SUMMARY
Physiology is regulated by interconnected cell and tissue circadian clocks. Disruption of the rhythms generated by the concerted activity of these clocks is associated with metabolic disease. Here we tested the interactions between clocks in two critical components of organismal metabolism – liver and skeletal muscle – by rescuing clock function either in each organ separately, or in both organs simultaneously, in otherwise clock-less mice. Experiments showed that individual clocks are partially sufficient for tissue glucose metabolism, yet the connections between both tissue clocks coupled to daily feeding rhythms maximize systemic glucose tolerance. This synergy relies in part on local transcriptional control of the glucose machinery, feeding-responsive signals such as insulin, and metabolic cycles that connect the muscle and liver. We posit that spatiotemporal mechanisms of muscle and liver play an essential role in the maintenance of systemic glucose homeostasis, and that disrupting this diurnal coordination can contribute to metabolic disease.
INTRODUCTION
Circadian clocks are defining features of bacteria, plants, and animals1,2. In mammals, the clock impinges broadly on physiology and drives rhythms of behavior, organ system function, and cellular pathways, thereby aligning them with the solar cycle. This process is linked with overall health when robust but with disease when disrupted3,4. Studies in humans and model organisms have made it clear that metabolism is a central aspect of circadian control. In mice, genetic and environmental circadian disruption impairs lipid and carbohydrate metabolism, induces hallmarks of metabolic syndrome, and leads to diabetes-like phenotypes5–7.
Circadian control originates in virtually all cells of the body, propagates anatomically, and sets layers of regulation. Pioneering studies using tissue explants and cell-type-specific clock mutant mice have established the existence of peripheral clocks (non-neuronal) that are functionally significant and that interact with circadian cues produced by central clocks in the brain8–11. The liver, a highly rhythmic and metabolic tissue, has autonomous clock functionality but requires inputs from central clocks, for example daily feeding and fasting behavior, to express more of its rhythmic potential and maintain local homeostasis12–15. Considering that the liver interacts with many partner tissues (e.g., muscle), each of which contains its own local clock, we sought to test whether peripheral-to-peripheral clock interactions are critical, much like brain-to-peripheral connections are. Determining such mechanisms will provide a better understanding of how metabolism is regulated in time and space.
Here, we investigated the interaction between clocks in liver and skeletal muscle, two organs that are connected metabolically through the exchange of metabolites and regulatory signals, but whose coordinated daily actions are poorly understood. By manipulating endogenous Bmal1, the main transcriptional activator of the molecular clock, we rescued clock function in liver, in muscle, or in both tissues, in otherwise clock-less mice. Through analyses of transcriptomics and metabolism, we defined muscle clock autonomy and teased apart key spatiotemporal aspects of glucose homeostasis. Our major finding is that systemic glucose tolerance can be achieved from the concerted actions of three circadian system components – the liver clock, the muscle clock, and a feeding-fasting rhythm, yet no component alone is sufficient. We propose that these spatiotemporal aspects of glucose homeostasis contribute to the relationship between circadian disruption and metabolic disease.
RESULTS
Characterization of muscle and liver+muscle clock reconstituted mice
To interrogate liver and muscle clocks, we used Bmal1-stopFL mice, in which endogenous expression of the critical clock component Bmal116 can be reconstituted exclusively in Cre recombinase-expressing tissues in otherwise Bmal1-deficient mice15,17. Bmal1-stopFL mice were crossed with hepatocyte-driven Alfp-Cre and skeletal muscle-driven Hsa-Cre lines to generate the following genotypes: wild-type (WT; all clocks); full Bmal1 knockout (KO; no clocks); hepatocyte Bmal1-reconstituted (Liver-RE; only liver clock) 13,15; skeletal muscle Bmal1-reconstituted (Muscle-RE; only skeletal muscle clock); and both hepatocyte and skeletal muscle Bmal1-reconstituted (Liver+Muscle-RE; only liver and skeletal muscle clocks) (Figure 1A). Western blotting and qPCR confirmed the expected expression of BMAL1 across genotypes (Figures S1A–S1E). Even though total food intake was similar across genotypes, KO mice exhibited a lower body weight and a higher fat-to-lean mass ratio than WT mice, and the presence of liver and/or muscle clocks did not alter these phenotypes (Figures 1B–1D). As reported previously for Liver-RE mice13,15,18, KO and all RE mice lacked robust 24 h rhythms of food intake and displayed severely blunted dark phase locomotor activity under 12 h light/12 h dark conditions with ad libitum (AL) feeding (Figure 1E). Only WT mice had consistent and robust diurnal metabolic cycles, as shown through respiratory exchange ratio (RER) and energy expenditure (Figures 1F and S1F). Only six of the 23 KO and RE mice had significant rhythms of both parameters (Figure S1F). Compared to KO, mice with liver and/or muscle clocks did not show significant differences in RER (Figure 1F).
Figure 1. Characterization of muscle or liver+muscle clock reconstituted mice.
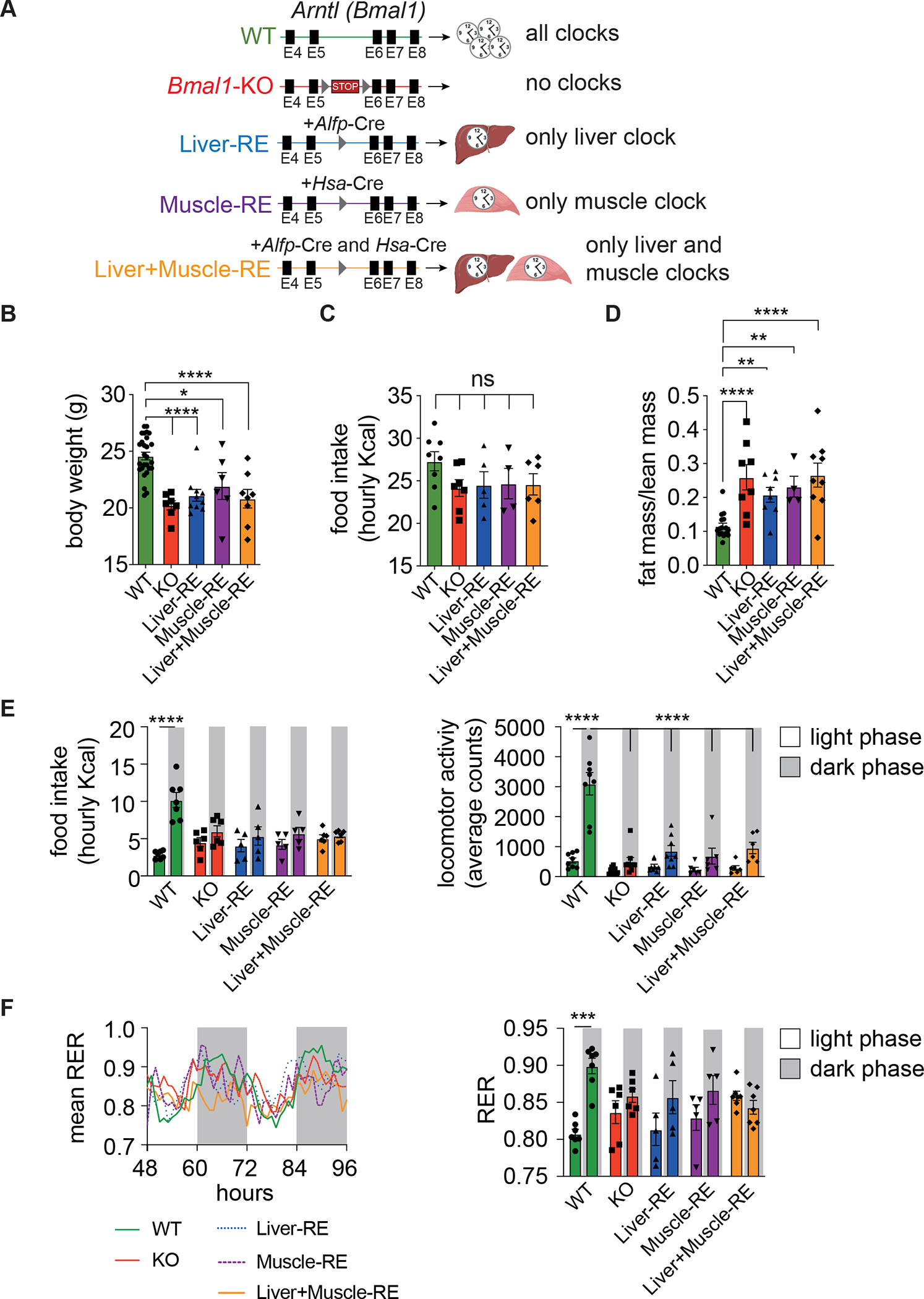
(A) Genetic scheme for tissue-specific clock reconstitution.
(B) Body weight, 8-week-old, n = 6–28 (WT-Alfp-Cre = 13; WT-Hsa-Cre = 4; WT-Alfp+Hsa-Cre = 10). One-way ANOVA, Fisher’s LSD post-hoc test, *p < 0.05, ****p < 0.0001.
(C) Total daily food intake in average Kcals per h, n = 4–8 (WT-Alfp-Cre = 2; WT-Hsa-Cre = 1; WT-Alfp+Hsa-Cre = 5). One-way ANOVA, Fisher’s LSD post-hoc test, ns – not significant.
(D) Fat-to-lean mass ratio. n = 4–16 (WT-Alfp-Cre = 4; WT-Hsa-Cre = 1; WT-Alfp+Hsa-Cre = 11). One-way ANOVA, Fisher’s LSD post-hoc test, **p < 0.01, ****p < 0.0001.
(E) Left, light vs dark phase food intake, n = 5–7 (WT-Alfp-Cre = 2; WT-Hsa-Cre = 1; WT-Alfp+Hsa-Cre = 4). Right, light vs dark phase locomotor activity, n = 6–9 (WT-Alfp+Hsa-Cre = 8). Two-way ANOVA, Bonferroni post-hoc test, ****p < 0.0001.
(F) Left, mean respiratory exchange ratio (RER), n = 5–7 (WT-Alfp-Cre = 2; WT-Hsa-Cre = 1; WT-Alfp+Hsa-Cre = 4). Right, average light vs dark phase RER. Two-way ANOVA, Bonferroni post-hoc test, ***p < 0.001.
Data are presented as mean ± SEM. See also Figure S1. Figure 1A created using Biorender.com.
Liver and muscle core clocks oscillate in an independent manner
To assess muscle clock autonomy, and probe potential muscle-liver connections, we performed RNA-sequencing (RNA-seq) on livers and gastrocnemius muscles (predominantly fast-twitch fibers19) harvested every 4 h over the diurnal cycle. For comparisons with Liver-RE, we used data from our previous study featuring mice of identical age, sex, diet, and conditions13. We used JTK_CYCLE20 to calculate peak phases and amplitudes for core clock genes and the differential rhythmicity algorithm dryR21 to compare genotypes (Tables S1 and S2). Since both single and double Cre-positive WT mice (i.e., Hsa-Cre, Alfp-Cre, or Hsa+Alfp-Cre) were similar according to dryR models, double Cre-positive WTs are shown in the figures.
In muscle, loss of Bmal1 disrupted clock gene oscillations as expected (Figures 2A, 2B, S2A and S2C). Even with highly efficient Cre-mediated recombination (Figure S1C), reconstitution of muscle Bmal1 led to only a partial rescue, with clear differences between Muscle-RE and WT. On average, amplitudes of clock genes in Muscle-RE were 56.4% lower (Figure S2A). Reflecting a lack of robustness, dryR assigned most clock genes to a model of genes oscillating with advanced phase and reduced amplitude in Muscle-RE or Liver+Muscle-RE mice and indicated that Cry1 did not oscillate (Figure 2A and S2C). In Liver-RE mice, nearly all clock genes were phase advanced with reduced amplitude (Figures 2C, 2D, S2B and S2D), as we have shown previously13; exceptions to this were Per1 and Bhlhe41.
Figure 2. Liver and muscle core clocks oscillate in an independent manner.
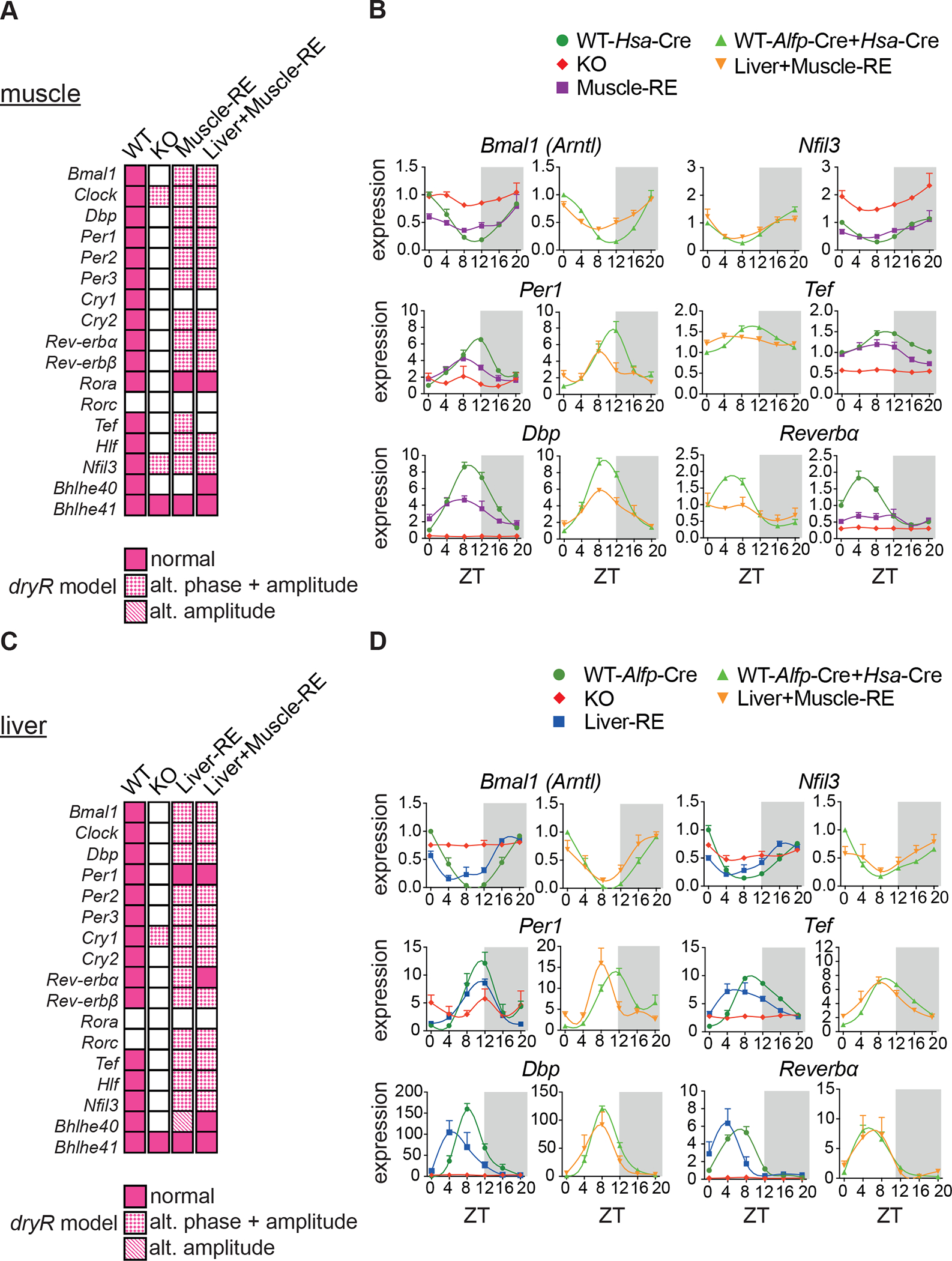
(A–D) RNA-seq of gastrocnemius muscle and liver harvested around the clock, 12 h light/12 h dark conditions, n = 3. ZT, zeitgeber time; ZT0, lights on; ZT12, lights off. No mature Bmal1 mRNA or protein was detected in KO mice. See also Figure S1.
(A, C) Oscillatory behavior of clock genes as captured by dryR.
(B, D) Relative expression of clock genes normalized to WT ZT0.
Data are presented as mean ± SEM. See also Figure S2 and Tables S1 and S2.
Next, we considered that liver and muscle molecular clocks may influence each other. The additional rescue of liver Bmal1 had little effect on muscle clock genes (Figure 2A and 2B). Like Muscle-RE mice, clock gene amplitudes remained lower in Liver+Muscle-RE mice as compared to WT mice (Figure S2A). In Liver+Muscle-RE mice, dryR models indicated that one clock gene, Bhlhe40, gained oscillation in muscle in the presence of the liver clock, whereas another gene, Tef, lost oscillation (Figure 2A–B). Inversely, dual clock rescue had little impact on liver clock genes. Nonetheless, dryR models indicated that the oscillation of Rev-Erbα and Bhlhe40 in liver was fully rescued in double but not in either single RE mouse (Figure 2C and 2D). Together, these data show that liver and muscle clocks are largely independent of one another.
Muscle Bmal1 is partially sufficient to drive the local diurnal transcriptome
Previous rescue experiments suggested that the muscle clock can have important effects on behavior22,23. However, whether endogenous muscle Bmal1 is sufficient to drive rhythmic functions within the muscle has not been defined. To explore this question, we assessed the state of clock-controlled output genes, focusing on genes that oscillated in both single and double Cre-positive WT mice. Considering the top 10 dryR models, which contained the greatest number of genes, we found sizable differences in the oscillatory nature liver and muscle. About three-times more genes oscillated in WT liver (3,750) as in WT muscle (1,150) (Figure 3A and 3B). The ability of each local clock to drive diurnal transcription was also different; reconstitution of Bmal1 induced oscillation of ~1,000 genes in liver but only of 250 genes in muscle (Figure 3A and 3B).
Figure 3. Local clocks in muscle and liver drive distinct autonomous outputs.
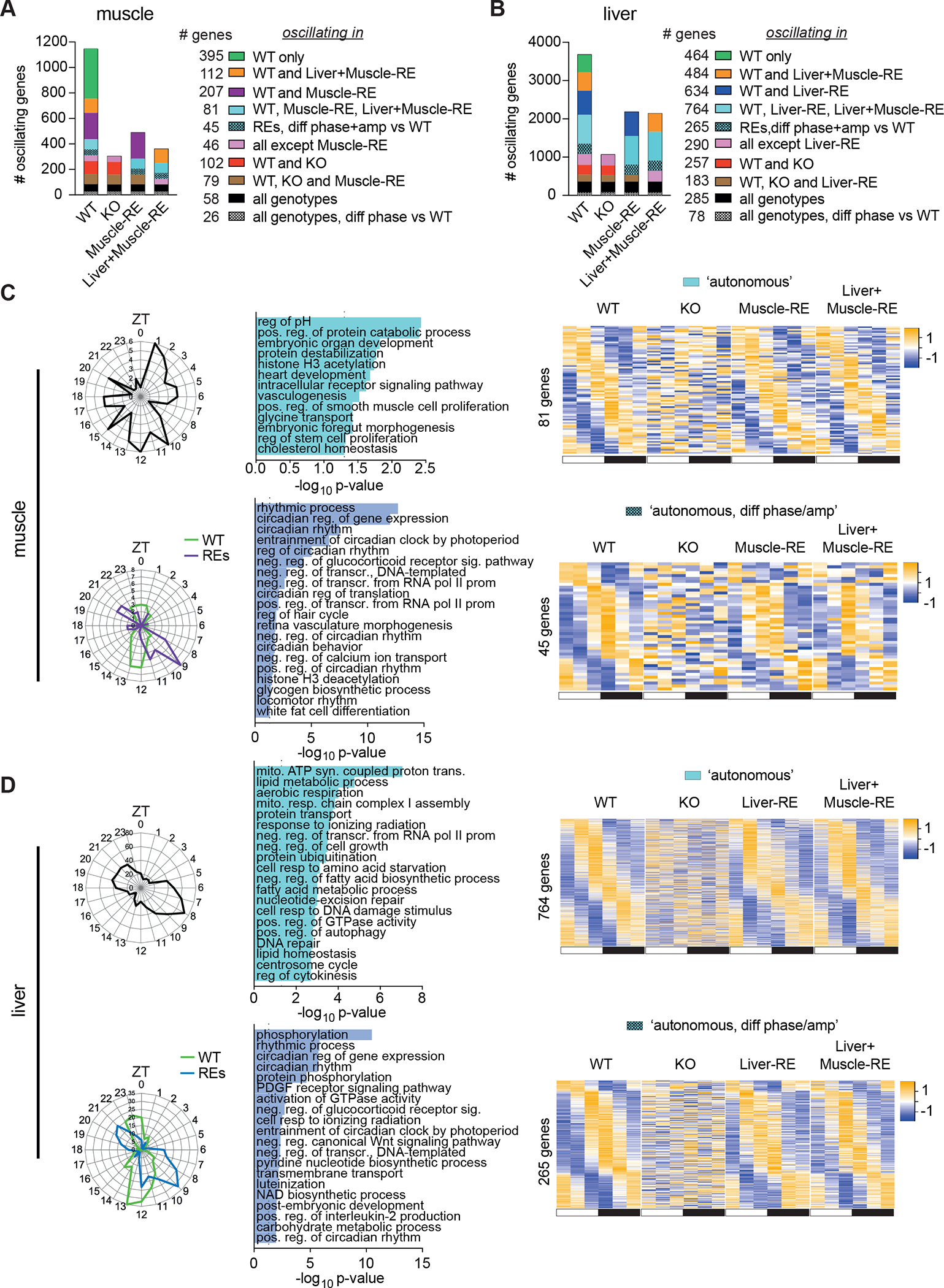
(A–D) RNA-seq of gastrocnemius muscle and liver harvested around the clock, 12 h light/12 h dark conditions, n = 3. ZT, zeitgeber time; ZT0, lights on; ZT12, lights off.
(A, B) Top 10 dryR models.
(C and D) Oscillating genes in muscle (C) and liver (D) by dryR. Left, polar histogram, peak phases. Middle, DAVID biological process enrichments, some pathway names abbreviated. Right, heatmaps of group averages sorted by WT peak phase. Top, autonomously oscillating genes. Bottom, autonomously oscillating genes with difference phase and amplitude in RE mice.
Next, we identified two dryR models representing autonomous genes. We defined autonomous as a loss of oscillation in KO mice with the reappearance of oscillation in single and double RE mice. One model contained 81 genes that oscillated identically in WT, Muscle-RE and Liver+Muscle-RE mice, peaking mainly around ZT1–5 and ZT10–12 (Figures 3A and 3C). These genes were involved in a range of biological processes, including development, protein catabolism, and intracellular signalling. Notably, the presence of muscle Bmal1 was sufficient to drive oscillation of Myod1, a principal regulator of the skeletal muscle lineage24 (Figure S3A). Another model, which included most core clock genes, featured 45 genes that oscillated in the three genotypes but with advanced phase and reduced amplitude in Muscle-RE and Liver+Muscle-RE mice (Figures 3A, 3C, and S3G). These genes were enriched for pathways involving transcription, glycogen metabolic process, phosphorylation, and Wnt signaling, among others.
Regulation of glucose metabolism is a central function of the muscle clock25–27. Here, we observed differential expression of many glucose-related genes. Although the oscillation of Slc2a4 (Glut4), the primary insulin-dependent glucose transporter, was rescued in muscle from Muscle-RE mice, average expression remained considerably lower than WT (Figure S3A). Other notable examples were Pfkfb3, encoding a glycolysis-promoting enzyme, and Pdp1, encoding a phosphatase that activates pyruvate dehydrogenase (PDH) phosphatase (Figure S3A).
In liver, genes that oscillated identically in WT, Liver-RE and Liver+Muscle-RE mice mostly peaked between ZT6 and ZT10, with an additional peak around ZT20, and were enriched for metabolic pathways including mitochondrial function, lipid metabolism, amino acid metabolism, and transcription (Figures 3B and 3D). Like muscle, liver contained an additional set of genes that oscillated in all three genotypes but had advanced phase and reduced amplitude in RE mice (Figures 3B and 3D). Consistent with our previous reports13,15, dryR analysis showed that these liver genes were functionally enriched for circadian rhythm, NAD+ biosynthesis, and carbohydrate metabolism (Figure 3D).
Manipulation of Bmal1 may also shape tissue phenotypes by altering the expression of non-rhythmic genes. We used dryR mean models, which test for differences in average daily expression, to assess the expression of non-rhythmic WT genes. In liver and muscle, the largest model contained genes with identical expression between WT, KO, and RE mice (Tables S3 and S4). The next largest model contained genes that were deregulated in KO and restored to WT levels in RE mice, representing 24.65% and 30.54% of non-rhythmic genes in muscle and liver, respectively (Figures S3B and S3D). The top enrichments of these genes were pathways involving immune function in liver and mitochondrial genes in muscle, and carbohydrate metabolic processes were also enriched in muscle (including Hk3, Ldhb, Pgk1, and Pgm1) (Figures S3C and S3E). The next largest model represented genes deregulated in KO and RE as compared to WT mice (i.e., not rescued). The smallest model contained genes that were differentially expressed between all three genotypes (Figures S3C and S3E). Overall, these analyses suggests that changes in the expression of non-rhythmic genes may also contribute to the state of metabolism in KO and RE mice.
Liver and muscle diurnal transcriptomes are influenced by Bmal1 function in distal tissues
Even though liver and muscle core clocks are largely independent, we considered that they may still influence clock-controlled output genes across the two tissues. Generally, heightened rhythmicity was not observed with dual versus single reconstitution, as evidenced by a similar number of oscillating genes in single and double RE mice (Figures 3A and 3B). Fewer genes oscillated in muscles of Liver+Muscle-RE as compared to Muscle-RE mice, showing a minor inhibitory effect of liver Bmal1 on oscillation of the muscle. However, sets of genes that oscillated exclusively in WT and Liver+Muscle-RE mice were among the top dryR models in liver (484 genes) and muscle (112 genes) (Figure 3A and 3B), indicating a degree of cross-tissue regulation. In muscle, genes rescued by dual reconstitution were confined to the dark phase and were related to intracellular signal transduction, phosphorylation, proliferation, and development (Figure 4A). In liver, the genes had more widely distributed phases and pertained to pathways involving ribosomes and NF kappa b activity (Figure 4B).
Figure 4. Liver and muscle diurnal transcriptomes are influenced by Bmal1 function in distal tissues.
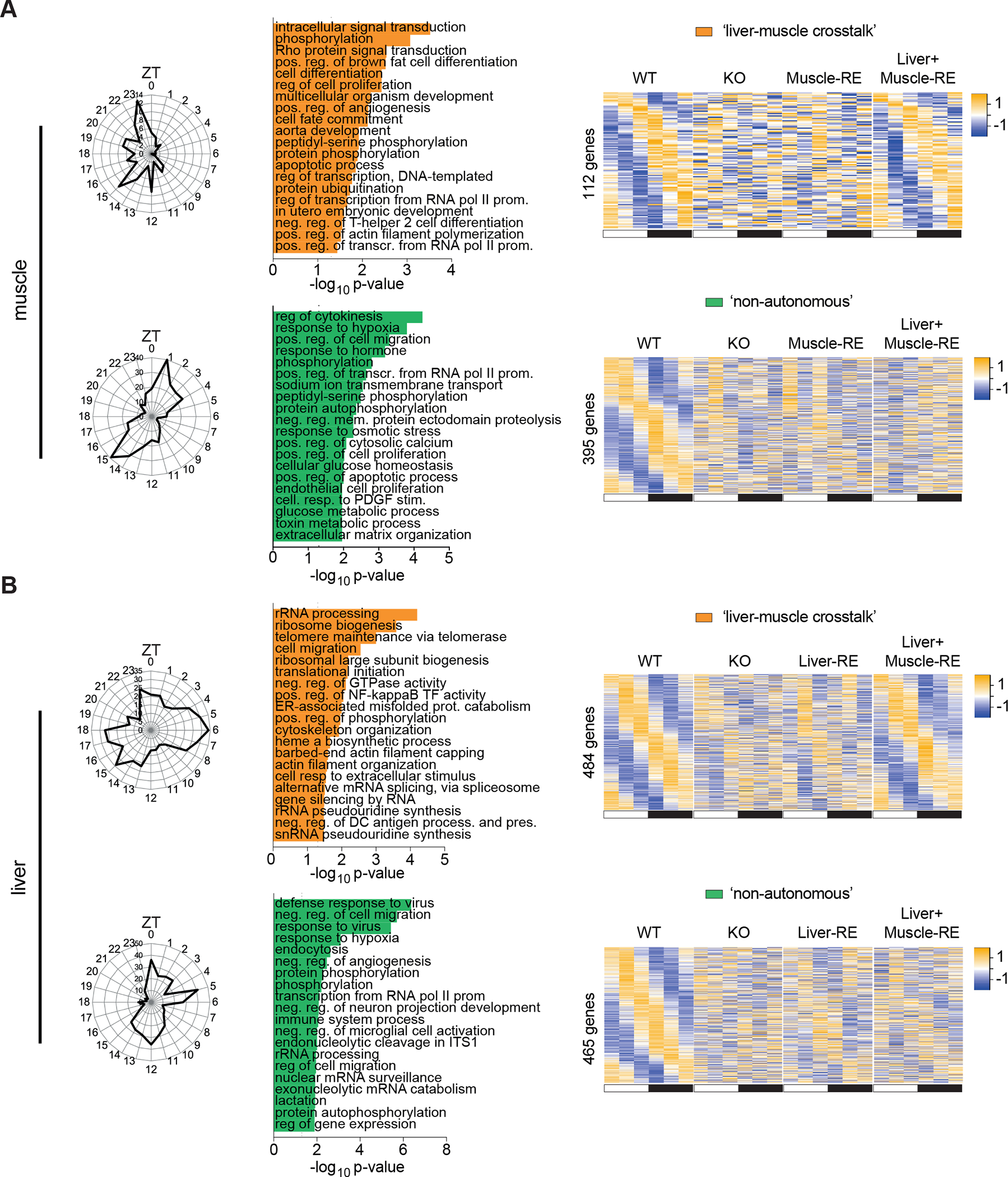
(A and B) dryR analysis of RNA-seq of gastrocnemius muscle (A) and liver (B) harvested around the clock, 12 h light: 12 h dark conditions, n = 3. ZT, zeitgeber time; ZT0, lights on; ZT12, lights off. Left, polar histogram, peak phases. Middle, DAVID biological process enrichments, some pathway names abbreviated. Right, heatmaps of group averages sorted by WT peak phase. Top, genes oscillating exclusively in WT and Liver+Muscle-RE. Bottom, genes oscillating exclusively in WT.
To complement dryR analysis, we performed two-way ANOVA genome-wide using Nitecap28. Mirroring dryR analysis, we observed a similar number of significant changes in single and double RE tissues as compared to WT (Figure S3F). This analysis also indicated a greater similarity between Muscle-RE and WT mice than Liver+Muscle-RE and WT mice.
We next interrogated the dryR model containing genes that oscillated exclusively in WT mice, which we referred to as genes that oscillate non-autonomously. These genes require a function of Bmal1 in additional tissue(s), for example the timed-release of a soluble signalling molecule. Interestingly, in muscle, response to hypoxia, response to hormone, and cellular glucose homeostasis were enriched pathways of genes oscillating non-autonomously (Figure 4A). Notable genes included Tbc1d1, which aids in GLUT4 trafficking to the plasma membrane29,30, and Hk2, which phosphorylates glucose in the first step of glycolysis (Figure S3A). Genes oscillating non-autonomously in liver were also enriched in response to hypoxia and pathways related to protein phosphorylation (Figure 4B), which is linked with extrinsic signalling events.
Additional dryR models contained a subset of genes that was rescued Liver-RE or Muscle-RE mice but then lost in double RE mice (Figures 3A, 3B, S4A and S4B). While the biological significance of this subset of genes is unclear, many functional enrichments in liver and muscle were distinct from other models (Figure S4A and S4B). dryR also constructed models with genes that oscillated irrespective of the presence of Bmal1 (Figures 3A, 3B, S4C, and S4D). Together, these data show that liver and muscle clocks influence clock-controlled output genes across the two tissues in specific ways.
Liver and muscle clocks are partially sufficient for control of glucose metabolism at the tissue but not systemic level
The metabolism of glucose within muscle and liver helps achieve whole-body glucose homeostasis31–33. Muscle is the primary site of postprandial glucose disposal, and the liver is the main producer of glucose34–36. As previous studies have shown that BMAL1 is necessary for maximal muscle glucose uptake25,27, we asked whether it is sufficient. We performed isotope tracing with U-13C-glucose to follow the metabolism of glucose into tissues (Figure 5A).
Figure 5. Liver and muscle clocks are partially sufficient for control of glucose metabolism at the tissue but not systemic level.
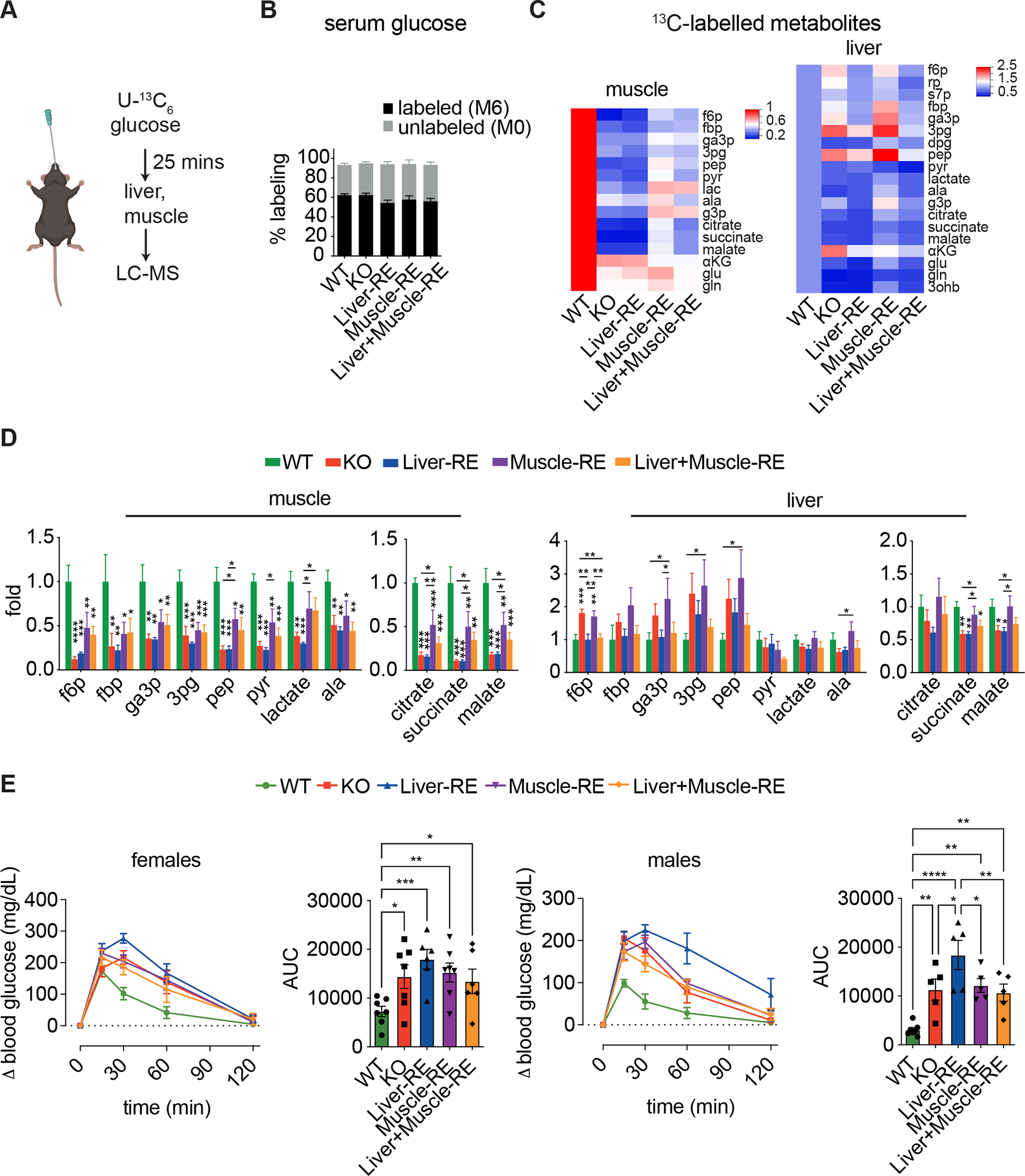
(A) Experimental design of isotope tracing, n = 5–6 (WT-Alfp-Cre = 2; WT-Hsa-Cre = 2; WT-Alfp+Hsa-Cre = 2). Data are 13C-labeled ion counts relative to WT.
(B) Labeling of circulating glucose.
(C) Heatmaps of downstream 13C metabolites. 3ohb, 3-hydroxybutyrate; 3pg, 3-phosphoglycerate; aKG, alpha-ketoglutarate; ala, alanine; dpg, 2,3 diphosphoglycerate; f6p, fructose 6-phosphate; fbp, fructose 1,6-bisphosphate; g3p, glycerol 3-phosphate; ga3p, glyceraldehyde 3-phosphate; glu, glutamate; gln, glutamine; lac, lactate; pep, phosphoenolpyruvate; pyr, pyruvate; rp, ribose phosphate; s7p, sedoheptulose 7-phosphate.
(D) Labelling of glycolysis and TCA cycle metabolites. One-way ANOVA, Fisher’s LSD post-hoc test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
(E) Oral glucose tolerance tests in female (left, n=6–7) and male (right, n=5–7) mice. WTs = WT-Alfp+Hsa-Cre. Bar graph area under the curve (AUC) – one-way ANOVA, Fisher’s LSD post-hoc test, *p < 0.05, **p < 0.01, ***p < 0.001, ns = not significant.
Data are presented as mean ± SEM. See also Figure S5. Figure 5A created using Biorender.com.
Gavage of 1.5 g/kg, a typical dose of a glucose tolerance test, sufficiently labeled serum glucose (Figure 5B). Compared to WT muscles, KO muscles showed substantially lower labeling of glycolytic and TCA cycle intermediates, indicating impaired glucose uptake and oxidation (Figures 5C and 5D). The presence of muscle BMAL1 in Muscle-RE mice restored labeling of these metabolites, but labelling was still considerably lower than in WT, highlighting the requirement of other tissue clocks for muscle glucose metabolism. Liver clock reconstitution did not alter muscle glucose metabolism, as Muscle-RE and Liver+Muscle-RE labeling patterns were similar (Figures 5C and 5D). While the liver does take up glucose during feeding to support anabolic pathways (e.g., glycogen and lipid synthesis), it also takes up muscle-derived metabolites of glucose (e.g., lactate and alanine) for oxidation or recycling37,38. In contrast to muscle, liver from KO mice displayed increased labeling of glycolytic intermediates and a modestly reduced labeling of TCA cycle intermediates (Figures 5C and 5D). In Liver-RE mice, we observed normalized labeling of glycolytic but not TCA cycle intermediates, suggesting disparate regulatory mechanisms (Figure 5D). Taken together, these data show partial rescue of tissue glucose metabolism by autonomous clocks in muscle and liver, with no evidence of improved metabolism with both clocks in tandem.
By balancing the consumption and production of glucose during periods of feeding and fasting, liver and muscle help maintain blood glucose within a physiological range32,34. To determine whether any RE mice had better glycemic control than KO mice, we performed oral glucose tolerance tests. Remarkably, glucose tolerance was similarly impaired in all genotypes as compared to WT, in both male and female mice (Figures 5E, S5A, and S5B). Our results demonstrate that despite partially rescuing tissue glucose metabolism, BMAL1 function in liver and muscle is insufficient to improve the glucose intolerance of KO mice.
Feeding rhythms and autonomous clocks bolster time-dependent gene expression in liver and muscle
Next, we considered which additional diurnal signals might invigorate liver and muscle clocks and enable improvement of glucose homeostasis at the systemic level. WT mice, but not KO and RE mice, exhibit rhythmic feeding-fasting behavior (Figure 1E), which impacts local clocks and transcript oscillations in peripheral organs13,14,39–43. We and others have demonstrated that feeding-fasting behavior bolsters the phase and output of the liver clock including glucose pathways13. To determine the effect of feeding rhythms on the muscle-autonomous clock, we performed experiments under night feeding (NF) conditions, giving mice access to food only during their active phase (the 12 h dark period; ZT12-ZT24/0) for 2 weeks. As expected, NF induced rhythms of feeding and fasting, as well as RER, in KO and RE mice (Figures 6A, S6B, and S6C). Only small changes in body weight were observed under NF (WT = +2.14%; KO and RE = −3.48% to −6.81%) (Figure S6A).
Figure 6. Feeding rhythms and autonomous clocks bolster time-dependent gene expression in liver and muscle.
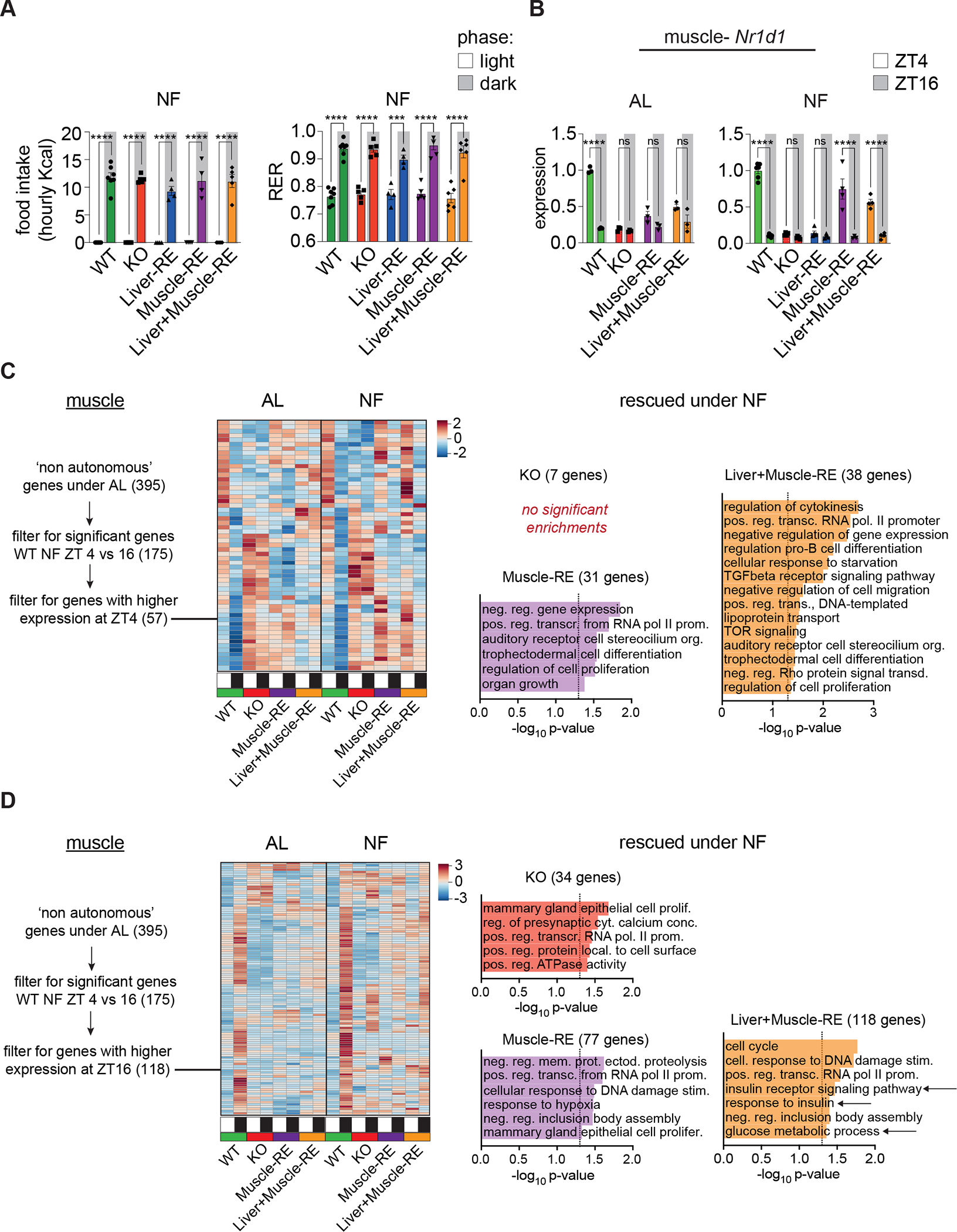
(A) Left, light vs dark phase food intake under night feeding (NF) conditions. n = 4–7 (WT-Alfp-Cre = 2; WT-Hsa-Cre = 1; WT-Alfp+Hsa-Cre = 4). Two-way ANOVA, Bonferroni’s post-hoc test, ****p < 0.0001. Right, average light and dark phase respiratory exchange ratio (RER), n = 5–7 (WT-Alfp-Cre = 2; WT-Hsa-Cre = 1; WT-Alfp+Hsa-Cre = 4). Two-way ANOVA, Bonferroni’s post-hoc test, ***p < 0.001.
(B) RNA-seq data from ad libitum (AL) and NF mice at ZT4 and ZT16, n = 3–6. For NF, WT-Alfp-Cre = 2; WT-Hsa-Cre = 2; WT-Alfp+Hsa-Cre = 2. Two-way ANOVA, Bonferroni’s post-hoc test, **p < 0.01, ****p < 0.0001. ns – not significant.
(C and D) Left, heatmaps of AL non-autonomously oscillating genes peaking at ZT4 (C) or ZT16 (D), DEseq2 FDR <0.05. Right, DAVID biological process enrichment for genes not significantly different vs WT at either time point (i.e., rescued by NF) (DE-seq2 FDR <0.05), some pathway names abbreviated. Arrows = glucose related pathways.
Data are presented as mean ± SEM. See also Figures S6–12.
We performed bulk RNA-seq on each tissue at ZT4 and ZT16 under NF. Using Liver-RE mice, we previously showed that NF corrects the phase and amplitude of core clock oscillations13; thus, we now focused on muscle. Assessing day-night differences, we found that NF made the expression profile of Rev-Erbɑ more similar to WT in both Muscle-RE and Liver+Muscle-RE mice (Figures 6B and S6D). While we cannot infer rhythmicity from two timepoint data, several other clock genes showed significant day-night changes under NF (Figures S6D) and these changes likely reflect enhancement of oscillatory phase and/or amplitude. Notably, for liver and muscle, we observed no additional benefit of dual reconstitution on clock gene expression (Figures S6D). These data indicate that like in liver, imposed feeding-fasting rhythms bolster clock gene expression in muscle.
Next, we assessed the genome-wide effect of NF. In liver, Liver-RE and Liver+Muscle-RE mice were most similar to WT mice (Figures S7A and S7B). In muscle, Liver+Muscle-RE showed the least number of changes versus WT at ZT4 and exhibited significant changes versus Muscle-RE mice (Figures S7A and S8A). Changes included genes involved in lipid and glucose metabolism and are supportive of a limited yet significant impact of the liver clock on the muscle at this time point (S8B). We also considered that reconstitution of Bmal1 in liver could alter gene expression in muscles (e.g., livers of Muscle-RE mice), and vice versa. However, we observed that without local Bmal1 also present, the cross-tissue impact was negligible (Figure S7A and S7B).
We then asked whether feeding-fasting rhythms rescued the temporal expression of genes that oscillated non-autonomously in WT mice under AL conditions. From 57 genes that exhibited higher expression at ZT4 vs ZT16 in WT NF mice, only 3 were significantly different in KO, and there was no further benefit of muscle clock reconstitution (Figure 6C and 6D). 118 genes exhibited lower expression at ZT4 vs ZT16 in WT NF mice. Of these, only 18 were significantly different in KO. Again, we observed no further benefit of muscle clock reconstitution on this set of genes (Figure 6C and 6D). Next, we functionally annotated genes that showed no significant difference vs WT (i.e., rescued genes) at ZT4 and ZT16. Although we observed relatively weak enrichments, many functions were identified, including those relevant to the metabolic phenotypes at hand. Notably, at ZT16, glucose metabolic process and two terms related to insulin signalling were identified as rescued specifically in Liver+Muscle-RE muscle (Figures 6C and 6D), suggesting an interaction of the two clocks on this process. Furthermore, identification of these pathways did not depend on the use of a specific algorithm; calling AL non-autonomously oscillating genes with JTK_CYCLE also led to identification of similar gene classes as rescued in Liver+Muscle-RE muscles (Figures S9). Further inspection of glucose genes revealed that NF also led to small improvements in the expression of Glut4 and Pfkm when the muscle clock was intact (Figure S10). These data show that NF improves the temporal profiles of gene expression in muscle, an effect that depends on Bmal1 in the muscle, and involves certain genes involved in glucose metabolism.
Analyses of liver agreed with our previous report13. Considering genome-wide day-night differences within genotypes, we observed 2- to 3-times the number of changes in Liver-RE and Liver+Muscle-RE, respectively, compared to KO liver (Figure S11). Like in muscle, there was still a considerably sized subset of genes whose expression was significantly different than in WT liver, indicating that there are other inputs besides feeding that maximize temporal gene expression. This was particularly evident at ZT4. Genes rescued by NF in Liver-RE and Liver+Muscle-RE at ZT4 were modestly enriched for actin cytoskeleton, immune function, and MAPK signaling, among others (Figure S11). Of note, some genes involved in mitochondrial translation were rescued in KO and Liver-RE mice at ZT16, showing that feeding-fasting cycles are sufficient to modulate this pathway. In addition, genes involved in autophagy were uniquely rescued in Liver+Muscle-RE mice, suggesting that an interaction of the two clocks is important for this function. Finally, using JTK_CYCLE to define AL non-autonomously oscillating genes, we found that genes involved in glucose metabolism were rescued by NF at ZT16 in Liver-RE and Liver+Muscle-RE mice (Figures S12).
Overall, these data show that feeding-fasting rhythms enhance the temporal expression of clock genes and sets of clock-controlled output genes in liver and muscle. Similar to AL conditions, there was no major benefit of dual clock reconstitution under NF. However, rescue of certain genes involved in glucose metabolism was observed in Liver+Muscle-RE mice, indicating a potential role for the interaction between feeding rhythms and liver and muscle clocks in driving this process from a transcriptional standpoint.
Synergy between feeding rhythms and liver and muscle clocks enables systemic glucose homeostasis
Next, we asked whether tissue clock function bolstered by feeding rhythms would improve serum glucose responses. Indeed, different glucose tolerances were uncovered in KO and RE mice under NF. The initial peak of glucose was blunted in Muscle-RE as compared to KO, suggesting that muscle Bmal1 alone was able to improve glucose clearance (Figures 7A and S13A). More strikingly, however, blood glucose curves of WT and Liver+Muscle-RE mice were indistinguishable; blood glucose peaked 30 min post gavage in Liver-RE and Muscle-RE, yet it peaked just 15 min post gavage in Liver+Muscle-RE mice (Figure 7A), indicating a more rapid rate of glucose clearance in the presence of both liver and muscle clocks. Comparison of areas under the curve revealed improved glucose tolerance in Liver+Muscle-RE mice as compared to KO mice (Figure 7A). These results demonstrate a functional connection between liver and muscle clocks for systemic glucose metabolism in the presence of a feeding-fasting rhythm.
Figure 7. Synergy between feeding rhythms and liver and muscle clocks enables systemic glucose homeostasis.
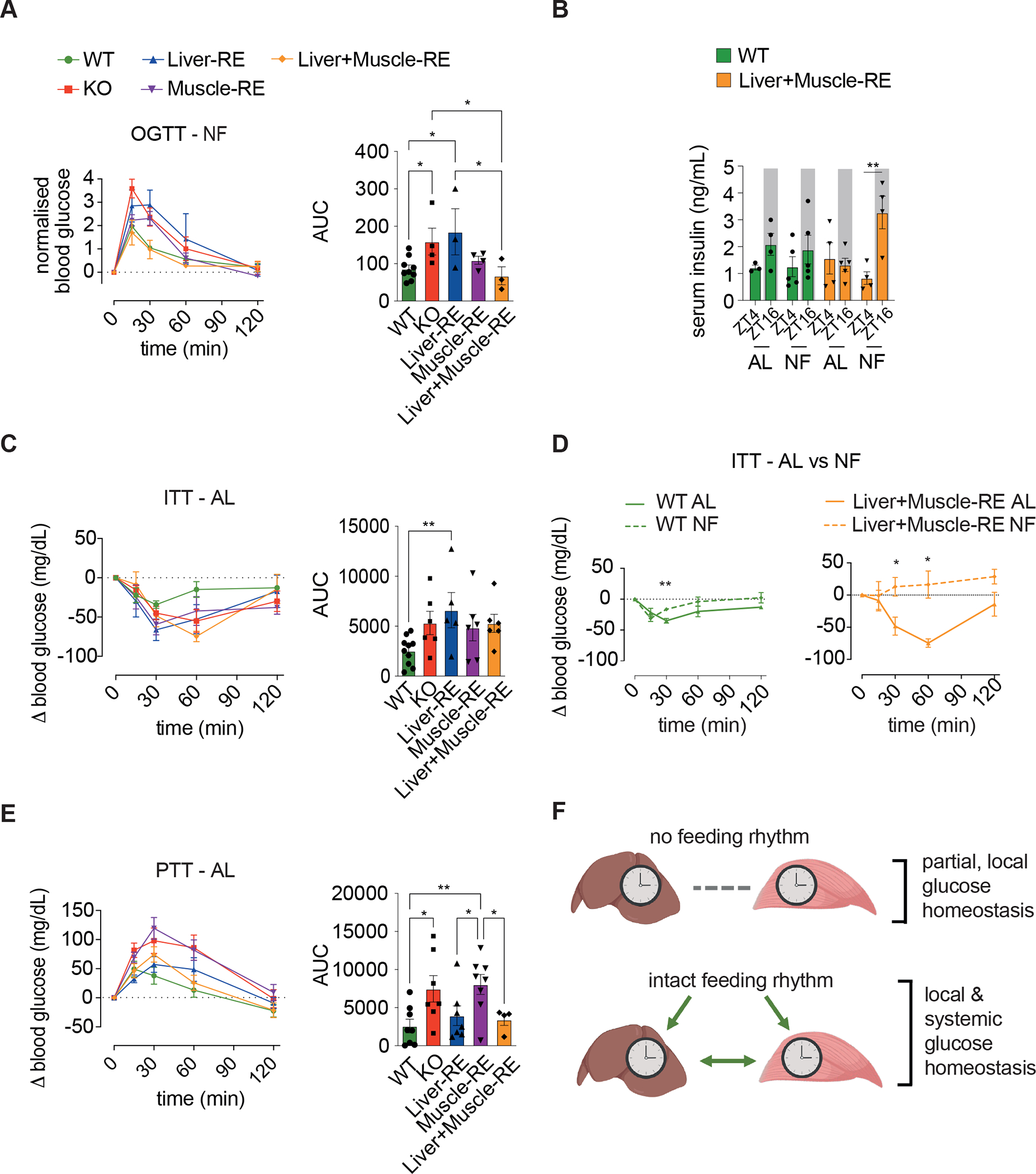
(A) Oral glucose tolerance test (OGTT) under night feeding (NF), n = 3–9 (WT-Alfp-Cre = 1; WT-Hsa-Cre = 3; WT-Alfp+Hsa-Cre = 4). Bar graph area under the curve (AUC) – one-way ANOVA, Fisher’s LSD post-hoc test, *p < 0.05, **p < 0.01, ***p < 0.001.
(B) Serum insulin levels under ad libitum (AL) or NF conditions, n = 4–5 (WTs = WT-Alfp+Hsa-Cre). Two-way ANOVA, Bonferroni post-hoc test, **p < 0.01.
(C) Insulin tolerance test (ITT) under AL conditions, n = 5–10 (WT-Alfp+Hsa-Cre = 7; WT-Hsa-Cre = 3). Bar graph AUC negative peaks – one-way ANOVA, Fisher’s LSD post-hoc test, **p < 0.01.
(D) Insulin tolerance test as in (C) vs NF conditions, NF n = 3–6 (WT-Alfp-Cre = 1; WT-Hsa-Cre = 3; WT-Alfp+Hsa-Cre = 2). Two-way repeated measures ANOVA, Fisher’s LSD post-hoc test. *p < 0.05, **p < 0.01.
(E) Pyruvate tolerance test (PTT) under AL conditions, n = 4–8 (WT = WT-Alfp+Hsa-Cre). Bar graph area under the curve – one-way ANOVA, Fisher’s LSD post-hoc test, *p < 0.05, **p < 0.01.
(F) Model of central findings.
Data are presented as mean ± SEM. See also Figure S13. Figure 7F created using Biorender.com.
During feeding, most glucose is cleared via insulin-induced uptake into tissues35,44–46. NF induced a rhythm of serum insulin in Liver+Muscle-RE mice, evidenced by a peak during feeding at ZT16 and a trough during fasting at ZT4 (Figure 7B). We have previously demonstrated serum insulin rhythms under NF in KO and Liver-RE mice13. Serum insulin is noteworthy because it can act as a synchronizing cue for peripheral oscillators, inhibit glucose production in liver, and drive glucose uptake into muscle27,29,47–50. Bmal1-null mice have a defect in glucose-stimulated insulin secretion and are hypersensitive to insulin26,51–53. Here, from insulin tolerance tests, we observed that RE mice are similarly hypersensitive to insulin under AL, although not all genotypes were statistically significant (Figures 7C and S13B). Interestingly, NF reduced insulin sensitivity similarly in KO and RE mice (Figures 7D, S13C, and S13F). Thus, we concluded that while insulin levels and action contributed to glucose tolerances under NF, the two factors did not explain why glucose tolerance was rescued specifically in Liver+Muscle-RE mice.
Because muscle is the main site of post-prandial glucose clearance35,45,46, improved glucose tolerance in Liver+Muscle-RE mice likely reflects faster and/or more efficient muscle oxidation of glucose. Moreover, since improved glucose tolerance was maximal in the presence of both liver and muscle Bmal1, liver Bmal1 must aid in glucose clearance. Glycolytic activity in muscle converts glucose to lactate and alanine, which are subsequently released, taken up by the liver, and recycled to glucose (i.e., the Cori and Cahil cycles) 37,38. Recent studies show that these cycles continue during feeding and that there are additional fates of lactate in the liver. As a preferred TCA cycle substrate, lactate can be fully oxidized or generate TCA cycle-derived amino acids. Lactate can also contribute to glycogen storage34,35,44. Consistent with reports that muscle-specific Bmal1 KO mice have normal glycogen content over the diurnal cycle54, we observed relatively small changes in the expression of glycogen synthase 1 (Gys1) and glycogen phosphorylase (Pygm) in KO or Muscle-RE mice vs WT (Figure S13E). We have shown previously that rhythms of glycogen synthesis and breakdown are restored to a significant degree in Liver-RE mice15 (Figure S13E). However, the contribution of glycogen synthesis to glucose clearance is estimated at ~6% and thus is not likely to explain our results44. Instead, oxidation of carbohydrate is responsible for ~90% of post-prandial glucose clearance44,45.
To assess these mechanisms, we leveraged pyruvate tolerance tests. Classically, the pyruvate tolerance test has been used as a measure of liver gluconeogenesis, yet current studies show that it also reflects liver lactate oxidation, as injected pyruvate is rapidly reduced to lactate in the blood55,56. We observed a larger and more sustained increase in blood glucose in response to injected pyruvate/lactate in KO mice, whereas the curves of Liver-RE and Liver+Muscle-RE mice were similar to WT (Figures 7E and S13D). These data suggest that liver Bmal1 is sufficient to limit gluconeogenic flux and/or promote oxidation of pyruvate/lactate. Taken together, our results suggest that improved glucose tolerance in Liver+Muscle-RE mice under NF feeding relies on the coordinated actions of muscle BMAL1 to metabolize incoming glucose, aided by higher insulin levels, and of liver BMAL1 to promote the clearance of muscle-derived metabolites of glucose (Figure 7F).
DISCUSSION
Interconnected molecular clocks in cells of the brain and body regulate metabolism. Here, we tested the interactions between clocks in liver and muscle, revealing that rescuing clock function in liver, muscle, or both tissues in combination, was not sufficient for glucose tolerance, a phenotype used to assess impairment of glucose homeostasis in rodents and humans. This insufficiency was observed despite well-established roles for liver and muscle clocks in the regulation of glucose pathways25,27,32,43,57,58. We reconciled this discrepancy by showing that a daily feeding-fasting rhythm is a key input to liver and muscle clocks that enables synergy between the two tissues for control of glucose metabolism at the systemic level.
Analysis of transcriptomes and metabolism revealed that the observed synergy among the three circadian system components was multi-faceted. Genes involved in glycolysis, gluconeogenesis, glucose oxidation, and related pathways, are driven daily by clock transcription factors, including Bmal115,27,31,32. This basal transcriptional state, which results in a partial ability to metabolize glucose, is further shaped by the interaction between clock transcription factors and feeding-fasting rhythms12–14. We have previously shown that integration of feeding signals by the autonomous liver clock enhances glucose homeostasis in the liver13. Here, we show a similar response in muscle. Feeding rhythms also overcame the defect in glucose-stimulated insulin release observed for mice deficient for pancreatic clock function51,59–61, including both KO and RE mice, by boosting serum insulin. Insulin, in turn, drives glucose uptake into muscle, wherein local Bmal1 temporally primes the machinery to process it. Rescue of muscle Bmal1 alone modestly improved glucose clearance, yet simultaneous actions involving Bmal1-dependent transcription in liver were required for maximal clearance. Liver takes up muscle-derived metabolites of glucose, including lactate and alanine, thereby clearing glucose-derived carbons from the circulation34,35,37,38,62, conceivably freeing muscle to take up more glucose. The synchronised engagement of these events presents a plausible explanation for improved glucose tolerance. Additionally, reconstitution of Bmal1 in the suprachiasmatic nucleus restores a feeding rhythm and partially restores glucose tolerance63. Thus, the central clock in the brain is also a critical piece of the glucose homeostasis framework, even in the absence of intact clocks in liver and muscle. The synergy between liver and muscle clocks may be aided by additional mechanisms, such as soluble factors or extracellular vesicles released by muscle and liver64–72. These mechanisms remain to be investigated.
This study also sheds light on the contributions of different cell types and systemic signals to circadian physiology in skeletal muscle. Single nuclei sequencing experiments show that ~70% of muscle nuclei are myonuclei, which constitute the differentiated muscle fibers, while the additional ~30% is comprised of fibrogenic progenitors, tenocytes, stem cells, endothelial cells and immune cells73. Here, we used Hsa-Cre to reconstitute Bmal1 expression specifically in the myofiber lineage74. Interestingly, whole tissue analysis revealed weaker oscillation of clock genes in this model, indicating that either additional cell types and/or additional systemic signals contribute significantly to the tissue ensemble rhythm. It is unlikely that this is a technical artifact of the model since we observed high Cre-mediated recombination efficiency. In addition, our analysis showed that the temporal expression pattern of clock genes, such as Rev-Erbα, was restored under NF conditions, meaning that lack of this systemic signal, rather than Rev-Erbα expression in other cell types, was responsible for the lack of Rev-Erbα oscillation under AL conditions. Still, not all clock genes were rescued. Another systemic signal of importance to muscle is daily periods of locomotion or exercise. Exercise can entrain the muscle clock and impact its functional outputs 75–77. As KO and RE mice do not exhibit the robust locomotor activity of WT, restoring this additional behavior is likely important for further restoration of muscle rhythms in RE mice. Nevertheless, Bmal1 is expressed in other muscle-based cell types, for example muscle stem cells; therefore, non-myofiber lineage cell types will also contribute. A more thorough interrogation at the single-cell level is warranted.
Limitations of the study
Factors that define systemic glucose metabolism, such as insulin secretion and sensitivity, the release of peptides from the gut, and neuronal circuits that control energy expenditure, are temporally regulated by the clock system31,61,78,79. Accordingly, studies report that glucose tolerance displays circadian rhythmicity in rodents79–81. A limitation of this study is that tolerance tests were performed at a single diurnal time point. Therefore, results may not necessarily represent glucose responses at all diurnal time points. Because fasting is required for glucose tolerance tests (to ensure the clearance of acute food intake from the gut), we sought to align this experimental fast with the natural fasting period of the mouse (i.e., the light phase). With this set-up, we avoided subjecting mice to starvation-like states associated with long fasts, and results were more reflective of physiological mechanisms82. In addition, the metabolic activities of liver and muscle are relevant to both light and dark phase glucose control. For example, shuttling of glucose-derived metabolites from muscle to liver (including recycling of lactate back to glucose) occurs in both the fed and fasted states34,35,44. Therefore, it is relevant to interrogate liver-muscle cross talk in the light phase.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kevin Koronowski (Koronowski@uthscsa.edu)
Materials availability
Mouse lines generated in this study are available from the lead contact with a completed materials transfer agreement.
Data and code availability
RNA-sequencing data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-BMAL1 | Abcam | ab93806; RRID: AB_10675117 |
| Anti-p84 | Genetex | GTX70220; RRID: AB_372637 |
| Anti-RFP | Rockland | 600–401-379 |
| Goat Anti-Rabbit IgG H&L (Alexa Fluor® 647) | Abcam | ab150079 |
| Anti-Tubulin | Sigma | T6199 |
| Chemicals, peptides, and recombinant proteins | ||
| U-13C6 glucose | Cambridge Isotope Laboratories | CLM-1396-PK |
| Critical commercial assays | ||
| Ultra Sensitive Mouse Insulin ELISA Kit | Crystal Chem | 90080 |
| Deposited data | ||
| RNA sequencing – WT and double RE liver and muscle | This paper | GSE197455 |
| RNA sequencing – WT, Bmal1 KO, and Muscle-RE muscle | This paper | GSE197726 |
| RNA sequencing – Night feeding dataset, all genotypes, liver and muscle | This paper | GSE228389 |
| Experimental models: Organisms/strains | ||
| Mouse: Wild Type (WT) – 1.Bmal1wt/wt, Alfp-cretg/0; 2. Bmal1wt/wt, Hsa-cretg/0; 3. Bmal1wt/wt, Alfp-cretg/0, Hsa- cretg/0 | S.A. Benitah lab, P.S. Welz, This study | |
| Mouse: Bmal1 knockout (KO) – Bmal1stopFL/stopFL | S.A. Benitah lab, P.S. Welz, This study | |
| Mouse: Bmal1 hepatocyte-reconstituted (Liver- RE) – Bmal1stopFL/stopFL; Alfp-cretg/0 | S.A. Benitah lab, P.S. Welz, This study | |
| Mouse: Bmal1 skeletal muscle-reconstituted (Muscle- RE) – Bmal1stopFL/stopFL; Hsa-cretg/0 | S.A. Benitah lab, P.S. Welz, This study | |
| Mouse: Bmal1 hepatocyte + skeletal muscle- reconstituted (Liver+Muscle-RE) – Bmal1stopFL/stopFL; Alfp- cretg/0; Hsa-cretg/0 | S.A. Benitah lab, P.S. Welz, This study | |
| Oligonucleotides | ||
| BMAL1 For - GCAGTGCCACTGACTACCAAGA | This study | |
| BMAL1Rev - TCCTGGACATTGCATTGCAT | This study | |
| Rn18S For - CGCCGCTAGAGGTGAAATTC | This study | |
| Rn18S Rev - CGAACCTCCGACTTTCGTTCT | This study | |
| Software and algorithms | ||
| dryR | Weger et al., 2021, PNAS | |
| JTK_CYCLE | Hughes et al., 2010, J. Biol. Rhythms. | |
| DAVID Bioinformatics Resource | Huang, Sherman and Lempicki, 2009, Nat. Protoc. | |
| Nitecap | Brooks et al., 2022, J. Biol. Rhythms. | |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Animals
Mice were bred and housed at the University of California Irvine vivarium, in accordance with the guidelines of the local Institutional Animal Care and Use Committee (IACUC). Animal experiments conformed to institutional and national regulatory standards and were performed considering ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines. Bmal1-stopFL mice were generated on the C57BL/6J background as previously described15,17 (see also Figure 1). Liver reconstituted (Liver-RE; hepatocyte-specific reconstitution of Bmal1) mice were generated by crossing Bmal1-stopFL with Alfp-Cre mice as previously described15. Skeletal muscle reconstituted (Muscle-RE; myocyte-specific reconstitution of Bmal1) mice were generated by crossing Bmal1-stopFL with mice expressing cre recombinase driven by the human alpha-skeletal actin (Hsa; Acta1) promotor (Hsa-Cre). Crosses of Bmal1-stopFL with both Cre lines generated Liver+Muscle reconstituted (Liver+Muscle-RE) mice. Experimental genotypes were: wild type (WT) – 1. Bmal1wt/wt, Alfp-cretg/0; 2. Bmal1wt/wt, Hsa-cretg/0; 3. Bmal1wt/wt, Alfp-cretg/0, Hsa-cretg/0. Bmal1 knockout (KO) – Bmal1stopFL/sto-FL; Bmal1 hepatocyte-reconstituted (Liver-RE) – Bmal1stopFL/stopFL; Alfp-cretg/0. Bmal1 skeletal muscle-reconstituted (Muscle-RE) – Bmal1stopFL/stopFL; Hsa-cretg/0. Bmal1 hepatocyte + skeletal muscle-reconstituted (Liver+Muscle-RE) – Bmal1stopFL/stopFL; Alfp-cretg/0; Hsa-cretg/0. The specific genotypes of WT controls for each experiment are listed in the figure legends. All experiments utilized 8- to 12-week-old mice entrained to a 12 h light/12 h dark cycle and fed ad libitum or placed under night feeding (NF) wherein mice were given ad libitum access to food during the dark period from zeitgeber time (ZT)12 to ZT24/0. For feasibility purposes, males were used for transcriptomic analyses and females for functional experiments. Key functional experiments were confirmed in both male and female mice.
METHOD DETAILS
Western blot
Frozen tissue was homogenized in lysis buffer (RIPA - 50 mM Tris-HCl [pH 8], 150 mM NaCl, 5 mM EDTA, 15 mM MgCl2 and 1% NP-40) supplemented with Protease Inhibitor Cocktail (Roche, Basel, Switzerland), 500 μM PMSF (serine protease inhibitor), 10 mM nicotinamide (Sirtuin inhibitor) and 330 nM TSA (Class I and II HDAC inhibitor). Samples were lysed on ice with periodic mixing for 30 min. Next, samples were sonicated (5 sec on, 5 sec off, 4 cycles, 60% amp.) and centrifuged at 13,200 rpm at 4°C for 15 min and the supernatant was collected. We used the Braford method to determine protein concentration, using Protein Assay Dye (BioRad). 20 μg protein from each sample was separated on 6% or 8% gels by SDS-PAGE and subsequently transferred to a nitrocellulose membrane. Membranes were then blocked with 5% milk in TBS-T (0.1% Tween-20, TBS) at room temperature for 2 h. Primary antibodies were diluted in 5% milk TBS-T and incubated with blots at 4°C overnight (BMAL1, Abcam – ab93806; ACTIN, Abcam – ab3280, P84, Genetex – GTX70220). Blots were incubated with HRP-conjugated secondary antibodies (Mouse IgG-HRP conjugate, EMD Millipore – AP160P; Rabbit IgG-HRP linked, EMD Millipore – 12–348) in 5% milk in TBS-T at room temperature for 1 h and visualized with Immobilon Western chemiluminescent HRP substrate (Millipore, Burlington, MA). Finally, blots were developed on HyBlot CL autoradiography film (Denville Scientific, Holliston, MA).
mCHERRY Immunostaining analysis
Freshly harvested gastrocnemius muscle was fixed for 120 minutes in 2% PFA, then washed 2 x with PBS, before incubation in 1:1 sucrose/OCT for 30 minutes, then frozen in OCT in tissue blocks using liquid-nitrogen cooled isobutane. 10 μm transverse gastrocnemius muscle sections were incubated in 5% w/v BSA 5% w/v normal goat serum 5.0% v/v Triton in PBS for 2 hours, washed 3 times with PBS then incubated with anti-RFP antibody (ROCKLAND, 600–401-379, 1:250, anti-RFP antibodies detect mCHERRY) in 5% w/v BSA 5% w/v normal goat serum 5.0% v/v Triton in PBS overnight at 4°C. Slides were washed 3 times in PBS then incubated in Alexa Fluor 647 anti- rabbit secondary antibody (Abcam ab150079) with DAPI 1/2000 in PBS for 2 hours at RT, washed 3 times in PBS then cover slips mounted using Fluoromount G. Images were acquired using Cell Observer (Zeiss) and analysed using FIJI. Fibers were traced manually using FIJI and each assigned as a ROI, integrated density was then calculated for each. A minimum of 100 ROI and 3 fields of view were used for each mouse.
Indirect calorimetry, food intake, and body composition
Metabolic parameters were measured using the Phenomaster metabolic cage system (TSE Systems Inc., Bad Homburg, Germany). The climate chamber was set to 21°C and 50% humidity with a 12h:12h light–dark cycle. Mice were maintained on standard chow (Teklad 2020x) ad libitum or time-restricted during the 12 hr dark phase as indicated, singly housed, and acclimated for 48h prior to data collection. VO2, VCO2, respiratory exchange ratio (RER), energy expenditure and food intake were monitored for 3 min per cage and measured every 39 min for four to six consecutive days. Body composition (lean mass and fat mass) was measured in non-anesthetized mice via MRI (QNMR EchoMRI100; Echo Medical Systems) prior to recordings. Food intake values were calculated by multiplying average grams eaten per hour by the kcal/g of the chow diet.
Locomotor activity
Mice were individually housed in home cages equipped with optical beam motion detection capability (Starr Life Sciences). Data were collected using VitalView 5.0 software and analyzed using Clocklab software (Actimetrics).
RNA-sequencing for ad libitum samples
Total RNA was extracted by Trizol for both liver and muscle samples, with muscle samples further purified using Direct-Zol columns (Zymo). RNA was monitored for quality control using the Agilent Bioanalyzer Nano RNA chip and Nanodrop absorbance ratios for 260/280nm and 260/230nm. Library construction was performed according to the Illumina TruSeq Total RNA stranded protocol. rRNA was depleted using ribo-zero rRNA gold removal kit (human/mouse/rat). rRNA-depleted RNA was chemically fragmented for three minutes. First strand synthesis was completed using random primers and reverse transcriptase to make cDNA. After second strand synthesis, cDNA was purified using AMPure XP beads, end-repaired and 3’ ends were adenylated. Illumina unique dual indexed adapters were ligated on the ends and the adapter-ligated fragments were enriched by nine cycles of PCR. The resulting libraries were validated by qPCR and sized by Agilent Bioanalyzer DNA high sensitivity chip. The concentrations for the libraries were normalized and then multiplexed together. The multiplexed libraries were sequenced using paired-end 100 cycles chemistry on the NovaSeq 6000.
RNA-Sequencing for NF samples
NGS experiments for NF samples were performed in the Genomics Unit of the CNIC. RNA extractions on liver samples were performed using RNeasy columns (Qiagen) and Direct-Zol columns (Zymo) were used for muscle samples. Quality control monitoring was performed as above for ad libitum samples, then single read sequencing was performed using the Illumina NextSeq 2000 System to achieve a minimum of 25 million reads per sample.
RNA-sequencing analysis for ad libitum samples
Sequencing results arrived in FastQ format. The pair-end reads from each replicate experiment were aligned to the reference genome assembly mm10 and the corresponding transcriptome using the Tuxedo protocol. Reads uniquely aligned to known exons, or splice junctions, extracted with no more than two mismatches were included in the transcriptome. Gene expression levels were directly computed from the read alignment results for each replicate. Standard FPKM (fragments per kilobase of exon per million mapped reads) values were extracted for each gene covered by the sequencing data and each replicate used in this study. Note, ad libitum RNA-sequencing data from livers of WT-Alfp-Cre, KO and Liver-RE is from our previous publication13 and was reanalysed by dryR21 alongside newly produced RNA-sequencing data of livers from WT-Alfp+Hsa-Cre and Liver+Muscle-RE mice. Fresh RNA extractions from tissue were performed on ZT4 and ZT16 liver samples of WT-Alfp-Cre, KO and Liver-RE night-fed mice collected for our previous study13, to minimise extraction and sequencing batch effects, and allow high confidence comparisons with WT-Alfp-Cre, KO, and Liver+Muscle-RE samples. Where shown, single RE or KO mice were normalized to the Alfp-Cre- or Hsa-Cre- positive WT control, double RE mice were normalized to the Alfp+Hsa-Cre-positive WT control, and data are displayed relative to WT controls within that feeding condition.
RNA-Sequencing data analysis for NF samples
Output FASTQ files from the sequencer as well as the quality of the reads were assessed using FASTQC v0.11.9 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Then, adapters and low quality bases were removed from the reads during a sequence-trimming step by means of BBDuk v35.85 (https://jgi.doe.gov/data-and-tools/software-tools/bbtools/bb-tools-user-guide/bbduk-guide/). Reads with less than 35 bp and less than a quality score of 25 were discarded.
High quality reads were mapped against the Mus musculus genome (GRCm38/Ensembl release 101 reference genome and gene annotation) employing STAR v2.7.1a83. Subsequently, FeatureCounts v 1.5.184 was used to quantify gene expression in terms of raw fragment counts. The latter information was displayed as an expression matrix that was loaded into R v4.0.3. Uninformative genes were filtered out from the matrix using HTSFilter package v1.30.185. Then, filtered raw counts were normalized by applying the TMM method. The differential gene expression analysis was conducted by the DESeq2 package v3.1686 in order to identify upregulated/downregulated genes. Multiple testing correction was performed with the FDR method and significance level was set at FDR < 0.05.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) terms were obtained performing hypergeometric tests (R phyper function), where FDR correction (FDR < 0.05) was applied to each individual pathway/term. Gene expression with respect to functional pathways was displayed using the pathview package version 1.30.1087.
For AL vs NF comparisons, non-autonomously oscillating genes were further narrowed down to those that exhibited a significant difference in expression between ZT4 and ZT16 in WT mice under NF.
qPCR
For liver and muscle, 500 ng-1 μg of Trizol-extracted RNA was reverse transcribed using Thermo Scientific Maxima Reverse Transcriptase and qPCR was performed using Syber Green Master Mix on a ThermoFisher Quantstudio 3. All gene expression data was normalized to 18S ribosomal RNA. Primer sequences are listed in the key resources table.
Rhythmicity and pathway analysis
To identify oscillating transcripts, transcriptomic data for each tissue was processed via dryR21. We used 5 group comparisons for each tissue. For liver: WT-Alfp-Cre, WT-Alfp+Hsa-Cre, KO, Liver-RE, Liver+Muscle-RE. For muscle: WT-Hsa-Cre, WT-Alfp+Hsa-Cre, KO, Muscle-RE, Liver+Muscle-RE. Genes with a BICW < 0.25 and amplitude < 0.25 were excluded. Only genes rhythmic in both WTs (single and double cre) were included, though WT-Alfp+Hsa-Cre is shown in heatmaps for clarity (see also Tables S1–2). Genes within each heatmap were normalised within each genotype due to observed batch effect in regards to overall levels of mRNA expression for muscle samples harvested in either batch 1 (WT-Hsa-Cre, KO, Muscle-RE) or batch 2 (WT-Alfp+Hsa-Cre, Liver+Muscle-RE). As such, for both muscle and liver, 3 group dryR was used to compare mean levels within batch 1 for muscle (WT-Hsa-Cre, KO, Muscle) and within batch 1 for liver (WT-Alfp-Cre, KO, Liver-RE). Gene ontology analysis was performed using DAVID88 for biological process for genes classifed into different dryR models. We performed select 2 group comparisons using Nitecap28 to identify genes with a significant change in the shape of their profile as determined by two-way ANOVA p<0.01.
Glucose, insulin, and pyruvate tolerance tests
Blood glucose was measured via the tail at 0, 15, 30, 60 and 120 min following gavage or intraperitoneal injection using an Accu-Chek Aviva Plus Blood Glucose Monitoring System. Glucose tolerance curves were calculated using current standards82; baseline was subtracted from raw blood glucose values at each time point to give change in glucose concentration (delta) over time. Raw data is shown in the corresponding supplementary figures. Area under the curve was calculated using standard parameters. For oral glucose tolerance tests, mice were fasted from ZT0 to ZT6 and then gavaged with 1.5 g/kg glucose. For pyruvate tolerance tests, mice were fasted from ZT0 to ZT8 and then injected I.P. with 1.5 g/kg sodium pyruvate. For insulin tolerance tests, mice were fasted from ZT0 to ZT3 and then injected I.P. with 0.3 U/kg insulin.
Glucose tracing
Glucose tracing was performed identically to oral glucose tolerance tests, except gavaged glucose was uniformly labeled with carbon 13 (U-13C6, Cambridge Isotope Laboratories, CLM-1396-PK). Tissues were harvested 25–30 min after gavage.
Glucose tracing sample preparation and metabolite extraction.
Serum (5 μl) was mixed with 150 μl 4°C 40:40:20 methanol:acetonitrile:water (extraction solvent), vortexed and immediately centrifuged at 16,000g for 10 min at 4°C. The supernatant (~ 100 μl) was collected for liquid chromatography-mass spectrometry (LC-MS) analysis. Frozen tissue samples were ground at liquid nitrogen temperature with a CryoMill (Retsch). To minimize data variation due to tissue heterogeneity, entire tissues were collected and ground. The resulting tissue powder (approximately 20 mg) was weighed and then extracted by adding 4°C extraction solvent, vortexed and centrifuged at 16,000g for 10 min at 4°C. The volume of the extraction solution (μl) was 40x the weight of tissue (mg) to make an extract of 25 mg of tissue per ml of solvent. The supernatant (40 μl) was collected for LC-MS analysis.
Metabolite measurements using LC-MS.
A quadrupole orbitrap mass spectrometer (Q Exactive; Thermo Fisher Scientific) operating in negative or positive ion mode was coupled to a Vanquish UHPLC system (Thermo Fisher Scientific) with electrospray ionization and used to scan from m/z 70 to 1,000 at 1 Hz, with a 140,000 resolution. LC separation was achieved on an XBridge BEH Amide column (2.1 × 150 mm2, 2.5 μm particle size, 130 Å pore size; Waters Corporation) using a gradient of solvent A (95:5 water: acetonitrile with 20 mM of ammonium acetate and 20 mM of ammonium hydroxide, pH 9.45) and solvent B (acetonitrile). Flow rate was 150 μl/min. The LC gradient was: 0 min, 85% B; 2 min, 85% B; 3 min, 80% B; 5 min, 80% B; 6 min, 75% B; 7 min, 75% B; 8 min, 70% B; 9 min, 70% B; 10 min, 50% B; 12 min, 50% B; 13 min, 25% B; 16 min, 25% B; 18 min, 0% B; 23 min, 0% B; 24 min, 85% B; and 30 min, 85% B. The autosampler temperature was 5°C and the injection volume was 3 μl. Data were analyzed using the MAVEN software (build 682, http://maven.princeton.edu/index.php). Natural isotope correction was performed with AccuCor R code (https://github.com/lparsons/accucor).
Insulin measurements
Collected blood was placed on ice and subsequently centrifuged at 3,000 rpm for 10 min at 4° C. The supernatant was stored at −80° C. All samples were thawed and used as starting material for the Ultra-Sensitive Mouse Insulin ELISA Kit (Cystal Chem 90080), which was utilized according to the manufacturer’s instructions.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data are displayed as mean ± S.E.M. unless otherwise noted. For each experiment, the number of biological replicates (n-value), statistical test and significance threshold can be found in the figure legend or main text. Complex statistical analyses of large-scale datasets are described within the corresponding methods section. Data were analyzed in Prism 6.0 software (GraphPad). The suitability of parametric vs non-parametric tests was determined by data distribution analysis tools in the software. Sample size was determined by referencing literature standards for studies of circadian rhythms89. Data were checked for outliers with Grubbs’ test.
Supplementary Material
Table S4. 3 group DryR mean expression analysis liver.
Table S3. 3 group DryR mean expression analysis muscle.
Table S2. 5 group DryR analysis liver.
Table S1. 5 group DryR analysis muscle.
ACKNOWLEDGEMENTS
This paper is dedicated to Paolo Sassone-Corsi, a hugely inspiring scientist and mentor and who remains an important influence on our work. Special thanks to our animal technician S. Sato and laboratory manager W. Orquiz. J.S. was supported by Zymo-CEM Postdoctoral Fellowship (Zymo Research) awarded at the University of California, Irvine. K.B.K. is supported by NIH, NIDDK F32 Fellowship - DK121425. T.S. was supported by a Japan Society for the Promotion of Science (JSPS) fellowship. C.M.G. was supported by the National Cancer Institute of the National Institutes of Health (NIH) under award number T32CA009054 and by the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement 749869. P.P. was funded by The Wenner-Gren Foundations, The Foundation Blanceflor Boncompagni Ludovisi, née Bildt and Tore Nilsson Foundation for Medical Science. V.S. was supported by a Marie -Curie grant- agreement 895390. A.V. was supported by the Hitachi-Nomura postdoctoral fellowship awarded through the Department of Biological Chemistry at the University of California, Irvine. The work of S.C., M.S., and P.B. was in part supported by NIH grant GM123558 C.J. was supported by the AASLD Foundation Pinnacle Research Award in Liver Disease, the Edward Mallinckrodt, Jr. Foundation Award, and NIH/NIAAA R01 AA029124. P.S.W. is supported by grant RYC2019026661-I funded by MCIN/AEI/10.13039/501100011033 and by “ESF Investing in your future”. Financial support for the S.M. Laboratory is provided through the NIH/NCI (Grants: R01CA244519, R01CA259370, K22CA212045). Research in the S.A.B. lab is supported partially by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (Grant agreement No. 787041), the Government of Cataluña (SGR grant), the Government of Spain (MINECO), the La Marató/TV3 Foundation, the Foundation Lilliane Bettencourt, the Spanish Association for Cancer Research (AECC) and The Worldwide Cancer Research Foundation (WCRF). The IRB Barcelona is a Severo Ochoa Center of Excellence (MINECO award SEV-2015-0505). P.M.C. acknowledges funding from MICINN-RTI2018-096068, ERC-2016-AdG-741966, LaCaixa-HEALTH-HR17-00040, MDA, UPGRADE-H2020-825825, AFM, DPP-Spain, Fundació La MaratóTV3– 80/19-202021, MWRF, and María-de-Maeztu Program for Units of Excellence to UPF (MDM-2014-0370) and the Severo- Ochoa Program for Centers of Excellence to CNIC (SEV2015-0505). Work in the P.S.-C. laboratory supported by NIH grants R21DK114652 and R21AG053592, a Challenge Grant from the Novo Nordisk Foundation (NNF202585), KAUST funding (OSR-2019-CRG8-URF/1/4042), and via access to the Genomics High Throughput Facility Shared Resource of the Cancer Center Support Grant (CA-62203) and the UCI and NIH-shared instrumentation grants 1S10RR025496-01, 1S10OD010794-01, and 1S10OD021718-01.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research
Footnotes
DECLARATION OF INTERESTS
SAB is a co-founder and scientific advisor of ONA Therapeutics.
REFERENCES
- 1.Rosbash M (2009). The implications of multiple circadian clock origins. PLoS Biol 7, e62. 10.1371/journal.pbio.1000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass J, and Lazar MA (2016). Circadian time signatures of fitness and disease. Science 354, 994–999. 10.1126/science.aah4965. [DOI] [PubMed] [Google Scholar]
- 3.Guan D, and Lazar MA (2021). Interconnections between circadian clocks and metabolism. J Clin Invest 131. 10.1172/JCI148278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cederroth CR, Albrecht U, Bass J, Brown SA, Dyhrfjeld-Johnsen J, Gachon F, Green CB, Hastings MH, Helfrich-Forster C, Hogenesch JB, et al. (2019). Medicine in the Fourth Dimension. Cell Metab 30, 238–250. 10.1016/j.cmet.2019.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fishbein AB, Knutson KL, and Zee PC (2021). Circadian disruption and human health. J Clin Invest 131. 10.1172/JCI148286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi SQ, Ansari TS, McGuinness OP, Wasserman DH, and Johnson CH (2013). Circadian disruption leads to insulin resistance and obesity. Curr Biol 23, 372–381. 10.1016/j.cub.2013.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kettner NM, Voicu H, Finegold MJ, Coarfa C, Sreekumar A, Putluri N, Katchy CA, Lee C, Moore DD, and Fu L (2016). Circadian Homeostasis of Liver Metabolism Suppresses Hepatocarcinogenesis. Cancer Cell 30, 909–924. 10.1016/j.ccell.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schibler U, and Sassone-Corsi P (2002). A web of circadian pacemakers. Cell 111, 919–922. 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 9.Kornmann B, Schaad O, Bujard H, Takahashi JS, and Schibler U (2007). System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5, e34. 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, and Schibler U (2004). Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119, 693–705. 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. (2004). PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A 101, 5339–5346. 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan D, Xiong Y, Trinh TM, Xiao Y, Hu W, Jiang C, Dierickx P, Jang C, Rabinowitz JD, and Lazar MA (2020). The hepatocyte clock and feeding control chronophysiology of multiple liver cell types. Science. 10.1126/science.aba8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greco CM, Koronowski KB, Smith JG, Shi J, Kunderfranco P, Carriero R, Chen S, Samad M, Welz PS, Zinna VM, et al. (2021). Integration of feeding behavior by the liver circadian clock reveals network dependency of metabolic rhythms. Sci Adv 7, eabi7828. 10.1126/sciadv.abi7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenwell BJ, Trott AJ, Beytebiere JR, Pao S, Bosley A, Beach E, Finegan P, Hernandez C, and Menet JS (2019). Rhythmic Food Intake Drives Rhythmic Gene Expression More Potently than the Hepatic Circadian Clock in Mice. Cell Rep 27, 649–657 e645. 10.1016/j.celrep.2019.03.064. [DOI] [PubMed] [Google Scholar]
- 15.Koronowski KB, Kinouchi K, Welz PS, Smith JG, Zinna VM, Shi J, Samad M, Chen S, Magnan CN, Kinchen JM, et al. (2019). Defining the Independence of the Liver Circadian Clock. Cell 177, 1448–1462 e1414. 10.1016/j.cell.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, and Bradfield CA (2000). Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009–1017. 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welz PS, Zinna VM, Symeonidi A, Koronowski KB, Kinouchi K, Smith JG, Guillen IM, Castellanos A, Furrow S, Aragon F, et al. (2019). BMAL1-Driven Tissue Clocks Respond Independently to Light to Maintain Homeostasis. Cell 177, 1436–1447 e1412. 10.1016/j.cell.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinturel F, Gos P, Petrenko V, Hagedorn C, Kreppel F, Storch KF, Knutti D, Liani A, Weitz C, Emmenegger Y, et al. (2021). Circadian hepatocyte clocks keep synchrony in the absence of a master pacemaker in the suprachiasmatic nucleus or other extrahepatic clocks. Genes Dev 35, 329–334. 10.1101/gad.346460.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sher J, and Cardasis C (1976). Skeletal muscle fiber types in the adult mouse. Acta Neurol Scand 54, 45–56. 10.1111/j.1600-0404.1976.tb07619.x. [DOI] [PubMed] [Google Scholar]
- 20.Hughes ME, Hogenesch JB, and Kornacker K (2010). JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms 25, 372–380. 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weger BD, Gobet C, David FPA, Atger F, Martin E, Phillips NE, Charpagne A, Weger M, Naef F, and Gachon F (2021). Systematic analysis of differential rhythmic liver gene expression mediated by the circadian clock and feeding rhythms. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2015803118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDearmon EL, Patel KN, Ko CH, Walisser JA, Schook AC, Chong JL, Wilsbacher LD, Song EJ, Hong HK, Bradfield CA, and Takahashi JS (2006). Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science 314, 1304–1308. 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehlen JC, Brager AJ, Baggs J, Pinckney L, Gray CL, DeBruyne JP, Esser KA, Takahashi JS, and Paul KN (2017). Bmal1 function in skeletal muscle regulates sleep. Elife 6. 10.7554/eLife.26557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrews JL, Zhang X, McCarthy JJ, McDearmon EL, Hornberger TA, Russell B, Campbell KS, Arbogast S, Reid MB, Walker JR, et al. (2010). CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci U S A 107, 19090–19095. 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harfmann BD, Schroder EA, Kachman MT, Hodge BA, Zhang X, and Esser KA (2016). Muscle-specific loss of Bmal1 leads to disrupted tissue glucose metabolism and systemic glucose homeostasis. Skelet Muscle 6, 12. 10.1186/s13395-016-0082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, and Fitzgerald GA (2004). BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol 2, e377. 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dyar KA, Ciciliot S, Wright LE, Bienso RS, Tagliazucchi GM, Patel VR, Forcato M, Paz MI, Gudiksen A, Solagna F, et al. (2014). Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol Metab 3, 29–41. 10.1016/j.molmet.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brooks TG, Mrcela A, Lahens NF, Paschos GK, Grosser T, Skarke C, FitzGerald GA, and Grant GR (2022). Nitecap: An Exploratory Circadian Analysis Web Application. J Biol Rhythms 37, 43–52. 10.1177/07487304211054408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.An D, Toyoda T, Taylor EB, Yu H, Fujii N, Hirshman MF, and Goodyear LJ (2010). TBC1D1 regulates insulin- and contraction-induced glucose transport in mouse skeletal muscle. Diabetes 59, 1358–1365. 10.2337/db09-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szekeres F, Chadt A, Tom RZ, Deshmukh AS, Chibalin AV, Bjornholm M, Al-Hasani H, and Zierath JR (2012). The Rab-GTPase-activating protein TBC1D1 regulates skeletal muscle glucose metabolism. Am J Physiol Endocrinol Metab 303, E524–533. 10.1152/ajpendo.00605.2011. [DOI] [PubMed] [Google Scholar]
- 31.Kalsbeek A, la Fleur S, and Fliers E (2014). Circadian control of glucose metabolism. Mol Metab 3, 372–383. 10.1016/j.molmet.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamia KA, Storch KF, and Weitz CJ (2008). Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A 105, 15172–15177. 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gachon F, Loizides-Mangold U, Petrenko V, and Dibner C (2017). Glucose Homeostasis: Regulation by Peripheral Circadian Clocks in Rodents and Humans. Endocrinology 158, 1074–1084. 10.1210/en.2017-00218. [DOI] [PubMed] [Google Scholar]
- 34.Hui S, Cowan AJ, Zeng X, Yang L, TeSlaa T, Li X, Bartman C, Zhang Z, Jang C, Wang L, et al. (2020). Quantitative Fluxomics of Circulating Metabolites. Cell Metab 32, 676–688 e674. 10.1016/j.cmet.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, Esparza LA, Reya T, Le Z, Yanxiang Guo J, et al. (2017). Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118. 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeFronzo RA, Jacot E, Jequier E, Maeder E, Wahren J, and Felber JP (1981). The effect of insulin on the disposal of intravenous glucose. Results from indirect calorimetry and hepatic and femoral venous catheterization. Diabetes 30, 1000–1007. 10.2337/diab.30.12.1000. [DOI] [PubMed] [Google Scholar]
- 37.Cori CF (1981). The glucose-lactic acid cycle and gluconeogenesis. Curr Top Cell Regul 18, 377–387. [PubMed] [Google Scholar]
- 38.Felig P, Pozefsky T, Marliss E, and Cahill GF Jr. (1970). Alanine: key role in gluconeogenesis. Science 167, 1003–1004. 10.1126/science.167.3920.1003. [DOI] [PubMed] [Google Scholar]
- 39.Petrus P, Smith JG, Koronowski KB, Chen S, Sato T, Greco CM, Mortimer T, Welz PS, Zinna VM, Shimaji K, et al. (2022). The central clock suffices to drive the majority of circulatory metabolic rhythms. Sci Adv 8, eabo2896. 10.1126/sciadv.abo2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, and Schibler U (2000). Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14, 2950–2961. 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, and Panda S (2009). Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A 106, 21453–21458. 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Goede P, Sen S, Oosterman JE, Foppen E, Jansen R, la Fleur SE, Challet E, and Kalsbeek A (2018). Differential effects of diet composition and timing of feeding behavior on rat brown adipose tissue and skeletal muscle peripheral clocks. Neurobiol Sleep Circadian Rhythms 4, 24–33. 10.1016/j.nbscr.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manella G, Sabath E, Aviram R, Dandavate V, Ezagouri S, Golik M, Adamovich Y, and Asher G (2021). The liver-clock coordinates rhythmicity of peripheral tissues in response to feeding. Nat Metab 3, 829–842. 10.1038/s42255-021-00395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.TeSlaa T, Bartman CR, Jankowski CSR, Zhang Z, Xu X, Xing X, Wang L, Lu W, Hui S, and Rabinowitz JD (2021). The Source of Glycolytic Intermediates in Mammalian Tissues. Cell Metab 33, 367–378 e365. 10.1016/j.cmet.2020.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeFronzo RA, and Tripathy D (2009). Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care 32 Suppl 2, S157–163. 10.2337/dc09-S302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merz KE, and Thurmond DC (2020). Role of Skeletal Muscle in Insulin Resistance and Glucose Uptake. Compr Physiol 10, 785–809. 10.1002/cphy.c190029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fougeray T, Polizzi A, Regnier M, Fougerat A, Ellero-Simatos S, Lippi Y, Smati S, Lasserre F, Tramunt B, Huillet M, et al. (2022). The hepatocyte insulin receptor is required to program the liver clock and rhythmic gene expression. Cell Rep 39, 110674. 10.1016/j.celrep.2022.110674. [DOI] [PubMed] [Google Scholar]
- 48.Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, and Spiegelman BM (2003). Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 423, 550–555. 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 49.Crosby P, Hamnett R, Putker M, Hoyle NP, Reed M, Karam CJ, Maywood ES, Stangherlin A, Chesham JE, Hayter EA, et al. (2019). Insulin/IGF-1 Drives PERIOD Synthesis to Entrain Circadian Rhythms with Feeding Time. Cell 177, 896–909 e820. 10.1016/j.cell.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamajuku D, Inagaki T, Haruma T, Okubo S, Kataoka Y, Kobayashi S, Ikegami K, Laurent T, Kojima T, Noutomi K, et al. (2012). Real-time monitoring in three-dimensional hepatocytes reveals that insulin acts as a synchronizer for liver clock. Sci Rep 2, 439. 10.1038/srep00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petrenko V, Saini C, Giovannoni L, Gobet C, Sage D, Unser M, Heddad Masson M, Gu G, Bosco D, Gachon F, et al. (2017). Pancreatic alpha- and beta-cellular clocks have distinct molecular properties and impact on islet hormone secretion and gene expression. Genes Dev 31, 383–398. 10.1101/gad.290379.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jouffe C, Weger BD, Martin E, Atger F, Weger M, Gobet C, Ramnath D, Charpagne A, Morin-Rivron D, Powell EE, et al. (2022). Disruption of the circadian clock component BMAL1 elicits an endocrine adaption impacting on insulin sensitivity and liver disease. Proc Natl Acad Sci U S A 119, e2200083119. 10.1073/pnas.2200083119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. (2010). Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466, 627–631. 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dyar KA, Hubert MJ, Mir AA, Ciciliot S, Lutter D, Greulich F, Quagliarini F, Kleinert M, Fischer K, Eichmann TO, et al. (2018). Transcriptional programming of lipid and amino acid metabolism by the skeletal muscle circadian clock. PLoS Biol 16, e2005886. 10.1371/journal.pbio.2005886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patgiri A, Skinner OS, Miyazaki Y, Schleifer G, Marutani E, Shah H, Sharma R, Goodman RP, To TL, Robert Bao X, et al. (2020). An engineered enzyme that targets circulating lactate to alleviate intracellular NADH:NAD(+) imbalance. Nat Biotechnol 38, 309–313. 10.1038/s41587-019-0377-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rabinowitz JD, and Enerback S (2020). Lactate: the ugly duckling of energy metabolism. Nat Metab 2, 566–571. 10.1038/s42255-020-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chaix A, Lin T, Le HD, Chang MW, and Panda S (2019). Time-Restricted Feeding Prevents Obesity and Metabolic Syndrome in Mice Lacking a Circadian Clock. Cell Metab 29, 303–319 e304. 10.1016/j.cmet.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doi R, Oishi K, and Ishida N (2010). CLOCK regulates circadian rhythms of hepatic glycogen synthesis through transcriptional activation of Gys2. J Biol Chem 285, 22114–22121. 10.1074/jbc.M110.110361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perelis M, Marcheva B, Ramsey KM, Schipma MJ, Hutchison AL, Taguchi A, Peek CB, Hong H, Huang W, Omura C, et al. (2015). Pancreatic beta cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science 350, aac4250. 10.1126/science.aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brown MR, Sen SK, Mazzone A, Her TK, Xiong Y, Lee JH, Javeed N, Colwell CS, Rakshit K, LeBrasseur NK, et al. (2021). Time-restricted feeding prevents deleterious metabolic effects of circadian disruption through epigenetic control of beta cell function. Sci Adv 7, eabg6856. 10.1126/sciadv.abg6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sadacca LA, Lamia KA, deLemos AS, Blum B, and Weitz CJ (2011). An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia 54, 120–124. 10.1007/s00125-010-1920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jang C, Hui S, Zeng X, Cowan AJ, Wang L, Chen L, Morscher RJ, Reyes J, Frezza C, Hwang HY, et al. (2019). Metabolite Exchange between Mammalian Organs Quantified in Pigs. Cell Metab 30, 594–606 e593. 10.1016/j.cmet.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petrus P, Smith JG, Koronowski KB, Chen S, Sato T, Greco CM, Mortimer T, Welz PS, Zinna VM, Shimaji K, et al. (2022). The central clock suffices to drive the majority of circulatory metabolic rhythms. Sci Adv 8, eabo2896. 10.1126/sciadv.abo2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seldin MM, Peterson JM, Byerly MS, Wei Z, and Wong GW (2012). Myonectin (CTRP15), a novel myokine that links skeletal muscle to systemic lipid homeostasis. J Biol Chem 287, 11968–11980. 10.1074/jbc.M111.336834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seldin MM, Lei X, Tan SY, Stanson KP, Wei Z, and Wong GW (2013). Skeletal muscle-derived myonectin activates the mammalian target of rapamycin (mTOR) pathway to suppress autophagy in liver. J Biol Chem 288, 36073–36082. 10.1074/jbc.M113.500736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Severinsen MCK, and Pedersen BK (2020). Muscle-Organ Crosstalk: The Emerging Roles of Myokines. Endocr Rev 41. 10.1210/endrev/bnaa016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bookout AL, de Groot MH, Owen BM, Lee S, Gautron L, Lawrence HL, Ding X, Elmquist JK, Takahashi JS, Mangelsdorf DJ, and Kliewer SA (2013). FGF21 regulates metabolism and circadian behavior by acting on the nervous system. Nat Med 19, 1147–1152. 10.1038/nm.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma X, Zhou Z, Chen Y, Wu Y, and Liu Y (2016). RBP4 functions as a hepatokine in the regulation of glucose metabolism by the circadian clock in mice. Diabetologia 59, 354–362. 10.1007/s00125-015-3807-1. [DOI] [PubMed] [Google Scholar]
- 69.Chen S, Feng M, Zhang S, Dong Z, Wang Y, Zhang W, and Liu C (2019). Angptl8 mediates food-driven resetting of hepatic circadian clock in mice. Nat Commun 10, 3518. 10.1038/s41467-019-11513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Priest C, and Tontonoz P (2019). Inter-organ cross-talk in metabolic syndrome. Nat Metab 1, 1177–1188. 10.1038/s42255-019-0145-5. [DOI] [PubMed] [Google Scholar]
- 71.Whitham M, Parker BL, Friedrichsen M, Hingst JR, Hjorth M, Hughes WE, Egan CL, Cron L, Watt KI, Kuchel RP, et al. (2018). Extracellular Vesicles Provide a Means for Tissue Crosstalk during Exercise. Cell Metab 27, 237–251 e234. 10.1016/j.cmet.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 72.Perrin L, Loizides-Mangold U, Skarupelova S, Pulimeno P, Chanon S, Robert M, Bouzakri K, Modoux C, Roux-Lombard P, Vidal H, et al. (2015). Human skeletal myotubes display a cell-autonomous circadian clock implicated in basal myokine secretion. Mol Metab 4, 834–845. 10.1016/j.molmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dos Santos M, Backer S, Aurade F, Wong MM, Wurmser M, Pierre R, Langa F, Do Cruzeiro M, Schmitt A, Concordet JP, et al. (2022). A fast Myosin super enhancer dictates muscle fiber phenotype through competitive interactions with Myosin genes. Nat Commun 13, 1039. 10.1038/s41467-022-28666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miniou P, Tiziano D, Frugier T, Roblot N, Le Meur M, and Melki J (1999). Gene targeting restricted to mouse striated muscle lineage. Nucleic Acids Res 27, e27. 10.1093/nar/27.19.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sato S, Basse AL, Schonke M, Chen S, Samad M, Altintas A, Laker RC, Dalbram E, Barres R, Baldi P, et al. (2019). Time of Exercise Specifies the Impact on Muscle Metabolic Pathways and Systemic Energy Homeostasis. Cell Metab 30, 92–110 e114. 10.1016/j.cmet.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 76.Small L, Altintas A, Laker RC, Ehrlich A, Pattamaprapanont P, Villarroel J, Pillon NJ, Zierath JR, and Barres R (2020). Contraction influences Per2 gene expression in skeletal muscle through a calcium-dependent pathway. J Physiol 598, 5739–5752. 10.1113/JP280428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolff G, and Esser KA (2012). Scheduled exercise phase shifts the circadian clock in skeletal muscle. Med Sci Sports Exerc 44, 1663–1670. 10.1249/MSS.0b013e318255cf4c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gil-Lozano M, Mingomataj EL, Wu WK, Ridout SA, and Brubaker PL (2014). Circadian secretion of the intestinal hormone GLP-1 by the rodent L cell. Diabetes 63, 3674–3685. 10.2337/db13-1501. [DOI] [PubMed] [Google Scholar]
- 79.Ding G, Li X, Hou X, Zhou W, Gong Y, Liu F, He Y, Song J, Wang J, Basil P, et al. (2021). REV-ERB in GABAergic neurons controls diurnal hepatic insulin sensitivity. Nature 592, 763–767. 10.1038/s41586-021-03358-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.la Fleur SE, Kalsbeek A, Wortel J, Fekkes ML, and Buijs RM. (2001). A daily rhythm in glucose tolerance: a role for the suprachiasmatic nucleus. Diabetes 50, 1237–1243. 10.2337/diabetes.50.6.1237. [DOI] [PubMed] [Google Scholar]
- 81.Zhong LX, Li XN, Yang GY, Zhang X, Li WX, Zhang QQ, Pan HX, Zhang HH, Zhou MY, Wang YD, et al. (2019). Circadian misalignment alters insulin sensitivity during the light phase and shifts glucose tolerance rhythms in female mice. PLoS One 14, e0225813. 10.1371/journal.pone.0225813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Virtue S, and Vidal-Puig A (2021). GTTs and ITTs in mice: simple tests, complex answers. Nat Metab 3, 883–886. 10.1038/s42255-021-00414-7. [DOI] [PubMed] [Google Scholar]
- 83.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 85.Rau A, Gallopin M, Celeux G, and Jaffrezic F (2013). Data-based filtering for replicated high-throughput transcriptome sequencing experiments. Bioinformatics 29, 2146–2152. 10.1093/bioinformatics/btt350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Luo W, and Brouwer C (2013). Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830–1831. 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang da W, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 89.Hughes ME, Abruzzi KC, Allada R, Anafi R, Arpat AB, Asher G, Baldi P, de Bekker C, Bell-Pedersen D, Blau J, et al. (2017). Guidelines for Genome-Scale Analysis of Biological Rhythms. J Biol Rhythms 32, 380–393. 10.1177/0748730417728663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S4. 3 group DryR mean expression analysis liver.
Table S3. 3 group DryR mean expression analysis muscle.
Table S2. 5 group DryR analysis liver.
Table S1. 5 group DryR analysis muscle.
Data Availability Statement
RNA-sequencing data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-BMAL1 | Abcam | ab93806; RRID: AB_10675117 |
| Anti-p84 | Genetex | GTX70220; RRID: AB_372637 |
| Anti-RFP | Rockland | 600–401-379 |
| Goat Anti-Rabbit IgG H&L (Alexa Fluor® 647) | Abcam | ab150079 |
| Anti-Tubulin | Sigma | T6199 |
| Chemicals, peptides, and recombinant proteins | ||
| U-13C6 glucose | Cambridge Isotope Laboratories | CLM-1396-PK |
| Critical commercial assays | ||
| Ultra Sensitive Mouse Insulin ELISA Kit | Crystal Chem | 90080 |
| Deposited data | ||
| RNA sequencing – WT and double RE liver and muscle | This paper | GSE197455 |
| RNA sequencing – WT, Bmal1 KO, and Muscle-RE muscle | This paper | GSE197726 |
| RNA sequencing – Night feeding dataset, all genotypes, liver and muscle | This paper | GSE228389 |
| Experimental models: Organisms/strains | ||
| Mouse: Wild Type (WT) – 1.Bmal1wt/wt, Alfp-cretg/0; 2. Bmal1wt/wt, Hsa-cretg/0; 3. Bmal1wt/wt, Alfp-cretg/0, Hsa- cretg/0 | S.A. Benitah lab, P.S. Welz, This study | |
| Mouse: Bmal1 knockout (KO) – Bmal1stopFL/stopFL | S.A. Benitah lab, P.S. Welz, This study | |
| Mouse: Bmal1 hepatocyte-reconstituted (Liver- RE) – Bmal1stopFL/stopFL; Alfp-cretg/0 | S.A. Benitah lab, P.S. Welz, This study | |
| Mouse: Bmal1 skeletal muscle-reconstituted (Muscle- RE) – Bmal1stopFL/stopFL; Hsa-cretg/0 | S.A. Benitah lab, P.S. Welz, This study | |
| Mouse: Bmal1 hepatocyte + skeletal muscle- reconstituted (Liver+Muscle-RE) – Bmal1stopFL/stopFL; Alfp- cretg/0; Hsa-cretg/0 | S.A. Benitah lab, P.S. Welz, This study | |
| Oligonucleotides | ||
| BMAL1 For - GCAGTGCCACTGACTACCAAGA | This study | |
| BMAL1Rev - TCCTGGACATTGCATTGCAT | This study | |
| Rn18S For - CGCCGCTAGAGGTGAAATTC | This study | |
| Rn18S Rev - CGAACCTCCGACTTTCGTTCT | This study | |
| Software and algorithms | ||
| dryR | Weger et al., 2021, PNAS | |
| JTK_CYCLE | Hughes et al., 2010, J. Biol. Rhythms. | |
| DAVID Bioinformatics Resource | Huang, Sherman and Lempicki, 2009, Nat. Protoc. | |
| Nitecap | Brooks et al., 2022, J. Biol. Rhythms. | |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


