Abstract
Background
B vitamins such as folate (B9), B6, and B12 are key in one carbon metabolism, which generates methyl donors for DNA methylation. Several studies have linked differential methylation to self-reported intakes of folate and B12, but these estimates can be imprecise, while metabolomic biomarkers can offer an objective assessment of dietary intakes. We explored blood metabolomic biomarkers of folate and vitamins B6 and B12, to carry out epigenome-wide analyses across up to three European cohorts. Associations between self-reported habitual daily B vitamin intakes and 756 metabolites (Metabolon Inc.) were assessed in serum samples from 1064 UK participants from the TwinsUK cohort. The identified B vitamin metabolomic biomarkers were then used in epigenome-wide association tests with fasting blood DNA methylation levels at 430,768 sites from the Infinium HumanMethylation450 BeadChip in blood samples from 2182 European participants from the TwinsUK and KORA cohorts. Candidate signals were explored for metabolite associations with gene expression levels in a subset of the TwinsUK sample (n = 297). Metabolomic biomarker epigenetic associations were also compared with epigenetic associations of self-reported habitual B vitamin intakes in samples from 2294 European participants.
Results
Eighteen metabolites were associated with B vitamin intakes after correction for multiple testing (Bonferroni-adj. p < 0.05), of which 7 metabolites were available in both cohorts and tested for epigenome-wide association. Three metabolites — pipecolate (metabolomic biomarker of B6 and folate intakes), pyridoxate (marker of B6 and folate) and docosahexaenoate (DHA, marker of B6) — were associated with 10, 3 and 1 differentially methylated positions (DMPs), respectively. The strongest association was observed between DHA and DMP cg03440556 in the SCD gene (effect = 0.093 ± 0.016, p = 4.07E−09). Pyridoxate, a catabolic product of vitamin B6, was inversely associated with CpG methylation near the SLC1A5 gene promoter region (cg02711608 and cg22304262) and with SLC7A11 (cg06690548), but not with corresponding changes in gene expression levels. The self-reported intake of folate and vitamin B6 had consistent but non-significant associations with the epigenetic signals.
Conclusion
Metabolomic biomarkers are a valuable approach to investigate the effects of dietary B vitamin intake on the human epigenome.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-023-01578-7.
Keywords: DNA methylation, Metabolomics, Biomarkers, B vitamins, Folate, Diet
Background
DNA methylation (DNAm) is an important epigenetic mechanism in development and over the lifecourse. In mammals, DNAm typically occurs through the transfer of methyl groups from S-adenosylmethionine (SAM) to cytosine residues at CpG dinucleotides. SAM is the product of one carbon metabolism, which includes the folate (B9) and methionine cycles and utilizes nutrients as substrates [1, 2]. SAM and DNAm levels are influenced by diet and the intake of nutrients [2], particularly B vitamins. Vitamins B6 and B12 are cofactors involved in the regulation of the catalytic activity of enzymes from the folate cycle where folate is the main substrate [3]. DNA methyltransferases (DNMTs) convert SAM into S-Adenosylhomocysteine (SAH), a metabolic precursor of homocysteine (hcy), and folate and vitamins B6 and B12 in the diet can reduce serum homocysteine (hcy-s) levels and promote its re-methylation to methionine [4]. In contrast, hcy accumulation and hyper-homocysteinemia can arise from nutritional deficiencies of B vitamins and lead to DNMT inhibition and DNA hypomethylation [5, 6].
Recently, two large-scale studies explored evidence for epigenome-wide association between self-reported B vitamin intakes and blood-based DNA methylation profiles. Chamberlain et al. [7] explored differential methylation with dietary intakes estimated from food frequency questionnaires (FFQs) in 5186 adult participants from the Melbourne Collaborative Cohort Study, reporting one association with B2 intake. Mandaviya et al. [8] also explored methylation associations with B vitamin intakes estimated from FFQs in a meta-analysis of 5841 participants across 10 European and North American cohorts, identifying multiple differentially methylated positions (DMPs) and regions associated with folate and B12 intakes. All but one signal showed an inverse correlation between folate intakes and whole blood DNAm levels. Overall, there is relatively modest overlap across the results from the two studies in adults [9], which may in part be attributed to differences in study design and methodology, or weak association between dietary intake of B vitamins and DNA methylation. A further study by Joubert et al. [10] reported associations between maternal intake of B vitamins in pregnancy and blood based methylation in newborns, but these results are not replicated in samples from adult participants.
FFQs are commonly used to assess habitual nutrient intakes in epidemiological studies due to their practicality for regular assessment of diets over time [11], but they also have limitations. By design, FFQs include a finite list of food items and portion sizes, and have limited specificity on food preparation and types of food [12]. Moreover, food intake greatly depends on ethnic, social, and cultural background and FFQs need to be well-tailored to the study population [11]. FFQs also suffer from social desirability bias where participants omit specific foods and beverages, therefore misreporting can occur. Further imprecision in the estimation of nutrient intakes from FFQ derived food intake estimates originate from the application of food composition databases [12].
Inaccurate dietary assessments may limit our understanding of the impact of B vitamins on DNAm. In contrast, biochemical markers may provide more accurate measures of specific aspects of dietary intake for the time point of biospecimen collection [13]. Metabolomics—the global assessment of all metabolites present in a biological sample—has major value for biomarker discovery in nutrition. Multiple cohort and intervention studies have identified metabolomic biomarkers of dietary patterns, foods and beverages such as tea, coffee, wine, cocoa, citrus fruits, fish, red meat, whole-grain products and more [14, 15]. Recently, Posma et al. [16] showed that urinary metabotypes collected three weeks apart are more stable than 24h dietary recalls, and that up to 67 nutrients, including folate and vitamin B6, can influence the urinary metabotype of participants.
In this study we aimed to use blood metabolomic biomarkers to investigate the effects of dietary B vitamins on blood DNAm variation. We first identified blood metabolomic biomarkers of folate and vitamins B6 and B12 dietary intakes in population-based samples from the UK, and subsequently explored their metabolomic-epigenetic associations in European cohorts. The metabolomic-epigenetic associations were compared with epigenetic associations of self-reported dietary B vitamin estimates in the current study and from previous work.
Results
Serum metabolites related to the intake of B vitamins were identified for use in downstream epigenetic association analyses aiming to detect differentially methylated signals related to B vitamin intakes in > 2000 participants of European ancestry (Fig. 1).
Fig. 1.
Data analysis workflow and main results from this study
B vitamin metabolomic biomarker discovery
Discovery of blood metabolomic biomarkers related to the intake of folate and vitamins B6 and B12 was conducted in 1064 samples from the TwinsUK cohort (Additional file 1: Table S1). Thirty-one metabolites were associated with the intake of one or more B vitamins (Bonferroni-adjusted p < 0.05; Additional file 1: Table S2). Of these, 18 metabolites were annotated to a known biochemical compound and were explored in downstream analyses.
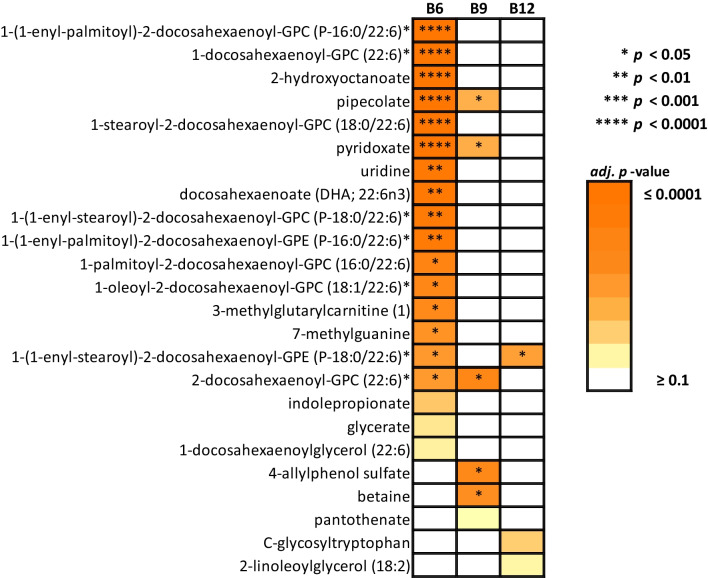
Among the 18 metabolites identified, 16, 5 and 1 metabolites were associated with the intakes of folate and vitamins B6 and B12, respectively (Bonferroni-adj. p < 0.05; Fig. 2). Compounds 2-docosahexaenoyl-GPC (22:6)*, pyridoxate and pipecolate were associated with both folate and vitamin B6, and 1-(1-enyl-stearoyl)-2-docosahexaenoyl-GPE (P-18:0/22:6)* was associated with both vitamins B6 and B12. Other compounds were associated with one B vitamin alone after multiple testing correction. Pyridoxate and pipecolate levels were not correlated with the levels of other biomarkers identified and fatty acid molecules had varying degrees of direct correlation with each other (Additional file 2: Figure S1).
Fig. 2.
Peak metabolomic associations for folate (B9) and vitamins B6 and B12 habitual intakes in the Metabolon platform (Bonferroni-adj. p < 0.1)
The strongest metabolomic associations were observed for vitamin B6 with 1-(1-enyl-palmitoyl)-2-docosahexaenoyl-GPC (P-16:0/22:6)* (b = 0.335 ± 0.058, adj. p = 5.09E−06, R2c = 0.591), followed by 1-docosahexaenoyl-GPC (22:6)* (b = 0.317 ± 0.058, adj. p = 4.88E−05, R2c = 0.480), 2-hydroxyoctanoate (b = − 0.313 ± 0.060, adj. p = 1.55E−04, R2c = 0.451) and pipecolate (b = 0.296 ± 0.059, adj. p = 4.76E−04, R2c = 0.489). The strongest association identified for folate was with 2-docosahexaenoyl-GPC (22:6)* (b = 0.001 ± 0.0003, adj. p = 0.012, R2c = 0.400), and the only compound associated with B12 was 1-(1-enyl-stearoyl)-2-docosahexaenoyl-GPE (P-18:0/22:6)* (b = 0.055 ± 0.013, adj. p = 0.030, R2c = 0.599). Docosahexaenoate (DHA; 22:6n3) was associated with vitamin B6 (b = 0.268 ± 0.057, adj. p = 2.53E-03, R2c = 0.519).
Of the 18 metabolites identified in our main analysis, 11 were annotated to the lipid superpathway with 5 of them playing a role in phospholipid metabolism and two belonging to the plasmalogen or lysolipid subpathways (Additional file 2: Figure S2). Other metabolic pathways found in our results included nucleotide and amino acid pathways. Pyridoxate — associated with vitamin B6 (b = 0.290 ± 0.059, adj. p = 8.34E-04, R2c = 0.407) and to a lesser extent with folate (b = 0.0013 ± 0.0003, adj. p = 0.039, R2c = 0.388) intakes — was annotated to the vitamin B6 metabolism pathway.
Sensitivity analyses
Three sensitivity analyses were carried out to assess the specificity of the 18 blood metabolic biomarkers of B vitamin intakes. First, we explored whether total energy intake and overall diet quality, estimated using the ‘AHEI-2010’ diet score [17], affected the biomarker results.
All 18 metabolomic associations reported in the main analysis remained nominally significant after adjusting for diet quality and energy intake (p < 0.05) (Additional file 1: Table S3), and 10 metabolomic associations remained significant after multiple testing correction, including 1-docosahexaenoyl-GPC (22:6)*, pyridoxate, uridine and DHA metabolites later on used in the downstream epigenome-wide association analysis. New associations were also identified in the sensitivity analysis, including folate intake associated with theanine and vitamin B12 associated with five other metabolites (Additional file 1: Table S3).
Second, we assessed the specificity of the 18 metabolite biomarkers of B vitamin intakes, by testing their association with the intake of 38 other nutrients estimated from FFQs (see Methods). Metabolomic associations with other nutrients were identified (Additional file 1: Table S4), and all B vitamin metabolomic biomarkers that were assessed in the downstream epigenome-wide association analysis below (1-docosahexaenoyl-GPC (22:6)*, 7-methylguanine, betaine, pipecolate, pyridoxate, uridine and DHA) were associated other nutrients. Pipecolate and DHA had the largest number of associations reported in this analysis with 13 and 16 other nutrient associations found for each metabolite at a Bonferroni-adj. p < 0.05 threshold extrapolated from the full 756 metabolite panel (Additional file 1: Table S5).
The final sensitivity analysis explored if the identified B vitamin intake biomarkers could be validated by assessing their association with the levels of folate, vitamin B12 and hcy in plasma, and hcy in serum (hcy-s), which were available in sample subsets for 473–729 individuals in the TwinsUK cohort (Additional file 1: Table S1). Pyridoxate and betaine were associated with levels of folate after multiple testing correction (Bonferroni-adj. p < 0.05) and 1-(1-enyl-stearoyl)-2-docosahexaenoyl-GPE (P-18:0/22:6)* was associated with levels of vitamin B12 nominally (p < 0.05; Additional file 1: Table S6). Directions of effect for circulating folate and vitamin B12 matched the directions of effect of the main analysis (Additional file 1: Table S2 and S6).
Of the 18 metabolomic B vitamin biomarkers, 10 were nominally associated with either hcy, hcy-s, or both (p < 0.05; Additional file 1: Table S6). Compounds 1-docosahexaenoyl-GPC (22:6)*, 7-methylguanine, pyridoxate and DHA, used in the downstream epigenome-wide association analysis, were associated with both. 7-methylguanine and DHA had the strongest associations with hcy and/or hcy-s in this sensitivity analysis (Bonferroni-adj. p < 0.05). As expected, all significant associations between homocysteine and blood metabolites showed the opposite direction of association effect to that observed between the intake of B vitamin and their respective blood metabolite biomarker (e.g., DHA levels increase with B12 intake and hcy levels lower with increased DHA in blood; Additional file 1: Tables S1 and S6). This result was expected, because folate and vitamins B6 and B12 break down homocysteine to methionine.
Extending these results to a metabolome-wide analysis, we observed 14, 15, 23 and 47 metabolites associated with the circulating levels of folate, vitamin B12, hcy and hcy-s, respectively (Bonferroni-adj. p < 0.05; Additional file 1: Table S7). Here, strongest signals were identified between vitamin B12 and pantothenate (vitamin B5), and homocysteine (hcy/hcy-s) and pseudouridine. The strongest signal for folate remained pyridoxate metabolome-wide (b = 0.050 ± 0.006, adj. p = 7.36E−13).
B vitamin metabolomic biomarker epigenome-wide meta-analysis
The 18 metabolomic biomarkers identified for folate and vitamins B6 and B12 intakes were explored for associations with blood DNAm levels at 430,768 autosomal probes in the TwinsUK and KORA F4 cohorts (n = 2182; Additional file 1: Table S8). The KORA F4 dataset included 7 of the 18 metabolites under investigation, therefore subsequent analyses focused on the 7 common metabolomic biomarkers of B vitamin intakes: 1-docosahexaenoyl-GPC (22:6)*, 7-methylguanine, betaine, pipecolate, pyridoxate, uridine and DHA. Epigenetic analyses were carried out within each cohort, taking into account cohort-specific confounders, and results were meta-analysed.
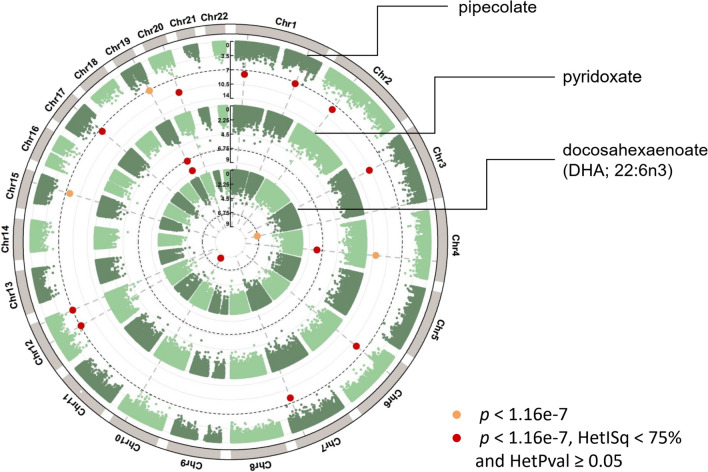
Three of the 7 metabolites in the epigenome-wide association meta-analysis showed significant differential DNAm levels in whole blood (Fig. 3). Pipecolate, pyridoxate and DHA were associated with 10, 3 and 1 DMPs each, respectively (Bonferroni-adj. p < 0.05, HetISq < 75% and HetPval ≥ 0.05). Pipecolate and pyridoxate are potential biomarkers of vitamin B6 and folate, while DHA is a potential metabolite biomarker of vitamin B6 intake alone, although they also show associations with other nutrient intakes.
Fig. 3.
Circular Manhattan plot of the B vitamin metabolomic biomarker results from the TwinsUK and KORA F4 epigenetic association meta-analysis (n = 2182)
Thirteen of the 14 DMPs identified in the meta-analysis had inverse directions of effect with DNAm (Table 1), that is, decreased DNAm levels with increased metabolite biomarker levels in blood. The strongest meta-analysis signals were observed for pipecolate and DMPs cg20732160 in the body of the PFKFB4 gene (b = − 0.029 ± 0.004, p = 1.68E−11), and cg10589813 located 751 bp upstream from CEBPB (b = − 0.029 ± 0.004, p = 3.49E−11). Pyridoxate was inversely associated with increased DNAm in SLC1A5 (cg02711608 and cg22304262) and SLC7A11 (cg06690548), with the strongest signal observed for DMP cg02711608 (b = − 0.038 ± 0.006, p = 1.63E−09) and the strongest effect size observed for DMP cg06690548 (b = − 0.073 ± 0.013, p = 1.65E−08). The association between DHA and DMP cg03440556 in the SCD gene was the one direct (positive) association identified in our analysis and had the strongest effect size overall (b = 0.093 ± 0.016, p = 4.07E−09).
Table 1.
B vitamin metabolomic biomarker results from the TwinsUK and KORA F4 epigenetic association meta-analysis (Bonferroni-adj. p < 0.05, HetISq < 75 and HetPVal ≥ 0.05; n = 2182)
| DMP | UCSC gene(s) (± 10 kb) |
Compound name |
Effect size | SE | p value | Direction* | HetISq | HetPVal |
|---|---|---|---|---|---|---|---|---|
| cg20732160 ( ~) | PFKFB4/UCN2 | Pipecolate | − 2.91E−02 | 4.32 E−03 | 1.68 E−11 | – – | 0.00 | 0.945 |
| cg10589813 (@) | CEBPB | Pipecolate | − 2.93 E−02 | 4.43 E−03 | 3.49 E−11 | – – | 0.00 | 0.825 |
| cg18120259 ( ~) | LOC100132354 | Pipecolate | − 2.67 E−02 | 4.36 E−03 | 9.18 E−10 | – – | 0.00 | 0.510 |
| cg11800635 ( ~) | LOXL3/DOK1/M1AP | Pipecolate | − 3.26 E−02 | 5.61 E−03 | 6.35 E−09 | – – | 0.00 | 0.385 |
| cg08616943 (@) |
LOC646329/MIR29A/ MIR29B1 |
Pipecolate | − 2.18 E−02 | 3.76 E−03 | 6.94 E−09 | – – | 0.00 | 0.942 |
| cg03523740 (^) | KPNA6/TXLNA | Pipecolate | − 1.69 E−02 | 2.97 E−03 | 1.14 E−08 | – – | 0.00 | 0.708 |
| cg12054453 ( ~) |
VMP1/MIR21/ DM119512/U6 |
Pipecolate | − 6.60 E−02 | 1.18 E−02 | 2.26 E−08 | – – | 46.60 | 0.171 |
| cg26841068 ($) | PRELP/OPTC | Pipecolate | − 2.26 E−02 | 4.17 E−03 | 5.66 E−08 | – – | 0.00 | 0.587 |
| cg13442969 (~ , #) | DYRK2 | Pipecolate | − 2.87 E−02 | 5.34 E−03 | 7.84 E−08 | – – | 57.50 | 0.125 |
| cg27180443 (^) | SCARB1 | Pipecolate | − 1.72 E−02 | 3.23 E−03 | 1.05 E−07 | – – | 0.00 | 0.570 |
| cg02711608 (~ , #, %) | SLC1A5 | Pyridoxate | − 3.76 E−02 | 6.23 E−03 | 1.63 E−09 | – – | 2.30 | 0.312 |
| cg06690548 ( ~) | SLC7A11 | Pyridoxate | − 7.33 E−02 | 1.30 E−02 | 1.65 E−08 | – – | 71.50 | 0.061 |
| cg22304262 (~ , #) | SLC1A5 | Pyridoxate | − 4.10 E−02 | 7.64 E−03 | 8.27 E−08 | – – | 0.00 | 0.444 |
| cg03440556 ( ~) | SCD | DHA | 9.25 E−02 | 1.57 E−02 | 4.07 E−09 | + + | 0.00 | 0.366 |
Location: ~ gene body, ^ TSS1500, $ 3’UTR, # 5’UTR, % 1st exon, @ intergenic region
*Directions of effect in TwinsUK and KORA, respectively
Cg03523740 (TXLNA) and cg27180443 (SCARB1) are located in the gene promoter (TSS1500) and were associated with pipecolate (Table 1). The pyridoxate-associated DMPs in SLC1A5 (cg02711608 and cg22304262) are in the upstream (5’) shelf of the same CpG island (chr19:47290585-47291983; Additional file 1: Table S9). Relative to SLC1A5, cg02711608 and cg22304262 are located on the 5’UTR, body or 1st exon of the gene depending on splicing.
In the individual cohort epigenome-wide analyses the three metabolite biomarkers associated with DMPs in the meta-analysis also displayed consistent direction of association, and further signals were detected albeit in smaller subsamples. In the KORA sample alone (n = 1673), pipecolate, pyridoxate and DHA were respectively associated with 9, 4 and 1 DMPs (Bonferroni-adj. p < 0.05; Additional file 1: Table S10). These include the three DMPs identified with pyridoxate in the meta-analysis (cg06690548, cg02711608, cg22304262), the 1 DMP associated with DHA (cg03440556), and 4 of the 10 DMPs associated with pipecolate (Additional file 1: Table S9). In the TwinsUK sample (n = 509), associations did not surpass epigenome-wide multiple testing correction (Bonferroni-adj. p < 0.05). However, at a more relaxed threshold (FDR = 10%) the individual cohort analyses also detected 15 DMPs for betaine in TwinsUK (lowest p = 4.76E−07 for cg08960352 in the body of the DYRK2 gene; Additional file 1: Table S11), and 126 DMPs for pipecolate in the KORA sample (lowest p = 5.85E−15 for cg06690548; data not shown).
If meta-analysis results were not filtered for heterogeneity among samples, pipecolate and DHA associated respectively with 3 and 1 further DMPs each (Additional file 1: Table S9; Fig. 3). Two of the high-heterogeneity DMPs associated with pipecolate included cg02711608 and cg06690548 identified for pyridoxate in the main results of our meta-analysis (Table 1).
B vitamin intake epigenome-wide meta-analysis
Pipecolate, pyridoxate and DHA were identified as potential metabolomic biomarkers of folate and vitamin B6 (Fig. 2) and showed evidence for association with 14 DNAm signals (Fig. 3). As a follow-up validation analysis, these 14 DMPs identified in our main analysis were also tested for association with diet FFQ-derived intakes of folate and vitamin B6 in the TwinsUK, KORA FF4 and LLS cohorts (n = 2294; Additional file 1: Table S12). Of the 14 DMPs, only cg10589813, upstream the CEBPB gene and associated with pipecolate, reached borderline nominal significance with habitual vitamin B6 intake (b = − 0.023 ± 0.013, p = 0.06). The directions of association, while not always consistent across cohorts, were often overall consistent with the results from the biomarker EWAS meta-analysis (Additional file 1: Table S13). Of the 10 DMPs associated with pipecolate, 6 and 7 had respective consistent inverse directions of effect with habitual vitamin B6 and folate. Of the 3 DMPs associated with pyridoxate, 2 and 1 had inverse directions of effect with B6 and folate. The main DMP result coming from DHA had a consistent positive direction of effect with dietary B6. Although the metabolomic biomarkers with DMPs were associated only with folate and vitamin B6 (Figs. 1 and 2), we tested if any of the 14 DMPs were significantly associated with vitamin B12 intake as well. Only cg20732160, located in the PFKFB4 gene and previously associated with pipecolate (a marker of folate and vitamin B6), was borderline significant with B12 (b = − 0.004 ± 0.002, p = 0.06). Nine of the 14 DMPs under study had consistent directions of effect between B12 intake and the three metabolites under study despite the metabolites marking only folate and vitamin B6 (Fig. 1).
Furthermore, an epigenome-wide association meta-analysis of FFQ-derived B vitamin intakes was performed across altogether 393,223 autosomal probes in the TwinsUK, KORA FF4 and LLS cohorts. No significant results were identified for vitamin B6 (lowest p = 2.68E−06) and folate (lowest p = 5.13E−06) and only 1 borderline significant result was found for vitamin B12: cg03473640 in the body of the MYO5A gene (b = − 0.013 ± 0.002, p = 1.30E−07), after multiple testing correction (Bonferroni-adj. p = 0.051).
Associated gene expression results
Using previously-published results from the BIOS consortium [18] we explored whether there were expression quantitative trait methylation signals among the 14 DMPs (Table 1). Overall, methylation levels at three DMPs—cg11800635, cg12054453 and cg02711608—were associated with the expression of genes annotated to them (Additional file 1: Table S14). Methylation levels at cg11800635 and cg12054453 were inversely associated with the expression of LOXL3/DOK1/M1AP and VMP1 genes, and methylation at cg02711608 was directly associated with the expression of SLC1A5 in a BIOS subsample of 2101 individuals.
The 14 DMPs identified in this study were in or within 10kb of 23 genes, of which 15 genes had whole blood gene expression data in a sample from the TwinsUK cohort (n = 297; mean age = 63.59 ± 7.59 and mean BMI = 25.96 ± 4.63 kg/m2). These included SLC1A5 and SLC7A11 genes (associated with pyridoxate in EWAS), SCD gene (associated with DHA in EWAS), and 12 genes with DMPs for pipecolate. Using the 15 candidate genes identified, we explored the association between gene expression and metabolomic biomarker levels. We observed one nominally significant association between TXLNA expression and pipecolate in blood (b = − 0.136 ± 0.108, p = 0.015; Additional file 1: Table S15), but no signals surpassed multiple testing correction.
Discussion
Our study identified 18 blood metabolite biomarkers of habitual folate and vitamins B6 and B12 intakes. Of these, three metabolomic biomarkers of folate and vitamin B6 showed a blood based epigenetic signature including signals in amino acid transporter genes SLC1A5 and SLC7A11, and in the stearoyl-CoA desaturase gene SCD. These signals may give insights into mechanisms involved in B vitamin uptake and regulation within the one-carbon metabolism pathway.
The B vitamins pyridoxine (B6), folate (B9) and cobalamin (B12) are essential soluble micronutrients that influence metabolism, physiology, immunity and development in living organisms through their roles in the one-carbon metabolism pathway—a biochemical network, which produces methyl groups for DNA synthesis and methylation. B6 and B12 function as enzymatic cofactors that facilitate reactions in the folate and methionine cycles in one-carbon metabolism; folate feeds into one-carbon metabolism as the principal substrate in the folate cycle. The conversion of hcy to methionine is particularly important as circulating hcy levels have been linked to several conditions, specifically, cardiovascular disease, diabetes, cancer and cognitive function. The B vitamins are proposed to have protective effects on human health through their influences on DNAm and levels of circulating hcy [19–22].
In this study we aimed to identify metabolomic biomarkers of folate and vitamins B6 and B12 to explore in downstream epigenome-wide association analysis towards identifying DNAm signatures of B vitamin intakes. Eighteen metabolites were identified as potential biomarkers of folate and vitamins B6 and B12, with one of the profiled metabolites—pyridoxate—acting within the vitamin B6 metabolic pathway. Sensitivity analyses showed that metabolite associations were non-specific. The non-specificity was expected since foods are composed of different nutrients and there will be a correlation of intakes according to an individual’s dietary choices. However, diet quality and total energy intake were not major confounders of our analysis. In line with our results, Posma et al. (2020) also identified associations between intakes of B vitamins with levels of betaines and fatty acids in urine [16]. Posma et al. identified direct correlations between folate, B6 and proline betaine/4-hydroxyproline betaine, and inverse correlations between folate, B6 and C5-C10 fatty acids in general [16]. Here, we identified direct correlations between folate and betaine, but the fatty acids identified had distinct directions of effect in blood depending on the molecule under study.
Metabolomic biomarker findings for intakes of folate and vitamin B12 were confirmed against their corresponding circulating levels in plasma, where direction of effect matched results based on self-assessed habitual dietary data and nominal significance was achieved for 4/6 metabolomic biomarkers. Hcy and hcy-s levels were both nominally associated with 8/18 metabolomic biomarkers identified for folate, and vitamins B6 and B12. Biomarkers with positive direction of association with folate and vitamins B6 and B12 had negative directions of association with circulating hcy levels in blood, and vice-versa. This matches current knowledge that plasma concentrations of hcy are inversely related to the intake of folate, B6 and B12, and nongenetic determinants of hcy concentrations in blood include inadequate concentrations of B vitamins [5, 6, 19–22].
Using epigenetic and metabolomic data from the TwinsUK and KORA F4 cohorts we were able to meta-analyse epigenome-wide associations for 7/18 metabolite biomarkers identified. Pipecolate (a marker of folate and vitamin B6), pyridoxate (a marker of folate and vitamin B6) and DHA (a marker of vitamin B6) were respectively associated with 10, 3 and 1 DMPs.
Of the 3 blood metabolomic biomarkers identified with DMPs epigenome-wide, pyridoxate has the most immediate link to B vitamins. Pyridoxate, or 4-pyridoxic acid, is the main catabolic product of vitamin B6 metabolism, and is formed from pyridoxal in the liver [23]. Pyridoxate is excreted into urine and its concentration in plasma is directly correlated with vitamin B6 intake [24]. Its use as a biomarker of vitamin B6 had mixed results in previous studies, however, and other forms of vitamin B6 have been encouraged in clinic [24]. In this study, we observed a strong positive correlation between the intake of dietary vitamin B6 and pyridoxate measured using Metabolon Inc. Pyridoxate was the only metabolite of the subpathway of vitamin B6 metabolism in our Metabolon panel of 756 metabolites, and therefore we suggest its use as a potential biomarker of vitamin B6 intake.
DHA is an essential omega-3 fatty acid from diet that needs phosphatidylcholine for circulation in the plasma and distribution to peripheral tissues [25]. As a consequence, it takes part in one-carbon metabolism, where methyl groups are transferred from SAM during the conversion of phosphatidylethanolamine-DHA to phosphatidylcholine-DHA [25]. Folate and vitamins B6 and B12 concentrations in plasma have been previously associated with DHA in blood in a cohort of European adolescents, likely due to their role in the maintenance of the levels of SAM [26]. DHA status has itself also been associated with B vitamin supplementation, where individuals with higher levels of DHA in plasma could gain more from supplementing their diet with vitamin B12 and folic acid in order to lower their hcy levels, which are associated with aging cognitive decline [27]. Pipecolate, or pipecolic acid, is a metabolite of lysine degradation in human physiological fluids, including the blood, urine and brain, with plasma pipecolate originating from both the bacterial catabolism of dietary lysine in the intestine and the direct dietary intake of plants with high levels of pipecolic acid [28, 29]. Pipecolate levels have been associated with pyridoxine-dependent epilepsy, but direct association of B6 deficiency and pipecolic acid metabolism is unlikely [30]. Indeed, we observed a positive correlation between B6 intake and pipecolate measured in plasma in our study.
The directions of effect for the DMPs identified from pyridoxate, pipecolate and DHA were often consistent with results obtained directly from FFQ-derived B vitamin intakes in the TwinsUK, KORA FF4 and LLS cohorts. Mandaviya et al. [8] identified associations between dietary folate and 6 DMPs (cg23465990, cg11832534, cg03249011, cg14398883, cg00826902, cg14145338), but these were not among those identified for pipecolate and pyridoxate in our main analysis. Dietary folate was associated with hypomethylation at single sites in Mandaviya et al. [8]; we observed the same trend here for pipecolate and pyridoxate. Previously Petersen et al. [31] reported an epigenome-wide analysis of serum metabolites in the KORA F4 cohort [31]. Petersen et al. [31] reported that methylation at 2 CpG sites—cg16936953 and cg12054453—was significantly negatively associated with pipecolate levels in blood. In line with this result, our meta-analysis identified DMP cg12054453 as peak signal for pipecolate. DMP cg16936953 was borderline significant (Bonferroni-adj. p = 0.052), but did not pass heterogeneity filters in a meta-analysis of results with TwinsUK. Overall, the predominantly inverse directions of effects identified epigenome-wide in Mandaviya et al. [8] for dietary folate, in Petersen et al. [31] for pipecolate, and in our study for pipecolate and pyridoxate suggest that population-wide differences in B vitamin intake within the normal reference values can affect one-carbon metabolism homeostasis with higher B vitamin linked to lower levels of methylation. This is particularly apparent for vitamin B6, which is a cofactor in the transsulfuration pathway that converts hcy to cysteine, and lowers the production of methionine available for DNA methylation. In our study, pipecolate and pyridoxate were markers of both vitamin B6 and folate, but had stronger associations with vitamin B6. Moreover, vitamin B6 and folate intakes were highly correlated in our data (Pearson’s r = 0.62 for folate and B6, while r = 0.04 for folate and B12, and r = 0.16 for vitamins B6 and B12). It is thus possible that we are primarily observing the effects of vitamin B6 in one-carbon metabolism in the inverse associations reported.
The genes annotated to the DMPs identified in our meta-analysis varied in function. Pipecolate was associated with decreased methylation in genes with important roles in cellular metabolism and homeostasis. Specifically, PFKFB4 is crucial in regulating the concentration of the glycolytic byproduct fructose-2,6-bisphosphate, while SCARB1 is a plasma membrane receptor for high-density lipoprotein and cholesterol trafficking between cells [32, 33]. DNAm in the PFKFB4 gene has been previously associated with the regulation of glycolytic potential in skeletal muscle [34]. DHA was associated with increased DNAm in the SCD gene, which encodes the Stearoyl CoA Desaturase-1 enzyme that converts saturated fatty acids into monounsaturated fatty acids and plays a role in obesity and insulin resistance. Decreased promoter methylation of the SCD gene has been previously linked to obesity [35].
Pyridoxate was associated with hypomethylation in amino acid transporter genes SLC1A5 and SLC7A11. SLC7A11 encodes a cysteine/glutamate antiporter system, a critical modulator of intracellular redox balance that mediates the exchange of intracellular glutamate for extracellular cystine, an essential precursor for glutathione synthesis [36, 37]. Vitamin B6-dependent enzymes also catalyse most reactions of the transsulfuration pathway, which drives homocysteine to cysteine and further into glutathione peroxidase proteins [38]. In our study pipecolate (direct marker of B6) was associated with hypomethylation in cg06690548, suggesting that vitamin B6-dependent hypomethylation in SLC7A11 may be related to processes implicated in cysteine homeostasis and oxidative stress. Hypermethylation of cg06690548 has also recently been associated with downregulation of SLC7A11 in Parkinson’s disease [39].
SLC1A5 is a sodium-dependent amino acid transporter with broad substrate specificity and preference for glutamine [40]. Consequently, SLC1A5 is expressed in highly proliferative cells such as inflammatory, stem and cancer cells to meet their augmented glutamine demand. Differential methylation of cg02711608 (located in the 5’UTR region of SLC1A5) has been linked to alcohol consumption and BMI [41–43]. Hypomethylation of cg02711608 and cg22304262 (also in the 5’UTR region of SLC1A5) has been linked to higher blood pressure [44]. A putative causal effect has further been demonstrated for DMP cg22304262 in the context of incident coronary heart disease [45], as recently reviewed by us [46]. Folate intake and supplementation have been associated with improved endothelial function [47], lower systolic and diastolic blood pressure [47, 48], and overall lower risk of incident hypertension [49]. DNAm could thus fulfil a mechanistic role in the mediation of B vitamin intake and determinants of cardiovascular risk. Pyridoxate was a stronger marker of B6 than folate in our results (Fig. 2, Additional file 1: Table S2). This could partially explain why pyridoxate-associated hypomethylation of cg02711608 and cg22304262 (Table 1)—linked to high blood pressure [44]—was found in the context of this study. Cg22304262 was hypermethylated with the intake of folate measured directly from FFQs (Additional file 1: Table S13), but diet cohort results were heterogeneous and lacked the consistency of the metabolomic results.
The functional relevance of our main results was explored in the BIOS consortium and in a subsample from TwinsUK to explore methylation-expression and metabolomic-expression associations. Overall, methylation levels at cg11800635 (associated with pipecolate in EWAS), cg12054453 (associated with pipecolate in EWAS) and cg02711608 (associated with pyridoxate in EWAS) were associated with the expression levels of genes in the BIOS consortium. Furthermore, DMP cg03523740 for pipecolate is located in the promoter region of the TXLNA gene and in TwinsUK TXLNA expression changed nominally with pipecolate levels.
As metabolomic platforms become more ubiquitously used in cohort studies, we aimed to identify metabolomic biomarkers of B vitamin intake in order to circumvent limitations of accuracy associated with habitual diet measurement. Moreover, using habitual dietary data to identify B vitamin-associated metabolites resulted in larger sample sizes and more power, in comparison to using folate and B12 data measured directly from plasma in TwinsUK (n > 1000 for habitual diet and n < 730 for folate and B12 in plasma).
Both the habitual diet and blood levels of B vitamins used in this study are within the normal ranges expected for humans. As such, in future a stratified analysis of the levels of B vitamins could reveal additional metabolic and epigenetic signatures of interest. Additionally, the discovery phase of our study included only UK females and the results may not reflect biomarkers in males or in individuals of non-European ancestry. Another limitation of our study was the small overlap between the blood metabolomic data available in TwinsUK and KORA F4. We were only able to meta-analyse epigenome-wide results for 7 of the 18 blood metabolomic biomarkers initially identified. It remains unknown whether, in addition to pipecolate, pyridoxate and DHA, other B vitamin metabolomic biomarkers identified in the discovery phase of our study have epigenome-wide effects in the DNA methylome. The B vitamins intake metabolomic biomarker identified with most confidence in our study was pyridoxate, because pyridoxate is the end product of vitamin B6 metabolism before excretion from the body. However, overall the non-specificity of the metabolomic biomarkers identified, while expected due to the high correlation of nutrients in food, also limits their application in nutritional assessment.
Mandaviya et al. [8] reported 6 DMPs from a stratified analysis of folate intake. Unlike Mandaviya et al. we were unable to identify DMPs for dietary folate after correcting for multiple testing. This was probably due to differences in our approach and much lower number of samples in the EWASs of our habitual diet meta-analysis (n = 2294) compared to Mandaviya et al. [8] (n = 5841). Instead, our findings identified 14 epigenome-wide signals for metabolomic biomarkers of B vitamins in a more modest sample size (n = 2182), suggesting that blood metabolites may offer not only an unbiased, but also more powerful approach over self-assessed reports of dietary intakes.
Conclusion
Using metabolomics and self-assessed dietary data we were able to identify blood metabolomic biomarkers of B vitamins with epigenome-wide association effects in whole blood DNAm. Pyridoxate—a catabolic product of the vitamin B6 metabolism—stands out as a potential blood metabolomic biomarker of B6 with noticeable epigenome-wide effects on DNAm. Significant epigenome-wide associations were observed from metabolomics data that were not observed with a similar sample size directly from self-reported dietary data. Metabolomic biomarkers of B vitamins are exact tools that can unveil novel differentially methylated signals of dietary intakes in the human epigenome.
Methods
Cohort information
TwinsUK. The TwinsUK registry is ongoing since 1992 and includes over 15,000 research volunteer twin participants from the United Kingdom [50]. Volunteers are monozygotic and dizygotic same-sex twins, predominately female (82%), middle-aged (mean age of 59 years) and over 18 years-old. Volunteers were recruited without selecting for disease and are mostly of European descent. Information on participants has been obtained through numerous questionnaire responses and comprehensive phenotyping over the years, with the particular application of several 'omic' technologies for a range of sample types. In this study we used epigenetic, transcriptomic and metabolomic profiling in TwinsUK, together with questionnaire level data from the twins.
KORA. The KORA (Cooperative Health Research in the Region of Augsburg) study is an ongoing registry of Southern German citizens with baseline recruiting dating back to 1999 (KORA S4). Selection of citizens was random with equal strata by sex and age and included 4261 subjects aged 25–74 years. Of these, KORA F4 (2006–2008) and KORA FF4 (2013/14), respectively the first and second follow-up to the S4 baseline, carried out with 3080 and 2279 participants each [51, 52]. In this study participants from the F4 follow-up were selected to explore methylation signal changes by metabolomic biomarkers, and participants from the FF4 follow-up were selected to explore methylation signal changes in response to diet, according to availability of data.
LLS. The Leiden Longevity Study (LLS) is a multigenerational study that recruited nonagenarian siblings of European descent and their offspring. Altogether 944 long-lived proband siblings (mean age of 94 years), 1671 offspring (mean age of 60 years) and 744 controls (the offspring spouses, mean age of 60 years) were recruited at baseline (between 2002 and 2006). Members of long-lived families are very similar to control groups with whom they likely share similar environment, lifestyle, and age, but have more favourable morbidity and mortality outcomes [53]. Members of long-lived families were analysed as one cohort of middle-aged people and the current study was restricted to unrelated individuals in epigenetic analyses.
Data collection and processing
Habitual B vitamin intakes
The habitual intakes of folate and vitamins B6 and B12 of participants was measured using food frequency questionnaires (FFQs) in the TwinsUK and LLS cohorts, and a blended approach comprising repeated 24h food lists and an FFQ in the KORA FF4 cohort.
TwinsUK. Food frequency questionnaires used in the TwinsUK study comprised 131 food and drink items from the EPIC Norfolk study [54]. Processing of these data has previously been described [55], and data were available for 3157 female twins. The daily intake of each item was calculated in g/day using the FETA software [56] and the default nutritional database based on the McCance and Widdowson’s The Composition of Foods (5th edition) [57]. The residual method was used to obtain B vitamin intake estimates independent of total energy intake [58]. In addition to B vitamins, the daily intakes of 38 other nutrients was estimated for use in sensitivity analysis of B vitamin intake associations. The 38 other nutrients quantified included altogether 16 macronutrients (i.e. total protein, total fat, total carbohydrates, starch, total sugars, glucose, fructose, sucrose, maltose, lactose, non‐starch polysaccharides, saturated fats, monounsaturated fats, polyunsaturated fats, trans fats and cholesterol), 11 minerals (i.e. sodium, potassium, calcium, magnesium, phosphorus, iron, copper, zinc, chloride, manganese and iodine), and 11 other vitamins/vitamin nutrient precursors (i.e. retinol, carotene, vitamin C, vitamin D, vitamin E, thiamine, riboflavin, niacin, tryptophan, pantothenate and biotin). The overall diet quality of the TwinsUK participants was calculated using the Alternate Healthy Eating Index 2010 (AHEI-2010) diet score [17], which ranges 0–10 and scores positively the intake of healthy foods (e.g., whole grains and healthy fats) and scores negatively the intake of unhealthy foods (e.g., red and processed meats). The AHEI-2010 accounts for the participants alcohol intake and was calculated here for the sensitivity of overall diet quality.
KORA FF4. Repeated 246-item 24-h food lists derived from the NAKO Health study [59] and 148-item FFQs adapted from the German version of the multilingual European Food Propensity Questionnaire [60] were used in the KORA FF4 study. The processing of these data was first described elsewhere [61], and data was available for 1602 participants. Classification of dietary intakes in KORA was performed with the EPIC-Soft software [62] and B vitamin intake data was calculated based on the German food composition database Bundeslebensmittelschlüssel, version 3.01 [63]. Like in TwinsUK, the residual method was used to get B vitamin intake estimates independent of energy intake in KORA FF4.
LLS. Food frequency questionnaires used in the LLS study included 218 items constructed from the 104-item VetExpress FFQ combined with the Dutch National Food Survey [64]. B vitamin intake data was estimated in grams per day using the NEVO table 2011 [65] as reference panel. A weighted average was calculated for the nutrient composition of a food item, based on the consumption of each NEVO product included in the food item according to the Dutch National Food Consumption Survey 2010. Dietary intake data in grams per day was collected from 1716 individuals.
The energy-adjusted intakes of folate and vitamins B6 and B12 were used in the discovery phase of our study to identify metabolomic biomarkers of B vitamins in participants from TwinsUK. Energy-adjusted intakes from TwinsUK, LLS and KORA FF4 were used for the epigenome-wide association meta-analysis of habitual B vitamin intakes. B vitamin outlier values were removed across analyses in similar fashion, where outliers 3 standard deviations away from the mean were excluded from the subsamples.
Blood levels of folate, vitamin B12 and homocysteine
Measured blood levels of folate (ng/mL) and B12 (ng/L) were available in the TwinsUK cohort for a subset of participants with metabolomics data. Homocysteine levels (µmol/L) were also available in plasma and serum. Overall, and after removing outliers 3 standard deviations away from the mean, a total of 729, 718, 473, and 707 individuals had circulating folate, vitamin B12, hcy and hcy-s levels measured within 2 years of metabolomics profiling, respectively.
Whole blood metabolome
Blood metabolites used in this study were profiled in the TwinsUK and KORA F4 cohorts using the Metabolon platform. Metabolon is a chromatography mass spectrometry platform that produces semiquantitative data where standards are used to determine the retention time and relative intensity of metabolites.
TwinsUK. Fasting blood serum samples were collected from female participants and profiled using the Metabolon platform (Metabolon, Inc., Durham, NC). The processing of samples has previously been described [66]. Metabolomic data were median-normalised by dividing metabolite concentrations by the day median of that metabolite and then rank inverse-normalised. Metabolites with more than 20% of missing values were excluded and minimum run day measures were imputed to the missing values. A total of 756 metabolites were kept for analysis from a total of 6196 samples taken from 2069 female twins spanning several years. Of the 756 metabolites, 591 (78%) are annotated and fall into the broad metabolic groups of amino acids, carbohydrates, cofactors and vitamins, energy, lipid, nucleotide, peptide, and xenobiotics. One of the profiled metabolites, pyridoxate, is known to act within the vitamin B6 metabolic pathway. A subset of 1063 (for folate and vitamin B6) and 1064 (for vitamin B12) female twins had a blood metabolomic profile within 2 years of FFQ. These twins were used for biomarker discovery in the TwinsUK sample.
KORA F4. Fasting blood serum samples were collected from participants of the KORA F4 (Cooperative Health Research in the Region of Augsburg) study population and profiled using the Metabolon platform (Metabolon, Inc., Durham, NC). The processing of samples was previously described [67, 68]. Like in TwinsUK, metabolomics data in KORA F4 was median-normalised by dividing metabolite concentrations by the day median due to fluctuations in the data caused by instrument maintenances that are day-dependent. Then in KORA each metabolite data was multiplied with their overall median values and log transformed. To match the TwinsUK outcome variables and for the purpose of meta-analysis, KORA data was normalised by rank-based inverse normal transformation in this study. Overall, and after quality control, 276 metabolites in human serum were profiled from 1768 participants of the KORA F4 population.
Whole blood DNA methylation
TwinsUK. Fasting whole blood DNAm of 990 individuals was profiled using the Infinium HumanMethylation450 BeadChip (Illumina Inc, San Diego, CA). DNAm was assessed at > 450,000 sites and processing of methylation signals was performed with R Bioconductor software [69]. Briefly, the ENmix package [70] was used for quality control of the data, and the minfi package [71] was used to exclude samples with median methylated and unmethylated signal ratio < 10.5. Background correction, dye bias correction and quantile normalization were performed with ENmix as previously described [72]. Underperforming probes and outlier samples were identified using standard parameter values and signals with detP > 0.000001 and nbead < 3 were excluded from the analysis. Maximum probe and sample missingness were set to 5%. Methylation beta-values (ranging 0–1 for un- to fully-methylated) were estimated with ENmix while adjusting for array probe type bias with the Regression on Correlated Probes (RCP) method [73]. Methylation beta-values were converted to methylation M-values with the lumi package [74] prior to downstream analysis for better statistical validity of the models. A total of 487 and 509 females had DNAm measures within 2 years of FFQ and 5 years of metabolomic profiling, respectively. The two subsamples were used in downstream analyses.
KORA. Fasting whole blood DNAm was available in the KORA F4 and FF4 waves used in this study for metabolomic and habitual diet intake analysis, respectively. KORA F4. Whole blood DNAm was measured with the HumanMethylation450 BeadChip and processing of data was previously described [75]. Briefly, the methylation data was extracted through Illumina’s Genome Studio (version 2011.1) methylation module (v1.9.0) and processed with the CPACOR pipeline [76]. Background correction was performed with minfi [71] and bad signals were excluded if detP > 0.01. Maximum sample missingness was set to 5% and methylation beta-values were estimated after quantile normalisation of the data. KORA FF4. Whole blood DNAm was measured with the Infinium MethylationEPIC BeadChip, which assesses methylation at > 850,000 sites of the human genome. Quality control of this data was previously described [77] and processed in similar fashion to DNAm in the KORA F4 population (i.e. following the CPACOR pipeline). KORA F4 and FF4 methylation data was converted to M-values prior to analysis in this study. A total of 1673 and 1322 participants respectively of KORA F4 and FF4 had a metabolomic profile and FFQ collected in the same wave as whole blood DNAm and were used in downstream analysis.
LLS. Fasting whole blood DNAm was available for 732 individuals of the LLS cohort. Processing and normalization of the data were done as described in the DNAmArray workflow (https://molepi.github.io/DNAmArray_workflow/). Briefly, methylation data was extracted using the minfi package [71] and sample-level quality control was performed using MethylAid [78]. Signal exclusion was performed based on detP > 0.01, nbead < 3 and zero values for intensity. Functional normalization of the data was performed using five principal components extracted using the control probes. Maximum sample missingness was set to 5% and methylation beta-values were converted to M-values to match other cohorts in this study. A total of 485 long-lived participants of the LLS study had DNAm and FFQ and were used in this study for the habitual B vitamin intake epigenetic meta-analysis.
Across cohorts, only autosomal probes were kept for analysis in this study. Polymorphic or probes that mapped to multiple locations in the genome were also removed. Altogether a total of 430,768 and 393,223 probes were identified in TwinsUK/KORA F4 and TwinsUK/KORA FF4/LLS cohort groups, respectively, and kept for the biomarker and habitual diet epigenetic meta-analyses.
Whole blood gene expression
Gene expression data used in this study was profiled in the TwinsUK cohort. Fasting whole blood transcriptomic data was obtained using Illumina RNA-Seq technologies (Illumina, Inc., San Diego, CA). There data and processing have previously been described [79]. Briefly, the STAR software v2.4.0.1 [80] was used to align reads to the hg19 reference genome and only uniquely mapped properly paired reads were kept after alignment. GENCODE annotation v19 gene counts were obtained with featurecounts [81], and then standardised with trimmed mean of M-values (TMM)-adjusted counts per million (CPMs) and inverse-normalised prior to downstream analysis. Only genes with at least 0.5 CPM expressed in 90% of samples were kept in the original data. A total of 23 genes were manually annotated to the 14 DMPs identified using the UCSC genome browser (hg19) selecting for genes ± 10 kb away from the CpG site. Fifteen out of the 23 genes from our main analysis were present in the data. A total of 297 female twins had gene expression data profiled within 5 years of metabolomic profiling. The 15 genes and 297 female twins identified were used for follow-up gene expression analysis in the TwinsUK cohort.
Statistical analyses
Discovery phase
Metabolomic data collected within 2 years of FFQ were used for the discovery of B vitamin metabolite biomarkers in the TwinsUK cohort. A total of 1063 (for folate and vitamin B6) and 1064 (for vitamin B12) female twins were included in this analysis after removing outliers. Twins were either monozygotic or dizygotic, and zygosity was included as a factor in the model to account for the level of shared genetic variation (i.e. MZ share approximately 100% while DZ share 50% of genetic variation). Twins with their co-twin in a twin pair missing were reclassified as unrelated individuals. Metabolome-wide associations between folate and vitamins B6 and B12 were separately undertaken for the 756 metabolites from Metabolon. Linear regression mixed-effects models were applied using the lme4 package [82]. Models were adjusted for the participants’ age and BMI, the time interval between food questionnaire and metabolomic sample collection, and the family and zygosity of participants as random effects. In this instance the energy-adjusted intake of a B vitamin was the predictor and the inverse-normalised signal of a metabolite was the outcome. Slight variations in final sample sizes were due to missing metabolomic data although each metabolite was profiled in n > 1000 in most cases (n < 1000 for 3 metabolites; lowest n = 976). Multiple testing adjustment of each B vitamin result was applied using Bonferroni correction (p = 0.05/756 tests = 6.61E−05, for Bonferroni-adj. p < 0.05). Structurally unidentified metabolites (unknowns) were discarded. Metabolites with single asterisk were annotated based on in silico prediction. A total of 18 metabolites were kept for downstream analyses.
Sensitivity analyses
Three sensitivity analyses were performed on the 18 putative biomarker metabolites identified during the biomarker discovery phase.
Total energy intake and diet quality. To assess their putative impact on the identification of biomarker metabolites, the biomarker discovery model described immediately above was extended to further include the total energy intake and overall diet quality of the participants as covariates. Here, the AHEI-2010 diet score [17]and total energy consumed (in kcal/day) were included as fixed effect variables. Multiple testing significance was presented with Bonferroni correction (p = 0.05/756 tests, Bonferroni-adj. p < 0.05).
Other nutrients. To determine the specificity of our findings, a panel of 38 other nutrients common from habitual diet were used in associations with the 18 biomarker metabolites identified in our main analysis. In this instance the other nutrient (e.g. glucose, iron, vitamin C, etc.) replaced the predictor variable in the original model, and the metabolite biomarker remained as the outcome. Predictor outliers were removed as previously described (n > 1000 in all instances). Only associations for the 18 metabolite biomarkers were performed per nutrient, but Bonferroni thresholds used to determine the significance of these results were set metabolome-wide as previously described (i.e. for each nutrient analysis adj. p = 0.05/756 tests).
Blood chemistry. To evaluate if B vitamin findings from habitual diet could be validated using the plasma or serum levels of B vitamins and homocysteine, associations were performed in subsamples of 473–729 twins with available folate, vitamin B12, hcy and hcy-s data. Circulating folate, vitamin B12 and homocysteine levels were used as predictors and associations were performed for the metabolites identified as the outcome. The 5 and 1 metabolites associated with dietary folate and vitamin B12 were used here in associations with folate and B12 levels in plasma, respectively. All 18 metabolites identified overall in our main analysis were used for the hcy and hcy-s associations. Linear mixed effects models were adjusted for age, BMI, time interval between blood metabolomics and other blood chemistry data, and family and zygosity of the twin sample. The significance of each metabolite result was determined with the Bonferroni correction threshold extrapolated previously from the full 756 metabolite panel (p = 0.05/756 tests).
Epigenome-wide association meta-analyses
B vitamin metabolite biomarkers. An epigenome-wide association study (EWAS) was performed in the TwinsUK cohort for each of the 18 metabolite biomarkers identified in the discovery phase of our study. Seven of the 18 metabolites identified in TwinsUK (i.e. 1-docosahexaenoyl-GPC (22:6)*, 7-methylguanine, betaine, pipecolate, pyridoxate, uridine and DHA) were also represented in the KORA F4 Metabolon data and EWASs results were meta-analysed between TwinsUK (n = 509) and KORA (n = 1673; total n = 2182). Epigenome-wide association studies were performed using linear regression mixed effects models where DNAm M-values were the outcome and inverse-normalised metabolite levels in blood were predictors. Metabolite levels 3 standard deviations away from the mean were excluded and models were adjusted for age, BMI, trichotomous smoking (0: never smoker; 1: ex-smoker; 2: current smoker), blood cell proportions (lymphocytes, granulocytes and monocytes), time interval between blood metabolomics and methylation profiling, and technical and cohort-specific variables such as family and zygosity in TwinsUK, and sex (0: female; 1: male) in the KORA F4 cohort. In TwinsUK, blood cell proportions were estimated with the Houseman method [83] using Horvath’s DNA Methylation Age Calculator [84], with lymphocytes corresponding to the sum of CD8T, CD4T, NK and B cell type proportions. Associations were performed across 430,768 autosomal probes and results were meta-analysed with METAL [85]. Here, the effect sizes and standard errors obtained in TwinsUK and KORA were used to conduct a fixed-effects inverse variance weighted meta-analysis. The heterogeneity of results was analysed with METAL across the two cohorts. The significance of results was established based on the Bonferroni correction (p = 0.05/430,768 tests = 1.17E−07, for Bonferroni-adj. p < 0.05) and sample heterogeneity (HetISq < 75% and HetPval ≥ 0.05). The false discovery rate method was used to explore results further in each cohort with q < 0.1. Genes of DMPs identified were manually annotated using the UCSC genome browser (hg19) selecting for genes ± 10 kb away from the CpG site.
B vitamin intakes. An EWAS of the habitual intake of folate and vitamins B6 and B12 was performed in the TwinsUK (n = 487), KORA FF4 (n = 1322) and LLS (n = 485) cohorts and meta-analysed in this study (total n = 2294). Analyses were performed using methylation M-values as the outcome variable and the energy-adjusted habitual diet intake of B vitamins as the predictor. B vitamin intakes 3 standard deviations away from the mean were excluded prior to analysis and models were adjusted for age, BMI, trichotomous smoking, blood cell proportions, and technical and cohort-specific variables. Meta-analysis of results was performed with METAL as described above (p = 0/393,223 tests = 1.27E−07, Bonferroni-adj. p < 0.05). Pipecolate, pyridoxate and DHA were identified as metabolomic biomarkers of folate and vitamin B6, and DMPs associated with these compounds were explored in further detail in context of habitual diet.
Gene expression follow-up
Methylation-expression associations for the DMPs identified in this study were explored in previously-published data from the BIOS (Biobank-Based Integrative Omics Studies) consortium in The Netherlands [18]. Cis expression quantitative trait methylation signals captured at FDR = 5% across 2101 samples were extracted from the “2015_09_02_cis_eQTMsFDR0.05-CpGLevel.txt” file hosted in the BIOS QTL browser (https://molgenis26.gcc.rug.nl/downloads/biosqtlbrowser/).
A targeted metabolomic-gene expression follow-up analysis of 15 genes with DMPs was performed in the TwinsUK cohort (n = 297). Levels of PFKFB4, CEBPB, DOK1, LOXL3, M1AP, LOC646329, MIR29B1, TXLNA, KPNA6, VMP1, DYRK2 and SCARB1 expression were tested for association with levels of pipecolate. Levels of SLC1A5 and SLC7A11 expression were tested for association with pyridoxate, and SCD expression levels were tested in association to DHA. Linear regression mixed effect models were implemented where inverse-normalised metabolite levels were included as the predictor and the inverse-normalised TMM-adjusted gene counts were included as the outcome. Models were adjusted for age, BMI, trichotomous smoking, time interval between blood metabolomics and gene expression profiling, and other technical covariates (fixed: insert-size median and mean GC content; random: primer index, date of sequencing and RNA extraction batch) used previously [86]. Family and zygosity were included as random effects. Multiple testing correction was applied as previously described.
Supplementary Information
Additional file 1: Table S1. Cohort characteristics for the B vitamin metabolomic biomarker discovery. Table S2. Blood metabolites associated with the dietary intake of folate (B9) and vitamins B6 and B12 (metabolome-wide Bonferroni-adj. p < 0.05). Table S3. B vitamin metabolomic biomarker associations adjusted for diet quality (AHEI-2010) and total energy intake (metabolome-wide Bonferroni-adj. p < 0.05). Table S4. B vitamin metabolomic biomarker associations with other nutrient intakes (metabolome-wide Bonferroni-adj. p < 0.05). Table S5. Summary of nutrients associated with the B vitamin metabolomic biomarkers identified (metabolome-wide Bonferroni-adj. p < 0.05). Table S6. B vitamin metabolomic biomarker associations with circulating blood levels of folate (B9), vitamin B12, hcy and hcy-s. Table S7. Metabolite associations with circulating blood levels of folate (B9), vitamin B12, hcy and hcy-s (metabolome-wide Bonferroni-adj. p < 0.05). Table S8. Cohort characteristics for the B vitamin metabolomic biomarker epigenome-wide meta-analysis. Table S9. Annotation of the epigenome-wide association meta-analysis of folate (B9) and vitamins B6 and B12 biomarker metabolites (epigenome-wide Bonferroni-adj. p < 0.05). Table S10. Epigenome-wide association of folate (B9) and vitamins B6 and B12 biomarker metabolites in the KORA F4 cohort (epigenome-wide Bonferroni-adj. p < 0.05). Table S11. Epigenome-wide association of folate (B9) and vitamins B6 and B12 biomarker metabolites in the TwinsUK cohort (epigenome-wide FDR = 10%). Table S12. Cohort characteristics for the B vitamin intake epigenome-wide meta-analysis. Table S13. DMP results in DMP results in the epigenome-wide association meta-analysis of folate (B9) and vitamin B6 dietary intakes. Table S14. Cis expression quantitative trait methylation signals from the BIOS consortium for the DMP identified in this study. Table S15. Gene expression results for the genes annotated in the biomarker metabolite EWAS meta-analysis.
Additional file 2: Figure S1. Correlation matrix of the serum levels of the metabolomic biomarkers identified. Figure S2. Pathway annotations of the 18 metabolomic biomarkers identified for folate and vitamins B6 and B12 (Bonferroni-adj. p < 0.05).
Acknowledgements
The authors thank all research volunteers who participated in the study. The authors acknowledge use of the research computing facilities at King’s College London, CREATE (https://doi.org/10.18742/rnvf-m076) and Rosalind (https://rosalind.kcl.ac.uk), which are delivered in partnership with the National Institute for Health Research (NIHR) Biomedical Research Centres at South London & Maudsley and Guy’s & St. Thomas’ NHS Foundation Trusts, and part-funded by capital equipment grants from the Maudsley Charity (award 980) and Guy’s & St. Thomas’ Charity (TR130505). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, King’s College London, or the Department of Health and Social Care. The contribution of all participants of the TwinsUK, KORA and Leiden Longevity Study is gratefully acknowledged.
Abbreviations
- AHEI-2010
Alternate healthy eating index 2010
- BMI
Body mass index
- DHA
Docosahexaenoate
- DMP
Differentially methylated position
- DNMT
DNA methyltransferase
- EWAS
Epigenome-wide association study
- FDR
False discovery rate
- FFQ
Food frequency questionnaire
- Hcy
Homocysteine measured in plasma
- Hcy-s
Homocysteine measured in serum
- R2c
Conditional R squared
- SAH
S-adenosylhomocysteine
- SAM
S-adenosylmethionine
Author contributions
R.C. and J.T.B. supervised the study and J.T.B. outlined the main conceptual ideas. R.C. and L.E. performed the main analyses. R.W., F.H. and L.S. performed analyses in the KORA or LLS cohorts. X.Y. performed the follow-up gene expression analysis in TwinsUK. C.C., S.V., O.M.M. and P-C.T. contributed to the data analysis. M.W., J.L., K.S.S., P.E.S., B.T.H., A.P., C.G., G.K., K.S., C.M. and M.M. contributed to student supervision and overall curation and processing of the data. M.B. and D.v.H. contributed to data acquisition in the LLS cohort. S.E.B., C.M. and O.M.M. helped with data interpretation. R.C., L.E. and J.T.B. wrote the manuscript, and R.C and L.E. prepared the visualisation of results. All authors reviewed and approved the manuscript.
Funding
This project was supported by the European HDHL Joint Programming Initiative funding scheme DIMENSION project (BBSRC BB/S020845/1 and BB/T019980/1 to J.T.B.). K.S.S. acknowledges funding from the Medical Research Council (MR/M004422/1 and MR/R023131/1). X.Y. is funded by King's-China Scholarship Council PhD Scholarship. C.M. is funded by the Chronic Disease Research Foundation. The TwinsUK study is funded by the Wellcome Trust, Medical Research Council, Versus Arthritis, European Union Horizon 2020, Chronic Disease Research Foundation (CDRF), ZOE LIMITED, and the National Institute for Health Research (NIHR) Clinical Research Network (CRN) and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London. The project DIMENSION, partnering site Helmholtz Munich, got financial support by a grant of the European HDHL Joint Programming Initiative funding scheme, administered by the Federal Ministry of Research in Germany, Grant No. 01EA1902A (R.W., M.W., C.G.). The project DIMENSION, partnering site LMU Munich, got financial support by a grant of the European HDHL Joint Programming Initiative funding scheme, administered by the Federal Ministry of Research in Germany, Grant No. 01EA1902B (F.H., J.L.). The Leiden Longevity Study has received funding from the European Union’s Seventh Framework Programme (FP7/2007–2011) under grant agreement number 259679, the Innovation-Oriented Research Program on Genomics (SenterNovem IGE05007), the Centre for Medical Systems Biology and the Netherlands Consortium for Healthy Ageing (Grant 050-060-810), all in the framework of the Netherlands Genomics Initiative, Netherlands Organization for Scientific Research (NWO), BBMRI-NL, a Research Infrastructure financed by the Dutch government (NWO 184.021.007 and 184.033.111).
Availability of data and materials
Many of the blood data analysed in TwinsUK is available through GEO GSE62992 and GSE121633 for methylation and EGA EGAD00001001088 for gene expression. Additional TwinsUK individual-level data are not permitted to be shared or deposited due to the original consent given at the time of data collection. However, access to these data can be applied for through the TwinsUK data access committee. For information on access and how to apply http://twinsuk.ac.uk/resources-for-researchers/access-our-data/. The informed consents given by KORA study participants do not cover data posting in public databases. However, data are available upon request from KORA Project Application Self-Service Tool (https://helmholtz-muenchen.managed-otrs.com/external/). Data requests can be submitted online and are subject to approval by the KORA Board. LLS DNA methylation data are available upon request via the BIOS consortium (https://www.bbmri.nl/acquisition-use-analyze/bios). FFQ data is available upon request.
Declarations
Ethics approval and consent to participate
TwinsUK. Ethical approval was granted by the National Research Ethics Service London-Westminster, the St Thomas’ Hospital Research Ethics Committee (EC04/015 and 07/H0802/84). All research participants have signed informed consent prior to taking part in any research activities. KORA F4 and FF4. The KORA cohort ethical approval was granted by the ethics committee of the Bavarian Medical Association (REC reference numbers: F4: #06068, FF4: #06068) and all were carried out in accordance with the principles of the Declaration of Helsinki. This covers consent for the use of biological material, including genetics. All research participants have signed informed consent prior to taking part in any research activities. The KORA data protection procedures were approved by the responsible data protection officer of the Helmholtz Munich. LLS. In accordance with the Declaration of Helsinki, we obtained written informed consent from all participants prior to their entering the study. Good clinical practice guidelines were maintained. The study protocol was approved by the ethical committee of the Leiden University Medical Center before the start of the study (P01.113).
Consent for publication
All authors have read and approved the manuscript for publication.
Competing interests
S.E.B. receives payments and options as a consultant to ZOE LIMITED. The other authors have no competing interests to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ricardo Costeira, Email: ricardo.de_oliveira_costeira@kcl.ac.uk.
Jordana T. Bell, Email: jordana.bell@kcl.ac.uk
References
- 1.Mentch SJ, Locasale JW. One-carbon metabolism and epigenetics: understanding the specificity. Ann N Y Acad Sci. 2016;1363:91–98. doi: 10.1111/nyas.12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serefidou M, Venkatasubramani AV, Imhof A. The impact of one carbon metabolism on histone methylation. Front Genet. 2019;10:1–7. doi: 10.3389/fgene.2019.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang N. Epigenetic modulation of DNA methylation by nutrition and its mechanisms in animals. Anim Nutr. 2015;1:144–151. doi: 10.1016/j.aninu.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An Y, Feng L, Zhang X, Wang Y, Wang Y, Tao L, et al. Dietary intakes and biomarker patterns of folate, vitamin B6, and vitamin B12 can be associated with cognitive impairment by hypermethylation of redox-related genes NUDT15 and TXNRD1. Clin Epigenetics. 2019;11:1–19. doi: 10.1186/s13148-019-0741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barroso M, Handy DE, Castro R. The link between hyperhomocysteinemia and hypomethylation. J Inborn Errors Metab Screen. 2017;5:232640981769899. [Google Scholar]
- 6.Ganguly P, Alam SF. Role of homocysteine in the development of cardiovascular disease. Nutr J. 2015;14:1–10. doi: 10.1186/1475-2891-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamberlain JA, Dugué PA, Bassett JK, Hodge AM, Brinkman MT, Joo JHE, et al. Dietary intake of one-carbon metabolism nutrients and DNA methylation in peripheral blood. Am J Clin Nutr. 2018;108:611–621. doi: 10.1093/ajcn/nqy119. [DOI] [PubMed] [Google Scholar]
- 8.Mandaviya PR, Joehanes R, Brody J, Castillo-fernandez JE, Dekkers KF, Do AN, et al. Association of dietary folate and vitamin B-12 intake with genome-wide DNA methylation in blood: a large-scale epigenome-wide association analysis in 5841 individuals. Am J Clin Nutr. 2019;110:437–450. doi: 10.1093/ajcn/nqz031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dugué PA, Chamberlain JA, Bassett JK, Hodge AM, Brinkman MT, Joo JHE, et al. Overall lack of replication of associations between dietary intake of folate and vitamin B-12 and DNA methylation in peripheral blood. Am J Clin Nutr. 2020;111:228–230. doi: 10.1093/ajcn/nqz253. [DOI] [PubMed] [Google Scholar]
- 10.Joubert BR, Den Dekker HT, Felix JF, Bohlin J, Ligthart S, Beckett E, et al. Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nat Commun. 2016;7:10577. doi: 10.1038/ncomms10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hafizah YN, Ang LC, Yap F, Najwa WN, Cheah WL, Ruzita AT, et al. Validity and reliability of a food frequency questionnaire (FFQ) to assess dietary intake of preschool children. Int J Environ Res Public Health. 2019;16:4722. doi: 10.3390/ijerph16234722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subar AF, Freedman LS, Tooze JA, Kirkpatrick SI, Boushey C, Neuhouser ML, et al. Addressing current criticism regarding the value of self-report dietary data. J Nutr. 2015;145:2639–2645. doi: 10.3945/jn.115.219634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shim J-S, Oh K, Kim HC. Dietary assessment methods in epidemiologic studies. Epidemiol Health. 2014;36:e2014009. doi: 10.4178/epih/e2014009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang A, Sun H, Yan G, Wang P, Wang X. Metabolomics for biomarker discovery: moving to the clinic. Biomed Res Int. 2015 doi: 10.1155/2015/354671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guasch-Ferre M, Bhupathiraju SN, Hu FB. Use of metabolomics in improving assessment of dietary intake. Clin Chem. 2018;64:82–98. doi: 10.1373/clinchem.2017.272344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Posma JM, Garcia-Perez I, Frost G, Aljuraiban GS, Chan Q, Van Horn L, et al. Nutriome–metabolome relationships provide insights into dietary intake and metabolism. Nat Food. 2020;1:426–436. doi: 10.1038/s43016-020-0093-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiuve SE, Fung TT, Rimm EB, Hu FB, McCullough ML, Wang M, et al. Alternative dietary indices both strongly predict risk of chronic disease. J Nutr. 2012;142:1009–1018. doi: 10.3945/jn.111.157222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet. 2017;49:131–138. doi: 10.1038/ng.3721. [DOI] [PubMed] [Google Scholar]
- 19.Yoshii K, Hosomi K, Sawane K, Kunisawa J. Metabolism of dietary and microbial vitamin b family in the regulation of host immunity. Front Nutr. 2019;6:1–12. doi: 10.3389/fnut.2019.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mascolo E, Vernì F. Vitamin B6 and diabetes: relationship and molecular mechanisms. Int J Mol Sci. 2020;21:3669. doi: 10.3390/ijms21103669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellmann H, Mooney S. Vitamin B6: A molecule for human health? Molecules. 2010;15:442–459. doi: 10.3390/molecules15010442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyon P, Strippoli V, Fang B, Cimmino L. B vitamins and one-carbon metabolism: implications in human health and disease. Nutrients. 2020;12:1–24. doi: 10.3390/nu12092867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obeid R, Geisel J, Nix WA. 4-Pyridoxic acid/pyridoxine ratio in patients with type 2 diabetes is related to global cardiovascular risk scores. Diagnostics. 2019;9:1–12. doi: 10.3390/diagnostics9010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulenad PM, Ulvik A, Rios-Avila R, Gregory JA. Direct and functional biomarkers of vitamin B6 status. Annu Rev Nutr. 2018;35:33–70. doi: 10.1146/annurev-nutr-071714-034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulkarni A, Dangat K, Kale A, Sable P, Chavan-Gautam P, Joshi S. Effects of altered maternal folic acid, vitamin B12 and docosahexaenoic acid on placental global DNA methylation patterns in wistar rats. PLoS ONE. 2011;6:1–7. doi: 10.1371/journal.pone.0017706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iglesia I, Huybrechts I, González-Gross M, Mouratidou T, Santabárbara J, Chajès V, et al. Folate and Vitamin B12 concentrations are associated with plasma DHA and EPA fatty acids in European adolescents: the healthy lifestyle in Europe by nutrition in adolescence (HELENA) study. Br J Nutr. 2017;117:124–133. doi: 10.1017/S0007114516004414. [DOI] [PubMed] [Google Scholar]
- 27.de Soest APM, van de Rest O, Witkamp RF, Cederholm T, de Groot LCPGM. DHA status influences effects of B-vitamin supplementation on cognitive ageing: a post-hoc analysis of the B-proof trial. Eur J Nutr. 2022;61:3731–3739. doi: 10.1007/s00394-022-02924-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hallen A, Cooper AJL. Reciprocal control of thyroid binding and the pipecolate pathway in the brain. Neurochem Res. 2017;42:217–243. doi: 10.1007/s11064-016-2015-9. [DOI] [PubMed] [Google Scholar]
- 29.Fujita T, Hada T, Higashino K. Origin of D- and L-pipecolic acid in human physiological fluids: a study of the catabolic mechanism to pipecolic acid using the lysine loading test. Clin Chim Acta. 1999;287:145–156. doi: 10.1016/s0009-8981(99)00129-1. [DOI] [PubMed] [Google Scholar]
- 30.Plecko B, Hoeger H, Jakobs C, Struys E, Stromberger C, Leschnik M, et al. Pipecolic acid concentrations in brain tissue of nutritionally pyridoxine-deficient rats. J Inherit Metab Dis. 2005;28:689–693. doi: 10.1007/s10545-005-0071-4. [DOI] [PubMed] [Google Scholar]
- 31.Petersen AK, Zeilinger S, Kastenmüller G, Werner RM, Brugger M, Peters A, et al. Epigenetics meets metabolomics: an epigenome-wide association study with blood serum metabolic traits. Hum Mol Genet. 2014;23:534–545. doi: 10.1093/hmg/ddt430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng Y, Akmentin W, Connelly MA, Lund-Katz S, Phillips MC, Williams DL. Scavenger receptor BI (SR-BI) clustered on microvillar extensions suggests that this plasma membrane domain is a way station for cholesterol trafficking between cells and high-density lipoprotein. Mol Biol Cell. 2004;15:384–396. doi: 10.1091/mbc.E03-06-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ros S, Santos CR, Moco S, Baenke F, Kelly G, Howell M, et al. Functional metabolic screen identifies 6-phosphofructo-2-kinase/fructose-2, 6-biphosphatase 4 as an important regulator of prostate cancer cell survival. Cancer Discov. 2012;2:328–343. doi: 10.1158/2159-8290.CD-11-0234. [DOI] [PubMed] [Google Scholar]
- 34.Shen L, Du J, Xia Y, Tan Z, Fu Y, Yang Q, et al. Genome-wide landscape of DNA methylomes and their relationship with mRNA and miRNA transcriptomes in oxidative and glycolytic skeletal muscles. Sci Rep. 2016;6:1–11. doi: 10.1038/srep32186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morcillo S, Martín-Núñez GM, Garciá-Serrano S, Gutierrez-Repiso C, Rodriguez-Pacheco F, Valdes S, et al. Changes in SCD gene DNA methylation after bariatric surgery in morbidly obese patients are associated with free fatty acids. Sci Rep. 2017;7:1–8. doi: 10.1038/srep46292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim JKM, Delaidelli A, Minaker SW, Zhang HF, Colovic M, Yang H, et al. Cystine/glutamate antiporter xCT (SLC7A11) facilitates oncogenic RAS transformation by preserving intracellular redox balance. Proc Natl Acad Sci. 2019;116:9433–9442. doi: 10.1073/pnas.1821323116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewerenz J, Hewett SJ, Huang Y, Lambros M, Gout PW, Kalivas PW, et al. The cystine/glutamate antiporter system xc- in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid Redox Signal. 2013;18:522–555. doi: 10.1089/ars.2011.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dalto DB, Matte JJ. Pyridoxine (Vitamin B6) and the glutathione peroxidase system; a link between one-carbon metabolism and antioxidation. Nutrients. 2017;9:1–13. doi: 10.3390/nu9030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallerga CL, Zhang F, Fowdar J, McRae AF, Qi T, Nabais MF, et al. Analysis of DNA methylation associates the cystine–glutamate antiporter SLC7A11 with risk of Parkinson’s disease. Nat Commun. 2020;11:1–10. doi: 10.1038/s41467-020-15065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scalise M, Pochini L, Console L, Losso MA, Indiveri C. The Human SLC1A5 (ASCT2) amino acid transporter: from function to structure and role in cell biology. Front Cell Dev Biol. 2018;6:1–17. doi: 10.3389/fcell.2018.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu C, Marioni RE, Hedman AK, Pfeiffer L, Tsai PC, Reynolds LM, et al. A DNA methylation biomarker of alcohol consumption. Mol Psychiatry. 2018;23:422–433. doi: 10.1038/mp.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perrier F, Viallon V, Ambatipudi S, Ghantous A, Cuenin C, Chajès V, et al. Association of leukocyte DNA methylation changes with dietary folate and alcohol intake in the EPIC study. Clin Epigenetics. 2019;11:1–13. doi: 10.1186/s13148-019-0637-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendelson MM, Marioni RE, Joehanes R, Liu C, Hedman ÅK, Aslibekyan S, et al. Association of body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: a Mendelian randomization approach. PLoS Med. 2017;14:1–30. doi: 10.1371/journal.pmed.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richard MA, Huan T, Ligthart S, Gondalia R, Jhun MA, Brody JA, et al. DNA methylation analysis identifies loci for blood pressure regulation. Am J Hum Genet. 2017;101:888–902. doi: 10.1016/j.ajhg.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westerman K, Sebastiani P, Jacques P, Liu S, Demeo D, José M. DNA methylation modules associate with incident cardiovascular disease and cumulative risk factor exposure. Clin Epigenetics Clin Epigenetics. 2019;11:1–14. doi: 10.1186/s13148-019-0705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia Y, Brewer A, Bell JT. DNA methylation signatures of incident coronary heart disease: findings from epigenome-wide association studies. Clin Epigenetics. 2021;13:1–16. doi: 10.1186/s13148-021-01175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McRae MP. High-dose folic acid supplementation effects on endothelial function and blood pressure in hypertensive patients: a meta-analysis of randomized controlled clinical trials. J Chiropr Med. 2009;8:15–24. doi: 10.1016/j.jcm.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asbaghi O, Salehpour S, Rezaei Kelishadi M, Bagheri R, Ashtary-Larky D, Nazarian B, et al. Folic acid supplementation and blood pressure: a GRADE-assessed systematic review and dose-response meta-analysis of 41,633 participants. Crit Rev Food Sci Nutr. 2021 doi: 10.1080/10408398.2021.1968787. [DOI] [PubMed] [Google Scholar]
- 49.Forman JP, Rimm EB, Stampfer MJ, Curhan GC. Folate intake and the risk of incident hypertension among US women. JAMA. 2005;293:320–329. doi: 10.1001/jama.293.3.320. [DOI] [PubMed] [Google Scholar]
- 50.Verdi S, Abbasian G, Bowyer RCE, Lachance G, Yarand D, Christofidou P, et al. TwinsUK: the UK adult twin registry update. Twin Res Hum Genet. 2019;22:1–7. doi: 10.1017/thg.2019.65. [DOI] [PubMed] [Google Scholar]
- 51.Huth C, Beuerle S, Zierer A, Heier M, Herder C, Kaiser T, et al. Biomarkers of iron metabolism are independently associated with impaired glucose metabolism and type 2 diabetes: the KORA F4 study. Eur J Endocrinol. 2015;173:643–653. doi: 10.1530/EJE-15-0631. [DOI] [PubMed] [Google Scholar]
- 52.Kowall B, Rathmann W, Stang A, Bongaerts B, Kuss O, Herder C, et al. Perceived risk of diabetes seriously underestimates actual diabetes risk: the KORA FF4 study. PLoS ONE. 2017;12:69–75. doi: 10.1371/journal.pone.0171152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westendorp RGJ, Van Heemst D, Rozing MP, Frölich M, Mooijaart SP, Blauw GJ, et al. Nonagenarian siblings and their offspring display lower risk of mortality and morbidity than sporadic nonagenarians: the Leiden longevity study. J Am Geriatr Soc. 2009;57:1634–1637. doi: 10.1111/j.1532-5415.2009.02381.x. [DOI] [PubMed] [Google Scholar]
- 54.Day N, Oakes S, Luben R, Khaw KT, Bingham S, Welch A, et al. EPIC-Norfolk: study design and characteristics of the cohort. Br J Cancer. 1999;80:95–103. [PubMed] [Google Scholar]
- 55.Teucher B, Skinner J, Skidmore PML, Cassidy A, Fairweather-Tait SJ, Hooper L, et al. Dietary patterns and heritability of food choice in a UK female twin cohort. Twin Res Hum Genet. 2007;10:734–748. doi: 10.1375/twin.10.5.734. [DOI] [PubMed] [Google Scholar]
- 56.Mulligan AA, Luben RN, Bhaniani A, Parry-Smith DJ, O’Connor L, Khawaja AP, et al. A new tool for converting food frequency questionnaire data into nutrient and food group values: FETA research methods and availability. BMJ Open. 2014;4:e004503. doi: 10.1136/bmjopen-2013-004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holland B, Welch A, Unwin D, Buss D, Paul A, Southgate D. McCance and Widdowson’s the composition of foods. 5. Cambridge: The Royal Society of Chemistry; 1992. [Google Scholar]
- 58.Willett WC, Howe R. Adjustmentfor total energyintake in epidemiologic studies. Am J Clin Nutr. 1997;65:1220S–S1228. doi: 10.1093/ajcn/65.4.1220S. [DOI] [PubMed] [Google Scholar]
- 59.Freese J, Feller S, Harttig U, Kleiser C, Linseisen J, Fischer B, et al. Development and evaluation of a short 24-h food list as part of a blended dietary assessment strategy in large-scale cohort studies. Eur J Clin Nutr. 2014;68:324–329. doi: 10.1038/ejcn.2013.274. [DOI] [PubMed] [Google Scholar]
- 60.Illner AK, Harttig U, Tognon G, Palli D, Salvini S, Bower E, et al. Feasibility of innovative dietary assessment in epidemiological studies using the approach of combining different assessment instruments. Public Health Nutr. 2010;14:1055–1063. doi: 10.1017/S1368980010003587. [DOI] [PubMed] [Google Scholar]
- 61.Mitry P, Wawro N, Six-Merker J, Zoller D, Jourdan C, Meisinger C, et al. Usual dietary intake estimation based on a combination of repeated 24-H food lists and a food frequency questionnaire in the KORA FF4 cross-sectional study. Front Nutr. 2019;6:145. doi: 10.3389/fnut.2019.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Slimani N, Deharveng G, Charrondière RU, Van Kappel AL, Ocké MC, Welch A, et al. Structure of the standardized computerized 24-h diet recall interview used as reference method in the 22 centers participating in the EPIC project. Comput Methods Progr Biomed. 1999;58:251–266. doi: 10.1016/s0169-2607(98)00088-1. [DOI] [PubMed] [Google Scholar]
- 63.Bundeslebensmittelschlüssel: BLS-Version 3.02 [Internet]. Max-Rubner Inst. [cited 2021 Apr 21]. Available from: https://www.blsdb.de/
- 64.Streppel MT, De Vries JH, Meijboom S, Beekman M, De Craen AJ, Slagboom PE, et al. Relative validity of the food frequency questionnaire used to assess dietary intake in the Leiden Longevity Study. Nutr J. 2013;12:1–8. doi: 10.1186/1475-2891-12-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.NEVO-tabel, Nederlands Voedingsstoffenbestand 2011. Den Haag: Voedingscentrum; 2011.
- 66.Pallister T, Haller T, Thorand B, Altmaier E, Cassidy A, Martin T, et al. Metabolites of milk intake: a metabolomic approach in UK twins with findings replicated in two European cohorts. Eur J Nutr. 2017;56:2379–91. doi: 10.1007/s00394-016-1278-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wägele B, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–62. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Altmaier E, Fobo G, Heier M, Thorand B, Meisinger C, Römisch-Margl W, et al. Metabolomics approach reveals effects of antihypertensives and lipid-lowering drugs on the human metabolism. Eur J Epidemiol. 2014;29:325–336. doi: 10.1007/s10654-014-9910-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:1–16. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu Z, Niu L, Li L, Taylor JA. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res. 2016;44:1–6. doi: 10.1093/nar/gkv907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Christiansen C, Tomlinson M, Eliot M, Nilsson E, Costeira R, Xia Y, et al. Adipose methylome integrative-omic analyses reveal genetic and dietary metabolic health drivers and insulin resistance classifiers. Genome Med. 2022;14:1–22. doi: 10.1186/s13073-022-01077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niu L, Xu Z, Taylor JA. RCP: a novel probe design bias correction method for Illumina Methylation BeadChip. Bioinformatics. 2016;32:2659–63. doi: 10.1093/bioinformatics/btw285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 75.Gomez-Alonso MDC, Kretschmer A, Wilson R, Pfeiffer L, Karhunen V, Seppälä I, et al. DNA methylation and lipid metabolism: an EWAS of 226 metabolic measures. Clin Epigenetics. 2021;13:1–19. doi: 10.1186/s13148-020-00957-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lehne B, Drong AW, Loh M, Zhang W, Scott WR, Tan ST, et al. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 2015;16:1–12. doi: 10.1186/s13059-015-0600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hellbach F, Sinke L, Costeira R, Baumeister SE, Beekman M, Louca P, et al. Pooled analysis of epigenome-wide association studies of food consumption in KORA, TwinsUK and LLS. Eur J Nutr. 2022 doi: 10.1007/s00394-022-03074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Iterson M, Tobi EW, Slieker RC, Den Hollander W, Luijk R, Slagboom PE, et al. MethylAid: visual and interactive quality control of large Illumina 450k datasets. Bioinformatics. 2014;30:3435–3437. doi: 10.1093/bioinformatics/btu566. [DOI] [PubMed] [Google Scholar]
- 79.Glastonbury CA, Couto Alves A, El-Sayed Moustafa JS, Small KS. Cell-Type heterogeneity in adipose tissue is associated with complex traits and reveals disease-relevant cell-specific eQTLs. Am J Hum Genet. 2019;104:1013–24. doi: 10.1016/j.ajhg.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 82.Bates D, Mächler M, Bolker BM, Walker SC. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67.
- 83.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:1–16. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:1–20. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Christiansen C, Castillo-Fernandez JE, Domingo-Relloso A, Zhao W, El-Sayed Moustafa JS, Tsai PC, et al. Novel DNA methylation signatures of tobacco smoking with trans-ethnic effects. Clin Epigenetics. 2021;13:1–13. doi: 10.1186/s13148-021-01018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Cohort characteristics for the B vitamin metabolomic biomarker discovery. Table S2. Blood metabolites associated with the dietary intake of folate (B9) and vitamins B6 and B12 (metabolome-wide Bonferroni-adj. p < 0.05). Table S3. B vitamin metabolomic biomarker associations adjusted for diet quality (AHEI-2010) and total energy intake (metabolome-wide Bonferroni-adj. p < 0.05). Table S4. B vitamin metabolomic biomarker associations with other nutrient intakes (metabolome-wide Bonferroni-adj. p < 0.05). Table S5. Summary of nutrients associated with the B vitamin metabolomic biomarkers identified (metabolome-wide Bonferroni-adj. p < 0.05). Table S6. B vitamin metabolomic biomarker associations with circulating blood levels of folate (B9), vitamin B12, hcy and hcy-s. Table S7. Metabolite associations with circulating blood levels of folate (B9), vitamin B12, hcy and hcy-s (metabolome-wide Bonferroni-adj. p < 0.05). Table S8. Cohort characteristics for the B vitamin metabolomic biomarker epigenome-wide meta-analysis. Table S9. Annotation of the epigenome-wide association meta-analysis of folate (B9) and vitamins B6 and B12 biomarker metabolites (epigenome-wide Bonferroni-adj. p < 0.05). Table S10. Epigenome-wide association of folate (B9) and vitamins B6 and B12 biomarker metabolites in the KORA F4 cohort (epigenome-wide Bonferroni-adj. p < 0.05). Table S11. Epigenome-wide association of folate (B9) and vitamins B6 and B12 biomarker metabolites in the TwinsUK cohort (epigenome-wide FDR = 10%). Table S12. Cohort characteristics for the B vitamin intake epigenome-wide meta-analysis. Table S13. DMP results in DMP results in the epigenome-wide association meta-analysis of folate (B9) and vitamin B6 dietary intakes. Table S14. Cis expression quantitative trait methylation signals from the BIOS consortium for the DMP identified in this study. Table S15. Gene expression results for the genes annotated in the biomarker metabolite EWAS meta-analysis.
Additional file 2: Figure S1. Correlation matrix of the serum levels of the metabolomic biomarkers identified. Figure S2. Pathway annotations of the 18 metabolomic biomarkers identified for folate and vitamins B6 and B12 (Bonferroni-adj. p < 0.05).
Data Availability Statement
Many of the blood data analysed in TwinsUK is available through GEO GSE62992 and GSE121633 for methylation and EGA EGAD00001001088 for gene expression. Additional TwinsUK individual-level data are not permitted to be shared or deposited due to the original consent given at the time of data collection. However, access to these data can be applied for through the TwinsUK data access committee. For information on access and how to apply http://twinsuk.ac.uk/resources-for-researchers/access-our-data/. The informed consents given by KORA study participants do not cover data posting in public databases. However, data are available upon request from KORA Project Application Self-Service Tool (https://helmholtz-muenchen.managed-otrs.com/external/). Data requests can be submitted online and are subject to approval by the KORA Board. LLS DNA methylation data are available upon request via the BIOS consortium (https://www.bbmri.nl/acquisition-use-analyze/bios). FFQ data is available upon request.