Summary
The emerging field of liquid biopsy stands at the forefront of novel diagnostic strategies for cancer and other diseases. Liquid biopsy allows minimally invasive molecular characterization of cancers for diagnosis, patient stratification to therapy, and longitudinal monitoring. Liquid biopsy strategies include detection and monitoring of circulating tumor cells, cell-free DNA, and extracellular vesicles. In this review, we address the current understanding and the role of existing liquid-biopsy-based modalities in cancer diagnostics and monitoring. We specifically focus on the technical and clinical challenges associated with liquid biopsy and biomarker development being addressed by the Liquid Biopsy Consortium, established through the National Cancer Institute. The Liquid Biopsy Consortium has developed new methods/assays and validated existing methods/technologies to capture and characterize tumor-derived circulating cargo, as well as addressed existing challenges and provided recommendations for advancing biomarker assays.
Graphical abstract
This review led by Batool et al. represents a critical appraisal of the work done in liquid biopsy. The authors identify and evaluate the challenges and potential approaches specific to each disease. Clinical implantation of liquid biopsy remains an unmet goal. With this review, the authors hope to provide recommendations for future studies.
Introduction
Cancer remains the second leading cause of death in the United States, with more than 1.8 million new cancer diagnoses and more than 0.6 million cancer deaths each year.1 With such an overwhelming burden on society, early cancer diagnosis is imperative to increasing survival rates and reducing care costs. For example, to date, as a result of population screening with invasive modalities such as colonoscopy, mammogram, and pap smear, overall cancer mortality has decreased by more than 25% from 1990 to 2015 in the United States, with a significant decline in mortality rates for colon cancer and breast cancer.2
Although imaging (breast) and some blood-based screenings (colorectal, prostate cancer) are available for certain cancers, these modalities do not provide insights into the genetic profiles of those cancers. Currently, the mainstay for diagnosis is a traditional tissue biopsy to confirm the mutational profile and guide treatment strategies. There remains a great need for minimally invasive, early detection methods that can provide a molecular diagnosis to allow timely patient stratification to appropriate therapies. Emerging evidence over the past two decades has given rise to the field of liquid biopsy, which involves the study of a sample of a biofluid (blood, cerebrospinal fluid [CSF], urine, saliva, amniotic fluid, ascitic fluid), examining cancer-derived circulating tumor cells (CTCs), circulating nucleic acids including cell-free DNA (cfDNA), cell-free RNA including mRNA, long non-coding RNAs (lncRNAs) and microRNA (miRNA), extracellular vesicles (EVs), tumor-educated platelets, proteins, and metabolites.3 However, most of the literature reports findings from single-center population studies lacking reproducibility and robust verification and validation studies.
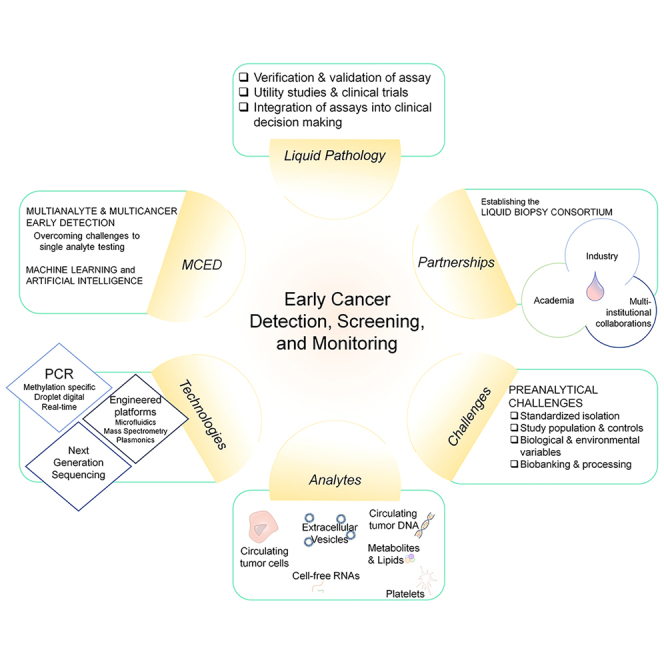
Therefore, large-scale, multi-institutional collaboration is essential for the development of reliable and reproducible liquid biopsy-based assays with clinical utility. The National Cancer Institute’s (NCI) Division of Cancer Prevention has initiated and sponsored an academic/industrial partnership program designed to advance and validate liquid biopsy technologies specifically targeted for early-stage cancer detection. The resulting Liquid Biopsy Consortium (LBC) has developed, tested, and cross-validated new methods and assays to capture and characterize tumor-derived circulating cargo for minimally invasive early diagnosis of cancer. Additionally, the consortium provides recommendations and best practices for developing liquid biopsy biomarkers with translational potential. The Precompetitive Collaboration on Liquid Biopsy for Early Cancer Assessment Consortium also works on non-invasive liquid biopsy methods to distinguish cancer from benign disease and aggressive from indolent cancers. Projects from six funded sites, described below, focus on the development of new tools/methods/assays and/or validations of existing technologies involving the capture of DNA, RNA, or EVs in circulating body fluids.
In this commentary, we highlight the need for advanced liquid biopsy-based tests for the early diagnosis of cancers. We review the current understanding of existing liquid biopsy-based modalities in early cancer diagnosis and monitoring, with a special focus on technical and clinical challenges associated with the development of clinically relevant liquid biopsy assays. We also elaborate on the need for the establishment and adoption of best practices to bridge these hurdles along the biomarker discovery pipeline to enhance clinical translation. Finally, we outline the mission, aims, structure, and function of the NCI’s LBC.
The potential of liquid biopsy-based early detection of cancers
The stimulus for liquid biopsy rests on several key facts. Although imaging and blood test screening for cancer detection are available for cancers of breast, cervix, colorectal, lung, and prostate, screening is not uniformly applied.4 Compliance reflects the availability, tolerance, and morbidity of the screening tool.5,6
For oncologists, the characterization of tumor-derived analytes within biofluids has emerged as a powerful method to identify tumor-specific genetic amplifications and aberrations. The aim is to achieve a rapid, non-invasive, and cost-efficient diagnosis of cancer, monitoring of disease status, and treatment response. Biofluid-based assays have been developed for diagnostics of large populations screened for cancer “in general” without speciation of organ type, early detection of cancer in asymptomatic individuals who are “at risk,” stratification of treatment cohorts in clinical trials, provision of molecular-based tumor stages, measurement of post-treatment minimal residual disease, and as indicators of response to therapy or post-treatment disease recurrence.
Tissue biopsy only provides a single snapshot of the tumor, rather than providing insights into its complex genomic landscape and intratumoral heterogeneity. This lack of information limits the clinician’s ability to determine an optimal therapeutic course. Although multiple biopsies from the tumor are sometimes performed, this procedure is limited by inaccessibility, potential surgical complications, morbidity, mortality, and economic considerations.7 Liquid biopsy-based assessment of mutations can overcome this challenge, as has been shown for clinically available BCR-ABL testing for chronic myelogenous leukemia8 and potentially for EGFRvIII9 and TERT10 testing in gliomas.
Liquid biopsy has the potential to offer advantages over tissue biopsies and imaging approaches. It provides profound cost and morbidity reductions when compared with surgical excision of tissues and costly computed tomographic (CT), positron emission, and magnetic resonance (MRI) studies. The analysis is relatively rapid compared with histologic examinations, providing results in minutes to hours. These analyses also provide mutation assessment across the histologically heterogeneous tumor tissues. Furthermore, it serves as a potential platform to address the common deficiencies and challenges in early detection modalities for cancers.
Challenges with current screening modalities
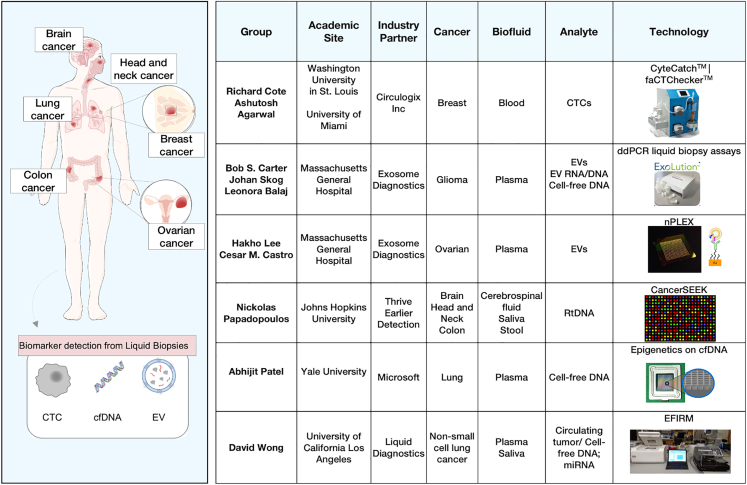
Cancer is a diverse condition that can originate from essentially all cell types and organs of a human body and has over a hundred distinct entities with diverse risk factors and epidemiologic features. Breast, prostate, lung, bronchus, and colorectal cancers are the most high-incident cancers in the United States (Figure S1A). Coincidentally, they contribute to the highest mortality as well (Figure S1B). The current screening programs are focused on identifying precursor lesions or malignancies when cancer is most treatable to plan the most effective treatment regimens.11 However, these screening modalities are often associated with certain challenges. Liquid biopsy has the potential to mitigate these deficiencies and supplement the screening tests to provide a more efficient diagnostic platform. The LBC comprises different groups working on detection of common cancers using novel platforms (Figure 1).
Figure 1.
Overview of the Liquid Biopsy Consortium and the different technologies investigated
(A) A summary of the academic-industrial partnership collaborations is provided, highlighting the leading principal investigators, target analytes, various liquid biopsy technologies used, and cancers of interest.
Liquid biopsy approach to mitigating deficiencies in early detection for cancers addressed by the consortium
Breast cancer is the most common cancer and the second most common cause of death from cancer in women. Currently, mammography is used in early-stage screening5 with definitive diagnosis made by pathology assessment. Technologies that isolate and analyze CTCs can be used for early detection of breast cancer. One example is the microfilter technology CyteCatch developed by Dr. Cote (Washington University in St. Louis) in collaboration with Circulogix to collect, quantify, and analyze CTCs and tumor microenvironment cells such as cancer-associated fibroblasts in early-stage breast cancer patients from blood.
Lung cancer manifests as 2 million new cases and 1.7 million deaths annually. Currently, low-dose spiral computed tomography is used for early detection of lung cancer.12 This, however, is associated with high false-positive rates, radiation exposure, and patient distress during ambiguous follow-ups. There is potential to harness the utility of circulating tumor DNA (ctDNA) for detection of common biomarkers. For instance, a research team led by Dr. Wong (University of California Los Angeles) has developed the electric field-induced release and measurement (EFIRM) liquid biopsy technology to detect the 10 most common DNA mutations in ctDNA and miRNA in the plasma and saliva of non-small cell lung carcinoma patients. The EFIRM technology allows for rapid and non-invasive detection of molecular targets directly in body fluids without prior extraction. Additionally, the Wong group has investigated the utility of this technology to detect an ultrashort (<60 bp) fragment of ctDNA. The group has partnered with the Liquid Diagnostics biotech company to develop and validate this technology and transition it to clinical application.
Ovarian cancer is the second-most diagnosed gynecological malignancy in women. Currently, there are no recommended screening tests for early detection of ovarian cancer.13 The current serum biomarker (CA-125) has low predictive value (5%) and high false-positive rates. Liquid biopsy-based assessment of EVs can be a new approach to early detection and treatment monitoring of this tumor, improving overall survival. An example of this effort is the Ovarian Cancer Exosomal Analysis with Nanoplasmonics project (led by Dr. Lee), which focuses on standardization of EV isolation methods (e.g., ExoLution,14 dual mode chromatography14) and proteomic analysis of EVs via a high-throughput nanoplasmonic platform (nPLEX).14
Liquid biopsy may also be beneficial in the diagnostics of inaccessible cancers, such as brain tumors. Globally, 3.7 and 2.6 out of 100,000 males and females, respectively, are annually diagnosed with primary malignant brain tumors.15 CT and MRI scans are used to detect cancerous lesions, while needle biopsies are used to detect tumor tissue in sensitive or hard-to-reach areas.16 Non-blood-based liquid biopsies have been recently developed as a useful tool for detecting and monitoring brain tumors.17 In the presence of inconclusive findings on imaging, the presence of mutant DNA/RNA in the CSF or blood may support a diagnosis of neoplasia or identify residual disease post-surgery. Targeted sequencing of CSF or blood analytes (EV RNA/DNA, CTCs, cfDNA) may also circumvent the need for craniotomy procedures to obtain a tissue biopsy to identify actionable genetic alterations. Finally, the minimally invasive nature of biofluid sampling can allow longitudinal monitoring and surveillance.18 Two leading investigators, Dr. Balaj and Dr. Carter, have developed assays for the detection of tumor-specific mutations in patients with glioma using blood and CSF.19
Other efforts within the consortium include sensitive detection of ctDNA via analysis of copy number variations using next-generation sequencing (NGS) led by Dr. Papadopoulos (Johns Hopkins University) for detection of colorectal, head and neck, and brain cancers. Additionally, computational models have been developed to integrate epigenomic and molecular signatures of ctDNA to differentiate benign lung nodules from malignant lesions in a work led by Dr. Patel (Yale University).
Liquid biopsy analytes
A range of biomarkers have been investigated for liquid biopsy. Each marker provides varying levels of information about the genetic landscape of a tumor. Disease-specific biomarkers can be detected at varying sensitivity and specificity in different biofluids (i.e., blood, plasma, saliva, urine, CSF).20 Frequently investigated biomarkers include ctDNA, EVs, and CTCs.
ctDNA represents fragments of tumor DNA released into biofluids.21 They are attractive analytes to study tumor-specific mutations, CNVs, methylation changes, or integrated viral sequences.22 CtDNA can be isolated from any biofluid, and certain biofluids are ideal for a given cancer due to anatomic proximity. Emerging evidence suggests that ctDNA was detectable in >75% of patients with advanced pancreatic, ovarian, colorectal, bladder, gastroesophageal, breast, melanoma, hepatocellular, and head and neck cancers but in less than 50% of primary brain, renal, prostate, or thyroid cancers.23 This potentially indicates the variability in overall ctDNA yield from different tumor lesions.
The concentration of isolated ctDNA from biofluids is low, and detection of rare mutant events is even more challenging.24 The concentration of ctDNA is negligible in healthy people. Higher sample volumes are sometimes used to isolate sufficient absolute quantities of ctDNA for detection of tumor-specific variants. The choice of biofluid is also an important variable. Use of alternative biofluids can potentially allow for earlier detection of premalignant lesions that are otherwise difficult to detect in the blood. This is especially true for CNS tumors that are separated from the systemic circulation via a blood-brain barrier. Additionally, ctDNA is highly fragmented, sometimes as low as 50 bp in length, requiring highly sensitive technologies to detect and quantify tumor-specific genomic alterations amid the background of cfDNA released by non-cancerous cells.
CTCs are intact cancer cells, sometimes found as clusters, that release the primary tumors into biofluids and are considered to have metastatic potential.25 It is not known whether CTCs are only detached subpopulations of the tumor or rather pulled randomly to represent the entire tumor. CTCs are exceedingly rare, occurring at a frequency of 1 per 106 to 107 leukocytes and are challenging to isolate. The yield may also vary as the tumor evolves, with low sensitivity in early-stage disease and more abundant CTCs isolated in late-stage disease.26 One potential way to overcome this is use of large volumes of biofluids to achieve optimal yield. Positive selection by antibody-based separation techniques is used to capture CTCs. These include flow cytometry, immunomagnetic, or adhesion-based methods. However, the captured CTC population only represents the subpopulation with a particular membrane protein. Other methods such as negative selection by depletion of unwanted components of whole blood using erythrocyte lysis, density gradient stratification, and magnetic bead-based removal of leukocytes have been explored.24 Advanced methodologies such as dielectric mobility and photoacoustic and microfluidic separation have been studied.27 Washington University in St. Louis (principal investigator, Dr. Cote), University of Miami School of Medicine, and Caltech are developing platforms to collect, quantify, and analyze CTCs and tumor microenvironment cells such as cancer-associated fibroblasts from early-stage breast cancer patients. The microfilter technology (CyteCatch), along with the automated fluidic platform faCTChecker, captures and isolates these rare cells of interest from blood and other biofluids. The captured cells are analyzed on-chip through image analysis where these images are being used initially as training sets to train deep learning algorithms that later will be used to automate the process. This group’s industry partners are Circulogix for the microfilter technology and Google for the deep-learning algorithm.
EVs are membranous structures released from the cells, including tumor cells, and contain nucleic acids (DNA, mRNA, miRNA, non-coding RNA), proteins, lipids, or other metabolites. The enclosed cargo closely reflects the cell of origin. The term “EV” covers a wide array of secreted vesicles, including microvesicles, exosomes, ectosomes, and oncosomes.28 EVs are actively released by cells and can interact with surrounding cells, and depending on their molecular content, they can exacerbate or ameliorate a cancerous phenotype. They have also been shown to modulate tumor proliferation, reprogram metabolic activity, induce angiogenesis, escape immune surveillance, acquire drug resistance, and undergo invasion.29 The disease-specific cargo, long half-life, and physical stability in collected biofluids advocates for their role as promising diagnostic and prognostic biomarkers for multiple diseases including tumor.
Use of blood-derived EVs as a diagnostic tool has been explored in multiple cancers including non-small-cell lung cancer, breast cancer, pancreatic cancer, colorectal cancer, ovarian cancer, and nasopharyngeal carcinoma.30 While EVs can be isolated from multiple biofluids, plasma and serum are most frequently utilized for EV detection and analysis. The heterogeneity of luminal content in different biofluids is closely linked to the site of disease. A study reported unique mRNA profiles of EVs isolated from serum and urine in patients with cholangiocarcinoma.31 Significantly higher diagnostic accuracy was achieved with EVs isolated from urine.31 Similar differences have also been reported for EV membrane analysis with differences in proteome profiling (saliva vs. serum) in patients with lung cancer.32 Further work is required to determine whether the heterogeneity in membranous and luminal contents of EVs isolated from different biofluids is significant and linked to the disease of interest. Current methods and technologies reported to date have mostly utilized a bulk EV approach. This provides information about the ensemble of the heterogeneous EV populations. However, analysis at the single-EV level would be of tremendous value in identifying disease-derived potential biomarkers that are uniquely enriched relative to the background host cell-derived biomarkers. There is a need for development of sensitive and robust technologies to reliably investigate the low-input nucleic acid (RNA, DNA) extracted from purified EVs.
Overall, the main utility of EVs is their role in disease diagnosis and monitoring. A vast majority of literature has utilized this analyte to develop sensitive and specific mutation detection assays. In addition, more work is now focused on developing a multianalyte approach. Studies have compared and investigated the feasibility of EVs versus other analytes individually and in combination (ctDNA, CTCs, tumor-educated platelets) in liquid biopsy (Table S1).
Liquid pathology
A new generation of targeted therapeutics and the advent of personalized oncology practice has transformed many types of cancer from diseases associated with acute mortality to more chronic conditions that require continuous monitoring to optimize disease remission and extend the quality of life for cancer patients. As such, cancer diagnostics will require approaches and testing platforms similar to those used to screen for and manage other chronic diseases such as diabetes and cardiovascular disease (i.e., low-cost, minimally invasive, point-of-care testing that is sufficiently accurate to guide reflexive testing using more extensive diagnostics).
Pathology has a long history of analysis of blood and other body fluids. Traditionally, this focused on chemical (pre-1980) and immunochemical (post-1980) analysis.33 The development of multiplexed (multi-marker) but single-analyte-type platforms has improved sensitivity, specificity, and throughput of assays.34 Malignant progression is a multifaceted process, and therefore, simultaneous and integrative analyses of biomarkers that contribute to that process (host genome, cancer genome, methylation programming, protein, EVs, and circulating tumor cell populations) are likely to create the most specific and sensitive assays for cancer predisposition, initiation, progression, and response to therapy.35 Recently, analyses that once were restricted to tissues have been increasingly performed from minimally invasive peripheral blood sampling, taking advantage of enormous advances in NGS technology,36 sophisticated proteomics, and the recognition that exosomes shed into blood may be exploited for diagnostic applications.37 Outstanding examples of this include tumor typing to determine possible therapeutic targets.38 These advances have led to a burgeoning field of liquid-based diagnoses, in what we have come to call “liquid biopsy.” This includes the detection of therapeutic efficacy based on blood tests that can report results much earlier than standard clinical methods, detection of early recurrence/minimal residual disease, and most recently, exciting work that may allow us to detect lethal cancers that currently have screening methods at an earlier, curable stage.39
A summary of the major analytes investigated in different cancers is provided in Table 1. In parallel with these analytic methods to evaluate shed components of cells have come new ways to evaluate cancer cells themselves that are in circulation (CTCs). While the original focus was restricted to the detection and quantification of CTCs, new technologies have allowed us to perform much deeper analysis of the cells, including their cellular and molecular compositions.10 We are also entering a new age of morphologic analysis of cells, where sophisticated advances in microscopy, image acquisition, and image analysis using deep learning/artificial intelligence provide ways of analyzing cells that extend far beyond our eyes and comprehension that could only be dreamed of a few years ago. These methods have created a paradigm shift in how we evaluate cancer, as well as a host of inherited diseases. While liquid pathology has long been an essential component of modern medical diagnosis, we are clearly entering a new and exciting era, where we can do and learn more, intervene sooner and more effectively, less invasively, and with greater safety, with a small blood sample.
Table 1.
Summary of reported liquid biopsy-based analytical techniques in brain, breast, lung, colon, and ovarian cancers
| Cancer | Biofluid | Analyte | Potential analytical techniques | Reference |
|---|---|---|---|---|
| Brain | plasma CSF |
EVs ctDNA CTCs |
ddPCR RT-PCR methylation-based PCR chip-based proteomic analysis CTC-iCHIP microfluidics density gradient centrifugation targeted sequencing deep sequencing NGS mass spectrometry imaging flow cytometry flow cytometry microfluidic nuclear magnetic resonance (μNMR) assay ELISA |
Figueroa, Lavon, Chen, Batool, Mouliere, Müller, Sullivan, Gao, Krol, Akers et al.6,19,40,41,42,43,44,45,46,47 |
| Breast | plasma | ctDNA CTCs cfmiRNA |
BEAMing ddPCR qRT-PCR TEC-seq personalized and ultra-deep sequencing large NGS panels nanotube-CTC chip CTC-iChip microarray |
Bettegowda, Phallen, Garcia-Murillas, Riva, Asaga, Roth, Coombes, Zhang, Kwan, Matamala, Hamam et al.9,23,48,49,50,51,52,53,54,55,56 |
| Lung | plasma pleural fluid |
EVs CTCs ctDNA miRNA |
PCR qRT-PCR ARMS-PCR CAPP-seq EFIRM NGS methylation-specific RT-PCR ISET CellSearch nano-quantum dots microarray |
Zhao, Uchida, Wan, Wei, Newman, Guo, Chen, Ponomaryova, Powrózek, Konecny, Powrózek, Ren, Allard, Tanaka, Hofman, Hofman, Hofman, Ilie, Dorsey, Heegaard, Sozzi, Shen, Wang, Montani, Xing, Wang Li, Fan, Razzak et al.57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85 |
| Colon | plasma serum saliva |
ctDNA CTCs EV RNA EV proteins TEPs |
CellMax biomimetic platform (CMx) CellSearch Safe-SeqS NGS digital PCR (BEAMing) ddPCR qPCR ALU-qPCR electrochemical sensing |
Tsai, Bork, Tsai, Musella, Krebs, Tie, Sun, Tie, Tie, Grasselli, Khan, Sun et al.86,87,88,89,90,91,92,93,94,95,96,97 |
| Ovarian | – | CTCs ctDNA EVs |
immunomagnetic bead capture microfluidic isolation and immunofluorescent staining CellSearch methylation-specific PCR microarray reduced representation bisulfite sequencing targeted deep sequencing whole-genome sequencing pyrosequencing qPCR nanoparticle tracking analysis nanoplasmonics |
Taylor, Hu, Liu, Nelson, Kim, Jaiswal, Salvi, Mari, Bronkhorst, Chen, Yokoi, Elias, Kim et al.98,99,100,101,102,103,104,105,106,107,108,109,110 |
Abbreviations: NGS, next-generation sequencing; cfmiRNA, circulating-free miRNA; TEC-seq, targeted error correction sequencing; ARMS-PCR, amplification refractory mutation system-based PCR; CAPP-seq, cancer personalized profiling by deep sequencing; EFIRM, electric field-induced release and management; ISET, isolation by size of epithelial tumor cell; TEP, tumor-educated platelets.
Multianalyte and multi-cancer early detection (MCED)
Despite the utility of single-analyte detection via liquid biopsy, barriers to high sensitivity and specificity remain challenging to the efficacy of these tests. As such, development of multianalyte early detection tests in this context has gained significant popularity over the last decade. By concurrently identifying multiple biomarkers, studies have presented an improved assay sensitivity for the detection of mutations as well as the potential to unveil therapy-resistant genes.48,111
Clinical inclusion of MCED blood-based tests will allow the detection of multiple cancers originating from different tissues. This promising, one-step testing has the potential to diagnose tumors at an early and more treatable stage. Examples of clinically applicable tests include Galleri, CancerSEEK, and OneTest. Galleri is a multi-cancer screening test for anyone over the age of 50 years when the risk of cancer is higher. It is based on cell-free DNA and detects tumor-specific methylation patterns to predict the tissue of origin. It can use the signal to potentially screen for more than 50 cancer types originating in multiple organ systems (kidney, lung, adrenal glands, gastrointestinal tract, oral cavity, etc.). Another platform, CancerSEEK, has been developed to detect eight unique proteins and oncogenic mutations using circulating DNA isolated from blood and screens up to nine cancers. Lastly, OneTest uses an integrated algorithm of machine learning and proteomics to screen for up to six cancers using well-established biomarkers. Importantly, these screening tests have been validated in large-scale, multi-institutional cohorts across countries, comprising different population types, and they represent an important milestone in achieving screening of cancers at an early, treatable stage.
A combination of these platforms may provide a more comprehensive window into the status of the tumors. As these diagnostic tests are developed, however, there remains a gap for inconsistencies to arise. Minimizing false positives is important to differentiate between low-prevalence marker signals and noise. Additionally, analysis and interpretation of the results need to be standardized and reproducible. Advances in machine learning and artificial intelligence will aid further development and advancement of multianalyte blood tests to predict progression.112
Addressing the challenges of biomarker-based liquid biopsy
Although a plethora of potential biomarkers to be used in clinical diagnostics is regularly published, fewer than 1% of these biomarkers successfully reach clinical practice.113 This can be attributed to several bottlenecks along different phases of biomarker discovery to clinical application (Table 2). Common setbacks include limited reproducibility of published findings, preanalytical errors, and post-analytical errors.
Table 2.
Common preanalytical issues and recommendations
| Preanalytical issue | Challenges | Recommendations |
|---|---|---|
| Isolation of disease-specific analytes from biofluids (CTCs, ctDNA, EVs) | lack of standardized protocols for processing of biofluids poor consistency among studies of reported kits employed for analyte isolation |
need to establish standard sample handling and collection protocols use of extensively tested and validated commercially available kits for optimal analyte isolation |
| Study population selection | convenience sampling commonly used to validate proposed mutation detection assays suboptimal control population |
multi-institutional collaborations to design a comprehensive patient population for improved generalizability of reported results selection of appropriate controls (healthy, benign disease of the same organ) |
| Confounding biological and environmental variables | influence of pre-sampling factors on quality of isolated analytes (circadian rhythm, fasting, metabolic disorders, hypertension, pregnancy, lactation) | inclusion of recommendations in sample collection protocols to improve rigor and reproducibility |
| Long-term sample storage conditions and biobanking | inconsistent data on decay rates reported with the use of different storage methods freeze-thaw cycles and thawing procedure can influence target nucleic acid concentration and integrity |
need to determine optimal storage conditions (time, temperature) for cell-free matrices and extracted nucleic acid evaluation of appropriate thawing procedure (duration, i.e., fast vs. slow, temperature) and sample storage volumes |
| Inclusion of study design specific controls | limited understanding of biological variation in individual samples as well as patients’ own biofluids over the course of disease | use of internal synthetic, known standards, to better account for biological variation in the biofluids and technical variation in reported techniques |
Study population selection is an important step in study design. Deficiencies in initial study design can introduce bias in reported findings. In addition to extensive cross-validation, the sensitivity and specificity should be evaluated in certain groups of patients with varying disease severity. Comorbidities or bias in the original design or lack of testing in heterogeneous disease groups can lead to differences in early- vs. late-stage testing results. Due to the rigorous nature of biobanking, many studies use “samples of convenience” rather than samples representative of the target population. Controls may vary in demographics and other biological parameters. These scenarios highlight the importance of a “fit-for-purpose” approach to biomarker discovery and validation to ensure results and applications are designed to clearly define and meet the intended use of the data.
Next, it is important to determine and evaluate the potential role of biomarkers in disease diagnosis and management. Some newly discovered biomarkers may be specific to the tumor but lack usefulness in actual clinical practice. This could be due to a limited role in clinical decision-making or minimal effect on treatment decisions. Understanding results in the context of disease is crucial to achieve wider implementation. Even more important is inclusion of efforts to train the multi-disciplinary teams involved in patient care to understand the implications of “positive” versus “negative” results.
The preanalytical phase represents an important stage of the liquid biopsy workflow. However, lack of standardized protocols can lead to variable findings. Key preanalytical variables include specimen collection, isolation methods, sample processing, downstream detection platforms, and biobanking for short- and long-term storage.
Finally, it is imperative to address sources of bias in data analysis, including systemic bias resulting from improper selection of controls, and statistical error. Inappropriate statistical methods can lead to incorrect conclusions. Furthermore, multiple comparisons using pre-defined training and validation datasets help to understand and to identify the variations in study populations. This is especially important when analyzing strong false-positive signals. These problems tend to arise more frequently when large data is applied to small cohorts or looking for a low, true positive signal. One of the most comprehensive study designs to eliminate bias is the prospective-specimen-collection, retrospective-blinded-evaluation (ProBe).114 This design focuses on four steps to reduce variation and bias: clinical application, outcome, case-control status, and selection. The comprehensive nature of ProBe reflects proper standards of research; it reduces the “exploratory” factor of biomarker study, effectively targets areas to reduce unintentional false discovery, and maintains high standards, potentially leading to more applicable biomarker discovery.
To improve the prospects of liquid biopsy implementation in clinical practice, more attention needs to be drawn to the numerous challenges in the study design and testing phase. This approach in combination with rigorous external and internal validation testing is an important next step in future biomarker development.
Partnership with industry
The commercialization of liquid biopsy-based diagnostics has garnered widespread attention. The market opportunity they represent is estimated to range between at least $30B and $130B in the United States alone. Large players have included GOOGLE Verily, Roche Holdings, Guardant Health, Illumina, and FoundationOne. The market remains under-penetrated particularly for orphan diseases, sensitive novel approaches, and academic-commercial interactions that emphasize clinically the benefits of synergistic efforts. These academic-commercial partnerships have been emphasized by the NCI, Division of Cancer Prevention, in the creation of the LBC (RFA CA17-029).
By and large, most companies have developed interrogative approaches using blood or its plasma: GRAIL, THRIVE, Bioprognos, DELPFI, EARLYDx, Helio/LAM, Nucleix, and Volition. However, academic centers have explored biofluids likely in close proximity to organs that are afflicted: saliva and sputum (mouth and pharyngeal cancers),115 sputum (lung and gastrointestinal cancers),116 urine (genitourinary),117 CSF (brain tumors and degenerative neurologic diseases),18,118 and seminal fluid.119,120 Academic-commercial collaborations have been more novel and represent the wellspring of the LBC. These include analyses of tumor-derived EVs, cfDNA, derived from CTCs, and circulating cell-free RNA. These analyses, combined with unique attention to specimen collection and study design, are complemented by a Consortium Biorepository and shared sample understanding as well as cross-laboratory validations.
Academic-industrial partnership can fuel discovery by validating promising biomarkers leading to rapid translation to a clinical setting. Academic labs can drive technological development and biomarker discovery, improve their performance, and define target populations for tests. The industry, in turn, can drive process development, cost reduction, automation, pre-analytics, analytical validation, and large trials. Thus, the combination of knowledge and scientific flexibility of academia with the organization and capabilities of the industry is aimed at streamlining the liquid biopsy-based biomarker development pipeline.
Other notable efforts in the field
Given the clinical utility of liquid biopsy in disease detection and surveillance, a number of international societies and consortiums have been initiated to standardize key variables identified in target analyte isolation and downstream analysis. Targeted international collaborations are an important step to achieving standard integrated protocols. European Liquid Biopsy Academy, funded under the European Commission’s Marie Sklodowska-Curie Programme, is focused at developing blood-based assays for the detection of stage I–IV non-small-cell lung cancer. Additionally, it covers the development and validation of technologies for analysis of four different types of analytes (CTCs, EVs, ctDNA, tumor-educated platelets) individually and in combination. These efforts will potentially enable a more robust large-scale validation and allow patient selection for targeted therapeutics. The International Society of Liquid Biopsy and International Liquid Biopsy Standardization Alliance Collaborative Community are some of the earlier organizations to coordinate efforts from multiple partners with the aim of standardizing the liquid biopsy workflow by addressing and identifying crucial preanalytical and analytical factors. BLOODPAC represents another effort as part of the White House Cancer Moonshot program to accelerate the development and clinical translation of proposed liquid biopsy assays. Early Detection Research Network (EDRN), founded by NIH, is a consortium of more than 300 academic and industry partners. There are four components of the EDRN (biomarker development, biomarker reference, clinical validation, and data management) that work synergistically to advance the development of biomarkers for early-stage cancer detection.
Vision for the future
Liquid biopsy will likely revolutionize how we detect and treat cancer in the future. As described in this commentary, several analytes such as DNA, proteins, metabolites in cfDNA, CTCs, and circulating EVs need to be incorporated into the liquid biopsy assays. Future research is needed to identify biological questions as to which cancer type can benefit from liquid biopsy-based assay on known etiology, type, and extent or foliation of cfDNA or ctDNA and mechanism behind the foliation. In addition to previously reported mechanisms (apoptosis, necrosis) of ctDNA release, active secretion leading to different patterns of ctDNA fragmentation has also been implicated. Within the nucleosome core formed by histone proteins, ctDNA is protected against cleavage by nucleases. However, the remaining linker ctDNA sequence located between nucleosomes is highly vulnerable. This therefore explains why different regions show high vs. low frequency of biological fragmentation. Such phenomenon can provide a biased representation of ctDNA sequences and unbalanced read coverage. We need to explore technologies that can be added to existing technologies to provide better sampling, improving the level of detection within a smaller sample (currently 5- to 10-mL samples are required), and improving the signal-noise ratio (e.g., chromatin immunoprecipitation contributes to mutational background noise to mutational-based liquid biopsy assays.
The major impediment to the use of liquid biopsy in clinical settings is the lack of prospective longitudinal cohorts for validating liquid biopsy technologies. We need to survey the existing cohorts in relation to quality and amount of sample availability and suitability of sample for genomic, proteomic, and epigenomic analysis and to assess quality control in place for these cohorts for maintaining the sample quality over years and the relevant acquisition guidelines (e.g., many cohorts have a process in place to request samples through their steering committees, working groups, review groups, etc.). There is a need to address the multiple preanalytical variables that have a crucial impact on standardization of reported liquid biopsy modalities (Table 2). Concurrently, we need to think creatively about a prospective study design(s) that can address the use of liquid biopsy in improving cancer-stage shift, minimize unnecessary invasive diagnostic workups, improve detection efficiency, and provide information comparatively with the standard of care. The objective of the liquid biopsy technologies should be to produce cost-effective, rigorous diagnostic tests that are complementary to the marketplace. Because the liquid biopsy technologies continue to be developed and refined, it is prudent to work with health-care professionals, including primary care doctors and the public to manage expectations.
The LBC is addressing some of the challenges in collaboration with several stakeholders, including foundations, industry, and academia. Our intention is not to adhere to a single approach but to promote the synergy of multiple circulating biomarkers with multiple analytes for multiple cancers. Our goal is to identify the combinations of markers that signal the presence of cancer. The consortium is considering circulating tumor biomarkers using a variety of approaches, including those that can be considered for monitoring tumor microenvironment and immune cells that could potentially be useful in developing targeted therapies.
Acknowledgments
This work is supported by the Division of Cancer Prevention, National Cancer Institute grants U01 CA230697 (B.S.C., L.B.), P01 CA069246 (B.S.C., L.B.), R01CA239078 (B.S.C., H.L.), CA237500 (B.S.C., L.B.), U01CA233360 (H.L., C.M.C., J.S.), and R01CA229777 (H.L., J.S.).
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101198.
Supplemental information
References
- 1.Website U.S. Cancer Statistics Working Group. U.S. Cancer Statistics Data Visualizations Tool, Based on 2019 Submission Data (1999-2017): U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute; www.cdc.gov/cancer/dataviz, released in June 2020.
- 2.Byers T., Wender R.C., Jemal A., Baskies A.M., Ward E.E., Brawley O.W. The American Cancer Society challenge goal to reduce US cancer mortality by 50% between 1990 and 2015: Results and reflections. CA A Cancer J. Clin. 2016;66:359–369. doi: 10.3322/caac.21348. [DOI] [PubMed] [Google Scholar]
- 3.Domínguez-Vigil I.G., Moreno-Martínez A.K., Wang J.Y., Roehrl M.H.A., Barrera-Saldaña H.A. The dawn of the liquid biopsy in the fight against cancer. Oncotarget. 2018;9:2912–2922. doi: 10.18632/oncotarget.23131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu X., Weiser E., Jacobson D.J., Griffin J.M., Limburg P.J., Finney Rutten L.J. Provider-perceived barriers to patient adherence to colorectal cancer screening. Prev. Med. Rep. 2022;25:101681. doi: 10.1016/j.pmedr.2021.101681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ACS Breast Cancer Early Detection Recommendations https://www.cancer.org/cancer/breast-cancer/screening-tests-and-early-detection/american-cancer-society-recommendations-for-the-early-detection-of-breast-cancer.html.
- 6.Mouliere F., Chandrananda D., Piskorz A.M., Moore E.K., Morris J., Ahlborn L.B., Mair R., Goranova T., Marass F., Heider K., et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 2018;10:eaat4921. doi: 10.1126/scitranslmed.aat4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah K.S.V., Ethunandan M. Tumour seeding after fine-needle aspiration and core biopsy of the head and neck--a systematic review. Br. J. Oral Maxillofac. Surg. 2016;54:260–265. doi: 10.1016/j.bjoms.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Ross T.S., Mgbemena V.E. Re-evaluating the role of BCR/ABL in chronic myelogenous leukemia. Mol. Cell. Oncol. 2014;1:e963450. doi: 10.4161/23723548.2014.963450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X., Zhao W., Wei W., You Z., Ou X., Sun M., Yin Y., Tang X., Zhao Z., Hu C., et al. Parallel Analyses of Somatic Mutations in Plasma Circulating Tumor DNA (ctDNA) and Matched Tumor Tissues in Early-Stage Breast Cancer. Clin. Cancer Res. 2019;25:6546–6553. doi: 10.1158/1078-0432.CCR-18-4055. [DOI] [PubMed] [Google Scholar]
- 10.Vasseur A., Kiavue N., Bidard F.-C., Pierga J.-Y., Cabel L. Clinical utility of circulating tumor cells: an update. Mol. Oncol. 2021;15:1647–1666. doi: 10.1002/1878-0261.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loud J.T., Murphy J. Cancer Screening and Early Detection in the 21 st Century. Semin. Oncol. Nurs. 2017;33:121–128. doi: 10.1016/j.soncn.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 13.Perrone M.G., Luisi O., De Grassi A., Ferorelli S., Cormio G., Scilimati A. Translational Theragnosis of Ovarian Cancer: where do we stand? Curr. Med. Chem. 2020;27:5675–5715. doi: 10.2174/0929867326666190816232330. [DOI] [PubMed] [Google Scholar]
- 14.Enderle D., Spiel A., Coticchia C.M., Berghoff E., Mueller R., Schlumpberger M., Sprenger-Haussels M., Shaffer J.M., Lader E., Skog J., Noerholm M. Characterization of RNA from Exosomes and Other Extracellular Vesicles Isolated by a Novel Spin Column-Based Method. PLoS One. 2015;10:e0136133. doi: 10.1371/journal.pone.0136133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bondy M.L., Scheurer M.E., Malmer B., Barnholtz-Sloan J.S., Davis F.G., Il’yasova D., Kruchko C., McCarthy B.J., Rajaraman P., Schwartzbaum J.A., et al. Brain Tumor Epidemiology: Consensus from the Brain Tumor Epidemiology Consortium (BTEC) Cancer. 2008;113:1953–1968. doi: 10.1002/cncr.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brain Cancer Screening https://moffitt.org/cancers/brain-cancer/diagnosis/screening/.
- 17.De Mattos-Arruda L., Mayor R., Ng C.K.Y., Weigelt B., Martínez-Ricarte F., Torrejon D., Oliveira M., Arias A., Raventos C., Tang J., et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat. Commun. 2015;6:8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller A.M., Shah R.H., Pentsova E.I., Pourmaleki M., Briggs S., Distefano N., Zheng Y., Skakodub A., Mehta S.A., Campos C., et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565:654–658. doi: 10.1038/s41586-019-0882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Figueroa J.M., Skog J., Akers J., Li H., Komotar R., Jensen R., Ringel F., Yang I., Kalkanis S., Thompson R., et al. Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro Oncol. 2017;19:1494–1502. doi: 10.1093/neuonc/nox085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yekula A., Muralidharan K., Kang K.M., Wang L., Balaj L., Carter B.S. From Laboratory to Clinic: Translation of Extracellular Vesicle Based Cancer Biomarkers. Methods. 2020;177:58–66. doi: 10.1016/j.ymeth.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothwell D.G., Ayub M., Cook N., Thistlethwaite F., Carter L., Dean E., Smith N., Villa S., Dransfield J., Clipson A., et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat. Med. 2019;25:738–743. doi: 10.1038/s41591-019-0380-z. [DOI] [PubMed] [Google Scholar]
- 22.Nassiri F., Chakravarthy A., Feng S., Shen S.Y., Nejad R., Zuccato J.A., Voisin M.R., Patil V., Horbinski C., Aldape K., et al. Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes. Nat. Med. 2020;26:1044–1047. doi: 10.1038/s41591-020-0932-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N., Bartlett B.R., Wang H., Luber B., Alani R.M., et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shankar G.M., Balaj L., Stott S.L., Nahed B., Carter B.S. Liquid biopsy for brain tumors. Expert Rev. Mol. Diagn. 2017;17:943–947. doi: 10.1080/14737159.2017.1374854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mader S., Pantel K. Liquid Biopsy: Current Status and Future Perspectives. Oncol. Res. Treat. 2017;40:404–408. doi: 10.1159/000478018. [DOI] [PubMed] [Google Scholar]
- 26.Young R., Pailler E., Billiot F., Drusch F., Barthelemy A., Oulhen M., Besse B., Soria J.-C., Farace F., Vielh P. Circulating tumor cells in lung cancer. Acta Cytol. 2012;56:655–660. doi: 10.1159/000345182. [DOI] [PubMed] [Google Scholar]
- 27.Palmirotta R., Lovero D., Cafforio P., Felici C., Mannavola F., Pellè E., Quaresmini D., Tucci M., Silvestris F. Liquid biopsy of cancer: a multimodal diagnostic tool in clinical oncology. Ther. Adv. Med. Oncol. 2018;10 doi: 10.1177/1758835918794630. 1758835918794630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becker A., Thakur B.K., Weiss J.M., Kim H.S., Peinado H., Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30:836–848. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yekula A., Yekula A., Muralidharan K., Kang K., Carter B.S., Balaj L. Extracellular Vesicles in Glioblastoma Tumor Microenvironment. Front. Immunol. 2019;10:3137. doi: 10.3389/fimmu.2019.03137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., Chen Y., Pei F., Zeng C., Yao Y., Liao W., Zhao Z. Extracellular Vesicles in Liquid Biopsies: Potential for Disease Diagnosis. BioMed Res. Int. 2021;2021:6611244. doi: 10.1155/2021/6611244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lapitz A., Arbelaiz A., O’Rourke C.J., Lavin J.L., Casta A.L., Ibarra C., Jimeno J.P., Santos-Laso A., Izquierdo-Sanchez L., Krawczyk M., et al. Patients with Cholangiocarcinoma Present Specific RNA Profiles in Serum and Urine Extracellular Vesicles Mirroring the Tumor Expression: Novel Liquid Biopsy Biomarkers for Disease Diagnosis. Cells. 2020;9 doi: 10.3390/cells9030721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Y., Liu S., Qiao Z., Shang Z., Xia Z., Niu X., Qian L., Zhang Y., Fan L., Cao C.-X., Xiao H. Systematic comparison of exosomal proteomes from human saliva and serum for the detection of lung cancer. Anal. Chim. Acta. 2017;982:84–95. doi: 10.1016/j.aca.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Whicher J.T., Warren C., Chambers R.E. Immunochemical Assays for Immunoglobulins. Ann. Clin. Biochem. 1984;21:78–91. doi: 10.1177/000456328402100202. [DOI] [PubMed] [Google Scholar]
- 34.Hoyt C.C. Multiplex Immunofluorescence and Multispectral Imaging: Forming the Basis of a Clinical Test Platform for Immuno-Oncology. Front. Mol. Biosci. 2021;8:674747. doi: 10.3389/fmolb.2021.674747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dagogo-Jack I., Shaw A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 36.Neumann M.H.D., Bender S., Krahn T., Schlange T. ctDNA and CTCs in Liquid Biopsy - Current Status and Where We Need to Progress. Comput. Struct. Biotechnol. J. 2018;16:190–195. doi: 10.1016/j.csbj.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee Y.T., Tan Y.J., Oon C.E. Molecular targeted therapy: Treating cancer with specificity. Eur. J. Pharmacol. 2018;834:188–196. doi: 10.1016/j.ejphar.2018.07.034. [DOI] [PubMed] [Google Scholar]
- 38.Dong J., Li B., Lin D., Zhou Q., Huang D. Advances in Targeted Therapy and Immunotherapy for Non-small Cell Lung Cancer Based on Accurate Molecular Typing. Front. Pharmacol. 2019;10:230. doi: 10.3389/fphar.2019.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu C., Xiang X., Han S., Lim H.Y., Li L., Zhang X., Ma Z., Yang L., Guo S., Soo R., et al. Blood-based liquid biopsy: Insights into early detection and clinical management of lung cancer. Cancer Lett. 2022;524:91–102. doi: 10.1016/j.canlet.2021.10.013. [DOI] [PubMed] [Google Scholar]
- 40.Lavon I., Refael M., Zelikovitch B., Shalom E., Siegal T. Serum DNA can define tumor-specific genetic and epigenetic markers in gliomas of various grades. Neuro Oncol. 2010;12:173–180. doi: 10.1093/neuonc/nop041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen W.W., Balaj L., Liau L.M., Samuels M.L., Kotsopoulos S.K., Maguire C.A., Loguidice L., Soto H., Garrett M., Zhu L.D., et al. BEAMing and Droplet Digital PCR Analysis of Mutant IDH1 mRNA in Glioma Patient Serum and Cerebrospinal Fluid Extracellular Vesicles. Mol. Ther. Nucleic Acids. 2013;2:e109. doi: 10.1038/mtna.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Batool S.M., Muralidharan K., Hsia T., Falotico S., Gamblin A.S., Rosenfeld Y.B., Khanna S.K., Balaj L., Carter B.S. Highly sensitive EGFRvIII detection in circulating extracellular vesicle RNA of glioma patients. Clin. Cancer Res. 2022;28:4070–4082. doi: 10.1158/1078-0432.CCR-22-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Müller C., Holtschmidt J., Auer M., Heitzer E., Lamszus K., Schulte A., Matschke J., Langer-Freitag S., Gasch C., Stoupiec M., et al. Hematogenous dissemination of glioblastoma multiforme. Sci. Transl. Med. 2014;6:247ra101. doi: 10.1126/scitranslmed.3009095. [DOI] [PubMed] [Google Scholar]
- 44.Sullivan J.P., Nahed B.V., Madden M.W., Oliveira S.M., Springer S., Bhere D., Chi A.S., Wakimoto H., Rothenberg S.M., Sequist L.V., et al. Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov. 2014;4:1299–1309. doi: 10.1158/2159-8290.CD-14-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao F., Cui Y., Jiang H., Sui D., Wang Y., Jiang Z., Zhao J., Lin S. Circulating tumor cell is a common property of brain glioma and promotes the monitoring system. Oncotarget. 2016;7:71330–71340. doi: 10.18632/oncotarget.11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krol I., Castro-Giner F., Maurer M., Gkountela S., Szczerba B.M., Scherrer R., Coleman N., Carreira S., Bachmann F., Anderson S., et al. Detection of circulating tumour cell clusters in human glioblastoma. Br. J. Cancer. 2018;119:487–491. doi: 10.1038/s41416-018-0186-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akers J.C., Ramakrishnan V., Kim R., Skog J., Nakano I., Pingle S., Kalinina J., Hua W., Kesari S., Mao Y., et al. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): a platform for glioblastoma biomarker development. PLoS One. 2013;8:e78115. doi: 10.1371/journal.pone.0078115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phallen J., Sausen M., Adleff V., Leal A., Hruban C., White J., Anagnostou V., Fiksel J., Cristiano S., Papp E., et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017;9:eaan2415. doi: 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia-Murillas I., Schiavon G., Weigelt B., Ng C., Hrebien S., Cutts R.J., Cheang M., Osin P., Nerurkar A., Kozarewa I., et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 2015;7:302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 50.Riva F., Bidard F.-C., Houy A., Saliou A., Madic J., Rampanou A., Hego C., Milder M., Cottu P., Sablin M.-P., et al. Patient-Specific Circulating Tumor DNA Detection during Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer. Clin. Chem. 2017;63:691–699. doi: 10.1373/clinchem.2016.262337. [DOI] [PubMed] [Google Scholar]
- 51.Asaga S., Kuo C., Nguyen T., Terpenning M., Giuliano A.E., Hoon D.S.B. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin. Chem. 2011;57:84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- 52.Roth C., Rack B., Müller V., Janni W., Pantel K., Schwarzenbach H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12:R90. doi: 10.1186/bcr2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coombes R.C., Page K., Salari R., Hastings R.K., Armstrong A., Ahmed S., Ali S., Cleator S., Kenny L., Stebbing J., et al. Personalized Detection of Circulating Tumor DNA Antedates Breast Cancer Metastatic Recurrence. Clin. Cancer Res. 2019;25:4255–4263. doi: 10.1158/1078-0432.CCR-18-3663. [DOI] [PubMed] [Google Scholar]
- 54.Kwan T.T., Bardia A., Spring L.M., Giobbie-Hurder A., Kalinich M., Dubash T., Sundaresan T., Hong X., LiCausi J.A., Ho U., et al. A Digital RNA Signature of Circulating Tumor Cells Predicting Early Therapeutic Response in Localized and Metastatic Breast Cancer. Cancer Discov. 2018;8:1286–1299. doi: 10.1158/2159-8290.CD-18-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matamala N., Vargas M.T., González-Cámpora R., Miñambres R., Arias J.I., Menéndez P., Andrés-León E., Gómez-López G., Yanowsky K., Calvete-Candenas J., et al. Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin. Chem. 2015;61:1098–1106. doi: 10.1373/clinchem.2015.238691. [DOI] [PubMed] [Google Scholar]
- 56.Hamam R., Ali A.M., Alsaleh K.A., Kassem M., Alfayez M., Aldahmash A., Alajez N.M. microRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection. Sci. Rep. 2016;6:25997. doi: 10.1038/srep25997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao X., Han R.B., Zhao J., Wang J., Yang F., Zhong W., Zhang L., Li L.-Y., Wang M.-Z. Comparison of epidermal growth factor receptor mutation statuses in tissue and plasma in stage I-IV non-small cell lung cancer patients. Respiration. 2013;85:119–125. doi: 10.1159/000338790. [DOI] [PubMed] [Google Scholar]
- 58.Uchida J., Kato K., Kukita Y., Kumagai T., Nishino K., Daga H., Nagatomo I., Inoue T., Kimura M., Oba S., et al. Diagnostic Accuracy of Noninvasive Genotyping of EGFR in Lung Cancer Patients by Deep Sequencing of Plasma Cell-Free DNA. Clin. Chem. 2015;61:1191–1196. doi: 10.1373/clinchem.2015.241414. [DOI] [PubMed] [Google Scholar]
- 59.Wan Y., Liu B., Lei H., Zhang B., Wang Y., Huang H., Chen S., Feng Y., Zhu L., Gu Y., et al. Nanoscale extracellular vesicle-derived DNA is superior to circulating cell-free DNA for mutation detection in early-stage non-small-cell lung cancer. Ann. Oncol. 2018;29:2379–2383. doi: 10.1093/annonc/mdy458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei F., Strom C.M., Cheng J., Lin C.-C., Hsu C.-Y., Soo Hoo G.W., Chia D., Kim Y., Li F., Elashoff D., et al. Electric Field-Induced Release and Measurement Liquid Biopsy for Noninvasive Early Lung Cancer Assessment. J. Mol. Diagn. 2018;20:738–742. doi: 10.1016/j.jmoldx.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Newman A.M., Bratman S.V., To J., Wynne J.F., Eclov N.C.W., Modlin L.A., Liu C.L., Neal J.W., Wakelee H.A., Merritt R.E., et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo N., Lou F., Ma Y., Li J., Yang B., Chen W., Ye H., Zhang J.-B., Zhao M.-Y., Wu W.-J., et al. Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci. Rep. 2016;6:33519. doi: 10.1038/srep33519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen K.-Z., Lou F., Yang F., Zhang J.-B., Ye H., Chen W., Guan T., Zhao M.-Y., Su X.-X., Shi R., et al. Circulating Tumor DNA Detection in Early-Stage Non-Small Cell Lung Cancer Patients by Targeted Sequencing. Sci. Rep. 2016;6:31985. doi: 10.1038/srep31985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ponomaryova A.A., Rykova E.Y., Cherdyntseva N.V., Skvortsova T.E., Dobrodeev A.Y., Zav’yalov A.A., Bryzgalov L.O., Tuzikov S.A., Vlassov V.V., Laktionov P.P. Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer. 2013;81:397–403. doi: 10.1016/j.lungcan.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 65.Powrózek T., Krawczyk P., Kucharczyk T., Milanowski J. Septin 9 promoter region methylation in free circulating DNA-potential role in noninvasive diagnosis of lung cancer: preliminary report. Med. Oncol. 2014;31:917. doi: 10.1007/s12032-014-0917-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Konecny M., Markus J., Waczulikova I., Dolesova L., Kozlova R., Repiska V., Novosadova H., Majer I. The value of SHOX2 methylation test in peripheral blood samples used for the differential diagnosis of lung cancer and other lung disorders. Neoplasma. 2016;63:246–253. doi: 10.4149/210_150419N208. [DOI] [PubMed] [Google Scholar]
- 67.Powrózek T., Krawczyk P., Nicoś M., Kuźnar-Kamińska B., Batura-Gabryel H., Milanowski J. Methylation of the DCLK1 promoter region in circulating free DNA and its prognostic value in lung cancer patients. Clin. Transl. Oncol. 2016;18:398–404. doi: 10.1007/s12094-015-1382-z. [DOI] [PubMed] [Google Scholar]
- 68.Ren M., Wang C., Sheng D., Shi Y., Jin M., Xu S. Methylation analysis of SHOX2 and RASSF1A in bronchoalveolar lavage fluid for early lung cancer diagnosis. Ann. Diagn. Pathol. 2017;27:57–61. doi: 10.1016/j.anndiagpath.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 69.Allard W.J., Matera J., Miller M.C., Repollet M., Connelly M.C., Rao C., Tibbe A.G.J., Uhr J.W., Terstappen L.W.M.M. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 70.Tanaka F., Yoneda K., Kondo N., Hashimoto M., Takuwa T., Matsumoto S., Okumura Y., Rahman S., Tsubota N., Tsujimura T., et al. Circulating tumor cell as a diagnostic marker in primary lung cancer. Clin. Cancer Res. 2009;15:6980–6986. doi: 10.1158/1078-0432.CCR-09-1095. [DOI] [PubMed] [Google Scholar]
- 71.Hofman V., Bonnetaud C., Ilie M.I., Vielh P., Vignaud J.M., Fléjou J.F., Lantuejoul S., Piaton E., Mourad N., Butori C., et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin. Cancer Res. 2011;17:827–835. doi: 10.1158/1078-0432.CCR-10-0445. [DOI] [PubMed] [Google Scholar]
- 72.Hofman V., Ilie M.I., Long E., Selva E., Bonnetaud C., Molina T., Vénissac N., Mouroux J., Vielh P., Hofman P. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay™ and the isolation by size of epithelial tumor cell method. Int. J. Cancer. 2011;129:1651–1660. doi: 10.1002/ijc.25819. [DOI] [PubMed] [Google Scholar]
- 73.Hofman V., Long E., Ilie M., Bonnetaud C., Vignaud J.M., Fléjou J.F., Lantuejoul S., Piaton E., Mourad N., Butori C., et al. Morphological analysis of circulating tumour cells in patients undergoing surgery for non-small cell lung carcinoma using the isolation by size of epithelial tumour cell (ISET) method. Cytopathology. 2012;23:30–38. doi: 10.1111/j.1365-2303.2010.00835.x. [DOI] [PubMed] [Google Scholar]
- 74.Ilie M., Hofman V., Long-Mira E., Selva E., Vignaud J.-M., Padovani B., Mouroux J., Marquette C.-H., Hofman P. Sentinel” circulating tumor cells allow early diagnosis of lung cancer in patients with chronic obstructive pulmonary disease. PLoS One. 2014;9:e111597. doi: 10.1371/journal.pone.0111597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dorsey J.F., Kao G.D., MacArthur K.M., Ju M., Steinmetz D., Wileyto E.P., Simone C.B., Hahn S.M., Hahn S.M. Tracking viable circulating tumor cells (CTCs) in the peripheral blood of non-small cell lung cancer (NSCLC) patients undergoing definitive radiation therapy: pilot study results. Cancer. 2015;121:139–149. doi: 10.1002/cncr.28975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heegaard N.H.H., Schetter A.J., Welsh J.A., Yoneda M., Bowman E.D., Harris C.C. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int. J. Cancer. 2012;130:1378–1386. doi: 10.1002/ijc.26153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sozzi G., Boeri M., Rossi M., Verri C., Suatoni P., Bravi F., Roz L., Conte D., Grassi M., Sverzellati N., et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J. Clin. Oncol. 2014;32:768–773. doi: 10.1200/JCO.2013.50.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shen J., Liao J., Guarnera M.A., Fang H., Cai L., Stass S.A., Jiang F. Analysis of MicroRNAs in sputum to improve computed tomography for lung cancer diagnosis. J. Thorac. Oncol. 2014;9:33–40. doi: 10.1097/JTO.0000000000000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang P., Yang D., Zhang H., Wei X., Ma T., Cheng Z., Hong Q., Hu J., Zhuo H., Song Y., et al. Early Detection of Lung Cancer in Serum by a Panel of MicroRNA Biomarkers. Clin. Lung Cancer. 2015;16:313–319.e1. doi: 10.1016/j.cllc.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 80.Montani F., Marzi M.J., Dezi F., Dama E., Carletti R.M., Bonizzi G., Bertolotti R., Bellomi M., Rampinelli C., Maisonneuve P., et al. miR-Test: a blood test for lung cancer early detection. J. Natl. Cancer Inst. 2015;107:djv063. doi: 10.1093/jnci/djv063. [DOI] [PubMed] [Google Scholar]
- 81.Xing L., Su J., Guarnera M.A., Zhang H., Cai L., Zhou R., Stass S.A., Jiang F. Sputum microRNA biomarkers for identifying lung cancer in indeterminate solitary pulmonary nodules. Clin. Cancer Res. 2015;21:484–489. doi: 10.1158/1078-0432.CCR-14-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang C., Ding M., Xia M., Chen S., Van Le A., Soto-Gil R., Shen Y., Wang N., Wang J., Gu W., et al. A Five-miRNA Panel Identified From a Multicentric Case-control Study Serves as a Novel Diagnostic Tool for Ethnically Diverse Non-small-cell Lung Cancer Patients. EBioMedicine. 2015;2:1377–1385. doi: 10.1016/j.ebiom.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li W., Wang Y., Zhang Q., Tang L., Liu X., Dai Y., Xiao L., Huang S., Chen L., Guo Z., et al. MicroRNA-486 as a Biomarker for Early Diagnosis and Recurrence of Non-Small Cell Lung Cancer. PLoS One. 2015;10:e0134220. doi: 10.1371/journal.pone.0134220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fan L., Qi H., Teng J., Su B., Chen H., Wang C., Xia Q. Identification of serum miRNAs by nano-quantum dots microarray as diagnostic biomarkers for early detection of non-small cell lung cancer. Tumour Biol. 2016;37:7777–7784. doi: 10.1007/s13277-015-4608-3. [DOI] [PubMed] [Google Scholar]
- 85.Razzak R., Bédard E.L.R., Kim J.O., Gazala S., Guo L., Ghosh S., Joy A., Nijjar T., Wong E., Roa W.H. MicroRNA expression profiling of sputum for the detection of early and locally advanced non-small-cell lung cancer: a prospective case-control study. Curr. Oncol. 2016;23:e86–e94. doi: 10.3747/co.23.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsai W.-S., Nimgaonkar A., Segurado O., Chang Y., Hsieh B., Shao H.-J., Wu J.-C., Lai J.-M., Javey M., Watson D., Mei R. Prospective clinical study of circulating tumor cells for colorectal cancer screening. J. Clin. Oncol. 2018;36:556. [Google Scholar]
- 87.Bork U., Rahbari N.N., Schölch S., Reissfelder C., Kahlert C., Büchler M.W., Weitz J., Koch M. Circulating tumour cells and outcome in non-metastatic colorectal cancer: a prospective study. Br. J. Cancer. 2015;112:1306–1313. doi: 10.1038/bjc.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsai W.-S., Chen J.-S., Shao H.-J., Wu J.-C., Lai J.-M., Lu S.-H., Hung T.-F., Chiu Y.-C., You J.-F., Hsieh P.-S., et al. Circulating Tumor Cell Count Correlates with Colorectal Neoplasm Progression and Is a Prognostic Marker for Distant Metastasis in Non-Metastatic Patients. Sci. Rep. 2016;6:24517. doi: 10.1038/srep24517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Musella V., Pietrantonio F., Di Buduo E., Iacovelli R., Martinetti A., Sottotetti E., Bossi I., Maggi C., Di Bartolomeo M., de Braud F., et al. Circulating tumor cells as a longitudinal biomarker in patients with advanced chemorefractory, RAS-BRAF wild-type colorectal cancer receiving cetuximab or panitumumab. Int. J. Cancer. 2015;137:1467–1474. doi: 10.1002/ijc.29493. [DOI] [PubMed] [Google Scholar]
- 90.Krebs M.G., Renehan A.G., Backen A., Gollins S., Chau I., Hasan J., Valle J.W., Morris K., Beech J., Ashcroft L., et al. Circulating Tumor Cell Enumeration in a Phase II Trial of a Four-Drug Regimen in Advanced Colorectal Cancer. Clin. Colorectal Cancer. 2015;14:115–122.e1-2. doi: 10.1016/j.clcc.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 91.Tie J., Wang Y., Tomasetti C., Li L., Springer S., Kinde I., Silliman N., Tacey M., Wong H.-L., Christie M., et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016;8:346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun X., Huang T., Cheng F., Huang K., Liu M., He W., Li M., Zhang X., Xu M., Chen S., Xia L. Monitoring colorectal cancer following surgery using plasma circulating tumor DNA. Oncol. Lett. 2018;15:4365–4375. doi: 10.3892/ol.2018.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tie J., Kinde I., Wang Y., Wong H.L., Roebert J., Christie M., Tacey M., Wong R., Singh M., Karapetis C.S., et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015;26:1715–1722. doi: 10.1093/annonc/mdv177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tie J., Cohen J., Wang Y., Lee M., Wong R., Kosmider S., Ananda S., Cho J.H., Faragher I., McKendrick J.J., et al. Serial circulating tumor DNA (ctDNA) analysis as a prognostic marker and a real-time indicator of adjuvant chemotherapy (CT) efficacy in stage III colon cancer (CC) J. Clin. Oncol. 2018;36:3516. [Google Scholar]
- 95.Grasselli J., Elez E., Caratù G., Matito J., Santos C., Macarulla T., Vidal J., Garcia M., Viéitez J.M., Paéz D., et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann. Oncol. 2017;28:1294–1301. doi: 10.1093/annonc/mdx112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Khan K., Rata M., Cunningham D., Koh D.-M., Tunariu N., Hahne J.C., Vlachogiannis G., Hedayat S., Marchetti S., Lampis A., et al. Functional imaging and circulating biomarkers of response to regorafenib in treatment-refractory metastatic colorectal cancer patients in a prospective phase II study. Gut. 2018;67:1484–1492. doi: 10.1136/gutjnl-2017-314178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun J., Fei F., Zhang M., Li Y., Zhang X., Zhu S., Zhang S. The role of SEPT9 in screening, diagnosis, and recurrence monitoring of colorectal cancer. BMC Cancer. 2019;19:450. doi: 10.1186/s12885-019-5663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taylor D.D., Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 99.Hu X., Feng Y., Zhang D., Zhao S.D., Hu Z., Greshock J., Zhang Y., Yang L., Zhong X., Wang L.-P., et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu E., Liu Z., Zhou Y. Carboplatin-docetaxel-induced activity against ovarian cancer is dependent on up-regulated lncRNA PVT1. Int. J. Clin. Exp. Pathol. 2015;8:3803–3810. [PMC free article] [PubMed] [Google Scholar]
- 101.Nelson N.J. Circulating tumor cells: will they be clinically useful? J. Natl. Cancer Inst. 2010;102:146–148. doi: 10.1093/jnci/djq016. [DOI] [PubMed] [Google Scholar]
- 102.Kim J.H., Chung H.H., Jeong M.S., Song M.R., Kang K.W., Kim J.S. One-step detection of circulating tumor cells in ovarian cancer using enhanced fluorescent silica nanoparticles. Int. J. Nanomed. 2013;8:2247–2257. doi: 10.2147/IJN.S45059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jaiswal S., Jamieson C.H.M., Pang W.W., Park C.Y., Chao M.P., Majeti R., Traver D., van Rooijen N., Weissman I.L. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138:271–285. doi: 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Salvi S., Gurioli G., De Giorgi U., Conteduca V., Tedaldi G., Calistri D., Casadio V. Cell-free DNA as a diagnostic marker for cancer: current insights. OncoTargets Ther. 2016;9:6549–6559. doi: 10.2147/OTT.S100901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mari R., Mamessier E., Lambaudie E., Provansal M., Birnbaum D., Bertucci F., Sabatier R. Liquid Biopsies for Ovarian Carcinoma: How Blood Tests May Improve the Clinical Management of a Deadly Disease. Cancers. 2019;11:774. doi: 10.3390/cancers11060774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bronkhorst A.J., Wentzel J.F., Aucamp J., van Dyk E., du Plessis L., Pretorius P.J. Characterization of the cell-free DNA released by cultured cancer cells. Biochim. Biophys. Acta. 2016;1863:157–165. doi: 10.1016/j.bbamcr.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 107.Chen H., Sun L.-Y., Zheng H.-Q., Zhang Q.-F., Jin X.-M. Total serum DNA and DNA integrity: diagnostic value in patients with hepatitis B virus-related hepatocellular carcinoma. Pathology. 2012;44:318–324. doi: 10.1097/PAT.0b013e328353a24c. [DOI] [PubMed] [Google Scholar]
- 108.Yokoi A., Yoshioka Y., Hirakawa A., Yamamoto Y., Ishikawa M., Ikeda S.-I., Kato T., Niimi K., Kajiyama H., Kikkawa F., Ochiya T. A combination of circulating miRNAs for the early detection of ovarian cancer. Oncotarget. 2017;8:89811–89823. doi: 10.18632/oncotarget.20688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Elias K.M., Fendler W., Stawiski K., Fiascone S.J., Vitonis A.F., Berkowitz R.S., Frendl G., Konstantinopoulos P., Crum C.P., Kedzierska M., et al. Diagnostic potential for a serum miRNA neural network for detection of ovarian cancer. Elife. 2017;6:e28932. doi: 10.7554/eLife.28932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim M.-Y., Oskarsson T., Acharyya S., Nguyen D.X., Zhang X.H.-F., Norton L., Massagué J. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hofmann L., Sallinger K., Haudum C., Smolle M., Heitzer E., Moser T., Novy M., Gesson K., Kroneis T., Bauernhofer T., El-Heliebi A. A Multi-Analyte Approach for Improved Sensitivity of Liquid Biopsies in Prostate Cancer. Cancers. 2020;12:2247. doi: 10.3390/cancers12082247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang Z., LaRiviere M.J., Ko J., Till J.E., Christensen T., Yee S.S., Black T.A., Tien K., Lin A., Shen H., et al. A Multianalyte Panel Consisting of Extracellular Vesicle miRNAs and mRNAs, cfDNA, and CA19-9 Shows Utility for Diagnosis and Staging of Pancreatic Ductal Adenocarcinoma. Clin. Cancer Res. 2020;26:3248–3258. doi: 10.1158/1078-0432.CCR-19-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kern S.E. Why your new cancer biomarker may never work: recurrent patterns and remarkable diversity in biomarker failures. Cancer Res. 2012;72:6097–6101. doi: 10.1158/0008-5472.CAN-12-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zheng Y. Study Design Considerations for Cancer Biomarker Discoveries. J. Appl. Lab. Med. 2018;3:282–289. doi: 10.1373/jalm.2017.025809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang Z., Li F., Rufo J., Chen C., Yang S., Li L., Zhang J., Cheng J., Kim Y., Wu M., et al. Acoustofluidic Salivary Exosome Isolation: A Liquid Biopsy Compatible Approach for Human Papillomavirus-Associated Oropharyngeal Cancer Detection. J. Mol. Diagn. 2020;22:50–59. doi: 10.1016/j.jmoldx.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mathai R.A., Vidya R.V.S., Reddy B.S., Thomas L., Udupa K., Kolesar J., Rao M. Potential Utility of Liquid Biopsy as a Diagnostic and Prognostic Tool for the Assessment of Solid Tumors: Implications in the Precision Oncology. J. Clin. Med. 2019;8:373. doi: 10.3390/jcm8030373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee B., Mahmud I., Marchica J., Dereziński P., Qi F., Wang F., Joshi P., Valerio F., Rivera I., Patel V., et al. Integrated RNA and metabolite profiling of urine liquid biopsies for prostate cancer biomarker discovery. Sci. Rep. 2020;10:3716. doi: 10.1038/s41598-020-60616-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yoo Y.K., Lee J., Kim H., Hwang K.S., Yoon D.S., Lee J.H. Toward Exosome-Based Neuronal Diagnostic Devices. Micromachines. 2018;9:634. doi: 10.3390/mi9120634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dong W.-W., Li H.-M., Qing X.-R., Huang D.-H., Li H.-G. Identification and characterization of human testis derived circular RNAs and their existence in seminal plasma. Sci. Rep. 2016;6:39080. doi: 10.1038/srep39080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rolland A.D., Lavigne R., Dauly C., Calvel P., Kervarrec C., Freour T., Evrard B., Rioux-Leclercq N., Auger J., Pineau C. Identification of genital tract markers in the human seminal plasma using an integrative genomics approach. Hum. Reprod. 2013;28:199–209. doi: 10.1093/humrep/des360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.