Abstract
Antimicrobial susceptibility testing (AST) remains the cornerstone of effective antimicrobial selection and optimization in patients. Despite recent advances in rapid pathogen identification and resistance marker detection with molecular diagnostics (e.g., qPCR, MALDI-TOF MS), phenotypic (i.e., microbial culture-based) AST methods - the gold standard in hospitals/clinics - remain relatively unchanged over the last few decades. Microfluidics-based phenotypic AST has been growing fast in recent years, aiming for rapid (i.e., turnaround time <8 h), high-throughput, and automated species identification, resistance detection, and antibiotics screening. In this pilot study, we describe the application of a multi-liquid-phase open microfluidic system, named under-oil open microfluidic systems (UOMS), to achieve a rapid phenotypic AST. UOMS provides an open microfluidics-based solution for rapid phenotypic AST (UOMS-AST) by implementing and recording a pathogen’s antimicrobial activity in micro-volume testing units under an oil overlay. UOMS-AST allows free physical access (e.g., by standard pipetting) to the system and label-free, single-cell resolution optical access. UOMS-AST can accurately and rapidly determine antimicrobial activities [including susceptibility/resistance breakpoint and minimum inhibitory concentration (MIC)] from nominal sample/bacterial cells in a system aligned with clinical laboratory standards where open systems and optical microscopy are predominantly adopted. Further, we combine UOMS-AST with a cloud lab data analytic technique for real-time image analysis and report generation to provide a rapid (<4 h) sample-to-report turnaround time, shedding light on its utility as a versatile (e.g., low-resource setting and manual laboratory operation, or high-throughput automated system) phenotypic AST platform for hospital/clinic use.
Introduction
Bacterial infection continues to be a major global health threat that has been exacerbated by the emergence and spread of antibiotic-resistant species.1 The need for rapid (with the turnaround time less than a single work shift, i.e., <8 h), reliable AST is crucial for reducing the empiric use of broad-spectrum antimicrobials while ensuring that patients receive timely and adequate treatment.2–4 The development and use of molecular methods, e.g., quantitative polymerase chain reaction (qPCR),5,6 matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (MS),7 and Raman spectroscopy,8 in clinical microbiology laboratories have revolutionized the speed of bacterial identification and antimicrobial resistance detection. However, molecular methods are useful when antimicrobial resistance genes or molecules are known, are currently limited to a few select pathogens and antimicrobials, and do not always predict phenotypic resistance.9
Phenotypic methods that directly and functionally screen for bacterial susceptibility/resistance by observing and quantifying the growth or inhibition event of bacteria after exposure to antibiotics provide clinically relevant results and thus remain the gold standard used in clinical settings.9 The traditional (i.e., manual) standard phenotypic AST methods (including disc diffusion and broth microdilution) require intensive labor and a long detection turnaround time of 1 or 2 days following bacterial identification. Several automated phenotypic AST systems have been approved by the U.S. Food and Drug Administration (FDA) and commercialized. Compared to the manual standard AST, the automated systems use standardized, custom, microdilution trays that are hydrated with the bacterial inoculum. Integrated software interprets the AST results and allows real-time report generation and integration into the electronic medical record. These automated systems significantly reduce labor and time (3.5–16 h) of data collection and quantification of antibiotic susceptibility/resistance, and today these are used in most large clinical microbiology laboratories. While improving upon the historical standards to provide more rapid AST results, reporting errors and discrepant results among the systems have been reported.10,11 In addition, access to these automated systems and platforms can be limited for new antibiotics and settings12 with low laboratory resources such as research labs, outpatient clinics, point of care, and middle- and low-income countries.
In the past decade, we have seen fast growing interest, effort, and investment in microfluidics-based phenotypic AST.13–17 Compared to the standard AST systems aforementioned (Table 1), microfluidics-based phenotypic AST allows versatile manipulation/organization of small-volume bacterial niches (e.g., micro- and sub-micro liter) on a device with small footprint, improved and high-resolution optical access, high throughput and flexible detection panel design. Recently, Idelevich, E. A. et al. developed a method named MALDI-TOF-MS-based direct-on-target microdroplet growth assay (DOT-MGA) allowing rapid phenotypic AST (<6 to 8 h) independently of the species-antibiotic agent combination as well as testing of different combinations in parallel.18,19 This method combines the advantages of microfluidics-based phenotypic AST, i.e., high-throughput compartmentalized micro bacterial niches, and fast turnaround time in detecting bacterial biomass by MS. Kandavalli, V. and Karempudi, P. et al. reported an important progress in microfluidics-based phenotypic AST to perform rapid phenotypic screening at the single-cell level in tandem with genotyping by in situ FISH.20 This method makes it possible to determine the susceptibility profile for each species in a mixed sample in 2 h.
Table 1.
A comparison matrix showing the novel features of UOMS-AST in microfluidics-based phenotypic AST compared to the standard phenotypic AST approaches/systems.
| Phenotypic AST | Small volume (μL to pL) | Micro & independent bacterial niche control | Label-free, single-cell detection/imaging | Antibiotics panel customization | Throughput (>100 units/test) & scalability | Real-time image analysis & report generation | Ease of adoption | ||
|---|---|---|---|---|---|---|---|---|---|
| Microfluidics-based AST | Open system | UOMSAST | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ |
| Single-liquid phase | ↓ | ↓ | ↓ | ↑ | ↑ | ↑ | ↑ | ||
| Closed system | ↑ | ↔ | ↑ | ↑ | ↑ | ↑ | ↔ | ||
|
Standard AST
(manual assay a & automated systemb) |
↓ | ↓ | ↓ | ↔ a | ↓ | ↔ b | ↔ b | ||
↑ - Available
↓ - Technically challenging/not available
↔ - Depends on specific method/system
In microfluidics, there has been renewed interest in the development of a fundamental branch in multi-liquid-phase open microfluidics, named UOMS.21–34 Different from closed-chamber/-channel microfluidics or single-liquid phase open microfluidics, cell culture in UMOS is implemented with the culture media and cells contained under an oil overlay, separating the cell culture/detection microenvironment from the ambient with an immiscible liquid (i.e., oil) rather than the closed chambers/channels of traditional microfluidic devices (that inflicts limited physical access to the systems) or open chambers/channels directly exposed to air (that causes media loss via evaporation, airborne sample contamination). Uniquely, UOMS allows: i) free physical access to the system with minimized evaporation and sample contamination, ii) high-resolution optical access with various microscopic techniques (e.g., bright-field,22–25,27 epifluorescence,23,25,29 confocal,31 multi-photon30), and iii) low adoption barrier (i.e., ease to make/use, cost efficiency). When combined with automated data analytics and reporting, UOMS provides an open microfluidics-based solution for rapid phenotypic AST (i.e., UOMS-AST) with advantages over the current standard AST methods (Table 1) and a natural alignment with application toward standard clinical laboratory settings where open-fluid handling (e.g., pipetting) and optical microscopy are typically adopted.
In this pilot study, we demonstrate the ability of UOMS-AST to detect antimicrobial activities including susceptibility/resistance breakpoint and MIC accurately and sensitively with manual laboratory operations. Further integrated with a cloud lab technique UOMS-AST allows real-time image analysis and report generation for rapid phenotypic AST with a sample-to-report turnaround time in 4 h. We also briefly discuss the high-throughput capacity and the development of an automated UOMS-AST system at the end but not the focus of this pilot study that aims to introduce the basic principles and workflow of UOMS-AST.
Results
The UOMS-AST platform
Since the 1980s, microfluidics and micro total analysis systems have rapidly grown and are now used in many commercial products.35,36 Compared to closed-chamber/-channel microfluidic systems or single-liquid phase open microfluidics, UOMS (Fig. 1A) utilizes surface chemistry contrast [i.e., free of solid and physical structures (Fig. 1B, Fig. S1)] and an oil barrier to confine aqueous media and cellular/molecular samples (Methods).25,29 Compared to microtiter plates, e.g., 384, 1536, or higher panel capacity, that confine and compartmentalize culture niches with physical walls, here we propose to use a chambered coverglass with a pre-patterned, open surface and virtual energy barriers (Fig. 1B) that spontaneously and robustly contain the media and cells on a device in the designated areas under an oil overlay.
Fig. 1.
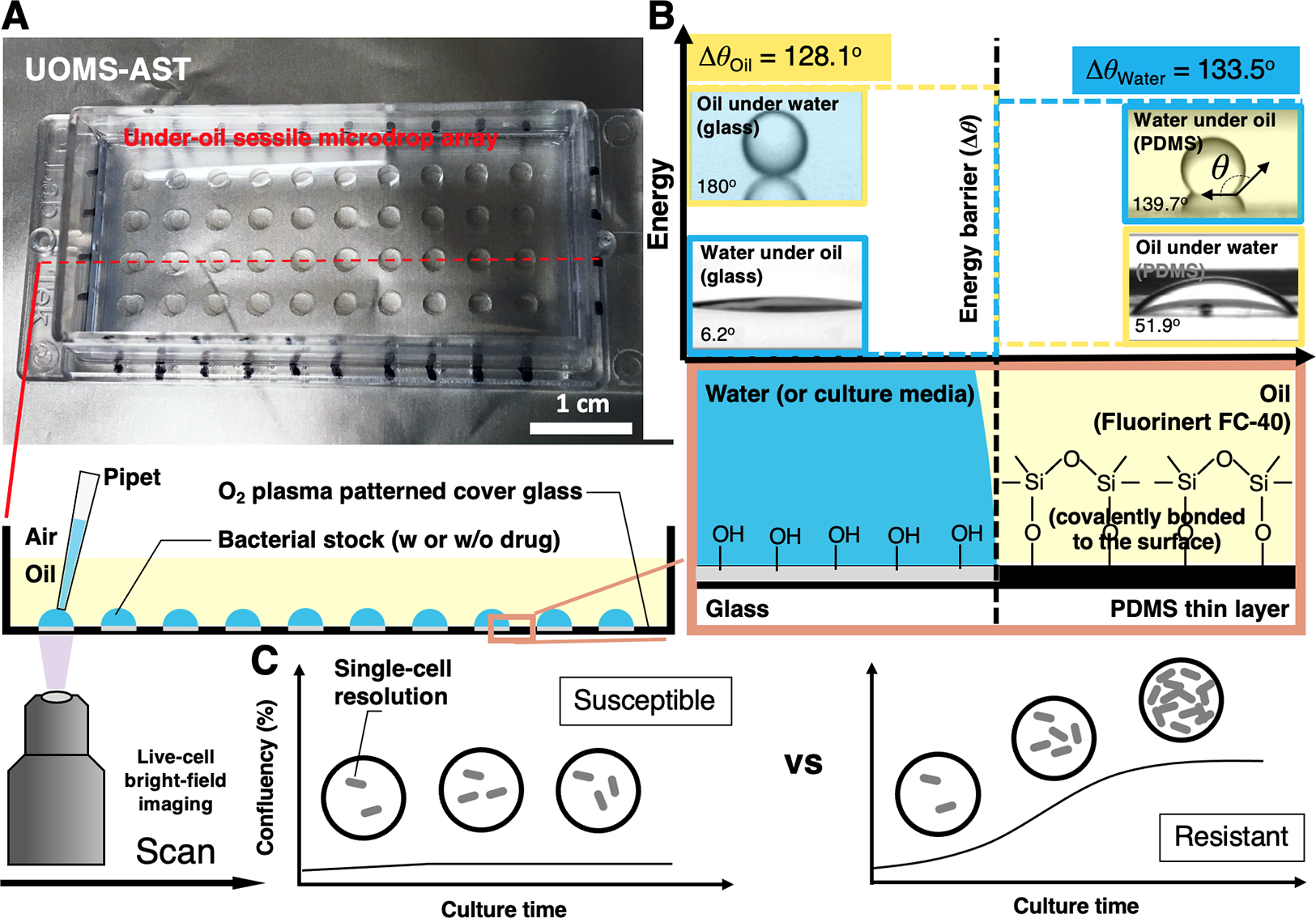
The configuration and physics of UOMS-AST. (A) A UOMS-AST device fabricated with a 1-well chambered coverglass (Nunc Lab-Tek - borosilicate glass #1.0 - medical grade silicone adhesive, 6 cm × 2.4 cm, 0.13–0.17 mm in bottom thickness). The glass surface was patterned for an array (4 × 10) of underoil (Fluorinert FC-40, or FC40, 1.5 mL per 1-well chambered coverglass) sessile (i.e., surface-attached) microdrops (2 mm in diameter). 2 μL of bacterial stock with or without antimicrobial was inoculated to each spot under oil by regular single-channel pipetting. (B) A schematic shows the surface chemistry contrast for liquid confinement with the energy (or virtual) barrier. The glass surface was modified by a monolayer of covalently bonded, polydimethylsiloxane (PDMS)-silane molecules and then selectively patterned by oxygen plasma (Fig. S1). (C) A schematic shows the live-cell, bright-field imaging in UOMS-AST and the detection of antimicrobial susceptibility/resistance based on confluency of bacterial cells in the field of view.
While UOMS can be performed with a coverglass-bottom microtiter plate (i.e., The plastic-bottom microtiter plate comes with a significantly compromised optical access to the bacterial cells especially under bright field.), the testing sample needs to be loaded to a well first followed by adding an oil overlay on top. The reversed order of reagent loading (i.e., oil first and then testing sample) on a microtiter plate leads to inconsistent oil-media distribution in the wells due to the lack of surface chemistry contrast, especially when the well size is small (e.g., 384, 1536) and surface tension becomes dominant over inertial forces. Moreover, to avoid media loss via evaporation and airborne contamination, loading the testing sample and oil on a microtiter plate needs to be done alternately through the wells one by one, which significantly adds time to device/sample preparation. While it might be possible to load the testing sample and then oil to the wells all together, a specially designed multi-channel pipet is required, which requires much larger sample volume for multi-channel pipetting (compared to single-channel pipetting) and increases the adoption barrier due to the limited access to those expensive liquid-handling instruments. Using foil rather than oil overlay to seal a microtiter plate comes with several limitations including 1) operation difficulty - The operator must seal a well right after liquid loading, otherwise, media loss via evaporation still occurs if the foil is applied after all liquid loading especially when the volume per well is small, e.g., a few microliters. 2) compromised bright-field imaging - Aluminum foil is completely opaque. Even if clear sealing film is used, the bright-field imaging will be compromised to some extent. 3) limited physical access to the wells - Once the plate is sealed by a foil, it blocks the physical access to the wells. While the foil can be removed and then put back to the plate, it adds extra steps and evaporation is inevitable during the liquid/sample operation.
By contrast, the chambered coverglass method proposed in this study allows loading and re-collection of the testing samples by single-channel pipetting directly under oil with minimized evaporation/airborne contamination), better oil compatibility [e.g., The coverglass-bottom microtiter plates typically use silicone adhesive to glue the plastic piece onto the glass substrate, which makes it not compatible with silicone oil due to the swelling and leakage issues. In comparison, the chambered coverglass uses either silicone adhesive (#1.0, compatible with fluorinated oil but not silicone oil) or acrylic adhesive (#1.5, compatible with both fluorinated oil and silicone oil) (Methods).], higher flexibility of culture pattern/testing panel customization, and lower cost (e.g., chambered coverglass, ~$10/piece versus 384-well plate, >$20/piece, or 1536-well plate, >$50/piece).
Specifically in the under-oil approach, we use fluorinated oil and/or silicone oil as the oil barrier, which minimizes oil extraction of lipophilic molecules [see the ultra-performance liquid chromatography-tandem mass spectrometer (UPLC-MS) characterization in our previous publication].31 The volume of oil required in UOMS-AST is small as long as enough to cover the sessile droplets on a device. For example, to cover the whole surface and sessile droplets in the 1-well chambered coverglass, we only need 1 to 2 mL of the oil. The oil cost per 1-well chambered coverglass is $1.20 or lower for silicone oil ($600 per liter), or $3.20 or lower for fluorinated oil ($1600 per liter). The testing spots (i.e., plasma-treated areas) are hydrophilic [with an under-oil (fluorinated oil) water contact angle (CA) θ = 6.2°] and the untreated background hydrophobic (with an under-oil water CA θ = 139.7°). Similarly, the untreated background is oleophilic (with an under-water oil CA θ = 51.9°) and the plasma-treated areas oleophobic (with an under-water oil CA θ = 180°). To each liquid, the CA differential (Δθwater = 133.5°, Δθoil = 128.1°) provides the energy barrier that holds and stabilizes the liquid on its preferred surface and thus enables a robust liquid-liquid boundary on the patterned, non-textured surface. Small volume (e.g., 2 μL in this work) of bacterial cells with or without antibiotic can be directly inoculated to the device under oil by regular pipetting. The coverglass bottom of the UOMS-AST device allows label-free (i.e., bright-field) optical access to the bacterial cells with single-cell resolution (Fig. 1C, Fig. S2, Movie S1 to Movie S5). These configurations make the UOMS-AST highly sensitive to pathogen’s antimicrobial activity.
Validation of UOMS-AST
To validate the UOMS-AST method, we benchmarked it against standard phenotypic AST and antimicrobial assessments, i.e., broth microdilution-based growth and time-based killing assay (Methods). In this test, we used Pseudomonas aeruginosa (strain PA01) in a standardized inoculum (5 × 105 cfu/mL) as a model human pathogen against four antibiotics with diverse mechanisms of action (Fig. 2A). The antibiotics were applied individually with different concentrations below and above their MIC. As shown, UOMS-AST was able to capture a full spectrum of different growth curves of the bacterium and displayed inhibition of growth at 0.5 μg/mL, consistent with the ciprofloxacin MIC in PA01 (Fig. 2B).
Fig. 2.
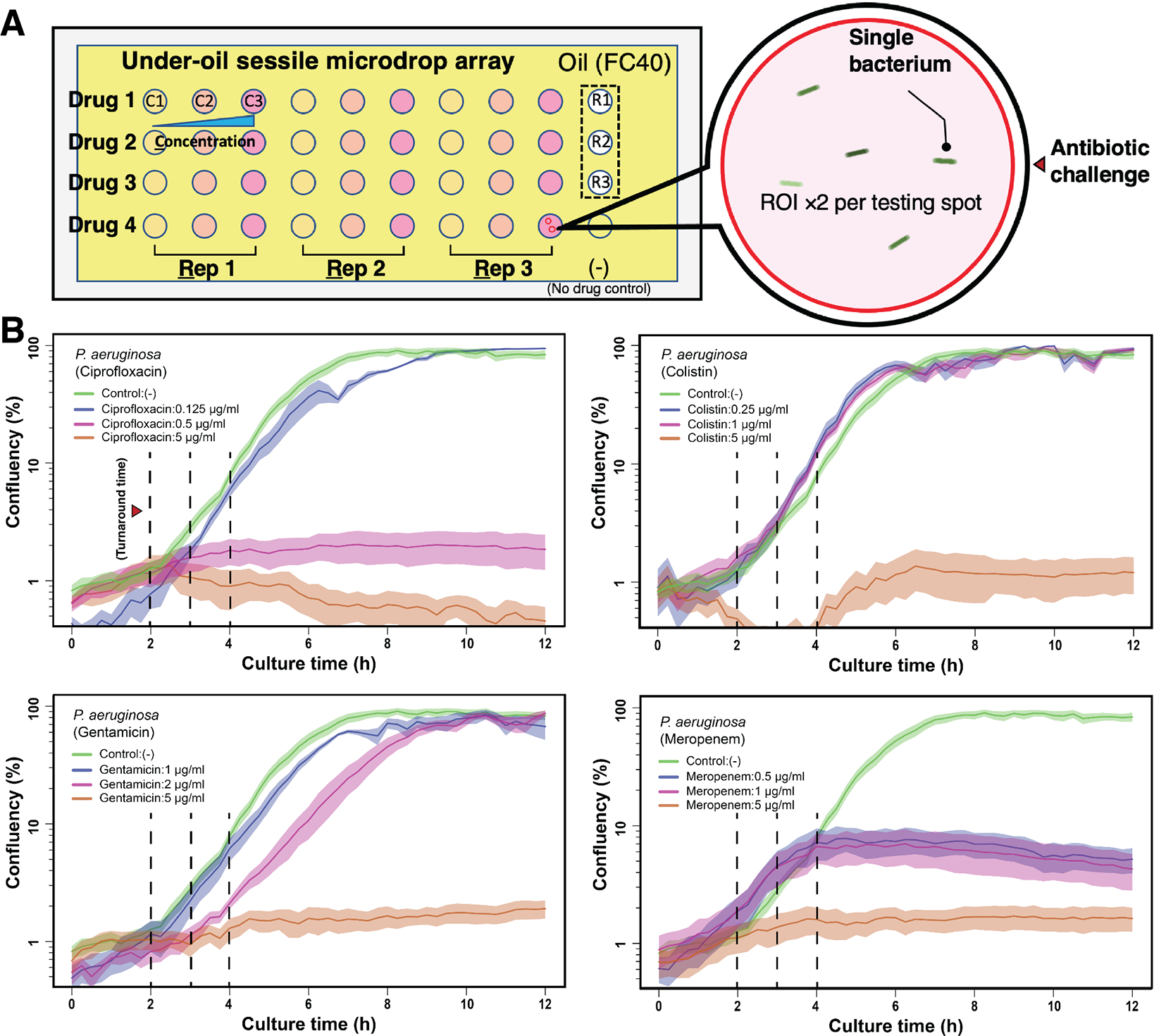
Results from UOMS-AST with P. aeruginosa (PA01). (A) A schematic shows the layout of UOMSAST on a chambered coverglass (Nunc Lab-Tek - borosilicate glass #1.0 - medical grade silicone adhesive, 6 cm × 2.4 cm, 0.13–0.17 mm in thickness) with 2 mm (in diameter) testing spots. Bacteria from 3 biological replicates (i.e., Rep 1 or R1, Rep 2 or R2, and Rep 3 or R3) were inoculated on the testing spots against 4 antibiotics (i.e., Drug 1, Drug 2, Drug 3, and Drug 4) at 3 different concentrations (i.e., C1, C2, and C3) and imaged under bright field [60× magnification, ROI ×2 (the red circles) per testing spot] in a time lapse. (B) The growth curves (i.e., confluency versus time, see Methods) of P. aeruginosa PA01 against four antibiotics. The antimicrobial activity and the difference across the conditions can be detected with a turnaround time of 2 to 4 h (vertical dashed lines). The solid line shows the mean of the biological replicates (×3) with the ROIs (×2) and the standard deviation (s.d.) is represented by the envelope on the plots. The microscopic images of bacteria are shown in Fig. S2.
To compare the results from UOMS-AST against the Clinical and Laboratory Standards Institute (CLSI) approved standard, we performed broth microdilution susceptibility testing with strain PA01 against the same four antibiotics. PA01 MICs were ciprofloxacin 0.25–0.5 μg/mL, gentamicin 2 μg/mL, colistin 1–2 μg/mL, and meropenem 0.5 μg/mL. In comparison with the confluency results of the UOMS-AST (Fig. 2B, Fig. S2), the antibiotic MIC susceptibility, determined at 16–20 h post incubation, aligned with the UOMS-AST data within the 2–4 h window (Fig. 2B, vertical dashed lines). To further compare UOMSAST to the killing activity of antibiotics, we performed time-kill curve analysis as a standard assay for determining antimicrobial dynamic killing over time. Using the same organism and antibiotic sets, the time-kill assay (Fig. 3) replicates the UOMS-AST results for ciprofloxacin activity across the concentrations tested (0.125–5 μg/mL). Similar comparability was noted with colistin. There were some notable differences with gentamicin and meropenem. Based on the gentamicin MIC of 2 μg/mL, we observed unexpected growth with 2 μg/mL gentamicin but no growth at 5 ug/ml. Meropenem also differed in MIC versus the UOMS-AST results, displaying UOMS confluency at 2–4 h with meropenem at 1 μg/mL despite MIC at 0.5 μg/mL. We posit that beta-lactam (i.e., meropenem) heterogeneous susceptibility in PA01 may contribute to these differences. Indeed the CLSI provides an acceptable range of antibiotic susceptibility for many pathogens, including P. aeruginosa.37 Further, MICs may vary among different clinically-approved platforms.38 Therefore, the droplet-specific characteristics and behavior of the UOMS may explain these differences versus standard testing. This will continue to be evaluated as the system is optimized and additional strains are tested. These results indicate that UOMS-AST has potential utility for not only MIC phenotypic testing, but also for bactericidal concentration determination of antibiotics. Although some antibiotics tested had turnaround times of 2–4 hours, the UOMS-AST will be further evaluated to identify the optimal turnaround time window for detection.
Fig. 3.
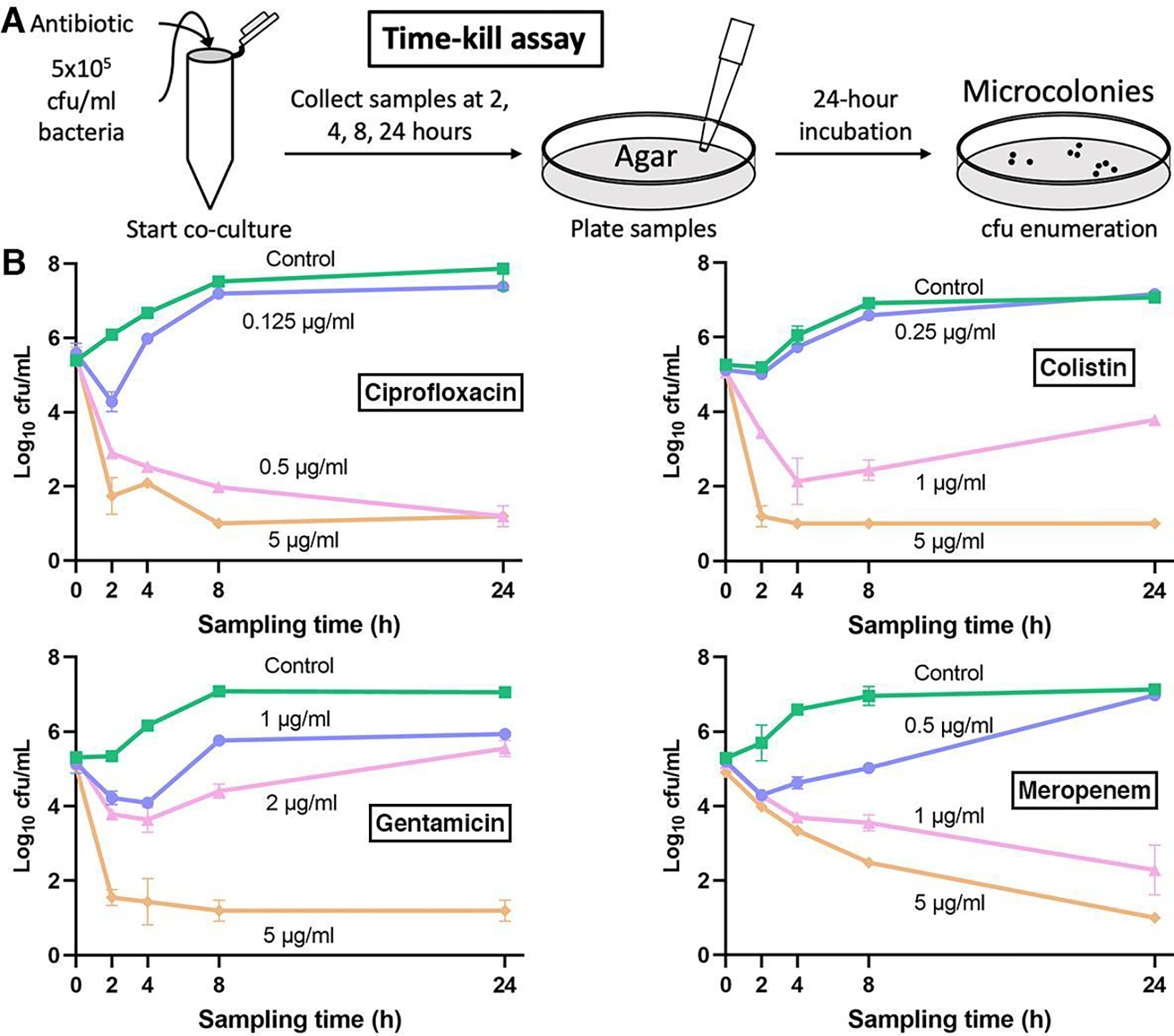
Standard time-killing curve results of Pseudomonas aeruginosa PA01. (A) A schematic showing the time-kill assay workflow. In this assay, the bacteria were collected from each broth culture at a series of time points (i.e., the sampling times) and then streaked/cultured (up to 24 h) on an agar plate to obtain cfu count. (B) The time-kill assay results of PA01 against the same antibiotics. Data were pooled and averaged with 3 replicates in each condition. Error bars, mean ± s.d.
Integration with cloud lab for real-time image analysis and report generation
Recently, various microfluidic platforms have been introduced to the field of phenotypic AST.20,39–42 Distinct from the bulk-scale (i.e., agar or microtiter plate-based) phenotypic AST, microfluidics-based phenotypic AST comes with the capability of handling and processing ultra-small volume (microliter to picoliter) of samples, performing high throughput antimicrobial screening, and single-cell level antimicrobial detection sensitivity. However, it must be noted that often the reported turnaround time in microfluidics-based phenotypic AST does not account for the time required for data analysis and report generation following data collection. Especially considering the data size (hundreds of GB to TB per run) from single-cell high throughput screening, the interpretation and quantification of the AST results can take days or weeks if done manually. An automated analytic and reporting mechanism in microfluidics-based phenotypic AST that can be implemented with clinical laboratory standards is required to achieve clinically relevant, rapid sample-to-report turnaround times.
Here we demonstrate the combined use of a cloud lab-based, live cell imaging system in UOMS-AST (Fig. 4A, Fig. S3, Methods). The system is equipped with a 10× magnification, high-resolution camera mounted on a motorized xy actuator and can be used in a standard cell culture incubator with controlled atmosphere (e.g., O2, CO2) and relative humidity. The UOMS-AST device in the workspace (i.e., a scanning area for the size of a standard microtiter plate) was scanned through a time course (Fig. 4B). A router transferred the images to cloud space during the scan. The image information in the defined region of interest (ROI) was analyzed with a selected algorithm (Fig. S3), e.g., confluency in this work, and the report was generated in real time. Lab and healthcare personnel would have access to the recorded images and plots via the client installed on a personal computer or mobile device (e.g., smart phone).
Fig. 4.
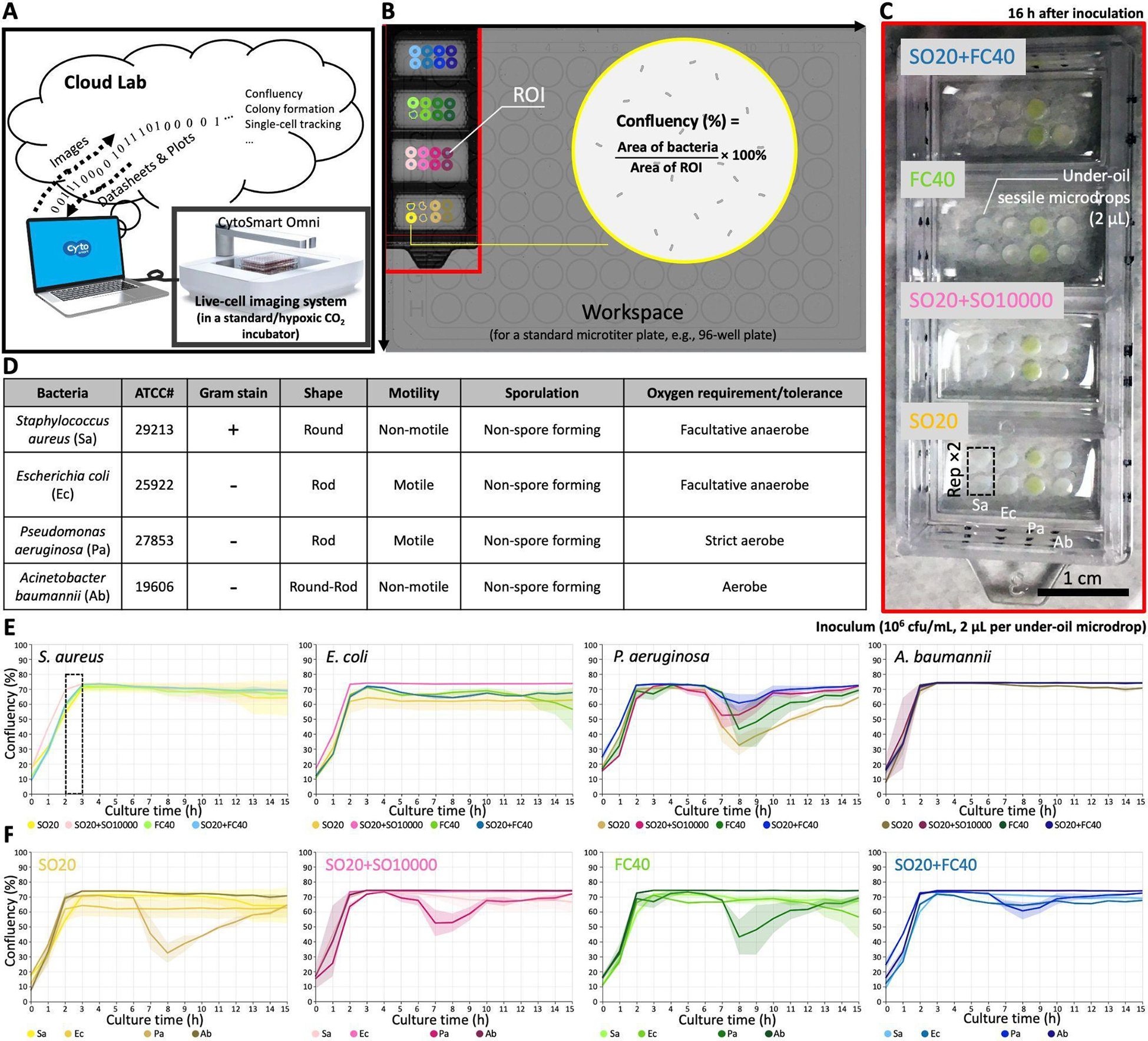
UOMS-AST integrated with cloud lab for real-time image analysis and report generation. (A) A schematic shows the data flow of cloud lab. (B) The workspace (recorded by the 10× magnification camera) on CytoSmart Omni (Fig. S3) can accommodate up to 6 pieces of (6 cm × 2.4 cm) chambered coverglass. Confluency of bacterial cells is automatically analyzed with the defined ROIs. (C) A camera picture of a 4-well chambered coverglass device (Nunc Lab-Tek II - borosilicate glass #1.5 - biocompatible acrylic adhesive, 6 cm × 2.4 cm, 0.16–0.19 mm in bottom thickness) with bacteria and different oil overlays. Each condition (i.e., per bacteria species per oil condition) has two replicates (i.e., Rep ×2). (D) A table shows the information of the four tested bacterial species. (E) The growth curves (i.e., confluency versus time) of each bacterial species against the four different oil conditions. (F) The growth curves of the bacteria species in each oil condition. The solid line shows the mean of the replicates and the s.d. is represented by the envelope on the plots.
We tested the capability of capturing the growth curve of four human pathogens - including Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter baumannii - that cause bacteremia and against four oil overlay conditions - including silicone oil (with the viscosity of 20 cSt, i.e., SO20), SO20+SO10000, FC40, and SO20+FC40 (Fig. 4C,D). We added silicone oil to this test because it provides three unique functions in UOMS: i) exclusive liquid repellency (ELR), where a liquid (e.g., culture media) is inherently and completely repelled by a solid surface (i.e., θ = 180°) when exposed to a secondary, immiscible liquid (e.g., oil) (see details in our previous publications23,25,29), ii) under-oil sweep distribution, where thousands of microdrops with a volume ranging from microliter to picoliter can be arrayed using automated or manual pipetting in a minute by dragging (or the so-called sweeping) a hanging drop of culture media (+ cells and/or drugs) across a patterned surface with double-ELR (i.e., under-oil water ELR + under-water oil ELR) (Fig. 5, and see details in our previous publications),25,29 and iii) autonomously regulated oxygen microenvironments (AROM), where cells spontaneously set up, regulate, and respond to the oxygen kinetics via a supply-demand balance as seen in vivo (see details in our previous publication).31 Studies have shown the broad impact of hypoxia on microbial infection and pathogenesis.43,44 The double-oil conditions (i.e., one oil plus another oil) showed the flexibility of adjusting the oil overlay by combining the properties of two oil types, e.g., different diffusion coefficients of vital gases (e.g., O2 and/or CO2),31 or under-oil media evaporation/loss rate.23 Specifically, SO20 allows smooth and robust under-oil sweep distribution due to its low viscosity and SO10000 can significantly reduce under-oil media evaporation/loss rate due to its ultra-high viscosity.45 In addition, no visible oil-oil interface is generated when two silicone oils are used together in a system. The combined use of SO20 and FC40 allows both reliable under-oil sweep distribution and minimized under-oil media evaporation/loss rate due the ultra-low diffusivity and solubility of water molecules in fluorinated oil compared to silicone oil.46
Fig. 5.
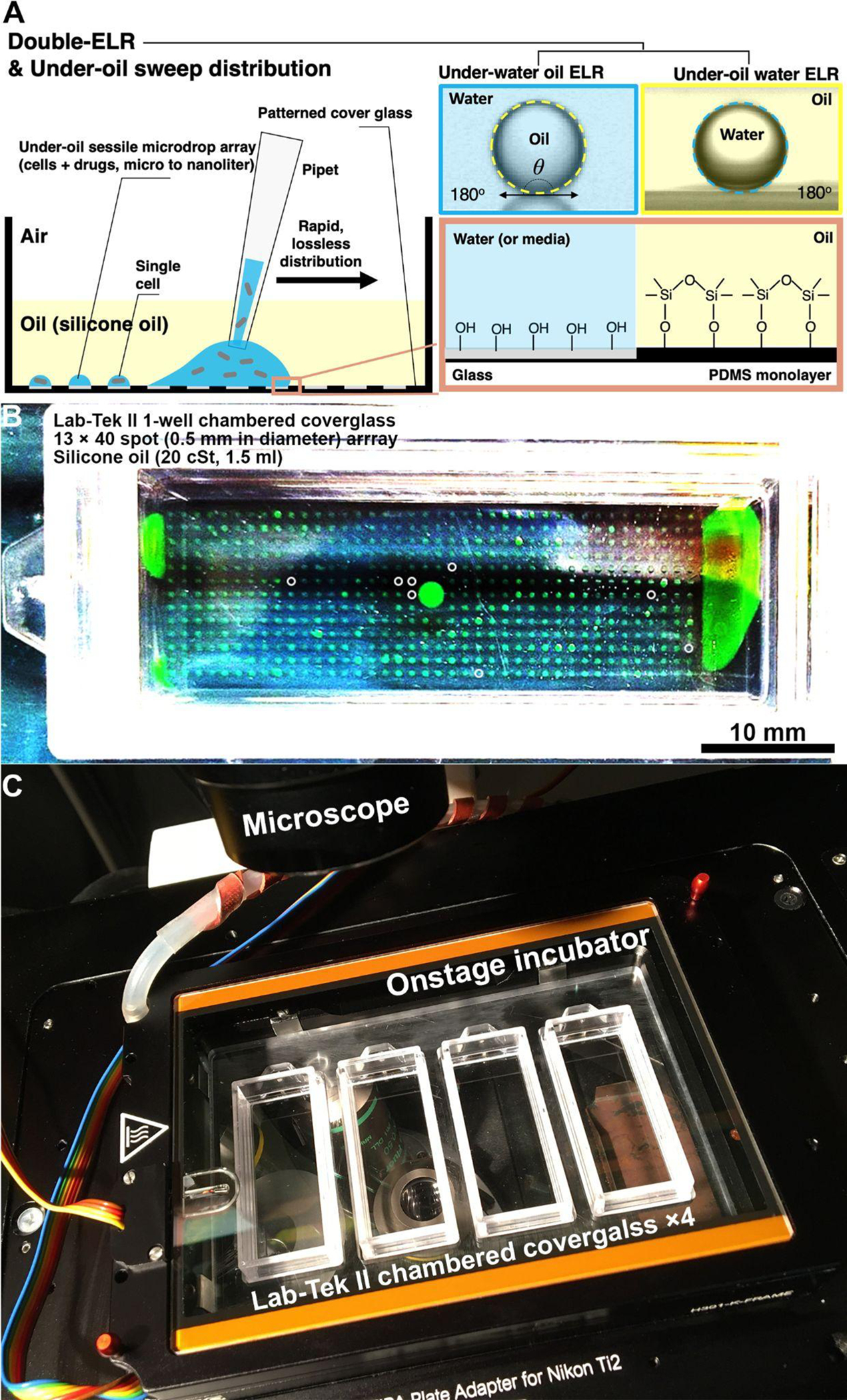
High-throughput UOMS-AST enabled by double-ELR and under-oil sweep distribution. (A) A schematic shows the physics of double-ELR and the operation of under-oil sweep distribution. (B) A camera picture shows an array of 520 under-oil spots (0.5 mm in diameter, 4 nL per spot) prepared by under-oil sweep distribution under silicone oil (20 cSt). The device is a 1-well chambered coverglass device (Nunc Lab-Tek II - borosilicate glass #1.5 - biocompatible acrylic adhesive, 6 cm × 2.4 cm, 0.16–0.19 mm in bottom thickness) compatible with silicone oil. The liquid is DMEM culture media + 10% fetal bovine serum (FBS) with 1 mM FITC for visualization. Dry spots were marked out with white circles. Different species and/or antibiotic type/concentration can be applied to the local domains on a device by sweeping a sample on a designated area. (C) A camera picture shows a home-made sample holder that houses 4 pieces of chambered coverglass in an onstage incubator on a scope.
As shown in the cloud lab results (Fig. 4E), the growth of the tested bacterial species was successfully recorded and quantified with real-time image analysis and report generation. All the four bacterial species reached confluence under oil within 3 h with a standardized inoculum (1 × 106 cfu/mL). The four oil overlay conditions showed high consistency on the growth curves against each bacterial species (Fig. 4F).
Conclusions
Patients with serious bacterial infections (e.g., bacteremia) are at significant risk of complications, including antibiotic treatment failure.47 Many studies have demonstrated that the most critical intervention to improve outcomes in patients with a severe infectious disease is early pathogen identification along with initiation of timely and effective antimicrobial therapy.48,49 This “window of opportunity” is optimized in the first 24 h, e.g., in septic patients, and within 48–72 h in non-septic (but still severe) bacteremia patients. Importantly, studies have found that patients who are switched to appropriate therapy after receiving inappropriate initial therapy for bacteremia and sepsis are still at higher risk of poor outcomes.49,50 Thus, rapid and accurate diagnosis is critical for all patients. Additionally, improved AST is a public health imperative to reduce unnecessary antibiotic use and hinder the risk of emerging antibiotic resistance.
Phenotypic AST provides functional readouts of antimicrobial activity and thus direct clinical guidance. Efforts have been seen in the development of next-generation phenotypic AST, striving to meet the following criteria: i) direct AST using the original clinical isolates (e.g., from blood, sputum, urine, abscess) to eliminate expansion culture that is time-consuming and introduces artificial passaging,9,51,52 ii) rapid AST, i.e., fast sample-to-answer/report turnaround time, iii) comprehensive test coverage including anaerobes,53,54 multispecies communities,55,56 and novel phenotypes such as heteroresistance (i.e., resistant mutants within the wild type population),57 and iv) lower adoption/implementation barriers (e.g., alignment with clinical laboratory standards, small footprint, minimal personnel training, operation, and maintenance).58
Microfluidics-based phenotypic AST has shown advantages over the traditional, bulk-scale methods regarding detection sensitivity, speed, and throughput. Compared to the reported microfluidic platforms in this field, UOMS is naturally aligned with the standard tools (e.g., pipette, microtiter plate/chambered coverglass, inverted microscope) and lab automation (e.g., robotic liquid handler, 2D/3D cell printer) in biology and laboratory medicine22–25,27,28 where open systems are traditionally adopted. Due to the oil protection, small-volume (microliter to picoliter)25 culture niches and/or testing units can be readily implemented and manipulated with minimized evaporation and sample contamination. The small scale allows facile (i.e., only running xy scan without z stack), label-free (i.e., bright-field), live-cell imaging with single-cell resolution. Label-free, single-cell detection is critical in next-generation phenotypic AST because it minimizes the risk of introducing random artifacts/pre-selection from fluorescent transfection of the isolated pathogens and maximizes the detection sensitivity and thus time efficiency. Importantly, the streamlined operation and small footprint of UOMS-AST make it suitable for translational applications in locations with limited space and access to infrastructures, e.g., mobile clinics and point-of-care settings.
Specifically, we highlight several unique features of UOMS-AST for its potential of commercialization. i) Low fabrication/material cost. The fabrication of UOMS devices is building upon room temperature (RT) chemical vapor deposition (CVD)23 and oxygen plasma surface patterning25,29 (Methods). The RT-CVD consumes minimal amounts of the surface-modifying reagents and energy. Both the surface modification and patterning are readily upscalable for mass production. Material-wise and compared to closed-chamber/-channel microfluidic devices, UOMS completely removes the top and sides (e.g., plastic or elastomer) that are required in closed channel designs. Moreover, the PDMS stamps for oxygen plasma surface patterning25,29 are reusable and long lasting. The PDMS stamps in this work have been used for 2 to 4 years without showing any significant decay. ii) Easy storage and distribution. The UOMS-AST devices with a designated pattern can be prepared in a specialized laboratory and sent to the users via a regular supply chain. UOMS-AST devices can be stored in a sealed plastic foil package filled with a small volume of deionized (DI) water that covers the patterned surface or under vacuum. Based on the tests we have run in our lab, the shelf life with the DI water or vacuum storage method can be several months or longer. From the DI water storage, the device is dried in nitrogen gas and ready for use. From the vacuum storage, the device can be used directly out of the package. iii) Low operating/environmental cost. Most microfluidic devices are designed or limited for one-time use. By contrast, the oil used in UOMS can be recycled, purified (e.g., by filtration), and reused. This will save the budget for the end users and in the meanwhile reduce the environmental burden of medical waste.
UOMS-AST, further combined with the three unique functions - ELR,23,25,29 under-oil sweep distribution,25,29 and AROM31 as described above in Results, provides a “less-is-more” strategy that leads to a next-generation phenotypic AST. The low adoption/implementation barriers give easy access to a significantly broadened group of end users, especially if commercialized. Importantly, the natural compatibility with clinical laboratory standards reserves the maximum lab resource efficiency and flexibility for different screening needs and tasks. Further, integrated with cloud lab techniques, automated, rapid, high-throughput (Fig. 5) antimicrobial detection and screening can be readily achieved. To better fit the use in hospitals and clinics with maximal efficiency/consistency and minimal human operation, we have been working on the development of an automated UOMS-AST system, with which sample preparation and loading, imaging, sample collection/disposal, device/oil recycling, image analysis and report generation will be all done automatically in a compact system.
Methods
Preparation of the UOMS devices
Detailed protocol was described in our previous publications.25,29 Briefly, the process includes i) surface modification, ii) surface patterning, and iii) under-oil sample loading. Surface modification introduces a monolayer of covalently bonded PDMS-silane (1,3-dichlorotetramethylsiloxane, Gelest, SID3372.0) molecules by RT-CVD (Fig. S1). The following surface patterning step is to transfer a designated pattern from a PDMS stamp (or mask) to the PDMS-silane grafted surface by selective oxygen plasma treatment. At last, oil is added to the device and cellular/molecular samples can be loaded under-oil by regular pipetting (the approach used in this work) or under-oil sweep distribution. The two types of chambered coverglass, Nunc Lab-Tek - borosilicate glass #1.0 - medical grade silicone adhesive, 6 cm × 2.4 cm, 0.13–0.17 mm in thickness and Nunc Lab-Tek II - borosilicate glass #1.5 - biocompatible acrylic adhesive, 6 cm × 2.4 cm, 0.16–0.19 mm in bottom thickness, were purchased from Thermo Fisher Scientific.
Bacteria growth conditions and antibiotic susceptibility testing
Bacterial strains stored at −80°C were plated on Mueller-Hinton agar (MHA, BD Difco, BD, Franklin Lakes, NJ USA) and incubated at 37°C overnight prior to use. Time-kill assays were completed for Pseudomonas aeruginosa PA01 and the antibiotics colistin, meropenem, ciprofloxacin, and gentamicin (Sigma-Aldrich, St. Louis, MO, USA). The inoculum was prepared from a 0.5 McFarland standard and diluted 1:100 to obtain a 106 cfu/mL starting bacterial concentration. The bacterial solution was aliquoted into microcentrifuge tubes and antibiotics were added at various concentrations. After 2, 4, 8, and 24 hours in a 37°C shaking incubator, a sample from each condition was collected, serially diluted, and spot-plated on MHA for cfu enumeration per mL. MICs were determined by broth microdilution, as described by CLSI.59
Imaging and time lapse
We used Nikon Eclipse Ti (40× objective with 1.5× tube lens, i.e., 60× magnification) to acquire the bright-field images, and run the time lapse (15 min interval for 12 h) (Fig. S2). The UOMS-AST device was kept at 37 °C, 21% O2, 5% CO2, 95% relative humidity (RH) via an on-stage incubator (Bold Line, Okolab) during imaging.
Batch-process image analysis and data visualization
We developed a custom image analysis workflow for objectively batch processing the time-lapse videos in JEX,60,61 an open-source image analysis software that uses well-established libraries from ImageJ. Briefly, raw masks identifying bacterial cells were generated in two ways to robustly accommodate variation in bright-field imaging. In the first approach, bright-field images were gamma adjusted (ɣ = 0.7), inverted, background subtracted, Gaussian-mean filtered, and thresholded. In the second, images were background subtracted, inverted, Gaussian-mean filtered, and thresholded. The two masks were then combined using an OR operation to form the final mask. The surface area contributed by the bacterial objects were then extracted in JEX and then exported to.csv for analysis in R/RStudio. For data visualization, plots with confluency (%) over time were smoothed using the ‘smooth.spline’ function of the R ‘stats’ package with a smoothing parameter spar = 0.4.
Cloud lab
The live-cell imaging system (CytoSmart Omni) adopted in the work is designed for use in a standard cell culture incubator with controlled temperature, atmosphere, and humidity (Fig. S3). The system comes with 10× high-resolution camera mounted on a motorized xy actuator. The workspace is designed for a standard microtiter plate. It can take up to 6 pieces of the chambered coverglass used in this work at a time. The camera scans the workspace every hour. ROIs on the device can be set after the first scan or the whole-time lapse scan. Image information from the ROIs will be analyzed by an algorithm (e.g., confluency) selected from the cloud lab. The user can have real-time access to the quantified results and plots with the client installed in a computer or mobile device.
Cell line authentication
The bacterial cultures used in this study were purchased from ATCC. Pseudomonas aeruginosa Pa01 is a standard model organism for laboratory analysis, and strains S. aureus ATCC 29213, E. coli ATCC 25922, P. aeruginosa ATCC 27853, and A. baumannii 19606 are standard strains for CLSI antimicrobial susceptibility testing. For this study, these strains were authenticated for genus and species by selecting a single colony from overnight growth on solid agar and analyzing by MALDI-TOF MS according to clinical protocols and manufacturer’s instructions for identification (MALDI Biotyper, Bruker Corp., Billerica, MA). All five organisms were confirmed as correct.
Statistical analysis
Raw data were directly used in statistical analysis with no data excluded. Data were present as mean ± s.d.. The replicate number was specified in the figure legends.
Supplementary Material
Acknowledgements
This work was supported by NSF EFRI-1136903-EFRI-MKS, NIH R01 CA247479, NIH R01 AI154940, NIH R01 EB010039, NIH R01 CA185251, NIH R01 CA186134, NIH R01 CA181648, NIH R01AI132627, NIH P30CA014520, EPA H-MAP 83573701, and American Cancer Society IRG-15-213-51. We thank the following individuals for their assistance with this work: Dr. Taylor Fleet for sample collection, Dr. Brittney Hynes for bacterial cell line authentication, and Dr. Duane S. Juang for introducing the live-cell imaging system.
Footnotes
Conflicts of interest
D.J.B. holds equity in BellBrook Labs LLC, Tasso Inc. Stacks to the Future LLC, Lynx Biosciences LLC, Onexio Biosystems LLC, Turba LLC, Flambeau Diagnostics LLC, and Salus Discovery LLC. The remaining authors declare no competing financial interests.
Code availability
The JEX workflows for batch-process image analysis and the R/R studio codes for data visualization are available upon request.
Data availability
All study data are included in the article and/or supporting information. The data that support the findings of this study are available from the corresponding authors upon reasonable request.
Notes and references
- 1.Ventola CL, P T, 2015, 40, 277–283. [PMC free article] [PubMed] [Google Scholar]
- 2.Paul M, Shani V, Muchtar E, Kariv G, Robenshtok E and Leibovici L, Antimicrob. Agents Chemother, 2010, 54, 4851–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonine NG, Berger A, Altincatal A, Wang R, Bhagnani T, Gillard P and Lodise T, Am. J. Med. Sci, 2019, 357, 103–110. [DOI] [PubMed] [Google Scholar]
- 4.Vasala A, Hytönen VP and Laitinen OH, Front. Cell. Infect. Microbiol, 2020, 10, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Athamanolap P, Hsieh K, Chen L, Yang S and Wang T-H, Anal. Chem, 2017, 89, 11529–11536. [DOI] [PubMed] [Google Scholar]
- 6.Jr WMD, Michael Dunne W Jr, Jaillard M, Rochas O and Van Belkum A, Expert Rev. Mol. Diagn, 2017, 17, 257–269. [DOI] [PubMed] [Google Scholar]
- 7.Hrabák J, Chudácková E and Walková R, Clin. Microbiol. Rev, 2013, 26, 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang K, Li H-Z, Zhu X, Su J-Q, Ren B, Zhu Y-G and Cui L, Anal. Chem, 2019, 91, 6296–6303. [DOI] [PubMed] [Google Scholar]
- 9.van Belkum A, Burnham C-AD, Rossen JWA, Mallard F, Rochas O and Dunne WM Jr, Nat. Rev. Microbiol, 2020, 18, 299–311. [DOI] [PubMed] [Google Scholar]
- 10.Zhou M, Wang Y, Liu C, Kudinha T, Liu X, Luo Y, Yang Q, Sun H, Hu J and Xu Y-C, Infect. Drug Resist, 2018, 11, 1347–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sader HS, Fritsche TR and Jones RN, J. Clin. Microbiol, 2006, 44, 1101–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Humphries RM and Hindler JA, Clin. Infect. Dis, 2016, 63, 83–88. [DOI] [PubMed] [Google Scholar]
- 13.Boedicker JQ, Li L, Kline TR and Ismagilov RF, Lab Chip, 2008, 8, 1265–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Churski K, Kaminski TS, Jakiela S, Kamysz W, Baranska-Rybak W, Weibel DB and Garstecki P, Lab Chip, 2012, 12, 1629–1637. [DOI] [PubMed] [Google Scholar]
- 15.Jiang L, Boitard L, Broyer P, Chareire A-C, Bourne-Branchu P, Mahé P, Tournoud M, Franceschi C, Zambardi G, Baudry J and Bibette J, Eur. J. Clin. Microbiol. Infect. Dis., 2016, 35, 415–422. [DOI] [PubMed] [Google Scholar]
- 16.Postek W, Pacocha N and Garstecki P, Lab Chip, 2022, 22, 3637–3662. [DOI] [PubMed] [Google Scholar]
- 17.Cao J, Chande C and Köhler JM, Lab Chip, 2022, 22, 2600–2623. [DOI] [PubMed] [Google Scholar]
- 18.Idelevich EA, Sparbier K, Kostrzewa M and Becker K, Clin. Microbiol. Infect, 2018, 24, 738–743. [DOI] [PubMed] [Google Scholar]
- 19.Idelevich EA, Nix ID, Busch JA, Sparbier K, Drews O, Kostrzewa M and Becker K, Diagnostics (Basel), , DOI: 10.3390/diagnostics11101803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kandavalli V, Karempudi P, Larsson J and Elf J, Nat. Commun, 2022, 13, 6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Boban M and Tuteja A, Lab Chip, 2017, 17, 1436–1441. [DOI] [PubMed] [Google Scholar]
- 22.Walsh EJ, Feuerborn A, Wheeler JHR, Tan AN, Durham WM, Foster KR and Cook PR, Nat. Commun, 2017, 8, 816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C, Yu J, Schehr J, Berry SM, Leal TA, Lang JM and Beebe DJ, ACS Appl. Mater. Interfaces, 2018, 10, 17065–17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soitu C, Feuerborn A, Tan AN, Walker H, Walsh PA, Castrejón-Pita AA, Cook PR and Walsh EJ, Proc. Natl. Acad. Sci. U. S. A, 2018, 115, E5926–E5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C, Yu J, Paine P, Juang DS, Berry SM and Beebe DJ, Lab Chip, 2018, 18, 2710–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng W, Chai Y, Forth J, Ashby PD, Russell TP and Helms BA, Nat. Commun, 2019, 10, 1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soitu C, Feuerborn A, Deroy C, Castrejón-Pita AA, Cook PR and Walsh EJ, Sci. Adv, 2019, 5, eaav8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li C, Niles DJ, Juang DS, Lang JM and Beebe DJ, SLAS Technol, 2019, 24, 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li C, Hite Z, Warrick JW, Li J, Geller SH, Trantow VG, McClean MN and Beebe DJ, Sci. Adv, 2020, 6, eaay9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Hurley A, Hu W, Warrick JW, Lozano GL, Ayuso JM, Pan W, Handelsman J and Beebe DJ, Nat. Commun, 2021, 12, 5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, Humayun M, Walker GM, Park KY, Connors B, Feng J, Pellitteri Hahn MC, Scarlett CO, Li J, Feng Y, Clark RL, Hefti H, Schrope J, Venturelli OS and Beebe DJ, Adv. Sci, 2022, 9, e2104510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juang DS, Berry SM, Li C, Lang JM and Beebe DJ, Anal. Chem, 2019, 91, 11848–11855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juang DS, Juang TD, Dudley DM, Newman CM, Accola MA, Rehrauer WM, Friedrich TC, O’Connor DH and Beebe DJ, Nat. Commun, 2021, 12, 4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juang DS, Lang JM and Beebe DJ, Lab Chip, 2022, 22, 286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Convery N and Gadegaard N, Micro Nano Eng, 2019, 2, 76–91. [Google Scholar]
- 36.Ortseifen V, Viefhues M, Wobbe L and Grünberger A, Front. Bioeng. Biotechnol, 2020, 8, 589074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin X, Zhou C, Zerr DM, Adler A, Addetia A, Yuan S and Greninger AL, mSphere, DOI: 10.1128/mSphere.00615-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sapino B, Mazzuccato S, Solinas M, Gion M and Grandesso S, New Microbiol, 2012, 35, 491–494. [PubMed] [Google Scholar]
- 39.Choi J, Yoo J, Lee M, Kim E-G, Lee JS, Lee S, Joo S, Song SH, Kim E-C, Lee JC, Kim HC, Jung Y-G and Kwon S, Sci. Transl. Med, 2014, 6, 267ra174. [DOI] [PubMed] [Google Scholar]
- 40.Baltekin Ö, Boucharin A, Tano E, Andersson DI and Elf J, Proc. Natl. Acad. Sci. U. S. A, 2017, 114, 9170–9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jalali F, Ellett F and Irimia D, Technology, 2017, 5, 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang K, Qin S, Wu S, Liang Y and Li J, Chem. Sci, 2020, 11, 6352–6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaffer K and Taylor CT, FEBS J, 2015, 282, 2260–2266. [DOI] [PubMed] [Google Scholar]
- 44.Chung H and Lee Y-H, Front. Microbiol, 2020, 11, 1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hofmeister AM, Measurements, Mechanisms, and Models of Heat Transport, 2019, 181–199. [Google Scholar]
- 46.Riess JG, Artif. Cells Blood Substit. Immobil. Biotechnol, 2005, 33, 47–63. [DOI] [PubMed] [Google Scholar]
- 47.Despotovic A, Milosevic B, Milosevic I, Mitrovic N, Cirkovic A, Jovanovic S and Stevanovic G, Am. J. Infect. Control, 2020, 48, 1211–1215. [DOI] [PubMed] [Google Scholar]
- 48.Kumar A, Ellis P, Arabi Y, Roberts D, Light B, Parrillo JE, Dodek P, Wood G, Kumar A, Simon D, Peters C, Ahsan M, Chateau D and Cooperative Antimicrobial Therapy of Septic Shock Database Research Group, Chest, 2009, 136, 1237–1248. [DOI] [PubMed] [Google Scholar]
- 49.Bassetti M, Rello J, Blasi F, Goossens H, Sotgiu G, Tavoschi L, Zasowski EJ, Arber MR, McCool R, Patterson JV, Longshaw CM, Lopes S, Manissero D, Nguyen ST, Tone K and Aliberti S, Int. J. Antimicrob. Agents, 2020, 56, 106184. [DOI] [PubMed] [Google Scholar]
- 50.Zilberberg MD, Shorr AF, Micek ST, Mody SH and Kollef MH, Chest, 2008, 134, 963–968. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Q, Lambert G, Liao D, Kim H, Robin K, Tung C-K, Pourmand N and Austin RH, Science, 2011, 333, 1764–1767. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Q, Robin K, Liao D, Lambert G and Austin RH, Mol. Pharm, 2011, 8, 2063–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veloo ACM, Baas WH, Haan FJ, Coco J and Rossen JW, Clin. Microbiol. Infect, 2019, 25, 1156.e9–1156.e13. [DOI] [PubMed] [Google Scholar]
- 54.Marteyn B, Scorza FB, Sansonetti PJ and Tang C, Cell. Microbiol, 2011, 13, 171–176. [DOI] [PubMed] [Google Scholar]
- 55.Stubbendieck RM, Vargas-Bautista C and Straight PD, Front. Microbiol, 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Connell JL, Ritschdorff ET, Whiteley M and Shear JB, Proc. Natl. Acad. Sci. U. S. A, 2013, 110, 18380–18385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nicoloff H, Hjort K, Levin BR and Andersson DI, Nat. Microbiol, 2019, 4, 504–514. [DOI] [PubMed] [Google Scholar]
- 58.Jorgensen JH and Ferraro MJ, Clin. Infect. Dis, 2009, 49, 1749–1755. [DOI] [PubMed] [Google Scholar]
- 59.Ferraro MJ, Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically: Approved Standard, 2003. [Google Scholar]
- 60.Warrick JW, Timm A, Swick A and Yin J, PLoS One, 2016, 11, e0145081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warrick JW, Young EWK, Schmuck EG, Saupe KW and Beebe DJ, Integr. Biol, 2013, 5, 720–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. The data that support the findings of this study are available from the corresponding authors upon reasonable request.


