Abstract
Acute myeloid leukemia (AML), the most frequent leukemia in adults, is driven by recurrent somatically acquired genetic lesions in a restricted number of genes. Treatment with tyrosine kinase inhibitors has demonstrated that targeting of prevalent FMS-related receptor tyrosine kinase 3 (FLT3) gain-of-function mutations can provide significant survival benefits for patients, although the efficacy of FLT3 inhibitors in eliminating FLT3-mutated clones is variable. We identified a T cell receptor (TCR) reactive to the recurrent D835Y driver mutation in the FLT3 tyrosine kinase domain (TCRFLT3D/Y). TCRFLT3D/Y-redirected T cells selectively eliminated primary human AML cells harboring the FLT3D835Y mutation in vitro and in vivo. TCRFLT3D/Y cells rejected both CD34+ and CD34− AML in mice engrafted with primary leukemia from patients, reaching minimal residual disease-negative levels, and eliminated primary CD34+ AML leukemia-propagating cells in vivo. Thus, T cells targeting a single shared mutation can provide efficient immunotherapy toward selective elimination of clonally involved primary AML cells in vivo.
Subject terms: T-cell receptor, Cytotoxic T cells, Immunization, Acute myeloid leukaemia, Cancer
Olweus and colleagues identify a T cell receptor reactive against the recurrent D835Y driver mutation in FLT3 in acute myeloid leukemia and show that engineered T cells against this neoantigen provide efficient immunotherapy.
Main
Neoantigens represent an attractive group of targets in cancer immunotherapy, as they are tumor specific and can be recognized by T cells as foreign in the context of major histocompatibility complex (MHC)1–4. Neoantigenic burden is an important determinant of clinical success upon checkpoint inhibition5–7, and case reports have demonstrated that neoantigen-reactive T cells can mediate clinical responses8–10. This has brought hope that T cells genetically modified to express TCRs derived from neoantigen-reactive T cells could provide efficient adoptive cell therapy. Patient T cells do, however, spontaneously recognize only 1–2% of candidate neoantigens predicted to be expressed and presented on human leukocyte antigen (HLA) in solid cancer11. As the large majority is unique to the individual patient, therapeutic targeting of neoantigens generally becomes a highly personalized effort12. In addition to being resource demanding, such a strategy might not benefit patients in time. By contrast, public neoantigens derived from recurrent oncogenic mutations have the advantage that a single TCR with off-the-shelf availability could target larger patient groups. Although shared mutations resulting in neoantigens presented on frequently expressed HLA alleles are rare in solid cancer, promising results were recently shown with T cells engineered to express a TCR targeting mutant KRAS in a patient with pancreatic adenocarcinoma13 and mutant p53 in a patient with breast cancer14.
AML is, in contrast to solid tumors, characterized by recurrent driver mutations in a restricted number of genes15, many of which are screened for in routine diagnostics. The only curative therapeutic option for many patients with AML today is allogeneic hematopoietic stem cell transplantation (allo-HSCT). Allo-HSCT remains, however, associated with high relapse rates as well as transplant-related morbidity and mortality resulting from donor T cells attacking healthy recipient cells, causing severe graft-versus-host disease16, and often a suitable donor cannot be identified. This has prompted development of alternative cellular therapies17,18. Successful targeting of the lymphoid-specific molecule CD19 in chimeric antigen receptor (CAR) T cell therapy of acute lymphocytic leukemia, has led to attempts at also directing CARs to myeloid cell surface antigens, including CD33 (ref. 19), CD123 (ref. 20) and FLT3 (ref. 21), overexpressed on AML cells. However, these molecules are also highly expressed on normal myeloid progenitor cells and even on hematopoietic stem cells, representing a major challenge22–24. Thus, CAR T cell therapies targeting these molecules are associated with toxicities and a need for transplantation to rescue normal hematopoiesis after a short period of treatment (for example, NCT03126864) (ref. 25). It is also unclear whether all AML-propagating cells express these antigens and therefore whether targeting has curative potential26.
Cytotoxic cells modified to express TCRs provide an opportunity to also target intracellular antigens with restricted expression in normal tissues, such as the specialized DNA polymerase terminal deoxynucleotidyl transferase (TdT)27, dramatically broadening the repertoire of potential targets. To this end, TCRs recognizing Wilms tumor 1 (WT1) were recently shown to reduce relapses in patients with AML after transplantation28. A TCR recognizing the recurrent neoantigen nucleophosmine 1 (NPM1) presented on HLA-A*02:01 (ref. 29,30) and T cell clones targeting the fusion core-binding factor subunit β (CBFB)–myosin heavy chain 11 (MYH11) in the context of HLA-B*40:01 (ref. 31) showed some efficacy in patient-derived xenograft (PDX) mouse models with low AML engraftment. However, no neoantigen-specific TCR has yet been shown to efficiently target primary human AML in in vivo models in which high and increasing leukemic burden is observed.
Recurrent driver mutations in FLT3 occur in approximately one-third of patients with de novo AML as internal tandem duplications (ITD) in the juxtamembrane domain or point mutations in the activation loop of the tyrosine kinase domain (Fig. 1a), which are screened for in routine diagnostics of AML32. Although, in most cases, FLT3 mutations are secondary events in leukemogenesis33, they are associated with accelerated clonal expansion and disease progression, and treatment with the tyrosine kinase inhibitor (TKI) midostaurin in patients with FLT3 mutations receiving standard induction chemotherapy does significantly increase long-term survival34. This indicates that FLT3-mutated AML clones have a survival advantage during standard therapy, expanding to drive relapse unless effectively eradicated, but the efficacy of TKIs in eliminating FLT3-mutated clones remains variable35–37.
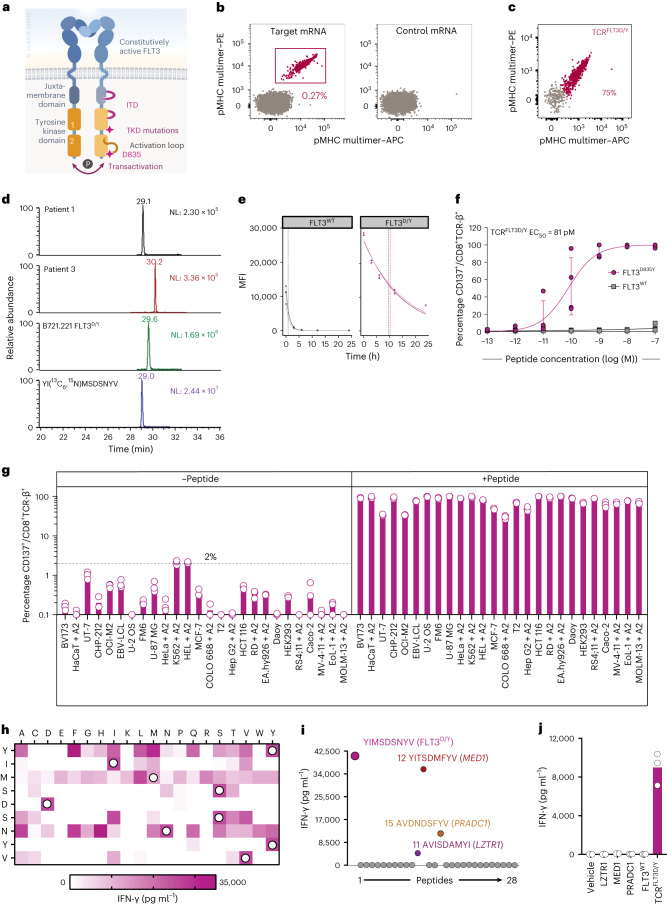
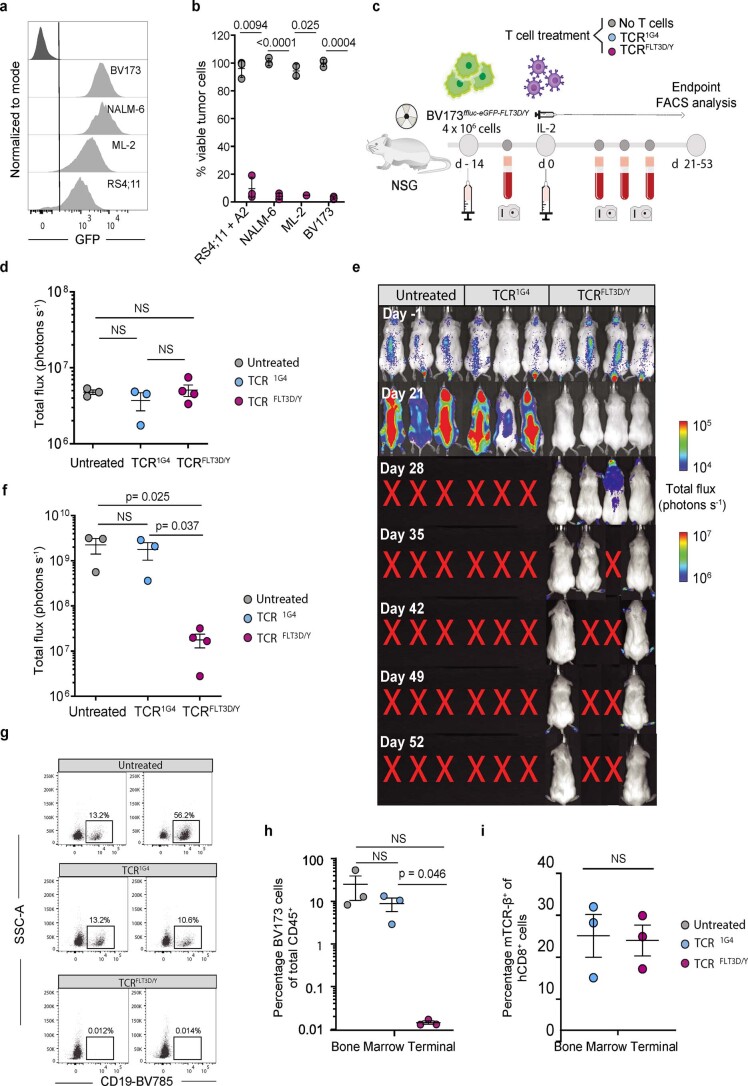
Fig. 1. TCRFLT3D/Y cells specifically recognize mutated peptide with high sensitivity in an HLA-A2-restricted manner and do not show off-target reactivity.
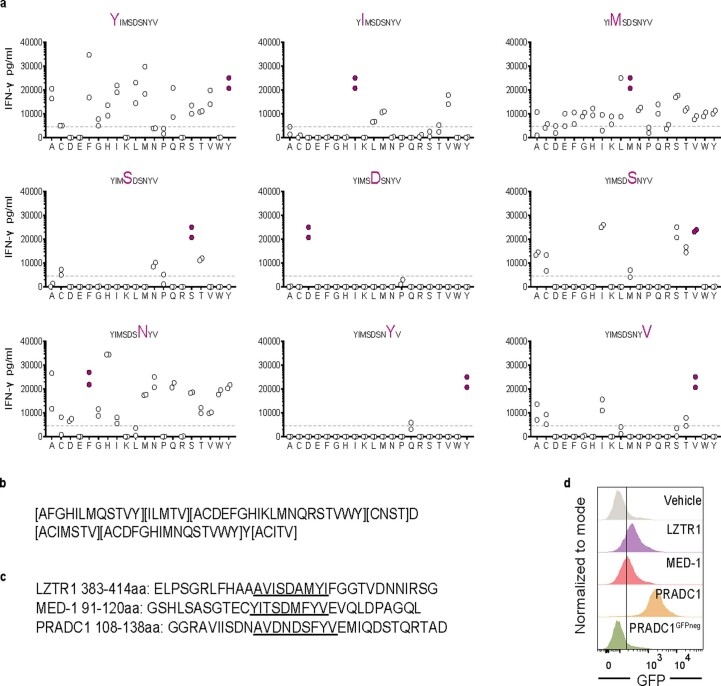
a, Schematic illustration of FLT3. TKD, tyrosine kinase domain. b, Naive CD8+ T cells co-cultured with autologous HLA-A2+ mRNA-transfected moDCs stained with FLT3D/Y pMHC multimers. c, CD8+ T cells transduced to express TCRFLT3D/Y stained with FLT3D/Y pMHC multimers (Gating strategy in Extended Data Fig. 3b). d, Parallel reaction-monitoring analysis, targeting the FLT3D835Y peptide (m/z = 1,091.43891+) in primary AML cells from two patient samples and the B721.221 cell line transduced to express FLT3D835Y and HLA-A2. NL = normalization level. e, Off-rates for FLT3WT or FLT3D/Y peptide binding to HLA-A2 measured by flow cytometry. Vertical lines indicate calculated half-lives in each experiment. Dots represent mean fluorescence intensity (MFI) values of intact pMHC complexes on fluorescent particles at the indicated time points (h) (one replicate per experiment, n = 3 independent experiments). f, Activation of TCRFLT3D/Y cells (CD137+) co-incubated with peptide-pulsed K562 cells. Data points are from n = 4 donors transduced to express TCR in n = 3 independent experiments, with each circle representing the mean of three technical replicates per donor, shown as mean ± s.e.m. g, Activation of CD8+ TCRFLT3D/Y cells co-incubated with HLA-A2+ cell lines with or without FLT3D/Y peptide. Results are from one experiment representative of n = 4 (BV173, CHP-212, EBV-LCL, K562, Daoy, RS4;11), n = 3 (HaCaT, U-2 OS, FM6, U-87 MG, HeLa, MV-4-11, EoL-1, MOLM-13) or, for the remaining cell lines, n = 2 independent experiments using different T cell donors; data points represent n = 3 technical replicates. The suffix + A2 denotes that cell lines were transduced with HLA-A*02:01, whereas remaining cell lines naturally express it. Connecting lines in f and bars in g show mean. The dashed line in g shows the highest level of activation by cell lines alone. h–j, IFN-γ produced by TCRFLT3D/Y cells co-incubated with K562 cells loaded with peptides from the mimotope library (h) or pulsed with the peptides that were predicted as potentially cross-reactive from the in silico search (i) or transfected with mRNA constructs encoding 30–32-mer peptides with the candidate cross-reactive peptide inducing reactivity (shown in i) in the middle, flanked by its naturally occurring sequence, or transfected with mRNA encoding the FLT3D/Y epitope or FLT3WT (j). White circles in h, amino acids of the FLT3D/Y peptide. Positive reaction for IFN-γ, 5,000–35,000 pg ml−1. LZTR1, leucine zipper-like post-translational regulator 1; MED1, mediator complex subunit 1; PRADC1, protease-associated domain-containing protein 1. Data in h–j are from one of n = 2 independent experiments, and individual data points represent one (h,i) or three (j) technical replicates.
Because of varying length, ITDs do not encode shared neoantigens. Mutations in the D835 position of FLT3 represent the most frequent point mutations in FLT3 (7–10% of patients with AML), with aspartic acid (D) to tyrosine (Y) being by far the most common amino acid substitution, although other substitutions can also occur32. The effect of TKIs on the D835Y mutation (FLT3D835Y) is limited by this mutation conferring primary and secondary resistance to first- and second-generation type II TKIs35–37.
We recently demonstrated that healthy donor T cells provide a rich source of neoantigen-reactive TCRs38,39. Here, we used this technology to identify a TCR reactive to a peptide from FLT3D835Y. The TCR (TCRFLT3D/Y) is restricted by HLA-A*02:01 (hereafter HLA-A2), expressed in approximately 50% of people of European, Middle Eastern or North African ancestry. We demonstrate that T cells redirected with TCRFLT3D/Y (TCRFLT3D/Y cells) specifically and efficiently eliminate primary human AML cells harboring FLT3D835Y in vitro and in vivo, while sparing cells expressing wild-type (WT) FLT3.
Results
Inducing FLT3D835Y-reactive T cells from healthy donor T cells
Naive T cells from a large number of HLA-A2-positive healthy blood donors (n = 16) were co-cultured with autologous monocyte-derived dendritic cells (moDCs) electroporated with mRNA encoding a 9-mer and a 10-mer peptide with the FLT3D835Y mutation in position 1, predicted to be strong binders to HLA-A2 (Extended Data Fig. 1a). The sequence was flanked by 9–11 amino acids at the N-terminal and C-terminal end (target mRNA, encoding KICDFGLARYIMSDSNYVVRGNVRLARLP) or control mRNA. After 10 d of co-culture, cells were stained with dual-color peptide–MHC (pMHC) multimers complexed with the nonameric or decameric peptide in which the FLT3D/Y mutation was in position 1. Multimer+CD8+T cells reactive with the 9-mer were detected in only one donor and only in the culture primed with the target mRNA and were sorted. Only one functional TCR sequence (TCRFLT3D/Y) was identified from two clones and from 55 sorted, sequenced single cells (Fig. 1b and Extended Data Fig. 1b,c).
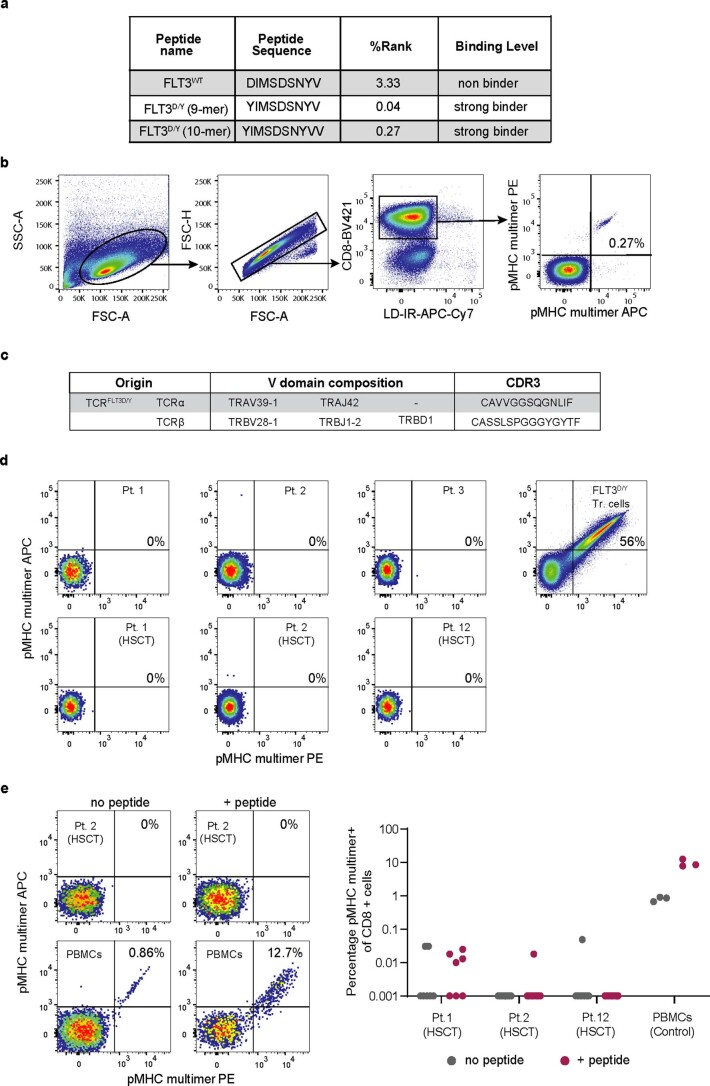
Extended Data Fig. 1. T cells reactive to FLT3D/Y HLA-A2 can be induced by culture of naïve healthy donor T cells but are not identified among memory T cells from AML patients in diagnostic samples or following HSCT.
(a) Predicted binding affinity of FLT3D/Y and FLT3WT peptides using the NetMHC-4.0 algorithm. Peptides with affinity <50 nM (EL%Rank <0.500) classify as strong binders, 50 nM<affinity<500 nM (EL%Rank <2.000) as weak binders and affinity >500 nM as non- binders85. (b) Gating strategy to identify CD8+ T cells staining as double positive events for pMHC multimers (APC- and PE-conjugated) complexed with the FLT3D/Y peptide. The three left plots show gates used for FSC/SSC, singlets and Live/Dead Fixable Near- IRneg/CD8+ events. The plot to the right shows multimer positive events in co-culture of naïve healthy donor CD8+ T cells with autologous moDCs transfected with target mRNA. (c) The TCRFLT3D/Y sequence. (d) Staining of samples from AML patients taken at point of diagnosis (top row, left three panels) and post allo-HSCT (bottom row) with pMHC multimers. TCRFLT3D/Y transduced T cells were used as a positive control for multimer staining (top right). (e) Staining of samples obtained from patient 2 after allo-HSCT following 5 days of in vitro expansion in absence (left) and presence (right) of the FLT3D/Y peptide (top panels). As positive control for in vitro expansion of memory T cells, TCRFLT3D/Y T cells were spiked into autologous healthy PBMCs under the same conditions (bottom panels). Inset numbers represent the percentage of pMHC multimer+ cells out of CD8+ cells. The graph to the right shows data for all three patients and positive controls. Data shown are from one experiment with each dot representing a technical replicate (n = 7 for pt 1, n = 12 for pt 2, n = 8 for pt 12 and n = 3 for PBMC healthy donor).
By contrast, memory cytotoxic T lymphocytes reactive to FLT3D835Y HLA-A2 were not identified among peripheral blood (PB) mononuclear cells (PBMCs) from diagnostic AML samples (n = 3) or from samples after allo-HSCT with HLA-matched donors (n = 3), directly after thawing or following a short 5-d in vitro culture in the presence of 100 nM of the FLT3D/Y mutant peptide to expand memory T cells (Extended Data Fig. 1d,e).
TCRFLT3D/Y recognizes mutant peptide with high peptide–HLA stability
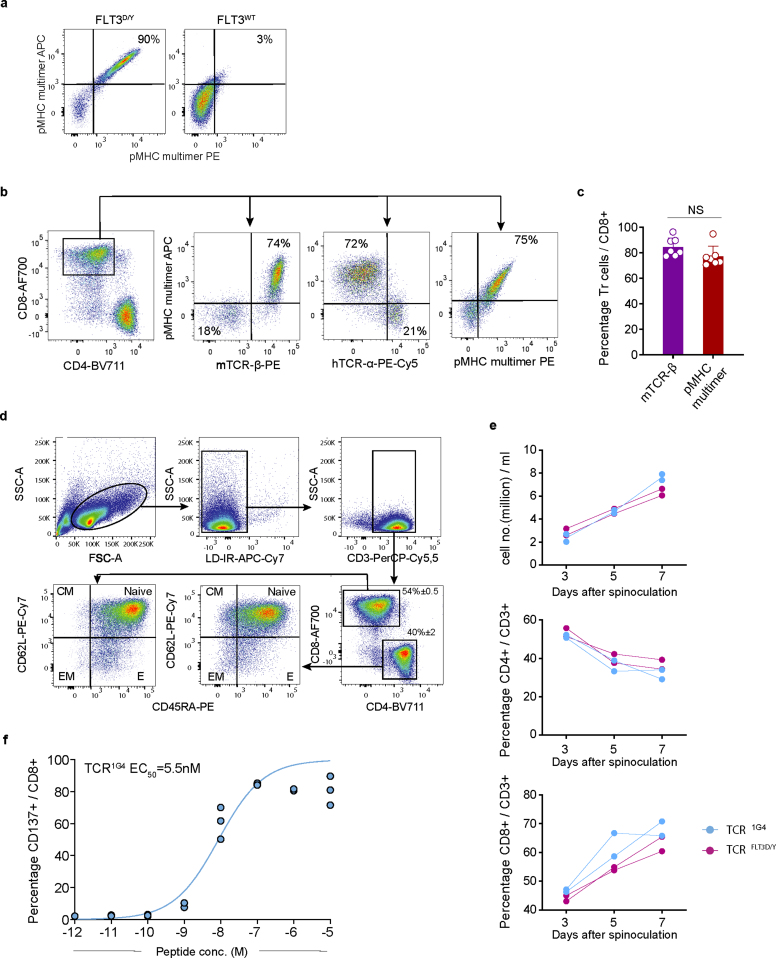
The TCRFLT3D/Y sequence was efficiently expressed in third-party PB T cells by retroviral transduction (Fig. 1c). Binding to HLA-A2 as well as endogenous processing and presentation of the peptide was demonstrated by immunopeptidomics and targeted mass spectrometry analysis of primary AML cells from two patients, using a monoallelic cell line overexpressing the FLT3D835Y mutation as a positive control (Fig. 1d, with ion chromatograms extracted using the precursor → b2, b3, b4, b5, b6, b7 and b8 fragment ion transactions, and Extended Data Fig. 2). We previously demonstrated that peptide–HLA stability is strongly predictive of neoantigen immunogenicity38, and the complex of HLA-A2 and the mutated peptide had an almost tenfold longer half-life than that of the WT peptide complexed with HLA-A2 (mean of 9.9 versus 0.83 h, n = 3) (Fig. 1e). In agreement with this, TCRFLT3D/Y cells stained brightly with pMHC multimers presenting mutant, but not WT, peptide (Extended Data Fig. 3a). A similar fraction of TCRFLT3D/Y-redirected CD8+ T cells stained positively with FLT3D/Y pMHC multimers and anti-mouse TCR-β–PE (reactive to the mouse constant region introduced into the TCR40) and negatively with anti-human TCR-α, indicating preferential pairing of the introduced TCR-α and TCR-β chains and suppression of the endogenous TCR (Extended Data Fig. 3b,c). Both CD8+ and CD4+ T cells exhibited a predominantly naive profile and expanded similarly to T cells transduced to express a control TCR in vitro, the clinically applied NY-ESO1-specific 1G4 (TCR1G4) (ref. 41), indicating lack of fratricide (Extended Data Fig. 3d,e).
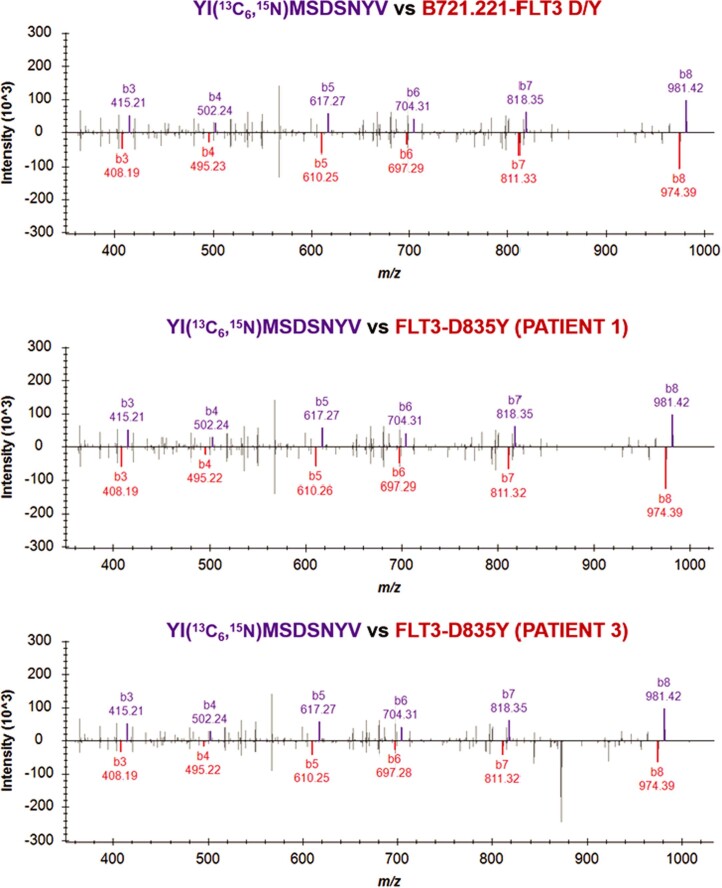
Extended Data Fig. 2. Tandem mass spectra of endogenous vs isotopically labelled peptide.
Mirror image plots comparing the fragmentation spectra of endogenous FLT3D/Y peptide to its isotopically labelled counterpart YI(13C6,15 N)MSDSNYV. Spectra obtained from immunoprecipitated HLA derived from the B721.221 cell line retrovirally transduced with a minigene encoding the D835Y mutation, and from patient 1 and patient 3, are displayed.
Extended Data Fig. 3. TCRFLT3D/Y cells maintain a predominantly naïve phenotype following expansion.
(a) Staining of TCRFLT3D/Y cells with pMHC multimers complexed with either the FLT3D/Y or the FLT3WT peptide (each multimer conjugated to both APC and PE, gating strategy shown in Extended Data Fig. 1b. Data shown is from one representative donor out of two stained in one experiment. (b) Gating strategy for TCRFLT3D/Y cells. Panels show gating on: CD8+ cells labeled with the APC pMHC multimer and antibodies reactive to mouse TCR-β or human TCR-α (middle panels), and CD8+ cells staining positively for APC and PE-labeled pMHC multimers complexed with the FLT3D/Y peptide (right plot). (c) Percentage of mTCR-β+ cells or pMHC multimer+ cells among CD8+ cells following transduction with the TCRFLT3D/Y. Each data point represents a different HLA-A2pos donor (n = 7 donors from 3 independent experiments). Data are analyzed by unpaired, two-tailed Student’s t-test and p < 0.05 was considered statistically significant. (d) Gating strategy to identify differentiation stages of TCR-transduced T cells three days after spinoculation. Top panels show gating on FSC/SSChi, Live/Dead Fixable Near-IRneg, CD3+ events. Bottom panels show gating on CD4+ or CD8+ populations and subsequent identification of naïve, central memory (CM), effector (E) and effector memory (EM) cells as defined by expression of CD45RA and CD62L analyzed in two donors in one experiment. (e) Expansion of HLA-A2pos PB T cells from two donors transduced in parallel with the TCRFLT3D/Y or TCR1G4 following indicated days after retroviral transduction. Data are from one experiment and dots represent one technical replicate for each HLA-A2pos PB T cell donor. (f) Activation of TCR1G4 cells measured as upregulation of CD137 after co-incubation with peptide-pulsed (NY-ESO-1 peptide SLLMWITQC) BV173 cells (K562 cells were not used, in contrast to Fig. 1f, as they express NY-ESO-1). EC50 = half maximal effective concentration (linear curve fitting). Data shown are from one experiment with one (out of two) T cell donors with individual data points representing technical replicates (n = 3).
TCRFLT3D/Y cells show high peptide sensitivity and specificity
TCRFLT3D/Y cells recognized K562 target cells transduced to express HLA-A2 and pulsed with picomolar concentrations of the mutant peptide (half-maximal effective concentration (EC50) = 81 pM) and displayed no reactivity against the corresponding WT peptide (Fig. 1f). By comparison, TCR1G4 cells recognized the cognate NY-ESO1 peptide with an EC50 of 5.5 nM (Extended Data Fig. 3f), in agreement with previous data showing an EC50 of 7.7 nM42. Furthermore, TCRFLT3D/Y cells showed no or negligible reactivity to a panel of 26 HLA-A2+ cell lines of different tissue origins, unless preloaded with mutant peptide, suggesting a high degree of peptide and HLA specificity (Fig. 1g).
We next mapped the fine specificity of TCRFLT3D/Y. TCRFLT3D/Y cells were screened for interferon (IFN)-γ production in response to target cells loaded with each peptide in a library of 161 peptides representing single-amino acid-substituted variants of the cognate peptide (Fig. 1h and Extended Data Fig. 4a). Peptide motifs harboring any ‘permitted’ alternative amino acids in each position were queried in the human proteome databases UniProtKB/Swiss-Prot by employing the ScanProsite tool (https://prosite.expasy.org/scanprosite/) (Extended Data Fig. 4b), identifying 28 additional 9-mers in the human proteome potentially recognized by TCRFLT3D/Y cells (Supplementary Table 1). However, only three of these peptides (peptide 11, AVISDAMYI, derived from LZTR1; peptide 12, YITSDMFYV, derived from MED1; and peptide 15, AVDNDSFYV, derived from PRADC1) activated TCRFLT3D/Y cells (Fig. 1i). The three genes encoding proteins harboring the potentially cross-reactive peptide sequences are ubiquitously expressed according to the HPA database (https://www.proteinatlas.org/). A lack of reactivity observed in response to the panel of 26 HLA-A2+ cell lines (Fig. 1g) therefore suggested that the peptides are not processed and presented. To further exclude this, we generated mRNA constructs encoding 30-mer peptides with the cross-reactive peptide in the middle flanked by the natural respective protein sequences and a green fluorescent protein (GFP) reporter (Extended Data Fig. 4c). Co-cultures demonstrated that TCRFLT3D/Y cells did not react to K562 HLA-A2+ target cells electroporated with the mRNA constructs encoding peptides derived from LZTR1, MED1 or PRADC1 (Fig. 1j and Extended Data Fig. 4d), indicating that these peptides are not naturally processed and presented on HLA-A2. In agreement with this, none of the 28 candidate peptides were found in the HLA ligand database43.
Extended Data Fig. 4. Mapping of peptide specificity does not reveal unintended targets to which the TCRFLT3D/Y cells cross-react.
(a) Graphs depicting IFN-γ response of TCRFLT3D/Y cells to K562 cells loaded with individual peptides from mimotope library containing a total of 161 nine-mers for the FLT3D/Y peptide, at a concentration of 10−8 M. Purple dots in each graph represent response to the FLT3D/Y peptide. Substituted amino acid in the original peptide is highlighted. IFN-γ concentration range for positive reactions was 5000–35000 pg/mL (cut-off indicated by horizontal lines). Graphs show results from two independent experiments that were performed, one technical replicate (dot) per peptide and experiment. (b) Peptide reactivity motifs for FLT3D/Y that were queried in the ScanProsite search tool against human proteome databases. Amino acids in square brackets [] indicate alternatives that are allowed for the given position in the peptide motifs. (c) mRNA-encoded amino acid sequences(30-32mers) derived from the sequence of the human proteins LZTR1, MED1 and PRADC1, with the candidate cross-reactive peptide identified in the ScanProsite database underlined. (d) Expression of mRNA encoding LZTR1, MED1 and PRADC1 sequences following electroporation of HLA-A2pos K562 cells as measured by GFP-reporter fluorescence using flow cytometry.
TCRFLT3D/Y cells kill primary FLT3D835Y AML cells in vitro
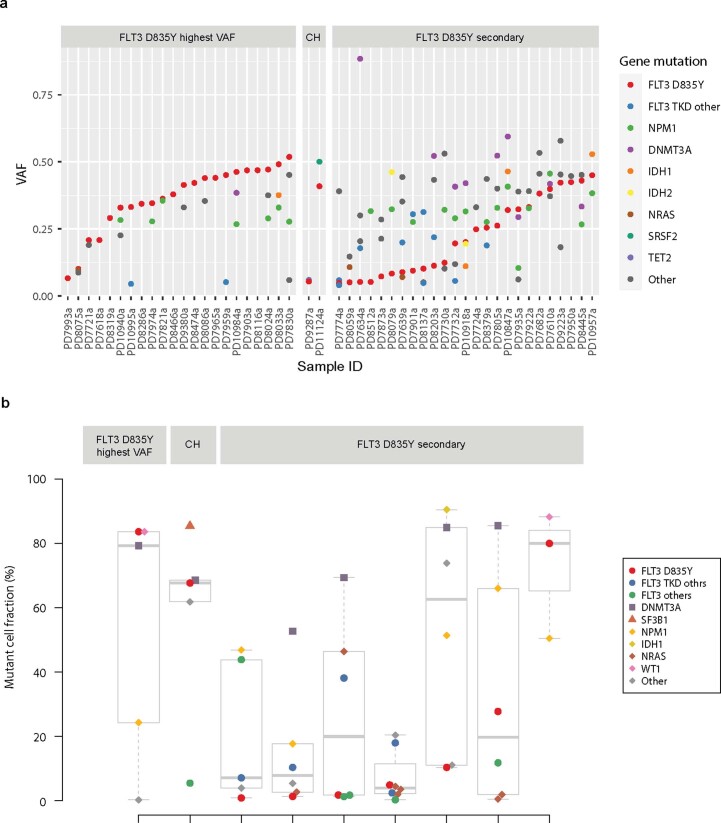
Reanalysis of publicly available DNA sequencing data of 49 patients with AML and the FLT3D835Y mutation15 demonstrated that the FLT3D835Y-mutated clone is frequently dominant (Extended Data Fig. 5a). The FLT3D835Y mutation had the highest variant allele frequency (VAF) among identified recurrent driver single-nucleotide variants (SNVs) and indels in 22 patients (45%). In two additional patients, it was only preceded by a recurrent SRSF2 and TET2 mutation, known to occur in normal individuals with clonal hematopoiesis (CH) and requiring additional driver mutations to transform to AML44. Similarly, reanalysis of single-cell DNA-sequenced AMLs showed in two of nine patients with the FLT3D835Y mutation that it had the highest VAF or was secondary only to CH-associated mutations45 (Extended Data Fig. 5b). In sum, this analysis of 58 FLT3D835Y-mutated patients with AML indicates that FLT3D835 is frequently clonal and that it might constitute an AML-initiating or transforming mutation.
Extended Data Fig. 5. Re-analysis of published mutation data for 58 FLT3 D835Y positive AML patient samples shows high VAF for FLT3 D835Y in a large fraction of patients.
(a) Mutation data (SNVs and indels) from AML patients reported in Papaemmanuil et al, 2016 NEJM were downloaded from https://www.cbioportal.org/. VAF was estimated from reported alternative allele reads divided by sequencing depth for the position. Patients harboring a FLT3 D835Y mutation were selected for in-depth analysis and displayed here. Patients are sorted from right to left within each subsection in descending order of FLT3 D835Y VAF. The following genes were considered as initiating events in clonal hematopoiesis (CH): DNMT3A, TET2, ASXL1, PPM1D, JAK2, SF3B1, SRSF2, TP53, GNAS and GNB1. (b) Mutation data from FLT3 D835Y positive AML patients (n = 9) reported in Morita et al, 2020 Nat Com were downloaded. The fraction of cells with mutations (mutant cell fraction) in each AML patient with mutations are plotted in indicated colors. Patients were categorized into 3 groups; FLT3 D835Y largest, only preceded by mutations in DNMT3A, TET2, and/or ASXL1 (DTA) and/or splicing factor mutations (SF3B1, SRAF2, and U2AF1) which are closely related with clonal hematopoiesis ‘CH’, and the rest of cases with FLT3 D835Y mutation according to the cell fraction of mutations. Interquartile range (IQR) and median values are shown. The dashed lines indicate 1.5xIQR and the dots indicate outliers.
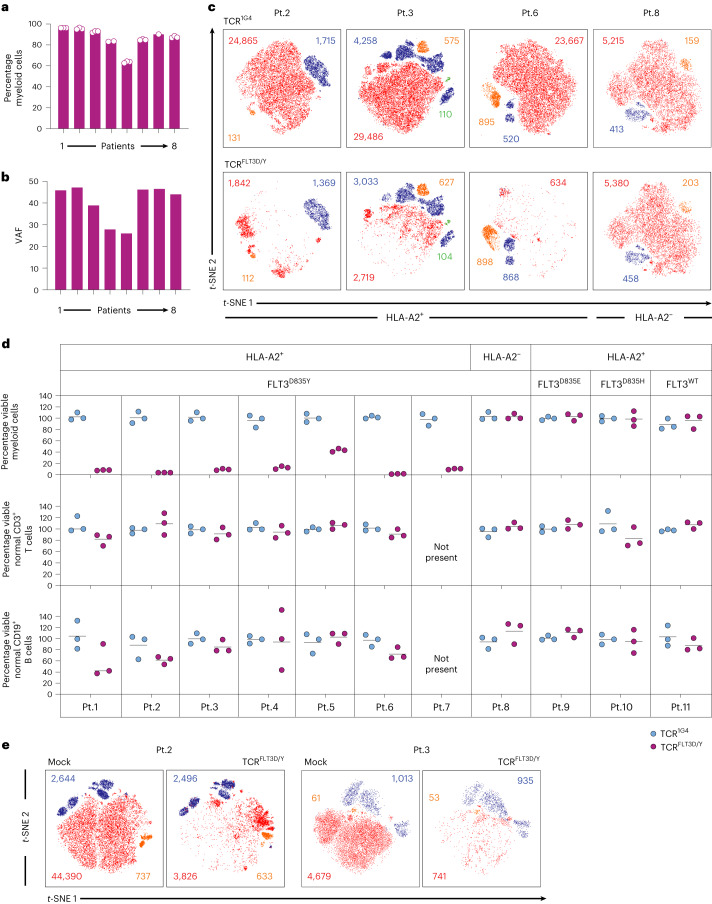
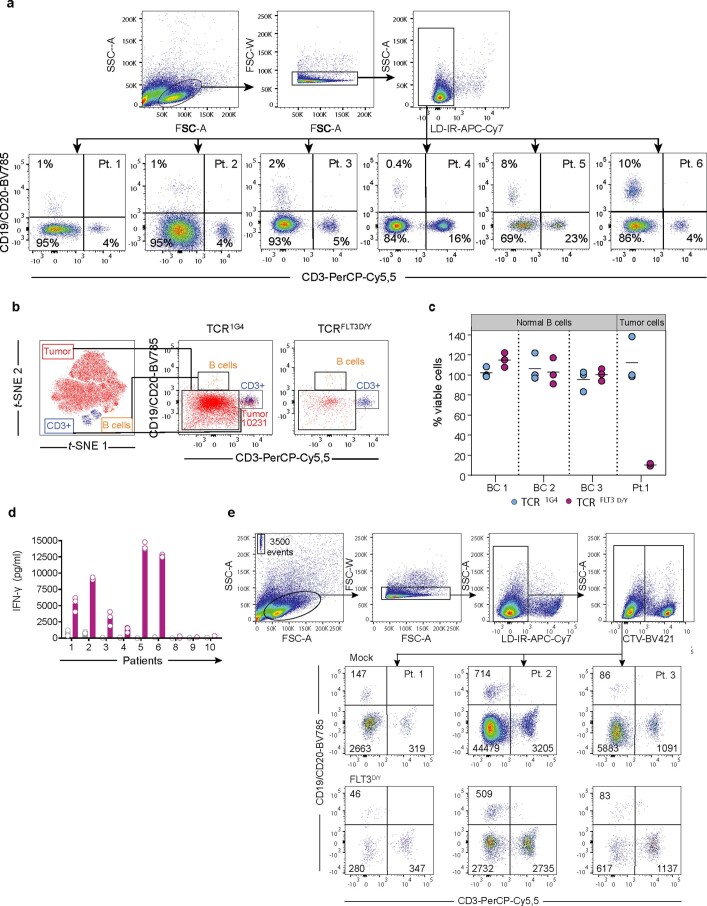
We next explored the specificity and efficacy with which TCRFLT3D/Y cells killed AML cells from 11 patients (Supplementary Table 2). DNA sequencing of mononuclear samples dominated by myeloid cells (Fig. 2a and Extended Data Fig. 6a) showed high FLT3D835Y clonal involvement in the eight FLT3D835Y+ patients (Fig. 2b and Supplementary Table 3). TCRFLT3D/Y cells killed myeloid cells from patients 1–7 with AML effectively at effector:target (E:T) ratios as low as 1:2 (mean, 87%; range, 53.3–98.7%; Fig. 2c,d). CD3+ T cells and CD19+CD20+ B cells were not significantly affected (Fig. 2c,d and Extended Data Fig. 6b), as expected in view of low clonal involvement46. However, B cell counts were very low (Extended Data Fig. 6a,b), and we therefore also investigated the effect of TCRFLT3D/Y cells on isolated B cells from healthy donors, which demonstrated a lack of killing (Extended Data Fig. 6c). The HLA restriction of TCRFLT3D/Y was confirmed by lack of killing of myeloid HLA-A2− FLT3D835Y patient cells (patient 8; Fig. 2c,d). Specificity for the D835Y substitution of FLT3 was demonstrated by lack of recognition of AML samples with alternative amino acid substitutions in the D835 position (patients 9 and 10) or expressing FLT3WT (patient 11) (Fig. 2d and Supplementary Table 2). Robust and specific IFN-γ production was observed upon co-incubation of TCRFLT3D/Y cells with HLA-A2+ FLT3D835Y patient cells (patients 1–6; Extended Data Fig. 6d). Finally, TCRFLT3D/Y cells derived from patients with AML killed autologous leukemia cells with similar efficacy and selectivity as third-party T cells, mimicking the clinical setting (Fig. 2e and Extended Data Fig. 6e). In most cases, two to four experiments were performed per patient sample, with only one experiment performed due to a limited amount of material in a few instances, as described in the figure legends.
Fig. 2. TCRFLT3D/Y cells efficiently kill primary AML cells harboring the FLT3D835Y mutation in vitro but spare normal lymphoid cells.
a, Percentage myeloid cells of live leukocytes for patients (Pt.) 1–8 with AML; gating strategy is shown in Extended Data Fig. 6a. Dots represents technical replicates from one representative experiment as described in d. b, PB or BM FLT3D/Y VAF for patients 1–8 as determined by next-generation sequencing. c, Representative t-distributed stochastic neighbor embedding (t-SNE) plots showing live primary myeloid cells (CD3−CD19−CD20− events) in red, T cells (CD3+) in blue, B cells (CD19+CD20+) in orange and normal CD34+lin− progenitor cells in green from n = 3 representative HLA-A2+ FLT3D/Y patients (patients 2, 3 and 6) with AML and one HLA-A2− FLT3D/Y patient (patient 8) following 72 h of co-culture with TCR1G4 (negative control, top) or TCRFLT3D/Y cells (E:T ratio, 1:2) as quantified by flow cytometry. Cells transduced to express TCR were excluded from analysis as CellTrace Violet (CTV)-positive events. d, Diagnostic samples from 11 patients with AML and the FLT3D/Y (patients 1–8), FLT3D/E (patient 9) or FLT3D/H (patient 10) mutation or FLT3WT (patient 11) (all HLA-A2+ except patient 8), analyzed as described in c. Each dot represents the fraction of live myeloid cells, B cells or T cells after co-culture with TCRFLT3D/Y cells (purple) in percent mean of the corresponding numbers in cultures treated with TCR1G4 cells (blue). Data points represent n = 3 technical replicates, and horizontal lines show means. Data shown are from one experiment representative of two to four experiments performed for each patient sample (n = 1 only for patient 7). e, t-SNE plots of PB diagnostic samples from patients 2 and 3 with AML showing live myeloid, T and B cells (color coded as in Fig. 2c) after 72 h of co-culture with autologous T cells transduced to express TCRFLT3D/Y or the mock control. Inset numbers in c,e denote absolute event counts of the indicated cell populations. The gating strategy is shown in Extended Data Fig. 6e.
Extended Data Fig. 6. TCRFLT3D/Y cells are activated by, and efficiently kill, primary AML cells expressing FLT3D/Y.
(a) Flow cytometry plots showing gating strategy to identify cell subsets in AML patient samples from PB (patient 2-6) or BM (patient 1). Cells are gated on FSC/SSChi, single, Live/Dead Fixable Near-IR events (top row), showing the fractions of CD3+ T cells, CD19+CD20+ B cells and myeloid cells, defined as CD3−CD19−CD20− events in the bottom row. (b) Populations were overlaid on a t- SNE plot (patient 1) with designated colors as indicated. Inset numbers show event counts for myeloid cells after co-culture with TCR1G4 or TCRFLT3D/Y cells, quantified as shown in e. (c) Quantification of normal CD19+ B cells isolated from n = 3 healthy blood donors (Buffy coat (BC) 1 - BC 3) and tumor cells from patient 1 (positive control) after performing the flow cytometry-based cytotoxicity assay for 72 h. Data points represent n = 3 technical replicates from one experiment and horizontal lines show mean. (d) Bar graph showing IFN-γ response of Mock (gray) and TCRFLT3D/Y cells (purple) after 24 h co-culture with HLA-A2pos patient cells expressing the FLT3 D835Y (Pt.1-6), FLT3 D835E (Pt.9) or FLT3 D835H (Pt. 10), or Pt.8 cells expressing FLT3 D835Y but being HLA-A2neg. Data points represent n = 3 technical replicates and bars show mean. Data shown are from one experiment representative of at least two performed for each patient sample. (e) Gating strategy for flow cytometry cytotoxicity assay to quantify viable cells in subpopulations from AML patients 1-3 after 72 h of co-culture with autologous T cells either expressing TCRFLT3D/Y (bottom row) or mock-transduced. Transduced T cells were labeled with cell-trace violet (shown in upper right plot) prior to co-culture with AML cells to distinguish them from T cells in the AML samples. Patient cell subsets are gated on FSC/SSC, singlets, Live/Dead Fixable Near-IRneg, CTVneg events. Numbers indicate absolute counts for CD3+, CD19+/CD20+ and myeloid cells after co-culture, as determined by addition of fluorescent beads (10,000) into each well and where 3,500 beads were acquired for flow cytometry analysis (shown in upper left plot).
TCRFLT3D/Y cells efficiently target human leukemia cells in vivo
We initially investigated whether TCRFLT3D/Y cells recognized endogenously presented antigen in a small experiment using an in vivo xenograft mouse model engrafted with a leukemia cell line. Because FLT3D835Y leukemic cell lines are not commercially available, we introduced the mutation into different leukemia cell lines and demonstrated that TCRFLT3D/Y cells killed >95% of leukemic cells in 24 h at low E:T ratios (1:2) (Extended Data Fig. 7a,b). BV173 FLT3D835Y-expressing cells (cell line origin, B cell precursor leukemia) were next transplanted into NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Extended Data Fig. 7c,d). All mice in the control groups (untreated (n = 3) and TCR1G4 cells (n = 3)) were killed after 21 d due to high leukemic burden, at which time leukemic cells were undetectable in the TCRFLT3D/Y cell-treated group (n = 4) (Extended Data Fig. 7e,f). Among four mice receiving the therapeutic TCR, two survived for the duration of the experiment (53 d) and one remained free of leukemia but was killed on day 41 due to graft-versus-host disease, whereas one relapsed on day 28 and was found dead on day 32 (Extended Data Fig. 7e). Upon termination, no leukemic cells were detected in the bone marrow (BM) of the two surviving TCRFLT3D/Y cell-treated mice (Extended Data Fig. 7g,h), while transduced T cells in the BM persisted (Extended Data Fig. 7i).
Extended Data Fig. 7. TCRFLT3D/Y cells efficiently target leukemia in a xenograft mouse model.
(a) Flow cytometry histograms showing expression of FLT3D/Y as measured by GFP- reporter fluorescence in transduced AML and B-ALL cell lines. Negative control (non-transduced BV173 cells) in top histogram. (b) Remaining viable FLT3D/Y-transduced, HLA-A2+ cells (purple dots) after 24 h co-culture with TCRFLT3D/Y cells (E:T ratio of 1:2), in percent of corresponding numbers following treatment with mock-transduced T cells (grey dots), quantified by flow cytometry. +A2 denotes that HLA-A2 was introduced by transduction. Data points are from n = 3 independent experiments with each dot representing the mean of n = 3 technical replicates in each experiment and are shown as mean ± s.e.m. Gating strategy and quantification as shown in Extended Data Fig. 6e. (c) Schematic overview of the BV173D835Y in vivo model. (d) Bioluminescence imaging (BLI) analysis of NSG mice day 13 after BV173D835Y cell injection, one day prior to T-cell therapy. Data shown are from one experiment, with mice grouped into untreated (n = 3 mice), TCR1G4 (n = 3 mice) and TCRFLT3D/Y (n = 4 mice) cell treated. (e) BLI of BV173D835Y engrafted leukemic cells in mice on indicated days relative to treatment with TCR1G4 or TCRFLT3D/Y cells, or no treatment. (f) Quantification of BV173D835Y engrafted leukemic cells in mice, 21 days after treatment with TCR1G4 or TCRFLT3D/Y cells or left untreated. (g) Flow cytometry plots showing BM tumor burden in two untreated and two TCR1G4 cell-treated mice at time of sacrifice (d 21), as well as two TCRFLT3D/Y cell treated mice sacrificed at end of experiment (d 53). (h) Percentage of bone marrow BV173D835Y leukemic cells in mice treated with TCR1G4 or TCRFLT3D/Y cells, or left untreated, out of total mouse and human CD45+ cells (21-53 days after treatment start). (i) Percentage of TCR-β+ cells out of human CD8+ cells in the BM of mice analyzed at point of sacrifice. Data in d, f, h, i are presented as mean ± s.e.m, are from one experiment with each dot representing an individual mouse, and are analyzed by unpaired, two-tailed Student’s t-test. P values are shown and p < 0.05 was considered statistically significant.
TCRFLT3D/Y cells efficiently eliminate primary AML in PDX models
To investigate the in vivo efficacy of TCRFLT3D/Y cells in disease-relevant models, we next treated mice engrafted with primary AML cells in different PDX models.
Model 1
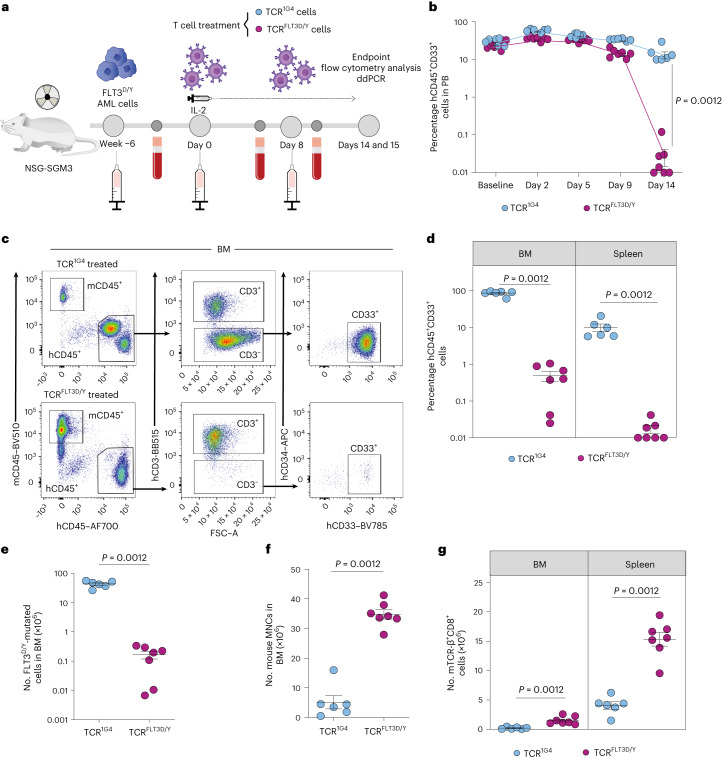
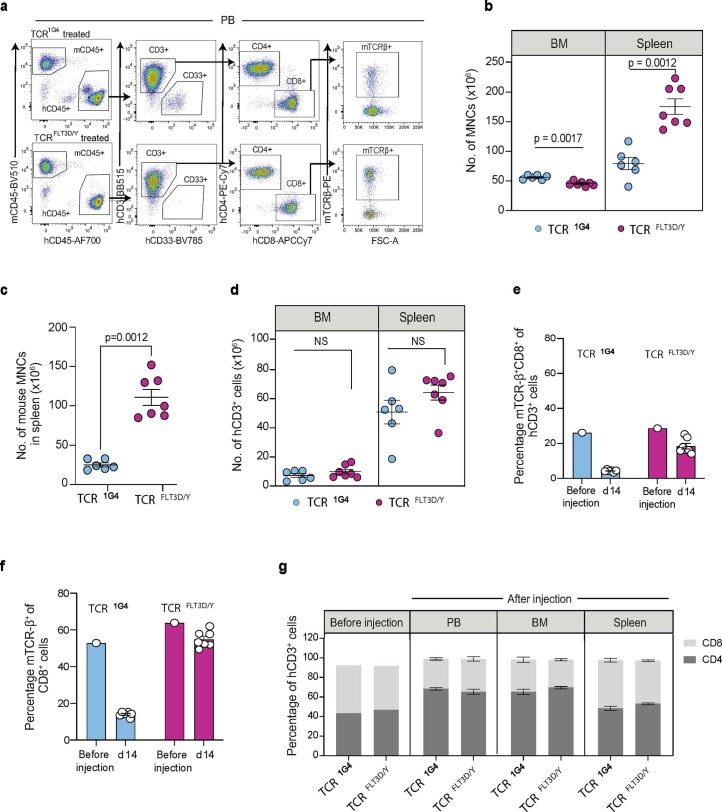
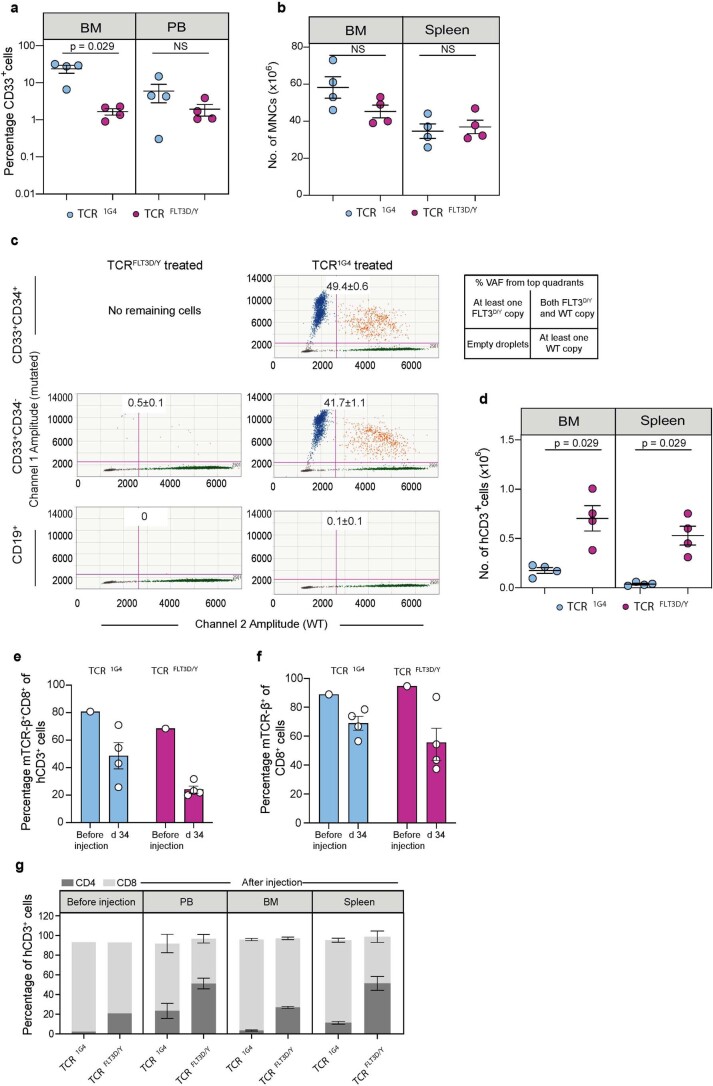
NSG-SGM3 mice highly engrafted with primary FLT3D835Y AML cells from patient 7 (24.5% ± 2.2% human CD33+ cells in PB) (Fig. 3a,b and Supplementary Tables 2 and 3) were treated with TCRFLT3D/Y (n = 7) or control TCR1G4 (n = 6) cells. Serial analysis of PB demonstrated a maintained high CD33+ engraftment in TCR1G4-treated mice, whereas CD33+ cells were virtually eliminated by day 14 in all TCRFLT3D/Y-treated mice (Fig. 3b and Extended Data Fig. 8a). Mice were closely monitored for potential allo-reactivity mediated by endogenous TCRs of the T cells transduced to express TCR derived from a third-party donor, which could otherwise confound the anti-tumor reactivity mediated by TCRFLT3D/Y. Thus, terminal analysis of all TCR1G4-treated and TCRFLT3D/Y-treated mice was performed on day 15 after T cell infusion when tumor burden started to decline slightly also in control mice (Fig. 3b). BM analysis demonstrated high CD33+ AML engraftment in TCR1G4-treated mice (mean, 86.9% ± 5.3%), which was reduced to a mean of 0.5% ± 0.2% in TCRFLT3D/Y-treated mice, with a similar reduction in the spleen (Fig. 3c,d and Supplementary Table 4). An almost complete elimination of FLT3D/Y AML cells in the BM was confirmed by droplet digital PCR (ddPCR) analysis, quantifying clonally involved cells (Fig. 3e and Supplementary Table 5). Importantly, efficient targeting of human AML cells in TCRFLT3D/Y-treated mice resulted in recovery of mouse hematopoiesis, which was severely suppressed by high leukemic burden in TCR1G4-treated mice (Fig. 3f and Extended Data Fig. 8b,c). T cells transduced to express TCR were detected in the BM, spleen and PB of all mice throughout the experiment (Fig. 3g and Extended Data Fig. 8d–g).
Fig. 3. TCRFLT3D/Y cells efficiently target primary AML in mice with high leukemic burden.
a, Schematic overview of the PDX in vivo model with FLT3D835Y-mutated primary AML cells from patient 7. b, Percentage of human hCD45+CD33+ cells in PB at baseline (1 d before T cell infusion) and on the indicated days after infusion with TCR1G4 (n = 6 mice) or TCRFLT3D/Y (n = 7 mice) cells. Numbers were adjusted for hCD3+ T cells. c, Representative flow cytometry plots of viable single BM mononuclear cells (MNCs) from TCR1G4 (top) and TCRFLT3D/Y (bottom) cell-treated NSG-SGM3 mice stably engrafted with primary AML FLT3D/Y cells from patient 7. d, Percentage of hCD45+CD33+ cells in the BM and spleen at terminal analysis 15 d after T cell infusion. Numbers were adjusted for hCD3+ T cells. e, Number of FLT3D835Y-mutated BM hCD45+CD3− cells determined by ddPCR. f,g, Number of mouse (m)CD45+ cells in BM (f) and mTCR-β+CD8+ cells in the BM and spleen (g) at the endpoint. All data are presented as mean ± s.e.m. and were generated from one experiment including six mice treated with TCR1G4 cells and seven mice treated with TCRFLT3D/Y cells. Each dot represents one mouse, and statistical analysis was performed with two-tailed Mann–Whitney test. P values are shown, and P < 0.05 was considered statistically significant.
Extended Data Fig. 8. TCRFLT3D/Y cells persist in vivo and mediate recovery of mouse hematopoiesis in patient 7 PDX model.
(a) Representative FACS plots of viable single PB MNCs from TCR1G4 (top) and TCRFLT3D/Y (bottom) T cell-treated NSG-SGM3 mice stably engrafted with primary AML FLT3 D835Y cells from patient 7 at 14 days post T cell infusion. Equivalent gating as shown was also used for spleen. (b) Number of total MNCs in BM and spleen of TCR T cell treated mice. (c) Number of mouse MNCs (mCD45+ cells) in spleen. (d) Number of total hCD3+ cells in BM and spleen. (e-f) Percentage of mTCR-β+CD8+ cells of hCD3+ cells (e) and percentage of mTCR-β+ cells of hCD8+ cells (f) in T cell samples used for injection and at indicated days after T cell injection, in PB. (g) Distribution (%) of CD4 and CD8 cells within the human CD3+ T cell samples used for injection, and in PB, BM and spleen 14-15 days after injection into mice. All data are from terminal analysis 15 days post T-cell infusion if not otherwise stated. The data are presented as mean ± s.e.m. of n = 6 individual mice treated with TCR1G4 cells and of n = 7 individual mice treated with TCRFLT3D/Y cells, measured in one experiment. Each dot represents one mouse and statistical analysis was performed with two-tailed Mann-Whitney test. P values are shown and p < 0.05 was considered statistically significant.
Model 2
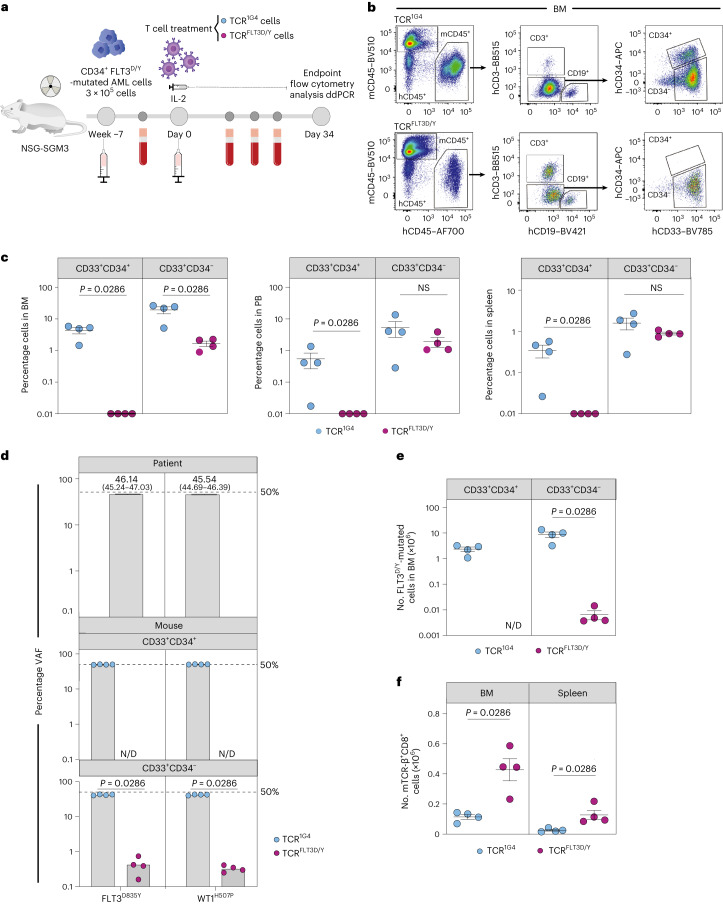
Primary xenografts from a second FLT3D835Y-mutated AML sample engrafted in NSG-SGM3 mice (patient 1, Supplementary Table 2) were investigated for response to TCRFLT3D/Y cells (Fig. 4a). Unlike patient 7 (Fig. 3c), this AML co-expressed CD34, a stem and progenitor marker characteristic of CD34+ AMLs47, for which CD34+ cells in contrast to CD34− cells have been shown to possess leukemia-propagating activity48,49. Control TCR-treated mice had relatively low levels of PB human CD33+ engraftment 34 d after T cell injection (mean, 6.0% ± 3.1% human (h)CD45+CD33+ cells), whereas BM engraftment was much higher (mean, 23.7% ± 5.8%) (Fig. 4b, Extended Data Fig. 9a,b and Supplementary Table 6). In TCRFLT3D/Y-treated mice, this was reduced to 1.9% ± 0.7% in PB and 1.7% ± 0.3% in the BM (Extended Data Fig. 9a,b). Terminal BM analysis on day 34 after T cell injection revealed distinct populations of CD33+CD34− (mean, 19.4% ± 4.8%) and CD33+CD34+ cells (mean, 4.4% ± 1.0%) in control TCR1G4-treated mice (Fig. 4b,c). In TCRFLT3D/Y-treated mice, some CD33+CD34− cells persisted in the BM although at greatly reduced levels (mean 1.7% ± 0.3%), whereas CD33+CD34+ cells were completely eliminated. Similar findings were observed in PB and the spleen (Fig. 4c and Supplementary Table 6).
Fig. 4. TCRFLT3D/Y cells eliminate primary CD34+ AML in vivo.
a, Schematic overview of the PDX in vivo model with FLT3D835Y-mutated primary AML cells from patient 1. b, Representative flow cytometry plots of BM from TCR1G4 (top) and TCRFLT3D/Y (bottom) cell-treated NSG-SGM3 mice stably engrafted with primary AML FLT3D/Y cells from patient 1. Equivalent gating was also used for PB and the spleen. c, Percentage of hCD45+CD33+CD34+ and hCD45+CD33+CD34− cells in the BM, PB and spleen at the endpoint (day 34 after T cell infusion) of TCR1G4 (n = 4 mice) or TCRFLT3D/Y (n = 4 mice) cell-treated mice. Numbers were adjusted for hCD3+ T cells. NS, not significant. d, Percentage VAF determined by ddPCR of FLT3D835Y and WT1H507P driver mutations in primary BM cells from patient 1 (top) and hCD45+CD33+CD34+ (middle) and hCD45+CD33+CD34− (bottom) cells from TCR cell-treated mice. N/D, not analyzed due to insufficient hCD45+CD33+CD34+ cells. Numbers show VAF and 95% confidence intervals. The dashed line at 50% indicates 100% clonality unless loss of heterozygosity. No significant differences in VAFs of the FLT3D835Y and WT1H507P mutations were observed. e, Number of FLT3D835Y-mutated hCD45+CD33+CD34+ and hCD45+CD33+CD34− cells in the BM as determined by ddPCR. N/D, not detected due to lack of hCD45+CD33+CD34+ cells. f, Number of mTCR-β+CD8+ cells in the BM and spleen at the endpoint. All data are presented as mean ± s.e.m. from terminal analysis 34 d after T cell infusion from one experiment including four mice treated with TCR1G4 cells and another four mice treated with TCRFLT3D/Y cells. Each dot represents one mouse, and statistical analysis was performed with two-tailed Mann–Whitney test. P values are shown, and P < 0.05 was considered statistically significant.
Extended Data Fig. 9. TCRFLT3D/Y cells eliminate FLT3D835Y leukemia cells and persist in vivo in patient 1 PDX model.
(a) Percentage of CD33+ cells in BM and PB at endpoint (day 34 post T cell infusion) of TCR1G4 cell (n = 4) or TCRFLT3D/Y cell (n = 4) treated mice. Numbers adjusted for hCD3+ T cells. (b) Number of total MNCs in BM and spleen of TCR T cell treated mice. (c) Representative ddPCR plots of BM from TCR cell-treated mice. Numbers in quadrants represent %VAF (mean ± s.e.m.) of the FLT3D/Y in BM among hCD45+CD33+CD34+, hCD45+CD33+CD34− and hCD45+CD19+ cells from TCRFLT3D/Y cell-treated (left) and TCR1G4 cell-treated (right) mice. ‘No remaining cells’ due to elimination of hCD45+CD33+CD34+ cells in TCRFLT3D/Y cell-treated mice. (d) Number of total hCD3+ cells in BM and spleen. (e-g) Percentage of mTCR-β+CD8+ cells of hCD3+ cells (e) and percentage of mTCR-β+ cells of hCD8+ cells (f) in sample used for injection and at indicated days after T cell infusion in PB. (g) Distribution (%) of CD4 and CD8 cells within the human CD3+ T cell samples analyzed prior to injection, and in PB, BM and spleen of individual mice at endpoint. All data are from terminal analysis 34 days post T-cell infusion unless otherwise stated. The data are presented as mean ± s.e.m. of n = 4 individual mice treated with TCR1G4 cells and of n = 4 mice treated with TCRFLT3D/Y cells, measured in one experiment. Each dot represents one mouse and statistical analysis was performed with two-tailed Mann-Whitney test. P values are shown and p < 0.05 was considered statistically significant.
DNA sequencing confirmed that cells engrafted in NSG-SGM3 mice were represented by the same exonic mutations as those detected in AML blasts from the BM of patient 1, with FLT3D835Y and WT1H507P representing the only detected recurrent driver mutations (Supplementary Table 7). Further quantification by ddPCR demonstrated that FLT3D835Y and WT1H507P mutations represented the dominating clone, with a VAF > 45% in patient 1 as well as in engrafted NSG-SGM3 mice (Fig. 4d). Notably, among the remaining CD33+CD34− BM cells of TCRFLT3D/Y-treated mice, the fraction of FLT3D835Y-mutated cells was reduced to only 0.9% ± 0.3% as compared with 83.4% ± 2.1% in TCR1G4-treated mice. This translates into more than a 1,300-fold reduction in CD33+CD34− FLT3D/Y AML cells in TCRFLT3D/Y-treated mice (Fig. 4d,e, Extended Data Fig. 9c and Supplementary Table 8). Human T cells including T cells transduced to express TCR were detected in the BM, spleen and PB of all treated mice throughout the course of the experiment (Fig. 4f and Extended Data Fig. 9d–g). Cells from patient 1 also reconstituted CD19+ B lymphocytes in engrafted mice (Fig. 4b), and, in agreement with previous studies46, these were not part of the FLT3D/Y leukemic clone (Extended Data Fig. 9c and Supplementary Table 8).
Model 3
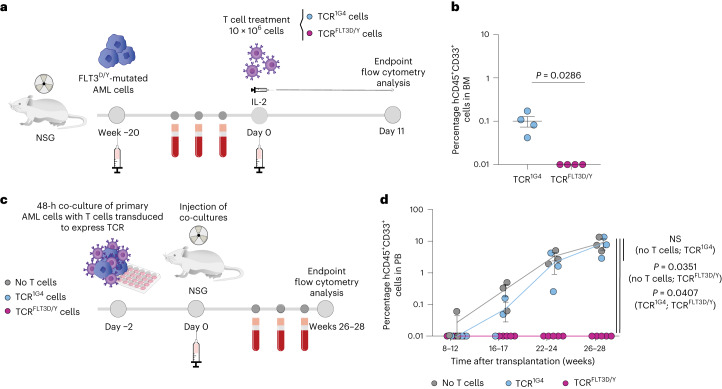
To establish a PDX model mimicking minimal residual disease (MRD)50, secondary transplantations of mice engrafted with AML cells from patient 1 were performed into NSG mice, a mouse strain that, in contrast to NSG-SGM3 mice, does not specifically enhance human myeloid lineages. In accordance with results from the NSG-SGM3 model, we observed efficient elimination of FLT3D/Y primary AML cells also in this MRD setting (Fig. 5a,b and Supplementary Table 9).
Fig. 5. TCRFLT3D/Y cells efficiently kill primary AML in an MRD setting and eliminate leukemia-propagating cells.
a, Schematic overview of the MRD PDX in vivo model with FLT3D835Y-mutated primary AML cells from patient 1. b, Percentage of hCD45+CD33+ cells in the BM of NSG mice engrafted with low levels of AML after treatment with TCR1G4 (n = 4 mice) or TCRFLT3D/Y (n = 4 mice) cells 11 d after T cell infusion. Numbers were adjusted for hCD3+ T cells. Data are presented as mean ± s.e.m. and were generated from one experiment. Each dot represents one mouse, and statistical analysis was performed with two-tailed Mann–Whitney test. c, Schematic overview of the PDX in vivo model with FLT3D835Y-mutated primary AML cells from patient 1 after in vitro targeting with TCR1G4 or TCRFLT3D/Y cells. d, Percentage of hCD45+CD33+ cells in PB at the indicated time after transplantation of primary AML cells from patient 1 following 48 h of co-culture without T cells (n = 3 mice) or with TCR1G4 (n = 3 mice) or TCRFLT3D/Y (n = 5 mice) cells. Data are presented as mean ± s.e.m. and were generated from two independent experiments. Each dot represents one mouse, and statistical analysis was performed by multilevel linear regression using the R package ‘lmerTest’ (further described in the Methods). P values are shown, and P < 0.05 was considered statistically significant.
Model 4
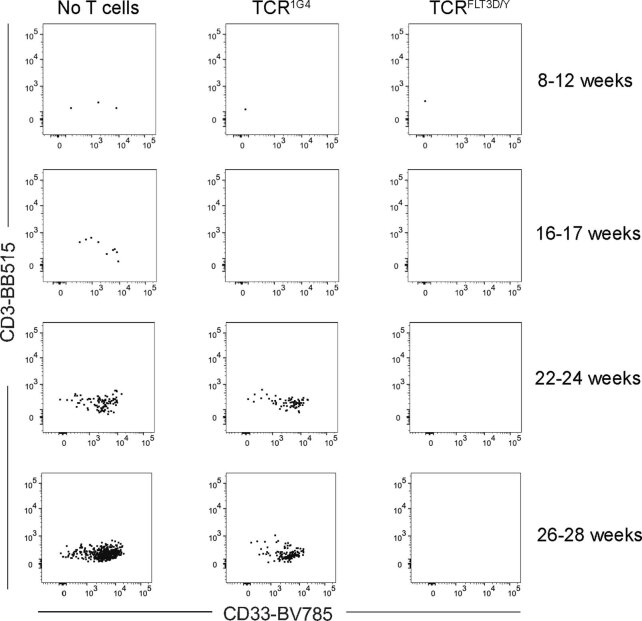
TCRFLT3D/Y treatment resulted in undetectable levels of CD34+ FLT3D/Y AML cells in the PB, spleen and BM of all mice engrafted with AML cells from patient 1, compatible with complete elimination of FLT3D/Y AML-propagating cells in vivo through specific TCRFLT3D/Y targeting. More definitive support for this would, however, require longer follow-up of mice to allow for potential outgrowth of rare and therapy-resistant AML stem cells that might have escaped T cell recognition. A long follow-up time was not possible in models 1–3 due to the continued presence of TCR T cells having endogenous TCR repertoires that cause allo-reactivity and xenoreactivity over time. Moreover, to establish whether all AML-propagating cells have been eliminated by TCRFLT3D/Y cells requires assessment in the absence of TCRFLT3D/Y T cells. To circumvent these limitations, AML cells from patient 1 were cultured with TCRFLT3D/Y or TCR1G4 cells or without T cells in vitro for 48 h before transplantation into NSG mice (Fig. 5c). The very few human T cells to which the AML cells were exposed in vitro did not persist in vivo (Extended Data Fig. 10). The mice did not receive interleukin (IL)-2 infusions. Potential AML development could therefore be followed for 28 weeks. During this time, mice injected with AML cells from control cultures without T cells or with control TCR1G4 cells showed progressive and high leukemic engraftment. By contrast, no detectable engraftment was observed at any time in any mice transplanted with AML cells co-cultured with TCRFLT3D/Y cells (Fig. 5d and Extended Data Fig. 10). Collectively, these experiments demonstrate that TCRFLT3D/Y cells can efficiently target and eliminate FLT3D835Y-mutated in vivo AML-propagating cells.
Extended Data Fig. 10. Lack of in vivo T-cell expansion upon transplantation of primary AML cells co-cultured with TCR1G4 or TCRFLT3D/Y T cells.
Representative FACS profiles of engrafted human CD45+ cells in the PB of mice analyzed at indicated weeks after transplantation of all cells remaining after 48 hours in vitro culture of AML cells without T cells (left), AML cells co-cultured with TCR1G4 cells (middle) or AML cells co-cultured with TCRFLT3D/Y cells (right). Profiles are from one mouse per treatment group from one experiment out of two performed.
Discussion
In this study, we identify a TCR recognizing an epitope from the recurrent driver mutation D835Y in the tyrosine kinase domain of FLT3 in AML, presented on the prevalent HLA-A2 allele. This TCR, selected from healthy donor T cell repertoires, mediated highly specific and efficient killing of primary AML cells harboring the FLT3D835Y mutation in vitro and in vivo. Different clinically relevant aspects of therapeutic effects were demonstrated in different in vivo mouse PDX models. This included an almost complete removal of mutated CD34− AML in model 1 with high engraftment and complete removal of CD34+ AML in model 2 with lower levels of reconstitution. In a third PDX model, TCRFLT3D/Y cells rejected patient AML cells in a setting resembling MRD. TCRFLT3D/Y cells thus efficiently eliminated leukemia with both high and low disease burden. AML cells that are CD34+ have previously been shown to propagate leukemia in vivo49. Here, we demonstrated, in a fourth PDX model followed for 7 months after AML injection, that TCRFLT3D/Y cells in vitro can specifically and efficiently target and kill leukemia-propagating CD34+ FLT3D835Y-mutated AML cells. Studies in humans would, however, be required to confirm that TCRFLT3D/Y cells could achieve this also in vivo. In sum, this suggests that a therapeutic TCR targeting a single, shared neoantigen has the potential of eliminating the reservoir of leukemia stem cells in AML.
The prospect of treating large patient groups with cytotoxic T cells expressing a single TCR targeting public neoantigens expressed by prevalent HLA molecules has emerged as an attractive therapeutic possibility51. However, although a large number of recurrent hotspot mutations are known, it has proven difficult to identify HLA-bound peptides from tumor samples by immunopeptidomics52,53. One reason might be that driver mutations that are poorly presented to the immune system provide a survival advantage. Another possibility is that mass spectrometry has insufficient sensitivity for detection of many neoantigenic peptides. We were, however, able to identify the FLT3D/Y neoepitope in two AML patient samples by targeted mass spectrometry, starting with a large number of leukemia cells. T cells are, on the other hand, the most sensitive tools available for detection of pMHC complexes54. Our technology using mRNA-transduced dendritic cells to prime naive T cells from healthy donors provides the means to identify neoepitopes and reactive TCRs in a single assay38,39. Presentation of the FLT3D/Y neoepitope on leukemic cells was confirmed by efficient killing of primary HLA-A2+ AML cells expressing FLT3D835Y, but not the less frequent FLT3D835E or FLT3D835H mutations or FLT3WT, by TCRFLT3D/Y cells, further demonstrating TCR specificity.
To date, most efforts to isolate neoantigen-specific TCRs employ strategies in which memory T cell responses in patient material are interrogated11,55,56. Identification of TCRs with sufficient affinity is essential for clinical impact41,57. A hurdle might be induction of T cell tolerance due to long-standing co-evolution between cancer and the immune system in the absence of inflammation and lack of sufficient priming. This might contribute to the low spontaneous reactivity that tumor-infiltrating T cells show to the large repertoire of predicted neoantigens in melanoma11. In support of this, immune responses to neoantigen vaccines seem to be dominated by de novo responses induced in naive T cells58–60. The possibility of inducing neoantigen-specific responses from naive patient-derived T cells is, however, limited by the amount of blood that can be collected and by T cell repertoires that might be reduced by foregoing therapy. In an earlier study, we showed that healthy donor-derived T cells recognized fivefold more neoantigens than tumor-infiltrating lymphocytes of patients with melanoma38. Here, we used the same approach to identify TCRFLT3D/Y, which recognizes peptide at picomolar concentrations and mediates efficient cytotoxicity against primary AML cells in vitro and in vivo. High antigen sensitivity, indicated by EC50 values below 10 nM, appears to be an important factor for TCRs in effectively killing cancer cells, as previously demonstrated by us27 and others9,41,61,62 Similarly, Foy et al., in their publication in Nature, discussed the potential association between low EC50 values observed for personalized neoantigen-reactive TCRs in their study and the limited clinical efficacy observed12.
We primed T cells from 16 healthy donors to identify T cells recognizing the YIMSDSNYV peptide presented by HLA-A2. This indicates low immunogenicity of the peptide, consistent with lack of recognition by the investigated AML patient T cells, although a limited number of cells were screened for reactivity. In sum, this demonstrates the advantage of accessing large, healthy donor T cell repertoires. We previously demonstrated that peptide–HLA stability is an important predictor of neoantigen immunogenicity38. Here, we showed that the off-rate for the FLT3835–843 peptide bound to HLA-A2 was significantly slower when substituting the WT amino acid D in position 1 with a Y. This is consistent with other studies showing that increased stability of the neoantigen relative to the corresponding WT peptide leads to sustained antigen presentation and increased T cell recognition63,64. Testing for potential off-target reactivity is essential to detect possible safety concerns before clinical use65. Mapping of TCR fine specificity using a library of single-amino acid-substituted peptides followed by a bioinformatic screen did not identify cross-recognized peptides, although this does not completely exclude potential recognition of peptide sequences that are unrelated to the cognate peptide. TCRFLT3D/Y cells did not, however, react to a panel of 26 HLA-A2+ cell lines of different tissue origins and spared normal blood cells and AML cells expressing the FLT3WT sequence or alternative FLT3D835 mutations.
Treatment with TCRFLT3D/Y cells would be limited to the 3–4% of patients that, in addition to expressing the mutation also express HLA-A2 in the European-descended population32. While recurrent FLT3 mutations are frequently known to represent secondary and accelerating AML mutations15,33,66, our reanalysis of 58 published FLT3D835Y-mutated AML patient samples15,45 demonstrated that this FLT3 mutation frequently is clonal and, in some cases, also might be the initiating mutation or secondary only to a CH mutation. These data are in agreement with the high VAF for FLT3D835Y in all included patients with AML harboring this mutation, where the only selection criterion upfront was the presence of the mutation. In sum, this highlights the therapeutic and potentially curative potential of eradicating FLT3D835Y-mutated AML clones. Furthermore, in cases in which TKIs effectively target FLT3 ITD but select for AML subclones with FLT3 point mutations, including FLT3D835Y, resistant to first-generation as well as second-generation TKIs35–37, FLT3D835Y-mutation-specific TCR treatment could be combined with TKIs. As part of a paradigm in which combination regimens are tailored based on tumor molecular profiles, TCR therapies targeting specific recurrent point mutations provide a potential means for highly efficacious eradication of specific tumor clones.
TCRs can access recurrent mutations currently inaccessible to CARs. Moreover, TCRs might have inherent advantages relative to CARs that improve T cell persistence and antigen sensitivity67. Today, TCR-based therapies are mostly applied in the form of genetically modified T cells, although soluble bispecific TCR engagers have emerged as new opportunities for off-the-shelf therapies at lower cost68. The results presented here show that a TCR targeting a single shared neoantigen generated from a healthy donor can provide highly efficacious and specific cancer treatment in vivo in multiple disease-relevant models, paving the way for future off-the-shelf, tumor-specific immunotherapies.
Methods
This study was approved by the Regional Committee for Medical and Health Research Ethics South-East Norway (2018/879, 2018/1246 and 2015/2357), the Institutional Review Board and the Data Protection Officer, Oslo University Hospital, the Swedish Ethical Review Authority, Stockholm (EPN 2017/2085-31/2) and the Ethical Committee in Central Denmark Region (1-45-70-88-21) and was performed in accordance with the Declaration of Helsinki. The Norwegian Food Safety Authority (application ID 17500) and Stockholms Djurförsöksetiska nämnd (17978-2018) approved all animal experiments.
Primary patient cells, healthy blood donor cells and cell lines
Buffy coats (PBMCs) from healthy donors were provided by the blood bank of Oslo University Hospital, and PB or BM MNCs from patients with leukemia were isolated from cryopreserved, biobanked material (ethical approvals 2018/879 and 2018/1246). MNCs derived by density-gradient centrifugation (Axis-Shield) were stained to determine HLA-A2 expression by flow cytometry. To confirm the presence of FLT3D835Y, genomic DNA was extracted (QIAGEN DNeasy purification kit) from patient primary cells, and samples were sequenced using the TruSight Myeloid panel (Illumina). Information regarding the patients’ sex (reported in Supplementary Table 2) was not considered during the design of the study but was part of the overall clinical information obtained at the hospital. No sex- or gender-based analysis was performed because each patient is shown individually in the study.
Epstein–Barr virus-transformed lymphoblastoid cell lines (EBV-LCL) were generated from HLA-A2+ and HLA-A2− PBMCs as described previously27. All other cell lines were gifted or obtained from the American Type Culture Collection (ATCC) or the German Collection of Microorganisms and Cell Cultures (DSMZ) as indicated in the Nature Portfolio Reporting Summary. Authentication was performed by short tandem repeat DNA profiling by Labcorp DNA Identification Lab (formerly Genetica, https://celllineauthentication.com/). Cell line cultures were grown in humidified cell incubators containing 5% CO2 at 37 °C using media according to provider guidelines and were tested frequently for potential mycoplasma contamination.
Minigene design
For generation of T cell responses, a minigene was designed to encode predicted epitopes (https://services.healthtech.dtu.dk/service.php?NetMHC-4.0) containing the FLT3D835Y mutation, codon optimized and synthesized by GenScript. Subsequently, it was cloned into the pCIpA102 vector for in vitro mRNA transcription using the RiboMAX Large Scale RNA production system (Promega), as previously described69,70. The minigene encoded the FLT3 amino acid sequence KICDFGLARYIMSDSNYVVRGNVRLARLP (FLT3D835Y 9-mer and 10-mer are underlined and the mutation in position 1 is shown in bold).
Induction of antigen-specific T cells
Monocytes from HLA-A2+ healthy donors were isolated on day −4 using CD14-reactive microbeads and the autoMACS Pro Separator (Miltenyi Biotec). The CD14− PBMC fraction was cryopreserved for later use. The monocytes were then cultured for 3 d in CellGro GMP DC medium (CellGenix) with 1% (vol/vol) human serum (HS; Trina biotech), 1% (vol/vol) penicillin–streptomycin (P/S; Sigma-Aldrich), 50 IU ml−1 IL-4 (PeproTech) and 800 IU ml−1 GM-CSF (Genzyme). Subsequently, moDCs were matured for 14–16 h by adding lipopolysaccharide (Sigma-Aldrich) and IFN-γ (PeproTech) to final concentrations of 10 ng ml−1 and 100 IU ml−1, respectively. On day −1, naive CD8+ T cells were isolated from the autologous CD14− cryopreserved PBMCs by use of the autoMACS Pro Separator and a CD8+ T cell-isolation kit, into which CD45RO- and CD57-reactive beads (Miltenyi Biotec) were added. On day 0, moDCs were collected, electroporated with mRNA and co-cultured with naive T cells in DC–T cell medium with 30 ng ml−1 IL-21 (PeproTech) at a DC:T cell ratio of 1:4. After 10 d, co-cultures were screened for the presence of FLT3D/Y pMHC multimer-reactive CD8+ T cells. pMHC multimers labeled with PE and APC were prepared in house as described previously71,72. Viable CD8+pMHC+ T cells (double positive for PE- and APC-conjugated pMHC multimers) were sorted by flow cytometry.
Expansion of memory T cells reactive to FLT3D/Y in samples from patients with AML following HSCT
For in vitro expansion of potential memory T cells reactive to the FLT3D/Y-mutant peptide in patients with AML that had undergone HSCT, cryopreserved PB samples were thawed and resuspended in Iscove’s Modified Dulbecco’s Medium (IMDM) with 20% (vol/vol) FCS (Trina biotech) and 0.1 mg ml−1 DNase. Viable cells were resuspended at a concentration of 1 M ml−1 and pulsed with the FLT3D/Y peptide at 100 ng ml−1 for 2 h at 37 °C. Cells were washed and resuspended at 3.75 million cells per ml in IMDM with 5% HS, 1× P/S and 20 U ml−1 IL-2 before culturing for 5 d. Identification of T cells reactive to the peptide was performed by staining with pMHC multimers as described above.
Sorting and cloning of pMHC multimer+CD8+ T cells
PBMCs from three healthy donors were mixed at an equal ratio (1:1:1) and irradiated with 35 Gy, washed and resuspended in X-VIVO 20 medium (Lonza, BioNordika) with 5% HS and 1% P/S (T cell medium). A total of 0.2 × 106 irradiated cells (feeders) were placed into tissue culture-treated 96-well plates and were supplemented with 100 μl T cell medium containing 2 μg ml−1 phytohemagglutinin (Remel Thermo Scientific), 80 ng ml−1 IL-2 (R&D Systems) and 4 ng ml−1 IL-15 (PeproTech). FLT3D/Y co-cultures were then collected and stained with LIVE/DEAD Fixable Near-IR, anti-CD3 antibody, anti-CD8a antibody and PE- and APC-conjugated pMHC multimers, and, using the FACSAria II (BD Biosciences) cell sorter, CD8+, pMHC-double-positive multimer populations were sorted as single cells into 96-well plates containing feeders. After 7 d, cultures were supplied with fresh T cell medium containing 1,750 U ml−1 IL-2 and 4 ng ml−1 IL-15, and expanding clones were identified by microscopy. On day 14, growing clones were restimulated with feeder cells prepared as described above and stained with FLT3D/Y pMHC multimers. To assess functionality after expansion, clones were stimulated with K562 cells pulsed with FLT3D/Y or FLT3WT peptides and assessed for CD137 upregulation.
TCR sequencing
The sequences for paired TCR-α and TCR-β chains from two clones and 55 single cells reactive to FLT3D/Y pMHC multimers were amplified as previously described but modified and adapted for the targeted amplification of transcripts encoding TCR-α and TCR-β (refs. 27,38,55). The MiXCR script was used to analyze sequencing data, and an in-house Python script TCR primer was used to reconstruct full-length TCR chains as described previously55,73. The output was manually verified using IMGT/V-QUEST74. For identified TCRs, codon-optimized sequences for TCR-α and TCR-β variable fragments were synthesized and cloned by GenScript.
Gene transfer to human PBMCs and cell lines
HLA-A2+ healthy donor-derived and patient-derived PBMCs were transduced to express FLT3D/Y- and NY-ESO-1 (1G4)-specific TCRs, as detailed in ref. 27. Briefly, 2 × 106 PBMCs per ml in CellGro GMP DC medium with 5% (vol/vol) HS, IL-7 and IL-15 (5 ng ml−1 each, PeproTech) were added to antibody-coated plates (anti-CD3 clone OKT3, eBioscience and anti-CD28 clone CD28.6, eBioscience) and incubated at 37 °C with 5% CO2 for 72 h. Retroviral supernatants were generated as described previously27. PBMCs were collected, resuspended in CellGro GMP DC medium with 5% HS, IL-7 and IL-15, mixed with retroviral supernatant, placed in non-tissue culture-treated six-well plates precoated with RetroNectin (20 μg ml−1, Takara) and spinoculated at 900g for 60 min twice on consecutive days. Transduction efficiency was determined after 3 d by staining with anti-mouse TCR-β chain antibody and/or the pMHC multimer followed by flow cytometry. Before functional experiments, cells were cultured for 48–72 h in CellGro GMP DC medium containing low concentrations of cytokines (0.5 ng ml−1 IL-7 and IL-15). Alternatively, cells were frozen for later experiments.
BV173, ML-2, RS4;11 and NALM-6 cell lines were transduced as described above using retroviral supernatant containing the FLT3D/Y minigene. For in vivo experiments, the BV173 cell line was stably transduced to express the FLT3D/Y minigene, firefly luciferase and GFP (hereafter, BV173D835Y). Complementary DNA encoding FLT3D/Y and HLA-A2 was cloned into the pCIpA102 vector for production of mRNA, as previously described70,71
Immunoprecipitation-targeted mass spectrometry analysis of FLT3 peptides presented on HLA
Monoallelic B721.221 cells expressing HLA-A2 were transduced to express the mutant FLT3 amino acid sequence VLVTHGKVVKICDFGLARYIMSDSNYVVRGNARLPVK. One hundred million cells from the B721.221 line and 400 million cells from patient samples (patients 1 and 3) were lysed in PBS containing 1% lauryl maltoside, 0.5 mM EDTA, 1 mM PMSF and Sigma protease inhibitors (1:200) for 1 h at 4 °C (1 ml lysis buffer per 100 million cells). The clarified cell lysates were then added to 200 µl of AminoLink Plus bead slurry (Thermo Fisher Scientific) coated with pan-HLA class I-specific antibody (W6/32, BioXCell) to enrich for HLA peptides27. The HLA-bound peptides were then sequentially eluted three times, each with 1 ml of 1% TFA. Peptide elutions were pooled and desalted using the Discovery DSC-C18 SPE column. The peptides were vacuum concentrated and dissolved in 25 μl of 3% acetonitrile containing 0.1% TFA, following spike-in with 200 pg of heavy isotope-labeled peptide (YI(13C6,15N)MSDSNYV). The peptide solution (5 μl) was analyzed using an EASY-nLC 1000 system (Thermo Fisher Scientific) connected to a Q Exactive HF mass spectrometer (Thermo Electron) equipped with a nano-electrospray ion source. For liquid chromatography separation, an EASY-Spray ES902 column (C18, 2-µm beads, 100 Å, 75-μm inner diameter) capillary with a bed length of 25 cm was used. A flow rate of 300 nl min−1 was employed with a solvent gradient of 7–35% B in 55 min to 90% B in 3 min. Solvent A was 0.1% formic acid, and solvent B was 0.1% formic acid–90% acetonitrile. The mass spectrometer was operated in parallel reaction-monitoring mode to specifically target the presence of endogenous FLT3 mutant (m/z = 1,091.47381+) and spiked-in isotope-labeled peptide (m/z = 1,098.49281+), eluting within a retention time window of 27–31 min, as determined using a synthetic analog. The MS/MS spectra using higher-energy collision-induced dissociation were acquired with a resolution of R = 15,000 after accumulation to a target of 1 × 105. The normalized collision energy was set to NCE 27, and the isolation window was m/z = 2.0. The maximum allowed ion accumulation for the MS/MS spectrum was 120 ms. Raw data were analyzed using Xcalibur software and Skyline (MacCoss Lab Software).
pMHC-stability assay
HLA-A2 molecules were prepared in house, as previously described71,75,76. The pMHC-stability assay was performed as previously described38 with minor modifications. UV-mediated peptide-exchange reactions were performed for 1 h, followed by incubation of the resulting product at 4 °C. The next day, streptavidin-coated beads were washed twice with PBS–1% Tween. The peptide–HLA monomers were coupled with the washed beads for 10 min at room temperature. After coupling, the beads were washed twice with PBS–1% Tween and resuspended in 200 μl. An aliquot of 20 μl beads was set aside for the 0-h time point, while the remaining beads were incubated at 37 °C. Twenty microliters of beads were collected at 3, 6, 12 and 24 h of incubation. After collecting at each time point, the beads were stained with 30 μl of anti-HLA-A2–PE antibody (343305, BioLegend, 1:100 dilution) for 10 min at room temperature. Samples from all time points were analyzed immediately after staining on a BD LSR II Flow Cytometer.
Antibodies, dyes and flow cytometry
For surface antibody staining of human PB and BM cells, antibodies were added to cells for 15–20 min at 4 °C, followed by washing steps. For intracellular staining, cells were suspended in Cytofix/Cytoperm (BD Biosciences) solution for 20 min, washed with Perm/Wash buffer (BD Biosciences) and then stained with antibodies. For mouse PB, BM and spleen, cells were processed into a single-cell suspension as previously described77 and Fc receptor blocked (human, Miltenyi Biotec; mouse, produced by mouse hybridoma cell line clone 2.4 G2, ATCC, HB-197) for 10 min at 4 °C before staining with antibodies for 15–20 min at 4 °C. All fluorescently conjugated antibodies are described in Supplementary Table 10 and the Nature Portfolio Reporting Summary. In PDX mice, the percentage of myeloid cells was determined as a fraction of combined mouse and human leukocytes (mCD45+hCD45+), subtracting hCD3+ events accounting for infused T cells. Flow cytometry analysis was performed on the BD LSR II flow cytometer or the BD LSRFortessa machine (both BD Biosciences), while cell sorting was performed on the FACSAria Fusion cell sorter (BD Biosciences). Data were analyzed using FlowJo (TreeStar) or FACSDiva (BD Biosciences) software. To visually display flow cytometry data, we used an unsupervised nonlinear dimensionality-reduction algorithm such as t-SNE by using FlowJo (TreeStar) software.
T cell-activation assays
T cells transduced to express TCR were co-cultured with cell lines or primary patient tumor cells at an E:T cell ratio of 1:2 (100,000:200,000 cells per well), and reactivity was investigated by measuring CD137 upregulation or IFN-γ release. When indicated, target cells were pulsed with FLT3D/Y or FLT3WT peptide (purities >90%) or 161 single-amino acid-substituted variants of the FLT3D/Y peptide (purity >70%) (GenScript Biotech) for 1–2 h or electroporated with mRNA encoding either FLT3WT or the FLT3D/Y minigene. Cells were placed in round- or flat-bottom 96-well plates and, after 18–20 h of co-incubation, were centrifuged at 700g for 2 min. Supernatants were collected for measurement of IFN-γ levels by ELISA, while cells were washed and stained with anti-CD137 antibody. In some experiments, transduced T cells were labeled with 0.75 μM CTV to distinguish them from target cells. Reagents for the IFN-γ ELISA were acquired from BD Pharmingen or R&D Systems: mouse anti-human IFN-γ capture antibody (NIB42), Biotin Mouse Anti-Human IFN-γ-detection antibody (4S.B3), streptavidin–HRP, stabilized tetramethylbenzidine and hydrogen peroxide as substrate solutions, sulfuric acid as the stop solution and recombinant human IFN-γ protein as the standard. The assay was performed according to the manufacturer’s instructions.
Flow cytometry-based cytotoxicity assay using cell lines as targets
Transduced cell lines stably expressing the FLT3D/Y minigene were co-cultured with CTV-labeled T cells transduced to express TCR at an E:T cell ratio of 1:2 (75,000:150,000 cells per well) for 48 h in round-bottom 96-well plates in triplicates. Following co-culture, cells were collected, washed and stained with human anti-CD3, anti-CD8 and anti-CD4 antibodies and LIVE/DEAD NIR for 15–20 min. Cells were then washed and resuspended in FACS buffer containing 10,000 CountBright Absolute Counting Beads (Thermo Fisher). An equal number of bead events (3,500) were recorded from every well. Normalized data were reported as percentage of the mean of the number of viable tumor cells acquired from three parallel wells co-cultured with TCR1G4 or TCRFLT3D/Y cells from each donor.
Flow cytometry-based assays for T cell activation and cytotoxicity using primary human samples
PB or BM samples from patients were thawed and resuspended in IMDM with 20% (vol/vol) FCS (Trina biotech) and 0.1 mg ml−1 DNase. Cells were centrifuged at 200g for 15 min at room temperature and transferred to round-bottom 96-well plates for assays measuring CD137 upregulation on T cells transduced to express TCR or cytotoxicity on target cells. Normal CD19+ B cells were isolated from healthy donor buffy coat MNCs using CD19-reactive microbeads and the autoMACS Pro Separator (Miltenyi Biotec) and transferred to round-bottom 96-well plates for assays measuring cytotoxicity on target cells. Individualized antibody panels and gating strategies to identify malignant blasts and normal leukocyte populations were designed after reviewing diagnostic phenotyping available in the hospital records. Allogeneic or, for patients 1–3, also autologous patient-derived T cells transduced to express TCRs, were used in the experiments. Cells transduced to express TCR were prelabeled with CTV dye to distinguish them from target cells. For cytotoxicity assays, 75,000 T cells per well were co-incubated with 150,000 target cells in three parallel wells per condition for 72 h and then stained with individualized antibody panels for flow cytometry. CountBright Absolute Counting Beads were used as described above. Examples of the gating strategy used to identify live tumor cells in patients are shown in Extended Data Fig. 6a. For patients 1–6 and 8, myeloid cells were identified as CD3−CD19−CD20−, normal T cells as CD3+ and normal B cells as CD19+CD20+. Cells from patient 7 were obtained from Jackson Laboratory (stock ID J000106565), and the patient-specific phenotypic markers CD33 and CD19 were used to identify leukemia cells based on the characterization profile from the provider.
TCRFLT3D/Y cell activity in the xenograft leukemia cell line model
This study was approved by the Norwegian Food Safety Authority (application ID 17500) and performed in compliance with institutional guidelines and the 2010/63/EU directive. Mice were observed for clinical signs of tumor spreading and were killed if they developed >20% weight loss, hunched posture, ruffled fur, limb paralysis or enlarged spleens. Maximum tumor burden was not exceeded. Experiments were terminated 2 months after T cell injection to avoid graft-versus-host disease, and surviving mice were killed humanely by cervical dislocation. Six mice were housed per cage in Eurostandard Type III cages (macrolone) with a light cycle from 7 a.m. to 7 p.m. at 22 ± 1 °C with 62 ± 5% humidity. Female (8–10-week-old) NSG (Jackson Laboratory) mice, bred in house, were sublethally irradiated (2.5 Gy, MultiRad225 X-ray, RPS Services) on day −15, and 4 × 106 cells of the human B-ALL cell line BV173D835Y were injected on day −14 through the tail vein. After leukemia was confirmed by BLI on day −1, mice were treated with 107 T cells transduced to express TCR1G4 or TCRFLT3D/Y. A group of control mice did not receive T cell injections. All mice were injected intraperitoneally (i.p.) daily with 2,500 IU IL-2 (R&D Systems). BLI imaging (by the IVIS Spectrum in vivo imaging system; analysis with Living Image software version 4.5.2, PerkinElmer) and blood analysis by flow cytometry were performed continuously. BM was collected at the endpoint and processed for flow cytometry to analyze the presence of T cells and tumor cells.
Activity of TCRFLT3D/Y cells in four primary AML PDX models
Experiments were approved by Stockholms Djurförsöksetiska nämnd (17978-2018). The maximal tumor burden permitted was defined by the impact on the animal’s health. Mice engrafted with leukemic cells were continuously monitored according to Karolinska Institutet’s health assessment, and no animal exceeded the humane endpoint. The housing conditions were 21 °C and 45–50% humidity. Two to five mice were housed per cage in IVC-Mouse GM500 cages with a light cycle from 4 a.m. to 4 p.m. (patient 7) or from 6 a.m. to 6 p.m. (patient 1). BM was collected from femur, tibia and crista from both hind legs at terminal analysis. Mainly female mice were used in this study, but, when both sexes were used, they were equally distributed within the treatment groups. No sex-based analysis was thus performed.
Model 1
Female NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV IL3,CSF2,KITLG)1Eav/MloySzJ (NSG-SGM3) mice stably engrafted with FLT3D835Y-expressing HLA-A2+ AML cells from patient 7 (Supplementary Table 2 and 3) at 5 weeks of age were obtained from Jackson Laboratory (stock ID J000106565). Upon arrival (5.5 weeks after transplantation), PB myeloid engraftment was confirmed by flow cytometry, and mice were allocated to treatment groups (TCRFLT3D/Y or TCR1G4 cells), resulting in a similar mean human myeloid engraftment between the groups before infusion with T cells. Cryopreserved T cells transduced to express TCR were thawed and cultured in X-VIVO 20 medium (Lonza) with 5% HS, 1% penicillin–streptavidin and 5 ng ml−1 IL-7 and IL-15 for 3–4 d before treatment. T cells containing 5 × 106 CD8+mTCR-β+ T cells were injected through the lateral tail vein, and all mice received daily i.p. injections of 2,500 IU human IL-2 (R&D Systems). At day 8 after T cell infusion, half of the mice received a second dose of 5 × 106 CD4-depleted (Miltenyi Biotec) TCRFLT3D/Y or TCR1G4 T cells. As no differences were observed between the mice receiving one or two doses of T cells, the data from these mice were pooled. The effect of the T cells was monitored in serially collected PB and at termination 15 d after T cell treatment in the BM and spleen through detailed flow cytometry analysis.
Model 2
BM CD34+ cells from patient 1 (FLT3D/Y and HLA-A2+; Supplementary Tables 2 and 3) were obtained by CD34 magnetic bead enrichment (Miltenyi Biotec) according to the manufacturer’s instructions and as previously described78. A total of 3 × 105 CD34+ cells per mouse were intrafemorally injected into sublethally irradiated (3.3 Gy, X-ray source) female and male NSG-SGM3 mice (Jackson Laboratory, stock 013062) 9 weeks of age. Upon confirmation of stable PB myeloid engraftment 7 weeks after transplantation, mice were allocated to treatment groups (TCRFLT3D/Y or TCR1G4 cells) based on their engraftment levels as described for patient 7. T cells containing 5 × 106 CD8+mTCR-β+ T cells were infused into each mouse by lateral tail vain injections, and all mice received daily i.p. injections of 2,500 IU human IL-2 for 2 weeks, followed by less frequent injections. Mice were monitored using serially collected PB and at termination 34 d after T cell treatment with the BM and spleen.
Model 3
To mimic an MRD setting, secondary intrafemoral transplantation of BM cells from NSG mice (Jackson Laboratory, stock 005557) engrafted with material from patient 1 was performed into sublethally irradiated (2.5 Gy, X-ray source) female NSG mice 12–13 weeks of age. Following confirmation of low but stable leukemic engraftment 20 weeks after transplantation, mice were treated with TCRFLT3D/Y or TCR1G4 CD4-depleted (Miltenyi Biotec) T cells (10 × 106 T cells, containing 90% CD8+ and 10% CD4+ T cells). All mice were injected i.p. daily with 2,500 IU human IL-2. The effect of the T cells was evaluated in the BM 11 d after T cell treatment.
Model 4
Following secondary transplantation of BM from mice engrafted with patient 1 cells, BM from three NOD.Cg-Prkdcscid Il2rgtm1Sug Tg(CMV-IL2)4-2Jic/JicTac (NOG-hIL2, Taconic) mice was isolated and cultured for 48 h in CellGro GMP DC medium with 5% HS, IL-7 and IL-15 either with no T cells or with TCR1G4 or TCRFLT3D/Y cells (E:T cell ratio, 1:2). After 48 h, the contents of each well were collected and intrafemorally injected into sublethally irradiated (2.25–2.5 Gy) female NSG mice 8–13 weeks of age. Engraftment levels were monitored in serially collected PB samples by flow cytometry. Data were pooled from two individual experiments. Differences in engraftment dynamics between mice engrafted with AML cells precultured with TCRFLT3D/Y or TCR1G4 cells or without T cells was assessed by multilevel linear regression using the R package ‘lmerTest’. Sampling time points and groups are treated as an interaction term with random effects for individual mice. Because the engraftment at the first time point was 0 for all mice except one, we fixed the intercept as 0. Estimated marginal means of models were obtained and compared between three groups using the R package ‘emmeans’. Multiple tests were corrected using the Benjamini–Hochberg method. Only mice that could be followed until the end of the experiment were included in the analysis.
Whole-exome sequencing of cells from patient 1 and PDX mice
DNA was isolated from AML cells purified with the FACSAria Fusion cell sorter (BD Biosciences) from patient 1 using the Maxwell RSC Cultured Cells DNA Kit (Promega), including primary BM AML blasts (CD3−CD19−) and T cells (CD3+CD8+CD19−CD33− and CD3+CD4+CD19-CD33-) and BM mCD45−hCD45+CD3−CD19− cells from one untreated and one TCR1G4-treated PDX mouse from model 2. Whole-exome sequencing libraries were prepared with the Lotus DNA Library Prep Kit (IDT). Exon regions were captured using xGen Exome Hyb Panel v2 and the xGen Hybridization and Wash Kit (IDT) on 4 July 2022 and 5 July 2022. After mixing with 1% PhiX, prepared libraries were sequenced using the NextSeq high-output kit (300 cycles) with settings ‘Read1, 151; Index1, 8; Index2, 8; Read2, 151’. Reads were aligned to the human (GRCh37) genome reference using Burrows–Wheeler Aligner version 0.7.17 with default parameter settings. PCR duplicates were marked with biobambam version 2.0.87. Reads were subjected to indel realignment and base quality score recalibration using GATK3 (version 3.8) and recalculation of MD/NM tags using SAMtools version 1.9. Errors associated with enzymatic fragmentation during library preparation were removed using FADE version 0.5.5, resulting in an average depth of 364 (331–439) in the final bam files79.
Mutations calling was performed using GenomonFisher (https://github.com/Genomon-Project/GenomonFisher) with the following parameters: (1) mapping quality score ≥20, (2) base quality score ≥15, (3) number of total reads ≥8, (4) number of variant reads ≥5, (5) VAF ≥ 0.05, (6) VAF in paired T cells <0.1, (7) VAF in other non-paired normal controls <0.05 in all and average <0.01, (8) strand ratio in tumor ≠0 or 1, (9) VAF by base counts divided by VAF by read count ≥0.5 and ≤2, (10) P value by Fisher <0.1, (11) P value by EBFilter80 <0.001, (12) mutation on exons, (13) not on the repeat regions.
Mutations were annotated by ANNOVAR81. Copy number analysis was performed using CNACS82. The WT1H507P mutation was, together with the FLT3D835Y mutation, the only identified potential driver mutation in AML15,83.
Droplet digital PCR
ddPCR was performed to quantify clonal involvement. DNA from patient 1 with AML was isolated as explained above, and DNA from AML PDX mice was isolated through flow cytometry sorting of mCD45−mTer119−hCD45+CD3− cells (PDX patient 7) or hCD45+CD33+CD34−, hCD45+CD33+CD34+ and hCD45+CD19+ cells (PDX patient 1) and subjected to whole-genome DNA amplification using the REPLI-g Single Cell Kit (QIAGEN) according to the manufacturer’s instructions. Samples with <90 cells were excluded. Briefly, a 20-µl PCR reaction mixture containing 1× ddPCR supermix for probes (no dUTP) (Bio-Rad), 1× primer-probe assay (FLT3D835Y, dHsaMDV2010047; WT1H507P, dHsaMDS871718945; Bio-Rad) and 60 ng DNA was mixed with Droplet Generation Oil for Probes (Bio-Rad). Droplets were prepared according to the manufacturer’s instructions on a QX200 droplet generator (Bio-Rad) and subjected to PCR (Bio-Rad): 95 °C for 10 min, 40 cycles of 94 °C for 30 s and 55 °C for 60 s and a 10-min incubation at 98 °C. Plates were read on a QX200 droplet reader (Bio-Rad) and analyzed using QuantaSoft version 1.5.38.1118 software (Bio-Rad) to calculate VAFs and 95% confidential intervals. The numbers of FLT3D835Y cells in the AML PDX mice were determined by combining the information from the fraction of mutated cells identified by ddPCR, and the frequency of phenotypically defined subsets (hCD45+CD3− or hCD45+CD33+CD34− and hCD45+CD33+CD34+, respectively, for patients 7 and 1) in the BM as determined by flow cytometry, and the total number of MNCs in the BM was counted with Sysmex.
External AML-targeted sequencing data analysis
Mutation data (SNVs and indels) from AML patients reported by Papaemmanuil et al.15 were downloaded from https://www.cbioportal.org. VAF was estimated from reported alternative allele reads divided by sequencing depth for the position. Patients harboring a FLT3D835Y mutation were selected for in-depth analysis.
The order of FLT3 mutations in AML using VAF and the mutated cell fraction was determined from the publicly available mutation list identified by targeted DNA sequencing by Morita et al.45.
Statistical analysis and reproducibility
Statistical analysis was performed in GraphPad Prism versions 6–8 (GraphPad Software). Comparison of mean values between two experimental groups was conducted with unpaired, two-tailed Student’s test. The ordinary ANOVA test with adjustment for multiple comparisons with Tukey’s post hoc test was employed for comparisons of more than two experimental groups. To determine differences between in vivo treatment groups in the PDX mouse models, Kruskal–Wallis ANOVA with Dunn’s multiple-comparison test and two-tailed Mann–Whitney test were performed. P values < 0.05 were considered statistically significant. The investigators were not blinded to allocation during experiments or to outcome assessment. PDX models 1–3 were performed once each, and data for PDX model 4 were generated from two independent experiments with mice from all treatment groups represented in both experiments. No statistical method was used to predetermine sample size, and experiments were not randomized. Sample sizes were estimated based on preliminary experiments and are similar to those previously reported27,84. Due to the nature of the other in vitro experiments, blinding was not possible and is not generally performed in the field as the data acquisition is quantitative (flow cytometry or MS) rather than qualitative and therefore less influenced by observer bias. Data distribution was assumed to be normal, but this was not formally tested. Data distribution is shown as individual data points.
Illustrations
Fig. 1a was generated by Science Shaped (https://scienceshaped.com/). All mouse illustrations were generated using Adobe Illustrator 2022 version 26.0.3.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Supplementary Tables 1–10. Supplementary Table 1: List of candidate off-target peptides reactive to TCRFLT3D/Y cells.Amino acid sequence of all potentially cross-reactive peptides predicted by the ScanProsite algorithm based on positive reactivities of the TCRFLT3D/Y cells against the peptide mimotope library of 161 peptides, shown in Fig. 1h. Supplementary Table 2: Clinical information for all leukemia patients included in the study.M, male; F, female; AML, acute myeloid leukemia; AMML, acute myelomonocytic leukemia; APL, acute promyelocytic leukemia. Cytogenetic characteristics from clinical diagnostics: TKD, tyrosine kinase domain; CFBbeta-MYH11 fusion inv(16)(p13,q22) is an inversion of chromosome 1–6, which results in a fused transcript between MYH11 (16p13.1) and CBFB (16q22.1); ITD, Internal tandem duplication; del(6q), deletion in long arm of chromosome 6. Allo-HSCT; allogeneic hematopoietic stem cell transplantation. Supplementary Table 3: Sequencing data for all FLT3D/Y positive leukemia patients.PT ID, Patient ID; Chr = chromosome; VAF, Variant allele frequency. Data for patients 16 was acquired using the TruSight™ Myeloid Panel. Data for patient 7 was acquired using the DFCI Rapid Heme Panel v2.0 (JAX Laboratories). Data for patient 8 was acquired by a custom-designed targeted DNA sequencing panel86. Supplementary Table 4: Number of events recorded by flow cytometry in PB, BM and spleen at terminal analysis for PDX models.Total event count is defined as single, viable, MNCs (mCD45+ cells + hCD45+ cells) adjusted for T cells by subtraction of hCD3+ events. Supplementary Table 5: ddPCR analysis of the FLT3 D835Y mutation in BM from micePD–X model with AML patient 7.Mouse ID; Population; Treatment; Number of cells analyzed; %VAF, variant allele frequency; CI, confidence interval; min, minimum %VAF of 95% CI; max, maximum %VAF of 95% CI; %CD33+ cells of hCD45+CD3- cells. Supplementary Table 6: Number of events recorded by flow cytometry in PB, BM and spleen at terminal analysis for PDX models.Total event count is defined as single, viable, MNCs (mCD45+ cells + hCD45+ cells) adjusted for T cells by subtraction of hCD3+ events. Supplementary Table 7: Somatic mutations in AML patient 1 BM blasts identified by whole exome sequencing.Somatic mutations identified by whole exome sequencing of blasts (CD3−CD19−) from BM of patient 1 were also detected in the two analyzed PDX mice transplanted with patient 1 blasts (one TCR1G4 cell treated mouse and one untreated mouse, both mCD45−hCD45+CD3−CD19). The FLT3D/Y mutation and a WT1 mutation (bold) were selected for further ddPCR analysis, shown in Fig. 4d and Extended Data Fig. 9c. Supplementary Table 8: ddPCR analysis of FLT3 D835Y and WT1 H507P mutations in BM from AML patient 1 and in transplanted PDX mice.Sample ID; Abbreviations; VAF, variant allele frequency; CI, confidence interval; min, mimimum VAF of 95% CI; max, maximum VAF of 95% CI. *Not enough cells for quantification. Supplementary Table 9: Number of events recorded by flow cytometry in PB, BM and spleen at terminal analysis for PDX models.Total event count is defined as single, viable, MNCs (mCD45+ cells + hCD45+ cells) adjusted for T cells by subtraction of hCD3+ events. Supplementary Table 10: Complete list of all antibodies used in the study.
Source data
Statistical source data for Fig. 1.
Statistical source data for Fig. 2.
Statistical source data for Fig. 3.
Statistical source data for Fig. 4.
Statistical source data for Fig. 5.
Statistical source data for Extended Data Fig. 1.
Statistical source data for Extended Data Fig. 3.
Statistical source data for Extended Data Fig. 4.
Statistical source data for Extended Data Fig. 6.
Statistical source data for Extended Data Fig. 7.
Statistical source data for Extended Data Fig. 8.
Statistical source data for Extended Data Fig. 9.
Acknowledgements
We thank the Oslo University Hospital (OUH) flow cytometry core facility for excellent technical assistance. This work was supported by the Research Council of Norway through its Centres of Excellence scheme (332727) and through grant 316060, the Norwegian Cancer Society grant 216135-2020, South-Eastern Regional Health Authority Norway (2021074), the Norwegian Childhood Cancer Foundation, Stiftelsen Kristian Gerhard Jebsen, the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 865805), the University of Oslo and Oslo University Hospital and the Novo Nordisk Foundation (to J.O.), by the Knut and Alice Wallenberg Foundation (KAW 2016.0105 to S.E.W.J. and KAW 2015.0195 to P.S.W.), the Swedish Research Council (538-2013-8995 to S.E.W.J. and 2015-03561 to P.S.W.) and by funding to S.E.W.J.: the Tobias Foundation (4-1122/2014), the Torsten Söderberg Foundation, the Center for Innovative Medicine at Karolinska Institutet (613/06) and the UK Medical Research Council (MC_UU_12009/5).
Extended data
Author contributions
E.G.: conceptualization, methodology, acquisition, analysis and interpretation of data, investigation, writing (original draft), writing (review and editing). M. Lehander and S.V.C.: methodology, acquisition, analysis and data interpretation, writing (original draft), writing (review and editing). W.Y., Y.L., T.K., M.M.N., R.C.B., T.T.T., T.Y., E.H.R., K.D., M. Laos, J.S.H., A.K.B., S.M., D.W.L.C.: acquisition, analysis and data interpretation. T.J.G., M.D.-S., A.H.: project administration, resources. M.A., A.M.: methodology. M.B., M.G., T.G.-D., S.L.: resources. S.E.W.J.: conceptualization, methodology, analysis and interpretation of data, investigation, supervision, funding acquisition, writing (original draft), writing (review and editing). P.S.W.: conceptualization, methodology, acquisition, analysis and interpretation of data, investigation, supervision, resources, writing (original draft), writing (review and editing). J.O.: conceptualization, methodology, supervision, analysis and interpretation of data, investigation, funding acquisition, writing (original draft), writing (review and editing).
Peer review
Peer review information
Nature Cancer thanks Saar Gill, Alexandre Harari and Claude Perreault for their contribution to the peer review of this work.
Data availability
The data that support the findings of this study are included in the text and in the Supplementary Information. Additional datasets used in the study are: the UniProt Homo sapiens database, using Mascot version 2.2.07 (https://www.matrixscience.com), curated human proteome databases UniProtKB/Swiss-Prot and the Protein Data Bank using the ScanProsite tool (https://prosite.expasy.org/scanprosite/), the pMHC class I binding prediction algorithm NetMHC version 4.0 (http://www.cbs.dtu.dk/services/NetMHC/), data from Papaemmanuil et al.15 (https://www.cbioportal.org) and data from Morita et al.45. Exome sequencing data have been deposited at the European Genome–Phenome Archive, which is hosted by the EBI and the CRG, under accession number EGAS00001007467 and can be shared according to institutional guidelines at Oslo University Hospital upon reasonable request. Mass spectrometry data have been deposited at the Proteomics Identification Database under accession number PXD043908. All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
Competing interests
A patent application has been filed by the Oslo University Hospital institutional technology transfer office Inven2 protecting the TCR sequence (J.O. and E.G. are inventors). J.O. is on the scientific advisory board of Asgard Therapeutics and is a cofounder of T-Rx therapeutics, a company that aims to develop TCR T cell therapies. The other co-authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sten Eirik W. Jacobsen, Petter S. Woll.
Contributor Information
Sten Eirik W. Jacobsen, Email: sten.eirik.jacobsen@ki.se
Petter S. Woll, Email: petter.woll@ki.se
Johanna Olweus, Email: johanna.olweus@medisin.uio.no.
Extended data
is available for this paper at 10.1038/s43018-023-00642-8.
Supplementary information
The online version contains supplementary material available at 10.1038/s43018-023-00642-8.
References
- 1.Coulie PG, et al. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc. Natl Acad. Sci. USA. 1995;92:7976–7980. doi: 10.1073/pnas.92.17.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolfel T, et al. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 3.Matsushita H, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castle JC, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72:1081–1091. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 5.van Rooij N, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J. Clin. Oncol. 2013;31:e439–e442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran E, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tran E, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med. 2016;375:2255–2262. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zacharakis N, et al. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat. Med. 2018;24:724–730. doi: 10.1038/s41591-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karpanen T, Olweus J. The potential of donor T-cell repertoires in neoantigen-targeted cancer immunotherapy. Front. Immunol. 2017;8:1718. doi: 10.3389/fimmu.2017.01718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foy SP, et al. Non-viral precision T cell receptor replacement for personalized cell therapy. Nature. 2023;615:687–696. doi: 10.1038/s41586-022-05531-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leidner R, et al. Neoantigen T-cell receptor gene therapy in pancreatic cancer. N. Engl. J. Med. 2022;386:2112–2119. doi: 10.1056/NEJMoa2119662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SP, et al. Adoptive cellular therapy with autologous tumor-infiltrating lymphocytes and T-cell receptor-engineered T cells targeting common p53 neoantigens in human solid tumors. Cancer Immunol. Res. 2022;10:932–946. doi: 10.1158/2326-6066.CIR-22-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papaemmanuil E, et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Socie G, Blazar BR. Acute graft-versus-host disease: from the bench to the bedside. Blood. 2009;114:4327–4336. doi: 10.1182/blood-2009-06-204669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mardiana S, Gill S. CAR T cells for acute myeloid leukemia: state of the art and future directions. Front. Oncol. 2020;10:697. doi: 10.3389/fonc.2020.00697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daver N, Alotaibi AS, Bucklein V, Subklewe M. T-cell-based immunotherapy of acute myeloid leukemia: current concepts and future developments. Leukemia. 2021;35:1843–1863. doi: 10.1038/s41375-021-01253-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenderian SS, et al. CD33-specific chimeric antigen receptor T cells exhibit potent preclinical activity against human acute myeloid leukemia. Leukemia. 2015;29:1637–1647. doi: 10.1038/leu.2015.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gill S, et al. Preclinical targeting of human acute myeloid leukemia and myeloablation using chimeric antigen receptor-modified T cells. Blood. 2014;123:2343–2354. doi: 10.1182/blood-2013-09-529537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jetani H, et al. CAR T-cells targeting FLT3 have potent activity against FLT3−ITD+ AML and act synergistically with the FLT3-inhibitor crenolanib. Leukemia. 2018;32:1168–1179. doi: 10.1038/s41375-018-0009-0. [DOI] [PubMed] [Google Scholar]
- 22.Olweus J, et al. Dendritic cell ontogeny: a human dendritic cell lineage of myeloid origin. Proc. Natl Acad. Sci. USA. 1997;94:12551–12556. doi: 10.1073/pnas.94.23.12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim MY, et al. Genetic inactivation of CD33 in hematopoietic stem cells to enable CAR T cell immunotherapy for acute myeloid leukemia. Cell. 2018;173:1439–1453. doi: 10.1016/j.cell.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haubner S, et al. Coexpression profile of leukemic stem cell markers for combinatorial targeted therapy in AML. Leukemia. 2019;33:64–74. doi: 10.1038/s41375-018-0180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tambaro FP, et al. Autologous CD33-CAR-T cells for treatment of relapsed/refractory acute myelogenous leukemia. Leukemia. 2021;35:3282–3286. doi: 10.1038/s41375-021-01232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laszlo GS, Estey EH, Walter RB. The past and future of CD33 as therapeutic target in acute myeloid leukemia. Blood Rev. 2014;28:143–153. doi: 10.1016/j.blre.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Ali, M. et al. T cells targeted to TdT kill leukemic lymphoblasts while sparing normal lymphocytes. Nat. Biotechnol.40, 488–498 (2021). [DOI] [PMC free article] [PubMed]
- 28.Chapuis AG, et al. T cell receptor gene therapy targeting WT1 prevents acute myeloid leukemia relapse post-transplant. Nat. Med. 2019;25:1064–1072. doi: 10.1038/s41591-019-0472-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Lee DI, et al. Mutated nucleophosmin 1 as immunotherapy target in acute myeloid leukemia. J. Clin. Invest. 2019;129:774–785. doi: 10.1172/JCI97482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie G, et al. CAR-T cells targeting a nucleophosmin neoepitope exhibit potent specific activity in mouse models of acute myeloid leukaemia. Nat. Biomed. Eng. 2021;5:399–413. doi: 10.1038/s41551-020-00625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biernacki MA, et al. CBFB–MYH11 fusion neoantigen enables T cell recognition and killing of acute myeloid leukemia. J. Clin. Invest. 2020;130:5127–5141. doi: 10.1172/JCI137723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daver N, Schlenk RF, Russell NH, Levis MJ. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia. 2019;33:299–312. doi: 10.1038/s41375-018-0357-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daver N, et al. Secondary mutations as mediators of resistance to targeted therapy in leukemia. Blood. 2015;125:3236–3245. doi: 10.1182/blood-2014-10-605808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stone RM, et al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N. Engl. J. Med. 2017;377:454–464. doi: 10.1056/NEJMoa1614359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith CC, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485:260–263. doi: 10.1038/nature11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith CC, Lin K, Stecula A, Sali A, Shah NP. FLT3 D835 mutations confer differential resistance to type II FLT3 inhibitors. Leukemia. 2015;29:2390–2392. doi: 10.1038/leu.2015.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith CC, et al. Heterogeneous resistance to quizartinib in acute myeloid leukemia revealed by single-cell analysis. Blood. 2017;130:48–58. doi: 10.1182/blood-2016-04-711820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stronen E, et al. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science. 2016;352:1337–1341. doi: 10.1126/science.aaf2288. [DOI] [PubMed] [Google Scholar]
- 39.Ali M, et al. Induction of neoantigen-reactive T cells from healthy donors. Nat. Protoc. 2019;14:1926–1943. doi: 10.1038/s41596-019-0170-6. [DOI] [PubMed] [Google Scholar]
- 40.Cohen CJ, Zhao Y, Zheng Z, Rosenberg SA, Morgan RA. Enhanced antitumor activity of murine–human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer Res. 2006;66:8878–8886. doi: 10.1158/0008-5472.CAN-06-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robbins PF, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J. Clin. Oncol. 2011;29:917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bethune MT, et al. Isolation and characterization of NY-ESO-1-specific T cell receptors restricted on various MHC molecules. Proc. Natl Acad. Sci. USA. 2018;115:E10702–E10711. doi: 10.1073/pnas.1810653115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcu A, et al. HLA Ligand Atlas: a benign reference of HLA-presented peptides to improve T-cell-based cancer immunotherapy. J. Immunother. Cancer. 2021;9:e002071. doi: 10.1136/jitc-2020-002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science. 2019;366:eaan4673. doi: 10.1126/science.aan4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morita K, et al. Clonal evolution of acute myeloid leukemia revealed by high-throughput single-cell genomics. Nat. Commun. 2020;11:5327. doi: 10.1038/s41467-020-19119-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shlush LI, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goardon N, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19:138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 48.Lapidot T, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 49.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 50.Schuurhuis GJ, et al. Minimal/measurable residual disease in AML: a consensus document from the European LeukemiaNet MRD Working Party. Blood. 2018;131:1275–1291. doi: 10.1182/blood-2017-09-801498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pearlman AH, et al. Targeting public neoantigens for cancer immunotherapy. Nat. Cancer. 2021;2:487–497. doi: 10.1038/s43018-021-00210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bassani-Sternberg M, Coukos G. Mass spectrometry-based antigen discovery for cancer immunotherapy. Curr. Opin. Immunol. 2016;41:9–17. doi: 10.1016/j.coi.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 53.Loffler MW, et al. Multi-omics discovery of exome-derived neoantigens in hepatocellular carcinoma. Genome Med. 2019;11:28. doi: 10.1186/s13073-019-0636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sykulev Y, Joo M, Vturina I, Tsomides TJ, Eisen HN. Evidence that a single peptide–MHC complex on a target cell can elicit a cytolytic T cell response. Immunity. 1996;4:565–571. doi: 10.1016/S1074-7613(00)80483-5. [DOI] [PubMed] [Google Scholar]
- 55.Scheper W, et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat. Med. 2019;25:89–94. doi: 10.1038/s41591-018-0266-5. [DOI] [PubMed] [Google Scholar]
- 56.Lowery FJ, et al. Molecular signatures of antitumor neoantigen-reactive T cells from metastatic human cancers. Science. 2022;375:877–884. doi: 10.1126/science.abl5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagarsheth NB, et al. TCR-engineered T cells targeting E7 for patients with metastatic HPV-associated epithelial cancers. Nat. Med. 2021;27:419–425. doi: 10.1038/s41591-020-01225-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sahin U, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 59.Ott PA, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547:217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lang, F., Schrors, B., Lower, M., Tureci, O. & Sahin, U. Identification of neoantigens for individualized therapeutic cancer vaccines. Nat. Rev. Drug Discov.21, 261–282 (2022). [DOI] [PMC free article] [PubMed]
- 61.Johnson LA, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin BY, et al. Engineered T cells targeting E7 mediate regression of human papillomavirus cancers in a murine model. JCI Insight. 2018;3:e99488. doi: 10.1172/jci.insight.99488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duan F, et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J. Exp. Med. 2014;211:2231–2248. doi: 10.1084/jem.20141308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chandran SS, et al. Immunogenicity and therapeutic targeting of a public neoantigen derived from mutated PIK3CA. Nat. Med. 2022;28:946–957. doi: 10.1038/s41591-022-01786-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Foldvari Z, Knetter C, Yang W, et al. A systematic safety pipeline for selection of T-cell receptors to enter clinical use. NPJ Vaccines10.1038/s41541-023-00713-y (2023). [DOI] [PMC free article] [PubMed]
- 66.Jan M, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci. Transl. Med. 2012;4:149ra18. doi: 10.1126/scitranslmed.3004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mansilla-Soto J, et al. HLA-independent T cell receptors for targeting tumors with low antigen density. Nat. Med. 2022;28:345–352. doi: 10.1038/s41591-021-01621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dolgin E. First soluble TCR therapy opens ‘new universe’ of cancer targets. Nat. Biotechnol. 2022;40:441–444. doi: 10.1038/s41587-022-01282-6. [DOI] [PubMed] [Google Scholar]
- 69.Abrahamsen IW, et al. Targeting B cell leukemia with highly specific allogeneic T cells with a public recognition motif. Leukemia. 2010;24:1901–1909. doi: 10.1038/leu.2010.186. [DOI] [PubMed] [Google Scholar]
- 70.Kumari S, et al. Alloreactive cytotoxic T cells provide means to decipher the immunopeptidome and reveal a plethora of tumor-associated self-epitopes. Proc. Natl Acad. Sci. USA. 2014;111:403–408. doi: 10.1073/pnas.1306549111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toebes M, et al. Design and use of conditional MHC class I ligands. Nat. Med. 2006;12:246–251. doi: 10.1038/nm1360. [DOI] [PubMed] [Google Scholar]
- 72.Hadrup SR, et al. Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nat. Methods. 2009;6:520–526. doi: 10.1038/nmeth.1345. [DOI] [PubMed] [Google Scholar]
- 73.Linnemann C, et al. High-throughput identification of antigen-specific TCRs by TCR gene capture. Nat. Med. 2013;19:1534–1541. doi: 10.1038/nm.3359. [DOI] [PubMed] [Google Scholar]
- 74.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toebes, M., Rodenko, B., Ovaa, H. & Schumacher, T. N. Generation of peptide MHC class I monomers and multimers through ligand exchange. Curr. Protoc. Immunol.87, 18.16.1–18.16.20 (2009). [DOI] [PubMed]
- 76.Sikorski K, et al. A high-throughput pipeline for validation of antibodies. Nat. Methods. 2018;15:909–912. doi: 10.1038/s41592-018-0179-8. [DOI] [PubMed] [Google Scholar]
- 77.Carrelha J, et al. Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature. 2018;554:106–111. doi: 10.1038/nature25455. [DOI] [PubMed] [Google Scholar]
- 78.Woll PS, et al. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell. 2014;25:794–808. doi: 10.1016/j.ccr.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 79.Gregory T, et al. Characterization and mitigation of fragmentation enzyme-induced dual stranded artifacts. NAR Genom. Bioinform. 2020;2:lqaa070. doi: 10.1093/nargab/lqaa070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shiraishi Y, et al. An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res. 2013;41:e89. doi: 10.1093/nar/gkt126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoshizato T, et al. Genetic abnormalities in myelodysplasia and secondary acute myeloid leukemia: impact on outcome of stem cell transplantation. Blood. 2017;129:2347–2358. doi: 10.1182/blood-2016-12-754796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rampal R, Figueroa ME. Wilms tumor 1 mutations in the pathogenesis of acute myeloid leukemia. Haematologica. 2016;101:672–679. doi: 10.3324/haematol.2015.141796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jain N, et al. TET2 guards against unchecked BATF3-induced CAR T cell expansion. Nature. 2023;615:315–322. doi: 10.1038/s41586-022-05692-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jurtz V, et al. NetMHCpan-4.0: improved peptide–MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang M, et al. Validation of risk stratification models in acute myeloid leukemia using sequencing-based molecular profiling. Leukemia. 2017;31:2029–2036. doi: 10.1038/leu.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables 1–10. Supplementary Table 1: List of candidate off-target peptides reactive to TCRFLT3D/Y cells.Amino acid sequence of all potentially cross-reactive peptides predicted by the ScanProsite algorithm based on positive reactivities of the TCRFLT3D/Y cells against the peptide mimotope library of 161 peptides, shown in Fig. 1h. Supplementary Table 2: Clinical information for all leukemia patients included in the study.M, male; F, female; AML, acute myeloid leukemia; AMML, acute myelomonocytic leukemia; APL, acute promyelocytic leukemia. Cytogenetic characteristics from clinical diagnostics: TKD, tyrosine kinase domain; CFBbeta-MYH11 fusion inv(16)(p13,q22) is an inversion of chromosome 1–6, which results in a fused transcript between MYH11 (16p13.1) and CBFB (16q22.1); ITD, Internal tandem duplication; del(6q), deletion in long arm of chromosome 6. Allo-HSCT; allogeneic hematopoietic stem cell transplantation. Supplementary Table 3: Sequencing data for all FLT3D/Y positive leukemia patients.PT ID, Patient ID; Chr = chromosome; VAF, Variant allele frequency. Data for patients 16 was acquired using the TruSight™ Myeloid Panel. Data for patient 7 was acquired using the DFCI Rapid Heme Panel v2.0 (JAX Laboratories). Data for patient 8 was acquired by a custom-designed targeted DNA sequencing panel86. Supplementary Table 4: Number of events recorded by flow cytometry in PB, BM and spleen at terminal analysis for PDX models.Total event count is defined as single, viable, MNCs (mCD45+ cells + hCD45+ cells) adjusted for T cells by subtraction of hCD3+ events. Supplementary Table 5: ddPCR analysis of the FLT3 D835Y mutation in BM from micePD–X model with AML patient 7.Mouse ID; Population; Treatment; Number of cells analyzed; %VAF, variant allele frequency; CI, confidence interval; min, minimum %VAF of 95% CI; max, maximum %VAF of 95% CI; %CD33+ cells of hCD45+CD3- cells. Supplementary Table 6: Number of events recorded by flow cytometry in PB, BM and spleen at terminal analysis for PDX models.Total event count is defined as single, viable, MNCs (mCD45+ cells + hCD45+ cells) adjusted for T cells by subtraction of hCD3+ events. Supplementary Table 7: Somatic mutations in AML patient 1 BM blasts identified by whole exome sequencing.Somatic mutations identified by whole exome sequencing of blasts (CD3−CD19−) from BM of patient 1 were also detected in the two analyzed PDX mice transplanted with patient 1 blasts (one TCR1G4 cell treated mouse and one untreated mouse, both mCD45−hCD45+CD3−CD19). The FLT3D/Y mutation and a WT1 mutation (bold) were selected for further ddPCR analysis, shown in Fig. 4d and Extended Data Fig. 9c. Supplementary Table 8: ddPCR analysis of FLT3 D835Y and WT1 H507P mutations in BM from AML patient 1 and in transplanted PDX mice.Sample ID; Abbreviations; VAF, variant allele frequency; CI, confidence interval; min, mimimum VAF of 95% CI; max, maximum VAF of 95% CI. *Not enough cells for quantification. Supplementary Table 9: Number of events recorded by flow cytometry in PB, BM and spleen at terminal analysis for PDX models.Total event count is defined as single, viable, MNCs (mCD45+ cells + hCD45+ cells) adjusted for T cells by subtraction of hCD3+ events. Supplementary Table 10: Complete list of all antibodies used in the study.
Statistical source data for Fig. 1.
Statistical source data for Fig. 2.
Statistical source data for Fig. 3.
Statistical source data for Fig. 4.
Statistical source data for Fig. 5.
Statistical source data for Extended Data Fig. 1.
Statistical source data for Extended Data Fig. 3.
Statistical source data for Extended Data Fig. 4.
Statistical source data for Extended Data Fig. 6.
Statistical source data for Extended Data Fig. 7.
Statistical source data for Extended Data Fig. 8.
Statistical source data for Extended Data Fig. 9.
Data Availability Statement
The data that support the findings of this study are included in the text and in the Supplementary Information. Additional datasets used in the study are: the UniProt Homo sapiens database, using Mascot version 2.2.07 (https://www.matrixscience.com), curated human proteome databases UniProtKB/Swiss-Prot and the Protein Data Bank using the ScanProsite tool (https://prosite.expasy.org/scanprosite/), the pMHC class I binding prediction algorithm NetMHC version 4.0 (http://www.cbs.dtu.dk/services/NetMHC/), data from Papaemmanuil et al.15 (https://www.cbioportal.org) and data from Morita et al.45. Exome sequencing data have been deposited at the European Genome–Phenome Archive, which is hosted by the EBI and the CRG, under accession number EGAS00001007467 and can be shared according to institutional guidelines at Oslo University Hospital upon reasonable request. Mass spectrometry data have been deposited at the Proteomics Identification Database under accession number PXD043908. All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.