Abstract
Homeodomain transcription factors control a variety of essential cell fate decisions during development. To understand the developmental regulation by these transcription factors, we describe here the molecular analysis of paired-like CVC homeodomain protein (PLC-HDP) trafficking. Complementary experimental approaches demonstrated that PLC-HDP family members are exported by the Crm1 pathway and contain an evolutionary conserved leucine-rich nuclear export signal. Importantly, inactivation of the nuclear export signal enhanced protein stability, resulting in increased transactivation of transfected reporters and decreased extracellular secretion. In addition, PLC-HDPs harbor a conserved active nuclear import signal that could also function as a protein transduction domain. In our study, we characterized PLC-HDPs as mobile nucleocytoplasmic shuttle proteins with the potential for unconventional secretion and intercellular transfer. Nucleocytoplasmic transport may thus represent a conserved control mechanism to fine-tune the transcriptional activity of PLC-HDPs prerequisite for regulating and maintaining the complex expression pattern during development.
Ordered development depends on the activity of transcription factors in a controlled manner. One defining feature of eukaryotic cells is their spatial and functional division into the nucleus and the cytoplasm by the nuclear envelope. Thus, among other mechanisms, regulated subcellular localization provides an attractive way to control the activity of transcription factors which has been demonstrated for several key players of signal transduction cascades (reference 6 and references therein). This type of regulation requires a specific and selective transport machinery for the controlled transport of macromolecules between both compartments. Nucleocytoplasmic transport takes place through the nuclear pore (29) and is regulated by specific signals and transport receptors. In general, active nuclear import requires energy and is mediated by short stretches of basic amino acids, termed nuclear localization signals (NLS), which interact with specific import receptors (reviewed in references 3 and 13). In contrast, signal-mediated nuclear export pathways (31) are less understood. The best-characterized nuclear export signals (NES) consist of a short leucine-rich stretch of amino acids, interact with the export receptor Crm1 (references 3 and 13 and references therein), and depend on the RanGTP/GDP axis. Leucine-rich NES have been identified in an increasing number of cellular and viral proteins executing heterogeneous biological functions. These include transcription control (6, 35), cell cycle control (43), and RNA transport (8). Proteins containing both NLS and NES have the capacity for continuous shuttling between the cytoplasm and the nucleus.
Homeodomain proteins (HDPs) have been shown to exert key developmental functions throughout the metazoa since defects in the evolutionary conserved homeobox genes were shown to cause many human disorders and aberrant animal phenotypes (reference 57 and references therein). Homeobox-containing genes encode transcription factors and are characterized by the homeodomain (HD), a motif that directs specific DNA binding to regulate the expression of target genes. Homeobox genes are grouped into several subclasses according to the primary structure of their homeodomain and its flanking sequences (reference 12 and references therein). Among the paired-like subclass, the paired-like CVC (PLC)-HDPs are characterized by a conserved CVC domain and can be grouped into the Vsx-1 and Vsx-2 family (32, 40), containing orthologs from several species. PLC-HDPs appear to play a particular role in ocular development (references 7, 21, 38, and 41 and references therein) and execute their functions by binding to the conserved locus control region (LCR), located upstream of the transcription initiation site of the red opsin gene, and thus specify the development and differentiation of cone photoreceptors and a subset of retinal inner nuclear layer bipolar cells (references 16 and 49 and references therein). The observation that null mutations in Chx10 cause congenital microphthalmia, including small eyes, cataracts, iris coloboma, and blindness in humans (42), mice (5), and zebra fish (2), underscores the importance of the PLC-HDP gene family for retinogenesis. Thus, a precise control of PLC-HDP functions is clearly critical for ordered development and homeostasis.
In concordance with their role as transcriptional regulators, homeoproteins localize predominantly to the nucleus, although several reports characterize them also as nucleocytoplasmic shuttle proteins, e.g., Extradenticle (1), Otx1 (56), and Engrailed (34). Since regulated subcellular localization has been reported for several transcription factors (e.g., p53, STATs, NF-κB, etc.) (6), we investigated the intracellular trafficking of PLC-HDPs and analyzed its consequences for PLC-HDP function as transcriptional regulators. As representatives of the Vsx-1 and Vsx-2 group, we studied the zebra fish Vsx1 and the murine Chx10 protein in detail. Nucleocytoplasmic transport was investigated by interspecies heterokaryon assays, microinjection of recombinant transport substrates, and the use of chemical transport inhibitors. We could demonstrate that PLC-HDPs contain a Crm1-dependent NES, previously described as the “octapeptide.” Nuclear export influenced PLC-HDP transcriptional activation by enhancing proteasomal protein degradation and by facilitating extracellular secretion. The predominant nuclear steady-state localization of PLC-HDPs is mediated by the presence of an active nuclear import signal. This NLS can function also as a protein transduction domain (PTD), explaining the evolutionary conservation of this signal. The integrity of both NES and NLS/PTD appears to be prerequisite for PLC-HDPs to function as mobile nucleocytoplasmic shuttle proteins with the potential for intercellular transfer.
MATERIALS AND METHODS
Plasmids.
Plasmids pc3-DrVsx1-green fluorescent protein (GFP) and pc3-MmChx10-GFP encode a zebra fish Vsx1-GFP or a mouse Chx10-GFP fusion protein, respectively. The coding regions of the genes were amplified by PCR with pSTT91zVsx-1 (27) and pT7tagNChx10 (42) as templates and appropriate primers containing BamHI and NheI restriction sites. The PCR products were subsequently cloned into the vector pc3-GFP as described previously (25). Likewise, truncated forms of various GFP fusion proteins were constructed by the same cloning strategy. To generate NES-deficient GFP fusion proteins critical residues were changed into alanines by mutagenesis as described previously (25). The MmChx10-responsive luciferase reporter pLCR-R-luc was constructed by PCR amplification of the luciferase gene with pHH-luc as the template (23), and appropriate primers containing SpeI/NotI-restriction sites and subsequent cloning into pcDNA3 (Invitrogen). Subsequently, the LCR, together with the red pigment promoter (48), was inserted into this construct by PCR amplification and subsequent cloning by SpeI restriction digest, thereby replacing the cytomegalovirus promoter. Potential nuclear export or import signals were cloned into the bacterial expression vector pGEX-GFP as described previously (45). pGEX-MmChx10 encodes a glutathione S-transferase (GST)-mouse Chx10 fusion protein. Plasmid p3-Crm1-HA, pGEX-RanQ69L, and pSV40-Gal were already described (18, 23).
Cells, transfection, microscopy, and microinjection.
Vero cells, the microglia cell line CRL-2540, 293 cells, NIH 3T3 cells, and HeLa cells were maintained under conditions recommended by the American Type Culture Collection and were prepared for microinjection or transfected as described previously (18). Microinjection, observation, and image analysis in living or fixed cells were performed as described previously (18). Cells were observed and analyzed by using the appropriate fluorescence filters as described previously (19), and 12-bit black and white images were captured by using a digital Axiocam CCD camera (Zeiss). Quantitation, image analysis, and presentation was performed by using IPLab Spectrum (Scanalytics) and Axiovision software (Zeiss). The total cellular GFP signal was measured by calculating the integrated pixel intensity in the imaged cell multiplied by the area of the cell. The nuclear signal was similarly obtained by measuring the pixel intensity in the nucleus. The cytoplasmic signal was calculated by subtracting the nuclear signal from the total cellular signal. All pixel values were measured below the saturation limits, and the background signal in an area with no cells was subtracted from all values. To determine the average intracellular localizations of the respective proteins, at least 200 fluorescent cells in three independent experiments were examined, and the standard deviations were determined.
Transactivation assays.
For transactivation assays, HeLa cells were transfected with 0.5 μg of the pLCR-R-luc reporter plasmid and the indicated amounts of the MmChx10 expression constructs, together with 0.1 μg of pSV40-Gal, and the cells were assayed for luciferase and β-galactosidase (β-Gal) activity as described previously (23). To analyze intercellular transactivation, 5 × 105 293 cells were transfected with either 3 μg of the indicated MmChx10 expression construct or with 1 μg of pLCR-R-luc and 0.1 μg of pSV40-Gal. At 12 h later MmChx10 transfected and pLCR-R-luciferase transfected cells were mixed at a ratio of 2:1 and assayed for luciferase and β-Gal activity 36 h later. Luciferase activity was normalized to β-Gal expression, and all measurements were conducted in duplicates in three independent experiments.
Purification of recombinant GST fusion proteins.
GST-GFP hybrid proteins were expressed and purified as described previously (45). Removal of GST by proteolytic cleavage using factor Xa protease (Roche) was performed according to the manufacturer's recommendations.
Immunoblotting, immunofluorescence, and antibodies.
Immunoblotting and immunofluorescence were carried out according to standard procedures, as previously described (18). Purified mouse Chx10 fused to GST was used for immunization of rabbits by using standard protocols (10). The immunoglobulin G fraction was purified by protein A chromatography and used at a 1:500 dilution for immunofluorescence.
Protein transduction assay.
Exponentially growing Vero cells were incubated with 1 μM concentrations of the corresponding recombinant GFP fusion proteins in phosphate-buffered saline (PBS) for 2 h. Subsequently, cells were extensively washed with PBS before incubation with trypsin (1 mg/ml) for 2 min to remove unspecifically bound protein from the cell surface. After removal of trypsin cells were cultured in medium for 3 h, washed with PBS, fixed with ice-cold methanol for 15 min and rehydrated in PBS prior to analysis by fluorescence microscopy.
Heterokaryon assay.
HeLa cells were transfected with the indicated plasmids and 12 h later seeded with untransfected mouse NIH 3T3 cells at a ratio of 1:3. Cells were cultured and fused 8 h later by using polyethylene glycol (Gibco) in the presence of cycloheximide as described previously (50). To discriminate between human donor and mouse acceptor nuclei, staining with Hoechst 33258 was performed as described previously (50). A total of 50 heterokaryons were chosen at random, and the percentage of fusion events positive for internuclear transfer was calculated in three independent experiments, and the standard deviations were determined.
Treatment with chemical export inhibitors.
Cells transfected with the indicated plasmids were treated with 10 nM leptomycin B (LMB; Sigma-Aldrich) or 5 nM Ratjadone A (Alexis Biochemicals) as described previously (25).
Crm1 pull-down assays and in vitro translation.
Coupled transcription-translation was performed by using the TNT reticulocyte lysate system (Promega) supplemented with [35S]methionine (Amersham) and the plasmid p3-Crm1-HA as a template. Crm1 pull-down assays with the specific recombinant GST-GFP substrates, Ran-GTP and nuclear extracts were performed as described previously (18). Care was taken to ensure equal input levels of labeled Crm1 protein into the binding reactions.
Secretion assay.
A total of 2 × 106 293 cells were transfected with the indicated plasmids and incubated for 8 h. Subsequently, cells were cultured in methionine-free medium supplemented with [35S] methionine (50 μCi) for additional 12 h. To block classical protein secretion, brefeldin A (BFA; Sigma-Aldrich) at 10 μg/ml was added to the cultures. Culture supernatants were collected and cleared by centrifugation (10,000 × g, 1 h, 4°C). Analysis of whole-cell lysates and immunoprecipitation of GFP fusion proteins from culture supernatants and cellular lysates by using a polyclonal anti-GFP antibody (BD Biosciences), as well as analysis of the complexes by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography, were performed as described previously (18, 28).
Pulse-chase experiments.
A total of 5 × 105 HeLa cells were transfected with pc3DrVsx1-GFP or pc3DrVsx1_NESmut-GFP, followed by incubation for 16 h. Subsequently, cells were incubated for 2 h in Dulbecco’s modified Eagle medium lacking methionine and pulse-labeled with 50 μCi of [35S]methionine (Amersham) for 2 h. Unlabeled methionine was then added to a final concentration of 100 mM. At the indicated time points, cells were washed with cold PBS, and whole-cell lysates were prepared as described previously (28). To prevent proteasomal degradation, cells were treated with the proteasome inhibitors MG-132 and hemin (Sigma-Aldrich; 50 μM final concentration). The total radioactivity in each sample was determined by trichloroacetic acid precipitation, and sample volumes were adjusted to represent equal amounts of radioactivity. Immunoprecipitation was done by using a polyclonal anti-GFP antibody (Clontech), and the complexes were resolved by SDS-PAGE as described previously (18). Band intensities were quantified by using a phosphorimager (Bio-Rad).
RESULTS
PLC-HDPs are active shuttle proteins and nuclear export is mediated by the Crm1 pathway.
To study PLC-HDPs localization and trafficking in live cells, we expressed the complete zebra fish (Dr) Vsx1 (amino acids [aa] 1 to 344) and murine (Mm) Chx10 (aa 1 to 380) as GFP fusion proteins. Fluorescence microscopy revealed that DrVsx1-GFP and MmChx10-GFP were predominantly nuclear. However, a significant amount of the respective protein was detectable also in the cytoplasm following transient expression in human (HeLa and 293) and rodent (NIH 3T3) cell lines (Fig. 1A and E and data not shown), indicating their potential for nucleocytoplasmic transport. Indirect immunofluorescence revealed a similar intracellular localization for the endogenous MmChx10 in the microglia cell line CRL-2540 (Fig. 1D) thereby excluding the possibility that the observed localization was due to the ectopic expression of GFP-tagged fusion proteins. Antiserum specificity was confirmed by staining MmChx10-GFP expressing HeLa cells (data not shown).
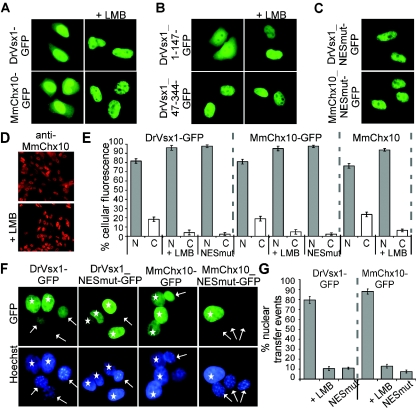
FIG. 1.
PLC-HD proteins are nucleocytoplasmic shuttle proteins. (A) HeLa cells were transfected with the indicated plasmids and analyzed by fluorescence microscopy. In living cells, DrVsx1-GFP and MmChx10-GFP localized predominantly to the nucleus. Significant amounts of the proteins were also detectable in the cytoplasm and accumulated completely in the nucleus after LMB treatment. (B) DrVsx1_1-147-GFP still responded to LMB treatment, whereas the construct lacking the first 47 aa (DrVsx1_47-344-GFP) displayed an exclusively nuclear localization. (C) Inactivation of the NES by mutating critical residues into alanines (DrVsx1_NESmut-GFP, aa37FAITDLLGL45 → 37AAITDLAGA45; MmChx10_NESmut-GFP, aa32FGIQEILGL40 → 32AGIQEIAGA40) resulted in complete nuclear localization. (D) Endogenous MmChx10 protein in the microglia cell line CRL-2540 displayed a similar intracellular localization and LMB responsiveness as observed for the MmChx10-GFP protein. MmChx10 was visualized by indirect immunofluorescence with a polyclonal anti-MmChx10 antiserum. (E) To determine the average intracellular localizations of the respective proteins, at least 200 fluorescent cells in three independent experiments were examined, and the standard deviations were determined. (F) DrVsx1- and MmChx10-GFP are capable of nucleocytoplasmic trafficking in a heterokaryon assay. Upon polyethylene glycol fusion of DrVsx1-GFP- and MmChx10-GFP-expressing HeLa donor cells with untransfected NIH 3T3 acceptor cells, DrVsx1-GFP and MmChx10-GFP were exported from the donor (marked by asterisks) and imported into the mouse acceptor nuclei (marked by arrows) 60 min after fusion. In contrast, NES-deficient mutants (DrVsx1_NESmut-GFP and MmChx10_NESmut-GFP) were not exported. (G) To quantify the number of transfer events, 50 heterokaryons were chosen at random, and the percentage of fusion events positive for internuclear transfer was calculated in three independent experiments with standard deviations. Scale bars: 10 μm (A, B, C, and F) and 100 μm (D).
To examine whether nuclear export was mediated via the Crm1 pathway, we used the export inhibitors LMB and Ratjadone A. These substances bind to Crm1, thereby preventing the interaction with leucine-rich NES (24, 55). LMB or Ratjadone A treatment not only resulted in exclusive nuclear accumulation of DrVsx1- and MmChx10-GFP in transfected HeLa and 293 cells but also blocked export of the endogenous MmChx10 protein (Fig. 1A/D/E and data not shown).
To further address whether DrVsx1-GFP and MmChx10-GFP were capable of nucleocytoplasmic trafficking, we performed heterokaryon assays in the presence of cycloheximide to prevent de novo protein synthesis. Upon fusion of DrVsx1-GFP and MmChx10-GFP expressing HeLa donor cells with untransfected NIH 3T3 acceptor cells, both PLC-HDPs were exported from the donor and imported into the mouse acceptor nuclei 60 min after fusion (Fig. 1F and G). As a control, incubation of the fused cells at 4°C (data not shown) or in the presence of LMB did not result in detectable accumulation of GFP fusion proteins in the acceptor nuclei, indicative for active transport (Fig. 1G). Since the cytoplasm of donor and acceptor cells are fused in the heterokaryon assay the low amount of cytoplasmic DrVsx1-GFP or MmChx10-GFP protein, respectively, initially present in the donor cells was diluted and not detectable postfusion. Of note, the level of nuclear fluorescence in the acceptor nuclei increased, and the fluorescence signal in the donor nuclei decreased over time, excluding the formal possibility that the observed nuclear transfer events resulted from the import of the cytoplasmic GFP fusion proteins present prior to fusion.
PLC-HDPs contain a highly conserved NES previously described as the “octapeptide.”
To identify domains directing nuclear export, we first expressed N- and C-terminal deletion mutants of DrVsx1 and MmChx10 as GFP hybrids. As indicated in Fig. 1B, only fusion proteins containing the first 47 aa responded to LMB treatment, indicating the presence of an active NES. Database searches identified potential NES in the PLC-HDPs, matching the still loosely defined consensus sequence for leucine-rich NES (18, 20). Because predicted signals need to be verified experimentally, we tested the activity of the potential NES in a highly stringent system that allows the observation and quantification of nuclear export in living cells, independent of drug treatment, nuclear import, and passive diffusion (45). Signals (Fig. 2A) were expressed as fusions with GST and GFP (GST-NES-GFP) and tested by microinjection. Due to the size of the fusion proteins (54 kDa, as a monomer) the localization of the microinjected autofluorescent transport substrate is not flawed by passive diffusion, and the protein remains at the site of injection for up to 24 h (45). We observed that only substrates containing active PLC-HDP NES were quantitatively exported into the cytoplasm within 16 h after microinjection into the nucleus of Vero (Fig. 2B) and microglia CRL-2540 cells (data not shown). As a stringent control, a signal in which essential residues were replaced by alanines was inactive under identical experimental conditions (Fig. 2A and B). Likewise, treatment with LMB completely prevented export (data not shown). Interestingly, analysis of NES representative for all known PLC-HDP family members revealed that these NES mediated export with comparable kinetics (Fig. 2A and B). Approximately 100 cells were injected and analyzed, and representative examples are shown. These results were confirmed in two independent experiments (data not shown). The evolutionary conservation of the NES strongly argues that nuclear export is critical for the biological function of PLC-HDPs. Since nuclear export had been proposed also for other members of the homeoprotein family (34), we included the proposed NES of the Engrailed homeoprotein (Fig. 2A; GgEn2 NES) in our study. In contrast to the PLC-HDP NES, the GgEn2 NES was not active in our assay (data not shown).
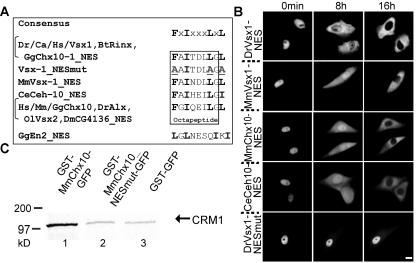
FIG. 2.
PLC-HDPs contain evolutionarily conserved active NES and interact with Crm1 in vitro. (A) Alignment of the tested PLC-HDPs export signals from different species with the NES consensus motif (18) and the inactive Engrailed “NES.” (B) Indicated GST-NES-GFP substrates were microinjected into the nuclei of Vero cells, and nuclear export was recorded in living cells by fluorescence microscopy after various time points. Approximately 100 cells were injected and representative examples are shown. Panels: left, t = 0 min; middle, t = 8 h; right, t = 16 h. Nuclear export was completed after 16 h. Inactivation of the NES by mutating critical residues into alanines (DrVsx1_NESmut) completely blocked export. (C) MmChx10 interacts with Crm1 in a GST pull-down assay. In vitro-translated 35S-labeled Crm1 protein was incubated with equal amounts of immobilized full-length GST-MmChx10-GFP, GST-MmChx10_NESmut-GFP, or GST-GFP in the presence of GST-RanQ69L and nuclear extracts. The specific binding of Crm1 to GST-MmChx10-GFP (lane 1) was abolished by mutating the NES (lane 2). GST-GFP served to control for unspecific binding (lane 3). Homo sapiens (Hs), Mus musculus (Mm), Bos taurus (Bt), Danio rerio (Dr), Carassius auratus (Ca), Oryzias latipes (Ol), Gallus gallus (Gg), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm). Scale bar, 10 μm.
NES inactivation prevents export of PLC-HDPs.
To verify the functionality of the export signals also in the context of the full-length proteins in vivo, we mutated critical residues of the NES into alanines (DrVsx1_NESmut-GFP, aa37FAITDLLGL45 → 37AAITDLAGA45; MmChx10_NESmut-GFP, aa32FGIQEILGL40 → 32AGIQEIAGA40). In contrast to the wild-type proteins DrVsx1_NESmut-GFP and MmChx10_NESmut-GFP displayed a complete nuclear localization after transient transfection (Fig. 1C). Likewise, the NES-deficient mutants were not exported in the heterokaryon assays (Fig. 1F and G), excluding the presence of additional NES or the possibility that export was mediated by shuttling interaction partners in trans.
PLC-HDP export signals interact with Crm1 in vitro.
If the defined PLC-HDPs are exported via the Crm1 pathway, these proteins should interact with the export receptor also in a cell-free system. We therefore performed in vitro interaction assays to biochemically verify the Crm1 interaction. Figure 2C demonstrates that recombinant GST-MmChx10-GFP significantly bound to Crm1 in the presence of Ran-GTP and nuclear extracts in contrast to inactive GST-MmChx10_NESmut-GFP or GST-GFP alone. Similar results were obtained for GST-DrVsx1-GFP or the other GST-NES-GFP fusion proteins, respectively (data not shown).
Nuclear export facilitates intracellular degradation of DrVsx1-GFP.
Kurtzman et al. (26) reported the polyubiquitination and degradation of DrVsx1 by the ubiquitin/proteasome pathway. Because the proteasome degradative pathway appears to operate predominantly in the cytoplasm (51), we investigated whether nuclear export influences indirectly the intracellular stability of DrVsx1. HeLa cells transiently expressing DrVsx1-GFP or DrVsx1_NESmut-GFP, respectively, were metabolically pulse-labeled, followed by a chase with cold methionine for 0, 30, 60, and 120 min. Subsequently, GFP fusion proteins were immunoprecipitated with anti-GFP antiserum and resolved by SDS-PAGE (Fig. 3A). Band intensities from two independent experiments were quantified by using a phosphorimager; this showed that both DrVsx1-GFP and DrVsx1_NESmut-GFP were degraded over time and that degradation could be reduced by treatment with proteasomal inhibitors. Interestingly, preventing nuclear export resulted in a significantly increased half-live for DrVsx1_NESmut-GFP (Fig. 3A and B), suggesting that nuclear export is continuously supplying substrate for the proteasomal degradation machinery. Similar results were obtained for MmChx10 (data not shown).
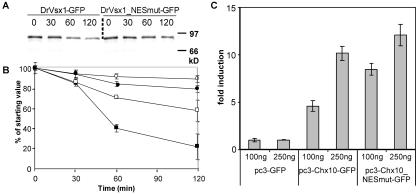
FIG. 3.
Nuclear export affects PLC-HD protein stability and transcriptional activation. (A) Preventing nuclear export increases the intracellular stability of DrVsx1-GFP. HeLa cells were transiently transfected with expression plasmids encoding DrVsx1-GFP (2 μg) or DrVsx1-NESmut-GFP (2 μg). After 16 h, methionine-starved cells were pulsed with [35S]methionine for 2 h and chased with excess methionine for the indicated times. Proteins were immunoprecipitated with anti-GFP antibody, resolved by SDS-PAGE and detected by fluorography. Whereas DrVsx1-GFP and DrVsx1-NESmut-GFP were degraded over time, inactivation of the NES resulted in a significantly increased half live of DrVsx1-NESmut-GFP. (B) Band intensities from two independent experiments (including the gel in panel A) were quantified by using a phosphorimager and graphed with standard errors for DrVsx1-GFP (▪), DrVsx1-GFP + proteasomal inhibitors (PI) (•), DrVsx-1_NESmut-GFP (□), and DrVsx-1_NESmut-GFP + PI (○). (C) Luciferase assays after cotransfection of HeLa cells with pSV40-Gal, an MmChx10-responsive luciferase reporter and different amounts of expression plasmids for GFP, MmChx10-GFP, and MmChx10_NESmut. Expression of wild-type MmChx10-GFP resulted in higher transcriptional activation compared to the export-deficient mutant. Luciferase activity was normalized to β-Gal expression. Error bars indicate the standard deviations.
Nuclear export influences MmChx10-mediated transactivation.
To investigate the effect of export-enhanced degradation on the transcriptional activity of MmChx10, we tested the ability of the export-defective MmChx10 to transactivate a MmChx10-responsive luciferase reporter plasmid in transient transfections. These experiments revealed a good correlation between dose-dependent MmChx10 mediated transactivation and protein stability since expression of MmChx10_NESmut resulted in increased stimulation of gene expression (Fig. 3C).
Nuclear export facilitates unconventional secretion of PLC-HD proteins.
Having demonstrated that PLC-HDPs are nucleocytoplasmic shuttle proteins, we investigated their potential for intercellular trafficking. In general, intercellular transfer requires both internalization and secretion. To analyze secretion of DrVsx1-GFP and to investigate the influence of nuclear export on secretion, we attempted to recover metabolically labeled DrVsx1-GFP or DrVsx1_NESmut-GFP protein, respectively, from the culture supernatant of transfected 293 cells. Figure 4A illustrates that DrVsx1-GFP, but not DrVsx1_NESmut-GFP could be immunoprecipitated by anti-GFP antibodies from the supernatant. Transfected cells were controlled by microscopic observation for cytotoxic effects caused by the expression of the respective GFP fusion proteins prior to lysate preparation to minimize unspecific protein release due to cell death. To also exclude the possibility that the observed result reflects differences in protein expression, equal expression levels of the GFP fusion proteins were verified by Western blot analysis of cellular extracts (Fig. 4B). Similar results were obtained for the MmChx10-GFP or MmChx10_NESmut-GFP protein, respectively (data not shown). Of note, we could not recover a NES-GFP fusion protein (DrVsx1 NES-GFP) from the supernatant of transfected 293 cells (data not shown). Thus, the continuous supply of cytoplasmic DrVsx1 from the nuclear pool by active nuclear export appears to facilitate secretion, but the NES itself does not represent an unconventional secretion signal. To determine whether protein secretion was mediated via the classical endoplasmic reticulum/Golgi-dependent pathway or by unconventional secretion, we attempted to inhibit secretion by treatment of the transfected cells with BFA. However, BFA treatment did not interfere with MmChx10-GFP release in support of secretion by the unconventional pathway, as also reported for several other proteins (39; data not shown).
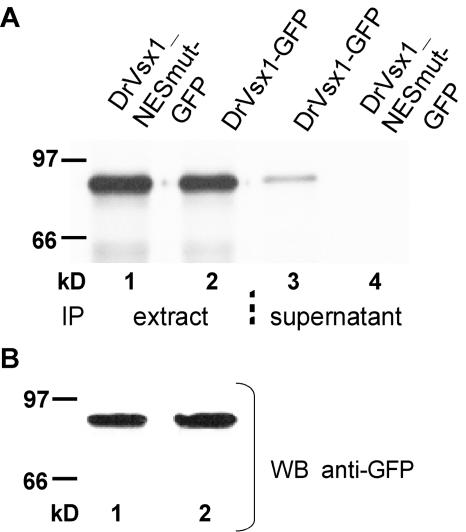
FIG. 4.
Nuclear export affects DrVsx1-GFP protein secretion. (A) 293 cells were transfected with the indicated plasmids (4 μg) and cultured in methionine-free medium supplemented with [35S]methionine. GFP fusion proteins were immunoprecipitated from the culture supernatants and from whole-cell lysates with an anti-GFP antibody. Although DrVsx1_NESmut-GFP and DrVsx1-GFP could be immunoprecipitated equally from cellular lysates (lanes 1 and 2), only DrVsx1-GFP could be recovered from the supernatants (lanes 3 and 4). (B) Equal expression levels of the GFP fusion proteins were verified by Western blot analysis of cellular lysates using a polyclonal anti-GFP antiserum.
PLC-HDPs contain a highly conserved active nuclear import signal which can function as a PTD.
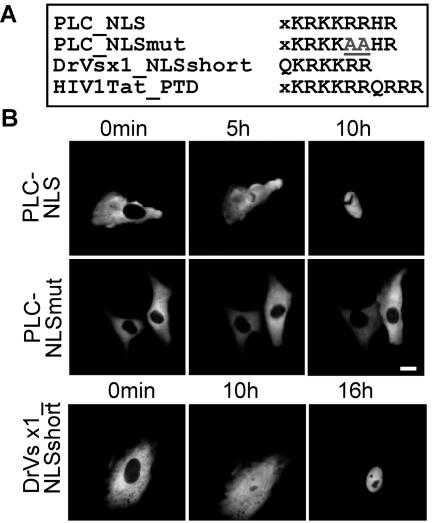
For continuous signal-mediated shuttling between the cytoplasm and the nucleus proteins require both NES and NLS. Thus, we next sought to determine whether the predominant nuclear steady-state localization of PLC-HDPs is the result of active nuclear import or is mediated by nuclear retention. The database comparisons revealed that the motif KRKKRRHR located at the beginning of the homeodomain is 100% conserved in all known PLC-HDPs (Fig. 5A). In contrast to GFP alone, a KRKKRRHR-GFP fusion protein localized to the nucleus (data not shown). However, because even a GFP-GFP fusion protein (54 kDa) can enter the nucleus by passive diffusion (45), we investigated whether this motif can function not only as nuclear retention but also as an active nuclear import signal. Microinjection experiments with recombinant GST-GFP fusion proteins (Fig. 5B) demonstrated that the tested signal mediated nuclear import and can therefore be considered as a bona fide nuclear import signal for PLC-HDs (PLC_NLS). Import activity was lost by replacing two conserved arginines by alanines (Fig. 5A and B, PLC_NLSmut). Approximately 100 cells were injected and analyzed, and representative examples are shown. These results were confirmed in two independent experiments (data not shown). Our observations are supported by the report of Kurtzman and Schechter (27), who demonstrated that a DrVsx1 mutant lacking the sequence QKRKKRR no longer accumulated in the nucleus. Of note, GST-QKRKKRR-GFP (Vsx1_short) was less active in mediating import (Fig. 5B).
FIG. 5.
PLC-HD proteins contain a highly conserved active nuclear import signal. (A) Sequence alignment of the NLS conserved in all PLC-HDP members, the inactive NLS mutant, Vsx1_short, and the HIV1Tat_PTD. (B) GST-NLS-GFP fusion protein were microinjected into the cytoplasm of Vero cells, and nuclear import was observed directly by fluorescence microscopy. Approximately 100 cells were injected, and representative examples are shown. Nuclear import of GST-PLC_NLS-GFP was completed after 10 h (upper panel). Import activity was lost by replacing two conserved arginines by alanines (GST-PLC_NLSmut-GFP, middle panel). In contrast, Vsx1_short was less active in mediating import (GST-Vsx1_NLSshort-GFP, lower panel). Scale bars, 10 μm.
Interestingly, the KRKKRRHR motif displayed a high homology to the widely used PTD KRKKRRQRRR of the human immunodeficiency virus type 1 (HIV-1) Tat protein (Fig. 5A) (52). To test the potential of the PLC-HD_NLS to also traverse intact cellular membranes, human cells were incubated with recombinant GFP fusion proteins, followed by treatment with trypsin to remove unspecifically bound protein. Fluorescence microscopy revealed that PLC-HD_NLS-GFP displayed a similar protein transduction activity as the positive control, HIV1Tat_PTD-GFP, and localized to the cytoplasm and nucleus of the treated cells (Fig. 6A). In contrast, GFP fusion proteins containing the QKRKKRR motif (Vsx1-short-GFP), the mutated NLS, or GFP alone could not mediate protein transduction under identical experimental conditions, arguing against PTD-independent cellular entry (Fig. 6A). These results provide a rational for the evolutionary conservation of the bifunctional KRKKRRHR motif. Importantly, recombinant full-length DrVsx1-GFP protein was also able to enter cells, although less efficiently, most likely due to its larger size since GST-HIV1Tat_PTD-GFP or GST-PLC-HD_NLS-GFP, respectively, also displayed a diminished transduction activity (data not shown).
FIG. 6.
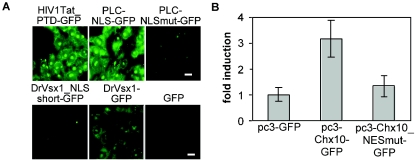
PLC-HDPs have the capacity for intercellular transport. (A) The PLC-HD NLS can function as a PTD. Vero cells were incubated with 1 μM concentrations of the indicated GFP fusion proteins for 2 h and treated as described in Materials and Methods. Fluorescence microscopy indicated that DrVsx1_NLS-GFP, HIV1Tat_PTD-GFP and, to a lesser extent, DrVsx1-GFP were able to enter the cells. In contrast, DrVsx1_NLSmut-GFP, DrVsx1_short-GFP or GFP could not mediate cellular entry. Scale bars, 10 μm. (B) Inhibition of nuclear export interferes with intercellular transactivation. Luciferase assays after cocultivation of 293 cells expressing MmChx10-GFP, MmChx10_NESmut-GFP, or GFP, together with 293 cells transfected with the MmChx10-responsive luciferase reporter and pSV40-Gal. Luciferase activity was normalized to β-Gal expression. Error bars indicate the standard deviations.
PLC-HD proteins have the potential for intercellular transport.
Having demonstrated that PLC-HDPs are nucleocytoplasmic shuttle proteins, can be secreted, and contain a PTD, we investigated their potential for intercellular trafficking. Although described for the Engrailed protein (22), we could not visually detect the spread of DrVsx1-GFP or MmChx10-GFP from transfected to untransfected HeLa cells by fluorescence microscopy upon cocultivation for up to 72 h (data not shown). This might be due to the low amount of secreted and internalized protein which could be below the detection level. However, since even low concentrations of transcription factors are sufficient to trigger biological relevant responses in vivo, we used an intercellular transactivation assay to investigate intercellular trafficking. 293 cells transfected with MmChx10-GFP, MmChx10_NESmut-GFP, or GFP alone were cocultivated with 293 cells transfected with the MmChx10 responsive pLCR-R-luc reporter plasmid. Intercellular transport of MmChx10 should result in enhanced luciferase activity that should be abolished by NES inactivation. Figure 6B indicates that the wild-type but not the export-deficient MmChx10 protein was able to activate reporter gene expression, supporting the potential of CVC-HDPs for intercellular trafficking.
DISCUSSION
Transcriptional networks ensure the ordered development of complex multifunctional organs as exemplified by the spinal cord (17) or the retina (9). In particular, homeodomain proteins represent transcription factors exerting key developmental functions. The paired-like CVC-HDPs play an essential role in ocular development and, therefore, a precise control of PLC-HDPs functions is critical for ordered development and homeostasis. In eukaryotic cells, the nuclear envelope generates two distinct cellular compartments that separate transcription and DNA replication from protein biosynthesis. Among other mechanisms, regulated subcellular localization provides an attractive way to control the activity of PLC-HDPs. We demonstrated for two representatives of the PLC-HDP family that endogenous MmChx10, as well as ectopically expressed MmChx10-GFP and DrVsx1-GFP proteins, did not exclusively localize to the nucleus. A similar, nonexclusive nuclear localization was recently reported for the MmChx10 protein (46). This could either be due to the retention of newly synthesized protein in the cytoplasm or to its continuous nucleocytoplasmic transport. We showed that PLC-HDs can shuttle between the nucleus and the cytoplasm by using the heterokaryon assay. Furthermore, we characterized the previously described “octapeptide” as part of an evolutionary conserved active leucine-rich NES present in all members of the PLC-HDP family. Nuclear export of PLC-HDPs was mediated by the Crm1 pathway, as supported by several lines of evidence. First, Crm1 antagonists caused nuclear accumulation of DrVsx1 and MmChx10, were able to block nuclear export in the heterokaryon assay, and prevented export of recombinant PLC-HDP-NES transport substrates. Second, DrVsx1-GFP, MmChx10-GFP, and PLC-HDP-NES bound to Crm1 in vitro, and these interactions could be prevented by mutating critical residues in the NES which also blocked export of the full-length proteins in vivo. The NES of PLC-HDPs fit the still loosely defined consensus sequence for leucine-rich export signals and are evolutionary conserved in all known PLC-HDPs from human, mouse, rat, chicken, and zebra fish (see Fig. 3A). Interestingly, the tested PLC-HDP NES were equally active in microinjection experiments and displayed a similar activity, as observed for the NES from other transcriptional regulators such as p53 or Mdm2 (19). As demonstrated in our previous work (18) and by others (20), it appears that the distance between the critical LxL motif and the next hydrophobic residue should not exceed 3 aa in the proposed NES consensus sequence (see Fig. 3). We are not aware of any functional NES breaking this rule. The 4-aa spacer in the suggested Engrailed export signal (34) marks this sequence as nonfunctional explaining the lack of activity observed in our study. To date, nuclear export has been proposed for a growing list of proteins, including also several homeodomain proteins, e.g., Extradenticle (1), Otx1 (56), and Engrailed (34). However, the numerous reports on nuclear export sometimes lead to conflicting results. To standardize the definition for active, Crm1-mediated nuclear export mediated by a “classical” leucine-rich NES, we propose the following quality criteria. (i) Nuclear export of a protein, as assayed by transfection and heterokaryon assay, should be blocked by Crm1 inhibitors. (ii) The export signal should be active also in the context of a heterologous system in trans and should interact with Crm1 in vitro. (iii) Mutation of critical residues in the NES should inactivate its export activity also in the context of the full-length protein. According to our knowledge, the present study is the first to demonstrate the nuclear export of homeodomain proteins fulfilling all of these criteria.
As transcription factors, PLC-HDPs have to access the nucleus to execute their function. Theoretically, the size of ca. 34 kDa allows PLC-HDPs to enter the nucleus also by passive diffusion. However, even smaller proteins are transported by active, signal-mediated mechanisms, most likely because active transport is more efficient and amendable to specific control mechanisms (3, 13). The transfection and/or microinjection experiments indicated that the conserved KRKKRRHR motif can not only function as a nuclear retention signal but also represents a bona fide monopartite nuclear import signal for PLC-HDPs in which the underlined arginines are critical for function and efficiency. Although Ubc9 has been suggested to mediate the nuclear localization of Vsx1 (27), we are currently investigating in detail whether Vsx1 is directly imported via the transportin 13/Ubc9 axis (37) or can also use alternative import pathways. Although the PLC-HDPs NLS is less active compared to the classical SV40 NLS (45), the rate of import still exceeds the rate of export, resulting in the observed dynamic but predominantly nuclear steady-state localization of PLC-HDPs.
The activity of transcriptional regulators can be modulated at various levels. As shown for other transcription factors (6, 44), these include posttranscriptional modifications in the nucleus or the cytoplasm, e.g., phosphorylation (35), sumoylation (10), and interactions with other proteins (14). The detailed molecular pathways regulating the activity of PLC-HDPs are currently under intense investigation. Here, we provided evidence that PLC-HDPs are dynamic transcription factors that have the capability to shuttle between the nucleus and the cytoplasm. Consequently, PLC-HDPs might be subjected to regulatory control mechanism homing in these specific compartments. Degradation by the ubiquitin/proteasome pathway is crucial to control protein homeostasis and was described for several transcription factors, including the DrVsx1 protein (26). In the present study, we found that inactivation of the nuclear export activity of DrVsx1 and MmChx10 resulted in increased protein stability and thus in increased transcriptional activation. Since the ubiquitin/proteasome pathway appears to act predominantly in the cytoplasm (47), our transactivation result indicate that regulating nucleocytoplasmic transport can indirectly influence the intracellular protein levels and the biological activity of PLC-HDPs by the proteasome pathway. A similar model was proposed for the IκB/NF-κB axis in which the nuclear export activity of IκB regulates the intracellular localization, degradation, and transcriptional activity of NF-κB (see references 30 and 33 and references therein). In contrast, Rehberg et al. reported that the inactivation of the NES in the Sox10 protein resulted in decreased transactivation by an unknown mechanism (44). Although it is less likely, we cannot formally rule out the possibility that the mutations introduced to generate the export deficient DrVsx1 protein directly resulted in enhanced degradation resistance. The increased stability observed would thus be due to conformational changes and not caused by blocking export.
We found that the continuous supply of cytoplasmic DrVsx1 or MmChx10 from the nuclear pool by active export facilitated also extracellular release of the proteins. PLC-HDPs in general lack a canonical secretion signal and appear not to be targeted into the endoplasmic reticulum by a cotranslational mechanism. In addition, secretion could not be inhibited by treatment with BFA, an inhibitor of the classical endoplasmic reticulum/Golgi-dependent secretion pathway. Thus, extracellular release appears to be mediated by the unconventional secretion pathway reported also for several viral and cellular proteins (see reference 39 and references therein). In this context, Julian Huxley's term “growth gradient” may be relevant for the biological function of PLC-HDPs. In the morphogen gradient model, the local concentration of a diffusible molecule can determine cells' rates of proliferation and differentiation as a continuous function of concentration (4, 11, 36). This model is made particularly attractive by our finding that PLC-HDPs also harbor a highly conserved PTD with a similar activity as the widely used HIV-1 Tat PTD (52). The mechanism of transduction has been studied extensively for a variety of proteins, including the Antennapedia and PDX-1 homeodomain transcription factors (15). Recent evidence suggests that transduction occurs via a multistep mechanism involving endocytosis and macropinocytosis (54). Although we could not visually monitor the spread of DrVsx1-GFP or MmChx10-GFP from expressing donor to untransfected acceptor cells as described for the Engrailed protein (22), the results of our intercellular transactivation assays support the intercellular trafficking of PLC-HDPs, as demonstrated for other PTD-containing transcription factors (53). However, to fully understand the biological relevance of transduction in vivo, the activity of PTD-deficient PLC-HDP mutants has to be investigated in adequate animal models.
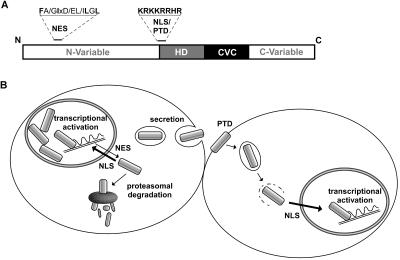
In summary, our report provides novel insights into the functional domain organization of PLC-HDPs (Fig. 7A). Based on our findings, we propose a model in which the continuous nucleocytoplasmic shuttling of PLC-HDPs contributes to the optimal and flexible execution of their transcriptional activities (Fig. 7B). The evolutionarily conserved combination of a PTD, together with active nuclear export and import signals, may allow the fine-tuning of intracellular protein levels by the proteasome pathway and, in addition, also permits intercellular transfer. The overlapping complex patterns of homeobox gene expression in the embryonic retina requires a complex regulatory network of transcription factors that specifies differentiation of competent retinal progenitors. Although transcriptional regulation of PLC-HDPs is an important control mechanism, PLC-HDPs may have additional unexpected paracrine activity, thereby influencing and maintaining the complex expression pattern during development. To ultimately gain profound insight into the detailed role of PLC-HDPs' nucleocytoplasmic shuttling for ordered development has to await transgenic mouse knock-in models in which nucleocytoplasmic transport of Chx10 is selectively abolished. Currently, we are pursuing this strategy in our laboratory.
FIG. 7.
(A) Organization of evolutionary conserved domains in PLC-HD proteins regulating cellular transport. (B) Model linking nucleocytoplasmic transport with PLC-HDP activity. The predominantly nuclear localization of PLC-HD proteins is the net result of import exceeding the rate of export due to the different activities of NES and NLS. Nuclear export allows PLC-HD protein levels to be regulated by the proteasomal degradation pathway and continuously supplies cargo for extracellular unconventional secretion. Intercellular transport and transactivation could be mediated by the protein transduction domain/NLS.
Acknowledgments
We thank B. Groner for support and N. Schechter, J. Nathans, and E. F. Percin for materials.
This study was supported by the Deutsche Forschungsgemeinschaft (Sta 598/1-2) and the Studienstiftung des Deutschen Volkes (S.K.K.).
REFERENCES
- 1.Affolter, M., T. Marty, and M. A. Vigano. 1999. Balancing import and export in development. Genes Dev. 13:913-915. [DOI] [PubMed] [Google Scholar]
- 2.Barabino, S. M., F. Spada, F. Cotelli, and E. Boncinelli. 1997. Inactivation of the zebrafish homologue of Chx10 by antisense oligonucleotides causes eye malformations similar to the ocular retardation phenotype. Mech. Dev. 63:133-143. [DOI] [PubMed] [Google Scholar]
- 3.Bednenko, J., G. Cingolani, and L. Gerace. 2003. Nucleocytoplasmic transport: navigating the channel. Traffic 4:127-135. [DOI] [PubMed] [Google Scholar]
- 4.Briscoe, J., Y. Chen, T. M. Jessell, and G. Struhl. 2001. A hedgehog-insensitive form of patched provides evidence for direct long-range morphogen activity of sonic hedgehog in the neural tube. Mol. Cell 7:1279-1291. [DOI] [PubMed] [Google Scholar]
- 5.Burmeister, M., J. Novak, M. Y. Liang, S. Basu, L. Ploder, N. L. Hawes, D. Vidgen, F. Hoover, D. Goldman, V. I. Kalnins, T. H. Roderick, B. A. Taylor, M. H. Hankin, and R. R. McInnes. 1996. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat. Genet. 12:376-384. [DOI] [PubMed] [Google Scholar]
- 6.Cartwright, P., and K. Helin. 2000. Nucleocytoplasmic shuttling of transcription factors. Cell Mol. Life Sci. 57:1193-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chow, R. L., B. Volgyi, R. K. Szilard, D. Ng, C. McKerlie, S. A. Bloomfield, D. G. Birch, and R. R. McInnes. 2004. Control of late off-center cone bipolar cell differentiation and visual signaling by the homeobox gene Vsx1. Proc. Natl. Acad. Sci. USA 101:1754-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen, B. R. 2003. Nuclear RNA export. J. Cell Sci. 116:587-597. [DOI] [PubMed] [Google Scholar]
- 9.Dyer, M. A. 2003. Regulation of proliferation, cell fate specification and differentiation by the homeodomain proteins Prox1, Six3, and Chx10 in the developing retina. Cell Cycle 2:350-357. [PubMed] [Google Scholar]
- 10.Endter, C., J. Kzhyshkowska, R. H. Stauber, H. Wolf, and T. Dobner. 2001. SUMO-1 modification required for transformation by adenovirus type 5 early 1B 55-kDa oncoprotein. Proc. Natl. Acad. Sci. USA 98:11312-11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franceschi, R. T. 1999. The developmental control of osteoblast-specific gene expression: role of specific transcription factors and the extracellular matrix environment. Crit. Rev. Oral Biol. Med. 10:40-57. [DOI] [PubMed] [Google Scholar]
- 12.Galliot, B., C. de Vargas, and D. Miller. 1999. Evolution of homeobox genes: Q50 paired-like genes founded the Paired class. Dev. Genes Evol. 209:186-197. [DOI] [PubMed] [Google Scholar]
- 13.Görlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 14.Green, E. S., J. L. Stubbs, and E. M. Levine. 2003. Genetic rescue of cell number in a mouse model of microphthalmia: interactions between Chx10 and G1-phase cell cycle regulators. Development 130:539-552. [DOI] [PubMed] [Google Scholar]
- 15.Green, I., R. Christison, C. J. Voyce, K. R. Bundell, and M. A. Lindsay. 2003. Protein transduction domains: are they delivering? Trends Pharmacol. Sci. 24:213-215. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi, T., J. Huang, and S. S. Deeb. 2000. RINX(VSX1), a novel homeobox gene expressed in the inner nuclear layer of the adult retina. Genomics 67:128-139. [DOI] [PubMed] [Google Scholar]
- 17.Hedlund, E., S. L. Karsten, L. Kudo, D. H. Geschwind, and E. M. Carpenter. 2004. Identification of a Hoxd10-regulated transcriptional network and combinatorial interactions with Hoxa10 during spinal cord development. J. Neurosci. Res. 75:307-319. [DOI] [PubMed] [Google Scholar]
- 18.Heger, P., J. Lohmaier, G. Schneider, K. Schweimer, and R. H. Stauber. 2001. Qualitative highly divergent nuclear export signals can regulate export by the competition for transport cofactors in vivo. Traffic 555:544-555. [DOI] [PubMed] [Google Scholar]
- 19.Heger, P., O. Rosorius, J. Hauber, and R. H. Stauber. 1999. Titration of cellular export factors, but not heteromultimerization, is the molecular mechanism of trans-dominant HTLV-1 Rex mutants. Oncogene 18:4080-4090. [DOI] [PubMed] [Google Scholar]
- 20.Henderson, B. R., and A. Eleftheriou. 2000. A comparison of the activity, sequence specificity, and CRM1 dependence of different nuclear export signals. Exp. Cell Res. 256:213-224. [DOI] [PubMed] [Google Scholar]
- 21.Heon, E., A. Greenberg, K. K. Kopp, D. Rootman, A. L. Vincent, G. Billingsley, M. Priston, K. M. Dorval, R. L. Chow, R. R. McInnes, G. Heathcote, C. Westall, J. E. Sutphin, E. Semina, R. Bremner, and E. M. Stone. 2002. VSX1: a gene for posterior polymorphous dystrophy and keratoconus. Hum. Mol. Genet. 11:1029-1036. [DOI] [PubMed] [Google Scholar]
- 22.Joliot, A., A. Maizel, D. Rosenberg, A. Trembleau, S. Dupas, M. Volovitch, and A. Prochiantz. 1998. Identification of a signal sequence necessary for the unconventional secretion of Engrailed homeoprotein. Curr. Biol. 8:856-863. [DOI] [PubMed] [Google Scholar]
- 23.Kino, T., A. Gragerov, J. B. Kopp, R. H. Stauber, G. N. Pavlakis, and G. P. Chrousos. 1999. The HIV-1 virion-associated protein Vpr is a coactivator of the human glucocorticoid receptor. J. Exp. Med. 189:51-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koster, M., S. Lykke-Andersen, Y. A. Elnakady, K. Gerth, P. Washausen, G. Hofle, F. Sasse, J. Kjems, and H. Hauser. 2003. Ratjadones inhibit nuclear export by blocking CRM1/exportin 1. Exp. Cell Res. 286:321-331. [DOI] [PubMed] [Google Scholar]
- 25.Krätzer, F., O. Rosorius, P. Heger, N. Hirschmann, T. Dobner, J. Hauber, and R. H. Stauber. 2000. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53, and Mdm2. Oncogene 19:850-857. [DOI] [PubMed] [Google Scholar]
- 26.Kurtzman, A. L., L. Gregori, A. L. Haas, and N. Schechter. 2000. Ubiquitination and degradation of the zebra fish paired-like homeobox protein VSX-1. J. Neurochem. 75:48-55. [DOI] [PubMed] [Google Scholar]
- 27.Kurtzman, A. L., and N. Schechter. 2001. Ubc9 interacts with a nuclear localization signal and mediates nuclear localization of the paired-like homeobox protein Vsx-1 independent of SUMO-1 modification. Proc. Natl. Acad. Sci. USA 98:5602-5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kzhyshkowska, J., H. Schutt, M. Liss, E. Kremmer, R. Stauber, H. Wolf, and T. Dobner. 2001. Heterogeneous nuclear ribonucleoprotein E1B-AP5 is methylated in its Arg-Gly-Gly (RGG) box and interacts with human arginine methyltransferase HRMT1L1. Biochem. J. 358:305-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamond, A. I., and J. E. Sleeman. 2003. Nuclear substructure and dynamics. Curr. Biol. 13:R825-R828. [DOI] [PubMed] [Google Scholar]
- 30.Lee, S. H., and M. Hannink. 2001. The N-terminal nuclear export sequence of IκBalpha is required for RanGTP-dependent binding to CRM1. J. Biol. Chem. 23:23. [DOI] [PubMed] [Google Scholar]
- 31.Lei, E. P., and P. A. Silver. 2002. Protein and RNA export from the nucleus. Dev. Cell 2:261-272. [DOI] [PubMed] [Google Scholar]
- 32.Levine, E. M., M. Passini, P. F. Hitchcock, E. Glasgow, and N. Schechter. 1997. Vsx-1 and Vsx-2: two Chx10-like homeobox genes expressed in overlapping domains in the adult goldfish retina. J. Comp. Neurol. 387:439-448. [PubMed] [Google Scholar]
- 33.Magnani, M., R. Crinelli, M. Bianchi, and A. Antonelli. 2000. The ubiquitin-dependent proteolytic system and other potential targets for the modulation of nuclear factor-κB (NF-κB). Curr. Drug Targets 1:387-399. [DOI] [PubMed] [Google Scholar]
- 34.Maizel, A., O. Bensaude, A. Prochiantz, and A. Joliot. 1999. A short region of its homeodomain is necessary for engrailed nuclear export and secretion. Development 126:3183-3190. [DOI] [PubMed] [Google Scholar]
- 35.McBride, K. M., and N. C. Reich. 2003. The ins and outs of STAT1 nuclear transport. Sci. STKE 195:RE13. [DOI] [PubMed] [Google Scholar]
- 36.Megason, S. G., and A. P. McMahon. 2002. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development 129:2087-2098. [DOI] [PubMed] [Google Scholar]
- 37.Mingot, J. M., S. Kostka, R. Kraft, E. Hartmann, and D. Gorlich. 2001. Importin 13: a novel mediator of nuclear import and export. EMBO J. 20:3685-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mintz-Hittner, H. A., E. V. Semina, L. J. Frishman, T. C. Prager, and J. C. Murray. 2004. VSX1 (RINX) mutation with craniofacial anomalies, empty sella, corneal endothelial changes, and abnormal retinal and auditory bipolar cells. Ophthalmology 111:828-836. [DOI] [PubMed] [Google Scholar]
- 39.Nickel, W. 2003. The mystery of nonclassical protein secretion: a current view on cargo proteins and potential export routes. Eur. J. Biochem. 270:2109-2119. [DOI] [PubMed] [Google Scholar]
- 40.Ohtoshi, A., M. J. Justice, and R. R. Behringer. 2001. Isolation and characterization of Vsx1, a novel mouse CVC paired-like homeobox gene expressed during embryogenesis and in the retina. Biochem. Biophys. Res. Commun. 286:133-140. [DOI] [PubMed] [Google Scholar]
- 41.Ohtoshi, A., S. W. Wang, H. Maeda, S. M. Saszik, L. J. Frishman, W. H. Klein, and R. R. Behringer. 2004. Regulation of retinal cone bipolar cell differentiation and photopic vision by the CVC homeobox gene Vsx1. Curr. Biol. 14:530-536. [DOI] [PubMed] [Google Scholar]
- 42.Percin, E. F., L. A. Ploder, Y. J. J., K. Arici, D. J. Horsford, A. Rutherford, B. Bapat, D. W. Cox, A. M. V. Duncan, V. I. Kalnins, A. Kocak-Altintas, J. C. Sowden, E. Trabousli, M. Sarfarazi, and R. R. McInnes. 2000. Human microphthalmia associated with mutations in the retinal homeobox gene CHX10. Nat. Genet. 25:397-401. [DOI] [PubMed] [Google Scholar]
- 43.Porter, L. A., and D. J. Donoghue. 2003. Cyclin B1 and CDK1: nuclear localization and upstream regulators. Prog. Cell Cycle Res. 5:335-347. [PubMed] [Google Scholar]
- 44.Rehberg, S., P. Lischka, G. Glaser, T. Stamminger, M. Wegner, and O. Rosorius. 2002. Sox10 is an active nucleocytoplasmic shuttle protein, and shuttling is crucial for Sox10-mediated transactivation. Mol. Cell. Biol. 22:5826-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosorius, O., P. Heger, G. Stelz, N. Hirschmann, J. Hauber, and R. H. Stauber. 1999. Direct observation of nucleo-cytoplasmic transport by microinjection of GFP-tagged proteins in living cells. BioTechniques 27:350-355. [DOI] [PubMed] [Google Scholar]
- 46.Rowan, S., and C. L. Cepko. 2004. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev. Biol. 271:388-402. [DOI] [PubMed] [Google Scholar]
- 47.Sakamoto, K. M. 2002. Ubiquitin-dependent proteolysis: its role in human diseases and the design of therapeutic strategies. Mol. Genet. Metab. 77:44-56. [DOI] [PubMed] [Google Scholar]
- 48.Smallwood, P. M., B. P. Olveczky, G. L. Williams, G. H. Jacobs, B. E. Reese, M. Meister, and J. Nathans. 2003. Genetically engineered mice with an additional class of cone photoreceptors: implications for the evolution of color vision. Proc. Natl. Acad. Sci. USA 100:11706-11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smallwood, P. M., Y. Wang, and J. Nathans. 2002. Role of a locus control region in the mutually exclusive expression of human red and green cone pigment genes. Proc. Natl. Acad. Sci. USA 99:1008-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stauber, R. H., and G. N. Pavlakis. 1998. Intracellular trafficking and interactions of the HIV-1 Tat protein. Virology 252:126-132. [DOI] [PubMed] [Google Scholar]
- 51.Ulrich, H. D. 2002. Natural substrates of the proteasome and their recognition by the ubiquitin system. Curr. Top. Microbiol. Immunol. 268:137-174. [DOI] [PubMed] [Google Scholar]
- 52.Vives, E., J. P. Richard, C. Rispal, and B. Lebleu. 2003. TAT peptide internalization: seeking the mechanism of entry. Curr. Protein Pept. Sci. 4:125-132. [DOI] [PubMed] [Google Scholar]
- 53.Wadia, J. S., and S. F. Dowdy. 2003. Modulation of cellular function by TAT mediated transduction of full length proteins. Curr. Protein Pept. Sci. 4:97-104. [DOI] [PubMed] [Google Scholar]
- 54.Wadia, J. S., R. V. Stan, and S. F. Dowdy. 2004. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat. Med. 10:310-315. [DOI] [PubMed] [Google Scholar]
- 55.Yashiroda, Y., and M. Yoshida. 2003. Nucleo-cytoplasmic transport of proteins as a target for therapeutic drugs. Curr. Med. Chem. 10:741-748. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, Y. A., A. Okada, C. H. Lew, and S. K. McConnell. 2002. Regulated nuclear trafficking of the homeodomain protein otx1 in cortical neurons. Mol. Cell Neurosci. 19:430-446. [DOI] [PubMed] [Google Scholar]
- 57.Zhao, Y., and H. Westphal. 2002. Homeobox genes and human genetic disorders. Curr. Mol. Med. 2:13-23. [DOI] [PubMed] [Google Scholar]