Abstract
Purpose:
Targeting the PD-1/PD-L1 interaction has led to durable responses in fewer than half of patients with mismatch repair-deficient (MMR-d) advanced colorectal cancers (CRC). Immune contexture, including spatial distribution of immune cells in the tumor microenvironment, may predict immunotherapy outcome.
Patients and Methods:
Immune contexture and spatial distribution, including cell-to-cell distance measurements, were analyzed by multiplex immunofluorescence in primary CRCs with d-MMR (N=33) from patients treated with anti-PD-1 antibodies. By digital image analysis, density, ratio, intensity, and spatial distribution of PD-L1, PD-1, CD8, CD3, CD68, LAG3, TGFβR2, MHC-I, CD14, B2M, and pan-cytokeratin were computed. Feature selection was performed by regularized Cox regression with LASSO, and a proportional hazards model was fitted to predict progression-free survival (PFS).
Results:
For predicting survival among patients with MMR-d advanced CRC receiving PD-1 blockade, cell-to-cell distance measurements, but not cell densities or ratios, achieved statistical significance univariately. By multivariable feature selection, only mean number of PD-1+ cells within 10μm of a PD-L1+ cell was significantly predictive of progression-free survival (PFS). Dichotomization of this variable revealed that those with high versus low values had significantly prolonged PFS [median not reached (>83 months) vs 8.5 months (95% CI: 4.7-NR)] with a median PFS of 28.4 months for all patients [HRadj= 0.14, 95% CI: 0.04, 0.56; p=0.005]. Expression of PD-1 was observed on CD8+ T-cells; PD-L1 on CD3+ and CD8+ T-lymphocytes, macrophages (CD68+) and tumor cells.
Conclusions:
In d-MMR CRCs, PD-1+ to PD-L1+ receptor to ligand proximity is a potential predictive biomarker for the effectiveness of PD-1 blockade.
Introduction
Anti-programmed cell death (PD-1) antibodies directly target PD-1 and inhibit the interaction of PD-1 with its ligand PD-L1 to restore the anti-tumor immune response (1). PD-1 is predominantly expressed by activated T cells as well as by natural killer cells, myeloid cells, and antigen-presenting cells (APC). Interaction of PD-1 with its ligand PD-L1 results in suppression of the anti-tumor immune response (1). Targeting the PD-1/PD-L1 axis has led to frequent and durable responses in patients with mismatch repair deficient (MMR-d) metastatic colorectal cancer (mCRC), (2–5) and is now the standard first-line treatment for these patients (6,7). Nearly 15% of CRCs have MMR-d and show microsatellite-instability–high (MSI-H) which results in hypermutation and abundant neoantigens (8,9). CRCs with MMR-d display heterogeneity in the density of tumor-infiltrating lymphocytes (TILs), and their TIL density has been shown to prognostically stratify patients with stage III MMR-d colon cancer (10). Despite often remarkable responses to PD-1 blockade, nearly half of patients fail to respond due to poorly understood mechanisms of resistance, (11) and these antibodies have potential toxicities as well as high cost (6). To date, PD-L1 remains the only target for companion diagnostics for immune checkpoint inhibitors (ICIs). However and in contrast to multiple other solid tumors, PD-L1 expression by immunohistochemistry has not been predictive of outcome of ICIs in CRC (4,12). Accordingly, there is an unmet need for a predictive biomarker to enable selection of patients who are most likely to benefit from ICIs.
Within the tumor microenvironment (TME), the presence, spatial distribution and functional status of immune cell types including lymphocytes, (13,14) macrophages, (15) and monocytes (16) may influence responsiveness or resistance to PD-1 blockade. Resistance can arise from genetic variation in antigen presentation machinery (APM) that may diminish capacity for neoantigen presentation (17). In this regard, CRCs with MMR-d frequently lose HLA-class-I-mediated antigen presentation due to silencing of HLA class I genes or other defects in the APM, (18,19) such as loss of beta2 microglobulin (B2M) which can render tumor cells resistant to CD8+ T-cell-mediated immunity (20). Prolonged stimulation with tumor antigens may lead to T-cell dysfunction or exhaustion mediated by co-inhibitory immune checkpoint proteins such as PD-1 and LAG-3 (21,22). Transforming growth factor-β (TGF-β) can cause tumor immunosuppression along with the WNT/βcatenin signaling pathway, which have been associated with reduced responsiveness to ICIs (23). Importantly, spatial distribution of immune cells relative to one another and to cancer cells can impact cellular interactions, and the interaction of PD-1 with PD-L1 is the target of anti-PD-1 drugs. Spatial organization of TME components (24) including PD-1/PD-L1 have been shown to influence immunotherapy responsiveness and/or outcome in Merkel cell carcinoma, (25) melanoma (26,27) and non–small cell lung cancer (24,28).
In this study, we examined multiple immune cell types and tumor cells including their spatial relationships in the TME of primary tumors from patients with MMR-d mCRC treated with anti-PD-1 therapy. Multiplex immunofluorescence (mIF) and digital image analysis enabled concurrent visualization and measurement of cell surface proteins and cytokeratin in whole slide images of tumor sections with preservation of tissue architecture and morphology (29). Associations of immune marker densities, compartment localization, and spatial distribution with patient progression-free survival (PFS) and tumor response were determined. PFS was chosen as the primary endpoint since analysis of PFS begins at initiation of immunotherapy until progression or death. We hypothesized that cell-to-cell proximity may be important for interactions that can influence clinical benefit from PD-1 blockade.
Methods:
Cohort characteristics
Primary CRCs from patients with MMR-d mCRC who were treated with anti-PD-1 antibodies at Mayo Clinic Comprehensive Cancer Center (RST, FLA, ARZ) and had available primary tumors were included (n=33). These patients were identified from the cohort patients with MMR-d mCRC treated by immunotherapy (n= 95). Patients received PD-1 blockade with pembrolizumab (n=32) or nivolumab (n=1) monotherapy. Median patient age was 62 years (IQR of 49, 74). 16 (48.4%) were female. Patient data were reviewed and recorded from the electronic health record (EPIC, Madison, WI) that included baseline characteristics of patient age (years) at initiation of ICI treatment, race (white vs other) primary tumor location [right (proximal to splenic flexure) vs left], tumor grade (G1, G2 and G3), number of tumor metastasis and involved metastatic sites, and number of prior regimens. Computed tomographic (CT) scans performed prior to the first dose of anti-PD-1 antibody were compared to those obtained nearest to the date of last follow-up for response assessment. Disease status was determined according to RECIST criteria (version 1.1) as follows: complete response (CR); partial response (PR); stable disease (SD); progressive disease (PD).
Primary tumors had been analyzed for DNA MMR proteins by immunohistochemistry or for microsatellite instability (MSI) by polymerase-chain-reaction (PCR). Data for KRAS (wild type vs mutated) and BRAFV600E (present vs absent) were recorded from somatic mutation profiling by next-generation sequencing (NGS) where available.
Multiplexed immunofluorescence staining
Formalin-fixed paraffin-embedded (FFPE) blocks from primary CRCs were sectioned (4–6 μm-thickness) and tissue from each case was placed on TOMO® slides for multiplex immunofluorescent (mIF) staining. Tumor enriched blocks were selected without knowledge of any patient information. Three immune biomarker panels were used for multiplexed IF staining consisting of immune response (panels 1,2) and immune resistance (panel 3) biomarkers (Figure 1). The panels include the following biomarkers: PD-L1, PD-1, CD8, CD3, CD68, LAG3, TGFβR2, MHC-I, CD14, B2M, DAPI and pan-cytokeratin. All slides subjected to digital image analysis met predefined quality control criteria.
Figure 1.
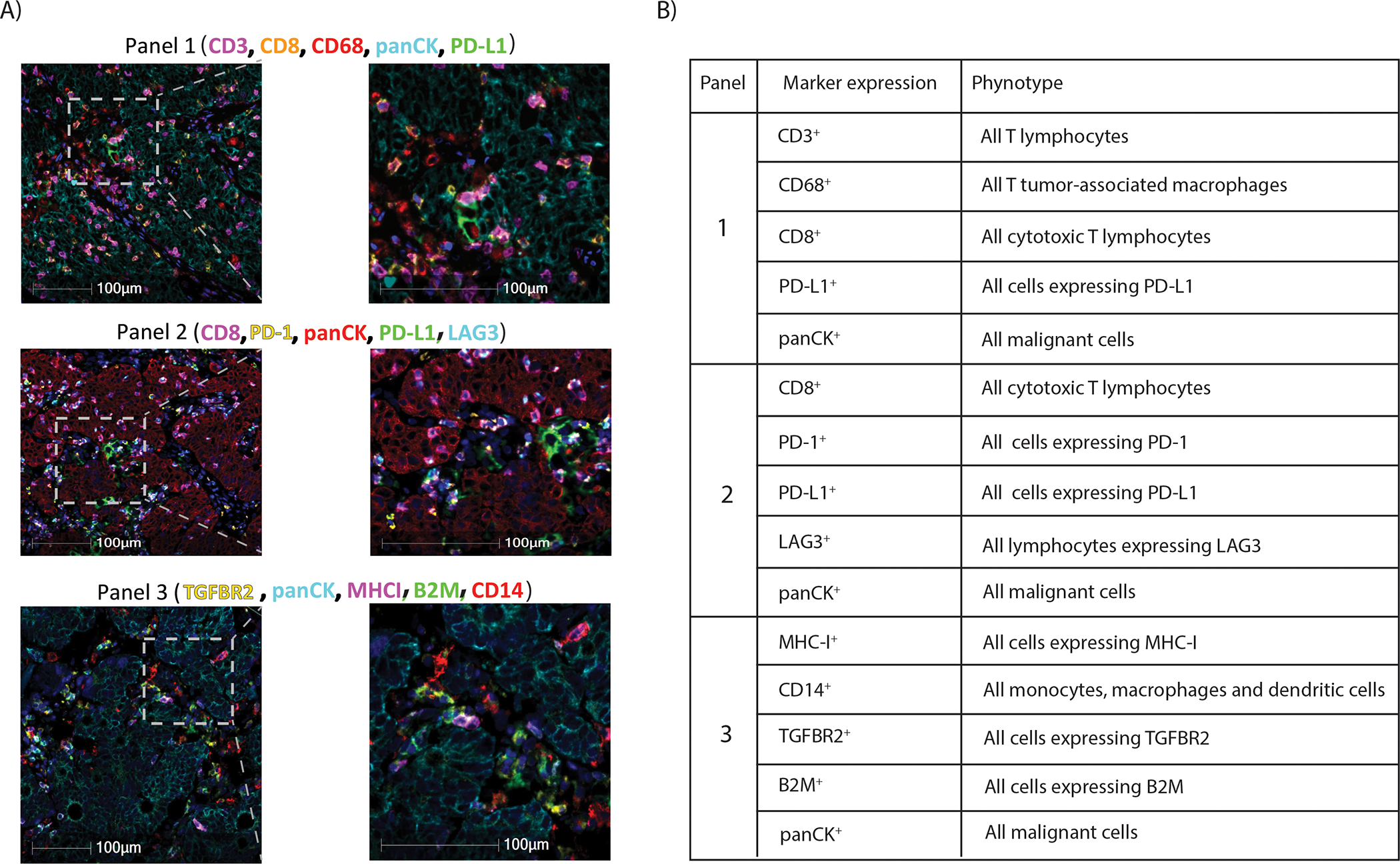
(A) Representative images of multiplex immunofluorescence panels in human colorectal cancers (Left); High power microscopic images of regions delineated by insets (Right) and (B) their observed phenotypes (Table).
Reagents
Rabbit monoclonal antibody CD8 (Clone #SP239, Abcam Cat#: ab178089, 0.7ug/ml), rabbit monoclonal antibody CD3 (Clone #: SP162, Abcam Cat#: ab135372, 0.05ug/ml), rabbit monoclonal antibody CD68 (Clone# SP251, Abcam Cat#: ab192847, 0.5ug/ml), rabbit monoclonal antibody PD-L1 (clone # SP263, Ventana Cat #790–4905/740–4907, ~1.6ug/ml), mouse monoclonal antibody pan-cytokeratin AE1/AE3/PCK26 cocktail, Ventana Cat# 760–2595/ 760–2135, 1:50 dilution from dispenser), rabbit monoclonal antibody LAG3 (Clone #EPR4392(2), Abcam Cat#: ab180187, 1:100 dilution from stock), PD-1 rabbit monoclonal antibody, clone #E18664, Abcam Cat# ab224774, 1:25 dilution from the stock), mouse monoclonal antibody MHC-I (clone #HCA2, Acris/Origene Cat#: AM03095PU-N, 1:2500 dilution from stock), mouse monoclonal antibody B2M (clone #B2M-02, Abcam Cat#: 27588, 0.5ug/ml), rabbit antibody CD14 (Clone# EPR3653, Ventana Cat#: 760–4523, 1:2 dilution), rabbit polyclonal antibody TGFBR2 (My BioSource Cat#MBS2400063, 1:400 dilution from the stock) were used.
Immunofluorescent multiplexed IHC assays were performed on a BenchMark ULTRA automated staining instrument (Ventana Medical Systems), using VENTANA reagents except as noted, according to the manufacturer’s instructions (29). Slides were de-paraffinized using EZ Prep solution (cat # 950–102) for 16 min at 72 °C. Epitope retrieval was accomplished with CC1 solution (cat #950–224) at high temperature (eg, 95–100 °C) for a period of time (e.g., 32–92 min) that is suitable for a specific tissue type. The epitope targets were sequentially detected with a thermochemical step in between to deactivate potential cross-reactivity of antibodies from the same species. For the 1st epitope detection, 1° antibodies was incubated for 8–32 min with an anti-species 2°Ab conjugated with horseradish peroxidase (GaR-HRP 2° antibody) from ultraView SISH DNP Detection Kit (cat # 760–098). After two rinses with reaction buffer, a Ty-fluor was introduced and incubated for 4 min, followed with the application of 0.01% H2O2 (DISCOVERY reagent cat # 760–244). The HRP in the 1° antibodies/2° antibody complex was then allowed to react with the Ty-fluor for 16–24 min, leading to oxidation and subsequent covalent binding of Ty-fluor to tyrosine residues in the vicinity of the antigens. The next four epitope targets were sequentially detected by following the above steps, with a > 16min 98°C incubation with CC2 as deactivation step to reduce the potential cross-reactivity. Five Ty-fluor conjugates were prepared in-house as research reagents: 31μM carboxyrhodamine-6G-Tyramide (Ty-R6G, DISCOVERY reagent cat #760–244), 8.6μM sulforhodamine101-tyramide (Ty-Red610 (Ty-R610), 17μM diethylaminocoumarin-tyramide (Ty-DCC), 32.6μM SulphoCy5-Tyramide (Ty-Cy5) and 52μM fluorescein isothiocyanate -Tyramide (Ty-FAM). The slides were counterstained with QD DAPI (DISCOVERY reagent cat #760–4196), and coverslipped with ProLong Diamond Antifade Mountant with DAPI (ThermoFisher Scientific, cat #P36962). Slides were coverslipped and subsequently scanned at 20× magnification using a Zeiss Axio Scan.Z1 and viewed with Zen 3 imaging software. We did not encounter sample loss.
Image acquisition and analysis
We examined immune marker IF data for undistinguished cell types by digital image analysis in five tissue compartments (overall tumor, tumor epithelia, tumor stroma, peritumor inside vs outside) in each case. For the peritumor compartment and using a computer automated system, regions were defined that 1) extends a pre-defined distance beyond the edge of the tumor region (peritumor outside); or 2) a pre-defined distance inside the tumor region (peritumor inside). Computed features within each compartment included cell density, fraction of positive cells, intensity of positive cells. We also examined spatial distribution between distinct markers on undistinguished cell types with distances measured using image analysis software [HighPlex FL module (version 4.1.3) in HALO (version: 3.3.2541.409)]. Identification of specific cell types with immune marker expression and their relative abundance were calculated for each case. Digital image analysis was performed with blinding to patient outcomes.
Statistical Analysis
For each computed feature from mIF digitized images, the mean, median, standard deviation, and quartiles were examined as potential predictive features. When insufficient cell counts prevented feature calculation, a value of not available (NA) was used. During data preprocessing, features which had greater than 10% NA values across subjects were dropped. For the remaining features, NA values were imputed using the median value across subjects. To reduce colinearty among features, the correlation between mean and median readouts for the same underlying feature was assessed. Where correlation exceeded 99%, only the median feature was kept.
Assessment of the association between individual mIF-derived features and patient PFS was performed by fitting univariable Cox proportional hazard models for each feature. The association between multiple mIF features and PFS was determined using a multivariable LASSO regularized Cox regression (30). The regularization parameter was chosen based on minimization of the partial-likelihood deviance (31). A stable estimate of the regularization parameter was achieved using 1000 repeated runs of 5-fold cross-validation, followed by computing the median optimal regularization parameter across runs. This median parameter was used to perform a LASSO regularized Cox regression for final feature selection. The resulting feature(s) were used to fit a Cox proportional hazards model for prediction of PFS. Survival curves for PFS were constructed using the Kaplan-Meier estimator. Adjusted hazard ratios (HRadj) and 95% confidence intervals (CI) are reported. For dichotomization of desired variables, a range of possible feature cutpoints and corresponding hazard ratios for prediction of PFS were examined. This procedure considered all possible continuous variables values as potential cutpoints. Two-sided P-values are reported and P-values <0.05 were considered to indicate statistical significance.
The association between individual mIF features and tumor response was performed by fitting a univariable logistic model for each feature. In this analysis, objective tumor response was considered as complete response (CR) or partial response (PR). For each model, the likelihood ratio (LLR) and resulting P-value are reported. Analyses were performed using R version 4.1.1 (Boston, MA, USA).
Written informed consents were obtained from the patients, and the studies were conducted in accordance with Declaration of Helsinki. This study was approved by the Mayo Clinic Institutional Review Board.
Data availability
The data generated in this study are available from the corresponding author upon reasonable request.
Results
Patient Characteristics
Among 33 patients with MMR-d mCRCs, 16 (48.4%) were female, 10 (30.3%) received first-line ICI therapy, and 23 (69.7%) had received ≥1 chemotherapy regimen prior to anti-PD-1 treatment. Median patient age was 62 years (IQR of 49, 74); 8 (28%) tumors harbored BRAFV600E and 9 (31%) had mutant KRAS. Patient baseline characteristics are shown in Supplementary Table S1. Patient median PFS was 28.4 months (95% CI: 10.4, NR) with 20 outcome events. At the date of last follow-up, there were 11 (33%) complete responses (CR) to anti-PD-1 treatment, 3 (9%) partial responses (PR), 1 (3%) stable disease, and 18 (55%) patients with progressive disease.
Multiplex Immunofluorescence
Multiplex IF was performed in tumor sections from each patient and analyzed by digital image analysis of 3 immune biomarker panels of which 2 panels evaluated biomarkers of immune response (panels 1, 2) and the third examined immune resistance (panel 3). Photomicrographs of IF staining for each panel and their related phenotype are shown in Figure 1. Panel 1: CD3, CD8, CD68, panCK, PD-L1; Panel 2: CD8, PD-1, panCK, PD-L1, LAG3; Panel 3: TGFBR2, panCK, MHC-I, B2M, CD14).
Association with Outcomes of Anti-PD-1 Treatment
Immune marker immunofluorescence (IF) data was examined for undistinguished cell types by digital image analysis (DIA) in five tissue compartments [overall tumor, tumor epithelia, tumor stroma, peritumor inside vs outside (see Methods)]. A total of 2283 features were computed using DIA. After preprocessing (see Methods), 509 features were retained in the dataset for use in subsequent analyses. Univariable Cox analysis found that only cell-to-cell distance readouts for immune markers or their relationship to panCK achieved statistical significance for their association with patient PFS (Supplementary Table S2). From panel 1, the distance measurement of a panCK+ tumor cell to the nearest CD8+ T-cell was significantly associated with PFS. From panel 2, the distance from PD-1+ to PD-L1+, CD8+PD-1+ to CD8+PD-L1+, CD8+PD-1+ to PD-L1+, and CD8+PD-1+ to panCK+PD-L1+ cell were each significantly associated with patient PFS (see Supplementary Table S2). Analysis using immune marker panel 3, however, did not reveal any features that were significantly associated with PFS.
Examination of TME features in relationship to response to anti-PD-1 therapy was also performed. Analysis of data from panel 1 revealed that the distance from a panCK+ cell to CD8+ cell, PD-L1+ panCK+ cell to CD8+ cell, and density of CD3+ cells in stroma were each significantly associated with objective tumor response by univariable analysis (Supplementary Table S3). Analysis of data from panel 2 revealed that the distance of PD-1+ cell to PD-L1+ cell, CD8+PD-1+ cell to CD8+PD-L1+ cell, and the density of the following cells were each significantly associated with anti-PD-1 response (CD3+ cells, CD8+ cells, CD3+/CD8+/PD-L1− cells, CD8+/panCK−, panCK+/PD-1+ cells, and panCK−/LAG3+ cells (Supplementary Table S3).
We found that the distance between undistinguished cells expressing PD-1 and PD-L1 in the overall tumor was the only feature that was significantly associated with patient PFS (p=0.005) by LASSO-derived feature selection. Next, we explored the distance measurement between PD-1+ and PD-L1+ cells and their associated hazard ratios (HR) for PFS. We found that a distance greater than 10μm converges to an HR of 1 indicating a decreasing prognostic impact (Figure 2A). However, the mean number of PD-1+ cells within 10μm of a PD-L1+ cell in the overall tumor was significantly associated with PFS (HR=6.2e-07, 95% CI: 4.1e-11, 0.009; p=2e-04). Next, we performed a multivariable Cox regression with adjustment for age, sex, histological grade, primary tumor sidedness, number of metastatic sites, presence of liver metastasis, number of prior regimes, and KRAS/BRAF status. After adjustment for these covariates, the mean number of PD-1+ cells within 10μm of a PD-L1+ cell in the overall tumor remained significantly associated with PFS (HRadj= 2.9e-10, 95% CI: 4.9e-17, 0.002; p=0.006). Our data indicate that any distance of PD-1+ within 10μm of PD-L1+ will be associated with a significant hazard ratio. In contrast to distance measurements, the ratio of PD-1+/PD-L1+ cells was not predictive of PFS (HR=1, 95% CI: 0.96,1.1; p= 0.47). We also analyzed tumor response data and found that responders (CR, PR) vs non responders to anti-PD-1 treatment had a higher mean number of PD-1+ cells within 10μm of a PD-L1+ cell in the overall tumor compared to nonresponders [median (IQR); 0.08 (0.04–0.17) vs 0.03 (0.002–0.09)].
Figure 2.
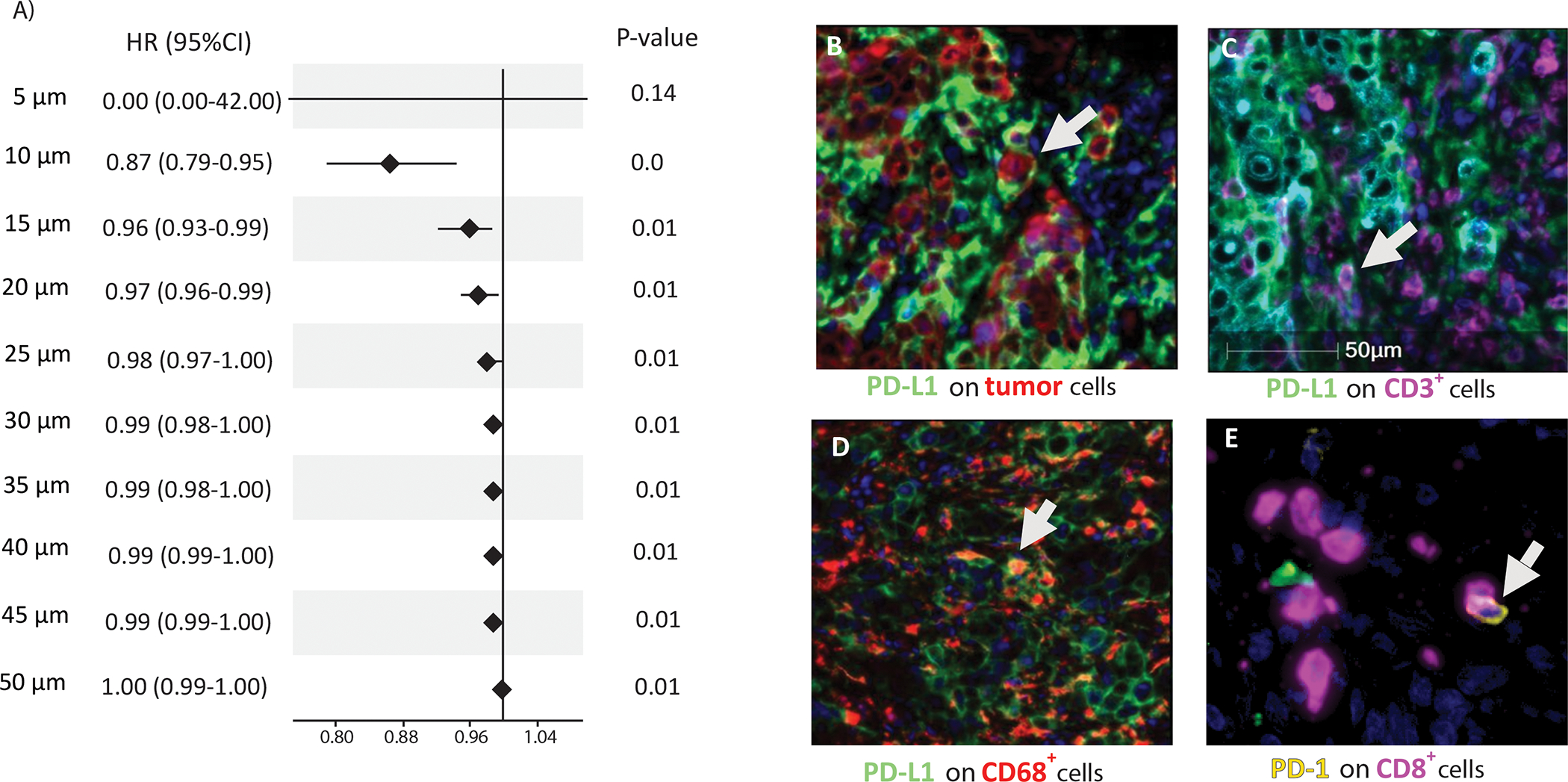
(A) Evaluation of PD-1-PD-L1 proximity cutpoints for prediction of progression-free survival (PFS). Forest plot of univariable Cox model hazard ratios and their 95% confidence intervals for relationship of PD-1 to PD-L1 proximity (5–50 μm) with patient PFS in colorectal cancers (N=33). PD-L1 was expressed on (B) tumor cells (panCK); (C) CD3+ T lymphocytes, and (D) CD68+ macrophages; (E) PD-1 expression on CD8+ T lymphocytes was also observed.
Regarding cellular localization of immune markers, we found that PD-L1 was expressed on CD3+ and CD8+ T lymphocytes, CD68+ macrophages, and tumor cells (Figure 2B–E, Supplementary Figure S1). Calculation of the relative abundance of immune markers for each case was performed to determine the dominant cell type with PD-L1 expression. PD-L1 expression was most commonly observed on T-cells (52%) followed by macrophages (34%) and tumor cells (14%).
To make our finding more interpretable, we converted the chosen continuous variable (mean number of PD-1+ cells within 10μm of PD-L1+ cells) to a categorical variable which defined high vs low value subgroups. Distribution of the average number of PD-1+ cells within 10μm of a PD-L1+ cell in the overall tumor and range of possible cutoff values and corresponding hazard ratios for prediction of PFS are shown in Supplementary Figure S2. A cutpoint of 0.044 for the chosen variable was selected due to maximization of hazard ratio at this cutpoint (Supplementary Figure S2). Using the cutoff value of 0.044, cases were dichotomized into high (n=20) vs low (n=13) value subgroups (Figure 3). We found that a high vs low PD-1 to PD-L1 proximity value was significantly associated with significantly longer PFS (HRadj= 0.14, 95% CI: 0.04, 0.56; p=0.005; [median: not reached (>83) vs 8.5 (95%CI: 4.7-NR) months]; Figure 4).
Figure 3.
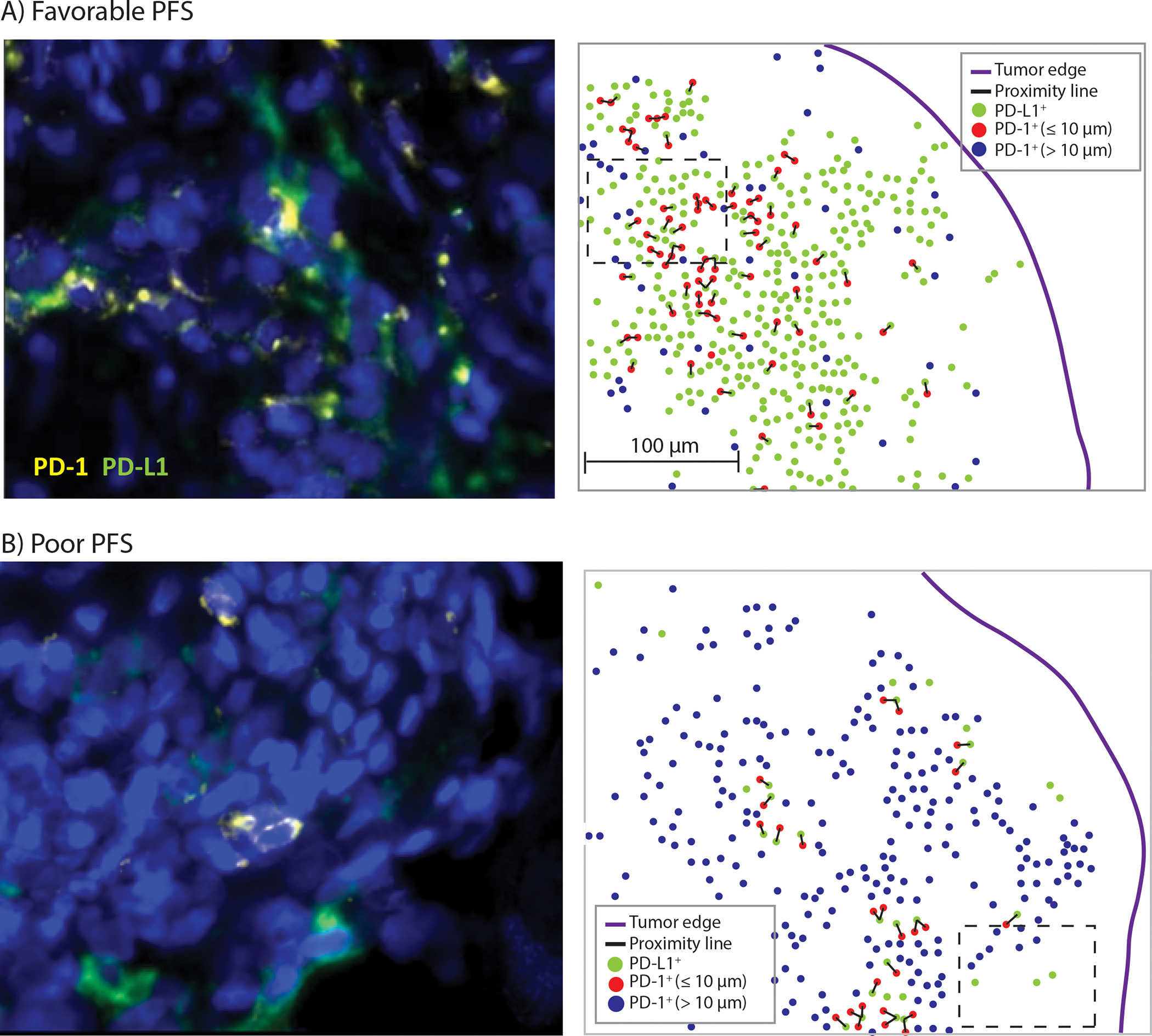
Representative multiplex immunofluorescence images of colorectal cancers with (A) Favorable PFS (high value) and (B) Poor PFS (low value) from anti-PD-1 treatment. High power microscopic images of regions delineated by insets (Left panels). The location of PD-1+ cells and PD-L1+ cells are shown within the overall tumor area (Right panels). After detection of PD-L1+ cells (green dots), distances between these cells and the neighboring PD-1+ cells were measured [proximity line (black color)]. Those PD-1+ cells that were within 10μm of a PD-L1+ cells are shown in blue color and those that are more than 10μm away from a PD-L1+ cells are shown in red color. For each PD-L1+ cell, the number of PD-1+ cells that were within 10μm of it was extracted and the mean of these values was calculated to generate the final variable “mean number of PD-1+ cells within 10μm of PD-L1+ cell” for each case.
Figure 4.
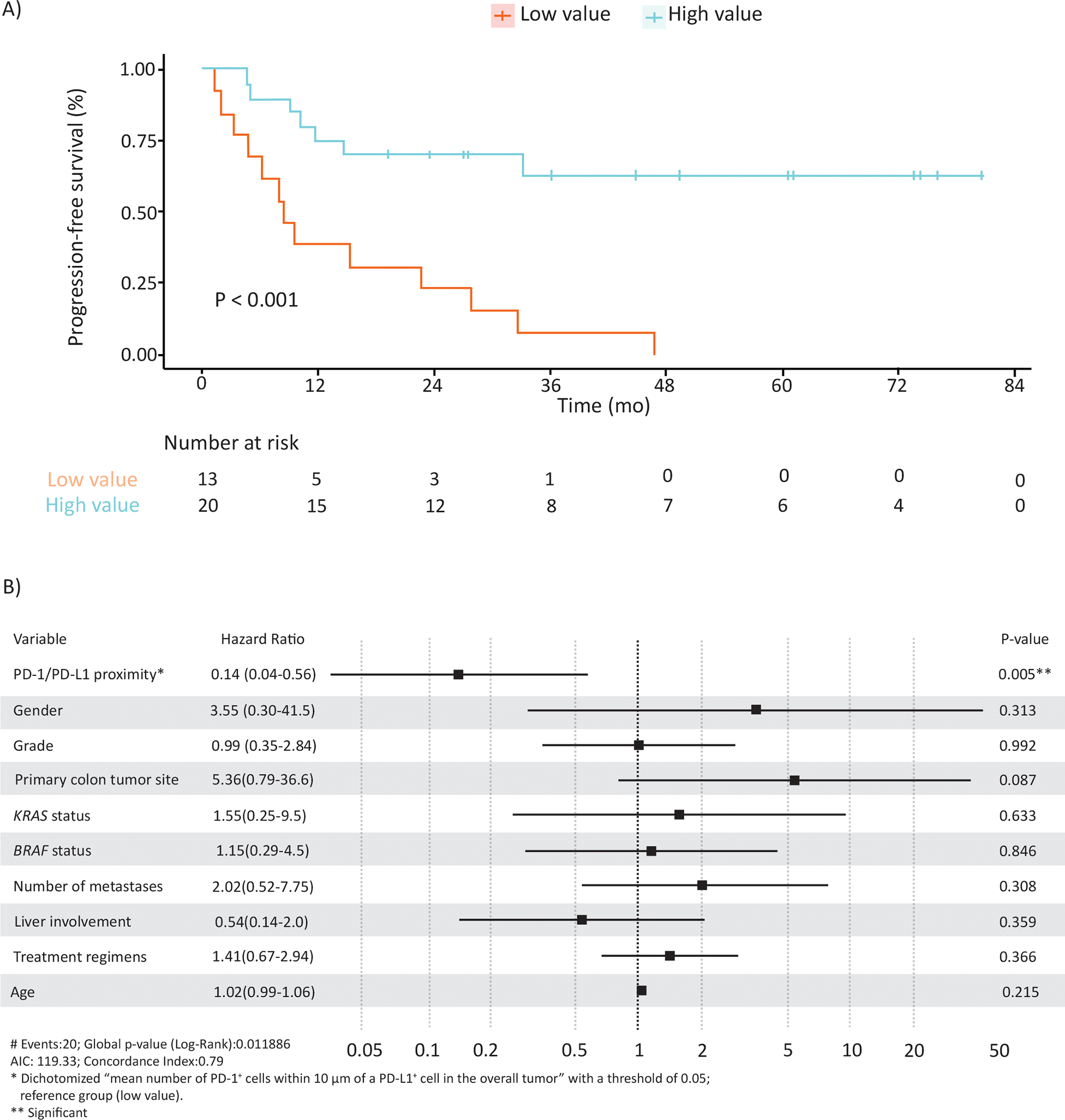
(A) Kaplan–Meier estimates of the association of PD-1/PD-L1 proximity value (mean number of PD-1+ cells within 10μm of PD-L1+ cell) with progression free survival. (B) Forest plot based on results of multivariable analysis of the association of PD-1/PD-L1 proximity value with progression free survival.
Discussion
In patients with advanced MMR-d CRC treated with an anti-PD-1, quantitative immunofluorescence revealed that a higher mean number of PD-1+ cells within 10μm of PD-L1+ cells using was associated with significantly better PFS and tumor response. Evaluation of a dichotomized variable (high vs low) also demonstrated that the mean number of PD-1+ cells within 10μm of PD-L1+ cells was significantly associated with PFS. Specifically, patients with high versus low values had a prolonged median PFS of >83 months vs 8.5 months that achieved statistical significance. Our results indicate that a quantitative threshold for PD-1 to PD-L1 proximity is important for the ability of PD-1 inhibitors to effectively block their interaction. Furthermore, the finding that PD-1-to-PD-L1 distances beyond 10μm are associated with shorter PFS suggests a mechanism of resistance to PD-1 blockade. Importantly, the predictive utility of PD-1 to PD-L1 proximity was found irrespective of the specific cell type expressing these checkpoint proteins. In contrast to proximity measurement, cell densities and their ratios, including PD-1/PD-L1, or other immune markers failed to predict patient outcomes. As expected, PD-L1 expression alone lacked predictive utility as has been reported in patients with MMR-d advanced CRCs receiving first-line ICI treatment (Checkmate 142 study) (32) and in proficient MMR CRCs (33). Our data demonstrate that PD-1 to PD-L1 proximity is a distinct biomarker which was shown to predict the outcome of PD-1 blockade in MMR-d tumors. In studies in advanced non-small cell lung cancer (NSCLC) and metastatic melanomas, a PD-1/PD-L1 co-localization score determined by immunofluorescence was shown to be associated with PFS and tumor response after treatment with ICIs (24). This score represents expression of PD-1 and PD-L1 in the same pixels (about 0.5 μm resolution) whereas our data delineate a critical threshold for PD-1 to PD-L1 proximity required to achieve clinical benefit from PD-blockade (24). In another study in advanced melanoma, a proximity ligation assay was used to assess the interaction of PD-1 with PD-L1 and whose dichotomized score was associated with patient outcome after PD-1 blockade (34). In contrast to these studies in other solid tumors, (24,34) our findings in MMR-d CRC define the minimum distance between PD-1 and PD-L1 proteins that appears necessary to enable effective PD-1 blockade which translates into clinical benefit. Studies in lung cancer and melanoma were limited by heterogeneous ICI treatment and use of tumor biopsies or tissue microarrays (24) whereas all patients in our study received PD-1 blockade and mIF was performed on whole slide images from all tumors.
PD-L1 is known to be expressed on the surface of many immune cell subtypes as well as cancer cells. Using immunofluorescence microscopy, we detected PD-1 expression on CD8+ T lymphocytes whereas PD-L1 was expressed on CD3+ and CD8+ T-cells, CD68+ macrophages, and tumor cells suggesting the potential for multiple cell types to influence the effectiveness of anti-PD-1 treatment. In contrast to melanoma and non-small cell lung cancer cells, CRC cell expression of PD-L1 is relatively infrequent (15) although it is expressed at a higher level in MMR-d tumor cells (35) and can be upregulated by BRAFV600E (36). Analysis of a subset of MMR-d mCRCs from the KEYNOTE- 177 study demonstrated that those tumors responding to PD-1 blockade were enriched in cytotoxic and proliferating PD-1+CD8+ T-cells interacting with PD-L1+ antigen-presenting macrophages (CD68+CD74+). These data were interpreted to suggest that PD-1 blockade releases the PD-1-PD-L1 interaction between CD8+ T-cells and macrophages to promote anti-tumor activity (15). Consistent with our findings, these studies underscore the importance of the PD-1-PD-L1 interaction in predicting ICI response or resistance. Relevant to its use as a biomarker, our data demonstrate that the proximity of PD-1 and PD-L1 appears most important for prediction of the outcome of PD-1 blockade.
We also analyzed tissue biomarkers in relationship to anti-PD-1 treatment response. We found that the distance of PD-1+ cell to PD-L1+ cell was significantly associated with objective tumor response which further supports the validity of our biomarker measurement. However, the categorical nature of the tumor response and our modest study sample size limited the ability to evaluate predictive accuracy by multivariable analysis. Strengths of our study include patients from a multi-site academic practice with long-term clinical follow-up and response data that were determined using established RECIST criteria. All patients received PD-1 blockade such that treatment was homogenous. Quantitative mIF was performed in whole slide images from surgical resection specimens. Study limitations include the retrospective nature of our analysis and need for validation in an independent patient cohort. We were unable to formally determine the predictive utility of PD-1/PD-L1 proximity with outcome given that our study was not randomized with a control arm that did not receive PD-1 blockade.
In conclusion, we found that the mean number of PD-1+ cells within 10μm of a PD-L1+ cell was significantly and independently associated with patient PFS after PD-1 blockade. These data suggest a quantitative threshold for receptor and ligand proximity for the effectiveness of PD-1 blockade. Examination of a dichotomized PD-1 to PD-L1 proximity variable was also shown to stratify patients for PFS, suggesting a potential predictive biomarker for ICI effectiveness with clinical applicability.
Supplementary Material
Translational Relevance.
Nearly half of patients with d-MMR metastatic CRC do not receive clinical benefit from anti-PD-1 treatment which underscores the need for a predictive biomarker. Using multiplex immunofluorescence, the mean number of PD-1+ cells within 10μm of a PD-L1+ cell was significantly and independently associated with patient progression-free survival after PD-1 blockade, suggesting the requirement for receptor and ligand proximity for anti-PD-1 effectiveness. This biologically important relationship of the PD-1/PD-L1 axis with delineation of a critical threshold suggest a potential predictive biomarker for anti-PD-1 therapy with potential clinical applicability.
Funding
This study was supported, in part, by NCI R01 CA210509 (to FAS) and by Roche Diagnostics (EA, SM, CW, AH, WZ, AM, DY, KS).
Footnotes
Author Disclosures
EA, SM, CW, AH, WZ, AM, DY, KS are employees of Ventana Medical Systems, Inc./Roche Tissue Diagnostics and hold ownership interest (including patents) in Roche Holding AG. FAS holds patents with Ventana Medical Systems, Inc./Roche Tissue Diagnostics. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Chen S, Crabill GA, Pritchard TS, McMiller TL, Wei P, Pardoll DM, et al. Mechanisms regulating PD-L1 expression on tumor and immune cells. J Immunother Cancer 2019;7(1):305 doi 10.1186/s40425-019-0770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357(6349):409–13 doi 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le DT, Kim TW, Van Cutsem E, Geva R, Jäger D, Hara H, et al. Phase II Open-Label Study of Pembrolizumab in Treatment-Refractory, Microsatellite Instability-High/Mismatch Repair-Deficient Metastatic Colorectal Cancer: KEYNOTE-164. J Clin Oncol 2020;38(1):11–9 doi 10.1200/jco.19.02107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372(26):2509–20 doi 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol 2017;18(9):1182–91 doi 10.1016/s1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.André T, Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt C, et al. Pembrolizumab in Microsatellite-Instability-High Advanced Colorectal Cancer. N Engl J Med 2020;383(23):2207–18 doi 10.1056/NEJMoa2017699. [DOI] [PubMed] [Google Scholar]
- 7.Casak SJ, Marcus L, Fashoyin-Aje L, Mushti SL, Cheng J, Shen YL, et al. FDA Approval Summary: Pembrolizumab for the First-line Treatment of Patients with MSI-H/dMMR Advanced Unresectable or Metastatic Colorectal Carcinoma. Clin Cancer Res 2021;27(17):4680–4 doi 10.1158/1078-0432.Ccr-21-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muzny DM, Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487(7407):330–7 doi 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015;5(1):43–51 doi 10.1158/2159-8290.Cd-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoon HH, Shi Q, Heying EN, Muranyi A, Bredno J, Ough F, et al. Intertumoral Heterogeneity of CD3(+) and CD8(+) T-Cell Densities in the Microenvironment of DNA Mismatch-Repair-Deficient Colon Cancers: Implications for Prognosis. Clin Cancer Res 2019;25(1):125–33 doi 10.1158/1078-0432.Ccr-18-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Z, Sinicrope FA. Mismatch Repair-Deficient Colorectal Cancer: Building on Checkpoint Blockade. J Clin Oncol 2022:Jco2102691 doi 10.1200/jco.21.02691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, et al. Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. J Clin Oncol 2018;36(8):773–9 doi 10.1200/jco.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 13.Rakaee M, Adib E, Ricciuti B, Sholl LM, Shi W, Alessi JV, et al. Association of Machine Learning-Based Assessment of Tumor-Infiltrating Lymphocytes on Standard Histologic Images With Outcomes of Immunotherapy in Patients With NSCLC. JAMA Oncol 2023;9(1):51–60 doi 10.1001/jamaoncol.2022.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park S, Ock CY, Kim H, Pereira S, Park S, Ma M, et al. Artificial Intelligence-Powered Spatial Analysis of Tumor-Infiltrating Lymphocytes as Complementary Biomarker for Immune Checkpoint Inhibition in Non-Small-Cell Lung Cancer. J Clin Oncol 2022;40(17):1916–28 doi 10.1200/JCO.21.02010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bortolomeazzi M, Keddar MR, Montorsi L, Acha-Sagredo A, Benedetti L, Temelkovski D, et al. Immunogenomics of Colorectal Cancer Response to Checkpoint Blockade: Analysis of the KEYNOTE 177 Trial and Validation Cohorts. Gastroenterology 2021;161(4):1179–93 doi 10.1053/j.gastro.2021.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grauers Wiktorin H, Nilsson MS, Kiffin R, Sander FE, Lenox B, Rydstrom A, et al. Histamine targets myeloid-derived suppressor cells and improves the anti-tumor efficacy of PD-1/PD-L1 checkpoint blockade. Cancer Immunol Immunother 2019;68(2):163–74 doi 10.1007/s00262-018-2253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbott CW, Boyle SM, Pyke RM, McDaniel LD, Levy E, Navarro FCP, et al. Prediction of Immunotherapy Response in Melanoma through Combined Modeling of Neoantigen Burden and Immune-Related Resistance Mechanisms. Clin Cancer Res 2021;27(15):4265–76 doi 10.1158/1078-0432.Ccr-20-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dierssen JWF, de Miranda NFCC, Ferrone S, van Puijenbroek M, Cornelisse CJ, Fleuren GJ, et al. HNPCC versus sporadic microsatellite-unstable colon cancers follow different routes toward loss of HLA class I expression. BMC Cancer 2007;7(1):33 doi 10.1186/1471-2407-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ijsselsteijn ME, Petitprez F, Lacroix L, Ruano D, van der Breggen R, Julie C, et al. Revisiting immune escape in colorectal cancer in the era of immunotherapy. Br J Cancer 2019;120(8):815–8 doi 10.1038/s41416-019-0421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. New England Journal of Medicine 2016;375(9):819–29 doi 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tu L, Guan R, Yang H, Zhou Y, Hong W, Ma L, et al. Assessment of the expression of the immune checkpoint molecules PD-1, CTLA4, TIM-3 and LAG-3 across different cancers in relation to treatment response, tumor-infiltrating immune cells and survival. Int J Cancer 2020;147(2):423–39 doi 10.1002/ijc.32785. [DOI] [PubMed] [Google Scholar]
- 22.Ma J, Li J, He N, Qian M, Lu Y, Wang X, et al. Identification and validation of a novel survival prediction model based on the T-cell phenotype in the tumor immune microenvironment and peripheral blood for gastric cancer prognosis. J Transl Med 2023;21(1):73 doi 10.1186/s12967-023-03922-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554(7693):544–8 doi 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gavrielatou N, Liu Y, Vathiotis I, Zugazagoitia J, Aung TN, Shafi S, et al. Association of PD-1/PD-L1 Co-location with Immunotherapy Outcomes in Non-Small Cell Lung Cancer. Clin Cancer Res 2022;28(2):360–7 doi 10.1158/1078-0432.Ccr-21-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giraldo NA, Nguyen P, Engle EL, Kaunitz GJ, Cottrell TR, Berry S, et al. Multidimensional, quantitative assessment of PD-1/PD-L1 expression in patients with Merkel cell carcinoma and association with response to pembrolizumab. J Immunother Cancer 2018;6(1):99 doi 10.1186/s40425-018-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515(7528):568–71 doi 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson DB, Bordeaux J, Kim JY, Vaupel C, Rimm DL, Ho TH, et al. Quantitative Spatial Profiling of PD-1/PD-L1 Interaction and HLA-DR/IDO-1 Predicts Improved Outcomes of Anti-PD-1 Therapies in Metastatic Melanoma. Clin Cancer Res 2018;24(21):5250–60 doi 10.1158/1078-0432.Ccr-18-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sánchez-Magraner L, Miles J, Baker CL, Applebee CJ, Lee DJ, Elsheikh S, et al. High PD-1/PD-L1 Checkpoint Interaction Infers Tumor Selection and Therapeutic Sensitivity to Anti-PD-1/PD-L1 Treatment. Cancer Res 2020;80(19):4244–57 doi 10.1158/0008-5472.Can-20-1117. [DOI] [PubMed] [Google Scholar]
- 29.Zhang W, Hubbard A, Jones T, Racolta A, Bhaumik S, Cummins N, et al. Fully automated 5-plex fluorescent immunohistochemistry with tyramide signal amplification and same species antibodies. Lab Invest 2017;97(7):873–85 doi 10.1038/labinvest.2017.37. [DOI] [PubMed] [Google Scholar]
- 30.Tibshirani R The lasso method for variable selection in the Cox model. Stat Med 1997;16(4):385–95 doi . [DOI] [PubMed] [Google Scholar]
- 31.Simon N, Friedman JH, Hastie T, Tibshirani R. Regularization Paths for Cox’s Proportional Hazards Model via Coordinate Descent. Journal of Statistical Software 2011;39(5):1 – 13 doi 10.18637/jss.v039.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenz HJ, Van Cutsem E, Luisa Limon M, Wong KYM, Hendlisz A, Aglietta M, et al. First-Line Nivolumab Plus Low-Dose Ipilimumab for Microsatellite Instability-High/Mismatch Repair-Deficient Metastatic Colorectal Cancer: The Phase II CheckMate 142 Study. J Clin Oncol 2022;40(2):161–70 doi 10.1200/jco.21.01015. [DOI] [PubMed] [Google Scholar]
- 33.Kim ST, Klempner SJ, Park SH, Park JO, Park YS, Lim HY, et al. Correlating programmed death ligand 1 (PD-L1) expression, mismatch repair deficiency, and outcomes across tumor types: implications for immunotherapy. Oncotarget 2017;8(44):77415–23 doi 10.18632/oncotarget.20492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Girault I, Adam J, Shen S, Roy S, Brard C, Faouzi S, et al. A PD-1/PD-L1 Proximity Assay as a Theranostic Marker for PD-1 Blockade in Patients with Metastatic Melanoma. Clin Cancer Res 2022;28(3):518–25 doi 10.1158/1078-0432.Ccr-21-1229. [DOI] [PubMed] [Google Scholar]
- 35.Zhao P, Li L, Jiang X, Li Q. Mismatch repair deficiency/microsatellite instability-high as a predictor for anti-PD-1/PD-L1 immunotherapy efficacy. Journal of Hematology & Oncology 2019;12(1):54 doi 10.1186/s13045-019-0738-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng D, Qin B, Pal K, Sun L, Dutta S, Dong H, et al. BRAFV600E-induced, tumor intrinsic PD-L1 can regulate chemotherapy-induced apoptosis in human colon cancer cells and in tumor xenografts. Oncogene 2019;38(41):6752–66 doi 10.1038/s41388-019-0919-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available from the corresponding author upon reasonable request.


