Abstract
In contrast to CD4+ T cells, human immunodeficiency virus type 1 (HIV-1)-infected macrophages typically resist cell death, support viral replication, and consequently, may facilitate HIV-1 transmission. To elucidate how the virus commandeers the macrophage's intracellular machinery for its benefit, we analyzed HIV-1-infected human macrophages for virus-induced gene transcription by using multiple parameters, including cDNA expression arrays. HIV-1 infection induced the transcriptional regulation of genes associated with host defense, signal transduction, apoptosis, and the cell cycle, among which the cyclin-dependent kinase inhibitor 1A (CDKN1A/p21) gene was the most prominent. p21 mRNA and protein expression followed a bimodal pattern which was initially evident during the early stages of infection, and maximum levels occurred concomitant with active HIV-1 replication. Mechanistically, viral protein R (Vpr) independently regulates p21 expression, consistent with the reduced viral replication and lack of p21 upregulation by a Vpr-negative virus. Moreover, the treatment of macrophages with p21 antisense oligonucleotides or small interfering RNAs reduced HIV-1 infection. In addition, the synthetic triterpenoid and peroxisome proliferator-activated receptor γ ligand, 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO), which is known to influence p21 expression, suppressed viral replication. These data implicate p21 as a pivotal macrophage facilitator of the viral life cycle. Moreover, regulators of p21, such as CDDO, may provide an interventional approach to modulate HIV-1 replication.
T lymphocytes and macrophages expressing CD4 and theseven-transmembrane-domain chemokine coreceptorsCXCR4 and CCR5 are susceptible to human immunodeficiency virus type 1 (HIV-1) infection (7). Infection in macrophages is also facilitated by the phospholipid binding protein annexin II (36). In contrast to CD4+ lymphocytes, HIV-1-infected macrophages typically resist cell death, in spite of the hostile environment generated by the virus (52, 64). Viruses budding from macrophage intracellular membranes may escape immune surveillance, allowing the macrophage to serve as a reservoir and a source of virus for infections of additional cells (30, 52, 64). The persistence of HIV-1 during highly active antiviral therapy and the poor susceptibility of macrophages to antiviral therapy (19, 30) have intensified interest in characterizing the mechanisms underlying infection and replication in this cell population. In addition to being viral hosts, macrophages also contribute to HIV-1 pathogenesis as incubators for multiple opportunistic infections (41). Moreover, increased HIV-1 replication occurs in macrophages which are coinfected with Mycobacterium avium, exacerbating both bacterial and viral infections and underscoring the importance of this population as a therapeutic target (41, 62).
Although macrophages express the requisite CD4 and chemokine coreceptors, which make them susceptible targets, and although R5 viral variants are preferentially transmitted, it remains a challenge to identify HIV-1-positive macrophages early after viral exposure in mucosal tissues (49) or in the absence of copathogens (41, 63). When exposed to HIV-1, monocyte-derived macrophages bind and internalize the virus, but the consequences of that interaction are ill defined. Since macrophages are triggered by this encounter to modify their phenotypic and functional repertoire, it is important to define the early stages when HIV-1 is gaining a foothold on the immune system and to identify key signals which not only promote permissiveness for infection but also enhance viral replication. To characterize the temporal events associated with the initial virus-macrophage encounter leading to viral replication, we monitored virus production by using multiple parameters, including RNA, the p24 antigen (Ag), and the ultrastructural detection of viral particles. In parallel, macrophage changes in gene expression subsequent to virus-receptor interactions were compared to gene expression in uninfected cells by use of cDNA expression arrays. An analysis of ∼1,200 genes at multiple intervals, from initial HIV-1 binding through levels of massive replication (10 to 14 days), revealed a profile of gene modulation which favored the virus life cycle and could potentially influence the recruitment and infection of additional HIV-1 host cells. One gene that was differentially expressed following virus binding and again at the peak of HIV-1 replication was the p21 gene, also known as the Cip1 (Cdk interacting protein) or Waf1 (wild-type p53-activated fragment) gene, which is associated with cell cycle regulation, antiapoptotic responses, and differentiation (16). The particular gene expression pattern for p21 led us to examine whether modulation of this transcript affects the HIV-1 viral life cycle. Our data demonstrate that the modulation of p21 in vitro results in a reduction in viral replication, implicating this cellular protein as an interventional target.
MATERIALS AND METHODS
HIV-1 infection and treatment of monocytes.
Human peripheral blood mononuclear cells obtained by leukapheresis from healthy volunteers were enriched for monocytes by elutriation (62), plated in T-75 flasks at 7.5 × 106 cells/ml, in six-well plates (Corning Costar Corporation) at 6 × 106 cells/well, or in glass chamber slides (Lab-Tek) at 1.5 × 106 cells/chamber in Dulbecco's modified Eagle's medium with 2 mM l-glutamine and 10 μg of gentamicin (BioWhittaker)/ml, allowed to adhere (4 to 6 h at 37°C and 5% CO2), supplemented with 10% fetal bovine serum (Invitrogen), and then differentiated into macrophages for 7 days. The macrophages were infected with pelleted R5 HIV-1BaL purified virions (50% tissue culture infective dose = 500 to 5,000) (Advanced BioTechnologies Inc.), the laboratory-adapted isolate ADA, or the primary viral isolate clade B 92US727 (NIH AIDS Research and Reference Reagent Program) as previously described (36, 62). For experiments with HIV Vpr mutants, pNLAD8 (NL4-3 with the CXCR4-tropic Env protein replaced with AD8.1 CCR5-tropic Env) and pNLAD8 delta-R (EcoRI fill-in plasmid that expresses the first 37 amino acids of Vpr) were obtained from Eric Freed (National Cancer Institute-Frederick, Frederick, Md.) (18). pNLAD8 Vpr− was constructed by introducing an A-to-T mutation at nucleotide 5559 (1), changing the methionine codon to a leucine, and mutating nucleotide 5557 from A to T to maintain an arginine codon in the Vif reading frame. Viral supernatants were produced by transfection of 293T human embryonic kidney cells by use of the Transit 293 transfection reagent (Mirus, Madison, Wis.). Viruses were titrated in a single-round lacZ Tat complementation assay using JC53BL cells (68). Briefly, a six-well tissue culture cluster was seeded with 5 × 105 JC53BL cells the day before infection. The cells were infected with virus dilutions, and the assay was developed for β-galactosidase activity with a 5-bromo-4-chloro-3-indolyl-β-galactosidase stain at 48 h postinfection. Positive, blue-staining cells were counted to score the number of infection events. Virions were isolated by centrifugation through a 20% sucrose pad in an SW41.1Ti rotor at 37,000 rpm at 4°C for 1 h. Immunoblotting for p24 was performed by use of a Bio-Rad Immuno-Star HRP substrate kit (Hercules, Calif.). Blots were developed by exposure to Lumifilm (Roche Applied Science, Indianapolis, Ind.). An antiserum against p24 was produced by the AIDS Vaccine Program, NCI-Frederick. In order to overcome the inherent block of viral infection by Vpr-negative virus in nondividing cells, we infected macrophages with the mutant virus at multiplicities of infection (MOIs) of >6 blue CFU per cell, while the wild-type NLAD8 virus was used at an MOI of 3.
Every 3 to 4 days, half of the culture medium was removed, analyzed for viral replication by a p24-specific enzyme-linked immunosorbent assay (ELISA; Perkin-Elmer Life Sciences), and replaced with fresh medium for up to 2 weeks. Control macrophages from each donor were mock infected, cultured, and refed in parallel. Infections were monitored by p24 ELISA, RNA (62), nested PCR, and transmission electron microscopy (TEM) (21). Adherent macrophages were also incubated with full-length synthetic viral protein R (Vpr) (26) at the indicated concentrations for 3 h, the total mRNA was isolated by use of an RNeasy mini kit (QIAGEN), and cell protein lysates were generated by use of a lysis buffer containing 20 mM Tris-HCl (pH 7.5), 1% Nonidet P-40, 150 mM NaCl, 10 mM NaF, 10 mM NaPPi, 2.5 mM EDTA, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 1× complete protease inhibitor cocktail (Boehringer Mannheim), 0.2 mM 3,4-dichloroisocoumarin, and 100 μg of chymostatin/ml. Elutriated T lymphocytes were blasted with phytohemagglutinin (10 μg/ml) (Sigma), infected with HIV-1 IIIB (50% tissue culture infective dose = 104) (Advanced Biotechnologies Inc.) for 6 h, washed, and cultured for 7 days, with supernatants being collected every 2 to 3 days for p24 ELISA.
Molecular analysis of p21 transcription.
The total cellular RNA was extracted at intervals from adherent control or infected macrophages from 6 h to 14 days by use of an RNeasy mini kit (QIAGEN) and then analyzed by Northern blotting (62) with a full-length HIV-1 probe (NIH AIDS Research and Reference Reagent Program) and with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (GIBCO BRL) to monitor RNA loading. For RNase protection assays (RPAs), 3 μg of RNA was evaluated by use of the hStress template Riboquant Multi-Probe RPA system (BD Pharmingen) (21). The gels were exposed to phosphor screens and analyzed with a phosphorimager. Band densities were normalized to that of the GAPDH gene by the use of ImageQuant (Molecular Dynamics) (21). For reverse transcription-PCR (RT-PCR), 1 μg of total RNA was reverse transcribed by use of an oligodeoxythymidylic acid primer (Invitrogen), and the resulting cDNA (0.5 to 1 μl) was amplified by PCR. The primer set for p21 was 5′-GACAGCAGAGGAAGACCAT-3′ (forward) and 5′-TGGAGTGGTAGAAATCTGTCAT-3′ (reverse). For GAPDH, the primer set was 5′-GAAGGTGAAGGTCGGAGTC-3′ and 5′-GAAGATGGTGATGGGATTTC-3′. PCRs were performed with 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphates, a 0.6 μM concentration of each primer, and 1 U of Taq polymerase (Sigma). cDNAs were amplified for 25 cycles with the following settings: 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s.
Nested PCR with viral DNA.
For analyses of newly synthesized viral DNAs (37), DNase-treated HIV-1BaL (200 μl; 104/ml) was added for 2 h to macrophages (6 × 106 cells/well) that had been pretreated with a p21 small interfering RNA (siRNA) or a control siRNA. The cultures were washed three times with phosphate-buffered saline (PBS), treated with trypsin-EDTA (0.05% trypsin, 0.53 mmol of EDTA/liter) for 5 min to remove noninternalized virus particles, washed, and incubated for 18 to 48 h. DNAs were extracted for nested PCR as described previously (37). PCR products from the second amplification were visualized by ethidium bromide staining after agarose gel electrophoresis.
cDNA expression array.
The total cellular RNAs were extracted from uninfected control and virus-infected macrophages by use of an RNeasy mini kit (QIAGEN). Hybridization to an Atlas human cDNA expression array (1.2 I; Clontech) was performed with 5 μg of DNase-digested total RNA as previously described (21). After normalization to housekeeping genes by the use of AtlasImage 1.01a (Clontech), gene expression in infected cells was compared with that in uninfected cells from the same donor at the same time interval and expressed as a ratio (fold change). Genes that were differentially up-regulated in four, five, or six of six donors at 6 h (day 0.25) with an average ≥2-fold increase above parallel uninfected control donors are reported here. For 14-day kinetic studies, RNAs were obtained from three donors and assessed for gene transcription. For some donors, variability in gene transcription was noted, based in part on background levels of activation, gene expression, and/or response to the virus. Nonetheless, the interarray variability was assessed by hybridizing the same sample to two different array membranes, which yielded a correlation coefficient (R2) of 0.95 (21). As indicated, the expression of selected genes was confirmed by multiple parameters, including RPA, PCR, immunofluorescence, and/or protein analysis (21). For the day 0 cDNA array (6 h), statistical significance between uninfected and infected cells was calculated by the nonparametric Wilcoxon signed rank procedure. The paired t test was rejected due to the small sample size and likely nonnormal distribution of responses. Genes with P values of ≤0.05 were considered significant. Additional statistical analyses of kinetic array data were performed by analysis of variance with repeated measures by the use of Partek Pro software, with P values of ≤0.05 considered significant.
Immunofluorescence microscopy.
Infected and control macrophages were cultured for 10 to 12 days, washed with PBS, fixed with 2% paraformaldehyde, washed, and incubated with 100 mM glycine for 20 min followed by 0.5% Triton X-100 for 10 min. The cells were incubated with 5% blocking serum for 30 min before the addition of a rabbit anti-p21 antibody (Santa Cruz) for 1 h, washed, and incubated with an Alexa fluor 594-conjugated secondary antibody (Molecular Probes) at 25°C. The nonspecific background was determined by use of an isotype control antibody and the secondary antibody alone. Images were captured with a Leica TCS-4D confocal microscope system with a Kr-Ar laser and a 40×, 1.0-numerical-aperture objective. Fluorescence was quantified with Metamorph analysis software (Universal Imaging).
Flow cytometry.
Adherent macrophages were detached by the use of cell dissociation buffer (Invitrogen), washed, and resuspended in PBS containing 2% fetal bovine serum and 0.01% sodium azide. The cells were stained with mouse anti-human phycoerythrin-CD4 and CCR5-fluorescein isothiocyanate (CCR5-FITC) or corresponding isotype controls (BD Pharmingen) for flow cytometry analysis.
Immunoprecipitation and Western blotting.
Cell lysates were generated, and p21 was immunoprecipitated from equal amounts of protein lysates with an anti-p21 conjugated agarose antibody (Santa Cruz) and incubated with constant rotation at 4°C for 2 h. Immunoprecipitates were washed, resuspended in sodium dodecyl sulfate sample buffer (New England Biolabs), electrophoresed in Tricine gels (Invitrogen), transferred to nitrocellulose membranes, and immunoblotted with anti-p21 (BD Pharmingen). Immunoblots were developed by enhanced chemiluminescence with the Super-Signal substrate (Pierce).
Suppression of p21 expression.
Cells were treated with p21 antisense phosphorothioate oligonucleotides conjugated to FITC and Penetratin (Q-Biogene). The sequences for the two p21 oligonucleotides and a negative control oligonucleotide were 5′-TGTCAGGCTGGTCTGCCTCC-3′ (oligo 1), 5′-ACATCACCAGGATTGGACAT-3′ (oligo 2), and 5′-TGGATCCGACATGTCAGA-3′ (oligo 3) (33). Oligonucleotides were added at 50 nM 60 min prior to HIV-1 infection and replenished at the time of medium replacement. Gene silencing was performed with SMARTpool siRNA duplexes (Dharmacon) specific for p21 (1 to 10 nM). A nonspecific siRNA control (Dharmacon) was utilized in parallel, and transfection was accomplished by the use of Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. In some experiments, cells were pretreated with 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO) or a CDDO analog (di-CDDO) (55) at 0.01 to 1.0 μM for 45 min prior to or concomitant with exposure to HIV-1. Cell viability was examined by use of a fluorescein-FragEL DNA fragmentation detection kit (Oncogene Research Products).
RESULTS
Kinetics of HIV-1 replication.
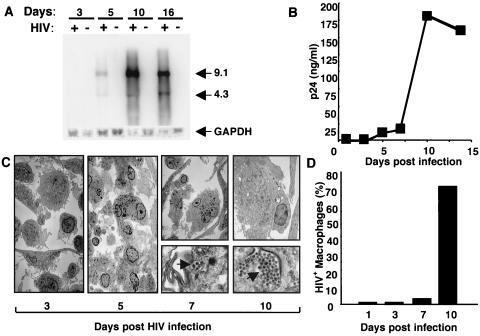
Elutriated monocytes were adhered for 7 days, exposed to R5 HIV-1BaL for 2 h, and washed, and the kinetics of cellular and viral changes were monitored. By Northern analysis, HIV-1 RNA was detected within 5 to 7 days after infection and reached a maximal level by 10 to 16 days (Fig. 1A). In parallel, the p24 Ag appeared within 5 days, then increased dramatically, and finally plateaued after 10 days (Fig. 1B). Consistent with the presence of viral RNA and p24, virus was detected by TEM around day 7, with ≥70% of the cells typically harboring large numbers of virions by day 10 (Fig. 1C and D). Virions were particularly evident within intracellular vacuoles as well as along convoluted macrophage membranes (Fig. 1C). Nonetheless, once the majority of cells were infected with large numbers of virions within and on the cell surface, p24 levels plateaued, independent of the concentration of the viral inoculum (not shown) and likely influenced by host factors.
FIG. 1.
Kinetics of HIV-1 infection in monocyte-derived adherent macrophages. (A) Cells were exposed to HIV-1 for the indicated intervals, and mRNAs were extracted and examined by Northern blotting. Bands of 9.1 and 4.3 kb correspond to viral gag/pol and env mRNAs, respectively. (B) Supernatants were collected from infected cultures (days 1 to 15) for p24 ELISA. (C) Cells were incubated for 3 to 10 days after infection and processed for TEM. Original magnification, ×10,000. Ultrastructure analysis revealed detectable virions (C and D) in macrophages by 5 to 7 days postinfection, with increasing virus numbers per cell (C) and numbers of infected macrophages (D) being most evident at or after day 10, as quantified by counting ≥200 cells/time point. The data shown correspond to a representative experiment (n ≥ 4).
Initial gene expression in infected macrophages.
To examine potential host factors underlying viral propagation, we examined transcriptional pathways activated downstream of CD4-HIV-1 coreceptor binding and signaling. Compared with the case in uninfected macrophages, an early and transient gene expression profile occurred, followed by a period of relatively quiescent gene expression and a subsequent delayed pattern that emerged in association with viral replication. Although substantial heterogeneity in the macrophage response to HIV-1 was observed (data not shown), which may reflect different levels of constitutive activation and differentiation of the uninfected macrophages and/or susceptibility to viral infection for each individual donor, the data shown represent genes that were differentially upregulated ≥2 fold in the majority of donors. Within 6 h, many upregulated genes (nearly 130 of ∼1,200 genes analyzed) were associated with signal transduction (24%) and transcription (26%) (Table 1). Components of the G protein receptor pathway which participate in signaling, such as GNAS, GNB1, GRB2, Rac1, and RhoA, were augmented subsequent to the interaction of HIV-1 with CD4 and the G-protein-coupled receptor CCR5. Genes corresponding to the mitogen-activated protein kinase (MAPK) family were also increased, including p38 MAPK, MAPKAP-K1, and MAPKAP-K2. Another signal transduction gene that was significantly upregulated was the gene for LIMK-1, a serine/threonine kinase that has been shown to participate in the regulation of actin cytoskeletal reorganization downstream of Rho family GTPases (4, 56, 71). Among the transcription factors influenced by the virus-macrophage encounter was the host Tat binding protein (TBP-1), known to interact with viral Tat (40); the cellular coactivator PC4, which has been identified as an HIV Tat-interacting protein (27); and RPL6, which binds the Tax-responsive element of human T-cell leukemia virus type 1 (39). In addition to signal transduction molecules, genes associated with the cell cycle, such as the cyclin-dependent kinase inhibitor 1A (CDKN1A/p21) gene, were significantly enhanced within hours, in parallel with the proliferating cell nuclear antigen (PCNA) (Table 1), which interacts with p21 (16). Although gene expression for caspases 3 and 4 was increased, genes encoding factors that contribute to cellular resistance to apoptosis, including IEX-1L and bcl-x (2, 70), were also elevated. The enhanced transcription of genes involved in cellular recruitment, including genes for chemokines (interleukin-8 and MCP-1) and MRP14 as well as surface adhesion molecules, may favor host cell accumulation and syncytium formation (23, 59). The metabolic pathway genes for dioxin-inducible cytochrome P450 (Table 1), which is associated with enhanced HIV-1 gene expression and the progression of AIDS (72), and heme oxygenase-1 (HO-1) (Table 1), a protein which is increased in the peripheral blood mononuclear cells of AIDS patients (34), were typically upregulated. In addition to HSP90 and HSP27, host molecules that have been implicated in the HIV-1 viral cycle (60, 65), HIF-1α, a transcription factor that participates in the regulation of genes involved in angiogenesis, glucose metabolism, cell survival, and cancer (44), was also upregulated. During this immediate early response, HIV-1 enhanced more genes than it suppressed in the subset of genes examined. Only tripeptidyl peptidase I, a lysosomal serine protease responsible for cleaving tripeptides from the N termini of oligopeptides (58), was reproducibly suppressed (data not shown) and may influence protein turnover.
TABLE 1.
Early HIV-1-induced gene expression in macrophagesa
| GenBank accession no. and functional group | Gene product | Average fold increase |
|---|---|---|
| Signal transduction | ||
| M36430 | GNB1 | 5.0 |
| X15014 | RalA | 3.3 |
| D26309 | LIMK-1 | 3.1* |
| M14631 | GNAS | 3.0 |
| L35253 | MAP kinase p38 | 3.0 |
| L29511 | GRB2 | 2.7 |
| AF068920 | SHOC 2 | 2.5 |
| L25080 | Ras homolog A (RhoA) | 2.5 |
| AF055581 | LNK adaptor | 2.4 |
| X17576 | NCK melanoma cytoplasmic Src homolog | 2.3 |
| M98343 | Cortactin (ems-1) | 2.3 |
| M65066 | PRKAR1B | 2.3 |
| U10550 | Gem (Ras family) | 2.2 |
| U78576 | PI4P5 kinase alpha | 2.2 |
| U12779 | MAPKAP kinase 2 | 2.2* |
| M19922 | INT2 | 2.1 |
| X15219 | SnoN | 2.1 |
| M29870 | Rac1 | 2.1 |
| M28213 | Rab2 | 2.1 |
| U24166 | EB1 | 2.1 |
| L20321 | Serine/threonine kinase NRK2 | 2.1 |
| M34181 | PKC beta | 2.1 |
| X60957 | Tyrosine kinase receptor Tie-1 | 2.0 |
| L05624 | MAPKK1 | 2.0 |
| X03484 | Raf1 protooncogene | 2.0 |
| L22075 | G13 | 2.0 |
| X94991 | Zyxin 2 | 2.0 |
| M63960 | PPIalpha | 2.0 |
| X06318 | PKC beta 1 | 2.0 |
| M77234 | Fte-1 | 2.0 |
| X08004 | Rap1b | 2.0 |
| Transcription | ||
| U10323 | NF45 | 3.6 |
| L19871 | Activating factor 3 (ATF3) | 3.1 |
| U12979 | Activated RNA polymerase II transcriptional coactivator p15 (PC4) | 2.9* |
| M81601 | Transcription elongation factor SII | 2.8 |
| M29038 | Stem cell protein | 2.8 |
| D90209 | Activating factor 4 (ATF4) | 2.8 |
| M34079 | TAT binding protein (TBP-1) | 2.7 |
| L34587 | RNA polymerase II elongation factor SIII p15 subunit | 2.6 |
| L04282 | CACCC-box DNA binding protein | 2.6 |
| U22431 | Hypoxia-inducible factor 1 alpha | 2.5* |
| L23959 | E2F dimerization partner 1 (DP1) | 2.5 |
| M83234 | NSEP | 2.5 |
| S40706 | GADD153 | 2.4 |
| M96824 | Nucleobindin precursor (NUC) | 2.4 |
| M36717 | Ribonuclease/angiogenin inhibitor (RAI) | 2.4 |
| D26156 | SW1/SNF-related actin-dependent regulator of chromatin | 2.3 |
| X69391 | 60S ribosomal protein (RPL6) | 2.3* |
| X59738 | Zinc finger X-chromosomal protein | 2.3 |
| M59079 | CBF-B | 2.2 |
| M96944 | PAX5 | 2.2 |
| AF084199 | PRD1-BF1 (transcription repressor protein) | 2.2 |
| M97796 | Inhibitor of DNA binding 2 (ID2) | 2.2 |
| U07418 | MutL protein homolog 1 (MLH1) | 2.2 |
| AF060222 | DNase II | 2.2 |
| U58198 | Interleukin enhancer binding factor (ILF) | 2.1 |
| Z36715 | Elk-3 | 2.1 |
| AF032119 | CASK | 2.1 |
| M97935 | STAT1 alpha/beta | 2.1 |
| Z30094 | Basic transcription factor 2 (BTF2p44) | 2.1 |
| J04111 | Jun protooncogene, AP-1 | 2.1 |
| D26155 | Transcriptional activator (hsnF2a) | 2.0 |
| M80397 | DNA polymerase delta catalytic subunit | 2.0 |
| AF076974 | Transformation/transcription domain-associated protein | 2.0 |
| Cell cycle and apoptosis | ||
| U13737 | Caspase 3 | 2.2 |
| L29222 | CDC-like kinase (CLK1) | 2.1 |
| AF071596 | IEX-1L anti-death protein | 2.1 |
| M15796 | Proliferating cyclic nuclear antigen (PCNA) | 2.1 |
| X96586 | FAN protein | 2.1 |
| U28014 | Caspase-4 | 2.1* |
| Z23115 | Bcl-x | 2.0 |
| U09579 | Cyclin-dependent kinase inhibitor 1A (CDKN1A) | 2.0* |
| Adhesion molecules and receptors | ||
| M14648 | Vitronectin receptor alpha (VNRA) | 3.3 |
| J03132 | Intercellular adhesion molecule 1 (ICAM1) | 3.1 |
| M81695 | CD11c antigen | 2.5 |
| X06256 | Fibronectin receptor alpha (FNRA) | 2.5 |
| D84657 | Photolyase/blue-light receptor homolog | 2.4 |
| X07979 | Fibronectin receptor beta (FNRB) | 2.7 |
| D13866 | Alpha 1 catenin | 2.4 |
| X72304 | Corticotropin releasing factor receptor 1 | 2.4 |
| M59911 | Integrin alpha 3 (ITGA3) | 2.3 |
| M37722 | Fibroblast growth factor receptor 1 | 2.2 |
| L25851 | Integrin alpha E (ITGAE) | 2.1 |
| M27492 | IL-1 receptor type I | 2.0 |
| J04536 | Leukosialin | 2.0 |
| X01057 | IL-2R alpha | 2.0 |
| M59040 | CD44 antigen | 2.0 |
| Chemokines and cytokines | ||
| Y00787 | IL-8 | 9.7 |
| M65291 | IL-12 alpha | 5.0 |
| M24545 | Monocyte chemotactic protein 1 (MCP-1) | 4.5 |
| X06233 | Migration inhibitory factor-related protein 14 (MRP14) | 3.8 |
| M92381 | Thymosin beta 10 | 3.7 |
| M17733 | Thymosin beta 4 | 3.6 |
| X01394 | Tumor necrosis factor alpha | 3.4 |
| X53655 | Neurotrophin-3 precursor | 2.6 |
| M21121 | Small inducible protein A5 (SCYA5) | 2.5 |
| M86492 | Glia maturation factor beta | 2.2 |
| M31145 | Insulin-like growth factor binding protein 1 | 2.2 |
| M27288 | Oncostatin M (OSM) | 2.1 |
| U13699 | IL-1 beta converting enzyme (ICE) | 2.1 |
| U16296 | T-lymphoma invasion and metastasis inducing (TIAM1) | 2.0 |
| M25667 | Neuromodulin | 2.0 |
| X02530 | Interferon gamma-induced protein (IP-10) | 2.0 |
| Proteases and protease inhibitors | ||
| M11233 | Cathepsin D | 3.2 |
| J05070 | Matrix metalloproteinase 9 (MMP9) | 3.1 |
| X56692 | C-reactive protein | 2.9 |
| AF059244 | Cystatin-related protein | 2.8 |
| X05562 | Procollagen alpha 2 | 2.5 |
| L23808 | Matrix metalloproteinase 12 | 2.2 |
| D00762 | Proteasome C8 | 2.1 |
| Z81326 | Protease inhibitor 12 | 2.0 |
| L40377 | Cytoplasmic antiprotease 2 (CAP2) | 2.0 |
| M23254 | Calpain 2 | 2.0 |
| X04106 | Calpain | 2.0 |
| Metabolism | ||
| U03688 | Dioxin-inducible cytochrome P450 (CYP1A1) | 4.5 |
| X06985 | Heme oxygenase 1 (HO-1) | 4.2 |
| U34683 | Glutathione synthetase | 3.0 |
| X07270 | 90-kDa heat shock protein A | 2.7* |
| U29091 | Selenium binding protein | 2.7 |
| L14595 | Neural amino acid transporter A (SATT) | 2.6 |
| D00099 | Na+/K+-transporting ATPase alpha 1 | 2.4 |
| M74524 | Ubiquitin-conjugating enzyme (UBE2A) | 2.3 |
| X91247 | Thioredoxin reductase | 2.3 |
| X54079 | 27-kDa heat shock protein | 2.2* |
| M11717 | 70-kDa heat shock protein 1 | 2.1 |
| L20046 | Xeroderma pigmentosum group G complementing protein | 2.0 |
| Y00264 | Alzheimer's disease amyloid A4 protein | 2.0* |
Total mRNA was extracted from uninfected and HIV-1-infected macrophages and analyzed by cDNA expression array. Values were normalized to those for housekeeping genes, and the data are presented as n-fold increases (≥2-fold) of infected cells compared to those in mock-infected control cultures. *, P ≤ 0.05.
Kinetics of HIV-1-induced gene expression.
The initial pattern of gene expression following binding of HIV-1 was not sustained, and interestingly, there was a reduced transcriptional response evident 3 to 5 days after infection, preceding the evidence of viral replication (Fig. 2). However, concomitant with evidence of the HIV-1 replicative cycle at 5 to 7 days postinfection (Fig. 1), a resurgence of gene expression began to manifest (Fig. 2). In addition to the reexpression of genes that were turned on by the initial HIV-macrophage interaction (i.e., MAPK, adhesion molecules, and p21), additional genes which were not differentially expressed during initial viral binding were upregulated at the peak of viral replication, emerging as potential regulatory host cell molecules for the viral life cycle. For example, increased transcription for the high-mobility-group protein I (HMG-I), one of a class of nonhistone DNA-binding proteins that modulate chromatin structure (8), and MutL protein homolog 1 (MLH1), a component of the DNA mismatch repair pathway (38), were evident during the progression of infection (Fig. 2). Furthermore, altered transcriptional profiles of apoptosis inhibitors and cell cycle regulators in infected cells implicated their involvement in viral permissiveness.
FIG. 2.
HIV-1-induced alterations in macrophage transcriptome. The figure shows changes in gene expression in HIV-1-treated macrophages compared to the gene expression levels in mock-infected macrophages from the same donor at intervals, from 0.25 to 14 days (mean values; n = 3 to 6). Increased or decreased gene transcription is represented in red and green, respectively. Genes shown in black indicate no change in transcriptional activity. *, P ≤ 0.05.
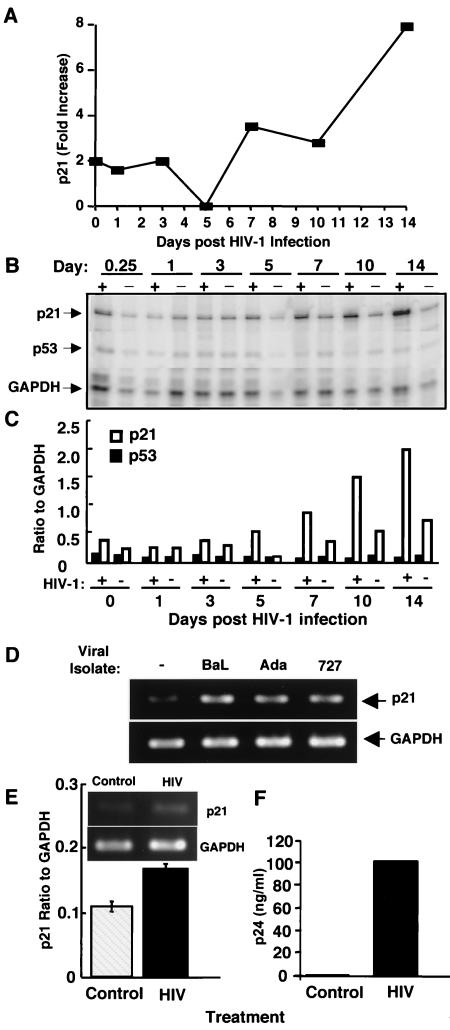
Increased p21 expression in infected macrophages.
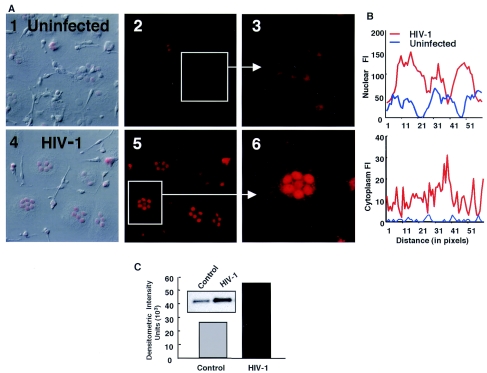
One of the intriguing genes that was significantly upregulated as an immediate early gene and then reexpressed at maximum levels during HIV-1 replication was the gene for p21, a cell cycle regulator (Table 1; Fig. 2 and 3A). Of the 1,200 genes studied, p21 was also the most upregulated transcript (up to eightfold) at the peak of viral replication. To further confirm the expression of this gene as a potential host cell regulator of viral production, we performed RPAs and confirmed the rapid early induction of p21 (Fig. 3B and C), which was followed by striking expression concomitant with viral replication, but without corresponding changes in another cell cycle-related gene, p53. To determine whether other HIV-1 viral isolates modulate p21, we infected cells with two additional isolates and found that p21 gene induction was not only evident after infection with another laboratory-adapted viral isolate, ADA, but importantly, was evident after infection with the primary clinical isolate 727 (Fig. 3D). Furthermore, enhanced p21 transcription correlated with increased protein levels in infected macrophages. Immunofluorescence assessment of the p21 protein revealed increased nuclear and cytoplasmic p21 staining in infected macrophages compared with that in mock-infected cells (Fig. 4A and B), consistent with enhanced protein expression detected in cell lysates by Western blotting (Fig. 4C). In contrast, infection of T lymphocytes with HIV-1 resulted in a modest increase in p21 transcription (Fig. 3E), despite elevated levels of p24 Ag (Fig. 3F).
FIG. 3.
Increased p21 gene expression in infected macrophages. (A) Kinetic profile of p21 expression from days 0.25 to 14 after infection (n = 3). (B) RPA analysis of uninfected (−) and HIV-1 infected (+) macrophages confirmed the enhanced gene expression for p21, with a minimal effect on p53 (data shown are for a representative donor; n = 2). (C) Densitometric analysis of RPA results for the p21 and p53 genes (shown in panel B), normalized to GAPDH. (D) Macrophages were infected with HIV-1BAL, the laboratory viral isolate ADA, or the primary isolate 727 and then washed, and the total RNA was collected after 12 days for analysis of p21 transcription by PCR. (E and F) Phytohemagglutinin-blasted T lymphocytes were infected with HIV-1 (IIIB), and day 6 supernatants were examined for p24 Ag. Total mRNA (6 h) was analyzed for p21 transcription by PCR.
FIG. 4.
Infected macrophages express more p21 protein. (A) Overlay confocal images from differential interference contrast (1 and 4) and immunofluorescence labeling for p21 in uninfected (1, 2, and 3) and virus-infected (4, 5, and 6) cells (original magnification, ×400). (B) Fluorescence intensity (FI) analysis (Metamorph) confirmed the enhanced nuclear and cytoplasmic p21 protein, as represented by the signal across equal line segments of nuclear or cytoplasmic areas (data shown are from a representative experiment; n = 3). (C) Increased p21 protein in infected macrophages (12 days) by immunoprecipitation, as quantified by densitometric analysis, relative to that in uninfected cells (n = 3).
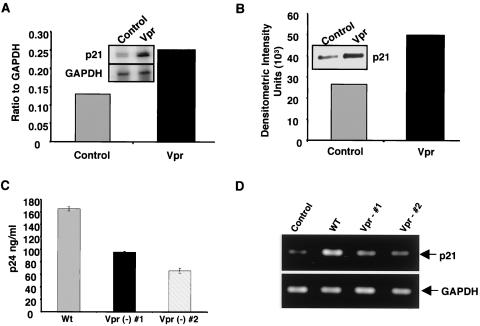
Since Vpr facilitates viral replication in nondividing cells (26, 54) and is required for efficient HIV-1 production during the late stages of replication in tissue macrophages (46), we assessed the potential contribution of Vpr to the mediation of p21 expression. Macrophages treated with Vpr for 3 h, but not with HIV-1 gp120, not only exhibited enhanced p21 transcription (Fig. 5A) but also had a corresponding increase in p21 protein (Fig. 5B). To further link Vpr to the modulation of p21 expression, we infected macrophages with Vpr mutant viruses at a high MOI (>6). Our studies with two different Vpr-negative viruses showed reduced viral replication (Fig. 5C), with little or no enhancement of p21 transcription (Fig. 5D), compared to the wild-type Vpr+ virus. Collectively, these studies implicate Vpr as one potential mechanism utilized by HIV-1 to drive p21 transcription.
FIG. 5.
Induction of p21 gene and protein expression by Vpr. Cells treated with Vpr (6 μg/ml) for 3 h showed increased gene transcription (A) and protein expression (B) for p21. (C) Macrophages were treated with control supernatants from uninfected or mock-transfected cells or from 293T cells infected with the wild-type virus clone pNLAD8 or the pNLAD8 Vpr-negative (#1) or pNLAD8-delta R (#2) R5 macrophage-tropic virus, and 12-day supernatants were analyzed by p24 ELISA. (D) The total RNA was analyzed for p21 and GAPDH by PCR. The data shown are from a representative experiment (n = 2).
Effect of p21 inhibitors on HIV-1 replication.
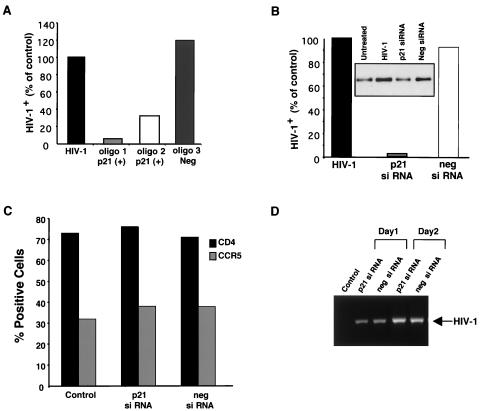
To determine whether increased p21 contributed to driving the viral life cycle, we treated macrophages with two separate p21 antisense oligonucleotides to suppress p21 expression in cells exposed to HIV-1. Both p21-specific oligonucleotides reduced viral replication, as assessed by p24 levels, whereas negative control oligonucleotides had no effect on p24 (Fig. 6A). In additional experiments, the suppression of p21 by a p21-specific siRNA, but not a control nonspecific siRNA, also inhibited HIV-1 replication (Fig. 6B), confirming the essential role of HIV-induced p21 expression in the viral life cycle. We have established that the effect of siRNA treatment resulting in a blockade of p21 and consequent reduced viral replication did not affect cell viability or CCR5 or CD4 cell surface expression (Fig. 6C). Consistent with the absence of alterations in cell surface recognition and binding receptors on the macrophages, we determined that the inhibition of p21 did not influence HIV-1 internalization or early RT (Fig. 6D), but rather acted at a later stage in the viral life cycle.
FIG. 6.
Inhibition of p21 reduces HIV-1 replication. (A) p21-specific oligonucleotides (1 and 2, 50 nM), but not a control oligonucleotide (3), inhibit HIV-1 growth in replicate cultures, as determined by p24 levels (data for day 12 are shown and are percentages of the positive HIV control with no oligonucleotide). (B) Macrophages treated with p21 siRNA duplexes (5 nM) 5 days prior to HIV infection (% of positive HIV control with no siRNA treatment) (data shown are from a representative experiment; n = 3). Percentages of HIV-1 infection were determined by comparing the p24 levels in untreated versus oligonucleotide- or siRNA-treated macrophages. The inset is a Western blot for p21 from day 14. (C) Cells treated with p21 and negative control siRNAs (5 days) were analyzed by flow cytometry for CD4 and CCR5 cell surface expression. (D) Nested PCR to detect proviral DNA on days 1 and 2 after HIV-1BaL infection of macrophages treated with p21 siRNA or negative control siRNA. The control represents uninfected cells.
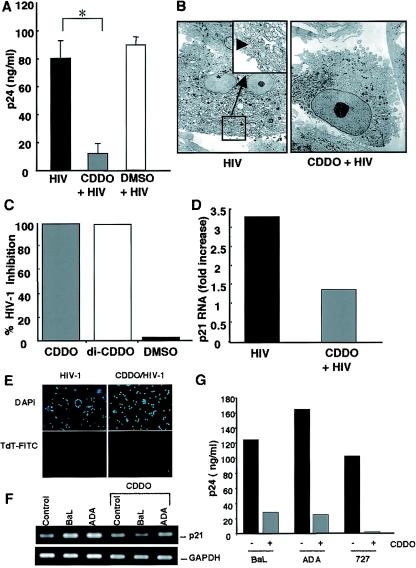
The ability of p21 antisense oligonucleotides and siRNA to block HIV-1 replication prompted an exploration of potential therapeutically relevant mechanisms of modulating this host cell target to inhibit HIV-1. It has been reported that PPARγ ligands, one of which includes the synthetic triterpenoid, CDDO, can modulate p21 activity (66, 67). The treatment of macrophages with CDDO, which had been added 45 min before (Fig. 7A to C), concomitant with infection, or at the onset of detectable viral replication (data not shown), reduced the levels of detectable virus when compared to untreated or dimethyl sulfoxide-treated control cultures. Similar results were observed when cells were treated with the CDDO analog di-CDDO (Fig. 7C). Supernatant p24 levels were inhibited ≥80%, and viral particles were rarely seen ultrastructurally in CDDO-treated macrophages (Fig. 7B and C). Demonstrating a further correlation between p21 and HIV, the CDDO-treated macrophages which exhibited a reduction in HIV-1 also showed reduced mRNA levels for p21 (Fig. 7D). No negative effects on cell viability were evident, as determined by terminal deoxynucleotidyl transferase and DAPI (4′,6′-diamidino-2-phenylindole) staining (Fig. 7E). In addition to HIV-1BaL, CDDO suppressed p21 expression and replication of both the laboratory-adapted viral isolate ADA and the clinical isolate 727 (Fig. 7F and G), confirming the participation of this pathway in macrophage HIV replication.
FIG. 7.
Inhibition of HIV-1 replication by CDDO. (A) CDDO-treated cells (as described in Materials and Methods) showed reduced viral replication, as quantitated by p24 levels, compared with dimethyl sulfoxide-treated control cells and untreated cells (day 10) (n = 3; *, P = 0.01 by one-tail paired t test). (B and C) By TEM, reduced numbers of infected cells were observed after CDDO or di-CDDO treatment. Analyses of ≥200 cells/treatment condition revealed the absence or near absence of detectable virions. (D) CDDO-treated cells infected with HIV-1 demonstrated reduced p21 transcription, as determined by RPA (data from day 12 postinfection are shown and are mean values from a representative experiment; n = 2). (E) Terminal deoxynucleotidyl transferase-FITC (apoptotic) and DAPI (nuclear) staining of cultures that were infected with HIV-1 and/or treated with CDDO. (F) Macrophages were infected with HIV-1BaL or ADA, treated with CDDO (0.1 μM) or left untreated, and analyzed by PCR for p21 and GAPDH. (G) Supernatants (12 days) collected from HIV-1BaL-, ADA-, or 727-infected cells that were treated with CDDO or left untreated were analyzed for viral replication by p24 ELISA.
DISCUSSION
Retroviruses rely on host cell molecules for replication and survival. We provide evidence of a novel role of the cyclin-dependent kinase inhibitor 1A (p21) in successful HIV-1 replication in macrophage hosts. Intact, infectious R5 HIV-1 induces reproducible alterations in immediate early gene transcription in primary macrophages. Consistent with viral binding to the CD4 and CCR5 seven-transmembrane-domain G-protein-coupled receptors, virus-initiated signal transduction induced transcriptional changes similar to and distinct from those observed in previous studies with the viral envelope gp120 (10, 42). While what functional significance can be assigned to each of the >100 genes that are upregulated after viral binding has yet to be established, our data support an initial burst of transcriptional activity followed by a quiescent phase and then a resurgence of new genes which is temporally associated with maximal viral replication. HIV-1-enhanced gene expression and/or phosphorylation of p38 MAPK and MAPKAP-K1 and -K2, which are important in the early postentry and late stages of HIV-1 infection (14, 45), may contribute to altered host cell receptiveness, as well as chemokine expression and the recruitment of new viral hosts (31). Furthermore, the induction of LIMK-1 and members of the Ras and Rho GTPase family, which are involved in the regulation of actin rearrangement, may be involved in transducing signals to the cellular cytoskeletal networks. In our analysis of the early transcriptional program, there was clear evidence that HIV-1 enhanced more genes than it suppressed. However, until we can document functional consequences of gene repression, it remains unclear if this confers a survival advantage on the virus.
In contrast to T cells, macrophages can coexist in vivo as well as in vitro with the virus for a prolonged time, during which they contribute to the pathogenesis of AIDS, acting as viral reservoirs and/or transmitting HIV-1 to neighboring cells. Although proapoptotic genes were upregulated in macrophages within hours after infection, the antiapoptotic genes encoding bcl-x, DAD1, and IEX-1L (2, 28, 70) were also increased. However, in T cells, an increased expression of proapoptotic transcripts and an inhibition of mitochondrial and DNA repair genes are observed (13). The differential gene expression and cell-specific modulation of host protein functions as a result of HIV-1 infection in these cell populations may underlie HIV-1-induced apoptosis in T cells (11, 13), while allowing macrophages to sustain a prolonged viral burden. A comparison of genes that are upregulated by HIV-1 in T lymphocytes (13, 20) with those we identified in macrophage hosts also revealed an early increase in cellular defense, transcription, and signaling genes in both populations.
After the initial HIV-1-induced burst of gene expression (6 to 24 h), an additional increase in transcriptional activity occurred concomitant with the onset of detectable viral replication (days 7 to 14). The lack of induction of new host molecules during the interim period may allow the infected cells to escape immune surveillance while the virus surreptitiously initiates its life cycle. Once replication commences, new transcription may be essential to facilitate the replicative process. One of the immediate early genes that was reexpressed within days after infection is MMP9, which facilitates the migration of HIV-1-infected monocytes across the vascular endothelium (15) and has been detected in HIV-1-infected patients (50). Enhanced transcription of other inflammatory mediators associated with increased viral replication and the pathophysiology of HIV-1 (31) was also represented in the transcriptional profile postinfection. Cell homeostasis and genomic stability influenced by HO-1, PCNA, HMG-I, and MLH1 may provide a receptive intracellular environment. The ability of PCNA to interact with MLH1 (22) suggests a link between mismatch repair and viral growth. The expression of PCNA independent of cell proliferation has been found in macrophage populations and identified as a potential factor contributing to susceptibility to simian immunodeficiency virus (SIV) infection (69). HMG-I, which participates in the integration of viral cDNA (17), may play a role in viral expression by modulating the interaction of transcription factors (25).
Among the genes expressed in a biphasic fashion is p21, which is increased after the initial HIV-1 interaction with macrophage receptors, followed by a striking increase in association with viral growth within intracellular compartments of the macrophage. Because of its unique profile of expression in infected cells, we focused on dissecting its potential contribution to macrophage vulnerability to infection. p21 is a CDK inhibitor induced by a p53-dependent pathway following DNA damage as well as by p53-independent pathways (16). A progressive upregulation of p21 mRNA and protein has been associated with the maturation of myeloid progenitor cells (51), but its connection with HIV-1 replication in macrophages has not been previously reported. Increased p21 in skin lesions of human papillomavirus was found to be further enhanced by HIV-1 coinfection (3), and the upregulation of p21 in macrophages infected with M. avium (21) may also be linked to their increased susceptibility to HIV replication (62).
While the initial increase in p21 gene expression likely represents a downstream consequence of CCR5/G protein signaling, the subsequent rise in gene transcription may be due to either intracellular or extracellular viral signals. Increased p21 protein in both the nuclear and cytoplasmic compartments of HIV-1-infected macrophages may generate a permissive environment and prevent the death of the host cells. The presence of p21 in the nucleus has been related to its cell cycle functions (16), whereas the cytoplasmic localization of this protein has been implicated in controlling the apoptosis of alveolar macrophages and during monocytic differentiation (5, 57). Originally described as a cell cycle inhibitor, p21 has also been associated with the modulation of apoptosis, the cytoplasmic regulation of nuclear import, and transcriptional regulation by acting as a transcriptional adaptor molecule (12). The transcriptional coactivator p300, which is essential for efficient viral replication through its interaction with the cyclic AMP response element binding protein (CREB) and HIV-1 Tat (29), can be stimulated by the coexpression of p21 through a novel transcriptional repression domain on p300 (48). An increase in TBP-1 may represent a viral strategy to ensure efficient regulation of transcription and reproduction. The induction of the transcription factors PC4 and RPL6 by the virus represents a potential pathway to maximize virus survival, while the transcription factor HIF-1, known to target p21 (44), may ensure the presence of this CDK during infection. Our studies support a causal relationship between HIV-1 and induced p21 expression, which in turn drives the viral life cycle in macrophages. Our data also demonstrate that Vpr independently enhances p21 transcription, similar to that reported in a replicating T-cell line (9), connecting this accessory protein with the p21-dependent infectious process (26, 46).
Targeting p21 with antisense oligonucleotides or a siRNA demonstrates the important role of this host molecule as a requisite regulator of subsequent viral replication in infected macrophages. The precise mechanism by which p21 contributes to HIV-1 replication requires further investigation, but the participation of p21 may be pivotal for the regulation of other host molecules that are required for successful viral replication, and their activities may be diminished by the blockade of p21. CDDO, a potent differentiating, antiproliferative, and anti-inflammatory compound (55) and a potential chemotherapeutic agent for cancer, was recently identified as a member of a new class of nuclear PPARγ ligands (66, 67). PPARγ is a nuclear hormone receptor involved in the gene regulation of lipid and glucose metabolism, cellular differentiation, and the control of macrophage inflammatory molecules (6). Agonists of PPARγ have recently been shown to influence retroviral replication (24, 47), and we now provide insight into a molecular target, since the PPARγ ligand CDDO, which inhibits p21, also inhibits HIV in parallel. Whether the antiviral effect of CDDO is totally mediated through this receptor by its effect on p21 (43) or also involves the inhibition of NF-κB (53), modulation of p38 MAPK (32), and/or production of cytokines that regulate cellular and viral components, such as transforming growth factor beta (35, 61), is still unresolved and is currently under intense investigation.
By using multiple parameters, we have documented that p21 contributes to the HIV-1 infection process in macrophage hosts. The increased expression accompanying infection, but most importantly, the ability to inhibit p21 by antisense oligonucleotides, siRNA, and CDDO and the suppression of HIV replication all point to the causal link between p21 and HIV. Since the macrophage represents a key target for HIV-1 infection and one of the major obstacles to eradicating the virus, even during highly active antiretroviral therapy (19, 30, 63), our study of the effect of HIV-1 on the macrophage transcriptome reveals important insights into the pattern of host gene expression underlying viral success in this population. Continued exploration of p21 and other virus-regulated macrophage genes that are critical for HIV-1 may disclose mechanisms by which this reservoir can be targeted and/or may serve as prognostic markers of disease progression. Finally, since anti-HIV-1 therapy is limited by the side effects that have accompanied conventional antiretroviral drugs and the constant emergence of drug-resistant virus, CDDO may be considered a candidate drug to target HIV-1 through a host cell factor, in conjunction with current antiviral therapy, to suppress replication sequelae in infected hosts.
Acknowledgments
We are grateful to Carl Wild from Panacos Pharmaceuticals Inc. for providing the ADA isolate and the primary isolate clade B 92US727. We also thank Eric Freed at National Cancer Institute for the NLAD8 and delta R constructs and Lori Coren, Alice Goodwin, and Wenwen Jin for their technical assistance. We thank Albert Kingman for his expertise with statistical analysis.
D. E. Ott’s contribution to this project has been funded by NCI, NIH, under contract number N01-CO-12400.
REFERENCES
- 1.Adachi, A., S. Koenig, H. E. Gendelman, D. Daugherty, S. Gattoni-Celli, A. S. Fauci, and M. A. Martin. 1987. Productive, persistent infection of human colorectal cell lines with human immunodeficiency virus. J. Virol. 61:209-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonsson, B., and J. C. Martinou. 2000. The Bcl-2 protein family. Exp. Cell Res. 256:50-57. [DOI] [PubMed] [Google Scholar]
- 3.Arany, I., A. Yen, and S. K. Tyring. 1997. p53, WAF1/CIP1 and mdm2 expression in skin lesions associated with human papillomavirus and human immunodeficiency virus. Anticancer Res. 17:1281-1285. [PubMed] [Google Scholar]
- 4.Arber, S., F. A. Barbayannis, H. Hanser, C. Schneider, C. A. Stanyon, O. Bernard, and P. Caroni. 1998. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393:805-809. [DOI] [PubMed] [Google Scholar]
- 5.Asada, M., T. Yamada, H. Ichijo, D. Delia, K. Miyazono, K. Fukumuro, and S. Mizutani. 1999. Apoptosis inhibitory activity of cytoplasmic p21(Cip1/WAF1) in monocytic differentiation. EMBO J. 18:1223-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bar-Tana, J. 2001. Peroxisome proliferator-activated receptor gamma (PPARgamma) activation and its consequences in humans. Toxicol. Lett. 120:9-19. [DOI] [PubMed] [Google Scholar]
- 7.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 8.Bustin, M., and R. Reeves. 1996. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acids Res. Mol. Biol. 54:35-100. [DOI] [PubMed] [Google Scholar]
- 9.Chowdhury, I. H., X. F. Wang, N. R. Landau, M. L. Robb, V. R. Polonis, D. L. Birx, and J. H. Kim. 2003. HIV-1 Vpr activates cell cycle inhibitor p21/Waf1/Cip1: a potential mechanism of G2/M cell cycle arrest. Virology 305:371-377. [DOI] [PubMed] [Google Scholar]
- 10.Cicala, C., J. Arthos, S. M. Selig, G. Dennis, Jr., D. A. Hosack, D. Van Ryk, M. L. Spangler, T. D. Steenbeke, P. Khazanie, N. Gupta, J. Yang, M. Daucher, R. A. Lempicki, and A. S. Fauci. 2002. HIV envelope induces a cascade of cell signals in non-proliferating target cells that favor virus replication. Proc. Natl. Acad. Sci. USA 99:9380-9385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark, E., F. Santiago, L. Deng, S. Chong, C. de La Fuente, L. Wang, P. Fu, D. Stein, T. Denny, V. Lanka, F. Mozafari, T. Okamoto, and F. Kashanchi. 2000. Loss of G(1)/S checkpoint in human immunodeficiency virus type 1-infected cells is associated with a lack of cyclin-dependent kinase inhibitor p21/Waf1. J. Virol. 74:5040-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coqueret, O. 2003. New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol. 13:65-70. [DOI] [PubMed] [Google Scholar]
- 13.Corbeil, J., D. Sheeter, D. Genini, S. Rought, L. Leoni, P. Du, M. Ferguson, D. R. Masys, J. B. Welsh, J. L. Fink, R. Sasik, D. Huang, J. Drenkow, D. D. Richman, and T. Gingeras. 2001. Temporal gene regulation during HIV-1 infection of human CD4+ T cells. Genome Res. 11:1198-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Corno, M., Q. H. Liu, D. Schols, E. de Clercq, S. Gessani, B. D. Freedman, and R. G. Collman. 2001. HIV-1 gp120 and chemokine activation of Pyk2 and mitogen-activated protein kinases in primary macrophages mediated by calcium-dependent, pertussis toxin-insensitive chemokine receptor signaling. Blood 98:2909-2916. [DOI] [PubMed] [Google Scholar]
- 15.Dhawan, S., B. S. Weeks, C. Soderland, H. W. Schnaper, L. A. Toro, S. P. Asthana, I. K. Hewlett, W. G. Stetler-Stevenson, S. S. Yamada, K. M. Yamada, et al. 1995. HIV-1 infection alters monocyte interactions with human microvascular endothelial cells. J. Immunol. 154:422-432. [PubMed] [Google Scholar]
- 16.Dotto, G. P. 2000. p21(WAF1/Cip1): more than a break to the cell cycle? Biochim. Biophys. Acta 1471:M43-M56. [DOI] [PubMed] [Google Scholar]
- 17.Farnet, C. M., and F. D. Bushman. 1997. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell 88:483-492. [DOI] [PubMed] [Google Scholar]
- 18.Freed, E. O., G. Englund, and M. A. Martin. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 69:3949-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garbuglia, A. R., M. Zaccarelli, S. Calcaterra, G. Cappiello, R. Marini, and A. Benedetto. 2001. Dynamics of viral load in plasma and HIV DNA in lymphocytes during highly active antiretroviral therapy (HAART): high viral burden in macrophages after 1 year of treatment. J. Chemother. 13:188-194. [DOI] [PubMed] [Google Scholar]
- 20.Geiss, G. K., R. E. Bumgarner, M. C. An, M. B. Agy, A. B. van't Wout, E. Hammersmark, V. S. Carter, D. Upchurch, J. I. Mullins, and M. G. Katze. 2000. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology 266:8-16. [DOI] [PubMed] [Google Scholar]
- 21.Greenwell-Wild, T., N. Vazquez, D. Sim, M. Schito, D. Chatterjee, J. M. Orenstein, and S. M. Wahl. 2002. Mycobacterium avium infection and modulation of human macrophage gene expression. J. Immunol. 169:6286-6297. [DOI] [PubMed] [Google Scholar]
- 22.Gu, L., Y. Hong, S. McCulloch, H. Watanabe, and G. M. Li. 1998. ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Res. 26:1173-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo, M. M., and J. E. Hildreth. 1995. HIV acquires functional adhesion receptors from host cells. AIDS Res. Hum. Retrovir. 11:1007-1013. [DOI] [PubMed] [Google Scholar]
- 24.Hayes, M. M., B. R. Lane, S. R. King, D. M. Markovitz, and M. J. Coffey. 2002. Peroxisome proliferator-activated receptor gamma agonists inhibit HIV-1 replication in macrophages by transcriptional and post-transcriptional effects. J. Biol. Chem. 277:16913-16919. [DOI] [PubMed] [Google Scholar]
- 25.Henderson, A., M. Bunce, N. Siddon, R. Reeves, and D. J. Tremethick. 2000. High-mobility-group protein I can modulate binding of transcription factors to the U5 region of the human immunodeficiency virus type 1 proviral promoter. J. Virol. 74:10523-10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henklein, P., K. Bruns, M. P. Sherman, U. Tessmer, K. Licha, J. Kopp, C. M. de Noronha, W. C. Greene, V. Wray, and U. Schubert. 2000. Functional and structural characterization of synthetic HIV-1 Vpr that transduces cells, localizes to the nucleus, and induces G2 cell cycle arrest. J. Biol. Chem. 275:32016-32026. [DOI] [PubMed] [Google Scholar]
- 27.Holloway, A. F., F. Occhiodoro, G. Mittler, M. Meisterernst, and M. F. Shannon. 2000. Functional interaction between the HIV transactivator Tat and the transcriptional coactivator PC4 in T cells. J. Biol. Chem. 275:21668-21677. [DOI] [PubMed] [Google Scholar]
- 28.Hong, N. A., M. Flannery, S. N. Hsieh, D. Cado, R. Pedersen, and A. Winoto. 2000. Mice lacking Dad1, the defender against apoptotic death-1, express abnormal N-linked glycoproteins and undergo increased embryonic apoptosis. Dev. Biol. 220:76-84. [DOI] [PubMed] [Google Scholar]
- 29.Hottiger, M. O., and G. J. Nabel. 1998. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J. Virol. 72:8252-8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Igarashi, T., C. R. Brown, Y. Endo, A. Buckler-White, R. Plishka, N. Bischofberger, V. Hirsch, and M. A. Martin. 2001. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): implications for HIV-1 infections of humans. Proc. Natl. Acad. Sci. USA 98:658-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kedzierska, K., and S. M. Crowe. 2001. Cytokines and HIV-1: interactions and clinical implications. Antivir. Chem. Chemother. 12:133-150. [DOI] [PubMed] [Google Scholar]
- 32.Kim, J. Y., J. A. Choi, T. H. Kim, Y. D. Yoo, J. I. Kim, Y. J. Lee, S. Y. Yoo, C. K. Cho, Y. S. Lee, and S. J. Lee. 2002. Involvement of p38 mitogen-activated protein kinase in the cell growth inhibition by sodium arsenite. J. Cell Physiol. 190:29-37. [DOI] [PubMed] [Google Scholar]
- 33.Lawson, B. R., R. Baccala, J. Song, M. Croft, D. H. Kono, and A. N. Theofilopoulos. 2004. Deficiency of the cyclin kinase inhibitor p21(WAF-1/CIP-1) promotes apoptosis of activated/memory T cells and inhibits spontaneous systemic autoimmunity. J. Exp. Med. 199:547-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levere, R. D., R. Staudinger, G. Loewy, A. Kappas, S. Shibahara, and N. G. Abraham. 1993. Elevated levels of heme oxygenase-1 activity and mRNA in peripheral blood adherent cells of acquired immunodeficiency syndrome patients. Am. J. Hematol. 43:19-23. [DOI] [PubMed] [Google Scholar]
- 35.Li, C. Y., L. Suardet, and J. B. Little. 1995. Potential role of WAF1/Cip1/p21 as a mediator of TGF-beta cytoinhibitory effect. J. Biol. Chem. 270:4971-4974. [DOI] [PubMed] [Google Scholar]
- 36.Ma, G., T. Greenwell-Wild, K. Lei, W. Jin, J. Swisher, N. Hardegen, C. T. Wild, and S. M. Wahl. 2004. Secretory leukocyte protease inhibitor (SLPI) binds to annexin II, a cofactor for macrophage HIV-1 infection. J. Exp. Med. 200:1337-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNeely, T. B., D. C. Shugars, M. Rosendahl, C. Tucker, S. P. Eisenberg, and S. M. Wahl. 1997. Inhibition of human immunodeficiency virus type 1 infectivity by secretory leukocyte protease inhibitor occurs prior to viral reverse transcription. Blood 90:1141-1149. [PubMed] [Google Scholar]
- 38.Modrich, P. 1997. Strand-specific mismatch repair in mammalian cells. J. Biol. Chem. 272:24727-24730. [DOI] [PubMed] [Google Scholar]
- 39.Morita, T., T. Sato, H. Nyunoya, A. Tsujimoto, J. Takahara, S. Irino, and K. Shimotohno. 1993. Isolation of a cDNA clone encoding DNA-binding protein (TAXREB107) that binds specifically to domain C of the tax-responsive enhancer element in the long terminal repeat of human T-cell leukemia virus type I. AIDS Res. Hum. Retrovir. 9:115-121. [DOI] [PubMed] [Google Scholar]
- 40.Nelbock, P., P. J. Dillon, A. Perkins, and C. A. Rosen. 1990. A cDNA for a protein that interacts with the human immunodeficiency virus Tat transactivator. Science 248:1650-1653. [DOI] [PubMed] [Google Scholar]
- 41.Orenstein, J. M., C. Fox, and S. M. Wahl. 1997. Macrophages as a source of HIV during opportunistic infections. Science 276:1857-1861. [DOI] [PubMed] [Google Scholar]
- 42.Popik, W., and P. M. Pitha. 2000. Exploitation of cellular signaling by HIV-1: unwelcome guests with master keys that signal their entry. Virology 276:1-6. [DOI] [PubMed] [Google Scholar]
- 43.Scott, M. T., N. Morrice, and K. L. Ball. 2000. Reversible phosphorylation at the C-terminal regulatory domain of p21(Waf1/Cip1) modulates proliferating cell nuclear antigen binding. J. Biol. Chem. 275:11529-11537. [DOI] [PubMed] [Google Scholar]
- 44.Semenza, G. L. 2003. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3:721-732. [DOI] [PubMed] [Google Scholar]
- 45.Shapiro, L., K. A. Heidenreich, M. K. Meintzer, and C. A. Dinarello. 1998. Role of p38 mitogen-activated protein kinase in HIV type 1 production in vitro. Proc. Natl. Acad. Sci. USA 95:7422-7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman, M. P., C. M. de Noronha, L. A. Eckstein, J. Hataye, P. Mundt, S. A. Williams, J. A. Neidleman, M. A. Goldsmith, and W. C. Greene. 2003. Nuclear export of Vpr is required for efficient replication of human immunodeficiency virus type 1 in tissue macrophages. J. Virol. 77:7582-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skolnik, P. R., M. F. Rabbi, J. M. Mathys, and A. S. Greenberg. 2002. Stimulation of peroxisome proliferator-activated receptors alpha and gamma blocks HIV-1 replication and TNFalpha production in acutely infected primary blood cells, chronically infected U1 cells, and alveolar macrophages from HIV-infected subjects. J. Acquir. Immune Defic. Syndr. 31:1-10. [DOI] [PubMed] [Google Scholar]
- 48.Snowden, A. W., L. A. Anderson, G. A. Webster, and N. D. Perkins. 2000. A novel transcriptional repression domain mediates p21(WAF1/CIP1) induction of p300 transactivation. Mol. Cell. Biol. 20:2676-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spira, A. I., P. A. Marx, B. K. Patterson, J. Mahoney, R. A. Koup, S. M. Wolinsky, and D. D. Ho. 1996. Cellular targets of infection and route of viral dissemination after an intravaginal inoculation of simian immunodeficiency virus into rhesus macaques. J. Exp. Med. 183:215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sporer, B., R. Paul, U. Koedel, R. Grimm, M. Wick, F. D. Goebel, and H. W. Pfister. 1998. Presence of matrix metalloproteinase-9 activity in the cerebrospinal fluid of human immunodeficiency virus-infected patients. J. Infect. Dis. 178:854-857. [DOI] [PubMed] [Google Scholar]
- 51.Steinman, R. A., J. Huang, B. Yaroslavskiy, J. P. Goff, E. D. Ball, and A. Nguyen. 1998. Regulation of p21(WAF1) expression during normal myeloid differentiation. Blood 91:4531-4542. [PubMed] [Google Scholar]
- 52.Stevenson, M. 2003. HIV-1 pathogenesis. Nat. Med. 9:853-860. [DOI] [PubMed] [Google Scholar]
- 53.Straus, D. S., G. Pascual, M. Li, J. S. Welch, M. Ricote, C. H. Hsiang, L. L. Sengchanthalangsy, G. Ghosh, and C. K. Glass. 2000. 15-Deoxy-delta 12,14-prostaglandin J2 inhibits multiple steps in the NF-kappa B signaling pathway. Proc. Natl. Acad. Sci. USA 97:4844-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subbramanian, R. A., A. Kessous-Elbaz, R. Lodge, J. Forget, X. J. Yao, D. Bergeron, and E. A. Cohen. 1998. Human immunodeficiency virus type 1 Vpr is a positive regulator of viral transcription and infectivity in primary human macrophages. J. Exp. Med. 187:1103-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suh, N., Y. Wang, T. Honda, G. W. Gribble, E. Dmitrovsky, W. F. Hickey, R. A. Maue, A. E. Place, D. M. Porter, M. J. Spinella, C. R. Williams, G. Wu, A. J. Dannenberg, K. C. Flanders, J. J. Letterio, D. J. Mangelsdorf, C. F. Nathan, L. Nguyen, W. W. Porter, R. F. Ren, A. B. Roberts, N. S. Roche, K. Subbramanian, and M. B. Sporn. 1999. A novel synthetic oleanane triterpenoid, 2-cyano-3,12-dioxoolean-1,9-dien-28-oic acid, with potent differentiating, antiproliferative, and anti-inflammatory activity. Cancer Res. 59:336-341. [PubMed] [Google Scholar]
- 56.Sumi, T., K. Matsumoto, Y. Takai, and T. Nakamura. 1999. Cofilin phosphorylation and actin cytoskeletal dynamics regulated by rho- and Cdc42-activated LIM-kinase 2. J. Cell Biol. 147:1519-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomita, K., G. Caramori, S. Lim, K. Ito, T. Hanazawa, T. Oates, I. Chiselita, E. Jazrawi, K. F. Chung, P. J. Barnes, and I. M. Adcock. 2002. Increased p21(CIP1/WAF1) and B cell lymphoma leukemia-x(L) expression and reduced apoptosis in alveolar macrophages from smokers. Am. J. Respir. Crit. Care Med. 166:724-731. [DOI] [PubMed] [Google Scholar]
- 58.Tomkinson, B. 1999. Tripeptidyl peptidases: enzymes that count. Trends Biochem. Sci. 24:355-359. [DOI] [PubMed] [Google Scholar]
- 59.Tremblay, M. J., J. F. Fortin, and R. Cantin. 1998. The acquisition of host-encoded proteins by nascent HIV-1. Immunol. Today 19:346-351. [DOI] [PubMed] [Google Scholar]
- 60.Vendeville, A., F. Rayne, A. Bonhoure, N. Bettache, P. Montcourrier, and B. Beaumelle. 2004. HIV-1 Tat enters T cells using coated pits before translocating from acidified endosomes and eliciting biological responses. Mol. Biol. Cell 15:2347-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wahl, S. M., J. B. Allen, N. McCartney-Francis, M. C. Morganti-Kossmann, T. Kossmann, L. Ellingsworth, U. E. Mai, S. E. Mergenhagen, and J. M. Orenstein. 1991. Macrophage- and astrocyte-derived transforming growth factor beta as a mediator of central nervous system dysfunction in acquired immune deficiency syndrome. J. Exp. Med. 173:981-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wahl, S. M., T. Greenwell-Wild, G. Peng, H. Hale-Donze, T. M. Doherty, D. Mizel, and J. M. Orenstein. 1998. Mycobacterium avium complex augments macrophage HIV-1 production and increases CCR5 expression. Proc. Natl. Acad. Sci. USA 95:12574-12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wahl, S. M., T. Greenwell-Wild, G. Peng, G. Ma, J. M. Orenstein, and N. Vazquez. 2003. Viral and host cofactors facilitate HIV-1 replication in macrophages. J. Leukoc. Biol. 74:726-735. [DOI] [PubMed] [Google Scholar]
- 64.Wahl, S. M., J. M. Orenstein, and P. D. Smith. 1996. Macrophage function in HIV infection, p. 303-336. In S. Gupta (ed.), Immunology of HIV infection. Plenum Medical Book Co., New York, N.Y.
- 65.Wainberg, Z., M. Oliveira, S. Lerner, Y. Tao, and B. G. Brenner. 1997. Modulation of stress protein (hsp27 and hsp70) expression in CD4+ lymphocytic cells following acute infection with human immunodeficiency virus type-1. Virology 233:364-373. [DOI] [PubMed] [Google Scholar]
- 66.Wakino, S., U. Kintscher, Z. Liu, S. Kim, F. Yin, M. Ohba, T. Kuroki, A. H. Schonthal, W. A. Hsueh, and R. E. Law. 2001. Peroxisome proliferator-activated receptor gamma ligands inhibit mitogenic induction of p21(Cip1) by modulating the protein kinase Cdelta pathway in vascular smooth muscle cells. J. Biol. Chem. 276:47650-47657. [DOI] [PubMed] [Google Scholar]
- 67.Wang, Y., W. W. Porter, N. Suh, T. Honda, G. W. Gribble, L. M. Leesnitzer, K. D. Plunket, D. J. Mangelsdorf, S. G. Blanchard, T. M. Willson, and M. B. Sporn. 2000. A synthetic triterpenoid, 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO), is a ligand for the peroxisome proliferator-activated receptor gamma. Mol. Endocrinol. 14:1550-1556. [DOI] [PubMed] [Google Scholar]
- 68.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williams, K., A. Schwartz, S. Corey, M. Orandle, W. Kennedy, B. Thompson, X. Alvarez, C. Brown, S. Gartner, and A. Lackner. 2002. Proliferating cellular nuclear antigen expression as a marker of perivascular macrophages in simian immunodeficiency virus encephalitis. Am. J. Pathol. 161:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu, M. X., Z. Ao, K. V. Prasad, R. Wu, and S. F. Schlossman. 1998. IEX-1L, an apoptosis inhibitor involved in NF-kappaB-mediated cell survival. Science 281:998-1001. [DOI] [PubMed] [Google Scholar]
- 71.Yang, N., O. Higuchi, K. Ohashi, K. Nagata, A. Wada, K. Kangawa, E. Nishida, and K. Mizuno. 1998. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 393:809-812. [DOI] [PubMed] [Google Scholar]
- 72.Yao, Y., A. Hoffer, C. Y. Chang, and A. Puga. 1995. Dioxin activates HIV-1 gene expression by an oxidative stress pathway requiring a functional cytochrome P450 CYP1A1 enzyme. Environ. Health Perspect. 103:366-371. [DOI] [PMC free article] [PubMed] [Google Scholar]