Abstract
The alphaproteobacterium Wolbachia pipientis infects arthropod and nematode species worldwide, making it a key target for host biological control. Wolbachia-driven host reproductive manipulations, such as cytoplasmic incompatibility (CI), are credited for catapulting these intracellular bacteria to high frequencies in host populations. Positive, perhaps mutualistic, reproductive manipulations also increase infection frequencies, but are not well understood. Here, we identify molecular and cellular mechanisms by which Wolbachia influences the molecularly distinct processes of germline stem cell (GSC) self-renewal and differentiation. We demonstrate that wMel infection rescues the fertility of flies lacking the translational regulator mei-P26 and is sufficient to sustain infertile homozygous mei-P26-knockdown stocks indefinitely. Cytology revealed that wMel mitigates the impact of mei-P26 loss through restoring proper pMad, Bam, Sxl, and Orb expression. In Oregon R files with wild-type fertility, wMel infection elevates lifetime egg hatch rates. Exploring these phenotypes through dual-RNAseq quantification of eukaryotic and bacterial transcripts revealed that wMel infection rescues and offsets many gene expression changes induced by mei-P26 loss at the mRNA level. Overall, we show that wMel infection beneficially reinforces host fertility at mRNA, protein, and phenotypic levels, and these mechanisms may promote the emergence of mutualism and the breakdown of host reproductive manipulations.
Wolbachia bacterial symbionts are being harnessed as biological control agents by exploiting their manipulation of host fertility. This study reveals that the wMel strain can have a beneficial impact on fruit fly development in both mutant and wild-type flies; this phenotype is essential to the long-term use of these bacteria for host population control.
Introduction
Endosymbiotic bacteria have evolved diverse strategies for infecting and manipulating host populations [1,2], which are now being leveraged for biological control applications [3]. Many of these bacteria reside within host cells and navigate female host development to colonize offspring, thus linking their fitness to that of their hosts through vertical transmission [4]. Inherited endosymbionts with reproductive manipulator capabilities go a step further by altering host development in ways that rapidly increase the frequency of infected, reproductive females in the host population. Reproductive manipulator strategies include cytoplasmic incompatibility (CI), where infected sperm require rescue by infected eggs, male-killing, feminization, and parthenogenesis [5,6]. These manipulations can be highly effective at driving bacterial symbionts into host populations, regardless of costs to the host individual or population [7].
Despite the success of parasitic reproductive manipulation, natural selection favors symbionts that increase the fertility of infected mothers, even if these variants reduce the efficacy of the parasitic mechanisms that initially drove the infection to high frequency [8]. In associations between arthropods and strains of the alphaproteobacterium Wolbachia pipientis, this scenario may be common. For example, measured fecundity in populations of Drosophila simulans infected with the wRi strain of Wolbachia swung from −20% to +10% across a 20-year span following wRi’s CI-mediated sweep across California in the 1980s [9]. This transition from fitness cost to benefit coincided with weakening of CI strength over the same time frame [10]. Fertility-enhancing mechanisms may be at work in other strains of Wolbachia that have reached high infection frequencies in their native hosts, yet do not currently exhibit evidence of parasitic reproductive manipulation [11]. Importantly, these fitness benefits may have also played a role in the early stages of population infection, when infected hosts are at too low of frequencies for CI to be effective [12].
Currently, the wMel strain of Wolbachia and its encoded CI mechanism are successfully being used to biologically control non-native hosts [3]. In Aedes aegypti mosquitoes, CI causes nearly 100% mortality of offspring born to uninfected mothers [13]. However, in its native host, the fruit fly Drosophila melanogaster, wMel exhibits CI that rarely exceeds 50% mortality and is extremely sensitive to paternal age [14–16], as well as grandmother age because titer increases with age [17,18]. Despite weak CI in its native host, wMel is found at moderate to high infection frequencies in populations worldwide [19,20]. Other data suggest that these frequencies may be explained by some emergent beneficial function that increases host fitness [21–25]. The molecular basis for these beneficial functions could be related to the loss of CI efficacy in D. melanogaster. Given that wMel’s use in non-native hosts relies on strong and efficient CI, it is essential that we learn the basis for its beneficial functions that could ultimately undermine CI function.
Host germline stem cell (GSC) maintenance and differentiation pathways are powerful targets for Wolbachia-mediated reproductive manipulation. Wolbachia strains have strong affinities for host germline tissues [26,27], positioning them at the right place to manipulate and enhance host fertility. In the strains that form obligate associations with Brugia filarial nematodes [28] and Asobara wasps [29], Wolbachia is required in the germline to prevent premature differentiation and achieve successful oogenesis (reviewed in [30]). In the facultative wMel-D. melanogaster association, the wMel strain can partially rescue select loss of function alleles of the essential germline maintenance and differentiation genes sex lethal (sxl) and bag-of-marbles (bam) in female flies [31–33]. It is known that wMel encodes its own factor, toxic manipulator of oogenesis (TomO), that partially recapitulates Sxl function in the GSC through derepression and overexpression of the translational repressor Nanos (Nos) [34,35]. However, Nos expression is negatively correlated with Bam expression in the early germarium [36]. Therefore, Bam’s function in cyst patterning and differentiation in wMel-infected mutant flies cannot be explained by shared mechanisms with sxl rescue or TomO’s known functions.
In a previous screen, our lab identified the essential fertility gene meiotic-P26 (mei-P26) as a host factor that influences wMel infection intensity in D. melanogaster cell culture, potentially through modulating protein ubiquitination [37]. Mei-P26 is a Trim-NHL protein that confers a wide range of functions through its multiple domains: its protein-binding NHL and B-box domains allow it to act as an adapter for multiple translational repressor complexes (e.g., Sxl, Nanos (Nos), Argonaute-1 (AGO1), see S1 to S2A Figs and S1 to S2 Tables). Its E3-ubiquitin ligase domain likely enables it to modulate proteolysis [38]. Given mei-P26’s in vitro role in infection [37] and its in vivo role in GSC maintenance [38], differentiation [39,40], and meiosis [40,41], we selected it as a candidate gene for modulating wMel–host interactions.
We report that D. melanogaster flies infected with the wMel strain of Wolbachia partially rescue mei-P26 mutations and exhibit reinforced fertility. In flies homozygous for hypomorphic mei-P26, infection with wMel elevates fertility in both males and females to a level sufficient to maintain a stable stock, whereas the uninfected stock is unsustainable. Infection rescues GSC maintenance and cyst differentiation by mitigating the downstream effects of perturbed mei-P26 function on other protein and mRNA expression. Specifically, wMel infection restores a wild-type-like expression profile for phosphorylated Mothers against decapentaplegic (pMad), Sxl, Bam, and Oo18 RNA-binding (Orb) proteins, as well as tumorous testis (tut) and benign gonadal cell neoplasm (bgcn) mRNAs. We find evidence of wMel’s beneficial reproductive manipulator abilities in Oregon R (OreR) wild-type flies, illustrating how bacterially mediated developmental resilience may be selected for in nature. These results are essential to understanding how wMel reaches high frequencies in natural D. melanogaster populations and these beneficial functions may be able to be harnessed for biological control applications.
Results
Here, we explore the effects of wMel Wolbachia infection on D. melanogaster’s essential fertility gene mei-P26 through dosage knockdown with an RNAi construct, disrupted function with a hypomorphic allele, mei-P26[1], and full knockdown with a null allele, mei-P26[mfs1]. Using fly fecundity assays, immunocytochemistry, and dual-RNAseq of both host and bacterial transcripts, we show that wMel can compensate for the loss of mei-P26 to significantly rescue host fertility.
Infection with wMel rescues female D. melanogaster fertility in mei-P26 deficient flies
Infection with wMel significantly rescued fertility defects induced by the loss of mei-P26 function relative to the OreR wild-type strain, as measured by offspring produced per female D. melanogaster per day (Figs 1A–1E and S3; p< = 2.2e-16 to 2.9e-2, Wilcoxon rank sum test; see S2–S6 Figs). Offspring production requires successful maintenance of the GSC, differentiation of a GSC daughter cell into a fully developed and fertilized egg, embryogenesis, and hatching into a first-instar larva. Breaking offspring production down into egg lay and egg hatch components (Fig 1B and 1C) revealed that as allelic strength increased, from nos-driven RNAi to hypomorphic and null alleles, fertility impacts shifted from those that impacted differentiation/development (egg hatch) to those that also impacted egg production (egg lay).
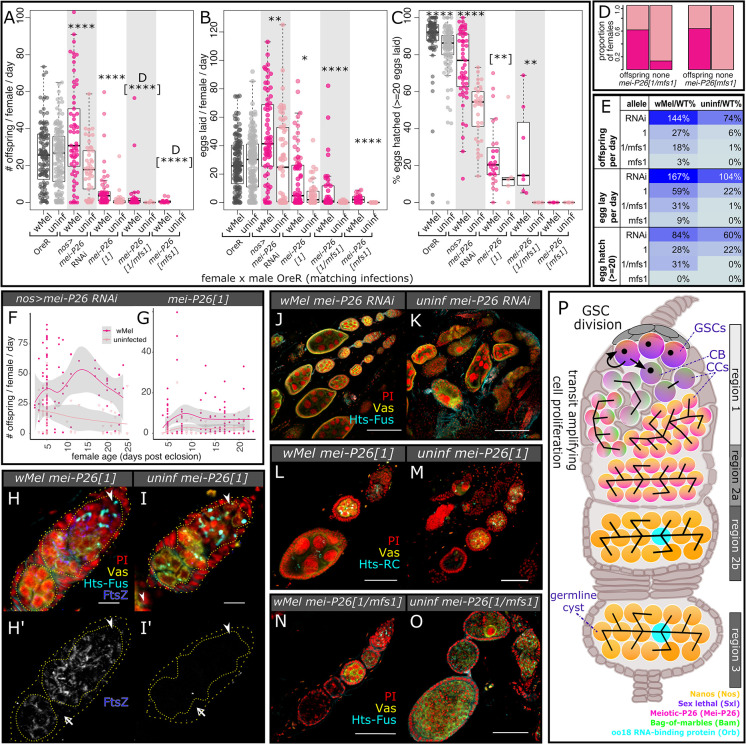
Fig 1. wMel infection rescues the loss of essential host fertility gene mei-P26 in females (and males, see S4E–S4G Fig).
(A–C) Beeswarm boxplots of (A) overall offspring production, broken into (B) eggs laid and (C) eggs hatched, from single female RNAi, hypomorphic, and null mei-P26 knockdown female flies crossed to single OreR males of the same infection state and age (3–7 days old). The null allele mei-P26[mfs1] reduced egg lay rates in both mutant heterozygotes and homozygotes below an average of 20 for both infected and uninfected females, necessitating an analysis of females with and without offspring (see D). Wilcoxon rank sum test *p< = 0.05, **< = 0.01, ***< = 0.001, ****< = 1e-4. Fisher’s exact test [**]p< = 0.01, [****]< = 1e-4. (D–E) (D) Barplots of the proportion of female samples of the indicated genotypes with offspring and without offspring, infected and uninfected with Wolbachia. (E) Table listing the percentage of wild-type fecundity demonstrated by wMel-infected (left) or uninfected (right) mei-P26 mutants in order of increasing severity, from top to bottom (see also S4–S6 Tables). Cell values from low to high proportions are colored from light to dark blue, respectively. (F, G) Knockdown mei-P26 D. melanogaster fecundity versus female age, fit with a local polynomial regression (dark gray bounds = 95% confidence intervals). Infection with wMel increases offspring production over the first 25 days of the (F) mei-P26 RNAi and (G) mei-P26[1] hypomorphic female lifespan. (H–O) Confocal mean projections of D. melanogaster ovarioles and germaria (3–7 days old). (H–I’) Intracellular wMel FtsZ localizes to germline (yellow, Vas+, inner dotted outline) and somatic (Vas-, space between inner dotted and outer dashed outlines) cells at high titers, with low background in uninfected flies. Bacteria are continuously located in the mei-P26[1] germline, starting in the GSC (arrowhead near Hts-bound spectrosome). Somatic cell regions are indicated with empty arrowheads. (J–O) Cytology reveals that wMel-infection rescues (J, K) mei-P26 RNAi, (L, M) mei-P26[1] (see also S5 Fig), and (N, O) mei-P26[1/mfs1]-induced oogenesis defects. Fluorescence channels were sampled serially and overlaid as indicated on each set of images as follows: cyan = anti-Hu Li Tai Shao (Hts) ring canal isoform (-RC) or fusome isoform (-FUS), yellow = anti-Vasa (Vas), blue = wMel anti-Filamenting temperature-sensitive mutant Z (FtsZ), and red = PI DNA staining. Scale bars: H–I’ = 10 μm, L–O = 50 μm, J, K = 100 μm. (P) Diagram of a wild-type D. melanogaster germarium, highlighting key events and cell types by region as in [42]. Somatic cells in beige, GSC-derived cells are brightly colored gradients leading to differentiated cyst cells in yellow, with specified oocytes in blue. The black structure that originates in the GSC is the spectrosome, which becomes a branching fusome as the CB is formed, moves away from the niche, and divides into CCs. The data underlying this figure can be found in S3 Table and on Dryad at doi.org/10.7291/D1DT2C. CB, cystoblast; CC, cystocyte; GSC, germline stem cell; OreR, Oregon R; PI, propidium iodide.
Infection with wMel mediated a significant degree of fertility rescue for all allelic strengths and nos:GAL4>UAS-driven RNAi knockdowns, suggesting a robust bacterial rescue mechanism that compensates for loss of protein dosage and function. Rescue was more than complete for nos-driven mei-P26 RNAi depletion (from 26% deficient offspring production in uninfected flies): wMel-infected mei-P26 RNAi females produced 44% more offspring than infected OreR wild-type females (37 versus 25 larvae/female/day, p< = 1.15E-02 Wilcoxon rank sum test, S4 Table and Fig 1A and 1E) due to a higher rate of egg production (p< = 6.99E-05 Wilcoxon rank sum test, S5 Table and Fig 1B and 1E), opposed to a higher hatch rate (Fig 1C and 1E and S6 Table). This may suggest a synergistic function between wMel and low-dose mei-P26 in the GSC. In uninfected and wMel-infected mei-P26[1] hypomorphic females, offspring production decreased 94% and 73%, respectively, compared to OreR wild-type of the matching infection status (p< = 2.26E-14 and 3.45E-12 Wilcoxon rank sum test, respectively, S4 Table and Fig 1A and 1E). Comparing uninfected and wMel-infected female mei-P26[1] fecundity revealed that wMel infection increases the number of offspring produced per day (1.6 versus 7 larvae/female/day, p< = 9.37E-05 Wilcoxon rank sum test, S4 Table and Fig 1A) through increasing both the rate of egg lay (p< = 2.82E-02 Wilcoxon rank sum test, S5 Table and Fig 1B) and egg hatch (p< = 2.55E-03 Wilcoxon rank sum test, S6 Table and Fig 1C). In uninfected and wMel-infected mei-P26[1]/mei-P26[mfs1] trans-heterozygous females, offspring production decreased 99% and 82%, respectively, relative to OreR wild-type (p< = 3.40E-04 and 5.01E-10 Wilcoxon rank sum test, S4 Table and Fig 1A and 1D). Comparing uninfected and wMel-infected trans-heterozygous flies showed that infection elevates offspring production (0.2 versus 4.6 larvae/female/day, p< = 1.03E-05 Fisher’s exact test, S4 Table and Fig 1A) through increasing the rate females lay eggs (p< = 4.71E-05 Wilcoxon rank sum test; Fig 1B) and the rate those eggs hatch (p< = 4.20E-03 Wilcoxon rank sum test; S6 Table and Fig 1C). In females homozygous for the most severe allele, mei-P26[mfs1], offspring production decreased 100% and 97% in uninfected and wMel-infected flies relative to wild-type, respectively (p< = 5.14E-06 Fisher’s exact test, S4 Table and Fig 1A). Offspring production was marginally rescued in mei-P25[mfs1] flies by wMel infection (0 (uninfected) versus 0.76 (infected) larvae/female/day, p< = 5.14E-06 Fisher’s exact test, S4 Table, Fig 1A), in part due to infected flies laying eggs at a higher rate than uninfected flies (p< = 3.04E-05 Wilcoxon rank sum test, Fig 1B). However, no female laid 20 or more eggs in one day, precluding an estimation hatch rate rescue (Fig 1C). See S2–S6 Tables and S3 Fig for a full description of the fecundity assays and data.
wMel-mediated mei-P26 rescue is robust and sufficient to rescue this gene in a stable stock. Infection with wMel elevates the number of offspring produced per day across the fly lifespan in mei-P26[1] hypomorphic and nos-driven RNAi-knockdown females (p< = 1.5e-11 to 3.4e-2 Kolmogorov–Smirnov test, S7 Table and Fig 1F and 1G). However, the underlying number of eggs laid per day and the hatch rate do vary considerably with age for both infection states and genotypes (S4A–S4D Fig). The wMel rescue mechanism is not specific to females, as the weaker impacts of mei-P26 loss on male fertility are mitigated by wMel-infection (S4E–S4G Fig; p< = 6.1e-6 to 2.9e-2 Wilcoxon rank sum test, S3–S5 Tables). Thus, we were able to establish a homozygous stock of mei-P26[1] flies infected with wMel Wolbachia. In contrast, the uninfected stock only lasted a few generations without balancer chromosomes (S4H and S4I Fig). Given the severe, yet significantly rescuable nature of the mei-P26[1] allele (Figs 1A–1C, S2B and S2D), we proceeded with this genotype for many of our subsequent assays for specific immunocytological rescue phenotypes.
Female germline morphology is rescued by wMel infection in mei-P26-deficient flies
In the D. melanogaster germarium, wMel is continuously present at high titers in both germline and somatic cells (Fig 1H–1H’, compared to control in Fig 1I–1I’), consistent with previous reports [26,27,43]. The bacteria localize more strongly to the germline-derived cells than the somatic cells (co-localization of FtsZ and Vas in Fig 1H–1H’). Intracellular wMel can be identified in the GSC, the cystoblast (CB), the cystocytes (CCs), and the developed cyst. This positioning puts wMel in all of the critical cell types and stages that mei-P26 is active, enabling the bacterium to compensate for mei-P26’s developmental functions in GSC maintenance and differentiation.
Comparing the cytology of mei-P26 knockdown oocytes infected and uninfected with Wolbachia confirmed that wMel-infected ovarioles exhibit far fewer developmental defects than uninfected, more closely resembling OreR wild-type (Figs 1J–1O and S5 versus Fig 1P; Vasa (Vas) = germline [44], Hu Li Tai Shao (Hts) = cytoskeletal spectrosome/fusome [45], propidium iodide (PI) = DNA). Specifically, wMel-infected mei-P26-knockdown ovarioles exhibited more normally formed cysts, consisting of somatically derived follicle cells surrounding germline-derived nurse cells and an oocyte, than uninfected ovarioles. Aberrant phenotypes included cysts lacking the follicle cell exterior (Fig 1K and 1M), the germline-derived interior (Fig 1M and 1N), or a normal number of differentiated nurse cells (Fig 1O).
These comparative data for a range of mei-P26 alleles, RNAi-knockdowns, and host ages indicate that wMel rescues mei-P26’s developmental functions in early host oogenesis. In contrast, wMel does not rescue mei-P26’s role in meiosis. Segregation defects were not rescued by wMel infection, as indicated by elevated X-chromosome nondisjunction (NDJ) rates in both infected and uninfected mei-P26[1] homozygous hypomorphs (NDJ = 7.7% and 5.6% by experiment, respectively, S6A and S6B Fig). In the following sections, we analyze wMel’s ability to rescue mei-P26 function at each of the critical time points in early oogenesis.
Host GSCs are maintained at higher rates in wMel-infected than uninfected mei-P26-deficient flies
GSCs were quantified by immunofluorescence staining of the essential GSC markers pMad and Hts in “young” 4- to 7-day-old (Fig 2) and “aged” 10- to 13-day-old females (S7A–S7C Fig). Positive pMad staining indicates the cell is responding to quiescent signals from the surrounding somatic GSC niche cells. Positive Hts staining requires localization to a single punctate spectrosome, opposed to a branching fusome [46].
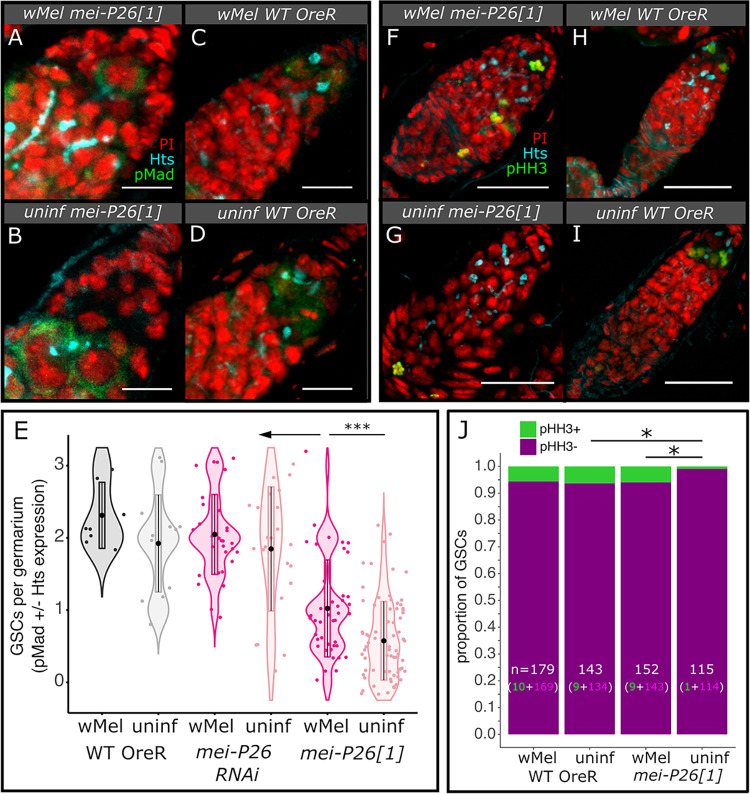
Fig 2. wMel infection rescues mei-P26-induced defects in female GSC maintenance.
(A–E) Infection with wMel confers higher numbers of GSCs per germarium in mei-P26[1] germaria than in uninfected flies. (A–D) Confocal mean projections of D. melanogaster germaria stained with antibodies against Hts and pMad. (E) Violin plots of the number of GSCs per germarium. As fully functional GSCs express pMad and have Hts-labeled spectrosomes, each was weighted by half, allowing for partial scores. Wilcoxon rank sum test * = p<0.05, ** = 0.01, *** = 0.001, **** = 1e-4. (F–J) Mitotic GSCs detected by pHH3 expression revealed that wMel restored mitosis in hypomorphic mei-P26 GSCs relative to uninfected GSCs. (F–I) Confocal mean projections of D. melanogaster germaria stained with antibodies against Hts and pHH3. (J) Stacked bar chart of the proportion of GSCs found expressing pHH3 in female germaria. Fisher’s exact test * = p < 0.05. Fluorescence channels were sampled serially and overlaid as indicated on each set of figs as follows: cyan = anti-Hts, yellow = anti-Vas, red = PI, green = anti-pMad in (A–D) and anti-pHH3 in (F–I). Scale bars A–D = 10 μm and F–I = 25 μm. The data underlying this figure can be found on Dryad at doi.org/10.7291/D1DT2C. GSC, germline stem cell; pHH3, phospho-Histone H3.
Staining revealed an increase in the average number of GSCs per mei-P26[1] germarium with wMel infection, relative to uninfected germaria in young, 4- to 7-day-old flies (infected/hypomorph: 1.0 versus uninfected/hypomorph: 0.58, p< = 2.9e-4 Wilcoxon rank sum test, S8 Table), but GSC maintenance did not reach OreR wild-type rates (infected/WT: 2.3 and uninfected/WT: 1.9, p< = 1.2e-4 to 2.8e-4 Wilcoxon rank sum test, Fig 2A–2E and S8 Table). wMel-infected mei-P26[1] germaria also had more cells manifesting other GSC properties, such as a large cytoplasm and physical attachment to the cap cells of the somatic niche [46], than uninfected germaria. In contrast to the mei-P26[1] allele, RNAi knockdown of mei-P26 did not significantly affect the numbers of GSCs per germarium (infected/RNAi: 2.1 and uninfected/RNAi: 1.9, S7A–S7C Fig and S8 Table), suggesting that the modest reductions in mei-P26 dosage have a stronger impact on differentiation than GSC maintenance. OreR wild-type germaria did not exhibit different numbers of GSCs due to wMel infection (Fig 2C–2E and S8 Table).
The number of GSCs per germarium in wMel-infected mei-P26[1] females converges on OreR wild-type values in aged 10- to 13-day-old germaria (S7C Fig). As wild-type flies age, the number of GSCs per germarium declines naturally [47], even in wMel-infected flies (aged/infected/WT: 1.6 versus young/infected/WT: 2.3, p< = 0.019 Wilcoxon rank sum test, Figs 2C–2E and S7C and S8 Table). Both infected and uninfected mei-P26[1] females appear to retain some of their GSCs as they age (aged/infected/hypomorph: 1.5 and aged/uninfected/hypomorph: 0.94, p< = 0.013–0.014 Wilcoxon rank sum test, S7C Fig and S8 Table). While OreR wild-type infected and uninfected flies lose from 3% to 31% of their GSCs in the first 2 weeks, infected and uninfected mei-P26[1] flies increase their GSC abundance by 43% and 63% in this time (S7C Fig), potentially through recruiting differentiated GSCs [48]. Taking both age-dependent trajectories in GSC retention into account, by the time wMel-infected mei-P26[1] germaria are 10 to 13 days old, they contain the same average number of GSCs per germarium as wild-type flies and significantly more GSCs than uninfected mei-P26[1] flies (p< = 0.015 Wilcoxon rank sum test, S7C Fig and S8 Table). Therefore, Wolbachia enhances stem cell maintenance in both young and old mei-P26[1] females.
Infected mei-P26[1] GSCs are functional, as indicated by their ability to divide by mitosis (Fig 2F–2J and S9 Table). We identified nuclei in mitosis by positive anti-phospho-histone H3 serine 10 (pHH3) staining, a specific marker of Cdk1 activation and mitotic entry [49]. Significantly fewer GSCs were in mitosis in uninfected mei-P26[1] germaria than infected mei-P26[1] and uninfected OreR wild-type germaria (0.09% versus 6%, p = 4.7e-2 and 4.6e-2 Fisher’s exact test, Fig 2F–2J and S9 Table). There was no difference in the frequency of GSCs in mitosis between uninfected and infected wild-type germaria (5% to 6%, Fig 2H–2J and S9 Table). Mitotic cystoblasts and cystocytes were only significantly enriched in infected mei-P26[1] germaria compared to infected wild-type germaria (p< = 1.3e-2 Wilcoxon rank sum test, S7D Fig and S10 Table), despite over-replication during transit-amplifying (TA) mitosis being a common phenotype in uninfected mei-P26[1] germaria (discussed below and [50]).
Regulation of host Sxl expression is rescued in wMel-infected germaria
Proper Sxl expression is required for GSC maintenance and differentiation [51]. High levels of Sxl in the GSC maintains stem cell quiescence [52]. When the GSC divides and the CB moves away from the niche, Sxl cooperates with Bam and Mei-P26 to bind to the nos 3′ untranslated region (UTR) and down-regulate Nos protein levels to promote differentiation [50,53].
We quantified Sxl localization patterns in whole ovaries stained with anti-Sxl antibodies by confocal microscopy of whole oocytes, revealing that Sxl dysregulation due to mei-P26 loss is mitigated by wMel infection (Fig 3). Less Sxl is expressed in uninfected mei-P26[1] germarium region 1 and more Sxl is expressed across regions 2a and 2b, relative to OreR wild-type (p< = 5.7e-10 to 5.3e-5 Wilcoxon rank sum test, Fig 3A–3B’ versus 3F–3H’ and 3I, S7E Fig, and S11 Table). Infection with wMel increased Sxl expression in germarium region 1, had no impact on region 2a, and suppressed expression in region 2b relative to uninfected mei-P26[1], replicating an expression pattern similar to that seen in wild-type germaria (p< = 5.7e-10 to 8.4e-3 Wilcoxon rank sum test, Figs 3A–3B’ versus 3C–3E’ and 3I and S7E and S11 Table). Interestingly, wMel infection may impact Sxl expression in OreR germaria, as the protein is significantly elevated in region 2a of wMel-infected relative to uninfected germaria (Fig 3I). Thus, Sxl dysregulation due to mei-P26 loss can be partially rescued by the presence of wMel, suggesting that wMel’s mechanism for mei-P26 rescue may underlie how Nos regulation in wMel-infected Sxl mutants [31,34].
Fig 3. Infection with wMel mitigates the consequences of dysfunctional mei-P26 on Sxl expression in the germarium.
(A–H’) Confocal mean projections of D. melanogaster germaria stained with antibodies against Sxl and pMad. Three phenotypes for each mei-P26[1] infection state are shown, (left) one as similar to wild-type as possible, (middle) one representative of GSC loss, pMad-negative, and (right) one representative of a developmentally altered phenotype retaining pMad-positive GSCs. We sampled the fluorescence channels serially and overlaid them as indicated on each set of images: cyan = anti-Sxl, red = PI, green = anti-pMad. Scale bars = 25 μm. (I) Bar-scatter plots of relative Sxl fluorescence expression levels across the germarium, by region (yellow outlines over Sxl-channel images in A’–H’). Colors as labeled in Figs 1A–1D and 2E, from left to right: dark gray = wMel-infected OreR, light gray = uninfected OreR, dark pink = wMel-infected mei-P26[1], light pink = uninfected mei-P26[1]. Wilcoxon rank sum test * = p<0.05, ** = 0.01, *** = 0.001, **** = 1e-4. (J) Model of wMel Sxl expression rescue in mei-P26[1] germaria. Green dashed line represents wMel’s up-regulation of pMad expression in the GSC (Fig 2A–2E). Purple dashed line represents wMel’s up-regulation of Sxl in the GSC/CB and down-regulation of Sxl by germarium region 2b. Arrows with + symbols indicate wMel’s action on gene expression. The data underlying this figure can be found on Dryad at doi.org/10.7291/D1DT2C. CB, cystoblast; GSC, germline stem cell; OreR, Oregon R; PI, propidium iodide.
Bam expression is partially restored in wMel-infected mei-P26 mutant germaria
In wild-type female flies, Bam expression begins immediately after the cystoblast daughter cell moves away from its undifferentiated sister, which remains bound to the GSC niche ([50,54] and illustrated in Fig 1P). Bam binds to the fusome and stabilizes CyclinA to promote TA mitosis, producing 16 cyst cells from a single CB [55]. Immediately following these 4 mitoses, Bam expression is down-regulated in wild-type ovaries to limit germline cysts to 16 germline-derived cells.
Through anti-Bam immunofluorescence confocal imaging and quantification, we determined that wMel-infection mitigates aspects of Bam dysregulation in mei-P26[1] germaria (Fig 4). Loss of Mei-P26 deregulates and extends Bam expression past germarium region 1 (Fig 4A–4B” versus 4C–4F” and 4G; S7F Fig and S12 Table), producing excess nurse-like cells [50]. Infection with wMel enables Bam up-regulation at the CB-to-16-cell stage in mei-P26[1] germaria relative to uninfected germaria (p< = 0.016, Fig 4G). Bam may also be down-regulated in stage 2a and 2b wMel-infected mei-P26[1] germaria because uninfected germaria express significantly more Bam than wild-type (p< = 0.019 Wilcoxon rank sum test, Fig 4G and S12 Table), whereas wMel-infected germaria do not (Fig 4G and S12 Table). Unfortunately, variance in mei-P26[1] Bam fluorescence intensity among samples is too large to resolve whether uninfected and infected germaria differ in Bam expression after the 16-cell stage with this dataset.
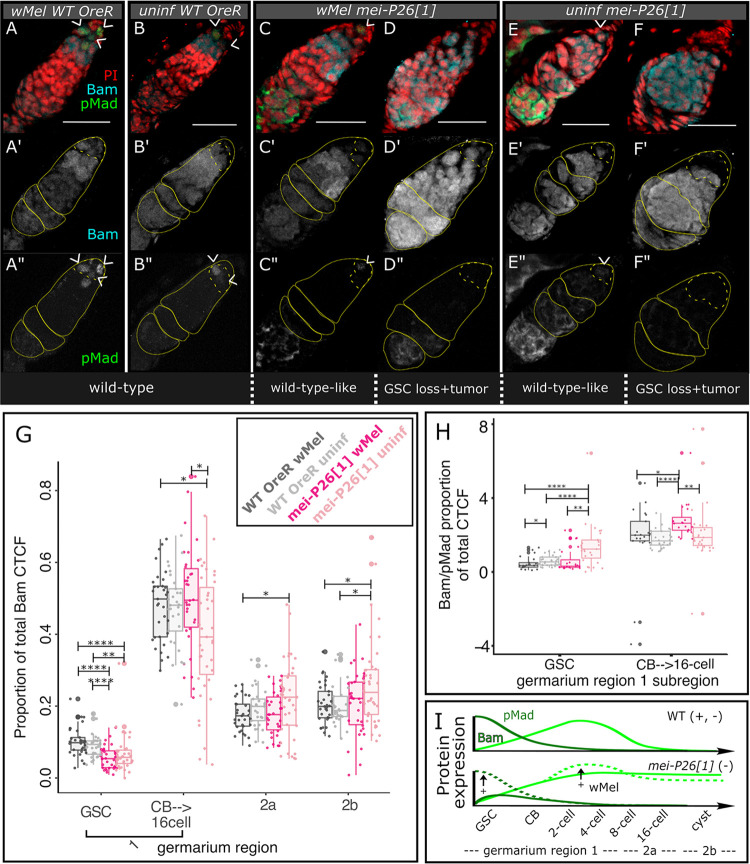
Fig 4. Infection with wMel mitigates the consequences of dysfunctional mei-P26 on Bam expression in the germarium.
(A–F”) Confocal mean projections of D. melanogaster germaria stained with antibodies against Bam and pMad. Two phenotypes for each mei-P26[1] infection state are shown, (left) one as similar to wild-type as possible and (right) one representative of developmentally altered phenotypes. (G) Bar-scatter plots of relative Bam fluorescence expression levels across the germarium, by region (yellow outlines over Bam-channel images in A’–F’). We subdivided region 1 into GSC and CB/cystocyte subsections because Bam expression ramps up in the CB. (H) Bar-scatter plot of the ratio of relative Bam to pMad fluorescence. As pMad and Bam expression are mutually exclusive in the GSC and CB/cystocytes, respectively, OreR wild-type values are near zero. Values significantly higher than this reflect dysregulation of the stem cell state. (I) Model of wMel rescue of Bam expression in mei-P26[1] germaria. Colors as labeled in Figs 1A–1C, 2E and 3I, from left to right: dark gray = wMel-infected OreR, light gray = uninfected OreR, dark pink = wMel-infected mei-P26[1], light pink = uninfected mei-P26[1]. Wilcoxon rank sum test * = p<0.05, ** = 0.01, **** = 1e-4. Scale bars = 25 μm. The data underlying this figure can be found on Dryad at doi.org/10.7291/D1DT2C. CB, cystoblast; GSC, germline stem cell; OreR, Oregon R.
Infection with wMel alters Bam expression in the GSC and CB to achieve an expression profile similar to and more extreme than wild-type germaria, respectively (Fig 4H and 4I). Both infected and uninfected mei-P26[1] germaria exhibit lower Bam expression than wild-type in the GSC niche (Fig 4G). This contrasts with expectations from the literature: bone morphogenic protein (BMP) signaling from the somatic niche induces pMad expression in the GSC, which directly represses Bam transcription, preventing differentiation [56]. Loss of mei-P26 function in the GSC should derepress Brat, resulting in pMad repression and inappropriate Bam expression [38]. However, we see lower Bam expression in both wMel-infected and uninfected mei-P26[1] GSCs relative to OreR wild-type (p< = 1.3e-6 to 1.4e-3 Wilcoxon rank sum test, Fig 4G and S13 Table). The elevated relative Bam/pMad expression ratio in uninfected mei-P26[1] GSCs (p< = 2.5e-4 to 4.0e-4 Wilcoxon rank sum test, Fig 4H and 4I and S13 Table) may explain this departure from expectations. Although GSC Bam expression is lower than in wild-type, Bam expression is clearly dysregulated and up-regulated relative to pMad expression, as expected for germline cells that have lost their stem cell identity [57,58]. Normalizing against pMad expression also reinforces the finding that wMel elevates Bam expression even higher than wild-type levels in mei-P26[1] hypomorphs at the CB-to-16-cell stage (p< = 1.2e-6 to 2.0e-2 Wilcoxon rank sum test, Fig 4H and 4I and S13 Table).
wMel rescues normal germline cyst and oocyte development impaired by mei-P26 knockdown
Mei-P26-deficient flies over-proliferate nurse cells and partially differentiate GSCs to produce tumorous germline cysts ([38,39] and Fig 5A–5D versus 5E–5H, Fig 5I and S14 Table). In RNAi knockdown ovaries, the formation of tumorous germline cysts is significantly mitigated by wMel infection (35.3% versus 16.3% of cysts with more than 15 nurse cells, respectively; p = 4.5e-3 Fisher’s exact test, Fig 5E and 5F versus 5G and 5H and S14 Table). In contrast, germline cyst tumors found in mei-P26[1] allele germaria were not rescued by wMel infection, despite the bacterium’s ability to rescue the rate at which these eggs hatch into progeny (Fig 5I versus Fig 1A and 1C), but consistent with their dysregulation of Bam expression in germarium regions 2a and 2b (Fig 4G). The stronger nature of the mei-P26[1] allele relative to the nos-driven RNAi knockdown likely underlies the difference in the number of tumorous cysts. Lack of tumor rescue indicates that either tumorous cysts produce normal eggs at some rate or wMel infection rescues tumors later in oogenesis, perhaps through some somatic mechanism [59–62].
Fig 5. wMel infection rescues cyst tumors and restores oocyte specification in mei-P26 mutants.
(A–I) Loss and overexpression of Mei-P26 induces the formation of tumorous cysts, which contain more than 15 nurse cells. wMel infection mitigates nos>mei-P26 RNAi-knockdown and exacerbates nos>mei-P26 OE phenotypes. (A–H) Confocal max projections of approximately 25 μm-thick sections of D. melanogaster oocyte cysts stained with PI. (I) Stacked bar chart of the proportion of germline cysts containing an abnormal number of nurse cells (less than or greater than 15 nurse cells). (J–R) Hypomorphic mei-P26[1] cysts infected with wMel significantly restored Orb translational regulation relative to uninfected cysts. Oocyte specification occurs in regions 2b-3 of the germarium (see diagram in Fig 1P), when Orb expression becomes restricted to the nascent oocyte. (J–Q’) Confocal mean projections of D. melanogaster germaria stained with antibodies against Orb and Vas. (R) Stacked bar chart of the proportion of germline cysts containing oocyte-specific Orb staining. Sample sizes are written on the bar charts. N = no, M = maybe, and Y = yes. Fisher’s exact test ** = p<1e-2, **** = p<1e-4. Scale bars = 50 μm. The data underlying this figure can be found on Dryad at doi.org/10.7291/D1DT2C. OE, overexpression; PI, propidium iodide.
Overexpression of mei-P26 produces tumors in both uninfected and infected ovaries; however, infected ovaries exhibit significantly more tumors than uninfected ovaries (p = 1.6e-3 Fisher’s exact test, Fig 5I and S14 Table). Finding that an excess of mei-P26 recapitulates the tumorous phenotype induced by a lack of mei-P26 indicates that this gene exerts a concentration-dependent dominant negative effect on its own function. Given that infection with wMel makes this dominant negative phenotype more severe, we hypothesize that wMel may make a factor that acts to enhance Mei-P26 function, potentially through a direct interaction or redundant function.
Infection with wMel increases the rate of oocyte-specific Orb localization in mei-P26[1] germline cysts (Figs 5J–5R and S7G and S15 Table), suggesting that wMel rescues functional cyst formation. Oocyte specification in region 2a of the germarium (see diagram in Fig 1P) is inhibited by the loss of mei-P26 through the derepression of orb translation [38]. Mei-P26 binds to the orb 3′ UTR and prevents its translation. Following cyst patterning and development, one of the 16 germline-derived cells is designated as the oocyte through specific oo18 RNA-binding protein (Orb) expression [63]. In region 2a, the germline cyst cell fated to become the oocyte activates orb translation [38]. Loss of mei-P26 results in orb derepression and nonspecific expression [38]. Infection with wMel restores oocyte-specific Orb expression in mei-P26[1] cysts relative to uninfected cysts (p = 5.9e-9 Fisher’s exact test, Fig 5L–5N’ versus 5O–5Q’ and S15 Table). We observed oocyte-specific Orb staining 93% to 94% of the time in wild-type cysts, whereas mei-P26[1] reduced this frequency to 25%, and wMel infection recovered mei-P26[1] cysts to 61% (counts in S15 Table). Oocyte-specific expression of Orb in wMel-infected cysts is robust, even in those that are developmentally aberrant (e.g., the double oocyte in Fig 5M–5M’). These results suggest that wMel rescues oocyte-specific Orb translation enhancing or duplicating Mei-P26’s function in orb translational repression.
OreR wild-type host fertility is beneficially impacted by wMel infection
Consistent with wMel’s ability to rescue the partial to complete loss of essential germline maintenance genes encompassing a range of functions, we discovered that this intracellular symbiont can reinforce fertility in D. melanogaster stocks displaying full wild-type fertility (Figs 1C and S8A–S8C and S4–S6 Tables). We analyzed egg lay, egg hatch, and overall offspring production rates independently to detect wMel effects on multiple components of fertility. Overall offspring production per female per day was elevated in wMel-infected nos:Gal4/CyO females relative to uninfected females by 49% (27 (uninfected) versus 40 (infected) offspring/female/day, p< = 2.2e-3 Wilcoxon rank sum test, S8A Fig and S4 Table). Infection with wMel increased the number of eggs laid per female per day for the nos:Gal4/CyO balancer stock by 49% (42 versus 28 eggs/female/day, p< = 2.2e-3 Wilcoxon rank sum test, S8B Fig and S5 Table), but did not have any detectable effect on egg lay rates in other genotypes with wild-type fertility. Following embryogenesis, infection with wMel elevated the rate that OreR and nos:Gal4/CyO wild-type eggs hatch into L1 stage larvae by 5.2% and 2.5%, respectively (88% and 91% (infected) versus 83% and 89% (uninfected) of eggs hatched, p< = 1e-4 to 2.1e-3 Wilcoxon rank sum test, Figs 1C and S8C and S6 Table). These results suggest that beneficial impacts of wMel infection are evident, yet variable among D. melanogaster stocks with wild-type fertility.
We measured OreR fertility across its lifetime (approximately 50 days) and found that wMel may produce higher lifetime fecundity relative to uninfected flies (Fig 6A–6C and S7 Table). OreR females were aged as indicated along the x-axis of Fig 6A–6C and mated to 3 to 7 days old males of matching infection status. wMel-infection maintains elevated OreR egg hatch rates as flies age (p< = 7.1e-9 Kolmogorov–Smirnov test, Fig 6C and S7 Table), while maintaining similar levels of egg production as uninfected flies (Fig 6B and S7 Table). This culminates in age ranges in which infected flies may produce an excess of offspring relative to uninfected flies (see gap between the regression confidence intervals when flies were approximately 20 to 30 days old, Fig 6A). The efficiency gains imparted on hosts by infection-induced elevated hatch rates could potentially accumulate for higher lifetime fitnesses, as resources are more efficiently converted to offspring.
Fig 6. The wMel strain of Wolbachia reinforces OreR wild-type fertility.
(A–C) D. melanogaster OreR fecundity versus female age, fit with a local polynomial regression (dark gray bounds = 95% confidence intervals). Infection with wMel may elevate (A) offspring and (B) egg offspring production at time points during the host lifespan through continuously elevating (C) hatch rate. (D–F’) Confocal mean projections of D. melanogaster (D–D’) infected and (E–E’) uninfected germaria and (F–F’) an infected ovariole. Intracellular wMel FtsZ localizes to germline (Vas+, outlined in dashed yellow) and somatic (Vas-) cells at high titers in infected OreR wild-type flies, with low background in uninfected flies. Bacteria in the GSC (* near the Hts-bound spectrosome) are indicated with arrows. Fluorescence channels were sampled serially and overlaid as indicated on each set of images as follows: cyan = anti-Hts, yellow = anti-Vas, blue = anti-FtsZ, and red = PI. Scale bars F–G = 10 μm, H = 50 μm. The data underlying this figure can be found in S3 Table and on Dryad at doi.org/10.7291/D1DT2C. GSC, germline stem cell; OreR, Oregon R; PI, propidium iodide.
Immunostaining of anti-FtsZ confirms that wMel exhibits high titers in the OreR wild-type germarium and concentrates in the oocyte in early cysts (Fig 6D–6F’). The intracellular bacteria are continuously located in the germline, starting in the GSC and proceeding through the end of embryogenesis and fertilization. Thus, wMel are located in the right place at the right time to reinforce female host fertility through interactions with the GSC and oocyte cyst, as well as to correct for perturbations caused by male CI-related alterations [64].
The wMel strain of Wolbachia in its native D. melanogaster host exhibits weak CI that decreases with age (S9A–S9D Fig), suggesting low maintenance of CI mechanisms [8]. Mating wMel-infected OreR males to infected (a “rescue cross”) and uninfected OreR virgin females (a “CI cross”) revealed that wMel reduces uninfected egg hatch by 26% when males are 0 to 1 day old (p< = 4.6e-4 Wilcoxon rank sum test, S9A Fig and S6 Table), but this moderate effect is eliminated by the time males are 5 days old, on average (S9C Fig and S6 Table). Only young males significantly impact offspring production (p< = 1.7E-2 Wilcoxon rank sum test, S9B and S9D Fig and S4 Table). In contrast, the Wolbachia wRi strain that naturally infects Drosophila simulans (the Riv84 line) and induces strong CI [10,65], reduces uninfected egg hatch by 95% when males are 0 to 1 day old (p< = 5.6e-8 Wilcoxon rank sum test, S9E Fig and S6 Table) and only loses some of this efficacy by 5 days (75% hatch reduction, p< = 3.6e-4, Wilcoxon rank sum test; S9G Fig, S6 Table). This significantly impacts overall offspring production (p< = 6.5E-7 and 5.0E-2 Wilcoxon rank sum test, S9F and S9H Fig and S4 Table). Paternal grandmothers of CI offspring were 3 to 7 days old (i.e., relatively young), contributing to weak CI in the D. melanogaster crosses [17]. Overall, these results suggest that wMel’s beneficial reproductive manipulations may affect its fitness more than its negative reproductive manipulations in nature.
wMel-mediated mei-P26[1] rescue may be enabled through rescuing and perturbing host transcription
Exploring wMel’s effect on host gene expression in OreR and mei-P26[1] ovaries with dual-eukaryotic and bacterial RNA sequencing (dual-RNAseq) revealed that infection significantly alters host gene expression in a variety of ways (Fig 7A–7C and S16–S19 Tables), which indicate a few potential mechanisms for wMel-mediated fertility rescue. Of the 35,344 transcripts in the D. melanogaster genome, 10,720 were expressed at high enough levels in at least 5 of 6 samples of each genotype+infection group to be included in the analysis. Of the 1,286 genes in the wMel genome, 663 were expressed in D. melanogaster ovaries (Fig 7D and S20 Table).
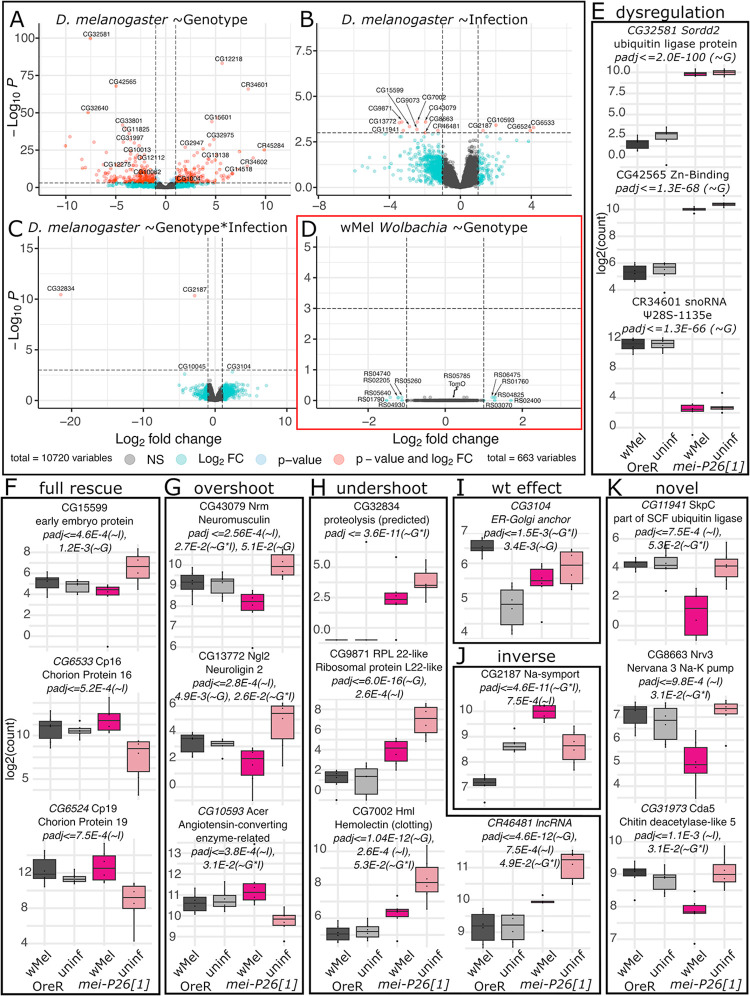
Fig 7. Differential expression analysis of wMel and D. melanogaster genes from host ovaries reveal that mei-P26 knockdown and infection influence global expression patterns.
(A–D) Volcano plots of log2-fold change in gene expression versus log10-FDR adjusted p-value (padj) for the test performed in each panel. (E–K) Normalized transcript count plots for (E) the 3 top genotype-associated (~G) Wald Test hits by padj, and (F–K) D. melanogaster genes significantly associated (padj< = 1e-3) with infection (~I) or jointly, genotype and infection (~G*I), organized by the differential expression pattern. See S10–S15 Figs for other significant and insignificant count plots. In all barplots, dark gray = wMel-infected OreR, light gray = uninfected OreR, dark pink = wMel-infected mei-P26[1], light pink = uninfected mei-P26[1]. The data underlying this figure can be found at NCBI, under BioProject number PRJNA1007602. OreR, Oregon R.
Both uninfected and wMel-infected hypomorphic mei-P26[1] ovaries exhibited a 5-fold depletion in mei-P26 expression relative to OreR ovaries (S2B and S2D Fig) and significant dysregulation of 1,044 other genes due to the mei-P26[1] allele after FDR correction (Wald test FDR-adjusted p-value (padj)< = 0.05, Figs 7A, 7E and S10A and S17 Table). The changes in gene expression due to the loss and dysfunction of mei-P26 far outnumber and outpace the changes in gene expression due to infection, in either D. melanogaster genotype, and highlight how important mei-P26 is as a global regulator of gene expression. This number is well in excess of the 366 genes thought to be involved in GSC self-renewal [66], underscoring how many processes mei-P26 is involved in, from germline differentiation [50] to somatic development [61]. Across biological process, cell component, and molecular function GO categories, loss of mei-P26 impacted processes involving chromatin, recombination, protein–protein interactions, and muscle cell differentiation (S10B–S10G Fig), supporting mei-P26’s known role in genetic repression, differentiation, and meiosis [38,40,50].
Infection with wMel significantly alters the expression of hundreds of D. melanogaster genes to either restore or compensate for mei-P26-regulated gene expression. We tested for genes significantly associated with infection state in DESeq2 under the full model: ~genotype + infection + infection*genotype, revealing 422 significantly differentially expressed genes (Wald test padj< = 0.05, Fig 7B and S18 Table). Testing for an effect of the interaction between infection and genotype yielded another 20 significantly differentially expressed genes (Wald test padj< = 0.05; Fig 7C and S19 Table). The most significantly differentially expressed genes are featured in Fig 7F–7K and other significant genes are plotted in Figs 8, S11 and S12. Binning gene expression patterns revealed that there are at least 6 main rescue gene expression phenotypes induced by wMel-infection in mutant ovaries: full rescue, overshoot, undershoot, wild-type effect, inverse, and novel (Figs 7F–7K, 8A, 8C, S11A, S11B, S12A and S12B). In “full rescue” phenotypes (Figs 7F and S11A), mei-P26[1] wMel gene expression is indistinguishable from OreR levels and significantly different from uninfected mei-P26[1] levels. “Overshoot” phenotypes (Fig 7G) exhibit expression that is significantly in the opposite direction of the uninfected mei-P26[1] knockdown effect relative to OreR expression. Oppositely, “undershoot” phenotypes (Fig 7H) are partial corrections towards OreR expression in mei-P26[1] wMel ovaries. The one “wild-type effect” gene, an ER and Golgi cytoskeletal membrane anchor protein, demonstrated differential expression only between wMel-infected and uninfected OreR ovaries (Fig 7I). Five genes exhibit “inverse” differential expression phenotypes, such as a sodium symporter (Fig 7J) and the transcription factor Chronophage (S12A Fig), in which mei-P26[1] and OreR ovaries show opposite changes in differential expression relative to the uninfected genotype. In total, only these 6 “wild-type effect” and “inverse” genes were differentially expressed due to infection in OreR ovaries. Lastly, “novel” genes (Fig 7K) are those that only exhibit differential expression in wMel-infected mei-P26[1] ovaries, suggesting a novel rescue or compensation pathway. For example, suppression of retinal degeneration disease 1 upon overexpression 2 (sordd2) ubiquitin ligase mRNA expression is up-regulated, without rescue, in both infected and uninfected mei-P26[1] ovaries (Fig 7E). One of the novel rescue candidates, SKP1-related C (skpC), is a down-regulated component of SCF ubiquitin ligase (Fig 7K), which might compensate for the up-regulation of sordd2.
Fig 8. Mei-P26 interactions predicted from the literature are supported by our ovary dual-RNAseq results.
(A, B) Predicted mei-P26 (A) protein and mRNA physical interactions and (B) genetic interactions. Symbols near gene names indicate significant DE interactions: * = significance by genotype; r = significance by infection, indicating a rescue phenotype; _ = significance by uncorrected p-values only. Red “X”s in A indicate genes that were not expressed in our dataset (CG6304 and mir-137). (C) Normalized transcript count plots for the genes designated with symbols in A and B. In all barplots, dark gray = wMel-infected OreR, light gray = uninfected OreR, dark pink = wMel-infected mei-P26[1], light pink = uninfected mei-P26[1]. (D) Cellular component GO terms for the 87 (~G*I) DE genes reveal an enrichment for early germline cytoskeletal structures and membrane proteins. Wald test association for listed padj and p-values are given in parentheses, as in Fig 7: genotype (~G), infection (~I), genotype*infection (~G*I). The data underlying this figure can be found at NCBI, under BioProject number PRJNA1007602. OreR, Oregon R.
GO terms associated with infection and the interaction between infection and genotype (Figs S11C–S11G, S12C–S12G and 8D) were consistent with wMel’s known ability to associate with host actin [67], microtubules, motor proteins [68–70], membrane-associated proteins [71], and chromatin [72]. Infection-associated genes were enriched in GO terms suggesting a cytoskeletal developmental function with chromatin interactions (S11C–S11G Fig). Across biological processes, cellular compartment, and molecular function categories, genes were enriched in terms suggesting interactions with actin, myosin, contractile fibers, and muscle cell differentiation. Chromatin interactions were indicated by associations with condensin complexes, centromeres, and mitotic chromosomes. Genotype-by-infection interaction-associated genes were also enriched in cytoskeletal and chromatin-interaction GO terms, in addition to plasma membrane/cell junction-associated terms such as cell junction and synapse structure maintenance, cell cortex, and lipid and calmodulin binding (S12C–S12G Fig).
Interestingly, 2 chorion proteins (Fig 7F), an ecdysteroid 22-kinase associated protein (S11A Fig), and an estrogen-related receptor (ERR) (S12A Fig), which are likely involved in chorion formation [73], were rescued by wMel. Considering that we have found mei-P26[1] embryos to have weaker corions then OreR wild-type embryos when treated with bleach, this finding suggests a novel chorion-associated function for mei-P26 and a novel rescue phenotype for wMel.
None of the 663 transcribed wMel genes were significantly differentially expressed between the OreR and mei-P26[1] host genotypes, after p-values were FDR-corrected (Figs 7D, S13 and S14 and S20 Table), suggesting host regulation by wMel’s constitutively expressed genes. On average, wMel transcriptomes were sequenced at 7.8 to 42.4× depth of coverage and D. melanogaster transcriptomes were sequenced at 25.9 to 35.9× (S16 Table). While these coverage ranges overlap, the 663 wMel genes detected only spanned −1.5-fold depletion to 1.3-fold up-regulation (S20 Table). In contrast, the 10,720 expressed D. melanogaster genes spanned an order of magnitude more differential expression, with −10.041-fold depletion to 9.866-fold up-regulation (S17–S19 Tables). Thus, wMel genes are weakly differentially transcribed compared to D. melanogaster genes, suggesting that wMel may alter host phenotypes through downstream impacts of constitutively expressed proteins. To explore the constitutive effects of wMel expression, we tested for GO pathway enrichment in the expressed transcriptome (S13 Fig). Pathways for DNA replication, protein expression, membrane transport, and heterotrophy (e.g., oxidative phosphorylation) were detected in abundance. While many of these genes are likely involved in normal bacterial cell maintenance and replication, a significant enrichment in pathways for oxidative phosphorylation, translation, transcription, and membrane functions (S13A Fig) suggest that some of these genes could be co-opted for host manipulation or processing host resources.
Among the 1,044 significantly differentially regulated D. melanogaster genes impacted by mei-P26 knockdown and infection were 6 genes (of 24) predicted in the literature (padj< = 0.05 Wald test and 9 of 24 by p-value< = 0.05; Figs 8 and S1 and S1 and S2 Tables). Loss of mei-P26 function caused dysregulation in oo18 RNA-binding protein (orb), gawky (gw), sex-lethal (sxl), benign gonial cell neoplasm (bgcn), tumorous testis (tut), and bantam (mir-ban) (padj< = 0.05 Wald test, Fig 8C and * in Fig 8A and 8B). Bam, Pumilio (pum), and Justice Seizure (jus) expression may be altered by mei-P26 knockdown, but additional samples are needed to resolve high infection variance (e.g., mir-ban) and low differential expression change (e.g., gw) (p-value< = 0.05 Wald test, Fig 8C and “*” in Fig 8A and 8B). Two of 6 genes exhibited evidence of wMel rescue with an “undershoot” rescue phenotype (“r” in Fig 8A and 8B): bgcn and tut by padjust (Fig 8C). If we consider genes with significant unadjusted p-values, gw may also exhibit evidence of “undershoot” rescue (Fig 8C, “r” in Fig 8A). Bantam or mir-ban expression suppression may be rescued as well, but high variance among wMel mei-P26[1] ovaries precludes DE rescue detection with this dataset (Fig 8C). In contrast, orb, sxl, bam, and nos transcript levels are not altered by mei-P26 knockdown or wMel infection, and therefore must be regulated posttranscriptionally. In total, these differential expression results reinforce and expand our understanding of how wMel rescues and reinforces host reproduction at transcriptional and posttranscriptional levels.
Discussion
The wMel strain of Wolbachia is a successful and widespread symbiont: naturally in D. melanogaster populations [19,20] and novelly through lab-generated trans-infections of wMel into non-native hosts used for biological control [3]. However, we know remarkably little about the processes that shape how symbiont and host coevolve and find mutually beneficial mechanisms for their reproduction. In this work, we confirm that wMel’s CI strength is typically weak in D. melanogaster, (Fig 9A–9D, [14–18]) and show that wMel confers benefits on host fertility through reinforcing host GSC maintenance and gamete differentiation (Figs 1–8).
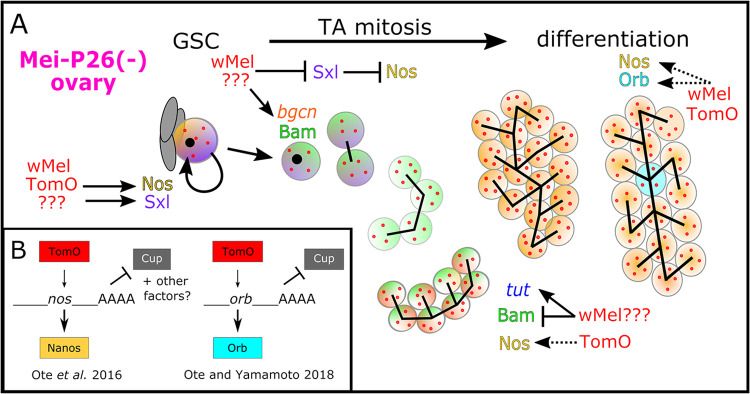
Fig 9. Model of wMel’s interactions with essential host genes in early GSC maintenance and germline cyst formation.
(A) In mei-P26 mutants, wMel restores normal Bam, Sxl, and Orb expression through various mechanisms, most of which are unknown (??? = unknown wMel factors). Protein and mRNA expression rescue correlates with cytological rescue from GSC maintenance through germline cyst differentiation. (B) Sxl rescue in the GSC is mediated through TomO’s interaction with RNPs containing nos mRNA, up-regulating Nos translation. Later, in stage 8 of cyst oogenesis, TomO was found to bind orb mRNA, displacing the translational repressor Cup, and up-regulating Orb translation [34,35]. Dashed arrow in A indicates TomO’s predicted function in enabling Orb derepression in early cyst differentiation. The factor that restores mei-P26 repression of Orb translation in mei-P26 mutants has not been identified. GSC, germline stem cell; RNP, ribonucleoprotein; TomO, toxic manipulator of oogenesis.
Here, we demonstrate that wMel infection can reinforce host fertility in both OreR wild-type and mei-P26 mutants by correcting perturbed gene expression patterns, and we shed light on the long-standing mystery of how wMel beneficially impacts host reproduction. Previously, wMel was shown to rescue the partial loss of Sxl [31] and Bam [32], 2 essential genes for 2 very different stages of early oogenesis: GSC maintenance and CB differentiation and TA-mitosis, respectively. These findings left unresolved how wMel suppresses these mechanistically and temporally distinct cellular processes and suggested that mei-P26 was not involved [31]. We show with an array of alleles and developmental assays that infection with wMel rescues defects associated with mutant mei-P26, a gene required for both GSC maintenance and differentiation. Infection with wMel rescues all stages of oogenesis in mei-P26 RNAi knockdowns, as well as hypomorphic and null alleles (Figs 1–5, 7, 8, S4–S7 and S10–S12 and S4–S15 and S18 and S19 Tables). Mei-P26 is a TRIM-NHL protein that regulates gene expression via mRNA translational inhibition through the Nos mRNA-binding complex, interactions with the RNA-induced silencing complex (RISC), and protein ubiquitination [38,74]. Importantly, Mei-P26 interacts with Sxl in the GSC and CB and Bam in the CB and cystocytes [36,50]. These interactions suggest that the mechanism wMel employs to rescue mei-P26 function may also be responsible for rescuing aspects of Sxl-dependent GSC maintenance and CB differentiation and Bam-dependent cyst differentiation.
In this in-depth molecular and cellular analysis of wMel-mediated manipulation of GSC maintenance and differentiation, we discovered that wMel rescues mei-P26 germline defects through correcting perturbed pMad, Sxl, Bam, and Orb protein expression and bgcn and tut mRNA expression towards OreR wild-type levels (diagrammed in Fig 9A). Infection with wMel partially rescued GSC-specific pMad expression relative to wild-type flies (Fig 2A–2E), suggesting that the bacteria restored BMP signaling from the somatic GSC niche [38]. Finding that these cells are also able to divide (Fig 2F–2J), further supports their functionality and stemness. Given that mei-P26 is likely involved in Drosophila Myc (dMyc) regulation, and overexpression of dMyc induces competitive GSCs [75], wMel’s GSC rescue mechanism may involve dMyc up-regulation. Although, our transcriptomic data indicate that dMyc is not significantly repressed at the transcriptional level (S15 Fig). In the germline cells that left the niche, wMel recapitulated the properly timed changes in pMad, Sxl, and Bam expression that are normally influenced by mei-P26 and are required during cystoblast differentiation to form the 16-cell germline cyst [39,40,50,74] (Figs 3, 4, and S7E–S7F). While Bam down-regulation is not fully rescued by wMel (Fig 4), finding that tut mRNA expression is partially reduced in wMel-infected mei-P26[1] germaria (Fig 8) suggests that there may be an unreported regulatory interaction between Tut, Bam, and Mei-P26 in females (as in males [76]), which could rescue the overexpression of Bam in late germline cyst development.
This is the first time wMel has been found to rescue oocyte differentiation post-cyst formation, suggesting that wMel has pervasive impacts throughout oogenesis that may reinforce embryogenesis. We confirmed the downstream consequences of Bam expression rescue in wMel-infected mei-P26[1] ovaries by finding fewer germline cyst tumors in infected flies (Fig 5A–5I). These germline cysts appeared to be functional, based on restored oocyte-specific expression of the essential oocyte differentiation protein Orb in infected mei-P26[1] cysts, relative to uninfected cysts (Fig 5J–5R). Finding that wMel rescues Cp16 and Cp19 chorion protein, tut, bgcn, and other transcript expression in ovary tissues (Figs 7F, 8, S11 and S12) further supports the conclusion that wMel can rescue the downstream impacts of mei-P26 loss. Future work is needed to explore how the overshoot and novel DE categories mitigate the loss of mei-P26, which may occur through some alternate or compensatory pathway. In addition to the mechanism discussed previously for ubiquitin ligase rescue, the loss of Ionotropic receptor 76a (Ir76a) or nicotinic Acetylcholine Receptor α5 (nAChRα5) expression (S10A Fig) could be rescued by the novel up-regulation of the Wunen-2 lipid phosphate phosphatase as a receptor or membrane transporter in wMel-infected mei-P26[1] ovaries (S12B Fig). Functions like these could underlie wMel-induced developmental resilience in OreR embryogenesis, as demonstrated by elevated hatch rates in wMel-infected OreR flies (Figs 1C and 6C).
The wMel strain’s rescue of D. melanogaster mei-P26 is likely independent of the Wolbachia TomO protein. Prior work on TomO indicates that the bacterial protein’s primary mode of action is through destabilizing ribonucleoprotein (RNP) complexes [35] (diagrammed in Fig 9B). This is how TomO elevates Nos expression in the GSC [34] and how TomO inhibits the translational repressor Cup from repressing Orb translation in mid-oogenesis (stage 8) [35,77]. In contrast, Mei-P26 interacts with Ago1 RISC to inhibit Orb translation through binding the orb ′3-UTR miRNA-binding sites [38]. Although Mei-P26 and TomO produce opposite outcomes for Orb expression and TomO is not significantly up-regulated in mei-P26[1] ovaries (S14C Fig), TomO-orb binding may underlie wMel’s ability to rescue Orb repression, if wMel produces other factors that modulate the interaction. Alternatively, wMel may make another RNA-binding protein. Given that Sxl is also repressed through the sxl 3′-UTR by Bruno [78], such a protein could be involved in restoring Sxl regulation in mei-P26 mutants. Neither orb nor sxl expression is rescued at the mRNA level by wMel infection (Fig 8C), lending further support for rescue at the translational or protein interaction/modification level.
Additional bacterial factors besides TomO must be involved in reinforcing host fecundity in wild-type flies and fertility mutants. First, Mei-P26 encodes 3 functional domains that confer at least 3 different functional mechanisms for modulating gene expression (mRNA-binding, miRNA-mediated, and ubiquitination). In a bacterial genome, these domains/functions are likely to be encoded by more than 1 protein [79]. Second, TomO rescues Sxl’s GSC maintenance functions by binding nos mRNA, increasing levels of Nos translation, but it cannot fully rescue Sxl loss [34]. Immediately after the GSC divides to produce the CB, Sxl interacts with Bam, Mei-P26, Bgcn, and Wh to inhibit Nos translation [36,74]. Thus, a wMel factor that inhibits Nos translation, counteracting TomO’s function to destabilize Nos translational repressors through RNA binding [34,35], remains to be identified. Furthermore, Nos up-regulation cannot explain Bam rescue because Nos and Bam exhibit reciprocal expression patterns during TA mitosis [36]. Third, exacerbation of the tumor phenotype induced by nos:Gal4-driven Mei-P26 overexpression by wMel infection (Fig 5I) suggests that wMel synthesizes a protein that directly mimics Mei-P26’s functions in differentiation and can reach antimorphic levels. Misregulated expression of a mei-P26-like factor could explain why wMel-infected mei-P26 RNAi females produce more eggs and offspring than OreR wild-type flies (Fig 1A and 1B). While TomO may recapitulate some of Mei-P26’s functions, such as orb-binding, at least one other factor is needed to recapitulate proper Orb, Nos, and Bam regulation.
Understanding how Wolbachia interacts with host development at the molecular level is essential to deploying these bacteria in novel hosts and relying on vertical transmission to maintain them in host populations. CI has been a powerful tool for controlling host populations [80] and driving Wolbachia to high infection frequencies [3]. The continuous presence of bacterial reproductive manipulators in host GSCs opens the opportunity for compensatory developmental functions to evolve, similar to what we have shown for mei-P26. While it is not known if any of the naturally occurring mei-P26 alleles [32,81] confer loss of function that is remedied by the presence of Wolbachia, natural variation in developmental genes theoretically could have provided the selection pressure for wMel to evolve its beneficial reproductive functions. Given that selection will favor the loss of CI if conflicting beneficial impacts on host fertility are realized [8], it is imperative that we understand the mechanisms underlying beneficial reproductive manipulations such as fertility reinforcement.
Materials and methods
Russell and colleagues Wolbachia endosymbionts manipulate GSC self-renewal and differentiation to enhance host fertility
Candidate gene selection
Leveraging what is known about wMel’s abilities to rescue essential maintenance and differentiation genes [31,32] with empirical data on protein–protein and protein–mRNA interactions among these genes (esyN [82] networks in Figs S1A–S1D and 8; references in S1 Table) and data on interactions between host genes and wMel titers [37], we identified the essential germline translational regulator meiotic P26 (mei-P26) as a potential target of bacterial influence over GSC maintenance and differentiation pathways (S1A and S1B Fig). Loss of mei-P26 in uninfected flies produces mild to severe fertility defects through inhibiting GSC maintenance, meiosis, and the switch from germline cyst proliferation and differentiation [38–40].
Drosophila stocks and genetic crosses
Flies were maintained on white food prepared according to the Bloomington Drosophila Stock Center (BDSC) Cornmeal Food recipe (aka “white food,” see https://bdsc.indiana.edu/information/recipes/bloomfood.html). The wMel strain of Wolbachia was previously crossed into 2 D. melanogaster fly stocks, one carrying the markers and chromosomal balancers w[1]; Sp/Cyo; Sb/TM6B, Hu and the other carrying the germline double driver: P{GAL4-Nos.NGT}40; P{GAL4::VP16-Nos.UTR}MVD1. These infected double balanced and ovary driver stocks were used to cross wMel into the marked and balanced FM7cB;;;sv[spa-pol], null/hypomorphic mutants, and UAS RNAi TRiP lines to ensure that all wMel tested were of an identical genetic background. The D. melanogaster strains obtained from the BDSC at the University of Indiana were: Oregon-R-C (#5), w1118; P{UASp-mei-P26.N}2.1 (#25771), y[1] w[*] P{w[+mC] = lacW}mei-P26-1 mei-P26[1]/C(1)DX, y[1] f[1]/Dp(1;Y)y[+]; sv[spa-pol] (#25716), y[1] w[1] mei-P26[mfs1]; Dp(1;4)A17/sv[spa-pol] (#25919), y[1] sc[*] v[1] sev[21]; P{y[+t7.7] v[+t1.8] = TRiP.GL01124}attP40 (#36855). We obtained the UASp-mRFP/CyO (1-7M) line from Manabu Ote at the Jikei University School of Medicine. Drosophila simulans w[–] stocks infected with the Riv84 strain of wRi and cured with tetracycline were sourced from Sullivan Lab stocks [83,84]. All fly stocks and crosses were maintained at room temperature or 25°C on white food (BDSC Cornmeal Food) because the sugar/protein composition of host food affects Wolbachia titer [84].
Wild-type fertility stocks: Paired wMel-infected (OreR_wMelDB) and uninfected Oregon R (OreR_uninf) stocks were made by crossing males of Oregon-R-C to virgin females from paired infected and uninfected balancer stocks of the genotype w[1];CyO/Sp;Hu/Sb. To ensure that no differences arose between these 2 D. melanogaster genotypes during the balancer cross or subsequently, we backcrossed males of the OreR_wMelDB stock to females of the OreR_uninf stock for 10 generations and repeated egg lay and hatch assays (F10_OreR_uninf). Paired infected and uninfected nosGal4/CyO and nosGal4/Sb flies were made by crossing to paired infected and uninfected balancer stocks, as described above.
The genomic and phenotypic consequences of the mei-P26 alleles studied here and previously (e.g., [31]) have been characterized [40]. The hypomorphic mei-P26[1] allele was created by the insertion of a P{lacW} transposon in the first intron of mei-P26, which codes for the RING domain. The mostly null mei-P26[fs1] allele tested by Starr and Cline in 2002 was generated by a second P{lacW} insertion in mei-P26[1]’s first insertion. The fully (male and female) null mei-P26[mfs1] allele arose by deleting the P{lacW} insertion from mei-P26[1], along with approximately 2.5 kb of flanking sequence. According to Page and colleagues (2000), these 3 alleles form a series, with increasing severity: mei-P26[1] < mei-P26[fs1] (and other fs alleles) < mei-P26[mfs1].
Fecundity crosses
Overview
SLR performed the 3,002 fecundity crosses continuously between October 2020 and January 2022, in batches based on their eclosion date (Table 1). Infected and uninfected OreR lines were maintained continuously to (1) control the age of grandmothers of CI males; (2) regularly have OreR virgins for mei-P26 and CI fecundity assays; (3) collect and age WT females for the aged fertility assay; and (4) obtain large sample sizes from mei-P26 fertility mutants. The full fecundity dataset is contained in S2 Table and plotted in S3 Fig.
Table 1. Fecundity cross genotypes, sexes, ages, and control crosses.
| Fecundity cross category | Female age | Male age | Parallel control crosses |
|---|---|---|---|
| wMel-infected mei-P26 rescue | 3–7 days old | 3–7 days | Uninfected and wild-type |
| wMel-infected OreR wild-type | 3–7 days old | 3–7 days | Uninfected OreR wild-type and other “wild-type” fertility stocks (e.g., nos:Gal4/CyO) |
| Aged wMel-infected mei-P26 rescue | 2–25 days old | 3–7 days | Uninfected and wild-type |
| Aged wMel-infected OreR wild-type | 2–45 days old | 3–7 days | Uninfected OreR wild-type |
| CI in wMel-D. melanogaster OreR | 3–7 days old | 0 or 3–7 days (from 3- to 7-day-old mothers) | Rescue, reciprocal, and uninfected |
| CI in wRi-D. simulans w[–] | 3–7 days old | 0 or 3–7 days (from 3- to 7-day-old mothers) | Rescue |
CI, cytoplasmic incompatibility; OreR, Oregon R.
Cross conditions
Food (vials of Bloomington’s white food recipe), laying media (grape food), and incubation conditions were made in-house in large batches monthly. Grape spoons: Approximately 1.5 mL of grape agar media (1.2× Welch’s Grape Juice Concentrate with 3% w/v agar and 0.05% w/v tegosept/methylparaben, first dissolved at 5% w/v in ethanol) was dispensed into small spoons, allowed to harden, and stored at 4°C until use. Immediately prior to use in a cross, we added ground yeast to the surface of each spoon.
Preparation of flies for fecundity crosses
Paired wMel-infected and uninfected genetic crosses were performed in parallel for the 4 mei-P26 mutants (RNAi, [mfs1], [1], [mfs1/1]) to produce homozygous virgin females of both infection states for parallel fecundity crosses. Mutant genetic crosses were performed multiple (3 or more) times across the 14 months to produce homozygotes from many different parents and matings. CI crosses were performed with males from infected grandmothers that were 3 to 7 days old. Males were aged either 0 (collected that day) or 5 days, depending on the cross. We collected males from vials over multiple days, so the majority of males were not the first-emerged.
Virgin female flies collected for fecundity crosses were transferred to fresh food and aged an average of 5 days at room temperature (from 3 to 7), or longer for the aged OreR, mei-P26 RNAi, and mei-P26[1] fecundity crosses. Males and virgin female flies were stored separately, and males were aged 0 or 3 to 7 days. Long-term aged virgin females were kept as small groups in vials of fresh white food. Every few days, the vials were inspected for mold and flies were moved on to fresh food.
Fecundity cross protocol
In the afternoon of the first day of each cross, 1 male and 1 female fly were knocked out on a CO2 pad, added to a vial containing a grape spoon, allowed to recover, and then transferred to a 25°C constant humidity incubator on a 12 light/dark cycle to allow for courting and mating. The following day, each spoon was replaced with a fresh spoon, and the vials were returned to 25°C for more mating. On the third day, we removed the flies from the vials, counted the number of eggs laid on the spoon’s grape media, and replaced the spoon in the vial at 25°C for 2 days. After approximately 40 h, we counted the number of hatched and unhatched eggs. The exact times and dates for all steps in all crosses were recorded (S2 Table) and plotted to show consistent results across the time frame (S3 Fig).
Fecundity cross analysis
Fecundity data were parsed using perl scripts and plotted in R. Individual crosses were treated as discrete samples. Hatch rates were calculated from samples that laid 20 or more eggs (for 3- to 7-day-old female plots) and 10 or more eggs (for age versus hatch rate plots). Egg lay and offspring production rates were calculated from raw counts, divided by the fraction of days spent laying (metadata in S2 Table). Crosses with 0 laid eggs (and offspring) were not rejected, as the fertility mutants often laid 0 to few eggs. Our cross conditions were highly consistent and run daily, lending confidence to these zero-lay/offspring samples (S3 Fig). Drosophila fecundity versus female age data were fit in R using the stat_smooth loess method, formula y~x, with 0.95 level confidence interval.
Non-disjunction assays
We placed females homozygous for the mei-P26[1] allele bearing the yellow mutation (y[1] w[*] mei-P26[1];;; sv[spa-pol]) in vials with males bearing a wild-type yellow allele fused to the Y chromosome (y[1] w[1] / Dp(1;Y)y+). We made 7 or 8 vials of 1 to 10 females mated to a count-matched 1 to 10 males for infected and uninfected mei-P26 hypomorphs, respectively. After eclosion, we screened for non-disjunction in the progeny by the presence of yellow males (XO) and normally colored females (XXY). Rates of non-disjunction (NDJ) were calculated to account for the inviable progeny (XXX and YO) with the equation NDJ rate = 2 *NDJ offspring/all offspring = (2XXY + 2X0)/(XX+XY+2XXY+2X0), as in [41].
Ovary fixation and immunocytochemistry
Within a day or 2 or eclosion, flies were transferred to fresh food and aged 3 to 7 days, or longer as indicated in the text. For long aging experiments, flies were transferred to fresh food every week and investigated for mold every few days. We dissected the ovaries from approximately 10 flies from each cross in 1× PBS and separated the ovarioles with pins. Ovaries were fixed in 600 μl heptane mixed with 200 μl devitellinizing solution (50% v/v paraformaldehyde and 0.5% v/v NP40 in 1× PBS), mixed with strong agitation, and rotated at room temperature for 20 min. Oocytes were then washed 5× in PBS-T (1% Triton X-100 in 1× PBS) and treated with RNAse A (10 mg/ml) overnight at room temperature.
After washing 6 times in PBS-T, we blocked the oocytes in 1% bovine serum albumin in PBS-T for 1 h at room temperature, and then incubated the oocytes in the primary antibodies diluted in PBS-T overnight at 4°C (antibodies and dilutions listed below). The following day, we washed the oocytes 6 times in PBS-T and incubated them in secondary antibodies diluted 1:500 in PBS-T overnight at 4°C. On the final day, we washed the oocytes a final 6 times in PBS-T and incubated them over 2 nights in PI mounting media (20 μg/ml PI (Invitrogen #P1304MP) in 70% glycerol and 1× PBS) at 4°C. Overlying PI medium was replaced with clear medium, and oocytes were mounted on glass slides. Slides were stored immediately at −20°C and imaged within a month. Infected and uninfected, as well as experimental and control samples were processed in parallel to minimize batch effects. Paired wMel-infected and uninfected OreR stocks were used as wild-type controls.
Primary monoclonal antibodies from the Developmental Studies Hybridoma Bank (University of Iowa) were used at the following dilutions in PBS-T: anti-Hts 1:20 (1B1) [45], anti-Vas 1:50 [44], anti-Orb 1:20 (4H8) [63], anti-Bam 1:5 [36], and anti-Sxl 1:10 (M18) [85]. Primary monoclonal antibodies from Cell Signaling Technology were used at the following dilutions: anti-Phospho-Smad1/5 (Ser463/465) (41D10) 1:300 (#9516S) and anti-Phospho-Histone H3 (Ser10) Antibody 1:200 (#9701S). Anti-Wolbachia FtsZ primary polyclonal antibodies were used at a 1:500 dilution (provided by Irene Newton). Secondary antibodies were obtained from Invitrogen: Alexa Fluor 405 Goat anti-Mouse (#A31553), Alexa Fluor 488 Goat anti-Rabbit (#A12379), and Alexa Fluor 647 Goat anti-Rat (#A21247).
Confocal imaging
Oocytes were imaged on a Leica SP5 confocal microscope with a 63× objective. Optical sections were taken at the Nyquist value for the objective, every 0.38 μm, at variable magnifications, depending on the sample. Most germaria were imaged at 4× magnification and ovarioles and cysts were imaged at a 1.5× magnification. Approximately 10 μm (27 slices) were sampled from each germarium and 25 μm (65 slices) were sampled from each cyst for presentation and analysis.
PI was excited with the 514 and 543 nm lasers, and emission from 550 to 680 nm was collected. Alexa 405 was imaged with the 405 nm laser, and emission from 415 to 450 nm was collected. Alexa 488 was imaged with the 488 laser, and emission from 500 to 526 nm was collected. Alexa 647 was imaged with the 633 laser, and emission from 675 to 750 nm was collected.
Image analysis
Germaria and ovarioles were 3D reconstructed from mean projections of approximately 10 μm-thick nyquist-sampled confocal images (0.38 μm apart) in Fiji/ImageJ. Germline cyst developmental staging followed the standard conventions established by Spradling [86] and the criteria described below. We also scored and processed the confocal z-stacks for analysis as follows in the sections below.
GSC quantification
GSCs were scored by the presence of pMad and an Hts-labeled spectrosome in cells with high cytoplasmic volumes adjacent to the somatic germ cell niche and terminal filament [56]. When antibody compatibility prevented both pMad and Hts staining (e.g., with anti-pHH3 staining), only one was used for GSC identification. The spectrosome was distinguished from the fusome by its position anterior of the putative GSC nucleus and the presence of a posterior fusome that continues into a posteriorly dividing cystoblast. We calculated the number of GSCs per germarium as the average number of pMad and Hts-spectrosome-expressing cells. Non-whole increments of GSCs indicate situations where a putative GSC expresses one attribute, but not the other.
Mitotic GSC quantification
Putative GSCs were identified as described above and scored for the presence of anti-pHH3 staining. The number of pHH3-positive cystocytes in region 1 was also quantified across germaria.
Oocyte cyst tumor quantification
Using a 10× objective, we manually counted the number of nurse cells in each stage 6 to 10b cyst. Each count was repeated 3 times consistently and the cyst tallied as having less than 15, 15, or more than 15 nurse cells per cyst.
Bam and Sxl expression measured by fluorescence
We summed 27-slice nyquist-sampled z-stacks of each germarium in Fiji/ImageJ. Fluorescence intensity was measured by setting ImageJ to measure: AREA, INTEGRATED DENSITY, and MEAN GRAY VALUE. Germarium regions were delimited as in [42] using Vas expression to indicate the germline. Three representative background selections were measured for subtraction. The corrected total cell fluorescence (CTCF) was calculated for each region of the oocyte as follows: CTCF = Integrated Density–Area of selected cell * Mean fluorescence of background readings. We controlled for staining intensity within a germarium and compared relative values among germaria.
Orb expression
Anti-Orb-stained oocyte cysts were manually scored for stage and Orb oocyte-staining in confocal z-stacks ImageJ.
Transcriptomics
We collected OreR and hypomorphic mei-P26[1] flies uninfected and infected with wMel and wMel-infected nos:Gal4>UAS:mei-P26 RNAi and nos:Gal4/CyO flies for RNAseq. Flies were collected after eclosion and moved to fresh food until they were 3 to 5 days old. Ovary dissections were performed in 1× PBS, in groups of 20 to 30 flies to obtain at least 10 mg of ovary tissue for each sample. After careful removal of all non-ovary somatic tissue, ovaries were promptly moved to RNAlater at room temperature, and then transferred to −80°C within 30 min for storage. Frozen tissue was shipped on dry ice to Genewiz Azenta Life Sciences for RNA extraction, cDNA synthesis, Illumina library preparation, and Illumina sequencing. The RNAi and control samples—both nos:Gal4>UAS:mei-P26 RNAi and nos:Gal4/CyO genotypes—were processed to cDNA with T-tailed primers, recovering only host transcripts. The mei-P26[1] and OreR samples were processed to cDNA using random hexamers and ribosomal sequences were depleted with sequential eukaryotic and bacterial rRNA depletion kits (Qiagen FastSelect). Illumina dual-indexed libraries were made from these cDNAs and sequenced as 2 × 150 bp reads.
We processed and analyzed RNAseq datasets for differential expression using standard computational approaches and custom perl parsing scripts. Briefly, following demultiplexing, we trimmed adapter fragments from the RNAseq reads with Trimmomatic [87]. To generate sitewise coverage data to examine read alignments directly, we used the STAR aligner [88] and samtools [89], and plotted read depths across samples in R. We quantified transcripts by pseudoalignment with Kallisto [90]. The choice of reference transcriptome was key to recovering the maximum number of alignments: we merged the NCBI RefSeq assemblies for the wMel reference genome CDSs and RNAs from genomic (accession GCF_000008025.1) and the D. melanogaster reference genome RNAs from genomic (accession GCF_000001215.4; Release_6_plus_ISO1_MT) to obtain alignments against the full, non-redundant host-symbiont transcriptome. Simultaneous mapping to both genomes was performed to avoid cross-species mismapping [91]. This reference transcriptome was indexed at a kmer length of 31 in Kallisto (version 0.45.1) [90] and reads were pseudoaligned against this reference with the “kallisto quant” command and default parameters. Host and symbionts have distinct transcriptome distributions [92], necessitating the separation of the 2 transcriptomes prior to transcript normalization and quantification in DESeq2 [93], which we performed with a custom perl script.
We imported the subset D. melanogaster and wMel Kallisto transcriptome quantifications separately into R with Tximport [94] for DESeq2 analysis [93]. Transcript-level abundances were mapped to gene IDs to estimate gene-level normalized counts. For each transcriptome, we filtered out low-count/coverage genes across samples by requiring at least 5 samples to have a read count of 10 or more. We modeled interactions among our experimental groups as a function of genotype, infection, and the interaction between genotype and infection (~genotype + infection + genotype*infection) and performed Wald Tests to detect differential expression. Briefly, the maximum likelihood gene model coefficient for each gene’s expression count was calculated and divided by its standard error to generate a z-statistic for each gene under the full and reduced models. These z-statistics were compared to the values obtained under standard normal distribution for p-value calculations. FDR/Benjamini–Hochberg corrections were performed on these p-values to reduce the number of false positives. Normalized counts for each gene were output with the plotCounts() function in DESeq2 for plotting in R.
We manually investigated the top hits for each test for evidence of germline expression in the Fly Cell Atlas project [95] through Flybase. GO categories for differentially expressed genes returned from each DESeq2 Wald Test and tested for significance against the reference transcriptome set of IDs with ShinyGO0.77 [96].
Transcriptomic data generated in this study are available through NCBI BioProject number PRJNA1007602.
Plotting and statistical analysis
Fecundity data were plotted, analyzed, and statistics were calculated in R. Differences in lay, hatch, and offspring production rates were evaluated with the nonparametric Wilcoxon rank sum test. When this was infeasible because non-zero samples were so few (e.g., null alleles), we compared the binomially distributed categorical groups of females who laid or did-not-lay eggs with the Fisher’s Exact test. All sample sizes, means, and p-values are presented in S2 Table. Fecundity was plotted across fly ages and fitted with local polynomial regression. Single age pots were made with base R and the beeswarm [97] package and fecundity-vs-time plots were made with the ggplot2 [98] package.
Confocal micrograph fluorescence intensities were analyzed and plotted, and statistics were calculated in R. Relative fluorescence intensities between oocyte genotypes were compared with the nonparametric Wilcoxon rank sum test. Counts of GSCs, mitotic GSCs, tumorous germline cysts, and orb-specific cysts were compared with Fisher Exact tests. Plots were made with the ggplot2 [98] package.
We analyzed and plotted the dual-RNAseq results from Kallisto and DESeq2 in R. Bar and scatter plots were made with the ggplot2 package [98] and volcano plots were made with the EnhancedVolcano package (release 3.17) [99].
Supporting information
(A) Diagram of the GSC niche illustrating a subset of the genes that shift in expression in the cystoblast following GSC mitosis. (B) Model of the relative levels of protein expression during germline cyst development. (C, D) esyN interaction networks for (C) sxl and (D) bam gene products (supporting references in S1 Table).
(TIF)
(A) Genomic map and gene model for mei-P26 and the studied alleles. The insertion of a P{lacW} transposon in the first intron of mei-P26[1] impacts the RING domain. The mei-P26[mfs1] allele was generated by deletion of this insertion and 0.7–1.6 kb of DNA flanking each side of the insertion site. (B, C) mei-P26 transcript coverage and (D, E) Kallisto Kallisto normalized transcript counts for D. melanogaster mei-P26 transcripts from (B, D) mei-P26[1] and OreR wMel-infected vs. uninfected ovaries and (C, E) nos:Gal4>UAS:meiP26RNAi vs. OreR wMel-infected ovaries. The data underlying this figure can be found at NCBI, under BioProject number PRJNA1007602.
(TIF)
(A, B) Female mei-P26 RNAi, (C, D) Female mei-P26[1], and (E, F) CI assays. Both (A, C, E) egg lay rates and (B, D, F) hatch rates were consistent over time, across genetic crosses, and across fecundity crosses. A factor of 0.1 was added to the y-axis values as an offset to see zero-lay and zero-percent hatch data points.
(TIF)
(A, B) Hypomorphic mei-P26[1] and (C, D) nos:Gal4>mei-P26RNAi D. melanogaster female fecundity vs. age, fit with a local polynomial regression (dark gray bounds = 95% confidence intervals). Infection with wMel elevates offspring production across the female lifespan through increasing the number of eggs laid and the proportion of those eggs that hatch. (E–G) Male mei-P26 rescue: wMel infection produced significantly higher rates of (D) overall offspring production, broken into (F) egg lay and (F) egg hatch, in RNAi, hypomorphic, and null mei-P26[1] knockdown male flies mated to wild-type females of the same age and infection status. Wilcoxon rank sum * = p < 0.05, ** = 0.01, **** = 1e-4. The data underlying this figure can be found on Dryad at doi.org/10.7291/D1DT2C. (H, I) Homozygous hypomorphic mei-P26[1] stocks (H) infected with wMel Wolbachia or (I) uninfected. Mold growth (green food vs. tan/brown food) is uninhibited in the uninfected stocks due to embryo and larval death, which both feeds and fails to stop mold. Infection enables stable robust stock persistence because larval production outruns mold growth. Both stocks were started at the same time (see 12/28 on the label). The wMel-infected stock never needed any adults added, whereas the uninfected stock produced too few offspring and had to be supplemented at every vial flip to keep the stock going artificially. We ended this after a few months and the uninfected stock fully died out.
(TIFF)
Red = PI DNA staining, yellow = anti-Vas staining, and cyan = anti-Hts staining. Scale bars A, D, G, H, I, J–L, N, O, Q, R = 50 μm; B, C, E, F, M, P = 25 μm.
(TIF)
(A) Table containing X-chromosome nondisjunction experimental data. (B) Beeswarm boxplot of the rate of X-chromosome non-disjunction (NDJ) in each experiment. There was no significant difference between infected and uninfected mei-P26[1] females.
(TIF)
(A, B) Confocal mean projections of D. melanogaster germaria stained with antibodies against Hts and pMad. RNAi knockdown of mei-P26 does not affect GSC maintenance (Fig 2E). (C) Violin plots of the number of GSCs per germarium in 10- to 13-day-old females. As fully functional GSCs express pMad and have Hts-labeled spectrosomes, each was weighted by half and allows for partial scores. Wilcoxon rank sum * = p < 0.05, ** = 0.01. (D) F-K) Violin plots of the number of mitotic cystocytes per germarium detected by pH3 expression. (E, F) Bar-scatter plots of total (E) Sxl and (F) Bam fluorescence expression levels across the germarium, by region. (G) 1D barplots of oocyte-specific Orb staining among germline cysts, distributed across cyst developmental stages. The data underlying this figure can be found on Dryad at doi.org/10.7291/D1DT2C.
(TIF)
(A–C) Beeswarm boxplots showing that wMel infection elevates wild-type D. melanogaster fertility relative to uninfected flies of the same genotype. (A) Overall offspring production, (B) egg lay, and (C) egg hatch were variably impacted in different “wild-type” genotypes. (D) D. melanogaster eggs laid per female per day plot against female age, fit with a local polynomial regression (dark gray bounds = 95% confidence intervals). The data underlying this figure can be found on Dryad at doi.org/10.7291/D1DT2C.
(TIF)
(A–D) Beeswarm box plots of (A, C) egg hatch rate and (B, D) offspring production of uninfected and wMel-infected D. melanogaster OreR females mated to (A, B) zero-day-old and (C, D) 5-day-old wMel-infected males. (E–H) Beeswarm box plots of (E, G) egg hatch rate and (F, G) offspring production in uninfected and wRi-infected D. simulans females mated to (A, B) zero-day-old and (C, D) 5-day-old wRi-infected males. Wilcoxon rank sum * = p < 0.05, ** = 0.01, *** = 0.001, **** = 1e-4. The data underlying this figure can be found on Dryad at doi.org/10.7291/D1DT2C.
(TIF)
(A) Kallisto normalized transcript counts for D. melanogaster genes (top 15 hits (including Fig 7E) padj< = 2.0E-30; see Figs 7 and S2 for other plots). Barplots are colored by group: dark gray = wMel-infected OreR, light gray = uninfected OreR, dark pink = wMel-infected mei-P26[1], light pink = uninfected mei-P26[1]. (B–G) GO analysis for mei-P26-associated DE genes reveal an enrichment for processes involving chromatin, recombination, protein–protein interactions, and muscle cell differentiation. GO enrichment (B–D) category plots and (E–G) term interaction networks for the categories of (B, E) biological process, (C, F) cellular component, and (D, G) molecular function. The data underlying this figure can be found at NCBI, under BioProject number PRJNA1007602.
(TIF)
(A, B) Kallisto normalized transcript counts for D. melanogaster genes exhibiting (A) rescue and (B) overshoot of OreR expression levels (padj< = 0.002; see Fig 7 for other plots). Barplots are colored by group: dark gray = wMel-infected OreR, light gray = uninfected OreR, dark pink = wMel-infected mei-P26[1], light pink = uninfected mei-P26[1]. (C–G) GO analysis for infection DE genes reveal an abundance of cytoskeletal and chromatin components. GO enrichment (C–E) category plots and (F, G) term interaction networks for the categories of (C, F) biological process, (D, G) cellular component, and (E) molecular function (no network for a single term). The data underlying this figure can be found at NCBI, under BioProject number PRJNA1007602.
(TIF)
(A–C) Kallisto normalized transcript counts for D. melanogaster genes exhibiting (A) inverse, (B) undershoot, and (C) novel regulation of OreR expression levels (padj< = 0.02; see Fig 7 for other plots). Barplots are colored by group: dark gray = wMel-infected OreR, light gray = uninfected OreR, dark pink = wMel-infected mei-P26[1], light pink = uninfected mei-P26[1]. (D–G) GO analysis for infection DE genes reveal cytoskeletal and membrane factors. GO enrichment (C–E) category plots and (F, G) term interaction networks for the categories of (C, G) biological process, (D) cellular component (see Fig 8D for network), and (E, F) molecular function. The data underlying this figure can be found at NCBI, under BioProject number PRJNA1007602.
(TIF)
GO enrichment (A) category plot, (B) hierarchical clustering tree, and (C) term interaction network the biological process category. The data underlying this figure can be found at NCBI, under BioProject number PRJNA1007602.
(TIFF)
Barplots are colored by group: dark gray = wMel-infected OreR, light gray = uninfected OreR, dark pink = wMel-infected mei-P26[1], light pink = uninfected mei-P26[1]. Wald test genotype-association p-values and adjusted p-values (padj). The data underlying this figure can be found at NCBI, under BioProject number PRJNA1007602.
(TIF)
Barplots are colored by group: dark gray = wMel-infected OreR, light gray = uninfected OreR, dark pink = wMel-infected mei-P26[1], light pink = uninfected mei-P26[1]. The data underlying this figure can be found at NCBI, under BioProject number PRJNA1007602.
(TIF)
(PDF)
(PDF)
Experimental genotypes, infection statuses, and sexes are listed. The mate for each cross was OreR, of the same infection status, and of the opposite sex as the experimental fly. Males were aged 3–6 days, except for the young male CI crosses, which were aged zero days (distinguished with “-0d” and “-5d” labels). P-values <0.01 are in light green and <0.05 are in dark green for clarity.
(PDF)
Experimental genotypes, infection statuses, and sexes are listed. The mate for each cross was OreR, of the same infection status, and of the opposite sex as the experimental fly. Males were aged 3–6 days, except for the young male CI crosses, which were aged zero days (distinguished with “-0d” and “-5d” labels). P-values <0.01 are in light green and <0.05 are in dark green for clarity.
(PDF)
Experimental genotypes, infection statuses, and sexes are listed. The mate for each cross was OreR, of the same infection status, and of the opposite sex as the experimental fly. Males were aged 3–6 days, except for the young male CI crosses, which were aged zero days (distinguished with “-0d” and “-5d” labels). Sample counts (n1, n2) in parentheses are for Fisher exact tests (samples with hatched eggs vs. no hatched eggs, opposed to % hatch for samples with 20 or more eggs laid). P-values <0.01 are in light green and <0.05 are in dark green for clarity.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Normal cysts contain 16 germline-derived cells, 15 nurse cells and 1 oocyte. Cysts containing greater or less than 15 nurse cells were scored as tumorous or abnormal, respectively.
(PDF)
(PDF)
Data deposited under NCBI BioProject number PRJNA1007602.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank the UCSC Life Sciences Microscopy Center (RRID:SCR_021135) and Ben Abrams for training and the use of their microscopes. We thank Irene Newton for aliquots of anti-Wolbachia FtsZ antibodies and Manabu Ote for a stock of UASp-mRFP/CyO (1-7M) flies. We thank Russ Corbett-Detig for his comments and feedback throughout this project.
Abbreviations
- BDSC
Bloomington Drosophila Stock Center
- BMP
bone morphogenic protein
- CB
cystoblast
- CC
cystocyte
- CI
cytoplasmic incompatibility
- CTCF
corrected total cell fluorescence
- ERR
estrogen-related receptor
- GSC
germline stem cell
- NDJ
non-disjunction
- OreR
Oregon R
- pHH3
phospho-Histone H3
- PI
propidium iodide
- RISC
RNA-induced silencing complex
- RNP
ribonucleoprotein
- TA
transit-amplifying
- TomO
toxic manipulator of oogenesis
- UTR
untranslated region
Data Availability
All relevant data are contained within the paper, Supporting Information files, NCBI BioProject number PRJNA1007602 or on Dryad: https://doi.org/10.7291/D1DT2C.
Funding Statement
This work was supported by the UC Santa Cruz Chancellor’s Postdoctoral Fellowship and the NIH (R00GM135583 to SLR; R35GM139595 to WTS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Leftwich PT, Edgington MP, Chapman T. Transmission efficiency drives host–microbe associations. Proc R Soc B Biol Sci. 2020;287:20200820. doi: 10.1098/rspb.2020.0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bright M, Bulgheresi S. A complex journey: transmission of microbial symbionts. Nat Rev Microbiol. 2010;8:218–230. doi: 10.1038/nrmicro2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Utarini A, Indriani C, Ahmad RA, Tantowijoyo W, Arguni E, Ansari MR, et al. Efficacy of Wolbachia-Infected Mosquito Deployments for the Control of Dengue. N Engl J Med. 2021;384:2177–2186. doi: 10.1056/NEJMoa2030243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drown DM, Zee PC, Brandvain Y, Wade MJ. Evolution of transmission mode in obligate symbionts. Evol Ecol Res. 2013;15:43. [PMC free article] [PubMed] [Google Scholar]
- 5.Doremus MR, Hunter MS. The saboteur’s tools: Common mechanistic themes across manipulative symbioses. Adv Insect Physiol. Elsevier. 2020:317–353. doi: 10.1016/bs.aiip.2020.03.003 [DOI] [Google Scholar]
- 6.Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969 [DOI] [PubMed] [Google Scholar]
- 7.Jansen VAA, Turelli M, Godfray HCJ. Stochastic spread of Wolbachia. Proc R Soc B Biol Sci. 2008;275:2769–2776. doi: 10.1098/rspb.2008.0914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turelli M. Evolution of Incompatibility-Inducing Microbes and Their Hosts. Evolution. 1994;48:1500. doi: 10.1111/j.1558-5646.1994.tb02192.x [DOI] [PubMed] [Google Scholar]
- 9.Weeks AR, Turelli M, Harcombe WR, Reynolds KT, Hoffmann AA. From Parasite to Mutualist: Rapid Evolution of Wolbachia in Natural Populations of Drosophila. Keller L, editor. PLoS Biol. 2007;5: e114. doi: 10.1371/journal.pbio.0050114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrington LB, Lipkowitz JR, Hoffmann AA, Turelli M. A Re-Examination of Wolbachia-Induced Cytoplasmic Incompatibility in California Drosophila simulans. Cordaux R, editor. PLoS ONE. 2011;6:e22565. doi: 10.1371/journal.pone.0022565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zug R, Hammerstein P. Bad guys turned nice? A critical assessment of Wolbachia mutualisms in arthropod hosts: Wolbachia mutualisms in arthropods. Biol Rev. 2015;90:89–111. doi: 10.1111/brv.12098 [DOI] [PubMed] [Google Scholar]
- 12.Kriesner P, Hoffmann AA, Lee SF, Turelli M, Weeks AR. Rapid sequential spread of two Wolbachia variants in Drosophila simulans. Charlat S, editor. PLoS Pathog. 2013;9:e1003607. doi: 10.1371/journal.ppat.1003607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross PA, Axford JK, Yang Q, Staunton KM, Ritchie SA, Richardson KM, et al. Heatwaves cause fluctuations in wMel Wolbachia densities and frequencies in Aedes aegypti. Kohl A, editor. PLoS Negl Trop Dis. 2020;14:e0007958. doi: 10.1371/journal.pntd.0007958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann AA. Partial cytoplasmic incompatibility between two Australian populations of Drosophila melanogaster. Entomol Exp Appl. 1988;48:61–67. doi: 10.1111/j.1570-7458.1988.tb02299.x [DOI] [Google Scholar]
- 15.Hoffmann AA, Hercus M, Dagher H. Population Dynamics of the Wolbachia Infection Causing Cytoplasmic Incompatibility in Drosophila melanogaster. Genetics. 1998;11. doi: 10.1093/genetics/148.1.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds KT, Hoffmann AA. Male age, host effects and the weak expression or non-expression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genet Res. 2002:80. doi: 10.1017/s0016672302005827 [DOI] [PubMed] [Google Scholar]
- 17.Layton EM, On J, Perlmutter JI, Bordenstein SR, Shropshire JD. Paternal Grandmother Age Affects the Strength of Wolbachia -Induced Cytoplasmic Incompatibility in Drosophila melanogaster. Dubilier N, editor. MBio. 2019;10:e01879–19, /mbio/10/6/mBio.01879-19.atom. doi: 10.1128/mBio.01879-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solignac M, Vautrin D, Rousset F. Widespread occurrence of the proteobacteria Wolbachia and partial cytoplasmic incompatibility in Drosophila melanogaster. Comptes Rendus Acad Sci—Ser. 1994;III(317):461–470. [Google Scholar]
- 19.Verspoor RL, Haddrill PR. Genetic Diversity, Population Structure and Wolbachia Infection Status in a Worldwide Sample of Drosophila melanogaster and D. simulans Populations. Welch JJ, editor. PLoS ONE. 2011;6:e26318. doi: 10.1371/journal.pone.0026318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kriesner P, Conner WR, Weeks AR, Turelli M, Hoffmann AA. Persistence of a Wolbachia infection frequency cline in Drosophila melanogaster and the possible role of reproductive dormancy: PERSISTENT WOLBACHIA FREQUENCY CLINE. Evolution. 2016;70:979–997. doi: 10.1111/evo.12923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fry AJ, Palmer MR, Rand DM. Variable fitness effects of Wolbachia infection in Drosophila melanogaster. Heredity. 2004;93:379–389. doi: 10.1038/sj.hdy.6800514 [DOI] [PubMed] [Google Scholar]
- 22.Harcombe W, Hoffmann AA. Wolbachia effects in Drosophila melanogaster: in search of fitness benefits. J Invertebr Pathol. 2004;87:45–50. doi: 10.1016/j.jip.2004.07.003 [DOI] [PubMed] [Google Scholar]
- 23.Serga SV, Maistrenko OM, Matiytsiv NP, Vaiserman AM, Kozeretska IA. Effects of Wolbachia infection on fitness-related traits in Drosophila melanogaster. Symbiosis. 2021;83:163–172. doi: 10.1007/s13199-020-00743-3 [DOI] [Google Scholar]
- 24.Strunov A, Lerch S, Blanckenhorn WU, Miller WJ, Kapun M. Complex effects of environment and Wolbachia infections on the life history of Drosophila melanogaster hosts. J Evol Biol. 2022;35:788–802. doi: 10.1111/jeb.14016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serga S, Maistrenko O, Rozhok A, Mousseau T, Kozeretska I. Fecundity as one of possible factors contributing to the dominance of the wMel genotype of Wolbachia in natural populations of Drosophila melanogaster. Symbiosis. 2014;63:11–17. doi: 10.1007/s13199-014-0283-1 [DOI] [Google Scholar]
- 26.Frydman HM, Li JM, Robson DN, Wieschaus E. Somatic stem cell niche tropism in Wolbachia. Nature. 2006;441:509–512. doi: 10.1038/nature04756 [DOI] [PubMed] [Google Scholar]
- 27.Toomey ME, Panaram K, Fast EM, Beatty C, Frydman HM. Evolutionarily conserved Wolbachia-encoded factors control pattern of stem-cell niche tropism in Drosophila ovaries and favor infection. Proc Natl Acad Sci U S A. 2013;110:10788–10793. doi: 10.1073/pnas.1301524110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foray V, Perez-Jimenez MM, Fattouh N, Landmann F. Wolbachia control stem cell behavior and stimulate germline proliferation in filarial nematodes. Dev Cell. 2018;45:198–211. doi: 10.1016/j.devcel.2018.03.017 [DOI] [PubMed] [Google Scholar]
- 29.Pannebakker BA, Loppin B, Elemans CP, Humblot L, Vavre F. Parasitic inhibition of cell death facilitates symbiosis. Proc Natl Acad Sci U S A. 2007;104:213–215. doi: 10.1073/pnas.0607845104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell SL, Castillo JR. Trends in Symbiont-Induced Host Cellular Differentiation. In: Kloc M, editor. Symbiosis: Cellular, Molecular, Medical and Evolutionary Aspects. Cham: Springer International Publishing; 2020. p. 137–176. doi: 10.1007/978-3-030-51849-3_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starr DJ, Cline TW. A host–parasite interaction rescues Drosophila oogenesis defects. Nature. 2002;418:76. doi: 10.1038/nature00843 [DOI] [PubMed] [Google Scholar]
- 32.Flores HA, Bubnell JE, Aquadro CF, Barbash DA. The Drosophila bag of marbles gene interacts genetically with Wolbachia and shows female-specific effects of divergence. PLoS Genet. 2015;11:e1005453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bubnell JE, Fernandez-Begne P, Ulbing CKS, Aquadro CF. Diverse wMel variants of Wolbachia pipientis differentially rescue fertility and cytological defects of the bag of marbles partial loss of function mutation in Drosophila melanogaster. G3. 2021. [cited 2021 Oct 2]. doi: 10.1093/g3journal/jkab312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ote M, Ueyama M, Yamamoto D. Wolbachia protein TomO targets nanos mRNA and restores germ stem cells in Drosophila sex-lethal mutants. Curr Biol. 2016;26:2223–2232. doi: 10.1016/j.cub.2016.06.054 [DOI] [PubMed] [Google Scholar]
- 35.Ote M, Yamamoto D. The Wolbachia protein TomO interacts with a host RNA to induce polarization defects in Drosophila oocytes. Arch Insect Biochem Physiol. 2018;99:e21475. doi: 10.1002/arch.21475 [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Minor NT, Park JK, McKearin DM, Maines JZ. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc Natl Acad Sci U S A. 2009;106:9304–9309. doi: 10.1073/pnas.0901452106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White PM, Serbus LR, Debec A, Codina A, Bray W, Guichet A, et al. Reliance of Wolbachia on high rates of host proteolysis revealed by a genome-wide RNAi screen of Drosophila cells. Genetics. 2017:genetics.116.198903. doi: 10.1534/genetics.116.198903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Maines JZ, Tastan OY, McKearin DM, Buszczak M. Mei-P26 regulates the maintenance of ovarian germline stem cells by promoting BMP signaling. Development. 2012;139:1547–1556. doi: 10.1242/dev.077412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neumüller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, et al. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008;454:241–245. doi: 10.1038/nature07014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Page SL, McKim KS, Deneen B, Van Hook TL, Hawley RS. Genetic Studies of mei-P26 Reveal a Link Between the Processes That Control Germ Cell Proliferation in Both Sexes and Those That Control Meiotic Exchange in Drosophila. Genetics. 2000;155:1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekelsky JJ, McKim KS, Messina L, French RL, Hurley WD, Arbel T, et al. Identification of novel Drosophila meiotic genes recovered in a P-element screen. Genetics. 1999;152:529–542. doi: 10.1093/genetics/152.2.529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waghmare I, Page-McCaw A. Wnt Signaling in Stem Cell Maintenance and Differentiation in the Drosophila Germarium. Genes. 2018;9:127. doi: 10.3390/genes9030127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fast EM, Toomey ME, Panaram K, Desjardins D, Kolaczyk ED, Frydman HM. Wolbachia Enhance Drosophila Stem Cell Proliferation and Target the Germline Stem Cell Niche. Science. 2011;334:990–992. doi: 10.1126/science.1209609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aruna S, Flores HA, Barbash DA. Reduced Fertility of Drosophila melanogaster Hybrid male rescue (Hmr) Mutant Females Is Partially Complemented by Hmr Orthologs From Sibling Species. Genetics. 2009;181:1437–1450. doi: 10.1534/genetics.108.100057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaccai M, Lipshitz HD. Differential distributions of two adducin-like protein isoforms in the Drosophila ovary and early embryo. Zygote. 1996;4:159–166. doi: 10.1017/S096719940000304X [DOI] [PubMed] [Google Scholar]
- 46.Hinnant TD, Merkle JA, Ables ET. Coordinating Proliferation, Polarity, and Cell Fate in the Drosophila Female Germline. Front Cell Dev Biol. 2020;8:19. doi: 10.3389/fcell.2020.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao R, Xuan Y, Li X, Xi R. Age-related changes of germline stem cell activity, niche signaling activity and egg production in Drosophila. Aging Cell. 2008;7:344–354. doi: 10.1111/j.1474-9726.2008.00379.x [DOI] [PubMed] [Google Scholar]
- 48.Kai T, Spradling A. Differentiating germ cells can revert into functional stem cells in Drosophila melanogaster ovaries. Nature. 2004;428:564–569. doi: 10.1038/nature02436 [DOI] [PubMed] [Google Scholar]
- 49.Su TT, Sprenger F, DiGregorio PJ, Campbell SD, O’Farrell PH. Exit from mitosis in Drosophila syncytial embryos requires proteolysis and cyclin degradation, and is associated with localized dephosphorylation. Genes Dev. 1998;12:1495–1503. doi: 10.1101/gad.12.10.1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Zhang Q, Carreira-Rosario A, Maines JZ, McKearin DM, Buszczak M. Mei-P26 Cooperates with Bam, Bgcn and Sxl to Promote Early Germline Development in the Drosophila Ovary. Singh SR, editor. PLoS ONE. 2013;8:e58301. doi: 10.1371/journal.pone.0058301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chau J, Kulnane LS, Salz HK. Sex-lethal Facilitates the Transition From Germline Stem Cell to Committed Daughter Cell in the Drosophila Ovary. Genetics. 2009;182:121–132. doi: 10.1534/genetics.109.100693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tseng C-Y, Kao S-H, Wan C-L, Cho Y, Tung S-Y, Hsu H-J. Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche. Kim SK, editor. PLoS Genet. 2014;10:e1004888. doi: 10.1371/journal.pgen.1004888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chau J, Kulnane LS, Salz HK. Sex-lethal enables germline stem cell differentiation by down-regulating Nanos protein levels during Drosophila oogenesis. Proc Natl Acad Sci U S A. 2012;109:9465–9470. doi: 10.1073/pnas.1120473109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joly W, Chartier A, Rojas-Rios P, Busseau I, Simonelig M. The CCR4 Deadenylase Acts with Nanos and Pumilio in the Fine-Tuning of Mei-P26 Expression to Promote Germline Stem Cell Self-Renewal. Stem Cell Rep. 2013;1:411–424. doi: 10.1016/j.stemcr.2013.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ji S, Li C, Hu L, Liu K, Mei J, Luo Y, et al. Bam-dependent deubiquitinase complex can disrupt germ-line stem cell maintenance by targeting cyclin A. Proc Natl Acad Sci U S A. 2017;114:6316–6321. doi: 10.1073/pnas.1619188114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie T, Spradling AC. A Niche Maintaining Germ Line Stem Cells in the Drosophila Ovary. Science. 2000;290:328–330. doi: 10.1126/science.290.5490.328 [DOI] [PubMed] [Google Scholar]
- 57.Chen D, Mckearin D. Dpp Signaling Silences bam Transcription Directly to Establish Asymmetric Divisions of Germline Stem Cells. Curr Biol. 2003;13:1786–1791. doi: 10.1016/j.cub.2003.09.033 [DOI] [PubMed] [Google Scholar]
- 58.Song X, Wong MD, Kawase E, Xi R, Ding BC, McCarthy JJ, et al. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development. 2004;131:1353–1364. doi: 10.1242/dev.01026 [DOI] [PubMed] [Google Scholar]
- 59.Glasscock E. The mei-P26 Gene Encodes a RING Finger B-box Coiled-Coil-NHL Protein That Regulates Seizure Susceptibility in Drosophilia. Genetics. 2005;170:1677–1689. doi: 10.1534/genetics.105.043174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herranz H, Hong X, Pérez L, Ferreira A, Olivieri D, Cohen SM, et al. The miRNA machinery targets Mei-P26 and regulates Myc protein levels in the Drosophila wing. EMBO J. 2010;29:1688–1698. doi: 10.1038/emboj.2010.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferreira A, Boulan L, Perez L, Milán M. Mei-P26 Mediates Tissue-Specific Responses to the Brat Tumor Suppressor and the dMyc Proto-Oncogene in Drosophila. Genetics. 2014;198:249–258. doi: 10.1534/genetics.114.167502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Santoso CS, Meehan TL, Peterson JS, Cedano TM, Turlo CV, McCall K. The ABC Transporter Eato Promotes Cell Clearance in the Drosophila melanogaster Ovary. G3. 2018;8:833–843. doi: 10.1534/g3.117.300427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lantz V, Chang JS, Horabin JI, Bopp D, Schedl P. The Drosophila orb RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes Dev. 1994;8:598–613. doi: 10.1101/gad.8.5.598 [DOI] [PubMed] [Google Scholar]
- 64.Kaur R, Leigh BA, Ritchie IT, Bordenstein SR. The Cif proteins from Wolbachia prophage WO modify sperm genome integrity to establish cytoplasmic incompatibility. Malik HS, editor. PLoS Biol. 2022;20:e3001584. doi: 10.1371/journal.pbio.3001584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turelli M, Hoffmann AA. Cytoplasmic incompatibility in Drosophila simulans: dynamics and parameter estimates from natural populations. Genetics. 1995;140:1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan D, Neumüller RA, Buckner M, Ayers K, Li H, Hu Y, et al. A Regulatory Network of Drosophila Germline Stem Cell Self-Renewal. Dev Cell. 2014;28:459–473. doi: 10.1016/j.devcel.2014.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sheehan KB, Martin M, Lesser CF, Isberg RR, Newton ILG. Identification and characterization of a candidate Wolbachia pipientis type IV effector that interacts with the actin cytoskeleton. MBio. 2016;7:e00622–e00616. doi: 10.1128/mBio.00622-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Russell SL, Lemseffer N, White P, Sullivan WT. Wolbachia and host germline components compete for kinesin-mediated transport to the posterior pole of the Drosophila oocyte. McGraw EA, editor. PLoS Pathog. 2018;14:e1007216. doi: 10.1371/journal.ppat.1007216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Serbus LR, Sullivan W. A cellular basis for Wolbachia recruitment to the host germline. PLoS Pathog. 2007;3:e190. doi: 10.1371/journal.ppat.0030190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferree PM, Frydman HM, Li JM, Cao J, Wieschaus E, Sullivan W. Wolbachia utilizes host microtubules and dynein for anterior localization in the Drosophila oocyte. PLoS Pathog. 2005;1:e14. doi: 10.1371/journal.ppat.0010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.White PM, Pietri JE, Debec A, Russell SL, Patel B, Sullivan W. Mechanisms of horizontal cell-to-cell transfer of Wolbachia spp. in Drosophila melanogaster. Appl Environ Microbiol. 2017;83:e03425–e03416. doi: 10.1128/AEM.03425-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Albertson R, Casper-Lindley C, Cao J, Tram U, Sullivan W. Symmetric and asymmetric mitotic segregation patterns influence Wolbachia distribution in host somatic tissue. J Cell Sci. 2009;122:4570–4583. doi: 10.1242/jcs.054981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hackney JF, Pucci C, Naes E, Dobens L. Ras signaling modulates activity of the ecdysone receptor EcR during cell migration in the drosophila ovary. Dev Dyn. 2007;236:1213–1226. doi: 10.1002/dvdy.21140 [DOI] [PubMed] [Google Scholar]
- 74.Rastegari E, Kajal K, Tan B-S, Huang F, Chen R-H, Hsieh T-S, et al. WD40 protein Wuho controls germline homeostasis via TRIM-NHL tumor suppressor Mei-p26 in Drosophila. Development. 2020;147:dev182063. doi: 10.1242/dev.182063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rhiner C, Díaz B, Portela M, Poyatos JF, Fernández-Ruiz I, López-Gay JM, et al. Persistent competition among stem cells and their daughters in the Drosophila ovary germline niche. Development. 2009;136:995–1006. doi: 10.1242/dev.033340 [DOI] [PubMed] [Google Scholar]
- 76.Chen D, Wu C, Zhao S, Geng Q, Gao Y, Li X, et al. Three RNA Binding Proteins Form a Complex to Promote Differentiation of Germline Stem Cell Lineage in Drosophila. Fuller MT, editor. PLoS Genet. 2014;10:e1004797. doi: 10.1371/journal.pgen.1004797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ote M, Yamamoto D. Impact of Wolbachia infection on Drosophila female germline stem cells. Curr Opin Insect Sci. 2020;37:8–15. doi: 10.1016/j.cois.2019.10.001 [DOI] [PubMed] [Google Scholar]
- 78.Wang Z, Lin H. Sex-lethal is a target of Bruno-mediated translational repression in promoting the differentiation of stem cell progeny during Drosophila oogenesis. Dev Biol. 2007;302:160–168. doi: 10.1016/j.ydbio.2006.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ye Y, Godzik A. Comparative Analysis of Protein Domain Organization. Genome Res. 2004;14:343–353. doi: 10.1101/gr.1610504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Beebe NW, Pagendam D, Trewin BJ, Boomer A, Bradford M, Ford A, et al. Releasing incompatible males drives strong suppression across populations of wild and Wolbachia -carrying Aedes aegypti in Australia. Proc Natl Acad Sci U S A. 2021;118:e2106828118. doi: 10.1073/pnas.2106828118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anderson JA, Gilliland WD, Langley CH. Molecular Population Genetics and Evolution of Drosophila Meiosis Genes. Genetics. 2009;181:177–185. doi: 10.1534/genetics.108.093807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bean DM, Heimbach J, Ficorella L, Micklem G, Oliver SG, Favrin G. esyN: Network Building, Sharing and Publishing. Promponas VJ, editor. PLoS ONE. 2014;9:e106035. doi: 10.1371/journal.pone.0106035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Casper-Lindley C, Kimura S, Saxton DS, Essaw Y, Simpson I, Tan V, et al. Rapid Fluorescence-Based Screening for Wolbachia Endosymbionts in Drosophila Germ Line and Somatic Tissues. Appl Environ Microbiol. 2011;77:4788–4794. doi: 10.1128/AEM.00215-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Serbus LR, White PM, Silva JP, Rabe A, Teixeira L, Albertson R, et al. The impact of host diet on Wolbachia titer in Drosophila. PLoS Pathog. 2015;11:e1004777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bopp D, Bell LR, Cline TW, Schedl P. Developmental distribution of female-specific Sex-lethal proteins in Drosophila melanogaster. Genes Dev. 1991;5:403–415. doi: 10.1101/gad.5.3.403 [DOI] [PubMed] [Google Scholar]
- 86.Spradling A. Developmental genetics of oogenesis. Dev Drosoph Melanogaster. 1993. [Google Scholar]
- 87.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- 91.Chung M, Bruno VM, Rasko DA, Cuomo CA, Muñoz JF, Livny J, et al. Best practices on the differential expression analysis of multi-species RNA-seq. Genome Biol. 2021;22:121. doi: 10.1186/s13059-021-02337-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marsh JW, Hayward RJ, Shetty AC, Mahurkar A, Humphrys MS, Myers GSA. Bioinformatic analysis of bacteria and host cell dual RNA-sequencing experiments. Brief Bioinform. 2017. [cited 2023 Feb 10]. doi: 10.1093/bib/bbx043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. 2016. doi: 10.12688/f1000research.7563.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li H, Janssens J, De Waegeneer M, Kolluru SS, Davie K, Gardeux V, et al. Fly Cell Atlas: A single-nucleus transcriptomic atlas of the adult fruit fly. Science. 2022;375:eabk2432. doi: 10.1126/science.abk2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Valencia A, editor. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eklund A. beeswarm. 2023. Available from: https://github.com/aroneklund/beeswarm. [Google Scholar]
- 98.Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer; 2009. doi: 10.1007/978-0-387-98141-3 [DOI] [Google Scholar]
- 99.Blighe K, Rana S, Turkes E, Ostendorf B, Grioni A, Lewis M. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. Bioconductor version: Release. 2023;(3.17). doi: 10.18129/B9.bioc.EnhancedVolcano [DOI] [Google Scholar]