Abstract
Chimerism happens rarely among most mammals but is common in marmosets and tamarins, a result of fraternal twin or triplet birth patterns in which in utero connected circulatory systems (through which stem cells transit) lead to persistent blood chimerism (12–80%) throughout life. The presence of Y-chromosome DNA sequences in other organs of female marmosets has long suggested that chimerism might also affect these organs. However, a longstanding question is whether this chimerism is driven by blood-derived cells or involves contributions from other cell types. To address this question, we analyzed single-cell RNA-seq data from blood, liver, kidney and multiple brain regions across a number of marmosets, using transcribed single nucleotide polymorphisms (SNPs) to identify cells with the sibling’s genome in various cell types within these tissues. Sibling-derived chimerism in all tissues arose entirely from cells of hematopoietic origin (i.e., myeloid and lymphoid lineages). In brain tissue this was reflected as sibling-derived chimerism among microglia (20–52%) and macrophages (18–64%) but not among other resident cell types (i.e., neurons, glia or ependymal cells). The percentage of microglia that were sibling-derived showed significant variation across brain regions, even within individual animals, likely reflecting distinct responses by siblings’ microglia to local recruitment or proliferation cues or, potentially, distinct clonal expansion histories in different brain areas. In the animals and tissues we analyzed, microglial gene expression profiles bore a much stronger relationship to local/host context than to sibling genetic differences. Naturally occurring marmoset chimerism will provide new ways to understand the effects of genes, mutations and brain contexts on microglial biology and to distinguish between effects of microglia and other cell types on brain phenotypes.
Introduction
Chimerism, in which an organism contains cells from genetically distinct animals, happens rarely among mammals. Chimerism is common, however, among marmosets and their close relatives the tamarins: in these primate species, animals usually give birth to dizygotic twins or trizygotic triplets whose blood contains cells from siblings. During development, the siblings share circulation in utero, allowing the exchange of hematopoietic stem cells (Gengozian et al., 1969; Wislocki, 1939). Most marmosets then exhibit blood chimerism throughout life: their blood-derived DNA is a mixture of both twins’ genomes, with the twin’s genome contributing 12% to 80% of the DNA in the blood (Niblack et al., 1977; The Marmoset Genome Sequencing and Analysis Consortium, 2014). This indicates that the twins’ hematopoietic stem cells establish permanent residency in one another’s bodies and contribute to blood cell populations throughout life.
A longstanding mystery involves whether other tissues and organisms also harbor chimerism. Beyond the blood, Y-chromosome DNA has been detected in the brain and other organs of female marmosets with male twins (Ross et al., 2007; Sweeney et al., 2012), eliciting much speculation about how chimerism might have shaped behavior and natural selection in marmosets. However, it is still not known what cell types harbor this sibling DNA; such observations could in principle be explained by the presence of blood cells within these organs.
Here, we analyze chimerism in the marmoset brain, liver, kidney and blood, using single-nucleus RNA-seq (snRNA-seq) to infer cell types and using combinations of transcribed SNPs (visible in the snRNA-seq data) to determine which marmoset sibling is the source of each cell. This approach makes it possible to determine whether chimerism arises from blood cells, resident immune cells, or other cell types, and to explore what chimerism can teach us about cellular migrations and population dynamics.
Results
Marmoset chimerism can be characterized at single-cell resolution
To identify which individual cells have the genome of the host marmoset, and which have the genome of the host’s birth sibling, we used combinations of many transcribed SNPs that were visible in the snRNA-seq data for each nucleus (Wells et al., 2023).
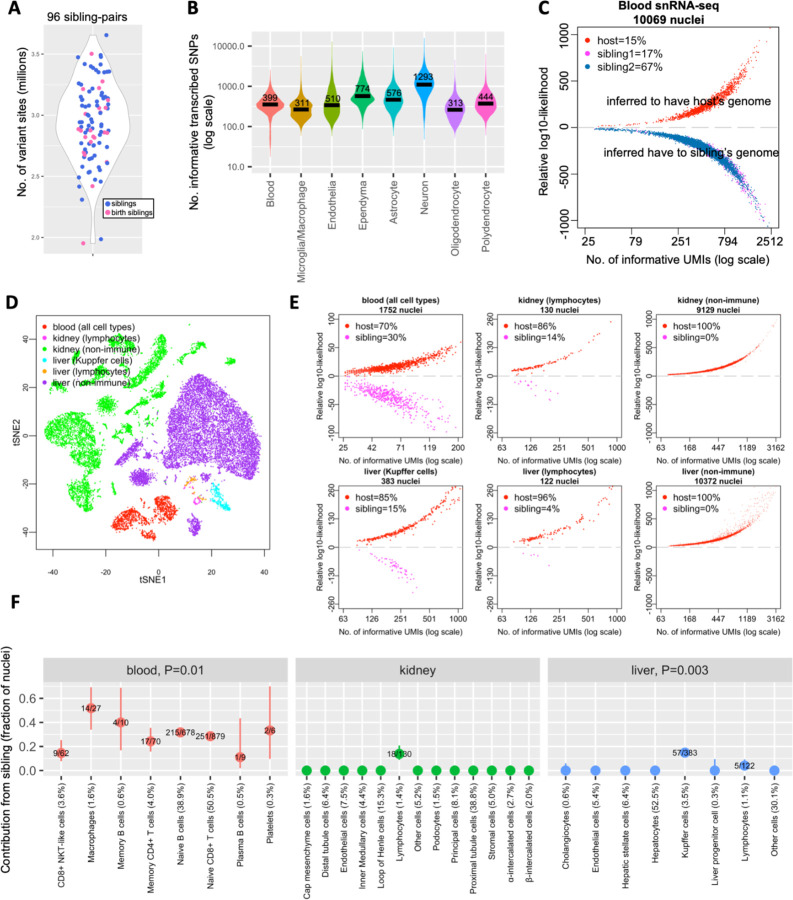
We first determined whether marmosets have sufficient sequence variation to enable the distinction between host and sibling cells. From whole-genome sequences of 123 marmosets, we identified 13 million polymorphic bi-allelic SNPs in the marmoset genome, with individual marmosets harboring 2.3 to 3.8 million (average 3.4 million) heterozygous sites – comparable to levels of heterozygosity in humans. For sibling comparisons, we detected a large number of sites at which any two siblings’ genomes differed, ranging from 2.0 to 3.7 million sites (average 2.9 million sites) across 96 sibling comparisons (Fig. 1A). To determine how many of these sites were visible in snRNA-seq data, we analyzed snRNA-seq data for several marmoset tissues. The results varied by cell type, reflecting that different cell types’ nuclei harbored different quantities of RNA. The hundreds of transcribed variant sites that differed between siblings (median>311 per nucleus) suggested ample power to distinguish between siblings in all cell types (Wells et al., 2023) (Fig. 1B).
Figure 1. Sibling chimerism analysis at single-cell resolution.
(A) Numbers of variant sites at which marmoset sibling genomes differ, for 96 sibling-pairs. Each dot is a sibling-pair; birth siblings are colored in pink. (B) Numbers of transcribed SNPs (per nucleus) visible in various cell types from blood and brain snRNA-seq datasets of marmoset CJ028. x-axis: snRNA-seq library; y-axis: number of ascertained, transcribed SNPs for which host and sibling have different genotypes; black horizontal lines: median values per cell type. (C) Donor-of-origin assignment of each nucleus in blood snRNA-seq of a marmoset. The host marmoset (CJ028) was born with two siblings; each nucleus was assigned to either the host or to one of the two birth siblings. x-axis: number of unique molecular identifiers (UMI; a measure of transcript abundance) that contains SNPs for which the host and sibling’s genomes differ, in log scale; y-axis: inferred likelihood that the cell has host genome minus likelihood that the cell has sibling genome (log10). (D) Two-dimensional visualization (tSNE plot) of snRNA-seq data from a marmoset’s (CJ026) blood, kidney and liver. (E) Donor-of-origin assignment in marmoset CJ026’s blood, kidney and liver. axes: same as in (C). (F) Levels of chimerism in each cell type ascertained in blood, kidney and liver snRNA-seq of marmoset CJ026. y-axis: fraction of sibling cells in each cell type; numbers in fraction: number of sibling nuclei over total nuclei in the cell type; percentage in x-axis labels: cell type representation in the tissue; vertical bars: binomial confidence interval (95%); P-values: test of heterogeneity (Chi-square) across immune cell types of a tissue (to test for differences in contribution of sibling across immune cell types).
We next evaluated whether the genome variation visible in snRNA-seq reads was sufficient to distinguish between host and sibling cells. For this we used Dropulation, which identifies the donors of individual cells (from a set of genome-sequenced candidate donors) by using combinations of the transcribed SNPs visible on the snRNA-seq reads of the individual cells (Wells et al., 2023). We first analyzed the blood cells by snRNA-seq of a marmoset (CJ028) born with two birth siblings and used Dropulation (Wells et al., 2023) to assign individual cells to the correct sibling (Fig. 1C). The relative likelihoods of the siblings could be strongly differentiated (relative likelihoods of 103 to 1023) for >99% of the nuclei (Fig. 1C). We found a high level of chimerism: 84% of all nuclei sampled in this marmoset’s blood appeared to contain the genome of one (67% - sibling #1) or the other (17% - sibling #2) of its two birth siblings (Fig. 1C), consistent with the wide range of chimerism found in previous studies: 4–82% in marmoset T cells and B cells (Niblack et al., 1977); 13–37% in marmoset whole blood (“The Marmoset Genome Sequencing and Analysis Consortium”, 2014).
Apparent liver and kidney chimerism arises from infiltrating monocytes
Earlier studies have identified Y-chromosome-derived DNA sequences in the organs of female marmosets with male birth siblings, suggesting that these organs harbor chimerism (Sweeney et al., 2012). However, such observations could also in principle arise from blood or from blood-derived immune cells that are present in those organs (Sweeney et al., 2012).
We performed snRNA-seq analysis of the blood (1,741 nuclei), liver (10,877 nuclei) and kidney (9,262 nuclei) of a marmoset (CJ026) with one birth sibling (Fig. 1D). The snRNA-seq profiles clustered into groups that were readily recognized (based on the RNAs expressed) as the principal cell types of each organ; we determined the identity of each cluster using scType, a cell-type identification tool that uses a database of known marker genes (Ianevski et al., 2022).
In kidney and liver, the only clearly twin-derived cells were cells of hematopoietic origin: the resident macrophages in liver (Kupffer cells), lymphocytes in liver, and lymphocytes in kidney (Fig. 1E,F). All non-hematopoietic cell types in liver and kidney appeared to contain only the host marmoset’s own genome. Chimerism levels for the two chimeric liver immune cell types appeared to diverge, with sibling-derived cells accounting for 15% of Kupffer cells and just 4% of lymphocytes (5/122 vs 57/383; Chi-square test P-value=0.003). In the blood, chimerism levels varied across the various cell types: the most abundant cell types, the Naive B cells and Naive CD8+ T cells, were respectively 29% and 32% sibling-derived, while the less-abundant CD8+ NKT-like cells were 15% sibling-derived (Fig. 1F, Chi-square test P-value=0.01).
These results indicate that, in this animal’s liver and kidney, apparent DNA chimerism likely arose from infiltrating immune cells rather than other cell types. These results also indicate that cells with siblings’ genomes can differ in their tendency to acquire specific hematopoietic cell fates and in their tendency to infiltrate into organs.
Marmoset brain microglia and macrophages exhibit abundant chimerism
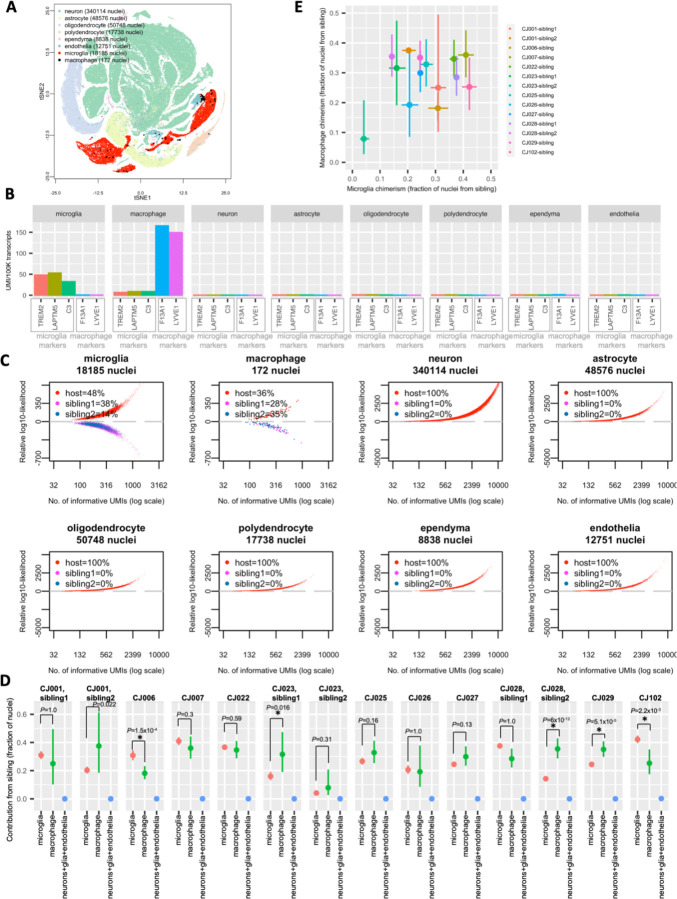
To characterize chimerism in the marmoset brain, we utilized a large snRNA-seq data set being generated for a marmoset brain cell atlas (Krienen et al., 2022). We first analyzed 497,000 single-nucleus RNA-expression profiles from the neocortex, thalamus, striatum, hippocampus, basal forebrain, hypothalamus and amygdala of an adult marmoset with two birth siblings (marmoset CJ028). We clustered the cell types using gene expression similarities (Fig. 2A) and identified brain cell types as in earlier work (Krienen et al., 2020), identifying neurons, astrocytes, oligodendrocytes, ependymal cells, endothelial cells, microglia and macrophages (Fig. 2B). Microglia (which expressed markers TREM2, LAPTM5 and C3) and macrophages (which expressed LYVE1 and F13A1) were a small fraction of all nuclei analyzed (about 3.6%), but due to the large number of nuclei we profiled (53 brain tissue dissections, 497,000 nuclei), we were able to ascertain sufficient numbers of microglia (18,175 nuclei) and to a lesser extent macrophages (172 nuclei) for many downstream analyses. We found microglia and macrophages in snRNA-seq data from 10 additional marmosets with different genetic backgrounds from 3 different colonies (sample information for 11 marmosets in Supplementary Table 1; a total of eleven marmoset brains were analyzed and all are unrelated except for CJ006 and CJ007 which are birth siblings, and CJ025 and CJ026 which are (non-birth) siblings; only CJ026 was assessed for liver and kidney, and 3 marmosets were assessed for blood: CJ026, CJ027 and CJ028). Brain snRNA-seq of all 11 marmosets showed consistently the presence of these two myeloid cell types in the brain (Supplementary Fig. 1; number of microglia and macrophages in Supplementary Table 2).
Figure 2. Microglia and macrophages, but not neurons and glia, are chimeric in the marmoset brain.
(A) Two-dimensional visualization (tSNE plot) of marmoset CJ028’s snRNA-seq profiles from 8 brain regions. (B) Expression of markers of microglia and macrophages in microglia, macrophages, neurons, glia (astrocytes, oligodendrocytes, polydendrocytes, ependymal cells) and endothelial cells. y-axis: expression levels, measured as numbers of detected transcripts (unique molecular identifiers (UMIs), a measure of transcript abundance) from that gene per 100,000 total. Microglia markers: TREM2, LAPTM5, C3; macrophage markers: F13A1, LYVE1. Data from all CJ028’s brain regions. (C) Donor assignment of each nucleus to one of three possible donors (host, sibling1 or sibling2) of marmoset CJ028’s brain snRNA-seq data. x-axis: number of UMI that contains SNPs for which the host and sibling’s genomes differ, in log10 scale; y-axis: inferred likelihood that the cell has host genome minus likelihood that the cell has sibling genome (log10). Data from all of CJ028’s brain regions. (D) Fractions of cells with a sibling’s genome among microglia, macrophages, and other brain cell types (neurons, astrocytes, oligodendrocytes, polydendrocytes, ependyma, endothelial) from 11 marmosets. CJ001, CJ023 and CJ028 were part of a triplet litter and the chimerism fraction of each birth sibling is shown in separate panels. y-axis: fraction of sibling-cells in the cell type. vertical bars: binomial confidence interval (95%). P-values: Chi-square test from comparison of chimerism fractions in microglia and macrophage; * (P-value<0.05). (E) Sibling contributions to microglial (x-axis) and macrophage (y-axis) populations in the same animals. Dots and bars are from (D). Pearson correlation R=0.32, 95% confidence interval (−0.26 to 0.73), P-value=0.27.
Donor-of-origin analysis of snRNA-seq data from 2.2 million nuclei sampled from 137 brain tissue samples from these 11 marmosets showed a clear and consistent pattern: microglia and macrophages, but not neurons, glia or endothelial cells, harbored chimerism (Fig. 2C, Supplementary Fig. 2). Microglia exhibited abundant chimerism – across the 11 marmosets, the total fraction of cells with the sibling’s genome ranged from 20% to 52% (for triplets, sum of two siblings; Fig. 2D). Macrophages exhibited a similarly wide range of sibling fractions across marmosets (18% to 64%, Fig. 2D).
The quantitative extent of microglial chimerism varied across individuals (Supplementary Fig. 3A; test of heterogeneity P-value<2.2×10−16), as did that of macrophage chimerism (Supplementary Fig. 3B; test of heterogeneity P-value=1×10−4). We asked whether microglial and macrophage chimerism were correlated. Intriguingly, only a modest correlation of chimerism levels across 14 host-sibling pairs was observed between the microglia and macrophages (Fig. 2E; Pearson correlation 0.31), suggesting that in addition to the cell’s genome, other factors such as local host environment play a role in differential recruitment or survival of sibling cells. Neither of these two myeloid cell types showed consistently higher chimerism than the other cell type did (Fig. 2D).
Sibling contributions in blood vs. brain
Though microglia (like macrophages) are myeloid cells that derive from hematopoietic stem cells, the ontogenies of microglia and brain macrophages are distinct from those of bone-marrow-derived peripheral blood mononuclear cells (Perdiguero & Geissmann, 2016). As such, differences in the developmental and migration histories of these cell populations could in principle have caused their chimerism fractions to diverge in a systematic way.
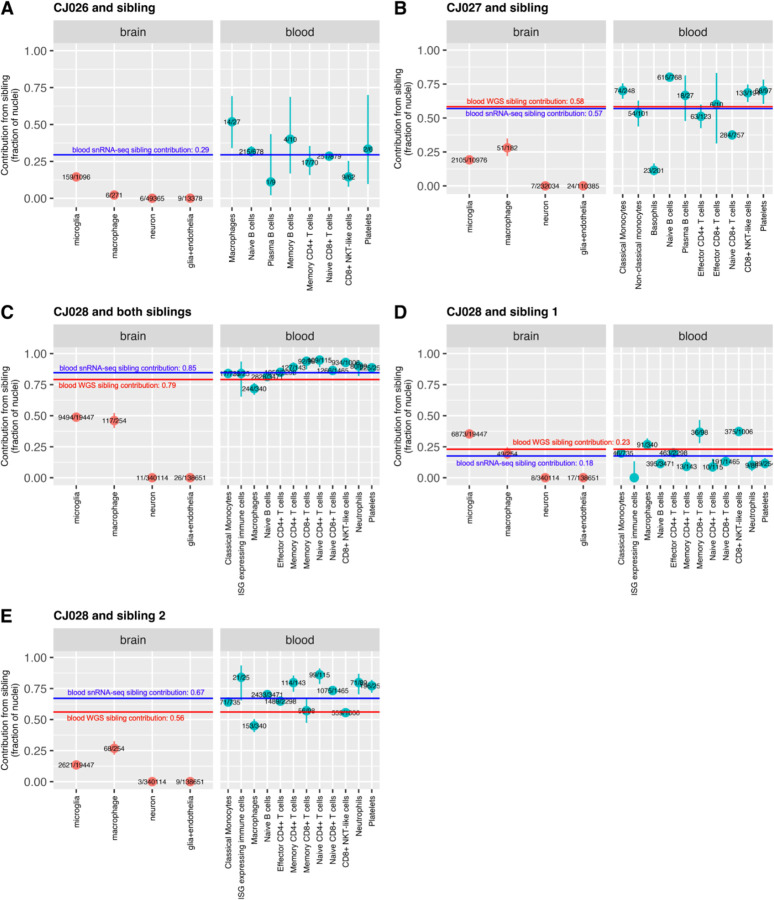
We analyzed three marmosets for which snRNA-seq was performed on both blood and brain tissues. Sibling contributions to microglia and brain macrophages were in general quite different from those in blood (Fig. 3).
Figure 3. Sibling contributions to hematopoiesis-derived cells diverge between blood and brain.
(A)-(E) Chimerism fractions in brain and blood of three animals. CJ028 was part of a triplet litter, and the contribution of each sibling is shown in a separate panel (D,E). Red horizontal lines: twin contribution to blood cells as ascertained from whole-genome sequencing of whole-blood-derived genomic DNA (Census-seq). Blue horizontal lines: total twin contribution to blood cells as estimated from PBMC snRNA-seq (all cells). Vertical bars: binomial confidence interval (95%). Numbers in fraction: number of sibling cells over total cells in the cell type. glia+endothelia: astrocytes, oligodendrocytes, polydendrocytes, ependymal cells, endothelial cells.
Marmoset CJ028’s chimerism (involving two birth siblings) provided a setting in which cells with three different genomes shared the same environment through development until adulthood (to two years of age). Among CJ028’s microglia, the fraction of cells from sibling 1 (35%) was greater than that from sibling 2 (13%) (two-sided test of proportionality P-value<2.2−16), while in blood, the opposite was true (fraction of cells from sibling 1 across all blood cell types was 18%, fraction of cells from sibling 2 across all blood cell types was 67%; two-sided test of proportionality P-value<2.2−16) (Fig. 3D,E, Supplementary Table 3).
Microglia chimerism fraction varies across brain regions
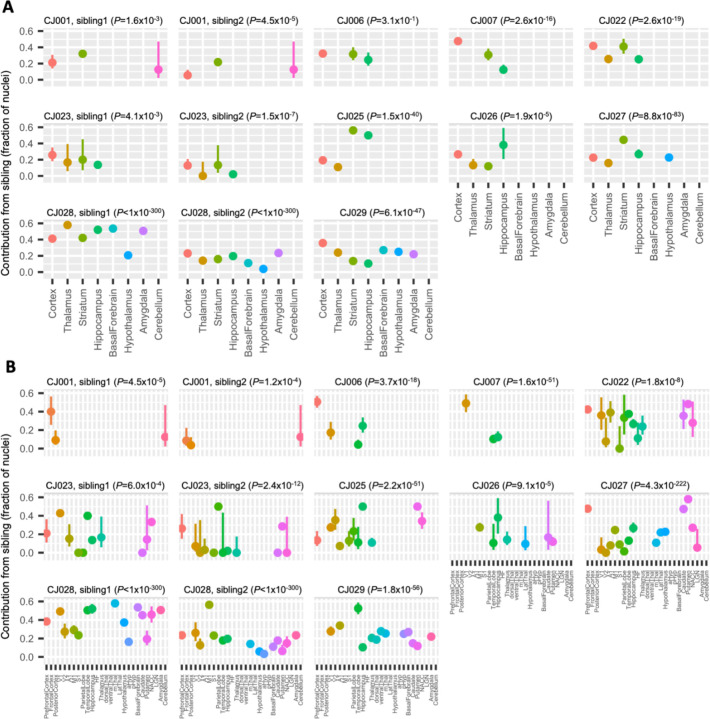
Sibling contributions to the microglial population could in principle be shaped by effects that are local to specific brain areas, including differential response of sibling microglia to local recruitment or proliferation cues. To evaluate whether the sibling contribution to the microglial population varied across brain areas within individual marmosets, we performed chimerism analysis for each of the brain regions profiled in the snRNA-seq datasets: neocortex, thalamus, striatum, hippocampus, basal forebrain, hypothalamus and amygdala (Krienen et al., 2020, 2022). Within each marmoset, the fraction of microglia with a sibling’s genome diverged across a marmoset’s brain regions (Fig. 4A). For example, in marmoset CJ025, sibling contributions to microglial populations ranged from 11% (21/193) in the thalamus to 56% (174/310) in the striatum (P-value=1.1×10−23, Chi-square test of thalamus vs striatum; P-value=1.5×10−40, Chi-square test across all 4 brain regions). For marmoset brains profiled with at least 300,000 nuclei, CJ027, CJ028 and CJ029, tests of heterogeneity P-values were even more significant: 8.8×10−83, <1×10−300, and 6.1×10−47, respectively. Analysis of finer brain substructures showed a similar result (Fig. 4B). None of the brain regions exhibited consistently higher or lower chimerism levels, suggesting that these divergences did not result from differential physical access of host and sibling microglia to different brain areas (Fig. 4).
Figure 4. Sibling contributions to brain microglial populations vary across an animal’s brain areas.
Contributions of sibling(s) to the microglial populations ascertained in principal brain areas (A), and finer-scale brain substructures (B). CJ001, CJ023 and CJ028 were part of a triplet litter and the chimerism contribution of each twin is shown in a separate panel. CJ102 was profiled in only one brain region and hence was not included in the analysis. y-axis: fraction of twin cells. Vertical bars: binomial confidence interval (95%); P-values: test of heterogeneity across an animal’s brain regions.
Gene-expression comparisons of host-to sibling-derived microglia
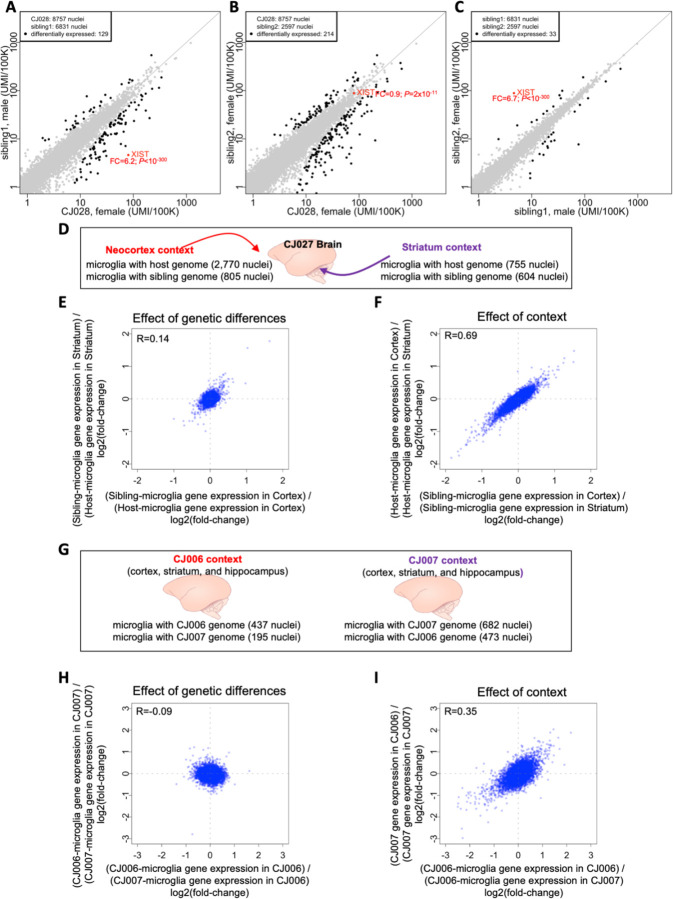
Chimerism provides the unusual opportunity to compare cells with different genomes in a shared in vivo biological context. We compared RNA expression between host- and sibling-derived microglia of a female marmoset with two birth siblings. Sex differences among the siblings (the host (CJ028) was a female and one of the two siblings was a male) allowed a natural control: the XIST gene encodes a non-coding RNA involved in silencing one copy of chromosome X in females and thus exhibits sex-specific expression due to cell-autonomous mechanisms. We found that (as expected) XIST transcripts were detected at far higher levels in the snRNA-seq profiles of microglia with the female twin’s genome relative to the male twin’s (Fig. 5A,C). By contrast, XIST transcripts were detected at similar levels in two microglial populations with the genomes of female twins (Fig. 5B).
Figure 5. Utilizing natural chimerism to distinguish cell-autonomous from non-cell-autonomous effects on gene expression, and to compare the effects context and genetic variation in shaping gene expression.
(A-C) Comparisons of RNA expression between microglial populations within host animal CJ028, who had two birth siblings. Comparisons of gene expression between microglia with the genomes of (A) the female host and male sibling, (B) the female host and female sibling, and (C) the two siblings (male and female). Each point represents a gene; its location on the plot represents the level of expression of that gene among microglia with two different genomes in the same animal. x- and y-axes: normalized gene expression levels (number of transcripts per 100,000 transcripts). FC: fold-change of gene expression, female/male for XIST. Fold-change and P-values were calculated using edgeR. Differentially expressed genes (black dots) were defined as: FDR Q-value<0.05 and fold-change>1.5 or less than 1/1.5 and the gene must be expressed in at least 10% of one of the microglia sets. (D-I) Higher effect of context than genetic differences in shaping gene expression. (D) In the brain of marmoset CJ027, the neocortex and striatum are two contexts where two sets of microglia with different genomes reside. (E) Effect of genetic differences. x-axis: log2-fold-change of cortical microglia gene expression between host and sibling cells; y-axis: log2-fold-change of striatal microglia gene expression between host and sibling cells. (F) Effect of context. x-axis: log2-fold-change of the sibling’s gene expression between the two brain regions; y-axis: log2-fold-change of the host’s gene expression between the two brain regions. (G-I) The brains (cortex, striatum and hippocampus) of two birth siblings provide biological contexts in which populations of microglia with two sibling genomes reside. The effect of genetic differences (H) and effect of animal context (I) are compared, for the same brain areas (combined data from cortex, striatum and hippocampus). x- and y-axes: log2-fold-changes of the gene expression between two sets of microglia (the sets being compared are indicated in the axis labels). R: Spearman correlation.
Gene-expression differences between host- and sibling-derived microglia in the same brain could in principle arise from asymmetries in their developmental histories (which would be shared across host animals) or from genomic differences (which would vary from host animal to host animal). In all eleven individual marmosets, analysis identified anywhere from six to hundreds of genes whose differential expression distinguished microglia with host vs. sibling genomes, but aside from sex differences (XIST gene) we did not find any gene that consistently distinguished host from sibling microglia across the sibling comparisons (Supplementary Fig. 4, Fig. 5A–C).
Brain context vs. genetic differences as determinants of microglial gene expression
Chimerism presents interesting opportunities to distinguish between cell-autonomous and contextual effects on biology, and to compare the magnitudes of such effects.
We first considered the difference in contexts provided by pairs of brain areas by analyzing snRNA-seq data from the neocortex and striatum of marmoset CJ027; the resident microglial populations with different genomes make it possible to compare contextual to genetic effects on microglial gene expression (Fig. 5D). Genetic effects appeared to elicit very many small-magnitude gene-expression differences; these differences were shared between cortical and striatal microglia (Fig. 5E). Brain-area context elicited much larger-magnitude gene-expression differences, which were experienced in common by microglia with both genotypes (Fig. 5F). We obtained similar results for all pairs of brain areas analyzed (52 context vs genetic effect from brain snRNA-seq of 6 marmosets with at least 60 cells available for analysis in each context; Supplementary Table 4; Supplementary Fig. 5).
We next considered the difference in contexts provided by the same brain area in different marmosets. Two of the marmosets profiled, CJ006 and CJ007, were birth siblings who passed away as neonates (the only birth siblings in our dataset), and thus provided the additional opportunity to distinguish genetic from contextual effects by analyzing the two sibling microglial populations in the cortex, striatum and hippocampus of both marmosets (Fig. 5G). The effects of context (host marmoset) in microglia from all three brain areas appeared to be far larger than the cell-autonomous effects of genetic differences (Fig. 5H,I).
Discussion
A longstanding debate concerns the extent of chimerism in marmosets and tamarins. Chimerism in these species has been detected in diverse organs but arises from unknown cell types (Ross et al., 2007; Sweeney et al., 2012). Here we found that chimerism in the brain, liver and kidney is present but appears to arise entirely from cells of the myeloid and lymphoid lineages, including infiltrating macrophages, monocytes, and microglia.
Cells of the myeloid and lymphoid lineages derive developmentally from hematopoietic stem cells. We found no strong evidence of chimerism among 2.2 million non-hematopoietic cells in the liver (from one marmoset), kidney (from one marmoset) or brain (from 11 marmosets). Thus, while marmosets share a circulation in utero, we found no evidence that non-hematopoietic stem cells or progenitors had been shared via this route in any appreciable number. However, we found that in the marmoset brain, the microglia and macrophages, which also derive from this lineage, routinely harbor abundant chimerism, with 10–50% of a marmoset’s microglia containing the genome(s) of birth sibling(s).
Organs in the same marmoset (liver, kidney, brain) differed markedly in the sibling contribution to resident macrophage and monocyte populations, with microglial chimerism fraction (the fraction of cells contributed by siblings) varying by as much as 40 percentage points across a marmoset’s brain areas. This phenomenon has more than one potential explanation. First, cells from the host and sibling could in principle respond differently to recruitment or proliferation cues that vary spatially; if this is the case, marmoset chimerism could provide a model for studying the effects of mutations and natural sequence variation on cell migration and recruitment. (Although we found that genetic effects were smaller than contextual effects in shaping microglial gene expression at any moment in time, genetic effects were clear (Fig. 5E), and even small effects on proliferation rates would tend to have effects that increase exponentially over time.) Second, beyond such recruitment effects, it is also possible that these differences suggest a substantial role of clonal expansions and population bottlenecks in shaping local microglial and macrophage populations.
We found that the cellular contribution of birth siblings to myeloid cell populations was significantly different in blood than in brain in the modest number of marmosets analyzed (Fig. 3). Unlike the differences among brain areas, the blood-brain differences tended to be directional, with more-modest sibling contributions in the brain than in the blood. Though this would need to be confirmed in many more marmosets to be definitive on its own, it is plausibly connected to this aspect of marmoset fetal development: sharing of a blood circulation between the two fetuses occurs during a temporal window that is more temporally extended than the waves of colonization of the brain by microglia, potentially allowing for greater exchange in the centers of blood hematopoiesis (the liver and then the bone marrow). In microglia and macrophages, due to the defined waves of hematopoiesis in the yolk sac and migration patterns of microglia and macrophages to the developing brain, the opportunity for a progenitor cell from a twin to colonize a host’s brain may need to occur during a more-restricted temporal window.
Comparisons of gene expression between microglial cells with host and sibling genomes in a shared brain context may provide many future opportunities to distinguish the cell-autonomous from the non-cell autonomous genetic effects of genetic differences and engineered mutations. Such analyses could become especially useful scientifically as genome editing increasingly enables the utilization of marmosets as a model organism in translational neuroscience (Aida & Feng, 2020; Feng et al., 2020). Our pilot analysis of host-sibling microglial gene expression differences in the brains of two co-twins revealed a large role of animal context (relative to genetic differences) in shaping microglial gene expression. This result points to an important principle: the ability to isolate the effects of a mutation will be greatly strengthened by the ability to make within-animal (rather than just between-animal) comparisons of cells with different genotypes.
A long history of innovation in genetics involves elaborate ways to create mosaics in mice, C. elegans and other laboratory organisms in order to distinguish cell-autonomous from non-cell autonomous genetic effects. Natural chimerism in marmosets may enable many straightforward ways to pursue such kinds of studies. Natural chimerism may also make it possible to determine when microglia or macrophages, as opposed to other cell types, mediate the effect of a mutation on an animal’s phenotype.
Microglia perform essential roles in the development and regulation of the central nervous system (CNS), including by sculpting or “pruning” neuronal circuits (Hammond et al., 2018; Schafer et al., 2012; Schafer & Stevens, 2015), and are implicated in or hypothesized to contribute to a wide range of brain disorders and diseases, including Alzheimer’s disease, Parkinson’s disease, autism spectrum disorder, and schizophrenia. Marmoset microglial chimerism will enable many new ways of studying microglia and the effects of genes and alleles upon brain biology.
Methods
Ethical compliance
Marmoset experiments were approved by and in accordance with Massachusetts Institute of Technology IACUC protocol number 051705020.
Nucleus Drop-seq library preparation and sequencing
Nucleus suspensions were prepared from frozen tissue and used for nucleus Drop-seq following the protocol we have described at https://doi.org/10.17504/protocols.io.2srged6. Drop-seq libraries were prepared as previously described (Macosko et al., 2015), with modifications, quantification and quality control as described in a previous study (Saunders et al., 2018), as well as the following modifications optimized for nuclei: in the Drop-seq lysis buffer, 8 M guanidine hydrochloride (pH 8.5) was substituted for water, nuclei were loaded into the syringe at a concentration of 176 nuclei/μl, and cDNA amplification was performed using around 6,000 beads per reaction, 15 PCR cycles. Raw sequencing reads were aligned to the calJac3 marmoset reference genome assembly and reads that mapped to exons or introns of each assembly were assigned to annotated genes (https://github.com/broadinstitute/Drop-seq). Drop-seq libraries are indicated in Supplementary Table 1.
Nucleus 10X Chromium library preparation and sequencing
Single-nucleus suspensions from frozen tissue were generated as for Drop-seq; GEM generation and library preparation followed the manufacturer’s protocol (protocol versions #CG00052 Chromium Single Cell 3’ v2 and #CG000183 Chromium Single Cell3′ v3 UG_Rev-A). Raw sequencing reads were processed and aligned using the same method for aligning Drop-seq reads. 10X Chromium libraries are indicated in Supplementary Table 1.
Clustering of cells using Independent Component Analysis
Nuclei from intact cells were identified and clustered into cell types using a method that we have previously described (Krienen et al., 2020; Saunders et al., 2018). Briefly, nuclei with less than 400 detected genes were not used in the analysis. A digital gene expression matrix was created for a set of libraries from the same animal that were to be co-analyzed (Supplementary Table 5), and independent component analysis using the fastICA package in R was used after normalization and variable gene selection as previously described (Krienen et al., 2020; Saunders et al., 2018). A Louvaine-based clustering algorithm was performed on the top 60 independent components. Due to the large number of nuclei profiled in some marmosets (CJ027, CJ028, CJ029), memory requirements exceeded machine limits and for these marmosets, we divided the clustering analysis into two or three batches (Supplementary Table 5). The brain of marmoset CJ022 was profiled using both Drop-seq and 10X and a separate clustering was done for each snRNA-seq method (Supplementary Table 5). We ran the clustering algorithm 12 times using 3 nearest neighbor parameters (10,20,30) and 4 resolution parameters (0.3, 0.5, 0.1, 1.0). Markers for each cluster were identified using differential gene expression analysis (Krienen et al., 2020; Saunders et al., 2018). We inspected each clustering result and chose the one which yielded separate clusters for microglia and macrophages (Supplementary Table 5).
Identification of cell types
For the brain datasets, the microglia and macrophage clusters were identified by the markers TREM2, C3, LAPTM5 for microglia and F13A1, LYVE1 for macrophages. The other brain cell types were identified using cell type markers for neurons, astrocytes, oligodendrocytes, polydendrocytes and endothelial cells that we used as before (Krienen et al., 2020). Cell types in blood, liver and kidney were identified using the ScType method (Ianevski et al., 2022).
Donor-of-origin analysis and detection of host-sibling doublets (Dropulation)
We used the Dropulation suite to calculate a donor likelihood for each cell (Wells et al., 2023) (software available at https://github.com/broadinstitute/Drop-seq). The host and birth sibling genotypes were provided as input to Dropulation’s AssignCellsToSamples tool, together with the snRNA-seq BAM file and a list of cell barcodes that were identified to be intact cells. To generate chimerism-free reference genotypes, we cultured fibroblasts and performed whole genome sequencing (WGS) on the resulting DNA. We found that the difference in likelihoods between host and sibling increase with the number of UMI of the cell, and hence we imposed a minimum number of UMI for each marmoset’s cells (Supplementary Table 5). We also performed doublet detection using Dropulation’s DetectDoublets to obtain a likelihood of a cell having a mix of transcripts from the host and sibling(s). Doublets lie between the host and sibling curves (Supplementary Fig. 6) and for each marmoset we empirically obtained a threshold for the Dropulation test statistic to identify them and were discarded in all analyses (Supplementary Fig. 6, Supplementary Table 5). The likelihoods plotted in Figures 1C,1E,2C and Supplementary Fig. 2 are from cells that have been filtered for minimum UMI and doublets.
Marmosets CJ006 and CJ007 were born in a triplet litter (tri-zygotic) that all died shortly after birth. We did not have access to any tissue from the third sibling and were not able to perform whole genome sequencing on it. For Dropulation analysis of CJ006 and CJ007’s brains, we provided only the genotypes of marmosets CJ006 and CJ007. Nuclei that contain the genome of the third unknown sibling will mostly be identified as doublets and were discarded in our analysis.
Additional filtering for microglia and macrophage clusters
We performed additional filtering of microglia and macrophage cells. When we compared the gene expression of host microglia and sibling microglia using cell-types from first-round clustering (and with UMI and doublet filtering), we found an abundance of genes that have higher expression in host than in the sibling (Supplementary Fig. 7, see panels B, C, G, I and K). The asymmetry could arise from neuronal cells mis-classified as microglia or macrophages. To filter out these mis-classified cells, we sub-clustered the microglia and macrophage cell types of each marmoset using the same fastICA and Louvaine-based clustering used in the first round of clustering. We found that some sub-clusters were not chimeric, indicating that they were not cells of hematopoietic origin, and that discarding these cells improved the symmetry between host and sibling gene expression (Supplementary Fig. 7).
Whole genome sequencing
Illumina libraries from fibroblast, blood, brain and buccal cells (Supplementary Table 6) were created as follows. An aliquot of genomic DNA (150ng in 50μL) is used as the input into DNA fragmentation (aka shearing). Shearing is performed acoustically using a Covaris focused-ultrasonicator, targeting 385bp fragments. Following fragmentation, additional size selection is performed using a SPRI cleanup. Library preparation is performed using a commercially available kit provided by KAPA Biosystems (KAPA Hyper Prep with Library Amplification Primer Mix, product KK8504), and with palindromic forked adapters using unique 8-base index sequences embedded within the adapter (purchased from Roche). The libraries are then amplified by 10 cycles of PCR. Following sample preparation, libraries are quantified using quantitative PCR (kit purchased from KAPA Biosystems) with probes specific to the ends of the adapters. This assay is automated using Agilent’s Bravo liquid handling platform. Based on qPCR quantification, libraries are normalized to 2.2nM and pooled into 24-plexes. Sample pools are combined with NovaSeq Cluster Amp Reagents DPX1, DPX2 and DPX3 and loaded into single lanes of a NovaSeq 6000 S4 flowcell cell using the Hamilton Starlet Liquid Handling system. Cluster amplification and sequencing occur on NovaSeq 6000 Instruments utilizing sequencing-by-synthesis kits to produce 151bp paired-end reads. Output from Illumina software is processed by the Picard data-processing pipeline to yield CRAM or BAM files containing demultiplexed, aggregated aligned reads. All sample information tracking is performed by automated LIMS messaging. All samples were sequenced to 30X coverage.
Variant site detection and genotyping from whole genome sequencing
Illumina paired-end reads were aligned to the calJac3 reference marmoset genome assembly using bwa (Li & Durbin, 2010) with command “bwa mem”. Duplicate reads were marked using Picard Markduplicates and for each chromosome, the GATK Haplotype Caller (McKenna et al., 2010) was run in genotype discovery GVCF mode. For each chromosome, the GVCFs of all samples analyzed in this study (from fibroblasts, blood, buccal cells, skin, brain and hair), were combined into a single GVCF file using GATK CombineGVCFs. To obtain the highest sensitivity in calling SNPs, we included in the GVCF additional fibroblasts whole genome sequences from the colony, yielding a total of 113 marmosets for multi-sample variant calling. The GVCF of each chromosome was genotyped using GATK GenotypeGVCFs. Only bi-allelic SNPs were used in the analysis and the following filters were used: QD<4.0 | FS>60.0 | MQ<40.0 | MQRankSum<−12.5 | ReadPosRankSum<−8.0 | MAF<0.01 | QUAL<500. SNP calls from all chromosomes were combined into one VCF file and additional filtering was performed to discard heterozygous sites that exhibited extreme allelic imbalance, i.e., the fraction of non-reference allele (from all samples) is less than 0.2 or greater than 0.8, and furthermore, sites in copy number variant regions were discarded (copy number variant regions were obtained by running Genome STRiP (Handsaker et al., 2015) on whole genome sequencing data from 113 fibroblast samples).
Dropulation analysis using sibling genotypes from whole genome sequencing of buccal cells
For four marmosets in our dataset (CJ022, CJ025, CJ026, CJ102; all born with one sibling), only the buccal cells (from cheek swabs) of their siblings were available for whole genome sequencing (the siblings are, CJ106, CJ104, CJ105 and CJ103, respectively). Using a method that quantifies chimerism from whole genome sequencing data (Census-seq; software available at https://github.com/broadinstitute/Drop-seq) (Mitchell et al., 2020), we estimated the chimerism fraction in buccal cells as follows: CJ106: 10%, CJ104: 24%, CJ105: 24% and CJ103: 9%. Thus, the genotypes we obtained for these marmosets will include errors and those genotyping errors could subsequently affect Dropulation (donor-of-origin) analysis that was used to estimate chimerism. To empirically estimate how sibling genotypes obtained from a chimeric tissue affect Dropulation analysis, we selected a host-sibling pair whose genome sequencing were both obtained from fibroblast cultures: CJ027 and its birth sibling CJ140. To simulate DNA contamination, we fixed the sequencing coverage of CJ140 to 40X, and replaced between 1% to 60% of the reads from CJ027’s sequencing reads (random subsampling using ‘samtools view -s’). We genotyped CJ140’s “chimeric” bam files using GATK’s “genotype given alleles” mode and compared the genotypes with CJ140’s true genotypes from its pure fibroblast WGS. We found that the sensitivity at heterozygous sites remains constant with different contamination levels, while the false positive rate increases. The false positive calls at heterozygous sites come from homozygous sites incorrectly genotyped as heterozygous (Supplementary Fig. 8A–F). Next, we re-analyzed donor-of-origin on brain snRNA-seq of CJ027 sibling (CJ140) genotypes from simulated contaminated DNA (300,000 nuclei; CJ027 genotypes from pure fibroblast WGS, CJ140 genotypes from WGS with various contamination levels). We found that chimerism in CJ140’s WGS resulted in doublets being assigned to the twin (Supplementary Fig. 8G–L), which subsequently causes a slight increase in chimerism estimates (Supplementary Fig. 8M–U). The contamination levels in buccal cells were from 9% to 24%, which we estimate will result in an overestimation in microglia chimerism of up to 3.5 percentage points and 4.5 percentage points in macrophage chimerism. Our results will not be affected by this over-estimation of chimerism in 4 marmosets since the conclusions were made from the analysis of all 11 marmosets (including 7 marmosets whose siblings were genotyped from fibroblast cultures).
Gene expression analysis of host and sibling meta cells
For each cell type, the host and sibling “meta cells” were calculated from the sum of UMI counts per gene across cells and were scaled to counts per 100,000 transcripts. The fold-changes and P-values of differentially expressed genes were identified using the binomTest method from the edgeR package (Robinson et al., 2010).
Supplementary Material
Acknowledgements
This work was supported by the NIH BRAIN Initiative U01MH114819, the Stanley Center for Psychiatric Research at the Broad Institute of MIT and Harvard, the James and Patricia Poitras Center for Psychiatric Disorders Research at MIT and the Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT.
Footnotes
Software availability
All software used in the analysis are publicly available. Drop-seq (analysis of snRNA-seq data, clustering, marker genes), Census-seq (estimation of chimerism in whole-genome sequencing data) and Dropulation analysis (estimation of chimerism in snRNA-seq data): https://github.com/broadinstitute/Drop-seq; alignment and variant detection of Illumina whole genome sequencing data: bwa (https://github.com/lh3/bwa), GATK (https://gatk.broadinstitute.org), BCFtools (https://github.com/samtools/bcftools), samtools (http://www.htslib.org/download), Picard Tools (https://broadinstitute.github.io/picard); R environment (https://www.rstudio.com/products/rstudio/download/, https://www.r-project.org); cell type identification scType (https://github.com/IanevskiAleksandr/sc-type).
Data availability
Brain snRNA-seq of 6 marmosets (CJ022, CJ023, CJ025, CJ026, CJ027, CJ028) were generated as part of the NIH’s Brain Initiative Cell Census Network (BICCN) project, while brain snRNA-seq of 5 marmosets (CJ001, CJ006, CJ007, CJ023, CJ102), and all blood, liver and kidney snRNA-seq were generated for this project. All snRNA-seq datasets are available in the BICCN NeMO portal (https://assets.nemoarchive.org/dat-hsgdsgu and https://assets.nemoarchive.org/dat-1je0mn3). All whole genome sequencing datasets will be provided in a manuscript (in preparation) that will describe naturally occurring genome variation in captive marmosets.
References
- Aida T., & Feng G. (2020). The dawn of non-human primate models for neurodevelopmental disorders. Current Opinion in Genetics & Development, 65, 160–168. 10.1016/j.gde.2020.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G., Jensen F. E., Greely H. T., Okano H., Treue S., Roberts A. C., Fox J. G., Caddick S., Poo M.-M., Newsome W. T., & Morrison J. H. (2020). Opportunities and limitations of genetically modified nonhuman primate models for neuroscience research. Proceedings of the National Academy of Sciences of the United States of America, 117(39), 24022–24031. 10.1073/pnas.2006515117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengozian N., Batson J. S., Greene C. T., & Gosslee D. G. (1969). Hemopoietic chimerism in imported and laboratory-bred marmosets. Transplantation, 8(5), 633–652. 10.1097/00007890-196911000-00009 [DOI] [PubMed] [Google Scholar]
- Hammond T. R., Robinton D., & Stevens B. (2018). Microglia and the Brain: Complementary Partners in Development and Disease. Annual Review of Cell and Developmental Biology, 34, 523–544. 10.1146/annurev-cellbio-100616-060509 [DOI] [PubMed] [Google Scholar]
- Handsaker R. E., Van Doren V., Berman J. R., Genovese G., Kashin S., Boettger L. M., & McCarroll S. A. (2015). Large multiallelic copy number variations in humans. Nature Genetics, 47(3), 296–303. 10.1038/ng.3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianevski A., Giri A. K., & Aittokallio T. (2022). Fully-automated and ultra-fast cell-type identification using specific marker combinations from single-cell transcriptomic data. Nature Communications, 13(1), 1246. 10.1038/s41467-022-28803-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krienen F. M., Goldman M., Zhang Q., Del Rosario R C. H., Florio M., Machold R., Saunders A., Levandowski K., Zaniewski H., Schuman B., Wu C., Lutservitz A., Mullally C. D., Reed N., Bien E., Bortolin L., Fernandez-Otero M., Lin J. D., Wysoker A., … McCarroll S. A. (2020). Innovations present in the primate interneuron repertoire. Nature, 586(7828), 262–269. 10.1038/s41586-020-2781-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krienen F. M., Levandowski K. M., Zaniewski H., del Rosario R. C. H., Schroeder M. E., Goldman M., Lutservitz A., Zhang Q., Li K. X., Beja-Glasser V. F., Sharma J., Shin T. W., Mauermann A., Wysoker A., Nemesh J., Kashin S., Vergara J., Chelini G., Dimidschstein J., … Feng G. (2023). A marmoset brain cell census reveals persistent influence of developmental origin on neurons. Sciences Advances, 9(41) DOI: 10.1126/sciadv.adk3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., & Durbin R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics, 26(5), 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko E. Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A. R., Kamitaki N., Martersteck E. M., Trombetta J. J., Weitz D. A., Sanes J. R., Shalek A. K., Regev A., & McCarroll S. A. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell, 161(5), 1202–1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., & DePristo M. A. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research, 20(9), 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J. M., Nemesh J., Ghosh S., Handsaker R. E., Mello C. J., Meyer D., Raghunathan K., de Rivera H., Tegtmeyer M., Hawes D., Neumann A., Nehme R., Eggan K., & McCarroll S. A. (2020). Mapping genetic effects on cellular phenotypes with “cell villages.” bioRxiv, 2020.06.29.174383. 10.1101/2020.06.29.174383 [DOI] [Google Scholar]
- Niblack G. D., Kateley J. R., & Gengozian N. (1977). T-and B-lymphocyte chimerism in the marmoset. Immunology, 32(2), 257–263. https://www.ncbi.nlm.nih.gov/pubmed/139360 [PMC free article] [PubMed] [Google Scholar]
- Perdiguero E. G., & Geissmann F. (2016). The development and maintenance of resident macrophages. Nature Immunology, 17(1), 2–8. 10.1038/ni.3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., & Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26(1), 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C. N., French J. A., & Orti G. (2007). Germline chimerism and paternal care in marmosets (Callithrix kuhlii). Proceedings of the National Academy of Sciences of the United States of America, 104(15), 6278–6282. 10.1073/pnas.0607426104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A., Macosko E. Z., Wysoker A., Goldman M., Krienen F. M., de Rivera H., Bien E., Baum M., Bortolin L., Wang S., Goeva A., Nemesh J., Kamitaki N., Brumbaugh S., Kulp D., & McCarroll S. A. (2018). Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell, 174(4), 1015–1030 e16. 10.1016/j.cell.2018.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D. P., Lehrman E. K., Kautzman A. G., Koyama R., Mardinly A. R., Yamasaki R., Ransohoff R. M., Greenberg M. E., Barres B. A., & Stevens B. (2012). Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron, 74(4), 691–705. 10.1016/j.neuron.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D. P., & Stevens B. (2015). Microglia Function in Central Nervous System Development and Plasticity. Cold Spring Harbor Perspectives in Biology, 7(10), a020545. 10.1101/cshperspect.a020545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney C. G., Curran E., Westmoreland S. V., Mansfield K. G., & Vallender E. J. (2012). Quantitative molecular assessment of chimerism across tissues in marmosets and tamarins. BMC Genomics, 13, 98. 10.1186/1471-2164-13-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- “The Marmoset Genome Sequencing and Analysis Consortium”, (2014). The common marmoset genome provides insight into primate biology and evolution. Nature Genetics, 46(8), 850–857. 10.1038/ng.3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells M. F., Nemesh J., Ghosh S., Mitchell J. M., Salick M. R., Mello C. J., Meyer D., Pietilainen O., Piccioni F., Guss E. J., Raghunathan K., Tegtmeyer M., Hawes D., Neumann A., Worringer K. A., Ho D., Kommineni S., Chan K., Peterson B. K., … McCarroll S. A. (2023). Natural variation in gene expression and viral susceptibility revealed by neural progenitor cell villages. Cell Stem Cell, 30(3), 312–332.e13. 10.1016/j.stem.2023.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wislocki G. B. (1939). Observations on twinning in marmosets. The American Journal of Anatomy, 64(3), 445–483. 10.1002/aja.1000640305 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.