Abstract
Background
The predictive efficacy of current biomarker of immune checkpoint inhibitors (ICIs) is not sufficient. This study investigated the causality between radiomic biomarkers and immunotherapy response status in patients with stage IB–IV non-small cell lung cancer (NSCLC), including its biological context for ICIs treatment response prediction.
Methods
CT images from 319 patients with pretreatment NSCLC receiving immunotherapy between January 2015 and November 2021 were retrospectively collected and composed a discovery (n=214), independent validation (n=54), and external test cohort (n=51). A set of 851 features was extracted from tumorous and peritumoral volumes of interest (VOIs). The reference standard is the durable clinical benefit (DCB, sustained disease control for more than 6 months assessed via radiological evaluation). The predictive value of combined radiomic signature (CRS) for pathological response was subsequently assessed in another cohort of 98 patients with resectable NSCLC receiving ICIs preoperatively. The association between radiomic features and tumor immune landscape on the online data set (n=60) was also examined. A model combining clinical predictor and radiomic signatures was constructed to improve performance further.
Results
CRS discriminated DCB and non-DCB patients well in the training and validation cohorts with an area under the curve (AUC) of 0.82, 95% CI: 0.75 to 0.88, and 0.75, 95% CI: 0.64 to 0.87, respectively. In this study, the predictive value of CRS was better than programmed cell death ligand-1 (PD-L1) expression (AUC of PD-L1 subset: 0.59, 95% CI: 0.50 to 0.69) or clinical model (AUC: 0.66, 95% CI: 0.51 to 0.81). After combining the clinical signature with CRS, the predictive performance improved further with an AUC of 0.837, 0.790 and 0.781 in training, validation and D2 cohorts, respectively. When predicting pathological response, CRS divided patients into a major pathological response (MPR) and non-MPR group (AUC: 0.76, 95% CI: 0.67 to 0.81). Moreover, CRS showed a promising stratification ability on overall survival (HR: 0.49, 95% CI: 0.27 to 0.89; p=0.020) and progression-free survival (HR: 0.43, 95% CI: 0.26 to 0.74; p=0.002).
Conclusion
By analyzing both tumorous and peritumoral regions of CT images in a radiomic strategy, we developed a non-invasive biomarker for distinguishing responders of ICIs therapy and stratifying their survival outcome efficiently, which may support the clinical decisions on the use of ICIs in advanced as well as patients with resectable NSCLC.
Keywords: immunotherapy, tumor biomarkers, non-small cell lung cancer
WHAT IS ALREADY KNOWN ON THIS TOPIC.
Current biomarkers for immunotherapy such as programmed cell death ligand-1 rely on invasive examination, and their accuracy is limited. Previous studies reported CT-based machine learning strategy could predict immunotherapy response for patients with advanced non-small cell lung cancer (NSCLC). However, the comparison of the peritumoral signatures and extensive application for earlier stage patients have not been explored.
WHAT THIS STUDY ADDS
This multicentered study developed the radiomic model, an efficient and non-invasive method, for immunotherapy response prediction for patients with stage IB–IV NSCLC. This is the first study to explore the application of radiomic signature for immune checkpoint inhibitors (ICIs) response prediction in patients with both advanced and earlier stage NSCLC, from which the importance of peritumoral features is highlighted.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
This study provides insights into the application of radiomic biomarker from advanced to earlier stage NSCLC. It confirms the potential of radiomics to identify patients with NSCLC most likely to respond to ICIs-based therapies and underlying prognostic value. The radiomics model was not influenced by tumor volume or pulmonary lesion choice strategy and could be widely used to aid clinicians in making decisions of ICIs-based therapies for patients with NSCLC.
Introduction
Immune checkpoint inhibitors (ICIs) for programmed cell death protein 1 (PD-1)/programmed cell death ligand-1 (PD-L1) have become the first-line standard therapy for advanced non-small cell lung cancer (NSCLC).1 Inspiringly, PD-1-based neoadjuvant therapy has been approved for patients with resectable NSCLC.2 Abnormalities in the immune checkpoint pathway prompt tumor cells to escape immune surveillance and proliferate continuously. ICIs can reactivate the immunity to cancer cells and have been adopted in the therapy of malignant tumors because of their remarkable tumor-killing effects and sustaining clinical benefits.3 Unfortunately, the responder rate is heterogeneous in current clinical trials of immunotherapy, with 20–50%4–6 among advanced NSCLC and 24%7 in resectable NSCLC.
To filter potential responders, improve clinical outcome, save medical cost and provide insight into tumor immune evasion mechanisms, it is imperative to develop predictive biomarker for immunotherapy response to stratify patients of different clinical outcomes. For this reason, many biomarkers have been explored, such as PD-L1 expression,8 tumor mutational load9 and tumor infiltrate immune cells.10 However, due to intra-patient tumor heterogeneity and the sampling bias brought by intrinsic tumor heterogeneity, they are still insufficient to provide promising predictive efficiency.11
Radiomics is the science which converts medical images into high-dimensional minable data and analyzes them by applying bioinformatics algorithms.12 It has several advantages, including (1) non-invasive; (2) standard image acquisition widely adopted in clinical imaging; and (3) less sampling bias for acquisition of information for the whole tumor. Some investigators have revealed that the radiomic models could effectively identify patients with advanced NSCLC sensitive to immunotherapy and implied the associations between radiomic features and immunotherapy response.13–15 However, limited by the therapy background of advanced NSCLC, practically no routine pathological response could be evaluated and thereby pathological validation was usually absent in their findings. In addition, tumor immune microenvironment played a crucial role in the cause of ICIs treatment failure,16 thereby the radiomic characteristics of peritumoral area may reveal some patterns of microenvironment and reflect the local immune response. In many cases, angiogenesis can be observed near the tumor due to a pathological response to hypoxia.17 The disorganization of blood vessels around the tumor will generate anoxic areas in the tumor microenvironment, thus reducing the efficacy of drugs.18 Therefore, peritumoral pulmonary parenchyma probably might give substantial value for the pretreatment prediction of immunotherapy response in radiomic analysis, which was often ignored in previous studies.19–22
In this work, we investigated the correlation between response of immunotherapy and radiomic features and constructed series radiomic models based on different scales of peritumoral and tumorous area. We sought to explore whether radiomic features of both tumorous and peritumoral areas could identify the responders of IB–IV stage NSCLC from ICIs therapy. The radiomic signatures were subsequently validated in the external validation cohort and pathological validation cohort. The associations of these radiomic features with overall survival (OS) and disease-free survival in patients with advanced NSCLC were evaluated. Finally, the utility of the model coupled with clinical variables and the biological basis of radiomic signature were explored.
Materials and methods
Data sets and patient enrollment
In this multicohort study, five separated cohorts of patients diagnosed with NSCLC were retrospectively included (online supplemental figure S1). The discovery data set D1, the test data set D2 and pathological validation data set D4 included patients receiving ICIs therapy in Guangdong Provincial People’s Hospital (GPPH) and external validation data set D3 was from Jiangxi Cancer Hospital (JXCH); and the bio-validation data set D5 was downloaded from the databases of The Cancer Imaging Archive (TCIA) and The Cancer Genome Atlas (TCGA).
jitc-2023-007369supp001.pdf (197.9KB, pdf)
jitc-2023-007369supp002.pdf (10.7KB, pdf)
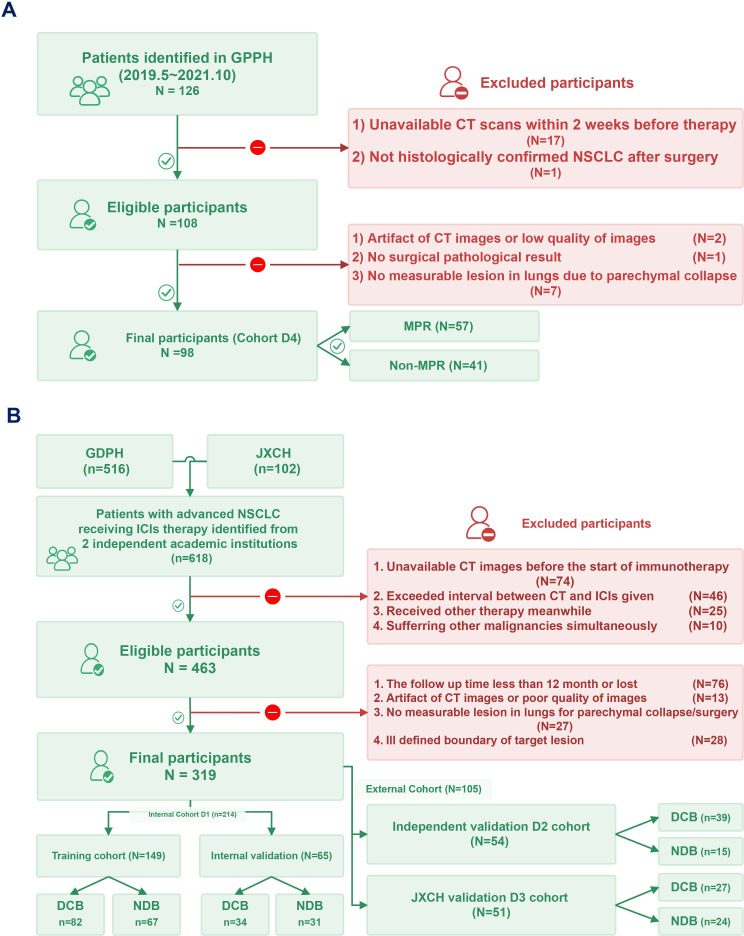
To develop the discovery data set D1, we identified 516 patients with advanced NSCLC admitted between January 2015 and June 2020 at GPPH, which were treated using PD-L1 or PD-1 single agent, or the combination of ICIs with platin-based regimen based on patient eligibility (figure 1). The major inclusion criteria were as follows (online supplemental appendix S1 for details): (1) above 18 years old; (2) patients receiving PD-1/PD-L1 blockade and had pathologically confirmed NSCLC (stage III and IV); (3) the data of chest thin-slice (≤5 mm) CT within 30 days before the first dose of immunotherapy were available. Patients with poor CT quality or whose pulmonary lesions were poorly discriminated from other lesions or adjacent tissues were excluded. Clinical statistics were obtained through manual abstraction from electronic medical records and patients whose follow-up period less than 6 months were excluded. Finally, 214 patients were included and randomly divided into training (n=149) and internal validation (n=65) cohorts for developing and validating the radiomic signature, respectively.
Figure 1.
The patient eligibility of entire cohorts. The training cohort comprising clinical data and the corresponding extracted imaging data of the retrospective patients were used to train the clinics-radiomics signature and OS Cox models, which were further validated using an external validation cohort from Jiangxi Cancer Hospital enrolled according to the inclusion and exclusion criteria. DCB, durable clinical benefit; GPPH, Guangdong Provincial People’s Hospital; ICIs, immune checkpoint inhibitors; JXCH, Jiangxi Cancer Hospital; MPR, major pathological response; NDB, non-durable clinical benefit; NSCLC, non-small cell lung cancer; OS, overall survival.
Four independent data sets were used to validate the radiomic signature. The same inclusion and exclusion criteria (details seen in online supplemental appendix S1) were applied to test data set D2 and external validation data set D3, retrospectively enrolling 54 patients in GPPH during July 2020 and June 2021, and 51 patients in JXCH during March 2019 and November 2021 respectively.
The pathological validation data set D4 consisted of 98 patients with stage IB–III NSCLC receiving radical resection of lung carcinoma after ICIs therapy between May 2019 and October 2021 at GPPH. This data set was to evaluate the prediction potential of the radiomic signature for the pathological response from ICIs and explore the concordance of the radiologic response with pathological response.
The biological validation data set D5 was extracted from the TCIA with available CT image data and corresponding transcriptomic data from TCGA. TCGA-NSCLC data set has 66 samples with matching pretreatment CT imaging and transcriptomic data, of which 6 samples were excluded due to unsatisfactory image quality. This TCGA-NSCLC data set with 60 patients was applied to validate the concordance between the radiomic signature and tumor microenvironmental characteristics.
Clinical endpoints evaluation
The primary predictor was the response status, durable clinical benefit (DCB) from immunotherapy, defined as partial response (PR) or sustained disease (SD) stabilization lasting longer than 6 months based on Response Evaluation Criteria in Solid Tumor (RECIST) (V.1.1) criteria. Besides, OS was the secondary endpoint for long-term efficacy, defined as from initiation of therapy to the date of last follow-up or date of death. The response outcome for the pathological validation data set is major pathological response23 (MPR) defined as less than 10% of a viable tumor in the primary tumor bed, and no viable tumor was observed in both tumor bed and lymph nodes were defined as a pathologic complete response (pCR). And the preoperative objective response rate (ORR) according to radiologic assessment adhering to RECIST (V.1.1) criteria was also recorded for further evaluation. Besides, the clinical stage of carcinoma was assessed based on the eighth TNM (tumor-node-metastasis) system and any discrepancy about the diagnosis of an illness was settled by consensus through discussion.
CT image acquisition and radiomic features extraction
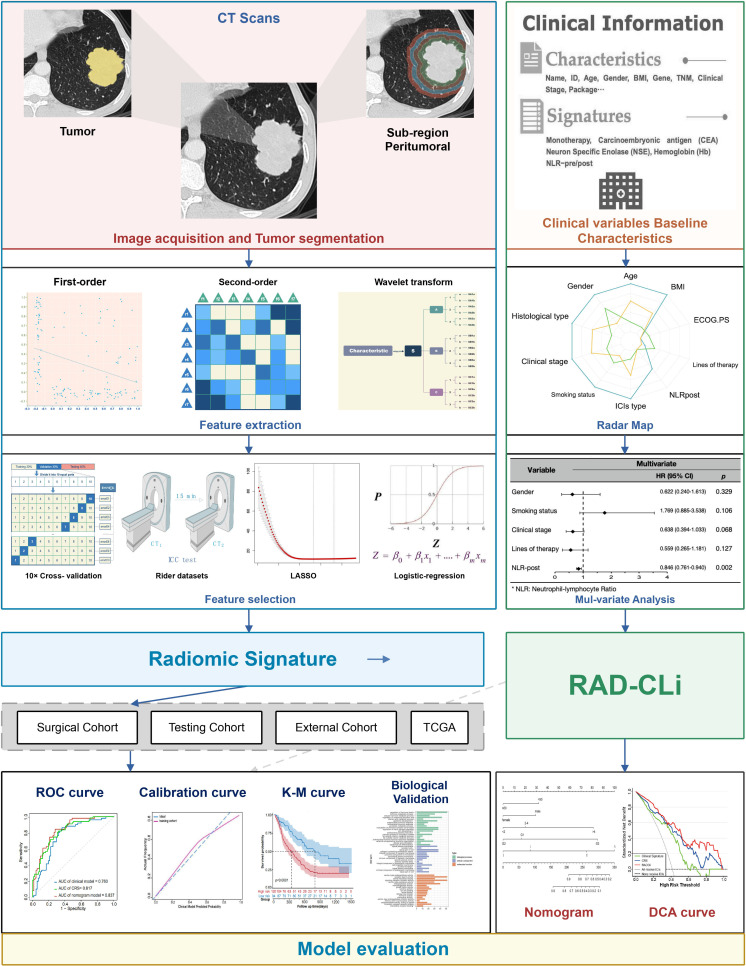
Through the Picture Archiving and Communication System, contrast-enhanced thoracic CT completed within 1 month of the initiation of ICIs therapy were obtained from two medical centers (details seen in online supplemental appendix S3). After quality and timing verification, these slices were segmented semi-manually with 3D Slicer (http://www.slicer.org) software by two experienced radiologists, SW (with 14 years of experience) and GW (with 18 years of experience) who were blind to the response status. After applying “draw”, “level tracing” and “smoothing” methods to outline the primary tumors in three orthogonally oriented planes, the three-dimensional (3D) volumes of interest (VOIs) of tumors were generated (details seen in online supplemental appendix S4). Radiologist LL (with 23 years of experience) assessed all tumor segmentations. Any disagreements were resolved by discussions between the three radiologists mentioned and three thoracic surgeons (QG, D-KZ, and HY).
The primary or largest pulmonary lesion was defined as the target lesion and 851 radiomic features were extracted from both VOIs of the tumor and peritumoral regions by applying the SlicerRadiomics package on the 3D Slicer platform. The effect of different lesion choice strategy was evaluated (details seen in online supplemental appendix S7.3). A two-way random intraclass correlation coefficient (ICC) model was applied to evaluate the impact of interobserver variability of the radiomic features between two radiologists (SW and GW) image segmentation and feature extraction process. Peritumoral regions were created by automated 20 mm dilation of tumor region, resulting in four annular rings of 5 mm each (figure 2).
Figure 2.
The workflow for the development of the integrated model to predict the responders from patients with NSCLC. AUC, area under the curve; BMI, body mass index; CRS, combined radiomic signature; DCA, decision curve analysis; ECOG PS, Eastern Cooperative Oncology Group performance status; ICC, intraclass correlation coefficient; ICIs, immune checkpoint inhibitors; K-M, Kaplan-Meier; NLR, neutrophil-to-lymphocyte ratio; NLRpost, NLR after first cycle of therapy; RAD-CLi, radiomic-clinical prediction model; ROC, receiver-operator characteristic; TCGA, The Cancer Genome Atlas; TNM, tumor-node-metastasis.
Radiomic features selection and radiomic signatures building
To guarantee the stability of the signature, non-reproducible or non-robust radiomic features were filtered on the Reference Image Database to Evaluate Therapy Response24 (RIDER, details seen in online supplemental appendix S5) data set using the ICC test. After filtered the highly-correlated features via the Spearman correlation analysis, the least absolute shrinkage and selection operator (LASSO) and a 10-fold cross-validation method with optimal λ were applied to filter out redundant features further. We applied LASSO and stepwise logistic regression as definite classifier because this configuration reached the highest performance with the least amount of overfitting (online supplemental appendix S8 and table S7). To predict response status, radiomic signatures were built based on predictive significant features of different regions of peritumoral areas using a binary stepwise logistic regression method. The performance and predictive efficacy of radiomic signatures were compared. Independent validations of the optimal radiomic signature were applied for the optimal radiomic signature in data sets D2 and D3 respectively.
jitc-2023-007369supp018.pdf (73.2KB, pdf)
Survival analysis
We performed log-rank statistical tests and Cox proportional hazard model analysis to evaluate the univariable discriminative ability of the variables and estimate the prognostic value of radiomic signature on OS. The discriminative ability was compared with traditional biomarkers like PD-L1 expression in the PD-L1 expression available subset (n=138) derived from data set D1 and D2, where the independent prognostic advantage of the radiomic signature was evaluated. The effect of tumor volume on combined radiomic signature (CRS and long-term survival was also evaluated (online supplemental appendix S6).
The association between features of radiomic signature on the molecular pathway
The R package “limma” was applied to identify differentially expressed genes (DEGs) between the high-risk group and low-risk group based on the radiomic signature. The functional annotation was analyzed using the Gene Ontology (GO) functional analysis while the pathway enrichment analysis of DEGs were investigated through the Kyoto Encyclopedia of Genes and Genomes pathway analysis. In terms of the potential functional biological pathway between the two groups, Gene Set Enrichment Analysis (GSEA) was applied with annotated gene collection “c2.cp.v2022.1.Hs.symbols.gmt” for canonical biological pathways and “c7.immunesigdb.v2022.1.Hs.symbols.gmt” for immunologic signature genes through the Molecular Signatures Database (http://www.gsea-msigdb.org/gsea/msigdb/). A gene set of 28 immune cells were imported to a single sample GSEA to quantify the infiltration level of immune cells in the tumor microenvironment by R package “GSVA”. Tumor Immune Dysfunction and Exclusion (TIDE, http://tide.dfci.harvard.edu/) score was estimated, indicating the potential condition of tumor immune exclusion and dysfunction. Responders of immune checkpoint inhibitors could be predicted by the TIDE algorithm as well. Correlations of radiomic signature and tumor immune landscape were calculated by spearmen correlation analysis. All the analysis tasks and visualization were processed in R software.
Development of a radiomic-clinical prediction model
Based on clinical information (gender, histologic type, clinical stage, etc) collected from the subset of patients, we developed a clinical signature after recognizing significant risk variables via stepwise regression method. The radiomic signature and clinical signature were combined to form a nomogram using logistic regression based on the D1 set. To assess the generalization ability of radiomic-clinical prediction model (RADCli), independent validation was applied to data sets D2 and D3.
Statistical analysis
P value<0.05 was considered statistically significant in all two-tailed tests. All statistical analyses were performed using R Project V.4.1.3 (http://www.r-project.org/) and SPSS V.23.0 (IBM, Armonk, New York, USA). For categorical data, we adopted Fisher’s exact test, Mann-rank Whitney’s sum test, and analysis of variance test for continuous variables and differences for the clinical covariates between responder and non-responders were evaluated. Model efficiencies were evaluated quantitatively using the area under the receiver-operator characteristic where sensitivity, specificity, and accuracy were compared among every signature. The Hosmer-Lemeshow test was used to assess model goodness of fit, drawing calibration curves. In addition, a two-way random ICC test was applied to assess the agreement between two times VOIs from the RIDER database and features whose ICC larger than 0.75 remained as stable features. We used the Akaike information criterion to select the optimal signature. The clinical utility was evaluated on the training and validation cohort using DCA.25 The R packages applied in this investigation are summarized in online supplemental appendix S2.
Result
Clinicopathologic characteristics of cohorts
A total of 744 patients with NSCLC receiving ICIs therapy were identified and 409 patients were eligible for analysis (online supplemental figure S1). The number of participants in the training, validation, test, external validation, and pathological validation cohorts were 149, 65, 54, 51, and 98, respectively. The baseline characteristics of the patients in this investigation are summarized in table 1 and online supplemental table S1. None of these characteristics differed significantly between the training and validation cohorts (p>0.05). The advanced NSCLC cohorts (D1, D2, D3), comprising 319 patients in total, included 262 (82.1%) men and 214 adenocarcinomas (67.1%), with a mean age of 59.3 years. Most of the participants received the anti-PD-1-based-regime (87.8%) and about one-half showed durable clinical benefit (52.4%). An unbalanced baseline characteristic was observed between D1 and D2 cohorts due to an independent cohort design. In the discovery data set (table 1), gender, clinical stage, smoking status, lines of therapy (LOT), and neutrophil-to-lymphocyte ratio after the first cycle of therapy (NLRpost) were identified as significant predictors for DCB (p<0.05).
Table 1.
Clinicopathological characteristics of the discovery data set
| Non-responder (N=113) | Responder (N=101) | P value | |
| Gender | |||
| Male | 85 (75.2%) | 91 (90.1%) | 0.007 |
| Female | 28 (24.8%) | 10 (9.9%) | |
| Age | |||
| Mean (SD) | 58.7 (12.5) | 57.5 (14.6) | 0.543 |
| BMI | |||
| Mean (SD) | 21.4 (3.01) | 21.8 (2.95) | 0.430 |
| Missing | 3 (2.7%) | 1 (1.0%) | |
| ECOG | |||
| 0–1 | 99 (87.6%) | 96 (95.0%) | 0.08984 |
| ≥2 | 14 (12.4%) | 5 (5.0%) | |
| Smoking status | |||
| Never smoked | 63 (55.8%) | 36 (35.6%) | 0.002 |
| Current or former smoker | 46 (40.7%) | 63 (62.4%) | |
| Missing | 4 (3.5%) | 2 (2.0%) | |
| Smoking package year | |||
| Mean (SD) | 15.0 (22.3) | 18.9 (24.4) | 0.226 |
| Missing | 1 (0.9%) | 1 (1.0%) | |
| Tumor histologic type | |||
| LUAD | 86 (76.1%) | 79 (78.2%) | 0.747 |
| LUSC | 27 (23.9%) | 22 (21.8%) | |
| Pathological stage | |||
| III | 9 (8.0%) | 12 (11.9%) | 0.004 |
| IVA | 30 (26.5%) | 47 (46.5%) | |
| IVB | 74 (65.5%) | 42 (41.6%) | |
| LOT | |||
| First line | 23 (20.4%) | 42 (41.6%) | < 0.001 |
| Second line or more | 90 (79.6%) | 59 (58.4%) | |
| ICIs | |||
| Anti PD-1 | 98 (86.7%) | 80 (79.2%) | 0.149 |
| Anti PD-L1 | 15 (13.3%) | 21 (20.8%) | |
BMI, body mass index; ECOG, Eastern Cooperative Oncology Group; ICIs, immune checkpoint inhibitors; LOT, line of therapy; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PD-1, programmed death-1; PD-L1, programmed cell death-ligand 1.
jitc-2023-007369supp012.pdf (79.4KB, pdf)
In the pathological validation data set (table 2), most of them were men (86.7%) and non-smokers (54.1%), with a mean age of 59.7 years. The majority of histological type was squamous carcinoma (62.2%) and the predominant pathological stage was stage III (70.4%). Most participants received the carboplatin-based regime (81.6%) and nivolumab is the majority for ICIs choice (55.1%).
Table 2.
Clinicopathological characteristics of the surgical data set D4
| Non-MPR (N=41) | MPR (N=57) | Total (N=98) | P value | |
| Gender | ||||
| Male | 33 (80.5%) | 52 (91.2%) | 85 (86.7%) | 0.141 |
| Female | 8 (19.5%) | 5 (8.8%) | 13 (13.3%) | |
| Age | ||||
| Mean (SD) | 58.4 (9.85) | 60.7 (9.39) | 59.7 (9.60) | 0.249 |
| BMI | ||||
| Mean (SD) | 23.2 (2.88) | 22.6 (2.78) | 22.9 (2.82) | 0.363 |
| Missing | 1 (2.4%) | 2 (3.5%) | 3 (3.1%) | |
| Smoking | ||||
| Never smoked | 26 (63.4%) | 27 (47.4%) | 53 (54.1%) | 0.151 |
| Current or former smoker | 15 (36.6%) | 30 (52.6%) | 45 (45.9%) | |
| ECOG | ||||
| 0 | 36 (87.8%) | 55 (96.5%) | 91 (92.9%) | 0.126 |
| 1 | 5 (12.2%) | 2 (3.5%) | 7 (7.1%) | |
| Pathological type | ||||
| LUAD | 20 (48.8%) | 11 (19.3%) | 31 (31.6%) | 0.002 |
| LUSC | 17 (41.5%) | 44 (77.2%) | 61 (62.2%) | |
| NOS | 4 (9.8%) | 2 (3.5%) | 6 (6.1%) | |
| T stage | ||||
| T1 | 6 (14.6%) | 5 (8.8%) | 11 (11.2%) | 0.661 |
| T2 | 17 (41.5%) | 21 (36.8%) | 38 (38.8%) | |
| T3 | 11 (26.8%) | 17 (29.8%) | 28 (28.6%) | |
| T4 | 7 (17.1%) | 14 (24.6%) | 21 (21.4%) | |
| N stage | ||||
| N0 | 7 (17.1%) | 13 (22.8%) | 20 (20.4%) | 0.598 |
| N1 | 13 (31.7%) | 22 (38.6%) | 35 (35.7%) | |
| N2 | 19 (46.3%) | 21 (36.8%) | 40 (40.8%) | |
| N3 | 2 (4.9%) | 1 (1.8%) | 3 (3.1%) | |
| M stage | ||||
| M0 | 40 (97.6%) | 56 (98.2%) | 96 (98.0%) | 1.000 |
| M1 | 1 (2.4%) | 1 (1.8%) | 2 (2.0%) | |
| Clinical stage | ||||
| I | 1 (2.4%) | 4 (7.0%) | 5 (5.1%) | 0.706 |
| II | 8 (19.5%) | 14 (24.6%) | 22 (22.4%) | |
| III | 31 (75.6%) | 38 (66.7%) | 69 (70.4%) | |
| IV | 1 (2.4%) | 1 (1.8%) | 2 (2.0%) | |
| PD-L1 TPS | ||||
| <1% | 5 (12.2%) | 8 (14.0%) | 13 (13.3%) | 0.968 |
| 1–49% | 12 (29.3%) | 14 (24.6%) | 26 (26.5%) | |
| ≥50% | 9 (22.0%) | 13 (22.8%) | 22 (22.4%) | |
| Could not be evaluated | 15 (36.6%) | 22 (38.6%) | 37 (37.8%) | |
| ICIs | ||||
| Camrelizumab | 2 (4.9%) | 6 (10.5%) | 8 (8.2%) | 0.103 |
| Durvalumab | 2 (4.9%) | 0 (0%) | 2 (2.0%) | |
| Nivolumab | 26 (63.4%) | 28 (49.1%) | 54 (55.1%) | |
| Pembrolizumab | 5 (12.2%) | 13 (22.8%) | 18 (18.4%) | |
| Sintilimab | 3 (7.3%) | 1 (1.8%) | 4 (4.1%) | |
| Tislelizumab | 3 (7.3%) | 9 (15.8%) | 12 (12.2%) | |
| Course of preoperative therapy | ||||
| Two or less | 6 (14.6%) | 8 (14.0%) | 14 (14.3%) | 0.650 |
| Three | 31 (75.6%) | 39 (68.4%) | 70 (71.4%) | |
| Four or more | 4 (9.8%) | 10 (17.5%) | 14 (14.3%) | |
| Type of platinum therapy | ||||
| Not platinum-based regime | 2 (4.9%) | 4 (7.0%) | 6 (6.1%) | 0.888 |
| Carboplatin | 33 (80.5%) | 47 (82.5%) | 80 (81.6%) | |
| Cisplatin | 3 (7.3%) | 4 (7.0%) | 7 (7.1%) | |
| Lobaplatin | 3 (7.3%) | 2 (3.5%) | 5 (5.1%) | |
BMI, body mass index; ECOG, Eastern Cooperative Oncology Group; ICIs, immune checkpoint inhibitors; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MPR, major pathological response; NOS, not otherwise specified; PD-L1, programmed cell death-ligand 1; TPS, tumor proportion score.
Radiomic signature construction and validation
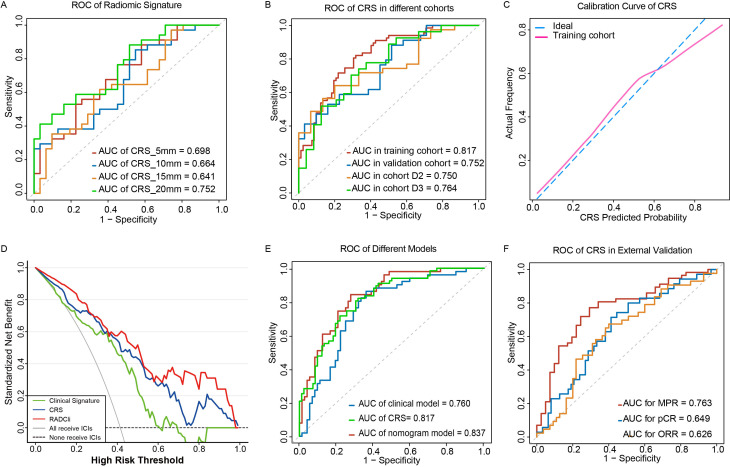
There were 4255 features extracted from five VOIs and 3630 features remaining with ICC>0.75 after the stability test. Next, they were processed by the LASSO algorithm to filter redundant and non-predictive features (online supplemental figure S2). Five radiomic signatures extracted from tumorous and four peritumoral areas, respectively, were constructed based on a backward stepwise logistic regression algorithm. The ability to clarify responders and non-responders of these four peritumoral radiomic predictors were compared in the discovery cohort (figure 3A; online supplemental table S2). The predictor of 20 mm peritumoral area outperformed others with superior goodness of fit (online supplemental table S3) and generalizability in internal validation cohort in accordance with the calibration curve analysis (figure 3C; online supplemental figure S3). In addition, combined with tumorous radiomic features, the CRS showed preferable prediction efficacy and robustness in D2 (area under the curve (AUC)=0.750, 95% CI: 0.616 to 0.8848) and D3 (AUC=0.764, 95% CI: 0.6327 to 0.8951) (figure 3B). A total of nine radiomic features selected for the radiomic score formula was displayed as in table 3. All radiomic features included exhibited good to excellent inter-rater reliability (details seen in online supplemental appendix S7.5 and table S8).
Figure 3.
Performance of clinical and radiomic signatures for predicting the responders to chemoimmunotherapy. (A) ROC of radiomic signatures in internal validation cohort. (B) ROC of CRS in different cohorts. (C) Calibration curve of CRS. (D) Decision curve analysis for the RADCli nomogram (red), CRS signature (blue), and clinical model (green). The analysis was performed across the most range of threshold probabilities at which a patient would be selected to receive immunotherapy. The y-axis indicates the net benefit; the x-axis indicates threshold probability, the value of the lowest suspected probability of DCB where the physician still may advise the patient to receive immunotherapy after balancing the benefit and harmfulness. The gray line represents the assumption that all patients receive immunotherapy. The black dotted line represents the hypothesis that no patient received immunotherapy. (E) ROC of RADCli nomogram (red), CRS model (green), and clinical model (blue). (F) ROC of CRS for ORR/pCR/MPR prediction in the surgical validation cohort. AUC, area under the curve; CRS, combined radiomic signature; DCB, durable clinical benefit; ICIs, immune checkpoint inhibitors; MPR, major pathological response; ORR, objective response rate; pCR, pathologic complete response; RADCli, radiomic-clinical nomogram; ROC, receiver-operator characteristic.
Table 3.
The features list of radiomic signature and the coefficients, respectively
| Feature | Location | Feature name | Coefficients |
| (Intercept) | 0.2656 | ||
| T710 | Tumorous | wavelet-HHL_glrlm-LowGrayLevelEmphasis | 0.2026 |
| T55 | Tumorous | wavelet-HHL_glrlm-LowGrayLevelRunEmphasis | 0.1385 |
| A457 | Peritumoral 0–5 mm | wavelet-LLH_firstorder-Skewness | 0.7839 |
| A730 | Peritumoral 0–5 mm | wavelet-HHL_glcm-Imc2 | 0.7238 |
| A765 | Peritumoral 0–5 mm | wavelet-HHL_glrlm-LongRunLowGrayLevelEmphasis | 0.978 |
| X2A194 | Peritumoral 5–10 mm | wavelet-HLL_firstorder-Mean | 0.5813 |
| X2A752 | Peritumoral 5–10 mm | wavelet-HHL_firstorder-Mean | 0.7193 |
| X3A287 | Peritumoral 10–15 mm | wavelet-LHL_firstorder-Mean | 0.4865 |
| X4A193 | Peritumoral 15–20 mm | wavelet-LHL_firstorder-Kurtosis | 0.3795 |
jitc-2023-007369supp003.pdf (134.7KB, pdf)
jitc-2023-007369supp013.pdf (114.2KB, pdf)
jitc-2023-007369supp014.pdf (10KB, pdf)
jitc-2023-007369supp004.pdf (140.6KB, pdf)
jitc-2023-007369supp019.pdf (104.9KB, pdf)
In DCA curve analysis we found that the CRS model would benefit in identifying responders and non-responders at most given threshold probability (figure 3D). Besides, both CRS and RADCli always showed more net-benefit than the clinical signature in predicting the probability of DCB. In data set D4, MPR recorded are not always consistent with ORR in D4 and 17 of 43 participants with radiological PR or SD did not achieve MPR. The ability of CRS to predict MPR to immunotherapy was shown to have an AUC of 0.763 (95% CI: 0.666 to 0.861; figure 3F) and the ability for ORR and pCR stratification were relatively ordinary with AUC of 0.626 and 0.649, respectively (figure 3F).
Construction of clinical signature
Clinical variables including gender, clinical stage, smoking status, LOT and, NLRpost were significantly associated with patients’ response status in univariate analysis (p<0.05; online supplemental table S5). These five characteristics were introduced to develop a clinical signature via stepwise regression and performed with AUC of 0.76 (95% CI: 0.674 to 0.846) in the training cohort and 0.66 (95% CI: 0.509 to 0.811) in the validation cohort in receiver-operator characteristic (ROC) analysis (online supplemental figure S4). Despite the clinical signature showing acceptable goodness of fit in both the training cohort (p=0.121) and validation cohort (p=0.153), it was still a limited predictor due to a relatively low accuracy of prediction.
jitc-2023-007369supp016.pdf (93.6KB, pdf)
jitc-2023-007369supp005.pdf (263.8KB, pdf)
Nomogram RADCli construction and validation
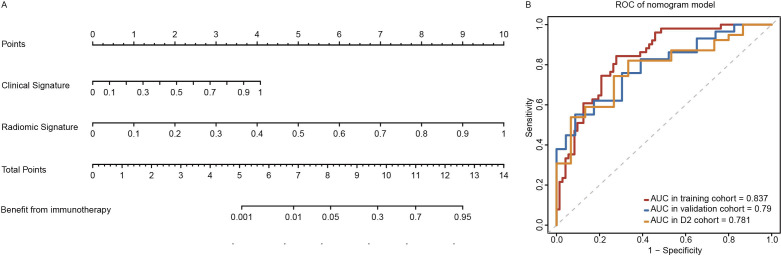
To improve the performance of predictive signatures further and develop a clinically adaptable tool for the identification of responders from ICIs therapy, we established a nomogram RADCli which included both radiomic and clinical signatures (figure 4). In multivariate logistic regression analysis, both radiomic (p<0.001) and clinical signatures (p=0.001) could serve as significant than any other signatures independent predictor. RADCli displayed a superior predictive performance (figure 3E), with an AUC of 0.837 (95% CI: 0.768 to 0.907) in the training cohort, 0.790 (95% CI: 0.669 to 0.911) in the internal validation cohort and 0.781 (95% CI: 0.651 to 0.911) in D2 data set (online supplemental table S4). In both training and validation cohorts, the RADCli model showed good agreement with the actual observations and classifications (online supplemental figure S5). As a result of the Hosmer-Lemeshow test, neither the training cohort (p=0.436) nor the validation cohort (p=0.826) achieved statistical significance. In DCA analysis, we calculated the net benefit of our models and it indicated that both RADCli and CRS had a higher net benefit in predicting patients for receiving PD-L1/PD-1 treatment across most threshold probability values (details seen in online supplemental appendix S7.2).
Figure 4.
Nomogram of RADCli and the ROC of RADCli in cohorts. (A) nomogram and (B) ROC analysis in several cohorts. RADCli displayed a superior predictive performance, with an AUC of 0.837 (95% CI: 0.768 to 0.907) in the training cohort, 0.790 (95% CI: 0.669 to 0.911) in the internal validation cohort and 0.781 (95% CI: 0.651 to 0.911) in an independent validation set D2. AUC, area under the curve; RADCli, radiomic-clinical nomogram; ROC, receiver-operator characteristic.
jitc-2023-007369supp015.pdf (41.8KB, pdf)
jitc-2023-007369supp006.pdf (119.4KB, pdf)
Evaluation of radiomic and clinical characteristics on long-term survival
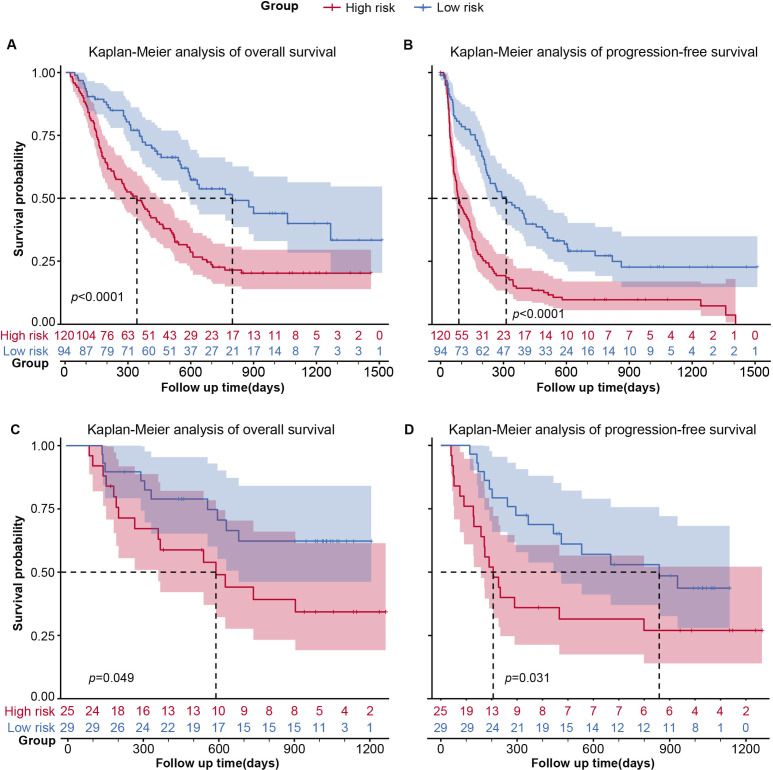
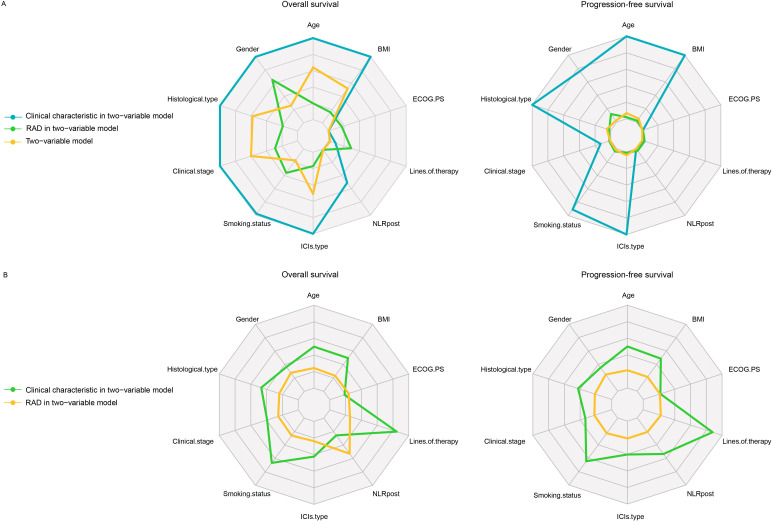
The median PFS and OS of the D1 set are 192 days (95% CI: 143 to 245 days) and 500 days (95% CI: 390 to 520 days) with an 849-day median follow-up. The two groups classified by CRS showed significantly different survival outcomes in discovery data set D1(figure 5A,B) and data set D2(figure 5C,D). The median PFS of outcomes of the high-risk and low-risk groups are 87 and 313 days, respectively. And the median OS of outcomes of the high-risk and low-risk groups are 340 and 796 days, respectively. We conducted a univariate analysis for every clinical variable for survival analysis (online supplemental table S4). We found CRS showed a promising stratification ability on OS (HR: 0.49, 95%CI: 0.27-0.89; p=0.020) and PFS (HR: 0.43, 95%CI: 0.26-0.74; p=0.002). LOT, gender and NLRpost could stratify both PFS and OS (p<0.05) alone, respectively. And we combine each characteristic with CRS for a two-variate analysis on OS and PFS separately based on Cox proportional hazard model (figure 6). Interestingly, only the combination of LOT and CRS could improve stratification of survival outcome, and both of them remained statistically significant for both OS (RAD: p=0.029; LOT: p<0.01) and PFS (RAD: p<0.01; LOT: p<0.01). Gender did not have significant performance for stratifying PFS (p=0.162) or OS (p=0.076) anymore after combination and another two variables clinical stage (clinical stage: p=0.013; RAD: p<0.01, figure 6A) and NLRpost (NLRpost: p<0.01; RAD: p<0.01, figure 6A) also have a good stratification performance for PFS after combination. The effect of tumor volume on CRS and long-term survival was insignificant (online supplemental table S6).
Figure 5.
The Kaplan-Meier survival analysis for CRS. Kaplan-Meier survival curves for PFS and OS on cohort D1 (n=214) for CRS. CRS, combined radiomic signature; OS, overall survival; PFS, progression-free survival.
Figure 6.
Clinicopathological characteristics associated with CRS in immunotherapy responder prediction. Evaluation of CRS and various clinicopathological characteristics. (A) The p value of each clinical characteristic and CRS in two-variable analysis by Cox regression and the log-rank p value of its model for OS and PFS. (B) HRs of CRS and clinical characteristics in each two-variable model. BMI, body mass index; CRS, combined radiomic signature; ECOG PS, Eastern Cooperative Oncology Group performance status; ICIs, immune checkpoint inhibitors; NLRpost, neutrophil-to-lymphocyte ratio after the first cycle of therapy; RAD, radiomic features.
jitc-2023-007369supp017.pdf (76.1KB, pdf)
Stratified PD-L1 expression not associated with OS
PD-L1 expression score was available for 138 participants in GPPH data sets. It was negative in 33 of 138. To compare the effectiveness of CRS to that of PD-L1 in identifying responders and OS prediction, a subset analysis was carried out in these 138 patients. The median survival of this subset is 592 days (95% CI: 514 to 874 days). ROC was drawn based on the prediction efficiency of the PD-L1 expression for responders and AUC of 0.594 with an optimal cut-off of 50.8% (sensitivity=0.759, specificity=0.424). Although different cut-off values were selected (1%/10%/50%), none of them have significantly different OS in patients with corresponding low and high PD-L1 expression, according to the related Kaplan-Meier survival curves (online supplemental figure S6A,B). In contrast, the CRS prediction stratifies two groups of participants with significantly different survival outcomes in this subset (p=0.00032, online supplemental figure S6C,D).
jitc-2023-007369supp007.pdf (397.8KB, pdf)
Biological validation of the radiomic biomarker
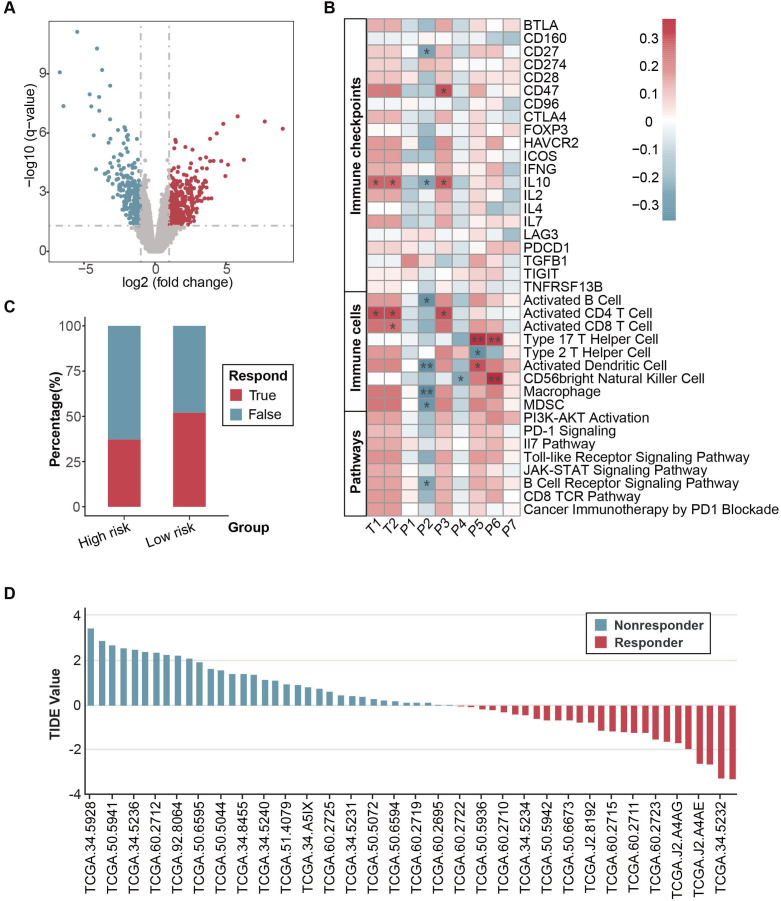
A total of 60 patients with NSCLC with matching RNA sequencing (RNA-seq) and CT imaging were collected from GSE58661. The image and corresponding clinical data were downloaded from TCIA. This biological validation cohort was separated into relatively high-risk (n=35) and low-risk groups (n=25) according to the stratified via radiomic signature. DEGs were compared between high-risk and low-risk groups, with 321 upregulated genes and 369 downregulated genes significantly (figure 7A). Kaplan-Meier survival analysis suggested survival differences found among the two groups, suggesting better survival benefits in the low-risk group, although they did not reach statistical significance (online supplemental figure S7). We then investigated the association between combined radiomic signature and oncological and biological molecular pathways. GO analysis showed dysregulated genes enriched in chemokine receptor binding (online supplemental figure S8). Correlation analysis showed a correlation between radiomic signatures from tumor and peritumor region and immune checkpoints, immune cells, and immune-related pathways (figure 7B). Peritumor signature P2 wavelet-HHL_glcm showed a negative correlation with the whole immune landscapes, especially for CD27, interleukin (IL)10, activated B cells, activated dendritic cells, macrophage, myeloid-derived suppressor cells (MDSC), and B cell receptor for signaling pathway significantly. Tumor signature T1 wavelet-HHL_glrlm, T2 wavelet-HHL_glrlm, P5 wavelet-HHL_firstorder, and P6 wavelet-LHL_firstorder showed a positive correlation with immune landscape biomarkers, especially with IL10, activated CD4 cells, activated dendritic cells, type 17 T helper cells and CD56-bright natural killer cells. For the prediction of immunotherapy, we use the TIDE algorithm to predict the responders according to RNA-seq data. Low-risk groups showed a higher percentage of predicted responders than high-risk groups (figure 7C). For the distribution of TIDE score, the predicted non-responders of immunotherapy have higher TIDE scores than responders, which indicates a higher level of tumor immune dysfunction in non-responders (figure 7D).
Figure 7.
Biological validation of CRS in TCGA-NSCLC cohort. (A) The volcano plot indicates differentially expressed genes in the TCGA-NSCLC cohort. (B) Correlation analysis between radiomic features with immune checkpoints, immune cells, and immune-related pathways. (C) Predicted responders of immunotherapy in TCGA-NSCLC cohort through transcriptomic data by TIDE algorithm. (D) Distribution of TIDE value TCGA-NSCLC cohort. CRS, combined radiomic signature; NSCLC, non-small cell lung cancer; TCGA, The Cancer Genome Atlas; TIDE, Tumor Immune Dysfunction and Exclusion.
jitc-2023-007369supp008.pdf (30KB, pdf)
Discussion
The early prediction of durable responders among patients who receive PD-L1/PD-1 inhibitors treatment is important for optimizing patient benefit and reducing social medical cost. We constructed multiple radiomic signatures based on pretreatment CT in this multicenter study. The optimal model combining predictive features of intratumoral and peritumoral areas demonstrated better predictive efficacy by applying a comparative strategy we applied in previous studies,26 27 and it was further validated in independent cohorts. We also found that the CRS has the potential prognostic ability for the long-term survival of advanced NSCLC and showed good stratification performance on MPR identification in resectable participants. In addition, mechanism analysis of RNA-seq data indicates a potential correlation between radiomic signature and immune checkpoints, immune cells, and immune-related pathways.
Nine radiomic features were incorporated in CRS, including two tumorous features and seven peritumoral features. Only hand-crafted features were extracted concerning its better expandability and generalization ability in a small-sample setting compared with deep learning algorithms. Among four higher-order features (glrlm and glcm parameters), LongRunLowGrayLevelEmphasis_HHL, one of the wavelet features derived from LongRunLowGrayLevelEmphasis, exhibited the strongest coefficient and had been found promising predictive efficacy in neoadjuvant chemoradiotherapy in esophageal cancer response and vascular endothelial growth factor expression prediction.28 29 The first-order variable of wavelet-LHL-kurtosis had shown significance in the identification of vessels around hepatocarcinoma, which is related to micro-metastasis.30 The dominant component of peritumoral features in the model also suggested the prime role in tumorous microenvironmental characteristics description and therapy response prediction.
As the biomarker approved for immunotherapy, PD-L1 did not achieve promising sensitivity or specificity11 yet and had inconsistent stratification performance for immunotherapy response and survival in previous clinical trials.31 32 In this article, we did not observe a significant association between PD-L1 expression and OS, and the stratified efficacy of PD-L1 for DCB identification was compromised compared with CRS. We optimized the model further by combining the CRS with clinical variables. RADCli nomogram were constructed and outperformed in immunotherapy responder prediction than any other signatures constructed. As we know, it is the first investigation to incorporate laboratory results like NLR with CRS for the early prediction of immunotherapy. Because CRS also stratified patients by OS significantly, especially combined with LOT, it might also identify patients who are likely to derive long-term survival benefits from therapy beyond objective responses.
Genetic level analysis of matching CT imaging and RNA-seq data TCIA database showed a correlation between radiomic signature and immune cells, suggesting the clustering of immune cells in the tumor immune microenvironment and the peritumoral region could be captured by radiomic analysis of CT image. Lung cancers with CD8+, CD3+, and PD-L1+ tumor-infiltrating cells display a solid texture on radiologic lesions and surrounding parenchyma. High PD-L1 levels were correlated with wavelet radiomic features, while Non-Uniformity-related radiomic features in CT imaging such as Gray Level Nonuniformity were highly expressed in CD8+ T-cell-rich tumor microenvironment.33 The relationship between radiomic features and genetic pathways has been explored in this work. Radiomic features of soft tissue sarcomas were associated with dysregulation of Hedgehog and Hippo signaling pathways.34 Hypoxia pathway could also be predicted in ovarian cancer by CT-based radiomic biomarkers.35 Hence, it is accessible and promising to validate the biological functions, related pathways, and mechanisms through functional annotations and pathway analysis for the explanation and understanding of radiomic signature. Concerning the underlying association, it is promising to adopt the deep learning algorithm for multi-module features integration and model training in larger-scale research.
Previous studies have revealed the significant association between radiomic features and immunotherapy response, but they seldom performed further radiomic feature extraction and analysis on peritumoral regions,19 22 which limited the further improvement of the model to some extent. The biological significance of immune cell distribution in the peritumoral region has already been emphasized36 and Khorrami et al incorporated the peritumoral region of 30 mm into their model in immunotherapy response prediction but they did not compare the predictive performance of different radiuses.15 This multicenter study was different from previous studies in these aspects as following. First, we incorporated the radiomic features extracted from both tumorous and annular peritumoral regions and the predictive value of the peritumoral area with different radii in this work was then investigated in a comparative strategy. Second, we tested the generalizability of CRS with two independent test cohorts (D2 and D3), which were from two different institutions across different immunotherapy agents. Even for participants in D3 treated with different ICIs agents (camrelizumab and sintilimab), CRS still yielded promising predictive accuracy. Third, we attempt to explore the extended value of CRS for pathological response prediction in a resectable NSCLC cohort in the neoadjuvant setting. Notably, MPR was predicted by the signature with better performance than ORR and pCR, suggesting the underlying compatible association between DCB and MPR. They were both closely correlated with survival benefits. And DCB disciplined clinical benefit on a chronologic scale while MPR did in a spatial scale, which is adopted by clinicians in the different therapy settings.
Despite the considerable diagnostic efficiency of CRS and RADCli applying both tumorous and peritumoral features, several limitations should be acknowledged for this study. First, limited sample size and selection bias are noted due to the retrospective nature of this investigation, although the characteristic of cohorts was basically balanced. Larger-scale studies in a prospective design are still warranted. Second, NLR post-therapy included in the clinical signature may limit the practical application of the nomogram in a pretreatment prediction setting although it is a stable independent factor for response status. More pretreatment variables or delta variables should be investigated further. Third, for the surgical cohort, it remains unclear whether the radiomic features are associated with survival outcomes due to the earliest admitted patients being treated in 2019, which would be evaluated in future. Finally, the strategy of pulmonary lesion selection disabled the direct evaluation of extrapulmonary metastatic status and disease burden, although this strategy decreased the heterogeneity brought by various metastatic lesion selection for metastatic disease and increased the feasibility and practicality of the model. More advanced feature extraction and auto-segmentation algorithm were guaranteed for this purpose.
Conclusion
The study constructed and verified CRS and integrated model for early non-invasive prediction of immunotherapy responders for patients with advanced NSCLC. It showed the potential association between radiomic features and pathological response of immunotherapy and laid the practical foundation of the radiomic predictive value in the neoadjuvant setting.
jitc-2023-007369supp009.pdf (57.8KB, pdf)
jitc-2023-007369supp010.pdf (421.6KB, pdf)
jitc-2023-007369supp011.pdf (72.1KB, pdf)
Acknowledgments
The results published here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. We thanked Professor Lu-Cheng from the Department of Radiology, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, Guangzhou, China, for his review of the design of this study.
Footnotes
Contributors: HZ is responsible for the overall content as guarantor. HZ and SWu conceived and designed the project. LL, SWa and GW contributed to CT segmentation and assessment. HZ, SWu, WZ, LL, LY, HY and DX contributed to the design of the study, writing the protocol, data preparation, analysis, and interpretation. HZ, SWu and LL drafted the manuscript. QL, ZH, HY, HZ, BX, LH and GL contributed to data collection and assessment. D-KZ, GQ and SWa performed the quality assessment and revised the manuscript. All authors have read and approved the submitted version.
Funding: This work supported by the International Science and Technology Cooperation Program of Guangdong (2022A0505050048), the Natural Science Foundation of Guangdong (2021A1515010838), Science and Technology Program of Guangzhou (201903010028), the Beijing Xisike Clinical Oncology Research Foundation (Y-HS202102-0038).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. The data that support the findings of this study are available from the corresponding authors with a signed data access agreement. The raw image and follow-up data are not publicly available because they contain sensitive information that could compromise patient privacy.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was conducted in accordance with the provisions of the Declaration of Helsinki and was approved by the ethics committee of Guangdong Provincial People’s Hospital (GDREC2020198H) and Jiangxi Cancer Hospital (2021ky039).
References
- 1. Reck M, Remon J, Hellmann MD. First-line Immunotherapy for non-small-cell lung cancer. J Clin Oncol 2022;40:586–97. 10.1200/JCO.21.01497 [DOI] [PubMed] [Google Scholar]
- 2. NCCN . NCCN clinical practice guidelines in oncology non-small cell lung cancer (version 3.2022–March 16,2022. 2022. Available: http://www.nccn.org/ [DOI] [PubMed]
- 3. Marin-Acevedo JA, Kimbrough EO, Lou Y. Next generation of immune Checkpoint inhibitors and beyond. J Hematol Oncol 2021;14:45. 10.1186/s13045-021-01056-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mok TSK, Wu Y-L, Kudaba I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet 2019;393:1819–30. 10.1016/S0140-6736(18)32409-7 [DOI] [PubMed] [Google Scholar]
- 5. Herbst RS, Giaccone G, de Marinis F, et al. Atezolizumab for first-line treatment of PD-L1-selected patients with NSCLC. N Engl J Med 2020;383:1328–39. 10.1056/NEJMoa1917346 [DOI] [PubMed] [Google Scholar]
- 6. Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med 2016;375:1823–33. 10.1056/NEJMoa1606774 [DOI] [PubMed] [Google Scholar]
- 7. Forde PM, Spicer J, Girard N. Neoadjuvant Nivolumab plus chemotherapy in lung cancer. reply. N Engl J Med 2022;387:572–3. 10.1056/NEJMc2208133 [DOI] [PubMed] [Google Scholar]
- 8. Wu Y-L, Zhang L, Fan Y, et al. Randomized clinical trial of Pembrolizumab vs chemotherapy for previously untreated Chinese patients with PD-L1-positive locally advanced or metastatic non-small-cell lung cancer: KEYNOTE-042 China study. Int J Cancer 2021;148:2313–20. 10.1002/ijc.33399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Truesdell J, Miller VA, Fabrizio D. Approach to evaluating tumor mutational burden in routine clinical practice. Transl Lung Cancer Res 2018;7:678–81. 10.21037/tlcr.2018.10.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang L, Chen Y, Wang H, et al. Massive PD-L1 and Cd8 double positive Tils characterize an immunosuppressive Microenvironment with high mutational burden in lung cancer. J Immunother Cancer 2021;9:e002356. 10.1136/jitc-2021-002356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu S, Stein JE, Rimm DL, et al. Comparison of biomarker modalities for predicting response to PD-1/PD-L1 Checkpoint blockade: A systematic review and meta-analysis. JAMA Oncol 2019;5:1195–204. 10.1001/jamaoncol.2019.1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278:563–77. 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun R, Limkin EJ, Vakalopoulou M, et al. A Radiomics approach to assess tumour-infiltrating Cd8 cells and response to anti-PD-1 or anti-PD-L1 Immunotherapy: an imaging biomarker, retrospective Multicohort study. Lancet Oncol 2018;19:1180–91. 10.1016/S1470-2045(18)30413-3 [DOI] [PubMed] [Google Scholar]
- 14. Mu W, Jiang L, Shi Y, et al. Non-invasive measurement of PD-L1 status and prediction of Immunotherapy response using deep learning of PET/CT images. J Immunother Cancer 2021;9:e002118. 10.1136/jitc-2020-002118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Khorrami M, Prasanna P, Gupta A, et al. Changes in CT Radiomic features associated with lymphocyte distribution predict overall survival and response to Immunotherapy in non-small cell lung cancer. Cancer Immunol Res 2020;8:108–19. 10.1158/2326-6066.CIR-19-0476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kubli SP, Berger T, Araujo DV, et al. Beyond immune Checkpoint blockade: emerging immunological strategies. Nat Rev Drug Discov 2021;20:899–919. 10.1038/s41573-021-00155-y [DOI] [PubMed] [Google Scholar]
- 17. Abou Khouzam R, Brodaczewska K, Filipiak A, et al. Tumor hypoxia regulates immune escape/invasion: influence on angiogenesis and potential impact of hypoxic biomarkers on cancer therapies. Front Immunol 2020;11:613114. 10.3389/fimmu.2020.613114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pietrobon V, Marincola FM. Hypoxia and the phenomenon of immune exclusion. J Transl Med 2021;19:9. 10.1186/s12967-020-02667-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He B, Dong D, She Y, et al. Predicting response to Immunotherapy in advanced non-small-cell lung cancer using tumor mutational burden Radiomic biomarker. J Immunother Cancer 2020;8:e000550. 10.1136/jitc-2020-000550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dercle L, Fronheiser M, Lu L, et al. Identification of non-small cell lung cancer sensitive to systemic cancer therapies using Radiomics. Clin Cancer Res 2020;26:2151–62. 10.1158/1078-0432.CCR-19-2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trebeschi S, Drago SG, Birkbak NJ, et al. Predicting response to cancer Immunotherapy using noninvasive Radiomic biomarkers. Ann Oncol 2019;30:998–1004. 10.1093/annonc/mdz108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mu W, Tunali I, Gray JE, et al. Radiomics of 18F-FDG PET/CT images predicts clinical benefit of advanced NSCLC patients to Checkpoint blockade Immunotherapy. Eur J Nucl Med Mol Imaging 2020;47:1168–82. 10.1007/s00259-019-04625-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hellmann MD, Chaft JE, William WN, et al. Pathological response after Neoadjuvant chemotherapy in Resectable non-small-cell lung cancers: proposal for the use of major pathological response as a Surrogate Endpoint. Lancet Oncol 2014;15:e42–50. 10.1016/S1470-2045(13)70334-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Armato SG, Meyer CR, Mcnitt-Gray MF, et al. The reference image database to evaluate response to therapy in lung cancer (RIDER) project: A resource for the development of change-analysis software. Clin Pharmacol Ther 2008;84:448–56. 10.1038/clpt.2008.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vickers AJ, Cronin AM, Elkin EB, et al. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak 2008;8:53. 10.1186/1472-6947-8-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liao G, Huang L, Wu S, et al. Preoperative CT-based peritumoral and Tumoral Radiomic features prediction for tumor spread through air spaces in clinical stage I lung adenocarcinoma. Lung Cancer 2022;163:87–95. 10.1016/j.lungcan.2021.11.017 [DOI] [PubMed] [Google Scholar]
- 27. Huang L, Lin W, Xie D, et al. Development and validation of a preoperative CT-based Radiomic Nomogram to predict pathology Invasiveness in patients with a solitary pulmonary Nodule: a machine learning approach, multicenter, diagnostic study. Eur Radiol 2022;32:1983–96. 10.1007/s00330-021-08268-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun Z, Li Y, Wang Y, et al. Radiogenomic analysis of vascular endothelial growth factor in patients with diffuse gliomas. Cancer Imaging 2019;19:68.:68. 10.1186/s40644-019-0256-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Beukinga RJ, Hulshoff JB, van Dijk LV, et al. Predicting response to Neoadjuvant Chemoradiotherapy in Esophageal cancer with Textural features derived from pretreatment 18F-FDG PET/CT imaging. J Nucl Med 2017;58:723–9. 10.2967/jnumed.116.180299 [DOI] [PubMed] [Google Scholar]
- 30. Fan Y, Yu Y, Wang X, et al. Texture analysis based on GD-EOB-DTPA-enhanced MRI for identifying vessels Encapsulating tumor clusters (VETC)-Positive hepatocellular carcinoma. J Hepatocell Carcinoma 2021;8:349–59. 10.2147/JHC.S293755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garassino MC, Gadgeel S, Esteban E, et al. Patient-reported outcomes following Pembrolizumab or placebo plus Pemetrexed and platinum in patients with previously untreated, metastatic, non-squamous non-small-cell lung cancer (KEYNOTE-189): a Multicentre, double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol 2020;21:387–97. 10.1016/S1470-2045(19)30801-0 [DOI] [PubMed] [Google Scholar]
- 32. Paz-Ares L, Vicente D, Tafreshi A, et al. A randomized, placebo-controlled trial of Pembrolizumab plus chemotherapy in patients with metastatic squamous NSCLC: protocol-specified final analysis of KEYNOTE-407. J Thorac Oncol 2020;15:1657–69. 10.1016/j.jtho.2020.06.015 [DOI] [PubMed] [Google Scholar]
- 33. Mazzaschi G, Milanese G, Pagano P, et al. Integrated CT imaging and tissue immune features disclose a radio-immune signature with high Prognostic impact on surgically Resected NSCLC. Lung Cancer 2020;144:30–9. 10.1016/j.lungcan.2020.04.006 [DOI] [PubMed] [Google Scholar]
- 34. Crombé A, Bertolo F, Fadli D, et al. Distinct patterns of the natural evolution of soft tissue Sarcomas on pre-treatment Mris captured with Delta-Radiomics correlate with gene expression profiles. Eur Radiol 2022;33:1205–18. 10.1007/s00330-022-09104-8 [DOI] [PubMed] [Google Scholar]
- 35. Feng S, Xia T, Ge Y, et al. Computed tomography imaging-based Radiogenomics analysis reveals hypoxia patterns and immunological characteristics in ovarian cancer. Front Immunol 2022;13:13. 10.3389/fimmu.2022.868067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beig N, Khorrami M, Alilou M, et al. Perinodular and Intranodular Radiomic features on lung CT images distinguish adenocarcinomas from Granulomas. Radiology 2019;290:783–92. 10.1148/radiol.2018180910 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-007369supp001.pdf (197.9KB, pdf)
jitc-2023-007369supp002.pdf (10.7KB, pdf)
jitc-2023-007369supp018.pdf (73.2KB, pdf)
jitc-2023-007369supp012.pdf (79.4KB, pdf)
jitc-2023-007369supp003.pdf (134.7KB, pdf)
jitc-2023-007369supp013.pdf (114.2KB, pdf)
jitc-2023-007369supp014.pdf (10KB, pdf)
jitc-2023-007369supp004.pdf (140.6KB, pdf)
jitc-2023-007369supp019.pdf (104.9KB, pdf)
jitc-2023-007369supp016.pdf (93.6KB, pdf)
jitc-2023-007369supp005.pdf (263.8KB, pdf)
jitc-2023-007369supp015.pdf (41.8KB, pdf)
jitc-2023-007369supp006.pdf (119.4KB, pdf)
jitc-2023-007369supp017.pdf (76.1KB, pdf)
jitc-2023-007369supp007.pdf (397.8KB, pdf)
jitc-2023-007369supp008.pdf (30KB, pdf)
jitc-2023-007369supp009.pdf (57.8KB, pdf)
jitc-2023-007369supp010.pdf (421.6KB, pdf)
jitc-2023-007369supp011.pdf (72.1KB, pdf)
Data Availability Statement
Data are available upon reasonable request. The data that support the findings of this study are available from the corresponding authors with a signed data access agreement. The raw image and follow-up data are not publicly available because they contain sensitive information that could compromise patient privacy.