Abstract
The contribution of regulatory genes to fluoroquinolone resistance was studied with clinical Escherichia coli strains bearing mutations in gyrA and parC and with different levels of fluoroquinolone resistance. Expression of marA and soxS was evaluated by Northern blot analysis of isolates that demonstrated increased organic solvent tolerance, a phenotype that has been linked to overexpression of marA, soxS, and rob. Among 25 cyclohexane-tolerant strains detected by a screen for increased organic solvent tolerance (M. Oethinger, W. V. Kern, J. D. Goldman, and S. B. Levy, J. Antimicrob. Chemother. 41:111–114, 1998), we found 5 Mar mutants and 4 Sox mutants. A further Mar mutant was detected among 11 fluoroquinolone-resistant, cyclohexane-susceptible E. coli strains used as controls. Comparison of the marOR sequences of clinical Mar mutants with that of E. coli K-12 (GenBank accession no. M96235) revealed point mutations in marR in all mutants which correlated with loss of repressor function as detected in a marO::lacZ transcriptional assay. We found four other amino acid changes in MarR that did not lead to loss of function. Two of these changes, present in 20 of the 35 sequenced marOR fragments, identified a variant genotype of marOR. Isolates with the same gyrA and parC mutations showed increased fluoroquinolone resistance when the mutations were accompanied by overexpression of marA or soxS. These data support the hypothesis that high-level fluoroquinolone resistance involves mutations at several chromosomal loci, comprising structural and regulatory genes.
A large number of studies of fluoroquinolone resistance among clinical Escherichia coli isolates has shown that mutations in the structural genes gyrA and parC are important mechanisms of resistance (13, 14, 21, 29, 37, 42). However, it has also become evident that additional mutations, such as mutations in one of the regulatory genes marRAB (10, 12, 16), soxRS (2, 44), and robA (4) and in other yet unidentified genes, potentially contribute to the resistance phenotype (15, 25). These regulatory genes, when overexpressed, confer low-level resistance to a number of structurally unrelated compounds, including quinolones (4, 12, 18).
The mar operon in E. coli consists of two divergently positioned transcriptional units that flank the operator marO (for a review, see reference 1). Transcriptional unit 2 comprises marRAB, which encodes the Mar repressor MarR, the activator MarA, and a putative small protein, MarB, of unknown function (10). In the absence of an inducer, MarR represses transcription of marRAB by binding to marO, thus negatively controlling expression of other genes on the chromosome by the activator MarA (1, 40). Upon induction of the marRAB operon by a variety of compounds, including tetracycline, chloramphenicol, and salicylate, MarR repression of marRAB is alleviated. Constitutive expression of marRAB also occurs when the repressor is rendered inactive by marR mutations (1). Mar mutants exist among clinical, fluoroquinolone-resistant isolates of E. coli (25).
In this study, we investigated the relative contributions of regulatory gene and structural gene changes to fluoroquinolone resistance in E. coli, using a large number of well-characterized clinical isolates with different levels of susceptibility to fluoroquinolones and with known mutations in the regions that determine quinolone resistance in gyrA (13) and parC (14).
(Part of this study was presented at the 97th General Meeting of the American Society for Microbiology, Miami Beach, Fla., 4 to 8 May 1997 [34].)
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
A total of 138 independently obtained clinical E. coli isolates were studied (33). The fluoroquinolone-resistant isolates (ofloxacin MICs, ≥8 μg/ml) comprised 21 bloodstream isolates from different cancer centers across Europe and the Middle East (E strains), 17 bloodstream isolates cultured from hematologic-oncologic patients at Ulm University Hospital, Ulm, Germany (HO strains), and 19 strains which comprised predominantly urinary tract isolates obtained from nonhematologic patients admitted to surgical services of Ulm University Hospital (NH strains). These strains differed by pulsed-field gel electrophoresis and PCR-randomly amplified polymorphic DNA analysis (32) and were gyrA double mutants with either one or two additional mutations in parC (13, 14). DNA sequencing of gyrA and parC provided information from both strands for the regions from nucleotides 123 to 366 (Leu41 through Tyr122) of gyrA (35) and from nucleotides 145 to 492 (Lys39 through Gln138) of parC (23). The present study also included 24 E. coli isolates with intermediate levels of fluoroquinolone susceptibility (M strains; ofloxacin MICs, 0.5 to 4 μg/ml) which were obtained from nonneutropenic patients at Ulm University Hospital and comprised predominantly urinary tract isolates. Finally, 57 fluoroquinolone-susceptible isolates (S strains; ofloxacin MICs, ≤0.25 μg/ml) from cancer patients at Ulm University Hospital, 18 of which were bloodstream isolates, served as a control group. The laboratory strains and plasmids used in this study are described in Table 1. Stock cultures were kept frozen at −80°C in the Microbank system (Mast, Germany) or in 20% glycerol.
TABLE 1.
Laboratory strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Reference(s) |
|---|---|---|
| Strains | ||
| AG100 | Wild-type E. coli K-12 strain | 16 |
| AG102 | marR1 mutant of AG100, selected on tetracycline | 10, 16 |
| ASS121 | SPC105 bearing a marOII::lacZ transcriptional fusion and a 1.24-kbp BspHI marRAB operon-specific deletion; Ampr | 11, 40 |
| GC4468 | Wild-type E. coli K-12 strain (F− Δlac-4169 rpsL) | 8 |
| DJ901 | GC4468, soxRSΔ901::Tn10Kmr | 18 |
| JTG1078 | GC4468, soxR105 zjc-2204::Tn10Kmr | 17 |
| Plasmids | ||
| pMAK-TU1&TU2 | Derivative of pMAK705; temperature-sensitive cloning vector with a 2.4-kb insertion containing marRAB and marC (bp 163 to 2592) and with a marR5 mutation; low copy number; Cmlr | 43 |
| pSPOK | Derivative of pSPORT1 (Gibco/BRL) in which the bla gene was replaced with a kanamycin resistance gene; used for cloning marOR from clinical strains; high copy number; IPTG inducible | 25 |
| pSXS | Derivative of pSE380 (Invitrogen, San Diego, Calif.) containing the soxS gene on a 432 bp-fragment; high copy number; IPTG inducible | 2 |
Susceptibility testing.
The MICs of selected antimicrobial agents were determined by a standard broth microdilution procedure with cation-adjusted Mueller-Hinton broth and a final inoculum of 5 × 105 CFU/ml according to National Committee for Clinical Laboratory Standards performance and interpretive guidelines (30). Microtiter plates were purchased from Merlin Diagnostics (Bornheim, Germany). Control strains included E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853.
RNA extraction and Northern blot analysis.
Overnight cultures were diluted 100-fold in fresh Luria-Bertani (LB) broth (per liter, 10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl) and grown to the mid-logarithmic phase at 30°C with shaking. In order to induce the marRAB operon, sodium salicylate (final concentration, 5 mM) was added to half the culture for 45 min before the cells were harvested by centrifugation (11). Similarly, the sox regulon was induced by the superoxide-generating agent paraquat (final concentration, 1.3 mM; Sigma, St. Louis, Mo.) in the presence of oxygen (18). Total RNA was extracted from a 50-ml culture by a cesium chloride (CsCl) method (6), with some modifications. Cells were washed in TE buffer (50 mM Tris-HCl, 50 mM EDTA [pH 8]) and lysed in the same buffer with 3.4% sodium dodecyl sulfate (SDS). CsCl (Cabot, Revere, Pa.) was added to 67% (wt/vol), and the preparation was centrifuged for 10 min at 14,000 × g. The supernatant was loaded on a cushion of 5.7 M CsCl–100 mM EDTA and centrifuged overnight at 150,000 × g at 20°C. The RNA pellet was treated with acid-phenol–chloroform, ethanol precipitated, resuspended in water, and stored at −80°C. The concentration of total RNA in the samples was determined spectrophotometrically by a ribonucleotide assay (39) based on the orcinol reaction (Sigma). Hybridization of radiolabelled DNA probes to the membrane-bound RNA (20 μg/lane) was performed at 65°C overnight according to the specifications of the membrane manufacturer (Amersham, Arlington Heights, Ill.).
The marA probe was a 387-bp PCR fragment containing the complete marA gene amplified from AG100 chromosomal DNA. The 432-bp soxRS probe was obtained from plasmid pSXS, kindly provided by B. Demple (2), by double digestion with EcoRI and HindIII (Gibco/BRL, Gaithersburg, Md.); it contained the complete soxS gene. After agarose gel electrophoresis, probes were purified with a QIAEXII gel extraction kit (Qiagen, Chatsworth, Calif.) and labelled with [α-32P]dCTP by using a Boehringer Mannheim (Indianapolis, Ind.) random primer labeling kit. RNA blots were washed twice with 2× SSPE (0.36 M NaCl, 0.02 M sodium phosphate, 0.002 M EDTA [pH 7.7]–0.1% SDS at room temperature and twice with 1× SSPE–0.1% SDS at 65°C. Washed membranes were air dried and exposed on a PhosphorImager screen (Molecular Dynamics, Sunnyvale, Calif.), and signals were visualized with ImageQuant software (Molecular Dynamics). Northern blot analysis was performed on RNAs from at least two independent extractions.
DNA manipulations.
Total chromosomal DNA was prepared as described previously (9). The marOR region was amplified from bp 1311 to 1858 (10) in a DNA thermocycler (model 480; Perkin-Elmer Cetus, Norwalk, Conn.) with the primer pair ORAB2 and RK3, which included PstI and EcoRI restriction sites, respectively, to allow directional cloning of the PCR fragment into plasmid pSPOK (25). After purification, both DNA strands were cycle sequenced by the Tufts University DNA Sequencing Facility with the same primers. In special cases, sequencing was repeated with a different PCR DNA batch to check for errors introduced during PCR. Recombinant DNA techniques, transformation, and restriction enzyme digestions were performed by standard techniques (38). Transformation of pMAK-TU1&TU2 (Table 1) into the clinical isolate NH10 was performed by electroporation with a gene pulser apparatus (Bio-Rad, Richmond, Calif.).
Test for MarR function with a marO::lacZ fusion.
Overnight cultures of ASS121 strains bearing pSPOK, with or without different cloned marOR sequences, were diluted 1:100 in fresh LB broth with 100 μg of ampicillin per ml, 50 μg of kanamycin per ml, and 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Alexis Corporation, Läufelfingen, Switzerland). Following growth at 30°C to late logarithmic phase, the cells were assayed in triplicate for β-galactosidase activity with a chemiluminescence assay kit (Tropix, Bedford, Mass.). Cells were solubilized with chloroform-SDS and diluted 103-fold. Twenty microliters of each dilution was added to 200 μl of reaction buffer containing the substrate Galacton (Tropix) and incubated for 60 min at room temperature. After the reaction was terminated by the addition of Emerald enhancer, the chemiluminescent signal was measured in a OptocompI luminometer (MGM Instruments, Hamden, Conn.). Data were expressed as relative light units/A530 and referred to the chemiluminescence of ASS121 bearing SPOK without the insert as a percentage of the control.
RESULTS
Identification of point mutations in marR that are causes for marA overexpression in clinical isolates of E. coli.
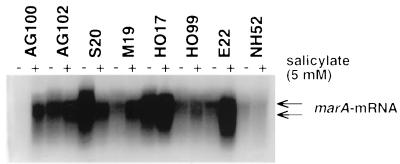
Since all known Mar mutants studied thus far have been shown to be cyclohexane tolerant (5, 43), we screened our collection of 138 clinical E. coli isolates for cyclohexane tolerance. We detected increased organic solvent tolerance in 1 of 57 fluoroquinolone-susceptible, in 3 of 24 low-level fluoroquinolone-resistant, and in 21 of 57 high-level fluoroquinolone-resistant E. coli clinical isolates (33). Northern blot analysis of RNAs harvested from these 25 cyclohexane-tolerant and 11 cyclohexane-susceptible, fluoroquinolone-resistant strains (some of the latter strains carried mutations in gyrA and parC which were identical to mutations in certain cyclohexane-tolerant strains) showed that six strains constitutively expressed marA (Fig. 1). One Mar mutant (NH52) was an exception in that it was cyclohexane susceptible (Table 2). Sequencing data were compared with the marOR sequence derived from AG100 (accession no. M96235 [10]) and showed six missense mutations (three transversions and three transitions) and two nonsense mutations among the six clinical E. coli isolates overexpressing marA (Table 2). The latter mutations were insertion of an early stop codon in strain M19 and a frameshift mutation in strain NH52. Since the sequences of NH52, HO99, and E22 differed by only one nucleotide from that of wild-type marOR, we repeated PCRs from the original frozen stocks of these strains and obtained identical results upon resequencing.
FIG. 1.
Northern blot analysis of marRAB mRNAs prepared from clinical E. coli strains incubated without (−) and with (+) 5 mM sodium salicylate for 45 min. RNA samples were transferred to Hybond-N+ membranes and probed with radioactively labelled marA. Arrows point to prominent transcripts of ∼1.1 and ∼0.9 kbp.
TABLE 2.
Sequences of mutant marR genes in six clinical Mar mutants with different ofloxacin susceptibilities
| Straina | Ofloxacin MIC (μg/ml) | Cyclo- hexane toleranceb | Mutation in marR (nucleotide changed) at amino acidc:
|
|||||
|---|---|---|---|---|---|---|---|---|
| Glu31 | Ile49 | Leu78 | Arg94 | Gly103 | Tyr137 | |||
| S20 | 0.25 | + | Ser (1724C→A) | Ser (1751G→A) | His (1853T→C) | |||
| M19 | 2 | + | Stop codon inserted (1535G→T) | Ser (1751G→A) | His (1853T→C) | |||
| NH52 | 8 | − | Frameshift (Δ1751d) | |||||
| HO17 | 32 | + | His (1725G→A) | Ser (1751G→A) | His (1853T→C) | |||
| HO99 | 32 | + | Met (1676C→A) | |||||
| E22 | 64 | + | Ser (1590T→G) | |||||
MICs indicating levels of fluoroquinolone resistance ranged from 0.25 through 64 μg/ml among the Mar mutants (Table 2). Strain S20 carried no gyrA and parC mutations, strain M19 had a Ser83→Leu mutation in gyrA, and strain NH52 had a Ser83→Leu and an Asp87→Asn mutation in gyrA and a Ser80→Ile mutation in parC. Likewise, strains HO17, HO99, and E22 were gyrA double mutants and had an additional parC mutation (13) (see Table 4).
TABLE 4.
Overexpression of marA and soxS in clinical fluoroquiolone-resistant E. coli strains with identical mutations in the regions determining quinolone resistance in gyrA and parC
| Straina | Ofloxacin MIC (μg/ml) | Cyclohexane toleranceb | Mutation inc:
|
Overexpression of marA | Overexpression of soxS | |||
|---|---|---|---|---|---|---|---|---|
|
gyrA at amino acid:
|
parC at amino acid:
|
|||||||
| Ser83 | Asp87 | Ser80 | Glu84 | |||||
| E5 | 8 | − | Leu | Asn | Lys | − | − | |
| E10d | 64 | − | Leu | Asn | Lys | − | − | |
| NH1 | 8 | − | Leu | Gly | Ile | − | − | |
| HO17 | 32 | + | Leu | Gly | Ile | + | − | |
| HO12 | 8 | − | Leu | Asn | Ile | − | − | |
| HO13d | 32 | − | Leu | Asn | Ile | − | − | |
| E7 | 32 | + | Leu | Asn | Ile | − | − | |
| HO99 | 32 | + | Leu | Asn | Ile | + | − | |
| E3 | 64 | + | Leu | Asn | Ile | − | + | |
| E19 | 64 | + | Leu | Asn | Ile | − | + | |
| E22 | 64 | + | Leu | Asn | Ile | + | − | |
The strains, including HO strains, are genotypically unrelated (32), and they are all bloodstream isolates except for NH1, which is a urinary tract isolate.
Cyclohexane tolerance of strains was tested on LB agar overlaid by the organic solvent and grown for 24 h at 30°C (33).
See reference 10.
Increased fluoroquinolone resistance not associated with cyclohexane tolerance.
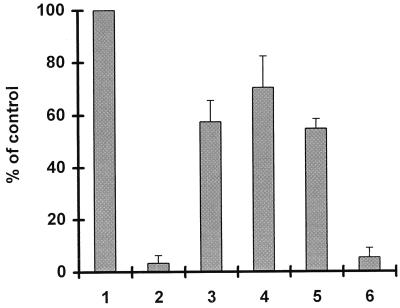
We cloned the marOR fragments of three of the Mar mutants (HO17, NH52, and M19) in an IPTG-inducible vector and tested the inhibitory function of their gene products on a transcriptional marO::lacZ fusion in the Δmar strain ASS121 (Table 1). The control strain (ASS121 with pSPOK without the insert) showed high β-galactosidase activity, which was defined as 100% activity (Fig. 2, bar 1). Introduction of wild-type marOR, derived from AG100, into ASS121 inhibited β-galactosidase activity almost completely (≤5% residual activity) (Fig. 2, bar 2). In contrast, suppression by marOR fragments from clinical Mar mutants was weak and yielded between 57 and 70% residual β-galactosidase activity (Fig. 2, bars 3 through 5), confirming low repressor activity. Of importance, the comparison of HO4 with HO17 showed that the R94→H mutation decreased the function of MarR but that the mutations at amino acids (aa) 103 and 137 did not.
FIG. 2.
Reporter gene assay for MarR function. The host strain ASS121, which lacks the mar and lac loci, carries a chromosomal marO::lacZ transcriptional fusion (40). The effect of the introduction of pSPOK carrying different marR genes on β-galactosidase activity was determined. Cells were grown for 5 h at 30°C in the presence of IPTG, and β-galactosidase activity was measured in triplicate cultures. Results are expressed as percentages of values determined for the control (ASS121 bearing pSPOK without the insert) and are the means and standard deviations of results from three to five consecutive assays. The origins of the cloned marR genes were as follows: bar 1, none; bar 2, AG100 (wild type); bar 3, HO17 (R94→H, G103→S, Y137→H); bar 4, M19 (G31 stop codon); bar 5, NH52 (G103 frameshift); and bar 6, HO4 (G103→S, Y137→H).
High frequency of strains with nucleotide sequences of marOR divergent from sequences in the GenBank database.
Twelve of 35 sequenced strains had a marOR nucleotide sequence identical to that of the E. coli K-12 strain AG100 (Table 3). In contrast, 20 of the 35 strains, including the three Mar mutants S20, M19, and HO17 (Table 2), had nucleotide changes which always occurred in combination: 1332A→C in marO and 1751G→A (Gly103→Ser) and 1853T→C (Tyr137→His) in marR (Tables 2 and 3). These mutations by themselves did not interfere with the wild-type function of the repressor MarR, since marA overexpression was not seen by Northern blot analysis in several strains carrying the mutations. When the function of the divergent MarR (Gly103→Ser and Tyr137→His) was studied in the above-described reporter gene assay, the gene products of two representative strains (HO4 and E1) were able to suppress β-galactosidase activity to a residual level of about 5% (e.g., Fig. 2, bar 6). Hence, we conclude that the observed marOR sequences represent a variant genotype of E. coli without loss of MarR function.
TABLE 3.
Sequences of functionally active marR genes in 29 clinical fluoroquinolone-resistant E. coli
| No. of strains | Mutation in marR (nucleotide changed) at amino acida
|
|||
|---|---|---|---|---|
| Ser3 | Ala53 | Gly103 | Tyr137 | |
| 12 | ||||
| 15 | Ser (1751G→A) | His (1853T→C) | ||
| 1 | Asn (1452G→A) | Ser (1751G→A) | His (1853T→C) | |
| 1 | Glu (1602C→A) | Ser (1751G→A) | His (1853T→C) | |
See reference 10.
Among strains with the variant genotype were five strains with one additional point mutation in MarR (Table 2). We infer that in the three Mar mutants S20, M19, and HO17, Arg94→Ser, Glu31→stop codon, and Arg94→His, respectively, were responsible for the loss of MarR function. In contrast, two other amino acid changes, Ser3→Asn and Ala53→Glu, were seen in strains with wild-type levels of expression of marA (Table 3).
Clinical isolates of E. coli overexpressing soxS.
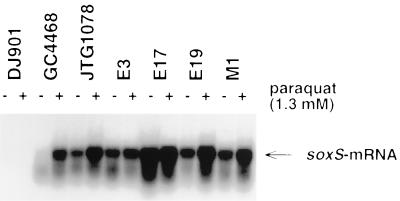
Expression of soxS mRNA in the presence of oxygen by vigorous shaking was investigated for all 25 cyclohexane-resistant strains and for the 11 cyclohexane-susceptible strains. RNA from the soxRS-deleted strain DJ901 and from the soxS-overexpressing strain JTG1078 served as negative and positive controls, respectively (Table 1; Fig. 3). Northern blot analysis demonstrated overexpression of soxS in four of the clinical isolates relative to expression in the control strains (Fig. 3). All Sox mutants were cyclohexane resistant. For three strains, ofloxacin MICs were high (64 μg/ml). gyrA and parC data are shown in Table 4 for strains E3 and E19. Strain E17 was a gyrA double mutant (Ser83→Leu, Asp87→Asn) with a Ser80→Arg mutation in parC. Strain M1, which is intermediately resistant to fluoroquinolone (ofloxacin MIC, 4 μg/ml), carried a Ser80→Leu mutation in gyrA.
FIG. 3.
Northern blot analysis of soxS mRNAs prepared from clinical E. coli strains incubated without (−) and with (+) 1.3 mM paraquat for 45 min. RNA samples were transferred to Hybond-N+ membranes and probed with radioactively labelled soxS. The arrow designates the ∼400-bp hybridizing band.
Expression of marA or soxS among fluoroquinolone-resistant strains with identical mutations in gyrA and parC.
For strains with the same mutations in gyrA and parC, oflaxacin MICs were nevertheless quite different (Table 4). For five strains overexpressing marA (HO17, HO99, and E22) or soxS (E3 and E19), ofloxacin MICs were four- to eightfold higher than those for strains matched for their gyrA and parC mutations but with wild-type levels of expression of these regulatory genes (NH1 and HO12). Three strains (E10, HO13, and E7) showed increased fluoroquinolone resistance without overexpressing marA or soxS. Two of these strains, E10 and HO13, were also cyclohexane susceptible. The basis of the increased fluoroquinolone resistance in these three strains is still unknown. The contribution of marA and soxS to the higher level of fluoroquinolone resistance in the topoisomerase mutants was not directly testable because of a lack of available antibiotic resistance markers for inactivation of the genes in these strains. However, transformation of the clinical topoisomerase mutant, non-Mar strain NH10 with pMAK-TU1&TU2, which specifies marA overexpression in trans, resulted in a twofold increase in the MIC of ofloxacin (16 versus 8 μg/ml) and a fourfold increase in the MIC of pefloxacin (64 versus 16 μg/ml) in conjunction with a newly observed cyclohexane tolerance. Thus, overexpression of marA can result in two- to fourfold increased resistance to fluoroquinolone compared with that mediated by topoisomerase mutations.
DISCUSSION
Resistance to the fluoroquinolones in E. coli is principally caused by mutations in the structural genes for topoisomerase II (gyrA and gyrB) and topoisomerase IV (parC and parE) (13, 14, 19, 21, 29, 37, 42) and by mutations affecting regulatory genes, namely, marA (10, 12), soxS (2, 44), and rob (4). In a previous study of 46 ofloxacin-resistant clinical E. coli strains, we noted a wide range of ofloxacin MICs for strains with identical mutations in gyrA and parC (13, 14). Similarly, among another 36 E. coli strains recently described, as many as 22 (61%) had higher levels of fluoroquinolone resistance than was expected from the mutations in the topoisomerase genes alone (15).
We adopted a screening approach to investigate the possible involvement of the regulatory genes marA and soxS in fluoroquinolone resistance in clinical E. coli isolates. Since overexpression of these loci has been linked to increased organic solvent tolerance as well as fluoroquinolone resistance (5, 27, 28, 43), we tested the strains for cyclohexane tolerance. This phenotype was significantly more frequent among the fluoroquinolone-resistant strains than among the fluoroquinolone-susceptible controls and was also associated with a higher level of resistance (33). Among 25 cyclohexane-tolerant strains detected by the screen (33) and 11 cyclohexane-susceptible strains used as controls, we found 6 Mar mutants and 4 Sox mutants in which the level of fluoroquinolone resistance was higher than was attributable to mutations in the structural genes gyrA and parC. We infer that overexpression of these regulatory genes enhanced fluoroquinolone resistance. However, other possibilities include mutations in the structural genes for the second subunits of gyrase, gyrB (29, 36, 45), and topoisomerase V, parE (7), although these appear to be rare events and would not explain the organic solvent tolerance.
Unexpectedly, one of the Mar mutants was found by its cyclohexane tolerance among clinical fluoroquinolone-susceptible E. coli strains. Although the strain was classified as fluoroquinolone-susceptible E. coli, the ofloxacin MIC of 0.25 μg/ml for this strain was already beyond the MIC at which 90% of fluoroquinolone-susceptible strains are inhibited and its antibiotic profile was characteristic of a Mar mutant, such as AG102 (16). Sequencing of gyrA and parC revealed no mutations in the quinolone-resistance-determining regions. We conclude that the small increase in fluoroquinolone resistance was due to overexpression of marA, as has been shown for the laboratory strain AG102 (12). Further studies are needed to determine whether the observed frequency of about 2% Mar mutants among apparent fluoroquinolone-susceptible E. coli strains cultured from hospital inpatients corresponds to the background level of Mar mutants.
Overexpression of marA in the clinical Mar mutants was due exclusively to point mutations in marR. Our previous study (25) described deletions in all three Mar mutants, accompanied by single amino acid changes in two of the mutants. Of interest, the point mutations found in the present study affected amino acids at highly conserved positions (aa 49 and 78) or a completely conserved position (aa 94) in the newly recognized family of MarR homologs (1, 26). In addition to the mutations that alleviated marR repression, we identified a small deletion in marO (from nucleotides 1369 to 1373) and four amino acid changes in marR without loss of repressor activity (Ser3→Asn, Ala53→Glu, Gly103→Ser, and Tyr137→His). Two of the last mutations, Gly103→Ser and Ser3→Asn, had previously been reported for two clinical Mar mutants (KM-D and J28, respectively) along with nucleotide deletions (25). It now appears that the nucleotide deletions and not the single amino acid changes are the cause of the Mar phenotype in these strains. The high frequency of the amino acid changes Gly103→Ser and Tyr137→His in the clinical strains tested suggests that the underlying nucleotide changes are genotypic variations without a change of phenotype.
The two-component soxRS regulatory system is involved in the adaptive response of E. coli to superoxide stress (2, 18, 41, 44). SoxR acts as the sensor and transcriptional activator of SoxS, which in turn activates a number of superoxide stress as well as antibiotic resistance genes (2, 31, 44). Four clinical E. coli strains displayed high constitutive expression of soxS, which was increased even further by paraquat induction (Fig. 3). Since constitutive expression of the sox locus follows mutations in soxR (2, 41, 44), we suspect that overexpression of soxS in the clinical strains is linked to mutations in soxR. That none of the clinical strains were both Mar and Sox mutants may relate to the fact that both regulatory systems control the expression of overlapping sets of target genes (3, 26).
Recent data suggest that a double mutation in gyrA plus a mutation at parC confers a ciprofloxacin MIC of 8 μg/ml (14, 15, 42). This finding indicates that additional mechanisms contribute to the fluoroquinolone resistance phenotype in about a third of fluoroquinolone-resistant E. coli isolates (14, 15, 42). In our study, half of the strains for which fluoroquinolone MICs were unexplainably high were Mar or Sox mutants (Table 4). The results of previous studies (22, 25) and the increase in fluoroquinolone MICs by marA overexpression in trans in the clinical strain NH10 are consistent with the hypothesis that removal of marA decreases fluoroquinolone MICs for the Mar strains to the lower level seen in the topoisomerase mutants which have a wild-type marRAB. Our conclusion is corroborated by recent work of Heisig and Wiedemann (20), who investigated the quinolone-resistance-determining region of KM-F, one of our previously reported Mar mutants (25). They found one mutation each in gyrA, gyrB, and parC (20). Upon deletion of mar by a kanamycin cassette (25), the ciprofloxacin MIC dropped by two dilutional steps, from 32 to 8 μg/ml (20). However, since the clinical strains of the present study were not isogenic (32), one cannot exclude the possibility that other differences between strains contributed to the level of fluoroquinolone resistance. Mutations in acrAB (24, 43) or rob (4), for instance, would affect both drug and organic solvent resistance and may account for the increased fluoroquinolone resistance in some of the other strains.
The Mar mutant NH52 deserves attention since it was cyclohexane susceptible despite overexpressing marA. The strain may be defective in one of the structural genes, such as acrAB, that is involved in mar-mediated cyclohexane tolerance (43). Alternatively, its wild-type tolerance to organic solvents, without overexpressing marA, might have been low for other reasons. Two of the 138 strains studied here were E. coli strains which do not grow with hexane (33). Both hypotheses would fit the observation that strain NH52 has wild-type levels of susceptibility to chloramphenicol and tetracycline.
The exact incidence of Mar or Sox mutants among clinical isolates of E. coli remains unknown. We may have missed some mutants like NH52 in the organic solvent screen, although this number would be very small. It appears, however, that mutations in the regulatory genes marA and soxS play a role in about 10 to 15% of clinical fluoroquinolone-resistant E. coli strains, which is in line with the results of our previous study (25). Mar and Sox mutants which have a higher level of fluoroquinolone resistance than expected from mutations in the structural genes gyrA and parC will likely be found among other strains. Our data support the hypothesis that chromosomal loci other than gyrA and parC contribute to fluoroquinolone resistance in a substantial number of clinical E. coli isolates.
ACKNOWLEDGMENTS
This study was supported in part by research grants from the U.S. Public Health Service (GM 51661 to S.B.L.), the Deutsche Forschungsgemeinschaft (Oe 195/1-1 to M.O.), and the University of Ulm (P172/1994 to M.O. and W.V.K.).
We thank Bruce Demple for helpful discussion of results and Laura McMurry and Patrick McDermott for invaluable comments during the preparation of the manuscript. The expert help of A. S. Ritter is greatly appreciated.
REFERENCES
- 1.Alekshun M N, Levy S B. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amábile-Cuevas C F, Demple B. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ariza R R, Cohen S P, Bachhawat N, Levy S B, Demple B. Repressor mutations in the marRAB operon that activate oxidative stress genes and multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1994;176:143–148. doi: 10.1128/jb.176.1.143-148.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariza R R, Li Z, Ringstad N, Demple B. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J Bacteriol. 1995;177:1655–1661. doi: 10.1128/jb.177.7.1655-1661.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asako H, Nakajima H, Kobayashi K, Kobayashi M, Aono R. Organic solvent tolerance and antibiotic resistance increased by overexpression of marA in Escherichia coli. Appl Environ Microbiol. 1997;63:1428–1433. doi: 10.1128/aem.63.4.1428-1433.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel R M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1996. pp. 4.4.1–4.4.7. [Google Scholar]
- 7.Breines D M, Ouabdesselam S, Ng E Y, Tankovic J, Shah S, Soussy C J, Hooper D C. Quinolone resistance locus nfxD of Escherichia coli is a mutant allele of the parE gene encoding a subunit of topoisomerase IV. Antimicrob Agents Chemother. 1997;41:175–179. doi: 10.1128/aac.41.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W, Kuo T. A simple and rapid method for the preparation of gram-negative genomic DNA. Nucleic Acids Res. 1993;21:2260. doi: 10.1093/nar/21.9.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen S P, Hächler H, Levy S B. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen S P, Levy S B, Foulds J, Rosner J L. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol. 1993;175:7856–7862. doi: 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen S P, McMurry L M, Hooper D C, Wolfson J S, Levy S B. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob Agents Chemother. 1989;33:1318–1325. doi: 10.1128/aac.33.8.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conrad S, Oethinger M, Kaifel K, Klotz G, Marre R, Kern W V. gyrA mutations in high-level fluoroquinolone-resistant Escherichia coli clinical isolates. J Antimicrob Chemother. 1996;38:443–455. doi: 10.1093/jac/38.3.443. [DOI] [PubMed] [Google Scholar]
- 14.Conrad S, Scheit L, Oethinger M, Klotz G, Marre R, Kern W V. Abstracts of the 36th Interscience Conference of Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1996. gyrA and parC mutations in high-level fluoroquinolone-resistant Escherichia coli clinical isolates, abstr. C9; p. 35. [Google Scholar]
- 15.Everett M J, Jin Y F, Ricci V, Piddock L V. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob Agents Chemother. 1996;40:2380–2386. doi: 10.1128/aac.40.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.George A M, Levy S B. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol. 1983;155:531–540. doi: 10.1128/jb.155.2.531-540.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenberg J T, Chou J H, Monach P A, Demple B. Activation of oxidative stress genes by mutations at the soxQ/cfxB/marA locus in Escherichia coli. J Bacteriol. 1991;173:4433–4439. doi: 10.1128/jb.173.14.4433-4439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg J T, Monach P, Chou J H, Josephy P D, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heisig P. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 1996;40:879–885. doi: 10.1128/aac.40.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heisig P, Wiedemann B. Abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. In vitro activity of the new quinolone Bay 12-8039 against defined mutants of Escherichia coli and Staphylococcus aureus, abstr. F-140; p. 169. [Google Scholar]
- 21.Hooper D C, Wolfson J S, Ng E Y, Swartz M N. Mechanisms of action of and resistance to ciprofloxacin. Am J Med. 1987;82:12–20. [PubMed] [Google Scholar]
- 22.Hüllen V, Heisig P, Wiedemann B. Bedeutung des marR-Genes für die klinische Resistenz von E. coli gegenüber fluorierten Chinolonen, abstr. Sy 9.2. Chemotherapie J. 1997;15:12. [Google Scholar]
- 23.Kato J-I, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 24.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 25.Maneewannakul K, Levy S B. Identification of mar mutants among quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1996;40:1695–1698. doi: 10.1128/aac.40.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller P F, Sulavik M C. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol Microbiol. 1996;21:441–448. doi: 10.1111/j.1365-2958.1996.tb02553.x. [DOI] [PubMed] [Google Scholar]
- 27.Nakajima H, Kobayashi K, Kobayashi M, Asako H, Aono R. Overexpression of the robA gene increases organic solvent tolerance and multiple antibiotic and heavy metal ion resistance in Escherichia coli. Appl Environ Microbiol. 1995;61:2302–2307. doi: 10.1128/aem.61.6.2302-2307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakajima H, Kobayashi M, Negishi T, Aono R. soxRS gene increased the level of organic solvent tolerance in Escherichia coli. Biosci Biotechnol Biochem. 1995;59:1323–1325. doi: 10.1271/bbb.59.1323. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura S, Nakamura M, Kojima T, Yoshida H. gyrA and gyrB mutations in quinolone-resistant strains of Escherichia coli. Antimicrob Agents Chemother. 1989;33:254–255. doi: 10.1128/aac.33.2.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing; fifth informational supplement. M100-S5. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1994. [Google Scholar]
- 31.Nunoshiba T, Hidalgo E, Amábile-Cuevas C F, Demple B. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J Bacteriol. 1992;174:6054–6060. doi: 10.1128/jb.174.19.6054-6060.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oethinger M, Conrad S, Kaifel K, Cometta A, Bille J, Klotz G, Glauser M P, Marre R, Kern W V the International Antimicrobial Therapy Cooperative Group of the European Organization for Research and Treatment of Cancer. Molecular epidemiology of fluoroquinolone-resistant Escherichia coli bloodstream isolates from patients admitted to European cancer centers. Antimicrob Agents Chemother. 1996;40:387–392. doi: 10.1128/aac.40.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oethinger M, Kern W V, Goldman J D, Levy S B. Association of organic solvent tolerance and fluoroquinolone resistance in clinical isolates of Escherichia coli. J Antimicrob Chemother. 1998;41:111–114. doi: 10.1093/jac/41.1.111. [DOI] [PubMed] [Google Scholar]
- 34.Oethinger M, Kern W V, Podglajen I, Levy S B. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. The multiple antibiotic resistance (mar) locus in fluoroquinolone-resistant blood isolates of E. coli, abstr. A-71; p. 13. [Google Scholar]
- 35.Oram M, Fisher L M. 4-Quinolone resistance mutations in the DNA gyrase of Escherichia coli clinical isolates identified by using the polymerase chain reaction. Antimicrob Agents Chemother. 1991;35:387–389. doi: 10.1128/aac.35.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouabdesselam S, Hooper D C, Tankovic J, Soussy C J. Detection of gyrA and gyrB mutations in quinolone-resistant clinical isolates of Escherichia coli by single-strand conformational polymorphism analysis and determination of levels of resistance conferred by two different single gyrA mutations. Antimicrob Agents Chemother. 1995;39:1667–1670. doi: 10.1128/aac.39.8.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piddock L J V. Mechanisms of resistance to fluoroquinolones: state-of-the-art 1992–1994. Drugs. 1995;49:29–35. doi: 10.2165/00003495-199500492-00006. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Schneider W C. Determination of nucleic acids in tissues by pentose analysis. Methods Enzymol. 1957;3:680–684. [Google Scholar]
- 40.Seoane A S, Levy S B. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J Bacteriol. 1995;177:3414–3419. doi: 10.1128/jb.177.12.3414-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsaneva I R, Weiss B. soxS, a locus governing a superoxide response regulon in Escherichia coli K-12. J Bacteriol. 1990;172:4197–4205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vila J, Ruiz J, Marcos A, Jimenez de Anta T. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1996;40:491–493. doi: 10.1128/aac.40.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White D G, Goldman J D, Demple B, Levy S B. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, and robA in Escherichia coli. J Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J, Weiss B. Two divergently transcribed genes, soxR and soxS, control a superoxide response regulon of Escherichia coli. J Bacteriol. 1991;173:2864–2871. doi: 10.1128/jb.173.9.2864-2871.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida H, Bogaki M, Nakamura M, Yamanaka L, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob Agents Chemother. 1991;35:1647–1650. doi: 10.1128/aac.35.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]