Abstract
Mutation of the survival motor neurons 1 (SMN1) gene causes motor neuron apoptosis and represents the major cause of spinal muscular atrophy in humans. Biochemical studies have established that the SMN protein plays an important role in spliceosomal small nuclear ribonucleoprotein (snRNP) biogenesis and that the SMN complex can interact with the zinc finger protein ZPR1. Here we report that targeted ablation of the Zpr1 gene in mice disrupts the subcellular localization of both SMN and spliceosomal snRNPs. Specifically, SMN localization to Cajal bodies and gems was not observed in cells derived from Zpr1−/− embryos and the amount of cytoplasmic snRNP detected in Zpr1−/− embryos was reduced compared with that in wild-type embryos. We found that Zpr1−/− mice die during early embryonic development, with reduced proliferation and increased apoptosis. These effects of Zpr1 gene disruption were confirmed and extended in studies of cultured motor neuron-like cells using small interfering RNA-mediated Zpr1 gene suppression; ZPR1 deficiency caused growth cone retraction, axonal defects, and apoptosis. Together, these data indicate that ZPR1 contributes to the regulation of SMN complexes and that it is essential for cell survival.
Spinal muscular atrophy (SMA) is an autosomal recessive disease in humans that is characterized by degeneration of the α-motor neurons of the spinal cord anterior horn. Infants born with SMA exhibit progressive muscle atrophy and paralysis that leads to respiratory failure and early death. Severe forms of SMA are caused by mutation of the survival motor neurons 1 (SMN1) gene (11, 24, 28, 51).
Two copies of the human SMN gene (SMN1 and SMN2) that are located in an inverted repeat on chromosome 5q13 have been identified. These two genes are very similar, but transcripts from the SMN2 gene undergo alternative splicing due to a translationally silent nucleotide difference (C → T, codon 280) in exon 7 (31, 36). In severe forms of SMA, the SMN1 gene is deleted and the SMN2 gene predominantly expresses a truncated SMN protein that lacks sequences derived from exon 7. The expression of low levels of full-length SMN protein causes motor neuron degeneration and SMA.
SMN has been implicated in the growth, development, and survival of spinal cord motor neurons (34, 42, 44). Biochemical analysis demonstrates that SMN plays an essential role in the assembly and maturation of spliceosomal small nuclear ribonucleoproteins (snRNPs) (35, 43, 53). After transcription, the Sm class of snRNAs (U1, U2, U4, and U5) are exported to the cytoplasm, where they are assembled with seven Sm proteins (SmB/B', SmD1 to SmD3, SmE, SmF, and SmG) to form Sm-core. The SMN protein complex is required for the specific assembly of Sm-core complexes on U snRNAs. This is mediated by SMN interactions with the U snRNA (52) and with the Arg/Gly-rich COOH tails of SmB, SmD1, and SmD3 (9). High-affinity interactions with SMN require that these Sm proteins be modified to contain symmetrical dimethyl-arginine (4, 10).
SMN plays a second role in the maturation of snRNPs following the assembly of snRNAs with Sm proteins. The Sm-core undergoes hypermethylation to form the 2′,2′,7′-trimethylguanosine (TMG) 5′-cap that is required for the recruitment of import receptors necessary for the translocation of snRNPs into the nucleus (50). Trimethylguanosine synthase 1 (TSG1), the enzyme that is responsible for the formation of the TMG 5′-cap, interacts with SMN (37). The hypermethylated 5′-cap of U snRNA recruits snurportin 1 to the snRNP complex (21) and snurportin 1 is able to bind both SMN (39) and importin β (21) to facilitate nuclear import of mature snRNP complexes.
Recent studies have demonstrated that the zinc finger protein ZPR1 represents a new component of SMN complexes (16). ZPR1 is part of a cytoplasmic snRNP complex that contains SMN, Sm proteins, U snRNA, snurportin 1, and importin β (39). The binding partner of ZPR1 in the SMN complex has not yet been identified, but it has been established that ZPR1 does not directly bind SMN (16). ZPR1 colocalizes with SMN in the nucleus, where both proteins accumulate in gems and Cajal bodies. It is likely that the binding of ZPR1 to SMN complexes is significant because SMN mutations that are associated with SMA disease disrupt the association of ZPR1 with SMN complexes (16). Furthermore, it is established that SMA patients express low levels of ZPR1 (20).
The purpose of this study was to examine the role of ZPR1 in mouse development. The effect of ZPR1 deficiency was investigated by disruption of the Zpr1 gene by using homologous recombination and also by Zpr1 gene silencing using RNA interference. We report that ZPR1 deficiency caused reduced growth and increased apoptosis. The effects of ZPR1 deficiency were associated with defects in the subcellular localization of snRNPs.
MATERIALS AND METHODS
Mice.
The murine Zpr1 gene was isolated from a 129/SvJ mouse λ Fix II genomic library (Stratagene), using the mouse Zpr1 cDNA as a probe. Sequence analysis confirmed that the clone carried the Zpr1 gene. A targeting vector was designed to replace exon 1 with a Neor cassette (see Fig. 1A). A thymidine kinase cassette was included for negative selection. The targeting vector was linearized with NotI, electroporated into TC1 embryonic stem cells (strain 129SvEv), and subsequently selected with G418 and ganciclovir. Targeted clones (four) were identified by Southern blot analysis, and two clones were used to create chimeric mice by blastocyst injection. Both clones transmitted the disrupted Zpr1 gene through the germ line. The mice were backcrossed to the C57BL/6J strain (The Jackson Laboratory) and maintained by mating Zpr1−/+ mice. Genomic DNA was isolated from the mouse tails with a genomic DNA isolation kit (Lamda Biotech) and used for PCR and Southern blot analysis. The animals were housed in a facility accredited by the American Association for Laboratory Animal Care, and the animal studies were approved by the Institutional Animal Care and Use Committee of the University of Massachusetts Medical School.
FIG. 1.
Targeted disruption of mouse Zpr1. (A) Exon-intron structure (coding and noncoding exons are numbered and represented by solid and open boxes, respectively) and restriction map of the mouse Zpr1 gene, structure of the targeting vector, and structure of the mutated allele after homologous recombination. (B) The mutated Zpr1 allele was detected by Southern blot analysis and PCR. Genomic DNA from targeted embryonic stem cells was digested with EcoRI and detected with cDNA probe (exons 10 to 14; bp 954 to 1379). The 17.5-kb fragment corresponding to the wild-type (WT) allele and the 9.0-kb fragment corresponding to the mutated (knockout [KO]) allele are indicated. (C) Genomic DNA samples were also examined by PCR; the wild-type (WT) allele (250 bp) and mutated (KO) allele (444 bp) are shown. (D) Genomic DNA prepared from E3.5 embryos cultured in vitro for 96 h was genotyped by PCR.
Blastocyst culture.
Heterozygous (−/+) mice were intercrossed to generate homozygous (−/−), heterozygous (−/+), and wild-type (+/+) mouse embryos. Blastocysts on day 3.5 postcoitum (E3.5) were flushed out with Dulbecco modified Eagles medium (DMEM) (Invitrogen) supplemented with 15% fetal bovine serum, β-mercaptoethanol (100 μM), glutamine (2 mM), and penicillin-streptomycin. Individual blastocysts were cultured for 96 h in 96-well tissue culture plates or on coverslips coated with 0.01% poly-l-lysine in either the presence or the absence of 103 U of LIF (Chemicon) per ml. Embryo growth and morphology were monitored at 24-h intervals. Embryos were washed with phosphate-buffered saline, (PBS), and genomic DNA was extracted by incubation at 55°C for 3 h in 20 μl of PCR lysis buffer (10 mM Tris-HCl [pH 9.0], 50 mM KCl, 2.5 mM MgCl2, 0.1% [vol/vol] Triton X-100) containing proteinase K (100 μg/ml). Lysates were boiled for 10 min and used for genotyping.
Genotyping.
Genomic DNA isolated from embryonic stem cell clones and mouse tails was used for determination of genotypes. Southern blot analysis was performed using a random-primed 32P-labeled 425-bp probe prepared by PCR using the primers 5′-CTCAGACATGACCAGAGACC-3′ and 5′-CCGCTGTGGGGCCAGGCCCG-3′ and the mouse Zpr1 cDNA as the template. Genotyping by PCR was performed to detect the wild-type allele (250 bp) using the primers 5′-CCCTCAGCGCCGAGGATGAG-3′ plus 5′-GCAGGAAAAGGAGCTCACGA-3′ and to detect the disrupted Zpr1 allele (444 bp) using the primers 5′-CTGCGGCCTCTTTAAGAAGG-3′ plus 5′-CTTCTTGACGAGTTCTTCTG-3′.
Culture of NSC-34 cells.
NSC-34 cells were maintained in DMEM supplemented with 10% fetal bovine serum, glutamine (2 mM), and penicillin-streptomycin (6). Differentiated cells with properties resembling those of motor neurons were obtained by culturing on coverslips in DMEM with 10% fetal bovine serum (2 days) and then in medium comprising 1:1 DMEM plus Ham's F12, 1% fetal bovine serum, penicillin-streptomycin, and 1% modified Eagle's medium nonessential amino acids. The medium was changed after 48 h and subsequently at 72-h intervals for 2 weeks.
Gene silencing with siRNA.
Motor neurons were transfected with 100 nM SMARTpool small interfering RNA (siRNA) (Dharmacon) designed to silence the mouse Zpr1 gene or Scramble II (Control; 5′-GCGCGCTTTGTAGGATTCG-3′). The SMARTpool of siRNA contained four double-stranded RNAs with target sequences (ZPR1.1, 5′-GATAATGCCTTGGTGATCA-3′; ZPR1.2, 5′-TAGAAGGACTGCTGAAAGA-3′; ZPR1.3, 5′-TCCCAGAGCTTGAGTTTGA-3′; and ZPR1.4, 5′-ACAGTTATCTGCAGAATGT-3′). Similar results were obtained using either the SMARTpool or the siRNA that targeted the sequences ZPR1.1 or ZPR1.3 alone. Transfection was performed with Oligofectamine (Invitrogen) by standard procedures. Cy3-labeled Luciferase GL2 duplex (Dharmacon) was used as a transfection control. Cells were harvested 72 h post-transfection, and the level of ZPR1 expression was examined by immunoblot analysis. Cover slips were processed for immunofluorescence analysis at 48 and 72 h post-transfection.
Immunofluorescence analysis.
Embryos (E3.5) cultured for 96 h on coverslips were incubated without and with 50 μM 5′-bromo-2′-deoxyuridine (BrdU) for 1 h at 37°C. Embryos and motor neurons were fixed with 4% paraformaldehyde in PBS for 15 min at room temperature and permeabilized with 0.2% (vol/vol) Triton X-100 in PBS for 5 min at room temperature or fixed with methanol (5 min) and acetone (2 min) at −20°C. Coverslips with blastocysts or motor neurons were blocked with 3% bovine serum albumin in PBS with 0.5% Tween 20 (PBS-T) for 30 min at 25°C. Coverslips were labeled with primary antibodies: anti-ZPR1 (clone LG1) (16), anti-Sm (clone Y12, LabVision); anti-TMG (Ab-1; Oncogene Research Products), anti-splicing factor SC-35 (Sigma), or anti-β-tubulin (clone TUJI; Covance) and detected with secondary antibodies coupled with fluorophores (Molecular Probes). Actin was labeled with phalloidin conjugated with Alexa 546 (Molecular Probes). Apoptosis assays were performed by detection of activated caspases using a caspase detection kit (Oncogene Research Products) by labeling with fluorescein isothiocyanate (FITC)-VAD-fmk for 1 h in culture at 37°C.
Double labeling (ZPR1-SMN) was carried out as described previously (16) by sequential incubations (1 h) with anti-SMN (clone 2B1) (30), with Alexa 546-conjugated anti-mouse immunoglobulin G (IgG) secondary antibody (Molecular Probes), and then with Alexa 488-conjugated anti-ZPR1 (clone LG1) at 25°C. Double labeling (ZPR1-p80 coilin) was carried out by sequential incubations (1 h) with rabbit anti-p80 coilin (no. R288) (1) and Alexa 546-conjugated anti-rabbit IgG secondary antibody and then with Alexa 488-conjugated LG1 (anti-ZPR1) at 25°C.
Triple labeling (ZPR1-SMN-BrdU) was carried out by sequential incubations (1 h) with anti-SMN (clone 2B1), Cy5-conjugated anti-mouse IgG secondary antibody (Jackson Immunoresearch), FITC-conjugated antibody to BrdU (BD Pharmingen), and Alexa 546-conjugated antibody to ZPR1 (clone LG1) at 25°C.
Processed coverslips were mounted on slides with mounting medium (Vectashield) containing 4′,6-diamino-2-phenylindole (DAPI). Fluorescence microscopy was performed using a Ziess inverted microscope (Axiovert M200) or a confocal laser scanning microscope (Leica TCS SP2) equipped with a 405-nm diode laser. Fluorescence images were quantitated using software obtained from Leica. Differential interference contrast images were obtained using the Ziess inverted microscope.
Transmission electron microscopy.
Mouse embryos (E3.5) were cultured for 96 h in 35-mm dishes (Nunc), washed with 0.5 M sodium cacodylate-HCl buffer (pH 7.0), and fixed with 2 ml of 1.25% gluteraldehyde for 30 min at 30°C and overnight at 4°C with 2 ml of 2.5% glutaraldehyde in cacodylate buffer. The embryos were postfixed (1 h) in 1% (wt/vol) osmium tetraoxide in 0.1 M phosphate buffer (pH 7.2). The fixed embryos were then washed with buffer and dehydrated through a graded ethanol series and transferred through a series of 100% ethanol and epoxy resin mixtures (LX 112/Araldite 502 epoxy resin) starting with 2 parts ethanol and 1 part epoxy resin, followed by a mixture of 1 part ethanol and 2 parts epoxy resin, and finally three changes of full-strength epoxy resin and were polymerized overnight at 70°C. The epoxy blocks were cut and mounted on blank epoxy stubs with a drop of Super Glue, and ultrathin sections were cut on a Reichart-Jung ultramicrotome using a diamond knife. The sections were collected and mounted on copper support grids in serial order, contrasted with lead citrate and uranyl acetate, and examined on a Philips CM 10 transmission electron microscope at 80-kV accelerating voltage.
Scanning electron microscopy.
Mouse embryos were fixed as described for transmission electron microscopy, dehydrated in graded alcohol (30, 50, 70, and 95% ethanol for 20 min each and then 100% ethanol twice for 20 min), and then critical-point dried in liquid CO2. The dried embryo samples were mounted onto aluminum stubs with silver conductive paste and sputter coated with gold-palladium alloy (60:40). The specimens were examined using an ETEC autoscan scanning electron microscope at 20-kV accelerating voltage.
RESULTS
Targeted disruption of the murine Zpr1 gene.
To define the physiological role of ZPR1, we examined the effect of disruption of the murine Zpr1 gene. A targeting vector was designed to replace exon 1 of the Zpr1 gene with a Neor cassette (Fig. 1A). This vector was electroporated into embryonic stem cells. Four correctly targeted cell clones were identified by Southern blot analysis (Fig. 1B). Two clones were injected into C57BL/6J blastocysts to obtain chimeric mice that were crossed with C57BL/6J mice. Chimeric mice derived from both of these clones transmitted the disrupted Zpr1 allele through the germ line (Fig. 1C).
Comparison of wild-type and heterozygous Zpr1−/+ mice did not lead to the detection of obvious morphological differences. Breeding Zpr1−/+ mice yielded wild-type and heterozygous mice at weaning, but no homozygous Zpr1−/− mice were found (Table 1). Similarly, genotyping of postimplantation embryos did not indicate the presence of Zpr1−/− embryos. We therefore examined preimplantation embryos at the blastocyst stage at E3.5. This analysis led to the identification of wild-type, heterozygous, and homozygous knockout embryos (Fig. 1D) in the expected Mendelian ratios (Table 1). These data indicate that ZPR1 deficiency caused a lethal defect in early development.
TABLE 1.
Progeny of heterozygous Zpr1−/+ mice intercrosses
| Stage | No. of Zpr1 mice that were:
|
Total no. | |||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | Untyped | ||
| Born live | 28 | 53 | 0 | 0 | 81 |
| E13.5 | 8 | 18 | 0 | 0 | 26 |
| E9.5 | 10 | 21 | 0 | 0 | 31 |
| E3.5 | 18 | 28 | 14 | 9 | 69 |
ZPR1 deficiency causes defective embryonic growth.
To determine the cause of embryonic death due to loss of ZPR1, we examined the growth and development of E3.5 blastocysts in vitro. All embryos appeared to be similar when visualized under differential interference contrast light microscopy at E3.5 (0 h; Fig. 2A). The blastocysts were cultured and observed at 24-h intervals. Both wild-type (Fig. 2A) and heterozygous Zpr1−/+ (data not shown) embryos showed normal growth, spreading of trophoblast giant cells, and growth of the inner cell mass surrounded by trophectoderm. In contrast, Zpr1−/− embryos were delayed in growth and failed to form normal trophectoderm and expand the inner cell mass (Fig. 2A). Addition of leukemia inhibitory factor stimulated the growth of the inner cell mass in wild-type and heterozygous Zpr1−/+ embryos but did not affect the proliferation of these cells in Zpr1−/− embryos (data not shown). Culture for 120 to 144 h resulted in substantial growth of wild-type and heterozygous embryos, but very few viable cells derived from Zpr1−/− embryos were detected (data not shown).
FIG. 2.
Mutation in Zpr1 results in cell death and early embryonic death in mice. (A) Differential interference contrast images of wild-type (WT) and mutant (KO) blastocysts (E3.5) at 0 h and after 96 h of culture in vitro. The inner cell mass (ICM) is surrounded by trophoblast giant (TG) cells. Scale bar, 100 μm. (B) Scanning electron micrographs of WT and mutant embryos (KO) cultured in vitro for 96 h. The ICM is growing on the trophectoderm (TE) formed by trophoblast giant cells in WT embryos. The ICM and TE of mutant embryo (KO) display defective growth (top; magnification, ×200; scale bar, 100 μm). Microvilli (MV), projections at the apical surface of cells from wild-type embryos, are absent in mutant (KO) embryos (bottom; magnification, ×1,000; scale bar, 10 μm). (C) In situ detection of caspase activation, using FITC-VAD-fmk and confocal laser-scanning microscopy. Activated caspase (green) in WT and mutant (KO) embryos was observed. Nuclei were stained with DAPI (blue). The mutant embryo exhibited a large amount of caspase activation. Scale bar, 40 μm. (D) Transmission electron micrographs of trophoblast giant cells (top) and cells from the ICM (bottom). The mutant embryo (KO) shows apoptotic bodies (AB) phagocytosed by the trophoblast giant cells, as well as aggregation of heterochromatin and accumulation at nuclear membrane (bottom). Arrowheads show nucleolar accessory bodies (Cajal bodies). Scale bar, 1.0 μm.
To further characterize the growth of embryos, we examined cultured embryos by scanning electron microscopy. Wild-type embryos contained a large inner cell mass surrounded by trophectoderm formed by trophoblast giant cells (Fig. 2B, top). Mutant embryos were significantly smaller. The inner cell mass of the mutant embryos contained only a few cells that were surrounded by a thin layer of irregular trophectoderm (Fig. 2B, top). Furthermore, higher magnification of these embryos showed marked differences in the apical surface of embryonic cells between wild-type and mutant embryos (Fig. 2B, bottom). The surface of the cells that form the trophectoderm and inner cell mass of wild-type embryos contained a high density of specialized projections (microvilli) that increase cell surface area and are involved in transport-secretion, enzymatic activity, and signal transduction. In contrast, cells of mutant embryos showed a smooth surface and were devoid of microvilli (Fig. 2B). These data demonstrate that ZPR1 deficiency caused defects in the apical surface of embryonic cells, including the loss of microvilli.
ZPR1 deficiency causes increased apoptosis.
To determine the cause of the developmental defect observed in Zpr1−/− embryos, we performed biochemical analysis of mouse embryos cultured in vitro. The impaired growth of Zpr1−/− embryos (Fig. 2A and B) suggested that ZPR1 deficiency might cause increased cell death. To test this hypothesis, we examined the activation of apoptotic caspases in cultured blastocysts by incubation with FITC-VAD-fmk. Confocal laser-scanning microscopy detected very little activated caspase in wild-type embryos (Fig. 2C). In contrast, a marked increase in the amount of activated caspase was detected in Zpr1−/− embryos. This observation suggests that ZPR1-deficient cells may exhibit increased apoptosis. Indeed, ultrastructural analysis by transmission electron microscopy demonstrated the presence of engulfed apoptotic bodies within the cytoplasm of the trophoblast giant cells of Zpr1−/− embryos but not wild-type embryos (Fig. 2D). Analysis of the inner cell mass of Zpr1−/− embryos demonstrated the presence of cells with an early apoptotic morphology (chromatin condensation and nuclear folding; Fig. 2D) and also cells undergoing late-stage apoptosis (data not shown). In contrast, no apoptosis within the inner cell mass of wild-type embryos was detected (Fig. 2D). Together, these data indicate that increased apoptosis contributes to the poor development of Zpr1−/− embryos.
ZPR1 is required for proliferation.
In addition to increased apoptosis (Fig. 2), the poor development of Zpr1−/− embryos may result from reduced cell proliferation. We therefore investigated DNA synthesis in wild-type and Zpr1−/− embryos by examining the incorporation of BrdU into DNA during the S phase of the cell cycle (Fig. 3A). Immunofluorescence analysis demonstrated that trophoblast giant cells did not incorporate BrdU in this assay. In contrast, marked incorporation of BrdU by the inner cell mass of wild-type embryos was observed. However, experiments using Zpr1−/− embryos indicated that BrdU incorporation was restricted to only a few cells in the inner cell mass (Fig. 3A). Together, these data demonstrated that Zpr1−/− embryos exhibit a severe defect in proliferation. It is likely that this reduced proliferation (Fig. 3A) together with increased apoptosis (Fig. 2C and D) accounts, in part, for the death of Zpr1−/− embryos.
FIG. 3.
ZPR1 deficiency results in defective embryo growth and causes mislocalization of SMN and coilin. (A) Blastocysts (E3.5) cultured in vitro were labeled with 50 μM BrdU for 1 h at 37°C. Wild-type (WT) and mutant (KO) embryos were stained with mouse monoclonal antibodies to ZPR1 (red), SMN (purple), and BrdU (green) and examined by indirect-immunofluorescence microscopy. Nuclei were stained with DAPI (blue). Scale bar, 100 μm. (B) Colocalization of ZPR1 and coilin in mouse embryos. Blastocysts were stained with antibodies to coilin (red) and ZPR1 (green). Colocalization of coilin (red) and ZPR1 (green) is represented by yellow. Nuclei were stained with DAPI (blue). Scale bar, 8.0 μm. (C) Colocalization of SMN and ZPR1 in mouse embryos. Blastocysts (E3.5) cultured for 96 h were stained with antibodies to SMN (red) and ZPR1 (green) and examined by confocal laser-scanning immunofluorescence microscopy. Colocalization of SMN (red) and ZPR1 (green) is represented by yellow. Scale bar, 8.0 μm.
The absence of BrdU incorporation into the trophoblast giant cells in this assay most probably reflects the observation that these cells undergo several rounds of endoreduplication during early development prior to cell cycle arrest. To examine whether this early endoreduplication of trophoblast giant cells might be affected by ZPR1 deficiency, we quantitated the nuclear DNA by measurement of DAPI fluorescence (Fig. 3A). The nuclear DNA content of ZPR1-deficient trophoblast giant cells was 52% ± 11% (mean ± standard deviation SD; n = 10) of the nuclear DNA of wild-type trophoblast giant cells (Fig. 3A). The cross-sectional area of the nucleus of ZPR1-deficient trophoblast giant cells was also smaller than that of wild-type cells (63% ± 4%; n = 10).
Together, these data indicate that ZPR1 deficiency causes reduced proliferation of the inner cell mass and reduced endoreduplication of the trophoblast giant cells.
ZPR1 deficiency causes defects in Cajal bodies.
It has been established in previous studies that nuclear ZPR1 localizes to gems and Cajal bodies (16). Cajal bodies were observed by transmission electron microscopy in both trophoblast giant cells and the inner cell mass of wild-type embryos (Fig. 2D). In contrast, electron microscopy examination of serial sections of cells derived from Zpr1−/− embryos failed to detect Cajal bodies (Fig. 2D). These data suggest that ZPR1 deficiency causes major defects in the electron-dense structure of Cajal bodies. To test this hypothesis, we examined the subnuclear distribution of p80 coilin, a cytological marker for Cajal bodies (1). In wild-type embryos, a small amount of coilin was diffusely distributed within the nucleoplasm and the majority of coilin was found to accumulate in Cajal bodies (Fig. 3B). In marked contrast, coilin was diffusely localized to the nucleoplasm in Zpr1−/− embryos (Fig. 3B). Interestingly, the total immunofluorescent staining of coilin in Zpr1−/− cells was markedly increased compared to that in wild-type cells (Fig. 3B). This increased staining may be caused by epitope unmasking when coilin redistributes from Cajal bodies to the nucleoplasm in Zpr1−/− cells, since studies of Zpr1 gene silencing caused similar changes in the subnuclear localization of coilin in the absence of increased coilin expression detected by immunoblot analysis (data not shown).
The ZPR1-associated protein SMN is also a marker for gems and Cajal bodies (16, 30). We therefore examined the localization of SMN in wild-type and Zpr1−/− embryos. Immunofluorescence analysis of wild-type embryos demonstrated the presence of punctate intranuclear staining that colocalized with ZPR1 (Fig. 3C). In contrast, punctate intranuclear localization of SMN was not observed in Zpr1−/− embryos (Fig. 3C).
Together, these data indicate that ZPR1 deficiency causes mislocalization of proteins (e.g., coilin and SMN) that normally accumulate in Cajal bodies and gems. This conclusion is consistent with the observation that electron microscopy detected Cajal bodies in wild-type embryos but not in Zpr1−/− embryos (Fig. 2D). The loss of Cajal bodies in Zpr1−/− embryos is likely to be biologically significant because these subnuclear bodies have been implicated in multiple aspects of RNA biogenesis (14, 40). Interestingly, it has been previously established that the association of coilin with Cajal bodies depends on active transcription since treatment of cells with actinomycin D causes relocalization of coilin from Cajal bodies to the nucleolar periphery (5, 47).
ZPR1 is required for the normal subcellular distribution of spliceosomal snRNPs.
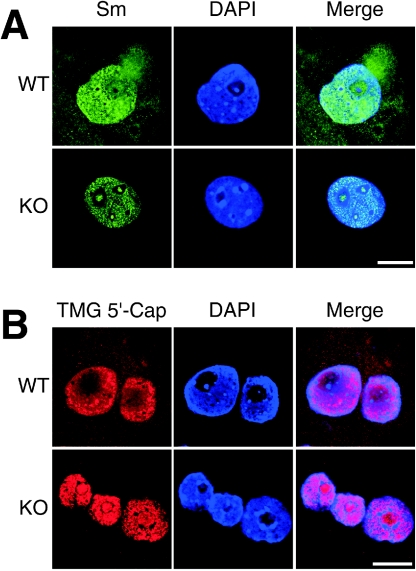
It has been established in previous studies that SMN plays an important role in the assembly of the Sm-core domain of snRNPs in the cytoplasm (35, 43, 53). In addition, cytoplasmic SMN can form a preimport snRNP complex with snurportin 1, importin β, and ZPR1 (39). ZPR1 deficiency may therefore cause changes in the subcellular localization of snRNPs. To test this hypothesis, we examined wild-type and Zpr1−/− embryos by immunofluorescence microscopy by staining with antibodies to Sm proteins (Fig. 4A) and to the TMG 5′-cap of U snRNA (Fig. 4B). No marked differences between the distribution of Sm proteins and the TMG 5′-cap were observed, indicating that these epitopes primarily identify similar groups of snRNP complexes.
FIG. 4.
ZPR1 deficiency causes defects in snRNP localization. (A) Loss of ZPR1 results in nuclear and nucleolar accumulation of Sm proteins. Wild-type (WT) and mutant (KO) blastocysts (E3.5) were cultured in vitro for 96 h and stained with antibodies to Sm proteins (green). All stained samples were examined by confocal laser-scanning immunofluorescence microscopy. Nuclei were stained with DAPI (blue). Scale bar, 20 μm. (B) Deficiency of ZPR1 causes nuclear and nucleolar accumulation of snRNPs. Embryos were stained with antibodies to TMG 5′-cap of U snRNA for detection of snRNPs (red) in wild-type and mutant embryos. Scale bar, 20 μm.
Immunofluorescence analysis demonstrated that ZPR1 deficiency caused major changes in the subcellular distribution of snRNPs (detected as Sm proteins or TMG 5′-cap). In wild-type embryos, snRNPs accumulated in the nucleus and were reproducibly detected in the cytoplasm. The location of snRNPs in both the nuclear and cytoplasmic compartments of wild-type cells is consistent with the established role of nuclear export and nuclear reimport during snRNP biogenesis (50). In marked contrast, snRNPs were detected only at very low levels in the cytoplasm of ZPR1-deficient embryos (Fig. 4). In addition, ZPR1 deficiency caused a marked change in the subnuclear localization of snRNPs because snRNPs were partially excluded from the nucleolus in wild-type embryos but accumulated within the nucleolus of Zpr1−/− embryos (Fig. 4). Interestingly, the effect of ZPR1 deficiency in causing localization of snRNPs within the nucleolus (Fig. 4) is similar to the effect of treatment of cells with the protein phosphatase inhibitor okadaic acid (32, 47).
To further characterize the effect of ZPR1 deficiency on the subcellular localization of Sm proteins and snRNPs, we performed quantitative analysis of images obtained by confocal immunofluorescence miscoscopy. The cytoplasmic and nuclear Sm protein staining in wild-type cells was 33% ± 2% and 67% ± 2% (n = 10), respectively. In contrast, ZPR1-deficient cells showed decreased cytoplasmic ZPR1 staining (2% ± 1%) and increased nuclear staining (98% ± 1%; n = 10). Quantitation of the staining of snRNPs indicated cytoplasmic and nuclear staining of 37% ± 3% and 63% ± 3% (n = 10), respectively. ZPR1 deficiency caused decreased staining of snRNPs in the cytoplasm (11% ± 2%) and increased staining in the nucleus (89% ± 2%; n = 10). These quantitative data confirm the conclusion that ZPR1 deficiency caused decreased cytoplasmic accumulation and increased nuclear accumulation of both Sm proteins and snRNPs.
Together, these data indicate that ZPR1 deficiency causes major defects in the subcellular localization of snRNPs. This observation suggested that ZPR1 deficiency may alter the normal function of spliceosomal snRNPs. Previous studies have demonstrated that the SC35 splicing factor can be detected in “nuclear speckles” and that these speckles aggregate when pre-mRNA splicing is disrupted (41). We therefore examined the subnuclear localization of the SC35 splicing factor. Immunofluorescence analysis demonstrated aggregation of the SC35 “nuclear speckles” in ZPR1-deficient embryos (data not shown). Together, these data indicate that ZPR1 deficiency influences snRNP complexes, the major target of SMN activity.
RNA interference-mediated suppression of Zpr1 gene expression causes mislocalization of SMN and snRNPs.
SMA is caused by the degeneration of α-motor neurons of the spinal cord. Motor neurons therefore represent a cell type that is relevant to SMA disease. Since Zpr1−/− mice die during early embryonic development, we were not able to study Zpr1−/− motor neurons. Consequently, we used an alternative approach, Zpr1 gene silencing by siRNA, to examine the effect of ZPR1 deficiency. These studies were performed using the NSC-34 mouse cell line, which can be induced to differentiate into cells that have motor neuron-like properties. Control studies demonstrated that these cells express both ZPR1 and SMN (Fig. 5A). Immunofluorescence analysis demonstrated extensive colocalization of ZPR1 and SMN in the soma and growth cones. The amount of nuclear accumulation of SMN and ZPR1 in these cells was small, but SMN and ZPR1 were found to colocalize in gems and Cajal bodies (Fig. 5A and B). Zpr1 siRNA, but not scambled siRNA, caused decreased expression of ZPR1 detected by immunoblot analysis (see Fig. S1 in the supplemental material). Similarly, immunofluorescence analysis demonstrated that Zpr1 siRNA, but not scrambled siRNA (Control), caused decreased expression of ZPR1 (Fig. 5B). Interestingly, both ZPR1 and SMN were detected in gems and Cajal bodies in control cells, but this localization was not detected following Zpr1 gene silencing (Fig. 5B). Similarly, coilin was localized to Cajal bodies in differentiated NSC-34 cells but was found to be diffusely distributed in the nucleoplasm following Zpr1 gene silencing (data not shown). These data confirm that ZPR1 is essential for the localization of SMN to gems and Cajal bodies.
FIG. 5.
Loss of ZPR1 causes mislocalization of SMN and snRNPs in differentiated NSC-34 cells. (A) Colocalization of SMN with ZPR1 in differentiated NSC-34 cells that resemble motor neurons. The cells were stained with antibodies to SMN and ZPR1 and examined by confocal laser-scanning immunofluorescence microscopy. Colocalization of SMN (red) with ZPR1 (green) is represented by yellow in nuclear bodies (top, see inset; scale bar, 40 μm) and in the neuronal growth cone (bottom, see inset; scale bar, 8.0 μm). (B) A low level of ZPR1 results in the loss of nuclear bodies and mislocalization of nuclear SMN in motor neurons. Motor neurons were transfected with scrambled siRNA (Control) and ZPR1-specific siRNA (siRNA) and cultured for 48 h. Cellular distribution of SMN (red) and ZPR1 (green) were examined using antibodies to SMN and ZPR1. Scale bar, 16 μm. (C) Loss of ZPR1 causes nuclear accumulation of snRNPs. Cells were transfected with scrambled siRNA (Control) and ZPR1-specific siRNA (siRNA), were cultured for 72 h, and stained with antibodies to ZPR1 (green) and TMG 5′-cap (red). Scale bar, 8.0 μm. Nuclei were stained with DAPI (blue).
To investigate whether ZPR1 influences snRNPs in NSC-34 cells, we examined the localization of snRNPs by immunofluorescence analysis using an antibody to the TMG 5′-cap of U snRNA that is assembled into snRNP complexes (Fig. 5C). Differentiated NSC-34 cells transfected with scrambled siRNA (Control) demonstrated the presence of snRNP in both the cytoplasm and the nucleus. In contrast, no cytoplasmic snRNPs were detected in cells following Zpr1 gene silencing. Furthermore, snRNPs were excluded from the nucleolus of control motor neurons but were present in the nucleolus of ZPR1-depleted cells.
Together, these data demonstrate that ZPR1 deficiency causes defects in the localization of both SMN and spliceosomal snRNPs in early embryos (Fig. 3 and 4) and cultured NSC-34 cells (Fig. 5).
ZPR1 deficiency causes axonal defects.
Axonal defects in motor neurons as a result of SMN deficiency have been described, and it is likely that such defects contribute to the pathogenesis of SMA (34, 44). We therefore examined whether ZPR1 deficiency might cause axonal defects in NSC-34 cells. Morphology was assessed by immunofluorescence analysis using an antibody to tubulin and phalloidin to stain actin (Fig. 6). Neurons transfected with scrambled siRNA (Control) demonstrated the presence of tubulin in axons and actin in both the soma and growth cones. Zpr1 gene silencing caused retraction of growth cones and disruption of the structural integrity of axonal microtubules. Control axons were long and straight, whereas the axons of ZPR1-deficient neurons were kinked and short. These data indicate that ZPR1 is important for the maintenance of motor neuron axons and growth cones.
FIG. 6.
Deficiency of ZPR1 causes defects in axons of differentiated NSC-34 cells. (A) Loss of ZPR1 causes axonal defects in differentiated NSC-34 cells that resemble motor neurons. Cells transfected with scrambled siRNA (Control) and ZPR1-specific siRNA (siRNA) were cultured for 72 h and stained with antibodies to tubulin (red) and ZPR1 (green). The arrowhead indicates the kinks in the axon of motor neuron after knockdown of ZPR1 (siRNA). Scale bar, 20 μm. (B) ZPR1 deficiency results in the loss of growth cones of differentiated NSC-34 cells. ZPR1 was stained with antibodies to ZPR1 (green), and actin was stained with phalloidin conjugated with Alexa 546 (red). Nuclei were stained with DAPI (blue). All samples were examined by confocal laser-scanning fluorescence microscopy. Scale bar, 20 μm.
ZPR1 is essential for cell survival.
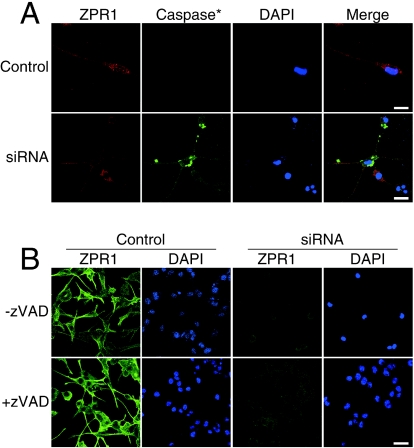
The apoptotic degeneration of motor neurons in SMA patients causes progressive muscle weakness. Since these patients express low levels of ZPR1 (20), we considered the possibility that ZPR1 deficiency might contribute to motor neuron death. To test this hypothesis, we investigated the effect of Zpr1 gene silencing on caspase activation in differentiated NSC-34 cells. Activated caspase was detected by immunofluorescence analysis after incubation of the motor neurons with FITC-VAD-fmk. Cells transfected with scrambled siRNA (Control) did not exhibit caspase activation (Fig. 7A). In contrast, caspase activation was observed in cultures of neurons transfected with Zpr1 siRNA. The most prominent staining of activated caspase appeared to correspond to apoptotic bodies derived from dead cells that were adherent to viable cells (Fig. 7A).
FIG. 7.
Loss of ZPR1 results in death of differentiated NSC-34 cells. (A) Low levels of ZPR1 causes caspase activation and death of differentiated NSC-34 cells that resemble motor neurons. Cells transfected with scrambled siRNA (Control) and ZPR1-specific siRNA (siRNA) were cultured for 72 h and labeled with FITC-VAD-fmk. Motor neurons stained with antibodies to ZPR1 (red) and activated caspase (green) were examined by confocal laser-scanning microscopy. Scale bar, 20 μm. (B) Inhibition of death of motor neurons that lack ZPR1 by zVAD. Cells were left untreated (−zVAD) or treated (+zVAD) with zVAD (10 μM) during transfection with scrambled siRNA (Control) and ZPR1-specific siRNA (siRNA). Samples were stained with antibodies to ZPR1 and examined by confocal laser-scanning microscopy. Nuclei were stained with DAPI (blue). Scale bar, 40 μm.
The observation that ZPR1 deficiency is associated with caspase activation suggests that the loss of ZPR1 might cause apoptosis. To test this hypothesis, we examined the effect of transfecting cultures of NSC-34 cells with scrambled siRNA or Zpr1 siRNA. The cells were cultured for 72 h, and the number of surviving neurons was examined by staining nuclear DNA with DAPI (Fig. 7B). Zpr1 gene silencing caused marked decreases in both the level of ZPR1 expression and the number of surviving neurons. The decreased neuronal survival was prevented by incubation with the caspase inhibitor zVAD-fmk (Fig. 7B). Together, these data demonstrate that ZPR1 is critical for sustaining cell survival.
DISCUSSION
The results of this study establish that Zpr1 is an essential gene in mice (Table 1) and that ZPR1 is required for the maintenance of motor neurons (Fig. 6 and 7). The ZPR1 protein interacts with SMN and is a component of cytoplasmic snRNP complexes (16,39). The observation that ZPR1 deficiency causes defects in the subcellular localization of snRNPs and SMN (Fig. 3 to 5) indicates that the interaction between ZPR1 and SMN is biologically significant. Interestingly, SMN mutations that are associated with the inherited disease SMA disrupt the interaction of ZPR1 with SMN (16). Furthermore, human patients with SMA express low levels of ZPR1 (20). In mice, ZPR1 deficiency prevents the localization of SMN to gems and Cajal bodies (Fig. 3 and 5); this may be significant for SMA because it is established that the severity of SMA negatively correlates with the number of SMN-positive gems and Cajal bodies (7, 29). Together, these data indicate that ZPR1 may contribute to SMN-associated disease.
ZPR1 and SMN prevent apoptosis.
SMN functions as a component of a protein complex (17) that includes the zinc finger protein ZPR1 (16, 39). SMN mutations that cause SMA disrupt the interaction of SMN with several components of this protein complex, including ZPR1 (16). The correlation between the interaction of ZPR1 with SMN and motor neuron degeneration in SMA suggests that ZPR1 and SMN may represent components of a common functional pathway.
It is established that loss of function, or mutation, of SMN can induce caspase-mediated apoptosis (22, 26, 45, 49). Similarly, mutation or silencing of the murine Zpr1 gene causes casapse activation and apoptosis (Fig. 2 and 7). The mechanism of apoptosis caused by Smn and Zpr1 mutations may be related, since it is known that SMN is essential for splicesomal snRNP assembly and maturation (35, 43, 53) and that ZPR1 deficiency causes defects in the subcellular localization of both SMN and snRNPs (Fig. 4 and 5). Interestingly, these effects of ZPR1 deficiency on the subcellular distribution of snRNPs and apoptosis resemble the effects of decreased expression of the SMN-interacting protein Gemin2 (23). Together, these data indicate that both ZPR1 and the SMN protein complex can function to sustain cell survival.
Nucleocytoplasmic trafficking and function of ZPR1.
ZPR1 redistributes between the cytoplasm and the nucleus (12, 13, 15). Genetic analysis indicates that the nuclear export of ZPR1 requires the peptidylprolyl isomerase activity of cyclophilin A (2), while biochemical studies suggest that snurportin 1 may contribute to the nuclear import of ZPR1 (39). This nucleocytoplasmic trafficking of ZPR1 resembles the trafficking of SMN (16), although it should be noted that the nuclear import of ZPR1 and SMN appear to be mediated by independent mechanisms (38) and it is unclear whether the mechanisms of ZPR1 and SMN export from the nucleus are related. Nevertheless, SMN and ZPR1 are both detected in the cytoplasm and the nucleus. The cytoplasmic function of ZPR1 may be to collaborate with SMN in the assembly and maturation of snRNPs to form preimport complexes with snurportin 1 (39). The effect of ZPR1 deficiency to disrupt the cytoplasmic localization of snRNP complexes is consistent with this hypothesis (Fig. 4 and 5).
In the nucleus, SMN and ZPR1 are found in the nucleoplasm, but these proteins also accumulate in gems and Cajal bodies (16, 30). These subnuclear bodies have been implicated in several aspects of RNA biogenesis (14, 33, 40). For example, recent studies have established that the guide RNA molecules that are required for site-specific modification of snRNAs are localized to Cajal bodies (27) and it has been demonstrated that the synthesis of 2-O-methylated nucleotides and pseudouridine in snRNAs occurs following nuclear import of snRNPs within Cajal bodies (25). Interestingly, Cajal bodies were not detected in Zpr1−/− embryos (Fig. 2D and 3B). Thus, ZPR1 may play an essential role in the maintenance of nuclear gems and Cajal bodies. Since these subnuclear bodies are dynamic structures that can exchange their protein components with the nucleoplasm (8), there are several possible mechanisms that could account for the requirement of ZPR1 to maintain these structures. Nevertheless, it appears that in addition to a cytoplasmic function that may contribute to the assembly of snRNPs and formation of preimport snRNP complexes (39), ZPR1 may contribute to the final maturation of imported snRNP complexes within Cajal bodies.
Previous studies have established coilin as a cytological marker for Cajal bodies (1). The coilin NH2-terminal region is able to self-associate (18), and the COOH terminus is able to recruit SMN by binding to symmetrical dimethylarginine residues located within an Arg-Gly-rich domain (3, 19). A major difference between cells that display gems and those that do not is that the COOH-terminal region of coilin is hypomethylated in cells containing gems and Cajal bodies and is fully methylated in cells containing only Cajal bodies (19). Interestingly, the COOH-terminal region of coilin can regulate the number of Cajal bodies (46), and deletion of this region prevents the recruitment of both SMN and snRNP complexes to Cajal bodies (48). In Zpr1−/− embryos, Cajal bodies were not detected by electron microscopy (Fig. 2D) and the nuclear distribution of coilin was profoundly altered (Fig. 3B). Consistent with the established role of coilin to recruit SMN, no accumulation of SMN in Cajal bodies was observed in Zpr1−/− embryos. It is possible that ZPR1 deficiency results in the formation of residual gems or Cajal bodies that lack coilin and SMN. Nevertheless, these data demonstrate that ZPR1 is essential for the normal morphology of gems and Cajal bodies.
Conclusions.
Our studies of Zpr1 gene silencing and Zpr1 gene disruption demonstrate that ZPR1 is an essential protein. ZPR1 deficiency causes defects in both the cytoplasmic and nuclear populations of spliceosomal snRNPs. Specifically, cytoplasmic snRNPs are not detected in Zpr1−/− embryos. Furthermore, ZPR1 deficiency markedly disrupts the subnuclear structures (gems and Cajal bodies) where final maturation of snRNP complexes is observed. These data are consistent with a collaborative role of ZPR1 in the function of SMN to mediate snRNP assembly/maturation and cell survival. The low level of ZPR1 expression in patients with spinal muscular atrophy may therefore contribute to disease progression.
Supplementary Material
Acknowledgments
We thank Neil Cashman for providing motor neurons, Eng Tan for providing the coilin antibody, Gideon Dreyfuss for providing the SMN antibody, Linda Evalgelista for performing the ES cell culture, John Nunnari and Greg Hendricks for performing electron microscopy, Stephen Jones for performing blastocyst injections, Brooke Barbara for contributing expert technical assistance, and Kathy Gemme for providing administrative assistance.
This work was supported, in part, by the Muscular Dystrophy Association (to L.G.), The Families of SMA (to L.G.), NIH/NINDS (to R.J.D.), and the Diabetes and Endocrinology Research Center (DERC) of the University of Massachusetts (NIH/NIDDK). R.A.F. and R.J.D. are investigators of the Howard Hughes Medical Institute.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andrade, L. E., E. M. Tan, and E. K. Chan. 1993. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc. Natl. Acad. Sci. USA 90:1947-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansari, H., G. Greco, and J. Luban. 2002. Cyclophilin a peptidyl-prolyl isomerase activity promotes ZPR1 nuclear export. Mol. Cell. Biol. 22:6993-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boisvert, F. M., J. Cote, M. C. Boulanger, P. Cleroux, F. Bachand, C. Autexier, and S. Richard. 2002. Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J. Cell Biol. 159:957-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brahms, H., L. Meheus, V. de Brabandere, U. Fischer, and R. Luhrmann. 2001. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B' and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 7:1531-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carmo-Fonseca, M., R. Pepperkok, M. T. Carvalho, and A. I. Lamond. 1992. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J. Cell Biol. 117:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cashman, N. R., H. D. Durham, J. K. Blusztajn, K. Oda, T. Tabira, I. T. Shaw, S. Dahrouge, and J. P. Antel. 1992. Neuroblastoma x spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev. Dyn. 194:209-221. [DOI] [PubMed] [Google Scholar]
- 7.Coovert, D. D., T. T. Le, P. E. McAndrew, J. Strasswimmer, T. O. Crawford, J. R. Mendell, S. E. Coulson, E. J. Androphy, T. W. Prior, and A. H. Burghes. 1997. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 6:1205-1214. [DOI] [PubMed] [Google Scholar]
- 8.Dundr, M., M. D. Hebert, T. S. Karpova, D. Stanek, H. Xu, K. B. Shpargel, U. T. Meler, K. M. Neugebauer, A. G. Matera, and T. Misteli. 2004. In vivo kinetics of Cajal body components. J. Cell Biol. 164:831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friesen, W. J., and G. Dreyfuss. 2000. Specific sequences of the Sm and Sm- like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN). J. Biol. Chem. 275:26370-26375. [DOI] [PubMed] [Google Scholar]
- 10.Friesen, W. J., S. Massenet, S. Paushkin, A. Wyce, and G. Dreyfuss. 2001. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell 7:1111-1117. [DOI] [PubMed] [Google Scholar]
- 11.Frugier, T., S. Nicole, C. Cifuentes-Diaz, and J. Melki. 2002. The molecular bases of spinal muscular atrophy. Curr. Opin. Genet. Dev. 12:294-298. [DOI] [PubMed] [Google Scholar]
- 12.Galcheva-Gargova, Z., L. Gangwani, K. N. Konstantinov, M. Mikrut, S. J. Theroux, T. Enoch, and R. J. Davis. 1998. The cytoplasmic zinc finger protein ZPR1 accumulates in the nucleolus of proliferating cells. Mol. Biol. Cell 9:2963-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galcheva-Gargova, Z., K. N. Konstantinov, I. H. Wu, F. G. Klier, T. Barrett, and R. J. Davis. 1996. Binding of zinc finger protein ZPR1 to the epidermal growth factor receptor. Science 272:1797-1802. [DOI] [PubMed] [Google Scholar]
- 14.Gall, J. G. 2000. Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 16:273-300. [DOI] [PubMed] [Google Scholar]
- 15.Gangwani, L., M. Mikrut, Z. Galcheva-Gargova, and R. J. Davis. 1998. Interaction of ZPR1 with translation elongation factor-1alpha in proliferating cells. J. Cell Biol. 143:1471-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gangwani, L., M. Mikrut, S. Theroux, M. Sharma, and R. J. Davis. 2001. Spinal muscular atrophy disrupts the interaction of ZPR1 with the SMN protein. Nat. Cell Biol. 3:376-383. [DOI] [PubMed] [Google Scholar]
- 17.Gubitz, A. K., W. Feng, and G. Dreyfuss. 2004. The SMN complex. Exp. Cell Res. 296:51-56. [DOI] [PubMed] [Google Scholar]
- 18.Hebert, M. D., and A. G. Matera. 2000. Self-association of coilin reveals a common theme in nuclear body localization. Mol. Biol. Cell 11:4159-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hebert, M. D., K. B. Shpargel, J. K. Ospina, K. E. Tucker, and A. G. Matera. 2002. Coilin methylation regulates nuclear body formation. Dev. Cell 3:329-337. [DOI] [PubMed] [Google Scholar]
- 20.Helmken, C., Y. Hofmann, F. Schoenen, G. Oprea, H. Raschke, S. Rudnik- Schoneborn, K. Zerres, and B. Wirth. 2003. Evidence for a modifying pathway in SMA discordant families: reduced SMN level decreases the amount of its interacting partners and Htra2-beta1. Hum. Genet. 114:11-21. [DOI] [PubMed] [Google Scholar]
- 21.Huber, J., U. Cronshagen, M. Kadokura, C. Marshallsay, T. Wada, M. Sekine, and R. Luhrmann. 1998. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 17:4114-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ilangovan, R., W. L. Marshall, Y. Hua, and J. Zhou. 2003. Inhibition of apoptosis by Z-VAD-fmk in SMN-depleted S2 cells. J. Biol. Chem. 278:30993-30999. [DOI] [PubMed] [Google Scholar]
- 23.Jablonka, S., B. Holtmann, G. Meister, M. Bandilla, W. Rossoll, U. Fischer, and M. Sendtner. 2002. Gene targeting of Gemin2 in mice reveals a correlation between defects in the biogenesis of U snRNPs and motoneuron cell death. Proc. Natl. Acad. Sci. USA 99:10126-10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jablonka, S., W. Rossoll, B. Schrank, and M. Sendtner. 2000. The role of SMN in spinal muscular atrophy. J. Neurol. 247(Suppl. 1):I37-I42. [DOI] [PubMed] [Google Scholar]
- 25.Jady, B. E., X. Darzacq, K. E. Tucker, A. G. Matera, E. Bertrand, and T. Kiss. 2003. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J. 22:1878-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerr, D. A., J. P. Nery, R. J. Traystman, B. N. Chau, and J. M. Hardwick. 2000. Survival motor neuron protein modulates neuron-specific apoptosis. Proc. Natl. Acad. Sci. USA 97:13312-13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiss, T. 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145-148. [DOI] [PubMed] [Google Scholar]
- 28.Lefebvre, S., L. Burglen, S. Reboullet, O. Clermont, P. Burlet, L. Viollet, B. Benichou, C. Cruaud, P. Millasseau, M. Zeviani, et al. 1995. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80:155-165. [DOI] [PubMed] [Google Scholar]
- 29.Lefebvre, S., P. Burlet, Q. Liu, S. Bertrandy, O. Clermont, A. Munnich, G. Dreyfuss, and J. Melki. 1997. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 16:265-269. [DOI] [PubMed] [Google Scholar]
- 30.Liu, Q., and G. Dreyfuss. 1996. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 15:3555-3565. [PMC free article] [PubMed] [Google Scholar]
- 31.Lorson, C. L., E. Hahnen, E. J. Androphy, and B. Wirth. 1999. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA 96:6307-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyon, C. E., K. Bohmann, J. Sleeman, and A. I. Lamond. 1997. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp. Cell Res. 230:84-93. [DOI] [PubMed] [Google Scholar]
- 33.Matera, A. G. 1999. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 9:302-309. [DOI] [PubMed] [Google Scholar]
- 34.McWhorter, M. L., U. R. Monani, A. H. Burghes, and C. E. Beattie. 2003. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J. Cell Biol. 162:919-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meister, G., C. Eggert, and U. Fischer. 2002. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 12:472-478. [DOI] [PubMed] [Google Scholar]
- 36.Monani, U. R., C. L. Lorson, D. W. Parsons, T. W. Prior, E. J. Androphy, A. H. Burghes, and J. D. McPherson. 1999. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 8:1177-1183. [DOI] [PubMed] [Google Scholar]
- 37.Mouaikel, J., U. Narayanan, C. Verheggen, A. G. Matera, E. Bertrand, J. Tazi, and R. Bordonne. 2003. Interaction between the small-nuclear-RNA cap hypermethylase and the spinal muscular atrophy protein, survival of motor neuron. EMBO Rep. 4:616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narayanan, U., T. Achsel, R. Luhrmann, and A. G. Matera. 2004. Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol. Cell 16:223-234. [DOI] [PubMed] [Google Scholar]
- 39.Narayanan, U., J. K. Ospina, M. R. Frey, M. D. Hebert, and A. G. Matera. 2002. SMN, the spinal muscular atrophy protein, forms a pre-import snRNP complex with snurportin1 and importin beta. Hum. Mol. Genet. 11:1785-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogg, S. C., and A. I. Lamond. 2002. Cajal bodies and coilin—moving towards function. J. Cell Biol. 159:17-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Keefe, R. T., A. Mayeda, C. L. Sadowski, A. R. Krainer, and D. L. Spector. 1994. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 124:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pagliardini, S., A. Giavazzi, V. Setola, C. Lizier, M. Di Luca, S. DeBiasi, and G. Battaglia. 2000. Subcellular localization and axonal transport of the survival motor neuron (SMN) protein in the developing rat spinal cord. Hum. Mol. Genet. 9:47-56. [DOI] [PubMed] [Google Scholar]
- 43.Paushkin, S., A. K. Gubitz, S. Massenet, and G. Dreyfuss. 2002. The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 14:305-312. [DOI] [PubMed] [Google Scholar]
- 44.Rossoll, W., S. Jablonka, C. Andreassi, A. K. Kroning, K. Karle, U. R. Monani, and M. Sendtner. 2003. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of β-actin mRNA in growth cones of motoneurons. J. Cell Biol. 163:801-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schrank, B., R. Gotz, J. M. Gunnersen, J. M. Ure, K. V. Toyka, A. G. Smith, and M. Sendtner. 1997. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. USA 94:9920-9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shpargel, K. B., J. K. Ospina, K. E. Tucker, A. G. Matera, and M. D. Hebert. 2003. Control of Cajal body number is mediated by the coilin C-terminus. J. Cell Sci. 116:303-312. [DOI] [PubMed] [Google Scholar]
- 47.Sleeman, J., C. E. Lyon, M. Platani, J. P. Kreivi, and A. I. Lamond. 1998. Dynamic interactions between splicing snRNPs, coiled bodies and nucleoli revealed using snRNP protein fusions to the green fluorescent protein. Exp. Cell Res. 243:290-304. [DOI] [PubMed] [Google Scholar]
- 48.Tucker, K. E., M. T. Berciano, E. Y. Jacobs, D. F. LePage, K. B. Shpargel, J. J. Rossire, E. K. Chan, M. Lafarga, R. A. Conlon, and A. G. Matera. 2001. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J. Cell Biol. 154:293-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vyas, S., C. Bechade, B. Riveau, J. Downward, and A. Triller. 2002. Involvement of survival motor neuron (SMN) protein in cell death. Hum. Mol. Genet. 11:2751-2764. [DOI] [PubMed] [Google Scholar]
- 50.Will, C. L., and R. Luhrmann. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13:290-301. [DOI] [PubMed] [Google Scholar]
- 51.Wirth, B. 2002. Spinal muscular atrophy: state-of-the-art and therapeutic perspectives. Amyotroph. Lateral Scler. Other Motor Neuron Disord. 3:87-95. [DOI] [PubMed] [Google Scholar]
- 52.Yong, J., T. J. Golembe, D. J. Battle, L. Pellizzoni, and G. Dreyfuss. 2004. snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol. Cell. Biol. 24:2747-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yong, J., L. Wan, and G. Dreyfuss. 2004. Why do cells need an assembly machine for RNA-protein complexes? Trends Cell Biol. 14:226-232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.