ABSTRACT
Neural crest cells generate numerous derivatives, including pigment cells, and are a model for studying how fate specification from multipotent progenitors is controlled. In mammals, the core gene regulatory network for melanocytes (their only pigment cell type) contains three transcription factors, Sox10, Pax3 and Mitf, with the latter considered a master regulator of melanocyte development. In teleosts, which have three to four pigment cell types (melanophores, iridophores and xanthophores, plus leucophores e.g. in medaka), gene regulatory networks governing fate specification are poorly understood, although Mitf function is considered conserved. Here, we show that the regulatory relationships between Sox10, Pax3 and Mitf are conserved in zebrafish, but the role for Mitf is more complex than previously emphasized, affecting xanthophore development too. Similarly, medaka Mitf is necessary for melanophore, xanthophore and leucophore formation. Furthermore, expression patterns and mutant phenotypes of pax3 and pax7 suggest that Pax3 and Pax7 act sequentially, activating mitf expression. Pax7 modulates Mitf function, driving co-expressing cells to differentiate as xanthophores and leucophores rather than melanophores. We propose that pigment cell fate specification should be considered to result from the combinatorial activity of Mitf with other transcription factors.
Keywords: Paired-type homeobox, Chromatophore, Melanocyte, Pigmentation, CRISPR/Cas9
Summary: Unique and sequential functions of Pax3 and Pax7, in combination with Mitf, allow the diversification of pigment cell types in teleost.
INTRODUCTION
All body pigment cells in vertebrates are considered to derive from neural crest cells (NCCs), a fascinating population of multipotent stem cells (Le Douarin and Kalcheim, 1999; Tang and Bronner, 2020). These cells form an interesting class of NCC derivatives, in part because traditionally all pigment cell types have been believed to develop from partially restricted NCC-derived progenitor cells, so called chromatoblasts (Bagnara et al., 1979; Schartl et al., 2016), although we have recently challenged that view, showing that neural crest cells appear to retain much broader multipotency, even in differentiating pigment cells (Kelsh et al., 2021; Subkhankulova et al., 2023). Regardless of their exact nature, these multipotent progenitor cells undergo a process of fate specification to one or more cell types, before differentiation and eventual commitment to individual specialized cell types. This process is regulated by a complex gene regulatory network (GRN) organized around a core set of transcription factors, which ultimately directs cells to differentiate, stably expressing a transcriptome appropriate to the specific cell type.
Mammals and birds have only one type of pigment cell, called the melanocyte (Sommer, 2011; Wegner, 2005). The core GRN that controls melanocyte differentiation consists of three transcription factors: the Sry-related HMG-type transcription factor Sox10, the paired-type homeodomain-containing Pax transcription factor Pax3 and the basic helix-loop-helix (bHLH) transcription factor Mitf (microphthalmia-associated transcription factor). Sox10 and Pax3 cooperatively promote melanocyte development through activating expression of Mitf, which in turn drives melanin-synthesizing enzymes, e.g. dopachrome tautomerase (Dct) (Jiao et al., 2004; Lang et al., 2005; Ludwig et al., 2004; Tsukamoto et al., 1992). Mutations in SOX10, PAX3 or MITF similarly cause Waardenburg syndrome in humans (Bondurand et al., 2000; Potterf et al., 2000; Tassabehji et al., 1992), which manifests as patchy pigmentation due to (partial) loss of melanocytes in the skin. Mitf is considered to be a master regulator of melanocytes in mammals as MITF/Mitf mutations result in absence of melanocytes, and MITF target genes are numerous and include at least the majority of melanocyte markers (Arnheiter, 2010; Goding and Arnheiter, 2019; Hemesath et al., 1994; Kawakami and Fisher, 2017; Opdecamp et al., 1997; Steingrimsson et al., 1994).
In contrast to mammals and birds, other vertebrates basically have three different types of pigment cell: the melanophore (equivalent to the mammalian melanocyte), the iridophore (iridescent cell) and the xanthophore (yellow to red pigment cell). A few species of teleosts, including the Japanese endemic fish medaka, have a fourth type, the leucophore (white cell), but these are absent from the zebrafish except for a few cells (which may or may not be homologous) in adults (Hashimoto, 2021; Lewis et al., 2019). A more complex GRN must be required to generate this diversity of pigment cell types. Here, we address this question of defining the GRN that achieves fate specification and differentiation of the multiple pigment cells in ectothermic vertebrates. We test the hypothesis that the core GRN is conserved in both endothermic and ectothermic vertebrates, but achieves added complexity through some additional components specific to ectothermic vertebrates (secondarily lost in the endothermic taxa). Among the three core transcription factors, Sox10 and Mitf have been shown to have highly conserved roles in melanocyte/melanophore development, whereas the function of Pax3 is still poorly understood in other vertebrate species (Greenhill et al., 2011; Hou et al., 2006).
Sox10 plays central roles in all non-ectomesenchymal fates of NCCs, in both fish and mammals, with its loss resulting in absence or severe depletion of all NCC-derived neuronal, glial and pigment cell types (Britsch et al., 2001; Dutton et al., 2001; Kelsh and Eisen, 2000; Nagao et al., 2018; Southard-Smith et al., 1998). Evidence to date suggests that development of each of these derivatives fails at the earliest stages of specification from the NCCs (Dutton et al., 2001; Elworthy et al., 2003, 2005; Greenhill et al., 2011; Kelsh, 2006; Lopes et al., 2008; Petratou et al., 2021, 2018). Thus, in zebrafish and medaka, loss of Sox10 function results in an almost-complete lack of melanophores (except for the brain-derived pigmented retinal epithelium), xanthophores and iridophores (Tsunogai et al., 2021), and in mice it results in complete lack of melanocytes (Britsch et al., 2001; Southard-Smith et al., 1998).
As in mammals, the zebrafish MITF homologue, Mitfa, plays a crucial role in melanophore development as zebrafish mitfaw2/w2 mutants (also known as nacre) lack melanophores throughout life (Lister et al., 1999). A crucial role in melanophore fate specification and differentiation is indicated by the extensive absence of melanophore markers in zebrafish mitfa mutants and by the role of Mitfa in directly driving dct transcription and melanin pigmentation in the melanophore lineage in zebrafish (Greenhill et al., 2011; Lister et al., 1999), as in mice (Jiao et al., 2004; Ludwig et al., 2004; Tsukamoto et al., 1992). Furthermore, expression of mitfa in the NCCs of zebrafish sox10 mutants is sufficient to rescue full melanophore differentiation (Elworthy et al., 2003), consistent with the master regulator model proposed for mammalian MITF. However, although xanthophores are formed in zebrafish mitfaw2/w2 mutants, their development has been noted as abnormal, suggesting that Mitfa may have a partially redundant role in xanthophore formation (Lister et al., 1999). Nevertheless, the exact role of Mitfa in xanthophore development remains unknown.
Pax3 functions as a cooperative partner of Sox10, and thus is also required for melanocyte specification in mammals (Kubic et al., 2008). Pax3 mutations result in loss of coat colour in the Splotch mutant mice (Conway et al., 1997; Tremblay et al., 1995), and also in splashed white horses (Hauswirth et al., 2012). In teleosts, however, the role of Pax3 is not clear. An antisense morpholino-mediated knockdown study in zebrafish showed that loss of Pax3 resulted in defective specification of xanthophores, but not of melanophores or iridophores (Minchin and Hughes, 2008).
Pax7 is closely related to Pax3 and has been suggested to have similar activity to Pax3. These two transcription factors play important roles in a variety of stem cells, including the NCCs, such as regulating fate decision and differentiation (Buckingham and Relaix, 2015; Nord et al., 2022; Relaix et al., 2004). The similarity is considered to reflect their emergence from a common ancestral gene through duplication (Robson et al., 2006). Pax7 knockdown by morpholino in the chick indicates that Pax7 is important for induction of the neural crest from the ectoderm, but not specifically required for melanocyte differentiation (Basch et al., 2006). To date, no PAX7-associated pigmentation disorders have been reported in humans and mice, although Pax7-expressing cells give rise to NCCs (Murdoch et al., 2012). Thus, Pax7 does not appear to play a major role in mammalian or avian melanocyte development.
Meanwhile, in teleosts, Pax7 seems to be involved in the GRN for pigment cell development. Zebrafish pax7a; pax7b double-homozygous mutants exhibit complete absence of xanthophores, but not of melanophores nor iridophores (Nord et al., 2016), suggesting an essential and specific role of Pax7 in xanthophore formation. However, in medaka, in which functional Pax7b has been evolutionarily lost, pax7a homozygous mutants exhibit a complete absence of xanthophores and leucophores. As a consequence, xanthophores and leucophores have been proposed to share a bipotent progenitor, specification of which requires Pax7a function in medaka (Kimura et al., 2014; Nagao et al., 2014, 2018). Although that conclusion may need revision in the context of our recent zebrafish observations (Kelsh et al., 2021; Subkhankulova et al., 2023), it remains clear that Pax7 function is necessary for both xanthophore and leucophore development.
Here, we used a comparative genetic approach to assess the roles for these key pigment cell transcription factors. We addressed the following key questions: (1) Is Pax3 one of the core transcription factors in the melanophore GRN in medaka and zebrafish?; (2) Do Pax3 and Pax7 control specification of other pigment cell types?; and (3) Does Mitfa have a role in the development of other pigment cell types in medaka? Our results suggest that Sox10, Pax3 and Mitf have a conserved role as components of the core GRN driving melanophore fate, but that they also work alongside Pax7 to promote xanthophore and leucophore fates in teleosts.
RESULTS
Two paralogous pax3 genes, pax3a and pax3b, are identified in medaka and zebrafish genomes
Vertebrate pax3 and pax7 are descendants of a common ancestral gene (named pax37 in ascidians), resulting from the second-round whole genome duplication in vertebrate evolution (Kusakabe and Kuratani, 2007; Wada et al., 1996). In teleosts, two paralogues for each of pax3 and pax7 were generated as a result of another round of whole-genome duplication (Amores et al., 1998; Postlethwait et al., 2004). Medaka and zebrafish have pax3a, pax3b, pax7a and pax7b in their genomes, although medaka pax7b lacks three exons (exons 6-8) and appears to be a non-functional pseudogene (see Fig. S1).
pax3 is expressed in neural crest cells prior to pax7
We observed expression of only pax3b and pax7a mRNAs in medaka NCCs. In situ hybridization analyses showed that pax3b-expressing cells were present in dorsal parts of the midbrain, hindbrain and spinal cord, dorsal somites and premigratory NCCs at early somite stages (Fig. 1A-A″). In contrast, pax7a expression was detected in both premigratory and laterally migrating NCCs (Fig. 1B-B″). At mid-somite stages, pax3b-expressing NCCs were seen in the posterior premigratory region, but not in the lateral migratory region (Fig. 1C,C′), whereas pax7a-expressing NCCs were observed in both premigratory and migrating regions (Fig. 1D-D″). By late somite stages, pax3b expression was no longer detectable in the NCCs, but pax7a expression was still detected in migratory NCCs (Fig. 1E-E″) and maintained until almost the hatching stage (Watakabe et al., 2018). These results indicate that expression of pax3b in medaka is transient, restricted to premigratory NCCs, and precedes that of pax7a, which is maintained into migrating NCCs. These expression patterns are similar to those previously reported for zebrafish pax3 (pax3a and pax3b) and pax7 (pax7a and pax7b) (Minchin and Hughes, 2008).
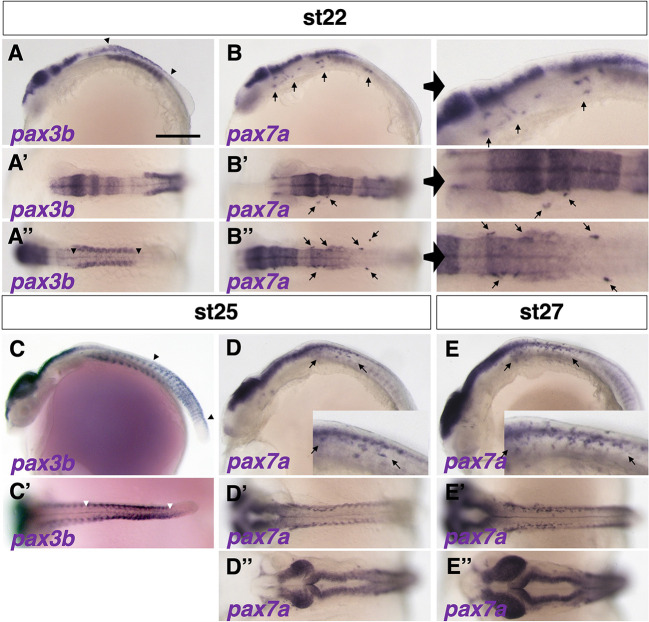
Fig. 1.
Expression patterns of pax3 and pax7 in medaka. (A-E″) Expression of pax3b mRNA was detected in NCCs at stage 22 (9 somite embryo) (A-A″, between arrowheads), whereas pax7a expression was observed in migrating NCCs anteroventral to the pax3b-expressing premigratory NCCs (B-B″; panels on the right show enlarged views; pax7a-expressing cells are indicated by arrows). The pax3b expression had shifted posteriorly at stage 25 (18-19 somite embryo) (C,C′, between arrowheads) and became undetectable by stage 27 (24 somite embryo) (not shown). pax7a mRNA was continuously observed in migrating NCCs from stage 25 to stage 27 (indicated by arrows; see also Fig. S2). pax3b and pax7a mRNAs were expressed in similar regions of neural tissue and somites (A-E). Expression of pax3b was observed in several anterior somites at stage 22 (A), shifted to posterior somites at stage 25 (C; see also Fig. S2), and appeared to precede that of pax7a in somites (B,D). The time windows of pax3b and pax7a expression overlapped during stages 22-25. (A-E) Lateral views. (A′-E′,A″,B″,D″,E″) Dorsal views. Images are representative of more than ten embryos. Scale bar: 250 µm.
The above results were confirmed by observing fluorescently labelled pax3b- and pax7a-expressing cells in a newly established stable transgenic medaka Tg(pax3b-hs:GFP) and previously reported Tg(pax7a-hs:GFP) lines (Fig. S2A) (Watakabe et al., 2018). pax7a-GFP remained detectable in NCCs until a late somite stage (Fig. S2D,F,H), much longer than the pax3b-GFP (Fig. S2B,C,E,G). We also tested whether the expression of pax3b and pax7a overlapped, by using Tg(pax3b-hs:GFP) crossed with the previously reported TgBAC(pax7a:DsRed) (Nagao et al., 2018). A portion, but not all, of the cells positive for pax3b-GFP were also positive for pax7a-DsRed (Fig. S2I-K), indicating that these cells express both Pax3b and Pax7a. We observed pax3b-GFP-positive but pax7a-DsRed-negative cells, which may be cells that have not yet reached the state at which they are ready to express pax7a, but which may indicate that pax3b-expressing cells give rise to pigment cell populations other than pax7a-expressing xanthophore/leucophore progenitors.
Loss of pax3 function results in delayed formation of xanthophores in zebrafish and medaka, and also of leucophores in medaka
To investigate the role of pax3 and pax7, we generated mutations in pax3a and pax3b in medaka (Fig. S1) and pax3a, pax3b, pax7a and pax7b in zebrafish using transcription activator-like effector nucleases (TALENs) or CRISPR/Cas9 (Fig. S3). In addition, we used medaka leucophore free-2 (lf-2) as a pax7a mutant, which was previously reported as a loss-of-function mutation (Fig. S1) (Kimura et al., 2014).
We examined pigment cell phenotypes of the mutants. The medaka pax3b homozygous mutants showed a delay in the formation of xanthophores and leucophores (Fig. 2A-D, Fig. S4), which was detected as a reduction in the number of these cell types in hatchlings (Fig. 2G,H). This phenotype was much milder than that of the pax7a mutant, in which these two cell types were completely lost (Fig. 2E,F, Fig. S4) (Kimura et al., 2014). Medaka pax3b mutant fish became apparently normal in pigmentation as they grew, with xanthophores and leucophores restored (Fig. S4).
Fig. 2.
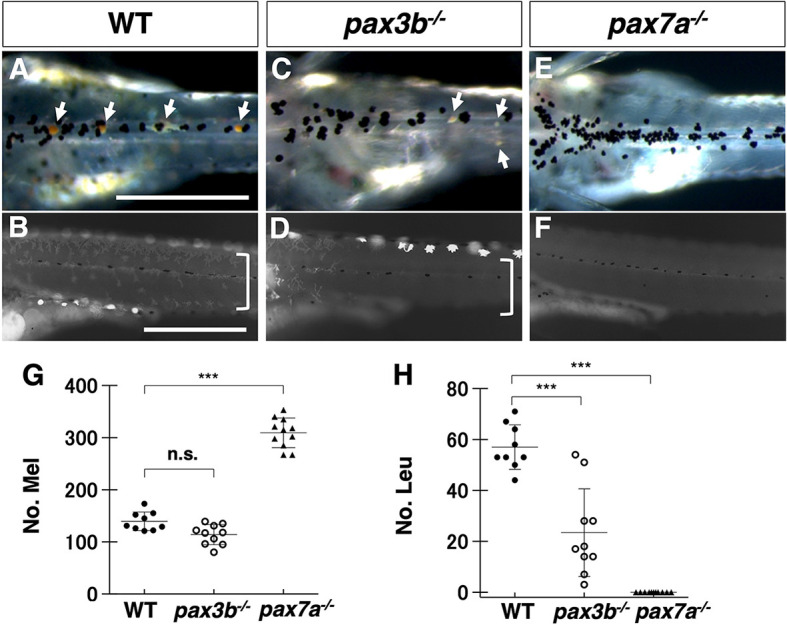
Phenotypes of medaka pax3 and pax7 mutants. (A-F) Medaka 9 dpf hatchlings. Comparison of pigment cell phenotypes between WT (A,B), pax3b−/− (C,D) and pax7a−/− (lf-2) (E,F). (A,C,E) Dorsal views of the trunk in dark field. (B,D,F) Lateral views of the trunk under UV light. Images are representative of more than ten embryos. Scale bars: 250 µm. (G,H) Quantification of the number of melanophores (G) and leucophores (H) on the dorsal surface of the trunk. Note that leucophores are not always white but often yellow or orange, and have strong fluorescence under UV light. See ‘Microscopy’ in the Materials and Methods section. The highly fluorescent cells in the ventral edge in B and in the dorsal edge in D are leucophores. Xanthophores and leucophores were severely reduced in number in medaka pax3b−/− mutant hatchlings (leucophores indicated by arrows in A and C; xanthophores, which are autofluorescent and dendritic under UV light, indicated by square brackets in B and D), and were completely absent in medaka pax7a−/− mutants (E,F,H). The number of melanophores was unaltered in pax3b−/− mutants (A,C,G), but were significantly increased in pax7a−/− mutants (E,G). Significant difference was determined by Kruskal–Wallis test. ***P<0.05. n=8 for WT, 10 for pax3b, and 11 for pax7a in G,H. n.s., not significant. Error bars represent s.d.
Similarly, zebrafish pax3a homozygous mutant hatchlings (Fig. S5) showed a delay in the formation of xanthophores, whereas xanthophores were completely and persistently lost in the pax7a; pax7b double mutant (Figs S5, S6). Melanophore number was not significantly altered in the absence of Pax3, but was increased in the absence of Pax7 in both medaka and zebrafish (Fig. 2G, Fig. S5J). The iridophore phenotype was faint and ambiguous in both medaka and zebrafish (Figs S4B,D,F, S5C,F,I,K).
The mutation in the paralogous gene, pax3a in medaka and pax3b in zebrafish, did not affect pigment cell development, or even enhance the phenotypes owing to the single paralogous mutation (medaka pax3b−/− and zebrafish pax3a−/−) (Figs S7 and S8).
In summary, the phenotypes due to loss of Pax3, by pax3b in medaka and pax3a in zebrafish, are most evident in xanthophores/leucophores and not significant or less severe in other cell types; notably, they do not include obvious melanophore phenotypes. Our results suggest that Pax3 is involved in the specification of xanthophores and leucophores in medaka, and xanthophores in zebrafish (which lack leucophores).
Loss of Pax3 affects the expression of early xanthophore/leucophore markers
To investigate the early phenotypes of pigment cells in the absence of Pax3, we examined the expression pattern of specification markers. We used GTP cyclohydrolase 2 (gch2) as a marker for xanthophore and leucophore progenitors (Pelletier et al., 2001), dct as a marker for melanophore progenitors (Kelsh et al., 2000) and purine nucleoside phosphorylase (pnp4a) as a marker for iridophore progenitors (Kimura et al., 2017; Petratou et al., 2021, 2018).
In medaka, expression of gch2 in xanthophore/leucophore progenitors was severely reduced in the pax3b mutant (Fig. 3A,B) and completely absent in the pax7a mutant (Fig. 3C). Melanophore progenitors expressing dct appeared to be slightly reduced in number in the pax3b mutant (Fig. 3E), whereas they were unaltered or even increased in number and expression level compared with wild type (WT) in the pax7a mutant (Fig. 3D,F). Iridophore progenitors expressing pnp4a appeared to be reduced in the yolk sac, but not in the eyes in the pax3b mutant (Fig. 3G,H), whereas they are unaltered in the pax7a mutant (Fig. 3I). We obtained roughly similar results from zebrafish pax3a and pax7a; pax7b mutants (Fig. S9), except that dct-expressing melanophore progenitors were little changed by the loss of Pax3 (Fig. S9D,E) and pnp4a-expressing iridophore progenitors appeared to be slightly increased in the pax7a; pax7b mutant (Fig. S9G-I).
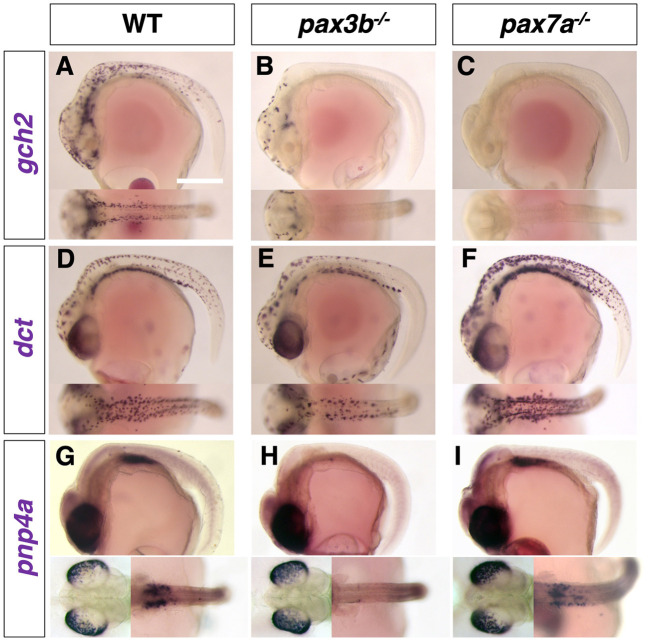
Fig. 3.
Pigment cell progenitors in medaka pax3b and pax7a mutant embryos. (A-I) Lateral views at top and dorsal views at bottom at stage 29. The gch2-expressing progenitors of xanthophore and leucophore were severely decreased in the pax3b mutant (A,B), with only a few remaining anteriorly, and completely absent in the pax7a mutant (C), which is consistent with the severity of the phenotypes in these mutants at later (hatching) stages. The dct-expressing progenitors of melanophore were slightly decreased in number in pax3b mutant (D,E), but unaltered or even increased in number and expression level in pax7a mutant compared with WT (F). The pnp4a-expressing progenitors of iridophore were not altered in the eyes in both mutants (G-I, bottom left), but the putative progenitors of peritoneum iridophores were decreased in the pax3b mutant (G,H), but not in the pax7a mutant (I, bottom right). Images are representative of more than ten embryos. Scale bar: 250 µm.
Like the phenotypes of pigmented cells in hatchlings, those of early specification markers in embryos suggest that the defects due to loss of Pax3 (Pax3a in zebrafish and Pax3b in medaka) are most manifest in the formation of xanthophores/leucophores.
pax3 functions upstream of pax7
To investigate the genetic relationship between pax3b and pax7a, we examined the expression patterns of both pax3b and pax7a in the pax7a and pax3b mutants. In situ hybridization analyses showed that, whereas pax3b-expressing cells were unaffected in the pax7a mutant medaka (Fig. 4A,B), pax7a-expressing NCCs were severely reduced from the trunk region in pax3b mutant medaka embryos (Fig. 4C,D). Similarly, in zebrafish, both pax7a and pax7b expression was severely reduced in the pax3a mutant (Fig. 4E-J). These results suggest that, consistent with their expression timings, in the NCCs pax3b functions upstream of pax7a in medaka, that pax3a functions upstream of pax7a and pax7b in zebrafish, and also that pax7 expression is partially dependent on pax3.
Fig. 4.
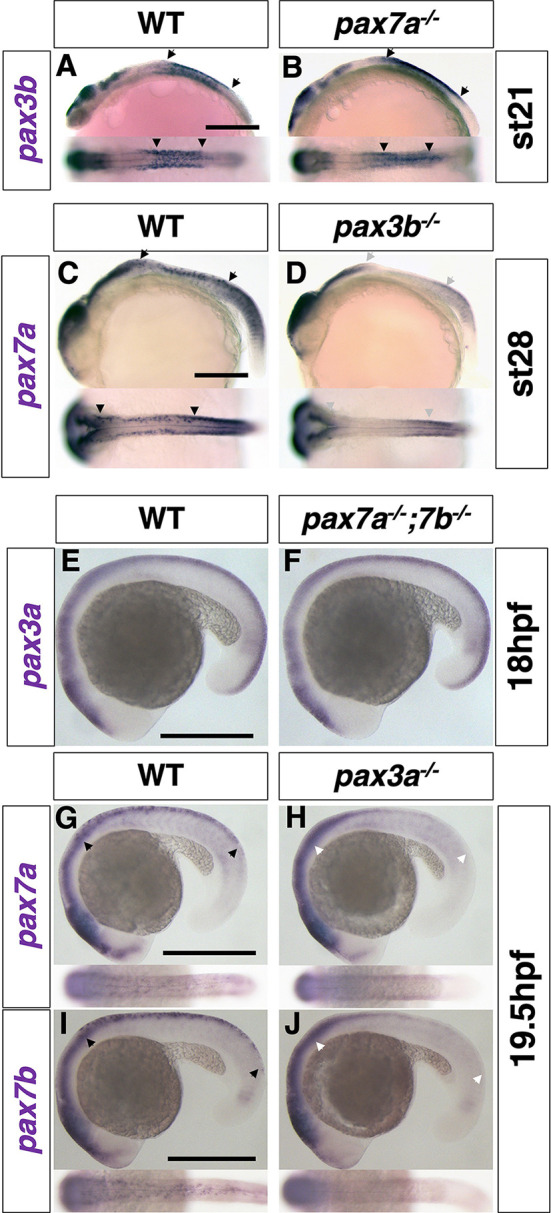
pax3 functions upstream of pax7. (A-D) Medaka. (A,B) pax3b expression at stage 21 (6 somite). (C,D) pax7a expression at stage 28 (30 somite). (E-J) Zebrafish. (E,F) pax3a expression at 18 hpf (18 somite). (G,H) pax7a expression at 19.5 hpf (21 somite). (I,J) pax7b expression at 19.5 hpf (21 somite). In medaka, pax3b expression was similar in WT and pax7a mutant embryos (A,B), whereas pax7a expression was largely reduced in NCCs in pax3b mutant embryos compared with WT (C,D). Similarly, in zebrafish, pax3a expression was not altered in pax7a; pax7b double-mutant embryos (E,F), but pax7a and pax7b expression was largely reduced in NCCs in pax3a mutant embryos compared with WT (G-J). Lateral views at top and dorsal views at bottom. Arrowheads indicate the anterior and posterior ends of the expression in the neural crest. Grey and white arrowheads indicate the absence of expression. Images are representative of more than ten embryos. Scale bars: 250 µm.
The expression of mitf is dependent on pax3
The dct expression patterns in zebrafish and medaka Pax3 mutants are intriguing, because they suggest that the core role for Pax3 in melanocyte development in mammals has been conserved in medaka, but perhaps not in zebrafish, melanophores. To test directly the idea that the core GRN consisting of the transcription factors Sox10, Pax3 and Mitf plays a central role in melanophore development in teleosts, we assessed whether mitf expression might be dependent on Pax3 in medaka and zebrafish, similar to its role in mammals (Lang et al., 2005), by examining mitfa expression in medaka pax3b and zebrafish pax3a mutant embryos (Fig. 5). In comparison with medaka WT embryos (Fig. 5A), the mitfa-expressing cells were severely reduced in pax3b mutants (Fig. 5B), whereas they were comparable or rather increased in pax7a mutant embryos (Fig. 5C). Similarly, in zebrafish at 19.5 hpf, early mitfa expression was partially lost in pax3a mutant (Fig. 5D,E), but not in pax7a; pax7b double-mutant embryos (Fig. 5F). The expression pattern in zebrafish in particular indicates that the Pax3 mutation might cause delayed upregulation of mitfa, whereas the loss of Pax7 activity might allow precocious upregulation of mitfa. Overall, and importantly, these results indicate that loss of Pax3 causes a reduction in mitfa expression in medaka and zebrafish, suggesting that mitfa transcription is partially dependent on Pax3 activity.
Fig. 5.
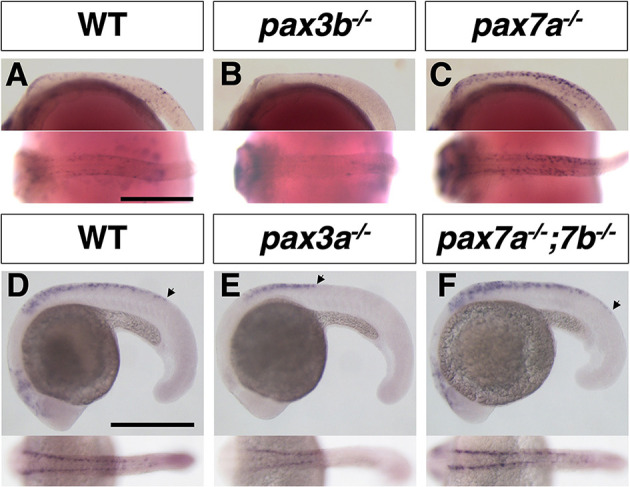
Loss of pax3 leads to a decrease in mitfa expression. (A-C) Medaka embryos at stage 28 (30 somite). (D-F) Zebrafish embryos at 19.5 hpf (21 somite). Arrowheads indicate the posterior ends of mitfa expression in the neural crest (D-F). Lateral views at top and dorsal views at bottom. The mitfa-expressing cells were largely lost in the absence of Pax3 in medaka (A,B) and in zebrafish (D,E). Those cells appeared to be normal or rather increased in the medaka pax7a mutant (C) and in the zebrafish pax7a; pax7b double mutant (F). Images are representative of more than ten embryos. Scale bars: 250 µm.
Cooperative function of Pax3 and Sox10 can promote mitf expression
To investigate whether Pax3 directly controls mitf transcription in teleosts, we performed a Pax3 overexpression experiment by synthetic RNA injection into zebrafish embryos. Working on the assumption that Pax3 cooperates with Sox10 to activate mitf transcription, as in mice, we injected synthetic RNAs of pax3a and/or sox10 into one-cell-stage embryos and examined whether mitfa mRNA was ectopically expressed in the injected embryos at 6 h post-fertilization (hpf), when mitfa mRNA is not yet endogenously expressed in zebrafish (Fig. 6A,A′).
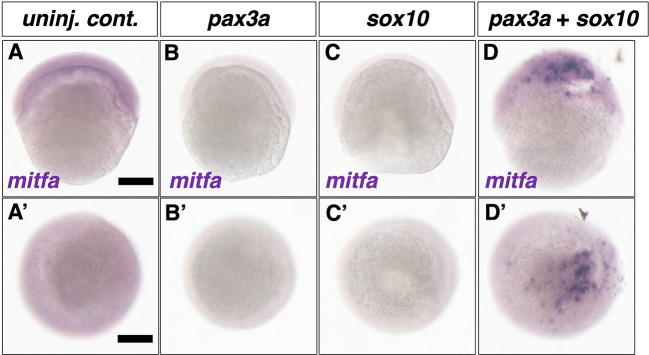
Fig. 6.
Overexpression of pax3a and sox10 induces ectopic expression of mitfa in zebrafish. (A-D′) mitfa expression at 6 hpf in control (A,A′), pax3a synthetic RNA-injected (B,B′), sox10 synthetic RNA injected (C,C′) and pax3a and sox10 synthetic RNA co-injected (D,D′) embryos. (A-D) Lateral views. (A′-D′) Animal pole views. mitfa mRNA was not expressed endogenously in 6 hpf embryo (A). Whereas overexpression of pax3a or sox10 by synthetic RNA injection into 1- to 2-cell-stage embryos failed to induce mitfa expression at 6 hpf, simultaneous overexpression of pax3a and sox10 can induce ectopic expression of mitfa. Scale bars: 200 µm. Fractions were 23/23 for pax3a injection, 16/16 for sox10 injection and 30/38 for pax3a and sox10 co-injection.
Whereas mitfa mRNA was not detected in situ when Pax3a (Fig. 6B,B′) or Sox10 (Fig. 6C,C′) were overexpressed alone, simultaneous overexpression of these transcription factors ectopically induced mitfa expression at a high rate (Fig. 6D,D′). Our results suggest that the cooperative action of Pax3 and Sox10 to promote mitf/Mitf expression is conserved between zebrafish and mice (Lang et al., 2005).
Mitfa can drive the progenitor markers for melanophore and xanthophore
The results above suggest that the GRN consisting of Sox10, Pax3 and Mitf is conserved in zebrafish, where Mitfa plays an essential role in melanophore differentiation (Greenhill et al., 2011). However, loss of Pax3 has the most severe effect on xanthophore formation in zebrafish and medaka. Given that Mitf mediates Pax3 function, we hypothesized that Mitf is involved in regulating, not only melanophore differentiation, but also xanthophore differentiation in zebrafish. To test this, we examined whether mitfa overexpression could induce ectopic expression of markers for melanophore (dct) and xanthophore (gch2) progenitors in zebrafish embryos (Fig. 7). The dct and gch2 mRNAs were not expressed endogenously in control 6 hpf embryos (Fig. 7A,D). Injection of mitfa synthetic RNA induced ectopic mRNA expression of dct (Fig. 7B), but also of gch2 (Fig. 7E). Thus, it is likely that Mitfa can promote both xanthophore and melanophore fates in zebrafish when overexpressed.
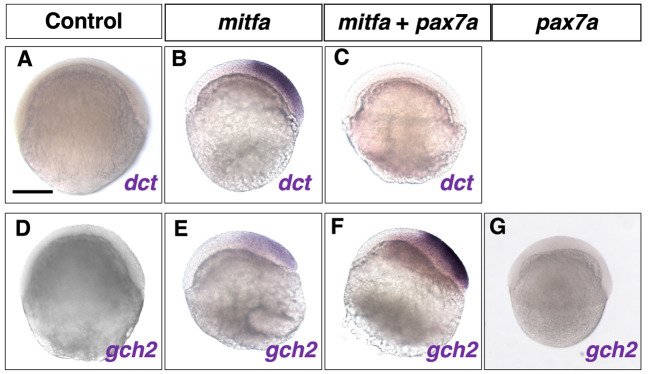
Fig. 7.
Overexpression of mitfa induces ectopic expression of progenitor markers for melanophore and xanthophore. (A-G) dct (A-C) and gch2 (D-G) expression in 6 hpf control (A,D), mitfa synthetic RNA-injected (B,E), mitfa and pax7a synthetic RNA-injected (C,F) and pax7a synthetic RNA-injected (G) embryos. Overexpression of mitfa alone can strongly induce ectopic dct expression (A,B; 22/23). Simultaneous overexpression of pax7a suppresses the ectopic dct expression by mitfa (C; 18/18). Ectopic expression of gch2 mRNA is induced by overexpression of mitfa alone (D,E; 20/24) and more strongly induced by overexpression of mitfa and pax7a in combination (F; 26/31). Overexpression of pax7a alone is unable to induce ectopic gch2 expression (G). Scale bar: 250 µm.
Pax7 can suppress Mitf action to drive Gch2 transcription
What determines the fate choice of the mitfa-expressing progenitors to either melanophore or xanthophore fate? To address this, we focused on the previous report in mice that Pax3 can inhibit Mitf-mediated transcriptional activation of Dct after activating Mitf transcription in cooperation with Sox10 (Lang et al., 2005). We hypothesized that after Pax3 has activated mitfa expression, Pax7 instead of Pax3 might play the inhibitory role against Mitfa-driven transcription of dct in zebrafish. To test this hypothesis, we examined whether Pax7 could affect the ectopic expression of dct and gch2, when co-overexpressed with Mitfa in zebrafish embryos. The ectopic expression of dct, which was detected in the mitfa-injected embryos, was lost in the 6 hpf embryos co-injected with mitfa and pax7a RNAs (Fig. 7C). By contrast, pax7a co-injection enhanced the ectopic expression of gch2 in the mitfa-injected 6 hpf embryos (Fig. 7F). Interestingly, injection of pax7a synthetic RNA alone was unable to activate ectopic gch2 expression (Fig. 7G).
Mitf specifies the fate of melanophore and xanthophore in medaka
To determine whether Mitf activity is required not only for melanophore fate but also for xanthophore fate, we attempted to generate loss-of-function mutants for Mitf in medaka. We found two mitf paralogous genes, mitfa and mitfb, in the medaka genome, as in zebrafish (Lister et al., 2001). The expression pattern of these genes, examined by in situ hybridization, showed that both appeared to be expressed in the NCCs (Fig. S10). We therefore induced mutations in mitfa and mitfb using CRISPR/Cas9, targeting the region encoding the bHLH domain (Fig. S11).
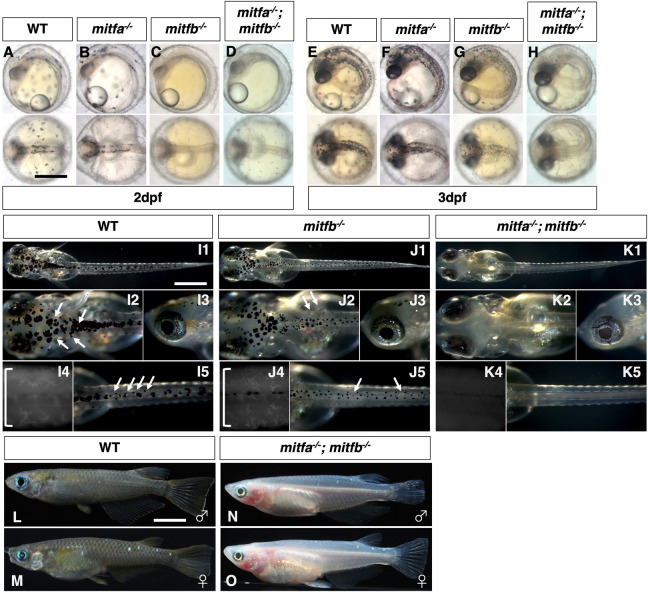
We successfully generated mutations in the mitfa and mitfb genes in medaka (Fig. S11). Single loss of mitfb resulted in a defect in melanophore formation, being delayed at 2 days post-fertilization (dpf) (Fig. 8A,C) and partially restored at 3 dpf (Fig. 8E,G) and at hatching (Fig. 8I,J), whereas loss of mitfa did not (Fig. 8B,F), suggesting that mitfb, but not mitfa, plays the more central role in melanophore development in medaka. The double loss of mitfa and mitfb resulted in complete absence of not only melanophores but also xanthophores and leucophores throughout embryogenesis (Fig. 8D,H,K), and even later during adulthood (Fig. 8L-O). Iridophore formation appeared normal throughout life (Fig. 8K2,K3,N,O). These data suggest that Mitfa and Mitfb are redundant in the NCCs and, together, are essential for the development of melanophores, xanthophores and leucophores in medaka.
Fig. 8.
Phenotypes of medaka mitfa and mitfb double homozygotes. (A-O) Phenotypes of wild-type (WT), mitfa single, mitfb single and mitfa and mitfb double mutants in 2 dpf (A-D) and 3 dpf (E-H) embryos, 7 dpf hatchlings (I-K) and 4-month-old adult male (L,M) and female (N,O) fish. (I4,J4,K4) Autofluorescence images showing xanthophores under UV light. (A-H) Upper panels are dorsal views; lower panels are lateral views. (G-K) Panels 1, 2 and 5 are dorsal views; panels 3 and 4 are lateral views. (L-O) Lateral views. Melanophores first appear on the head, the anterior body and the yolk at 2 dpf (A) and increase in number to become distributed throughout the body at 3 dpf (E) in the WT embryo. The mitfa mutant embryo looks normal at this time (B,F), whereas the mitfb mutant does not have melanophores at 2 dpf (C), but shows their delayed formation at 3 dpf (G). The mitfa; mitfb double mutant completely lacks melanophores during this period and thereafter (D,H). At the hatching stage, all the four types of pigment cells are differentiated (pigmented) in WT (I1-I5). The mitfb mutant looks normal except that melanophores are relatively small and few leucophores are found (arrows in J2,J5) compared with those in WT (I1-I5). The mitfa; mitfb double mutant completely lacks not only melanophores (K1,K2,K5) but also xanthophores (K4) and leucophores (K1,K2,K5), but retains iridophores in the eyes and on the yolk (K1,K2,K3). Square brackets indicate xanthophores on the lateral surface of the body (I4,J4). In adulthood, compared with WT (L,M), it is obvious that the mitfa; mitfb double mutant lacks all visible pigmentation except for that of iridophores in the skin and the iris (N,O). Scale bars: 0.5 mm (in A for A-H, in I1 for I1-K1); 5 mm (in L for L-O).
In situ hybridization analyses support the idea that Mitfs are not only required for melanophore fate but also for specification of xanthophore/leucophore fate (Fig. 9). Expression of gch2 and dct was absent from the body surface in the mitfa; mitfb double homozygotes (Fig. 9A-D). In contrast, pnp4a mRNA was expressed in the eyes and on the yolk similarly in WT and the mitfa; mitfb double mutant (Fig. 9E,F). These results are fully consistent with the above phenotype that the double mutant has only iridophores but lacks the other three types of pigment cells.
Fig. 9.
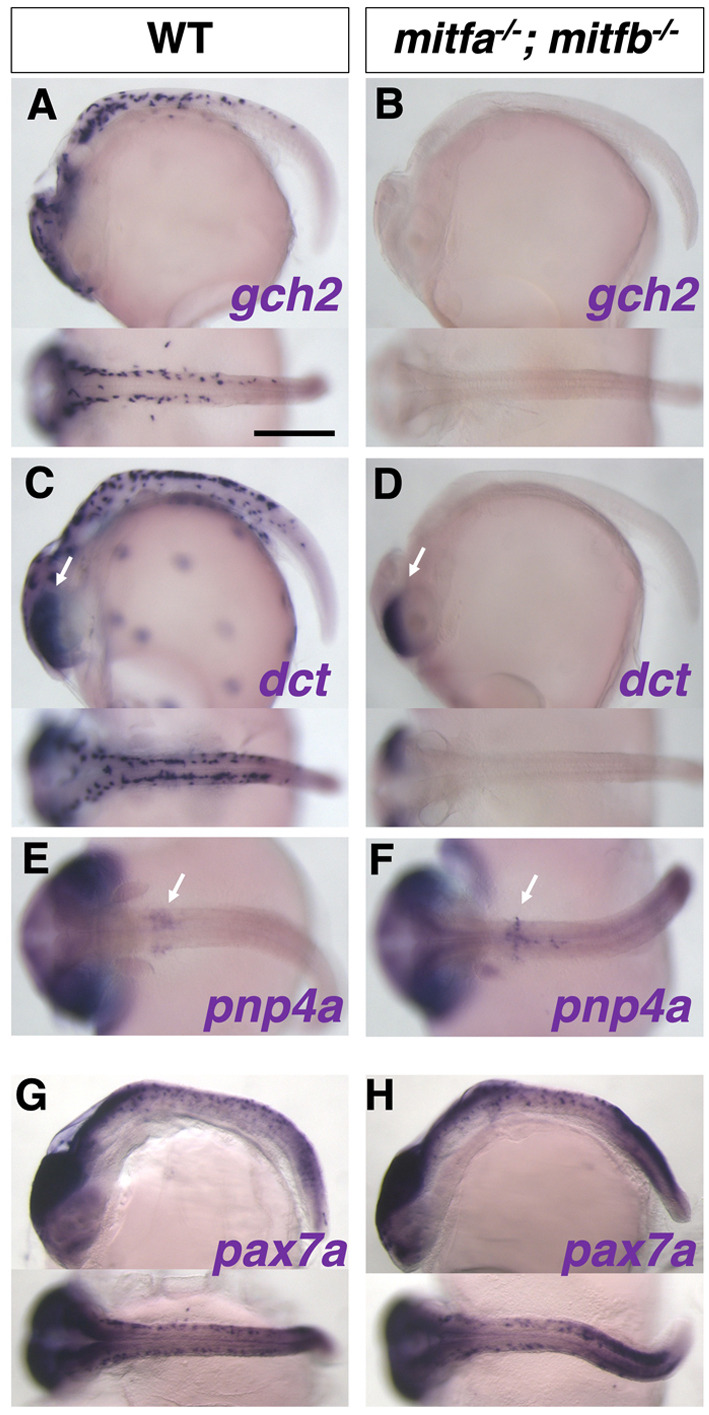
In situ analyses of medaka mitfa−/−; mitfb−/− double mutants with pigment cell markers. Results of in situ analyses with specification markers, dct for melanophore, gch2 for xanthophore/leucophore, and pnp4a for iridophore, are consistent with the phenotypes of differentiated pigment cells. The gch2-expressing xanthophore/leucophore progenitors are lost in the double mutant (A,B). The dct-expressing melanophore progenitors are absent from the body surface, but not from the eyes (retinal pigment epithelium, RPE) in the double-mutant embryo (C,D; arrows indicate the signal in RPE), consistent with the phenotype that the mutant retains melanized RPE (see Fig. 8K1,K3). The pnp4a-expressing iridophore progenitors appear unchanged in the double mutant compared with WT (E,F; arrows indicate the signal on the yolk). The mitfa−/−; mitfb−/− double mutant shows normal expression pattern of pax7a mRNA compared with WT (G,H), suggesting that the defect in xanthophore and leucophore formation in the double mutant is not mediated by Pax7a function. Lateral views at top and dorsal views at bottom. (A-D,G,H) Stage 28. (E,F) Stage 29. Scale bar: 250 µm.
To exclude the possibility that Mitfs regulate pax7a expression to activate the transcription of dct and gch2, we examined the expression of pax7a mRNA in the mitfa; mitfb double homozygotes. The pax7a-expressing cells were not notably altered in the double mutant (Fig. 9H) compared with WT (Fig. 9G), suggesting that Mitfs directly activate the transcription of dct and gch2, and that, consistent with the overexpression results in Fig. 7, Pax7a alone is not sufficient to drive gch2 transcription.
Finally, to assess whether the function of Mitf is conserved among fish species, we generated zebrafish mitfa−/−; mitfb−/− double mutants by additionally disrupting the mitfb gene by CRISPR/Cas9 in the mitfanacre background (Fig. S12) and observed the double-mutant phenotype (Fig. 10). Compared with a single mitfa mutant, mitfa−/−; mitfb−/− double mutants showed a slightly more severe reduction of xanthophores, with fewer fluorescent xanthophores on the trunk (Fig. 10A3,B3). However, unlike the medaka mitfa; mitfb double homozygotes, the zebrafish double mutants retained xanthophores on the head (Fig. 10A1,B1). Thus, the results suggest that in zebrafish xanthophore development may be partially dependent on Mitf function, but that some other factor(s) may compensate for the loss of Mitfs.
Fig. 10.
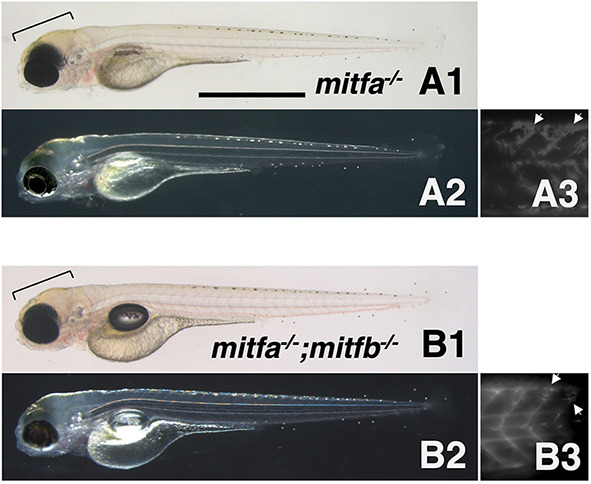
Phenotypes of zebrafish mitfa and mitfb double homozygotes. (A1-B3) 4 dpf mitfa−/− (nacre) hatchling (A1-A3) and 4 dpf mitfa−/−; mitfb−/− hatchling (B1-B3) under normal transmission optics (A1,B1), dark-field epi-illumination optics (A2,B2) and autofluorescence images showing xanthophores under UV light (A3,B3). Yellowish-pigmented xanthophores are observed in the dorsal head of the mitfa−/− hatchling (A1,A2). Autofluorescence emitted by xanthophores is clearly visible in the trunk of mitfa−/− (A3). Similarly, the mitfa−/−; mitfb−/− hatchling has xanthophores in the head (B1,B2) and autofluorescent cells in the trunk (B3). Brackets indicate pigmented xanthophores and arrows indicate autofluorescent (possibly immature) xanthophores. Scale bar: 1 mm.
DISCUSSION
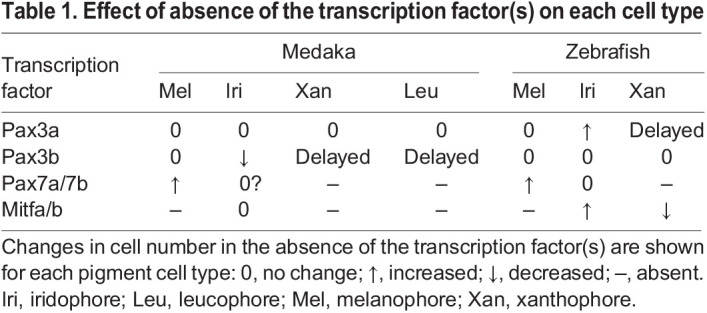
We established medaka and zebrafish mutants of transcription factors that we expected to be involved in the GRN of pigment cell-fate specification. Their phenotypes are summarized in Table 1.
Table 1.
Effect of absence of the transcription factor(s) on each cell type
Although Pax7 has been considered a requisite for xanthophore differentiation in medaka and zebrafish and additionally for leucophore differentiation in medaka (Kimura et al., 2014; Minchin and Hughes, 2008; Nord et al., 2016), Pax3, paralogous to Pax7, has not yet been extensively studied in the context of pigment cell development. In this study, we elucidated the function of Pax3 in pigment cell development and proposed that the set of transcription factors Sox10, Pax3 and Mitf regulate the melanophore and xanthophore fates (plus leucophore fate in medaka). Furthermore, we found that the choice of melanophore versus xanthophore/leucophore fate appears to be regulated by the interaction of Mitf and Pax7. In the absence of Pax7, Mitf strongly drives the melanophore progenitor gene dct, whereas Mitf and Pax7 cooperatively drive the xanthophore/leucophore progenitor gene gch2.
Partially overlapping but sequential function of Pax3 and Pax7
Pax3 is expressed and functions before Pax7 during NCC development, and appears to be temporally restricted to the initial phase of pigment cell progenitor formation. Pax7 expression occurs slightly later than, but overlaps with, Pax3 expression, and is long maintained until a later stage of cell differentiation (Fig. 1, Fig. S2). Assuming that Pax3 and Pax7 are highly homologous and have similar activities (Pax3a versus Pax7a/7b= 96.8% similarity; Pax7a versus Pax7b=100% in homeodomain of zebrafish proteins), loss of Pax3 might be partially compensated by Pax7 at early stages, but loss of Pax7 cannot be compensated by Pax3 at later stages because Pax3 is no longer expressed. The mutant phenotypes support this idea: xanthophores and leucophores are partially formed in the absence of Pax3, whereas these pigment cell types are completely lost in the absence of Pax7, in medaka (Fig. 2, Fig. S4). This is also the case for xanthophores in zebrafish (Figs S5, S6).
The Sox10, Pax3 and Mitf GRN in teleosts
Pax3 is required for activation of mitf expression, as shown in the medaka pax3b mutant and zebrafish pax3a mutant with delayed mitfa expression (Fig. 5). Overexpression of Pax3 together with Sox10 resulted in ectopic expression of mitfa in zebrafish embryos (Fig. 6). As a previous report using human cultured cells showed that Pax3 functions with Sox10 to activate Mitf expression (Lang et al., 2005), we propose that a conserved role of Pax3 in vertebrates is to form a core melanophore GRN with Sox10 to regulate Mitf in the NCCs.
The medaka and zebrafish pax3 mutants only showed ambiguously the melanophore-related phenotypes. Although loss of Pax3 appeared to substantially decrease mitf expression, it eventually resulted in only partially or barely reduced melanophore formation in both medaka and zebrafish (Fig. 2, Figs S4-S6). We reason this can be explained as mitf expression is delayed in the absence of Pax3 and perhaps subsequently compensated by some other transcription factors. In contrast, loss of Pax7 resulted in significant increase of mitfa-expression in progenitors and pigmented melanophores in both species (Fig. 2, Figs S4-S6). As Pax7, and presumably Pax3, inhibit Mitf protein from promoting melanophore fate (see below), loss of Pax3 or Pax7 would tend to increase melanophore formation (Fig. 2, Fig. S5). These conflicting actions may explain the ambiguous melanophore phenotypes in pax3 mutants.
Pax7 interacts with Mitf to segregate pigment cell lineages
In mice, Mitf directly activates Dct expression and drives the cells to differentiate into melanocytes (Lang et al., 2005; Ludwig et al., 2004). In teleosts, Mitf also drives dct expression, reflecting a broader role in driving melanophore differentiation (Elworthy et al., 2003; Greenhill et al., 2011; Lister et al., 1999). However, here we show that co-expression of Pax7 can inhibit Mitf from driving dct transcription, which we suggest has physiological relevance when pax7 is expressed in a subset of the mitfa-expressing NCC population (Fig. 7). In these cells, gch2 is activated by the cooperative action of Pax7 and Mitfa instead of dct (Fig. 7), presumably reflecting a broader role for the combinatorial impact of these transcription factors, which together drive differentiation into xanthophores or leucophores. If pax7 is not co-expressed, the mitfa-expressing cells would differentiate into melanophores, as in mice. We still do not know how the mitfa-expressing progenitor population is divided into two, one expressing Pax7 and the other not.
Model
We propose that a more complex GRN than in mammals controls the fate specification of melanophore progenitors and xanthophore/leucophore progenitors in medaka (Fig. 11). Pax3 and Sox10 activate mitf expression in the NCC-derived multipotent progenitors for pigment cells. The fate of the mitf-expressing progenitor cells is determined by whether or not they express Pax7. These progenitors, if expressing Pax7, give rise to xanthophores and leucophores in medaka (to xanthophores in zebrafish), or, if not expressing Pax7, they give rise to melanophores.
Fig. 11.
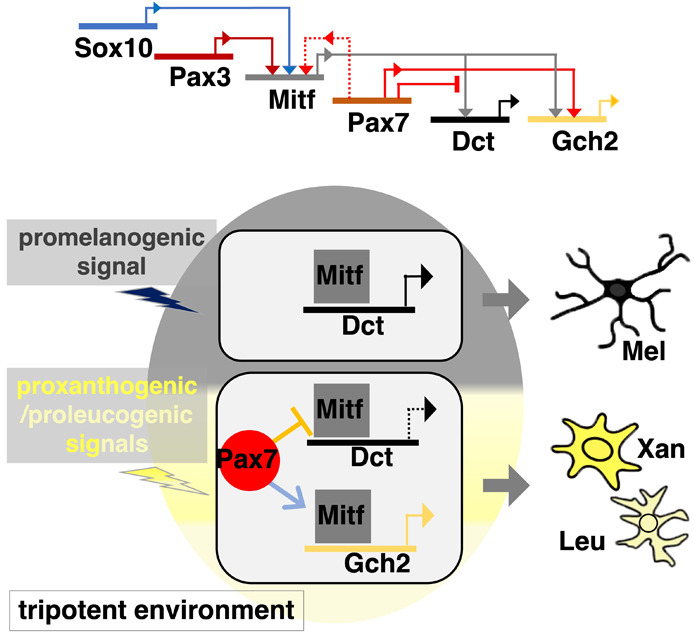
Model of a gene regulatory network. Our results lead us to postulate a GRN that controls the development of melanophores with Dct expression and xanthophores/leucophores with Gch2 expression. In medaka NCCs, Sox10 (Sox10a and Sox10b in medaka) and Pax3 (Pax3b in medaka) activate Mitf (Mitfa and possibly Mitfb) expression. Mitf can drive transcription of Dct and Gch2. Pax7 represses transcription of Dct by inhibiting Mitf, and promotes transcription of Gch2 cooperatively with Mitf. Pax7 may compensate for loss of Pax3 by activating Mitf expression (dotted line). Expression of Mitf and Pax7 are dependent upon promelanogenic and proxanthogenic/proleucogenic signals, respectively (the cell resides in a tripotent environment). A fraction of the Mitf-expressing cells, if expressing Pax7a (lower cell), would differentiate into the Gch2-expressing xanthophore/leucophore lineage because Mitf and Pax7 cooperatively activate Gch2 expression, whereas Pax7 represses Dct by inhibiting Mitf. Another cell, not expressing Pax7 (upper cell), would differentiate into the Dct-expressing melanophore lineage where Mitf activates Dct expression. Leu, leucophore; Mel, melanophore; Xan, xanthophore.
Previously, we and others have explained pigment cell development from NCCs in terms of a progressive fate restriction model, postulating partially restricted intermediates, such as a bipotent progenitor of melanophores and iridophores and a bipotent progenitor of xanthophores and leucophores. Recently, based upon detailed evaluation of pigment cell development in the zebrafish, we have proposed an alternative view, the cyclical fate restriction (CFR) model. Under CFR, we predict that pigment cell development proceeds directly and dynamically from a highly multipotent state (Kelsh et al., 2021; Subkhankulova et al., 2023). We consider that this view can be readily applied to the situation in medaka too. The key is to replace the concept of a bipotent or tripotent progenitor with that of a bipotent or tripotent environment – one in which the broadly multipotent cell is exposed to two or three key fate-determining signals, which limit the options available to the multipotent cell in that region. In this context, we suggest that a pigment cell progenitor expresses mitf in response to a promelanogic environment. But, in addition, if the cell receives unknown signals that are xanthogenic and leucogenic (a tripotent environment), and when these are strong or prolonged enough, pax7 expression becomes dominant, and the cell adopts a xanthophore or leucophore fate. If the signals are too transient/weak, the cell would differentiate into a melanophore.
To our knowledge, Pax7 has not been implicated in pigment cell development in mammals and birds, which lack xanthophores, whereas Pax3 plays a key role in melanocyte development in mammals (Conway et al., 1997; Tassabehji et al., 1992). It is conceivable that the sequential activity of earlier Pax3 and later Pax7 is key to the development of xanthophores and leucophores in vertebrates, and also that loss of Pax3-dependent Pax7 expression may contribute to the evolutionary loss of xanthophore fate from the endothermic vertebrates.
Mitf is a key player in the formation of both melanophore and xanthophore progenitors
Overexpression of Mitfa alone can ectopically activate the expression of not only dct but also gch2 in zebrafish embryos. This suggests an unexpected role of Mitfa in promoting the xanthophore fate. Indeed, our genetic study in medaka showed that Mitfs (Mitfa and Mitfb) are essential for the formation of, not only melanophores, but also xanthophores and leucophores (Figs 8, 9).
Mitf has long been believed to be the master regulator of melanophores in teleosts as in mammals (Arnheiter, 2010) because zebrafish mitfa mutants lack melanophores completely (Lister et al., 1999). At the same time, however, xanthophore pigmentation becomes reduced in the mitfa mutant, implying that Mitf is involved in xanthophore formation and that, perhaps, Mitfb compensates for the loss of Mitfa. Lister et al. generated mitfa; mitfb double mutants, but did not describe whether they lacked xanthophores (Lister et al., 2001).
Our study clearly shows that loss of Mitfa and Mitfb results in complete absence of melanophores, xanthophores and leucophores throughout life in medaka (Fig. 8), suggesting that Mitfs (Mitfa and Mitfb) are required for fate specification of, not only melanophores, but also xanthophores and leucophores in medaka. In contrast, zebrafish mitfa; mitfb double mutants retain xanthophores, as we confirmed by using the mitfb allele we generated in combination with the mitfanacre allele (Fig. S12, Fig. 10). It remains unclear why Mitf loss-of-function phenotypes differ between zebrafish and medaka with respect to xanthophore formation. However, the slightly enhanced reduction of xanthophores by loss of Mitfb in the zebrafish mitfa/nacre mutant suggests that Mitf activity is also involved in xanthophore development in zebrafish (Fig. 10). We speculate that Tfec, a transcription factor homologous to Mitf, which is required for iridophore specification, might partially compensate for the loss of Mitfa and Mitfb to form xanthophores on the head in zebrafish. Indeed, Tfec and Mitfs are partially redundant for pigment cell development in zebrafish (Petratou et al., 2021; Subkhankulova et al., 2023). We speculate that the degree of redundancy of Mitf and Tfec may differ between medaka and zebrafish, so that the formation of xanthophores in these species shows differential regulation by Mitf and Tfec.
In conclusion, the core GRN consisting of Sox10, Pax3 and Mitf to regulate pigment cell development is conserved across teleosts and mammals. We propose that the core GRN governs melanophore and xanthophore/leucophore potentials in teleosts. Recruitment of Pax7 to the GRN allows teleosts to create cell diversity, e.g. xanthophores/leucophores, implying an evolutionary aspect of cell diversification in vertebrates. Finally, although Mitf is considered to be a master regulator of the melanocyte phenotype, this picture may be rather misleading outside of a mammalian context; instead, we note that our results and previous studies make it clear that pigment cell fate determination in fish likely reflects the combinatorial activity of multiple transcription factors, including those of the Mitf/Tfec, Pax and Sox families. Further work will be required to define fully the transcription factor complements responsible for each cell type.
MATERIALS AND METHODS
Ethics
The animal work in this study was approved by the Nagoya University Animal Experiment Committee and was conducted in accordance with the Regulations on Animal Experiments at Nagoya University.
Strains and fish husbandry
The Nagoya and d-rR strains of the medaka fish Oryzias latipes were used as the WT (Nagao et al., 2014; Yamamoto, 1953). Medaka pax3a and pax3b mutant strains were generated by TALENs in the d-rR strain and maintained in the d-rR background, designated as d-rR; pax3aex2del5 and d-rR; pax3bex2del11. The d-rR; pax3bex2del11 strain was crossed with the Nagoya strain to obtain Nagoya; pax3bex2del11 having melanized melanophores (see Figs S4, S7). The pax3aex2del5/ex2del5 and pax3bex2del11/ex2del11 mutants are considered null mutants and are therefore referred to as pax3a−/− and pax3b−/−, respectively. The medaka leucophore free-2 strain, previously described (Kimura et al., 2014), was used as pax7a null mutant. The medaka mitfaex6del1 and mitfbex6del7 mutation was generated by CRISPR/Cas9 on the Nagoya background. In this work, the homozygous mutants are designated as mitfa−/− and mitfb−/−.
The AB strain of the zebrafish Danio rerio was used as the WT. Zebrafish pax3a, pax3b and pax7a mutant strains were generated by TALENs and pax7b mutant by CRISPR/Cas9. The alleles isolated in this study are pax3aex2del14, pax3bex2del11, pax3b ex2del16, pax7aex2del19 and pax7bex1del10. All the mutant alleles are considered null alleles, and thus these zebrafish mutants are designated as pax3a−/−, pax3b−/−, pax7a−/− and pax7b−/−.
Genotyping
Mutations in medaka and zebrafish were detected using PCR fragment length polymorphism by polyacrylamide gel electrophoresis (PAGE), as previously described (Nagao et al., 2018). PCR primer sets were: medaka pax3aex2del5, 5′-AGGTCTCTGGATTTTTCTAACCTAAACCCG-3′ and 5′-TGGTGCGCCATCTCCACGATCTTATG-3′; medaka pax3bex2del11, 5′-TGCTGTGATTGAACGCAGTGTCCACCCCGC-3′ and 5′-GTCTCCTGGTACCGGCACAGGATTTTGGAC-3′; zebrafish pax3aex2del14, 5′-CGCTGACTTTTCCTCTTTTGT-3′ and 5′-GCGGATGTGATTGGGCAAAGG-3′; zebrafish pax3bex2del11 or ex2del16, 5′-TGTCAACTCCGATGGGTCAG-3′ and 5′-AGAGACTCTGAGCTGGCGGG-3′; zebrafish pax7aex2del19, 5′-TCTCAACACCTCTGGGTCAAG-3′ and 5′-CCATTTCCACTATTTTGTGTC-3′; zebrafish pax7bex1del10, 5′-GTAGAATGTCATCCTTACCGG-3′ and 5′-CTTCCAAGGGGAATCCGGTGC-3′; medaka mitfaex6del1 5′-AGGTTCAACATTAATGACCGCATT-3′ (WT allele-specific sense) and 5′-AGGTTCAACATTAATGACCGCATA-3′ (mutant allele-specific sense) and 5′-TGATTGCAGATGATTTGCCCATC-3′ (common antisense); medaka mitfbex6del7, 5′-GCATTGAAGCATTTCTTCCAG-3′ and 5′-AAAATGGCGGGATATACGTAC-3′. PCR products were separated by PAGE to detect the deletion in size or by agarose gel to check for presence or absence (for mitfaex6del1).
TALEN- and CRISPR/Cas9-mediated mutagenesis
Mutagenesis was carried out according to methods described previously, e.g. for TALEN (Ansai et al., 2013) and for CRISPR/Cas9 (Ansai and Kinoshita, 2014). The internal spacer sequences of a pair of TALEN targets were: medaka pax3a, 5′-CAACCAGCTCGGCGG-3′; medaka pax3b, 5′-TTGCCTAACCATATCCG-3′; zebrafish pax3a, 5′-GGGCCGCGTGAACCA-3′; zebrafish pax3b, 5′-TCTGCCCAACCACATTC-3′; zebrafish pax7a, 5′-TTTTCATCAACGGA-3′.
The target sequences of CRISPR/Cas9 were: zebrafish pax3b, 5′-ATCACGGAATCCGGCCCT-3′; zebrafish pax7b, 5′-TACCGCGAATGATGCGAC-3′. We purchased crRNA for targeting medaka mitfa and mitfb: mitfa, 5′-AGGTTCCTAGTTCCTTAATGCGG-3′; mitfb, 5′-GAAGGTTCAACATCAACGATCGG-3′ (Integrated DNA Technologies), and injected each with tracrRNA and Cas9 protein (Integrated DNA Technologies) into one-cell embryos.
Transcript detection in whole-mount embryos
Whole-mount in situ hybridization was performed as previously described (Nagao et al., 2014). The digoxigenin (Roche Diagnostics GmbH)-labelled antisense riboprobe was synthesized from a plasmid containing the full- or partial-length open reading frame of the cDNAs using SP6 or T7 polymerase (Promega) after restriction enzyme digestion (New England Bio Labs).
Microscopy
Melanophores were examined under a stereomicroscope (MZ APO, Leica Microsystems) using a combination of bright- and darkfield illumination. Leucophores and iridophores were identified in dark field. Xanthophores were identified by detecting their autofluorescence under UV light with a DAPI filter (Imager.D1, Carl Zeiss).
Note that larval leucophores appear orange to varying degrees and are clearly visible under darkfield illumination, whereas xanthophores appear yellowish but are ambiguous under brightfield illumination. Under UV light, leucophores show stronger autofluorescence than xanthophores, but iridophores and melanophores do not. Leucophores and xanthophores can be distinguished by their location: leucophores are located along the dorsal and ventral midlines, whereas xanthophores are located laterally across the body surface in the larva.
A Zeiss LSM700 confocal microscope was used to observe GFP fluorescent cells in the transgenic embryos.
Constructs and microinjection of synthetic RNAs
The full-length open reading frame of the cDNA for each gene was subcloned into a pCS2 plasmid (Turner and Weintraub, 1994). Artificial mRNA (synthetic RNA) was transcribed in vitro using SP6 polymerase (Promega) after NotI digestion (New England Bio Labs).
Supplementary Material
Acknowledgements
The authors thank Dr Masato Kinoshita for gifts of TALEN and CRISPR/Cas9 tools. We thank Yumiko Takayanagi for managing fish care.
Footnotes
Author contributions
Conceptualization: H.H.; Investigation: M.M., H.T., A.S., I.W., H.K., Y.N., K.N., S.H., T.S., H.H.; Resources: T.K.; Writing - original draft: H.H.; Writing - review & editing: R.N.K., M.H., H.H.; Visualization: H.H.; Supervision: M.H.; Project administration: H.H.; Funding acquisition: H.H.
Funding
This study was supported by a Grant-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science (20K06757 to H.H.). Open access funding provided by Nagoya University. Deposited in PMC for immediate release.
Data availability
All relevant data can be found within the article and its supplementary information.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/lookup/doi/10.1242/dev.202114.reviewer-comments.pdf
References
- Amores, A., Force, A., Yan, Y. L., Joly, L., Amemiya, C., Fritz, A., Ho, R. K., Langeland, J., Prince, V., Wang, Y.-L.et al. (1998). Zebrafish hox clusters and vertebrate genome evolution. Science 282, 1711-1714. 10.1126/science.282.5394.1711 [DOI] [PubMed] [Google Scholar]
- Ansai, S. and Kinoshita, M. (2014). Targeted mutagenesis using CRISPR/Cas system in medaka. Biol. Open 3, 362-371. 10.1242/bio.20148177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansai, S., Sakuma, T., Yamamoto, T., Ariga, H., Uemura, N., Takahashi, R. and Kinoshita, M. (2013). Efficient targeted mutagenesis in medaka using custom-designed transcription activator-like effector nucleases. Genetics 193, 739-749. 10.1534/genetics.112.147645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnheiter, H. (2010). The discovery of the microphthalmia locus and its gene, Mitf. Pigment. Cell Melanoma. Res. 23, 729-735. 10.1111/j.1755-148X.2010.00759.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnara, J. T., Matsumoto, J., Ferris, W., Frost, S. K., Turner, W. A., Jr., Tchen, T. T. and Taylor, J. D. (1979). Common origin of pigment cells. Science 203, 410-415. 10.1126/science.760198 [DOI] [PubMed] [Google Scholar]
- Basch, M. L., Bronner-Fraser, M. and Garcia-Castro, M. I. (2006). Specification of the neural crest occurs during gastrulation and requires Pax7. Nature 441, 218-222. 10.1038/nature04684 [DOI] [PubMed] [Google Scholar]
- Bondurand, N., Pingault, V., Goerich, D. E., Lemort, N., Sock, E., Le Caignec, C., Wegner, M. and Goossens, M. (2000). Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum. Mol. Genet. 9, 1907-1917. 10.1093/hmg/9.13.1907 [DOI] [PubMed] [Google Scholar]
- Britsch, S., Goerich, D. E., Riethmacher, D., Peirano, R. I., Rossner, M., Nave, K. A., Birchmeier, C. and Wegner, M. (2001). The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 15, 66-78. 10.1101/gad.186601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham, M. and Relaix, F. (2015). PAX3 and PAX7 as upstream regulators of myogenesis. Semin. Cell Dev. Biol. 44, 115-125. 10.1016/j.semcdb.2015.09.017 [DOI] [PubMed] [Google Scholar]
- Conway, S. J., Henderson, D. J. and Copp, A. J. (1997). Pax3 is required for cardiac neural crest migration in the mouse: evidence from the splotch (Sp2H) mutant. Development 124, 505-514. 10.1242/dev.124.2.505 [DOI] [PubMed] [Google Scholar]
- Dutton, K. A., Pauliny, A., Lopes, S. S., Elworthy, S., Carney, T. J., Rauch, J., Geisler, R., Haffter, P. and Kelsh, R. N. (2001). Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development 128, 4113-4125. 10.1242/dev.128.21.4113 [DOI] [PubMed] [Google Scholar]
- Elworthy, S., Lister, J. A., Carney, T. J., Raible, D. W. and Kelsh, R. N. (2003). Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development. Development 130, 2809-2818. 10.1242/dev.00461 [DOI] [PubMed] [Google Scholar]
- Elworthy, S., Pinto, J. P., Pettifer, A., Cancela, M. L. and Kelsh, R. N. (2005). Phox2b function in the enteric nervous system is conserved in zebrafish and is sox10-dependent. Mech. Dev. 122, 659-669. 10.1016/j.mod.2004.12.008 [DOI] [PubMed] [Google Scholar]
- Goding, C. R. and Arnheiter, H. (2019). MITF-the first 25 years. Genes Dev. 33, 983-1007. 10.1101/gad.324657.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhill, E. R., Rocco, A., Vibert, L., Nikaido, M. and Kelsh, R. N. (2011). An iterative genetic and dynamical modelling approach identifies novel features of the gene regulatory network underlying melanocyte development. PLoS Genet. 7, e1002265. 10.1371/journal.pgen.1002265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, H., Goda, M. and Kelsh, R. N. (2021). Pigment cell development in teleosts. In Pigments, Pigment Cells and Pigment Patterns (ed. Hashimoto H., Goda M., Futahashi R., Kelsh R. N. and Akiyama T.), pp. 209-246. Springer. [Google Scholar]
- Hauswirth, R., Haase, B., Blatter, M., Brooks, S. A., Burger, D., Drogemuller, C., Gerber, V., Henke, D., Janda, J., Jude, R.et al. (2012). Mutations in MITF and PAX3 cause “splashed white” and other white spotting phenotypes in horses. PLoS Genet. 8, e1002653. 10.1371/journal.pgen.1002653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemesath, T. J., Steingrimsson, E., Mcgill, G., Hansen, M. J., Vaught, J., Hodgkinson, C. A., Arnheiter, H., Copeland, N. G., Jenkins, N. A. and Fisher, D. E. (1994). microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 8, 2770-2780. 10.1101/gad.8.22.2770 [DOI] [PubMed] [Google Scholar]
- Hou, L., Arnheiter, H. and Pavan, W. J. (2006). Interspecies difference in the regulation of melanocyte development by SOX10 and MITF. Proc. Natl. Acad. Sci. USA 103, 9081-9085. 10.1073/pnas.0603114103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, Z., Mollaaghababa, R., Pavan, W. J., Antonellis, A., Green, E. D. and Hornyak, T. J. (2004). Direct interaction of Sox10 with the promoter of murine Dopachrome Tautomerase (Dct) and synergistic activation of Dct expression with Mitf. Pigment Cell Res. 17, 352-362. 10.1111/j.1600-0749.2004.00154.x [DOI] [PubMed] [Google Scholar]
- Kawakami, A. and Fisher, D. E. (2017). The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab. Invest. 97, 649-656. 10.1038/labinvest.2017.9 [DOI] [PubMed] [Google Scholar]
- Kelsh, R. N. (2006). Sorting out Sox10 functions in neural crest development. BioEssays 28, 788-798. 10.1002/bies.20445 [DOI] [PubMed] [Google Scholar]
- Kelsh, R. N. and Eisen, J. S. (2000). The zebrafish colourless gene regulates development of non-ectomesenchymal neural crest derivatives. Development 127, 515-525. 10.1242/dev.127.3.515 [DOI] [PubMed] [Google Scholar]
- Kelsh, R. N., Schmid, B. and Eisen, J. S. (2000). Genetic analysis of melanophore development in zebrafish embryos. Dev. Biol. 225, 277-293. 10.1006/dbio.2000.9840 [DOI] [PubMed] [Google Scholar]
- Kelsh, R. N., Camargo Sosa, K., Farjami, S., Makeev, V., Dawes, J. H. P. and Rocco, A. (2021). Cyclical fate restriction: a new view of neural crest cell fate specification. Development 148, dev176057. 10.1242/dev.176057 [DOI] [PubMed] [Google Scholar]
- Kimura, T., Nagao, Y., Hashimoto, H., Yamamoto-Shiraishi, Y., Yamamoto, S., Yabe, T., Takada, S., Kinoshita, M., Kuroiwa, A. and Naruse, K. (2014). Leucophores are similar to xanthophores in their specification and differentiation processes in medaka. Proc. Natl. Acad. Sci. USA 111, 7343-7348. 10.1073/pnas.1311254111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, T., Takehana, Y. and Naruse, K. (2017). pnp4a Is the Causal Gene of the Medaka Iridophore Mutant guanineless. G3 7, 1357-1363. 10.1534/g3.117.040675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubic, J. D., Young, K. P., Plummer, R. S., Ludvik, A. E. and Lang, D. (2008). Pigmentation PAX-ways: the role of Pax3 in melanogenesis, melanocyte stem cell maintenance, and disease. Pigment. Cell Melanoma. Res. 21, 627-645. 10.1111/j.1755-148X.2008.00514.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe, R. and Kuratani, S. (2007). Evolutionary perspectives from development of mesodermal components in the lamprey. Dev. Dyn. 236, 2410-2420. 10.1002/dvdy.21177 [DOI] [PubMed] [Google Scholar]
- Lang, D., Lu, M. M., Huang, L., Engleka, K. A., Zhang, M., Chu, E. Y., Lipner, S., Skoultchi, A., Millar, S. E. and Epstein, J. A. (2005). Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature 433, 884-887. 10.1038/nature03292 [DOI] [PubMed] [Google Scholar]
- Le Douarin, N. and Kalcheim, C. (1999). The Neural Crest, 2nd edn. Cambridge, NY, USA: Cambridge University Press. [Google Scholar]
- Lewis, V. M., Saunders, L. M., Larson, T. A., Bain, E. J., Sturiale, S. L., Gur, D., Chowdhury, S., Flynn, J. D., Allen, M. C., Deheyn, D. D.et al. (2019). Fate plasticity and reprogramming in genetically distinct populations of Danio leucophores. Proc. Natl. Acad. Sci. USA 116, 11806-11811. 10.1073/pnas.1901021116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister, J. A., Robertson, C. P., Lepage, T., Johnson, S. L. and Raible, D. W. (1999). nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development 126, 3757-3767. 10.1242/dev.126.17.3757 [DOI] [PubMed] [Google Scholar]
- Lister, J. A., Close, J. and Raible, D. W. (2001). Duplicate mitf genes in zebrafish: complementary expression and conservation of melanogenic potential. Dev. Biol. 237, 333-344. 10.1006/dbio.2001.0379 [DOI] [PubMed] [Google Scholar]
- Lopes, S. S., Yang, X., Muller, J., Carney, T. J., Mcadow, A. R., Rauch, G. J., Jacoby, A. S., Hurst, L. D., Delfino-Machin, M., Haffter, P.et al. (2008). Leukocyte tyrosine kinase functions in pigment cell development. PLoS Genet. 4, e1000026. 10.1371/journal.pgen.1000026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, A., Rehberg, S. and Wegner, M. (2004). Melanocyte-specific expression of dopachrome tautomerase is dependent on synergistic gene activation by the Sox10 and Mitf transcription factors. FEBS Lett. 556, 236-244. 10.1016/S0014-5793(03)01446-7 [DOI] [PubMed] [Google Scholar]
- Minchin, J. E. and Hughes, S. M. (2008). Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest. Dev. Biol. 317, 508-522. 10.1016/j.ydbio.2008.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch, B., Delconte, C. and Garcia-Castro, M. I. (2012). Pax7 lineage contributions to the mammalian neural crest. PLoS One 7, e41089. 10.1371/journal.pone.0041089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao, Y., Suzuki, T., Shimizu, A., Kimura, T., Seki, R., Adachi, T., Inoue, C., Omae, Y., Kamei, Y., Hara, I.et al. (2014). Sox5 functions as a fate switch in medaka pigment cell development. PLoS Genet. 10, e1004246. 10.1371/journal.pgen.1004246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao, Y., Takada, H., Miyadai, M., Adachi, T., Seki, R., Kamei, Y., Hara, I., Taniguchi, Y., Naruse, K., Hibi, M.et al. (2018). Distinct interactions of Sox5 and Sox10 in fate specification of pigment cells in medaka and zebrafish. PLoS Genet. 14, e1007260. 10.1371/journal.pgen.1007260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord, H., Dennhag, N., Muck, J. and Von Hofsten, J. (2016). Pax7 is required for establishment of the xanthophore lineage in zebrafish embryos. Mol. Biol. Cell 27, 1853-1862. 10.1091/mbc.e15-12-0821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord, H., Kahsay, A., Dennhag, N., Pedrosa Domellof, F. and Von Hofsten, J. (2022). Genetic compensation between Pax3 and Pax7 in zebrafish appendicular muscle formation. Dev. Dyn. 251, 1423-1438. 10.1002/dvdy.415 [DOI] [PubMed] [Google Scholar]
- Opdecamp, K., Nakayama, A., Nguyen, M. T., Hodgkinson, C. A., Pavan, W. J. and Arnheiter, H. (1997). Melanocyte development in vivo and in neural crest cell cultures: crucial dependence on the Mitf basic-helix-loop-helix-zipper transcription factor. Development 124, 2377-2386. 10.1242/dev.124.12.2377 [DOI] [PubMed] [Google Scholar]
- Pelletier, I., Bally-Cuif, L. and Ziegler, I. (2001). Cloning and developmental expression of zebrafish GTP cyclohydrolase I. Mech. Dev. 109, 99-103. 10.1016/S0925-4773(01)00516-0 [DOI] [PubMed] [Google Scholar]
- Petratou, K., Subkhankulova, T., Lister, J. A., Rocco, A., Schwetlick, H. and Kelsh, R. N. (2018). A systems biology approach uncovers the core gene regulatory network governing iridophore fate choice from the neural crest. PLoS Genet. 14, e1007402. 10.1371/journal.pgen.1007402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petratou, K., Spencer, S. A., Kelsh, R. N. and Lister, J. A. (2021). The MITF paralog tfec is required in neural crest development for fate specification of the iridophore lineage from a multipotent pigment cell progenitor. PLoS One 16, e0244794. 10.1371/journal.pone.0244794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait, J., Amores, A., Cresko, W., Singer, A. and Yan, Y. L. (2004). Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 20, 481-490. 10.1016/j.tig.2004.08.001 [DOI] [PubMed] [Google Scholar]
- Potterf, S. B., Furumura, M., Dunn, K. J., Arnheiter, H. and Pavan, W. J. (2000). Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Hum. Genet. 107, 1-6. 10.1007/s004390000328 [DOI] [PubMed] [Google Scholar]
- Relaix, F., Rocancourt, D., Mansouri, A. and Buckingham, M. (2004). Divergent functions of murine Pax3 and Pax7 in limb muscle development. Genes Dev. 18, 1088-1105. 10.1101/gad.301004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson, E. J., He, S. J. and Eccles, M. R. (2006). A PANorama of PAX genes in cancer and development. Nat. Rev. Cancer 6, 52-62. 10.1038/nrc1778 [DOI] [PubMed] [Google Scholar]
- Schartl, M., Larue, L., Goda, M., Bosenberg, M. W., Hashimoto, H. and Kelsh, R. N. (2016). What is a vertebrate pigment cell? Pigment. Cell Melanoma. Res. 29, 8-14. 10.1111/pcmr.12409 [DOI] [PubMed] [Google Scholar]
- Sommer, L. (2011). Generation of melanocytes from neural crest cells. Pigment. Cell Melanoma. Res. 24, 411-421. 10.1111/j.1755-148X.2011.00834.x [DOI] [PubMed] [Google Scholar]
- Southard-Smith, E. M., Kos, L. and Pavan, W. J. (1998). Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat. Genet. 18, 60-64. 10.1038/ng0198-60 [DOI] [PubMed] [Google Scholar]
- Steingrimsson, E., Moore, K. J., Lamoreux, M. L., Ferre-D'amare, A. R., Burley, S. K., Zimring, D. C., Skow, L. C., Hodgkinson, C. A., Arnheiter, H., Copeland, N. G.et al. (1994). Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat. Genet. 8, 256-263. 10.1038/ng1194-256 [DOI] [PubMed] [Google Scholar]
- Subkhankulova, T., Camargo Sosa, K., Uroshlev, L. A., Nikaido, M., Shriever, N., Kasianov, A. S., Yang, X., Rodrigues, F., Carney, T. J., Bavister, G.et al. (2023). Zebrafish pigment cells develop directly from persistent highly multipotent progenitors. Nat. Commun. 14, 1258. 10.1038/s41467-023-36876-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W. and Bronner, M. E. (2020). Neural crest lineage analysis: from past to future trajectory. Development 147, dev193193. 10.1242/dev.193193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassabehji, M., Read, A. P., Newton, V. E., Harris, R., Balling, R., Gruss, P. and Strachan, T. (1992). Waardenburg's syndrome patients have mutations in the human homologue of the Pax-3 paired box gene. Nature 355, 635-636. 10.1038/355635a0 [DOI] [PubMed] [Google Scholar]
- Tremblay, P., Kessel, M. and Gruss, P. (1995). A transgenic neuroanatomical marker identifies cranial neural crest deficiencies associated with the Pax3 mutant Splotch. Dev. Biol. 171, 317-329. 10.1006/dbio.1995.1284 [DOI] [PubMed] [Google Scholar]
- Tsukamoto, K., Jackson, I. J., Urabe, K., Montague, P. M. and Hearing, V. J. (1992). A second tyrosinase-related protein, TRP-2, is a melanogenic enzyme termed DOPAchrome tautomerase. EMBO J. 11, 519-526. 10.1002/j.1460-2075.1992.tb05082.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunogai, Y., Miyadai, M., Nagao, Y., Sugiwaka, K., Kelsh, R. N., Hibi, M. and Hashimoto, H. (2021). Contribution of sox9b to pigment cell formation in medaka fish. Dev. Growth Differ. 63, 516-522. 10.1111/dgd.12760 [DOI] [PubMed] [Google Scholar]
- Turner, D. L. and Weintraub, H. (1994). Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 8, 1434-1447. 10.1101/gad.8.12.1434 [DOI] [PubMed] [Google Scholar]
- Wada, H., Holland, P. W. and Satoh, N. (1996). Origin of patterning in neural tubes. Nature 384, 123. 10.1038/384123a0 [DOI] [PubMed] [Google Scholar]
- Watakabe, I., Hashimoto, H., Kimura, Y., Yokoi, S., Naruse, K. and Higashijima, S. I. (2018). Highly efficient generation of knock-in transgenic medaka by CRISPR/Cas9-mediated genome engineering. Zool. Lett. 4, 3. 10.1186/s40851-017-0086-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner, M. (2005). Secrets to a healthy Sox life: lessons for melanocytes. Pigment Cell Res. 18, 74-85. 10.1111/j.1600-0749.2005.00218.x [DOI] [PubMed] [Google Scholar]
- Yamamoto, T.-O. (1953). Artificially induced sex-reversal in genotypic males of the medaka (Oryzias latipes). J. Exp. Zool. 123, 571-594. 10.1002/jez.1401230309 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.