SUMMARY
OSCA/TMEM63s form mechanically activated (MA) ion channels in plants and animals, respectively. OSCAs and related TMEM16s and TMCs form homo-dimers with two pores. Here, we uncover an unanticipated monomeric configuration of TMEM63 proteins. Structures of TMEM63A and TMEM63B (referred to as TMEM63s) revealed a single highly restricted pore. Functional analyses demonstrated TMEM63s are bona fide mechanosensitive ion channels, characterized by small conductance and high thresholds. TMEM63s possess evolutionary variations in the intracellular linker IL2, which mediates dimerization in OSCAs. Replacement of OSCA1.2 IL2 with TMEM63A IL2 or mutations to key variable residues resulted in monomeric OSCA1.2 and MA currents with significantly higher thresholds. Structural analyses revealed substantial conformational differences in the mechano-sensing domain IL2 and gating helix TM6 between TMEM63s and OSCA1.2. Our studies reveal that mechanosensitivity in OSCA/TMEM63 channels is affected by oligomerization and suggest gating mechanisms that may be shared by OSCA/TMEM63, TMEM16 and TMC channels.
Keywords: TMEM63, OSCA, IL2, TM6, mechanosensitive, ion channel, high-threshold, oligomerization, gating, TMEM16, TMC, hearing
In brief
Zheng et al. reveal that mammalian TMEM63 proteins form uniquely monomeric mechanosensitive ion channels, in contrast to dimeric OSCA channels in plants. The monomeric configurations of TMEM63s induce less favorable conformations of mechano-sensing domain IL2 and gating helix TM6 for channel activation, endowing high-threshold mechanosensitivties.
INTRODUCTION
Mechanosensitivity, the ability to detect and respond to mechanical stimuli, is fundamental to many physiological processes in all living organisms 1. Mechanically activated (MA) ion channels convert external or internal mechanical cues into electrical signals, enabling cells to respond rapidly to their physical environment 2–5. MA ion channels are widely expressed in all domains of life including bacteria, plants and animals and play critical roles in diverse biological processes. The bacterial MscS and MscL proteins were the first identified MA ion channels and are implicated in osmotic stress responses 6. In carnivorous plants, FLYC1 and FLYC2 are expressed in mechanosensory cells within touch-sensitive hairs triggered for capturing prey 7. In invertebrates, such as Drosophila melanogaster, NompC, a member of the transient receptor potential (TRP) channel family, is involved in touch sensation and sound detection 8–10. In mammals, the K2P potassium channel family may contribute to mechanical nociception 11–13, while the Piezo family of MA channels is implicated in a wide range of physiological processes, including gentle touch sensation, blood pressure regulation and vascular development 5,14,15. Lastly, TMC1 from the transmembrane-channel like (TMC) family forms the pore of the mechanosensory transduction complex in inner ear hair cells and is critical for hearing 16–18.
Despite progress in identifying MA ion channels, the largest family of MA ion channels OSCA/TMEM63 was only recently identified 19,20. OSCA channels exist in plants with 15 paralogs identified in Arabidopsis thaliana 19–21. OSCA1.1 mutations in A. thaliana lead to impaired rapid osmotic stress-induced Ca2+ elevation, suggesting a role of OSCAs as osmosensors in plants 20,21. Electrophysiological studies demonstrated that OSCAs are bona fide pore-forming mechanosensitive ion channels with preferred selectivity for cations 19. Cryo-electron microscopy (Cryo-EM) analysis of OSCA1.1, OSCA1.2 and OSCA3.1 22–25 revealed that OSCA channels form homo-dimers with one ion permeation pathway in each monomer. The OSCA monomer can be divided into a transmembrane domain (TMD), containing 11 transmembrane (TM) segments labeled TM0 to TM10, and an intracellular domain (ICD). The ICD is mainly composed of a long intracellular linker between TM2 and TM3, known as IL2, and a C-terminal tail. A notable feature of IL2 is that it contains two long amphipathic α-helices with an intervening hook domain inserted into the membrane, which was proposed to sense membrane tension 22. Sequence analysis indicated that OSCA/TMEM63 family is distantly related to the TMEM16/anoctamin and TMC families 26. Structural analysis of TMEM16 proteins, such as mTMEM16A (calcium-activated chloride channel in mouse) 27,28 and nhTMEM16 (lipid scramblase in fungus Nectria haematococca) 29, and TMC-1 in C. elegans 30 revealed that they all share a dimeric architecture with TM domain arrangements similar to OSCA channels.
TMEM63 proteins, including A, B, and C members, are animal orthologues of the plant OSCAs, with recently identified physiological functions and genetic pathologies. TMEM63A heterozygous variants have been identified in young human patients with hypo-myelinating leukodystrophies, characterized by myelin deficits, and global developmental delays with seizures 31–34. Ten distinct TMEM63B heterozygous variants were recently reported in 16 unrelated human patients with a broad range of brain function abnormities, including severe early onset developmental and epileptic encephalopathy, intellectual disability, and severe motor and cortical visual impairment 35. Biallelic variants of TMEM63C were also identified in individuals with hereditary spastic paraplegias 36. These clinical reports and genetic studies have demonstrated critical roles for TMEM63s in normal central nervous system development and neurodegenerative diseases. TMEM63B deficiency in mice was also reported to lead to deafness and it was suggested to act as an osmo-sensor in outer hair cells of the inner ear 37. When expressed in heterologous cells, TMEM63A and TMEM63B evoke small MA currents while no current was detected for TMEM63C 19. This raised fundamental questions as to whether TMEM63s are bona fide pore-forming MA ion channels. Furthermore, the structures and mechano-gating mechanisms of the entire TMEM63 family remain unexplored.
Here, we reveal an unexpected monomeric configuration of TMEM63s in vitro and in vivo, which contrasts with the dimeric assembly of OSCAs, TMEM16s and TMC1. We also present cryo-EM structures of human TMEM63A and TMEM63B with nominal resolutions of 3.8 Å and 3.6 Å, respectively. Structural and biochemical analysis reveal that the monomeric configuration of TMEM63s is governed by the IL2 domain. Electrophysiological recordings reveal that TMEM63A and TMEM63B are pore-forming MA ion channels with small conductance and high-threshold mechanosensitivity, compared to dimeric OSCA1.2. Moreover, by leveraging monomeric OSCA1.2 mutants, we find that the two protomers in an OSCA dimer are mechanically gated in an independent manner, and monomeric OSCA1.2 exhibits higher thresholds than dimeric OSCA1.2. Last, structural analysis uncovers distinct conformations of mechano-sensing domain IL2 and gating helix TM6 between monomeric TMEM63s and dimeric OSCA1.2. Based on these data, we propose a mechanistic model that explains how protein oligomerization affects mechanosensitivity of the OSCA/TMEM63 channels.
RESULTS
TMEM63s are monomeric proteins
We first examined the oligomeric configuration of overexpressed human TMEM63 proteins in HEK293T cell lysates using fluorescent size-exclusion chromatography (FSEC) (Figure 1A). With four membrane proteins as controls, we generated a standard curve, in which the oligomeric molecular weight (MW, kDa) showed a linear function of the retention time T (min) (MW = −242.6 × T + 2391, Figure 1A). Compared to A. thaliana OSCA1.2, human TMEM63A and TMEM63B exhibited right-shifted peak profiles, with calculated MWs of 114 kDa and 130 kDa, respectively (Figure 1B), consistent the size of glycosylated monomers (Figure S2B and S3B). The monomeric nature of TMEM63s was further supported by cross-linking assays in cell lysates (Figure 1C and S1A) and intact cells (Figure 1D), and by blue native polyacrylamide gel electrophoresis (PAGE) (Figure 1C). Human TMEM63C protein also displayed a monomeric configuration. Formation of hetero-dimers among TMEM63A, B and C proteins was not observed (Figure S1B). These data showed that overexpressed TMEM63s in HEK cells exist as monomers.
Figure 1. TMEM63A and TMEM63B exist as monomers.
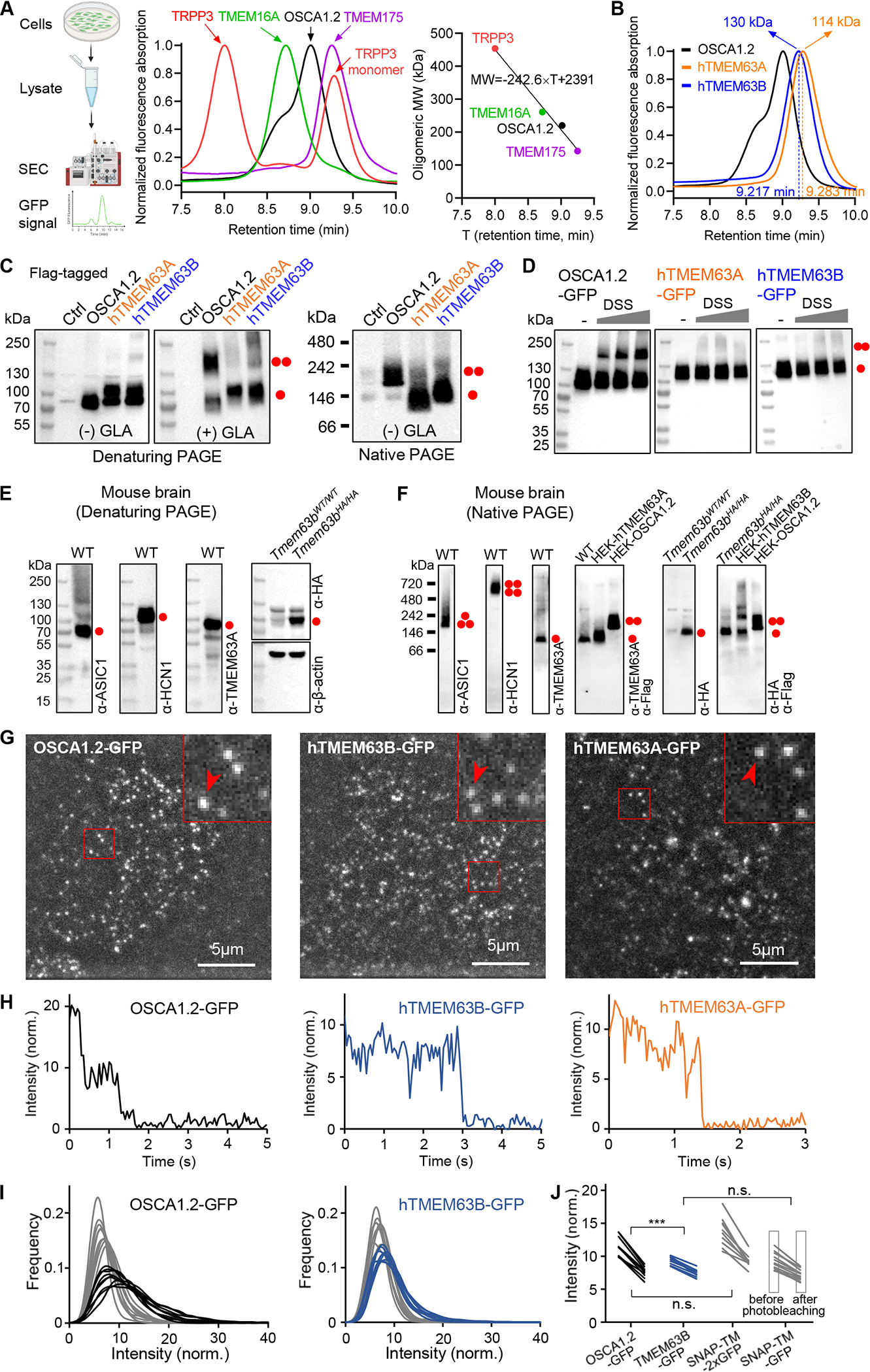
(A) Left, schematic diagram of FSEC; middle, normalized FSEC traces of GFP-tagged membrane proteins; right, oligomeric MW (with GFP) versus retention time fit deduced from middle panel data.
(B) Normalized FSEC traces of OSCA1.2, human TMEM63A and TMEM63B. Calculated oligomeric MWs (without GFP) are shown.
(C) Representative denaturing and blue native PAGE of HEK293T cell lysates without (Ctrl) or with indicated protein overexpression. The cell lysate was treated without (−) or with (+) 10 mM glutaraldehyde (GLA). Monomeric and dimeric protein bands are indicated.
(D) Representative western blots of HEK293T cell lysates with overexpression of GFP-tagged proteins. Cells were treated without or with membrane permeable disuccinimidyl suberate (DSS) (in mM: 0.0312, 0.0625, 0.125) before lysate preparation.
(E) Representative denaturing PAGEs of whole brain tissue of Tmem63bWT/WT (WT) or Tmem63bHA/HA knock-in mice.
(F) Representative blue native PAGEs of membrane fraction of mouse brain tissues. HEK293T cell lysates with overexpression of Flag-tagged proteins were also included.
(G) Single-molecule images from the first frames of CHO cells expressing OSCA1.2-GFP, hTMEM63B-GFP and hTMEM63A-GFP.
(H) Intensity time courses of marked spots from panel G.
(I) Intensity distribution before (black, blue) and after (gray) photobleaching for OSCA1.2-GFP and hTMEM63B-GFP.
(J) Mean intensity values before and after photobleaching for OSCA1.2-GFP, hTMEM63B-GFP and the control proteins containing one or two GFP tags.
See also Figure S1.
Next, we assessed oligomeric status of endogenous TMEM63s in mouse brain using denaturing and blue native PAGE. As controls, acid-gated ASIC1 channels and voltage-gated HCN1 channels were first confirmed to assemble as trimers and tetramers, respectively (Figure 1E and 1F). In contrast, TMEM63A forms monomers with an apparent MW close to 100 kDa, consistent with overexpressed TMEM63A in HEK cells (Figure 1E and 1F). Due to the lack of specific antibodies against TMEM63B, we evaluated the oligomeric status of TMEM63B by employing a Tmem63bHA/HA knock-in mouse model, in which an HA tag was fused to the N-terminus of TMEM63B 37. Similar to TMEM63A, TMEM63B also exists as monomers (Figure 1E and 1F). As such, endogenous TMEM63A and TMEM63B in the brain are monomeric.
Finally, we assessed the oligomeric state of TMEM63s in situ in the cell surface membrane. Overexpressed TMEM63s or OSAC1.2 with a C-terminal GFP tag in mammalian CHO cells was subjected to single molecule analysis using TIRF microscopy. For TMEM63A-GFP, we observed diffusing fluorescent spots, but also considerable amount of background (Figure 1G, Movie S1). In contrast, OSCA1.2-GFP and TMEM63B-GFP showed less background (Figure 1G, Movie S2 and S3), allowing more reliable single-molecule analysis. We observed that many fluorescent spots of OSCA1.2-GFP exhibited two-step photobleaching while TMEM63A-GFP or TMEM63B-GFP exclusively showed only a single bleaching step (Figure 1H), indicating a monomeric nature of TMEM63s. We then assessed the oligomeric states of OSCA1.2-GFP and TMEM63B-GFP by quantifying distribution of their fluorescence intensities before and after photobleaching. For OSCA1.2-GFP, the intensity histograms before photobleaching showed a peak for a single GFP, but with a pronounced shoulder at about twice intensity (Figure 1I). After 7.5 s, the shoulder disappeared, supporting the notion that a majority of the GFP tags had bleached and one tag at most was left (spots with both tags bleached are invisible and therefore do not contribute to the histogram) (Figure 1I). For TMEM63B-GFP, the shoulder at higher intensities was not visible (Figure 1I). These observations suggested that OSCA1.2 forms dimers, but TMEM63B remains monomeric. We made similar observations for control proteins containing a monomeric transmembrane domain with either one or two C-terminal GFP tags (Figure S1C) 38. While the mean intensities of the spots from OSCA1.2-GFP or the control protein with two GFP tags decreased by factors of 1.48 ± 0.05 (N = 10 cells) and 1.53 ± 0.04 (N = 9), respectively, the mean intensities from TMEM63B-GFP and the control with one GFP both decreased by 1.27 ± 0.02 (N = 12, N = 9) (Figure 1J). Taken together, these data support that TMEM63s are monomers in living cells while OSCA1.2 forms a dimer.
Structures of TMEM63A and TMEM63B
To understand the monomeric nature of TMEM63s, we determined cryo-EM structures of TMEM63A (3.8 Å) and TMEM63B (3.6 Å) in lipid nanodiscs and lauryl maltose neopentyl glycol (LMNG)/cholesteryl hemisuccinate (CHS), respectively (Figure S2 and S3, Table S1). Purified TMEM63A and TMEM63B proteins are monomeric (Figure S1D) and are glycosylated as confirmed by PNGase F treatment (Figure S2B and S3B). The high-resolution maps of the TMDs of both proteins allowed us to build an atomistic model unambiguously (Figure S2J, S3J, and S4). We were also able to identify extra densities of conserved N-linked glycosylation motif in both TMEM63A (38NST40 and 450NVT452) and TMEM63B (462NVT464) (Figure S4). In contrast, only secondary structures could be distinguished from the relatively low-resolution density map of cytosolic IL2 (Figure S2J, S3J, and S4), indicating its flexibility or multiple conformational states. By docking the AlphaFold2 39 model of the IL2 domain into cryo-EM densities, we were able to build complete models of TMEM63A and TMEM63B.
Different from the dimeric assembly of OSCAs 22–25, both TMEM63A and TMEM63B (hereafter referred as TMEM63s due to their similar structures) are monomers without any symmetry (Figure 2A–2D). The TMD of TMEM63s contains 11 transmembrane (TM) helices with an overall arrangement resembling OSCAs, TMEM16s, and TMC1 (Figure 2C, 2E and S5A). The ICD of TMEM63s is mainly composed of the IL2 domain that contains 4 stranded antiparallel β-sheet (IL2β1- IL2β4) and five α-helices (IL2H1-IL2H5) (Figure 2C). Notably, IL2H2 and IL2H3, two long amphipathic helices shared by TMEM63s and OSCAs, are parallel to the membrane with an intervening loop, known as the “hook” domain, inserted into the membrane. These features may allow IL2 to sense membrane tension.
Figure 2. Cryo-EM structures of human TMEM63A and TMEM63B.
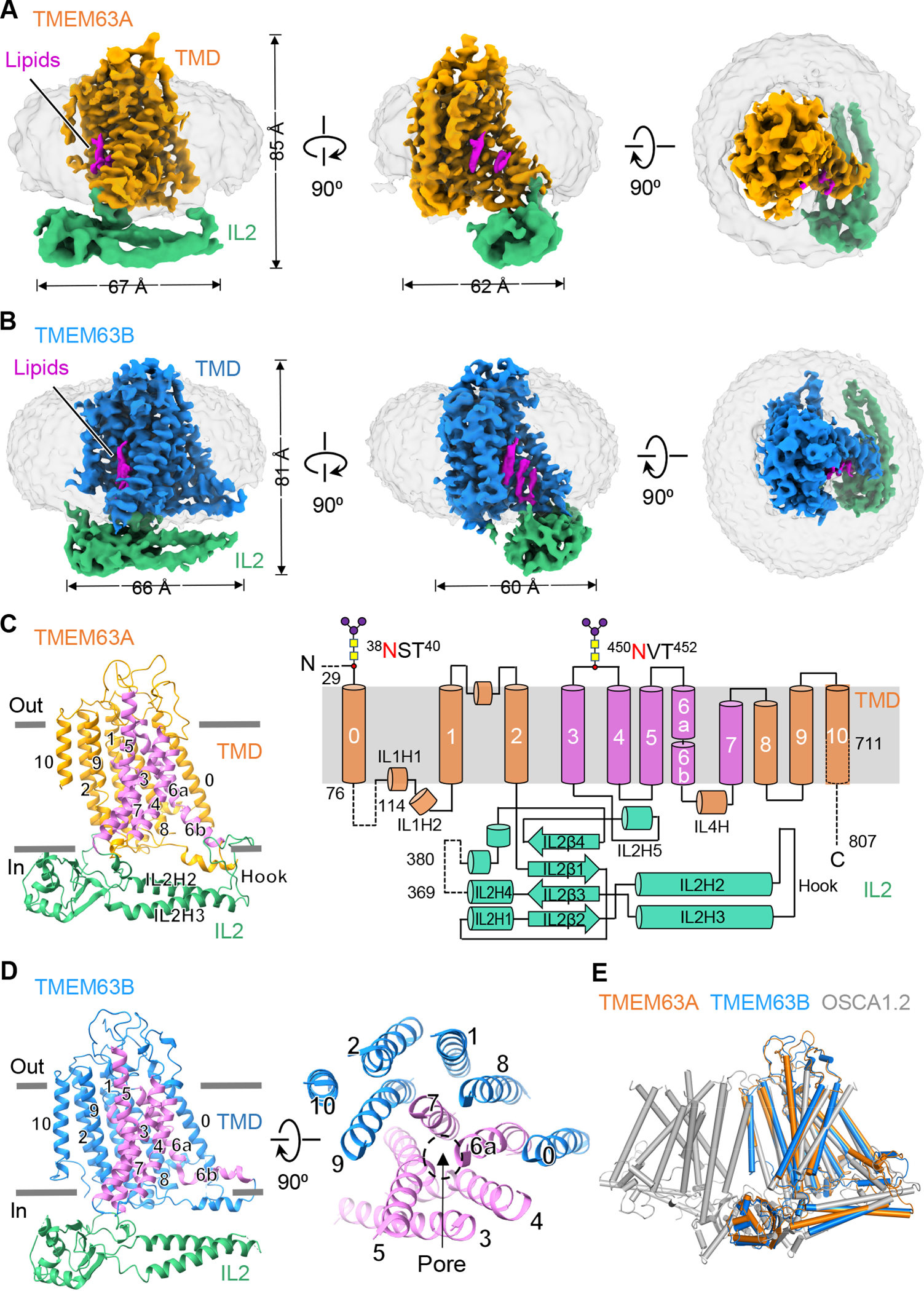
(A and B) Side and top views of EM density maps of TMEM63A in nanodisc (A) and TMEM63B in detergent LMNG (B). Densities for the transmembrane domain (TMD) are colored in either orange (TMEM63A) or blue (TMEM63B). Densities of intracellular linker 2 (IL2) are colored green. Densities of nanodisc and detergent are grey. Lipid densities are highlighted in magenta.
(C) Left, side view of TMEM63A in ribbon representation, with the pore-lining helices (TM3-TM7) colored in magenta. Right, topology diagram of TMEM63A with major structural components highlighted. Dashed lines denote regions excluded in model building due to missing density. Two N-linked glycosylation sites were predicted based on the protruding densities near N38 and N450 (Figure S4).
(D) Left, side view of TMEM63B with the pore-lining helices (TM3-TM7) colored in magenta. Right, top view of the TM segments of TMEM63B with the location of the pore indicated.
(E) Structural alignments of monomeric human TMEM63A and TMEM63B with one subunit of dimeric OSCA1.2 (PDB: 6MGV).
Additionally, non-protein densities within a cavity of TMDs of TMEM63s were observed and tentatively assigned as lipids (Figure 2A and 2B). Give the critical roles of lipids in regulation/gating of mechanosensitive channels such as TRAAK, Piezo1, and MscS 40,41, it is possible that lipids may also affect functional aspects of TMEM63s.
Ion permeation pores of TMEM63s
Like OSCAs, TMEM16s and TMC1, we identified an ion permeation pore in TMEM63s at a similar position formed by TM3-TM7 (Figure 3A and S5A) 42. Compared with OSCA1.2, the putative pore in TMEM63s exhibited two remarkable features. First, the TMEM63 pore contains a drastically elongated constriction region, known as the ‘neck’, which extends over 50 Å (Figure 3A and 3B). Comparison of pore domains between OSCA1.2 and TMEM63s revealed noticeable differences in the pore-lining TM4 and TM6 (Figure S5B). Although an open pore conformation has not been resolved, it is highly likely that TMEM63s possess a more constricted pore than OSCA1.2 in the open state. Second, an N-terminal domain (NTD) in TMEM63s (S30-V45 in TMEM63A and P19-L36 in TMEM63B), absent in OSCAs, folds into a hairpin on top of the putative pore (Figure 2A, S5C and S5D). Collectively, these pore features may significantly limit ion flux, leading to a low channel conductance of TMEM63s.
Figure 3. Ion permeation pores in TMEM63s.
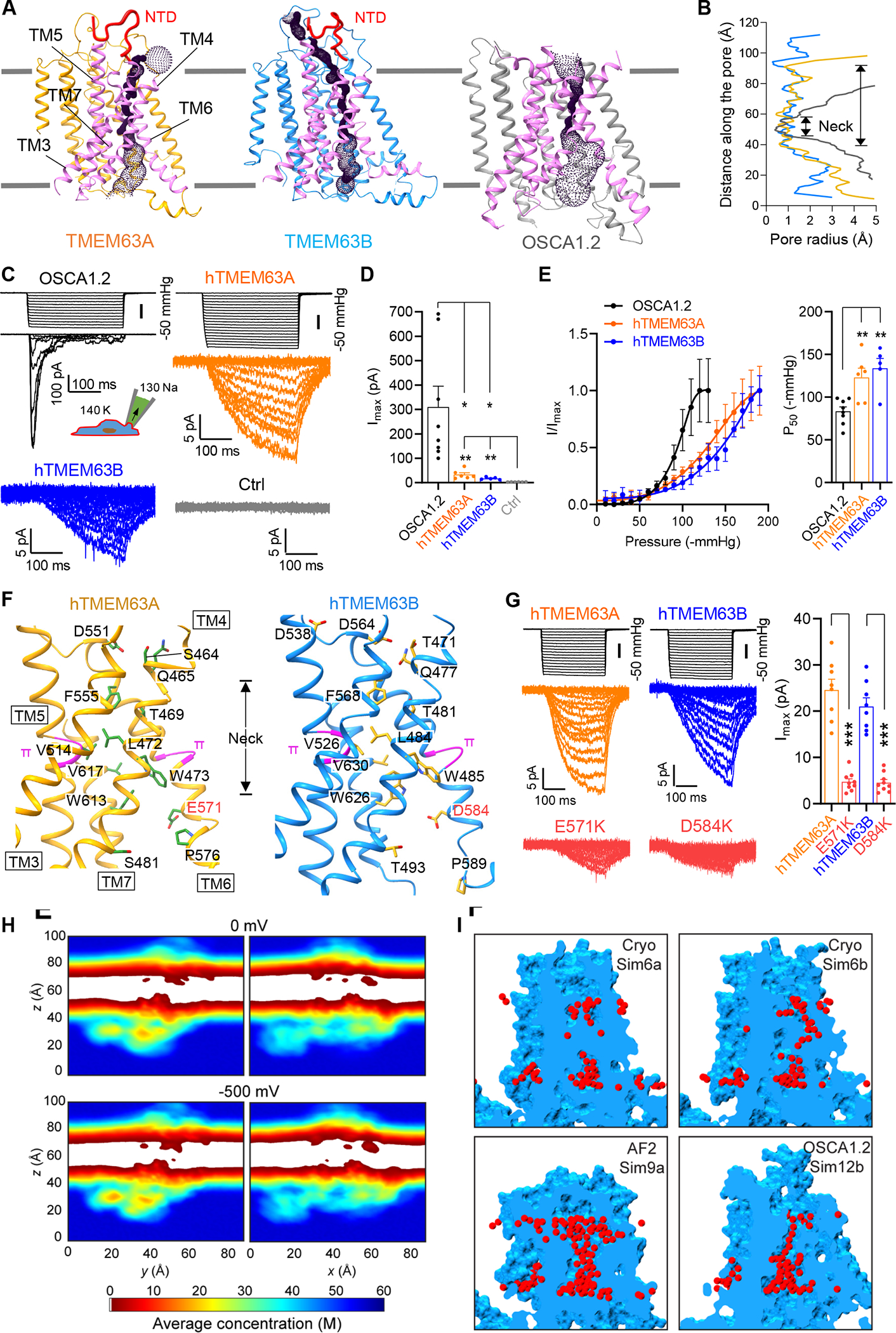
(A) Predicted location of the pore in TMEM63A, TMEM63B and OSCA1.2 (PDB: 6MGV). Pore-lining TMs are colored in magenta and N-terminal domains (NTDs) in TMEM63A and TMEM63B are accentuated by thickening line and colored in red.
(B) Van der Waals pore radii against distance along the pore, with neck regions indicated.
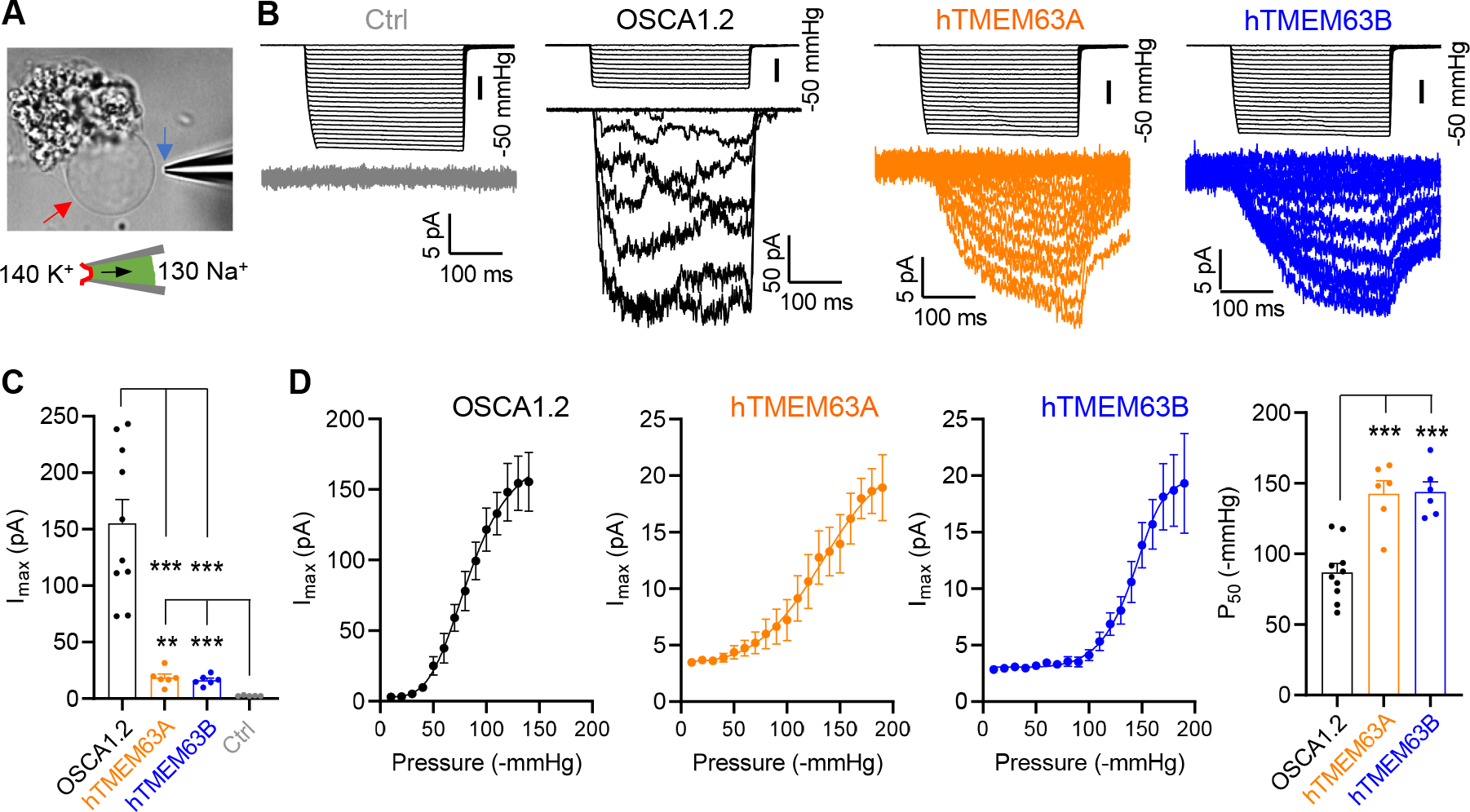
(C) Representative traces of stretch-activated currents (−80 mV) from HEK293T cells without (Ctrl) or with indicated protein overexpression. A diagram was shown to indicate main cations (mM) used in the bath and pipette solutions.
(D) Quantification of maximal current amplitude (Imax) from individual patches. *p<0.05, **p<0.01
(E) Left, Average and normalized pressure-response current curves fitted with a Boltzmann equation. OSCA1.2 (N = 8), hTMEM63A (N = 6), hTMEM63B (N = 5). Right, quantification of P50 values. **p<0.01.
(F) Pore-lining residues in TMEM63A (Left) and TMEM63B (Right) from HOLE analysis.
(G) Left, representative traces of stretch-activated currents from HEK293T cells expressing hTMEM63A, hTMEM63B or indicated mutants; Right, quantification of Imax. ***p<0.001.
(H) Two-dimensional slices showing time- and space-averaged water concentrations for different all-atom MD simulations (Sim1a – top; Sim1b – bottom; see also Figure S7).
(I) Examples of transient water channels formed in simulations of our experimentally derived TMEM63B model (Sim6a and Sim6b), of an AlphaFold2 (AF2) TMEM63B model (Sim9a), and of an experimentally derived model of OSCA1.2.
In line with the structural observations, electrophysiological recordings revealed small stretch-activated currents in heterologous HEK293T cells expressing TMEM63A (Imax = 33.5 ± 7.5 pA; N=6) or TMEM63B (Imax = 17.4 ± 2.3 pA; N=5), whereas OSCA1.2 were 10-fold larger (Imax = 310.2 ± 85.1 pA; N=8) (Figure 3C and 3D). The TMEM63A currents were dramatically reduced by Gd3+ (76% reduction in Imax; Figure S1E), which excluded the possibility of pressure-induced leak. In addition, robust single-channel currents were evident in OSCA1.2-transfected cells (with a conductance of 129 ± 3 pS, see Figure 5L and 5M), but we were not able to resolve single-channel currents of TMEM63s, which were probably too small to be detected. Of note, although TMEM63s had small current amplitudes, they lacked inactivation (Figure 3C and S1F), which allows them to conduct similar numbers of charges/ions to OSCA1.2 and Piezo1 under sustained mechanical stimuli (Figure S1G).
Figure 5. The oligomeric configuration affects mechanosensitivity of OSCA/TMEM63s.
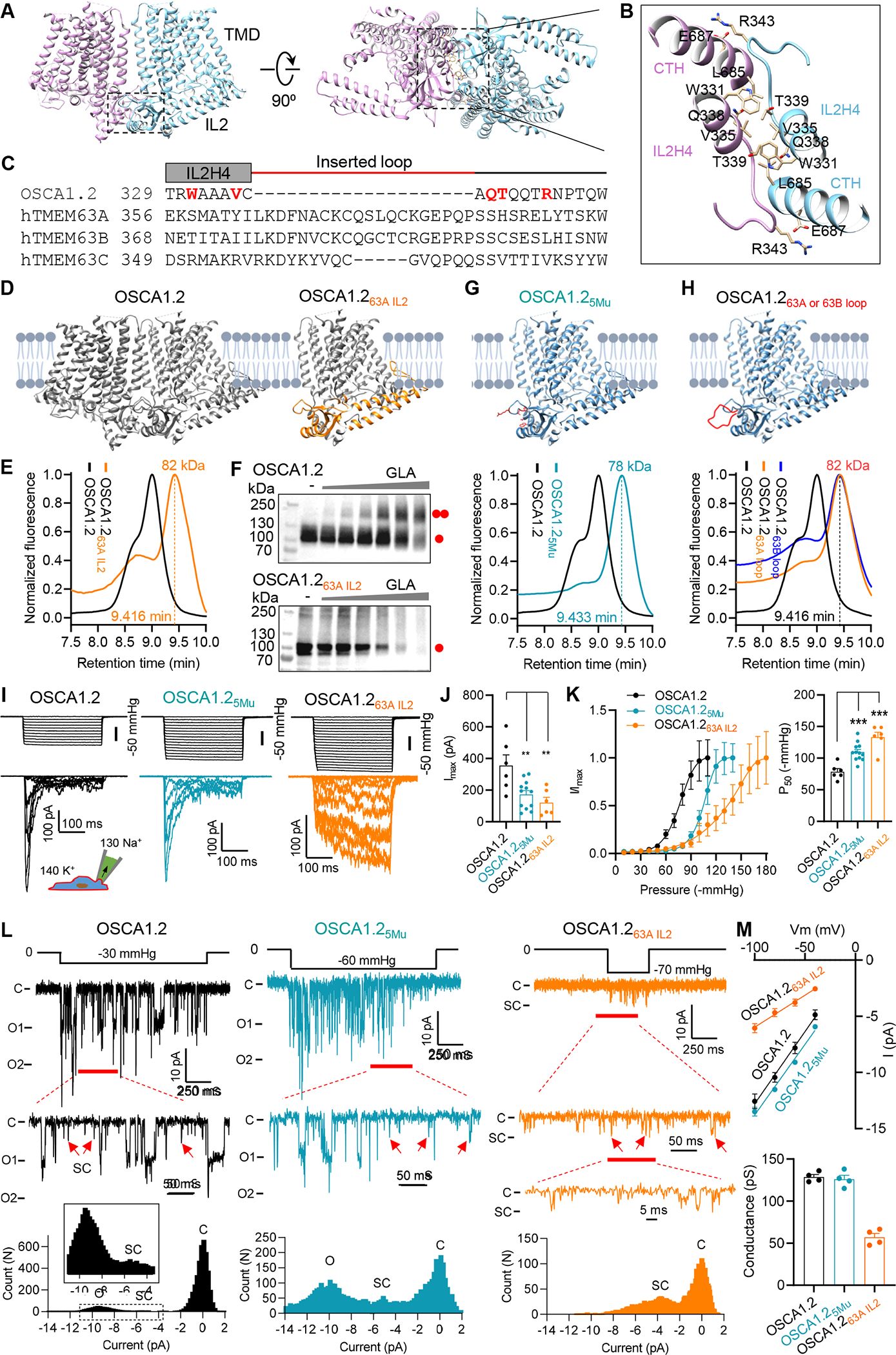
(A) Side and bottom views of dimeric OSCA1.2 (PDB: 6MGV). The rectangle indicates the dimerization interface in IL2.
(B) Enlarged dimerization interface in OSCA1.2, with interacting residues are shown. CTH, C-terminal helix (L685-R698).
(C) Sequence alignment of the region within IL2 that mediates the dimerization of OSCA1.2. Residues that directly mediate OSCA1.2 dimerization are highlighted in red. The inserted loop in TMEM63s is indicated by the red line.
(D) Illustrations of dimeric OSCA1.2 and monomeric chimera protein OSCA1.263A IL2.
(E) Normalized FSEC traces of GFP-tagged OSCA1.2 and OSCA1.263A IL2, with MW of the OSCA1.263A IL2 (without GFP) indicated.
(F) Representative western blots of GFP-tagged OSCA1.2 (upper) or OSCA1.263A IL2 (lower). Cell lysates were treated without or with GLA at various concentrations (in mM: 0.3125, 0.625, 1.25, 2.5, 5, 10).
(G) Illustration of monomeric OSCA1.25Mu mutant containing 5 mutations, W331G, V335G, Q338G, T339G R343A (Upper), and normalized FSEC traces (Lower).
(H) Illustration of monomeric OSCA1.2 with inserted 20-aa loop from hTMEM63A (OSCA1.263A loop) or hTMEM63B (OSCA1.263B loop) (Upper) and normalized FSEC traces (Lower).
(I and J) Representative traces of stretch-activated currents recorded from HEK293T cells (I) and quantification of Imax (J). **p<0.01.
(K) Left, average and normalized pressure-response current curves fitted with a Boltzmann equation for OSCA1.2 (N = 6), OSCA1.25Mu (N = 12), OSCA1.263A IL2 (N = 6); Right, quantification of P50 from individual patches. ***p<0.001.
(L) Single-channel currents recorded at −80 mV. The closed (C) and fully open (O) states are indicated. Red arrows mark the assumed sub-conductance (SC) state. Current amplitude histogram for each trace is shown at the bottom.
(M) Upper, averaged current-voltage relationship. OSCA1.2 (N = 4), OSCA1.25Mu (N = 4), OSCA1.263A IL2 (N = 4); Lower, the mean single-channel conductance from individual patches.
See also Figure S9.
Pore-lining residues in TMEM63s are mostly hydrophobic within the neck region and hydrophilic at either end of the neck (Figure 3F). In particular, the acidic residues at both extracellular and intracellular sides (D551/E571 in TMEM63A and D538/D564/D584 in TMEM63B) of the neck may provide an attractive electrostatic environment for cations. Positively charged residues were previously found at an equivalent pore location of TMEM16A to account for a selectivity for anions 27,28. As most pore-lining residues in the neck region are located in TM4 and TM6 (Figure 3F), the pore opening of TMEM63s probably involves movement of TM4, TM6 or both. Like OSCA1.2, TMEM63s possess a pair of π-helices in TM5 and TM6, respectively (Figure 3F). The π-helix is energetically unstable and the transition between π- and α-helix in pore-lining helices has been implicated in gating of many ion channels, including TRPs and TMEM16A 28,43. To experimentally investigate the channel pore, we replaced the putative pore-lining acidic residues in TM6 of TMEM63A (E571) and TMEM63B (D584) with positively charged lysines. These substitutions had minimal effect on protein surface expression (Figure S1H), but dramatically reduced the maximal stretch-activated currents for both TMEM63A and TMEM63B (Figure 3G), suggesting involvement of this acidic residue in cation permeation in TMEM63s. A similar substitution in OSCA1.2 (E531K) also drastically reduced single-channel current (see Figure S9F), consistent with a previous claim that E531 contributes to the ion permeation pathway of OSCA1.2 22.
To further characterize the ion permeation pore of TMEM63s, we carried out equilibrium (0 mV) and non-equilibrium (−500 mV) all-atom molecular dynamics (MD) simulations 44 on models based on our structure of TMEM63B (Figure S5E), on an AlphaFold2 model of TMEM63B, and on the structure of OSCA1.2 (Table S2). Our structural model of TMEM63B was simulated with or without the NTD (Figure S5F) and incorporated missing regions from AlphaFold2 prediction including two conformations (C1 and C2) for the first intracellular linker (IL1) region (Figure S5G). The AlphaFold2 model contains two loops connecting TM3/TM4 and TM5/TM6, respectively, that show prominently different conformations from our experimentally derived models (Figure S5H). Equilibrium simulations (200 ns each) revealed a stable TMD for all systems simulated (root-mean-squared deviation (RMSD) < 2 Å; Figure S6), while IL2 domains were more dynamic. Analysis of water density throughout equilibrium simulations did not reveal persistent hydration of the putative pore in TMEM63B models, regardless of NTD presence, IL1 conformation, or membrane composition (Figure 3H and S7). Nevertheless, we were able to identify potential permeation pathways that match those visualized with the HOLE program using the static structures (Figure 3I). Transient hydration was more readily observed for the AlphaFold2 model of TMEM63B and for the OSCA1.2 channel (Figure 3I). At −500 mV, we did not observe any ion permeation events over 100 ns of simulation for each system. A pore with a conductance of ~120 pS, as expected for OSCA1.2, should conduct up to ~37 ions under the simulated conditions. These data supported that our resolved structures of TMEM63s, as well as the published OSCA1.2 model 22, represent closed states, and that the pore of TMEM63s is substantially constricted and of low conductance.
TMEM63s form high-threshold mechanosensitive ion channels
Stretch-activated currents evoked by TMEM63 expression in HEK293T cells exhibited not only small amplitudes, but also high thresholds. As shown in Figure 3E, the pressure required for half-maximal activation (P50) of human TMEM63A (−123.3 ± 11.2 mmHg) or TMEM63B (−134.2 ± 11.4 mmHg) was significantly larger than that for OSCA1.2 (−85.9 ± 5.2 mmHg). To examine whether monomeric TMEM63s are bona fide pore-forming mechanosensitive ion channels with high thresholds, we performed electrophysiological recordings in proteoliposomes (Figure 4A). Reconstitution of purified OSCA1.2 proteins into liposomes induced robust macroscopic stretch-activated currents (Imax = 155.4 ± 20.9 pA; N=10) (Figure 4B and 4C), confirming that OSCA1.2 is an intrinsically mechanosensitive ion channel 19. Single-channel current recordings revealed a conductance of 133 pS (Figure S1I), consistent with measurements from HEK293T cells. As a negative control, little current was detected in the empty liposomes (Imax = 2.6 ± 0.2 pA; N=5) (Figure 4B and 4C). Using the established protocol, we next detected small but significant stretch-activated currents for both TMEM63A (Imax = 18.7 ± 3.1 pA; N=6) and TMEM63B (Imax = 16.3 ± 1.8 pA; N=6) (Figure 4B and 4C). No single-channel current was resolved, in accordance with our assumption of small conductance of TMEM63s. Akin to recordings in HEK293T cells, the P50 of TMEM63A (−142.6 ± 9.1 mmHg) or TMEM63B (−143.9 ± 7.3 mmHg) was substantially higher than OSCA1.2 (−86.9 ± 6.4 mmHg) (Figure 4D), supporting the characteristic high-threshold mechanosensitivity for monomeric TMEM63s. Furthermore, TMEM63s from other species, including mouse and fruit fly, also adopted a monomeric configuration (Figure S1J and S1K) and exhibited high-threshold mechanosensitivities in HEK293T cells (Figure S1L and S1M).
Figure 4. TMEM63A and TMEM63B form high-threshold mechanosensitive ion channels.
(A) Representative image showing a proteoliposome, indicated by a red arrow, grown from the lipid cloud. A recording pipette, indicated by a blue arrow, is also shown for excised patch recording.
(B) Representative traces of stretch-activated currents from unilamellar liposomes reconstituted without (Ctrl) or with indicated purified proteins.
(C) Quantification of Imax. OSCA1.2 (N = 10), hTMEM63A (N = 6), hTMEM63B (N = 6) or Ctrl (N = 5). **p<0.01, ***p<0.001.
(D) Average pressure-response current curves fitted with a Boltzmann equation for OSCA1.2 (N = 10), hTMEM63A (N = 6) and hTMEM63B (N = 6). The right panel shows quantification of P50 values. ***p<0.001.
See also Figure S1.
Oligomeric configuration affects mechanosensitivity of OSCA/TMEM63s
We hypothesized that the differential mechanosensitivity between TMEM63s and OSCA1.2 is a result of their distinct oligomeric configuration. To examine this hypothesis, we first sought to unveil the structural basis underlying their distinct oligomerization. In OSCA1.2, the dimerization is solely mediated by the IL2 domain (Figure 5A and 5B). Sequence alignments revealed that residues mediating interactions within the dimerization interface of OSCA1.2 are variable among TMEM63s (Figure 5C and S8). Moreover, TMEM63s contain an additional inserted fragment of 20 amino acid (aa) in the middle of dimerization region (Figure 5C). This 20-aa fragment was not resolved in our structures and probably forms a flexible loop. Conceivably, this 20-aa loop would clash with the dimerization interface.
To verify the importance of IL2 in protein oligomerization, we first generated a chimeric protein, OSCA1.263A IL2, in which the IL2 domain of OSCA1.2 (S194-P367) was replaced with that of TMEM63A (N224-Q414) (Figure 5D). FSEC and cross-linking assays revealed a monomeric configuration of OSCA1.263A IL2 (Figure 5E and 5F), suggesting that the IL2 domain is a key determinant of dimerization. Of note, dimeric TMEM63A was not achieved by replacement with OSCA1.2 IL2 or further with OSCA1.2 C-terminus (Figure S9A and S9B), so OSCA1.2 IL2 itself is not sufficient to induce a dimerization. Furthermore, replacements of 5 dimerization interfacial residues in OSCA1.2 (W331G, V335G, Q338G, T339G and R343A) lead to a monomeric mutant OSCA1.25Mu (containing these five mutations together) (Figure 5G and S9C). Additionally, engineering the 20-aa inserted loop of TMEM63s into OSCA1.2 led to a monomeric configuration (Figure 5H). Collectively, these data support that the genetic variances in the IL2 domain lead to distinct oligomerization status between OSCAs and TMEM63s.
To examine whether the oligomeric configuration affects the mechanosensitivity of OSCA/TMEM63s, we functionally characterized the OSCA1.2 monomeric variants, OSCA1.25Mu and OSCA1.263A IL2. In HEK293T cells, OSCA1.25Mu expression evoked robust stretch-activated currents (Imax = 171.6 ± 25.5 pA) with rapid activation and fast inactivation, akin to OSCA1.2 (Figure 5I and 5J), demonstrating that monomeric OSCA1.2 preserves mechanosensitive channel function. Notably, compared to OSCA1.2, OSCA1.25Mu showed a substantially higher P50 (−109.8 ± 4.1 mmHg, Figure 5K), suggesting that switching from a dimer to monomer leads to high-threshold mechanosensitivity. Similarly, monomeric OSCA1.263A IL2 expression also induced high-threshold (P50 = −133.1 ± 7.7 mmHg) stretch-activated currents (Imax = 120.8 ± 33.2 pA) (Figure 5I–5K). Unlike OSCA1.25Mu, OSCA1.263A IL2 showed little inactivation (Figure 5I), even with prolonged application of pressures (Figure S1F). Of note, OSCA1.25Mu and OSCA1.263A IL2 exhibited smaller Imax than OSCA1.2 (Figure 5J), at least partially due to lower total and cell surface protein expression (Figure S9D). Together, these data demonstrate the influence of oligomeric status on mechanosensitivity of OSCA/TMEM63s.
We next examined the influence of the oligomeric configuration on single-channel behavior. In HEK293T cells, OSCA1.2 induced two types of stretch-activated unitary currents, a dominant fully open current with a conductance of 129 ± 3 pS, and a transient smaller current with approximately half amplitude (Figure 5L and 5M). This transient current was suggested to be the single-pore current, and the fully open current was thought to arise from the simultaneous opening of two pores 19,22. Thus, the two subunits in OSCA1.2 were assumed to function cooperatively. However, our recordings of monomeric OSCA1.25Mu showed a similar fully open current with a conductance of 126 ± 5 pS and a small current of half amplitude (Figure 5L and 5M), which indicated that the fully open current observed in OSCA1.2 is from a single-pore and two subunits in OSCA1.2 function in a largely independent manner. The half-amplitude current likely represents a sub-conductance state, i.e., a partial open state, of a single pore. This presumption of independent gating of OSCA1.2 two subunits is further supported by single-channel recordings of a OSCA1.2 concatemer, containing two protomers of different conductance in tandem (E531K mutant and WT) (Figure S9E). This OSCA1.2 concatemer induced a mix of two levels of unitary current of WT (11 pA) and E531K mutant (3 pA), instead of one averaged level (7 pA) (Figure S9F). Our recordings from monomeric OSCA1.263A IL2 only showed currents resembling the assumed sub-conductance currents, with a conductance of 57 ± 5 pS (Figure 5L and 5M). One possible explanation is that the IL2 domain from TMEM63A only induces activation of the OSCA1.2 pore to a partially open state. Comparison of current traces between OSCA1.2 and OSCA1.25Mu revealed that the mean open-dwell time of the pore in OSCA1.2 (τ = 3.5 ± 0.3 ms) is longer than that in OSCA1.25Mu (τ = 1.8 ± 0.2 ms) (Figure 5L and S9G). Therefore, the oligomeric status of OSCA/TMEM63s barely affects the pore conductance, but has an influence on the pore open-dwell time.
Oligomeric configuration affects conformation of IL2 and TM6
To mechanistically understand how the oligomeric configuration affects mechanosensitivity of OSCA/TMEM63, we compared structures of monomeric TMEM63s and dimeric OSCA1.2. While the TMD exhibited large similarity between TMEM63s and OSCA1.2, we observed prominent conformational differences of the IL2 domain which exhibited more intimate contact with the membrane in OSCA1.2 (Figure 6A). Isolated IL2 domains were superimposed between TMEM63s and OSCA1.2, we thus speculated that the dimerization in OSCA1.2 induces a rigid-body movement of IL2 domain towards the membrane (Figure 6A) and stabilizes the IL2 domain in such conformation to allow membrane association of hypothetic mechano-sensing elements, including amphipathic helices IL2H2/IL2H3 and the hook domain. (Figure 6A). We deleted the hook domain in OSCA1.2 (Q272-K287), TMEM63A (L298-W317) and TMEM63B (M310-Q329) and found that the deletion had little effect on the mechanosensitivity of these channels (Figure S9H), demonstrating that the hook domain is dispensable for mechano-sensation. We thus propose that the amphipathic helices IL2H2/IL2H3 may serve as the mechano-sensor, given the prevalence of the amphipathic helix in other mechanosensitive ion channels 1,30,45. Compared with OSCA1.2, the IL2 domain in TMEM63s is largely mobile (Figure S6) and mainly adopts a conformation in which IL2H2/IL2H3 is farther away from the membrane surface (Figure 6A), which could confer high-threshold mechanosensitivity.
Figure 6. The oligomeric configuration affects conformations of mechano-gating elements, including IL2 and TM6, in OSCA/TMEM63 family.
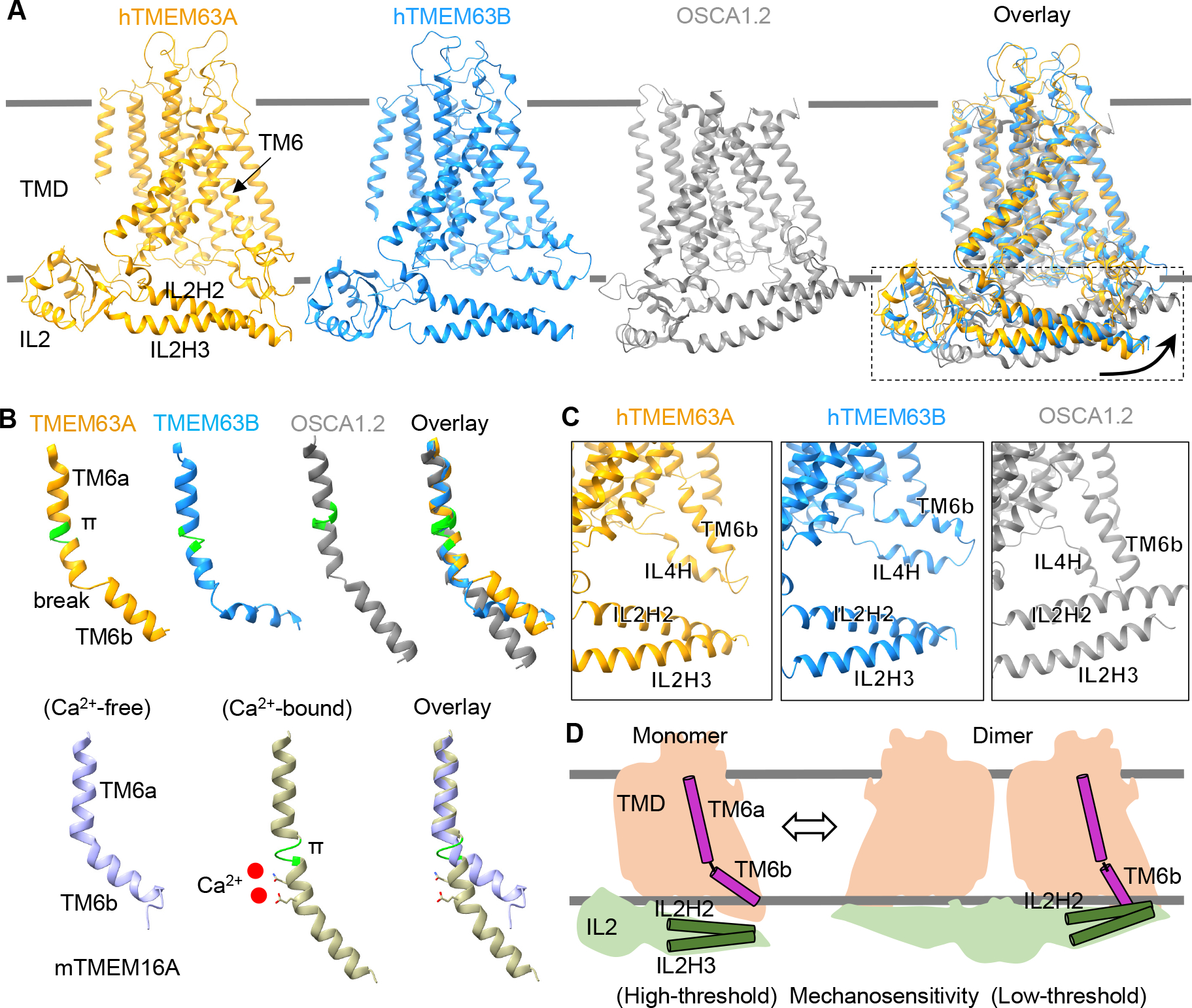
(A) Structural comparisons among hTMEM63A, hTMEM63B and OSCA1.2 (one subunit, PDB: 6MGV) based on the alignment of TMDs. Arrows indicate the movement of IL2 domain from the conformation in hTMEM63A or hTMEM63B to that in OSCA1.2.
(B) Upper, ribbon representations of TM6 from hTMEM63A, hTMEM63B and OSCA1.2 (PDB: 6MGV). A π-helix in the TM6 is colored in green and a break point in the TM6 is indicated. Lower, TM6 from mouse TMEM16A in Ca2+-free (PDB: 5OYG) and Ca2+-bound (PDB: 5OYB) conformations. The π-helix and Ca2+ ions in the Ca2+-bound state are shown.
(C) Structural comparisons showing the relative positions of IL2H2/IL2H3 to TM6b/IL4H in hTMEM63A, hTMEM63B and OSCA1.2. The hook domain is not shown for clarity.
(D) Schematic diagram showing distinct conformations of IL2 and TM6 in monomeric and dimeric OSCA/TMEM63 proteins.
The pore opening (gating) of a mechanosensitive ion channel involves both sensation of mechanical force and transduction of the force to open the pore. In the Ca2+-activated chloride channel TMEM16A, the pore-lining TM6 directly binds with Ca2+ and undergoes a dramatic rearrangement upon Ca2+ binding (Figure 6B) 27,28. Thus, TM6 was proposed to act as a gating helix to bridge Ca2+ binding and pore opening in TMEM16A 46. Interestingly, a similar conformational rearrangement in TM6 was observed in the structural alignment of TMEM63A, TMEM63B and OSCA1.2 (Figure 6B). The cytoplasmic half of TM6, named TM6b, in dimeric OSCA1.2 is oriented to establish stable physical contact with IL2H2 (Figure 6C), whereas such a physical link was not observed in TMEM63s (Figure 6C). This conformation of TM6b in OSCA1.2 places it in a favorable position to transduce force sensed by IL2H2/IL2H3 to promote pore opening, while in TMEM63A and TMEM63B, pore opening may require greater force. Interestingly, transient physical contact between TM6b and IL2H2 in TMEM63B was observed in our MD simulation studies (Figure S10), implicating the IL2-TM6 interaction in TMEM63 gating. We propose that these features of monomeric TMEM63s have evolved to fill a functional niche, namely as high-threshold mechanosensitive ion channels (Figure 6D).
DISCUSSION
Like thermosensation in mammals, which is mediated by various TRP channels detecting a wide range of temperatures 47,48, mechanosensation may also involve distinct molecules with sensitivities that span a broad physiological range of mechanical forces encountered in the biological world. To date, Piezo proteins (piezo1 and piezo2) 5,49 and TMC1 18 are the predominant mechanosensitive ion channels identified in mammals with low thresholds. We propose that TMEM63s evolved to function as high-threshold mechanosensitive ion channels, complementary to Piezo and TMC1 channels. Human patients with TMEM63B variants exhibit chronic haemolytic anaemia 35, characterized by dysmorphic red blood cells (RBCs) due to dehydration. Interestingly, Piezo1 mutations are known to cause similar symptoms in humans with dehydrated hereditary stomatocytosis (DHS) 50. RBCs are highly deformable cells that experience a broad range of mechanical stimuli when circulating through blood vessels of variable sizes. It is plausible that Piezo1 and TMEM63B may work complementarily in RBCs to sense mild to intense mechanical force, thereby ensuring effective mechanosensory responses of RBCs. TMEM63B has also been suggested to function as an osmosensor in cochlear outer hair cells and TMEM63B deficiency in mice leads to outer hair cell death and deafness 37. Outer hair cells act as mechanical actuators capable of amplifying sound signals in the ear through a process known as cochlear amplification 51. The mechanical actuator protein, prestin, is densely packed in the outer hair cell basolateral membrane and its electromotility activity likely evokes significant mechanical force within the basolateral membrane of outer hair cells. Therefore, the high-threshold activation of TMEM63B may be beneficial and could allow the channel to remain in a closed state during normal electromotility function of outer hair cells. Opening of TMEM63B would only be triggered when outer hair cells experience severe osmotic stress. Alternatively, it is plausible that prestin activity may generate sufficient force to activate TMEM63B directly. Since prestin is voltage-dependent it is also plausible that TMEM63B activation may provide feedback to modulate prestin activity, outer hair cell electromotility and cochlear amplification.
Unlike well-established ion channels containing one central pore formed by multiple subunits, OSCA, TMEM16 and TMC channels are dimers with one pore in each subunit (Figure S5A). Since each subunit of the dimeric channels contains a complete ion permeation pathway, several fundamental questions arise. Can the monomeric protein function as a channel? If yes, why is dimerization needed? Our data, recorded from TMEM63A and TMEM63B in HEK293T cells and liposomes (Figure 3C and 4B), demonstrate that monomeric proteins can form functional channels, which is further supported by the recordings from monomeric OSCA1.25Mu and OSCA1.263A IL2 (Figure 5I). Comparison of single-channel currents between OSCA1.2 and monomeric OSCA1.25Mu (Figure 5L) and recordings from a OSCA1.2 concatemer (Figure S9E and S9F) further suggest that the two subunits in the dimeric OSCA1.2 function in a largely independent manner. Independent gating of TMEM16A subunits while in a dimeric complex was previously deduced from measurement of Ca2+-induced activation in a fused protein with two subunits of different Ca2+ sensitivities in tandem 52,53. Independent function may extend to other members in this superfamily, including TMCs. Structural comparisons between monomeric TMEM63s and dimeric OSCA1.2 revealed distinct conformations of the mechano-sensing domain IL2 and the gating helix TM6 (Figure 6A and 6B). We suggest that dimerization may affect the conformation of key structural elements (Figure 6D), thus leading to new functional characteristics. For example, the dimeric OSCA1.2 showed mechanosensitivity with a lower threshold, compared to monomeric OSCA1.25Mu, OSCA1.263A IL2 and TMEM63s (Figure 3E and 5K). Therefore, we suggest that monomeric proteins in OSCA/TMEM63, TMEM16 and TMC families are functional and the two subunits in the dimeric protein function independently; dimerization could lead to distinct biophysical properties, thereby diversifying functionalities in this superfamily of ion channel proteins.
TMEM63 structures displayed similar TMD architectures with OSCA1.2, TMEM16A and TMC-1 (Figure S5A). It is unclear whether they share a common gating mechanism. Structural studies 28 in mouse TMEM16A revealed that Ca2+ binding induced a dramatic rearrangement in TM6 (Figure 6B), highlighting a critical role of TM6 in the gating of TMEM16A. Indeed, mutations in TM6 of TMEM16A resulted in significant alterations in Ca2+ sensitivity 46. A recent functional study 54 in mouse TMC1 also showed that mutations in TM6 can affect the mechanosensitivity of sensory hair cells in the inner ear, which implies an involvement of TM6 in the gating process of TMC1. Interestingly, our structural studies revealed a conformational difference in TM6 between monomeric TMEM63s and OSCA1.2, akin to the Ca2+-induced conformational change in TM6 of TMEM16A (Figure 6B). Given the mechanosensitivity difference between TMEM63s and OSCA1.2 (Figure 3E and 4D), it is highly likely that TM6 functions as a gating helix in the OSCA/TMEM63 family. In support of this hypothesis, pathogenic variants in humans have been identified in TM6 of both TMEM63A and TEM63B 31–35 (Figure 7). In particular, variants of the glycine residue in the π-helix of TM6 (G567 in TMEM63A and G580 in TMEM63B) were reported in high frequency. Based on this evidence, it is plausible that TM6 is a common gating helix among OSCA/TMEM63, TMEM16 and TMC families. How TM6 couples stimulus-induced local conformational changes to pore opening may be diverse in this ion channel superfamily and remains to be elucidated. The density map of TMEM63A in nanodisc suggested a membrane distortion near the IL2H2/IL2H3 and TM6b. Similar distortion has been observed in TMEM16F, a scramblase with ion channel function, and has been shown to be related to TMEM16F scramblase activity 55. Whether TMEM63A possesses scramblase activity is yet to be determined. It will also be interesting to see whether this membrane distortion is involved in the mechano-gated pore opening process of TMEM63s.
Figure 7. Disease-associated variants in humans identified in TMEM63A and TMEM63B.
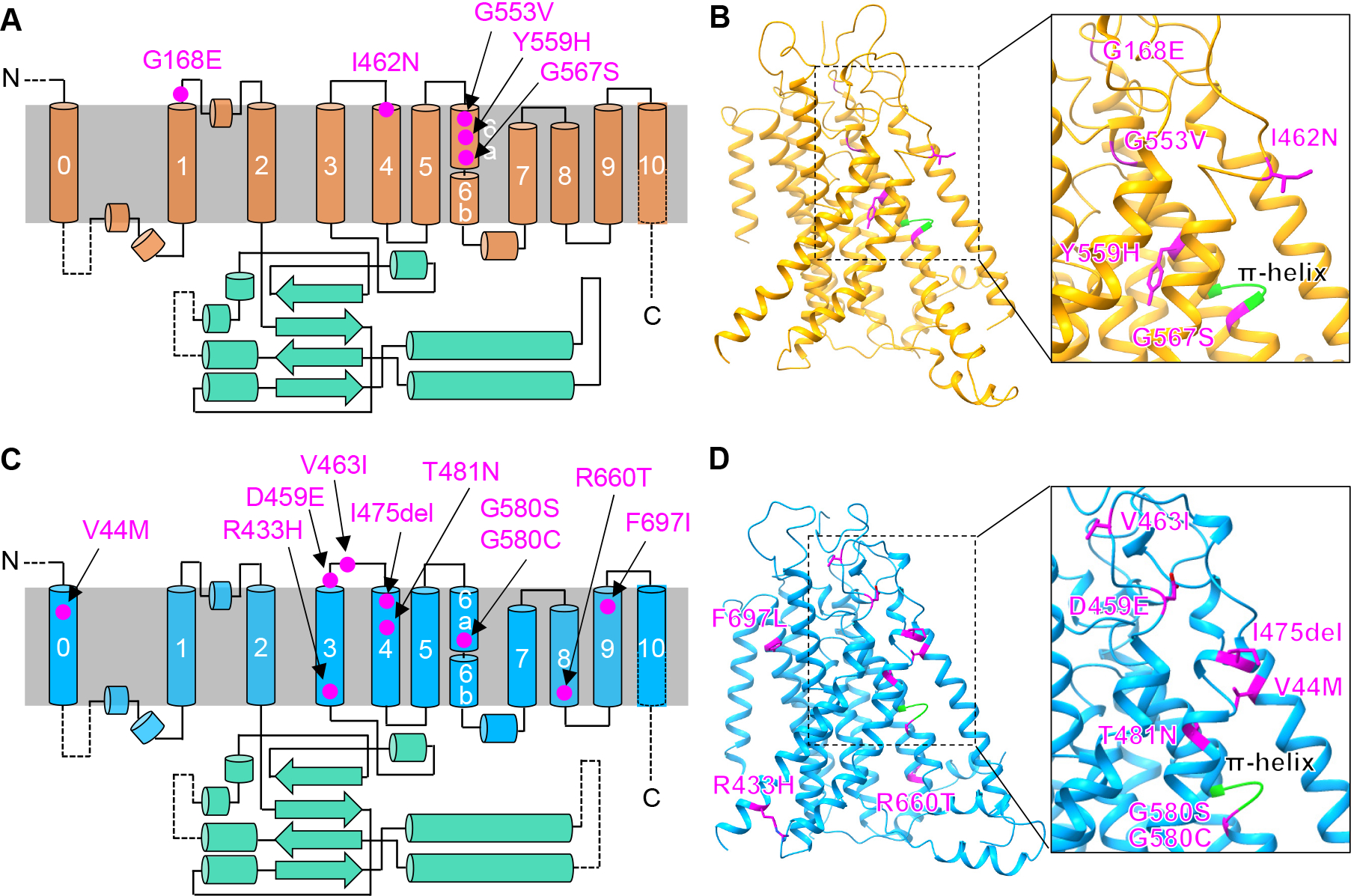
(A and C) Location of five mutations of TMEM63A (A) 31–34 and 10 mutations of TMEM63B (C) 35 on a topology diagram.
(B and D) Mutations mapped to structures of TMEM63A (B) and TMEM63B (D).
See also Figure S8.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Jeffrey R. Holt (Jeffrey.holt@childrens.harvard.edu).
Materials availability
Plasmids generated in this study are available upon request.
Data and code availability
The cryo-EM volumes and coordinates have been deposited in EMDB and PDB and are publicly available as of the data of publication. Accession numbers are listed in the key resources table. All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-GFP | Thermo Fisher Scientific | Cat#A-31852; RRID: AB_162553 |
| Rabbit polyclonal anti-Flag | Sigma-Aldrich | Cat#F7425; RRID: AB_439687 |
| Mouse monoclonal anti-β-actin | Santa Cruz Biothechnology | Cat#sc-47778 HRP; RRID: AB_2714189 |
| Mouse monoclonal anti-HCN1 | Addgene | Cat#180092; RRID: AB_2750810 |
| Mouse monoclonal anti-ASIC1 | Addgene | Cat#184198 |
| Rabbit polyclonal anti-TMEM63A | Sigma-Aldrich | Cat#HPA068918; RRID: AB_2686057 |
| Rabbit polyclonal anti-HA | Abcam | Cat#ab9110; RRID: AB_307019 |
| Bacterial and Virus Strains | ||
| XL10-Gold Ultracompetent E. coli | Agilent Technologies | Cat#200315 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Glutaraldehyde | Sigma-Aldrich | Cat#G5882 |
| Disuccinimidyl suberate | Thermo Fisher Scientific | Cat#A39267 |
| n-Dodecyl-β-D-Maltopyranoside (DDM) | Anatrace | Cat#D310-CH210 |
| lauryl maltose neopentyl glycol (LMNG) | Anatrace | Cat#NG310-CH210 |
| Lipofectamine 3000 | Thermo Fisher Scientific | Cat#L3000008 |
| Expi293 Expression Medium | Thermo Fisher Scientific | Cat#A1435102 |
| Polyethylenimine Max 40k | Polysciences | Cat#24765–1 |
| tris(2-carboxyethyl)phosphine (TCEP) | Sigma-Aldrich | Cat#646547 |
| Protease inhibitor cocktail | Sigma-Aldrich | Cat#S8830 |
| POPC:POPE:POPG lipid mix (3:1:1) | Anatrace | Cat#P516:P416:P616 MIX |
| 3C protease | Sigma-Aldrich | Cat#SAE0045 |
| MSP2N2 scaffold protein | Sigma-Aldrich | Cat#MSP12 |
| Critical Commercial Assays | ||
| Superose 6 Increase 10/300 GL | GE Healthcare | Cat#29–0915-96 |
| Quikchange Site-Directed Mutagenesis | Agilent Technologies | Cat#210515 |
| Deposited Data | ||
| Coordinates of human TMEM63A in Nanodiscs | This paper | PDB: 8EHW |
| Cryo-EM map of human TMEM63A in Nanodiscs | This paper | EMDB: EMD-28153 |
| Coordinates of human TMEM63B in LMNG | This paper | PDB: 8EHX |
| Cryo-EM map of human TMEM63B in LMNG | This paper | EMDB: EMD-28154 |
| Experimental Models: Cell Lines | ||
| Human: HEK293T cells | ATCC | CRL-3216 |
| Human: Expi293F suspended cells | Thermo Fisher Scientific | Cat#A14527 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Tmem63bHA/HA knock-in | Du et al. 37 | N/A |
| Recombinant DNA | ||
| cDNA: Human TMEM63A | Genscript | Accession number NM_014698.3 |
| cDNA: Human TMEM63B | Genscript | Accession number NM_018426.3 |
| cDNA: Human TMEM63C | Genscript | Accession number NM_020431.4 |
| Plasmid: pEG BacMam | Goehring et al. 56 | N/A |
| Plasmid: OSCA1.2-PP-EGFP | Jojoa-Cruz et al. 22 | N/A |
| Software and Algorithms | ||
| Serial EM | Mastronarde 57 | https://bio3d.colorado.edu/SerialEM |
| MotionCor2 | Zheng et al. 58 | https://emcore.ucsf.edu/cryoem-software |
| CTFFIND4 | Rohou and Grigorieff 59 | https://grigoriefflab.umassmed.edu/ctf_estimation_ctffind_ctftilt |
| Relion 3.0 | Scheres 60 | https://www3.mrc-lmb.cam.ac.uk/relion/ |
| crYOLO | Wagner et al. 61 | https://cryolo.readthedocs.io/en/stable/ |
| cryoSPARC V2 | Punjani et al. 62 | https://cryosparc.com/ |
| SBGrid | Morin et al. 63 | https://sbgrid.org/ |
| UCSF Chimera | Pettersen et al. 64 | https://www.cgl.ucsf.edu/chimera/download.html |
| UCSF ChimeraX | Pettersen et al. 65 | https://www.cgl.ucsf.edu/chimerax/download.html |
| PyMOL | Schrodinger, LLC | http://www.pymol.org/ |
| COOT | Emsley et al. 66 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PHENIX | Adams et al. 75 | https://phenix-online.org/download/ |
| HOLE | Smart et al. 42 | http://www.holeprogram.org/ |
| pClamp 10 | Molecular Devices | https://www.moleculardevices.com/products/axon-patch-clamp-system/acquisition-and-analysis-software/pclamp-software-suite |
| Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| Quantifoil R1.2/1.3 400-mesh gold grid | Electron Microscopy Sciences | Cat#Q3100AR1.3 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cultured cell lines
Suspended Expi293F cells were grown in Expi293 Expression Medium with 8% CO2 environment at a rotation speed of 100 rpm. When the cell density reached 1–2 × 106 cells/mL, constructs expressing TMEM63A or TMEM63B were transiently transfected with polyethylenimine Max 40k. For 1L of cell culture, 1 mg construct and 3 mg PEI Max 40k was used. Adherent HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium plus 10% fetal bovine serum and 10 μg/mL ciprofloxacin hydrochloride, with 5% CO2 environment. Transient transfections were performed with lipofectamine 3000 based on the manufacturer’s instructions.
Mice
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Oregon Health & Science University (protocol IP00002957). The Tmem63bHA/HA line has been maintained on a C57BL/6 background and is genotyped using the following primers: Forward- TCA ACA GCA GCA ACC CGAAG, Reverse- CAC ATG AAG TCC AGA GCCAG. Brain tissue from Tmem63bHA/HA knock-in or littermate Tmem63bWT/WT mice was collected at post-natal day 21 (P21), and snap-frozen in liquid nitrogen before use. Tissues were collected from animals of both sexes.
METHOD DETAILS
cDNA constructs and mutagenesis
The cDNA constructs of human TMEM63A (OHu08349C, Accession #: NM_014698.3), TMEM63B (OHu13152C, Accession #: NM_018426.3) and TMEM63C (OHu27782C, Accession #: NM_020431.4) were purchased from Genscript (Piscataway, NJ). The TMEM63A, B or C coding sequence was then subcloned into pEG BacMam vector 56, a kind gift from Eric Gouaux (OHSU Vollum Institute, OR). An EGFP tag was placed at the C-terminus of TMEM63 proteins with a 3C protease cleavage site in the middle. A TwinStrep tag was added to the EGFP C-terminus for affinity purification. The OSCA1.2-PP-EGFP and mouse TMEM63A-GFP constructs 22 was kindly provided by Ardem Patapoutian (Scripps Research Institute, CA). Flag-tagged OSCA1.2, human TMEM63A or TMEM63B constructs were generated by cloning the coding sequence to pEG BacMam vector with a C-terminal Flag tag. Drosophila TMEM63 cDNA was purchased from Addgene (Cat # 136598) and cloned into pEG BacMam with a C-terminal GFP tag. pcDNA3.1(+) construct expressing mouse TMEM63B cDNA (OMu53492C, Accession#: XM_006524138.3) with a GFP tag in the C-terminus was purchased from Genscript. The TMEM16A-GFP construct 67 was provided by Lily Yeh Jan (University of California at San Francisco, CA). Constructs expressing TRPP3-GFP and TMEM175-GFP were descripted previously 68,69. Chimeric constructs were generated with NEBuilder HiFi DNA Assembly Cloning Kit (New England Biolabs, Ipswich, MA). The OSCA1.2 concatemer was generated by cloning the coding sequence of OSCA1.2 E531K mutant into the upstream of OSCA1.2-PP-EGFP with a linker fragment in the middle that contains a glycophorin A helix, as described before 70. Point mutations, insertions or deletions were introduced with QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, La Jolla, CA). All constructs were confirmed by sequencing.
Fluorescence size-exclusion chromatography (FSEC)
Constructs expressing GFP-tagged proteins were transfected into HEK293T cells in 6-well plates with lipofectamine™ 3000 (Thermo Fisher Scientific). Around 30 h following the transfection, cells were washed twice with ice-cold PBS buffer (pH 7.4) and lysed in Tris buffer, supplemented with 1% DDM (Anatrace) and Protease Inhibitor Cocktail (Thermo Fisher Scientific), for 30 min in cold room. The cell lysates were then cleared by centrifugation at maximum speed and 40 μL samples were applied to FSEC (Xbridge Protein BEH SEC Column, 450 Å, 3.5 μm, 7.8 mm X 150 mm, SKU176003598, Waters Corporation, Milford, MA) in Tris buffer supplemented with 0.05% DDM (Anatrace) and 1 mM Tris (2-carboxyethyl) phosphine (TCEP, P1020–25, Ubiquitin-Proteasome Biotechnologies, Dallas, TX).
Cross-linking assay
For the cross-linking of purified proteins, 7 μg of purified OSCA1.2-GFP, TMEM63A-GFP or TMEM63B-GFP protein was used in 30 μL reaction buffer (50 mM HEPES, 150 mM NaCl, pH 7.4) supplemented with 0.05% DDM (Anatrace). The cross-linker glutaraldehyde (Cat# G5882, Sigma-Aldrich, St. Louis, MO) was then added to the final concentration of 10 mM. The mixture was incubated at room temperature for 5 min. The reaction was then quenched by adding 3 μL of 1 M Tris (pH 7.4). The proteins were then subjected to SDS-PAGE and stained with Coomassie Brilliant Blue R-250 (Bio-Rad, Hercules, CA)
For the cross-linking of over-expressed proteins in HEK293T cell lysates, cells in a 6-well plate were first washed with PBS buffer (pH 8.0) twice. The cell lysates were then prepared with 500 μL reaction buffer supplemented with 1% DDM and Halt™ Protease Inhibitor Cocktail (Thermo Fisher Scientific, Waltham, MA). Of note, DDM is used for all cell lysate preparations in our study unless otherwise specified. Cell lysates (30 μL) were incubated with glutaraldehyde (Sigma-Aldrich) at indicated concentrations for 5 min at room temperature. 3 μL 1 M Tris (pH 7.4) was then added to quench the reaction. The mixture was then subjected to western blot with antibodies against GFP (A-31852, Thermo Fisher Scientific), Flag (F7425, Sigma-Aldrich) and β-actin (sc-47778, Santa Cruz Biotechnology, Dallas, TX).
For cross-linking in intact cells, HEK293T cells expressing indicated proteins in 6-well plates were washed three times with PBS buffer (pH 8.0). At the last wash, the cross-linker disuccinimidyl suberate (DSS, A39267, Thermo Fisher Scientific) was added to a variety of concentrations, as indicated. The reaction mixture was incubated for 10 min at room temperature. 1 M Tris (pH 7.5) solution was then added to a final concentration of 50 mM and incubated for 10 min at room temperature to quench the reaction. The cells were then washed twice with Tris buffer (20 mM Tris, 150 mM NaCl, pH 7.4) and lysed in the same buffer supplemented with 1% DDM (Anatrace) and Protease Inhibitor Cocktail (Thermo Fisher Scientific). The cell lysates were then subjected to western blot with anti-GFP antibody (A-31852, Thermo Fisher Scientific).
Blue native PAGE
Blue native PAGE followed by immunoblotting was performed as described 68 with modifications of sample preparation. Cell lysates from HEK293T cells expressing OSCA1.2, TMEM63A or TMEM63B were prepared with NativePAGE Sample Prep Kit (Thermo Fisher Scientific) according to the manufacturer’s manual. To prepare samples of mouse brain tissue for blue native PAGE, the whole brain of Tmem63bWT/WT or Tmem63bHA/HA knock-in mice was first washed with ice-cold PBS solution (pH 8). The brain tissue was then homogenized on ice in 4 ml NativePAGE 1X sample buffer contained in the NativePAGE Sample Prep Kit. The mixture was next centrifuged at a maximal speed to collect the pellet (cell membrane fraction). Wash the pellet with 2 ml NativePAGE 1X sample buffer twice. The washed pellet was then solubilized in 2 ml sample buffer supplemented with 1% DDM (Anatrace) and Protease Inhibitor Cocktail (Thermo Fisher Scientific) at cold room for 2 hrs, followed by centrifugation at a maximal speed. The supernatant was collected and diluted 5 folds. When preparing brain tissue sample for denaturing western blot, the whole brain was directly homogenized and solubilized in 2 ml CelLytic MT lysis buffer (Sigma-Aldrich) with 1% DDM and Protease Inhibitor Cocktail. To run blue native gel, Coomassie blue G-250 was added to the supernatants from HEK293T cells or mouse brain tissue at 4:1 of detergent:G-250 ratio. Protein complexes were separated at 150 V for 100 min at room temperature using NuPAGE Novex 4–16% Bis-Tris protein gels (Thermo Fisher Scientific). For immunoblotting, separated proteins were transferred to polyvinylidene difluoride membrane using NuPAGE Novex transfer buffer (Thermo Fisher Scientific). The membrane was stained with ponceau red to visualize the protein ladder, which was then marked with ballpoint pen. Next, the membrane was blocked with 3% skimmed mild in PBS buffer with 0.1% tween-20 for 30 min and then incubated with antibodies against Flag (F7425, Sigma-Aldrich), HCN1 (180092, Addgene), ASIC1 (184198, Addgene), TMEM63A (HPA068918, Sigma-Aldrich) or HA (ab9110, Abcam, Cambridge, UK).
Live-cell single-molecule imaging and tracking
Single-molecule imaging was done in transfected CHO cells using Total Internal Reflection (TIRF) Microscopy at an Olympus IX71 microscope equipped with a 100x NA1.7 objective and a 488nm laser. Movies of 200 frames were recorded at 20 Hz, a power density of 110 W/cm2 (hTMEM63A-GFP was recorded at 33 Hz and a power density of 360 W/cm2 for a better quality) and 100 nm pixel size with an EMCCD camera (Andor iXon DV-897 BV). Tracking was done with the particle tracking tool from the MOSAIC suite in ImageJ, and intensities were extracted from the identified spots after background subtraction. Intensity time courses were normalized to the different frame rates and power densities to yield comparable brightness values in Figure 1H–1J and Figure S1C.
Protein expression and purification
The human TMEM63A or TMEM63B in the pEG BacMam vector was expressed in Expi293F cells by transient transfection using PEI Max 40k (polysciences). 36–48 hours after transfection, cells from 1 liter culture were collected for protein purification. The cell pellet was washed with ice cold Tris buffer (50 mM Tris, 150 mM NaCl, pH 7.6) and was then resuspended and sonicated in 50 mL Tris buffer plus 1 mM tris(2-carboxyethyl)phosphine (TCEP) and protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). The membrane fraction was collected via ultracentrifugation for 30 min at 40,000 rpm. To purify proteins in detergent LMNG, the membrane fraction was first homogenized in 60 mL Tris buffer supplemented with protease inhibitor cocktail, 1 mM TCEP and 0.7% LMNG/0.07% CHS (Anatrace, Maumee, OH). The homogenized mixture was then incubated at 4 °C for 2.5 h on a nutating mixer, followed by ultracentrifugation at 40,000 rpm for 1 h. The supernatant was then incubated with 1 mL Strep-Tactin XT 4Flow resin (IBA Lifesciences, Gottingen, Germany) at 4 °C for 3 h. The resin was washed with 10 column volumes of buffer W (IBA Lifesciences) plus 1 mM TCEP and 0.005% LMNG/0.0005% CHS. The GFP-tagged human TMEM63A or TMEM63B was eluted with buffer BXT (IBA Lifesciences) plus 1 mM TCEP and 0.005% LMNG/0.0005% CHS. The eluate was concentrated with a 100 kDa MWCO Amicon Ultra Centrifugal filter (MilliporeSigma, Burlington, MA) and the GFP tag was removed by treatment with 3C protease (Sigma-Aldrich, 1:100 in molar ratio) overnight at 4 °C. The TMEM63A or B protein was then further purified by size-exclusion chromatography (SEC) using a Superose 6 Increase column (GE Healthcare, Chicago, IL) equilibrated with Tris buffer supplemented with 1 mM TCEP and 0.005% LMNG/0.0005% CHS. Fractions corresponding to the TMEM63A or TMEM63B were pooled and concentrated to around 2.5 mg/mL. The OSCA1.2 protein was expressed and purified as described previously 22 with the exception that Expi293F cells were used.
To obtain purified proteins in lipid nanodiscs, the TMEM63A or TMEM63B was first purified in detergent n-Dodecyl-β-D-maltoside (DDM, Anatrace) using the same purification procedures with LMNG replaced by DDM. In the membrane fraction solubilization, the 1% DDM/0.1% CHS was used, while in other steps, 0.05% DDM/0.005% CHS was used. After elution and concentration, purified TMEM63A or TMEM63B with GFP tag was mixed with MSP2N2 scaffold protein and lipid (POPC:POPE:POPG in a 3:1:1 ratio, Anatrace) at a molar ratio of TMEM63 monomer:MSN2P2:lipid = 1:4:30. The mixture was incubated on ice for 1 hour. To remove the DDM detergent, around 10–20 mg Bio-Beads SM2 (Bio-Rad, Hercules, CA) were added to the mixture, which was then incubated at 4 °C with constant rotation for 1 h. Another batch of Bio-Beads SM2 was added to the system for 1 hour rotation at 4 °C. A third batch of Bio-Beads SM2 together with 3C protease was added and the mixture was incubated overnight at 4 °C. The Bio-Beads SM2 were then removed and the sample was applied to SEC with a Superose 6 Increase column equilibrated with Tris buffer plus 1 mM TCEP. Fractions corresponding to the TMEM63A or TMEM63B nanodiscs were collected and reapplied to the SEC. The nanodisc fractions were then pooled and concentrated to around 2 mg/mL.
Cryo-EM sample preparation and data collection
A 3 μL drop of purified TMEM63A or TMEM63B protein in either detergent LMNG or nanodisc was applied to a glow-discharged Quantifoil R1.2/1.3 400-mesh gold grid (Electron Microscopy Sciences, Hatfield, PA). The grid was then blotted for 6 s in 100% humidity at 6 °C with a wait time of 3 s. The blotted grid was plunged into nitrogen-cooled liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). All grids were screened using a FEI Talos Arctica microscope at the Harvard Cryo-EM Center for Structural Biology. The data was then collected using a 300-kV Titan Krios microscope (FEI) equipped with a K3 direct electron detector with Gatan Quantum Image Filter (Gatan) at the Harvard Cryo-EM Center. For TMEM63A in nanodisc, images were collected in a counting mode at a nominal magnification of 105,000x with a pixel size of 0.825 Å. 50 frames were collected per movie for a total accumulated dose of 50.7 electrons per Å using a defocus range of −1.0 to −2.0 μm. For TMEM63A in LMNG, images were collected with a 200 kV Talos Arctica at a nominal magnification of 36,000x with a pixel size of 1.1 Å. 47 frames were collected per movie for a total accumulated dose of 52 electrons per Å with a defocus range of −1.4 to −2.5 μm. For TMEM63B in either nanodisc or LMNG, images were collected in a counting mode at a nominal magnification of 105,000x with a pixel size of 0.825 Å. 50 frames were collected per movie for a total accumulated dose of 53 electrons per Å with a defocus range of −0.8 to −2.1 μm.
Cryo-EM imaging processing
The image processing for both TMEM63A and TMEM63B is shown in Figure S2G and S3G, respectively. In short, dose fractionated images were motion corrected with MotionCor2 58 (v1.2.6) followed by CTF estimation using CTFFIND4 59. Particle picking was carried out using crYOLO 61 resulting in 4,552,788 and 2,433,802 particles for TMEM63A and TMEM63B respectively. Classification steps are carried out in RELION 60 (v3.0) unless otherwise noted. TMEM63A particles were subjected to multiple rounds of 3D classification until a clear secondary structure in the transmembrane region was observed, resulting in 198,144 particles being selected. CTF parameters were re-estimated for these particles using patch CTF estimation within cryoSPARC prior to non-uniform refinement 71 in cryosparc, resulting in a 3.8Å reconstruction.
TMEM63B particles then underwent 2D classification resulting in 1,994,464 particles before a single round of full particle 3D classification giving 550,379 well aligning particles. A subsequent masked classification excluding the micelle was carried out giving a single class of 64,399 with high resolution features. These particles were polished in RELION 72 and CTF parameters were re-estimated in cryoSPARC patch CTF refinement resulting in a reconstruction at 3.64Å from cryoSPARC non-uniform refinement. Map quality and resolution was subsequently slightly improved using cryosparc local refinement excluding the micelle giving a final map at 3.62Å.
Structural biology applications other than cryosparc used in this project were compiled and configured by SBGrid 63.
Cryo-EM model building and refinement
The sharpened EM density maps obtained from cryoSPARC 62 were used for atomic model building. For both TMEM63A and TMEM63B, the Alphafold2 39 -predicted model served as an initial template. The model was first fitted into the density map in UCSF ChimeraX 65. Then, each individual residue was manually examined and adjusted to fit the map in Coot 66. To build a model for the IL2 domain, the IL2 structure predicted by Alphafold2 was docked into the density map with rigid body fitting and adjusted to fit the density map in secondary structural level using UCSF ChimeraX. Due to the missing densities, several fragments in TMEM63A (M1-D29, W76-C114, A369-E380, C711-A807) and TMEM63B (M1-N18, R71-F125, V307-V330, V381-E392, Y741-Q832) were excluded from final models. Generated models were subsequently refined in PHENIX using phenix.real_space_refine 73,74 and manually inspected in Coot for several iterations. The model geometry was evaluated with the MolProbity Web Server 75. The ion permeation pore was predicted using the HOLE program 42. Figures were generated with Pymol (Schrödinger), UCSF ChimeraX or Chimera 64.
Molecular dynamics simulations
A cryo-EM model that lacked IL1 (residues R71-F125) was used to build a complete human TMEM63B model for simulation studies. This model had a RMSD Cα of ~0.8 Å with respect to the deposited structure (PDB: 8EHX) and retained a IL2 generated during refinement. A trimmed TMEM63B sequence (residues P19-N740) was submitted to AlphaFold2 v2.1.0 39, which made 50 predictions without relaxation in both monomer and multimer mode 76. The top-ranking structures differed in the number of helical elements in IL1 (Figure S5G): multimer mode predicted two helices (conformation C1; ipTM+pTM 0.18) while monomer mode predicted one (conformation C2; mean pLDDT 79.19). Residues V65-G70 and C126 were removed from the cryo-EM structure and then IL1 of each top-ranked prediction was positioned using a MatchMaker alignment of 20 residues flanking IL1 in ChimeraX 65. TMEM63B systems based on the cryo-EM structure were built with and without residues P19-L36 (+/− NTD; see Figure S5F). Additional systems using the complete AF2-monomer prediction described above were also built. All TMEM63B systems have a N-terminal protonated proline and C-terminal methylamidation, as well as disulfide bonds involving residues S320-S327 and S384-S89. Systems with the NTD (+NTD) have an additional disulfide bond involving residues S23-S559. A monomeric OSCA1.2 was modeled using a 3.1 Å resolution structure in nanodisc (PDB: 6MGV). All non-terminal missing loops (residues P51-M70, V123-Y155, I402-K419, and N491-I501) were replaced based on an AlphaFold2-multimer prediction of full-length dimeric OSCA1.2 (ipTM+pTM 0.83). Here, loops were placed using a MatchMaker alignment of 10 flanking residues.
After selecting neutral histidine protonation states to favor evident hydrogen bonding, our initial protein models were oriented and positioned in the membrane plane using the PPM 3.0 webserver (https://opm.phar.umich.edu/ppm_server3). Each protein was embedded in either pure POPC (VMD Membrane Builder 77) or a more-realistic soybean membrane comprising seven types of phospholipids and two plant sterols (https://user.eng.umd.edu/~jbklauda/memb.html). In all cases, the initial patch was patterned in a regular array, trimmed to a final x- and y-extent of 130 Å, and lipids with head groups within 1.4 Å of protein were removed. The protein-lipid system was solvated with 18 Å box padding in the z-direction using VMD solvate. The systems were neutralized, and ions were added to achieve a final concentration of 150 mM KCl with VMD autoionize.
All MD simulations were performed using NAMD 2.14 78 and CHARMM36 force fields with CMAP correction and the TIP3P model for water 79. A van der Waals cutoff of 12 Å (with a switching function starting at 10 Å) was used with periodic boundary conditions. The particle mesh Ewald method was used to calculate long range electrostatic interactions without cutoff and with a grid point density >1 Å3. An integration timestep of 2 fs was used with the SHAKE algorithm. Electrostatic interactions were computed every other time step.
Equilibrium simulations to obtain a well-packed and disordered membrane bilayer were carried out in steps that included a minimization of 1,000 steps followed by 0.5 ns of free dynamics with everything fixed except for lipid tails, followed by a second minimization of 1,000 steps and 0.5 ns of free dynamics with only the protein constrained (k = 1 kcal/mol/Å2). This step was followed by a 0.5 ns-long equilibration with the protein constrained except for IL1 and a final step with 0.5 ns of free dynamics with no constraints. All these steps used a Langevin damping coefficient of γ = 1 ps−1. The rest of the equilibrium and voltage simulations, lasting up to 200 ns and 100 ns respectively, were performed using γ = 0.1 ps−1. Langevin dynamics was used to maintain the temperature of the system at 310 K. The NpT ensemble at 1 atm was simulated using a hybrid Nosé-Hoover Langevin piston method with a decay period of 200 fs and 50 fs damping time constant. Simulations with a normalized applied electric field to generate a −500 mV membrane potential were carried out following Gumbart et al 80.
To obtain water density plots, systems were centered according to the protein position at each frame and sequentially unwrapped and wrapped using VMD PBCTools. The number density of water oxygen atoms throughout each trajectory was computed and converted to concentration using VMD’s volmap plugin with a resolution of 1 Å using frames saved every 50 ps. Density plots show averages computed using a box (90 × 90 × 100 Å3) centered at the protein’s center-of-mass. MATLAB surfaces were generated by a custom script with (Figure 3H) or without (Figure S7) interpolated shading. RMSD values were calculated every saved frame (5 ps) in VMD for a given selection after aligning that same selection to its initial conformation. Selections for TMEM63B include Cα atoms of transmembrane helices (residues 40–66, 148–178, 201–226, 427–458, 477–501, 506–545, 566–585, 620–638, 643–667, 674–702, and 707–727) and IL2 (residues 240–412). Same regions were defined for OSCA1.2 based on a structural alignment. Contacts between IL2 and TM6b/IL4H were quantified using buried surface area (BSA) computed every 5 ps. To compute the BSA between selections A and B, the VMD measure sasa command was used to calculate . Selections for TMEM63B were residues 589–614 (TM6b/ILH4) and residues 278 to 354 (IL2). Molecular figures from simulations were rendered using VMD.
Surface protein biotinylation assay
The biotinylation assay for detecting proteins in cell surface membrane was performed as described previously 81. In brief, HEK293T cells in 6-well plates were transfected with OSCA1.2 constructs using lipofectamine 3000 (Thermo Fisher Scientific). After 48 hours, cells were washed twice with ice-cold PBS solution (pH 8), followed by incubation with sulfo-NHS-SS-Biotin (0.5 mg/mL in 500 μL PBS, Piece, Rockford, IL) at room temperature for 15 min. The excess nonreacted biotin was then quenched with NH4Cl. Next, cells were washed once with PBS and lysed using CelLytic M lysis buffer (Sigma-Aldrich) supplemented with proteinase inhibitor (Thermo Fisher Scientific). Cell lysates were then incubated with 50 μL Streptavidin bead (Pierce) for 4 h at 4 °C. The captured surface proteins were washed with PBS buffer plus 0.05% DDM (Anatrace) and were then subjected to western blot and detected with GFP antibody (Thermo Fisher Scientific) and β-actin antibody (sc-47778 HRP, Santa Cruz Biotechnology).
Proteoliposome reconstitution
Liposome reconstitution of purified membrane proteins were performed with dehydration/rehydration (D/R) method as previously described 82. Specifically, 20 mg azolectin (P5638, Sigma-Aldrich) was first dissolved in 1 mL chloroform in a glass test tube. The chloroform was then completely removed by nitrogen gas (around 15 min). We then added 2 mL D/R buffer (5 mM HEPES, 200 mM KCl, pH 7.2) to the glass tube to make a final concentration of 10 mg/mL for azolectin. To dissolve the azolectin, the mixture was vortexed for 10 min, followed by 30 min of bath sonication (around 6 cycles of 5 min sonication with 2 min off) until the mixture looked translucent. We took 200 μL of the resulting lipid solution to a 1.5 mL Eppendorf tube and added fresh purified protein (in DDM detergent) with a desired weight ratio (1:500 for OSCA1.2 and 1:200 for TMEM63A or TMEM63B). The mixture was then rotated gently for 1 h at room temperature. Next, the DDM detergent was removed by incubating with BioBeads SM2 (Bio-Rad) for 3 h at room temperature. After removal of the BioBeads, the mixture was centrifuged at 40,000 rpm for 45 min at 4 °C (MLS 50 rotor, Optima MAX Ultracentrifuge, Beckman Coulter Brea, CA). The supernatant was discarded and the pellet was resuspended in 80 μL D/R buffer. Four drops with each 20 μL of the resuspension were placed on a clean glass slide and were kept in a vacuum desiccator overnight for dehydration. Next day, 20 μL D/R buffer was added to each dehydrated spot and kept it at 4 °C (cold room) without disturbance for 24 h before carrying out electrophysiological recordings.
Electrophysiological recordings
Proteoliposome recordings were adapted based on previous description 82. We added 5 μL of the rehydrated drop to the bath solution (in mM): 140 KCl, 10 HEPES, 1 MgCl2, 10 glucose, pH 7.3 adjusted with KOH) in the recording chamber and waited for 5–10 min to allow the lipids to settle down. The unilamellar liposome membrane would then be visible under the microscope and ready to be recorded. The recording pipette was made from borosilicate glass with an outer diameter of 1.5 mm (Warner Instruments, Hamden, CT) using a micropipette puller (model P-2000, Sutter Instruments, Novato, CA) and polisher (ALA Scientific Instruments). The polished pipette was back-filled with internal solution containing (in mM): 130 NaCl, 5 KCl, 10 HEPES, 10 TEA-Cl, 1 CaCl2, 1 MgCl2, pH 7.3 (with NaOH). The pipette resistance was in the range of 3–5 MΩ when filled with the internal solution. The stretch-activated current was evoked in excised configuration with stepwise, 300 ms negative pressure pulses (Δ10 mmHg with 2 s between stimuli) using a high-speed pressure clamp system (HSPC-1, ALA Scientific Instruments, Farmingdale, NY). The membrane potential inside the patch was held at −80 mV. Currents were recorded using an Axopatch 200B patch-clamp amplifier (Molecular Devices, Union City, CA), filtered at 10 kHz through a low-pass analog filter, and sampled at a rate of 20 kHz with a digidata 1322A digitizer (Molecular Devices). The pClamp 10 software (Axon Instruments, Union City, CA) was used for data acquisition and analysis. Recordings were not corrected for liquid junction potential. For single-channel recordings, macroscopic currents were first recorded to find out the pressure threshold for stretch-induced activation. The single-channel openings were then resolved with the pressure threshold using the same conditions described above.
Electrophysiological recordings on cultured cells were performed as previously described 83. HEK293T cells in 6-well plates were transfected with indicated constructs using lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Cells were seeded 24–48 h following transfection onto matrigel-coated coverslips (BD Bioscience, Billerica, MA) and used for recordings. 4–12 h later, the stretch-activated macroscopic or single-channel currents were recorded with cell-attached configurations. Experimental conditions were similar to what was described above for proteoliposome recordings. The stretch-activated peak current (I) against pressure (P) was fitted with a Boltzmann equation I = Imin + (Imax−Imin)/(1+exp(−Z*(P−P50)), where the Imin is the minimal stretch-activated current and Imax is the maximal stretch-activated current; P50 is the pressure required for half-maximal activation and Z represents the slope of the activation curve.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were analyzed and plotted using Clampfit and GraphPad Prism 8.2.1 (GraphPad Software Inc., La Jolla, CA) and expressed as means ± SEM (standard error of the mean) as noted in the text and figure legends. Number (N) of patches recorded is indicated for each mean value in the text and figure legends. Statistical analyses were carried out using Student’s t-tests when comparing two groups or one-way or two-way ANOVA for three or more groups, with corrections for multiple comparisons. A probability value (p) of less than 0.05, 0.01, and 0.001 was considered statistically significant and indicated by *, **, and ***, respectively. The investigators were not blinded to the experimental groups.
Supplementary Material
Movie S1. Live cell imaging of TMEM63A-GFP molecules overexpressed in CHO cells, related to Figure 1.
Movie S2. Live cell imaging of OSCA1.2-GFP molecules overexpressed in CHO cells, related to Figure 1.
Movie S3. Live cell imaging of TMEM63B-GFP molecules overexpressed in CHO cells, related to Figure 1.
Highlights.
TMEM63s form monomeric mechanosensitive ion channels
Variations in IL2 affect oligomeric configuration of OSCA/TMEM63 family
Oligomerization dictates the threshold of mechanosensitivity in OSCA/TMEM63 family
Mechano-sensing IL2 and gating helix TM6 adopt distinct conformations in OSCA/TMEM63s
ACKNOWLEDGEMENTS
We thank members of the Holt/Géléoc laboratory for their contributions and discussions throughout the project. We thank Destinee Semidey for helping with genotyping mice. Cryo-EM data were collected with the assistance of Megan Mayer, Sarah Sterling and Richard Walsh at the Harvard Medical School cryo-Electron Microscopy Center supported by grants from the NIH National Institute of General Medical Sciences (GM103310). Simulations were performed using the Stampede2 supercomputer at the Texas Advanced Computing Center (XSEDE grant MCB140226 to M.S.). This work is supported by NIH/NIDCD RO1 DC01352 (JRH).
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kefauver JM, Ward AB, and Patapoutian A (2020). Discoveries in structure and physiology of mechanically activated ion channels. Nature 587, 567–576. 10.1038/s41586-020-2933-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox CD, Bavi N, and Martinac B (2019). Biophysical Principles of Ion-Channel-Mediated Mechanosensory Transduction. Cell Rep 29, 1–12. 10.1016/j.celrep.2019.08.075. [DOI] [PubMed] [Google Scholar]
- 3.Douguet D, and Honore E (2019). Mammalian Mechanoelectrical Transduction: Structure and Function of Force-Gated Ion Channels. Cell 179, 340–354. 10.1016/j.cell.2019.08.049. [DOI] [PubMed] [Google Scholar]
- 4.Jin P, Jan LY, and Jan YN (2020). Mechanosensitive Ion Channels: Structural Features Relevant to Mechanotransduction Mechanisms. Annu Rev Neurosci 43, 207–229. 10.1146/annurev-neuro-070918-050509. [DOI] [PubMed] [Google Scholar]
- 5.Murthy SE, Dubin AE, and Patapoutian A (2017). Piezos thrive under pressure: mechanically activated ion channels in health and disease. Nat Rev Mol Cell Biol 18, 771–783. 10.1038/nrm.2017.92. [DOI] [PubMed] [Google Scholar]
- 6.Booth IR, and Blount P (2012). The MscS and MscL families of mechanosensitive channels act as microbial emergency release valves. J Bacteriol 194, 4802–4809. 10.1128/jb.00576-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Procko C, Murthy S, Keenan WT, Mousavi SAR, Dabi T, Coombs A, Procko E, Baird L, Patapoutian A, and Chory J (2021). Stretch-activated ion channels identified in the touch-sensitive structures of carnivorous Droseraceae plants. Elife 10. 10.7554/eLife.64250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker RG, Willingham AT, and Zuker CS (2000). A Drosophila mechanosensory transduction channel. Science 287, 2229–2234. 10.1126/science.287.5461.2229. [DOI] [PubMed] [Google Scholar]
- 9.Yan Z, Zhang W, He Y, Gorczyca D, Xiang Y, Cheng LE, Meltzer S, Jan LY, and Jan YN (2013). Drosophila NOMPC is a mechanotransduction channel subunit for gentle-touch sensation. Nature 493, 221–225. 10.1038/nature11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Yan Z, Jan LY, and Jan YN (2013). Sound response mediated by the TRP channels NOMPC, NANCHUNG, and INACTIVE in chordotonal organs of Drosophila larvae. Proc Natl Acad Sci U S A 110, 13612–13617. 10.1073/pnas.1312477110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brohawn SG (2015). How ion channels sense mechanical force: insights from mechanosensitive K2P channels TRAAK, TREK1, and TREK2. Ann N Y Acad Sci 1352, 20–32. 10.1111/nyas.12874. [DOI] [PubMed] [Google Scholar]
- 12.Natale AM, Deal PE, and Minor DL Jr. (2021). Structural Insights into the Mechanisms and Pharmacology of K(2P) Potassium Channels. J Mol Biol 433, 166995. 10.1016/j.jmb.2021.166995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plant LD (2012). A Role for K2P Channels in the Operation of Somatosensory Nociceptors. Front Mol Neurosci 5, 21. 10.3389/fnmol.2012.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagriantsev SN, Gracheva EO, and Gallagher PG (2014). Piezo proteins: regulators of mechanosensation and other cellular processes. J Biol Chem 289, 31673–31681. 10.1074/jbc.R114.612697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young M, Lewis AH, and Grandl J (2022). Physics of mechanotransduction by Piezo ion channels. J Gen Physiol 154. 10.1085/jgp.202113044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan B, Akyuz N, Liu XP, Asai Y, Nist-Lund C, Kurima K, Derfler BH, Gyorgy B, Limapichat W, Walujkar S, et al. (2018). TMC1 Forms the Pore of Mechanosensory Transduction Channels in Vertebrate Inner Ear Hair Cells. Neuron 99, 736–753 e736. 10.1016/j.neuron.2018.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan B, Geleoc GS, Asai Y, Horwitz GC, Kurima K, Ishikawa K, Kawashima Y, Griffith AJ, and Holt JR (2013). TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron 79, 504–515. 10.1016/j.neuron.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng W, and Holt JR (2021). The Mechanosensory Transduction Machinery in Inner Ear Hair Cells. Annu Rev Biophys 50, 31–51. 10.1146/annurev-biophys-062420-081842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murthy SE, Dubin AE, Whitwam T, Jojoa-Cruz S, Cahalan SM, Mousavi SAR, Ward AB, and Patapoutian A (2018). OSCA/TMEM63 are an Evolutionarily Conserved Family of Mechanically Activated Ion Channels. Elife 7. 10.7554/eLife.41844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan F, Yang H, Xue Y, Kong D, Ye R, Li C, Zhang J, Theprungsirikul L, Shrift T, Krichilsky B, et al. (2014). OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 514, 367–371. 10.1038/nature13593. [DOI] [PubMed] [Google Scholar]
- 21.Hou C, Tian W, Kleist T, He K, Garcia V, Bai F, Hao Y, Luan S, and Li L (2014). DUF221 proteins are a family of osmosensitive calcium-permeable cation channels conserved across eukaryotes. Cell Res 24, 632–635. 10.1038/cr.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jojoa-Cruz S, Saotome K, Murthy SE, Tsui CCA, Sansom MS, Patapoutian A, and Ward AB (2018). Cryo-EM structure of the mechanically activated ion channel OSCA1.2. Elife 7. 10.7554/eLife.41845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Wang J, and Sun L (2018). Structure of the hyperosmolality-gated calcium-permeable channel OSCA1.2. Nat Commun 9, 5060. 10.1038/s41467-018-07564-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maity K, Heumann JM, McGrath AP, Kopcho NJ, Hsu PK, Lee CW, Mapes JH, Garza D, Krishnan S, Morgan GP, et al. (2019). Cryo-EM structure of OSCA1.2 from Oryza sativa elucidates the mechanical basis of potential membrane hyperosmolality gating. Proc Natl Acad Sci U S A 116, 14309–14318. 10.1073/pnas.1900774116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang M, Wang D, Kang Y, Wu JX, Yao F, Pan C, Yan Z, Song C, and Chen L (2018). Structure of the mechanosensitive OSCA channels. Nat Struct Mol Biol 25, 850–858. 10.1038/s41594-018-0117-6. [DOI] [PubMed] [Google Scholar]
- 26.Medrano-Soto A, Moreno-Hagelsieb G, McLaughlin D, Ye ZS, Hendargo KJ, and Saier MH Jr. (2018). Bioinformatic characterization of the Anoctamin Superfamily of Ca2+-activated ion channels and lipid scramblases. PLoS One 13, e0192851. 10.1371/journal.pone.0192851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dang S, Feng S, Tien J, Peters CJ, Bulkley D, Lolicato M, Zhao J, Zuberbuhler K, Ye W, Qi L, et al. (2017). Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature 552, 426–429. 10.1038/nature25024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulino C, Kalienkova V, Lam AKM, Neldner Y, and Dutzler R (2017). Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM. Nature 552, 421–425. 10.1038/nature24652. [DOI] [PubMed] [Google Scholar]
- 29.Brunner JD, Lim NK, Schenck S, Duerst A, and Dutzler R (2014). X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature 516, 207–212. 10.1038/nature13984. [DOI] [PubMed] [Google Scholar]
- 30.Jeong H, Clark S, Goehring A, Dehghani-Ghahnaviyeh S, Rasouli A, Tajkhorshid E, and Gouaux E (2022). Structures of the TMC-1 complex illuminate mechanosensory transduction. Nature 610, 796–803. 10.1038/s41586-022-05314-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukumura S, Hiraide T, Yamamoto A, Tsuchida K, Aoto K, Nakashima M, and Saitsu H (2022). A novel de novo TMEM63A variant in a patient with severe hypomyelination and global developmental delay. Brain Dev 44, 178–183. 10.1016/j.braindev.2021.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Tonduti D, Mura E, Masnada S, Bertini E, Aiello C, Zini D, Parmeggiani L, Cantalupo G, Talenti G, Veggiotti P, et al. (2021). Spinal cord involvement and paroxysmal events in “Infantile Onset Transient Hypomyelination” due to TMEM63A mutation. J Hum Genet 66, 1035–1037. 10.1038/s10038-021-00921-1. [DOI] [PubMed] [Google Scholar]
- 33.Yan H, Helman G, Murthy SE, Ji H, Crawford J, Kubisiak T, Bent SJ, Xiao J, Taft RJ, Coombs A, et al. (2019). Heterozygous Variants in the Mechanosensitive Ion Channel TMEM63A Result in Transient Hypomyelination during Infancy. Am J Hum Genet 105, 996–1004. 10.1016/j.ajhg.2019.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan H, Ji H, Kubisiak T, Wu Y, Xiao J, Gu Q, Yang Y, Xie H, Ji T, Gao K, et al. (2021). Genetic analysis of 20 patients with hypomyelinating leukodystrophy by trio-based whole-exome sequencing. J Hum Genet 66, 761–768. 10.1038/s10038-020-00896-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vetro A, Balestrini S, Pelorosso C, Masi A, Hambleton S, Argilli E, Conti V, Giubbolini S, Barrick R, Bergant G, et al. (2022). Stretch-activated ion channel TMEM63B associates with developmental and epileptic encephalopathies and progressive neurodegeneration. medRxiv, 2022.2011.2022.22282283. 10.1101/2022.11.22.22282283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabara LC, Al-Salmi F, Maroofian R, Al-Futaisi AM, Al-Murshedi F, Kennedy J, Day JO, Courtin T, Al-Khayat A, Galedari H, et al. (2022). TMEM63C mutations cause mitochondrial morphology defects and underlie hereditary spastic paraplegia. Brain 145, 3095–3107. 10.1093/brain/awac123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du H, Ye C, Wu D, Zang YY, Zhang L, Chen C, He XY, Yang JJ, Hu P, Xu Z, et al. (2020). The Cation Channel TMEM63B Is an Osmosensor Required for Hearing. Cell Rep 31, 107596. 10.1016/j.celrep.2020.107596. [DOI] [PubMed] [Google Scholar]
- 38.Lan C, Stulz A, Barthes NPF, Lauw S, Salavei P, Jung M, Heerklotz H, and Ulbrich MH (2021). Designed membrane protein heterodimers and control of their affinity by binding domain and membrane linker properties. Nanoscale 13, 20692–20702. 10.1039/d1nr06574b. [DOI] [PubMed] [Google Scholar]
- 39.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romero LO, Massey AE, Mata-Daboin AD, Sierra-Valdez FJ, Chauhan SC, Cordero-Morales JF, and Vasquez V (2019). Dietary fatty acids fine-tune Piezo1 mechanical response. Nat Commun 10, 1200. 10.1038/s41467-019-09055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Battle AR, Ridone P, Bavi N, Nakayama Y, Nikolaev YA, and Martinac B (2015). Lipid-protein interactions: Lessons learned from stress. Biochim Biophys Acta 1848, 1744–1756. 10.1016/j.bbamem.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 42.Smart OS, Neduvelil JG, Wang X, Wallace BA, and Sansom MS (1996). HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph 14, 354–360, 376. 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 43.Zubcevic L, and Lee SY (2019). The role of pi-helices in TRP channel gating. Curr Opin Struct Biol 58, 314–323. 10.1016/j.sbi.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karplus M, and Petsko GA (1990). Molecular dynamics simulations in biology. Nature 347, 631–639. 10.1038/347631a0. [DOI] [PubMed] [Google Scholar]
- 45.Jeong H, Clark S, Goehring A, Dehghani-Ghahnaviyeh S, Rasouli A, Tajkhorshid E, and Gouaux E (2022). Structure of C. elegans TMC-1 complex illuminates auditory mechanosensory transduction. bioRxiv, 2022.2005.2006.490478. 10.1101/2022.05.06.490478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peters CJ, Gilchrist JM, Tien J, Bethel NP, Qi L, Chen T, Wang L, Jan YN, Grabe M, and Jan LY (2018). The Sixth Transmembrane Segment Is a Major Gating Component of the TMEM16A Calcium-Activated Chloride Channel. Neuron 97, 1063–1077.e1064. 10.1016/j.neuron.2018.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dhaka A, Viswanath V, and Patapoutian A (2006). Trp ion channels and temperature sensation. Annu Rev Neurosci 29, 135–161. 10.1146/annurev.neuro.29.051605.112958. [DOI] [PubMed] [Google Scholar]
- 48.Kashio M, and Tominaga M (2022). TRP channels in thermosensation. Curr Opin Neurobiol 75, 102591. 10.1016/j.conb.2022.102591. [DOI] [PubMed] [Google Scholar]
- 49.Delmas P, Parpaite T, and Coste B (2022). PIEZO channels and newcomers in the mammalian mechanosensitive ion channel family. Neuron 110, 2713–2727. 10.1016/j.neuron.2022.07.001. [DOI] [PubMed] [Google Scholar]
- 50.Albuisson J, Murthy SE, Bandell M, Coste B, Louis-Dit-Picard H, Mathur J, Feneant-Thibault M, Tertian G, de Jaureguiberry JP, Syfuss PY, et al. (2013). Dehydrated hereditary stomatocytosis linked to gain-of-function mutations in mechanically activated PIEZO1 ion channels. Nat Commun 4, 1884. 10.1038/ncomms2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fettiplace R (2017). Hair Cell Transduction, Tuning, and Synaptic Transmission in the Mammalian Cochlea. Compr Physiol 7, 1197–1227. 10.1002/cphy.c160049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeng G, Aggarwal M, Yu WP, and Chen TY (2016). Independent activation of distinct pores in dimeric TMEM16A channels. J Gen Physiol 148, 393–404. 10.1085/jgp.201611651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim NK, Lam AK, and Dutzler R (2016). Independent activation of ion conduction pores in the double-barreled calcium-activated chloride channel TMEM16A. J Gen Physiol 148, 375–392. 10.1085/jgp.201611650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akyuz N, Karavitaki KD, Pan B, Tamvakologos PI, Brock KP, Li Y, Marks DS, and Corey DP (2022). Mechanical gating of the auditory transduction channel TMC1 involves the fourth and sixth transmembrane helices. Sci Adv 8, eabo1126. 10.1126/sciadv.abo1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng S, Dang S, Han TW, Ye W, Jin P, Cheng T, Li J, Jan YN, Jan LY, and Cheng Y (2019). Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling. Cell Rep 28, 567–579 e564. 10.1016/j.celrep.2019.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goehring A, Lee CH, Wang KH, Michel JC, Claxton DP, Baconguis I, Althoff T, Fischer S, Garcia KC, and Gouaux E (2014). Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc 9, 2574–2585. 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51. 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 58.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221. 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530. 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wagner T, Merino F, Stabrin M, Moriya T, Antoni C, Apelbaum A, Hagel P, Sitsel O, Raisch T, Prumbaum D, et al. (2019). SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol 2, 218. 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 63.Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, and Sliz P (2013). Collaboration gets the most out of software. Elife 2, e01456. 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 65.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 67.Tien J, Lee HY, Minor DL Jr., Jan YN, and Jan LY (2013). Identification of a dimerization domain in the TMEM16A calcium-activated chloride channel (CaCC). Proc Natl Acad Sci U S A 110, 6352–6357. 10.1073/pnas.1303672110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng W, Hussein S, Yang J, Huang J, Zhang F, Hernandez-Anzaldo S, Fernandez-Patron C, Cao Y, Zeng H, Tang J, and Chen XZ (2015). A novel PKD2L1 C-terminal domain critical for trimerization and channel function. Sci Rep 5, 9460. 10.1038/srep09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng W, Shen C, Wang L, Rawson S, Xie WJ, Nist-Lund C, Wu J, Shen Z, Xia S, Holt JR, et al. (2022). pH regulates potassium conductance and drives a constitutive proton current in human TMEM175. Sci Adv 8, eabm1568. 10.1126/sciadv.abm1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Last NB, Kolmakova-Partensky L, Shane T, and Miller C (2016). Mechanistic signs of double-barreled structure in a fluoride ion channel. Elife 5. 10.7554/eLife.18767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Punjani A, Zhang H, and Fleet DJ (2020). Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat Methods 17, 1214–1221. 10.1038/s41592-020-00990-8. [DOI] [PubMed] [Google Scholar]
- 72.Zivanov J, Nakane T, and Scheres SHW (2019). A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17. 10.1107/S205225251801463X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, and Adams PD (2018). Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544. 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21. 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Evans R, O’Neill M, Pritzel A, Antropova N, Senior A, Green T, Žídek A, Bates R, Blackwell S, Yim J, et al. (2022). Protein complex prediction with AlphaFold-Multimer. bioRxiv, 2021.2010.2004.463034. 10.1101/2021.10.04.463034. [DOI] [Google Scholar]
- 77.Humphrey W, Dalke A, and Schulten K (1996). VMD: visual molecular dynamics. J Mol Graph 14, 33–38, 27–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 78.Phillips JC, Hardy DJ, Maia JDC, Stone JE, Ribeiro JV, Bernardi RC, Buch R, Fiorin G, Henin J, Jiang W, et al. (2020). Scalable molecular dynamics on CPU and GPU architectures with NAMD. J Chem Phys 153, 044130. 10.1063/5.0014475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buck M, Bouguet-Bonnet S, Pastor RW, and MacKerell AD Jr. (2006). Importance of the CMAP correction to the CHARMM22 protein force field: dynamics of hen lysozyme. Biophys J 90, L36–38. 10.1529/biophysj.105.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gumbart J, Khalili-Araghi F, Sotomayor M, and Roux B (2012). Constant electric field simulations of the membrane potential illustrated with simple systems. Biochim Biophys Acta 1818, 294–302. 10.1016/j.bbamem.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zheng W, Yang X, Hu R, Cai R, Hofmann L, Wang Z, Hu Q, Liu X, Bulkley D, Yu Y, et al. (2018). Hydrophobic pore gates regulate ion permeation in polycystic kidney disease 2 and 2L1 channels. Nat Commun 9, 2302. 10.1038/s41467-018-04586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nikolaev YA, Cox CD, Ridone P, Rohde PR, Cordero-Morales JF, Vasquez V, Laver DR, and Martinac B (2019). Mammalian TRP ion channels are insensitive to membrane stretch. J Cell Sci 132. 10.1242/jcs.238360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zheng W, Gracheva EO, and Bagriantsev SN (2019). A hydrophobic gate in the inner pore helix is the major determinant of inactivation in mechanosensitive Piezo channels. Elife 8. 10.7554/eLife.44003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Live cell imaging of TMEM63A-GFP molecules overexpressed in CHO cells, related to Figure 1.
Movie S2. Live cell imaging of OSCA1.2-GFP molecules overexpressed in CHO cells, related to Figure 1.
Movie S3. Live cell imaging of TMEM63B-GFP molecules overexpressed in CHO cells, related to Figure 1.
Data Availability Statement
The cryo-EM volumes and coordinates have been deposited in EMDB and PDB and are publicly available as of the data of publication. Accession numbers are listed in the key resources table. All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-GFP | Thermo Fisher Scientific | Cat#A-31852; RRID: AB_162553 |
| Rabbit polyclonal anti-Flag | Sigma-Aldrich | Cat#F7425; RRID: AB_439687 |
| Mouse monoclonal anti-β-actin | Santa Cruz Biothechnology | Cat#sc-47778 HRP; RRID: AB_2714189 |
| Mouse monoclonal anti-HCN1 | Addgene | Cat#180092; RRID: AB_2750810 |
| Mouse monoclonal anti-ASIC1 | Addgene | Cat#184198 |
| Rabbit polyclonal anti-TMEM63A | Sigma-Aldrich | Cat#HPA068918; RRID: AB_2686057 |
| Rabbit polyclonal anti-HA | Abcam | Cat#ab9110; RRID: AB_307019 |
| Bacterial and Virus Strains | ||
| XL10-Gold Ultracompetent E. coli | Agilent Technologies | Cat#200315 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Glutaraldehyde | Sigma-Aldrich | Cat#G5882 |
| Disuccinimidyl suberate | Thermo Fisher Scientific | Cat#A39267 |
| n-Dodecyl-β-D-Maltopyranoside (DDM) | Anatrace | Cat#D310-CH210 |
| lauryl maltose neopentyl glycol (LMNG) | Anatrace | Cat#NG310-CH210 |
| Lipofectamine 3000 | Thermo Fisher Scientific | Cat#L3000008 |
| Expi293 Expression Medium | Thermo Fisher Scientific | Cat#A1435102 |
| Polyethylenimine Max 40k | Polysciences | Cat#24765–1 |
| tris(2-carboxyethyl)phosphine (TCEP) | Sigma-Aldrich | Cat#646547 |
| Protease inhibitor cocktail | Sigma-Aldrich | Cat#S8830 |
| POPC:POPE:POPG lipid mix (3:1:1) | Anatrace | Cat#P516:P416:P616 MIX |
| 3C protease | Sigma-Aldrich | Cat#SAE0045 |
| MSP2N2 scaffold protein | Sigma-Aldrich | Cat#MSP12 |
| Critical Commercial Assays | ||
| Superose 6 Increase 10/300 GL | GE Healthcare | Cat#29–0915-96 |
| Quikchange Site-Directed Mutagenesis | Agilent Technologies | Cat#210515 |
| Deposited Data | ||
| Coordinates of human TMEM63A in Nanodiscs | This paper | PDB: 8EHW |
| Cryo-EM map of human TMEM63A in Nanodiscs | This paper | EMDB: EMD-28153 |
| Coordinates of human TMEM63B in LMNG | This paper | PDB: 8EHX |
| Cryo-EM map of human TMEM63B in LMNG | This paper | EMDB: EMD-28154 |
| Experimental Models: Cell Lines | ||
| Human: HEK293T cells | ATCC | CRL-3216 |
| Human: Expi293F suspended cells | Thermo Fisher Scientific | Cat#A14527 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Tmem63bHA/HA knock-in | Du et al. 37 | N/A |
| Recombinant DNA | ||
| cDNA: Human TMEM63A | Genscript | Accession number NM_014698.3 |
| cDNA: Human TMEM63B | Genscript | Accession number NM_018426.3 |
| cDNA: Human TMEM63C | Genscript | Accession number NM_020431.4 |
| Plasmid: pEG BacMam | Goehring et al. 56 | N/A |
| Plasmid: OSCA1.2-PP-EGFP | Jojoa-Cruz et al. 22 | N/A |
| Software and Algorithms | ||
| Serial EM | Mastronarde 57 | https://bio3d.colorado.edu/SerialEM |
| MotionCor2 | Zheng et al. 58 | https://emcore.ucsf.edu/cryoem-software |
| CTFFIND4 | Rohou and Grigorieff 59 | https://grigoriefflab.umassmed.edu/ctf_estimation_ctffind_ctftilt |
| Relion 3.0 | Scheres 60 | https://www3.mrc-lmb.cam.ac.uk/relion/ |
| crYOLO | Wagner et al. 61 | https://cryolo.readthedocs.io/en/stable/ |
| cryoSPARC V2 | Punjani et al. 62 | https://cryosparc.com/ |
| SBGrid | Morin et al. 63 | https://sbgrid.org/ |
| UCSF Chimera | Pettersen et al. 64 | https://www.cgl.ucsf.edu/chimera/download.html |
| UCSF ChimeraX | Pettersen et al. 65 | https://www.cgl.ucsf.edu/chimerax/download.html |
| PyMOL | Schrodinger, LLC | http://www.pymol.org/ |
| COOT | Emsley et al. 66 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PHENIX | Adams et al. 75 | https://phenix-online.org/download/ |
| HOLE | Smart et al. 42 | http://www.holeprogram.org/ |
| pClamp 10 | Molecular Devices | https://www.moleculardevices.com/products/axon-patch-clamp-system/acquisition-and-analysis-software/pclamp-software-suite |
| Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| Quantifoil R1.2/1.3 400-mesh gold grid | Electron Microscopy Sciences | Cat#Q3100AR1.3 |


