SUMMARY
Formation of either an ovary or testis during human embryonic life is one of the most important sex-specific events leading to the emergence of secondary sexual characteristics and sex assignment of babies at birth. Our study focused on the sex-specific and sex-indifferent characteristics of the prenatal ovarian stromal cells, cortical cords and germline, with the discovery that the ovarian mesenchymal cells of the stroma are transcriptionally indistinguishable from the mesenchymal cells of the testicular interstitium. We found that first wave pre-granulosa cells emerge at W7 from early supporting gonadal cells with stromal identity and are spatially defined by KRT19 levels. We also identified rare transient state f0 Spermatogonia cells within the ovarian cords between W10 and W16. Taken together, our work illuminates a unique plasticity of the embryonic ovary during human development.
eTOC BLURB
To explore human ovarian development, Wamaitha and Nie et al. analyze prenatal ovaries aged 7–21 weeks and map ovarian stroma and cord formation. They identify sex-specific and sex-indifferent progenitors during gonadal establishment and detect transient fetal spermatogonia-like germ cells within ovarian cortical cords, highlighting the plasticity of human prenatal gonads.
Graphical Abstract
INTRODUCTION
Increasing knowledge of human ovary development and follicle formation has implications for understanding and treating infertility, disorders of sex development, germ cell tumor formation, and other endocrine diseases. Germline specification and early gonadal development are challenging to study in humans, so model organisms such as the mouse, cow and non-human primates have been critical thus far as key developmental events are common between species, albeit on different timelines (Figure 1A).
Figure 1: Single-cell transcriptome profile of ovarian cells from 7 to 16 weeks post-fertilization.
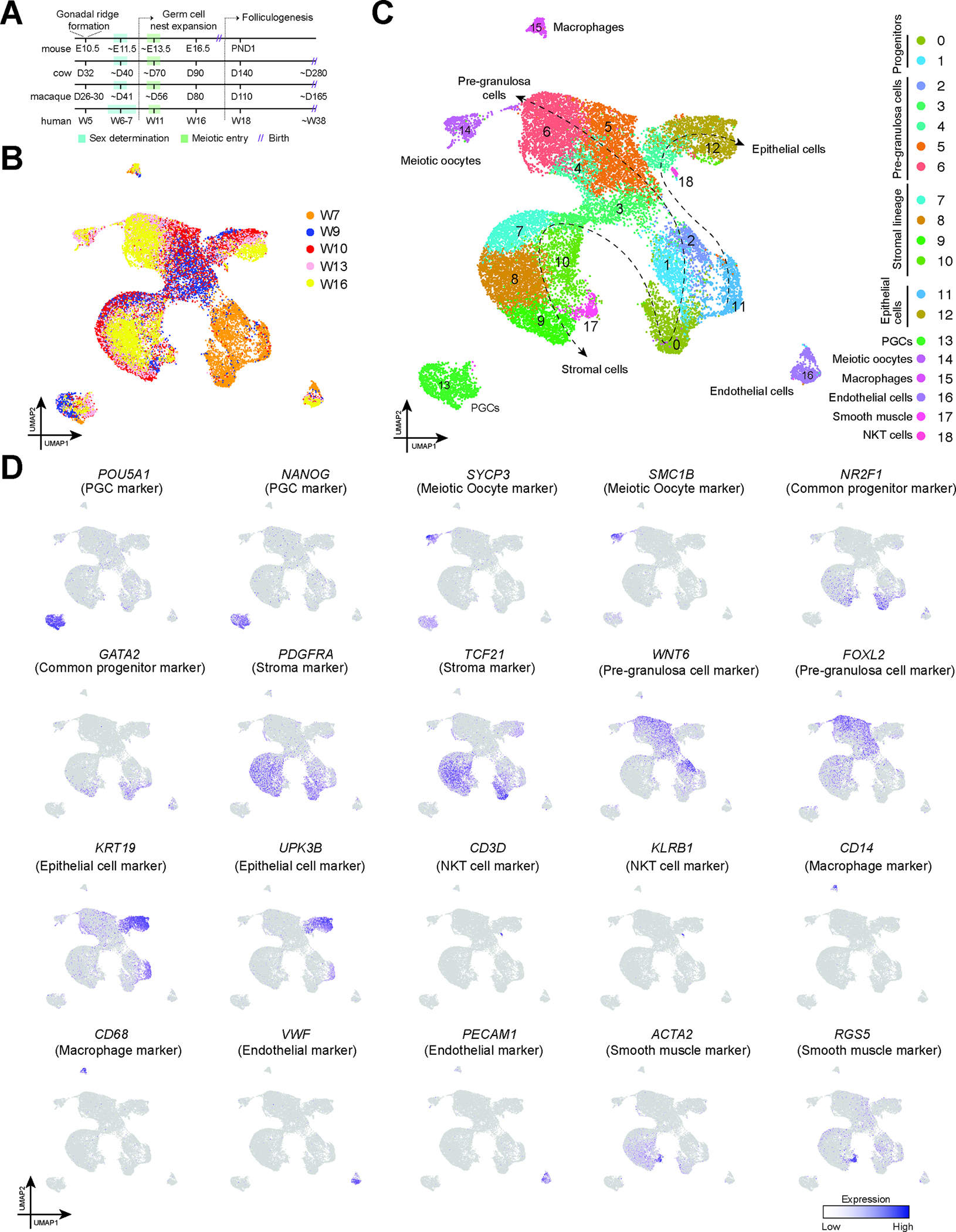
A: Illustration comparing the timing of selected ovarian development events identified in mouse, bovine, primate (rhesus or cynomolgus macaque) and human studies.
B: UMAP plot showing the distribution of single cells collected from fetal ovaries between 7 and 16 weeks of development (W7 – W16). Cells are colored based on their donor of origin.
C: UMAP plot from Fig 1A colored according to the cell types. The dashed lines show the lineage trajectory.
D: Expression of markers identifying major ovarian cell types cast on the UMAP plot from Fig 1A. Normalized expression is plotted on a high-to-low scale (purple-grey).
In the mouse embryo, the gonads (ovary and testis) develop from bilateral gonadal ridge epithelia located on the surface of the mesonephros. The gonadal epithelium becomes specified at embryonic (E) day 10.5 due to the expression of key transcription factors including LHX9, GATA4, WT1 and SF1 1,2. Twenty-four to forty-eight hours after the gonadal ridge is specified, supporting cells called pre-granulosa cells begin to develop from what appears to be two separate developmental origins; WNT4+ bipotential precursors which are localized in the center of the embryonic ovary, and KRT19+/LGR5+ ovarian surface epithelial cells 3–7. In the testis, activation of sex-determining region on the Y chromosome (SRY) and its target FGF9 represses WNT4 in these bipotential precursors, giving rise to Sertoli cells 8,9. In the ovary, WNT4 represses FGF9, resulting in formation of LGR5-/FOXL2+ pre-granulosa cells from the bipotential progenitors (Kim et al., 2006). In contrast, the LGR5+/KRT19+ pre-granulosa cells originating from the surface epithelium initiate FOXL2 expression after birth 6. The current rodent model follows that the first-wave FOXL2+ pre-granulosa cells ultimately contribute to medullary follicles, which mature prior to puberty onset 4,5. Conversely, the second-wave LGR5+ pre-granulosa cells originating from the KRT19+ ovarian surface epithelium contribute to the cortical follicles, which ultimately constitute the primordial follicles that form the long-term ovarian reserve in mice 5. The progenitor cells of the embryonic ovarian stroma also arise from SF1 positive gonadal progenitors, and by single cell RNA-Seq (scRNA-Seq) are reported to exhibit similar characteristics to embryonic testicular interstitial cells 10.
In the bovine embryo, the gonadal ridge develops around D30, with sex determination occurring about a week later 11. In contrast to the mouse, recent evidence from bovine studies predicts that pre-granulosa cells in cattle develop from somatic precursors known as gonadal ridge epithelial-like (GREL) cells (which are KRT19+), that proliferate to generate both the ovarian surface epithelium and the pre-granulosa cells 12. In this hypothesis, penetration by stromal cells from the mesonephros sequesters these GREL cell descendants to their locations either at the surface or within the cortical cords 12. This is different from the mouse, where the ovarian stroma originates from the coelomic epithelium 10.
In the human embryo, the gonadal ridge is specified at W5, with the somatic cells of the bilateral gonad formed by cells from both the genital ridge epithelium and stromal cells from the mesonephros 13. Sex determination begins at around W6, coincident with the expression of SRY in the human testis and organization into the testis cords 13–19. In the ovary, rudimentary primary sex cords develop from the gonadal ridge epithelium between W5–6, which then separate from the surface epithelium and become re-localized to the medulla to form the so-called rete ovarii 20–22. A second wave of cord formation occurs between W6–7, giving rise to the nascent ovarian cortical cords 18,21. Once established, the cortical cords expand, invaginating as pillar-like projections into the gonadal stroma 18,20,21,23.
FOXL2 is one of the earliest markers of pre-granulosa cell development in humans 17. In a recent comprehensive scRNA-Seq analysis, FOXL2 expression was first localized to bipotential early supporting gonadal cells (ESGCs) at W7, and subsequently to first-wave pre-granulosa cell-I (preGC-I) cells identified at W8 24. The ESGCs, marked by TSPAN8/LGR5, are also reported to express gonadal interstitial cell markers that are repressed as FOXL2 is expressed. However, further evidence for this interstitial identity remains to be confirmed. A second wave of cortical pre-granulosa cells called preGC-II appears from W8, with preGC-IIa emerging first in the outer cortex, expressing low levels of FOXL2, and preGC-IIb appearing second in the inner cortex, expressing high levels of FOXL2 24. Discovery of these multiple pre-granulosa sub populations is speculated to represent a similar scenario to the two waves of pre-granulosa cells reported in mice 24.
Although pre-granulosa cell specification in the human ovary has been thoroughly described 24, the human ovarian stroma remains poorly defined, particularly the mesenchymal cells of the ovarian stroma which have a low similarity index to the equivalent cells in mice 24. Given this dearth of knowledge on the mesenchyme of ovarian stroma (herein called stromal cells), we carried out un-biased profiling of the developing human ovary using a 10X Genomics dataset 25. Our analysis uncovered insights into human ovarian stromal cell development, leading to a re-evaluation of the gonadal supporting cells originating from ESGCs, and revealing plasticity in the sex-specific differentiation of the human germline.
RESULTS
scRNA-seq transcriptome profiling of human fetal ovaries from W7–16.
To examine prenatal ovarian development and identify cell populations in the ovarian stroma, we analyzed a total of 23248 single cells from W7-W16 prenatal human ovaries, with an average of 3115 genes in each cell. Red blood cell genes were found to contaminate cells in the embryonic stage, so the cluster of red blood cells, as well as their marker genes (Supplemental Table 1), were removed from the dataset. We applied uniform manifold approximation and projection (UMAP) to visualize the distribution of single cells with the ovarian samples (Figure 1B). Delineation of the samples by age (Figure S1A) revealed a dramatic developmental shift in the transcriptome of embryonic ovarian somatic cells from W7-W9. Cluster analysis revealed that clusters 0, 1, 2, and 11 derive in large part from W7, suggesting their identity as undefined precursor populations. In contrast, other clusters could be annotated based on previously published identifying markers 5,12 (Figure 1C and D). Dot plots were also generated to illustrate the percentage of cells in each cluster that expressed a given marker gene (Figure S1B). In particular, cluster 13 corresponds to PGCs expressing canonical markers such as POU5F1 and NANOG, whereas cluster 14 corresponds to meiotic germ cells and primordial oocytes, as identified by expression of SMC1B, SYCP3, and ZP3 respectively. Clusters described as stromal (mesenchymal) lineage were identified based on the expression of stromal markers TCF21 and PDGFRA 26,27. Clusters 11 and 12 expressed epithelial markers including KRT19 and UPK3B, genes previously associated with gonadal ridge epithelial cells 12, while clusters 1–6 expressed markers associated with pre-granulosa cells (FOXL2, WNT6). Finally, clusters 15, 16, 17, and 18 corresponded to endothelial cells (VWF+, PECAM1+), macrophages (CD14+, CD68+), NKT cells (CD3D+, KLRB1+) and smooth muscle cells (ACTA2+, RGS5+) respectively. As anticipated, we did not detect any cells with a thecal identity during this window of development 17.
Mesenchymal cells of the prenatal ovary and testis have an equivalent transcriptome
In order to explore sex-specific differences in the somatic cells of the ovary and testis, we first combined the sequencing data from prenatal ovaries (clusters 0–10, Figure 1C) with previous scRNA-seq data collected from testicular somatic cells from W6 post-fertilization to 1 year of age 28 (Figure 2A and 2B). Cell identity within these compartments was then assigned based on expression of key marker genes (Figure 2C). Progenitor cells (expressing GATA2, NR2F1, and TCF21) from ovarian and testicular samples clustered together indicating a similar transcriptome, with GATA2 expression consistent with a mesonephric origin of the stromal compartment 24. Delineation of the samples by age highlighted the progenitor cluster as being composed of cells from the W6 and W7 timepoints (Figure S1C). As expected, the supporting sex-specific pre-granulosa cells (FOXL2) and Sertoli cells (SOX9) clustered separately from each other, as did the fetal Leydig cells (HSD3B2) of the testis. In contrast, the mesenchymal stromal and interstitial cells of the ovary and testis respectively clustered together, indicating these cells are transcriptionally sex-indifferent (Figure 2A and 2C).
Figure 2: Gonadal somatic cell lineage specification during sex determination.
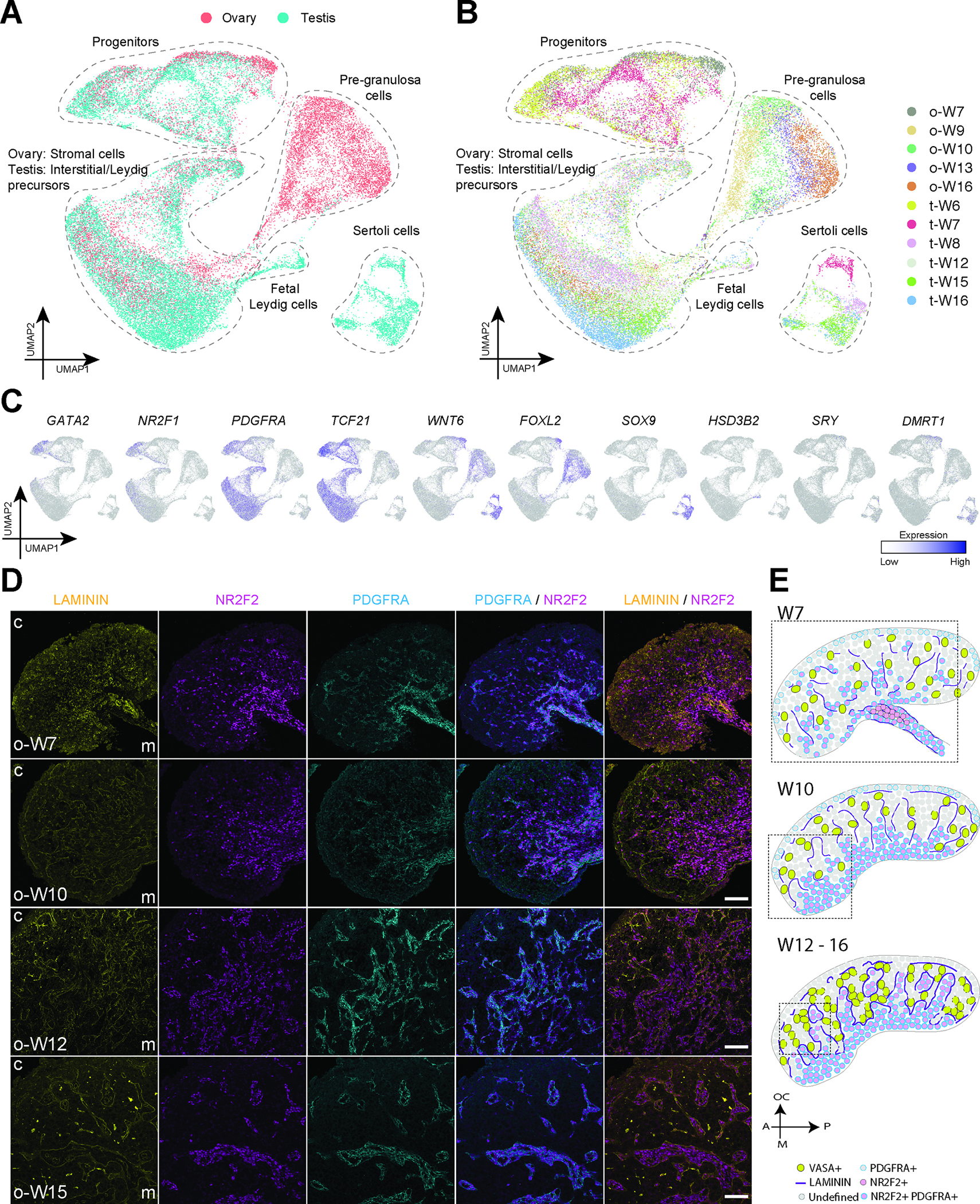
A: UMAP plot showing common progenitor-derived somatic cell lineages from fetal ovaries and testes at W7–16. See also Figure S1.
B: UMAP plot from Fig. 2A colored according to embryonic and fetal age.
C: Expression of markers identifying major cell types cast on the UMAP plot from Fig. 2A. Normalized expression is plotted on a high-to-low scale (purple-grey).
D: Immunofluorescence for selected sex-indifferent markers in fetal ovaries at W7–16 – Laminin (yellow), PDGFRA (cyan), NR2F2 (magenta). Nuclei were counterstained with DAPI (grey). Cortex (c) and medulla (m) labels included to show section image orientation. Scale bars 50 μm.
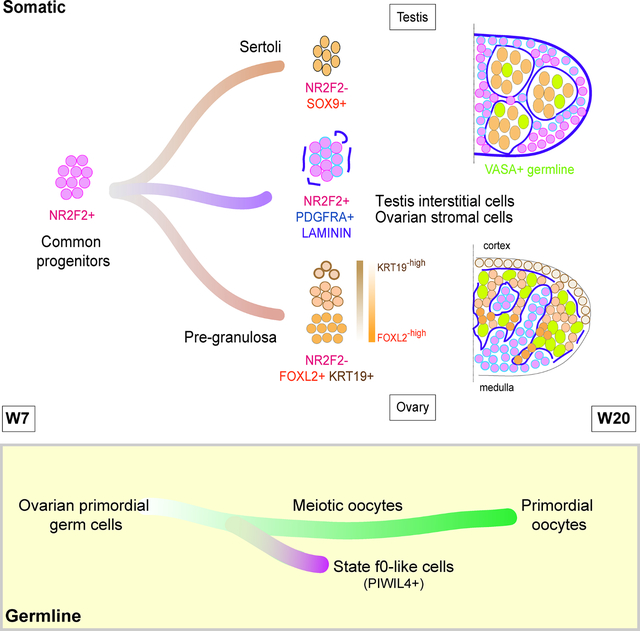
E: Model of ovarian development between W7–16.
Although 10X Genomics provides a unique opportunity to predict cell relationships at unprecedented scale, examining spatial relationships during organogenesis is equally important. Therefore, to investigate the architecture of the ovarian stroma and testicular interstitium during cord formation, we conducted immunofluorescence in gonads from W7–16 (Figure 2D; Figure S2A–C). The orphan nuclear receptor NR2F2 (COUP-TFII) was used to mark the stromal/interstitial cells 5,6,29,30. This strategy successfully delineated stroma/interstitium from VASA+ germ cells in the cords as early as W7 (Figure S2A).
To evaluate establishment of basement membranes during stromal infiltration and cord formation, we next performed immunofluorescence at W7 for LAMININ. Our results show that LAMININ protein is expressed by both the NR2F2 positive stromal streams, and NR2F2 negative cells of the cords, with discontinuous deposition of LAMININ between the cords and stroma (Figure 2D). Starting at W10, continuous LAMININ basement membranes separate the NR2F2 stromal compartment from the cords (Figure 2D). Co-staining LAMININ with PDGFRA, a gene identified as marking gonadal stromal/interstitial cells and progenitors (Figure 2C) reveals that PDGFRA largely overlaps with NR2F2 in the stroma from W7 as predicted (Figure 2D). However, we could detect background levels of PDGFRA expression by cells within the cortical cords, which may represent cord cells in the process of repressing PDGFRA. A simplified illustration of the ovarian structural changes outlined here is shown in Figure 2E.
Similar to the developing ovary at W7, PDGFRA was expressed at high levels in NR2F2+ interstitial cells in the W7 testis, with low but detectable levels of PDGFRA in the cords (Figure S2B). Discontinuous LAMININ deposition was also detected around the testis cords at W7, which resolved to continuous LAMININ basement membranes by W10 (Figure S2B). A polyclonal LAMININ antibody was used in this analysis, however previous reports indicate that LAMA1 is the predominant LAMININ expressed in the prenatal testis from W6 onwards 15. By W12, our data revealed that the NR2F2 interstitial cells around the cords are strongly positive for PDGFRA (Figure S2B). Based on their morphology and location, these cells may represent testicular interstitial progenitor precursors of peritubular myoid cells, smooth muscle cells that make up the outer cell layer of the seminiferous tubules 31,32. A simplified illustration of the testicular structural changes outlined here is shown in Figure S2D.
Given the prediction that supporting cells of the ovary and testis originate from ESGCs with stromal/interstitial cell identity 24, we conducted immunofluorescence for NR2F2 with FOXL2 or SOX9 in the ovary and testis respectively (Figure S2C). In the W7-W12 ovary, FOXL2 and NR2F2 were mostly mutually exclusive, however clusters of NR2F2+ FOXL2+ expressing cells were identified (arrows) (Figure S2C). This co-expression pattern is consistent with previous observations noting FOXL2 expression in some stromal cells within human and bovine fetal ovaries 12,33. In the prenatal testis from W7–12, we did not observe NR2F2+ expression within the SOX9+ cells of the cords; instead we identified rare double positive cells in the interstitium (FigureS2C; arrows). The presence of these rare cells co-expressing stromal/interstitial markers with FOXL2 or SOX9 is consistent with an origin from ESGCs that co-express stromal/interstitial markers prior to sex-determination.
Evidence for TSPAN8/LGR5 ESGCs at W6 of human ovarian development
Given the hypothesis that SOX9/NR2F2 and FOXL2/NR2F2 cells are likely descendants of bipotential progenitor ESGCs 24, we next set out to identify ESGCs in the current data set. To achieve this, we conducted a focused analysis of W7 ovarian somatic cells (clusters 0, 1, and 2, Figure 1B) combined with W6 and W7 testis somatic cell precursor populations 28. As anticipated, Sertoli and pre-granulosa cells clustered separately from each other, while conversely, bipotential cells and common progenitors from the prenatal testis and ovary clustered together (Figure 3A). Differential gene expression analysis on the populations from Figure 3A (minus the Sertoli cells) revealed that the common progenitor cells were enriched for GO terms for cell division, embryonic morphogenesis, and mesodermal cell differentiation (Figure 3B; Figure S3A). The analysis also identified an additional sub-population of bipotential cells that shared similarities with pre-granulosa cells; we termed these bipotential transitional cells. Bipotential and bipotential transitional cells were enriched for GO terms for cellular response to stress and actin cytoskeleton organization (Figure 3B). Finally, pre-granulosa cells showed enrichment for GO terms related to signal transduction, cell junction organization, and regulation of hormone levels and expression of WNT (Figure 3B; Figure S3C).
Figure 3: Gene expression dynamics in ovarian somatic cells.
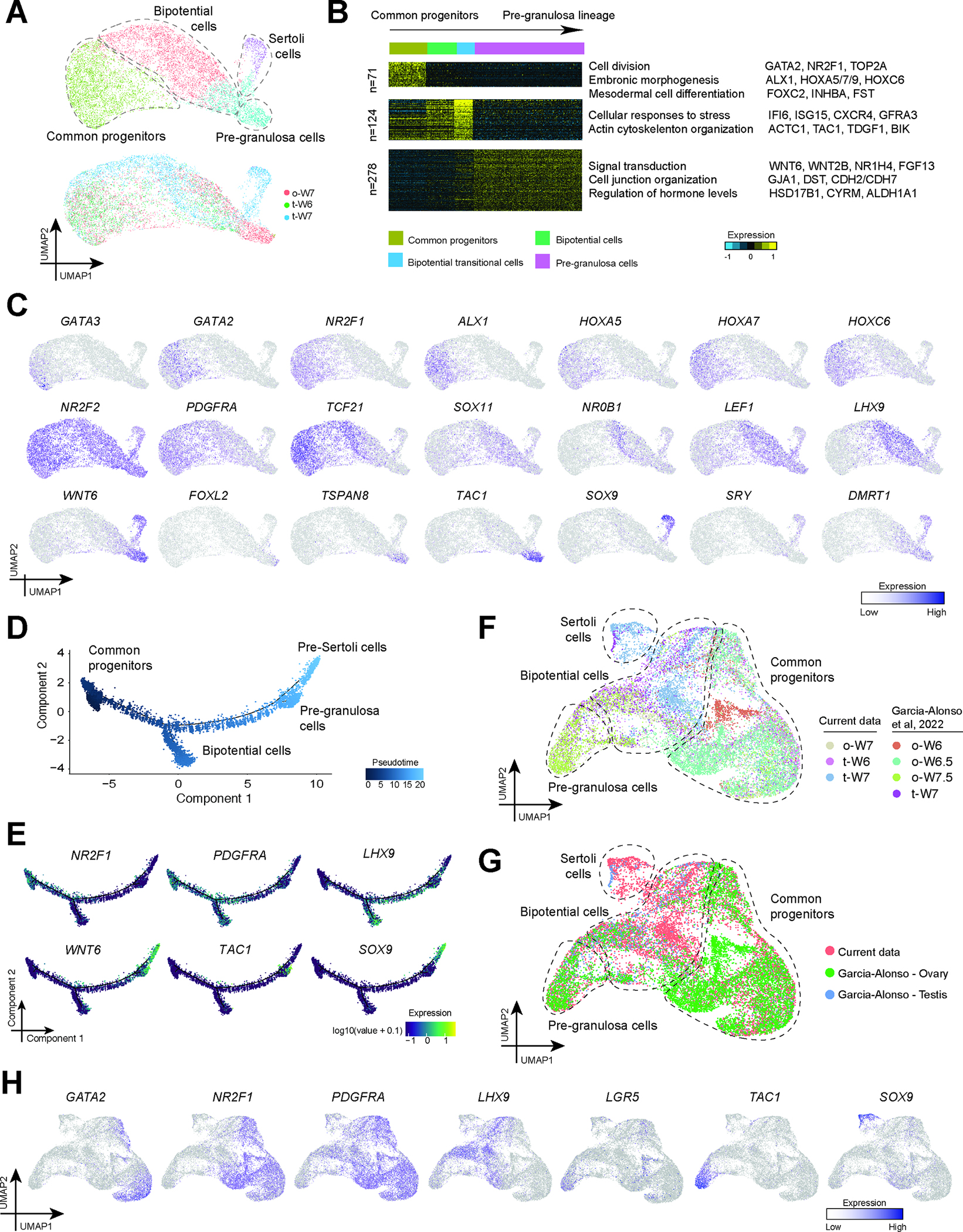
A: UMAP plot showing focused analysis of the progenitor cluster annotated in Fig. 2A. Top figure labeled with cell identities and colored by Seurat analysis-designated cell groups; bottom figure colored by donors of origin.
B: K-means clustering of differentially expressed genes (DEGs) (n =473) across common progenitor-derived somatic cell lineages. Note: Each row represents a gene, and each column represents a single cell, with columns/cells placed by developmental trajectory and depicted by a thick colored line at the top. Scaled gene expression levels are colored according to Z-score (color key is at the top right). Corresponding GO terms enriched in the DEGs are listed with example genes on the right of the heatmap.
C: Expression of transcription factors or markers identifying the cell types cast on the UMAP plot from Fig. 4A. Normalized expression is plotted on a high-to-low scale (purple-grey).
D: Pseudotime trajectory of common progenitors, bipotential cells and supporting cells from ovary and testis samples at W6–7 analyzed by Monocle.
E: Expression of selected markers of common progenitor (NR2F1), bipotential (LHX9), pre-granulosa (WNT6, TAC1) and pre-Sertoli (WNT6, SOX9) cells plotted on the pseudotime trajectory. PDGFRA expression is gradually downregulated with the highest expression in the common progenitors. Normalized expression is plotted on a high-to-low scale (purple-green-yellow).
F: UMAP plot showing combined analysis of common progenitors, bipotential cells and supporting cells from ovary and testis samples at W6–7 (progenitor cluster annotated in Fig. 2A) and corresponding cells from W6–7 samples collected in Garcia-Alonso et al. 24. Corresponding cells were parsed out with the Scanpy package using key cell markers along with their identity labeled in the paper (see Methods). The figure is labeled with cell identities and colored by Seurat analysis-designated cell groups.
G: UMAP plot from 3F colored according to dataset of origin.
H: Expression of selected markers cast on the UMAP plot from Fig. 3F. Normalized expression is plotted on a high-to-low scale (purple-grey).
See also Figure S3.
Bipotential cells at W6 and W7 were enriched for LHX9, an early gonadal ridge marker in the mouse 1, and the WNT pathway regulator LEF1 that is eventually silenced in Sertoli cells but active in the ovary 34–36. The stromal/interstitial markers NR2F2, PDGFRA and TCF21 were expressed in both common progenitors and bipotential cells, while SRY was expressed in some bipotential cells and in Sertoli cells (Figure 3C). SOX9 was also enriched in Sertoli cells as expected, while TAC1, a member of the tachykinin peptide family implicated in ovarian function 37,38, was enriched in bipotential transitional cells and pre-granulosa cells (Figure 3C). WNT6 was expressed in bipotential cells, Sertoli and pre-granulosa cells, as was DMRT1, consistent with previous work in humans and mice showing they are initially expressed in both sexes before becoming testis specific (Figure 3C) 39,40. We also performed computational pseudotime analysis of the common progenitor lineage including common progenitors, bipotential cells, and supporting cells using Monocle (Figure 3D). PDGFRA and NR2F1 expression were strongest in the common progenitors, and were gradually downregulated over the trajectory. LHX9 expression was mainly enriched in bipotential and pre-granulosa cells later in the trajectory, while SOX9, TAC1 and WNT6 were enriched in either Sertoli, pre-ganulosa or both cell types (Figure 3E), in line with the prior UMAP plots
This analysis also revealed expression of TSPAN8 (Figure 3C), a specific marker of ESGCs in pre-granulosa cells at W7, consistent with previous reports 24. Analysis of LGR5 (Figure S3B), another marker of ESGCs revealed some rare scattered cells in the TSPAN8 cluster (Figure S3B). In order to increase the power of our analysis, we integrated four additional W6–7.5 samples 24 with the three samples above, and re-analyzed the seven samples together (Figure 3F). As three of the four additional samples were embryonic ovaries, this data set contributed most of the pre-granulosa cells (Figure 3F–G). The combined analysis identified clusters of common progenitors, bipotential cells, pre-granulosa and Sertoli cells as before. UMAP visualizations of selected genes showed GATA2, NR2F1 expression in the common progenitor cluster, LHX9 in the bipotential cluster, and TAC1 and SOX9 in pre-granulosa and Sertoli cells respectively (Figure 3H). PDGFRA expression was strong in progenitor and bipotential cells confirming a stromal/interstitial-like identity to the progenitors in the W6–7 ovary and testis of both data sets. Rare LGR5+ cells were detected in the pre-granulosa cluster with TAC1 (Figure 3G; Figure 3H). Additional LGR5+ cells were identified in the common progenitor cluster, which may represent an earlier state of this stem-like population (Figure 3H).
To investigate possible sexual dimorphism at W6–7, we performed differential gene expression analysis on common progenitor and bipotential populations. Using a fold change of 2 and P-adjust value lower than 0.05 we identified only one sex chromosome-linked gene, RPS4Y1, listed as differentially expressed in the common progenitor data (Supplemental Table 2). Adjusting our analysis to use a lower threshold (fold change of 1.2 and P-adjust value lower than 0.05) still only returned a limited number of differentially expressed genes (DEGs) (Supplemental Table 2). Similar results were obtained for the bipotential cells (Supplemental Table 3). We conclude that any sexual dimorphism in these early cell populations may be subtle. Expression of TSPAN8 and LGR5 in pre-granulosa cells at this stage suggests the ESGC program is not yet fully repressed, whereas the absence of TSPAN8 or LGR5 in Sertoli cells of the testis at this age indicates more rapid repression of the progenitor program in testis supporting cells.
Pre-granulosa cells are spatially defined by KRT19 from at least W7
In humans, two waves of pre-granulosa cells were recently identified, and hypothesized to be analogous to the two-waves that develop in mice 24. In mice, KRT19+ cells mark the LGR5+ population originating from the surface epithelium 3–7. At the early timepoints (W6–7), we noticed KRT19 expression in some cells within the common progenitor, bipotential and pre-granulosa cell clusters (Figure S3B). Therefore, to characterize the relationship between FOXL2 and KRT19, we performed immunofluorescence from W7–W15 (Figure S3C). In this analysis at W7, KRT19 was strongly enriched in the surface epithelium and the underlying FOXL2+ cells in the ovarian cortex, with FOXL2+ single positive cells observed in the interior of the ovary (Figure S3C). By W15, most FOXL2+ cells were KRT19+, with more KRT19 localized to those FOXL2+ cells under the surface epithelium (Figure S3C). Analysis of the cortex of W6 ovaries indicates that KRT19 is expressed first by cells of the surface epithelium and underlying cortical cells, with FOXL2 turning on between W6-W7 (Figure S3D). Taken together, the gradient in KRT19 expression from ovarian surface epithelium to medulla suggests spatial patterning of the FOXL2+ pre-granulosa cells during embryonic ovary development.
To investigate the relationship between the KRT19+ and FOXL2+ cells, we returned to our scRNA-seq analysis and focused on the pre-granulosa populations from W9-W16 (clusters 3–6 and 12; Figure 1C). We identified three co-existing pre-granulosa cell populations (Figure 4A and 4B). One population (designated as “Epithelial”) was enriched for KRT19 and LHX9 but not FOXL2 or WNT6, and specifically expressed UPK3B (Figure 4A and 4C). This profile appears similar to the epithelial cell population from Clusters 11 and 12 in Figure 1B. The other two populations (designated as pre-granulosa 1 and 2, “PG1” and “PG2”) expressed FOXL2, LHX9, WNT6 and KITLG (Figure 4A and 4C). KRT19+ cells were present in both PG1 and PG2, with slightly more enrichment in PG2. Based on the immunofluorescence analysis, we propose that PG2 reflects the population of KRT19+FOXL2+ pre-granulosa cells situated closer to the surface epithelium, with PG1 reflecting the FOXL2+ pre-granulosa cells found closer to the ovarian medulla, where occasional KRT19 protein expression can be detected (Figure S3C). The proliferation marker Ki-67 was not enriched in either PG1 or PG2, consistent with observations in mouse and bovine fetal ovaries that FOXL2+ cells are not cycling (Figure S3E). 4,12.
Figure 4: Characterizing pre-granulosa cells in human fetal ovaries.
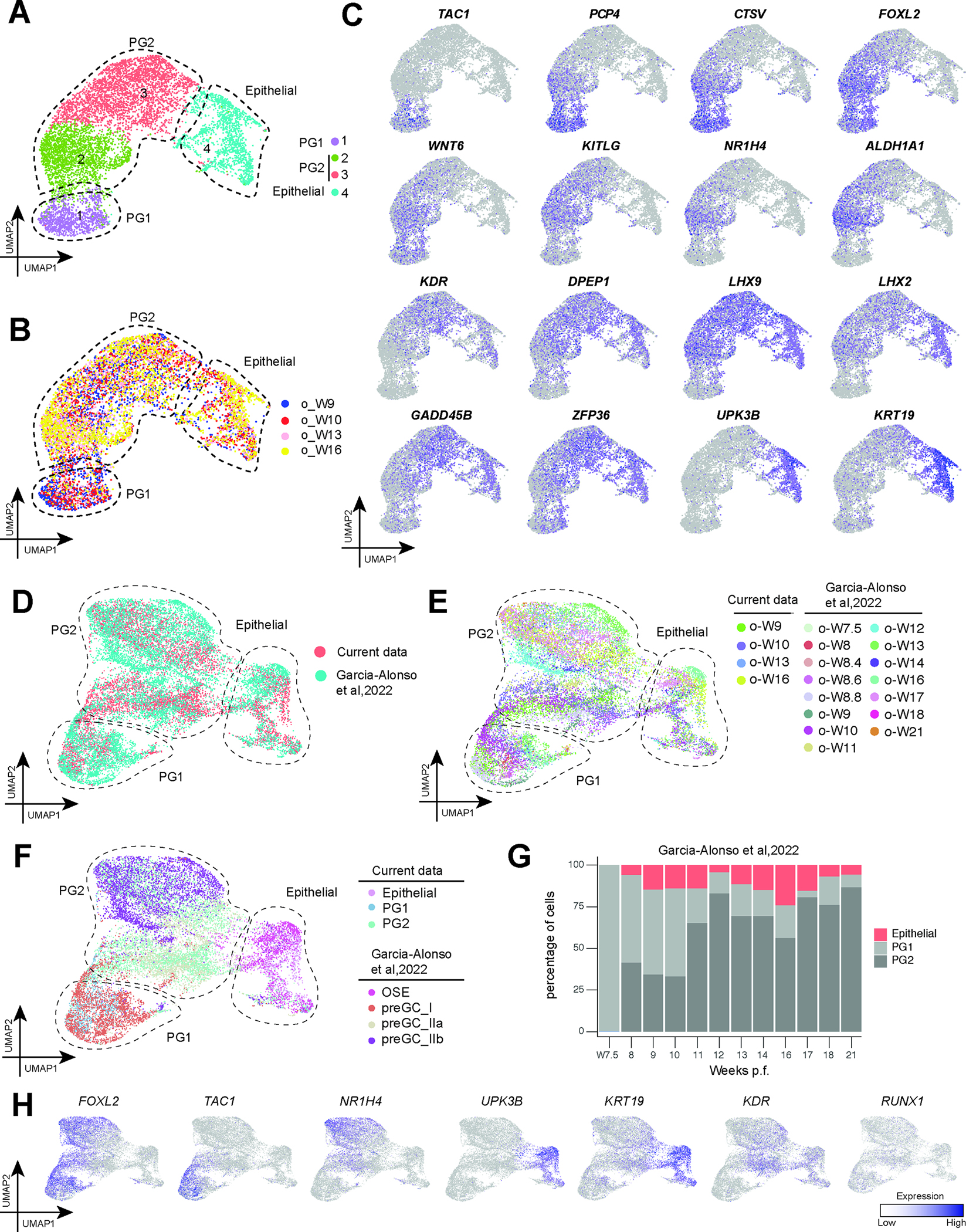
A: UMAP plot showing focused analysis of epithelial cells and pre-granulosa cells from W9–16 (cluster 12 and 3–6 annotated in Fig. 1A). The figure is labeled with cell identities and colored by Seurat analysis-designated cell groups.
B: UMAP plot from Fig 4B colored by donors of origin.
C: Expression of markers of pre-granulosa cells (PG1, PG2) or epithelial cells cast on the UMAP plot from Fig. 4B. Normalized expression is plotted on a high-to-low scale (purple-grey).
D: UMAP plot showing combined analysis of epithelial cells and pre-granulosa cells from W9–16 (cluster 12 and 3–6 annotated in Fig. 1A) and W7.5–21 24. The figure is labeled with cell identities and colored by Seurat analysis-designated cell groups.
E: UMAP plot from Fig 4D colored by donors of origin.
F: UMAP plot from Fig 4D colored by granulosa cell identity clusters identified in Fig 4A or in Garcia-Alonso et al. 24.
G: Bar graph illustrating the percentage of PG1 (dark grey), PG2 (light grey) and epithelial cells (red) present at each timepoint in the additional W7.5- W21 fetal samples from Garcia-Alonso et al. 24. The four timepoints at W8 (W8, W8.4, 8.6, 8.8) were condensed into one bar.
H: Expression of selected markers cast on the UMAP plot from Fig. 4D. Normalized expression is plotted on a high-to-low scale (purple-grey).
See also Figure S3.
Given that PG1 is first observed at W7 in the embryonic ovary (Figure 3C and H), we predicted that PG1 may correlate to first-wave preGC-I identified in Garcia-Alonso et al. 24 (Figure 4C). To compare the pre-granulosa cell populations in our dataset to those in Garcia-Alonso et al., we re-analyzed our data with fifteen additional ovarian samples from W7–21 (Figure 4D). We could once again identify three populations of cells (PG1, PG2, and Epithelial) in the combined dataset (Figure 4D, 4E). As predicted, PG1 overlaps with the preGC_I population identified in the Garcia-Alonso et al., 2022 study, while PG2 overlaps with the preGC_IIa and preGC_IIb populations (Figure 4F) 24. The Epithelial cluster overlaps with the ovarian surface epithelial (OSE) cluster (Figure 4F) 24. We also investigated the expression of RUNX1, a transcription factor, whose expression was previously noted in mouse and human fetal ovaries via bulk RNA sequencing 41. However, our analysis found RUNX1 expression is relatively low and rare in all three populations of pre-granulosa cells (Figure 4H). In order to evaluate the proportion of first wave PG1 (equivalent to preGC-1) and second wave PG2 (equivalent to pre-GC-IIa/b) over time, we measured the proportion of each population from 7.5W to 21W and showed that PG1/preCG-I is found exclusively at W7.5, whereas from W8, PG1 and PG2 pre-granulosa cells exist simultaneously in the ovary with progressively more PG2 over time (Figure 4G). Altogether, we find evidence for diverse pre-granulosa cell populations in the human embryonic and fetal ovary with ESGC-derived PG1/preGC-I co-existing with PG2/preGC-IIa/b through to at least 21 weeks.
Emergence of cells resembling State fetal 0 (f0) Spermatogonia within the ovary.
Given that our data set captures both sex-indifferent progenitors and sex-specific supporting cells during the initial stages of gonad establishment, we were next interested in the timing of sex-specific differences emerging in the human germline. To achieve this, we combined the sequencing data from ovarian germline cells collected from W7-W16 (clusters 13 and 14, Figure 1C) with scRNA-seq data collected from testicular germ cells from W6 post-fertilization to 1 year of age 25,28 (Figure S4A).
Ovarian germ cells were divided into four stages based on transcriptome analysis and UMAP clustering: PGCs (identified by expression of POU5F1, SOX17 and TFAP2C), retinoic acid (RA) responsive MGCs identified by (ZGLP1, STRA8), MGCs in prophase 1 (SPO11, SYCP1), and primordial oocytes (FIGLA, ZP3) (Figure S4A–C). As previously reported 25, at W7 PGCs were the only germ cells present in the ovary, with rare RA-responsive MGCs emerging at W9. MGCs were first identified in fetal ovaries at W10, and primordial oocytes at W13, with a large fraction of ovarian germline cells still composed of PGCs (Figure S4D). In the testis, very rare RA-responsive MGCs were detected at W8, which were mostly lost by W12. State f0 spermatogonia were first identified in the testis at W12, with almost all germ cells at W16 corresponding to state f0 Spermatogonia (Figure S4D). When Monocle was used to calculate the pseudotime trajectory of all germ cells, divergence of testis germ cells towards state f0 Spermatogonia was noted from W12, while ovarian germ cells diverged towards primordial oocytes from W9–10 (Figure 5A).
Figure 5: Rare State f0 Spermatogonia-like cells are found in fetal ovaries.
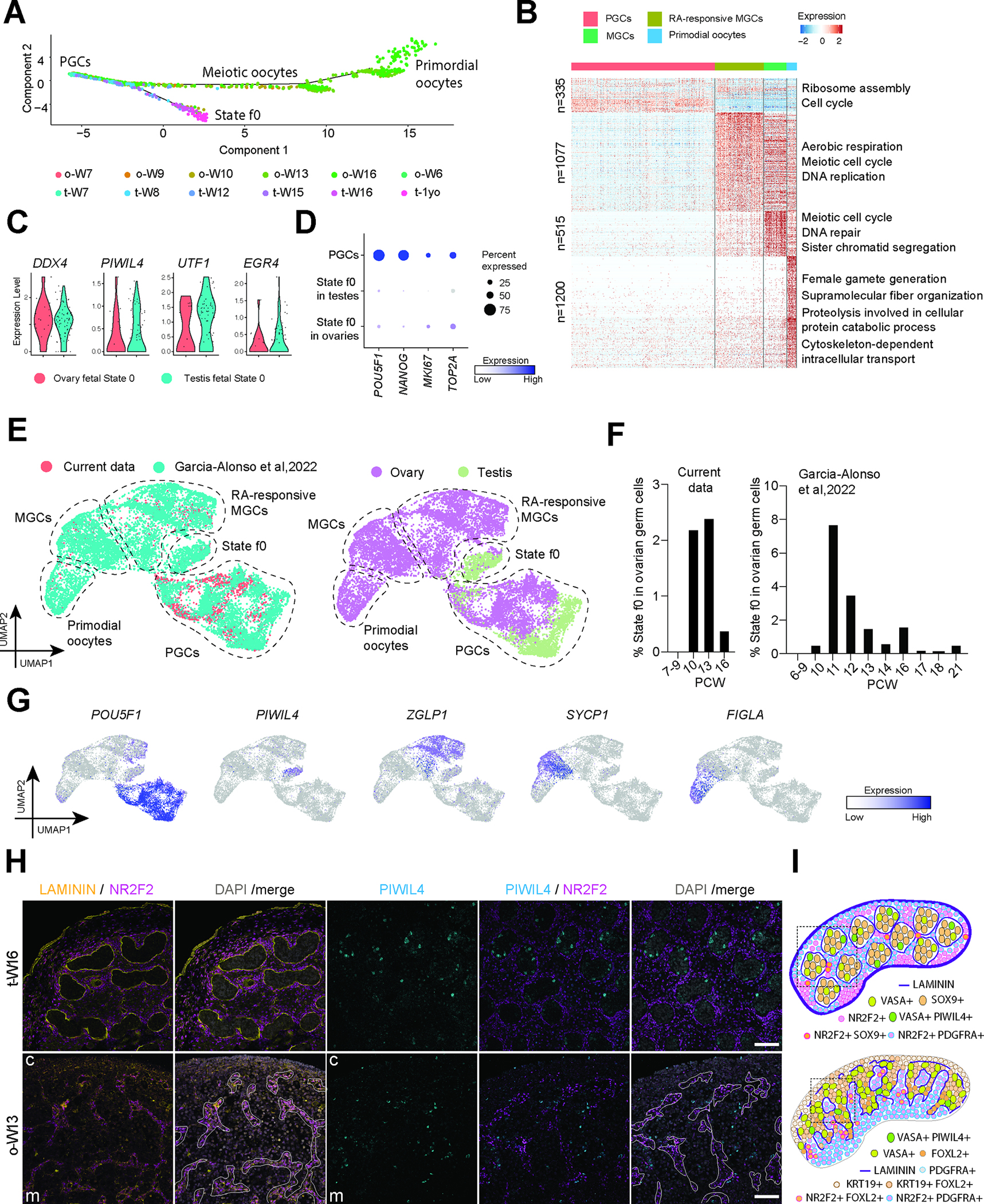
A: Pseudotime trajectory of both testis and ovary germ cells from W7–1 year old analyzed by Monocle. Cells are colored by donor origin.
B: Heatmap showing differentially expressed genes (DEGs) for each germ cell stage. Scaled gene expression levels are colored according to Z-score (color key is at the top right). Corresponding GO terms enriched in the DEGs are listed on the right of the heatmap. Ovarian f0 cells not included, too few and corresponding column is not visible
C: Violin plots showing equivalent expression of selected marker genes of germ cells or state f0 Spermatogonia cells in both ovary and testis.
D: Dotplot comparing percentage expression of selected pluripotency (POU5F1, NANOG) and proliferation (MKI67, TOP2A) markers in PGCs and state f0 cells. Normalized expression is plotted on a high-to-low scale (purple-grey), with dot size increasing with the percentage of cells within a cluster (0% – 75%) that express a given gene.
E: UMAP plot showing combined analysis of germ cells from W7–1year old and W7.5–21 from Garcia-Alonso et al. 24. The figure is labeled with cell identities and colored by study origin (left) or by ovarian or testicular origin (right). The low expression of these genes in the ovarian state f0 Spermatogonia cells indicates they are not carcinoma cells
F: Percentage of state f0 cells plotted across W7–16 in the current data (left) or W6–21 from Garcia-Alonso et al. 24 (right).
G: Expression of selected markers cast on the UMAP plot from Fig. 3E. Normalized expression is plotted on a high-to-low scale (purple-grey).
H: Immunofluorescence for LAMININ (yellow) NR2F2 (magenta) and PIWIL4 (cyan) / NR2F2 (magenta) in the W16 testis and W13 ovary, to situate PIWIL4+ cells relative to the cords. Nuclei counterstained with DAPI (grey) in merged image. Testis cords are defined by the thick LAMININ membrane. For the ovary panels where the LAMININ membranes are thinner at this stage, NR2F2+ stromal clusters are outlined in white line – the ovarian cords are defined in opposition to this. Scale bars 50 μm.
I: Condensed illustration of ovarian and testis development.
Comparing testicular and ovarian PGCs at W7 prior to sex-specific PGC differentiation revealed subtle transcriptional shifts based on biological sex, with W7 ovarian PGCs clustering slightly apart from testicular PGCs (Figure S4A). Notable expression differences driving this sex-based difference include genes associated with maternal imprinting and X-chromosome dampening in the ovarian PGCs, with Y-chromosome expressed genes upregulated in testicular PGCs (Figure S4E). In agreement with previous studies, we observed dynamic transcriptome changes as PGCs develop into meiotic oocytes, including downregulation of pluripotency genes and upregulation of meiotic genes (Figure 5B, S4F, S4G) 25,42.
Early studies aimed at identifying meiosis inducing factors showed that PGCs situated outside of seminiferous tubules enter into meiosis 43. The inductive factor was later identified as RA, which drives STRA8-mediated meiotic entry in prenatal ovaries and postnatal testis 44,45. We therefore speculate that rare MGCs identified in the fetal testis at W8 represent testicular germ cells not fully protected from RA by Sertoli cells (Figure S4D). Moreover, surprisingly, we identified a transient population of germ cells in W10-W16 fetal ovaries that clustered with state f0 (PIWIL4+) testicular cells in cluster 6 (Figure 5C, S4A, S4B, S4D). This result suggests that rare germ cells in the human fetal ovary are capable of initiating a state f0 Spermatogonia program.
Further analysis of cluster 6 from Figure S4A demonstrates a strikingly similar transcriptome between these rare prenatal ovarian germ cells and the testicular state f0 cells, particularly in the expression pattern of DDX4, PIWIL4, UTF1, and EGR4 (Figure 5C). Pseudotime analysis of the W10 and W13 ovarian germ cells again reveals a small proportion of cells progressing along the lower trajectory (Figure S5A). Our lineage trajectory analysis predicts that these cells do not correct themselves – rather, these cells differentiate from PGCs in the ovary and do not enter into a meiotic state. In order to rule out the hypothesis that these cells represent the initiation of germ cell carcinomas in the ovary, we investigated the expression of proliferation (MKI67 and TOP2A) and pluripotency markers (POU5F1 and NANOG) with PGCs as controls (Figure 5D). The low expression of these genes in the ovarian state f0 Spermatogonia cells indicates they are likely not carcinoma cells.
To evaluate whether ovarian state f0 Spermatogonia are found in other prenatal ovarian data sets, we examined 15 additional ovarian samples from W7-W21 24. As before, the combined data could be segregated into PGCs, MGCs, RA-responsive MGCs and primordial oocytes (Figure 5E). PIWIL4+ state f0 cells formed a distinct cluster in the combined dataset, comprising cells from both ovary and testis samples (Figure 5E, 5G). The number of PIWIL4+ ovarian cells reached its peak around W11–13 and declined from W14 onward (Figure 5F). Differential gene expression analysis found no DEGs between PIWIL4+ state f0 Spermatogonia in the ovary and those in the testis. To rule out the identification of these cells as being associated with droplet based sequencing technology, we also re-analyzed data from the plate-based fetal germ cell (FGC) dataset 42, and identified state f0 cells in both ovaries and testes (Figure S5C). Taken together, using three independently generated scRNA-Seq data sets across two different technologies we consistently identify transient state f0 Spermatogonia in the developing human ovary.
To confirm this finding at the protein level, we conducted immunofluorescence of prenatal ovaries from W10-W16 and identified rare PIWIL4+ VASA+ germ cells in five independent ovarian samples representing W10–15 (Figure S5D). To situate the PIWIL4+ cells either within or outside the cords of the developing ovary, we performed immunofluorescence for LAMININ and NR2F2 at W13, when PIWIL4+ cells are readily detectable and LAMININ basement membranes are continuous (Figure 5H). As a positive control, we also stained the fetal testis at W16 for PIWIL4, LAMININ and NR2F2. In the testis, PIWIL4+ f0 Spermatogonia are only present within the LAMININ-bordered cords and not within the NR2F2+ interstitium (Figure 5H). Similarly, within the fetal ovary, we observed that the PIWIL4+ cells were located within the cortical ovarian cords rather than in the NR2F2+ stromal streams (Figure 5H). In general, we find that PIWIL4+ cells tend to be found in small clusters in the cords positioned closer to the medulla of the ovary, though occasional single cells can be observed closer to the surface epithelium in the cord. Altogether, our data indicates that the major lineage trajectory for ovarian germline cells involves the progression of PGCs into meiosis, with a small transient population of VASA+ germ cells within and outside the ovarian cords expressing PIWIL4 and becoming presumptive f0 spermatogonia that rapidly disappear. A summary of heterogenous cell populations in the prenatal ovary and testis are shown in Figure 5I.
DISCUSSION
Early histological and EM studies of the developing human prenatal ovary proposed that the ovarian cords make up the majority of the developing organ and are connected directly with the surface epithelia separated by minimal stroma 16,20–22,46–48. Here we show that cord formation in the embryonic ovary begins before W7. We identified PDGFRA and TCF21 mRNA expression in progenitor cells of the embryonic ovary and testis during a time window when sex-specific differentiation of supporting cells within the cords has commenced and stromal infiltration from the mesonephros can be observed. At the protein level, similar to the dynamics revealed by the scRNA-seq results, we found that PDGFRA is largely repressed in the supporting cells of the ovary and testis cords, and instead is restricted to stromal/interstitial cells by W7 in both organs. However, rare supporting cells harboring a dual stromal-like and supporting-cell identity can be detected in both organs between W7-W12, suggesting that the origin of supporting cord cells and stroma/interstitium are related.
Previous work in humans has implicated the developmental origins of ovarian and testicular supporting cells as ESGC progenitors that express TSPAN8/LGR5+ 24. We identified highly specific TSPAN8 expression at W7 in the progenitors that give rise to pre-granulosa cells, consistent with the ESGC hypothesis for ovarian supporting cell development 24. LGR5 expression by contrast was more elusive in our dataset, and appeared to mark not only the TSPAN8+ cells, but also a subpopulation of stromal-like common progenitor cells that express PDGFRA and NR2F1. The stromal-like identity of LGR5 progenitors does not necessarily imply that ESGCs originate in the stroma. Instead, we hypothesize that the early embryonic progenitors of the ovary and testis have a transcriptional program that at later stages becomes highly restricted to the stroma/interstitium of the gonads and remains largely sex-indifferent until steroidogenic and peritubular myoid cells are specified.
Lineage tracing experiments in the mouse has led to the classification of two distinct pre-granulosa cell populations prior to birth, one that is FOXL2+ but KRT19-/LGR5-negative, and another that is KRT19+/LGR5+ but FOXL2-negative. The importance of this observation is that this second pre-granulosa population (KRT19+/LGR5+) only express FOXL2 after birth, and generate the granulosa cells of primordial follicles that make up the ovarian reserve. A similar two-wave paradigm for pre-granulosa cell differentiation during embryonic and fetal development was recently shown using a comprehensive scRNA-Seq analysis of human ovaries 24. The two waves of pre-granulosa cells were coined preGC-I and preGC-IIa/b, with preGC-1 arising first by W7, and preGC-II arising second by W8 24. Using a different computational pipeline with our data set, we also identified two granulosa cells populations in the developing ovary that we called PG-1 and PG-2. Combining our data with that from Garcia-Alonso et al. 24 revealed that PG-1 and PG-2 correspond to preGC-I and preGC-IIa/b respectively, therefore confirming through an alternate approach that two major types of pre-granulosa cells develop between W7-W21. However, our data supports a model whereby the two waves of pre-granulosa cell differentiation have also diverged from the dynamics reported in mouse ovaries as the PGC-2/pre-GCIIa/b pre-granulosa cells appear to initiate FOXL2 expression before birth during cord establishment. In the mouse, this is a postnatal event more closely aligned with cord breakdown.
Further analysis of pre-granulosa cell positioning in the W7-W15 human ovary reveals that KRT19 expression spatially discriminates pre-granulosa cell populations. Specifically, our data shows that FOXL2+ cells closer to the surface epithelium express high levels of KRT19, while FOXL2+ cells towards the center initially express no or low levels of KRT19 at W7, with increasing KRT19 expression detected by the FOXL2+ cells towards the center over time. We speculate this may be due to either the FOXL2+ cells acquiring KRT19 expression later, or perhaps the specification of KRT19+/FOXL2+ cells at the surface ingressing inwards as development progresses.
In the bovine model, KRT19+ gonadal ridge epithelial (GREL) cells are speculated to give rise to both pre-granulosa cells and the surface epithelium; GREL cells share a number of markers with surface epithelium cells but are a distinct cell type 12. We did not identify KRT19+ sub-populations with a unique enough identity to be called GREL cells in this study but at the current time we cannot rule out whether this may be due to the rarity of GREL cells, which were therefore not captured in the 10X genomics data set, or whether a GREL cell population is not present in in the developing human ovary per se. Recent studies further characterizing GREL cell identity in the bovine fetal ovary may help future comparisons in this regard 49,50. It does not appear that the GREL cell in bovine is equivalent to the ESGCs identified in human ovaries, as ESGCs do not express KRT19.
Establishment of basement membranes during organogenesis is a crucial process providing structure, elasticity, and a source of signaling between tissue layers. We discovered that the initial specification of ovarian and testicular supporting cells at W7 precedes the establishment of continuous LAMININ-containing basement membranes. In contrast, continuous LAMININ staining occurs after the emergence of the second wave of granulosa cells in the ovary suggesting a potential relationship between these two events.
Surprisingly, we identified rare and transient ovarian germ cells with a very similar identity to state f0 Spermatogonia during the late embryonic to early fetal transition. These were detected in both our scRNA-seq dataset and the scRNA-seq dataset compiled by Garcia-Alonso et al. 24, as well as the SMART-Seq dataset of fetal germ cells developed by 42. The fate of these ovarian state f0 spermatogonia-like germ cells is not clear, though one hypothesis is that they ultimately degenerate as development progresses. It is unclear what cues (or missing cues) are responsible for a rare population of ovarian cortical cord germ cells entering state f0 and expressing PIWIL4 rather than entering the meiotic pathway. However, the presence of these cells in the fetal ovary, and within the cortical cords during a time window where the gonadal progenitor program is not fully repressed, supports the concept that ovarian niche and the germ cells themselves are still relatively pliable.
In summary, the formation of the ovary and testis during human embryonic life is one of the most important sex-specific events leading to the emergence of secondary sexual characteristics and sex assignment of babies at birth. In the current study, we show that embryonic and fetal gonadal progenitors and interstitial cells within the prenatal ovary and testis have largely overlapping transcriptional signatures for an extended period during the late first and early second trimester of prenatal life, with the germ cells retaining some plasticity that allows transient state f0 spermatogonia-like cells to be identified in the fetal ovary. This stands in contrast to mice where rapid mutual antagonism of the testis and ovary pathways leads to the maintenance and stabilization of their respective transcriptional programs prior to birth and sex-specific differentiation of ovarian germ cells into meiotic germ cells.
LIMITATIONS OF STUDY
This study involved the analysis of human ovary and testis samples from week 7–21 post fertilization. Given the challenges in accurately assessing human embryo and fetal age, together with the low number of samples across this prenatal time window, future work with additional samples will likely refine the temporal aspects of our findings. Furthermore, in order to accurately decipher the earliest embryonic progenitors associated with human gonad formation, human samples at week 5 post-fertilization will be needed. Given the limitations in using human embryonic and fetal tissue, future studies in non-human primate models will be needed to fill gaps in knowledge before 7 weeks and after 21 weeks post-fertilization.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr Amander Clark (clarka@ucla.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
All software tools can be found online (see Key Resources Table). The accession numbers for the sequencing data reported in this paper are GEO:GSE143380, GEO:GSE161617 25 and GEO:GSE86146 42. Any additional information will be available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-COUP-TFII | R&D Systems | RRID: AB_2155627 |
| Anti-FOXL2 | Novus Biologicals | RRID: AB_2106188 |
| Anti-FOXL2 | Abcam | RRID: AB_869813 |
| Anti-LAMININ | Abcam | RRID: AB_298179 |
| Anti-Ki67 | BD Biosciences | RRID: AB_396287 |
| Anti-KRT19 | Abcam | RRID: AB_2281020 |
| Anti-KRT19 | Novus Biologicals | RRID: AB_2737392 |
| anti-NANOG | Abcam | RRID: AB_10863442 |
| Anti-PDGFRA | R&D Systems | RRID: AB_354459 |
| Anti-PIWIL4 | ThermoFisher | RRID: AB_2548922 |
| Anti-SOX9 | Millipore | RRID: AB_2239761 |
| Anti-VASA (MVH) | R&D Systems | RRID: AB_2277369 |
| Anti-VASA (MVH) | Abcam | RRID: AB_443012 |
| donkey anti-goat IgG AF488 | Jackson Labs | RRID:AB_2340430 |
| donkey anti-goat IgG AF647 | ThermoFisher | RRID:AB_2340437 |
| goat anti-mouse IgG2a AF488 | ThermoFisher | RRID: AB_2535771 |
| donkey anti-mouse IgG AF594 | Jackson Labs | RRID:AB_2340854 |
| donkey anti-mouse IgG2b AF594 | ThermoFisher | RRID: AB_2535781 |
| donkey anti-rabbit IgG AF488 | Jackson Labs | RRID:AB_2313584 |
| donkey anti-rabbit IgG AF647 | Jackson Labs | RRID:AB_2492288 |
| Deposited Data | ||
| Human ovary samples | GEO database | GSE143380 |
| Human testis samples | GEO database | GSE161617 |
| Human fetal gonads (SMART-Seq) | GEO database | GSE86146 |
| Software and Algorithms | ||
| Fiji | Schindelin et al., 2012 51 | RRID:SCR_002285 |
| Imaris | Bitplane | RRID: SCR_007370 |
| Seurat (version: 3.0.0) | Satija et al., 2015 52 | RRID:SCR_016341 |
| Metascape | Zhou et al., 2019 57 | RRID:SCR_016620 |
| Monocle 2 | Trapnell et al., 2014; Qiu et al., 2017 53,54 | RRID:SCR_016339 |
| Cluster 3.0 | de Hoon et al., 2004 56 | http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm |
| Scanpy | Wolf et al., 2018 55 | RRID:SCR_018139 |
| Treeview 3.0 | Keil et al., 2016 58 | https://bitbucket.org/TreeView3Dev/treeview3/src/master/ |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Human fetal tissues
All human fetal tissue was obtained following informed consent. Donated human fetal tissue sent to UCLA did not carry any personal identifiers. No payments were made to donors and the donors knowingly and willingly consented to provide research materials without restrictions for research and for use without identifiers. Prenatal gonads (W5–W16) were obtained from either the University of Washington Birth Defects Research Laboratory (BDRL) or the Karolinska Institutet. At the BDRL, prenatal gonads were obtained with regulatory oversight from the University of Washington IRB approved Human Participants protocol, combined with a Certificate of Confidentiality from the Federal Government. Developmental age was documented by the BDRL as days post fertilization using a combination of prenatal intakes and Carnegie staging. At the Karolinska Institutet, the Regional Human Ethics Committee, Stockholm, Sweden, approved the collection (Dnr 2007/1477–31 with complementary permissions 2011/1101–32 and 2013/564–32) and gave ethical approval to perform the gonadal studies: Dnr 2013/457–31/4). Developmental age was documented by Karolinska Institutet as days post fertilization by the examination of anatomical landmarks such as nervous system, limb, eye and gonadal development according to the atlas of England.
METHOD DETAILS
Immunostaining
Gonads were fixed in 4% PFA at room temperature for 2 hours on a platform rocker, and then washed thrice with PBS and placed into paraffin blocks (Histogel,Thermo Scientific HG4000012) for sectioning and mounting on glass slides. For immunofluorescence staining, sections were deparaffinized and rehydrated using Xylene followed by a graded ethanol series (100%, 95%, 70%, 50%, water, PBS) respectively. Antigen retrieval was performed in a hot water bath (95°C) for 40 minutes using Tris-EDTA solution (pH 9.0). Sections were then washed in PBS, 0.2% Tween-20 (PBS-T) and subsequently permeabilized in PBS, 0.05% Triton X-100 for 10 minutes. Sections were blocked with 10% normal donkey serum for 30 minutes and then incubated with primary antibodies diluted in PBS-T. The primary antibodies, anti-NR2F2 (PP-H7147–00, 1:200, RRID: AB_2155627), anti-FOXL2 (NB100–1277; 1:100, RRID: AB_2106188) (ab246511; 1:100, RRID: AB_869813), anti-GATA4 (sc-25310, RRID: AB_627667), anti-KRT19 (NBP215186; 1:200, RRID: AB_2737392), anti-KRT19 (ab52625; 1:200, RRID: AB_2281020), anti-LAMININ (ab11575, 1:100, RRID:AB_298179); anti-PDGFRA (AF-307, 1:100, RRID: AB_354459); anti-PIWIL4 (PA531448, 1:100, RRID: AB_2548922); anti-SOX9 (ab5535, 1:100, RRID: AB_354459); anti-VASA (AF2030; 1:200, RRID: AB_2277369) (ab13840, 1:200, RRID: AB_443012) were incubated overnight at 4 °C. Sections were then rinsed thrice in PBS-T before incubation at room temperature for one hour in secondary antibodies diluted in PBS-T (PIWIL4 1:1000, all others 1:400; see Key Resources Table). Slides were treated with TruBlack reagent as per kit instrutions. DAPI was then added for ten minutes followed by Prolong Gold antifade mountant (Invitrogen P10144). For sections immunostained for presence of PIWIL4 protein, antigen retrieval was performed with sodium citrate (ph 6.0) and Superblock blocking buffer (Thermo Scientific 37580) and SignalBoost Immunoreaction Enhancer Kit (Millipore 407207) were used.
Microscopy and image analysis
Gonadal sections were examined on an LSM 880 (Carl Zeiss) controlled by Zen Black software with a Plan-Apochromat 20×/0.8 NA and a Plan-Apochromat 40×/1.4 NA M27 oil immersion objective at room temperature. Acquired images were processed using IMARIS 8.1 (Bitplane, RRID: SCR_007370) or Fiji 51 (RRID:SCR_002285) image analysis software.
QUANTIFICATION AND STATISTICAL ANALYSIS
scRNA-seq data analysis
We analyzed the scRNA-seq data of human ovaries using the Seurat package 52 (R package, v.3, RRID:SCR_016341). Unique molecular identifier (UMI) count tables for each ovary were loaded into R using the Read10X function. After adding their sample information, the tables were merged to create a Seurat object. Only cells with >800 genes/cell and < 20% of mitochondrial genome reads were retained. The Seurat object was normalized by default setting and underwent linear dimensional reduction via principle component analysis (PCA). The top 5000 highly variable genes and 1–40 principle components (PCs) were used in the cluster analysis. The uniform manifold approximation and projection (UMAP) technique was used to visualize the dataset by choosing 3–8 PCs in the cluster analysis. We then identified cell types by plotting marker genes with the function FeaturePlot. Contaminating red blood cell genes were detected in the embryonic stage, so the cluster of red blood cells, as well as their marker genes (Supplemental Table 1), were removed from the dataset.
The Seurat object of the ovary and testis somatic cells (Fig. 2A) was generated by reciprocal PCA via Seurat 3. Briefly, to create a combined Seurat object, the UMI count tables of the human ovary and testis somatic cells were merged together, and red blood cell genes (Supplemental Table 1) and some sex-chromosome-linked genes (RPS4Y1, RPS4X, XIST, XIAP) were removed. The combined Seurat object was split by SplitObject function, and PCA was run individually on each dataset (the ovary/testis). This was followed by identifying anchors to integrate the two datasets together via FindIntegrationAnchors and IntegrateData function. Finally, the standard workflow for visualization and clustering was applied.
When performing focused analysis on germ cells or pre-granulosa cells, cell barcodes of the target cells were extracted and the corresponding UMI count table from the total matrix was used to create a new Seurat object. The fetal germ cell and somatic cell dataset from Li et al. 42 was analyzed similarly to the fetal ovary data with the same standard for filtering, normalization, and optimized parameters. The Seurat object of germ cells was converted to a CellDataSet object for importing into the Monocle package 53,54 (v2.10.1, RRID:SCR_016339). Pseudotime analysis was performed according to the default setting.
For the combined analysis of data from Garcia-Alonso et al. 24 and the current manuscript, data was downloaded from the “Reproductive Cell Atlas” (https://www.reproductivecellatlas.org/gonads.html). Corresponding cells from Garcia-Alonso et al. were parsed out based on key cell markers along with their identity labeled in the paper using the Scanpy package 55 (RRID:SCR_018139). The gene matrix and metadata were extracted from the original Scanpy objects and merged for conversion into Seurat objects. The new Seurat object was then loaded and reciprocal PCA employed (Seurat package) to integrate their data with ours. This is a conservative method that removes the batch effect and cells in different states/types are not likely to mix together after integration. Note: For germ cells and common progenitor-bipotential supporting cell analysis, all the corresponding cells from Garcia-Alonso et al. 24 were extracted. For pre-granulosa cell lineage study, the cells from Garcia-Alonso et al. 24 were subset to 10,000 cells to make the two datasets comparable.
Differentially expressed genes (DEGs) between ovaries and testes were identified by the FindMarkers function in Seurat. DEGs among ovarian common progenitor-derived somatic cells or the pre-granulosa lineage were identified by the FindAllMarkers function in Seurat. The genes with expression level difference greater than 1.2 and padjust values less than 0.01 were set as the threshold. DEGs in Fig S4E were clustered by k-means clustering using Cluster 3.0 56 with an optimized clustering number. GO analysis of the DEGs was performed using the online tool Metascape 57 (RRID:SCR_016620).
scRNAseq data can be explored at (https://germline.mcdb.ucla.edu/) and (https://www.reproductivecellatlas.org/gonads.html).
Supplementary Material
HIGHLIGHTS.
Human ovarian stroma and cords arise from stromal-like progenitors before W7.
KRT19 expression spatially discriminates pre-granulosa cells in the cords by W7
Continuous basement membranes between cords and stroma are established by W10.
Spermatogonia-like cells arise transiently within the fetal ovary between W10-W16.
ACKNOWLEDGEMENTS
We are grateful to the donors and their families, who made this work possible. We are grateful for the human fetal tissue procurement supported by an NIH research grant to Ian Glass at the University of Washington Birth Defects Laboratory (5R24HD000836). We thank the UCLA Translational Pathology Core Laboratory and the microscopy core at the UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research Center (BSCRC). This project was funded by NIH Grant HD079546 (ATC), with computational analysis funded by NIH Grant R01AG069725 (BRC). SEW was supported by a UCLA Presidents Postdoctoral Fellowship. ECP was supported by UPLIFT: UCLA Postdocs’ Longitudinal Investment in Faculty (Award # K12 GM106996). BRC was supported by the Howard Hughes Medical Institute. JG was supported by a National Key Research & Development Grant (2022YFA1104100). This work was supported, in part, by the Bill & Melinda Gates Foundation [INV-003385], Ovarian Contraceptive Discovery Initiative (ATC). Under the grant conditions of the Foundation, a Creative Commons Attribution 4.0 Generic License has already been assigned to the author accepted manuscript version that might arise from this submission.
INCLUSION and DIVERSITY
We support inclusive, diverse and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
Amander Clark is on the Board of the International Society for Stem Cell Research. Bradley Cairns and Jingtao Guo are founders of Paterna Biosciences Inc and members of its scientific advisory board. Xichen Nie is an employee and shareholder of Paterna Biosciences Inc. Paterna Biosciences Inc currently focuses on male (primarily adult) germ cell growth and differentiation, which does not overlap significantly with this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Birk OS, Casiano D, Wassif CA, Cogliati T, Zhao L, Zhao Y, Grinberg A, Huang S, Kreidberg JA, Parker KL, et al. (2000). The LIM homeobox gene Lhx9 is essential for mouse gonad formation. Nature 403, 909–913. 10.1038/35002622. [DOI] [PubMed] [Google Scholar]
- 2.Hu YC, Okumura LM, and Page DC (2013). Gata4 is required for formation of the genital ridge in mice. PLOS Genetics 9(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harikae K, Miura K, Shinomura M, Matoba S, Hiramatsu R, Tsunekawa N, Kanai-Azuma M, Kurohmaru M, Morohashi K-I, and Kanai Y (2013). Heterogeneity in sexual bipotentiality and plasticity of granulosa cells in developing mouse ovaries. Journal of Cell Science 126, 2834–2844. [DOI] [PubMed] [Google Scholar]
- 4.Mork L, Maatouk DM, McMahon JA, Guo JJ, Zhang P, McMahon AP, and Capel B (2012). Temporal Differences in Granulosa Cell Specification in the Ovary Reflect Distinct Follicle Fates in Mice1. Biology of Reproduction 86, 37, 31–39. 10.1095/biolreprod.111.095208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niu W, and Spradling A, C. (2020). Two distinct pathways of pregranulosa cell differentiation support follicle formation in the mouse ovary. Proceedings of the National Academy of Sciences 117, 20015–20026. 10.1073/pnas.2005570117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rastetter RH, Bernard P, Palmer JS, Chassot A-A, Chen H, Western PS, Ramsay RG, Chaboissier M-C, and Wilhelm D (2014). Marker genes identify three somatic cell types in the fetal mouse ovary. Developmental Biology 394, 242–252. 10.1016/j.ydbio.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Zheng W, Zhang H, Gorre N, Risal S, Shen Y, and Liu K (2014). Two classes of ovarian primordial follicles exhibit distinct developmental dynamics and physiological functions. Human Molecular Genetics 23, 920–928. 10.1093/hmg/ddt486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim Y, Kobayashi A, Sekido R, DiNapoli L, Brennan J, Chaboissier M-C, Poulat F, Behringer RR, Lovell-Badge R, and Capel B (2006). Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol 4, e187–e187. 10.1371/journal.pbio.0040187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y-H, Chen T-M, Huang B-M, Yang S-H, Wu C-C, Lin Y-M, Chuang J-I, Tsai S-J, and Sun HS (2020). FGF9 is a downstream target of SRY and sufficient to determine male sex fate in ex vivo XX gonad culture. Biology of Reproduction 103, 1300–1313. 10.1093/biolre/ioaa154. [DOI] [PubMed] [Google Scholar]
- 10.Stévant I, Kühne F, Greenfield A, Chaboissier M-C, Dermitzakis ET, and Nef S (2019). Dissecting Cell Lineage Specification and Sex Fate Determination in Gonadal Somatic Cells Using Single-Cell Transcriptomics. Cell Reports 26, 3272–3283.e3273. 10.1016/j.celrep.2019.02.069. [DOI] [PubMed] [Google Scholar]
- 11.Wrobel KH, and Süß F (1998). Identification and temporospatial distribution of bovine primordial germ cells prior to gonadal sexual differentiation. Anatomy and Embryology 197, 451–467. 10.1007/s004290050156. [DOI] [PubMed] [Google Scholar]
- 12.Hummitzsch K, Irving-Rodgers HF, Hatzirodos N, Bonner W, Sabatier L, Reinhardt DP, Sado Y, Ninomiya Y, Wilhelm D, and Rodgers RJ (2013). A New Model of Development of the Mammalian Ovary and Follicles. PLOS ONE 8, e55578. 10.1371/journal.pone.0055578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wartenberg H (1982). Development of the early human ovary and role of the mesonephros in the differentiation of the cortex. Anatomy and Embryology 165, 253–280. [DOI] [PubMed] [Google Scholar]
- 14.Hanley NA, Ball SG, Clement-Jones M, Hagan DM, Strachan T, Lindsay S, Robson S, Ostrer H, Parker KL, and Wilson DI (1999). Expression of steroidogenic factor 1 and Wilms’ tumour 1 during early human gonadal development and sex determination. Mechanisms of Development 87, 175–180. 10.1016/S0925-4773(99)00123-9. [DOI] [PubMed] [Google Scholar]
- 15.Kurek M, Åkesson E, Yoshihara M, Oliver E, Cui Y, Becker M, Alves-Lopes JP, Bjarnason R, Romerius P, Sundin M, et al. (2021). Spermatogonia Loss Correlates with LAMA 1 Expression in Human Prepubertal Testes Stored for Fertility Preservation. Cells 10, 241. 10.3390/cells10020241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurilo LF (1981). Oogenesis in antenatal development in man. Human Genetics 57, 86–92. [DOI] [PubMed] [Google Scholar]
- 17.Mamsen LS, Ernst EH, Borup R, Larsen A, Olesen RH, Ernst E, Anderson RA, Kristensen SG, and Andersen CY (2017). Temporal expression pattern of genes during the period of sex differentiation in human embryonic gonads. Scientific Reports 7, 15961. 10.1038/s41598-017-15931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satoh M (1991). Histogenesis and organogenesis of the gonad in human embryos. J Anat 177, 85–107. [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Workman S, and Wilson MJ (2019). The molecular pathways underlying early gonadal development. Journal of Molecular Endocrinology 62, R47–R64. 10.1530/JME-17-0314. [DOI] [PubMed] [Google Scholar]
- 20.Konishi I, Fujii S, Okamura H, Parmley T, and Mori T (1986). Development of interstitial cells and ovigerous cords in the human fetal ovary: an ultrastructural study. J Anat 148, 121–135. [PMC free article] [PubMed] [Google Scholar]
- 21.Motta PM, Makabe S, and Notttola SA (1997). The ultrastructure of human reproduction. 1. The natural history of the female germ cell: origin, migration and differentiation inside the developing ovary. Human Reproduction Update 3, 281–297. 10.1093/humupd/3.3.281. [DOI] [PubMed] [Google Scholar]
- 22.Wenzel JG, and Odend’hal S (1985). The mammalian rete ovarii: a literature review. Cornell Veterinarian 75, 411–425. [PubMed] [Google Scholar]
- 23.Heeren AM, van Iperen L, Klootwijk DB, de Melo Bernardo A, Roost MS, Gomes Fernandes MM, Louwe LA, Hilders CG, Helmerhorst FM, van der Westerlaken LAJ, and Chuva de Sousa Lopes SM (2015). Development of the follicular basement membrane during human gametogenesis and early folliculogenesis. BMC Developmental Biology 15, 4. 10.1186/s12861-015-0054-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Alonso L, Lorenzi V, Mazzeo CI, Alves-Lopes JP, Roberts K, Sancho-Serra C, Engelbert J, Marečková M, Gruhn WH, Botting RA, et al. (2022). Single-cell roadmap of human gonadal development. Nature 607, 540–547. 10.1038/s41586-022-04918-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chitiashvili T, Dror I, Kim R, Hsu F-M, Chaudhari R, Pandolfi E, Chen D, Liebscher S, Schenke-Layland K, Plath K, and Clark A (2020). Female human primordial germ cells display X-chromosome dosage compensation despite the absence of X-inactivation. Nature Cell Biology 22, 1436–1446. 10.1038/s41556-020-00607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green CD, Ma Q, Manske GL, Shami AN, Zheng X, Marini S, Moritz L, Sultan C, Gurczynski SJ, Moore BB, et al. (2018). A Comprehensive Roadmap of Murine Spermatogenesis Defined by Single-Cell RNA-Seq. Developmental cell 46, 651–667.e610. 10.1016/j.devcel.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen Y-C, Shami AN, Moritz L, Larose H, Manske GL, Ma Q, Zheng X, Sukhwani M, Czerwinski M, Sultan C, et al. (2021). TCF21(+) mesenchymal cells contribute to testis somatic cell development, homeostasis, and regeneration in mice. Nature communications 12, 3876–3876. 10.1038/s41467-021-24130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo J, Sosa E, Chitiashvili T, Nie X, Rojas EJ, Oliver E, Plath K, Hotaling JM, Stukenborg J-B, Clark AT, and Cairns BR (2021). Single-cell analysis of the developing human testis reveals somatic niche cell specification and fetal germline stem cell establishment. Cell Stem Cell 28, 764–778.e764. 10.1016/j.stem.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lottrup G, Nielsen JE, Maroun LL, Møller LMA, Yassin M, Leffers H, Skakkebæk NE, and Rajpert-De Meyts E (2014). Expression patterns of DLK1 and INSL3 identify stages of Leydig cell differentiation during normal development and in testicular pathologies, including testicular cancer and Klinefelter syndrome. Human Reproduction 29, 1637–1650. 10.1093/humrep/deu124. [DOI] [PubMed] [Google Scholar]
- 30.Sato Y, Suzuki T, Hidaka K, Sato H, Ito K, Ito S, and Sasano H (2003). Immunolocalization of Nuclear Transcription Factors, DAX-1 and COUP-TF II, in the Normal Human Ovary: Correlation with Adrenal 4 Binding Protein/ Steroidogenic Factor-1 Immunolocalization during the Menstrual Cycle. The Journal of Clinical Endocrinology & Metabolism 88, 3415–3420. 10.1210/jc.2002-021723. [DOI] [PubMed] [Google Scholar]
- 31.Ostrer H, Masch RJ, and Shapiro E (2007). A cellular study of human testis development. Sexual Development 1, 286–292. 10.1159/000108930. [DOI] [PubMed] [Google Scholar]
- 32.Potter SJ, and DeFalco T (2017). Role of the testis interstitial compartment in spermatogonial stem cell function. Reproduction 153, R151–R162. 10.1530/REP-16-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duffin K, Bayne RAL, Childs AJ, Collins C, and Anderson RA (2009). The forkhead transcription factor FOXL2 is expressed in somatic cells of the human ovary prior to follicle formation. Mol Hum Reprod 15, 771–777. 10.1093/molehr/gap065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Behrens J, von Kries JP, Kühl M, Bruhn L, Wedlich D, Grosschedl R, and Birchmeier W (1996). Functional interaction of β-catenin with the transcription factor LEF-1. Nature 382, 638–642. 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Moreno SA, Lin Y-T, Futtner CR, Salamone IM, Capel B, and Maatouk DM (2019). CBX2 is required to stabilize the testis pathway by repressing Wnt signaling. PLOS Genetics 15, e1007895. 10.1371/journal.pgen.1007895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jameson SA, Lin Y-T, and Capel B (2012). Testis development requires the repression of Wnt4 by Fgf signaling. Developmental biology 370, 24–32. 10.1016/j.ydbio.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Debeljuk L (2006). Tachykinins and ovarian function in mammals. Peptides 27, 736–742. 10.1016/j.peptides.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 38.García-Ortega J, Pinto FM, Prados N, Bello AR, Almeida TA, Fernández-Sánchez M, and Candenas L (2016). Expression of Tachykinins and Tachykinin Receptors and Interaction with Kisspeptin in Human Granulosa and Cumulus Cells1. Biology of Reproduction 94, 124, 121–110. 10.1095/biolreprod.116.139881. [DOI] [PubMed] [Google Scholar]
- 39.Jørgensen A, Nielsen JE, Blomberg Jensen M, Græm N, and Rajpert-De Meyts E (2012). Analysis of meiosis regulators in human gonads: a sexually dimorphic spatio-temporal expression pattern suggests involvement of DMRT1 in meiotic entry. Mol Hum Reprod 18, 523–534. 10.1093/molehr/gas030. [DOI] [PubMed] [Google Scholar]
- 40.Lei N, Hornbaker KI, Rice DA, Karpova T, Agbor VA, and Heckert LL (2007). Sex-Specific Differences in Mouse DMRT1 Expression Are Both Cell Type- and Stage-Dependent During Gonad Development1. Biology of Reproduction 77, 466–475. 10.1095/biolreprod.106.058784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicol B, Grimm SA, Chalmel F, Lecluze E, Pannetier M, Pailhoux E, Dupin-De-Beyssat E, Guiguen Y, Capel B, and Yao HHC (2019). RUNX1 maintains the identity of the fetal ovary through an interplay with FOXL2. Nature Communications 10, 5116. 10.1038/s41467-019-13060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L, Dong J, Yan L, Yong J, Liu X, Hu Y, Fan X, Wu X, Guo H, Wang X, et al. (2017). Single-Cell RNA-Seq Analysis Maps Development of Human Germline Cells and Gonadal Niche Interactions. Cell Stem Cell 20, 858–873.e854. 10.1016/j.stem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 43.McLaren A, and Southee D (1997). Entry of Mouse Embryonic Germ Cells into Meiosis. Developmental Biology 187, 107–113. 10.1006/dbio.1997.8584. [DOI] [PubMed] [Google Scholar]
- 44.Bowles J, Knight D, Smith C, Wilhelm D, Richman J, Mamiya S, Yashiro K, Chawengsaksophak K, Wilson Megan J, Rossant J, et al. (2006). Retinoid Signaling Determines Germ Cell Fate in Mice. Science 312, 596–600. 10.1126/science.1125691. [DOI] [PubMed] [Google Scholar]
- 45.Koubova J, Menke Douglas B, Zhou Q, Capel B, Griswold Michael D, and Page David C (2006). Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proceedings of the National Academy of Sciences 103, 2474–2479. 10.1073/pnas.0510813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Byskov AG (1986). Differentiation of mammalian embryonic gonad. Physiological Reviews 66, 71–117. 10.1152/physrev.1986.66.1.71. [DOI] [PubMed] [Google Scholar]
- 47.Gruenwald P (1942). The Development of the Sex Cords in the Gonads of Man and Mammals. American Journal of Anatomy 70, 359–397. 10.1002/aja.1000700303. [DOI] [Google Scholar]
- 48.Pinkerton JH, McKay DG, Adams EC, and Hertig AT (1961). Development of the human ovary--a study using histochemical technics. Obstetrics & Gynecology 18, 152–181. [PubMed] [Google Scholar]
- 49.Hummitzsch K, Hatzirodos N, Irving-Rodgers HF, Hartanti MD, Perry VEA, Anderson RA, and Rodgers RJ (2019). Morphometric analyses and gene expression related to germ cells, gonadal ridge epithelial-like cells and granulosa cells during development of the bovine fetal ovary. PLOS ONE 14, e0214130. 10.1371/journal.pone.0214130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu M, Hummitzsch K, Bastian NA, Hartanti MD, Wan Q, Irving-Rodgers HF, Anderson RA, and Rodgers RJ (2022). Isolation, culture, and characterisation of bovine ovarian fetal fibroblasts and gonadal ridge epithelial-like cells and comparison to their adult counterparts. PLOS ONE 17, e0268467. 10.1371/journal.pone.0268467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nature Biotechnology 33, 495–502. 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu X, Hill A, Packer J, Lin D, Ma Y-A, and Trapnell C (2017). Single-cell mRNA quantification and differential analysis with Census. Nature Methods 14, 309–315. 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature Biotechnology 32, 381–386. 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biology 19, 15. 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Hoon MJL, Imoto S, Nolan J, and Miyano S (2004). Open source clustering software. Bioinformatics 20, 1453–1454. 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 57.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Communications 10, 1523. 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keil C, Leach RW, Faizan SM, Bezawada S, Parsons L, and Baryshnikova A (2016). Treeview 3.0 (beta 1) - Visualization and analysis of large data matrices. Zenodo. 10.5281/zenodo.1303402. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All software tools can be found online (see Key Resources Table). The accession numbers for the sequencing data reported in this paper are GEO:GSE143380, GEO:GSE161617 25 and GEO:GSE86146 42. Any additional information will be available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-COUP-TFII | R&D Systems | RRID: AB_2155627 |
| Anti-FOXL2 | Novus Biologicals | RRID: AB_2106188 |
| Anti-FOXL2 | Abcam | RRID: AB_869813 |
| Anti-LAMININ | Abcam | RRID: AB_298179 |
| Anti-Ki67 | BD Biosciences | RRID: AB_396287 |
| Anti-KRT19 | Abcam | RRID: AB_2281020 |
| Anti-KRT19 | Novus Biologicals | RRID: AB_2737392 |
| anti-NANOG | Abcam | RRID: AB_10863442 |
| Anti-PDGFRA | R&D Systems | RRID: AB_354459 |
| Anti-PIWIL4 | ThermoFisher | RRID: AB_2548922 |
| Anti-SOX9 | Millipore | RRID: AB_2239761 |
| Anti-VASA (MVH) | R&D Systems | RRID: AB_2277369 |
| Anti-VASA (MVH) | Abcam | RRID: AB_443012 |
| donkey anti-goat IgG AF488 | Jackson Labs | RRID:AB_2340430 |
| donkey anti-goat IgG AF647 | ThermoFisher | RRID:AB_2340437 |
| goat anti-mouse IgG2a AF488 | ThermoFisher | RRID: AB_2535771 |
| donkey anti-mouse IgG AF594 | Jackson Labs | RRID:AB_2340854 |
| donkey anti-mouse IgG2b AF594 | ThermoFisher | RRID: AB_2535781 |
| donkey anti-rabbit IgG AF488 | Jackson Labs | RRID:AB_2313584 |
| donkey anti-rabbit IgG AF647 | Jackson Labs | RRID:AB_2492288 |
| Deposited Data | ||
| Human ovary samples | GEO database | GSE143380 |
| Human testis samples | GEO database | GSE161617 |
| Human fetal gonads (SMART-Seq) | GEO database | GSE86146 |
| Software and Algorithms | ||
| Fiji | Schindelin et al., 2012 51 | RRID:SCR_002285 |
| Imaris | Bitplane | RRID: SCR_007370 |
| Seurat (version: 3.0.0) | Satija et al., 2015 52 | RRID:SCR_016341 |
| Metascape | Zhou et al., 2019 57 | RRID:SCR_016620 |
| Monocle 2 | Trapnell et al., 2014; Qiu et al., 2017 53,54 | RRID:SCR_016339 |
| Cluster 3.0 | de Hoon et al., 2004 56 | http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm |
| Scanpy | Wolf et al., 2018 55 | RRID:SCR_018139 |
| Treeview 3.0 | Keil et al., 2016 58 | https://bitbucket.org/TreeView3Dev/treeview3/src/master/ |


