts and invest in long-term longitudinal studies to link immunological patterns with epidemiologic outcomes.
Identifying determinants of disease severity and novel therapies during pandemics
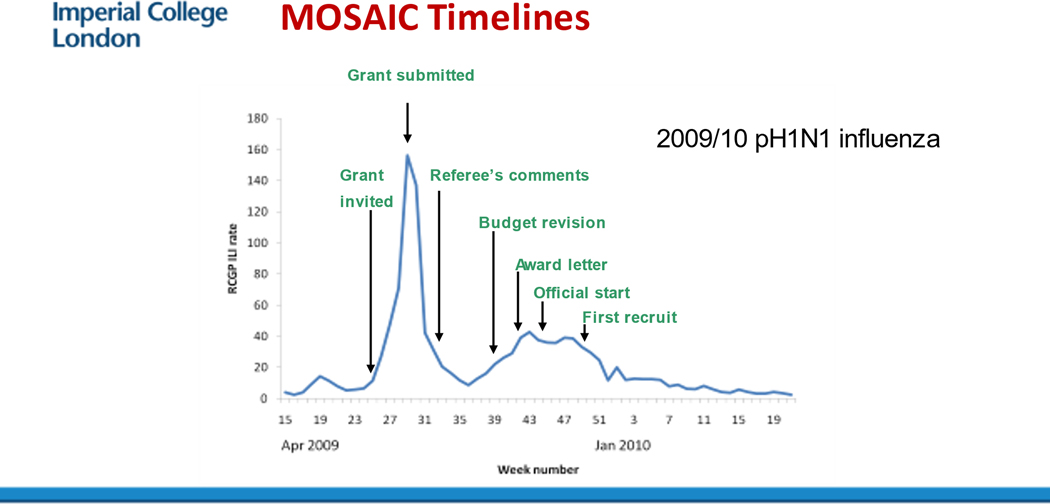
Peter Openshaw from Imperial College London (ICL) discussed efforts to study ongoing pandemics in the UK, understand the factors that determine disease severity for common respiratory viruses (including SARS-CoV-2), and identify effective interventions. The need for long-term research investment is clear: the Centre for Respiratory Infections was funded in 2008, shortly before the 2009 H1N1 influenza pandemic. Without this fortunate event, the comprehensive study of factors involved in severe disease that included virologic, genomic, transcriptomic, and immunologic analyses of more than 250 patients with severe influenza-like illness could not have been launched. One of the early results of this collaboration (the MOSAIC study, Figure 1) was discovery of the association between a polymorphism in IFITM3 with severe illness.29 Transcriptomics also revealed a signature characteristic of an inflammatory response to bacterial infection that occurred in late phases of influenza virus infection, indicating the time-based changes30 that are now evident in other conditions such as COVID-10. Openshaw stressed that long-term investments in facilities are vital if such large, coordinated studies are to be performed as well as healthcare data linkage. When COVID-19 hit UK the UK was able to launch the International Severe Acute Respiratory and emerging Infection Consortium (ISARIC-4C) observational cohort which gathered extensive data on more than 340,000 patients hospitalized with COVID-19 and serial biological sampling on more than 2000.31 Initial findings from the study have identified an association between several plasma biomarkers, including IL-6 and D-dimer as well as markers of endothelial inflammation, with disease severity.32 In addition many gene variants have been identified that are associated with disease severity1, including polymorphisms that reduce JAK signaling.33 These data enabled researchers to prioritize the JAK inhibitor baricitinib in the RECOVERY trial, which established that baricitinib reduced mortality among hospitalized adults.34 Rapid integration of research trials was transformative during the COVID pandemic.
Figure 1.
The delay that occurred in setting up the MOSAIC consortium in 2009 following the traditional path of applying for a grant and getting it funded. This occurred despite all parties recognising that speed was essential.
Tracking B cell maturation following vaccination
Ali Ellebedy from Washington University School of Medicine in St. Louis presented work on tracking B cell maturation in response to COVID-19 vaccines. Ellebedy’s group uses fine needle aspiration to sample the draining lymph nodes located near the armpits following vaccination. This is the site of germinal center activity where responding B cells undergo affinity maturation to enhance their binding to immunizing antigens. The cells eventually become either memory B cells or long-lived bone marrow plasma cells (BMPC), which are important for long-term immunity. Ellebedy showed that they can detect vaccine-induced germinal center responses after influenza virus vaccination and track antigen-specific germinal center B cells over time.35 They also used this method to monitor the B cell response to the Pfizer’s mRNA COVID-19 vaccine in humans. Sampling the draining lymph nodes showed that vaccination induced a strong spike-specific germinal center response that persisted for at least six months. Unlike the response to the influenza virus vaccine, COVID-19 vaccination often engaged multiple lymph nodes. Bone marrow sampling confirmed that vaccination induced spike-specific long-lived BMPCs.36,37 Ellebedy showed that the antibodies produced by the BMPCs, which had had months to mature, were of higher quality than those produced in the blood by plasmablasts immediately following vaccination. These later antibodies had higher avidity and higher neutralization activity than earlier antibodies.38 Ellebedy’s work shows that the persistent germinal center reaction induced by mRNA COVID-19 vaccination leads to affinity-matured long-term antibody responses with potent neutralization activity.
The negative impact of interferon in lung epithelial repair
Andreas Wack from the Francis Crick Institute discussed the impact of interferons (IFN) on epithelial repair following viral infection. Interferons have important antiviral activity, but they can also have antiproliferative or proapoptotic effects. Working in a mouse model of influenza infection, Wack showed that IFN levels peak early in infection and decline as AT2 cell proliferation increases. However, there is a window during which IFN levels are relatively high and AT2 cells are proliferating to repair the recovering lung. He showed that IFNs can negatively impact epithelial repair during viral infection. Exogenous exposure to IFN ɑ, β, and λ reduced epithelial proliferation during viral recovery without impacting viral load. Likewise, knocking out genes required for endogenous IFN signaling increased lung epithelial proliferation and differentiation.39,40 Transcriptome analyses of IFN-treated airway epithelial cells indicated that short-term exposure to IFN induces genes involved in the antiviral response while long-term exposure also arrests cell development and increases antiproliferative effects through p53. Additional work confirmed that IFNs induce p53 expression in vivo in the lung epithelium after viral infection. Knocking out IFN-ƛ receptors improved epithelial recovery and barrier function.39 Wack’s work shows that while IFNs play important roles in mediating the antiviral response, they can also negatively impact proliferation and differentiation of the epithelium, which can impair barrier integrity and increase the risk of severe disease and secondary bacterial infections.
New Approaches to Studying Respiratory Viruses
Developing effective therapies and vaccines against respiratory viruses will require novel methods and techniques. Speakers discussed new insights into viral biology and host immune responses, including how viruses co-opt host cell resources to expand their genomes, the impact of host cell composition on viral dynamics, and novel inflammatory signaling mechanisms, and viral protein-host protein interactions. They also presented techniques to measure virus burst size and to surveil clinical samples for early detection of novel, emerging viruses.
Expanding the translatable regions of viral genomes
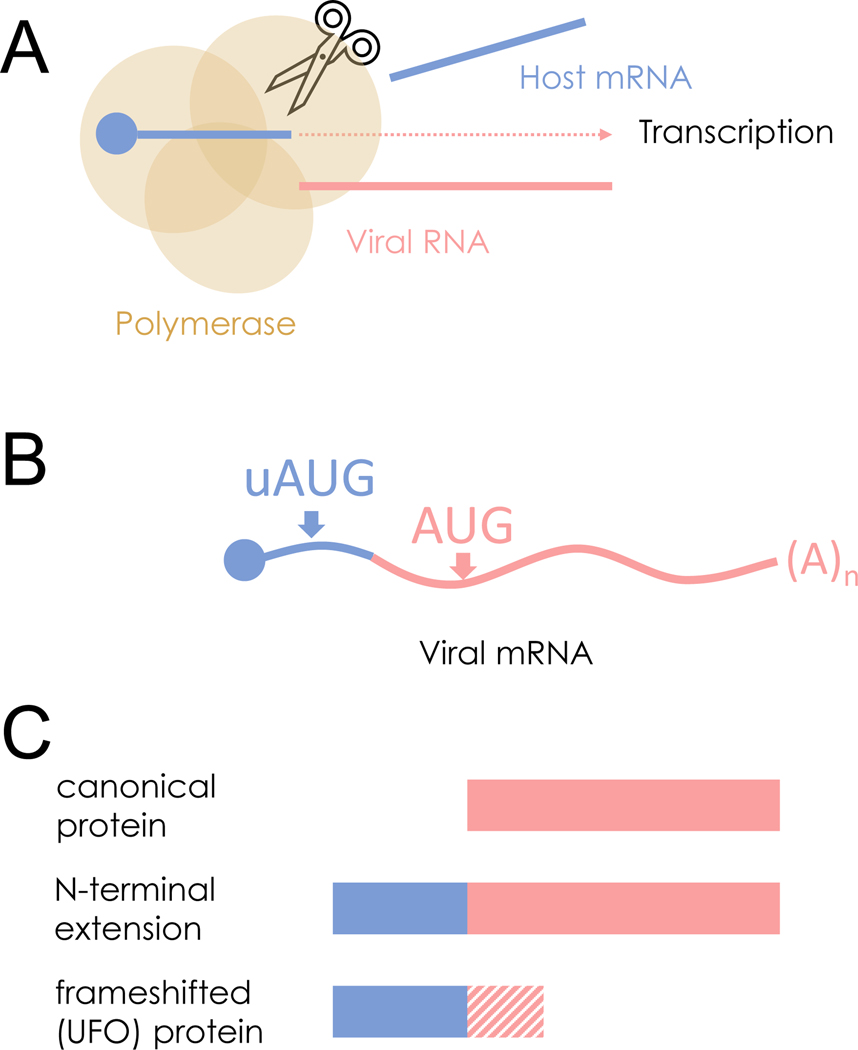
Ed Hutchinson from the University of Glasgow discussed the impact of chimeric human-virus genes in influenza virus infections – a concept that had originally been investigated independently by his group and the group of Ivan Marazzi from the Icahn School of Medicine at Mount Sinai, until the discovery of each other’s preprints led the groups to combine their work into a single and more detailed study. Viruses have devised a range of strategies to compress a lot of information into their small genomes.41 Hutchinson and his colleagues realized that the segmented negative strand RNA viruses (sNSVs), a large group of viruses including major pathogens such as the influenza viruses, would have an additional way to do this. The sNSVs transcribe their genes through a process known as cap-snatching. In this process, the virus polymerase binds to host mRNA and cleaves it shortly downstream of the 5′ cap, using the cleaved product as a starting point when transcribing a viral gene. The result is a hybrid mRNA containing both host sequences with a 5′ cap that can be recognized by the ribosome for translation, as well as viral sequences. Since many host untranslated regions (UTR) have upstream, noncanonical start codons42,43, Hutchinson’s group was interested if these host-derived start codons allow the translation of novel viral proteins (“start snatching”). They showed that host sequences in the cap-snatched mRNA contain start codons that can be recognized by ribosomes. These alternative start codons create upstream viral open reading frames (ORF) that result in either N-terminal extensions in the viral protein (if the ORF is in-frame with the viral ORF) or in novel proteins (if the upstream ORF is frameshifted). About half of influenza genome segments have N-terminal extensions encoded into canonical genes, suggesting that this phenomenon is relatively common. Hutchinson showed that the proteins resulting from viral upstream ORFs can be detected in infected cells. While interrupting these ORFs did not impact viral replication in cells, it did impact disease in mice, indicating that these proteins may have functions in infections.44–46 Hutchinson’s work shows that upstream viral ORFs expand the translatable regions of the viral genome beyond those that contain a viral start codon (Figure 2).
Figure 2.
Expanding the translatable regions of viral genomes. (A) Segmented negative strand RNA viruses (sNSVs) such as the influenza viruses transcribe their genes by ‘cap snatching’ from host mRNAs. (B) Cap snatching results in a hybrid mRNA containing host and viral sequences. If the host mRNA contains upstream start codons (uAUGs) this can provide additional opportunities to translate viral proteins through ‘start snatching.’ (C) Start snatching expands the accessory proteome of sNSVs by allowing the translation of proteins with N-terminal extensions (from in-frame uAUGs) or novel upstream frameshifted open reading frames (from out-of-frame uAUGs).
A high-throughput method to measure viral burst size
Mallory Thomas from Connie Chang’s lab at Montana State University presented a high-throughput, single-cell method to quantify viral burst size—the number of virions released from a single cell and an important indicator of viral fitness. Current methods to measure burst size either average across the population, thus masking heterogeneity across infected cells47, or are low throughput.48 These methods also only measure infectious viruses; however, the majority of influenza particles are not fully infectious.49 Chang’s group has developed a drop-based microfluidic platform to quantify burst size from single-cell infections. In this method, single cells are isolated within a droplet and infected with influenza virus. The viral progeny are then isolated from the droplets and quantified by measuring matrix protein (M) gene abundance.50 Thomas showed unpublished data in which she used this method to quantify the burst size of H3N2 and H1N1 influenza viruses to understand the diversity of burst size across cells and between viruses.
The impact of cell composition on influenza virus dynamics
Ryan Langlois from the University of Minnesota presented work on influenza virus tropism in the human airway. Langlois’s group uses flow cytometry to identify and quantify cell types in human tracheal bronchial epithelial cells grown at the air-liquid interface, which supports the growth of ciliated, secretory, basal, and other cell populations.51,52 Langlois presented unpublished data on how the ratio of these cell populations within cell culture impacts influenza virus replication dynamics and immune responses.
An antiviral role for IL-6 trans signaling
Daniel Lingwood from Harvard Medical School presented work on understanding the role of interleukin 6 (IL-6) trans signaling in viral immunity. IL-6 is a cytokine that activates pro-inflammatory signaling pathways via binding to the IL-6 receptor (IL-6R) in complex with gp130 on the cell membrane. It can also initiate signaling in cells that do not express IL-6R by binding to soluble IL-6R (sIL-6R) and forming a complex with gp130 on the cell membrane in a process known as trans-signaling.53 Lingwood showed that IL-6 signaling is regulated by a blood buffer system that controls levels of free IL-6. Normally, relatively high levels of sIL-6R and soluble gp130 (sgp130) combined with the tight binding affinity between IL-6, sIL-6R, and sgp130 mean that the majority of circulating IL-6 is complexed with sIL-6R and sgp130 and therefore unable to initiate downstream signaling. In mice, IL-6 levels increase following influenza infection. This is accompanied by an increase in sIL-6R and a decrease in sgp130. The result is an increase in IL-6 trans signaling, with antiviral effects.54
Proviral and antiviral activities of a host protein
Steven Baker from the Lovelace Biomedical Research Institute showed how mitochondrial enoyl-CoA/ACP reductase (MECR) can have both antiviral and proviral effects as a result of alternative splicing. Baker and co-authors used mass spectrometry (MS), to identify host proteins that interact with the influenza virus polymerase. One of these hits, MECR, was found to have antiviral effects when knocked down with siRNA. Baker showed full-length MECR is generally proviral. It is involved in mitochondrial fatty acid synthesis, which is important for host cell metabolism and consequent virus growth. However, a truncated form of MECR, cMECR, exhibits antiviral effects by binding to the influenza polymerase and inhibiting RNP assembly.55 Baker noted that this is an interesting case of how this dual functionality may safeguard antiviral host proteins from virus antagonism.
Extracellular vesicles and RSV replication
Melanie Folkins from David Marchant’s group at the University of Alberta presented unpublished data on the role of extracellular vesicles in protecting RSV. The work is based on previous work by Folkins demonstrating that amoebas can harbor RSV virions without negatively affecting their infectivity.56,57 Combined with the fact that RSV uses vesicle trafficking throughout its replication cycle58, Folkins is investigating the role of extracellular vesicles in viral replication.
Screening for emerging respiratory viruses
Ellen Foxman from Yale University presented work on leveraging the airway innate immune response to screen for unexpected or emerging respiratory viruses. Many hospitals use multiplex respiratory polymerase chain reaction (PCR) panels to test for respiratory infections; however, this approach only identifies viruses included in the panel. Comprehensive testing of samples that test negative via these panels to identify new or emerging viruses is both costly and time-consuming. Foxman’s group has previously shown that the IFN response is highly upregulated in the nasopharynx following respiratory virus infection.59,60 They are investigating the potential of using the IFN response as a sensitive indicator of viral respiratory infection. Prior to the COVID-19 pandemic, Foxman’s group screened over 200 nasopharyngeal samples that tested virus-negative by PCR for elevated CXCL10. Subsequent analyses of these samples found that many were infected with seasonal coronaviruses, and one case of influenza C was identified. Transcriptome analyses suggest that it may be possible to use additional aspects of the host response to identify the type of infection a patient has. In the early days of the pandemic, before PCR tests were widely available, Foxman’s group screened 375 nasopharyngeal samples that tested virus-negative via conventional respiratory panels and identified four undiagnosed cases of COVID-19. Viral genome analyses showed that these four cases were genetically distinct, representing separate introductions of the virus. This work demonstrates practical applications of host response-based screening in virus discovery and surveillance.61
Novel Insights into Respiratory Virus Biology
Monitoring the antibody response via structural biology
Andrew Ward from The Scripps Research Institute presented work on using structural biology to monitor the antibody response to influenza and coronaviruses. Typically, structural biology is one of the last steps when investigating the antiviral immune response. Ward’s group has developed new methods to move structural analyses further upstream in the process so that they can be done in parallel with serum and repertoire analyses. The method, EM-based polyclone epitope mapping (EMPEM) enables comprehensive epitope mapping within days using electron microscopy.62 Ward’s group has used EMPEM to map the serum antibody response over time in people given an H5N1 influenza vaccine. He showed that most vaccinees had pre-existing stem antibodies, which makes sense since the stem domain is highly conserved. Antibodies against the less conserved H5N1 head domain dominated approximately four weeks after vaccination and subsequently wanted while the response against the stem domain persisted.63 They have also used EMPEM to identify a new type of stem broadly neutralizing antibodies elicited from the seasonal flu vaccine. These antibodies target a conserved epitope and are cross-reactive against several influenza subgroups.64 Ward’s group has adapted EMPEM to incorporate cryo-EM data (CryoEMPEM), which provides molecular details of the antibody-epitope interaction in a relatively high-throughput manner.65,66 He showed how they used cryoEMPEM to map the antibody response to seasonal coronaviruses using stabilized spike proteins. They found that most individuals had pre-existing antibodies against the N-terminal domain (NTD) while immunity against the more conserved S2 region was limited. They identified different antibody classes based on their epitope, including those that block the sialic acid receptor binding site, those that bind the C-terminal domain loop, and those that bind across protein subunit interfaces, likely preventing the conformation change from the prefusion to postfusion forms.67 Ward hopes that insights like these can inform vaccine design.
Impairments in the interferon response and susceptibility to SARS-CoV-2
Martin Schwemmle from the University of Freiburg presented work on what drives the age-dependent severity of COVID-19. Both age and impaired IFN response correlate with disease severity in those with COVID-19.68–70 Schwemmle’s group has established a mouse model to investigate whether there is a connection between age-dependent disease severity and an impaired IFN response. He showed that the enhanced susceptibility of aged mice to SARS-CoV-2-induced disease correlated with an increased viral load compared to adult mice, even though lung pathology was similar between the two age groups. Aged mice exhibited a delayed, diminished, and dysregulated immune response. In particular, Schwemmle showed that impairments in the IFN-γ response may be responsible for age-dependent disease severity. In adult mice deficient in type I and type III IFN signaling, disease severity was similar to that of aged mice. Knocking out the IFN-γ receptor increased disease severity, while knocking out the IFN-ɑ and -γ receptors sensitized adult animals to disease. Therapeutic administration of IFN-λ was able to reduce disease severity and mortality in adult mice with the IFN-ɑ receptor knocked out but did not have a dramatic effect as either a prophylactic or therapeutic treatment in aged mice. However, supplementing IFN-λ with IFN-γ in aged mice reversed the age-dependent disease phenotype. Together, these data demonstrate a complex relationship between IFN signaling and COVID-19 disease severity, clarifying the nonredundant antiviral roles of type I, II, and III IFNs. They also demonstrate a potential therapeutic role for IFNs.71
An artificial library-based approach to studying defective viral genomes
Alistair Russell from the University of California, San Diego presented work on analyzing defective influenza A viral genomes. Defective or nonstandard viral genomes often arise as a result of large internal deletions as the virus replicates. These deletions reduce the overall fitness of the virus population, both by competing for resources with intact viral genomes and by activating the host innate immune response. Studying these deletions has been difficult. They occur stochastically, so teasing out the impact of a specific deletion versus defective viral genomes as a whole can be difficult. In addition, methods that depend on PCR enrichment can distort the abundance of deletions. Russell’s group is overcoming these barriers using an artificial library-based approach that uses barcoding to sample the library in an unbiased fashion. He showed that deletions display a fitness optimum between 300 and 600 nucleotides, which is consistent with the diversity seen in nature. In addition, stimulation of the IFN response did not correlate with the length of the defective genomes, indicating that other properties are important for the host immune response.72
Insights from human challenge studies
Christopher Chiu from Imperial College London gave an overview of how human challenge studies can provide important, unique insights on respiratory virus infections and determinants of disease. Human infection challenge studies offer an opportunity to study viral infection with a known strain, constant dose, and detailed assessment of clinical and immunologic readouts. Unlike most field studies, challenge studies enable researchers to sample pre-infection, early infection, and asymptomatic infection to tease out host susceptibility and protective factors.73 Chiu showed how human challenge studies of RSV infection have provided information on viral load dynamics and symptoms. Serum antibody levels against RSV have been a poor indicator of protection; however, challenge studies showed that nasal antibody levels correlate more strongly with infection, although they do not completely explain why some individuals are protected against infection while others are not.74 However, more recent studies in healthy older adults, who had both higher infection rates and viral loads than younger individuals, showed that nasal IgA responses were impaired with aging with systemic IgG potentially compensating.75 RNA sequencing of nasal tissue samples has revealed differences in transcription prior to infection and during the virus incubation period that correlate with the development of symptomatic infection. For example, individuals with higher expression of genes associated with neutrophilic inflammation prior to infection were more susceptible to infection. Work in a mouse model to understand the mechanism showed that pre-infection neutrophils enhance the recruitment of immunopathogenic CD8+ T cells.76 This work is another example of a common theme throughout the meeting―CD8+ T cells can be a double-edged sword when it comes to infection. While they are important for viral clearance, they can also have pathologic effects if dysregulated. Chiu also gave an overview of the first SARS-CoV-2 human challenge study, which was conducted during the pandemic. In a small study of 34 seronegative individuals, the infection rate was approximately 50%. Uninfected individuals did not develop a neutralizing antibody response despite transient viral detections in a proportion. Data from infected individuals revealed viral kinetics, including exact assessments of incubation period and viral shedding.77 Now that the majority of the population has been vaccinated and/or exposed to SARS-CoV-2, this study cannot be easily replicated but these data long with human challenge studies of breakthrough infection may be useful to understand factors associated with protection against primary and breakthrough infection as well as in trials of new vaccines and other interventions.
Influenza reassortment in animal hosts
Ketaki Ganti from Emory University presented work on understanding influenza A reassortment in swine. Swine is an important intermediate host between human and non-human animals for influenza. The potential for reassortment within such a host would increase viral diversity and potentially increase viral fitness. Ganti, a research scientist in Anice Lowen’s group, is interested in understanding whether the within-host dynamics of influenza A infection in swine are conducive to reassortment. They quantified reassortment in swine using well-matched parental strains of human pandemic H1N1 influenza virus. Ganti showed that reassortment yielded modest viral diversity in the nasal tract. Overall, there was limited viral mixing between anatomic sites, and viral replication was highly compartmentalized within the respiratory tract. These data suggest that the concept of swine as a viral mixing vessel may be more a result of permissiveness to infection than of high reassortment.78
Prevention and Treatment Strategies: Vaccines and Antivirals
The symposium ended with a session on novel prevention and treatment strategies. Speakers gave an update on the clinical results for Pfizer’s RSV F protein vaccine, discussed challenges and strategies regarding seasonal and pandemic influenza virus vaccines, reviewed efforts to create broad-spectrum coronavirus antivirals to increase pandemic preparedness, and shared preclinical insights on the immunogenicity of COVID-19 boosters in a population that has largely been exposed to SARS-CoV-2.
Clinical data on a prefusion RSV F protein vaccine
Iona Munjal from Pfizer gave an overview of the company’s progress on different approaches to developing an RSV vaccine for infants and the elderly. Nearly everyone is exposed to RSV as a child; however, exposure does not lead to durable, lifelong protection, and RSV disease contributed to high morbidity and mortality among infants and the elderly.79 Early efforts to create an RSV vaccine focused on a post-fusion conformation of the viral F protein and led to enhanced disease. Newer efforts, including Pfizer’s RSV vaccine, have leveraged structural studies of the F protein to stabilize it in its prefusion conformation.80,81 Munjal described Pfizer’s maternal vaccination program to protect young infects from RSV. In a phase 2b trial, the RSVpreF vaccine was shown to boost neutralizing antibody titers when administered to healthy pregnant women. High titers of neutralizing antibodies were also detectable in the cord blood after birth, indicating that protective antibodies are efficiently transferred across the placenta. A post-hoc analysis showed that RSVpreF prevented lower respiratory tract infections (LRTI) in infants whose mothers were vaccinated.82 A larger, phase 3 global study is underway to confirm the efficacy of RSVpreF in infants. A separate program in older adults showed that RSVpreF elicits a robust immune response with a good safety and tolerability profile. 83,84 While it is unclear whether the antibody response is sufficient to afford protection, data from a phase 1/2 challenge study in healthy adults showed that RSVpreF was highly efficacious against symptomatic and asymptomatic mild-to-moderate infection and reduced infectious viral shedding.85 A global phase 3 trial of RSVpreF is underway in individuals 60 years of age and older, including those at risk for severe disease.
Challenges and solutions to improving influenza virus vaccines
Kanta Subbarao from the WHO Collaborating Centre for Reference and Research on Influenza discussed various challenges and strategies to improve seasonal and pandemic influenza vaccines. Currently, developing seasonal influenza vaccines takes months. By the time vaccines are available, the predominant strains may have already mutated, rendering the vaccine less effective.86 Subbarao noted the importance of modeling to predict which viral clades will dominate. Modeling is especially important following the COVID-19 pandemic as low influenza activity since early 2020 means that there are limited data on which strains are circulating. Seasonal vaccines also are less effective in some high-risk populations, such as the elderly. Subbarao stressed the importance of research efforts to increase immunogenicity, potentially with higher doses or adjuvants, to protect vulnerable populations. Finally, vaccine production in embryonated chicken eggs can induce mutations in HA that may impact the antibody response. Establishing new manufacturing methods, such as cell-grown vaccines and recombinant HA vaccines, will be key to avoiding this complication. In terms of a pandemic influenza vaccine, Subbarao stressed the need to consider breadth. Current vaccines are very strain specific.87 Creating an effective pandemic vaccine with this conventional approach would therefore require intense monitoring of viral genetic and antigenic drift, identifying strains that are likely to cross the species barrier into humans, and preparing vaccine candidates against that specific strain. Enhancing the breadth of influenza vaccines, likely by starting with a subtype-specific vaccine, would help countries be more prepared when a zoonotic influenza virus emerges. Strategies such as whole-virion vaccines and multivalent vaccines may provide more breadth. Ultimately, however, the end goal is to develop a universal influenza vaccine that targets all influenza A viruses. The NIH has established criteria for a universal influenza vaccine87, but technical, regulatory, and logistical challenges remain.
Broad-spectrum coronavirus antivirals
Timothy Sheahan from the University of North Carolina presented work on developing broad-spectrum antivirals against coronaviruses. The past decade has demonstrated the propensity for coronaviruses to jump from animals to humans and cause severe morbidity and mortality around the world. Having broad-spectrum coronavirus antivirals on hand could mitigate the impact of potential future pandemics. Sheahan discussed the potential of nucleoside analogs as antiviral drugs. They study the potential of various agents in primary human airway epithelial cell cultures, which recapitulate the complexity and structure of the human airway. Prior to the pandemic, Sheahan showed that the nucleoside analog remdesivir had broad-spectrum activity against multiple coronaviruses, including MERS-CoV and SARS-CoV-1.88–91 Remdesivir would later become the first treatment approved for COVID-19.92 Sheahan showed that in a transgenic MERS mouse model93–95, remdesivir treatment reduces viral load and mitigates acute lung injury.91 His lab has also developed replication and disease mouse models of SARS-CoV-296,97, which have been instrumental in the preclinical assessment of several agents.96,98,99 These studies demonstrated the importance of treatment timing. In mice, remdesivir was only able to mitigate symptoms if given prior to or early in infection, even though later treatment reduced viral titers.99 Later trials in humans have similarly shown mixed results with remdesivir treatment, potentially due to differences in when patients receive therapy.100 Sheahan’s group has also conducted preclinical studies of molnupiravir, another nucleoside analog with activity against several coronaviruses as well as Ebola virus, influenza virus, and Venezuelan equine encephalitis virus.101 In a phase 2a study, molnupiravir was shown to reduce infectious virus shedding among non-hospitalized, unvaccinated adults with COVID-19.102 A subsequent phase 3 study showed that treatment reduced the risk of hospitalization and death by 30%103, suggesting that there is still a need to improve current treatments and develop novel agents.
The impact of pre-existing immunity on COVID-19 vaccine boosters
Pablo Penaloza-MacMaster from Northwestern University presented work on the effect of pre-existing immunity on mRNA vaccine boosters. In a cohort of individuals primed with an mRNA COVID-19 vaccine, Penaloza-MacMaster’s group found an inverse correlation between pre-boost antibody titers and fold increase after boosting. In other words, it appeared that the humoral response elicited by mRNA vaccination abrogated subsequent de novo antibody responses. Similar effects were seen in mice. Penaloza-MacMaster’s group showed that prior immunization with mRNA-LNPs generated antigen-specific antibodies that accelerate the clearance of vaccine antigen upon booster immunization, via antibody effector mechanisms. As new variants dominate, one of the key questions has been whether booster doses should be updated to reflect current circulating viruses. Penaloza-MacMaster showed that in mice primed with an ancestral vaccine, boosting with a monovalent Omicron-specific vaccine was not substantially superior to boosting with an ancestral vaccine. The ancestral boost elicited higher titers of ancestral-specific antibodies and similar titers of Omicron-specific antibodies as the monovalent Omicron boost. 104 Studies like this are key to informing future vaccination strategies as the virus evolves.
The neonatal T cell response to hMPV infection
Taylor Eddens from John Williams’s group at the University of Pittsburgh presented work on understanding the neonatal CD8+ T cell response in human metapneumovirus (hMPV) infection. Williams’s group has investigated the signaling between dendritic cells and CD8+ T cells in adult mice in response to hMPV, including the impact of PD-1/L1 signaling, which typically downregulates T cell function.105–107 However, the immune system within the neonate lung has distinct differences from the adult lung that may impact its response to viral infection. In particular, the immune system within the neonate lung has a more anti-inflammatory and tolerant phenotype.108 Eddens presented unpublished data on the role of PD-1/L1 signaling in a neonate model of hMPV infection, including its impact on CD8+ T cell function, viral clearance, and disease pathology.
Acknowledgments
Work in Andreas Wack’s lab was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (CC2085), the UK Medical Research Council (CC2085), and the Wellcome Trust (CC2085).
Christopher Chiu is supported by the UK Vaccine Taskforce, the Wellcome Trust (087805/Z/08/Z), the Kwok Foundation, MRC EMINENT Network (MR/R502121/1), which is co-funded by GSK, and by the Biomedical Research Centre award to Imperial College Healthcare National Health Service (NHS) Trust, Imperial’s Health Protection Research Unit in Respiratory Infections, the Comprehensive Local Research Networks, and the MRC HIC-Vac network (MR/R005982/1). The views expressed are those of the author and not necessarily those of the NHS, the NIHR, or the Department of Health and Social Care.
Work in the Hutchinson group is funded by the UK Medical Research Council (MR/N008618/1, MR/V035789/1 and MC_PC_19026). We thank Dr Léa Meyer for assistance in figure preparation.
The Ellebedy laboratory is supported by NIH grants U01AI141990, 1U01AI150747, 5U01AI144616-02, and R01AI168178-01.
The Ellebedy laboratory received funding from Emergent BioSolutions, AbbVie and Moderna that are unrelated to the data presented in the current study. AHE received consulting and speaking fees from InBios International, Inc, Fimbrion Therapeutics, RGAX, Mubadala Investment Company, Moderna, Pfizer, GSK, Danaher, Third Rock Ventures, Goldman Sachs and Morgan Stanley. AHE is the founder of ImmuneBio Consulting. AHE is a recipient of a licensing agreement with Abbvie that is unrelated to the data presented in the current study.
Footnotes
Sci Immunol. 2021 Jan 8;6(55). doi: 10.1126/sciimmunol.abb6852.
Competing Interests
J. Sun is a consultant for the Teneofour company.
References
- 1.Narasimhan H, Wu Y, Goplen NP, et al. 2022. Immune determinants of chronic sequelae after respiratory viral infection. Sci. Immunol 7: eabm7996. [DOI] [PubMed] [Google Scholar]
- 2.Lin W-HW, Kouyos RD, Adams RJ, et al. 2012. Prolonged persistence of measles virus RNA is characteristic of primary infection dynamics. Proc. Natl. Acad. Sci. U. S. A 109: 14989–14994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao X-H, He Z-C, Li T-Y, et al. 2020. Pathological evidence for residual SARS-CoV-2 in pulmonary tissues of a ready-for-discharge patient. Cell Res. 30: 541–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pebody RG, McLean E, Zhao H, et al. 2010. Pandemic Influenza A (H1N1) 2009 and mortality in the United Kingdom: risk factors for death, April 2009 to March 2010. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull 15: 19571. [PubMed] [Google Scholar]
- 5.Zheng Y, Liu X, Le W, et al. 2020. A human circulating immune cell landscape in aging and COVID-19. Protein Celi 11: 740–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goplen NP, Wu Y, Son YM, et al. 2020. Tissue-resident CD8+ T cells drive age-associated chronic lung sequelae after viral pneumonia. Sci. Immunol 5: eabc4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheon IS, Li C, Son YM, et al. 2021. Immune signatures underlying post-acute COVID-19 lung sequelae. Sci. Immunol 6: eabk1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiner AI, Zhao G, Zayas HM, et al. 2022. ΔNp63 drives dysplastic alveolar remodeling and restricts epithelial plasticity upon severe lung injury. 2022.02.23.481695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendel SK, Kellermann L, Hausmann A, et al. 2022. Tuft Cells and Their Role in Intestinal Diseases. Front. Immunol 13: 822867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barr J, Gentile ME, Lee S, et al. 2022. Injury-induced pulmonary tuft cells are heterogenous, arise independent of key Type 2 cytokines, and are dispensable for dysplastic repair. eLife 11: e78074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melms JC, Biermann J, Huang H, et al. 2021. A molecular single-cell lung atlas of lethal COVID-19. Nature 595: 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia GL, Valenzuela A, Manzoni T, et al. 2020. Distinct Chronic Post-Viral Lung Diseases upon Infection with Influenza or Parainfluenza Viruses Differentially Impact Superinfection Outcome. Am. J. Pathol 190: 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vignuzzi M. & López CB 2019. Defective viral genomes are key drivers of the virus-host interaction. Nat. Microbiol 4: 1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genoyer E. & López CB 2019. The Impact of Defective Viruses on Infection and Immunity. Annu. Rev. Virol 6: 547–566. [DOI] [PubMed] [Google Scholar]
- 15.Xu J, Sun Y, Li Y, et al. 2017. Replication defective viral genomes exploit a cellular pro-survival mechanism to establish paramyxovirus persistence. Nat. Commun 8: 799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felt SA, Sun Y, Jozwik A, et al. 2021. Detection of respiratory syncytial virus defective genomes in nasal secretions is associated with distinct clinical outcomes. Nat. Microbiol 6: 672–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davenport FM & Hennessy AV 1956. A serologic recapitulation of past experiences with influenza A; antibody response to monovalent vaccine. J. Exp. Med 104: 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davenport FM, Hennessy AV, Stuart-Harris CH, et al. 1955. Epidemiology of influenza; comparative serological observations in England and the United States. Lancet Lond. Engl 269: 469–474. [DOI] [PubMed] [Google Scholar]
- 19.Fonville JM, Wilks SH, James SL, et al. 2014. Antibody landscapes after influenza virus infection or vaccination. Science 346: 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henry C, Zheng N-Y, Huang M, et al. 2019. Influenza Virus Vaccination Elicits Poorly Adapted B Cell Responses in Elderly Individuals. Cell Host Microbe 25: 357–366.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gostic KM, Ambrose M, Worobey M, et al. 2016. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 354: 722–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gostic KM, Bridge R, Brady S, et al. 2019. Childhood immune imprinting to influenza A shapes birth year-specific risk during seasonal H1N1 and H3N2 epidemics. PLoS Pathog. 15: e1008109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arevalo P, McLean HQ, Belongia EA, et al. 2020. Earliest infections predict the age distribution of seasonal influenza A cases. eLife 9: e50060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gouma S, Kim K, Weirick ME, et al. 2020. Middle-aged individuals may be in a perpetual state of H3N2 influenza virus susceptibility. Nat. Commun 11: 4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linderman SL, Chambers BS, Zost SJ, et al. 2014. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013–2014 influenza season. Proc. Natl. Acad. Sci. U. S. A 111: 15798–15803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skowronski DM, Zou M, Sabaiduc S, et al. 2020. Interim estimates of 2019/20 vaccine effectiveness during early-season co-circulation of influenza A and B viruses, Canada, February 2020. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull 25:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kissling E, Pozo F, Buda S, et al. 2019. Low 2018/19 vaccine effectiveness against influenza A(H3N2) among 15–64-year-olds in Europe: exploration by birth cohort. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull 24:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oidtman RJ, Arevalo P, Bi Q, et al. 2021. Influenza immune escape under heterogeneous host immune histories. Trends Microbiol. 29: 1072–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Everitt AR, Clare S, Pertel T, et al. 2012. IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunning J, Blankley S, Hoang LT, et al. 2018. Progression of whole-blood transcriptional signatures from interferon-induced to neutrophil-associated patterns in severe influenza. Nat. Immunol 19: 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Docherty AB, Harrison EM, Green CA, et al. 2020. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: prospective observational cohort study. BMJ 369: m1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thwaites RS, Sanchez Sevilla Uruchurtu A, Siggins MK, et al. 2021. Inflammatory profiles across the spectrum of disease reveal a distinct role for GM-CSF in severe COVID-19. Sci. Immunol 6: eabg9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pairo-Castineira E, Clohisey S, Klaric L, et al. 2021. Genetic mechanisms of critical illness in COVID-19. Nature 591: 92–98. [DOI] [PubMed] [Google Scholar]
- 34.Marconi VC, Ramanan AV, de Bono S, et al. 2021. Efficacy and safety of baricitinib for the treatment of hospitalised adults with COVID-19 (COV-BARRIER): a randomised, double-blind, parallel-group, placebo-controlled phase 3 trial. Lancet Respir. Med 9: 1407–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner JS, Zhou JQ, Han J, et al. 2020. Human germinal centres engage memory and naive B cells after influenza vaccination. Nature 586: 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner JS, O’Halloran JA, Kalaidina E, et al. 2021. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature 596: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner JS, Kim W, Kalaidina E, et al. 2021. SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Nature 595: 421–425. [DOI] [PubMed] [Google Scholar]
- 38.Kim W, Zhou JQ, Horvath SC, et al. 2022. Germinal centre-driven maturation of B cell response to mRNA vaccination. Nature 604: 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Major J, Crotta S, Llorian M, et al. 2020. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science 369: 712–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Broggi A, Ghosh S, Sposito B, et al. 2020. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science 369: 706–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinto RM, Lycett S, Gaunt E, et al. 2021. Accessory Gene Products of Influenza A Virus. Cold Spring Harb. Perspect. Med 11: a038380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H, Wang Y. & Lu J. 2019. Function and Evolution of Upstream ORFs in Eukaryotes. Trends Biochem. Sci 44: 782–794. [DOI] [PubMed] [Google Scholar]
- 43.Elfakess R, Sinvani H, Haimov O, et al. 2011. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res. 39: 7598–7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clohisey S, Parkinson N, Wang B, et al. 2020. Comprehensive Characterization of Transcriptional Activity during Influenza A Virus Infection Reveals Biases in Cap-Snatching of Host RNA Sequences. J. Virol 94: e01720–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rialdi A, Hultquist J, Jimenez-Morales D, et al. 2017. The RNA Exosome Syncs IAV-RNAPII Transcription to Promote Viral Ribogenesis and Infectivity. Cell 169: 679–692.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho JSY, Angel M, Ma Y, et al. 2020. Hybrid Gene Origination Creates Human-Virus Chimeric Proteins during Infection. Cell 181: 1502–1517.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacobs NT, Onuoha NO, Antia A, et al. 2019. Incomplete influenza A virus genomes occur frequently but are readily complemented during localized viral spread. Nat. Commun 10: 3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heldt FS, Kupke SY, Dorl S, et al. 2015. Single-cell analysis and stochastic modelling unveil large cell-to-cell variability in influenza A virus infection. Nat. Commun 6: 8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brooke CB 2017. Population Diversity and Collective Interactions during Influenza Virus Infection. J. Virol 91: e01164–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loveday EK, Sanchez HS, Thomas MM, et al. 2022. Single-Cell Infection of Influenza A Virus Using Drop-Based Microfluidics. Microbiol. Spectr e0099322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fiege JK, Thiede JM, Nanda HA, et al. 2021. Single cell resolution of SARS-CoV-2 tropism, antiviral responses, and susceptibility to therapies in primary human airway epithelium. PLoS Pathog. 17: e1009292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonser LR, Koh KD, Johansson K, et al. 2021. Flow-Cytometric Analysis and Purification of Airway Epithelial-Cell Subsets. Am. J. Respir. Cell Mol. Biol 64: 308–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka T. & Kishimoto T. 2014. The biology and medical implications of interleukin-6. Cancer Immunol. Res 2: 288–294. [DOI] [PubMed] [Google Scholar]
- 54.Yousif AS, Ronsard L, Shah P, et al. 2021. The persistence of interleukin-6 is regulated by a blood buffer system derived from dendritic cells. Immunity 54: 235–246.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker SF, Meistermann H, Tzouros M, et al. 2022. Alternative splicing liberates a cryptic cytoplasmic isoform of mitochondrial MECR that antagonizes influenza virus. 2020.11.09.355982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Folkins MA, Dey R. & Ashbolt NJ 2020. Interactions between Human Reovirus and Free-Living Amoebae: Implications for Enteric Virus Disinfection and Aquatic Persistence. Environ. Sci. Technol 54: 10201–10206. [DOI] [PubMed] [Google Scholar]
- 57.Dey R, Folkins MA & Ashbolt NJ 2021. Extracellular amoebal-vesicles: potential transmission vehicles for respiratory viruses. NPJ Biofilms Microbiomes 7: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krzyzaniak MA, Zumstein MT, Gerez JA, et al. 2013. Host cell entry of respiratory syncytial virus involves macropinocytosis followed by proteolytic activation of the F protein. PLoS Pathog. 9: e1003309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Landry ML & Foxman EF 2018. Antiviral Response in the Nasopharynx Identifies Patients With Respiratory Virus Infection. J. Infect. Dis 217: 897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheemarla NR, Watkins TA, Mihaylova VT, et al. 2021. Dynamic innate immune response determines susceptibility to SARS-CoV-2 infection and early replication kinetics. J. Exp. Med 218: e20210583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheemarla NR, Hanron A, Fauver JR, Bishai J, Watkins TA, Brito AF, Zhao D, Alpert T, Vogels CBF, Ko AI, Schulz WL, Landry ML, Grubaugh ND, van Dijk D, Foxman EF. Nasal host response-based screening for undiagnosed respiratory viruses: a pathogen surveillance and detection study. Lancet Microbe. 2023. Jan;4(1):e38–e46. doi: 10.1016/S2666-5247(22)00296-8. PMID: 36586415.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bianchi M, Turner HL, Nogal B, et al. 2018. Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization. Immunity 49: 288–300.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han J, Schmitz AJ, Richey ST, et al. 2021. Polyclonal epitope mapping reveals temporal dynamics and diversity of human antibody responses to H5N1 vaccination. Cell Rep. 34: 108682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guthmiller JJ, Han J, Utset HA, et al. 2022. Broadly neutralizing antibodies target a haemagglutinin anchor epitope. Nature 602: 314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Antanasijevic A, Sewall LM, Cottrell CA, et al. 2021. Polyclonal antibody responses to HIV Env immunogens resolved using cryoEM. Nat. Commun 12: 4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Antanasijevic A, Bowman CA, Kirchdoerfer RN, et al. 2022. From structure to sequence: Antibody discovery using cryoEM. Sci. Adv 8: eabk2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bangaru S, Antanasijevic A, Kose N, et al. 2022. Structural mapping of antibody landscapes to human betacoronavirus spike proteins. Sci. Adv 8: eabn2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bastard P, Rosen LB, Zhang Q, et al. 2020. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370: eabd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Q, Bastard P, Liu Z, et al. 2020. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370: eabd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bastard P, Gervais A, Le Voyer T, et al. 2021. Autoantibodies neutralizing type I IFNs are present in ~4% of uninfected individuals over 70 years old and account for ~20% of COVID-19 deaths. Sci. Immunol 6: eabl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beer J, Crotta S, Breithaupt A, et al. 2022. Impaired immune response drives age-dependent severity of COVID-19. J. Exp. Med 219: e20220621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mendes M. & Russell AB 2021. Library-based analysis reveals segment and length dependent characteristics of defective influenza genomes. PLoS Pathog. 17: e1010125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roestenberg M, Hoogerwerf M-A, Ferreira DM, et al. 2018. Experimental infection of human volunteers. Lancet Infect. Dis 18: e312–e322. [DOI] [PubMed] [Google Scholar]
- 74.Habibi MS, Jozwik A, Makris S, et al. 2015. Impaired Antibody-mediated Protection and Defective IgA B-Cell Memory in Experimental Infection of Adults with Respiratory Syncytial Virus. Am. J. Respir. Crit. Care Med 191: 1040–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ascough S, Dayananda P, Kalyan M, et al. 2022. Divergent age-related humoral correlates of protection against respiratory syncytial virus infection in older and young adults: a pilot, controlled, human infection challenge model. Lancet Healthy Longev. 3: e405–e416. [DOI] [PubMed] [Google Scholar]
- 76.Habibi MS, Thwaites RS, Chang M, et al. 2020. Neutrophilic inflammation in the respiratory mucosa predisposes to RSV infection. Science 370: eaba9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Killingley B, Mann AJ, Kalinova M, et al. 2022. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat. Med 28: 1031–1041. [DOI] [PubMed] [Google Scholar]
- 78.Ganti K, Bagga A, Carnaccini S, et al. 2022. Influenza A virus reassortment in mammals gives rise to genetically distinct within-host sub-populations. 2022.02.08.479600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Y, Wang X, Blau DM, et al. 2022. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in children younger than 5 years in 2019: a systematic analysis. Lancet Lond. Engl 399: 2047–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Graham BS 2017. Vaccine development for respiratory syncytial virus. Curr. Opin. Virol 23: 107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Graham BS, Gilman MSA & McLellan JS 2019. Structure-Based Vaccine Antigen Design. Annu. Rev. Med 70: 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simões EAF, Center KJ, Tita ATN, et al. 2022. Prefusion F Protein-Based Respiratory Syncytial Virus Immunization in Pregnancy. N. Engl. J. Med 386: 1615–1626. [DOI] [PubMed] [Google Scholar]
- 83.Walsh EE, Falsey AR, Scott DA, et al. 2022. A Randomized Phase 1/2 Study of a Respiratory Syncytial Virus Prefusion F Vaccine. J. Infect. Dis 225: 1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baber J, Arya M, Moodley Y, et al. 2022. A Phase 1/2 Study of a Respiratory Syncytial Virus Prefusion F Vaccine With and Without Adjuvant in Healthy Older Adults. J. Infect. Dis jiac189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schmoele-Thoma B, Zareba AM, Jiang Q, et al. 2022. Vaccine Efficacy in Adults in a Respiratory Syncytial Virus Challenge Study. N. Engl. J. Med 386: 2377–2386. [DOI] [PubMed] [Google Scholar]
- 86.Chen J-R, Liu Y-M, Tseng Y-C, et al. 2020. Better influenza vaccines: an industry perspective. J. Biomed. Sci 27: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Erbelding EJ, Post DJ, Stemmy EJ, et al. 2018. A Universal Influenza Vaccine: The Strategic Plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis 218: 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sheahan TP, Sims AC, Graham RL, et al. 2017. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med 9: eaal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Agostini ML, Andres EL, Sims AC, et al. 2018. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 9: e00221–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brown AJ, Won JJ, Graham RL, et al. 2019. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res. 169: 104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sheahan TP, Sims AC, Leist SR, et al. 2020. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun 11: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Commissioner O. of the. 2020.FDA October 22, 2020 Accessed October 14, 2022. https://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-covid-19.
- 93.Peck KM, Cockrell AS, Yount BL, et al. 2015. Glycosylation of mouse DPP4 plays a role in inhibiting Middle East respiratory syndrome coronavirus infection. J. Virol 89: 4696–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cockrell AS, Yount BL, Scobey T, et al. 2016. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat. Microbiol 2: 16226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Douglas MG, Kocher JF, Scobey T, et al. 2018. Adaptive evolution influences the infectious dose of MERS-CoV necessary to achieve severe respiratory disease. Virology 517: 98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dinnon KH, Leist SR, Schäfer A, et al. 2020. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586: 560–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Leist SR, Dinnon KH, Schäfer A, et al. 2020. A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell 183: 1070–1085.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schäfer A, Muecksch F, Lorenzi JCC, et al. 2021. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med 218: e20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martinez DR, Schäfer A, Leist SR, et al. 2021. Prevention and therapy of SARS-CoV-2 and the B.1.351 variant in mice. Cell Rep. 36: 109450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee TC, Murthy S, Del Corpo O, et al. 2022. Remdesivir for the treatment of COVID-19: a systematic review and meta-analysis. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis 28: 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sheahan TP, Sims AC, Zhou S, et al. 2020. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med 12: eabb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fischer WA, Eron JJ, Holman W, et al. 2022. A phase 2a clinical trial of molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus. Sci. Transl. Med 14: eabl7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jayk Bernal A, Gomes da Silva MM, Musungaie DB, et al. 2022. Molnupiravir for Oral Treatment of Covid-19 in Nonhospitalized Patients. N. Engl. J. Med 386: 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dangi T, Sanchez S, Lew MH, et al. 2022. Pre-existing immunity modulates responses to mRNA boosters. 2022.06.27.497248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Erickson JJ, Gilchuk P, Hastings AK, et al. 2012. Viral acute lower respiratory infections impair CD8+ T cells through PD-1. J. Clin. Invest 122: 2967–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Erickson JJ, Rogers MC, Hastings AK, et al. 2014. Programmed death-1 impairs secondary effector lung CD8+ T cells during respiratory virus reinfection. J. Immunol. Baltim. Md 1950 193: 5108–5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Erickson JJ, Rogers MC, Tollefson SJ, et al. 2016. Multiple Inhibitory Pathways Contribute to Lung CD8+ T Cell Impairment and Protect against Immunopathology during Acute Viral Respiratory Infection. J. Immunol. Baltim. Md 1950 197: 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Eddens T, Parks OB & Williams JV 2022. Neonatal Immune Responses to Respiratory Viruses. Front. Immunol 13: 863149. [DOI] [PMC free article] [PubMed] [Google Scholar]