Abstract
Phosphatase and tensin homolog (PTEN) loss associates to adverse outcomes in prostate cancer and can be measured via immunohistochemistry (IHC). The purpose of the study was to establish the clinical application of an in-house developed artificial intelligence (AI) image analysis workflow for automated detection of PTEN loss on digital images for identifying patients at risk of early recurrence and metastasis. Post-surgical tissue microarray sections from the Canary Foundation (n=1264) stained with anti-PTEN antibody were evaluated independently by pathologist conventional visual scoring (cPTEN) and an automated AI-based image analysis pipeline (AI-PTEN). The relationship of PTEN evaluation methods with cancer recurrence and metastasis was analyzed using multivariable Cox proportional hazard and decision curve models. Both cPTEN scoring by pathologist and quantification of PTEN loss by AI (High-Risk AI-qPTEN) were significantly associated to shorted MFS in univariable analysis (cPTEN HR: 1.54, CI:1.07–2.21, p=0.019; AI-qPTEN HR: 2.55, CI:1.83,3.56), p<0.001). In multivariable analyses, AI-qPTEN showed a statistically significant association with shorter metastasis-free survival (MFS) (HR:2.17, CI:1.49–3.17, p<0.001) and recurrence-free survival (HR:1.36, CI:1.06–1.75, p=0.016) when adjusting for relevant post-surgical clinical nomogram (CAPRA-S) while cPTEN does not show a statistically significant association (HR:1.33, CI:0.89–2, p=0.2 and HR:1.26, CI:0.99–1.62, p=0.063, respectively) when adjusting for CAPRA-S risk stratification. More importantly, AI-qPTEN was associated with shorter MFS in patients with favorable pathological stage and negative surgical margins (HR: 2.72, CI:1.46–5.06, p=0.002). Workflow also demonstrated enhanced clinical utility in decision curve analysis, more accurately identifying men who might benefit from adjuvant therapy post-surgery. This study demonstrates the clinical value of an affordable and fully automated AI-powered PTEN assessment for evaluating the risk of developing metastasis or disease recurrence after radical prostatectomy. Adding AI-qPTEN assessment workflow to clinical variables may affect post-operative surveillance or management options, particularly in low-risk patients.
INTRODUCTION
The incidence rates of advanced and metastatic prostate cancer (PCa) are steadily increasing, likely due to a decline in routine prostate cancer screening1,2. Although the 10-year survival for patients with localized PCa remains excellent (>95%), prostate cancer-specific death is almost exclusive to patients with advanced or metastatic disease, where survival rates drop to approximately 30%2. Identifying men at risk of developing metastatic PCa is crucial for management decisions, as these at-risk individuals could potentially benefit from intensified adjuvant therapies3. Several risk stratification strategies (Partin tables, Kattan and MSKCC nomogram, NCCN risk classification, CAPRA scores) have been created to improve disease prognostication; however, their impact on treatment management is debatable in clinical practice4,5. To improve the accuracy of risk prediction, molecular biomarker tests were introduced in pre- and post-operative settings as additive tests to clinical-pathological variables6,7. Despite the promise, their routine use has been hindered by high financial costs and feasibility4. As a result, an unmet need to develop cost-effective and accurate tools that could better predict clinically meaningful endpoints (i.e., metastasis, death or recurrence)8,9.
PTEN (Phosphatase and tensin homolog), a tumor suppressor gene which many studies advocate using its loss (assessed by immunohistochemical (IHC) assay) in clinical practice to improve risk assessment10–12. The IHC scoring method is qualitative, however it is quite challenging to interpret the heterogeneity seen in PTEN expression, further leading to misclassification and confounding associations with the outcomes10,13. Precision medicine aims to develop and implement unbiased, quantitative approaches that have been proven to improve disease prognostication and drug response prediction.
The development of digital biomarkers and artificial intelligence (AI) has a tremendous role in this regard14–16. We recently demonstrated that digital pathology and semi-automated quantitative assessment of PTEN loss (qPTEN) using a commercial digital image analysis (DIA) software could further improve the post-surgical risk stratification tool (CAPRA-S17) and better identify men at risk of experiencing shorter biochemical recurrence13. However, this type of image analysis still requires substantial manual effort from expert pathologists, including identifying the regions of interest and, therefore, is not fully automated18,19. We subsequently showed that AI-based workflow (AI-PTEN) could reliably detect PTEN loss in prostate cancer tissue cores, offering a streamlined objective biomarker assessment system20.
The objective of the current study was to evaluate the clinical utility of a fully automated PTEN loss detection workflow and investigate its value for the prediction of clinically significant endpoints such as disease recurrence- (RFS) and metastases-free survival (MFS) to improve risk stratification.
METHODS
Cohort Description
The Canary retrospective Prostate Cancer Tissue Microarray (TMA) cohort, described previously21, comprises cases from six participating sites (Stanford University, University of California San Francisco, University of Washington, University of British Columbia, University of Texas Health San Antonio, Eastern Virginia Medical School). The cohort is well suited for tissue-based biomarker validation studies. It reflects population diversity in North America, long clinical follow-up and a relatively high prevalence of events such as disease recurrence and metastasis21. At initial cohort development, 1264 patients who underwent radical prostatectomy (RP) between 1995 and 2004 were included on TMAs (3 cancer cores and 1 benign core for each case). Cancer grading was centrally re-reviewed (JKM) following ISUP 2014 and WHO 2016 classification22,23. Detailed clinical and pathological characteristics are provided in Supplemental Table S2.1.
Immunohistochemical staining, slide scanning, and visual assessment
Evaluation of PTEN IHC in this cohort has been previously reported from different tissue levels (i.e. sections within a TMA core block) and different antibody11. IHC in this study was performed on an automated Discovery XT (Ventana Medical System, Inc, Tucson, AZ) platform using the validated staining protocol and monoclonal anti-PTEN antibody (clone 138G6, Cell Signaling)12,13,24,25. Stained TMA (n=33) were scanned at 0.4948 microns per pixel (effective magnification ×20) on an Aperio scanner (Leica Biosystems; .svs file format) with JPEG2000 compression (Compression Quality 30, Compresion Ratio 29.54).
Conventional assessment of PTEN expression (cPTEN) was performed by a pathologist (TJ) using previously established criteria for manual scoring12,13,24,25. Quality assessment of TMA images was also performed visually (TJ) for the tissue, staining, and scanning quality artifacts as they can significantly impact the findings of these studies. Hence, the slides and/or cases with unacceptable artifacts were removed from study analyses. cPTEN assessment was performed by an expert GU pathologist (TJ) and PTEN loss was found in 24% (n=246/1025) of cases (Supplemental Table S2.1). All equivocal cores by visual reading were reviewed by a second reader (PP). A detailed description of quality assessment and exclusion criteria is provided in Supplemental Figure S1.1.
AI-based image analysis automated AI-based image analysis
The automated image analysis pipeline included few modifications as detailed in the Supplemental Figure 1.2, which further improves previously published AI algorithms for PTEN loss detection/classification on TMA core images (AI-PTEN)20. The core-based pipeline includes the following: TMA core extraction (step 1), stain normalization (step 2) and prediction of the following: cancer containing regions (step 3), presence or absence of PTEN loss at a core-level using a multi-resolution PTEN classification method (step 4), and areas with PTEN loss within cancer regions using a pixel-level segmentation approach (step 5). In the end, the pipeline generates the following quantitative measures for each core: binary prediction of PTEN status (loss vs no loss), area (μm2) of cancer, and area (μm2) of PTEN loss. In the remaining steps of the workflow, core-based image analysis outputs are utilized for patient-level statistical modeling of clinical endpoints, as described in the following sections.
Statistical Analysis
Optimizing selection of cancer containing cores
Patient-level statistical analysis requires the aggregation of AI predictions from all cores of a given patient. For this goal, the selection of sufficient cancer-containing cores has been introduced (Supplemental Figure 1.2, Step 6). Briefly, logistic regression models were trained using k-fold (k=7) cross-validation where all patients from one center were repetitively excluded from the model for further validation. The optimal thresholds for cancer detection for both core and patient-level analysis were selected as described in the Supplemental Methods and were used for downstream analysis (Supplemental Table S2.3). Finally, the total AI-based quantitative PTEN loss (AI-qPTEN) is represented on the patient-level as the AI-measured proportion of total PTEN loss within the total cancer area, AI − qPTEN = ∑ PTEN loss (um2)/∑ cancer (um2), as summarized in Supplemental Figure 1.2. Throughout the manuscript, any patient-level workflow is referred to as AI-qPTEN and any core-level analysis is referred to as AI-PTEN.
Outcome analysis
To evaluate the association of AI-qPTEN with patient outcomes, MFS and RFS were used as co-primary endpoints in this study. Recurrence-free intervals were defined as the time from surgery to either 1) a single serum prostate-specific antigen (PSA) level of > 0.2 ng/mL more than 8 weeks after RP or 2) receipt of salvage or secondary therapy after RP or 3) clinical or radiological evidence of metastatic disease after RP.
K-fold (where k=7) cross-validation was used to identify clinically relevant AI-qPTEN thresholds for the most significant association with MFS and RFS as described in the Supplemental Methods. Patients with higher than median threshold values of AI-qPTEN were considered to be at higher risk of experiencing recurrence and/or metastasis (High-Risk AI-qPTEN). Cox’s proportional hazards models were used for univariable and multivariable analysis to assess an association of cPTEN and AI-qPTEN with RFS and MFS after adjusting for standard clinicopathological features. All survival analysis was performed using maxstat and survival R packages26–28. Decision curve analysis was completed using dcurve R package29–32 to evaluate the net benefit of the combined CAPRA-S/AI-qPTEN model compared to CAPRA-S alone.
Core-based performance of AI-PTEN was reported by sensitivity, specificity, and accuracy and was compared to cPTEN as a reference. Here true positive is the correct classification for PTEN loss, and true negative is the correct classification for PTEN intact as predicted by AI. Fisher’s exact and Chi-square tests were utilized to evaluate the association of cPTEN with available clinical and pathological variables using SPSS Statistics v24 (Armonk, NY). All statistical tests were two-sided, with P values below 0.05 considered statistically significant.
RESULTS
Cohort characteristics
The final cohort consisted of 1025 patients. The complete summary of clinical and demographic information is presented in Table 1. Distribution of Grade Groups (GG) was as follows: GG=1, n=400/1025 (39%); GG=2, n=381/1025 (37%); GG≥3, n=223/1025 (22%). Almost half of the patients developed recurrence (n=457/1025, 45%). The median RFS was 5yr (interquartile range [IQR]: 1.1–7.0). The rate of metastasis was 14% (n=141/1025) with a median MFS of 6.4yrs (IQR: 4.7–8.4) (Table 1). As expected, there was a significant association of cPTEN with adverse clinicopathological variables except for pre-operative serum prostate-specific antigen (PSA) levels20 (Supplemental Table S2.1). AI-PTEN showed high sensitivity (86%) and specificity (86%) for detecting PTEN loss compared to cPTEN as a reference on the core-level (Supplemental Table S2.2) and AI-qPTEN achieved AUC=0.92 compared to cPTEN as a reference on the patient-level.
Table 1.
Description of clinicopathological factors in the Canary prostate cancer cohort.
| Characteristic | N = 10251 |
|---|---|
| Grade Group (GG) | |
| 1 | 400 (40%) |
| 2 | 381 (38%) |
| ≥3 | 223 (22%) |
| Unknown | 21 |
| Seminal Vesicle Invasion (SVI) | |
| Absent | 930 (93%) |
| Present | 71 (7.1%) |
| Unknown | 24 |
| Extraprostatic Extension (EPE) | |
| Absent | 700 (70%) |
| Present | 302 (30%) |
| Unknown | 23 |
| Surgical Margin | |
| Absent | 666 (68%) |
| Present | 312 (32%) |
| Unknown | 47 |
| Lymph Node Invasion | |
| Absent | 658 (95%) |
| Present | 32 (4.6%) |
| Unknown | 335 |
| Pre-Op PSA (ng/ml) | |
| Median | 6.4 |
| IQR | 4.8 – 9.5 |
| Unknown | 112 |
| Metastasis | |
| Absent | 875 (86%) |
| Present | 141 (14%) |
| Unknown | 9 |
| Metastasis-free Survival (yrs) | |
| Median | 6.4 |
| IQR | 4.7 – 8.4 |
| Unknown | 30 |
| Recurrence-free Survival (yrs) | 5.0 |
| Median | 5 |
| IQR | 1.1 – 7 |
| Unknown | 27 |
| Recurrence | |
| Absent | 559 (55%) |
| Present | 457 (45%) |
| Unknown | 9 |
Statistics presented: n (%); IQR: Interquartile Range
Prognostic association of AI-qPTEN with MFS
In k-fold cross-validation, an optimal AI-qPTEN threshold of 0.04 was identified as a predictor for shorter MFS (i.e., High-Risk AI-qPTEN) using log-rank statistics (Supplemental Table S2.4). In univariable analysis, High-Risk AI-qPTEN had a higher hazard associated with shorter MFS compared to cPTEN (HR:2.55, CI:1.83–3.56, p<0.001 vs HR:1.54, CI:1.07–2.21, p=0.019) (Table 2). Similar risk was observed for complete loss vs partial loss by stratified cPTEN analysis (HR:1.55, CI:1.00–2.41, p=0.049 vs HR:1.52, CI: 0.91–2.53, p=0.11, respectively)(Table 2). In Kaplan-Meir analysis, High-Risk AI-qPTEN better-stratified men at risk of experiencing a significantly shorter MFS compared to cPTEN (Figure 2), especially men classified with low and intermediate CAPRA-S risk scores (Supplemental Figure S2.1). In addition, AI-qPTEN showed a strong association with shorter MFS in CAPRA-S low and intermediate-risk patients with 7-years post-surgical follow-up (Supplemental Figure S2.3).
Table 2.
Univariable Cox’s proportional hazards analysis of AI-qPTEN and other post-surgical variables with recurrence- and metastasis-free survival. cPTEN: Conventional visual scoring, AI: Artificial Intelligence, AI-qPTEN: AI-based quantitative PTEN loss, CAPRA-S: Cancer of the Prostate Risk Assessment (CAPRA) post-surgical score, HR: Hazard Ratio, CI: Confidence Interval
| Recurrence-free Survival | Metastasis-free Survival | |||||||
|---|---|---|---|---|---|---|---|---|
| N | HR1 | 95% CI1 | p-value | N | HR1 | 95% CI1 | p-value | |
| Grade Group (GG) | 986 | 983 | ||||||
| 1 | — | — | — | — | ||||
| 2 | 1.36 | 1.09, 1.71 | 0.007 | 2.42 | 1.52, 3.85 | <0.001 | ||
| ≥3 | 2.39 | 1.89, 3.03 | <0.001 | 5.03 | 3.18, 7.95 | <0.001 | ||
| preopPSA | 895 | 1.04 | 1.03, 1.04 | <0.001 | 892 | 1.02 | 1.01, 1.03 | <0.001 |
| Seminal Vesicle Invasion (SVI) | 983 | 981 | ||||||
| Absent | — | — | — | — | ||||
| Present | 2.98 | 2.25, 3.96 | <0.001 | 2.32 | 1.44, 3.72 | <0.001 | ||
| Surgical Margin | 960 | 959 | ||||||
| Absent | — | — | — | — | ||||
| Present | 1.92 | 1.58, 2.33 | <0.001 | 1.24 | 0.87, 1.76 | 0.2 | ||
| Extraprostatic Extension (EPE) | 984 | 981 | ||||||
| Absent | — | — | — | — | ||||
| Present | 1.98 | 1.64, 2.40 | <0.001 | 1.56 | 1.11, 2.18 | 0.01 | ||
| Lymph Node Invasion | 676 | 673 | ||||||
| Absent | — | — | — | — | ||||
| Present | 4.15 | 2.81, 6.14 | <0.001 | 3.69 | 2.11, 6.43 | <0.001 | ||
| cPTEN - Binary | 998 | 995 | ||||||
| Intact | — | — | — | — | ||||
| Loss | 1.66 | 1.36, 2.03 | <0.001 | 1.54 | 1.07, 2.21 | 0.019 | ||
| cPTEN - Stratified | 998 | 995 | ||||||
| Intact | — | — | — | — | ||||
| Loss - Partial | 1.38 | 1.02,1.86 | 0.035 | 1.52 | 0.91, 2.54 | 0.11 | ||
| Loss- Complete | 1.89 | 1.49,2.39 | <0.001 | 1.55 | 1.00, 2.41 | 0.049 | ||
| AI-qPTEN - Continuous | 998 | 2.62 | 1.79, 3.82 | <0.001 | 995 | 4.12 | 2.28, 7.44 | <0.001 |
| AI-qPTEN - Categorical | 998 | 995 | ||||||
| Ref | — | — | — | — | ||||
| High-Risk (>0.04) | 1.71 | 1.39, 2.11 | <0.001 | 2.55 | 1.83, 3.56 | <0.001 | ||
| CAPRA-S | 594 | 593 | ||||||
| Low | — | — | — | — | ||||
| Intermediate | 2.63 | 1.98, 3.50 | <0.001 | 3.78 | 2.35, 6.08 | <0.001 | ||
| High | 4.76 | 3.53, 6.44 | <0.001 | 3.21 | 1.89, 5.44 | <0.001 | ||
HR = Hazard Ratio, CI = Confidence Interval
Figure 2.
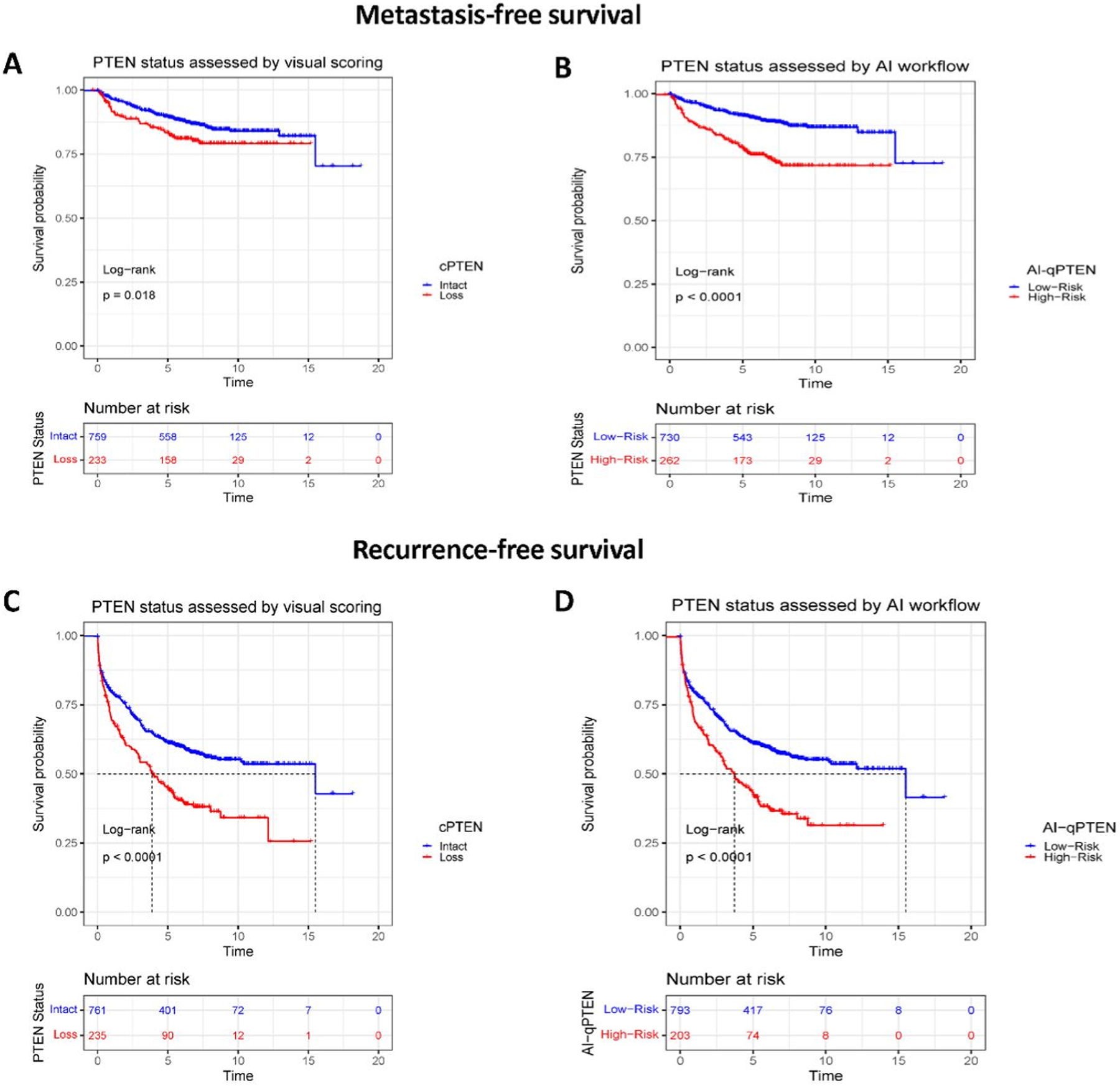
Evaluation of PTEN loss assessment methods (cPTEN and AI-qPTEN) and their association to metastasis- (Panels A and B) and recurrence-free survival (Panels C and D) in Kaplan-Meir analysis. Patients with AI-qPTEN >0.04 and >0.13 were considered as “High-risk” and the rest as “Ref” or “Low-Risk” for MFS and RFS endpoints. Patients stratified as having PTEN Loss or as High-risk by AI-qPTEN demonstrate statistically significant decreased median MFS and RFS for both cPTEN and AI-qPTEN. Log-rank test was performed to assess differences in survival. RFS: Recurrence-free survival, MFS: Metastasis-free survival, cPTEN: Conventional visual scoring, AI: Artificial intelligence, AI-qPTEN: AI-based quantitative PTEN loss
In a multivariable analysis involving post-surgery clinic-pathological variables, High-Risk AI-qPTEN was identified as a significant independent predictor of metastasis-free survival (HR:1.82, CI:1.24–2.66, p=0.002). In contrast, cPTEN did not achieve statistical significance when adjusting for clinical features (HR:1.12, CI:0.74–1.69, p=0.6) (Supplemental Table S2.6). Similar results were found when both PTEN assessment methods were tested with CAPRA-S in the multivariable models. The latter demonstrated added clinical utility of High-Risk AI-qPTEN to CAPRA-S (HR:2.2, CI:1.49–3.17 p <0.001) at 7-years after prostatectomy (Table 3) and where all available patients were included for all years of follow-up (Supplemental Table S2.5). More importantly, in the subset analysis of patients with favorable pathologic stage and negative surgical margins, AI-qPTEN identified men at risk of experiencing shorter metastasis-free survival (HR:2.72, CI:1.46–5.06, p=0.002) (Table 4).
Table 3:
Multivariable Cox proportional hazard analysis involving CAPRA-S and AI-qPTEN with metastasis- and recurrence-free survival at 7 and 5 years. Patients are stratified into low, intermediate, and high-risk CAPRA-S groups. Similarly, patients are stratified as High-risk if AI-qPTEN results in >0.04 proportion of cancer burden contained PTEN loss by AI assessment. For cPTEN analysis, patients were stratified as PTEN Intact (PTEN loss=0%) or PTEN Loss (PTEN loss>0%). cPTEN: Conventional visual scoring, AI: Artificial Intelligence, AI-qPTEN: AI-based quantitative PTEN loss, CAPRA-S: Cancer of the Prostate Risk Assessment (CAPRA) post-surgical score, HR: Hazard Ratio, CI: Confidence Interval
|
Metastasis-free survival at 7 year
(n=340) |
Recurrence-free survival at 5 year
(n=535) |
|||||||||||||||||
| No PTEN | With cPTEN | With AI-qPTEN | No PTEN | With cPTEN | With AI-qPTEN | |||||||||||||
| Characteristic | HR 1 | HR 1 | HR 1 | HR 1 | HR 1 | HR 1 | ||||||||||||
| CAPRA-S | ||||||||||||||||||
| Low | — | — | — | — | — | — | ||||||||||||
| Intermediate | 3.13 | 3.1 | 2.95 | 2.56 | 2.53 | 2.51 | ||||||||||||
| High | 2.62 | 2.45 | 2.14 | 4.67 | 4.55 | 4.45 | ||||||||||||
| cPTEN | ||||||||||||||||||
| Intact | — | — | ||||||||||||||||
| Loss | 1.33 | 1.26 | ||||||||||||||||
| AI-qPTEN | ||||||||||||||||||
| Ref | — | — | ||||||||||||||||
| High-Risk | 2.17 | 1.36 | ||||||||||||||||
| Concordance Index | ||||||||||||||||||
HR = Hazard Ratio, CI = Confidence Interval, Ref = Reference
Table 4:
Multivariable cox proportional hazard analysis for PTEN assessment (cPTEN and AI-qPTEN) with metastasis-free survival in patients with favorable pathologic stage and negative surgical margins (n=295). Patients are stratified as High-Risk if AI-qPTEN results in >0.04 proportion of cancer burden contained PTEN loss by AI assessment. For cPTEN analysis, patients were stratified as PTEN Intact (PTEN loss=0%) or PTEN Loss (PTEN loss>0%). cPTEN: Conventional visual scoring, AI: Artificial Intelligence, AI-qPTEN: AI-based quantitative PTEN loss, CAPRA-S: Cancer of the Prostate Risk Assessment (CAPRA) post-surgical score, HR: Hazard Ratio, CI: Confidence Interval
| With cPTEN | With AI-qPTEN | |||||
|---|---|---|---|---|---|---|
| Characteristic | HR1 | 95% CI1 | p-value | HR1 | 95% CI1 | p-value |
| Grade Group (GG) | ||||||
| 1 | — | — | — | — | ||
| 2 | 2.9 | 1.22, 6.92 | 0.016 | 2.62 | 1.10, 6.25 | 0.03 |
| 3+ | 4.8 | 1.89, 12.2 | <0.001 | 4.26 | 1.68, 10.8 | 0.002 |
| pre-Op PSA | 1.07 | 1.03, 1.11 | <0.001 | 1.07 | 1.03, 1.11 | <0.001 |
| cPTEN | ||||||
| Intact | — | — | ||||
| Loss | 1.43 | 0.72, 2.82 | 0.3 | |||
| AI-qPTEN | ||||||
| Ref | — | — | ||||
| High-Risk (>0.04) | 2.72 | 1.46, 5.06 | 0.002 | |||
| Concordance Index | 0.713 | 0.743 | ||||
HR = Hazard Ratio, CI = Confidence Interval
Decision curve analysis of combined CAPRA-S and High-Risk AI-qPTEN showed an increase in net benefit across several threshold probabilities, especially for patients who were classified as low-risk according to CAPRA-S (Figure 3). High-Risk AI-qPTEN demonstrated significant added value in accurately identifying men who were initially classified as low-risk according to CAPRA-S due to favorable pathologic stage and negative surgical margins (Figure 3). More specifically, High-Risk AI-qPTEN successfully reclassified 25% (68/269) of CAPRA-S low-risk men who experience significantly shorter MFS (Supplemental Figure S2.1).
Figure 3.
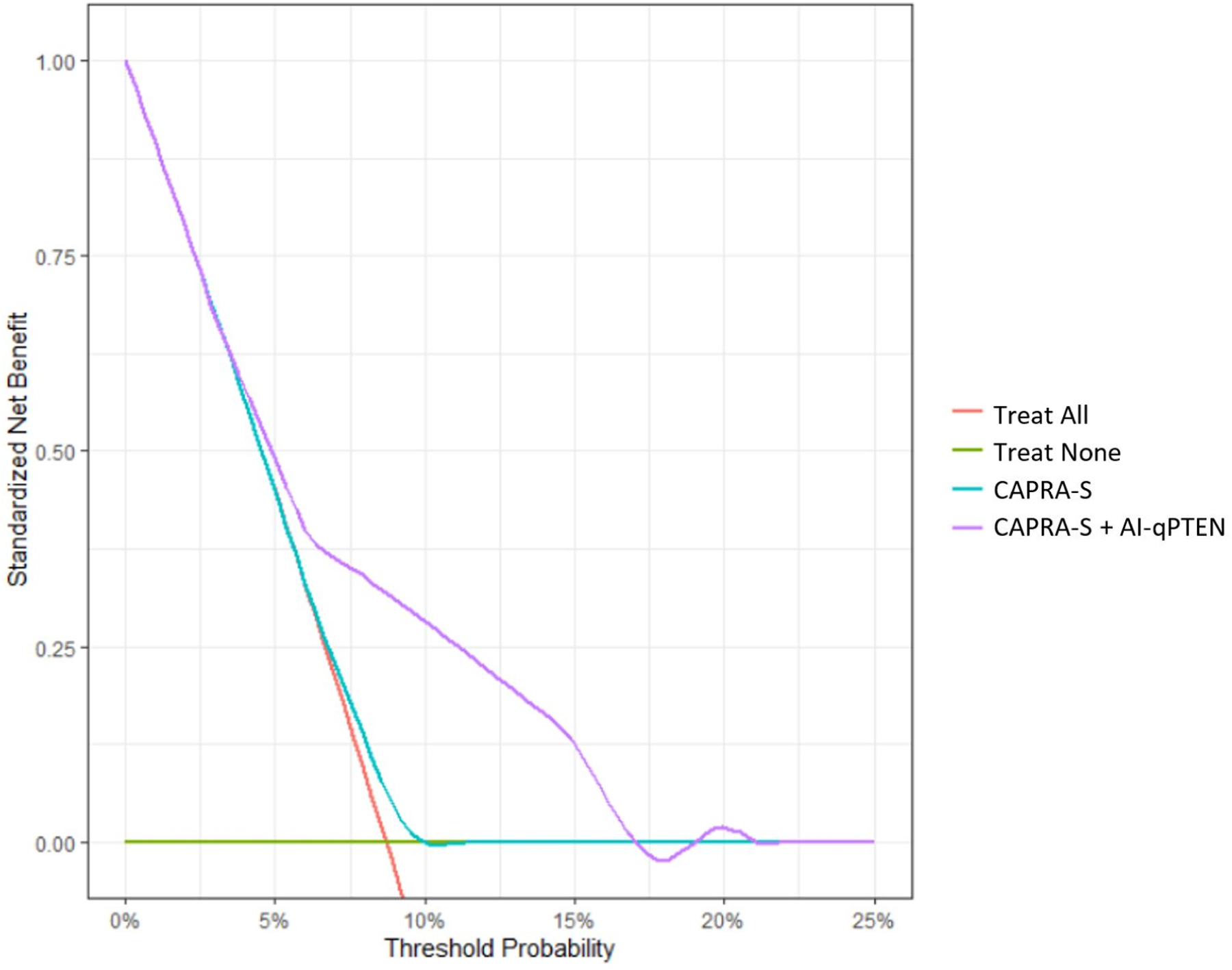
Decision curve analysis assessing the added benefit of including High-Risk AI-qPTEN (>0.04) in patients classified as low risk by CAPRA-S. Net increase in the proportion of men appropriately identified for therapeutic intervention post-surgery is shown on the y-axis and the associated probabilities on the x-axis. The green line represents a strategy of treating no men, assuming none of them experience metastasis. The orange line on the other hand assumes that all men will develop metastasis and therefore all men are treated. Multivariable models of CAPRA-S with and without High-Risk AI-qPTEN are shown as teal and purple lines, respectively. A combined model involving CAPRA-S and High-Risk AI-qPTEN showed greater net benefit across all threshold probabilities, signifying the ability to appropriately identify patients initially diagnosed with low risk PCa for additional therapeutic interventions after surgery. PCa: Prostate Cancer, CAPRA-S: Cancer of the Prostate Risk Assessment (CAPRA) post-surgical score, cPTEN: Conventional visual scoring, AI: Artificial intelligence, AI-qPTEN: AI-based quantitative PTEN loss
Discordant cases between AI-qPTEN and cPTEN results are highlighted in Supplemental Figure S2.5 and S2.6. Stratified cPTEN scoring, distinguishing between partial and complete PTEN loss by pathologist visual assessment is shown in Supplemental Figure S2.7. Multivariable analysis adjusting for CAPRA-S risk stratification additionally did not demonstrate differential prognostic significance for cPTEN loss stratified as complete or partial by visual assessment (Supplemental Table S2.11).
Prognostic association of AI-qPTEN with RFS
Using a similar 7-fold cross-validation strategy described above, 0.13 was selected as an optimal threshold as it showed the best association with shorter recurrence-free survival (Supplemental Table S2.4). In univariable analysis, men with >0.13 AI-qPTEN (High-Risk AI-qPTEN) had a statistically significant association with shorter RFS (HR: 1.71, CI:1.39–2.11, p<0.001) (Table 2). In Kaplan-Meir analysis, the cases assessed by AI-qPTEN and (cPTEN) demonstrated similar median recurrence-free survival (Figure 2). Unlike observations in MFS, stratified cPTEN assessment by complete vs partial loss demonstrated differential RFS risk (HR:1.89, CI: 1.49–2.39, p<0.001 vs. HR:1.38, CI:1.02–1.86, p=0.035, respectively)(Table 2), as additionally demonstrated in Supplemental Figure S2.7.
Interestingly, in multivariable analysis, AI-qPTEN was not significantly associated with recurrence-free survival (Supplemental Table S2.7). Although, multivariable analyses adjusted for CAPRA-S showed superior prognostic performance of High-Risk AI-qPTEN (HR:1.36, CI: 1.06–1.75, p=0.016) compared to cPTEN (Table 3 and Supplemental Table S2.8). Similar to MFS, ~20% of men diagnosed with low-risk CAPRA scores (50/270) were better classified according to their risk of shorter RFS when using AI-qPTEN (Supplemental Figure S2.2 and S2.4). It is additionally worth noting in multivariable analysis adjusting for CAPRA-S risk stratification, only complete cPTEN loss by visual assessment was significantly associated with RFS (Supplemental Table S2.11).
Cox’s proportional hazard analysis without regression models (i.e., cancer core selection and prognostically relevant AI-qPTEN) did not show any significant association with either MFS (Supplemental Table S2.9) or RFS (Supplemental Table S2.10), emphasizing the need to integrate these components into the overall workflow.
DISCUSSION
Precision medicine focuses on measuring clinically meaningful biomarkers that could improve treatment management and survival for cancer patients. Therefore, an accurate understanding of recurrence and metastasis risks after initial cancer treatment is critical. Over the past decade, various clinical risk assessment systems were created to better identify men at risk of developing recurrent or metastatic prostate cancer4,5, however, it is common to underestimate the “true risk” when using models composed only of clinicopathological variables33.
This multi-center study demonstrates the potential of improving post-surgical risk stratification of prostate cancer patients using a single molecular biomarker in combination with existing stratification tools such as CAPRA-S. In our prior work using commercially available image analysis software (HALO, Indica Labs) has shown that the degree of PTEN loss has prognostic importance in identifying the patients at higher risk of early biochemical recurrence than conventional (visual and dichotomous) assessment (cPTEN)13. In this study, we further demonstrate the clinical value of our recently published20 fully automated AI-based PTEN assessment for predicting clinically meaningful endpoints such as disease recurrence and metastasis-free survival. The workflow reported here includes the following components: AI-based stain normalization of digital images, AI-based prostate cancer detection, and statistical modeling for selecting sufficient cancer cores to detect clinically relevant thresholds for predicting MFS and RFS (Supplemental Figure S1.2). This multi-center retrospective validation study demonstrates the robust generalization of AI-based algorithms with balanced sensitivity and specificity of 86%/86% compared to conventional assessment of PTEN loss (cPTEN). Interestingly, we showed that only AI-qPTEN remained statistically predictive of both MFS and RFS in multivariable analysis. More importantly, AI-qPTEN provided additional stratification power for the cases classified as low-risk for recurrence according to post-surgical CAPRA-S (Supplemental Figure S2.1, S2.2, S2.3, S2.4) and held clinical relevance for the group of patients without adverse post-surgical clinical features (i.e. lymph node invasion, extra prostatic extension, seminal vesicle invasion and positive surgical margin) (Table 4). Adding this workflow to conventional risk stratification tools may better inform physicians which patients may benefit from intensified surveillance or post-operative interventions (Figure 3). Nevertheless, the study did not discover a notable correlation between PTEN loss and the condition under the stratified cPTEN criteria, which included categories such as intact, partial, or complete loss. This finding provides additional support for using the AI-qPTEN approach for improved accuracy and reliability inassessing PTEN loss (Table S2.11, Figure S2.7).
In this context, we believe that AI-qPTEN workflow should be tested in the future prospective clinical trials evaluating the benefits of adjuvant vs salvage therapies for prostate cancer patients and studying the importance of biomarker-guided interventional trial designs.
In addition to the clinical importance of AI-qPTEN, the workflow has an advantage of being cost-effective34 and affordable over commercial molecular tests (Decipher, Oncotype Dx Prostate, ProMark, and Prolaris)4,5,7,35, as their utilization in clinical practice is limited due to their high cost, especially in low-income countries4,5,36–38. In line with our findings, Leapman et al. (2018) elegantly showed superior prognostic power of using a single biomarker model (PTEN loss) in conjunction with the CAPRA-S over a commercially available test (Prolaris) for assessing the risk of metastasis and cancer-specific death39. Although this study did not utilize threshold-based quantitative PTEN assessment, which further improves risk-stratification power of the models13,40. Evaluation of PTEN IHC in this CANARY cohort has been previously reported from different tissue levels (i.e. sections within a TMA core block) and different antibody, which prohibits direct comparison of AI-qPTEN results in this study to prior published study11; however, similar observations of PTEN loss by conventional assessment in univariable analysis were observed in this study.
The use of a large multi-institutional cohort for workflow validation, along with meaningful survival endpoints (i.e., recurrence and metastasis) within the group of localized prostate cancer, and long-term follow-up is the biggest strength of this study. In addition, successful validation of AI-PTEN workflow on different antibody clones (Clone 138G6-current study vs Clone D4.3-previous study, Cell Signaling) shows generalizability of the algorithm for any CLIA-certified labs using various clones of validated PTEN antibodies. However, there are a few limitations which need to be considered in future: 1) Evaluation of algorithms should be performed on whole slide images in independent cohort enriched with desirable endpoints (i.e., metastasis) before deployment in clinical practice. Tissue artifacts were rigorously assessed by labor-intensive visual inspection of individual TMA cores, resulting in a large number of cores removed from analysis due to poor quality. Incorporating automated quality assessment (QA) of digital images could be considered for additional improvements. The CANARY cohort was constructed21 with oversampling of patients with recurrent Gleason 6 disease and non-recurrent Gleason 8–10 patients therefore associations of PTEN loss with histology grades requires additional validation studies.
In conclusion, our findings strongly support the use of PTEN loss as a prognostic biomarker test to further improve the risk-stratification of clinically localized low-risk cancers. Our novel AI-based unbiased, standardized, quantitative high-throughput PTEN loss assessment workflow offers the possibility of assessing the risk of recurrence and metastasis using digital pathology images of the prostate cancer surgical tissue. This cost-efficient deep-learning algorithm has the potential to be integrated into the clinical labs’ digital pathology workflow as an LDT (Laboratory-developed Test) to offer accessible, affordable personalized cancer care for prostate cancer patients worldwide.
Supplementary Material
Figure 1. Image Analysis Pipeline.
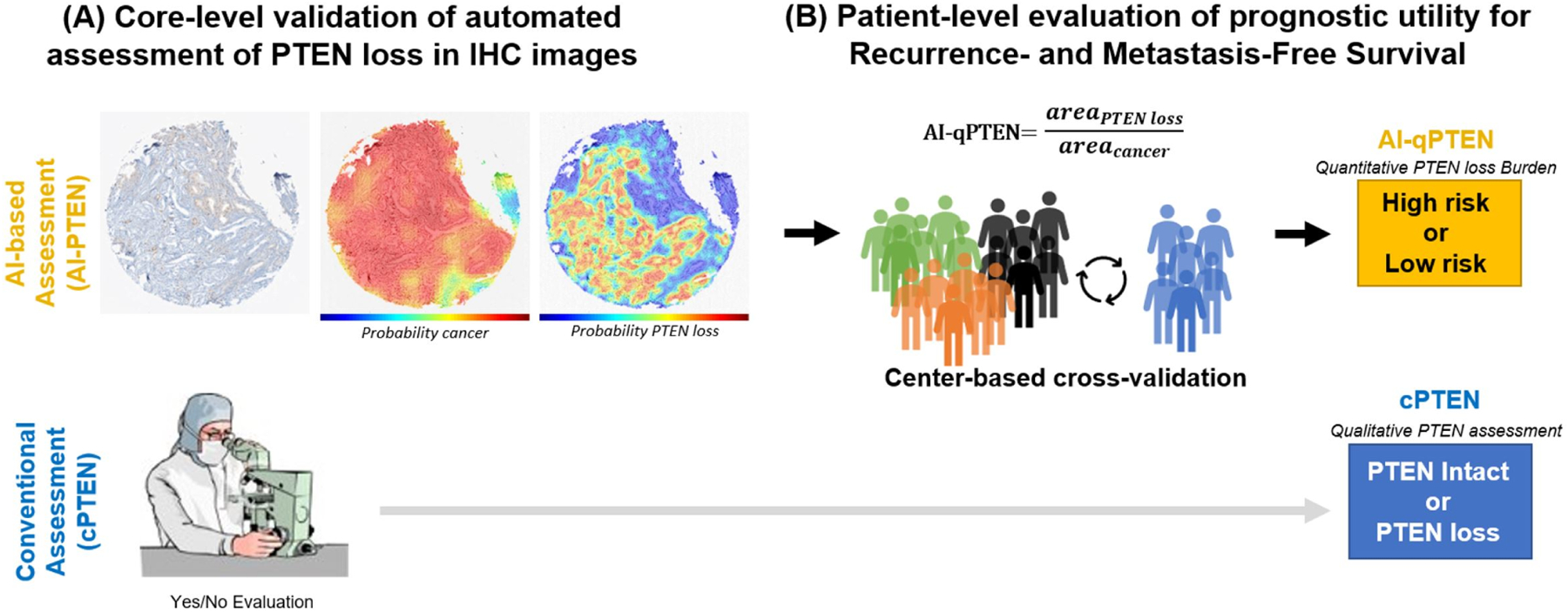
(A) Core-level Analysis: Each TMA core is assessed independently, undergoing cancer detection and PTEN loss identification (classifier followed by pixel-based segmentation). The final output is an area estimate for PTEN loss area and cancer area. A representative core is shown with partial PTEN loss, i.e. not all cancer areas exhibiting loss. An expert pathologist performs qualitative assessment for each core. (B) Patient-level Analysis: All cores from an individual patient are aggregated and selected by total tumor content based on cross-validation analysis before a patient-level estimate of PTEN loss relative to cancer burden (AI-qPTEN) is calculated. Clinically relevant cut-points for an association of AI-qPTEN with patient outcomes are identified through a modified CV partitioned by the institution/center. Conventional assessment (cPTEN) is determined by qualitative review across all cores. AI: Artificial intelligence, AI-qPTEN: AI-based quantitative PTEN loss, TMA: Tissue Micro-Array, CV: cross-validation
ACKNOWLEDGEMENT
This work was supported by the Canary Foundation (Palo Alto, CA), contributing multi-center Canary Prostate Cancer Tissue Microarray (CPCTM) validation cohort; the National Cancer Institute Early Detection Research Network; The Department of Defense W81XWH-09-LCRP-CTRA.
We would like to thank MICCAI (https://gleason2019.grand-challenge.org/) and PANDA challenge for publicly available H&E datasets (https://panda.grand-challenge.org/), that were used for training our cancer detection algorithms.
Tamara Jamaspishvili was supported by a Transformative Pathology Fellowship funded by the Ontario Institute for Cancer Research through funding provided by the government of Ontario. This project is in part supported by the Intramural Research Program of the Center for Cancer Research, NCI. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This work utilized the computational resources of the NIH HPC Biowulf cluster.
FUNDING
The author(s) received no specific funding for this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors declare no competing financial or non-financial Interests
ETHICS APPROVAL / CONSENT TO PARTICIPATE
Tissue specimens and corresponding clinical data were collected at each of the participating sites with central IRB approval, which included sharing of de-identified data. Authors of this publication received de-identified data under a data sharing agreement with Canary Foundation.
DATA AVAILABILITY
The data that support the findings of this study are available from Canary Foundation, but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of Canary Foundation.
REFERENCES
- 1.Jemal A, Culp MB, Ma J, Islami F, Fedewa SA. Prostate Cancer Incidence 5 Years After US Preventive Services Task Force Recommendations Against Screening. J Natl Cancer Inst. 2021;113(1):64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel DA, O’Neil ME, Richards TB, Dowling NF, Weir HK. Prostate Cancer Incidence and Survival, by Stage and Race/Ethnicity - United States, 2001–2017. MMWR Morb Mortal Wkly Rep. 2020;69(41):1473–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shevach J, Chaudhuri P, Morgans AK. Adjuvant therapy in high-risk prostate cancer. Clin Adv Hematol Oncol HO. 2019;17(1):45–53. [PubMed] [Google Scholar]
- 4.Eggener SE, Rumble RB, Armstrong AJ, et al. Molecular Biomarkers in Localized Prostate Cancer: ASCO Guideline. J Clin Oncol Off J Am Soc Clin Oncol. 2020;38(13):1474–1494. [DOI] [PubMed] [Google Scholar]
- 5.Clinckaert A, Devos G, Roussel E, Joniau S. Risk stratification tools in prostate cancer, where do we stand? Transl Androl Urol. 2021;10(1):12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooperberg MR, Simko JP, Cowan JE, et al. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J Clin Oncol Off J Am Soc Clin Oncol. 2013;31(11):1428–1434. [DOI] [PubMed] [Google Scholar]
- 7.Klein EA, Cooperberg MR, Magi-Galluzzi C, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol. 2014;66(3):550–560. [DOI] [PubMed] [Google Scholar]
- 8.Kornberg Z, Cowan JE, Westphalen AC, et al. Genomic Prostate Score, PI-RADS™ version 2 and Progression in Men with Prostate Cancer on Active Surveillance. J Urol. 2019;201(2):300–307. [DOI] [PubMed] [Google Scholar]
- 9.Brooks MA, Thomas L, Magi-Galluzzi C, et al. GPS Assay Association With Long-Term Cancer Outcomes: Twenty-Year Risk of Distant Metastasis and Prostate Cancer-Specific Mortality. JCO Precis Oncol. 2021;5:PO.20.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jamaspishvili T, Berman DM, Ross AE, et al. Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol. 2018;15(4):222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lotan TL, Wei W, Morais CL, et al. PTEN Loss as Determined by Clinical-grade Immunohistochemistry Assay Is Associated with Worse Recurrence-free Survival in Prostate Cancer. Eur Urol Focus. 2016;2(2):180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lotan TL, Wei W, Ludkovski O, et al. Analytic validation of a clinical-grade PTEN immunohistochemistry assay in prostate cancer by comparison with PTEN FISH. Mod Pathol Off J U S Can Acad Pathol Inc. 2016;29(8):904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jamaspishvili T, Patel PG, Niu Y, et al. Risk Stratification of Prostate Cancer Through Quantitative Assessment of PTEN Loss (qPTEN). J Natl Cancer Inst. 2020;112(11):1098–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartl D, de Luca V, Kostikova A, et al. Translational precision medicine: an industry perspective. J Transl Med. 2021;19(1):245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aeffner F, Zarella MD, Buchbinder N, et al. Introduction to Digital Image Analysis in Whole-slide Imaging: A White Paper from the Digital Pathology Association. J Pathol Inform. 2019;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lujan G, Quigley JC, Hartman D, et al. Dissecting the Business Case for Adoption and Implementation of Digital Pathology: A White Paper from the Digital Pathology Association. J Pathol Inform. 2021;12:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooperberg MR, Hilton JF, Carroll PR. The CAPRA-S score: A straightforward tool for improved prediction of outcomes after radical prostatectomy. Cancer. 2011;117(22):5039–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acs B, Rantalainen M, Hartman J. Artificial intelligence as the next step towards precision pathology. J Intern Med. 2020;288(1):62–81. [DOI] [PubMed] [Google Scholar]
- 19.Chang HY, Jung CK, Woo JI, et al. Artificial Intelligence in Pathology. J Pathol Transl Med. 2019;53(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harmon SA, Patel PG, Sanford TH, et al. High throughput assessment of biomarkers in tissue microarrays using artificial intelligence: PTEN loss as a proof-of-principle in multi-center prostate cancer cohorts. Mod Pathol Off J U S Can Acad Pathol Inc. 2021;34(2):478–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawley S, Fazli L, McKenney JK, et al. A model for the design and construction of a resource for the validation of prognostic prostate cancer biomarkers: the Canary Prostate Cancer Tissue Microarray. Adv Anat Pathol. 2013;20(1):39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am J Surg Pathol. 2016;40(2):244–252. [DOI] [PubMed] [Google Scholar]
- 23.Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur Urol. 2016;70(1):93–105. [DOI] [PubMed] [Google Scholar]
- 24.Troyer DA, Jamaspishvili T, Wei W, et al. A multicenter study shows PTEN deletion is strongly associated with seminal vesicle involvement and extracapsular extension in localized prostate cancer. The Prostate. 2015;75(11):1206–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lotan TL, Carvalho FL, Peskoe SB, et al. PTEN loss is associated with upgrading of prostate cancer from biopsy to radical prostatectomy. Mod Pathol Off J U S Can Acad Pathol Inc. 2015;28(1):128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hothorn T, Hothorn MT, Suggests TH. Package ‘Maxstat.’ Citeseer; 2017. [Google Scholar]
- 27.Therneau TM. A Package for Survival Analysis in R. New York, NY, USA; 2019. [Google Scholar]
- 28.Therneau TM, Grambsch PM, Therneau TM, Grambsch PM. The Cox Model. Springer; 2000. [Google Scholar]
- 29.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Mak Int J Soc Med Decis Mak. 2006;26(6):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfeiffer RM, Gail MH. Estimating the decision curve and its precision from three study designs. Biom J Biom Z. 2020;62(3):764–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sjoberg DD. Dcurves: Decision Curve Analysis for Model Evaluation, 2021. [Google Scholar]
- 33.Basourakos SP, Tzeng M, Lewicki PJ, et al. Tissue-Based Biomarkers for the Risk Stratification of Men With Clinically Localized Prostate Cancer. Front Oncol. 2021;11:676716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Djuric U, Zadeh G, Aldape K, Diamandis P. Precision histology: how deep learning is poised to revitalize histomorphology for personalized cancer care. NPJ Precis Oncol. 2017;1(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuzick J, Swanson GP, Fisher G, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12(3):245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erho N, Crisan A, Vergara IA, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PloS One. 2013;8(6):e66855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knezevic D, Goddard AD, Natraj N, et al. Analytical validation of the Oncotype DX prostate cancer assay - a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics. 2013;14:690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spratt DE, Zhang J, Santiago-Jiménez M, et al. Development and Validation of a Novel Integrated Clinical-Genomic Risk Group Classification for Localized Prostate Cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2018;36(6):581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leapman MS, Nguyen HG, Cowan JE, et al. Comparing Prognostic Utility of a Single-marker Immunohistochemistry Approach with Commercial Gene Expression Profiling Following Radical Prostatectomy. Eur Urol. 2018;74(5):668–675. [DOI] [PubMed] [Google Scholar]
- 40.Cyll K, Kleppe A, Kalsnes J, et al. PTEN and DNA Ploidy Status by Machine Learning in Prostate Cancer. Cancers. 2021;13(17). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from Canary Foundation, but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of Canary Foundation.


