Abstract
Nuclear receptor coactivator 2 (Ncoa2) is a member of the Ncoa family of co-activators, and we previously showed that Ncoa2 regulates the differentiation of induced regulatory T cells. However, it remains unknown if Ncoa2 plays a role in CD8+ T-cell function. Here, we show that Ncoa2 promotes CD8+ T cell-mediated immune responses against tumors by stimulating T-cell activation via upregulating PGC-1α expression to enhance mitochondrial function. Mice deficient in Ncoa2 in T cells (Ncoa2fl/fl/CD4Cre) displayed defective immune responses against implanted MC38 tumors, which associated with significantly reduced tumor-infiltrating CD8+ T cells and decreased IFNγ production. Consistently, CD8+ T cells from Ncoa2fl/fl/CD4Cre mice failed to reject tumors after adoptive transfer into Rag1−/− mice. Further, in response to TCR stimulation, Ncoa2fl/fl/CD4Cre CD8+ T cells failed to increase mitochondrial mass, showed impaired oxidative phosphorylation, and had lower expression of PGC-1α, a master regulator of mitochondrial biogenesis and function. Mechanically, T cell activation-induced phosphorylation of CREB triggered the recruitment of Ncoa2 to bind to enhancers, thus, stimulating PGC-1α expression. Forced expression of PGC-1α in Ncoa2fl/fl/CD4Cre CD8+ T cells restored mitochondrial function, T-cell activation, IFNγ production, and anti-tumor immunity. This work informs the development of Ncoa2-based therapies that modulate CD8+ T cell-mediated anti-tumor immune responses.
Keywords: Ncoa2, PGC-1α, mitochondria, CD8+ T cells, tumor, immune response
Introduction
Cytotoxic CD8+ T cells mediate protective immune responses against viral/bacterial infections and cancer (1, 2). Infiltration of functional CD8+ T cells into tumors positively correlates with survival of cancer patients and the success of cancer immunotherapy (3, 4). Naïve CD8+ T cells are not able to attack targets until differentiation into effector T cells. Engagement of the T-cell receptor (TCR) by antigen initiates T-cell activation, which leads to the proliferation and differentiation of a very limited number of antigen-specific naïve CD8+ T cells into a large population of antigen-specific effector cytotoxic T cells (i.e., clonal expansion) capable of killing target cells expressing the antigen.
Naïve, resting T cells are small and inactive, with minimum metabolic activity. However, the engagement of the TCR by antigen initiates a cascade of signaling events that boost metabolism, including protein synthesis, DNA replication, ATP production, and gene transcription required for proliferating and differentiating into larger effector T cells. Mitochondria are the bioenergetic and biosynthetic organelles that play an important role in T-cell function (5-7). In metabolically quiescent, resting T cells, mitochondria efficiently couple oxidative phosphorylation (OXPHOS) to the tricarboxylic acid (TCA) cycle in order to produce the ATP required for T-cell survival and maintenance. Upon TCR stimulation, mitochondrial biogenesis is stimulated, resulting in significantly increased mitochondrial mass and a functional switch (8). The major function of mitochondria in activated T cells switches to generate essential building blocks for synthesis of the macromolecules required for T-cell proliferation (9), and activated T cells rely on less efficient anaerobic glycolysis to generate ATP (10) as well as some building blocks (10, 11). Peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1α) is a critical regulator of mitochondrial biogenesis and activates the expression of many mitochondrial genes (12). Deletion of PGC-1α leads to mitochondrial dysfunction and cellular defects (13, 14), and forced expression of PGC-1α stimulates T cell-mediated immune responses, including anti-tumor immunity, by enhancing mitochondrial function (15, 16). PGC-1α is thus a critical molecule for regulating mitochondrial function. However, little is known about how PGC-1α is regulated in T cells.
Nuclear receptor co-activator 2 (Ncoa2) belongs to a family of Ncoa co-activators consisting of three members, Ncoa1 (or SRC1), Ncoa2 (or SRC2/TIF2/GRIP1), and Ncoa3 (or SRC3/pCIP/ACTR/AIB1). Ncoas do not directly bind to target DNA, they function as co-activators for steroid nuclear receptors and other transcription factors to regulate gene transcription (17). As such, Ncoas are believed to orchestrate transcription programs critical for multiple cellular functions (17). The function of Ncoas in the immune system is not fully known. Our previous study illustrated that Ncoa1 can reciprocally regulate the differentiation of inflammatory T-helper 17 (Th17) cells and inhibitory regulatory T cells (Tregs) (18). Similar to Ncoa1, another study shows that Ncoa3 promotes Th17 differentiation (19), which was also demonstrated by an independent study that further indicated a selective role of Ncoa3 in the differentiation of pathogenic Th17 cells (20). Therefore, Ncoa1 and Ncoa3 have a nonredundant function in stimulating Th17 differentiation. Regarding Tregs, a recent report using germline Ncoa3−/− mice and a Ncoa3 inhibitor suggested a possible function of Ncoa3 in Tregs (21). However, Ncoa3 was found to be dispensable for Treg differentiation (20). We recently discovered Ncoa2’s critical role in the differentiation of induced Tregs (iTregs) (22). However, the function of Ncoa2 in CD8+ T cells remains unknown.
In the present study, we show that Ncoa2 promotes CD8+ T cell-dependent immune responses against tumors via stimulating PGC-1α expression to boost mitochondrial function. CD8+ T cells deficient in Ncoa2 were not effective at mediating tumor rejection, which associated with their reduced proliferation and cytokine production, as well as defective mitochondrial function including reduced mitochondrial mass and oxidative phosphorylation. Consistently, PGC-1α, which controls the expression of many mitochondrial genes, was downregulated in Ncoa2fl/fl/CD4Cre CD8+ T cells and forced expression of PGC-1α in Ncoa2fl/fl/CD4Cre CD8+ T cells rescued mitochondrial function, T-cell activation, and CD8+ T cell-mediated rejection of tumors. Therefore, our results demonstrate a previously unknown function of Ncoa2 in CD8+ T-cell activation by promoting PGC-1α-dependent mitochondrial function.
Materials and Methods
Mice
C57BL/6J (IMSR_JAX:000664), OT1 (IMSR_JAX:003831), CD4Cre (IMSR_JAX:022071), Rag1−/− (IMSR_JAX:002216), CRISPR/Cas9-EGFP (IMSR_JAX:028555), and CD45.1+ (IMSR_JAX:002014) mice were purchased from the Jackson Laboratory. Ncoa2fl/fl mice were gifted by Dr. Jianming Xu (Baylor College of Medicine). Ncoa2fl/fl and CD4Cre mice were crossed to generate the Ncoa2fl/fl/CD4Cre mouse strain, which was further crossed with OT1 mice (gift from Dr. Jianhua Yu, City of Hope) to generate the OT1/Ncoa2fl/fl/CD4Cre strain. All mice were bred on the C57BL/6j background and maintained in a pathogen-free animal facility at the City of Hope. Eight-to-twelve-week-old male mice were used for implanting MC38 or MC38-Ova tumors (sex-matched) as described below; age-matched littermates were used. Both male and female mice were used for in vitro CD8+ T-cell assays as specified. All animal experiments were conducted per the protocols approved by the Institutional Animal Care and Use Committee at the City of Hope (IACUC #07023).
Vectors
MIGR1 retroviral vector was gifted by Dr. Warren S. Pear (University of Pennsylvania). The coding sequences of murine Ncoa2 transcript variant a (NM_008678.3), Ppargc1a (PGC-1α) transcript variant 1 (NM_008904.3), and Creb1 (CREB) transcript variant a (NM_133828.2) were PCR-amplified from the cDNA of wild-type (WT) CD8+ T cells (see sections below). The reverse transcription of mRNA to cDNA was done using Tetro™ cDNA Synthesis Kit (BIO 65043, Meridian Bioscience) and oligo (dT)18 primer, which was included in the kit. Additional 3xFlag tag sequence (ATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAG) was added to the 5’ of PGC-1α coding sequence. For measuring PGC-1α-mediated increases in mitochondrial mass using MitoTracker Green (Thermo Fisher Scientific), the EGFP sequence of the MIGR1-3xFlag-PGC-1α vector was replaced with TagBFP sequence. The TagBFP sequence was PCR-amplified from pMSCV-U6sgRNA(Bbs I)PGKpuro2ABFP (Addgene, 102796) to avoid the channel overlapping during flow cytometric analysis. The CREBS133A mutant vector was generated by PCR-amplification of 5’ and 3’ fragments with S133A mutation, and additional BsaI type II restriction sites were incorporated into primers. Subsequently, the two fragments were ligated using Golden gate assembly method. Further PCR-amplified product was used to replace WT CREB in MIGR1 vector. Lentiviral VSg-GFP transfer plasmid (equivalent to pHR’), VSVg envelope plasmid, and gag-pol packaging plasmid were obtained from Dr. Inder M. Verma (Salk Institute for Biological Studies). The Ova sequence was amplified from the cDNA of B16-F10-Ova cancer cells (gift from Jianhua Yu, City of Hope, California) and first inserted into MIGR1 vector. The Ova-IRES-EGFP sequence was then cut from this vector between BglII and SalI restriction sites and inserted into VSg-GFP vector digested with BamHI and SalI restriction enzymes, replacing the original EGFP sequence. PGL3-Basic and pGL3-SV40-Renilla luciferase vectors were purchased from (Promega, Madison, WI). The PGC-1α luciferase vector (P-E1) containing a 2-kb promoter in front of the luciferase coding sequence was obtained from Addgene (plasmid # 8887). PGL3-TK was cloned by inserting the core sequence of thymidine kinase (TK) promoter between BglII and HindIII sites of PGL3-basic vector. A DNA sequence of approximately 1.5 kb, expanding the Rgn5 region, was inserted upstream TK core promoter of the PGL3-TK luciferase vector (I-E2-TK). Vectors containing truncated enhance elements (P-ΔE1 and I-ΔE2-TK) were cloned by PCR-amplification of 5’ and 3’ residue fragments of deleted regions and ligation of two fragments using additional introduced BbsI restriction sites through the Golden gate assembly method. pMSCV-U6sgRNA(Bbs I)PGKpuro2ABFP was used for Cas9-mediated genomic gene deletion with single-guide RNAs (sgRNAs) and a MIGR1 backboned vector with dual sgRNA cassettes and one phosphoglycerate kinase (PGK) promoter–TagBFP cassette was previously generated for large gene fragment deletion (21).
Cell lines
MC38 colon cancer cells were obtained from Dr. Mingye Feng (City of Hope, California), and HEK293T cells were gifted by Dr. Saul Priceman (City of Hope, California). The Ova-expressing MC38 cell line was generated by lentiviral transduction of Ova to MC38 cells as described above. Cells were cultured in Dulbecco’s Modified Eagle Medium (Corning Inc.) supplemented with 10% fetal bovine serum (Corning Inc.) and 1% penicillin-streptomycin (Gibco).
Antibodies
The following antibodies against mouse proteins were purchased from BioLegend: CD3 (145-2C11), CD28 (37.51), TruStain FcX™ (anti-mouse CD16/32, clone 93), CD45 (clone 30-F11), CD4 (RM4-5), CD8 (53-6.7), IFNγ (XMG-1.2), PD-1 (29F.1A12), Lag3 (C9B7W), Tim3 (RMT3-23), TNFα (MP6-XT22), CD69 (H1.2F3), and CD25 (PC61). The antibody against Foxp3 (FJK-16s) was obtained from eBioscience, and anti-PGC-1α (NBP1-04676) was bought from Novus Biologicals. The H-2K(b) SIINFEKL tetramer (Ova tetramer) was provided by the National Institute of Health (NIH) Tetramer Core Facility. Rabbit IgG (P120-101) and anti-Ncoa2 (A300-346A) were purchased from Bethyl Laboratories. CREB (48H2) and phospho-CREB (Ser133) (87G3) antibodies were obtained from Cell Signaling Technology.
Viral transduction
The generation of retrovirus was done by transfection of empty or the PGC-1α vector into Platinum-E (Plat-E; Cell Biolabs) packaging cells using BioT transfection reagent (Bioland Scientific). For lentivirus, the expression vector (Ova), packaging vector (gag-pol), and VSV-g envelope vector were mixed at a 3:2:1 ratio and transfected to HEK293T cells using the BioT reagent. Fresh medium was changed 10 hours post-transfection, and the virus-containing medium was collected at 24 and 48 hours thereafter. Retroviral or lentiviral supernatants were filtered with a 0.45 μm polyvinylidene difluoride (PVDF) syringe filter (Millipore) and supplemented with 10 μg/mL polybrene (Sigma-Aldrich), 10 ng/mL IL2 (Biolegend), and 50 μM β-mercaptoethanol (Gibco). Activated CD8+ T cells (4 x 105; see “T-cell in vitro culture” section below) were suspended in the retroviral medium and centrifuged in a 24-well plate at 2500 rpm for 2 hours, followed by additional incubation at 37 °C for 3 hours and expansion in fresh culture medium (see “Acute and chronic T-cell activation” section).
CRISPR-Cas9-mediated genomic DNA deletion
To perform genomic gene deletion using CRISPR-Cas9, single-guide RNA (sgRNA) sequences targeting coding regions of Ncoa1 and Ncoa3 genes were obtained from a library (Addgene, library 67988) and cloned into the blue fluorescent protein (BFP)-expressing pMSCV-U6sgRNA(Bbs I)PGKpuro2ABFP vector. For the deletion of the Ppargc1a promoter or intronic elements, two respective gRNAs were designed using an online tool (CRISPOR, http://crispor.tefor.net/) to target the 5’ and 3’ ends of the desired regions. Subsequently, the gRNAs for each Ppargc1a gene regions were cloned into a previously validated vector containing two U6 promoter-sgRNA cassettes and a phosphoglycerate kinase (PGK) promoter-BFP cassette (21). The sgRNAs were delivered to Cas9/EGFP-expressing cells via retroviral transduction. BFP+/GFP+ cells were analyzed using BD LSRFortessa or sorted using FACS Aria Fusion II (BD Biosciences) to validate gene deletion and measure mRNA levels. The “regular” PCR primers for determining gene deletion are listed in Table S1, and gRNA sequences are listed in Table S2.
Luciferase assay
PGC-1α luciferase vector containing a 2-kb promoter in front of the luciferase coding sequence was obtained from Addgene (plasmid # 8887). A DNA sequence of approximately 1.5 kb, expanding the Rgn5 region, was inserted upstream of the thymidine kinase (TK) core promoter previously placed in the basic PGL3 luciferase vector. The Rgn2 in the promoter element and Rgn5 in the intronic element were truncated by ligating the 5’ and 3’ residue fragments via Golden Gate assembly. The resulting PCR product was then amplified and inserted into the PGL3 basic vector to replace the full-length promoter or intronic sequence.
To measure the promoter or intronic enhancer activity, the PGL3-basic vector (200 ng), PGC-1α promoter or intronic enhancer luciferase vector alone (200 ng), or PGC-1α promoter or intronic enhancer luciferase vector together with the Ncoa2 or CREB vector (2 μg) was delivered to 4 x 105 HEK293T cells seeded in a 6-well plate. To assess the promotive effect of Noca2 on CREB-mediated luciferase expression, a low amount of CREB or Ncoa2 vector (500 ng each) was used to observe any synergistic effects. Rinella luciferase vector (100 ng) was co-transfected to cells in each group for normalizing different transfection efficiencies. An empty vector was used to adjust the total plasmid DNA to the same amount. Luciferase activity was measured in Dual-Luciferase® Reporter Assay System (Promega, Madison, WI) per the manufacturer’s instruction at 1-day post-transfection in a Synergy HTX multi-mode reader (Agilent, Santa Clara, CA). To determine luciferase activity, a previously reported calculation method was used (23). Briefly, after background subtraction, Firefly luciferase activities were normalized to Renilla luciferase values, followed by an additional normalization of all Firefly to Renilla ratios to the PGL-basic group.
Primary T-cell isolation
Spleens from WT, Ncoa2fl/fl/CD4Cre, OT1, OT1/Ncoa2fl/fl/CD4Cre or CRISPR/Cas9-EGFP naïve mice were harvested and smashed in a 40-μm cell strainer using RPMI 1640 medium (Corning Inc.) to make single-cell suspensions. Splenocytes were further treated with red blood cell lysis buffer (Invitrogen) and then resuspended in Robo buffer (STEMCELL Technologies). Total splenocytes were sequentially labeled with antibody cocktails and magnetic beads from the Naive CD4+ T-cell Isolation Kit (130-104-453, Miltenyi Biotec) or Naive CD8+ T-cell Isolation Kit (130-096-543, Miltenyi Biotec) and passed through LS columns (130-042-401, Miltenyi Biotec) placed in a QuadroMACS™ separator (130-091-051, Miltenyi Biotec) to negatively select naïve CD4+ or CD8+ T cells via magnetic cell sorting (MACS). The purity of isolated cells was higher than 95%. Thymus and inguinal lymph nodes of naïve WT and Ncoa2fl/fl/CD4Cre mice were harvested and homogenized through a 40-μm cell strainer using RPMI 1640 medium (Corning Inc.). Single-cell suspensions were then subjected to flow cytometry to examine normal T-cell development, as described below.
T-cell in vitro culture
For in vitro assays and PGC-1α transduction, 4 x 105 WT, Ncoa2fl/fl/CD4Cre, OT1, or OT1/Ncoa2fl/fl/CD4Cre naïve CD8+ T cells (see “Primary T-cell isolation” section above) were suspended in RPMI 1640 medium (Corning Inc) containing 10% FBS (Corning Inc.), 2 mM L-glutamine, 100 U/ml penicillin, 100 mg/ml streptomycin (10378-016, Gibco) and 50 μM β-mercaptoethanol and activated in a 24-well plate coated with anti-CD3 (2 μg/mL, Biolegend) and anti-CD28 (5 μg/mL, Biolegend) for 48 hours. Murine recombinant IL2 (10 ng/mL, Biolegend) was supplemented at 24 hours and during further expansion as described below. To determine mitochondria-dependent T-cell activation, naïve WT and Ncoa2fl/fl/CD4Cre CD8+ T cells were activated in the presence of 2.5 ng/mL oligomycin (Sigma-Aldrich) for 12 hours, followed by the measurement of CD69 and CD25 expression by flow cytometry as described below.
Acute and chronic T-cell activation
Following initial activation for 2 days, WT or Ncoa2fl/fl/CD4Cre CD8+ T cells were cultured at 1 x 106 cells/mL in RPMI 1640 medium supplemented with 10% FBS, 2 mM L-glutamine, 100 U/mL penicillin, 100 mg/mL streptomycin, 50 μM β-mercaptoethanol, and 10 ng/mL IL2, with continuous activation by plate-bound anti-CD3 (2 μg/mL) for additional 8 days (chronic activation). Cells were passaged to a new plate coated with anti-CD3 every 2 days. For acute activation, CD8+ T cells were passaged into plates without pre-coated anti-CD3. With forced expression of PGC-1α, CD8+ T cells were retrovirally transduced with MIGR1 empty vector or MIGR1-3xFlag-PGC-1α vector at day 2 after initial activation and subjected to acute or chronic activation conditions until day 8, at which point cells were used for experiments.
Tumor engraftment
MC38 or MC38-Ova cells (number of cells in figure legends) were suspended in 100 μL PBS solution containing 25% Matrigel (Corning Inc.). For engraftment to WT or Ncoa2fl/fl/CD4Cre mice, 2 x 105 MC38 or 3 x 106 MC38-Ova tumor cells were subcutaneously injected. For demonstrating the effects of Ncoa2 deficiency in CD8+ T cells specifically, 0.5 x 106 naïve WT or Ncoa2fl/fl/CD4Cre CD8+ T cells (from OT1 mice) were mixed with an equal number of WT CD4+ T cells and then adoptively transferred to MC38-Ova tumor-bearing Rag1−/− mice on day 5 post tumor engraftment. Tumor size was measured every two days, and mice were euthanized on day 15 for analysis as described below. For measuring cell proliferation in vivo, isolated naïve OT1 CD8+ T cells were stained with cell trace violet dye (Thermo Fisher Scientific) and then adoptively transferred to MC38-Ova tumor-bearing Rag1−/− mice on day 3 post-implantation. Proliferation was then assessed 3 days-post via flow cytometry as specified below. For adoptive transfer models, (i) 5 x 105 MC38-Ova cells were engrafted to Rag1−/− mice and OT1 or OT1/Ncoa2fl/fl/CD4Cre naïve CD8+ T cells and WT naïve CD4+ T cells were adoptively co-transferred, or (ii) 1 x 105 MC38-Ova cells were implanted with OT1 or OT1/Ncoa2fl/fl/CD4Cre CD8+ T cells that were in vitro-activated and retrovirally transduced with EV or PGC-1α alone (no helper T cells). Tumor volumes were measured using a caliper every 2 days and tumor volume was calculated by volume = (length X width2)/2. For RNA-seq analysis, total CD8+ T cells from OT1 and OT1/Ncoa2fl/fl/CD4Cre mice were sorted using a FACS Aria Fusion II (BD Biosciences) from single-cell suspensions of splenocytes collected from mice engrafted with MC38-Ova cells on day 3 post-implantation.
Tumor digestion and cell isolation
Tumors were weighed for enzymatic digestion using a tumor dissociation kit (Miltenyi Biotech, 130-096-730) per the manufacturer’s instructions (1 g per reaction). Briefly, tumors were minced into small pieces and transferred to a gentleMACS™ C Tube (130-093-334) containing 2.35 mL RPMI 1640 medium (Corning Inc.) supplemented with Enzyme A, D, and R. The mixture was incubated at 37 °C with continuous and gentle rotation for 40 minutes using a gentleMACS Dissociator with heaters (37C_m_TDK_1 program) and rotated at 160 rpm in a rotator shake (New Brunswick) at 37 °C for another 40 minutes. Tissues were further dissociated at a relatively high-speed using a build-in program (m_impTumor_02) with the gentleMACS Dissociator. Single-cell suspensions were passed through a 70 μm cell strainer and washed 2-3 times using PBS, followed by the removal of erythrocytes using red blood cell lysis buffer. Cells were then resuspended in a Robo buffer (STEMCELL Technologies) and subjected to direct flow cytometric analysis or cell sorting for measuring mitochondrial oxygen consumption rate (OCR) as indicated below.
In vitro killing assay
MC38-Ova cells were treated with 1 ng/mL IFNγ (Biolegend) overnight to induce the expression of MHC-I. The tumor cells were then plated in a 6-well plate (2 × 105 cells per well). The same number of OT1 or OT1/Ncoa2fl/fl/CD4Cre CD8+ T cells (see “T-cell in vitro culture” section above) was added to the MC38-Ova culture at 6 hours and co-incubated for 16 hours in the presence of a low concentration of IL2 (1 ng/mL). Floating cells (OT1 cells and dead cells) and trypsinized cancer cells (GFP+) were collected and mixed with 1 x 105 GFP-negative counting cells (MC38) for subsequent flow cytometric analysis as specified below. Dead cells and CD8+ T cells were gated out to show the relative percentage of surviving cancer cells and counting cells. Cell survival was calculated as the percentage of surviving cells in experimental groups to the control group without OT1 cells. The killing capacity was calculated as 100% - cell survival percentage.
Flow cytometry
Single-cell suspensions were stained with LIVE/DEAD Fixable Near IR dye (Thermo Fisher Scientific) in PBS at room temperature for 5 minutes before staining. Before staining surface proteins, cells were first incubated with TruStain FcX™ (anti-mouse CD16/32) in RoboSepTm buffer (STEMCELL Technologies) at 4 °C for 10 minutes to block non-specific binding. Surface proteins were stained with fluorophore-conjugated antibodies at 4 °C for 15-30 minutes. For detecting cytokine production, cells were suspended in RPMI 1640 medium (Corning Inc) containing 10% FBS, 2 mM L-glutamine, 100 U/mL penicillin, 100 mg/mL streptomycin, and 50 μM β-ME and stimulated with 50 ng/mL PMA (Sigma-Aldrich) and 750 ng/mL ionomycin (Sigma-Aldrich) in the presence of Golgistop™ (BD Bioscience) at 37 °C for 4 hours. For staining intracellular cytokines, cells were fixed in CytoFix/CytoPerm buffer (BD Bioscience) for 20 minutes, washed using 1x Perm/Wash buffer (BD Bioscience), and stained with antibodies in the Perm/Wash buffer at 4 °C for 15 minutes. For staining transcriptional factors or other intracellular proteins, cells were fixed in TF Fix/Perm buffer (BD Bioscience) at 4 °C for 20 minutes, washed once with 1x TF Perm/Wash buffer (BD Bioscience), and stained with antibodies against target proteins in the TF Perm/Wash buffer at 4 °C for 15 minutes. To measure cell proliferation, naïve CD8+ T cells were stained with CellTrace™ Violet dye (Thermo Fisher Scientific) in PBS (1:5000) at room temperature for 20 minutes. These cells were then subjected to in vitro stimulation with anti-CD3/CD28 or adoptive transfer to MC38-Ova bearing Rag1−/− mice as described above. The flow cytometric analysis was done on day 2 and day 3, respectively. For measuring mitochondrial mass or membrane potential, cells were stained with 100 μM MitoTracker™ Green FM (Mito-Green, Thermo Fisher Scientific) and 10 μM MitoTracker™ Deep Red FM (MDR, Thermo Fisher Scientific) in PBS at room temperature for 20 minutes before the surface staining. Subsequent analysis was performed in the BD LSRFortessa flow cytometer.
Metabolism assay
For in vitro activated CD8+ T cells, naïve cells were activated using plate-bound anti-CD3/CD28 for 2 days as described. For tumor-infiltrating CD8+ T cells, cells were sorted from single-cell suspension of MC38 tumors engrafted to WT or Ncoa2fl/fl/CD4Cre mouse (see “Tumor digestion and cell isolation” section above) using FACS Aria Fusion II (BD Biosciences) and were subsequently stimulated with plate-bound anti-CD3/CD28 antibodies for 2 days. Cells were resuspended with XF DMEM medium (Agilent Technologies) supplemented with 10 mM glucose, 2 mM L-glutamine, and 1 mM sodium pyruvate and plated in a Seahorse XF 96-well plate (Agilent Technologies) at 2 x 105 cells per well. The OCR was measured using the Seahorse XF Cell Mito Stress Test Kit (103015-100, Agilent Technologies) under basal conditions and after sequential injection of oligomycin (2.5 mM), carbonyl cyanide-4-trifluoromethoxyphenylhydrazone (FCCP, 1.0 μM), and rotenone/antimycin (0.5 μM). The extracellular acidification rate (ECAR) was measured using the Seahorse XF Glycolysis Stress Test Kit (103020-100, Agilent Technologies) under glucose-depleted conditions and sequential injection of glucose (10 mM), oligomycin (1.0 μM), and 2-deoxy-D-glucose (50 mM). Data were analyzed using the Seahorse Wave software. To measure mitochondrial mass or membrane potential, cells were respectively stained with Mito-Green and MDR, then analyzed using flow cytometry, as described above.
Chromatin immunoprecipitation (ChIP)
A public Ncoa2 ChIP-seq dataset (GSE53039) of mouse liver from GEO database was used to predict Ncoa2 binding sites in the Ppargc1a gene locus. Reads were analyzed using Partek Flow through alignment to the mm10 mouse reference genome using the Burrow-Wheeler aligner (BWA). Peaks were identified with the model-based analysis of ChIP-seq 2 (MACS2) tool (version 2.1.1) and quantified with a minimum region size of 50 bp. Primers were designed to detect an Ncoa2 non-binding site in the Ppargc1a promoter (Rgn1), a potential binding site within the promoter (Rgn2), and three intronic sites showing potential binding (Rgn3-5).
In vitro-cultured CD8+ T cells (2 x 107 cells/per reaction) were fixed in 1% formaldehyde at room temperature for 10 minutes to cross-link proteins with chromatin. The reaction was stopped by incubating in glycine for 5 minutes. Genomic DNA was fragmented with an enzyme cocktail (ChIP-IT Express Enzymatic kit, Active Motif) for 10 minutes as directed. Cell lysates were centrifuged at 15,000 rpm for 10 minutes to remove debris, and the supernatant was used for immunoprecipitation after quantification following the manufacture’s protocol. The following steps were performed using components from ChIP-IT High Sensitivity kit (#103930, Active Motif) as per the manufacture’s guide. Briefly, an equal amount of DNA (30 μg/210 μL per reaction) was incubated with 5 μg of IgG control or anti-Ncoa2 overnight, followed by precipitation with protein G agarose beads. Beads complexed with DNA fragments were extensively washed 5 times using ChIP buffer in ChIP filtration columns, and DNA was eluted with 100 μL elution buffer followed by reverse cross-linking through an incubation with 20 μg proteinase K at 55 °C for 30 minutes, followed by an additional 2 hours at 80 °C. Five volumes of DNA purification biding buffer (500 μL) was added to the reaction mix, and the pH was adjusted by adding 5 μL of 3 M sodium acetate. Subsequent DNA purification was performed using DNA columns included in the kit. The recovered DNA was subjected to RT-qPCR with 1/25 of total amount as DNA template using primers listed in Supplementary Table S1 and was performed using the PowerUp™ SYBR™ Green Master Mix (Applied Biosystems) in a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific). Gene expression was calculated using 2−ΔΔCt method and normalized to hemoglobin beta chain complex (Hbb) gene. Subsequently, the values were further normalized to the IgG control group. Data from three independent experiments were collected for analysis.
RNA-seq analysis
WT or Ncoa2fl/fl/CD4Cre OT1 mice were challenged with MC38-Ova cancer cells for 3 days. CD8+ T cells from the spleens were sorted as described above for RNA extraction with the RNeasy® mini kit (QIAGEN). RNA quality and quantity were assessed using a BioAnalyser Systems (Agilent Technologies) at Novogene. Selected samples with RNA Integrity Number (RIN) higher than 8.0 were subjected to sequencing library preparation. For RNA-seq data analysis, sequence reads passing quality check were preprocessed for adaptor and polyA removal using trimmomatic v.0.39 and fastp v.0.23.2, followed by mapping to the mouse reference genome mm10, with an index build containing the transcript/gene annotations using STAR v.2.7.9a. Paired-end fragment counts of each gene annotated in RefGene were calculated by the featureCounts function in the Subread package v.1.6.4. To promote the accuracy of the gene expression estimates, a total of 13561 genes with counts per million (CPM) > 0.1 in at least three samples were selected for downstream analysis. Bioconductor package edgeR v.3.32.1 was applied with a design matrix for testing differential expression between Ncoa2fl/flCD4Cre and WT samples based on quasi-likelihood F-tests. Statistical p-values were adjusted by Benjamini and Hochberg method. Genes below the false discovery rate of 0.05 were considered significant.
Gene set enrichment analysis (GSEA).
Preranked Gene Set Enrichment Analysis was conducted for functional enrichment analysis with Gene Ontology (GO) resource obtained from mouse Molecular Signatures Database (MSigDB) collections v2022.1. Genes from edgeR results were ranked by the sign of logFC in combination with −log10 of the p-value for this analysis.
Statistical analysis
Data are presented as mean ± SEM unless otherwise specified. Statistical analyses were performed using Student’s t-test (two-tailed, unpaired). The P values were represented as follows: *P< 0.05, **P< 0.01, ***P< 0.001; ns, not significant. A P value less than 0.05 was considered statistically significant.
Data and materials availability
The reagents, including DNA construct and mice, will be provided by Z.S. pending scientific review and a completed material transfer agreement. Requests for such reagents should be submitted to mta@coh.org. All data needed to evaluate the paper’s conclusions are present in the paper and/or the Supplementary Materials. The RNA sequencing data from this paper are available in the GEO database (accession code: GSE224019).
Results
Ncoa2-deficient mice exhibit defective anti-tumor immune responses
To determine the function of Ncoa2 in conventional T cells, Ncoa2fl/fl mice were crossed to CD4Cre mice (Ncoa2fl/fl/CD4Cre) to conditionally delete Ncoa2 in T-cell compartments, which was confirmed by the lack of Ncoa2 mRNA (Supplementary Fig. S1A) (22). The body weight of Ncoa2fl/fl/CD4Cre mice was comparable to that of wildtype (WT) mice at different ages (Supplementary Fig. S1B). No obvious defects in thymic cellularity and distribution of thymocyte populations (CD4−CD8− double negative (DN), CD4+CD8+ double positive (DP), and CD4+ or CD8+ single positive (SP) cells) were observed in Ncoa2fl/fl/CD4Cre mice (Supplementary Fig. S1C-D). Further, WT and Ncoa2fl/fl/CD4Cre mice showed comparable spleen cellularity (Supplementary Fig. S1E), which was supported by a comparable percentage of CD4+ and CD8+ T cells in the spleens (Supplementary Fig. S1F). Equivalent percentages of CD4+ or CD8+ T cells were also observed in lymph nodes (Supplementary Fig. S1G). These results suggest normal thymic T-cell development and migration of developed T cells into the periphery.
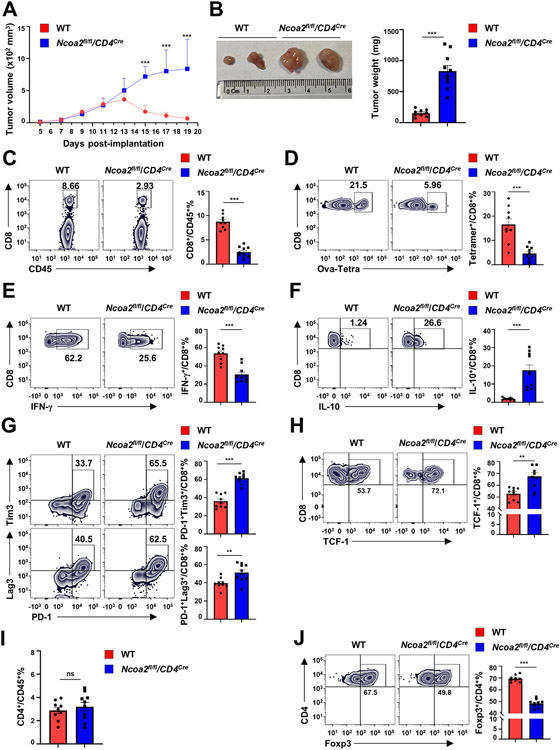
To determine the in vivo function of Ncoa2 in immune responses, we used a subcutaneous MC38 colorectal adenocarcinoma model, which induces CD8+ T cell-mediated anti-cancer responses (24, 25). The implanted ovalbumin (Ova)-expressing MC38 tumor cells exhibited significantly greater growth in Ncoa2fl/fl/CD4Cre mice compared to that in Ncoa2fl/fl (WT) mice, as evidenced by larger tumor volume (Fig. 1A), visually larger tumor size (Fig. 1B, left panel), and increased tumor weight (Fig. 1B, right panel). Because tumor-infiltrating lymphocytes (TILs), especially IFNγ-producing CD8+ T cells, are biomarkers of anti-tumor immune responses (26-28), we analyzed tumor lymphocyte infiltration. Consistent with the higher tumor burden observed in Ncoa2fl/fl/CD4Cre mice, there was a significant decrease in CD8+ T cells in TILs (Fig. 1C; Supplementary Fig. S1H for gating strategy), whereas their survival was comparable to WT CD8+ T cells (Supplementary Fig. S1I). Further analysis using tetramers that recognized the Ova-specific TCR also confirmed significantly reduced Ova-tetramer (Ova-Tetra)+ CD8+ T cells (Fig. 1D). Additionally, the Ova-specific CD8+ population in TILs from Ncoa2fl/fl/CD4Cre mice produced much less IFNγ (Fig. 1E; Supplementary Fig. S1J for IFNγ staining control) and skewed to IL10-producing cells (Fig. 1F; Supplementary Fig. S1K for gating strategy), suggesting CD8+ T-cell dysfunction in the absence of Ncoa2. Consistent with this finding, significant upregulation of exhaustion markers, PD-1, Lag3, Tim3 (Fig. 1G), and TCF-1 (Fig. 1H), in CD8+ TILs was observed. In contrast to reduced TIL CD8+ T cells in Ncoa2fl/fl/CD4Cre mice, CD4+ T cells were comparable between WT and Ncoa2fl/fl/CD4Cre mice (Fig. 1I). Consistent with our previous report (22), Ncoa2fl/fl/CD4Cre mice also showed reduced Tregs (Fig. 1J), which cannot account for the impaired anti-cancer immune responses. Similar defects in immune responses against implanted MC38 tumor (without expression of Ova) were also observed in Ncoa2fl/fl/CD4Cre mice, indicated by reduced tumor size and weight (Supplementary Fig. S1L), decreased TIL CD8+ but not CD4+ T cells (Supplementary Fig. S1M), reduced IFNγ (Supplementary Fig. S1N), increased IL10-producing (Supplementary Fig. S1O) CD8+ T cells, upregulated PD-1, Lag3, and Tim3 exhaustion markers on CD8+ T cells (Supplementary Fig. S1P), and reduced Tregs (Supplementary Fig. S1Q). Therefore, Ncoa2fl/fl/CD4Cre mice are defective in anti-tumor immune responses and this defect associates with reduced IFNγ+CD8+ T-cell infiltration.
Figure 1. Ncoa2 deficient mice are defective in anti-tumor immune responses.
WT and Ncoa2fl/fl/CD4Cre mice were subcutaneously implanted with 3 x 106 MC38-ovalbumin (Ova) cancer cells and euthanized on day 19 post-implantation for assessment (n=9-10, 2 independent experiments). (A-B) Tumor volume (A), size (B, left panel), and weight (B, right panel) of indicated mice. (B) Tumor size and weight were measured at day 19 post-engraftment when mice were eventually euthanized. (C) Flow cytometric analysis of the percentage of CD8+ T cells among CD45+ tumor-infiltrating lymphocytes (TILs) from indicated mice. (D) Representative flow cytometric analysis and percentage of H-2K(b) Ova257-264 tetramer+ (Ova-Tetra+) among tumor infiltrated CD8+ T cells from indicated mice (E-H) Representative flow cytometric analysis (left panels) and percentage (right panel) of IFN-γ+ (E), IL-10+ (F), PD-1+Tim3+ (G, top panels), PD-1+Lag3+ (G, bottom panels), and TCF-1+ (H) cells among tumor-infiltrating CD8+/Ova-Tetra+ T cells from indicated mice. (I-J) Flow cytometric analysis of the percentage of CD4+/CD45+ (I) and Fopx3+/CD4+ in tumors shown in (A). Data are shown as mean ± SEM. **P<0.01; ***P<0.001 (two-tailed unpaired Student’s t-test).
Ncoa2 is essential for CD8+ T cell-mediated tumor rejection
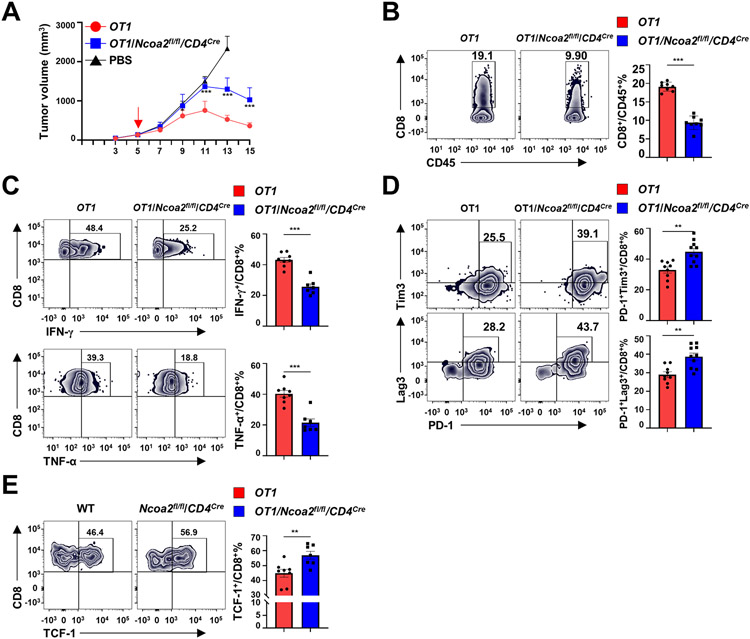
Ncoa2fl/fl/CD4Cre mice lack Ncoa2 in T-cell compartments, including both CD4+ and CD8+ T cells. To specifically determine Ncoa2 function in CD8+ T cells, we crossed Ncoa2fl/fl/CD4Cre mice with OT1 transgenic mice to generate OT1/Ncoa2fl/fl/CD4Cre mice that expressed a TCR recognizing the Ova peptide 257-264 (Ova257-264) presented within the context of MHC-I. OT1 or OT1/Ncoa2fl/fl/CD4Cre donor CD8+ T cells and WT CD4+ T cells were adoptively co-transferred into immunodeficient recipient Rag1−/− mice bearing Ova-expressing MC38 tumor cells (Supplementary Fig. S2A-C). Rag1−/− recipients with OT1/Ncoa2fl/fl/CD4Cre CD8+ T cells showed higher tumor burden compared to recipients with WT OT1 CD8+ T cells (Fig. 2A). As expected, control Rag1−/− mice transferred with PBS (no CD8+ T cells) had the highest tumor burden due to CD8+ T-cell deficiency and thus a lack of CD8+ T cell-driven anti-tumor responses. Compared to Rag1−/− recipients with WT OT1 CD8+ T cells, recipients with OT1/Ncoa2fl/fl/CD4Cre donor cells had a reduced infiltrating CD8+ T-cell responses, including significantly reduced IFNγ- and TNFα-producing CD8+ T-cell infiltrates (Fig. 2B-C; Supplementary Fig. S2D for gating strategy). Additionally, we also observed upregulated expression of exhaustion markers PD-1, Lag3, Tim3 (Fig. 2D), and TCF-1 (Fig. 2E) on CD8+ T cells in recipients treated with OT1/Ncoa2fl/fl/CD4Cre donor cells, further indicating CD8+ T-cell dysfunction. As expected, there were no obvious differences in CD4+ T-cell numbers among CD45+ TILs and Tregs between Rag1−/− mice reconstituted with CD8+ T cells from OT1 versus OT1/Ncoa2fl/fl/CD4Cre mice (Supplementary Fig. S2E-F), excluding the role of Tregs in differential anti-tumor immunity. Overall, these results demonstrated the essential function of Ncoa2 in CD8+ T cell-mediated anti-tumor immune responses.
Figure 2. Ncoa2 is essential for CD8+ T cell-mediated tumor rejection.
(A) Quantification of tumor volume in Rag1−/− recipient mice implanted with 5 x 105 MC38-Ova cancer cells and then adoptively transferred with PBS (n=4) or indicated CD8+ (5 x 105) and WT CD4+ (5 x 105) naïve T cells at day 5 post-engraftment (n=8; 3 independent experiments). (B-F) Representative flow cytometric analysis (left panels) and percentage of CD8+/CD45+ lymphocytes (B), IFN-γ+/CD8+ (C, top panels), TNF-α+/CD8+ (C, bottom panels), PD-1+Tim3+/CD8+ (D, top panels), PD-1+Lag3+/CD8+ (D, bottom panels), TCF1+/CD8+ (E) cells in tumors from Rag1−/− mice shown in (A) on day 15 post-engraftment. Data are shown as mean ± SEM. **P<0.01; ***P<0.001 (two-tailed unpaired Student’s t-test).
Ncoa2 plays an essential role in CD8+ T-cell activation
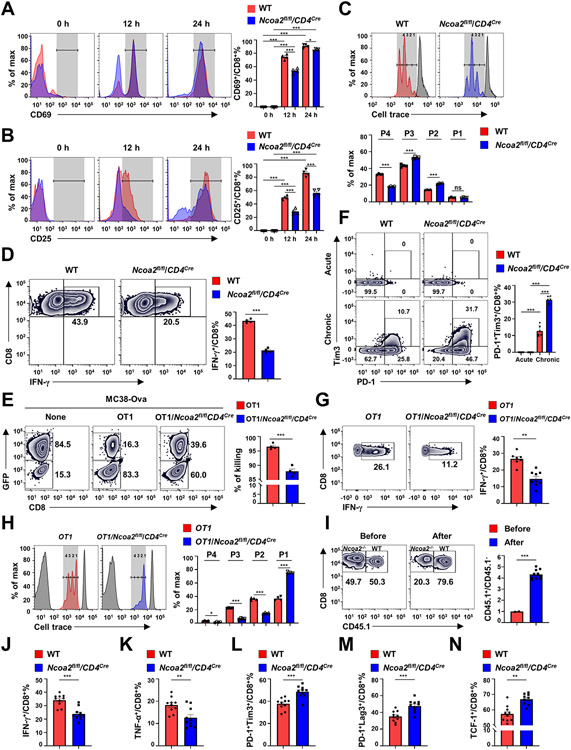
T-cell activation is the first step to initiate adaptive immune responses, so we next determined the function of Ncoa2 in CD8+ T-cell activation. We found impaired activation of CD8+ T cells from Ncoa2fl/fl/CD4Cre mice in response to anti-CD3/CD28 stimulation, as exhibited by significantly reduced expression of activation markers, CD69 and CD25 (Fig. 3A-B). Indeed, Ncoa2fl/fl/CD4Cre CD8+ T cells were defective in activation-induced proliferation and IFNγ production (Fig. 3C-D), but not survival (Supplementary Fig. S3A), compared to WT cells. Activated CD8+ T cells exhibit cytotoxic activity towards targets expressing antigens recognized by the TCR. Thus, an in vitro tumor cell killing assay was performed to determine the cytotoxic capacity of OT1 CD8+ T cells towards co-cultured tumor cells (Supplementary Fig.S3B). OT1/Ncoa2fl/fl/CD4Cre CD8+ T cells had significantly reduced killing capacity compared to WT OT1 CD8+ T cells (Fig. 3E; Supplementary Fig. S3C for gating strategy), indicating Ncoa2 requirement for CD8+ T-cell cytotoxic activity. Chronic stimulation of CD8+ T cells with anti-CD3 for eight days in vitro after initial activation with anti-CD3/CD28 was done to mimic continuous activation by tumor antigens in the tumor microenvironment (Supplementary Fig. S3D) (29, 30). Compared to WT controls, acute stimulation of CD8+ T cells in vitro after initial activation did not upregulate exhaustion markers; however, chronic activation significantly upregulated the exhaustion markers PD-1 and Tim3 on Ncoa2fl/fl/CD4Cre CD8+ T cells (Fig. 3F). This further confirmed CD8+ T-cell dysfunction in the absence of Ncoa2. To observe T-cell activation in vivo, naïve CD8+ T cells from OT1 or OT1/Ncoa2fl/fl/CD4Cre mice were labeled with cell trace dye and adoptively transferred to Rag1−/− recipients, followed by challenge with Ova-expressing MC38 cells. Transplanted OT1/Ncoa2fl/fl/CD4Cre CD8+ T cells consistently showed defects in IFNγ production and proliferation (Fig. 3G-H). The defective proliferation was even more dramatic compared to in vitro-stimulated Ncoa2fl/fl/CD4Cre CD8+ T cells. To compare the activation of WT and Ncoa2fl/fl/CD4Cre CD8+ T cells in the same recipient, naïve CD8+ T cells from CD45.1-expressing WT and CD45.2-expressing Ncoa2fl/fl/CD4Cre mice were mixed at 50:50 ratio and adoptively transferred to Rag1−/− mice (Fig. 3I). The percentage of WT Ova-Tetramer+CD8+ T cells increased to about 80%, whereas Ncoa2fl/fl/CD4Cre Ova-Tetramer+CD8+ T cells were reduced to 20%, and no difference in survival was observed (Supplementary Fig. S3E), confirming defective proliferation in the absence of Ncoa2. Consistently, cells from Ncoa2fl/fl/CD4Cre mice showed reduced production of IFNγ and TNFα (Fig. 3J-K), and upregulated exhaustion markers PD-1, Tim3, Lag3, and TCF-1 (Fig. 3L-N). Altogether, Ncoa2 is required for CD8+ T cell activation-induced proliferation, cytokine production, and cytotoxicity.
Figure 3. Ncoa2 plays an essential role in CD8+ T-cell activation.
(A-B) Representative flow cytometric analysis (left panels) and percentage (right panels) of activation markers CD69 (A) and CD25 (B) among indicated CD8+ T cells in vitro activated by α-CD3/28 antibodies for 0, 12, and 24 h (n=4). (C) Flow cytometric analysis of the proliferation of indicated CD8+ T cells labeled with cell trace dye and stimulated with α-CD3/28 antibodies (top panel). Plots were overlayed with a peak of non-divided cells (gray). The bottom panel is the percentage of indicated cells in each peak (P1-4) shown on the top panel (n=4). (D) Representative flow cytometric analysis (left panel) and quantification (right panel) of IFN-γ+ cells among indicated CD8+ T cells in vitro activated with α-CD3/28 antibodies for 48 h (n=4). (E) Representative flow cytometric analysis (left panel) and quantification (right panel) of cytotoxic activity against MC38-Ova cells by indicated OT1 CD8+ T cells at 16 h post co-culture (n=4 independent experiments). Left panel: Plots show relative percentages between GFP+ targeting cells and GFP− counting cells with dead cells and OT1 cells gated out. (F) Representative flow cytometric analysis (left panels) and percentage (right panel) of PD-1+Tim3+ cells among indicated CD8+ T cells that were acutely (top panels) or chronically (bottom panels) activated with plate-bound α-CD3 antibody for consecutive eight days (n=6; 3 independent experiments). (G) Representative flow cytometric analysis (left panel) and percentage (right panel) of IFN-γ+ among indicated CD8+ T cells adoptively transferred into Rag1−/− mice challenged with Ova-MC38 for 3 days (n=7-9). (H) Representative flow cytometric analysis of in vivo proliferation of indicated OT1 CD8+ T cells labeled with cell trace dye and adoptively transferred to MC38-Ova-bearing Rag1−/− mice (n=4; 2 independent experiments). Peaks of unstained control and non-divided cells were overlayed (gray). The right panel is the percentage of indicated cells in each peak (P1-4) shown on the left panel. (I) Flow cytometric analysis of the percentage of CD45.1 WT and CD45.2 Ncoa2fl/flCD4Cre CD8+ T cells (0.5 x 106) mixed at 50:50 ratio before or after adoptively transferred to Rag1−/− mice which were challenged with 0.1 x 106 MC38-Ova cancer cells (2 independent experiments). (J-F) Percentage of IFN-γ+ (J), TNF-α+ (K), PD-1+Tim3+ (L), PD-1+Lag3+ (M) or TCF-1+ cells among indicated CD45.1 WT or CD45.2 Ncoa2fl/flCD4Cre Ova-Tera+CD8+ T cells from TILs 19 day post tumor engraftment shown in I. Data shown are mean ± SEM. *P<0.05; **P<0.01; ***P<0.001; ns, not significant (two-tailed unpaired Student’s t-test).
Ncoa2 is required for the upregulation of PGC-1α, which is critical for mitochondrial function
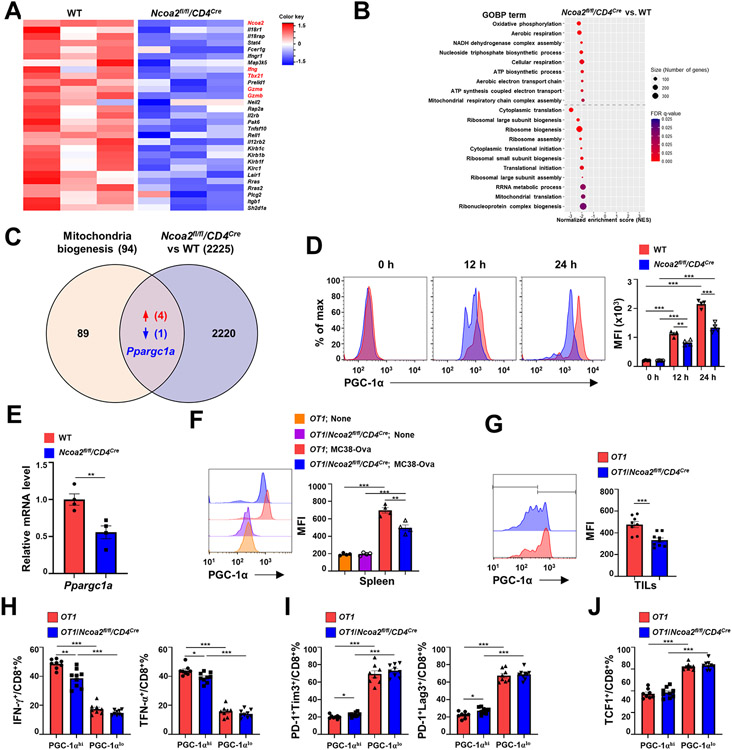
Because Ncoa2 is a transcription co-activator that regulates cellular function via controlling gene expression, we performed RNA-sequencing (RNA-seq) to determine Ncoa2-regulated gene expression. The computational principal component analysis (PCA) revealed that six RNA-seq samples were divided into WT or Ncoa2fl/fl/CD4Cre group (Supplementary Fig. S4A), indicating reproducible gene expression patterns within each group. Of the 860 differentially expressed genes (DEGs) identified, 368 were upregulated and 492 were downregulated between WT versus Ncoa2fl/fl/CD4Cre CD8+ T cells (Supplementary Fig. S4B). Ncoa2 was among the most downregulated genes in Ncoa2fl/fl/CD4Cre CD8+ T cells (Fig. 4A), confirming the deletion of this gene. Additionally, genes critical for CD8+ T-cell activation, including Ifng and cytotoxicity Gzma and Gzmb, were downregulated in Ncoa2fl/fl/CD4Cre CD8+ T cells (Fig. 4A), consistent with the observed defective T-cell activation and cytotoxicity (Fig. 3A-E). Gene set enrichment analysis (GSEA) identified dysregulated gene ontology biological processes (GOBP) in Ncoa2fl/fl/CD4Cre CD8+ T cells (Fig. 4B). A major cellular process strongly affected by Ncoa2 deficiency was mitochondrial metabolism, including oxidative phosphorylation, aerobic respiration, NADH dehydrogenase complex assembly, cellular respiration, ATP synthetic process, aerobic electron transport chain, ATP synthesis-coupled electron transport, and mitochondrial respiration chain complex assembly (Fig. 4B; Supplementary Fig. S4C). Another cellular process affected was protein translation, including translation initiation and ribosome biogenesis and assembly (Fig. 4B). Because mitochondrial activity not only provides energy but also building blocks, amino acids, for translation (9), T-cell activation stimulates mitochondrial biogenesis and energy production that is critical for T-cell proliferation, and impaired mitochondrial function prevents T-cell activation (5-7) as observed in Ncoa2fl/fl/CD4Cre CD8+ T cells. We next cross-examined the genes critical for mitochondrial biogenesis/function (REACTOME_MITOCHONDRIAL_BIOGENESIS) and DEGs with less strict criteria to include more potential upstream regulators (2225 genes) and identified peroxisome proliferator-activated receptor γ coactivator 1-α (Ppargc1a), encoding PGC-1α, a master regulator of mitochondrial biogenesis, which was the only downregulated DEG (Fig. 4C) (12-14). Additionally, we also identified four upregulated genes, Camk4, Crebbp, Ppargc1b, and Perm1, which cannot explain the observed mitochondrial defects (Fig. 4C). Thus, we evaluated PGC-1α expression during CD8+ T-cell activation. Naïve T cells expressed very low PGC-1α, whereas in vitro-activated T cells upregulated PGC-1α, presumably to meet the requirements for increased mitochondrial biogenesis and function (Fig. 4D). However, Ncoa2fl/fl/CD4Cre CD8+ T cells failed to upregulate PGC-1α protein and mRNA expression to that observed in WT controls, indicating an impaired mitochondrial function in the absence of Ncoa2 (Fig. 4D-E). The reduced expression of PGC-1α was also detected in vivo in CD8+ T cells obtained from spleens (Fig. 4F), lymph nodes (Supplementary Fig. 4D), and tumors (Fig. 4G) of OT1/Ncoa2fl/fl/CD4Cre mice versus WT OT1 mice challenged with Ova-expressing MC38. Ncoa2 is thus required for the upregulation of PGC-1α, which is critical for mitochondrial function.
Figure 4. Ncoa2 is required for the upregulation of PGC-1α critical for mitochondrial function.
(A-B) Computational analysis of RNA sequencing (RNA-seq) data of CD8+ T cells sorted from the spleen of OT1 and OT1/Ncoa2fl/fl/CD4Cre mice that were challenged with MC38-Ova cancer cells for 3 days (n=3). (A) Heatmap of CD8 T cell activation-related genes. (B) Gene set enrichment analysis (GSEA) plots of DEGs involved in dysregulated mitochondrial metabolism in Ncoa2fl/fl/CD4Cre CD8+ T cells. (C) Venn diagram of gene overlapping between 94 mitochondrial biogenesis-function-related genes (Reactome database) and 2225 DEGs (including additional genes with fc>2 or fc<−2 in addition to 860 DEGs). (D) Representative flow cytometric analysis (left panels) and mean fluorescent intensity (MFI, right panel) of PGC-1α in CD8+ T cells stimulated by α-CD3/CD28 antibodies for 0, 12, and 24 h. (E) RT-qPCR analysis of PGC-1α mRNA level in indicated CD8+ T cells stimulated by α-CD3/CD28 antibodies for 24 h (n=4). (F-G) Representative flow cytometric analysis (left panels) and MFI of PGC-1α in CD8+ T cells from the spleen (F, n=4) or tumor infiltrated CD8+ T cells (G, n=8-9) of indicated OT1 mice at day 3 (F) or 14 (G) post-challenge with MC38-Ova cells. (H-J) Percentages of IFN-γ+ and TNF-α+ (H), PD-1+Tim3+ and PD-1+Lag3+ (I), and TCF1+ (J) CD8+ T cells in PGC-1αhi or PGC-1αlo populations shown in G. MFI: mean fluorescence intensity. (D-H) Data shown are mean ± SEM. *P<0.05; **P<0.01; ***P<0.001; ns, not significant (two-tailed unpaired Student’s t-test).
We further compared cytokine production and exhaustion markers on PGC-1αhi and PGC-1αlo populations of WT and Ncoa2fl/fl/CD4Cre CD8+ T cells and showed reduced IFNγ and TNFα production in PGC-1αhi but not PGC-1αlo CD8+ T cells from Ncoa2fl/fl/CD4Cre CD8+ mice (Fig. 4H). PGC-1αhi cells have abundant mitochondrial activity and therefore can maintain high T-cell activity, whereas PGC-1αlo cells are relatively inactive due to insufficient mitochondrial activity. Consistent with this finding, both WT and Ncoa2fl/fl/CD4Cre CD8+ cells expressed higher PD-1, Tim3, Lag3, and TCF-1 in PGC-1αlo populations compared to that in PGC-1αhi populations (Fig. 4I-J). Therefore, the defects observed in Ncoa2fl/fl/CD4Cre CD8+ T cells are largely attributed to the higher percentage of PGC-1αlo populations. Again, these results thus support defective T-cell activation in Ncoa2fl/fl/CD4Cre CD8+ T cells due to impaired mitochondrial function.
Ncoa2, recruited by CREB, binds to the Ppargc1a locus to stimulate gene expression
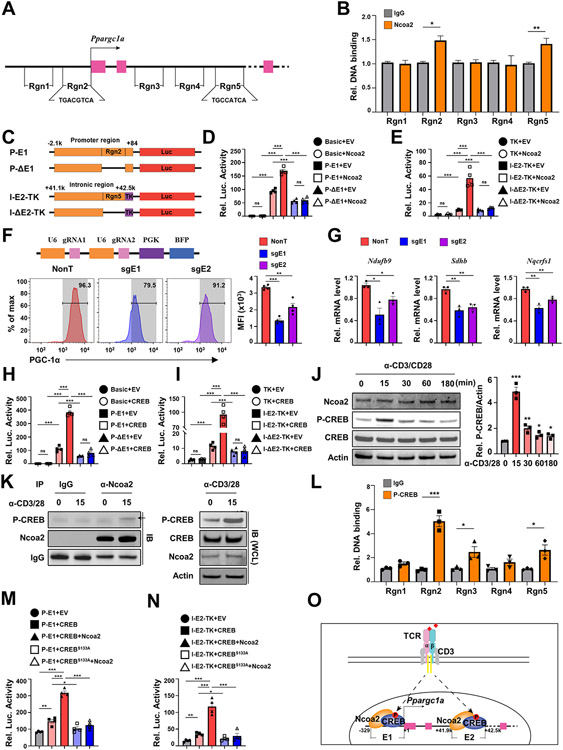
Because activated Ncoa2fl/fl/CD4Cre CD8+ T cells expressed lower PGC-1α, and Ncoa2 is a transcription co-activator, Ncoa2 may transcriptionally activate PGC-1α expression. By analyzing Ncoa2 chromatin immunoprecipitation sequencing (ChIP-seq) data obtained from the liver (GSE53039), we found several possible Ncoa2-binding sites at the Ppargc1a gene locus, including three intronic sites [region 3 (Rgn3)-Rgn5] and one at the promoter region (Rgn2) close to the transcriptional start site (Supplementary Fig. S5A). We designed four pairs of primers to detect those peaks with an additional pair of primers targeting a promoter region without the Ncoa2-binding site (Rgn1) as a negative control (Fig. 5A; Supplementary Fig. S5A). ChIP analysis of activated CD8+ T cells detected enhanced Ncoa2 DNA-binding signals at the Rgn2 (enhancer 1, E1) and Rgn5 (enhancer 2, E2) at the Ppargc1a locus (Fig. 5B). To validate that Ncoa2 stimulated transcription through E1 and E2, we designed four luciferase reporters; two reporters were driven by a 2-kb Ppargc1a promoter element with E1 (P-E1) or without E1 (P-ΔE1); the other two reporters were driven by a basic thymidine kinase promoter (TK) fused downstream of a 1.5-kb intronic element with E2 (I-E2-TK) or without E2 (I-ΔE2-TK) (Fig. 5C). Ncoa2 stimulated the activity of a luciferase reporter driven by P-E1, but failed to stimulate a reporter driven by P-ΔE1 or without a promoter (basic reporter) (Fig. 5D). Also, Ncoa2 stimulated the reporter activity in the presence of E2 (I-E2-TK) but not in the absence of E2 (I-ΔE2-TK) or in the TK promoter only reporter (TK) (Fig. 5E), supporting the notion that Ncoa2 stimulates PGC-1α enhancer activity via E1 and E2. To determine whether E1 and E2 regulated endogenous Ppargc1a expression, the two enhancer regions were deleted individually by CRISPR-Cas9 in the presence of two simultaneously expressed sgRNAs (Fig. 5F, top panel), with the non-targeting (NonT) sgRNAs as the negative control, which was confirmed by PCR analysis (Supplementary Fig. S5B). Deletion of E1 (sgE1) or E2 (sgE2) region reduced the percentage of PGC-1α+ cells, as well as mean fluorescence intensity (MFI) of PGC-1α compared to NonT control (Fig. 5F). Reduced PGC-1α after E1 or E2 deletion also led to decreased expression of known PGC-1α target genes, Ndufb9, Sdhb, and Nqcrfs1 (Fig. 5G). Activated WT CD8+ T cells expressed relatively lower Ncoa1, whereas higher expression of Ncoa2 and Ncoa3, and Ncoa2fl/fl/CD4Cre CD8+ T cells had slightly increased Ncoa1 and Ncoa3 levels, likely due to compensation for lack of Ncoa2 (Supplementary Fig. S5C). In contrast to the deletion of Ncoa2, deletion of Ncoa1 or Ncoa3 did not obviously affect the expression of PGC-1α (Supplementary Fig. S5D), PGC-1α downstream target genes, Ndufb9, Sdhb, and Nqcrfs1 (Supplementary Fig. S5E), and CD8+ T-cell proliferation (Supplementary Fig. S5F), indicating the selective role of Ncoa2 in regulating PGC-1α expression in CD8+ T cells.
Figure 5. Ncoa2 recruited by CREB binds to the Ppargc1a locus to stimulate gene expression.
(A) Scheme of Ppargc1a gene locus. Region 1 (Rgn1) to Rgn5 are the five pairs of PCR primers covered regions. Pink boxes are exons. Arrow is the transcription start site. CREB-binding DNA sequences are identified in Rgn2 and Rgn5. (B) Chromatin immunoprecipitation (ChIP) signals with control IgG or anti-Ncoa2 antibody (Ncoa2) in indicated Rgn from CD8+ T cells activated in vitro (n=3 independent experiments). (C) schematic representation of indicated luciferase reporter constructs. P, Ppargc1a promoter; E1, enhancer 1 at Rgn2; I-E2, intronic enhance 2 at Rgn5; TK, minimal thymidine kinase gene promoter; ΔE, deleted enhancer; Luc, luciferase. (D-E) Relative luciferase activity from indicated reporter transfected into 293T cells together with expression plasmid for Ncoa2 or control empty plasmid (EV). Basic is a promoter-less reporter (n=4-6). (F) Flow cytometric analysis of PGC-1α expression in activated Cas9-expressing CD8+ T cells retrovirally transduced with non-targeting (NonT) sgRNA controls or dual sgRNAs that target to delete Rgn2 containing E1 or Rgn5 containing E2 (n=4). (G) qPCR analysis of indicated PGC-1α downstream mitochondrial genes in control or E1 or E2 deleted CD8+ T cells shown in F. (H-J) relative luciferase activity from indicated reporter transfected into 293T cells together with expression plasmid for CREB or control empty plasmid (EV) (n=4). (K) Immunoblot analysis of indicated proteins in in vitro activated CD8+ T cells by α-CD3/28 antibodies for different times. P-CREB, ser133 phosphorylated CREB. (L) Immunoblot (IB) analysis of P-CREB in control IgG or anti-Ncoa2 antibody immunoprecipitated complexes (IP) from CD8+ T cells stimulated with α-CD3/28 antibodies for 0 or 15 min (n=3). (L) ChIP signals with control IgG or anti-P-CREB antibody in indicated Rgn from CD8+ T cells activated in vitro (n=3 independent experiments). (M-N) Relative luciferase activity from indicated reporter transfected into 293T cells together with expression plasmid for CREB or GREBS133A or control empty plasmid (EV) (M) and Ncoa2 (N). (O) Scheme of the Ncoa2-mediated regulation of Ppargc1a gene expression through recruitment by CREB to E1 and E2. Data shown are mean ± SEM. *P<0.05; **P<0.01; ***P<0.001 (two-tailed unpaired Student’s t-test).
Because Ncoa2 does not have DNA-binding activity, Ncoa2 needs to be recruited by other transcription factors to regulate gene expression. We searched E1 and E2 sequences for transcription factor-binding sites and found conserved CREB-binding sites within both E1 and E2 (Fig. 5A). Indeed, CREB stimulated a reporter driven by P-E1 but not the reporter driven by P-ΔE1 (Fig. 5H). CREB also stimulated a reporter driven by the I-E2-TK but not I-ΔE2-TK or TK promoter alone (Fig. 5I), supporting that CREB, the same as Ncoa2, can stimulate Ppargc1a expression via E1 and E2. Upon T-cell activation, CREB is phosphorylated at serine 133 (31), which activates CREB to stimulate the expression of the genes required for T-cell activation. Mutation of serine 133 to alanine (CREBS133A) to prevent phosphorylation inhibits T-cell activation (32). Indeed, obvious Ser133-phosphorylated CREB was detected 15 mins. After T-cell activation (Fig. 5J; Supplementary Fig. S5G for full blot images), whereas protein levels of both Ncoa2 and CREB were not obviously changed before and after T-cell activation. Phosphorylated CREB was detected in anti-Ncoa2-immunoprecipitated complexes (Fig. 5K; Supplementary Fig. S5H for full blot images), indicating that T cell activation-induced phosphorylation of CREB triggers the recruitment of Ncoa2. A ChIP assay detected P-CREB-binding to E1 and E2 regions in activated CD8+ T cells (Fig. 5L). Further, CREB but not CREBS133A stimulated a reporter driven by P-E1 but not P-ΔE1, and a reporter driven by I-E2-TK but not I-ΔE2-TK or TK promoter alone (Supplementary Fig. 5I). Ncoa2 could significantly further enhance CREB- but not CREBS133A-stimulated luciferase reporter driven by P-E1 or I-E2-TK (Fig. 5M-N). Our results thus support a model whereby TCR-induced phosphorylation of CREB triggers the recruitment of Ncoa2 to stimulate Ppargc1a expression via E1 and E2 (Fig. 5O).
CD8+ T cells deficient in Ncoa2 display defective mitochondrial function
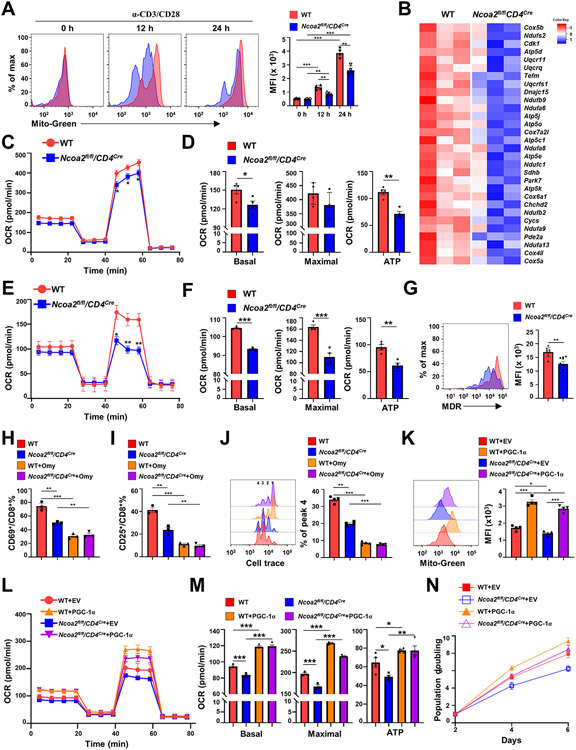
The lower levels of PGC-1α in the absence of Ncoa2 indicated abnormal mitochondrial function, we thus determined how lack of Ncoa2 affected mitochondrial function. T-cell activation stimulates mitochondrial biogenesis to increase mitochondrial mass, which is essential for cell proliferation via PGC-1α upregulation (8). Therefore, we monitored the mitochondrial mass using MitoTracker green (Mito-Green) during T-cell activation. No changes in mitochondrial mass were observed in naïve CD8+ T cells between WT and Ncoa2fl/fl/CD4Cre groups (Fig. 6A). As expected, T-cell activation corresponded with an increase in mitochondrial mass compared to the naïve state; albeit activated Ncoa2fl/fl/CD4Cre CD8+ T cells showed lower levels of mitochondrial mass versus activated WT cells, which was further confirmed by lower levels of mitochondrial DNA (Supplementary Fig. S6A), suggesting defective mitochondrial biogenesis in activated Ncoa2fl/fl/CD4Cre CD8+ T cells. Our GSEA analysis of the DEGs indicated reduced mitochondrial respiration and oxidative phosphorylation in Ncoa2fl/fl/CD4Cre CD8+ T cells due to the downregulation of genes in this pathway (Fig. 4B, Fig. 6B; Supplementary Fig. S4C). Indeed, oxygen consumption rate (OCR), the indicator for mitochondrial respiration, was decreased in Ncoa2fl/fl/CD4Cre CD8+ T cells activated in vitro (Fig. 6C-D) and CD8+ T-cell tumor infiltrates from OT1/Ncoa2fl/fl/CD4Cre mice challenged with Ova-expressing MC38 (Fig. 6E-F). Extracellular acidification rate (ECAR) was increased in Ncoa2fl/fl/CD4Cre CD8+ T cells, likely due to compensation for decreased OCR (Supplementary Fig. S6B). Moreover, mitochondrial membrane potential detected by MitoTracker Deep Red (MDR) was also reduced in tumor-infiltrating Ncoa2fl/fl/CD4Cre CD8+ T cells (Fig. 6G), again indicating impaired mitochondrial function. The defective mitochondrial membrane potential in Ncoa2fl/fl/CD4Cre CD8+ T cells compared to WT cells was further confirmed by staining with tetramethylrhodamine, ethyl ester (TMRE) (Supplementary Fig. S6C). Further, an inhibitor of mitochondrial ATPases, oligomycin (Omy), inhibited the activation of both WT and Ncoa2fl/fl/CD4Cre CD8+ T cells, indicated by reduced upregulation of CD69 (Fig. 6H) and CD25 (Fig. 6I) and decreased T-cell proliferation (Fig. 6J). The differences in CD8+ T-cell activation between WT and Ncoa2fl/fl/CD4Cre CD8+ cells were diminished in the presence of Omy. These results suggest that T-cell activation depends on mitochondrial oxidative phosphorylation and that the differences observed in WT and Ncoa2fl/fl/CD4Cre CD8+ cell activation is largely related to mitochondrial dysfunction. To determine whether downregulated PGC-1α is responsible for the impaired mitochondrial function, PGC-1α was overexpressed in Ncoa2fl/fl/CD4Cre CD8+ T cells by retroviral transduction. Forced expression of PGC-1α increased mitochondrial mass (Fig. 6K), respiration (Fig. 6L-M), and cell proliferation (Fig. 6N) of Ncoa2fl/fl/CD4Cre CD8+ T cells. Therefore, Ncoa2 stimulates CD8+ T-cell activation by directly controlling mitochondrial function in a PGC-1α-dependent manner.
Figure 6. CD8+ T cells deficient in Ncoa2 display defective mitochondrial function.
(A) Representative flow cytometric analysis (left panels) and mean fluorescence intensity (MFI) (right panels) of the mitochondrial mass by MitoTracker Green (Mito-Green) of indicated CD8+ T cells activated in vitro for 0, 12, and 24 h (n=4). (B) Expression heatmap of genes regulating oxidative phosphorylation in indicated CD8+ T cells. Negative Z-score indicates downregulation. (C-F) Seahorse analysis of oxygen consumption rate (OCR) detected over 80 minutes in CD8+ T cells activated in vitro (C and D, n=6-7) or activated in vivo from tumor-infiltrated lymphocytes (E and F, n=8-9). Bar graphs show basal or maximal OCR, and ATP production (D and F). (G) Representative flow cytometric analysis (left panels) and MFI of mitochondrial member potential by MitoTracker Deep Red (MDR) in indicated CD8+ T cells activated in vitro (n=8-9). (H-I) The percentage of CD69+ (H) or CD25+(I) among indicated CD8+ cells in vitro activated in the absence or presence of oligomycin (Omy). (J) Proliferation detected by Cell Trace dye (quantification was done to peak 4) of indicated CD8+ T cells activated by plate α-CD3/28. (K) Representative flow cytometric analysis (merged, left panels) and MFI (right panels) of the mitochondrial mass by Mito-Green of indicated CD8+ T cells retrovirally expressing GFP alone (EV, empty virus) or with PGC-1α and activated in vitro (n=4). (L-M) Seahorse analysis of mitochondrial OCR detected over 80-minutes (L, n=5-6) of indicated CD8+ T cells retrovirally expressing GFP alone (EV) or with PGC-1α and activated in vitro. (M) The basal (left panel), maximal (middle panel) OCR and ATP production calculated from L. (N) Doubling of cell number over 6 days post-in vitro stimulation of indicated CD8+ T cells retrovirally expressing GFP alone (EV) or with PGC-1α. Data shown are mean ± SEM. *P<0.05; **P<0.01; ***P<0.001 (two-tailed unpaired Student’s t-test).
Forced PGC-1α expression restores Ncoa2fl/fl/CD4Cre CD8+ T cell-mediated anti-tumor immunity
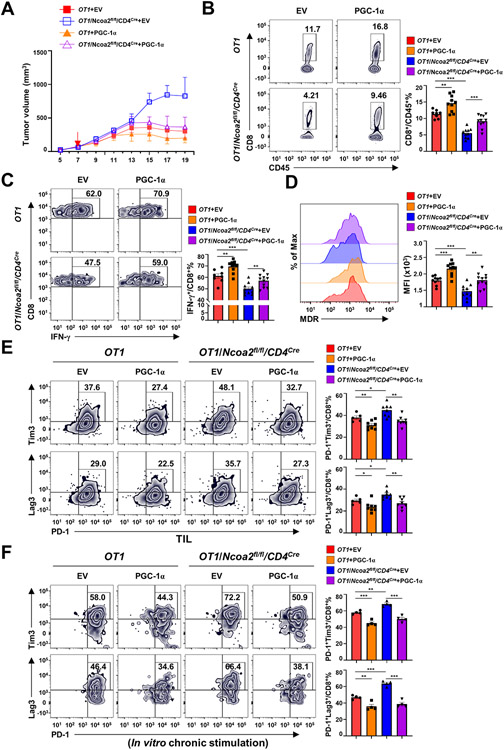
To determine whether Ncoa2-regulated PGC-1α was required for CD8+ T cell-mediated immune responses, we monitored tumor growth in Rag1−/− mice adoptively transferred with sorted OT1 or OT1/Ncoa2fl/fl/CD4Cre CD8+ T cells forcedly expressing PGC-1α (Supplementary Fig. S7A-B). Although tumors grew fast in Rag1−/− mice with OT1/Ncoa2fl/fl/CD4Cre CD8+ T cells expressing GFP alone (empty virus, EV), forced expression of PGC-1α in OT1/Ncoa2fl/fl/CD4Cre CD8+ T cells significantly reduced tumor growth to the levels observed in tumor-bearing Rag1−/− mice with WT OT1 CD8+ T cells, suggesting the restoration of Ncoa2fl/fl/CD4Cre CD8+ T cell-mediated anti-tumor immune responses (Fig. 7A). Consistently, forced expression of PGC-1α increased tumor-infiltrating CD8+ T cells including those producing IFNγ (Fig. 7B-C) and restored mitochondrial function, as indicated by the increased mitochondrial membrane potential (Fig. 7D; Supplementary Fig. S7C; Supplementary Fig. S7D for gating strategy). In addition, forced expression of PGC-1α in OT1/Ncoa2fl/fl/CD4Cre CD8+ cells reduced exhaustion markers PD-1, Lag3, and Tim3 on tumor-infiltrating OT1/Ncoa2fl/fl/CD4Cre CD8+ T cells in vivo, as well as in vitro, in chronically stimulated OT1/Ncoa2fl/fl/CD4Cre CD8+ T cells, indicating rescue of defects due to lack of Ncoa2 (Fig. 7E-F). To rule out the possibility that the observed rescue was due to overexpression of PGC-1α that is higher than WT levels, we adjusted the titer of the retrovirus to express PGC-1α in Ncoa2fl/fl/CD4Cre CD8+ T cells at the levels similar as the WT cells (Supplementary Fig. S7E). We still observed rescue of PGC-1α downstream target genes, Ndufb9, Sdhb, and Uqcrfs1 (Supplementary Fig. S7F), cell proliferation (Supplementary Fig. S7G), and IFNγ production (Supplementary Fig. S7H) in Ncoa2fl/fl/CD4Cre CD8+ T cells. In summary, these results show that Ncoa2 regulates CD8+ T cell-mediated anti-tumor immunity by activating PGC-1α-dependent mitochondrial function.
Figure 7. Forced expression of PGC-1α restores Ncoa2fl/fl/CD4Cre CD8+ T cell-mediated anti-tumor immunity.
(A) Quantification of the volume of tumors in Rag1−/− mice implanted with 5 x 105 MC38-Ova cancer cells and then adoptively transferred with indicated CD8+ (5 x 105) expressing GFP alone (EV) or with PGC-1α for 7 days (n=8-11; 2 independent experiments). (B-E) Representative flow cytometric analysis (left panels) and percentage (B, C, E, and F) or MFI (C) of CD8+/CD45+ cells (B), IFN-γ+/CD8+ cells (C), mitochondrial membrane potential by MDR (D), and PD-1+Tim3+/CD8+ cells (E, top panels) and PD-1+Lag3+/CD8+ cell (E, bottom panels) percentage (E) in tumor infiltrated lymphocytes 19 days post tumor implantation shown in (A). (F) Representative flow cytometric analysis (left panel) and percentage of PD-1+Tim3+/CD8+ (top panels) and PD-1+Lag3+/CD8+ (bottom panels) cells retrovirally expressing GFP alone (EV) or with PGC-1α and chronically activated with plate-bound α-CD3 antibody for 8 days. (B-F) Data are shown as mean ± SEM. *P<0.05; **P<0.01; ***P<0.001 (two-tailed unpaired Student’s t-test).
Discussion
We previously showed that mice deficient of Ncoa2 in Tregs (Ncoa2fl/fl/Foxp3YFP-Cre) had overwhelming T cell-mediated immune responses, leading to severe autoimmunity due to a reduced number of induced Tregs (22). However, mice deficient of Ncoa2 in T-cell compartments (Ncoa2fl/fl/CD4Cre) display impaired T cell-dependent anti-tumor cytotoxicity, which cannot be explained by reduced number and/or impaired function of Tregs. These results led us to explore the function of Ncoa2 in CD8+ T cells since tumor rejection is CD8+ T cell-dependent (33). Here, we uncovered an essential function for Ncoa2 in CD8+ T cells that is separate from its function in Tregs. Our results show that: (1) Rag1−/− mice reconstituted with CD8+ T cells from Ncoa2fl/fl/CD4Cre mice have a defective tumor rejection response, and (2) upon TCR stimulation, there is reduced proliferation and IFNγ production of activated Ncoa2fl/fl/CD4Cre CD8+ T cells both in vitro and in vivo.
Naïve T cells express low PGC-1α, which correlates with relatively low mitochondrial mass and respiration. However, in response to TCR stimulation, T cells upregulate PGC-1α to stimulate the increase of mitochondrial mass and oxidative respiration to meet the demands for activation-induced proliferation and cytokine production (8). In contrast to resting naïve T cells that depend on mitochondrial oxidative respiration to generate the principal source of energy ATP, the other important function of mitochondria in activated T cells is to generate precursor substrates for the biosynthesis of macromolecules required for cell proliferation. For example, the TCA cycle intermediate citrate is exported from mitochondria to the cytosol where it is converted to oxaloacetate and acetyl-CoA (6). While oxaloacetate is used to synthesize aspartate, a precursor for the synthesis of protein and nucleotide purines and pyrimidines (34, 35) and acetyl-CoA is required for lipid synthesis. Pharmacological inhibition of mitochondrial oxidative respiration diminishes T cell activation-induced proliferation (36, 37), partially due to a lack of mitochondrial metabolism-mediated synthesis of macromolecules required for cell proliferation. In this study, we revealed a previously unknown function of Ncoa2 in the regulation of mitochondrial function during CD8+ T-cell activation. In response to TCR stimulation, CD8+ T cells deficient in Ncoa2 had lower mitochondrial activity, indicated by reduced mitochondrial mass and oxidative respiration, which associated with reduced proliferation and IFNγ production compared to WT cells. This result suggests that lower mitochondrial activity is not sufficient to support the demands for activation-induced cell proliferation in the absence of Ncoa2. Our RNA-seq analysis showed that Ncoa2fl/fl/CD4Cre CD8+ T cells expressed lower PGC-1α, critical for mitochondrial function, and forced expression of PGC-1α restored Ncoa2fl/fl/CD4Cre CD8+ T-cell activation and immune responses responsible for tumor rejection. Because PGC-1α expression is fine-tuned to reflect the demands of cellular activity, our research reveals Ncoa2 as a critical regulator for boosting the mitochondrial activity required for CD8+ T-cell activation via upregulation of PGC-1α expression.
PGC-1α is subject to both transcriptional and post-transcriptional regulation (38). We suggest that Ncoa2 transcriptionally promotes PGC-1α expression by interaction with its promoter because Ncoa2 binds to the PGC-1α promoter and activates the PGC-1α-promoter-driven luciferase reporter expression. Given that Ncoa2 lacks direct DNA-binding capacity, it needs to be recruited to the PGC-1α promoter by other transcription factors. Indeed, we identified CREB as the transcription factor that recruits Ncoa2 to the Ppargc1a gene locus, as CREB binds to the same two enhancer regions as Ncoa2 at the Ppargc1a locus to stimulate gene expression. Moreover, TCR stimulation induced CREB phosphorylation that triggered the interaction between CREB and Ncoa2, whereas the non-phosphorylatable form of CREB was not able to stimulate the two-enhancer activity of Ppargc1a. Our results thus illustrate the previously unknown mechanisms responsible for Ncoa2-regulated Ppargc1a expression.
We detected upregulation of some exhaustion markers on Ncoa2fl/fl/CD4Cre CD8+ T cells from the tumor microenvironment or under chronically stimulated conditions. However, we don’t think these are the traditionally defined irreversibly exhausted T cells because they failed to be fully activated initially, suggesting that their dysfunction primarily derived from defective early activation events. As a result, subsequent events, such as T cell effector function and exhaustion, did not occur normally. For example, TCF-1 is typically downregulated in terminally exhausted T cells (39). However, we found higher TCF-1 in Ncoa2fl/fl/CD4Cre CD8+ T cells, which exhibited elevated levels of exhaustion markers Tim3 and PD-1. TCF-1 is highly expressed in naïve T cells and downregulated in response to T-cell activation (39-41). Thus, the relatively higher TCF-1 observed in Ncoa2fl/fl/CD4Cre CD8+ T cells may be attributed to failed downregulation of TCF-1 or other dysregulated pathways resulting from defective initial T-cell activation. Therefore, due to the defective initial T-cell activation, investigating exhaustion in this study was challenging.
Inappropriate activation of T cells leads to various types of autoimmune diseases such as multiple sclerosis, type 1 diabetes, and inflammatory bowel disease (42, 43). Conversely, failed T-cell activation in response to pathogens results in severe immunodeficiency and cancer growth. Therefore, drugs are developed to either boost T-cell activation such as checkpoint inhibitors for cancer treatment or to inhibit T-cell activation such as cyclosporin A for preventing transplantation rejection. Our results suggest that targeting Ncoa2 is a potential treatment for controlling T-cell function. Members of the Ncoa family are considered oncogenes, as they play roles in tumorigenesis in different types of cancers (44, 45). Small-molecule Ncoa inhibitors have already been developed for the treatment of cancers (46, 47). It would be interesting to test whether Ncoa2 inhibitors can prevent CD8+ T cell-mediated autoimmunity. However, the clinical application of Ncoa2 inhibitors is complicated by the fact that Ncoa2 also regulates Tregs. We have shown that Ncoa2 is required for the generation of Tregs (22) and deletion of Ncoa2 in Tregs leads to the development of auto-inflammation. However, the function of Tregs is to inhibit CD4+ and CD8+ T-cell activation via multiple mechanisms including contact-dependent via surface inhibitory molecules such as CTLA-4 and contact-independent mechanisms via secreting inhibitory cytokines such as TGF-β (48). Without the normal function of CD4+ and CD8+ T cells, impaired Treg function may not be sufficient to boost immune responses, which was clearly demonstrated by the failed tumor rejection in mice deficient in Ncoa2. Our observation that CD8+ T cell-mediated immune responses were impaired even though the number of Tregs was significantly reduced in the absence of Ncoa2 supports the feasibility of the application of Ncoa2 inhibitors in treating autoimmunity. In conclusion, our study revealed an important mechanism for Ncoa2-regulated T-cell activation and the future therapeutic potential for targeting Ncoa2 in cancer and autoimmunity.
Supplementary Material
Synopsis.
Nuclear receptor coactivator 2 (Ncoa2) is identified as a critical regulator for boosting the mitochondrial activity during CD8+ T-cell activation via upregulation of PGC-1α expression. The data highlight the potential of targeting Ncoa2 as a cancer therapy.
Acknowledgments
The authors thank Dr. Mingye Feng (City of Hope) for kindly sharing MC38 colon cancer cells and Warren S. Pear (University of Pennsylvania) for the gift of the MIGR1 retroviral vector. We thank NIH Tetramer Core Facility for providing H-2K(b) SIINFEKL (Ova) tetramers for detecting antigen-specific CD8+ T cells. This work was supported by grants from the NIH (R01-AI109644 and R21-AI163256 to Z.S.), institutional pilot funding, Jackie and Bruce Barrow Cancer Research Scholars’ Program, and Caltech-CoH Biomedical Initiative. Research reported in this publication included work performed in the animal, genomic, and flow cytometry cores supported under NIH grant P30CA033572. We thank Dr. Chathurani S. Jayasena for critically reviewing and editing the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- 1.Harty JT, Tvinnereim AR, White DW, CD8+ T cell effector mechanisms in resistance to infection. Annu Rev Immunol 18, 275–308 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Philip M, Schietinger A, CD8+ T cell differentiation and dysfunction in cancer. Nat Rev Immunol 22, 209–223 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenberg SA, Restifo NP, Adoptive cell transfer as personalized immunotherapy for human cancer. Science 348, 62–68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raskov H, Orhan A, Christensen JP, Gogenur I, Cytotoxic CD8+ T cells in cancer and cancer immunotherapy. Br J Cancer 124, 359–367 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinberg SE, Sena LA, Chandel NS, Mitochondria in the regulation of innate and adaptive immunity. Immunity 42, 406–417 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desdin-Mico G, Soto-Heredero G, Mittelbrunn M, Mitochondrial activity in T cells. Mitochondrion 41, 51–57 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Peng G, Mitochondria orchestrate T cell fate and function. Nat Immunol 22, 276–278 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Ron-Harel N et al. , Mitochondrial biogenesis and proteome remodeling promote one-carbon metabolism for T cell activation. Cell Metab 24, 104–117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bantug GR, Galluzzi L, Kroemer G, Hess C, The spectrum of T cell metabolism in health and disease. Nat Rev Immunol 18, 19–34 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Vander Heiden MG, Cantley LC, Thompson CB, Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganeshan K, Chawla A, Metabolic regulation of immune responses. Annu Rev Immunol 32, 609–634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Z et al. , Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98, 115–124 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Leone TC et al. , PGC-1α deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol 3, e101 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J et al. , Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell 119, 121–135 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Dumauthioz N et al. , Enforced PGC-1α expression promotes CD8 T cell fitness, memory formation and antitumor immunity. Cell Mol Immunol 18, 1761–1771 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scharping NE et al. , The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 45, 701–703 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Walsh CA, Qin L, Tien JC, Young LS, Xu J, The function of steroid receptor coactivator-1 in normal tissues and cancer. Int J Biol Sci 8, 470–485 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sen S et al. , SRC1 promotes TH17 differentiation by overriding Foxp3 suppression to stimulate RORγt activity in a PKC-θ-dependent manner. Proc Natl Acad Sci U S A 115, E458–E467 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Z et al. , SRC3 is a cofactor for RORγt in TH17 differentiation but not thymocyte development. J Immunol 202, 760–769 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka K et al. , Regulation of pathogenic T helper 17 cell differentiation by steroid receptor coactivator-3. Cell Rep 23, 2318–2329 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Nikolai BC et al. , Steroid receptor coactivator 3 (SRC-3/AIB1) is enriched and functional in mouse and human Tregs. Sci Rep 11, 3441 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W et al. , Steroid nuclear receptor coactivator 2 controls immune tolerance by promoting induced Treg differentiation via up-regulating Nr4a2. Sci Adv 8, eabn7662 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong X et al. , Decoupling the role of RORγt in the differentiation and effector function of TH17 cells. Sci Adv 8, eadc9221 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juneja VR et al. , PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med 214, 895–904 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang H et al. , Facilitating T cell infiltration in tumor microenvironment overcomes resistance to PD-L1 blockade. Cancer Cell 30, 500 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Mahmoud SM et al. , Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol 29, 1949–1955 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Hwang WT, Adams SF, Tahirovic E, Hagemann IS, Coukos G, Prognostic significance of tumor-infiltrating T cells in ovarian cancer: a meta-analysis. Gynecol Oncol 124, 192–198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galon J et al. , Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313, 1960–1964 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Scharping NE et al. , Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat Immunol 22, 205–215 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vardhana SA et al. , Impaired mitochondrial oxidative phosphorylation limits the self-renewal of T cells exposed to persistent antigen. Nat Immunol 21, 1022–1033 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wen AY, Sakamoto KM, Miller LS, The role of the transcription factor CREB in immune function. J Immunol 185, 6413–6419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barton K et al. , Defective thymocyte proliferation and IL-2 production in transgenic mice expressing a dominant-negative form of CREB. Nature 379, 81–85 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Durgeau A, Virk Y, Corgnac S, Mami-Chouaib F, Recent advances in targeting CD8 T-cell immunity for more effective cancer immunotherapy. Front Immunol 9, 14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birsoy K et al. , An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan LB et al. , Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162, 552–563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang CH et al. , Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sena LA et al. , Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 38, 225–236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez-Marcos PJ, Auwerx J, Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr 93, 884S–890 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Lyu T, Cao Y, Feng H, Role of TCF-1 in differentiation, exhaustion, and memory of CD8+ T cells: A review. FASEB J 35, e21549 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Willinger T et al. , Human naive CD8 T cells down-regulate expression of the WNT pathway transcription factors lymphoid enhancer binding factor 1 and transcription factor 7 (T cell factor-1) following antigen encounter in vitro and in vivo. J Immunol 176, 1439–1446 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Zhao DM et al. , Constitutive activation of Wnt signaling favors generation of memory CD8 T cells. J Immunol 184, 1191–1199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Z, Xie H, Wang R, Sun Z, Retinoid-related orphan receptor γ t is a potential therapeutic target for controlling inflammatory autoimmunity. Expert Opin Ther Targets 11, 737–743 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Zhu J, Yamane H, Paul WE, Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol 28, 445–489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rohira AD et al. , Targeting SRC coactivators blocks the tumor-initiating capacity of cancer stem-like cells. Cancer Res 77, 4293–4304 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suresh S et al. , SRC-2-mediated coactivation of anti-tumorigenic target genes suppresses MYC-induced liver cancer. PLoS Genet 13, e1006650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song X et al. , Development of potent small-molecule inhibitors to drug the undruggable steroid receptor coactivator-3. Proc Natl Acad Sci U S A 113, 4970–4975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y et al. , Small molecule inhibition of the steroid receptor coactivators, SRC-3 and SRC-1. Mol Endocrinol 25, 2041–2053 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakaguchi S, Yamaguchi T, Nomura T, Ono M, Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.