Abstract
During the investigation of lignicolous freshwater fungi in plateau lakes in Yunnan Province, China, eight Lentitheciaceae species were collected from five lakes viz. Luguhu, Qiluhu, Xingyunhu, Cibihu, and Xihu lake. Based on morphological characters and phylogenetic analysis of combined ITS, LSU, SSU, and tef 1-α sequence data, a new genus Paralentithecium, two new species (Paralentithecium suae, and Setoseptoria suae), three new records (Halobyssothecium phragmitis, H. unicellulare, and Lentithecium yunnanensis) and three known species viz. Halobyssothecium aquifusiforme, Lentithecium pseudoclioninum, and Setoseptoria bambusae are reported.
Keywords: three new taxa, lignicolous freshwater fungi, plateau lake, phylogeny, taxonomy
1. Introduction
Lignicolous freshwater fungi are those fungi that grow on submerged woody debris in freshwater habitats, including lentic (e.g., lakes, ponds, swamps, and pools), lotic (e.g., rivers, streams, creeks, brooks), and other habitats (e.g., cooling tower, tree holes) [1,2,3]. They play an important role in the material and energy cycle of freshwater ecosystems [4,5,6,7,8]. Lignicolous freshwater fungi are a highly diverse group, with the majority belonging to Dothideomycetes and Sordariomycetes (Ascomycota), and a few species in Eurotiomycetes and Orbiliomycetes [3,9,10,11,12]. Lignicolous freshwater fungi have been investigated worldwide, but mainly in lotic habitats of tropical, subtropical, and temperate regions [10,13,14,15,16], with a few from lentic habitats [17]. Those fungi in lentic habitats are poorly studied. This study collects submerged decaying wood from plateau lakes in Yunnan, China, to investigate the species diversity of lignicolous freshwater fungi in the lakes.
Yunnan Province is in the southwest of China, it is a low-latitude, high-altitude inland province, and is one of the biodiversity hotspots in the Yunnan–Guizhou Plateau [18]. Yunnan has three climatic zones, tropical (southwest, south, and southeast borders), subtropical (west, middle, and east), and temperate regions (high-elevation area in the northwest) [19]. The special geographical location and climatic conditions endow Yunnan with abundant natural resources. There are plateau cold-resistant biomes in the west and tropical biomes in the south and southwest. Plateau lakes are an important part of terrestrial lakes and an important part of regional water cycling. They are distributed at higher altitudes, have a large number, and have a wide basin area. They are sensitive to climate change and have made outstanding contributions to coping with global climate change and shaping regional biodiversity patterns [20,21]. There have been several biological studies conducted on plateau lakes in Yunnan, such as waterbirds [22,23], invasive fish [24], water plants [25,26,27], and microorganisms [2,17,28,29,30,31,32]. Yunnan has abundant lignicolous freshwater fungi resources, and from 1986 to 2021, a total of 281 lignicolous freshwater fungi taxa have been reported. These species were mainly reported in lotic habitats (rivers, streams), with only 53 (19%) species from plateau lakes [12].
Species of Lentitheciaceae from freshwater habitats are mainly in Halobyssothecium, Lentithecium, Setoseptoria, and Tingoldiago [14,15,33,34,35,36,37]. The family was introduced by Zhang et al. [35] to accommodate those lentitheciaceous taxa that have narrow peridia, fusiform to broadly cylindrical pseudoparaphyses, hyaline ascospores with 1–3-transverse septa and containing refractive globules, surrounded by a mucilaginous sheath or extended appendage-like sheaths and asexual morphs are stagonospora-like or dendrophoma-like [14,38,39,40]. Currently, more than 100 species are reported in Lentitheciaceae. The last treatment of Lentitheciaceae was provided by Wijayawardene et al. [41] with acceptance of 18 genera: Crassoascoma [42], Darksidea [43], Groenewaldia [44], Halobyssothecium [45], Katumotoa [46], Keissleriella [47], Lentithecium [35], Murilentithecium [40], Neolentithecia [48], Neoophiosphaerella [34], Phragmocamarosporium [49], Pleurophoma [50,51], Poaceascoma [52], Pseudokeissleriella [53], Pseudomurilentithecium [54], Setoseptoria [37], Tingoldiago [55], and Towyspora [56].
We are investigating the diversity of lignicolous freshwater fungi from plateau lakes in Yunnan Province, and 13 collections of lentitheciaceous-like taxa were obtained. Based on morphological and multigene phylogenetic analysis, a new genus Paralentithecium is introduced to accommodate P. aquaticum, and a new taxon P. suae, Setoseptoria suae sp. nov. and new records Halobyssothecium and Lentithecium are described and illustrated. The sexual morph of Halobyssothecium phragmitis is also introduced.
2. Materials and Methods
2.1. Samples Collection
The fresh samples were submerged in lake water with a diameter of less than 2 cm and a length of more than 20 cm, including tree trunks, branches, twigs, and rotten branches of grasses. The specimens in this study were collected from Dali City (Cibihu, Xihu, and Erhai Lakes), Lijiang City (Luguhu Lake), and Yuxi City (Xingyunhu and Qiluhu Lakes) in Yunnan. The collected samples were placed in plastic ziplock bags and were taken back to the laboratory for processing.
2.2. Sample Processing and Cultivation
The samples were brought to the laboratory in ziplock bags to avoid moisture loss and then trimmed to 15 cm in length with pruning scissors. Each sample with a label number that is attached to the end of the sample with a thumbtack (Figure 1a). In addition, plastic boxes with the size of 24 cm × 16 cm × 6 cm were prepared. First, rinse the inside of the plastic box with sterile water, then wipe the entire plastic box with 75% alcohol. After drying, put two layers of sterilized tissues on the bottom of the box, lay three sterilized straws on the tissues to prevent the sample from directly touching the sterilized tissues, and add an appropriate amount of sterile water (the water soaks sterile tissues, but accumulates at the bottom), and then arrange the processed samples horizontally on the straw, ten samples in each plastic box, and label the boxes in obvious places (Figure 1b,c). The samples were placed on a culture rack and incubated at room temperature for one week (Figure 1d).
Figure 1.
(a) Sample with a label (arrow indicates sample number); (b) Samples in the plastic box; (c) Plastic box with labels (arrows indicate labels documenting detailed sampling sites and sample order); (d) The samples were incubated on the culture rack.
2.3. Morphological Studies and Isolation
Macromorphological characters of samples were observed using Optec SZ 760 compound stereomicroscope (Chongqing Optec Instrument Co., Ltd., Chongqing, China). The temporarily prepared microscope slide was placed under a Nikon ECLIPSE Ni-U compound stereomicroscope (Nikon, Tokyo, Japan) for observation and microscopic morphological photography. The morphology of colonies on native substrates was photographed with a Nikon SMZ1000 stereo-zoom microscope. Indian ink was used to reveal the presence of a gelatinous sheath around the ascospores or conidia. The measurements of photomicrographs were obtained using Tarosoft (R) Image Frame Work version 0.9.7. Images were edited with Adobe Photoshop CS5 Extended version 12.0.0.0 software (Adobe Systems, San Jose, CA, USA).
Single spore isolations were performed as follows: the tip of a sterile toothpick dipped in sterile water was used to capture the conidia of the target colony directly from the specimen; the conidia were then streaked on the surface of water agar (WA, Composition: Agar 20 g/L, Chloramphenicol 0.1 g/L) or potato dextrose agar (PDA, CM123, Composition: Potato infusion 5.0 g/L, Dextrose 20 g/L, Agar 20 g/L, Chloramphenicol 0.1 g/L, from Beijing Bridge Technology Co., Ltd., Beijing, China) and incubated at room temperature overnight. The single germinated conidia were transferred to fresh PDA medium and incubated at room temperature. A few of the remaining germinated spores in the media plate were separated along with agar using a needle and transferred onto water-mounted glass slides for photographs to capture the germination position of the germ tubes.
After finalizing the observation and isolation, the specimens were dried under natural light, wrapped in absorbent paper, and placed in a ziplock bag with mothballs. Specimens were deposited in the herbarium of Kunming Institute of Botany, Academia Sinica (KUN-HKAS). The living cultures were deposited in the China General Microbiological Culture Collection Center (CGMCC) and Kunming Institute of Botany Culture Collection (KUNCC). MycoBank numbers are registered in the MycoBank database (https://www.mycobank.org/Registration%20home (accessed on 4 August 2023)). Entries will be added to the Greater Mekong Subregion database [57].
2.4. DNA Extraction, PCR Amplification and Sequencing
DNA extraction, PCR amplification, sequencing, and phylogenetic analysis were done following the methods of Dissanayake et al. [58]. Mycelia for DNA extraction from each isolate was grown on PDA for 3–4 weeks at room temperature. Total genomic DNA was extracted from 100 to 300 mg axenic mycelium via scraping from the edges of the growing culture using a sterile scalpel and transferred to a 1.5 mL microcentrifuge tube using sterilized inoculum needles. The mycelium was ground to a fine powder with liquid nitrogen or quartz sand to break the cells for DNA extraction. When the cultures could not be maintained with some of the collected samples, fruiting structures (20–50 mg) were removed from the natural substrate using a sterile scalpel placed on sterile paper and then transferred to a 1.5 mL microcentrifuge tube. DNA was extracted with the TreliefTM Plant Genomic DNA Kit (TSP101) following the manufacturer’s guidelines.
Four gene regions, ITS, LSU, SSU, and tef 1-α were amplified using ITS5/ITS4 [59], LR0R/LR5 [60], NS1/NS4 [59], and EF1-983F/EF1-2218R [61] primer pairs, respectively. The PCR mixture contained 12.5 µL of 2× Power Taq PCR MasterMix (a premix and ready-to-use solution, including 0.1 Units/µL Taq DNA Polymerase, 500 µm dNTP Mixture each (dATP, dCTP, dGTP, dTTP), 20 mm Tris–HCl pH 8.3, 100 Mm KCl, 3 mM MgCl2, stabilizer, and enhancer), 1 µL of each primer including forwarding primer and reverse primer (10 µm), 1 µL template DNA extract and 9.5 µL deionized water. The PCR thermal cycling conditions of ITS and SSU were as follows: 94 °C for s min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 56 °C for 50 s, elongation at 72 °C for 1 min, and a final extension at 72 °C for 10 min; LSU and tef 1-α were as follows: 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 50 s, elongation at 72 °C for 1 min, and a final extension at 72 °C for 10 min. PCR products were then purified using minicolumns, purification resin, and buffer according to the manufacturer’s protocols (Amersham product code: 27-9602-01). The sequences were carried out at Beijing Tsingke Biological Engineering Technology and Services Co., Ltd. (Beijing, China).
2.5. Phylogenetic Analyses
ITS, LSU, SSU, and tef 1-α sequence data used for phylogenetic analysis are selected based on the preliminary identification results and the related publications [14,15]. The sequences were aligned using MAFFT online service: multiple alignment program MAFFT v.7 (http://mafft.cbrc.jp/alignment/server/index.html (accessed on 30 August 2023)) [62], and sequence trimming was performed with trimAl v1.2 for Windows, and all parameters were set by default (http://trimal.cgenomics.org for specific operation steps (accessed on 30 August 2023)) [63]. The sequence dataset was combined using SquenceMatrix v.1.7.8 [64]. FASTA alignment formats were changed to PHYLIP and NEXUS formats by the website: ALignment Transformation EnviRonment (ALTER) (http://sing.ei.uvigo.es/ALTER/ (accessed on 30 August 2023)) [65]. The alignments and phylogenetic trees were deposited in TreeBASE (http://www.treebase.org/ (accessed on 31 August 2023), accession number: 30729-30733).
The single-gene phylogenetic tree was obtained based on maximum likelihood (ML) only, and the multigene phylogenetic tree was obtained based on maximum likelihood (ML) and Bayesian criterion (BI). ML tree and BI tree were run on the CIPRES Science Gateway portal [66,67,68,69]. MrModeltest v. 2.3 [70] was run under the AIC (Akaike Information Criterion) implemented in PAUP v. 4.0b10. to evaluate the best-fit model in both ML and BI analyses. ML analyses for the datasets were performed with RAxML-HPC2 on XSEDE v. 8.2.10 [66] using the determined best-fit substitution model with 1000 bootstrap iterations. The BI analysis was computed with MrBayes v. 3.2.6 [69]. Six simultaneous Markov chains were run with a suitable number of generations, and trees were sampled every 100th generation, ending the run automatically when the standard deviation of split frequencies dropped below 0.01. Alignment gaps were treated as missing characters in the analysis of the combined data set, where they will occur in relatively conserved regions. Trees were inferred using the heuristic search option with 1000 random sequence additions, with maxtrees set at 1000. Phylogenetic trees were visualized using FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 31 August 2023)), editing and typesetting using Adobe Illustrator (AI) (Adobe Systems Inc., San Jose, CA, USA). The new sequences were submitted in GenBank, and the strain information used in this paper is provided in Table 1.
Table 1.
Taxa used in the phylogenetic analyses and their corresponding GenBank accession numbers.
| Species | Strain/Voucher Number | GenBank Accession Number | |||
|---|---|---|---|---|---|
| LSU | SSU | ITS | tef 1-α | ||
| Bambusicola bambusae | MFLUCC 11–0614 T | JX442035 | JX442039 | NR_121546 | KP761722 |
| Bambusicola irregulispora | MFLUCC 11–0437 T | JX442036 | JX442040 | NR_121547 | KP761723 |
| Bambusicola massarinia | MFLUCC 11–0389 T | JX442037 | JX442041 | JX442033 | KP761725 |
| Bambusicola splendida | MFLUCC 11–0439 T | JX442038 | JX442042 | NR121549 | KP761726 |
| Crassoascoma potentillae | UESTCC 21.0010 | OK161254 | OK161233 | OK161237 | OK181165 |
| Crassoascoma potentillae | UESTCC 21.0011 | OK161255 | OK161234 | OK161238 | OK181166 |
| Crassoascoma potentillae | UESTCC 21.0012 | OK161256 | OK161235 | OK161239 | OK181167 |
| Crassoascoma potentillae | CGMCC 3.20483 T | OK161257 | OK161236 | OK161240 | OK181168 |
| Darksidea alpha | CBS 135650 T | KP184019 | KP184049 | NR_137619 | KP184166 |
| Darksidea beta | CBS 135637 T | KP184023 | KP184074 | NR_137957 | KP184189 |
| Darksidea delta | CBS 135638 T | KP184024 | KP184069 | NR_137075 | KP184184 |
| Darksidea epsilon | CBS 135658 T | KP184029 | KP184070 | NR_137959 | KP184186 |
| Darksidea gamma | CBS 135634 T | KP184031 | KP184073 | NR_137587 | KP184188 |
| Darksidea zeta | CBS 135640 T | KP184013 | KP184071 | NR_137958 | KP184191 |
| Halobyssothecium aquifusiforme | GZCC 20–0481 T | OP377925 | OP378010 | OP377825 | OP473005 |
| Halobyssothecium aquifusiforme | MFLUCC 19–0305 | OP377929 | OP378014 | OP377829 | OP473008 |
| Halobyssothecium aquifusiforme | KUNCC 22–12665 | OR335346 | OR335329 | OR335289 | OR367662 |
| Halobyssothecium bambusicola | MFLUCC 20–0226 T | MT068489 | MT068494 | MN833419 | MT477868 |
| Halobyssothecium cangshanense | DLUCC 0143 T | KU991149 | KU991150 | – | – |
| Halobyssothecium caohaiense | GZCC 19–0482 T | MW133831 | MW134611 | OP377841 | OP473019 |
| Halobyssothecium carbonneanum | CBS 144076 T | MH069699 | – | MH062991 | – |
| Halobyssothecium estuariae | MFLUCC 19–0386 T | MN598871 | MN598868 | MN598890 | MN597050 |
| Halobyssothecium estuariae | MFLUCC 19–0387 T | MN598872 | MN598869 | MN598891 | MN597051 |
| Halobyssothecium kunmingense | KUMCC 19–0101 T | MN913732 | MT864313 | MT627715 | MT954408 |
| Halobyssothecium obiones | 20AV2566 | – | – | KX263862 | – |
| Halobyssothecium obiones | 27AV2385 | – | – | KX263864 | – |
| Halobyssothecium obiones | MFLUCC 15–0381 T | MH376744 | MH376745 | MH377060 | MH376746 |
| Halobyssothecium phragmitis | MFLUCC 20–0223 T | MT068486 | MT068491 | MT232435 | MT477865 |
| Halobyssothecium phragmitis | MFLUCC 20–0225 | MT068488 | MT068493 | MT232437 | MT477867 |
| Halobyssothecium phragmitis | HKAS 127181 | OR506189 | OR506192 | OR506177 | OR513794 |
| Halobyssothecium thailandica | MFLUCC 21–0062 T | MZ433248 | MZ429435 | MZ429434 | – |
| Halobyssothecium unicellulare | MD129 | KX505375 | KX505373 | – | – |
| Halobyssothecium unicellulare | KUNCC 22–12413 | OR335347 | OR335330 | OR335290 | |
| Halobyssothecium unicellulare | MD6004 T | KX505376 | KX505374 | – | – |
| Halobyssothecium versicolor | MFLUCC 20–0222 T | MT068485 | MW346047 | MT232434 | MT477864 |
| Halobyssothecium voraginesporum | CBS H-22560 T | KX499520 | KX499519 | – | – |
| Kalmusia scabrispora | KT2202 | AB524594 | AB524453 | LC014576 | AB539107 |
| Karstenula rhodostoma | CBS 690.94 | GU301821 | GU296154 | – | GU349067 |
| Katumotoa bambusicola | KT 1517a T | AB524595 | AB524454 | LC014560 | AB539108 |
| Keissleriella bambusicola | KUMCC 18–0122 T | MK995880 | MK995878 | MK995881 | MN213156 |
| Keissleriella breviasca | KT 581 | AB807587 | AB797297 | AB811454 | AB808566 |
| Keissleriella breviasca | KT 649 T | AB807588 | AB797298 | AB811455 | AB808567 |
| Keissleriella camporesiana | MFLUCC 15–0029 T | MN401741 | MN401743 | MN401745 | MN397907 |
| Keissleriella camporesii | MFLUCC 15–0117 T | MN252886 | MN252907 | MN252879 | – |
| Keissleriella caraganae | KUMCC 18–0164 T | MK359439 | MK359444 | MK359434 | MK359073 |
| Keissleriella cirsii | MFLUCC 16–0454 T | KY497780 | KY497782 | KY497783 | KY497786 |
| Keissleriella cladophila | CBS 104.55 T | GU301822 | GU296155 | MH857391 | GU349043 |
| Keissleriella culmifida | KT2308 | AB807591 | AB797301 | LC014561 | AB808570 |
| Keissleriella culmifida | KT2642 | AB807592 | AB797302 | LC014562 | AB808571 |
| Keissleriella dactylidicola | MFLUCC 13–0866 T | KT315506 | KT315505 | – | KT315507 |
| Keissleriella dactylidis | MFLUCC 13–0751 T | KP197668 | KP197666 | KP197667 | KP197669 |
| Keissleriella genistae | CBS 113798 | GU205222 | GU205242 | – | – |
| Keissleriella gloeospora | KT829 | AB807589 | AB797299 | LC014563 | AB808568 |
| Keissleriella linearis | IFRD2008 | FJ795435 | FJ795478 | – | – |
| Keissleriella linearis | MFLUCC 19–0410 | MN598873 | MN598870 | MN598892 | MN607978 |
| Keissleriella linearis | MFLUCC 20–0224 | MT068487 | MT068492 | MT232436 | MT477866 |
| Keissleriella phragmiticola | CPC 33249 | MT223903 | – | MT223808 | MT223715 |
| Keissleriella phragmiticola | MFLUCC 17–0779 T | MG829014 | – | MG828904 | – |
| Keissleriella poagena | CBS 136767 | KJ869170 | – | KJ869112 | – |
| Keissleriella quadriseptata | KT2292 T | AB807593 | AB797303 | AB811456 | AB808572 |
| Keissleriella rara | CBS 118429 | GU479791 | GU479757 | – | – |
| Keissleriella rosacearum | MFLUCC 15–0045 T | MG829015 | MG829123 | – | – |
| Keissleriella rosae | MFLUCC 15–0180 T | MG829016 | MG922549 | – | – |
| Keissleriella rosarum | MFLUCC 15–0089 T | MG829017 | MG829124 | MG828905 | – |
| Keissleriella sparticola | MFLUCC 14–0196 T | KP639571 | – | – | – |
| Keissleriella tamaricicola | MFLUCC 14–0168 T | KU900300 | – | KU900328 | – |
| Keissleriella taminensis | KT571 | AB807595 | AB797305 | LC014564 | AB808574 |
| Keissleriella taminensis | KT594 | AB807596 | AB797306 | – | – |
| Keissleriella taminensis | KT678 | AB807597 | AB797307 | LC014565 | AB808575 |
| Keissleriella trichophoricola | CBS 136770 T | KJ869171 | – | KJ869113 | – |
| Keissleriella yonaguniensis | HHUF 30138 T | AB807594 | AB797304 | AB811457 | AB808573 |
| Keissleriella sp. | KT895 | AB807590 | AB797300 | – | AB808569 |
| Latorua caligans | CBS 576.65 T | MH870362 | – | MH858723 | – |
| Latorua grootfonteinensis | CBS 369.72 T | MH877741 | – | – | – |
| Lentithecium clioninum | KT1149A T | AB807540 | AB797250 | LC014566 | AB808515 |
| Lentithecium clioninum | KT1220 | AB807541 | AB797251 | LC014567 | AB808516 |
| Lentithecium fluviatile | CBS 122367 | FJ795451 | FJ795493 | – | GU349074 |
| Lentithecium fluviatile | CBS 123090 | FJ795450 | FJ795492 | – | – |
| Lentithecium pseudoclioninum | KT1113 T | AB807544 | AB797254 | AB809632 | AB808520 |
| Lentithecium pseudoclioninum | GZCC 19–0483 | MW133832 | MW134612 | OM692194 | – |
| Lentithecium pseudoclioninum | KUNCC 22–12414 | OR335348 | OR335331 | OR335291 | – |
| Lentithecium pseudoclioninum | KUNCC 22–12415 | OR335349 | OR335331 | OR335291 | – |
| Lentithecium yunnanensis | KUNCC 22–10776 T | ON227127 | ON227123 | ON227126 | ON228074 |
| Lentithecium yunnanensis | KUNCC 22–10777 | ON227124 | ON227122 | ON227125 | ON228075 |
| Lentithecium yunnanensis | KUNCC 22–12420 | OR335350 | OR335333 | OR335293 | OR367664 |
| Lentithecium yunnanensis | KUNCC 22–12421 | OR335351 | OR335334 | OR335294 | OR367665 |
| Lentithecium yunnanensis | KUNCC 22–12422 | OR335352 | OR335335 | OR335295 | OR367666 |
| Longipedicellata aptrootii | MFLUCC 10–0297 T | KU238894 | KU238895 | KU238893 | KU238892 |
| Longipedicellata aptrootii | MFLUCC 18–0988 | MN913744 | – | MT627733 | – |
| Macrodiplodiopsis desmazieri | CBS 140062 T | KR873272 | – | KR873240 | – |
| Massarina cisti | CBS 266.62 | FJ795447 | FJ795490 | LC014568 | AB808514 |
| Massarina eburnea | CBS 139697 | AB521735 | AB521718 | LC014569 | AB808517 |
| Massarina eburnea | CBS 473.64 | GU301840 | GU296170 | AF383959 | GU349040 |
| Multiseptospora thailandica | MFLUCC 11–0183 T | KP744490 | KP753955 | KP744447 | KU705657 |
| Murilentithecium clematidis | MFLUCC 14–0561 | KM408758 | KM408760 | KM408756 | KM454444 |
| Murilentithecium clematidis | MFLUCC 14–0562 T | KM408759 | KM408761 | KM408757 | KM454445 |
| Murilentithecium lonicerae | MFLUCC 18–0675 T | MK214373 | MK214376 | MK214370 | MK214379 |
| Murilentithecium rosae | MFLUCC 15–0044 T | MG829030 | MG829137 | MG828920 | – |
| Neolentithecia changchunensis | CCMJ10012 T | MZ518790 | MZ518820 | MZ519071 | – |
| Neoophiosphaerella sasicola | KT1706 T | AB524599 | AB524458 | LC014577 | AB539111 |
| Parabambusicola thysanolaenae | KUMCC 18–0147 T | MK098199 | MK098205 | MK098190 | MK098209 |
| Parabambusicola thysanolaenae | KUMCC 18–0148 | MK098198 | MK098202 | MK098193 | MK098211 |
| Paraconiothyrium brasiliense | CBS 100299 T | JX496124 | AY642523 | JX496011 | – |
| Paraphaeosphaeria michotii | MFLUCC 13–0349 T | KJ939282 | KJ939285 | KJ939279 | – |
| Paraphaeosphaeria minitans | CBS 122788 | EU754173 | EU754074 | – | GU349083 |
| Phragmocamarosporium hederae | MFLUCC 13–0552 T | KP842915 | KP842918 | – | – |
| Phragmocamarosporium platani | MFLUCC 14–1191 T | KP842916 | KP842919 | – | – |
| Phragmocamarosporium rosae | MFLUCC 17–0797 T | MG829051 | MG829156 | – | MG829225 |
| Pleomonodictys descalsii | CBS 142298 T | KY853522 | – | KY853461 | – |
| Pleomonodictys capensis | CBS 968.97 T | KY853521 | – | KY853460 | – |
| Pleurophoma ossicola | CBS139905 T | KR476769 | – | KR476736 | – |
| Pleurophoma ossicola | CPC24985 | KR476770 | – | KR476737 | – |
| Pleurophoma pleurospora | CBS130329 T | JF740327 | – | – | – |
| Poaceascoma aquaticum | MFLUCC 14–0048 T | KT324690 | KT324691 | – | – |
| Poaceascoma halophila | MFLUCC 15–0949 T | MF615399 | MF615400 | – | – |
| Poaceascoma helicoides | MFLUCC 11–0136 T | KP998462 | KP998463 | KP998459 | KP998461 |
| Poaceascoma taiwanense | MFLUCC 18–0083 T | MG831567 | MG831568 | MG831569 | – |
| Paralentithecium aquaticum | CBS 123099 T | GU301823 | GU296156 | NR_160229 | GU349068 |
| Paralentithecium suae | CGMCC 3.24265 T | OQ732683 | OQ875040 | OQ874972 | OR367672 |
| Pseudokeissleriella bambusicola | CGMCC 3.20950 T | ON614138 | ON614096 | ON614135 | ON639623 |
| Pseudokeissleriella bambusicola | UESTCC 22.0028 | ON614137 | ON614095 | ON614134 | ON639622 |
| Setoseptoria arundelensis | MFLUCC 17–0759 T | MG829073 | MG829173 | MG828962 | – |
| Setoseptoria arundinacea | CBS 123131 | GU456320 | GU456298 | – | GU456281 |
| Setoseptoria arundinacea | CBS 619.86 | GU301824 | GU296157 | – | – |
| Setoseptoria arundinacea | MAFF 239460 | AB807574 | AB797284 | LC014594 | AB808550 |
| Setoseptoria arundinacea | MAFF 243842 T | AB807575 | AB797285 | LC014595 | AB808551 |
| Setoseptoria bambusae | GZCC 17–0044 | OP377919 | OP378004 | OP377820 | OP472999 |
| Setoseptoria bambusae | KUNCC 22–12416 | OR335353 | OR335336 | OR335296 | OR367667 |
| Setoseptoria bambusae | KUNCC 22–12417 | OR335354 | OR335337 | OR335297 | OR367668 |
| Setoseptoria bambusae | KUNCC 22–12418 | OR335355 | OR335338 | OR335298 | OR367669 |
| Setoseptoria englandensis | MFLUCC 17–0778 T | MG829074 | MG829174 | MG828963 | – |
| Setoseptoria lulworthcovensis | MFLU 18–0110 T | MG829075 | MG829175 | – | – |
| Setoseptoria magniarundinacea | KT1174 | AB807576 | AB797286 | LC014596 | AB808552 |
| Setoseptoria phragmitis | CBS 114802 T | KF251752 | – | KF251249 | KF253199 |
| Setoseptoria phragmitis | CBS 114966 | KF251753 | – | KF251250 | KF253200 |
| Setoseptoria scirpi | MFLUCC 14–0811 T | KY770982 | KY770980 | MF939637 | KY770981 |
| Setoseptoria suae | CGMCC 3.24266 T | OQ874972 | OQ875041 | OQ874972 | OR367673 |
| Splanchnonema platani | CBS 221.37 | MH867404 | – | MH855894 | DQ677908 |
| Splanchnonema platani | CBS 222.37 | KR909316 | KR909318 | KR909311 | KR909319 |
| Tingoldiago clavata | MFLUCC 19–0495 | MN857180 | MN857188 | MN857184 | – |
| Tingoldiago clavata | MFLUCC 19–0496 T | MN857178 | MN857186 | MN857182 | – |
| Tingoldiago clavata | MFLUCC 19–0498 | MN857179 | MN857187 | MN857183 | – |
| Tingoldiago graminicola | KH155 | AB521745 | AB521728 | LC014599 | AB808562 |
| Tingoldiago graminicola | KH68 T | AB521743 | AB521726 | LC014598 | AB808561 |
| Tingoldiago graminicola | KT891 | AB521744 | AB521727 | LC014600 | AB808563 |
| Tingoldiago hydei | MFLUCC 19–0499 T | MN857177 | – | MN857181 | – |
| Towyspora aestuari | MFLUCC 15–1274 T | KU248852 | KU248853 | NR_148095 | – |
Notes: The ex-type cultures are indicated using “T” after strain numbers; newly generated sequences are indicated in bold. “–” stands for no sequence data in GenBank.
3. Results
3.1. Phylogenetic Analysis
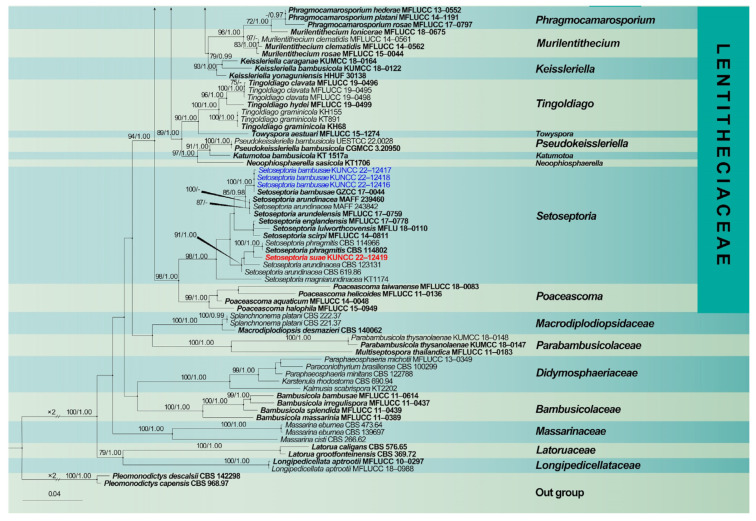
The combined ITS, LSU, SSU, and tef 1-α dataset comprises 147 taxa, including nine genera of Lentitheciaceae, with Pleomonodictys capensis (CBS 968.97) and P. descalsii (CBS 142298) as outgroup taxa (Figure 2). The dataset comprised 3777 characters (LSU: 1285 bp; SSU: 1021 bp; ITS: 539 bp; tef 1-α: 932 bp, including gaps). Maximum likelihood (ML) analysis and Bayesian analysis produced similar topologies that were consistent across the major clades. The likelihood of the final tree is evaluated and optimized under GAMMA. The best RAxML tree with a final likelihood value of −31,318.755060 is presented (Figure 2). The matrix had 1636 distinct alignment patterns, with 27.52% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.239720, C = 0.248808, G = 0.272414, T = 0.239058; substitution rates AC = 1.212047, AG = 2.534776, AT = 1.388124, CG = 1.249521, CT = 7.002685, GT = 1.000000, α = 0.226056, Tree-Length: 3.286461. Bayesian analyses generated 4412 trees (average standard deviation of split frequencies: 0.009960) from which 3310 were sampled after 25% of the trees were discarded as burn-in. The alignment contained a total of 1441 unique site patterns. Bootstrap support values with an ML greater than 75%, and Bayesian posterior probabilities (PP) greater than 0.97 are given above the nodes.
Figure 2.
Maximum likelihood (ML) tree is based on combined LSU, SSU, ITS, and tef 1-α sequence data. Bootstrap support values with an ML greater than 70% and Bayesian posterior probabilities (PP) greater than 0.97 are given above the nodes, shown as “ML/PP”. The tree is rooted to Pleomonodictys capensis (CBS 968.97) and P. descalsii (CBS 142298). New species are indicated in red bold, new strains are indicated in blue, and type strains are in black bold.
The multigene phylogenetic analyses showed that the 13 fresh collections clustered within Lentitheciaceae. Five known species, Halobyssothecium aquifusiforme (KUNCC 22-12665), H. phragmitis (KUN-HKAS 127181), H. unicellulare (KUNCC 22-12413), Lentithecium pseudoclioninum (KUNCC 22-12414 and KUNCC 22-12415), L. yunnanensis (KUNCC 22-124201, KUNCC 22-12421 and KUNCC 22-12422) and Setoseptoria bambusae (KUNCC 22-12416, KUNCC 22-12417 and KUNCC 22-12418) clustered with their ex-type strains, respectively. Paralentithecium suae (KUNCC 22-12412) clustered sister to P. aquaticum (CBS 123099) in an independent clade within Lentitheciaceae. Setoseptoria suae (KUNCC 22-12419) was placed sister to S. phragmitis (CBS 114804 and CBS 114966). Single-gene phylogenies are shown as Supplemental Materials (Figures S1–S4) because they resulted in being less informative and resolutive than those based on the four-loci concatenated tree.
3.2. Taxonomy
Halobyssothecium aquifusiforme J. Yang, Jian K. Liu & K.D. Hyde, Fungal Diversity 119: 39 (2023). Figure 3.
Figure 3.
Halobyssothecium aquifusiforme (KUN-HKAS 1124599). (a,b) Appearance of ascomata on the host; (c,d) Sections of ascomata; (e) Ostiole; (f,g) Section of peridium; (h) Pseudoparaphyses; (i–n) Asci; (o–u) Ascospores; (v) Germinated conidium; (w,x) Colony on MEA, obverse (w) and reverse (x); Scale bar: (c,d) = 150 µm; (e,i–n) = 30 µm; (f–h) = 20 µm; (o–v) = 10 µm.
Index Fungorum number: IF559450; Facesoffungi number: FoF12783.
Saprobic on submerged decaying wood in a freshwater lake. Asexual morph: Undetermined. Sexual morph: Ascomata 354–382 µm high, 328–366 µm wide, immersed, clustered, sometimes solitary, scattered, subglobose or ellipsoidal, dark brown to black, carbonaceous, uniloculate, ostiolate. Ostiolar neck central, 86–114 µm long, 138–168 µm wide, papillate, rounded, short, dark brown, composed of several layers of pseudoparenchymatous cells. Peridium 22–35 µm thick, composed of several layers of pseudoparenchymatous cells, an outer layer composed of black cells, arranged in a textura angularis, inner layer composed of hyaline, flattened cells, arranged in a textura angularis. Pseudoparaphyses about 2 µm wide, branched, septate, hyaline, filamentous, anastomosing above the asci. Asci 97–129 × 13–16 µm ( = 113 × 14 µm, n = 20) µm, 8-spored, clavate to subcylindrical, bitunicate, fissitunicate, apex rounded, short pedicellate, with an ocular chamber. Ascospores (20–)24–27 × 7–8 µm ( = 25 × 8 µm, n = 40), overlapping, uniseriate to biseriate, central cells are brown to dark brown, 1-septate when young, 3-septate when mature, constricted at the septa, slightly curved, fusiform, guttulate, conical and narrowly rounded at the ends, one cell on the central septum side is swollen, lacking gelatinous sheaths or appendages.
Culture characteristics: Ascospore germinating on PDA within 12 h. Colonies on PDA reaching 3 cm diameter in 6 weeks at room temperature. Mycelium superficial, initially white, later becoming brown to black, with pale brown dense aerial mycelium on the surface, mastoid, marginal mycelium smooth, sparse, brown to black; from below, light brown at the center, dark brown at the margin.
Material examined: China, Yunnan Province, Dali City, Eryuan County, Cibihu Lake, 26°09′59″ N, 99°55′27″ E (2050 m), on unknown submerged decaying wood, 21 July 2021, S.P. Huang and L.L. Li, L788 (KUN-HKAS 124599), living cultures (KUNCC 22-12665).
Known host and distribution: China, Guizhou Province, Anshun City, Gaodang village, 26.071° N, 105.698° E, Suoluo River, on decaying wood submerged in a freshwater stream HKAS 112638, (holotype), HKAS 112641 (paratype) [15].
Notes: The phylogenetic analysis showed that our new strain, KUNCC 22-12665 clustered sister to Halobyssothecium aquifusiforme (GZCC 20-0481 and MFLUCC 19-0305) with 99% ML/1.00 PP supports (Figure 2). Our species is similar to H. aquifusiforme in having immersed, subglobose ascomata, and fusiform, guttulate, septate ascospores which are constricted at the septum [15]. We, therefore, identified our new collection as H. aquifusiforme and provided detailed descriptions and illustrations for it. Halobyssothecium aquifusiforme is an aquatic species that was collected on submerged decaying wood in a freshwater stream in Guizhou, China. Our two new collections were collected from a plateau lake in Yunnan.
Halobyssothecium phragmitis M.S. Calabon, E.B.G. Jones, S. Tibell & K.D. Hyde, Mycological Progress 20: 711 (2021). Figure 4.
Figure 4.
Halobyssothecium phragmitis (KUN-HKAS 124600, new geographic record). (a,b) Appearance of ascomata on the host; (c,d) Sections of ascomata; (e,f) Section of peridium; (g) Pseudoparaphyses; (h,i) Asci; (j–n) Ascospores. Scale bar: (c,d) = 200 µm; (e–g) = 20 µm; (h,i) = 40 µm; (j–n) = 10 µm.
Index Fungorum number: IF558090; Facesoffungi number: FoF 09431.
Saprobic on submerged decaying wood in a freshwater lake. Sexual morph: Ascomata 529–566 µm high, 545–691 µm wide, immersed or semi-immersed, solitary to gregarious, scattered, subglobose or ellipsoidal, dark brown, subcarbonaceous or coriaceous, uniloculate, with indistinct ostiolate. Ostiolar neck 172–265 µm high, 184–320 µm wide, central, papillate, rounded, short, dark brown, composed of several layers of pseudoparenchymatous cells. Peridium 26–77 µm thick, composed of several layers of pseudoparenchymatous cells, an outer layer composed of brown cells, arranged in a textura angularis and textura globulosa, and an inner layer composed of hyaline, flattened cells, arranged in a textura angularis. Pseudoparaphyses 2–3 µm wide, septate, hyaline, filiform, branched, anastomosing above the asci. Asci (102–)111–130(–137) × 10–12 µm ( = 121 × 11 µm, n = 30), 8-spored, clavate to subcylindrical, bitunicate, fissitunicate, short pedicellate with an ocular chamber. Ascospores 22–27 × 5–6 µm ( = 25 × 6 µm, n = 40), overlapping, uniseriate to biseriate, fusiform with narrow ends, cells swollen nearly central septum and gradually narrow toward ends, slightly curved, pale brown to dark brown and lightening from central cells to the end cells, 1-septate when young, 5-septate when mature, and constricted at the septa, lacking gelatinous sheaths or appendages. Asexual morph: Coelomycetes [14].
Material examined: China, Yunnan Province, Dali City, Eryuan County, Xihu Lake, 26°00′33″ N, 100°03′35″ E (1970 m), on unknown submerged decaying wood, 8 May 2021, S.P. Huang and L.L Li, L783 (KUN-HKAS 127181).
Known host and distribution: SWEDEN, Gotland, Kappelshamnsviken, on dead Phragmites culm (Poaceae), MFLU 20–0550 (holotype); ibid., Sudersand, on dead Phragmites (Poaceae) stem, MFLU 20–0552 (paratype) [14].
Notes: Halobyssothecium phragmitis was introduced by Calabon et al. [14], and only the asexual morph is known. This species was collected on Phragmites (Poaceae) culm in Europe. Phylogenetic analysis combined with ITS, LSU, SSU, and tef 1-α sequence data showed that our new collection (KUN-HKAS 127181) clustered with two strains of H. phragmitis (MFLUCC 20–0223 and MFLUCC 20–0225). The comparison of ITS, LSU, SSU, and tef 1-α sequences between our new collection (KUN-HKAS 127181) and the ex-type of H. phragmitis (MFLUCC 20–0226) showed 8 bp, 1 bp, 3 bp, and 3 bp differences, respectively. Morphologically, our new collection is similar to other sexual members of Halobyssothecium in having immersed or semi-immersed, subglobose or ellipsoidal, dark brown, subcarbonaceous or coriaceous ascomata, clavate to subcylindrical, bitunicate asci and 3-septate, fusiform ascospores [14,15,45,71]. Based on phylogenetic analysis and morphological evidence, we identified our new collection as H. phragmitis, and described its asexual morph. This is the first report of this species in China [14].
Halobyssothecium unicellulare (Abdel-Aziz) M.S. Calabon, K.D. Hyde & E.B.G. Jones, Mycological Progress 20: 715 (2021). Figure 5.
Figure 5.
Halobyssothecium unicellulare (KUN-HKAS 124589, new geographic record). (a,b) Appearance of conidiomata on the host. (c,d) Sections of conidiomata. (e,f) Conidiomatal wall. (g,h) Developing conidia attach to conidiogenous cells. (i–l) Conidia. (m) Germinated conidium. (n) Colony on PDA, obverse (upper) and reverse (lower). Scale bar: (c,d) = 40 µm, (e,f) = 20 µm, (g–m) = 10 µm.
Index Fungorum number: IF558094; Facesoffungi number: FoF 09437
Saprobic on submerged decaying wood in a freshwater lake. Sexual morph: Undetermined. Asexual morph: Coelomycetes. Conidiomata 135–178 µm high, 205–242 µm wide, immersed to semi-immersed, most immersed, clustered, sometimes solitary, scattered, subglobose or ellipsoidal, uniloculate, dark brown to black, carbonaceous, short ostiolate, papillate, rounded. Conidiomatal walls 14–31 µm thick, composed of several layers of hyaline to black–brown cells of textura angularis. Conidiophores are reduced to conidiogenous cells. Conidiogenous cells 5–12 × 3–5 µm ( = 8 × 4 µm, n = 30), hyaline, thin-walled, holoblastic, smooth, subglobose to pear-shaped, swollen at the base, sometimes one conidiogenous cell producing two conidia. Conidia 9–11 × 4–5 µm ( = 10 × 5 µm, n = 60), subglobose, ovate, clavate, ellipsoid, allantoid or irregular, hyaline, aseptate, several small to one big guttulate, smooth-walled.
Culture characteristics: Conidia germinating on PDA within 12 h and germ tubes produced from one end of the conidia. Colonies on PDA, circular, reaching 5 cm in one month at room temperature, flat surface, pale brown to brown in PDA medium. Mycelium superficial, white to brown, hairy, effuse with wavy edge, dense, circular, raised, undulate to filiform with age; reverse light brown in the middle, with a dark brown deposit on the outside.
Material examined: China, Yunnan Province, Dali City, Eryuan County, Xihu Lake, 26°00′33″ N, 100°03′35″ E (1970 m), on unknown submerged decaying wood, 8 May 2021, S.P. Huang and L.L Li, L412 (KUN-HKAS 124589), living cultures, KUNCC 22-12413.
Known host and distribution: EGYPT, Sohag City, on decayed wood submerged in the River Nile, CBS H-22674 (holotype) [72].
Notes: The multigene phylogenetic analysis showed that our new collection (KUNCC 22-12413) clustered with the ex-type strain of Halobyssothecium unicellulare (MD 6004) with 91% ML/1.00 PP support (Figure 2). Morphologically, our new collection fits well with the original description of H. unicellulare [72]. The nucleotide comparison of LSU and SSU sequence data between our new collection (KUNCC 22-12413) and H. unicellulare (MD 6004) revealed 2 bp (including one gap) and 1 bp (including one gap) differences, respectively. We therefore identified it as H. unicellulare and it was reported from China for the first time.
Lentithecium pseudoclioninum Kaz. Tanaka & K. Hiray, Studies in Mycology 82: 99 (2015). Figure 6.
Figure 6.
Lentithecium pseudoclioninum (KUN-HKAS 124590). (a) Appearance of ascomata on the host; (b,c) Sections of ascomata; (d) Ostiole; (e,f) Section of peridium; (g) Pseudoparaphyses; (h,i) Asci; (j–o) Ascospores; (p) Germinated conidium; (q) Colony on PDA, obverse (upper) and reverse (lower). Scale bar: (b,c) = 100 µm; (d) = 50 µm; (e–g) = 20 µm; (h,i) = 30 µm; (j–p) = 10 µm.
Index Fungorum number: IF811309; Facesoffungi number: FoF12785.
Saprobic on submerged decaying wood in a freshwater lake. Asexual morph: Undetermined. Sexual morph: Ascomata 201–310 µm high, 227–274 µm wide, black, semi-immersed, gregarious, erumpent, globose or subglobose, uniloculate, ostiolate. Ostiolar neck central, papillate, 92–110 µm long, 100–107 µm wide. Peridium 20–32 µm, thick-walled, brown to dark brown cells, composing several layers of pseudoparenchymatous cells of textura angularis, outer layers heavily pigmented, inner layers hyaline to pale brown, flattened. Pseudoparaphyses 2–3 µm wide, filamentous, branched septate. Asci 98–118 × 14–16 µm ( = 108 × 15 µm, n = 30), 8-spored, bitunicate, fissitunicate, cylindric-clavate, slightly curved, pedicellate, apex rounded with a minute ocular chamber. Ascospores 28–32 × 8–10 µm ( = 30 × 9 µm, n = 30), overlapping uni- to biseriate, narrowly fusiform, with a nearly median primary septum, constricted at the septum, hyaline, guttulate, usually with 2–4 larger guttules, asymmetrical, broadly fusiform, narrowly rounded at the ends, with a mucilaginous sheath.
Culture characteristics: Ascospore germinating on PDA within 12 h and germ tubes produced from the ends of the spore. Colonies on PDA, circular, reaching 5 cm in one month at room temperature, smooth surface, papillae, brown to dark brown. Mycelium superficial, hairy, smooth, circular, reverse grayish; reverse pale to brown, crack at the middle, flocculent at the edge.
Material examined: China, Yunnan Province, Dali City, Eryuan County, Xihu Lake, 26°00′33″ N, 100°03′35″ E (1970 m), on unknown submerged decaying wood, 8 May 2021, S.P. Huang and L.L Li, L413 (KUN-HKAS 124590), living cultures (KUNCC 22-12414); ibid., Erhai Lake, 26°00′32″ N, 100°03′35″ E (1970 m), on unknown submerged decaying wood, 01 April 2021, Z.Q. Zhang, L445 (KUN-HKAS 124593), living cultures (KUNCC 22-12415)
Known host and distribution: JAPAN, Aomori, Hirosaki, Aoki, Mohei pond, on submerged twigs of woody plant, KT 1113 (holotype) and KT 1111 (paratype); China, Guizhou Province, Weining City, Caohai National Nature Reserve, near 26.817° N, 104.217° E, on submerged decaying aquatic plants in Caohai lake, GZAAS 20-0378 [15].
Notes: Our two new collections are morphologically consistent with the holotype of Lentithecium pseudoclioninum [34]. In addition, phylogenetic analysis revealed that these two collections clustered with L. pseudoclioninum (Figure 2). Based on morphological and phylogenetic evidence, we identified our new collection as L. pseudoclioninum. Lentithecium pseudoclioninum has been collected on submerged twigs of woody plants in China and Japan [15,34]. Our two specimens were collected from a freshwater plateau lake in Yunnan, China.
Lentithecium yunnanensis W.H. Lu, Karun. & Tibpromma, Phytotaxa 554: 108 (2022). Figure 7.
Figure 7.
Lentithecium yunnanensis (KUN-HKAS 124597, new habitat records). (a–c) Appearance of ascomata on the host; (d,e) Sections of ascomata; (f) Section of peridium; (g) Pseudoparaphyses; (h) Ascomata wall with hypha; (i–k) Asci; (l–p) Ascospores; (q) Germinated conidium; (r) Colony on PDA, obverse (upper) and reverse (lower). Scale bar: (d,e) = 100 µm; (f,g) = 20 µm; (i–k) = 30 µm; (h,l–q) = 10 µm.
Index Fungorum number: IF559622; Facesoffungi number: FoF 10778.
Saprobic on submerged decaying wood in a freshwater lake. Asexual morph: Undetermined. Sexual morph: Ascomata 246–285 µm high, 179–229 µm wide, immersed to semi-immersed, clustered, sometimes solitary, scattered, subglobose or ellipsoidal, dark brown to black, carbonaceous, uni- to bi-loculate, with indistinct ostiolate. Ostiolar neck central, papillate, 127–156 µm long, 96–110 µm wide. Peridium 11–21 µm thick, composed of several layers of pseudoparenchymatous cells, outer layer composed of back brown to brown cells, arranged in textura angularis, inner layer composed of hyaline cells, arranged in textura angularis. Pseudoparaphyses about 2 µm wide, hyaline, filamentous, branched, septate, globose to subglobose swollen at the apex, sometimes swollen at the septum, anastomosing at the apex, embedded in a hyaline gelatinous matrix. Asci 98–117 × 14–15 µm ( = 108 × 15 µm, n = 15), 8-spored, clavate to subcylindrical, bitunicate, apex rounded, short pedicellate with an ocular chamber. Ascospores 27–30 × 5–6 µm ( = 28 × 6 µm, n = 30), overlapping, uniseriate to biseriate, hyaline, 1-septate, smooth, constricted at the septa, slightly curved, guttulate, lacking gelatinous sheaths or appendages.
Culture characteristics: Ascospore germinating on PDA within 12 h and germ tubes produced from both ends of the spore. Colonies on PDA, circular, reaching 6 cm in 45 days at room temperature, smooth surface, papillae, brown in PDA medium. Mycelium superficial, brown to dark brown, hairy, smooth, circular; reverse brown to dark brown, crack at the middle, flocculent at the edge.
Material examined: China, Yunnan Province, Dali City, Eryuan County, Xihu Lake, 26°17′37″ N, 99°58′33″ E (2100 m), on unknown submerged decaying wood, 22 July 2021, L.L. Li, L680 (KUN-HKAS 124598), living cultures (KUNCC 22-12420 = KUNCC 22-12422); ibid., 26°17′24″ N, 99°57′56″ E (2100 m), on unknown submerged decaying wood, 22 July 2021, X.J. Yuan, L679 (KUN-HKAS 124597), living culture (KUNCC 22-12421).
Known host and distribution: China, Yunnan, Kunming, Songhua Dam Reservoir, on dead culms of Artemisia sp., HKAS 123192 (holotype) [73].
Notes: Lentithecium yunnanensis is a terrestrial species introduced by Lu et al. [73] that occurs on dead culms of Artemisia sp. near humid places. We collected two Lentithecium-like collections from decaying wood submerged in Xihu Lake, Dali, Yunnan Province. Phylogenetic analysis showed that our two new collections clustered with two strains of L. yunnanensis (KUNCC 22-10776 and KUNCC 22-10776). In addition, the morphology of our two collections is similar to the holotype of L. yunnanensis in having semi-immersed to immersed, subglobose to globose ascomata with short ostioles, and hyaline, clavate to fusiform, septate ascospores. Therefore, the two new collections were identified as L. yunnanensis, which was reported from the freshwater habitat for the first time.
Paralentithecium H.W. Shen, K.D. Hyde & Z.L. Luo gen. nov.
MycoBank number: 849738.
Etymology: referring to the comparable morphological characters to that of Lentithecium.
Saprobic on submerged decaying wood in a freshwater lake. Asexual morph: Undetermined. Sexual morph: Ascomata immersed to semi-immersed, clustered, sometimes solitary, scattered, subglobose or ellipsoidal, dark brown to black, carbonaceous, uni- to bi-loculate, with indistinct ostiolate. Peridium thick, composed of several layers of pseudoparenchymatous cells, an outer layer composed of back brown to brown cells, arranged in textura angularis, and an inner layer composed of hyaline cells, arranged in textura angularis. Pseudoparaphyses thick, hyaline, filamentous, branched, septate, globose to subglobose swollen at the apex, sometimes swollen at the septum, anastomosing at the apex, embedded in a hyaline gelatinous matrix. Asci 8-spored, clavate to subcylindrical, bitunicate, apex rounded, short pedicellate with an ocular chamber. Ascospores overlapping, uniseriate to biseriate, hyaline, 1-septate, smooth, constricted at the septa, slightly curved, with gelatinous sheaths.
Type species: Paralentithecium aquaticum (Yin. Zhang, J. Fourn. & K.D. Hyde) H.W. Shen & Z.L. Luo.
Paralentithecium aquaticum (Yin. Zhang, J. Fourn. & K.D. Hyde) H.W. Shen & Z.L. Luo, comb. nov.
MycoBank number: MB 512791.
Basionym: Lentithecium aquaticum Yin. Zhang, J. Fourn. & K.D. Hyde, Fungal Diversity 38: 234 (2009).
Known host and distribution: FRANCE, Ariège, Rimont, Peyrau, on submerged wood of Fraxinus excelsior; on submerged wood of Alnus glutinosa; Le Baup brook, along D 18, on submerged wood of Platanus sp. [35].
Notes: Lentithecium aquaticum was introduced by Zhang et al. [35] based on phylogenetic analysis and morphological characteristics. The placement of this species was not stable and has been changed by several studies [10,45,71,74]. Previous phylogenetic analyses indicated that Lentithecium aquaticum did not cluster with other Lentithecium species, and it formed an individual lineage basal to Darksidea, Halobyssothecium and Lentithecium [10,14,34,41,48,71]. Furthermore, phylogenetic studies of Dayarathne et al. [45] and Devadatha et al. [71] showed that L. aquaticum clustered within Setoseptoria. Several other studies excluded L. aquaticum from Lentithecium [10,74]. The latest phylogenetic analysis based on combined ITS, LSU, SSU, and tef 1-α genes showed that L. aquaticum formed a separate lineage outside of Lentithecium [48]. Our phylogenetic analysis shows that L. aquaticum clusters with our new collection KUNCC 22-12412 and forms a distinct lineage within Lentitheciaceae with 100 ML/1.00 PP support (Figure 2). Therefore, we propose a new genus, Paralentithecium to accommodate Paralentithecium aquaticum (Lentithecium aquaticum) and a new species P. suae.
Paralentithecium suae H.W. Shen, K.D. Hyde & Z.L. Luo sp. nov. Figure 8.
Figure 8.
Paralentithecium suae (KUN-HKAS 124587, holotype). (a–c) Appearance of ascomata on the host; (d) Sections of ascomata; (e,f) Section of peridium; (g,h) Pseudoparaphyses; (i–l) Asci; (m–r) Ascospores; (s) Germinated conidium; (t,u) Colony on PDA, obverse (t) and reverse (u). Scale bar: (d) = 100 µm; (e–l) = 40 µm; (m–s) = 20 µm.
MycoBank number: 849739; Facesoffungi number: FoF 14876.
Etymology: “suae” (Lat.) in memory of the Chinese mycologist Prof. Hong-Yan Su (4 April 1967–3 May 2022).
Holotype: KUN-HKAS 124587.
Saprobic on submerged decaying wood in a freshwater lake. Asexual morph: Undetermined. Sexual morph: Ascomata 212–253 µm high, 175–204 µm wide, immersed to semi-immersed, clustered, sometimes solitary, scattered, subglobose or ellipsoidal, dark brown to black, carbonaceous, uni- to bi-loculate, with indistinct ostiolate. Peridium 17–32 µm thick, composed of several layers of pseudoparenchymatous cells, outer layer composed of bark brown to brown cells, arranged in textura angularis, inner layer composed of hyaline cells, arranged in textura angularis. Pseudoparaphyses 2–3 µm wide, hyaline, filamentous, branched, septate, globose to subglobose swollen at the apex, sometimes swollen at the septum (6–10 µm wide), anastomosing at the apex, embedded in a hyaline gelatinous matrix. Asci 104–134 × 24–28 µm ( = 119 × 26 µm, n = 25), 8-spored, clavate to subcylindrical, bitunicate, apex rounded, short pedicellate with an ocular chamber. Ascospores 28–34 × 11–14 µm ( = 31 × 13 µm, n = 40), overlapping, uniseriate to biseriate, hyaline, 1-septate, broadly fusiform, smooth, constricted at the septa, slightly curved, guttulate, with gelatinous sheaths.
Culture characteristics: Ascospore germinating on PDA within 12 h and germ tubes produced from both ends of the spore. Colonies on PDA, circular, reaching 4–5 cm in one month at room temperature, smooth surface, papillae, brown to dark brown, olive green in PDA medium. Mycelium superficial, brown to dark brown, hairy, smooth, circular; reverse dark brown, crack at the middle, flocculent at the edge, dark brown with greenish.
Material examined: China, Yunnan Province, Lijiang City, Ninglang County, Luguhu Lake, 27°44′15″ N, 100°45′16″ E (2700 m), on unknown submerged decaying wood, 5 March 2021, Z.Q. Zhang and L. Sha, L184 (KUN-HKAS 124587, holotype), ex-type living cultures (CGMCC 3.24265 = KUNCC 22–12412).
Notes: In our phylogenetic analysis, Paralentithecium suae clustered with P. aquaticum with 100% ML/1.00 PP support (Figure 2). Comparison of ITS, LSU, SSU, and tef 1-α sequences between Paralentithecium suae and P. aquaticum revealed 11 bp, 4 bp, 4 bp, and 22 bp differences, respectively. Paralentithecium suae resembles P. aquaticum in having hyaline, 1-septate, broadly fusiform ascospores with gelatinous sheaths [35]. However, P. suae is distinct from P. aquaticum in having globose to subglobose pseudoparaphyses that are swollen at the apex and sometimes swollen at the septum. In contrast, the pseudoparaphyses of P. aquaticum are not swollen. In addition, ascospores of P. aquaticum contain four refractive globules, while P. suae has ascospores with many small guttules [35]. Therefore, we introduce P. suae as a new species.
Setoseptoria bambusae J. Yang, Jian K. Liu & K.D. Hyde, Fungal Diversity 119: 44 (2022). Figure 9.
Figure 9.
Setoseptoria bambusae (KUN-HKAS 124592). (a–c) Appearance of ascomata on the host; (d,e) Sections of ascomata; (f,g) Section of peridium; (h) Pseudoparaphyses; (i–k) Asci; (l–o) Ascospores; (p,q) Ascospore stained in Indian ink; (r) Germinated conidium; (s) Colony on PDA, obverse (left) and reverse (right). Scale bar: (d,e) = 150 µm; (f–k) = 30 µm; (h,i) = 30 µm; (l–r) = 20 µm.
Index Fungorum number: IF559452; Facesoffungi number: FoF12786.
Saprobic on submerged decaying wood in a freshwater lake. Asexual morph: Undetermined. Sexual morph: Ascomata 245–375 µm high, 194–296 µm wide, black, superficial to semi-immersed, gregarious, fully or partly erumpent, globose, uniloculate, ostiolate. Peridium 26–39 µm wide, thick, multi-layered, outer layer most heavily pigmented, comprising blackish to dark brown amorphous layer, middle layer heavily pigmented, inner layer, pale brown to hyaline, cells towards the inside lighter, flattened, thick-walled. Pseudoparaphyses 2–3 µm wide, filamentous, branched septate. Asci 113–128 × 15–19 µm ( = 120 × 17 µm, n = 30), 8-spored, bitunicate, fissitunicate, clavate to cylindric-clavate, pedicellate, apex rounded with a minute ocular chamber. Ascospores 32–40 × 6–8 µm ( = 36 × 7 µm, n = 20), overlapping uni- to biseriate, narrowly fusiform, with a nearly median primary septum, deeply constricted at the septum, hyaline, guttulate, asymmetrical, conical, and narrowly rounded at the ends.
Culture characteristics: Ascospore germinating on PDA within 12 h and germ tubes produced from one end of the spore. Colonies on PDA, circular, reaching 6 cm in 45 days at room temperature, smooth surface, papillae, pale brown in PDA medium. Mycelium superficial, grayish-brown to brown, hairy, smooth, circular; reverse pale brown at the edges, dark brown in the middle, flocculent at the edge.
Material examined: China, Yunnan Province, Yuxi City, Jiangchuan District, Xingyunhu Lake, 24°23′05″ N, 102°48′22″ E (1720 m), on unknown submerged decaying wood, 10 July 2021, H.W. Shen, L511 (KUN-HKAS 124592), living culture (KUNCC 22–12417); ibid., 24°23′05″ N, 102°48′22″ E (1720 m), on the submerged stem of Phragmites sp. (Poaceae), 10 July 2021, S. Luan, L579 (KUN-HKAS 124596), living culture (KUNCC 22–12418); ibid., on submerged stem of Phragmites sp. (Poaceae), 10 July 2021, Y.K. Jiang, L474 (KUN-HKAS 124591), living culture (KUNCC 22–12416).
Known host and distribution: China, Guizhou Province, Anshun City, Gaodang Village, 26.071° N, 105.698° E, Suoluo River, on decaying bamboo culms submerged in a freshwater stream, HKAS 112629 (holotype) [15].
Notes: Setoseptoria bambusae was introduced by Yang et al. [15] to accommodate two collections, GZCC 17–0044 (ex-type strain) and IFRD500-013 (previously identified as S. arundinaceae, without description). In this study, our four new collections clustered with the ex-type strain of S. bambusae with 100% ML/1.00 PP statistical support (Figure 2). Furthermore, our collections fit the morphological characteristics of S. bambusae except for the size of asci and ascospores, our isolate has shorter asci (113–128 vs. 130–180 µm) and longer ascospores (32–40 vs. 28–37 µm). Therefore, we identified them as S. bambusae. Our four new collections were collected from lentic freshwater habitats. The holotype was collected from lotic habitats.
Setoseptoria suae H.W. Shen, K.D. Hyde & Z.L. Luo sp. nov. Figure 10.
Figure 10.
Setoseptoria suae (KUN-HKAS 124595, holotype). (a–c) Appearance of conidiomata on the host; (d) Sections of conidiomata; (e,f) Section of peridium; (g,h) Conidiomata and conidiogenous cells; (i–o) Conidia; (p) Germinated conidium; (q) Colony on PDA, obverse (left) and reverse (right). Scale bar: (d) = 100 µm; (e,f) = 30 µm; (g,h) = 10 µm; (h,i) = 30 µm; (i–p) = 10 µm.
MycoBank number: 849740; Facesoffungi number: FoF 14877.
Etymology: “suae” (Lat.) in memory of the Chinese mycologist Prof. Hong-Yan Su (4 April 1967–3 May 2022).
Holotype: KUN-HKAS 124595.
Saprobic on submerged decaying wood in a freshwater lake. Sexual morph: Undetermined. Asexual morph: Conidiomata 383–512 µm high, 173–196 µm wide, solitary, scattered, semi-immersed to immersed in the host, pycnidial, subglobose to ellipsoidal, unilocular, black, ostiolate, apapillate. Ostiole short, centrally located. Conidiomatal wall 33–55 µm wide, thickening at the upper zone, thick-walled, composed of several layers of textura angularis, an outer layer comprising brown to dark brown cells, pigmented; inner layer comprising hyaline cells. Conidiophores reduced to conidiogenous cells. Conidiogenous cells (4–)7–15(–26) × 4–6 µm ( = 11 × 5 µm, n = 25), arising from the inner layers of conidiomata, hyaline, enteroblastic, phialidic, determinate, ampuliform, subcylindrical to lageniform. Conidia 33–43 × 4–6 µm ( = 38 × 5 µm, n = 50), subcylindrical, with obtuse to subobtuse ends, straight or slightly curved, hyaline, (1–)3-septate, euseptate, mostly with one large central guttule per cell when young, with many small guttules in each cell at maturity, slightly constricted at the septum, smooth-walled.
Culture characteristics: Conidia germinated on PDA within 12 h and germ tubes produced from the ends of the spore. Colonies on PDA, circular, reaching 6 cm in one month at room temperature, brown to dark brown. Mycelium superficial, brown to dark brown, hairy, smooth, circular; dark brown from below.
Material examined: China, Yunnan Province, Yuxi City, Tonghai County, Qiluhuhu Lake, 24°08′37″ N, 102°46′24″ E (1800 m), on submerged stem of Phragmites sp. (Poaceae), 11 July 2021, H.W. Shen, L570 (KUN-HKAS 124595, holotype), ex-type living cultures (CGMCC 3.24266 = KUNCC 22–12419).
Notes: Phylogenetic analysis showed that Setoseptoria suae clustered with S. phragmitis with 100% ML/0.99 PP statistical support (Figure 2). The comparison of ITS and LSU sequences between S. suae and S. phragmitis shows that the similarities are 96.9% (538/555 bp) and 99.9% (826/827 bp), respectively. Setoseptoria suae resembles S. phragmitis in having immersed, globose conidiomata, hyaline, subcylindrical, smooth, guttulate, (1–)3-septate conidia [37]. However, Setoseptoria suae can be distinguished from S. phragmitis by its larger conidia (33–43 × 4–6 µm vs. (19–)25–35(–38) × (3.5–)4 µm). In addition, the conidia of S. phragmitis mostly have one large central guttule per cell, while Setoseptoria suae has conidia with many small guttules in each cell. We, therefore, introduce S. suae as a new species.
4. Discussion
Yunnan, located on the Yunnan–Guizhou Plateau, is one of the global biodiversity hotspots with rich biological resources [18,19,75]. In recent years, research on lignicolous freshwater fungi in Yunnan has developed rapidly, and a large number of new species have been reported from lotic freshwater habitats such as streams and rivers [10,13,76,77,78,79,80,81]. A few studies have reported lignicolous freshwater fungi from lentic habitats in Yunnan Province. For example, Cai et al. [17] and Luo et al. [2] investigated lignicolous freshwater fungi in Fuxianhu and Dianchi Lakes, respectively. However, freshwater fungi in lentic habitats have not been updated recently. In this study, we investigate the freshwater fungi in Cibihu, Luguhu, Qiluhu, Xihu, and Xingyunhu lakes in Yunnan Province, one new genus, two new species, and three new records are reported, the results indicate that high undiscovered diversity of lignicolous freshwater fungi in lentic habitats.
Zhang et al. [36] provided the first multigene phylogenetic analysis of Pleosporales and introduced the family Lentitheciaceae which accepted the genera Lentithecium, Katumotoa, and Keissleriella. Dong et al. [10] treated the family with ten genera and this was followed by Wijayawardene et al. [41]. Previous studies based on morphology and phylogenetic analyses showed that the classification of Lentithecium, Keissleriella, and Setoseptoria is confusing as the placement of several taxa was problematic and has been transferred to different genera. For example, Suetrong et al. [82] transferred Keissleriella rara to Lentithecium as L. rarum; however, later studies showed that L. rarum clustered with K. trichophoricola in Keissleriella [14]. Similarly, Zhang et al. [35] transferred Keissleriella linearis to Lentithecium as L. lineare, Singtripop et al. [83] re-examined the type specimen of K. linearis (L. lineare) and transferred it to Keissleriella based on LSU phylogenetic analysis, and this was confirmed by subsequent phylogenetic studies [14,34,72]. The placements of Lentithecium species have been revised in recent years based on multigene phylogenetic studies [14,34,84]. Calabon et al. [14] transferred several Lentithecium species with brown and versicolored ascospores without sheaths and hyaline conidia to Halobyssothecium, including L. cangshanense, L. carbonneanum, L. kunmingense, L. unicellulare, and L. voraginesporum. Currently, 13 species are accepted in Halobyssothecium. In the present study, we report the sexual morph of H. phragmitis and provide detailed morphological descriptions for its sexual morph.
Acknowledgments
Hong-Wei Shen thanks Long-Li Li, Qiu-Xia Yang, Sha Luan, and Si-Ping Huang for their help with sample collection, DNA extraction, and PCR amplification. Thanks to Rong-Ju Xu for his help in specimens and culture preservation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof9100962/s1.
Author Contributions
Conceptualization, Z.-L.L., S.B. and K.D.H.; methodology, H.-W.S. and X.-J.S.; formal analysis, H.-W.S.; investigation, H.-W.S., D.-F.B., X.-J.S. and X.-G.T.; resources, Z.-L.L. and S.B.; data curation, H.-W.S.; writing—original draft preparation, H.-W.S. and X.-G.T.; writing—review and editing, D.-F.B., S.B., X.-J.S., K.D.H. and Z.-L.L.; funding acquisition, Z.-L.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequences generated in this study were submitted to GenBank database.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the National Natural Science Foundation of China (Project ID: 32060005) and the Yunnan Fundamental Research Project (202201AW070001).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Wong M.K.M., Goh T.K., Hodgkiss I.J., Hyde K.D., Ranghoo V.M., Tsui C.K.M., Ho W.H., Wong W.S.W., Yuen T.K. Role of Fungi in Freshwater Ecosystems. Biodivers. Conserv. 1998;7:1187–1206. doi: 10.1023/A:1008883716975. [DOI] [Google Scholar]
- 2.Luo J., Yin J.F., Cai L., Zhang K.Q., Hyde K.D. Freshwater fungi in Lake Dianchi, a heavily polluted lake in Yunnan, China. Fungal Divers. 2004;16:93–112. [Google Scholar]
- 3.Hyde K.D., Fryar S., Tian Q., Bahkali A.H., Xu J.C. Lignicolous freshwater fungi along a north-south latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function? Fungal Ecol. 2016;19:190–200. doi: 10.1016/j.funeco.2015.07.002. [DOI] [Google Scholar]
- 4.Zare-Maivan H., Shearer C.A. Extracellular enzyme production and cell wall degradation by freshwater lignicolous fungi. Mycologia. 1988;80:365–375. doi: 10.1080/00275514.1988.12025551. [DOI] [Google Scholar]
- 5.Yuen T.K., Hyde K.D., Hodgkiss I.J. Physiological growth parameters and enzyme production in tropical freshwater fungi. Mater. Org. (Berl) 1998;32:2–16. [Google Scholar]
- 6.Abdel-Raheem A., Shearer C.A. Extracellular enzyme production by freshwater ascomycetes. Fungal Divers. 2002;11:1–19. [Google Scholar]
- 7.Bucher V.V.C., Hyde K.D., Pointing S.B., Reddy C.A. Production of wood decay enzymes, mass loss and lignin solubilization in wood by marine ascomycetes and their anamorphs. Fungal Divers. 2004;15:14. [Google Scholar]
- 8.Duarte S., Fernandes I., Nogueira M.J., Cássio F., Pascoal C. Temperature alters interspecific relationships among aquatic fungi. Fungal Ecol. 2013;6:187–191. doi: 10.1016/j.funeco.2013.02.001. [DOI] [Google Scholar]
- 9.Dong W., Hyde K.D., Doilom M., Yu X.D., Bhat D.J., Jeewon R., Boonmee S., Wang G.N., Nalumpang S., Zhang H. Pseudobactrodesmium (Dactylosporaceae, Eurotiomycetes, Fungi) a novel lignicolous genus. Front. Microbiol. 2020;11:456. doi: 10.3389/fmicb.2020.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong W., Wang B., Hyde K.D., McKenzie E.H.C., Raja H.A., Tanaka K., Abdel-Wahab M.A., Abdel-Aziz F.A., Doilom M., Phookamsak R., et al. Freshwater Dothideomycetes. Fungal Divers. 2020;105:319–575. doi: 10.1007/s13225-020-00463-5. [DOI] [Google Scholar]
- 11.Calabon M.S., Hyde K.D., Jones E.B.G., Luo Z.L., Dong W., Hurdeal V.G., Gentekaki E., Rossi W., Leonardi M., Thiyagaraja V., et al. Freshwater fungal numbers. Fungal Divers. 2022;114:3–235. doi: 10.1007/s13225-022-00503-2. [DOI] [Google Scholar]
- 12.Shen H.W., Bao D.F., Bhat D.J., Su H.Y., Luo Z.L. lignicolous freshwater fungi in Yunnan Province, China: An overview. Mycology. 2022;13:119–132. doi: 10.1080/21501203.2022.2058638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo Z.L., Hyde K.D., Liu J.K., Bhat D.J., Bao D.F., Li W.L., Su H.Y. Lignicolous freshwater fungi from china ii: Novel Distoseptispora (Distoseptisporaceae) species from northwestern Yunnan Province and a suggested unified method for studying lignicolous freshwater fungi. Mycosphere. 2018;9:444–461. doi: 10.5943/mycosphere/9/3/2. [DOI] [Google Scholar]
- 14.Calabon M.S., Jones E.B.G., Hyde K.D., Boonmee S., Tibell S., Tibell L., Pang K.L., Phookamsak R. Phylogenetic assessment and taxonomic revision of Halobyssothecium and Lentithecium (Lentitheciaceae, Pleosporales) Mycol. Prog. 2021;20:701–720. doi: 10.1007/s11557-021-01692-x. [DOI] [Google Scholar]
- 15.Yang J., Liu L.L., Jones E.B.G., Hyde K.D., Liu Z.Y., Bao D.F., Liu N.G., Li W.L., Shen H.W., Yu X.D., et al. Freshwater fungi from karst landscapes in China and Thailand. Fungal Divers. 2023;119:1–212. doi: 10.1007/s13225-023-00514-7. [DOI] [Google Scholar]
- 16.Tsui C.K.M., Hyde K.D., Hodgkiss I.J. Biodiversity of fungi on submerged wood in hong kong streams. Aquat. Microb. Ecol. 2000;21:289–298. doi: 10.3354/ame021289. [DOI] [Google Scholar]
- 17.Cai L., Tsui C.K.M., Zhang K., Hyde K.D. Aquatic fungi from lake Fuxian, Yunnan, China. Fungal Divers. 2000;14:57–70. [Google Scholar]
- 18.Xu Y., Shen Z.H., Ying L.X., Wang Z.H., Huang J.H., Zang R.G., Jiang Y.X. Hotspot analyses indicate significant conservation gaps for evergreen broadleaved woody plants in China. Sci. Rep. 2017;7:1859. doi: 10.1038/s41598-017-02098-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian L.S., Chen J.H., Deng T., Sun H. Plant diversity in yunnan: Current status and future directions. Plant Divers. 2020;42:281–291. doi: 10.1016/j.pld.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S.R., He X.Y., Yang W.S., Ren G.P., Li Y.P., Zhou J., Cai Q.H., Xiao W. Spatial distribution and significance of high mountain micro-waterbodies in northwestern Yunnan, China. J. Hydroecol. 2017;38:18–23. doi: 10.1016/j.foodhyd.2016.10.006. [DOI] [Google Scholar]
- 21.Liu S.R., De Yang D., Li X.F., Tan L., Sun J., He X.Y., Yang W.S., Ren G.P., Fornacca D., Cai Q.H., et al. Diversity in benthic and environmental characteristics on alpine micro-waterbodies and stream ecosystems in northwest Yunnan. Biodivers. Sci. 2019;27:1298–1308. [Google Scholar]
- 22.Wang R.X., Yang X.J. Waterbird composition and changes with wetland park construction at lake Dianchi, Yunnan-Guizhou plateau. Mt. Res. Dev. 2021;41:R29–R37. doi: 10.1659/MRD-JOURNAL-D-19-00055.1. [DOI] [Google Scholar]
- 23.Lu B., Pan M., Li B., Yang B., Song R.B., Li Y. Morphological characteristics and habitat demand analysis of common water birds in Dianchi Lake. Environ. Sci. Surv. 2022;41:15–19. [Google Scholar]
- 24.Xiao Q.Z., Chen L.J., Qiu Y.P., Chen G.Z. The alien fish Largemouth bass, Micropterus salmoides, found in Dianchi basin, Yunnan, China. Chin. J. Zool. 2020;55:834–835. [Google Scholar]
- 25.Wu Y., Li L., Zheng L., Dai G., Ma H., Shan K., Wu H., Zhou Q., Song L. Patterns of succession between bloom-forming cyanobacteria Aphanizomenon flos-aquae and Microcystis and related environmental factors in large, shallow Dianchi Lake, China. Hydrobiologia. 2016;765:1–13. doi: 10.1007/s10750-015-2392-0. [DOI] [Google Scholar]
- 26.Yang W., Deng D.G., Meng X.L., Zhang S. Temporal and spatial variations of phytoplankton community structure in lake Erhai, a chinese plateau lake, with reference to environmental factors. Russ. J. Ecol. 2019;50:352–360. doi: 10.1134/S1067413619040179. [DOI] [Google Scholar]
- 27.Wang H., Wen Z., Zhang Z.H., Zhang X.L., Fu H., Cao Y., Ni L.Y., Cao T., Li K.Y. Environmental vs. spatial drivers of submerged macrophyte community assembly in different seasons and water depths in a mesotrophic bay of Erhai Lake, China. Ecol. Indic. 2020;117:106696. doi: 10.1016/j.ecolind.2020.106696. [DOI] [Google Scholar]
- 28.Han L., Li Z.Y., Guo X.F., Tan J.L., He S.Z., Cui X.L., Li S.L. Hannaella dianchiensis sp. nov., a basidiomycetous yeast species isolated from lake water. Int. J. Syst. Evol. Microbiol. 2017;67:2014–2018. doi: 10.1099/ijsem.0.001908. [DOI] [PubMed] [Google Scholar]
- 29.Liu X.Y. Master’s Thesis. Dali University; Dali, China: 2016. Studies of Aquatic Fungal Diversity in Erhai Lake and Morphology, Molecular Phylogenetic Systematics of Minimelanlocus. [Google Scholar]
- 30.Han L. Ph.D. Dissertation. Yunnan University; Kunming, China: 2018. Diversity and Spatial Distribution of Fungi in Dianchi Lake of Yunnan plateau. [Google Scholar]
- 31.Chen Z.B., Xu S.G., Yu L., Liu J.N., Su Y., Shao X.D., Ruan Y.N., Wang D.K. Bacterial diversity of surface sediment at Lake Dian in summer and winter seasons. Int. J. Agric. Biol. 2020;23:559–565. [Google Scholar]
- 32.Zhang Y., Zuo J.E., Wang S.K., Salimova A., Li A.J., Li L.L. Spatial distribution of nitrogen metabolism functional genes of Eubacteria and Archaebacteria in Dianchi Lake. Huanjing Kexue/Environ. Sci. 2020;41:2908–2917. doi: 10.13227/j.hjkx.201909196. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka K., Hatakeyama S., Harada Y. Three new freshwater ascomycetes from rivers in Akkeshi, Hokkaido, northern Japan. Mycoscience. 2005;46:287–293. doi: 10.1007/S10267-005-0248-6. [DOI] [Google Scholar]
- 34.Tanaka K., Hirayama K., Yonezawa H., Sato G., Toriyabe A., Kudo H., Hashimoto A., Matsumura M., Harada Y., Kurihara Y., et al. Revision of the Massarineae (Pleosporales, Dothideomycetes) Stud. Mycol. 2015;82:75–136. doi: 10.1016/j.simyco.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Wang H.K., Fournier J., Crous P.W., Jeewon R., Pointing S.B., Hyde K.D. Towards a phylogenetic clarification of Lophiostoma/Massarina and morphologically similar genera in the Pleosporales. Fungal Divers. 2009;38:225–251. [Google Scholar]
- 36.Zhang Y., Schoch C.L., Fournier J., Crous P.W., de Gruyter J., Woudenberg J.H.C., Hirayama K., Tanaka K., Pointing S.B., Spatafora J.W., et al. Multi-locus phylogeny of Pleosporales: A taxonomic, ecological and evolutionary re-evaluation. Stud. Mycol. 2009;64:85–102. doi: 10.3114/sim.2009.64.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quaedvlieg W., Verkley G.J.M., Shin H.D., Barreto R.W., Alfenas A.C., Swart W.J., Groenewald J.Z., Crous P.W. Sizing up Septoria. Stud. Mycol. 2013;75:307–390. doi: 10.3114/sim0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y., Crous P.W., Schoch C.L., Hyde K.D. Pleosporales. Fungal Divers. 2012;53:1–221. doi: 10.1007/s13225-011-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hyde K.D., Jones E.B.G., Liu J.K., Ariyawansa H., Boehm E., Boonmee S., Braun U., Chomnunti P., Crous P.W., Dai D.Q., et al. Families of Dothideomycetes. Fungal Divers. 2013;63:1–313. doi: 10.1007/s13225-013-0263-4. [DOI] [Google Scholar]
- 40.Wanasinghe D.N., Jones E.B.G., Camporesi E., Boonmee S., Ariyawansa H.A., Wijayawardene N.N., Mortimer P.E., Xu J., Yang J.B., Hyde K.D. An exciting novel member of Lentitheciaceae in Italy from Clematis vitalba. Cryptogam. Mycol. 2014;35:323–337. doi: 10.7872/crym.v35.iss4.2014.323. [DOI] [Google Scholar]
- 41.Wijayawardene N., Hyde K., Dai D., Sánchez-García M., Goto B., Saxena R., Erdoğdu M., Selçuk F., Rajeshkumar K., Aptroot A., et al. Outline of fungi and fungus-like taxa—2021. Mycosphere. 2022;13:53–453. doi: 10.5943/mycosphere/13/1/2. [DOI] [Google Scholar]
- 42.Liu Z.P., Zhang S.N., Cheewangkoon R., Zhao Q., Liu J.K. Crassoascoma gen. nov. (Lentitheciaceae, Pleosporales): Unrevealing microfungi from the Qinghai-Tibet plateau in China. Diversity. 2022;14:15. doi: 10.3390/d14010015. [DOI] [Google Scholar]
- 43.Knapp D.G., Kovács G.M., Zajta E., Groenewald J.Z., Crous P.W. Dark septate endophytic pleosporalean genera from semiarid areas. Persoonia. 2015;35:87–100. doi: 10.3767/003158515X687669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajeshkumar K.C., Varma R.K., Sruthi O.P., Gautam A.K., Crous P.W. Groenewaldia (Lentitheciaceae), a new corticolous fungal genus from India. Mycol. Prog. 2023;22:43. doi: 10.1007/s11557-023-01888-3. [DOI] [Google Scholar]
- 45.Dayarathne M.C., Wanasinghe D.N., Jones E.B.G., Chomnunti P., Hyde K.D. A novel marine genus, Halobyssothecium (Lentitheciaceae) and epitypification of Halobyssothecium obiones comb. nov. Mycol. Prog. 2018;17:1161–1171. doi: 10.1007/s11557-018-1432-3. [DOI] [Google Scholar]
- 46.Tanaka K., Harada Y. Bambusicolous fungi in Japan (6): Katumotoa, a new genus of phaeosphaeriaceous ascomycetes. Mycoscience. 2005;46:313–318. doi: 10.1007/S10267-005-0251-Y. [DOI] [Google Scholar]
- 47.von Höhnel F.X.R. Fragmente Zur Mykologie XXIII. Mitteilung, Nr. 1154 bis 1188. [(accessed on 16 February 2023)];Sitz. Kais. Akad. Wiss. Math.-Nat. Kl. Abt. I. 1919 128:535–625. Available online: https://archive.org/details/sbaww_128_0535-0625/mode/2up. [Google Scholar]
- 48.Hyde K.D., Suwannarach N., Jayawardena R.S., Manawasinghe I.S., Liao C., Doilom M., Cai L., Zhao P., Buyck B., Phukhamsakda C., et al. Mycosphere notes 325–344—Novel species and records of fungal taxa from around the world. Mycosphere. 2021;12:1101–1156. doi: 10.5943/mycosphere/12/1/14. [DOI] [Google Scholar]
- 49.Wijayawardene N.N., Hyde K.D., Bhat D.J., Goonasekara I.D., Nadeeshan D., Camporesi E., Schumacher R.K., Wang Y. additions to brown spored coelomycetous taxa in Massarinae, Pleosporales: Introducing Phragmocamarosporium gen. nov. and Suttonomyces gen. nov. Cryptogam. Mycol. 2015;36:213–224. doi: 10.7872/crym/v36.iss2.2015.213. [DOI] [Google Scholar]
- 50.de Gruyter J., Aveskamp M.M., Woudenberg J.H.C., Verkley G.J.M., Groenewald J.Z., Crous P.W. Molecular phylogeny of Phoma and allied anamorph genera: Towards a reclassification of the Phoma complex. Mycol. Res. 2009;113:508–519. doi: 10.1016/j.mycres.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 51.Crous P.W., Wingfield M.J., Guarro J., Sutton D.A., Acharya K., Barber P.A., Boekhout T., Dimitrov R.A., Dueñas M., Dutta A.K., et al. Fungal Planet description sheets: 320–370. Persoonia. 2015;34:167–266. doi: 10.3767/003158515X688433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phookamsak R., Manamgoda D.S., Li W.J., Dai D.Q., Singtripop C., Hyde K.D. Poaceascoma helicoides gen et sp. nov., a new genus with scolecospores in Lentitheciaceae. Cryptogam. Mycol. 2015;36:225–236. doi: 10.7872/crym/v36.iss2.2015.225. [DOI] [Google Scholar]
- 53.Yang Y., Zhang S.N., Yu X.D., Liu J.K. Pseudokeissleriella bambusicola gen. et sp. nov. (Lentitheciaceae, Pleosporales) from bamboos in Sichuan Province, China. Phytotaxa. 2022;560:263–273. doi: 10.11646/phytotaxa.560.3.1. [DOI] [Google Scholar]
- 54.Hyde K.D., Dong Y., Phookamsak R., Jeewon R., Bhat D.J., Jones E.B.G., Liu N.G., Abeywickrama P.D., Mapook A., Wei D., et al. Fungal Diversity Notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020;100:5–277. doi: 10.1007/s13225-020-00439-5. [DOI] [Google Scholar]
- 55.Hirayama K., Tanaka K., Raja H.A., Miller A.N., Shearer C.A. A molecular phylogenetic assessment of Massarina ingoldiana sensu lato. Mycologia. 2010;102:729–746. doi: 10.3852/09-230. [DOI] [PubMed] [Google Scholar]
- 56.Li G.J., Hyde K.D., Zhao R.L., Hongsanan S., Abdel-Aziz F.A., Abdel-Wahab M.A., Alvarado P., Alves-Silva G., Ammirati J.F., Ariyawansa H.A., et al. Fungal diversity notes 253–366: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;78:1–237. doi: 10.1007/s13225-016-0366-9. [DOI] [Google Scholar]
- 57.Chaiwan N., Gomdola D., Wang S., Monkai J., Tibpromma S., Doilom M., Wanasinghe D.N., Mortimer P.E., Lumyong S., Hyde K.D. https://gmsmicrofungi.org: An online database providing updated information of microfungi in the Greater Mekong Subregion. Mycosphere. 2021;12:1513–1526. doi: 10.5943/mycosphere/12/1/19. [DOI] [Google Scholar]
- 58.Dissanayake L.S., Samarakoon M.C., Mortimer P.E., Lu Y.Z., Li Q.R., Hyde K.D., Kang J.C. Morpho-molecular characterization of two novel amphisphaeriaceous species from Yunnan, China. Phytotaxa. 2020;446:144–158. doi: 10.11646/phytotaxa.446.3.1. [DOI] [Google Scholar]
- 59.White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal rna genes for phylogenetics. PCR Protoc. 1990;18:315–322. [Google Scholar]
- 60.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal dna from several cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rehner S. Primers for Elongation Factor 1-α (EF1-α) Insect Biocontrol Laboratory USDA, ARS, PSI; Beltsville, MD, USA: 2001. p. 4. [Google Scholar]
- 62.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2018;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. TrimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vaidya G., Lohman D.J., Meier R. Cladistics multi-gene datasets with character set and codon information. Cladistics. 2011;27:171–180. doi: 10.1111/j.1096-0031.2010.00329.x. [DOI] [PubMed] [Google Scholar]
- 65.Glez-Peña D., Gómez-Blanco D., Reboiro-Jato M., Fdez-Riverola F., Posada D. ALTER: Program-oriented conversion of dna and protein alignments. Nucleic Acids Res. 2010;38:14–18. doi: 10.1093/nar/gkq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stamatakis A., Hoover P., Rougemont J. A rapid bootstrap algorithm for the raxml web servers. Syst. Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 67.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 68.Miller M.A., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic Trees; Proceedings of the 2010 Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010. [Google Scholar]
- 69.Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nylander J.A.A. Modeltest v2. Program Distributed by the Author. Evolutionary Biology Centre, Uppsala University; Uppsala, Sweden: 2004. [Google Scholar]
- 71.Devadatha B., Calabon M.S., Abeywickrama P.D., Hyde K.D., Jones E.B.G. Molecular data reveals a new holomorphic marine fungus, Halobyssothecium estuariae, and the asexual morph of Keissleriella phragmiticola. Mycology. 2020;11:167–183. doi: 10.1080/21501203.2019.1700025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hyde K.D., Hongsanan S., Jeewon R., Bhat D.J., McKenzie E.H.C., Jones E.B.G., Phookamsak R., Ariyawansa H.A., Boonmee S., Zhao Q., et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;80:1–270. doi: 10.1007/s13225-016-0373-x. [DOI] [Google Scholar]
- 73.Lu W.H., Dai D.Q., Lu L., Liu X.F., Wei X.M., Karunarathna S.C., Tibpromma S. Additions to Microfungi in China: Lentithecium yunnanensis sp. nov. Phytotaxa. 2022;554:103–121. doi: 10.11646/phytotaxa.554.2.1. [DOI] [Google Scholar]
- 74.Crous P.W., Wingfield M.J., Burgess T.I., Hardy G.E.S.J., Gené J., Guarro J., Baseia I.G., García D., Gusmão L.F.P., Thangavel R., et al. Fungal Planet description sheets: 716–784. Persoonia. 2018;40:240–393. doi: 10.3767/persoonia.2018.40.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wijayawardene N.N., Dissanayake L.S., Li Q.R., Dai D.Q., Xiao Y., Wen T.C., Karunarathna S.C., Wu H.X., Zhang H., Tibpromma S. Yunnan–Guizhou Plateau: A mycological hotspot. Phytotaxa. 2021;523:1–31. doi: 10.11646/phytotaxa.523.1.1. [DOI] [Google Scholar]
- 76.Su H.Y., Hyde K.D., Maharachchikumbura S.S.N., Ariyawansa H.A., Luo Z.L., Promputtha I., Tian Q., Lin C.G., Shang Q.J., Zhao Y.C., et al. The families Distoseptisporaceae fam. nov., Kirschsteiniotheliaceae, Sporormiaceae and Torulaceae, with new species from freshwater in Yunnan Province, China. Fungal Divers. 2016;80:375–409. doi: 10.1007/s13225-016-0362-0. [DOI] [Google Scholar]
- 77.Bao D.F., McKenzie E.H.C., Bhat D.J., Hyde K.D., Luo Z.L., Shen H.W., Su H.Y. Acrogenospora (Acrogenosporaceae, Minutisphaerales) appears to be a very diverse genus. Front. Microbiol. 2020;11:1606. doi: 10.3389/fmicb.2020.01606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bao D.F., Luo Z.L., Liu J.K., Bhat D.J., Sarunya N., Li W.L., Su H.Y., Hyde K.D. Lignicolous freshwater fungi in china iii: Three new species and a new record of Kirschsteiniothelia from northwestern Yunnan Province. Mycosphere. 2018;9:755–768. doi: 10.5943/mycosphere/9/4/4. [DOI] [Google Scholar]
- 79.Luo Z.L., Hyde K.D., Bhat D.J., Jeewon R., Maharachchikumbura S.S.N., Bao D.F., Li W.L., Su X.J., Yang X.Y., Su H.Y. Morphological and molecular taxonomy of novel species Pleurotheciaceae from freshwater habitats in Yunnan, China. Mycol. Prog. 2018;17:511–530. doi: 10.1007/s11557-018-1377-6. [DOI] [Google Scholar]
- 80.Luo Z.L., Hyde K.D., Liu J.K., Maharachchikumbura S.S.N., Jeewon R., Bao D.F., Bhat D.J., Lin C.G., Li W.L., Yang J., et al. Freshwater Sordariomycetes. Fungal Divers. 2019;99:451–660. doi: 10.1007/s13225-019-00438-1. [DOI] [Google Scholar]
- 81.Dong W., Hyde H.D., Jeewon R., Doilom M., Yu X.D., Wang G.N., Liu N.G., Hu D.M., Nalumpang S., Zhang H. Towards a natural classification of annulatascaceae-like taxa ⅱ: Introducing five new genera and eighteen new species from freshwater. Mycosphere. 2021;12:1–88. doi: 10.5943/mycosphere/12/1/1. [DOI] [Google Scholar]
- 82.Suetrong S., Schoch C.L., Spatafora J.W., Kohlmeyer J., Volkmann-Kohlmeyer B., Sakayaroj J., Phongpaichit S., Tanaka K., Hirayama K., Jones E.B.G. Molecular systematics of the marine Dothideomycetes. Stud. Mycol. 2009;64:155–173. doi: 10.3114/sim.2009.64.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Singtripop C., Camporesi E., Ariyawansa H.A., Wanasinghe D.N. Keissleriella dactylidis, sp. nov., from Dactylis glomerata and its phylogenetic placement chonticha. Sci. Asia. 2015;41:295–304. doi: 10.2306/scienceasia1513-1874.2015.41.295. [DOI] [Google Scholar]
- 84.Tibell S., Tibell L., Pang K.L., Calabon M., Jones E.B.G. Marine fungi of the Baltic Sea. Mycology. 2020;11:195–213. doi: 10.1080/21501203.2020.1729886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences generated in this study were submitted to GenBank database.