Abstract
By selecting the R5 human immunodeficiency virus type 1 (HIV-1) strain JR-CSF for efficient use of a CCR5 coreceptor with a badly damaged amino terminus [i.e., CCR5(Y14N)], we previously isolated variants that weakly utilize CCR5(Δ18), a low-affinity mutant lacking the normal tyrosine sulfate-containing amino-terminal region of the coreceptor. These previously isolated HIV-1JR-CSF variants contained adaptive mutations situated exclusively in the V3 loop of their gp120 envelope glycoproteins. We now have weaned the virus from all dependency on the CCR5 amino terminus by performing additional selections with HeLa-CD4 cells that express only a low concentration of CCR5(Δ18). The adapted variants had additional mutations in their V3 loops, as well as one in the V2 stem (S193N) and four alternative mutations in the V4 loop that eliminated the same N-linked oligosaccharide from position N403. Assays using pseudotyped viruses suggested that these new gp120 mutations all made strong contributions to use of CCR5(Δ18) by accelerating a rate-limiting CCR5-dependent conformational change in gp41 rather than by increasing viral affinity for this damaged coreceptor. Consistent with this interpretation, loss of the V4 N-glycan at position N403 also enhanced HIV-1 use of a different low-affinity CCR5 coreceptor with a mutation in extracellular loop 2 (ECL2) [i.e., CCR5(G163R)], whereas the double mutant CCR5(Δ18,G163R) was inactive. We conclude that loss of the N-glycan at position N403 helps to convert the HIV-1 envelope into a hair-trigger form that no longer requires strong interactions with both the CCR5 amino terminus and ECL2 but efficiently uses either site alone. These results demonstrate a novel functional role for a gp120 N-linked oligosaccharide and a high degree of adaptability in coreceptor usage by HIV-1.
The human immunodeficiency virus type 1 (HIV-1) infection pathway involves a sequential process whereby reversible binding to the CD4 receptor induces a conformational change in the viral gp120-gp41 trimeric complexes that exposes or induces formation of previously inaccessible epitopes in both gp120 and gp41 and dramatically enhances gp120 affinity for a coreceptor (46, 57, 65, 69, 71, 73, 75). Energetic studies have suggested that the gp120 subunit has a high entropy or conformational flexibility that inhibits binding of antibodies or other ligands and that binding to CD4 substantially reduces this flexibility, thus enhancing the subsequent binding of gp120 to additional ligands by an energetic (entropic) mechanism (36, 47). The sequential reversible binding of gp120 to CD4 and to a coreceptor is believed to lower the activation energy for irreversible conformational changes in the metastable gp41 subunits. These conformational changes pull the virus tightly onto the cell surface and induce fusion of the viral and cellular membranes (9, 24).
The gp120 glycoprotein contains an inner core of conserved sequences and several variable loops that are heavily N-glycosylated and that form a protective surface shell (8, 32, 41, 55, 60, 74, 77, 78). X-ray crystallographic studies have revealed the structural organization of the gp120 core complexed with a CD4 amino-terminal domain and the monoclonal antibody (MAb) 17b, which occludes a portion of the coreceptor binding region (37, 38). This gp120 core is folded into inner and outer domains connected by a bridging sheet, with discontinuous residues from these domains forming the CD4 binding site (36-38, 61, 62). Residues in the bridging sheet consisting of conserved region 4 (C4), the V1/V2 base, and the V3 loop comprise a coreceptor binding site (36-38, 45, 61, 62, 70, 77, 78), with the highly variable V3 loop controlling the coreceptor specificity (13, 14, 68, 75).
Coreceptors are members of a large family of G-protein-coupled chemokine receptors, with CCR5 and CXCR4 being the principal coreceptors used by HIV-1 in vivo (2, 13, 18, 19, 21, 29). Typically, HIV-1 transmission involves an R5 virus that uses CCR5 (40, 58, 64), whereas X4 variants that can use CXCR4 often form during disease progression (15, 20, 30, 44).
Mutagenesis and biochemical studies have demonstrated that the tyrosine-rich and highly acidic amino terminus of CCR5 is essential for binding R5 gp120s and for R5 HIV-1 infections (22, 25, 35, 56, 63). The tyrosines at positions 10, 14, and 15, which are modified posttranslationally by sulfation, appear to be most critical (22, 25, 27, 34, 35). Indeed, sulfated peptides corresponding to the CCR5 amino terminus competitively block R5 HIV-1 infections of cells that contain CCR5 (16, 28) and restore HIV-1 infectivity for cells that express CCR5(Δ18), an amino-terminal deletion mutant of CCR5 lacking amino acids 2 to 19 (26). Additional studies have implicated other regions of CCR5 in coreceptor function, with extracellular loop 2 (ECL2) being most clearly shown (3, 5, 34, 35, 49, 63, 67). MAbs specific for the CCR5 amino terminus block gp120 binding but do not prevent HIV-1 infections, whereas MAbs such as 2D7 that are specific for ECL2 have smaller effects on gp120 binding yet are potent inhibitors of infection (39, 48). In addition, we recently analyzed a G163 residue near the transmembrane 4/ECL2 junction that is critical for HIV-1 infections (34, 67). A G163R substitution, which occurs naturally in African green monkey CCR5, has no effect on simian immunodeficiency virus infections or on CCR5 signaling but potently inhibits binding of R5 HIV-1 gp120 molecules and HIV-1 infections. In agreement with these conclusions, we found that variants of the R5 HIV-1 strain JR-CSF that were selected for replication in HeLa-CD4/CCR5(Y14N) cells were weakly able to use CCR5(Δ18) [or CCR5(Δ16), which lacks amino acids 2 to 17] and were more sensitive than the wild-type virus to inhibition by the G163R mutation in ECL2 (51). These results suggested that the adapted viruses, which contained mutations situated exclusively in the gp120 V3 loop, had a reduced dependency on the CCR5 amino terminus and a correspondingly increased reliance on ECL2 (51). A further implication is that these sites in CCR5 have additive or collaborative roles in infection, so that an absence or defect at one site can be compensated for by increased dependency on the other.
We have extended our previous studies by selecting HIV-1JR-CSF variants that efficiently replicate in HeLa-CD4/CCR5(Δ18) or HeLa-CD4/CCR5(Δ16) cell clones that express relatively low concentrations of these mutant coreceptors. The adapted viruses obtained from these stringent selections remained dependent on CCR5 and contained functionally adaptive mutations not only in the V3 loop but also in the V2 base and in a site for N-linked glycosylation in the V4 region of gp120. Surprisingly, removal of this same V4 N-glycan caused HIV-1JR-CSF to become less sensitive rather than more sensitive to the CCR5 ECL2 mutation G163R. Our results suggest that loss of this V4 N-glycan converts HIV-1JR-CSF into a hair-trigger form that is no longer reliant on both the CCR5 amino terminus and ECL2. Rather, the adapted viruses can use the CCR5 amino terminus very efficiently although they are no longer dependent on it.
MATERIALS AND METHODS
Cells and viruses.
HeLa-CD4 cells (clone HI-J), derivative cell clones that express distinct amounts of wild-type CCR5 or of the mutants CCR5(Y14N) and CCR5(G163R), and HEK293T cells were maintained as previously described (31, 34, 53). HeLa-CD4 cells expressing CCR5(Δ16) or CCR5(Δ18), which contained deletions of amino acids 2 to 17 or 2 to 19, respectively, were created by transduction with either the pSFF-Rd16 or pSFF-Rd18 retroviral vector (51). The transduced cell population was labeled with the anti-CCR5 MAb 2D7 (BD Biosciences PharMingen, San Diego, Calif.) followed by fluorescein isothiocyanate-conjugated rabbit anti-mouse immunoglobulin (MP Biomedicals), and positive cells were pooled by fluorescence-activated cell sorting. Cell clones were isolated from the positive population by limiting dilution and expansion. Coreceptor expression levels in the clones were determined by quantitative flow cytometry with the Dako (Carpinteria, Calif.) Qifikit (34, 51). HIV-1JR-CSF isolates adapted to grow in HeLa-CD4 cells expressing small amounts of the CCR5(Y14N) virus were created as described previously (51). The wild-type HIV-1JR-CSF isolate was grown from an infectious molecular clone, pYK-JRCSF, that had been obtained from the AIDS Research and Reference Reagent Program (ARRRP), Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, and was contributed by I. Chen and Y. Koyanagi. HIV-gpt virions pseudotyped with wild-type or adapted JR-CSF envelopes were also generated as described previously (51).
Adaptation of HIV-1 JR-CSF for growth in HeLa-CD4 cell clones expressing CCR5(Δ16) or CCR5(Δ18).
We independently selected HIV-1JR-CSF in cell clones that expressed relatively low concentrations of CCR5(Δ16) [cell clone R5d16.8 with 2.3 ×104 CCR5(Δ16) molecules per cell] or CCR5(Δ18) [cell clone R5d18.2 with 2.7 × 104 CCR5(Δ18) molecules per cell]. These selections were initiated by using an HIV-1JR-CSF isolate that had been adapted to grow in HeLa-CD4 cells expressing small amounts of CCR5(Y14N) [clone JYN.4 with 6 × 104 CCR5(Y14N) molecules per cell] (34, 35, 51). Passaging of virus was performed as previously described except that 106 cells per T-75 filter-capped flask were seeded 24 h prior to passage. Initially, we observed no syncytium formation; however, after four passages, syncytia began to appear. We passaged the virus two more times at more dilute concentrations and harvested genomic DNA from cells infected with passage 6 virus.
Cloning, sequencing, and expression of envelope genes from adapted viruses.
Envelopes from CCR5(Δ16)- or CCR5(Δ18)-adapted viruses were cloned, amplified, and sequenced as described previously (51). HIV-1JR-CSF env mutants were created by using the QuickChange (Stratagene, La Jolla, Calif.) mutagenesis method according to the manufacturer's recommendations, using as our templates JR-CSF rev/env genes cloned into pcDNA3.0 (51). HIV-1JR-CSF envelopes containing CCR5(ΔNt)-adapted mutations were created in the following manner. The S298N, N300Y, T315P V3 mutant was created by introducing the T315P mutation into JR-CSF env already containing the S298N and N300Y mutations (51) by using the primers TP FWD (5′GGGAGAGCATTTTATCCAACAGGAGAAATAATAG3′) and TP REV (5′CTATTATTTCTCCTGTTGGATAAAATGCTCTCCC3′). The S298N, N300Y, I307M, F313L V3 mutant was created by amplifying the JR-CSF env construct containing the mutations S298N, N300Y, and F313L (51) with the forward primer IM FWD (5′GAAAAAGTATACATATGGGACCAGGGAGAG3′) and the reverse primer IM REV (5′CTCTCCCTGGTCCCATATGTATACTTTTTC3′). We created an envelope mutant containing the S298N, N300Y, I307M, F313L, and T315P V3 mutations by PCR amplifying JR-CSFrev/env with the S298N, N300Y, I307 M, and F313L V3 mutations with the primer set NYMLP FWD (5′GGGAGAGCATTGTATCCAACAGGAGAAATAGG3′) and NYMLP REV (5′CCTATTTCTCCTGTTGGATACAATGCTCTCCC3′). The V356A C3 mutation was added to an envelope construct containing the S298N, N300Y, and T315P V3 mutations by amplification with the primers VA FWD (5′CAATTTAATAATAAAACAATAGCCTTTACTCACTCCTCAGGAGGGG3′) and VA REV (5′CCCCTCCTGAGGAGTGAGTAAAGGCTATTGTTTTATTATTAAATTG3′). TheV2 (S193N), and V4 N-glycan deletion (N403S) mutations were placed (singly or together) into JR-CSF rev/env expression vectors having either wild-type or CCR5(ΔNt)-adapted V3 sequences by using the following primers for the S193N mutation: V2SN FWD (5′CCAAATATAGGTTAATAAATTGTAACACCTCAGTCATTACACAAGCC3′) and V2SN REV (5′GGCTTGTGTAATGACTGAGGTGTTACAATTTATTAACCTATATTTGG3′). To destroy the glycosylation site in V4, we designed the following primers to change 403N to 403S: V4NS FWD (5′GGCACTGAAGGAAGTGACACCATCATACTC3′) and V4NS REV (5′GAGTATGATGGTGTCACTTCCTTCAGTGCC3′). Underlined bases indicate point mutations introduced in order to generate the mutant envelopes.
CCR5 mutagenesis.
A CCR5 double mutant containing both the 18-amino-acid N-terminal deletion and the G163R mutation in ECL2 was generated by using the QuickChange (Stratagene) method. The expression vector pcDNA3.0-CCR5(G163R) was used as the template to perform the PCR with mutagenic primers (51) that delete amino acids 2 to 19.
Infectivity assays.
Infections were performed with infectious replication-defective HIV-gpt viruses pseudotyped with wild-type or mutant JR-CSF envelopes as previously described (51, 52), with the following alterations. Pseudotyped HIV-gpt virions were harvested from HEK293T cells that had been seeded at 2.4 × 106 cells/100-mm-diameter plate and 24 h later transfected with PolyFect (Qiagen, Valencia, Calif.) according to the manufacturer's recommendations. Viruses were harvested at 48 h posttransfection and used to infect HeLa-CD4 cells expressing wild-type or mutant CCR5 that had been seeded at 2.5 × 104 cells/well in 12-well plates 24 h earlier. Infectious titers were determined by placing the cells in selective medium and then fixing, staining, and counting the colonies that grew 7 to 10 days later (31, 52). Some infectivity assays were performed in the presence of various concentrations of the anti-CCR5 MAb 2D7 (BD Biosciences), the small-molecule CCR5 antagonist TAK-779 (obtained from the ARRRP) (4), or the peptide inhibitor T-20 (obtained from the ARRRP, contributed by Roche).
Infections with wild-type HIV-1JR-CSF and variants that had been adapted to use mutant CCR5 coreceptors were performed as previously described (53). Target cells were HeLa-CD4 cells stably or transiently expressing wild-type or mutant coreceptors (33-35, 53). Infectious titers for replication-competent HIV-1JR-CSF were obtained by the focal infectivity method of Chesebro et al. (10, 11). Briefly, at 72 h postinfection we fixed cells in 95% ethanol and then rinsed them in a physiological saline solution containing 1 mM EDTA. Foci of infection were visualized by immunoperoxidase staining as previously described (10, 11), using as primary antibody supernatant from the anti-HIV p24 hybridoma 183-H12-5C (12, 72), which had been obtained from the ARRRP and was contributed by Bruce Chesebro and Hardy Chen.
Infection kinetic assays.
Kinetics of infection were measured for HIV-gpt viruses pseudotyped with a CCR5(ΔNt)-adapted envelope with or without the V4 N-glycan deletion mutation. Briefly, this assay entails binding virus to target cells at 4°C by spinoculating at 188 × g for 2 h. The spinoculated cells are then rinsed in the cold and warmed to 37°C, and a completely inhibitory concentration (25 μM) of TAK-779 is then added at specific time points. The following day, the cells are placed in selective medium as described previously (31, 52). For more details, see the accompanying paper (50).
Fusion assays.
The syncytium-inducing ability of CCR5(ΔNt)-adapted envelopes was measured by coculturing envelope-transfected HEK293T cells with HeLa-CD4 target cells expressing wild-type or mutant CCR5 coreceptors as described previously (51).
RESULTS
Selection of HIV-1JR-CSF variants that efficiently replicate in HeLa-CD4/CCR5(ΔNt) cells.
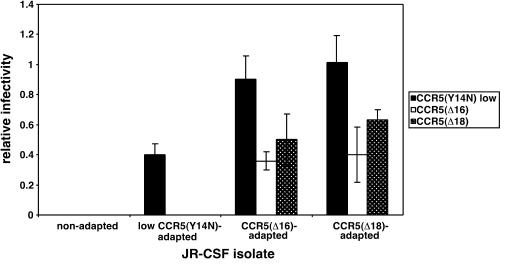
Wild-type HIV-1JR-CSF is unable to use CCR5(Δ16) or CCR5(Δ18). However, we previously isolated HIV-1JR-CSF variants that were adapted to replicate in HeLa-CD4/CCR5(Y14N) cells that contain a severe mutation in the CCR5 amino terminus (51). An initial selection in cells that contain a large amount of CCR5(Y14N) yielded an adapted variant that contained the V3 loop mutations S298N and F313L. This variant was further selected with cells that contained a low concentration of CCR5(Y14N), resulting in the additional V3 loop mutation N300Y. Interestingly, this adapted HIV-1JR-CSF virus was weakly able to infect our clones of HeLa-CD4/CCR5(Δ16) (clone R5d16.8) and HeLa-CD4/CCR5(Δ18) (clone R5d18.2) that express small amounts of these severely disabled coreceptors (i.e., the titers were approximately 0.04% of the titer in the JC.53 cells, which express a large optimal quantity of wild-type CCR5). Initially, we observed no cytopathic effect of the virus in these HeLa-CD4/CCR5(Δ16) and HeLa-CD4/CCR5(Δ18) cells, but abundant syncytia were seen during the fourth passages. After the sixth passage, the adapted virus population was highly syncytium inducing, and focal infectivity assays indicated approximately 3,500- and 1,000-fold increases in relative infectivities for these CCR5(Δ16)- and CCR5(Δ18)-adapted virus populations compared to the initial virus (Fig. 1). For the remainder of this paper, we refer to these CCR5(Δ16)- and CCR5(Δ18)-adapted viruses collectively as CCR5(ΔNt)-adapted viruses. These adapted virus populations were unable to infect HeLa-CD4 cells that lacked CCR5. Specifically, CCR5(ΔNt)-adapted viruses had large titers on HeLa-CD4/CCR5 cells (ca. 5 × 106 focus-forming units/ml) but had titers of zero on HeLa-CD4 cells, indicating that they had not gained the ability to use CXCR4.
FIG. 1.
Infectivity of wild-type and mutant CCR5-adapted HIV-1JR-CSF isolates in HeLa-CD4/CCR5 cell clones. Infections were performed with either wild-type HIV-1JR-CSF or uncloned virus populations that had been adapted to grow in HeLa-CD4 cell clones that expressed either low concentrations of CCR5(Y14N) [6.0 × 104 CCR5(Y14N) molecules/cell], low concentrations of CCR5(Δ16) [clone R5d16.8, 2.3 × 104 CCR5(Δ16) molecules/cell], or low concentrations of CCR5(Δ18) [clone R5d18.2, 2.7 × 104 CCR5(Δ18) molecules/cell]. Relative infectivities were determined by normalizing titers in each target cell to titers obtained in HeLa-CD4 cells expressing low concentrations of wild-type CCR5 (2.7 × 104 molecules/cell). The means from three independent assays are displayed. Error bars are standard errors of the means.
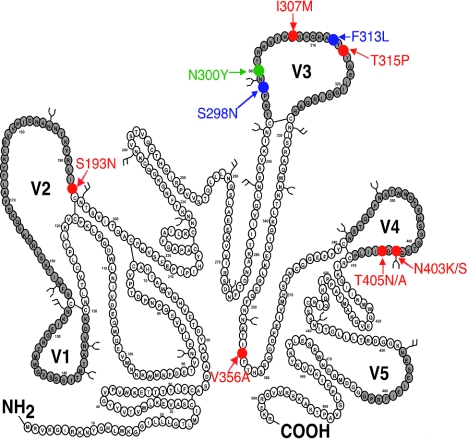
We then cloned and sequenced the entire env genes of the adapted viruses. Table 1 summarizes the mutations that we found in the env clones and the numbers of clones that contained each sequence. The env mutations were all situated within gp120 rather than gp41. As summarized in Fig. 2, the CCR5(ΔNt)-adapted viruses contained several new V3 loop mutations in addition to those that formed in the previous selections. Moreover, the CCR5(ΔNt)-adapted viruses contained four alternative V4 mutations (N403S, N403K, T405A, and T405N), all of which eliminate the same NDT consensus site for N-linked glycosylation. We also observed less frequent mutations at the base of the V2 loop (S193N) and in the C3 region of gp120 (V356A).
TABLE 1.
Adaptive mutations in HIV-1JR-CSF grown in HeLa-CD4/CCR5(ΔNt) cells
| Adaptive mutationsa | No. of clones |
|---|---|
| S298N, N300Y, T315P, N403S | 2 |
| S298N, N300Y, T315P, N403K | 1 |
| S298N, N300Y, T315P, T405N | 1 |
| S298N, N300Y, T315P, V356A, N403S | 9 |
| S298N, N300Y, I307M, F313L | 1 |
| S298N, N300Y, I307M, F313L, N403S | 1 |
| S298N, N300Y, I307M, F313L, N403K | 2 |
| S298N, N300Y, T315P, N403S | 2 |
| S193N, S298N, N300Y, T315P, N403K | 1 |
| S298N, N300Y, I307M, F313L | 4 |
| S193N, S298N, N300Y, I307M, F313L | 2 |
| S193N, S298N, N300Y, I307M, F313L, T405N | 1 |
| S298N, N300Y, I307M, F313L, N403K | 2 |
| S298N, N300Y, I307M, F313L, T405A | 2 |
| S298N, N300Y, I307M, F313L, T405N | 3 |
| S298N, N300Y, I307M, F313L, N403S | 1 |
We used a CCR5(Y14N)-adapted HIV-1JR-CSF virus population that could weakly replicate in HeLa-CD4/CCR5(ΔNt) cells to obtain our CCR5(Nt)-adapted mutants. Approximately 20% of the input virus had the NYL mutation that we had previously shown confers partial independence from the CCR5 N terminus. All of the adaptive mutations occurred in gp120.
FIG. 2.
Adaptive mutations generated during the passage of HIV-1JR-CSF in HeLa-CD4 cell clones expressing mutant coreceptors. The amino acid sequence of HIV-1JR-CSF is depicted. The variable loops are shaded gray, and N-linked glycosylation sites are indicated by the branched structures. Blue residues specify adaptive mutations (S298N and F313L) that arose during selection in HeLa-CD4 cells expressing high concentrations of CCR5(Y14N) [1.7 × 105 CCR5(Y14N) molecules/cell]. The single green residue indicates an additional mutation (N300Y) that arose when the high-CCR5(Y14N)-adapted viruses were then passaged in HeLa-CD4 cells expressing low concentrations of CCR5(Y14N) [6.0 × 104 CCR5(Y14N) molecules/cell]. Red residues depict adaptive mutations generated when CCR5(Y14N)-adapted viruses were grown in HeLa-CD4 cell clones expressing CCR5(Δ16) or CCR5(Δ18). These novel adaptive mutations are in V2(S193N), V3 (I307 M, T315P), and C3 (V356A). Multiple, alternative mutations in V4 destroy the NDT consensus sequence for N-linked glycosylation at residue 403.
We made env cDNAs that contain these mutations alone and in different combinations, and we transfected them into 293T cells to produce pseudotyped HIV-gpt virions for infectivity assays. Thus, as shown in Fig. 3, we analyzed the effects of the V2 loop and V4 N-glycan mutations in the contexts of different combinations of the V3 loop mutations. These diverse envelope glycoproteins and the wild-type glycoprotein were produced in equal amounts when their expression vectors were transfected into replica cultures of 293T cells, and they were also processed with similar efficiencies (results not shown). In addition, the pseudotyped viruses that were prepared in parallel all had similar titers (e.g., ranging from 4 × 105 to 8 × 105 CFU/ml) in HeLa-CD4/CCR5 cells that express wild-type CCR5. Thus, the wild-type CCR5 amino terminus had not become inhibitory for these adapted viruses.
FIG. 3.
Infectivity of HIV-gpt viruses pseudotyped with mutant CCR5-adapted HIV-1JR-CSF envelopes. HIV-gpt virions pseudotyped with the envelopes described in the bottom portion of the figure were used to infect HeLa-CD4 cell clones expressing similar levels of CCR5(Δ16) or CCR5(Δ18). Relative infectivities were determined by normalizing titers in the CCR5(ΔNt) cells to titers obtained in a HeLa-CD4 clone expressing similar levels of wild-type CCR5. The envelopes are grouped according to the particular mutations present in the V3 loop. The presence or absence of the V2 mutation (S193N) and the V4 N-glycan deletion mutation (N403S) is indicated by + and −, respectively. The data are averages from three or four independent assays, and error bars are standard errors of the means.
Figure 3 shows the titers of these pseudotyped viruses in our HeLa-CD4/CCR5(Δ16) and HeLa-CD4/CCR5(Δ18) cell clones (termed R5d16.8 and R5d18.2, respectively), which express approximately 2.7 × 104 molecules of these mutant coreceptors per cell. To control for differences between independently made virus preparations, we normalized the titer of each virus relative to its titer in a clone of HeLa-CD4/CCR5 cells that expresses the same amount of wild-type CCR5. As shown in Fig. 3, viruses with the wild-type gp120 V3 loop had background titers in both the R5d16.8 and R5d18.2 cells, regardless of the presence of the S193N V2 mutation and/or the V4 N-glycan deletion mutation. In contrast, viruses with CCR5(ΔNt)-adapted V3 loops had increased relative infectivities compared to the wild-type and to the starting CCR5(Y14N)-adapted viruses. Thus, single mutations in the V3 crown of I307M in the S298N, N300Y, F313L context or of T315P in the S298N, N300Y context contribute substantially to use of CCR5(Δ16) and CCR5(Δ18).
We also tested the adaptive contributions of the V2 and V4 mutations in the contexts of the CCR5(ΔNt)-adapted V3 loops (Fig. 3). In these contexts, the V2 loop mutation alone did not enhance utilizations of CCR5(Δ16) or CCR5(Δ18), whereas the V4 N-glycan deletion mutation alone strongly enhanced these utilizations. However, the greatest enhancements occurred when these mutations were present together. Thus, the V2 base mutation contributed to utilization of CCR5(Δ16) and CCR5(Δ18) only in the presence of the V4 N-glycan mutation. It should be noted that enhancing effects of the V2 and V4 loop mutations were most clearly evident when the efficiencies of infection were relatively low and were more difficult to discern in the context of the most highly adapted V3 loop, which contained all five substitutions. These results support the idea that the effects of the adaptive mutations are additive or synergistic and that their individual contributions are most easily detectable when the infectivities are suboptimal. For gp120s that function at nearly the same efficiency on the cells with CCR5(ΔNt) as on the cells with wild-type CCR5, as indicated by relative infectivities close to unity, additional mutations do not increase the efficiencies of infection. Because the conservative V356A mutation in the gp120 C3 region had only a modest effect on coreceptor utilization (data not shown), it was not investigated further.
Fusogenicities of CCR5(ΔNt)-adapted viruses with and without the V4 N-glycan at position 403.
We cocultured 293T cells that were transiently expressing the wild-type or mutant envelope glycoproteins with HeLa-CD4 target cells containing wild-type CCR5, CCR5(Δ16), or CCR5(Δ18). The tested envelope glycoproteins had roughly equal syncytium-inducing abilities with target cells expressing wild-type CCR5 (Fig. 4). In contrast, only the CCR5(ΔNt)-adapted envelope glycoproteins formed syncytia with target cells that contained CCR5(Δ16) or CCR5(Δ18). Removal of the V4 N-glycan at position N403 was associated with a strong enhancement in fusogenicity in these assays. Thus, both the V3 loop mutations and the V4 N-glycan loss mutation make important contributions to HIV-1 use of the CCR5(ΔNt) coreceptors.
FIG. 4.
Syncytium induction mediated by wild-type and mutant CCR5-adapted envelopes. HEK293T cells transiently expressing wild-type HIV-1JR-CSF envelope, CCR5(Y14N)-adapted envelope, or CCR5(ΔNt)-adapted envelopes with (+) or without (−) the V4 403 N-glycan were cocultured with HeLa-CD4 cells expressing similar levels of wild-type CCR5, CCR5(Δ16), or CCR5(Δ18). The envelopes are grouped according to the V3 loop sequence. Relative fusogenicity values were obtained for each envelope construct by normalizing fusogenicity with a given target cell to the fusion of wild-type HIV-1JR-CSF envelope with cells expressing wild-type CCR5. The means from one representative experiment performed in triplicate are displayed; error bars are standard errors of the means.
Removal of the N403 N-glycan in gp120 accelerates a postassembly step in the entry pathway.
Because the tyrosine sulfate-containing amino-terminal region of CCR5 is a major site for association with gp120 (22, 25, 27, 34, 35), HIV-1 affinities for CCR5(Δ16) and CCR5(Δ18) are greatly reduced compared to their affinities for wild-type CCR5 (see below). Consequently, we initially anticipated that the adaptive viral mutations that facilitate highly efficient use of these CCR5(ΔNt) coreceptors would enhance viral affinities for these severely damaged coreceptors. However, this hypothesis was difficult to reconcile with the fact that the N-glycan at position N403 is in the V4 loop, which is on the silent face of gp120 far removed from the presumptive site of CCR5 interaction with gp120 (37, 38, 78). In addition, we found that the adaptive mutations that contribute to CCR5(ΔNt) utilization also caused large increases in efficiencies of CCR5(Y14N) utilization but had negligible effects on viral affinities for CCR5(Y14N) as seen by the titers of the adapted viruses in our previously characterized panel of HeLa-CD4/CCR5(Y14N) cell clones, which contain different amounts of CCR5(Y14N). Thus, the amounts of cell surface CCR5(Y14N) required for optimal infections (i.e., the 50% effective concentrations in these assays) were only marginally altered by these adaptive viral mutations (results not shown) (51). These results raised the interesting possibility that the adaptive viral mutations were increasing the efficiencies of CCR5(ΔNt) usage without significantly enhancing the viral affinities for the damaged coreceptors.
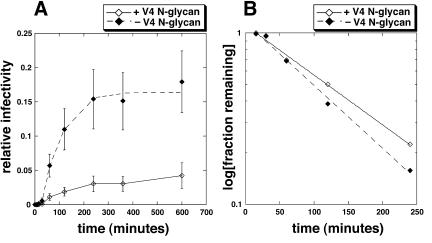
To further investigate this issue, we used our recently described kinetic method (50). Specifically, we compared the entry kinetics into HeLa-CD4/CCR5(Δ18) cells of HIV-gpt viruses pseudotyped with two CCR5(Δ18)-adapted envelope constructs, one containing the V3 loop mutations S298N, N300Y, and T315P and the other containing this same V3 loop but also lacking the V4 N-glycan at residue 403. Briefly, we bound the viruses to the cells at 4°C, washed the cultures with fresh medium at 4°C, and then initiated the infections by warming to 37°C. At different subsequent times, we added fully inhibitory concentrations of the small-molecule CCR5 antagonist TAK-779 to block infections that had not progressed beyond the CCR5-dependent steps of the entry pathway. Relative infectivity values (i.e., the titer of the virus in each culture was normalized to the final 10-h titer in cells that contain wild-type CCR5) were plotted versus time. Figure 5A shows the infection kinetics of the two pseudotyped viruses in the HeLa-CD4/CCR5(Δ18) cells. As described elsewhere, salient features of the entry kinetics include a lag phase preceding the completion of any infectious virus entry and a plateau phase when the entry has reached completion (50). The length of the lag phase is influenced by the concentration of receptor on the cells and its affinity for the virus, and it is consequently prolonged by reducing the CCR5 concentration, by mutations in CCR5 or in the virus that reduce their affinity of association, and by competitive entry inhibitors that bind to CCR5 and thereby reduce the pool available to the virus (50). Thus, the lag is required for the concentration-dependent assembly of viral complexes with CCR5 that are competent to undergo further steps of infection. However, the assemblages are reversible because competitive CCR5 inhibitors such as TAK-779 block entry efficiently even when added after the lag has been completed. In contrast, the subsequent rates of entry and the final relative infectivity values at the plateaus are determined principally by a kinetic competition between two opposing processes, one involving a rate-limiting CCR5-dependent conformational change in gp41 that results in infectious virus entry and the other being a constitutive process of viral inactivation that probably involves its endocytosis and degradation in lysosomes (43, 66). In agreement with the data in Fig. 3, the pseudotyped virus with the CCR5(ΔNt)-adapted envelope lacking the V4 N-glycan is more infectious for these CCR5(Δ18)-containing cells than the virions that have the wild-type V4 loop sequence. However, both viruses have the same lag times. Mathematical analysis of the infectivity data according to the equations in the accompanying paper (50) confirmed that the lag phases are identical for the adapted viruses (these lags are seen as the intercepts on the upper axis in Fig. 5B). These results strongly suggest that the V4 N-glycan has no effect on the affinity or kinetics of association of the virus with CCR5(Δ18). In addition, the difference in the slopes of the lines in Fig. 5B demonstrates that the CCR5(ΔNt)-adapted virus lacking the V4 N-glycan enters the cells at a higher rate than the adapted virus that has the wild-type V4 N-glycan at position N403. Because these viruses do not differ in their degrees of association with CCR5(Δ18), as described above, the slope difference in Fig. 5B implies that the V4 N-glycan increases the activation energy barrier for the rate-limiting CCR5-dependent postassembly step in entry and that its removal therefore lowers this activation energy barrier and enables the CCR5(Δ18) coreceptor to function more efficiently. Specifically, according to our kinetic model, these slopes are equal to −(αki + k2)/2.3, where α is a measure of virus-coreceptor association, ki is the rate constant for cellular entry of fully assembled virus-coreceptor complexes, and k2 is the rate constant for constitutive virus inactivation (50). Since α and k2 are the same for both viruses in this study, we conclude that the slope difference must be due to a difference in ki. As described elsewhere, ki is limited principally by a CCR5-dependent slow conformational change in gp41 (50). These conclusions were supported by additional evidence described below.
FIG. 5.
Infection kinetics mediated by CCR5(Δ18)-adapted envelopes in HeLa-CD4 cells expressing CCR5(Δ18). (A) Infection kinetics of HIV-gpt viruses pseudotyped with CCR5(Δ18)-adapted envelopes (V3 loop sequence S298N, N300Y, T315P) with or without the V4 N-glycan. Infections were terminated at each time point upon the addition of 25 μM TAK-779. Relative infectivity values were obtained by normalizing titers for each virus in CCR5(Δ18) cells at each time point to titers in HeLa-CD4 cells expressing a large amount of wild-type CCR5 at the final time point (10 h). The data were generated from the averages of three independent experiments. Error bars are standard errors of the means. (B) Mathematical analysis of the infectivity data. Infectivity data were analyzed according to the equations in reference 50. The x intercept denotes the lag phase and is the same for both viruses (∼15 min). The adapted virus lacking the V4 N-glycan has the steeper slope, indicating that it enters CCR5(Δ18) cells with a greater rate constant than the adapted virus with wild-type V4.
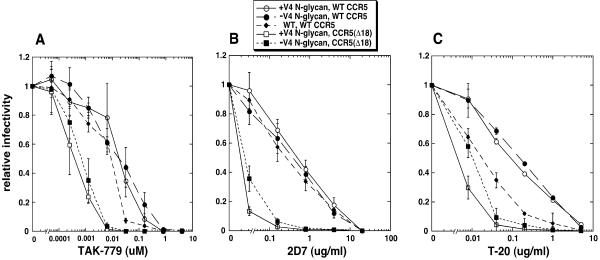
Sensitivities of CCR5(ΔNt)-adapted viruses to entry inhibitors.
To gain a further understanding of the mechanism by which adaptive mutations in V3 and the N-glycan loss in V4 enhance use of CCR5(ΔNt) coreceptors, we assayed the sensitivities of HIV-gpt viruses pseudotyped with these CCR5(ΔNt)-adapted envelopes to the entry inhibitors TAK-779, 2D7, and T-20. TAK-779 is a small-molecule CCR5 antagonist that competitively inhibits gp120 binding by interacting with a hydrophobic pocket within CCR5 (4, 23); 2D7 is a monoclonal antibody that binds to an epitope in ECL2 of CCR5 and prevents 125I-gp120 binding and HIV-1 infections (76); and T-20 binds to the three-stranded prehairpin conformational intermediate formed by gp41, thus inhibiting the fusion reaction (24). We analyzed the inhibitor sensitivities of HIV-gpt viruses pseudotyped with the wild-type JR-CSF envelope and two CCR5(ΔNt)-adapted envelopes that both had the V3 loop mutations S298N, N300Y, and T315P and with one of these also lacking the V4 loop N403 N-glycan. The target cells used in these assays were HeLa-CD4 cells expressing either wild-type CCR5 or CCR5(Δ18), both of which expressed similar amounts of coreceptor (∼2.7 × 104 molecules/cell). Results for these inhibitor studies are shown in Fig. 6. Figure 6A shows the TAK-779 dose response. The TAK-779 sensitivities of the wild-type and mutant viruses in HeLa-CD4 cells expressing wild-type CCR5 are all similar. In light of recent evidence that gp120 molecules with greater affinity for CCR5 have decreased sensitivity to TAK-779 (59), the results in Fig. 6A suggest that the CCR5(ΔNt)-adapted viruses do not have altered affinities for wild-type CCR5 compared to the wild-type virus. Interestingly, the CCR5(ΔNt)-adapted viruses with and without the V4 N-glycan both had approximately 30-fold-increased TAK-779 sensitivities when assayed on cells with CCR5(Δ18) (50% inhibitory concentration, ∼0.001 μM) compared to cells with wild-type CCR5 (50% inhibitory concentration, ∼0.03 μM). These results confirm that the CCR5(ΔNt)-adapted viruses bind weakly to CCR5(Δ18) and that tight coreceptor binding is not a mechanism used by these adapted viruses to circumvent inefficient coreceptor functioning. The TAK-779 sensitivities of the CCR5(ΔNt)-adapted viruses were unaffected by the V4 N-glycan, indicating that removal of this N-glycan does not enhance utilization of CCR5(Δ18) by increasing viral affinity for this damaged coreceptor, in agreement with our kinetic data in Fig. 5. Figure 6B shows the effects of the 2D7 MAb. The overall pattern we observe is similar to the results shown in Fig. 6A [i.e., CCR5(ΔNt)-adapted viruses have increased inhibitor sensitivities in cells with CCR5(Δ18) compared to cells with wild-type CCR5, and they have inhibitor sensitivities similar to those of the wild-type virus in cells expressing wild-type CCR5], supporting the conclusion that these adapted viruses do not assemble more efficiently with coreceptors. We do observe a very slight increase in 2D7 resistance in cells with CCR5(Δ18) of the adapted virus that lacks the V4 N-glycan. This result may reflect a mechanism of inhibition by 2D7 that is only partially competitive (50), and it suggests that removal of this N-glycan may also enhance a postassembly step in the utilization of this coreceptor. In contrast to the interpretation in a previous report (1), we also found that removal of the V4 N-glycan in the context of the wild-type JR-CSF envelope also had no effect on the viral 2D7 sensitivity (data not shown).
FIG. 6.
Inhibitor sensitivities of HIV-gpt virions pseudotyped with wild-type and CCR5(Δ18)-adapted envelopes. HIV-gpt viruses pseudotyped with wild-type (WT) or CCR5(Δ18)-adapted envelopes (V3 loop sequence S298N, N300Y, T315P) with (+) or without (−) the V4 N-glycan at position 403 were used to infect HeLa-CD4 cells expressing similar amounts of wild-type CCR5 or CCR5(Δ18) in the absence or presence of various amounts of inhibitors. Relative infectivity values were calculated for each virus in a given cell line by dividing by the titers obtained in the absence of inhibitor in the same cell line. (A) TAK-779; (B) 2D7; (C) T-20. Error bars are standard errors of the means; n = 3 to 6.
Figure 6C displays the T-20 dose-response curves of the wild-type and adapted viruses. In contrast to curves generated with competitive inhibitors such as TAK-779 or 2D7, these curves show that the CCR5(ΔNt)-adapted viruses have substantially reduced sensitivities to T-20 compared to wild-type viruses when assayed in cells expressing wild-type CCR5. Since T-20 targets the gp41 prehairpin structure, the resolution of which is required for fusion, we conclude that the prehairpin intermediate is resolved more rapidly in adapted viruses than in wild-type viruses. The T-20 sensitivities are identical in adapted viruses with and without the V4 N-glycan in cells expressing wild-type CCR5 because this coreceptor is functioning maximally. However, when these adapted viruses use a coreceptor that functions suboptimally, such as CCR5(Δ18), they become much more sensitive to T-20, and in this circumstance, removal of the V4 N-glycan significantly decreases this sensitivity. These results suggest that deletion of the CCR5 amino terminus lowers virus-coreceptor affinity (Fig. 6A and B) and reduces the rate of the T-20-sensitive conformational change in gp41 (Fig. 6C). The adaptive mutations in the V3 loop and in the V4 N403 site counteract these limitations by increasing the rate of a specific postassembly step, the resolution of the T-20-sensitive prehairpin intermediate. Thus, the adaptive mutations may lower the activation energy barrier for this conformational change and thereby make this limiting step more easily triggered by a weak coreceptor.
Effects of the V4 N-glycan mutation on viral utilization of a mutant CCR5 with a damaged extracellular loop 2.
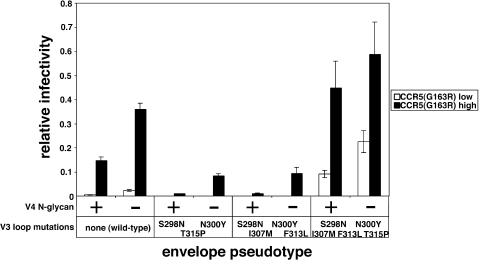
Based on the above evidence that the viral mutations that facilitate use of CCR5(ΔNt) may make the virus more hair-trigger by lowering the activation energy barrier required for a rate-limiting conformational change in gp41, we attempted to learn whether these mutations would have a global effect on mutant CCR5 use. Consequently, we tested the ability of our CCR5(ΔNt)-adapted viruses with and without the V4 N-glycan to use the ECL2 mutant CCR5(G163R). As shown in Fig. 7, removal of the V4 N-glycan at position N403 substantially increased viral utilizations of CCR5(G163R) in the contexts of all tested V3 loops. The viruses with the mutant V3(S298N, N300Y, T315P) loop or the V3(S298N, N300Y, I307 M, F313L) loop were less efficient at using the CCR5(G163R) coreceptor than the wild-type virus, in agreement with previous evidence that the N300Y mutation confers extreme sensitivity to the G163R mutation (51). However, the CCR5(ΔNt)-adapted envelope with all five V3 mutations was able to use CCR5(G163R) relatively efficiently, indicating that the I307M and T315P mutations overcome the inhibitory effect of the N300Y mutation on CCR5(G163R) use. Considered together, these results strongly suggest that the adaptive mutations that facilitate use of CCR5(Δ18) also enhance use of CCR5(G163R). This is especially true of the V4 mutation that eliminates the N-glycan at position N403, but it is also the case for the I307 M and T315P mutations in the V3 loop.
FIG. 7.
Infectivity of wild-type and CCR5(Δ18)-adapted viruses in HeLa-CD4 cells expressing CCR5(G163R). HIV-gpt viruses pseudotyped with wild-type or CCR5(ΔNt)-adapted envelopes, with (+) or without (−) the V4 N-glycan, were used to infect HeLa-CD4 cells expressing low (1.9 × 104 molecules/cell) or high (9.8 × 104 molecules/cell) concentrations of CCR5(G163R) (34, 67). Relative infectivities were determined by normalizing titers in each target cell to titers obtained in HeLa-CD4 cells expressing large amounts of wild-type CCR5. The averages from three independent assays are shown, and error bars are standard errors of the means.
These results could possibly be interpreted in two ways. One is that the CCR5(ΔNt)-adapted envelopes may have become reliant on a region of CCR5 other than either the amino terminus or the G163 region. Alternatively, they may be able to efficiently and interchangeably use either the amino terminus or the G163 region without requiring both sites. In contrast, the wild-type virus functions efficiently only if both CCR5 sites are present. To discern between these two possibilities, we tested our CCR5(ΔNt)-adapted virus populations for their abilities to use the CCR5(Δ18,G163R) double mutant. Immunofluorescence microscopy revealed that CCR5(Δ18,G163R) was highly expressed at the cell surface (data not shown). As shown in Fig. 8, the wild-type and the CCR5(Y14N)-adapted viruses were able to use CCR5(G163R) much more efficiently than they used CCR5(Δ18) or the double mutant. In contrast, the CCR5(ΔNt)-adapted viruses were able to efficiently use CCR5(Δ18), although they were also unable to use the double mutant. These results strongly suggest that the CCR5(ΔNt)-adapted viruses have not become reliant on another region of CCR5. Rather, they can use the CCR5 amino-terminal region or the G163 ECL2 regions efficiently and interchangeably.
FIG. 8.
Infectivities of wild-type and CCR5(ΔNt)-adapted HIV-1 in HeLa-CD4 cells expressing mutant coreceptors. Replication competent preparations of wild-type JR-CSF, CCR5(Δ16)-adapted virus, or CCR5(Δ18)-adapted virus populations were used to infect HeLa-CD4 cells transiently expressing CCR5(G163R), CCR5(Δ18), or CCR5(Δ18,G163R). Relative infectivity values were obtained by dividing titers obtained in each transfected cell line by titers generated in HeLa-CD4 cells transfected with wild-type CCR5. The high relative infectivity values for CCR5(G163R)-transfected cells exhibited by these replication-competent viruses are caused by the high titers of the replication-competent viruses and the high expression level of CCR5(G163R) in this transient assay (34, 51). The averages from two or three independent experiments are shown, and error bars are the ranges or standard errors of the means.
DISCUSSION
Isolation of adapted HIV-1JR-CSF variants that efficiently use CCR5(Δ16) and CCR5(Δ18).
Previously, we reported the isolation of HIV-1JR-CSF variants that efficiently replicate in HeLa-CD4 cells that express a low concentration of CCR5(Y14N) (51). The Y14N mutation causes severe damage to the amino-terminal region of CCR5 by eliminating an important site for tyrosine sulfation (17, 25, 27, 28) and by creating an NYT consensus site for N-linked glycosylation (34). The adapted HIV-1JR-CSF variants that resulted from the latter selections had a reduced dependency on the CCR5 amino terminus and an increased reliance on the CCR5 ECL2, as revealed by their weak but improved ability to use CCR5(Δ16) and CCR5(Δ18) and by their increased sensitivity to inhibition by the proximal ECL2 mutation G163R (51). The adaptations found in this previous study were all within the V3 loop of gp120 and consisted of S298N, N300Y, and F313L (51). These mutations caused only minimal increases in viral affinities for CCR5(Y14N), and their major effects were to increase the efficiencies of a rate-limiting CCR5-dependent step of infection that follows assembly of the essential virus-CD4-CCR5(Y14N) complexes (50, 51). Specifically, these adaptive mutations decreased the viral sensitivities to T-20, implying that they lower the activation energy barrier that retards the conversion of the three-stranded coil prehairpin conformation of gp41 into a six-helix bundle (50).
In this investigation, we have further weaned HIV-1JR-CSF from all dependency on the CCR5 amino terminus by selecting for viral replication in HeLa-CD4/CCR5(Δ16) and HeLa-CD4/CCR5(Δ18) cell clones that express relatively low concentrations of these mutant coreceptors. Thus, our present results establish that HIV-1JR-CSF can abandon its reliance on the tyrosine-sulfated amino-terminal region of CCR5 by a surprisingly small number of adaptive substitutions that seem to occur entirely within the variable loop regions of gp120. Interestingly, these new adaptive mutations have additive or synergistic effects on utilization of CCR5(Δ16) and CCR5(Δ18), and they occur not only in the crown of the V3 loop (i.e., I307M and T315P) but also in the V2 base (S193N) and in a functionally important site for N-linked glycosylation in the V4 loop (N403K/S or T405N/A). The locations of these new mutations within gp120 are indicated in the schematic diagram in Fig. 2. The V3 loop and V2 stem substitutions are consistent with previous evidence that these sites occur in the coreceptor recognition region of gp120. However, the V4 N-glycan mutations are surprising because they are distant from the coreceptor interaction region (38, 77, 78), because they have such a prominent and strong effect on utilization of the CCR5(Δ16) and CCR5(Δ18) coreceptors (Fig. 3), and for the mechanistic reasons discussed below.
Functions of the V4 N-glycan at position 403.
We believe that the CCR5(ΔNt) adaptive mutations, including the mutations that eliminate the V4 N-glycan, have important mechanistic implications concerning HIV-1 infections. As discussed above, the N300Y V3 loop mutation that contributed to HIV-1JR-CSF utilization of CCR5(Y14N) caused a reduced dependency of the virus on the CCR5 amino terminus and a correspondingly increased reliance on the ECL2 region (51). Such shifts in dependency between different CCR5 regions imply that the virus makes functionally redundant, collaborative contacts with different sites in the coreceptor, so that a weakening in one contact site can be compensated for by increases at another site. The V4 N-glycan role is somewhat unexpected from this perspective because it occurs far from the coreceptor interaction region of gp120 (38, 77, 78) and because the V2 base substitution S193N seems to contribute toward use of CCR5(Δ18) only in the absence of the V4 N-glycan (Fig. 3). Furthermore, consistent with its position far from the coreceptor binding region of gp120, our results suggest that removal of the V4 N-glycan does not increase the affinity of gp120 for CCR5(Δ18) (Fig. 5 and 6). This idea is supported by both kinetic and inhibitor data. Specifically, our kinetic assays reveal that the CCR5(ΔNt)-adapted mutants with and without the V4 N-glycan have identical lag phases in HeLa-CD4/CCR5(Δ18) cells, indicating that the V4 N-glycan does not alter the virus-coreceptor assembly process despite its strong inhibitory effect on the rate and efficiency of infection (Fig. 5). These results suggest that removal of the V4 N-glycan increases a rate-limiting CCR5-dependent postassembly step in the entry pathway. Furthermore, CCR5(ΔNt)-adapted viruses both in the presence and absence of the V4 mutation had identical sensitivities to TAK-779 on CCR5(Δ18) cells and on cells expressing wild-type CCR5, further indicating that this mutation does not affect CCR5 affinity (Fig. 6A). Moreover, we find that removal of the V4 N-glycan enhances use of an additional mutant coreceptor, CCR5(G163R), that is damaged in ECL2 (Fig. 6A). Thus, loss of the V4 N-glycan appears to globally enhance the use of inefficiently functioning mutant coreceptors that are damaged at different sites.
We can envision two general models to explain this result. First, loss of the V4 N-glycan could convert the gp120-gp41 complexes into hair-trigger variants that adopt a fusogenic conformation readily, even in the absence of any strong coreceptor interactions. Thus, according to this model, we envision that loss of this N-glycan could lower the activation energy barrier for a slow, irreversible, rate-limiting CCR5-dependent change in gp41 conformation (50). Thereby, the gp120-gp41 complexes would become less dependent on making multiple contacts with CCR5. By this mechanism, the virus would become “ambidextrous,” able to interchangeably use one or another of several alternative sites in CCR5. A second model would be that loss of the V4 N-glycan might increase viral affinity for a region of CCR5, such as ECL3, distinct from the amino terminus or ECL2. We strongly favor the first model because the V4 loop is distant from the coreceptor binding region, which makes it difficult to imagine that it would influence virus binding to a specific region of CCR5 (38, 77, 78), and because loss of the V4 N-glycan does not appear to alter virus-CCR5 affinity, as discussed above (Fig. 5 and 6). In addition, our adapted viruses were unable to use a double mutant, CCR5(Δ18,G163R), indicating that they have not become adapted to rely on alternative regions of CCR5, such as ECL1 or ECL3. Furthermore, our kinetic, fusion, and T-20 inhibition assays indicate that removal of the V4 N-glycan increases the efficiency of postassembly steps. Specifically, elimination of this N-glycan causes the gp41 prehairpin intermediate to be resolved more rapidly, and this ultimately results in more syncytium formation in HeLa-CD4 cells expressing CCR5(ΔNt) (Fig. 4 and 6C). Based on all of these considerations, we propose that the V4 N-glycan strongly influences the activation energy barrier for a limiting step involving conversion of the gp120-gp41 complexes into a fusogenic conformation. An N-glycan in this area of V4 is highly conserved in gp120, although it is not universally present and its precise position is subject to some variations.
Our evidence suggests that the adaptive V3 loop mutations also may affect this same slow step in viral entry without significantly influencing the virus-CCR5(Δ18) affinities. This is implied in part by the inability of these V3 mutations to significantly alter the viral sensitivities to TAK-779 or to the 2D7 monoclonal antibody in cells that contain wild-type CCR5 (Fig. 6A). In contrast, in these same cells the adaptive V3 loop mutations strongly reduce the viral sensitivities to inhibition by T-20, which binds to the prehairpin intermediate gp41 conformation (Fig. 6C). These V3 loop mutations also increase viral fusogenicities (Fig. 4) and contribute to the more efficient use of both CCR5(Δ18) and CCR5(G163R) (Fig. 7). Thus, these V3 loop mutations in conjunction with the other adaptive mutations allow the virus to more efficiently use single interaction sites in CCR5.
Interestingly, several other N-glycans influence coreceptor usages (32, 42, 54, 55). Most notably, the V3 N-glycan at N301 contributes to the switch between CCR5 and CXCR4 specificity (54). Furthermore, removal of the N-glycan at the amino terminus of CXCR4 enables it to be used by R5 strains of HIV-1 (7). Recent evidence indicates that variably situated gp120 N-glycans also form a critically important glycan shield that protects HIV-1 from neutralizing antibodies (8, 32, 41, 55, 60, 74, 77, 78). The highly neutralizing 2G12 monoclonal antibody recognizes a series of N-glycans that are situated on the “silent face” of gp120 (6), near the functional N403 N-glycan analyzed in this report.
Finally, we emphasize that the functional effects of important adaptive changes in gp120 can be discerned only under specific assay conditions. For example, we were able to readily detect strong effects of the V2 base mutation and of the V4 N-glycan mutations on infections of HeLa-CD4/CCR5(Δ18) cells that express a relatively low concentration of this mutant coreceptor. In contrast, as described above, the effects of these mutations were more difficult to detect in situations in which the gp120-CCR5 complexes functioned more efficiently. Our results suggest that removal of the V4 N-glycan and changes in the V3 loop lower the activation energy for a limiting step in the membrane fusion reaction, thus increasing the rate and efficiency of infection. However, this effect would be difficult to observe in cells with wild-type CCR5, because in that circumstance the gp120 is already functioning in a rapid and efficient manner. In this context it is notable that late in AIDS progression the pool of susceptible cells changes substantially, with cells containing CXCR4 and small amounts of CCR5 becoming more predominant. Similar to our CCR5(Δ18)-adapted viruses, the variants that use CXCR4 are less reliant on the coreceptor amino terminus and are believed to be more syncytium inducing. However, after infection of a new host, the absence of these selection pressures would be expected to lead to their loss in the viral population, as is typically observed.
Acknowledgments
This research was supported by NIH grant CA67358.
We thank our colleagues Kristine Rose, Susan Kozak, Mariana Marin, Sheetal Golem, and James Durnin for encouragement and helpful suggestions.
REFERENCES
- 1.Aarons, E. J., S. Beddows, T. Willingham, L. Wu, and R. A. Koup. 2001. Adaptation to blockade of human immunodeficiency virus type 1 entry imposed by the anti-CCR5 monoclonal antibody 2D7. Virology 287:382-390. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 3.Atchison, R. E., J. Gosling, F. S. Monteclaro, C. Franci, L. Digilio, I. F. Charo, and M. A. Goldsmith. 1996. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science 274:1924. [DOI] [PubMed] [Google Scholar]
- 4.Baba, M., O. Nishimura, N. Kanzaki, M. Okamoto, H. Sawada, Y. Iizawa, M. Shiraishi, Y. Aramaki, K. Okonogi, Y. Ogawa, K. Meguro, and M. Fujino. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. USA 96:5698-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieniasz, P. D., R. A. Fridell, I. Aramori, S. S. Ferguson, M. G. Caron, and B. R. Cullen. 1997. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 16:2599-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065-2071. [DOI] [PubMed] [Google Scholar]
- 7.Chabot, D. J., H. Chen, D. S. Dimitrov, and C. C. Broder. 2000. N-linked glycosylation of CXCR4 masks coreceptor function for CCR5-dependent human immunodeficiency virus type 1 isolates. J. Virol. 74:4404-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chackerian, B., L. M. Rudensey, and J. Overbaugh. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 71:7719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 10.Chesebro, B., and K. Wehrly. 1988. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J. Virol. 62:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chesebro, B., K. Wehrly, J. Metcalf, and D. E. Griffin. 1991. Use of a new CD4-positive HeLa cell clone for direct quantitation of infectious human immunodeficiency virus from blood cells of AIDS patients. J. Infect. Dis. 163:64-70. [DOI] [PubMed] [Google Scholar]
- 12.Chesebro, B., K. Wehrly, J. Nishio, and S. Perryman. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 66:6547-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 14.Cocchi, F., A. L. DeVico, A. Garzino-Demo, A. Cara, R. C. Gallo, and P. Lusso. 1996. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat. Med. 2:1244-1247. [DOI] [PubMed] [Google Scholar]
- 15.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cormier, E. G., M. Persuh, D. A. Thompson, S. W. Lin, T. P. Sakmar, W. C. Olson, and T. Dragic. 2000. Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 97:5762-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cormier, E. G., D. N. Tran, L. Yukhayeva, W. C. Olson, and T. Dragic. 2001. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J. Virol. 75:5541-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 19.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. C. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149-1158. [DOI] [PubMed] [Google Scholar]
- 20.Douek, D. C., L. J. Picker, and R. A. Koup. 2003. T cell dynamics in HIV-1 infection. Annu. Rev. Immunol. 21:265-304. [DOI] [PubMed] [Google Scholar]
- 21.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 22.Dragic, T., A. Trkola, S. W. Lin, K. A. Nagashima, F. Kajumo, L. Zhao, W. C. Olson, L. Wu, C. R. Mackay, G. P. Allaway, T. P. Sakmar, J. P. Moore, and P. J. Maddon. 1998. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J. Virol. 72:279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dragic, T., A. Trkola, D. A. Thompson, E. G. Cormier, F. A. Kajumo, E. Maxwell, S. W. Lin, W. Ying, S. O. Smith, T. P. Sakmar, and J. P. Moore. 2000. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. USA 97:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 25.Farzan, M., H. Choe, L. Vaca, K. Martin, Y. Sun, E. Desjardins, N. Ruffing, L. Wu, R. Wyatt, N. Gerard, C. Gerard, and J. Sodroski. 1998. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J. Virol. 72:1160-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farzan, M., S. Chung, W. Li, N. Vasilieva, P. L. Wright, C. E. Schnitzler, R. J. Marchione, C. Gerard, N. P. Gerard, J. Sodroski, and H. Choe. 2002. Tyrosine-sulfated peptides functionally reconstitute a CCR5 variant lacking a critical amino-terminal region. J. Biol. Chem. 277:40397-40402. [DOI] [PubMed] [Google Scholar]
- 27.Farzan, M., T. Mirzabekov, P. Kolchinsky, R. Wyatt, M. Cayabyab, N. P. Gerard, C. Gerard, J. Sodroski, and H. Choe. 1999. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96:667-676. [DOI] [PubMed] [Google Scholar]
- 28.Farzan, M., N. Vasilieva, C. E. Schnitzler, S. Chung, J. Robinson, N. P. Gerard, C. Gerard, H. Choe, and J. Sodroski. 2000. A tyrosine-sulfated peptide based on the N terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry. J. Biol. Chem. 275:33516-33521. [DOI] [PubMed] [Google Scholar]
- 29.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 30.Hazenberg, M. D., D. Hamann, H. Schuitemaker, and F. Miedema. 2000. T cell depletion in HIV-1 infection: how CD4+ T cells go out of stock. Nat. Immunol. 1:285-289. [DOI] [PubMed] [Google Scholar]
- 31.Kabat, D., S. L. Kozak, K. Wehrly, and B. Chesebro. 1994. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J. Virol. 68:2570-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolchinsky, P., E. Kiprilov, P. Bartley, R. Rubinstein, and J. Sodroski. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 75:3435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozak, S. L., E. J. Platt, N. Madani, F. E. Ferro, Jr., K. Peden, and D. Kabat. 1997. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J. Virol. 71:873-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhmann, S. E., E. J. Platt, S. L. Kozak, and D. Kabat. 2000. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J. Virol. 74:7005-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuhmann, S. E., E. J. Platt, S. L. Kozak, and D. Kabat. 1997. Polymorphisms in the CCR5 genes of African green monkeys and mice implicate specific amino acids in infections by simian and human immunodeficiency viruses. J. Virol. 71:8642-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 37.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure Fold. Des. 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 38.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, B., M. Sharron, C. Blanpain, B. J. Doranz, J. Vakili, P. Setoh, E. Berg, G. Liu, H. R. Guy, S. R. Durell, M. Parmentier, C. N. Chang, K. Price, M. Tsang, and R. W. Doms. 1999. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J. Biol. Chem. 274:9617-9626. [DOI] [PubMed] [Google Scholar]
- 40.Liu, R., W. A. Paxton, S. Choe, D. Ceradini, S. R. Martin, R. Horuk, M. E. MacDonald, H. Stuhlmann, R. A. Koup, and N. R. Landau. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367-377. [DOI] [PubMed] [Google Scholar]
- 41.Lue, J., M. Hsu, D. Yang, P. Marx, Z. Chen, and C. Cheng-Mayer. 2002. Addition of a single gp120 glycan confers increased binding to dendritic cell-specific ICAM-3-grabbing nonintegrin and neutralization escape to human immunodeficiency virus type 1. J. Virol. 76:10299-102306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malenbaum, S. E., D. Yang, L. Cavacini, M. Posner, J. Robinson, and C. Cheng-Mayer. 2000. The N-terminal V3 loop glycan modulates the interaction of clade A and B human immunodeficiency virus type 1 envelopes with CD4 and chemokine receptors. J. Virol. 74:11008-11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marechal, V., F. Clavel, J. M. Heard, and O. Schwartz. 1998. Cytosolic Gag p24 as an index of productive entry of human immunodeficiency virus type 1. J. Virol. 72:2208-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCune, J. M. 2001. The dynamics of CD4+ T-cell depletion in HIV disease. Nature 410:974-979. [DOI] [PubMed] [Google Scholar]
- 45.Moore, J. P., and J. Binley. 1998. HIV. Envelope's letters boxed into shape. Nature 393:630-631. [DOI] [PubMed] [Google Scholar]
- 46.Moore, J. P., B. A. Jameson, R. A. Weiss, and Q. J. Sattentau. 1993. The HIV-cell fusion reaction, p. 234-289. In J. Bentz (ed.), Viral fusion mechanisms. CRC Press, Boca Raton, Fla.
- 47.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olson, W. C., G. E. Rabut, K. A. Nagashima, D. N. Tran, D. J. Anselma, S. P. Monard, J. P. Segal, D. A. Thompson, F. Kajumo, Y. Guo, J. P. Moore, P. J. Maddon, and T. Dragic. 1999. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J. Virol. 73:4145-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Picard, L., G. Simmons, C. A. Power, A. Meyer, R. A. Weiss, and P. R. Clapham. 1997. Multiple extracellular domains of CCR-5 contribute to human immunodeficiency virus type 1 entry and fusion. J. Virol. 71:5003-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Platt, E. J., J. P. Durnin, and D. Kabat. 2005. Kinetic factors control efficiencies of cell entry, efficacies of entry inhibitors, and mechanisms of adaptation of human immunodeficiency virus. J. Virol. 79:4347-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Platt, E. J., S. E. Kuhmann, P. P. Rose, and D. Kabat. 2001. Adaptive mutations in the V3 loop of gp120 enhance fusogenicity of human immunodeficiency virus type 1 and enable use of a CCR5 coreceptor that lacks the amino-terminal sulfated region. J. Virol. 75:12266-12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Platt, E. J., N. Madani, S. L. Kozak, and D. Kabat. 1997. Infectious properties of human immunodeficiency virus type 1 mutants with distinct affinities for the CD4 receptor. J. Virol. 71:883-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pollakis, G., S. Kang, A. Kliphuis, M. I. Chalaby, J. Goudsmit, and W. A. Paxton. 2001. N-linked glycosylation of the HIV-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 co-receptor utilization. J. Biol. Chem. 276:13433-13441. [DOI] [PubMed] [Google Scholar]
- 55.Polzer, S., M. T. Dittmar, H. Schmitz, and M. Schreiber. 2002. The N-linked glycan g15 within the V3 loop of the HIV-1 external glycoprotein gp120 affects coreceptor usage, cellular tropism, and neutralization. Virology 304:70-80. [DOI] [PubMed] [Google Scholar]
- 56.Rabut, G. E., J. A. Konner, F. Kajumo, J. P. Moore, and T. Dragic. 1998. Alanine substitutions of polar and nonpolar residues in the amino-terminal domain of CCR5 differently impair entry of macrophage- and dualtropic isolates of human immunodeficiency virus type 1. J. Virol. 72:3464-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raja, A., M. Venturi, P. Kwong, and J. Sodroski. 2003. CD4 binding site antibodies inhibit human immunodeficiency virus gp120 envelope glycoprotein interaction with CCR5. J. Virol. 77:713-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rana, S., G. Besson, D. G. Cook, J. Rucker, R. J. Smyth, Y. Yi, J. D. Turner, H. H. Guo, J. G. Du, S. C. Peiper, E. Lavi, M. Samson, F. Libert, C. Liesnard, G. Vassart, R. W. Doms, M. Parmentier, and R. G. Collman. 1997. Role of CCR5 in infection of primary macrophages and lymphocytes by macrophage-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the delta ccr5 mutation. J. Virol. 71:3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 61.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retroviruses 16:741-749. [DOI] [PubMed] [Google Scholar]
- 62.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 63.Rucker, J., M. Samson, B. J. Doranz, F. Libert, J. F. Berson, Y. Yi, R. J. Smyth, R. G. Collman, C. C. Broder, G. Vassart, R. W. Doms, and M. Parmentier. 1996. Regions in β-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell 87:437-446. [DOI] [PubMed] [Google Scholar]
- 64.Samson, M., F. Libert, B. J. Doranz, J. Rucker, C. Liesnard, C. M. Farber, S. Saragosti, C. Lapoumeroulie, J. Cognaux, C. Forceille, G. Muyldermans, C. Verhofstede, G. Burtonboy, M. Georges, T. Imai, S. Rana, Y. Yi, R. J. Smyth, R. G. Collman, R. W. Doms, G. Vassart, and M. Parmentier. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722-725. [DOI] [PubMed] [Google Scholar]
- 65.Sattentau, Q. J. 1992. CD4 activation of HIV fusion. International J. Cell Cloning 10:323-332. [DOI] [PubMed] [Google Scholar]
- 66.Schaeffer, E., R. Geleziunas, and W. C. Greene. 2001. Human immunodeficiency virus type 1 Nef functions at the level of virus entry by enhancing cytoplasmic delivery of virions. J. Virol. 75:2993-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siciliano, S. J., S. E. Kuhmann, Y. Weng, N. Madani, M. S. Springer, J. E. Lineberger, R. Danzeisen, M. D. Miller, M. P. Kavanaugh, J. A. DeMartino, and D. Kabat. 1999. A critical site in the core of the CCR5 chemokine receptor required for binding and infectivity of human immunodeficiency virus type 1. J. Biol. Chem. 274:1905-1913. [DOI] [PubMed] [Google Scholar]
- 68.Speck, R. F., K. Wehrly, E. J. Platt, R. E. Atchison, I. F. Charo, D. Kabat, B. Chesebro, and M. A. Goldsmith. 1997. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J. Virol. 71:7136-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sullivan, N., Y. Sun, Q. Sattentau, M. Thali, D. Wu, G. Denisova, J. Gershoni, J. Robinson, J. Moore, and J. Sodroski. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thali, M., C. Furman, E. Helseth, H. Repke, and J. Sodroski. 1992. Lack of correlation between soluble CD4-induced shedding of the human immunodeficiency virus type 1 exterior envelope glycoprotein and subsequent membrane fusion events. J. Virol. 66:5516-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toohey, K., K. Wehrly, J. Nishio, S. Perryman, and B. Chesebro. 1995. Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology 213:70-79. [DOI] [PubMed] [Google Scholar]
- 73.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 74.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 75.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 76.Wu, L., G. LaRosa, N. Kassam, C. J. Gordon, H. Heath, N. Ruffing, H. Chen, J. Humblias, M. Samson, M. Parmentier, J. P. Moore, and C. R. Mackay. 1997. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J. Exp. Med. 186:1373-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 78.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]