Abstract
To understand the genomic basis accounting for the phenotypic differences between human and apes, we compare the matrices consisting of the cis-element frequencies in the proximal regulatory regions of their genomes. One such frequency matrix is represented by a robust singular value decomposition. For each singular value, the negative and positive ends of the sorted motif eigenvector correspond to the dual ends of the sorted gene eigenvector, respectively, comprising a dual eigen-module defined by cis-regulatory element frequencies (CREF). The CREF eigen-modules at levels 1, 2, 3, and 6 are highly conserved across humans, chimpanzees, and orangutans. The key biological processes embedded in the top three CREF eigen-modules are reproduction versus embryogenesis, fetal maturation versus immune system, and stress responses versus mitosis. Although the divergence at the nucleotide level between the chimpanzee and human genome was small, their cis-element frequency matrices crossed a singularity point, at which the fourth and fifth singular values were identical. The CREF eigen-modules corresponding to the fourth and fifth singular values were reorganized along the evolution from apes to human. Interestingly, the fourth sorted gene eigenvector encodes the phenotypes unique to human such as long-term memory, language development, and social behavior. The number of motifs present on Alu elements increases substantially at the fourth level. The motif analysis together with the cases of human-specific Alu insertions suggests that mutations related to Alu elements play a critical role in the evolution of the human-phenotypic gene eigenvector.
Keywords: cis-element frequency, dual eigenmodule, polarized eigenvector, Alu element, cognition
Introduction
Many efforts have been made to explore the origin of humans, in particular, to elucidate the evolutionary history from apes to humans. Phylogenetic studies based on DNA sequences infer that the chimpanzee is our closest living relative. The whole genome comparison indicates that the divergence rate between humans and chimpanzees at the nucleotide level is as little as 1.23% (Waterson et al. 2005). King and Wilson (1975) proposed that cues for biological differences might be located in the noncoding DNA making up 98.5% of the human genome. Indeed, evidences from various species support the claim that changes in cis-regulatory sequences constitute a key genetic basis for adaptation (Wray 2007).
A key problem in the study of the evolution of regulatory sequences is the gain and loss of cis-regulatory elements, which has been explained by turnover to some extent (Stone and Wray 2001; Doniger and Fay 2007), and by recombination and selection (Paixao and Azevedo 2010). Insertions of transposon elements can also bring in cis-regulatory elements. For example, Alu elements have been suggested to be carriers of cis-regulatory elements (Polak and Domany 2006). The farther away from the transcription starting sites, the more Alu elements insertion events were observed (Tsirigos and Rigoutsos 2009). Alu retrotransposition events are estimated to occur at a 2.2-fold higher rate in humans compared with chimpanzees and bonobos (Hormozdiari et al. 2013). Overall, insertion/deletions between the human and chimpanzee genome are estimated to comprise 3% of both genomes (Waterson et al. 2005).
In this study, we investigated the genome-wide frequency distribution of 1,403 vertebrate cis-motifs in the proximal regulatory regions flanking transcription starting sites (TSS) of three hominidae genomes: human, chimpanzee, and orangutan. We constructed a cis-regulatory element frequency (CREF) matrix for each species, using the motif position weight matrices (PWMs) of their transacting factors collected from the TRANSFAC database (Wingender et al. 1996). One such CREF matrix was decomposed into dual eigen-modules. Cross-species comparisons enabled us to identify conserved CREF eigen-modules at levels one, two, three, and six, as well as divergent eigen-modules at levels four and five. Compared with apes’, the human eigen-modules at levels four and five were reorganized. Surprisingly, the gene module at level 4 encodes the human-specific phenotypes such as biological processes key to long-term memory, language development, and social behavior.
Results
Cis-Element Frequency Matrices
This study focuses on the cis–trans regulation around the proximal regulatory regions. Rather than the condition-specific binding data from ChIP-seq experiments, we would consider the cis–trans binding strength that involves solely DNA sequences. We count the occurrences of a cis-element in the proximal regulatory region of a protein-coding gene, and define it as the binding strength of the corresponding transfactor and the gene target. The definition is motivated by two perspectives. First, intuitively more occurrences of a cis-element would increase its chance to be bound by the partner factor. We take a probability model for the binding of a regulator and a DNA target (see Supplementary Material online), and show that the chance of at least one successful binding is roughly proportional to the occurrences of the cis-element (Feng et al. 2019). Second, as reported (Xu et al. 2015), the number of mRNA molecules in the vicinity of the transcription is, within a certain range, roughly proportional to the number of bound transcription factors, which can be approximated by the number of the binding sites. Thus, the frequency of a cis-element measures a kind of capacity of the transcriptional regulation.
Specifically, we perform a systematic search for potential binding sites of transcription factors whose binding motifs are available in the TRANSFAC database. The motif of a cis-element is represented by its PWM (Lawrence and Reilly 1990; Lawrence et al. 1993). The motif search is carried out within the region from −1,000 to +500 bp of the annotated most upstream TSSs of protein-coding genes in the human, chimpanzee, and orangutan genomes. The TSS positions are retrieved from the Ensembl database (Zerbino et al. 2018). The collection of the cis-element frequencies of all genes can be arranged in a CREF matrix whose rows and columns are the respective genes and motifs for each species. These three species–specific matrices are the basis of the dual eigen-analysis.
Dual Eigen-modules of CREF Matrices
To dissect the underlying structure of the high-dimensional CREF matrices, we adopt the dual eigen-analysis as illustrated in figure 1, which detects functional modules in three major steps.
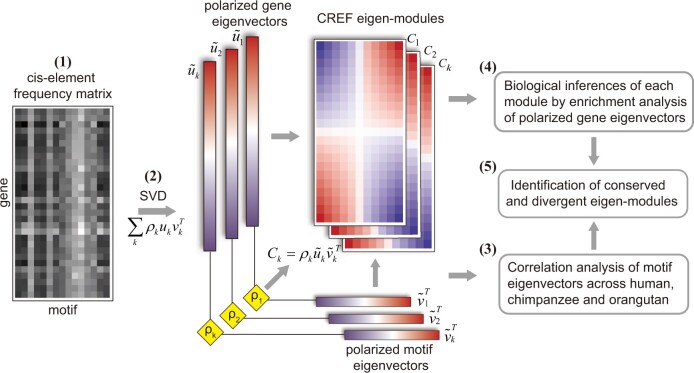
Fig. 1.
The illustration of the CREF cis-regulatory element frequency (CREF) eigen-modules. (1) The matrix of CREFs in the proximal regulatory sequences represents the cis-trans binding strength across all genes. (2) The singular value decomposition of the CREF matrix. Each singular value , together with the polarized gene/motif eigenvector arranged by sorting their loadings comprise a CREF eigen-module; each species has one such CREF matrix together with its SVD. (3-5) The biological analysis of the CREF eigen-modules.
First, we compute the robust singular value decompositions (SVD) of species-specific CREF matrices (Golub and Loan 1996; Cand et al. 2011). Several eigen-components are thus derived, each of which is comprised a singular value and a pair of eigenvectors: one vector for the motifs and the other for the protein-coding genes. The motif eigenvector, denoted by v, is composed of the weights of all cis-element motifs, referred to as the motif loadings, whereas the gene eigenvector, denoted by u, is composed of the weights of all genes, referred to as the gene loadings. The top eigen-components of SVDs are expected to capture the principal information of the genome-wide cis–trans binding strengths. Supplementary table S1, Supplementary Material online, shows the top six singular values and the percentages of their contributions in each species.
Second, we sort the loadings of each eigenvector. The eigenvectors are thus polarized with a positive and a negative pole (fig. 1(2)). Each pair of polarized gene and motif eigenvectors, together with the singular value comprise an eigen-module of CREF. Unlike the original cis–trans binding matrix, CREF eigen-modules stratify the cis-element frequencies into different levels. At each level, the most prominent genes and their corresponding motifs can be found near the two poles. Naturally the decomposition defines regulatory modules by the combinations of the motifs sitting at one pole. The singular value of one module reflects its frequency level.
Third, on the one hand, we compare each CREF eigen-module across humans, chimpanzees, and orangutans by correlation analysis of their polarized motif eigenvectors. On the other hand, we make inferences regarding the biological relevance of each CREF eigen-module using gene enrichment analysis of its polarized gene eigenvector. Finally, we identify the conserved and divergent modules (fig. 1(3–5)).
Comparison of CREF Dual Eigen-modules between Humans and Apes
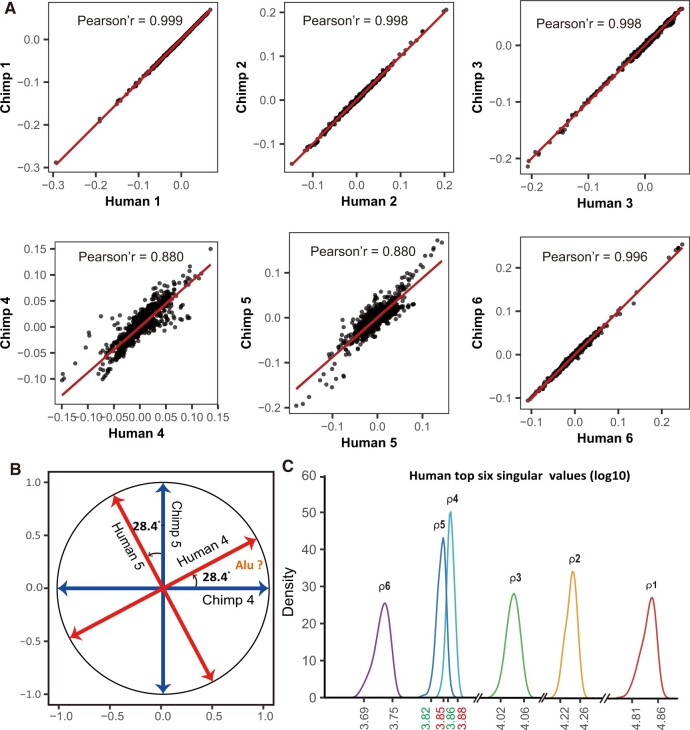
The nature of the dual eigen-modules allows us to compare them across species along either the gene eigenvectors or the motif eigenvectors. Consequently, the conservation of CREF eigen-modules can be evaluated from either the motif eigenvector or the gene eigenvector. However, the gene annotations of the three genomes are not of the same quality. We first consider the Pearson correlation coefficients between the motif eigenvectors of two species. When the motif correlation coefficient of one CREF eigen-module is beyond a threshold, which is taken to be 0.99 hereafter, then the module is defined to be conserved. As shown in figure 2A, the Pearson correlations between the top three and the sixth human–chimpanzee motif eigenvectors are >0.99, thus, they are conserved. In contrast, the middle two motif eigenvectors, the fourth and fifth, are less correlated. A similar pattern (supplementary fig. S1A, Supplementary Material online) is observed when we compare the loadings of orangutans’ top motif eigenvectors versus humans’. Notably, all top six motif eigenvectors, including the fourth and fifth, are conserved between chimpanzees and orangutans (supplementary fig. S1B, Supplementary Material online). Thus, we postulate that the first three and the sixth CREF eigen-modules were conserved in hominidae, whereas significant evolution occurred in the fourth and fifth CREF eigen-modules from the two apes to humans. Furthermore, by projecting the human fourth and fifth motif eigenvectors onto the chimpanzee’s, we discover that the 2D eigen-space spanned by the fourth and fifth motif eigenvectors is almost identical, from chimpanzee to human. Interestingly, these two eigen-directions of human, are rotated ∼28° from those of chimpanzee’s (fig. 2B). The eigenvector rotation is explained in details in the section “Multiplicity of a Singular Value and Its Eigen-space” (see Materials and Methods).
Fig. 2.
(A) The scatter plots of human’s top six motif eigenvector loadings versus chimpanzee’s. The top three pairs and the sixth pair are highly correlated (Pearson correlation >0.99), suggesting these four modules are conserved. In contrast, the fourth and fifth pairs are less correlated, suggesting divergence occurred in these two modules. (B) Rotation between the fourth and fifth motif eigenvectors from apes to humans. In the 2D eigensubspace spanned by the fourth and fifth motif eigenvectors, human’s two eigen-directions were rotated ∼28.4° from those of chimpanzee’s. The motif analysis suggests that mutations related to Alu elements are likely to be a key driving factor of the rotation. (C) The sampling distributions of human’s top six singular values are shown by their densities from right to left. The sampling distributions were obtained from 100 random submatrices containing 80% of the motifs. The sampling distributions of the top three and the sixth singular values are well separated from adjacent ones, whereas the fourth and the fifth overlap by a large portion, suggesting a possible fusion event in the history, namely, a 2D eigen-space of a common singular value.
Stability of CREF Eigen-modules
The general eigenvalue and vector perturbation problem has been studied extensively in numerical linear algebra. A basic mathematical result of the perturbation analysis states that if an eigenvalue is well separated from other eigenvalues, then its associated eigenvector is fairly stable with respect to small perturbations to the matrix (Stewart and Sun 1990). But when an eigenvalue approaches its neighboring eigenvalue(s), its eigenvector will become more sensitive to perturbations. In the case of the human CREF matrix, the top three and the sixth singular values are well separated from others, whereas the relative distance between the fourth and fifth singular values is much closer (supplementary table S1, Supplementary Material online). This suggests that the top three and the sixth CREF eigen-modules are more stable, whereas the fourth and fifth CREF eigen-modules are less so. According to the expansions of the eigenvectors (see Materials and Methods), the two neighboring vectors interfere more with each other, or become wobbly, when their eigenvalues are closer.
Moreover, as the fourth and fifth singular values are well separated from the third and sixth ones, the matrix perturbation theory claims that the invariant subspace composed of the fourth and fifth eigenvectors, either on the gene side or on the motif side, is more stable than the single eigenvectors themselves are.
Other than the mathematical stability, we are also concerned with the sensitivity of the distances between adjacent singular values of the human CREF matrix, with respect to the set of motifs selected in the study. To examine this, we introduced perturbations by random sampling and checked the resulting sample distributions (see Supplementary Material online). As shown in figure 2C, the sampling distributions of the top three and the sixth singular values are well separated from adjacent ones, whereas those of the fourth and fifth singular values overlap somewhat. We also notice that their sampling distributions overlap to some extent too in the case of chimpanzees, but no overlap is observed in the case of orangutans (supplementary fig. S3, Supplementary Material online) because the distance between the fourth and fifth singular values for the latter is sufficiently large.
The Biological Relevance of Principal CREF Eigen-modules
Next, our focus turns from the mathematical structure of CREF eigen-modules to their biological relevance. The CREF eigen-modules are defined by the SVD of the CREF matrix. The SVD stratified the frequency matrix into orthogonal components by their specific frequency levels from high to low. The first component captures the largest portion of global motif frequencies; the second component, which is orthogonal to the previous one, captures the next largest portion of motif frequencies, and so on and so forth. Within a certain principal component, those genes with heavy and similar loadings around one pole correspond to the cis-motifs around the dual pole of the coupling vector. This fact leads us to hypothesize that within one eigen-component, the transcription of the principal genes via the corresponding cis-motifs was selected over time to achieve a certain kind of efficacy. In other words, when transcriptional regulations were stratified into different levels, they could be realized in a more synergistic way.
To figure out the biological activities embedded at each level, we collected predefined gene subsets from gene ontology (GO) (Ashburner et al. 2000; Carbon et al. 2017), KEGG (Kanehisa and Goto 2000; Kanehisa et al. 2016, 2017), and REACTOME (Fabregat et al. 2018), and applied the Wilcoxon scoring method (Cheng et al. 2007) to see what gene subsets were enriched at the two ends of the polarized gene eigenvectors. To simply and clearly characterize the biological relevance of each eigen-module, we organized the significant subsets according to three rules (see Supplementary Material online). Figure 3 summarizes the results of enrichment analysis for the top six polarized gene eigenvectors of humans and chimpanzees (for more details, see supplementary tables S3–S18, Supplementary Material online).
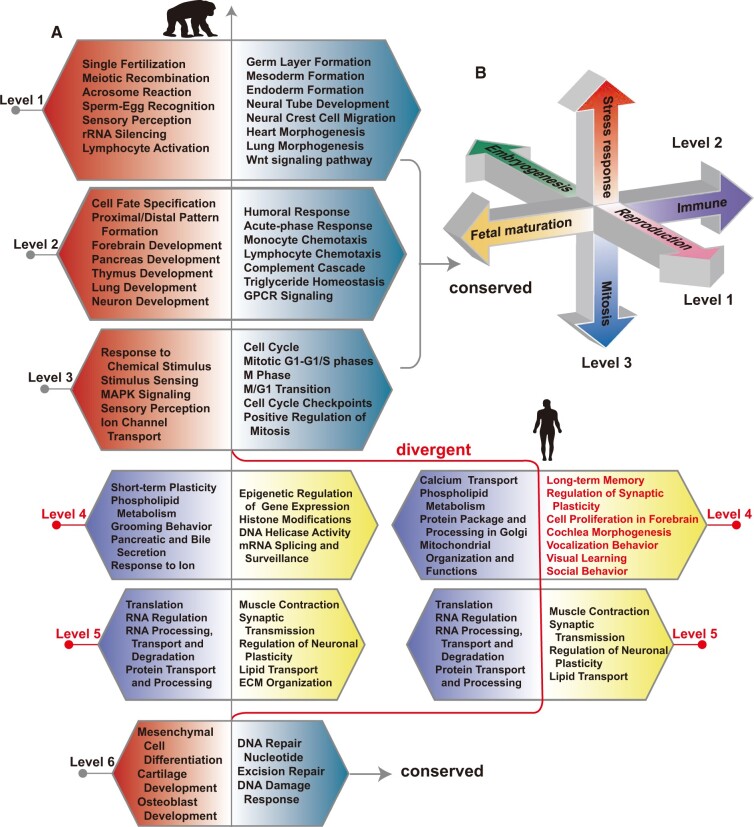
Fig. 3.
(A) Summarization of the biological processes enriched at the two ends of the top six polarized gene eigenvectors for humans and chimpanzees. Each box, whose poles are marked in two different colors, corresponds to one polarized gene eigenvector. From top to bottom, the six polarized gene eigenvectors are arranged in the descending order of their singular values. The enrichment results of the top three and the sixth gene eigenvectors are highly conserved in consistency with the results of the motif eigenvectors, therefore, only the results for human are shown. Significant divergences in the fourth and fifth eigenvectors are observed between humans and chimpanzees. Notably, the GO categories of regulation of synaptic plasticity, social behavior, vocalization behavior, visual learning, and long-term memory are enriched at the human fourth gene eigenvector, but are not so at that of chimpanzee. (B) The key functions of the top three CREF eigen-modules are represented by three polarized axes. They are highly conserved in humans, chimpanzees, and orangutans. Reproduction and embryogenesis lie at the two poles at the top level. Fetal maturation faces immune system at level 2. Stress responses and mitosis stay opposite in the level 3 axis. They are supported primarily by the enrichment results of the polarized gene eigenvectors, and are also supported by the known cis–trans regulation such as those in embryogenesis and mitosis due to the duality.
Top Three CREF Modules Are Highly Conserved
At a certain frequency level, if a gene subset is significantly enriched in both human and chimpanzee, the enrichment is conserved. We evaluated the overall conservation of enrichment results of one CREF eigen-modules using the portion of subsets shared by both human and chimpanzee. The significance threshold is taken to be 0.01 and 0.05, respectively, for humans and chimpanzees, as the gene subset annotations of the former are adopted for both species and the number of genes available for the latter in the same subset is usually smaller. Considering the quality of gene subset annotations, we first choose the KEGG pathways for the conservation analysis. In line with the conservation of the top three and the sixth motif eigenvectors, their dual gene eigenvectors share >75% of enriched KEGG pathways. In contrast, the portion of pathways shared by the fourth gene eigenvectors of the two species drops to only 46%. Similar results for the GO biological processes are shown in supplementary table S2, Supplementary Material online. Nevertheless, the portion of KEGG pathways shared by the fifth gene eigenvectors of the two species is as high as 90%. It turns out that a large number of KEGG pathways enriched at the fifth level are basal biological processes such as RNA/protein processing and transport (fig. 3 and supplementary tables S13–S16, Supplementary Material online), which are supposed to be relatively conserved across these close species. Later, we will show that the corresponding cis-regulatory motifs at level 5 did change somewhat from chimpanzee to human.
The major biological processes enriched near the two poles of the top three polarized gene eigenvector are highly conserved across humans, chimpanzees, and orangutans (fig. 3A and B). As a matter of fact, they comprise the key processes of life. By and large, reproduction and embryogenesis lie at the two ends of the top level. The former is indicated by processes such as meiosis and fertilization; whereas the latter is indicated by early embryonic events such as germ layer formation and neural tube development. At level 2, fetal maturation, characterized by organ development, lies at one end, whereas the immune system lies at the other end. Cell–environment and cell–cell interactions such as stress responses, and mitosis stay in the opposite direction at the third level. Due to the duality, the enriched biological processes in the gene eigenvector are supposed to correspond to the cis-elements near the two ends of the polarized motif eigenvector. However, it should be noted that the cis-regulatory mechanisms of these biological processes are only partially known. In the following, we describe two known examples of regulators and target relationships, which provide support to the dual eigen-analysis.
Dual Eigen-analysis Identifies the Regulators Involved in Early Embryogenesis at the First Level
With a close look at the top polarized motif eigenvector, we find a list of high-ranking motifs at the pole coupled with the embryogenesis pole of the gene eigenvector. From them, we select three representatives shown in figure 4A. One such is ZIC3_05, ranking in the third place; its binding factor ZIC3 plays a critical role in embryogenesis. The ZIC3 null embryos exhibit a huge range of phenotypes including primitive streak dysgenesis, incorrect body axis establishment, and neural tube defect (Houtmeyers et al. 2013). Moreover, two binding elements of other members of the ZIC family, ZIC1_01 and ZIC2_05, also rank among the top ten. In addition to ZIC proteins, the other two examples are MRG2_01 and TBX5_Q5, ranking 47 and 15, respectively. The binding factor of MRG2_01, MEIS3, directs neural anteroposterior patterning as a direct target of canonical WNT signaling (Elkouby et al. 2010). TBX5 plays an important role in heart morphogenesis, and its dysregulation leads to abnormal cardiac morphogenesis and congenital heart diseases (Horb and Thomsen 1999). Other high-ranking cis-elements are listed in supplementary table S20, Supplementary Material online, where the literature of their functions in embryogenesis can be found.
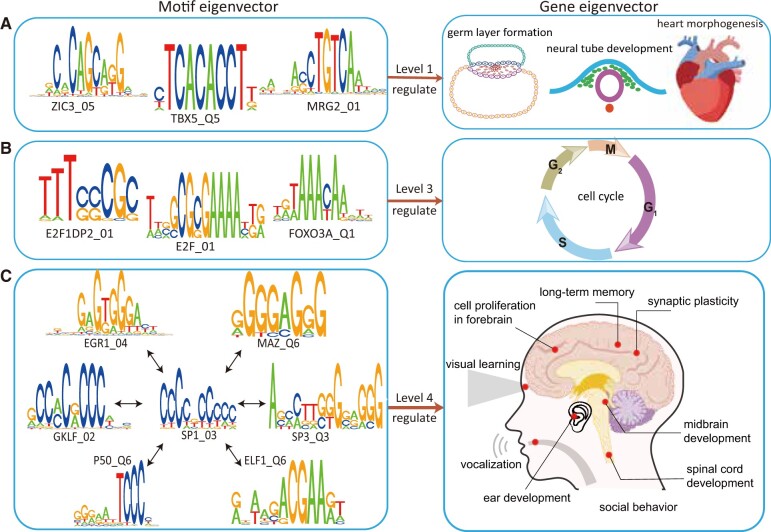
Fig. 4.
Examples of the regulatory correspondence between the polarized motif- and gene eigenvectors. The examples are from the human CREF modules, and can find experimental support in the literature. In each row, certain major biological processes enriched near one end of a polarized gene eigenvector are shown in the right box, whereas their cis-regulatory elements are shown in the left box. (A) Embryogenesis at level 1 and its three cis-elements present at the dual end of the first motif eigenvector; (B) mitosis at level 3 and its three cis-elements present at the dual end of the third motif eigenvector; (C) the cognition-language system enriched at level 4, and some of their cis-elements present at the dual end of the fourth motif eigenvector. The shown cis-elements except for ELF_Q6 are boosted from the chimpanzee’s fifth motif eigenvector. They center around SP1, whose binding motif is an MPA. Among them, EGR1_04, P50_Q6 are, respectively, the binding motifs of EGR1 and NFKB1, two key regulators of long-term memory.
Cell Cycle Regulators Stand around One Pole of the Third Polarized Motif Eigenvector
The motifs captured by the third motif eigenvector are binding elements of cell cycle factors experimentally validated by previous studies. The phase-specific motifs and their bind factors are illustrated in figure 4B and supplementary table S21, Supplementary Material online. Among them, E2F1, a member of the E2F family transcription factors, is a well-known cell cycle regulator and primarily functions as an activator of transcription in cell cycle control (Lam and La Thangue 1994; Ren et al. 2002). Moreover, E2F1 protein can pair with DP2 protein as a heterodimer to efficiently activate cell cycle regulated transcription (Lam and La Thangue 1994; Wu et al. 1995). In addition to the E2F family, the dual eigen-analysis suggests that several members of the forkhead family play important roles in the cell cycle regulation. It has been reported that FOXO3 contributes to G2-M progression by directly activating the transcription of GADD45 (growth arrest and DNA damage-inducible protein). Ectopic expression of FOXO1, FOXO3, or FOXO4 blocks cell cycle at the G1 phase by up-regulating the P27KIP1 level (Medema et al. 2000). It is somewhat surprising that the binding elements of three major reprograming factors, NANOG, SOX2, and OCT4, are present at the mitosis end as well (Takahashi et al. 2007; Yu et al. 2007).
Distance between the Fourth and Fifth Frequency Levels
To measure the relative distance between two adjacent levels, we take the ratio of the difference of their singular values over the larger one. Two observations are noted. On the one hand, the distance between the fourth and fifth levels is far less than that between other adjacent pairs in both humans and apes (supplementary table S1, Supplementary Material online). On the other hand, when comparing the distance between the fourth and fifth levels across species, we find that it is as small as 3.0% in humans, 4.0% in chimpanzees, and 8.2% in orangutans. We further examine the sensitivity of the distance with respect to the selection of motifs. By randomly sampling 80% of the motifs, we generated matched distances in all three species, then repeated the sampling. We observed that, in 74% of the cases, the distances did decrease in the order of orangutan, chimpanzee, and human. The conclusion is further supported by statistical tests that compare the sampling distributions (supplementary fig. S4A, Supplementary Material online). This suggests that the distance decreases from the two apes to human regardless of motif selection. This kind of comparison of the relative distances between the fourth and fifth CREF eigen-modules across species might provide some clues to the evolution of regulation in hominidae.
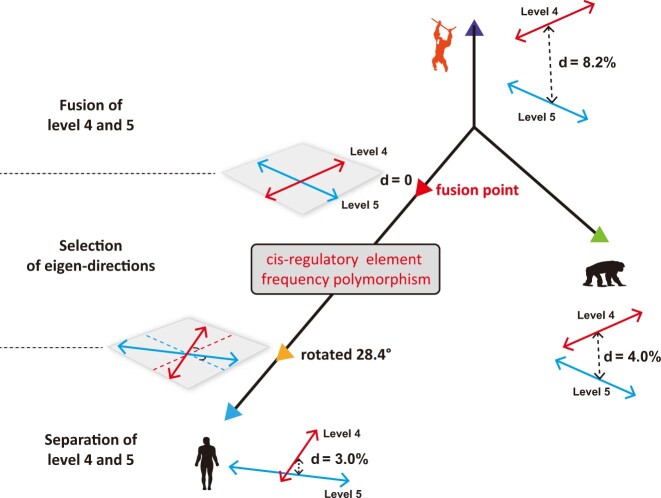
A Hypothetical Evolution of Hominidae Genomes in the Fourth and Fifth CREF Eigen-modules
Putting together the mathematical facts and computational results, we propose one continuous evolutionary path of the fourth and fifth CREF eigen-modules (fig. 5). They might have undergone a genomic evolution consisting of three major stages. In the first stage, after the common ancestor split from the orangutan lineage, the distance between the fourth and fifth levels decreased more quickly in the human lineage than in the chimpanzee lineage. In the former case, the fourth and fifth levels approached each other, whereas the eigen-directions remained unchanged. That is, the two levels during this period were still distinct so that their corresponding eigenvectors were orthogonal to each other, forming two 1D eigen-spaces. At one point, the two levels became identical and consequently the two 1D eigenvectors fused into a 2D eigen-space. The fusion of eigen-modules introduced a new type of options for selection.
Fig. 5.
A hypothetical evolution of the fourth and fifth CREF eigen-modules from the apes to humans. In the ape genomes, the distances between the fourth and fifth singular values are statistically significant, therefore, their corresponding eigenvectors are orthogonal to each other. Along the evolution, the two singular values approached, likely driven by the mutations related to Alu elements together with other mechanisms. At one point, the two levels became identical and consequently the two 1D eigenvectors fused into a 2D eigen-space, in which any direction was an eigenvector. This 2D eigen-space formed a space of the CREF-polymorphisms. Then selection could occur. In comparison to the fourth and fifth motif eigenvectors of the chimpanzee, the human’s ones were rotated 28.4°. We compare the deviance of the distance between the fourth and fifth levels in each species from that generated at its fusion point. The results support that the distances to the fusion point is increasing in the order of humans, chimpanzees, and orangutans.
One key selection is supposed to occur in the second stage. We refer to the 2D eigen-space at the fusion point as a space of CREF-polymorphisms, because any linear combination of the original eigen-directions is an eigenvector (see Materials and Methods). The eigen-directions in the 2D space can be quantified by a continuous angle of rotation with respect to the eigenvectors in the chimpanzee genome. According to this setting, only the eigen-direction with the highest fitness was eventually selected in the human lineage. As shown later by the enrichment analysis, the human-specific phenotypes such as long-term memory, language development, and social behaviors were indeed selected in the human’s fourth gene eigenvector.
The final stage is the separation of the two levels after the fixation of the eigen-direction in the human lineage. The distance between the fourth and fifth levels is only 3.0% in humans. We further use random perturbations to test if humans are indeed the closest to the fusion point (see Supplementary Material online).
Genes Specific to Cognition and Language Development Cluster near One Pole of the Fourth Gene Eigenvector in Human
Corresponding to the rotation or reorganization of the motif eigenvectors, the enriched biological processes and functions were modified too. The modification along the fourth gene eigenvector was phenomenal, whereas that along the fifth one is less so. It is found that a group of gene subsets representing human-specific phenotypes are enriched at the positive pole of the human gene eigenvector, but not along the fourth gene eigenvectors of the two apes (fig. 6B and supplementary fig. S5B and table S19, Supplementary Material online). Next, we elaborate on two aspects that laid the foundation for our human uniqueness.
Fig. 6.
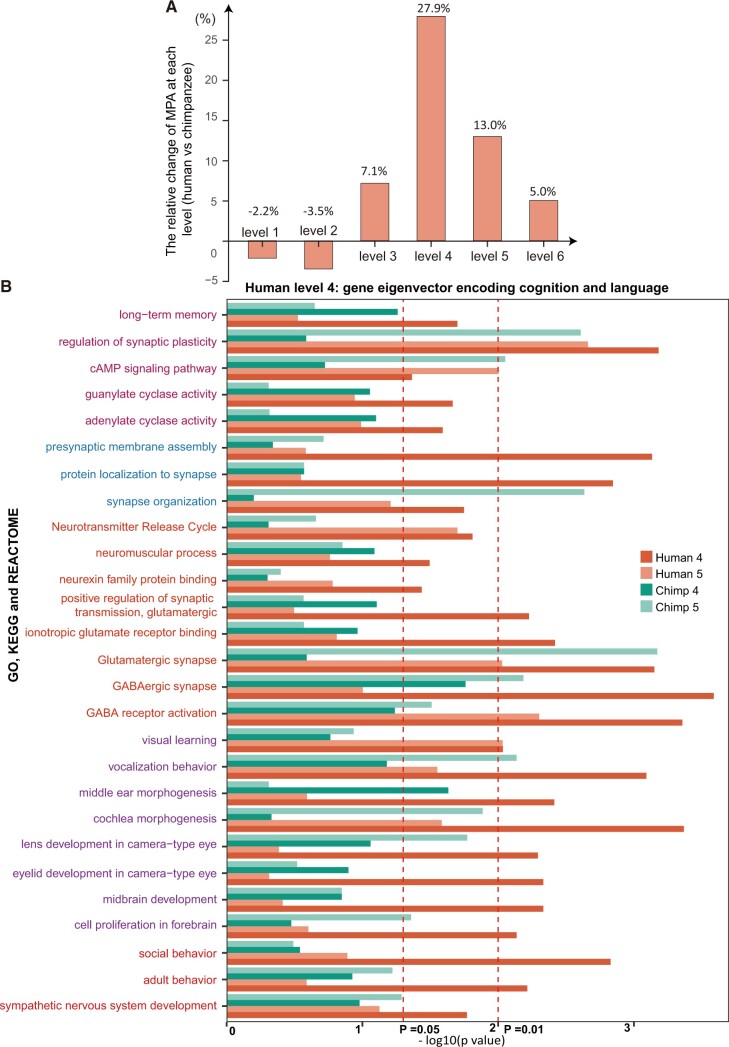
(A) The relative change of motifs present on Alu (MPAs) in percentages at each level from chimpanzee to human. The number of MPAs increases most significantly at level 4 by 27.9%, followed by 13.0% at the fifth level. (B) Shown are gene subsets that are significantly enriched near the positive end of the fourth polarized gene eigenvector of humans, but are not or less significantly enriched along that of chimpanzees. Their significances at the human fourth and fifth, chimpanzee fourth and fifth eigenvectors are represented by the dark orange and light orange, dark green, and light green bars, respectively. The left red-dashed line marks the P value 0.05, whereas the right one marks the P value 0.01. According to their biological relevance, the enriched gene subsets are grouped by memory, synaptic activity, organ development, and behaviors related to language, and complex behavior, shown in different colors.
First, long-term memory, as an integral part of our existence, is uniquely present in humans. Long-term memory helps individuals accumulate experiences, thereby enhancing their ability to live in more complicated conditions. Indeed, long-term memory is enriched in the human fourth gene eigenvector. Several specific biological pathways related to long-term memory are shown in the upper part of figure 6B. They include the neurochemical process synaptic plasticity, which is thought to be indispensable for memory consolidation, together with much more active glutamatergic and GABAergic synaptic transmission. In addition, adenylyl and guanylate cyclase activity are significantly enhanced. Calcium-stimulated adenylyl cyclase activity produces the cAMP signals, which are necessary for late phase long-term potentiation and long-term memory. Guanylate cyclase is also reported to regulate the activity of dopamine neurons in the midbrain, thereby controlling diverse behavioral processes (Gong et al. 2011). Other pathways contributing to synapse connectivity include presynaptic membrane assembly and neurexin family protein binding. Neuroligin–neurexin interaction can trigger adhesion between dendrites and axons bidirectionally (Dean and Dresbach 2006).
Second, the human language is the most powerful communication system on account of its complexity, productivity, and grammaticality. The human language is also unique in adaptability. It has three forms: speech, sign, and writing. It is found that cochlea morphogenesis, middle ear morphogenesis, and vocalization behavior, which are the basis for oral language, are enhanced in the human fourth gene eigenvector. On the visual side, visual learning and camera-type eye development are enriched.
Other enriched annotated gene sets include social behavior, adult behavior, and development of sympathetic nervous system, spinal cord, dendrites, hippocampus, midbrain, and neuronal system. According to the stratification by SVD, these all belong to the same level. This fact is surprisingly reasonable because when parts needed in a system somehow match one another in quantity, a kind of efficiency can be achieved.
At the other pole of the same level, the pathways of phospholipid, lipids, and lipoproteins metabolism are enriched (supplementary table S10, Supplementary Material online). The KEGG pathway—steroid hormone biosynthesis is significant even after correction (P value = 4.37e-03). The most significant GO biological process is protein targeting to Golgi (P value = 1.32e-04). Related GO cell components include the integral component of Golgi membrane (P value =1.20e-02). It is interesting to note that the Golgi is fragmented in a variety of neurodegenerative diseases (Joshi et al. 2015). For example, Golgi fragmentation is highly related to the formation of extracellular amyloid plaques by secreted amyloid beta, which is the hall mark of Alzheimer’s disease. Thus, the genes at this end, among many tasks, are involved in the pathways supporting the brain development and activities.
Motifs Underlying the CREF Module Reorganization
Next, we look into the cis-elements at the end of the human fourth polarized motif eigenvector dual with the human cognitive gene eigenvector. When checking the motifs at the positive end of the human fourth polarized motif eigenvector, we discover that nearly one-quarter of them, 47 out of 200, are boosted from the fifth motif eigenvector of the common ancestor. In other words, they rank high at the fifth level but rank quite low at the fourth level in chimpanzees, but are now prominent at the fourth motif eigenvector in humans. We will refer to them as newcomers hereafter. Similarly, we refer to those which already rank at the top of the fourth polarized motif eigenvector of chimpanzees as old-timers.
The newcomers SP1_03 and LYF1_01 have been experimentally validated to be carried by Alu elements (Oei et al. 2004; Polak and Domany 2006). The two most abundant mobile elements in hominidae genomes are L1 and Alu elements (Deininger 2011); the former is more common in intergenic regions, whereas the latter is more common in intragenic regions. We shall now examine how many newcomer motifs are related to Alu elements.
To do this, we need a comprehensive list of the motifs present on Alu elements (MPA). The MPAs are defined as follows. First, we take the minFP (minimize false positives) option in TRANSFAC to search the motif occurrence in 60 Alu consensus sequences which cover three major Alu subfamilies in primates: AluJ, AluY, and AluS (Bao et al. 2015). This results in 25 MPAs. Second, we compare these with those reported in the literature (Polak and Domany 2006), part of which were experimentally validated. If one such reported motif was not included in the 25 MPAs, we obtain its TRANSFAC counterparts through its binding factor. In this way, the final list of MPAs is generated.
To quantitatively measure the impact of Alu elements on the cis–trans regulation, we count the number of MPA among the top and bottom 150 motifs at each level for both chimpanzee and human genomes. The number of MPA changes by −2.2%, −3.5%, 7.1%, 27.9%, 13.0%, and 5.0%, respectively, from levels one to six (fig. 6A). The same patterns are observed in orangutans (supplementary fig. S5A, Supplementary Material online). It is obvious that the number of MPAs increases substantially at levels four and five, implying that the Alu elements might account partially for the CREF module reorganization between the fourth and fifth motif eigenvectors.
Human-Specific Alu Insertions and the Fourth Gene Eigenvector
It is interesting to understand how Alu elements, of course along with other factors, drive the change of CREF profiles, thereby reorganizing the modules. In the following, we describe three such mechanisms.
First, human-specific Alu elements were inserted directly into the proximal regulatory regions. To have a rather complete account, we consider the 8,817 human-specific Alu elements that were identified recently by comparing the genomes of human and nine nonhuman primates including chimpanzees (Tang et al. 2018). We filtered out those falling out of the proximal regulatory regions, namely, −1,000 to 500 bp around TSS of the most upstream transcripts of humans, mostly principal ones. This resulted in 47 genes, each of which contains a clean human-specific Alu insertion. Of these, 25 genes rank higher in the fourth gene eigenvector of human than in that of chimpanzee; 15 genes rank lower; 7 genes lack annotations in chimpanzee. Among the 25 higher ranking genes, 21 are related to cognition, language, and brain development, whereas only 3 out of 15 lower ranking genes are so. Under the hypergeometric model, these numbers equate a P value of 3.91e-06. In addition, for each MPA, we compared its occurrences in each human-specific Alu insertion versus its frequency in the neighboring regulatory DNA sequence by a statistical test, and combined the 47 resulting P values by the Fisher’s method (Fisher 1925, 1948). All MPAs that are newcomers around the poles of the fourth polarized human motif eigenvector are significant (supplementary table S23, Supplementary Material online). The results show that the Alu insertions indeed play a significant role in the selection of human-specific cognition–language–behavior motif eigenvector.
A list of the 47 gene annotations is shown in supplementary table S24, Supplementary Material online. Among them, CEBPG can form stable heterodimers with CEBPB that is a key regulator of synaptic plasticity and memory formation; CTHRC1 functions in cochlea morphogenesis; GRK7 belongs to visual perception; VDAC3 is involved in synaptic transmission and learning; BSG plays a pivotal role in neural network formation; TRIB3 may play a role in programed neuronal cell death but does not appear to affect nonneuronal cells. SEMA4F is involved in axon guidance; mutation in MKS1 causes Meckel syndrome 1; mutation in CDT1 causes Meier–Gorlin syndrome 4.
Among the seven genes lacking annotated counterparts in chimpanzees, some are related to cognition as well. For example, OCLM, ranking 1,448 in the fourth gene eigenvector, is involved in visual perception. In addition, other human-specific Alu insertions than the above 8,817 ones may exist. One such example is ADAM10, which is involved in neuronal plasticity and cochlea development. A human-specific insertion AluJr4 is identified 700 bp upstream of its TSS (supplementary fig. S6, Supplementary Material online).
In the second mechanism, one or more Alu elements are present around the TSS of a human-specific transcript (supplementary table S25, Supplementary Material online). In the example of MYH14 that is involved in vocalization behavior and axon guidance, the TSS of the human principal transcript extends from that of chimpanzees toward the 5′ end. Two elements AluJo and AluYm1 are shown in its regulatory region (∼830 bp, ∼1,250 bp upstream of the TSS). In the case of DRD3 that is involved in social behavior, no Alu element is present in its proximal regulatory region of chimpanzee, whereas the most upstream transcript in humans starts at a different position, as shown in supplementary figure S7, Supplementary Material online. An AluJr, which is not a human-specific one, was located within 500 bp downstream from the TSS.
Finally, when the TSS of a gene changes, the regulatory DNA sequence changes accordingly, thereby changing its motif frequencies. We show that some changes of TSSs across species are accompanied by human-specific Alu insertions nearby (supplementary table S24, Supplementary Material online). The principal TSS of the gene PAX2, involved in cochlea morphogenesis and brain morphogenesis, shifts in human toward the 3′ end compared with that of chimpanzee. About 8 kb downstream of the regulatory sequence, a human-specific AluYa5 was inserted into an existing MIR element (supplementary fig. S8, Supplementary Material online). In the case of NRXN1 that is involved in social behavior, its principal TSS of human shifts ∼3 kb toward the 5′ end. About 16 kb upstream of the TSS is a human-specific insertion AluYg6. For another example, GABRP is one subunit of the receptor bound by the major inhibitory neurotransmitter GABA in the mammalian brain. Compared with the TSS of chimpanzees, several transcripts of humans start ∼20 kb farther toward the 5′ end. Nearby, 1.7 kb downstream of the most upstream TSS is an AluYa4 insertion that was reported to be missing in the human reference genome (GRCh36) yet fixed in humans (Hormozdiari et al. 2013).
Although the above evidence suggests that the Alu element is one of the genetic factors underlying the motif change in the human-specific eigen-module, other factors such point mutations, simple repeats, and repetitive elements other than Alu may contribute to the reorganization as well. A complete account of the mechanism is yet to be accomplished.
Gene Eigenvector Corresponding to Human-Specific Phenotypes and Its Cis–Trans Regulation
Among the biological processes enriched along the human cognitive gene eigenvector, the transcription factors involved in long-term memory have been studied extensively. Long-term memory depends on a temporally limited phase of RNA and protein synthesis (Davis and Squire 1984; Sutton and Schuman 2006). Synaptic plasticity is a mechanism closely related to memory formation. Based on certain models of synaptic plasticity and memory formation, scientists identified quite a few key transcription factors such as CREB, C/EBP, EGR1, AP-1, and NFKB1 (Alberini 2009). Among these, EGR1 is a marker of neuronal plasticity. Its binding cis-elements, EGR1_04 ranks 68, and is a newcomer. EGR2, another member of the EGR family, was reported to play a regulatory role in long-term memory (Alberini 2009). Its binding element EGR2_Q6 is a newcomer too with rank 114. C/EBP is a key regulator required for synaptic plasticity and memory formation. Its binding partner, CEBP_Q3 ranks 16, and is an old-timer. NFKB1 plays a critical role not only in the immune system but also in memory formation. Its binding partner, P50_Q6, is boosted from the position of rank 149 in chimpanzees to rank 81 in humans. However, TAXCREB_02, the binding element of the basal factor CREB, is found at the top of the other pole, ranking 112. It is both a newcomer and an MPA in the fourth polarized motif eigenvector of humans. Similarly, AP1_02, the binding element of AP-1, is a marker of neuronal activation, ranking 197 on the same pole as that of CREB. It is an MPA yet an old-timer motif. Notably, CREB and AP-1 are activated upon stimuli such as stress and growth factors; whereas C/EBP functions downstream of CREB.
We notice a common feature of several newcomer elements in the polarized motif eigenvector paired with the human cognitive gene eigenvector: they all interact with SP1, whose binding elements are boosted from the chimpanzee’s fifth motif eigenvector as well (fig. 4C). One such example is SP1_03, ranking 28; another is MAZ_Q6, ranking 35. Their binding factors SP1 and MAZ mediate enhancement of NR1 (NMDA receptor subunit type 1) promoter activity during neuronal differentiation (Okamoto et al. 2002). The expression of NR1 is also regulated by NFKB1, which involves interaction with SP1 (Liu et al. 2004). LRF (also known as ZBTB7A), whose binding element LRF_Q2 ranks 39 interacts with both NFKB1 and SP1 (Lee et al. 2002, 2005). KLF13, whose binding element BTEB3_Q5 ranks 67, represses transcription in competition with the activator SP1 (Kaczynski et al. 2001; Lee et al. 2002). KLF4, also known as GKLF with a binding element GKLF_Q4 ranking 32, inhibits SP1-mediated activation of the CYP1A1 promoter (Zhang et al. 1998). SP3, whose binding element SP3_Q3 ranks 140, interacts with SP1 in quite some regulatory situations. For example, SP1 and SP3 act as oxidative stress-induced transcription factors in cortical neurons, positively regulating neuronal survival (Ryu et al. 2003). SP1 interacts with some old-timer as well. One such example is ELF1, whose binding element ranks 38. ELF1 serves as a molecular cue to guide visual projection from the retina to the tectum both in vitro and in vivo (Nakamoto et al. 1996; Gunther et al. 2000). Among these cis-elements, SP1_03 is an MPA. We recall that the binding of SP1 on Alu has been verified experimentally (Oei et al. 2004).
Discussion
Rather than the protein-coding DNA sequences, we consider the evolution of hominidae from the perspective of the CREF profile in the proximal regulatory regions. The profile measures the gain and loss of cis-elements across genomes, and is analyzed by a systems biology approach (Wang et al. 2019). Mathematically the CREF profile of a species is stratified into dual modules at levels from high to low, leading to an understanding of the global organization of transcriptional regulation.
The dual eigen-analysis is different from the classical principal component analysis (PCA) or factor analysis. First, we choose SVD simply as a data-driven representation of cis-element frequencies without assumptions on their distributions or covariances. The dual eigen-modules that result from the representation are then aligned or compared across species, leading to a new discovery of evolution. Second, not only does the dual eigen-analysis look for linear combinations of cis-motifs and those of genes to explain the variations in the CREF matrix but also aims to unravel the correspondence between the motif- and gene eigenvectors by their coupling in SVD. Third, at each singular value level, the correspondence is established by sorting the motif- and gene eigenvector. The dual correspondence of the polarized motif- and gene eigenvectors is not known in PCA. We do notice that SVD has been adopted to summarize the tissue-specific expression profiles in the literature (Alter et al. 2000).
The comparative analysis of the dual CREF modules across humans, chimpanzees, and orangutans unravel a divergence between the fourth and fifth dual eigenvectors, specifically, an eigen-direction rotation. It is astonishing that the polarized gene eigenvector at the fourth level gathers almost all the physiological elements necessary for language development and for learning. These elements include ear development, vocalization, visual learning, long-term memory, brain development, and behavior. Thus, we demonstrate that the regulatory structure of the cognition-language system unique to humans can, by and large, be identified using only the cis-element frequencies, without the assistance of structures or sequences of proteins.
In this article, the dramatic influence of the motif frequency change, in consistency with dramatic change in traits, is described mathematically by the sensitivity of two adjacent eigenvectors as their eigenvalues approach a common value. This phenomenon is an analog to the phase transition in physics. For example, water becomes vapor upon heating to the boiling point. The boiling point in the CREF eigen-space situation is when the fourth and fifth singular values become identical. Behind the common level is a fused 2D eigen-space, in which any rotation is an eigen-direction. The degree of rotation is the biological polymorphism of regulation. Although whether a species at this fusion point really existed in the history is unknown, the three-stage scheme in figure 5 can guarantee the continuity of the CREF eigen-space evolution.
It is noticed that the definition of CREF modules hinges on the annotation of TSS. When multiple annotated transcripts are available, it is expected that the principal transcripts will be selected. The APPRIS system defines one kind of principal transcripts (Rodriguez et al. 2013). We calculated the CREF modules for humans and chimpanzees based on the principal transcript tagged as the main functional isoform by APPRIS. Module reorganization was observed too, and the rotation between the fourth and fifth level was ∼52° (supplementary fig. S9, Supplementary Material online). The enrichment analysis was, by and large, similar to that reported above (supplementary fig. S10, Supplementary Material online). However, the APPRIS annotation is not available for orangutans.
The CREF matrices rely on the criterion of motif search. The same criterion should be applied to all the species in the comparative genomic study so that any observed difference should be caused by their genomic difference. Two considerations are relevant in the criterion selection. First, the criterion is expected to have the power to tell apart the difference of genotype–phenotype association between species. Second, a fair fraction of the cis-element occurrences reported under a criterion are expected to be bound physically by the cognate transcription factors. We have tested the strict option “minimizing false positives” in TRANSFAC. It did not show any power of detecting CREF module differences among the three species. Besides, the strict criterion reported only half of the sites which were used to derive the motif position weight matrices (Zhang et al. 2006). In contrast, the more general criterion as adopted in this article did provide the discriminant power as shown in figures 2 and 3. Although this option is supposed to have more false positive cis–trans bindings, the precise fraction is hard to access at this point. It is noted that experimentally validated binding sites were obtained under certain cellular conditions, whereas in this article, we consider condition-free binding sites. In other words, we want to include binding sites under all possible conditions. Indeed, highly degenerate yet functional sites do exist as reported in the literature (Zhang et al. 2006). In addition, binding sites derived from transposable elements are far less conserved between species (Polavarapu et al. 2008). Definitely the false positive rate needs to be quantitatively evaluated in the future.
The reliability of our analysis was further strengthened by the following techniques. First, to rule out the effects of gene repertoire variations, we considered the matrices that consisted exclusively of motif frequencies in the proximal regulatory regions of the known orthologous genes. In spite of this, the reorganization between the human’s and apes’ CREF modules, as well as the conservation between the two apes’, was still observed (supplementary fig. S2, Supplementary Material online). Second, the random sampling was used to deal with the motif selection issue. Third, a robust version of SVD decomposition was used to address errors in the DNA sequences, and uncertainty in the motif PWMs. Finally, the robust Wilcoxon scoring method was adopted for the enrichment analysis.
To summarize, the dual eigen-analysis of the CREF profiles identified a gene eigenvector that encodes the phenotype of cognition, language development, and behavior unique to humans. The evidence from motif analysis and the cases of human-specific Alu insertions imply that mutations related to Alu elements play a critical role in the evolution of the human-specific gene phenotypic eigenvector.
Materials and Methods
Cis-Element Frequency Profile
The genomes and gene annotation were downloaded from the Ensembl database (release 84) (Zerbino et al. 2018) using R package BiomaRt (Durinck et al. 2009). The Ensembl human gene annotation merges the most recent manually curated annotations from Havana and the results from its automatic annotation pipeline (Aken et al. 2016). For nonhuman primate clade annotations, Ensembl uses a combination of protein-genome alignment, annotation projection from human GENCODE gene set, and RNA-seq alignment. We focused our analysis on protein-coding genes, including 19,763 human genes, 16,442 chimpanzee genes, and 15,994 orangutan genes. For a gene with multiple annotated transcripts, the start position of the most upstream transcript toward the 5′ end is chosen as the transcription start site (TSS). The proximal regulatory sequences between −1,000 and +500 bp with respect to the TSSs were extracted from the human genome (GRCh38.p5), the chimpanzee genome (CHIMP2.1.4), and the orangutan genome (PPYG2) through Ensembl REST API (Yates et al. 2015). About 1,403 vertebrate motifs together with their PWMs from the TRANSFAC database (Matys et al. 2006) were used in this study. Motif frequencies in the promoters were calculated using the MATCH program (Kel et al. 2003) with the minFN (minimize false negative) option.
Robust SVD
The first step of the dual eigen-analysis is the singular value decomposition of the CREF profile of each species. We denote one such matrix by , where ,is the total number of protein-coding genes, and is the total number of motifs. We note that motif frequencies can be influenced by the quality of the genome, the annotation, and motifs’ PWMs. To reduce the influence of the uncontrollable factors, we adopt a robust method consisting of two steps.
First, is represented as the sum of a low-rank matrix , which is expected to capture the principal genomic information of a cis-element regulatory profile, and a sparse matrix , which includes the unexpected and noisy variations. We reconstruct from by solving the following optimization problem:
where denotes the nuclear norm of a matrix, that is, the sum of its singular values, denotes the sum of the absolute values of the matrix entries, and λ is a positive penalizing parameter. To solve the above optimization problem, we choose the inexact augmented Lagrange multipliers (IALM) algorithm (Lin et al. 2010) with a fixed weighting parameter .
Second, we performed the conventional SVD on the low-rank matrix :
where are nonnegative, decreasing singular values, and their corresponding gene eigenvector and motif eigenvector are, respectively, of size and , are mutually orthogonal and so are .
Multiplicity of a Singular Value and Its Eigen-space
Consider the SVD of a CREF matrix , let the fourth and the fifth singular value approach each other. At the fusion point when they become identical, has a singular value with multiplicity two, that is, = . Their corresponding eigenvectors and span a 2D eigen-space. Any vector in the space can be written as , for some real number . It is easy to check that is still a motif eigenvector of :
where its coupling gene vector is . Accordingly, there are infinite number of eigenvector pairs in the 2D eigen-space.
Sensitivity of Eigenvalues and Eigenvectors
The theory of eigenvector sensitivity upon perturbation is helpful for understanding the rotation of the fourth and fifth eigen-directions. In fact, the singular values of are the square roots of the nonzero eigenvalues of :
The sensitivity of eigenvalues and eigenvectors can be rigorously analyzed under a variety of conditions (Stewart and Sun 1990). Here, we rather give one intuitive result. Assume that is a perturbation of and is symmetric too. Then the perturbed eigenvalues and eigenvectors have the following expansions,
where and denotes the 2-norm of a matrix, and the big notation indicates asymptotic upper bound as in mathematical analysis. According to the expansions, the interference of one eigenvector to another is roughly inversely proportional to the distance between the two eigenvalues. For example, when is approaching , the interaction between and becomes dominant, whereas those between others pairs are ignorable. After normalization of the vectors, the perturbation is approximately a rotation between and .
Supplementary Material
Acknowledgments
We owe many debts to two anonymous reviewers and an anonymous reader whose constructive suggestions have greatly enhanced this research and improved the organization of the material. We give our grateful thanks to Dr Weimou Zheng and Dr Aihui Zhou for discussions on stability of eigenvectors, and to Dr Lingan Wu and Ms Xinyi Cai for proofreading the article. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant Number XDB13040600), the National Natural Science Foundation of China (Grant Numbers 11871462 and 91530105), the National Center for Mathematics and Interdisciplinary Sciences of the Chinese Academy of Sciences, the Key Laboratory of Systems and Control of the Chinese Academy of Sciences, and the National Key Research and Development Program of China (Grant Number 2017YFC0908400).
Contributor Information
Liang Li, National Center of Mathematics and Interdisciplinary Sciences, Academy of Mathematics and Systems Science, Chinese Academy of Sciences, Beijing, China; University of Chinese Academy of Sciences.
Sheng Zhang, National Center of Mathematics and Interdisciplinary Sciences, Academy of Mathematics and Systems Science, Chinese Academy of Sciences, Beijing, China; University of Chinese Academy of Sciences.
Lei M Li, National Center of Mathematics and Interdisciplinary Sciences, Academy of Mathematics and Systems Science, Chinese Academy of Sciences, Beijing, China; University of Chinese Academy of Sciences; Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, Kunming, China.
References
- Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, Fernandez Banet J, Billis K, Garcia Giron C, Hourlier T, et al. 2016. The Ensembl gene annotation system. Database 2016:baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberini CM. 2009. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 89(1):121–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter O, Brown PO, Botstein D.. 2000. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci U S A. 97(18):10101–10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O.. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cand EJ, Li X, Ma Y, Wright J.. 2011. Robust principal component analysis? J ACM. 58:1–37. [Google Scholar]
- Carbon S, Chan J, Kishore R, Lee R, Muller H-M, Raciti D, Van Auken K, Sternberg P.. 2017. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45:D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Fabrizio P, Ge H, Wei M, Longo VD, Li LM.. 2007. Significant and systematic expression differentiation in long-lived yeast strains. PLoS One 2(10):e1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis HP, Squire LR.. 1984. Protein synthesis and memory: a review. Psychol Bull. 96(3):518–559. [PubMed] [Google Scholar]
- Dean C, Dresbach T.. 2006. Neuroligins and neurexins: linking cell adhesion, synapse formation and cognitive function. Trends Neurosci. 29(1):21–29. [DOI] [PubMed] [Google Scholar]
- Deininger P. 2011. Alu elements: know the SINEs. Genome Biol. 12(12):236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger SW, Fay JC.. 2007. Frequent gain and loss of functional transcription factor binding sites. PLoS Comput Biol. 3(5):e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, Huber W.. 2009. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 4(8):1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkouby YM, Elias S, Casey ES, Blythe SA, Tsabar N, Klein PS, Root H, Liu KJ, Frank D.. 2010. Mesodermal Wnt signaling organizes the neural plate via Meis3. Development 137(9):1531–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, et al. 2018. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 46(D1):D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Zhang S, Li L, Li LM.. 2019. The cis-trans binding strength defined by motif frequencies facilitates statistical inference of transcriptional regulation. BMC Bioinformatics 20(S7):201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. 1925. Statistical methods for research workers. Edinburgh (United Kingdom: ): Oliver and Boyd. [Google Scholar]
- Fisher RA. 1948. Questions and Answers. Am Stat. 2:30–31. [Google Scholar]
- Golub GH, Loan C.. 1996. Matrix computations. 3rd ed. Baltimore (MD): Johns Hopkins University Press. [Google Scholar]
- Gong R, Ding C, Hu J, Lu Y, Liu F, Mann E, Xu F, Cohen MB, Luo M.. 2011. Role for the membrane receptor guanylyl cyclase-C in attention deficiency and hyperactive behavior. Science 333(6049):1642–1646. [DOI] [PubMed] [Google Scholar]
- Gunther M, Laithier M, Brison O.. 2000. A set of proteins interacting with transcription factor Sp1 identified in a two-hybrid screening. Mol Cell Biochem. 210(1/2):131–142. [DOI] [PubMed] [Google Scholar]
- Horb ME, Thomsen GH.. 1999. Tbx5 is essential for heart development. Development 126(8):1739–1751. [DOI] [PubMed] [Google Scholar]
- Hormozdiari F, Konkel MK, Prado-Martinez J, Chiatante G, Herraez IH, Walker JA, Nelson B, Alkan C, Sudmant PH, Huddleston J, et al. 2013. Rates and patterns of great ape retrotransposition. Proc Natl Acad Sci U S A. 110(33):13457–13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtmeyers R, Souopgui J, Tejpar S, Arkell R.. 2013. The ZIC gene family encodes multi-functional proteins essential for patterning and morphogenesis. Cell Mol Life Sci. 70(20):3791–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi G, Bekier ME 2nd, Wang Y.. 2015. Golgi fragmentation in Alzheimer’s disease. Front Neurosci. 9:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczynski J, Zhang JS, Ellenrieder V, Conley A, Duenes T, Kester H, van Der Burg B, Urrutia R.. 2001. The Sp1-like protein BTEB3 inhibits transcription via the basic transcription element box by interacting with mSin3A and HDAC-1 co-repressors and competing with Sp1. J Biol Chem. 276(39):36749–36756. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K.. 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45(D1):D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S.. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M.. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44(D1):D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel MO, Wingender E.. 2003. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 31(13):3576–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M, Wilson A.. 1975. Evolution at two levels in humans and chimpanzees. Science 188(4184):107–116. [DOI] [PubMed] [Google Scholar]
- Lam EW, La Thangue NB.. 1994. DP and E2F proteins: coordinating transcription with cell cycle progression. Curr Opin Cell Biol. 6:859–866. [DOI] [PubMed] [Google Scholar]
- Lawrence CE, Altschul SF, Boguski MS, Liu JS, Neuwald AF, Wootton JC.. 1993. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science 262(5131):208–214. [DOI] [PubMed] [Google Scholar]
- Lawrence CE, Reilly AA.. 1990. An expectation maximization (EM) algorithm for the identification and characterization of common sites in unaligned biopolymer sequences. Proteins 7(1):41–51. [DOI] [PubMed] [Google Scholar]
- Lee DK, Kang JE, Park HJ, Kim MH, Yim TH, Kim JM, Heo MK, Kim KY, Kwon HJ, Hur MW.. 2005. FBI-1 enhances transcription of the nuclear factor-kappaB (NF-kappaB)-responsive E-selectin gene by nuclear localization of the p65 subunit of NF-kappaB. J Biol Chem. 280(30):27783–27791. [DOI] [PubMed] [Google Scholar]
- Lee DK, Suh D, Edenberg HJ, Hur MW.. 2002. POZ domain transcription factor, FBI-1, represses transcription of ADH5/FDH by interacting with the zinc finger and interfering with DNA binding activity of Sp1. J Biol Chem. 277(30):26761–26768. [DOI] [PubMed] [Google Scholar]
- Lin Z, Chen M, Ma Y.. 2010. The augmented Lagrange multiplier method for exact recovery of corrupted low-rank matrices. unpublished data. Available from: https://arxiv.org/abs/1009.5055v3. Accessed February 22, 2020.
- Liu A, Hoffman PW, Lu W, Bai G.. 2004. NF-kappaB site interacts with Sp factors and up-regulates the NR1 promoter during neuronal differentiation. J Biol Chem. 279(17):17449–17458. [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. 2006. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34(90001):D108–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema RH, Kops GJ, Bos JL, Burgering BM.. 2000. AFX-like forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404(6779):782–787. [DOI] [PubMed] [Google Scholar]
- Nakamoto M, Cheng H-J, Friedman GC, McLaughlin T, Hansen MJ, Yoon CH, O’Leary DDM, Flanagan JG.. 1996. Topographically specific effects of ELF-1 on retinal axon guidance in vitro and retinal axon mapping in vivo. Cell 86(5):755–766. [DOI] [PubMed] [Google Scholar]
- Oei SL, Babich VS, Kazakov VI, Usmanova NM, Kropotov AV, Tomilin NV.. 2004. Clusters of regulatory signals for RNA polymerase II transcription associated with Alu family repeats and CpG islands in human promoters. Genomics 83(5):873–882. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Sherman K, Bai G, Lipton SA.. 2002. Effect of the ubiquitous transcription factors, SP1 and MAZ, on NMDA receptor subunit type 1 (NR1) expression during neuronal differentiation. Brain Res Mol Brain Res. 107(2):89–96. [DOI] [PubMed] [Google Scholar]
- Paixao T, Azevedo RB.. 2010. Redundancy and the evolution of cis-regulatory element multiplicity. PLoS Comput Biol. 6:e1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Domany E.. 2006. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics 7(1):133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polavarapu N, Marino-Ramirez L, Landsman D, McDonald JF, Jordan IK.. 2008. Evolutionary rates and patterns for human transcription factor binding sites derived from repetitive DNA. BMC Genomics 9(1):226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD.. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16(2):245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JM, Maietta P, Ezkurdia I, Pietrelli A, Wesselink JJ, Lopez G, Valencia A, Tress ML.. 2013. APPRIS: annotation of principal and alternative splice isoforms. Nucleic Acids Res. 41(D1):D110–D117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Lee J, Zaman K, Kubilis J, Ferrante RJ, Ross BD, Neve R, Ratan RR.. 2003. Sp1 and Sp3 are oxidative stress-inducible, antideath transcription factors in cortical neurons. J Neurosci. 23(9):3597–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G, Sun J.. 1990. Matrix perturbation theory. Boston: Academic Press.
- Stone JR, Wray GA.. 2001. Rapid evolution of cis-regulatory sequences via local point mutations. Mol Biol Evol. 18(9):1764–1770. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM.. 2006. Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127(1):49–58. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S.. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131(5):861–872. [DOI] [PubMed] [Google Scholar]
- Tang W, Mun S, Joshi A, Han K, Liang P.. 2018. Mobile elements contribute to the uniqueness of human genome with 15,000 human-specific insertions and 14 Mbp sequence increase. DNA Res. 25(5):521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsirigos A, Rigoutsos I.. 2009. Alu and b1 repeats have been selectively retained in the upstream and intronic regions of genes of specific functional classes. PLoS Comput Biol. 5(12):e1000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Chen L, Wang W.. 2019. Genomic insights into ruminant evolution: from past to future prospects. Zool Res. 40(6):476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterson RH, Lander ES, Wilson RK, The Chimpanzee S, Analysis C.. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87. [DOI] [PubMed] [Google Scholar]
- Wingender E, Dietze P, Karas H, Knüppel R.. 1996. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 24(1):238–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 8(3):206–216. [DOI] [PubMed] [Google Scholar]
- Wu CL, Zukerberg LR, Ngwu C, Harlow E, Lees JA.. 1995. In vivo association of E2F and DP family proteins. Mol Cell Biol. 15(5):2536–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Sepulveda LA, Figard L, Sokac AM, Golding I.. 2015. Combining protein and mRNA quantification to decipher transcriptional regulation. Nat Methods. 12(8):739–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A, Beal K, Keenan S, McLaren W, Pignatelli M, Ritchie GR, Ruffier M, Taylor K, Vullo A, Flicek P.. 2015. The Ensembl REST API: Ensembl data for any language. Bioinformatics 31(1):143–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318(5858):1917–1920. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Giron CG, et al. 2018. Ensembl 2018. Nucleic Acids Res. 46(D1):D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Xuan Z, Otto S, Hover JR, McCorkle SR, Mandel G, Zhang MQ.. 2006. A clustering property of highly-degenerate transcription factor binding sites in the mammalian genome. Nucleic Acids Res. 34(8):2238–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Shields JM, Sogawa K, Fujii-Kuriyama Y, Yang VW.. 1998. The gut-enriched Kruppel-like factor suppresses the activity of the CYP1A1 promoter in an Sp1-dependent fashion. J Biol Chem. 273(28):17917–17925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.