Abstract
The weakened protective efficacy of COVID-19 vaccines and antibodies caused by SARS-CoV-2 variants presents a global health emergency, which underscores the urgent need for universal therapeutic antibody intervention for clinical patients. Here, we screened three alpacas-derived nanobodies (Nbs) with neutralizing activity from twenty RBD-specific Nbs. The three Nbs were fused with the Fc domain of human IgG, namely aVHH-11-Fc, aVHH-13-Fc and aVHH-14-Fc, which could specifically bind RBD protein and competitively inhibit the binding of ACE2 receptor to RBD. They effectively neutralized SARS-CoV-2 pseudoviruses D614G, Alpha, Beta, Gamma, Delta, and Omicron sub-lineages BA.1, BA.2, BA.4, and BA.5 and authentic SARS-CoV-2 prototype, Delta, and Omicron BA.1, BA.2 strains. In mice-adapted COVID-19 severe model, intranasal administration of aVHH-11-Fc, aVHH-13-Fc and aVHH-14-Fc effectively protected mice from lethal challenges and reduced viral loads in both the upper and lower respiratory tracts. In the COVID-19 mild model, aVHH-13-Fc, which represents the optimal neutralizing activity among the above three Nbs, effectively protected hamsters from the challenge of SARS-CoV-2 prototype, Delta, Omicron BA.1 and BA.2 by significantly reducing viral replication and pathological alterations in the lungs. In structural modeling of aVHH-13 and RBD, aVHH-13 binds to the receptor-binding motif region of RBD and interacts with some highly conserved epitopes. Taken together, our study illustrated that alpaca-derived Nbs offered a therapeutic countermeasure against SARS-CoV-2, including those Delta and Omicron variants which have evolved into global pandemic strains.
Keywords: COVID-19, SARS-CoV-2, Nanobody, Broad-spectrum, Therapeutic, Rodent models
Highlights
-
•
Alpaca-derived nanobodies showed a cross-neutralizing activity against SARS-CoV-2 variants.
-
•
aVHH-13-Fc provided complete protection in mild and severe COVID-19 rodent models.
-
•
The binding site between aVHH-13-Fc and the RBD variant of SARS-CoV-2 is partially located on the RBM.
1. Introduction
The pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in billions of confirmed cases and millions of deaths worldwide. Simultaneously, mutations have accumulated in SARS-CoV-2, of which several representative variants of SARS-CoV-2 have been noted as variants of concerns (VOCs) by World Health Organization (WHO). Since 2020, previously circulating VOCs including Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), and Delta (B.1.617.2) have been circulated and induced waves of prevalence (Janik et al., 2021). More recently, Omicron (B.1.1.529) and its sub-lineages have been actively replacing other VOCs to become the most dominant variant. Despite the milder symptoms and lower fatality rate reported in Omicron variants, immune escape caused by accumulated mutations in spike (S) protein has seriously hindered the performance of vaccines and therapeutic countermeasures against SARS-CoV-2 infection (Kustin et al., 2021; Altarawneh et al., 2022; Kuhlmann et al., 2022; Shuai et al., 2022).
Neutralization antibodies have achieved remarkable progress in both prophylactic and therapeutic performance against SARS-CoV-2. Although a variety of therapeutic antibody drugs, such as homologous or heterologous monoclonal antibodies (Chi et al., 2020; VanBlargan et al., 2021; Weinreich et al., 2021) or their cocktails (Baum et al., 2020), nanobodies (Nbs) (Xiang et al., 2020; Huo et al., 2021a; Koenig et al., 2021; Xu et al., 2021), humanized antibodies (Du et al., 2021) and chimeric antibodies (Fedry et al., 2021) have been licensed or approved to enter clinical trials, the current epidemic situation and the emergence of SARS-CoV-2 variants have prompted the acceleration of the development of therapeutic antibody drugs with broad-spectrum neutralizing activity (Baig, 2021; Smail et al., 2021). Among the antibodies mentioned above, particularly, camelid-derived single-domain antibody fragments, namely VHHs or Nbs, are minimal (< 15 kDa), monomeric, soluble and stable therapeutics that are able to reach cryptic epitopes otherwise inaccessible by conventional human antibody domains (Hamers-Casterman et al., 1993; Muyldermans, 2013; Li et al., 2021). These versatile properties in specificity, thermotolerance, stability, low immunogenicity, and ease of manipulation and production in cost-effective systems have made the Nbs a desirable candidate for next-generation interventions against SARS-CoV-2 variants infection (Scully et al., 2019).
Derived from llama immunized with MERS-CoV or SARS-CoV-1 S antigens, cross-reactivity has been reported between SARS-CoV-1 S-directed VHH and SARS-CoV-2 S. Crystal structures of these VHHs bound to their respective viral targets reveal two distinct epitopes, but both VHHs interfere with receptor binding. These data provided the molecular basis for the neutralization of pathogenic beta coronaviruses by VHHs (Hanke et al., 2020; Wrapp et al., 2020a). In further investigations, more Nbs were developed and proved to work by interfering with ACE2 binding, stabilizing the epitope-inaccessible/inactive form of S protein, or inhibiting membrane fusion mediated by SARS-CoV-2 S protein with target cells or premature inducing of the post-fusion conformation to preclude bona fide fusion events (Custodio et al., 2020; Haga et al., 2021; Huo et al., 2021b; Koenig et al., 2021; Yang et al., 2021; Wang et al., 2023). Although these VHHs were well established and were fully revealed in crystal structures in terms of neutralization of beta coronaviruses, development of therapeutic antibodies to confer broad and protective activity is still a global priority, particularly for vulnerable individuals of coronavirus disease 2019 (COVID-19). Of importance, a comprehensive evaluation of these Nbs in distinctive animal models, both mild and severe COVID-19 infection models, has not yet been completed, which hampered the fully application of Nbs between different groups of COVID-19 individuals in clinical settings.
In our study, to search for broad neutralization antibodies, we identified a total of 20 Nbs capable of binding to the receptor binding domain (RBD) of SARS-CoV-2 and neutralizing pseudovirus from hyper-immunized alpacas by utilizing a nanobody phage library. Three of them were characterized as potency against all tested VOCs, including SARS-CoV-2 D614G, Alpha, Beta, Gama, Delta and Omicron subvariants BA.2 and BA.4/5. The half maximal inhibitory concentration (IC50) against a panel of tested coronaviruses reached single digital μg/mL or less, representing broad and potent Nbs. Passive delivery of three of the representative Nbs, aVHH-11-Fc, aVHH-13-Fc, aVHH-14-Fc, protected mice from the lethal challenge of mouse-adapted SARS-CoV-2 strain BMA8 (Yan et al., 2022). Further evaluation of aVHH-13-Fc in golden hamsters confirmed the therapeutic efficacy of the nanobody. These results indicated that we have identified a broad-spectrum and protective nanobody that may provide potential benefits for COVID-19 patients with mild to severe symptoms.
2. Material and methods
2.1. Animals, viruses and cells
Two 3–4-year-old male Australian alpacas were purchased and fed at the Qifeng Mountain alpaca breeding plant in Taiyuan, Shanxi Province, China. 9-month-old female BALB/c mice and 8-week-old female golden hamsters were purchased from Changchun Yisi Experimental Animal Technology Center. BALB/c mouse-adapted SARS-CoV-2 (BMA8), SARS-CoV-2 prototype (human/CHN/Beijing-01/2020), Delta (CSTR.16698.06. NPRC6. CCPM-B-V-049-2105-6), Omicron BA.1 (Omicron CoV/human/CHN_CVRI-01/2022) and BA.2 (Omicron CoV/human/CHN_CVRI-04/2022) variants were stored in the Biosafety Level 3 Laboratory (BSL-3) of Changchun Veterinary Research Institute. BMA8 was obtained by passaging the SARS-CoV-2 prototype in aged BALB/c mice for eight generations, in which a mouse-adapted virus caused 100% death in aged BALB/c mice (Yan et al., 2022). Vero E6 cells (ATCC® CRL-1587™) were cultured at 37 °C in Dulbecco's modified Eagle medium (DMEM) (GIBCO, Grand Island, NY, USA) containing 10% fetal bovine serum (ThermoFisher, MA, USA) with 5% CO2 for BMA8 propagation and titration.
2.2. Immunization of alpaca, construction of phage display VHH library and isolation of VHH phage specific for SARS-CoV-2
Alpacas were simultaneously immunized with 150 μg SARS-CoV-2 S-trimer protein (SinoBiological, Pecking, China) and 1 mg DNA vaccine for 6 times. The DNA vaccine was constructed based on pcDNA3.1(+) backbone expressing the extracellular domain, aa 1–1208 of SARS-CoV-2 S protein. Briefly, 150 μg SARS-CoV-2 S-trimer protein was mixed with 15 μg of ODN and 1.5 mL aluminum adjuvant (Thermo Fisher, MA, USA) as a subunit vaccine. 1 mg DNA vaccine was mixed with 10 μg of ODN. Two vaccines were simultaneously immunized at a 3-week interval (week 0, week 3, week 6, week 9, week 12) via a three-point subcutaneous injection on the neck. Alpaca peripheral blood was collected on 7-day after final booster immunization. Peripheral blood mononuclear cells (PBMCs) from whole blood were isolated using a lymphocyte separation solution Ficoll (GE Healthcare, Uppsala, Sweden), and total RNA of PBMCs was extracted with Trizol (Ambion, Texas, USA). Reverse transcription was performed with SuperScript III reverse transcriptase to obtain cDNA according to the manufacturer's instruction (Takara, Dalian, China). The fragments of the VHH gene were amplified by two rounds of PCR and cloned into the phage plasmid pcomb3XSS vector (Hou et al., 2022). The upstream primer sequence was 5′-CTTGGTGGTCCTGGCTGC. The downstream primer sequence was GGTACGTGCTGTTGAACTGTTCC-3′. The recombinant plasmids were electrically transferred to E. coli TG1 competent cells (Agilent, California, USA). Collected ampicillin-resistant bacterial colonies were defined as bacterial libraries. Bacterial libraries containing the VHH library were infected with helper phage VCSM13 to produce phages displaying the encoded VHHs. Phages were purified and precipitated with PEG/NaCl to obtain a phage library. After three round panning by RBD protein (GenBank: MN908947) immobilized on immune tube, monoclonal phages were tested by phage enzyme-linked immunosorbent assay (Phage-ELISA) (Gauhar et al., 2021). Positive candidates screened by Phage-ELISA were inoculated from monoclonal plate to 2 mL 2× YT medium, cultured at 37 °C until OD600 to 0.8. The positive bacterial colonies were sequenced. The sequencing primer sequence was 5′-TGTGTGGAATTGTGAGCGGA-3′. Sequencing results were analyzed via https://www.imgt.org/IMGT_vquest/input.
2.3. Molecular cloning and expression of VHH
Three Nb monomers were selected for expression according to the results of phage ELISA and pseudovirus neutralization assay. The gene sequence of the antibodies was cloned to pcDNA3.1(+) vector, and pcDNA3.1(+)-Fc vector with human IgG1 Fc fusion, respectively. The recombinant plasmids were transfected into 293F cells to express the Nbs. A volume of 160 μL ExpiFectamine™ 293 Reagent was added into opti-MEM with a final volume of 3 mL 60 μg recombinant plasmid was taken and added into opti-MEM with a final volume of 3 mL, and placed at room temperature for 5 min. Plasmids was added to the transfection reagents drop by drop, placed at room temperature for 15 min. The density of 293F cells was adjusted to 3 × 106 cells/mL, and the mixture was added drop by drop. Enhancer I and enhancer II were added after cultured for 20 h (37 °C, 8% CO2, 125 rpm). At 4 days after transfection, the supernatant containing Nbs was purified by Histrap™ HP purification column (Cytiva, Danaher, USA) according to instructions. The supernatants containing Nbs fused with human IgG1 Fc were purified according to instructions by HiTrap™ MabSelect SuRe™ LX purification column (Cytiva, Danaher, USA).
2.4. Metabolic half-life of Nb in vivo
To investigate the metabolism of Nb in vivo, 8-week-old female BALB/c mice were intravenously injected with aVHH-13 or aVHH-13-Fc at a dose of 3 mg/kg into the tail vein. 50 μL blood samples were collected at pre-set time point 5 min, 10 min, 15 min, 30 min, 45 min, 1 h, 1.5 h and 2 h post-injection of aVHH-13 or PBS control. 50 μL blood samples were taken at 30 min, 2 h, 6 h, 1 day, 3 days, 5 days, 7 days, 9 days and 11 days post administration of aVHH-13-Fc. Metabolism of Nbs was detected by indirect ELISA, and the method in details was provided as below. Tmax was calculated by DAS (Drug And Statistics) 2.0 (software available at https://www.drugchina.net/). Through compartmental model, weight analysis and automatic compartmental judgment, F-test, AIC judgment, the optimal number of atrioventricular and weight value were determined.
2.5. SARS-CoV-2 pseudovirus-based neutralization assay
Neutralization titer was measured based on lentivirus pseudoviral packaging system according to the previous protocol (Donofrio et al., 2021). Briefly, pseudoviruses of SARS-CoV-2 Alpha, Beta, Delta, Gamma, Omicron sub-lineages BA.1, BA.2, BA.4 and BA.5 variants were prepared by co-transfection of SARS-CoV-2 S and lentivirus skeleton plasmids into HEK293 cells. Pseudoviruses were harvested 48 h post transfection. For pseudovirus-based neutralization assay, pseudoviruses were added in serial diluted serum samples or antibodies. After incubation for 1 h at 37 °C, HEK 293 cells were added to the mixture. After incubation for 48 h, luciferase was added and neutralization titers were presented as IC50.
2.6. Competitive ELISA
For competitive ELISA, 1 μg/mL recombinant SARS-CoV-2 RBD-his protein (SinoBiological, Pecking, China) was added in 96-well plates and incubated at 4 °C overnight. The plates were washed with PBST buffer and blocked with 3% BSA solution. The plates were washed three times with PBST buffer and then 40 ng/mL ACE2-Fc was added. 20 μg/mL purified Nbs were immediately added and incubated at 37 °C for 1 h. ACE2-Fc control well and blank well were set at the same time. After three times of washing, mouse monoclonal anti-human IgG Fc (Abcam, Cambridge, UK) antibody was added at 1:10,000 dilution and incubated at 37 °C for 1 h, After three times of washing, goat anti-mouse IgG (H + L) HRP (Bioworld, Minnesota, USA) antibody was added at 1:15,000 dilution and incubated for 1 h. Subsequently, TMB was added for color rendering. The reaction was stopped with adding 1 nmol/L H2SO4. Results were read at 450 nm absorbance. Percent activity (%) = [1 − (ODVHH − ODblank)/(ODACE2 − ODblank)] × 100%.
2.7. Indirect ELISA
SARS-CoV-2 RBD-his protein (SinoBiological, Pecking, China) was used as an antigen with a coating concentration of 1 μg/mL, 50 μL/well coated in 96-well plates at 4 °C overnight. Then the plates were blocked with 1% BSA solution and incubated for 2 h at 37 °C. The aVHH-11-Fc, aVHH-13-Fc and aVHH-14-Fc antibodies with initial concentration of 5 μg/mL were performed with a series of two-fold dilutions and incubated for 1.5 h at 37 °C. The plates were washed for five times with PBST buffer. Then 100 μL horseradish peroxidase (HRP) labeled rabbit anti-camelied VHH monoclonal antibody (GenScript, New Jersey, USA) was added at 1:15,000 dilution and incubated for 1 h at 37 °C. After five times of PBST buffer washing, 100 μL TMB (3,3′,3,5′-tetramethyl benzidine) was added for color rendering, and 100 μL/well H2SO4 solution was added to stop color rendering. Finally, the absorbance value was measured at 450 nm.
2.8. SARS-CoV-2 authentic virus-based neutralization assay
Neutralization titer was detected using SARS-CoV-2 BMA8, prototype, Delta, Omicron BA.1 and Omicron BA.2. The three strains of Nbs were diluted in a 96-well plate with a concentration of 50 μg/mL in the first well, followed by successive dilution in a two-fold scale. Subsequently, 50 μL SARS-CoV-2 were added into 96 well plates, with 100 median tissue culture infective dose (TCID50) per well. The mixture was incubated at 37 °C and 5% CO2 for 1 h. Subsequently, 100 μL Vero E6 cells were added to each well. Virus control and blank cell control wells were set. After 72 h, the results were obtained according to the CPE. The maximum antibody dilution concentrations that neutralize all viral CPE were defined as the neutralizing titer.
2.9. Mice and golden hamster challenge study
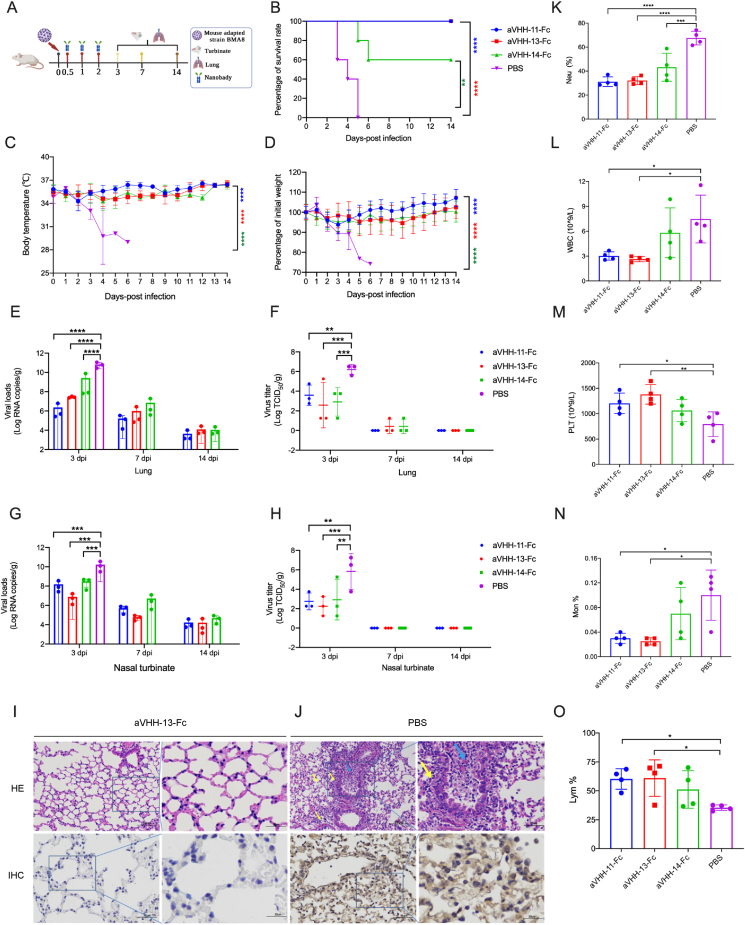
9-month-old female BALB/c mice were randomized into four groups (n = 20). Animals were infected with 10 median lethal dose (LD50) mice-adapted SARS-CoV-2 BMA8 through intranasal delivery route. The mice were given aVHH-11, aVHH-13 and aVHH-14 at a dose of 20 mg/kg at 0.5 h, 1 h and 2 h after infection through intranasal delivery route, respectively. All mice were monitored for clinical signs of disease, survival rate, weight change and temperature for 14 days. At 3 days post infection (dpi), 7 dpi and 14 dpi, three mice in each group were sacrificed, and lungs and nasal turbinates were harvested to test viral RNA copies, viral titers, histopathology and immunohistochemistry (IHC). Animals were euthanized if they lost 25% of their initial body weigh following the Institutional Animal Care and Use Committee (IACUC) regulation.
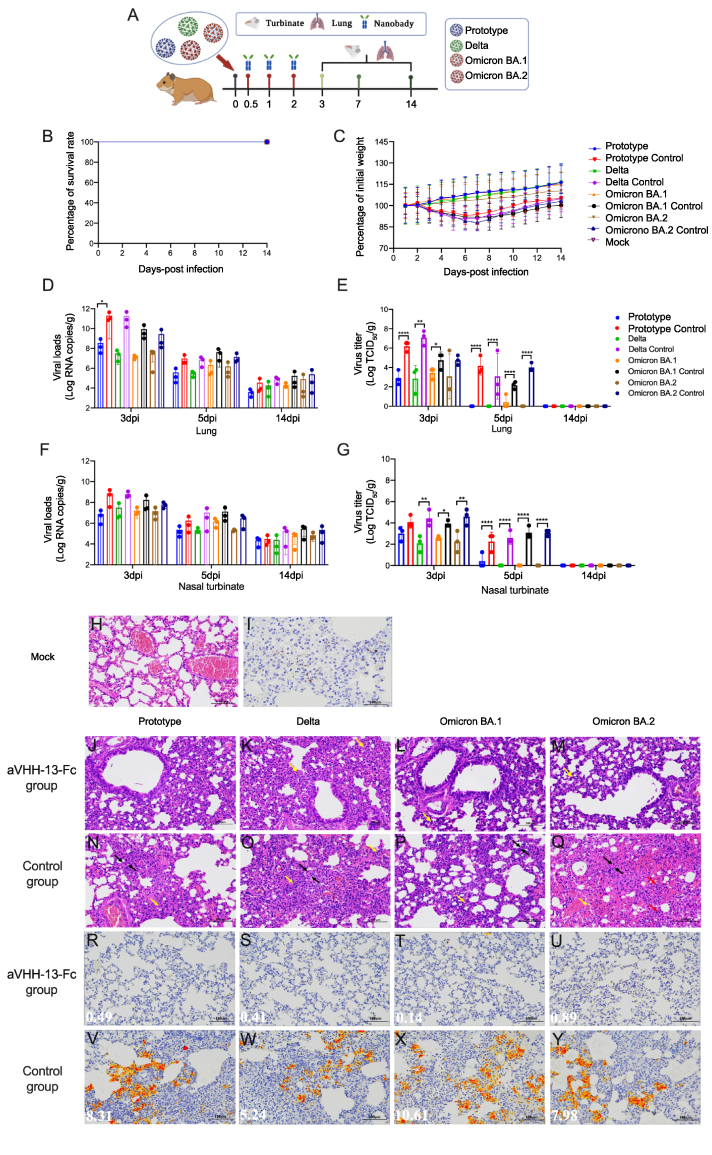
8-week-old female golden hamsters were randomized into eight groups (n = 14). The golden hamsters were challenged with 1000 TCID50 SARS-CoV-2 prototype, Delta, Omicron sub-lineages BA.1 and BA.2 through intranasal delivery route. The hamsters were given aVHH-13 three times at a dose of 20 mg/kg at 0.5 h, 1 h and 2 h after infection, respectively. Clinical signs and weight were monitored for 14 days. At 3 dpi, 7 dpi and 14 dpi, three hamsters in each group were euthanized. Referred to the mice challenge study, lungs and turbinates were harvested to test viral RNA copies, viral titers. Histopathology and IHC were performed in the lungs.
2.10. Viral load determination by RT-qPCR
Viral load determination was conducted according to the previous study (Wang et al., 2022; Yan et al., 2022). Briefly, viral RNA from homogenate samples was extracted with an RNA extraction Mini kit (TIANGEN, Pecking, CN) according to the manufacturer's protocol. Quantification of viral RNA was performed by RT-qPCR targeting the N gene of SARS-CoV-2. RT-qPCR was performed using Premix Ex Taq (Takara, Dalian, China) with the following primers and probes: NF (5′-GGGGAACTTCTCCTGCTAGAAT-3′), NR (5′-CAGACATTTTGCTCTCAAGCTG-3′) and NP (5′-FAM-TTGCTGCTGCTTGACAGATT-TAMRA-3′).
2.11. Viral load determination by TCID50
To determine the TCID50 titer, the tissue homogenate supernatant was serially diluted at a 10-fold interval. 100 μL of samples were added to 96-well cell culture plates that had been plated with Vero E6 cells. The plates were incubated at 37 °C for 1 h and then washed three times with sterile PBS buffer. 100 μL of DMEM medium (CORNING, New York, USA) containing 2% FBS and 1% penicillin streptomycin was added into 96-well plate. The TCID50 was calculated by observing the number of wells with CPE based on Reed and Muench method after incubation for 72 h.
2.12. Hematological test
To determine complete blood cell counts in mice, blood was collected through the orbital venous plexus into anticoagulant tubes containing EDTAK2. The blood cell count was carried out under the automatic blood analyzer (BC-5000 vet, Mindray, China), and the operating procedures were carried out according to the instrument operating instructions.
2.13. Histopathology and IHC
The procedures of the histopathology and IHC assays were conducted according to the previous study (Wang et al., 2022; Yan et al., 2022). Lung samples from the mice which were euthanized at 3 dpi were fixed in 10% buffered formalin and embedded in paraffin wax. Lung tissue was stained with hematoxylin and eosin (H&E) for histopathological assessment. For IHC, quenching of paraffin-embedded lung tissue was performed with 3% hydrogen peroxide in methanol for 10 min. Tissue was added with 500-fold dilution of mouse monoclonal anti-SARS-CoV-2 N antibody (SinoBiological, Pecking, China) at 4 °C overnight. After incubation with primary antibody, the sections were washed with PBS buffer for three times and species-matched secondary antibodies were applied for 2 h at 4 °C. The sections were observed under ZEISS Axio Vert. A1 microscope (ZEISS Axio Vert. A1, Germany). Positive regions were marked in brown and percentage of positive area was marked in white font.
2.14. Molecular docking
Crystal structure of the C-terminal domain of SARS-CoV-2 RBD proteins were applied in structural modeling (built at https://swissmodel.expasy.org/). The chemical structures of the aVHH-13 as MOL2 files were converted into three dimensional PDB files and energy minimization was conducted using MM2 force field. Energy minimized 3D structures were then converted into docking input file format (i.e. PDBQT). Autodock (version 4.2) (available at https://autodock.scripps.edu/download-autodock4/) was used for protein-ligand docking. The 3D-grid box was set to be with dimensions 23.75 × 7.54 × 21.50 Å in XYZ axes, with a grid point spacing of 0.587 Å. The binding poses were generated by the Lamarckian Genetic Algorithm. Ten binding poses were generated for each ligand. After docking, the 2D receptor-ligand complexes were constructed and visualized using PyMOL (available at https://pymol.org/2/).
2.15. Data analysis and processing
GraphPad Prism 8.0. Software (GraphPad Software Inc. San Diego, CA, USA) was used to analyze the data, which were expressed as mean ± standard error of mean (SEM). Significant differences between groups were determined using a one-way ANOVA analysis of variance. P < 0.05 was considered statistically significant. Significance levels were defined as ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 and ∗∗∗∗P < 0.0001.
3. Results
3.1. Alpaca immunization and neutralizing antibody (nAb) detection
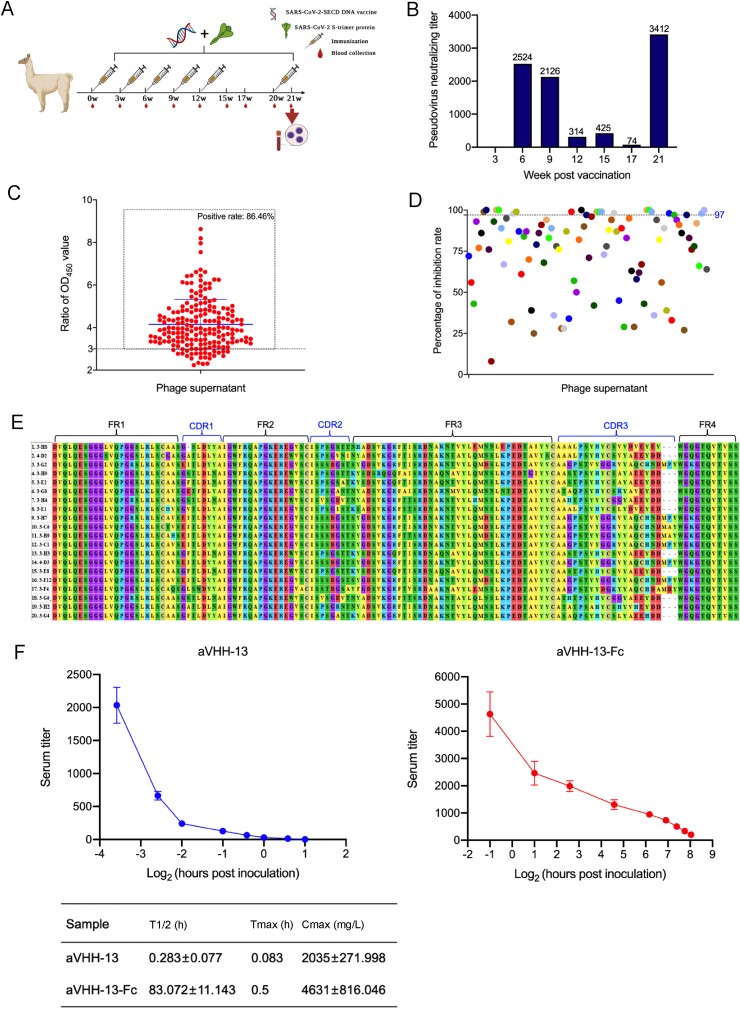
To enhance the immune response and determine the optimum time point for PBMCs isolation and phage library establishment, alpacas were immunized with DNA vaccine and S-trimer protein at a three-week interval (Fig. 1A). Correspondingly, nAb titers were measured. The results showed that nAb titer against SARS-CoV-2 D614G pseudovirus reached 2524 at 6th week post vaccination. Nevertheless, vaccination from week 9 to week 12 failed to strengthen nAb titers, which may due to immune tolerance. At week 20, another boosting dose was given, which aroused an upmost nAb titer of up to 3412 (Fig. 1B). Consequently, PBMCs were collected at this time point. Overall, after 6 doses of immunization, PMBCs were obtained when the nAb titer reached the upmost level at week 21.
Fig. 1.
Screening and characterization of SARS-CoV-2 nanobodies (Nbs). A Alpacas were immunized with SARS-CoV-2 S DNA vaccine and S-trimer protein for 6 doses at week 0, 3, 6, 9, 12, and 20, respectively. B Neutralizing titers of immunized alpaca serum was detected by SARS-CoV-2 D614G pseudovirus based on HIV pseudovirus system following vaccination. PBMCs were isolated from the whole blood of alpacas on week 21 for construction of phage library. C Positive rate of 192 monoclonal phage supernatants by phage-ELISA, the positive sample was defined as the ratio of OD450 value between the experiment group and negative control is greater than 3. D Percentage of inhibition of 192 monoclonal phage supernatants to SARS-CoV-2 D614G pseudovirus. According to the capture of luciferase report gene, Nbs with inhibition rates greater than or equal to 97% were extracted for sequencing and alignment. E Sequence alignment of 20 VHH gene sequences. FR1 to FR4 and CDR1 to CDR3 were marked. F Metabolic dynamics of Nbs. Antibody level of aVHH-13 and aVHH-13-Fc were measured at different time points post tail vein injection in mice. T1/2: the half-life of terminal elimination, the time required for the terminal phase blood drug concentration to decrease by half. Tmax: peak time, the time for the drug action to reach the peak. Cmax: peak concentration, the highest value of blood drug concentration after administration. Data were presented as mean with standard deviation.
3.2. Screening of SARS-CoV-2 Nbs
After RNA extraction and reverse transcription of isolated PBMCs, a phage library with a capacity of 8 × 1013 PFU/mL was established. The positive rate of phage library was 96%. To identify Nbs that specifically bind to the SARS-CoV-2 RBD protein, supernatants from 192 randomly selected monoclonal phages were detected by phage ELISA. Positive samples were defined by the OD450 ratio of the specific sample to negative control over 3. The result suggested that the positive rate of phage supernatants was 86.46% (166/192) (Fig. 1C). Meanwhile, 192 phage supernatants were tested by pseudovirus neutralization detection assay. Based on nanobody (Nb) screening results, 20 samples with inhibition rates greater than or equal to 97% were selected, of which inhibition rates of 7 samples were 100% (Fig. 1D). Sequence comparison of the CDR1, CDR2 and CDR3 regions of the Nb sequences showed that the longest CDR3 region sequence was 22 amino acids. The genetic Nbs of the same family shared a homologous CDR3 sequence but have a few different amino acids in their framework or CDR3 sequence (Fig. 1E).
To evaluate the in vivo half-life of Nbs, we injected the Nbs into mice through the tail vein, then collected the sera from the mice after different time intervals, and measured the VHH titer using indirect ELISA. To extend the half-life of Nbs, they were fused with Fc, termed aVHH-13-Fc. aVHH-13-Fc exhibited a significantly extended half-life of 71.929–94.215 h, while the half-life of aVHH-13 was 0.206–0.36 h. The time required to reach the highest concentration (drug peak concentration) on the concentration curve of antibody aVHH-13 and aVHH-13-Fc in mouse plasma after administration is 0.083 h and 0.5 h, respectively. The highest value of Nb concentrations in the blood after administration were 1763.002–2306.998 mg/L in aVHH-13 group and 3814.954–5447.046 mg/L in aVHH-13-Fc group (Fig. 1F). Thus, compared to monomeric Nb, Fc-fused Nb has a significantly extended in vivo half-life likely due to its dimeric structure.
3.3. Competition with ACE2 and binding ability with RBD of Nbs
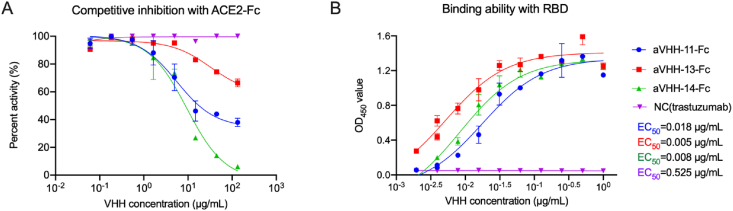
The functional competitive ability of the three Nbs to block RBD-ACE2 binding was tested by competitive ELISA. The results showed that aVHH-11-Fc, aVHH-13-Fc and aVHH-14-Fc had the ability to block ACE2 binding to RBD (Fig. 2A). As for binding capacity with RBD, aVHH-13-Fc exhibited the best binding activity with SARS-CoV-2 RBD protein, with an EC50 of 0.005 μg/mL (135.135 pmol/L), followed by aVHH-14-Fc of 0.008 μg/mL (216.216 pmol/L) and aVHH-11-Fc of 0.018 μg/mL (486.487 pmol/L) (Fig. 2B). This result showed that these three Nbs can preferentially combine with RBD and competitively inhibit the way that RBD combines with ACE2.
Fig. 2.
Competitive inhibition with ACE2 and binding ability with RBD of nanobodies (Nbs). A The competitive inhibition between Nbs and ACE2-Fc in SARS-CoV-2 RBD-binding, determined by competitive ELISA. Briefly, RBD-his protein was coated, following blocking, ACE2-Fc adding and Nbs adding, anti-human IgG Fc antibody and HRP labeled secondary antibody was added. After color rendering, results were read at 450 nm absorbance. B The binding affinity between Nbs and SARS-CoV-2 RBD determined by indirect ELISA. Binding capacity was expressed in half effect concentration (EC50). Data were presented as mean with standard deviation.
3.4. Nbs possess the broad-spectrum neutralizing capability against SARS-CoV-2 pseudovirus
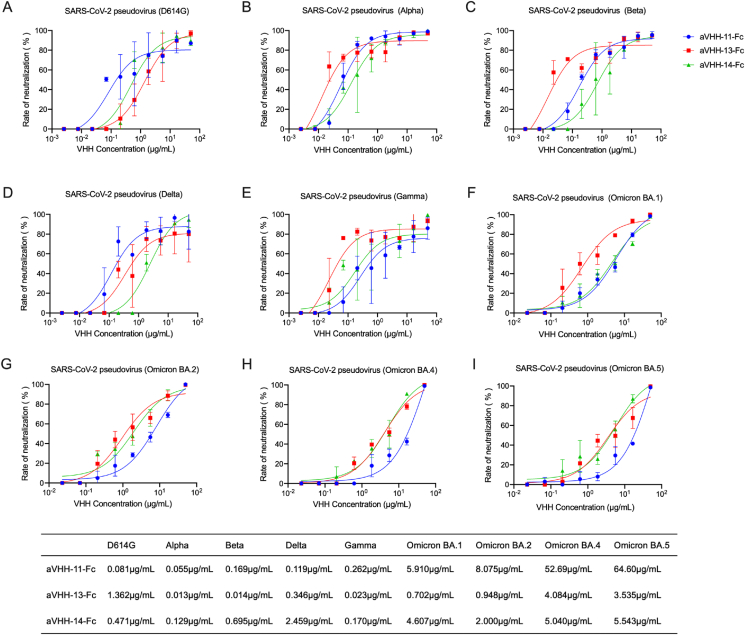
To verify the cross-neutralizing ability of the three Nbs against divergent SARS-CoV-2 variants, a lentivirus-based pseudovirus neutralizing assay was carried out. The aVHH-11-Fc exhibited the strongest inhibitory effect on SARS-CoV-2 D614G (IC50 = 0.081 μg/mL, 2.189 nmol/L) and Delta variant (IC50 = 0.119 μg/mL, 3.216 nmol/L) (Fig. 3A and D); aVHH-13-Fc exhibited the strongest inhibitory effect on Alpha variant (IC50 = 0.013 μg/mL, 351.351 pmol/L), Beta variant (IC50 = 0.014 μg/mL, 378.378 pmol/L), Gamma variant (IC50 = 0.023 μg/mL, 621.622 pmol/L) (Fig. 3B, C, 3E). aVHH-14-Fc could inhibit all variants despite lower efficacy than aVHH-11-Fc or aVHH-13-Fc (Fig. 3A–E). For the Omicron sub-lineage BA.1, BA.2, BA.4 and BA.5, aVHH-13-Fc showed the best neutralization coverage, followed by aVHH-14-Fc (Fig. 3F–I). These results suggested the three Nbs had cross neutralizing activity against all the tested major variants, including D614G, Alpha, Beta, Gamma, Delta and Omicron variants, although their inhibition ability to Omicron-related mutants had been weakened to some extent. These results suggest that aVHH-11-Fc, aVHH-13-Fc and aVHH-14-Fc represent different neutralizing capability on various SARS-CoV-2 variants, of which aVHH-13-Fc seems to provide a broad-spectrum neutralizing of SARS-CoV-2 VOCs.
Fig. 3.
Nanobodies (Nbs) possess broad-spectrum neutralizing capability against pseudoviral SARS-CoV-2 VOCs. A–I Measurement of the neutralizing potency of aVHH-11-Fc, aVHH-13-Fc and aVHH-14-Fc against SARS-CoV-2 D614G, Alpha, Beta, Delta, Gamma, Omicron BA.1, Omicron BA.2, Omicron BA.4, Omicron BA.5 pseudoviruses, respectively. Briefly, neutralization titer was measured based on lentivirus pseudoviral system. Pseudoviruses of SARS-CoV-2 strains were prepared by co-transfection of SARS-CoV-2 S and lentivirus skeleton plasmids, harvested and tittered. Then pseudoviruses were added in serial diluted serum samples or antibodies. After incubation and cells adding, luciferase was added and neutralization titers were presented as IC50. IC50 was determined by the serum dilution that inhibiting 50% concentration of luciferase. Data were presented as mean with standard deviation.
3.5. Nbs exert the broad-spectrum neutralizing capability against authentic SARS-CoV-2
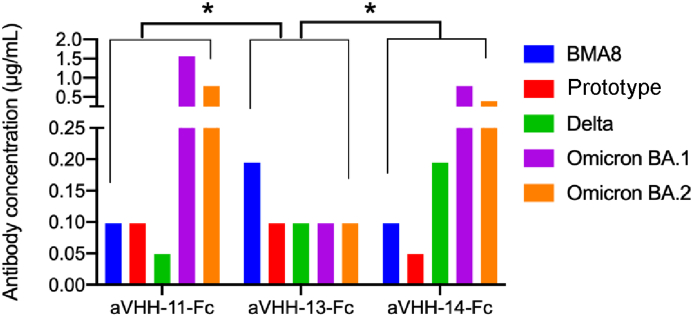
According to the results of the neutralizing assay against authentic SARS-CoV-2, for BMA8, the neutralization titer that neutralizes all viral CPE of aVHH-11-Fc (0.098 μg/mL, 2.649 nmol/L) and aVHH-14-Fc was the same, higher than that of aVHH-13-Fc (0.195 μg/mL, 5.270 nmol/L). For prototype, aVHH-14-Fc (0.049 μg/mL, 1.324 nmol/L) was stronger than aVHH-11-Fc (0.098 μg/mL, 2.649 nmol/L) and aVHH-13-Fc (0.098 μg/mL, 2.649 nmol/L). For Delta, aVHH-11-Fc (0.049 μg/mL, 1.324 nmol/L) was stronger than aVHH-13-Fc (0.098 μg/mL, 2.649 nmol/L) and aVHH-14-Fc (0.195 μg/mL, 5.270 nmol/L). For Omicron BA.1, aVHH-13-Fc (0.098 μg/mL, 2.649 nmol/L) was stronger than aVHH-11-Fc (1.563 μg/mL, 42.243 nmol/L) and aVHH-14-Fc (0.781 μg/mL, 21.108 nmol/L). For Omicron BA. 2, aVHH-13-Fc (0.098 μg/mL, 2.649 nmol/L) was stronger than aVHH-11-Fc (0.781 μg/mL, 21.108 nmol/L) and aVHH-14-Fc (0.391 μg/mL, 10.568 nmol/L). Compared with aVHH-11-Fc and aVHH-11-Fc, aVHH-13-Fc exhibit significantly higher neutralizating capacity against all the tested SARS-CoV-2 strains (P < 0.05). Corresponding to the results of pseudovirus-based neutralization assay, aVHH-13-Fc provided a better coverage of neutralization against SARS-CoV-2 VOCs (Fig. 4).
Fig. 4.
Broad-spectrum neutralization capability against authentic SARS-CoV-2. Neutralization antibody titers of aVHH-11-Fc, aVHH-13-Fc, and aVHH-14-Fc for SARS-CoV-2 BMA8, prototype, Delta, Omicron BA.1 and Omicron BA.2. Briefly, neutralization titer was detected by authentic SARS-CoV-2. Nbs were successively diluted dilution in a two-fold scale and 100 TCID50 SARS-CoV-2 were added. Post incubation, cells were added. The maximum antibody dilution concentrations that neutralize all viral CPE were defined as the neutralizing titer. Statistical analysis was performed by two-way ANOVA, ∗P < 0.05. Data were presented as mean with standard deviation.
3.6. Nbs protect mice against lethal challenge of mouse-adapted SARS-CoV-2
To investigate the therapeutic efficacy of the three Nbs, the previously established COVID-19 lethal mouse model was applied (Yan et al., 2022). Following intranasal challenges with 10LD50 of SARS-CoV-2 BMA8, the aged BALB/c mice were administrated with three doses of aVHH-11-Fc, aVHH-13-Fc or aVHH-14-Fc, respectively, at 0.5 h, 1 h and 2 h post-challenge through intranasal delivery route. PBS was served as a parallel control (Fig. 5A). During continuous monitoring for 14 days, 100% (20/20) animals in the aVHH-11-Fc and aVHH-13-Fc treatment group survived, while 60% of animals survived in the aVHH-14-Fc treatment groups. All animals succumbed to the SARS-CoV-2 BMA8 infection at 3–5 dpi in PBS control groups (Fig. 5B). Accordingly, temperature was well maintained in the Nb treatment groups and the weight of the surviving mice recovered to their original weight at the endpoint of the study, while the body temperature of the mice decreased continuously after SARS-CoV-2 infection in the PBS control group and all mice in the control group had obvious weight loss (Fig. 5C and D).
Fig. 5.
Protective efficacy of nanobodies (Nbs) against lethal infection of the mouse-adapted SARS-CoV-2 in mice. A Experimental schedule for nanobody therapeutics in mice. Groups of BALB/c mice (n = 20) were challenged with 50 LD50 of BMA8 via the intranasal route, followed by three doses successive intranasal administration with aVHH-11-Fc, aVHH-13-Fc or aVHH-14-Fc at 0.5, 1 and 2 h post infection. Nbs with a dose of 20 mg/kg were given to each mouse. The survival rate, weight change and body temperature of BALB/c mice were monitored daily after SARS-CoV-2 BMA8 infection. Lung and nasal turbinates were taken (n = 3) on 3 days post infection (dpi), 7 dpi and 14 dpi, respectively for viral loads determination and virus tittering. B–D Survival rate, weight change and body temperature during the 14 days successive monitoring post challenge. E, G Viral loads quantified by RT-qPCR at 3 dpi, 7 dpi and 14 dpi in lung and nasal turbinates. F, H Virus titer was conducted by TCID50 using Reed-Muench methods at 3 dpi, 7 dpi and 14 dpi in lung and nasal turbinates. I HE and IHC staining of lung tissue from nanobody treatment group (aVHH-13-Fc). No obvious pulmonary lesions or SARS-CoV-2 antigens were recorded. Scale bar = 100 μm. J HE and IHC staining of lung tissue from PBS control group. Pulmonary lesions including mucosal epithelial cell degeneration, necrosis and sloughing, inflammatory necrosis in some lumens (blue arrows), and inflammatory cell infiltration (yellow arrows) were observed. Abundant SARS-CoV-2 antigens were observed in IHC staining (yellow region). K–O The hematological values of BALB/c mice were analyzed at 3 days after SARS-CoV-2 BMA8 infection. K neutrophil (Neu) percentage. L White blood cell (WBC) count. M platelet (PLT). N Monocyte (Mno). O lymphocyte (LYM) percentage. Data are presented as the mean ± standard deviation (n = 4). Statistical analysis was performed by two-way ANOVA. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
Three animals from each group were sacrificed for viral load titration and pathogenesis analysis at 3 dpi, 7 dpi and 14 dpi, respectively. Among the three Nb treatment groups, viral loads in the lungs and nasal turbinates were significantly lower than that of the control group at 3 dpi. Viral RNA levels in the Nb treatment groups were 1.5–4.5 log lower than that in the PBS control group. (Fig. 5E and G). For viral titers, the Nb treatment groups were 2.4–3.9 log lower than the PBS control group in lung and nasal turbinates, respectively (Fig. 5F and H). At 7 dpi and 14 dpi, no significant differences were observed in these Nb treatment groups. The above results suggested that aVHH-11-Fc and aVHH-13-Fc fully protected BALB/c mice against BMA8 lethal challenge, highlighting their therapeutic potential in the severe COVID-19 cases. Besides, aVHH-13-Fc appears to be more effective than aVHH-11-Fc despite the differences of viral loads in the nasal turbinates and lungs were not significantly.
The gross pathology and histological assays were performed to check the pathological changes in the lungs at 3 dpi. In aVHH-13 treatment group, lung lesions were not observed, and Nb treatment significantly reduced viral infection in the lungs (Fig. 5I). In the PBS control group, extensive lung lesions were found, including mucosal epithelial cell degeneration, necrosis and sloughing, inflammatory necrosis in some lumens (blue arrows), and inflammatory cell infiltration (yellow arrows). Further immunostaining results showed that abundant SARS-CoV-2-positive regions could be detected in the PBS control group (Fig. 5J). These results proved that Nb prevented lethal challenge, limited viral replication, and avoided pulmonary lesions in severe COVID-19 model.
Hematological analysis revealed that BMA8-infected BALB/c mice in the control group showed a moderate increase in white blood cell (WBC) counts (Fig. 5L), with significant decreases in the percentage of monocytes (Mon%) (Fig. 5N) and lymphocytes (Lym%) (Fig. 5O) and a decrease in platelet count (PLT) (Fig. 5M). A significant increase in the percentage of neutrophils (Neu%) (Fig. 5K) was also observed in the control group rather than the treatment groups injected with Nbs, These results suggested that after Nb treatment, Neu%, WBC, Mon% were declined, and PLT and Lym% were elevated compared to PBS control (Chen et al., 2020).
3.7. aVHH-13-Fc protected hamster against infection of SARS-CoV-2
Based on the in vivo and in vitro study, aVHH-13-Fc was determined as the optimum Nb candidate. To further evaluate the therapeutic efficacy of this Nb in mild animal models, hamsters were challenged with 1000 TCID50 of SARS-CoV-2 prototype, Delta, Omicron BA.1 and BA.2, respectively through intranasal inoculation (n = 14). Subsequently, three doses of aVHH-13-Fc were given at 0.5 h, 1 h and 2 h post-challenge, respectively through intranasal inoculation. PBS served as a control (Fig. 6A). Following the successive monitoring for 14 days, animals in all groups survived from the challenge, as the golden hamsters served as a moderate COVID-19 animal model (Fig. 6B) (Wang et al., 2021). In all treatment groups, the weight showed a continuous growth trend. In the PBS control group, animals experienced temporary weight loss at 2–7 dpi and then gradually increased at 8–14 dpi (Fig. 6C).
Fig. 6.
Protective efficacy of nanobodies (Nbs) against SARS-CoV-2 VOCs in golden hamsters. A Experimental schedule for Nbs therapeutics in golden hamsters. Groups of golden hamsters (n = 14) were challenged with 1000 TCID50 of SARS-CoV-2 prototype, Delta, Omicron BA.1 or BA.2 via the intranasal route, followed by three doses successive intranasal administration of aVHH-11-Fc, aVHH-13-Fc or aVHH-14-Fc at 0.5, 1 and 2 h post infection. Nbs with a dose of 20 mg/kg were given to each hamster. The survival rate and weight change were monitored daily after challenge. Lung and nasal turbinates were taken on 3 days post infection (dpi), 7 dpi and 14 dpi, respectively for viral loads determination and virus tittering. B, C Survival rate and weight change during the 14 days monitoring post challenge. D, F Viral loads quantified by RT-qPCR at 3 dpi, 7 dpi and 14 dpi in lung and nasal turbinates. E, G Virus titers was conducted by TCID50 using Reed-Muench methods at 3 dpi, 7 dpi and 14 dpi in lung and nasal turbinates. Data are presented as the mean ± SD (n = 3). Statistical analysis was performed by two-way ANOVA (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001). J–M The basic structure of the lung tissues in treatment groups given purified aVHH-13-Fc post challenge of SARS-CoV-2 prototype, Delta, Omicron BA.1 and Omicron BA.2, respectively; Scale bar = 100 μm N–Q Abnormality of lung tissue structure were marked, represent as alveolar wall thickening (black arrow), mild bleeding (yellow arrow) and a small amount of lymphocyte and neutrophil infiltration (red arrow). R–U Viral antigens detected in the lung sections of purified aVHH-13-Fc treatment group post challenge of SARS-CoV-2 prototype, Delta, Omicron BA.1 and Omicron BA.2, respectively. V–Y Abundant viral antigens (yellow regions) detected in lung sections in control group. The figure showed immunohistochemistry (IHC) labeling against SARS-CoV-2 N, scale bar = 100 μm.
Three hamsters from each group were sacrificed for viral load and pathogenesis analysis at 3 dpi, 5 dpi and 14 dpi, respectively. Lung sections of challenged animals were stained with H&E and subjected to IHC. Compared with each control group, the animals in treatment group after infection of SARS-CoV-2 prototype, Delta, Omicron BA.1 and Omicron BA.2 had fewer viral RNAs in lungs by 588.84-, 6309.57-, 660.89-, 93.33-fold at 3 dpi, 25.70-, 23.44-, 20.42-, 9.33-fold at 5 dpi and 8.51-, 3.98-, 8.51-, 3.02-fold at 14 dpi, respectively (Fig. 6D). For viral titers, significantly higher virus titers in lung were detected in each control group in comparison to treatment group at 3 dpi. No infectious virus was present in all treatment groups at 5 dpi and 14 dpi except for one individual animal of Omicron BA.1 group (Fig. 6E). For detection of virus RNA copies in turbinate, in comparison to control group, all Nb-given hamsters had lower viral RNAs by 93.33-, 20.89-, 10.72-, 4.27-fold at 3 dpi, 2.34-, 48.98-, 8.91-, 14.45-fold at 5 dpi and 1.38-, 6.92-, 5.13-, 3.24-fold at 14 dpi, respectively (Fig. 6F). The trend of live virus titer detection in turbinate bone was consistent with what have been found in lung. Higher virus titers were observed in control group of Delta, Omicron BA.1 and BA.2 than that in treatment group at 3 dpi. Also, significant differences were observed in viral titer in nasal turbinate on 5 dpi between treatment group and control group in terms of prototype, delta omicron BA.1 and omicron BA.2 (Fig. 6G). The above results indicate that aVHH-13-Fc provided substantive therapeutic efficacy against parental SARS-CoV-2 prototype and SARS-CoV-2 VOCs including, Delta, Omicron BA.1 and BA.2.
In the PBS control group, more extensive lung lesions were confirmed, with more lung lobes showing activated inflammatory cells (red and yellow arrow) and alveolar septal thickening (black arrow) (Fig. 6N–Q). In contrast, moderate inflammatory cells were observed in aVHH-13-Fc treatment group (Fig. 6J–M). Consistently, immunostaining with the SARS-CoV-2 antigen showed aVHH-13-Fc treatment significantly reduced positive region of viral antigen in the lungs, while large amounts of SARS-CoV-2 antigen could be detected in the control group (red arrow) (Fig. 6R–U, 6V–6Y). These results demonstrated that aVHH-13-Fc inhibited SARS-CoV-2 replication in the upper and lower respiratory tracts and protected hamsters from lung lesions.
3.8. Molecular mechanism underlying the neutralizing activities of Nb
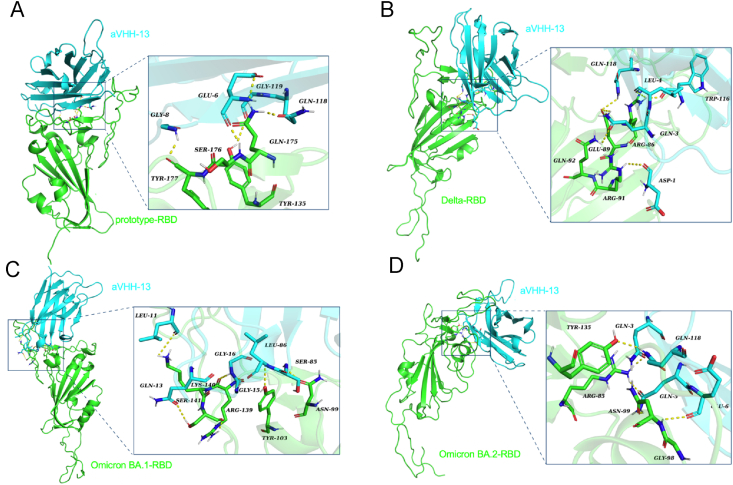
To understand the structural mechanism underlying the broad-spectrum neutralizing activities of aVHH-13, we examined the interactions between aVHH-13 and SARS-CoV-2 RBD of prototype, Delta, Omicron BA.1 and BA.2 using molecular docking (Fig. 7). The key binding sites between RBDs and aVHH-13 are shown in Table 1. Tyr135 (453Y), Gln175 (493Q), Ser176 (494S), and Tyr177 (495Y) in prototype -RBD play a crucial role in corresponding binding to Gly 119, Gln 118, Glu 6 and Gly 8 of aVHH-13, respectively (Fig. 7A). Arg86 (402R), Glu89 (405E), Arg91 (407R), and Gln92 (408Q) in Delta RBD play a crucial role in binding to aVHH-13, corresponding to Trp116, Leu4, Gln3, Gln118 and Asp1, respectively in aVHH-13 (Fig. 7B). Asn99 (414 N), Tyr103 (418Y), Arg139 (454R), Lys140 (455K), and Ser141 (456S) in Omicron BA.1-RBD play a crucial role in binding to aVHH-13, corresponding to Ser85, Leu86, Gly15, Gly16, Leu11 and Gln13, respectively in aVHH-13 (Fig. 7C). Arg85 (400R), Gly98 (413G), Asn99 (414Q), and Tyr135 (450Y) in Omicron BA.2-RBD play a crucial role in binding to aVHH-13, corresponding to Gln3, Gln5, Glu6 and Gln118, respectively in aVHH-13 (Fig. 7D).
Fig. 7.
Structure docking model of nanobodies bound to RBDs. Crystal structure of SARS-CoV-2 RBD proteins were applied in structural modeling with aVHH-13, energy minimization and energy minimized were then conducted. A Structural analysis of the combination of aVHH-13 and SASR-CoV-2 prototype-RBD. RBD is colored in green and aVHH-13 is colored in cyan (same color annotation below). B Structural analysis of the combination of aVHH-13 and SASR-CoV-2 Delta-RBD. C Structural analysis of the combination of aVHH-13 and SASR-CoV-2 Omicron BA.1-RBD. D Structural analysis of the combination of aVHH-13 and SASR-CoV-2 Omicron BA.2-RBD.
Table 1.
The key binding sites between aVHH-13 and RBDs.
| SARS-CoV-2 RBDs | Prototype-RBD | Delta-RBD | Omicron BA.1-RBD | Omicron BA.2-RBD |
|---|---|---|---|---|
| Binding sites | Tyr135, Gln175, Ser176, Tyr177 | Arg86, Glu89, Arg91, Gln92 | Asn99, Tyr103, Arg139, Lys140, Ser141 | Arg85, Gly98, Asn99, Tyr135 |
4. Discussion
Remarkable advances have been made in SARS-CoV-2 monoclonal antibodies based on sophisticated screening and structure elucidation techniques (Baum et al., 2020; Cao et al., 2020; Hansen et al., 2020; Pinto et al., 2020; Shi et al., 2020; Wu et al., 2020), which have enabled clear crystal structure, cross-neutralization among beta coronavirus and antibody cocktails targeting homologous or heterologous regions of S protein (Chi et al., 2020; Rogers et al., 2020; Wec et al., 2020). Whereas immune escape caused by VOCs, potential antibody-dependent enhancement (ADE) and high costs in antibody production compromised the development of nAbs. Due to the minimization in size, camelid-derived Nbs offer several advantages and may complement other monoclonal antibodies (Custodio et al., 2020; Hanke et al., 2020; Huo et al., 2020; Schoof et al., 2020; Tortorici et al., 2020; Wrapp et al., 2020a). Characteristics such as tissue penetration, hydrophilicity, stability, low heterogeneity in humans, and easy in manipulation and modification rendered Nbs infinite and broad application prospects. Our study fulfilled the pool of Nbs and reported isolation and characterization of single domain neutralizing antibodies from alpacas. These VHHs bound to the SARS-CoV-2 RBD proteins with high affinity, competitive competed with ACE2 and were capable of neutralizing SARS-CoV-2 VOCs in vitro.
In clinical, therapeutic countermeasures are particularly needed for vulnerable COVID-19 patients. In previous studies, mouse-adapted COVID-19 models were generated, which recapitulated the characterization and clinical manifestations of severe COVID-19 patients (Leist et al., 2020; Wang et al., 2020; Yan et al., 2022). In contrast, golden hamsters are naturally susceptible models of SARS-CoV-2, which represented mild to moderate COVID-19 infection (Boudewijns et al., 2020; Chan et al., 2020; Sia et al., 2020). Thus, the evaluation of Nbs in these rodent models would provide omni-directional therapeutic efficacy of nAbs in different groups of COVID-19 patients. In this study, therapeutic given of Nbs protected mice and golden hamsters in both the upper and lower respiratory tract against the challenge of SARS-CoV-2, which supports the reasonable application of Nbs in clinical. Owing to the promising results of aVHH-13 post intranasal administration in animals challenging studies, nebulized spray may be also practicable and particularly attractive in the case of respiratory pathogens because VHHs could theoretically be inhaled directly to the site of infection in an effort to maximize bioavailability and function (Wrapp et al., 2020b).
In the docking models between aVHH-13 and RBDs from different SARS-CoV-2 strains, the docking site of aVHH-13 was predicted to be located in the region from R402 to Y495 in corresponding S proteins, which largely lies in the receptor binding mortify (RBM) of S protein (aa 437–508) (Lan et al., 2020). Representatively, aVHH13 binds to Q493 and S494 of the RBD of SARS-CoV-2 prototype, which are key amino acid residues corresponding to the RBD-hACE2 interaction (Luan et al., 2020). Based on the structural comparisons and classification conducted by Barnes et al. (2020), aVHH13 may be classified as a class 1/2 antibody, which directly overlap with the binding footprint of ACE2. These may help explain the potent neutralizing ability of aVHH13 against SARS-CoV-2. The perennial substitutions accumulated in the RBD of SARS-CoV-2 S protein, particularly in RBM, compromised the efficacy of vaccines and therapeutic antibodies (Starr et al., 2021; Bowen et al., 2022; Liu et al., 2022). Nevertheless, a potent RBM-targeting antibody with broad neutralizing capacity across sarbecoviruses was also reported, which highlighted the potential conserved epitopes in RBD (Starr et al., 2021). In the docking model, aVHH-13 interacts with several highly conserved epitopes between SARS-CoV-2 strains and even SARS-CoV-1 (Li et al., 2022b), specifically, such as G413, Q414, etc. Further, crystal structure analysis is required to reveal the mechanism of broad neutralization.
To our knowledge, Nbs targeting two independent epitopes on RBD could synergistically prevent viral escape, which is also seen in antibody cocktail (Sun et al., 2021). These Nbs induce the premature activation of the viral S by an unusual mode of neutralization and yield insights into the mechanism of fusion (Koenig et al., 2021; Xu et al., 2021). In this study, diversity in the CDR3 region, neutralizing coverage of SARS-CoV-2 VOCs, participation in ACE2 competition, and performance in animal challenge studies may collectively reflect the independent epitopes involved in aVHH-11-Fc, aVHH-13-Fc, and aVHH-14-Fc. According to the results of alignment, the CDR1 and CDR2 regions of aVHH-11 and aVHH-14 are identical, with delicate differences in amino acid lying in the CDR3 region. In contrast, significant differences exist in CDR regions of aVHH-13 compared to the counterparts. As has been proved, the long CDR3 loop can increase the size of the antigen-binding loop and can bind to concave epitopes unaccessible by traditional antibodies (Muyldermans et al., 1994; Vu et al., 1997). Thus, bispecific Nbs targeting heterologous epitopes warrant further investigation.
Taken together, our current results illustrate novel Nbs screening from the phage library, which are functional in receptor binding, ACE2 competition-inhibition, and SARS-CoV-2 neutralization. Therapeutic efficacy of these broad-spectrum Nbs has been confirmed in both severe and mild to moderate rodent COVID-19 rodent models. In subsequent studies, double epitope antibodies and antibody cocktails could be prepared based on this Nb monomer to further optimize the neutralizing spectrum (Chi et al., 2022; Li et al., 2022a). What's more, intranasal delivered Nbs could be converted into aerosol-inhaled formulation, which enabled the in-depth and uniform distribution of Nbs to the unique circumstance in lungs, the original and extensive site of SARS-CoV-2 infection (Mettelman et al., 2022; Xu et al., 2022). These versatile Nbs warrant further study for rapid and cost-effective treatment against COVID-19.
5. Conclusions
In summary, we screened three alpacas-derived SARS-CoV-2 Nbs from a phage library, which could effectively neutralize pseudoviruses and authentic SARS-CoV 2 VOCs. What's more, they could also provide therapeutic efficacy in both mild and severe COVID-19 animal models. Among all Nbs, aVHH-13-Fc exhibited the best protective efficacy and bound to the RBM and conserved epitopes of SARS-CoV-2. Taken together, our study illustrated that alpaca-derived Nbs offered a therapeutic countermeasure against SARS-CoV-2.
Data availability
All of the data generated during the current study are included in the manuscript.
Ethics statement
The animal studies were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences. The animal protocols were approved by the IACUC of Changchun Veterinary Research Institute (approval number: AMMS-11-2022-010). Ethics approval number for the animal challenge study was IACUC-DWZX-2020-028.
Author contributions
Qiuxue Han: conceptualization, methodology, data curation, writing-original draft preparation. Shen Wang: data curation, writing-original draft preparation. Zhenshan Wang: methodology, data curation. Cheng Zhang: methodology. Xinyue Wang: methodology. Na Feng: conceptualization. Tiecheng Wang: conceptualization. Yongkun Zhao: conceptualization, data curation. Hang Chi: conceptualization, methodology. Feihu Yan: data curation, writing-reviewing and editing. Xianzhu Xia: conceptualization, writing-reviewing and editing.
Conflict of interest
The authors declare that there are no competing interests.
Acknowledgements
This work was supported by Jilin Province Youth Talent Support Project (grant number QT202115).
Contributor Information
Hang Chi, Email: ch_amms@163.com.
Feihu Yan, Email: yanfh1990@163.com.
Xianzhu Xia, Email: xiaxzh@cae.cn.
References
- Altarawneh H.N., Chemaitelly H., Ayoub H.H., Tang P., Hasan M.R., Yassine H.M., Al-Khatib H.A., Smatti M.K., Coyle P., Al-Kanaani Z., Al-Kuwari E., Jeremijenko A., Kaleeckal A.H., Latif A.N., Shaik R.M., Abdul-Rahim H.F., Nasrallah G.K., Al-Kuwari M.G., Butt A.A., Al-Romaihi H.E., Al-Thani M.H., Al-Khal A., Bertollini R., Abu-Raddad L.J. Effects of previous infection and vaccination on symptomatic omicron infections. N. Engl. J. Med. 2022;387:21–34. doi: 10.1056/NEJMoa2203965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig A.M. Targeting neuroinvasion by SARS-CoV-2: emerging trends in drug and antibody delivery to combat covid-19. ACS Chem. Neurosci. 2021;12:2555–2557. doi: 10.1021/acschemneuro.1c00372. [DOI] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., Robbiani D.F., Nussenzweig M.C., West A.P., Jr., Bjorkman P.J. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., Wei Y., Atwal G.S., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudewijns R., Thibaut H.J., Kaptein S.J.F., Li R., Vergote V., Seldeslachts L., Van Weyenbergh J., De Keyzer C., Bervoets L., Sharma S., Liesenborghs L., Ma J., Jansen S., Van Looveren D., Vercruysse T., Wang X., Jochmans D., Martens E., Roose K., De Vlieger D., Schepens B., Van Buyten T., Jacobs S., Liu Y., Martí-Carreras J., Vanmechelen B., Wawina-Bokalanga T., Delang L., Rocha-Pereira J., Coelmont L., Chiu W., Leyssen P., Heylen E., Schols D., Wang L., Close L., Matthijnssens J., Van Ranst M., Compernolle V., Schramm G., Van Laere K., Saelens X., Callewaert N., Opdenakker G., Maes P., Weynand B., Cawthorne C., Vande Velde G., Wang Z., Neyts J., Dallmeier K. Stat2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat. Commun. 2020;11:5838. doi: 10.1038/s41467-020-19684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen J.E., Addetia A., Dang H.V., Stewart C., Brown J.T., Sharkey W.K., Sprouse K.R., Walls A.C., Mazzitelli I.G., Logue J.K., Franko N.M., Czudnochowski N., Powell A.E., Dellota E., Jr., Ahmed K., Ansari A.S., Cameroni E., Gori A., Bandera A., Posavad C.M., Dan J.M., Zhang Z., Weiskopf D., Sette A., Crotty S., Iqbal N.T., Corti D., Geffner J., Snell G., Grifantini R., Chu H.Y., Veesler D. Omicron spike function and neutralizing activity elicited by a comprehensive panel of vaccines. Science. 2022;377:890–894. doi: 10.1126/science.abq0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., Su X.D., Du Z., Feng Y., Qin C., Qin C., Jin R., Xie X.S. Potent neutralizing antibodies against sars-cov-2 identified by high-throughput single-cell sequencing of convalescent patients' b cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., Chan W.M., Fan Z., Tsoi H.W., Wen L., Liang R., Cao J., Chen Y., Tang K., Luo C., Cai J.P., Kok K.H., Chu H., Chan K.H., Sridhar S., Chen Z., Chen H., To K.K., Yuen K.Y. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (covid-19) in golden syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., Chen Z., Guo Y., Zhang J., Li Y., Song X., Chen Y., Xia L., Fu L., Hou L., Xu J., Yu C., Li J., Zhou Q., Chen W. A neutralizing human antibody binds to the n-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Wang L., Liu C., Cheng X., Zheng H., Lv L., Tan Y., Zhang N., Zhao S., Wu M., Luo D., Qiu H., Feng R., Fu W., Zhang J., Xiong X., Zhang Y., Zu S., Chen Q., Ye Q., Yan X., Hu Y., Zhang Z., Yan R., Yin J., Lei P., Wang W., Lang G., Shao J., Deng Y., Wang X., Qin C. An engineered igg-vhh bispecific antibody against sars-cov-2 and its variants. Small Methods. 2022;6 doi: 10.1002/smtd.202200932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio T.F., Das H., Sheward D.J., Hanke L., Pazicky S., Pieprzyk J., Sorgenfrei M., Schroer M.A., Gruzinov A.Y., Jeffries C.M., Graewert M.A., Svergun D.I., Dobrev N., Remans K., Seeger M.A., McInerney G.M., Murrell B., Hallberg B.M., Low C. Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2. Nat. Commun. 2020;11:5588. doi: 10.1038/s41467-020-19204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donofrio G., Franceschi V., Macchi F., Russo L., Rocci A., Marchica V., Costa F., Giuliani N., Ferrari C., Missale G. A Simplified SARS-CoV-2 Pseudovirus Neutralization Assay. Vaccines; Basel: 2021. p. 9,389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Shi R., Zhang Y., Duan X., Li L., Zhang J., Wang F., Zhang R., Shen H., Wang Y., Wu Z., Peng Q., Pan T., Sun W., Huang W., Feng Y., Feng H., Xiao J., Tan W., Wang Y., Wang C., Yan J. A broadly neutralizing humanized ace2-targeting antibody against SARS-CoV-2 variants. Nat. Commun. 2021;12:5000. doi: 10.1038/s41467-021-25331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedry J., Hurdiss D.L., Wang C., Li W., Obal G., Drulyte I., Du W., Howes S.C., van Kuppeveld F.J.M., Forster F., Bosch B.J. Structural insights into the cross-neutralization of sars-cov and SARS-CoV-2 by the human monoclonal antibody 47d11. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abf5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauhar A., Privezentzev C.V., Demydchuk M., Gerlza T., Rieger J., Kungl A.J., Walsh F.S., Rutkowski J.L., Stocki P. Single domain shark vnar antibodies neutralize SARS-CoV-2 infection in vitro. Faseb. J. 2021;35 doi: 10.1096/fj.202100986RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K., Takai-Todaka R., Matsumura Y., Song C., Takano T., Tojo T., Nagami A., Ishida Y., Masaki H., Tsuchiya M., Ebisudani T., Sugimoto S., Sato T., Yasuda H., Fukunaga K., Sawada A., Nemoto N., Murata K., Morimoto T., Katayama K. Nasal delivery of single-domain antibody improves symptoms of SARS-CoV-2 infection in an animal model. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E.B., Bendahman N., Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- Hanke L., Vidakovics Perez L., Sheward D.J., Das H., Schulte T., Moliner-Morro A., Corcoran M., Achour A., Karlsson Hedestam G.B., Hällberg B.M., Murrell B., McInerney G.M. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020;11:4420. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., Chung K.M., Hermann A., Ullman E., Cruz J., Rafique A., Huang T., Fairhurst J., Libertiny C., Malbec M., Lee W-y, Welsh R., Farr G., Pennington S., Deshpande D., Cheng J., Watty A., Bouffard P., Babb R., Levenkova N., Chen C., Zhang B., Romero Hernandez A., Saotome K., Zhou Y., Franklin M., Sivapalasingam S., Lye D.C., Weston S., Logue J., Haupt R., Frieman M., Chen G., Olson W., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.N., Cai Y., Li W.H., He W.M., Zhao Z.Y., Zhu W.J., Wang Q., Mai X., Liu J., Lee H.C., Stjepanovic G., Zhang H., Zhao Y.J. A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of sarm1. Nat. Commun. 2022;13:7898. doi: 10.1038/s41467-022-35581-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N., Ren J., Zhou D., Harrison P.J., Weckener M., Clare D.K., Vogirala V.K., Radecke J., Moynie L., Zhao Y., Gilbert-Jaramillo J., Knight M.L., Tree J.A., Buttigieg K.R., Coombes N., Elmore M.J., Carroll M.W., Carrique L., Shah P.N.M., James W., Townsend A.R., Stuart D.I., Owens R.J., Naismith J.H. Neutralizing nanobodies bind SARS-CoV-2 spike rbd and block interaction with ace2. Nat. Struct. Mol. Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- Huo J., Bas A.L., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N., Ren J., Zhou D., Harrison P.J., Weckener M., Clare D.K., Vogirala V.K., Radecke J., Moynie L., Zhao Y., Gilbert-Jaramillo J., Knight M.L., Tree J.A., Buttigieg K.R., Coombes N., Elmore M.J., Carroll M.W., Carrique L., Shah P.N.M., James W., Townsend A.R., Stuart D.I., Owens R.J., Naismith J.H. Author correction: neutralizing nanobodies bind SARS-CoV-2 spike rbd and block interaction with ace2. Nat. Struct. Mol. Biol. 2021;28:326. doi: 10.1038/s41594-021-00566-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo J., Mikolajek H., Le Bas A., Clark J.J., Sharma P., Kipar A., Dormon J., Norman C., Weckener M., Clare D.K., Harrison P.J., Tree J.A., Buttigieg K.R., Salguero F.J., Watson R., Knott D., Carnell O., Ngabo D., Elmore M.J., Fotheringham S., Harding A., Moynié L., Ward P.N., Dumoux M., Prince T., Hall Y., Hiscox J.A., Owen A., James W., Carroll M.W., Stewart J.P., Naismith J.H., Owens R.J. A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the syrian golden hamster model of covid-19. Nat. Commun. 2021;12:5469. doi: 10.1038/s41467-021-25480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janik E., Niemcewicz M., Podogrocki M., Majsterek I., Bijak M. The emerging concern and interest SARS-CoV-2 variants. Pathogens. 2021;10:633. doi: 10.3390/pathogens10060633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig P.A., Das H., Liu H., Kummerer B.M., Gohr F.N., Jenster L.M., Schiffelers L.D.J., Tesfamariam Y.M., Uchima M., Wuerth J.D., Gatterdam K., Ruetalo N., Christensen M.H., Fandrey C.I., Normann S., Todtmann J.M.P., Pritzl S., Hanke L., Boos J., Yuan M., Zhu X., Schmid-Burgk J.L., Kato H., Schindler M., Wilson I.A., Geyer M., Ludwig K.U., Hallberg B.M., Wu N.C., Schmidt F.I. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021;371 doi: 10.1126/science.abe6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann C., Mayer C.K., Claassen M., Maponga T., Burgers W.A., Keeton R., Riou C., Sutherland A.D., Suliman T., Shaw M.L., Preiser W. Breakthrough infections with SARS-CoV-2 omicron despite mrna vaccine booster dose. Lancet. 2022;399:625–626. doi: 10.1016/S0140-6736(22)00090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustin T., Harel N., Finkel U., Perchik S., Harari S., Tahor M., Caspi I., Levy R., Leshchinsky M., Ken Dror S., Bergerzon G., Gadban H., Gadban F., Eliassian E., Shimron O., Saleh L., Ben-Zvi H., Keren Taraday E., Amichay D., Ben-Dor A., Sagas D., Strauss M., Shemer Avni Y., Huppert A., Kepten E., Balicer R.D., Netzer D., Ben-Shachar S., Stern A. Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in bnt162b2-mrna-vaccinated individuals. Nat. Med. 2021;27:1379–1384. doi: 10.1038/s41591-021-01413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ace2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Leist S.R., Dinnon K.H., Schäfer A., Tse L.V., Okuda K., Hou Y.J., West A., Edwards C.E., Sanders W., Fritch E.J., Gully K.L., Scobey T., Brown A.J., Sheahan T.P., Moorman N.J., Boucher R.C., Gralinski L.E., Montgomery S.A., Baric R.S. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell. 2020;183:1070–1085.e12. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Zhan W., Yang Z., Tu C., Hu G., Zhang X., Song W., Du S., Zhu Y., Huang K., Kong Y., Zhang M., Mao Q., Gu X., Zhang Y., Xie Y., Deng Q., Song Y., Chen Z., Lu L., Jiang S., Wu Y., Sun L., Ying T. Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody. Cell. 2022;185:1389–1401.e1318. doi: 10.1016/j.cell.2022.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Ren Y., Aw Z.Q., Chen B., Yang Z., Lei Y., Cheng L., Liang Q., Hong J., Yang Y., Chen J., Wong Y.H., Wei J., Shan S., Zhang S., Ge J., Wang R., Dong J.Z., Chen Y., Shi X., Zhang Q., Zhang Z., Chu J.J.H., Wang X., Zhang L. Broadly neutralizing and protective nanobodies against SARS-CoV-2 omicron subvariants ba.1, ba.2, and ba.4/5 and diverse sarbecoviruses. Nat. Commun. 2022;13:7957. doi: 10.1038/s41467-022-35642-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.F., He L., Deng Y.Q., Qi S.H., Chen Y.H., Zhang X.L., Hu S.X., Fan R.W., Zhao G.Y., Qin C.F. Generation and characterization of a nanobody against sars-cov. Virol. Sin. 2021;36:1484–1491. doi: 10.1007/s12250-021-00436-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., Yu J., Chik K.K., Yuen T.T., Yoon C., To K.K., Chen H., Yin M.T., Sobieszczyk M.E., Huang Y., Wang H.H., Sheng Z., Yuen K.Y., Ho D.D. Striking antibody evasion manifested by the omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- Luan J., Lu Y., Jin X., Zhang L. Spike protein recognition of mammalian ace2 predicts the host range and an optimized ace2 for SARS-CoV-2 infection. Biochem. Biophys. Res. Commun. 2020;526:165–169. doi: 10.1016/j.bbrc.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettelman R.C., Allen E.K., Thomas P.G. Mucosal immune responses to infection and vaccination in the respiratory tract. Immunity. 2022;55:749–780. doi: 10.1016/j.immuni.2022.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- Muyldermans S., Atarhouch T., Saldanha J., Barbosa J.A., Hamers R. Sequence and structure of vh domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. Protein Eng. 1994;7:1129–1135. doi: 10.1093/protein/7.9.1129. [DOI] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J.B., Chen R.E., Havenar-Daughton C., Snell G., Telenti A., Virgin H.W., Lanzavecchia A., Diamond M.S., Fink K., Veesler D., Corti D. Cross-neutralization of SARS-CoV-2 by a human monoclonal sars-cov antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W-t, Limbo O., Smith C., Song G., Woehl J., Yang L., Abbott R.K., Callaghan S., Garcia E., Hurtado J., Parren M., Peng L., Ramirez S., Ricketts J., Ricciardi M.J., Rawlings S.A., Wu N.C., Yuan M., Smith D.M., Nemazee D., Teijaro J.R., Voss J.E., Wilson I.A., Andrabi R., Briney B., Landais E., Sok D., Jardine J.G., Burton D.R. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof M., Faust B., Saunders R.A., Sangwan S., Rezelj V., Hoppe N., Boone M., Billesbølle C.B., Puchades C., Azumaya C.M., Kratochvil H.T., Zimanyi M., Deshpande I., Liang J., Dickinson S., Nguyen H.C., Chio C.M., Merz G.E., Thompson M.C., Diwanji D., Schaefer K., Anand A.A., Dobzinski N., Zha B.S., Simoneau C.R., Leon K., White K.M., Chio U.S., Gupta M., Jin M., Li F., Liu Y., Zhang K., Bulkley D., Sun M., Smith A.M., Rizo A.N., Moss F., Brilot A.F., Pourmal S., Trenker R., Pospiech T., Gupta S., Barsi-Rhyne B., Belyy V., Barile-Hill A.W., Nock S., Liu Y., Krogan N.J., Ralston C.Y., Swaney D.L., García-Sastre A., Ott M., Vignuzzi M., Walter P., Manglik A. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive spike. Science. 2020;370:1473–1479. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully M., Cataland S.R., Peyvandi F., Coppo P., Knöbl P., Kremer Hovinga J.A., Metjian A., de la Rubia J., Pavenski K., Callewaert F., Biswas D., De Winter H., Zeldin R.K. Caplacizumab treatment for acquired thrombotic thrombocytopenic purpura. N. Engl. J. Med. 2019;380:335–346. doi: 10.1056/NEJMoa1806311. [DOI] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W.J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F.S., Wang Q., Gao G.F., Yuan Z., Yan J. A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Shuai H., Chan J.F., Hu B., Chai Y., Yuen T.T., Yin F., Huang X., Yoon C., Hu J.C., Liu H., Shi J., Liu Y., Zhu T., Zhang J., Hou Y., Wang Y., Lu L., Cai J.P., Zhang A.J., Zhou J., Yuan S., Brindley M.A., Zhang B.Z., Huang J.D., To K.K., Yuen K.Y., Chu H. Attenuated replication and pathogenicity of SARS-CoV-2 b.1.1.529 omicron. Nature. 2022;603:693–699. doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R., Poon L.L.M., Nicholls J.M., Peiris M., Yen H.L. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smail S.W., Saeed M., Twana A., Khudhur Z.O., Younus D.A., Rajab M.F., Abdulahad W.H., Hussain H.I., Niaz K., Safdar M. Inflammation, immunity and potential target therapy of SARS-CoV-2: a total scale analysis review. Food Chem. Toxicol. 2021;150 doi: 10.1016/j.fct.2021.112087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Czudnochowski N., Liu Z., Zatta F., Park Y.J., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A.J., Marzi R., Glass W.G., Zhang I., Dingens A.S., Bowen J.E., Tortorici M.A., Walls A.C., Wojcechowskyj J.A., De Marco A., Rosen L.E., Zhou J., Montiel-Ruiz M., Kaiser H., Dillen J.R., Tucker H., Bassi J., Silacci-Fregni C., Housley M.P., di Iulio J., Lombardo G., Agostini M., Sprugasci N., Culap K., Jaconi S., Meury M., Dellota E., Jr., Abdelnabi R., Foo S.C., Cameroni E., Stumpf S., Croll T.I., Nix J.C., Havenar-Daughton C., Piccoli L., Benigni F., Neyts J., Telenti A., Lempp F.A., Pizzuto M.S., Chodera J.D., Hebner C.M., Virgin H.W., Whelan S.P.J., Veesler D., Corti D., Bloom J.D., Snell G. SARS-CoV-2 rbd antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Wang L., Feng R., Wang N., Wang Y., Zhu D., Xing X., Yang P., Zhang Y., Li W., Wang X. Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms. Cell Res. 2021;31:597–600. doi: 10.1038/s41422-021-00497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., Zatta F., De Marco A., Guarino B., Bianchi S., Lauron E.J., Tucker H., Zhou J., Peter A., Havenar-Daughton C., Wojcechowskyj J.A., Case J.B., Chen R.E., Kaiser H., Montiel-Ruiz M., Meury M., Czudnochowski N., Spreafico R., Dillen J., Ng C., Sprugasci N., Culap K., Benigni F., Abdelnabi R., Foo S.C., Schmid M.A., Cameroni E., Riva A., Gabrieli A., Galli M., Pizzuto M.S., Neyts J., Diamond M.S., Virgin H.W., Snell G., Corti D., Fink K., Veesler D. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBlargan L.A., Adams L.J., Liu Z., Chen R.E., Gilchuk P., Raju S., Smith B.K., Zhao H., Case J.B., Winkler E.S., Whitener B.M., Droit L., Aziati I.D., Bricker T.L., Joshi A., Shi P.Y., Creanga A., Pegu A., Handley S.A., Wang D., Boon A.C.M., Crowe J.E., Jr., Whelan S.P.J., Fremont D.H., Diamond M.S. A potently neutralizing SARS-CoV-2 antibody inhibits variants of concern by utilizing unique binding residues in a highly conserved epitope. Immunity. 2021;54:2399–2416. doi: 10.1016/j.immuni.2021.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu K.B., Ghahroudi M.A., Wyns L., Muyldermans S. Comparison of llama vh sequences from conventional and heavy chain antibodies. Mol. Immunol. 1997;34:1121–1131. doi: 10.1016/s0161-5890(97)00146-6. [DOI] [PubMed] [Google Scholar]
- Wang, Hu Y., Li B., Wang H., Shen J. Applications of nanobodies in the prevention, detection, and treatment of the evolving SARS-CoV-2. Biochem. Pharmacol. 2023;208 doi: 10.1016/j.bcp.2022.115401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Li L., Yan F., Gao Y., Yang S., Xia X. Covid-19 animal models and vaccines: current landscape and future prospects. Vaccines. 2021;9:1082. doi: 10.3390/vaccines9101082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Zhang C., Liang B., Wang W., Feng N., Zhao Y., Wang T., Guo Z., Yan F., Yang S., Xia X. Characterization of immune response diversity in rodents vaccinated with a vesicular stomatitis virus vectored covid-19 vaccine. Viruses. 2022;14:1127. doi: 10.3390/v14061127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Shuai L., Wang C., Liu R., He X., Zhang X., Sun Z., Shan D., Ge J., Wang X., Hua R., Zhong G., Wen Z., Bu Z. Mouse-adapted SARS-CoV-2 replicates efficiently in the upper and lower respiratory tract of balb/c and c57bl/6j mice. Protein & Cell. 2020;11:776–782. doi: 10.1007/s13238-020-00767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., Tse L.V., Johnson N.V., Hsieh C.-L., Wang N., Nett J.H., Champney E., Burnina I., Brown M., Lin S., Sinclair M., Johnson C., Pudi S., Bortz R., Wirchnianski A.S., Laudermilch E., Florez C., Fels J.M., O’Brien C.M., Graham B.S., Nemazee D., Burton D.R., Baric R.S., Voss J.E., Chandran K., Dye J.M., McLellan J.S., Walker L.M. Broad neutralization of sars-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., Perry C., Pan C., Hosain R., Mahmood A., Davis J.D., Turner K.C., Hooper A.T., Hamilton J.D., Baum A., Kyratsous C.A., Kim Y., Cook A., Kampman W., Kohli A., Sachdeva Y., Graber X., Kowal B., DiCioccio T., Stahl N., Lipsich L., Braunstein N., Herman G., Yancopoulos G.D., Trial I. Regn-cov2, a neutralizing antibody cocktail, in outpatients with covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Hoffmann M., Pöhlmann S., Graham B.S., Callewaert N., Schepens B., Saelens X., McLellan J.S. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1004–1015.e1015. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Team V.-C.C.-R., Hoffmann M., Pohlmann S., Graham B.S., Callewaert N., Schepens B., Saelens X., McLellan J.S. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1004–1015 e1015. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., Gong Y., Xiao H., Fan Z., Tan S., Wu G., Tan W., Lu X., Fan C., Wang Q., Liu Y., Zhang C., Qi J., Gao G.F., Gao F., Liu L. A noncompeting pair of human neutralizing antibodies block covid-19 virus binding to its receptor ace2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Nambulli S., Xiao Z., Liu H., Sang Z., Duprex W.P., Schneidman-Duhovny D., Zhang C., Shi Y. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science. 2020;370:1479–1484. doi: 10.1126/science.abe4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Wu S., Yi L., Peng S., Wang F., Si W., Hou L., Zhu T. Safety, mucosal and systemic immunopotency of an aerosolized adenovirus-vectored vaccine against SARS-CoV-2 in rhesus macaques. Emerg. Microb. Infect. 2022;11:438–441. doi: 10.1080/22221751.2022.2030199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Xu K., Jung S., Conte A., Lieberman J., Muecksch F., Lorenzi J.C.C., Park S., Schmidt F., Wang Z., Huang Y., Luo Y., Nair M.S., Wang P., Schulz J.E., Tessarollo L., Bylund T., Chuang G.Y., Olia A.S., Stephens T., Teng I.T., Tsybovsky Y., Zhou T., Munster V., Ho D.D., Hatziioannou T., Bieniasz P.D., Nussenzweig M.C., Kwong P.D., Casellas R. Nanobodies from camelid mice and llamas neutralize SARS-CoV-2 variants. Nature. 2021;595:278–282. doi: 10.1038/s41586-021-03676-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F., Li E., Wang T., Li Y., Liu J., Wang W., Qin T., Su R., Pei H., Wang S., Feng N., Zhao Y., Yang S., Xia X., Gao Y. Characterization of two heterogeneous lethal mouse-adapted SARS-CoV-2 variants recapitulating representative aspects of human covid-19. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.821664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Wang Y., Jin Y., Zhu Y., Wu Y., Li C., Kong Y., Song W., Tian X., Zhan W., Huang A., Zhou S., Xia S., Tian X., Peng C., Chen C., Shi Y., Hu G., Du S., Wang Y., Xie Y., Jiang S., Lu L., Sun L., Song Y., Ying T. A non-ace2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants. Signal Transduct. Targeted Ther. 2021;6:378. doi: 10.1038/s41392-021-00810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All of the data generated during the current study are included in the manuscript.