ABSTRACT
Human noroviruses (HuNoVs) are the leading cause of acute gastroenteritis. In immunocompetent hosts, symptoms usually resolve within 3 days; however, in immunocompromised persons, HuNoV infection can become persistent, debilitating, and sometimes life-threatening. There are no licensed therapeutics for HuNoV due to a near half-century delay in its cultivation. Treatment for chronic HuNoV infection in immunosuppressed patients anecdotally includes nitazoxanide, a broad-spectrum antimicrobial licensed for treatment of parasite-induced gastroenteritis. Despite its off-label use for chronic HuNoV infection, nitazoxanide has not been clearly demonstrated to be an effective treatment. In this study, we standardized a pipeline for antiviral testing using multiple human small intestinal enteroid lines representing different intestinal segments and evaluated whether nitazoxanide inhibits replication of five HuNoV strains in vitro. Nitazoxanide did not exhibit high selective antiviral activity against any HuNoV strain tested, indicating it is not an effective antiviral for HuNoV infection. Human intestinal enteroids are further demonstrated as a model to serve as a preclinical platform to test antivirals against HuNoVs to treat gastrointestinal disease.
Abstr
KEYWORDS: human norovirus, enteroids, intestinal organoids, nitazoxanide, antivirals, genotypes
INTRODUCTION
Human noroviruses (HuNoVs) pose a major public health burden worldwide. These positive-sense, single-stranded RNA viruses are the leading worldwide cause of acute and sporadic cases of gastroenteritis, causing an estimated $4 billion in direct health-care costs yearly (1 – 4). HuNoV-associated gastroenteritis is typically self-resolving within 3 days (5). However, patients who are immunocompromised can develop chronic HuNoV infection. In addition to continuous shedding of HuNoV, these patients can experience gastroenteritis symptoms that last for several months or years, resulting in severe morbidity and at times mortality (6).
HuNoVs are members of the diverse Norovirus genus, which includes at least 39 genotypes capable of infecting humans (7). Currently, no HuNoV vaccines have been approved (8, 9). Off-label therapies are used to treat chronic HuNoV infection based on anecdotal evidence and expert opinion, but many of these have proven inefficacious (6). Recently, two clinical trials to treat chronic HuNoV infection have been established, including a phase I trial for a biological treatment utilizing norovirus-specific T-cell therapy (NCT04691622) (10) and a phase II trial evaluating the broad-spectrum antimicrobial nitazoxanide (NTZ) (NCT03395405) (11).
NTZ is a Food and Drug Administration-approved drug for the treatment of diarrhea associated with Cryptosporidium parvum and Giardia lamblia. The mechanism by which NTZ inhibits these protozoa is by acting as a non-competitive inhibitor of pyruvate:ferredoxin/flavodoxin oxidoreductases that are involved in anaerobic metabolism (12, 13). NTZ has also been reported to have putative antiviral activity against influenza, rotavirus, Ebola, and several other viruses (14 – 17). However, the mechanism of NTZ against these viruses differs and is commonly believed to work through stimulation of the host’s innate immunity (16, 18, 19) or inhibition of viral protein maturation (20 – 23). NTZ was first evaluated for treating noroviral diarrhea in a small randomized double-blind clinical trial held in Egypt (24). The six HuNoV-infected but otherwise healthy patients who underwent NTZ treatment had symptom resolution significantly faster than those who received placebo (n = 7), although the median time to recovery in the placebo recipients was much longer than usual for previously healthy persons. Subsequent case reports and studies have demonstrated inconsistent efficacy of administering NTZ to resolve chronic norovirus-associated diarrhea (25 – 37). The reason for this inconsistency remains unclear. One possibility is variable efficacy of NTZ by virus strain, considering there are several differences in biological properties such as cell entry and sensitivity to host interferon pathways that are now known between some HuNoV genotypes (38 – 41). A large diversity of HuNoVs including a wide variety of genotypes have been reported in chronically infected patients in some studies (26, 42 – 46). However, the HuNoV genotype infecting chronically afflicted patients are reported infrequently in most clinical studies, possibly because most clinical assays lack the capability to fully classify viruses and distinguish genotypes beyond GI and GII genogroups (47). Another possibility is potential differences in disease severity caused by GII.4 HuNoV compared to other genotypes, which has been observed in pediatric populations (48).
The lack of an in vitro model for HuNoV replication for a near half-century since the virus’ discovery has been a major factor in the lack of antiviral development for these viruses. Surrogate models such as a genome-encoded plasmid replicon of the prototype HuNoV GI.1 in Huh7 liver cells were used to assess NTZ activity (19). NTZ and its active metabolite, tizoxanide, inhibited replication of the GI.1-based HuNoV replicon. In 2016, we established a HuNoV replication system using non-transformed human intestinal enteroids (HIEs) that support reproducible, robust and successful replication of at least 12 HuNoV genotypes (39, 49). Two studies tested the effect of NTZ on GII.4 HuNoV replication in HIEs and reported differing outcomes. In the first study, NTZ had no inhibitory activity against GII.4 Sydney [P31] virus in an adult jejunal HIE plated as monolayers. The HIE monolayer was intact at concentrations below 10 µM, indicating no major cytotoxicity unlike what was seen at 100-µM NTZ (35). In the second study, by contrast, NTZ and tizoxanide treatment of fetal ileum-derived three-dimensional (3D) HIEs reduced replication of three different GII.4 HuNoV isolates at a non-cytotoxic concentration (50). The basis for the conflicting results from these initial in vitro studies with NTZ is unclear.
We evaluated whether NTZ inhibits the replication of several HuNoV genotypes, using HIEs derived from different donors and different intestinal segments. Utilizing non-linear regression analyses to distinguish whether NTZ exhibits selective activity in vitro, we demonstrate that NTZ causes significant cytotoxicity to HIE monolayer cultures with no significant reduction in replication below cytotoxic levels among five different HuNoV strains tested, regardless of HIE small intestinal segment or donor derivation.
RESULTS
Establishment of a standardized antiviral pipeline for HuNoVs
We first determined the 50% tissue culture infectious dose (TCID50) for each virus genotype in the specific HIE lines used for antiviral testing. The TCID50 values vary for each virus and each HIE line (Table 1), suggesting the need for a standard dose of virus to effectively compare antiviral results between different genotypes. To standardize a testing pipeline for assessing antiviral activity across different samples and HIE lines, we used 100 TCID50 in antiviral assays (Fig. 1A). We also assessed cell viability in tandem with every antiviral assessment. The half maximal effective concentration and cytotoxic concentration (EC50 and CC50, respectively) were estimated for each antiviral to calculate the selective index (SI), a measure of the antiviral’s therapeutic window (51). We also estimated an effective concentration for 90% inhibition as this cutoff yielded less variability between results in HIEs for antibody neutralization (52).
TABLE 1.
List of HuNoV isolates used in this study and their infectivity to different HIE lines a
| Viral isolate | VP1 genotype | P-type | Viral titer (g.e./µL) | HIE line | g.e./TCID50 |
|---|---|---|---|---|---|
| BCM 723–100595 | GI.1 | GI.P1 | 1.3 × 105 | J4FUT2 | 7.3 × 103 |
| TCH 04–577 | GII.3 | GII.P21 | 2.7 × 106 | J4FUT2 | 6.8 × 102 |
| BCM 16–2 | GII.4 Sydney | GII.P31 | 6.3 × 105 | J2 | 5.0 × 103 |
| BCM 16–16 | GII.4 Sydney | GII.P16 | 4.3 × 106 | D2004 | 4.7 × 104 |
| BCM 16–16 | GII.4 Sydney | GII.P16 | 4.3 × 106 | J2 | 1.1 × 104 |
| BCM 16–16 | GII.4 Sydney | GII.P16 | 4.3 × 106 | J2004 | 8.5 × 103 |
| BCM 16–16 | GII.4 Sydney | GII.P16 | 4.3 × 106 | IL2004 | 5.3 × 103 |
| 1295–44 | GII.17 | GII.P13 | 9.3 × 106 | J4FUT2 | 7.5 × 103 |
Five different HuNoV isolates were used consisting of four different VP1 genotypes and five different polymerase types (P-types). The viral titer is reported as genome equivalents (g.e.). The TCID50 is the number of g.e. estimated to infect 50% of the HIE line in which it was tested.
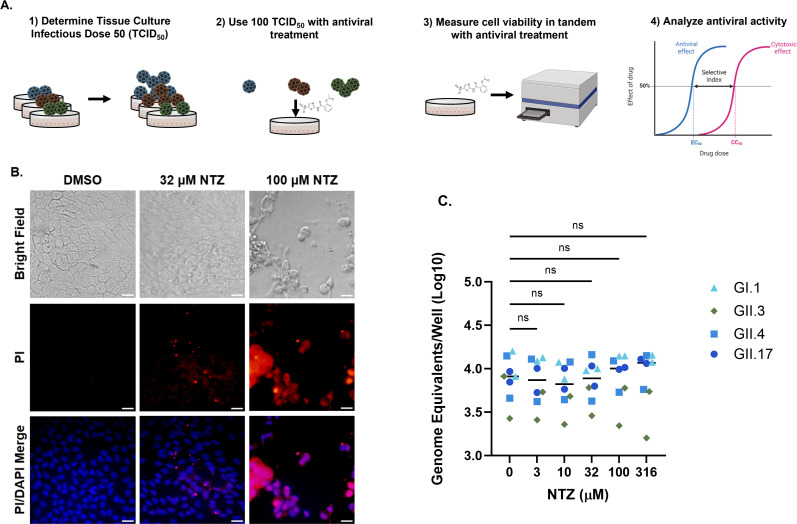
FIG 1.
Parameters measured to optimize assessment of antiviral treatment of HuNoV in HIEs. (A) Schematic representation of conditions used for standardizing HuNoV antiviral testing in HIEs. After determining the TCID50 for each genotype in each HIE line, 100 TCID50 of each virus was used to assess antiviral activity with different concentrations of compounds. Cytotoxicity was evaluated in tandem with each antiviral assay, and final antiviral activity was analyzed by determining the selective index. (B) Bright field and fluorescent microscopy images of vehicle (1% dimethyl sulfoxide (DMSO)) and 32- and 100-µM NTZ-treated J2 HIEs after 24 h. Cell nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI, blue) and dead cells were stained with propidium iodide (PI) (red). (C) Effect of NTZ on HuNoV adsorption to HIE monolayers. One hundred TCID50 of four HuNoV genotypes was added to either J2 (GII.4) or J4FUT2 (GI.1, GII.3, GII.17) HIEs for 1 or 2 h, and genome equivalents were quantitated. Horizontal bar = median. Images were taken at ×40 magnification. Scale bar = 200 µM. Data are compiled from n = 2 experiments. ns, not significant.
We then used this pipeline to test the effect of NTZ on HuNoV replication. In preliminary studies to determine dose range for infectivity studies, we performed cytotoxicity assessments using the CellTiter-Glo version 2.0 assay with different doses of NTZ. The highest concentrations in the original range of doses (3–316 µM) resulted in high cytotoxicity and detachment of the monolayer from the plate. While the monolayers were intact and had few dead or damaged cells as indicated by positive staining for propidium iodide (Fig. 1B) at 32-µM NTZ, treatment with 100-µM NTZ and above resulted in disassociation of the monolayer. There were few cells attached to the plate after staining and washing, indicating high cytotoxicity. Therefore, in our subsequent experiments with NTZ, we used drug concentrations below 100 µM. Standard replication and antiviral assays include HIE monolayers for binding (1–2 h) and replication (24 hpi). We assessed whether NTZ affected adsorption of four different HuNoV genotypes (GI.1, GII.3, GII.4, and GII.17) to HIEs. With increasing doses of NTZ, there was no significant difference in the number of viral genome equivalents detected after 1 or 2 h (GII.3, GII.4, and GI.1, GII.17, respectively), suggesting that NTZ does not affect binding of HuNoV to cells (Fig. 1C). Based on these data, we compared the effect of NTZ treatment on the number of genome equivalents detected at 24 hpi in all subsequent experiments.
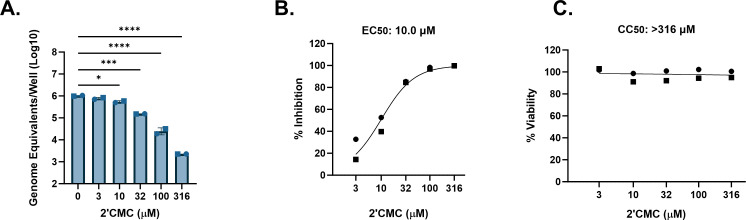
We then evaluated the capability of this model to measure antiviral activity. The degree of antiviral activity was evaluated by assessing the selective index (SI = CC50/EC50), with an SI of <2, 2 to <10, 10 to <50, and ≥50 being defined as having no, weak, moderate, and high antiviral activity, respectively (51). As a proof of principle, we initially evaluated a positive control compound, 2′-C-methylcytidine (2′CMC), previously reported to inhibit HuNoV replication in HIEs (50, 53). We tested the effect of 2′CMC on the replication of a GII.4 Sydney [P31] HuNoV strain in a jejunal J2 HIE line, given that GII.4 Sydney is the most widely circulating HuNoV strain (42, 54, 55) and has been shown to infect multiple HIEs. 2′CMC inhibited GII.4 Sydney replication in a dose-dependent manner with a range of 0.12–2.65 log10 reduction between 3 and 316 µM (Fig. 2A). This translated to a 23.5%–100.0% inhibition of replication (Fig. 2B), and an EC50 of 10.0 µM (Table 2). Using a luciferase-based commercial assay to determine viability, we found that 2′CMC was not cytotoxic to HIEs at any concentration tested (Fig. 2C). Therefore, an exact CC50 value could not be estimated and was presumed greater than the highest concentration tested (Table 2). Using these values, we determined the SI of 2′CMC to be >31.6, indicating selective activity for GII.4 Sydney [P31] HuNoV replication in HIEs and at least moderate antiviral activity. Due to variability in the percent inhibition at the lower concentrations around the EC50, we also estimated an effective concentration for 90% inhibition, as this cutoff has been shown to yield less variability between results in HIEs for antibody neutralization (52). The estimated EC90 of 2′CMC for GII.4 Sydney [P31] was 54.0 µM (Table 2).
FIG 2.
HIEs are useful models for antiviral studies with HuNoV. J2 HIEs were treated with five ascending doses of 2′CMC and the vehicle (2% H2O) for 24 h. (A) Replication after 24 h of GII.4 Sydney with 2′CMC or vehicle treatment (B) Percent inhibition by 24 h of GII.4 Sydney by 2′CMC. (C) Percent viability was assessed in the J2 HIE line after 24 h. Based on EC50 and CC50 (panels B and C, respectively), the SI was calculated as >31.6. *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001. Data are compiled from n = 2 experiments.
TABLE 2.
Summary of effectiveness of compounds against HuNoV in HIEs a
| Figure | Genotype | P-type | HIE line | Compound | EC90 (μM) | EC50 (μM) | CC50 (μM) | SI | Antiviral activity |
|---|---|---|---|---|---|---|---|---|---|
| 2 | GII.4 Sydney | GII.P31 | J2 | 2′CMC | 54.0 | 10.0 | >316 | >31.6 | At least moderate |
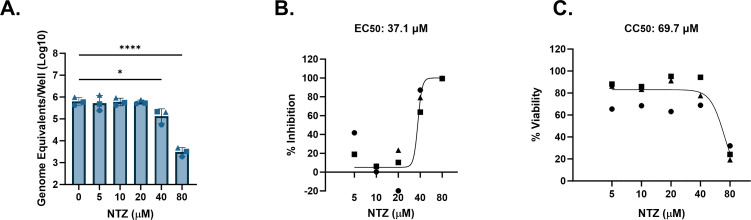
| 3 | GII.4 Sydney | GII.P31 | J2 | NTZ | 43.0 | 37.1 | 69.7 | 1.9 | Inactive |
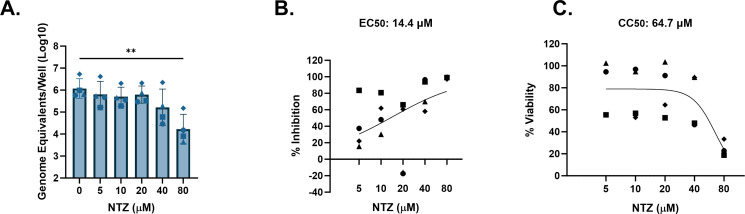
| 4 | GII.3 | GII.P21 | J4FUT2 | NTZ | >80 | 14.4 | 64.7 | 4.5 | Weak |
| 5 | GII.17 | GII.P13 | J4FUT2 | NTZ | >80 | 33.4 | 63.6 | 1.9 | Inactive |
| 6 | GI.1 | GI.P1 | J4FUT2 | NTZ | 76.3 | 27.4 | 73.3 | 2.7 | Weak |
| 7 | GII.4 Sydney | GII.P16 | J2 | NTZ | >80 | 12.9 | 51.2 | 4.0 | Weak |
| 7 | GII.4 Sydney | GII.P16 | D2004 | NTZ | >80 | 42.8 | 50.9 | 1.2 | Inactive |
| 7 | GII.4 Sydney | GII.P16 | J2004 | NTZ | >80 | 53.9 | 63.1 | 1.2 | Inactive |
| 7 | GII.4 Sydney | GII.P16 | IL2004 | NTZ | 46.0 | 40.0 | 57.0 | 1.4 | Inactive |
EC50 and EC90 were calculated on the percent inhibition data, and CC50 was calculated on the percent viability data presented in Fig. 2 to 7 using non-linear regression. Selective index (SI) was calculated using EC50 and CC50. Antiviral activity of each compound is denoted based on the SI: inactive = SI < 2; weak = 2 ≤ SI < 10; moderate = 10 ≤ SI < 50; high = SI ≥ 50.
Nitazoxanide does not significantly inhibit HuNoV replication in jejunal HIEs at non-cytotoxic concentrations
Using the same protocol as described in Fig. 1, we tested the antiviral activity of NTZ against several HuNoV strains in jejunal HIEs. A significant reduction in replication of GII.4 Sydney [P31] HuNoV in J2 HIEs was observed with 40- and 80-µM NTZ (0.67 and 2.31 log10, respectively; Fig. 3A), resulting in 76.7% and 99.5% inhibition (Fig. 3B). This yielded in an estimated EC50 of 37.1 µM (Table 2). However, there was also a reduction in viability at 80 µM (Fig. 3C), resulting in an estimated CC50 of 69.7 µM. The SI was 1.9 and is much lower than recommended SI ratios for antimicrobials being developed for clinical use (56, 57). In this case, NTZ would be considered to have no antiviral activity given that its SI is <2. The EC90 of NTZ against GII.4 Sydney [P31] was estimated to be 43.0 µM, similar to the CC50 of this compound.
FIG 3.
NTZ inhibition of GII.4-Sydney HuNoV replication in jejunal HIEs parallels its cytotoxicity. J2 HIEs were treated with five ascending doses of NTZ and the vehicle (1% DMSO) for 24 h. (A) Replication of GII.4 Sydney with NTZ treatment. (B) Percent inhibition of GII.4 Sydney by NTZ. (C) Percent viability was assessed in the J2 HIE line after 24 h. Based on EC50 and CC50 (panels B and C, respectively), the SI was calculated as 1.9. *P ≤ 0.05, ****P ≤ 0.0001. Data are compiled from n = 3 experiments.
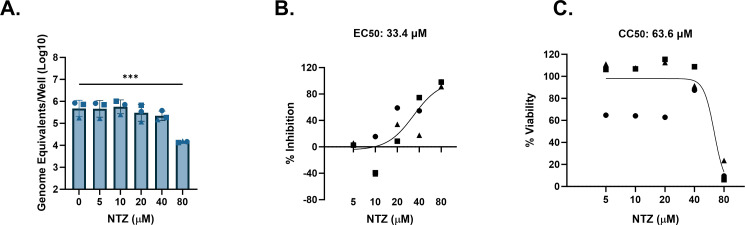
Other highly prevalent GII genotypes include GII.3 and GII.17 (55, 58). We previously showed that a genetically modified J4FUT2 HIE line supports better replication of several non-GII.4 HuNoV strains including GII.3 and GII.17 (59). We therefore tested cytotoxicity and antiviral activity of NTZ against these two genotypes in J4FUT2 HIEs. For GII.3[P21], significant reduction in replication by NTZ only occurred at 80 µM, but results were highly variable between experiments (Fig. 4A). The average percent inhibition of GII.3 by NTZ ranged from 39.7% to 55.2% between 5 and 20 µM and achieved 79.6%–98.8% inhibition at 40 and 80 µM (Fig. 4B). The EC50 was estimated to be 14.4 µM. On average, NTZ was cytotoxic at 40 and 80 µM (Fig. 4C) and resulted in a CC50 of 64.7 µM. The resulting SI was 4.5, indicating weak antiviral activity of NTZ against GII.3[P21]. Similar results were observed using GII.17[P13] in J4FUT2 HIEs. A significant (1.5 log10, 95.9%) reduction in replication occurred at 80-µM NTZ (Fig. 5A and B, respectively). The EC50 was estimated to be 33.4 µM. However, NTZ also significantly reduced cell viability at 80 µM (Fig. 5C) with an estimated CC50 of 63.6 µM, resulting in an SI of 1.9. These data indicate no antiviral activity of NTZ for GII.17[P13]. Although >90% inhibition was achieved by 80 µM, the calculated EC90 value was estimated to be >80 µM, possibly due to the variability of inhibition at the lower concentrations.
FIG 4.
NTZ minimally inhibits GII.3 HuNoV in jejunal HIEs. J4FUT2 HIEs were treated with five ascending doses of NTZ and the vehicle (1% DMSO) for 24 h. (A) Replication of GII.3 with NTZ treatment. (B) Percent inhibition of GII.3 by NTZ. (C) Percent viability was assessed in the J4FUT2 HIE line after 24 h. Based on EC50 and CC50 (panels B and C, respectively), the SI was calculated as 4.5. **P ≤ 0.01. Data are compiled from n = 4 experiments.
FIG 5.
NTZ inhibition of GII.17 HuNoV replication in jejunal HIEs parallels its cytotoxicity. J4FUT2 HIEs were treated with five ascending doses of NTZ and the vehicle (1% DMSO) for 24 h. (A) Replication of GII.17 with NTZ treatment. (B) Percent inhibition of GII.17 by NTZ. (C) Percent viability was assessed in the J4FUT2 HIE line after 24 h. Based on EC50 and CC50 (panels B and C, respectively), the SI was calculated as 1.9. ***P ≤ 0.001. Data are compiled from n = 3 experiments.
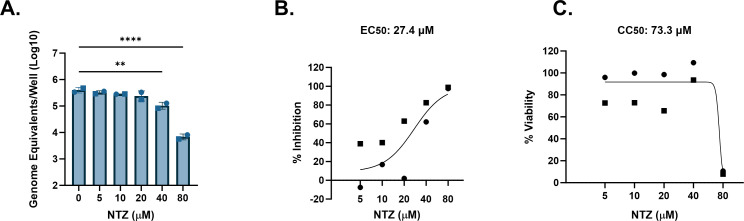
NTZ was previously reported to inhibit replication of viral RNA in a replicon model of HuNoV GI.1 (19). When using J4FUT2 HIEs, NTZ showed significant inhibition of GI.1[P1] HuNoV replication at 40 and 80 µM, resulting in 0.6–1.8 log10 reduction of genome equivalents (Fig. 6A). This corresponds to 72.3%–98.2% inhibition in replication (Fig. 6B). The EC50 for GI.1[P1] in J4FUT2 HIEs was estimated to be 27.4 µM. The average viability was steady up to 40 µM but significantly reduced at 80 µM. The CC50 was estimated to be 73.3 µM. Using these data, the SI ratio for NTZ was 2.7, indicating poor selective inhibition of GI.1 replication and designation as a weak antiviral. The EC90 value was 76.3 µM and was again similar to the CC50.
FIG 6.
NTZ inhibition of GI.1 HuNoV replication in jejunal HIEs parallels its cytotoxicity. J4FUT2 HIEs were treated with five ascending doses of NTZ and the vehicle (1% DMSO) for 24 h. (A) Replication of GI.1 with NTZ treatment. (B) Percent inhibition of GI.1 by NTZ. (C) Percent viability was assessed in the J4FUT2 HIE line after 24 h. Based on EC50 and CC50 (panels B and C, respectively), the SI was calculated as 2.7. **P ≤ 0.01, ****P ≤ 0.0001. Data are compiled from n = 2 experiments.
Nitazoxanide activity against HuNoV replication is similar in HIEs from different intestinal segments from single individuals
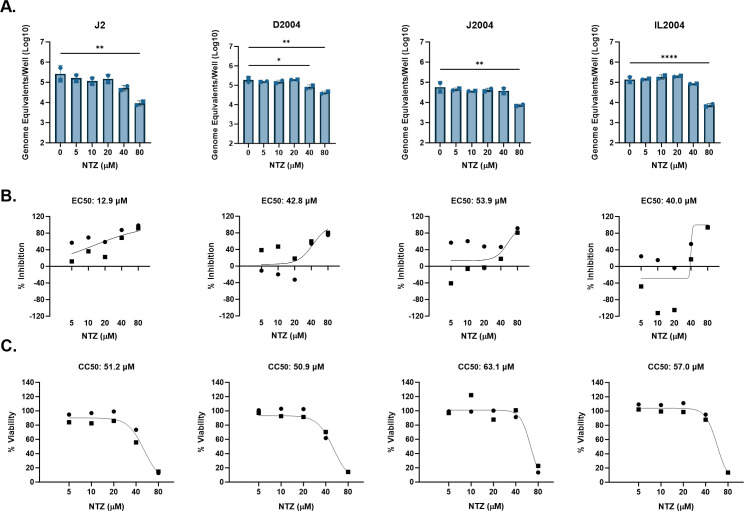
The data presented above failed to demonstrate significant NTZ activity against HuNoV replication in jejunal HIEs. However, HuNoVs can infect and replicate in all segments of the small intestine (39, 49). We therefore evaluated whether NTZ activity might be different in HIEs derived from different small intestinal segments. Replication of a GII.4 Sydney [P16] isolate was examined in duodenal, jejunal, and ileal HIEs derived from the same donor (D2004, J2004, and I2004, respectively) and in the well-characterized jejunal J2 HIE line used above.
Across the different HIE lines, 80-µM NTZ significantly inhibited GII.4 Sydney [P16] replication (Fig. 7A). NTZ was also significantly inhibitory at 40 µM in the duodenal D2004 HIEs. Based on the percent inhibition by NTZ, the EC50s for GII.4 Sydney [P16] in D2004, J2004, and IL2004 were estimated between 40.0–53.9 µM. In J2, NTZ activity was greater, with an EC50 of 12.0 µM. The CC50 values for J2, D2004, J2004, and IL2004 in these experiments ranged between 50.9 and 63.1 µM. The SIs for the duodenal, jejunal, and ileal lines derived from the same donor ranged from 1.2 to 1.4, demonstrating no antiviral activity. For the same virus isolate tested in the jejunal line J2, NTZ had low selective activity with an SI ratio of 4.0, indicating weak antiviral activity.
FIG 7.
NTZ inhibition of GII.4 HuNoV replication in small intestinal HIEs parallels its cytotoxicity. J2, D2004, J2004, and IL2004 HIEs were treated with five ascending doses of NTZ and the vehicle (1% DMSO) for 24 h. (A) Replication of GII.4 Sydney with NTZ treatment in each HIE line. (B) Percent inhibition of GII.4 Sydney by NTZ in each HIE line. (C) Percent viability was assessed in the J2, D2004, J2004, and IL2004 HIE lines after 24 h. Based on EC50 and CC50 (panels B and C, respectively), the following SI values were calculated: J2 = 4.0, D2004 = 1.2, J2004 = 1.2, and IL2004 = 1.4. *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001. Data are compiled from n = 2 experiments.
Nitazoxanide does not influence innate immunity for antiviral activity
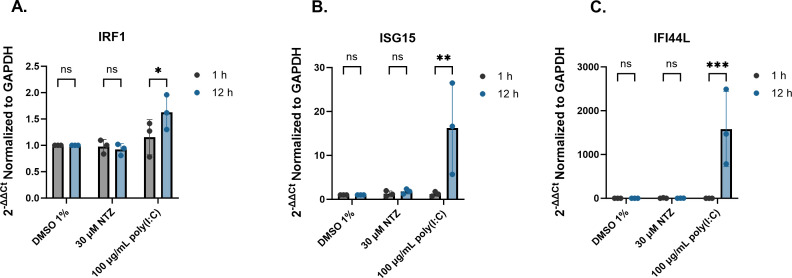
Dang et al. previously reported that NTZ-mediated inhibition of the HuNoV GI.1 replicon in Huh7 cells was due to stimulation of IRF-1 and other select interferon stimulated genes (ISGs) (19). Upon treating J2 HIEs with NTZ, no significant increase in expression was observed between 1 and 12 h for IRF1, ISG15, or IFI44L (Fig. 8). Treatment with poly(I:C), the positive control, for 12 h stimulated expression (1.6-fold) of IRF-1. Poly(I:C) also elicited expression (16- and 1582-fold) of ISG15 and IFI44L, respectively, compared to DMSO by 12 h. Treatment of HIEs with NTZ did not induce significant gene expression of ISGs.
FIG 8.
NTZ does not stimulate expression of ISGs in human intestinal enteroid cultures. J2 HIEs were treated with 1% DMSO, 30-µM NTZ, and 100-µg/mL poly(I:C) for 12 h. The fold change gene expression of (A) IRF-1, (B) ISG15, and (C) IFI44L relative to GAPDH were quantitated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). ns = not significant. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Data are compiled from n = 3 experiments.
DISCUSSION
HIEs are increasingly used to test antivirals as well as to assess neutralizing antibody titers, viral stability, and disinfectants for HuNoVs (35, 50, 52, 53, 60 – 66). However, differences in donor characteristics, HIE culture formats, virus inoculum, and cytotoxicity assays used confound comparisons between experiments and across studies. Our work establishes a standardized method to rigorously evaluate antivirals across a variety of host and virus genotypes and intestinal segments. While 2′CMC shows clear, at least moderate antiviral activity, we demonstrate that NTZ, a compound often prescribed off-label for chronic HuNoV infection, has weak to no antiviral activity across multiple virus genotypes and HIE lines (Table 2). We found that the SI for NTZ is not greater than 10 against any of the five HuNoV strains tested (Table 2), suggesting it is unlikely to be a clinically useful HuNoV antiviral. In most experiments, the EC50 was close in value to the CC50, demonstrating that inhibition of viral replication was largely attributed to its cytotoxicity. Our data are consistent with recent observational clinical studies reporting NTZ’s inefficacy for chronically infected HuNoV patients compared to other or no treatments (34, 36, 37). Thus, HIEs can serve as a valuable preclinical tool to robustly evaluate antivirals for HuNoV infection.
Our findings contrast with some previous in vitro reports of NTZ as a potential antiviral against HuNoVs. Dang et al. (19) reported NTZ inhibition of a GI.1 replicon in Huh7 cells, while our data show no inhibition of GI.1[P1] replication in HIEs. These differences could be attributed to either the cell type (transformed hepatocytes versus non-transformed intestinal epithelial cells) or the replication model used. Considering a comparison within HIEs, the report from Mirabelli et al., which described that NTZ inhibited three different GII.4 HuNoV isolates, utilized fetal ileum-derived HIEs in a 3D format (50), unlike our study that mostly utilized adult jejunal-derived HIEs in a monolayer format. The age of the donor from which the HIEs were derived or the format that the HIEs were plated could be an important distinction in the differences between the previously mentioned study and ours.
Instead, our results confirm and extend another report showing no significant inhibition of GII.4 Sydney [P31] infection in HIEs by NTZ (35). It is also important to note that this report utilized the same J2 HIEs and plated them as monolayers as is commonly used. The highest dose of NTZ tested by van Kampen et al. (35) that kept the cell monolayers intact was 10 µM, and testing was performed only against a single GII.4 Sydney 2012 strain. We did not observe any EC50 below 10 µM, which may be why van Kampen et al. (35) failed to detect any decrease in viral replication. Regardless, the GII.4 Sydney [P16] infection we examined was minimally inhibited by NTZ in J2 HIEs but not in the J2004 HIEs derived from another patient. Further, a different GII.4 Sydney [P31] isolate tested in the same J2 HIEs showed poor selective inhibition with NTZ. This suggests donor-specific and potentially virus strain-specific differences in response and leads to further questions on the potential effectiveness of this drug across the general population. While the isolates used in our study were both GII.4 Sydney viruses based on their capsid protein sequences, they have differing P-types; further investigation is needed to delineate whether differences in the P-type or other viral or host genetic attributes mediate NTZ antiviral activity.
HIEs are heterogeneous because they represent the genetics of the individual donors from whom they were established; this is reflected in RNAseq analyses showing that gene expression first segregates by HIE line in cultures plated in the same format (67, 68). HIEs can vary in their responses based on intestinal segment of origin, plating format, and differentiation status (68). In addition, HIEs generated from different donors vary in their permissiveness to support replication of different HuNoVs (39, 59). For example, HIE lines from individuals lacking a functional FUT2 gene, termed secretor negative, are resistant to infection by many HuNoV strains (39). These factors highlight important considerations for using HIEs as a model for evaluating antivirals for HuNoV. We implemented antiviral testing using secretor-positive HIEs known to be susceptible to multiple HuNoV genotypes, used a standard inoculum dose by determining the TCID50 for every genotype for each HIE line tested, and performed experiments with lines cultured in the same medium and plated in the same format (monolayers). We also used HIE lines that were optimal for replication of a given HuNoV genotype such as GI.1 and GII.17 in J4FUT2 rather than J2, which has been reported previously (59). Our TCID50 studies revealed that HIEs established from different intestinal segments have differing permissiveness to infection even when generated from the same donor. We observed that from the same donor, the TCID50 for GII.4 Sydney [P16] virus was at least five times higher for the duodenal HIE line compared to the jejunal and ileal lines, suggesting that GII.4 preferentially infects the jejunum and ileum. This distinction was also reported by Hosmillo et al. (69), who found their GII.4 HuNoV consistently replicated more efficiently in ileal organoids compared to duodenal HIEs and vice versa for GII.3 (69). Contrastingly, our group previously observed higher replication of GII.3 [P21] in ileal HIEs compared to duodenal HIEs from two donors, an effect not observed for GII.4 Sydney [P31] (49). The basis for variation between the results in this study and prior publications is not clear but may be due to differences in media used, HIE donor genotype and phenotype, or differences in the virus, all of which are factors that need to be explored further. To eliminate possible differences in replication efficiency and to standardize infections, our studies utilized 100 TCID50 for each virus and each tested HIE line, achieving similar levels of replication throughout. In addition to standardizing viral inoculum dose, and as required for standard antiviral testing, we assessed viability of drug treatments for every HIE line in tandem with every antiviral experiment. After taking these measures to reduce variability, we estimated EC50 and CC50 values to calculate the SI ratio, which is routinely used in the antiviral field to determine whether a drug is likely to be effective clinically (51, 56, 57).
Dang et al. attributed the activity of NTZ to inhibit the GI.1 replicon model to stimulating an endogenous innate immune response, namely, IRF-1 (19). They also tested expression of select innate immunity genes in HIE cultures and concluded that NTZ may be effective in HIEs. By contrast, we did not detect a significant increase in IRF-1 or ISG-15 in our studies, although these ISGs were induced in the HIEs by poly(I:C), a positive control (Fig. 8). We also evaluated IFI44L, another ISG routinely upregulated in HIEs by HuNoV infection and found its expression was stimulated by poly(I:C) but not by NTZ. Whether NTZ could modulate other host responses apart from any direct antiviral effect on replication remains to be determined. For example, it is possible NTZ could exert indirect antiviral activity via immune cells which are lacking in our current HIE culture model. However, our data are consistent with several observational clinical studies where no differences were reported between NTZ treatment and other or no treatments in chronically infected HuNoV patients (34, 36, 37). A recent clinical trial (NCT03395405) (11) aimed to assess NTZ treatment for persons chronically infected with HuNoV but failed to reach its targeted enrollment numbers and was concluded early in the COVID-19 pandemic; that said, reported outcomes of the 31 randomized patients suggest that NTZ did not improve clinical resolution of symptoms compared to placebo, with no effect on fecal viral load observed. The standard adult NTZ dose, one 500-mg tablet by mouth twice daily, results in maximal concentrations of the active metabolite tizoxanide of approximately 10 µg/mL (~38 µM) (70). Based on this, the range of doses tested in this study is relevant to achievable drug concentrations in sera.
While our study did not test virus strains representing the entire repertoire of genotypes that can replicate in HIEs (49), it included the most prevalent HuNoV genotypes and showed NTZ had weak to no antiviral activity for these clinically important genotypes. Our study does not support the use of NTZ as a clinical antiviral for HuNoVs. We have further provided important considerations to make future HuNoV antiviral studies more standardized and robust and provide more evidence that HIEs are an excellent and relevant model for screening HuNoV therapeutics.
MATERIALS AND METHODS
Maintenance and culture of HIEs
HIE cultures were obtained from the BCM organoid core; these cultures were originally derived from biopsy specimens obtained during bariatric surgery as described previously (39, 71) or from intestinal organ donations from a LifeGift organ procurement organization (K. Ettayebi and M. K. Estes, unpublished data). A genetically modified J4FUT2 line was used to obtain enhanced replication of some virus strains as described previously (59). 3D cultures of HIEs suspended in Matrigel were maintained in complete medium with growth factors (L-WRN media prepared from cell line ATCC CRL-3276 grown in DMEM-F-12 supplemented with 20% FBS) until processing into monolayer cultures. Processing and plating of monolayer cultures were performed and seeded into 96-well plates using Intesticult proliferation medium for 24 h and then differentiated for 5 days using Intesticult differentiation (INTd) medium prior to inoculation with the virus, as described previously (49).
HuNoVs and assessment of viral infectivity
Preparation of stool filtrates were described previously (49). The viruses and respective titers are listed in Table 1. Total RNA was extracted using a Kingfisher Flex machine and MagMAX-96 viral RNA isolation kit. Extracted RNA was used for RT-qPCR, and viral replication was quantitated relative to a standard curve as described previously (49). In brief, the primer pair and probe sets NIFG1F/NV1LCR/NIFG1P and COG2R/QNIF2d/QNIFS were used to detect GI and GII genotypes, respectively (64, 72). Reactions were first run at 50°C (15 min) and 95°C (5 min), followed by 40 cycles of 95°C (15 s) and 60°C (35 s). Standard curves were based on recombinant HuNoV GII.4 (Houston virus) or GI.1 (Norwalk virus) RNA. RNA in each infected inoculated well was evaluated with technical duplicates.
Determination of TCID50
TCID50 values were determined for viruses in the HIE line known to yield maximum virus replication (59). Virus filtrates were serially diluted twofold in complete media without growth factors (CMGF−) supplemented with 500-µM sodium glycochenodeoxycholate (GCDCA; Sigma, G0759) and inoculated onto three wells of HIE monolayers for 1 h (GII.4 and GII.3) or 2 h (GI.1 and GII.17). Following three washes with CMGF−, INTd media supplemented with GCDCA was added for 24 h (49). Viral RNA was extracted, and RT-qPCR was used to determine if each inoculated well was positive (above the limit of detection) or negative (below the limit of detection) for infection. RNA from each inoculated well was run in duplicate for RT-qPCR. The Reed-Muench method was used to determine the TCID50 (39, 73). The TCID50 was averaged from two experiments. The TCID50 values for each virus relative to the HIE line used are shown in Table 1.
Antiviral treatment of HIEs
United States Pharmacopeia-grade NTZ (Sigma, 1463960) was serially diluted twofold in ≥99.7% pure DMSO (Sigma, D2650). Pure 2′CMC of ≥95% (Sigma, M4949) was serially diluted in 0.5 log10 increments in Milli-Q H2O. Dilutions of compound in their vehicle were then added to 500-µM GCDCA supplemented CMGF− and INTd medium for a 1- to 2-h inoculation and 24-h incubation periods, respectively. NTZ was added to the media with a final concentration of 1% DMSO, and 2′CMC was added to the media with a final concentration of 2% H2O. One hundred TCID50 of each virus was added to the inoculation media and then added onto three wells of the optimal HIE line and incubated at 37°C. After the incubation period, the HIE monolayers were washed three times with CMGF−. The media was replaced with vehicle- or compound-prepared INTd media, and cells were incubated at 37°C for 24 h. Stocks of NTZ were stored at −20°C and were not stored longer than 2 weeks.
Assessment of HIE viability
For the initial dose range assessment studies, cell viability was determined by staining for propidium iodide. After 24 h of treatment with a compound, cells were stained with propidium iodide (Invitrogen, P1304MP) for 10 min to mark dead cells. Monolayers were washed three times with CMGF−. Four percent paraformaldehyde (Electron Microscopy Sciences) was then added to the monolayers for 15 min followed by three washes with phosphate-buffered saline (PBS). Monolayers were then permeated using 0.5% Triton-X 100 for 15 min, then washed with PBS and stained using DAPI (Invitrogen R37606). After incubating, monolayers were washed three times with PBS. Cells were imaged at ×40 magnification using an Olympus epifluorescent microscope. For all other studies, cell viability was determined using the CellTiter-Glo version 2.0 Cell Viability Assay (Promega, G9242). HIE cells were seeded onto black 96-well plates (Greiner Bio-One, 655086) and differentiated for 5 days as described above. Three wells of HIE monolayers were treated with vehicle and compound in incubation media without virus. After 24 h of treatment of a compound, the manufacturer’s protocol was used for the CellTiter-Glo version 2.0 Cell Viability Assay.
Measuring relative gene expression for innate immune genes
Fold changes in mRNA expression for host interferon-stimulated genes were determined by RT-qPCR using RNA extracted 12 h following the HIE inoculation and RNA extraction methods presented above. A positive control, poly(I:C) (Millipore, 528906) was diluted to 100 µg/mL (1% H2O vehicle) in INTd medium supplemented with GCDCA for 12-h inoculation of HIEs, a time point chosen based on high stimulation of ISGs in HIEs by this agonist. The delta-delta-Ct method was used to calculate relative values to DMSO-treated HIEs, after normalization to the housekeeping gene GAPDH. The TaqMan primer probes were of IRF1, ISG15, IFI44L, and GAPDH (ThermoFisher Scientific; Hs00971965_m1, Hs01921425_s1, Hs00915292_m1, and Hs99999905_m1, respectively).
Statistical analyses
Each treatment performed on HIEs had three technical replicate wells. Two technical replicates were performed on each well for RT-qPCR and then averaged. The estimated genome equivalents between the three technical replicate wells per condition were then averaged. Data from each averaged condition were then pooled among repeated experiments. n is represented as the number of experiments and is noted in the figure legends. Gene expression of ISGs between 1 and 12 h was done using a two-way analysis of variance (ANOVA) and Sidak post hoc multiple comparisons analysis. Percent inhibition was determined as
For determining the effect of 2′CMC and NTZ on HuNoV replication, one-way ANOVA and Dunnett’s post hoc multiple comparisons analyses were performed on 24-h replication data, and non-linear regression analysis was performed on percent inhibition and viability data to determine EC50, EC90, and CC50. The non-linear regression formula used was the [Agonist] versus response – Find ECanything in GraphPad Prism. For EC50 and EC90 calculations, the top value was constrained to 100. For CC50 calculations, the bottom value was constrained to 0. SI was calculated as a ratio of the CC50 over the EC50. Statistical analyses were performed using GraphPad Prism version 9.5.0.
ACKNOWLEDGMENTS
This work was funded in part by Public Health Service grants (U19 AI144297, M.K.E. and R.L.A.; and PO1-AI057788, M.K.E., R.L.A., and B.V.V.P.)
This work was also supported by a training fellowship to M.L. from the Gulf Coast Consortia on Training Interdisciplinary Pharmacology Scientists (grant no. T32GM120011 and T32GM139801). Figure 1A was created with some images from BioRender.com.
Contributor Information
Mary K. Estes, Email: mestes@bcm.edu.
Miguel Angel Martinez, IrsiCaixa Institut de Recerca de la Sida, Badalona, Barcelona, Spain .
REFERENCES
- 1. Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, Parashar UD, Koopmans M, Lopman BA. 2014. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis 14:725–730. doi: 10.1016/S1473-3099(14)70767-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ramani S, Atmar RL, Estes MK. 2014. Epidemiology of human noroviruses and updates on vaccine development. Curr Opin Gastroenterol 30:25–33. doi: 10.1097/MOG.0000000000000022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pires SM, Fischer-Walker CL, Lanata CF, Devleesschauwer B, Hall AJ, Kirk MD, Duarte ASR, Black RE, Angulo FJ. 2015. Aetiology-specific estimates of the global and regional incidence and mortality of diarrhoeal diseases commonly transmitted through food. PLoS One 10:e0142927. doi: 10.1371/journal.pone.0142927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartsch SM, Lopman BA, Ozawa S, Hall AJ, Lee BY. 2016. Global economic burden of norovirus gastroenteritis. PLoS One 11:e0151219. doi: 10.1371/journal.pone.0151219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, Graham DY. 2008. Norwalk virus shedding after experimental human infection. Emerg Infect Dis 14:1553–1557. doi: 10.3201/eid1410.080117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bok K, Green KY. 2012. Norovirus gastroenteritis in immunocompromised patients. N Engl J Med 367:2126–2132. doi: 10.1056/NEJMra1207742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chhabra P, de Graaf M, Parra GI, Chan M-W, Green K, Martella V, Wang Q, White PA, Katayama K, Vennema H, Koopmans MPG, Vinjé J. 2019. Updated classification of norovirus genogroups and genotypes. J Gen Virol 100:1393–1406. doi: 10.1099/jgv.0.001318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang M, Fu M, Hu Q. 2021. Advances in human norovirus vaccine research. Vaccines (Basel) 9:732. doi: 10.3390/vaccines9070732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tan M. 2021. Norovirus vaccines: current clinical development and challenges. Pathogens 10:1641. doi: 10.3390/pathogens10121641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Children's National Research Institute . 2023. Adoptive T lymphocyte administration for chronic norovirus treatment in immunocompromised hosts (ATLANTIC). Available from: https://clinicaltrials.gov/ct2/show/study/NCT04691622
- 11. NIAID . 2022. NNITS-nitazoxanide for norovirus in transplant patients study. Available from: https://clinicaltrials.gov/ct2/show/study/NCT03395405
- 12. Sisson G, Goodwin A, Raudonikiene A, Hughes NJ, Mukhopadhyay AK, Berg DE, Hoffman PS. 2002. Enzymes associated with reductive activation and action of nitazoxanide, nitrofurans, and metronidazole in Helicobacter pylori. Antimicrob Agents Chemother 46:2116–2123. doi: 10.1128/AAC.46.7.2116-2123.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoffman PS, Sisson G, Croxen MA, Welch K, Harman WD, Cremades N, Morash MG. 2007. Antiparasitic drug nitazoxanide inhibits the pyruvate oxidoreductases of Helicobacter pylori, selected anaerobic bacteria and parasites, and Campylobacter jejuni. Antimicrob Agents Chemother 51:868–876. doi: 10.1128/AAC.01159-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haffizulla J, Hartman A, Hoppers M, Resnick H, Samudrala S, Ginocchio C, Bardin M, Rossignol J-F, US Nitazoxanide Influenza Clinical Study Group . 2014. Effect of nitazoxanide in adults and adolescents with acute uncomplicated influenza: a double-blind, randomised, placebo-controlled, phase 2B/3 trial. Lancet Infect Dis 14:609–618. doi: 10.1016/S1473-3099(14)70717-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rossignol JF, Abu-Zekry M, Hussein A, Santoro MG. 2006. Effect of nitazoxanide for treatment of severe rotavirus diarrhoea: randomised double-blind placebo-controlled trial. Lancet 368:124–129. doi: 10.1016/S0140-6736(06)68852-1 [DOI] [PubMed] [Google Scholar]
- 16. Jasenosky LD, Cadena C, Mire CE, Borisevich V, Haridas V, Ranjbar S, Nambu A, Bavari S, Soloveva V, Sadukhan S, Cassell GH, Geisbert TW, Hur S, Goldfeld AE. 2019. The FDA-approved oral drug nitazoxanide amplifies host antiviral responses and inhibits Ebola virus. iScience 19:1279–1290. doi: 10.1016/j.isci.2019.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rossignol JF. 2014. Nitazoxanide: a first-in-class broad-spectrum antiviral agent. Antiviral Res 110:94–103. doi: 10.1016/j.antiviral.2014.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Trabattoni D, Gnudi F, Ibba SV, Saulle I, Agostini S, Masetti M, Biasin M, Rossignol J-F, Clerici M. 2016. Thiazolides elicit anti-viral innate immunity and reduce HIV replication. Sci Rep 6:27148. doi: 10.1038/srep27148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dang W, Xu L, Ma B, Chen S, Yin Y, Chang KO, Peppelenbosch MP, Pan Q. 2018. Nitazoxanide inhibits human norovirus replication and synergizes with ribavirin by activation of cellular antiviral response. Antimicrob Agents Chemother 62:e00707-18. doi: 10.1128/AAC.00707-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rossignol JF, La Frazia S, Chiappa L, Ciucci A, Santoro MG. 2009. Thiazolides, a new class of anti-influenza molecules targeting viral hemagglutinin at the post-translational level. J Biol Chem 284:29798–29808. doi: 10.1074/jbc.M109.029470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Elazar M, Liu M, McKenna SA, Liu P, Gehrig EA, Puglisi JD, Rossignol JF, Glenn JS. 2009. The anti-hepatitis C agent nitazoxanide induces phosphorylation of eukaryotic initiation factor 2α via protein kinase activated by double-stranded RNA activation. Gastroenterology 137:1827–1835. doi: 10.1053/j.gastro.2009.07.056 [DOI] [PubMed] [Google Scholar]
- 22. La Frazia S, Ciucci A, Arnoldi F, Coira M, Gianferretti P, Angelini M, Belardo G, Burrone OR, Rossignol J-F, Santoro MG. 2013. Thiazolides, a new class of antiviral agents effective against rotavirus infection, target viral morphogenesis, inhibiting viroplasm formation. J Virol 87:11096–11106. doi: 10.1128/JVI.01213-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Piacentini S, La Frazia S, Riccio A, Pedersen JZ, Topai A, Nicolotti O, Rossignol J-F, Santoro MG. 2018. Nitazoxanide inhibits paramyxovirus replication by targeting the fusion protein folding: role of glycoprotein-specific thiol oxidoreductase ERp57. Sci Rep 8:10425. doi: 10.1038/s41598-018-28172-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rossignol JF, El-Gohary YM. 2006. Nitazoxanide in the treatment of viral gastroenteritis: a randomized double-blind placebo-controlled clinical trial. Aliment Pharmacol Ther 24:1423–1430. doi: 10.1111/j.1365-2036.2006.03128.x [DOI] [PubMed] [Google Scholar]
- 25. Siddiq DM, Koo HL, Adachi JA, Viola GM. 2011. Norovirus gastroenteritis successfully treated with nitazoxanide. J Infect 63:394–397. doi: 10.1016/j.jinf.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Echenique IA, Stosor V, Gallon L, Kaufman D, Qi C, Zembower TR. 2016. Prolonged norovirus infection after pancreas transplantation: a case report and review of chronic norovirus. Transpl Infect Dis 18:98–104. doi: 10.1111/tid.12472 [DOI] [PubMed] [Google Scholar]
- 27. Kempf B, Edgar JD, Mc Caughey C, Devlin LA. 2017. Nitazoxanide is an ineffective treatment of chronic norovirus in patients with X-linked agammaglobulinemia and may yield false-negative polymerase chain reaction findings in stool specimens. J Infect Dis 215:486–487. doi: 10.1093/infdis/jiw497 [DOI] [PubMed] [Google Scholar]
- 28. Jurgens PT, Allen LA, Ambardekar AV, McIlvennan CK. 2017. Chronic norovirus infections in cardiac transplant patients. Prog Transplant 27:69–72. doi: 10.1177/1526924816679843 [DOI] [PubMed] [Google Scholar]
- 29. Avery RK, Lonze BE, Kraus ES, Marr KA, Montgomery RA. 2017. Severe chronic norovirus diarrheal disease in transplant recipients: clinical features of an under-recognized syndrome. Transpl Infect Dis 19. doi: 10.1111/tid.12674 [DOI] [PubMed] [Google Scholar]
- 30. Haubrich K, Gantt S, Blydt-Hansen T. 2018. Successful treatment of chronic norovirus gastroenteritis with nitazoxanide in a pediatric kidney transplant recipient. Pediatr Transplant 22:e13186. doi: 10.1111/petr.13186 [DOI] [PubMed] [Google Scholar]
- 31. Ghusson N, Vasquez G. 2018. Successfully treated norovirus- and sapovirus-associated diarrhea in three renal transplant patients. Case Rep Infect Dis 2018:6846873. doi: 10.1155/2018/6846873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wright S, Kleven D, Kapoor R, Kavuri S, Gani I. 2020. Recurring norovirus & sapovirus infection in a renal transplant patient. IDCases 20:e00776. doi: 10.1016/j.idcr.2020.e00776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ashton G, Shand A, Arnott I, Din S. 2021. Profound diarrhoea and weight loss in an immunocompromised patient. BMJ Case Rep 14:e236913. doi: 10.1136/bcr-2020-236913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hedvat J, Salerno DM, Kovac D, Scheffert JL, Corbo H, Chen JK, Choe JY, Jennings DL, Anamisis A, Liu EC, Lee JH, Shertel T, Lange NW. 2022. Nitazoxanide treatment for norovirus infection in solid organ transplant recipients. Clin Transplant 36:e14594. doi: 10.1111/ctr.14594 [DOI] [PubMed] [Google Scholar]
- 35. van Kampen JJA, Dalm VASH, Fraaij PLA, Oude Munnink BB, Schapendonk CME, Izquierdo-Lara RW, Villabruna N, Ettayebi K, Estes MK, Koopmans MPG, de Graaf M. 2022. Clinical and in vitro evidence favoring immunoglobulin treatment of a chronic norovirus infection in a patient with common variable immunodeficiency. J Infect Dis 226:1781–1789. doi: 10.1093/infdis/jiac085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nair SN, Bhaskaran A, Chandorkar A, Fontana L, Obeid KM. 2023. Noroviral diarrhea in solid organ transplant recipients: an analysis of interventions and outcomes. Clin Transplant 37:e14855. doi: 10.1111/ctr.14855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Soneji M, Newman AM, Toia J, Muller WJ. 2022. Metronidazole for treatment of norovirus in pediatric transplant recipients. Pediatr Transplant 26:e14390. doi: 10.1111/petr.14390 [DOI] [PubMed] [Google Scholar]
- 38. Lin SC, Qu L, Ettayebi K, Crawford SE, Blutt SE, Robertson MJ, Zeng XL, Tenge VR, Ayyar BV, Karandikar UC, Yu X, Coarfa C, Atmar RL, Ramani S, Estes MK. 2020. Human norovirus exhibits strain-specific sensitivity to host interferon pathways in human intestinal enteroids. Proc Natl Acad Sci U S A 117:23782–23793. doi: 10.1073/pnas.2010834117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng X-L, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK. 2016. Replication of human noroviruses in stem cell-derived human enteroids. Science 353:1387–1393. doi: 10.1126/science.aaf5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ayyar BV, Ettayebi K, Salmen W, Karandikar UC, Neill FH, Tenge VR, Crawford SE, Bieberich E, Prasad BVV, Atmar RL, Estes MK. 2023. CLIC and membrane wound repair pathways enable pandemic norovirus entry and infection. Nat Commun 14:1148. doi: 10.1038/s41467-023-36398-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nordgren J, Svensson L. 2019. Genetic susceptibility to human norovirus infection: an update. Viruses 11:226. doi: 10.3390/v11030226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Parra GI. 2019. Emergence of norovirus strains: a tale of two genes. Virus Evol 5:vez048. doi: 10.1093/ve/vez048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Davis A, Cortez V, Grodzki M, Dallas R, Ferrolino J, Freiden P, Maron G, Hakim H, Hayden RT, Tang L, Huys A, Kolawole AO, Wobus CE, Jones MK, Karst SM, Schultz-Cherry S. 2020. Infectious norovirus is chronically shed by immunocompromised pediatric hosts. Viruses 12:619. doi: 10.3390/v12060619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brown JR, Roy S, Tutill H, Williams R, Breuer J. 2017. Super-infections and relapses occur in chronic norovirus infections. J Clin Virol 96:44–48. doi: 10.1016/j.jcv.2017.09.009 [DOI] [PubMed] [Google Scholar]
- 45. Brown JR, Shah D, Breuer J. 2016. Viral gastrointestinal infections and norovirus genotypes in a paediatric UK hospital, 2014-2015. J Clin Virol 84:1–6. doi: 10.1016/j.jcv.2016.08.298 [DOI] [PubMed] [Google Scholar]
- 46. Green KY. 2014. Norovirus infection in immunocompromised hosts. Clin Microbiol Infect 20:717–723. doi: 10.1111/1469-0691.12761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kondapi DS, Ramani S, Estes MK, Atmar RL, Okhuysen PC. 2021. Norovirus in cancer patients: a review. Open Forum Infect Dis 8:fab126. doi: 10.1093/ofid/ofab126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Haddadin Z, Batarseh E, Hamdan L, Stewart LS, Piya B, Rahman H, Spieker AJ, Chappell J, Wikswo ME, Dunn JR, Payne DC, Vinjé J, Hall AJ, Halasa N. 2021. Characteristics of GII.4 norovirus versus other genotypes in sporadic pediatric infections in Davidson county, Tennessee, USA. Clin Infect Dis 73:e1525–e1531. doi: 10.1093/cid/ciaa1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ettayebi K, Tenge VR, Cortes-Penfield NW, Crawford SE, Neill FH, Zeng X-L, Yu X, Ayyar BV, Burrin D, Ramani S, Atmar RL, Estes MK. 2021. New insights and enhanced human norovirus cultivation in human intestinal enteroids. mSphere 6:e01136-20. doi: 10.1128/mSphere.01136-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mirabelli C, Santos-Ferreira N, Gillilland MG, Cieza RJ, Colacino JA, Sexton JZ, Neyts J, Taube S, Rocha-Pereira J, Wobus CE, López S. 2022. Human norovirus efficiently replicates in differentiated 3D-human intestinal enteroids. J Virol 96:e0085522. doi: 10.1128/jvi.00855-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Josset L, Textoris J, Loriod B, Ferraris O, Moules V, Lina B, N’guyen C, Diaz J-J, Rosa-Calatrava M. 2010. Gene expression signature-based screening identifies new broadly effective influenza A antivirals. PLoS One 5:e13169. doi: 10.1371/journal.pone.0013169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Atmar RL, Ettayebi K, Ayyar BV, Neill FH, Braun RP, Ramani S, Estes MK. 2020. Comparison of microneutralization and histo-blood group antigen-blocking assays for functional norovirus antibody detection. J Infect Dis 221:739–743. doi: 10.1093/infdis/jiz526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hayashi T, Murakami K, Hirano J, Fujii Y, Yamaoka Y, Ohashi H, Watashi K, Estes MK, Muramatsu M, Pasetti MF. 2021. Dasabuvir inhibits human norovirus infection in human intestinal enteroids. mSphere 6:e0062321. doi: 10.1128/mSphere.00623-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cannon JL, Barclay L, Collins NR, Wikswo ME, Castro CJ, Magaña LC, Gregoricus N, Marine RL, Chhabra P, Vinjé J, Tang Y-W. 2017. Genetic and epidemiologic trends of norovirus outbreaks in the United States from 2013 to 2016 demonstrated emergence of novel GII.4 recombinant viruses. J Clin Microbiol 55:2208–2221. doi: 10.1128/JCM.00455-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. van Beek J, de Graaf M, Al-Hello H, Allen DJ, Ambert-Balay K, Botteldoorn N, Brytting M, Buesa J, Cabrerizo M, Chan M, Cloak F, Di Bartolo I, Guix S, Hewitt J, Iritani N, Jin M, Johne R, Lederer I, Mans J, Martella V, Maunula L, McAllister G, Niendorf S, Niesters HG, Podkolzin AT, Poljsak-Prijatelj M, Rasmussen LD, Reuter G, Tuite G, Kroneman A, Vennema H, Koopmans MPG, NoroNet . 2018. Molecular surveillance of norovirus, 2005-16: an epidemiological analysis of data collected from the NoroNet network. Lancet Infect Dis 18:545–553. doi: 10.1016/S1473-3099(18)30059-8 [DOI] [PubMed] [Google Scholar]
- 56. FDA . 2014. Vaginal microbicides: development for the prevention of HIV infection. Available from: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/vaginal-microbicidesdevelopment-prevention-hiv-infection-pdf
- 57. Smee DF, Hurst BL, Evans WJ, Clyde N, Wright S, Peterson C, Jung KH, Day CW. 2017. Evaluation of cell viability dyes in antiviral assays with RNA viruses that exhibit different cytopathogenic properties. J Virol Methods 246:51–57. doi: 10.1016/j.jviromet.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cannon JL, Bonifacio J, Bucardo F, Buesa J, Bruggink L, Chan MC-W, Fumian TM, Giri S, Gonzalez MD, Hewitt J, Lin J-H, Mans J, Muñoz C, Pan C-Y, Pang X-L, Pietsch C, Rahman M, Sakon N, Selvarangan R, Browne H, Barclay L, Vinjé J. 2021. Global trends in norovirus genotype distribution among children with acute gastroenteritis. Emerg Infect Dis 27:1438–1445. doi: 10.3201/eid2705.204756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Haga K, Ettayebi K, Tenge VR, Karandikar UC, Lewis MA, Lin S-C, Neill FH, Ayyar BV, Zeng X-L, Larson G, Ramani S, Atmar RL, Estes MK, Griffin DE. 2020. Genetic manipulation of human intestinal enteroids demonstrates the necessity of a functional fucosyltransferase 2 gene for secretor-dependent human norovirus infection. mBio 11:e00251-20. doi: 10.1128/mBio.00251-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Costantini V, Morantz EK, Browne H, Ettayebi K, Zeng X-L, Atmar RL, Estes MK, Vinjé J. 2018. Human norovirus replication in human intestinal enteroids as model to evaluate virus inactivation. Emerg Infect Dis 24:1453–1464. doi: 10.3201/eid2408.180126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Estes MK, Ettayebi K, Tenge VR, Murakami K, Karandikar U, Lin SC, Ayyar BV, Cortes-Penfield NW, Haga K, Neill FH, Opekun AR, Broughman JR, Zeng XL, Blutt SE, Crawford SE, Ramani S, Graham DY, Atmar RL. 2019. Human norovirus cultivation in nontransformed stem cell-derived human intestinal enteroid cultures: success and challenges. Viruses 11:638. doi: 10.3390/v11070638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lindesmith LC, McDaniel JR, Changela A, Verardi R, Kerr SA, Costantini V, Brewer-Jensen PD, Mallory ML, Voss WN, Boutz DR, Blazeck JJ, Ippolito GC, Vinje J, Kwong PD, Georgiou G, Baric RS. 2019. Sera antibody repertoire analyses reveal mechanisms of broad and pandemic strain neutralizing responses after human norovirus vaccination. Immunity 50:1530–1541. doi: 10.1016/j.immuni.2019.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Randazzo W, Costantini V, Morantz EK, Vinjé J. 2020. Human intestinal enteroids to evaluate human norovirus GII.4 inactivation by aged-green tea. Front Microbiol 11:1917. doi: 10.3389/fmicb.2020.01917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ettayebi K, Salmen W, Imai K, Hagi A, Neill FH, Atmar RL, Prasad BVV, Estes MK. 2022. Antiviral activity of olanexidine-containing hand rub against human noroviruses. mBio 13:e0284821. doi: 10.1128/mbio.02848-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Desdouits M, Polo D, Le Mennec C, Strubbia S, Zeng X-L, Ettayebi K, Atmar RL, Estes MK, Le Guyader FS. 2022. Use of human intestinal enteroids to evaluate persistence of infectious human norovirus in seawater. Emerg Infect Dis 28:1475–1479. doi: 10.3201/eid2807.220219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hayashi T, Murakami K, Ando H, Ueno S, Kobayashi S, Muramatsu M, Tanikawa T, Kitamura M. 2023. Inhibitory effect of Ephedra herba on human norovirus infection in human intestinal organoids. Biochem Biophys Res Commun 671:200–204. doi: 10.1016/j.bbrc.2023.05.127 [DOI] [PubMed] [Google Scholar]
- 67. Saxena K, Simon LM, Zeng XL, Blutt SE, Crawford SE, Sastri NP, Karandikar UC, Ajami NJ, Zachos NC, Kovbasnjuk O, Donowitz M, Conner ME, Shaw CA, Estes MK. 2017. A paradox of transcriptional and functional innate interferon responses of human intestinal enteroids to enteric virus infection. Proc Natl Acad Sci U S A 114:E570–E579. doi: 10.1073/pnas.1615422114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Criss ZK, Bhasin N, Di Rienzi SC, Rajan A, Deans-Fielder K, Swaminathan G, Kamyabi N, Zeng X-L, Doddapaneni H, Menon VK, Chakravarti D, Estrella C, Yu X, Patil K, Petrosino JF, Fleet JC, Verzi MP, Christakos S, Helmrath MA, Arimura S, DePinho RA, Britton RA, Maresso AW, Grande-Allen KJ, Blutt SE, Crawford SE, Estes MK, Ramani S, Shroyer NF. 2021. Drivers of transcriptional variance in human intestinal epithelial organoids. Physiol Genomics 53:486–508. doi: 10.1152/physiolgenomics.00061.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hosmillo M, Chaudhry Y, Nayak K, Sorgeloos F, Koo B-K, Merenda A, Lillestol R, Drumright L, Zilbauer M, Goodfellow I. 2020. Norovirus replication in human intestinal epithelial cells is restricted by the interferon-induced JAK/STAT signaling pathway and RNA polymerase II-mediated transcriptional responses. mBio 11:e00215-20. doi: 10.1128/mBio.00215-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stockis A, De Bruyn S, Gengler C, Rosillon D. 2002. Nitazoxanide pharmacokinetics and tolerability in man during 7 days dosing with 0.5 g and 1 g b.i.d. Int J Clin Pharmacol Ther 40:221–227. doi: 10.5414/cpp40221 [DOI] [PubMed] [Google Scholar]
- 71. Saxena K, Blutt SE, Ettayebi K, Zeng X-L, Broughman JR, Crawford SE, Karandikar UC, Sastri NP, Conner ME, Opekun AR, Graham DY, Qureshi W, Sherman V, Foulke-Abel J, In J, Kovbasnjuk O, Zachos NC, Donowitz M, Estes MK, Sandri-Goldin RM. 2016. Human intestinal enteroids: a new model to study human rotavirus infection, host restriction, and pathophysiology. J Virol 90:43–56. doi: 10.1128/JVI.01930-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Miura T, Parnaudeau S, Grodzki M, Okabe S, Atmar RL, Le Guyader FS. 2013. Environmental detection of genogroup I, II, and IV noroviruses by using a generic real-time reverse transcription-PCR assay. Appl Environ Microbiol 79:6585–6592. doi: 10.1128/AEM.02112-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408 [DOI] [Google Scholar]