Abstract
When the electron transport chain (ETC) function is impaired, cancer cells rely on reductive carboxylation (RC) to convert α-ketoglutarate (αKG) to citrate for macromolecular synthesis, thereby promoting tumor growth. Currently, there is no viable therapy to inhibit RC for cancer treatment. In this study, we demonstrate that the mitochondrial uncoupler treatment effectively inhibits RC in cancer cells. Mitochondrial uncoupler treatment activates the ETC and increases the NAD+/NADH ratio. Using U-13C-glutamine and 1-13C-glutamine tracers, we show that mitochondrial uncoupling accelerates the oxidative TCA cycle and blocks RC under hypoxia, in von Hippel-Lindau (VHL) tumor suppressor-deficient kidney cancer cells, or under anchorage-independent growth condition. Together, these data demonstrate that mitochondrial uncoupling redirects α-KG from RC back to the oxidative TCA cycle, highlighting that the NAD+/NADH ratio is one key switch that determines the metabolic fate of α-KG. Inhibiting RC could be a key mechanism by which mitochondrial uncouplers inhibit tumor growth.
Implications:
Mitochondrial uncoupling is a novel strategy to target reductive carboxylation in cancer.
Keywords: Warburg effect, reductive carboxylation, mitochondrial uncoupler, NAD+/NADH, anchorage-independent growth
Introduction
A metabolic hallmark of all cancer cells is the diversion of glucose flux from mitochondrial oxidation to lactate production, known as the Warburg effect (1). This metabolic reprogramming is primarily caused by the impairment of mitochondrial respiration due to electron transport chain (ETC) inhibition. As a result, NADH oxidation is attenuated, leading to a decrease in the intracellular NAD+/NADH ratio, which induces reductive stress. This reductive stress not only triggers the Warburg effect but also drives other metabolic reprogramming events, such as reductive carboxylation, a sibling of the Warburg effect.
Reductive carboxylation is a process that enables cancer cells to convert glutamine-derived α-ketoglutarate (α-KG) to citrate, which subsequently produces acetyl-CoA and aspartate for macromolecular synthesis (2-4). Cancer cells reprogram the TCA cycle by switching the major carbon source from glucose to glutamine through reductive carboxylation, which is reported to be hyperactive in multiple scenarios such as hypoxia, loss of the von Hippel–Lindau (VHL) tumor suppressor and HIF stabilization, ETC protein deletion or inhibitor treatment, or anchorage-independent growth (3-8). Reductive carboxylation is a crucial anaplerosis metabolic pathway to support lipid and nucleotide synthesis (2,3), maintain redox homeostasis (8), and aerobic glycolysis (4) when ETC activity is inhibited. Thus, reductive carboxylation may therefore be a metabolic vulnerability that can be targeted for cancer therapy. However, there are currently no specific and effective therapeutic interventions to target reductive carboxylation in cancer cells. Novel therapeutic approaches are therefore urgently required.
Because reductive carboxylation is driven by ETC inhibition and a low NAD+/NADH ratio (4,5,7,9), we proposed that activating the ETC to oxidize NADH will eliminate the reductive stress and inhibit reductive carboxylation. In a recent study, we reported that mitochondrial uncouplers could activate the ETC, resulting in an increased NAD+/NADH ratio and dampened Warburg effect to induce neuroblastoma differentiation(10). Building on these findings, we now demonstrate that mitochondrial uncoupling could activate mitochondrial NADH oxidation to inhibit reductive carboxylation.
In this study, we used multiple mitochondrial uncouplers to activate mitochondrial respiration and ETC. By using U-13C-glutamine and 1-13C-glutamine tracers, we show that mitochondrial uncoupling significantly inhibited reductive carboxylation, particularly under hypoxia. In addition, mitochondrial uncoupling also inhibited reductive carboxylation in VHL-deficiency renal cancer cells or upon anchorage-independent growth condition. Moreover, mitochondrial uncoupling blocked spheroid formation and cell proliferation in anchorage-independent growth condition. Together, these results suggest the new therapeutic approach of using mitochondrial uncouplers to inhibit reductive carboxylation and tumor growth, in addition to its role in dampening the Warburg effect.
Materials and Methods
Cell culture and reagents
The neuroblastoma cell lines SK-N-BE(2) and NB16 were obtained from Dr. John M. Maris’ laboratory (Children’s Hospital of Philadelphia). A certificate of analysis is available from Dr. Maris’ group. Clear cell renal cell carcinoma (ccRCC) VHL deficient (786-O-VHL and RCC4-VHL) or VHL-reconstituted (786-O+VHL, RCC4+VHL) isogenic cell lines were obtained from Dr. Erinn Rankin’s laboratory (Stanford University). All cell lines were tested and found negative for mycoplasma (MycoAlert Mycoplasma Detection Kit, Lonza). The authors did not authenticate these cell lines. All cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM)/F12 medium (Caisson Labs, DFL15) supplemented with 1% penicillin-streptomycin (Gibco, 15140122), 10% fetal bovine serum (Sigma-Aldrich, F0926), and 1 mM glutamine (Gibco, 25030081). For spheroid culture, ultra-low attachment dishes/plates (Corning) were used with the same culture medium as for monolayer culture. Mitochondrial uncouplers NEN (#17118), FCCP (#15218), and BAM15 (#17811) were obtained from Cayman Chemicals.
Cell proliferation assay
2x103 cells (n=6) were plated in 96 well plate (Coring 3603). After 24 hours of incubation, the indicated treatment was added, and the plates were transferred to either a normoxia cell culture incubator or a hypoxia chamber for at least 72 hours. Once the control group cells under normoxia reached around 95% confluence, they were stained with 20 μM Hoechst 33342 (Thermo Scientific, 62249) for 10 minutes, and the cell count was automatically determined using Cytation5 (Agilent).
Spheroid formation assay
5x102 SK-N-BE(2) or NB16 cells were plated in 12-well ultra-low attachment plates and treated as indicated. After 12 days, the number of spheroids was counted and their morphology was assessed under a microscope (Leica DMi8). The spheroids were then dissociated into single cells using trypsin and counted using a hemocytometer.
Metabolic analysis
Metabolomics and isotope tracing analyses were conducted using an Agilent 1290 Infinity Liquid Chromatography (LC) System (Agilent) coupled to a Q-TOF 6545 mass spectrometer (MS, Agilent). Targeted analysis, isotopologues extraction, and natural isotope abundance correction were performed with Agilent Profinder B.10.00 (Agilent) as previously described (11).
Stable isotope tracing analysis
The cells were treated as specified and, during the final two or three hours, the medium was changed to DMEM/F-12 without glutamine (Gibco, 21331020) supplemented with 2 mM U-13C-glutamine (Cambridge Isotope Laboratories, CLM-1822-H) and 10% dialyzed FBS (Gibco, 26400044). For the tracing analysis under spheroid culture conditions, 3×103 SK-N-BE(2) or NB16 cells were plated in 6-well ultra-low attachment plates. The cells were incubated for 12 days with a medium change on day 6. After this incubation, treatments were added for three hours, and medium with tracers was added for two hours.
Statistics
Spheroid formation assays and LC-MS experiments were performed in at least biological triplicate. Results are presented as mean ± standard deviation. Statistical analysis was conducted using Student's t-tests or one-way ANOVA tests with post-hoc Tukey HSD tests to determine the significance of differences between groups (two-tailed; unequal variance). P < 0.05 was considered statistically significant, and P values are denoted as follows: *P < 0.05; **P <0.01; ***P < 0.001, ****P < 0.0001.
Result
Mitochondrial uncoupling activates NADH oxidation and inhibits reductive carboxylation under both normoxia and hypoxia
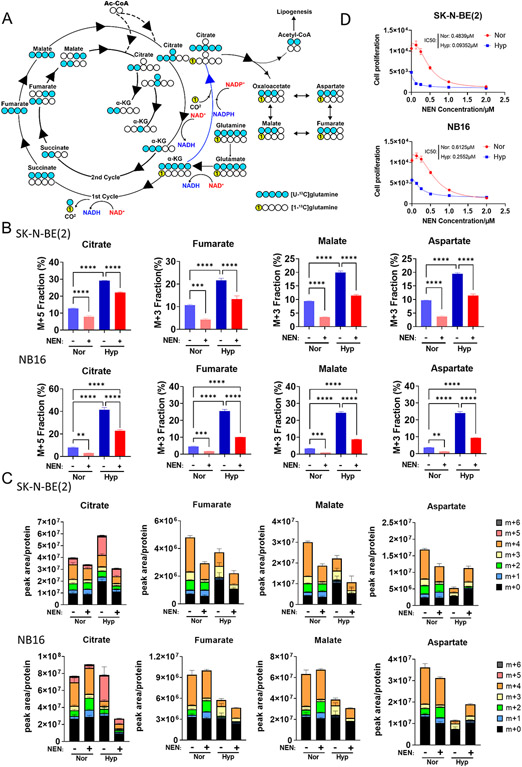
Mitochondrial uncoupling dissipates the proton gradient across the inner mitochondrial membrane, inhibiting ATP synthesis and activating the ETC to promote NADH oxidation (12,13). Consistent with our previous observation, the mitochondrial uncoupler NEN increased the intracellular NAD+/NADH ratio in both SK-N-BE(2) and NB16 cells (Supplemental Figure 1A) (10). Because the pyruvate/lactate ratio is coupled with the NAD+/NADH ratio (14,15), NEN treatment also increased the pyruvate/lactate ratio, indicating reversal of the Warburg effect (Supplemental Figure 1A). Since both the Warburg effect and reductive carboxylation are caused by ETC inhibition and a reduced NAD+/NADH ratio, we next tested whether mitochondrial uncoupling inhibited reductive carboxylation. Using U-13C-glutamine as the tracer, we measured the carbon flow from glutamine into TCA cycle intermediates (Figure 1A). The fraction labeling of M+5 citrate, M+3 fumarate, M+3 malate, and M+3 aspartate was significantly reduced by NEN treatment at both 5h and 24h, suggesting that mitochondrial uncoupling inhibits reductive carboxylation (Supplemental Figure 1B). In SK-N-BE(2) cells, the labeling fractions of glutamine, glutamate, and α-KG were not reduced by NEN treatment, while in NB16 cells, NEN treatment reduced the labeling of glutamate and α-KG slightly (Supplemental Figure 1C). To exclude the possibility that mitochondrial uncoupling inhibited reductive carboxylation through inhibition of glutamine metabolism, the ratios of reductive product to oxidative product from glutamine were calculated. Ratios of citrate (M+5/M+4), fumarate (M+3/M+4), malate (M+3/M+4), and aspartate (M+3/M+4) were significantly reduced by NEN treatment at 5h and 24h, confirming that mitochondrial uncoupling inhibited the reductive carboxylation and rediverted the glutamine carbon from the reductive pathway to the oxidative pathway (Supplemental Figure 1D). Consistent with this observation, the M+2 fractions of fumarate, malate, and aspartate, which are converted from the second cycle of oxidative TCA, were increased by NEN treatment (Supplemental Figure 2A and B). To confirm that the inhibition of reductive carboxylation by NEN treatment was due to mitochondrial uncoupling, we also tested two other mitochondrial uncouplers, FCCP and BAM15. In line with the results from the NEN treatment, both FCCP and BAM15 inhibited reductive carboxylation and diverted the carbon flux into the oxidative pathway (Supplemental Figure 3).
Figure 1. Mitochondrial uncoupling inhibits reductive carboxylation induced by hypoxia.
(A) Schematic of 13C-labeling patterns of TCA cycle metabolites from U-13C-glutamine and 1-13C-glutamine. Blue circles indicate 13C from U-13C-glutamine, yellow circles labelled “1” indicate 13C from 1-13C-glutamine, and hollow circles indicate 12C. (B, C) SK-N-BE(2) and NB16 cells were pretreated under normoxia or hypoxia (0.5% O2) for 16 hours, followed by DMSO or 1 μM NEN for 3 hours, then labeled with U-13C-glutamine for 2 hours. The relative isotopic labeling fraction of metabolites were measured using LC-MS (n=3). (D) SK-N-BE(2) and NB16 cells were plated and treated by different concentration of NEN under normoxia and hypoxia, followed by cell counting by Cytation5 (n=6).
Hypoxia, a common microenvironmental stress in solid tumors, significantly enhances reductive carboxylation flux, which is essential for tumor cell proliferation under hypoxia (3,5). Although substantial inhibition of reductive carboxylation by mitochondrial uncoupling was observed under normoxia, we wondered whether hypoxia-induced reductive carboxylation could also be inhibited by mitochondrial uncoupling. Employing the same isotope tracing method, we measured reductive carboxylation activity with or without NEN treatment under hypoxia (0.5% oxygen). In line with previous reports, reductive carboxylation was induced by hypoxia, determined by increased labeling fractions of M+5 citrate, M+3 fumarate, M+3 malate, and M+3 aspartate (Figure 1B, C; Supplemental Figure 4B). NEN treatment significantly inhibited the reductive carboxylation induced by hypoxia (Figure 1B, C; Supplemental Figure 4B). Ratios of citrate (M+5/M+4), fumarate (M+3/M+4), malate (M+3/M+4), and aspartate (M+3/M+4) were significantly increased under hypoxia and uniformly reduced by NEN treatment, confirming that hypoxia-induced reductive carboxylation can be inhibited by NEN treatment (Supplemental Figure 4A). In accordance with the increased dependence on RC in cancer cells under hypoxia, SK-N-BE(2) and NB16 cells exhibited higher sensitivity to NEN treatment under hypoxic conditions compared to normoxic condition (Figure 1D). To further validate that mitochondrial uncoupling inhibits reductive carboxylation, we used another tracing strategy developed by Metallo et al.(3). When using 1-13C-glutamine tracer, only metabolites generated from reductive carboxylation are labeled (M+1 citrate, M+1 fumarate, M+1 malate, and M+1 aspartate) since the heavy carbon is oxidized to form CO2 in the oxidative direction (Figure 1A). Consistent with the results from U-13C-glutamine tracing, NEN treatment inhibited the reductive carboxylation induced by hypoxia, judging by the full reduction of M+1 citrate, M+1 fumarate, M+1 malate, and M+1 aspartate (Supplemental 5A). Furthermore, the mitochondrial uncouplers FCCP and BAM15 also inhibited the hypoxia-induced reductive carboxylation (M+1 fractions) and diverted the carbon flux in the oxidative direction (Supplemental Figure 5B).
Mitochondrial uncoupling inhibits reductive carboxylation in VHL-deficient kidney cancer cells
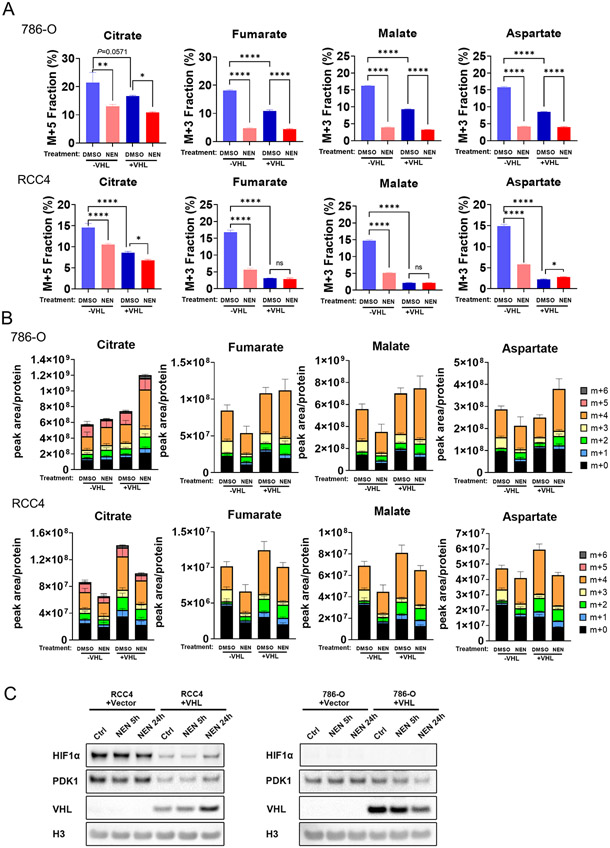
VHL loss is well-known to induce reductive carboxylation by activating HIF signaling (3,6). Previously, we reported that mitochondrial uncoupling repressed HIF accumulation under both normoxic and hypoxic conditions (10). Thus, we wondered whether mitochondrial uncoupling also inhibit reductive carboxylation in VHL-deficient clear cell renal cell carcinoma (ccRCC) cancer cells. The VHL-deficient 786-O and RCC4 cells had hyperactive reductive carboxylation even under normoxic conditions, with approximately 20%-15% of citrate, fumarate, malate, and aspartate generated from reductive carboxylation (Figure 2A). Consistently, VHL reconstitution significantly inhibited reductive carboxylation, determined by reduced labeling fractions of M+5 citrate, M+3 fumarate, M+3 malate, and M+3 aspartate (Figure 2A, B). However, the reduced fraction of these metabolites could be contributed by different mechanisms. For citrate, no significant change on M+5 citrate abundance was observed, the reduced fraction of M+5 citrate is due to the increased abundance of non–M+5 citrate. The reduced m+3 labeling fraction of fumarate, malate and aspartate was caused by both increased non–m+3 metabolites abundance and decreased M+3 metabolites abundance (Figure 2A, B). In contrast, NEN treatment significantly repressed reductive carboxylation activity by reducing both labeling fractions and abundance of M+5 citrate, M+3 fumarate, and M+ 3 malate in VHL-deficient cells (Figure 2A, B). In addition, NEN treatment re-directed the glutamine flux from reductive carboxylation to oxidative metabolism (Supplemental Figure 6A, B).
Figure 2. Mitochondrial uncoupling inhibits reductive carboxylation generation in VHL-null ccRCC cells.
(A, B) The ccRCC cells were pretreated with DMSO or 1 μM NEN for 3 hours, then labeled with U-13C-glutamine for 3 hours. Relative isotopic labeling fractions or ratios of metabolites were measured using LC-MS. (C) The RCC4 and 786-O cells were treated by DMSO or NEN for 5 or24 hours, followed by immunoblot to examine the protein level.
Interestingly, we found that 1μM NEN (the same concentration used in isotope tracing experiment) did not reduce the HIF-1α or its transcriptional target PDK1 expression in RCC4 cells (Figure 2C), and no HIF-1α expression was detected in 786-O cells (Figure 2C), suggesting that the inhibitory effect on reductive carboxylation by NEN was not caused by repressing HIF-1α signaling.
Mitochondrial uncoupling inhibits reductive carboxylation during anchorage-independent growth
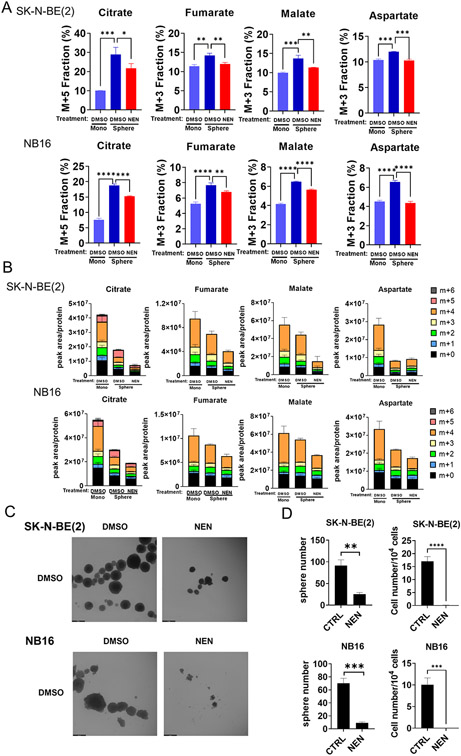
Cells growing in anchorage-independent conditions preferentially use reductive carboxylation to maintain redox homeostasis (8). Thus, we wondered whether mitochondrial uncoupling could inhibit reductive carboxylation during anchorage-independent growth. When cultured in low-attachment dishes, the neuroblastoma cells formed spheroids (Figure 3C). The labeling fraction of M+5 citrate, M+3 fumarate, malate, or aspartate significantly increased in spheroids compared to cells cultured in monolayer, meanwhile the total abundance of these metabolites were all significantly reduced in spheroid (Figure 3A, B). As expected, NEN treatment significantly reduced M+5 citrate, M+3 fumarate, malate, or aspartate labeling fraction and abundance in spheroids (Figure 3A, B; Supplemental Figure 7A). In addition, NEN treatment decreased spheroid formation and cell proliferation in spheroid culture conditions (Figure 3C, D). FCCP or BAM15 treatment also inhibited cell proliferation in 3D culture conditions (Supplemental Figure 8A).
Figure 3. Mitochondrial uncoupling inhibits reductive carboxylation during anchorage-independent growth.
(A, B) SK-N-BE(2) and NB16 cells in monolayer culture, or spheroid culture condition treated with DMSO or 1 μM NEN for 3 hours, were labeled with U-13C-glutamine for 3 hours. Relative isotopic labeling fractions or ratios of metabolites were measured using LC-MS. (C) Morphology of SK-N-BE(2) and NB16 cells treated with DMSO or 1 μM NEN for 12 days in low attachment dishes. (Scale bar: 500μm) (D) Quantitation of spheroid and cell number from samples shown in (C).
In addition to redirecting the carbon flux from RC to the oxidative TCA cycle, mitochondrial uncoupling also limits the ATP synthesis, resulting in a lower ATP/ADP level (10). To examine whether a low ATP synthesis rate also inhibits spheroid formation, the neuroblastoma cells were treated with rotenone, which is a complex I inhibitor that induces reductive carboxylation and reduces ATP synthesis (Supplemental Figure 9A) (9). Interestingly, rotenone treatment only reduced cell proliferation upon anchorage-independent condition, but did not reduce spheroid numbers (Supplemental Figure 9B, C). In addition, we performed the spheroid formation assay under hypoxia, a stress condition that also inhibits ETC activity and ATP synthesis. Surprisingly, hypoxia significantly increased the number of the spheroid in SK-N-BE(2) cells, while in NB16 cells, the spheroid number was not affected by hypoxia (Supplemental Figure 9D, E). In line with the smaller spheroid, hypoxia slow down the cell proliferation in SK-N-BE(2) and NB16 cells (Supplemental Figure 9D, E). NEN treatment eliminated spheroid formation and blocked cell proliferation in spheroid culture conditions under hypoxia (Supplemental Figure 9D, E), indicating that mitochondrial uncoupling also inhibits anchorage-independent growth under hypoxia.
Discussion
Upon the induction of the Warburg effect, pyruvate carbon is diverted away from the TCA cycle to lactate production, causing a shortage of citrate. To compensate for this, cancer cells utilize glutamine as an alternative carbon source to generate citrate through reductive carboxylation. Therefore, the induction of the Warburg effect is always associated with increased reductive carboxylation flux. Inhibition of pyruvate dehydrogenase kinase (PDK) activity by dichloroacetate (DCA) partially restores pyruvate entry into the TCA cycle and inhibits reductive carboxylation (3,8), indicating that pyruvate dehydrogenase (PDH) inhibition is a cause of reductive carboxylation. However, the concentrations of DCA needed to be at a high mM range to inhibit PDK and cell proliferation (11,16), which is difficult to reach in patients without causing toxicity. Therefore, it is essential to develop an alternative strategy to inhibit reductive carboxylation for cancer therapy.
It was shown that knocking down/out the NADP+-dependent enzymes IDH1 and/or IDH2 blocked reductive carboxylation (3,5,7,8). Because IDH activity is essential for cellular metabolism, inhibiting IDH with inhibitors will likely cause toxicity in normal cells. For instance, livers from Idh1 knockout (KO) mice displayed accelerated and more pronounced oxidative stress compared to wild-type (WT) livers. Furthermore, Idh1 KO mice demonstrated an increased vulnerability to death caused by lipopolysaccharide (LPS) and exhibited elevated serum levels of inflammatory cytokines (17). Notably, the concentrations of IDH inhibitors required to inhibit wild-type IDH1 are considerably higher than those needed to inhibit mutant IDH1 (18,19). It remains uncertain whether the existing IDH inhibitors can achieve sufficiently high concentrations within tumor tissues to effectively inhibit reductive carboxylation.
Because PDH uses NAD+ as a substrate, according to the law of mass action, the NAD+/NADH ratio is the key to driving pyruvate entry into the TCA cycle. The reduction of the NAD+/NADH ratio upon ETC inhibition is not only the trigger of the Warburg effect but also the driver of reductive carboxylation (4,9). Because NADPH can be produced from NADH through nicotinamide nucleotide transhydrogenase (NNT), it was proposed that a reduced NAD+/NADH ratio can drive reductive carboxylation by reducing the NADP+/NADPH ratio (5,7). Here we used mitochondrial uncouplers to restore NAD+/NADH ratios and inhibit reductive carboxylation in multiple systems, uncovering a novel application of mitochondrial uncouplers as therapeutic agents to target reductive carboxylation. Given that niclosamide is an FDA-approved anti-parasitic drug with minimal toxicity, we anticipate that its application in targeting reductive carboxylation could potentially provide a safe and effective therapeutic approach for cancer treatment.
Supplementary Material
Acknowledgments
This work was supported by a Stanford Maternal and Child Health Research Institute Research Scholar Award (2020), an American Cancer Society Research Scholar Grant (RSG-20-036-01), and an Agilent Solutions Innovation Research Award (SIRA) to J.Y. We thank Dr. Yuqin Dai (Stanford ChEM-H Institute Metabolomics Knowledge Center) for support and use of facilities, Dr. Erinn Rankin (Stanford University) for providing the ccRCC cell lines.
Footnotes
Competing interests
H.J., Y.L. and J.Y. submitted a patent application related to this manuscript.
Reference:
- 1.Warburg O. On the origin of cancer cells. Science 1956;123(3191):309–14 doi 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell 2015;162(3):540–51 doi 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2011;481(7381):380–4 doi 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaude E, Schmidt C, Gammage PA, Dugourd A, Blacker T, Chew SP, et al. NADH Shuttling Couples Cytosolic Reductive Carboxylation of Glutamine with Glycolysis in Cells with Mitochondrial Dysfunction. Mol Cell 2018;69(4):581–93 e7 doi 10.1016/j.molcel.2018.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A 2011;108(49):19611–6 doi 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gameiro PA, Yang J, Metelo AM, Perez-Carro R, Baker R, Wang Z, et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab 2013;17(3):372–85 doi 10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 2011;481(7381):385–8 doi 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 2016;532(7598):255–8 doi 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fendt S-M, Bell EL, Keibler MA, Olenchock BA, Mayers JR, Wasylenko TM, et al. Reductive glutamine metabolism is a function of the α-ketoglutarate to citrate ratio in cells. Nature communications 2013;4(1):1–11 doi 10.1038/ncomms3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang H, Greathouse RL, Tiche SJ, Zhao M, He B, Li Y, et al. Mitochondrial uncoupling induces epigenome remodeling and promotes differentiation in neuroblastoma. Cancer Research 2022. doi 10.1158/0008-5472.Can-22-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Gruber JJ, Litzenburger UM, Zhou Y, Miao YR, LaGory EL, et al. Acetate supplementation restores chromatin accessibility and promotes tumor cell differentiation under hypoxia. Cell Death Dis 2020;11(2):102 doi 10.1038/s41419-020-2303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alasadi A, Chen M, Swapna GVT, Tao H, Guo J, Collantes J, et al. Effect of mitochondrial uncouplers niclosamide ethanolamine (NEN) and oxyclozanide on hepatic metastasis of colon cancer. Cell Death Dis 2018;9(2):215 doi 10.1038/s41419-017-0092-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luengo A, Li Z, Gui DY, Sullivan LB, Zagorulya M, Do BT, et al. Increased demand for NAD(+) relative to ATP drives aerobic glycolysis. Mol Cell 2021;81(4):691–707 e6 doi 10.1016/j.molcel.2020.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patgiri A, Skinner OS, Miyazaki Y, Schleifer G, Marutani E, Shah H, et al. An engineered enzyme that targets circulating lactate to alleviate intracellular NADH:NAD(+) imbalance. Nat Biotechnol 2020;38(3):309–13 doi 10.1038/s41587-019-0377-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Hu Q, Cheng F, Su N, Wang A, Zou Y, et al. SoNar, a Highly Responsive NAD+/NADH Sensor, Allows High-Throughput Metabolic Screening of Anti-tumor Agents. Cell Metab 2015;21(5):777–89 doi 10.1016/j.cmet.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stockwin LH, Yu SX, Borgel S, Hancock C, Wolfe TL, Phillips LR, et al. Sodium dichloroacetate selectively targets cells with defects in the mitochondrial ETC. Int J Cancer 2010;127(11):2510–9 doi 10.1002/ijc.25499. [DOI] [PubMed] [Google Scholar]
- 17.Itsumi M, Inoue S, Elia AJ, Murakami K, Sasaki M, Lind EF, et al. Idh1 protects murine hepatocytes from endotoxin-induced oxidative stress by regulating the intracellular NADP(+)/NADPH ratio. Cell Death Differ 2015;22(11):1837–45 doi 10.1038/cdd.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okoye-Okafor UC, Bartholdy B, Cartier J, Gao EN, Pietrak B, Rendina AR, et al. New IDH1 mutant inhibitors for treatment of acute myeloid leukemia. Nat Chem Biol 2015;11(11):878–86 doi 10.1038/nchembio.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popovici-Muller J, Lemieux RM, Artin E, Saunders JO, Salituro FG, Travins J, et al. Discovery of AG-120 (Ivosidenib): A First-in-Class Mutant IDH1 Inhibitor for the Treatment of IDH1 Mutant Cancers. ACS Med Chem Lett 2018;9(4):300–5 doi 10.1021/acsmedchemlett.7b00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.