Abstract
The diversity and phylogeny of nodA and nifH genes were studied by using 52 rhizobial isolates from Acacia senegal, Prosopis chilensis, and related leguminous trees growing in Africa and Latin America. All of the strains had similar host ranges and belonged to the genera Sinorhizobium and Mesorhizobium, as previously determined by 16S rRNA gene sequence analysis. The restriction patterns and a sequence analysis of the nodA and nifH genes divided the strains into the following three distinct groups: sinorhizobia from Africa, sinorhizobia from Latin America, and mesorhizobia from both regions. In a phylogenetic tree also containing previously published sequences, the nodA genes of our rhizobia formed a branch of their own, but within the branch no correlation between symbiotic genes and host trees was apparent. Within the large group of African sinorhizobia, similar symbiotic gene types were found in different chromosomal backgrounds, suggesting that transfer of symbiotic genes has occurred across species boundaries. Most strains had plasmids, and the presence of plasmid-borne nifH was demonstrated by hybridization for some examples. The nodA and nifH genes of Sinorhizobium teranga ORS1009T grouped with the nodA and nifH genes of the other African sinorhizobia, but Sinorhizobium saheli ORS609T had a totally different nodA sequence, although it was closely related based on the 16S rRNA gene and nifH data. This might be because this S. saheli strain was originally isolated from Sesbania sp., which belongs to a different cross-nodulation group than Acacia and Prosopis spp. The factors that appear to have influenced the evolution of rhizobial symbiotic genes vary in importance at different taxonomic levels.
Rhizobia are soil bacteria that are capable of forming a nitrogen-fixing symbiosis with leguminous plants. All rhizobia belong to the alpha subgroup of the Proteobacteria, based on the sequences of the gene coding for small-subunit (16S) rRNA (23). There are three major rhizobial branches, the genus Bradyrhizobium, the genus Azorhizobium, and the fast-growing rhizobia. The latter organisms are currently split into three genera, the genera Rhizobium, Sinorhizobium, and Mesorhizobium, plus a fourth potential genus that so far is represented only by Rhizobium galegae (14, 24, 46). To form an effective symbiosis, rhizobia require several classes of specific genes. These include nod genes, which encode the production of Nod factors, which stimulate the plants to produce symbiotic nodules, and nif genes, which produce the nitrogen-fixing nitrogenase enzyme. In this study we explored the diversity of these genes in rhizobia from a particular group of hosts and the extent to which they may have a different evolutionary history than the rest of the bacterial genome.
The nod genes are unique to rhizobia, and the phylogenies of nodA, nodB, nodC, and nodD, which are found in all rhizobia, resemble each other but differ from the phylogeny of 16S rRNA (19). Indeed, it has been suggested that the phylogenies of nod genes may correlate with the host plant (6, 40, 42). Other studies, performed by using a variety of techniques, have reported various degrees of correlation between symbiotic genes and chromosomal genotypes, but in general the authors concluded that symbiotic genes appear to have been transferred between strains (16–18, 22, 29, 37, 39, 44, 47). Symbiotic genes are often located on plasmids, which probably increases the likelihood of gene transfer.
By contrast, nif genes are found in many bacteria besides rhizobia. It is not clear whether nif genes are, from an evolutionary point of view, part of the symbiotic genome or part of the “normal” bacterial genome (46). It has been reported that, for bacteria as a whole, the phylogeny of nifH closely resembles that of 16S rRNA genes, and thus these genes probably share a common evolutionary history (13, 43, 45); however, there is also evidence of phylogenetic discordance that could be due to lateral transfer of nif genes (7).
We have recently described the phenotypic and phylogenetic diversity of a fairly large group of fast-growing rhizobia that were isolated from Acacia senegal and Prosopis chilensis trees growing in Sudan and Kenya (11, 12, 26, 48). In the present study we selected a representative subset of these organisms and added a few Latin American strains with similar host ranges. The strains used represent two rhizobial genera commonly associated with these trees, namely, the genera Sinorhizobium (4) and Mesorhizobium (14, 46). The sinorhizobia have a range of 16S rRNA gene sequences and probably belong to a number of different species. We also included the type strains of two species from Africa, Sinorhizobium teranga and Sinorhizobium saheli, which are closely related to our isolates.
Our aim was to determine whether these rhizobia which belong to different species but have similar host ranges share the same set of symbiotic genes as a result of lateral transfer or whether they have converged to have similar host ranges but use different genes. As representatives of the nodulation and nitrogen fixation genes we chose nodA and nifH, respectively, because a considerable number of comparative sequences already are known and these genes have a number of other advantages. For nodA, only one copy has ever been detected, and although reiteration of nifH has been reported for several rhizobia, sequencing of the multiple copies has shown that they are identical or nearly so (1, 27, 31). An initial restriction fragment length polymorphism (RFLP) examination of the total DNA of our strains indicated that both genes probably are present only in a single copy. The two genes are also relatively small, and conserved primer sites allow PCR amplification of most of each gene. Both gene products have well-defined functions; NodA is a host-specific determinant of the transfer of fatty acids in Nod factor biosynthesis (2, 32, 33), while NifH is the Fe protein subunit of nitrogenase. The phylogeny of nifH has been explored by a number of authors (13, 28, 43, 45), and the phylogeny of nodA resembles the phylogenies of the other common nod genes (19). Debellé et al. (3) have recently suggested that the study of nodA and nodFE, which together specify Nod factors of many fast-growing rhizobia, combined with cloning and characterization of the Nod factor receptors of the host plants, could in the future provide a strong basis for the analysis of coevolution of the two partners.
In our study, after an initial RFLP analysis in which we used the total DNA of selected strains, we used restriction digestion of PCR-amplified nodA and nifH gene products from 54 isolates to compare the distributions of nod and nif types to each other and to the distribution of 16S rRNA gene sequence types and also to see whether these distributions correlated with a tree of isolation or geographical origin. Representative gene types were selected for DNA sequencing.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used are listed in Table 1. Chromosomal sequence types s1 to s12, based on the sequence of a 230-bp fragment of 16S rDNA, were determined previously, as indicated in Table 1. Sequence type s1 is identical to Sinorhizobium meliloti and Sinorhizobium fredii, sequence type s2 is identical to Sinorhizobium meliloti type B, now named Sinorhizobium medicae (34), sequence type s7 is identical to S. teranga, and sequence types s10 and s11 are identical to two alternative sequences of S. saheli (12). Sequence type s12 represents a group of mesorhizobia identical in the 230-bp region, i.e., Mesorhizobium huakuii, Mesorhizobium ciceri, Mesorhizobium tianshanense, Mesorhizobium mediterraneum, and cluster U of de Lajudie et al. (4). On the basis of their whole-cell protein patterns, the Brazilian mesorhizobia used in this study also belong to cluster U (4), as do the Sudanese mesorhizobia (26). Strains were maintained on yeast-mannitol agar plates supplemented with Congo red (1:400).
TABLE 1.
Strains used in this study and their origins of isolation, host trees, 16S rDNA sequence types, nodA and nifH restriction patterns and types, 16S rRNA-nod-nif combinations, and plasmid profiles
| Straina | Origin | Host tree | 16S rRNA sequence typeb | Source(s) of 16S rRNA sequencec |
nodA restriction patternsd
|
nod typee |
nifH restriction patternsd
|
nif typee | 16S rRNA-nod-nif combination | Plasmid profilef | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HhaI | HinfI | RsaI | AluI | HhaI | HinfI | RsaI | |||||||||
| Sinorhizobium strains | |||||||||||||||
| H1394 | Kenya | Prosopis chilensis | s1 | 1 | a | a | a | Ag | a | a | a | a | Ag | 1AA | NP |
| BR827 | Brazil | Leucaena leucocephala | s1 | 2 | b | b | b | Bg | b | b | a | a | Bg | 1BB | p1 |
| M6 | Mexico | Prosopis sp. | s1 | 3 | c | c | c | Cg | b | c | a | b | Cg | 1CC | p2h |
| H1396 | Kenya | Prosopis chilensis | s2 | 1 | a | d | d | D | a | d | a | a | D | 2DD | p3 |
| H1552 | Kosti, Sudan | Prosopis chilensis | s2 | 1 | d | d | d | Eg | c | d | a | c | Eg | 2EE | p4h |
| H1680 | Tendelti, Sudan | Prosopis chilensis | s2 | 1 | d | d | d | E | a | e | b | a | F | 2EF | p5h |
| H1700 | Khartum, Sudan | Acacia senegal | s2 | 1 | d | d | d | Eg | a | e | b | a | Fg | 2EF | p5 |
| H1704 | Khartum, Sudan | Acacia senegal | s2 | 1 | d | d | d | E | a | e | b | a | F | 2EF | p5 |
| H1706 | El Obeid, Sudan | Acacia senegal | rel. s2 | 4 | d | d | d | E | a | e | b | a | F | 2EF | p5 |
| H1707 | Kosti, Sudan | Prosopis chilensis | rel. s2 | 4 | d | d | d | E | a | e | b | a | F | 2EF | p5 |
| H1708 | Kosti, Sudan | Prosopis chilensis | rel. s2 | 4 | d | d | d | E | a | e | b | a | F | 2EF | p6 |
| H1624 | El Obeid, Sudan | Acacia senegal | s3 | 1 | d | d | d | E | a | e | b | a | F | 3EF | p7 |
| H1482 | El Fau, Sudan | Acacia senegal | s4 | 1 | a | a | a | A | c | e | a | a | G | 4AG | p8 |
| H1489 | Kosti, Sudan | Acacia senegal | s4 | 1 | a | a | a | Ag | c | e | a | a | Gg | 4AG | p8h |
| H1492 | Kosti, Sudan | Prosopis chilensis | s4 | 1 | a | a | a | A | c | e | a | a | G | 4AG | p8 |
| H1493 | Khartum, Sudan | Prosopis chilensis | s4 | 1 | a | a | a | A | c | e | a | a | G | 4AG | p8 |
| H1502 | Tendelti, Sudan | Acacia senegal | rel. s4 | 4 | a | a | a | A | c | e | a | a | G | 4AG | p8 |
| H1481 | El Fau, Sudan | Acacia senegal | rel. s4 | 4 | a | a | a | A | c | e | a | a | G | 4AG | p8 |
| H1484 | Kosti, Sudan | Prosopis chilensis | rel. s4 | 4 | a | a | a | A | c | e | a | a | G | 4AG | p8 |
| H1485 | Kosti, Sudan | Prosopis chilensis | rel. s4 | 4 | a | a | a | A | c | e | a | a | G | 4AG | p8 |
| H1486 | Kosti, Sudan | Prosopis chilensis | rel. s4 | 4 | a | a | a | A | c | e | a | a | G | 4AG | p8 |
| H1488 | Kosti, Sudan | Prosopis chilensis | rel. s4 | 4 | a | a | a | A | c | e | a | a | G | 4AG | p8 |
| H1491 | Kosti, Sudan | Prosopis chilensis | rel. s4 | 4 | a | a | a | A | c | e | a | a | G | 4AG | p8 |
| H1505 | Kosti, Sudan | Prosopis chilensis | rel. s4 | 4 | a | a | a | A | c | e | a | a | G | 4AG | p8 |
| H1504 | Kosti, Sudan | Prosopis chilensis | rel. s4 | 4 | a | a | a | A | c | e | a | a | G | 4AG | p8 |
| H1476 | Tendelti, Sudan | Prosopis chilensis | s4 | 1 | a | a | a | A | c | f | a | a | Hg | 4AH | p8 |
| H1679 | Tendelti, Sudan | Prosopis chilensis | s4 | 1 | a | a | a | A | c | f | a | a | H | 4AH | p8 |
| H1501 | Kosti, Sudan | Acacia senegal | s4 | 1 | e | d | d | F | c | f | a | a | H | 4FH | p9 |
| H1498 | Kosti, Sudan | Prosopis chilensis | s4 | 1 | e | d | d | F | c | f | b | a | I | 4FI | p8 |
| H1483 | Kosti, Sudan | Prosopis chilensis | rel. s4 | 4 | a | e | a | G | d | d | a | a | J | 4GJ | ND |
| H1392 | Kenya | Prosopis chilensis | s5 | 1 | f | f | e | H | a | e | b | a | F | 5HF | NP |
| H1698 | El Fau, Sudan | Acacia senegal | s6 | 1 | f | f | f | I | a | e | b | a | F | 6IF | p10h |
| H1703 | Tendelti, Sudan | Acacia senegal | s7 | 1 | f | g | f | J | a | e | b | a | F | 7JF | p11h |
| H1550 | El Obeid, Sudan | Acacia senegal | s7 | 1 | f | h | f | K | a | e | b | a | F | 7KF | p12h |
| ORS1009T | Senegal | Acacia laeta | s7 | 5 | f | h | f | Kg | a | e | b | a | Fg | 7KF | p13 |
| H1395 | Kenya | Acacia senegal | s8 | 1 | f | f | f | I | a | e | b | a | F | 8IF | p14 |
| H1551 | El Fau, Sudan | Acacia senegal | s9 | 1 | f | f | g | L | a | e | c | a | K | 9LK | p15 |
| DWO607 | Kenya | Acacia seyal | s10 | 6 | f | f | f | Ig | a | e | b | a | Fg | 10IF | p16h |
| H1393 | Kenya | Prosopis chilensis | s10 | 1 | a | i | h | Mg | d | d | a | a | J | 10MJ | p17 |
| BR4007 | Brazil | Prosopis juliflora | s10 | 2 | g | c | i | Ng | e | f | a | d | Lg | 10NL | NP |
| ORS609T | Senegal | Sesbania cannabina | s10+s11 | 5, 1 | h | j | j | Og | a | g | a | c | Mg | 10+11OM | p18 |
| H1478 | El Obeid, Sudan | Acacia senegal | s11 | 1 | e | d | d | F | a | d | a | a | Dg | 11FD | p19h |
| H1495 | Khartum, Sudan | Acacia senegal | s11 | 1 | e | d | d | F | a | e | b | a | F | 11FF | p20h |
| H1496 | El Fau, Sudan | Acacia senegal | s11 | 1 | e | d | d | Fg | a | e | b | a | F | 11FF | p20h |
| H1500 | El Fau, Sudan | Acacia senegal | s11 | 1 | a | f | k | P | a | e | b | a | F | 11PF | p21 |
| H1506 | El Obeid, Sudan | Acacia senegal | s11 | 1 | a | f | k | P | a | e | b | a | F | 11PF | p22 |
| H1480 | Kosti, Sudan | Acacia senegal | s11 | 1 | a | e | a | Gg | d | d | a | a | Jg | 11GJ | NP |
| H1499 | El Fau, Sudan | Acacia senegal | s1+s10 | 1 | e | d | d | Fg | c | d | a | a | Ng | 1+10FN | p23 |
| Mesorhizobium strains | |||||||||||||||
| H1487 | El Obeid, Sudan | Acacia senegal | s12 | 1 | i | c | l | Qg | b | h | a | a | Og | 12QO | NP |
| H260 | Tendelti, Sudan | Acacia senegal | s12 | 3 | j | k | l | Rg | b | h | a | a | O | 12RO | ND |
| DUS466 | Kenya | Acacia arenaria | s12 | 7 | k | l | l | Sg | b | h | a | a | O | 12SO | ND |
| DWO366 | Kenya | Acacia polyacantha | s12 | 6 | l | m | m | Tg | b | e | a | a | Pg | 12TP | p24 |
| BR3804 | Brazil | Chamaecrista ensiformis | s12 | 2 | m | n | l | Ug | f | i | a | a | Qg | 12UQ | p25 |
| INPA78B | Brazil | Leucaena diversifolia | s12 | 2 | n | o | n | Vg | f | j | a | e | Rg | 12VR | ND |
H, HAMBI Culture Collection of Division of Microbiology, University of Helsinki, Helsinki, Finland; BR, CNPBS/EMBRAPA collection, Seropedica, Rio de Janeiro, Brazil; M, CINVESTAV collection, Irapuato, Mexico; ORS,ORSTOM Collection, Dakar, Senegal; DWO, strain obtained from D. W. Odee, KFR, Kenya Forestry Research Institute Rhizobium culture collection, Nairobi, Kenya; DUS, Dundee University Strain Collection, Dundee, Scotland; INPA, National Institute of Amazonia Research, Manaus, Brazil. Strains ORS1009T and ORS609T are the type strains of S. teranga and S. saheli, respectively.
Sequence types s1 to s12 as described by Haukka et al. (12). rel. s2 and rel. s4, no sequence data were available, but data from other methods, such as pulsed-field gel electrophoresis (11), and data from Nick and Lindström (26), revealed close relatedness to the strains having sequence types s2 and s4, respectively.
1, Haukka et al. (12); 2, Moreira et al. (25); 3, new data; 4, data based on pulsed-field gel electrophoresis patterns (11) and data from Nick and Lindström (26); 5, de Lajudie et al. (4); 6, Odee et al. (28a); 7, McInroy et al. (24a).
The restriction patterns are the band patterns obtained with each endonuclease; identical patterns are designated by the same letter.
The combinations of restriction patterns were used to define nod and nif types; identical types are designated by the same letter.
NP, no plasmid detected after at least four attempts; ND, not determined conclusively.
Sequence determined in this study.
The nifH probe hybridized with one of the plasmids.
RFLP analysis of total DNA.
Total DNA was isolated by using a modified standard protocol, in which 5 ml of fresh culture was spun down, washed in 1 ml of TEN (TE containing 100 mM NaCl, pH 8.0), and resuspended in a solution containing 350 μl of TEN, 35 μl of 10% sodium dodecyl sulfate, and 50 μl of proteinase K (10 mg/ml). After about 1 h of lysis at 37°C, the DNA was sheared by using a 23-gauge needle and extracted three times with 400 μl of phenol-chloroform and twice with chloroform. The DNA was precipitated with ethanol and resuspended in 100 μl of H2O. Approximately 5 μl of DNA was digested with three enzymes, EcoRI, HindIII, and BamHI (all from Promega), by following the manufacturer’s instructions. Digests were electrophoresed slowly overnight in three separate 0.8 to 1% agarose gels, after which the DNA was transferred to nylon membranes by using standard Southern blotting (36). The membranes were hybridized successively with different probes by using either ECL direct nucleic acid labelling and detection systems (catalog no. RPN3000; Amersham Life Science) or a Gene Images random prime labelling module and a Gene Images CDP-Star detection module (catalog no. RPN3500; Amersham Life Science). Probes for nifH and nodA were prepared by following the manufacturer’s instructions from the PCR-amplified gene products (see below) of strain H1552.
PCR amplification.
For PCR a loopful of bacterial culture from a yeast-mannitol agar plate was suspended in water and boiled, and 25 μl was used to prepare a 50-μl PCR that also contained 1× reaction buffer, 2 mM MgCl2, and 1 U of Taq polymerase (all from Promega), as well as 0.05% Tween 20, 0.05% Nonidet P-40, each deoxynucleoside triphosphate at a concentration of 0.2 mM, and 6.25 pmol of each primer (final concentration, 125 mM). The following temperature cycle was used: 2 min at 93°C, 35 cycles consisting of 45 s at 93°C, 45 s at 62°C, and 2 min at 72°C, and finally 5 min at 72°C. nifH was amplified by using primers nifH-1 and nifH-2 (7), which amplify a 601-bp fragment between positions 256 and 856 in the S. meliloti sequence. Primers for nodA were designed by comparing known nodA sequences for S. meliloti, S. fredii, Sinorhizobium sp. strain NGR234, Rhizobium leguminosarum bv. trifolii, and R. galegae and were obtained from Cruachem Ltd., Glasgow, United Kingdom. Forward primer nodA-1 (5′-TGCRGTGGAARNTRNNCTGGGAAA-3′) starts from base 14 in nodA, and reverse primer nodA-2 (5′-GGNCCGTCRTCRAAWGTCARGTA-3′) ends in nodB 88 bases (in the S. meliloti sequence) after the end of nodA and amplifies a fragment of approximately 666 bp. The PCR conditions used for nodA were the same as those used for nifH, except that 20 pmol of each primer (final concentration, 400 nM) was used and the annealing step in the PCR was for 1 min at 49°C. For a few strains multiple nodA bands were obtained; in each of these cases the band of the expected size was excised from the agarose gel and transferred into 50 μl of sterile water for a few hours. After this 10 μl of water containing the eluted DNA was used as a template in a new PCR.
Restriction digestion of PCR products.
Both the nodA PCR products and the nifH PCR products of 54 strains were digested with endonucleases HhaI, HinfI, and RsaI; in addition, AluI was used for nifH. The reaction mixtures (15 μl) contained 5 to 12 μl of PCR product, 5 U of enzyme, and 1.5 μl of buffer specific for the enzyme (Promega). The restriction products were electrophoresed in 2.5% agarose gels at 80 V for about 2 h. Ethidium bromide-stained gels were photographed by using a UVP gel documentation system, and the band patterns were scored manually.
Direct sequencing of PCR products.
PCR products were purified by using Wizard PCR Preps columns (Promega) as recommended by the manufacturer. The purified products were electrophoresed in a 1% agarose gel to estimate the amount of DNA and then sequenced by using an ABI PRISM Ready Reaction dye terminator cycle sequencing kit with Amplitaq DNA polymerase FS (Perkin-Elmer). An estimated 100 ng of DNA was used for each reaction together with 1.6 pmol of primer, 4 μl of ready reaction mixture, and enough H2O so that the final volume was 10 μl. The primers used were the primers used for the PCR amplifications described above (nifH-1, nifH-2, nodA-1, and nodA-2). Cycle sequencing PCR amplification and subsequent DNA precipitation were performed by following the manufacturer’s instructions. Resuspended samples were examined with a Perkin-Elmer model 377 ABI PRISM DNA sequencer by using standard conditions recommended by the manufacturer.
Phylogenetic analysis.
Each PCR product was sequenced in both directions, and the sequences were assembled and checked with the AutoAssembler 1.4 program (Perkin-Elmer). The revised sequences were analyzed by using the SEQNET computer facility provided by the BBSRC Daresbury Laboratory, Genetics Computer Group, United Kingdom. Sequences were aligned by using the Pileup program of the Wisconsin GCG package (5). ClustalW (41) was used to construct phylogenetic trees from the Jukes-Cantor distances by using the neighbor-joining method (35). The trees were displayed by using TreeView (30).
Plasmid profiles.
Plasmid profiles were determined on horizontal agarose gels by using the in-well lysis method of Eckhardt (8), as modified by Hashem and Kuykendall (10), except that the strains were cultured in half-strength tryptone-yeast extract liquid medium until the late log phase. The cell cultures were then centrifuged slowly, washed with water, and centrifuged again, and only then was N-laurolylsarcosine added to the resulting pellets. Two gels were blotted onto nylon membranes and hybridized with the nifH probe as described above for the total DNA RFLP gels.
Nucleotide sequence accession numbers.
Our nifH sequences have been deposited in the EMBL data bank under accession no. Z95211 through Z95230; these sequences include the sequences of two strains, S. fredii USDA191 and Rhizobium sp. (Lonchocarpus sp.) strain BR6001 (25), which were determined but are not included in Table 1. The nodA sequences have been deposited in the EMBL data bank under accession no. Z95231 through Z95250.
RESULTS
Copy number and genomic arrangement of nodA and nifH genes.
Total DNAs isolated from 15 strains were restricted separately with three endonucleases, electrophoresed in agarose gels, and blotted, and the blots were hybridized successively with nifH and nodA probes (data not shown). The results obtained suggested that only a single copy of each gene was present, because each probe hybridized to only one band, except when there was an internal restriction site in the gene. The restriction sites were later verified from the sequences. The sizes of the hybridizing bands differed in distantly related strains.
RFLP analysis of PCR-amplified nodA and nifH genes.
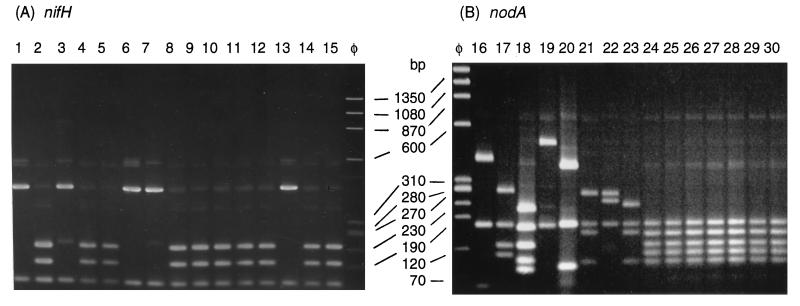
To compare the restriction patterns of symbiotic genes, nodA and nifH were amplified from 54 strains, as shown in Table 1. nifH amplification always resulted in a single band at 601 bp, but nodA primers, which are degenerate and require a low annealing temperature, in some cases gave faint bands in addition to the expected band at 666 bp. Generally, these extra bands did not hinder restriction and comparison of the brighter bands (Fig. 1). However, strains H1500, H1506, H1478, and H1501 had strong extra bands, so the expected band was excised from the gel and reamplified before restriction. The resulting restriction patterns for all strains were combined to define 22 nod types and 18 nif types, as shown in Table 1.
FIG. 1.
Typical gel showing restriction patterns obtained with HinfI. (A) nifH PCR products of strains. Lane 1, H1679; lane 2, H1550; lane 3, H1480; lane 4, H1496; lane 5, H1395; lane 6, H1476; lane 7, H1502; lane 8, H1680; lane 9, H1703; lane 10, H1706; lane 11, H1624; lane 12, H1707; lane 13, H1552; lane 14, H1708; lane 15, H1698; lane φ, marker φX174 HaeIII. (B) nodA PCR products of strains. Lane φ, marker φX174 HaeIII; lane 16, H1487; lane 17, DWO366; lane 18, BR3804; lane 19, INPA78B; lane 20, DUS466; lane 21, H1698; lane 22, H1703; lane 23, ORS1009; lane 24, H1488; lane 25, H1485; lane 26, H1486; lane 27, H1505; lane 28, H1481; lane 29, H1484; lane 30, H1490.
Correlated variation of the different genes.
The combinations of nodA and nifH restriction types and 16S rRNA gene sequence types representing chromosomal background were not random (Table 1). Sinorhizobia and mesorhizobia never had the same nod or nif types, and Latin American sinorhizobia had unique symbiotic gene types. Among the sinorhizobia from Sudan were two widespread clones, one having 16S rRNA gene-nod-nif combination 2EF and the other having 16S rRNA gene-nod-nif combination 4AG, but otherwise the pairing of nod and nif types was fairly free; e.g., nif type F could combine with nod types E, F, H, I, J, K, and P. The same nod-nif pair type was found in different chromosomal backgrounds in only three cases; type EF was found in sequence types s2 and s3, type GJ was found in sequence types s4 and s11, and type IF was found in sequence types s6, s8, and s10. However, even though similar restriction patterns and types indicate that the genes are closely related, the genes are not necessarily identical at the sequence level. For example, when nifH was sequenced from three strains having nif type F, strains ORS1009T and DWO607 differed from each other in only 9 bases (1.6%), but H1700 differed from both of these strains in 27 bases (4.8%).
Sequencing and phylogeny of nodA and nifH genes.
Based on the restriction patterns of PCR products, 20 nodA and 18 nifH products were chosen for sequencing (Table 1). All of the significantly different restriction types were included. We deduced that the remaining nif types, types I and K, as well as nod types D, H, J, L, and P, closely resembled other African sinorhizobia either on the basis the similarity of their restriction patterns or on the basis of the results of partial sequencing of the gene (data not shown). Sequences of nifH were determined nearly up to the primers; the 558-bp sequence that was used for further analysis started from the second base after the forward primer and ended two bases before the reverse primer. For nodA, the 525-bp sequence started 29 bases after the forward primer (base 67 in the entire nodA sequence) and ended at the stop codon, which, depending on the strain, is approximately 65 bases from the reverse primer.
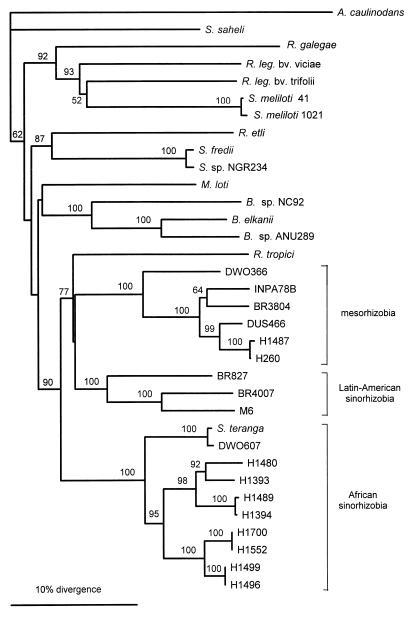
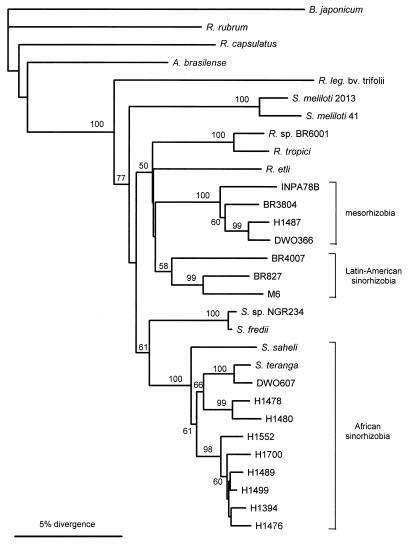
Phylogenetic analysis of the aligned 525-bp nodA and 558-bp nifH sequences resulted in the trees shown in Fig. 2 and 3. In both trees three separate clusters of tree rhizobia were evident, one consisting of African sinorhizobia, one consisting of Latin American sinorhizobia, and one consisting of mesorhizobia from both regions. High bootstrap values (from 1,000 resamplings) showed the robustness of these groups. In the nodA tree only Rhizobium tropici was found in a cluster formed by our rhizobia, whereas in the nifH tree the distances between strains were smaller and our tree rhizobia groups were intermingled with reference strains. Although the reference strains had quite different positions in the two trees, among our tree rhizobia the only major difference was the position of the S. saheli sequences. In the nifH tree S. saheli fell into the cluster of African sinorhizobia as expected, although it was on the outskirts of the cluster. However, most of the base substitutions in S. saheli nifH were in the third bases of codons, and thus in a tree based on protein translations of DNA sequences S. saheli was actually identical to H1476 and H1394. Otherwise, the trees based on protein translations of the sequences resembled the DNA trees closely (data not shown). The nodA sequence of S. saheli, on the other hand, stood on its own in both DNA and protein trees.
FIG. 2.
Phylogenetic tree based on the nodA sequences and constructed by using the neighbor-joining method. Only bootstrap values greater than 50% are shown. Sequences of the following strains were used (the numbers in parentheses are accession numbers): Azorhizobium caulinodans ORS571 (L18897), Rhizobium galegae HAMBI1174 (X87578), Rhizobium leguminosarum bv. viciae 248 (X01650), Rhizobium leguminosarum bv. trifolii ANU843 (X03721), Sinorhizobium meliloti 41 (X01649), Sinorhizobium meliloti 1021 (M11268), Rhizobium etli CE-3 (M58625), Sinorhizobium fredii USDA257 (M73699), Sinorhizobium sp. strain NGR234 (X73362), Mesorhizobium loti NZP2213 (L06241), Bradyrhizobium sp. strain NC92 (U33192), Bradyrhizobium elkanii USDA94 (U04609), Bradyrhizobium (Parasponia) sp. strain ANU289 (X03720), and Rhizobium tropici CFN299 (X98514). Abbreviations: A., Azorhizobium; R. leg., Rhizobium leguminosarum; S., Sinorhizobium; B., Bradyrhizobium; M., Mesorhizobium; R., Rhizobium.
FIG. 3.
Phylogenetic tree based on the nifH sequences and constructed by using the neighbor-joining method. Only bootstrap values greater than 50% are shown. Sequences of the following strains were used (the numbers in parentheses are accession numbers): Bradyrhizobium japonicum USDA110 (K01620), Rhodospirillum rubrum (M33774), Rhodobacter capsulatus SB1003 (X07866), Azospirillum brasilense Sp7 (X51500), Rhizobium leguminosarum bv. trifolii (K00490), Sinorhizobium meliloti 41 (J01781), Sinorhizobium meliloti CC2013 (M55232), Rhizobium tropici (M55225), Rhizobium etli CFN-42 (M15942), and Sinorhizobium sp. strain ANU240 (= NGR234Smr) (M26961). Abbreviations: B., Bradyrhizobium; R. rubrum, Rhodospirillum rubrum; R. capsulatus, Rhodobacter capsulatus; A., Azospirillum; R. leg., Rhizobium leguminosarum; R., Rhizobium; S., Sinorhizobium.
Plasmid profiles.
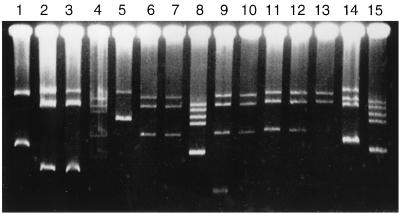
Comparison of plasmid numbers and sizes showed that the plasmid contents of the strains varied greatly and that only closely related strains had the same plasmid profile (Fig. 4 and Table 1). Profile p8 was shared by 16 closely related strains, and, in addition, profile p9 lacked only a small plasmid of profile p8 (Fig. 4). Likewise, profile p5 was shared by five strains, and profiles p4 and p6 closely resembled profile p5. The plasmids found in our tree rhizobia were generally very large, even larger than the 1.5-Mbp plasmid of S. meliloti that was used as a marker. All of the strains were examined at least once, but despite examining four or more gels for some strains, we never detected any plasmids in them (Table 1). Hybridization of the nifH probe with the Southern blots of some of the plasmid gels showed that at least 11 of the strains carried their symbiotic genes in one of the large plasmids (Table 1).
FIG. 4.
Plasmid profiles. The standards used were S. meliloti NZP4010, which has a megaplasmid of approximately 1,500 kbp, and R. leguminosarum bv. viciae 3841 (= 300 Strr [15]). Lane 1, S. meliloti; lane 2, H1495; lane 3, H1496; lane 4, H1478; lane 5, H1506; lane 6, H1481; lane 7, H1482; lane 8, R. leguminosarum; lane 9, H1484; lane 10, H1485; lane 11, H1486; lane 12, H1502; lane 13, H1501; lane 14, H1680; lane 15, R. leguminosarum.
DISCUSSION
Three symbiotic lineages with the same host specificity.
In our collection of rhizobia, which were isolated largely from members of the genera Acacia and Prosopis, the nodulation specificity gene nodA and the nitrogen fixation gene nifH show the same phylogenetic pattern. There are three major branches which correspond to taxonomic and geographic divisions: sinorhizobia from Africa, sinorhizobia from Latin America, and mesorhizobia from both regions. Rhizobia in all three groups have similar host ranges that include members of the genera Acacia, Prosopis, and Leucaena (21, 48). Our demonstration that there are three deep symbiotic lineages with similar host ranges raises some interesting questions about the relationships among symbiotic genes, bacterial species, and plant hosts and about the biogeographic pattern of this variation.
Introduced P. chilensis shares sinorhizobia with native African A. senegal.
When legume species are introduced into regions outside their natural areas of distribution (e.g., soybeans in North America or the genus Lotus in New Zealand), it has often proved necessary to inoculate them with the appropriate rhizobia in order to establish an effective symbiosis. Sometimes, however, the incoming legume may be able to adopt the symbionts of a native plant belonging to the same cross-inoculation group (24), and our results indicate that this is the case for P. chilensis in Africa. P. chilensis is a South American tree that has been introduced into Africa. In African soils it is nodulated by sinorhizobia that show some variation but can be identical to the bacteria found on the African tree A. senegal in 16S rRNA gene sequence, nodA and nifH restriction patterns, and plasmid profiles (Table 1), identical in nodA sequence (Fig. 3), and very similar in nifH sequence (Fig. 2). The similarity of strains from these two hosts is not so surprising because previous work has shown that these strains are capable of nodulating members of both of the host genera (48). Since A. senegal is native to Africa, we tentatively suggest that this is a natural host of these sinorhizobia, while the ability of the sinorhizobia to nodulate the introduced Prosopis species is fortuitous. To take this question further, we need to ask what nodulates P. chilensis in its native area, South America.
Latin American Prosopis sinorhizobia are different from African isolates.
Although we do not have strains obtained from P. chilensis in South America, we do have two isolates obtained from other Prosopis species in Latin America (strains BR4007 and M6). Both of these strains are Sinorhizobium strains, and they have similar nodA and nifH sequences despite the fact that one is from Brazil and the other is from Mexico. However, the nodA and nifH sequences are quite different from those of the African symbionts of P. chilensis (Fig. 2 and 3). This supports the idea that sinorhizobia have had a long history of separate evolution on Prosopis spp. and other Latin American hosts on the one hand and on African hosts, including Acacia spp., on the other hand. These lineages have overlapping host ranges, but there are substantial differences in their symbiotic genes. Of course, this hypothesis needs to be tested by more extensive sampling of Latin American strains. If the distinction between African and Latin American sinorhizobia is confirmed by further sampling, it would be worth investigating them for possible differences in environmental adaptation.
Mesorhizobia on African Acacia spp. are similar to mesorhizobia on Latin American trees.
In contrast to the Sinorhizobium situation, the Mesorhizobium strains from Acacia spp. in Africa are genetically similar to two isolates from Brazil, BR3804 isolated from Chamaecrista sp. and INPA78B isolated from Leucaena sp. (Table 1 and Fig. 2 and 3). While it is possible that this similarity might reflect recent human-mediated movement between the continents, there is no strong reason to suspect this and certainly no record of deliberate introduction. It could well be that these are cosmopolitan organisms that occur naturally in both Africa and South America. There are other rhizobia with wide distributions that are apparently natural (24). For example, similar strains of R. leguminosarum are found on native legumes in western North America and in Europe (38). The biogeography of bacteria is still very poorly understood, and it is quite possible that many bacteria are sufficiently mobile that their global distribution is determined more by the suitability of the habitat than by geographic proximity. For rhizobia the presence of host plants is, of course, a relevant habitat factor, but it does not readily explain the difference in the geographic distributions of the sinorhizobia and mesorhizobia found in this study because suitable hosts are present in all of the locations.
Sinorhizobia and mesorhizobia on African Acacia spp. do not share symbiotic determinants.
When we began this study, we had both Sinorhizobium and Mesorhizobium isolates from acacia plants grown in the same African soils. One question which we posed was whether these bacteria shared the same host because host range determinants had been transferred between them. Although the number of mesorhizobial strains in our study was quite small, the answer seems to be clearly negative; the bacteria belonging to the two genera have nodA and nifH genes that are very distinct (Fig. 2 and 3). Even if there is some diversity within each genus, this diversity is small compared to the difference between the genera. There is, therefore, no evidence for recent gene transfer between Sinorhizobium and Mesorhizobium species.
There may be gene transfer among African sinorhizobial species.
Although Sinorhizobium strains have only small differences in their 16S rRNA gene sequences, these differences are large enough to divide the strains into separate species, when these data are supported by other characteristics (4, 12, 26). The African isolates used in this study have diverse partial 16S rRNA gene sequences that match the sequences of all five of the Sinorhizobium species described so far, plus a range of intermediate sequences. This suggests that these organisms represent several species. Nevertheless, identical nodA or nifH restriction patterns, or even identical sequences in the case of nodA, are found in strains with different 16S rDNA sequences. Thus, within the group of African sinorhizobia it seems likely that the different species have exchanged genes by lateral transfer. The presence of plasmids in nearly all of these strains (Fig. 4 and Table 1) and our observation that nifH can be located on these plasmids (Table 1) suggest that plasmid-mediated gene transfer occurs.
Phylogeny of nifH.
The phylogenetic trees based on the nodA and nifH genes of the Acacia rhizobia reveal the same three major clusters. Correlations between nod and nif genes have also been reported by Urtz and Elkan (44), who used the RFLP technique to study the diversity among bradyrhizobia that nodulate peanuts. The similarity of the nod and nif trees seems incompatible with the view that nifH phylogeny closely follows the phylogeny of 16S rRNA (13, 24, 45), whereas the phylogeny of nod genes is more closely related to the phylogeny of host plants (6, 42). At a higher taxonomic level nifH and 16S rRNA phylogenies are similar, and, for example, the nifH genes of species in the alpha subdivision of the Proteobacteria are quite distinct from the nifH genes of species in the gamma subdivision (45). However, the phylogenetic tree based on nifH sequences (Fig. 3) shows that bacteria with a Sinorhizobium background do not necessarily carry similar nifH genes. Indeed, the nifH tree is not consistent with the tree based on 16S rRNA in many respects, with the genera Rhizobium and Sinorhizobium thoroughly interspersed. Eardly et al. (7) presented another relevant example. Thus, lateral transfer of nif genes may have a significant effect when the species or genus level is reached. It would not be surprising if nod and nif genes showed similar evolutionary patterns at this level, because they are often linked on the same plasmid. However, some data indicate that nod and nif genes have come together fairly recently (9). This possibility is also supported by the fact that some strains (e.g., S. saheli strains) have very different locations when the nifH and nodA trees are compared (Fig. 2 and 3).
S. saheli may have two biovars.
Lortet et al. (21) and Lorquin et al. (20) have recently shown that similar Nod factors are produced by S. teranga, mesorhizobia belonging to cluster U, and R. tropici, all of which nodulate A. senegal. In agreement with this, our results place all of these strains in the same nodA sequence branch, albeit it is a very deep branch (Fig. 2). However, the thin-layer chromatography results of Lortet et al. (21) showed that there could be two types of S. teranga Nod factors, depending on whether the strain was isolated from Acacia sp. or Sesbania sp.; the same symbiont hardly ever nodulated members of both genera. Since the Nod factor structure corresponded to the host specificity of strains, Lortet et al. proposed division of S. teranga into two biovars, S. teranga bv. acaciae and S. teranga bv. sesbaniae. In our study, a corresponding division based on host plants was found when we compared nodA sequences of two S. saheli strains. Strain H1496 was isolated from Acacia sp., but it has been found to be a typical S. saheli strain based on its partial 16S rRNA gene sequence (12) and other properties, such as the high DNA-DNA reassociation value characteristics and total proteins (26). This strain has a nodA sequence similar to the nodA sequences of other strains isolated from Acacia spp. and related species. By contrast, the type strain of S. saheli (ORS609T), which was isolated from Sesbania sp., has a very different nodA sequence (Fig. 2). Since NodA plays an important role in determining the host specificity of Nod factors (2, 32, 33), it seems only natural that analyzing NodA and Nod factors should lead to congruent results. Our results suggest that S. saheli, like S. teranga, should be divided into two biovars, S. saheli bv. acaciae and S. saheli bv. sesbaniae. A good correlation between nodA sequences and host range has recently been confirmed with more examples of rhizobia from Acacia and Sesbania species (1a), and, therefore, simple nodA PCR and restriction can be used as an easy screening technique in addition to, or instead of, Nod factor analysis.
Factors influencing evolution at different taxonomic levels.
As a way to summarize our findings, we considered those factors that appear to have influenced the evolution of rhizobial symbiotic genes, as reflected in the nod and nif phylogenetic trees. Five hierarchical levels were considered. At the first, highest level, rhizobia were considered one group in the context of all nitrogen-fixing bacteria; nifH phylogeny can be seen to follow 16S rRNA phylogeny, reflecting a common evolutionary history. Moving one level down, we considered the genera of rhizobia. Among Sinorhizobium species, for example, we observed in the phylogenies based on nod and nif some correlation with host plant range, for the temperate narrow-host-range symbiont S. meliloti, the broad-host-range symbiont S. fredii, and tropical tree rhizobia all form clusters of their own. At the next level, we concentrated on the tree rhizobia, the only group for which we had enough samples for a detailed analysis. Here it seems that the chromosomal background affected the evolution of symbiotic genes. This might be because the symbiotic system requires the correct chromosomal background in order to function optimally, or alternatively, the chromosomal and symbiotic genes may have simply coevolved together for a very long time without significant gene exchange. Down at the fourth level, geographical factors could also further differentiate the phylogenetic groups. This could be a consequence of either divergent evolution due to geographical separation or adaptation to different environmental conditions, which naturally also influence the range of legume hosts available. Finally, the fifth level was where we looked at the evolution of individual strains and found various combinations of nod and nif types in different 16S rRNA backgrounds. Thus, at this level the lateral transfer between strains seems to explain the differences in the various phylogenetic trees.
ACKNOWLEDGMENTS
We gratefully acknowledge receiving strains from F. M. Moreira, D. W. Odee, S. G. McInroy, and J. J. Peña-Cabriales. We also thank A. Mould for technical assistance with the ABI automated sequencer and A. Jumpponen for critical reading of the manuscript.
This work was funded mainly by an exchange grant from the Academy of Finland and the Royal Society to K.H. We also thank the Finnish Cultural Foundation and the University of York for financial support.
REFERENCES
- 1.Badenoch-Jones J, Holton T A, Morrison C M, Scott K F, Shine J. Structural and functional analysis of nitrogenase genes from the broad-host-range Rhizobium strain ANU240. Gene. 1989;77:141–153. doi: 10.1016/0378-1119(89)90368-5. [DOI] [PubMed] [Google Scholar]
- 1a.Boivin, C. 1997. Personal communication.
- 2.Debellé F, Plazanet C, Roche P, Pujol C, Savagnac A, Rosenberg C, Promé J C, Dénarié J. The NodA proteins of Rhizobium meliloti and Rhizobium tropici specify the N-acylation of Nod factors by different fatty acids. Mol Microbiol. 1996;22:303–314. doi: 10.1046/j.1365-2958.1996.00069.x. [DOI] [PubMed] [Google Scholar]
- 3.Debellé F, Yang G P, Ferro M, Truchet G, Promé J C, Dénarié J. Rhizobium nodulation factors in perspective. NATO Adv Study Inst Ser G Ecol Sci. 1997;G39:15–23. [Google Scholar]
- 4.de Lajudie P, Willems A, Pot B, Dewettinck D, Maestrojuan G, Neyra M, Collins M D, Dreyfus B, Kersters K, Gillis M. Polyphasic taxonomy of rhizobia: emendation of the genus Sinorhizobium and description of Sinorhizobium meliloti comb. nov., Sinorhizobium saheli sp. nov., and Sinorhizobium teranga sp. nov. Int J Syst Bacteriol. 1994;44:715–733. [Google Scholar]
- 5.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobert R C, Breil B T, Triplett E W. DNA sequence of the common nodulation genes of Bradyrhizobium elkanii and their phylogenetic relationships to those of other nodulating bacteria. Mol Plant Microbe Interact. 1994;7:564–572. doi: 10.1094/mpmi-7-0564. [DOI] [PubMed] [Google Scholar]
- 7.Eardly B D, Young J P W, Selander R K. Phylogenetic position of Rhizobium sp. strain Or 191, a symbiont of both Medicago sativa and Phaseolus vulgaris, based on partial sequences of the 16S rRNA and nifH genes. Appl Environ Microbiol. 1992;58:1809–1815. doi: 10.1128/aem.58.6.1809-1815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckhardt T. A rapid method for the identification of plasmid deoxyribonucleic acid in bacteria. Plasmid. 1978;1:584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- 9.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 10.Hashem F M, Kuykendall D. Plasmid DNA content of several agronomically important Rhizobium species that nodulate alfalfa, berseem clover, or Leucaena. In: Graham P H, Sadowsky M J, Vance C P, editors. Symbiotic nitrogen fixation. Proceedings of the 14th North American Conference on Symbiotic Nitrogen Fixation. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1993. pp. 181–188. [Google Scholar]
- 11.Haukka K, Lindström K. Pulsed-field gel electrophoresis for genotypic comparison of Rhizobium bacteria that nodulate leguminous trees. FEMS Microbiol Lett. 1994;119:215–220. [Google Scholar]
- 12.Haukka K, Lindström K, Young J P W. Diversity of partial 16S rRNA sequences among and within strains of African rhizobia isolated from Acacia and Prosopis. Syst Appl Microbiol. 1996;19:352–359. [Google Scholar]
- 13.Hennecke H, Kaluza K, Thöny B, Fuhrmann M, Ludwig W, Stackebrandt E. Concurrent evolution of nitrogenase genes and 16S rRNA in Rhizobium species and other nitrogen fixing bacteria. Arch Microbiol. 1985;142:342–348. [Google Scholar]
- 14.Jarvis B D W, van Berkum P, Chen W X, Nour S M, Fernandez M P, Cleyet-Marel J C, Gillis M. Transfer of Rhizobium loti, Rhizobium huakuii, Rhizobium ciceri, Rhizobium mediterraneum, and Rhizobium tianshanense to Mesorhizobium gen. nov. Int J Syst Bacteriol. 1997;47:895–898. [Google Scholar]
- 15.Johnston A W B, Hombrecher G, Brewin N J, Cooper M C. Two transmissible plasmids in Rhizobium leguminosarum strain 300. J Gen Microbiol. 1982;128:85–93. [Google Scholar]
- 16.Kaijalainen S, Lindström K. Restriction fragment length polymorphism analysis of Rhizobium galegae strains. J Bacteriol. 1989;171:5561–5566. doi: 10.1128/jb.171.10.5561-5566.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laguerre G, Geniaux E, Mazurier S I, Casartelli R R, Amarger N. Conformity and diversity among field isolates of Rhizobium leguminosarum bv. viciae, bv. trifolii, and bv. phaseoli revealed by DNA hybridization using chromosome and plasmid probes. Can J Microbiol. 1993;39:412–419. [Google Scholar]
- 18.Laguerre G, Mavingui P, Allard M-R, Charnay M-P, Louvrier P, Mazurier S-I, Rigottier-Gois L, Amarger N. Typing of rhizobia by PCR DNA fingerprinting and PCR-restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: application to Rhizobium leguminosarum and its different biovars. Appl Environ Microbiol. 1996;62:2029–2036. doi: 10.1128/aem.62.6.2029-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindström K, Paulin L, Roos C, Suominen L. Nodulation genes of Rhizobium galegae. In: Tikhonovich I A, Provorov N A, Romanov V I, Newton W E, editors. Nitrogen fixation: fundamentals and applications. Proceedings of the 10th International Congress on Nitrogen Fixation. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 365–370. [Google Scholar]
- 20.Lorquin J, Lortet G, Ferro M, Méar N, Promé J C, Boivin C. Sinorhizobium teranga bv. acaciae ORS1073 and Rhizobium sp. strain ORS1001, two distantly related Acacia-nodulating strains, produce similar Nod factors that are O-carbamoylated, N-methylated, and mainly sulfated. J Bacteriol. 1997;179:3079–3083. doi: 10.1128/jb.179.9.3079-3083.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lortet G, Méar N, Lorquin J, Dreyfus B, de Lajudie P, Rosenberg C, Boivin C. Nod factor TLC profiling as a tool to characterize symbiotic specificity of rhizobial strains: application to Sinorhizobium saheli, Sinorhizobium teranga and Rhizobium sp. strains isolated from Acacia and Sesbania. Mol Plant Microbe Interact. 1996;9:736–747. [Google Scholar]
- 22.Louvrier P, Laguerre G, Amarger N. Distribution of symbiotic genotypes in Rhizobium leguminosarum biovar viciae populations isolated directly from soils. Appl Environ Microbiol. 1996;62:4202–4205. doi: 10.1128/aem.62.11.4202-4205.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martínez-Romero E, Caballero-Mellado J. Rhizobium phylogenies and bacterial genetic diversity. Crit Rev Plant Sci. 1996;15:113–140. [Google Scholar]
- 24a.McInroy, S. G., K. Haukka, and J. P. W. Young. Unpublished data.
- 25.Moreira, F. M., K. Haukka, and J. P. W. Young. 1997. Unpublished data.
- 26.Nick, G., and K. Lindström. 1997. Unpublished data.
- 27.Norel F, Elmerich C. Nucleotide sequence analysis of the two nifH copies of Rhizobium ORS571. J Gen Microbiol. 1987;133:1563–1576. [Google Scholar]
- 28.Normand P, Bousquet J. Phylogeny of nitrogenase sequences in Frankia and other nitrogen-fixing microorganisms. J Mol Evol. 1989;29:436–447. doi: 10.1007/BF02602914. [DOI] [PubMed] [Google Scholar]
- 28a.Odee, D. W., K. Haukka, and J. P. W. Young. Unpublished data.
- 29.Paffetti D, Scotti C, Gnocchi S, Fancelli S, Bazzicalupo M. Genetic diversity of an Italian Rhizobium meliloti population from different Medicago sativa varieties. Appl Environ Microbiol. 1996;62:2279–2285. doi: 10.1128/aem.62.7.2279-2285.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Applic Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 31.Quinto C, de la Vega H, Flores M, Leemans J, Cevallos M A, Pardo M A, Azpiroz R, Girard M L, Calva E, Palacios R. Nitrogenase reductase: a functional multigene family in Rhizobium phaseoli. Proc Natl Acad Sci USA. 1985;82:1170–1174. doi: 10.1073/pnas.82.4.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritsema T, Wijfjes A H M, Lugtenberg B J J, Spaink H P. Rhizobium nodulation protein NodA is a host-specific determinant of the transfer of fatty acids in Nod factor biosynthesis. Mol Gen Genet. 1996;251:44–51. doi: 10.1007/BF02174343. [DOI] [PubMed] [Google Scholar]
- 33.Roche P, Maillet F, Plazanet C, Debellé F, Ferro M, Truchet G, Promé J C, Dénarié J. The common nodABC genes of Rhizobium meliloti are host-range determinants. Proc Natl Acad Sci USA. 1996;93:15305–15310. doi: 10.1073/pnas.93.26.15305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rome S, Fernandez M P, Brunel B, Normand P, Cleyet-Marel J-C. Sinorhizobium medicae sp. nov., isolated from annual Medicago spp. Int J Syst Bacteriol. 1996;46:972–980. doi: 10.1099/00207713-46-4-972. [DOI] [PubMed] [Google Scholar]
- 35.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:374–377. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Schofield P R, Gibson A H, Dudman W F, Watson J M. Evidence for genetic exchange and recombination of Rhizobium symbiotic plasmids in a soil population. Appl Environ Microbiol. 1987;53:2942–2947. doi: 10.1128/aem.53.12.2942-2947.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strain S R, Whittam T S, Bottomley P J. Analysis of genetic structure in soil populations of Rhizobium leguminosarum recovered from the USA and the UK. Mol Ecol. 1995;4:105–114. [Google Scholar]
- 39.Sullivan J T, Patrick H N, Lowther W L, Scott D B, Ronson C W. Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc Natl Acad Sci USA. 1995;92:8985–8989. doi: 10.1073/pnas.92.19.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas P M, Golly K F, Virginia R A, Zyskind J W. Cloning of nod gene regions from mesquite rhizobia and bradyrhizobia and nucleotide sequence of the nodD gene from mesquite rhizobia. Appl Environ Microbiol. 1995;61:3422–3429. doi: 10.1128/aem.61.9.3422-3429.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ueda T, Suga Y, Yahiro N, Matsuguchi T. Phylogeny of Sym plasmids of rhizobia by PCR-based sequencing of a nodC segment. J Bacteriol. 1995;177:468–472. doi: 10.1128/jb.177.2.468-472.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ueda T, Suga Y, Yahiro N, Matsuguchi T. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J Bacteriol. 1995;177:1414–1417. doi: 10.1128/jb.177.5.1414-1417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urtz B E, Elkan G H. Genetic diversity among Bradyrhizobium isolates that effectively nodulate peanut (Arachis hypogaea) Can J Microbiol. 1996;42:1121–1130. doi: 10.1139/m96-144. [DOI] [PubMed] [Google Scholar]
- 45.Young J P W. Phylogenetic classification of nitrogen-fixing organisms. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 43–86. [Google Scholar]
- 46.Young J P W, Haukka K E. Diversity and phylogeny of rhizobia. New Phytol. 1996;133:87–94. [Google Scholar]
- 47.Young J P W, Wexler M. Sym plasmid and chromosomal genotypes are correlated in field populations of Rhizobium leguminosarum. J Gen Microbiol. 1988;134:2731–2739. [Google Scholar]
- 48.Zhang X, Harper R, Karsisto M, Lindström K. Diversity of Rhizobium bacteria isolated from the root nodules of leguminous trees. Int J Syst Bacteriol. 1991;41:104–113. [Google Scholar]